Abstract
Here we report a modified peptide reagent useful for the rapid, native elution of protein complexes containing a Protein-A-tagged component. We tested this reagent for the elution of tagged endogenous protein complexes from yeast (Nup53p/Nup170p dimer; Nup1p/Kap95p/Kap60p trimer; pentameric GINS complex) and bacteria (RNAP holoenzyme). The majority of the affinity-isolated material is released within 15 minutes under mild conditions, and the elution reagent itself is readily depleted from the elution mixture by simple spin column gel filtration. This reagent is ideal for eluting protein complexes after Protein A/IgG affinity isolation when protease cleavage is not possible or not desirable and facile depletion of the elution reagent is needed.
Keywords: native elution, competitive elution, Protein A, SpA, PrA, affinity isolation, affinity capture
Affinity isolation methodologies have led the successful drive to capture endogenous protein complexes. Among these affinity systems, the S. aureus Protein A (SpA)/mammalian immunoglobulin G (IgG) interaction (1,2) is widely utilized. Moreover, tandem affinity purification (TAP)-tagged (3) yeast strains available from genome-wide tagging efforts (4,5) incorporate tandem repeats of the SpA-derived IgG binding Z-domain (6). Thus, a vast library of strains exist for the purification of diverse protein complexes via the SpA/IgG interaction.
To recover protein complexes in the most active and physiologically relevant state, it is desirable to work fast – minimizing the time between capture and assay. Native elution from SpA/IgG affinity systems generally follows two lines: competitive elution of the tagged bait protein and its coprecipitated interactors (7,8), or protease cleavage of the tag from the bait protein, releasing the bait and its coprecipitated interactors (3). The efficiency of release of different protein complexes from the stationary phase by protease cleavage is not uniform, and cleavage requires long incubation times ranging from hours to overnight (9,10). Improvement of these parameters, if at all possible, would likely require fundamental engineering of the protease. SpA binds to the hinge region on the Fc fragment of IgG; a bacteriophage display screen found a 13 amino acid peptide (FcIII peptide: DCAWHLGELVWCT) that bound competitively to the same region with high affinity (11). Our group previously developed a modified, higher solubility version of this peptide capable of competitively eluting protein complexes in their native state at high yield within ~2 h (biotinylated FcIII peptide, termed Bio-Ox) (8). We reasoned that a larger increase in the solubility of FcIII could reduce the time to achieve competitive elution and therefore sought to improve this approach further.
As the N terminus has previously been used to increase FcIII peptide solubility via conjugation to biotin (8), we tested several alternative N-terminal substitutions aimed at further solubility gains. Among our tested variants, a modification comprising a polyethylene glycol moiety of four units length (PEG[4]) proved to be the most effective at increasing and maintaining FcIII peptide solubility. Longer PEG modifications, up to eight units in length, exhibited problems with in-solution stability, precipitating out when held on ice. Coupling additional groups (e.g., biotin, arginine, poly-histidine) to the PEG moiety did not further improve performance (data shown for R-PEG[4], see below). The new reagent described here, termed PEGylOx, is an FcIII peptide N-terminally PEGylated with a four-unit polymer and cyclized by oxidation of the cysteines to cystine (Figure 1A).
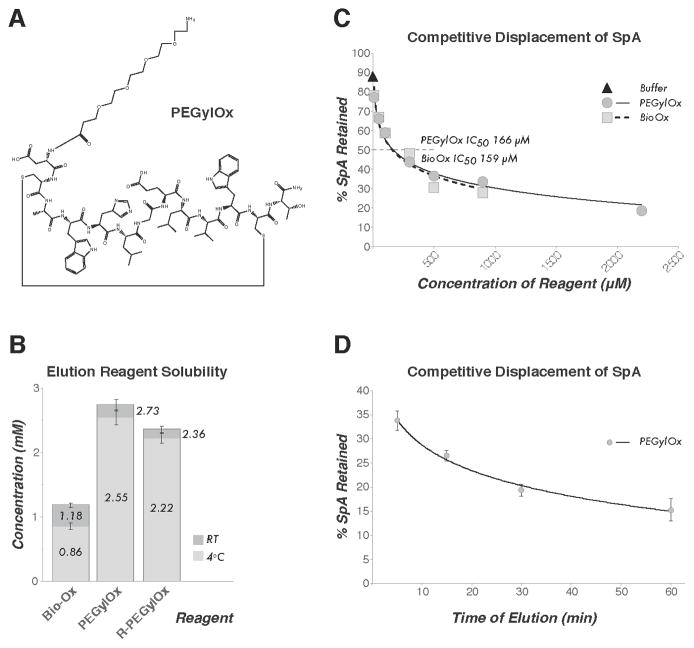
Figure 1. PEGylation of FcIII peptide increases solubility and maintains competitive displacement of S. aureus Protein A (SpA) from mammalian IgG.
(A) The chemical structure of PEGylOx. (B) The concentrations of Bio-Ox, PEGylOx, and R-PEGylOx at room temperature (RT) or 4°C, as determined by UV280 spectrophotometry on three independent aliquots of lyophilized peptide suspended in the described buffered solution. Error bars indicate the standard deviation (SD) (C) The percentage of SpA retained by immobilized rabbit IgG after treatment with differing concentrations of Bio-Ox or PEGylOx for 1 h at RT – as determined by image densitometry of Coomassie stained SDS-PAGE gels (detailed in Supplementary Section S3). for both reagents is indicated. All values are A buffer only control is included, and the IC50 the average of duplicate experiments. (D) The percentage of SpA retained by immobilized rabbit IgG after treatment with a saturated solution of PEGylOx for 5, 15, 30, or 60 min. All values are the average of triplicate experiments with SD indicated.
For this study, Bio-Ox was synthesized as previously described (8). PEGylated peptides were synthesized by standard Fmoc solid-phase synthesis methods. Incorporation of PEG spacers into the peptide sequence was accomplished using N-Fmoc-amido-(PEG) n-acid building blocks (Detailed in Supplementary Section S1, or obtained from 21st Century Biochemicals, Marlboro, MA). Prior to use, lyophilized peptides were resuspended in a 40 mM Tris-Cl buffered solution at pH 8.0, with 100 mM NaCl, 1 mM EDTA (pH 8.0), 0.01% v/v Tween 20, and 5% v/v ethanol. This solution was centrifuged at top speed in a bench-top microcentrifuge for 5 min at room temperature (RT) to remove any insoluble fraction. The resulting supernatant was assayed by UV spectrophotometry at a wavelength of 280 nm (UV280) to determine the final peptide concentration using an estimated molar extinction coefficient (12) (ε280) of 11125 M−1cm−1 (Supplementary Section S2A). We found that the addition of 5% v/v ethanol aided modified FcIII peptide solubility, increasing Bio-Ox solubility almost 3× (0.440 mM to 1.18 mM) (8), and exhibited no observable negative effect on the stability of any protein complex we have tested thus far (data not shown). PEGylOx was found to be 2.3× more soluble at saturation than Bio-Ox at RT and almost 3× more soluble at 4°C, demonstrating the advantage of the PEG[4] conjugate over biotin as a solubility enhancer. Although we reasoned that additional charged groups may provide further solubility gains, no such advantage was observed by the coupling of an R residue to the PEG[4] moiety (Figure 1B). Next, we conducted an assay for the competitive displacement of wild type (wt) SpA from immobilized IgG. Bio-Ox and PEGylOx each exhibited a similar (Figure 1C, Supplementary Section IC50 S2B) — consistent with the ability of both Bio-Ox and PEGylOx to effectively displace SpA, and the complete conservation of the mechanism of action of the peptide upon substitution of the biotin modification for PEG[4].
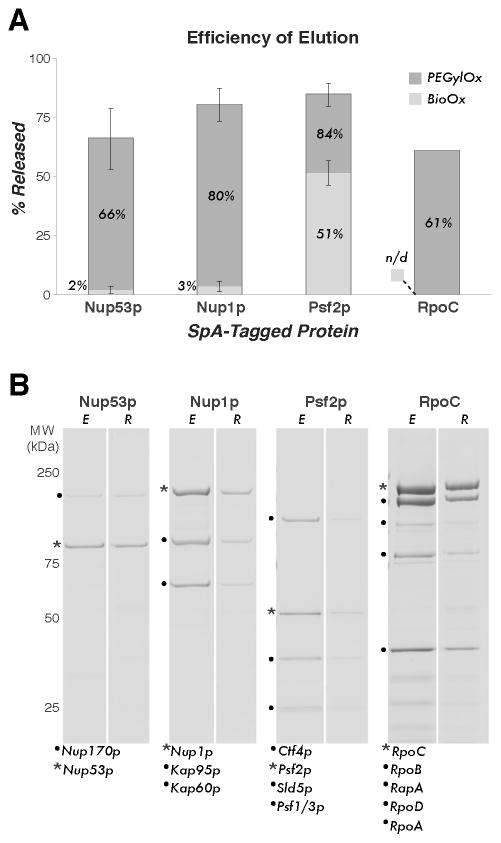
Macromolecular complexes tend to disintegrate with time after isolation; thus, to determine the shortest sensible time-scale of elution, we conducted a time course to displace SpA from immobilized IgG with a saturated PEGylOx solution. We found that > 70% of the SpA was released within 15 min (Figure 1D, Supplementary Section S2C). We then tested saturated solutions of PEGylOx and Bio-Ox for their ability to elute already well-defined SpA-tagged protein complexes (13–15) (Supplementary Section S4A) isolated from endogenous sources (Supplementary Protocol) and found that the saturated PEGylOx solution released numerous protein complexes with greater efficiency and uniformity between samples than the saturated Bio-Ox solution. While Bio-Ox is effective with longer incubation times (8), PEGylOx shows a distinct advantage at short elution times. Example protein complexes that we quantified exhibited approximately 60%–85% efficiency of release during a 15 minutes incubation with PEGylOx. In contrast, Bio-Ox was essentially unable to release two of the test complexes within this time, and one complex was released with only moderate efficacy (Figure 2). We conclude that PEGylOx is an improved, rapidly acting reagent for competitive native elution of SpA-tagged protein complexes.
Figure 2. Efficiency of elution of endogenous protein complexes by competitive displacement using Bio-Ox or PEGylOx.
(A) The percentage of S. aureus Protein A (SpA)-tagged protein released from immobilized rabbit IgG after treatment with Bio-Ox or PEGylOx for 15 min at room temperature (RT). At least three experimental replicates were averaged where error bars are shown, indicating the standard deviation (SD). The value for RpoC-SpA release was determined from a single experiment using PEGylOx only (n/d: Bio-Ox not done). All values determined by image densitometry of Coomassie blue stained SDS-PAGE gels (Supplementary Section S3). (B) Example Coomassie blue stained gels showing elution of protein complexes using PEGylOx for each of the proteins quantified in (A). The SpA tagged protein is marked with an asterisk (*), the eluted fractions are denoted E and the retained fractions are denoted R. Displayed are the S. cerevisiae Nup53p/Nup170p dimer (13), Nup1p/Kap95p/Kap60p trimer (13), and pentameric GINS complex (14), and the E. coli RNA polymerase holoenzyme (15). All complexes were prepared from ~0.5 g of cell material. Co-precipitating proteins are labeled based on the referenced work and MALDI-MS analyses (data not shown).
Finally, as with Bio-Ox (8), PEGylOx may be depleted from samples by spin-column-based gel filtration chromatography (Supplementary Protocol). We assayed the level of depletion for PEGylOx and found greater than 100× depletion of the peptide through the application of eluted samples to desalting/buffer exchange spin columns with a 40 kDa molecular weight cut-off (MWCO) as assayed by UV280 and Coomassie staining. We tested Bio-Rad (Hercules, CA) Bio-Gel P-30 (#732–6223) and Thermo (Waltham, MA) Zeba 40K MWCO (#87764) spin columns and achieved equivalent results (Supplemetary Section S5). Resins with lower MWCOs that we tested (Bio-Rad Bio-Gel P-6 #732–6221 and Thermo Zeba 7K MWCO #89877; UV280 data for P-6 in Supplementary Section S5) were only able to deplete PEGylOx by ~10–20×. In contrast, we were unable to significantly deplete 3xFLAG peptide in this manner (data not shown). Hence PEGylOx also exhibits the beneficial property of being depleted from samples by desalting/buffer exchange when needed for downstream applications.
Unfortunately, the optimization of affinity isolation/native elution often requires attention to a very broad range of parameters – pH, overall ionic strength, salt-type(s) and concentration, detergent-type(s) and concentration, time, and temperature, among others. Optimized parameters for the capture of a given complex from cell extract will often need to be determined empirically through trial and error – but some prior knowledge about the requirements of the affinity system can help provide boundaries for exploring new isolation conditions.
Protein affinity isolation is typically carried out at 4°C, which retards the disintegration of most protein complexes and helps reduce enzyme activity within the cell extract; this includes proteolysis – although an appropriate cocktail of protease inhibitors should also be included. Solvents for extracting protein complexes from cells typically employ a near physiological pH (~7.0–8.0 based on the pH of the cytosol), although there is good reason to vary this parameter for complexes believed to reside in cellular compartments of differing physiological pH. In this study, we have constrained our pH to a range we know works well for the SpA affinity tag. The canonical SpA interaction with Human IgG Fc occurs through an induced hydrophobic fit, which is weakened as the pH is shifted to acidic values. This pH sensitivity is exploited to fractionate differing IgG subclasses from one another on SpA affinity media (16–19). Hence, a slightly alkaline pH promotes the capture and retention of SpA-tagged proteins on IgG media while still remaining within the typical physiological range. This phenomenon bears a connection to the number of SpA domains present (18), and we have observed that the affinity of tandem repeats of the Z-domain for IgG is significantly diminished at pH ~6.0 and below (20). Moreover, since PEGylOx interacts with Fc through a highly conserved mechanism (11), its optimal binding conditions are likely to be very similar to those mentioned above. Along these lines, we have successfully achieved competitive displacement of protein complexes using PEGylOx in HEPES buffered solution at pH 7.4 and ammonium acetate, pH 7.0 – with results comparable to those presented here using Tris pH 8.0 (data not shown). Finally, keep in mind that the multiplicity and variety of IgG binding domains present in different SpA-tag configurations may factor into performance through avidity effects (2,18,21,22) (Supplementary Section S4B).
Through affinity isolation and subsequent washing, the complex of interest is purified away from the bulk cell extract and thus away from the majority of proteolytic activity. At this point, the importance of low temperature in maintaining the integrity of the protein complex may be somewhat diminished. For many of our complexes we carry out brief post-capture washes and a subsequent 15 min incubation with PEGylOx at RT (Supplementary Protocol). This is primarily a matter of convenience, although elution is expected to occur more rapidly as temperature increases (i.e., at RT versus 4°C). The precise extent of the difference in elution efficiency can vary from complex to complex (8), but going above RT could have the undesirable side effect of accelerating the disintegration of the complex of interest. In cases where we observe that complex components are labile when working at RT, we make every effort to work at between 0°–4°C. However, in such cases, we take this as an indicator that further optimization of the extraction solvent may provide greater stability during RT handling. For this work, the following extraction and washing solutions were used: Nup53p and Nup1p – 40 mM Tris-Cl buffer solution at pH 8.0 with 50 mM trisodium citrate, 300 mM NaCl, 0.1% v/v Tween 20, and 1 mM EDTA (pH 8.0); Psf2p – 20 mM HEPES-Na buffered solution at pH 7.4 with 150 mM NaCl, 2 mM MgCl2, 0.1% v/v Tween 20; RpoC – HEPES-Na buffered solution at pH 7.4 with 150 mM NaCl, 2 mM MgCl2, 0.1 mM CaCl2, and 0.1% v/v Tween 20, including the addition of DNase I to extracts.
Supplementary Material
Method summary.
To improve isolation of Protein-A tagged protein complexes, we modified our previously described competitive elution peptide (Bio-Ox) with the addition of a polyethylene glycol moiety of four unit lengths to increase solubility. This new peptide (PEGylOx) enables elution of affinity-isolated material in less than 15 minutes under mild reaction conditions.
Acknowledgments
This work was funded by the National Institutes of Health (NIH)/National Center for Research Resources (NCRR) grant no. U54 GM103511. We thank Zachary Quinkert for stimulating discussion; Lars Westblade, Seth Darst, and Robert Landick for providing the RpoC-SpA E. coli strain and useful information; and Brian Chait for providing the Psf2p-SpA strain, and for stimulating discussions.
Footnotes
To purchase reprints of this article, contact: biotechniques@fosterprinting.com
Supplementary material for this article is available at www.BioTechniques.com/article/114012.
Competing Interests
The authors declare no competing interests.
References
- 1.Lindmark R, Thorén-Tolling K, Sjöquist J. Binding of immunoglobulins to protein A and immunoglobulin levels in mammalian sera. J Immunol Methods. 1983;62:1–13. doi: 10.1016/0022-1759(83)90104-7. [DOI] [PubMed] [Google Scholar]
- 2.Moks T, Abrahmsén L, Nilsson B, Hellman U, Sjöquist J, Uhlén M. Staphylococcal protein A consists of five IgG-binding domains. Eur J Biochem. 1986;156:637–643. doi: 10.1111/j.1432-1033.1986.tb09625.x. [DOI] [PubMed] [Google Scholar]
- 3.Rigaut G, Shevchenko A, Rutz B, Wilm M, Mann M, Seraphin B. A generic protein purification method for protein complex characterization and proteome exploration. Nat Biotechnol. 1999;17:1030–1032. doi: 10.1038/13732. [DOI] [PubMed] [Google Scholar]
- 4.Ghaemmaghami S, Huh WK, Bower K, Howson RW, Belle A, Dephoure N, O’Shea EK, Weissman JS. Global analysis of protein expression in yeast. Nature. 2003;425:737–741. doi: 10.1038/nature02046. [DOI] [PubMed] [Google Scholar]
- 5.Gavin AC, Aloy P, Grandi P, Krause R, Boesche M, Marzioch M, Rau C, Jensen LJ, et al. Proteome survey reveals modularity of the yeast cell machinery. Nature. 2006;440:631–636. doi: 10.1038/nature04532. [DOI] [PubMed] [Google Scholar]
- 6.Nilsson B, Moks T, Jansson B, Abrahmsén L, Elmblad A, Holmgren E, Henrichson C, Jones TA, Uhlén M. A synthetic IgG-binding domain based on staphylococcal protein A. Protein Eng. 1987;1:107–113. doi: 10.1093/protein/1.2.107. [DOI] [PubMed] [Google Scholar]
- 7.Nilsson J, Nilsson P, Williams Y, Pettersson L, Uhlén M, Nygren PA. Competitive elution of protein A fusion proteins allows specific recovery under mild conditions. Eur J Biochem. 1994;224:103–108. doi: 10.1111/j.1432-1033.1994.tb20000.x. [DOI] [PubMed] [Google Scholar]
- 8.Strambio-de-Castillia C, Tetenbaum-Novatt J, SImai B, Chait BT, Rout MP. A method for the rapid and efficient elution of native affinity-purified protein A tagged complexes. J Proteome Res. 2005;4:2250–2256. doi: 10.1021/pr0501517. [DOI] [PubMed] [Google Scholar]
- 9.Polayes DA, Parks TD, Johnston SA, Dougherty WG. Application of TEV Protease in Protein Production. In: Reischl U, editor. Methods Mol Med. Vol. 13. 1998. pp. 169–83. [DOI] [PubMed] [Google Scholar]
- 10.Puig O, Caspary F, Rigaut G, Rutz B, Bouveret E, Bragado-Nilsson E, Wilm M, Séraphin B. The tandem affinity purification (TAP) method: a general procedure of protein complex purification. Methods. 2001;24:218–229. doi: 10.1006/meth.2001.1183. [DOI] [PubMed] [Google Scholar]
- 11.DeLano WL, Ultsch MH, de Vos AM, Wells JA. Convergent solutions to binding at a protein-protein interface. Science. 2000;287:1279–1283. doi: 10.1126/science.287.5456.1279. [DOI] [PubMed] [Google Scholar]
- 12.Grimsley GR, Pace CN. Spectrophotometric determination of protein concentration. In: Taylor G, editor. Curr Protoc Protein Sci. Wiley; 2004. p. 3.1. [DOI] [PubMed] [Google Scholar]
- 13.Alber F, Dokudovskaya S, Veenhoff LM, Zhang W, Kipper J, Devos D, Suprapto A, Karni-Schmidt O, Williams R, Chait BT, et al. The molecular architecture of the nuclear pore complex. Nature. 2007;450:695–701. doi: 10.1038/nature06405. [DOI] [PubMed] [Google Scholar]
- 14.Gambus A, van Deursen F, Polychronopoulos D, Foltman M, Jones RC, Edmondson RD, Calzada A, Labib K. A key role for Ctf4 in coupling the MCM2-7 helicase to DNA polymerase alpha within the eukaryotic replisome. EMBO J. 2009;28:2992–3004. doi: 10.1038/emboj.2009.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Westblade LF, Minakhin L, Kuznedelov K, Tackett AJ, Chang EJ, Mooney RA, Vvedenskaya I, Wang QJ, Fenyö D, Rout MP, et al. Rapid isolation and identification of bacteriophage T4-encoded modifications of Escherichia coli RNA polymerase: a generic method to study bacteriophage/host interactions. J Proteome Res. 2008;7:1244–1250. doi: 10.1021/pr070451j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Deisenhofer J. Crystallographic refinement and atomic models of a human Fc fragment and its complex with fragment B of protein A from Staphylococcus aureus at 2.9- and 2.8-Å resolution. Biochemistry. 1981;20:2361–2370. [PubMed] [Google Scholar]
- 17.Tashiro M, Tejero R, Zimmerman DE, Celda B, Nilsson B, Montelione GT. High-resolution solution NMR structure of the Z domain of staphylococcal protein A. J Mol Biol. 1997;272:573–590. doi: 10.1006/jmbi.1997.1265. [DOI] [PubMed] [Google Scholar]
- 18.Brown NL, Bottomley SP, Scawen MD, Gore MG. A study of the interactions between an IgG-binding domain based on the B domain of staphylococcal protein A and rabbit IgG. Mol Biotechnol. 1998;10:9–16. doi: 10.1007/BF02745859. [DOI] [PubMed] [Google Scholar]
- 19.Schwarz A. Affinity purification of monoclonal antibodies. In: Bailon P, Ehrlich GK, Fung WJ, Berthold W, editors. Methods Mol Biol. Humana Press; 2000. pp. 49–56. [Google Scholar]
- 20.LaCava J, Houseley J, Saveanu C, Petfalski E, Thompson E, Jacquier A, Tollervey D. RNA degradation by the exosome is promoted by a nuclear polyadenylation complex. Cell. 2005;121:713–724. doi: 10.1016/j.cell.2005.04.029. [DOI] [PubMed] [Google Scholar]
- 21.Ljungberg UK, Jansson B, Niss U, Nilsson R, Sandberg BEB, Nilsson B. The interaction between different domains of staphylococcal protein A and human polyclonal IgG, IgA, IgM and F(ab′)2: Separation of affinity from specificity. Mol Immunol. 1993;30:1279–1285. doi: 10.1016/0161-5890(93)90044-c. [DOI] [PubMed] [Google Scholar]
- 22.Jansson B, Uhlén M, Nygren PA. All individual domains of staphylococcal protein A show Fab binding FEMS. Immunol Med Microbiol. 1998;20:69–78. doi: 10.1111/j.1574-695X.1998.tb01112.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.