Abstract
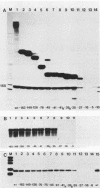
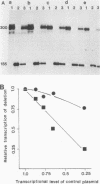
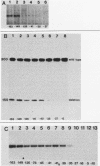
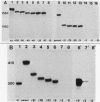
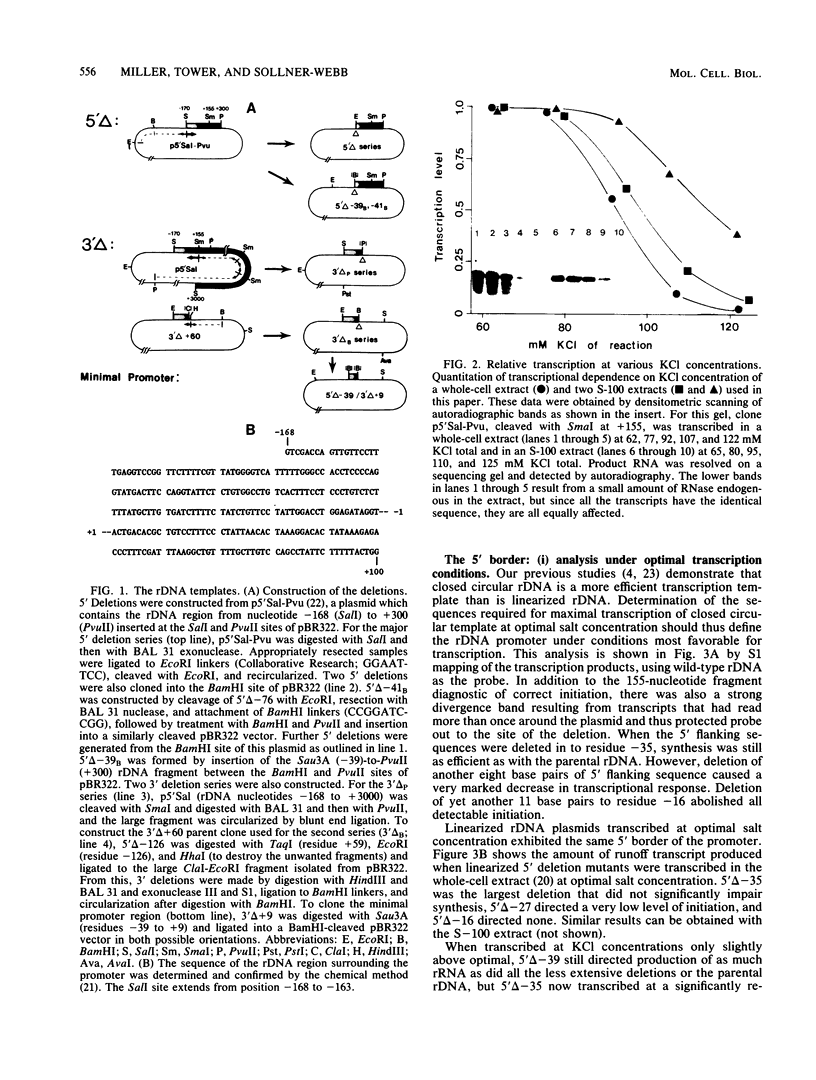
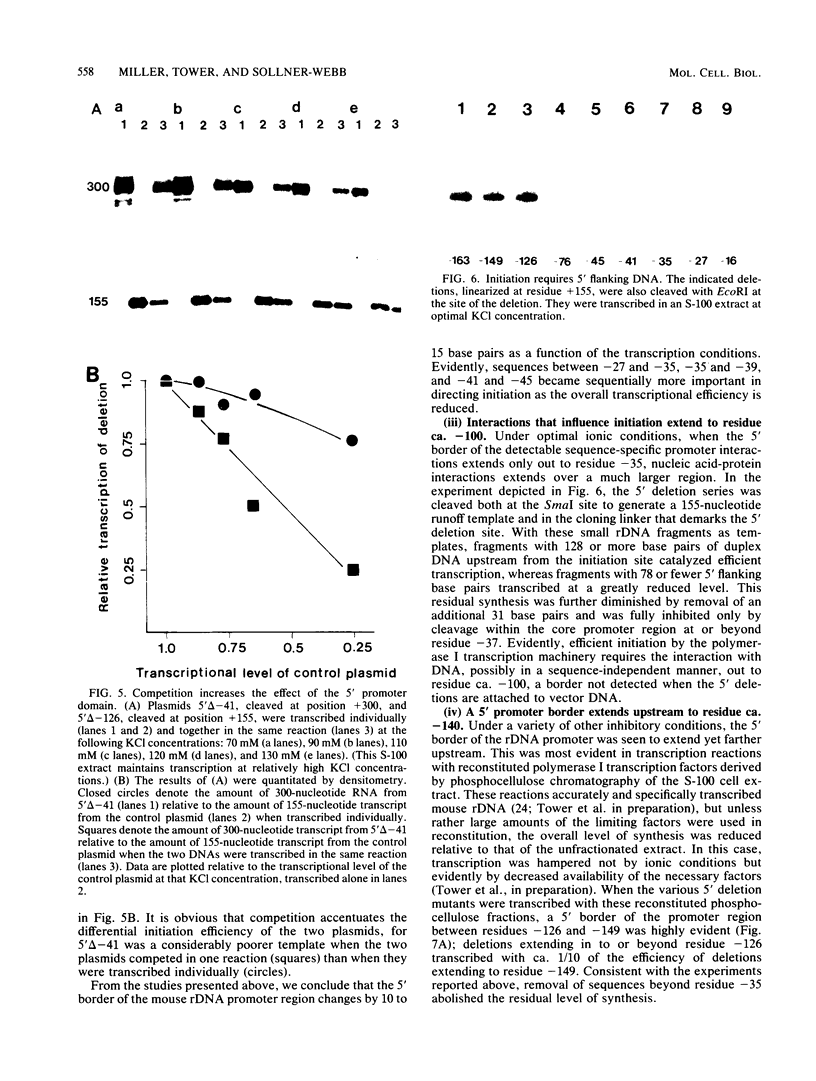
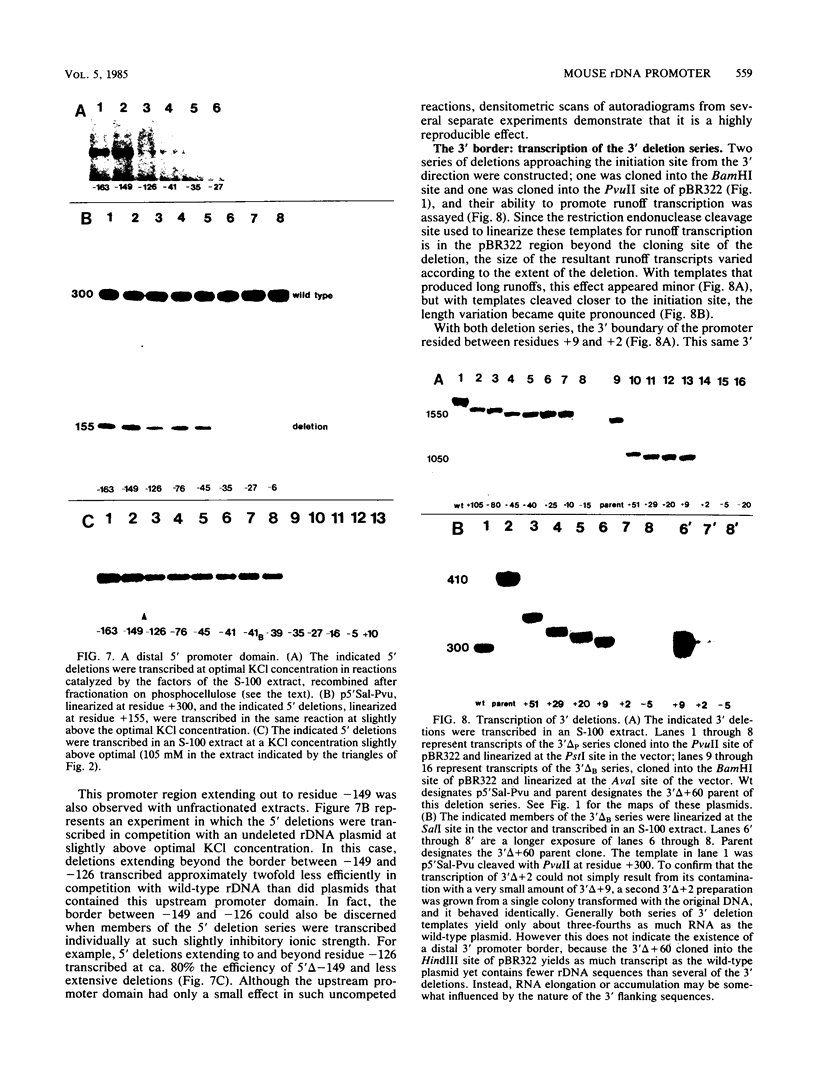
To determine the size and location of the mouse rDNA promoter, we constructed systematic series of deletion mutants approaching the initiation site from the 5' and 3' directions. These templates were transcribed in vitro under various conditions with S-100 and whole-cell extracts. Surprisingly, the size of the rDNA region that determines the level of transcription differed markedly, depending on the reaction conditions. In both kinds of cell extracts, the apparent 5' border of the promoter was at residue ca. -27 under optimal transcription conditions, but as reaction conditions became less favorable, the 5' border moved progressively out to residues -35, -39, and -45. The complete promoter, however, extends considerably further, for under other nonoptimal conditions, we observed major effects of promoter domains extending in the 5' direction to positions ca. -100 and -140. In contrast, the apparent 3' border of the mouse rDNA promoter was at residue ca. +9 under all conditions examined. We also show that the subcloned rDNA region from -39 to +9 contains sufficient information to initiate accurately and that the region between +2 and +9 can influence the specificity of initiation. These data indicate that, although the polymerase I transcription factors recognize and accurately initiate with only the sequences downstream of residue -40, sequences extending out to residue -140 greatly favor the initiation reaction; presumably, this entire region is involved in rRNA transcription in vivo.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bach R., Grummt I., Allet B. The nucleotide sequence of the initiation region of the ribosomal transcription unit from mouse. Nucleic Acids Res. 1981 Apr 10;9(7):1559–1569. doi: 10.1093/nar/9.7.1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogenhagen D. F., Sakonju S., Brown D. D. A control region in the center of the 5S RNA gene directs specific initiation of transcription: II. The 3' border of the region. Cell. 1980 Jan;19(1):27–35. doi: 10.1016/0092-8674(80)90385-2. [DOI] [PubMed] [Google Scholar]
- Bogenhagen D. F., Wormington W. M., Brown D. D. Stable transcription complexes of Xenopus 5S RNA genes: a means to maintain the differentiated state. Cell. 1982 Feb;28(2):413–421. doi: 10.1016/0092-8674(82)90359-2. [DOI] [PubMed] [Google Scholar]
- Cizewski V., Sollner-Webb B. A stable transcription complex directs mouse ribosomal RNA synthesis by RNA polymerase I. Nucleic Acids Res. 1983 Oct 25;11(20):7043–7056. doi: 10.1093/nar/11.20.7043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davison B. L., Egly J. M., Mulvihill E. R., Chambon P. Formation of stable preinitiation complexes between eukaryotic class B transcription factors and promoter sequences. Nature. 1983 Feb 24;301(5902):680–686. doi: 10.1038/301680a0. [DOI] [PubMed] [Google Scholar]
- Dierks P., van Ooyen A., Cochran M. D., Dobkin C., Reiser J., Weissmann C. Three regions upstream from the cap site are required for efficient and accurate transcription of the rabbit beta-globin gene in mouse 3T6 cells. Cell. 1983 Mar;32(3):695–706. doi: 10.1016/0092-8674(83)90055-7. [DOI] [PubMed] [Google Scholar]
- Engelke D. R., Ng S. Y., Shastry B. S., Roeder R. G. Specific interaction of a purified transcription factor with an internal control region of 5S RNA genes. Cell. 1980 Mar;19(3):717–728. doi: 10.1016/s0092-8674(80)80048-1. [DOI] [PubMed] [Google Scholar]
- Financsek I., Mizumoto K., Mishima Y., Muramatsu M. Human ribosomal RNA gene: nucleotide sequence of the transcription initiation region and comparison of three mammalian genes. Proc Natl Acad Sci U S A. 1982 May;79(10):3092–3096. doi: 10.1073/pnas.79.10.3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green M. R., Maniatis T., Melton D. A. Human beta-globin pre-mRNA synthesized in vitro is accurately spliced in Xenopus oocyte nuclei. Cell. 1983 Mar;32(3):681–694. doi: 10.1016/0092-8674(83)90054-5. [DOI] [PubMed] [Google Scholar]
- Grosschedl R., Birnstiel M. L. Delimitation of far upstream sequences required for maximal in vitro transcription of an H2A histone gene. Proc Natl Acad Sci U S A. 1982 Jan;79(2):297–301. doi: 10.1073/pnas.79.2.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grummt I. Mapping of a mouse ribosomal DNA promoter by in vitro transcription. Nucleic Acids Res. 1981 Nov 25;9(22):6093–6102. doi: 10.1093/nar/9.22.6093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grummt I. Nucleotide sequence requirements for specific initiation of transcription by RNA polymerase I. Proc Natl Acad Sci U S A. 1982 Nov;79(22):6908–6911. doi: 10.1073/pnas.79.22.6908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grummt I., Roth E., Paule M. R. Ribosomal RNA transcription in vitro is species specific. Nature. 1982 Mar 11;296(5853):173–174. doi: 10.1038/296173a0. [DOI] [PubMed] [Google Scholar]
- Grummt I. Specific transcription of mouse ribosomal DNA in a cell-free system that mimics control in vivo. Proc Natl Acad Sci U S A. 1981 Feb;78(2):727–731. doi: 10.1073/pnas.78.2.727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu S. L., Manley J. L. DNA sequence required for initiation of transcription in vitro from the major late promoter of adenovirus 2. Proc Natl Acad Sci U S A. 1981 Feb;78(2):820–824. doi: 10.1073/pnas.78.2.820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohorn B. D., Rae P. M. A component of Drosophila RNA polymerase I promoter lies within the rRNA transcription unit. Nature. 1983 Jul 14;304(5922):179–181. doi: 10.1038/304179a0. [DOI] [PubMed] [Google Scholar]
- Kohorn B. D., Rae P. M. Localization of DNA sequences promoting RNA polymerase I activity in Drosophila. Proc Natl Acad Sci U S A. 1983 Jun;80(11):3265–3268. doi: 10.1073/pnas.80.11.3265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Learned R. M., Smale S. T., Haltiner M. M., Tjian R. Regulation of human ribosomal RNA transcription. Proc Natl Acad Sci U S A. 1983 Jun;80(12):3558–3562. doi: 10.1073/pnas.80.12.3558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Learned R. M., Tjian R. In vitro transcription of human ribosomal RNA genes by RNA polymerase I. J Mol Appl Genet. 1982;1(6):575–584. [PubMed] [Google Scholar]
- Manley J. L., Fire A., Cano A., Sharp P. A., Gefter M. L. DNA-dependent transcription of adenovirus genes in a soluble whole-cell extract. Proc Natl Acad Sci U S A. 1980 Jul;77(7):3855–3859. doi: 10.1073/pnas.77.7.3855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- McKnight S. L., Gavis E. R., Kingsbury R., Axel R. Analysis of transcriptional regulatory signals of the HSV thymidine kinase gene: identification of an upstream control region. Cell. 1981 Aug;25(2):385–398. doi: 10.1016/0092-8674(81)90057-x. [DOI] [PubMed] [Google Scholar]
- Miller K. G., Sollner-Webb B. Transcription of mouse rRNA genes by RNA polymerase I: in vitro and in vivo initiation and processing sites. Cell. 1981 Nov;27(1 Pt 2):165–174. doi: 10.1016/0092-8674(81)90370-6. [DOI] [PubMed] [Google Scholar]
- Mishima Y., Financsek I., Kominami R., Muramatsu M. Fractionation and reconstitution of factors required for accurate transcription of mammalian ribosomal RNA genes: identification of a species-dependent initiation factor. Nucleic Acids Res. 1982 Nov 11;10(21):6659–6670. doi: 10.1093/nar/10.21.6659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakonju S., Bogenhagen D. F., Brown D. D. A control region in the center of the 5S RNA gene directs specific initiation of transcription: I. The 5' border of the region. Cell. 1980 Jan;19(1):13–25. doi: 10.1016/0092-8674(80)90384-0. [DOI] [PubMed] [Google Scholar]
- Sollner-Webb B., Wilkinson J. A., Roan J., Reeder R. H. Nested control regions promote Xenopus ribosomal RNA synthesis by RNA polymerase I. Cell. 1983 Nov;35(1):199–206. doi: 10.1016/0092-8674(83)90222-2. [DOI] [PubMed] [Google Scholar]
- Urano Y., Kominami R., Mishima Y., Muramatsu M. The nucleotide sequence of the putative transcription initiation site of a cloned ribosomal RNA gene of the mouse. Nucleic Acids Res. 1980 Dec 20;8(24):6043–6058. doi: 10.1093/nar/8.24.6043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wandelt C., Grummt I. Formation of stable preinitiation complexes is a prerequisite for ribosomal DNA transcription in vitro. Nucleic Acids Res. 1983 Jun 11;11(11):3795–3809. doi: 10.1093/nar/11.11.3795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasylyk B., Derbyshire R., Guy A., Molko D., Roget A., Téoule R., Chambon P. Specific in vitro transcription of conalbumin gene is drastically decreased by single-point mutation in T-A-T-A box homology sequence. Proc Natl Acad Sci U S A. 1980 Dec;77(12):7024–7028. doi: 10.1073/pnas.77.12.7024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weil P. A., Segall J., Harris B., Ng S. Y., Roeder R. G. Faithful transcription of eukaryotic genes by RNA polymerase III in systems reconstituted with purified DNA templates. J Biol Chem. 1979 Jul 10;254(13):6163–6173. [PubMed] [Google Scholar]
- Wilkinson J. A., Miller K. G., Sollner-Webb B. Dinucleotide primers facilitate convenient identification of the mouse ribosomal DNA transcription initiation site. A general method for analysis of transcription by RNA polymerases I and III. J Biol Chem. 1983 Nov 25;258(22):13919–13928. [PubMed] [Google Scholar]
- Yamamoto O., Takakusa N., Mishima Y., Kominami R., Muramatsu M. Determination of the promoter region of mouse ribosomal RNA gene by an in vitro transcription system. Proc Natl Acad Sci U S A. 1984 Jan;81(2):299–303. doi: 10.1073/pnas.81.2.299. [DOI] [PMC free article] [PubMed] [Google Scholar]