SUMMARY
The methylcytosine hydroxylase Tet2 has been implicated in hematopoietic differentiation and the formation of myeloid malignancies when mutated. An ideal system to study the role of Tet2 in myelopoeisis is C/EBPa induced transdifferentiation of pre-B cells into macrophages. Here we found that C/EBPa binds to upstream regions of Tet2 and that the gene becomes activated. Tet2 knockdowns impaired the upregulation of macrophage markers as well as phagocytic capacity, suggesting that the enzyme is required for both early and late stage myeloid differentiation. A slightly weaker effect was seen in primary cells with a Tet2 ablation. Expression arrays of transdifferentiating cells with Tet2 knockdowns permitted the identification of a small subset of myeloid genes whose upregulation was blunted. Activation of these target genes was accompanied by rapid increases of promoter hydroxy-methylation. Our observations indicate that Tet2 helps C/EBPa rapidly de-repress myeloid genes during the conversion of pre-B cells into macrophages.
Keywords: Epigenetics, Transdifferentiation, DNA methylation, DNA hydroxymethylation
INTRODUCTION
The methylation of cytosines is a well-studied mechanism to stably repress gene expression (Reik, 2007). While the enzymes involved in the establishment and maintenance of methylated cytosines (MeC) have been studied for many years, enzymes with the potential to remove or modify the mark were only recently discovered (Wu and Zhang, 2010). For example, activation-induced cytidine deaminase (AID) has been shown to remove the methyl-cytosine mark through an intermediate involving the DNA mismatch repair machinery (Rai et al., 2008). In contrast, the Ten-eleven translocation (Tet) family of dioxygenase enzymes can hydroxylate the methyl group, resulting in hydroxymethylated CpGs (OH-MeC) (Tahiliani et al., 2009). Of the three members of the Tet family, Tet2 is highly expressed in the hematopoietic system and peaks in macrophages (Ko et al., 2010). Tet2 mutations have been described in several types of hematopoietic disorders (Tefferi, 2010), including acute myeloid leukemia (AML), chronic myelomonocytic leukemia (CMML), and myelodysplastic syndrome (MDS) (Langemeijer et al., 2009; Tefferi et al., 2009). Several of these mutations have been directly linked to Tet2 loss of function with coordinated reductions in genome-wide hydroxymethylation (Figueroa et al., 2010; Ko et al., 2010). Studies of Tet2−/− mice revealed expansion of the hematopoietic stem cell compartment, biased differentiation down the myelomonocytic lineage, as well as myeloid cell transformation resembling MDS (Moran-Crusio et al., 2011; Quivoron et al., 2011). Therefore, Tet2 plays important functions in the steady-state maintenance of hematopoietic stem cells as well as in commitment to the myeloid lineage. However, the mechanism by which Tet2 functions remains largely elusive and target genes have not yet been described.
Several hypotheses have been proposed to explain the function of OH-MeC residues. First, they might neutralize the silencing potential associated with MeCs since the modification prevents association with methyl-binding domains (MBDs) (Jin et al., 2010; Valinluck et al., 2004). Second, a role for the mark in regulating the steady-state fidelity of DNA methylation has been proposed (Williams et al., 2011). Third, OH-MeC might serve as a docking site itself to recruit additional transcription regulators (Yildirim et al., 2011). Finally, the modification might represent an intermediate to complete demethylation, such as mediated by DNA damage repair mechanisms (Wu and Zhang, 2010). Evidence in support of each of these possibilities has been reported, with the majority of studies carried out in ES cells. However, if Tet2 mediated OH-MeC plays an important role in gene activation remains to be determined.
We have previously described the CCAAT/enhancer binding protein alpha (C/EBPa) induced conversion of pre-B cells into functional macrophages (Bussmann et al., 2009; Xie et al., 2004). Transdifferentiation occurs between homogenous populations of cells within a matter of days, with essentially 100% efficiency and involving a single cell division (Bussmann et al., 2009; Xie et al., 2004). Here we have used a beta-estradiol (bEst) inducible pre-B cell line (called C10 or C11) in a knockdown approach to address the function of Tet2 in myeloid cell fate decisions. We found that C/EBPa activates Tet2 and that expression of the gene is necessary for rapid and complete transdifferentiation. Gene expression analysis revealed the misregulation of nearly 100 genes of which the majority represented myeloid genes with impaired activation. Tet2 directly binds to the methylated promoters of its target genes, resulting in hydroxylation and correlating with the genes’ strong activation. Our findings suggest a mechanism by which repressed genes may become activated during transdifferentiation.
RESULTS
C/EBPa activates Tet2 during B cell to macrophage transdifferentiation and binds to sites upstream of the gene
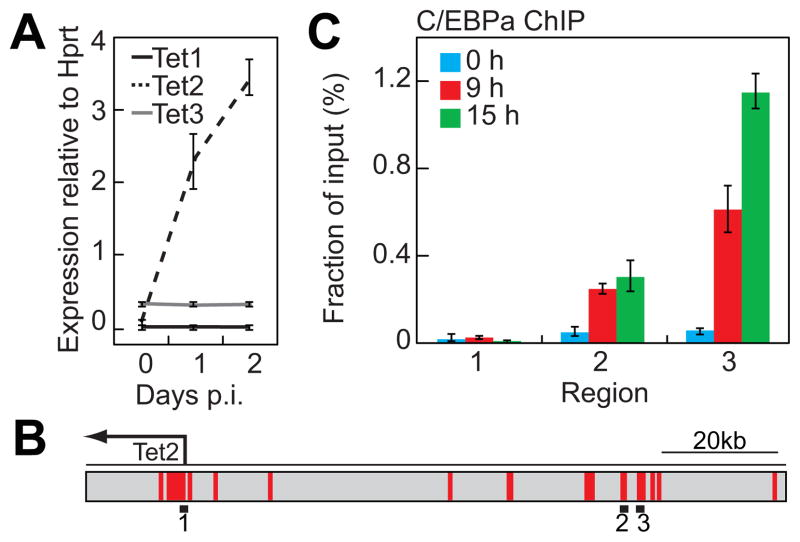
B cell to macrophage transdifferentiation occurs amidst a dynamic landscape of expression changes encompassing thousands of genes, including the upregulation of myeloid genes that are completely silenced in the starting cell population (Bussmann et al., 2009). In a previous study we found that the genome wide distribution of DNA methylation remained largely unaltered during transdifferentiation (Rodriguez-Ubreva et al., 2011). However, it seemed possible that a conversion of MeC to OH-MeC by Tet enzymes plays a role in the gene expression changes that accompany the lineage switch. We therefore decided to analyze the role of these enzymes during pre-B cell to macrophage transdifferentiation. Our gene expression array data (Bussmann et al., 2009) revealed that Tet2 is upregulated approximately 4 fold during bEst induced reprogramming of C10 cells (data not shown). This was validated by qRT-PCR confirming the rapid upregulation of Tet2 and revealing it to be the major Tet enzyme expressed in the system when compared to Tet1 and Tet3 (Figure 1A). We next investigated whether C/EBPa directly associates with Tet2 regulatory elements in transdifferentiating cells. For this purpose, we performed chromatin immunoprecipitation (ChIP) experiments in uninduced, 9h-induced and 15h-induced cells. We directed our analysis at the Tet2 promoter (Figure 1B, region 1), as well as two upstream regulatory elements that are highly conserved between mouse and human and have been previously implicated in Oct4-mediated Tet2 regulation in ES cells (Figure 1B, regions 2 and 3) (Koh et al., 2011). C/EBPa was not enriched at any of these sites in the uninduced pre-B cell line (Figure 1C, blue bars). However, at 9h post-induction (p.i.) C/EBPa bound to both enhancer regions and became further enriched at 15 hours at the region 3 (Figure 1C, red and green bars) while the promoter remained negative at all time points. These data show that Tet2 is upregulated during pre-B cell to macrophage transdifferentiation and suggests that C/EBPa directly mediates this through binding to upstream regulatory elements.
Figure 1. C/EBPa induces Tet2 expression during B cell to macrophage transdifferentiation and binds to the Tet2 enhancer.
A) qRT-PCR analysis of Tet1/2/3 expression levels in C10 cells. Data are presented relative to Hprt expression as mean ±SD of three biological replicates.
B) Schematic representation of the Tet2 locus indicating the amplicons used for ChIP analysis (black bars). Regions highlighted in red represent highly conserved sequences between mouse and human (>60% conservation).
C) ChIP assay demonstrating C/EBPa enrichment at Tet2 upstream regulatory regions during transdifferentiation. Data are presented as mean percentage of input ±SD of three biological replicates.
Knockdowns of Tet2 impair B cell to macrophage transdifferentiation
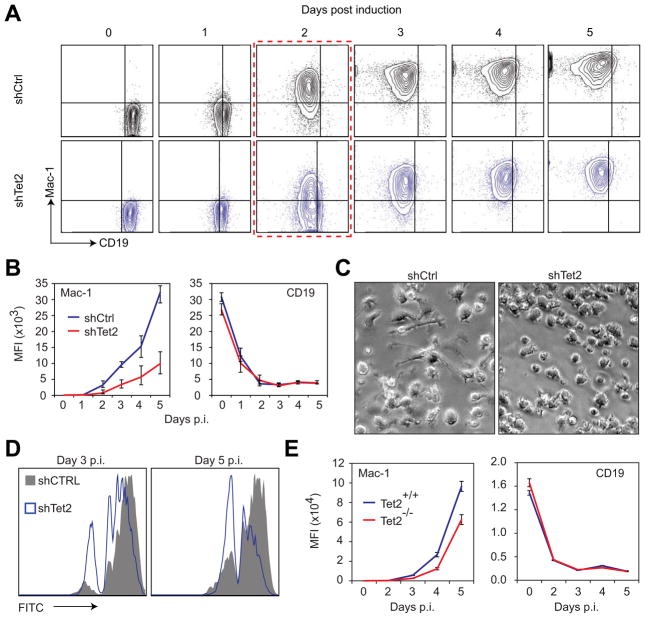
To assess the functional relevance of the C/EBPa-induced upregulation of Tet2 we infected C10 cells singly or in combination with two different knockdown constructs (shTet2kd1-puro and shTet2kd2-puro lentiviruses) followed by puromycin selection. Each shRNA construct alone resulted in an approximately 40% reduction of Tet2 mRNA levels whereas their combination led to an 80% reduction (Figure S1A) without affecting Tet1 and Tet3 (Figure S1B). Then we induced the cells with bEst and monitored the expression of Mac-1 and CD19 at 0h and 48h p.i. While the macrophage marker Mac-1 became expressed in about 90% of control cells after 48 hours, this proportion was reduced by approximately 35% in cells expressing either shTet2kd1 or shTet2kd2, and by 55% in cells with both constructs (Figure S1C, left panel). The observed complementation of the Tet2 knockdowns suggests that their effects do not represent an off target artifact. In contrast to Mac-1 upregulation, silencing of the B-cell marker CD19 was not affected (Figure S1C, right panel). To determine whether the effect on Mac-1 represents a block or a delay in macrophage marker expression, cells were induced with bEst and analyzed by FACS at 24-hour intervals (Figure 2A and S1D). These experiments again showed that the combined knockdown constructs (herein referred to as Tet2 kd) resulted in a striking reduction in Mac-1 expression levels that persisted throughout the five-day time course (Figure 2B, left panel) while CD19 downregulation was not affected (Figure 2B, right panel). The reduction of Mac-1 expression levels as determined by mean fluorescence intensity analysis was accompanied by an approximately 1-day delay of Mac-1 upregulation (Figure 2A & S1D), with the 48h time point showing the largest difference in Mac-1 positive versus Mac-1 negative cells (Figure 2A, red box). The kd cells also differed from control 5 day-induced cells by a more rounded morphology and fewer filopodia (Figure 2C). To test for macrophage functionality we performed a phagocytosis assay by incubating 3 day-induced and 5 day-induced C11 cells with or without Tet2 kd for 2 hours in the presence of green-fluorescent 1 micron beads, followed by removal of free beads and FACS analysis. Fewer of the kd cells were able to ingest the beads when compared to control induced cells at both time points assayed (Figure 2D) and similar observations were made with bacteria as a substrate (data not shown). Since we could not distinguish between cells expressing only one or both sh constructs it is possible that the more inhibited cells correspond to the sub-population containing both knockdown constructs. Our results show that Tet2 kds cause a partial inhibition of C/EBPa induced transdifferentiation by reducing myeloid marker upregulation as well as the ability to phagocytose bacteria in transdifferentiated cells.
Figure 2. Disruption of Tet2 impairs B cell to macrophage transdifferentiation. See also Figure S1.
A) FACS plots of CD19 (x-axis) and Mac-1 (y-axis) marker levels in control (shCtrl, black) and Tet2 kd (shTet2, blue) C10 cells at the indicated time points during transdifferentiation. Dotted red outline indicates the time-point where the percentage of Mac-1 positive cells was most inhibited in the Tet2 kd sample.
B) Mac-1 (left panel) and CD19 (right panel) mean fluorescence intensity (MFI) of control (shCtrl, blue) and Tet2 kd (shTet2, red) cells at the indicated time points p.i. of transdifferenitation. Data are presented as mean ±SD of three biological replicates.
C) Images of control (shCtrl) and Tet2 kd (shTet2) C10 cells taken 5 days after the induction of transdifferentiation.
D) Histogram plot representation of the phagocytic capacity of control (shCtrl, dark grey) and Tet2 kd (shTet2, blue) C10 cells at day 3 and day 5 of transdifferentiation to engulf green fluorescent beads as determined by FACS analysis. The peaks represent cells that have not ingested particles (far left) to cells that have ingested one or more particles (subsequent peaks to the right). Data presented are representative of three biological replicates.
E) Mac-1 (left panel) and CD19 (right panel) MFI as determined by FACS of wildtype (Tet2+/+, blue) and Tet2 null (Tet2−/−, red) cells at the indicated time p.i. of transdifferenitation. Data represent mean ±SD of three biological replicates.
To determine whether our Tet2 kd constructs affect myeloid differentiation, we sorted LinnegKitposSca-1posFlt3pos bone marrow progenitors (Adolfsson et al., 2005) and infected them with either shTet2kd2-egfp or sh control-egfp viruses. After one day in culture GFP+ cells were sorted and 60 single cells each were distributed into 96 well plates containing S17 stromal cells in media supplemented with GM-CSF, IL3, Flt3l, and IL7. At day 10 approximately 60% of the wells showed clonal expansion, revealing a significant increase upon Tet2 kd in the proportion of Gr-1/Mac-1 double negative cells (DN; Wilcoxon test P=0.01) as well as of immature myeloid cells (Mac-1lo; Wilcoxon test P=0.03) and a concomitant reduction in the proportion of mature macrophages (Mac-1hi, Wilcoxon test P=0.04). In addition, Gr-1/Mac-1 double positive cells, consisting of granulocytes and macrophages, showed a slight reduction (Figures S1E, F). Together, these observations indicate that a knockdown of Tet2 in hematopoietic progenitor cells results in a block of myeloid differentiation, supporting earlier studies (Figueroa et al., 2010; Moran-Crusio et al., 2011) and further validating the knockdown constructs used.
To unequivocally rule out off-target effects that may be associated with the knockdown approach, we performed transdifferentiation assays with Tet2−/− and wild type pre-B cells. To this end, CD19 positive cells were isolated from the bone marrow of knockout mice (Moran-Crusio et al., 2011) as well as from wild type littermates and infected with C/EBPaER-hCD4 retrovirus. Two days later CD19+hCD4+ cells were sorted, induced to transdifferentiate and monitored by FACS for the expression of surface markers. Similar to the kds described above, Tet2−/−B cells upregulated Mac-1 to lower levels than control cells with no appreciable differences in CD19 downregulation (Figure 2E). However, the effect of Tet2 loss of function was less dramatic as compared to the cell line. This may be attributed to compensation by Tet3, which is also expressed in the primary cells and becomes further upregulated following bEst induction (Figure S1G). Alternatively, it might be due to the increased DNA methylation associated with cultured cell lines (Weber et al., 2005). Our data reveal that Tet2 knockdowns partially inhibit the generation of the macrophage phenotype in a pre-B cell line induced to transdifferentiate by C/EBPa and show that primary cells with a complete knockdown also exhibit striking but smaller differences in Mac-1 upregulation.
Tet2 knockdowns impair activation of myeloid target genes
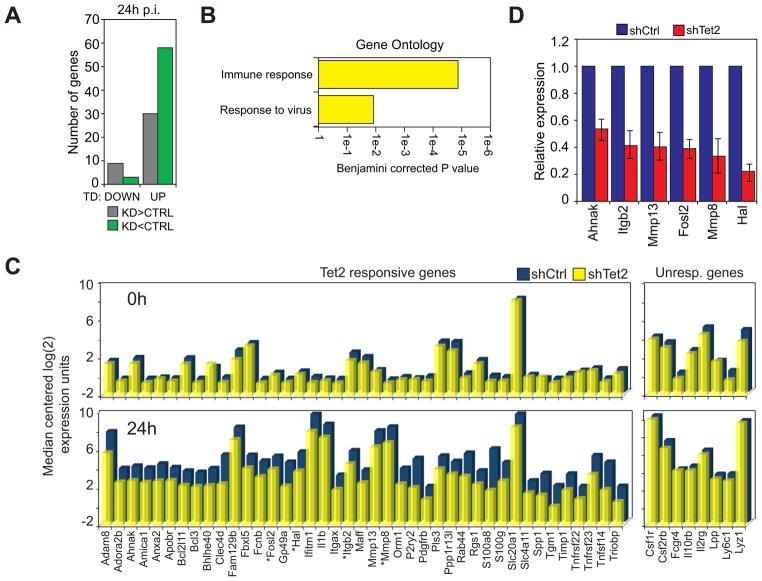
The observed effects of Tet2 disruption on immune cell transdifferentiation prompted us to search for gene expression defects during the process. We therefore performed a microarray analysis of RNAs derived from control and Tet2 kd cells at 0 and 24h p.i. To explore the effect of Tet2 knockdowns on genes that are specifically modulated during transdifferentiation we first selected microarray probes that were up or downregulated ≥2 fold in control cells at 24 hours compared to uninduced cells, revealing 1586 upregulated and 1546 downregulated genes (Data S1). Of the downregulated genes, only eleven were misregulated in Tet2 kd cells ≥1.5 fold at 24h p.i. (Fig. 3A, Data S1). In contrast, within the upregulated cohort 88 unique genes were affected by Tet2 kd at 24h p.i. Fifty-eight of these genes were less expressed in kd cells and 30 were more expressed compared to controls (Figure 3A). We decided to concentrate on the group of 58 genes whose expression was delayed because they were more strongly affected and represented the predominant subgroup. A gene ontology analysis revealed that the top enriched terms associated within this cohort of misregulated genes (designated as Tet2 responsive genes) were ‘Immune response’, and ‘Response to virus’ (Benjamini corrected P = 3.7e-5 and 1.2e-2, respectively)(Figure 3B). The median centered expression value of a representative selection of Tet2 responsive genes in control and Tet2 kd cells is shown for 0h and 24h p.i. in Figure 3C. The expression defects detected by microarray were confirmed by qRT-PCR in 20 of the 22 genes assayed, including Ahnak, Itgb2, Mmp13, Fosl2 (Fra-2), Mmp8, and Hal (Figure 3D, Figure S2A). Several of these genes are known to play a role in macrophages, such as integrin β2, which is the partner of the α chain of Mac-1; Fosl2, which is essential for the function of the macrophage-related osteoclasts (Bozec et al., 2008); MMP13, which has been shown to be produced by scar associated macrophages (Fallowfield et al., 2007); and MMP8, which is produced by myelomonocytic precursors (Prikk et al., 2001). In contrast, no macrophage functions have been described for Ahnak and Hal, a histidine ammonia-lyase. Of note, Tet2 kd did not impair the upregulation of several other genes known to be important for macrophage function, such as Csf1r, Lyz1 and Fcgr4 (Figure 3C, right panel). We refer to this group as ‘Tet2 unresponsive genes’. Together, our results indicate that Tet2 function is critical for the full activation of >50 genes that become upregulated during pre-B cell to macrophage transdifferentiation.
Figure 3. Disruption of Tet2 impairs the upregulation of myeloid specific genes. See also Figure S2.
A) Number of genes that are downregulated (DOWN) or upregulated (UP) during transdifferentiation and misexpressed at least 1.5 fold in Tet2 kd cells versus the control. Misexpressed genes are further subdivided into those that are expressed at higher levels in Tet2 kd cells (KD>CTRL, grey) and those that are expressed at lower levels in Tet2 kd cells (KD<CTRL, green).
B) Top Biological Process ontology terms associated with genes upregulated during transdifferentiation and expressed lower in Tet2 kd cells when compared to controls. X-axis represents the Benjamini-corrected P value of ontology term enrichment.
C) Bar plot of genes from the microarray that show a decreased expression upon Tet2 kd (Tet2 responsive genes) compared to selected genes not affected by Tet2 kd (Unresponsive genes). Median centered expression values are presented for the indicated gene (x-axis) in uninduced (0h) and 24h induced control cells (shCtrl, blue bars) and Tet2 knockdown cells (shTet2, yellow bars). Asterisks denote verified direct Tet2 target genes.
D) qRT-PCR validation of several genes identified by microarray at 24h p.i.. Samples were normalized against Hprt and expression in Tet2 kd cells (shTet2, red bars) was set relative to that in control cells (shCtrl, blue bars). Data are presented as the mean ±SD of three biological replicates.
Tet2 binds to the promoter of genes impaired by the knockdowns
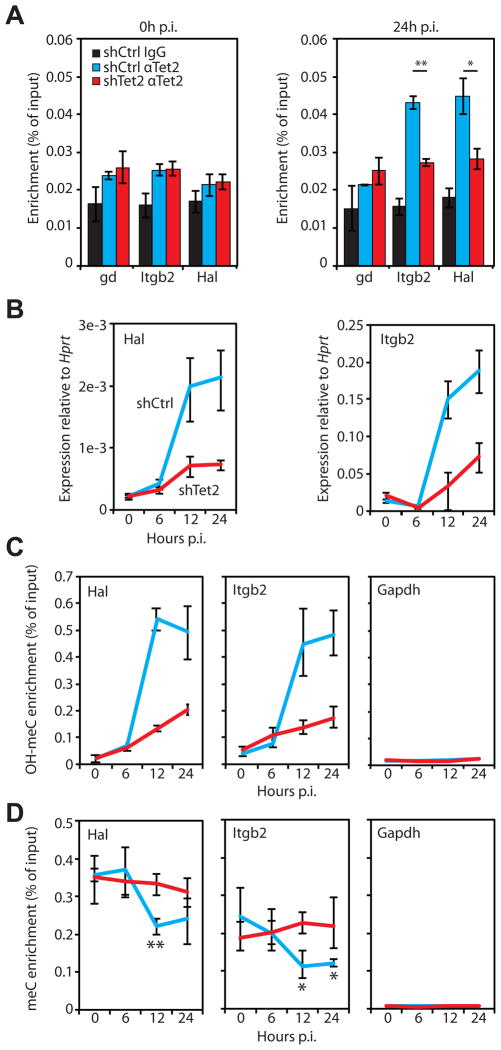
To address whether the genes displaying incomplete activation upon Tet2 kd are direct targets of Tet2, we performed ChIP experiments with control and Tet2 kd C10 cells induced for 24 hours. After immunoprecipitation with anti-Tet2 antibody, qPCR was performed using primers specific for the promoters of several Tet2 responsive genes. A gene desert region and the promoter of a Tet2 unresponsive gene (Rasgrf1) were used as negative controls. These experiments revealed specific Tet2 enrichment at the promoters of Itgb2, Fosl2, Mmp8 and Hal (Figure 4A, Figure S3A, blue bars). In contrast, we did not observe Tet2 binding at the gene desert control nor at the promoters of Rasgrf1 and the Tet2 responsive genes Ahnak and Mmp13. Perhaps in the latter Tet2 binding is at regions outside of the proximal promoters assayed here or they might be indirectly affected. Tet2 binding to the four target genes was found by ChIP assay to be significantly reduced in Tet2 kd cells induced for 24 hours. (Student’s t-test: Hal, P=0.006; Itgb2, P=0.00016; Fosl2, P=0.0012; Mmp8, P=0.0052)(Figure 4A, Figure S3A, red bars) while no Tet2 signal above background was detected in uninduced pre-B cells (Figure 4A). In addition, the four target genes also exhibited decreased expression in pre-B cells from Tet2 knockout animals induced to transdifferentiate for 24 hours compared to Tet2+/+ cells (Figure S3B). These results show that Tet2 binds directly to the promoters of several of the Tet2 responsive genes studied.
Figure 4. Tet2 binding to the promoters of selected genes results in OH-MeC accumulation. See also Figure S3.
A) Tet2 binding at target gene promoters in 0h and 24h p.i. cells was analyzed by chromatin immunoprecipitation (ChIP) followed by qPCR. The experiment was carried out in control (shCtrl, blue bars) and Tet2 kd (shTet2, red bars) cells. Local background was determined through IgG experiments carried out in shCtrl samples (IgG, black bars). Data presented as mean percentage of input ±SD of three biological replicates. Student’s t-test P value between shCtrl and shTet2 replicates < 0.01 (*), < 0.001 (**). Gene desert region, gd.
B) qRT-PCR analysis of Hal (left panel) and Itgb2 (right panel) gene expression in control (shCtrl, blue lines) and Tet2 kd (shTet2, red lines) cells induced to transdifferentiate for the time indicated on the x-axis. Data presented as mean expression relative to Hprt ±SD of three biological replicates.
C) Hydroxymethyl-DIP (αOH-MeC) assays followed by qPCR were performed at the indicated gene promoters (Hal, Itgb2, Gapdh) in control and Tet2 kd (colored as in B) cells at 0, 6, 12, and 24h p.i. of transdifferentiation. Data presented as in A.
D) Methyl-DIP (αMeC) assays followed by qPCR were performed at the indicated gene promoters (Hal, Itgb2, Gapdh) in control and Tet2 kd (colored as in B) cells at 0, 6, 12, and 24h p.i. of transdifferentiation. Data presented as in A. Asterisks indicate Student’s t-test P value between 0h and the indicated time in shCtrl cells < 0.05 (*), < 0.01 (**).
Promoters of upregulated Tet2 target genes are enriched for OH-MeC
If the enzymatic activity of Tet2 played a role in the activation of myeloid target genes we would expect to see an enrichment of OH-MeC within their promoters after induction of transdifferentiation. To assay for the appearance of OH-MeC at Tet2 targets, we performed OH-MeC DNA immunoprecipitation (OH-meDIP) experiments followed by qPCR, using the same primers as for the Tet2 binding assays. DNA was extracted from uninduced and 24-hour induced control and Tet2 kd cells, immunoprecipitated using an antibody specific for the OH-MeC modification (Figure S3C), and the samples analyzed by qPCR. As a background control we included primers that recognize the promoter region of the Gapdh gene. We observed a 2 to 4-fold enrichment of OH-MeC at target gene promoters in cells induced to transdifferentiate for 24 hours compared to uninduced cells (Figure S3D). This enrichment was Tet2 dependent as it was significantly reduced in Tet2 kd cells (Student’s t-test: Itgb2, P=0.012; Fosl2, P=0.036; Mmp8, P=0.009; Hal, P=0.021) (Figure S3E). In contrast, in uninduced cells the Tet2 target gene promoters showed little OH-MeC enrichment compared to Gapdh (Figure S3D). These data demonstrate that promoter hydroxymethylation accompanies the Tet2 dependent activation of myeloid target genes.
To investigate the timing of gene upregulation relative to Tet2 binding and OH-MeC enrichment, we chose Hal and Itgb2 since these two Tet2 target genes gave the highest level of OH-MeC enrichment compared to background controls in OH-meDIP experiments (Figure S3D). For this analysis, control and Tet2 kd C10 cells were induced to transdifferentiate and cell samples were collected at 0, 6, 12, and 24h p.i. At each time point RNA and DNA was extracted and subjected to qRT-PCR and OH-meDIP, respectively. qRT-PCR analysis revealed a dramatic upregulation of both genes between 6h and 12h and this was impaired by the kd construct (Figure 4B). OH-meDIP analysis showed a remarkably similar pattern of enrichment of OH-meC during upregulation that was inhibited in Tet2 kd cells (Figure 4C). The Gapdh promoter, used as a negative control, showed low enrichment through all time points (Figure 4C). In parallel with the OH-meDIP experiments we also performed meDIP experiments with an antibody specific for methylated cytosine residues (Figure S3C). This experiment revealed an enrichment of meC signal at both the Hal and Itgb2 promoters relative to the control Gapdh promoter at all time points analyzed. In addition, there was a slight depletion of the meC signal at both Tet2 target genes at 12h p.i. (Student’s t-test: Hal, P=0.001; Itgb2, P=0.043) not seen in Tet2 kd cells (Figure 4D). In summary, Tet2 function is required for promoter hydroxymethylation and full gene activation during C/EBPa induced transdifferentiation. Furthermore promoters show a partial depletion of methylation at 12h p.i..
Tet2 function during transdifferentiation requires DNA methylation
The above results suggest that Tet2 is required to relieve the silencing effects of MeC at the promoters of a subset of upregulated genes. Supporting this finding, analysis of upregulated genes revealed that Tet2 responsive genes are expressed at a significantly lower level in uninduced cells than Tet2 unresponsive genes (Kolmogorov-Smirnov test P=1.029e-12)(Figure 5A). In addition, analyses of a methyl-binding domain (MBD) pull down experiment (MethylCAP-chip) performed in uninduced cells (Rodriguez-Ubreva et al., 2011) revealed enrichment of MeC within the promoters of Tet2 responsive genes when compared to unresponsive genes (Figure 5B). If Tet2 was indeed necessary to relieve MeC-mediating silencing, then removal of the modification prior to induction of transdifferentiation should preclude its necessity, thus rescuing the phenotype in Tet2 kd cells. Pretreatment of the inducible cell line with the DNA methylation inhibitor 5-azacytidine (azaC) resulted in about 30% global demethylation at 24h p.i as assessed by cytosine extension assay (Figure S4A,B) (Fujiwara and Ito, 2002; Pogribny et al., 1999). Similar levels of gene specific demethylation were observed at the Hal gene promoter, as assessed by bisulfite sequencing analysis (Figure S4C). Furthermore, drug treatment partially rescued Mac-1 upregulation (Figure 5C) after bEst induction of Tet2 kd cells and also rescued the impaired upregulation of the Tet2 target genes Itgb2, Fosl2, Mmp8, and Hal at 24 hours p.i. (Figure 5D).
Figure 5. DNA methylation contributes to Tet2 target gene silencing. See also Figure S4.
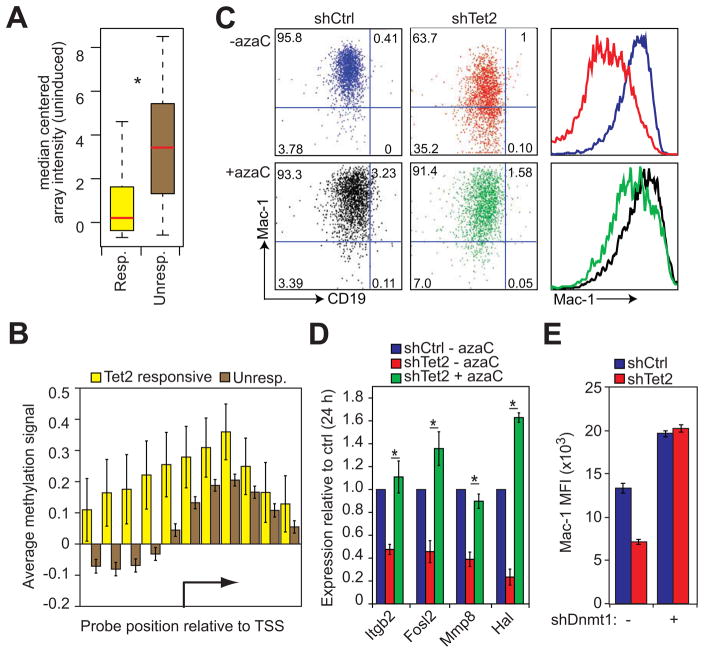
A) Box plot representation of median-centered gene expression (log2 expression units) in uninduced control cells for genes that are upregulated during normal transdifferentiation and either respond to Tet2 kd (Resp., yellow, n=58) or do not respond to kd (Unresp., brown, n=1447). Red line indicates the median expression value of each cohort. Asterisk denotes Kolmogorov-Smirnov test P=1.029e-12.
B) Bar plot of average DNA methylation at the transcription start sites (TSS) of Tet2 responsive (yellow bars, n=58) and unresponsive genes (brown bars, n=1182) as determined by MethylCAP-chip assay. Data are presented as the mean normalized signal from a methyl-DNA binding domain pull down assay hybridized to promoter tiling arrays. Error bars represent ±SD between aligned probes within a given group.
C) FACS plots of 48h p.i. pre-B cell to macrophage transdifferentiation of control (shCtrl) and Tet2 kd (shTet2) cells carried out with or without treatment with 5-azacytidine (+azaC, -azaC). Quadrant gates are set with respect to uninduced control cells. Mac-1 histogram plots (right panels) colored with respect to FACS plots. Data presented are representative of three biological replicates.
D) qRT-PCR analysis of Tet2 target genes 24h p.i. in control cells (shCtrl – azaC, blue) and Tet2 kd cells with or without 5-azacytidine pretreatment (shTet2 + azaC, green; shTet2 – azaC, red). Data represented as mean ±SD of Hprt normalized expression set relative to shCtrl levels of three biological replicates. Asterisks indicate Student’s t-test P value < 0.05 between shTet2 kd +/− azaC.
E) Bar plots of Mac-1 expression in control (shCtrl, blue bars) and Tet2 knockdown (shTet2, red bars) cells superinfected or not with a lentivirus containing an shDnmt1 knockdown construct. Data presented as mean ±SD of three biological replicates. Representative FACS plots are presented in Figure S4C.
We next tested whether DNA methyltransferase 1 (Dnmt1) knockdown could likewise rescue the phenotypes associated with Tet2 kd since Dnmt1 is responsible for maintenance of DNA methylation in mammalian cells (Chen and Li, 2006) and loss of function results in a dramatic decrease of MeC genome-wide (Chen et al., 2007). To this end, the inducible pre-B cell line C11 was infected with shTet2 lentivirus, selected in puromycin and then super-infected with an shDnmt1 lentivirus harboring GFP (Ventura et al., 2004). Doubly infected cells were FACS sorted and subjected to bisulfite sequencing to verify gene specific demethylation (Figure S4D). Separate aliquots of cells were then induced to transdifferentiate for 48 hours. FACS analysis revealed knockdown of Dnmt1 not only enhanced Mac-1 upregulation in control cells (Figure 5E, blue bars; Figure S3E) but also completely rescued the Mac-1 activation defect in Tet2 kd cells (Figure 5E; Figure S3E). Together, these results indicate that DNA methylation is indeed required for the ability of Tet2 to enhance the de-repression of macrophage genes, consistent with the idea that methylated residues are a substrate of the enzyme.
Promoters of selected Tet2 target genes may become demethylated following transcriptional activation
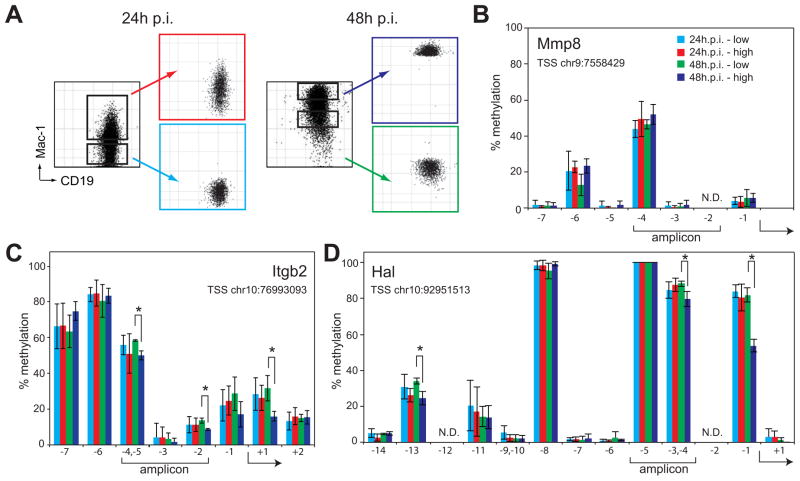
Previous reports have implicated methyl cytosine hydroxylation as an intermediate to complete demethylation through DNA repair pathways (Guo et al., 2011; He et al., 2011; Ito et al., 2011). Therefore, we investigated the timing of demethylation of promoters at the direct Tet2 target genes Hal, Itgb2 and Mmp8. For this purpose C10 cells were treated with bEst and cells sorted based on the extent of their reprogramming as measured by Mac-1 upregulation, generating Mac-1 low and Mac-1 high cells at 24 and 48h p.i. (Figure 6A). DNA was extracted from these four cell fractions and subjected to Sequenom MassARRAY analysis, a sensitive and quantitative method to measure CpG methylation (Ehrich et al., 2005). This analysis revealed heavily methylated cytosine residues within the promoters of all three genes studied. However, while there were no significant differences in the promoter DNA methylation of the 24h p.i. samples (Figure 6B-D light blue and red bars), in the fast responding 48 hour sample we found evidence that specific cytosine residues within the Itgb2 and Hal promoters showed a decrease in methylation of the more rapidly transdifferentiating cell fraction (Figure 6C and D). Notably this was not apparent in the Mmp8 promoter (Figure 6B). These data show that the upregulation of the Tet2 target genes Hal, Itgb2, and Mmp8 correlate with hydroxylation of methylated cytosine residues at the corresponding promoters and that their sustained upregulation may be followed by demethylation, with all but one promoter showing relatively small changes in DNA methylation.
Figure 6. Cytosines within selected Tet2 target gene promoters may become demethylated after induced transdifferentiation.
A) FACS plots of C10 cells induced for 24 or 48h prior and after sorting into Mac-1 low and Mac-1 high populations.
B–D) Percentage of individual modified cytosines at the Mmp8 (B), Itgb2 (C) and Hal (D) promoters as determined by Sequenom MassARRAY analysis at the indicated positions. X-axis depicts the position of analyzed cytosines with respect to the TSS of the gene and y-axis the percentage of methylation. Data represent mean ±SD of three biological replicates. Asterisks, Student’s t-test P value < 0.05 between Mac-1 high and Mac-1 low populations 48h p.i.; Brackets below the x-axis, position of the qPCR amplicons used for DNA immunoprecipitation and ChIP experiments in Fig. 4. The genomic coordinate of each TSS is given in the graph and the arrow indicates the relative location within the individual CpGs analyzed. CpGs that could not be analyzed, N.D. Color corresponds to the FACS plots shown in A. Chromosome position of the assayed region: Mmp8, chr9:7557999-7558511; Itgb2, chr10:76992548-76993258; Hal, chr10:92950958-92951420. Position data is relative to mouse genome build mm9.
DISCUSSION
Two recent studies with Tet2 knockout mice have reported an increase in the number of myelomonocytic cells and an impairment of Mac-1 expression in a portion of the myeloid cells, leading to the hypothesis that the enzyme functions to inhibit premature differentiation of the myeloid lineage (Ko et al., 2011; Moran-Crusio et al., 2011). Our Tet2 knockdowns also induce a Mac-1 low phenotype in transdifferentiated macrophages as well as in normal progenitors but we attribute this to a requirement of Tet2 for the de-repression of a subset of macrophage genes. The fact that we do not see an expansion of the myeloid pool could be because the process is direct and does not involve the formation of early progenitors (Di Tullio et al., 2011). It will be interesting to determine whether the Mac-1 low populations described in Tet2−/− mice are also impaired in their phagocytic capacity, as observed here. Annotation of the 58 genes exhibiting defective activation reveals no genes that are overtly involved in phagocytosis. However, 19 genes encode proteins integral or closely associated with the plasma membrane, including Mmp8, Mmp13, Ahnak, and Itgb2, and may be contributing to the altered phenotype.
It has been reported that HSCs from mice with reduced Dnmt1 activity cannot suppress key myelo-erythroid regulators and exhibit a decrease in lymphoid progeny (Broske et al., 2009). Together with our observations, these data suggest that both the functional neutralization of DNA methylation by hydroxymethylation as well as demethylation are key events controlling macrophage specification. This hypothesis is supported by our observation that the promoters of two out of three Tet2 target genes studied begin to show evidence of demethylation after Tet2 mediated hydroxylation during the cell conversion process.
The idea that Tet2 alleviates the silencing effect of MeC is supported by several lines of evidence: i) Tet2 responsive genes are promoter-hypermethylated in the starting pre-B cells. ii) AzaC induced-demethylation or Dnmt1 kd prior to induction of the lineage switch precludes the necessity for Tet2 function during the process. iii) Tet2 target gene promoters become hydroxymethylated at a time when the effect of the kd on myeloid gene expression is fully obvious yet before demethylation is observed.
The observation that at later time points during transdifferentiation target gene promoters become demethylated supports previous findings suggesting that Tet mediated hydroxylation serves as an intermediate to complete demethylation through DNA repair pathways (Guo et al., 2011; Ito et al., 2011). This multi-step/multi-enzyme mechanism of complete demethylation may not be rapid enough for the immediate activation of gene transcription as a response to external signaling that may induce cell fate transitions. It is therefore reasonable to assume that cells have two separate but interconnected mechanisms of activation of MeC-silenced genes: a fast one involving hydroxymethylation and a slow one involving complete demethylation. Whether this complete demethylation is active, such as through the thymine-DNA glycosylase repair pathway or passive through DNA replication is unclear. However, the fact that during pre-B cell to macrophage transdifferentiation cells divide once within 24h p.i. and that we only observe demethylation a day later in the fastest switching cell subpopulation suggests that demethylation involves an active mechanism.
Perhaps surprisingly, only about 6% of all genes that become upregulated during C/EBPa induced transdifferentiation are affected by the Tet2 kd. Also, the majority of the Tet2 responsive genes are upregulated to some extent even when Tet2 is knocked down. This is consistent with the finding that there is very little change in the genome-wide distribution of MeC upon induction of transdifferentiation (Rodriguez-Ubreva et al., 2011) and implies that C/EBPa is able to overcome the silencing effects of DNA methylation at most myeloid genes. Alternatively, methylation may not be important for the silencing of these genes in pre-B cells. However, the observed and relatively subtle block of macrophage differentiation in Tet2 kd cells might be sufficient to confer a selective growth advantage to Tet2 deficient cells during the development of AML.
Collectively, our data show that the expression of myeloid genes in hematopoietic precursors and in B lineage cells is subjected to two layers of regulation at the level of DNA modifications. The first one is well-studied and consists in the addition of methyl groups to cytosines by DNA methyltransferases and is responsible for long-term repression (also known as ‘epigenetic memory’ (Reik, 2007)). The second layer, postulated here, consists in the Tet2 mediated hydroxy-methylation of methylated cytosine residues. This layer may afford the cells with the capacity to rapidly respond to lineage instructive transcription factors activated by internal or external cues. Such a regulator could be PU.1, an obligatory partner of C/EBPa in the activation of myeloid genes (Bussmann et al., 2009; Feng et al., 2008; Laiosa et al., 2006; Xie et al., 2004) that is upregulated by M-CSF during the induction of macrophage fate in granulocyte/macrophage progenitors (Rieger et al., 2009; Sarrazin et al., 2009) or C/EBPa itself. The two layers of epigenetic gene regulation may thus maximize the cell’s flexibility to adapt to changing requirements during differentiation. It will now be interesting to determine how Tet2 is recruited to its target genes and whether Tet-mediated hydroxymethylation plays a more general role in transcription factor induced transdifferentiation in other cell systems.
EXPERIMENTAL PROCEDURES
Cell culture and transdifferentiation induction
C10 cells and C11 cells (maintained in RPMI 1640 media (Lonza BE12-918F) supplemented with 20mM L-Glutamine (Gibco 25030), 10%(v/v) FBS and 20μM 2-mercaptoethanol) were cultured using standard methods. Transdifferentiation assays were performed as in (Bussmann et al., 2009). CD19 positive primary B-cells were obtained from bone marrow of Tet2+/+ and Tet2−/− littermates (Moran-Crusio et al., 2011) using MACS separation (Miltenyi Biotec). Primary cells were induced to transdifferentiate as described (Di Tullio et al., 2011).
FACS analysis and phagocytosis assay
Cells were washed in PBS+10%(v/v) FBS, incubated with Fc Block and antibody as outlined in the Supplementary Table 1, washed in PBS+10% FBS, analyzed with an LSRII or FACScanto machine or sorted on a FACSAria (all BD Biosciences). The data were analyzed using FloJo version 8.8.7. Phagocytosis assays were performed with green fluorescent carboxylated 1μ beads (Polysciences, Inc. 17154) as described in (Bussmann et al., 2009).
Generation of knockdown cells
Lentiviral shRNA-puromycin constructs were obtained from Sigma-Aldrich (shTet2kd1, TRC0000250893; shTet2kd2, TRC0000250896; shControl, SHC002). shRNA-EGFP plasmids were constructed by standard PCR cloning methods which replaced puroR with EGFP amplified from pET15-EGFP. shDnmt1 plasmid was constructed by cloning a hairpin (Ventura et al., 2004) downstream of the U6 promoter in a lentiviral construct (He et al., 2008) with a PGK promoter driven EGFP transgene. For virus production details see the supplement. Tet2 knock-down and control C10 cells were generated through spinoculation of lentivirus for 45 min at 1000×g and 32°C. Cells were kept at 37°C for 12–16 hrs prior to washing and beginning selection in 1μg/ml puromycin or subjected to FACS sorting for GFP expression.
Azacytidine experiments
C10 cells were pre-incubated in growth medium containing 0.5μM 5-azacytidine (Sigma A3656) for 3h prior to induction of transdifferentiation and maintained in drug supplemented medium throughout the time-course.
qRT-PCR, qPCR, and microarrays
RNA was isolated from cells using the RNeasy Mini Kit (Qiagen 74106) as per manufacturer’s instructions and quality was assessed on a Bioanalyzer 2100 (Agilent) prior to use. For reverse transcription, 1μg of RNA was processed using High Capacity RNA to cDNA Master Mix (Applied Biosystems 4390712) following the manufacturer’s instructions. For quantitative PCR, cDNA or ChIP DNA was mixed with appropriate primers (Table S2) and 2X Power SYBR Green (Applied Biosystems 4367659), the reactions were run on an ABI7900 Real Time PCR machine and data was imported into Microsoft Excel for processing. Microarray experiments and data analysis are described in the supplement.
DNA extraction and DNA immunoprecipitation (DIP)
DNA extraction and IP were carried out by a previously published protocol (http://www.epigenome-noe.net/, PROT33) with the following alterations. Genomic DNA was fragmented to a mean size of 350 bp using a Bioruptor (Diagenode) for 7×30sec cycles. DNA (resuspended in 1X IP buffer) was incubated with 5μg of anti-OHMeC antibody or 8μg of anti-MeC (Table S1) at 4°C for 2h and 40μl of a 1:1 mixture of BSA-blocked Protein A (Millipore 16–125) and Protein G (Millipore 16–266) agarose beads were added, followed by an additional 2h incubation at 4°C. After washing 3 times in 1x IP buffer, immune complexes were released by incubation with 70μg Proteinase K, and DNA was extracted once with phenol, extracted once with chloroform and recovered by EtOH precipitation for qPCR analysis.
MethylCAP-chip array analysis
MethylCAP-chip data of uninduced C10 cells were generated in a previous study (Rodriguez-Ubreva et al., 2011). Raw data were analyzed as outlined in the supplement.
Sequenom analysis
For a detailed protocol of Sequenom analysis see the supplement. Briefly, genomic DNA from sorted cell populations was purified using Qiagen DNeasy columns (Qiagen 69506), bisulfite treated (Zymo Research) and subjected to methylation analysis using Sequenom MassARRAY technology by a protocol similar to that published (Ehrich et al., 2005). PCR primers for amplification of the promoter regions of the Hal, Itgb2 and Mmp8 genes were designed by using Epidesigner (Sequenom). Primer sequences are available in the Table S2.
Chromatin immunoprecipitation (ChIP)
1mg of crosslinked and sonicated chromatin (prepared as in (Morey et al., 2012)) was incubated with 10μg of αTet2 antibody, 2μg of αC/EBPa (Santa Cruz) or an equal amount of purified rabbit IgG overnight at 4°C in IP buffer. After a 2-hour incubation with BSA-blocked protein A/protein G agarose, the beads were washed 3x in low salt buffer (50mM Hepes pH 7.5, 140 mM NaCl, 1% Triton X-100), once in high salt buffer (50mM Hepes pH 7.5, 500 mM NaCl, 1% Triton X-100), reverse crosslinked at 65°C for 3 hours, purified over columns (Qiagen #28706), and subjected to qPCR using primers outlined in Table S2.
Statistical Analysis
Parametric statistical testing in Figs. 4, 5 & 6 was carried out in Microsoft Excel using two-tailed, homoscedastic Student’s t test. Nonparametric statistical testing was carried out in R v2.11.1 using Wilcoxon rank sum test (Fig. S1F) or the Kolmogorov-Smirnov test (Fig. 5A).
Supplementary Material
HIGHLIGHTS.
C/EBPa induces Tet2 upregulation during pre-B cell to macrophage transdifferentiation
Tet2 is required for efficient myeloid transdifferentiation
Tet2 knockdown during transdifferentiation permitted the identification of target genes
Tet2 target gene activation correlates with hydroxy-methylation of methylated promoters
Acknowledgments
We would like to thank Almudena Ramiro, Pia Cosma, Min Ye, Matthias Stadtfeld, Chris van Oevelen and Salvador Aznar Benitah for comments and Graf lab members for discussions. This work has been supported by grants from the Spanish Ministry of Science and Innovation (MICINN) (SAF2007-63058, TG; PI081346, EB), the CONSOLIDER Epigenetica (CSD2006-49, EB and TG), AGAUR (SGR768, TG). I.A. is a Howard Hughes Medical Institute Early Career Investigator supported by the National Health Institutes (RO1CA133379, RO1CA105129, RO1CA149655) and the V Foundation for Cancer Research. EMK is supported by an EMBO long-term fellowship. KH was supported by grants from the Danish National Research Foundation and the Danish Cancer Society.
Footnotes
ACCESSION NUMBERS
AUTHOR CONTRIBUTIONS:
EMK initiated the project, carried out the work and co-wrote the manuscript. TG directed the project and co-wrote the manuscript. JRU and EB performed the MBD-array experiments. JC and KH provided the Tet2 antibody. LC and IA provided Tet2−/− bone marrow samples.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adolfsson J, Mansson R, Buza-Vidas N, Hultquist A, Liuba K, Jensen CT, Bryder D, Yang L, Borge OJ, Thoren LA, et al. Identification of Flt3+ lympho-myeloid stem cells lacking erythro-megakaryocytic potential a revised road map for adult blood lineage commitment. Cell. 2005;121:295–306. doi: 10.1016/j.cell.2005.02.013. [DOI] [PubMed] [Google Scholar]
- Bozec A, Bakiri L, Hoebertz A, Eferl R, Schilling AF, Komnenovic V, Scheuch H, Priemel M, Stewart CL, Amling M, et al. Osteoclast size is controlled by Fra-2 through LIF/LIF-receptor signalling and hypoxia. Nature. 2008;454:221–225. doi: 10.1038/nature07019. [DOI] [PubMed] [Google Scholar]
- Broske AM, Vockentanz L, Kharazi S, Huska MR, Mancini E, Scheller M, Kuhl C, Enns A, Prinz M, Jaenisch R, et al. DNA methylation protects hematopoietic stem cell multipotency from myeloerythroid restriction. Nat Genet. 2009;41:1207–1215. doi: 10.1038/ng.463. [DOI] [PubMed] [Google Scholar]
- Bussmann LH, Schubert A, Vu Manh TP, De Andres L, Desbordes SC, Parra M, Zimmermann T, Rapino F, Rodriguez-Ubreva J, Ballestar E, et al. A robust and highly efficient immune cell reprogramming system. Cell Stem Cell. 2009;5:554–566. doi: 10.1016/j.stem.2009.10.004. [DOI] [PubMed] [Google Scholar]
- Chen T, Hevi S, Gay F, Tsujimoto N, He T, Zhang B, Ueda Y, Li E. Complete inactivation of DNMT1 leads to mitotic catastrophe in human cancer cells. Nat Genet. 2007;39:391–396. doi: 10.1038/ng1982. [DOI] [PubMed] [Google Scholar]
- Chen T, Li E. Establishment and maintenance of DNA methylation patterns in mammals. Current topics in microbiology and immunology. 2006;301:179–201. doi: 10.1007/3-540-31390-7_6. [DOI] [PubMed] [Google Scholar]
- Di Tullio A, Manh TP, Schubert A, Mansson R, Graf T. CCAAT/enhancer binding protein {alpha} (C/EBP{alpha})-induced transdifferentiation of pre-B cells into macrophages involves no overt retrodifferentiation. Proc Natl Acad Sci U S A. 2011;108:17016–17021. doi: 10.1073/pnas.1112169108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrich M, Nelson MR, Stanssens P, Zabeau M, Liloglou T, Xinarianos G, Cantor CR, Field JK, van den Boom D. Quantitative high-throughput analysis of DNA methylation patterns by base-specific cleavage and mass spectrometry. Proc Natl Acad Sci U S A. 2005;102:15785–15790. doi: 10.1073/pnas.0507816102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fallowfield JA, Mizuno M, Kendall TJ, Constandinou CM, Benyon RC, Duffield JS, Iredale JP. Scar-associated macrophages are a major source of hepatic matrix metalloproteinase-13 and facilitate the resolution of murine hepatic fibrosis. J Immunol. 2007;178:5288–5295. doi: 10.4049/jimmunol.178.8.5288. [DOI] [PubMed] [Google Scholar]
- Feng R, Desbordes SC, Xie H, Tillo ES, Pixley F, Stanley ER, Graf T. PU.1 and C/EBPalpha/beta convert fibroblasts into macrophage-like cells. Proc Natl Acad Sci U S A. 2008;105:6057–6062. doi: 10.1073/pnas.0711961105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueroa ME, Abdel-Wahab O, Lu C, Ward PS, Patel J, Shih A, Li Y, Bhagwat N, Vasanthakumar A, Fernandez HF, et al. Leukemic IDH1 and IDH2 mutations result in a hypermethylation phenotype, disrupt TET2 function, and impair hematopoietic differentiation. Cancer Cell. 2010;18:553–567. doi: 10.1016/j.ccr.2010.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara H, Ito M. Nonisotopic cytosine extension assay: a highly sensitive method to evaluate CpG island methylation in the whole genome. Analytical biochemistry. 2002;307:386–389. doi: 10.1016/s0003-2697(02)00053-2. [DOI] [PubMed] [Google Scholar]
- Guo JU, Su Y, Zhong C, Ming GL, Song H. Hydroxylation of 5-methylcytosine by TET1 promotes active DNA demethylation in the adult brain. Cell. 2011;145:423–434. doi: 10.1016/j.cell.2011.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He J, Kallin EM, Tsukada Y, Zhang Y. The H3K36 demethylase Jhdm1b/Kdm2b regulates cell proliferation and senescence through p15(Ink4b) Nature structural & molecular biology. 2008;15:1169–1175. doi: 10.1038/nsmb.1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He YF, Li BZ, Li Z, Liu P, Wang Y, Tang Q, Ding J, Jia Y, Chen Z, Li L, et al. Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science. 2011;333:1303–1307. doi: 10.1126/science.1210944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito S, Shen L, Dai Q, Wu SC, Collins LB, Swenberg JA, He C, Zhang Y. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science. 2011;333:1300–1303. doi: 10.1126/science.1210597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin SG, Kadam S, Pfeifer GP. Examination of the specificity of DNA methylation profiling techniques towards 5-methylcytosine and 5-hydroxymethylcytosine. Nucleic Acids Res. 2010;38:e125. doi: 10.1093/nar/gkq223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko M, Bandukwala HS, An J, Lamperti ED, Thompson EC, Hastie R, Tsangaratou A, Rajewsky K, Koralov SB, Rao A. Ten-Eleven-Translocation 2 (TET2) negatively regulates homeostasis and differentiation of hematopoietic stem cells in mice. Proc Natl Acad Sci U S A. 2011;108:14566–14571. doi: 10.1073/pnas.1112317108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko M, Huang Y, Jankowska AM, Pape UJ, Tahiliani M, Bandukwala HS, An J, Lamperti ED, Koh KP, Ganetzky R, et al. Impaired hydroxylation of 5-methylcytosine in myeloid cancers with mutant TET2. Nature. 2010;468:839–843. doi: 10.1038/nature09586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh KP, Yabuuchi A, Rao S, Huang Y, Cunniff K, Nardone J, Laiho A, Tahiliani M, Sommer CA, Mostoslavsky G, et al. Tet1 and Tet2 regulate 5-hydroxymethylcytosine production and cell lineage specification in mouse embryonic stem cells. Cell Stem Cell. 2011;8:200–213. doi: 10.1016/j.stem.2011.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laiosa CV, Stadtfeld M, Xie H, de Andres-Aguayo L, Graf T. Reprogramming of committed T cell progenitors to macrophages and dendritic cells by C/EBP alpha and PU.1 transcription factors. Immunity. 2006;25:731–744. doi: 10.1016/j.immuni.2006.09.011. [DOI] [PubMed] [Google Scholar]
- Langemeijer SM, Kuiper RP, Berends M, Knops R, Aslanyan MG, Massop M, Stevens-Linders E, van Hoogen P, van Kessel AG, Raymakers RA, et al. Acquired mutations in TET2 are common in myelodysplastic syndromes. Nat Genet. 2009;41:838–842. doi: 10.1038/ng.391. [DOI] [PubMed] [Google Scholar]
- Moran-Crusio K, Reavie L, Shih A, Abdel-Wahab O, Ndiaye-Lobry D, Lobry C, Figueroa ME, Vasanthakumar A, Patel J, Zhao X, et al. Tet2 loss leads to increased hematopoietic stem cell self-renewal and myeloid transformation. Cancer Cell. 2011;20:11–24. doi: 10.1016/j.ccr.2011.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morey L, Pascual G, Cozzuto L, Roma G, Wutz A, Benitah SA, Di Croce L. Nonoverlapping functions of the Polycomb group Cbx family of proteins in embryonic stem cells. Cell Stem Cell. 2012;10:47–62. doi: 10.1016/j.stem.2011.12.006. [DOI] [PubMed] [Google Scholar]
- Pogribny I, Yi P, James SJ. A sensitive new method for rapid detection of abnormal methylation patterns in global DNA and within CpG islands. Biochemical and biophysical research communications. 1999;262:624–628. doi: 10.1006/bbrc.1999.1187. [DOI] [PubMed] [Google Scholar]
- Prikk K, Maisi P, Pirila E, Sepper R, Salo T, Wahlgren J, Sorsa T. In vivo collagenase-2 (MMP-8) expression by human bronchial epithelial cells and monocytes/macrophages in bronchiectasis. J Pathol. 2001;194:232–238. doi: 10.1002/path.849. [DOI] [PubMed] [Google Scholar]
- Quivoron C, Couronne L, Della Valle V, Lopez CK, Plo I, Wagner-Ballon O, Do Cruzeiro M, Delhommeau F, Arnulf B, Stern MH, et al. TET2 inactivation results in pleiotropic hematopoietic abnormalities in mouse and is a recurrent event during human lymphomagenesis. Cancer Cell. 2011;20:25–38. doi: 10.1016/j.ccr.2011.06.003. [DOI] [PubMed] [Google Scholar]
- Rai K, Huggins IJ, James SR, Karpf AR, Jones DA, Cairns BR. DNA demethylation in zebrafish involves the coupling of a deaminase, a glycosylase, and gadd45. Cell. 2008;135:1201–1212. doi: 10.1016/j.cell.2008.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reik W. Stability and flexibility of epigenetic gene regulation in mammalian development. Nature. 2007;447:425–432. doi: 10.1038/nature05918. [DOI] [PubMed] [Google Scholar]
- Rieger MA, Hoppe PS, Smejkal BM, Eitelhuber AC, Schroeder T. Hematopoietic cytokines can instruct lineage choice. Science. 2009;325:217–218. doi: 10.1126/science.1171461. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Ubreva J, Ciudad L, Gomez-Cabrero D, Parra M, Bussmann LH, di Tullio A, Kallin EM, Tegner J, Graf T, Ballestar E. Pre-B cell to macrophage transdifferentiation without significant promoter DNA methylation changes. Nucleic Acids Res. 2011 doi: 10.1093/nar/gkr1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarrazin S, Mossadegh-Keller N, Fukao T, Aziz A, Mourcin F, Vanhille L, Kelly Modis L, Kastner P, Chan S, Duprez E, et al. MafB restricts M-CSF-dependent myeloid commitment divisions of hematopoietic stem cells. Cell. 2009;138:300–313. doi: 10.1016/j.cell.2009.04.057. [DOI] [PubMed] [Google Scholar]
- Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, Agarwal S, Iyer LM, Liu DR, Aravind L, et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tefferi A. Novel mutations and their functional and clinical relevance in myeloproliferative neoplasms: JAK2, MPL, TET2, ASXL1, CBL, IDH and IKZF1. Leukemia. 2010;24:1128–1138. doi: 10.1038/leu.2010.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tefferi A, Lim KH, Levine R. Mutation in TET2 in myeloid cancers. N Engl J Med. 2009;361:1117. doi: 10.1056/NEJMc091348. author reply 1117–1118. [DOI] [PubMed] [Google Scholar]
- Valinluck V, Tsai HH, Rogstad DK, Burdzy A, Bird A, Sowers LC. Oxidative damage to methyl-CpG sequences inhibits the binding of the methyl-CpG binding domain (MBD) of methyl-CpG binding protein 2 (MeCP2) Nucleic Acids Res. 2004;32:4100–4108. doi: 10.1093/nar/gkh739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ventura A, Meissner A, Dillon CP, McManus M, Sharp PA, Van Parijs L, Jaenisch R, Jacks T. Cre-lox-regulated conditional RNA interference from transgenes. Proc Natl Acad Sci U S A. 2004;101:10380–10385. doi: 10.1073/pnas.0403954101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber M, Davies JJ, Wittig D, Oakeley EJ, Haase M, Lam WL, Schubeler D. Chromosome-wide and promoter-specific analyses identify sites of differential DNA methylation in normal and transformed human cells. Nat Genet. 2005;37:853–862. doi: 10.1038/ng1598. [DOI] [PubMed] [Google Scholar]
- Williams K, Christensen J, Pedersen MT, Johansen JV, Cloos PA, Rappsilber J, Helin K. TET1 and hydroxymethylcytosine in transcription and DNA methylation fidelity. Nature. 2011;473:343–348. doi: 10.1038/nature10066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu SC, Zhang Y. Active DNA demethylation: many roads lead to Rome. Nat Rev Mol Cell Biol. 2010;11:607–620. doi: 10.1038/nrm2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie H, Ye M, Feng R, Graf T. Stepwise reprogramming of B cells into macrophages. Cell. 2004;117:663–676. doi: 10.1016/s0092-8674(04)00419-2. [DOI] [PubMed] [Google Scholar]
- Yildirim O, Li R, Hung JH, Chen PB, Dong X, Ee LS, Weng Z, Rando OJ, Fazzio TG. Mbd3/NURD complex regulates expression of 5-hydroxymethylcytosine marked genes in embryonic stem cells. Cell. 2011;147:1498–1510. doi: 10.1016/j.cell.2011.11.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.