Abstract
The molecular genetic alterations underlying the development and diversity of salivary gland carcinomas are largely unknown. To characterize these events, comparative genomic hybridization analysis was performed, using a single-nucleotide polymorphism microarray platform, of 60 fresh-frozen specimens that represent the main salivary carcinoma types: mucoepidermoid carcinoma (MEC), adenoid cystic carcinoma (ACC), and salivary duct carcinoma (SDC). The results were correlated with the clinicopathologic features and translocation statuses to characterize the genetic alterations. The most commonly shared copy number abnormalities (CNAs) in all types were losses at chromosomes 6q23-26 and the 9p21 region. Subtype-specific CNAs included a loss at 12q11-12 in ACC and a gain at 17q11-12 in SDC. Focal copy number losses included 1p36.33-p36-22 in ACC, 9p13.2 in MEC, and 3p12.3-q11-2, 6q21-22.1, 12q14.1, and 12q15 in SDC. Tumor-specific amplicons were identified at 11q23.3 (PVRL1) in ACC, 11q13.3 (NUMA1) in MEC, and 6p21.1 (CCND3), 9p13.2 (PAX5), 12q15 (CNOT2/RAB3IP), 12q21.1 (GLIPR1L1), and 17q12 (ERBB2/CCL4) in SDC. A comparative CNA analysis of fusion-positive and fusion-negative ACCs and MECs revealed relatively lower CNAs in fusion-positive tumors than in fusion-negative tumors in both tumor types. An association between CNAs and high grade and advanced stage was observed in MECs only. These findings support the pathogenetic segregation of these entities and define novel chromosomal sites for future identification of biomarkers and therapeutic targets.
Salivary gland carcinomas (SGCs) comprise numerous morphological, biological, and clinically diverse entities that sometimes have overlapping diagnostic and management difficulties.1,2 The heterogeneity of these tumors has largely been linked to their derivation from different segments of the ductal-acinar unit of the salivary glands. In that context, tumors derived from the terminal duct are composed of dual epithelial and myoepithelial cells and are less aggressive than those derived from purely epithelial-lined ductal segments. These behavioral differences have been, at least in part, attributed to the suppressive role of myoepithelial cells.3–5 Although numerous subtypes have been recognized, mucoepidermoid carcinoma (MEC), adenoid cystic carcinoma (ACC), and salivary duct carcinoma (SDC) are the most common salivary malignancies, with a combined incidence of 75%. In addition to representing the major histopathological categories, they manifest disparate intertumoral phenotypic, genetic, and clinical characteristics.6–13
Surgical resection with postoperative radiotherapy is the primary treatment for SGCs, but patients with nonresectable, recurrent, and metastatic disease have limited therapeutic options. Several chemotherapeutic and targeted agent trials have been conducted in patients with advanced SGCs, with minimal success.6,7,14–18 However, the results of these trials were disappointing and complicated by the inclusion of different phenotypes, patients with variable clinicopathologic characteristics, and small sample sizes. These shortcomings, together with the lack of progress in understanding the events associated with development of salivary cancers, highlight the need for novel biomarker-based trial concepts. Identifying the genetic and molecular events associated with SGC evolution and clinical diversity is central to future progress in their management.16–18
High-resolution, microarray-based, comparative genomic hybridization and single-nucleotide polymorphism (SNP) array platforms have greatly advanced the detection of genomic alterations and have led to the localization of critical genetic aberrations that are associated with major solid tumors.18–29 Furthermore, the ability to detect focal DNA loss as a result of homozygous deletion, hemizygosity, and/or copy number neutral loss of heterozygosity (CNN-LOH) allows for further in-depth identification of previously unknown events.30,31 Using these tools, investigators were able to characterize and define the genetic alteration in several solid human malignant tumors.21,22,26,27
We hypothesized that early molecular genetic alterations are shared among all SGCs and that the subsequent acquisition of type-specific events underlies their morphological and biological diversity. To test this hypothesis and to characterize molecular genetic alterations of SGCs, we used an oligonucleotide SNP array platform and in-depth informatics programs.
Materials and Methods
Tumors and DNA Sample Processing
We searched the head and neck pathology database at The University of Texas M. D. Anderson Cancer Center (Houston, TX) to identify all patients with SGCs who had been treated surgically from 1995 to 2008. Twenty cases from each tumor type, ACCs, MECs, and SDCs were selected based on the availability of at least 1.0 g of fresh-frozen tumor tissues. Seventeen salivary gland tissues (10 from matching tumor cases and seven from neck resections from patients with other diseases) were used as control (Supplemental Table S1). The 20 ACCs had previously been included in our earlier comparative genomic hybridization analysis.32 The tissues had been harvested immediately after resection by experienced head and neck pathologists (A.E.N.), placed in liquid nitrogen, and stored at −80°C until use. Frozen sections of each tissue specimen were evaluated for tumor content and phenotypic viability. Specimens with >20% of host nonneoplastic elements underwent macrodissection to remove host elements to enrich tumor content to >80%. One ACC tumor was disqualified because of excessive stroma; of the 59 tumor specimens, 14 had undergone macrodissection. All tissues were obtained according to an Institutional Review Board–approved protocol for nonmucosal head and neck cancer.
DNA Extraction
Fresh-frozen tissue was processed using the Gentra Puregene tissue kit (Qiagen, Valencia, CA). In brief, normal and tumor tissues were carefully dissected, and proteinase K was immediately added for cell digestion. After protein precipitation, DNA was precipitated using isopropanol, followed by a 70% ethanol washing procedure. Purified DNA was dissolved and eluted in DNA hydration solution and stored at −80°C. The quality of DNA was assessed using electrophoresis and nanodrop measurement. A 260/280 wavelength ratio of at least 1.8 was used to qualify specimens for analysis.
Microarray Analysis
We used the Affymetrix 250k Nsp SNP array (Affymetrix, Inc., Santa Clara, CA) to survey the genotype and DNA copy number abnormality (CNA) data for 59 tumor specimens and 17 healthy salivary gland specimens. Each array contains 261,563 SNP sites, which are approximately uniformly distributed throughout the genome. The SNP site mapping information was provided by Affymetrix, Inc., using human genome sequence version (National Center for Biotechnology Information) NCBI36/hg18. Two samples were repeatedly sampled to assess data reproducibility.
RT-PCR Analysis of Chimeric Fusions
Screening for the t(6;9)/MYB-NFIB and t(11;19)/CTRC1-MAML2 fusion transcripts was performed as previously described.3,12,13
FISH Analysis
We performed fluorescence in situ hybridization (FISH) analysis on touch preparations from fresh tumor fragments using standard, commercially available, spectrum orange– and spectrum green–labeled probes for ERBB2 amplification (PathVysion HER-2 DNA probe kit; Abbott Laboratories, Des Plaines, IL). Known amplified samples and normal parotid tissue were used as the positive and negative controls, respectively. Tumor samples were evaluated for amplification patterns; we also determined the number of centromeric probe signals of the corresponding chromosomes, with at least 100 cells evaluated per slide. Amplification was defined as the presence of >10 copies per tumor cell in ≥20% of cells. We also performed an FISH analysis on paraffin sections of selected cases to verify 1p35-36 deletions. We used the bacterial artificial chromosome clones, RP11-219 C24 and RP-11-163 M9, and a control clone, RP11-201 K10 (1q21), as previously described.32 For PAX5 gene amplification, we used an orange-labeled bacterial artificial chromosome (BAC) clone (RP11-243F8) and a green-labeled centromeric chromosome 9 probe on case SDC10 (PAX5 amplification) and a control negative tumor.
IHC Staining
We performed immunohistochemical (IHC) staining for human epidermal growth factor receptor 2 (HER-2) using the automated BOND MAX strainer (Vision Biosystems Waverley, Australia) on a paraffin section (4 μm thick) prepared from the five tumors with gene amplification, according to a previously published protocol.33 The anti–HER-2 monoclonal antibody (clone e2-4001, mouse, 1:300 was used; Thermo Fisher Scientific, Fremont, CA) and the secondary conjugate antibody, applied and counterstained with H&E. HER-2 membrane staining was evaluated on the basis of previously described criteria.33 The IHC staining for cyclin D3 (CCND3) was performed using anti-CCND3 polyclonal antibody (clone C-16, rabbit, 1:100; Santa Cruz Biotechnology, Inc., Santa Cruz, CA) after citrate buffer antigen retrieval for 30 minutes.
Statistical Analysis
The data analyses were performed using Partek and R software packages. We used Partek Genomics Suite software (St. Louis, MO) to extract the copy number and B-allele fraction data from the CEL files of the Affymetrix SNP array raw data. The subsequent steps of the analyses were performed using R. A scanning window of 509 SNPs and the median was used to represent the copy number of the DNA segment enclosed in the data was applied. The raw data CEL files, the R-scripts, and the processed DNA copy number data used in the raw data analyses are available from the Gene Expression Omnibus database (http://www.ncbi.nlm.nih.gov/geo; accession number GSE44434).
Broad CNA Analysis
To characterize broad CNAs, we used the running median smoothing method to denoise the copy number data, with a scanning window of 509 SNP probes (approximately 5.5 million bp of the genome) was applied. Accordingly, gains and losses less than half the window size (2.75 million bp) were excluded from the broad CNA analysis. The five-σ rule was used to determine whether the gain or loss was significant; σ represents the estimated dispersion of the copy number measurement for a given chromosomal segment enclosed by a scanning window. This was calculated as follows:
| (1) |
in which mad stands for the median absolution deviation and CNi − CNi+1 stands for the differential between nonoverlapping consecutive scanning windows. If the copy number profile can be regarded as a piecewise constant plus gaussian noise, this formulation will provide a robust estimate of the SD. The gain or loss of a chromosomal segment in the scanning window was defined as the median copy number differing significantly from two.
Focal CNA Analysis
To identify focal CNAs, a double-filtering strategy to process copy number profiles was used with a median smoothing method to denoise the copy number data with a scanning window width of 25 SNP probes. To evaluate the effects of broad CNAs, we reapplied the running median smoothing, with a scanning window the width of 1625 SNP probe sets. The twice-smoothed copy number data were then subtracted from the once-smoothed copy number data to identify the focal CNA profile; the resulting CNA profile included an aberrant segment of <2.7 million bp. Supplemental Figure S1 illustrates the relationship between focal and broad CNAs. Double filtering caused focal alterations to become prominent peaks and valleys; the copy number profile of most of the genome appears flat, with perturbations from a small amount of noise. To evaluate the statistical significance of the focal CNAs, we used the five-σ rule. The procedure was similar to that used to evaluate the broad CNAs, except that the estimated dispersion of the focal copy number profile was calculated as follows:
| (2) |
in which mad represents the median absolute deviation and FCNi − FCNi+1 denotes the difference in copy numbers between nonoverlapping, consecutive scanning windows. The procedure resembles that used to evaluate broad CNAs, except for a smaller scanning window size. Because smaller scanning windows involve fewer SNP probes, the focal CNA profile contains a higher level of noise than does the broad CNA profile. Therefore, σf is higher than σ.
Because focal gain or loss involves numerous SNPs, we grouped focal CNAs as discrete events on the basis of the defined focal CNA profile maximum across samples.
Allele-Specific Copy Number Analysis
To compute the allele-specific copy number for a genomic segment, we first collected the B-allele fraction data from the segment:
| (3) |
where R is a B-allele fraction data point and β is a threshold value set to be 0.15. This threshold was estimated from the frequency of heterozygous SNP loci probed on the array. Approximately 85% of the probed sites on the array are homozygous. The estimate of the R value of the segment was computed as follows:
| (4) |
in which the mode value is the most common. The two allele-specific copy numbers were obtained as follows:
| (5) |
| (6) |
The sum of CNa and CNb is the total copy number that reflects the broad-scale CNA profile. However, the allele-specific CNA analysis could not identify localized CNAs that spanned less than half the size of the scanning window, indicating that localized CNAs that were <2.7 million bp in the chromosomes were ignored in the broad CNA profiles described in this study.
Results
Fifty-nine tumors qualified for the analysis [one ACC sample did not meet the minimum DNA quality requirement (see Materials and Methods)]. Patients' clinical and pathological characteristics are presented in Supplemental Table S1. The cohort was composed of 25 women and 34 men, aged 21 to 91 years (mean, 56 years). Thirty-nine tumors were located in major glands and 22 in minor glands; they ranged from 1.0 to 15.0 cm (median, 4.7 cm).
We characterized large-scale broad and focal (narrow) copy number DNA changes at different chromosomal regions in all tumor specimens.
Broad CNA Analysis
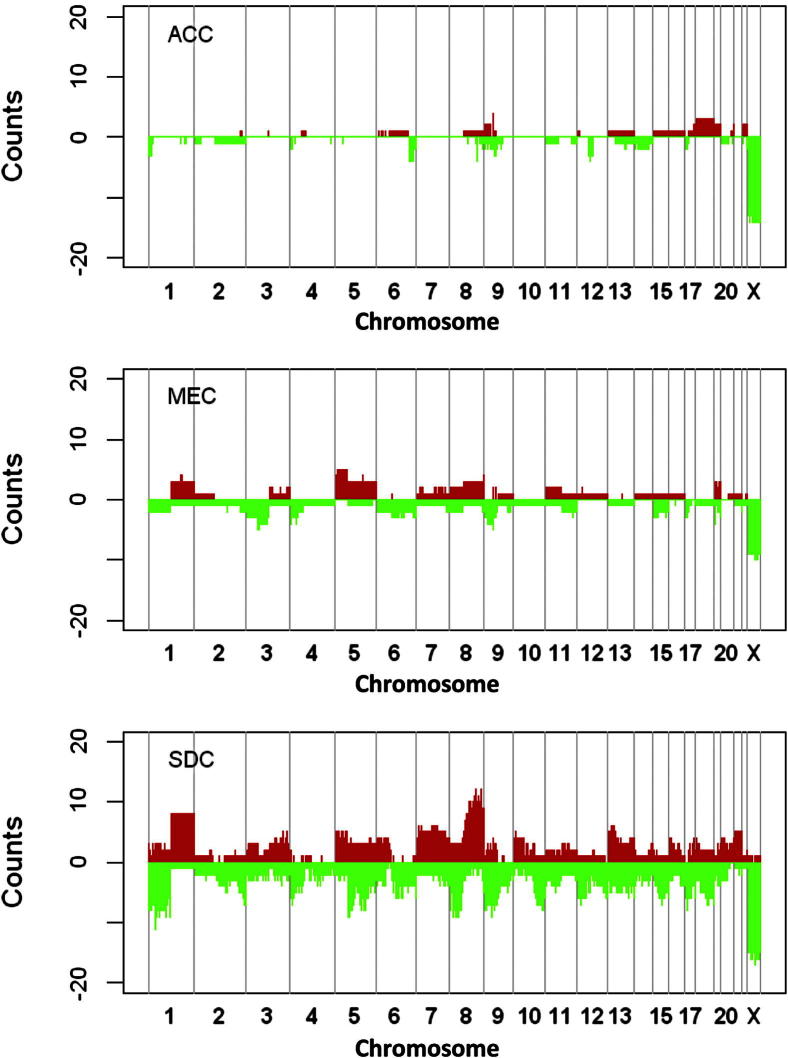
Figure 1 and Supplemental Table S2 show the distributions of broad gains and losses in ACCs, MECs, and SDCs. Gains and losses of >2.7 million bp (half the window size) were included and counted as broad CNAs in the analysis. The results revealed distinct differences among the subtypes: ACC had the lowest level of aberrations, followed by MEC and SDC (Figure 1). Common broad losses at chromosomes 1p36, 6q24, and 9p21 were found in tumors from all subtypes. Preferential broad CNAs included four deletions at the 1p36, 6q27, and 12p14 and gains at the 9p22 and 18p11 regions in ACCs only. In SDCs, broad CNAs that nearly included entire arms or chromosomes were observed and included gains at the 1q and 8q regions; chromosomes 7, 13, 15, and 21; and 1p, 2q, 4p, 5p, and 5q, 6q, 8p, 9p, 10q, 17p, and 8q regions. In general, broad CNAs in MECs and SDCs had considerable overlap.
Figure 1.
Distribution of CNAs across the human genome in ACCs, MECs, and SDCs. The heights of the red regions illustrate the number of samples with copy number gains; the x axis shows the genomic order of the probed segments. The heights of the green regions represent the number of copy number losses. CNAs in chromosome X are affected by sex and were not counted.
Focal CNA Analysis
Table 1 shows the distribution of and lists the significant focal CNAs found in at least three tumors. The most commonly altered locus in tumors of different types was at 9p21.3; this was found in one ACC, one MEC, and four SDCs. One MEC sample also appeared to have a gain at the CDKN2A gene. In addition, a focal gain at chromosome Xp21.3 (CD99 gene) region in tumors from men was noted; because a copy of the CD99 gene is also located on chromosome Y, the possibility of a pseudoautosomal gain cannot be excluded. We also identified alterations at loci on chromosomes 1p36.33, 14q11.2, 15q11.2, 17p21.31, and Xp22.3 in both tumors and normal tissue (data not shown). CNAs at these loci have previously been reported in healthy individuals.34 A comparison of broad and focal CNA in one sample is illustrated in Supplemental Figure S1. The focal gain at 6q21, in which the FYN gene (a membrane-associated tyrosine kinase) is located, can be seen on the focal, but not the broad, CNA profile.
Table 1.
Recurrent Focal CNAs in Major SGCs
| SNP ID | Chromosome | Cytoband | Position | ACC (%) |
MEC (%) |
SDC (%) |
Gene | |||
|---|---|---|---|---|---|---|---|---|---|---|
| G | L | G | L | G | L | |||||
| 2131660 | 1 | p36.33 | 1145994 | 11 | 0 | 10 | 10 | 10 | 0 | TP73 |
| 1988527 | 6 | q27 | 168176083 | 16 | 16 | 0 | 20 | 5 | 5 | FGFR1OP |
| 1822252 | 7 | p22.1 | 5642226 | 5 | 0 | 5 | 15 | 10 | 5 | RNF216 |
| 1996220 | 9 | p21.3 | 21763222 | 0 | 5 | 5 | 10 | 0 | 10 | CDKN2A |
| 2191519 | 9 | p21.3 | 22115503 | 0 | 5 | 0 | 5 | 0 | 20 | CDKN2A |
| 2185617 | 10 | q11.22 | 47062478 | 11 | 5 | 10 | 15 | 10 | 0 | ANAX8 |
| 1822332 | 13 | q31.1 | 83036907 | 5 | 5 | 0 | 5 | 5 | 0 | SLITRK1 |
| 1942128 | 14 | q11.2 | 19417144 | 32 | 32 | 35 | 45 | 40 | 50 | APEX1 |
| 4204773 | 15 | q11.2 | 19163125 | 21 | 37 | 45 | 25 | 35 | 25 | POTEB |
| 1943770 | 15 | q11.2 | 19912458 | 32 | 32 | 40 | 25 | 45 | 20 | OR4N4 |
| 2249545 | 16 | p13.2 | 7092171 | 5 | 0 | 5 | 5 | 0 | 0 | A2BP1 |
| 1822964 | 16 | p11.2 | 34307201 | 16 | 5 | 5 | 25 | 10 | 0 | TP53TG3 |
| 2200283 | 17 | p13.3 | 285707 | 0 | 0 | 0 | 0 | 15 | 10 | RPH3AL |
| 2264661 | 17 | q12 | 31489488 | 11 | 11 | 5 | 10 | 20 | 10 | CCL3L1 |
| 2005155 | 17 | q12 | 34949761 | 0 | 0 | 0 | 0 | 25 | 0 | ERBB2 |
| 2029077 | 17 | q21.31 | 41602401 | 16 | 26 | 50 | 40 | 20 | 35 | MAPT |
| 4222078 | 18 | p11.32 | 1738722 | 5 | 5 | 5 | 0 | 5 | 5 | None |
| 4240882 | 20 | p12.1 | 14798101 | 0 | 5 | 0 | 5 | 0 | 5 | MACROD2 |
| 1919387 | 22 | q11.21 | 18703211 | 0 | 5 | 0 | 20 | 5 | 0 | GGT2 |
| 2192475 | 22 | q11.22 | 20825481 | 0 | 11 | 15 | 0 | 0 | 5 | VPREB1 |
| 4207761 | X | p22.33 | 2581725 | 26 | 0 | 10 | 0 | 15 | 0 | CD99 |
| 2170987 | X | q22.3 | 103915401 | 5 | 5 | 0 | 0 | 0 | 0 | IL1RAPL2 |
The genomic loci were identified from SNP probe positions in which the variance focal CNA profile peaked across samples. Loci with more than three significant focal CNAs are listed. A complete list can be found in Supplemental Table S2.
G, gain; L, loss.
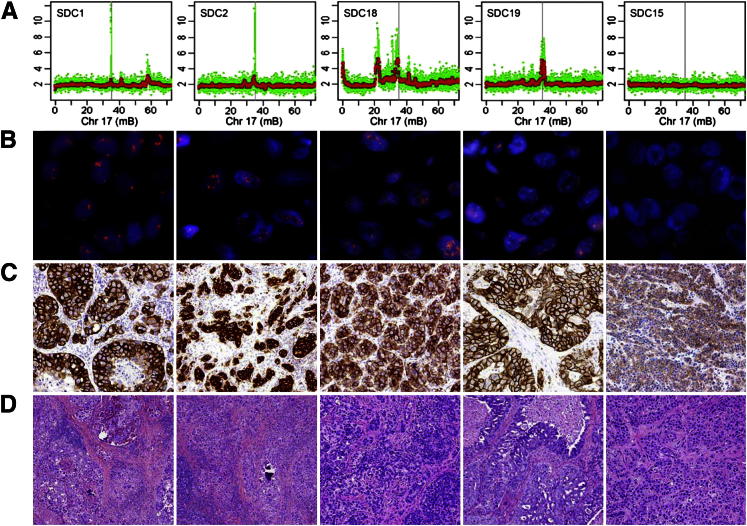
In this study, focal gains with a mean copy number higher than six were considered to represent amplicons (Table 2). Based on this definition, several amplicons in different tumor types were identified. These included one amplicon each at 11q13 and 11q23 in MECs and ACCs, respectively, and four at the 17q12 locus in four different SDC tumors (Figure 2A).
Table 2.
Genomic Regions with CNN-LOH/UPD in Major SGCs
| Chromosome | Cytoband | Position | ACC (%) | MEC (%) | SDC (%) |
|---|---|---|---|---|---|
| 1 | p36.33–p36.22 | 775852–9341877 | 16 | 0 | 0 |
| 1 | q43–q43 | 236829721–240601698 | 0 | 10 | 5 |
| 2 | q11.2–q12.2 | 100704414–105985632 | 0 | 5 | 10 |
| 2 | q14.2–q14.3 | 119250745–124670233 | 5 | 5 | 5 |
| 3 | p24.3–p24.2 | 21618434–25295115 | 0 | 5 | 10 |
| 3 | p12.3–q11.2 | 78686977–96257804 | 0 | 0 | 15 |
| 5 | q33.2–q35.3 | 153512108–180629495 | 0 | 5 | 10 |
| 6 | q21–q22.1 | 109415645–114741718 | 0 | 0 | 15 |
| 9 | p24.3–p24.1 | 140524–6300829 | 0 | 10 | 5 |
| 9 | p22.3–p22.1 | 15942197–18968625 | 11 | 5 | 0 |
| 9 | p21.2–p21.1 | 27800651–31371316 | 11 | 0 | 0 |
| 9 | p21.1–p13.2 | 31373712–37537256 | 5 | 10 | 0 |
| 9 | p13.2–q21.13 | 37574687–73460374 | 0 | 15 | 0 |
| 9 | q22.33–q31.1 | 99537571–104645839 | 0 | 10 | 5 |
| 9 | q33.1–q33.1 | 117901921–121619119 | 5 | 5 | 5 |
| 9 | q33.3–q34.3 | 128519158–140147760 | 5 | 5 | 5 |
| 10 | q21.1–q21.1 | 57418162–61122758 | 5 | 5 | 10 |
| 10 | q21.1–q21.3 | 61177020–66136170 | 0 | 5 | 10 |
| 11 | p15.1–p14.3 | 20031569–23207906 | 5 | 0 | 10 |
| 11 | p13–p13 | 31282908–35998329 | 5 | 0 | 10 |
| 11 | p11.2–q11 | 44781630–56339353 | 0 | 0 | 10 |
| 11 | q12.3–q13.4 | 62942205–74605123 | 0 | 5 | 10 |
| 11 | q14.1–q14.3 | 84276650–88310071 | 0 | 5 | 10 |
| 12 | q14.1–q14.3 | 58492903–64094393 | 0 | 0 | 15 |
| 12 | q14.3–q15 | 64124796–68546087 | 0 | 0 | 10 |
| 12 | q15–q21.1 | 68546631–73477497 | 0 | 0 | 15 |
| 12 | q21.2–q21.31 | 78005428–83047419 | 0 | 0 | 10 |
| 12 | q21.31–q21.33 | 83065065–88560841 | 0 | 5 | 10 |
| 14 | q12–q21.1 | 29399863–39750495 | 5 | 0 | 10 |
| 18 | q12.2–q12.3 | 34364676–39279784 | 0 | 5 | 10 |
Data show the number of CNN-LOH events that occurred in ACCs, MECs, and SDCs. The table lists only those genomic regions that had at least three events. The events were identified using allele-specific CNAs that were significantly different from the reference level, whereas the total CNA was not significantly different in a scanning window (for details, see Materials and Methods).
UPD, uniparental disomy.
Figure 2.
Validation of 17q12 amplification in SDCs. Composite illustration of chromosome (Chr) 17q12 DNA (A) and corresponding ERBB2 gene amplification by FISH (B) and the expression by IHC analysis (C) in the four SDCs with amplicons at this location (SDCs 1, 2, 18, and 19) and case 15, an SDC with no amplicons at this site, is far right (SDC15). Note the lack of ERBB2 gene amplification and the low expression of this gene. The y axis shows copy number data (in green). The red points were obtained by median smoothing (window size, 25). The x axis shows the position on chromosome 17. The position of the ERBB2 gene is marked by the gray vertical bars. C: The results of ERBB2 (HER-2) expression, which suggest that copy number gains contribute to elevated gene expression. D: Corresponding light microscopic images (H&E stain) of each tumor.
Allele-Specific Analysis
CNN-LOH represents loss of one allele and the duplication of the other allele. Our search revealed multiple recurrent CNN-LOH in ACCs, MECs, and SDCs (Table 3); the most common were alterations at 12q14-15 in eight SDCs. Supplemental Figure S1 shows an example of a CNN-LOH in which no aberration on broad CNA data was noted; compensatory CNN-LOH alterations were noted at both alleles of chromosomes 5 and 9. We also found multiple CNN-LOH alterations at centromeric 9p regions in tumors of all subtypes (two ACCs, eight MECs, and one SDC). Interestingly, an MEC specimen (MEC3) had no broad CNAs and only CNN-LOH in chromosomes 2, 3, 5, 9, and 18. Paradoxically, CNN-LOH alterations were found within focal and broad copy number loss regions in several tumors; these included loss at 1p36 in three ACC samples. Surprisingly, concurrent alterations in nontumorous and tumor samples (case ACC5) at the 4q23-q25 region (Chr4:99350642-109772330, spanning approximately 10 million bp) were detected in multiple samples (Supplemental Figure S2). Because normal tissues were obtained from matched tissue specimens, the germline nature of this event cannot be excluded.30,31
Table 3.
Amplicons at Different Chromosomal Regions in Major SGCs
| Sample | Cytoband | Position∗ | CNA | Gene |
|---|---|---|---|---|
| ACC1 | 11q23.3–24.1 | 118706909–122013373 | 9.9 | PVRL1 |
| MEC1 | 11q13.3–q13.4 | 70322842–71448250 | 7.2 | NUMA1 |
| SDC1 | 17q12 | 35084426–35264341 | 10.8 | ERBB2 |
| SDC10 | 12q15 | 68689573–69077882 | 7.0 | CNOT2, RAB3IP |
| SDC10 | 9p13.2 | 36791146–36905407 | 7.1 | PAX5 |
| SDC18† | 17q12 | 31473221–31501499 | 6.9 | CCL4 |
| SDC19 | 17q12 | 35130293–35264341 | 6.6 | ERBB2 |
| SDC2 | 17q12 | 34786442–35130293 | 12.0 | ERBB2 |
| SDC4 | 6p21.1 | 41772099–42368678 | 6.0 | CCND3 |
| SDC7 | 12q21.1–q21.2 | 73790896–74456589 | 6.9 | GLIPR1L1 |
A segment was defined as an amplicon if the mean copy number was higher than six.
The genomic intervals of the segments are shown.
The amplicon at 17q12 in sample SDC18 encodes no known gene. The nearest gene, CCL4, is shown instead.
Relationship between CNAs and Gene Fusion
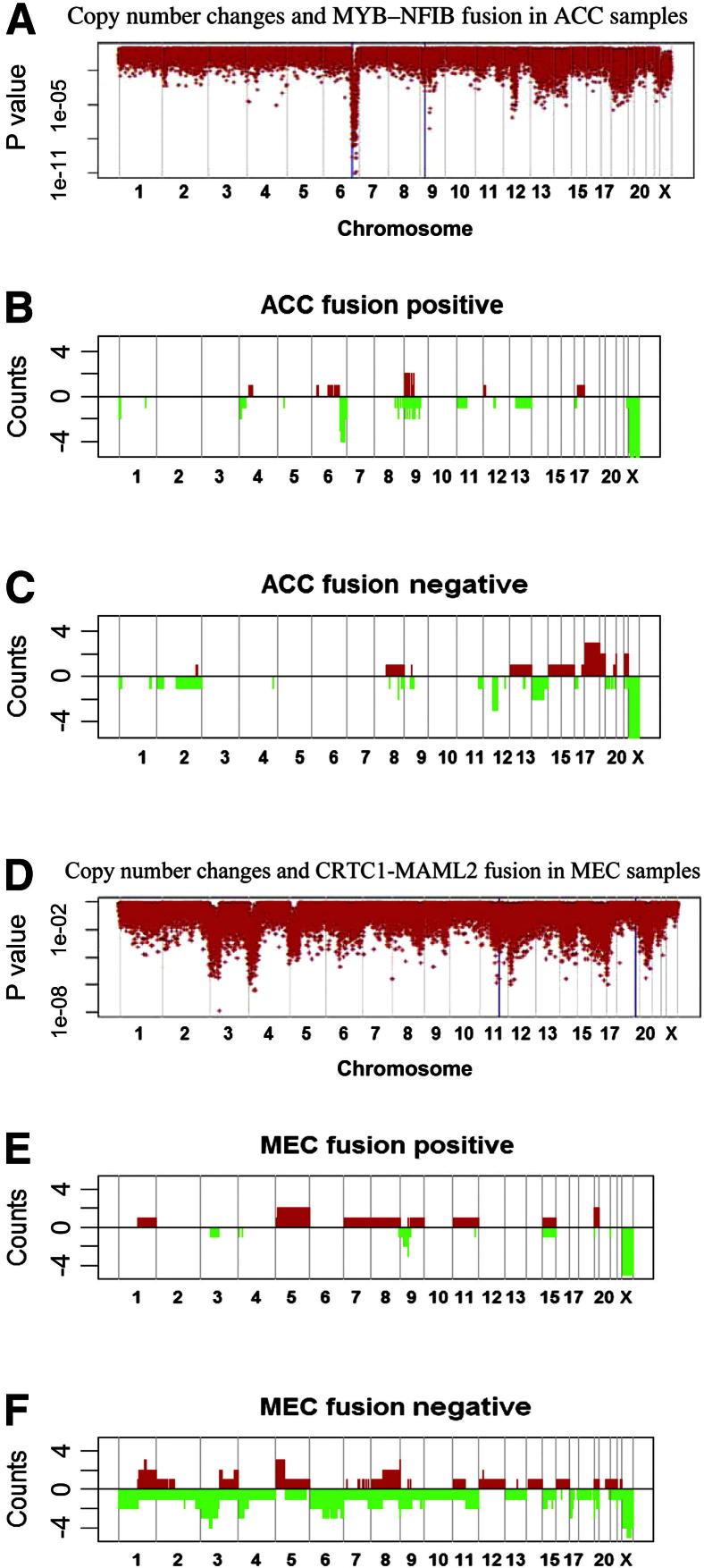
To determine whether CNAs differ on the basis of the presence or absence of fusion genes, we performed t-tests to scan for the mean copy number in fusion-positive and fusion-negative tumors. The P values shown in Figure 3A reflect the comparison of MYB-NFIB fusion-positive ACCs and fusion-negative ACCs. Highly significant associations were seen at 6q25.2 (P = 1.1 × 10−11) and 9p13.3 (P = 9.4 ×10−8) near the loci encoding MYB and NFIB. Figure 3, B and C, shows CNA distributions stratified by fusion status. Detailed copy number data at chromosomes 9 and 6 from fusion-positive and fusion-negative ACCs are shown in Supplemental Figure S3. Fusion-positive ACCs had fewer alterations than did fusion-negative tumors (Figure 3, B and C), with exclusive loss at 4p and 6q and gain only at 9q. Loss at 12q and chromosome 14 and gains at chromosomes 18 and 22 characterized fusion-negative ACCs. Interestingly, of the eight fusion-positive ACC samples, four had breakpoints within 0.5 Mb of the MYB sites (Supplemental Figure S3).
Figure 3.
Association between CNAs and gene fusion in ACCs and MECs. The x axis is ordered by chromosomal location. The vertical gray lines show the boundaries of the chromosomes. In A and B, the y axis shows log10 (P values), which were obtained from t-tests using copy number data for fusion-positive and fusion-negative samples. The blue lines and asterisks indicate the sites involved in gene fusion. Results regarding chromosome X should be ignored because they were affected by sex differences. C and D: The distribution of CNAs in fusion-positive and fusion-negative ACC samples. Gains are shown as red bars; losses are shown as green bars. Similarly, E and F show the distribution of CNAs in fusion-positive and fusion-negative samples of MECs, respectively.
Similarly, Figure 3, D–F, illustrates the association between CNAs and CRTC1-MAML2 fusion status in MECs. Judging from the t-test P values, a significant CNA difference was found at 11p15.1 (P = 2.6 ×10−4) near CRTC1 but not near MAML2. There also appear to be significant loss (P < 2 ×10−6) loci at 3p22.1, 4p15.1, 12p12.3, and 17p12. Overall, fusion-negative tumors had more CNAs than did fusion-positive tumors (Figure 3, E and F). Losses of 1p, 2q, 4q, 6q, 10, and 13 and gains of chromosomes 12, 14, 16, and 18 were mainly found in fusion-negative MECs. In both MECs and ACCs, fusion-positive tumors had relatively fewer CNAs than did fusion-negative tumors.
Validation Findings
We validated the CNA findings at the 1p36, 9p13.2, and 17q12 regions by FISH or IHC analysis. Examples of ERBB2 in SDC cases (Figure 2B) and the PAX5 gene in one SDC case (SDC10), as identified by the validation FISH, are shown in Supplemental Figure S4. In ERBB2- and PAX5-amplified cases (>8 copy numbers), the increase was proportional to the ploidy level of the tumor (Table 2). In addition, FISH analysis using control probe RP11-201K10 (1q21) and 1p36 target probes (RP11-219C24, RP11-163M9) were performed on seven tumors with 1p36 loss and configured the loss of one or two copies of 1p36 target probes.32 An IHC analysis using antibodies specific to CCND3 (6p21.1) revealed high nuclear expression of tumor cells in amplified cases (SDC4) and lack of nuclear staining in nonamplified cases (SDC10), as shown in Supplemental Figure S5.
Discussion
In this study, we characterized common and distinctive broad and focal genetic alterations in major SGCs. In general, fewer chromosomal aberrations were found in ACCs than in MECs and SDCs, underlining their perceived histological and clinical differences. The most common broad CNAs shared in all subtypes were located at chromosomes 6q23-26 and the 9p21 regions. Subtype-limited broad alterations were found in ACC and included losses at the 6q and 12q22,35,36 regions and gains at chromosomes 9p and 18. Although common loss at the 1p36 chromosome region was found in both ACC and SDC tumors, different loci within this region are associated with ACC and SDC.32 These findings, along with evidence of accelerated tumor growth in mouse models with deletion at this chromosomal region, highlight the potential importance of this region in oncogenesis.37–41 Moreover, these broad findings are in agreement with those previously reported in individual studies of these tumors.42–48
Our results highlight the localized chromosomal gains and losses among and between different neoplasm types. Surprisingly, multiple instances of localized gains and losses at chromosome 14q11.4, 15q11.2, and 17q21.3 regions were also observed in tumors and matched nonneoplastic salivary tissues. These regions were previously reported to be altered in North American population, presumably as common polymorphic sites.34 More important, the finding of common loss at the 9p21 locus, where the CDKN2A gene resides, in all three tumor types. This finding lends further evidence for the involvement of this gene in the early evolution of salivary tumors.47 We also noted a restricted loss of the 22q11.21 locus that houses the RANBP1 gene in several MECs. The role of this gene as a regulator of mitotic microtubule function and organization in MEC tumorigenesis, however, is unknown.49 Paradoxically, focal gain at the 17q12 region and amplification of the ERBB2 gene were found in several SDCs.
Our more localized CNN-LOH analysis revealed multiple restricted and shared alterations in several tumor subtypes. CNN-LOH at chromosome 1p36.33 was found in ACC, suggesting potential association with their ACC tumorigenesis.32 Strikingly, we noted concurrent CNN-LOH and focal amplifications at chromosome 12q15 and 12q21 regions in several SDCs. The findings indicate that these loci may either contain critical genes or represent focal regions of genomic instability. Interestingly, a recent study showed that an average of six focal recurrent deletions per tumor occur in human carcinomas, and these may result from a loss of tumor suppressor gene, genomic instability (fragile site), hemizygosity, and/or CNN-LOH.50 These findings, together with evidence for an association between homologous recombination and these events at sites with no known tumor-suppressor genes, lend further credence to our findings.50–52
Surprisingly, multiple mutually exclusive amplicons were identified in different tumor types; these included one amplicon in an ACC and one in an MEC at different sites on the 11p regions and multiple nonoverlapping amplicons at chromosomes 6p, 12q, and 17q in several SDCs. Not surprisingly, some of the amplicons have also been identified in high-grade mammary ductal carcinoma.31,53 Most of these amplicons occurred at regions that lacked copy number gains, suggesting that a limited number of oncogenes reside at these locations. Genes within these amplicons (Table 2), including CCND3, NUMA1, PAX5, CNOT2, RAB3IP, GLIPR1L1, ERBB2, CCL4, and PVRL1, have been linked to cell growth and apoptosis in several tumor types.54–56 Of particular interest is the identification of amplicons at the 17q12 region, where the ERBB2 gene resides, in several SDCs. Although high ERBB2 expression has been reported, a thorough validation was needed by our group and others. Our validation efforts confirm the amplification of known oncogenes within these amplicons and the likelihood of their involvement in the development and progression of some of these tumors.33,56–59
A comparative analysis of fusion-positive and fusion-negative ACCs and MECs revealed fewer CNAs in fusion-positive than fusion-negative tumors (Supplemental Figure S3). Interestingly, fusion-positive ACCs commonly had copy number loss at or near the 6q25 and 9p23 translocation sites, whereas fusion-negative ACCs had no alterations at these loci. In contrast, no discernible CNA differences were found at the 11p or 19q fusion sites in fusion-negative or fusion-positive MECs. Together, these results suggest that a genomic difference exists between tumors on the basis of fusion status; tumors with fusions are most likely driven by this event, but studies of human malignancies with fusion genes have revealed a similar association.13,20 Fusion-negative tumors arise by a different mechanism, such as genomic instability. Our clinicopathologic analysis in this relatively small cohort revealed no association between CNA and clinicopathologic features in patients with ACC and SDC; however, high CNAs were much more common in high-stage, high-grade, and clinically aggressive MECs. Collectively, the specificity of the reciprocal translocations in MECs and ACCs, along with the histogenetic and biological differences, strongly supports their segregation into distinct clinicopathologic entities in future targeted therapy trials.
In conclusion, we identified several CNA differences between SGC subtypes, supporting their genetic phenotypic classification for treatment.
Acknowledgments
We thank Jie Li and Irina I. Makovetskaya for technical assistance and Yan Cai, Deborah A. Rodriguez, and Stella U. Njoku for tissue retrieval and data collection.
Footnotes
Supported in part by the National Institute of Dental and Craniofacial Research, the Office of Rare Diseases Research grant U01DE019765, the Head and Neck SPORE (Specialized Programs of Research Excellence) program grant P50 CA097007, The Kenneth D. Muller Professorship, and National Cancer Institute grant CA-16672.
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Cancer Institute or the NIH.
Contributor Information
Li Zhang, Email: lzhangli@mdanderson.org.
Adel K. El-Naggar, Email: anaggar@mdanderson.org.
Supplemental Data
A: Representative plot of focal CNAs. The light blue points show the raw broad copy number data (top panel). The red points represent the denoised copy number data using a scanning window size of 25 SNP probes (bottom panel). The y axis shows the copy number values in a log2 scale of the sample. Note the focal alteration at chromosome 6 that was not visible on broad analysis. Focal gain identified on chromosome 6q in one MEC (bottom panel), but note corresponding raw CNAs (top panel). B: Representative plot of CNN-LOH. Copy number profile of sample MEC16 (top panel). The sum of allele-specific copy numbers (ASCN) at each genomic location equals that in the bottom panel. In chromosomes 5 and 9, no apparent copy number change was found, but the allele-specific copy number reveals compensatory changes in chromosomes 5 and 9.
LOH on chromosome 4 in ACC5 and its matching normal tissue (N3). No copy number change was found on chromosome 4 in the normal and matching tumor samples. This finding is consistent with both germline and somatic events that lead to uniparental disomy. The y axis shows the b-allelle fraction (BAF) values. The absence of values in the middle section indicates LOH.
Copy number data of ACC samples on chromosome 6. The x axis shows the physical positions of the SNP probes on chromosome 6. The red bars show the locus encoding the MYB gene. The y axis shows the copy number values. The sample name is indicated in the top right corner of each panel. The fusion-positive samples are ACC2, ACC3, ACC6, ACC10, ACC17, ACC18, ACC19, and ACC23. Copy number data of ACC samples on chromosome 9. The x axis shows the physical positions of the SNP probes on chromosome 9. The red bars show the locus encoding the NFIB gene. The y axis shows the copy number values. The sample name is indicated in the top right corner of each panel. The fusion-positive samples are ACC2, ACC3, ACC6, ACC10, ACC17, ACC18, ACC19, and ACC23.
Validation of 9p13.2 amplification in SDCs. FISH using PAX5 BAC clones (RP11-243F8, red) and centromeric chromosome 9 (green) probe; note PAX5 amplification in SDC10 (Table 2).
Validation of 6p21.1 amplification in SDCs. A: CCND3 nuclear-positive staining in tumor cells (SDC4, Table 2). B: CCND3 case with no amplifications showing no nuclear staining was detected in SDC 10 (Table 2).
References
- 1.Spiro R.H. Salivary neoplasms: overview of a 35-year experience with 2,807 patients. Head Neck Surg. 1986;8:177–184. doi: 10.1002/hed.2890080309. [DOI] [PubMed] [Google Scholar]
- 2.Goode R.K., Auclair P.L., Ellis G.L. Mucoepidermoid carcinoma of the major salivary glands: clinical and histopathologic analysis of 234 cases with evaluation of grading criteria. Cancer. 1998;82:1217–1224. doi: 10.1002/(sici)1097-0142(19980401)82:7<1217::aid-cncr2>3.0.co;2-c. [DOI] [PubMed] [Google Scholar]
- 3.Mitani Y., Li J., Rao P.H., Zhao Y.J., Bell D., Lippman S.M., Weber R.S., Caulin C., El-Naggar A.K. Comprehensive analysis of the MYB-NFIB gene fusion in salivary adenoid cystic carcinoma: incidence, variability, and clinicopathologic significance. Clin Cancer Res. 2010;16:4722–4731. doi: 10.1158/1078-0432.CCR-10-0463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Luna M.A., Batsakis J.G., Ordonez N.G., Mackay B., Tortoledo M.E. Salivary gland adenocarcinomas: a clinicopathologic analysis of three distinctive types. Semin Diagn Pathol. 1987;4:117–135. [PubMed] [Google Scholar]
- 5.Batsakis J.G., Regezi J.A., Luna M.A., El-Naggar A. Histogenesis of salivary gland neoplasms: a postulate with prognostic implications. J Laryngol Otol. 1989;103:939–944. doi: 10.1017/s0022215100110552. [DOI] [PubMed] [Google Scholar]
- 6.Murrah V.A., Batsakis J.G. Salivary duct carcinoma. Ann Otol Rhinol Laryngol. 1994;103:244–247. doi: 10.1177/000348949410300315. [DOI] [PubMed] [Google Scholar]
- 7.Chomette G., Auriol M., Tranbaloc P., Vaillant J.M. Adenoid cystic carcinoma of minor salivary glands: analysis of 86 cases: clinico-pathological, histoenzymological and ultrastructural studies. Virchows Arch A Pathol Anat Histol. 1982;395:289–301. doi: 10.1007/BF00429355. [DOI] [PubMed] [Google Scholar]
- 8.Ross D.A., Huaman J.A., Barsky S.H. A study of the heterogeneity of the mucoepidermoid tumor and the implication for future therapies. Arch Otolaryngol Head Neck Surg. 1992;118:1172–1178. doi: 10.1001/archotol.1992.01880110040010. [DOI] [PubMed] [Google Scholar]
- 9.El-Naggar A.K., Huvos A.G. In: Adenoid Cystic Carcinoma. Barnes L., Eveson J.W., Reichart P., Sidransky D., editors. IARC Press; Lyon, France: 2005. [Google Scholar]
- 10.Goode R.K., El-Naggar A.K. In: Mucoepidermoid Carcinoma. Barnes L.E.J., Reichert P., Sidransky D., editors. IARC Press; Lyon, France: 2005. [Google Scholar]
- 11.Tonon G., Modi S., Wu L., Kubo A., Coxon A.B., Komiya T., O'Neil K., Stover K., El-Naggar A., Griffin J.D., Kirsch I.R., Kaye F.J. t(11;19)(q21;p13) Translocation in mucoepidermoid carcinoma creates a novel fusion product that disrupts a Notch signaling pathway. Nat Genet. 2003;33:208–213. doi: 10.1038/ng1083. [DOI] [PubMed] [Google Scholar]
- 12.Tirado Y., Williams M.D., Hanna E.Y., Kaye F.J., Batsakis J.G., El-Naggar A.K. CRTC1/MAML2 fusion transcript in high grade mucoepidermoid carcinomas of salivary and thyroid glands and Warthin's tumors: implications for histogenesis and biologic behavior. Genes Chromosomes Cancer. 2007;46:708–715. doi: 10.1002/gcc.20458. [DOI] [PubMed] [Google Scholar]
- 13.Persson M., Andren Y., Mark J., Horlings H.M., Persson F., Stenman G. Recurrent fusion of MYB and NFIB transcription factor genes in carcinomas of the breast and head and neck. Proc Natl Acad Sci U S A. 2009;106:18740–18744. doi: 10.1073/pnas.0909114106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Glisson B., Colevas A.D., Haddad R., Krane J., El-Naggar A., Kies M., Costello R., Summey C., Arquette M., Langer C., Amrein P.C., Posner M. HER2 expression in salivary gland carcinomas: dependence on histological subtype. Clin Cancer Res. 2004;10:944–946. doi: 10.1158/1078-0432.ccr-03-0253. [DOI] [PubMed] [Google Scholar]
- 15.Haddad R., Colevas A.D., Krane J.F., Cooper D., Glisson B., Amrein P.C., Weeks L., Costello R., Posner M. Herceptin in patients with advanced or metastatic salivary gland carcinomas: a phase II study. Oral Oncol. 2003;39:724–727. doi: 10.1016/s1368-8375(03)00097-6. [DOI] [PubMed] [Google Scholar]
- 16.Locati L.D., Perrone F., Losa M., Mela M., Casieri P., Orsenigo M., Cortelazzi B., Negri T., Tamborini E., Quattrone P., Bossi P., Rinaldi G., Bergamini C., Calderone R.G., Liberatoscioli C., Licitra L. Treatment relevant target immunophenotyping of 139 salivary gland carcinomas (SGCs) Oral Oncol. 2009;45:986–990. doi: 10.1016/j.oraloncology.2009.05.635. [DOI] [PubMed] [Google Scholar]
- 17.Nashed M., Casasola R.J. Biological therapy of salivary duct carcinoma. J Laryngol Otol. 2009;123:250–252. doi: 10.1017/S0022215108002314. [DOI] [PubMed] [Google Scholar]
- 18.Mitelman F., Johansson B., Mertens F. The impact of translocations and gene fusions on cancer causation. Nat Rev Cancer. 2007;7:233–245. doi: 10.1038/nrc2091. [DOI] [PubMed] [Google Scholar]
- 19.Mani R.S., Chinnaiyan A.M. Triggers for genomic rearrangements: insights into genomic, cellular and environmental influences. Nat Rev Genet. 2010;11:819–829. doi: 10.1038/nrg2883. [DOI] [PubMed] [Google Scholar]
- 20.Stenman G. Fusion oncogenes and tumor type specificity: insights from salivary gland tumors. Semin Cancer Biol. 2005;15:224–235. doi: 10.1016/j.semcancer.2005.01.002. [DOI] [PubMed] [Google Scholar]
- 21.Albertson D.G., Collins C., McCormick F., Gray J.W. Chromosome aberrations in solid tumors. Nat Genet. 2003;34:369–376. doi: 10.1038/ng1215. [DOI] [PubMed] [Google Scholar]
- 22.Taylor B.S., Barretina J., Maki R.G., Antonescu C.R., Singer S., Ladanyi M. Advances in sarcoma genomics and new therapeutic targets. Nat Rev Cancer. 2011;11:541–557. doi: 10.1038/nrc3087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rutherford S., Hampton G.M., Frierson H.F., Moskaluk C.A. Mapping of candidate tumor suppressor genes on chromosome 12 in adenoid cystic carcinoma. Lab Invest. 2005;85:1076–1085. doi: 10.1038/labinvest.3700314. [DOI] [PubMed] [Google Scholar]
- 24.Bernheim A., Toujani S., Saulnier P., Robert T., Casiraghi O., Validire P., Temam S., Menard P., Dessen P., Fouret P. High-resolution array comparative genomic hybridization analysis of human bronchial and salivary adenoid cystic carcinoma. Lab Invest. 2008;88:464–473. doi: 10.1038/labinvest.2008.18. [DOI] [PubMed] [Google Scholar]
- 25.Morio T., Morimitsu Y., Hisaoka M., Makishima K., Hashimoto H. DNA copy number changes in carcinoma in pleomorphic adenoma of the salivary gland: a comparative genomic hybridization study. Pathol Int. 2002;52:501–507. doi: 10.1046/j.1440-1827.2002.01388.x. [DOI] [PubMed] [Google Scholar]
- 26.Persson F., Winnes M., Andren Y., Wedell B., Dahlenfors R., Asp J., Mark J., Enlund F., Stenman G. High-resolution array CGH analysis of salivary gland tumors reveals fusion and amplification of the FGFR1 and PLAG1 genes in ring chromosomes. Oncogene. 2008;27:3072–3080. doi: 10.1038/sj.onc.1210961. [DOI] [PubMed] [Google Scholar]
- 27.Amatschek S., Koenig U., Auer H., Steinlein P., Pacher M., Gruenfelder A., Dekan G., Vogl S., Kubista E., Heider K.H., Stratowa C., Schreiber M., Sommergruber W. Tissue-wide expression profiling using cDNA subtraction and microarrays to identify tumor-specific genes. Cancer Res. 2004;64:844–856. doi: 10.1158/0008-5472.can-03-2361. [DOI] [PubMed] [Google Scholar]
- 28.Liu C.J., Lin S.C., Chen Y.J., Chang K.M., Chang K.W. Array-comparative genomic hybridization to detect genomewide changes in microdissected primary and metastatic oral squamous cell carcinomas. Mol Carcinog. 2006;45:721–731. doi: 10.1002/mc.20213. [DOI] [PubMed] [Google Scholar]
- 29.Myllykangas S., Himberg J., Bohling T., Nagy B., Hollmen J., Knuutila S. DNA copy number amplification profiling of human neoplasms. Oncogene. 2006;25:7324–7332. doi: 10.1038/sj.onc.1209717. [DOI] [PubMed] [Google Scholar]
- 30.Tuna M., Knuutila S., Mills G.B. Uniparental disomy in cancer. Trends Mol Med. 2009;15:120–128. doi: 10.1016/j.molmed.2009.01.005. [DOI] [PubMed] [Google Scholar]
- 31.Smith A.C., Shuman C., Chitayat D., Steele L., Ray P.N., Bourgeois J., Weksberg R. Severe presentation of Beckwith-Wiedemann syndrome associated with high levels of constitutional paternal uniparental disomy for chromosome 11p15. Am J Med Genet A. 2007;143A:3010–3015. doi: 10.1002/ajmg.a.32030. [DOI] [PubMed] [Google Scholar]
- 32.Rao P.H., Roberts D., Zhao Y.J., Bell D., Harris C.P., Weber R.S., El-Naggar A.K. Deletion of 1p32-p36 is the most frequent genetic change and poor prognostic marker in adenoid cystic carcinoma of the salivary glands. Clin Cancer Res. 2008;14:5181–5187. doi: 10.1158/1078-0432.CCR-08-0158. [DOI] [PubMed] [Google Scholar]
- 33.Williams M.D., Roberts D.B., Kies M.S., Mao L., Weber R.S., El-Naggar A.K. Genetic and expression analysis of HER-2 and EGFR genes in salivary duct carcinoma: empirical and therapeutic significance. Clin Cancer Res. 2010;16:2266–2274. doi: 10.1158/1078-0432.CCR-09-0238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zogopoulos G., Ha K.C., Naqib F., Moore S., Kim H., Montpetit A., Robidoux F., Laflamme P., Cotterchio M., Greenwood C., Scherer S.W., Zanke B., Hudson T.J., Bader G.D., Gallinger S. Germ-line DNA copy number variation frequencies in a large North American population. Hum Genet. 2007;122:345–353. doi: 10.1007/s00439-007-0404-5. [DOI] [PubMed] [Google Scholar]
- 35.Harada T., Chelala C., Bhakta V., Chaplin T., Caulee K., Baril P., Young B.D., Lemoine N.R. Genome-wide DNA copy number analysis in pancreatic cancer using high-density single nucleotide polymorphism arrays. Oncogene. 2008;27:1951–1960. doi: 10.1038/sj.onc.1210832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kasamatsu A., Endo Y., Uzawa K., Nakashima D., Koike H., Hashitani S., Numata T., Urade M., Tanzawa H. Identification of candidate genes associated with salivary adenoid cystic carcinomas using combined comparative genomic hybridization and oligonucleotide microarray analyses. Int J Biochem Cell Biol. 2005;37:1869–1880. doi: 10.1016/j.biocel.2005.04.004. [DOI] [PubMed] [Google Scholar]
- 37.Bergamaschi A., Hjortland G.O., Triulzi T., Sorlie T., Johnsen H., Ree A.H., Russnes H.G., Tronnes S., Maelandsmo G.M., Fodstad O., Borresen-Dale A.L., Engebraaten O. Molecular profiling and characterization of luminal-like and basal-like in vivo breast cancer xenograft models. Mol Oncol. 2009;3:469–482. doi: 10.1016/j.molonc.2009.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bagchi A., Mills A.A. The quest for the 1p36 tumor suppressor. Cancer Res. 2008;68:2551–2556. doi: 10.1158/0008-5472.CAN-07-2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thompson P.M., Gotoh T., Kok M., White P.S., Brodeur G.M. CHD5, a new member of the chromodomain gene family, is preferentially expressed in the nervous system. Oncogene. 2003;22:1002–1011. doi: 10.1038/sj.onc.1206211. [DOI] [PubMed] [Google Scholar]
- 40.Garnis C., Campbell J., Davies J.J., Macaulay C., Lam S., Lam W.L. Involvement of multiple developmental genes on chromosome 1p in lung tumorigenesis. Hum Mol Genet. 2005;14:475–482. doi: 10.1093/hmg/ddi043. [DOI] [PubMed] [Google Scholar]
- 41.Ragnarsson G., Eiriksdottir G., Johannsdottir J.T., Jonasson J.G., Egilsson V., Ingvarsson S. Loss of heterozygosity at chromosome 1p in different solid human tumours: association with survival. Br J Cancer. 1999;79:1468–1474. doi: 10.1038/sj.bjc.6690234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bagchi A., Papazoglu C., Wu Y., Capurso D., Brodt M., Francis D., Bredel M., Vogel H., Mills A.A. CHD5 is a tumor suppressor at human 1p36. Cell. 2007;128:459–475. doi: 10.1016/j.cell.2006.11.052. [DOI] [PubMed] [Google Scholar]
- 43.Vekony H., Ylstra B., Wilting S.M., Meijer G.A., van de Wiel M.A., Leemans C.R., van der Waal I., Bloemena E. DNA copy number gains at loci of growth factors and their receptors in salivary gland adenoid cystic carcinoma. Clin Cancer Res. 2007;13:3133–3139. doi: 10.1158/1078-0432.CCR-06-2555. [DOI] [PubMed] [Google Scholar]
- 44.El-Rifai W., Rutherford S., Knuutila S., Frierson H.F., Jr., Moskaluk C.A. Novel DNA copy number losses in chromosome 12q12–q13 in adenoid cystic carcinoma. Neoplasia. 2001;3:173–178. doi: 10.1038/sj.neo.7900158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Freier K., Flechtenmacher C., Walch A., Ohl S., Devens F., Burke B., Hassfeld S., Lichter P., Joos S., Hofele C. Copy number gains on 22q13 in adenoid cystic carcinoma of the salivary gland revealed by comparative genomic hybridization and tissue microarray analysis. Cancer Genet Cytogenet. 2005;159:89–95. doi: 10.1016/j.cancergencyto.2004.09.007. [DOI] [PubMed] [Google Scholar]
- 46.Yu Y., Baras A.S., Shirasuna K., Frierson H.F., Jr., Moskaluk C.A. Concurrent loss of heterozygosity and copy number analysis in adenoid cystic carcinoma by SNP genotyping arrays. Lab Invest. 2007;87:430–439. doi: 10.1038/labinvest.3700536. [DOI] [PubMed] [Google Scholar]
- 47.Cerilli L.A., Swartzbaugh J.R., Saadut R., Marshall C.E., Rumpel C.A., Moskaluk C.A., Frierson H.F., Jr. Analysis of chromosome 9p21 deletion and p16 gene mutation in salivary gland carcinomas. Hum Pathol. 1999;30:1242–1246. doi: 10.1016/s0046-8177(99)90044-8. [DOI] [PubMed] [Google Scholar]
- 48.Bergamaschi A., Kim Y.H., Wang P., Sørlie T., Hernandez-Boussard T., Lonning P.E., Tibshirani R., Børresen-Dale A.L., Pollack J.R. Distinct patterns of DNA copy number alteration are associated with different clinicopathological features and gene-expression subtypes of breast cancer. Genes Chromosomes Cancer. 2006;45:1033–1040. doi: 10.1002/gcc.20366. [DOI] [PubMed] [Google Scholar]
- 49.Rensen W.M., Roscioli E., Tedeschi A., Mangiacasale R., Ciciarello M., Di Gioia S.A., Lavia P. RanBP1 downregulation sensitizes cancer cells to taxol in a caspase-3-dependent manner. Oncogene. 2009;28:1748–1758. doi: 10.1038/onc.2009.24. [DOI] [PubMed] [Google Scholar]
- 50.Solimini N.L., Xu Q., Mermel C.H., Liang A.C., Schlabach M.R., Luo J., Burrows A.E., Anselmo A.N., Bredemeyer A.L., Li M.Z., Beroukhim R., Meyerson M., Elledge S.J. Recurrent hemizygous deletions in cancers may optimize proliferative potential. Science. 2012;337:104–109. doi: 10.1126/science.1219580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Reliene R., Bishop A.J., Schiestl R.H. Involvement of homologous recombination in carcinogenesis. Adv Genet. 2007;58:67–87. doi: 10.1016/S0065-2660(06)58003-4. [DOI] [PubMed] [Google Scholar]
- 52.Cox C., Bignell G., Greenman C., Stabenau A., Warren W., Stephens P., Davies H., Watt S., Teague J., Edkins S., Birney E., Easton D.F., Wooster R., Futreal P.A., Stratton M.R. A survey of homozygous deletions in human cancer genomes. Proc Natl Acad Sci U S A. 2005;102:4542–4547. doi: 10.1073/pnas.0408593102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.van Dartel M., Hulsebos T.J. Amplification and overexpression of genes in 17p11.2 ∼ p12 in osteosarcoma. Cancer Genet Cytogenet. 2004;153:77–80. doi: 10.1016/j.cancergencyto.2004.03.007. [DOI] [PubMed] [Google Scholar]
- 54.Roijer E., Nordkvist A., Strom A.K., Ryd W., Behrendt M., Bullerdiek J., Mark J., Stenman G. Translocation, deletion/amplification, and expression of HMGIC and MDM2 in a carcinoma ex pleomorphic adenoma. Am J Pathol. 2002;160:433–440. doi: 10.1016/S0002-9440(10)64862-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Harris T., Pan Q., Sironi J., Lutz D., Tian J., Sapkar J., Perez-Soler R., Keller S., Locker J. Both gene amplification and allelic loss occur at 14q13.3 in lung cancer. Clin Cancer Res. 2011;17:690–699. doi: 10.1158/1078-0432.CCR-10-1892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pruneri G., Valentini S., Fabris S., Del Curto B., Laszlo D., Bertolini F., Martinelli G., Leocata P., Viale G., Neri A. Cyclin D3 immunoreactivity in follicular lymphoma is independent of the t(6;14)(p21.1;q32.3) translocation or cyclin D3 gene amplification and is correlated with histologic grade and Ki-67 labeling index. Int J Cancer. 2004;112:71–77. doi: 10.1002/ijc.20354. [DOI] [PubMed] [Google Scholar]
- 57.Santarius T., Shipley J., Brewer D., Stratton M.R., Cooper C.S. A census of amplified and overexpressed human cancer genes. Nat Rev Cancer. 2010;10:59–64. doi: 10.1038/nrc2771. [DOI] [PubMed] [Google Scholar]
- 58.Rabbitts T.H., Stocks M.R. Chromosomal translocation products engender new intracellular therapeutic technologies. Nat Med. 2003;9:383–386. doi: 10.1038/nm0403-383. [DOI] [PubMed] [Google Scholar]
- 59.Lips E.H., de Graaf E.J., Tollenaar R.A., van Eijk R., Oosting J., Szuhai K., Karsten T., Nanya Y., Ogawa S., van de Velde C.J., Eilers P.H., van Wezel T., Morreau H. Single nucleotide polymorphism array analysis of chromosomal instability patterns discriminates rectal adenomas from carcinomas. J Pathol. 2007;212:269–277. doi: 10.1002/path.2180. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A: Representative plot of focal CNAs. The light blue points show the raw broad copy number data (top panel). The red points represent the denoised copy number data using a scanning window size of 25 SNP probes (bottom panel). The y axis shows the copy number values in a log2 scale of the sample. Note the focal alteration at chromosome 6 that was not visible on broad analysis. Focal gain identified on chromosome 6q in one MEC (bottom panel), but note corresponding raw CNAs (top panel). B: Representative plot of CNN-LOH. Copy number profile of sample MEC16 (top panel). The sum of allele-specific copy numbers (ASCN) at each genomic location equals that in the bottom panel. In chromosomes 5 and 9, no apparent copy number change was found, but the allele-specific copy number reveals compensatory changes in chromosomes 5 and 9.
LOH on chromosome 4 in ACC5 and its matching normal tissue (N3). No copy number change was found on chromosome 4 in the normal and matching tumor samples. This finding is consistent with both germline and somatic events that lead to uniparental disomy. The y axis shows the b-allelle fraction (BAF) values. The absence of values in the middle section indicates LOH.
Copy number data of ACC samples on chromosome 6. The x axis shows the physical positions of the SNP probes on chromosome 6. The red bars show the locus encoding the MYB gene. The y axis shows the copy number values. The sample name is indicated in the top right corner of each panel. The fusion-positive samples are ACC2, ACC3, ACC6, ACC10, ACC17, ACC18, ACC19, and ACC23. Copy number data of ACC samples on chromosome 9. The x axis shows the physical positions of the SNP probes on chromosome 9. The red bars show the locus encoding the NFIB gene. The y axis shows the copy number values. The sample name is indicated in the top right corner of each panel. The fusion-positive samples are ACC2, ACC3, ACC6, ACC10, ACC17, ACC18, ACC19, and ACC23.
Validation of 9p13.2 amplification in SDCs. FISH using PAX5 BAC clones (RP11-243F8, red) and centromeric chromosome 9 (green) probe; note PAX5 amplification in SDC10 (Table 2).
Validation of 6p21.1 amplification in SDCs. A: CCND3 nuclear-positive staining in tumor cells (SDC4, Table 2). B: CCND3 case with no amplifications showing no nuclear staining was detected in SDC 10 (Table 2).