Abstract
Troponin T (TnT) plays a major role in striated muscle contraction. We recently demonstrated that the fast skeletal muscle TnT3 isoform is localized in the muscle nucleus, and either its full-length or COOH-terminus leads to muscle cell apoptosis. Here, we further explored the mechanism by which it enters the nucleus and promotes cytotoxicity. Amino acid truncation and substitution showed that its COOH-terminus contains a dominant nuclear/nucleolar localization sequence (KLKRQK) and the basic lysine and arginine residues might play an important role in the nuclear retention and nucleolar enrichment of KLKRQK-DsRed fusion proteins. Deleting this domain or substituting lysine and arginine residues (KLAAQK) resulted in a dramatic loss of TnT3 nuclear and nucleolar localization. In contrast, the GATAKGKVGGRWK domain-DsRed construct localized exclusively in the cytoplasm, indicating that a nuclear exporting sequence is possibly localized in this region. Additionally, we identified a classical DNA-binding Leucine Zipper Domain (LZD) which is conserved among TnT isoforms and species. Deletion of LZD or KLKRQK sequence significantly reduced cell apoptosis compared to full-length TnT3. We conclude that TnT3 contains both a nuclear localization signal and a DNA binding domain, which may mediate nuclear/nucleolar signaling and muscle cell apoptosis.
Keywords: Troponin T3, nuclear/nucleolar localization, leucine zipper domain, apoptosis
INTRODUCTION
Muscle fiber depolarization leads to sarcoplasmic reticulum calcium release (Tang et al. 2011; Wang et al. 2012), which causes conformational changes in the troponin (Tn) complex to expose binding sites for myosin on the actin filaments, and initiating the interaction between myosin and actin, causing muscle contraction (Geeves and Holmes 1999; Jimenez-Moreno et al. 2008; Jimenez-Moreno et al. 2010). The Tn complex is composed of three subunits: the calcium-binding TnC; the inhibitory TnI; and the Tm-binding TnT. Thin-filament regulatory proteins Tn and tropomyosin (Tm) sense changes in intracellular calcium concentration (Gordon et al. 2000; Szczesna and Potter 2002; Tobacman 1996), and bind to the myofilaments; this classic Tn regulatory function takes place on thin filaments in the myoplasm. However, growing evidence indicates that Tn complex and Tm may also localize in the cell nucleus. For instance, actin-binding proteins including noncardiac tropomyosin have been identified in the nuclei of cultured mammalian nonmuscle cells (Dingova et al. 2009); Drosophila TnI and tropomyosin enter the nuclei and regulate chromosomal stability (Sahota et al. 2009). Note that cardiac TnI and TnT are associated with the nucleus in cardiac myocytes of adult humans (Bergmann et al. 2009; Kajstura et al. 2010). The Tn complex was recently found in the cardiac nucleus of neonatal mice (Asumda and Chase 2012), indicating a possible function in nuclear signaling.
We recently reported that the fast skeletal muscle TnT3 and its fragments are located in the myofiber nucleus as well as the cytoplasm, and their relative abundance changes with aging. We also showed that various TnT3 regions have different apoptotic effects when overexpressed in the mouse muscle cell line C2C12 (Zhang et al. 2011), which is consistent with reports from the Jin laboratory indicating that nonmyofilament-associated TnT and its fragments are somehow toxic and lead to apoptosis when transiently overexpressed in C2C12 or NIH3T3 cells (Jeong et al. 2009). Since we reported that TnT3 cytotoxicity may be related to its nuclear/nucleolar localization (Zhang et al. 2011), here, we examine the mechanisms of TnT3 subcellular localization and cytotoxicity.
The nuclear localization signal (NLS) of TnT and other Tn subunits has been predicted but not verified (Bergmann et al. 2009). A functional overlap between NLS and nucleolar localization signal (NoLS) has been shown (Scott et al. 2010). In this study, we sought to identify the peptide sequences needed to regulate TnT3 nuclear/nucleolar targeting and to define the TnT3 nuclear or nucleolar amino acid sequence that induces cytotoxicity. We mapped the NLS/NoLS at the COOH-terminus of TnT3 by sequential deletion and site-directed mutagenesis and, using flow cytometry, found a Leucine Zipper Domain (LZD) that mediates cell apoptosis. This study is the first evidence that TnT3 contains genuine NLSs/NoLSs and mediates DNA binding through its LZD to function as a transcription factor.
MATERIALS AND METHODS
Cell culture and transfection
The C2C12 cell line was cultured as described (Zhang et al. 2009). Only undifferentiated C2C12 myoblasts, which do not exhibit endogenous myofilaments, were used for transient exogenous protein expression. Briefly, undifferentiated C2C12 myoblasts were plated on tissue culture dishes or glass coverslips coated with 0.5% gelatin in growth medium (GM) consisting of Dulbecco’s modified Eagle Medium (DMEM, 1 g/l glucose) containing 10% FBS (Atlanta Biologicals, Atlanta, GA) and 2 mM Glutamax (Invitrogen, Carlsbad, CA). To induce C2C12 differentiation, cells about 80% confluent were switched to differentiation medium consisting of Dulbecco’s modified Eagle Medium (DMEM, 1 g/l glucose) containing 2% horse serum (Atlanta Biologicals, Atlanta, GA) and 2 mM Glutamax (Invitrogen). Lipofactamine 2000 (Invitrogen) was used for cell transfection.
Construction of expression plasmids
The plasmids for expressing different NH2-terminally DsRed-fused TnT3 COOH-terminal fragments (designated as TnCT-MD/, -BD/, -KL/, -YD/, -DQ/, and -GA/DsRed) were constructed according to our previous report (Zhang et al. 2011). Briefly, we constructed several truncation mutants for mapping TnT3 NLS/NoLS using specific PCR primers with HindIII/SacII or XhoI/NotI restriction enzyme digestion sites flanking the 5’ end of the forward and reverse primers, respectively. PCR was performed using a full-length TnT3/DsRed plasmid as a template. Table 1 lists primer sequences. Sequencing confirmed all constructs (DNA Sequencing Service, GenScript, Piscataway, NJ).The amplified product was digested by HindIII/SacII or XhoI/NotI, followed by ligation into DsRed2-N1 backbone, generated by HindIII/SacII digestion of pDsRed2-N1 vector (Clontech, PaloAlto, CA) or XhoI/NotI digestion of TnFL/DsRed vector, resulting in the various TnT3 COOH-terminus fragments conjugated with DsRed at the COOH terminus of each fragment. To construct TnFL-GFP and -DsRed double fluorescent conjugated fusion protein (GFP/TnFL/DsRed), GFP cDNA was cloned using designed primers (see Table 1) and pEYFP-N1as a PCR template. The PCR product was XhoI/HindIII, digested and ligated into the XhoI/HindIII digested TnFL/DsRed vector as described (Zhang et al. 2011).
Table 1. Primers designed for TnT3 constructs.
Restriction enzyme recognition sites are underlined, and the sequences in red were sites for mutagenesis into alanine.
| Primers for truncation of TnT3 COOH terminus | |
| TnCTMD-F | 5’-GCTAAAGCTTGCCGCCATGCTGAACATTGACCATCTTAGC-3’ (HindIII) |
| TnCTBD-F | 5’-GCTAAAGCTTGCCGCCATGGCCAAGGAACTCTGGGATACC-3’ (HindIII) |
| TnCTKL-F | 5’-GCTACTCGAGGCCGCCATGAAGCTGAAACGTCAGAAA-3’ (XhoI) |
| TnCTYD-F | 5’-GCTACTCGAGGCCGCCATGTATGATATCACCACCCTC-3’ (XhoI) |
| TnCTDQ-F | 5’-GCTACTCGAGGCCGCCATGGACCAAGCCCAGAAGCAC-3’ (XhoI) |
| TnCTGA-F | 5’-GCTAAAGCTTGCCGCCATGGGTGCCACAGCCAAGG-3’ (HindIII) |
| TnCT-R1 | 5’-GCTACCGCGGCTTCCAGCGCCCACCGAC-3’ (SacII) |
| TnCT-R2 | 5’-GCTAGCGGCCGCTACAGGAACAGGTGGTG-3’ (NotI) |
| Primers for amino acid substitution by site-directed mutagenesis | |
| M1-F | |
| M1-R | 5’-CAATGTTCAGAGGTGCTGCTGCTTCGGCAAGAATC-3’ |
| M2-F | |
| M2-R | 5’-CCCAGAGTTCTGCGGCTGCGTCTGCCAGTGCGTCATCGCTAAG-3’ |
| M3-F | |
| M3-R | 5’-GATATCATATTTCTGTGCTGCCAGCTTCTCCCCAAAC-3’ |
| M4-F | |
| M4-R | 5’-CGAATTTGTCAGTCTCTGCTTGGTACAAGGTATCCCATGCTTCCTTGGCC-3’ |
| Primers for construction of deletion plasmids | |
| TnFLΔBD-F | 5’-AAGATTCTTGCCGAAGCCAAGGAACTCTGG-3’ |
| TnFLΔBD-R | 5’-CCAGAGTTCCTTGGCTTCGGCAAGAATCTT-3’ |
| TnFLΔLZD-F | 5’-GAGACTGACAAATTCG-3’ |
| TnFLΔLZD-F | 5’-TTCCTTGGCCTTGTCC-3’ |
| TnFLΔNLS-F | 5’-TATGATATCACCACCC-3’ |
| TnFLΔNLS-F | 5’-CTCCCCAAACTCGAA-3’ |
| Primers for BD or NLS DsRed fusion protein-expressing plasmids | |
| BD-Red/F | 5’-GCTACTCGAGGCCGCCATGAGGCGCAAGCCTCTGAACATTGACCATCTTAGCGATGACAAGCTGAGGGACAAGATGGCCTCCTCCGAGAACGT-3’ (XhoI) |
| NLS-Red/F | 5’-GCTACTCGAGGCCGCCATGAAGCTGAAACGTCAGAAAATGGCCTCCTCCGAGAACGT -3’ (XhoI) |
| BD/NLS-Red/R | 5’-GCTAGCGGCCGCTACAGGAACAGGTGGTG-3’ (NotI) |
| Primers for construction of GFP/TnFL/DsRed fusion protein-expressing plasmids | |
| TnT3GR-F | 5’-GCTACTCGAGGCCGCC ATGGTGAGCAAGGGCGAGGAGC-3’ (XhoI) |
| TnT3GR-R | 5’-GCTAAGCTTCTTGTACAGCTCGTCCATGCCG-3’ (HindIII) |
Construction of BD/ or NLS/DsRed fusion protein expression plasmids
To further verify if the predicted bipartite domain (BD, RRKPLNIDHLSDDKLRDK) and the manually identified subdomain (KLKRQK) contain NLS/NoLS, we performed PCR using designed primers (Table 1) to incorporate the BD or KLKRQK coding sequence into the NH2-terminus of DsRed2-N1 expression vectors. The amplified product was treated with XhoI/NotI and ligated into the DsRed2-N1 backbone treated with the same restriction enzymes, resulting in BD/DsRed or NLS/DsRed, respectively.
Construction of deletion plasmids
BD deletion, part of the predicted LZD (LWDTLYQL) or the manually identified KLKRQK domain in the TnFL/DsRed plasmid, was performed using PCR and described primer sets (Table 1). The amplified products were treated with either DpnI and directly transformed into Oneshot Top10 competent cells (Invitrogen) to produce TnFL-ΔBD /DsRed (Figure 5) or with DpnI and then T4 polynucleotide kinase (Promega, Madison, WI) before self-ligation and transformation to produce TnFL-ΔNLS/DsRed or TnFL-ΔLZD/DsRed (Figs. 5, 10).
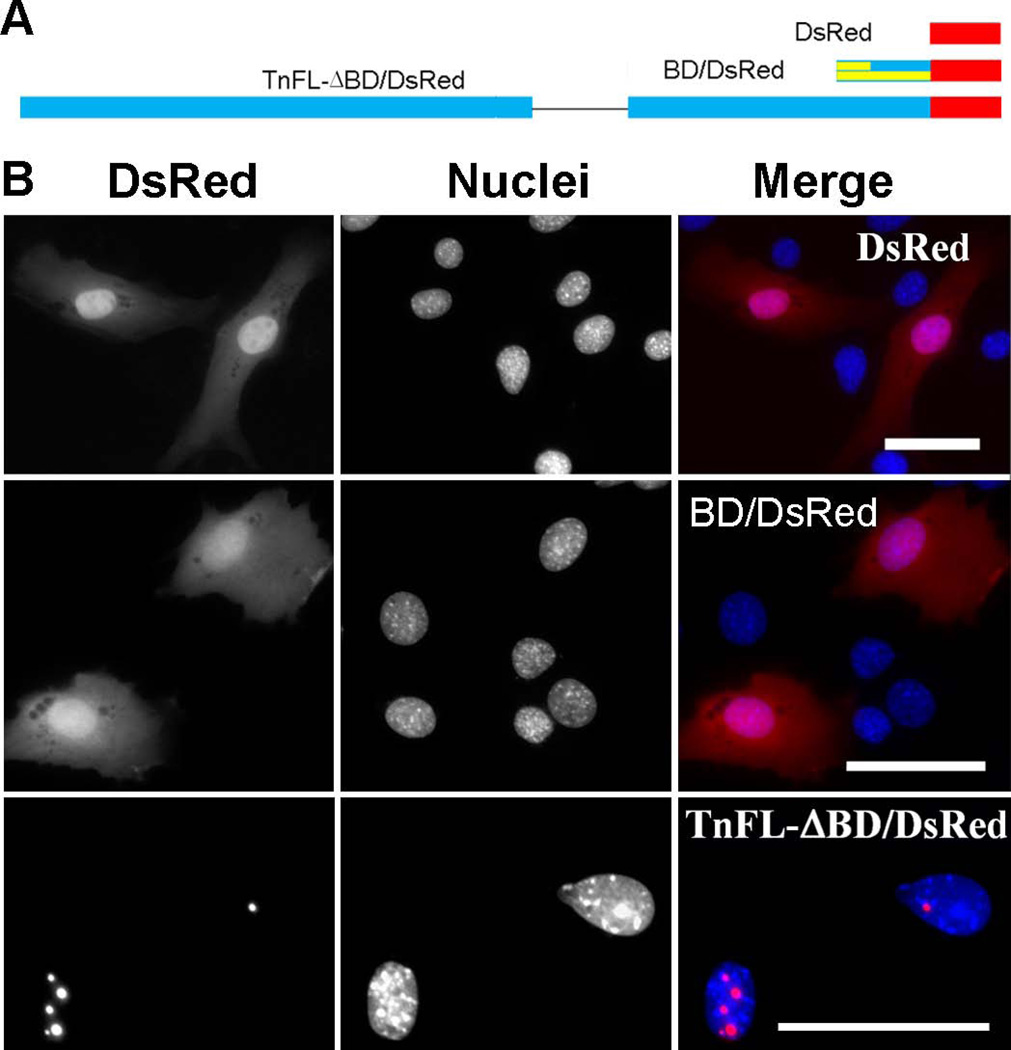
Figure 5. The predicted bipartite NLS does not play a major role in TnT3 nuclear localization.
(A) Schematic representation of a DsRed-fused bipartite NLS alone (BD/DsRed) and bipartite NLS-deleted full-length TnT3 (TnFL-ΔBD/DsRed). (B) DsRed alone is localized throughout the C2C12 cell with no signs of nucleolar enrichment. BD/DsRed shows a subcellular localization similar to that of DsRed alone, with weak nucleolar enrichment. TnFL-ΔBD/DsRed shows an exclusive nucleolar enrichment like that described for wild-type TnFL/DsRed (Zhang et al. 2011). Nuclei (blue, Hoechst 33342), TnT3-DsRed fusion protein (red). Scale bar, 50 µm.
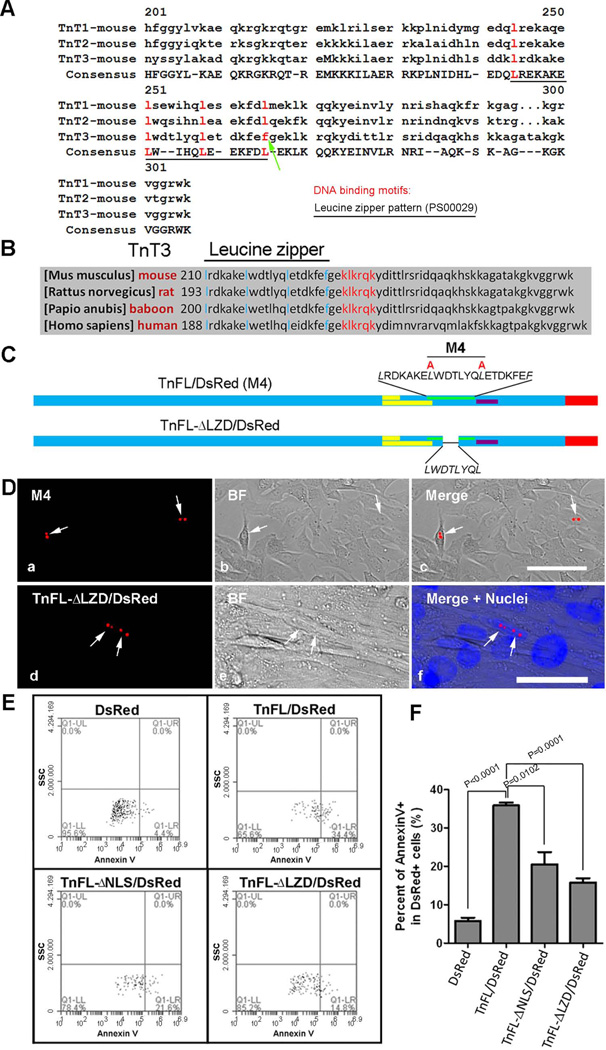
Figure 10. TnT3 Leucine Zipper Domain (LZD) is related to C2C12 cytotoxicity but not required for nuclear/nucleolar localization.
(A) In silico identification of an LZD in TnT1 and TnT2 and sequence alignment showing that F replaces the 4th L in TnT3 (arrow). (B) Alignment of TnT3 COOH-terminal sequences showing that LZD is conserved across mammalian species. (C) Schematic representation of LZD partially deleted from full-length TnT3 (TnFL-DLZD/DsRed) and constructs with leucine mutated into alanine (M4). (D) LZD mutation (a-c) or truncation (d-f) does not affect nuclear localization and nucleolar enrichment in C2C12 myoblasts and myotubes. Nuclei (blue, Hoechst 33342), TnT3-DsRed fusion protein (red), brightfield (BF). Scale bars, 50 µm. (E) Representative analyses of Annexin V staining after DsRed pre-gating. C2C12 cells transfected with various TnFL/DsRed constructs or DsRed alone for 48 hours were analyzed by flow cytometry. Data were collected on at least 50,000 freshly stained cells and at least 1000 DsRed positive cells and analyzed for staining with the cell death marker Annexin V (n=3). SSC, side scatter. (F) Percent of Annexin V+ in C2C12 cells expressing DsRed, TnFL/DsRed, TnFL-DNLS/DsRed, and TnFL-DLZD/DsRed.
Amino acids substitution by site-directed mutagenesis
Alanine substitution for lysine (K) or arginine (R) residues was performed as described (Zhang et al. 2009). Briefly, amino acid substitution was performed by PCR using Pfu DNA polymerase (Stratagene, La Jolla, CA) and the designed primers listed in Table 1. The following PCR protocol was used: initial denaturation at 95°C for 1 minute; three-step cycling, with 18 cycles consisting of denaturation at 95°C for 50 seconds, annealing at 60°C for 50 seconds, and extension at 68°C for 7 minutes, and a final extension at 68°C for 7 minutes. PCR products were treated with DpnI restriction enzyme (Promega) before transformation into Oneshot Top 10 competent cells (Invitrogen). Mutations were confirmed by gene sequencing (GenScript, Piscataway, NJ).
Microscopy and image analysis
Cells cultured on coverslips were fixed with 4% paraformaldehyde (15 minutes at room temperature), and their membrane permeabilized at room temperature with 0.5% Triton X-100 in PBS buffer for 5 minutes. After three washes with PBS, they were incubated in blocking buffer (PBS with 10% normal goat serum [Sigma, St. Louis, MO]) at room temperature for 1 hour and labeled at room temperature with primary and secondary antibodies for 2 and 1 hour, respectively (Birbrair et al. 2011; Birbrair et al. 2012a; Birbrair et al. 2012b).
For cells permeabilized prior to fixation, we followed a described protocol (Asumda and Chase 2012) that allowed extraction of cytoplasmic components. Briefly, cells were grown on glass cover slips to subconfluency, washed with cold PBS and cytoskeleton (CSK) buffer (10 mmol/l HEPES-KOH, pH 7.4, 300 mmol/l sucrose, 100 mmol/l NaCl, 3 mmol/l MgCl2), and extracted on ice with 0.5% TritonX-100 in CSK buffer with protease inhibitors for 5 minutes. They were then fixed with 2% paraformaldehyde at room temperature for 15 minutes and washed extensively, first with PBS and then with 0.5% NP-40. They were washed with 0.5% Tween-20 plus 3% BSA prior to incubation with primary antibodies at 4°C overnight and then appropriate secondary antibodies at 37°C for 1 hour. Controls for both procedures involved incubation with secondary antibodies only.
After cells were counterstained with Hoechst 33342 (Invitrogen) and mounted in DAKO fluorescent mounting medium (DAKO, Carpinteria, CA), widefield immunofluorescence images were taken on an inverted motorized fluorescent microscope (Olympus, IX81, Tokyo, Japan) with an Orca-R2 Hamamatsu CCD camera (Hamamatsu, Japan). Camera driver and image acquisition were controlled by a MetaMorph Imaging System (Olympus). Digital image files were transferred to Photoshop 7.0 to assemble montages. Images represent three independent experiments.
Annexin V staining and flow cytometry
To examine apoptosis/necrosis after transfection with various constructs, C2C12 cells were cultured in GM for 48 hours and stained with FITC Annexin V (BD Bioscience, San Jose, CA), following the manufacturer’s protocol. Briefly, the cells were harvested by trypsinization, washed twice with PBS, and stained with 2.5 µg/ml FITC Annexin V in 1× binding buffer (10 mM Hepes [pH 7.4], 140 mM NaCl, 2.5 mM CaCl2) at room temperature for 15 minutes. Cellular DsRed and FITC were analyzed by flow cytometry (with an Accuri C6 Flow Cytometer; BD Biosciences, San José, CA), which allowed us to discriminate viable (AnnexinV −) and apoptotic/necrotic (Annexin V +) cells. The samples were acquired immediately after staining, and the value in the dot-plot indicates the percent of Annexin V + cells after DsRed pregating. DsRed fluorescence was detected with the FL2 channel; Annexin V FITC fluorescence with the FL1 channel. To quantify only the transfected DsRed-positive cells for Annexin V staining, background fluorescence of (1) unstained nontransfected cells, (2) nontransfected cells stained for FITC Annexin V, and (3) transfected DsRed-positive cells was used to set up gates, photomultiplier tube voltages, and amperage gain for each channel. Data were collected on freshly stained cells and analyzed using BD Accuri™ C6 CFlowPlus software.
Antibodies and reagents
Rabbit anti-TnT3 polyclonal antibody (ARP51287_T100) was purchased from Aviva Systems Biology (San Diego, CA), and Hoechst 33342 and Alexa 568-conjugated anti rabbit IgG from Invitrogen.
Statistical analysis
Experimental groups were analyzed using analysis of variance (ANOVA) followed by Student’s t-test. Data are expressed as means ± SD. Statistical analyses were made with Prism 5.0a (Graphpad Software, Inc., La Jolla, CA). P < 0.05 was considered significant.
RESULTS
Endogenous TnT3 is also located in the nucleus of C2C12 cells
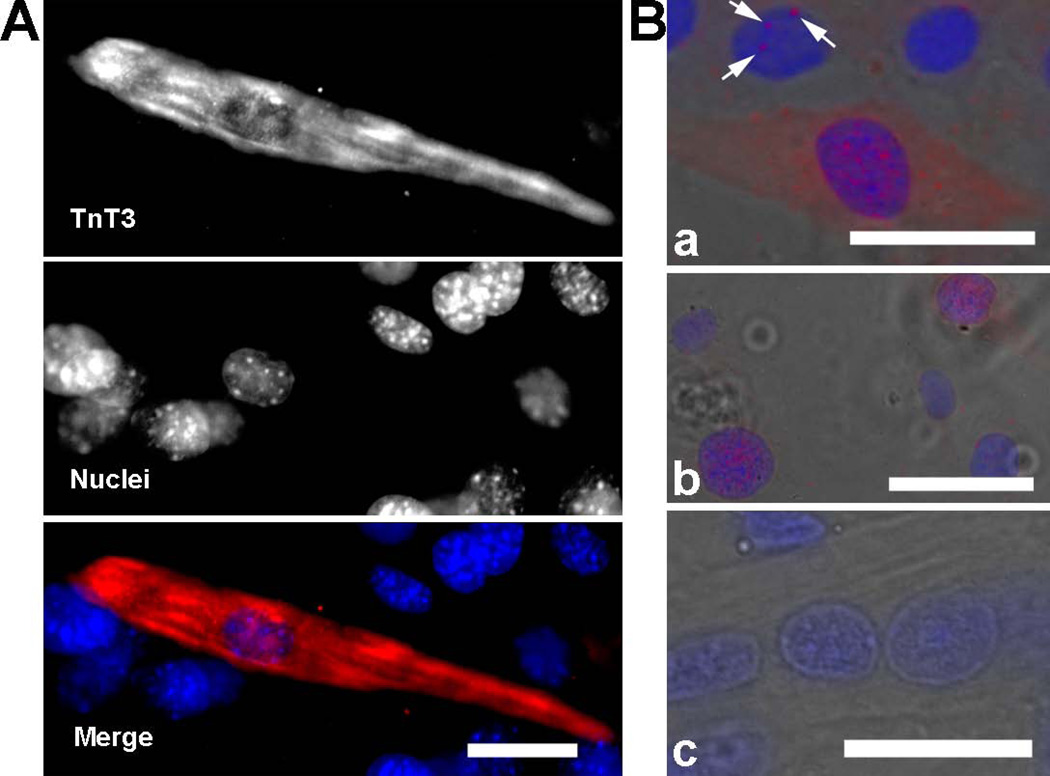
We showed that endogenous TnT3 is located in the cytoplasm and nucleus of adult myofibers by immunoblots or expressing DsRed-tagged intact TnT3(TnFL/DsRed) or its COOH-terminus (TnCT/DsRed) (Zhang et al. 2011). Both constructs were closely related to the nucleolus (Zhang et al. 2011). Here, we detected endogenous TnT3 in the nucleus of C2C12 cells by immunofluorescence staining (Figure 1A). TnT3 was not detected in most undifferentiated myoblasts, but became noticeable at very early stages of differentiation. In addition to the abundant cytoplasmic pool, a small fraction of endogenous TnT3 was observed in the nucleus of elongated myocytes (Figure 1A). After removing most of the cytoplasmic TnT3 before cell fixation (see Materials and Methods), nuclear TnT3 was apparent in nuclear foci as distinctive punctates (Figure 1B). Endogenous TnT proteins were not detected in NIH3T3 fibroblasts using the same procedure (data not shown), which rules out any unspecific staining.
Figure 1. Endogenous nuclear TnT3 in cultured C2C12 early in differentiation.
C2C12 cells were immunostained with TnT3 antibody following two procedures: (A) Fixed prior to permeabilization. An elongated mononucleated myocyte was positively stained with TnT3 in both cytoplasmic and nuclear areas. Neighboring cells were not stained. (B) Permeabilized prior to fixation to allow extraction of cytoplasmic components. TnT3 was detected in some nuclear areas as isolated foci (a, arrows) or abundant punctates (a and b); a control, using only the secondary antibody, shows no positive staining (c). Nuclei (blue, Hoechst 33342) and TnT3 (red), and brightfield images are merged. Scale bars, 50 µm.
Mapping the nuclear/nucleolar targeting sequence of TnT3
TnT3 was found to have several predicted nuclear targeting sequences (monopartite or bipartite) using PSORT II (http://psort.hgc.jp/form2.html) (Figure 2). As only DsRed-tagged, intact TnT3 and COOH-terminus showed strong nuclear/nucleolar localization (Zhang et al. 2011), we focused the TnT3 NLS/NoLS mapping strategy on the COOH-terminus.
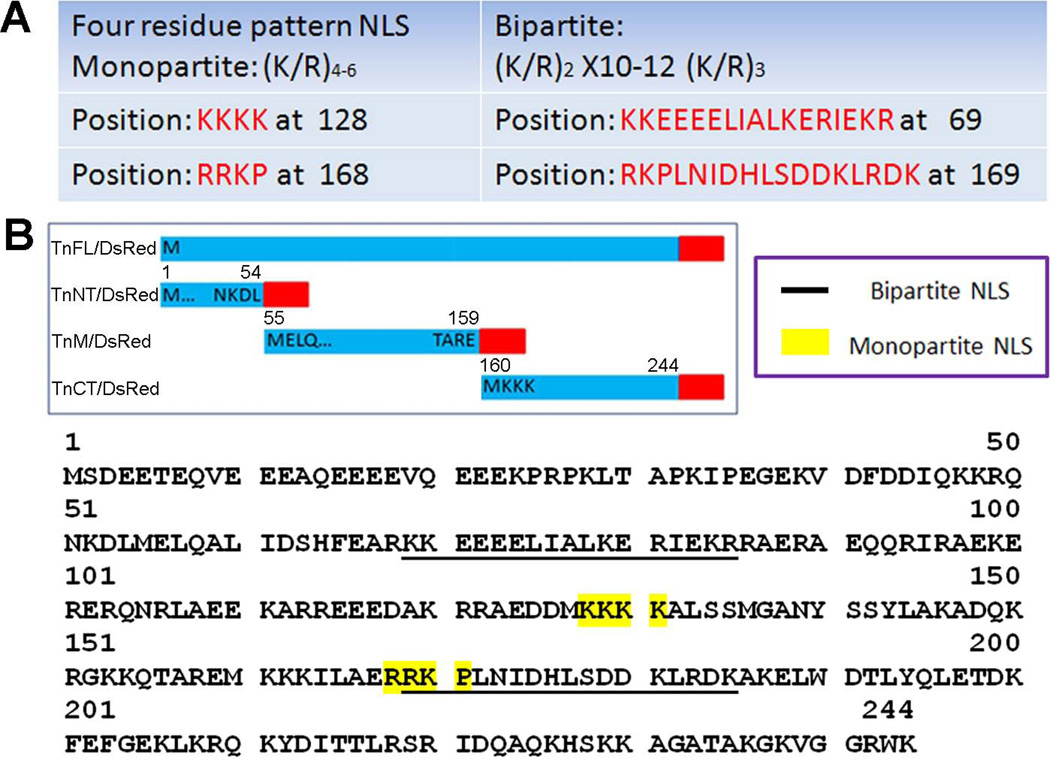
Figure 2. Prediction of TnT3 nuclear localization signal (NLS).
(A) Predicted monopartite and bipartite NLS sequence and positions. (B) Predicted NLS distribution along TnT3. Monopartite NLS is highlighted in yellow, and bipartite is underlined; one of each is localized in the TnM/DsRed and TnCT/DsRed constructs. The analysis used PSORT II.
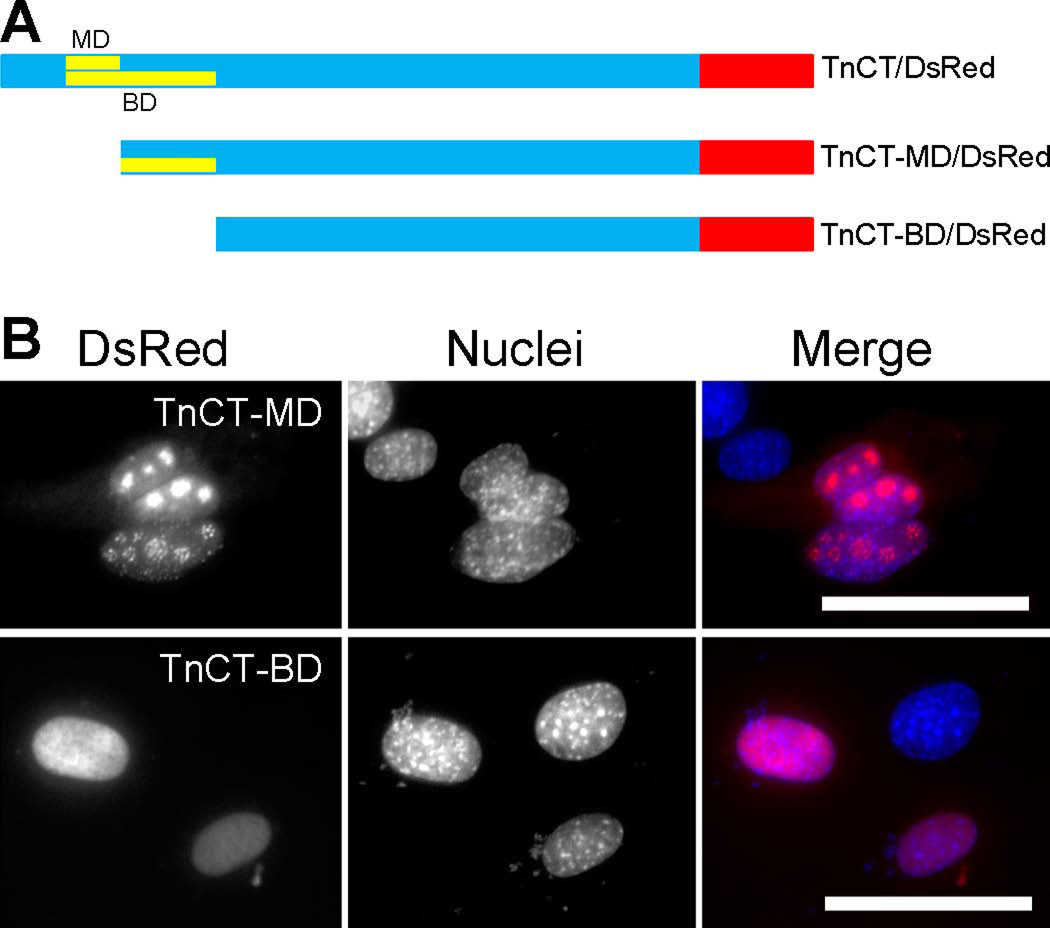
Note that deleting 171 amino acids from the NH2-terminus (TnCT-MD), which included the predicted monopartite domain (MD), had no apparent effect on the nuclear/nucleolar localization of TnT3 COOH-terminus (Figure 3). To our surprise, further deletion of the predicted bipartite domain (BD) failed again to prevent TnT3 COOH-terminus nuclear localization, although nucleolar enrichment of the COOH terminus was largely reduced (Figure 3). This observation implies that TnT3’s NoLS may be located, at least partially, in its bipartite domain, while the NLS is most probably located downstream of the BD.
Figure 3. Removal of the predicted monopartite and bipartite NLSs from TnCT/DsRed does not prevent its nuclear localization.
(A) MD or BD-deleted TnT3 COOH-terminus fused to DsRed. (B) C2C12 cells expressing MD or BD deletion constructs, which both localized exclusively in the nucleus, but enrichment in the nucleolar area was reduced, particularly for TnCT-BD/DsRed. Nuclei (blue, Hoechst 33342), TnT3-DsRed fusion protein (red). Scale bar, 50 µm.
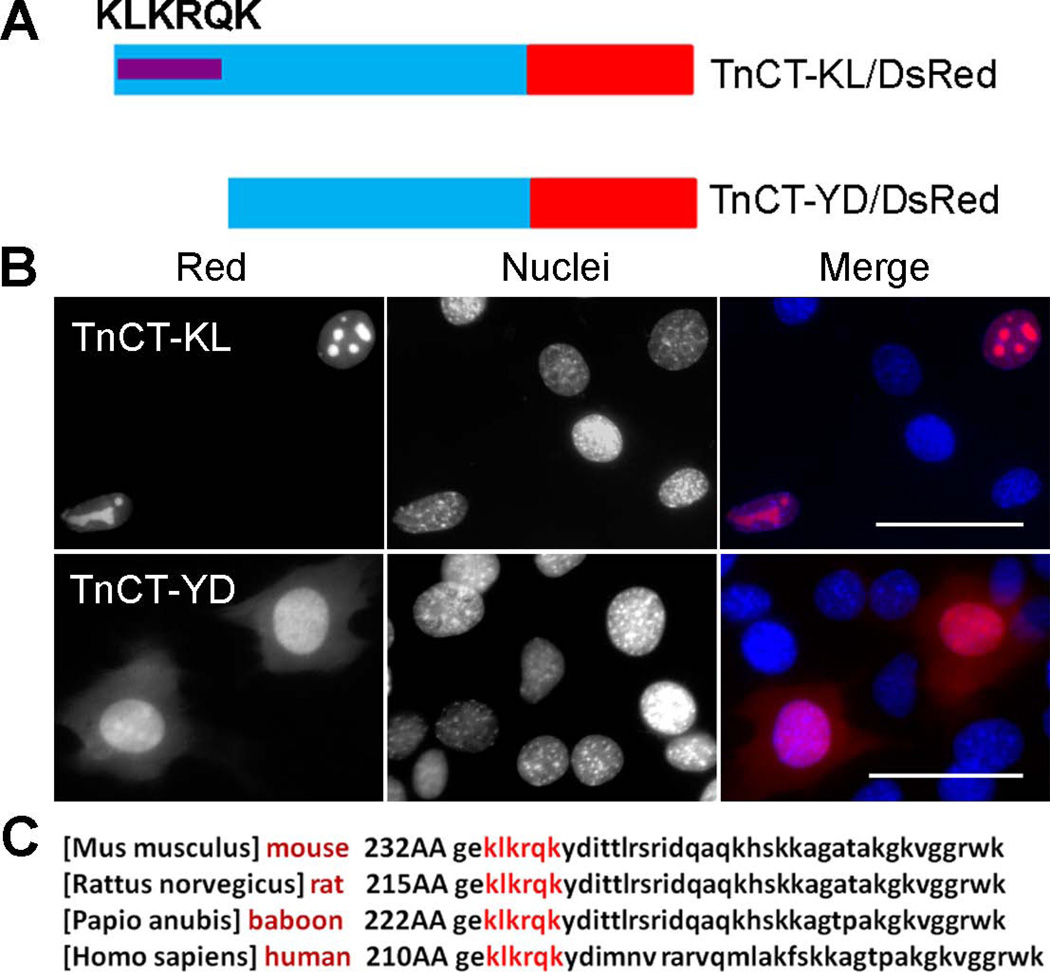
To determine the NLS, we further analyzed the TnT3 COOH-terminus. We manually identified a sequence (KLKRQK), which is similar to the reported KxKxK motif that works as a nuclear targeting sequence (Banfic et al. 2009). We constructed two shorter TnCT/DsRed constructs, including (TnCT-KL/DsRed) or excluding (TnCT-YD/DsRed) the KLKRQK motif (Figure 4A). The TnCT-KL/DsRed construct still went exclusively into the nucleus, with an apparent enrichment in the nucleolar area when expressed in C2C12 myoblasts (Figure 4B). In contrast, the TnCT-YD/DsRed localized in both cytoplasm and nucleus (Figure 4B) and showed a distribution pattern similar to control DsRed (Fig. 5B), which strongly indicates that the NLS is most possibly localized in the KLKRQK region and conserved among many species (Figure 4C) as expected, given that TnT isoforms have highly conserved sequences in the middle region and the COOH terminus.
Figure 4. KLKRQK is needed for TnCT nuclear localization and is conserved among species.
(A) Schematic representation of DsRed-fused TnT3 COOH-terminus mutant with the KLKRQK sequence retained (TnCT-KL/DsRed) or removed (TnCT-YD/DsRed). (B) C2C12 cells expressing the deletion constructs. TnCT-KL/DsRed is localized exclusively in the nucleus with an enriched nucleolar area. In contrast, TnCT-YD/DsRed is present throughout the cell, with marked reduction in nucleolar enrichment. Nuclei (blue, Hoechst 33342), TnT3-DsRed fusion protein (red). Scale bar, 50 µm. (C) The Tnnt3 KLKRQK sequence is conserved across mammalian species.
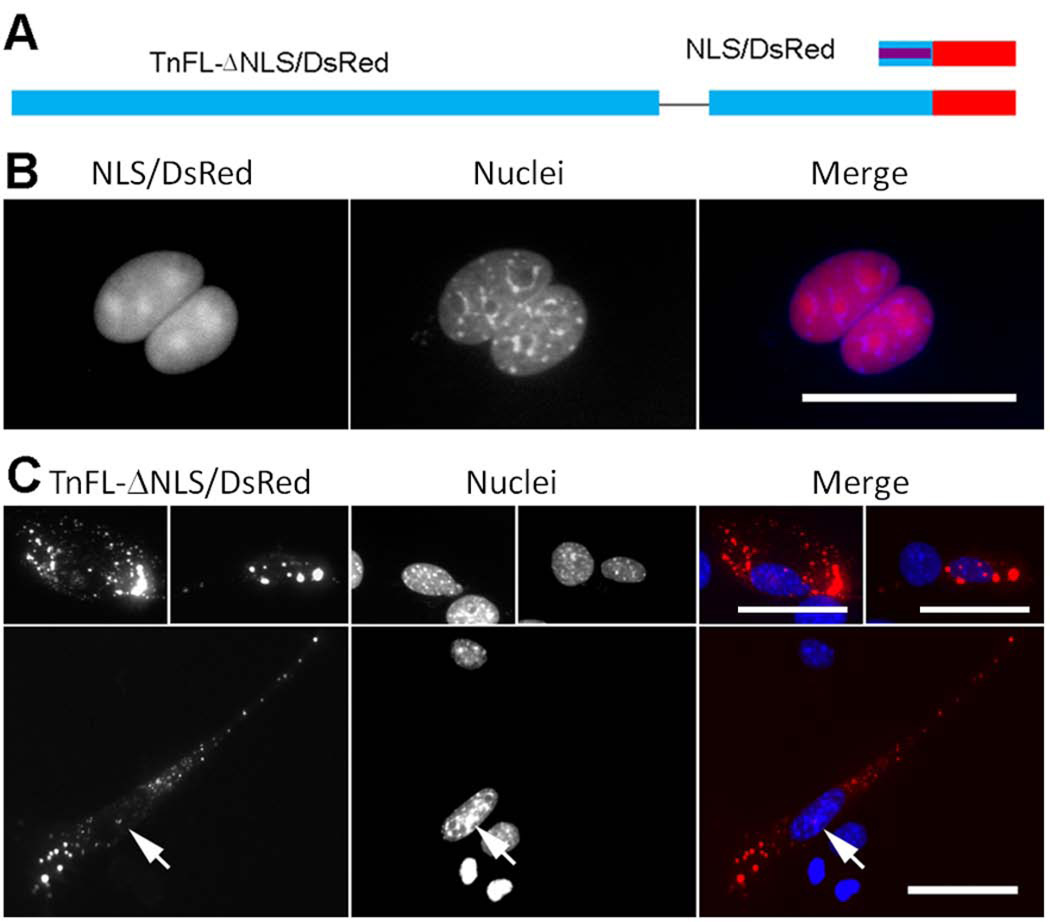
To further verify the identity of TnT3 NoLS and NLS, we subcloned the BD or the KLKRQK peptide into the DsRed2-N1 vector (BD/DsRed or NLS/DsRed). NH2-terminally DsRed-tagged BD or NLS fusion protein expressed in C2C12 myoblasts showed different subcellular localizations (Figs. 5 and 6). These findings confirm that KLKRQK but not BD is a strong signal for TnT3 nuclear/nucleolar targeting, supported by the TnFL/DsRed deletion of either the BD (TnFL-ΔBD/DsRed) or the NLS domains (TnFL-ΔNLS/DsRed), respectively (Figs. 5 and 6).
Figure 6. KLKRQK is a genuine NLS and plays a major role in TnT3 nuclear localization.
(A) Schematic representation of DsRed-fused KLKRQK alone (NLS/DsRed) and full-length TnT3 devoid of this sequence (TnFL-ΔNLS/DsRed). (B) NLS/DsRed shows an exclusive nuclear localization with an enriched nucleolar area in C2C12 cells. (C) TnFL-ΔNLS/DsRed shows exclusion from the C2C12 nucleus, and only a minor fraction is localized in the nucleolar area (arrows). Nuclei (blue, Hoechst 33342), TnT3-DsRed fusion protein (red). Scale bar, 50 µm.
The positively charged amino acids lysine (K) and arginine (R) in the NLS/NoLS are required for TnT3 nuclear targeting
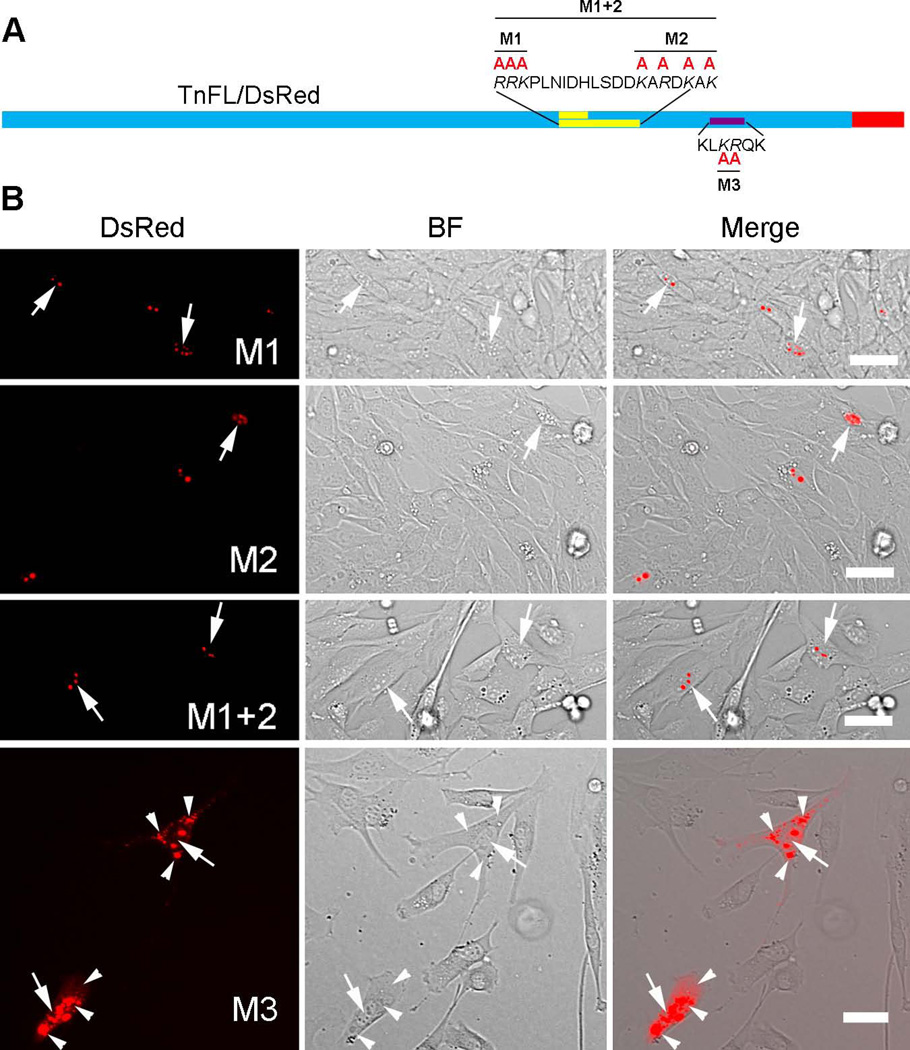
The positively charged basic amino acids located in the nuclear targeting sequences are responsible for their function (Dingwall and Laskey 1991). Therefore, we mutated K and R into the neutral amino acid A and found that the mutation in the NLS domain (M3) but not the BD region (M1, M2, M1+2) strongly impaired nuclear/nucleolar enrichment of TnFL/DsRed (Figure 7), similar to the deletion reported for NLS or BD and shown in Figures 5 and 6.
Figure 7. Basic residues in the KLKRQK sequence play a critical role in TnT3 nuclear localization.
(A) Schematic representation of DsRed, full-length TnT3 fusion protein with K or R amino acid substitution by A in the MD, BD, and NLS regions. (B) Only mutating the NLS region greatly reduced its nuclear localization in C2C12 cells (M3). Arrows indicate the nucleus while arrowheads the cytoplasm. BF, brightfield. Scale bar, 50 µm.
The TnT3 nuclear export signal (NES) is localized in the COOH-terminus
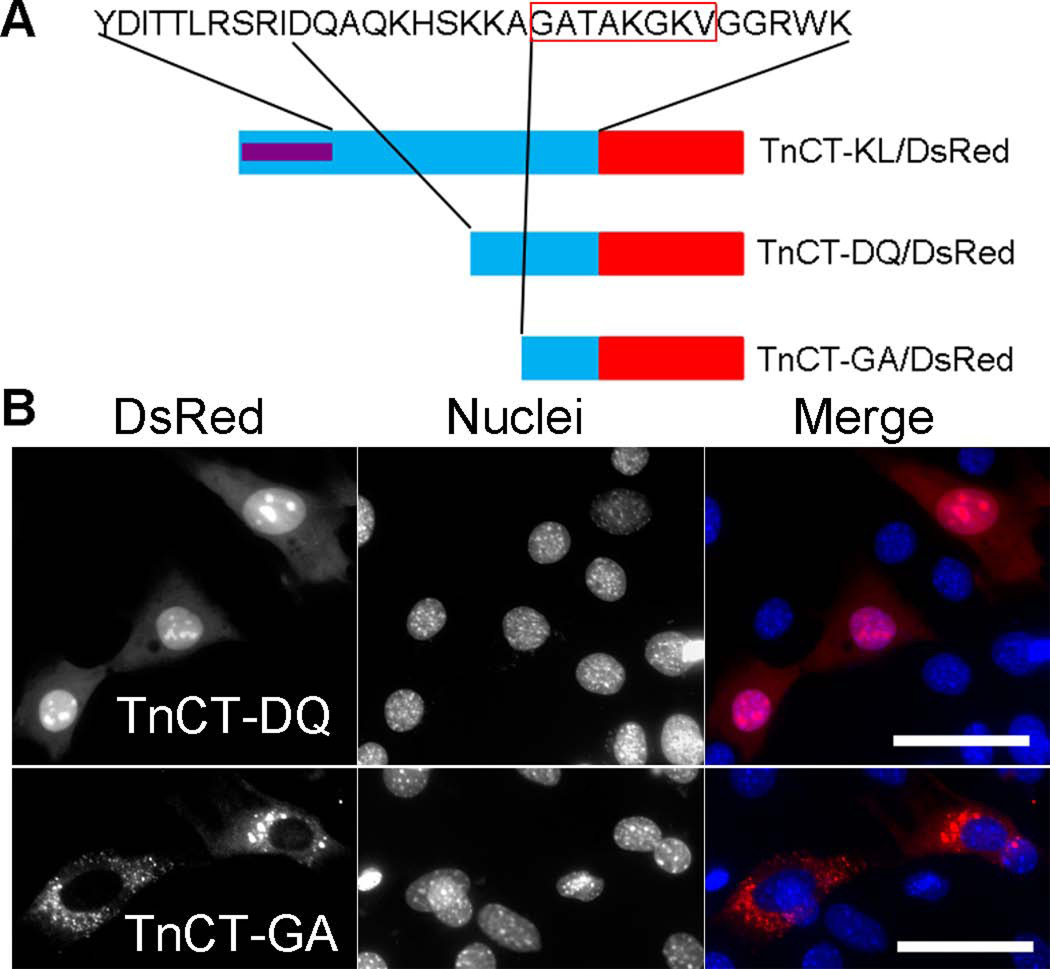
Since TnCT-YD/DsRed showed less nuclear enrichment than TnCT-KL/DsRed, we tested whether TnT3 has a NES in its COOH-terminus by examining the truncation constructs TnCT-DQ/DsRed and TnCT-GA/DsRed (Figure 8A). TnCT-GA/DsRed showed stronger cytoplasmic localization than all other deletion constructs and the DsRed fluorescent protein alone (Figures 5B and 8B). We found that TnT3 has a predicted nuclear exporting sequence in its COOH-terminus (Figure S1) by using the NetNES1.1 server at www.cbs.dtu.dk/services/NetNES/. The overlap between TnCT-GA/DsRed and the predicted NES region (GATAKGKV) is possibly TnT3 NES.
Figure 8. NESs and NLSs in the TnT3 COOH-terminus.
(A) Schematic representation of DsRed-fused TnT3 COOH-terminus deletion constructs. (B) TnCT-DQ/DsRed is localized throughout the C2C12 cell with an enriched nucleolar area, indicating a NLS/NoLS. In contrast, TnCT-GA/DsRed shows a dramatic exclusion from the nucleus, indicating a possible NES in the GATAKGKVGGRWK region, where the sequence in the red box overlaps with the predicted NES (Figure S1). Nuclei (blue, Hoechst 33342), TnT3-DsRed fusion protein (red). Scale bar, 50 µm.
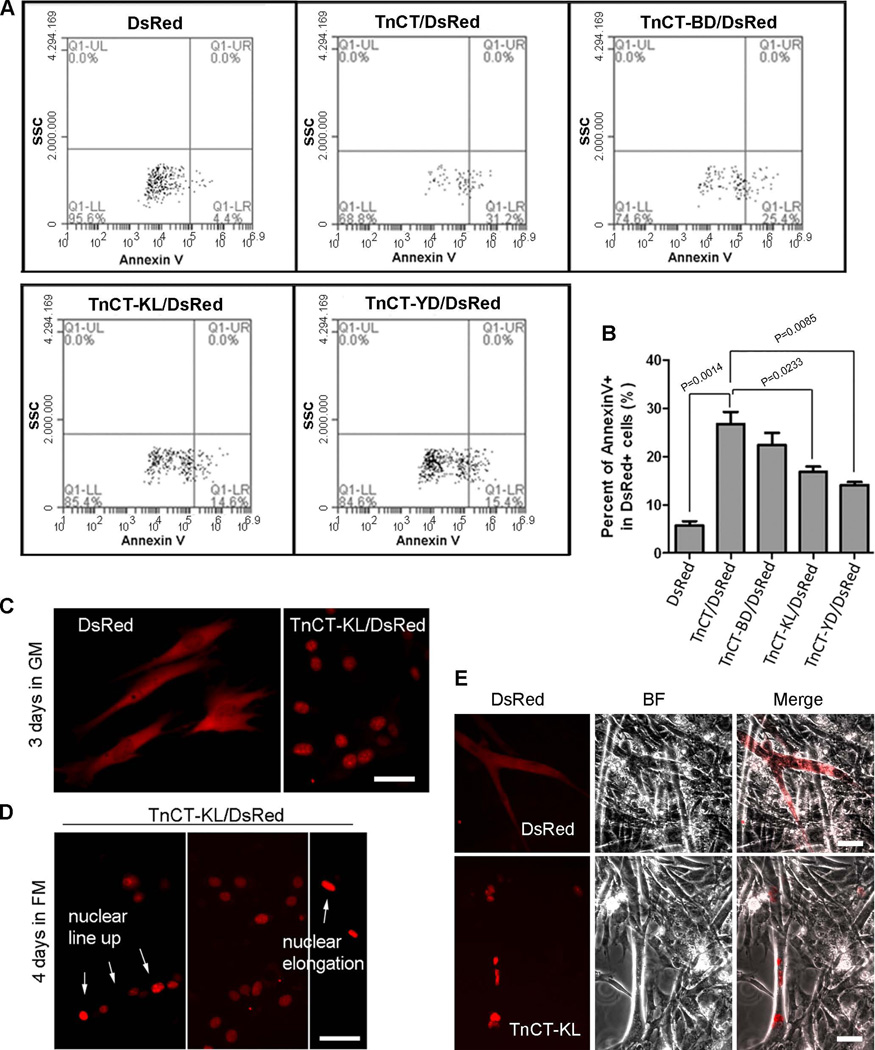
Mutants with various TnT3 COOH-terminus lengths showed differential cytotoxicity
We showed that expressing TnCT/DsRed in C2C12 or NIH3T3 cells leads to apoptosis (Zhang et al. 2011). Similar cytotoxicity for nonmyofilament-associated TnT fragments, especially the COOH-terminus, has been reported (Jeong et al. 2009). To determine whether TnCT cytotoxicity is related to its nuclear/nucleolar localization, we performed Annexin V staining to detect apoptotic and necrotic cells with COOH-terminus DsRed fusion proteins of different lengths and DsRed alone. As expected, compared to DsRed alone, TnCT/DsRed increased apoptosis and necrosis, and truncating the COOH-terminus dramatically reduced cytotoxicity (Figure 9, A and B). Surprisingly, cells transfected with TnCT-KL/DsRed survived and proliferated as well as those expressing DsRed alone, which could also fuse into myotubes in differentiation medium (Figure 9, C–E). As expected, TnCT-GA/DsRed-expressing cells showed fusion competency (Figure S2). We hypothesize that a domain localized upstream of the TnT3 NLS region plays a major role in mediating cytotoxicity.
Figure 9. TnT3 cytotoxicity is gradually reduced by truncating the TnT3 COOH-terminus.
C2C12 cells transfected with various TnCT/DsRed constructs or DsRed alone for 48 hours were analyzed by flow cytometry. Data were collected on at least 50,000 freshly stained cells and at least 1000 DsRed positive cells and analyzed for staining with the cell death marker Annexin V (n=3). (A) Representative analyses of Annexin V staining after DsRed pregating. SSC, side scatter. (B) The percent of total apoptotic and necrotic cells (Annexin V+) was compared among different constructs. (C) A representative fluorescence image showing that C2C12 cells expressing TnCT-KL/DsRed or DsRed remain healthy in extended GM cultures and survive and fuse into myotubes in differentiation medium. Arrows indicate nuclear line up or elongation (D). (E) Brightfield merged with DsRed image showing that TnCT-KL/DsRed is localized in myotube nuclei, while DsRed alone is expressed throughout the myotube. Scale bars, 50 µm.
Deleting the LZD reduced TnT3 cytotoxicity
TnT1 and TnT2 were found to have a classical LZD (Vinson et al. 1989) in the COOH-terminus using PDBsum (www.ebi.ac.uk/thornton-srv/databases/pdbsum/index.html), a program designed to predict the 3D structure of proteins and nucleic acids. Although TnT3 does not exhibit a classic LZD, we noticed that the fourth leucine in the TnT3 candidate LZD is replaced with a phenylalanine (Phe) to produce the so-called “relaxed leucine repeat” (Figure 10A), which may increase Fos DNA homodimerization and enhance its DNA binding efficiency (O'Shea et al. 1992; O'Shea et al. 1989). Note that this LZD is just upstream of the identified NLS/NoLS and conserved among species (Figure 10B).
Considering that the LZD is an important motif for regulating transcription factor DNA binding (Vinson et al. 1989), we tested whether removing or mutating it would affect TnFL/DsRed’s cytotoxicity. The partially removed LZD construct, TnFL-ΔLZD/DsRed, and the LZD mutant (L into A), M4, were expressed. They still localized mainly in the nucleus, but cells survived longer, and they were even found in the nuclei of fully differentiated C2C12 myotubes (Figure 10, C and D). To examine whether TnFL cytotoxicity was due to its LZD, we performed Annexin V staining to detect apoptosis and necrotic cells with various TnFL DsRed fusion proteins or DsRed alone. Compared to DsRed alone, TnFL/DsRed did increase cytotoxicity, while TnFL-ΔLZD/DsRed dramatically reduced it, as with TnFL-ΔNLS/DsRed (Figure 10, E and F). Taken together, these results indicate that TnT3 cytotoxicity is probably mediated by its nuclear localization and an LZD-mediated nuclear signaling pathway.
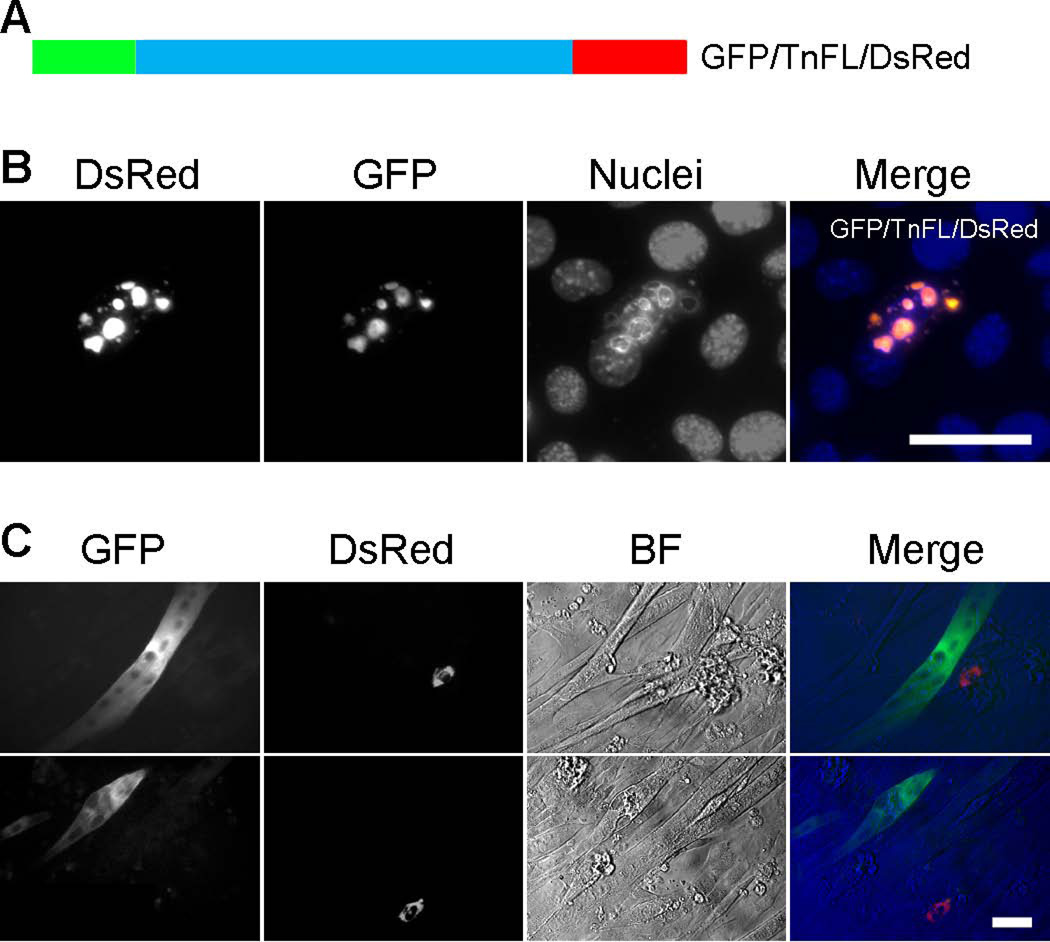
To support this concept, we constructed a double fluorescent protein tagged TnFL expressing plasmid, with GFP fused to the NH2-terminus and DsRed fused to the COOH-terminus of TnT3 (GFP/TnFL/DsRed) (Figure 11A). Expressing GFP/TnFL/DsRed in C2C12 myoblasts showed the expected nuclear/nucleolar localization of overlapped GFP and DsRed fluorescence, indicating that TnT3 was intact (Figure 11B). When cells were cultured in differentiation medium for more than 5 days, very few fluorescence-positive cells survived, and most contained either GFP signal (in myoblasts or myotubes) or DsRed signal (in myoblasts) (Figure 11C). This finding indicates that TnT3 toxicity is reduced by fragmentation and the consequent exclusion from the nucleus, as shown for GFP-tagged NH2-terminal fragments, where NLS was supposedly removed from the COOH-terminus, or by removing the LZD, as seen in the DsRed-tagged COOH-terminal fragments. At this stage, the TnT cleavage site is unknown and merits further investigated.
Figure 11. Fragmented GFP/TnFL/DsRed showed decreased cytotoxicity.
(A) Schematic representation of GFP (NH2-terminal) and DsRed (COOH-terminal) full-length TnT3 double fluorescence construct (GFP/TnFL/DsRed). C2C12 cells cultured in GM for 2 days showed nucleolar GFP and DsRed fluorescence overlap, indicating nucleolar localization of intact TnT3 (B). No double fluorescence-positive cells survived culture in differentiation medium yet most showed either GFP (in myoblasts or myotubes) or DsRed (in myoblasts) fluorescence mainly in the cytoplasm (C). Scale bars, 50 µm.
DISCUSSION
We demonstrated that endogenous TnT3 is present in the nuclei of mature myofibers and that full-length DsRed-tagged TnT3 or its COOH-terminal enters the nucleus when expressed in C2C12 cells (Zhang et al. 2011). However, the mechanism by which this occurs is unknown. The current study shows that endogenous TnT3 is present in the nucleus of C2C12 cells at an early stage of differentiation. Using a combined software prediction and amino acid truncation and substitution strategy, we establish that it contains both NLS/NoLS and NES at its COOH-terminus. Our finding is consistent with a recent report showing that NoLSs localize predominantly at the ends of proteins, which are easily accessible to interacting partners (Scott et al. 2010). We also identify an LZD that may be involved in the cytotoxic effect of nonmyofilament-associated TnT3 and its fragments. TnT3 may modulate the nuclear signaling pathway when its LZD interacts with DNA or nucleolar RNA.
TnT3 exhibits a NLS/NoLS and NES
Although the software predicted several NLS candidates along the full-length TnT3, we focused our search on the COOH-terminus, based on our previous experimental evidence that the DsRed-tagged TnT3 COOH-terminus (TnCT/DsRed) localizes exclusively in the nucleus, while the NH2-terminus and the middle region do not (Zhang et al. 2011). Surprisingly, removing the predicted monopartite (TnCT-MD/DsRed) and bipartite (TnCT-BD/DsRed) domains in the COOH-terminus failed to prevent TnCT nuclear localization, which strongly indicates that other NLSs downstream of the predicted bipartite domain exist. Further constructs retaining (TnCT-KL/DsRed) or truncating (TnCT-YD/DsRed) the KLKRQK sequence in TnCT showed different cellular localizations, with TnCT-KL/DsRed localized exclusively in the nucleus, and the TnCT-YD/DsRed throughout the cells. These findings suggest that the KLKRQK sequence is essential for TnT3 nuclear localization. Fusing this sequence to the DsRed NH2-terminus consistently enriched nuclear/nucleolar localization, while depleting it from full-length TnT3 (TnFL-ΔNLS/DsRed) inhibited its nuclear localization. We conclude that KLKRQK is a genuine NLS for TnT3. Given that both NLS/DsRed and TnCT-KL/DsRed showed nucleolar enrichment in addition to their nucleoplasmic localization, KLKRQK may function as a joint NLS-NoLS region (Scott et al. 2010).
TnCT-GA/DsRed, with a calculated molecular weight smaller than 50 kDa, should enter the nucleus passively, as DsRed alone does, yet, compared to other TnT3 COOH-terminal DsRed fusion proteins, it is largely excluded, strongly supporting the concept that it may contain a NES, as the software predicts.
TnT3 may have many NLSs/NoLSs either upstream or downstream of KLKRQK, based on the fact that removing MD or BD (TnCT-MD/ or TnCT-BD/DsRed) reduced nucleolar enrichment. The MD and BD upstream of KLKRQK might be working as a NoLS cooperatively with others, particularly with the KLKRQK sequence, which works as the major NLS/NoLS. That BD/DsRed alone and full-length TnT3 with BD depletion (TnFL-ΔBD/DsRed) did not show nucleolar enrichment support this concept. Similarly, regions downstream of KLKRQK may also contain NLS/NoLS since we noticed that shorter truncations (TnCT-DQ/DsRed) were still somewhat enriched in nucleus/nucleolus but not the TnCT-GA/DsRed that seems to contain a NES. We conclude that KLKRQK is the most likely TnT3 NLS/NoLS, and its basic residues play a major role since mutating K and R into neutral residue A in the TnFL/DsRed (M3) could diminish its nuclear localization. In contrast, mutations in the MD and BD regions (M1, M2) did not. KLKRQK is conserved among many species, including humans and primates, strongly supporting a role for TnT3 nuclear signaling in mammalian skeletal muscle.
Although dimerization of some transcription factors through the LZD is found to be a prerequisite for their nuclear localization (Nagoshi and Yoneda 2001), partial depletion or mutation of leucine into alanine (M4) in this domain in the TnFL/DsRed construct (TnFL-ΔLZD/DsRed) did not inhibit its nuclear/nucleolar enrichment, indicating that the LZD is not necessary for TnT3 nuclear/nucleolar localization. That the TnT3 COOH-terminal forms an alpha-helical coiled-coil with TnI to mediate its nuclear localization is unlikely as our experiments were carried out in undifferentiated C2C12 myoblasts, differentiation stage at which the expression of endogenous myofilaments is not obvious.
Some transcription factors are known to have many NLSs and NESs that may allow multilevel regulation. For example, ATF2 contains two NLSs and two NESs, and P53 contains three NLSs and two NESs (Hsu and Hu 2012; Shaulsky et al. 1990; Zhang and Xiong 2001). The fact that TnT3 may contain many NLSs/NoLSs and NESs and the NH2-terminus could affect TnT conformation and function; TnT3 may work as a multilevel regulated transcription factor. Future work should further map TnT3 NES, which will elucidate the mechanisms underlying the regulation of TnT3 transport between the nucleus and cytoplasm.
Regulation of nonmyofilament-associated TnT3 cytotoxicity
The LZD is a well-known protein-protein interacting domain present in the B-ZIP (basic region leucine zipper) class of eukaryotic transcription factors (Vinson et al. 1989). Identification of an LZD in all three TnT isoforms is additional evidence that TnT functions as a transcription factor by either leucine zipper-mediated direct DNA binding or binding and regulating other LZD containing transcription factors.
Replacing Leucines in LZD with other hydrophobic amino acids (Ile, Val, Phe, Met) may enhance transcription activity. For example, Fos LZD variants were shown to have increased association capacity and transcription activity (Porte et al. 1995). Substituting the fourth leucine in the TnT3 LZD with a Phe residue may increase TnT3’s capacity in forming heterodimers with other LZD-containing proteins (Pearlman et al. 1994). Considering that apoptotic transcription factors (Fos and Jun) form heterodimers through LZDs (O'Shea et al. 1989), this domain may enable TnT3’s direct or indirect participation in the apoptotic signaling pathway. In support of this concept, removal or LZD (TnCT-KL/, -YD/DsRed and TnFL-ΔLZD/DsRed) or mutation of leucine into alanine (M4) showed greatly reduced cytotoxicity. Similarly, TnT3 fragments excluded from the nucleus (TnCT-GA/DsRed) or generated from endogenous cleavage of GFP/TnFL/DsRed also showed greatly reduced cytotoxicity. Given that all three TnT isoforms contain an LZD, and TnT2 and TnT3 isoforms are present in the muscle nucleus (Asumda and Chase 2012; Zhang et al. 2011), TnT2 may also have a conserved function in the nuclear signaling pathway.
The nucleolus is a large nuclear domain directly involved in ribosome biosynthesis (Sirri et al. 2008). Compelling evidence shows that in addition to ribogenesis, nucleolar proteins are tightly linked to cell-cycle regulation, proliferation, senescence, and apoptosis (Ma et al. 2008; Pederson and Tsai 2009). Considering that TnT3 and its fragments tested here showed variable nucleolar enrichment, we cannot rule out the possibility that this process is also involved in muscle cytotoxicity directly or indirectly by interacting with nuclear actins (Dopie et al. 2012).
Conclusions and future directions
Here, we conclude that TnT3 contains at least one NLS/NoLS, one NES, and an LZD, which enables its nuclear localization and possible function as a transcription factor. We also verified that not all TnT3 COOH-terminal fragments are similarly toxic, only those containing both an NLS and LZD. Further work will focus on the genes TnT3 regulates and the nuclear signaling pathway leading to muscle cell apoptosis. Transgenic mice with TnT3 KO or the LZD truncation may help us to understand the physiological function of endogenous nuclear TnT3. Elucidating these events may finally define TnT3 as a potential therapeutic target for interventions in muscle aging and injury/repair. Since an LZD is present in all three TnT isoforms, and the TnT middle and COOH-terminal region are highly conserved, the cardiac TnT2 will be another important isoform to investigate.
Supplementary Material
ACKNOWLEDGMENTS
This study was supported by grants from the National Institutes of Health/National Institute on Aging AG13934, AG15820, and FIRCA-BB TW008091to Osvaldo Delbono and the Wake Forest Claude D. Pepper Older Americans Independence Center (P30-AG21332).
Abbreviations
- TnT
Troponin T
- NLSs/NoLSs
nuclear/nucleolar localization signals
- NES
nuclear export signal
- LZD
Leucine Zipper Domain
- BD
Bipartite domain
- MD
Monopartite domain
- TnFL
full-length TnT
- TnNT
TnT NH2-terminus
- TnCT
TnT COOH-terminus
- TnM
TnT middle region
REFERENCES
- Asumda FZ, Chase PB. Nuclear cardiac troponin and tropomyosin are expressed early in cardiac differentiation of rat mesenchymal stem cells. Differentiation. 2012;83(3):106–115. doi: 10.1016/j.diff.2011.10.002. [DOI] [PubMed] [Google Scholar]
- Banfic H, Visnjic D, Mise N, Balakrishnan S, Deplano S, Korchev YE, Domin J. Epidermal growth factor stimulates translocation of the class II phosphoinositide 3-kinase PI3K-C2beta to the nucleus. Biochem J. 2009;422(1):53–60. doi: 10.1042/BJ20090654. [DOI] [PubMed] [Google Scholar]
- Bergmann O, Bhardwaj RD, Bernard S, Zdunek S, Barnabe-Heider F, Walsh S, Zupicich J, Alkass K, Buchholz BA, Druid H, et al. Evidence for cardiomyocyte renewal in humans. Science. 2009;324(5923):98–102. doi: 10.1126/science.1164680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birbrair A, Wang ZM, Messi ML, Enikolopov GN, Delbono O. Nestin-GFP transgene reveals neural precursor cells in adult skeletal muscle. PLoS One. 2011;6(2):e16816. doi: 10.1371/journal.pone.0016816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birbrair A, Zhang T, Wang ZM, Messi ML, Enikolopov GN, Mintz A, Delbono O. Skeletal muscle neural progenitor cells exhibit properties of NG2-glia. Exp Cell Res. 2012a doi: 10.1016/j.yexcr.2012.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birbrair A, Zhang T, Wang ZM, Messi ML, Enikolopov GN, Mintz A, Delbono O. Skeletal muscle pericyte subtypes differ in their differentiation potential. Stem Cell Res. 2012b;10(1):67–84. doi: 10.1016/j.scr.2012.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dingova H, Fukalova J, Maninova M, Philimonenko VV, Hozak P. Ultrastructural localization of actin and actin-binding proteins in the nucleus. Histochem Cell Biol. 2009;131(3):425–434. doi: 10.1007/s00418-008-0539-z. [DOI] [PubMed] [Google Scholar]
- Dingwall C, Laskey RA. Nuclear targeting sequences--a consensus? Trends Biochem Sci. 1991;16(12):478–481. doi: 10.1016/0968-0004(91)90184-w. [DOI] [PubMed] [Google Scholar]
- Dopie J, Skarp KP, Rajakyla EK, Tanhuanpaa K, Vartiainen MK. Active maintenance of nuclear actin by importin 9 supports transcription. Proc Natl Acad Sci U S A. 2012;109(9):E544–E552. doi: 10.1073/pnas.1118880109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geeves MA, Holmes KC. Structural mechanism of muscle contraction. Annu Rev Biochem. 1999;68:687–728. doi: 10.1146/annurev.biochem.68.1.687. [DOI] [PubMed] [Google Scholar]
- Gordon AM, Homsher E, Regnier M. Regulation of contraction in striated muscle. Physiol Rev. 2000;80(2):853–924. doi: 10.1152/physrev.2000.80.2.853. [DOI] [PubMed] [Google Scholar]
- Hsu CC, Hu CD. Critical role of N-terminal end-localized nuclear export signal in regulation of activating transcription factor 2 (ATF2) subcellular localization and transcriptional activity. J Biol Chem. 2012;287(11):8621–8632. doi: 10.1074/jbc.M111.294272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong EM, Wang X, Xu K, Hossain MM, Jin JP. Nonmyofilament-associated troponin T fragments induce apoptosis. American Journal of Physiology-Heart and Circulatory Physiology. 2009;297(1):H283–H292. doi: 10.1152/ajpheart.01200.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimenez-Moreno R, Wang ZM, Gerring RC, Delbono O. Sarcoplasmic reticulum Ca2+ release declines in muscle fibers from aging mice. Biophys J. 2008;94(8):3178–3188. doi: 10.1529/biophysj.107.118786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimenez-Moreno R, Wang ZM, Messi ML, Delbono O. Sarcoplasmic reticulum Ca2+ depletion in adult skeletal muscle fibres measured with the biosensor D1ER. Pflugers Arch. 2010;459(5):725–735. doi: 10.1007/s00424-009-0778-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajstura J, Urbanek K, Perl S, Hosoda T, Zheng H, Ogorek B, Ferreira-Martins J, Goichberg P, Rondon-Clavo C, Sanada F, et al. Cardiomyogenesis in the adult human heart. Circ Res. 2010;107(2):305–315. doi: 10.1161/CIRCRESAHA.110.223024. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Ma L, Chang N, Guo S, Li Q, Zhang Z, Wang W, Tong T. CSIG inhibits PTEN translation in replicative senescence. Mol Cell Biol. 2008;28(20):6290–6301. doi: 10.1128/MCB.00142-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagoshi E, Yoneda Y. Dimerization of sterol regulatory element-binding protein 2 via the helix-loop-helix-leucine zipper domain is a prerequisite for its nuclear localization mediated by importin beta. Mol Cell Biol. 2001;21(8):2779–2789. doi: 10.1128/MCB.21.8.2779-2789.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Shea EK, Rutkowski R, Kim PS. Mechanism of specificity in the Fos-Jun oncoprotein heterodimer. Cell. 1992;68(4):699–708. doi: 10.1016/0092-8674(92)90145-3. [DOI] [PubMed] [Google Scholar]
- O'Shea EK, Rutkowski R, Stafford WF, 3rd, Kim PS. Preferential heterodimer formation by isolated leucine zippers from fos and jun. Science. 1989;245(4918):646–648. doi: 10.1126/science.2503872. [DOI] [PubMed] [Google Scholar]
- Pearlman JA, Powaser PA, Elledge SJ, Caskey CT. Troponin-T Is Capable of Binding Dystrophin Via a Leucine-Zipper. Febs Letters. 1994;354(2):183–186. doi: 10.1016/0014-5793(94)01119-2. [DOI] [PubMed] [Google Scholar]
- Pederson T, Tsai RY. In search of nonribosomal nucleolar protein function and regulation. J Cell Biol. 2009;184(6):771–776. doi: 10.1083/jcb.200812014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porte D, Oertel-Buchheit P, Granger-Schnarr M, Schnarr M. Fos leucine zipper variants with increased association capacity. J Biol Chem. 1995;270(39):22721–22730. doi: 10.1074/jbc.270.39.22721. [DOI] [PubMed] [Google Scholar]
- Sahota VK, Grau BF, Mansilla A, Ferrus A. Troponin I and Tropomyosin regulate chromosomal stability and cell polarity. J Cell Sci. 2009;122(Pt 15):2623–2631. doi: 10.1242/jcs.050880. [DOI] [PubMed] [Google Scholar]
- Scott MS, Boisvert FM, McDowall MD, Lamond AI, Barton GJ. Characterization and prediction of protein nucleolar localization sequences. Nucleic Acids Res. 2010;38(21):7388–7399. doi: 10.1093/nar/gkq653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaulsky G, Goldfinger N, Ben-Ze'ev A, Rotter V. Nuclear accumulation of p53 protein is mediated by several nuclear localization signals and plays a role in tumorigenesis. Mol Cell Biol. 1990;10(12):6565–6577. doi: 10.1128/mcb.10.12.6565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirri V, Urcuqui-Inchima S, Roussel P, Hernandez-Verdun D. Nucleolus: the fascinating nuclear body. Histochem Cell Biol. 2008;129(1):13–31. doi: 10.1007/s00418-007-0359-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szczesna D, Potter JD. The role of troponin in the Ca(2+)-regulation of skeletal muscle contraction. Results Probl Cell Differ. 2002;36:171–190. doi: 10.1007/978-3-540-46558-4_13. [DOI] [PubMed] [Google Scholar]
- Tang S, Wong HC, Wang ZM, Huang Y, Zou J, Zhuo Y, Pennati A, Gadda G, Delbono O, Yang JJ. Design and application of a class of sensors to monitor Ca2+ dynamics in high Ca2+ concentration cellular compartments. Proc Natl Acad Sci U S A. 2011;108(39):16265–16270. doi: 10.1073/pnas.1103015108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tobacman LS. Thin filament-mediated regulation of cardiac contraction. Annu Rev Physiol. 1996;58:447–481. doi: 10.1146/annurev.ph.58.030196.002311. [DOI] [PubMed] [Google Scholar]
- Vinson CR, Sigler PB, McKnight SL. Scissors-grip model for DNA recognition by a family of leucine zipper proteins. Science. 1989;246(4932):911–916. doi: 10.1126/science.2683088. [DOI] [PubMed] [Google Scholar]
- Wang ZM, Tang S, Messi ML, Yang JJ, Delbono O. Residual sarcoplasmic reticulum Ca2+ concentration after Ca2+ release in skeletal myofibers from young adult and old mice. Pflugers Arch. 2012;463(4):615–624. doi: 10.1007/s00424-012-1073-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T, Birbrair A, Wang ZM, Taylor J, Messi ML, Delbono O. Troponin T nuclear localization and its role in aging skeletal muscle. Age (Dordr) 2011 doi: 10.1007/s11357-011-9368-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T, Zaal KJ, Sheridan J, Mehta A, Gundersen GG, Ralston E. Microtubule plus-end binding protein EB1 is necessary for muscle cell differentiation, elongation and fusion. J Cell Sci. 2009;122(Pt 9):1401–1409. doi: 10.1242/jcs.039255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Xiong Y. A p53 amino-terminal nuclear export signal inhibited by DNA damage-induced phosphorylation. Science. 2001;292(5523):1910–1915. doi: 10.1126/science.1058637. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.