Abstract
Soft tissue sarcomas are a heterogeneous group of tumors associated with poor clinical outcome. Although a subset of soft tissue sarcomas is characterized by simple karyotypes and recurrent chromosomal translocations, the mechanisms driving cytogenetically complex sarcomas are largely unknown. Clinical evidence led us to partially inactivate Pten and Tp53 in the smooth muscle lineage of mice, which developed high-grade undifferentiated pleomorphic sarcomas, leiomyosarcomas, and carcinosarcomas that widely recapitulate the human disease, including the aberrant karyotype and metastatic behavior. Pten was found haploinsufficient, whereas the wild-type allele of Tp53 invariably gained point mutations. Gene expression profiles showed up-regulated Notch signaling in PtenΔ/+Tp53Δ/+ tumors compared with Pten+/+Tp53Δ/+ tumors. Consistently, Pten silencing exacerbated the clonogenic and invasive potential of Tp53-deficient bone marrow–derived mouse mesenchymal stem cells and tumor cells and activated the Notch pathway. Moreover, the increased oncogenic behavior of PtenΔ/+Tp53Δ/+ and shPten-transduced Pten+/+Tp53Δ/+ tumor cells was counteracted by treatment with a γ-secretase inhibitor, suggesting that the aggressiveness of those tumors can be attributed, at least in part, to enhanced Notch signaling. This study demonstrates a cooperative role for Pten and Tp53 suppression in complex karyotype sarcomas while establishing Notch as an important functional player in the cross talk of these pathways during tumor progression. Our results highlight the importance of molecularly subclassifying patients with high-grade sarcoma for targeted treatments.
Soft tissue sarcomas (STSs) are rare but represent some of the most aggressive adult and childhood malignancies. Believed to originate from as-yet poorly defined mesenchymal stem/progenitor cells, they can arise in connective tissue (eg, tendons, fat, and fascia), muscle, nerve, or blood vessel.1 However, regardless of the tissue of origin, STSs share an overall poor prognosis.2 One reason is that they are frequently discovered at more advanced stages, but the deeper problem is that there is limited understanding of their pathogenesis and, thus, little guidance for developing targeted therapies. Although some STSs harbor characteristic chromosomal translocations or simple genetic mutations, the rest show more pronounced genomic instability, with a complex karyotype and multiple chromosomal aberrations that are not consistent among tumors within a particular subtype.1,3 This larger category, for which there have been no driver mutations identified, includes high-grade undifferentiated pleomorphic sarcomas (HGUPSs; the most common sarcomas diagnosed in the extremities), leiomyosarcomas (LMSs; which arise in smooth muscle), and carcinosarcomas (CSs; a mixed epithelial/connective tissue malignancy).1,3
Despite the heterogeneity of STSs, there are hints that some alterations are shared among multiple tumor types. Individuals with Li-Fraumeni syndrome, caused by a germ-line TP53 mutation, have an increased tendency to develop STSs, and several different sarcomas have shown disruptions in the Tp53 tumor-suppressor pathway.1,3,4 Individuals with neurofibromatosis type 1, who carry NF1 mutations leading to up-regulation of Ras signaling, are also predisposed to develop STS. Mouse models of these diseases replicate the broad spectrum of neoplasia seen in human patients, but it has been difficult to engineer in vivo models that develop nonsyndromic forms of sarcoma, particularly HGUPS and LMS. There may be a requirement for multiple genetic hits (eg, intramuscular delivery of an adenovirus expressing the Cre recombinase in mice with conditional mutations in both Kras and Tp53 initiated high-grade sarcomas with myofibroblastic differentiation).5 Similarly, uterine LMSs that arose after Tp53 and BRCA1 were conditionally deleted under the anti-müllerian hormone type II receptor (Amhr2),6 and simultaneous inactivation of both genes significantly accelerated tumor progression. Despite their usefulness, these models likely represent only a portion of all HGUPSs, LMSs, and/or CSs, given the molecular heterogeneity of these tumors. In fact, to our knowledge, there is no mouse model of CS.
We have, therefore, sought to generate mouse models based on genetic defects widely found in tumor tissue from patients with STS in the hope that these models would allow us to dissect molecular pathways frequently involved in sarcoma formation and metastasis and to test the efficacy of new therapies. Herein, we describe novel conditional mice heterozygous for Tp53 and/or Pten in the smooth muscle (SM) lineage. These mice develop HGUPS, LMS, and CS, which recapitulate the histological and biological features of the corresponding human neoplasia, including their metastatic behavior, and provide insight into the molecular mechanisms involved in these tumors.
Materials and Methods
Generation of Knockout Mice, Animal Husbandry, and Genotyping
PtenL/L7,8 and Tp53L/L9 (National Cancer Institute mouse repository, Bethesda, MD) were intercrossed, and the progeny were crossed with Tagln-cre23 [Tg(Tagln-cre23)1Her/J; Jackson Labs, Bar Harbor, ME] mice for smooth muscle–specific deletion of one or two alleles of Pten and Tp53. The mouse cohort followed up for overall survival up to 27 months included 90 animals: 13 Tagln-cre+/PtenΔ/+Tp53Δ/+, 28 Tagln-cre+/PtenΔ/+Tp53+/+, 16 Tagln-cre+/Pten+/+Tp53Δ/+, 5 Tagln-cre+/PtenΔ/ΔTp53+/+, and 28 control mice (including 2 Tagln-cre−/PtenΔ/+Tp53Δ/+, 7 Tagln-cre−/PtenΔ/+Tp53+/+, 1 Tagln-cre−/Pten+/+Tp53Δ/+, 3 Tagln-cre−/PtenΔ/ΔTp53+/+, and 15 Tagln-cre−/Pten+/+Tp53+/+). Another mouse cohort was maintained to increase the amount of tumors isolated and explanted. B6;129-Gt(Rosa)26Sortm2Sho/J mice (Rosa26-LSL-EGFP, number 004077; Jackson Labs10) and B6.129S4-Gt(Rosa)26Sortm1Sor/J (Rosa26-LSL-LacZ, number 003474; Jackson Labs11) were obtained. All mice were handled according to New York University (NYU) Institutional Animal Care and Use Committee approved protocols (numbers 061108-03 and 100108-01). For genotyping, tail DNA was subjected to polymerase chain reaction analysis using the primers and protocols described.7,8,10,11
Mouse Histopathological and IHC Characteristics
Mouse tissue samples collected were fixed in 10% neutral-buffered formalin, processed, and embedded in paraffin, according to standard protocols. Sections (5 μm thick) were prepared for antibody detection and H&E staining. Immunohistochemistry (IHC) was conducted following the standard avidin-biotin immunoperoxidase staining procedure. Diaminobenzidine was used as the chromogen, and hematoxylin was used to counterstain nuclei. Antibodies used were desmin (DER11, number 2530; Ventana Medical Systems, Tucson, AZ) and proliferating cell nuclear antigen (PCNA; PC10, number MS106; Neomarkers, Fremont, CA).
Generation of Cell Lines and Culture Conditions
Tumor fragments were minced in HBSS with antibiotics/antimycotics (Hyclone Thermo, South Logan, UT) and dissociated by incubation on 100 U/mL collagenase V (Sigma, St. Louis, MO) for 30 minutes at 37°C. Twenty-four hours later, media were changed to mouse mesenchymal stem cell medium that consists of Iscove’s modified Dulbecco’s medium (Gibco, Grand Island, NY) with 2 mmol/L l-glutamine (Hyclone Thermo), 9% fetal bovine serum (Cell Gro, Manassas, VA), and 9% donor horse serum (Cell Gro).
RNA Extraction, RT-PCR, and Real-Time PCR Analyses
Total RNA was extracted using Qiazol (Qiagen, Valencia, CA), followed by RNeasy Mini Kit (Qiagen). Semiquantitative RT-PCR was performed by reverse transcribing 500 ng of total RNA with a Taqman Assay kit (Applied Biosystems, Austin, TX), followed by standard PCR using RNA-specific primers. Quantitative real-time RT-PCR was performed using FastStart SYBR Green MasterMix (Roche, Basel, Switzerland) and BioRad iCycler equipment (Hercules, CA).
Expression Analysis
Expression profiling of PtenΔ/+Tp53Δ/+ (n = 4) and Pten+/+Tp53Δ/+ (n = 5) murine HGUPS was performed using the Affymetrix Genechip system (Affymetrix, Santa Clara, CA). Total RNA was extracted with Qiazol (Qiagen), and quality and quantity were determined using a Bioanalyzer (Agilent Technologies, Santa Clara, CA) and Nanodrop ND-1000 (Thermo Scientific, Waltham, MA). Total RNA (100 ng) was used to prepare cDNA following the Affymetrix 3′IVT Express Kit labeling protocol (Affymetrix). Standardized array processing procedures recommended by Affymetrix were performed, including hybridization, fluidics processing, and scanning of the Affymetrix Mouse Genome-430 2.0 arrays. The raw data (.cel files) were normalized for probe-level summarization by the robust multichip average and analyzed by using Pavlidis template matching (P < 0.05). For these steps, GeneSpring software version GX11 (Agilent Technologies) was used. Gene set enrichment analysis (GSEA)12,13 was used to identify significantly enriched gene expression patterns underlying PtenΔ/+Tp53Δ/+ HGUPS, by querying the C2 (curated pathways) and C5 (Gene Ontologies) categories of the GSEA MSigDB, version 3.0 (Broad Institute, Boston, MA). The raw array data were deposited at the National Center for Biotechnology Information Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo; accession number GSE42103).
DNA Extraction, Loss of Heterozygosity, and Sequencing Analyses
For genomic DNA extraction, formalin-fixed, paraffin-embedded (FFPE) tissues (10 μm thick) were cut and placed on uncharged glass slides. Tissues were deparaffinized with xylene, washed in ethanol, and digested with proteinase K. DNA was extracted using the Qiagen FFPE genomic DNA isolation kit. Total DNA quantity and quality were quantitated using Nanodrop ND-1000 (Thermo Scientific). Primer sequences used for DNA amplification are listed in Table 1. For sequencing, PCR fragments were cloned into pCRII-TOPO vector, and >20 colonies were sequenced using T7 and M13R primers.
Table 1.
Primers Used for Sequencing, Genotyping, and Expression Array Validation
| Gene | No. | Sequence |
|---|---|---|
| Tp53 | 11 Forward | 5′-ATGACTGCCATGGAGGAGTCACA-3′ |
| Tp53 | 11 Reverse | 5′-TCAGTCTGAGTCAGGCCCCA-3′ |
| Tp53 | 1 Forward | 5′-CACAAAAACAGGTTAAACCCAG-3′ |
| Tp53 | 1 Reverse | 5′-AGCACATAGGAGGCAGAGAC-3′ |
| Tp53 | 10 Reverse | 5′-AAGGGGTATGAGGGACAAGG-3′ |
| Pten | 2 Forward | 5′-CATCTCTCTCCTCCTTTTTCTTCA-3′ |
| Pten | 4 Reverse | 5′-TTCATGGTATTTTATCCCTCTTGA-3′ |
| Pten | 2 Reverse | 5′-CAAACATCATCTTGTGAAACAGC-3′ |
| Pten | 3 Forward | 5′-GGCAAATTTTTAAAGGCACAAG-3′ |
| Pten | 3 Reverse | 5′-AAGTTGAACTGCTAGCCTCTGG-3′ |
| Pten | 4 Forward | 5′-ACTTTGAGTTCCCTCAGCCA-3′ |
| Pten | 5 Reverse | 5′-AAAAGTTCCCCTGCTGATGATTTGT-3′ |
| Pten | 6 Forward | 5′-TGTTTTTGACCAATTAAAGTAGGCTGTG-3′ |
| Pten | 7 Forward | 5′-TTCTCTTGAGCACTGTTTCACAGGC-3′ |
| Hes1 | Forward | 5′-AAAGCCTATCATGGAGAAGAGGCG-3′ |
| Hes1 | Reverse | 5′-GGAATGCCGGGAGCTATCTTTCTT-3′ |
| Notch1 | Forward | 5′-ACACTGACCAACAAATGGAGG-3′ |
| Notch1 | Reverse | 5′-GTGCTGAGGCAAGGATTGGA-3′ |
| Jak1 | Forward | 5′-CTCTCTGTCACAACCTCTTCGC-3′ |
| Jak1 | Reverse | 5′-TTGGTAAAGTAGAACCTCATGCG-3′ |
| Ripk1 | Forward | 5′-GAAGACAGACCTAGACAGCGG-3′ |
| Ripk1 | Reverse | 5′-CCAGTAGCTTCACCACTCGAC-3′ |
| R18s1 | Forward | 5′-TTGTACACACCGCCCGTCGC-3′ |
| R18s1 | Reverse | 5′-CTTCTCAGCGCTCCGCCAGG-3′ |
| Gapdh | Forward | 5′-ACCGCCGTTATGAAATCTTG-3′ |
| Gapdh | Reverse | 5′-CACATTGGGGGTAGGAACAC-3′ |
Western Blot Analysis
Cell pellets were lysed using radioimmunoprecipitation (RIPA) assay buffer (Thermo Scientific), supplemented with protease and phosphatase inhibitors (Roche). Tumors were lysed with the same buffer, adding 0.1 mg/mL aprotinin, 0.1 mg/mL leupeptin, and 0.5 μg/mL pepstatin. Protein (30 μg) was resolved on 4% to 20% Tris-glycine SDS-PAGE gels (Invitrogen, Carlsbad, CA). Proteins were transferred to polyvinylidene difluoride (PVDF) membranes, blocked with 1% bovine serum albumin (BSA) in Tris-buffered saline (TBS)-Tween 0.01% for 1 hour, and incubated overnight at 4°C with primary antibody [PTEN, number 9552 (Cell Signaling, Danvers, MA) and tubulin, number T9026 (Sigma)]. Membranes were incubated with horseradish peroxidase–conjugated secondary antibodies for 1 hour before development using Enhanced Chemoluminescence Plus (ECL Plus), a Western blot detection kit (GE Healthcare, Piscataway, NJ).
Bone Marrow Extraction
Bone marrow (BM) was collected by flushing the long bones of murine tibias and femurs with an insulin syringe. BM was washed with HBSS twice. Cells were plated in a 75-mm2 flask in complete isolation media, which consisted of RPMI 1640 medium (Gibco, Carlsbad, CA), 9% fetal bovine serum (Cell Gro), 9% horse serum (HyClone Thermo), 100 U/mL penicillin/streptomycin (Thermo Scientific), and 2 mmol/L l-glutamine (Invitrogen). After 48 hours, adherent cells were washed with PBS and fresh complete isolation medium was added every 3 to 4 days. After 2 weeks in culture, cells were lifted with 0.25% trypsin (Gibco) and plated for different experiments in complete expansion medium (mouse mesenchymal stem cell medium).
Adenovirus Infection
BM-derived cells were infected in low serum with Ad5–CMV–Cre–green fluorescent protein (GFP). Adenoviruses were purchased from Baylor College of Medicine (Houston, TX). At 7 hours after infection, mouse mesenchymal stem cell medium was added and changed every 2 days. After 5 days, cells were plated.
Lentiviral Infection
Lentiviruses were propagated using previously described methods.14 GIPZ short-hairpin (shPten) and empty vector were purchased from Open Biosystems (Huntsville, AL). Cell lines were infected 3× and selected with 1 μg/mL puromycin 48 hours after transfection to generate stable cell lines.
Clonability
A total of 0.5, 1, or 2 × 103 cells were seeded in 6-well plates. Medium was replaced every 3 days, and after 5 to 10 days, cells were fixed and stained with 1% crystal violet. After extensive washing, colonies were counted. When indicated, cells were treated with 500 nmol/L Compound E (ALX-270-415-C250; Enzo Life Sciences, Farmingdale, NY).
Growth Curves
A total of 1 × 103 cells were plated in 96-well plates. Every 2 days, cells were fixed and stained with 1% crystal violet. After extensive washing, 15% acetic acid was used to dissolve the stain, and color intensity was quantified at 595 nm. Relative growth is represented as the percentage of growth compared with the control. When indicated, cells were treated with 500 nmol/L Compound E.
Fibronectin Transwell Invasion Assay
A suspension of 8 × 103 cells was added to cell culture inserts (Falcon, Austin TX) containing a polycarbonate filter with 8-μm-diameter pores coated with 100 μg/mL fibronectin and 2.5% bovine serum albumin. Cells were incubated for 19 hours under standard culture conditions. Tumor cells remaining on the topside of the membrane were removed, and cells that had migrated to the underside were fixed and stained with 1% crystal violet. Five preset fields per insert were imaged and scored. When indicated, cells were treated 1 day before seeding and during the experiment with 500 nmol/L Compound E.
β-Galactosidase Staining
Tissues were fixed in 1% paraformaldehyde/0.2% glutaraldehyde in PBS containing 2 mmol/L MgCl2, 5 mmol/L EGTA, and 0.02% NP-40 at 4°C for 2 hours. After rinsing in PBS and staining with X-Gal for 4 hours, tissues were dehydrated with isopropanol and embedded in paraffin. Tissue was divided into sections (5 μm thick) and counterstained with eosin.
Fluorescence-Activated Cell Sorter Analysis
Fluorescence-activated cell sorting of primary BM cells was performed on a BD LSR II flow cytometer. Antibodies were purchased from eBioscience (San Diego, CA): c-kit APC number 17-1171-82, Stem cell antigen-1 (Sca) PE-Cy7 number 25-5981-82, Myeloid differentiation antigen (Gr1) APC-Cy7 number 47-5931-82, Macrophage-1 antigen (Mac1) APC-Cy7 number 47-0112-82, TER-119 APC-Cy7 number 47-5921-82, B220 APC-Cy7 number 47-0452-82, CD4 APC-Cy7 number 47-0042-82, CD8 APC-Cy7 number 47-0081-82, and IL7R APC-Cy7 number 47-1271-82. Data were analyzed using FlowJo software version X (10.0.0) (Tree Star, Ashland, OR).
Karyotyping
Subconfluent cultures were treated with 0.05 μg/mL KaryoMAX Colcemid (Invitrogen) for 40 to 60 minutes before harvesting. Cells were trypsinized to a single-cell suspension, pelleted at 180 × g for 8 minutes, and resuspended in warm 0.075 mol/L KCl. After an 8-minute incubation at 37°C, the hypotonic solution was diluted with three parts of Carnoy's fixative (3:1 methanol/glacial acetic acid) and gently mixed, and the cells were pelleted as before. The supernatant was removed, and the cell pellet was loosened by gently flicking the base of the tube. The cells were then fixed in three changes of fixative. Fixed cell suspensions were stored at −20°C. Fixed metaphase preparations were dropped onto dry slides, and the quality of spreading was assessed by phase microscopy. Spreading was adjusted by altering the drying time, increasing local humidity (water bath), or applying heat (hotplate). Slides were then air dried and aged (at 37°C for several days or at 60°C for several hours). Aged slides were immersed in 0.08 μg/mL DAPI in 2× standard saline citrate for 3 minutes, rinsed, air dried, mounted in antifade solution (Vectashield; Vector Labs, Burlingame, CA), and stored at 4°C. DAPI-stained slides were scanned using a Nikon E800 epifluorescence microscope (Melville, NY) equipped with a digital imaging system (Applied Spectral Imaging, Vista, CA). Metaphase images were inverted to resemble conventional G-banding and karyotyped using BandView software (Applied Spectral Imaging, ASI; Carlsbad, CA). Where possible, a minimum of 20 metaphases was examined for each sample. To avoid confusion, abnormal karyotypes were described according to the International System for Human Cytogenetic Nomenclature, rather than the Standardized Genetic Nomenclature for Mice, because the experimental systems are intended to model human disease.
Patients and Tumor Collection
Paraffin blocks of the tumor specimens were obtained from the NYU Pathology Department at Bellevue Hospital Center (H10457-014; New York, NY) and Northwestern University (STU7147; Chicago, IL).
Extraction of Tumor Genomic DNA
For FFPE samples, tissue sections from blocks were placed into microcentrifuge tubes (five sections, 10 μm thick), and consecutive H&E-stained slides were obtained from each block. Tumor-enriched areas were identified, and core punches were taken from the corresponding region of the block. DNA was extracted from FFPE cores using a Qiagen DNeasy kit, according to the manufacturer’s directions.
Mass Spectrometric Genotyping
A two-step process was used as previously described.15 DNA extracted from FFPE blocks was quantified using picogreen analysis. Whole genome amplified DNA was used as input for multiplex PCR using primers from OncoMap 3 and OncoMap 3 Extended (Dana-Farber Cancer Institute, Boston, MA), which together compose 1047 independent assays interrogating 983 unique mutations across 116 genes. Mass spectrometric genotyping using iPLEX technology (Sequenom Inc, San Diego, CA) was performed, and candidate mutations identified were subsequently subject to a second round of homogeneous Mass-Extend, using independent primers and probes. Candidate calls were classified as aggressive or conservative, depending on their apparent robustness. Sample quality was considered adequate for analysis if >80% of the attempted genotypes resulted in identifiable products.
Statistical Analysis
Statistical significance was determined using GraphPad Prism Software version 5 (La Jolla, CA). Overall survival was analyzed by Kaplan-Meier curves, and the log-rank (Mantel-Cox) test was used to determine differences in survival. Statistical significance was defined as P < 0.05. Two-tailed Student’s t-tests were used to compare time points.
Results
PTEN and TP53 Loci Are Concurrently Deleted in Many Human LMS and HGUPS Tumors
We performed mutational analysis of 40 human LMS samples and 7 normal myometria using OncoMap3 Core and Extended panels,15 assessing 1047 mutations from 116 genes (Supplemental Table S1 and Supplemental Table S2). Independent validation of 73 candidate mutations on 44 genes only confirmed six mutations in two genes, EGFR and TP53, none of which co-occurred in the same tumor sample (Supplemental Figure S1A). Specifically, three (7.5%) of the LMSs harbored a low-frequency germ-line single-nucleotide polymorphism in EGFR (S703F), and another three cases (7.5%) carried mutations in TP53 (one V157F and two R273C amino acid substitutions). Overall, the incidence of hotspot mutations in the most commonly altered oncogenes and tumor suppressors was extremely low in our LMS cohort, in accordance with a recent analysis covering fewer candidate mutations.16
We then searched for chromosomal aberrations in several tumor suppressor loci using published single-nucleotide polymorphism array analysis17 of DNA from 27 LMSs and three MFHs/HGUPs paired with their corresponding normal tissue (Supplemental Figure S1B). (HGUPS, alias undifferentiated sarcoma, has, at times, been coextensive with MFH or malignant fibrous histiocytoma; a history of the changes in subclassification is given by Matushansky et al.18) The most frequent alteration observed was partial deletion of chromosome 10 at the PTEN locus, in approximately 50% to 70% of cases. Moreover, 85% of LMSs showed a reduced copy number of PTEN and/or TP53. Strikingly, although 11% of LMS tumors displayed only TP53 loss, 67% harbored concomitant reduction of both PTEN and TP53 copy numbers (Supplemental Figure S1B).
In agreement with the genomic findings, data mining of several independent expression profiles revealed that human HGUPS (undifferentiated/MFH) and LMS had significantly lower expression of PTEN and TP53 than either liposarcomas (Supplemental Figure S1C)19–21 or other STSs (Supplemental Figure S1D),22 and 33% of HGUPS and LMS had concomitantly reduced mRNA levels of both PTEN and TP53.19
In summary, concurrent reduction of PTEN and TP53 levels, resulting from genetic deletions or chromosomal losses, is characteristic of a large subgroup of human HGUPS and LMS.
Partial Inactivation of Pten and Tp53 in the SM Lineage Leads to Various Sarcoma Subtypes with Ability to Metastasize
These findings encouraged us to investigate the differential effect of reduced gene dosage of Pten, Tp53, or combinations of both in early progenitors of the SM lineage. We, therefore, crossbred PtenL/LTp53L/L (PtenL/L7,8 and Tp53L/L9) and Tagln(transgelin)-cre23 mice to generate all of the possible genetic combinations of mice defective in one or both alleles of these two tumor suppressors. Immunostaining of Tagln-cre/Rosa26-LSL-LacZ reporter mouse11 tissues revealed diffuse LacZ signal in the myometrium and in the SM layers of the bladder (Supplemental Figure S2A), confirming cre-mediated recombination in the SM of Tagln-cre mice. The progeny of the Tagln-cre x PtenL/LTp53L/L crosses followed an almost mendelian distribution, except for Tagln-cre+/PtenΔ/ΔTp53Δ/Δ and Tagln-cre+/PtenΔ/ΔTp53Δ/+ mice, which were deemed less viable. Tagln-cre+/PtenΔ/ΔTp53+/+ mice were smaller at birth and presented complex pathological characteristics, which resulted in a dramatically reduced lifespan (average, 68.2 ± 9.5 days). This is consistent with our previous report24 showing that these mice develop SM hyperplasia and incipient abdominal leiomyosarcomas.
Only 4 (10%) of 40 Tagln-cre+/PtenΔ/+Tp53+/+ (hereafter, PtenΔ/+Tp53+/+) mice developed sarcomas (Table 2), suggesting that heterozygous Pten works as a strong tumor suppressor in this context. Sarcoma penetrance in Tagln-cre+/Pten+/+Tp53Δ/+ (hereafter, Pten+/+Tp53Δ/+) mice was higher, affecting 6 (22%) of 27 mice. Concomitant inactivation of a Tp53 and a Pten allele led to the highest sarcoma incidence, affecting 14 (38%) of 37 Tagln-cre+/PtenΔ/+Tp53Δ/+ mice (hereafter, PtenΔ/+Tp53Δ/+), and the shortest overall survival (244.1 ± 29 days) compared with Pten heterozygous mice (376.9 ± 33 days; P = 0.0024) and Tp53 heterozygous mice (546.4 ± 36 days; P < 0.0001) (Figure 1A). Partial loss of Pten, thus, synergizes with Tp53 heterozygosity in sarcomagenesis by accelerating tumorigenesis, progression, or both.
Table 2.
Incidence of HGUPS, LMS, and CS per Genotype
| Genotype |
Incidence∗ | Tumor distribution∗ |
||||
|---|---|---|---|---|---|---|
| Tagln-cre | Pten | Tp53 | HGUPS | LMS | CS | |
| + | +/+ | +/+ | 0/28 | 0/0 | 0/0 | 0/0 |
| + | Δ/+ | Δ/+ | 14/37 (38) | 9/14 (64.3) | 3/14 (21.4) | 2/14 (14.3) |
| + | Δ/+ | +/+ | 4/40 (10) | 2/4 (50) | 1/4 (25) | 1/4 (25) |
| + | +/+ | Δ/+ | 6/27 (22.2) | 2/6 (33.3) | 3/6 (50) | 1/6 (22.2) |
+, wild-type allele; Δ, floxed allele; CS, carcinosarcomas; HGUPS, high-grade undifferentiated pleomorphic sarcomas; LMS, leiomyosarcomas.
Data are given as number/total (percentage).
Figure 1.
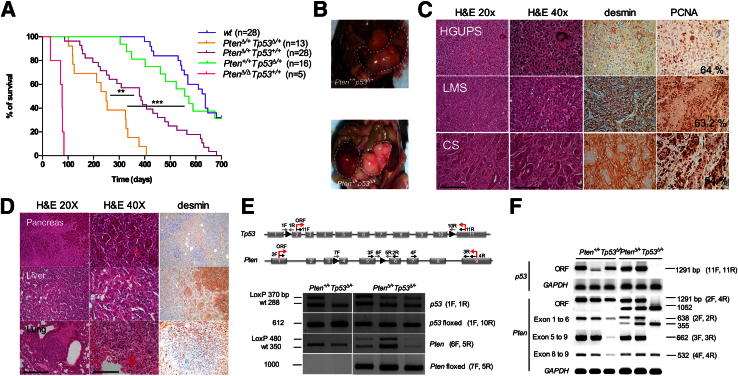
Conditional Pten and Tp53 heterozygous deletion in the SM lineage leads to HGUPS, LMS, and CS. A: Survival plot for mice with the indicated genotypes as a function of days. A statistically significant decrease in lifespan was found for PtenΔ/+Tp53Δ/+ mice compared with PtenΔ/+Tp53+/+ mice (∗∗P = 0.0024) and Pten+/+Tp53Δ/+ mice (∗∗∗P < 0.0001 ). B: Macroscopic images of a uterine LMS in a Pten+/+Tp53Δ/+ mouse (top panel) and an abdominal HGUPS in a PtenΔ/+Tp53Δ/+ mouse (bottom panel). C: H&E and IHC stainings of sections of the three sarcoma subtypes found on PtenΔ/+Tp53Δ/+ mice with antibodies against desmin and PCNA. The percentage of PCNA-positive cells is indicated. Scale bars, ×20 (250 μm); ×40 (125 μm). D: H&E and IHC staining for desmin in metastases found in the indicated organs. The top two rows correspond to a Pten+/+Tp53Δ/+ LMS, and the bottom panel depicts a PtenΔ/+Tp53Δ/+ CS. E: Top panel, scheme of primers used for Pten and Tp53 PCR-based assays to genotype or sequence for mutations and/or deletions. ORF, open reading frame. Bottom panel, electrophoresis of PCR products from genomic DNA isolated from tumor tissue using the indicated primer pairs. Floxed denotes the recombined allele after cre-mediated recombination. F: Gel electrophoresis of PCR amplification products for Tp53 (top two lanes) and Pten (bottom five lanes) in DNA from short-term cultures of the corresponding murine tumors. In parenthesis, primer pairs used for detection. GAPDH is used as a loading control.
Histopathological evaluation of PtenΔ/+Tp53Δ/+ and Pten+/+Tp53Δ/+ tumors led to the diagnosis of a variety of lesions (Table 2), including HGUPS, LMS, and CS, predominantly affecting the genitourinary tract and the retroperitoneum (Figure 1B). HGUPS displayed cellular atypia, hypercromatic nuclei, numerous aberrant mitosis, and local areas of necrosis (Figure 1C). LMSs were characterized by spindle cells with cigar-shaped nuclei, arranged in intersecting bundles and positive for SM markers, such as desmin (Figure 1C) or smooth muscle actin (data not shown). CSs were composed of their characteristic mixed areas of mesenchymal and epithelial origin, as shown by positivity for desmin (Figure 1C) and cytokeratins (data not shown), respectively. The proliferative index as per PCNA immunostaining was similar in HGUPS, LMS, and CS (approximately 60%). Pten+/+Tp53Δ/+ sarcomas included 50% LMS and 33.3% HGUPS, whereas PtenΔ/+Tp53Δ/+ mice had a higher incidence of undifferentiated sarcomas (64% HGUPS and 21% LMS). CS incidence was 22% and 14%, respectively (Table 2). Both mouse lines also developed nonmesenchymal tumors, as summarized in Supplemental Table S3.
Some PtenΔ/+Tp53+/+, Pten+/+Tp53Δ/+, and PtenΔ/+ Tp53Δ/+ tumors (particularly the CSs) were able to colonize distant organs, including liver, pancreas, or lungs (Figure 1D), but metastases were detected much earlier in the life of PtenΔ/+Tp53Δ/+ mice. This observation underlines the ability of the PtenΔ/+Tp53Δ/+ model to recapitulate the highly aggressive behavior of human sarcomas, which normally display hematogeneous spread through the portal system to reach distal organs.
Pten But Not Tp53 Is Haploinsufficient for Sarcoma Tumor Suppression in PtenΔ/+Tp53Δ/+ Mice
We asked whether sarcoma formation associates with loss of heterozygosity of the remaining Pten and/or Tp53 wild-type alleles in PtenΔ/+Tp53Δ/+ and Pten+/+Tp53Δ/+ mice. The wild-type allele of Pten was occasionally found partially deleted (3 of 10 PtenΔ/+Tp53Δ/+and 0 of 4 Pten+/+Tp53Δ/+), and sequencing did not reveal point mutations (Figure 1, E and F, and Supplemental Table S4). The remaining Tp53 allele, however, was invariably found mutated in short-term cultures derived from resected murine tumors (six of six cases), recurrently in the DNA-binding domain (Supplemental Table S4). This suggests that Tp53 must be fully inactivated to allow sarcomagenesis initiated by Pten heterozygosity to proceed. We conclude that Pten is generally haploinsufficient for sarcoma tumor suppression, whereas there is strong selective pressure to completely abrogate Tp53 function to allow sarcoma development.
Interestingly, tumor cells from PtenΔ/+Tp53Δ/+ and Pten+/+Tp53Δ/+ mice showed numerous structural and numerical changes (Supplemental Figure S3). These tumors, thus, recapitulate the chromosomal instability characteristic of the corresponding human lesions.
Pten Down-Regulation in Pten+/+Tp53Δ/+ Bone Marrow–Derived Mesenchymal Stem Cells and in Tumor Cells Increases Their Clonogenic and Invasive Capacity
PtenΔ/+Tp53Δ/+ tumors progress much more rapidly than Pten+/+Tp53Δ/+ lesions. To determine which advantage Pten suppression confers on Tp53-deficient murine tumors, we compared the growth properties of BM-derived mesenchymal stem cells (MSCs) from these mice, as a surrogate of the plausible cell of origin of HGUPS and LMS.17,25,26 We first confirmed that a fraction of BM-MSC and their progeny are targeted in our mouse model on Cre recombination using fluorescence-activated cell sorter analysis of Tagln-cre/Rosa26-LSL-EGFP mice BM. This analysis showed 6.46% ± 0.52% (n = 4) of Lin−ScaI+c-kit− cells, which encompass the MSC pool, as positive for GFP (Supplemental Figure S2B). Then, BM-MSCs were isolated from PtenL/+Tp53L/+ and Pten+/+Tp53L/+ Cre− mice, and recombination was acutely triggered by Ad5-CMV-Cre-GFP infection in vitro. We observed that after infection, PtenΔ/+Tp53Δ/+ MSCs displayed significantly higher clonogenic potential (Figure 2A) and proliferative capacity (Figure 2B) than Pten+/+Tp53Δ/+ or wild-type MSCs. These results were confirmed in MSCs isolated from the corresponding PtenΔ/+Tp53Δ/+ and Pten+/+Tp53Δ/+ mice, in which recombination had occurred genetically, in vivo (data not shown).
Figure 2.
Pten silencing in Tp53 heterozygous mesenchymal stem cells and tumor cells exacerbates their oncogenic behavior. Murine BM-MSCs were infected with Ad5-CMV-Cre-GFP and subjected to colony formation (A) and proliferation assays (B). Relative proliferation at day 6 after infection is shown. C: Western blot analysis of two murine Pten+/+Tp53Δ/+ HGUPS cell lines infected with shPten (sh) or nonsilencing control (V) and parental (C) cells. Colony formation assay (quantitated in the adjacent histogram) (D) and growth curve (E) of shPten-infected HGPUS cells compared with nonsilencing control. F: Representative images of a transwell invasion assay on two HGUPS cell lines transduced with nonsilencing control or shPten lentivirus. Graphs indicate relative number of cells per field. B and D–F: Representative of three independent experiments ± SD. ∗P < 0.05, ∗∗P < 0.01, and ∗∗∗P < 0.001.
To determine whether the enhanced oncogenic properties of PtenΔ/+Tp53Δ/+ tumor cells were caused by Pten down-regulation, we stably infected two cell lines derived from Pten+/+Tp53Δ/+ HGUPS with a lentivirus carrying a short-hairpin RNA for Pten (shPten). In both cell lines, reduced Pten expression (Figure 2C) was accompanied by a significant increase in colony formation (Figure 2D) and cell invasion (Figure 2F), without major effects in proliferation (Figure 2E). These in vitro properties are consistent with the more aggressive tumor behavior and reduced overall survival observed in PtenΔ/+Tp53Δ/+mice; they further indicate that the differences between PtenΔ/+Tp53Δ/+and Pten+/+Tp53Δ/+ tumor behavior can be directly attributed to lower Pten levels.
Notch Signaling Is Increased in PtenΔ/+Tp53Δ/+ Murine HGUPS Compared with Pten+/+Tp53Δ/+ Tumors
We compared the gene expression profiles of PtenΔ/+Tp53Δ/+ (n = 4) and Pten+/+Tp53Δ/+ (n = 5) murine HGUPS tissues to identify transcriptional changes that could account for their distinct phenotypic behavior. Functional annotation (DAVID)27,28 and GSEA12,13 revealed that the most significantly up-regulated pathways in PtenΔ/+Tp53Δ/+ tumors were Notch (P = 0.012), Akt (P = 0.011), mitogen-activated protein kinase (MAPK; P < 0.0001), and NF-κB (P < 0.0001) (Figure 3, A and B). Further analyses of individual genes within those pathways showed Notch1, Pdk1, Jak1, Tradd, Map3k9, Pak2, and Ripk1 as significantly up-regulated (Figure 3C). Array data were validated by quantitative real-time PCR for Notch1, Jak1, and Ripk1 in PtenΔ/+Tp53Δ/+ and Pten+/+Tp53Δ/+ tumor-derived cell lines (Figure 3D). These data demonstrate that partial loss of Pten in a Tp53 heterozygous context results in a distinct molecular profile, which includes up-regulation of Notch signaling.
Figure 3.
The Notch signaling pathway is up-regulated in PtenΔ/+Tp53Δ/+ murine HGUPS. A: Histogram of pathways up-regulated in PtenΔ/+Tp53Δ/+ tumors according to DAVID.27,28B: GSEA performed on genes differentially expressed (fold change of >1.25 and P < 0.05) in PtenΔ/+Tp53Δ/+ compared with Pten+/+Tp53Δ/+. C: Selected differentially expressed genes found in relevant pathways. D: Validation of the RNA array data by quantitative real-time RT-PCR comparing PtenΔ/+Tp53Δ/+ and Pten+/+Tp53Δ/+ murine HGUPS cell lines. The fold change (FC) is shown for different genes. GAPDH was used for normalization. Error bars represent SD between two experimental replicates. ∗P < 0.05, ∗∗P < 0.01.
Increased Notch Signaling Partially Accounts for the Greater Invasive and Clonogenic Potential of PtenΔ/+Tp53Δ/+ Murine HGUPS
Because the Notch pathway exerts key roles in tumorigenesis,29 maintains MSCs in an undifferentiated state,30 and can be targeted pharmacologically,31 we selected it for further analyses. We sought to determine whether Notch signaling was responsible for the increased invasive and clonogenic potential of PtenΔ/+Tp53Δ/+ tumor cells compared with Pten+/+Tp53Δ/+ cells. Indeed, treatment with the γ-secretase inhibitor (GSI) Compound E, which suppressed Hes1 levels (Figure 4A), reduced the invasive capacity of PtenΔ/+Tp53Δ/+ cells (Figure 4B), suggesting that these tumors particularly rely on Notch signaling for their aggressive behavior. Surprisingly, treatment of Pten+/+Tp53Δ/+ cells had the opposite effect (Figure 4B). Furthermore, treatment with this GSI counteracted the ability of shPten, which induced Hes-1 levels (Figure 4C), to enhance the clonogenic and invasive potential of Pten+/+Tp53Δ/+ sarcoma cells (Figure 4, D and E). These results demonstrate that Notch activation subsequent to Pten silencing in a Tp53-deficient background contributes to the increased aggressiveness of double-heterozygous tumors. In sum, higher Notch signaling is characteristic of HGUPS that have dual Pten/Tp53 suppression, and it may contribute to a faster disease course in patients with this genetic makeup.
Figure 4.
Increased Notch signaling partially accounts for the higher invasive and clonogenic potential of PtenΔ/+Tp53Δ/+ sarcoma. A: Relative expression of Hes1 by quantitative real-time RT-PCR of PtenΔ/+Tp53Δ/+ and Pten+/+Tp53Δ/+ HGUPS murine sarcoma cell lines, with (+) or without (−) 500 nmol/L Compound E. Relative values referred to R18S1, used as a housekeeping gene. B, Left panel: Transwell invasion assay for PtenΔ/+Tp53Δ/+ and Pten+/+Tp53Δ/+ HGUPS sarcoma cell lines, with (+) or without (−) 500 nmol/L Compound E. Data are given as means ± SD. Right panel: Representative images of the invasion assay. C: Relative expression of Hes1 in Pten+/+Tp53Δ/+ HGUPS transduced with shPten or vector (v), in the presence (+) or absence (−) of 500 nmol/L Compound E. Clonability (D) and transwell invasion assay (E) in Pten+/+Tp53Δ/+ HGUPS transduced with shPten or vector (v), with (+) or without (−) 500 nmol/L Compound E. Data are given as means ± SD. ∗P < 0.05, ∗∗P < 0.01, and ∗∗∗P < 0.001.
Discussion
The extremely low frequency of point mutations in conventional oncogenes, highlighted by our extensive analysis of tumor samples, has typically limited our ability to model LMS and HGUPS in mice. Herein, we demonstrate that concurrent chromosomal deletions encompassing the Pten and Tp53 loci, which are common hallmarks of human HGUPS and LMS,16 act synergistically to initiate these tumors in our genetically engineered mice.
Similar genomic alterations in some LMS and HGUPS32 suggest that they may share common pathways of transformation. Our large-scale mutation profiling of human LMS using the Oncomap platform found nucleotide changes in TP53 and EGFR in 6 of 40 patients; other rare mutations previously reported for HGUPS and LMS were not found in our cohort, including those in PTEN and HRAS.33–36 These findings underline the low incidence of recurrent point mutations in classic oncogenes and tumor suppressors characteristic of these tumors. In contrast, published single-nucleotide polymorphism and gene expression arrays reveal frequent concurrent 10q22-23 genomic losses and down-regulation of PTEN37 and TP53,38–42 providing a rationale for genetically engineering these lesions in mice to model HGUPS and LMS.
Genetic alterations of TP53 and PTEN are among the most frequent causal events in many cancers, and their combined inactivation has shown strong oncogenic potential in other mouse models. Loss of Tp53 alone was sufficient to initiate sarcomas in our model, although tumor progression was dramatically accelerated in conjunction with Pten inactivation. Conditional inactivation of both Pten and Tp53 in other mouse models has been shown to cause metastatic bladder cancer,43 glioblastoma,44 and invasive, but not metastatic, prostate cancer.7 In contrast to our findings, those studies showed Tp53 depletion to be generally insufficient for transformation: both alleles of Tp53 and Pten needed to be inactivated to generate tumors that recapitulate the biological behavior of the corresponding human lesions. These differences in results suggest that there are context-dependent tumor suppressor roles for Tp53 and Pten in epithelial and mesenchymal transformation and tumorigenesis.
Complete Pten inactivation has been shown to trigger a prosenescence response (Pten-initiated cellular senescence), which acts as a potent antitumoral barrier in vivo7 that can be bypassed by Tp53 genetic silencing.7,45 In our model, partial Pten inactivation was accompanied by Tp53 point mutations in both PtenΔ/+Tp53+/+ (data not shown) and PtenΔ/+Tp53Δ/+ murine tumors. This could explain why human sarcomas with complex genetics do not select for complete loss of PTEN—partial loss has been reported to occur in 39% of 111 LMS and HGUPS33—and emphasize the significance of PTEN haploinsufficiency for sarcoma initiation and progression.
Partial Pten/Tp53 inactivation in our mice led to multiple histological phenotypes (HGUPS, LMS, and CS), suggesting a common molecular32 and a common cellular origin for these tumors. The appearance of multiple histological phenotypes characterized by various degrees of differentiation might be explained by targeting of different cells of origin that represent various stages of differentiation along the SM lineage, each producing a specific histological phenotype. Alternatively, it could be that a specific cell of origin, perhaps a progenitor, subsequently differentiates into multiple tumor phenotypes through the accumulation of stochastic secondary events. In fact, the variety of tumors observed, ranging from HGUPS to well-differentiated LMS, may also reflect how transforming events affect the ability of a single precursor cell to differentiate.
In recent years, there has been much debate over the cell of origin for various sarcoma subtypes, including synovial, Ewing, and HGUPS. Xenografted and genetically engineered mouse models25,46,47 have revealed some of these tumors to be derived from mesenchymal stem cells or progenitors, rather than somatic, differentiated cell types. We propose that cre-mediated inactivation of Pten or Tp53 target MSC-like cells committed toward the SM lineage. The fact that there were no other tumors of mesenchymal origin (ie, liposarcomas, osteosarcomas, or chondrosarcomas) supports this hypothesis. It is also possible, however, that other mesenchymal lineages, such as the adipocytic or chondrocytic, are not as susceptible to transformation triggered by Tp53 and/or Pten defects. The appearance of carcinosarcomas is particularly puzzling: they might originate from a common epithelial/mesenchymal precursor within the uterus,48 or the carcinomatous component might result from transgene expression in some epithelial cells of the uterus, supporting a combined tumor model.49 A third possibility is that transformation of the epithelium could be non–cell autonomous, resulting from aberrant signaling from the surrounding sarcoma cells.
One of the salient characteristics of our genetically engineered mouse model is its spontaneous development of metastases. We found metastases in both Pten+/+Tp53Δ/+ and PtenΔ/+Tp53Δ/+ mice, although the combined inactivation of Tp53 and Pten defines a subgroup of tumors with much faster dissemination. This suggests a particularly aggressive course of Pten/Tp53-inactivated tumors, as supported by our in vitro data.
Pten suppression in a Tp53 heterozygous background strongly promoted the clonogenic capacity of MSCs and the invasive potential of established murine tumor cell lines. Faster proliferation was also apparent in MSCs in response to partial Pten/Tp53 conditional inactivation, but not in already established Pten+/+Tp53Δ/+ tumors, perhaps because of additional hits acquired during tumor progression, which are partly redundant with the proproliferative effects of Pten suppression. Nonetheless, the suppression of Pten in a Tp53-inactivated background induced more malignant behavior and more rapid disease course.
Expression profiling of PtenΔ/+Tp53Δ/+ and Pten+/+Tp53Δ/+ tumors revealed an up-regulation of the MAPK pathway on Pten inactivation. Aberrant MAPK signaling occurs in many human malignancies, where it promotes tumor proliferation and metastasis. In particular, Ras, a central signal transduction mediator, has been previously involved in sarcomagenesis.5 Therefore, selective inhibition of the MAPK pathway, currently possible through highly specific compounds being tested in the clinic (ie, BRAF and MEK inhibitors), may serve as a viable therapeutic approach against double Pten/Tp53-defective HGUPS.
In addition, the microarray profile showed a significant up-regulation of the Notch signaling pathway in Pten/Tp53 heterozygous tumors compared with Pten wt/Tp53 heterozygous tumors. Furthermore, the increased invasive and clonogenic potentials conferred by Pten suppression (or shPten) were largely dependent on Notch signaling. The highly conserved Notch pathway plays critical roles in development and cancer in a cell-specific manner, leading to oncogenic or tumor-suppressive effects in different contexts.29 Tumor-promoting effects of Notch signaling have been described in human osteosarcoma,50 Ewing sarcoma,51 and rhabdomyosarcoma,52 and they have recently been implicated in the regulation of self-renewal in HGUPS.53 A link between NOTCH and PTEN deregulation has been reported in human melanoma, in which tumors lacking PTEN show hyperactivated phosphatidylinositol 3-kinase/AKT signaling and NOTCH1 up-regulation via NF-κB.54 Interestingly, the NF-κB pathway was found significantly up-regulated in PtenΔ/+Tp53Δ/+ mice compared with Pten+/+Tp53Δ/+ mice, according to our array data (Figure 3). Conversely, in other tumor models, such as T-ALL, phosphatidylinositol 3-kinase–AKT has been shown to contribute to NOTCH1-induced transformation, with the PTEN status determining the cellular response to GSI.55 In our system, treatment of Pten/Tp53 heterozygous HGUPSs with a GSI strongly inhibited their in vitro oncogenic properties, in contrast to Tp53 heterozygous HGUPSs that paradoxically showed opposite effects. These data lend support to the notion that context is important for determining the effects of Notch signaling and inhibition.
At present, most HGUPSs and LMSs are treated with similar chemotherapy regimens, despite their histological and molecular heterogeneity.18,56 Previous observations in a Pten-null murine LMS model24 and data not shown revealed antiproliferative effects of mTor inhibition in both Pten-wt and Pten-het murine HGUPS. However, inhibition of mTOR has been deemed incapable of achieving durable therapeutic responses in patients with sarcoma,57 suggesting the possibility of feedback and compensatory effects, such as those shown in other tumor types.58 Our observation that Pten het/Tp53 het murine tumors are particularly sensitive to Notch inhibition raises the possibility that HGUPS or LMS patients bearing this genetic makeup might be successfully treated with NOTCH inhibitors either alone or in combination with mTOR inhibitors. In contrast, this treatment might be harmful for PTEN wt/TP53 het cases, according to our preclinical studies.
These observations suggest that classifying HGUPS and LMS patients based on tumor genetic and expression criteria could improve patient outcomes by predicting disease progression and providing a decision-making tool for designing individualized treatments (as proposed in Supplemental Figure S4).
Acknowledgments
We thank Martha Vega for genotyping, Elizabeth Charytonowicz and Xavier Jirau-Serrano for supervising the mouse colony, members of the Aifantis laboratory (Camille Loubry, Jasper Mullenders, and Beatriz Aranda-Orgilles) for reagents and discussions, and Jordi Barretina and Emmanuelle Palescandolo (Broad Institute) for technical advice.
Footnotes
Supported by American Cancer Society grant RSG-08-161-01-DDC, Edna’s Foundation of Hope, Spanish Ministerio de Educacion y Ciencia postdoctoral fellowship (M.V.G.), a National Cancer Center fellowship (M.V.G.), members of the NYU Cancer Institute Histopathology and Immunohistochemistry cores for NYUCI Center Support grant NIH/National Cancer Institute5 P30CA16087-31, and the Molecular Cytogenetics core facility at Memorial Sloan-Kettering.
Current address of J.Z., World Health Organization/International Agency for Research on Cancer, Lyon, France.
Supplemental Data
Downregulation of PTEN and TP53 is characteristic of human high-grade undifferentiated pleomorphic sarcomas (HGUPS)/indifferentiated sarcoma (UND)/malignant fibrous histiocytoma (MFH), and leiomyosarcomas (LMS). A: Frequency of mutations of specific genes in LMS patient cohort (n = 40). N indicates number of mutated cases per total of samples analyzed. Percentage from the total is shown in parentheses. B: Status of PTEN and TP53 genomic loci in LMS (n = 27) and HGUPS/MFH (n = 3) paired with normal tissue from the same patient. The corresponding gene locus is highlighted (blue, loss; red: gain; each column indicates one sample). PTEN and TP53 relative array expression values (number of cases: HGUPS/UND=71; LMS=52 and LPS=44; C) and (SYS=16; MLPS=19; WLPS=3; DLPS=15; MPNST=3; MFBS=15; FBS=4; LMS=6; MFH/UND=71; D). DLPS, dedifferentiated liposarcoma; FBS, fibrosarcoma; LPS/LIP, liposarcoma; MFBS: myxoid fibrosarcoma; MLPS, myxoid liposarcoma; MPNST, malignant peripheral nerve sheath tumor; SYS, synovial sarcoma; WLPS, well-differentiated liposarcoma. (*P < 0.05; **P < 0.01; ***P < 0.001.
A: LacZ staining of smooth muscle in bladder and myometrium of the uterus in Tagln-cre crossed with the Rosa26-LacZ reporter mice. Scale bars: ×10 (125 μm); ×20 (250μm). B: Representative FACS analysis of Lin--c-kit--Sca1+-GFP+ in BM cells of Tagln-cre mice crossed with Rosa26-EGFP reporter mice. Average of GFP+ cells in cre- (n = 3) and cre+ (n = 4) is shown.
Examples of karyotypes of PtenΔ/+Tp53Δ/+ and Pten+/+Tp53Δ/+ murine sarcoma cell lines. Red arrows indicate presence of the alteration in most cells; black arrow, chromosomal aberration present in a single cell. Chromosome analysis was performed on a minimum of 20 metaphases.
Model proposed for signaling pathway activation downstream of PtenΔ/+Tp53Δ/+ and Pten+/+Tp53Δ/+ murine sarcomas, and targeted therapies impacting in specific components of those pathways.
References
- 1.Taylor B.S., Barretina J., Maki R.G., Antonescu C.R., Singer S., Ladanyi M. Advances in sarcoma genomics and new therapeutic targets. Nat Rev Cancer. 2011;11:541–557. doi: 10.1038/nrc3087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Agrawal S., Payal Y.S., Sharma J.P., Meher R., Varshney S. Montgomery T-tube: anesthetic management. J Clin Anesth. 2007;19:135–137. doi: 10.1016/j.jclinane.2006.06.007. [DOI] [PubMed] [Google Scholar]
- 3.Demicco E.G., Lazar A.J. Clinicopathologic considerations: how can we fine tune our approach to sarcoma? Semin Oncol. 2011;38(Suppl 3):S3–S18. doi: 10.1053/j.seminoncol.2011.09.001. [DOI] [PubMed] [Google Scholar]
- 4.Mulligan L.M., Matlashewski G.J., Scrable H.J., Cavenee W.K. Mechanisms of p53 loss in human sarcomas. Proc Natl Acad Sci U S A. 1990;87:5863–5867. doi: 10.1073/pnas.87.15.5863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kirsch D.G., Dinulescu D.M., Miller J.B., Grimm J., Santiago P.M., Young N.P., Nielsen G.P., Quade B.J., Chaber C.J., Schultz C.P., Takeuchi O., Bronson R.T., Crowley D., Korsmeyer S.J., Yoon S.S., Hornicek F.J., Weissleder R., Jacks T. A spatially and temporally restricted mouse model of soft tissue sarcoma. Nat Med. 2007;13:992–997. doi: 10.1038/nm1602. [DOI] [PubMed] [Google Scholar]
- 6.Xing D., Scangas G., Nitta M., He L., Xu X., Ioffe Y.J., Aspuria P.J., Hedvat C.Y., Anderson M.L., Oliva E., Karlan B.Y., Mohapatra G., Orsulic S. A role for BRCA1 in uterine leiomyosarcoma. Cancer Res. 2009;69:8231–8235. doi: 10.1158/0008-5472.CAN-09-2543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen Z., Trotman L.C., Shaffer D., Lin H.K., Dotan Z.A., Niki M., Koutcher J.A., Scher H.I., Ludwig T., Gerald W., Cordon-Cardo C., Pandolfi P.P. Crucial role of p53-dependent cellular senescence in suppression of Pten-deficient tumorigenesis. Nature. 2005;436:725–730. doi: 10.1038/nature03918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Trotman L.C., Niki M., Dotan Z.A., Koutcher J.A., Di Cristofano A., Xiao A., Khoo A.S., Roy-Burman P., Greenberg N.M., Van Dyke T., Cordon-Cardo C., Pandolfi P.P. Pten dose dictates cancer progression in the prostate. PLoS Biol. 2003;1:E59. doi: 10.1371/journal.pbio.0000059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Donehower L.A., Harvey M., Slagle B.L., McArthur M.J., Montgomery C.A., Butel J., Bradley A. Mice deficient for p53 are developmentally normal but susceptible to spontaneous tumours. Nature. 1992;356:215–221. doi: 10.1038/356215a0. [DOI] [PubMed] [Google Scholar]
- 10.Mao X., Fujiwara Y., Chapdelaine A., Yang H., Orkin S.H. Activation of EGFP expression by Cre-mediated excision in a new ROSA26 reporter mouse strain. Blood. 2001;97:324–326. doi: 10.1182/blood.v97.1.324. [DOI] [PubMed] [Google Scholar]
- 11.Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat Genet. 1999;21:70–71. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- 12.Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S., Mesirov J.P. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mootha V.K., Lindgren C.M., Eriksson K.F., Subramanian A., Sihag S., Lehar J., Puigserver P., Carlsson E., Ridderstrale M., Laurila E., Houstis N., Daly M.J., Patterson N., Mesirov J.P., Golub T.R., Tamayo P., Spiegelman B., Lander E.S., Hirschhorn J.N., Altshuler D., Groop L.C. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet. 2003;34:267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- 14.Naldini L., Blomer U., Gallay P., Ory D., Mulligan R., Gage F.H., Verma I.M., Trono D. In vivo gene delivery and stable transduction of nondividing cells by a lentiviral vector. Science. 1996;272:263–267. doi: 10.1126/science.272.5259.263. [DOI] [PubMed] [Google Scholar]
- 15.MacConaill L.E., Campbell C.D., Kehoe S.M., Bass A.J., Hatton C., Niu L., Davis M., Yao K., Hanna M., Mondal C., Luongo L., Emery C.M., Baker A.C., Philips J., Goff D.J., Fiorentino M., Rubin M.A., Polyak K., Chan J., Wang Y., Fletcher J.A., Santagata S., Corso G., Roviello F., Shivdasani R., Kieran M.W., Ligon K.L., Stiles C.D., Hahn W.C., Meyerson M.L., Garraway L.A. Profiling critical cancer gene mutations in clinical tumor samples. PLoS One. 2009;4:e7887. doi: 10.1371/journal.pone.0007887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Murray S., Linardou H., Mountzios G., Manoloukos M., Markaki S., Eleutherakis-Papaiakovou E., Dimopoulos M.A., Papadimitriou C.A. Low frequency of somatic mutations in uterine sarcomas: a molecular analysis and review of the literature. Mutat Res. 2010;686:68–73. doi: 10.1016/j.mrfmmm.2010.01.019. [DOI] [PubMed] [Google Scholar]
- 17.Barretina J., Taylor B.S., Banerji S., Ramos A.H., Lagos-Quintana M., Decarolis P.L. Subtype-specific genomic alterations define new targets for soft-tissue sarcoma therapy. Nat Genet. 2010;42:715–721. doi: 10.1038/ng.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Matushansky I., Charytonowicz E., Mills J., Siddiqi S., Hricik T., Cordon-Cardo C. MFH classification: differentiating undifferentiated pleomorphic sarcoma in the 21st century. Expert Rev Anticancer Ther. 2009;9:1135–1144. doi: 10.1586/era.09.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chibon F., Lagarde P., Salas S., Perot G., Brouste V., Tirode F., Lucchesi C., de Reynies A., Kauffmann A., Bui B., Terrier P., Bonvalot S., Le Cesne A., Vince-Ranchere D., Blay J.Y., Collin F., Guillou L., Leroux A., Coindre J.M., Aurias A. Validated prediction of clinical outcome in sarcomas and multiple types of cancer on the basis of a gene expression signature related to genome complexity. Nat Med. 2010;16:781–787. doi: 10.1038/nm.2174. [DOI] [PubMed] [Google Scholar]
- 20.Baird K., Davis S., Antonescu C.R., Harper U.L., Walker R.L., Chen Y., Glatfelter A.A., Duray P.H., Meltzer P.S. Gene expression profiling of human sarcomas: insights into sarcoma biology. Cancer Res. 2005;65:9226–9235. doi: 10.1158/0008-5472.CAN-05-1699. [DOI] [PubMed] [Google Scholar]
- 21.Henderson S.R., Guiliano D., Presneau N., McLean S., Frow R., Vujovic S., Anderson J., Sebire N., Whelan J., Athanasou N., Flanagan A.M., Boshoff C. A molecular map of mesenchymal tumors. Genome Biol. 2005;6:R76. doi: 10.1186/gb-2005-6-9-r76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nakayama R., Nemoto T., Takahashi H., Ohta T., Kawai A., Seki K., Yoshida T., Toyama Y., Ichikawa H., Hasegawa T. Gene expression analysis of soft tissue sarcomas: characterization and reclassification of malignant fibrous histiocytoma. Mod Pathol. 2007;20:749–759. doi: 10.1038/modpathol.3800794. [DOI] [PubMed] [Google Scholar]
- 23.Lepore J.J., Cheng L., Min Lu M., Mericko P.A., Morrisey E.E., Parmacek M.S. High-efficiency somatic mutagenesis in smooth muscle cells and cardiac myocytes in SM22alpha-Cre transgenic mice. Genesis. 2005;41:179–184. doi: 10.1002/gene.20112. [DOI] [PubMed] [Google Scholar]
- 24.Hernando E., Charytonowicz E., Dudas M.E., Menendez S., Matushansky I., Mills J., Socci N.D., Behrendt N., Ma L., Maki R.G., Pandolfi P.P., Cordon-Cardo C. The AKT-mTOR pathway plays a critical role in the development of leiomyosarcomas. Nat Med. 2007;13:748–753. doi: 10.1038/nm1560. [DOI] [PubMed] [Google Scholar]
- 25.Matushansky I., Hernando E., Socci N.D., Mills J.E., Matos T.A., Edgar M.A., Singer S., Maki R.G., Cordon-Cardo C. Derivation of sarcomas from mesenchymal stem cells via inactivation of the Wnt pathway. J Clin Invest. 2007;117:3248–3257. doi: 10.1172/JCI31377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Danielson L.S., Menendez S., Attolini C.S., Guijarro M.V., Bisogna M., Wei J., Socci N.D., Levine D.A., Michor F., Hernando E. A differentiation-based microRNA signature identifies leiomyosarcoma as a mesenchymal stem cell-related malignancy. Am J Pathol. 2010;177:908–917. doi: 10.2353/ajpath.2010.091150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dennis G., Jr., Sherman B.T., Hosack D.A., Yang J., Gao W., Lane H.C., Lempicki R.A. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4:P3. [PubMed] [Google Scholar]
- 28.Huang da W., Sherman B.T., Lempicki R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 29.Lobry C., Oh P., Aifantis I. Oncogenic and tumor suppressor functions of Notch in cancer: it’s NOTCH what you think. J Exp Med. 2011;208:1931–1935. doi: 10.1084/jem.20111855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hilton M.J., Tu X., Wu X., Bai S., Zhao H., Kobayashi T., Kronenberg H.M., Teitelbaum S.L., Ross F.P., Kopan R., Long F. Notch signaling maintains bone marrow mesenchymal progenitors by suppressing osteoblast differentiation. Nat Med. 2008;14:306–314. doi: 10.1038/nm1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Purow B. Notch inhibition as a promising new approach to cancer therapy. Adv Exp Med Biol. 2012;727:305–319. doi: 10.1007/978-1-4614-0899-4_23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Derre J., Lagace R., Nicolas A., Mairal A., Chibon F., Coindre J.M., Terrier P., Sastre X., Aurias A. Leiomyosarcomas and most malignant fibrous histiocytomas share very similar comparative genomic hybridization imbalances: an analysis of a series of 27 leiomyosarcomas. Lab Invest. 2001;81:211–215. doi: 10.1038/labinvest.3780229. [DOI] [PubMed] [Google Scholar]
- 33.Gibault L., Ferreira C., Pérot G., Audebourg A., Chibon F., Bonnin S., Lagarde P., Vacher-Lavenu M.C., Terrier P., Coindre J.M., Aurias A. From PTEN loss of expression to RICTOR role in smooth muscle differentiation: complex involvement of the mTOR pathway in leiomyosarcomas and pleomorphic sarcomas. Mod Pathol. 2012;25:197–211. doi: 10.1038/modpathol.2011.163. [DOI] [PubMed] [Google Scholar]
- 34.Saito T., Oda Y., Kawaguchi K., Takahira T., Yamamoto H., Tamiya S., Tanaka K., Matsuda S., Sakamoto A., Iwamoto Y., Tsuneyoshi M. PTEN/MMAC1 gene mutation is a rare event in soft tissue sarcomas without specific balanced translocations. Int J Cancer. 2003;104:175–178. doi: 10.1002/ijc.10918. [DOI] [PubMed] [Google Scholar]
- 35.Rieske P., Bartkowiak J., Szadowska A., Debiec-Rychter M. Malignant fibrous histiocytomas and H-ras-1 oncogene point mutations. Mol Pathol. 1999;52:64–67. doi: 10.1136/mp.52.2.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kawaguchi K., Oda Y., Saito T., Takahira T., Yamamoto H., Tamiya S., Iwamoto Y., Tsuneyoshi M. Genetic and epigenetic alterations of the PTEN gene in soft tissue sarcomas. Hum Pathol. 2005;36:357–363. doi: 10.1016/j.humpath.2005.01.017. [DOI] [PubMed] [Google Scholar]
- 37.Gibault L., Perot G., Chibon F., Bonnin S., Lagarde P., Terrier P., Coindre J.M., Aurias A. New insights in sarcoma oncogenesis: a comprehensive analysis of a large series of 160 soft tissue sarcomas with complex genomics. J Pathol. 2011;223:64–71. doi: 10.1002/path.2787. [DOI] [PubMed] [Google Scholar]
- 38.Sandberg A.A. Updates on the cytogenetics and molecular genetics of bone and soft tissue tumors: leiomyoma. Cancer Genet Cytogenet. 2005;158:1–26. doi: 10.1016/j.cancergencyto.2004.08.025. [DOI] [PubMed] [Google Scholar]
- 39.Das P., Kotilingam D., Korchin B., Liu J., Yu D., Lazar A.J., Pollock R.E., Lev D. High prevalence of p53 exon 4 mutations in soft tissue sarcoma. Cancer. 2007;109:2323–2333. doi: 10.1002/cncr.22680. [DOI] [PubMed] [Google Scholar]
- 40.Yoo J., Lee H.K., Kang C.S., Park W.S., Lee J.Y., Shim S.I. p53 Gene mutations and p53 protein expression in human soft tissue sarcomas. Arch Pathol Lab Med. 1997;121:395–399. [PubMed] [Google Scholar]
- 41.Ito M., Barys L., O’Reilly T., Young S., Gorbatcheva B., Monahan J., Zumstein-Mecker S., Choong P.F., Dickinson I., Crowe P., Hemmings C., Desai J., Thomas D.M., Lisztwan J. Comprehensive mapping of p53 pathway alterations reveals an apparent role for both SNP309 and MDM2 amplification in sarcomagenesis. Clin Cancer Res. 2011;17:416–426. doi: 10.1158/1078-0432.CCR-10-2050. [DOI] [PubMed] [Google Scholar]
- 42.Perot G., Chibon F., Montero A., Lagarde P., de The H., Terrier P., Guillou L., Ranchere D., Coindre J.M., Aurias A. Constant p53 pathway inactivation in a large series of soft tissue sarcomas with complex genetics. Am J Pathol. 2010;177:2080–2090. doi: 10.2353/ajpath.2010.100104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Puzio-Kuter A.M., Castillo-Martin M., Kinkade C.W., Wang X., Shen T.H., Matos T., Shen M.M., Cordon-Cardo C., Abate-Shen C. Inactivation of p53 and Pten promotes invasive bladder cancer. Genes Dev. 2009;23:675–680. doi: 10.1101/gad.1772909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zheng H., Ying H., Yan H., Kimmelman A.C., Hiller D.J., Chen A.J., Perry S.R., Tonon G., Chu G.C., Ding Z., Stommel J.M., Dunn K.L., Wiedemeyer R., You M.J., Brennan C., Wang Y.A., Ligon K.L., Wong W.H., Chin L., DePinho R.A. p53 And Pten control neural and glioma stem/progenitor cell renewal and differentiation. Nature. 2008;455:1129–1133. doi: 10.1038/nature07443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xue W., Zender L., Miething C., Dickins R.A., Hernando E., Krizhanovsky V., Cordon-Cardo C., Lowe S.W. Senescence and tumour clearance is triggered by p53 restoration in murine liver carcinomas. Nature. 2007;445:656–660. doi: 10.1038/nature05529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Haldar M., Hancock J.D., Coffin C.M., Lessnick S.L., Capecchi M.R. A conditional mouse model of synovial sarcoma: insights into a myogenic origin. Cancer Cell. 2007;11:375–388. doi: 10.1016/j.ccr.2007.01.016. [DOI] [PubMed] [Google Scholar]
- 47.Tirode F., Laud-Duval K., Prieur A., Delorme B., Charbord P., Delattre O. Mesenchymal stem cell features of Ewing tumors. Cancer Cell. 2007;11:421–429. doi: 10.1016/j.ccr.2007.02.027. [DOI] [PubMed] [Google Scholar]
- 48.Armstrong A.B., Wang M., Eble J.N., MacLennan G.T., Montironi R., Tan P.H., Lopez-Beltran A., Zhang S., Baldridge L.A., Spartz H., Cheng L. TP53 mutational analysis supports monoclonal origin of biphasic sarcomatoid urothelial carcinoma (carcinosarcoma) of the urinary bladder. Mod Pathol. 2009;22:113–118. doi: 10.1038/modpathol.2008.176. [DOI] [PubMed] [Google Scholar]
- 49.Wada H., Enomoto T., Fujita M., Yoshino K., Nakashima R., Kurachi H., Haba T., Wakasa K., Shroyer K.R., Tsujimoto M., Hongyo T., Nomura T., Murata Y. Molecular evidence that most but not all carcinosarcomas of the uterus are combination tumors. Cancer Res. 1997;57:5379–5385. [PubMed] [Google Scholar]
- 50.Engin F., Bertin T., Ma O., Jiang M.M., Wang L., Sutton R.E., Donehower L.A., Lee B. Notch signaling contributes to the pathogenesis of human osteosarcomas. Hum Mol Genet. 2009;18:1464–1470. doi: 10.1093/hmg/ddp057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schaefer K.L., Eisenacher M., Braun Y., Brachwitz K., Wai D.H., Dirksen U., Lanvers-Kaminsky C., Juergens H., Herrero D., Stegmaier S., Koscielniak E., Eggert A., Nathrath M., Gosheger G., Schneider D.T., Bury C., Diallo-Danebrock R., Ottaviano L., Gabbert H.E., Poremba C. Microarray analysis of Ewing’s sarcoma family of tumours reveals characteristic gene expression signatures associated with metastasis and resistance to chemotherapy. Eur J Cancer. 2008;44:699–709. doi: 10.1016/j.ejca.2008.01.020. [DOI] [PubMed] [Google Scholar]
- 52.Roma J., Masia A., Reventos J., Sanchez de Toledo J., Gallego S. Notch pathway inhibition significantly reduces rhabdomyosarcoma invasiveness and mobility in vitro. Clin Cancer Res. 2011;17:505–513. doi: 10.1158/1078-0432.CCR-10-0166. [DOI] [PubMed] [Google Scholar]
- 53.Wang C.Y., Wei Q., Han I., Sato S., Ghanbari-Azarnier R., Whetstone H., Poon R., Hu J., Zheng F., Zhang P., Wang W., Wunder J.S., Alman B.A. Hedgehog and Notch signaling regulate self-renewal of undifferentiated pleomorphic sarcomas. Cancer Res. 2012;72:1013–1022. doi: 10.1158/0008-5472.CAN-11-2531. [DOI] [PubMed] [Google Scholar]
- 54.Bedogni B., Warneke J.A., Nickoloff B.J., Giaccia A.J., Powell M.B. Notch1 is an effector of Akt and hypoxia in melanoma development. J Clin Invest. 2008;118:3660–3670. doi: 10.1172/JCI36157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Palomero T., Sulis M.L., Cortina M., Real P.J., Barnes K., Ciofani M., Caparros E., Buteau J., Brown K., Perkins S.L., Bhagat G., Agarwal A.M., Basso G., Castillo M., Nagase S., Cordon-Cardo C., Parsons R., Zúñiga-Pflücker J.C., Dominguez M., Ferrando A.A. Mutational loss of PTEN induces resistance to NOTCH1 inhibition in T-cell leukemia. Nat Med. 2007;13:1203–1210. doi: 10.1038/nm1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Beck A.H., Lee C.H., Witten D.M., Gleason B.C., Edris B., Espinosa I., Zhu S., Li R., Montgomery K.D., Marinelli R.J., Tibshirani R., Hastie T., Jablons D.M., Rubin B.P., Fletcher C.D., West R.B., van de Rijn M. Discovery of molecular subtypes in leiomyosarcoma through integrative molecular profiling. Oncogene. 2010;29:845–854. doi: 10.1038/onc.2009.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Quek R., Wang Q., Morgan J.A., Shapiro G.I., Butrynski J.E., Ramaiya N., Huftalen T., Jederlinic N., Manola J., Wagner A.J., Demetri G.D., George S. Combination mTOR and IGF-1R inhibition: phase I trial of everolimus and figitumumab in patients with advanced sarcomas and other solid tumors. Clin Cancer Res. 2011;17:871–879. doi: 10.1158/1078-0432.CCR-10-2621. [DOI] [PubMed] [Google Scholar]
- 58.O’Reilly K.E., Rojo F., She Q.B., Solit D., Mills G.B., Smith D., Lane H., Hofmann F., Hicklin D.J., Ludwig D.L., Baselga J., Rosen N. mTOR inhibition induces upstream receptor tyrosine kinase signaling and activates Akt. Cancer Res. 2006;66:1500–1508. doi: 10.1158/0008-5472.CAN-05-2925. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Downregulation of PTEN and TP53 is characteristic of human high-grade undifferentiated pleomorphic sarcomas (HGUPS)/indifferentiated sarcoma (UND)/malignant fibrous histiocytoma (MFH), and leiomyosarcomas (LMS). A: Frequency of mutations of specific genes in LMS patient cohort (n = 40). N indicates number of mutated cases per total of samples analyzed. Percentage from the total is shown in parentheses. B: Status of PTEN and TP53 genomic loci in LMS (n = 27) and HGUPS/MFH (n = 3) paired with normal tissue from the same patient. The corresponding gene locus is highlighted (blue, loss; red: gain; each column indicates one sample). PTEN and TP53 relative array expression values (number of cases: HGUPS/UND=71; LMS=52 and LPS=44; C) and (SYS=16; MLPS=19; WLPS=3; DLPS=15; MPNST=3; MFBS=15; FBS=4; LMS=6; MFH/UND=71; D). DLPS, dedifferentiated liposarcoma; FBS, fibrosarcoma; LPS/LIP, liposarcoma; MFBS: myxoid fibrosarcoma; MLPS, myxoid liposarcoma; MPNST, malignant peripheral nerve sheath tumor; SYS, synovial sarcoma; WLPS, well-differentiated liposarcoma. (*P < 0.05; **P < 0.01; ***P < 0.001.
A: LacZ staining of smooth muscle in bladder and myometrium of the uterus in Tagln-cre crossed with the Rosa26-LacZ reporter mice. Scale bars: ×10 (125 μm); ×20 (250μm). B: Representative FACS analysis of Lin--c-kit--Sca1+-GFP+ in BM cells of Tagln-cre mice crossed with Rosa26-EGFP reporter mice. Average of GFP+ cells in cre- (n = 3) and cre+ (n = 4) is shown.
Examples of karyotypes of PtenΔ/+Tp53Δ/+ and Pten+/+Tp53Δ/+ murine sarcoma cell lines. Red arrows indicate presence of the alteration in most cells; black arrow, chromosomal aberration present in a single cell. Chromosome analysis was performed on a minimum of 20 metaphases.
Model proposed for signaling pathway activation downstream of PtenΔ/+Tp53Δ/+ and Pten+/+Tp53Δ/+ murine sarcomas, and targeted therapies impacting in specific components of those pathways.