This work summarizes recent progress in the use of small molecules for the expansion and generation of desirable lineage-restricted stem and progenitor cells in vitro and for selectively controlling cell fate of lineage-restricted stem and progenitor cells in vivo, thereby facilitating stem cell-based clinical applications. All of the examples listed suggest that small molecules can be used to facilitate the generation and expansion of desirable lineage-restricted stem and progenitor cells for various purposes, and selectively control the differentiation of lineage-restricted stem and progenitor cells in vitro and in vivo for therapeutics purposes.
Keywords: Stem/progenitor cell, Differentiation, Hematopoietic stem cells, Neural stem cell, Stem cell expansion, T cell, Induced pluripotent stem cells, Mesenchymal stem cells, Self-renewal, Cell fate conversion
Abstract
Generation and manipulation of lineage-restricted stem and progenitor cells in vitro and/or in vivo are critical for the development of stem cell-based clinical therapeutics. Lineage-restricted stem and progenitor cells have many advantageous qualities, including being able to efficiently engraft and differentiate into desirable cell types in vivo after transplantation, and they are much less tumorigenic than pluripotent cells. Generation of lineage-restricted stem and progenitor cells can be achieved by directed differentiation from pluripotent stem cells or lineage conversion from easily obtained somatic cells. Small molecules can be very helpful in these processes since they offer several important benefits. For example, the risk of tumorigenesis is greatly reduced when small molecules are used to replace integrated transcription factors, which are widely used in cell fate conversion. Furthermore, small molecules are relatively easy to apply, optimize, and manufacture, and they can more readily be developed into conventional pharmaceuticals. Alternatively, small molecules can be used to expand or selectively control the differentiation of lineage-restricted stem and progenitor cells for desirable therapeutics purposes in vitro or in vivo. Here we summarize recent progress in the use of small molecules for the expansion and generation of desirable lineage-restricted stem and progenitor cells in vitro and for selectively controlling cell fate of lineage-restricted stem and progenitor cells in vivo, thereby facilitating stem cell-based clinical applications.
Introduction
The breakthrough of induced pluripotent stem cell (iPSC) technology holds great promise for personalized cell therapy [1, 2]. However, iPSCs or even embryonic stem cells (ESCs), representing a very early developmental stage, cannot be directly applied to patients, where functional tissue-specific cell types are needed. Furthermore the use of iPSCs/ESCs poses a high risk of tumor formation [1]. Great efforts have been made toward stepwise differentiation of ESCs or iPSCs into desirable tissue-specific cell types, such as hematopoietic stem cells (HSCs), dopaminergic neuronal cells, cardiomyocytes, and pancreatic islet β cells [3–6]. However, these pluripotent cell-derived differentiated cells have some important limitations: (a) the differentiation usually results in a heterogeneous mixture of cells that are often very difficult to expand and maintain in vitro, making it difficult to derive a sufficient amount of functional cells, and (b) these cells engraft poorly upon transplantation [2]. Therefore, advances must be made in the differentiation of pluripotent stem cells toward suitable cell fates before they can be generally useful for therapy.
On the other hand, endogenous lineage-restricted stem and progenitor cells reside in the body in special microenvironments called niches and can each differentiate into several tissue-specific cell types [7, 8]. Some cells and the tissues they populate, owing to ample stores of stem cells, can readily regenerate after injury, such as skin cells and the cells that line the digestive tract. However, other tissues, perhaps because of low numbers of the tissue-specific stem cells or inadequate activity of the niche cells (supporting stem cells), are very difficult to regenerate after injury, such as pancreatic islet β-cells, hepatocytes, and cardiomyocytes [1, 3–8]. This represents an underlying mechanism of many degenerative diseases or poor recovery after tissue injury. Lineage-restricted stem and progenitor cells are well suited for cell replacement: they efficiently engraft and differentiate into desirable cell types in vivo after transplantation and are much less tumorigenic than pluripotent cells or their derivatives [2]. Some lineage-restricted stem and progenitor cells can be expanded in vitro when cultured under special conditions [9], but some are refractory to expansion. Therefore, developing methods to obtain large amounts of lineage-restricted stem cells represents a critical step in the realization of stem cell-based therapeutics [2, 9]. Generally speaking, there are three methods to obtain these stem cells: (a) expansion of stem cells directly isolated from a donor, (b) stepwise differentiation from ESCs/iPSCs, and (c) lineage conversion of one tissue-specific cell type into another lineage-restricted stem cell.
Stem cells have the ability to go through numerous cycles of cell division resulting in expansion of stem cells while maintaining their intact state or keeping all of their original potential, and this is called self-renewal, an important feature for stem cells. The self-renewal of these lineage-restricted stem cells is strictly controlled by their own transcriptional network and the signaling in their niches to maintain a homeostatic balance of having enough but not an overabundance of these cells; therefore their numbers are usually very low [7, 9]. Because of this, it is often very difficult to isolate them in sufficient quantity for cell-based transplantation therapy [9], which would likely require a large amount of cells. However, endogenous lineage-restricted stem and progenitor cells are an ideal source for cell replacement because they are fully functional and show higher engraftment efficiency after transplantation than those generated by stepwise differentiation from ESCs/iPSCs or by lineage conversion from easily obtained somatic cells with transcription factors. The use of small molecules together with cytokines/growth factors to spur the expansion of these lineage-restricted stem cells represents a practical strategy to obtain these cells in sufficient quantities. Therefore, screening small molecules that can expand stem cells in vitro is an important aspect of stem cell research. Recent progress has clearly shown that small molecules can facilitate the expansion of lineage-restricted stem cells in vitro, and these expanded cells can retain their functional characteristics upon transplantation [9].
Although much effort has gone into the directed differentiation of ESCs/iPSCs to desirable cell types, often these target cell types are terminally differentiated somatic cells with limited expansion potential [2]. Another, perhaps more therapeutically relevant strategy is to differentiate ESCs/iPSCs into lineage-restricted stem cells and allow for their capture and expansion using defined culture conditions [2, 10, 11]. The use of small molecules has been demonstrated to be very powerful tool to maintain the self-renewal and allow for the expansion of ESC/iPSC-derived lineage-restricted stem and progenitor cells [10, 11].
In an alternative strategy for differentiating cells from pluripotent cells, the desired cells can also be generated by lineage conversion from other somatic cell types using various factors. For example, the forced expression of MyoD in fibroblasts is able to convert fibroblasts into myocytes [12], and Bcl11b deficiency in pre-T cells causes these cells to become NK-like cells [13]. A similar strategy, often employing more transcription factors, has been used to convert adult somatic cells into lineage-restricted stem and progenitor cells in vitro [14–16]. Significant progress has been made; however, there are still many issues that need resolution. For example, the efficiency of these conversions is usually very low, and virus-based methods permanently introduce ectopic genes into cells. Small molecules have been used to enhance conversion efficiency and in some cases can even replace transcription factors [17], raising the possibility that conversion of adult cells into lineage-restricted stem cells could be accomplished without the introduction of ectopic genes in the future.
Although there are many promising prospects with culturing various lineage-restricted stem and progenitor cells in vitro for cellular replacement therapy, another attractive strategy to stem cell-based medicine is to manipulate the resident endogenous stem cell population in vivo with conventional pharmaceuticals. Because lineage-restricted stem cells have the potential to differentiate into several types of cells, each of which has different functions, selective manipulation of the differentiation of these lineage-restricted stem and progenitor cells in vivo can have desirable therapeutic effects. For example, selective promotion of the differentiation of mesenchymal stem cells (MSCs) into chondrocytes (cartilage cells) can ameliorate osteoarthritis (OA) by helping to repair the damaged cartilage characteristic of this disease [8]. Carefully designed assays have been used to screen for small molecules that can selectively control the differentiation of lineage-restricted stem and progenitor cells into a suitable lineage for desired therapeutic effects. Such small molecules enable the precise control of the differentiation of lineage-restricted stem and progenitor cells in vivo to ameliorate the diseases and have the potential to be developed into traditional pharmaceutical therapeutics [8, 18–20].
Small molecules, which target signaling pathways, transcription factors, nuclear receptors, or epigenetic enzymes, have many advantages compared with genetic methods for controlling the fate and function of stem cells. For example, small molecules provide high temporal control over protein function, including rapid and reversible activation and inhibition; their effect can be adjusted by their concentration; they can target more than one protein simultaneously; and they have the potential to be developed into traditional therapeutics for clinical application [1]. Additionally, the use of small molecules to direct stem cell fate avoids the use of exogenously delivered transgenes, which may have potential to cause cancer or other diseases [1]. In this review, we will focus on how small molecules can be used to facilitate the expansion and generation of desirable lineage-restricted stem and progenitor cells for various purposes and how small molecules can be used to selectively control the differentiation of lineage-restricted stem and progenitor cells in vitro and in vivo.
Expansion of Lineage-Restricted Stem Cells In Vitro With Small Molecules
Lineage-restricted stem cells are usually quite susceptible to the loss of their identity when cultured in vitro [2]. Moreover, these cells exist in vivo in very limited amounts. Therefore, it is a formidable challenge to obtain these cells from donors in sufficient quantities for therapeutic purposes. However, because of their high degree of function, efficient engraftment potential, and the low tumor risk they pose, they are an ideal cell source for cell replacement therapy. Moreover, in some cases, lineage-restricted stem cells provide the only way to treat many devastating diseases [9, 21–23]. For example, HSC transplantation is currently the only curative option for many patients with leukemia or lymphoma malignances. HSCs used for transplantation are obtained from bone marrow, from HSC-mobilized peripheral blood, or from human umbilical cord blood. Human cord blood is becoming increasingly popular because it has many advantages compared with HSC-mobilized peripheral blood and bone marrow cells from a donor, including no risk of graft-versus-host disease if the cells come from the patient's own banked cord blood. However, the amount of HSCs obtained from human cord blood is far less than is needed for transplantation into an adult patient [21, 22]. Expansion of functional HSCs in vitro is urgently needed to make HSC transplantation more accessible. Finding small molecules that can expand functional HSCs in vitro is an important goal for the field. Boitano et al. [9] used CD34 and CD133 as markers to screen small molecules that can promote in vitro expansion of CD34+ human hematopoietic stem/progenitor cells. After screening 100,000 small molecules, they found that StemRegenin 1 (SR1) can promote HSC expansion (Table 1), increasing the number of CD34+ cells 50-fold compared with controls after 21 days in culture. More importantly, the expanded CD34+ cells are enriched in HSCs that have increased ability to functionally engraft in immunodeficient mice. Culture of cord blood-derived CD34+ cells with this compound increased the number of functional HSCs by more than 17-fold compared with uncultured CD34+ cells. Mechanistic studies show that SR1 acts by antagonizing the aryl hydrocarbon receptor [9], a nuclear receptor that has been implicated in pathway-regulating hematopoiesis with the direct role unclear. This compound may prove useful in the clinic to extend the utility of our current methods for obtaining therapeutic HSCs. These studies clearly show that expansion of functional lineage-restricted stem cell in vitro with small molecules is possible, and such small molecules will accelerate stem cell-based clinical applications.
Table 1.
Small molecules used in generation and manipulation of lineage-restricted stem and progenitor cells
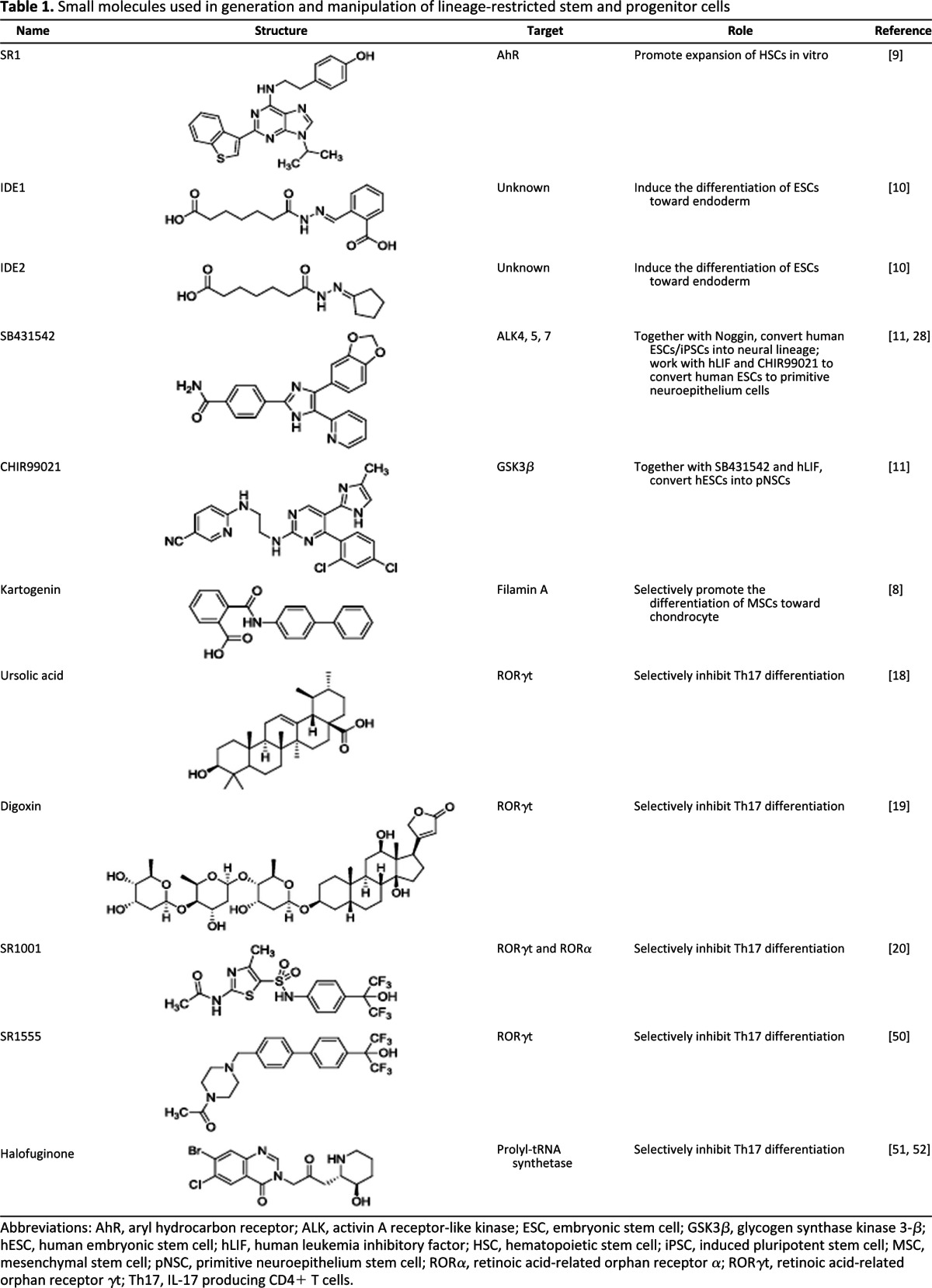
Abbreviations: AhR, aryl hydrocarbon receptor; ALK, activin A receptor-like kinase; ESC, embryonic stem cell; GSK3β, glycogen synthase kinase 3-β; hESC, human embryonic stem cell; hLIF, human leukemia inhibitory factor; HSC, hematopoietic stem cell; iPSC, induced pluripotent stem cell; MSC, mesenchymal stem cell; pNSC, primitive neuroepithelium stem cell; RORα, retinoic acid-related orphan receptor α; RORγt, retinoic acid-related orphan receptor γt; Th17, IL-17 producing CD4+ T cells.
Directed Differentiation/Cell Fate Conversion to Lineage-Restricted Stem and Progenitor Cells
As discussed above, lineage-restricted stem and progenitor cells are invaluable cells that are able to differentiate into the entire repertoire of a certain cell lineage, maintain tissue homeostasis, and mediate tissue/organ repair and regeneration in vivo. An alternative strategy to obtain these cells from tissue donors is to generate stably expandable lineage-restricted stem and progenitor cells in vitro by either directed differentiation from pluripotent stem cells or lineage conversion from easily obtained somatic cell sources, such as fibroblasts, for use in therapeutic transplantation. Here we will highlight some developments and strategies for obtaining desirable cell types, particularly neural stem cells, using directed ESC differentiation or somatic cell transdifferentiation by chemical approaches, which may prove useful for clinical regenerative therapy.
Directed Differentiation of Embryonic Stem Cells to Neural Stem Cells
ESCs possess the ability to differentiate into diverse cell types in vitro and in vivo, including lineage-restricted stem and progenitor cells. However, the paramount challenge for the directed differentiation of ESCs is the efficient and long-term expansion of desirable cell types as a homogenous population. Protocols for stepwise differentiation of ESCs into desirable cell types in vitro have been developed that recapitulate embryonic development in vivo. This principle has also been applied to screen for chemical inducers of differentiation. For example, by using the expression of a fluorescent reporter, dTomato, under control of the Sox17 promoter as an indicator, Borowiak et al. [10] screened a collection of 4,000 compounds and identified two compounds, IDE1 and IDE2, that are able to induce up to 80% of mouse ESCs to definitive endoderm-like cells (Table 1), a higher efficiency than is produced with conventional growth factor treatment. These definitive endoderm-like cells induced by IDE1 and IDE2 express characteristic markers of definitive endoderm, are able to integrate in vivo into the endodermal epithelium of the developing mouse embryo, and can form pancreatic progenitors.
Improved understanding of signaling requirements for development guides the design of directed differentiation methods. An excellent example is that the dual inhibition of SMAD signaling is able to convert human ESCs/iPSC into the neural lineage. Receptor-regulated SMADs act as substrates for transforming growth factor (TGF)-β family of receptors and consequently regulate downstream cellular processes. In brief, SMADs 1, 5, and 8 serve principally as substrates for bone morphogenetic proteins (BMPs), and SMADs 2 and 3 serve principally as substrates for the TGF-β, activin A, and Nodal receptors. Based on the knowledge that BMP inhibitors served as a critical neural inducer in frog [23–26] and that SB431542 (activin A receptor-like kinase [ALK] 4, 5, and 7 inhibitor) was shown to enhance neural induction in an embryoid body-based hESC neural induction protocol [27], Chambers et al. [28] rationalized and found that combined blockade of SMAD signaling using Noggin (BMP inhibitor) and SB431542 was sufficient to achieve highly efficient neural conversion (Table 1). By monitoring the expression of Pax6, an early marker of neuroectoderm differentiation, the dual SMAD inhibition led to the rapid conversion of hESCs to the neural lineage with over 80% efficiency. This dual inhibition of activin A and BMP pathways prevents the differentiation of hESCs toward endoderm and mesoderm, respectively, and enables neutralization of primitive ectoderm. Actually, in this dual inhibition case, SB431542 inhibits the activity of activin A-mediated TGF-β signaling pathway, which is required for hESC self-renewal. One of the downstream targets of activin A is Nanog [29], a key part of the pluripotency network. Therefore, this inhibition abolishes the pluripotency network and primes the hESCs to differentiate. The important implication revealed by this study is that the desirable cell type, such as neuroectoderm here, could be obtained by combining the disruption of pluripotency and blockade of the undesirable differentiation signaling.
Another elegant study illustrates the successful capture of stage-specific neural precursors from hESCs by small molecule inhibitors. The basic fibroblast growth factor (bFGF)-responsive definitive neural stem cells (NSCs) first appear on embryonic day 8.5 (E8.5) in the mouse embryo [30]. These cells can be grown under typically used conditions (bFGF/epidermal growth factor) but during expansion and passaging gradually transit into glial-restricted precursors, which are much less neurogenic [31]. At an earlier stage (E5.5–7.5), Tropepe et al. [32] found that there exist primitive neuroepithelium stem cells (pNSCs) that are generated in the presence of leukemia inhibitory factor (LIF), which is normally used to maintain ESCs in an undifferentiated state. However, these pNSCs could not be maintained in culture. To this end, Li et al. [11] successfully developed a chemical approach that is able to convert hESCs into homogeneous pNSCs with combined treatment of human LIF and two additional small molecule inhibitors, CHIR99021 and SB431542 (Table 1). CHIR99021, a small molecule inhibitor to glycogen synthase kinase 3-β (GSK3β), is able to mediate the activation of canonical Wnt signaling and other downstream pathways [33] to promote stem cell self-renewal [34], and SB431542 inhibits TGF-β pathway, which has been implicated in the mesenchymal-to-epithelial transition and mouse somatic cell reprogramming by other reports [35, 36]. Interestingly, treatment of hESCs with these small molecules cocktail did not safeguard the pluripotent status but instead led to efficient conversion to pNSCs. Importantly, in contrast to the bFGF-responsive NSCs, these pNSCs can be expanded long term in this defined condition without losing their highly neurogenic propensity and responsiveness to morphogenic signals to be patterned into region-specific neuronal subtypes. In addition, the converted pNSCs exhibit in vivo engraftment potential, suggesting potential for clinical application.
This study overcomes crucial obstacles to clinical trials for transplantation of neuronal cells. This chemically defined condition efficiently directs hESCs to a homogenous pNSC population. Because this condition results in a very pure and expandable cell population that can give rise to various neural cell types, it is an ideal method to generate neural cells for therapeutic cell replacement. For example, these cells could be used to treat diseases and injuries in which neuronal cells are lost such as neurodegenerative diseases, for example, Alzheimer's and Parkinson's diseases, as well as head injuries and strokes.
Lineage Conversion of Somatic Cells Into Neural Stem Cells
The recent breakthrough of iPSC technology established the paradigm of ectopically overexpressing multiple target cell-specific transcription factors (TFs) in starting somatic cells to achieve their conversion to the target cell type. Using this strategy, a diversity of cell types have been generated from fibroblasts, such as neurons [37–39], hepatocytes [40, 41], and cardiomyocytes [42–44], by ectopic expression of lineage-specific TFs. One of the excellent representatives is that by using a combination of three developmentally relevant transcription factors, Gata4, Mef2c, and Tbx5, Ieda et al. [42] and Qian et al. [44] have succeeded in rapid and efficient reprogramming of postnatal cardiac or dermal fibroblast into functional cardiomyocyte-like cells in vitro and in vivo, respectively. Several recent studies advanced this concept by converting the somatic cells into tissue-specific stem/progenitor cells, such as hematopoietic progenitors [45] and neural progenitor cells [14–16, 46], thus overcoming the challenge of cell expansion. However, the efficiency of the TF-mediated cell fate conversion is still low. To address this, researchers have turned to small molecules in attempts to increase the efficiency.
TF-directed conversion of fibroblasts to neuron-like cells has been widely reported both in human and mouse cells. Ladewig et al. [17] recently reported a method that combines overexpression of two factors (Ascl1 and Ngn2) with small molecule inhibitors of GSK3β and TGF-β receptor. This condition was shown to efficiently convert human fibroblasts into functional neuron-like cells with high yields and neuronal purities. Without the small molecule treatment, neuron-like cells comprised <5% of the total population 23 days after transgene induction; however, when treated with small molecule cocktails, a 4- to 17-fold increase was detected, highlighting the functional value of small molecules in this process. This illustrates the importance of small molecules in advancing the utility of converting cells from other easily obtained somatic cells.
Small Molecules Can Selectively Regulate the Differentiation of Lineage-Restricted Stem Cells In Vivo
Lineage-restricted stem/progenitor cells have the ability to differentiate into several cell types, each having different, sometimes opposing, functions. For example, MSCs, which reside in bone marrow and many adult tissues and are widely used as a cell source in cell therapy to treat autoimmune and many other diseases, have self-renewal ability and can differentiate into osteoblasts (bone cells), chondrocytes (cartilage cells), and adipocytes (fat cells) [47]. Another example is the CD4 T-cell lineage: CD4 naive T cells can differentiate, under different cytokine conditions, into Th1, Th2, Th17, and regulatory T cells, each of which has different functions [48]. Th1 cells can promote the proliferation of cytotoxic CD8+ T cells and secrete interferon γ to induce the differentiation of type 1 macrophage and clear intracellular pathogens. Th2 cells can stimulate B cells to proliferate, to switch antibody class, to increase neutralizing antibody production, and to clear extracellular pathogen. Th17 cells provide antimicrobial immunity at epithelial/mucosal barriers and fight against fungal infection. Th17 cells have also been shown to play a critical role in autoimmune disease such as multiple sclerosis, psoriasis, rheumatoid arthritis, and Crohn's disease. Regulatory T cells can modulate immune response, maintain tolerance to self-antigens, and protect from autoimmune diseases [48, 49]. Since each lineage (or cell type) has unique functions, selective inhibition or promotion of (a) particular lineage(s) by small molecules while minimizing the effects on other lineages could be very useful for conventional pharmaceutical therapies.
The promotion of the differentiation of one lineage from a progenitor could help to treat some intractable diseases. In OA, a degenerative joint disease affecting more than 70% of Americans between 55 and 70 years old, chondrocytes are progressively lost, and cartilage degenerates. Therefore, it may represent a fertile approach to find small molecules that can be used to repair and regenerate cartilage. Johnson et al. [8] found that kartogenin can selectively promote differentiation of human MSC into chondrocytes by screening 22,000 small molecules using image-based high throughput screening (HTS) assay. The authors showed that kartogenin was effective in the collagenase VII-induced chronic joint injury model and the acute surgical model of OA (Table 1). Mechanistic studies revealed that kartogenin can bind filamin A, disrupting its interaction with the transcription factor core-binding factor β subunit (CBFβ). This allows CBFβ to translocate to the nucleus where it can bind Runx1 to promote chondrocyte differentiation [8]. Further development of such molecules may lead to novel treatment for OA patients.
On the other hand, inhibition of differentiation of a cell type from a progenitor, for example Th17 from naïve CD4 T cells, could also be therapeutically useful. Xu et al. [18] screened more than 2,000 known bioactive compounds in human naïve CD4 T cells using high throughput fluorescence-activated cell sorting to identify small molecules that can selectively inhibit Th17 cells differentiation in vitro. Interestingly, ursolic acid (a natural product rich in many kinds of fruits and a Food and Drug Administration-approved drug to treat cancer) was identified as a specific inhibitor of Th17 cell differentiation (Table 1). Mechanistic studies showed that ursolic acid can selectively bind to the ligand binding domain of retinoic acid-related orphan receptor γt (RORγt), the master transcription factor for Th17 cells, inhibiting its transcriptional activity, while not affecting retinoic acid-related orphan receptor α (RORα), another ROR family nuclear receptor important for Th17 differentiation. In a mouse experimental autoimmune encephalomyelitis (EAE) model (a mice model for human multiple sclerosis), it was shown that ursolic acid can effectively ameliorate this disease [18]. Ursolic acid has been used clinically for cancer treatment with very few side effects; therefore it may have a potential to be developed into a therapeutic for autoimmune diseases involving Th17 cells. Similarly, Huh et al. [19] have independently identified digoxin as an inhibitor of Th17 cell differentiation by antagonizing RORγt (Table 1). However, since digoxin has many other targets and is highly toxic to human cells, a modified, less toxic, digoxin analog was developed and remains to be further tested in animal models. Furthermore, Solt et al. [20] have identified SR1001, a synthetic compound, as a specific inhibitor for Th17 cell differentiation by antagonizing RORγt and RORα (Table 1). Considering that RORα plays a critical role in circadian rhythm and metabolism, there are safety concerns for the use of SR1001. Recently, a derivative of SR1001 has been developed, but much effort is needed to evaluate the effect of this modified compound in the EAE model [50]. Moreover, Sundrud and colleagues [51, 52] found that halofuginone selectively inhibits mouse and human Th17 differentiation by activating the amino acid starvation response through inhibiting prolyl-tRNA synthetase activity (Table 1).
It must be noted that choosing suitable markers or signature genes is very important for cell-based chemical screening. In rare cases, a single marker may work reasonably well for a primary HTS, but in many cases, a single marker is insufficient for this purpose and leads to biased and inappropriate hit selection. Therefore, it is highly desirable to use a cohort of gene expression to define a particular cellular phenotype. Although technologies, such as Luminex and Fluidigm, can partially provide the solution, it is still a significant challenge to perform a medium- to large-scale chemical screen using multiplexed methods. As DNA sequencing becomes more affordable, RASL-seq technology (RNA Annealing, Selection, and Ligation, followed by next generation sequencing) provides a cost-effective and true high-throughput, multiplexed platform for cell-based chemical screening. Briefly, two short probes per target mRNA (DNA oligonucleotides) that can match two adjacent exons are incubated together with cell lysate. After annealing, selection, and ligation, purified ligated DNA oligonucleotides are amplified and bar-coded by polymerase chain reaction before pooling for parallel sequencing [53]. The quantity of ligated oligonucleotides can be determined by read frequencies during sequencing and can reflect the relative expression of target mRNAs. This new technology will greatly accelerate identification of new chemicals that can regulate self-renewal, differentiation, and reprogramming of lineage-restricted cells in vitro and in vivo.
All of the examples listed above suggest that carefully designed screening assays can be used to screen small molecules that selectively modulate the differentiation of tissue-specific progenitor cells, and these small molecules can function in vivo to ameliorate disease phenotypes involving the progenitor and its differentiated cell types. These kinds of small molecules could be further developed into pharmaceuticals to control cell fate in vivo with therapeutic effects.
Conclusion
Substantial advances toward regenerative medicine have been made in the past several years, especially since the discovery of iPSCs. With the growing understanding of the mechanisms of cell fate commitment, including programming and reprogramming, effective approaches that exert precise control of cell fate, behavior, and cellular function have emerged as the prerequisite for successful clinical therapy. ESCs/iPSCs, although possessing the potential to develop into all cell types in vitro and in vivo, exhibit significant risk of tumorigenesis. The two alternative strategies are acquisition of expandable lineage-restricted stem and progenitor cells in vitro for transplantation and control of the cell fate switch, especially the lineage-restricted stem and progenitor cells, for existing cell populations in vivo.
Although much progress has been made toward these goals, regenerative biology and medicine is still in its early stage. Efficacious cell replacement requires the generation of correct cell type, which is tightly controlled by their intrinsic regulators and external cues. Chemical approaches are highly attractive for such manipulation of cell fate. However, more potent and functionally specific small molecules are required for precise control of certain biological processes, such as signaling pathways or enzymatic activity. Additionally, optimization of the specific timing, dose, and combination of small molecules is essential to bring about the desired outcome. Notably, delivery of the small molecules to the desirable target tissue in vivo is still challenging. Therefore, continuous identification and characterization of small molecules, as well as comprehensive understanding of the detailed mechanisms, particularly context-dependent behavior, would no doubt be of great interest for both chemists and biologists. The development of this promising area would definitely facilitate the mechanistic study of stem cell biology, drug discovery, and, more importantly, the development of clinical therapy of human diseases.
Acknowledgments
S.D. is supported by funding from the National Institute of Child Health and Human Development, National Heart, Lung, and Blood Institute, National Eye Institute, and National Institute of Mental Health (NIH), California Institute for Regenerative Medicine, Prostate Cancer Foundation, and the Gladstone Institute. The authors apologize to all of the scientists whose research could not be properly discussed and cited in this review because of space limitations.
Author Contributions
T.X. and M.Z.: conception and design, manuscript writing; T.L. and M.X.: manuscript writing; S.D.: conception and design, manuscript writing, provision of study material, financial support, final approval of manuscript.
Disclosure of Potential Conflicts of Interest
The authors declare no potential conflicts of interest.
References
- 1.Xu Y, Shi Y, Ding S. A chemical approach to stem-cell biology and regenerative medicine. Nature. 2008;453:338–344. doi: 10.1038/nature07042. [DOI] [PubMed] [Google Scholar]
- 2.Kim J, Ambasudhan R, Ding S. Direct lineage reprogramming to neural cells. Curr Opin Neurobiol. 2012;22:1–7. doi: 10.1016/j.conb.2012.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zambidis ET, Peault B, Park TS, et al. Hematopoietic differentiation of human embryonic stem cells progresses through sequential hematoendothelial, primitive, and definitive stages resembling human yolk sac development. Blood. 2005;106:860–870. doi: 10.1182/blood-2004-11-4522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kriks S, Shim JW, Piao J, et al. Dopamine neurons derived from human ES cells efficiently engraft in animal models of Parkinson's disease. Nature. 2011;480:547–551. doi: 10.1038/nature10648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lian X, Hsiao C, Wilson G, et al. Robust cardiomyocyte differentiation from human pluripotent stem cells via temporal modulation of canonical Wnt signaling. Proc Natl Acad Sci USA. 2012;109:E1848–E1857. doi: 10.1073/pnas.1200250109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.D'Amour KA, Bang AG, Eliazer S, et al. Production of pancreatic hormone-expressing endocrine cells from human embryonic stem cells. Nat Biotechnol. 2006;24:1392–1401. doi: 10.1038/nbt1259. [DOI] [PubMed] [Google Scholar]
- 7.Orkin SH, Zon LI. Hematopoiesis: An evolving paradigm for stem cell biology. Cell. 2008;132:631–644. doi: 10.1016/j.cell.2008.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Johnson K, Zhu S, Tremblay MS, et al. A stem cell-based approach to cartilage repair. Science. 2012;336:717–721. doi: 10.1126/science.1215157. [DOI] [PubMed] [Google Scholar]
- 9.Boitano AE, Wang J, Romeo R, et al. Aryl hydrocarbon receptor antagonists promote the expansion of human hematopoietic stem cells. Science. 2010;329:1345–1348. doi: 10.1126/science.1191536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Borowiak M, Maehr R, Chen S, et al. Small molecules efficiently direct endodermal differentiation of mouse and human embryonic stem cells. Cell Stem Cell. 2009;4:348–358. doi: 10.1016/j.stem.2009.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li W, Sun W, Zhang Y, et al. Rapid induction and long-term self-renewal of primitive neural precursors from human embryonic stem cells by small molecule inhibitors. Proc Natl Acad Sci USA. 2011;108:8299–8304. doi: 10.1073/pnas.1014041108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Graf T, Enver T. Forcing cells to change lineages. Nature. 2009;462:587–594. doi: 10.1038/nature08533. [DOI] [PubMed] [Google Scholar]
- 13.Li P, Burke S, Wang J, et al. Reprogramming of T cells to natural killer-like cells upon Bcl11b deletion. Science. 2010;329:85–89. doi: 10.1126/science.1188063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ring K, Tong L, Balestra M, et al. Direct reprogramming of mouse and human fibroblasts into multipotent neural stem cells with a single factor. Cell Stem Cell. 2012;11:100–109. doi: 10.1016/j.stem.2012.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Han D, Tapia N, Hermann A, et al. Direct reprogramming of fibroblasts into neural stem cells by defined factors. Cell Stem Cell. 2012;10:465–472. doi: 10.1016/j.stem.2012.02.021. [DOI] [PubMed] [Google Scholar]
- 16.Thier M, Worsdorfer P, Lakes YB, et al. Direct conversion of fibroblasts into stably expandable neural stem cells. Cell Stem Cell. 2012;10:473–479. doi: 10.1016/j.stem.2012.03.003. [DOI] [PubMed] [Google Scholar]
- 17.Ladewig J, Mertens J, Kesavan J, et al. Small molecules enable highly efficient neuronal conversion of human fibroblasts. Nat Methods. 2012;9:575–578. doi: 10.1038/nmeth.1972. [DOI] [PubMed] [Google Scholar]
- 18.Xu T, Wang X, Zhong B, et al. Ursolic acid suppresses interleukin-17 (IL-17) production by selectively antagonizing the function of RORgamma t protein. J Biol Chem. 2011;286:22707–22710. doi: 10.1074/jbc.C111.250407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huh JR, Leung MW, Huang P, et al. Digoxin and its derivatives suppress TH17 cell differentiation by antagonizing RORgammat activity. Nature. 2011;472:486–490. doi: 10.1038/nature09978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Solt LA, Kumar N, Nuhant P, et al. Suppression of TH17 differentiation and autoimmunity by a synthetic ROR ligand. Nature. 2011;472:491–494. doi: 10.1038/nature10075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hofmeister CC, Zhang J, Knight KL, et al. Ex vivo expansion of umbilical cord blood stem cells for transplantation: Growing knowledge from the hematopoietic niche. Bone Marrow Transplant. 2007;39:11–23. doi: 10.1038/sj.bmt.1705538. [DOI] [PubMed] [Google Scholar]
- 22.Goessling W, Allen RS, Guan X, et al. Prostaglandin E2 enhances human cord blood stem cell xenotransplants and shows long-term safety in preclinical nonhuman primate transplant models. Cell Stem Cell. 2011;8:445–458. doi: 10.1016/j.stem.2011.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sasai Y, Lu B, Steinbeisser H, et al. Xenopus chordin: A novel dorsalizing factor activated by organizer-specific homeobox genes. Cell. 1994;79:779–790. doi: 10.1016/0092-8674(94)90068-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hemmati-Brivanlou A, Kelly OG, Melton DA. Follistatin, an antagonist of activin, is expressed in the Spemann organizer and displays direct neuralizing activity. Cell. 1994;77:283–295. doi: 10.1016/0092-8674(94)90320-4. [DOI] [PubMed] [Google Scholar]
- 25.Smith WC, Harland RM. Expression cloning of noggin, a new dorsalizing factor localized to the Spemann organizer in Xenopus embryos. Cell. 1992;70:829–840. doi: 10.1016/0092-8674(92)90316-5. [DOI] [PubMed] [Google Scholar]
- 26.Valenzuela DM, Economides AN, Rojas E, et al. Identification of mammalian noggin and its expression in the adult nervous system. J Neurosci. 1995;15:6077–6084. doi: 10.1523/JNEUROSCI.15-09-06077.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Smith JR, Vallier L, Lupo G, et al. Inhibition of activin/nodal signaling promotes specification of human embryonic stem cells into neuroectoderm. Dev Biol. 2008;313:107–117. doi: 10.1016/j.ydbio.2007.10.003. [DOI] [PubMed] [Google Scholar]
- 28.Chambers SM, Fasano CA, Papapetrou EP, et al. Highly efficient neural conversion of human ES and iPS cells by dual inhibition of SMAD signaling. Nat Biotechnol. 2009;27:275–280. doi: 10.1038/nbt.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xu RH, Sampsell-Barron TL, Gu F, et al. NANOG is a direct target of TGFbeta/activin-mediated SMAD signaling in human ESCs. Cell Stem Cell. 2008;3:196–206. doi: 10.1016/j.stem.2008.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hitoshi S, Seaberg RM, Koscik C, et al. Primitive neural stem cells from the mammalian epiblast differentiate to definitive neural stem cells under the control of Notch signaling. Genes Dev. 2004;18:1806–1811. doi: 10.1101/gad.1208404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Qian X, Shen Q, Goderie SK, et al. Timing of CNS cell generation: A programmed sequence of neuron and glial cell production from isolated murine cortical stem cells. Neuron. 2000;28:69–80. doi: 10.1016/s0896-6273(00)00086-6. [DOI] [PubMed] [Google Scholar]
- 32.Tropepe V, Hitoshi S, Sirard C, et al. Direct neural fate specification from embryonic stem cells: A primitive mammalian neural stem cell stage acquired through a default mechanism. Neuron. 2001;30:65–78. doi: 10.1016/s0896-6273(01)00263-x. [DOI] [PubMed] [Google Scholar]
- 33.Ring DB, Johnson KW, Henriksen EJ, et al. Selective glycogen synthase kinase 3 inhibitors potentiate insulin activation of glucose transport and utilization in vitro and in vivo. Diabetes. 2003;52:588–595. doi: 10.2337/diabetes.52.3.588. [DOI] [PubMed] [Google Scholar]
- 34.Sato N, Meijer L, Skaltsounis L, et al. Maintenance of pluripotency in human and mouse embryonic stem cells through activation of Wnt signaling by a pharmacological GSK-3-specific inhibitor. Nat Med. 2004;10:55–63. doi: 10.1038/nm979. [DOI] [PubMed] [Google Scholar]
- 35.Li W, Wei W, Zhu S, et al. Generation of rat and human induced pluripotent stem cells by combining genetic reprogramming and chemical inhibitors. Cell Stem Cell. 2009;4:16–19. doi: 10.1016/j.stem.2008.11.014. [DOI] [PubMed] [Google Scholar]
- 36.Tojo M, Hamashima Y, Hanyu A, et al. The ALK-5 inhibitor A-83–01 inhibits Smad signaling and epithelial-to-mesenchymal transition by transforming growth factor-beta. Cancer Sci. 2005;96:791–800. doi: 10.1111/j.1349-7006.2005.00103.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vierbuchen T, Ostermeier A, Pang ZP, et al. Direct conversion of fibroblasts to functional neurons by defined factors. Nature. 2010;463:1035–1041. doi: 10.1038/nature08797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pang ZP, Yang N, Vierbuchen T, et al. Induction of human neuronal cells by defined transcription factors. Nature. 2011;476:220–223. doi: 10.1038/nature10202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kim J, Efe JA, Zhu S, et al. Direct reprogramming of mouse fibroblasts to neural progenitors. Proc Natl Acad Sci USA. 2011;108:7838–7843. doi: 10.1073/pnas.1103113108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Huang P, He Z, Ji S, et al. Induction of functional hepatocyte-like cells from mouse fibroblasts by defined factors. Nature. 2011;475:386–389. doi: 10.1038/nature10116. [DOI] [PubMed] [Google Scholar]
- 41.Sekiya S, Suzuki A. Direct conversion of mouse fibroblasts to hepatocyte-like cells by defined factors. Nature. 2011;475:390–393. doi: 10.1038/nature10263. [DOI] [PubMed] [Google Scholar]
- 42.Ieda M, Fu JD, Delgado-Olguin P, et al. Direct reprogramming of fibroblasts into functional cardiomyocytes by defined factors. Cell. 2010;142:375–386. doi: 10.1016/j.cell.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Efe JA, Hilcove S, Kim J, et al. Conversion of mouse fibroblasts into cardiomyocytes using a direct reprogramming strategy. Nat Cell Biol. 2011;13:215–222. doi: 10.1038/ncb2164. [DOI] [PubMed] [Google Scholar]
- 44.Qian L, Huang Y, Spencer CI, et al. In vivo reprogramming of murine cardiac fibroblasts into induced cardiomyocytes. Nature. 2012;485:593–598. doi: 10.1038/nature11044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Szabo E, Rampalli S, Risueno RM, et al. Direct conversion of human fibroblasts to multilineage blood progenitors. Nature. 2010;468:521–526. doi: 10.1038/nature09591. [DOI] [PubMed] [Google Scholar]
- 46.Lujan E, Chanda S, Ahlenius H, et al. Direct conversion of mouse fibroblasts to self-renewing, tripotent neural precursor cells. Proc Natl Acad Sci USA. 2012;109:2527–2532. doi: 10.1073/pnas.1121003109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Le Blanc K, Mougiakakos D. Multipotent mesenchymal stromal cells and the innate immune system. Nat Rev Immunol. 2012;12:383–396. doi: 10.1038/nri3209. [DOI] [PubMed] [Google Scholar]
- 48.Steinman L. A brief history of T(H)17, the first major revision in the T(H)1/T(H)2 hypothesis of T cell-mediated tissue damage. Nat Med. 2007;13:139–145. doi: 10.1038/nm1551. [DOI] [PubMed] [Google Scholar]
- 49.Littman DR, Rudensky AY. Th17 and regulatory T cells in mediating and restraining inflammation. Cell. 2010;140:845–858. doi: 10.1016/j.cell.2010.02.021. [DOI] [PubMed] [Google Scholar]
- 50.Solt LA, Kumar N, He Y, et al. Identification of a selective RORγ ligand that suppresses T(H)17 cells and stimulates T regulatory cells. ACS Chem Biol. 2012;7:1515–1519. doi: 10.1021/cb3002649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sundrud MS, Koralov SB, Feuerer M, et al. Halofuginone inhibits TH17 cell differentiation by activating the amino acid starvation response. Science. 2009;324:1334–1338. doi: 10.1126/science.1172638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Keller TL, Zocco D, Sundrud MS, et al. Halofuginone and other febrifugine derivatives inhibit prolyl-tRNA synthetase. Nat Chem Biol. 2012;8:311–317. doi: 10.1038/nchembio.790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li H, Zhou H, Wang D, et al. Versatile pathway-centric approach based on high-throughput sequencing to anticancer drug discovery. Proc Natl Acad Sci USA. 2012;109:4609–4614. doi: 10.1073/pnas.1200305109. [DOI] [PMC free article] [PubMed] [Google Scholar]


