Abstract
Immune-mediated damage to tumor vessels is a potential means of preventing solid tumor progression. Antiangiogenic cancer vaccines capable of inducing this kind of damage include formulations comprised of endothelial cell-specific antigens. Identification of antigens capable of eliciting efficient vaccination is difficult because the endothelial cell phenotype is affected by surrounding tissues, including angiogenic stimuli received from surrounding tumor cells. Therefore, phenotype endothelial cell variations (heterogeneity) were examined in the context of the development of an efficient vaccine using mass spectrometry-based cell surface profiling. This approach was applied to primary human microvascular endothelial cell (HMEC) cultures proliferated under growth stimuli provided by either normal tissues (growth supplement from human hypothalamus) or cancer cells (MCF-7, LNCap and HepG2). It was found that tumors induced pronounced, tumor type-dependent changes to HMEC surface targets that in an in vitro model of human antiangiogenic vaccination directly facilitated HMEC escape from cytotoxic T cell-mediated cell death. Furthermore, it was found that tumors influenced the HMEC phenotype unidirectionally and that HMEC imunogenicity was reciprocal to the intensity of tumor-induced changes to the HMEC surface. These findings provide data for the design of tumor-specific endothelial cell based vaccines with sufficient immunogenicity without posing a risk to the elicitation of autoimmunity if administered in vivo.
Keywords: antiangiogenic cancer vaccine, microvascular endothelial cells, cell surface profiling, cell heterogeneity, vaccine design, cell proteomic footprinting
Introduction
Despite tremendous progress in basic research, effective treatments for most types of cancers are still lacking. Therefore, development of novel, effective therapies, specifically the development of anti-cancer vaccine-based therapies designed to specifically prevent cancer represent a promising treatment option.1 Tumor vaccine formulations comprised of tumor antigens seem to be a promising cancer preventive approach,2 however, targeting tumor endothelium with antiangiogenic vaccines has advantages over targeting tumor cells. Tumor endothelium (in comparison with tumor cells) is genetically stable with a low probability of developing acquired resistance to drugs.3 Furthermore, given that the endothelial to tumor cell ratio can vary between 1:50 and 1:100 the number of cells targeted is much smaller.4 Furthermore, destruction of a small number of endothelial cells can lead to vascular obstruction resulting in arrest of tumor growth or its destruction since vascular integrity is essential to growth and metastasis.5-8
Among the various approaches used to direct immune response against tumor endothelial antigens, active immunization using endothelial cells is most promising compared with immunotherapies targeting specific epitopes since cell-based vaccines can target multiple autologous target cell antigens, most of which have not been isolated or characterized.9 Previously, this approach was shown to inhibit the growth of experimental tumors in mouse models.10-15 However, additional considerations, such as endothelial cell heterogeneity, need to be considered before this approach can be developed.
Endothelial cell heterogeneity has been described at the level of cell morphology, function, gene expression and antigen composition.16,17 Endothelial cell phenotypes vary between different organs, as well as between different tissues of the same organ. In addition to the tissue of origin, the gene expression profile of tumor endothelial cells can be sufficiently influenced by the tumor.18-20 Previous studies have shown that conditioned medium from tumor cells resulted in the change of gene expression by cultured endothelial cells.21-23 Other studies have described differences in gene expression profiles in isolated tumor endothelial cells compared with endothelial cells harvested from matched tissues.24,25 These data showed that endothelial cell heterogeneity should be investigated in the context of the efficient design of endothelial cell-based vaccines. To this end, human microvascular endothelial cell (HMEC) primary cultures were established and tumor type-specific changes induced by culturing HMEC in the presence of tumor-conditioned medium were examined. Changes were studied using mass spectrometry-based approach for cell surface profiling. This approach identified tumor-induced heterogeneity in cell surface antigen expression profiles associated with HMEC escape from cytotoxic T cell-mediated cell death and provides a novel means of selecting cells for the development of a cellular cancer vaccine.
Results
Primary HMEC cultures
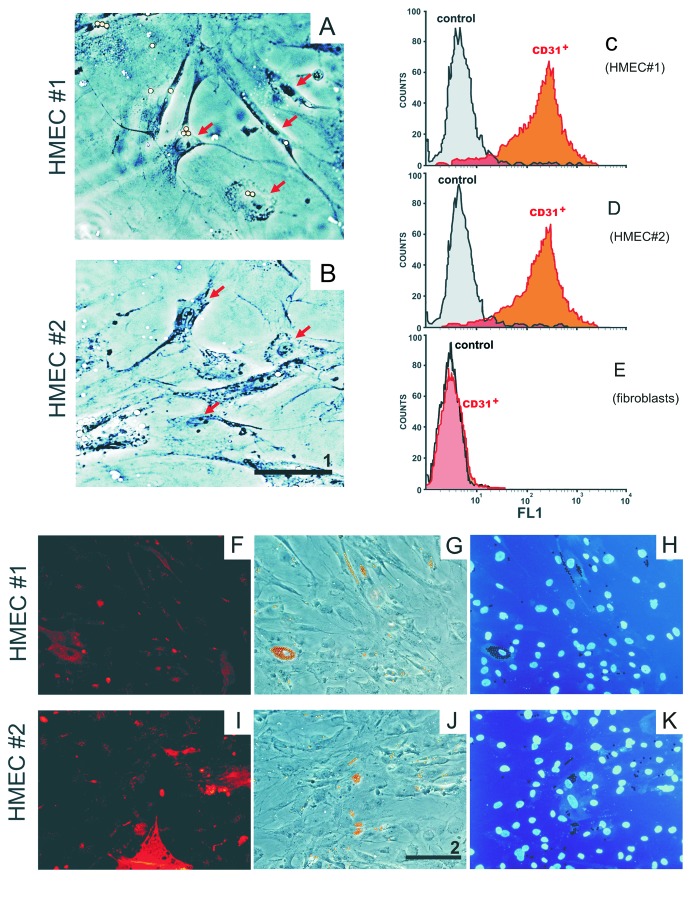
Anti-CD31 beads were used to isolate HMEC from a thoracic fat biopsy. Figure 1A and B show endothelial cells isolated from fat biopsies obtained from donors 1 and 2, respectively. HMEC presented with morphology typical for adipose-derived microvascular ECs with numerous cytoplasmic extensions and/or a cobblestone-like morphology26 that formed net-like structures when incubated in the presence of tumor-conditioned medium. Immunofluorescent staining (Fig. 1F and I) revealed that an endothelial cell marker CD31 is associated with almost 90% of cells during the first passage following isolation. No overgrowth of contaminating fibroblasts or mesothelial cells was detected, demonstrating that primary HMEC cultures were successfully established. FACS analysis of HMEC cultures confirmed that CD31+ comprised 89.6% and 87.4% of cells obtained from the fat of donors 1 and 2, respectively (Fig. 1C and D). Non-specific binding of the primary antibody to negative control cells (fibroblasts) did not exceed 1.5% (Fig. 1E).
Figure 1. Primary HMEC cultures. A representative HMEC from donor 1 (A) and donor 2 (B). HMEC have numerous cytoplasmic extensions and/or cobblestone-like morphology (arrows) typical for adipose-derived microvascular endothelial cells.26 Flow cytometric analysis of HMEC cultures from donor 1 (C) and donor 2 (D). Cells were stained with mouse anti-human CD31 antibody (biotinylated horse anti-mouse IgG as secondary antibody and streptavidin-RPE as a fluorescent label). Fluorescently stained cells are labeled as “CD31+.” Baseline florescence was determined using the same cells exposed only to the biotinylated secondary antibody and streptavidin-RPE (labeled as “control”). Immunofluorescent staining of CD31+ cells (F and I) correspond to donors 1 and 2, respectively. Microscopic images (G and J) and DAPI staining of nuclei from cells (H and K) depicted in (F and I). Images were obtained using a Leiса DM5000B microscope (scale bar #1, 20 µm; scale bar #2, 50 µm). Adult skin fibroblasts were used as negative anti-C31-binding control (E).
HMEC surface profiles
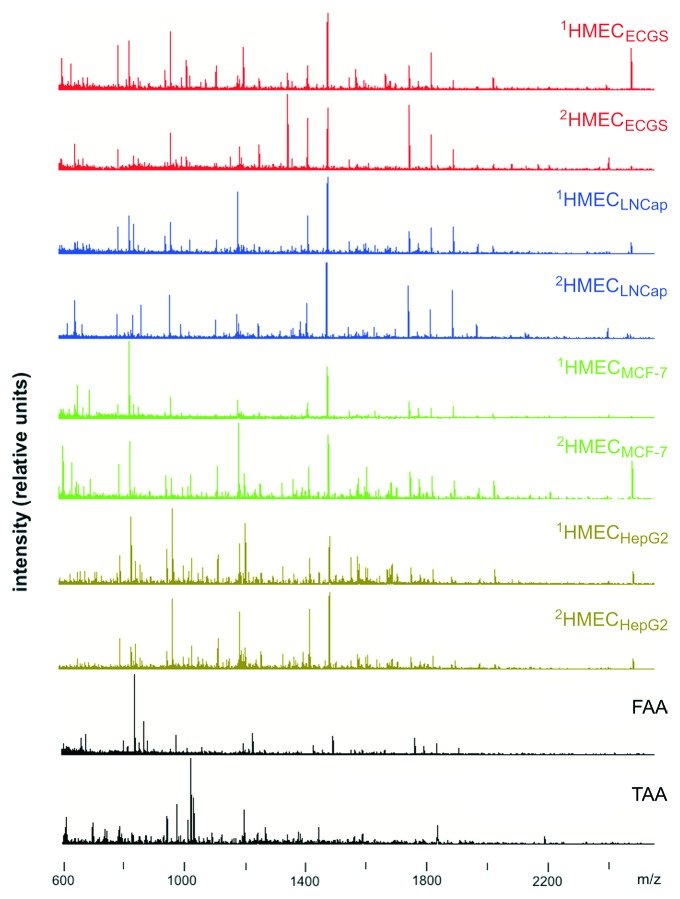
Mass spectrometry analysis of proteolytically-cleaved cell surface targets, i.e., endothelial cell-associated antigens (EAA), tumor-associated antigens (TAA) and fibroblast-associated antigens (FAA), resulted in the detection of 164 ± 29 (mean ± standard deviation) positively charged (glyco)peptides ions per sample. Representative mass spectra used to generate cell surface profiles are shown in Figure 2.
Figure 2. Mass spectra used to generate cell surface profiles. Representative spectra of EAA of HMEC obtained from donor 1 and donor 2 stimulated using endothelial cell growth supplement (1HMECECGS and 2HMECECGS), conditioned medium from MCF-7 cells (1HMECMCF-7 and 2HMECMCF-7), LNCap cells (1HMECLNCap and 2HMECLNCap), or HepG2 cells (1HMECHepG2 and 2HMECHepG2). Superscript numbers correspond to donor 1 or 2, respectively. The mass spectra of fibroblast-associated antigens (FAA) and tumor(MCF-7 cells)-associated antigens (TAA) are also shown.
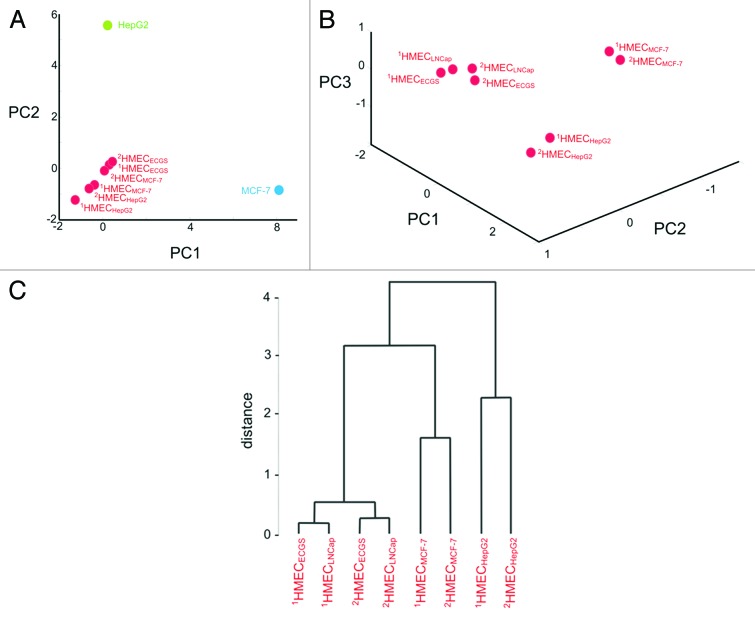
Principal component analysis (PCA) plot of cell surface profiles (Fig. 3A) revealed a relationship between HMEC of different donors stimulated to grow in the presence of endothelial cell growth supplement (ECGS) or tumor-conditioned medium (cancer cells were cultured to generate the conditioned medium). The 2-dimensional plot depicted covers 92.5% of variance contained in these profiles and, therefore, accurately reflects the relationship between them. All profiles on a plot related to the HMEC grouped together and were distinct from cancer cell surface profiles that in turn were also unrelated to each other.
Figure 3. Principal component analysis (PCA) of cell surface profiles. PCA of cell surface profiles obtained from HMEC and cancer cells that were projected in the space of the first two principal components (A). PCA of cell surface profiles obtained only for HMEC projected into the space of the first 3 principal components (B) and the dendrogram depicting the distances measured between points (C). “1HMECECGS” and “2HMECECGS” - cell surface profile of HMEC stimulated to grow in the presence of endothelial cell growth supplement; “1HMECMCF-7” and “2HMECMCF-7” - cell surface profile of HMEC stimulated to grow in the presence of MCF-7 cell conditioned medium; “1HMECLNCap” and “2HMECLNCap” - cell surface profiles of HMEC stimulated to grow in the presence of LNCap cell conditioned medium; “1HMECHepG2” and “2HMECHepG2” - cell surface profiles of HMEC stimulated to grow in the presence of HepG2 cell conditioned medium; the “MCF-7” - point corresponds to the cell surface profile obtained from MCF-7 cells; the “HepG2” - point corresponds to the cell surface profile obtained from HepG2 cells.
PCA analysis of HMEC surface profiles alone demonstrated that they projected into the space of the 3 principal components (Fig. 3B) with the majority of the variability (83.0%) contained in these profiles. Therefore, 3-dimentianal PCA plot accurately reflects the relationship between HMEC profiles and shows that points related to respective HMEC profiles grouped according to the growth stimuli provided. Moreover, points correspondent to HMEC following growth in the presence of conditioned medium from HepG2 or MCF-7 cells were far from points observed following growth in the presence of ECGS. Points resulting from HMEC stimulation with LNCap conditioned medium were close to these points.
Cytotoxicity assays
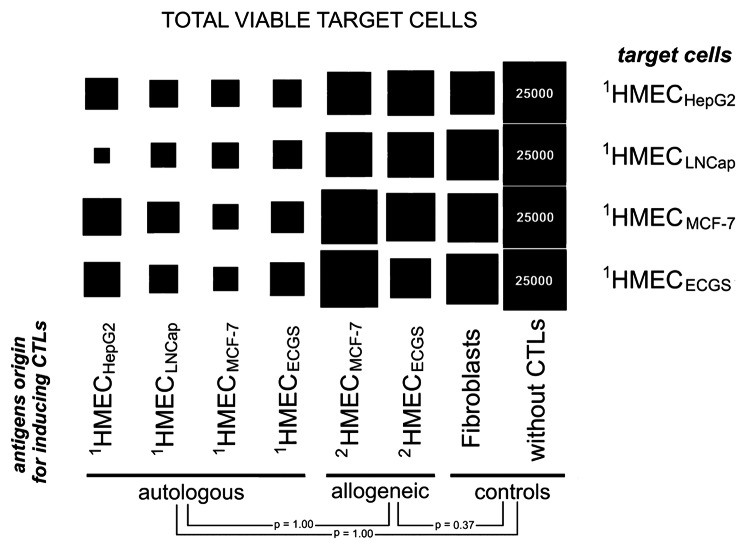
The immunologic properties of respective HMEC were evaluated by loading dendritic cells (DCs) with EAA as a means of activating and stimulating human cytotoxic T lymphocytes (CTLs) against target HMEC. CTLs stimulated with unloaded DCs or FAA-loaded DCs incubated in the presence of HMEC were used as negative controls. On day 3, surviving target HMEC cells were detected using trypan blue exclusion (Fig. 4). A slight increase in cytotoxic activity was observed when CTLs were stimulated with FAA-loaded or allogeneic EAA-loaded DCs. Only DCs loaded with autologous EAA induced effective immune responses measured by high death rates of target HMEC (Fig. 4).
Figure 4. Cytotoxicity of effector CTLs against HMEC. Target HMEC cells (3.7 × 104 cells/well) were incubated in the presence of effector CTLs at a 1:8 ratio. After 3 d, floating CTLs were removed, target cells carefully washed with HBSS, attached HMEC were trypsinized and cell viability determined. Square size reflects viable target cell counts. Data was scaled to bring all controls (target cells incubated without CTLs) to equal values (25,000 cells). Data represent the mean value of 3 independent measurements. The standard deviations for the number of viable cells were in the 1,200–3,400 cell range. P values determined by t-test.
CTLs stimulated with EAA-loaded DCs from 1HMECMCF-7 were most effective against the same 1HMECMCF-7 target cells. 1HMECHepG2 cells were most efficiently killed by CTLs stimulated with EAA-loaded DCs from 1HMECMCF-7 or 1HMECLNCap. Target 1HMECLNCap were killed more effectively by CTLs stimulated by EAA-loaded DCs from 1HMECHepG2.
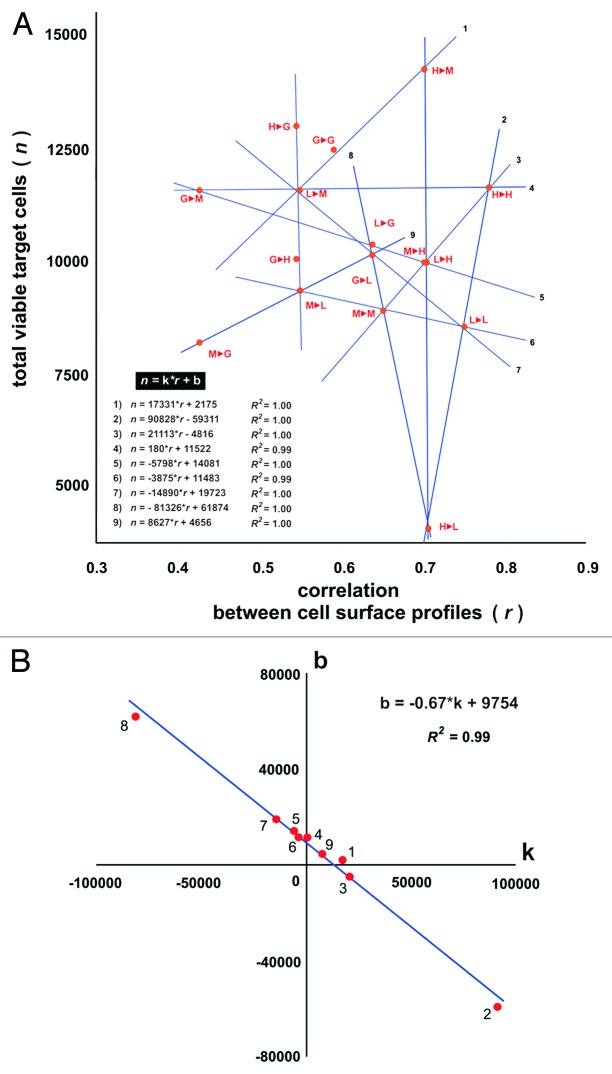
To further demonstrate the impact on immune response efficiency elicited by respective target cell surface antigen profiles, the number of viable target cells surviving cytotoxicity assays was plotted against profile correlation values calculated pairwise for target HMEC and HMEC used to generate antigens (EAA) for eliciting immune response in the cytotoxicity assays. As shown on Figure 5, some points on this plot can be linearly approximated with R2 ≥ 0.99. Moreover, all points on the plot were located at the intersection of respective lines intersected.
Figure 5. Cytotoxicity of effector CTLs against target HMEC plotted vs. correlation of cell surface profiles (A) and plotted coefficients of linear equations that describe the dependence of cytotoxicity from the correlation of cell surface profiles (B). (A) Autologous EC antigens were used to induce CTLs. The average number of viable target cells in 3 wells is presented. Accuracy of liner approximation (R2) was achieved using coordinates of 3 respective points. Equations for linear approximations are shown on the plot. Correlation values (coefficient r) were calculated for HMEC surface profiles used to generate antigens (EAA) for eliciting immune response and the surface profile of target HMEC used in same cytotoxicity assay. “1►2” – 1st letter corresponds to HMEC used to generate antigens (EAA) for eliciting immune response in the cytotoxicity assays; 2nd letter corresponds to the target HMEC used in the same cytotoxicity assay. “G” – 1HMECECGS; “M” – 1HMECMCF-7; “L” – 1HMECLNCap; “H” – 1HMECHepG2. (B) “k” and “b” values correspond to coefficients of linear equations showed on plot (A). Equation of linear approximation of the coordinates and accuracy of this approximation (R2) are shown on the plot.
Discussion
It is known that endothelial cells have tissue-specific phenotypes. In addition to tissue type specificity, the phenotype of tumor endothelial cells can be influenced by tumor cell-specific growth stimuli. Therefore, endothelial cell heterogeneity should be investigated in the context of designing efficient endothelial cell-based vaccines. The simplest way to do this is to model tumor-endothelium interactions in vitro by culturing primary HMEC cultures with tumor-conditioned medium. Tumor cells release growth factors in culture medium that affect HMEC and support their proliferation. As a control, HMEC stimulated by normal tissues should be performed despite the limited methods available for carrying this out. Conditioned media from untransformed cells possesses limited capacity to support cell cultures due to lack of growth factors and therefore cannot sustain endothelial cell growth. Therefore the most effective means of providing growth stimuli from normal tissue is the endothelial cell growth supplement (ECGS) prepared from brain gland tissue.27 Unfortunately, commercially available growth supplements are all xenogeneic (bovine origin), therefore ECGS was prepared from human hypothalamus, thereby providing the control needed for the described experiments.
The use of HMEC in this model system is based on previous studies that characterized the type of endothelial cells involved in mediating tumor angiogenesis that identified endothelial cells of microvascular origin. In addition, HMEC exhibit a number of functional differences compared with large vessel-derived endothelial cells,28,29 including responses to stimulators30,31 and their extracellular protein expression pattern.32-34 The use of primary cultures (rather than cell lines) is essential to investigating natural endothelial cell phenotypes and the respective responses of endothelial cells to growth stimuli. These parameters are difficult to assess when using immortalized cell lines that have intrinsic proliferative properties resulting from virus transfection and not due to responses to growth stimuli. The use of HMEC derived from subcutaneous fat tissues in our experiments resulted from the observation that HMEC lining tumor vessels grow up from surrounding tumor tissues. However, the location of the primary tumors, as well as sites of metastasis can occur in many different tissues throughout the body. Therefore, it was rational to derive HMEC for these experiments from abundant and easily accessible tissues such is subcutaneous fat.
Although isolation and culture of microvascular endothelial cells is difficult due to potential contamination by other cell types, including fibroblasts and mesothelial cells,35-37 selection of CD31+ cells using magnetic beads38 (followed by confirmatory immunofluorescent staining for CD31 and FACS analysis) was used in this study to isolate HMEC.37,39 CD31 is expressed on all endothelial cells but not on most other cell types including fibroblasts and mesothelial cells. Therefore positive selection of CD31+ cells is a common means of isolating endothelial cells from different tissues (excluding whole blood or bone marrow since monocytes and other cells expressing CD31 are present in these tissues). Furthermore, the purity of established HMEC primary cultures was confirmed by FACS analysis performed using anti-CD31 antibodies. In addition, non-specific binding of the anti-human CD31 antibody was ruled out by using fibroblasts as negative anti-C31-binding controls since fibroblasts are the main cause of primary culture contamination. Other means of confirming HMEC purity were not tested.
Figure 1 confirms that the 2 primary HMEC used in the described experiments were successfully established without overgrowth of contaminating cells. The ~90% purity of the primary HMEC cultures was considered high40 and corresponded to the purity observed for commercially available high-grade primary cultures.41 However, it should be noted that we did not attempt to obtain HMEC cultures of higher purity since it would have required repeated CD31+ cell enrichment steps combined with cell culture steps. Since the experiments described in this report required cells to be maintained in culture for short periods of time (since the molecular phenotype of isolated cells tends diverge significantly in a short period of time42) the extended culture times required to achieve CD31+ cultures with > 90% purity would not have been justified.
Design and development of cell-based vaccines focuses the immune response against target cells following immunization with these cells as a source of native antigens.43-45 An advantage of applied mass spectrometry-based approach for the profiling of cell surface targets (the set of prioritized antigens for cancer vaccine design46,47) that can be used as immunogens (rather than using whole cells) is that even though cell-based vaccines express the antigen profile of interest, whole cells also contain abundant amounts of intracellular antigens which are ubiquitous to all mammalian cells that could elicit untoward immune responses to self antigens.42 Access to cell surface targets (recognized by either antibodies or cytotoxic immune cells) means they are accessible to proteases and therefore can be isolated in vitro following proteolytic cleavage.48-50 Recently, it was shown that proteolytically-cleaved cell-surface targets could be directly analyzed using mass spectrometry, a technique known as cell proteomic footprinting51 which already has been successfully applied in the design of cellular cancer vaccines.50 In this study, the HMEC heterogeneity induced with either tumor-conditioned medium or growth factors from normal tissue was performed using this approach.
The degree of change to the HMEC surface antigen expression profile following incubation in the presence of tumor-conditioned medium is shown in Figure 3A. The PCA plot shows that these changes could be considered as deviations from the typical HMEC phenotype. HMEC profiles were grouped close together and distant from cancer cell profiles. Figure 3B shows the relationship between surface profiles within the HMEC groups and Figure 3C represents dendrogram where the length of dendrogram branches reflects similarity between profiles. From these observations, 3 conclusions were made: First, HMEC cells from the same tissue had the same surface antigen profile based on the high similarity between HMEC surface profiles obtained from adipose tissues from different donors. Second, tumors induced reproducible tumor type-specific changes to the HMEC surface antigen profile which can range from relatively insignificant (for example 1HMECLNCap and 2HMECLNCap) to pronounced (for example 1HMECHepG2 and 2HMECHepG2). Third, pronounced changes to the HMEC surface antigen profile were less reproducible. For example, 1HMECHepG2 and 2HMECHepG2 profiles were more significantly divergent from each other than 1HMECLNCap and 2HMECLNCap profiles.
The effect of the HMEC surface change on the ability to escape from the immune response was determined with cytotoxicity assays, as an in vitro model of immune response following cell-based anti-cancer vaccination. It should be noted that cell viability in these cytotoxicity assays was determined using trypan blue exclusion. Although more laborious, this cell counting/cell viability assay was favored over the 51Cr release assay (as well as MTT assay) since this method allowed for direct assessment of viable cell numbers, unlike methods using indirect measures of radioactivity release. Only direct counting allowed assessment of changes to the number of surviving cells.52 Moreover, with direct cell counting, lower effector-to-target cell ratios and longer incubation times could be employed that better reflected likely in vivo scenarios, thereby providing a more sensitive and appropriate assay that better allowed the objectives of this study to be performed.53 Therefore, using an in vitro model of human antiangiogenic therapies (where human CTLs were incubated in the presence of endothelial cell targets) DCs loaded with autologous antigens were found to stimulate cytotoxic activity more effectively than DCs loaded with allogeneic antigens (Fig. 4). The most efficient targeting of 1HMECMCF-7 was the result of stimulation by 1HMECMCF-7 antigens. However, 1HMECHepG2 were more efficiently targeted by autologous 1HMECMCF-7 or 1HMECLNCap and 1HMECLNCap were more efficiently targeted by autologous 1HMECHepG2.
Even though these data suggested that autologous antigens provided the most immunogenic stimulus (and represent the most promising prospect for cancer prevention) the rules for selecting autologous HMEC for the elicitation of immune responses against tumor endothelium as well as establishing a connection between cell surface heterogeneity and immune evasion remained unclear. Thus, statistically significant differences between autologous antigens for the elicitation of immune responses was not observed. These observations resulted in the design of additional experiments developed to examine the immune responses targeting by different autologous HMEC. If the cell surface profiles of the target HMEC and the HMEC used as a vaccine were plotted against viable target cell counts (Fig. 5) groups of points on this plot may be linearly approximated with R2 values equal or almost equal to 1, suggesting that cytotoxicity of CTLs was directly predefined by cell surface profiles and is described by following equation: n =k*r + b
where n is a number of total viable target cell in cytotoxicity assays and represents target cell escape (the reciprocal value of the observed CTL-mediated immune response); r is the correlation of target cell profile and the profile of cells used for targeting the immune response; b represents the coefficient which contributes to the immune response independent from the correlation of target cell profile and the profile of cells used for targeting the immune response; k represents the coefficient which defines immune response intensity directly from this correlation.
Therefore, it was rational to suggest that k reflects the intensity of tumor-induced changes at the cell surface, b reflects the immunogenicity of cell surface targets associated with these changes. Moreover, all points on the plot were located at the intersection of respective lines, suggesting that k and b varied dependently on each other. Indeed, when linear equations were built for respective lines and all k and b values were defined, it was found that b values were linearly dependent on k values according to the following equation (see also Fig. 5B):
| b = -0.67*k + 9754 (R2 of linear approximation is 0.99) |
Thus, the immunogenicity of HMEC was inversely proportional to the intensity of tumor-induced changes at the HMEC surface. From this observation it was concluded that HMEC heterogeneity was the result of the unidirectional influence of tumor cells, i.e., this influence was not specific for the tumor type and HMEC heterogeneity was a result of differences in strength of this influence. More significant influences lead to more pronounced changes in HMEC surfaces and simultaneously lead to loss of HMEC immunogenicity. Consequently, in cytotoxicity assays the observed efficacy of CTLs in killing of target cells was directly defined by the similarity between surface profiles of target HMEC and HMEC used for targeting immune responses and by the actual immunogenicity of these cells.
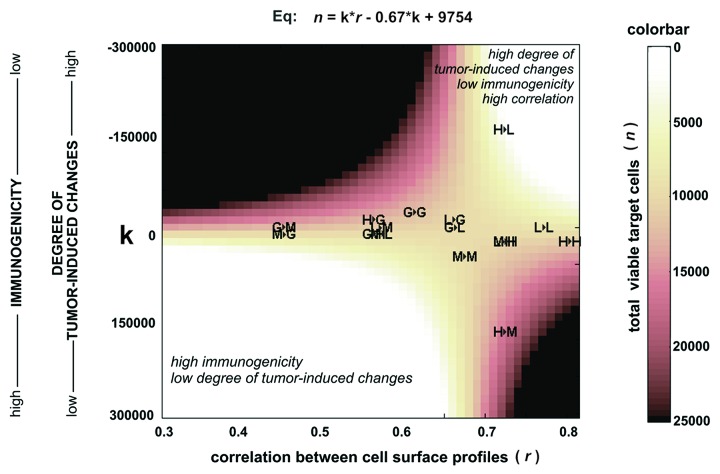
As a means of better understanding to what degree these results could impact vaccine design, the dependence of total viable target cells in cytotoxicity assays (n) from the intrensity of tumor-induced changes at the cell surface (k) and correlation of cell surface profiles (r) was plotted (Fig. 6). This plot shows that tumor endothelial cells were efficiently killed by vaccines projected into two areas of this plot.
Figure 6. Cytotoxicity of effector CTLs against target HMEC plotted according to equation (Eq) that describes the dependence of cytotoxicity from the correlation of cell surface profiles (r) and intensity of tumor-induced changes (k) at the HMEC surface. Autologous EC antigens were used to induce CTLs. Correlation values (coefficient r) were calculated for HMEC surface profiles used to generate antigens (EAA) for eliciting immune response and the surface profile of target HMEC used in same cytotoxicity assay. Results of cytotoxicity assays are projected on the plot. “1►2” -1st letter corresponds to HMEC used to generate antigens (EAA) for eliciting immune response in the cytotoxicity assays; 2nd letter corresponds to the target HMEC used in the same cytotoxicity assay. “G” -1HMECECGS; “M” -1HMECMCF-7; “L”- 1HMECLNCap; “H” -1HMECHepG2. The equation was developed from equations described in the “Discussion.”
The first area is located to the lower left corner corresponding to vaccines with endothelial cell-derived antigens slightly modified by the tumor. These data suggest that the tumor vasculature can be destroyed by autoimmune reactions with the potential of killing endothelial cells in healthy tissues. Such vaccines are less acceptable for medical use since they involve the elicitation of autoimmune responses that limit the practical implementation of these vaccines.54-59
The second area located at the right upper corner corresponds to vaccines with antigens sufficiently modified by the tumor, suggesting that these vaccines are likely safe based on the difference of these antigens relative to antigens associated with endothelial cells in healthy tissues. However, based on the plot it is likely that these vaccines would also be less immunogenic, requiring efficient targeting of immune responses by using endothelial cells well matched with endothelial cells in tumors. Figure 6 clearly describes conditions needed for the design of such efficient antiangiogenic vaccines. The most safe and efficient vaccines should be located to the right upper corner of this plot.
Among pairs of antigens and target cells involved in cytotoxicity assays, projection of antigens from 1HMECHepG2 and target cells 1HMECLNCap (see “H►L” in Fig. 6) is located more closely to the right upper corner, identifying the most efficient target cell killing. These observations can be explained by the specific composition of growth factors released by HepG2 and LNCap cells, however, from a vaccine design perspective and findings described in this report, the observed high killing rate at “H►L” was the result of the most preferable combinations of r and k for this cell pair, due to the killing rate of target cells that is a function of these variables (see equation in Fig. 6).
The next antigens:target pair demonstrating a high target cell killing rate was “M►L.” This antigens:target pair was located near the lower left corner of the plot corresponding to vaccines with relatively high immunogenicity and a low degree of tumor-induced changes at the cell surface. So pair “M►L” describes a condition where target cell killing in vivo was expected to be accompanied with autoimmune reactions resulting in the destruction of vessels in normal tissues.
Finally, one additional feature of this study should be discussed. Besides target cells and antigens (autologous and allogeneic in relation to target cells), monocyte-derived DCs and CTLs were used in an in vitro model of antiangiogenic vaccination. For consistency, these monocyte-derived cells were obtained from one donor and therefore were allogeneic in relation to target cells used in cytotoxicity assays. Utilizing monocyte-derived cells and target HMEC from the same person (i.e., autologous DCs and CTLs) in in vitro experiments further strengthened the findings reported and may provide additional insights for antiangiogenic, anti-cancer vaccinations.
Conclusion
This study showed that tumors induce pronounced, tumor type-dependent changes to HMEC surface targets using an in vitro model of human antiangiogenic vaccination that facilitated HMEC escape from CTL-mediated cell death. Previously, animal and human studies corroborated the capability of in vitro induced specific CTLs to mediate in vivo protection against tumor challenge.60,61 Therefore, data obtained in this study can be directly used for design of endothelial-based cancer vaccines that can be applied in the development of in vivo studies. A direct dependence between CTL killing efficacy and target cell surface profiles, and cells used for targeting immune responses allowed for the accurate design of vaccines matched to their target cells. A direct influence of tumors on the HMEC phenotype was also established in addition to demonstrating a reciprocal dependence between the intensity of tumor-induced changes in HMEC and the immunogenicity of these cells. This information will provide useful information for researchers for the efficient design of vaccines. Specifically, vaccines with antigen compositions divergent from antigens expressed by normal endothelial cells can be designed to avoid the elicitation of autoimmune reactions. Evidence for undesired autoimmune reactions following anti-cancer vaccination has accumulated from work in animal models, as well as clinical trials.54-59 Furthermore, by controlling the intensity of tumor-induced changes in HMEC (e.g., measured by cell surface profiling), it is possible to avoid developing vaccines with low immunogenicity—one of the main reasons cancer vaccines are ineffective.62,63
Materials and Methods
Cell cultures
Two thoracic subcutaneous adipose tissue biopsies were obtained from male patients (49 and 41 y old) undergoing open-thoracic surgical procedures at the National Medico-Surgical Center. The protocol was approved by the Research Ethics Committee and the patients provided their written informed consent. The biopsy specimens were transported to the laboratory in Ringer solution (transport time 45 min), and after removal of the visible fibrous tissue, the fat was finely minced and incubated for 45 h at 37°C in digest solution (0.5 mg/mL collagenase IA (Sigma-Aldrich) prepared with Hank’s Balanced Salt Solution (HBSS) at a ratio of 4:1 (v/v).64 The digested material was then intensely shaken for 2 min then centrifuged (300 × g for 10 min) to separate adipocytes and free oil from the stromovascular components. The stromovascular pellet was resuspended in HBSS and washed 3× by centrifugation (600 × g for 5 min). The resulting pellet was incubated in 0.25% trypsin (activity 300 U/mg, PanEco, Russia) containing 1 mM EDTA for 15 min at RT, followed by 3 washes with PBS containing 0.1% bovine serum albumin (BSA) by centrifugation (600 × g for 5 min). Endothelial cells were isolated using Dynabeads CD31 Endothelial Cells (Invitrogen, San Diego, CA) according to the manufacturer’s protocol and seeded in 6-well tissue culture plates in the presence of endothelial cell growth selection medium [DMEM with d-valine (PanEco, Russia)], 20% fetal bovine serum (FBS, PAA Laboratories), 100 U penicillin, 100 μg/mL streptomycin, 2 mM L-glutamine, 12 U/mL heparin, 30 μL/mL endothelial cell growth supplement from the human hypothalamus) at 37°C in 7.5% CO2. Culture media was changed every 2–3 d and after the first passage cells were grown to 65% confluence and used in future experiments following stimulation with endothelial cell growth supplement (1HMECECGS and 2HMECECGS for HMEC from donor 1 and donor 2, respectively. Superscript numbers correspond to donor number). To provide allogeneic growth stimulus from normal tissues endothelial cell growth supplement (ECGS) was obtained from the human hypothalamus as described.27 Briefly, hypothalamuses harvested at autopsy from the 2 male (46 and 48 y old) donors who died accidentally were kindly provided by Dr. Sirotkin V.I. (The Russian State Medical University) in accordance with Federal law Nº8-FL and Order Nº82 from the Ministry of Health of the Russian Federation. ECGS was prepared by homogenization of approximately 10 g of human hypothalamus on ice in 1.5 mL of cold (4°C) 0.1 M NaCl. The homogenate was stirred at 40°C for 2 h, centrifuged at 13,800 × g for 40 min and the supernatants recovered and fractionated with streptomycin sulfate (Invitrogen) in order to remove soluble lipids. Supernatants were extracted with 0.5% streptomycin sulfate at pH 7.0 for at least 1 h after which the extract was centrifuged at 13,800 × g for 40 min. Supernatants were then collected and used as ECGS.
To obtain HMEC with tumor-induced phenotypes, cell cultures were incubated for 5 d with conditioned medium collected from MCF-7 human breast adenocarcinoma (1HMECMCF-7 and 2HMECMCF-7), LNCap human prostate adenocarcinoma (1HMECLNCap and 2HMECLNCap) and HepG2 human hepatocellular carcinoma (1HMECHepG2 and 2HMECHepG2) cells (ATCC) as described by Folkman et al.65 Briefly, growth medium was aspirated and replaced with the medium to be conditioned. After 48 h, medium was collected, centrifuged for 10 min at 600 × g, filter sterilized (0.2 μm) and added to cell cultures with endothelial cell growth medium at a 1:2 ratio.
MCF-7 cells, LNCap and HepG2 cells were grown to 65% confluence in DMEM (Invitrogen) supplemented with 10% FBS (Invitrogen). Cells were washed 5 times with Hank’s Balanced Salt Solution (HBSS) and used for preparation of tumor-associated antigens (TAA).
Primary fibroblast culture was established from adult skin biopsy (45 y old woman; donor provided written informed consent) as described by Rittie and Fisher.66 Primary culture was cultured in DMEM, 10% FBS at 5% CO2 at 37°C and 3rd passage cells used to obtain fibroblast-associated antigens (FAA).
Immunofluorescent staining and FACS analysis
Endothelial cells grown on the gelatin-coated glass coverslips were fixed for 10 min in 4% paraformaldehyde in PBS at RT. After blocking with casein, cells were incubated with a monoclonal mouse anti-human CD31 (PECAM-1) antibody (clone JC70A; Cat.# M0823) that were visualized following incubation with a biotinylated horse anti-mouse IgG antibody (Vector Labs; Cat.#BA-2000) and streptavidin conjugated with R-phycoerythrin (streptavidin-RPE; Cat.#R0438). The preparations were then stained with DAPI (1 μg/mL in PBS; Sigma) and mounted in Moviol. Samples were visualized using a Leiса DM5000B microscope.
For FACS analysis, endothelial cell cultures were washed once with PBS containing 0.04% EDTA and incubated with 1 mL of cell dissociation solution (Sigma-Aldrich, USA) to release the cells. Cells were washed once with DMEM, 10% FBS, blocked in TBS with 1% BSA (Sigma-Aldrich) for 20 min on ice, and incubated with monoclonal mouse anti-human CD31 antibody (clone JC70A; Cat. #M0823) prepared in TBS, 1% BSA at 2 μg/mL for 30 min on ice. Following incubation, cells were washed with TBS, 1% BSA, then incubated for 30 min on ice with a biotinylated horse anti-mouse IgG secondary antibody (Vector Labs; Cat. #BA-2000). Сells were washed with TBS, 1% BSA, then incubated with streptavidin conjugated with R-phycoerythrin (streptavidin-RPE, Dako; Cat.#R0438) for 30 min on ice. Stained cells were washed with TBS, 1% BSA and resuspended in 0.5 mL TBS, 1% BSA and then analyzed using a FACS Calibur flow cytometer and Cell Quest software (Becton Dickinson). In addition, non-specific binding of the primary anti-human CD31 antibody was ruled out by using adult skin fibroblasts (prepared as described in “Cell culture” section) as a negative control cell line.
Preparation of EAA, TAA and FAA
HMEC, cancer cells (MCF-7 and HepG2) and fibroblasts grown to 65% confluence were washed 5 × with HBSS before being treated with 0.2 µg/mL trypsin (15,000 U/mg, Promega) in HBSS. One mL of trypsin solution was added to each 25 cm2 flask, incubated for 20 min at 37°C in saturated humidity, then collected again and centrifuged (600 × g for 5 min). The resulting supernatant contained cell surface targets and was considered as solutions of endothelial cell-associated antigens (EAA), tumor-associated antigens (TAA) and fibroblast-associated antigens (FAA) in case of HMEC, cancer cells and fibroblasts, respectively.
Cell surface profiling
The obtained antigen solutions were desalted by using ZipTipC18 (Millipore Corp.) according to the manufacturer’s protocol. MALDI samples were prepared using a standard “dried droplet” method with 2,5-dihydroxybenzoic acid (DHB) as the matrix. All mass spectra were acquired on an AutoFLEX MALDI-TOF mass spectrometer (Bruker Daltonik) in linear positive ion mode. The mass spectrometer was set up for priority detection of ions with the m/z range from 600 to 3,500 at a mass accuracy of 80–100 ppm. Mass peak lists were created manually. All peaks above noise level were selected to generate peak lists that represent the cell surface profiles (also known as cell proteomic footprints51).
Cell surface profile processing
Resultant lists of peak intensities from each cell culture (cell surface profiles) were pooled using Matlab software (Mathworks). Two peaks were considered to be related to the same ion if their mass difference did not exceed 0.2 Da. Pooled intensities were processed by principal component analysis (PCA) using the princomp function of the Matlab program. Projections of peak intensities on the first three principal components were used for visualization of the similarity/divergence among cell surface profiles. The distances between profiles on the PCA plot were also depicted as a dendrogram using the Matlab program.
Preparation of EAA-loaded DCs
Monocyte-derived DCs were generated as described previously.67 Briefly, fresh peripheral blood mononuclear cells (PBMCs) from healthy donors were isolated using Ficoll-Hypaque (PanEco, Russia) gradient centrifugation and were then allowed to adhere to culture flasks for 1 h. Non-adherent cells were collected and centrifuged, and cell pellets were mixed with autologous serum containing 10% DMSO and stored in liquid nitrogen. Cryopreserved, non-adherent PBMCs were later used as a source of effector cells (cytotoxic T lymphocytes, CTLs) for cytotoxicity assays. The adherent cell fraction was cultured in RPMI-1640 (PanEco) supplemented with 10% FBS (PAA Laboratories) in the presence of 1,000 U/mL granulocyte macrophage colony-stimulating factor (Sigma-Aldrich) and 1,000 U/mL interleukin-4 (Sigma-Aldrich). After 6 d in culture EAA (1 mL) or FAA (1 mL) were combined with an equal volume (1 mL) of immature DCs. Antigen loading was allowed to occur for 3 h, then DCs were matured with 1,000 U/mL tumor necrosis factor-α (Sigma-Aldrich) for 48 h. Matured, EAA-loaded and FAA-loaded DCs were then used to stimulate CTLs. For successive cycles of CTLs stimulation, aliquots of matured and loaded DCs were cryopreserved and thawed as needed. The freezing method used has been previously described.68
Stimulation of CTLs
EAA-loaded DCs (4.5 × 104) were combined with 9 × 105 autologous non-adherent PBMCs (1:20) in 4 mL of RPMI-1640 medium supplemented with 30 U/mL of clinical grade human interleukin-2 (Ronkoleukin, Russia) and 10% FBS. The culture medium supplemented with interleukin-2 was replaced every third day. Additional antigen-loaded DCs were added to PBMCs seven days after stimulation. After incubation for five days, the non-adherent PBMCs containing stimulated CTLs were washed by centrifugation and used as effector CTLs in cytotoxicity assays.
Cytotoxicity assays
HMEC (2 × 104 cells/well) were seeded into 48-well plates, which yielded 3.7 × 104 cells/well after 48 h. Effector CTLs were then added to HMEC at an effector:target ratio of 8:1. On the 3rd day, target HMEC were washed to remove CTLs and were photographed using an inverted phase contrast microscope. In duplicate wells, attached HMEC were trypsinized and viability was detected using trypan blue exclusion.69 Cell counts were averaged over three measurements and the number of HMEC in the absence of effector CTLs, as well as with CTLs that had been stimulated with FAA-loaded DCs, were used as controls.
Acknowledgments
We would like to thank Dr. A.N. Dobrovolskaya (National Medico-Surgical Center, Moscow, Russia) for kindly providing the adipose tissue biopsy samples and Dr. V.I. Sirotkin (Russian State Medical University, Moscow, Russia) for the autopsy hypothalamuses.
Sources of support
This work was funded by ZAO BioBohemia (Moscow, Russia).
Glossary
Abbreviations:
- EAA
endothelial-associated antigens
- TAA
tumor-associated antigens
- FAA
fibroblast-associated antigens
- ECGS
endothelial cell growth supplement
- HMEC
human microvascular endothelial cells
- DCs
dendritic cells
- PBMCs
peripheral blood mononuclear cells
- PCA
principal component analysis
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/vaccines/article/22828
References
- 1.Lollini PL, Cavallo F, Nanni P, Forni G. Vaccines for tumour prevention. Nat Rev Cancer. 2006;6:204–16. doi: 10.1038/nrc1815. [DOI] [PubMed] [Google Scholar]
- 2.Trial watch : Progress for Phase III cancer vaccines. Nat Rev Drug Discov. 2008;7:966–7. doi: 10.1038/nrd2766. No_authors_listed. [DOI] [PubMed] [Google Scholar]
- 3.Boehm T, Folkman J, Browder T, O’Reilly MS. Antiangiogenic therapy of experimental cancer does not induce acquired drug resistance. Nature. 1997;390:404–7. doi: 10.1038/37126. [DOI] [PubMed] [Google Scholar]
- 4.Bussolino F, Arese M, Audero E, Giraudo E, Marchiò S, Mitola S, et al. Biological aspects of tumour angiogenesis. Cancer Modelling and Simulation. London: Chapman and Hall/CRC; 2003. p. 1–22. [Google Scholar]
- 5.Folkman J. Tumor angiogenesis: therapeutic implications. N Engl J Med. 1971;285:1182–6. doi: 10.1056/NEJM197111182852108. [DOI] [PubMed] [Google Scholar]
- 6.Folkman J. What is the evidence that tumors are angiogenesis dependent? J Natl Cancer Inst. 1990;82:4–6. doi: 10.1093/jnci/82.1.4. [DOI] [PubMed] [Google Scholar]
- 7.Pluda JM. Tumor-associated angiogenesis: mechanisms, clinical implications, and therapeutic strategies. Semin Oncol. 1997;24:203–18. [PubMed] [Google Scholar]
- 8.Carmeliet P, Jain RK. Angiogenesis in cancer and other diseases. Nature. 2000;407:249–57. doi: 10.1038/35025220. [DOI] [PubMed] [Google Scholar]
- 9.Copier J, Dalgleish A. Overview of tumor cell-based vaccines. Int Rev Immunol. 2006;25:297–319. doi: 10.1080/08830180600992472. [DOI] [PubMed] [Google Scholar]
- 10.Wei YQ, Wang QR, Zhao X, Yang L, Tian L, Lu Y, et al. Immunotherapy of tumors with xenogeneic endothelial cells as a vaccine. Nat Med. 2000;6:1160–6. doi: 10.1038/80506. [DOI] [PubMed] [Google Scholar]
- 11.Scappaticci FA, Nolan GP. Induction of anti-tumor immunity in mice using a syngeneic endothelial cell vaccine. Anticancer Res. 2003;23(2B):1165–72. [PubMed] [Google Scholar]
- 12.Corsini E, Gelati M, Calatozzolo C, Alessandri G, Frigerio S, De Francesco M, et al. Immunotherapy with bovine aortic endothelial cells in subcutaneous and intracerebral glioma models in rats: effects on survival time, tumor growth, and tumor neovascularization. Cancer Immunol Immunother. 2004;53:955–62. doi: 10.1007/s00262-004-0529-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Okaji Y, Tsuno NH, Kitayama J, Saito S, Takahashi T, Kawai K, et al. Vaccination with autologous endothelium inhibits angiogenesis and metastasis of colon cancer through autoimmunity. Cancer Sci. 2004;95:85–90. doi: 10.1111/j.1349-7006.2004.tb03175.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen XY, Zhang W, Zhang W, Wu S, Bi F, Su YJ, et al. Vaccination with viable human umbilical vein endothelial cells prevents metastatic tumors by attack on tumor vasculature with both cellular and humoral immunity. Clin Cancer Res. 2006;12:5834–40. doi: 10.1158/1078-0432.CCR-06-1105. [DOI] [PubMed] [Google Scholar]
- 15.Okaji Y, Tsuno NH, Saito S, Yoneyama S, Tanaka M, Nagawa H, et al. Vaccines targeting tumour angiogenesis--a novel strategy for cancer immunotherapy. Eur J Surg Oncol. 2006;32:363–70. doi: 10.1016/j.ejso.2006.01.016. [DOI] [PubMed] [Google Scholar]
- 16.Aird WC. Phenotypic heterogeneity of the endothelium: I. Structure, function, and mechanisms. Circ Res. 2007;100:158–73. doi: 10.1161/01.RES.0000255691.76142.4a. [DOI] [PubMed] [Google Scholar]
- 17.Aird WC. Phenotypic heterogeneity of the endothelium: II. Representative vascular beds. Circ Res. 2007;100:174–90. doi: 10.1161/01.RES.0000255690.03436.ae. [DOI] [PubMed] [Google Scholar]
- 18.St Croix B, Rago C, Velculescu V, Traverso G, Romans KE, Montgomery E, et al. Genes expressed in human tumor endothelium. Science. 2000;289:1197–202. doi: 10.1126/science.289.5482.1197. [DOI] [PubMed] [Google Scholar]
- 19.Khodarev NN, Yu J, Labay E, Darga T, Brown CK, Mauceri HJ, et al. Tumour-endothelium interactions in co-culture: coordinated changes of gene expression profiles and phenotypic properties of endothelial cells. J Cell Sci. 2003;116:1013–22. doi: 10.1242/jcs.00281. [DOI] [PubMed] [Google Scholar]
- 20.Bhati R, Patterson C, Livasy CA, Fan C, Ketelsen D, Hu Z, et al. Molecular characterization of human breast tumor vascular cells. Am J Pathol. 2008;172:1381–90. doi: 10.2353/ajpath.2008.070988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hellebrekers DM, Castermans K, Viré E, Dings RP, Hoebers NT, Mayo KH, et al. Epigenetic regulation of tumor endothelial cell anergy: silencing of intercellular adhesion molecule-1 by histone modifications. Cancer Res. 2006;66:10770–7. doi: 10.1158/0008-5472.CAN-06-1609. [DOI] [PubMed] [Google Scholar]
- 22.Hellebrekers DM, Jair KW, Viré E, Eguchi S, Hoebers NT, Fraga MF, et al. Angiostatic activity of DNA methyltransferase inhibitors. Mol Cancer Ther. 2006;5:467–75. doi: 10.1158/1535-7163.MCT-05-0417. [DOI] [PubMed] [Google Scholar]
- 23.Hellebrekers DM, Melotte V, Viré E, Langenkamp E, Molema G, Fuks F, et al. Identification of epigenetically silenced genes in tumor endothelial cells. Cancer Res. 2007;67:4138–48. doi: 10.1158/0008-5472.CAN-06-3032. [DOI] [PubMed] [Google Scholar]
- 24.Unger RE, Oltrogge JB, von Briesen H, Engelhardt B, Woelki U, Schlote W, et al. Isolation and molecular characterization of brain microvascular endothelial cells from human brain tumors. In Vitro Cell Dev Biol Anim. 2002;38:273–81. doi: 10.1290/1071-2690(2002)038<0273:IAMCOB>2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 25.Bussolati B, Deambrosis I, Russo S, Deregibus MC, Camussi G. Altered angiogenesis and survival in human tumor-derived endothelial cells. FASEB J. 2003;17:1159–61. doi: 10.1096/fj.02-0557fje. [DOI] [PubMed] [Google Scholar]
- 26.Kern PA, Knedler A, Eckel RH. Isolation and culture of microvascular endothelium from human adipose tissue. J Clin Invest. 1983;71:1822–9. doi: 10.1172/JCI110937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maciag T, Cerundolo J, Ilsley S, Kelley PR, Forand R. An endothelial cell growth factor from bovine hypothalamus: identification and partial characterization. Proc Natl Acad Sci USA. 1979;76:5674–8. doi: 10.1073/pnas.76.11.5674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kumar S, West DC, Ager A. Heterogeneity in endothelial cells from large vessels and microvessels. Differentiation. 1987;36:57–70. doi: 10.1111/j.1432-0436.1987.tb00181.x. [DOI] [PubMed] [Google Scholar]
- 29.Lang I, Pabst MA, Hiden U, Blaschitz A, Dohr G, Hahn T, et al. Heterogeneity of microvascular endothelial cells isolated from human term placenta and macrovascular umbilical vein endothelial cells. Eur J Cell Biol. 2003;82:163–73. doi: 10.1078/0171-9335-00306. [DOI] [PubMed] [Google Scholar]
- 30.Shreeniwas R, Ogawa S, Cozzolino F, Torcia G, Braunstein N, Butura C, et al. Macrovascular and microvascular endothelium during long-term hypoxia: alterations in cell growth, monolayer permeability, and cell surface coagulant properties. J Cell Physiol. 1991;146:8–17. doi: 10.1002/jcp.1041460103. [DOI] [PubMed] [Google Scholar]
- 31.Hewett PW. Identification of tumour-induced changes in endothelial cell surface protein expression: an in vitro model. Int J Biochem Cell Biol. 2001;33:325–35. doi: 10.1016/S1357-2725(01)00020-6. [DOI] [PubMed] [Google Scholar]
- 32.Swerlick RA, Lee KH, Wick TM, Lawley TJ. Human dermal microvascular endothelial but not human umbilical vein endothelial cells express CD36 in vivo and in vitro. J Immunol. 1992;148:78–83. [PubMed] [Google Scholar]
- 33.Swerlick RA, Lee KH, Li LJ, Sepp NT, Caughman SW, Lawley TJ. Regulation of vascular cell adhesion molecule 1 on human dermal microvascular endothelial cells. J Immunol. 1992;149:698–705. [PubMed] [Google Scholar]
- 34.Lee KH, Lawley TJ, Xu YL, Swerlick RA. VCAM-1-, ELAM-1-, and ICAM-1-independent adhesion of melanoma cells to cultured human dermal microvascular endothelial cells. J Invest Dermatol. 1992;98:79–85. doi: 10.1111/1523-1747.ep12495643. [DOI] [PubMed] [Google Scholar]
- 35.Pötzsch B, Grulich-Henn J, Rössing R, Wille D, Müller-Berghaus G. Identification of endothelial and mesothelial cells in human omental tissue and in omentum-derived cultured cells by specific cell markers. Lab Invest. 1990;63:841–52. [PubMed] [Google Scholar]
- 36.Hewett PW, Murray JC, Price EA, Watts ME, Woodcock M. Isolation and characterization of microvessel endothelial cells from human mammary adipose tissue. In Vitro Cell Dev Biol Anim. 1993;29A:325–31. doi: 10.1007/BF02633961. [DOI] [PubMed] [Google Scholar]
- 37.Hull MA, Hewett PW, Brough JL, Hawkey CJ. Isolation and culture of human gastric endothelial cells. Gastroenterology. 1996;111:1230–40. doi: 10.1053/gast.1996.v111.pm8898637. [DOI] [PubMed] [Google Scholar]
- 38.Hewett PW, Murray JC. Immunomagnetic purification of human microvessel endothelial cells using Dynabeads coated with monoclonal antibodies to PECAM-1. Eur J Cell Biol. 1993;62:451–4. [PubMed] [Google Scholar]
- 39.Hewett PW, Murray JC. Human omental mesothelial cells: a simple method for isolation and discrimination from endothelial cells. In Vitro Cell Dev Biol. 1994;30:145–7. doi: 10.1007/BF02631436. [DOI] [PubMed] [Google Scholar]
- 40.Jin Y, Liu Y, Antonyak M, Peng X. Isolation and characterization of vascular endothelial cells from murine heart and lung. Methods Mol Biol. 2012;843:147–54. doi: 10.1007/978-1-61779-523-7_14. [DOI] [PubMed] [Google Scholar]
- 41.Jankowski R. A Comparison of Commercially-Available Human Skeletal Muscle Cells and Media for Research Applications. Nat Methods. 2011 [Application Notes] [Google Scholar]
- 42.Lokhov PG, Balashova EE. Cellular cancer vaccines: an update on the development of vaccines generated from cell surface antigens. J Cancer. 2010;1:230–41. doi: 10.7150/jca.1.230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Thompson PL, Dessureault S. In: Shurin MR, Smolkin YS, editors. Immune-Mediated Diseases From Theory to Therapy New York: Springer; 2007. p. 345-55. [Google Scholar]
- 44.de Gruijl TD, van den Eertwegh AJ, Pinedo HM, Scheper RJ. Whole-cell cancer vaccination: from autologous to allogeneic tumor- and dendritic cell-based vaccines. Cancer Immunol Immunother. 2008;57:1569–77. doi: 10.1007/s00262-008-0536-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chiang CL, Benencia F, Coukos G. Whole tumor antigen vaccines. Semin Immunol. 2010;22:132–43. doi: 10.1016/j.smim.2010.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cheever MA, Allison JP, Ferris AS, Finn OJ, Hastings BM, Hecht TT, et al. The prioritization of cancer antigens: a national cancer institute pilot project for the acceleration of translational research. Clin Cancer Res. 2009;15:5323–37. doi: 10.1158/1078-0432.CCR-09-0737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lang JM, Andrei AC, McNeel DG. Prioritization of cancer antigens: keeping the target in sight. Expert Rev Vaccines. 2009;8:1657–61. doi: 10.1586/erv.09.134. [DOI] [PubMed] [Google Scholar]
- 48.Balashova EE, Lokhov PG. Proteolytically-cleaved fragments of cell surface proteins stimulate a cytotoxic immune response against tumor-activated endothelial cells in vitro. J Cancer Sci Ther. 2010;2:126–31. doi: 10.4172/1948-5956.1000037. [DOI] [Google Scholar]
- 49.Balashova EE, Lokhov PG. Proteolytically-cleaved fragments of cell-surface proteins from live tumor cells stimulate anti-tumor immune response in vitro. Journal of Carcinogenesis & Mutagenesis. 2010;1:1–3. doi: 10.4172/2157-2518.1000103. [DOI] [Google Scholar]
- 50.Balashova EE, Dashtiev MI, Lokhov PG. Proteomic footprinting of drug-treated cancer cells as a measure of cellular vaccine efficacy for the prevention of cancer recurrence. Mol Cell Proteomics. 2012;11:M111–, 014480. doi: 10.1074/mcp.M111.014480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lokhov P, Balashova E, Dashtiev M. Cell proteomic footprint. Rapid Commun Mass Spectrom. 2009;23:680–2. doi: 10.1002/rcm.3928. [DOI] [PubMed] [Google Scholar]
- 52.Nolan KF, Yun CO, Akamatsu Y, Murphy JC, Leung SO, Beecham EJ, et al. Bypassing immunization: optimized design of “designer T cells” against carcinoembryonic antigen (CEA)-expressing tumors, and lack of suppression by soluble CEA. Clin Cancer Res. 1999;5:3928–41. [PubMed] [Google Scholar]
- 53.Yun CO, Nolan KF, Beecham EJ, Reisfeld RA, Junghans RP. Targeting of T lymphocytes to melanoma cells through chimeric anti-GD3 immunoglobulin T-cell receptors. Neoplasia. 2000;2:449–59. doi: 10.1038/sj.neo.7900108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ludewig B, Ochsenbein AF, Odermatt B, Paulin D, Hengartner H, Zinkernagel RM. Immunotherapy with dendritic cells directed against tumor antigens shared with normal host cells results in severe autoimmune disease. J Exp Med. 2000;191:795–804. doi: 10.1084/jem.191.5.795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dudley ME, Wunderlich JR, Robbins PF, Yang JC, Hwu P, Schwartzentruber DJ, et al. Cancer regression and autoimmunity in patients after clonal repopulation with antitumor lymphocytes. Science. 2002;298:850–4. doi: 10.1126/science.1076514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Phan GQ, Yang JC, Sherry RM, Hwu P, Topalian SL, Schwartzentruber DJ, et al. Cancer regression and autoimmunity induced by cytotoxic T lymphocyte-associated antigen 4 blockade in patients with metastatic melanoma. Proc Natl Acad Sci USA. 2003;100:8372–7. doi: 10.1073/pnas.1533209100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Maker AV, Phan GQ, Attia P, Yang JC, Sherry RM, Topalian SL, et al. Tumor regression and autoimmunity in patients treated with cytotoxic T lymphocyte-associated antigen 4 blockade and interleukin 2: a phase I/II study. Ann Surg Oncol. 2005;12:1005–16. doi: 10.1245/ASO.2005.03.536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Attia P, Phan GQ, Maker AV, Robinson MR, Quezado MM, Yang JC, et al. Autoimmunity correlates with tumor regression in patients with metastatic melanoma treated with anti-cytotoxic T-lymphocyte antigen-4. J Clin Oncol. 2005;23:6043–53. doi: 10.1200/JCO.2005.06.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Finn OJ. Cancer vaccines: between the idea and the reality. Nat Rev Immunol. 2003;3:630–41. doi: 10.1038/nri1150. [DOI] [PubMed] [Google Scholar]
- 60.Paczesny S, Shi H, Saito H, Mannoni P, Fay J, Banchereau J, et al. Measuring melanoma-specific cytotoxic T lymphocytes elicited by dendritic cell vaccines with a tumor inhibition assay in vitro. J Immunother. 2005;28:148–57. doi: 10.1097/01.cji.0000154247.97254.ef. [DOI] [PubMed] [Google Scholar]
- 61.Ossevoort MA, Feltkamp MC, van Veen KJ, Melief CJ, Kast WM. Dendritic cells as carriers for a cytotoxic T-lymphocyte epitope-based peptide vaccine in protection against a human papillomavirus type 16-induced tumor. J Immunother Emphasis Tumor Immunol. 1995;18:86–94. doi: 10.1097/00002371-199508000-00002. [DOI] [PubMed] [Google Scholar]
- 62.Bodey B, Bodey B, Jr., Siegel SE, Kaiser HE. Failure of cancer vaccines: the significant limitations of this approach to immunotherapy. Anticancer Res. 2000;20:2665–76. [PubMed] [Google Scholar]
- 63.Schreiber TH, Raez L, Rosenblatt JD, Podack ER. Tumor immunogenicity and responsiveness to cancer vaccine therapy: the state of the art. Semin Immunol. 2010;22:105–12. doi: 10.1016/j.smim.2010.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hutley LJ, Herington AC, Shurety W, Cheung C, Vesey DA, Cameron DP, et al. Human adipose tissue endothelial cells promote preadipocyte proliferation. Am J Physiol Endocrinol Metab. 2001;281:E1037–44. doi: 10.1152/ajpendo.2001.281.5.E1037. [DOI] [PubMed] [Google Scholar]
- 65.Folkman J, Haudenschild CC, Zetter BR. Long-term culture of capillary endothelial cells. Proc Natl Acad Sci USA. 1979;76:5217–21. doi: 10.1073/pnas.76.10.5217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rittié L, Fisher GJ. Isolation and culture of skin fibroblasts. Methods Mol Med. 2005;117:83–98. doi: 10.1385/1-59259-940-0:083. [DOI] [PubMed] [Google Scholar]
- 67.Romani N, Gruner S, Brang D, Kämpgen E, Lenz A, Trockenbacher B, et al. Proliferating dendritic cell progenitors in human blood. J Exp Med. 1994;180:83–93. doi: 10.1084/jem.180.1.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.John J, Dalgleish A, Melcher A, Pandha H. Cryopreserved dendritic cells for intratumoral immunotherapy do not require re-culture prior to human vaccination. J Immunol Methods. 2005;299:37–46. doi: 10.1016/j.jim.2004.12.014. [DOI] [PubMed] [Google Scholar]
- 69.Yun CO, Nolan KF, Beecham EJ, Reisfeld RA, Junghans RP. Targeting of T lymphocytes to melanoma cells through chimeric anti-GD3 immunoglobulin T-cell receptors. Neoplasia. 2000;2:449–59. doi: 10.1038/sj.neo.7900108. [DOI] [PMC free article] [PubMed] [Google Scholar]