Abstract
Kwashiorkor, an enigmatic form of severe acute malnutrition, is the consequence of inadequate nutrient intake plus additional environmental insults. To investigate the role of the gut microbiome, we studied 317 Malawian twin pairs during the first 3 years of life. During this time, half of the twin pairs remained well-nourished, while 43% became discordant and 7% manifested concordance for acute malnutrition. Both children in twin pairs discordant for kwashiorkor were treated with a peanut-based, ready-to-use therapeutic food (RUTF). Time-series metagenomic studies revealed that RUTF produced a transient maturation of metabolic functions in kwashiorkor microbiomes that regressed when RUTF was stopped. Previously frozen fecal communities from several discordant pairs were each transplanted into gnotobiotic mice. The combination of Malawian diet and kwashiorkor microbiome produced marked weight loss in recipient mice, accompanied by perturbations in amino acid, carbohydrate and intermediary metabolism that were only transiently ameliorated with RUTF. These findings implicate the gut microbiome as a causal factor in kwashiorkor.
Malnutrition is the leading cause of child mortality worldwide (1). Moderate acute malnutrition (MAM) refers to simple wasting with a weight-for-height Z score (WHZ) between two and three standard deviations below the median defined by World Health Organization (WHO) Child Growth Standards (2,3). Severe acute malnutrition (SAM) refers to either marasmus, which is extreme wasting with WHZ scores less than −3, or kwashiorkor, a virulent form of SAM characterized by generalized edema, hepatic steatosis, skin rashes and ulcerations, and anorexia (4,5). The cause of kwashiorkor remains obscure. Speculation regarding its pathogenesis has focused on inadequate protein intake and/or excessive oxidative stress, but substantial evidence to refute these hypotheses has come from epidemiologic surveys and clinical trials (6–9). Our comparative metagenomic study of the gut microbiomes of 531 healthy infants, children and adults, living in the USA, Venezuela, and Malawi revealed a maturational program where the proportional representation of genes encoding functions related to micro- and macronutrient biosynthesis and metabolism changes during postnatal development (10). Together, these observations give rise to the following testable hypotheses: (i) the gut microbiome provides essential functions needed for healthy postnatal growth and development; (ii) disturbances in microbiome assembly and function (e.g., those prompted by enteropathogen infection), affect the risk for kwashiorkor; and (iii) in a self-reinforcing pathogenic cascade, malnutrition affects gut microbiome functions involved in determining nutritional status, thus further worsening health status. A challenge is that there may be several gut microbiome configurations associated with kwashiorkor among different hosts and even within a given host over time. Moreover, microbiome configurations associated with kwashiorkor may be differentially affected by therapeutic food interventions and features that are reconfigured during treatment may not persist after withdrawal of treatment.
To address some of these hypotheses, we performed a longitudinal comparative study of the fecal microbiomes of monozygotic (MZ) and dizygotic (DZ) twin pairs born in Malawi who became discordant for kwashiorkor. Malawi has one of the highest infant mortality rates in the world (1,11). We reasoned that a healthy (well-nourished) co-twin in a discordant twin pair represented a very desirable control given his/her genetic relatedness to the affected co-twin, and their similar exposures to diet and microbial reservoirs in their shared early environment. Ready-to-use therapeutic food (RUTF) composed of peanut paste, sugar, vegetable oil and milk fortified with vitamins and minerals has become the international standard of treatment for SAM in community-based treatment programs (12). In Malawi, the standard of care for twins discordant for kwashiorkor is to treat both co-twins with RUTF to limit food sharing; this practice allowed us to compare and contrast their microbiomes prior to, during and after treatment. Following each child in a twin pair prospectively permitted each individual to serve as his or her own control. Moreover, if there are many different routes to disrupted microbiome structure/function, then each discordant twin pair could provide an illustration of underlying pathology. As an additional set of controls, we defined temporal variation of the fecal microbiomes in twin pairs who remained well-nourished, lived in the same geographic locations as discordant pairs, and never received RUTF.
A total of 317 twin pairs less than 3 years old were enrolled, regardless of their health status, from five villages in the southern region of Malawi. Children were followed until they were 36 months old. Zygosity testing (13) revealed that 46 (15%) twin pairs were monozygotic (MZ). Using WHO criteria (3), kwashiorkor was diagnosed based on the presence of bilateral pitting pedal edema, marasmus when a child had a WHZ less than −3, and MAM when the WHZ score was between −2 and −3 and bilateral pitting pedal edema was absent (2,3). SAM was treated with RUTF and MAM with a soy-peanut ready-to-use supplementary food (14). After diagnosis with SAM, anthropometry was assessed and a fecal sample was collected every two weeks until the child recovered (defined as WHZ score greater than −2 and no edema).
Fifty percent of twin pairs remained well-nourished throughout the study while 43% became discordant and 7% manifested concordance for acute malnutrition. The prevalence of discordant compared to concordant phenotypes was significantly different (p<10−15, Binomial and Chi-squared tests). MAM was significantly more frequent than SAM, affecting 81 (60%) of the 135 discordant twin pairs (p=0.02, Chi-square test) (table S1A). Of the 634 children in the study, 7.4% developed kwashiorkor, 2.5% marasmus, and 13.9% MAM; 10.7% had multiple episodes of malnutrition with the most frequent combination being marasmus and MAM (5.5% of the children, table S1B). There was no significant relationship between concordance for acute malnutrition and zygosity, nor did we find significant differences in the number of MZ versus dizygotic (DZ) twin pairs affected with kwashiorkor, marasmus, or MAM in our cohort (Chi-squared and Fisher’s exact tests). Taking all 135 discordant pairs into account, there was no statistically significant difference in the incidence of discordance for kwashiorkor, marasmus, or MAM in MZ versus DZ twin pairs (table S1A). In addition, we did not find any association between gender or geographic location and the type or incidence of malnutrition, or in the discordance rate for MAM or SAM among twin pairs (table S1B, C). For the current study, we chose to focus on children with kwashiorkor because survival of both members of a twin pair was significantly higher than with marasmus, onset occurred at a later age allowing us to better assess the state of functional development of the gut microbiome, the duration of RUTF treatment was more uniform than in cases of marasmus, and there was a lower incidence of relapse to SAM (see supplementary online text for additional details).
Microbiomes of healthy twins and pairs discordant for kwashiorkor
We selected nine same gender twin pairs who remained well-nourished in our study cohort and 13 of 19 same gender twin pairs who became discordant for kwashiorkor for metagenomic analyses of their microbiomes [n=5 MZ and 4 DZ healthy pairs; 7 MZ and 6 DZ pairs discordant for kwashiorkor; for 12 of the 13 discordant pairs, there was a single episode of kwashiorkor during the study period; see supplementary online text for the criteria used for their selection, and table S2A for additional information about subjects and their samples]. DNA prepared from fecal samples was subjected to multiplex shotgun pyrosequencing and the resulting reads were annotated by comparison to the KEGG database (table S2A) and to a database of 462 sequenced human gut microbes (table S3). Principal coordinates analysis (PCoA) of Hellinger distances computed from the KEGG enzyme commission number (EC) content of fecal microbiomes was used to visualize variation in this dataset (Fig. 1A and fig. S1). Principal coordinate 1 (PC1), which explained the largest amount of variation, was strongly associated with age and family membership (Fig. 1A). Since age encompasses a variety of metabolic and dietary changes, we used the positions of microbiomes along PC1 to assess functional development of the microbiomes of twin pairs who remained healthy and twin pairs who became discordant for kwashiorkor. When microbiomes from three consecutive time points from twins who remained healthy were plotted along PC1, there was a steady progression towards a configuration found in older children (Fig. 1B). A similar result was observed in healthy co-twins from discordant twin pairs. This was not the case for their siblings with kwashiorkor: their fecal microbiomes, sampled at the time of diagnosis, as well as during and following RUTF, did not show significant differences in their position in the ordination plot (Fig. 1C). We obtained the same results for KEGG Orthology (KO) groups. Comparing the representation of KEGG ECs between a healthy and malnourished co-twin within each family using Fischer’s exact test, we identified ECs that were significantly different in as few as one and in as many as six of the twin pairs (table S5).
Fig 1. Functional development of the gut microbiomes of Malawian twin pairs concordant for healthy status, and twin pairs who became discordant for kwashiorkor.
(A) PCoA of Hellinger distances between KEGG EC profiles. The position of each fecal microbiome along principal coordinate 1 (PC1), which describes the largest amount of variation (17%) in this dataset of 308 sequenced twin fecal microbiomes, is plotted against age. Each sphere represents a microbiome colored by the age of the human donor. PC1 is strongly associated with age (and with family membership) (linear mixed-effects model, table S4). We did not find significant associations between the positions of samples along other principal coordinates and the other host parameters presented in table S2A). On average the degree of intrapersonal variation in a co-twin was not smaller than the variation between co-twins (fig. S2). Similar to twins who remained healthy, the temporal variation within a co-twin member of a discordant twin pair was equal to the variation between co-twins, but still smaller compared to unrelated children (fig. S2). (B) Average ± SEM PC1 coordinate obtained from the data shown in panel A for microbiomes sampled at three consecutive time points from nine twin pairs who remained well-nourished (healthy) during the study (subjects surveyed between three weeks and 24.5 months of age). (C) Average ± SEM PC1 coordinate obtained from panel A for microbiomes sampled before, during and after RUTF treatment from co-twins discordant for kwashiorkor. *p<0.05, Friedman test with Dunn’s post-hoc test applied to data shown in panels B and C. Similar results were obtained using other distance metrics [Bray Curtis, Euclidian, and Kulzyncki (fig. S3)]. (See fig. S4 which shows how changes in the relative proportion of Actinobacteria parallel the patterns observed with the changes along PC1; children with kwashiorkor manifested a statistically significant decrease in Actinobacteria with introduction of RUTF, unlike their healthy co-twins).
These associations between the configurations of gut microbial communities and health status do not establish whether the microbiome is a causal factor in the pathogenesis of kwashiorkor. We reasoned that transplanting previously frozen fecal microbial communities, obtained from discordant twin pairs at the time one of the co-twins presented with kwashiorkor, into gnotobiotic mice would allow us to assess the degree to which donor phenotypes could be transmitted via their gut microbiomes, and to identify features of microbial community structure, metabolism and host-microbial co-metabolism associated with donor health status and diet. In the absence of a distinct and consistent taxonomic signature of kwashiorkor (supplementary online text), we selected pre-treatment fecal samples from three discordant twins based on the following criteria: all twin pairs were of similar age, and neither co-twin in any pair had diarrhea, vomiting, or were consuming antibiotics at the time that fecal samples were collected. The twins selected included DZ pair 196 (aged 16.5 months), DZ pair 56 (aged 18 months), MZ twin pair 57 (aged 21 months) (see fig. S5 and table S2A for clinical characteristics).
Transplantation of fecal microbial communities into gnotobiotic mice
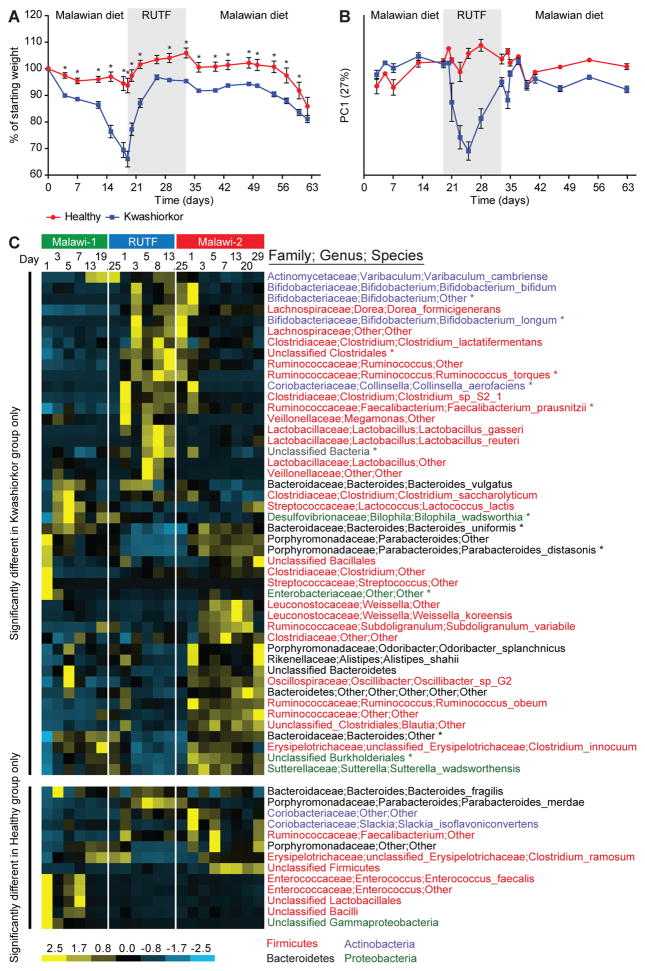
Fecal microbiota samples from the six selected human donors were each transplanted, with a single oral gavage, into separate groups of adult 8 week-old male C57BL/6J germ-free mice. Beginning one week prior to gavage, animals were fed, ad libitum, a sterilized diet based on the staple foods consumed by individuals living in rural southern Malawi (10) (see table S6 for the composition of this low caloric density, nutrient-deficient diet and fig. S6 for experimental design). In the case of two of the three discordant twin pairs (families 196 and 57), transplantation of the kwashiorkor co-twin’s microbiota resulted in significantly greater weight loss in recipient mice over the ensuing three weeks than those harboring their healthy sibling’s microbiota (Fig. 2A). This discordant weight loss phenotype was dependent upon the combination of Malawian diet and kwashiorkor microbiota; when separate groups of animals were placed on a standard mouse chow there were no significant differences between the weights of mice with a kwashiorkor compared to healthy co-twin microbiota (93.6±4% versus 93±11% of their starting weights after 21 days, respectively) or compared to mice with a healthy co-twin microbiota consuming a Malawian diet (102.4±11.1%).
Fig 2. Transplantation of fecal microbiota from kwashiorkor and healthy co-twins from family 196 into gnotobiotic mice fed Malawian and RUTF diets.
(A) Discordant weight loss in recipient mice, n=10 mice/group, *p<0.05, Student’s t-test. Data points are colored by recipient group; blue, kwashiorkor co-twin fecal microbiota recipients; red, healthy co-twin fecal microbiota recipients. (B) Average ± SEM PC1 coordinate obtained from the weighted UniFrac distances shown in fig. S9A, B for fecal microbiota sampled from mice over time. Same color key as in panel A. (C) Heatmap of phylotypes assigned to species-level taxa whose representation in the fecal microbiota of gnotobiotic mice change significantly (p<0.05, Students t-test with Bonferroni correction) as a function of donor microbiota and Malawian versus RUTF diets. An asterisk indicates taxa that changed significantly in both healthy and kwashiorkor microbiota transplant recipients. Species level taxa are colored by phylum: Firmicutes (red), Actinobacteria (blue), Bacteroidetes (black) and Proteobacteria (green). Switching from a Malawian diet to RUTF produces a rapid change in the configuration of the transplanted kwashiorkor microbiota. A bloom in Lactobacilli occurs early during treatment with RUTF but regresses by the end of this diet period and remains unchanged when animals are returned to the Malawian diet. Bifidobacterium spp also bloom early during administration of RUTF. Unlike the Lactobacilli, their increase is sustained into the early phases of the second Malawian diet period (M2) after which they diminish. Like the members of Bifidobacterium, R. torques increases its representation during RUTF and then rapidly diminishes when mice returned to a Malawian diet. The increase in F. prausnitzii is sustained into and through M2. The responses of the Bacteroidales were opposite to that of the other three groups: they decrease with RUTF and re-emerge with M2. The response of the Lactobacilli observed in the kwashiorkor transplant recipients is not seen in gnotobiotic mice containing the healthy co-twin’s microbiota. The pattern of change of the two Ruminococcus spp., B. uniformis, P. distasonis, B. longum and an unclassified Bifidobacterium taxon are shared by both recipient groups (healthy and kwashiorkor), although the Bifidobacterium response is more diminutive in the healthy microbiota treatment group. Parabacteroides merdae, an unclassified taxon from the genus Faecalibacterium, as well as a member of the Coriobacteriaceae are specifically elevated in the healthy co-twin’s microbiota when mice switch to a RUTF diet. Of these, only P. merdae does not persist when animals are returned to the Malawian diet (also see table S7A and table S8A).
Three weeks after gavage, mice consuming the Malawian diet were switched to the RUTF given to children with SAM. At the time of the diet switch, all recipients of kwashiorkor microbiota from families 196 and 57 had become severely anorectic. All mice in each treatment group rapidly gained weight while consuming RUTF. In the case of the most discordant set of recipients (from family 196), mice with the kwashiorkor co-twin’s microbiota did not achieve the same body weight as recipients of the healthy sibling’s microbiota, but did reach 96.8%±2.8% of their pre-gavage weight. After two weeks on RUTF, all mice in all treatment groups were returned to the Malawian diet. While all recipients of microbiota transplants lost weight, this re-exposure did not produce the profound weight loss that mice colonized with the kwashiorkor microbiota had experienced during their first exposure (Fig. 2A). These results indicate that the gut microbiota from two of the three discordant pairs are able to transmit a discordant malnutrition phenotype, manifested by weight loss, to recipient gnotobiotic mice. Given that the most discordant weight loss phenotype was produced by microbiota from twin pair 196, we initiated in-depth time-series analyses of the organismal, gene, and metabolite content of their transplanted microbial communities as a function of co-twin donor and diet (fig. S6 and table S2B, C).
Transplantation was efficient: (i) 62 of 72 species-level taxa present in the input community from the healthy co-twin and 58 of 67 species-level taxa from the kwashiorkor co-twin were detected in fecal microbiota collected from all transplant recipients across time points and diets (table S7A, B); (ii) 90.6% and 89.6% of the 859 ECs detected in each of the healthy and kwashiorkor input communities were identified in the fecal microbiota of transplant recipients after three weeks on the Malawian diet; and (iii) the proportional representation of ECs in input versus output fecal communities was highly correlated, (R2=0.893–0.936; fig. S7A, B). PCR-Luminex assays (15–18) for 22 common bacterial, parasitic, and viral enteropathogens in the input human microbiota as well as in recipient mouse fecal samples indicated that the markedly discordant weight loss phenotype in recipients of these microbiota was not due to transfer and/or subsistence of any of the surveyed pathogens (fig. S8A, B and supplementary online text).
Comparing the two groups of gnotobiotic recipients while they consumed a Malawian diet disclosed significant differences in the proportional representation of 37 species-level taxa. Organisms with the most statistically significant differences, and whose relative proportion were higher in mice with the kwashiorkor microbiota were Bilophila wadsworthia, a hydrogen-consuming, sulfite-reducing organism related to members of Desulfovibrio (phylum Proteobacteria) that has been linked to inflammatory bowel disease (IBD) in humans, and induces a pro-inflammatory T-helper 1 response in a mouse model of IBD (19), and Clostridium innocuum, a gut symbiont that can function as an opportunist in immunocompromised hosts (20) (table S8B). B. wadsworthia and members of the order Clostridiales were also overrepresented in the fecal microbiota of the kwashiorkor co-twin from family 196 compared to his healthy co-twin at the time he presented with kwashiorkor (table S8B).
Switching from a Malawian diet to RUTF produced a rapid change in configuration of the fecal microbiota that was most pronounced in recipients of the kwashiorkor co-twin’s community (Fig. 2B and fig. S9A, B). Thirty species-level taxa exhibited significant changes in their representation in kwashiorkor microbiota transplant recipients (Fig. 2C, tables S7B and S8A) with prominent increases in Bifidobacteria (B. longum, B. bifidum, plus another unclassified taxon), two Lactobacilli [L. reuteri and L. gasseri, which can produce bacteriocins and stimulate the innate immune system to inhibit the growth and eliminate various enteropathogens (21–23)], plus two members of Ruminococcus [R. torques, a mucus degrader (24), and Faecalibacterium prausnitzii, a member of the order Clostridiales that exhibits anti-inflammatory activity in a mouse model of colitis, and whose decreased representation is associated with increased risk of ileal Crohn’s disease (25)]. There were statistically significant decreases in the representation of members of the Bacteroidales (B. uniformis, Parabacteroides distasonis, plus an unclassified Parabacteroides taxon) (see Fig. 2C and legend for time courses). Twenty-eight bacterial species-level taxa also exhibited significant changes in their representation in gnotobiotic mice harboring the healthy co-twin’s microbiota in response to RUTF. The pattern of change of 13 different taxa including the two Ruminococcus spp., B. uniformis, P. distasonis, B. longum and an unclassified Bifidobacterium taxon were shared by both recipient groups (healthy and kwashiorkor), although the Bifidobacterium response was less pronounced in the healthy microbiota treatment group (Fig. 2C, table S7A, B and table S8A). These changes were representative of those that occurred in the human donors: the change in Bifidobacterium was unique to the kwashiorkor co-twin (table S9).
Metabolic profiles associated with kwashiorkor
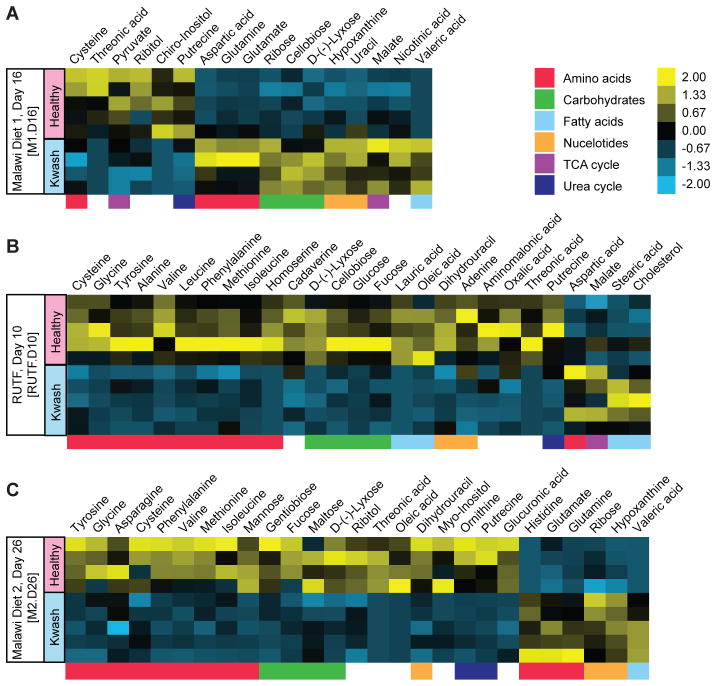
GC-MS analyses of short chain fatty acids (SCFA) and 69 other products of carbohydrate, amino acid, nucleotide and lipid (fatty acid) metabolism in cecal and fecal samples collected during the different diet periods (n=4–5 mice/family 196 microbiota donor) showed that levels of the majority of these metabolites increased when mice were switched to RUTF. In contrast, levels of several di- and monosaccharides (maltose, gentibiose and tagatose) decreased (fig. S10). There were a number of significant differences between the two groups of mice while they were consuming the different diets (Fig. 3). While switching to RUTF produced a significant increase in fecal levels of six essential amino acids (valine, leucine, isoleucine, methionine, phenylalanine, threonine) and three nonessential amino acids (alanine, tyrosine, serine) in both groups, the response was initially greater in the kwashiorkor group. Four weeks after returning to a Malawian diet, levels of six of these amino acids remained higher in the healthy microbiota recipient group than before RUTF but in the kwashiorkor group they fell to pre-RUTF treatment levels (fig. S11A, B). The same pattern of transient response was observed with urea cycle intermediates in the kwashiorkor group (fig. S11C). RUTF-associated increases in levels of propionate, butyrate, lactate and succinate were generally greater in mice harboring the healthy co-twin’s microbiota (fig. S12A). Similarly, acetate levels were elevated early on during RUTF in the healthy but not the kwashiorkor microbiota (fig. S12A). Increases in these end products of fermentation were accompanied by reductions in the levels of a number of mono- and disaccharides (fig. S12B). The observed differences in metabolic profiles were not attributable to differences in microbial community biomass: there were no statistically significant differences in fecal DNA content between recipients of the healthy and kwashiorkor co-twin microbiota when assayed at the midpoint of RUTF treatment [986.5±108.3 (mean±SEM) versus 903.2 ±97.7 ng DNA/mg feces, respectively; p=0.22, Student’s t-test] or 4 weeks after cessation of treatment (559.7±62.2 versus 651.6 ±98.9 ng DNA/mg feces, respectively; p=0.44; for characterization of gnotobiotic recipients of family 57 transplants, including taxonomic and metabolic responses to the different diets that they share with family 196 recipients, see supplementary online text, fig. S7C, D, table S7C, D, table S8C, D and table S9).
Fig. 3. Metabolites with significant differences in their fecal levels in gnotobiotic mice colonized with microbiota from discordant twin pair 196 as a function of diet.
Data are from fecal samples collected three days before the end of (A) the first period of consumption of the Malawian diet (M1, day 16; abbreviated M1.D16), (B) RUTF treatment (RUTF.D10), and (C) the second period of Malawian diet consumption (M2.D26). Significant differences are defined as p<0.05 according to Student’s t-test. Procrustes analysis of data obtained from the transplanted microbiota from discordant co-twins in family 196 (fig. S13) revealed significant correlation between metabolic and taxonomical profiles on each diet with an overall goodness of fit (M2 value) of 0.380 (p<0.0001; 1,000 Monte Carlo label permutations) for all diets and microbiota.
Microbial-host co-metabolism as a function of donor microbiota and host diet
Urine metabolite profiles were generated using standard 1H NMR spectroscopy (26) (Table 1, fig. S15, and supplementary online text). A pronounced metabolic shift seen in response to RUTF was not sustained on re-introduction of the Malawian diet; urinary metabolic profiles at the end of the second Malawian diet period (M2) resembled those from the first period (M1), with renewed differentiation between healthy and kwashiorkor microbiota transplant groups. The kwashiorkor microbiota-associated metabolic phenotype was not as distinctive in M2 as it was during M1. When the data were reanalyzed after excluding the RUTF samples, the metabolic differentiation was more apparent between the two Malawian dietary periods (fig. S15).
Table 1. Metabolite analysis of urine samples obtained from mice with transplanted healthy or kwashiorkor co-twin microbiota from family 196 at each diet phase.
R2X represents the variation in 1H-NMR spectral data explained by the O-PLD-DA model, where a value of 1 would indicate 100% of the variation in the spectral dataset is explained by the model. Q2Y represents the predictive ability of the model and is calculated by leaving a percentage of the data out (15%) while calculating the ability of the model to discriminate between classes in any pairwise comparison (e.g., kwashiorkor versus healthy) and indicates how robust or significant the metabolic differences between two classes are. The numbers in each column are obtained from O-PLS-DA models and represent the correlation values between the NMR data and a given class. Metabolites are colored according to their overrepresentation in a treatment group. (A) Urinary metabolites with differences in their levels between mice transplanted with the healthy co-twin versus the kwashiorkor co-twin microbiota within a given diet. Color code: blue, higher in kwashiorkor co-twin microbiota recipients; red, higher in healthy co-twin microbiota recipients; white, no significant difference between the two classes. (B) Urinary metabolites with differences in their representation in mice transplanted with healthy or kwashiorkor co-twin microbiota between diets. Color code: red, higher during the M1 diet phase relative to RUTF or relative to M2; orange, higher on RUTF relative to M1 or M2; blue, higher on M2 compared to RUTF or M1); white, no significant differences in the comparison.
| A | M1 | RUTF | M2 |
|---|---|---|---|
|
| |||
| R2X | 0.38 | 0.49 | 0.36 |
|
| |||
| Q2Y | 0.62 | 0.8 | 0.78 |
|
| |||
| 2-oxoadipate | 0.8747 | 0.9069 | 0.6603 |
| taurine | 0.8578 | 0.6236 | |
| lactate | 0.4618 | ||
| creatine | 0.9303 | ||
| creatinine | 0.7692 | ||
| methylamine | 0.6755 | 0.8338 | 0.7646 |
| dimethylamine | 0.7732 | ||
| trimethylamine | 0.8469 | ||
| trimethylamine N-oxide | |||
| phenylacetylglycine | 0.8522 | 0.8816 | |
| indoxyl sulfate | 0.6591 | 0.8138 | |
| hippurate | 0.6301 | 0.9485 | 0.9786 |
| allantoin | 0.7662 | ||
| B | Recipients of healthy co-twin donor microbiota | Recipients of kwashiorkor co-twin donor microbiota | ||||
|---|---|---|---|---|---|---|
|
| ||||||
| M1 vs RUTF | RUTF vs M2 | M1 vs M2 | M1 vs RUTF | RUTF vs M2 | M1 vs M2 | |
| R2X | 0.66 | 0.58 | 0.61 | 0.74 | 0.52 | 0.67 |
|
| ||||||
| Q2Y | 0.57 | 0.94 | 0.31 | 0.87 | 0.93 | 0.68 |
|
| ||||||
| 2-oxoglutarate | 0.7448 | 0.8538 | 0.9062 | 0.8761 | ||
| citrate | 0.7065 | 0.7921 | 0.827 | 0.7137 | ||
| succinate | 0.6794 | 0.6841 | 0.8348 | 0.846 | ||
| fumarate | 0.7439 | 0.7553 | ||||
| acetate | 0.7 | |||||
| 2-oxoadipate | 0.8382 | 0.6198 | ||||
| taurine | 0.632 | 0.7469 | 0.5998 | |||
| lactate | 0.5558 | 0.4733 | ||||
| creatine | 0.7432 | 0.9474 | 0.759 | 0.8781 | ||
| creatinine | 0.8297 | 0.7051 | 0.6838 | |||
| methylamine | 0.7562 | |||||
| dimethylamine | 0.7469 | |||||
| trimethylamine | 0.7149 | 0.6639 | 0.8534 | 0.878 | ||
| trimethylamine N-oxide | 0.799 | |||||
| phenylacetylglycine | 0.8447 | 0.5716 | 0.6616 | |||
| indoxyl sulfate | 0.8021 | 0.8699 | 0.9593 | |||
| hippurate | 0.8602 | |||||
| allantoin | 0.8203 | 0.9133 | 0.93 | 0.769 | ||
| 1-methylnicotinamide | 0.7554 | |||||
 Higher metabolite concentration in recipients of kwashiorkor co-twin microbiota
Higher metabolite concentration in recipients of kwashiorkor co-twin microbiota
 Higher metabolite concentration in recipients of healthy co-twin donor microbiota
Higher metabolite concentration in recipients of healthy co-twin donor microbiota
 Higher metabolite concentration during Malawian diet phase 1
Higher metabolite concentration during Malawian diet phase 1
 Higher metabolite concentration during RUTF
Higher metabolite concentration during RUTF
 Higher metabolite concentration during Malawian diet phase 2
Higher metabolite concentration during Malawian diet phase 2
Among the notable results, we found that levels of urinary taurine were affected by both donor microbiota and diet. Mice with a healthy donor microbiota excreted more taurine while consuming both the Malawian diet and RUTF compared to mice with a kwashiorkor microbiota; in both recipient groups, urinary taurine levels were higher when mice were consuming a Malawian diet (Table 1). B. wadsworthia grows well on bile acids and uses taurine from taurine-conjugated bile acids as a terminal electron acceptor, converting it to ammonia, acetate and sulfide (27). In agreement with this property, fecal levels of B. wadsworthia showed an inverse relationship with urinary taurine levels (table S7A, B). In addition to urinary taurine levels, fecal methionine and cysteine concentrations were significantly lower in mice harboring kwashiorkor compared to healthy co-twin microbiota when consuming a Malawian diet (Fig. 3). Dietary methionine and cysteine meet most of the human body’s needs for sulfur; these amino acids are more abundant in animal and cereal proteins than in vegetable proteins. The Malawian diet is deficient in total protein and in animal protein. Studies have found a decrease of serum methionine levels and urinary sulfate excretion in patients with kwashiorkor (28–33). Moreover, cysteine or methionine deficiency in experimental animals can produce weight loss (correctable by sulfate supplementation) (34–36). Our findings suggest that the combination of a kwashiorkor microbiota and Malawian diet may contribute to abnormal sulfur metabolism, thereby impacting the pathogenesis and manifestations of this form of SAM.
For mice containing the healthy co-twin’s microbiota, urinary excretion of tricarboxylic acid (TCA) cycle intermediates 2-oxoglutarate, citrate, succinate and fumarate were closely coupled together. These mitochondrial metabolites typically follow similar reabsorption control mechanisms in the renal tubule that are closely linked to tubular pH. In mice containing kwashiorkor microbiota, urinary fumarate excretion was effectively decoupled from the other TCA intermediates (fig. S15). Differential excretion rates of TCA intermediates can occur where there is selective enzymatic inhibition of the TCA cycle (37). The TCA cycle also appears to be disrupted in the kwashiorkor microbiota itself (3-fold increase in cecal levels of succinate (p<0.05, unpaired Student’s t-test), and an increased succinate to fumarate ratio (0.58 versus 0.23, p<0.05, unpaired Student’s t-test), suggesting inhibition of succinate dehydrogenase, the enzyme responsible for converting succinate to fumarate. Taken together, these observations suggest that the kwashiorkor microbiota examined in these gnotobiotic mice may generate chemical products that result in a selective inhibition of one or more TCA cycle enzymes, making energy metabolism a bigger challenge for these children when they are exposed to a micro- and macronutrient deficient, low calorie diet.
Prospectus
The discordance rate for kwashiorkor is high for both MZ and DZ twins in our study population. Our results illustrate the value of using twins discordant for nutritional phenotypes to characterize the interrelationship between the functional development of the gut microbiome in children and their nutritional status. Linking metagenomic analyses with dietary experiments in gnotobiotic mice that have received gut microbiome transplants from twins discordant for kwashiorkor allowed us to gain insights into pathogenesis by identifying transmissible features associated with healthy versus diseased donors. By replicating a human donor’s gut community in multiple recipient mice, we have been able to mimic a clinical intervention, and identify community characteristics, including differences in taxonomic composition and differences in taxonomic and metabolic responses to RUTF. The resulting data provide biomarkers of community metabolism and of microbial-host co-metabolism that delineate and discriminate diet and microbiota effects, including biomarkers indicative of the more labile, short-lived nature of the responses of microbiota from kwashiorkor donors to RUTF. The interrelationships between diet, microbiota, and many facets of host physiology can be explored in detail in these ‘personalized’ gnotobiotic mouse models. These models may be useful for developing new and more effective approaches for treatment and/or prevention. In addition, studies of other forms of malnutrition using an approach analogous to that described in this study, could also help provide insights about the contribution of the gut microbiome to this global health problem.
Supplementary Material
Acknowledgments
We thank Sabrina Wagoner, Jill Manchester and Marty Meier for superb technical assistance, Jill Manchester, Su Deng and Jessica Hoisington-López for assistance with DNA sequencing, Maria Karlsson, David O’Donnell and Sabrina Wagoner for help with gnotobiotic mouse husbandry, Barbara Mickelson (Teklad Diets) and Heidi Sandige for assistance with the design of the mouse diets, and members of the Gordon lab for valuable suggestions during the course of this work. This work was supported by grants from the Bill & Melinda Gates Foundation, and the NIH (DK30292, DK078669, T32-HD049338). M.I.S was the recipient of a post-doctoral fellowship from the St. Louis Children’s Discovery Institute (MD112009-201). J.V.L was the recipient of an Imperial College Junior Research Fellowship. Illumina V4-16S rRNA and 454 shotgun pyrosequencing datasets have been deposited in EBI (ERP001861, ERP001871, ERP001819, ERP001909).
Footnotes
Author contributions:
M.I.S., T.Y., and J.I.G designed the experiments; M.M. designed and implemented the clinical monitoring and sampling for the trial, R.M. and I.T. participated in patient recruitment, sample collection, sample preservation and clinical evaluations; M.I.S. performed experiments involving gnotobiotic mice while T.Y. characterized microbiota obtained from twins; M.I.S., T.Y., J.C., A.L.K., S.S.R., P.C., J.C.M., J.L., E.H., J.V.L., E.H., and J.N. generated data; M.I.S., T.Y., E.H., J.N., D.K, L.K.U, R.K. and J.I.G. analyzed the results; M.I.S., T.Y., and J.I.G. wrote the paper.
Supplementary Material: www.sciencemag.org
Materials and Methods
References and notes
- 1.UN Inter-Agency Group for Child Mortality Estimation (UNIGME) Levels & Trends in Child Mortality Report. 2011 www.childinfo.org/files/Child_Mortality_Report_2011.pdf.
- 2.World Health Organization. Multicentre Growth Reference Study Group (WHO MGRSG), WHO child growth standards based on length/height, weight and age. Acta Paediatr. 2006;(Suppl 450):76–85. [Google Scholar]
- 3.World Health Organization, United Nations Children’s Fund. WHO child growth standards and the identification of severe acute malnutrition in infants and children. 2009 www.who.int/nutrition/publications/severemalnutrition/9789241598163/en/index.html. [PubMed]
- 4.Williams CD, Oxon BM, Lond H. Kwashiorkor: a nutritional disease of children associated with a maize diet. Lancet. 1935;226:1151–1152. [PMC free article] [PubMed] [Google Scholar]
- 5.Ahmed T, Rahman S, Cravioto A. Oedematous malnutrition. Indian J Med Res. 2009;130:651–654. [PubMed] [Google Scholar]
- 6.Gopalan C. In: Calorie Deficiencies and Protein Deficiencies: Kwashiorkor and marasmus: evolution and distinguishing features. McCance RA, Widdowson EM, editors. Churchill; London, UK: 1968. pp. 48–58. [Google Scholar]
- 7.Golden MH. Protein deficiency, energy deficiency, and the oedema of malnutrition. Lancet. 1982;1:1261–1265. doi: 10.1016/s0140-6736(82)92839-2. [DOI] [PubMed] [Google Scholar]
- 8.Ciliberto H, et al. Antioxidant supplementation for the prevention of kwashiorkor in Malawian children: randomised, double blind, placebo controlled trial. Br Med J. 2005;330:1109–1114. doi: 10.1136/bmj.38427.404259.8F. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lin CA, et al. A prospective assessment of food and nutrient intake in a population of Malawian children at risk for kwashiorkor. J Pediatr Gastroenterol Nutr. 2007;44:487–493. doi: 10.1097/MPG.0b013e31802c6e57. [DOI] [PubMed] [Google Scholar]
- 10.Yatsunenko T, et al. Human gut microbiome viewed across age and geography. Nature. 2012;486:222–227. doi: 10.1038/nature11053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Black RE, et al. Maternal and child undernutrition: global and regional exposures and health consequences. Lancet. 2008;371:243–260. doi: 10.1016/S0140-6736(07)61690-0. [DOI] [PubMed] [Google Scholar]
- 12.World Health Organization, World Food Programme, United Nations System Standing Committee on Nutrition, United Nations Children’s Fund, Community-Based Management of Severe Acute Malnutrition. Community-based management of severe acute malnutrition. 2007 www.who.int/nutrition/topics/statement_commbased_malnutrition/en/index.html.
- 13.Materials and methods are available as supplementary material on Science Online.
- 14.LaGrone L, Cole S, Schondelmeyer A, Maleta K, Manary MJ. Locally produced ready-to-use supplementary food is an effective treatment of moderate acute malnutrition in an operational setting. Ann Trop Paed. 2010;30:103–108. doi: 10.1179/146532810X12703901870651. [DOI] [PubMed] [Google Scholar]
- 15.Liu J, et al. Simultaneous detection of six diarrhea-causing bacterial pathogens with an in-house PCR-luminex assay. J Clin Microbiol. 2012;50:98–103. doi: 10.1128/JCM.05416-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Taniuchi M, et al. Multiplex polymerase chain reaction method to detect Cyclospora, Cystoisospora, and Microsporidia in stool samples. Diagn Microbiol Infect Dis. 2011;71:386–390. doi: 10.1016/j.diagmicrobio.2011.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Taniuchi M, et al. High throughput multiplex PCR and probe-based detection with Luminex beads for seven intestinal parasites. Am J Trop Med Hyg. 2011;84:332–337. doi: 10.4269/ajtmh.2011.10-0461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Taniuchi M, et al. Development of a multiplex polymerase chain reaction assay for diarrheagenic Escherichia coli and Shigella spp. and its evaluation on colonies, culture broths, and stool. Diagn Microbiol Infect Dis. 2012;73:121–128. doi: 10.1016/j.diagmicrobio.2012.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Devkota S, et al. Dietary-fat-induced taurocholic acid promotes pathobiont expansion and colitis in Il10(-/-) mice. Nature. 2012;487:104–108. doi: 10.1038/nature11225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Crum-Cianflone N. Clostridium innocuum bacteremia in a patient with acquired immunodeficiency syndrome. Am J Med Sci. 2009;337:480–482. doi: 10.1097/MAJ.0b013e31819f1e95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Itoh T, Fujimoto Y, Kawai Y, Toba T, Saito T. Inhibition of food-borne pathogenic bacteria by bacteriocins from Lactobacillus gasseri. Lett Appl Microbiol. 21:137–141. doi: 10.1111/j.1472-765x.1995.tb01025.x. [DOI] [PubMed] [Google Scholar]
- 22.Fernández MF, Boris S, Barbés C. Probiotic properties of human lactobacilli strains to be used in the gastrointestinal tract. J Appl Microbiol. 2003;94:449–55. doi: 10.1046/j.1365-2672.2003.01850.x. [DOI] [PubMed] [Google Scholar]
- 23.Kato-Mori Y, et al. Fermentation metabolites from Lactobacillus gasseri and Propionibacterium freudenreichii exert bacteriocidal effects in mice. J Med Food. 2010;13:1460–1467. doi: 10.1089/jmf.2010.1137. [DOI] [PubMed] [Google Scholar]
- 24.Salyers AA, West SE, Vercellotti JR, Wilkins TD. Fermentation of mucins and plant polysaccharides by anaerobic bacteria from the human colon. Appl Environ Microbiol. 1977;34:529–33. doi: 10.1128/aem.34.5.529-533.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sokol H, et al. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn’s disease patients. Proc Natl Acad Sci USA. 2008;105:16731–16736. doi: 10.1073/pnas.0804812105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Beckonert O, et al. Metabolic profiling, metabolomic and metabonomic procedures for NMR spectroscopy of urine, plasma, serum and tissue extracts. Nature Protoc. 2007;2:2692–2703. doi: 10.1038/nprot.2007.376. [DOI] [PubMed] [Google Scholar]
- 27.Laue H, Denger K, Cook AM. Taurine reduction in anaerobic respiration of Bilophila wadsworthia RZATAU. Appl Environ Microbiol. 1997;63:2016–2021. doi: 10.1128/aem.63.5.2016-2021.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Edozien JC, Phillips EJ, Collis WFR. The free amino acids of plasma and urine in kwashiorkor. Lancet. 1960;1:615–618. doi: 10.1016/s0140-6736(60)90502-x. [DOI] [PubMed] [Google Scholar]
- 29.Whitehead RG, Dean RF. Serum amino acids in kwashiorkor. I. Relationship to clinical condition. Am J Clin Nutr. 1964;14:313–319. doi: 10.1093/ajcn/14.6.313. [DOI] [PubMed] [Google Scholar]
- 30.Awwaad S, Eisa EA, El-Essawy M. Methionine metabolism in kwashiorkor in Egyptian children. J Trop Med Hyg. 1962;65:179–181. [PubMed] [Google Scholar]
- 31.Arroyave G, Wilson D, Funes C, Behar M. The free amino acids in blood plasma of children with kwashiorkor and marasmus. Am J Clin Nutr. 1962;11:517–524. [Google Scholar]
- 32.Rlttyerah T, Pereira SM, Dumm ME. Serum amino acids of children on high and low protein intakes. Am J Clin Nutr. 1965;17:11–4. doi: 10.1093/ajcn/17.1.11. [DOI] [PubMed] [Google Scholar]
- 33.Ittyerah TR. Urinary excretion of sulfate in kwashiorkor. Clin Chim Acta. 1969;25:365–369. doi: 10.1016/0009-8981(69)90194-6. [DOI] [PubMed] [Google Scholar]
- 34.Baker DH. Utilization of isomers and analogs of amino acids and other sulfur- containing compounds. Prog Food Nutr Sci. 1986;10:133–178. [PubMed] [Google Scholar]
- 35.Orentreich N, Matias JR, Defelice A, Zimmerman JA. Low methionine ingestion by rats extends life span. J Nutr. 1993;123:269–274. doi: 10.1093/jn/123.2.269. [DOI] [PubMed] [Google Scholar]
- 36.Mori M, Manabe S, Uenishi K, Sakamoto S. Nutritional improvements of soy protein isolate by different levels of methionine supplementation in pregnant rats. Tokushima J Exp Med. 1993;40:35–42. [PubMed] [Google Scholar]
- 37.Nicholson JK, Timbrell JA, Sadler PJ. Proton NMR spectra of urine as indicators of renal damage. Mercury-induced nephrotoxicity in rats. Mol Pharmacol. 1985;27:644–651. [PubMed] [Google Scholar]
- 38.Manary MJ. Local production and provision of ready-to-use therapeutic food (RUTF) spread for the treatment of severe childhood malnutrition. Food Nutr Bull. 2006;27(3 Suppl):S83–S89. doi: 10.1177/15648265060273S305. [DOI] [PubMed] [Google Scholar]
- 39.McPeek MS, Sun L. Statistical tests for detection of misspecified relationships by use of genome-screen data. Am J Hum Genet. 2000;66:1076–1094. doi: 10.1086/302800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Turnbaugh PJ, et al. A core gut microbiome in obese and lean twins. Nature. 2009;457:480–484. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Caporaso JG, et al. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 2012;6:1621–1624. doi: 10.1038/ismej.2012.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Caporaso JG, et al. QIIME allows analysis of high-throughput community sequencing data. Nature Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McNulty NP, et al. The impact of a consortium of fermented milk strains on the gut microbiome of gnotobiotic mice and monozygotic twins. Science Transl Med. 2011;3:106ra106. doi: 10.1126/scitranslmed.3002701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pinheiro J, Bates D, DebRoy S, Sarkar D the R Development Core Team. nlme: Linear and Nonlinear Mixed Effects Models. R package version 3.1–104. 2012;2012 [Google Scholar]
- 45.Chen M, et al. Metabonomic study of aristolochic acid-induced nephrotoxicity in rats. J Proteome Res. 2006;5:995–1002. doi: 10.1021/pr050404w. [DOI] [PubMed] [Google Scholar]
- 46.Cheng J, Yuan C, Graham TL. Potential defense-related prenylated isoflavones in lactofen-induced soybean. Phytochemistry. 2011;72:875–881. doi: 10.1016/j.phytochem.2011.03.010. [DOI] [PubMed] [Google Scholar]
- 47.Veselkov KA, et al. Recursive segment-wise peak alignment of biological 1H NMR spectra for improved metabolic biomarker recovery. Anal Chem. 2009;81:56–66. doi: 10.1021/ac8011544. [DOI] [PubMed] [Google Scholar]
- 48.World Health Organization, United Nations Children’s Fund. Integrated management of childhood illnesses: caring for newborns and children in the community. 2011 whqlibdoc.who.int/publications/2011/9789241548045_Manual_eng.pdf.
- 49.Breiman L. Random forests. Machine Learning J. 2001;45:5–32. [Google Scholar]
- 50.Albert MJ, Faruque AS, Faruque SM, Sack RB, Mahalanabis D. Case-control study of enteropathogens associated with childhood diarrhea in Dhaka, Bangladesh. J Clin Microbiol. 1999;37:3458–3464. doi: 10.1128/jcm.37.11.3458-3464.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Taras D, Simmering R, Collins MD, Lawson PA, Blaut M. Reclassification of Eubacterium formicigenerans Holdeman and Moore 1974 as Dorea formicigenerans gen. nov., comb. nov., and description of Dorea longicatena sp. nov., isolated from human faeces. Int J Syst Evol Microbiol. 2002;52:423–428. doi: 10.1099/00207713-52-2-423. [DOI] [PubMed] [Google Scholar]
- 52.Wensinck F, van de Merwe JP, Mayberry JF. An international study of agglutinins to Eubacterium, Peptostreptococcus and Coprococcus species in Crohn’s disease, ulcerative colitis and control subjects. Digestion. 1983;27:63–69. doi: 10.1159/000198931. [DOI] [PubMed] [Google Scholar]
- 53.Subcommittee on Laboratory Animal Nutrition, Committee on Animal Nutrition, Board on Agriculture, National Research Council. Nutrient Requirements of Laboratory Animals, Fourth Revised Edition, 1995. 4. The National Academies Press; 1995. [PubMed] [Google Scholar]
- 54.Chaparro CM, Dewey KG. Use of lipid-based nutrient supplements (LNS) to improve the nutrient adequacy of general food distribution rations for vulnerable sub-groups in emergency settings. Matern Child Nutr. 2010;6(Suppl 1):1–69. doi: 10.1111/j.1740-8709.2009.00224.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.