Abstract
Viral emergence as a result of zoonotic transmission constitutes a continuous public health threat. Emerging viruses such as SARS coronavirus, hantaviruses and henipaviruses have wildlife reservoirs. Characterising the viruses of candidate reservoir species in geographical hot spots for viral emergence is a sensible approach to develop tools to predict, prevent, or contain emergence events. Here, we explore the viruses of Eidolon helvum, an Old World fruit bat species widely distributed in Africa that lives in close proximity to humans. We identified a great abundance and diversity of novel herpes and papillomaviruses, described the isolation of a novel adenovirus, and detected, for the first time, sequences of a chiropteran poxvirus closely related with Molluscum contagiosum. In sum, E. helvum display a wide variety of mammalian viruses, some of them genetically similar to known human pathogens, highlighting the possibility of zoonotic transmission.
Keywords: Virome, Bat, Megabat, Poxvirus, Viral emergence, Metagenomics, Adenovirus
Highlights
-
•
The first metagenomic study of a chiropteran (bat) suborder.
-
•
Demonstrates a novel and thorough bioinformatics pipeline for metagenomic studies.
-
•
Multiple novel, diverse viruses present in an urban African bat bushmeat species.
-
•
The study is supported with further molecular evidence and virus isolation.
-
•
The study contains the first evidence of chiropteran poxviruses and a novel bat adenovirus isolate.
Introduction
Zoonoses caused by unknown agents represent a significant proportion of the challenge of emerging infectious diseases (EIDs) (Morens et al., 2004). Viruses account for approximately 25–44% of all EIDs (Jones et al., 2008, Taylor et al., 2001) and studies suggest they are the pathogen class most likely to emerge (Cleaveland et al., 2007, Dobson and Foufopoulos, 2001). Hantaviruses, henipaviruses, SARS coronaviruses and filoviruses are all viruses of zoonotic origin. Nearly 80% of zoonotic EIDs originate from wildlife, and the overall contribution of wildlife pathogens to human EID events is increasing and represent an ongoing threat to global health (Cleaveland et al., 2007, Jones et al., 2008). For example, a novel coronavirus associated with acute respiratory disease was recently diagnosed in pneumonia patients in Saudi Arabia and London (Bermingham et al., 2012, Zaki et al., 2012). Analysis of the novel coronavirus genome suggests a possible bat origin (Bermingham et al., 2012).
South and East Asia, Eastern Europe, Latin America and tropical Africa constitute areas of increased relative risk for zoonotic emergence from wildlife (Jones et al., 2008, Morens et al., 2004). Numerous studies have successfully combined metagenomics with next generation sequencing to explore the viruses of different animal species, including: domestic pigs and turkeys (Day et al., 2010, Shan et al., 2011); Californian sea lions (Li et al., 2011); and rodents (Phan et al., 2011). Characterising the viruses of candidate reservoir species in high-risk geographical areas is an important step toward better understanding viral emergence.
Bats are the primary reservoirs for many viral zoonoses, including henipaviruses, filoviruses, some lyssaviruses and SARS-like coronaviruses (Halpin et al., 2000, Kuzmin et al., 2008, Li et al., 2005, Luby et al., 2009, Towner et al., 2007). Indeed, seminal work has been recently published on the role of bats as natural reservoirs of paramyxoviruses (Drexler et al., 2012). Detailed studies of the viruses of insectivorous bat species in both North America and China have been conducted (Donaldson et al., 2010, Ge et al., 2012, Li et al., 2010a, Wu et al., 2012). These studies found large numbers of insect and plant viruses, which were thought to reflect dietary inputs, as well phage sequences and mammalian viruses. The majority of the mammalian viruses identified in those studies were those previously identified in bats (often with high diversity being reported in individual populations) and include: Adenoviridae (Li et al., 2010c); Parvoviridae (Li et al., 2010b); Circoviridae (Ge et al., 2011); Coronaviridae (Tang et al., 2006, Woo et al., 2006) and Astroviridae (Chu et al., 2008). Papillomaviridae and Herpesvirdae sequences were also commonly found (Donaldson et al., 2010, Ge et al., 2012, Wu et al., 2012) and some studies also reported Picornaviridae, Flaviviridae and Retroviridae (Li et al., 2010a, Wu et al., 2012).
If we consider that the ∼1200 bat species constitute approximately 20% of the class Mammalia and that they are near-globally distributed, the benefits of expanding our knowledge of bat viruses on geographic and taxonomic levels become evident. Here we conducted a metagenomic study to detect viruses of E. helvum, a frugivorous African bat species that is widely-distributed and migratory throughout much of sub-Saharan Africa. The species is eaten as bushmeat, and the populations studied have ample opportunities for human contact, including a roost directly over a hospital in Accra, Ghana (Hayman et al., 2012).
Results
Bioinformatics analysis
Performance comparison among assemblers
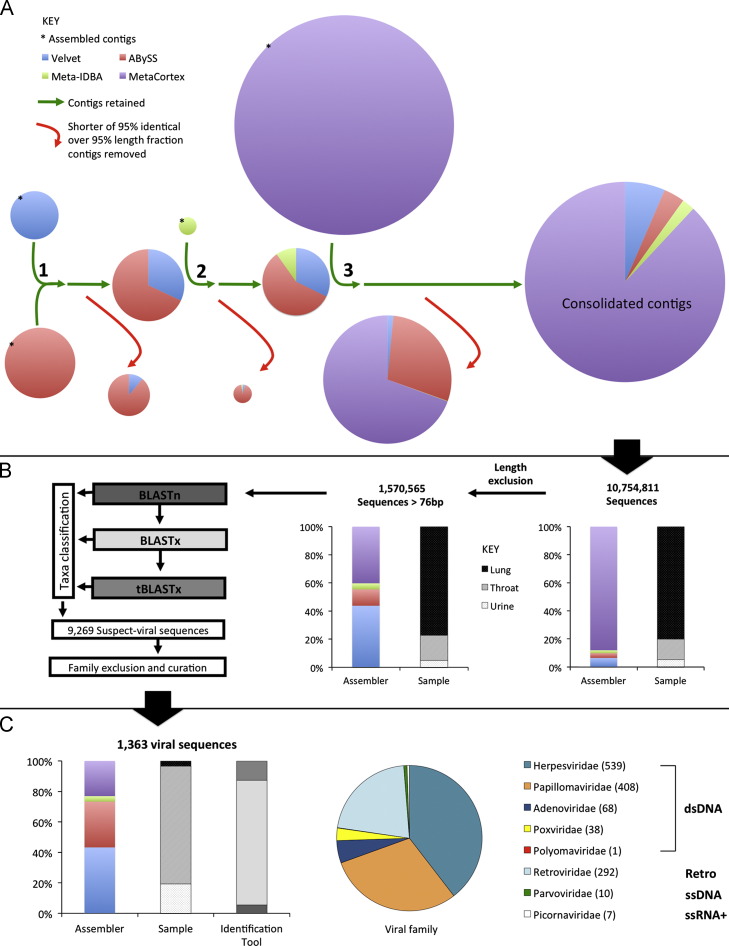
Here, we show important performance differences among four different assemblers (Velvet, ABySS, MetaIDBA and MetaCortex) and three sample types.
The assemblers generated different numbers of contigs, with MetaCortex and ABySS producing more sequences than Velvet and MetaIDBA for each sample type ( Fig. 1A). Sample type also affected the number of contigs, with increasing cellularity resulting in more contigs ( Table 1), except with MetaIDBA, where the sample allowed to iterate over a kmer size-range generated the most contigs. Contig length parameters also varied with assembler and sample type. Velvet generated contigs with the longest average length across sample types, while ABySS typically produced the longest contigs. Regarding sample type, the longest contigs were generated from the throat sample for each assembler (Table 1).
Fig. 1.
Bioinformatic analysis pipeline for 15,155,461 assembled contigs. (A) Contigs assembled from sequencing reads by different de novo assemblers (starred, denoted by colours) were consolidated by sequential comparisons (numbered, curved green arrows) and removal (red arrows) of duplicate sequences. The size of charts is proportional to the number of contigs. (B) Consolidated sequences subject to sequential BLAST comparison with automated taxonomic classification to identify suspect-viral sequences. Then sequences were excluded manually on the basis of related viral family and curation to identify a final set of viral sequences. Proportions of sequences assembled by each algorithm (coloured as in A) and from each sample type (coloured as in the inset key) before and after length-exclusion are shown in the stacked charts. (C) Proportions of 1,363 mammalian-virus related contigs by assembly algorithm, sample-type and identification algorithm (shaded as in B) are shown in the stacked chart. Proportions by viral family are shown in the chart on the right. Number of contigs related to members of each family are shown in parentheses after the viral family names, which are grouped by genome type. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Table 1.
Number and length distributions of contiguous sequences by assembler. Metrics for total assembled sequences are shown for each sample type.
| Sample type | Contiguous sequences | Assembler |
|||
|---|---|---|---|---|---|
| ABySS | Velvet | MetaIDBA | MetaCortex | ||
| Urine | Number | 157,518 | 28,886 | 4,597 | 819,081 |
| Average length (bp) | 91 | 222 | 94 | 79 | |
| Largest (bp) | 6,166 | 4,127 | 2,744 | 5,704 | |
| N50 (bp) | 119 | 244 | 78 | 76 | |
| Throat | Number | 621,363 | 113,317 | 217,483 | 1,917,401 |
| Average length (bp) | 67 | 160 | 100 | 79 | |
| Largest (bp) | 14,875 | 5,392 | 14,173 | 15,292 | |
| N50 (bp) | 75 | 175 | 78 | 76 | |
| Lung | Number | 1,496,223 | 706,515 | 14,007 | 9,358,870 |
| Average length (bp) | 69 | 118 | 86 | 77 | |
| Largest (bp) | 3,955 | 783 | 2,745 | 1,980 | |
| N50 (bp) | 76 | 116 | 73 | 76 | |
As well as generating contigs of differing length and number, the nucleotide composition of contigs varied among assemblers. Base composition of the total assembled contigs revealed important differences (Supp. Fig. 2A). Velvet and MetaCortex contigs had similar base compositions, while ABySS contigs incorporated non-ATCG notations (e.g. N, R, Y, Supp. Fig. 2A), and MetaIDBA contigs were primarily composed of adenine (though this was not true for MetaIDBA contigs included in final analyses (Supp. Fig. 2B)).
Consolidation of contigs combines the strengths of assembler approaches and reduces complexity
De novo assemblers have different contig-construction methods, resulting in strengths and weaknesses in different situations. By consolidating contigs from multiple assemblers, we combined the strengths, while reducing the computational complexity of analyzing contigs from assemblers separately. The proportion of contigs retained after consolidation differed among assemblers. For example, ≤22% of ABySS contigs but ≥94% of MetaIDBA contigs were retained into the final consolidated set for each sample type ( Table 2). The consolidation resulted in the discard of approximately 30% of the total assembled contigs (∼4.4 million sequences). Consistent with the observed assembler differences in contig generation, a variable proportion of sequences were also length-excluded per assembler, but these proportions were approximately equal among sample types (Fig. 1B, Table 2).
Table 2.
Number of contiguous sequences (by assembler and sample type) assembled, removed and retained during contig consolidation. The final four rows represent those sequences in the consolidated contiguous set, and the percentage of these >76 bp in length that underwent BLAST identification is shown in parentheses in the table.
| Number of contiguous sequences |
Assembler | Sample |
|||
|---|---|---|---|---|---|
| Process |
Outcome |
Urine |
Throat |
Lung |
|
| Assembly | Total generated (% in consolidated) | Velvet | 28,886 (60) | 113,317 (60) | 706,515 (87) |
| ABySS | 157,518 (22) | 621,363 (14) | 1,496,223 (16) | ||
| MetaIDBA | 4,597 (97) | 217,483 (94) | 14,007 (100) | ||
| MetaCortex | 819,081 (63) | 1,917,401 (63) | 9,358,870 (83) | ||
| Comparison 1 Velvet vs. ABySS | Removed | Velvet | 9,268 | 23,593 | 51,218 |
| ABySS | 46,253 | 165,256 | 439,302 | ||
| Retained | Velvet | 19,618 | 89,724 | 655,297 | |
| ABySS | 111,265 | 456,107 | 1,056,921 | ||
| Total retained | 130,883 | 545,831 | 1,712,218 | ||
| Comparison 2: Velvet/ABySS vs. MetaIDBA | Removed | Velvet | 550 | 13,315 | 877 |
| ABySS | 17,516 | 233,758 | 26,886 | ||
| MetaIDBA | 45 | 7,463 | 0 | ||
| Retained | Velvet | 19,068 | 76,409 | 654,420 | |
| ABySS | 93,749 | 222,349 | 1,030,035 | ||
| MetaIDBA | 4,552 | 210,020 | 14,007 | ||
| Total retained | 117,369 | 508,418 | 1,698,462 | ||
| Comparison 3: Velvet/ABySS/MetaIDBA vs. MetaCortex | Removed | Velvet | 1,776 | 8,648 | 41,201 |
| ABySS | 58,594 | 137,011 | 786,165 | ||
| MetaIDBA | 85 | 5,871 | 49 | ||
| MetaCortex | 303,863 | 701,171 | 1,620,716 | ||
| Retained (%>76 bp) | Velvet | 17,292 (100) | 67,761 (99) | 613,219 (98) | |
| Consolidated | ABySS | 35,155 (63) | 85,338 (42) | 243,870 (51) | |
| contigs | MetaIDBA | 4,467 (33) | 204,149 (30) | 13,958 (36) | |
| MetaCortex | 515,218 (7) | 1,216,230 (9) | 7,738,154 (6) | ||
| Total retained | 572,132 | 1,573,478 | 8,609,201 | ||
Eidolon helvum samples contain large numbers of viral sequences
To identify viral sequences in the consolidated contigs, we used multiple algorithms, which had different efficacies. While BLASTn identified 258 suspect-viral sequences among the sample types, BLASTx and tBLASTx identified a further 6448 and 2563 viral sequences, respectively. Manual exclusion and curation of these 9269 suspect-viral sequences was used to further focus analysis on viral sequences of interest (Fig. 1B). Here, we aimed to explore the viruses that likely infect E. helvum (for which they probably constitute a natural reservoir) not the viruses infecting their dietary inputs or bacterial flora. Consequently, 8095 suspect-viral sequences related to viral families not known to infect vertebrates were excluded from further analysis. Subsequent to close inspection of the remaining 1174 suspect-viral sequences, a further 11 sequences were removed due to incorrect classification in the database (not shown). This resulted in 1363 viral sequences related to eight mammalian-infecting viral families being identified. While the majority (77%) were related to viruses with double stranded DNA (dsDNA) genomes, 21% were related to retroviruses (classified separately as sequences may have derived from exogenous-RNA or proviral-DNA forms) and single-stranded DNA and positive-sense single-stranded RNA viruses were also present ( Table 3, Fig. 1C).
Table 3.
Identification of viral sequences (divided by viral family) according to sample type, assembler and identification algorithm.
| Sequence origin |
Related viral family |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Herpes | Papilloma | Adeno | Pox | Polyoma | Picorna | Retro | Parvo | Total (%) | ||
| Assembly algorithm | ABySS | 188 | 111 | 18 | 12 | 3 | 74 | 3 | 409 (30%) | |
| MetaCortex | 113 | 94 | 29 | 9 | 1 | 69 | 315 (23%) | |||
| Meta-IDBA | 27 | 14 | 2 | 4 | 2 | 49 (4%) | ||||
| Velvet | 211 | 189 | 21 | 15 | 1 | 3 | 145 | 5 | 590 (43%) | |
| Sample | Lung | 8 | 1 | 18 | 1 | 235 | 263 (19%) | |||
| Throat sw. | 531 | 405 | 18 | 38 | 1 | 53 | 8 | 1054 (77%) | ||
| Urine | 2 | 32 | 6 | 4 | 2 | 46 (4%) | ||||
| ID algorithm | BLASTn | 14 | 9 | 36 | 5 | 4 | 6 | 1 | 75 (6%) | |
| BLASTx | 454 | 343 | 22 | 30 | 1 | 3 | 258 | 4 | 1115 (82%) | |
| tBLASTx | 71 | 56 | 10 | 3 | 28 | 5 | 173 (13%) | |||
| Total (% of total) | 539 (40%) | 408 (30%) | 68 (5%) | 38 (3%) | 1 (0%) | 7 (0%) | 292 (21%) | 10 (1%) | 1363 (100%) | |
All sample types, assembly algorithms and BLAST comparison algorithms identified viral sequences (Table 3, Fig. 1C). By using multiple assembly and identification algorithms we generated more contigs and identified more viral sequences.
Analysis of viral sequences by family
Herpesviridae
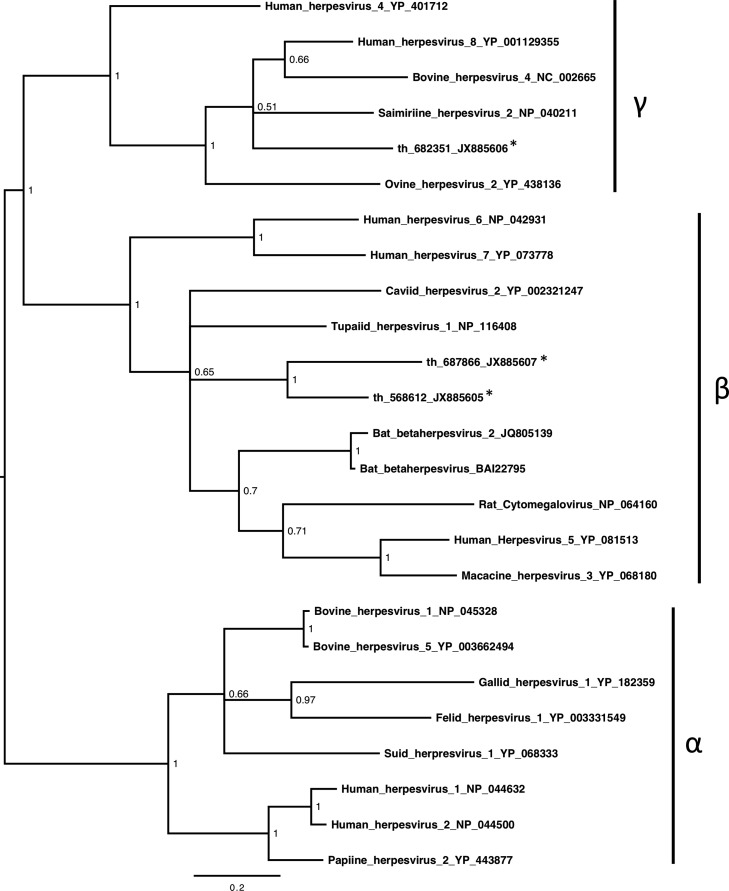
We identified 539 sequences related to herpesviruses, mostly from the throat sample(Table 3). Sequences related to a wide range of genes and proteins involved in diverse functions including gene regulation, nucleotide metabolism, DNA replication as well as envelope glycoproteins and other structural proteins. Most sequences related to members of the betaherpesvirinae (n=366) and gammaherpesvirinae (n=171), and only two sequences most closely related with alphaherpesvirinae (Supp. Table 1). Phylogenetic analysis of a region of the DNA polymerase showed the presence of distinct herpesviruses in the throat sample, including some related to other bat betaherpesviruses (Zhang et al., 2012) and a novel gammaherpesvirus ( Fig. 2). The presence of contig th_687866 in the throat sample was confirmed by PCR and sequencing (not shown).
Fig. 2.
Presence of diverse herpesviruses in throat swab samples fromEidolon helvum. Midpoint-rooted phylogenetic tree based on 166aa of the DNA polymerase protein encoded by three contigs (starred) and representatives from the Herpesviridae. Herpesvirinae subfamily clusters are shown. Posterior probability values are shown and the bar represents the expected number of amino acid substitutions per site.
Papillomaviridae
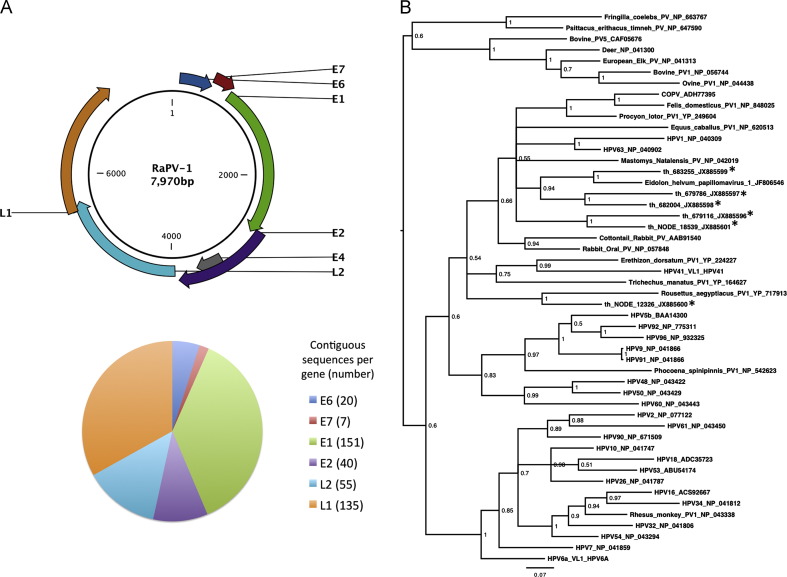
Most papillomavirus sequences derived from the throat sample (405 of 408), but the other sample types also contained papillomavirus sequences (Table 3). Sequences related to both early (E) and late (L) genes of viral replication in proportions approximate to gene length ( Fig. 3A). The sequences related to members of genetically-diverse genera within the Papillomaviridae (Supp. Table 1, Fig. 3B). Phylogenetic analysis of overlapping fragments showed two related sequences of novel papillomavirus(es), one (th_683255) with 76% aa ID with an incomplete, unpublished ‘Eidolon helvum papillomavirus 1’ and another (th_NODE_12326) with 64% aa ID with Rousettus aegyptiacus papillomavirus 1. We confirmed the presence of contig th_679786 by PCR and sequencing (not shown).
Fig. 3.
Gene origin and diversity of papillomavirus sequences present inE. helvum. (A) Top: schematic representation of Rousettus aegyptiacus papillomavirus 1 (RaPV-1) genome. Bottom: gene classification of 408 contigs identified as papillomaviruses. The pie chart shows the proportion of contigs related to each gene. The number of contigs is shown in brackets after the gene name. (B) Midpoint-rooted phylogenetic tree based on 157 aa of the major capsid (L1) protein encoded by six contigs (starred) and representatives from the Papillomaviridae. Posterior probability values are shown at each node and the bar represents the expected number of amino acid substitutions per site.
Adenoviridae
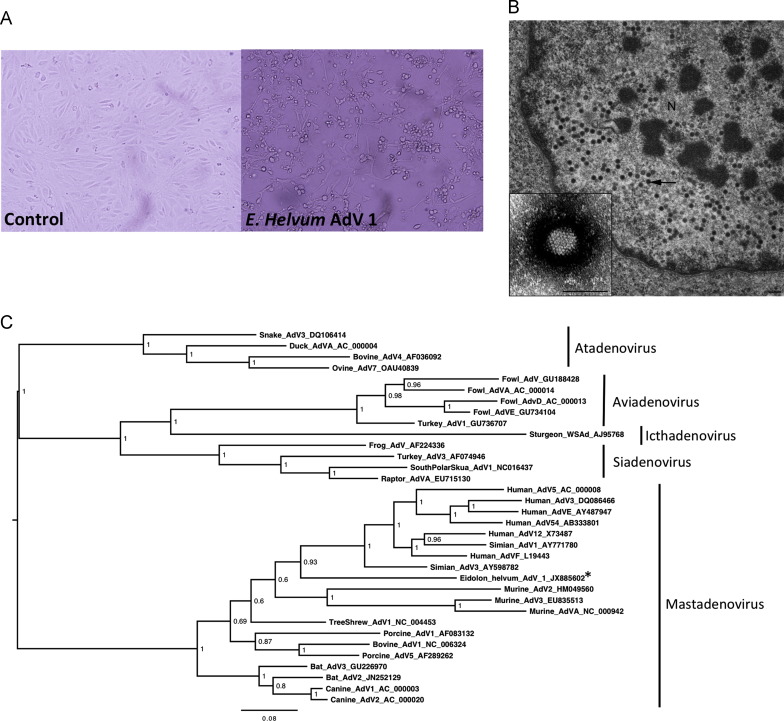
Sequences related to adenoviruses were present in all three sample types (Table 3). All sequences related to members of the mammalian-infecting genus Mastadenovirus. When aligned with the prototypic member of this genus (Human adenovirus C, NC_001405), the 68 adenovirus-like contigs displayed 65–100% aa identity with twelve proteins involved in capsid morphogenesis, DNA replication and encapsidation, and apoptosis (Supp. Table 1). We isolated an adenovirus (referred to as E. helvum adenovirus 1) from a urine sample obtained from this bat population ( Fig. 4A and B), and although phylogenetic analysis identified as a mastadenovirus, it was distinct from those previously described in bats (Fig. 4C). Notably, E. helvum adenovirus 1 clustered with human adenoviruses. Contigs from the throat (th_NODE_10144) and urine samples (ur_NODE_27579) shared 77% and 90% aa identity with the isolated virus’ hexon protein over distances of 58 and 63aa, respectively.
Fig. 4.
Isolation ofE. helvumadenovirus 1 (AdV1). (A) Pteropus alecto primary kidney cells showing cytopathic effect (and control) at day 6 post infection with E. helvum AdV1. Magnification is 200 X (B) Transmission electron micrograph of a Pteropus alecto cell infected with E. helvum AdV1. Arrow indicates adenovirus particles observed in the nucleus (N). Bar represents 200 nm. Insert adenovirus particle negative stained from supernatant of the same culture. Insert bar represents 100 nm. (C) Midpoint-rooted phylogenetic tree based on alignment of hexon proteins (915aa) of E. helvum AdV1 (starred) and representative adenoviruses. Genera are demarcated by adjacent lines and names. Posterior probability values are shown and the bar represents the expected number of amino acid substitutions per site.
Poxviridae
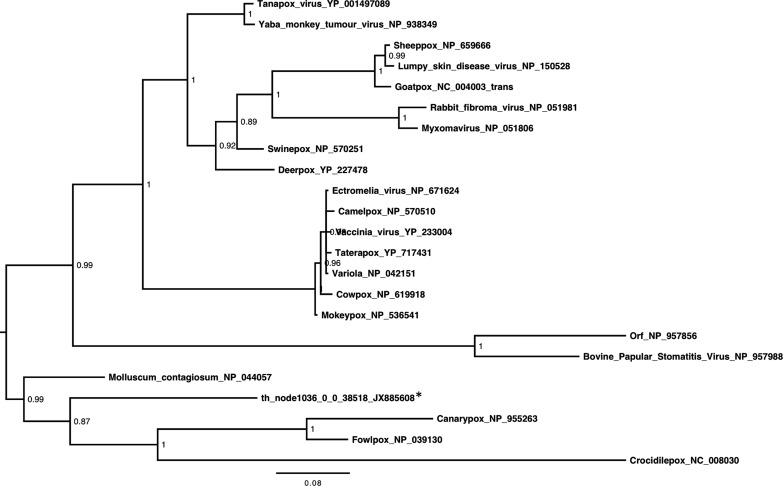
We detected 38 contigs related to poxviruses, all derived from the throat sample and related to chordate-infecting poxviruses. Most (n=25) were related to Molluscum contagiosum (MC), a human contagion (Supp. Table 1). We compared all poxvirus contigs against MC reference proteins (NC_001731) using BLASTx. The sequences shared 29–74% aa identity with 23 different MC proteins and had e-values of 3.45 e −111–8.03 e −4, showing that all sequences had significant similarity to MC. Sequences were related to proteins in the outer (variable) as well the inner (core) regions of the genome (Supp. Fig. 3), but no proteins unique to Molluscipox were detected. This relationship was exemplified by phylogenetic analysis of contig th_node1036_0_0_38518, related to the major core protein ( Fig. 5). We confirmed the presence of this contig in individual throat swabs by PCR. Five of the forty throat swabs (13% of samples, 95% CI 6–26%) contained this sequence. To further confirm the presence of poxvirus sequences in E. helvum, we aligned the MetaCortex contigs (of any length, from all three sample types) against the MC genome using BLAT (which reports alignment blocks of over 95% identity) and increased the number of poxviral related contigs to 12,845 (Supp. Fig. 3)
Fig. 5.
Phylogenetic analysis of anE. helvumpoxviral sequence. Midpoint-rooted phylogenetic tree based on 286 aa of the major core protein encoded by a contig from the throat sample (starred) and representatives from the Poxviridae. Posterior probability values are shown and the bar represents the expected number of amino acid substitutions per site.
Polyomaviridae
We assembled a sequence from the throat sample that was related to the VP1 capsid protein of polyomaviruses. This phylogenetically-clustered with primate polyomaviruses with low confidence, likely due to the short length of the sequence (Supp. Fig. 4).
Retroviridae
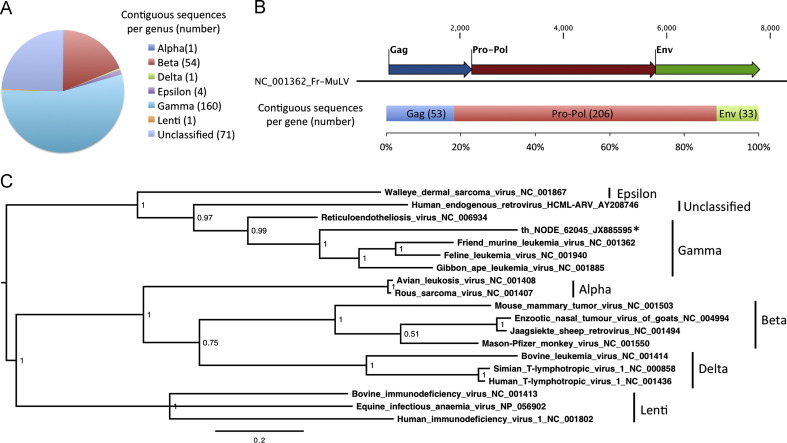
There were 292 sequences related to retroviruses, primarily derived from the lung sample, though sequences were also present in the urine and throat samples (Table 3). Retroviral sequences related primarily to gamma, beta and unclassified retroviruses ( Fig. 6A). The sequences related to all three canonical genes of retroviruses in proportions approximate to gene length (Fig. 6B). Translations of many retrovirus contigs contained stop codons within the region of BLAST alignment, suggesting that they derived from non-functional, endogenous retroviruses. The longest ORF of a partial polymerase protein sequence (th_NODE_62045) was phylogenetically related to, but distinct from, both avian and mammalian viruses (Fig. 6C).
Fig. 6.
Classification and phylogenetic analysis of retroviral sequences present inE. helvum. (A) The chart shows proportions of retroviral sequences related to different retroviral genera. The number of sequences related to each genus is shown in parentheses. (B) Retroviral sequences classified by the gene they encode. The top bar represents the genome organization of Friend-murine leukemia virus, with numbers representing genome nucleotide positions. The lower bar shows the proportion of 292 retroviral contigs that were related to each gene. The number of sequences related to each gene is shown in parentheses. (C) Midpoint-rooted phylogenetic tree based on 176aa of the Pro-Pol gene of representative retroviruses and a contig from the throat sample (starred), posterior probability values are shown at each node and the bar represents the expected number of amino acid substitutions per site.
Parvoviridae
Ten sequences derived from the throat (n=8) and urine (n=2) related to members of the Parvoviridae, from both the mammalian-infecting Parvovirinae subfamily and the invertebrate-infecting Densovirinae subfamily (Supp. Fig. 5). The analysed Parvovirinae-like sequence (th_node7292_0_0_7345) related to members of different genera (e.g. Erythrovirus and Betaparvovirus) and was distinct from a known E. helvum parvovirus (Supp. Fig. 5B).
Picornaviridae
Contigs from the urine (n=6) and lung (n=1) sample related to picornaviruses. Urine contigs related to the polyprotein of members of the genus Kobuvirus and the longest sequence, ur_181630, phylogenetically-clustered with human and canine kobuviruses (Supp. Fig. 6). Short sequence lengths precluded useful phylogenetic comparison of these Kobuvirus sequences with those detected in North American insectivorous bats (Li et al., 2010a), however, there was 50% identity over a 90aa overlapping region. The lung picornavirus sequence related to members of the genus Enterovirus, but was too short (79 bp) for useful phylogenetic analysis.
Discussion
Here we described the first detailed study of metagenomic viral sequences from a megachiropteran species. E. helvum have a wide geographical distribution and live in close contact with human populations. As such, this bat species is an ideal candidate reservoir host, also being a source of bushmeat in Ghana and likely being infected with henipaviruses, Lagos bat virus (lyssavirus) and Ebola virus (filovirus) in this location (Drexler et al., 2009, Drexler et al., 2012, Hayman et al., 2010, Hayman et al., 2008a, Hayman et al., 2008b, Wright et al., 2010). Our results highlight the utility of metagenomic studies to assess zoonotic risk in wildlife.
The impact of bioinformatics tools on metagenomic studies
Differences in the assembler efficacy manifested as differences in number, length-parameters and base-composition of the generated contigs. ABySS and Velvet are consensus assemblers designed to assemble a single genome from sequence reads. Contrastingly, MetaCortex and MetaIDBA are meta-assemblers specifically designed to address situations where multiple genomes would be expected. Generally, consensus assemblers adopt more stringent algorithms for error removal in order to build longer contigs, while meta-assemblers preserve sample variation. The consolidation of contigs from multiple assemblers generated a more robust contig set and reduced the number of sequences by one third, facilitating downstream processing.
The BLAST algorithm used also impacted the number of contigs identified as suspect-viral. BLASTn identified fewer sequences than tBLASTx against the same database with identical retention criteria. Similarly, BLASTx identified more suspect-viral sequences than BLASTn (although a different database was used), supporting the observation that protein-based comparisons are more effective than nucleotide-based comparisons where divergent sequences are expected (Kunin et al., 2008). The use of multiple identification algorithms here, enabled the detection of more viral sequences.
Viral sequences
The relative identification success for mammalian viral sequences in this metagenomic study compared with others, as well as among sample types and analytical tools, provide guidance on how best to approach such studies. Using the Illumina platform, and working with a frugivorous bat species, we found that 28% of viral sequences identified were of mammalian-origin, more than the ≤10% previously identified in insectivorous bat species (Donaldson et al., 2010, Ge et al., 2012, Li et al., 2010a, Wu et al., 2012). The sample type also affected the level of detection, with most viral sequences being derived from the throat sample (though differences attributable to colony differences from which the samples were collected cannot be ruled out). These discrepancies in detection show that the quantitative and qualitative success of viral metagenomic studies is determined partly by the study species, sample type, and molecular and bioinformatic tools used.
Here we aimed to identify viruses circulating in E. helvum that might have zoonotic potential. We detected novel, often diverse, viruses in many viral families, in samples collected over a short time frame from a small number of bats. The viral sequences were often distinct from those previously described in bats and often we saw diversity within the viral family (e.g. herpesvirus and retroviral sequences from different subfamilies, and at least six phylogenetically-distinct papillomaviruses from different genera). Clearly, a wide range of diverse and previously uncharacterized viruses circulate in E. helvum.
Given the proximity of this species with humans over a large geographical area, it is important to consider the zoonotic potential of these viruses. We detected poxvirus, adenovirus and polyomavirus sequences closely related with those from humans and primates (in the latter two cases, more closely related than with those viruses previously described in bats). We also isolated an adenovirus from urine collected directly underneath the colony, a sample type to which humans are regularly exposed. The relatedness of these viruses to human pathogens may indicate that these viruses are more likely to emerge (Antia et al., 2003). Given the relationship of these viruses with human pathogens and E. helvum's high rate of human contact, more extensive active surveillance such as molecular and serological studies of humans in contact with relevant bat populations seems appropriate.
Our study expands the known chiropteran viral profile. While many viral families detected here have previously been reported in bat populations, here we report the detection of poxvirus sequences in bats. Although Li et al. reported pox-related sequences derived from a circovirus (Li et al., 2010a), the viral sequences reported here are likely to derive from a true poxvirus. We detected sequences with a high degree of relatedness to 23 different proteins throughout the genome of M. contagiosum. The presence of this virus in 5% of the throat swabs suggests a high prevalence of poxvirus infection in this bat population. Given the relatedness of the virus to MC, the possibility of zoonotic transmission of poxviruses from bats to humans should be considered further, and in other geographical areas.
The commonalities between our findings and those in other metagenomic studies of bats provide insight on the relationship that bats may have with their natural pathogens. Of the eight viral families detected here, six have previously been detected during metagenomic studies of insectivorous bats (Donaldson et al., 2010, Ge et al., 2012, Li et al., 2010a, Wu et al., 2012) and one (the Polyomaviridae) was detected in Myotis spp. using consensus PCR (Misra et al., 2009). We worked with a host from a separate taxonomic suborder to those studies and still found similar viruses, suggesting that a common viral footprint may be present in all chiropteran species. Continued description of viral profiles of disparate host species and geographical locations will deepen our understanding of host-pathogen relationships in these important zoonotic reservoir species.
While these results represent an interesting development in the study of these reservoir hosts, the limitations of metagenomic methods should be acknowledged. Only partial sequence information was generated for each viral family under study, limiting analysis. Additionally, although some viral sequences were confirmed by PCR and virus isolation, making them likely-derived from true viruses, the same cannot be said of the retroviral sequences. The high proportion of retroviral sequences in the lung, as well as the presence of stop codons in a number of sequences indicate that they likely derive from endogenous proviruses. Furthermore, in common with other studies of this nature, most viral sequences here were derived from those with dsDNA genomes. Drexler et al. (2012) showed that metagenomic methods could not detect paramyxoviral RNA where consensus PCR was successful. Similarly, this population of E. helvum has been demonstrated to harbour a high prevalence and diversity of paramyxoviruses yet none were detected here (Baker et al., 2012). Although some progress is being made toward validating the scope of viral metagenomic studies (Sachsenroder et al., 2012), further work is needed to conclude whether this repeatedly-observed bias is biological (perhaps consequent to the long, sometimes latent, infection periods of dsDNA viruses) or if it constitutes a laboratory artifact. Due to these methodological limitations, it is likely that the viral sequences described here are not an exhaustive representation of the viruses of E. helvum.
Materials and methods
Ethics declaration
This study was approved by the Zoological Society of London's animal ethics committee.
Populations under study
Two colonial populations (250,000–1,000,000 bats each) of E. helvum in Ghana were sampled: one in Accra and one in Tano Sacred Grove (TSG, approx. 400 km North, Supp. Fig. 1). The Accra population is urban, roosting in trees over a city center hospital. The TSG population is rural, roosting in a protected forest area. The two populations comprise part of a metapopulation (Peel, 2012). Interspecies co-roosting of these populations was not observed in five years of field study.
Sample collection
Urine for metagenomic analysis was collected twice from beneath the Accra roost; in January and March 2009. Sterile cotton swabs were saturated with urine on plastic sheeting placed beneath the roost, and placed in 1 ml of virus transport medium (VTM: Hank's Balanced Salt Solution, 1% BSA [w/v], gentamicin 100 µg/ml and amphotericin B 2 µg/ml). Urine samples for virus isolation were collected in 2010, as previously described (Baker et al., 2012). Throat swabs were collected from individual, manually-restrained bats caught from TSG in March 2009. Swabs were placed in 1 ml of VTM. Lung tissue was collected from healthy, adult bats euthanized by anaesthetic overdose (ketamine/medetomidine), captured in Accra in March 2009. An individual piece of tissue (approx. 5 mm2) from each bat was used. Samples were frozen at −80 °C until further processing.
Sample pooling, enrichment and nucleic acid manipulation
This work was performed at the Department of Veterinary Medicine, University of Cambridge. Lung tissues (n=5) were disrupted in 1 ml PBS using sterile pestles before homogenisation and pooling. Urine samples (n=80) were pooled, with individual samples contributing 500 µl of VTM. Throat swabs in VTM (n=40) were pooled similarly. Sample pools were centrifuged (1500 g, 10 min, room temperature) and supernatants filtered (5 µm spin filters then 0.85 µm syringe filters). Filtrates were divided in two before overnight precipitation (at 4 °C) using polyethylene glycol (5% PEG, 0.15 M NaCl), and centrifugated (10,700 g, 1 h, 4 °C). Pellets resuspended in 175 µl PBS were DNAse (RNAse-free DNAse 1, 100 U, Ambion) (adapted from (Allander et al., 2001), and then RNAse (RNAse A, 0.5 U, Invitrogen) treated, both for 30 min at 37 °C.
Total nucleic acids were extracted (High Pure Viral Nucleic Acid Kit, Roche). Sequence-Independent Single-Primer Amplification (adapted from (Djikeng et al., 2008)) was performed as follows: cDNA was generated by reverse transcription (Superscript III, Invitrogen) followed by complementary-strand synthesis (DNA polymerase I large [Klenow] fragment, NEB) using primer FR26RV-N. PCR amplification was performed with FR20RV (Advantage 2 PCR kit, Clontech) with thermal profiling: 1 min at 95 °C, 25 cycles of 30 s at 95 °C, 1 min at 65 °C, 2 min 30 s at 68 °C, followed by 1 min at 68 °C. PCR products were size-fractionated (Chromospin 200 columns, Clontech) and visualized on a Bioanalyser DNAchip (Agilent Technologies). For each sample type, 1 µg of DNA was prepared for sequencing.
Next generation sequencing
This work was carried out at the Wellcome Trust Sanger Institute. The DNA library was sheared (Covaris AFA, Covaris) to 200–300 bp and purified (QIAquick spin columns, Qiagen), blunt-end repaired, and ligated to sequencing primers. Ligation products were purified (Agencourt Ampure SPRI beads, Beckman Coultor Genomics) and the library was 200 bp size-selected by agarose gel electrophoresis. Following purification (Gel extraction kit, Qiagen), the library was PCR amplified for 10 cycles (Phusion DNA polymerase) in triplicate using Illumina adaptor-specific primers. The primers were removed (Agencourt Ampure SPRI beads, Beckman Coultor Genomics), and libraries quantified by qPCR were diluted to 40 nM for cluster generation. Libraries were sequenced on an Illumina GAII (Illumina Inc) for 76 bp paired-end reads. Following data QA, QC and computational primer removal there were 5,218,132 sequence reads from the urine sample, 15,809,698 from the throat sample and 22,530,774 from the lung tissue sample.
Generation of consolidated contiguous sequences (contigs)
Sequences from each sample type were processed individually. Reads were de novo assembled using four assembly algorithms: Velvet 1.1.04 (Zerbino and Birney, 2008), ABySS 1.2.7 (Simpson et al., 2009), MetaIDBA 0.19 (Peng et al., 2011) and MetaCortex (Leggett and Caccamo, personal communication), a recently-developed variant of Cortex (Iqbal et al., 2012). These assemblers are based on de Bruijn graphs, which are constructed by dividing reads into smaller, overlapping sequences called kmers. For ABySS, Velvet and MetaCortex, a range of kmer sizes (21, 31, 41, 51, 61 and 71) were evaluated, with 31 proving optimal (providing the largest number of contigs ≥100 nt and the largest number of viral matches to the NCBI nt database) for Velvet and ABySS, and 61 being optimal for MetaCortex. MetaIDBA iterates over a kmer size range, and the throat sample was iterated over 21–71 nt. When assembling the urine and lung samples, MetaIDBA was unable to reach completion when allowed to iterate, so was fixed to 71 nt. The four contig sets for each sample type then underwent a consolidation process comprised of sequential BLAT alignments (Kent, 2002), followed by removal of shorter contigs that were 95% identical over a 95% length fraction (Fig. 1A). First, contigs generated by Velvet and ABySS were compared (Comparison 1). The retained contigs were then compared with MetaIDBA contigs (Comparison 2), and then those still retained were compared with MetaCortex contigs (Comparison 3, Fig. 1A). Consequently, sequences retained after Comparison 3 were a consolidated contig set with reduced redundancy.
Identification of suspect-viral sequences
Contigs≤76 nt in length (considered potentially-derived from single sequencing reads) were not analysed further (Fig. 1B). Remaining sequences underwent sequential BLAST comparison (Altschul et al., 1990) against NCBI databases (at November 25, 2011) and taxonomic classification queried from the NCBI taxonomy web service. Contigs underwent BLASTn comparison with the NCBI nt database. The taxonomic classification of the source organism of the reference sequence with which the contig ‘best’ aligned (i.e. the alignment with the lowest expect value [e-value]) was retrieved. If the reference sequence taxonomy was viral and aligned with the contig with an e-value of≤0.0001, the sequence was flagged as suspect-viral and retained for further analysis. Sequences not flagged as suspect-viral then proceeded to BLASTx comparison with the NCBI nr database, with the same retention criteria. Sequences still not retained underwent tBLASTx comparison with the NCBI nt database, and were similarly retained. Those not retained in this final comparison round were discarded.
Classification and curation of viral sequences
Suspect-viral sequences related to viral families not known to infect vertebrates were excluded. Remaining suspect-viral sequences were manually curated by examination of the region of database sequence that matched the contig in BLAST alignment. Where the database sequence providing the alignment with the lowest e-value appeared to be classified into the incorrect taxonomic group, all BLAST hits for the contig were examined and the majority taxa was determined to be the likely origin of the sequence (Fig. 1B). Remaining sequences and BLAST results were then grouped by viral family.
Analysis of viral sequences
Reference sequences were downloaded from NCBI, and global alignments with contigs generated using Clustal X (Version 2 (Thompson et al., 1994)) and Muscle 3.8.31 (Edgar, 2004). Gap-stripped alignments (columns with>50% gaps were removed) were then used to infer phylogenetic trees using MrBayes (Ronquist and Huelsenbeck, 2003), as previously described (Baker et al., 2012). Local BLAST comparisons and pairwise identities were performed using Genomics workbench (Vs 5.1, CLC Bio).
Amplification of viral sequences by PCR
Primers (sequences available on request) were designed to detect poxvirus, herpesvirus and papillomavirus contigs in the throat sample nucleic acids submitted for sequencing. Poxvirus PCR was also performed on nucleic acids extracted from the individual throat swabs. PCR (DreamTaqGreenPCR Mastermix, Fermentas) products were visualized by gel electrophoresis, purified (Gel extraction kit, QIAGEN) and sequenced.
Adenovirus isolation and characterization
An adenovirus causing cytopathic effect in Pteropus alecto primary kidney cells (Crameri et al., 2009) was isolated from sample U69 (Baker et al., 2012). Negative contrast electron microscopy (EM) was used to examine 6 day post-infection culture supernatant. Supernatant was adsorbed onto parlodion-filmed copper grids coated with carbon and stained with nano-W stain (Nanoprobes, Yaphank, NY, USA). Thin section EM was used to examine cells 5 days post-infection (as in (Weir et al., 2012) except using Sorenson's phosphate buffer (300 mosM/kg, pH 7.2)). The full-length hexon gene of this isolate was sequenced, as in (Zhang et al., 2012).
Nucleotide sequences
Viral sequences discussed here were deposited in GenBank (JX885594 – JX885611), except for the too-short polyomavirus sequence (Supp. Fig. 4). Sequencing data were deposited in the European Nucleotide Archive (ERP001979). Supplementary fasta files of all viral and suspect-viral sequences and comparison outputs are available online, and assembled contigs are available on request.
Acknowledgments
We would like to thank the Veterinary Services Directorate (Ghana), Professor Anthony Fooks at the Veterinary Laboratories Agency (UK). Huajun Zhang for assistance in isolation and preparation of adenoviral DNA for sequencing. The authors acknowledge the support of the Australian Microscopy and Microanalysis Research Facility to the microscopy facilities of the Australian Animal Health Laboratory. This study was funded by a Wellcome Trust research-training fellowship (KB) and the viral genomics group at the Wellcome Trust Sanger Institute (PK). NB and PRM are also funded by the Wellcome Trust. JLNW is supported by the Alborada Trust and the RAPIDD program of the Science and Technology Directorate, Department of Homeland Security. The authors also thank Professor Massimo Palmarini and the anonymous reviewers for their helpful comments on the manuscript.
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.virol.2013.03.014.
Appendix A. Supplementary materials
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material
References
- Allander T., Emerson S.U., Engle R.E., Purcell R.H., Bukh J. A virus discovery method incorporating DNase treatment and its application to the identification of two bovine parvovirus species. PNAS. 2001;98:11609–11614. doi: 10.1073/pnas.211424698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Antia R., Regoes R.R., Koella J.C., Bergstrom C.T. The role of evolution in the emergence of infectious diseases. Nature. 2003;426:658–661. doi: 10.1038/nature02104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker K.S., Todd S., Marsh G., Fernandez-Loras A., Suu-Ire R., Wood J.L., Wang L.F., Murcia P.R., Cunningham A.A. Co-circulation of diverse paramyxoviruses in an urban African fruit bat population. J. Gen. Virol. 2012;93:850–856. doi: 10.1099/vir.0.039339-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bermingham, A., Chand, M.A., Brown, C.S., Aarons, E., Tong, C., Langrish, C., Hoschler, K., Brown, K., Galiano, M., Myers, R., Pebody, R.G., Green, H.J., Boddington, N.L., Gopal, R., Price, N., Newsholme, W., Drosten, C., Fouchier, R.A., Zambon, M., 2012. Severe Respiratory Illness Caused by a Novel Coronavirus, in a Patient Transferred to the United Kingdom from the Middle East, September 2012. Eurosurveillance 17, Available online: 〈http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=20290〉. [PubMed]
- Chu D.K., Poon L.L., Guan Y., Peiris J.S. Novel astroviruses in insectivorous bats. J. Virol. 2008;82:9107–9114. doi: 10.1128/JVI.00857-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleaveland S., Haydon D.T., Taylor L. Overviews of pathogen emergence: which pathogens emerge, when and why? Curr. Top. Microbiol. Immunol. 2007;315:85–111. doi: 10.1007/978-3-540-70962-6_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crameri G., Todd S., Grimley S., McEachern J.A., Marsh G.A., Smith C., Tachedjian M., De Jong C., Virtue E.R., Yu M., Bulach D., Liu J.P., Michalski W.P., Middleton D., Field H.E., Wang L.F. Establishment, immortalisation and characterisation of pteropid bat cell lines. PLoS One. 2009;4:e8266. doi: 10.1371/journal.pone.0008266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day J.M., Ballard L.L., Duke M.V., Scheffler B.E., Zsak L. Metagenomic analysis of the turkey gut RNA virus community. Virol. J. 2010;7:313. doi: 10.1186/1743-422X-7-313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djikeng A., Halpin R., Kuzmickas R., Depasse J., Feldblyum J., Sengamalay N., Afonso C., Zhang X., Anderson N.G., Ghedin E., Spiro D.J. Viral genome sequencing by random priming methods. BMC Genomics. 2008;9:5. doi: 10.1186/1471-2164-9-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobson A., Foufopoulos J. Emerging infectious pathogens of wildlife. Philos. Trans. R. Soc. Lond B. Biol. Sci. 2001;356:1001–1012. doi: 10.1098/rstb.2001.0900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaldson E.F., Haskew A.N., Gates J.E., Huynh J., Moore C.J., Frieman M.B. Metagenomic analysis of the viromes of three North American bat species: viral diversity among different bat species that share a common habitat. J. Virol. 2010;84:13004–13018. doi: 10.1128/JVI.01255-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drexler J.F., Corman V.M., Gloza-Rausch F., Seebens A., Annan A., Ipsen A., Kruppa T., Muller M.A., Kalko E.K., Adu-Sarkodie Y., Oppong S., Drosten C. Henipavirus RNA in African bats. PLoS One. 2009;4:e6367. doi: 10.1371/journal.pone.0006367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drexler J.F., Corman V.M., Muller M.A., Maganga G.D., Vallo P., Binger T., Gloza-Rausch F., Rasche A., Yordanov S., Seebens A., Oppong S., Sarkodie Y.A., Pongombo C., Lukashev A.N., Schmidt-Chanasit J., Stocker A., Carneiro A.J., Erbar S., Maisner A., Fronhoffs F., Buettner R., Kalko E.K., Kruppa T., Franke C.R., Kallies R., Yandoko E.R., Herrler G., Reusken C., Hassanin A., Kruger D.H., Matthee S., Ulrich R.G., Leroy E.M., Drosten C. Bats host major mammalian paramyxoviruses. Nat. Commun. 2012;3:796. doi: 10.1038/ncomms1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R.C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge X., Li J., Peng C., Wu L., Yang X., Wu Y., Zhang Y., Shi Z. Genetic diversity of novel circular ssDNA viruses in bats in China. J. Gen. Virol. 2011 doi: 10.1099/vir.0.034108-0. [DOI] [PubMed] [Google Scholar]
- Ge X., Li Y., Yang X., Zhang H., Zhou P., Zhang Y., Shi Z. Metagenomic analysis of viruses from bat fecal samples reveals many novel viruses in insectivorous bats in China. J. Virol. 2012;86:4620–4630. doi: 10.1128/JVI.06671-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halpin K., Young P.L., Field H.E., MacKenzie J.S. Isolation of Hendra virus from pteropid bats: a natural reservoir of Hendra virus. J. Gen. Virol. 2000;81:1927–1932. doi: 10.1099/0022-1317-81-8-1927. [DOI] [PubMed] [Google Scholar]
- Hayman D.T., Emmerich P., Yu M., Wang L.F., Suu-Ire R., Fooks A.R., Cunningham A.A., Wood J.L. Long-term survival of an urban fruit bat seropositive for Ebola and Lagos bat viruses. PLoS One. 2010;5:e11978. doi: 10.1371/journal.pone.0011978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayman D.T., Fooks A.R., Horton D., Suu-Ire R., Breed A.C., Cunningham A.A., Wood J.L. Antibodies against Lagos bat virus in megachiroptera from West Africa. Emerg. Infect. Dis. 2008;14:926–928. doi: 10.3201/eid1406.071421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayman D.T., Suu-Ire R., Breed A.C., McEachern J.A., Wang L., Wood J.L., Cunningham A.A. Evidence of henipavirus infection in West African fruit bats. PLoS One. 2008;3:e2739. doi: 10.1371/journal.pone.0002739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayman D.T.S., McCrea R., Restif O., Suu-Ire R., Fooks A.R., Wood J.L.N., Cunningham A.A., Rowcliffe M.J. Demography of straw-colored fruit bats in Ghana. J. Mammal. 2012 doi: 10.1644/11-MAMM-A-270.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iqbal Z., Caccamo M., Turner I., Flicek P., McVean G. De novo assembly and genotyping of variants using colored de Bruijn graphs. Nat. Genet. 2012;44:226–232. doi: 10.1038/ng.1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones K.E., Patel N.G., Levy M.A., Storeygard A., Balk D., Gittleman J.L., Daszak P. Global trends in emerging infectious diseases. Nature. 2008;451:990–993. doi: 10.1038/nature06536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent W.J. BLAT—the BLAST-like alignment tool. Genome Res. 2002;12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunin V., Copeland A., Lapidus A., Mavromatis K., Hugenholtz P. A bioinformatician's guide to metagenomics. Microbiol. Mol. Biol. Rev. 2008;72:557–578. doi: 10.1128/MMBR.00009-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzmin I.V., Niezgoda M., Franka R., Agwanda B., Markotter W., Beagley J.C., Urazova O.Y., Breiman R.F., Rupprecht C.E. Lagos bat virus in Kenya. J. Clin. Microbiol. 2008;46:1451–1461. doi: 10.1128/JCM.00016-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Shan T., Wang C., Cote C., Kolman J., Onions D., Gulland F.M., Delwart E. The fecal viral flora of California sea lions. J. Virol. 2011;85:9909–9917. doi: 10.1128/JVI.05026-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Victoria J.G., Wang C., Jones M., Fellers G.M., Kunz T.H., Delwart E. Bat guano virome: predominance of dietary viruses from insects and plants plus novel mammalian viruses. J. Virol. 2010;84:6955–6965. doi: 10.1128/JVI.00501-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Shi Z., Yu M., Ren W., Smith C., Epstein J.H., Wang H., Crameri G., Hu Z., Zhang H., Zhang J., McEachern J., Field H., Daszak P., Eaton B.T., Zhang S., Wang L.F. Bats are natural reservoirs of SARS-like coronaviruses. Science. 2005;310:676–679. doi: 10.1126/science.1118391. [DOI] [PubMed] [Google Scholar]
- Li Y., Ge X., Hon C.C., Zhang H., Zhou P., Zhang Y., Wu Y., Wang L.F., Shi Z. Prevalence and genetic diversity of adeno-associated viruses in bats from China. J. Gen. Virol. 2010;91:2601–2609. doi: 10.1099/vir.0.020032-0. [DOI] [PubMed] [Google Scholar]
- Li Y., Ge X., Zhang H., Zhou P., Zhu Y., Zhang Y., Yuan J., Wang L.F., Shi Z. Host range, prevalence, and genetic diversity of adenoviruses in bats. J. Virol. 2010;84:3889–3897. doi: 10.1128/JVI.02497-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luby S.P., Hossain M.J., Gurley E.S., Ahmed B.N., Banu S., Khan S.U., Homaira N., Rota P.A., Rollin P.E., Comer J.A., Kenah E., Ksiazek T.G., Rahman M. Recurrent zoonotic transmission of Nipah virus into humans, Bangladesh, 2001–2007. Emerg. Infect. Dis. 2009;15:1229–1235. doi: 10.3201/eid1508.081237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misra V., Dumonceaux T., Dubois J., Willis C., Nadin-Davis S., Severini A., Wandeler A., Lindsay R., Artsob H. Detection of polyoma and corona viruses in bats of Canada. J. Gen. Virol. 2009;90:2015–2022. doi: 10.1099/vir.0.010694-0. [DOI] [PubMed] [Google Scholar]
- Morens D.M., Folkers G.K., Fauci A.S. The challenge of emerging and re-emerging infectious diseases. Nature. 2004;430:242–249. doi: 10.1038/nature02759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peel A.J. Department of Veterinary Medicine University of Cambridge; Cambridge: 2012. The Epidemiology of Lagos Bat Virus and Henipaviruses in Straw Coloured Fruit Bats (Eidolon helvum), Using Population Genetics to Infer Population Connectivity. pp. 309. [Google Scholar]
- Peng Y., Leung H.C., Yiu S.M., Chin F.Y. Meta-IDBA: a de Novo assembler for metagenomic data. Bioinformatics. 2011;27:i94–101. doi: 10.1093/bioinformatics/btr216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phan T.G., Kapusinszky B., Wang C., Rose R.K., Lipton H.L., Delwart E.L. The fecal viral flora of wild rodents. PLoS Pathog. 2011;7:e1002218. doi: 10.1371/journal.ppat.1002218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronquist F., Huelsenbeck J.P. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- Sachsenroder J., Twardziok S., Hammerl J.A., Janczyk P., Wrede P., Hertwig S., Johne R. Simultaneous identification of DNA and RNA viruses present in pig faeces using process-controlled deep sequencing. PLoS One. 2012;7:e34631. doi: 10.1371/journal.pone.0034631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan T., Li L., Simmonds P., Wang C., Moeser A., Delwart E. The fecal virome of pigs on a high-density farm. J. Virol. 2011;85:11697–11708. doi: 10.1128/JVI.05217-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson J.T., Wong K., Jackman S.D., Schein J.E., Jones S.J., Birol I. ABySS: a parallel assembler for short read sequence data. Genome Res. 2009;19:1117–1123. doi: 10.1101/gr.089532.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang X.C., Zhang J.X., Zhang S.Y., Wang P., Fan X.H., Li L.F., Li G., Dong B.Q., Liu W., Cheung C.L., Xu K.M., Song W.J., Vijaykrishna D., Poon L.L., Peiris J.S., Smith G.J., Chen H., Guan Y. Prevalence and genetic diversity of coronaviruses in bats from China. J. Virol. 2006;80:7481–7490. doi: 10.1128/JVI.00697-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor L.H., Latham S.M., Woolhouse M.E. Risk factors for human disease emergence. Philos. Trans. R Soc. Lond B Biol. Sci. 2001;356:983–989. doi: 10.1098/rstb.2001.0888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson J.D., Higgins D.G., Gibson T.J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towner J.S., Pourrut X., Albarino C.G., Nkogue C.N., Bird B.H., Grard G., Ksiazek T.G., Gonzalez J.P., Nichol S.T., Leroy E.M. Marburg virus infection detected in a common African bat. PLoS One. 2007;2:e764. doi: 10.1371/journal.pone.0000764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weir R.P., Moody N.J., Hyatt A.D., Crameri S., Voysey R., Pallister J., Jerrett I.V. Isolation and characterisation of a novel Bohle-like virus from two frog species in the Darwin rural area, Australia. Dis. Aquat. Org. 2012;99:169–177. doi: 10.3354/dao02472. [DOI] [PubMed] [Google Scholar]
- Woo P.C., Lau S.K., Li K.S., Poon R.W., Wong B.H., Tsoi H.W., Yip B.C., Huang Y., Chan K.H., Yuen K.Y. Molecular diversity of coronaviruses in bats. Virology. 2006;351:180–187. doi: 10.1016/j.virol.2006.02.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright E., Hayman D.T., Vaughan A., Temperton N.J., Wood J.L., Cunningham A.A., Suu-Ire R., Weiss R.A., Fooks A.R. Virus neutralising activity of African fruit bat (Eidolon helvum) sera against emerging lyssaviruses. Virology. 2010;408:183–189. doi: 10.1016/j.virol.2010.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z., Ren X., Yang L., Hu Y., Yang J., He G., Zhang J., Dong J., Sun L., Du J., Liu L., Xue Y., Wang J., Yang F., Zhang S., Jin Q. Virome analysis for identification of novel mammalian viruses in bat species from Chinese provinces. J. Virol. 2012 doi: 10.1128/JVI.01394-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaki A.M., van Boheemen S., Bestebroer T.M., Osterhaus A.D., Fouchier R.A. Isolation of a Novel Coronavirus from a Man with Pneumonia in Saudi Arabia. New England J. Med. 2012 doi: 10.1056/NEJMoa1211721. [DOI] [PubMed] [Google Scholar]
- Zerbino D.R., Birney E. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 2008;18:821–829. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Todd S., Tachedjian M., Barr J.A., Luo M., Yu M., Marsh G.A., Crameri G., Wang L.F. A novel bat herpesvirus encodes homologues of MHC classes I and II, C-type lectin, and a unique family of immune-related genes. J. Virol. 2012 doi: 10.1128/JVI.00723-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material