Abstract
NALCN is an intriguing, orphan ion channel among the 4x6TM family of related voltage-gated cation channels, sharing a common architecture of four homologous domains consisting of six transmembrane helices, separated by three cytoplasmic linkers and delimited by N and C-terminal ends. NALCN is one of the shortest 4x6TM family members, lacking much of the variation that provides the diverse palate of gating features, and tissue specific adaptations of sodium and calcium channels. NALCN’s most distinctive feature is that that it possesses a highly adaptable pore with a calcium-like EEEE selectivity filter in radially symmetrical animals and a more sodium-like EEKE or EKEE selectivity filter in bilaterally symmetrical animals including vertebrates. Two lineages of animals evolved alternative calcium-like EEEE and sodium-like EEKE / EKEE pores, spliced to regulate NALCN functions in differing cellular environments, such as muscle (heart and skeletal) and secretory tissue (brain and glands), respectively. A highly adaptable pore in an otherwise conserved ion channel in the 4x6TM channel family is not consistent with a role for NALCN in directly gating a significant ion conductance that can be either sodium ions or calcium ions. NALCN was proposed to be an expressible Gd3+-sensitive, NMDG+-impermeant, non-selective and ohmic leak conductance in HEK-293T cells, but we were unable to distinguish these reported currents from leaky patch currents (ILP) in control HEK-293T cells. We suggest that NALCN functions as a sensor for the much larger UNC80/UNC79 complex, in a manner consistent with the coupling mechanism known for other weakly or non-conducting 4x6TM channel sensor proteins such as Nax or Cav1.1. We propose that NALCN serves as a variable sensor that responds to calcium or sodium ion flux, depending on whether the total cellular current density is generated more from calcium-selective or sodium-selective channels.
Keywords: molecular evolution, sodium leak conductance channel, sodium channels, calcium channels, ion selectivity
A Brief History of NALCN Research
Hermann J. Muller, in the 1930s, came across the mutant “narrow abdomen” or “na” allele of what has been dubbed NALCN or “sodium leak channel, non-selective1 while cataloguing phials of fruit fly progeny derived from males subject to relatively high doses of radiation crossed with virgin females.2 Six decades later, Howard A. Nash rediscovered Muller’s narrow abdomen phenotype in mutagenized flies that were especially sluggish in climbing up the side of the fly phial in response to halothane anaesthetic.3 The hypersensitive fly mutants mapped to an unusual four repeat channel, whose rat homolog had been cloned by Dr. Edward Perez-Reyes’ lab at the University of Virginia, a year earlier in 1999.4 NALCN has been labeled also as “Voltage-Independent Sodium Channel 2.1” (NaVI2.1)5 and “Voltage-gated channel-like protein 1” (VGCNL1) in mammals,6 “Unknown- or U-type” (Dmα1U) in Drosophila7 and nematode cation channel (nca-1/nca-2) in C. elegans.8 NALCN (NA Leak Channel) received its official designation when one research group reported its function as a non-selective, sodium leak conductance channel, after countless others tried similar experiments for more than a decade.1 The features of the reported sodium leak currents are equally present in native HEK-293T cells without expressing a NALCN gene and its associated UNC79 or UNC80 subunits or SRC kinase.9 The reported ohmic, linear current is identical to a leak patch current from an imperfect whole cell patch clamp seal1 that can be corrected for by online P/N leak subtraction.10 The inward sodium current of a leaky patch has NALCN’s reported characteristic of disappearing when NMDG+ replaces external sodium ions and blockade of NALCN with 10 uM Gd3+9. Ten uM Gd3+ dramatically improves the membrane seals of native HEK-293T and other patched cells, mimicking the apparent block of a linear, non-selective conductance.9 If 10 uM Gd3+ targets a specific linear conductance through a membrane channel protein, it would be hard to distinguish this current from membrane seal changes in a whole cell patch.
NALCN is attractive as a sodium leak conductance because structurally NALCN appears as a cation channel, which is voltage-independent, and has the appearance of a non-selective type pore that is in between sodium and calcium channels. Furthermore, a missing sodium conductance is the simplest explanation for the hyperpolarized plasma membranes in animal mutants that lack NALCN channels such as C. elegans,8,11,12 Drosophila,3,13 snails10 and mammals.1 Many publications now have ascribed 10 uM Gd3+-sensitive sodium leak currents as confirmation of the presence of NALCN, on the assumption that 10 uM Gd3+ is a blocker of a NALCN conductance. We propose an alternative possibility, that NALCN is a weakly conducting or non-conducting channel. We glean clues as to the function of NALCN from the structural and evolutionary history of this intriguing ion channel and also collate all the common observations of NALCN mutants in C. elegans, Drosophila and mice. We come to a conclusion that NALCN might function as a variable calcium and sodium sensor protein.
Structural Features of NALCN, Calcium Channels and Sodium Channels
The origin of 4x6TM channels
The orphan NALCN and the 20 calcium and sodium channel mammalian genes belong to a structurally related superfamily of voltage-gated cation channels with 24 membrane spanning α helices. Each cation channel is a large single polypeptide of ~1700 to ~2900 amino acids of four homologous domains (DI,DII, DIII,DIV) chained together by cytoplasmic linkers, with each domain containing six transmembrane helices (4x6TM) (Fig. 1) that is typified to a subunit of a “Shaker”-type voltage-gated potassium channel gene (1x6TM), which forms tetramers of four different expressed subunits. The close kinship of Domains II and Domains IV, and Domains I and III, is evidence that all members of the 4x6TM family emerge from a likely common single domain ancestor, followed by a period of divergence of the domains and followed by another repeated duplication and period of divergence before speciation into the 21 4x6TM channels consisting of NALCN, sodium and calcium channels.14 4x6TM cation channels are found in photosynthetic green algae,15 and there is also a single calcium transporter in yeast16 but not found in bacteria, suggesting that the 4x6TM family may have arose in basal eukaryotes, perhaps more than once. Single-celled organisms can possess both an L-type channel and a sodium channel,17 but the earliest NALCN is found in multicellular organisms without a tissue level organization like sponges.18
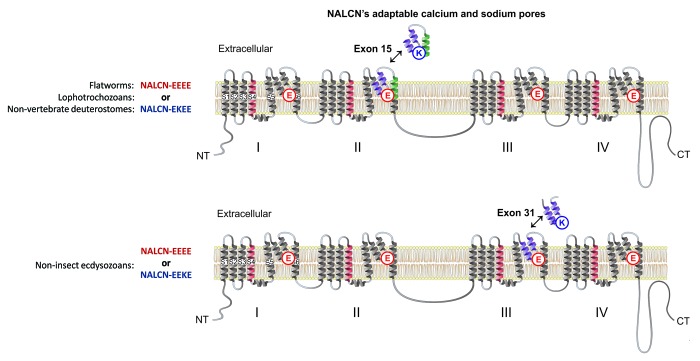
Figure 1. Alternative splicing in NALCN generates selectivity filters that resemble a calcium channel pore (EEEE) or sodium channel pore (EKEE/EEKE), by alternative splicing of either exon 15 or exon 31 in different animal lineages.
The voltage sensor
The 6TM domain contributes to one quadrant of the 4x6TM cation channel pore and consists of two interdependent but separate modules, a voltage-sensor domain (transmembrane segments S1 to S4) and a pore domain (segments S5 and S6). The voltage-sensing S4 helix has repeating positive charges (of Lys+ and Arg+ amino acid residues) and an amphipathic S4-S5 linker that couples the S4 helix to gating (opening/closing) of the pore domain in response to membrane voltage changes.19 All NALCN channels from simple multicellular organisms without a tissue level organization (sponge) to humans have a similar distribution of charges in S4 helices, albeit reduced compared with most sodium and calcium channels. The fewer positive changes in the S4 helix of NALCN resemble non-voltage gated channels such as yeast calcium transporter Cch1p16 and the non-conducting salt sensor, Nax16 (Fig. S1). It should be noted that not all S4 helices of all four domains contribute equally to voltage-sensitive activation gating, and the consequences to gating depends on both the domain and position of the missing charges in the S4 helices. NALCN also has conserved counter-charges in S2 and S3 segments of the voltage-sensor domain, and an amphipathic S4-S5 linker which is critical for the mobility of the voltage-sensor of calcium and sodium channels (Fig. S1). NALCN and salt sensor Nax may have altered voltage-sensitivity, but the level of conservation of cationic charges and countercharges in the voltage sensor domain suggest that NALCN and Nax possess a mobile S4 helix like conventional voltage-gated sodium and calcium channels. Overexpression of a gain-of-function NALCN mutation causes over-excitability, with the mutation located in a hotspot location for gain-of-function alleles associated with altered gating of voltage-gated channels, such as slowing of inactivation.8,11,20 The NALCN gain-of-function mutation is in the proximal I-II linker adjacent to the IS6 transmembrane domain (Domain I, segment 6), where gating modifiers such as β subunits of Cav1 and Cav2 calcium channels are coupled to a rigid post 1S6 helix,21 and where a “gating brake” of Cav3 calcium channels is located.22 The existence of this NALCN mutation within a hotspot region associated with gating behavior, which produces an over-excitability phenotype, leads us to speculate that NALCN may have a mobile voltage-sensor to mediate its functional activation.
Models for pore selectivity
The pore domain is formed by re-entrant cytoplasmic loops spanning the S5 and S6 membrane helices of all four domains, containing a signature selectivity filter that dictates the relative permeability to sodium, calcium or potassium ions. Contributing S6 helices of each domain line the inner pore below the selectivity filter and form a C-terminal helical bundle that meet like an inverted teepee serving as a channel gate to occlude ion passage at their cytoplasmic ends.23
X-ray structures from Roderick Mackinnon elegantly illustrate the concept of potassium channel permeation.24 The potassium selectivity filter is made up of four rigid amino acid backbones that together mimic the hydration shell oxygen atoms surrounding potassium ions in solution.24 Optimized arrangement of surrogate oxygen groups lining the pore selectivity filter provides passage that is only energetically favorable for dehydrated potassium ions to permeate.24 The selectivity of sodium and calcium channels and NALCN is much more ambiguous because it is governed by protruding and flexible, amino acid side chains of selectivity filter residues in the pore, which produce a wider and shorter pore than the potassium channel, according to the X-ray structures of NavAb, a 1x6TM bacterial sodium channel crystalized by Bill Catterall and colleagues.23 Also notably different from the potassium permeation through potassium channels is that sodium ions are expected to permeate the pore of sodium channels in a semi-hydrated23 or hydrated state.25
Selectivity filter residues of calcium and sodium pores
The selectivity filter residues for ion permeation are located at the most constrictive point of the “hourglass” re-entrant (Pore-) P-loops between segments 5 and segments 6 of all four domains (Fig. 1). The three lineages of 4x6TM channels (Cav, Nav and NALCN) appear to follow universal rules, almost without exception, in eight established pore configurations, including Ca2+-selective channels with negatively-charged glutamates and aspartates (EEEE, EDEE, EEDD, DEEA) and Na+-selective channels with a positively-charged lysine (K) in either the 2nd or 3rd domain (EKEE, EEKE, DKEA, DEKA) (Fig. 2). While these four residues play critical roles in governing sodium and calcium selectivity of 4x6TM channels, each re-entrant pore is not equal in its contribution as in symmetrical (homo-multimeric) potassium channels, and furthermore the divergent residues of the re-entrant P-loops outside of the selectivity filter residues also can influence ion selectivity.26,27
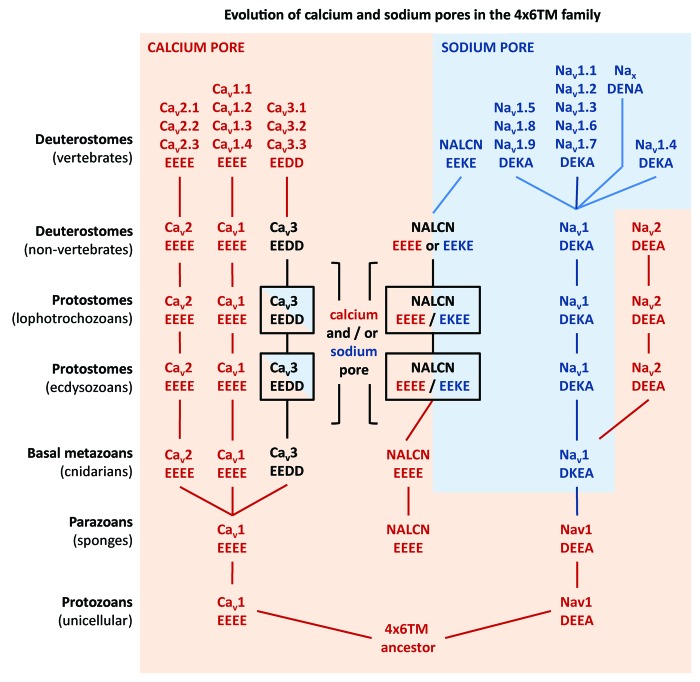
Figure 2. NALCN’s adaptable pore mimics the pores that differentiate the classes of calcium- and sodium-selective channels. NALCN is calcium (EEEE) pore when Cav and Nav channels are calcium-selective and adapts a sodium pore in animals that have acquired a classical Nav1 DEKA pore. Protostome invertebrates have promiscuous calcium (EEEE, DEEA, EEDD) and sodium (DEKA, EEKE, EKEE, EEDD) pores for all 4x6TM classes, the Cav, Nav and NALCN channels. NALCN is limited to a sodium (EEKE) pore in nematodes, insects and vertebrates but is a calcium (EEEE) pore, or can splice in alternative calcium (EEEE) or sodium (EKEE/EEKE) pore in other animals.
The selectivity filter consists of dipole pairs of characteristic negatively charged glutamate residues projecting into pore, providing an EEEE selectivity filter generating the highly calcium-selective filter ubiquitously featured in all Cav1 and Cav2 calcium channels.28,29 Invertebrates express a unique, calcium-selective Nav2 channel with a DEEA pore.30 A more sodium selective channel is generated by replacing the negatively charged glutamate residue with a lysine residue in either Domain II or Domain III.31 The lysine residue governs the highly sodium selective, DEKA configuration of the classical Nav1 sodium channels that generates the upstroke of the action potential,28,31 and what is considered a slightly less sodium-selective, DKEA configuration found in Nav1 channels of basal cnidarians.26 NALCN has indeterminate pores that can both resemble calcium channels or sodium channels, or both.9 Basal NALCN channels in radially symmetrical animals (sponge/Trichoplax/Cnidaria) all have calcium-like EEEE pores.9,18
Variability in calcium and sodium pores of 4x6TM channels
Different invertebrates groups appear to have independently evolved NALCN channels with alternative calcium-like EEEE and sodium-like EKEE or EEKE pores.9 Alternative EEEE and EKEE pores are generated by splicing of mutually-exclusive, exon 15a/exon 15b isoforms in lophotrochozoans (mollusks/annelids) and non-chordate deuterostomes (echinoderms/hemichordates) (Fig. 1).9 In a separate evolutionary event within particular arthropods (myriapods/chelicerates) alternative splicing of mutually-exclusive, exon 31a/exon 31b, generates alterative EEEE and EEKE pores for NALCN (Fig. 1).9 NALCN channels are exclusively the sodium-selective EEKE variant in vertebrates, as well, evidently in typical invertebrate model organisms, like Drosophila (insects) and C. elegans (nematodes).9
Notable NALCN features
NALCN evolved with the advent of multicellularity in animals
A NALCN gene is a required gene in mammals1 and present in every known animal species, beginning with the simplest multicellular organisms, such as sponge and the placozoan Trichoplax.9 Single-celled organisms closely related to animals lack NALCN, such as the ciliate paramecium, which has an EEEE L-type calcium channel that functions in turning movements,32 and protozoans that have a DEEA Nav1 type sodium channel.17 NALCN is in between sodium and calcium channels in overall structure and may have evolved from an ancestral channel resembling the 4x6TM calcium transporter Cch1 found in the sister group to the animals, the fungi.17,18
NALCN is linked to the interconnectivity between cells in multicellular animals
The emergence of NALCN in primitive multicellular organisms without organized tissues and not single-celled organisms may relate to a role for NALCN in the interconnectivity of cells. Interestingly, NALCN in invertebrates at least is functionally linked to a gap junction hemichannel, innexin (UNC7).11
NALCN evolved in a parallel lineage from ancestral multicellular organisms and did not emerge from a specialized branch of sodium channels or calcium channels
NALCN first appears in extant relatives of the most basal multicellular organisms without organs18,25 and thus evolved differently from the highly-specialized vertebrate-specific genes like the non-conducting Nax salt sensor for the subfornical (a brain circumventricular) organ33 and weakly conducting Cav1.1 channel for muscle.34 Nax and Cav1.1 adapted as sensor proteins for functions in vertebrates, losing a primary purpose in ion conduction that was present in their forbearers that were likely both voltage-gated and ion conducting, Nav1 and Cav1 channels in basal organisms respectively. NALCN is highly conserved in the simple multicellular organisms which lack organs, so its fundamental role likely is not associated within specialized intracellular signaling associated with the organ function.
NALCN has resisted gene duplication compared with other members of the 4x6TM family and is uniquely, a single copy gene in most animals
Although NALCN is required in all multicellular organisms, and it is noteworthy that NALCN is mostly a single copy gene in every animal, with exceptions such as sponge (Amphimedon), anthozoan cnidarian (Nematostella) and nematode (Caenorhabditis), which contain two mostly redundant NALCN gene copies.9 While NALCN has largely resisted increases in gene numbers, sodium and calcium channels expanded from five genes in invertebrates to 20 different mammalian genes. Many of the mammalian sodium and calcium channels are selectively expressed for different functions in different cells such as skeletal muscle (Cav1.1, Nav1.4), neurons (Cav2.1 and Cav2.2, Nav1.1, Nav1.2, Nav1.3, Nav1.6, Nav1.7, Nav1.8, and Nav1.9), retina and leukocytes (Cav1.4), heart muscle (Nav1.5), inner hair cells (Cav1.3) and subfornical organ (Nax).35,36
NALCN is a highly conserved ion channel between animal groups
NALCN is more highly conserved across the whole protein from end to end from sponges to humans than are other 4x6TM channels.9 Conversely, sodium and calcium channels have undergone an accelerated rate of evolution, associated with the rising complexity of cellular signaling in higher invertebrates and vertebrates. NALCN’s lack of variability suggests its general functions have little changed from its origins in simple multicellular organisms without significant tissue level organization such as sponge or the placozoan, Trichoplax.
NALCN protein is short and lacking of the typical variation in cytoplasmic regions common in other 4x6TM channels
4x6TM channels differ from the tetrameric 1x6TM channels in having, single N- and C-termini and inter-Domain I-II and II-III cytoplasmic linkers serving as protein interaction platforms adapted for different intracellular environments. NALCN is the second shortest 4x6TM channel in total size (1738 amino acids) because of the shortness of its cytoplasmic N- and C-terminal ends and the I-II and II-III linkers.9 The shortest full-length 4x6TM channel (1682 amino acids) is the highly specialized, non-conducting sodium channel Nax that serves as a sodium sensor in glial cells and couples to the α subunit of the Na+/K+-ATPase, to transduce a signal that regulates salt appetite in the subfornical organ.33 The difference between the largest (LCav3, 2,886 amino acids37) and smallest (Nax33 or NALCN9) 4x6TM channels is an extra ~70% of the whole channel length. The shortness and lack of variability in NALCN channel, specifically in the cytoplasmic regions, may suggest that NALCN has a more limited function like the sensor protein Nax.
Large UNC80/UNC79 proteins coupled to NALCN are unconventional compared with the small accessory proteins of most voltage-gated channels
Most of the 4x6TM channels outside of Cav3 T-type channels are known to be associated with accessory subunits. These subunits in vertebrate calcium channels (α2δ, β, γ)38 and sodium channels (β1, β2, β3, β4)39 are smaller than the conducting channel α subunits. Theses accessory subunits are promiscuous in being able to bind to several different, related cation channel types. They serve to facilitate membrane expression (and can prevent protein degradation40) and affect the biophysical features of the ion conducting channels in a manner similar to 1x6TM potassium channel β subunits.41 NALCN channels require accessory subunits for function too, but these are unlike the sodium and calcium channel accessory subunits. NALCN (1738 aa) is associated with cytoplasmic protein, UNC80, which is twice its size (3258 aa).8,42 UNC80 itself is coupled to UNC79, which is also larger than NALCN (2635 aa).43 NALCN, UNC80 and UNC79 are contiguous proteins, and the mutant phenotypes of these three proteins overlap.8,42-45 NALCN constitutes less than ¼ of the size of the NALCN/UNC80/UNC79 complex. The majority or ¾ of the size of the NALCN complex embodies large interconnected, cytoplasmic proteins UNC/80 and UNC79. A typical sodium channel β subunit in comparison is a miniscule, 1/10th of the size of a voltage-gated sodium channel. It seems more probable that NALCN is the external calcium or sodium ion sensor for the intracellular signaling mechanism performed by the much larger UNC80/UNC79 complex, than to conceive of a model where NALCN functions mostly as an independently conducting and signaling unit.
NALCN coupling to intracellular UNC80/UNC79 complex is reminiscent of known ion sensor proteins, Cav1.1 and Nax
The small NALCN channel intimately coupled to a larger UNC80-UNC79 intracellular protein complex is reminiscent to calcium channel Cav1.1, for example, which is adapted as a specialized voltage-sensing receptor of the T-tubule junction membrane.34 Cav1.1 (1873 aa) is bi-directionally coupled to a much larger intracellular sarcoplasmic reticulum’s ryanodine receptor subunit (~5000 aa).34 The calcium entry through Cav1.1 is minor, and the channel’s primary purpose is signaling voltage changes to the massive web of ryanodine receptor-gated, intracellular calcium, surrounding muscle fibers.34 Of all the 4x6TM channels, NALCN appears to have more in common with the non-conducting Nax salt sensor in its small size, lack of variability and reduced voltage sensor,33 and also similar to the highly specialized and weakly-conducting voltage sensor protein, Cav1.1 whose function is inseparable from the much larger, calcium conducting ryanodine receptor subunit.34
UNC80 and UNC79 have not provided insights into how NALCN works
So far, no functions are ascribed to NALCN’s partners UNC80 and UNC79. UNC80 and UNC79 are highly disordered proteins that likely bear flexible docking sites for multiple protein interactions. Neither UNC80 nor UNC79 are classified within known protein families.
A variable sodium and calcium pore is not consistent with NALCN being an ion conducting channel
The argument for NALCN gating a significant ion conductance on its own (i.e., ionotropic effect) is difficult to reconcile, when it has a variable pore for ion selectivity that can be calcium-selective or sodium-selective in different animal species.9 NALCN cannot be a ubiquitous sodium leak channel, when NALCN likely evolved in basal animals with a calcium-selective EEEE pore. These simple animals do not possess a NALCN pore that resembles a sodium channel EKEE or EEKE.9 While both sodium and calcium ions can depolarize membranes, these are not interchangeable conductances, where the impact of the relatively inert and highly abundant Na+ ion serves mostly an electrogenic role, while Ca2+ influx at the same levels is highly toxic to cells, and serves as an exquisitely-sensitive signaling molecule. Instead, variability in the pore suggests that ion permeation is somewhat inconsequential for NALCN, which has permitted unprecedented levels of variability in the selectivity filter without dramatically impacting the physiology of cells.
NALCN lacks the abundant alternative-splicing associated with most other 4x6TM channels
Each 4x6TM channel usually possesses significant (sometimes > 100 different) alternatively-spliced isoforms,46 and many of these are designed to fine-tune the ion channel gating and expression characteristics. Sodium and calcium channels are limited to specified voltage ranges, which shape their signal specificity. NALCN lacks the extensive alternative-splicing of other 4x6TM channels.
A key feature of NALCN is its variable and adaptive pore
Invertebrate NALCN from snail Lymnaea stagnalis possess no significant gene splicing other than the alternative exon that splices in a novel pore to change ion selectivity.9 The variable and adaptive pore is the distinctive feature of NALCN, as is the variable-splicing that generates channel gating changes in voltage-sensitive 4x6TM channels. Mutually-exclusive splicing to generate alternative EEEE calcium and EEKE or EKEE sodium pores, evolved twice independently, within the two major lineages of invertebrates, the lophotrochozoans - deuterostomes (mollusks/annelids - echinoderms/hemichordates) and the ecdysozoa (centipedes, arachnids) (Fig. 1).9 Evolutionary pressure was present to duplicate exons for generating alternatively-spliced pores but to also retain both alternatives pores in many animal groups including deuterostomes.
Dual, alternative NALCN calcium and sodium pores are adapted for differing tissue functions in invertebrates
Expression patterns of the EKEE sodium pore and the EEEE calcium pore are equally abundantly-expressed overall in the snail Lymnaea stagnalis.9 Snail NALCN with a sodium pore is most abundant in secretory brain and glandular tissue, which correlate with cells containing Nav1channels and sodium-dependent membrane fluxes generating action potentials.9 Snail calcium pore is associated with contractile heart and skeletal muscle, where there are greater requirements for general calcium-sensing.9 The heart of snails, for example, completely lack sodium-dependent action potentials and expression of Nav1 sodium channels.27 NALCN is thus likely working mostly as a calcium sensor in muscle, to sense the ion fluxes during contraction that are mostly carried by calcium ions.
NALCN could still be a regulated channel even if it were non-conducting or weakly-conducting
Structurally, NALCN has reduced S4 charges in the voltage-sensor relative to standard voltage-gated sodium and calcium channels, but this is as true for NALCN from the simplest basal animals, such as sponge, as it is for human NALCN. An overall level of conservation of cationic charges and countercharges in the voltage sensor domain, and conservation of an amphipathic S4-S5 linker (Fig. S1) suggests that NALCN is like other 4x6TM channels in containing mobile elements that are triggered upon activation.
Other known leak conductance channels are regulated leak channels
It is hard to imagine NALCN, the singleton sodium leak channel, as the depolarizing counterpart to the two-pore, KCNK potassium leak conductance channels. There are more than 50 different KCNK genes in animals, and at least 18 human KCNK genes.47 KCNK leak conductance channels are governed by fundamental cellular conditions such as membrane stretch, external pH and temperature, are variable in their conductances and are often rectifying channels.47 NALCN was first described as a linear, non-selective, ohmic conductance at all voltage ranges,1 but it has also been asserted that NALCN can be receptor-operated.42,48 A lack of gating mechanism would mean in essence, that NALCN is more like an unregulated hole in the membrane when open, draining membrane gradients at high metabolic cost to ATP driven pumps and dampening activity through shunting inhibition because of the membrane’s lowered input resistance.
NALCN with a calcium or sodium pore correlates with animals that rely on calcium or sodium ion flux, respectively
It appears that NALCN’s changeable calcium and sodium pores correlate with the changing roles of calcium and sodium ions as signaling molecules in animal evolution (Fig. 2). The first appearance of a 4x6TM channel with a lysine in the pore (DKEA), is in cnidarians, which are the most primitive animal group with a nervous system that contains sodium-dependent action potentials.49 Prior to the cnidarians, all calcium channels, sodium channels and NALCN have calcium pores (EEEE or DEEA). Invertebrate 4xTM channels then evolved both calcium pores (EEEE/EEDD/DEEA) and sodium pores (DEKA/ EKEE/DKEA/ /EEKE).9 Invertebrates have greater promiscuity between sodium and calcium selectivity within the major 4x6TM channels. They have both sodium-selective Nav1 (DEKA)49 and an invertebrate-specific calcium-selective sodium channel gene Nav2 (DEEA).30 Cav3 T-type channels in invertebrates have a differing sodium and calcium ion permeability through alternative splicing, which is not present in the more exclusively calcium-selective Cav3 channels in vertebrates.27 The greater flexibility in ion selectivity may relate to the presence of ~4× fewer numbers of 4x6TM channels to draw upon in invertebrate genomes for structural and functional diversification.
Many animal species have lost NALCN with a calcium pore
The calcium pore is lost in insects and nematodes as well as many parasitic species of flatworms, arachnids and annelids. Free-living forms, closely related to the parasites that have lost the calcium pore, maintain dual calcium and sodium pores. The loss of selective ion sensing in endoparasites may reflect that they live within the regulated environment of their host with a more limited requirement for NALCN sensing.9 In higher vertebrates, NALCN is mostly a brain protein but is also can be found in heart tissues (particularly atria) and glandular tissues, including the thyroid gland, the adrenal gland and the pancreas.1,48 Vertebrates lost the more muscle-specific, NALCN spliced calcium-sensing pore, which may relate to the decreasing reliance of plasma membrane calcium flux through L-type calcium channels in the progressive evolution of muscle excitation-contraction coupling.50
Mammalian NALCN with the sodium pore has been linked to sodium ion sensing
Interestingly, NALCN mice mutants have altered serum sodium levels, which is consistent with a sodium-sensing role for mammalian NALCN.51 A more adaptable calcium or sodium sensor in invertebrates may reflect the cellular content of regulated calcium and sodium fluxes derived from voltage-gated sodium and calcium channels. It seems more than coincidence that a NALCN sodium-sensing lysine pore, co-evolved in similar animals that also contained the first sodium-conducting channels, and that the radially-symmetrical animals with only calcium sensing NALCN pores, are devoid of Nav1 sodium currents across their cell membranes and limited mostly to calcium conductances through their sodium and calcium channels (Fig. 2).
NALCN is concentrated in cells with rapid ion flux rates
NALCN loss of function phenotypes are prominently associated with defective pacemaking in rhythmically oscillating neurons,1,8,12,13,44 and there are defects evident in secretion especially in cells with fast or continuous vesicle turnover.8,11-13 A potential adaptable sodium and calcium sensing pore of NALCN may play a role in particular cells with rapid ion fluxes such as pacemaker tissue, where there would be the greatest need to monitor and respond to changing calcium and sodium concentrations across cell membrane and between cells through gap junctions.
There are links between NALCN and energy metabolism
NALCN/UNC80/UNC79 knockouts do not result in drastic developmental defects in the embryo,52-54 suggesting that the NALCN complex does not have a critical role in development. NALCN appears to have an association with metabolism and mitochondrial function. Defects in the NALCN complex produce animals that are smaller,3,45 consume more energy45 and are more hypersensitive to alcohol and volatile anesthetics.55 It is tempting to speculate that NALCN coupled to UNC80/UNC79 relates to a defect in mitochondrial signaling, linking a sensing of rapid turnover of membrane cation flux with energy usage and generation.
Conclusions
Many different channel conductances active at low membrane potentials near rest are known to contribute to pacemaking (e.g., T-type channels and HCN channels), but these are highly-regulated and diverse and do not resemble a non-specific, linear, ohmic leak conductance. NALCN-UNC80 complexes influence membrane conductances, suggesting a potential leak channel role,56,57 but the difficulties and elusiveness of locating expressed NALCN currents are more suggestive that its major function might not be its ion conductance. Rather, NALCN’s distinctiveness is in its changeable pore, which mimics the pores that differentiates the classes of calcium- and sodium-selective channels. Appropriately, NALCN is in between sodium and calcium channels in overall structure and evolved after them in likely ancestors of extant multicellular organisms. NALCN’s adaptable pore switches with the changing dominance of calcium and sodium fluxes in primitive to more advanced animals and evolves a dual pore in two different lineages of animals to serve differing proportions of calcium and sodium fluxes required in different tissues of many animals. NALCN is a ubiquitous and likely critical gene in all multicellular animals, but it has resisted changes in gene numbers and diversity of form that is the hallmark of voltage-gated sodium and calcium channels. NALCN, with a similar overall structure as other 4x6TM channels, evolved from a calcium sensor in primitive, radially symmetrical animals, to dual calcium and sodium sensor in different tissues of more advanced bilaterally symmetrical animals9 to mostly serving as a sodium sensor in the mammalian brain.51 The animal knockdown of NALCN is perinatal lethal, not embryonic lethal in mammals, which may relate directly to a reduced electrical activity in spinal nerves innervating the diaphragm.
A future model of NALCN function will have to tie all the known attributes of NALCN together. NALCN possess an adaptable calcium and sodium sensing pore and is enriched in pacemaking cells with rapid ion fluxes. NALCN’s activation likely involves a mobile voltage-sensor domain that is coupled through a large, cytoplasmic UNC80/UNC79 complex to possible functions, which may include a homeostatic regulation of sodium or calcium ions, and energy utilization. Overall, there is no definitive proof that NALCN is an ion sensor, but at least one should consider this as an alternative possibility to the prevailing viewpoint that NALCN is a non-selective, ohmic leak conductance channel resembling the Gd3+-sensitive, leaky patch current (ILP) observable in control HEK-293T cells.
Supplementary Material
Acknowledgments
We thank Arnaud Monteil and Gerald Zamponi with helpful comments on this manuscript and an NSERC-MSFSS award for providing Adriano Senatore an opportunity to examine the functions of NALCN with the CNRS group including Dr. Monteil in Montpellier, France. We would like to thank the Heart and Stroke Foundation of Canada and NSERC Discovery Operating for operating grant funds to J.D.S.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/channels/article/23981
References
- 1.Lu B, Su Y, Das S, Liu J, Xia J, Ren D. The neuronal channel NALCN contributes resting sodium permeability and is required for normal respiratory rhythm. Cell. 2007;129:371–83. doi: 10.1016/j.cell.2007.02.041. [DOI] [PubMed] [Google Scholar]
- 2.Miller HM. Sex-linked mutant characters induced by X-ray dosage of 5000 r-u. Drosoph Inf Serv. 1934;2:9. [Google Scholar]
- 3.Nash HA, Scott RL, Lear BC, Allada R. An unusual cation channel mediates photic control of locomotion in Drosophila. Curr Biol. 2002;12:2152–8. doi: 10.1016/S0960-9822(02)01358-1. [DOI] [PubMed] [Google Scholar]
- 4.Lee JH, Cribbs LL, Perez-Reyes E. Cloning of a novel four repeat protein related to voltage-gated sodium and calcium channels. FEBS Lett. 1999;445:231–6. doi: 10.1016/S0014-5793(99)00082-4. [DOI] [PubMed] [Google Scholar]
- 5.Sharman JL, Mpamhanga CP. IUPHAR-DB: An Open-Access, Expert-Curated Resource for Receptor and Ion Channel Research. ACS Chem Neurosci. 2011;2:232–5. doi: 10.1021/cn200025w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Magrane M, Consortium U. UniProt Knowledgebase: a hub of integrated protein data. Database (Oxford) 2011;2011:bar009. doi: 10.1093/database/bar009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Littleton JT, Ganetzky B. Ion channels and synaptic organization: analysis of the Drosophila genome. Neuron. 2000;26:35–43. doi: 10.1016/S0896-6273(00)81135-6. [DOI] [PubMed] [Google Scholar]
- 8.Yeh E, Ng S, Zhang M, Bouhours M, Wang Y, Wang M, et al. A putative cation channel, NCA-1, and a novel protein, UNC-80, transmit neuronal activity in C. elegans. PLoS Biol. 2008;6:e55. doi: 10.1371/journal.pbio.0060055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Senatore A, Monteil A, van Minnen J, Smit AB, Spafford JD. NALCN ion channels have alternative selectivity filters resembling calcium channels or sodium channels. PLoS One. 2013;8:e55088. doi: 10.1371/journal.pone.0055088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lu TZ, Feng ZP. A sodium leak current regulates pacemaker activity of adult central pattern generator neurons in Lymnaea stagnalis. PLoS One. 2011;6:e18745. doi: 10.1371/journal.pone.0018745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bouhours M, Po MD, Gao S, Hung W, Li H, Georgiou J, et al. A co-operative regulation of neuronal excitability by UNC-7 innexin and NCA/NALCN leak channel. Mol Brain. 2011;4:16. doi: 10.1186/1756-6606-4-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jospin M, Watanabe S, Joshi D, Young S, Hamming K, Thacker C, et al. UNC-80 and the NCA ion channels contribute to endocytosis defects in synaptojanin mutants. Curr Biol. 2007;17:1595–600. doi: 10.1016/j.cub.2007.08.036. [DOI] [PubMed] [Google Scholar]
- 13.Lear BC, Lin JM, Keath JR, McGill JJ, Raman IM, Allada R. The ion channel narrow abdomen is critical for neural output of the Drosophila circadian pacemaker. Neuron. 2005;48:965–76. doi: 10.1016/j.neuron.2005.10.030. [DOI] [PubMed] [Google Scholar]
- 14.Strong M, Chandy KG, Gutman GA. Molecular evolution of voltage-sensitive ion channel genes: on the origins of electrical excitability. Mol Biol Evol. 1993;10:221–42. doi: 10.1093/oxfordjournals.molbev.a039986. [DOI] [PubMed] [Google Scholar]
- 15.Verret F, Wheeler G, Taylor AR, Farnham G, Brownlee C. Calcium channels in photosynthetic eukaryotes: implications for evolution of calcium-based signalling. New Phytol. 2010;187:23–43. doi: 10.1111/j.1469-8137.2010.03271.x. [DOI] [PubMed] [Google Scholar]
- 16.Locke EG, Bonilla M, Liang L, Takita Y, Cunningham KW. A homolog of voltage-gated Ca(2+) channels stimulated by depletion of secretory Ca(2+) in yeast. Mol Cell Biol. 2000;20:6686–94. doi: 10.1128/MCB.20.18.6686-6694.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liebeskind BJ, Hillis DM, Zakon HH. Evolution of sodium channels predates the origin of nervous systems in animals. Proc Natl Acad Sci U S A. 2011;108:9154–9. doi: 10.1073/pnas.1106363108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liebeskind BJ, Hillis DM, Zakon HH. Phylogeny unites animal sodium leak channels with fungal calcium channels in an ancient, voltage-insensitive clade. Mol Biol Evol. 2012;29:3613–6. doi: 10.1093/molbev/mss182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Catterall WA. Ion channel voltage sensors: structure, function, and pathophysiology. Neuron. 2010;67:915–28. doi: 10.1016/j.neuron.2010.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yeh E, Kawano T, Weimer RM, Bessereau JL, Zhen M. Identification of genes involved in synaptogenesis using a fluorescent active zone marker in Caenorhabditis elegans. J Neurosci. 2005;25:3833–41. doi: 10.1523/JNEUROSCI.4978-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vitko I, Shcheglovitov A, Baumgart JP, Arias-Olguín II, Murbartián J, Arias JM, et al. Orientation of the calcium channel beta relative to the alpha(1)2.2 subunit is critical for its regulation of channel activity. PLoS One. 2008;3:e3560. doi: 10.1371/journal.pone.0003560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Perez-Reyes E. Characterization of the gating brake in the I-II loop of CaV3 T-type calcium channels. Channels (Austin) 2010;4:453–8. doi: 10.4161/chan.4.6.12889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Payandeh J, Scheuer T, Zheng N, Catterall WA. The crystal structure of a voltage-gated sodium channel. Nature. 2011;475:353–8. doi: 10.1038/nature10238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Doyle DA, Morais Cabral J, Pfuetzner RA, Kuo A, Gulbis JM, Cohen SL, et al. The structure of the potassium channel: molecular basis of K+ conduction and selectivity. Science. 1998;280:69–77. doi: 10.1126/science.280.5360.69. [DOI] [PubMed] [Google Scholar]
- 25.McCusker EC, Bagnéris C, Naylor CE, Cole AR, D’Avanzo N, Nichols CG, et al. Structure of a bacterial voltage-gated sodium channel pore reveals mechanisms of opening and closing. Nat Commun. 2012;3:1102. doi: 10.1038/ncomms2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gur Barzilai M, Reitzel AM, Kraus JE, Gordon D, Technau U, Gurevitz M, et al. Convergent evolution of sodium ion selectivity in metazoan neuronal signaling. Cell Rep. 2012;2:242–8. doi: 10.1016/j.celrep.2012.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Senatore A, Guan W, Boone AN, Spafford JD. Cav3 T-type calcium channels become sodium-permeable using an alternate extracellular turret outside the selectivity filter. 2013 In press. [Google Scholar]
- 28.Heinemann SH, Terlau H, Stühmer W, Imoto K, Numa S. Calcium channel characteristics conferred on the sodium channel by single mutations. Nature. 1992;356:441–3. doi: 10.1038/356441a0. [DOI] [PubMed] [Google Scholar]
- 29.Tikhonov DB, Zhorov BS. Possible roles of exceptionally conserved residues around the selectivity filters of sodium and calcium channels. J Biol Chem. 2011;286:2998–3006. doi: 10.1074/jbc.M110.175406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhou W, Chung I, Liu Z, Goldin AL, Dong K. A voltage-gated calcium-selective channel encoded by a sodium channel-like gene. Neuron. 2004;42:101–12. doi: 10.1016/S0896-6273(04)00148-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lipkind GM, Fozzard HA. Voltage-gated Na channel selectivity: the role of the conserved domain III lysine residue. J Gen Physiol. 2008;131:523–9. doi: 10.1085/jgp.200809991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Eckert R, Brehm P. Ionic mechanisms of excitation in Paramecium. Annu Rev Biophys Bioeng. 1979;8:353–83. doi: 10.1146/annurev.bb.08.060179.002033. [DOI] [PubMed] [Google Scholar]
- 33.Shimizu H, Watanabe E, Hiyama TY, Nagakura A, Fujikawa A, Okado H, et al. Glial Nax channels control lactate signaling to neurons for brain [Na+] sensing. Neuron. 2007;54:59–72. doi: 10.1016/j.neuron.2007.03.014. [DOI] [PubMed] [Google Scholar]
- 34.Bannister RA, Beam KG. Ca(V)1.1: The atypical prototypical voltage-gated Ca(2+) channel. Biochim Biophys Acta. 2012 doi: 10.1016/j.bbamem.2012.09.007. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Catterall WA. Voltage-gated calcium channels. Cold Spring Harb Perspect Biol. 2011;3:a003947. doi: 10.1101/cshperspect.a003947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Catterall WA. Voltage-gated sodium channels at 60: structure, function and pathophysiology. J Physiol. 2012;590:2577–89. doi: 10.1113/jphysiol.2011.224204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Senatore A, Spafford JD. Gene transcription and splicing of T-type channels are evolutionarily-conserved strategies for regulating channel expression and gating. PLoS One. 2012;7:e37409. doi: 10.1371/journal.pone.0037409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dolphin AC. Calcium channel auxiliary α2δ and β subunits: trafficking and one step beyond. Nat Rev Neurosci. 2012;13:542–55. doi: 10.1038/nrn3317. [DOI] [PubMed] [Google Scholar]
- 39.Brackenbury WJ, Isom LL. Na Channel β Subunits: Overachievers of the Ion Channel Family. Front Pharmacol. 2011;2:53. doi: 10.3389/fphar.2011.00053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Altier C, Garcia-Caballero A, Simms B, You H, Chen L, Walcher J, et al. The Cavβ subunit prevents RFP2-mediated ubiquitination and proteasomal degradation of L-type channels. Nat Neurosci. 2011;14:173–80. doi: 10.1038/nn.2712. [DOI] [PubMed] [Google Scholar]
- 41.Pongs O, Schwarz JR. Ancillary subunits associated with voltage-dependent K+ channels. Physiol Rev. 2010;90:755–96. doi: 10.1152/physrev.00020.2009. [DOI] [PubMed] [Google Scholar]
- 42.Lu B, Su Y, Das S, Wang H, Wang Y, Liu J, et al. Peptide neurotransmitters activate a cation channel complex of NALCN and UNC-80. Nature. 2009;457:741–4. doi: 10.1038/nature07579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lu B, Zhang Q, Wang H, Wang Y, Nakayama M, Ren D. Extracellular calcium controls background current and neuronal excitability via an UNC79-UNC80-NALCN cation channel complex. Neuron. 2010;68:488–99. doi: 10.1016/j.neuron.2010.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pierce-Shimomura JT, Chen BL, Mun JJ, Ho R, Sarkis R, McIntire SL. Genetic analysis of crawling and swimming locomotory patterns in C. elegans. Proc Natl Acad Sci U S A. 2008;105:20982–7. doi: 10.1073/pnas.0810359105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Speca DJ, Chihara D, Ashique AM, Bowers MS, Pierce-Shimomura JT, Lee J, et al. Conserved role of unc-79 in ethanol responses in lightweight mutant mice. PLoS Genet. 2010;6:6. doi: 10.1371/journal.pgen.1001057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lipscombe D, Andrade A, Allen SE. Alternative splicing: Functional diversity among voltage-gated calcium channels and behavioral consequences. Biochim Biophys Acta. 2012 doi: 10.1016/j.bbamem.2012.09.018. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Enyedi P, Czirják G. Molecular background of leak K+ currents: two-pore domain potassium channels. Physiol Rev. 2010;90:559–605. doi: 10.1152/physrev.00029.2009. [DOI] [PubMed] [Google Scholar]
- 48.Swayne LA, Mezghrani A, Varrault A, Chemin J, Bertrand G, Dalle S, et al. The NALCN ion channel is activated by M3 muscarinic receptors in a pancreatic beta-cell line. EMBO Rep. 2009;10:873–80. doi: 10.1038/embor.2009.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Spafford JD, Spencer AN, Gallin WJ. A putative voltage-gated sodium channel alpha subunit (PpSCN1) from the hydrozoan jellyfish, Polyorchis penicillatus: structural comparisons and evolutionary considerations. Biochem Biophys Res Commun. 1998;244:772–80. doi: 10.1006/bbrc.1998.8332. [DOI] [PubMed] [Google Scholar]
- 50.Schredelseker J, Shrivastav M, Dayal A, Grabner M. Non-Ca2+-conducting Ca2+ channels in fish skeletal muscle excitation-contraction coupling. Proc Natl Acad Sci U S A. 2010;107:5658–63. doi: 10.1073/pnas.0912153107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sinke AP, Caputo C, Tsaih SW, Yuan R, Ren D, Deen PM, et al. Genetic analysis of mouse strains with variable serum sodium concentrations identifies the Nalcn sodium channel as a novel player in osmoregulation. Physiol Genomics. 2011;43:265–70. doi: 10.1152/physiolgenomics.00188.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lu B, Su Y, Das S, Liu J, Xia J, Ren D. The neuronal channel NALCN contributes resting sodium permeability and is required for normal respiratory rhythm. Cell. 2007;129:371–83. doi: 10.1016/j.cell.2007.02.041. [DOI] [PubMed] [Google Scholar]
- 53.Nakayama M, Iida M, Koseki H, Ohara O. A gene-targeting approach for functional characterization of KIAA genes encoding extremely large proteins. FASEB J. 2006;20:1718–20. doi: 10.1096/fj.06-5952fje. [DOI] [PubMed] [Google Scholar]
- 54.Speca DJ, Chihara D, Ashique AM, Bowers MS, Pierce-Shimomura JT, Lee J, et al. Conserved role of unc-79 in ethanol responses in lightweight mutant mice. PLoS Genet. 2010;6:6. doi: 10.1371/journal.pgen.1001057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Humphrey JA, Hamming KS, Thacker CM, Scott RL, Sedensky MM, Snutch TP, et al. A putative cation channel and its novel regulator: cross-species conservation of effects on general anesthesia. Curr Biol. 2007;17:624–9. doi: 10.1016/j.cub.2007.02.037. [DOI] [PubMed] [Google Scholar]
- 56.Lu TZ, Feng ZP. NALCN: a regulator of pacemaker activity. Mol Neurobiol. 2012;45:415–23. doi: 10.1007/s12035-012-8260-2. [DOI] [PubMed] [Google Scholar]
- 57.Ren D. Sodium leak channels in neuronal excitability and rhythmic behaviors. Neuron. 2011;72:899–911. doi: 10.1016/j.neuron.2011.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.