Abstract
Inositol pyrophosphates are highly energetic inositol polyphosphate molecules present in organisms from slime molds and yeast to mammals. Distinct classes of enzymes generate different forms of inositol pyrophosphates. The biosynthesis of these substances principally involves phosphorylation of inositol hexakisphosphate (IP6) to generate the pyrophosphate IP7. Initial insights into functions of these substances derived primarily from yeast, which contain a single isoform of IP6 kinase (yIP6K), as well as from the slime mold Dictyostelium. Mammalian functions for inositol pyrophosphates have been investigated by using cell lines to establish roles in various processes, including insulin secretion and apoptosis. More recently, mice with targeted deletion of IP6K isoforms as well as the related inositol polyphosphate multikinase (IPMK) have substantially enhanced our understanding of inositol polyphosphate physiology. Phenotypic alterations in mice lacking inositol hexakisphosphate kinase 1 (IP6K1) reveal signaling roles for these molecules in insulin homeostasis, obesity, and immunological functions. Inositol pyrophosphates regulate these processes at least in part by inhibiting activation of the serine-threonine kinase Akt. Similar studies of IP6K2 establish this enzyme as a cell death inducer acting by stimulating the proapoptotic protein p53. IPMK is responsible for generating the inositol phosphate IP5 but also has phosphatidylinositol 3-kinase activity—that participates in activation of Akt. Here, we discuss recent advances in understanding the physiological functions of the inositol pyrophosphates based in substantial part on studies in mice with deletion of IP6K isoforms. These findings highlight the interplay of IPMK and IP6K in regulating growth factor and nutrient-mediated cell signaling.
INTRODUCTION
Inositol phosphates have been recognized as biologically pertinent molecules since the identification in the early 20th century of inositol hexakisphosphate (IP6, also known as phytic acid) as an abundant constituent of plants that makes up their principal phosphate store. Interest in these substances escalated with the discovery in the mid-1980s of IP3 [inositol 1,4,5-trisphosphate, also abbreviated as Ins(1,4,5) P3] as a second messenger that releases calcium from intracellular stores (1, 2). Inositol polyphosphate derivatives with highly energetic diphosphates (inositol pyrophosphates) were identified in the mid-1990s (3, 4). The term “inositol polyphosphate” is often used to designate inositol with more than a single phosphate, whereas the energetic diphosphates are referred to as inositol pyrophosphates. Initial characterization of inositol pyrophosphate molecules revealed a rapid turnover, suggesting a role in cellular signaling (3–5).
New insights into the functions of the inositol pyrophosphates have been afforded by the identification of their synthetic enzymes (6), in particular by cloning of the cDNAs for these enzymes (7–9) and through their genetic deletion (10–14). Inositol pyrophosphates generated by IP6 kinase 1 (IP6K1) impact insulin homeostasis (10, 11) by inhibiting the serine-threonine protein kinase Akt (11). Mice lacking IP6K1 are protected against age- and high-fat diet–induced obesity (11) and display improved neutrophil phagocytosis (12). IP6K2 is a mediator of p53-determined cell death (13), and IP6K2-deleted mice are predisposed to aerodigestive (upper respiratory and upper digestive) tract carcinoma (14).
The isomer of IP7 generated by the three mammalian IP6Ks and the yeast isoform (Kcs1, designated yIP6K) is 5-IP7 (5-diphosphoinositol pentakisphosphate) (15). A distinct form of IP7, 1-/3-IP7 (1-/3-diphosphoinositol pentakisphosphate, which has an unquantifiable mixture of 1- and 3-diphosphoinositol pentakisphosphates) (16), synthesized in yeast by the enzyme Vip1 (17), promotes phosphate homeostasis (18). Characterization of PP-IP5K (diphosphoinositol pentakisphosphate kinase), the mammalian isoform of Vip1 (16, 19, 20), established that, in mammals, this enzyme is primarily responsible for IP8 formation (16, 19, 20).
IPMK (inositol polyphosphate multikinase) was first identified as Arg82, a transcriptional regulator in yeast (21, 22). IPMK generates inositol tetrakisphosphate (IP4) [both Ins(1,3,4,5)P4 and Ins(1,4,5,6) P4] and IP5 [Ins(1,3,4,5,6)P5] and thus acts upstream of the IP6Ks (7, 23, 24) (Fig. 1). IPMK is also a physiologically important PI 3-kinase that forms PIP3 [phosphatidylinositol (3,4,5)-trisphosphate] (25), which activates Akt (26). IPMK, in a catalytically independent fashion, activates mammalian target of rapamycin (mTOR) (27). Thus, IPMK and IP6K1 participate in a network regulating growth factor–and nutrient-mediated signaling.
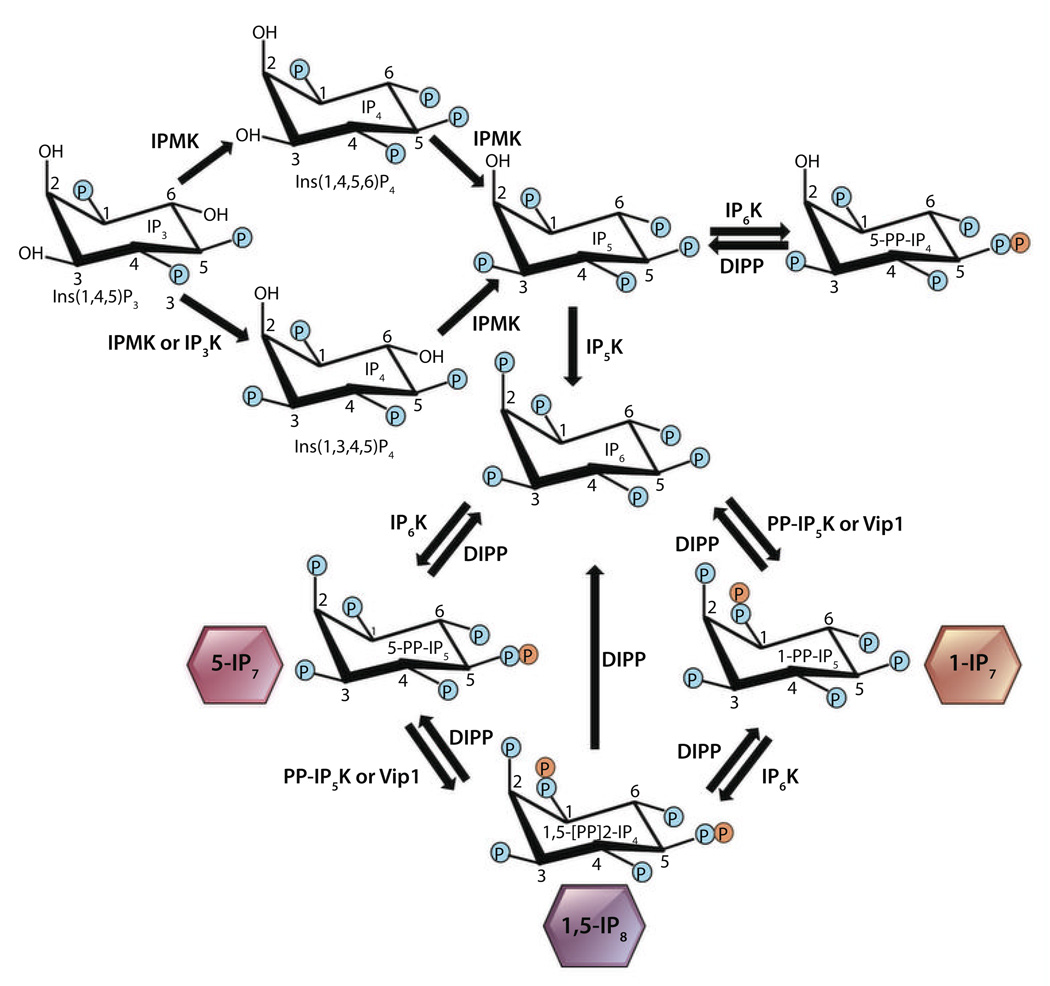
Fig. 1.
Inositol polyphosphate biosynthetic pathway. IP3K or IPMK phosphorylates IP3 to generate IP4 [Ins(1,3,4,5)P4 or Ins(1,4,5,6)P4]. Further sequential phosphorylation of IP4 by IPMK and IP5K yields IP6 (7, 23, 24, 33–36, 44). IP6Ks pyrophosphorylate the 5 position, generating 5-PP-IP4 from IP5, 5-PP-IP5 (5-IP7) from IP6, and 1-/3-,5-(PP)2-IP4 (1,5-IP8) from 1-/3-PP-IP5 (7, 17, 19, 20). 5-IP7 is the predominant inositol pyrophosphate in mammals. PP-IP5Ks, the human Vip1 isoforms, pyrophosphorylate the 1 or 3 position to yield 1-/3-PP-IP5 (depicted as 1-IP7 for simplicity) from IP6, or 1-/3-,5-(PP)2-IP4 (1,5-IP8) from 5-IP7, and physiologically, they are primarily involved in generating IP8. DIPPs dephosphorylate inositol pyrophosphates to IP6 or IP5 (32, 47).
Inositol Pyrophosphate Metabolism
The complex metabolic pathway generating the pyrophosphates has been extensively reviewed elsewhere and is beyond the scope of this review (28–32). Hormones or growth factors activate phospholipase C to generate IP3 from PIP2 [phosphatidylinositol (4,5)-bisphosphate] (28). IP3K [Ins(1,4,5) P3 3-kinase] converts IP3 to IP4 (33). In addition, IPMK sequentially converts IP3 to IP5, albeit with species-specific isomeric preference (7, 23, 24, 34–36) (Fig. 1). Cellular concentrations of IP5 and IP6, are substantially higher than those of IP3. Because the techniques for measuring endogenous concentrations of inositol phosphates are limited, most studies assess concentrations of inositol phosphates by examining the conversion by cultured cells of [3H]inositol into the relatively polar inositol phosphates, which can be separated by ion-exchange chromatography. The existence of pyrophosphates was initially postulated when several groups identified metabolites more polar than IP6 (37–40) and speculated that they might represent diphosphates incorporating energetic pyrophosphate bonds. Subsequently, definitive evidence emerged that these more polar agents are diphosphorylated (3, 4, 41) and characterized as 5-IP7 (15) and IP8 (4, 16, 19, 20, 41, 42). IP5 and IP6 are the best-characterized precursor molecules for the inositol pyrophosphates. Diphosphoinositol tetrakisphosphate (PP-IP4), synthesized by IP6Ks from IP5 (30–32, 43) occurs endogenously in most mammalian tissues. IP5K [Ins(1,3,4,5,6)P5 2-kinase] converts IP5 to IP6 (44), which is the substrate for IP6Ks and Vip1 to synthesize 5-IP7, 1-/3-IP7 and 1-/3-,5-IP8 (7, 17, 19, 20) (Fig. 1). Diphosphorylated derivatives of IP3 and IP4 have also been reported (45). The enzymatic formation in vitro of a triphosphate derivative of IP6 (43) raises the possibility that triphosphorylated IP6 might exist in vivo (46). Diphosphoinositol-poly-phosphate phosphohydrolases (DIPPs) dephosphorylate all inositol pyrophosphates including PP-IP4 (32, 47) (Fig. 1). The discovery of the DIPPs has provided a mechanism that accounts for the rapid turnover of inositol pyrophosphates (3–5).
Intracellular concentrations of IP7 in most mammalian tissues are about 1 to 5 µM (30, 31, 48–50), whereas those of IP6, the most abundant inositol phosphate, are 15 to 60 µM (28, 31, 51). The slime mold Dictyostelium discoideum has high concentrations of inositol pyrophosphates, about 10 µM in the disaggregated state and increasing to 100 to 250 µM during starvation and aggregation (52). The predominant form of IP7 in Dictyostelium is 6-IP7, which differs from yeast and mammalian isomers (15).
Mammalian inositol pyrophosphate concentrations are dynamic. IP8 concentrations increase 1000 to 2500% within 20 to 30 min after exposure to sorbitol-induced hyperosmotic conditions (53, 54) or heat shock (55). AICAR [N-(β-d-ribofuranosyl)-5-aminoimidazole-4-carboxamide], which is transformed in cells to an adenosine monophosphate (AMP) derivative, prevents the sorbitol-mediated increase in IP8 independently of its ability to stimulate AMP kinase (AMPK), the latter a well-characterized action of AMP (56). Various cell stressors, including the anticancer drug cisplatin and broad-spectrum kinase inhibitors, increase intracellular IP7 concentrations by activating IP6K2 (57–59).
One classic criterion for putative intracellular second messengers, exemplified by IP3 (2), is that their formation can be altered in response to intercellular stimuli such as neurotransmitters, hormones, and growth factors. Concentrations of IP6 and IP7 decline markedly after overnight serum starvation (11). Conversely, insulin-like growth factor 1 (IGF-1) stimulates IP6 and IP7 formation (11). IGF-1 increases the IP7-to-IP6 ratio in mouse embryonic fibroblasts (MEFs), an effect absent in MEFs lacking IP6K1, which indicates a meaningful “second-messenger response” that depends on IP6K1 activity. Similar alterations of inositol pyrophosphate concentrations occur in the hepatocellular carcinoma cell line HepG2 after IGF-1 or insulin treatment (11). IP7/IP6 ratios increase with age in primary hepatocytes. Moreover, IP7 concentrations increase markedly during NIH3T3-L1 adipogenic differentiation (11). Modest increases in IP8 but not IP7 were observed after epidermal growth factor (EGF) treatment of DDT1 MF-2 smooth muscle cells (53). Inositol pyrophosphates are also responsive to chemokine signaling. Formylmethionyl-leucyl-phenylalanine, a bacterial peptide that triggers chemotaxis, rapidly depletes IP7 in the human promyelocytic leukemia cell line HL60 (12).
Regulation of the pyrophosphates is also determined by factors that regulate the biosynthesis of their precursor inositol phosphates. Several groups have reported increases in total inositol phosphate concentrations after treatment with various growth factors like EGF, IGF-1, platelet-derived growth factor (PDGF), and the Wnt (for vertebrate Int and Drosophila Wingless) family (60–64). Concentrations of IP5 and IP6 increase after prolonged PDGF treatment of serum-deprived NIH3T3 cells (63). The Wnt family of signaling proteins participates in embryogenesis and adult tissue homeostasis with aberrations promoting degenerative diseases and cancer (65). Serum-starved F9 teratocarcinoma or human embryonic kidney–293 (HEK293) cells displayed increased IP5 concentrations within 15 min of Wnt 3a treatment (64). Thus, inositol polyphosphate concentrations alter dynamically in response to diverse stimuli.
Molecular Mechanisms of Inositol Pyrophosphate Signaling
There appear to be two principal mechanisms whereby inositol pyrophosphates affect other cellular constituents: binding and pyrophosphorylation. Inositol polyphosphates physiologically bind to several proteins (66–70). Inositol polyphosphates bind to the membrane-targeted PH domains of several proteins (68, 69, 71–76) (Table 1). PH domains (77) bind phospholipids such as PIP3 and PIP2 (78, 79) and so recruit signaling proteins to membranes. In Dictyostelium, PIP3 binds the PH domain of the protein CRAC (cytosolic regulator of adenylate cyclase), which mediates cyclic AMP (cAMP)–related chemotaxis (73). IP7 competes with PIP3 for binding to CRAC and prevents the chemotactic response. Deletion of IP6K depletes IP7 and thereby augments slime mold sensitivity to cAMP and, consequently, aggregation. A critical regulatory role for inositol pyrophosphates in the chemotactic response to cAMP is evident by the marked elevation of inositol pyrophosphates in the amoeba elicited by cAMP (73). In mammals, one of the best-characterized examples is the recruitment of the kinase Akt, a PH domain–containing protein, to plasma membranes (80) downstream of hormone- or growth factor–mediated activation of PI 3-kinase to form PIP3. 5-IP7 blocks PIP3-mediated activation of Akt (11). The other IP7 isomer, 1-/3-IP7, also exerts its physiological effects by binding and inhibiting the cyclin-cyclin dependent kinase (CDK) complex of yeast (18, 81).
Table 1.
PH domain–containing enzymes whose catalytic activity or membrane translocation, or both, are regulated by inositol phosphates or pyrophosphates. BTK, Bruton’s tyrosine kinase.
The pyrophosphate bond in IP7 is as energetic as those of ATP (ATP), which raises the possibility that IP7, like ATP, might phosphorylate proteins (4, 31, 82). Isolation and cloning of IP6Ks so as to generate [32P]IP7, with the label in the β-phosphate anticipated to be donated, led to the demonstration that [32P]IP7 phosphorylates various proteins (83). The process is magnesium dependent, like ATP phosphorylation. However, the phosphate transfer by IP7 is nonenzymatic and occurs in vitro without the intervention of protein kinases. IP7-mediated phosphorylation has been characterized both in yeast and mammals (83–85), with the most extensive work done in yeast. Proteins that are particularly good targets for this phosphorylation process tend to be rich in serines adjacent to acidic amino acids and close to a stretch of basic amino acids, as exemplified by nucleolar yeast proteins, which are particularly good substrates (83, 84). The importance of acidic amino acids is reminiscent of the consensus motif for phosphorylation by CK2 (casein kinase 2, an acidophilic serine-threonine kinase, discussed in detail in the cell death and development section).
Target proteins purified from bacteria or λ-protein phosphatase–treated proteins purified from mammalian systems are not phosphorylated by IP7 (83, 84). Although phosphorylation as a posttranslational modification in the bacterial proteome is increasingly appreciated (86), the relevant bacterial kinases differ markedly from their mammalian counterparts (86). These hints that something more than conventional phosphorylation was involved led to the discovery that IP7 can only phosphorylate a protein that had been previously phosphorylated on the same serine, principally by CK2. Hence, IP7 pyrophosphorylates proteins (84). Such a process provides a unique type of signaling event, distinguishing IP7 phosphorylation from that involving ATP. There are other distinctions. Unlike ATP-dependent phosphorylation, pyrophosphorylation by IP7 resists actions of most phosphatases while being more acid labile (84). Certain lines of evidence indicate that such protein pyrophosphorylation occurs in vivo. For instance, pyrophosphorylation in vitro of overexpressed nucleolar protein NSR1, a target of IP7 phosphorylation, is enhanced when purified from yeast lacking yIP6K, presumably because there is less endogenous pyrophosphorylation of the protein in the absence of yIP6K and endogenous IP7 (83). In mammals, pyrophosphorylation appears to inhibit physiological interactions between the β subunit of adaptor protein–3 (AP-3), a clathrin-associated protein, and the kinesin family motor protein Kif3A (85).
The energy of hydrolysis for inositol pyrophosphates is similar to that of AT P, which supports a physiologic role for inositol pyrophosphorylation (87). The pyrophosphorylating ability of 1-/3-IP7 (84) supports the pyrophosphorylation potential of other inositol pyrophosphates such as IP8.
Physiologic Functions of Inositol Pyrophosphates
1. Nuclear dynamics
Yeasts have been used extensively to characterize nuclear functions of inositol pyrophosphates. Overexpression of catalytically active yIP6K leads to shortened telomeres, whereas its deletion lengthens them (88, 89) (Table 2), which indicates that inositol pyrophosphates regulate telomere length. The ability of PP-IP4 to shorten telomeres (89) requires Tel1, the yeast version of ATM, the kinase that is mutated in ataxia telangiectasia (88, 89).
Table 2.
Physiological processes regulated by inositol pyrophosphate–synthesizing enzymes.
| Functions | Enzyme | Mouse | References |
|---|---|---|---|
| Insulin sensitivity and resistance to weight gain | IP6K1 | Mouse | (10, 11) |
| Insulin release | IP6K1 | Min6 cells | (50) |
| Neutrophil phagocytosis | IP6K1 | Mouse | (12) |
| HIV-VLP release | IP6K1 | HeLa, MEF cells | (85) |
| Dopamine release | IP6K1* | PC12 cells | (94) |
| Apoptosis and autophagy | IP6K2, yIP6K–Vip1 | Cell lines, yeast | (13, 57–59, 95–97, 102, 103) |
| Development | IP6K2 | Zebrafish | (104) |
| Telomere length maintenance | yIP6K | Yeast | (88, 89) |
| DNA hyperrecombination | yIP6K | Yeast | (90) |
| Vacuole biogenesis | yIP6K | Yeast | (91) |
| Endocytosis | yIP6K | Yeast | (92) |
| Cell wall integrity, filamentous growth | yIP6K, Asp1 | Yeast | (91, 93) |
| Phosphate homeostasis | Vip1 | Yeast | (18, 81) |
| Chemotaxis | IP6K | Slime mold | (73) |
Indicates action of IP6K1 independent of its catalytic activity.
Mutations of yeast protein kinase C (PKC) lead to hyperrecombination of chromosomes; this augmentation of DNA recombination is reversed by mutating yIP6K, which suggests a role for yIP6K in mediating this process (90). The enhanced recombination linked to PKC mutations is associated with DNA damage, augmented transcription, and cyclin mutation and leads to a prolongation of the S phase. Catalytically active yeast or mammalian IP6K both restore hyperrecombination (90).
Yeasts have evolved a complex system for regulating phosphate disposition during starvation. Phosphate starvation specifically increases the concentration of 1-/3-IP7 (18), which binds and stimulates Pho81-mediated inhibition of the Pho80-Pho85 cyclin-CDK complex. Cyclin-CDK complex inhibition leads to reduced phosphorylation and nuclear accumulation of the transcription factor Pho4, which results in enhanced expression of genes from the phosphate-responsive signaling pathway (PHO genes) required to maintain phosphate homeostasis (18, 81). Whether these findings are relevant to mammalian physiology is unclear.
2. Vacuole biogenesis, vesicular trafficking, and cellular morphology
Studies deleting yIP6K implicate inositol pyrophosphates in vesicular dynamics. Yeasts lacking yIP6K display prominent vacuolar abnormalities (46, 91). Yeasts with mutations of enzymes in the inositol pyrophosphate pathway display selective abnormalities of endocytic trafficking, which indicates that inositol pyrophosphates are physiologic enhancers of endocytosis (92). Generation of inositol pyrophosphates by yIP6K regulates cellular morphology (91, 93) and stimulates cell wall integrity in S. cerevisiae (92), whereas Asp1, a Vip1 homolog, enhances the dimorphic switch from single cell to filamentous invasive growth form in Schizosaccharomyces pombe (93).
Identification of GRAB [guanine nucleotide–exchange factor (GEF) for Rab3A] as an IP6K1-binding protein revealed a role for IP6K1 in synaptic vesicle exocytosis (94) (Table 2). GRAB inhibits neurotransmitter release via its GEF activity on Rab3A, which is a guanosine triphosphatase (GTPase) protein. IP6K1 competes with GRAB for binding to Rab3A and thus stimulates neurotransmitter release. Catalytically inactive IP6K1 also stimulates transmitter release (94).
Insulin release from pancreatic β cells involves exocytotic processes analogous to those mediating neurotransmitter release. Overexpression of the three isoforms of IP6K stimulates insulin release, as does exogenous 5-IP7 (50) (Fig. 2 and Table 2). Depletion of IP6K1 but not IP6K2 in pancreatic β cells inhibits exocytosis (50), and mice with targeted deletion of IP6K1 have reduced plasma levels of insulin (10, 11).
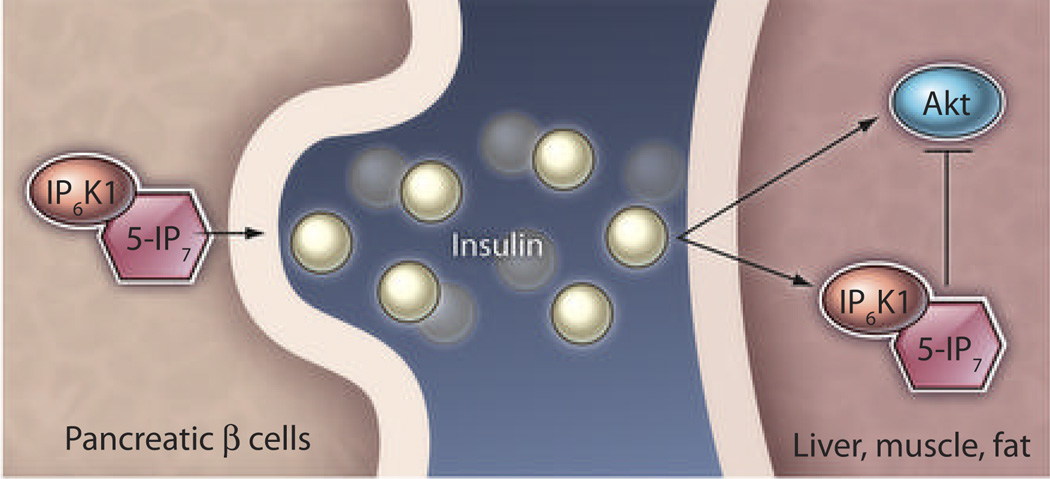
Fig. 2.
Inositol polyphosphates in insulin homeostasis. Inositol pyrophosphates (5-IP7) facilitate insulin secretion from pancreatic β cells (50). The secreted hormone signals through its receptors in insulin-responsive tissues and activates the protein kinase Akt through the PI 3-kinase pathway. Insulin also increases inositol pyrophosphate concentrations, which decreases Akt activity in liver, skeletal muscles, and adipose tissues (11) and thereby maintains homeostasis between insulin release and its signaling.
The β subunit of AP-3 (AP3β1) is a clathrin-associated protein required for HIV-1 release from infected cells by means of binding to the kinesin motor protein Kif3A. Pyrophosphorylation of AP3β1 decreases its interaction with Kif3A, which reduces the release of HIV-1 virus–like particles (VLPs) (85) (Fig. 3).
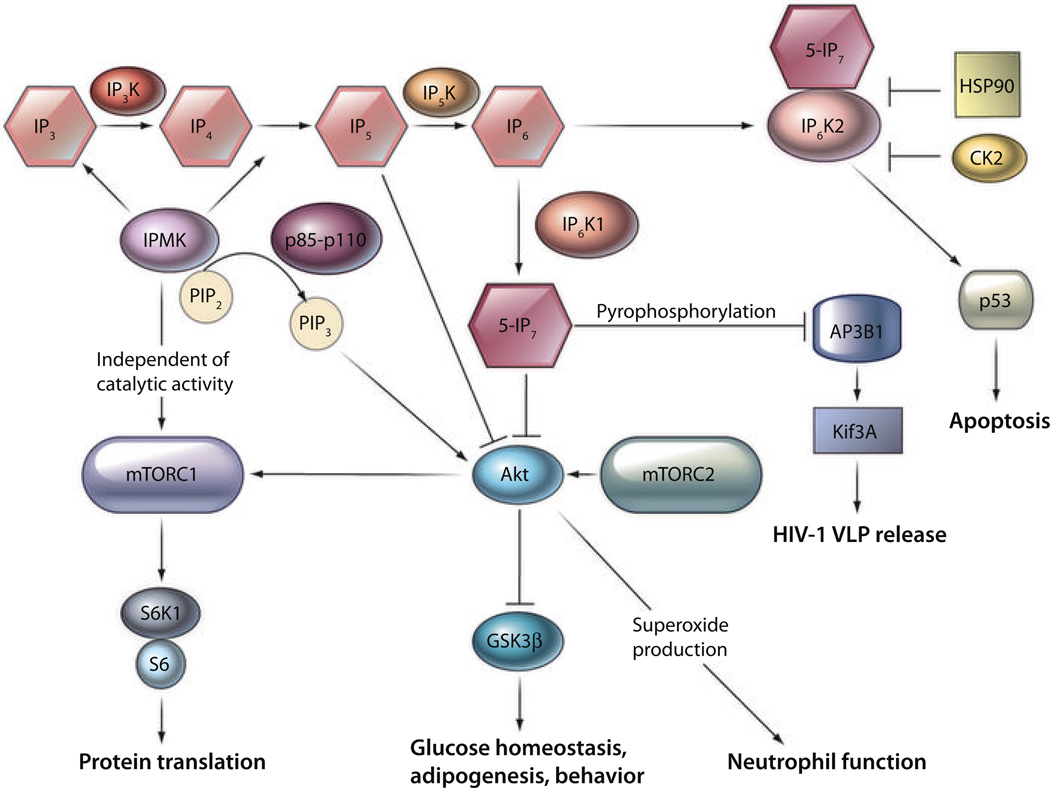
Fig. 3.
Inositol poly- and pyrophosphates in mammalian cellular signaling networks. The combined soluble inositol phosphate kinase activities of IP3K, IPMK, and IP5K generate IP5 and IP6 from IP3, which are precursors of inositol pyrophosphates. 5-IP7 inhibits Akt activity, which decreases mTORC1-mediated protein translation and increases GSK3β-mediated glycogenolysis, adipogene-sis (11), and behavior. IP5 also inhibits Akt signaling (124, 125). Inhibition by 5-IP7 of Akt diminishes neutrophil phagocytic functions (12). 5-IP7 regulates the release of virus like particles through its pyrophosphorylating activities (85). IPMK acts as a PI 3-kinase to activate Akt (25, 26). In a manner independent of its catalytic activity, IPMK binds, stabilizes, and activates the mTORC1 complex (27). Thus, IPMK enhances or inhibits Akt signaling, respectively, by means of its PI 3-kinase and soluble inositol phosphate kinase activities. IP6K2 is a proapoptotic protein regulated by HSP90, which binds and inactivates it (58), as well as by CK2, which phosphorylates IP6K2 to enhance its proteasomal degradation (59). Apoptotic stimuli stabilize, activate, and facilitate nuclear translocation of IP6K2, where it binds p53 and augments p53’s apoptotic actions (13).
3. Cell death and development
Hints of a selective role for IP6K2 in apoptosis came from studies of the mechanisms whereby interferon-β (IFN-β) suppresses proliferation of the ovarian cancer cell line OVCAR-3 by enhancing apoptosis (95, 96) (Table 2). These studies showed that the abundance of IP6K2, which was linked to cell death by antisense knockout (95), increased with IFN-β treatment. Moreover, overexpression of IP6K2 increased the growth-suppressive and apoptotic influences of IFN-β, whereas a dominant-negative mutant of the inositol phosphate–binding domain prevented the actions of IFN-β (96). Subsequent work extended the involvement of IP6K2 in apoptosis to other systems (57). Thus, transfection of IP6K2 in multiple cell lines substantially enhances the apoptotic actions of various cell stressors, which includes anticancer drugs such as cisplatin, etoposide, and staurosporine. Moreover, exposure to cisplatin enhanced IP7 generation up to 400% (57, 58). Overexpression of any one of the three isoforms of IP6K increased sensitivity to the apoptotic actions of hypoxia and staurosporine. However, knockdown of IP6K2 by RNA interference markedly reduced cell death, whereas knockdown of IP6K1 and IP6K3 did not (57).
Under basal conditions, IP6K2 is sequestered in the cytoplasm bound to the cytosolic heat shock protein 90 (HSP90). Cell stressors—such as cisplatin and novobiocin, another anticancer drug—disrupt this binding, which liberates and activates IP6K2 and leads to cell death (58). Single amino acid mutations of IP6K2 that selectively inhibit its binding to HSP90 increase its catalytic activity and apoptosis (58). Thus, a substantial part of the apoptotic actions of drugs such as cisplatin and novobiocin, which also inhibit the ATPase activity of HSP90, may reflect disruption of IP6K2-HSP90 binding.
IP6K2-mediated cytotoxicity was associated with the translocation of IP6K2 to mitochondria in HEK293 cells preincubated with staurosporine (57) but with its translocation to the nucleus in OVCAR-3 cells under IFN-β–treated conditions (97). Different cell types and other experimental conditions might explain these discrepancies.
In several mammalian cells, IP6K2 selectively augments p53-related cell death (13). Somatic cell gene disruption of IP6K2 virtually abolishes IP7 generation in HCT116 cells (an epithelial cell line derived from human colon carcinoma), which indicates that the IP6K2 isoform is primarily responsible for forming IP7 in these cells. Deletion of IP6K2 reduces apoptosis elicited by 5-fluorouracil, which acts through p53 but is not reduced by sulindac, an anti-inflammatory cyclooxygenase-2 inhibitor that elicits Bcl2-associated X protein (BAX)–dependent, but p53-independent, apoptosis (13). IP6K2 deletion causes cell cycle arrest associated with a marked increase in expression of proarrest genes such as p21 and 14-3-3σ. By contrast, there is no alteration in prodeath genes such as PUMA and NOXA. With moderate cell stress, p53 primarily activates proarrest genes, which enables the cell to repair damage, whereas, with grave insults, p53 activates proapoptosis programs. IP6K2 appears to act by inhibiting expression of proarrest genes so that the prodeath program is activated seemingly by default. Overexpression of IP6K2 markedly diminishes expression of proarrest genes without influencing that of prodeath ones (13).
How might IP6K2 regulate p53? IP6K2 binds directly to the p53 DNA binding core domain (13). This binding underlies IP6K2‘s apoptotic actions, because dominant-negative constructs of IP6K2, which prevent its binding to p53, block the proapoptotic influences of IP6K2. Accordingly, drugs that block IP6K2-p53 binding might prevent p53-mediated cell death and could diminish neuronal cell death in conditions such as stroke and neurodegenerative disease; IP6K2’s prodeath influences might also reflect its role in induction of the Apo2 ligand, Apo2L [TRAIL, tumor necrosis factor (TNF)–related apoptosis-inducing ligand] (97). Apo2L (TRAIL) is a member of the TNF superfamily of cytokines that preferentially induces apoptosis in cancer cells upon binding to death receptors 4 and 5 (DR4 and DR5) (98). Blocking antibodies to Apo2L (TRAIL) prevents the apoptotic influences of IFN-β–IP6K2 (97).
IP6K2’s prodeath actions might also involve antagonism of the trophic influences of CK2 (59). CK2 is a protein kinase with prominent prosurvival functions. Its abundance is increased in many cancers, and its inhibitors display antitumor activity (99, 100). CK2 stimulates angiogenesis to create a hospitable environment for tumor growth (101). The proapoptotic effects of CK2 inhibitors are reduced after IP6K2 deletion (59). CK2 phosphorylates IP6K2 to decrease its stability (59). Phosphorylation occurs at serines 347 and 356, whose mutations lead to marked increases in IP6K2 abundance. The clinically beneficial effects of CK2 inhibitors could involve stabilization of IP6K2 to augment its apoptotic actions.
Autophagy has been implicated in IP6K-mediated cell death (102). Overexpression of the IP6Ks increases the numbers of autophagosomes and proportionately enhances cell death (102), whereas depletion of IP6K2 by RNA interference diminishes both cell death and the numbers of autophagosomes.
Mice with targeted deletion of IP6K2 exhibit normal development, growth, and fertility (14). However, they display a 400% greater incidence of oral and esophageal tumors. Fibroblasts from these mice resist the antiproliferative effects of IFN-β (14).
IP8 has also been implicated in cell death (103). Thus, yeast with yIP6K or Vip1 deleted display increased resistance to cell death induced by hydrogen peroxide. Furthermore, hydrogen peroxide elicits a rapid decrease in cellular inositol pyrophosphate concentrations in vivo and inhibits yIP6K in vitro (103).
Inositol pyrophosphates have recently been linked to zebrafish development (104), where IP6K2 activates the Hedgehog developmental pathway by PP-IP4 generation. In zebrafish, deletion of IP6K2 impairs development of the craniofacial skeleton and muscle fibers, as well as the development and migration of neural crest cells (104). Similarly, IP6K2 depletion in NIH3T3 cells inhibits Hedgehog target gene expression, whereas overexpression of IP6K2 elicits stimulatory effects (104). One might speculate about links between the apoptotic actions of IP6K2 and its influences on Hedgehog signaling in mammalian nervous system development. Embryonic mammalian brain contains twice as many neurons as adult brain, as dropout of neurons is a prominent feature of brain ontogeny. Whether Hedgehog signaling interfaces with the neuronal dropout process and whether loss of neurons in development is related to IP6K2 are unanswered questions.
4. Growth factor and cytokine signaling
Studies with IP6K1 knockout mice have established a role for IP7 in regulating the growth factor–mediated Akt–mTOR–glycogen synthase kinase–3β (Akt-mTOR-GSK3β) signaling cascade (Table 2 and Fig. 3). Hints of the existence of this role came from evidence that IP7 interferes with the binding of PIP3 to the Akt PH domain (73). Akt enhances growth factor signaling, glucose uptake, glycogen synthesis, and protein synthesis (80). Akt influences critical regulatory proteins such as glucose transporter–4 (GLUT4), GSK3 α and β, and the mTORC1 complex (80). Akt overexpression in skeletal muscle leads to insulin sensitivity, skeletal muscle hypertrophy, and augmented hepatic fatty acid oxidation with reduced fat accumulation (105). Akt also exerts lipogenic effects. Akt1 and Akt2 double-knockout mice display reduced adipose mass and skeletal muscle atrophy (106). Conversely, Akt phosphorylates GSK3β, which inhibits its activity and which may affect lipogenesis, because GSK3β inhibition abolishes adipogenesis (107, 108). In organisms with insulin resistance and obesity, Akt and GSK3β activities are reciprocally regulated. Thus, Akt and mTOR signaling are diminished, and GSK3β activity is increased, in insulin-resistant tissues of aged and obese mice (109–111). These evidently complex relationships of Akt and inositol pyrophosphates with insulin homeostasis (11, 50) and lipid disposition (11) may reflect isoform- and tissue-specific influences (Fig. 2).
An initial link of IP6K1 to Akt signaling came from observations that growth factors such as IGF-1 markedly stimulate IP7 formation in serum-starved cells, with the increase abolished in IP6K1-depleted cells (11). The IP6K1 knockout cells display markedly augmented IGF-1 and insulin-stimulated Akt signaling, with enhanced phosphorylation of Akt downstream effectors such as GSK3β, tuberin, p70S6 kinase1, and S6 itself. These effects reflect loss of IP6K1 catalytic activity because they can be rescued by the wild-type enzyme but not by catalytically inactive IP6K1 (11).
How might IP7 regulate Akt? Inositol polyphosphate binding to various membrane-targeted proteins like Akt, phosphoinositide-dependent kinase–1 (PDK1), and interleukin 2–inducible T cell kinase (ITK) regulates their activity and membrane translocation (68, 69, 71–76) (Table 1). In the growth factor signaling cascade, extracellular receptor stimulation leads to activation of the p85-p110 PI 3-kinase, which generates the phospholipid PIP3. PIP3 activates Akt at the plasma membrane by facilitating its phosphorylation by the kinase PDK1. Growth factor stimulation of cells increases the membrane localization of Akt, with these effects substantially increased in cells lacking IP6K1. The IP6K inhibitor TNP {N2-[m-(trifluoromethyl)benzyl], N6-(p-nitrobenzyl) purine} (112) also increases growth factor stimulation of Akt, manifested as its phosphorylation at threonine-308, a process mediated by PDK1. 5-IP7 inhibits the phosphorylation of Akt by PDK1 in vitro. In the absence of added PIP3, IP7 inhibits Akt phosphorylation with a median inhibitory concentration of about 20 nM, substantially more potent than any other known action of IP7 (11). IP5 and IP6 are much less potent at inhibiting Akt phosphorylation by PDK1, whereas IP3 and IP4 are inactive (11). Inhibition is absent in Akt lacking the PH domain, which indicates that 5-IP7 exerts its inhibitory effect through the PH domain of Akt. This fits with evidence that IP7 interferes with PIP3 binding to PH domains of several proteins (73). IP7 inhibits PIP3’s activation of Akt-T308 phosphorylation more potently when added before PIP3 than when preparations are preincubated with PIP3 (11), consistent with observations that preincubation with PIP3 prevents IP7 from displacing PIP3 from Akt’s PH domain (113).
Detailed mechanisms whereby IP7 inhibits Akt are not fully resolved. IP7 competes with PIP3 for binding PH domains of Akt and other proteins with a potency similar to that of IP4 (73). However, IP7’s potency as an inhibitor of Akt phosphorylation is much greater (11), which suggests that its activity may depend on factors other than binding. Binding assays (73) used the Akt-PH domain, whereas the kinase assays (11) utilized full-length Akt, which may account in part for discrepant potencies. Similarly, full-length PDK1 binds phosphoinositides or inositol polyphosphates better than the PDK1-PH domain does (76). Variations in Akt conformation may contribute to the dual regulation of Akt by phosphoinositides and inositol pyrophosphates. Thus, PDK1-mediated phosphorylation depends on PH-and kinase domain–dependent conformational changes in Akt (114). In the PH-in conformation, the Akt activation loop is inaccessible to PDK1. Growth factor–mediated Akt binding to PIP3 converts Akt to the PH-out conformation, which is available for PDK1 phosphorylation. Moreover, PDK1 and Akt form a complex in vivo (114). In this complex mode of PDK1-Akt-PIP3-IP7 regulation, 5-IP7 appears to stabilize the inactive (PH-in) conformation of Akt, whereas PIP3 does the opposite. Whether IP7 influences this process by pyrophosphorylating Akt or PDK1 is unclear.
The increased negative charge of IP7 may explain, in part, why it is preferred over other inositol phosphates for mediating certain biologic actions. Thus, the specificity for binding of IP5 isomers to various PH domains depends on the number of basic residues in the domain. PH domains of Akt and pleckstrin bind more selectively to IP5 isomers than does the general receptor for phosphoinositides–1 (GRP1) PH domain, which reflects the fewer basic residues of the PH domains of Akt and pleckstrin (115).
The intracellular distribution of inositol pyrophosphate–synthesizing enzymes might affect inositol poly- and pyrophosphate-mediated PH domain regulation in vivo. Indeed, receptor-regulated compartmentalization of inositol pyrophosphate synthesis has been demonstrated (116). The mammalian IP8-synthesizing enzyme PP-IP5K1 has a cryptic PIP3-binding PH domain, which also binds IP6 to a lesser extent (116). PP-IP5K1 translocates to the plasma membrane in response to PDGF treatment (116). Accordingly, IP8 might influence the membranous disposition of Akt.
The enhanced Akt activation in response to insulin in IP6K1 knockout mice has notable physiologic consequences. Initial studies of IP6K1 knockout mice revealed reduced concentrations of blood insulin with normal plasma glucose, which implies insulin hypersensitivity (10). Enhanced glucose tolerance confirms the sustained insulin sensitivity of 10-month-old IP6K1 mutants (11). Moreover, glucose uptake into muscle and fat of the mutants is tripled. Consistent with the insulin hypersensitivity, IP6K1 knockout mice are resistant to obesity elicited by high-fat diets. These metabolic alterations in the IP6K1 knockouts appear to result from the increased Akt signaling, which leads to decreased GSK3β activity in consequence of phosphorylation by Akt of GSK3β. This also leads to increased glycogen abundance and reduced adipogenesis. Accordingly, IP6K1 inhibitors may be useful in treating type 2 diabetes and obesity. The likelihood of adverse effects from such inhibitors can be inferred from the phenotype of the IP6K1 knockouts. They weigh about 15% less than controls, owing to less fat deposition, and males have reduced numbers of sperm, but otherwise the mice appear normal. Because of Akt’s role in cell survival and proliferation (80, 117), one might anticipate that the increased Akt signaling of the IP6K1 mutants would facilitate tumor formation. However, no spontaneous tumors have been observed in 2-year-old IP6K1 knockout mice, which corresponds roughly to 75 to 80 years of human life.
PI 3-kinase activity and PIP3 signaling influence neutrophil chemotaxis and phagocytosis through NADPH (nicotinamide adenine dinucleotide phosphate) oxidase–mediated superoxide production (118–120). IP6K1 appears to regulate neutrophil function through Akt inhibition (12). Thus, IP6K1-depleted neutrophils display enhanced Akt activation and amplified superoxide production which leads to increased phagocytic and bactericidal activity (12).
IP6K1 signaling may have behavioral consequences. Genetic abnormalities leading to decreased Akt abundance have been linked to schizophrenia (121). The mood-stabilizing actions of lithium have been attributed to direct GSK3 inhibition (122). In an alternative model, lithium dissociates a β-arrestin–protein phosphatase–Akt complex, which leads to augmented Akt activity, which in turn inhibits GSK3β (123). Thus, IP6K1 inhibitors, by enhancing Akt signaling, might be therapeutic in diverse psychiatric diseases.
IPMK and IP6K: A Molecular Switch in Growth Factor Signaling?
An interface between IP6K1 and IPMK in regulating Akt is implied by the finding that IPMK has robust and selective PI 3-kinase activity in vitro (25). Initially, it was not clear how IPMK could contribute to cellular formation of PIP3, as one can deplete PIP3 with wortmannin, which inhibits PI 3-kinases but does not inhibit IPMK. The availability of IPMK-deleted cells permitted a resolution of this conundrum (26). Deletion of IPMK leads to a 50% decrease in PIP3 generation. Moreover, IPMK purified from cells treated with wortmannin displays a 70% reduction in catalytic activity. Thus, in intact cells, wortmannin inhibits IPMK with potency similar to that for inhibiting the p110 PI 3-kinase. Presumably, PIP3 formed by the p110 enzyme activates a kinase that phosphorylates IPMK to stimulate its catalytic activity. In support of this notion, dephosphorylation of IPMK with protein phosphatases reduces its catalytic activity to the same extent as wortmannin treatment of intact cells (26).
If IPMK is a physiologically important PI 3-kinase, then it should regulate Akt. Indeed, the stimulation of Akt by growth factors such as insulin, IGF-1, and EGF is substantially reduced in IPMK-deleted cells (26). IPMK’s stimulation of Akt is due to its PI 3-kinase rather than its inositol phosphate kinase activity. Thus, in IPMK-deleted cells the loss of Akt signaling is rescued by overexpressing wild-type IPMK but not an IPMK isoform from Arabidopsis that has inositol phosphate kinase, but not lipid kinase, activity (26). Akt is a major determinant of cell proliferation and, as expected, the rate of proliferation in IPMK-deleted mouse embryonic fibroblasts is reduced about 50%. In U87MG glioma cells, depletion of IPMK by RNA interference also substantially reduces proliferation, which is augmented by overexpression of IPMK (26).
IPMK thus appears to be a required link in growth factor signaling via PIP3. PI 3-kinases generate PIP3, which, by means of unknown kinases, elicits phosphorylation and activation of IPMK, whose PI 3-kinase activity also synthesizes PIP3 to stimulate Akt. Reasons for a cell to generate PIP3 in two sequential steps are not clear. One likely possibility is the amplifying effect attendant upon a series of enzymatic steps. It is also possible that the PIP3 that activates Akt derives in part from p110 and in part from IPMK. However, the 70% reduction in Akt activation after IPMK deletion suggests that p110 and IPMK act sequentially.
IPMK’s deletion abolishes the formation of IP5, IP6, and IP7, which indicates that it is rate limiting in the generation of inositol pyrophosphates (26). Like IP7, IP5 also inhibits Akt (11, 124), although to a lesser extent than IP7 (11), and IP5 derivatives display an-titumor activities (125). Because IP5 can be converted by IP6Ks to PP-IP4, IP5-mediated Akt inhibition is likely to involve generation of PP-IP4 in vivo. IP7 (and possibly IP5), are thus opposing the influences exerted by the lipid kinase activity of IPMK. Accordingly, IPMK may exert two divergent actions, as its lipid kinase activity promotes Akt signaling, whereas its inositol phosphate kinase activity exerts the opposite effect (Fig. 3).
All of the above signaling influences of the IP6K1-IPMK system involve the activation by growth factors of Akt and its downstream targets, such as mTOR. IPMK may be a key element regulating mTOR. mTOR was first isolated as a target for the immunosuppressive actions of the drug rapamycin (126, 127) and subsequently has been shown to act by means of a complex of mTOR-binding proteins to serve as a major stimulant of protein synthesis (128). There are two mTOR complexes differentiated by the association of mTOR with raptor in mTOR complex-1 (mTORC1) (129), which is targeted by IPMK, and rictor in mTORC2 (130). mTORC1 is the principal determinant of protein synthesis and cell growth in response to nutrient amino acids. IPMK is bound to mTOR and raptor in the mTORC1 complex (27). IPMK appears to be a critical component of the complex, because depletion of IPMK reduces mTOR binding to raptor and the GTPase Rag without influencing interactions of mTORC2 with rictor. IPMK is critical for mTOR signaling, because its deletion reduces mTOR signaling in response to amino acids by about 60%. Overexpression of IPMK rescues this deficiency. Remarkably, catalytically inactive IPMK is equally effective in restoring mTOR signaling. Thus, IPMK acts in a noncatalytic fashion to stabilize mTORC1 and to facilitate protein translation. This conclusion is supported by findings that a dominant-negative construct that selectively blocks the binding of IPMK to mTOR abrogates nutrient-elicited mTOR signaling (27).
IP6K1 and IPMK thus appear to be key regulators of metabolic signaling in response to growth factors and nutrients (Fig. 3). A fuller understanding of how IPMK switches between its lipid kinase and inositol phosphate kinase activities awaits identification of its regulation. We do not know whether an analogous regulation sequesters a pool of IPMK to act noncatalytically in stabilizing the mTORC1 complex for responses to altered amino acid availability. Indeed, it is not clear whether cells switch between devoting their signaling resources to coping with growth factors versus nutrients or whether the two activities occur in parallel without any crosstalk.
Conclusions
Delineation and genetic manipulation of key enzymes in the inositol pyrophosphate family have revealed major roles for these substances in diverse areas, especially metabolic regulation and the balance between cell growth and death. As intracellular messenger molecules, inositol pyrophosphates may well emerge as regulators comparable in importance to the cyclic nucleotides. The field is young and has been hampered by technical limitations. Thus, IP7 and IP8 are difficult to synthesize so that they are not readily available from commercial sources, although innovative approaches to their generation may overcome such hurdles (131, 132). A recently developed polyacrylamide gel procedure affords facile separation of inositol pyrophosphates, which may simplify IP7 purification and detection (133). Antibodies to the critical enzymes tend to lack adequate specificity for many purposes. It is hoped that these hurdles will be surmounted and unexploited areas addressed, such as roles of these substances in the brain and the function of IP6K3. Major advances have been facilitated by the development of mice with deletion of inositol pyrophosphate biosynthetic enzymes. Some of the recent insights may have therapeutic relevance, especially in treating diabetes and obesity. IP6K inhibitors such as TNP (112) may help clarify the functions of the inositol pyrophosphates and may also afford therapeutic benefit.
Acknowledgments
We thank M. Koldobskiy, R. Tyagi, K. Juluri, J. Xu, J. K. Werner, J. Cha, D. Maag, M. Maxwell, R. Xu, and F. Rao for valued contributions. Funding: This work was supported by U.S. Public Health Service Grants MH18501 and DA-000266 (to S.H.S.).
Footnotes
Competing interests: S.H.S. is on the Scientific Advisory Board of Cerecor, Inc., which may eventually do research related to inositol phosphates.
References and Notes
- 1.Streb H, Irvine RF, Berridge MJ, Schulz I. Release of Ca2+ from a nonmitochondrial intracellular store in pancreatic acinar cells by inositol-1,4,5-trisphosphate. Nature. 1983;306:67–69. doi: 10.1038/306067a0. [DOI] [PubMed] [Google Scholar]
- 2.Berridge MJ, Lipp P, Bootman DM. The versatility and universality of calcium signalling. Nat. Rev. Mol. Cell Biol. 2000;1:11–21. doi: 10.1038/35036035. [DOI] [PubMed] [Google Scholar]
- 3.Menniti FS, Miller RN, Putney JW, Jr, Shears SB. Turnover of inositol polyphosphate pyrophosphates in pancreatoma cells. J. Biol. Chem. 1993;268:3850–3856. [PubMed] [Google Scholar]
- 4.Stephens L, Radenberg T, Thiel U, Vogel G, Khoo KH, Dell A, Jackson TR, Hawkins PT, Mayr GW. The detection, purification, structural characterization, and metabolism of diphosphoinositol pentakisphosphate(s) and bisdiphosphoinositol tetrakisphosphate(s) J. Biol. Chem. 1993;268:4009–4015. [PubMed] [Google Scholar]
- 5.Glennon MC, Shears SB. Turnover of inositol pentakisphosphates, inositol hexakisphosphate and diphosphoinositol polyphosphates in primary cultured hepatocytes. Biochem. J. 1993;293:583–590. doi: 10.1042/bj2930583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Voglmaier SM, Bembenek ME, Kaplin AI, Dormán G, Olszewski JD, Prestwich GD, Snyder SH. Purified inositol hexakisphosphate kinase is an ATP synthase: Diphosphoinositol pentakisphosphate as a high-energy phosphate donor. Proc. Natl. Acad. Sci. U.S.A. 1996;93:4305–4310. doi: 10.1073/pnas.93.9.4305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Saiardi A, Erdjument-Bromage H, Snowman AM, Tempst P, Snyder SH. Synthesis of diphosphoinositol pentakisphosphate by a newly identified family of higher inositol polyphosphate kinases. Curr. Biol. 1999;9:1323–1326. doi: 10.1016/s0960-9822(00)80055-x. [DOI] [PubMed] [Google Scholar]
- 8.Schell MJ, Letcher AJ, Brearley CA, Biber J, Murer H, Irvine RF. PiUS (Pi uptake stimulator) is an inositol hexakisphosphate kinase. FEBS Lett. 1999;461:169–172. doi: 10.1016/s0014-5793(99)01462-3. [DOI] [PubMed] [Google Scholar]
- 9.Saiardi A, Nagata E, Luo HR, Snowman AM, Snyder SH. Identification and characterization of a novel inositol hexakisphosphate kinase. J. Biol. Chem. 2001;276:39179–39185. doi: 10.1074/jbc.M106842200. [DOI] [PubMed] [Google Scholar]
- 10.Bhandari R, Juluri KR, Resnick AC, Snyder SH. Gene deletion of inositol hexakisphosphate kinase 1 reveals inositol pyrophosphate regulation of insulin secretion, growth, and spermiogenesis. Proc. Natl. Acad. Sci. U.S.A. 2008;105:2349–2353. doi: 10.1073/pnas.0712227105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chakraborty A, Koldobskiy MA, Bello NT, Maxwell M, Potter JJ, Juluri KR, Maag D, Kim S, Huang AS, Dailey MJ, Saleh M, Snowman AM, Moran TH, Mezey E, Snyder SH. Inositol pyrophosphates inhibit Akt signaling, thereby regulating insulin sensitivity and weight gain. Cell. 2010;143:897–910. doi: 10.1016/j.cell.2010.11.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Prasad A, Jia Y, Chakraborty A, Li Y, Jain SK, Zhong J, Roy SG, Loison F, Mondal S, Sakai J, Blanchard C, Snyder SH, Luo HR. Inositol hexakisphosphate kinase 1 regulates neutrophil function in innate immunity by inhibiting phosphatidylinositol-(3,4,5)-trisphosphate signaling. Nat. Immunol. 2011;12:752–760. doi: 10.1038/ni.2052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Koldobskiy MA, Chakraborty A, Werner JK, Jr, Snowman AM, Juluri KR, Vandiver MS, Kim S, Heletz S, Snyder SH. p53-mediated apoptosis requires inositol hexakisphosphate kinase-2. Proc. Natl. Acad. Sci. U.S.A. 2010;107:20947–20951. doi: 10.1073/pnas.1015671107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Morrison BH, Haney R, Lamarre E, Drazba J, Prestwich GD, Lindner DJ. Gene deletion of inositol hexakisphosphate kinase 2 predisposes to aerodigestive tract carcinoma. Oncogene. 2009;28:2383–2392. doi: 10.1038/onc.2009.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Albert C, Safrany ST, Bembenek ME, Reddy KM, Reddy K, Falck J, Bröcker M, Shears SB, Mayr GW. Biological variability in the structures of diphosphoinositol polyphosphates in Dictyostelium discoideum and mammalian cells. Biochem. J. 1997;327:553–560. doi: 10.1042/bj3270553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lin H, Fridy PC, Ribeiro AA, Choi JH, Barma DK, Vogel G, Falck JR, Shears SB, York JD, Mayr GW. Structural analysis and detection of biological inositol pyrophosphates reveal that the family of VIP/diphosphoinositol pentakisphosphate kinases are 1/3-kinases. J. Biol. Chem. 2009;284:1863–1872. doi: 10.1074/jbc.M805686200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mulugu S, Bai W, Fridy PC, Bastidas RJ, Otto JC, Dollins DE, Haystead TA, Ribeiro AA, York JD. A conserved family of enzymes that phosphorylate inositol hexakisphosphate. Science. 2007;316:106–109. doi: 10.1126/science.1139099. [DOI] [PubMed] [Google Scholar]
- 18.Lee YS, Mulugu S, York JD, O’Shea EK. Regulation of a cyclin-CDK-CDK inhibitor complex by inositol pyrophosphates. Science. 2007;316:109–112. doi: 10.1126/science.1139080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Choi JH, Williams J, Cho J, Falck JR, Shears SB. Shears, Purification, sequencing, and molecular identification of a mammalian PP-InsP5 kinase that is activated when cells are exposed to hyperosmotic stress. J. Biol. Chem. 2007;282:30763–30775. doi: 10.1074/jbc.M704655200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fridy PC, Otto JC, Dollins DE, York JD. Cloning and characterization of two human VIP1-like inositol hexakisphosphate and diphosphoinositol pentakisphosphate kinases. J. Biol. Chem. 2007;282:30754–30762. doi: 10.1074/jbc.M704656200. [DOI] [PubMed] [Google Scholar]
- 21.Bercy J, Dubois E, Messenguy F. Regulation of arginine metabolism in Saccharomyces cerevisiae: expression of the three ARGR regulatory genes and cellular localization of their products. Gene. 1987;55:277–285. doi: 10.1016/0378-1119(87)90287-3. [DOI] [PubMed] [Google Scholar]
- 22.Amar N, Messenguy F, El Bakkoury M, Dubois E. ArgRll, a component of the ArgR-Mcm1 complex involved in the control of arginine metabolism in Saccharomyces cerevisiae, is the sensor of arginine. Mol. Cell. Biol. 2000;20:2087–2097. doi: 10.1128/mcb.20.6.2087-2097.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Odom AR, Stahlberg A, Wente SR, York JD. A role for nuclear inositol 1,4,5-trisphosphate kinase in transcriptional control. Science. 2000;287:2026–2029. doi: 10.1126/science.287.5460.2026. [DOI] [PubMed] [Google Scholar]
- 24.Frederick JP, Mattiske D, Wofford JA, Megosh LC, Drake LY, Chiou ST, Hogan BL, York JD. An essential role for an inositol polyphosphate multikinase, Ipk2, in mouse embryogenesis and second messenger production. Proc. Natl. Acad. Sci. U.S.A. 2005;102:8454–8459. doi: 10.1073/pnas.0503706102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Resnick AC, Snowman AM, Kang BN, Hurt KJ, Snyder SH, Saiardi A. Inositol poly-phosphate multikinase is a nuclear PI3-kinase with transcriptional regulatory activity. Proc. Natl. Acad. Sci. U.S.A. 2005;102:12783–12788. doi: 10.1073/pnas.0506184102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maag D, Maxwell MJ, Hardesty DA, Boucher KL, Choudhari N, Hanno AG, Ma JF, Snowman AS, Pietropaoli JW, Xu R, Storm PB, Saiardi A, Snyder SH, Resnick AC. Inositol polyphosphate multikinase is a physiologic PI3-kinase that activates Akt/PKB. Proc. Natl. Acad. Sci. U.S.A. 2011;108:1391–1396. doi: 10.1073/pnas.1017831108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim S, Kim SF, Maag D, Maxwell MJ, Resnick AC, Juluri KR, Chakraborty A, Koldobskiy MA, Cha SH, Barrow R, Snowman AM, Snyder SH. Amino acid signaling to mTOR mediated by inositol polyphosphate multikinase. Cell Metab. 2011;13:215–221. doi: 10.1016/j.cmet.2011.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Irvine RF, Schell MJ. Back in the water: The return of the inositol phosphates. Nat. Rev. Mol. Cell Biol. 2001;2:327–338. doi: 10.1038/35073015. [DOI] [PubMed] [Google Scholar]
- 29.Shears SB. How versatile are inositol phosphate kinases? Biochem. J. 2004;377:265–280. doi: 10.1042/BJ20031428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bennett M, Onnebo SM, Azevedo C, Saiardi A. Inositol pyrophosphates: Metabolism and signaling. Cell. Mol. Life Sci. 2006;63:552–564. doi: 10.1007/s00018-005-5446-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shears SB. Diphosphoinositol polyphosphates: Metabolic messengers? Mol. Pharmacol. 2009;76:236–252. doi: 10.1124/mol.109.055897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Barker CJ, Illies C, Gaboardi GC, Berggren PO. Inositol pyrophosphates: structure, enzymology and function. Cell. Mol. Life Sci. 2009;66:3851–3871. doi: 10.1007/s00018-009-0115-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Irvine RF, Letcher AJ, Heslop JP, Berridge MJ. The inositol tris/tetrakisphosphate pathway—demonstration of Ins(1,4,5)P3 3-kinase activity in animal tissues. Nature. 1986;320:631–634. doi: 10.1038/320631a0. [DOI] [PubMed] [Google Scholar]
- 34.Chattaway JA, Drobak BK, Watkins PAC, Dawson AP, Letcher AJ, Stephens LR, Irvine RF. An inositol 1,4,5-trisphosphate 6-kinase activity in pea roots. Planta. 1992;187:542–545. doi: 10.1007/BF00199975. [DOI] [PubMed] [Google Scholar]
- 35.Stevenson-Paulik J, Odom AR, York JD. Molecular and biochemical characterization of two plant inositol polyphosphate 6-/3-/5-kinases. J. Biol. Chem. 2002;277:42711–42718. doi: 10.1074/jbc.M209112200. [DOI] [PubMed] [Google Scholar]
- 36.Saiardi A, Caffrey JJ, Snyder SH, Shears SB. Inositol polyphosphate multikinase (ArgRIII) determines nuclear mRNA export in Saccharomyces cerevisiae. FEBS Lett. 2000;468:28–32. doi: 10.1016/s0014-5793(00)01194-7. [DOI] [PubMed] [Google Scholar]
- 37.Europe-Finner GN, Gammon B, Newell PC. Accumulation of [3H]-inositol into inositol polyphosphates during development of Dictyostelium. Biochem. Biophys. Res. Commun. 1991;181:191–196. doi: 10.1016/s0006-291x(05)81400-7. [DOI] [PubMed] [Google Scholar]
- 38.Stephens LR, Hawkins PT, Stanley AF, Moore T, Poyner DR, Morris PJ, Hanley MR, Kay RR, Irvine RF. myo-inositol pentakisphosphates. Structure, biological occurrence and phosphorylation to myo-inositol hexakisphosphate. Biochem. J. 1991;275:485–499. doi: 10.1042/bj2750485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Oliver KG, Putney JW, Jr, Obie JF, Shears SB. The interconversion of inositol 1,3,4,5,6-pentakisphosphate and inositol tetrakisphosphates in AR4-2J cells. J. Biol. Chem. 1992;267:21528–21534. [PubMed] [Google Scholar]
- 40.Wong NS, Barker CJ, Morris AJ, Craxton A, Kirk CJ, Michell RH. The inositol phosphates in WRK1 rat mammary tumour cells. Biochem. J. 1992;286:459–468. doi: 10.1042/bj2860459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mayr GW, Radenberg T, Thiel U, Vogel G, Stephens LR. Phosphoinositol diphosphates: Nonenzymic formation in vitro and occurrence in vivo in the cellular slime mold Dictyostelium. Carbohydr. Res. 1992;234:247–262. [Google Scholar]
- 42.Shears SB, Ali N, Craxton A, Bembenek ME. Synthesis and metabolism of bis-diphosphoinositol tetrakisphosphate in vitro and in vivo. J. Biol. Chem. 1995;270:10489–10497. doi: 10.1074/jbc.270.18.10489. [DOI] [PubMed] [Google Scholar]
- 43.Draskovic P, Saiardi A, Bhandari R, Burton A, Ilc G, Kovacevic M, Snyder SH, Podobnik M. Inositol hexakisphosphate kinase products contain diphosphate and triphosphate groups. Chem. Biol. 2008;15:274–286. doi: 10.1016/j.chembiol.2008.01.011. [DOI] [PubMed] [Google Scholar]
- 44.Verbsky JW, Wilson MP, Kisseleva MV, Majerus PW, Wente SR. The synthesis of inositol hexakisphosphate Characterization of human inositol 1,3,4,5,6-pentakisphosphate 2-kinase. J. Biol. Chem. 2002;277:31857–31862. doi: 10.1074/jbc.M205682200. [DOI] [PubMed] [Google Scholar]
- 45.Seeds AM, Bastidas RJ, York JD. Molecular definition of a novel inositol polyphosphate metabolic pathway initiated by inositol 1,4,5-trisphosphate 3-kinase activity in Saccharomyces cerevisiae. J. Biol. Chem. 2005;280:27654–27661. doi: 10.1074/jbc.M505089200. [DOI] [PubMed] [Google Scholar]
- 46.Saiardi A, Caffrey JJ, Snyder SH, Shears SB. The inositol hexakisphosphate kinase family Catalytic flexibility and function in yeast vacuole biogenesis. J. Biol. Chem. 2000;275:24686–24692. doi: 10.1074/jbc.M002750200. [DOI] [PubMed] [Google Scholar]
- 47.Safrany ST, Caffrey JJ, Yang X, Bembenek ME, Moyer MB, Burkhart WA, Shears SB. A novel context for the ‘MutT’ module, a guardian of cell integrity, in a diphosphoinositol polyphosphate phosphohydrolase. EMBO J. 1998;17:6599–6607. doi: 10.1093/emboj/17.22.6599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fisher DI, Safrany ST, Strike P, McLennan AG, Cartwright JL. Nudix hydrolases that degrade dinucleoside and diphosphoinositol polyphosphates also have 5-phosphoribosyl 1-pyrophosphate (PRPP) pyrophosphatase activity that generates the glycolytic activator ribose 1,5-bisphosphate. J. Biol. Chem. 2002;277:47313–47317. doi: 10.1074/jbc.M209795200. [DOI] [PubMed] [Google Scholar]
- 49.Ingram SW, Safrany ST, Barnes LD. Disruption and overexpression of the Schizosaccharomyces pombe aps1 gene, and effects on growth rate, morphology and intracellular diadenosine 5′,5 ’-P1,P5-pentaphosphate and diphosphoinositol polyphosphate concentrations. Biochem. J. 2003;369:519–528. doi: 10.1042/BJ20020733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Illies C, Gromada J, Fiume R, Leibiger B, Yu J, Juhl K, Yang SN, Barma DK, Falck JR, Saiardi A, Barker CJ, Berggren PO. Requirement of inositol pyrophosphates for full exocytotic capacity in pancreatic beta cells. Science. 2007;318:1299–1302. doi: 10.1126/science.1146824. [DOI] [PubMed] [Google Scholar]
- 51.Szwergold BS, Graham RA, Brown TR. Observation of inositol pentakis- and hexakisphosphates in mammalian tissues by 31P NMR. Biochem. Biophys. Res. Commun. 1987;149:874–881. doi: 10.1016/0006-291x(87)90489-x. [DOI] [PubMed] [Google Scholar]
- 52.Laussmann T, Pikzack C, Thiel U, Mayr GW, Vogel G. Diphospho-myo-inositol phosphates during the life cycle of Dictyostelium and Polysphondylium. Eur. J. Biochem. 2000;267:2447–2451. doi: 10.1046/j.1432-1327.2000.01264.x. [DOI] [PubMed] [Google Scholar]
- 53.Pesesse X, Choi K, Zhang T, Shears SB. Signaling by higher inositol polyphosphates Synthesis of bisdiphosphoinositol tetrakisphosphate (“InsP8”) is selectively activated by hyperosmotic stress. J. Biol. Chem. 2004;279:43378–43381. doi: 10.1074/jbc.C400286200. [DOI] [PubMed] [Google Scholar]
- 54.Safrany ST. Protocols for regulation and study of diphosphoinositol polyphosphates. Mol. Pharmacol. 2004;66:1585–1591. doi: 10.1124/mol.104.002667. [DOI] [PubMed] [Google Scholar]
- 55.Choi K, Mollapour E, Shears SB. Signal transduction during environmental stress: InsP8 operates within highly restricted contexts. Cell. Signal. 2005;17:1533–1541. doi: 10.1016/j.cellsig.2005.03.021. [DOI] [PubMed] [Google Scholar]
- 56.Choi K, Mollapour E, Choi JH, Shears SB. Cellular energetic status supervises the synthesis of bis-diphosphoinositol tetrakisphosphate independently of AMP-activated protein kinase. Mol. Pharmacol. 2008;74:527–536. doi: 10.1124/mol.107.044628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nagata E, Luo HR, Saiardi A, Bae BI, Suzuki N, Snyder SH. Inositol hexakisphosphate kinase-2, a physiologic mediator of cell death. J. Biol. Chem. 2005;280:1634–1640. doi: 10.1074/jbc.M409416200. [DOI] [PubMed] [Google Scholar]
- 58.Chakraborty A, Koldobskiy MA, Sixt KM, Juluri KR, Mustafa AK, Snowman AM, van Rossum DB, Patterson RL, Snyder SH. HSP90 regulates cell survival via inositol hexakisphosphate kinase-2. Proc. Natl. Acad. Sci. U.S.A. 2008;105:1134–1139. doi: 10.1073/pnas.0711168105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chakraborty A, Werner JK, Jr, Koldobskiy MA, Mustafa AK, Juluri KR, Pietropaoli J, Snowman AM, Snyder SH. Casein kinase-2 mediates cell survival through phosphorylation and degradation of inositol hexakisphosphate kinase-2. Proc. Natl. Acad. Sci. U.S.A. 2011;108:2205–2209. doi: 10.1073/pnas.1019381108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Blakeley DM, Corps AN, Brown KD. Bombesin and platelet-derived growth factor stimulate formation of inositol phosphates and Ca2+ mobilization in Swiss 3T3 cells by different mechanisms. Biochem. J. 1989;258:177–185. doi: 10.1042/bj2580177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tilly BC, van Paridon PA, Verlaan I, de Laat SW, Moolenaar WH. Epidermal-growth-factor-induced formation of inositol phosphates in human A431 cells Differences from the effect of bradykinin. Biochem. J. 1988;252:857–863. doi: 10.1042/bj2520857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Takasu N, Takasu M, Komiya I, Nagasawa Y, Asawa T, Shimizu Y, Yamada T. Insulin-like growth factor I stimulates inositol phosphate accumulation, a rise in cytoplasmic free calcium, and proliferation in cultured porcine thyroid cells. J. Biol. Chem. 1989;264:18485–18488. [PubMed] [Google Scholar]
- 63.Balla T, Sim SS, Baukal AJ, Rhee SG, Catt KJ. Inositol polyphosphates are not increased by overexpression of Ins(1,4,5)P3 3-kinase but show cell-cycle dependent changes in growth factor-stimulated fibroblasts. Mol. Biol. Cell. 1994;5:17–27. doi: 10.1091/mbc.5.1.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gao Y, Wang HY. Inositol pentakisphosphate mediates Wnt/beta-catenin signaling. J. Biol. Chem. 2007;282:26490–26502. doi: 10.1074/jbc.M702106200. [DOI] [PubMed] [Google Scholar]
- 65.Logan CY, Nusse R. The Wnt signaling pathway in development and disease. Annu. Rev. Cell Dev. Biol. 2004;20:781–810. doi: 10.1146/annurev.cellbio.20.010403.113126. [DOI] [PubMed] [Google Scholar]
- 66.Majerus PW. Inositol phosphate biochemistry. Annu. Rev. Biochem. 1992;61:225–250. doi: 10.1146/annurev.bi.61.070192.001301. [DOI] [PubMed] [Google Scholar]
- 67.Alcázar-Román AR, Wente SR. Inositol poly-phosphates: A new frontier for regulating gene expression. Chromosoma. 2008;117:1–13. doi: 10.1007/s00412-007-0126-4. [DOI] [PubMed] [Google Scholar]
- 68.Fukuda M, Kojima T, Kabayama H, Mikoshiba K. Mutation of the pleckstrin homology domain of Bruton’s tyrosine kinase in immunodeficiency impaired inositol 1,3,4,5-tetrakisphosphate binding capacity. J. Biol. Chem. 1996;271:30303–30306. doi: 10.1074/jbc.271.48.30303. [DOI] [PubMed] [Google Scholar]
- 69.Hirata M, Kanematsu T, Takeuchi H, Yagisawa H. Pleckstrin homology domain as an inositol compound binding module. Jpn. J. Pharmacol. 1998;76:255–263. doi: 10.1254/jjp.76.255. [DOI] [PubMed] [Google Scholar]
- 70.Ye W, Ali N, Bembenek ME, Shears SB, Lafer EM. Inhibition of clathrin assembly by high affinity binding of specific inositol polyphosphates to the synapse-specific clathrin assembly protein AP-3. J. Biol. Chem. 1995;270:1564–1568. [PubMed] [Google Scholar]
- 71.Huang YH, Grasis JA, Miller AT, Xu R, Soonthornvacharin S, Andreotti AH, Tsoukas CD, Cooke MP, Sauer K. Positive regulation of Itk PH domain function by soluble lP4. Science. 2007;316:886–889. doi: 10.1126/science.1138684. [DOI] [PubMed] [Google Scholar]
- 72.Jia Y, Subramanian KK, Erneux C, Pouillon V, Hattori H, Jo H, You J, Zhu D, Schurmans S, Luo HR. Inositol 1,3,4,5-tetrakisphosphate negatively regulates phosphatidylinositol-3,4,5-trisphosphate signaling in neutrophils. Immunity. 2007;27:453–467. doi: 10.1016/j.immuni.2007.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Luo HR, Huang YE, Chen JC, Saiardi A, Iijima M, Ye K, Huang Y, Nagata E, Devreotes P, Snyder SH. Inositol pyrophosphates mediate chemotaxis in Dictyostelium via pleckstrin homology domain-PtdIns(3,4,5)P3 interactions. Cell. 2003;114:559–572. doi: 10.1016/s0092-8674(03)00640-8. [DOI] [PubMed] [Google Scholar]
- 74.Takeuchi H, Kanematsu T, Misumi Y, Sakane F, Konishi H, Kikkawa U, Watanabe Y, Katan M, Hirata M. Distinct specificity in the binding of inositol phosphates by pleckstrin homology domains of pleckstrin, RAC-protein kinase, diacylglycerol kinase and a new 130 kDa protein. Biochim. Biophys. Acta. 1997;1359:275–285. doi: 10.1016/s0167-4889(97)00109-2. [DOI] [PubMed] [Google Scholar]
- 75.Frech M, Andjelkovic M, Ingley E, Reddy KK, Falck JR, Hemmings BA. High affinity binding of inositol phosphates and phosphoinositides to the pleckstrin homology domain of RAC/protein kinase B and their influence on kinase activity. J. Biol. Chem. 1997;272:8474–8481. doi: 10.1074/jbc.272.13.8474. [DOI] [PubMed] [Google Scholar]
- 76.Komander D, Fairservice A, Deak M, Kular GS, Prescott AR, Peter Downes C, Safrany ST, Alessi DR, van Aalten DM. Structural insights into the regulation of PDK1 by phosphoinositides and inositol phosphates. EMBO J. 2004;23:3918–3928. doi: 10.1038/sj.emboj.7600379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lemmon MA. Membrane recognition by phospholipid-binding domains. Nat. Rev. Mol. Cell Biol. 2008;9:99–111. doi: 10.1038/nrm2328. [DOI] [PubMed] [Google Scholar]
- 78.Fruman DA, Rameh LE, Cantley LC. Phosphoinositide binding domains: embracing 3-phosphate. Cell. 1999;97:817–820. doi: 10.1016/s0092-8674(00)80792-8. [DOI] [PubMed] [Google Scholar]
- 79.Di Paolo G, De Camilli P. Phosphoinositides in cell regulation and membrane dynamics. Nature. 2006;443:651–657. doi: 10.1038/nature05185. [DOI] [PubMed] [Google Scholar]
- 80.Manning BD, Cantley LC. AKT/PKB signaling: Navigating downstream. Cell. 2007;129:1261–1274. doi: 10.1016/j.cell.2007.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lee YS, Huang K, Quiocho FA, O’Shea EK. Molecular basis of cyclin-CDK-CKI regulation by reversible binding of an inositol pyrophosphate. Nat. Chem. Biol. 2008;4:25–32. doi: 10.1038/nchembio.2007.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Laussmann T, Eujen R, Weisshuhn CM, Thiel U, Vogel G. Structures of diphospho-myo-inositol pentakisphosphate and bisdiphospho-myo-inositol tetrakisphosphate from Dictyostelium resolved by NMR analysis. Biochem. J. 1996;315:715–720. doi: 10.1042/bj3150715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Saiardi A, Bhandari R, Resnick AC, Snowman AM, Snyder SH. Phosphorylation of proteins by inositol pyrophosphates. Science. 2004;306:2101–2105. doi: 10.1126/science.1103344. [DOI] [PubMed] [Google Scholar]
- 84.Bhandari R, Saiardi A, Ahmadibeni Y, Snowman AM, Resnick AC, Kristiansen TZ, Molina H, Pandey A, Werner JK, Jr, Juluri KR, Xu Y, Prestwich GD, Parang K, Snyder SH. Protein pyrophosphorylation by inositol pyrophosphates is a posttranslational event. Proc. Natl. Acad. Sci. U.S.A. 2007;104:15305–15310. doi: 10.1073/pnas.0707338104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Azevedo C, Burton A, Ruiz-Mateos E, Marsh M, Saiardi A. Inositol pyrophosphate mediated pyrophosphorylation of AP3B1 regulates HIV-1 Gag release. Proc. Natl. Acad. Sci. U.S.A. 2009;106:21161–21166. doi: 10.1073/pnas.0909176106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kobir A, Shi L, Boskovic A, Grangeasse C, Franjevic D, Mijakovic I. Protein phosphorylation in bacterial signal transduction. Biochim. Biophys. Acta. 2011 doi: 10.1016/j.bbagen.2011.01.006. [DOI] [PubMed] [Google Scholar]
- 87.Hand CE, Honek JF. Phosphate transfer from inositol pyrophosphates InsP5PP and InsP4(PP)2: A semi-empirical investigation. Bioorg. Med. Chem. Lett. 2007;17:183–188. doi: 10.1016/j.bmcl.2006.09.066. [DOI] [PubMed] [Google Scholar]
- 88.Saiardi A, Resnick AC, Snowman AM, Wendland B, Snyder SH. Inositol pyrophosphates regulate cell death and telomere length through phosphoinositide 3-kinase-related protein kinases. Proc. Natl. Acad. Sci. U.S.A. 2005;102:1911–1914. doi: 10.1073/pnas.0409322102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.York SJ, Armbruster BN, Greenwell P, Petes TD, York JD. Inositol diphosphate signaling regulates telomere length. J. Biol. Chem. 2005;280:4264–4269. doi: 10.1074/jbc.M412070200. [DOI] [PubMed] [Google Scholar]
- 90.Luo HR, Saiardi A, Yu H, Nagata E, Ye K, Snyder SH. Inositol pyrophosphates are required for DNA hyperrecombination in protein kinase c1 mutant yeast. Biochemistry. 2002;41:2509–2515. doi: 10.1021/bi0118153. [DOI] [PubMed] [Google Scholar]
- 91.Dubois E, Scherens B, Vierendeels F, Ho MM, Messenguy F, Shears SB. In Saccharomyces cerevisiae, the inositol polyphosphate kinase activity of Kcs1p is required for resistance to salt stress, cell wall integrity, and vacuolar morphogenesis. J. Biol. Chem. 2002;277:23755–23763. doi: 10.1074/jbc.M202206200. [DOI] [PubMed] [Google Scholar]
- 92.Saiardi A, Sciambi C, McCaffery JM, Wendland B, Snyder SH. Inositol pyrophosphates regulate endocytic trafficking. Proc. Natl. Acad. Sci. U.S.A. 2002;99:14206–14211. doi: 10.1073/pnas.212527899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Pöhlmann J, Fleig U. Asp1, a conserved 1/3 inositol polyphosphate kinase, regulates the dimorphic switch in Schizosaccharomyces pombe. Mol. Cell. Biol. 2010;30:4535–4547. doi: 10.1128/MCB.00472-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Luo HR, Saiardi A, Nagata E, Ye K, Yu H, Jung TS, Luo X, Jain S, Sawa A, Snyder SH. GRAB: A physiologic guanine nucleotide exchange factor for Rab3A, which interacts with inositol hexakisphosphate kinase. Neuron. 2001;31:439–451. doi: 10.1016/s0896-6273(01)00384-1. [DOI] [PubMed] [Google Scholar]
- 95.Morrison BH, Bauer JA, Kalvakolanu DV, Lindner DJ. Inositol hexakisphosphate kinase 2 mediates growth suppressive and apoptotic effects of interferon-beta in ovarian carcinoma cells. J. Biol. Chem. 2001;276:24965–24970. doi: 10.1074/jbc.M101161200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Morrison BH, Bauer JA, Hu J, Grane RW, Ozdemir AM, Chawla-Sarkar M, Gong B, Almasan A, Kalvakolanu DV, Lindner DJ. Inositol hexakisphosphate kinase 2 sensitizes ovarian carcinoma cells to multiple cancer therapeutics. Oncogene. 2002;21:1882–1889. doi: 10.1038/sj/onc/1205265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Morrison BH, Tang Z, Jacobs BS, Bauer JA, Lindner DJ. Apo2L/TRAIL induction and nuclear translocation of inositol hexakisphosphate kinase 2 during IFN-beta-induced apoptosis in ovarian carcinoma. Biochem. J. 2005;385:595–603. doi: 10.1042/BJ20040971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Mahalingam D, Szegezdi E, Keane M, de Jong S, Samali A. TRAIL receptor signalling and modulation: Are we on the right TRAIL? Cancer Treat. Rev. 2009;35:280–288. doi: 10.1016/j.ctrv.2008.11.006. [DOI] [PubMed] [Google Scholar]
- 99.Scaglioni PP, Yung TM, Cai LF, Erdjument-Bromage H, Kaufman AJ, Singh B, Teruya-Feldstein J, Tempst P, Pandolfi PP. A CK2-de-pendent mechanism for degradation of the PML tumor suppressor. Cell. 2006;126:269–283. doi: 10.1016/j.cell.2006.05.041. [DOI] [PubMed] [Google Scholar]
- 100.Ruzzene M, Pinna LA. Addiction to protein kinase CK2: A common denominator of diverse cancer cells? Biochim. Biophys. Acta. 2010;1804:499–504. doi: 10.1016/j.bbapap.2009.07.018. [DOI] [PubMed] [Google Scholar]
- 101.Ljubimov AV, Caballero S, Aoki AM, Pinna LA, Grant MB, Castellon R. Involvement of protein kinase CK2 in angiogenesis and retinal neovascularization. Invest. Ophthalmol. Vis. Sci. 2004;45:4583–4591. doi: 10.1167/iovs.04-0686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Nagata E, Saiardi A, Tsukamoto H, Satoh T, Itoh Y, Itoh J, Shibata M, Takizawa S, Takagi S. Inositol hexakisphosphate kinases promote autophagy. Int. J. Biochem. Cell Biol. 2010;42:2065–2071. doi: 10.1016/j.biocel.2010.09.013. [DOI] [PubMed] [Google Scholar]
- 103.Onnebo SM, Saiardi A. Inositol pyrophosphates modulate hydrogen peroxide signalling. Biochem. J. 2009;423:109–118. doi: 10.1042/BJ20090241. [DOI] [PubMed] [Google Scholar]
- 104.Sarmah B, Wente SR. Inositol hexakisphosphate kinase-2 acts as an effector of the vertebrate Hedgehog pathway. Proc. Natl. Acad. Sci. U.S.A. 2010;107:19921–19926. doi: 10.1073/pnas.1007256107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Izumiya Y, Hopkins T, Morris C, Sato K, Zeng L, Viereck J, Hamilton JA, Ouchi N, LeBrasseur NK, Walshs K. Fast/Glycolytic muscle fiber growth reduces fat mass and improves metabolic parameters in obese mice. Cell Metab. 2008;7:159–172. doi: 10.1016/j.cmet.2007.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Peng XD, Xu PZ, Chen ML, Hahn-Windgassen A, Skeen J, Jacobs J, Sundararajan D, Chen WS, Crawford SE, Coleman KG, Hay N. Dwarfism, impaired skin development, skeletal muscle atrophy, delayed bone development, and impeded adipogenesis in mice lacking Akt1 and Akt2. Genes Dev. 2003;17:1352–1365. doi: 10.1101/gad.1089403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Ross SE, Hemati N, Longo KA, Bennett CN, Lucas PC, Erickson RL, MacDougald OA. Inhibition of adipogenesis by Wnt signaling. Science. 2000;289:950–953. doi: 10.1126/science.289.5481.950. [DOI] [PubMed] [Google Scholar]
- 108.Tang QQ, Grønborg M, Huang H, Kim JW, Otto TC, Pandey A, Lane MD. Sequential phosphorylation of CCAAT enhancer-binding protein beta by MAPK and glycogen synthase kinase 3beta is required for adipogenesis. Proc. Natl. Acad. Sci. U.S.A. 2005;102:9766–9771. doi: 10.1073/pnas.0503891102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Funai K, Parkington JD, Carambula S, Fielding RA. Age-associated decrease in contraction-induced activation of downstream targets of Akt/mTor signaling in skeletal muscle. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2006;290:R1080–R1086. doi: 10.1152/ajpregu.00277.2005. [DOI] [PubMed] [Google Scholar]
- 110.Shao J, Yamashita H, Qiao L, Friedman JE. Decreased Akt kinase activity and insulin resistance in C57BL/KsJ-Leprdb/db mice. J. Endocrinol. 2000;167:107–115. doi: 10.1677/joe.0.1670107. [DOI] [PubMed] [Google Scholar]
- 111.Kaidanovich O, Eldar-Finkelman H. The role of glycogen synthase kinase-3 in insulin resistance and type 2 diabetes. Expert Opin. Ther. Targets. 2002;6:555–561. doi: 10.1517/14728222.6.5.555. [DOI] [PubMed] [Google Scholar]
- 112.Padmanabhan U, Dollins DE, Fridy PC, York JD, Downes CP. Characterization of a selective inhibitor of inositol hexakisphosphate kinases: Use in defining biological roles and metabolic relationships of inositol pyrophosphates. J. Biol. Chem. 2009;284:10571–10582. doi: 10.1074/jbc.M900752200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Downes CP, Gray A, Fairservice A, Safrany ST, Batty IH, Fleming I. The regulation of membrane to cytosol partitioning of signalling proteins by phosphoinositides and their soluble headgroups. Biochem. Soc. Trans. 2005;33:1303–1307. doi: 10.1042/BST0331303. [DOI] [PubMed] [Google Scholar]
- 114.Calleja V, Alcor D, Laguerre M, Park J, Vojnovic B, Hemmings BA, Downward J, Parker PJ, Larijani B. Intramolecular and intermolecular interactions of protein kinase B define its activation in vivo. PLoS Biol. 2007;5:e95. doi: 10.1371/journal.pbio.0050095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Jackson SG, Al-Saigh S, Schultz C, Junop MS. Inositol pentakisphosphate isomers bind PH domains with varying specificity and inhibit phosphoinositide interactions. BMC Struct. Biol. 2011;11:11. doi: 10.1186/1472-6807-11-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Gokhale NA, Zaremba A, Shears SB. Receptor-dependent compartmentalization of PPIP5K1, a kinase with a cryptic polyphosphoinositide binding domain. Biochem. J. 2011;434:415–426. doi: 10.1042/BJ20101437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Carpten JD, Faber AL, Horn C, Donoho GP, Briggs SL, Robbins CM, Hostetter G, Boguslawski S, Moses TY, Savage S, Uhlik M, Lin A, Du J, Qian YW, Zeckner DJ, Tucker-Kellogg G, Touchman J, Patel K, Mousses S, Bittner M, Schevitz R, Lai MH, Blanchard KL, E J, Thomas JE. A transforming mutation in the pleckstrin homology domain of AKT1 in cancer. Nature. 2007;448:439–444. doi: 10.1038/nature05933. [DOI] [PubMed] [Google Scholar]
- 118.Stephens L, Ellson C, Hawkins P. Roles of PI3Ks in leukocyte chemotaxis and phagocytosis. Curr. Opin. Cell Biol. 2002;14:203–213. doi: 10.1016/s0955-0674(02)00311-3. [DOI] [PubMed] [Google Scholar]
- 119.Olson TS, Ley K. Chemokines and chemokine receptors in leukocyte trafficking. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2002;283:R7–R28. doi: 10.1152/ajpregu.00738.2001. [DOI] [PubMed] [Google Scholar]
- 120.Hawkins PT, Davidson K, Stephens LR. The role of PI3Ks in the regulation of the neutrophil NADPH oxidase. Biochem. Soc. Symp. 2007;74:59–67. doi: 10.1042/BSS0740059. [DOI] [PubMed] [Google Scholar]
- 121.Emamian ES, Hall D, Birnbaum MJ, Karayiorgou M, Gogos JA. Convergent evidence for impaired AKT1-GSK3beta signaling in schizophrenia. Nat. Genet. 2004;36:131–137. doi: 10.1038/ng1296. [DOI] [PubMed] [Google Scholar]
- 122.Jope RS. Lithium and GSK-3: One inhibitor, two inhibitory actions, multiple outcomes. Trends Pharmacol. Sci. 2003;24:441–443. doi: 10.1016/S0165-6147(03)00206-2. [DOI] [PubMed] [Google Scholar]
- 123.Beaulieu JM, Marion S, Rodriguiz RM, Medvedev IO, Sotnikova TD, Ghisi V, Wetsel WC, Lefkowitz RJ, Gainetdinov RR, Caron MG. A beta-arrestin 2 signaling complex mediates lithium action on behavior. Cell. 2008;132:125–136. doi: 10.1016/j.cell.2007.11.041. [DOI] [PubMed] [Google Scholar]
- 124.Piccolo E, Vignati S, Maffucci T, Innominato PF, Riley AM, Potter BV, Pandolfi PP, Broggini M, Iacobelli S, Innocenti P, Falasca M. Inositol pentakisphosphate promotes apoptosis through the PI 3-K/Akt pathway. Oncogene. 2004;23:1754–1765. doi: 10.1038/sj.onc.1207296. [DOI] [PubMed] [Google Scholar]
- 125.Falasca M, Chiozzotto D, Godage HY, Mazzoletti M, Riley AM, Previdi S, Potter BV, Broggini M, Maffucci T. A novel inhibitor of the PI3K/Akt pathway based on the structure of inositol 1,3,4,5,6-pentakisphosphate. Br. J. Cancer. 2010;102:104–114. doi: 10.1038/sj.bjc.6605408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Heitman J, Movva NR, Hall MN. Targets for cell cycle arrest by the immunosuppressant rapamycin in yeast. Science. 1991;253:905–909. doi: 10.1126/science.1715094. [DOI] [PubMed] [Google Scholar]
- 127.Sabatini DM, Erdjument-Bromage H, Lui M, Tempst P, Snyder SH. RAFT1: a mammalian protein that binds to FKBP12 in a rapamycin-dependent fashion and is homologous to yeast TORs. Cell. 1994;78:35–43. doi: 10.1016/0092-8674(94)90570-3. [DOI] [PubMed] [Google Scholar]
- 128.Ma XM, Blenis J. Molecular mechanisms of mTOR-mediated translational control. Nat. Rev. Mol. Cell Biol. 2009;10:307–318. doi: 10.1038/nrm2672. [DOI] [PubMed] [Google Scholar]
- 129.Kim DH, Sarbassov DD, Ali SM, King JE, Latek RR, Erdjument-Bromage H, Tempst P, Sabatini DM. mTOR interacts with raptor to form a nutrient-sensitive complex that signals to the cell growth machinery. Cell. 2002;110:163–175. doi: 10.1016/s0092-8674(02)00808-5. [DOI] [PubMed] [Google Scholar]
- 130.Sarbassov DD, Ali SM, Kim D-H, Guertin DA, Latek RR, Erdjument-Bromage H, Tempst P, Sabatini DM. Rictor, a novel binding partner of mTOR, defines a rapamycin-insensitive and raptor-independent pathway that regulates the cytoskeleton. Curr. Biol. 2004;14:1296–1302. doi: 10.1016/j.cub.2004.06.054. [DOI] [PubMed] [Google Scholar]
- 131.Reddy KM, Reddy KK, Falck JR. Synthesis of 2- and 5-diphospho-myo-inositol pentakisphosphate (2- and 5-PP-InsP5), intracellular mediators. Tetrahedron Lett. 1997;38:4951–4952. [Google Scholar]
- 132.Zhang H, Thompson J, Prestwich GD. A scalable synthesis of the IP7 isomer, 5-PP-Ins(1,2,3,4,6) P5. Org. Lett. 2009;11:1551–1554. doi: 10.1021/ol900149x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Losito O, Szijgyarto Z, Resnick AC, Saiardi A. Inositol pyrophosphates and their unique metabolic complexity: Analysis by gel electrophoresis. PLoS ONE. 2009;4:e5580. doi: 10.1371/journal.pone.0005580. [DOI] [PMC free article] [PubMed] [Google Scholar]