Tbc1 is identified as the fission yeast orthologue of tubulin-folding cofactor C. In addition to its roles in tubulin folding, Tbc1 acts as a GAP in regulating the small GTPase Alp41/Arl2, which interacts with cofactor D Alp1. A model is given of the final stages of the tubulin cofactor pathway.
Abstract
Supplying the appropriate amount of correctly folded α/β-tubulin heterodimers is critical for microtubule dynamics. Formation of assembly-competent heterodimers is remarkably elaborate at the molecular level, in which the α- and β-tubulins are separately processed in a chaperone-dependent manner. This sequential step is performed by the tubulin-folding cofactor pathway, comprising a specific set of regulatory proteins: cofactors A–E. We identified the fission yeast cofactor: the orthologue of cofactor C, Tbc1. In addition to its roles in tubulin folding, Tbc1 acts as a GAP in regulating Alp41/Arl2, a highly conserved small GTPase. Of interest, the expression of GDP- or GTP-bound Alp41 showed the identical microtubule loss phenotype, suggesting that continuous cycling between these forms is important for its functions. In addition, we found that Alp41 interacts with Alp1D, the orthologue of cofactor D, specifically when in the GDP-bound form. Intriguingly, Alp1D colocalizes with microtubules when in excess, eventually leading to depolymerization, which is sequestered by co-overproducing GDP-bound Alp41. We present a model of the final stages of the tubulin cofactor pathway that includes a dual role for both Tbc1 and Alp1D in opposing regulation of the microtubule.
INTRODUCTION
Microtubules are dynamical protein structures that act as cellular scaffolding within the cell. The structures are involved in a large array of functions, ranging from fundamental activities such as cell polarity, mitosis, and cell division to intricate regulatory processes such as nuclear positioning. Owing to the importance of their cellular functions, study of microtubules is an expanding field of research. Several studies have aimed at deciphering the mechanisms of its various roles in regulation, such as those involving microtubule-associated proteins (MAPs), posttranslational modifications, and dynamical instability. Surprisingly, however, far less is known about the basics of microtubules, in particular tubulin biogenesis.
For proteins to fold into the necessary macromolecular structures, so-called helper proteins termed molecular chaperones assist proteins to achieve their functionally active conformation. Chaperonins are chaperones that depend on ATP hydrolysis. One group of important chaperonins makes up the chaperonin-containing TCP-1 (CCT; or c-cpn) complex, with which both tubulin (Llorca et al., 2000) and actin (Llorca et al., 1999) must interact to reach their native state before proper folding. This CCT complex, in combination with the molecular chaperone prefoldin, is the first step of the tubulin cofactor pathway (Yaffe et al., 1992).
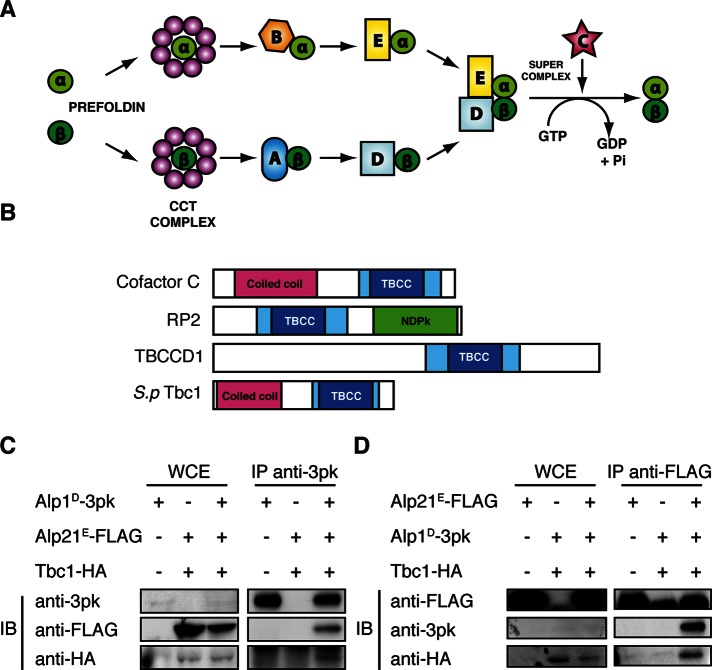
The tubulin cofactor pathway consists of a specific set of chaperones that aid the folding of α- and β-tubulin monomers into a functional heterodimer. The pathway was first established in vitro, in which folding assays showed that there were several proteins required to sufficiently fold tubulin (Gao et al., 1992, 1993, 1994). Since then, a full set of the required cofactor molecules have been identified as cofactors A–E (Tian et al., 1996, 1997, 1999; Lopez-Fanarraga et al., 2001), and a model pathway has been established that intricately assigns these cofactors to efficiently fold a functional heterodimer (schematically shown in Figure 1A). As mentioned earlier, the native α- and β-tubulin monomers are first captured by the CCT complex. After release, the α-tubulin monomer is captured by cofactor B, which then delivers it to cofactor E. In parallel, the β-tubulin monomer is captured by cofactor A, which is then replaced by cofactor D. The two parallel pathways converge at this stage, and with the introduction of cofactor C, GTP hydrolysis occurs, triggering the release of the newly folded α/β-tubulin heterodimer for addition to the plus end of the microtubule.
FIGURE 1:
Fission yeast Tbc1 is a conserved orthologue of tubulin-folding cofactor C. (A) Model of the canonical tubulin-folding pathway. The native α- and β-tubulin monomers are folded into functional heterodimers by a specific set of chaperones, cofactors A–E. (B) The conserved domains of the human TBCC domain containing proteins and fission yeast Tbc1. The TBCC domain is shown in blue, with the dark blue indicating two CARP domains usually found in CAP proteins. The coiled-coil regions shown in magenta suggest a protein–protein interaction domain, and the nucleotide diphosphate kinase (NDPK) shown in green is a phosphocarrier domain. (C, D) Immunoprecipitation of Alp1D-3Pk, Alp21E-3FLAG, and Tbc1–3HA. The immunoblot from IP against an anti-Pk antibody and an anti-FLAG antibody is shown in C and D, respectively. In both blots, the first lane shows the single-tagged strain and the second lane the doubly tagged strain. The third lane on the far right is the result from the triply tagged strain. For each IP 5 mg of protein was used, and 30 μg of whole-cell extract was loaded as a control.
After the in vitro studies of the tubulin cofactor pathway, we investigated the fission yeast orthologues of the cofactors to observe their in vivo function. Studies of the pathway in vivo had been limited due to the high degree of essentiality of these proteins, and therefore Schizosaccharomyces pombe was an ideal model organism in which to isolate mutants and functionally characterize the cofactors with regard to microtubules. The orthologues had been identified in a screen for temperature sensitivity, with defects in cell morphology (Hirata et al., 1998; Radcliffe et al., 1998) and mitochondria distribution (Yaffe et al., 1996). Among them, the cofactors were picked up as frequently mutated loci within these polarity mutants. In fission yeast, cofactors A, B, D, and E are Alp31A, Alp11B, Alp1D, and Alp21E, respectively. All of the fission yeast cofactor mutants showed similar bent morphology, with defective microtubules. Interaction studies showed consistency with the in vitro model, with Alp31A and Alp1D able to bind β-tubulin, and Alp11B and Alp21E able to bind α-tubulin. However, in the fission yeast system, there were some differences between the α- and β-tubulin pathways. Alp31A appeared to be nonessential and did not show similar properties to the parallel cofactor Alp11B on the α-tubulin arm of the pathway. Data also suggested that the cofactor D orthologue, Alp1D, might have increased importance in fission yeast. Alp1D was shown to be toxic to the cell when massively overproduced, but multicopy plasmids of Alp1D enabled rescue of the temperature sensitivity of the alp11B and alp21E mutants. This was not the case in the opposite situation, in which the overproduction of Alp11B or Alp21E could not rescue the other cofactor mutants (Hirata et al., 1998). These genetic data suggested that Alp1D was acting downstream of the other cofactors rather than in parallel with Alp21E. This indicated that it was likely that Alp1D was required not only for the β-tubulin pathway, but also for the α-tubulin pathway, suggesting its unique importance compared with the other cofactors.
The fission yeast pathway provided some interesting insights regarding the functional hierarchy of the cofactors. However, to complete the path, a player was missing—the orthologue of cofactor C had not yet been identified. In humans, cofactor C has two related proteins—retinitis pigmentosa 2 (RP2) and TBCCD1. RP2 is an extensively studied protein due to its significance in disease (Schwahn et al., 1998; Chapple et al., 2001). It acts as a GTPase-activating protein (GAP) for small GTPase Arl3 (Veltel et al., 2008a, b) and has functions related to ciliogenesis in photoreceptors, as well as other cilia-related functions in kidney (Schrick et al., 2006), Golgi apparatus (Evans et al., 2010), and G protein trafficking (Schwarz et al., 2012). The other related protein, TBCCD1, was shown in Chlamydomonas reinhardtii to have functions in the centriole, with mutants showing defective spindle orientation and positioning (Feldman and Marshall, 2009). Data from mammalian cells also show that human TBCCD1 is an integral centrosome component, with depletion resulting in defects in primary cilia and Golgi apparatus anchoring (Goncalves et al., 2010). Both of these related proteins are involved in microtubule function, showing their close functional conservation with cofactor C.
Moreover, cofactor C, RP2, and TBCCD1 have in common a conserved tubulin-folding cofactor C (TBCC) domain that is responsible for GAP activity. Structural studies showed that, in addition to the TBCC domain, these three proteins also share a CARP domain, which is found in cyclase-associated proteins (Bartolini, 2002), as shown in Figure 1B. In the fission yeast system, no orthologues of any of these proteins have been reported, making the identification and characterization of the cofactor C orthologue all the more important.
In this study, we identified the fission yeast orthologue of cofactor C—Tbc1—and isolated temperature-sensitive mutants to observe its in vivo function in fission yeast cells. It acts as a GAP for the small GTPase Alp41, which, when in its GDP-bound form, interacts with another cofactor in the pathway—the cofactor D orthologue Alp1D. This led us to propose a model of the final stages of the tubulin cofactor pathway in which a dual role of both Tbc1 and Alp1D is involved in opposing regulation of the microtubule.
RESULTS
Fission yeast Tbc1 is able to interact with the other cofactors in the pathway
We first identified the potential cofactor C orthologue from a screen carried out for the α-tubulin mutant atb2-983 (Asakawa et al., 2006). This gene was found as a multicopy suppressor of this mutant and named tbc1+ (tubulin-folding cofactor C 1). We confirmed that it was a true candidate by various means. First, we carried out a sequence alignment between human, Saccharomyces cerevisiae, and Drosophila melanogaster orthologues and RP2 (Schwahn et al., 1998) and TBCCD1 (Goncalves et al., 2010). This showed that the TBCC domain was moderately conserved in fission yeast Tbc1, along with the arginine finger (Ahmadian et al., 1997) and the surrounding regions crucial for GAP activity (Figure 1B and Supplemental Figure S1).
After sequence analysis, we confirmed its identity by observing its interaction partners. We assessed whether Tbc1 was able to interact with the other cofactors; as previously described, at the final stages of the cofactor pathway, cofactor D bound to β-tubulin and cofactor E bound to α-tubulin come together to form a supercomplex with cofactor C (Tian et al., 1996). We would therefore expect the orthologue of cofactor C to interact with these other two cofactors. We tagged the fission yeast cofactors D and E—Alp1D and Alp21E, respectively—at the C-terminus with 3pk and 3FLAG. We similarly tagged Tbc1 with 3x hemagglutinin (3HA). As shown in Figure 1C, both Alp21E-3FLAG and Tbc1-3HA coimmunoprecipitated with Alp1D-3pk. The reciprocal immunoprecipitation (IP) showed identical results (Figure 1D). From this we conclude that Tbc1 is the orthologue of cofactor C and is able to form a supercomplex with Alp1D and Alp21E.
Temperature-sensitive mutants of tbc1 show loss of microtubules
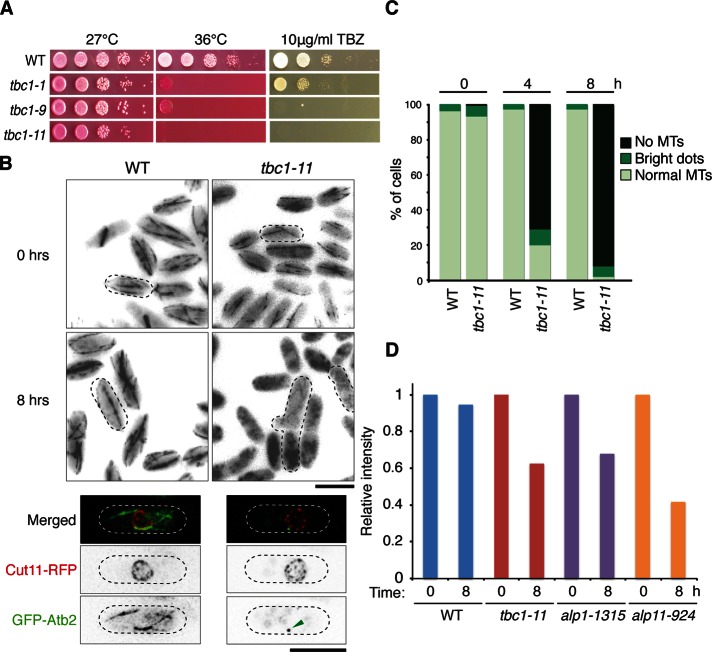
Tbc1 is essential in fission yeast (Kim et al., 2010). Therefore, to observe the defects caused by the loss of function of this gene, we isolated temperature-sensitive (ts) mutants of tbc1 by PCR-based random mutagenesis. From the mutants isolated, Figure 2A shows a selection of alleles spotted on plates containing phloxine B and 10 μg/ml thiabendazole (TBZ). Phloxine B stains dead cells red, and TBZ is a microtubule drug often used for observing mutants with microtubule defects. All three alleles shown are growth sensitive to both temperature and TBZ. From these, we used the mutant strain tbc1-11 for further analysis, as it showed stronger sensitivity to both conditions. When sequenced, this mutant was found to have a point mutation at F174S, which lay in the conserved GAP (CARP-TBCC) domain of Tbc1 (Supplemental Figure S1).
FIGURE 2:
Isolation and characterization of tbc1 ts mutants. (A) Serial dilution spot test of tbc1 ts mutants. Mutants were spotted onto rich media plates containing phloxine B or TBZ and incubated at either 27 or 36°C. Three representative alleles are shown. (B) Microtubule morphology. Wild-type and tbc1-11 mutant cells containing GFP-labeled Atb2 (α2-tubulin) were incubated at 36°C for 8 h and observed for microtubule defects. Bar, 10 μm. Bottom, single-cell images of wild-type and tbc1-11 mutant cells with an additional Cut11-RFP label (a marker for the nuclear envelope). The arrowhead indicates a bright GFP dot localizing in the nuclear periphery. Bar, 5 μm. (C) Quantification of microtubule phenotypes. Three hundred cells were counted and characterized into three classes: no microtubules, cells with no microtubules but with a bright GFP dot, and normal microtubule structures. Percentage of cells showing these phenotypes are shown for cells after 4 and 8 h of incubation at the restrictive temperature. (D) The α-tubulin concentration in various cofactor mutants. Protein was extracted from each wild-type/mutant strain after 0 and 8 h of incubation at 36°C. An immunoblot was performed using 30 μg of protein for each strain, and the intensity of each band was calculated using Photoshop CS3 normalized against a control band.
The TBZ sensitivity indicated that this mutant had microtubule defects. To further observe the microtubules in living cells, we used the mutant strain with α2-tubulin Atb2 tagged with green fluorescent protein (GFP) at the N-terminus under the SV40 promoter (Pardo and Nurse, 2005; Bratman and Chang, 2007). After 8 h of incubation at the restrictive temperature, the mutant showed severe morphological and microtubule defects (Figure 2B). Wild-type fission yeast cells showed an even rod shape with smooth and continuous microtubule structures that extended along the full length of the cell. In contrast, the mutant cells showed a characteristic bent or branched morphology, and almost all of the cells had no remaining microtubules. On quantification of cells (n = 300), after 4 h of incubation 71% of cells showed complete loss of microtubules and a further 9% showed no filamentous microtubule structures but only bright GFP dots within the cell (Figure 2C). The percentage of cells showing these phenotypes increased further after 8 h, with 92% of cells showing no microtubules and 6% showing the bright GFP dots. These bright dots of GFP often were located in close proximity to the nucleus, as seen localizing in the vicinity of the Cut11–red fluorescent protein (RFP) signal (Figure 2B, bottom), which marks the nuclear envelope (West et al., 1998). This suggests that the microtubules attempted to nucleate and failed to do so, resulting in local accumulation of tubulin. The remaining cells showed some residual microtubules, although they were broken or shorter than those in wild-type cells. Of interest, in the mutant cells, the microtubules already appeared more fragile even at the permissive temperature (Figure 2B). This loss of microtubules not only was consistent with other cofactor mutants such as Alp1D (Hirata et al., 1998), but it also confirmed Tbc1’s involvement in the tubulin cofactor pathway, as the loss of its function led to a clear decrease in microtubules.
The requirement of Tbc1 for microtubule biogenesis was further confirmed by assessing α-tubulin concentration within the mutant cells. Wild-type and mutant strains were grown up to 8 h at the restrictive temperature, protein extracted, and then blotted with an anti–α-tubulin antibody. The ts mutants for alp1 (alp1-1315) and alp11 (alp11-924) were used as positive controls that displayed reduced levels of α-tubulin (Hirata et al., 1998; Radcliffe and Toda, 2000). As expected, the tbc1-11 mutant showed a decrease in α-tubulin levels (Figure 2D), together with the other cofactor mutants. This further indicated that tubulin regulation had been disrupted in this mutant, possibly through a similar mechanism by which the other cofactor mutants showed defective microtubules.
Temperature-sensitive mutants of alp41 show a similar loss of microtubules
Having confirmed that Tbc1 is the orthologue of cofactor C and plays a role in the tubulin-folding pathway, we investigated the possible functions of Tbc1 in microtubule regulation. The conservation of the functional domains seen in human cofactor C and its related protein RP2 led us to hypothesize that Tbc1 might also have GAP function. We therefore searched for a G protein for which Tbc1 might be regulating as a GAP and identified Alp41, the orthologue of human Arl2, as a possible candidate. Previous studies of Alp41 found it to be essential, with lethal deletion mutants showing the characteristic branched and bent morphology similar to that of the other cofactor mutants (Radcliffe et al., 2000). However, the gene was not extensively studied, as no conditional mutants were available.
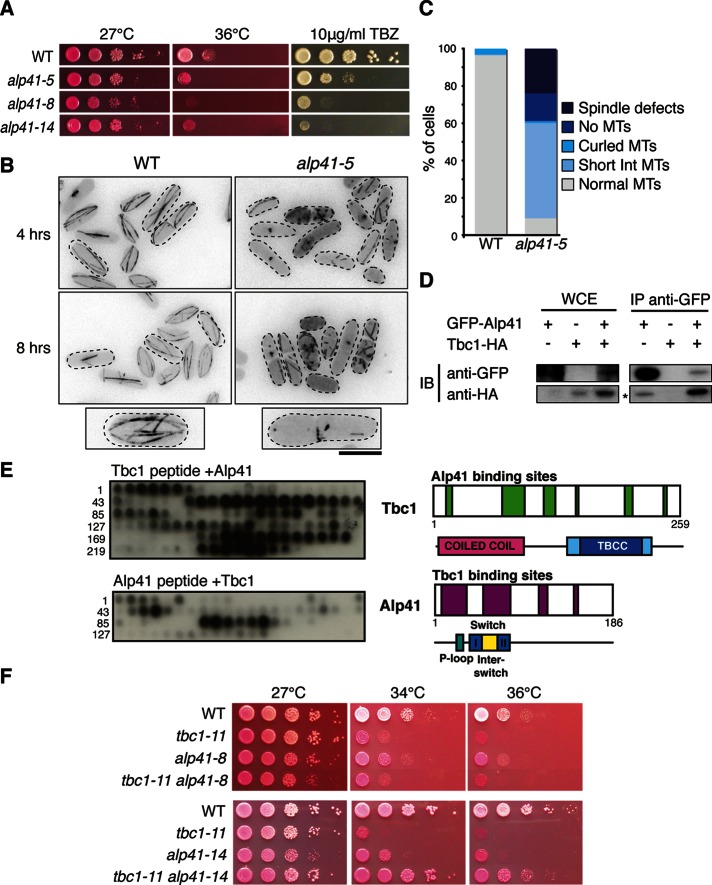
Therefore we isolated temperature-sensitive mutants of alp41 to observe the microtubule defects caused by loss of function. Figure 3A shows selected alleles spotted onto plates containing phloxine B or TBZ. Each allele showed sensitivity to both temperature and TBZ, albeit some slightly weaker than with the tbc1 mutants (Figure 2A). To observe the microtubule phenotype, we endogenously tagged α2-tubulin Atb2 with GFP for visualization. After 8 h of incubation at the restrictive temperature, we observed a similar morphological phenotype to that of the tbc1 mutants, with the cells appearing bent, short, or branched (Figure 3B). The microtubules within these mutant cells were either short or fragmented, with some cells showing a complete loss of microtubules. When quantified, the majority of the cells had defective microtubules of varied degrees. The phenotypes were put into four classes as shown in Figure 3C. The interphase microtubules were classified as being short or curled, some cells showed a complete loss of microtubules, and spindle defects were observed in some cells.
FIGURE 3:
Isolation and characterization of alp41 ts mutants and the physical interaction between Alp41 and Tbc1. (A) Serial dilution spot test of alp41 ts mutants. Mutants were spotted onto rich media plates containing phloxine B or TBZ and incubated at either 27 or 36°C. Three representative alleles are shown. (B) Microtubule morphology. Wild-type and alp41-5 mutant cells with GFP-labeled Atb2 were incubated at 36°C for 4 and 8 h and observed for microtubule defects. Bottom, representative cells from wild-type and mutant strains. Bar, 5 μm. (C) Quantification of microtubule phenotypes. Three hundred cells were quantified and characterized into five microtubule phenotypes: “spindle defects” showing malformed/dysfunctional mitotic spindle, no microtubules, curled microtubules, short interphase microtubules, or normal microtubules. (D) Immunoprecipitation of Alp41 and Tbc1. IPs were performed using a doubly tagged strain expressing GFP-Alp41 and Tbc1-3HA. First lane, control strain expressing GFP-Alp41 only; second lane, Tbc1-3HA only; third lane, doubly tagged strain. A 30-μg amount of whole-cell extract was loaded. Asterisk, unspecific band detected by antibody. (E) Peptide array assay showing interaction of Tbc1 and Alp41. Recombinant Alp41 and Tbc1 proteins (hexahistidine tagged) were purified from E. coli and incubated with synthesized peptides. Left, immunoblots against an anti-histidine antibody; right, schematic of the interacting regions with the known domains. Numbers on the blot mark the amino acid number at the beginning of the row. Top blot, cellulose membrane spotted with Tbc1 peptide incubated with Alp41 protein. Bottom blot, cellulose membrane spotted with Alp41 peptide incubated with Tbc1 protein. (F) Mutual rescue of tbc1 and alp41 ts mutants. Double mutants of tbc1 and alp41 were spotted onto rich media plates with added phloxine B. Plates were incubated at 27, 34, and 36°C. Two alleles of alp41 (-8 and -14) were crossed with tbc1-11.
Not only were these defective microtubule phenotypes consistent with the other cofactor mutants, but these results also showed that Alp41 must play a crucial role in microtubule regulation and formation. Previously fission yeast Alp41 and human Arl2 had not been named as “cofactors” in the pathway per se, but these data suggest that they might be involved very closely in its regulation. On sequence analysis, we found that all the mutants isolated had mutations within the N-terminus (Supplemental Figure S2), where the conserved domains such as the switch regions of the small GTPases lie (Kühnel et al., 2006). This strongly suggests that the mutations might affect the activity of Alp41 as a GTPase.
Fission yeast Tbc1 is a GAP for Alp41
Owing to structural conservation and their possible roles in the cofactor pathway, we hypothesized that Tbc1 and Alp41 are involved in the regulation of one another. To confirm this, we examined whether the two proteins were able to interact biochemically and genetically. Figure 3D shows the interaction of Tbc1 and Alp41 by immunoprecipitation using strains with 3HA-tagged Tbc1 and GFP-tagged Alp41. Tbc1-3HA coimmunoprecipitated with GFP-Alp41.
We investigated this interaction further by using an in vitro peptide array assay to determine whether the interaction was direct. The advantage of this system is that it uses purified recombinant proteins, which, in contrast to immunoprecipitation, means that there are no other associated proteins such as MAPs or tubulins that could allow them to interact indirectly. The peptide arrays that were synthesized covered the entire region of Tbc1 or Alp41. Twenty amino acid residues per spot were spotted onto a cellulose membrane, with two residue increments between the spots. Hexahistidine-tagged Alp41 and Tbc1, expressed in and purified from bacteria (see Materials and Methods), were incubated with the membrane and the interactions detected using an anti-histidine antibody by immunoblotting. As shown in Figure 3E, there is clear interaction between Tbc1 and Alp41. This also allowed us to determine the domains that are important for binding between the two proteins, which is shown schematically on the right of the figure. In the case of Alp41, the largest interacting region lies in the N-terminal of the protein, where the P-loop and the two switch regions lie. These are the regions important for GTPase activity, which would further strengthen our results showing that Tbc1 is a GAP for Alp41. The result for Tbc1 showed that the interaction regions span the entire Tbc1, with some regions lying at the end of the coiled-coil domain, as well as in the GAP/TBCC domain. Taken together, these results confirm that Tbc1 and Alp41 directly interact.
We also observed the genetic interaction of the two proteins by crossing the tbc1 and alp41 mutants to observe any rescue of temperature sensitivity. According to our hypothesis, Tbc1 is a GAP for Alp41. Therefore, if Tbc1 loses its GAP activity as in the tbc1-11 mutant, Alp41 will no longer be converted from its active, GTP-bound state to the inactive, GDP-bound state. This interferes with the balance between the GTP- and GDP-bound states of Alp41 in the cell. Therefore, by crossing the two mutants, the balance may be restored and consequently the cells may show a wild-type phenotype. Of interest, we found that when tbc1-11 was crossed with alp41-8, there was no rescue, and there was still strong temperature sensitivity, as shown in Figure 3F, top. However, as Figure 3F, bottom, shows, when tbc1-11 was crossed with alp41-14, there was a clear rescue. The precise reason for this allele specificity remains to be determined. Nonetheless, biochemical as well as genetic results indicate that Tbc1 is a GAP for the Alp41 GTPase.
Overproduction of GTP or GDP form of Alp41 is toxic to the cell
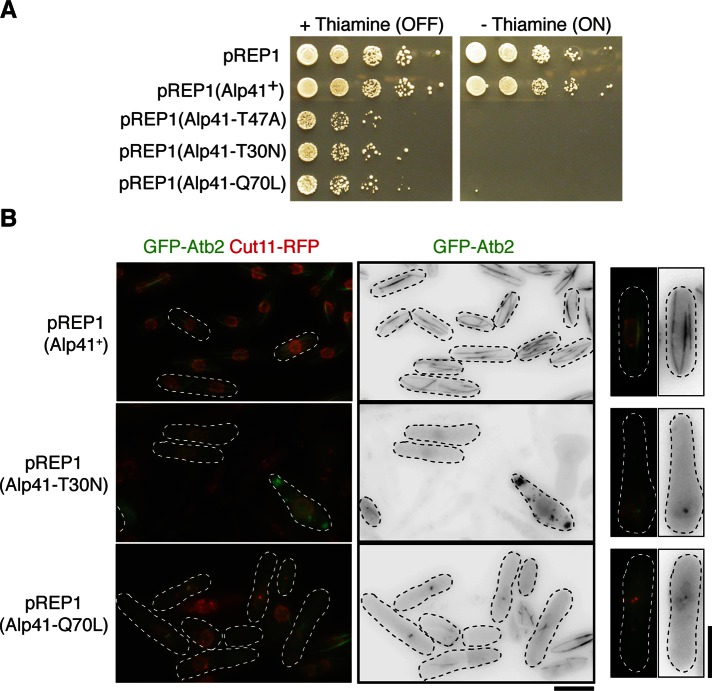
Having established that Tbc1 is a GAP for Alp41, we investigated further the effect of defective Alp41 regulation. Alp41’s identity as a small GTPase implies that it can be in either of two states: the active, GTP-bound or inactive, GDP-bound form. To determine the possible effect of the imbalance in the Alp41 nucleotide state, we introduced mutations into alp41 that would render it either constitutively GTP or GDP bound. These mutations, compiled from a range of literature on the related G proteins (Bhamidipati et al., 2000; Kühnel et al., 2006), included Alp41-Q70L, a GTPase-deficient form of Alp41 that renders it GTP bound; Alp41-T30N, which loses the ability to bind with GTP and is therefore constitutively GDP bound; and another GDP-bound form, Alp41-T47A, which induces a defect in the effector loop that inhibits the switching from GDP to GTP. The mutant forms of Alp41 were overproduced in wild-type cells under the inducible nmt1 promoter (Maundrell, 1990). These cells were spotted onto plates with or without thiamine, and the growth was observed. Figure 4A shows that under minus-thiamine conditions when the promoter is on, the cells overproducing either form of the nucleotide states of Alp41 showed severe toxicity, resulting in complete lack of growth. The overproduction of the wild-type Alp41, however, had no effect on cell growth.
FIGURE 4:
Phenotypic consequences from overproduction of constitutively GTP/GDP-bound forms of Alp41. (A) Spot test. Plasmids expressing constitutively GTP- or GDP-bound form of Alp41 were transformed into wild-type cells and transformants spotted onto minimal media plates with or without thiamine (promoter is OFF when thiamine is added, ON when without, as indicated). (B) Microtubule morphology. Strains containing GFP-tagged Atb2 and Cut11-RFP were transformed with the foregoing plasmids and incubated in minus-thiamine conditions (promoter ON) for 32 h. Far right, representative cell from each condition. Bars, 5 μm.
To examine whether the toxicity was caused by microtubule defects, we transformed the plasmids into strains with GFP-tagged Atb2 and monomeric RFP–tagged Cut11. After incubation in media lacking thiamine (the nmt1 promoter derepressed), there was a striking morphological phenotype and severe loss of microtubules in both the GTP-bound and GDP-bound states (Figure 4B). The phenotype was similar to that of the tbc1 and alp41 ts mutants, for which there were no microtubules at all, short, fragmented microtubules, or bright spots of GFP dots that accumulated around the nuclear envelope. Of interest and rather unexpectedly, identical phenotypes were seen in both GTP- and GDP-bound Alp41 overproduction strains, even though they are in completely opposite nucleotide states. These microtubule and morphological phenotypes were not seen when the plasmid containing the wild-type Alp41 was overexpressed within these cells, suggesting that in wild-type conditions the balance between the two nucleotide forms must be maintained to prevent toxicity. This highlights the importance of the continuous cycling between the two states of Alp41.
Alp41 GTPase interacts with cofactor D, Alp1D, only when bound to GDP
To determine the exact function of the Alp41 GTPase, we searched for its potential interactors. Previous studies in higher eukaryotes showed that the human orthologue Arl2 was able to interact with tubulin-folding cofactor D (Bhamidipati et al., 2000). We picked up Alp1D to be a potential interactor of Alp41 and carried out immunoprecipitation experiments to establish whether this interaction could be seen in fission yeast. Strains were constructed in which Alp1D and Alp41 were tagged with 3pk and GFP, respectively, and immunoprecipitation performed with beads coupled with GFP antibody. Surprisingly, we found that there was no interaction in either the control or the double-tagged strain (Figure 5A). The reciprocal immunoprecipitation showed identical results (unpublished data).
FIGURE 5:
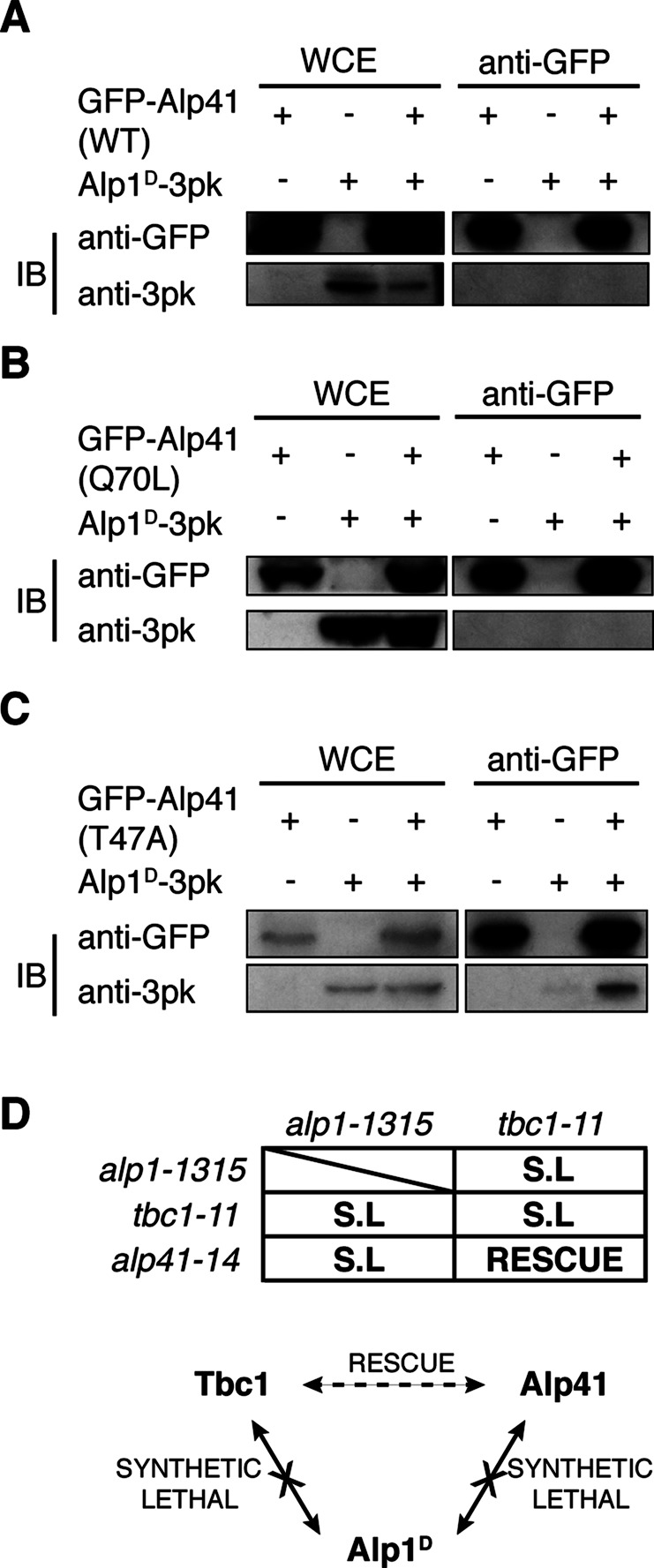
Alp41 interacts with Alp1D only when in its GDP-bound form. (A–C) Immunoprecipitation. IPs were performed using a doubly tagged strain expressing the mutated form of Alp41 tagged with GFP and Alp1D-3Pk. First lane, control strain expressing GFP-Alp41 only; second lane, Alp1D-3Pk only; third lane, the doubly tagged strain. A 30-μg amount of whole-cell extract was loaded as a control. (A) Wild-type Alp41, (B) GTP-bound Alp41-Q70L, and (C) GDP-bound Alp41-T47A. (D) Genetic interactions between Tbc1, Alp41, and Alp1D. Mutants of each gene were crossed and growth/lethality observed. SL, synthetic lethality.
Next we investigated whether the nucleotide state could determine the interaction. Strains with GFP-tagged, GTP/GDP-bound Alp41 were constructed (as in Figure 4) and the immunoprecipitation repeated. The result showed a striking difference between the immunoprecipitations carried out in the two states. Alp1D did not coimmunoprecipitate with the GTP-bound form, Alp41-Q70L (Figure 5B), but instead a clear interaction was seen for the GDP-bound form, Alp41-T47A (Figure 5C). This was consistent with the data from higher eukaryotes suggesting that the Alp41 orthologue, Arl2, interacted more readily with cofactor D in its GDP-bound state (Bhamidipati et al., 2000). Our data indicate that Alp1D is an interacting partner of Alp41 and that the interaction is specific to when Alp41 is in the inactive, GDP-bound form.
To confirm this regulation of Alp41 and Alp1D, we crossed the mutants of these two genes, respectively alp41-14 and alp1-1315 (Hirata et al., 1998), and examined them for possible genetic interaction. We found that the double mutants were all synthetically lethal (Figure 5D). Because the rescue between the tbc1 mutant tbc1-11 and alp41 mutants was allele specific, we crossed several alleles of alp41, but they all showed synthetic lethality with alp1-1315. Similarly, tbc1-11 also showed synthetic lethality with alp1-1315. Because alp1 and tbc1 are both cofactors in the same tubulin-folding pathway, this synthetic lethality was unsurprising, but its lethality with alp41 suggests possible relevance to its function in the pathway.
Alp1D overproduction causes disruption of microtubules
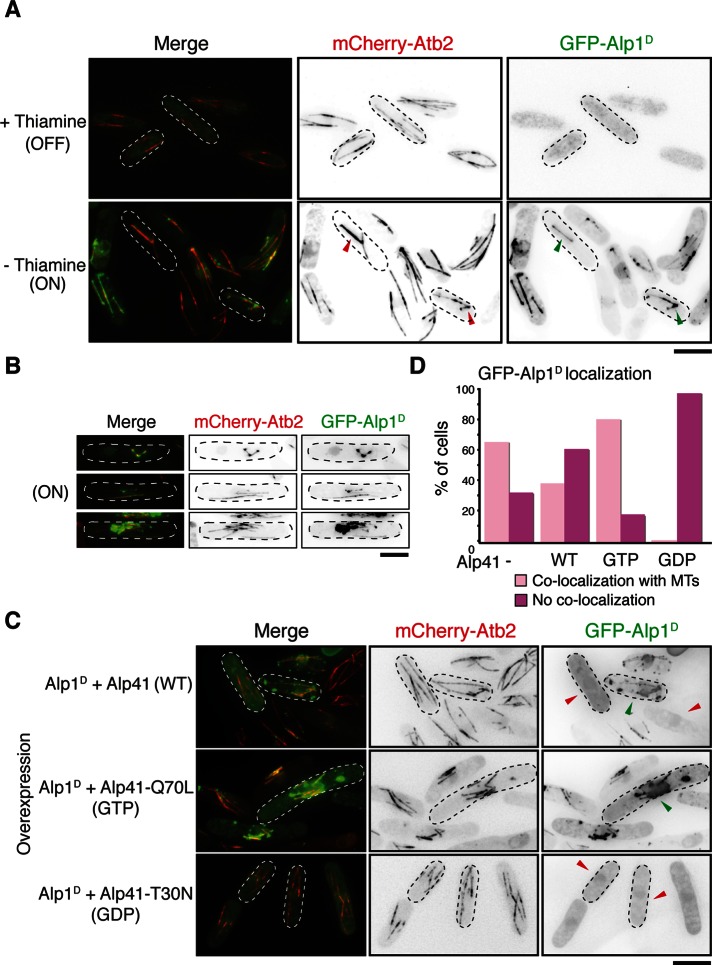
Alp41, when in its GDP-bound form, interacts with Alp1D, which is known to function in the tubulin-folding pathway. However, its interaction with specifically the GDP-bound form of Alp41 suggests that it might have additional roles. Previous studies in fission yeast showed that Alp1D seems to function most downstream of the other cofactors (Radcliffe et al., 1999). Considering that Alp1D interacts with Alp41 only when in the GDP-bound form, the interaction must be significant for the function of Alp1D. Therefore, to elucidate this function, we overproduced Alp1D within the cell to observe any effects. For this purpose, we overproduced GFP-tagged Alp1D in a strain containing Atb2 tagged with mCherry. As shown in Figure 6A, the cells were drastically elongated or misshapen, branched, or bent. In addition, there were severe microtubule defects: microtubules appeared short or fragmented or were completely lost. Most striking of all, the GFP-Alp1D signal strongly colocalized with the residual microtubules. The Alp1D signal appeared to strongly colocalize with, or “paint,” the whole of the microtubule structures at all cell cycle stages. Previous studies suggested that Alp1D behaves like a MAP biochemically (Hirata et al., 1998), but here we show for the first time such an extent of colocalization within live cells. On longer incubation, the Alp1D localization further accumulated on the microtubules, and eventually the microtubules became shorter and disassembled (Figure 6B and Supplemental Figure S3). We believe that this microtubule localization is specific to Alp1D, as no such localization has been reported for overproduced Tbc1 or Alp21 (Matsuyama et al., 2006). Regarding Alp41, we observed localization of the various forms of Alp41 (wild type, GTP bound, and GDP bound) when overproduced and found that each form exhibited cytosolic localization (Supplemental Figure S4). Of interest, we saw no microtubule localization when we expressed Alp1D under its endogenous promoter in any conditions (Supplemental Figure S5), suggesting that this disruptive function of Alp1D occurs only when it is present in excess. We conclude from this result that localization of overproduced Alp1D leads to disassembly of the microtubule.
FIGURE 6:
Overproduction of Alp1D causes disruption of microtubule structure, which can be rescued with co-overproduction of GDP-bound Alp41. (A) Cells transformed with an overexpression plasmid of Alp1D were cultured under minus-thiamine conditions for 20 h. Note that cells have not yet undergone complete microtubule disruption, by which colocalization of Alp1D with the remaining microtubules is visible (marked with arrowheads). (B) Single-cell images in which there is clear localization of the GFP-Alp1D on the microtubules. (C) Cells were double transformed with overexpression plasmids of Alp1D and a wild-type, GTP-bound (Q70L), or GDP-bound (T30N) form of Alp41. Green arrowheads indicate cells that show GFP-Alp1D colocalizing with microtubules. Red arrowheads indicate cells where this colocalization is not seen. (D) Quantification of GFP-Alp1D localization. Two hundred cells showing GFP-Alp1D overproduction were counted and categorized into cells showing colocalization of GFP-Alp1D signal on microtubules or cells showing no colocalization. Bars, 5 μm.
GDP-bound Alp41 sequesters overproduced Alp1D from microtubules and rescues abnormal microtubule structures
The results in Figure 6, A and B, suggest that microtubule localization of overproduced Alp1D causes disruption of microtubules. In addition, the interaction data in Figure 5 suggest that Alp41, when in the GDP-bound form, which is cytoplasmic (Supplemental Figure S4), is capable of binding to Alp1D. Taken these results together, we speculated that GDP-bound Alp41 absorbs the excess Alp1D within the cell. To address this, we co-overproduced Alp1D and one of the forms of Alp41 together to observe any change in localization of Alp1D. Strikingly, we found that when the GDP-bound form of Alp41 (T30N) was co-overproduced with Alp1D, there was no longer localization of Alp1D on the microtubules. Instead there was cytosolic localization, and the microtubules remained intact (Figure 6C). In contrast, when GTP-bound Alp41 was overproduced together with Alp1D, there was no change in localization, with Alp1D strongly painting the microtubules and causing disruption. The wild-type form of Alp41 showed intermediate results, with some cells showing localization on the microtubule and some not. This was clarified further after quantification, by which 200 cells showing Alp1D overproduction were categorized into those showing clear localization of Alp1D on the microtubules and those that did not (Figure 6D). Whereas 81% of cells co-overproducing GTP-bound Alp41 showed colocalization with microtubules, only 2% were seen in the cells co-overproducing GDP-bound Alp41. The overproduction of Alp1D only showed 66% of cells showing colocalization, which was reduced to 39% when wild-type Alp41 was co-overproduced. This clear dissociation of Alp1D from the microtubules when GDP-bound Alp41 is co-overproduced strengthens our proposal that GDP-bound Alp41, produced by the GAP activity of Tbc1, is able to suppress the toxic microtubule binding of Alp1D by physical association.
DISCUSSION
In this study we investigated the fundamentals of microtubule biogenesis by resolving the complete fission yeast tubulin-folding pathway. We identified Tbc1, the fission yeast orthologue of tubulin-folding cofactor C and found it to be a GAP for the small GTPase Alp41, the orthologue of human Arl2. We further showed that the cycling of Alp41 between its GTP- and GDP-bound forms was crucial for its functions in microtubule regulation. Finally, we found that only the GDP-bound form of Alp41 was able to bind Alp1D, the fission yeast orthologue of cofactor D. Analysis of Alp1D led us to the hypothesis that there are two roles of Alp1D: to fold heterodimers in the tubulin-folding pathway and to depolymerize microtubules, which is antagonized by its interaction with GDP-bound Alp41.
Tbc1 regulates Alp41 and the critical cycling between its GDP- and GTP-bound states
The fission yeast orthologues of the tubulin-folding cofactors were previously identified and studied (Hirata et al., 1998; Radcliffe et al., 1998, 1999, 2000; Radcliffe and Toda, 2000; Fedyanina et al., 2006), but cofactor C remained unidentified. Having identified fission yeast Tbc1, we found that the ts mutants showed loss of microtubules. In addition to its function in supercomplex formation, we speculated that it may have other roles as a GAP and found Alp41 to be the corresponding small GTPase. The alp41-mutant phenotype strongly suggested that it was essential for microtubule formation. As a small GTPase, Alp41 must exist in both the GTP-bound form and the GDP-bound form. To see the possible effects of restricting this behavior, we overproduced both the GTP-bound and GDP-bound form of Alp41 in fission yeast. Rather unexpectedly, we saw that when either of the two forms of Alp41 was overproduced in wild-type cells, they showed identical microtubule phenotypes. Given that the two forms are in complete opposite states, this was surprising and led us to propose that it is not the nucleotide state of Alp41 that is crucial in its role as a microtubule regulator but instead the continuous cycling between the two forms.
There are other examples of GTPases, such as the Ran GTPase, in which it is crucial for the balance of the GTP- and GDP-bound states to be maintained by continuous cycling (Matynia et al., 1996). Similar to this case, it could be the cycling of the GTP and GDP states of Alp41 that is crucial for at least its microtubule regulation and not which state in which it exists. This could also explain the similar phenotypes seen in the tbc1 mutant, in which Alp41 would be fixed in the GTP form, as its GAP function would be compromised. We conclude that the cycling of Alp41 between its GTP- and GDP-bound states is essential for its function in microtubule regulation. These results bring to light important questions: what is the effector of Alp41, and is there a guanine nucleotide exchange factor (GEF)? Given that the human orthologue Arl2 has several interactors, including BART (Sharer and Kahn, 1999; Zhang et al., 2009), PDEδ (Hanzal-Bayer et al., 2002), and HRG4 (Kobayashi et al., 2003), we envision that fission yeast Alp41, when bound to GTP, interacts with an effector molecule. The identification of this effector, and possibly a GEF, would be the next direction of the present study.
The dual roles of Alp1D in microtubule regulation
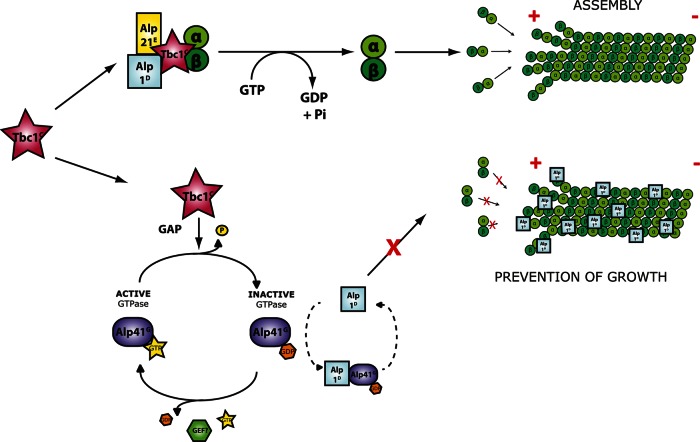
We showed that the cofactor D orthologue Alp1D interacts with Alp41 when in the GDP-bound state. This is consistent with previous data that showed that the human cofactor D preferentially bound to Arl2 when in its GDP form (Bhamidipati et al., 2000). To further understand the physiological significance of this interaction, we overproduced Alp1D in wild-type cells. A strong toxicity was seen, with cells showing severe defects in cell morphology with either short, fragmented microtubules or none at all. Remarkably, Alp1D colocalized with, or “painted,” the remaining microtubules, which eventually disappeared upon prolonged Alp1D overproduction. Not only did we confirm the previous result that Alp1D is a MAP (Hirata et al., 1998) in living cells, but it also indicated that the “painting” of microtubules by Alp1D caused this depolymerization. This proposes a dual role of Alp1D: processing tubulin for heterodimer formation, and acting in a contrary way in depolymerizing microtubules. It is of note that vertebrate cofactor D also displays microtubule-depolymerizing activities upon overproduction (Martin et al., 2000; Tian et al., 2010). Of interest, we found that this otherwise toxic behavior of excess Alp1D could be suppressed by the co-overproduction of GDP-bound Alp41. We propose that one of the functions of GDP-bound Alp41, produced by the GAP activity of Tbc1, is to bind with and absorb the excess Alp1D within the cell. Our model, given next, describes the situations that can lead to either polymerization or depolymerization of microtubules (Figure 7).
FIGURE 7:
Model of the final stages of the tubulin-folding pathway. The dual functions of Tbc1 are shown in two parallel pathways. Top pathway shows the last steps of the tubulin-folding pathway as shown in Figure 1A after Tbc1 forms a supercomplex together with Alp1D and Alp21E. Lower pathway shows the alternative function of Tbc1 as a GAP for Alp41. The subsequently GDP-bound Alp41 is now able to bind to Alp1D, thus preventing excess Alp1D from localizing on the microtubules, where they would otherwise cause dissociation of microtubules. The lack of interaction of Alp41 and Alp1D strongly suggests that the two arms of the pathway are independent of one another.
The interplay among Tbc1, Alp41, and Alp1D in microtubule biogenesis
After the processing of the α- and β-tubulin monomers by the CCT complex, they are captured by Alp11B and Alp31A, respectively, which are then replaced by Alp21E and Alp1D. In the final stage of the pathway, the α-tubulin–bound Alp21E and β-tubulin–bound Alp1D converge to form a supercomplex with Tbc1. Here two parallel situations can arise. If Tbc1 is readily available to form the supercomplex, the forward process ensues as normal, with GTP hydrolysis being triggered by the combined actions of the cofactors, releasing the newly folded α/β heterodimer for incorporation into the plus end of the microtubule (Figure 7, top path). However, in the alternative situation in which Tbc1 readily acts as a GAP for Alp41, there is excess Alp1D within the cell as supercomplex formation is restrained. This excess Alp1D, assuming that the β-tubulin monomer pool is steady, would be free and go on to “paint” the preexisting microtubules, preventing further polymerization and leading to eventual depolymerization of the microtubules (Figure 7, lower path, far right). To maintain and regulate microtubule dynamics, this must be inhibited. This is done by the action of Alp41, which is now GDP bound by the action of Tbc1 and able to bind and absorb the free Alp1D within the cell (Figure 7, lower path, center).
This model indicates that there are several competitive events within these final stages of the pathway. First there is competition regarding the protein to which Tbc1 is bound: Alp41 or the supercomplex. The ability of Alp1D to bind to Alp41 only when it is GDP bound shows that when in the GTP-bound form, Tbc1 is readily able to bind Alp41. There is also competition for Alp1D binding: if Tbc1 is not acting as a GAP for Alp41, it is able to form the supercomplex, which in turn absorbs the excess Alp1D within the cell. If there is excess, it is absorbed by GDP-bound Alp41. We believe that the two pathways are parallel and that Tbc1 does not perform both its functions as part of the same supercomplex. The lack of interaction between Alp41 and Alp1D (Figure 5A) supports this notion, together with previous results (Bhamidipati et al., 2000; Shern et al., 2003). This intricate balance of Alp1D, Alp41, and Tbc1 binding suggests a homeostatic mechanism to maintain the tubulin heterodimer pool and microtubule dynamics within the cell.
Physiological implications for microtubule dynamics
In this study we presented an in vivo model of the coordinated functions of Tbc1, Alp1D, and the Alp41 GTPase. The complex interplay among these proteins leads to an inhibition of the toxic effects caused by the cofactors themselves. The cofactors’ primary roles were initially believed to be the production of functional heterodimers from α- and β-tubulin monomers. However, studies gave evidence of their potentially destructive nature (Bhamidipati et al., 2000; Martin et al., 2000; Kortazar et al., 2006, 2007), indicating that there may be two conserved faces to these cofactors and how they contribute to microtubule regulation. Not only have the cofactors been shown to destabilize and disrupt the native tubulin heterodimer in vitro (Bhamidipati et al., 2000), but in addition in vivo the cofactors have been shown to destroy microtubule structures (Martin et al., 2000; Cunningham and Kahn, 2008; Fanarraga et al., 2010; Tian et al., 2010) (although there are slight discrepancies in the extent of disruption, depending on the species). We found that within the fission yeast system, the overproduction of the cofactor D orthologue Alp1D led to the complete depolymerization of microtubules within the cell. Although we did not assess the biochemical properties of Alp1D in comparison to its bovine or human counterparts, this localization pattern of Alp1D as a MAP shows its capability for microtubule depolymerization. Its regulation appears to be through a stoichiometric competition among Tbc1, Alp41, and Alp1D and thus highlights this dual role of cofactors and their possible functions within the cell. Evidence shows that together with the actions of cofactor B, cofactor E is also capable of depolymerizing microtubules (Feierbach et al., 1999; Radcliffe et al., 1999; Kortazar et al., 2006, 2007; Carranza et al., 2013), which is consistent with our work suggesting that there is potential that the other cofactors have roles aside from heterodimer folding.
Several reports link cofactor function to human disease. It has been suggested that cofactor C is involved in tumor cell regulation (Hage-Sleiman et al., 2010, 2011), in which an increased sensitivity to the S phase–targeting drug gemcitabine and microtubule-targeting drugs has been observed. Cofactor D has emerged as a possible centriolar protein, with its silencing leading to defective ciliogenesis and multiciliated cells (Fanarraga et al., 2010). These emerging roles of cofactors, which can lead to more widespread consequences, may increase the significance of these studies and shine new light on the tubulin-folding pathway.
MATERIALS AND METHODS
Fission yeast culture and genetics
All yeast strains used are described in Supplemental Table S1. Fission yeast cells were grown and maintained in standard conditions as described (Moreno et al., 1991). For temperature-sensitive mutants, cells were cultured in rich YE5S medium until mid–log phase at 27°C and subsequently shifted to the restrictive temperature of 36°C for further culture until observation. For overexpression using nmt1 series plasmids, cells were grown in Edinburgh Minimal Medium (EMM) with required amino acid supplements plus 15 μM thiamine overnight. Thiamine was then washed out by filtration pump and cultured in the same EMM minus thiamine conditions for ∼16–32 h as necessary.
Spot tests were carried out by first equalizing cells to a concentration of 2 × 107 cells/ml. Subsequently 10-fold dilutions were spotted onto plates. Plates were then incubated at the necessary temperature between 27 and 36°C. Plates contained rich YE5S medium, rich medium with added TBZ (10 μg/ml) or phloxine B (7.5 μg/ml), or EMM with added supplements with or without 15 μM thiamine.
Yeast strain construction
Temperature-sensitive mutants were isolated by random mutagenesis. tbc1 or alp41 was tagged with an HphR selection cassette and the fragment randomly mutagenized by error-prone PCR amplification (10× dGTP compared with other dNTPs) using Vent DNA polymerase (New England BioLabs, Ipswich, MA). Products were ethanol precipitated and transformed into a wild-type strain. Hph-resistant clones were selected by replica plating and temperature sensitivity assessed by further replica plating onto YE5S plates containing phloxine B. We screened 9000 and 10,000 clones for tbc1 and alp41, respectively. We isolated 11 mutants for tbc1 and 14 for alp41. These were backcrossed to ensure proper integration of the Hph-resistant marker by checking the cosegregation of the drug marker with temperature sensitivity and removing any off-site mutations. Mutants were sequenced (mutation sites of representative alleles are shown in Supplemental Figures S1 and S2).
The C- or N-terminus–tagged proteins were constructed by chromosomal integration of PCR-amplified fragments (Bahler et al., 1998; Sato et al., 2005). Proper tagging was confirmed by immunoblot and colony PCR.
Fluorescence microscopy
Live cells were imaged in an imaging chamber containing 800 μl of agarose sealed with a glass coverslip. The DeltaVision-SoftWoRx system (Olympus, Tokyo, Japan, and Applied Precision, Issaquah, WA) was used for obtaining all fluorescence microscopy data. The images were acquired using an Olympus IX71 wide-field inverted epifluorescence microscope in a temperature-controlled environmental chamber. The images were taken with the Olympus UPlan Sapo 63× or 100×/numerical aperture 1.4, oil immersion objective and captured with a CoolSNAP-HQ digital charge-coupled device camera (Photometrics, Tucson, AZ). Fifteen sections were taken along the z-axis at 0.2-μm intervals. The images were further processed using deconvolution and then projected into a single projection. Analysis, including counts and measurements, was done using MetaMorph (Molecular Devices, Sunnyvale, CA), and further processing and analysis used Excel (Microsoft, Redmond, WA) or Photoshop CS3 (Adobe, San Jose, CA).
Immunoprecipitation
Protein was extracted by mechanical shaking of cells in protein extraction buffer (50 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid [HEPES], 50 mM NaF, 50 mM Na-β-glycerophosphate, 5 mM ethylene glycol tetraacetic acid, 5 mM EDTA, and 0.2% Triton X-100 with added 1× protease inhibitor cocktail [PIC; Sigma-Aldrich, St. Louis, MO]) and 1 mM phenylmethylsulfonyl fluoride (PMSF) with acid-washed glass beads in a FastPrep FP120 apparatus (4 × 30 s; power, 6.0). Extract was cleared of debris by centrifugation for 1 min and subsequently 5 min at 13,000 × g, and the concentration was determined by Bradford assay (Bio-Rad, Hercules, CA). A range of 3–10 mg of protein was used per immunoprecipitation experiment, in which the extract was incubated with slow rotation for 1 h, 30 min with protein A Dynabeads coated with antibodies against Pk (AbD Serotec, Raleigh, NC), GFP (Invitrogen, Carlsbad, CA), FLAG (Sigma-Aldrich), or HA (BAbCO, Richmond, CA). The beads were then washed in wash buffer (50 mM Tris-HCl, pH 7.4, 1 mM EDTA, 150 mM NaCl, 0.05% NP-40, 10% glycerol, 1 mM dithiothreitol, and 1.5 mM p-nitrophenyl phosphate with added 1× PIC and 0.1 mM PMSF) and boiled in Laemmli buffer for 5 min. Protein extract was loaded and resolved on denaturing 4–12% gradient gels (Bio-Rad) and transferred by wet transfer onto polyvinylidene fluoride membrane. Membranes were blocked with 10% skimmed milk and blotted with anti-Pk (AbD Serotec), anti-GFP (Roche, Indianapolis, IN), anti-FLAG (Sigma-Aldrich), or anti-HA (BAbCO) antibodies at a dilution of 1:1000 in ImmunoShot solution 1 (2B Scientific, Upper Heyford, United Kingdom). After washing, blots were incubated in anti-mouse horseradish peroxidase–conjugated secondary antibody (GE Healthcare, Piscataway, NJ) in ImmunoShot Solution 2 (2B Scientific) at a dilution of 1:2000. The ECL chemiluminescence kit (GE Healthcare) was used for detection.
Expression and purification of recombinant proteins
cDNAs encoding Alp41 and Tbc1 were cloned into the pET28a vector containing the hexahistidine tag. The protein was expressed in Escherichia coli (BL20) by incubation at 19°C overnight upon 1 mM isopropyl-β-d-thiogalactoside induction in 2 l of culture. The cells were pelleted by centrifugation at 8000 × g for 30 min at 4°C, and lysis buffer (50 mM HEPES, 250 mM NaCl, 10% glycerol, 5 mM β-mercaptoethanol, 0.5% Triton X-100, 1 mM phenylmethylsulfonyl fluoride [PMSF], 1× PIC, 40 mM imidazole) was added. The sample was mixed vigorously and incubated at 4°C on ice for a further 1 h. After incubation the cells were sonicated for 7 s with 30-s intervals seven times or until the sample was no longer viscous. The supernatant was collected by centrifugation at 14,800 × g for 20 min at 4°C and added to a Ni-nitriloacetic acid agarose column (Qiagen, Valencia, CA). The column was rotated for 1 h and the flowthrough collected. After four washes with wash buffer (50 mM HEPES, 250 mM NaCl, 10% glycerol, 5 mM β-mercaptoethanol, 0.5% Triton X-100, 1 mM PMSF, 1× PIC, 60 mM imidazole), elution buffer (50 mM HEPES, 250 mM NaCl, 10% glycerol, 5 mM β-mercaptoethanol, 0.5% Triton X-100, 1 mM PMSF, 1× PIC, 250 mM Imidazole) was passed through the column and 10-ml fractions collected. Laemmli buffer was added and the sample boiled before loading onto an SDS–PAGE gel.
Peptide array assay
Peptides were synthesized and spotted onto a cellulose membrane at two–amino acid residue increments, 20 residues per spot. The membrane was activated by 1 h of incubation at room temperature in solution with 50% ethanol and 10% glacial acetic acid. It was then briefly washed, and 20 μg of protein in phosphate-buffered saline/Tween was added to incubate on a shaker overnight at 4°C. The membrane was then washed once and then blocked. After blocking, the membrane was handled for immunoblot as described for the immunoprecipitation using the anti-histidine antibody (Qiagen).
Supplementary Material
Acknowledgments
We thank Fred Chang and Paul Nurse for strains used in this study. We thank Kazuhiko Asakawa and Vicky Anderson for conducting the initial part of this project. We are grateful to Céline Bouchoux for technical advice on protein purification and the Cancer Research UK Peptide Synthesis Unit for preparation of the peptide arrays. This work was supported by Cancer Research UK (T.T.).
Abbreviations used:
- GAP
GTPase-activating protein
- IP
immunoprecipitation
- MAP
microtubule-associated protein
- PIC
protease inhibitor cocktail
- RP2
retinitis pigmentosa 2
- TBCC
tubulin-folding cofactor C
- TBZ
thiabendazole
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E12-11-0792) on April 10, 2013.
REFERENCES
- Ahmadian MR, Stege P, Scheffzek K, Wittinghofer A. Confirmation of the arginine-finger hypothesis for the GAP-stimulated GTP-hydrolysis reaction of Ras. Nat Struct Biol. 1997;4:686–689. doi: 10.1038/nsb0997-686. [DOI] [PubMed] [Google Scholar]
- Asakawa K, Kume K, Kanai M, Goshima T, Miyahara K, Dhut S, Tee WW, Hirata D, Toda T. The V260I mutation in fission yeast α-tubulin Atb2 affects microtubule dynamics and EB1-Mal3 localization and activates the Bub1 branch of the spindle checkpoint. Mol Biol Cell. 2006;17:1421–1435. doi: 10.1091/mbc.E05-08-0802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahler J, Wu JQ, Longtine MS, Shah NG, McKenzie A 3rd, Steever AB, Wach A, Philippsen P, Pringle JR. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast. 1998;14:943–951. doi: 10.1002/(SICI)1097-0061(199807)14:10<943::AID-YEA292>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- Bartolini F. Functional overlap between retinitis pigmentosa 2 protein and the tubulin-specific chaperone cofactor C. J Biol Chem. 2002;277:14629–14634. doi: 10.1074/jbc.M200128200. [DOI] [PubMed] [Google Scholar]
- Bhamidipati A, Lewis SA, Cowan NJ. ADP ribosylation factor-like protein 2 (Arl2) regulates the interaction of tubulin-folding cofactor D with native tubulin. J Cell Biol. 2000;149:1087–1096. doi: 10.1083/jcb.149.5.1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bratman SV, Chang F. Stabilization of overlapping microtubules by fission yeast CLASP. Dev Cell. 2007;13:812–827. doi: 10.1016/j.devcel.2007.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carranza G, et al. Autoinhibition of TBCB regulates EB1-mediated microtubule dynamics. Cell Mol Life Sci. 2013;70:357–371. doi: 10.1007/s00018-012-1114-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapple JP, Grayson C, Hardcastle AJ, Saliba RS, van der Spuy J, Cheetham ME. Unfolding retinal dystrophies: a role for molecular chaperones. Trends Mol Med. 2001;7:414–421. doi: 10.1016/s1471-4914(01)02103-7. [DOI] [PubMed] [Google Scholar]
- Cunningham LA, Kahn RA. Cofactor D functions as a centrosomal protein and is required for the recruitment of the γ-tubulin ring complex at centrosomes and organization of the mitotic spindle. J Biol Chem. 2008;283:7155–7165. doi: 10.1074/jbc.M706753200. [DOI] [PubMed] [Google Scholar]
- Evans RJ, Schwarz N, Nagel-Wolfrum K, Wolfrum U, Hardcastle AJ, Cheetham ME. The retinitis pigmentosa protein RP2 links pericentriolar vesicle transport between the Golgi and the primary cilium. Hum Mol Genet. 2010;19:1358–1367. doi: 10.1093/hmg/ddq012. [DOI] [PubMed] [Google Scholar]
- Fanarraga ML, Bellido J, Jaen C, Villegas JC, Zabala JC. TBCD links centriologenesis, spindle microtubule dynamics, and midbody abscission in human cells. PLoS One. 2010;5:e8846. doi: 10.1371/journal.pone.0008846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedyanina OS, Mardanov PV, Tokareva EM, McIntosh JR, Grishchuk EL. Chromosome segregation in fission yeast with mutations in the tubulin folding cofactor D. Curr Genet. 2006;50:281–294. doi: 10.1007/s00294-006-0095-9. [DOI] [PubMed] [Google Scholar]
- Feierbach B, Nogales E, Downing KH, Stearns T. Alf1p, a CLIP-170 domain-containing protein, is functionally and physically associated with α-tubulin. J Cell Biol. 1999;144:113–124. doi: 10.1083/jcb.144.1.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldman JL, Marshall WF. ASQ2 encodes a TBCC-like protein required for mother-daughter centriole linkage and mitotic spindle orientation. Curr Biol. 2009;19:1238–1243. doi: 10.1016/j.cub.2009.05.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y, Melki R, Walden PD, Lewis SA, Ampe C, Rommelaere H, Vandekerckhove J, Cowan NJ. A novel cochaperonin that modulates the ATPase activity of cytoplasmic chaperonin. J Cell Biol. 1994;125:989–996. doi: 10.1083/jcb.125.5.989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y, Thomas JO, Chow RL, Lee GH, Cowan NJ. A cytoplasmic chaperonin that catalyzes β-actin folding. Cell. 1992;69:1043–1050. doi: 10.1016/0092-8674(92)90622-j. [DOI] [PubMed] [Google Scholar]
- Gao Y, Vainberg IE, Chow RL, Cowan NJ. Two cofactors and cytoplasmic chaperonin are required for the folding of α- and β-tubulin. Mol Cell Biol. 1993;13:2478–2485. doi: 10.1128/mcb.13.4.2478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goncalves J, Nolasco S, Nascimento R, Lopez Fanarraga M, Zabala JC, Soares H. TBCCD1, a new centrosomal protein, is required for centrosome and Golgi apparatus positioning. EMBO Rep. 2010;11:194–200. doi: 10.1038/embor.2010.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hage-Sleiman R, Herveau S, Matera E-L, Laurier J-F, Dumontet C. Tubulin binding cofactor C (TBCC) suppresses tumor growth and enhances chemosensitivity in human breast cancer cells. BMC Cancer. 2010;10:135. doi: 10.1186/1471-2407-10-135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hage-Sleiman R, Herveau S, Matera EL, Laurier JF, Dumontet C. Silencing of tubulin binding cofactor C modifies microtubule dynamics and cell cycle distribution and enhances sensitivity to gemcitabine in breast cancer cells. Mol Cancer Ther. 2011;10:303–312. doi: 10.1158/1535-7163.MCT-10-0568. [DOI] [PubMed] [Google Scholar]
- Hanzal-Bayer M, Renault L, Roversi P, Wittinghofer A, Hillig RC. The complex of Arl2-GTP and PDEδ: from structure to function. EMBO J. 2002;21:2095–2106. doi: 10.1093/emboj/21.9.2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirata D, Masuda H, Eddison M, Toda T. Essential role of tubulin-folding cofactor D in microtubule assembly and its association with microtubules in fission yeast. EMBO J. 1998;17:658–666. doi: 10.1093/emboj/17.3.658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DU, et al. Analysis of a genome-wide set of gene deletions in the fission yeast Schizosaccharomyces pombe. Nat Biotechnol. 2010;28:617–623. doi: 10.1038/nbt.1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi A, Kubota S, Mori N, McLaren MJ, Inana G. Photoreceptor synaptic protein HRG4 (UNC119) interacts with ARL2 via a putative conserved domain. FEBS Lett. 2003;534:26–32. doi: 10.1016/s0014-5793(02)03766-3. [DOI] [PubMed] [Google Scholar]
- Kortazar D, Carranza G, Bellido J, Villegas JC, Fanarraga ML, Zabala JC. Native tubulin-folding cofactor E purified from baculovirus-infected Sf9 cells dissociates tubulin dimers. Protein Expr Purif. 2006;49:196–202. doi: 10.1016/j.pep.2006.03.005. [DOI] [PubMed] [Google Scholar]
- Kortazar D, Fanarraga ML, Carranza G, Bellido J, Villegas JC, Avila J, Zabala JC. Role of cofactors B (TBCB) and E (TBCE) in tubulin heterodimer dissociation. Exp Cell Res. 2007;313:425–436. doi: 10.1016/j.yexcr.2006.09.002. [DOI] [PubMed] [Google Scholar]
- Kühnel K, Veltel S, Schlichting I, Wittinghofer A. Crystal structure of the human retinitis pigmentosa 2 protein and its interaction with Arl3. Structure. 2006;14:367–378. doi: 10.1016/j.str.2005.11.008. [DOI] [PubMed] [Google Scholar]
- Llorca O, Martin-Benito J, Ritco-Vonsovici M, Grantham J, Hynes GM, Willison KR, Carrascosa JL, Valpuesta JM. Eukaryotic chaperonin CCT stabilizes actin and tubulin folding intermediates in open quasi-native conformations. EMBO J. 2000;19:5971–5979. doi: 10.1093/emboj/19.22.5971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llorca O, McCormack EA, Hynes G, Grantham J, Cordell J, Carrascosa JL, Willison KR, Fernandez JJ, Valpuesta JM. Eukaryotic type II chaperonin CCT interacts with actin through specific subunits. Nature. 1999;402:693–696. doi: 10.1038/45294. [DOI] [PubMed] [Google Scholar]
- Lopez-Fanarraga M, Avila J, Guasch A, Coll M, Zabala JC. Review: postchaperonin tubulin folding cofactors and their role in microtubule dynamics. J Struct Biol. 2001;135:219–229. doi: 10.1006/jsbi.2001.4386. [DOI] [PubMed] [Google Scholar]
- Martin L, Fanarraga ML, Aloria K, Zabala JC. Tubulin folding cofactor D is a microtubule destabilizing protein. FEBS Lett. 2000;470:93–95. doi: 10.1016/s0014-5793(00)01293-x. [DOI] [PubMed] [Google Scholar]
- Matsuyama A, et al. ORFeome cloning and global analysis of protein localization in the fission yeast Schizosaccharomyces pombe. Nat Biotechnol. 2006;24:841–847. doi: 10.1038/nbt1222. [DOI] [PubMed] [Google Scholar]
- Matynia A, Dimitrov K, Mueller U, He X, Sazer S. Perturbations in the spi1p GTPase cycle of Schizosaccharomyces pombe through its GTPase-activating protein and guanine nucleotide exchange factor components result in similar phenotypic consequences. Mol Cell Biol. 1996;16:6352–6362. doi: 10.1128/mcb.16.11.6352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maundrell K. nmt1 of fission yeast. A highly transcribed gene completely repressed by thiamine. J Biol Chem. 1990;265:10857–10864. [PubMed] [Google Scholar]
- Moreno S, Klar A, Nurse P. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 1991;194:795–823. doi: 10.1016/0076-6879(91)94059-l. [DOI] [PubMed] [Google Scholar]
- Pardo M, Nurse P. The nuclear rim protein Amo1 is required for proper microtubule cytoskeleton organisation in fission yeast. J Cell Sci. 2005;118:1705–1714. doi: 10.1242/jcs.02305. [DOI] [PubMed] [Google Scholar]
- Radcliffe P, Hirata D, Childs D, Vardy L, Toda T. Identification of novel temperature-sensitive lethal alleles in essential β-tubulin and nonessential α2-tubulin genes as fission yeast polarity mutants. Mol Biol Cell. 1998;9:1757–1771. doi: 10.1091/mbc.9.7.1757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radcliffe PA, Hirata D, Vardy L, Toda T. Functional dissection and hierarchy of tubulin-folding cofactor homologues in fission yeast. Mol Biol Cell. 1999;10:2987–3001. doi: 10.1091/mbc.10.9.2987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radcliffe PA, Toda T. Characterisation of fission yeast alp11 mutants defines three functional domains within tubulin-folding cofactor B. Mol Gen Genet. 2000;263:752–760. doi: 10.1007/s004380000252. [DOI] [PubMed] [Google Scholar]
- Radcliffe PA, Vardy L, Toda T. A conserved small GTP-binding protein Alp41 is essential for the cofactor-dependent biogenesis of microtubules in fission yeast. FEBS Lett. 2000;468:84–88. doi: 10.1016/s0014-5793(00)01202-3. [DOI] [PubMed] [Google Scholar]
- Sato M, Dhut S, Toda T. New drug-resistant cassettes for gene disruption and epitope tagging in Schizosaccharomyces pombe. Yeast. 2005;22:583–591. doi: 10.1002/yea.1233. [DOI] [PubMed] [Google Scholar]
- Schrick JJ, Vogel P, Abuin A, Hampton B, Rice DS. ADP-ribosylation factor-like 3 is involved in kidney and photoreceptor development. Am J Pathol. 2006;168:1288–1298. doi: 10.2353/ajpath.2006.050941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwahn U, et al. Positional cloning of the gene for X-linked retinitis pigmentosa 2. Nat Genet. 1998;19:327–332. doi: 10.1038/1214. [DOI] [PubMed] [Google Scholar]
- Schwarz N, Hardcastle AJ, Cheetham ME. Arl3 and RP2 mediated assembly and traffic of membrane associated cilia proteins. Vision Res. 2012;75:2–4. doi: 10.1016/j.visres.2012.07.016. [DOI] [PubMed] [Google Scholar]
- Sharer JD, Kahn RA. The ARF-like 2 (ARL2)-binding protein, BART. Purification, cloning, and initial characterization. J Biol Chem. 1999;274:27553–27561. doi: 10.1074/jbc.274.39.27553. [DOI] [PubMed] [Google Scholar]
- Shern JF, Sharer JD, Pallas DC, Bartolini F, Cowan NJ, Reed MS, Pohl J, Kahn RA. Cytosolic Arl2 is complexed with cofactor D and protein phosphatase 2A. J Biol Chem. 2003;278:40829–40836. doi: 10.1074/jbc.M308678200. [DOI] [PubMed] [Google Scholar]
- Tian G, Bhamidipati A, Cowan NJ, Lewis SA. Tubulin folding cofactors as GTPase-activating proteins. GTP hydrolysis and the assembly of the α/β-tubulin heterodimer. J Biol Chem. 1999;274:24054–24058. doi: 10.1074/jbc.274.34.24054. [DOI] [PubMed] [Google Scholar]
- Tian G, Huang Y, Rommelaere H, Vandekerckhove J, Ampe C, Cowan NJ. Pathway leading to correctly folded β-tubulin. Cell. 1996;86:287–296. doi: 10.1016/s0092-8674(00)80100-2. [DOI] [PubMed] [Google Scholar]
- Tian G, Lewis SA, Feierbach B, Stearns T, Rommelaere H, Ampe C, Cowan NJ. Tubulin subunits exist in an activated conformational state generated and maintained by protein cofactors. J Cell Biol. 1997;138:821–832. doi: 10.1083/jcb.138.4.821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian G, Thomas S, Cowan NJ. Effect of TBCD and its regulatory interactor Arl2 on tubulin and microtubule integrity. Cytoskeleton (Hoboken) 2010;67:706–714. doi: 10.1002/cm.20480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veltel S, Gasper R, Eisenacher E, Wittinghofer A. The retinitis pigmentosa 2 gene product is a GTPase-activating protein for Arf-like 3. Nat Struct Mol Biol. 2008a;15:373–380. doi: 10.1038/nsmb.1396. [DOI] [PubMed] [Google Scholar]
- Veltel S, Kravchenko A, Ismail S, Wittinghofer A. Specificity of Arl2/Arl3 signaling is mediated by a ternary Arl3-effector-GAP complex. FEBS Lett. 2008b;582:2501–2507. doi: 10.1016/j.febslet.2008.05.053. [DOI] [PubMed] [Google Scholar]
- West RR, Vaisberg EV, Ding R, Nurse P, McIntosh JR. cut11+: a gene required for cell cycle-dependent spindle pole body anchoring in the nuclear envelope and bipolar spindle formation in Schizosaccharomyces pombe. Mol Biol Cell. 1998;9:2839–2855. doi: 10.1091/mbc.9.10.2839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaffe MB, Farr GW, Miklos D, Horwich AL, Sternlicht ML, Sternlicht H. TCP1 complex is a molecular chaperone in tubulin biogenesis. Nature. 1992;358:245–248. doi: 10.1038/358245a0. [DOI] [PubMed] [Google Scholar]
- Yaffe MP, Harata D, Verde F, Eddison M, Toda T, Nurse P. Microtubules mediate mitochondrial distribution in fission yeast. Proc Natl Acad Sci USA. 1996;93:11664–11668. doi: 10.1073/pnas.93.21.11664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T, Li S, Zhang Y, Zhong C, Lai Z, Ding J. Crystal structure of the ARL2-GTP-BART complex reveals a novel recognition and binding mode of small GTPase with effector. Structure. 2009;17:602–610. doi: 10.1016/j.str.2009.01.014. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.