SUMMARY
KRAS is the most commonly mutated oncogene, yet no effective targeted therapies exist for KRAS mutant cancers. We developed a pooled shRNA-drug screen strategy to identify genes that, when inhibited, cooperate with MEK inhibitors to effectively treat KRAS mutant cancer cells. The anti-apoptotic BH3 family gene BCL-XL emerged as a top hit through this approach. ABT-263 (navitoclax), a chemical inhibitor that blocks the ability of BCL-XL to bind and inhibit pro-apoptotic proteins, in combination with a MEK inhibitor led to dramatic apoptosis in many KRAS mutant cell lines from different tissue types. This combination caused marked in vivo tumor regressions in KRAS mutant xenografts and in a genetically engineered KRAS-driven lung cancer mouse model, supporting combined BCL-XL/MEK inhibition as a potential therapeutic approach for KRAS mutant cancers.
INTRODUCTION
KRAS mutations occur in ~20% of all cancers, with particularly high frequency in pancreatic (~90%), colorectal (~40%), and lung (~30%) cancers (Malumbres and Barbacid, 2003; Montagut and Settleman, 2009). However, no effective therapies exist for KRAS mutant cancers, largely because KRAS itself has proven difficult to target directly with small molecules (Young et al., 2009). Targeting single KRAS effector pathways (e.g., MEK) has also failed to induce clinical responses (Adjei et al., 2008), likely because KRAS activates multiple critical effectors, such as the MEK-ERK, PI3K-AKT, and NF-κB pathways (Montagut and Settleman, 2009).
Investigators have identified potential therapeutic approaches for KRAS mutant cancers that are yet to be explored in the clinic, including inhibitors of TBK1, TAK1, and the GATA2 transcriptional network (Barbie et al., 2009; Singh et al., 2012; Kumar et al., 2012). Previously, our laboratory and others showed that simultaneous targeting of more than one KRAS effector pathway (specifically the MEK-ERK and PI3K-AKT pathways) induced responses in KRAS-driven mouse tumor models (Engelman et al., 2008; She et al., 2010). While these data support the promise of targeted combination strategies, toxicity has prevented dosing both inhibitors at or near their maximally tolerated doses when used in combination (LoRusso et al., 2012; Speranza et al., 2012). Thus, potent and continuous suppression of the MEK and PI3K pathways may not be possible in patients with currently available agents. Furthermore, this approach may be effective only in a subset of KRAS mutant cancers. Consequently, additional effective combination therapy strategies for KRAS mutant cancers are critically needed.
RESULTS
To enable rapid development of MEK inhibitor-based combination therapies for KRAS mutant cancers, we developed a pooled shRNA-drug screen strategy (Figure 1A) aimed at identifying genes that, when inhibited, cooperate with MEK inhibitors to inhibit the proliferation and survival of KRAS mutant tumor cells. This screen utilized a ~5000 shRNA library targeting ~1,200 “druggable” genes, such as kinases and regulators of cell proliferation and survival. Target cells infected with this library were cultured in the presence or absence of the allosteric MEK inhibitor selumetinib (AZD6244, ARRY-142886) for 7 days. Since lentiviral shRNA integrates into the genome of a target cell, if a given shRNA decreases cell viability, the relative abundance of that shRNA will decrease over the 7-day period. We can thus identify shRNAs that “drop out” specifically with MEK inhibitor treatment relative to vehicle. This screen differs from other recently performed synthetic lethal RNAi screens in KRAS mutant cancer cell lines because it specifically assays for genes that cooperate with MEK inhibitors to reduce cell viability (Barbie et al., 2009; Luo et al., 2009; Scholl et al., 2009). Furthermore, by selecting for shRNAs with decreased abundance in MEK inhibitor versus vehicle-treated cells, shRNAs that are universally toxic to cells are filtered out, since these shRNAs drop out in both conditions. While this screen can be readily modified to incorporate other inhibitors in future studies, MEK inhibitors were chosen as the backbone of potential combination strategies in this study because large-scale screening of >600 cell lines with >100 targeted compounds identified MEK inhibitors as the most effective agents in KRAS mutant cell lines (Garnett et al., 2012). MEK inhibitors have also led to stable disease in patients with KRAS mutant cancer (Infante et al., 2010).
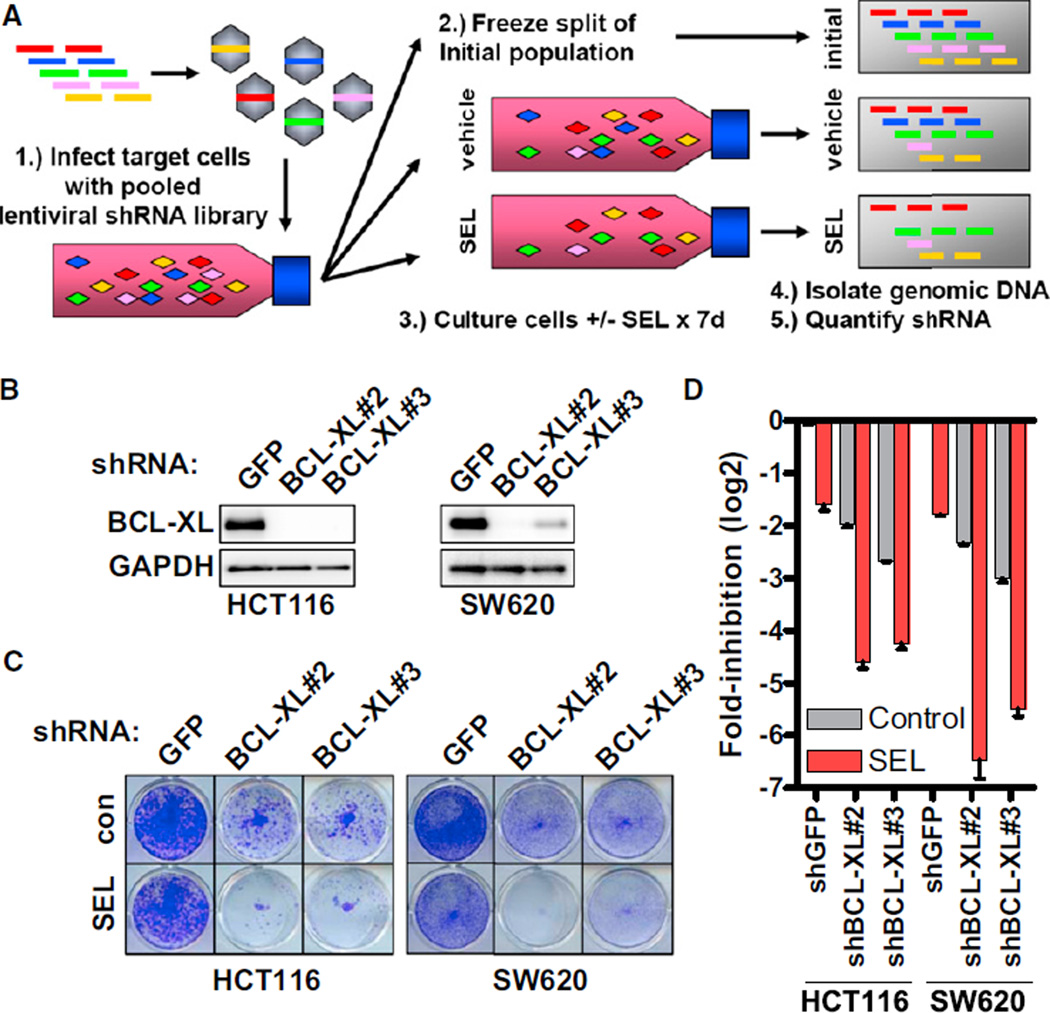
Figure 1. Identification of BCL-XL as a Potential Target for Combination Therapy with MEK Inhibitors in KRAS Mutant Cancers.
(A) Schematic of the pooled shRNA-drug screen approach. 1: Target cells are infected with a pooled lentiviral shRNA library. 2 and 3: Cells are aliquoted into three parts: one part is immediately frozen to represent the initial population, and the other two parts are treated with vehicle or 1 µM selumetinib (SEL) for 7 days. 4 and 5: Genomic DNA is isolated from cells, lentiviral cassettes are PCR-amplified, and individual shRNA abundance is quantified by deep sequencing.
(B) Western blot of cells infected with shRNAs targeting GFP or BCL-XL and lysates.
(C) Cells were infected with the indicated shRNAs. Following 48-hr puromycin selection, cells were cultured with or without 1 µM SEL for an additional 72 hr and stained with crystal violet.
(D) Quantification of crystal violet staining from cells in (C). Error bars represent SEM. See also Figure S1 and Tables S1 and S2.
We screened two KRAS mutant cell lines with different sensitivities to MEK/PI3K inhibition—HCT116 (sensitive) and SW620 (insensitive) (Figure S1A and S1B available online)—to identify combination strategies independent of MEK/PI3K sensitivity. Hits for each cell line were determined as described in Experimental Procedures, and we identified 17 hits common to both cell lines (Figure S1C; Tables S1 and S2). The anti-apoptotic BH3 family member BCL-XL (BCL2L1) emerged as the most promising hit in validation studies (Figure S1D). Knockdown of BCL-XL produced profound suppression of cell viability in the presence of selumetinib (Figures 1B–1D). ABT-263 (navitoclax) is a small molecule inhibitor that occupies the BH3 binding groove of BCL-XL and BCL-2, inhibiting their anti-apoptotic effects (Tse et al., 2008). ABT-263 does not effectively inhibit the anti-apoptotic proteins MCL-1 and BCL2-A1. The combination of ABT-263 and selumetinib caused significantly greater reduction in cell viability than either agent alone (Figure 2A). Combinations using other MEK inhibitors and another active BH3 mimetic produced similar efficacy, but a less active enantiomer of ABT-263 was not effective, suggesting that these effects were on-target (Figure S2A–S2D). These combinations led to an overall decrease in cell titer, relative to pretreatment starting cell titer, indicating induction of cell death. Indeed, ABT-263/selumetinib caused significantly more apoptosis than either agent alone (Figure 2B). Although this screen was not designed to identify combinations with efficacy specific for KRAS mutant versus wild-type cancers, lack of efficacy of ABT-263/selumetinib in an isogenic HCT116 cell line with wild-type KRAS suggests that KRAS mutations may indeed predispose to sensitivity to this combination (Figure S2E and S2F).
Figure 2. Pharmacologic Inhibition of BCL-XL and MEK in KRAS Mutant Cancer Cells.
(A) Cells were treated with vehicle (Control), 1 µM ABT-263 (ABT), 1 µM selumetinib (SEL), or the combination for 72 hr. Values represent the change in viable cell number relative to starting cell titer immediately before treatment. p < 0.01(*) or < 0.001(**) for ABT/SEL versus all other groups by one-way ANOVA with Tukey post-hoc test.
(B) Cells were treated as in (A) for 72 hr and the percentage of apoptotic cells was determined by Annexin V staining. All error bars represent SEM.
(C) Western blot of cells treated with ABT, SEL, or the combination for 24 hr.
(D) Cells were treated for 24 hr with ABT, SEL, or the combination. Left, immunoprecipitation was performed with immunoglobulin G control antibody or anti-BIM antibody, and the immunoprecipitate was probed with the indicated antibodies. Right, pre-immunoprecipitation lysates (L) and supernatants (S) following immunoprecipitation with anti-BIM antibody were probed with the indicated antibodies.
(E) Thirty KRAS mutant cell lines were treated with ABT, SEL, or the combination for 72 hr and the percentage of apoptotic cells was determined by Annexin V staining. For each cell line, the percent apoptosis is color-coded by quartile.
(F) The percentage of all cell lines exhibiting the indicated degree of apoptosis is shown.
See also Figure S2 and Table S3.
We investigated the mechanism by which ABT-263 and selumetinib cooperate to induce apoptosis in KRAS mutant cancer cells. Consistent with prior results, suppression of phosphorylated ERK (P-ERK) by selumetinib led to increased levels of the pro-apoptotic protein BIM, a well-known target of MAPK signaling (Figure 2C; Ley et al., 2003; Faber et al., 2009). The lack of marked apoptosis induced by selumetinib alone (Figures 2B and 2C) is consistent with previous studies demonstrating that induction of BIM alone is insufficient to cause apoptosis, but that concomitant suppression of one or more anti-apoptotic proteins is also needed (Faber et al., 2009; Rahmani et al., 2009). As expected, neither ABT-263 nor selumetinib led to a decrease in the levels of the anti-apoptotic proteins BCL-XL, BCL-2, or MCL-1 (Figure 2C). Immunoprecipitaion of BIM revealed that when BIM levels are induced by selumetinib, a proportionally increased amount of BCL-XL associates with BIM (Figure 2D), consistent with the notion that induction of BIM alone is not sufficient to induce marked apoptosis because it is bound and inhibited by pro-survival BH3 proteins, including BCL-XL. However, ABT-263 completely disrupted the association of BCL-XL with BIM under basal conditions and following BIM induction by selumetinib. In the presence of ABT-263, BIM was now able to complex with MCL-1, which has been shown to promote apoptosis by freeing apoptotic mediators, such as BAK and BAX, from inhibition by MCL-1 (Willis et al., 2007). Thus, ABT-263 may effectively combine with MEK inhibitors to promote apoptosis by blocking the ability of BCL-XL to bind and inhibit the increased levels of BIM protein induced by MEK inhibition, thereby “freeing” BIM to trigger an apoptotic response. When evaluated across a panel of 30 KRAS mutant cell lines (ten colorectal, ten lung, ten pancreatic; Table S3), ABT-263/selumetinib induced marked apoptosis in a large proportion of cell lines (Figures 2E and 2F), suggesting that this strategy could be effective in a significant proportion of KRAS mutant cancers of different tissue types.
To identify potential biomarkers predicting which KRAS mutant cancers might be most likely to respond to ABT-263/selumetinib therapy, we analyzed gene expression profiles from these KRAS mutant cell lines to identify genes whose expression correlated closely with the degree of apoptosis induced by ABT-263/selumetinib (Figure 3A). Gene set enrichment analysis (GSEA; Subramanian et al., 2005) revealed that four of the top ten gene sets enriched were related to epithelial versus mesenchymal differentiation (Table S4). Moreover, 25% of the genes identified were represented in a previously reported epithelialto-mesenchymal transition (EMT) gene signature (Figure 3A; Taube et al., 2010). Increased sensitivity to ABT-263/selumetinib also correlated with a previously identified “KRAS dependency” gene signature associated with epithelial differentiation (Figure 3B; Singh et al., 2009). Expression levels of E-cadherin protein (a marker of epithelial differentiation) also correlated with sensitivity (Figures 3C and S3A). Consistent with this hypothesis, shRNA-mediated knockdown of Zeb1, a master regulator of mesenchymal differentiation, in the mesenchymal KRAS mutant lung cancer cell line A549 induced an epithelial phenotype (increased E-cadherin and decreased vimentin) and increased sensitivity to ABT-263/selumetinib (Figures 3D and 3E). Thus, epithelial differentiation and/or EMT may be useful biomarkers to predict subsets of KRAS mutant cancers that are sensitive or resistant to this combination.
Figure 3. Epithelial Differentiation Predicts Sensitivity to the Combination of ABT-263 and Selumetinib.
(A) Heat map showing the top genes differentially expressed according to sensitivity to ABT-263/selumetinib as generated by the PAMR algorithm utilizing gene expression profiles from the KRAS mutant cell line panel. Red arrows represent genes that are upregulated and blue arrows represent genes that are down-regulated in a previously reported EMT gene signature. Vimentin (VIM) and E-cadherin (CDH1), markers of mesenchymal and epithelial differentiation, respectively, are indicated.
(B and C) Correlation of apoptosis induced by ABT-263/selumetinib with (B) a KRAS dependency gene signature and (C) E-cadherin protein expression. Each dot represents a single cell line. P values were generated by two-tailed t test.
(D) Western blot of A549 cells infected with shRNA targeting GFP (shGFP) or Zeb1 (shZeb1).
(E) A549 cells infected with shGFP or shZeb1 were treated with vehicle (CON), 1 µM ABT-263 (ABT), 1 µM selumetinib (SEL), or the combination for 72 hr. Values represent the change in viable cell number relative to starting cell titer immediately before treatment. The p values were determined by one-way ANOVA with Tukey post-hoc test. All error bars represent SEM.
See also Figure S3 and Table S4.
Given the broad efficacy of combined BCL-XL/MEK inhibition in KRAS mutant cancers in vitro, we evaluated the efficacy of ABT-263/selumetinib in KRAS mutant xenografts. Consistent with prior results, MEK inhibition alone slowed tumor growth relative to vehicle-treated control, but failed to induce tumor regressions (Figure 4A; She et al., 2010). Although ABT-263 alone had minimal effect on tumor growth, ABT-263/selumetinib led to marked tumor regressions in all 3 KRAS mutant xenografts (Figures 4A and 4B). Mice tolerated combined treatment well with no overt signs of toxicity (Figure S4A). Selumetinib alone led to robust suppression of P-ERK and tumor cell proliferation, but caused only a minimal increase in apoptosis (Figure 4C). However, ABT-263/selumetinib led to a dramatic induction of tumor cell apoptosis, consistent with the tumor regressions observed with this therapy.
Figure 4. In Vivo Efficacy of Combined BCL-XL and MEK Inhibition.
(A) KRAS mutant xenografts were treated with vehicle (CON), ABT-263 (ABT, 100 mg/kg daily), selumetinib (SEL, 25 mg/kg twice daily), or both drugs in combination. The mean percent change in tumor volume relative to initial tumor volume is shown. Error bars represent SEM.
(B) Waterfall plot showing the percent change in tumor volume (relative to initial volume) for individual tumors in the SEL and ABT/SEL groups following 21 days of treatment. P values were generated by two-tailed t test.
(C) Tumor tissue from HCT116 xenografts treated for 3 days with the indicated drug regimens was evaluated by immunohistochemistry for P-ERK, Ki67 (a marker of proliferation), or cleaved caspase 3 (a marker of apoptosis). Tumors were harvested 3 hr after dosing on day 3. Scale bar represents 100 µm.
(D and E) Established lung tumors in LSL-KRASG12D mice were treated with vehicle, ABT, SEL, or both drugs in combination as in (A). (D) MR images of two mice obtained pretreatment and following 1 week of treatment with the ABT/SEL combination. Red arrows indicate dense areas of lung tumor in the pretreatment images. (E) Mean percent tumor regression of lung tumors following 2 weeks of the indicated treatment. p represents ABT/SEL versus each treatment group by one-way ANOVA with Tukey post-hoc test. All error bars represent SEM.
See also Figure S4.
Our in vitro studies suggest that subsets of KRAS mutant cancers from multiple tissue types, including colorectal, lung, and pancreatic cancers, may be susceptible to this therapeutic approach. Thus, we assessed the efficacy of combined BCL-XL/MEK inhibition in established KRAS-driven lung tumors in the LSL-KRASG12D mouse model (Jackson et al., 2001; Engelman et al., 2008.) ABT-263/selumetinib led to significantly greater tumor regression than either agent alone, and led to near-complete regression of tumors in some cases (Figures 4D and 4E). In some mice selected for long-term treatment with ABT-263/selumetinib, durable tumor regressions lasting up to 7 weeks were observed (Figure S4B and S4C). This combination also led to regressions in a similar model also lacking p53 (Figure S4D). Overall, these data indicate that ABT-263/selumetinib has substantial preclinical in vivo efficacy in KRAS mutant cancer models from different tumor types. The marked tumor regressions observed support combined BCL-XL/MEK inhibition as a targeted therapy combination for evaluation in clinical trials in patients with KRAS mutant cancer.
Despite the marked in vivo efficacy observed with combined BCL-XL/MEK inhibition, our results suggest that this strategy is unlikely to be universally effective in all KRAS mutant cancers and that biomarkers predicting sensitivity and resistance are needed. Indeed, we observed that epithelial differentiation and EMT may help identify subsets of KRAS mutant cancers that are more or less likely to respond to this therapy (Figures 3A–3C). Interestingly, some, but not all, xenograft tumors harvested after long-term treatment with ABT-263/selumetinib showed loss of membrane expression of E-cadherin and increased vimentin expression, indicative of EMT (Figure S3B), further supporting the notion that cancers that have undergone EMT may be less sensitive to this combination. Though no acquired mutations were identified in the tumor cells that survived long-term treatment (see Supplemental Experimental Procedures), we observed that most residual tumors showed partial recovery of P-ERK, suggesting that failure to maintain full MAPK pathway suppression may contribute to the development of resistance to this combination (Figure S4E). With respect to EMT, analysis of KRAS mutant lung cancers from 25 patients revealed that 56% of patients showed features of epithelial differentiation, whereas 44% showed evidence of mesenchymal differentiation (Figure S3C). These results indicate that the epithelial/mesenchymal status of KRAS mutant cancers can be readily assessed in patients, and that a substantial percentage of KRAS mutant lung cancers retain an epithelial phenotype, which our data suggest may predict sensitivity to this therapy. Thus, the epithelial/ mesenchymal status of KRAS mutant cancers may be useful to evaluate in early clinical trials of combined BCL-XL/MEK inhibition.
DISCUSSION
Although KRAS is the most commonly mutated oncogene, KRAS mutant cancers have proven refractory to targeted therapies and remain a major clinical challenge. We identified combined BCL-XL and MEK inhibition as a therapeutic strategy that led to increased efficacy in KRAS mutant cancer cell lines from different tumor types and to in vivo tumor regressions in several KRAS mutant cancer models. These findings, along with prior reports (Engelman et al., 2008; She et al., 2010), provide further evidence that targeted therapy combinations may be an important avenue to generate therapeutic efficacy in KRAS mutant cancers.
Although MEK inhibitors were among the most effective agents in KRAS mutant cancer cell lines in a large-scale cell line screen (Garnett et al., 2012), MEK inhibition tends to have largely cytostatic effects in KRAS mutant cancers, causing <25% apoptosis in 90% of cell lines tested. The primarily cytostatic effects of MEK inhibitors may explain why they can slow tumor growth in vivo in KRAS mutant tumor xenografts, but rarely cause tumor regressions (Figures 4A and 4B; She et al., 2010). These findings are also consistent with the clinical experience with MEK inhibitors in KRAS mutant cancers, where stable disease is commonly observed, but true tumor regressions and/or responses are rarely seen (Adjei et al., 2008; Infante et al., 2010). However, the ability of MEK inhibitors to decrease proliferation and lead to stable disease in patients with KRAS mutant cancers suggests that MEK inhibitors may be good backbones for targeted therapy combinations. In particular, combination approaches that increase the cell death response to MEK inhibitors may be promising strategies to generate clinical responses in KRAS mutant cancers.
While MEK inhibition alone does not lead to pronounced apoptosis in KRAS mutant cancer cells, it may “prime” cells for death through induction of the pro-apoptotic protein BIM. Our results suggest that these increased levels of BIM are bound and inhibited by anti-apoptotic proteins, such as BCL-XL. Thus, BIM induction alone by MEK inhibitors is insufficient to cause apoptosis, but may leave KRAS mutant cancer cells primed for death by a second insult. Indeed, we found that ABT-263 could abrogate the inhibitory complex between BCL-XL and BIM, leading to robust apoptosis. In broad terms, this mechanism is consistent with prior findings that inhibition of another antiapoptotic protein, BCL-2, increases the efficacy of kinase inhibitors in HER2-amplified cancers, BRAF mutant melanomas, and acute myeloid leukemia cells (Milella et al., 2002; Konopleva et al., 2006; Cragg et al., 2008; Muranen et al., 2012). Thus, potentiators of apoptosis may be particularly effective when partnered with the appropriate targeted therapy in molecularly defined cancer subsets. Our results suggest that agents (e.g., ABT-263) that directly target BCL-XL or agents that decrease levels of BCL-XL by targeting upstream regulators may be particularly effective therapeutic combination partners with MEK inhibitors in KRAS mutant cancers.
EXPERIMENTAL PROCEDURES
Detailed experimental information is provided in Supplemental Experimental Procedures.
Pooled shRNA Screen and Analysis
Detailed pooled shRNA screen procedures are provided in Supplemental Experimental Procedures. Briefly, target cells were infected with lentivital pooled shRNA library and split into three aliquots. One aliquot was immediately frozen to represent the initial population. The remaining two aliquots were seeded into separate 15 cm plates. The next day, media containing DMSO or 1 µM selumetinib was added, and cells were cultured for 7 days. Genomic DNA was isolated from all three cell aliquots, and shRNA abundance was quantified by deep sequencing. The ratio of the abundance of each shRNA in MEK inhibitor-treated versus both the vehicle-treated and initial samples was calculated. For each cell line, a given shRNA was considered a “hit” if it showed a decrease in abundance of at least 2-fold relative to both the vehicle-treated and initial samples.
Mouse Treatment Studies
HCT116, SW620, or SW1463 cells were injected (5 × 106 cells per injection) into the flanks of athymic nude mice (Charles River Laboratories). Once tumors reached an average volume of ~100–200 mm3, mice were randomized into treatment arms and tumor volume was assessed by caliper measurements over a 21- to 28-day period. Lung tumors in LSL-KRASG12D mice were induced by inhalation of adenoviral Cre recombinase and monitored and measured by serial MRI scans as previously described (Engelman et al., 2008). All mouse studies were conducted through Institutional Animal Care and Use Committee (IACUC) approved animal protocols in accordance with institutional guidelines.
Patient Samples
Human tumor specimens were obtained from the Massachusetts General Hospital under Institutional Review Board-approved studies. All patients provided written, informed consent. KRAS mutation status was determined by the Massachusetts General Hospital Clinical Laboratory and Department of Pathology.
Statistical Analyses
One-way ANOVA with Tukey post-hoc test was used for Figures 2A, 2B, 3E, and 4E. A two-tailed t test was used for Figures 3B, 3C, and 4B. Statistical significance was established for p < 0.05.
Supplementary Material
Significance.
Although KRAS is the most commonly mutated oncogene in human cancer, KRAS has proven difficult to target pharmacologically, and no effective therapies exist for KRAS mutant cancers. Recently, there has been evidence that targeted therapy combinations inhibiting multiple downstream effectors of KRAS may be a promising approach for KRAS mutant cancers. Here, we report a pooled shRNA-drug screen designed to identify MEK inhibitor-based targeted therapy combinations for KRAS mutant cancers. Through this approach, we identified combined BCL-XL and MEK inhibition as a strategy with robust in vitro and in vivo efficacy in KRAS mutant cancer models. Thus, this targeted therapy combination may represent a potential therapeutic avenue for KRAS mutant cancers.
ACKNOWLEDGMENTS
This study was supported by grants from the NIH Gastrointestinal SPORE P50 CA127003 (to R.C. and J.E.); an American Cancer Society Institutional Research grant, a Damon Runyon Clinical Investigator award, a Conquer Cancer Foundation of ASCO Young Investigator award (to R.C.); and K08CA120060, R01CA137008, 1U01CA141457-01, the V Foundation, American Cancer Society RSG-06-102-01-CCE, the Ellison Foundation Scholar (all to J.E.); NIH Lung SPORE P50CA090578, R01CA140594 (to J.E. and K.W.); R01CA122794 and R01CA163896 (to K.W.); and 1U54HG006097-01 (to C.B.). Research support for this study was also provided to J.E. by AstraZeneca.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information includes four figures, four tables, and Supplemental Experimental Procedures and can be found with this article online at ttp://dx. doi.org/10.1016/j.ccr.2012.11.007.
REFERENCES
- Adjei AA, Cohen RB, Franklin W, Morris C, Wilson D, Molina JR, Hanson LJ, Gore L, Chow L, Leong S, et al. Phase I pharmacokinetic and pharmacodynamic study of the oral, small-molecule mitogen-activated protein kinase kinase 1/2 inhibitor AZD6244 (ARRY-142886) in patients with advanced cancers. J. in. Oncol. 2008;26:2139–2146. doi: 10.1200/JCO.2007.14.4956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbie DA, Tamayo P, Boehm JS, Kim SY, Moody SE, Dunn IF, Schinzel AC, Sandy P, Meylan E, Scholl C, et al. Systematic RNA interference reveals that oncogenic KRAS-driven cancers require TBK1. Nature. 2009;462:108–112. doi: 10.1038/nature08460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cragg MS, Jansen ES, Cook M, Harris C, Strasser A, Scott CL. Treatment of B-RAF mutant human tumor cells with a MEK inhibitor requires Bim and is enhanced by a BH3 mimetic. J. Clin. Invest. 2008;118:3651–3659. doi: 10.1172/JCI35437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelman JA, Chen L, Tan X, Crosby K, Guimaraes AR, Upadhyay R, Maira M, McNamara K, Perera SA, Song Y, et al. Effective use of PI3K and MEK inhibitors to treat mutant Kras G12D and PIK3CA H1047R murine lung cancers. Nat. Med. 2008;14:1351–1356. doi: 10.1038/nm.1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faber AC, Li D, Song Y, Liang MC, Yeap BY, Bronson RT, Lifshits E, Chen Z, Maira SM, García-Echeverría C, et al. Differential induction of apoptosis in HER2 and EGFR addicted cancers following PI3K inhibition. Proc. Natl. Acad. Sci. USA. 2009;106:19503–19508. doi: 10.1073/pnas.0905056106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garnett MJ, Edelman EJ, Heidorn SJ, Greenman CD, Dastur A, Lau KW, Greninger P, Thompson IR, Luo X, Soares J, et al. Systematic identification of genomic markers of drug sensitivity in cancer cells. Nature. 2012;483:570–575. doi: 10.1038/nature11005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Infante JR, Fecher LA, Nallapareddy S, Gordon MS, Flaherty KT, Cox DS, DeMarini DJ, Morris SR, Burris HA, Messersmith WA. Safety and efficacy results from the first-in-human study of the oral MEK 1/2 inhibitor GSK1120212. J. Clin. Oncol. 2010;28:15s. [Google Scholar]
- Jackson EL, Willis N, Mercer K, Bronson RT, Crowley D, Montoya R, Jacks T, Tuveson DA. Analysis of lung tumor initiation and progression using conditional expression of oncogenic K-ras. Genes Dev. 2001;15:3243–3248. doi: 10.1101/gad.943001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konopleva M, Contractor R, Tsao T, Samudio I, Ruvolo PP, Kitada S, Deng X, Zhai D, Shi YX, Sneed T, et al. Mechanisms of apoptosis sensitivity and resistance to the BH3 mimetic ABT-737 in acute myeloid leukemia. Cancer Cell. 2006;10:375–388. doi: 10.1016/j.ccr.2006.10.006. [DOI] [PubMed] [Google Scholar]
- Kumar MS, Hancock DC, Molina-Arcas M, Steckel M, East P, Diefenbacher M, Armenteros-Monterroso E, Lassailly F, Matthews N, Nye E, et al. The GATA2 transcriptional network is requisite for RAS oncogene-driven non-small cell lung cancer. Cell. 2012;149:642–655. doi: 10.1016/j.cell.2012.02.059. [DOI] [PubMed] [Google Scholar]
- Ley R, Balmanno K, Hadfield K, Weston C, Cook SJ. Activation of the ERK1/2 signaling pathway promotes phosphorylation and proteasome-dependent degradation of the BH3-only protein. Bim. J. Biol. Chem. 2003;278:18811–18816. doi: 10.1074/jbc.M301010200. [DOI] [PubMed] [Google Scholar]
- LoRusso P, Shapiro G, Pandya SS, Kwak EL, Jones C, Belvin M, Musib LC, de Crespigny A, McKenzie M, Gates MR, et al. A first-in-human phase Ib study to evaluate the MEK inhibitor GDC-0973, combined with the pan-PI3K inhibitor GDC-0941, in patients with advanced solid tumors. J. Clin. Oncol. 2012;30 (suppl; abstr 2566) [Google Scholar]
- Luo J, Emanuele MJ, Li D, Creighton CJ, Schlabach MR, Westbrook TF, Wong KK, Elledge SJ. A genome-wide RNAi screen identifies multiple synthetic lethal interactions with the Ras oncogene. Cell. 2009;137:835–848. doi: 10.1016/j.cell.2009.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malumbres M, Barbacid M. RAS oncogenes: the first 30 years. Nat. Rev. Cancer. 2003;3:459–465. doi: 10.1038/nrc1097. [DOI] [PubMed] [Google Scholar]
- Milella M, Estrov Z, Kornblau SM, Carter BZ, Konopleva M, Tari A, Schober WD, Harris D, Leysath CE, Lopez-Berestein G, et al. Synergistic induction of apoptosis by simultaneous disruption of the Bcl-2 and MEK/MAPK pathways in acute myelogenous leukemia. Blood. 2002;99:3461–3464. doi: 10.1182/blood.v99.9.3461. [DOI] [PubMed] [Google Scholar]
- Montagut C, Settleman J. Targeting the RAF-MEK-ERK pathway in cancer therapy. Cancer Lett. 2009;283:125–134. doi: 10.1016/j.canlet.2009.01.022. [DOI] [PubMed] [Google Scholar]
- Muranen T, Selfors LM, Worster DT, Iwanicki MP, Song L, Morales FC, Gao S, Mills GB, Brugge JS. Inhibition of PI3K/mTOR leads to adaptive resistance in matrix-attached cancer cells. Cancer Cell. 2012;21:227–239. doi: 10.1016/j.ccr.2011.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahmani M, Anderson A, Habibi JR, Crabtree TR, Mayo M, Harada H, Ferreira-Gonzalez A, Dent P, Grant S. The BH3-only protein Bim plays a critical role in leukemia cell death triggered by concomitant inhibition of the PI3K/Akt and MEK/ERK1/2 pathways. Blood. 2009;114:4507–4516. doi: 10.1182/blood-2008-09-177881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholl C, Fröhling S, Dunn IF, Schinzel AC, Barbie DA, Kim SY, Silver SJ, Tamayo P, Wadlow RC, Ramaswamy S, et al. Synthetic lethal interaction between oncogenic KRAS dependency and STK33 suppression in human cancer cells. Cell. 2009;137:821–834. doi: 10.1016/j.cell.2009.03.017. [DOI] [PubMed] [Google Scholar]
- She QB, Halilovic E, Ye Q, Zhen W, Shirasawa S, Sasazuki T, Solit DB, Rosen N. 4E-BP1 is a key effector of the oncogenic activation of the AKT and ERK signaling pathways that integrates their function in tumors. Cancer Cell. 2010;18:39–51. doi: 10.1016/j.ccr.2010.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh A, Greninger P, Rhodes D, Koopman L, Violette S, Bardeesy N, Settleman J. A gene expression signature associated with “K-Ras addiction“ reveals regulators of EMT and tumor cell survival. Cancer Cell. 2009;15:489–500. doi: 10.1016/j.ccr.2009.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh A, Sweeney MF, Yu M, Burger A, Greninger P, Benes C, Haber DA, Settleman J. TAK1 inhibition promotes apoptosis in KRAS-dependent colon cancers. Cell. 2012;148:639–650. doi: 10.1016/j.cell.2011.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speranza G, Kinders RJ, Khin S, Weil MK, Do KT, Horneffer Y, Juwara L, Allen D, Williams PM, Lih CJ, et al. Pharmacodynamic biomarker-driven trial of MK-2206, and AKT inhibitor, with AZD6244 (selumetinib), a MEK inhibitor, in patients with advanced colorectal carcinoma. J. Clin. Oncol. 2012;30 (suppl; abstr 3529) [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taube JH, Herschkowitz JI, Komurov K, Zhou AY, Gupta S, Yang J, Hartwell K, Onder TT, Gupta PB, Evans KW, et al. Core epithelial-to-mesenchymal transition interactome gene-expression signature is associated with claudin-low and metaplastic breast cancer subtypes. Proc. Natl. Acad. Sci. USA. 2010;107:15449–15454. doi: 10.1073/pnas.1004900107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tse C, Shoemaker AR, Adickes J, Anderson MG, Chen J, Jin S, Johnson EF, Marsh KC, Mitten MJ, Nimmer P, et al. ABT-263: a potent and orally bioavailable Bcl-2 family inhibitor. Cancer Res. 2008;68:3421–3428. doi: 10.1158/0008-5472.CAN-07-5836. [DOI] [PubMed] [Google Scholar]
- Willis SN, Fletcher JI, Kaufmann T, van Delft MF, Chen L, Czabotar PE, Ierino H, Lee EF, Fairlie WD, Bouillet P, et al. Apoptosis initiated when BH3 ligands engage multiple Bcl-2 homologs, not Bax or Bak. Science. 2007;315:856–859. doi: 10.1126/science.1133289. [DOI] [PubMed] [Google Scholar]
- Young A, Lyons J, Miller AL, Phan VT, Alarcón IR, McCormick F. Ras signaling and therapies. Adv. Cancer Res. 2009;102:1–17. doi: 10.1016/S0065-230X(09)02001-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.