Abstract
Peroxisome proliferator–activated receptor gamma (PPARγ) agonists have anticancer activity and influence cell differentiation. We examined the impact of the selective PPARγ agonist efatutazone on mammary cancer pathogenesis in a mouse model of BRCA1 mutation. Mice with conditional loss of full-length BRCA1 targeted to mammary epithelial cells in association with germline TP53 insufficiency were treated with efatutazone through the diet starting at age 4 months and were euthanized at age 12 months or when palpable tumor reached 1 cm3. Although treatment did not reduce percentage of mice developing invasive cancer, it significantly reduced prevalence of noninvasive cancer and total number of cancers per mouse and increased prevalence of well-differentiated cancer subtypes not usually seen in this mouse model. Invasive cancers from controls were uniformly estrogen receptor α negative and undifferentiated, whereas well-differentiated estrogen receptor α–positive papillary invasive cancers appeared in efatutazone-treated mice. Expression levels of phosphorylated AKT and CDK6 were significantly reduced in the cancers developing in efatutazone-treated mice. Efatutazone treatment reduced rates of mammary epithelial cell proliferation and development of hyperplastic alveolar nodules and increased expression levels of the PPARγ target genes Adfp, Fabp4, and Pdhk4 in preneoplastic mammary tissue. Intervention efatutazone treatment in mice with BRCA1 deficiency altered mammary cancer development by promoting development of differentiated invasive cancer and reducing prevalence of noninvasive cancer and preneoplastic disease.
The anticancer activities of peroxisome proliferator–activated receptor gamma (PPARγ) agonists include promotion of differentiation1–5 and apoptosis6 in addition to inhibition of cell proliferation,6,7 inflammation,3 and angiogenesis.8 The agent studied herein, efatutazone (CS-7017/RS5444), is a selective high-affinity thiazolidinedione (TZD)-class PPARγ agonist that can induce PPARγ-dependent transactivation but cannot activate either PPARα or PPARδ transactivation.9–11 Efatutazone restrains growth of human anaplastic thyroid and colon cancer cells in xenograft mouse models9,11,12 and inhibits formation and progression of azoxymethane-induced colonic adenomas in mice.13 In anaplastic thyroid cancer cells, efatutazone reduces cell proliferation through a PPARγ-dependent mechanism that affects activation of the Rho-related GTP-binding protein RhoB and CDK inhibitor 1 signaling pathways.11,12 A recent phase 1 trial in patients with advanced malignancy demonstrated acceptable toxicity with some evidence of disease control.14 The EC50 of 0.20 nmol/L reported for PPARγ promoter activation by efatutazone is estimated to be 1/50th of the EC50 of the more commonly used PPARγ agonist rosiglitazone.2,9 An appropriate in vivo dose for pathophysiologic experiments is known from published dose-ranging and efficacy studies previously performed, including studies in mouse models.9,11–13
PPARγ belongs to a family of nuclear receptors that bind to peroxisome proliferator hormone response elements located in the promoters of target genes.15,16 PPARγ binds to members of the retinoic X receptor (RXR) family as heterodimers. Ligands of PPARγ can stimulate target gene transcription, including adipose differentiation–related protein (Adfp), fatty acid binding protein 4 (Fabp4), and pyruvate dehydrogenase kinase isozyme 4 (Pdhk4).15,16 Therapeutic roles for TZD-class PPARγ agonists are under debate. Although there is evidence that this drug class reduces preneoplastic and cancer cell growth and differentiation4,5,17–22 with alterations in cell-cycle proteins,11,12,17–19,22 including RAC-α serine/threonine-protein kinase (AKT),20,21 in vivo changes in expression levels of cell-cycle proteins are not always accompanied by alterations in cell growth,16 and results from many studies using PPARγ agonists as single agents have not been promising.23,24 Moreover, three TZD-class drugs that were Food and Drug Administration approved25 for diabetes are currently restricted owing to an increased risk of cardiovascular events (rosiglitazone; http://www.fda.gov/Drugs/DrugSafety/PostmarketDrugSafetyInformationforPatientsandProviders/ucm143349.htm, last accessed April 7, 2013), under investigation for raising the risk of bladder cancer (pioglitazone; http://www.fda.gov/Drugs/DrugSafety/PostmarketDrugSafetyInformationforPatientsandProviders/ucm109136.htm, last accessed April 7, 2013), or withdrawn owing to hepatotoxicity (troglitazone; http://www.fda.gov/Safety/MedWatch/SafetyInformation/SafetyAlertsforHumanMedicalProducts/ucm173081.htm, last accessed April 7, 2013). Finally, off-target PPARγ-independent anticancer effects of some TZD-class drugs have been identified, complicating the interpretation of experiments showing reductions in cancer cell growth.26 Efatutazone was tested in this study because it is a selective PPARγ agonist with high affinity for PPARγ that showed acceptable toxicity in a phase 1 trial,14 because interest remains in developing PPARγ-modulating drugs27 for specific settings,23 and because the impact of TZD-class drugs on the development of mammary cancer initiated by loss of function of the genetic risk factor breast cancer 1, early onset (Brca1) was not defined.
Genetically engineered mice that undergo a mammary epithelial cell–targeted deletion of Brca1 exon 11 in somatic cells coupled with germline tumor protein p53 (TP53) haploinsufficiency are an established tool for studies of BRCA1 mutation–related breast cancer pathogenesis. These mice model the predilection for the development of poorly differentiated, triple-negative/basal-type mammary cancers found in human patients with BRCA1 mutation.28–35 The cell of origin for BRCA1 mutation–related human and mouse mammary cancer is reported to be a luminal estrogen receptor α (ERα)–negative mammary epithelial progenitor cell36 that can show a basal-like differentiation pattern.37 Inactivation of BRCA1 in ERα-negative cancer stem cells is hypothesized to push cancer development toward a basal-type phenotype.38 Loss of normal BRCA1 function in mammary epithelial cells alters cell differentiation and fate specification so that luminal cells are, molecularly speaking, more basal-like.39 In 7,12-dimethylbenz(a)anthracene–treated mice carrying two intact BRCA1 genes, another PPARγ agonist, GW7845, promotes the appearance of more differentiated ERα-positive mammary adenocarcinomas.10 However, it is also known that PPARγ activation can induce a stellate cell morphology corresponding to triple-negative pathobiology in three-dimensional cell culture in vitro.40 These investigations were initiated to assess the in vivo impact of a PPARγ agonist on mammary cancer subtype development in the setting of BRCA1 deficiency.
Intervention treatment with the PPARγ agonist efatutazone at age 4 months did not change the prevalence of cancers >1 cm3 by age 12 months but did significantly decrease the total number of cancers formed, promoted the development of well-differentiated cancer subtypes not found in the absence of drug treatment, and reduced the levels of phosphorylated AKT (pAKT) and CDK6 expression levels in the invasive cancers formed. This was accompanied by a decline in preneoplasia prevalence and decreased rates of mammary epithelial cell proliferation.
Materials and Methods
Mouse Model, Efatutazone Administration, and Necropsy
C57Bl/6 mice exhibiting loss of full-length BRCA1 expression in mammary epithelial cells through conditional Mouse Mammary Tumor Virus (MMTV)-Cre (Line D)41 transgene-mediated Brca1 floxed exon 11 (f11) deletion accompanied by loss of one germline copy of TP53 (Brca1f11/f11/p53+/-/MMTV-Cre)28–31,33,34,41 were identified using tail samples (Transnetyx Inc., Cordova, TN). Efatutazone was administered through the diet (F3028, rodent diet, grain-based, 1/2-in pellets; Bio-Serv, Frenchtown, NJ) at a 30-mg/kg concentration, a dose selected for its maximal efficacy (antitumor activity) and limited toxicity profile (bioavailability in fasted mice: 86.6%; Daiichi Sankyo, Tokyo, Japan),9,11–13 starting at 4 months of age. Control mice received the same diet without added efatutazone. Mice were necropsied at 12 months of age or when the largest palpable tumor size reached 1 cm3. Cohorts of control (n = 13) and efatutazone-treated (n = 13) Brca1f11/f11/p53+/-/MMTV-Cre mice were entered into the study. Two mice in the control group were excluded: one was sacrificed owing to an untreatable skin condition and one was found dead. Five mice in the efatutazone-treated group were excluded: one was sacrificed owing to an untreatable skin condition, three were found dead, and one developed lymphoma, resulting in n = 11 control and n = 8 efatutazone evaluable mice. Mammary gland cancers ≥1 cm3 (n = 10 control mice, n = 10 efatutazone-treated mice) were harvested and divided: half were fixed in 10% buffered formalin overnight at 4°C and embedded in paraffin and half were snap frozen in liquid nitrogen and stored at −80°C. One inguinal mammary gland was prepared for whole mount,29,31 and the other, as available, was fixed in 10% buffered formalin overnight at 4°C and was embedded in paraffin. One mammary gland was snap frozen in liquid nitrogen and stored at −80°C. Animal procedures were performed in accordance with federal guidelines and were approved by the Georgetown University Institutional Animal Care and Use Committee.
Whole-Mount Analysis, Immunohistochemical Analysis, and Pathologic Evaluation
Mammary gland whole mounts fixed in Carnoy’s solution and stained in carmine alum were evaluated for the presence or absence of dense lobular growth and number of hyperplastic alveolar nodules (HANs) per gland (n = 10 control mice; n = 8 efatutazone-treated mice). Images were obtained using a Nikon Eclipse E800M microscope equipped with a Nikon DXM1200 camera (Nikon Instruments Inc., Melville, NY).29,31 Five-micrometer sections of formalin-fixed, paraffin-embedded tissue were stained with H&E for histologic evaluation.42 The presence or absence of nonpalpable invasive and noninvasive cancer, hyperplasia, and stromal pathology or alteration with efatutazone treatment was scored by a board-certified pathologist (B. V.S.K.) on one H&E section taken from the center of one inguinal mammary gland of each mouse (n = 8 control mice, n = 8 efatutazone-treated mice). Cancer multiplicity was defined as the number of cancers detected by either external palpation (>1 cm3) or examination of one H&E section taken from the center of the inguinal mammary gland from each mouse with cancer. Serial 5-μm sections of mammary glands were used for the detection of protein expression by immunohistochemical (IHC) analysis performed using the Vectastain ABC kit (Vector Laboratories, Burlingame, CA) or Mouse on Mouse (M.O.M.) peroxidase kit (PK-2200; Vector Laboratories) as appropriate using the following primary antibodies: ERα (dilution 1:750; SC-542; Santa Cruz Biotechnology Inc., Santa Cruz, CA), progesterone receptor (PGR) (dilution 1:250; SC-538; Santa Cruz Biotechnology Inc.), Ki-67 protein (dilution 1:100; NCL-L-Ki-67-MM1; Novocastra, Newcastle on Tyne, UK), cyclin D1 (dilution 1:50; SP4; RM-9104-S; NeoMarkers, Thermo Scientific, Fremont, CA), cyclin E (dilution 1:80; SC-198; Santa Cruz Biotechnology Inc.), pAKT serine (ser473) (dilution 1:35; D9E; Cell Signaling Technology Inc., Danvers, MA), AKT (pan) (dilution 1:600; C67E7; Cell Signaling Technology Inc.), retinoblastoma-associated protein (RB) (dilution 1:25; sc-50; Santa Cruz Biotechnology Inc.), serine (ser807/811) phosphorylated RB (pRB) (dilution 1:100; 93083s; Cell Signaling Technology Inc.), CDK4 (dilution 1:2000; sc-260; Santa Cruz Biotechnology Inc.), CDK6 (dilution 1:750; SAB4300596; Sigma-Aldrich, St. Louis, MO), keratin, type II cytoskeletal 5 (CK5) (dilution 1:1000; PRB-160P; Covance Inc., Princeton, NJ), tumor protein 63 (p63) (dilution 1:1000; MS-107-P0; NeoMarkers), PPARγ (dilution 1:60; sc-7196; Santa Cruz Biotechnology Inc.), RXRα (1:300; sc-553; Santa Cruz Biotechnology Inc.), and PPARα (dilution 1:300; ab8934; Abcam Inc., Cambridge, MA) following either manufacturer instructions or as previously published.16,29,31,43–47 IHC analysis was performed on sections of cancer tissue (n = 10 control mice, n = 10 efatutazone-treated mice; exceptions are pRb: n = 8 control mice; and PPARα: n = 8 control mice and n = 3 efatutazone-treated mice) and mammary glands (n = 10 control mice, n = 8 efatutazone-treated mice; exceptions are PPARα: n = 5 control mice and n = 5 efatutazone-treated mice). Percentages of epithelial cells demonstrating nuclear-localized ERα, PGR, or Ki-67 were calculated by counting ≥500 cells per section. Cancers were designated as ERα or PGR positive if >10% of the cancer cells demonstrated nuclear-localized expression. The proliferation index was calculated as the percentage of epithelial cells with nuclear-localized Ki-67 in ≥500 epithelial cells per mammary gland section. Qualitative IHC scoring included measures of intensity (0 indicates no stain; 1, weak; 2, intermediate; and 3, strong) and proportion of cells stained (1 indicates ≤1/3 positive cells; 2, 1/3 to 2/3 positive cells; and 3, ≥2/3 positive cells). The average of the proportion and intensity scores was used to determine a final IHC score for pAKT Ser473, AKT (pan), pRB, RB, cyclin D1, cyclin E, CDK4, CDK6, PPARα, PPARγ, and RXRα as follows: 0 indicates none; 1, low; 2, medium; and 3, high. Presence and absence of staining in myoepithelial and cancer cells was evaluated for CK5 and p63 (n = 8 control mice, n = 7 efatutazone-treated mice). A board-certified academic pathologist (B.V.S.K.) blinded to the identity and treatment group of the samples read the mammary cancer, epithelial, and stromal pathology reports.
RNA Isolation and Real-Time RT-PCR
Total RNA was isolated using TRIzol reagent (Life Technologies, Grand Island, NY), and cDNA prepared from 2 μg of total RNA by a reverse transcription reaction. Three independent samples of mammary gland tissue from the efatutazone-treated and untreated cohorts were randomly selected for analysis. Taqman gene expression assays (Life Technologies) were used to detect Adfp (Mm00475794_m1), Fabp4 (Mm00445878_m1), Pdhk4 (Mm01166879_m1), and eukaryotic 18s rRNA (Hs99999901_s1). Reactions were performed according to the manufacturer's instructions using the ABI Prism 7700 sequence detector and ABI SDS 2.1 software version 2.1 (Life Technologies). Fold change in mRNA expression was calculated using the CT method (2-ΔΔCT method).48
Western Blot Analysis
Total protein samples (15 μg per lane) isolated from cancer tissue were electrophoresed on 4% to 12% gradient Bis-Tris gels (NP0335; Life Technologies), transferred to polyvinylidene difluoride membranes (EMD Millipore, Billerica, MA), and blotted using primary antibody against pAKT Ser473 (dilution 1:1000; D9E; Cell Signaling Technology Inc.), pAKT Threonine (Thr)308 (dilution 1:1000; C31E5; Cell Signaling Technology Inc.), AKT (pan) (dilution 1:1000; C67E7; Cell Signaling Technology Inc.), CDK6 (dilution 1:1000; DCS83; Cell Signaling Technology Inc.), and CDK4 (dilution 1:1000; DCS156; Cell Signaling Technology Inc.). The blot was incubated with Amersham ECL (GE Healthcare, Piscataway, NJ) and horseradish peroxidase–conjugated secondary antibody (dilution 1:10,000; GE Healthcare) and was visualized using the SuperSignal West Pico chemiluminescent substrate (Thermo Scientific) and Amersham Hyperfilm ECL (GE Healthcare), or the blot was incubated with Odyssey IRDye 680LT or 800CW (LI-COR Biosciences, Lincoln, NE) as appropriate and visualized using a LI-COR Odyssey infrared imager (LI-COR Biosciences). Protein markers (Bio-Rad Laboratories, Hercules, CA) were used as molecular standards. Western blots were quantified by measuring means ± SEM relative densities (Photoshop CS5; Adobe Systems Inc., San Jose, CA; or Odyssey Image Studio software version 3.0; LI-COR Biosciences). Expression levels of pAKT were normalized to AKT (pan). AKT (pan), CDK4, and CDK6 were normalized to actin. Samples were randomly selected from each group for quantitative Western blot analysis: pAKT Thr308: n = 3 control mice, n = 6 efatutazone-treated mice; pAKT Ser473, CDK6: n = 3 control mice, n = 4 efatutazone-treated mice; and AKT (pan), CDK4: n = 3 control mice, n = 3 efatutazone-treated mice.
Statistical Analysis
Student’s t-tests were used to compare age at tumor development, mean numbers of invasive and noninvasive cancers, number of HANs, percentage of mammary epithelial cells with nuclear-localized Ki-67, and protein expression levels (GraphPad Prism version 4.03 for Windows; GraphPad Software Inc., San Diego, CA). Proportions of mice with noninvasive cancer, papillary and squamous histologic features, and IHC scores were compared using Z-tests (McCallum Layton, Leeds, UK). U-tests were used to compare real-time RT-PCR data (GraphPad Prism). Fisher’s exact test was used to compare the prevalence of mammary glands demonstrating dense lobular growth and HANs (GraphPad Prism). Significance was assigned at P ≤ 0.05.
Results
Efatutazone treatment starting at age 4 months did not prevent the appearance of cancers ≥1 cm3 by age 12 months but reduced cancer multiplicity and promoted the appearance of differentiated cancer histologic types.
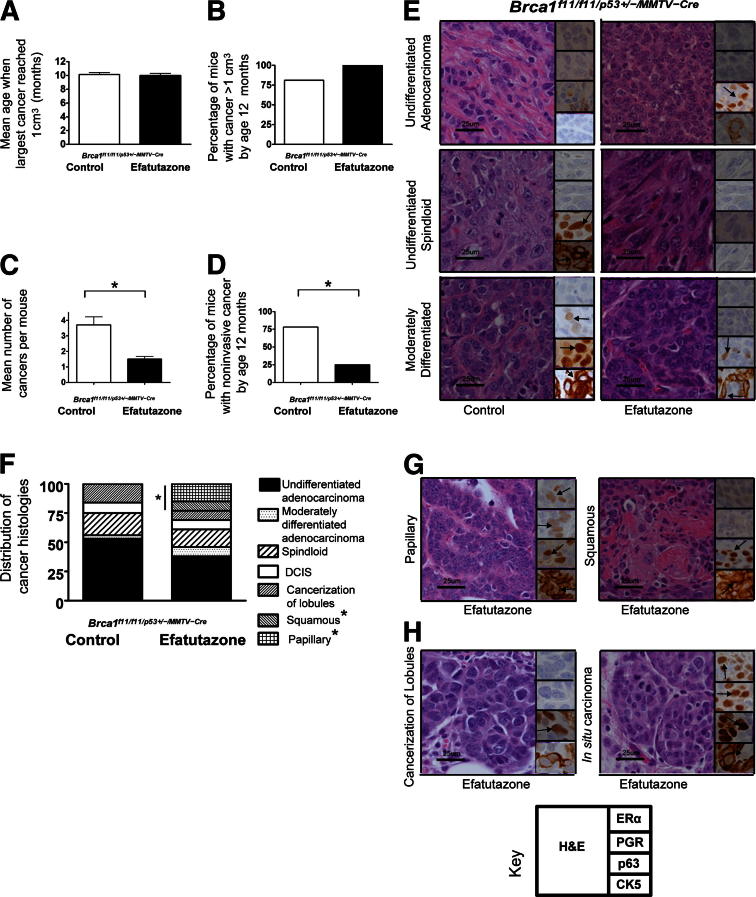
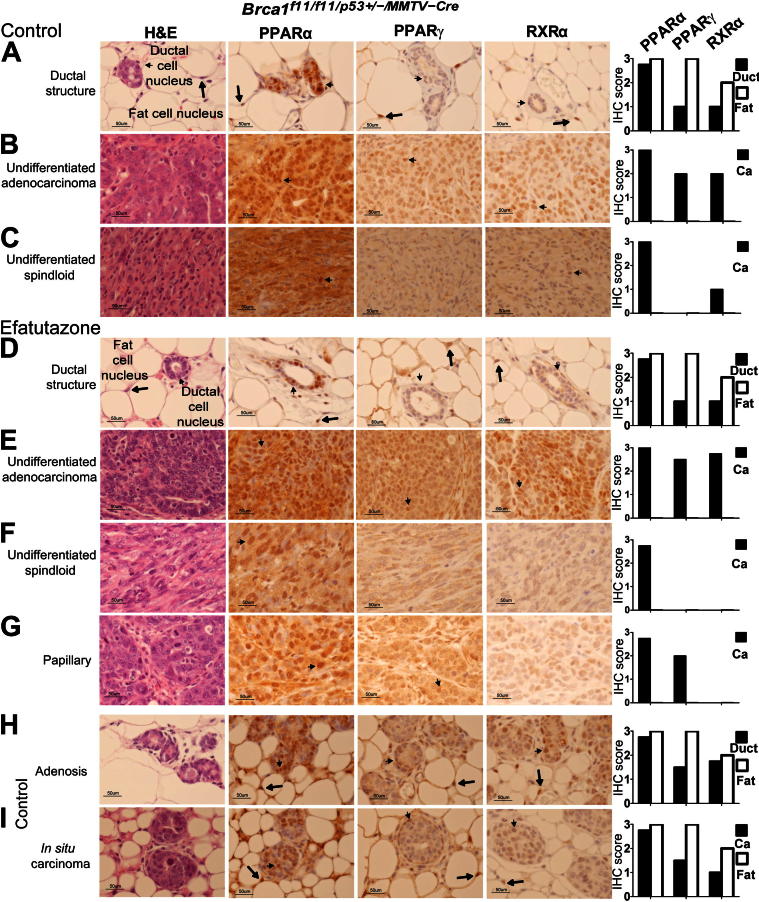
Efatutazone treatment did not significantly reduce the means ± SEM age at which tumors reached 1 cm3 [10.1 ± 0.3 months (control) versus 10.0 ± 0.3 months (efatutazone)] (Figure 1A) or the prevalence of cancers ≥1 cm3 [81% (control) versus 100% (efatutazone)] (Figure 1B). ERα-negative undifferentiated and moderately differentiated adenocarcinomas and undifferentiated spindloid cancers appeared in efatutazone-treated and control mice (Figure 1E). However, efatutazone treatment significantly reduced cancer multiplicity [3.7 ± 0.9 per mouse (control) versus 1.5 ± 0.3 per mouse (efatutazone); P < 0.05, Student’s t-test] (Figure 1C), decreased the percentage of mice with noninvasive cancers [78% (control) versus 25% (efatutazone); P < 0.05, Z-test] (Figure 1D), and altered the spectrum of cancer histologic types that developed (Figure 1F). Well-differentiated ERα-positive papillary and ERα-negative squamous cancers appeared only in the treated group [0% (control) versus 23% (efatutazone); P < 0.05, Z-test] (Figure 1, F and G). Noninvasive cancerization of lobules and in situ cancer were found in both groups (Figure 1, F and H). Expression of the myoepithelial cell proteins CK5 and p6349 were more prominent in the more differentiated cancers. Efatutazone treatment did not significantly alter expression patterns of PPARα, PPARγ, and RXRα when specific tissue types were compared (Figure 2).44–47 Prominent nuclear localization of all three proteins was observed in epithelial and fat cells, with the exception of one spindloid cancer that demonstrated appreciable cytoplasmic PPARα staining (Figure 2C). Relative PPARγ and RXRα expression levels were higher in fat cell nuclei (scores of 2 to 3) than in normal-appearing mammary duct cells (scores of 1) (Figure 2, A and D). Relatively higher expression levels of PPARγ and RXRα were found in adenocarcinomas (scores of 2 to 3) (Figure 2, B and E) than in spindloid cancers (scores of 0 to 1) (Figure 2, C and F). Papillary cancers exhibited expression of PPARα and PPARγ but not RXRα (Figure 2G). PPARγ and RXRα expression was slightly higher in adenosis and in situ cancer (Figure 2, H and I) than in the normal-appearing mammary ductal cells (Figure 2, A and D). Efatutazone treatment did not significantly change the appearance of the mammary stroma, which in mice is composed primarily of fat cells (Figure 2, A and D). Periductal fibrosis was found associated with cancerization of lobules, atypical hyperplasia and adenosis, and desmoplasia with some invasive cancers, without significant differences in appearance between the control and efatutazone-treated cohorts.
Figure 1.
Impact of efatutazone treatment on mammary cancer development in Brca1f11/f11/p53+/-/MMTV-Cre mice. A: Bar graphs comparing means ± SEM age in months when the largest cancer reached 1 cm3 in the control (10.1 ± 0.3) and efatutazone-treated (10.0 ± 0.3) cohorts. n = 11 control mice; n = 8 efatutazone-treated mice. B: Bar graphs comparing the percentage of mice with at least one cancer >1 cm3 by age 12 months in the control (81%) and efatutazone-treated (100%) cohorts. n = 11 control mice with 10 cancers >1 cm3; n = 8 efatutazone-treated mice with 10 cancers >1 cm3. C: Bar graphs comparing the means ± SEM cancer numbers in mice with cancer (>1 and <1 cm3) from control (3.7 ± 0.9) and efatutazone-treated (1.5 ± 0.3) cohorts. ∗P < 0.05, Student’s t-test, two-tailed. n = 32 control cancers; n = 13 efatutazone-treated cancers. D: Bar graphs comparing the percentage of mice with noninvasive cancers by age 12 months in the control (78%) and efatutazone-treated (25%) cohorts. ∗P < 0.05, Z-test. n = 11 control mice; n = 8 efatutazone-treated mice. E: Representative histologic types of invasive undifferentiated adenocarcinoma, undifferentiated spindloid cancers, and moderately differentiated adenocarcinomas from control and efatutazone-treated mice. H&E-stained sections are shown with insets illustrating representative ERα, PGR, p63, and CK5 IHC staining. F: Stacked bar graphs comparing the distribution of cancer phenotypes in the control versus efatutazone-treated groups: undifferentiated adenocarcinomas (54% versus 39%), moderately differentiated adenocarcinomas (3% versus 6%), spindloid s (20% versus 18%), ductal carcinoma in situ (DCIS) (7% versus7%), cancerization of lobules (16% versus 7%), squamous cancers (0% versus 7%), and papillary cancers (0% versus 16%). ∗P < 0.05, Z-test, squamous and papillary control (0%) versus efatutazone (23%). n = 32 control cancers; n = 13 efatutazone-treated cancers. G: Representative histologic features of papillary and squamous cancers found only in efatutazone-treated mice. H: Representative histologic features of in situ carcinoma and cancerization of lobules found in control and efatutazone-treated mice. The key illustrates where H&E and IHC ERα, PGR, p63, and CK5 images are positioned. Original magnification, ×60. Scale bars: 25 μm. Arrows indicate representative cells with positive staining.
Figure 2.
Expression patterns of PPARα, PPARγ, and RXRα in mammary tissue and cancers from control and efatutazone-treated mice. Representative images of H&E-stained sections and PPARα, PPARγ, and RXRα IHC sections for each tissue shown. Small arrows indicate mammary epithelial ductal cell nuclei and large arrows indicate fat cell nuclei. Bar graphs summarize relative IHC scores for ductal epithelial (duct), fat (Fat), and cancer (Ca) cells. A: Mammary ductal structure and surrounding stromal fat pad (control). B: Undifferentiated adenocarcinoma (control). C: Undifferentiated spindloid cancer (control). D: Mammary ductal structure and surrounding stromal fat pad (efatutazone treated). E: Undifferentiated adenocarcinoma (efatutazone treated). F: Undifferentiated spindloid cancer (efatutazone treated). G: Papillary cancer (efatutazone treated). H: Adenosis (control). I: In situ carcinoma (control). IHC scores: 0 indicates no stain; 1, weak; 2, intermediate; and 3, strong. n = 10 (PPARγ and RXRα) and 5 (PPARα) control cancers; n = 8 (PPARγ and RXRα) and 5 (PPARα) efatutazone-treated cancers. Original magnification, ×40. Scale bars: 50 μm.
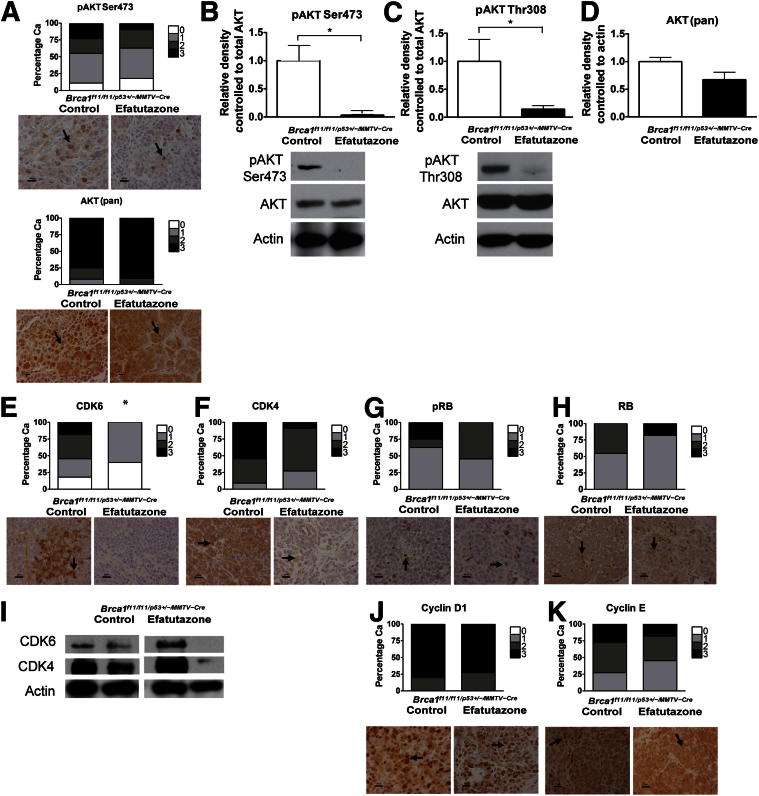
Relative Levels of pAKT Are Lower in Mammary Gland Tissue and Cancers of Efatutazone-Treated Mice
Significant reductions in means ± SEM relative expression levels of pAKT Ser473 [1.0 ± 0.3 (control) versus 0 0.04 ± 0.1 (efatutazone); P < 0.05, Student’s t-test] and pAKT Thr308 [1.0 ± 0.4 (control) versus 0.1 ± 0.1 (efatutazone); P < 0.05, Student’s t-test] but not total AKT [1.0 ± 0.1 (control) versus 0.7 ± 0.2 (efatutazone)] were found in the cancers that developed on efatutazone treatment as assessed by Western blot analysis (Figure 3, A–D). In some settings, PPARγ agonists can reduce expression of cell-cycle regulators,16–19,21,50 but CDK6 (Figure 3E) was the only cell-cycle regulator expressed at statistically significantly lower levels in cancers from efatutazone-treated mice as assessed by IHC analysis [scores of 0 to 3 (control) versus 0 to 1 (efatutazone); P < 0.05, Z-test]. Statistically nonsignificant trends toward lower CDK4 (Figure 3F) and pRB (Figure 3G) but not total RB (Figure 3H) levels were found using IHC analysis. Although some cancers showed decreased expression levels of CDK6 and CDK4 when evaluated by Western blot analysis, others did not, and the overall mean differences in relative expression levels were not statistically significantly different (Figure 3I). Efatutazone treatment did not significantly alter cyclin D1 and cyclin E expression patterns (Figure 3, J and K).
Figure 3.
Levels of pAKT and expression of CDK6 are significantly reduced in cancers from efatutazone-treated Brca1f11/f11/p53+/-/MMTV-Cre- mice. A: Stacked bar graphs summarizing IHC scores with representative IHC analysis images below for pAKT Ser473 and AKT (pan) in cancers from control and efatutazone-treated cohorts. B: Bar graphs comparing means ± SEM relative densities of pAKT Ser473 evaluated by Western blot analysis in cancers from control (1.0 ± 0.3) and efatutazone-treated (0 0.04 ± 0.1) cohorts. ∗P < 0.05, Student’s t-test, two-tailed. pAKT Ser473 levels were normalized to AKT (pan). AKT (pan) levels were normalized to actin. Representative Western blots of pAKT Ser473 and AKT (pan) are shown below the bar graphs. Actin is shown as a loading control. C: Bar graphs comparing means ± SEM relative densities of pAKT Thr308 evaluated by Western blot analysis in cancers from control (1.0 ± 0.4) and efatutazone-treated (0.1 ± 0.1) cohorts. ∗P < 0.05, Student’s t-test, two-tailed. pAKT Thr308 levels were normalized to AKT (pan). AKT (pan) levels were normalized to actin. Representative Western blots of pAKT Thr308 and AKT (pan) are shown below the bar graphs. D: Bar graphs comparing means ± SEM relative densities of AKT (pan) in cancers from the control (1.0 ± 0.1) and efatutazone-treated (0.7 ± 0.2) cohorts. AKT (pan) levels were normalized to actin. Stacked bar graphs summarizing IHC scores with representative IHC images below in cancers from control and efatutazone-treated cohorts for CDK6 (E), CDK4 (F), pRB (G), and RB (H). I: Representative Western blots demonstrating a range of CDK6 and CDK4 expression levels in cancers from control and efatutazone-treated cohorts. Actin is shown as a loading control. White bars indicate images isolated from different regions of the same blot. Stacked bar graphs summarizing IHC scores with representative IHC images below in cancers from the control and efatutazone-treated cohorts for cyclin D1 (J) and cyclin E (K). IHC scores: 0 indicates no stain; 1, weak; 2, intermediate; and 3, strong, represented by different shades: 0 indicates none (white); 1, low (light gray); 2, medium (dark gray); and 3, high (black). IHC analysis: n = 10 control mice, n = 10 efatutazone-treated mice. Western blot analysis: control: n = 3 [pAKT Ser473, pAKT Thr308, AKT (pan)]; efatutazone treated: n = 4 (pAKT Ser473 and CDK6), n = 6 (pAKT Thr308), n = 3 [AKT (pan) and CDK4]. Ca, cancer. IHC images: original magnification, ×40. Arrows indicate cells with representative staining for protein indicated. Scale bars: 20 μm, except for 10 μm for AKT (pan). ∗P < 0.05.
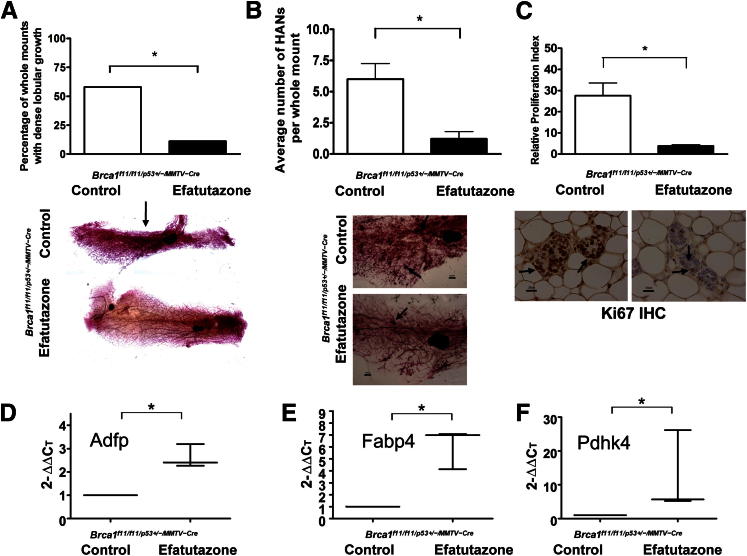
Efatutazone Treatment Reduces Dense Lobular Growth, HANs, and Mammary Epithelial Cell Proliferation in the Mammary Glands of Brca1f11/f11/p53+/-/MMTV-Cre Mice and Increases Expression of PPARγ Downstream Genes
Efatutazone treatment reduced the prevalence of dense lobular growth [58% (control) versus 11% (efatutazone); P < 0.05, Fisher’s exact test] (Figure 4A), means ± SEM number of HANs per gland [6.0 ± 1.2 (control) versus 1.2 ± 0.6 (efatutazone); P < 0.05, Student’s t-test] (Figure 4B), and means ± SEM rates of mammary epithelial cell proliferation [21.9% ± 4.5% (control) versus 3.9% ± 0.6% (efatutazone); P < 0.05, Student’s t-test] (Figure 4C). The percentage of mice demonstrating HANs was also significantly reduced by efatutazone [91.7% (control) versus 44.4% (efatutazone); P < 0.05, Fisher’s exact test]. No significant differences in the percentages of mammary epithelial cells demonstrating nuclear-localized ERα were found when comparing the two groups [5.4% ± 1.5% (control) versus 7.6% ± 1.4% (efatutazone)]. Significant fold increases in expression of the PPARγ target genes Adfp (2.3- to 3.2-fold), Fabp4 (4.1- to 7.1-fold), and Pdhk4 (5.2- to 26.1-fold) were found in mammary tissue of efatutazone-treated mice (P < 0.05, U-test) (Figure 4, D–F).
Figure 4.
Efatutazone treatment reduces lobular growth, preneoplasia, and cell proliferation and increases expression of PPARγ downstream genes in mammary tissue from Brca1f11/f11/p53+/-/MMTV-Cre mice. A: Bar graphs comparing the percentage of mammary gland whole mounts with dense lobular growth in control (58%) and efatutazone-treated (11%) mice. ∗P < 0.05, Fisher’s exact test. Representative mammary gland whole mounts are shown below the bar graphs. Arrow indicates area of dense lobular growth. n = 10 control mice; n = 8 efatutazone-treated mice. Original magnification, ×4. B: Bar graphs comparing means ± SEM number of HANs per mammary gland whole mount in control (6.0 ± 1.2) and efatutazone-treated (1.2 ± 0.6) mice. ∗P < 0.05, Student’s t-test, two-tailed. Representative whole mounts with arrows indicating HANs are shown below the bar graphs. n = 10 control mice; n = 8 efatutazone-treated mice. Original magnification: ×10. C: Bar graphs comparing the means ± SEM proliferation indices of mammary epithelial cells from control (21.9% ± 4.5%) and efatutazone-treated (3.9% ± 0.6%) mice determined from Ki-67 IHC analysis. ∗P < 0.05, Student’s t-test, two-tailed. Representative Ki-67 IHC images are shown below the bar graphs. Arrows indicate nuclear-localized Ki-67 staining. n = 10 control mice; n = 8 efatutazone-treated mice. Original magnification, ×40. D: Graph illustrating the median, minimum (2.3), and maximum (3.2) relative fold increases in RNA expression levels of Adfp in mammary tissue from efatutazone-treated compared with control mice. ∗P < 0.05, U-test. E: Graph illustrating the median, minimum (4.1) and maximum (7.1) relative fold increases in RNA expression levels of Fabp4 in mammary tissue from efatutazone-treated compared with control mice. ∗P < 0.05, U-test. F: Graph illustrating the median, minimum (5.2), and maximum (26.1) relative fold increases in RNA expression levels of Pdhk4 in mammary tissue from efatutazone-treated compared with control mice. ∗P < 0.05, U-test. D–F: n = 3 control mice and n = 3 efatutazone-treated mice. ∗P < 0.05. Scale bars: 1000 μm (B); 20 μm (C).
Discussion
The spectrum of mammary cancer subtypes initiated by loss of full-length Brca1 in association with Tp53 haploinsufficiency was modified by treatment with the PPARγ agonist efatutazone. The appearance of well-differentiated cancer histologic features with efatutazone treatment indicated that the histologic fate of cancer progenitor cells was not fixed in this model by 4 months of age, the time when drug treatment was initiated, and expands the number of histologic cancer subtypes found in this model.28,29,31,33 Recognition of an intervention that introduces an ERα-positive differentiated cancer subtype in this model opens the door to the identification of a cell(s) of origin for ERα-positive and ERα-negative cancer subtypes developing in the setting of BRCA1 deficiency.36,51
In contrast to the lack of effect on the appearance of cancers >1 cm3 by age 12 months in this model, efatutazone treatment markedly inhibited the development of smaller invasive and noninvasive cancers and decreased preneoplasia prevalence. It could be that the larger cancers formed from clinically unapparent efatutazone-resistant cancer or cancer progenitor cells52 that had progressed to a certain stage in the gland when efatutazone treatment was initiated at 4 months of age, whereas cancer stem or progenitor cells at an earlier stage or not yet formed were inhibited. It is known that in vitro exposure of MCF-7 and primary breast cancer cells to the PPARγ agonist pioglitazone reduces the formation of mammospheres.53 Alternatively, there could be different types of mammary stem or progenitor cells in the glands of Brca1f11/f11/p53+/-/MMTV-Cre mice, ones that were not inhibited by efatutazone and others that were. The appearance of differentiated ERα-positive papillary cancers not usually seen in this model raises the question of whether efatutazone treatment could have modified some early cancer or progenitor cells toward a more well-differentiated phenotype. Future studies that vary the timing of efatutazone exposure could determine whether there is a time point earlier than age 4 months that would more profoundly inhibit cancer development; explore whether the impact of efatutazone is reversible, that is, whether cancer development would be restored after discontinuation of the drug; and directly investigate the impact of efatutazone on cancer stem and progenitor cells.
Expression of PPARγ and RXRα were verified in the epithelial and stromal tissue compartments. It is not yet known whether the inhibitory action of efatutazone on mammary epithelial cell proliferation and development of hyperplasia and cancer is mediated by its action on only one tissue compartment or whether it is a combined effect on different tissue compartments. PPARα expression was examined because exposure to a PPARα agonist promotes mammosphere formation and it is up-regulated in MCF7 cells grown as mammospheres.53 PPARα expression was found in the normal-appearing mammary epithelial cells of the Brca1f11/f11/p53+/-/MMTV-Cre mice and in the cells composing adenosis, noninvasive, and invasive cancers. A previous report indicated that PPARα expression in normal mammary epithelial cells is low,54 but whether PPARα plays a specific role in cancer progression in mammary cells deficient in BRCA1 remains to be determined.
The reduced levels of pAKT and decreased rates of cell proliferation found herein with efatutazone treatment in vivo are consistent with previous in vitro studies of PPARγ agonists.1–4,10,19,21 Significantly, the decreased levels of pAKT did not translate into reduced cancer prevalence but did correlate with reduced numbers of cancers and lower rates of HAN development.
Although the use of PPARγ agonists for the treatment of diabetes is currently limited, thousands of women have been exposed. A challenging translational question would be to determine whether diabetic women exposed to a PPARγ agonist have different patterns of breast cancer development than unexposed women, an area that has not yet been addressed in publications, to our knowledge.
In conclusion, the PPARγ agonist efatutazone significantly modified cancer development and promoted the appearance of more differentiated mammary cancers in the setting of BRCA1 deficiency. The studies set a platform for further investigations of the cells of origin and the role of PPARγ in the generation of diverse breast cancer subtypes in different genetic backgrounds.
Acknowledgments
These studies were conducted with the Lombardi Comprehensive Cancer Center Histopathology and Tissue, Animal, and Genomics and Epigenetics Shared Resources.
Footnotes
Supported in part by Daiichi Sankyo Inc. (P.A.F.); National Cancer Institute, NIH grant RO1CA112176 (P.A.F.); Department of Defense grant W81XWH-11-1-0074 (R.E.N.); the World Class University program through the National Research Foundation of Korea funded by the Ministry of Education, Science, and Technology (R31-10069 to P.A.F.); NIH grant IG20 RR025828-01 (Rodent Barrier Facility Equipment), and NIH grant NCI 5P30CA051008 (Histology and Tissue, Genomics and Epigenomics, and Animal Shared Resources).
Disclosures: This work was partially supported by Daiichi Sankyo Inc., which manufactures efatutazone. An investigational new drug application for efatutazone is on file with the US Food and Drug Administration.
See related Commentary on page 1972.
References
- 1.Mueller E., Sarraf P., Tontonoz P., Evans R.M., Martin K.J., Zhang M., Fletcher C., Singer S., Spiegelman B.M. Terminal differentiation of human breast cancer through PPAR gamma. Mol Cell. 1998;1:465–470. doi: 10.1016/s1097-2765(00)80047-7. [DOI] [PubMed] [Google Scholar]
- 2.Chen L., Bush C.R., Necela B.M., Su W., Yanagisawa M., Anastasiadis P.Z., Fields A.P., Thompson E.A. RS5444, a novel PPARgamma agonist, regulates aspects of the differentiated phenotype in nontransformed intestinal epithelial cells. Mol Cell Endocrinol. 2006;251:17–32. doi: 10.1016/j.mce.2006.02.006. [DOI] [PubMed] [Google Scholar]
- 3.Chawla A., Barak Y., Nagy L., Liao D., Tontonoz P., Evans R.M. PPAR-gamma dependent and independent effects on macrophage-gene expression in lipid metabolism and inflammation. Nat Med. 2001;7:48–52. doi: 10.1038/83336. [DOI] [PubMed] [Google Scholar]
- 4.Tontonoz P., Singer S., Forman B.M., Sarraf P., Fletcher J.A., Fletcher C.D., Brun R.P., Mueller E., Altiok S., Oppenheim H., Evans R.M., Spiegelman B.M. Terminal differentiation of human liposarcoma cells induced by ligands for peroxisome proliferator-activated receptor gamma and the retinoid X receptor. Proc Natl Acad Sci U S A. 1997;94:237–241. doi: 10.1073/pnas.94.1.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Demetri G.D., Fletcher C.D., Mueller E., Sarraf P., Naujoks R., Campbell N., Spiegelman B.M., Singer S. Induction of solid tumor differentiation by the peroxisome proliferator-activated receptor-gamma ligand troglitazone in patients with liposarcoma. Proc Natl Acad Sci U S A. 1999;96:3951–3956. doi: 10.1073/pnas.96.7.3951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Elstner E., Müller C., Koshizuka K., Williamson E.A., Park D., Asou H., Shintaku P., Said J.W., Heber D., Koeffler H.P. Ligands for peroxisome proliferator-activated receptorgamma and retinoic acid receptor inhibit growth and induce apoptosis of human breast cancer cells in vitro and in BNX mice. Proc Natl Acad Sci U S A. 1998;95:8806–8811. doi: 10.1073/pnas.95.15.8806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Altiok S., Xu M., Spiegelman B.M. PPARgamma induces cell cycle withdrawal: inhibition of E2F/DP DNA-binding activity via down-regulation of PP2A. Genes Dev. 1997;11:1987–1998. doi: 10.1101/gad.11.15.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xin X., Yang S., Kowalski J., Gerritsen M.E. Peroxisome proliferator-activated receptor gamma ligands are potent inhibitors of angiogenesis in vitro and in vivo. J Biol Chem. 1999;274:9116–9121. doi: 10.1074/jbc.274.13.9116. [DOI] [PubMed] [Google Scholar]
- 9.Shimazaki N., Togashi N., Hanai M., Isoyama T., Wada K., Fujita T., Fujiwara K., Kurakata S. Anti-tumour activity of CS-7017, a selective peroxisome proliferator-activated receptor gamma agonist of thiazolidinedione class, in human tumour xenografts and a syngeneic tumour implant model. Eur J Cancer. 2008;44:1734–1743. doi: 10.1016/j.ejca.2008.04.016. [DOI] [PubMed] [Google Scholar]
- 10.Yin Y., Russell R.G., Dettin L.E., Bai R., Wei Z.-L., Kozikowski A.P., Kopelovich L., Kopleovich L., Glazer R.I. Peroxisome proliferator-activated receptor delta and gamma agonists differentially alter tumor differentiation and progression during mammary carcinogenesis. Cancer Res. 2005;65:3950–3957. doi: 10.1158/0008-5472.CAN-04-3990. [DOI] [PubMed] [Google Scholar]
- 11.Copland J.A., Marlow L.A., Kurakata S., Fujiwara K., Wong A.K.C., Kreinest P.A., Williams S.F., Haugen B.R., Klopper J.P., Smallridge R.C. Novel high-affinity PPARgamma agonist alone and in combination with paclitaxel inhibits human anaplastic thyroid carcinoma tumor growth via p21WAF1/CIP1. Oncogene. 2006;25:2304–2317. doi: 10.1038/sj.onc.1209267. [DOI] [PubMed] [Google Scholar]
- 12.Marlow L.A., Reynolds L.A., Cleland A.S., Cooper S.J., Gumz M.L., Kurakata S., Fujiwara K., Zhang Y., Sebo T., Grant C., McIver B., Wadsworth J.T., Radisky D.C., Smallridge R.C., Copland J.A. Reactivation of suppressed RhoB is a critical step for the inhibition of anaplastic thyroid cancer growth. Cancer Res. 2009;69:1536–1544. doi: 10.1158/0008-5472.CAN-08-3718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Su W., Necela B.M., Fujiwara K., Kurakata S., Murray N.R., Fields A.P., Thompson E.A. The high affinity peroxisome proliferator-activated receptor-gamma agonist RS5444 inhibits both initiation and progression of colon tumors in azoxymethane-treated mice. Int J Cancer. 2008;123:991–997. doi: 10.1002/ijc.23640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pishvaian M.J., Marshall J.L., Wagner A.J., Hwang J.J., Malik S., Cotarla I., Deeken J.F., He A.R., Daniel H., Halim A.-B., Zahir H., Copigneaux C., Liu K., Beckman R.A., Demetri G.D. A phase 1 study of efatutazone, an oral peroxisome proliferator-activated receptor gamma agonist, administered to patients with advanced malignancies. Cancer. 2012;118:5403–5413. doi: 10.1002/cncr.27526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rosen E.D., Spiegelman B.M. PPARgamma: a nuclear regulator of metabolism, differentiation, and cell growth. J Biol Chem. 2001;276:37731–37734. doi: 10.1074/jbc.R100034200. [DOI] [PubMed] [Google Scholar]
- 16.Cabrera M.C., Díaz-Cruz E.S., Kallakury B.V.S., Pishvaian M.J., Grubbs C.J., Muccio D.D., Furth P.A. The CDK4/6 Inhibitor PD0332991 reverses epithelial dysplasia associated with abnormal activation of the cyclin-CDK-Rb pathway. Cancer Prev Res (Phila) 2012;5:810–821. doi: 10.1158/1940-6207.CAPR-11-0532-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kawa S., Nikaido T., Unno H., Usuda N., Nakayama K., Kiyosawa K. Growth inhibition and differentiation of pancreatic cancer cell lines by PPAR gamma ligand troglitazone. Pancreas. 2002;24:1–7. doi: 10.1097/00006676-200201000-00001. [DOI] [PubMed] [Google Scholar]
- 18.Morrison R.F., Farmer S.R. Role of PPARgamma in regulating a cascade expression of cyclin-dependent kinase inhibitors, p18(INK4c) and p21(Waf1/Cip1), during adipogenesis. J Biol Chem. 1999;274:17088–17097. doi: 10.1074/jbc.274.24.17088. [DOI] [PubMed] [Google Scholar]
- 19.Wakino S., Kintscher U., Kim S., Yin F., Hsueh W.A., Law R.E. Peroxisome proliferator-activated receptor gamma ligands inhibit retinoblastoma phosphorylation and G1– >S transition in vascular smooth muscle cells. J Biol Chem. 2000;275:22435–22441. doi: 10.1074/jbc.M910452199. [DOI] [PubMed] [Google Scholar]
- 20.Kim K.Y., Kim S.S., Cheon H.G. Differential anti-proliferative actions of peroxisome proliferator-activated receptor-gamma agonists in MCF-7 breast cancer cells. Biochem Pharmacol. 2006;72:530–540. doi: 10.1016/j.bcp.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 21.Teresi R.E., Shaiu C.-W., Chen C.-S., Chatterjee V.K., Waite K.A., Eng C. Increased PTEN expression due to transcriptional activation of PPARgamma by lovastatin and rosiglitazone. Int J Cancer. 2006;118:2390–2398. doi: 10.1002/ijc.21799. [DOI] [PubMed] [Google Scholar]
- 22.Shao J., Sheng H., DuBois R.N. Peroxisome proliferator-activated receptors modulate K-Ras-mediated transformation of intestinal epithelial cells. Cancer Res. 2002;62:3282–3288. [PubMed] [Google Scholar]
- 23.Ondrey F. Peroxisome proliferator-activated receptor gamma pathway targeting in carcinogenesis: implications for chemoprevention. Clin Cancer Res. 2009;15:2–8. doi: 10.1158/1078-0432.CCR-08-0326. [DOI] [PubMed] [Google Scholar]
- 24.Robbins G.T., Nie D. PPAR gamma, bioactive lipids, and cancer progression. Front Biosci. 2012;17:1816–1834. doi: 10.2741/4021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Olefsky J.M. Treatment of insulin resistance with peroxisome proliferator-activated receptor gamma agonists. J Clin Invest. 2000;106:467–472. doi: 10.1172/JCI10843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wei S., Yang J., Lee S.-L., Kulp S.K., Chen C.-S. PPARgamma-independent antitumor effects of thiazolidinediones. Cancer Lett. 2009;276:119–124. doi: 10.1016/j.canlet.2008.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cariou B., Charbonnel B., Staels B. Thiazolidinediones and PPARγ agonists: time for a reassessment. Trends Endocrinol Metab. 2012;23:205–215. doi: 10.1016/j.tem.2012.03.001. [DOI] [PubMed] [Google Scholar]
- 28.Herschkowitz J.I., Simin K., Weigman V.J., Mikaelian I., Usary J., Hu Z., Rasmussen K.E., Jones L.P., Assefnia S., Chandrasekharan S., Backlund M.G., Yin Y., Khramtsov A.I., Bastein R., Quackenbush J., Glazer R.I., Brown P.H., Green J.E., Kopelovich L., Furth P.A., Palazzo J.P., Olopade O.I., Bernard P.S., Churchill G.A., Van Dyke T., Perou C.M. Identification of conserved gene expression features between murine mammary carcinoma models and human breast tumors. Genome Biol. 2007;8:R76. doi: 10.1186/gb-2007-8-5-r76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jones L.P., Li M., Halama E.D., Ma Y., Lubet R., Grubbs C.J., Deng C.-X., Rosen E.M., Furth P.A. Promotion of mammary cancer development by tamoxifen in a mouse model of Brca1-mutation-related breast cancer. Oncogene. 2005;24:3554–3562. doi: 10.1038/sj.onc.1208426. [DOI] [PubMed] [Google Scholar]
- 30.Xu X., Wagner K.U., Larson D., Weaver Z., Li C., Ried T., Hennighausen L., Wynshaw-Boris A., Deng C.X. Conditional mutation of Brca1 in mammary epithelial cells results in blunted ductal morphogenesis and tumour formation. Nat Genet. 1999;22:37–43. doi: 10.1038/8743. [DOI] [PubMed] [Google Scholar]
- 31.Jones L.P., Tilli M.T., Assefnia S., Torre K., Halama E.D., Parrish A., Rosen E.M., Furth P.A. Activation of estrogen signaling pathways collaborates with loss of Brca1 to promote development of ERalpha-negative and ERalpha-positive mammary preneoplasia and cancer. Oncogene. 2008;27:794–802. doi: 10.1038/sj.onc.1210674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Furth P.A., Cabrera M.C., Díaz-Cruz E.S., Millman S., Nakles R.E. Assessing estrogen signaling aberrations in breast cancer risk using genetically engineered mouse models. Ann N Y Acad Sci. 2011;1229:147–155. doi: 10.1111/j.1749-6632.2011.06086.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Diaz-Cruz E.S., Cabrera M.C., Nakles R., Rutstein B.H., Furth P.A. BRCA1 deficient mouse models to study pathogenesis and therapy of triple negative breast cancer. Breast Dis. 2010;32:85–97. doi: 10.3233/BD-2010-0308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bachelier R., Xu X., Li C., Qiao W., Furth P.A., Lubet R.A., Deng C.-X. Effect of bilateral oophorectomy on mammary tumor formation in BRCA1 mutant mice. Oncol Rep. 2005;14:1117–1120. [PubMed] [Google Scholar]
- 35.Atchley D.P., Albarracin C.T., Lopez A., Valero V., Amos C.I., Gonzalez-Angulo A.M., Hortobagyi G.N., Arun B.K. Clinical and pathologic characteristics of patients with BRCA-positive and BRCA-negative breast cancer. J Clin Oncol. 2008;26:4282–4288. doi: 10.1200/JCO.2008.16.6231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Molyneux G., Smalley M.J. The cell of origin of BRCA1 mutation-associated breast cancer: a cautionary tale of gene expression profiling. J Mammary Gland Biol Neoplasia. 2011;16:51–55. doi: 10.1007/s10911-011-9202-8. [DOI] [PubMed] [Google Scholar]
- 37.Proia T.A., Keller P.J., Gupta P.B., Klebba I., Jones A.D., Sedic M., Gilmore H., Tung N., Naber S.P., Schnitt S., Lander E.S., Kuperwasser C. Genetic predisposition directs breast cancer phenotype by dictating progenitor cell fate. Cell Stem Cell. 2011;8:149–163. doi: 10.1016/j.stem.2010.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Melchor L., Benítez J. An integrative hypothesis about the origin and development of sporadic and familial breast cancer subtypes. Carcinogenesis. 2008;29:1475–1482. doi: 10.1093/carcin/bgn157. [DOI] [PubMed] [Google Scholar]
- 39.Lindeman G.J., Visvader J.E. Cell fate takes a slug in BRCA1-associated breast cancer. Breast Cancer Res. 2011;13:306. doi: 10.1186/bcr2840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Han J., Chang H., Giricz O., Lee G.Y., Baehner F.L., Gray J.W., Bissell M.J., Kenny P.A., Parvin B. Molecular predictors of 3D morphogenesis by breast cancer cell lines in 3D culture. PLoS Comput Biol. 2010;6:e1000684. doi: 10.1371/journal.pcbi.1000684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wagner K.U., Wall R.J., St-Onge L., Gruss P., Wynshaw-Boris A., Garrett L., Li M., Furth P.A., Hennighausen L. Cre-mediated gene deletion in the mammary gland. Nucleic Acids Res. 1997;25:4323–4330. doi: 10.1093/nar/25.21.4323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Paik S., Kim C., Song Y., Kim W. Technology insight: application of molecular techniques to formalin-fixed paraffin-embedded tissues from breast cancer. Nat Clin Pract Oncol. 2005;2:246–254. doi: 10.1038/ncponc0171. [DOI] [PubMed] [Google Scholar]
- 43.Frech M.S., Halama E.D., Tilli M.T., Singh B., Gunther E.J., Chodosh L.A., Flaws J.A., Furth P.A. Deregulated estrogen receptor alpha expression in mammary epithelial cells of transgenic mice results in the development of ductal carcinoma in situ. Cancer Res. 2005;65:681–685. [PMC free article] [PubMed] [Google Scholar]
- 44.Nomoto K., Tsuneyama K., Abdel Aziz H.O., Takahashi H., Murai Y., Cui Z.-G., Fujimoto M., Kato I., Hiraga K., Hsu D.K., Liu F.-T., Takano Y. Disrupted galectin-3 causes non-alcoholic fatty liver disease in male mice. J Pathol. 2006;210:469–477. doi: 10.1002/path.2065. [DOI] [PubMed] [Google Scholar]
- 45.Sarruf D.A., Yu F., Nguyen H.T., Williams D.L., Printz R.L., Niswender K.D., Schwartz M.W. Expression of peroxisome proliferator-activated receptor-gamma in key neuronal subsets regulating glucose metabolism and energy homeostasis. Endocrinology. 2009;150:707–712. doi: 10.1210/en.2008-0899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ijpenberg A., Pérez-Pomares J.M., Guadix J.A., Carmona R., Portillo-Sánchez V., Macías D., Hohenstein P., Miles C.M., Hastie N.D., Muñoz-Chápuli R. Wt1 and retinoic acid signaling are essential for stellate cell development and liver morphogenesis. Dev Biol. 2007;312:157–170. doi: 10.1016/j.ydbio.2007.09.014. [DOI] [PubMed] [Google Scholar]
- 47.Childs A.J., Cowan G., Kinnell H.L., Anderson R.A., Saunders P.T.K. Retinoic acid signalling and the control of meiotic entry in the human fetal gonad. PLoS One. 2011;6:e20249. doi: 10.1371/journal.pone.0020249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schmittgen T.D., Livak K.J. Analyzing real-time PCR data by the comparative CT method. Nat Protoc. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 49.Walker R.A., Hanby A., Pinder S.E., Thomas J., Ellis I.O. Current issues in diagnostic breast pathology. J Clin Pathol. 2012;65:771–785. doi: 10.1136/jclinpath-2012-200733. [DOI] [PubMed] [Google Scholar]
- 50.Theocharis S., Margeli A., Vielh P., Kouraklis G. Peroxisome proliferator-activated receptor-gamma ligands as cell-cycle modulators. Cancer Treat Rev. 2004;30:545–554. doi: 10.1016/j.ctrv.2004.04.004. [DOI] [PubMed] [Google Scholar]
- 51.Kendrick H., Regan J.L., Magnay F.-A., Grigoriadis A., Mitsopoulos C., Zvelebil M., Smalley M.J. Transcriptome analysis of mammary epithelial subpopulations identifies novel determinants of lineage commitment and cell fate. BMC Genomics. 2008;9:591. doi: 10.1186/1471-2164-9-591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vassilopoulos A., Wang R.-H., Petrovas C., Ambrozak D., Koup R., Deng C.-X. Identification and characterization of cancer initiating cells from BRCA1 related mammary tumors using markers for normal mammary stem cells. Int J Biol Sci. 2008;4:133–142. doi: 10.7150/ijbs.4.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Papi A., Guarnieri T., Storci G., Santini D., Ceccarelli C., Taffurelli M., De Carolis S., Avenia N., Sanguinetti A., Sidoni A., Orlandi M., Bonafé M. Nuclear receptors agonists exert opposing effects on the inflammation dependent survival of breast cancer stem cells. Cell Death Differ. 2012;19:1208–1219. doi: 10.1038/cdd.2011.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yang Q., Kurotani R., Yamada A., Kimura S., Gonzalez F.J. Peroxisome proliferator-activated receptor alpha activation during pregnancy severely impairs mammary lobuloalveolar development in mice. Endocrinology. 2006;147:4772–4780. doi: 10.1210/en.2006-0437. [DOI] [PMC free article] [PubMed] [Google Scholar]