Abstract
To assess the extent and nature of somatic categorical selection of CDR-H3 content in peritoneal cavity (PerC) B cells, we analyzed the composition of VH7183DJCμ transcripts derived from sorted PerC B-1a, B-1b, and B-2 cells. We divided these sequences into those that contained N nucleotides (N+) and those that did not (N−), and then compared them to sequences cloned from sorted IgM+IgD+ B cells from neonatal liver and both wild-type and TdT deficient adult bone marrow. We found that the PerC B-1a N− repertoire is enriched for the signatures of CDR-H3 sequences present in neonatal liver and shares many features with the B-1b N− repertoire, whereas the PerC B-1a N+, B-1b N+ and B-2 N+ repertoires are enriched for adult bone marrow sequence signatures. However, we also found several sequence signatures that were not shared with other mature perinatal or adult B cell subsets; but were either unique or variably shared between the two or even among all three of the PerC subsets that we examined. These signatures included an increased number of sequences lacking N nucleotides in the B-2 population and an increased use of DH reading frame 2 which created CDR-H3s of greater average hydrophobicity. These findings provide support for both ontogenetic origin and shared antigen receptor-influenced selection as the mechanisms that shape the unique composition of the B-1a, B-1b and B-2 repertoires. The peritoneal cavity may thus serve as a general reservoir for B cells with antigen binding specificities that are uncommon in other mature compartments.
Keywords: B cells, Antibodies, Gene Rearrangement, Repertoire Development
Introduction
Complementarity determining region-3 of the heavy chain (CDR-H3) lies at the center of the antigen-binding site of the antibody, as classically defined. This central location allows CDR-H3 to often play a critical role in the recognition and subsequent immune response to an antigen (1, 2). CDR-H3 is also the focus for the initial diversity of the antibody repertoire. Each individual CDR-H3 is created de novo by the combinatorial rearrangement of VH, DH and JH gene segments in developing B cells of the fetal and neonatal liver (NL) and of the post-natal bone marrow (BM). In addition to the combinatorial diversity provided by arrays of V, D and J gene segments, the D→J and V→D junctions are further somatically diversified by the variable loss or palindromic (P) gain of terminal gene segment nucleotides (3) and, in post natal tissues, the random addition of N nucleotides (4, 5).
Although at first glance VDJ rearrangement and N addition would appear to permit unrestricted CDR-H3 diversity, the CDR-H3 repertoires expressed by specific B cell subsets often exhibit characteristic categorical constraints. These may include biases in VDJ gene segment usage, in DH reading frame preference, and in the number (i.e. length) and physicochemical properties (e.g. hydrophobicity or charge) of the encoded amino acids (6–8). Some of these categorical constraints are genetically predetermined (9–13). Others are progressively imposed by selective mechanisms as developing B cells pass through critical developmental checkpoints in both primary and secondary lymphoid tissues (6, 14–16). In either case, these biases create what appear to be preferred ranges of potential structures and antigen binding complementarity surfaces in each B cell subset. For example, in previous studies, we identified somatically imposed sequence signatures that distinguished the range of CDR-H3 repertoires expressed in the spleen by marginal zone (MZ) B cells from those expressed by follicular (FO) B cells (14).
In the present work, we have sought to test whether B cells residing in the peritoneal cavity (PerC) also display evidence of categorical repertoire selection. We specifically sought to compare the repertoires expressed by PerC B-1a and B-1b cells, both major sources of natural antibodies, to that of PerC B-2 cells, which have long been assumed to correspond to the conventional, recirculating mature B cell pool (17), as exemplified by the IgM+IgD+ B cells of the bone marrow. Some previous studies have identified alternative lineages of differing ontogenetic origin as the basis for B-1a, B-1b and B-2 cell repertoire diversity (18, 19), whereas others have found evidence of a role for somatic selection of the B cell receptor based on antigen engagement and signal strength (20, 21). Having recently performed an analysis of the pattern of categorical selection exhibited by the CDR-H3 repertoires expressed in BALB/c perinatal liver and in TdT deficient adult bone marrow (15, 16), we postulated that a comparative analysis of PerC B cell CDR-H3 repertoires might yield new insights into the derivation and selective pressures that give rise to the signature range of immunoglobulin repertoire diversity within these key PerC subsets.
Our analysis of the immunoglobulin H chain sequences reported in this work provides support for both the ontogenetic and the antigen driven somatic selection models of PerC B cell repertoire development. In support of the ontogenetic model, we found that each PerC B cell subset includes aspects of CDR-H3 signatures that are characteristic of their presumed ontogenetic origin, i.e. perinatal or adult. However, we also found support for the somatic selective model in that the CDR-H3 repertoires of B-1a, B-1b, and B-2 cells appear to share sequence characteristics that distinguish them from the repertoires expressed by mature B cells of either the perinatal liver or the post-natal bone marrow. The latter results raise the possibility that B cells expressing BCRs enriched for specific categories of CDR-H3 amino acid sequence content are favored for entry or survival in the PerC, or both; and that this selective advantage occurs regardless of ontogenetic origin or the expression of CD5.
Materials and Methods
Mice
The mice analyzed represent the progeny of a mixed 129/C57BL6 forbearer that had been backcrossed for ten generations onto BALB/cJ (Stock No. 000651; Jackson Laboratories, Bar Harbor, ME.) (6). Our initial analysis of bone marrow CDR-H3 repertoire development had been performed on four separate mice derived from the same breeding pool (6). Additional samples were independently obtained from five individual 8–10-wk-old BALB/cJ mice bred in our own mouse colony (6, 11, 12, 14). All studies were performed in accordance with University of Alabama at Birmingham Institutional Animal Care and Use Committee regulations.
Flow cytometry and cell sorting
Peritoneal B cells were isolated from two different mice. Single-cell suspensions were prepared by washing the peritoneal cavity with 10 mL of ice-cold FACS buffer (1x PBS with 2% heat-inactivated fetal calf serum). Cells were washed and resuspended in an appropriate volume of FACS buffer for counting and staining. Total peritoneal cells from individual mice were incubated in 100 μl of fluorescently labeled antibodies in FACS buffer. Sorting was performed on a MoFlo instrument (DakoCytomation, Ft. Collins, CO). The following mAbs were used to isolate peritoneal B cells into the B-1a, B-1b and B-2 subpopulations (Supplemental Fig. 1) anti-IgM (Cy5) (Jackson ImmunoResearch, West Grove, PA), anti-CD19 (Spectral Red) (Southern Biotechnology, Birmingham, AL), anti-Mac-1 (FITC) (BD Pharmingen, San Diego, CA) and anti-CD5 (PE) (BD Pharmingen, San Diego, CA).
Sorting, RNA preparation, RT-PCR and sequencing
For each peritoneal B cell population, 2×104 cells were sorted directly into RLT lysis buffer (RNeasy mini-kit; Qiagen, Valencia, CA). Because the lysis precluded reanalysis of the cells, we routinely used companion unlysed sorted cells to confirm the purity of the sorted populations. Based on “tight gating” for surface expression of IgM, CD19, Mac-1 and CD5, this purity of the populations was typically 99% for all peritoneal B cell subsets (data not shown).
RNA isolation, RT-PCR amplification, and sequencing were performed as previously described (6). A listing of the 383 unique, in-frame VH7183DJCμ sequences used for analysis in this work is provided in Supplemental Table I. All unique sequences have been placed in the GenBank database (accession numbers HM132462-HM132836). The sequences from adult bone marrow fraction F (BM F) used in this work were previously reported (6, 11, 12, 14). The sequences from neonatal liver fraction F (NL F) used for comparison were previously reported (15), as were the sequences from TdT deficient adult bone marrow (16).
Sequence analysis
CDR-H3 was identified as the region between (but not including) the 3′ terminal VH- TGT codon for the conserved Cys at Kabat (1) position 92 (IMGT 104) and the 5′ terminal JH TGG codon for the conserved Trp at Kabat position 103 (IMGT 118). CDR-H3 was separated into two components, the base [Kabat amino acids 93 and 94 (IMGT 105 and 106; typically alanine and arginine) and Kabat amino acids 100–102 (IMGT 115–117; typically phenylalanine, aspartic acid, and tyrosine] and the loop [the intervening amino acids].
Structural Analysis
We used the “H3” rules, as published by Shirai (22, 23) to predict structural features of the CDR-H3 base and loop, as previously described (14). Briefly, the structure of the CDR-H3 base (termed kinked, extra-kinked, or extended) can be predicted in sequences that contain a minimum of five amino acid residues, including IMGT positions 105–118 (Kabat positions 93–103). In approximately 25–30% of the sequences with a kinked or extra-kinked CDR-H3 base, the H3-rules can predict whether an intact hydrogen bond ladder may be formed within the loop of the CDR-H3 region or whether the hydrogen bond ladder is likely to be broken. For example, proline residues tend to inhibit formation of a stable hydrogen bond ladder, whereas the presence of a VH-encoded arginine at the amino terminus of CDR-H3 in conjunction with a JH-encoded aspartic acid at the C terminus permits formation of a salt bridge that stabilizes the base. Glycine residues permit greater flexibility (23).
Statistical analysis
Differences between populations were assessed by Student’s t-test, two tailed; Fisher’s exact test, two tailed; χ2-test; or Levene’s tests for the homogeneity of variance, as appropriate. Analysis was performed with JMP version 7.0 (SAS Institute, Inc., Cary, NC). Means are reported with the standard error of the mean.
Results
Creation of a PerC B-1a, B-1b and B2-derived VH7183DJCμ sequence database
Initial studies of PerC CDR-H3 sequences identified the extent of N nucleotide addition as a characteristic feature that could be used to distinguish between immunoglobulin sequences derived from PerC B-1a (IgMhiCD19+Mac-1−/+CD5+) B cells versus those obtained from the alternative B-2 (IgMloCD19+Mac-1− CD5−) population (24–26). B-1a cells characteristically demonstrate a paucity of N addition, whereas B-2 cells demonstrate an extensive array of N nucleotides in CDR-H3. These classic observations contributed greatly to the perception that many PerC B-1a cells were the progeny of fetal progenitors, which lack TdT activity; whereas B-2 cells were the progeny of post-natal bone marrow-derived progenitors. However, in the absence of a detailed comparative analysis of the CDR-H3 repertoires expressed by conventional bone marrow B cells; these initial studies left open the question of whether the PerC B-2 repertoire was unrestrictedly drawn from the conventional B cell repertoire, as represented by the mature, recirculating bone marrow IgM+IgD+ B cells (Hardy fraction F) (27); or whether the B-2 subset might also be subject to somatic PerC-specific categorical selection.
To distinguish between these possibilities, we used RT-PCR to randomly clone VH7183DJCμ transcripts from sorted peritoneal cavity B-1a, B-1b (IgMhiCD19+Mac-1+CD5−), and B-2 cells. We chose to focus our studies on the VH7183 family because it comprises 10% of the repertoire, it is a VH family that is expressed in both fetal and adult B cell progenitors, and it does not contain any of the iconic PerC sequences that are over-represented in transcripts using members of the VH11, VH12 or VHS107 families (28–30). Indeed, we have generated an extensive database of VH7183 expressed by multiple other tissues and B cell subsets that can be used as a standard for comparison. These comparison populations include mature CD19+sIgMlosIgDhi B cells from neonatal liver (NL F ) and adult bone marrow fraction F (BM F).
For the present study, we obtained a total of 379 in-frame, open, and unique sequences (See Supplemental Table I). Of these, 147 were derived from sorted B-1a cells, 118 from B-1b (CD19hiMac-1+CD5−) and 114 from B-2 (Supplemental Fig. 1). We compared these sequences as a population to 254 unique BM F sequences (6, 11, 12, 14) and 132 unique sequences from neonatal liver CD19+sIgMlosIgDhi fraction F (NL F) (15). Also available for comparison were 167 sequences from fraction B (pro-B), 313 from C (early pre-B), 229 sequences from D (late pre-B) and 203 from E (immature B) from wild-type TdT sufficient bone marrow; 73 unique sequences from TdT deficient bone marrow fraction F (BM F TdT−/−); and 102 unique CDR-H3 sequences from splenic transitional T1, 97 unique CDR-H3 sequences from splenic transitional T2, 97 unique CDR-H3 sequences from splenic follicular, and 151 unique CDR-H3 sequences from splenic marginal zone B cells (6, 11, 12, 14, 16).
Given that the absence of N addition in many B-1a CDR-H3 sequences is typically attributed to their fetal origin (31), we used N addition as an additional variable by grouping sequences into those that lacked N nucleotides (N−) and those that contained them (N+). These additional comparative groups included PerC B-1a, B-1b and B-2 N− and N+ sequences, NL F cells N− and N+ sequences, and BM F N+ sequences. Only five of the 254 (2%) adult BM F sequences from the TdT sufficient wild-type mice lacked N nucleotides, a number too small to permit a meaningful comparison. Thus, to control for the effect of an absence of N nucleotides on the adult bone marrow repertoire, we further included 62 N− sequences obtained from TdT deficient bone marrow (16), with a focus on mature fraction F (BM F TdT−/− N−).
N− sequences are more common in the PerC B-2 subset than in bone marrow fraction F
In accordance with previous observations (26), when compared to the B-1a CDR-H3 repertoire the B-2 CDR-H3 repertoire contained more sequences with N nucleotides (p<0.0001) (Fig. 1). However, when compared to the CDR-H3 repertoires of bone marrow immature B cells (Fraction E), mature BM F B cells or splenic FO B cells, the PerC B-2 repertoire proved to also be five to seven-fold enriched for CDR-H3s that lacked N regions (p<0.001, χ2). The increased prevalence (15%) of N− CDR-H3s in the B-2 repertoire lay intermediate between that of the PerC B-1b (23%) and the splenic MZ (10%) repertoires (p=0.15 and p=0.24, respectively; Fig 1). [The difference in N addition between B-1b and MZ achieved statistical significance at p=0.007, as did the difference between B-1a (39%) and B-1b (23%, p=0.005).]
Figure 1. Frequency of N addition at both VH-D and D-JH junctions among VH7183DJCμ transcripts isolated from 1-day liver, bone marrow, peritoneal cavity and splenic B cells.
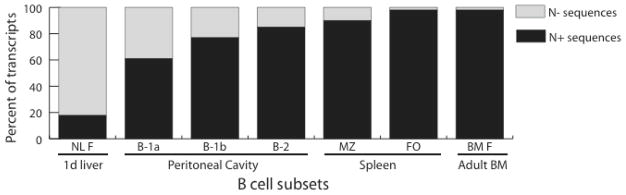
The frequency of sequences containing N additions is reported as the percent of the sequenced CDR-H3 (containing identifiable D gene) transcripts from 1-day liver Hardy fraction F (NL F) (15), adult bone marrow Hardy fraction F (BM F) (6, 27), peritoneal cavity (PerC) B-1a, B-1b and B-2 (Supplementary Figure 1), and spleen follicular (FO) and marginal zone (MZ) B cells (14). χ2 analyses demonstrate that the frequency of B-1a N− sequences differ significantly from B-1b (p=0.005) and all other B cell subsets (p<0.0001). The B-1b population differs from BM F, FO (p<0.0001) and MZ (p=0.007) but does not differ from B-2 (p=0.152). The B-2 population differs from BM F (p<0.0001) and FO (p=0.001), but does not differ from MZ (p=0.247).
Increased use of VH7183.18 in all three of the peritoneal B cell subsets examined
Differences in the immunoglobulin repertoires expressed by the PerC B cell subsets extended beyond CDR-H3 to the V domain as a whole, as represented by VH usage. The fetal liver H chain repertoire is heavily enriched for use of members of the DH-proximal VH7183 family (32–34). Within that family, which numbers 17 active members in the BALB/c IgMa haplotype, VH81X (VH7183.1) is the most highly expressed. For example, in our database of perinatal liver VH7183-containing sequences, VH81X-containing transcripts comprised 10% of the NL F B cell VH7183-containing clones (Fig. 2, top and (15)). When compared as a whole irrespective of N addition, only 2% (p<0.01) of the B-1a, B-1b or B-2 cloned cDNAs used VH81X. This matched the prevalence of this ‘fetal’ sequence in the CDR-H3 repertoire of mature adult B cells (BM F; Fig. 2, top).
Figure 2. VH usage of VH7183DJCμ transcripts isolated from sorted 1-day liver, peritoneal cavity and bone marrow mature B cells.
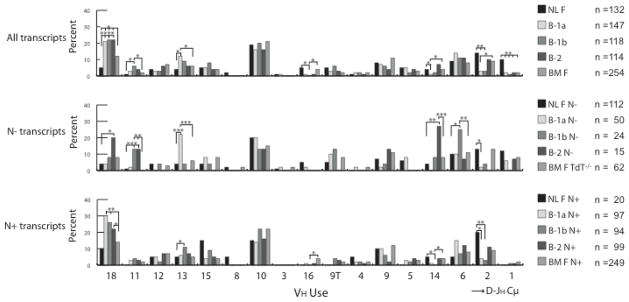
VH usage is reported as the percent of the sequenced population of unique, in-frame, open transcripts from neonatal liver fraction F (NL F - mature), peritoneal cavity (PerC) B-1a, B-1b and B-2 (Supplementary Figure 1), and bone marrow fraction F (BM F - mature, recirculating) CDR-H3s from either wild type or TdT−/− mice (BM F TdT−/−). The VH segments are arranged in germline order with the most DH proximal sequences to the right. The number of sequences analyzed is shown. (top) all VH7183 transcripts, (middle) VH7183 transcripts from PerC B cells CDR-H3 without N nucleotide addition (N−), (bottom) VH7183 transcripts from PerC B cell CDR-H3 containing N nucleotide addition (N+). All comparisons were made to fraction F, either from 1 day liver (NL F) or bone marrow (BM F) using χ2-test or Fisher’s exact test as appropriate. Significant differences between PerC B cell subsets and both fraction F are labelled with asterisks with *p≤0.05, **p≤0.01, *** p≤0.001, **** p≤0.0001.
In young adults, bone marrow fraction E (BM E) and BM F B cells preferentially express VH7183.10 (6)., and this VH continues to be over-expressed in both the FO and MZ populations of the spleen (6, 14). In the PerC B cell subsets, VH7183.10 usage was equivalent to that observed in BM E, BM F, splenic FO and splenic MZ (Fig. 2 and data not shown). Thus, when taken as a whole, all three PerC populations were marked by the two major VH usage characteristics most representative of the repertoires expressed by a large array of adult B cell subsets.
However, although the pattern of usage of VH81X and VH7183.10 was similar to BM F and other adult subsets, several other VH gene segment signatures were found that were shared by all three PerC subsets, but not by the bone marrow or spleen B cells. For example, the most prominent member of the VH7183 family expressed by the PerC B cells proved to be VH7183.18. It contributed to more than 20% of PerC B cell sequences (p<0.001 and p<0.05 when compared to NL F and BM F, respectively).
The usage of individual VH7183 gene segments in fetal versus adult B cell progenitors has been found to be heavily influenced by their physical location relative to the DH locus (35, 36). Fetal cells preferentially use DH proximal VH gene segments rather than DH distal ones. VH7183.18 is the most DH distal member of the VH7183 family, again suggesting an ‘adult’ influence on the repertoire of all three PerC B cell subsets. Intriguingly, the transitional T1 B cells of the spleen also exhibited a high prevalence of the use of this gene segment (17%, data not shown), suggesting that either there might be shared selection pressures acting on T1 and PerC B cells or that the transitional 1 stage is a branch point for selection (14).
VH7183 gene usage in N− B-1a transcripts followed the perinatal pattern
When the three PerC B cell repertoires were analyzed by the presence or absence of N nucleotides, the extent of correlation between ‘fetal’ and ‘adult’ usage was found to differ by subset. Among the B-1a-derived clones, the pattern of VH usage in the N− repertoire proved highly similar to NL F (Fig. 2, middle), demonstrating an increased use of VH81X and decreased use of VH7183.10. Indeed, only two of the other fifteen VH7183 gene segments diverged significantly in prevalence from that observed in NL F B cells. Use of VH7183.13, which is DH-distal, was uniquely significantly increased; whereas use of VH7183.2, which is DH-proximal, was significantly decreased but matched that of the N− B-1b and B-2 repertoires. Among the N+ compartment, use of VH81X and VH7183.10 followed the pattern expressed by adult BM F B cells; and enhanced use of DH-distal VH7183.18 was evident.
Unlike B-1a, the B-1b-derived N− repertoire followed an ‘adult’, rather than a ‘fetal’, pattern, with VH usage closely resembling that of adult BM F TdT−/− N− sequences. None of these N− sequences used VH81X, and use of VH7183.10, and VH7183.18 matched that of TdT− BM F N−. Two outliers in the N− subset were noted: VH7183.6 in B-1b and VH7183.14 in B-2, both of which are DH proximal. The N+ PerC B-1b-derived VH repertoire also resembled BM F more than NL F (Fig 2, middle). Among the sequence signatures that diverged from BM F, an increased use of DH distal VH7183.18 was prominent. DH distal VH7183.13 was also increased, whereas use of DH proximal VH7183.16 and VH7183.14 was decreased.
Among the B-2 N− derived sequences, the pattern of VH usage, including VH81X and VH7183.10, again largely matched that of BM F TdT−/− N−. Exceptions included increased use of DH distal VH7183.18 and VH7183.11. Not unexpectedly, VH usage among the B-2 N+ sequences largely matched WT BM F. However, the N+ repertoire was again enriched for the use of DH distal VH7183.18; and there was also a decrease in the use of DH proximal VH7183.16 (Fig. 2).
Altered patterns of DH and JH usage in B-2 cells
Signature differences in individual DH and JH gene segments are also apparent in the fetal and adult CDR-H3 repertoires (15). Perinatal B cells use the DQ52 gene segment at a greater frequency than adults; whereas adult B cells use DFL family gene segments more frequently than perinatal B cells.
We examined DH usage in the presence or absence of N nucleotides and found that both the B-1a and B-1b N− repertoires contained DQ52 more frequently than the B-1a N+ and B-1b N+ cells, respectively; whereas B-2 cells matched the pattern of adult bone marrow DQ52 usage irrespective of the presence or absence of N nucleotides (Fig. 3, middle). With regards to DFL gene segments, B-1a and B-1b B cells demonstrated an adult usage pattern, and B-2 cells used DFL gene segments more frequently than adult bone marrow again irrespective of the presence or absence of N nucleotides. The increased use of DFL family members in B-2 cells achieved statistical significance when compared to B-1a (p<0.01), B-1b (p<0.01), NL F (p<0.01) and BM F (p<0.05; Fig 3).
Figure 3. DH, DH reading frame and JH usage of VH7183DJCμ transcripts isolated from sorted 1-day liver, peritoneal cavity and bone marrow mature B cells.
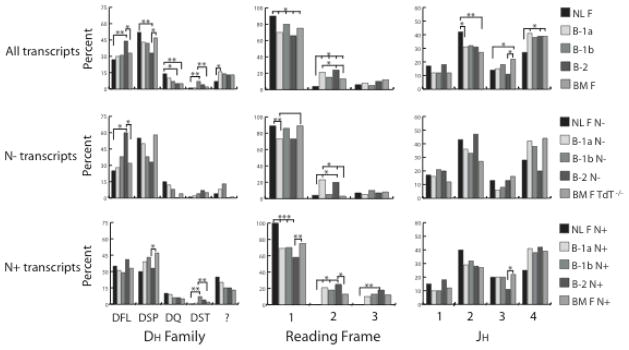
DH and JH usage is reported as the percent of the sequenced population of unique, in-frame, open transcripts from neonatal liver fraction F (NL F - mature), peritoneal cavity (PerC) B-1a, B-1b and B-2, and bone marrow fraction F (BM F - mature, recirculating) CDR-H3 from either wild type or TdT−/− mice (BM F TdT−/−) which use the particular DH or JH. DH reading frame usage is reported as the percent of the sequenced population of DFL- and DSP-containing transcripts from each B cell population that use the specified reading frame. (top) all VH7183 transcripts; (middle) VH7183 transcripts from PerC B cells CDR-H3 without N nucleotide addition (N−); (bottom) VH7183 transcripts from PerC B cell CDR-H3 containing N nucleotide addition (N+). All comparisons were made to fraction F, either from 1 day liver (NL F) or bone marrow (BM F) using χ2-test or Fisher’s exact test as appropriate. Significant differences between PerC B cell subsets and both fraction F are designated labelled with asterisks, with *p≤0.05, **p≤0.01, *** p≤0.001, **** p≤0.0001.
Within the JH locus, perinatal B cells demonstrate a relative preference for use of JH2, whereas adult cells use JH3 and JH4 more frequently. While B-1a N− sequences demonstrated an increased use of JH2 and a diminished use of JH3 and JH4 relative to adult, the differences in JH2 between B-1a and NLF were still significant at p < 0.05 (Fig 3, top). Among the N+ sequences, no differences in JH usage were observed between B-1a and BM F N+ or BM F N−. B-1b cells followed the same pattern, at the same level of significance. Among the B-2 cells, there was a trend for the N− sequences to be closer in JH usage to NL F than to BMF, but these trends did not achieve statistical significance. No differences were observed among the N+ repertoires.
DH reading frame usage also diverged from a direct fetal/adult paradigm. DH gene segments can theoretically be read in any one of six reading frames, three by deletion and three by inversion. However, a preference for use of reading frame 1 by deletion is a near universal facet of DH usage in all jawed vertebrates (6). This bias is greatest in the perinatal period (15). While the three PerC B cell subsets maintained the expected preference for RF1 by deletion, use of RF2 was unexpectedly enhanced (p<0.05; Fig 3, top), especially given the fact that use of RF2 was increased even in the absence of N addition, a situation where microhomology between the 3′ termini of the DH gene segments and the 5′ terimini of the JH gene segments facilitates RF1 rearrangement (Fig. 3) (24, 31, 37).
The global amino acid content of the CDR-H3 loops is largely dependent on N nucleotide additions
The CDR-H3 loop is biased for the use of tyrosine and against the use of highly charged or highly hydrophobic amino acids (7, 8). This bias is first established in developing BM B cells (6) and is most apparent in the perinatal liver (15). The bias for the use of tyrosine in the CDR-H3 loop was maintained in all three PerC B cell subsets when analyzed irrespective of N addition (Fig. 4, top). However, this bias was substantially diminished when compared to NL F (p<0.0001, Fig 4, top). B-1a cells were intermediate in their use of tyrosine between NL F (p=0.0009) and BM F (p<0.05), whereas use of tyrosine in B-1b and B-2 cells was statistically indistinguishable from BM F. PerC B-1a, B-1b and B-2 cells also used arginine (charged) and valine (hydrophobic) more frequently than NL F (p<0.0001; Fig. 4, top).
Figure 4. Distribution of amino acids in the CDR-H3 loops of the VH7183DJCμ transcripts isolated from sorted 1-day liver, peritoneal cavity and bone marrow mature B cells.

Amino acids use is reported as the percent of the sequenced population from neonatal liver Fraction F (NL F - mature); peritoneal cavity (PerC) B-1a, B-1b and B-2; and bone marrow fraction F (BM F - mature, recirculating) from either wild type or TdT−/− mice (BM F TdT−/−). The amino acids are arranged by relative hydrophobicity, as assessed by a normalized Kyte-Doolittle scale (39, 40). The number of loops analyzed is shown. (top) all VH7183 transcripts; (middle) VH7183 transcripts from PerC B cells CDR-H3 without N nucleotide addition (N−); (bottom) VH7183 transcripts from PerC B cell CDR-H3 containing N nucleotide addition (N+). All comparisons were made to fraction F, either from 1 day liver (NL F) or bone marrow (BM F) using χ2-test or Fisher’s exact test as appropriate. Significant differences between PerC B cell subsets and both fraction Fs are labelled with asterisks, with *p≤0.05, **p≤0.01, *** p≤0.001, **** p≤0.0001.
Although the difference in tyrosine usage was found to primarily reflect the contribution of N addition, each PerC B cell subset again demonstrated its own characteristic amino acid signature. PerC B cell sequences lacking N nucleotides from all three subsets had higher tyrosine content than BM F (p<0.05), and completely lacked lysine, proline, cysteine, and phenylalanine. This matched the pattern observed in both NL F and BM F TdT−/− (Fig. 4, middle). Among the sequences lacking N nucleotides, histidine was more common in B-1a N− CDR-H3s, serine in B-1b and B-2, and valine in B-2. The increased use of histidine in B-1a reflected a preference for a specific type of V→D overlap in terminal sequence at Kabat position 95 (38). The increased use of serine in B-1b and B-2 correlated with the increased use of DFL16.1, which is enriched for serine (11). The increased use of valine in B-2 reflected the increased use of RF-2 (13). Among the PerC N+ sequences amino acid content proved similar, but not identical, to BM F.
One invariant amino acid was glycine, which was present at the same frequency in all three PerC subsets regardless of the presence or absence of N nucleotides. The frequency of use of glycine matched that of both WT BM F and BM F TdT−/−, but WT BM F was significantly greater than that observed in NL F (p <0.05) (Fig. 4). These data would suggest that a specific level of glycine offers a selective advantage in an adult environment.
The average lengths of B-1a, B-1b and B-2 CDR-H3s lie between neonatal and adult CDR-H3s
During B cell development, average CDR-H3 length is adjusted to fit an apparently preferred set point specific for each developmental checkpoint (6, 14). For example, in adult bone marrow the average length of BM F CDR-H3 is 0.5 codons longer than BM E (6). This particular transition appears to occur in the periphery because the average length of splenic T1 CDR-H3s matches BM Fr. E, whereas the average length of follicular CDR-H3s matches BM Fr. F (14).
The lengths of CDR-H3 in PerC B cells in comparison to neonatal liver and bone marrow demonstrated a progressive hierarchy with NL F ≪B1a <B1b ≪B2 ≪BM F (Fig. 5, left). The average CDR-H3 length of NL F B cells was 0.8 and 0.9 codons shorter than B-1a and B-1b, respectively (p<0.005). The B-1a sequences averaged 0.1 codons less than B-1b, but this difference did not achieve statistical significance. In turn, the average lengths of B-1a and B-1b CDR-H3 were 0.9 and 0.8 codons shorter than B-2 (p<0.05), and B-2 was 0.6 codons shorter than BM F (p<0.05, Fig. 5, left). The average length of the B-2 CDR-H3s matched that of BM E and splenic T1 [data not shown, (14)].
Figure 5. Average CDR-H3 length of VH7183DJCμ transcripts isolated from sorted 1-day liver, peritoneal cavity and bone marrow mature B cells.
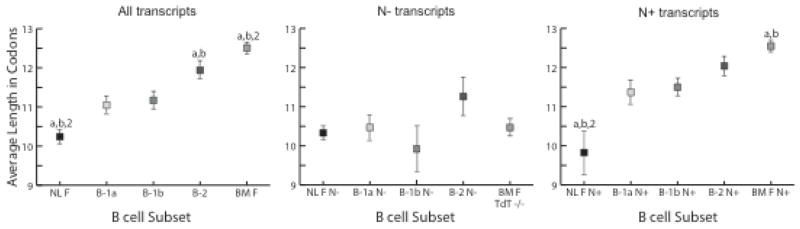
Average CDR-H3 codon lengths of the sequenced population from neonatal liver fraction F (NL F - mature); peritoneal cavity (PerC) B-1a, B-1b and B-2; and bone marrow fraction F (BM F - mature, recirculating) from either wild type or TdT−/− mice (BM F TdT−/−). Error bars depict the standard error of each mean. (left) all VH7183 transcripts; VH7183 transcripts from PerC B cells CDR-H3 without N nucleotide addition (N−); (right) VH7183 transcripts from PerC B cell CDR-H3 containing N nucleotide addition (N+). Statistical analyzes were made comparing to PerC B cell subsets using Student’s t-test, two tailed; the extent of statistical significance (p<0.05) is indicated with (a) compared to B-1a, (b) compared to B-1b and (2) compared to B-2.
The presence or absence of N nucleotides greatly affected CDR-H3 length. Among the N− sequences, the average lengths of NL F, B-1a, B-1b, B-2 (p=0.53) and BM F TdT−/− were statistically indistinguishable from each other. However, among the N+ sequences, the average length of all three PerC B cell sequences were easily distinguishable from NL F (p<0.0001; Fig. 5, right) and, for B-1a and B-1b, from BM F. [The average length of the B-2 N+ sequences was shorter than BM F but did not achieve statistical significance (Fig. 5, right)]. No statistically significant differences in average length were found among the N+ sequences from B-1a, B-1b or B2 (Fig. 5, right). Thus, the differences in average CDR-H3 length between the three PerC B cell subsets and NL F and BM F as a whole were mostly due to the sequences that contained N nucleotides.
In addition to changes in average length, we previously demonstrated that in the progression from fraction E to the intermediary populations in the spleen (T1 and FO) to recirculating fraction F the variance in CDR-H3 length narrowed (14). This decrease in variance was greatly influenced by the progressive loss of CDR-H3s containing fewer than nine codons (6, 14). However, in the peritoneal cavity sequences containing CDR-H3s of fewer than nine codons appeared to be retained in all three of the PerC subsets (Supplemental Fig. 2), including B-2 with 9% short sequences versus only 4% for BM F (p < 0.05). This enrichment for short sequences is especially notable when compared to TdT deficient BM F TdT−/−, which is devoid of CDR-H3s containing only 5 codons, whereas this length is still prevalent in B-1a and B-1b cells (Supplemental Fig. 2, center).
N nucleotide addition had a variable effect on the prevalence of short CDR-H3s. Of the B-1a CDR-H3s, one in five contained fewer than nine codons. N addition had no effect on the prevalence of CDR-H3s with fewer than nine codons, a prevalence that matched that of NL F. However, for the B-1b subset, one in four of the N− sequences were short, but only one in ten of the N+ sequences contained fewer than nine codons (Supplemental Fig. 2). Similarly, short CDR-H3s were more common among B-2 N− sequences (one in eight) than in N+ (one in twelve).
Enrichment for hydrophobic CDR-H3s in the PerC B cell subsets
The average hydrophobicities of each of the B-1a, B-1b and B-2 cell CDR-H3 repertoires were significantly more hydrophobic than NL F (p<0.01, p<0.01 and p<0.0001, respectively; Fig. 6, left). The average hydrophobicity of the B-2 CDR-H3 repertoire was also significantly more hydrophobic than BM F (p<0.05). This trend towards increased hydrophobicity was apparent among both the N+ and N− sequences (Fig 6).
Figure 6. Average CDR-H3 hydrophobicity of VH7183DJCμ transcripts isolated from sorted 1-day liver, peritoneal cavity and bone marrow mature B cells.
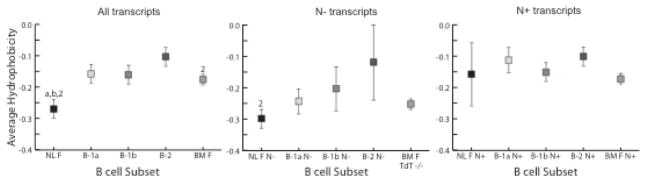
Average CDR-H3 loop hydrophobicity as assessed by a normalized Kyte-Doolittle scale (39, 40) from neonatal liver fraction F (NL F - mature); peritoneal cavity (PerC) B-1a, B-1b and B-2; and bone marrow fraction F (BM F - mature, recirculating) from either wild type or TdT−/− mice (BM F TdT−/−). Error bars depict the standard error of each mean. (left) all VH7183 transcripts; VH7183 transcripts from PerC B cells CDR-H3 without N nucleotide addition (N−); (right) VH7183 transcripts from PerC B cell CDR-H3 containing N nucleotide addition (N+). Statistical analyzes were made comparing to PerC B cell subsets using Student’s t-test, two tailed; the extent of statistical significance (p≤0.05) is indicated with (a) compared to B-1a, (b) compared to B-1b and (2) compared to B-2.
As with length, the variance in average hydrophobicity of CDR-H3 also normally decreases with development (6, 14). This decrease in variance is due, in part, to the loss of sequences at the extremes (6, 14). In BALB/c bone marrow Hardy fraction C (BM C), for example, there are a number of sequences with an average normalized Kyte–Doolittle hydrophobicity (39, 40) of less than −0.700 (charged) or greater than +0.600 (hydrophobic); whereas by BM F sequences with these characteristics largely no longer contribute to the repertoire [see Supplemental Fig. 3 and (6, 14)]. This focusing of the hydrophobicity of CDR-H3 is less apparent in fetal B cells, which are more dependent on the use of germline sequence and neutral DH RF1 (15).
In the PerC, the hydrophobicity distributions of the repertoires of all three of B cell subsets were more similar to that of NL F than BM F (Supplemental Fig. 3). In particular, the distribution of average CDR-H3 hydrophobicity for the B-2 subset demonstrated the same pattern of preservation of highly hydrophobic CDR-H3s as that of BM E and splenic T1 B cells [Supplemental Fig. 3 and (14)].
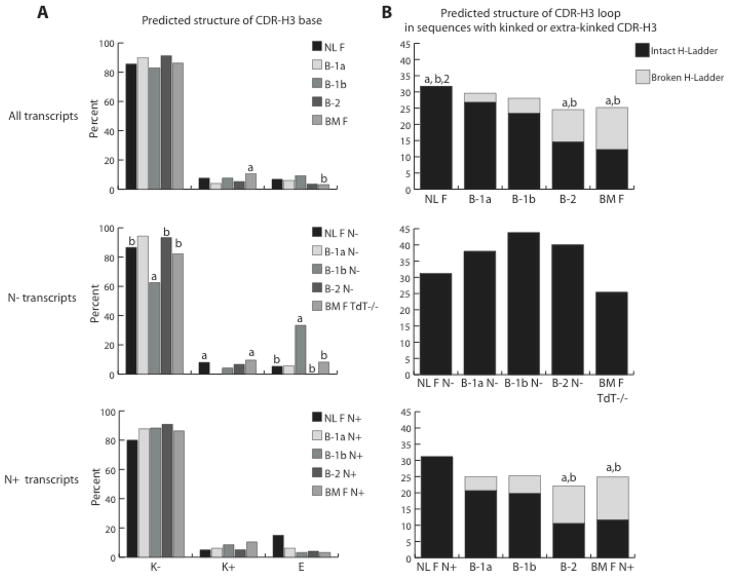
Predicted CDR-H3 base and loop structure
Given the great diversity of CDR-H3, it has proven difficult to definitively predict the structure generated by an individual sequence. However, using the “H3-rules” of Shirai (23) it is possible to gain insight into the likely structure of the base of the CDR-H3 loop and to predict whether sequences with a kinked or extra-kinked CDR-H3 base will be able to generate an intact or a broken hydrogen bond ladder. We found that the B-1a CDR-H3 repertoire was the least likely to include extra-kinked bases (p < 0.02, vs BM F); whereas the B-1b CDR-H3 repertoire was the most likely to create extended bases (p <0.05 vs BM F) (Fig. 6A). However, while the differences in likely base structures between the PerC B cell subsets appeared relatively minor when considered as a whole, striking differences were found among the sequences that lacked N nucleotides. The B-1a N− CDR-H3 repertoire was devoid of extra-kinked bases (p< 0.02 vs both NL F and BM F TdT−/−), whereas more than one-third of the B-1b N− CDR-H3 repertoire contained likely extended bases with a compensatory decrease in the likelihood of generating kinked bases.
When we evaluated likely loop structures, we found that the B-1a and B-1b structures were again similar to each other in the likely distribution of intact or broken hydrogen ladders when compared to NL F (p<0.05, p<0.01, respectively), B-2 (p<0.001, p<0.05, respectively), and BM F (p<0.0001, p<0.001, respectively), whereas the B-2 repertoire proved very similar in likely structures to BM F (Figure 6B). No broken ladders were predicted in the N− repertoires. Among the N+ repertoire the NL F clearly diverged from B-1a and B-1b, which shared a similar likely distribution of intact versus broken ladders; and both of these sets diverged from the B-2 and BM F repertoires, which again shared similar likelihoods. Thus among that subset of N+ sequences with kinked or extra-kinked bases, the B-1a and B-1b repertoires shared a similar pattern of loop structure distributions.
Discussion
The recent identification of a specific B-1 progenitor (19) has provided strong support for previous models proposing distinct developmental pathways for the peritoneal B cell subsets (18, 41, 42). In previous work we had shown that somatic selection for or against certain categories of immunoglobulin CDR-H3 sequence occurs during B cell development in the bone marrow of BALB/c mice (6), and that the sequence content of CDR-H3 appears to help shape the fate of the B cells in peripheral lymphoid tissues (14). In the present study, we analyzed the expressed VH7183-containing CDR-H3 repertoire in the peritoneal cavity (PerC) B cell subsets and compared them to repertoires expressed in neonatal liver and adult bone marrow. Because we were comparing equivalent repertories, these studies allowed us to test the extent to which specific categories of CDR-H3 sequence, including those that reflect ontogenetic origin (24, 25, 31), might be associated with populating PerC B cell niches. We identified key signatures that were variably shared among the PerC B-1a, B-1b, and B-2 subsets and IgM+IgD+ B cell subsets within the neonatal liver and the adult bone marrow with or without N addition (Fig. 8). We also identified key signatures that appeared to be specific for each individual PerC B cell subset as well as those that were shared between two or all three of these subsets.
Figure 8. Schematic representation of the shared and unique features of the PerC B cell CDR-H3 repertoire.

The key signatures shared between the PerC subsets and mature B cells within the neonatal liver and the adult bone marrow with (N+) or without (N−) N addition are represented. Also represented are the key signatures that appeared to be specific for each individual B cell subset as well as those that were shared between two or all three (PerC) of these subsets. Note that the CDR-H3 repertoire overlap is greater among the PerC N+ transcripts than in PerC N− transcripts. (
 ) NL F: features shared with neonatal liver fraction F; (
) NL F: features shared with neonatal liver fraction F; (
 ) BM F: features shared with adult bone marrow from WT; (
) BM F: features shared with adult bone marrow from WT; (
 ) TdT: features shared with adult bone marrow from TdT deficient mice; (
) TdT: features shared with adult bone marrow from TdT deficient mice; (
 ) B-1a: unique features of B-1a cells; (
) B-1a: unique features of B-1a cells; (
 ) B-1b: unique features of B-1b cells; (
) B-1b: unique features of B-1b cells; (
 ) B-2: unique features of B-2 cells. (Bold black) features shared between two or all three PerC B cell subsets.
) B-2: unique features of B-2 cells. (Bold black) features shared between two or all three PerC B cell subsets.
In a number of ways, our work confirms and extends previous observations by other investigators (24–26, 43). Our findings confirmed that the PerC B-1a N− repertoire is enriched for the signatures of CDR-H3 sequences present in neonatal liver, whereas the PerC B-2 N+ repertoire is enriched for adult bone marrow sequence signatures. Our analysis extended the structural significance of these findings by demonstrating that the predicted CDR-H3 loop structures in all the N− transcripts from the PerC B cell subsets also share neonatal signatures, such as the absence of broken ladder. Similarly, broken ladders were present only in N+ sequences, matching the adult pattern of BM F. These observations provide strong support for the role of ontogenetic origin in controlling the composition of the antibody repertoire expressed by individual PerC B cell subsets. In particular, our findings support the now classic view that the PerC B-1a population serves as a reservoir for B cells that can produce fetal-like immunoglobulins, which reinforces the role of these cells as a source of ontogenetically regulated natural antibodies (44).
Our findings also confirm that a significant subset of the B-1a repertoire includes CDR-H3s with extensive N region addition (24–26). Our extended findings that B-1a sequences containing N regions (N+) were more similar to BM F provide new support for the view that at least some B-1a cells, likely expressing CDR-H3s with N nucleotides, are derived from post-natal bone marrow (45, 46).
Our findings confirm previous reports (26) that the B-1b population shares a number of sequence characteristics with the B-1a population. Our extended findings that the B-1b N+ repertoire also exhibits many of the features that characterize the adult BM F repertoire provide new support for the view that the B-1b population is also a mixture of the progeny of the perinatal liver and the postnatal bone marrow (19, 42, 45).
Finally, our extended findings show that the N+ B-2 repertoire also shares many of the characteristics of the adult BM F repertoire, again providing new support for the view that B-2 cells whose antigen receptors contain N nucleotides are the product of post-natal B cell development.
However, in addition to these findings, which were to be expected if ontogenetic origin plays a key role in determining the fate of the B-1a, B-1b and B-2 repertoires, there were several unexpected findings which suggest that PerC-specific somatic selection is also playing a role in all three subsets. In particular, the PerC B-2 population, which is usually considered to be directly derived from the pool of conventional B cells that circulate through the blood, spleen and bone marrow (27), presented significantly fewer N regions than the other so called ‘conventional’ B cells. And, we identified several unique sequence signatures that were shared between the two or even among all three of the PerC subsets but not with other mature perinatal or adult compartments. This latter finding suggests that all three PerC subsets are enriched for B cells that have responded to a shared antigen receptor-based selective stimulus.
We considered three possible explanations for the increased prevalence of N− sequences in the B-2 population.
First, it is possible that the B-2 cells that express CDR-H3s without N nucleotides were generated in the perinatal period and are the product a PerC environment that permits or promotes long term survival. This view would be consistent with the results ofco-transfer studies of B220− cells sorted from both fetal liver and adult bone marrow in the same adoptive recipients showing that B-2 cells can be derived from both sources (47). However, IL-7−/− mice, which are populated primarily by B-lymphocytes of perinatal origin, show few cells with B-2 phenotype in the PerC (48). And, although a fetal origin for N− B-2 sequences is quite attractive, our analysis failed to demonstrate the same type of signature sequence similarity between the B-2 N− and NL F repertoires that was so evident for the B-1a N− sequences (Fig. 8).
Second, it is possible that while the current surface markers used to identify B-2 cells within the PerC also identify the conventional, recirculating mature bone marrow B cell pool, these PerC B-2 cells actually are either a separate B cell population for which adequate markers have not yet been identified or that these PerC B-2 cells represent a single temporal point in a continuing developmental pathway where B-2 cells can acquire a B-1b like phenotype in the peritoneal compartment as proposed by Rothstein and colleagues (49). However, given the divergence in VH-D-JH usage between B-1b and B-2 as documented by this and previous work by others (25, 26), and also the divergence in the predicted CDR-H3 base, mainly in N− transcripts, and loop between B-1b and B-2 it seems less likely that B-2 cells represent a single temporal point in the B-1b development. Deep repertoire sequencing of the major B cell subsets from mice co-transferred with fetal liver and bone marrow B220− cells into the same irradiated recipient may shed further light on this issue.
Third, antigen receptor-influenced selection against B cells with N-region containing CDR-H3 sequences or for B cells with CDR-H3s lacking N addition might be influencing entry or survival, or both (43). Such a selective process would be consistent with our current hypothesis that individual niches exert categorical selection of the B cell repertoire (14).
We recognize that all three of these possibilities are not necessarily mutually exclusive. Still, although all three PerC B cell subsets demonstrated several sequence signatures that were unique and differentiated them from other mature B cell subsets, the B-2 repertoire was the most marked of the three (Fig. 8). However, several of the sequence signatures that separated B-2 from BM F and NL F were shared with the B-1a and B-1b subsets. For example, we found a unique preference for VH7183.18 gene usage among all three PerC N+ B cell subsets, which might suggest that the cells bearing BCR encoded for VH7183.18 are derived from adult precursors. In support of this hypothesis, the splenic transitional 1 (T1) B cells also uses VH7183.18 at the same high frequency (14), suggesting that the VH content also influence entry into the peritoneal compartment, as we have shown for the spleen (14). The functional meaning of the enhanced frequency of the VH7183.18 gene in the PerC is unclear, and may require analysis of the specificity of PerC VH7183.18-containing antibodies. But at least in terms of B cell development, our data point to a close relation among the splenic T1 subset and PerC B-1a N+, B-1b N+ and B-2 cells irrespective of N addition, which would be compatible with a precursor-product relationship with selection based on the composition of the antigen receptor as the final force driving cells into each individual mature compartment. Support for such a precursor-product hypothesis comes from the observation that in the absence of a spleen PerC B-1a cell development is inhibited (50).
Evidence of enrichment for hydrophobic CDR-H3s in B-1a N+, B-1b N+, and B-2 cells irrespective of N addition is another new finding. In BM F and the splenic follicles, the CDR-H3 loop is normally biased for the expression of neutral, hydrophilic amino acids such as tyrosine, glycine, and serine, whereas use of hydrophobic and charged amino acids is minimized (8). This bias reflects, in part, evolutionary selection of D and JH sequence coupled with a preference for use of only one of the six potential D reading frames, the RF-1 (8, 51). However, there is precedent for alteration of this pattern in specific B cell subsets. In particular, marginal zone B cells are enriched for the presence of charged CDR-H3s (14). This increased use of RF-2, which typically creates hydrophobic patches within CDR-H3, is clearly one of the mechanisms that underlies the shift to a more hydrophobic average CDR-H3 hydrophobicity that we identified in all three PerC B cell subsets.
In previous studies, we have used mice with altered germline DH loci to study the effect of altering the pattern of hydrophobicity in CDR-H3. We have shown that whereas mice forced to express highly charged (ΔD-iD) CDR-H3s demonstrated a decrease in PerC B-1a and B-1b cells numbers (12), the mice forced to use hydrophobic (ΔD-DμFS) CDR-H3s did not (53). Combining our previous studies (6, 11–14, 53) with the present data showing the enrichment for hydrophobic sequences into the PerC of the WT mice, we now propose that the poor filling of the PerC B-1 compartment in ΔD-iD mice and the normal number of PerC B cells presented by ΔD-DμFS follow the same pattern of exclusion or inclusion of specific categories of CDR-H3 hydrophobicity observed in splenic MZ and follicular B cells and in the mature bone marrow B cells from WT mice (14). While suggestive, it remains unclear whether the low number of cells remaining in the PerC B cell compartment in the mice forced to express highly charged CDR-H3 (ΔD-iD) present the same CDR-H3 repertoire as imposed by the DH-altered segment or whether there is evidence for selection of alternative clones that best fit the categorical exigencies in each compartment. The composition of immunoglobulin CDR-H3 repertoires in the peripheral B cell compartments of those DH-limited mice is a current focus of investigation in our laboratory.
In summary, a detailed analysis of sequences expressed by PerC B cells has shown that while each PerC B cell subset retains a number of CDR-H3 sequence signatures that mark their respective developmental and ontogenetic origins (54), i.e. fetal/adult origins or both, the final result includes signature features that are unique to the peritoneal cavity. We interpret our findings to suggest that PerC B cells are the product of PerC-specific antigen-receptor mediated selective pressures, which may, in part, help regulate either entry or survival in the peritoneal compartment. PerC B-1a and B-1b cells have been credited as the source of the natural antibodies, the innate arm of the adaptive immune system (55, 56). Our studies would suggest that at least one factor that shapes this repertoire is permissiveness for hydrophobic CDR-H3s, which may exhibit unique antigen binding patterns. The fact that the CDR-H3 repertoire expressed by the B-2 compartment, which is presumed to be the source of adaptive immunity, appears to be shaped by the same antigen-receptor based selective pressures would suggest that this compartment may also be able to produce a unique range of antigen binding specificities that are uncommon or rare elsewhere, and thus serve as a reservoir for the diversity needed to respond to unusual or difficult antigens or vaccines.
Supplementary Material
Supplemental Figure 1: Representative gates used to identify and sort peritoneal B lineage cell populations.
Supplemental Figure 2: Distribution of CDR-H3 length of VH7183DJCμ transcripts isolated from sorted 1-day liver, peritoneal cavity and bone marrow mature B cells. Distribution of CDR-H3 lengths of the sequenced population from neonatal liver fraction F (NL F - mature); peritoneal cavity (PerC) B-1a, B-1b and B-2; and adult bone marrow fraction F (BM F - mature, recirculating) from TdT sufficient and deficient mice (BM F TdT−/−). (left) all VH7183 transcripts; VH7183 transcripts without N nucleotide addition (N−); (right) VH7183 transcripts containing N nucleotide addition (N+). Prevalence is reported as the percent of the sequenced population of unique, in-frame, open transcripts from each B cell subset. To facilitate visualization of the change in variance of the distribution, the vertical lines mark the preferred range of lengths in the bone marrow fraction F.
Supplemental Figure 3: Distribution of average CDR-H3 hydrophobicity of VH7183DJCμ transcripts isolated from sorted 1-day liver, peritoneal cavity and bone marrow mature B cells. Distribution of CDR-H3 loops hydrophobicity as assessed by a normalized Kyte-Doolittle scale (39, 40) from neonatal liver fraction F (NL F - mature); peritoneal cavity (PerC) B1a, B1b and B2; and adult bone marrow fraction F (BM F - mature, recirculating) from TdT sufficient and deficient mice (BM F TdT−/−). (left) all VH7183 transcripts; VH7183 transcripts CDR-H3 without N nucleotide addition (N−); (right) VH7183 transcripts containing N nucleotide addition (N+). Prevalence is reported as the percent of the sequenced population of unique, in-frame, open transcripts from each B cell subset. To facilitate visualization of the change in variance of the distribution, the vertical lines mark the preferred range of average hydrophobicity in the bone marrow fraction F.
Figure 7. Distribution of the predicted structures of the base and the loop of CDR-H3 from VH7183DJCμ transcripts isolated from sorted 1-day liver, peritoneal cavity and bone marrow mature B cells.
Structural properties were predicted according to Shirai’s (23) “H3-rules” deduced from the analysis of well-determined CDR-H3 crystal structures. These rules allow prediction of structural features from the primary CDR-H3 sequence based upon the location and hydrophobicity of amino acids, and size of the side chain. Frequencies are reported as percentage of all sequences analyzed. (A) Frequency of kinked (k−), extra- kinked (k+) and extended (E) CDR-H3 bases from neonatal liver fraction F (NL F - mature); peritoneal cavity (PerC) B-1a, B-1b and B-2; and bone marrow fraction F (BM F - mature, recirculating) from either wild type or TdT−/− mice (BM F TdT−/−). (B) Frequency of broken and intact hydrogen bond ladders within the CDR-H3 loop for those H chains that contain kinked or extra-kinked bases, depicted in A. (top) all VH7183 transcripts; (middle) VH7183 transcripts from NL F and PerC B cells CDR-H3 without N nucleotide addition (N−) and bone marrow fraction F from TdT deficient mice; (bottom) VH7183 transcripts from NL F, PerC B and BM F cells CDR-H3 containing N nucleotide addition (N+). Statistical analyzes were made comparing to PerC B cell subsets, using χ2-test or Fisher’s exact test as appropriate. The extent of statistical significance (p≤0.05) is indicated with (a) compared to B-1a, (b) compared to B-1b and (2) compared to B-2.
Table 1.
Cell numbers in the peritoneal cavity of normal and mutant mice (ΔD-DFL)
| N | Total Cells × 106 | CD19+ × 106 | Fraction
|
|||
|---|---|---|---|---|---|---|
| B1a × 105 | B1b × 105 | B2 × 105 | ||||
| wt/wt | 9 | 3.9 (0.1) | 1.3 (0.1) | 3.9 (0.3) | 1.1 (0.1) | 3.9 (0.3) |
| ΔD-DFL/ΔD-DFL | 9 | 4.1 (0.2) | 1.2 (0.1) | 3.5 (0.3) | 1.0 (0.1) | 3.3 (0.2) |
| Cell numbers in the peritoneal cavity of normal and mutant mice (ΔD-iD)
| ||||||
|---|---|---|---|---|---|---|
| N | Total Cells × 106 | CD19+ × 106 | Fraction
|
|||
| B1a × 105 | B1b × 105 | B2 × 105 | ||||
| wt/wt | ||||||
| ΔD-iD/ΔD-iD | ||||||
| Cell numbers in the peritoneal cavity of normal and mutant mice (ΔD-rf2)
| ||||||
|---|---|---|---|---|---|---|
| N | Total Cells × 106 | CD19+ × 106 | Fraction
|
|||
| B1a × 105 | B1b × 105 | B2 × 105 | ||||
| wt/wt | 10 | 3.8 (0.2) | 1.4 (0.1) | 2.3 (0.3) | 1.9 (0.2) | 4.2 (0.4) |
| ΔD-rf2/ΔD-rf2 | 10 | 4.0 (0.2) | 1.4 (0.1) | 2.2 (0.2) | 2.0 (0.1) | 4.5 (0.2) |
| Cell numbers in the peritoneal cavity of normal and mutant mice (C56/BL/6, sle2, ΔD-iD)
| ||||||
|---|---|---|---|---|---|---|
| N | Total Cells × 106 | CD19+ × 106 | Fraction
|
|||
| B1a × 105 | B1b × 105 | B2 × 105 | ||||
| wt/wt | 10 | 5.0 (0.2) | 1.3 (0.1) | 5.4 (0.5) | 2.7 (0.3) | 1.8 (0.2) |
| ΔD-iD/ΔD-iD | 10 | 4.9 (0.2) | 1.1 (0.1) | 4.6 (0.6) | 2.1(0.3) | 1.5 (0.1) |
Total nucleated cells that excluded trypan blue. Values shown are cell counts per first peritoneal cavity lavage (average cellularity of per peritoneal cavity collected from each experimental animal) of paired 8 week old homozygous ΔD-rf2 (ΔD-rf2) or homzogyous wild-type (WT) littermate progeny of heterozygous ΔD-rf2/WT BALB/c mice. The standard error of the mean is presented in parentheses. The number of cells in fractions B1a (CD19+ CD5+ Mac 1+), B1b (CD19+ CD5− Mac-1+), and B2 (CD19+ CD5− Mac-1−) was determined from the relative proportion of total cells.
p ≤ 0.05.
p ≤ 0.01,
p ≤ 0.005,
p ≤ 0.0001 versus BALB/c WT littermate.
Acknowledgments
The authors wish to thank P. Burrows, J. Kearney, A. Nóbrega, and E. Szymanska for their invaluable advice and support.
Abbreviations
- BM
bone marrow
- BM F
bone marrow mature IgM+IgD+ B cells
- BM F TdT−/−
bone marrow mature IgM+IgD+ B cells from TdT deficient mice
- CDR-H3
complementarity determining region 3 of the immunoglobulin heavy chain
- FO
splenic follicular B cells
- MZ
splenic marginal zone B cells
- N−
CDR-H3 sequences containing a recognizable DH but lacking evidence of N addition
- N+
CDR-H3 sequences that contain evidence of N addition
- NL
neonatal liver
- NL F
neonatal liver mature IgM+IgD+ B cells
- P
palindromic nucleotide gain
- PerC
peritoneal cavity
- RT-PCR
polymerase chain reaction amplification of transcripts cloned into cDNA by reverse transcription of mRNA
- TdT−/−
Terminal deoxynucleotidyl Transferase deficient mice
- WT
wild type
References
- 1.Kabat EA, Wu TT, Perry HM, Gottesman KS, Foeller C. Sequences of proteins of immunological interest. U.S. Department of Health and Human Services; Bethesda, Maryland: 1991. [Google Scholar]
- 2.Xu JL, Davis MM. Diversity in the CDR3 region of V(H) is sufficient for most antibody specificities. Immunity. 2000;13:37–45. doi: 10.1016/s1074-7613(00)00006-6. [DOI] [PubMed] [Google Scholar]
- 3.Lafaille JJ, DeCloux A, Bonneville M, Takagaki Y, Tonegawa S. Junctional sequences of T cell receptor gamma delta genes: implications for gamma delta T cell lineages and for a novel intermediate of V-(D)-J joining. Cell. 1989;59:859–870. doi: 10.1016/0092-8674(89)90609-0. [DOI] [PubMed] [Google Scholar]
- 4.Alt FW, Baltimore D. Joining of immunoglobulin heavy chain gene segments: Implications from a chromosome with evidence of three D-J heavy fusions. Proceedings of the National Academy of Sciences, USA. 1982;79:4118–4122. doi: 10.1073/pnas.79.13.4118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tonegawa S. Somatic generation of antibody diversity. Nature. 1983;302:575–581. doi: 10.1038/302575a0. [DOI] [PubMed] [Google Scholar]
- 6.Ivanov II, Schelonka RL, Zhuang Y, Gartland GL, Zemlin M, Schroeder HW., Jr Development of the expressed immunoglobulin CDR-H3 repertoire is marked by focusing of constraints in length, amino acid utilization, and charge that are first established in early B cell progenitors. Journal of Immunology. 2005;174:7773–7780. doi: 10.4049/jimmunol.174.12.7773. [DOI] [PubMed] [Google Scholar]
- 7.Zemlin M, Klinger M, Link J, Zemlin C, Bauer K, Engler JA, Schroeder HW, Jr, Kirkham PM. Expressed murine and human CDR-H3 intervals of equal length exhibit distinct repertoires that differ in their amino acid composition and predicted range of structures. Journal of Molecular Biology. 2003;334:733–749. doi: 10.1016/j.jmb.2003.10.007. [DOI] [PubMed] [Google Scholar]
- 8.Ivanov II, Link JM, Ippolito GC, Schroeder HW., Jr . Constraints on hydropathicity and sequence composition of HCDR3 are conserved across evolution. In: Zanetti M, Capra JD, editors. The Antibodies. Taylor and Francis Group; London: 2002. pp. 43–67. [Google Scholar]
- 9.Perlmutter RM, Kearney JF, Chang SP, Hood LE. Developmentally controlled expression of immunoglobulin VH genes. Science. 1985;227:1597–1607. doi: 10.1126/science.3975629. [DOI] [PubMed] [Google Scholar]
- 10.Alt FW, Blackwell TK, DePinho RA, Reth MG, Yancopoulos GD. Regulation of genome rearrangement events during lymphocyte differentiation. Immunological Reviews. 1986;89:5–30. doi: 10.1111/j.1600-065x.1986.tb01470.x. [DOI] [PubMed] [Google Scholar]
- 11.Schelonka RL, I, Ivanov I, Jung D, Ippolito GC, Nitschke L, Zhuang Y, Gartland GL, Pelkonen J, Alt FW, Rajewsky K, Schroeder HW., Jr A single D H gene segment is sufficient for B cell development and immune function. Journal of Immunology. 2005;175:6624–6632. doi: 10.4049/jimmunol.175.10.6624. [DOI] [PubMed] [Google Scholar]
- 12.Ippolito GC, Schelonka RL, Zemlin M, Ivanov II, Kobayashi R, Zemlin C, Gartland GL, Nitschke L, Pelkonen J, Fujihashi K, Rajewsky K, Schroeder HW., Jr Forced usage of positively charged amino acids in immunoglobulin CDR-H3 impairs B cell development and antibody production. Journal of Experimental Medicine. 2006;203:1567–1578. doi: 10.1084/jem.20052217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zemlin M, Schelonka RL, Ippolito GC, Zemlin C, Zhuang Y, Gartland GL, Nitschke L, Pelkonen J, Rajewsky K, Schroeder HW., Jr Regulation of repertoire development through genetic control of D H reading frame preference. Journal of Immunology. 2008;181:8416–8424. doi: 10.4049/jimmunol.181.12.8416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schelonka RL, Tanner J, Zhuang Y, Gartland GL, Zemlin M, Schroeder HW., Jr Categorical selection of the antibody repertoire in splenic B cells. Eur J Immunol. 2007;37:1010–1021. doi: 10.1002/eji.200636569. [DOI] [PubMed] [Google Scholar]
- 15.Schelonka RL, Szymanska E, Vale AM, Zhuang Y, Gartland GL, Schroeder HW., Jr DH and JH Usage in Murine Fetal Liver Mirrors that of Human Fetal Liver. Immunogenetics. 2010 doi: 10.1007/s00251-010-0469-5. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schelonka RL, Ivanov II, Vale AM, Szymanska E, Zemlin M, Gartland GL, Schroeder HW., Jr The CDR-H3 repertoire from TdT deficient adult bone marrow is a close, but not exact, homologue of the CDR-H3 repertoire from perinatal liver. Journal of Immunology. 2010 doi: 10.4049/jimmunol.1001419. in review. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Herzenberg LA, Tung JW. B cell lineages: documented at last! Nature Immunology. 2006;7:225–226. doi: 10.1038/ni0306-225. [DOI] [PubMed] [Google Scholar]
- 18.Hayakawa K, Hardy RR, Herzenberg LA. Progenitors for Ly-1 B cells are distinct from progenitors for other B cells. J Exp Med. 1985;161:1554–1568. doi: 10.1084/jem.161.6.1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Montecino-Rodriguez E, Leathers H, Dorshkind K. Identification of a B-1 B cell-specified progenitor. Nature Immunology. 2006;7:293–301. doi: 10.1038/ni1301. [DOI] [PubMed] [Google Scholar]
- 20.Haughton G, Arnold LW, Whitmore AC, Clarke SH. B-1 cells are made, not born. Immunol Today. 1993;14:84–87. doi: 10.1016/0167-5699(93)90064-R. discussion 87–91. [DOI] [PubMed] [Google Scholar]
- 21.Lam KP, Rajewsky K. B cell antigen receptor specificity and surface density together determine B-1 versus B-2 cell development. Journal of Experimental Medicine. 1999;190:471–477. doi: 10.1084/jem.190.4.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shirai H, Kidera A, Nakamura H. Structural classification of CDR-H3 in antibodies. FEBS Letters. 1996;399:1–8. doi: 10.1016/s0014-5793(96)01252-5. [DOI] [PubMed] [Google Scholar]
- 23.Shirai H, Kidera A, Nakamura H. H3-rules: identification of CDR-H3 structures in antibodies. FEBS Letters. 1999;455:188–197. doi: 10.1016/s0014-5793(99)00821-2. [DOI] [PubMed] [Google Scholar]
- 24.Gu H, Forster I, Rajewsky K. Sequence homologies, N sequence insertion and JH gene utilization in VH-D-JH joining: implications for the joining mechanism and the ontogenetic timing of Ly1 B cell and B-CLL progenitor generation. EMBO Journal. 1990;9:2133–2140. doi: 10.1002/j.1460-2075.1990.tb07382.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tornberg UC, Holmberg D. B-1a, B-1b and B-2 B cells display unique VHDJH repertoires formed at different stages of ontogeny and under different selection pressures. EMBO Journal. 1995;14:1680–1689. doi: 10.1002/j.1460-2075.1995.tb07157.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kantor AB, Merrill CE, Herzenberg LA, Hillson JL. An unbiased analysis of V(H)-D-J(H) sequences from B-1a, B-1b, and conventional B cells. Journal of Immunology. 1997;158:1175–1186. [PubMed] [Google Scholar]
- 27.Hardy RR, Hayakawa K. B cell development pathways. Annual Review of Immunology. 2001;19:595–621. doi: 10.1146/annurev.immunol.19.1.595. [DOI] [PubMed] [Google Scholar]
- 28.Reininger L, Ollier P, Poncet P, Kaushik A, Jaton JC. Novel V genes encode virtually identical variable regions of six murine monoclonal anti-bromelain-treated red blood cell autoantibodies. Journal of Immunology. 1987;138:316–323. [PubMed] [Google Scholar]
- 29.Pennell CA, Arnold LW, Haughton G, Clarke SH. Restricted Ig variable region gene expression among Ly-1+ B cell lymphomas. Journal of Immunology. 1988;141:2788–2796. [PubMed] [Google Scholar]
- 30.Feeney AJ. Predominance of the prototypic T15 anti-phosphorylcholine junctional sequence in neonatal pre-B cells. Journal of Immunology. 1991;147:4343–4350. [PubMed] [Google Scholar]
- 31.Feeney AJ. Lack of N regions in fetal and neonatal mouse immunoglobulin V-D-J junctional sequences. Journal of Experimental Medicine. 1990;172:1377–1390. doi: 10.1084/jem.172.5.1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yancopoulos GD, Desiderio SV, Paskind M, Kearney JF, Baltimore D, Alt FW. Preferential utilization of the most JH-proximal VH gene segments in pre-B cell lines. Nature. 1984;311:727–733. doi: 10.1038/311727a0. [DOI] [PubMed] [Google Scholar]
- 33.Malynn BA, Yancopoulos GD, Barth JE, Bona CA, Alt FW. Biased expression of JH-proximal VH genes occurs in the newly generated repertoire of neonatal and adult mice. Journal of Experimental Medicine. 1990;171:843–859. doi: 10.1084/jem.171.3.843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Viale AC, Coutinho A, Freitas AA. Differential expression of VH gene families in peripheral B cell repertoires of newborn or adult immunoglobulin H chain congenic mice. Journal of Experimental Medicine. 1992;175:1449–1456. doi: 10.1084/jem.175.6.1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Williams GS, Martinez A, Montalbano A, Tang A, Mauhar A, Ogwaro KM, Merz D, Chevillard C, Riblet R, Feeney AJ. Unequal VH gene rearrangement frequency within the large VH7183 gene family is not due to recombination signal sequence variation, and mapping of the genes shows a bias of rearrangement based on chromosomal location. Journal of Immunology. 2001;167:257–263. doi: 10.4049/jimmunol.167.1.257. [DOI] [PubMed] [Google Scholar]
- 36.Sen R, Oltz E. Genetic and epigenetic regulation of IgH gene assembly. Curr Opin Immunol. 2006;18:237–242. doi: 10.1016/j.coi.2006.03.008. [DOI] [PubMed] [Google Scholar]
- 37.Feeney AJ. Predominance of VH-D-JH junctions occurring at sites of short sequence homology results in limited junctional diversity in neonatal antibodies. Journal of Immunology. 1992;149:222–229. [PubMed] [Google Scholar]
- 38.Marshall AJ, Doyen N, Bentolila LA, Paige CJ, Wu GE. Terminal deoxynucleotidyl transferase expression during neonatal life alters D(H) reading frame usage and Ig-receptor-dependent selection of V regions. Journal of Immunology. 1998;161:6657–6663. [PubMed] [Google Scholar]
- 39.Kyte J, Doolittle RF. A simple method for displaying the hydropathic character of a protein. Journal of Molecular Biology. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 40.Eisenberg D. Three-dimensional structure of membrane and surface proteins. Annual Review of Biochemistry. 1984;53:595–623. doi: 10.1146/annurev.bi.53.070184.003115. [DOI] [PubMed] [Google Scholar]
- 41.Kantor AB, Herzenberg LA. Origin of murine B cell lineages. Annual Review of Immunology. 1993;11:501–538. doi: 10.1146/annurev.iy.11.040193.002441. [DOI] [PubMed] [Google Scholar]
- 42.Tung JW, Mrazek MD, Yang Y, Herzenberg LA. Phenotypically distinct B cell development pathways map to the three B cell lineages in the mouse. Proceedings of the National Academy of Sciences, USA. 2006;103:6293–6298. doi: 10.1073/pnas.0511305103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Carlsson L, Overmo C, Holmberg D. Selection against N-region diversity in immunoglobulin heavy chain variable regions during the development of pre-immune B cell repertoires. International Immunology. 1992;4:549–553. doi: 10.1093/intimm/4.5.549. [DOI] [PubMed] [Google Scholar]
- 44.Avrameas S. Natural autoantibodies: from ‘horror autotoxicus’ to ‘gnothi seauton’. Immunology Today. 1991;12:154–159. doi: 10.1016/S0167-5699(05)80045-3. [DOI] [PubMed] [Google Scholar]
- 45.Esplin BL, Welner RS, Zhang O, Borghesi LA, Kincade PW. A differentiation pathway for B1 cells in adult bone marrow. Proceedings of the National Academy of Sciences, USA. 2009;106:5773–5778. doi: 10.1073/pnas.0811632106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Holodick NE, Repetny K, Zhong X, Rothstein TL. Adult BM generates CD5+ B1 cells containing abundant N-region additions. Eur J Immunol. 2009;39:2383–2394. doi: 10.1002/eji.200838920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kantor AB, Stall AM, Adams S, Herzenberg La, Herzenberg LA. Differential development of progenitor activity for three B-cell lineages. Proceedings of the National Academy of Sciences, USA. 1992;89:3320–3324. doi: 10.1073/pnas.89.8.3320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Carvalho TL, Mota-Santos T, Cumano A, Demengeot J, Vieira P. Arrested B lymphopoiesis and persistence of activated B cells in adult interleukin 7(−/)− mice. J Exp Med. 2001;194:1141–1150. doi: 10.1084/jem.194.8.1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hastings WD, Tumang JR, Behrens TW, Rothstein TL. Peritoneal B-2 cells comprise a distinct B-2 cell population with B-1b-like characteristics. Eur J Immunol. 2006;36:1114–1123. doi: 10.1002/eji.200535142. [DOI] [PubMed] [Google Scholar]
- 50.Wardemann H, Boehm T, Dear N, Carsetti R. B-1a B cells that link the innate and adaptive immune responses are lacking in the absence of the spleen. J Exp Med. 2002;195:771–780. doi: 10.1084/jem.20011140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gu H, Kitamura D, Rajewsky K. DH reading frame bias: evolutionary selection, antigen selection or both? Evolutionary selection. Immunology Today. 1991;12:420–421. doi: 10.1016/0167-5699(91)90146-K. [DOI] [PubMed] [Google Scholar]
- 52.Ichihara Y, Hayashida H, Miyazawa S, Kurosawa Y. Only DFL16, DSP2, and DQ52 gene families exist in mouse immunoglobulin heavy chain diversity gene loci, of which DFL16 and DSP2 originate from the same primordial DH gene. European Journal of Immunology. 1989;19:1849–1854. doi: 10.1002/eji.1830191014. [DOI] [PubMed] [Google Scholar]
- 53.Schelonka RL, Zemlin M, Kobayashi R, Szalai A, Ippolito GC, Zhuang Y, Gartland GL, Fujihashi K, Rajewsky K, Schroeder HW., Jr Preferential use of D H reading frame 2 alters B cell development and antigen-specific antibody production. Journal of Immunology. 2008;181:8409–8415. doi: 10.4049/jimmunol.181.12.8409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tung JW, Herzenberg LA. Unraveling B-1 progenitors. Current Opinion in Immunology. 2007;19:150–155. doi: 10.1016/j.coi.2007.02.012. [DOI] [PubMed] [Google Scholar]
- 55.Baumgarth N, Herman OC, Jager GC, Brown L, Herzenberg LA. Innate and acquired humoral immunities to influenza virus are mediated by distinct arms of the immune system. Proceedings of the National Academy of Sciences, USA. 1999;96:2250–2255. doi: 10.1073/pnas.96.5.2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Baumgarth N, Herman OC, Jager GC, Brown LE, Herzenberg LA, Chen J. B-1 and B-2 cell-derived immunoglobulin M antibodies are nonredundant components of the protective response to influenza virus infection. Journal of Experimental Medicine. 2000;192:271–280. doi: 10.1084/jem.192.2.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1: Representative gates used to identify and sort peritoneal B lineage cell populations.
Supplemental Figure 2: Distribution of CDR-H3 length of VH7183DJCμ transcripts isolated from sorted 1-day liver, peritoneal cavity and bone marrow mature B cells. Distribution of CDR-H3 lengths of the sequenced population from neonatal liver fraction F (NL F - mature); peritoneal cavity (PerC) B-1a, B-1b and B-2; and adult bone marrow fraction F (BM F - mature, recirculating) from TdT sufficient and deficient mice (BM F TdT−/−). (left) all VH7183 transcripts; VH7183 transcripts without N nucleotide addition (N−); (right) VH7183 transcripts containing N nucleotide addition (N+). Prevalence is reported as the percent of the sequenced population of unique, in-frame, open transcripts from each B cell subset. To facilitate visualization of the change in variance of the distribution, the vertical lines mark the preferred range of lengths in the bone marrow fraction F.
Supplemental Figure 3: Distribution of average CDR-H3 hydrophobicity of VH7183DJCμ transcripts isolated from sorted 1-day liver, peritoneal cavity and bone marrow mature B cells. Distribution of CDR-H3 loops hydrophobicity as assessed by a normalized Kyte-Doolittle scale (39, 40) from neonatal liver fraction F (NL F - mature); peritoneal cavity (PerC) B1a, B1b and B2; and adult bone marrow fraction F (BM F - mature, recirculating) from TdT sufficient and deficient mice (BM F TdT−/−). (left) all VH7183 transcripts; VH7183 transcripts CDR-H3 without N nucleotide addition (N−); (right) VH7183 transcripts containing N nucleotide addition (N+). Prevalence is reported as the percent of the sequenced population of unique, in-frame, open transcripts from each B cell subset. To facilitate visualization of the change in variance of the distribution, the vertical lines mark the preferred range of average hydrophobicity in the bone marrow fraction F.