Abstract
Diatoms with symbiotic N2-fixing cyanobacteria are often abundant in the oligotrophic open ocean gyres. The most abundant cyanobacterial symbionts form heterocysts (specialized cells for N2 fixation) and provide nitrogen (N) to their hosts, but their morphology, cellular locations and abundances differ depending on the host. Here we show that the location of the symbiont and its dependency on the host are linked to the evolution of the symbiont genome. The genome of Richelia (found inside the siliceous frustule of Hemiaulus) is reduced and lacks ammonium transporters, nitrate/nitrite reductases and glutamine:2-oxoglutarate aminotransferase. In contrast, the genome of the closely related Calothrix (found outside the frustule of Chaetoceros) is more similar to those of free-living heterocyst-forming cyanobacteria. The genome of Richelia is an example of metabolic streamlining that has implications for the evolution of N2-fixing symbiosis and potentially for manipulating plant–cyanobacterial interactions.
 Cyanobacterial symbionts of marine diatoms can localize intracellularly or
externally to their host partners. Here Hilton et al. describe the genomes of two
diazotroph cyanobacterial symbionts of diatoms and show that the location of the symbiont
affects expression of nitrogen assimilation genes.
Cyanobacterial symbionts of marine diatoms can localize intracellularly or
externally to their host partners. Here Hilton et al. describe the genomes of two
diazotroph cyanobacterial symbionts of diatoms and show that the location of the symbiont
affects expression of nitrogen assimilation genes.
Cyanobacteria form partnerships with taxonomically diverse hosts that are usually multicellular, and these symbioses are ubiquitous in terrestrial and aquatic environments1. Cyanobacteria are autotrophic microorganisms and some can convert dinitrogen (N2) gas to ammonium. Two groups of understudied planktonic symbioses are the partnerships between marine diatoms and the heterocyst-forming cyanobacteria, Richelia intracellularis and Calothrix rhizosoleniae (Fig. 1a–c).
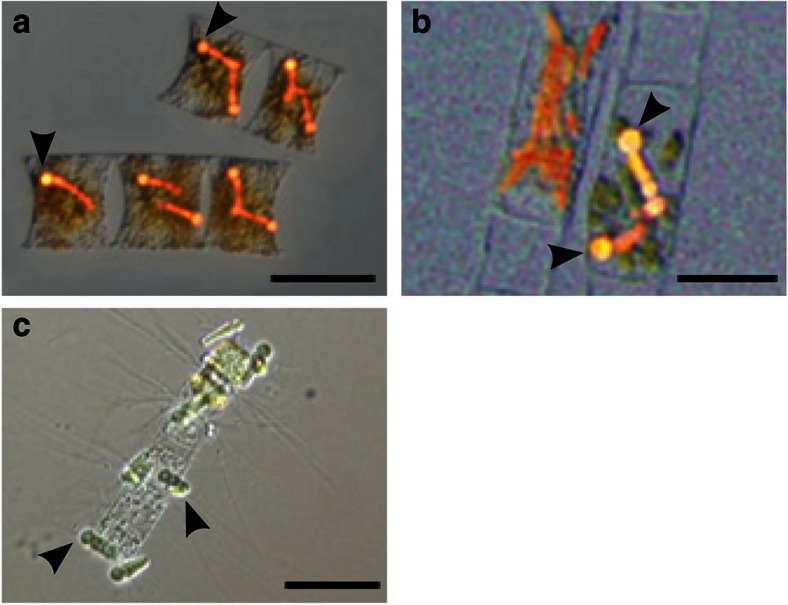
Figure 1. Cyanobacteria in symbiosis with diatoms.
Photomicrographs of cyanobacterial symbionts (denoted by arrows) representative of those sequenced in this study with host diatoms. Differential interference contrast bright field overlaid with blue light epifluorescence images of the diatoms H. membranaceus (a) and H. hauckii (b), with intracellular cyanobacterial symbionts. Bright-field microscopy image of epiphytic cyanobacterial symbiont C. rhizosoleniae SC01 attached to the host diatom Chaetoceros sp. (c). Scale bars, 50 μm.
Richelia and Calothrix species convert N2 and transfer the fixed N to their host2. Richelia and Calothrix associate with different hosts and also differ in cellular location (internal versus external), implying different life histories and mechanisms for nutrient exchanges with their partners. The Richelia symbionts of the diatom genera Rhizosolenia and Hemiaulus reside inside the diatom cell wall and are passed on to the next generation of the host3. The Rhizosolenia symbiont is outside the plasmalemma in the periplasmic space3; the Hemiaulus symbiont’s location is unknown. In contrast, Calothrix attaches externally to Chaetoceros spp. and can be cultured without the host diatom in N-deplete media4. Reports of free-living Richelia may be a result of broken diatoms5,6, whereas Calothrix have been observed as individual trichomes in the plankton7,8. The mechanism of formation of a Calothrix–Chaetoceros association and whether the symbiont is transmitted to the next generation is unknown.
We compared the genomes of two of the Richelia internal symbiont strains (R. intracellularis HH01, RintHH, symbiont of Hemiaulus hauckii and R. intracellularis HM01, RintHM, symbiont of H. membranaceus) with that of the external symbiont Calothrix rhizosoleniae SC01 (CalSC). We found that genome size and content, especially N metabolism genes, differed substantially, suggesting the cellular location (intracellular versus extracellular) has dictated varying evolutionary paths and resulted in different mechanisms involved in maintaining the symbiosis (Table 1).
Table 1. Nostocales genomes statistics.
| Cyanobacterium | Accession number | Symbiotic state | Size (Mb) | Percent GC | Percent coding | TCs |
|---|---|---|---|---|---|---|
| Anabaena variabilis ATCC 29413 | PRJNA10642 | Free-living | 7.1 | 41 | 82 | 570 |
| Nostoc punctiforme PCC 73102 | PRJNA216 | Facultative | 9.1 | 41 | 77 | 575 |
| Nostoc sp. PCC 7120 | PRJNA244 | Free-living | 7.2 | 41 | 82 | 559 |
| ‘Nostoc azollae’ 0708 | PRJNA30807 | Obligate | 5.5 | 38 | 52 | 286 |
| Raphidiopsis brookii D9a | PRJNA40111 | Free-living | 3.2 | 40 | 86 | 300 |
| Cylindrospermopsis raciborskii CS-505a | PRJNA40109 | Free-living | 3.9 | 40 | 85 | 344 |
| Nodularia spumigena CCY 9414a | PRJNA13447 | Free-living | 5.3 | 41 | 82 | 428 |
| Richelia intracellularis HH01a | PRJEA104979 | Obligate | 3.2 | 34 | 56 | 190 |
| Calothrix rhizosoleniae SC01a | PRJNA19291 | Facultative | 6.0 | 39 | 76 | 400 |
TC, transporter classification.
aGenome is in a draft state.Available genomes of the Order Nostocales.
Results
General features of the diatom symbiont genomes
On the basis of the 16 rRNA and ntcA gene sequences, the diatom symbionts cluster within the cyanobacterial Order Nostocales (Fig. 2), but their genome sizes vary greatly (RintHH, 3.2 Mb; CalSC, 6.0 Mb; Table 1). The percent coding information of the CalSC genome is only slightly lower than the free-living Nostocales members, whereas the RintHH genome percent coding is further reduced, similar to ‘Nostoc azollae’ 0708 (Table 1). Similarly, the RintHH genome GC content and transporter count are lower than any other genome in the Order, whereas the CalSC genome is a more characteristic Nostocales genome in each respect (Table 1).
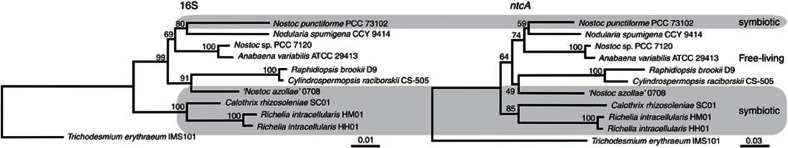
Figure 2. Nostocales phylogeny.
Neighbor-joining phylogenetic trees of 16S rRNA and ntcA sequences from seven previously sequenced cyanobacteria and three additional diatom symbionts from this study. The organisms observed in symbiotic relationships are shaded in grey. Both trees are rooted with T. erythraeum IMS101. Locus tags (16S, ntcA): ‘N. azollae’ 0708 (Aazo_R0008, Aazo_1065), R. brookii (CRD_01297,CRD_00550), C. raciborskii CS-505 (CRC_01246, CRC_00858), N. sp. PCC 7120 (allrr01, alr4392), N. spumigena CCY9414 (N9414_r17988, N9414_19492), N. punctiforme PCC 73102 (Npun_r020, Npun_F5511), A. variabilis ATCC 29413 (Ava_R0006, Ava_3283), T. erythraeum IMS101 (Tery_R0014, Tery_2023), C. rhizosoleniae SC01 (CSC01_11477, CSC01_6586), R. intracellularis HH01 (RintHH_r10, RintHH_12150), R. intracellularis HM01 (RintHM_3660, RintHM_9700).
The genome of RintHM, the symbiont of H. membranaceus, a diatom that is closely related to H. hauckii, is only 2.2 Mb and is lacking a number of sequences expected of a full genome (including transfer RNAs for four amino acids and several nitrogenase genes). Therefore, we believe it is a partial genome, likely due to low-sequencing coverage (average depth of coverage 13 × ). However, 16S rRNA and ntcA sequences confirm the morphologically similar symbionts are also related genetically (Fig. 2), as previously demonstrated by nifH and hetR sequences9,10. In addition, analysis of the contigs showed that there are no evident gene insertions/deletions or genome rearrangements between the two Hemiaulus sp. symbiont genomes. The 1,671 shared genes of the symbionts average 97.5% sequence identity (DNA) (Supplementary Fig. S1) and show no significant difference in the GC content of the genes sequenced.
Nitrogen metabolism of the diatom symbionts
Given its small size, the RintHH genome is highlighted by many gene deletions, including numerous genes important in N metabolism, such as the transporters for ammonium and nitrate, and the genes encoding nitrate and nitrite reductases (Fig. 3). The diatom symbiont genomes are each missing genes that encode urea transporters and urease, which are functional in all previously sequenced Nostocales genomes, except for the genome of ‘N. azollae’ 0708 (ref. 11).
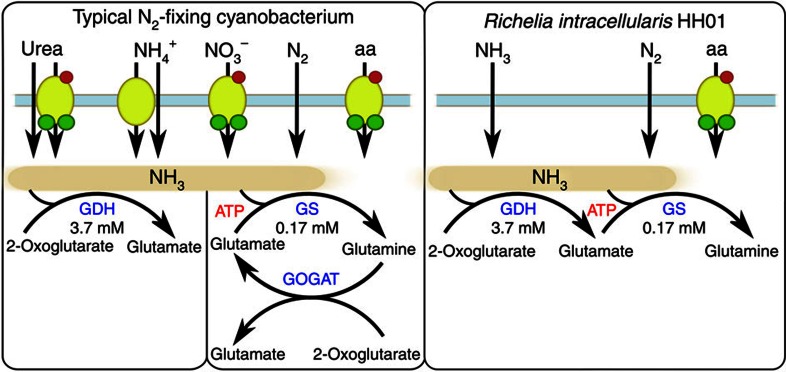
Figure 3. Overview of the limited N pathways of Richelia.
Nitrogen metabolism pathways common in N2-fixing cyanobacteria compared with R. intracellularis HH01. Redrawn from 33,34. Km-values for GDH and GS are for ammonia in each reaction from the N2-fixing cyanobacterium Synechocystis PCC 6803 (refs 18, 35).
The most unusual gene deletion in RintHH is the gene for an important enzyme in C and N metabolism, glutamate synthase, also known as glutamine:2-oxoglutarate aminotransferase (GOGAT). This enzyme is part of glutamine synthetase (GS)-GOGAT (GS-GOGAT), a generally universal pathway for high-affinity N assimilation (found in all other sequenced cyanobacterial genomes12, including CalSC and ‘N. azollae’ 0708), which uses glutamine, synthesized by GS, and a C skeleton, 2-oxoglutarate, to produce two glutamate molecules. The glutamate produced by GOGAT is then recycled for further ammonium assimilation by GS. The gene encoding GS is present and functional in each symbiont genome; however, they are each lacking a gene that encodes a GS-inactivating factor that is found in all previously sequenced Nostocales genomes (asl2329 in Nostoc sp. PCC 7120).
The multiple N metabolism genes missing from the RintHH genome are common to, and widely dispersed across, the genomes of all closed Nostocales genomes (Supplementary Fig. S2). Given this, and the high-sequencing coverage of the RintHH draft genome (average depth of coverage >40 × ), it is unlikely that the missing genes are actually present in the RintHH genome. The RintHH genome does contain a tRNA for each of the 20 amino acids, as expected from a complete genome. Other sequences expected to be present are also in the assembly, such as the previously studied genes hetR and nifH9,10, and genes responsible for known characteristics of RintHH, such as nitrogen fixation, heterocyst formation, and phycoerythrin and chlorophyll pigments. Moreover, to decrease any possible bias during the process, two RintHH samples were separately sorted, amplified and sequenced.
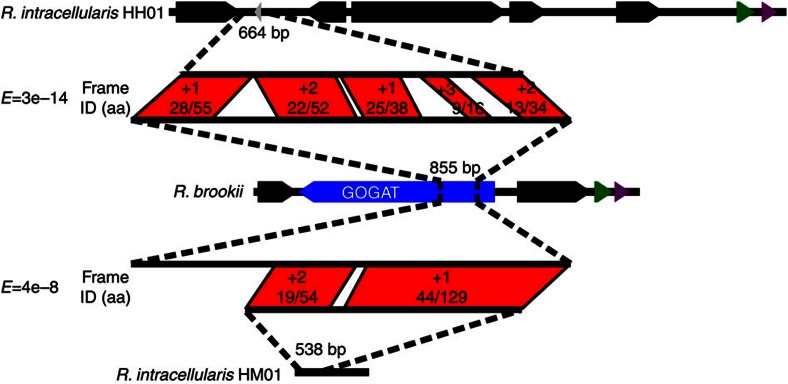
The majority of the N metabolism genes show no similarity to the RintHH genome. However, an intergenic sequence, which contains a small predicted hypothetical protein, has a top hit to the Raphidiopsis brookii GOGAT-encoding gene in the NCBI database of non-redundant (nr) protein sequences (BLASTx, E-value=3e−14; Fig. 4). The intergenic sequence covers less than 20% of the GOGAT gene and aligns in all three unidirectional frames. This intergenic region is found downstream of two genes that are part of a conserved region downstream of the GOGAT gene in Nostocales genomes. A single contig from the RintHM genome also shows similarity to the GOGAT gene in the same manner (Fig. 4).
Figure 4. GOGAT gene remnants in Richelia.
The intergenic space between two R. intracellularis HH01 ORFs, including an annotated 120 bp hypothetical protein (in grey, RintHH_15420) compared with the gene encoding GOGAT in Raphidiopsis brookii D9 (in blue, CRD_00957) and the genomic context of each. The middle alignment represents the top BLAST hit of the R. intracellularis HH01 intergenic space (1561, bp) in the nr database (BLASTx, E-value=3e−14). The green and purple genes encode a mannose-6-phosphate isomerase and a probable iron binding protein, respectively.
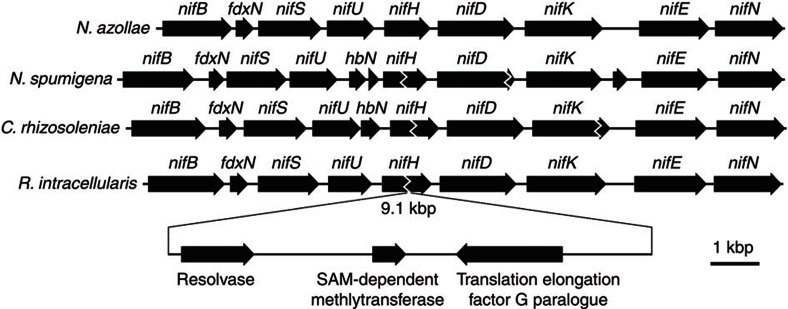
Gene interruptions on the diatom symbiont nif operon
The similarities with other heterocyst-forming cyanobacteria include the presence of insertion sequences in the middle of RintHH and CalSC N2-fixation genes13,14. The RintHH nifH gene is interrupted in this manner by a 9.1-kb sequence (Fig. 5). The CalSC nifH and nifK genes are each interrupted in the same manner by longer sequences (each >20 kb). The nifH interruptions in RintHH and CalSC appear to occur at the same location within the nifH gene; however, the CalSC nifH element is at least twice as long as that in the RintHH genome. Recombination genes found on each nifH elements show high similarity to each other (71% ID, protein) and are presumably the mechanisms for excision of the element during heterocyst formation.
Figure 5. Nitrogenase gene organization.
The nif operon and surrounding genes of R. intracellularis HH01 and C. rhizosoleniae SC01 compared with other representative Nostocales cyanobacteria ‘Nostoc azollae’ 0708 and Nodularia spumigena CCY9414, and the genes along the insertion element interrupting the R. intracellularis HH01 nifH gene.
Discussion
To date, the intracellular RintHH genome is the smallest N2-fixing, heterocyst-forming cyanobacteria genome sequenced. Within Nostocales, the R. brookii D9 genome is slightly smaller than that of RintHH, but it is unable to form heterocysts or fix N2 (ref. 15). In contrast, the CalSC genome is similar in size and content to the genomes of free-living organisms in this Order and N. punctiforme, a facultative, or opportunistic, symbiont.
The genome reduction in RintHH, marked by its size, percent coding and GC content, is similar to that of ‘N. azollae’ 0708, the obligate, or host-dependent, symbiont of the water fern Azolla filiculoides11. These features are commonly exhibited by genomes of obligate symbionts, indicating that RintHH is also dependent on its host. Obligate symbionts have more unnecessary genes than free-living or facultative symbiotic organisms due to metabolic redundancy encoded by the host genome and the lack of full exposure to the environment16. Examples of genes dispensable to obligate symbionts may be those absent or non-functional in both RintHH and ‘N. azollae’ 0708, but present in other heterocyst-forming cyanobacteria genomes (Supplementary Table S1). Decreased evolutionary pressure to keep functional genes leads to a lower percent coding and eventually to genome size reduction as non-functioning genes are deleted. The smaller genome leads to accelerated sequence evolution, increasing AT bias16. The lack of CalSC genome reduction may be taken as evidence that this organism is an opportunistic partner. This is consistent with the external location of CalSC on the diatom setae (spine-like projections) and the ability to maintain it in culture independent of the host diatom in filtered seawater-based media4. In contrast, RintHH lives inside the host diatom cell wall, and possibly even within the cytoplasm, with little or no exposure to the external environment, and thus the genome reduction is consistent with that of an obligate symbiont.
The numerous absent N metabolism genes appear to have been selectively deleted from multiple regions throughout the RintHH genome. The lack of ammonium transporters and enzymes required to take up and assimilate urea or nitrate limits the possible N sources for RintHH to amino acids, N2, and passive diffusion of ammonia in oceanic environments, where concentrations of amino acids and ammonium are extremely low. Therefore, deletions in N metabolism genes ensure N2 fixation within the partner diatom persists, and is likely important for maintaining the symbiotic partnership.
The lack of GOGAT, on the other hand, likely streamlines host–symbiont interactions and seems to be a more recent deletion than the other N metabolism genes, given the similarity between GOGAT genes and intergenic space in the RintHH genome. Without GOGAT, RintHH must use an alternate pathway for assimilation of N2-derived ammonium with glutamate dehydrogenase (GDH; Fig. 3), unless the host diatom provides glutamate for the symbiont. In contrast, GS-GOGAT is the main N assimilation pathway used by Anabaena azollae in obligate symbiosis with host Azolla caroliniana, and very little N is assimilated through GDH17. Given the high N2 fixation rates by the cyanobacterial symbiont when associated with the host diatom2, it is feasible that intracellular ammonium concentrations are elevated and facilitate assimilation by the low-affinity GDH enzyme18. However, an adequate concentration of 2-oxoglutarate would also be needed to support ammonium assimilation. If these C skeletons are provided by the host, as in the Nostoc–Gunnera symbiosis19, the symbiont may perceive the increase of intracellular C:N as N starvation20, causing continued N2 fixation by the cyanobacterium. Thus, the lack of GOGAT eliminates a common metabolic pathway and creates an N exchange pathway between host and symbiont that provides the host with a way to regulate the symbiont’s growth and activity.
The lack of a GS-inactivating factor streamlines N metabolism further in RintHH. GS catalyses the conversion of glutamate to glutamine, and without an inactivating factor it will maintain low intracellular glutamate concentrations. The subsequent increasing glutamine pool may indicate this amino acid is the form of N passed to the host. The absence of this regulator shows parallels between the Richelia–Hemiaulus and Calothrix–Chaetoceros associations, and separates the diatom symbionts from other heterocyst-forming cyanobacteria.
However, with regard to N metabolism, the similarities are minimal and the fundamental differences between the RintHH and CalSC genomes reflect the evolutionary selection of their metabolic interactions and cellular locations with the partner diatom. The extracellular CalSC symbiont is exposed to the open ocean environment at all times, and can therefore use a suite of dissolved inorganic nitrogen sources, albeit at low concentrations. Furthermore, the CalSC genome possesses a gene to encode GOGAT and, thus, the symbiont is capable of assimilating N through the high-affinity GS-GOGAT, in addition to GDH. However, a scenario for enhancing N2 fixation by C transfer from the diatom to the external symbiont CalSC, as hypothesized for the Richelia–Hemiaulus association, would require a direct host–symbiont transport system. Otherwise, the extracellular C would likely be diluted immediately and available to other microorganisms. Thus, the extracellular location of CalSC on Chaetoceros spp. likely requires different mechanisms for N metabolism and exchange than intracellular RintHH. The differences in genome content and metabolic potential reflect the differences between obligate and facultative symbionts.
Many heterocyst-forming cyanobacteria have DNA sequences interrupting N2 fixation-related genes in vegetative cells, which are excised during genome rearrangements coincident with heterocyst development21, but the functional significance and evolutionary origin of these elements are unknown. These interrupting sequences have been seen previously in several genes13,14,22, but the CalSC genome is the first example of a nifK element. The location of elements within nifH and high similarity between the genes likely responsible for the excision of the interrupting sequence are the only apparent similarities between the two nifH elements in these closely related cyanobacteria. Although their similarities indicate the nifH elements in each organism have the same evolutionary origin, there seems to be little evolutionary pressure on the contents and length of the element.
The characteristics of the genomes of symbiotic heterocyst-forming cyanobacteria reflect the differences in cellular location and host dependency. The absence of basic N metabolism enzymes and transporters in the RintHH genome streamline it, while maintaining the association and providing a mechanism for host regulation of the symbiont. In contrast, the genome of CalSC has few deletions relative to free-living heterocyst-forming cyanobacteria. The differences between genomes suggest mechanisms that may be important in defining facultative or obligate symbioses, with implications for the biology and ecology of these widespread symbiotic associations in the sea. Furthermore, differences in the genomic composition of morphologically and taxonomically similar microorganisms provides an important example of how one partner’s metabolic capabilities can evolve with a symbiosis. Finally, the genomes reported in this study, in addition to other recent discoveries of extensive metabolic streamlining in N2-fixing cyanobacteria23, yield the possibility of yet undiscovered plants or algae containing N2-fixing organelles.
Methods
H. hauckii and H. membranaceus symbiont DNA preparation
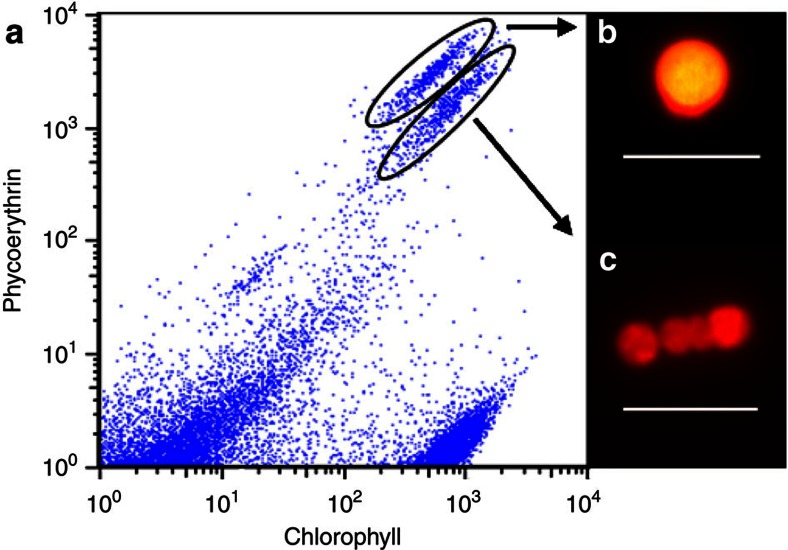
Stable Hemiaulus–Richelia cultures, isolated from the western Gulf of Mexico, were grown in N-free YBC-II medium at 25 °C (ref. 24), filtered on to a 3-μm pore size, 25-mm diameter polyester filter (Sterlitech) and frozen for storage. TE buffer (1 × ) was added, and once the filter thawed the cells were resuspended by vortexing for 1 min. The majority of diatoms were broken at this stage, releasing the symbionts in the process. Samples were then analysed on the Influx flow cytometer and cell sorter (BD Biosciences), and cyanobacteria cells were distinguished from other cells by their phycoerythrin pigmentation (Fig. 6). For the H. hauckii symbionts, the vegetative trichomes and heterocysts had separated during the sample preparation and the cells formed separate populations on the flow cytometer based on slightly different chlorophyll and phycoerythrin signatures (Fig. 6). Sorting gates, defined by relative pigment values, allowed for the isolation of vegetative cells from the rest of the sample. Two replicate sorts of 5,000 symbiont vegetative trichomes (3–5 cells per trichome) were sorted. Genomic DNA in each sample was amplified by multiple displacement amplification using the Repli-g Midi kit (Qiagen). The manufacturer’s protocol for 0.5 μl of cell material was followed with one exception: after buffer D2 was added, the samples were incubated for 5 min at 65 °C and then put on ice for 1 min, instead of 10 min on ice without a 65 °C incubation.
Figure 6. Flow cytometric analysis ofR. intracellularis–H. hauckii culture samples.
A cytogram displaying events gated based upon chlorophyll (692/40 nm) and phycoerythrin (572/27 nm) detection channels with the cyanobacteria symbiont populations easily separated (a). Insets are microscopy images under blue excitation of a vegetative trichome (b) and a heterocyst cell (c) representative of circled populations. Scale bars, 10 μm. The vegetative trichome population was sorted for genome sequencing samples.
To ensure uncontaminated samples, each amplified DNA sample was PCR-amplified using universal 16S rRNA primers 27F (5′-AGAGTTTGATCMTGGCTCAG-3′) and 1492R (5′-GGTTACCTTGTTACGACTT-3′)25. The PCR was carried out in 50 μl reactions consisting of 1 × PCR buffer, 2 mM MgCl2, 200 μM dNTPs, 0.2 μM of each primer and 1.5 U of Platinum Taq DNA polymerase (Invitrogen). A touchdown PCR was performed as follows: an initial denaturing step at 94 °C for 5 min, followed by 30 cycles of three 1-min steps (denaturation at 94 °C, annealing at 53–41 °C and elongation at 72 °C) and a final elongation step at 72 °C for 10 min. The first cycle annealing took place at 53 °C and was lowered by 0.4 °C for each cycle to reach 41 °C for the final cycle. Resulting products were run on a 1.2% agarose gel, the distinct bands of ~1,500 bp were excised and then recovered using the Zymoclean DNA Recovery Kit (Zymo Research). The recovered DNA was then ligated and plated for blue/white screening using the pGem-T and pGem-T Easy Vectors Systems (Promega). Twenty-four colonies per sample were picked and grown overnight at 37 °C in 2 × LB media with carbenicillin (200 μg ml−1). The Montáge Plasmid MiniprepHTS Kit (Millipore) was used following the manufacturer’s instructions for the full lysate protocol for plasmid DNA miniprep. Samples were sequenced at UC-Berkeley DNA Sequencing Facility and each sequence was subject to BLAST analysis against the nt database (BLASTn). All sequences were identical and had top hits to 16S rRNA sequences of heterocyst-forming cyanobacteria, confirming no contaminant genomes were present in the samples.
DNA concentration and quality were checked (Agilent 2100 Bioanalyzer, Agilent Technologies) before submission for 454 Titanium sequencing (Roche) at the UCSC Genome Technology Center.
The symbionts of H. membranaceus were processed in the same manner, but heterocysts and vegetative cells did not separate during sample preparation, and both cell types were present in the sorted samples. Moreover, we were confident from flow cytometry that the cell preparation was pure enough to determine the comparative features we were looking for, and that we would be able to distinguish between the closely related symbiont and the few bacteria that could be carried through by flow cytometry. Therefore, no contamination or DNA quality checks were performed in preparation of the RintHM samples.
H. hauckii and H. membranaceus symbiont genome assembly
A total of 433,028 reads were sequenced from RintHM samples. The reads assembled to nearly 8 Mb and the assembly contained four 16S rRNA sequences with low similarity to each other (<83% ID), indicating multiple DNA sources in the data. RintHM contigs were defined as those which had a better BLAST hit to RintHH than to any other organism in the nt database. The resulting 2,212,909 bp (941 contigs, coverage depth 13 × ) were made up of 77,324 reads averaging 380 bp each. An additional 31 contigs, totalling 97,821 bp, had a top hit in the nt database to a cyanobacterium other than RintHH, but none of those contigs contained any of the N metabolism genes of interest.
The two RintHH samples yielded a total 409,035 reads, averaging 344 bp each. The read data were pooled and assembled into 3,243,759 bp in 90 contigs (coverage depth 43 × ) and appeared to be non-contaminated. There are seven contigs longer than 100 kbp, an additional 32 contigs longer than 25 kbp (Supplementary Fig. S3) and 91% of bases with 15 × coverage or greater.
CalSC DNA preparation
CalSC genomic DNA was extracted from pelleted cells using a sucrose lysis protocol, including the optional back extraction26. The exceptions to this protocol were our use of 10% SDS in the lysate for Fraction B instead of 20% SDS and the 1-h incubation of Fraction B after adding the lysate was at 37 °C rather than 55 °C. The genomic DNAs from Fraction B were pooled and divided into three equal volume samples. The three genomic extracts were checked for purity and quantity (Agilent 2100 Bioanalyzer, Agilent Technologies), and the DNA concentrations ranged between 38.37 and 66.38 ng μl−1. Samples were then submitted to JCVI for 454 sequencing.
CalSC genome assembly
Once the read data from JCVI (2,477,040 reads, 968 MB) were assembled, the number of contigs (69,919) and size of the assembly (81.4 Mb) immediately suggested that more than one organism was in the sequencing samples. The longest contig of 1.2 MB in length contained a full-length rRNA operon predicted by RDP (Ribosomal Database Project) to be a Planctomycete, confirming the presence of organisms other than CalSC. A plot of the number of reads on each contig against the length of the contig showed strong linear relationships (Supplementary Fig. S4), representing defined clusters of coverage depth, based on the relative abundance of the each organism’s genome in the sample. Spot-checking the phylogeny of BLASTn results for long open reading frames (ORFs) on long contigs revealed that the contigs lying along the line marked in red (representing a coverage depth of 30 × ) were those that came from CalSC (Supplementary Fig. S4). Each predicted ORF >450 bp on contigs with depth of coverage 15–45 × was subject to BLAST analysis against the nt database. A contig was considered to be part of the CalSC genome if at least one of these ORFs on the contig had a top hit to a cyanobacterial sequence, and 471 contigs met this criterion (5,967,587 bp). One additional contig (5,416 bp) containing the rRNA operon was added. It had been overlooked initially due to its lack of ORFs and its relatively higher coverage depth (71 × , indicating it is present in two copies in the genome). The end result was a 5,973,003 bp genome composed of 472 contigs (30 × coverage depth).
Genomic analysis
After assembly and contamination screening, the genomes were submitted to RAST (Rapid Annotation using Subsystem Technology)27 for annotation.
The nitrogen metabolism genes not found in the RintHH genome were pulled from each Nostocales genome, and each gene was subject to BLAST analysis against a database of all 409,035 reads (tBLASTn, e<10). Two thousand seven hundred and twenty reads had hits at least 25% identical (AA) across at least 50% the length of the read or gene, whichever was shorter. A BLAST analysis of each of these reads against the nr database was performed (BLASTx, e<10). Twenty-one reads had a top hit to a GOGAT-encoding gene, and each of these reads is assembled into the intergenic region discussed below as likely GOGAT remnants in the RintHH genome. No other reads had a top hit in the nr database of a nitrogen metabolism-related gene.
Predicted ORFs in each genome with a BLAST hit in the Transporter Classification database28 (BLASTp, E-value <1E−19) were counted as transporter genes.
For the 16S rRNA and the ntcA phylogenetic trees, nucleotide sequences were acquired from DOE Joint Genome Institute for each of the seven previously sequenced Nostocales genomes and Trichodesmium erythraeum IMS101, and were aligned with the sequences from the three diatom symbiont genomes using Clustal W29 (1,421 bp, 16S rRNA; 646 bp, ntcA). Phylogenetic analyses were rendered in Mega5 (ref. 30) using the Neighbor-Joining method31. The Tamura–Nei test was run to detect the best models. Statistical support for nodes was based on 1,000 bootstrap replicates32.
Author contributions
J.A.H., R.A.F., J.P.Z. and T.A.V. designed the study and wrote the paper; T.A.V and R.A.F. grew and collected cultures; J.A.H. and B.J.C. sorted symbiont cells; J.A.H. and H.J.T. assembled and analysed the genomes.
Additional information
Accession codes: The genomes described in this study have been deposited at the European Nucleotide Archive (ENA) under accession numbers CAIY01000001 to CAIY01000090 (RintHH), CAIS01000001 to CAIS01000941 (RintHM) and SRX023670 (CalSC).
How to cite this article: Hilton, J. A. et al. Genomic deletions disrupt nitrogen metabolism pathways of a cyanobacterial diatom symbiont. Nat. Commun. 4:1767 doi: 10.1038/ncomms2748 (2013).
Supplementary Material
Supplementary Figures S1-S4 and Supplementary Table S1
Acknowledgments
This work was sponsored in part by the Gordon and Betty Moore Foundation (J.P.Z.), NSF (BIO OCE-0929015, R.A.F. and J.P.Z.; OCE-0726726, T.A.V.) and the NSF Center for Microbial Oceanography: Research and Education (J.P.Z.). The CalSC genome was sequenced through the Microbial Genome Sequencing Project managed by JCVI and was sponsored by the Gordon and Betty Moore Foundation. We thank J. Meeks for comments on the manuscript; M. Hogan, S. Bench and K. Turk for technical help and discussions; K. Karplus and J. Long for their effort on the CalSC genome assembly; and A. Worden and H. Wilcox for suggestions on CalSC culture DNA extraction methods.
References
- Usher K. M., Bergman B. & Raven J. A. Exploring cyanobacterial mutualisms. Ann. Rev. Ecol. Evol. Syst. 38, 255–273 (2007). [Google Scholar]
- Foster R. A. et al. Nitrogen fixation and transfer in open ocean diatom–cyanobacterial symbioses. ISME J. 5, 1484–1493 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villareal T. A. In Marine Pelagic Cyanobacteria: Trichodesmium and Other Diazotrophs eds Carpenter E. J., Capone D. G., Reuter J. 163–175Kluwer (1992). [Google Scholar]
- Foster R. A., Goebel N. L. & Zehr J. P. Isolation of Calothrix rhizosoleniae (cyanobacteria) strain SC01 from Chaetoceros (Bacillariophyta) spp. diatoms of the Subtropical North Pacific Ocean. J. Phycol. 46, 1028–1037 (2010). [Google Scholar]
- Lyimo T. J. Distribution and abundance of the cyanobacterium Richelia intracellularis in the coastal waters of Tanzania. J. Ecol. Nat. Environ. 3, 85–94 (2011). [Google Scholar]
- Zhang Y., Zhao Z., Sun J. & Jiao N. Diversity and distribution of diazotrophic communities in the South China Sea deep basin with mesoscale cyclonic eddy perturbations. FEMS Microbiol. Ecol. 78, 417–427 (2011). [DOI] [PubMed] [Google Scholar]
- Gomez F., Furuya K. & Takeda S. Distribution of the cyanobacterium Richelia intracellularis as an epiphyte of the diatom Chaetoceros compressus in the western Pacific Ocean. J. Plank. Res. 27, 323–330 (2005). [Google Scholar]
- White A. E., Prahl F. G., Letelier R. M. & Popp B. N. Summer surface waters in the Gulf of California: prime habitat for biological N2 fixation. Global Biogeochem. Cycles 21, GB2017 (2007). [Google Scholar]
- Janson S., Wouters J., Bergman B. & Carpenter E. J. Host specificity in the Richelia-diatom symbiosis revealed by hetR gene sequence analysis. Environ. Microbiol. 1, 431–438 (1999). [DOI] [PubMed] [Google Scholar]
- Foster R. A. & Zehr J. P. Characterization of diatom-cyanobacteria symbioses on the basis of nifH, hetR and 16S rRNA sequences. Environ. Microbiol. 8, 1913–1925 (2006). [DOI] [PubMed] [Google Scholar]
- Ran L. et al. Genome erosion in a nitrogen-fixing vertically transmitted endosymbiotic multicellular cyanobacterium. PLoS ONE 5, e11486 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsson J., Nylander J. & Bergman B. Genome fluctuations in cyanobacteria reflect evolutionary, developmental and adaptive traits. BMC Evol. Biol. 11, 187 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golden J. W., Robinson S. J. & Haselkorn R. Rearrangement of nitrogen fixation genes during heterocyst differentiation in the cyanobacterium Anabaena. Nature 314, 419–423 (1985). [DOI] [PubMed] [Google Scholar]
- Carrasco C. D., Buettner J. A. & Golden J. W. Programmed DNA rearrangement of a cyanobacterial hupL gene in heterocysts. Proc. Natl Acad. Sci. 92, 791–795 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stucken K. et al. The smallest known genomes of multicellular and toxic cyanobacteria: comparison, minimal gene sets for linked traits and the evolutionary implications. PLoS ONE 5, e9235 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran N. A. Tracing the evolution of gene loss in obligate bacterial symbionts. Curr. Opin. Microbiol. 6, 512–518 (2003). [DOI] [PubMed] [Google Scholar]
- Meeks J. C. et al. Assimilation of exogenous and dinitrogen-derived 13NH4+ by Anabaena azollae separated from Azolla caroliniana Willd. Arch. Microbiol. 142, 229–233 (1985). [Google Scholar]
- Florencio F., Marques S. & Candau P. Identification and characterization of a glutamate dehydrogenase in the unicellular cyanobacterium Synechocystis PCC 6803. FEBS Lett. 223, 37–41 (1987). [Google Scholar]
- Söderbäck E. & Bergman B. The Nostoc-Gunnera symbiosis: carbon fixation and translocation. Physiol. Plant 89, 125–132 (1993). [Google Scholar]
- Muro-Pastor M. I., Reyes J. C. & Florencio F. J. Cyanobacteria perceive nitrogen status by sensing intracellular 2-oxoglutarate levels. J. Biol. Chem. 276, 38320–38328 (2001). [DOI] [PubMed] [Google Scholar]
- Carrasco C. D., Ramaswamy K., Ramasubramanian T. & Golden J. W. Anabaena xisF gene encodes a developmentally regulated site-specific recombinase. Genes Dev. 8, 74–83 (1994). [DOI] [PubMed] [Google Scholar]
- Vintila S., Selao T., Norén A., Bergman B. & El-Shehawy R. Characterization of nifH gene expression, modification and rearrangement in Nodularia spumigena strain AV1. FEMS Microbiol. Ecol. 77, 449–459 (2011). [DOI] [PubMed] [Google Scholar]
- Thompson A. W. et al. Unicellular cyanobacterium symbiotic with a single-celled eukaryotic alga. Science 337, 1546–1550 (2012). [DOI] [PubMed] [Google Scholar]
- Pyle A. Light dependant growth and nitrogen fixation rates in the Hemiaulus hauckii and Hemiaulus membranaceus diatom-diazotroph associations. MS thesis, Dept. Marine Science, Austin, Texas 78pp (2011). [Google Scholar]
- Lane D. J. In Nucleic Acid Techniques in Bacterial Systematics 115–175John Wiley & Sons (1991). [Google Scholar]
- Cuvelier M. L. et al. Targeted metagenomics and ecology of globally important uncultured eukaryotic phytoplankton. Proc. Natl Acad. Sci. 107, 14679–14684 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aziz R. K. et al. The RAST Server: rapid annotations using subsystems technology. BMC Genomics 9, 75 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saier M. H. J., Yen M. R., Noto K., Tamang D. G. & Elkan C. The transporter classification database: recent advances. Nucleic Acids Res. 37, D274–D278 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson J. D., Higgins D. G. & Gibson T. J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673–4680 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K. et al. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitou N. & Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406–425 (1987). [DOI] [PubMed] [Google Scholar]
- Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39, 783–791 (1985). [DOI] [PubMed] [Google Scholar]
- Muro-Pastor M. I. & Florencio F. J. Regulation of ammonium assimilation in cyanobacteria. Plant Physiol. Biochem. 41, 595–603 (2003). [Google Scholar]
- Yan D. Protection of the glutamate pool concentration in enteric bacteria. Proc. Natl Acad. Sci. 104, 9475–9480 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mérida A., Leurentop L., Candau P. & Florencio F. J. Purification and properties of glutamine synthetases from the cyanobacteria Synechocystis sp. strain PCC 6803 and Calothrix sp. strain PCC 7601. J. Bacteriol. 172, 4732–4735 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures S1-S4 and Supplementary Table S1