Abstract
Lipopolysaccharide (LPS) from gram-negative bacteria activates B cells, enabling them to proliferate and differentiate into plasma cells. This response is critically dependent on the expression of TLR4, but other genes, such as RP105 and MHC class II, have also been shown to contribute to B cell LPS response. Here we have evaluated the role of genetic control of the B cell response to LPS at the single cell level by using limiting dilution analysis. This approach avoided the co-stimulatory and feeder-cell dependent effects that occur in bulk cultured B lymphocytes. We compared the response to LPS of peritoneal cavity (PEC) and splenic B cells on the BALB/c genetic background (LPS-low responder) to those on the C57BL/6J background (LPS-high responder) and their F1 progeny (B6xBc). Both PEC and splenic B cells from B6 exhibited 100% clonal growth in the presence of LPS, whereas BALB/c PEC and splenic B cells achieved only 50% and 23% clonal growth, respectively. Surprisingly, PEC B cells on the F1 (B6xBc) background behaved as high responders (close to 100 %, as in B6), while splenic B cells displayed a response that was closer to that of the low responder BALB/c strain (≈ 30 %), revealing a differential genetic control. Splenic follicular B cells on both the F1 and BALB/c backgrounds were also low responders, averaging 18 % and 25 % growth, respectively. Marginal zone B cells on the F1 and BALB/c backgrounds were moderately higher, achieving 45–50% growth. Adding CpG (the TLR9 ligand) to the LPS stimulus promoted pushed splenic B cell clonal growth in the low responder strain BALB/c up to 90%, showing that the non-response to LPS is a specific effect that cannot be attributed to a general deficiency of in vitro growth by the BALB/c cells. The data presented here reveals a previous unsuspected behavior in the genetic control of the B cell response to LPS, with an opposing impact in splenic versus peritoneal cavity B cells. These results suggest the existence of an as yet unidentified genetic factor exclusively expressed by coelomic B cells that contributes to the control the LPS signaling pathway in the B lymphocyte.
INTRODUCTION
Lipopolysaccharide (LPS) from the cell wall of gram-negative bacteria plays a fundamental role in the triggering of innate immunity. The action of LPS on distinct cell types of the immune system, and on somatic cells as well, depends on the expression of toll-like receptor four (TLR4) (Poltorak et al. 1998). The recognition of LPS by TLR4 triggers a cascade of intracellular events that leads to the translocation of NF-KB and AP-1 to the nucleus, resulting in the transcription of a series of cytokine genes, e.g. TNF-α, IL-1, IL-6, INF-β, which play a critical role in innate immune responses (Kawai 2007). Mice genetically deficient for TLR4 are both highly susceptible to gram-negative bacterial infections and resistant to LPS sepsis, confirming that this molecule plays a fundamental in the response to LPS (Hoshino et al. 1999; Kalis et al. 2003). The triggering of TLR4 by LPS depends on multiple factors such as LPS-binding protein (LBP), CD14, and MD2 that act in concert to promote the efficient engagement of TLR4 and its associated signaling machinery. It has been proposed that LBP binds to LPS to create an LBP:LPS complex that is then recognized by CD14 in the cell membrane. Subsequent association with the MD2/TLR4 complex then enables the recruitment of downstream signaling molecules (da Silva Correia et al. 2001).
The triggering of TLR4 in antigen presenting cells has a profound impact on the activation of T cells and the commitment of the adaptive immune response to TH1/TH2 phenotypes. The indirect action of LPS in T lymphocyte activation is thought to be mediated by TLR4-dependent up-regulation of the co-stimulatory molecules B7-1 and B7-2 on the surface of dendritic cells, together with secretion of cytokines (e.g. IL-12, TNF-α) (Medzhitov 2001). Elegant experiments have shown that, in the absence of TLR4, dendritic cells drive the proliferation of antigen specific T lymphocytes that are unable to acquire effector functions (Sporri and Reis e Sousa 2005).
B cell responses in the mouse are also affected by TLR4 ligands, not only because of their role as APCs in T cell dependent humoral immunity, but also because LPS has direct effects in B lymphocytes. LPS promotes the full activation of B cells, leading to proliferation and differentiation to plasmocytes (Andersson et al. 1977). The action of LPS in B cells seems to depend not only on TLR4, but also on another receptor from the TLR family, RP105 (Ogata et al. 2000). The genetic ablation of this element profoundly affects the B cell response to LPS and it has been proposed that RP105 associates with MD1 and cooperates with TLR4/MD2 to activate the B lymphocyte. It has also been shown that an MHC linked locus has a significant impact in the B cell response to LPS, possibly through the control of the expression of RP105 (Rodo et al. 2006).
Here we have studied the genetic control of the B cell response to LPS at the single cell level by limiting dilution analysis, which eliminates the co-stimulatory and feeder-cell dependent effects of LPS that occur in bulk cultures of B lymphocytes. Using the S17 stromal cell line as feeder cells (Collins and Dorshkind 1987), we show that in the frequency of B cells responding to LPS was close to 100 % in LPS-high responding C57BL/6 mice. This extensive response was observed in both peritoneal cavity and splenic B cells. The effect of LPS on B cell activation differed in BALB/c mice, which are known to be low responders to LPS. Activation achieved a ~50% response in PEC B cells and only a ~23% response in splenic B cells. To determine whether this effect was dominant, we examined LPS-driven B cell activation on the F1 (B6xBc) background. Surprisingly, B6xBc PEC B cells behaved as high responders (~100% as in B6), whereas B6xBc splenic B cells displayed a response similar to the low responder BALB/c strain (~30% activation). Concomitant triggering with both the TLR9 ligand CpG and LPS achieved ~90% activation BALB/c B cells, demonstrating that the LPS low response was a specific function of the LPS signaling pathway, rather than an inherent inability to induce B cell activation in the BALB/c cells. These data reveal a previous unsuspected behavior in the genetic control of the B cell response to LPS, with a diametrically opposing impact on splenic versus peritoneal B cells. No direct correlation between the response to LPS and the expression of R105 and MHC class II was observed, suggesting the existence of a yet unidentified genetic factor that contributes to the control the LPS signaling pathway in coelomic B cells.
MATERIAL AND METHODS
Animals
BALB/cJ, C57BL/6J, F1 (CB6F1/J, BALB/cJ Female x C57BL/6J Male), CB.17 and BC8 mice aged 8 to 12 weeks old were obtained from Instituto de Microbiologia, Universidade Federal do Rio de Janeiro, or from Department of Microbiology, University of Alabama at Birmingham. Rats were were obtained from Instituto de Microbiologia, Unversidade Federal do Rio de Janeiro. All experiments were approved by and performed in compliance with the regulations of the “Ethical Committee for Animal Studies” of the Instituto de Microbiologia, Universidade Federal do Rio de Janeiro and the IACUC of the University of Alabama at Birmingham.
Cells, flow-cytometry and cell sorting
Single cell suspensions were prepared from spleen by the gentle disruption of the organ in a Petri dish containing complete medium. PEC B cells were isolated from the peritoneal cavity by washing with 9 ml of complete RPMI 1640 medium (GIBCO) supplemented with 10% heat-inactivated FCS, 2 mM L-glutamine, 1mM Sodium Pyruvate, 50 μM 2-ME, 100 U penicillin, and 100 μg/ml streptomycin. Cells were washed and resuspended in an appropriate volume of RPMI for counting and staining. The percentage of B cells in each cell suspension was determined by flow cytometry analysis, gating on B220+ cells. For sorting, total peritoneal cavity cells were pooled and incubated at 2.5 × 108 cells/mL in RPMI medium containing the followings monoclonal Abs: anti-B220 (RA 3.6B2) PB (BD Pharmingen), anti-CD5 PE (53-7.3)(BD Pharmingen), and anti-Mac-1 FITC (M1/70) (BD Pharmingen). Splenic follicular B cells were sorted as B220+IgM+CD23+CD21−, and splenic marginal zone B cells were sorted as B220+IgM+CD23-CD21+. Anti-CD23 (B3B4), anti-CD21 (8D9), anti-IgM (II/41), anti-RP105 (RP/14) and anti-MHC II (M5/114.15.2) were from Ebioscience. Analysis and sorting were then performed on a MoFlo instrument (DakoCytomation) and the cells were collected directly in sterile tubes containing RPMI medium for further use in the cultures.
B cell culture and Limiting Dilution Assay (LDA)
B cells were cultured in 250μl of complete RPMI medium in 96 well flat-bottom in the presence of combinations of 30μg/mL of LPS (Salmonella typhimurium, Sigma-Aldrich), 1.5 μg/ml of CpG (ODN 1828, Invivogen), and 1.0 μg/ml of Pam3Cys (Invivogen). All cultures received 6 × 105 rat thymocytes or 3×103 S17 as growth-supporting cells/well. In the majority of the experiments the bone marrow-derived stromal cell line S17 was used as feeder cells (Collins and Dorshkind 1987). In the remaining minority of studies, thymocytes obtained from 4 weeks old Sprague–Dawley Rats were used as feeder cells instead. The preparation of S17 as feeder cell was done as follows. Briefly, one day before start the LDA cultures, plates were coated with 3 × 103 S17 per well and incubated overnight at 37°C with 5% CO2. The next day, the S17 culture plates were irradiated at 3000 rads and received the B cells at variable numbers in 48 replicates for each cell concentration, 18; 6; 2 and 0.66 B cell per well, to determine the frequency of IgM secreting clones by ELISA according to Poisson’s distribution (Andersson et al. 1977). Culture supernatants were typically harvested on the 9th day of culture.
ELISA
To determine the presence of IgM in the supernatants from cultures of secreting B cells, ELISA was performed using anti mouse IgM-specific reagents (Southern Biotechnology). Briefly, microplates (half-area, Costar, 96 well plate, #3690) were coated overnight at 4oC with 2.0 μg/ml of monoclonal anti-mouse IgM in PBS. The wells were washed with PBS and blocked with PBS 1% gelatin (MERCK, #345808) for 2 h at room temperature. Samples were diluted in PBS-1% gelatin/0.1% tween and incubated overnight at 4oC. The wells were then washed with PBS. Secondary antibody was diluted in PBS-1% gelatin/0.1% tween (SIGMA), and added for 2 h at room temperature. The wells were further washed with PBS and the reaction revealed with OPD substract (SIGMAFAST P9187). The reaction was stopped with 50 μl of HCl 1N and read at 490 nm.
RESULTS AND DISCUSSION
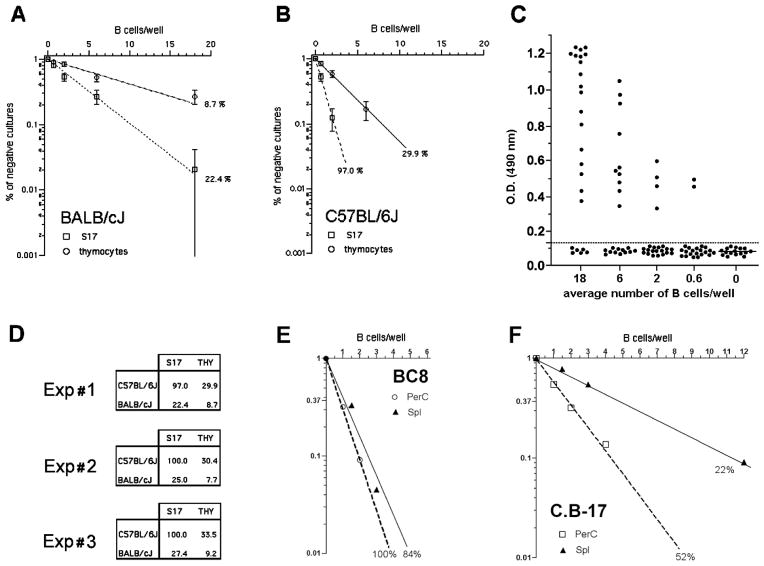
In vitro B cell activation by LPS depends not only on the direct triggering of B lymphocyte itself, but also on the presence of ill-characterized factors that support B cell growth and are provided by feeder cells. Indeed, splenic B cells do not grow in response to LPS stimulus if cultured below a critical density of close to 2000 cells/ml, which requires the presence of growth supporting feeder cells (Andersson et al. 1977). Thymocytes have been classically used as feeders cell types for low density B cell cultures (Andersson et al. 1977), but stromal cell lines (S17, OP9) are now routinely used in their place. These stromal cell lines show superior capacity to sustain B cell growth and differentiation. However, it is important to note that in higher density cell cultures the B cell itself may provide the feeder cell function. Therefore it is not possible to ascertain if differences in the response to LPS displayed by purified B cells from different mouse strains or cultured in bulk are exclusively due to genetic control on the B cell activation itself, or if it would also be due to the impact of a feeder cell effect. In order to eliminate this bias, we analyzed B cell response to LPS from high (C57BL/6) and low (BALB/c) responder mouse strains by culturing B lymphocytes under limiting dilution conditions down to single B cell/well using a common feeder cell type in all cultures. Figures 1A and 1B show the results obtained with splenic B cells from BALB/c and C57BL/6 mice that were cultured either with young rat thymocytes or S17 stroma cells as feeder cells. The B cell response was measured by the presence of significant amount of IgM in the supernatants in day 9 of culture (fig. 1C). In both cases, the frequency of B cells responding in the BALB/c strain was approximately threefold lower than in the B6 strain. However, in the presence of S17 feeders, close to 100 % of B6 B cells responded to LPS stimulus, demonstrating the major impact of the feeder cell factor. [The results obtained in three different experiments are shown in Figure 1D.] Thereafter S17 was chosen as feeder cells for all subsequent experiments described here.
FIGURE 1. Evaluation of feeder cell efficiency for single B cell cultures.
Splenic B cells were cultivated under limiting dilution conditions in the presence of LPS, 30μg/mL, using either rat thymocytes (6 × 105/well) or S17 stroma cell line (3×103/well) as feeder cells. The percentage of negative cultures is plotted against the B cell number/well, and frequencies of responding cells were calculated according to Poisson’s distribution. The growth of IgM secreting B cell clones was evaluated by the presence of IgM in the culture supernatant by ELISA from cells derived from (A) C57BL/6J or (B) BALB/c mice. OD values of individual culture wells are shown in (C). Culture wells were scored positive with OD values above the threshold line of 0.120 OD. Results from three independent experiments comparing the frequencies of response to LPS of C57BL/6J and BALB/c mice are shown in (D). (E) and (F) show the frequencies of response to LPS of congenic mouse strains BC8 and C.B-17.
As the parameter being used here to evaluate the response to LPS is the presence of secreted IgM, we thought it would be important to test if the variation in the heavy chain genes alone could modify the outcome of the limiting dilution assay. For that purpose, we evaluated the frequency of splenic B cells responding to LPS in the congenic mouse lineages BC8 (B6 background, congenic for the IgMa haplotype) and CB.17 (BALB/c background, congenic for the IgMb haplotype). We found that genetic variation in these loci had no effects on the frequencies of B lymphocytes responding to LPS. BALB/c and CB.17 showed similar lower frequencies of response, whereas C57Bl/6 and BC8 exhibited equivalent higher frequencies (fig. 1E). These results indicated that the differences in frequencies of B cell clonal growth to LPS are controlled by background genes outside the IgH loci.
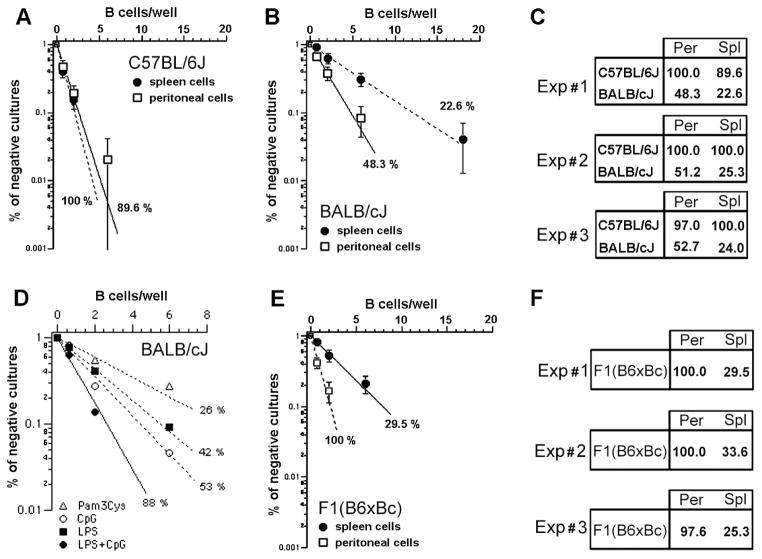
In short term B cell bulk cultures, the response to LPS varies according to different B cell populations, being maximal for MZ cells (Meyer-Bahlburg et al. 2008; Oliver et al. 1999). These data suggested that B cell populations might have distinct response to LPS in limiting dilution cultures as well. Our results confirm this hypothesis, with BALB/c PEC B cells demonstrating superior clonal growth when compared to their splenic counterpart (Figure 2A). Clonal growth also differed by genetic background with clonal growth close to 100 % in both PEC and splenic B cell populations from C57BL/6 mice (Figure 2B). This observation was confirmed in three independent experiments (Figure 2C). Concomitant administration of CpG, a TLR9 ligand, but not Pam3Cys, a TLR2 ligand, with LPS was able to nearly double the growth of B1a B cells from BALB/c, achieving a clonal expansion of ~90 % (Figure 2D). This latter experiment demonstrated that the absence of response to LPS is a specific effect that cannot be attributed to a general, inferior capacity of BALB/c B cells to grow in vitro.
FIGURE 2. Genetic control of LPS response in B cells from spleen or peritoneal cavity.
Splenic or peritoneal cavity B cells were cultivated under limiting dilution conditions in the presence of LPS, 30μg/mL, using S17 stroma cell line as feeder cells. The percentage of negative cultures is plotted against the B cell number/well, and frequencies of responding cells were calculated according to Poisson’s distribution. The results from three independent experiments comparing the frequencies of response to LPS of peritoneal and splenic B cells from (A) C57BL/6J and (B) BALB/c mice are shown in (C). Shown in (D) are the frequencies of response to different combinations of TLR ligands by sorted B1a cells. In (E), the frequencies of response to LPS of PEC and splenic B cells from F1 mice are depicted. (F) shows the results of three independent experiments with F1 B cells.
To begin to test the genetics behind the differences in response between the C57BL/6 and BALB/c backgrounds, we then compared the clonal growth of PEC versus splenic B cells in F1 (CB6F1/J, BALB/cJ Female x C57BL/6J Male) mice. We found that these populations suffered distinct and opposing genetic impacts (figure 2E). PEC B cells from the F1 mice exhibited near 100 % clonal growth, i.e. a response that was equal to the high responder B6 strain. In contrast, splenic B cells from the F1 heterozygotes displayed depressed clonal growth values close to those observed in the low responder BALB/c strain (Figure 2F).
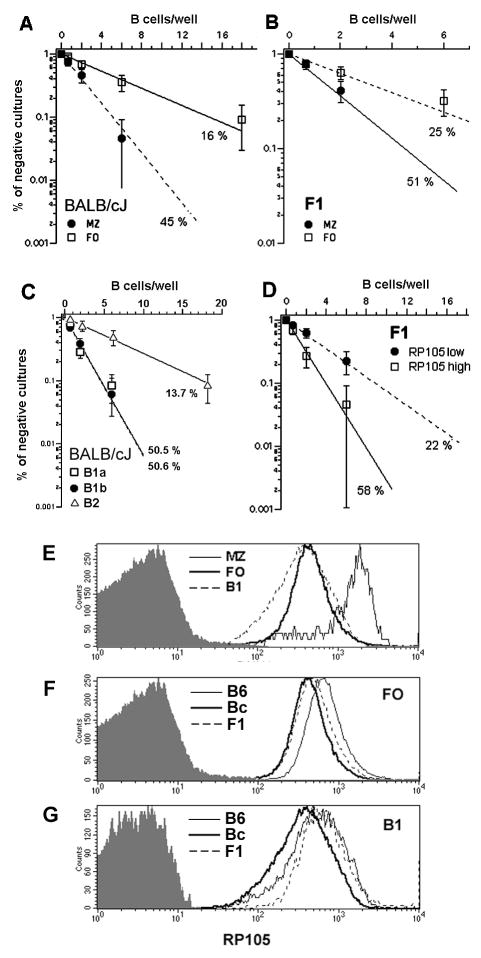
To better characterize the cells responding to LPS in BALB/c and F1 mice, we used flow cytometric cell sorting to isolate follicular (FO) and marginal zone (MZ) B cells from the spleen, and B1a, B1b and B2 were isolated from the peritoneal cavity. We then tested their clonal response to LPS. On a BALB/c background, B1a and B1b B cells demonstrated equivalent clonal growth (~50%) after stimulation, whereas B2 peritoneal cells displayed a clonal growth frequency similar to total splenic B cells (Figure 3A). From the spleen, MZ B cells responded better to LPS (~45 %) than FO B cells (~16 %) (Figure 3B). As FO B cells represent the majority of splenic B lymphocytes, these values are in agreement with the frequencies obtained for total splenic B cells (Figure 2). MZ B cells from F1 mice also responded better than FO B cells (Figure 3C), with frequencies similar to those observed for BALB/c.
FIGURE 3. Genetic control of LPS response in different B cell subpopulations from spleen and peritoneal cavity and the role of RP105.
Splenic or peritoneal cavity B cells from F1(B6xBc) were cultivated under limiting dilution conditions in the presence of LPS, 30μg/mL, using S17 stroma cell line as feeder cells. The percentage of negative cultures is plotted against the B cell number/well, and frequencies of responding cells were calculated according to Poisson’s distribution. (A) FO and MZ B cells from BALB/cJ; (B) FO and MZ B cells from F1; and (C) B1a, B1b and B2 peritoneal cavity B cells from BALB/cJ. In (D), FO B cells from F1 mice were sorted according to the expression of RP105, as RP105low and RP105high, and then evaluated for their clonal response to LPS. Expression levels of RP105 in the different B cell subpopulations was measured by flow-cytometry. (E) Acomparison of RP105 expression in FO, MZ and B1 from BALB/cJ. (F) A comparison of RP105 expression in FO from BALB/c, C57BL/6J and F1 mice. (G) A comparison of RP105 expression in B1 from BALB/c, C57BL/6J and F1 mice. The shaded histograms indicate the background staining with control antibody isotype.
Recently it has been shown that the MHC locus has a strong impact in the B cell response to LPS, probably through the regulation of the expression of RP105 (Rodo et al 2006). Expression of RP105 has been previously shown to be substantially augmented in MZ B cells (Nagai et al. 2005). We thus evaluated PEC B1 cells, and splenic FO to determine whether the surface expression of RP105 might vary between these populations as well. We found that PEC B1 and B2 B cells showed levels of RP105 that were equivalent to those expressed by splenic FO B cells (Figure 3E). This same pattern of expression was observed for the all mouse strains studied here. Between mouse strains, RP105 expression by splenic FO and MZ B cells were lower in BALB/c than C57BL/6, with levels in the F1 heterozygotes that were intermediate between the two parental strains (Figure 3F). In contrast, while RP105 expression in BALB/c PEC B1 B cells was also lower than C57BL/6, the F1 heterozygotes expressed RP105 at levels equivalent to the C57BL/6 parent (Figure 3G).
The much higher expression of RP105 in MZ B cells could contribute to their fast response to LPS in bulk cultures (Oliver et al. 1999). And, in support of this view, we found that splenic MZ B cells from BALB/c and F1 mice showed higher clonal frequencies of response to LPS than FO B cells (Figure 3A and 3B). However, in limiting dilution cultures both FO and MZ splenic B cells from C57BL/6 mice responded equally well to LPS, indicating that variation in the expression of RP105 did not affect the splenic B cell response in this mouse strain. To examine this issue further, we sorted FO B cells from F1 mice according to high or low expression of RP105. We found that RP105low FO B cells responded with a clonal frequency of 22 %, whereas RP105high FO B cells showed a higher clonal response frequency of 58% (Figure 3D), thus supporting a role for RP105 in the augmented response to LPS in limiting dilution cultures in these heterozygotes. However, in BALB/c, the MZ B cells responded at a ~50% rate to LPS, matching the pattern seen in B1 B cells even though the latter showed much lower expression of RP105. Indeed, the expression of RP105 was equivalent to that of FO B cells, which exhibited a lower clonal frequency response of ~ 15%. Among F1 mice, PEC B cells responded 100 % to LPS, although exhibiting lower expression of RP105 than MZ cells that scored 51 %. Thus, a general correlation between levels of expression of RP105 and clonal frequencies of response to LPS could not be established.
Although the response to LPS in limiting dilution assays in B1 cells did not correlate with the levels of expression of RP105, these PEC B cells exhibited a super response. This suggests that other factors may be be involved in the control of this response in PEC B cells. The opposite effect observed for peritoneal versus splenic B cells of F1 mice indicates a multifactorial and cell lineage specific genetic control of the response to LPS. The preferential activation of B1 cells by LPS is in line with the proposal that these cells would have been selected in evolution to participate in innate immunity through the generation of “natural antibodies” (Casali and Schettino 1996; Herzenberg 1989), which provide immediate protection against bacterial infections (Briles et al. 1981). The same argument may apply to MZ B cells, whose phenotypic and functional similarities to B1 cells have been described (Martin and Kearney 2002).
Peritoneal B1a cells have been shown to derive from a distinct B cell precursor that is abundant in the fetal liver and neonatal bone marrow but progressively disappears during ontogeny (Montecino-Rodriguez et al. 2006). It is interesting to note that the absence of molecules involved in the BCR signaling affects primarily the formation of the B1a cell population (Desiderio 1997; Sato et al. 1996), suggesting that BCR signaling machinery operates with different threshold in B1a cells. It seems reasonable to speculate that B1a cells may also respond better to LPS because of an altered threshold for their activation through TLR4/RP105. The same argument may apply to B1b peritoneal cells, whose hematopoietic origins are still unclear. The different embryonic origin of those lymphocytes may be related to their higher response to LPS through the expression of accessory molecules that participate in the signaling cascades triggered by LPS, or with their regulation by other genes, e.g. MHC II, as suggested (Rodo et al. 2006). Interestingly, fetal liver B cell precursors, which tend to be the progenitors of PEC B1a cells, totally lack the expression of MHC molecules, which are only expressed several days after the maturation of the precursor into a B lymphocyte (Lam and Stall 1994).
In conclusion, the data presented here revealed a previous unsuspected behavior in the genetic control of the B cell response to LPS, with a differential impact in splenic B cells versus peritoneal cavity B1 cells. These results suggest new experimental approaches to characterize the mechanisms controlling TLR4 signaling pathway in coelomic B cells.
Acknowledgments
This work was supported by CNPq, FAPERJ and FINEP and in part by AI048115 (HWS) and AI078449 (HWS). AMV was supported by a fellowship from CAPES and CNPq.
Footnotes
The authors declare that they have no competing financial interests.
References
- Andersson J, Coutinho A, Lernhardt W, Melchers F. Clonal growth and maturation to immunoglobulin secretion in vitro of every growth-inducible B lymphocyte. Cell. 1977;10:27–34. doi: 10.1016/0092-8674(77)90136-2. [DOI] [PubMed] [Google Scholar]
- Briles DE, Nahm M, Schroer K, Davie J, Baker P, Kearney J, Barletta R. Antiphosphocholine antibodies found in normal mouse serum are protective against intravenous infection with type 3 streptococcus pneumoniae. J Exp Med. 1981;153:694–705. doi: 10.1084/jem.153.3.694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casali P, Schettino EW. Structure and function of natural antibodies. Curr Top Microbiol Immunol. 1996;210:167–79. doi: 10.1007/978-3-642-85226-8_17. [DOI] [PubMed] [Google Scholar]
- Collins LS, Dorshkind K. A stromal cell line from myeloid long-term bone marrow cultures can support myelopoiesis and B lymphopoiesis. J Immunol. 1987;138:1082–7. [PubMed] [Google Scholar]
- da Silva Correia J, Soldau K, Christen U, Tobias PS, Ulevitch RJ. Lipopolysaccharide is in close proximity to each of the proteins in its membrane receptor complex. transfer from CD14 to TLR4 and MD-2. J Biol Chem. 2001;276:21129–35. doi: 10.1074/jbc.M009164200. [DOI] [PubMed] [Google Scholar]
- Desiderio S. Role of Btk in B cell development and signaling. Curr Opin Immunol. 1997;9:534–40. doi: 10.1016/s0952-7915(97)80107-0. [DOI] [PubMed] [Google Scholar]
- Herzenberg LA. Toward a layered immune system. Cell. 1989;59:953–4. doi: 10.1016/0092-8674(89)90748-4. [DOI] [PubMed] [Google Scholar]
- Hoshino K, Takeuchi O, Kawai T, Sanjo H, Ogawa T, Takeda Y, Takeda K, Akira S. Cutting edge: Toll-like receptor 4 (TLR4)-deficient mice are hyporesponsive to lipopolysaccharide: evidence for TLR4 as the Lps gene product. J Immunol. 1999;162:3749–52. [PubMed] [Google Scholar]
- Kalis C, Kanzler B, Lembo A, Poltorak A, Galanos C, Freudenberg MA. Toll-like receptor 4 expression levels determine the degree of LPS-susceptibility in mice. Eur J Immunol. 2003;33:798–805. doi: 10.1002/eji.200323431. [DOI] [PubMed] [Google Scholar]
- Kawai TaAS. TLR signaling. Semin Immunol. 2007;19:9. doi: 10.1016/j.smim.2006.12.004. [DOI] [PubMed] [Google Scholar]
- Lam KP, Stall AM. Major histocompatibility complex class II expression distinguishes two distinct B cell developmental pathways during ontogeny. J Exp Med. 1994;180:507–16. doi: 10.1084/jem.180.2.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin F, Kearney JF. Marginal-zone B cells. Nat Rev Immunol. 2002;2:323–35. doi: 10.1038/nri799. [DOI] [PubMed] [Google Scholar]
- Medzhitov R. Toll-like receptors and innate immunity. Nat Rev Immunol. 2001;1:135–45. doi: 10.1038/35100529. [DOI] [PubMed] [Google Scholar]
- Meyer-Bahlburg A, Andrews SF, Yu KO, Porcelli SA, Rawlings DJ. Characterization of a late transitional B cell population highly sensitive to BAFF-mediated homeostatic proliferation. J Exp Med. 2008;205:155–68. doi: 10.1084/jem.20071088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montecino-Rodriguez E, Leathers H, Dorshkind K. Identification of a B-1 B cell-specified progenitor. Nat Immunol. 2006;7:293–301. doi: 10.1038/ni1301. [DOI] [PubMed] [Google Scholar]
- Nagai Y, Kobayashi T, Motoi Y, Ishiguro K, Akashi S, Saitoh S, Kusumoto Y, Kaisho T, Akira S, Matsumoto M, Takatsu K, Miyake K. The radioprotective 105/MD-1 complex links TLR2 and TLR4/MD-2 in antibody response to microbial membranes. J Immunol. 2005;174:7043–9. doi: 10.4049/jimmunol.174.11.7043. [DOI] [PubMed] [Google Scholar]
- Ogata H, Su I, Miyake K, Nagai Y, Akashi S, Mecklenbrauker I, Rajewsky K, Kimoto M, Tarakhovsky A. The toll-like receptor protein RP105 regulates lipopolysaccharide signaling in B cells. J Exp Med. 2000;192:23–9. doi: 10.1084/jem.192.1.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver AM, Martin F, Kearney JF. IgMhighCD21high lymphocytes enriched in the splenic marginal zone generate effector cells more rapidly than the bulk of follicular B cells. J Immunol. 1999;162:7198–207. [PubMed] [Google Scholar]
- Poltorak A, He X, Smirnova I, Liu MY, Van Huffel C, Du X, Birdwell D, Alejos E, Silva M, Galanos C, Freudenberg M, Ricciardi-Castagnoli P, Layton B, Beutler B. Defective LPS signaling in C3H/HeJ and C57BL/10ScCr mice: mutations in Tlr4 gene. Science. 1998;282:2085–8. doi: 10.1126/science.282.5396.2085. [DOI] [PubMed] [Google Scholar]
- Rodo J, Goncalves LA, Demengeot J, Coutinho A, Penha-Goncalves C. MHC class II molecules control murine B cell responsiveness to lipopolysaccharide stimulation. J Immunol. 2006;177:4620–6. doi: 10.4049/jimmunol.177.7.4620. [DOI] [PubMed] [Google Scholar]
- Sato S, Ono N, Steeber DA, Pisetsky DS, Tedder TF. CD19 regulates B lymphocyte signaling thresholds critical for the development of B-1 lineage cells and autoimmunity. J Immunol. 1996;157:4371–8. [PubMed] [Google Scholar]
- Sporri R, Reis e Sousa C. Inflammatory mediators are insufficient for full dendritic cell activation and promote expansion of CD4+ T cell populations lacking helper function. Nat Immunol. 2005;6:163–70. doi: 10.1038/ni1162. [DOI] [PubMed] [Google Scholar]