Abstract
A complex network of transcription factors contributes to the establishment and maintenance of the osteoblastic phenotype. Although relatively few transcription factors, such as Runx2 and osterix, are essential to the process of osteoblastic differentiation, others serve the purpose of fine-tuning in response to various environmental and hormonal cues. The nuclear receptor (NR) superfamily of transcription factors are involved in numerous aspects of bone biology. In this study, we characterized the expression pattern of the entire NR superfamily in differentiating primary murine calvarial cells in order to identify novel NR regulatory patterns. Dynamic patterns of NR expression were observed throughout the differentiation process. Interestingly, retinoic acid receptor-related orphan receptor β(Rorβ) expression was markedly suppressed at later stages of differentiation. To gain further insight into the function of NRs in bone biology, the NR superfamily was also profiled in mouse bone marrow precursor cells isolated from either young (6 month) or aging, osteoporotic (18–22 month) mice. Of interest, Rorβ was potently overexpressed in the aged cohort. Collectively, these data provided evidence that Rorβ expression is inversely correlated with osteogenic potential, suggesting Rorβ may be an important and unexplored regulator of osteogenesis. To validate this hypothesis, a cell model stably expressing Rorβ in mouse osteoblastic MC3T3-E1 cells was produced (MC3T3-Rorβ). These cells displayed markedly suppressed bone nodule formation as well as reduced osteocalcin and osterix gene expression. Since these genes are Runx2 targets, we reasoned that Rorβ may interfere with Runx2 activity. Consistent with this, transient transfection analysis demonstrated that Rorβ inhibited Runx2-dependent activation of a Runx2-reporter construct. In summary, our data provide a comprehensive profile of NR expression during osteoblast differentiation and identify Rorβ as a novel regulator of osteogenesis and potentially of age-related bone loss through antagonism of Runx2 activity.
Keywords: Nuclear receptor, Rorβ, osteoblast, Runx2, mineralization, aging
Introduction
Osteoblast differentiation is a complex, multistep process involving the coordinate actions of numerous genes and proteins. It is initiated through the induction and secretion of a variety of extracellular matrix components that eventually become mineralized (1). Many of the genes controlling this process have been identified using molecular, cellular and animal models (2,3). Transcription factors such as Runx2 and osterix are considered master controller genes in bone, as deletion of either in mice results in a dramatic phenotype where the cartilaginous skeleton fails to ossify during embryonic development, leading to a complete lack of mineralized bone (4–7). Perturbations in the process of osteoblast differentiation lead to diseases such as osteoporosis, and understanding the molecular mechanisms that modulate this process will undoubtedly lead to the generation of more effective treatment modalities.
Nuclear receptors (NRs) are a large group of ligand-inducible transcription factors that regulate many facets of mammalian physiology. Alterations in NR signaling have been implicated in diseases such as diabetes, heart disease, cancer and osteoporosis (8–11). This family includes receptors for steroid and thyroid hormones, as well as retinoids and vitamin D. In addition, there exists a large group of NRs that lack identified ligands and are termed orphan receptors (12). NRs share several functional and structural domains: a highly variable N-terminal transactivation function, a central zinc finger DNA binding domain and a C-terminal ligand binding domain involved in ligand binding, dimerization and transactivation. The molecular function of NRs is typically achieved through the modulation of gene expression at the transcriptional level, through either direct protein-DNA interactions via NR response elements, or indirectly through protein-protein interactions with other DNA-bound transcription factors (13). Examination of the phenotypes in mouse models deficient for select members of the NR superfamily have revealed alterations in bone metabolism, establishing bone as an endocrine-sensitive organ. However, the complete gene expression patterns of the NR superfamily throughout normal osteoblast differentiation have not been extensively characterized.
In this report, we investigated the expression patterns of the entire NR superfamily during the process of osteoblastic differentiation in a primary mouse calvarial osteoblast system and in an aging mouse model of bone loss. The data reveal significant alterations in NR expression throughout osteoblast differentiation and with aging, as well as identify novel genes which may play important roles in bone metabolism. As a result of this analysis, we identify Rorβ as an important and previously unrecognized regulator of osteogenesis.
Materials and Methods
Cell culture and differentiation
Isolation of primary mouse calvarial osteoblasts and bone marrow stromal cells from C57BL/6 mice were as previously described (14). Cells were maintained in αMEM growth medium (Invitrogen, Carlsbad, CA) supplemented with 1X antibiotic/antimycotic (Invitrogen) and 10% (v/v) fetal bovine serum (Hyclone, Logan, UT). For the osteoblast differentiation assays, passage 4 cells were plated at a density of 104 cells/cm2 in 6-well plates (Corning Incorporated Life Sciences, Lowell, MA) in αMEM growth medium (n=4). At confluence the media was replaced with growth medium supplemented with 50 mg/L ascorbic acid and 10 mM β-glycerophosphate (Sigma-Aldrich, St. Louis, MO). MC3T3-E1 mouse osteoblasts were cultured and treated identically to the primary mouse calvarial osteoblasts except the osteoblast differentiation medium was supplemented with 100 μg/mL recombinant Bmp2 (R&D Systems, Minneapolis, MN) unless otherwise noted in the text. U2OS cells were cultured as previously described.
Alizarin red staining
Alizarin red staining was done as previously described (14). Briefly, cells were washed in 1X PBS and fixed in 3.75% paraformaldehyde overnight at room temperature. Following two 1X PBS washes, the cells were stained with 1.2% Alizarin red (v/v) (Sigma-Aldrich) pH 4.2 for 20 minutes. The cells were extensively washed with 1X PBS and scanned.
RNA isolation and quantitative RT-PCR (QPCR)
Total cellular RNA was harvested at the indicated times following induction of osteoblast differentiation using QIAzol Lysis Reagent and RNeasy Mini Columns (Qiagen, Valencia, CA). DNase treatment was performed to degrade potential contaminating genomic DNA using an on-column RNase-free DNase solution (Qiagen). Three μg of total RNA was used in a reverse transcriptase (RT) reaction using the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems by Life Technologies, Foster City, CA) according to manufacturer instructions. The RT reactions were diluted 1:5 and 1 μl used in a 10 μl total reaction volume for real-time quantitative PCR (QPCR) using the QuantiTect SYBR Green PCR Kit (Qiagen) and the ABI 7900HT Fast Real-Time PCR System (Applied Biosystems). All primers were designed using Primer Express® Software Version 3.0 (Applied Biosystems). The nuclear receptor, bone marker and reference gene primer sequences used in this study can be found in Supplemental Tables 1–3, respectively.
Normalization for variations in input RNA was performed using a panel of 9 reference genes [18S, glucose-6-phosphate dehydrogenase (G6pdh), glyceraldehyde-3-phosphate dehydrogenase (Gapdh), hypoxanthine guanine phosphoribosyl transferase (Hprt), ribosomal protein L13A (Rpl13A), polymerase (RNA) II (DNA directed) polypeptide A (Polr2a), TATA binding protein (Tbp), tubulin alpha 1a (Tubα1a) and β2-microglobulin (B2m)] using the geNorm algorithm to select the 3 most stable reference genes (15,16). The PCR Miner algorithm was used to correct for variations in amplification efficiencies (17). The median cycle threshold (Ct) for each gene in each sample was normalized to the geometric mean of the median Ct of the reference genes as determined by the geNorm algorithm using the formula: . The resulting ΔCt for each gene was used to calculate relative gene expression changes between samples. Analysis of gene expression in hematopoietic lineage negative (lin-) bone marrow cells, a mesenchymal-enriched cell fraction (18) from young (6 month) and aged (18–22 month) mice (Fig. 4), was performed using cDNA samples (n=7–8) derived from a previous study in our laboratory using the methods described therein (19).
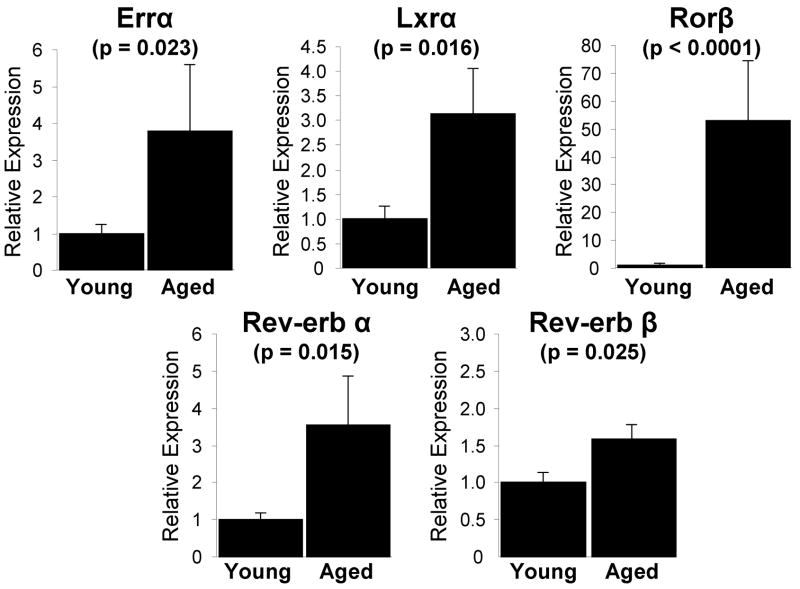
Fig. 4.
NR expression in bone marrow lin- cells in aged mice. Samples from a previous study (19), from which cDNAs from the lin- cell population in femoral bone marrow of young (6 month) and aged (18–22 month) mice were generated, were analyzed for expression of the NR superfamily using QPCR (n=7–8). Only those genes exhibiting statistically significant expression changes (p ≤ 0.05, Student’s t-test) are indicated. The bars represent fold-induction relative to the young mouse cohort. The data are presented as the mean ± SE and p-values are indicated.
Hierarchal clustering and tree analysis
Unsupervised cluster analysis was performed essentially as previously described (20). Briefly, the mean gene expression fold-changes for each gene were adjusted using log transformation to center the data around 0 and normalized to set the magnitude of the control value (day 0) for each gene to 1 using Gene Cluster 3.0 (http://rana.lbl.gov/eisen/). The adjusted data were clustered by calculating Pearson’s centered correlation coefficients followed by average linkage analysis in the Gene Cluster 3.0 program. Expression heatmaps, which visually describe the cluster results, were generated using TreeView (http://rana.lbl.gov/eisen/). The red shades represent genes upregulated and green shades represent genes downregulated relative to the control value (Supplemental Fig. 1).
Production of the MC3T3-GFP and –Rorβ-GFP cell models
A Rorβ expression construct, pCMV6-Rorβ (Origene, Rockville, MD), was cloned as an EcoRI/MluI fragment into a vector coexpressing GFP under the control of an internal ribosome entry sequence (data not shown). A vector expressing only GFP was used as a control. These vectors were electroporated into MC3T3-E1 cells using the Neon Transfection System (Invitrogen) and selected with 400 ug/mL G418 antiobiotic (Invitrogen). Following 2 weeks of cell selection and expansion, fluorescence-activated cell sorting was used to isolate the GFP-expressing population (essentially as described in (21)) and the cells were again expanded resulting in the MC3T3-GFP and MC3T3–Rorβ-GFP cell models.
Transient transfection
U2OS cells were plated in 6-well plates at a density of 2.6 × 104 cells/cm2 the day before transfection. Five-hundred (500) ng of pCMV6-Rorβ (Origene), p6OSE2-Luc (22), and pCMV-Cbfa1/Runx2 (4) constructs each were transiently transfected (n=6) using X-tremeGENE9 transfection reagent (Roche Diagnostics, Indianapolis, IN). Following incubation at 37°C for 48 hours, cells were harvested in 1X Passive Lysis Buffer and equal quantities of protein extracts were assayed using Luciferase Assay Reagent on a GloMax® 96 Microplate Luminometer (Promega, Madison, WI). Protein concentrations were determined using a BCA Protein Assay Kit (Thermo Scientific, Rockford, IL).
siRNA Studies
MC3T3-E1 cells were transfected in 24-well plates at a density of 2.6 × 104 cells/cm2 using HyperFect Transfection Reagent (Qiagen) with either a non-specific siRNA control (AllStars Negative Control siRNA) or a mouse-specific Rorβ siRNA (Qiagen) at a concentration of 33 nM according to the manufacturer’s protocol. Following incubation at 37°C for 48 hours, cells were collected and QPCR analysis was performed as described above.
Statistical Analyses
Calculations and statistical analyses were performed using Microsoft ® Office Excel 2003 (Microsoft Corporation, Redmond, WA). The data are presented as the mean ± standard error (SE) and comparisons between groups were performed on the log-transformed data using Student’s t-test. All p-values ≤ 0.05 were considered statistically significant.
Results
Osteoblastic differentiation of primary mouse calvarial cells is associated with increases in bone marker and osteocytic gene expression
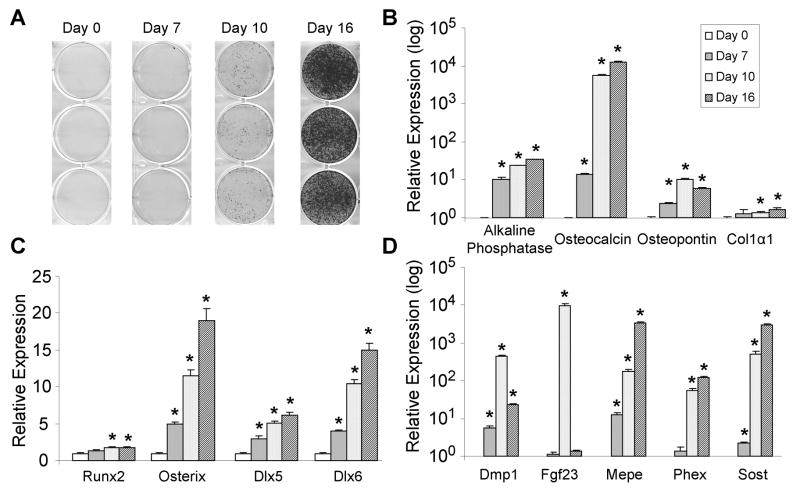
Primary mouse calvarial cells were chosen for this study since they represent a well known osteoblast model system that exhibits robust mineralization and increases in bone marker gene expression following induction of differentiation by ascorbate and β-glycerophosphate. Differentiation was induced at confluence and samples collected at day 0, 7, 10 and 16 and assayed for calcium deposition by Alizarin red staining and osteoblast gene expression by quantitative PCR (QPCR). Robust Alizarin red-positive nodule formation is observed at day 10 and 16 following the induction of differentiation (Fig. 1A), indicative of a highly mineralizing cell population. Gene expression profiles of classic osteoblast marker genes [(alkaline phosphatase, osteocalcin, osteopontin and collagen, type I, alpha (Col1α1)] involved in formation of the secreted glycoprotein matrix were increased compared to day 0 (Fig. 1B), confirming the production of a highly osteogenic cell population. Transcriptional regulation of osteoblastic differentiation is a tightly controlled process involving Runx2, osterix and Dlx5/6 (2,3,14), and as is observed in Fig. 1C, expression of these genes is increased at nearly all timepoints. Due to the high expression of these bone marker genes at day 16, we surmised these cells may be starting to exhibit an osteocytic phenotype. Remarkably, large increases in the well-established osteocytic markers dentin matrix protein 1 (Dmp1), fibroblast growth factor 23 (Fgf23), matrix extracellular phosphoglycoprotein (Mepe), phosphate regulating endopeptidase homolog, X-linked (Phex) and sclerostin (Sost) were observed at the later stages of differentiation (Fig. 1D). Collectively, these data indicate that differentiation of primary mouse calvarial cells leads to marked increases in mineralization, as well as activation of genes involved in osteoblast and osteocyte biology.
Fig. 1.
Bone-marker and osteocytic gene expression during differentiation of primary mouse calvarial osteoblasts. (A) Osteoblastic differentiation of primary mouse calvarial osteoblasts was induced at confluence and mineralization was determined by Alizarin red stain at the indicated timepoints. QPCR analyses of genes involved in, (B) forming the extracellular bone matrix, (C) osteoblastic transcriptional regulation, and (D) osteocyte biology were performed from samples harvested at the indicated timepoints (n=4). The bars represent fold-induction relative to the expression at day 0 for each gene. The data are presented as the mean ± SE and an asterisk (*) represents statistical significance of p ≤ 0.01 (Student’s t-test).
Expression of the NR superfamily during osteoblastic differentiation
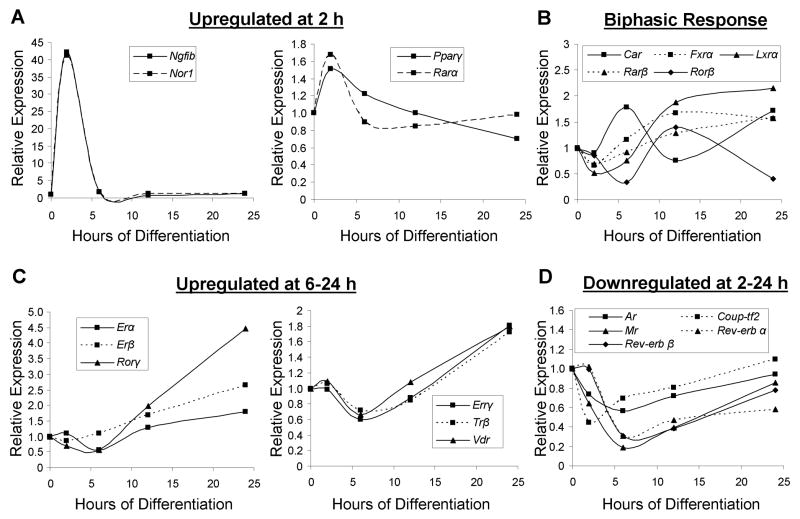
Previous analyses of NR superfamily expression have been performed on the adipocytic cell line 3T3-L1 during differentiation (23), on an array of mouse tissues (24), during macrophage activation (25) and on a differentiating ES cell population (20). As a first step to understanding the influence of osteoblastic differentiation on NR gene expression, we examined the mRNA expression patterns of the 49 NRs during the first 24 hours following addition of osteoblast differentiation media. Analysis of the entire NR superfamily revealed that 35 were expressed at either 2 or 24 hours following osteoblastic induction; whereas 14 were not expressed at any timepoint using the previously published threshold cutoff of Ct ≥ 33 (23). The expression patterns of the NRs were subjected to hierarchal cluster analysis and a heatmap was generated, further subdividing the genes as either upregulated or downregulated (Supplemental Fig. S1A). Those genes exhibiting statistical significance (p≤0.05) and a fold-change ≥ 1.5 compared to time 0 were reassayed using QPCR on an expanded timecourse (0, 2, 6, 12, and 24 hours) to generate a more detailed profile of gene expression. Comparison of the temporal patterns of NR gene expression revealed a group of four transiently upregulated genes at 2 hours, including nerve growth factor-induced gene B (Ngfib), neuron-derived orphan receptor 1 (Nor1), peroxisome proliferator-activated receptor gamma (Pparγ), and retinoic acid receptor alpha (Rarα) (Fig. 2A). It is of interest that Pparγ, the main controller of adipogenesis (26), is induced at 2 hours followed by downregulation by 24 hours of osteogenic media treatment. Another group of genes exhibited biphasic expression during the timecourse, which includes liver X receptor alpha (Lxrα), Rarβ, farnesoid X receptor (Fxrα), constitutive androstane receptor (Car) and retinoic acid receptor-related orphan receptor beta (Rorβ) that are suppressed at 2 hours and upregulated at 24 hours (Fig. 2B). Genes upregulated primarily at 24 hours were observed including vitamin D receptor (Vdr), Rorγ, estrogen-related receptor gamma (Errγ), thyroid hormone receptor beta (Trβ) and estrogen receptor (Er)-α and –β (Fig. 2C). The genes transiently downregulated at 2 hours include Rev-erb-α/β, chicken ovalbumin upstream promoter transcription factor 2 (Couptf2), as well as classic group 3 receptors such as androgen receptor (Ar) and mineralocorticoid receptor (Mr) (Fig. 2D). Collectively, these data demonstrate that significant NR gene expression changes occur very early in the process of osteoblastic differentiation that may set the stage for modulated sensitivity to specific hormonal or nutritional influences.
Fig. 2.
Expression of NRs early in osteoblastic differentiation. QPCR analysis of NRs regulated at a ≥ 1.5-fold threshold (upregulated or downregulated) and containing at least one statistically significant timepoint (p ≤ 0.05) was conducted in primary mouse calvarial osteoblasts collected at 0, 2, 6, 12, and 24 hours following induction of osteoblastic differentiation (n=4). The genes were grouped by their expression characteristics; (A) upregulated at 2 hours, (B) NRs displaying a biphasic response, (C) upregulated from 6 to 24 hours, and (D) downregulated from 2 to 24 hours. Figure legends within each graph describe the graph labels. Strictly for figure clarity, SEs and p-values have been omitted but the data conform to the above statistical standards (≥ 1.5-fold threshold and p ≤ 0.05, at least at one timepoint).
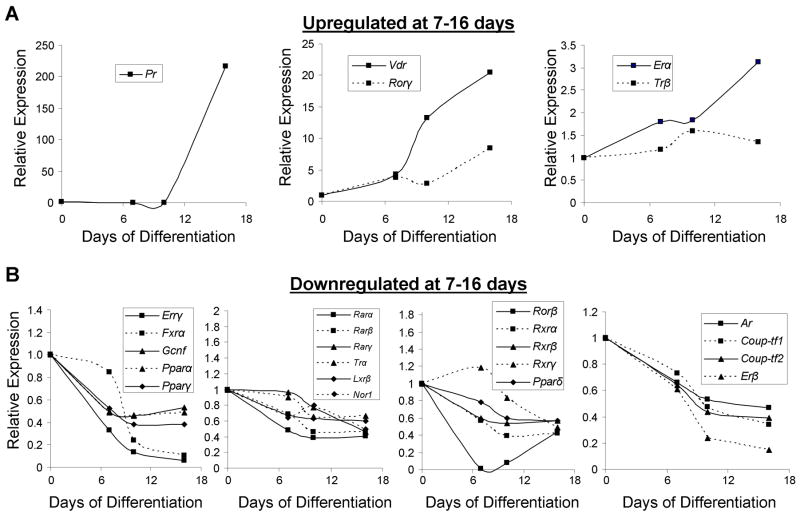
To characterize transcriptional regulation of NR expression at later stages of osteoblastic differentiation, the expression patterns of the NRs at 7, 10 and 16 days following osteoblastic induction were determined using QPCR and the data subjected to hierarchal cluster analysis and a heatmap generated (Supplemental Fig. S1B). Examination of the temporal expression pattern in late differentiating osteoblasts revealed that 36 were expressed late in osteoblastic differentiation, whereas 13 were not expressed. Further examination of the expressed NRs revealed that only 5 genes [Pr, Vdr, Rorγ, Erα, and Trβ] were significantly upregulated late in differentiation using the expanded timecourse (Fig. 3A). Pr was induced 217-fold at day 16 but was undetectable at all earlier timepoints, suggesting that Pr may have functions limited to the late-osteoblastic and/or osteocytic phenotype. Upregulation of Vdr (20-fold), Erα (3.1-fold) and Trβ (1.6-fold) which have reported roles in bone biology (27), was also observed late in osteoblastic differentiation. Rorγ, whose role in osteoblast differentiation is unknown, was upregulated 8.4-fold. Most remarkable is the observation that 78% (28/36) of the expressed genes were downregulated at the later timepoints including all members of the Rar (NR1B), Rxr (NR2B), and Ppar (NR1C) gene families, which have known roles in the support of adipogenesis (28,29). Strikingly, significant downregulation of Errγ (16-fold), Fxrα (9.5-fold), Ar (2.1-fold), and Erβ (6.7-fold) was observed, some of which have known functions in osteoblasts (Fig. 3B). Of particular interest, Rorβ expression drastically declined to undetectable levels at day 7–10 and returns to ~40% of control at day 16. A similar temporal Rorβ expression profile was observed in primary bone marrow stromal cells (Supplemental Fig. S2). Rorβ gene expression was not detectable in osteoclast cultures (data not shown). Collectively, these data clearly demonstrate that significant changes in NR expression occur during late osteoblastic differentiation, mostly downregulation, which may be important in osteoblast and/or osteocyte function.
Fig. 3.
Expression of NRs late in osteoblastic differentiation. Osteoblastic differentiation of primary mouse calvarial osteoblasts was induced at confluence and harvested at 0, 7, 10 and 16 days (n=4) and QPCR performed as in Fig. 2. The genes were grouped by their expression characteristics; (A) upregulated at 7–16 days or (B) downregulated at 7–16 days. Figure legends within each graph describe the graph labels. Strictly for figure clarity, SEs and p-values have been omitted but the data conform to the above statistical standards (≥ 1.5-fold threshold and p ≤ 0.05, at least at one timepoint).
Expression of the NR superfamily and in mouse hematopoietic lineage negative (lin-) bone marrow precursors derived from young and aged mice
Although characterization of NR gene expression during calvarial osteoblast differentiation yielded interesting and important results regarding the function of these receptors in this in vitro system, understanding which NRs are important in regulating osteogenesis in a more physiological system is imperative. We therefore characterized NR gene expression changes associated with age-related bone loss in mice. A previous study in our laboratory (19) analyzed the bone marker gene expression patterns of cells isolated from the lineage negative (lin-) population in femoral bone marrow of young (6 month) and aged (18–22 month) mice. In this study, the aged mice had marked reductions in bone mass and in osteoblast numbers on bone-surfaces, consistent with an age-related impairment in osteogenesis (19). The bone marrow lin- cells represent a population depleted of the hematopoietic cell lineage, which have been shown to be highly enriched for osteoprogenitor cells which mineralize in vitro, form bone in vivo, and express bone-related genes, thereby providing a useful cell population for evaluation of effects of aging on osteoblast progenitor cells (18). Therefore, we applied our QPCR methodology to these samples (n=7–8) and surprisingly found only 5 NRs with statistically significant gene expression changes in aged mice when compared to young mice (Fig 4). Significant upregulation of Errα (3.8-fold), Lxrα (3.1-fold), Rev-erbα (3.6-fold) and Rev-erbβ (1.6-fold) was observed. Interestingly, all these NRs are associated with either age-related bone loss or the support of adipogenesis (30–37). Rorβ, a gene originally thought to have actions limited to the central nervous system, retina and in circadian rhythms (38), was induced 53-fold.
Rorβ expression is inversely correlated with osteoblast differentiation
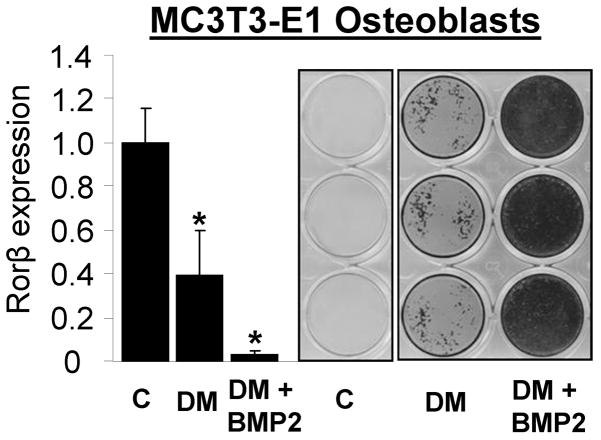
Examination of the calvarial osteoblast and aged mouse datasets revealed that Rorβ exhibits a gene expression pattern inversely correlated with osteogenic potential in both models. Therefore to verify this phenomenon in an independent system, the mouse MC3T3-E1 osteoblastic cell line was differentiated with or without 100 ng/mL Bmp2 (to strongly induce osteoblastic differentiation in this model) for 14 days. As seen in Fig. 5, Rorβ expression declines 2.5- and 20-fold with differentiation media (DM) or DM + Bmp2, respectively. It is interesting that the amount of mineral formed and the degree of Rorβ suppression are also inversely correlated (e.g. more Rorβ suppression associated with more robust mineralization).
Fig. 5.
Rorβ gene expression decreases with increasing mineralization of MC3T3-E1 mouse osteoblasts. Parallel sets of MC3T3-E1 cells were treated for 14 days with either growth media (C), standard osteoblast differentiation media (DM, described in “Materials and Methods”), or DM supplemented with 100 ng/μg bone morphogenetic protein (BMP)-2 (DM + BMP2). Bone nodule formation was determined using Alizarin red stain and Rorβ gene expression quantified using QPCR (n=4). The data are presented as the mean ± SE and an asterisk (*) represents statistical significance of p ≤ 0.01 (Student’s t-test) compared with growth media (C) alone.
Stable Rorβ expression in MC3T3-E1 cells results in impaired mineralization and suppressed Runx2 function
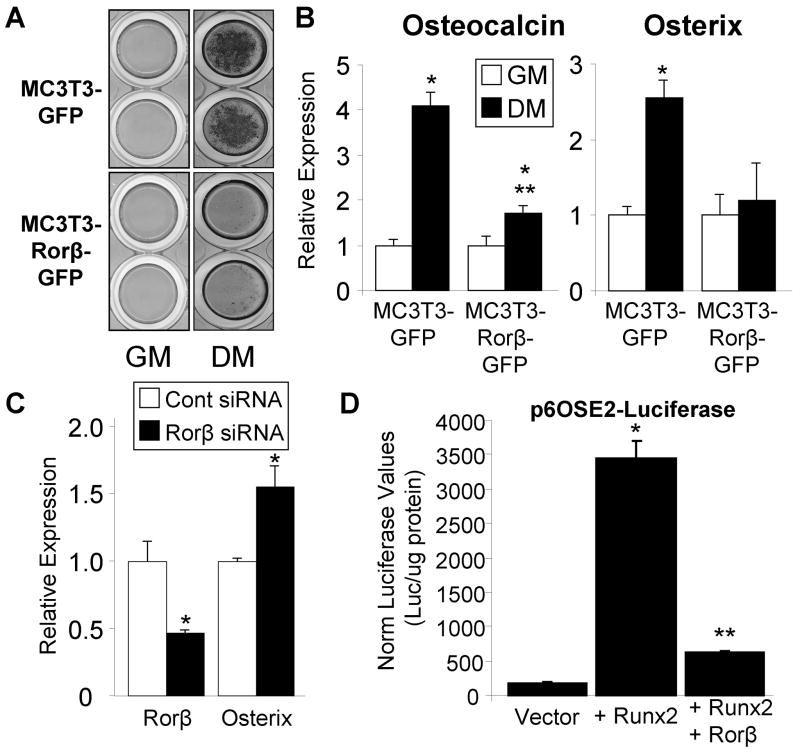
To directly test whether Rorβ inhibits cell mineralization, two cell models were produced in MC3T3-E1 mouse osteoblasts which stably express either GFP (control) or Rorβ-GFP. Following transfection, the cells were antibiotic selected and high GFP-expressing cells were isolated using fluorescence-activated cell sorting, resulting in two cell models: MC3T3-GFP (control) and MC3T3-Rorβ-GFP (Fig. 6A). QPCR analysis of these two cell models revealed a 2.5-fold overexpression of Rorβ in MC3T3-Rorβ-GFP cells (Fig. 6B), confirming stable genomic integration and expression of the Rorβ transgene. To determine the amount of Rorβ expression during osteoblastic differentiation in these models, MC3T3-GFP and MC3T3-Rorβ-GFP cells were treated with either growth or osteoblastic differentiation media for 14 days. QPCR analysis demonstrated that Rorβ expression declines 2.5-fold with osteoblastic differentiation medium in the control MC3T3-GFP cell model (Fig. 6C), confirming our previous data in other models (Fig. 3B and 5). The identical experiment in the MC3T3-Rorβ-GFP model resulted in increased Rorβ expression levels that do not decline during osteoblast differentiation (Fig. 6C), providing an ideal model to test whether decreased Rorβ expression is prerequisite for osteoblast differentiation in the absence of gross overexpression. Indeed, mineralization capacity of the MC3T3-Rorβ-GFP model following 14 days of differentiation is severely inhibited, as compared to the MC3T3-GFP control model (Fig. 7A). QPCR analysis of two classic bone marker genes, osteocalcin and osterix, revealed significant inhibition of differentiation-induced expression in the MC3T3-Rorβ-GFP model (Fig. 7B). Analysis of other bone marker genes, such as alkaline phosphatase, osteopontin, Runx2 and Col1α1, did not significantly change (data not shown), demonstrating that Rorβ-dependent inhibition of mineralization may affect a small, but important, subset of bone regulatory genes. RNA interference (RNAi) assays using a Rorβ-specific siRNA resulted in a 54% reduction of Rorβ mRNA and concomitantly increased expression of osterix by 55% (Fig. 7C). Contrary to the Rorβ overexpression data (Fig. 7B), osteocalcin expression was not significantly changed with the Rorβ siRNA (data not shown). Since osterix is a direct transcriptional target of Runx2 (6,22), we reasoned that Rorβ may antagonize Runx2 transcriptional activity. To test this possibility, cells were transiently transfected with the Runx2-dependent reporter construct p6OSE2-Luc (4) in the presence of Runx2 and/or Rorβ. As expected, Runx2 transfection alone resulted in a 17–fold increase in reporter activity (Fig. 7D). Cotransfection of Rorβ significantly attenuated the ability of Runx2 to activate the reporter, indicating that Rorβ-dependent inhibition of Runx2 function may contribute to the observed inhibition of mineralization in the MC3T3-Rorβ-GFP cell model.
Fig. 6.
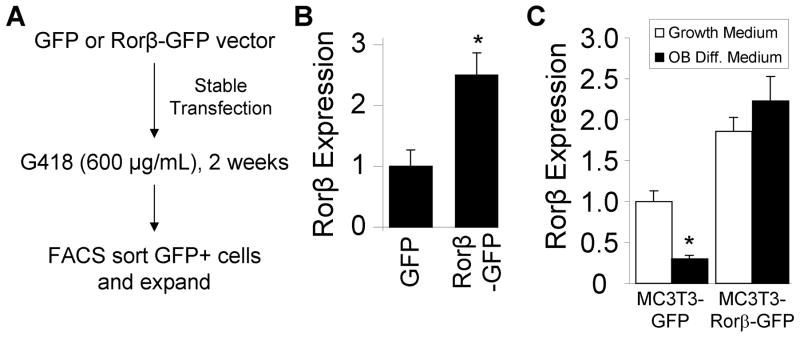
Development of a constitutive, Rorβ-expressing cell system in MC3T3-E1 mouse osteoblasts. (A) A GFP or Rorβ-GFP expression vectors were stably transfected into mouse MC3T3-E1 osteoblasts. Following two weeks of G418 antibiotic selection, the cells were sorted based on GFP positivity using fluorescence-activated cell sorting (FACS) and expanded. (B) MC3T3-GFP and MC3T3-Rorβ-GFP cells were assayed for Rorβ expression using QPCR (n=4). (C) MC3T3-GFP and MC3T3-Rorβ-GFP cells were plated and treated at confluence with either growth medium or osteoblast differentiation medium for 14 days and harvested. Rorβ expression was assayed using QPCR (n=4). The data are presented as the mean ± SE and an asterisk (*) represents statistical significance of p ≤ 0.05 (Student’s t-test).
Fig. 7.
Rorβ expression inhibits osteoblast differentiation through suppression of Runx2 function. (A) MC3T3-GFP and MC3T3-Rorβ-GFP cells were plated and treated at confluence with either growth medium (GM) or osteoblast differentiation medium (DM) for 14 days and bone nodule formation was determined using Alizarin red staining. (B) Identically treated cells were prepared and osteocalcin and osterix expression was assayed using QPCR (n=4). A single asterisk (*) represents significance of p ≤ 0.01 compared to GM within each cell model and double asterisks (**) represents significance of p ≤ 0.01 compared to MC3T3-GFP DM (Student’s t-test). (C) MC3T3-E1 cells were transfected with either a non-specific siRNA control (Cont) or a mouse-specific Rorβ siRNA (Rorβ) and cells harvested 48 hours later. Rorβ and osterix expression was assayed as in panel B. A single asterisk (*) represents significance of p ≤ 0.01 compared to the control siRNA (Student’s t-test). (D) U2OS cells were transiently transfected (n=6) with the indicated plasmids and harvested 72 hours later. Luciferase and protein assays were performed. The data are presented as the mean ± SE. A single asterisk (*) represents significance of p ≤ 0.01 compared to vector alone and double asterisks (**) represents significance of p ≤ 0.01 compared to Runx2 alone (Student’s t-test).
Discussion
Osteoblast differentiation involves the concerted action of large number of transcription factors and regulatory molecules (14). Osteoblasts originate from multipotent mesenchymal stem cells which commit to the osteoblastic lineage following the actions of Runx2 and osterix, as loss of either of these transcription factors inhibits osteoblastic mineralization resulting in the inability of the cartilaginous skeleton to ossify during embryonic development (4–7). However, other transcriptional regulatory networks are important in mediating endocrine and nutritional signals, which influence osteoblastic mineralization and therefore bone formation. Nuclear receptors (NRs) are a large class of intracellular proteins that act as sensors, translating environmental signals into meaningful transcriptional responses. Therefore, it is important to characterize the expression patterns of NRs during osteoblast differentiation in order to gain an understanding of the potential NRs involved, which may lead to drug targets to combat age-related bone loss and osteoporosis. We used a QPCR screening approach to identify NRs with unique expression patterns in both primary mouse calvarial cells and preosteoblastic bone marrow cells isolated from young (6 month) and aged (18–22) mice. Using this approach we identified Rorβ as a novel regulator of osteogenesis.
Primary mouse calvarial cells were chosen since they are a well known mineralizing osteoblast model system, thereby representing an excellent system to study NR gene regulation in the context of osteoblastic mineralization. These cells exhibited robust mineralization and stimulation of classic bone marker gene expression following the induction of differentiation. Interestingly, the osteocyte-associated genes Sost, Dmp1, Fgf23, Mepe and Phex were expressed at later stages of differentiation. Although expression of these genes was surprising, since calvarial osteoblasts are not considered an osteocytic model, it is not unprecedented. For example, Sost is expressed at later stages of differentiation coinciding with the expression of the osteogenic marker, osteocalcin (39). Furthermore, Mepe expression is low in proliferating MC3T3-E1 cells but is dramatically increased during the mineralization stage when stimulated with Bmp2 (40). Although the primary goal of this study was to characterize NR gene expression during osteoblastic differentiation, the upregulation of a few genes (e.g. Erα, Vdr and Pr) at the very late stages of osteoblastic differentiation may provide important clues into the function of the corresponding proteins in the osteocyte.
We have previously demonstrated decreased bone mass and a marked reduction in osteoblast numbers on bone surfaces in the mice from which we obtained the lin- bone marrow cells (19). In the current study, we observed statistical increases in relatively few NR genes: Errα, Lxrα, Rev-erbα/β, and Rorβ. It is interesting that 4 of these NRs (excluding Rorβ) are known to be involved in adipogenesis (31–34,36), suggesting that this may play a role in the decreased bone mass and increased fat acquisition associated with aging. In addition, a major finding of this study is the identification of Rorβ as a novel regulator of osteogenesis. We observed that Rorβ levels dramatically decreased during osteoblastic differentiation of both calvarial and MC3T3-E1 osteoblasts and that Rorβ levels were significantly increased (53-fold) in the preosteogenic lin- cells in the bone marrow of aged mice. This indicates that Rorβ expression is inversely correlated with osteogenic potential and further suggests that suppression of Rorβ may be prerequisite for osteoblastic mineralization to occur. Our data supports this hypothesis since stable Rorβ expression in MC3T3-E1 osteoblasts inhibited mineralization and expression of osteocalcin and osterix.
No data exists concerning the function of Rorβ in bone. Rorβ was originally shown to exhibit a restricted pattern of expression limited to certain regions of the brain and retina (41). Generation and analysis of a Rorβ-null mouse model demonstrated behavioral, motor, and visual defects, as well as decreased male fertility (42–45). Interestingly, characterization of Rorα-deficient mice uncovered several bone-related abnormalities, including thin, long bones (46). This provides precedence that some Ror family members may participate in bone physiology. However, the bone phenotype of Rorβ-null mice has not been studied. The suppression of Rorβ expression during osteoblast differentiation, coupled with marked increase with aging suggests an inhibitory role of Rorβ in bone; thus, examination of the skeletal phenotype of Rorβ null mice would be of interest.
Mechanistically, Rorβ shares a typical NR domain organization containing a DNA-binding domain, N- and C-terminal transactivation domains and ligand binding domain (47). It regulates gene transcription through direct binding to a specific DNA response element through its centrally located zinc-finger binding motif (48,49). Unlike most members of the NR family, Rorβ fails to dimerize with itself or the obligate heterodimer partner Rxr, and therefore acts as a monomer (50). The retinoid, all-trans retinoic acid (ATRA), was shown to specifically bind the Rorβ ligand-binding pocket (51) and shown to repress transcriptional activation of Rorβ, demonstrating that Rorβ has ligand-independent activities that are relieved through ATRA binding; however, this activity was shown to be cell type specific.
Our observation that Rorβ potently inhibits Runx2 transactivation suggests that it may influence bone biology at a fundamental level, however the precise mechanism used by Rorβ to mediate Runx2 inhibition is unknown. It is conceivable that it involves protein interactions between Rorβ and Runx2 and that these interactions inhibit normal Runx2 function. Examination of the literature for evidence of other nuclear receptors that interact with Runx2 revealed that ERα interacts with Runx2 through its DNA-binding domain, suppressing the transactivation potential of Runx2 (52). Whether Rorβ uses a similar mechanism will require studies to characterize the molecular relationship between Rorβ and Runx2. Furthermore, since Rorβ itself binds DNA it is also possible that Runx2-independent actions involving specific Rorβ-regulated genes may be an important component to its function in osteoblasts, however no data exists on Rorβ target genes in osteoblasts.
In further support that Rorβ may exert some of its anti-osteogenic function through Runx2 inhibition, we observed attenuated gene expression of the known Runx2-dependent genes osteocalcin and osterix in the MC3T3-Rorβ-GFP cell model during differentiation. Suppression of Rorβ using RNAi resulted in enhanced expression of osterix, consistent with Rorβ-dependent Runx2 antagonism, but surprisingly, osteocalcin gene expression was unaffected. This may be partially explained by the modest Rorβ knockdown (54%) or by the relatively short experimental timecourse (48 hours). However, this data is consistent with our observation that Rorβ overexpression incompletely inhibited osteocalcin while completely attenuating osterix gene expression, suggesting that osterix may be more sensitive to Rorβ levels than osteocalcin. Examination of the long-term effects of Rorβ suppression on both gene expression and differentiation will require development of stable shRNA cell lines or osteoblast-specific deletion of Rorβ in mice.
In summary, we have characterized the NR temporal gene expression profile in a differentiating osteoblast model system and in bone marrow osteoprogenitor cells of aged mice and identified Rorβ as a novel transcription factor important for the suppression of osteoblastic differentiation, possibly through the inhibition of Runx2 activity. This is the first demonstration of Rorβ expression and function in any bone-related system and therefore identifies a novel NR important for the regulation of bone formation. Further characterization of the function of Rorβ in bone may define it as a potential clinical target to combat age-related osteoporosis.
Supplementary Material
Global expression of the NR superfamily during osteoblastic differentiation. Osteoblastic differentiation of primary mouse calvarial osteoblasts was induced at confluence and samples harvested at either (A) 0, 2 and 24 hours or (B) 0, 7, 10 and 16 days (n=4 for each timepoint). QPCR was performed on the 49 members of the NR superfamily and hierarchal clustering software was used to generate expression heatmaps where the red colors represent upregulation (Up Reg.) and the green colors represent downregulation (Down Reg.) when compared with hour 0 or day 0, respectively. An asterisk within the heatmap represents statistical significance (p ≤ 0.05, Student’s t-test) compared with hour or day 0.
Rorβ expression in primary mouse bone marrow stromal cells (mBMSCs). Osteoblastic differentiation of mBMSCs was induced at confluence and samples harvested at 0, 7, 10 and 16 days (n=4 for each timepoint). (A) QPCR was performed using primers specific for Rorβ. The data are presented as the mean ± SE and an asterisk (*) represents statistical significance of p ≤ 0.05 (Student’s t-test). (B)) Mineralization was determined by Alizarin red stain at the indicated timepoints.
Acknowledgments
We would like to thank James M. Peterson for the QPCR data analyses, Drs. Patricia Ducy and Gerard Karsenty for the p6OSE2-Luc and pCMV-Cbfa1/Runx2 expression constructs, and Dr. Merry Jo Oursler for the osteoclastic cDNAs. This work was supported by a grant (P01-AG004875) from the National Institutes of Health (NIH) to DGM and SK. DGM, MMR and SK conceived the study.
DGM, MMR and GL performed the experiments. All authors were involved in interpretation of data and production of the final manuscript.
Funding: This work was funded by a grant (P01-AG004875) from the National Institutes of Health (NIH) to DGM and SK.
Footnotes
Disclosures
All authors state that they have no conflicts of interest.
Contributor Information
Matthew M. Roforth, Email: roforth.matthew@mayo.edu.
Gang Liu, Email: liu.gang@mayo.edu.
Sundeep Khosla, Email: khosla.sundeep@mayo.edu.
David G. Monroe, Email: monroe.david@mayo.edu.
References
- 1.Nefussi JR, Brami G, Modrowski D, Oboeuf M, Forest N. Sequential expression ob bone matrix proteins during rat calvaria osteoblast differentiation and bone nodule formation in vitro. J Histochem Cytochem. 1997;45:493–503. doi: 10.1177/002215549704500402. [DOI] [PubMed] [Google Scholar]
- 2.Karsenty G. Minireview: Transcriptional control of osteoblast differentiation. Endocrinology. 2001;142:2731–2733. doi: 10.1210/endo.142.7.8306. [DOI] [PubMed] [Google Scholar]
- 3.Komori T. Regulation of osteoblast differentiation by transcription factors. J Cell Biochem. 2006;99:1233–1239. doi: 10.1002/jcb.20958. [DOI] [PubMed] [Google Scholar]
- 4.Ducy P, Zhang R, Geoffroy V, Ridall AL, Karsenty G. Osf2/Cbfa1: a transcriptional activator of osteoblast differentiation. Cell. 1997;89:747–754. doi: 10.1016/s0092-8674(00)80257-3. [DOI] [PubMed] [Google Scholar]
- 5.Komori T, Yagi H, Nomura S, Yamaguchi A, Sasaki K, Deguchi K, Shimizu Y, Bronson RT, Gao YH, Inada M, Sato M, Okamoto R, Kitamura Y, Yoshiki S, Kishimoto T. Targeted disruption of Cbfa1 results in a complete lack of bone formation owing to maturational arrest of osteoblasts. Cell. 1997;89:755–764. doi: 10.1016/s0092-8674(00)80258-5. [DOI] [PubMed] [Google Scholar]
- 6.Nakashima K, Zhou X, Kunkel G, Zhang Z, Deng JM, Behringer RR, Crombrugghe BD. The novel zinc finger-containing transcription factor osterix is required for osteoblast differentiation and bone formation. Cell. 2002;108:17–29. doi: 10.1016/s0092-8674(01)00622-5. [DOI] [PubMed] [Google Scholar]
- 7.Otto F, Thornell AP, Crompton T, Denzel A, Gilmour KC, Rosewell IR, Stamp GWH, Beddington RSP, Mundlos S, Olsen BR, Selby PB, Owen MJ. Cbfa1, a candidate gene for cleidocranial dysplasia syndrome, is essential for osteoblast differentiation and bone development. Cell. 1997;89:765–771. doi: 10.1016/s0092-8674(00)80259-7. [DOI] [PubMed] [Google Scholar]
- 8.Altucci L, Gronemeyer H. Nuclear receptors in cell life and death. Trends Endocrinol Metab. 2001;12(10):460–8. doi: 10.1016/s1043-2760(01)00502-1. [DOI] [PubMed] [Google Scholar]
- 9.Chiang JY. Nuclear receptor regulation of lipid metabolism: potential therapeutics for dyslipidemia, diabetes, and chronic heart and liver diseases. Curr Opin Investig Drugs. 2005;6 (10):994–1001. [PubMed] [Google Scholar]
- 10.O’Malley BW, Kumar R. Nuclear receptor coregulators in cancer biology. Cancer Res. 2009;69(21):8217–22. doi: 10.1158/0008-5472.CAN-09-2223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang D, DuBois RN. Inflammatory mediators and nuclear receptor signaling in colorectal cancer. Cell Cycle. 2007;6(6):682–5. doi: 10.4161/cc.6.6.4030. [DOI] [PubMed] [Google Scholar]
- 12.Ranhotra HS. The mammalian orphan nuclear receptors: orphans as cellular guardians. J Recept Signal Transduct Res. 2011;31(1):20–25. doi: 10.3109/10799893.2010.538853. [DOI] [PubMed] [Google Scholar]
- 13.McKenna NJ, O’Malley BW. Nuclear receptors, coregulators, ligands, and selective receptor modulators: making sense of the patchwork quilt. Ann N Y Acad Sci. 2001;949:3–5. doi: 10.1111/j.1749-6632.2001.tb03997.x. [DOI] [PubMed] [Google Scholar]
- 14.Monroe DG, Hawse JR, Subramaniam M, Spelsberg TC. Retinoblastoma binding protein-1 (RBP1) is a Runx2 coactivator and promotes osteoblastic differentiation. BMC Musculoskelet Disord. 2010;11:104–113. doi: 10.1186/1471-2474-11-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Radonic A, Thulke S, Mackay IM, Landt O, Siegert W, Nitsche A. Guideline to reference gene selection for quantitative real-time PCR. Biochem Biophys Res Commun. 2004;313:856–862. doi: 10.1016/j.bbrc.2003.11.177. [DOI] [PubMed] [Google Scholar]
- 16.Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F. Accurate normalization of real-time quantitative RT-PCR data by geometeric averaging of multiple internal control genes. Genome Biol. 2002;3:research0034.1-0-34.11. doi: 10.1186/gb-2002-3-7-research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhao S, Fernald RD. Comprehensive algorithm for quantitative real-time polymerase chain reaction. J Comput Biol. 2005;12(8):1047–1064. doi: 10.1089/cmb.2005.12.1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Itoh S, Aubin JE. A novel purification method for multipotential skeletal stem cells. J Cell Biochem. 2009;108:368–377. doi: 10.1002/jcb.22262. [DOI] [PubMed] [Google Scholar]
- 19.Syed FA, Modder UI, Roforth M, Hensen I, Fraser DG, Peterson JM, Oursler MJ, Khosla S. Effects of chronic estrogen treatment on modulating age-related bone loss in female mice. J Bone Miner Res. 2010;25(11):2438–2446. doi: 10.1002/jbmr.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xie C-Q, Jeong Y, FUM, Bookout AL, Garcia-Barrio MT, Sun T, Kim B-H, Xie Y, Root S, Zhang J, Xu R-H, Chen E, Mangelsdorf DJ. Expression profiling of nuclear receptors in human and mouse embryonic stem cells. Mol Endocrinol. 2009;23(5):724–733. doi: 10.1210/me.2008-0465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nielsen R, Grontved L, Stunnenberg HG, Mandrup S. Peroxisome proliferator-activated receptor subtype- and cell-type-specific activation of genomic target genes upon adenoviral transgene delivery. Molecular & Cellular Biology. 2006;26(15):5698–714. doi: 10.1128/MCB.02266-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ducy P, Karsenty G. Two distinct osteoblast-specific cis-acting elements control expression of a mouse osteocalcin gene. Molecular & Cellular Biology. 1995;15(4):1858–69. doi: 10.1128/mcb.15.4.1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fu M, Sun T, Bookout AL, Downes M, Yu RT, Evans RM, Mangelsdorf DJ. A nuclear receptor atlas: 3T3-L1 adipogenesis. Mol Endocrinol. 2005;19:2437–2450. doi: 10.1210/me.2004-0539. [DOI] [PubMed] [Google Scholar]
- 24.Bookout AL, Jeong Y, Downes M, Yu RT, Evans RM, Mangelsdorf DJ. Anatomical profiling of nuclear receptor expression reveals a hierarchical transcriptional network. Cell. 2006;126:789–799. doi: 10.1016/j.cell.2006.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Barish GD, Downes M, Alaynick WA, Yu RT, Ocampo CB, Bookout AL, Mangelsdorf DJ, Evans RM. A Nuclear Receptor Atlas: macrophage activation. Mol Endocrinol. 2005;19 (10):2466–77. doi: 10.1210/me.2004-0529. [DOI] [PubMed] [Google Scholar]
- 26.Kawai M, Sousa KM, MacDougald OA, Rosen CJ. The many facets of PPARgamma: novel insights for the skeleton. Am J Physiol Endocrinol Metab. 2010;299(1):E3–E9. doi: 10.1152/ajpendo.00157.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bassett JH, Williams GR. The skeletal phenotypes of TRalpha and TR beta mutant mice. J Mol Endocrinol. 2009;42(4):269–282. doi: 10.1677/JME-08-0142. [DOI] [PubMed] [Google Scholar]
- 28.Tontonoz P, Spiegelman BM. Fat and beyond: the diverse biology of PPARgamma. Ann Rev Biochem. 2008;77:289–312. doi: 10.1146/annurev.biochem.77.061307.091829. [DOI] [PubMed] [Google Scholar]
- 29.Ziouzenkova O, Plutzky J. Retinoid metabolism and nuclear receptor responses: New insights into coordinated regulation of the PPAR-RXR complex. FEBS Letters. 2008;582(1):32–8. doi: 10.1016/j.febslet.2007.11.081. [DOI] [PubMed] [Google Scholar]
- 30.Delhon I, Gutzwiller S, Morvan F, Rangwala S, Wyder L, Evans G, Studer A, Kneissel M, Fournier B. Absence of estrogen receptor-related-alpha increases osteoblastic differentiation and cancellous bone mineral density. Endocrinology. 2009;150(10):4463–72. doi: 10.1210/en.2009-0121. [DOI] [PubMed] [Google Scholar]
- 31.Ijichi N, Ikeda K, Horie-Inoue K, Yagi K, Okazaki Y, Inoue S. Estrogen-related receptor alpha modulates the expression of adipogenesis-related genes during adipocyte differentiation. Biochem Biophys Res Commun. 2007;358(3):813–8. doi: 10.1016/j.bbrc.2007.04.209. [DOI] [PubMed] [Google Scholar]
- 32.Kubo M, Ijichi N, Ikeda K, Horie-Inoue K, Takeda S, Inoue S. Modulation of adipogenesis-related gene expression by estrogen-related receptor gamma during adipocytic differentiation. Biochim Biophys Acta. 2009;1789(2):71–7. doi: 10.1016/j.bbagrm.2008.08.012. [DOI] [PubMed] [Google Scholar]
- 33.Kumar N, Solt LA, Wang Y, Rogers PM, Bhattacharyya G, Kamenecka TM, Stayrook KR, Crumbley C, Floyd ZE, Gimble JM, Griffin PR, Burris TP. Regulation of adipogenesis by natural and synthetic REV-ERB ligands. Endocrinology. 2010;151(7):3015–25. doi: 10.1210/en.2009-0800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Laitinen S, Fontaine C, Fruchart JC, Staels B. The role of the orphan nuclear receptor Rev-Erb alpha in adipocyte differentiation and function. Biochimie. 2005;87(1):21–5. doi: 10.1016/j.biochi.2004.12.006. [DOI] [PubMed] [Google Scholar]
- 35.Robertson KM, Norgard M, Windahl SH, Hultenby K, Ohlsson C, Andersson G, Gustafsson JA. Cholesterol-sensing receptors, liver X receptor alpha and beta, have novel and distinct roles in osteoclast differentiation and activation. J Bone Miner Res. 2006;21 (8):1276–87. doi: 10.1359/jbmr.060503. [DOI] [PubMed] [Google Scholar]
- 36.Seo JB, Moon HM, Kim WS, Lee YS, Jeong HW, Yoo EJ, Ham J, Kang H, Park MG, Steffensen KR, Stulnig TM, Gustafsson JA, Park SD, Kim JB. Activated liver X receptors stimulate adipocyte differentiation through induction of peroxisome proliferator-activated receptor gamma expression. Mol Cell Biol. 2004;24(8):3430–44. doi: 10.1128/MCB.24.8.3430-3444.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Teyssier C, Gallet M, Rabier B, Monfoulet L, Dine J, Macari C, Espallergues J, Horard B, Giguere V, Cohen-Solal M, Chassande O, Vanacker JM. Absence of ERRalpha in female mice confers resistance to bone loss induced by age or estrogen-deficiency. PLoS ONE. 2009;4(11):e7942. doi: 10.1371/journal.pone.0007942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sumi Y, Yagita K, Yamaguchi S, Ishida Y, Kuroda Y, Okamura H. Rhythmic expression of ROR beta mRNA in the mice suprachiasmatic nucleus. Neuroscience Letters. 2002;320(1–2):13–6. doi: 10.1016/s0304-3940(02)00011-3. [DOI] [PubMed] [Google Scholar]
- 39.Li X, Zhang Y, Kang H, Liu W, Liu P, Zhang J, Harris SE, Wu D. Sclerostin binds to LRP5/6 and antagonizes canonical Wnt signaling. J Biol Chem. 2005;280(20):19883–7. doi: 10.1074/jbc.M413274200. [DOI] [PubMed] [Google Scholar]
- 40.Cho YD, Yoon WJ, Woo KM, Baek JH, Lee G, Cho JY, Ryoo HM. Molecular regulation of matrix extracellular phosphoglycoprotein expression by bone morphogenetic protein-2. J Biol Chem. 2009;284(37):25230–40. doi: 10.1074/jbc.M109.008391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jetten AM. Retinoid-related orphan receptors (RORs): critical roles in development, immunity, circadian rhythm, and cellular metabolism. Nuclear Receptor Signaling. 2009;7:e003. doi: 10.1621/nrs.07003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Andre E, Gawlas K, Becker-Andre M. A novel isoform of the orphan nuclear receptor RORbeta is specifically expressed in pineal gland and retina. Gene. 1998;216(2):277–83. doi: 10.1016/s0378-1119(98)00348-5. [DOI] [PubMed] [Google Scholar]
- 43.Masana MI, Sumaya IC, Becker-Andre M, Dubocovich ML. Behavioral characterization and modulation of circadian rhythms by light and melatonin in C3H/HeN mice homozygous for the RORbeta knockout. American Journal of Physiology-Regulatory Integrative & Comparative Physiology. 2007;292(6):R2357–67. doi: 10.1152/ajpregu.00687.2006. [DOI] [PubMed] [Google Scholar]
- 44.Jia L, Oh EC, Ng L, Srinivas M, Brooks M, Swaroop A, Forrest D. Retinoid-related orphan nuclear receptor RORbeta is an early-acting factor in rod photoreceptor development. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(41):17534–9. doi: 10.1073/pnas.0902425106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Srinivas M, Ng L, Liu H, Jia L, Forrest D. Activation of the blue opsin gene in cone photoreceptor development by retinoid-related orphan receptor beta. Molecular Endocrinology. 2006;20(8):1728–41. doi: 10.1210/me.2005-0505. [DOI] [PubMed] [Google Scholar]
- 46.Meyer T, Kneissel M, Mariani J, Fournier B. In vitro and in vivo evidence for orphan nuclear receptor RORalpha function in bone metabolism. Proc Natl Acad Sci USA. 2000;97(16):9197–202. doi: 10.1073/pnas.150246097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jetten AM, Kurebayashi S, Ueda E. The ROR nuclear orphan receptor subfamily: critical regulators of multiple biological processes. Progress in Nucleic Acid Research & Molecular Biology. 2001;69:205–47. doi: 10.1016/s0079-6603(01)69048-2. [DOI] [PubMed] [Google Scholar]
- 48.Carlberg C, Hooft van Huijsduijnen R, Staple JK, DeLamarter JF, Becker-Andre M. RZRs, a new family of retinoid-related orphan receptors that function as both monomers and homodimers. Molecular Endocrinology. 1994;8(6):757–70. doi: 10.1210/mend.8.6.7935491. [DOI] [PubMed] [Google Scholar]
- 49.Giguere V, Tini M, Flock G, Ong E, Evans RM, Otulakowski G. Isoform-specific amino-terminal domains dictate DNA-binding properties of ROR alpha, a novel family of orphan hormone nuclear receptors. Genes & Development. 1994;8(5):538–53. doi: 10.1101/gad.8.5.538. [DOI] [PubMed] [Google Scholar]
- 50.Stehlin C, Wurtz JM, Steinmetz A, Greiner E, Schule R, Moras D, Renaud JP. X-ray structure of the orphan nuclear receptor RORbeta ligand-binding domain in the active conformation. EMBO Journal. 2001;20(21):5822–31. doi: 10.1093/emboj/20.21.5822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stehlin-Gaon C, Willmann D, Zeyer D, Sanglier S, Van Dorsselaer A, Renaud JP, Moras D, Schule R. All-trans retinoic acid is a ligand for the orphan nuclear receptor ROR beta. Nature Structural Biology. 2003;10(10):820–5. doi: 10.1038/nsb979. [DOI] [PubMed] [Google Scholar]
- 52.Khalid O, Baniwal SK, Purcell DJ, Leclerc N, Gabet Y, Stallcup MR, Coetzee GA, Frenkel B. Modulation of Runx2 activity by estrogen receptor-alpha: implications for osteoporosis and breast cancer. Endocrinology. 2008;149(12):5984–95. doi: 10.1210/en.2008-0680. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Global expression of the NR superfamily during osteoblastic differentiation. Osteoblastic differentiation of primary mouse calvarial osteoblasts was induced at confluence and samples harvested at either (A) 0, 2 and 24 hours or (B) 0, 7, 10 and 16 days (n=4 for each timepoint). QPCR was performed on the 49 members of the NR superfamily and hierarchal clustering software was used to generate expression heatmaps where the red colors represent upregulation (Up Reg.) and the green colors represent downregulation (Down Reg.) when compared with hour 0 or day 0, respectively. An asterisk within the heatmap represents statistical significance (p ≤ 0.05, Student’s t-test) compared with hour or day 0.
Rorβ expression in primary mouse bone marrow stromal cells (mBMSCs). Osteoblastic differentiation of mBMSCs was induced at confluence and samples harvested at 0, 7, 10 and 16 days (n=4 for each timepoint). (A) QPCR was performed using primers specific for Rorβ. The data are presented as the mean ± SE and an asterisk (*) represents statistical significance of p ≤ 0.05 (Student’s t-test). (B)) Mineralization was determined by Alizarin red stain at the indicated timepoints.