A transcriptional regulator generated by alternative splicing uses a cryptic binding site to negatively regulate bHLH-type transcription factors that promote jasmonate responses.
Abstract
The plant hormone jasmonate (JA) activates gene expression by promoting ubiquitin-dependent degradation of jasmonate ZIM domain (JAZ) transcriptional repressor proteins. A key feature of all JAZ proteins is the highly conserved Jas motif, which mediates both JAZ degradation and JAZ binding to the transcription factor MYC2. Rapid expression of JAZ genes in response to JA is thought to attenuate JA responses, but little is known about the mechanisms by which newly synthesized JAZ proteins exert repression in the presence of the hormone. Here, we show in Arabidopsis (Arabidopsis thaliana) that desensitization to JA is mediated by an alternative splice variant (JAZ10.4) of JAZ10 that lacks the Jas motif. Unbiased protein-protein interaction screens identified three related basic helix-loop-helix transcription factors (MYC2, MYC3, and MYC4) and the corepressor NINJA as JAZ10.4-binding partners. We show that the amino-terminal region of JAZ10.4 contains a cryptic MYC2-binding site that resembles the Jas motif and that the ZIM motif of JAZ10.4 functions as a transferable repressor domain whose activity is associated with the recruitment of NINJA. Functional studies showed that the expression of JAZ10.4 from the native JAZ10 promoter complemented the JA-hypersensitive phenotype of a jaz10 mutant. Moreover, treatment of these complemented lines with JA resulted in the rapid accumulation of JAZ10.4 protein. Our results provide an explanation for how the unique domain architecture of JAZ10.4 links transcription factors to a corepressor complex and suggest how JA-induced transcription and alternative splicing of JAZ10 premessenger RNA creates a regulatory circuit to attenuate JA responses.
The small-molecule hormone jasmonate (JA) mediates plant responses to various environmental stresses and developmental cues. JA has been extensively characterized for its role in controlling reproductive development (McConn and Browse, 1996; Li et al., 2004; Nagpal et al., 2005; Browse, 2009) and defense responses to herbivores and pathogens (Howe et al., 1996; McConn et al., 1997; Kessler et al., 2004; Glazebrook, 2005; Howe and Jander, 2008). Increasing evidence indicates that this lipid-derived hormone also controls vegetative growth and cell differentiation responses (Staswick et al., 1992; Yan et al., 2007; Zhang and Turner, 2008; Yoshida et al., 2009; Chen et al., 2011; Yang et al., 2012). JA regulates gene expression by controlling the abundance of jasmonate ZIM domain (JAZ) transcriptional repressor proteins (Chini et al., 2007; Thines et al., 2007; Yan et al., 2007; Chung et al., 2009; Pauwels and Goossens, 2011). JAZs belong to the family of TIFY proteins that is defined by a highly conserved TIFYXG motif embedded within the ZIM domain (Vanholme et al., 2007). Low intracellular concentrations of the receptor-active form of JA, jasmonoyl-l-Ile (JA-Ile), permit JAZ proteins to accumulate in the nucleus, where they bind transcription factors (TFs) to actively repress JA-response genes. The best characterized targets of JAZ repressors are the basic helix-loop-helix TF MYC2 (also known as JASMONATE INSENSITIVE1 [JIN1]) and its closely related paralogs MYC3 and MYC4 (Lorenzo et al., 2004; Chini et al., 2007; Melotto et al., 2008; Cheng et al., 2011; Fernández-Calvo et al., 2011; Niu et al., 2011; Song et al., 2011; Kazan and Manners, 2013). In response to tissue damage or other conditions that elevate JA-Ile levels, JAZ proteins are recruited to the F-box protein CORONATINE INSENSITIVE1 (COI1), which determines the specificity of the E3 ubiquitin ligase SCFCOI1 for JAZ substrates (Thines et al., 2007; Katsir et al., 2008; Melotto et al., 2008; Fonseca et al., 2009; Koo and Howe, 2009; Yan et al., 2009; Sheard et al., 2010). Ubiquitin-dependent degradation of JAZ proteins relieves the repression of TFs, thereby allowing the expression of JA-response genes (Chini et al., 2007; Thines et al., 2007).
Recent studies have provided insight into the mechanisms by which TF-bound JAZ proteins repress transcription. NINJA (for novel interactor of JAZ) was identified as an adaptor protein that physically links the ZIM domain of JAZ to the corepressor TOPLESS (TPL) and TOPLESS-related (TPR) proteins. As is the case for other targets of TPL, NINJA contains an ERF-associated amphiphilic repression (EAR) motif that mediates interaction with TPL (Szemenyei et al., 2008; Pauwels et al., 2010). Interestingly, some JAZs contain an EAR motif and thus may interact directly with TPL (Kagale et al., 2010; Arabidopsis Interactome Mapping Consortium, 2011; Causier et al., 2012). It was recently demonstrated that the repressive function of JAZ8 depends on its TPL-interacting EAR motif (Shyu et al., 2012). Thus, whereas some JAZs repress transcription through NINJA (Pauwels et al., 2010), other JAZs such as JAZ8 may repress gene expression by a NINJA-independent mechanism in which corepressors are recruited directly to a TF-bound JAZ (Shyu et al., 2012).
Alternative splicing provides a mechanism to increase the repressive activity and functional diversity of JAZ proteins. The best example of this form of posttranscriptional regulation is splice variants of JAZ10 that are truncated in the C-terminal Jas motif, which mediates interaction with COI1 and MYC2 (Yan et al., 2007; Chung and Howe, 2009; Chung et al., 2010). Alternative splicing of JAZ10 pre-mRNA generates three protein variants that differentially interact with COI1. The full-length JAZ10.1 isoform binds strongly to COI1 in the presence of JA-Ile, whereas C-terminally truncated splice variants interact weakly (JAZ10.3) or not at all (JAZ10.4) with COI1. JAZ10.3 and JAZ10.4 are more stable than JAZ10.1 in JA-stimulated cells and, as a consequence, exert dominant repression in transgenic overexpression assays (Yan et al., 2007; Chung and Howe, 2009; Chung et al., 2010). A role for JAZ10 splice variants in attenuating JA responses is supported by the fact that jaz10 loss-of-function mutants (e.g. jaz10-1) are hypersensitive to JA and that JAZ10 expression is rapidly induced by JA (Yan et al., 2007; Chung et al., 2008; Demianski et al., 2012). JAZ10 may also play a role in attenuating JA-mediated defense processes under environmental conditions that prioritize plant growth over defense (Moreno et al., 2009; Cerrudo et al., 2012).
Initial insight into the role of JAZs as transcriptional repressors came from experiments showing that the expression of truncated JAZ proteins (referred to as JAZΔJas) lacking the Jas motif results in reduced sensitivity to JA (Chini et al., 2007; Thines et al., 2007; Yan et al., 2007). This observation is explained by the fact that the Jas motif harbors an instability element (i.e. degron) that interacts with COI1 in a hormone-dependent manner. Thus, removal of this element creates a stable repressor (Katsir et al., 2008; Melotto et al., 2008; Sheard et al., 2010). Because the Jas motif also mediates TF binding, however, the mechanism by which JAZ10.4 and other JAZΔJas proteins interact with TFs to repress JA responses has remained unclear (Chini et al., 2009; Wager and Browse, 2012). The strong JA-insensitive phenotype of JAZ10.4-overexpressing plants, together with the ability of JAZ10.4 to interact with MYC2 in yeast (Saccharomyces cerevisiae), suggest that sequence determinants outside the Jas motif promote binding to MYC2 (Chung and Howe, 2009). There is also evidence that JAZ1 can interact with MYC2 through sequences outside the Jas motif (Withers et al., 2012). Specific sequence determinants that mediate TF binding to JAZ10.4 and other JAZΔJas proteins have not been identified. In this study, we discovered a cryptic MYC2-binding site at the N terminus of JAZ10.4, providing a mechanism by which JAZ10.4 represses JA responses. We also describe a transgenic complementation assay to test the proposed function of JAZ10.4 in dampening JA responses. We use this system to show that the native JAZ10 promoter is sufficient to drive JAZ10.4 protein expression and complement jaz10 phenotypes in response to JA treatment. Our findings demonstrate how JA-induced transcription and alternative splicing of JAZ10 establish a negative feedback loop to attenuate JA responses.
RESULTS
JAZ10.4 Interacts with Basic Helix-Loop-Helix TFs and NINJA
As an unbiased approach to identify binding partners of JAZ10.4, we performed a high-throughput yeast two-hybrid (Y2H) screen for Arabidopsis (Arabidopsis thaliana) proteins that interact with JAZ10.4. Sequence information obtained from 295 positive clones (among 3.2 × 107 clones screened) produced a list of putative interacting proteins. Based on calculated confidence scores (Formstecher et al., 2005), five proteins were categorized as high-confidence interactors: MYC2, MYC3, MYC4, NINJA, and GENERAL REGULATING FACTOR6 (GRF6, also known as 14-3-3λ/GF14λ; Table I). GRF6 was not identified as a JAZ10.4-interacting protein in planta (see below) and thus was not characterized further. Sequencing of positive clones provided information on the minimal interaction domain of each prey protein (Formstecher et al., 2005). The results suggested that JAZ10.4 interacts with the N-terminal region of MYC2, MYC3, and MYC4 and the C-terminal region of NINJA (Table I). These findings are in agreement with previous studies showing that most full-length JAZs (including JAZ10.1) interact with the N terminus of MYC TFs (Cheng et al., 2011; Fernández-Calvo et al., 2011; Niu et al., 2011) and that JAZ1 interacts with the C-terminal region of NINJA (Pauwels et al., 2010).
Table I. List of high-confidence JAZ10.4-interacting proteins identified in a Y2H screen.
| Protein | Arabidopsis Genome Initiative Code | Length | No. of Clonesa | SIDb |
|---|---|---|---|---|
| MYC2 | At1g32640 | 623 | 11 | 51–272 |
| MYC3 | At5g46760 | 592 | 34 | 40–279 |
| MYC4 | At4g17880 | 589 | 100 | 46–246 |
| NINJA | At4g28910 | 425 | 22 | 229–425 |
| GRF6 | At5g10450 | 273 | 15 | 37–248 |
Number of positive clones for which sequence information was obtained. bThe Selected Interaction Domain (SID) defines the minimum interaction domain of the prey identified as the amino acid sequence shared by all prey fragments matching with the same reference protein.
As an alternative approach to identify JAZ10.4 interacting partners, we used mass spectrometry (MS) to identify proteins that copurify with JAZ10.4 expressed in cultured Arabidopsis T87 cells. The utility of this system for studying JAZ proteins was assessed by an analysis of cell lines expressing yellow fluorescent protein (YFP) fusions of each JAZ10 splice variant. Confocal microscopy and western-blot analysis showed that JAZ10.3-YFP and JAZ10.4-YFP accumulate to higher levels than JAZ10.1-YFP in both control and JA-treated T87 cells (Supplemental Fig. S1). This finding is consistent with previous studies showing that JAZ10.3 and JAZ10.4 are more stable than JAZ10.1 in JA-treated seedlings (Chung and Howe, 2009). MS-based analysis of coimmunoprecipitated JAZ10.4-YFP complexes (four independent replicates) identified NINJA as a major copurifying protein (Table II). MYC2, MYC3, and MYC4 were also identified as JAZ10.4-interacting partners in at least two of the four replicate experiments. We also identified JAZ12 and PEAPOD2 (PPD2) as components of the JAZ10.4 complex, which is in agreement with studies showing that JAZs form heteromeric complexes with other ZIM domain-containing proteins (Chini et al., 2009; Chung and Howe, 2009; Pauwels et al., 2010). These collective findings show that JAZ10.4 interacts in vivo with NINJA and the basic helix-loop-helix TFs MYC2, MYC3, and MYC4.
Table II. List of JAZ10.4-interacting proteins identified by immunoaffinity purification of JAZ10.4-YFP followed by MS.
nd, Not detected.
| Prey Protein | Arabidopsis Genome Initiative Code | Bait Proteina |
|||||
|---|---|---|---|---|---|---|---|
| JAZ10.4 |
JAZ10.4I107A |
||||||
| rep 1 | rep 2 | rep 3 | rep 4 | rep 1 | rep 2 | ||
| JAZ10b | At5g13220 | 25/17 | 42/10 | 34/12 | 56/9 | 28/7 | 47/6 |
| NINJA | At4g28910 | 21/10 | 22/10 | 39/11 | 34/11 | nd | 6/5 |
| MYC2 | At1g32640 | nd | nd | 3/2 | 16/7 | nd | nd |
| MYC3 | At5g46760 | 5/3 | nd | 17/6 | 18/8 | nd | 5/3 |
| MYC4 | At4g17880 | nd | nd | 3/1 | 4/4 | nd | 6/4 |
| PPD2 | At4g14720 | 4/4 | 8/5 | 2/2 | 8/5 | nd | nd |
| JAZ12 | At5g20900 | nd | 4/2 | 6/3 | 3/1 | nd | 3/2 |
JAZ10.4-containing complexes were purified from T87 Arabidopsis cell cultures expressing either JAZ10.4-YFP (JAZ10.4) or JAZ10.4I107A-YFP (JAZ10.4 I107A) as bait. Data are shown for four and two independent experiments (rep) with JAZ10.4 and JAZ10.4 I107A, respectively. Data entries denote the number of assigned spectra/number of unique peptides for each of the listed copurifying proteins. Only peptides with 95% or greater confidence are shown. None of the listed proteins were identified in control cultures expressing YFP. bIndicates the identification of the bait protein (JAZ10.4 or JAZ10.4 I107A) by MS/MS.
JAZ10.4 Contains a Cryptic MYC2-Interacting Domain That Is Required for the Repression of JA Responses
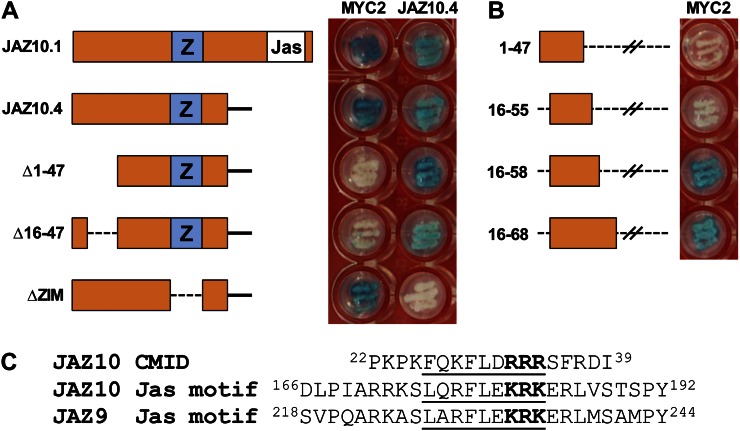
The ability of JAZ10.4 to interact with MYC2 was unexpected, because previous studies have shown that the Jas motif, which is not present in JAZ10.4, is necessary and sufficient for interaction with MYC2 (Katsir et al., 2008; Melotto et al., 2008; Chini et al., 2009). We used Y2H assays to map the MYC2-interacting domain of JAZ10.4. The strength of the JAZ10.4 interaction with MYC2 in this assay was comparable to that of the Jas motif-containing full-length splice variant, JAZ10.1 (Fig. 1A). Deletion of the ZIM domain abolished JAZ10.4 dimerization but did not affect JAZ10.4 interaction with MYC2, as reported previously (Chung and Howe, 2009). However, constructs lacking the N-terminal 47 amino acids (JAZ10.4Δ1-47) or lacking only amino acid residues 16 to 47 (JAZ10.4Δ16-47) failed to bind MYC2. Systematic deletion analysis identified a 43-amino acid fragment (JAZ10.416-58) that is sufficient for MYC2 interaction (Fig. 1B). Interestingly, this cryptic MYC2-interacting domain (CMID) contains a nine-amino acid sequence (FQKFLDRRR) that strongly resembles the core of the Jas motif of JAZ10 and other JAZ proteins, including a tribasic cluster of residues (Fig. 1C). In the Jas motif of JAZ9, the central R residue within the tribasic cluster (Fig. 1C) is essential for interaction with MYC2 (Withers et al., 2012).
Figure 1.
The N-terminal region of JAZ10.4 has a cryptic MYC2-interacting domain. A, Y2H assays of JAZ10.4 deletion constructs (DNA-binding domain bait fusions) with MYC2 and JAZ10.4 (activation domain prey fusions). Yeast strains coexpressing the indicated bait and prey proteins were plated on medium containing 5-bromo-4-chloro-3-indolyl-β-D-galactopyranoside. Blue-color formation in streaked yeast cells is indicative of protein-protein interaction. Photographic images were taken after 30 h of incubation at 30°C. JAZ10.1 is included as a positive control. The ZIM (Z) and Jas domains are indicated. The solid black line denotes the C-terminal region that is specific for the JAZ10.4 isoform. B, C-terminal deletion constructs of JAZ10.4 were tested for interaction with MYC2 as described in A. C, Sequence similarity between the conserved Jas motif of JAZ10 and JAZ9 and the CMID near the N terminus of JAZ10. The underlined sequence shows the region of conservation, which includes a tribasic motif (boldface).
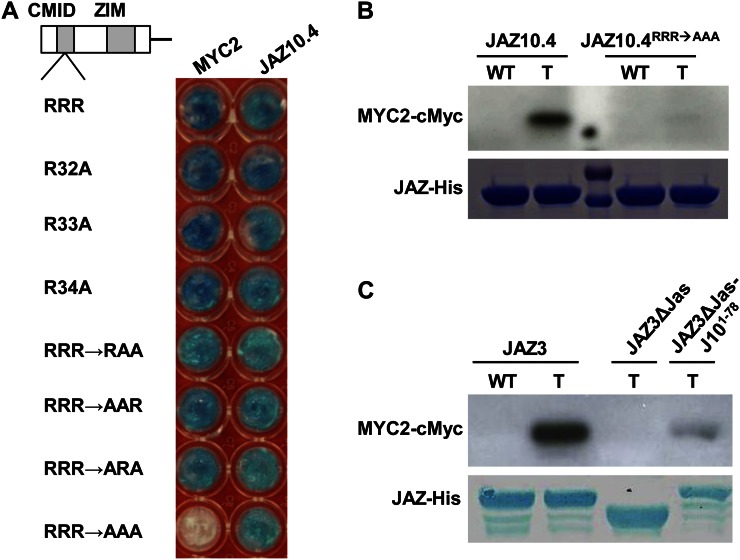
To assess whether the tribasic cluster (RRR32-34) in the JAZ10.4 CMID plays a role in MYC2 binding, we mutated each Arg residue, individually and in combination, to an Ala and tested the resulting constructs for interaction with MYC2. Single and double R→A substitutions had no effect on MYC2 binding in yeast. However, substitution of all three residues (RRR→AAA) abolished MYC2 binding but did not affect JAZ10.4 dimerization (Fig. 2A). We also expressed wild-type and mutant (JAZ10.4RRR→AAA) versions of JAZ10.4 in Escherichia coli as maltose-binding protein-JAZ-6xHis fusions and tested these proteins in pull-down assays for their ability to bind a cMyc-tagged derivative of MYC2 expressed in Arabidopsis. The results verified that the RRR32-34 motif of JAZ10.4 is required for MYC2 binding (Fig. 2B). To determine whether the CMID of JAZ10.4 is sufficient for MYC2 binding, we fused the N-terminal 78 amino acids of JAZ10.4 (JAZ101-78) to the C terminus of a JAZ3 derivative (JAZ3∆Jas) that lacks the entire Jas motif and thus does not interact with MYC2 (Chini et al., 2009). Pull-down assays showed that fusion of JAZ101-78 to the C terminus of JAZ3∆Jas partially restores interaction with MYC2 (Fig. 2C). Introduction of the RRR32-34→AAA mutation into the full-length JAZ10.1 isoform had no effect on MYC2 binding, presumably because JAZ10.1RRR→AAA contains an intact Jas motif (Supplemental Fig. S2).
Figure 2.
An RRR motif in the N-terminal region of JAZ10.4 is required for interaction with MYC2. A, Site-directed mutagenesis of the RRR motif within the CMID of JAZ10.4. The indicated R→A substitution mutants of JAZ10.4 were tested for interaction with MYC2 or JAZ10.4 as a positive control. Y2H assays were performed as described in Figure 1. B, In vitro pull-down assay of the JAZ10.4-MYC2 interaction. Assays were performed using the indicated JAZ10.4-His recombinant proteins and crude extracts from leaves of wild-type (WT) or 35S:cMyc-MYC2 transgenic (T) plants. Protein bound to JAZ-His was separated by SDS-PAGE and analyzed by immunoblotting (anti-cMyc antibody) for the presence of cMyc-MYC2. The Coomassie blue-stained gel shows total input protein as a loading control. C, The CMID of JAZ10.4 is sufficient for MYC2 binding. In vitro pull-down assays were performed as described in B using JAZ3, JAZ3ΔJas, or a chimeric JAZ (JAZ3ΔJas-J101-78) in which the CMID of JAZ10.4 was fused to the C terminus of JAZ3ΔJas. [See online article for color version of this figure.]
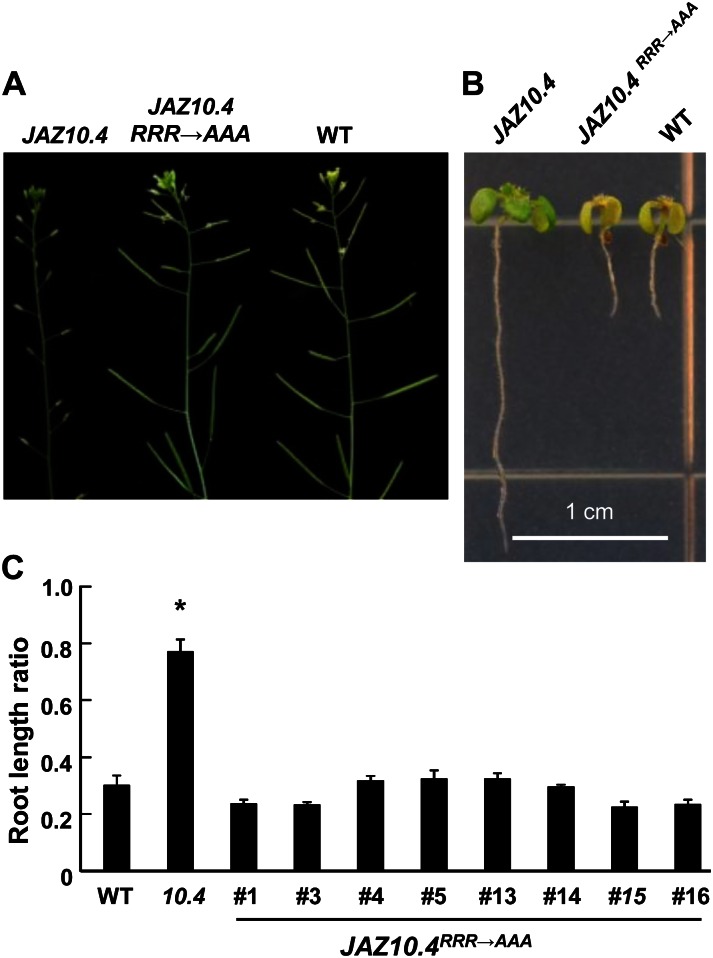
To determine whether the CMID is required for JAZ10.4 function in vivo, we compared JA responses in transgenic lines (35S:JAZ10.4-YFP and 35S:JAZ10.4RRR→AAA-YFP) that overexpress YFP fusions of JAZ10.4 and JAZ10.4RRR→AAA. In agreement with previous studies (Chung and Howe, 2009), 24% of all 35S:JAZ10.4-YFP plants tested (n = 73 independent T1 lines) exhibited hallmark characteristics of JA-associated male sterility, including short anther filaments and reduced anther dehiscence (Fig. 3A). Root growth inhibition assays also showed that 35S:JAZ10.4-YFP seedlings were strongly insensitive to JA (Fig. 3B). All 35S:JAZ10.4RRR→AAA-YFP lines (n = 50 independent T1 lines) were fully fertile and showed no signs of JA-associated male sterility. Experiments performed with T3 seedlings that are homozygous for the transgene further showed that the sensitivity of 35S:JAZ10.4RRR→AAA-YFP seedlings to JA is comparable to that of wild-type seedlings (Fig. 3, B and C). Confocal microscopy analysis showed that both JAZ10.4-YFP and JAZ10.4RRR→AAA-YFP localize to the nucleus of trichome cells (Supplemental Fig. S3). JAZ10.4RRR→AAA-YFP also showed nuclear localization in roots, although the signal was more diffuse (presumably cytosolic) than that of JAZ10.4-YFP. This suggests that the inability of JAZ10.4RRR→AAA-YFP to repress JA responses does not result from mislocalization of the JAZ fusion protein. Taken together, the results indicate that the N-terminal region of JAZ10.4 contains a cryptic MYC2-binding site that is required for JAZ10.4-mediated repression of JA responses.
Figure 3.
The RRR motif is required for the JAZ10.4-mediated repression of JA responses. A, The photograph shows silique development in the wild type (WT) and transgenic lines that overexpress JAZ10.4 or JAZ10.4RRR→AAA. B, The photograph shows seedlings of the indicated genotypes grown for 10 d on Murashige and Skoog medium containing 20 μm MeJA. C, Quantification of JA-induced root growth inhibition in the wild type, 35S:JAZ10.4 (10.4), and eight independent JAZ10.4RRR→AAA lines. Seedlings were grown for 10 d on Murashige and Skoog agar plates containing or not containing 20 µm MeJA. The root length ratio was calculated by dividing the average root length of MeJA-treated seedlings by the average root length of seedlings of the same genotype grown in the absence of MeJA. Data show means ± se (n = 12 seedlings per genotype). The asterisk denotes a significant difference (P < 0.05, Student’s t test) in comparison with the wild type. [See online article for color version of this figure.]
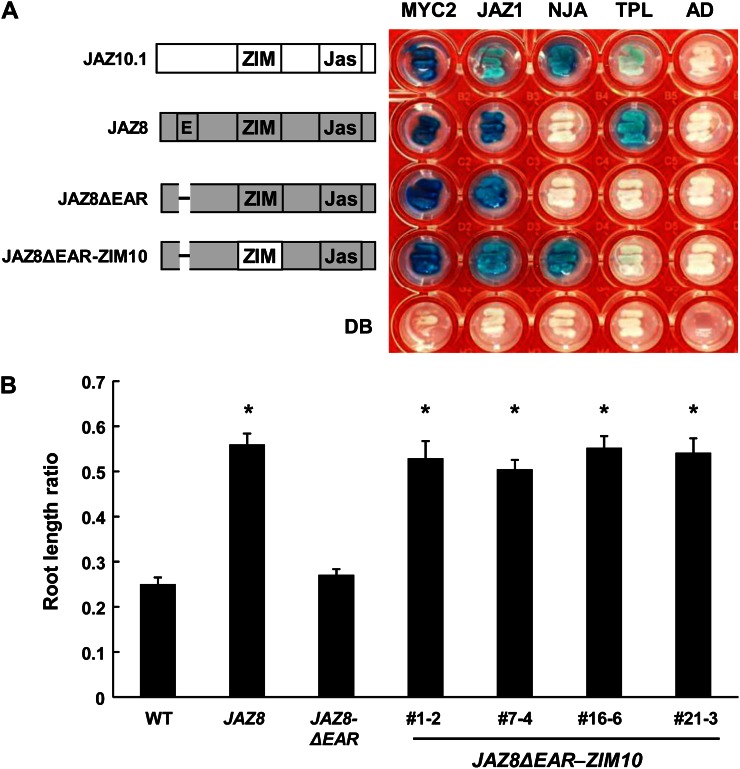
The ZIM Domain of JAZ10.4 Interacts with NINJA to Repress JA Responses
The identification of NINJA as a JAZ10.4-interacting protein (Tables I and II) suggests that repression of JA responses by JAZ10.4 involves recruitment of the NINJA-TPL corepressor complex to the ZIM domain of JAZ10.4 (Pauwels et al., 2010). To address this question, we tested whether the ZIM domain (residues 104–130) of JAZ10 is capable of conferring repressive activity on a JAZ that binds MYC2 but fails to associate with the NINJA-TPL corepressor complex. We took advantage of the fact that repression of JA responses by JAZ8 does not depend on its ZIM domain but rather relies on an EAR motif at the N terminus of JAZ8 that directly binds TPL. A mutant (JAZ8ΔEAR) of JAZ8 lacking the EAR motif does not interact with TPL and is unable to repress JA responses (Fig. 4A; Shyu et al., 2012). A chimeric protein (JAZ8ΔEAR-ZIM10) in which the ZIM domain of JAZ8ΔEAR was replaced with the ZIM domain of JAZ10 exhibited strong interaction with NINJA in yeast (Fig. 4A). In root growth assays, transgenic lines that express JAZ8ΔEAR-ZIM10 from the 35S promoter exhibited a JA-insensitive phenotype identical to that of JAZ8-overexpressing lines (Fig. 4B). Thus, the ZIM domain of JAZ10 restores repression activity to a JAZ variant (JAZ8ΔEAR) that binds MYC2 but is unable to recruit the corepressor complex.
Figure 4.
The ZIM domain of JAZ10 interacts with NINJA and is required for the repression of JA responses. A, Y2H assays depicting the interaction of JAZ10 or JAZ8 chimeric proteins (DNA binding-domain fusions [DB]) with MYC2, JAZ1, NINJA (NJA), and TPL (activation domain fusions [AD]). Empty vectors were included as negative controls. Sequence regions derived from JAZ10 and JAZ8 are shown in white and gray, respectively, together with various protein domains. E, EAR motif. Y2H assays were performed as described in Figure 1, except that the photographic image was taken after 48 h of incubation of yeast cells at 30°C. B, Overexpression of a JAZ8ΔEAR-ZIM10 chimeric protein confers insensitivity to JA in root growth assays. Seedlings were grown for 8 d on Murashige and Skoog agar plates containing or not containing 20 µm MeJA, and root length ratios were calculated as described in Figure 3C. Data show means ± se of at least 11 seedlings per genotype. Asterisks denote significant differences (P < 0.05, Student’s t test) in comparison with the wild type (WT).
The highly conserved Ile residue (Ile-107 in the TIFY motif) of the ZIM domain is required both for JAZ10.4 dimerization and the repression of JA responses by JAZ10.4 (Chung and Howe, 2009). Given that NINJA represses JA responses through its interaction with the ZIM domain of JAZ (Pauwels et al., 2010), we tested whether Ile-107 of JAZ10.4 may also be important for interaction with NINJA. Y2H assays showed that, whereas substitution of Ile-107 to Ala eliminates JAZ10.4 dimerization, this mutation reduced but did not abolish JAZ10.4 interaction with NINJA (Fig. 5A). A T106A mutation had no effect on JAZ10.4 binding to either JAZ10.4 or NINJA, suggesting that the effect is specific for Ile-107. In vitro pull-down assays confirmed that I107A eliminates JAZ10.4 dimerization and reduces the extent to which JAZ10.4 interacts with NINJA (Fig. 5, B and C). MS analysis of protein complexes immunopurified from T87 cells provided additional evidence that I107A disrupts the ability of NINJA to copurify with JAZ10.4 (Table II). Based on the number of assigned mass spectra corresponding to NINJA and JAZ10.4, we estimated that the amount of NINJA copurifying with JAZ10.4I107A was less than 20% of that copurifying with wild-type JAZ10.4 (Table II). These findings show that Ile-107 is not only critical for JAZ10.4 dimerization but also suggest a key role for this residue in stabilizing the JAZ10.4-NINJA interaction.
Figure 5.
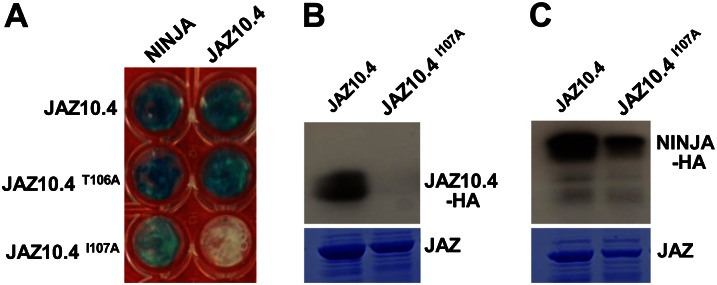
Ile-107 in the TIFY motif is involved in JAZ10.4 interaction with both JAZ10 and NINJA. A, Y2H assay depicting the role of Thr-106 and Ile-107 (within the TIFY motif) in JAZ10.4 dimerization and JAZ10.4 interaction with NINJA. Y2H assays were performed as described in Figure 1, except that the photographic image was taken after 24 h of incubation of yeast cells at 30°C. B, In vitro pull-down assay of the JAZ10.4-JAZ10.4 interaction. Assays were performed using the indicated wild-type (JAZ10.4) and mutant (JAZ10.4I107A) recombinant proteins and crude extract from a yeast strain expressing a JAZ10.4-HA fusion protein. Protein bound to JAZ-His was separated by SDS-PAGE and analyzed by immunoblotting with an anti-HA antibody. A Coomassie blue-stained gel is shown as a loading control. C, In vitro pull-down assay of the JAZ10.4-NINJA interaction. Assays were performed as described in B except for the use of a crude extract from a yeast strain expressing a NINJA-HA fusion protein. [See online article for color version of this figure.]
JA-Induced Expression of JAZ10.4 Attenuates JA Responses
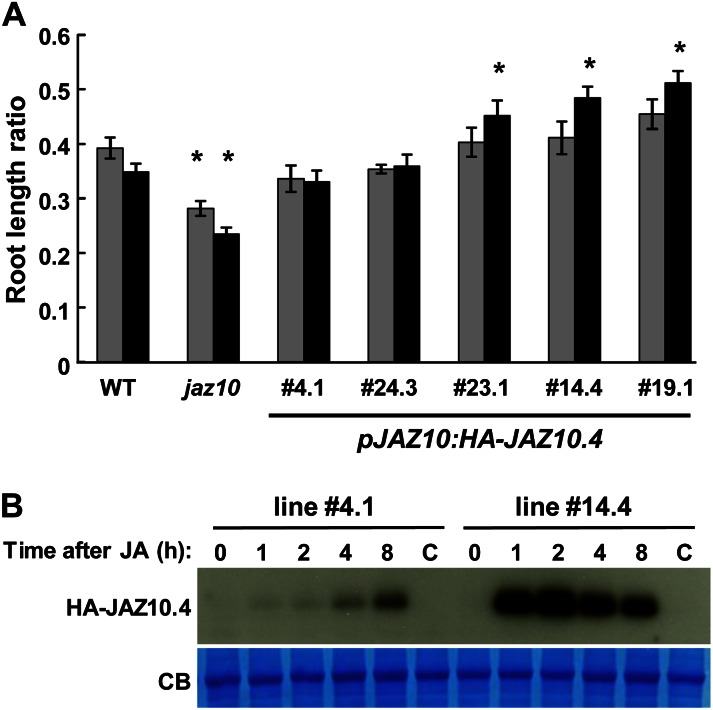
To further test the hypothesis that JAZ10.4 participates in a negative feedback loop to restrain JA responses, we tested whether expression of this splice variant from the native JAZ10 promoter is sufficient to attenuate JA responses. For these experiments, we used a 2.0-kb JAZ10 promoter fragment that confers JA-inducible expression of a GUS reporter gene (Sehr et al., 2010). A hemagglutinin (HA) epitope tag was added to the N terminus of JAZ10.4 to facilitate protein detection in plant tissues. The resulting pJAZ10:HA-JAZ10.4 transgene was introduced into the jaz10-1 mutant that is hypersensitive to JA-induced root growth inhibition (Demianski et al., 2012). Of eight lines that were confirmed to contain a single segregating copy of the transgene, all were significantly less sensitive to exogenous methyl jasmonate (MeJA; 20 μm) than jaz10-1 seedlings. Further analysis of five representative pJAZ10:HA-JAZ10.4 lines showed that all but one line (4.1) were also less sensitive than jaz10-1 to 10 μm MeJA (Fig. 6A). On medium containing 20 μm MeJA, root growth of some lines (e.g. 14.4) was significantly less sensitive than that of wild-type seedlings (Fig. 6A). This finding suggests that differences in the expression level of JAZ10.4 between transgenic lines may modulate sensitivity to JA.
Figure 6.
JA-induced expression of JAZ10.4 complements the JA-hypersensitive phenotype of jaz10-1. A, Root growth inhibition assay of the wild type (WT), jaz10-1 (jaz10), and five independent lines in which the pJAZ10:HA-JAZ10.4 transgene was introduced into the jaz10-1 mutant background. Root length was measured in 8-d-old seedlings grown on Murashige and Skoog medium containing 0, 10, or 20 µm MeJA. The root length ratio was calculated by dividing the average root length of seedlings grown in the presence of either 10 µm (gray bars) or 20 µm (black bars) MeJA by the average root length of seedlings of the same genotype grown in the absence of MeJA. Data show means ± se of 12 to 14 seedlings per genotype. Asterisks indicate significant differences in root length (P < 0.05, Student’s t test) in comparison between wild-type and jaz10 seedlings grown at the same concentration of MeJA or between wild-type and pJAZ10:HA-JAZ10.4 seedlings grown at the same concentration of MeJA. B, Accumulation of HA-JAZ10.4 protein in response to JA treatment. Ten-day-old liquid-grown seedlings of the indicated pJAZ10:HA-JAZ10.4 line were treated with 50 µm MeJA and harvested at various times (h) thereafter. As controls, seedlings were harvested immediately prior to treatment (0) and 8 h after mock treatment (C). Total protein was subjected to immunoblot analysis with an anti-HA antibody to detect HA-JAZ10.4. A Coomassie blue-stained gel of protein extracts was used as a loading control (CB). [See online article for color version of this figure.]
We used immunoblot analysis to test whether the level of JA-induced accumulation of JAZ10.4 in specific transgenic lines correlates with observed differences in root growth sensitivity to JA. Lines that exhibited either wild-type-like (line 4.1) or reduced (line 14.4) sensitivity to the hormone were grown in liquid Murashige and Skoog medium and treated with 50 μm MeJA. At various times after treatment, seedlings were harvested for western-blot analysis with an anti-HA antibody (Fig. 6B). We detected the accumulation of JAZ10.4 protein in both lines within 1 h of treatment. In the case of line 4.1, JAZ10.4 protein levels steadily increased during the 8-h time course. In the moderately JA-insensitive 14.4 line, however, JA treatment resulted in a much stronger accumulation of JAZ10.4, with protein levels peaking at 2 h and then declining slowly at later time points. These results show that JAZ10.4, when expressed from the native JAZ10 promoter, rapidly accumulates in response to JA treatment and that the strength of JAZ10.4 expression inversely correlates with the level of sensitivity to JA.
DISCUSSION
Domain Architecture of the JAZ10.4 Repressor
In our initial studies of JAZ10 alternative splicing, the strong repressive activity of JAZ10.4 was enigmatic because this isoform lacks the MYC2-interacting Jas motif (Chung and Howe, 2009). In this study, we show that repression by JAZ10.4 depends on a CMID located near the N terminus of JAZ10.4. The ability of JAZ10.4 to interact with MYC3 and MYC4 in yeast and in planta indicates that this JAZ splice variant likely targets the core MYC2/MYC3/MYC4 triad of TFs that control primary JA responses in leaves and roots (Cheng et al., 2011; Fernández-Calvo et al., 2011; Niu et al., 2011). The minimal CMID defined by deletion analysis contains a sequence (FQKFLDRRR) that has striking similarity to the central region of the conserved Jas motif (Fig. 1C). We show that a tribasic amino acid cluster (RRR) within this sequence is required both for MYC2 binding and dominant repressor function in planta. The RRR motif, therefore, may be functionally equivalent to the highly conserved KRK sequence of the Jas motif, which was recently shown to be critical for binding of JAZ9 to MYC2 and nuclear localization of JAZ9 (Withers et al., 2012). This conclusion is supported by the finding that the CMID restores, at least in part, the ability of JAZ3ΔJas to interact with MYC2. The apparent increase in partitioning of JAZ10.4RRR→AAA-YFP to the cytosol is consistent with the recent finding that MYC2 interaction correlates with the efficiency of the nuclear localization of JAZ1 and JAZ9 (Withers et al., 2012). Our results indicate, however, that the accumulation of JAZ10.4RRR→AAA-YFP in the nucleus is not strictly dependent on MYC2 interaction with the CMID. The fact that MYC2 interacts with the RRR-mutated form of JAZ10.1 (JAZ10.1RRR→AAA) suggests that two MYC-binding sites (Jas and CMID) on JAZ10.1 may act redundantly in the recruitment of TFs. It is also possible that these sites engage multiple TFs simultaneously to modulate the repressive activity of JAZ10.1 and JAZ10.3, which contain full-length and truncated Jas motifs, respectively. It is noteworthy that the CMID encompasses a sequence region (referred to as domain 1) that is weakly conserved between various members of the Arabidopsis JAZ family (Thines et al., 2007). Inspection of the domain 1 sequence indicates that the Jas motif-like sequence within the CMID is not well conserved in other Arabidopsis JAZ proteins. The presence of JAZ10 orthologs in other plant species, however, suggests that the production of JA-resistant truncated JAZs via alternative splicing may be a general mechanism to dampen JA responses.
Transcriptional repression by JAZ proteins is thought to involve the recruitment of TPL and TPR corepressors to JAZ-TF complexes bound to promoter regions of JA-response genes (Pauwels and Goossens, 2011). Genetic and biochemical evidence supports the existence of two mechanisms to explain how JAZs are linked to TPL/TPRs. One mechanism invokes interaction of the ZIM domain with NINJA, which contains an EAR motif that interacts with TPL/TPRs (Pauwels et al., 2010). Alternatively, and as exemplified by JAZ8, a subset of JAZs contain an EAR motif that allows direct binding to TPL independently of the ZIM domain and NINJA (Shyu et al., 2012). A third group of JAZ proteins (JAZ5 and JAZ6) have the potential to interact with corepressors via both NINJA-dependent and -independent mechanisms, but this remains to be confirmed (Kagale et al., 2010; Pauwels and Goossens, 2011). Our results indicate that JAZ10.4 represses JA responses through the former NINJA/ZIM-dependent pathway. In support of this conclusion, mutation of Ile-107 within the conserved TIFY motif abolished the ability of ectopically expressed JAZ10.4 to confer JA insensitivity (Chung and Howe, 2009; this study). Various protein-protein interaction assays, including MS analysis of JAZ10.4 protein complexes purified from Arabidopsis cells, showed that I107A strongly diminishes the ability of JAZ10.4 to interact with NINJA. An essential role for the ZIM domain in JAZ10.4 repression is also supported by domain-swap experiments showing that the ZIM domain of JAZ10 confers repressive activity on a modified JAZ (JAZ8ΔEAR) that binds MYC2 but fails to interact with NINJA. Overexpression of JAZ8ΔEAR-ZIM10 and JAZ8 has similar quantitative effects on JA-mediated root growth inhibition, suggesting that our engineered NINJA-dependent JAZ8 repressor is functionally equivalent to the wild-type JAZ8 repressor (Fig. 4). In contrast to JAZ10.4, however, overexpression of JAZ8ΔEAR-ZIM10 or JAZ8 does not result in male sterility. This finding suggests that JAZ10.4 may be more stable than JAZ8 or, alternatively, that JAZ10.4 and JAZ8 target different sets of TFs in their respective overexpressing lines. These findings highlight the modular nature of JAZ functional domains and provide proof of concept for the idea of using synthetic JAZ repressors to control specific JA responses.
An important feature of the ZIM domain is its ability to mediate JAZ interaction with both NINJA and other TIFY proteins (Vanholme et al., 2007; Chini et al., 2009; Chung and Howe, 2009). Using cell extracts from Arabidopsis T87 cells, we found that JAZ10.4 copurifies not only with MYC TFs and NINJA but also with JAZ12 and PPD2. This finding is in agreement with tandem affinity purification tagging screens that identified NINJA, MYC TFs, and various TIFY proteins as components of JAZ-containing multiprotein complexes (Pauwels et al., 2010; Fernández-Calvo et al., 2011). Our in vivo and in vitro studies of the JAZ10.4I107A mutant further showed that the TIFY motif is required for JAZ10.4 interaction with both NINJA and other JAZ proteins (Chung and Howe, 2009). Although the MS data indicate that JAZ10.4I107A retains the ability to interact with MYC TFs, we cannot exclude the possibility that this mutation alters the stability of JAZ10.4-TF complexes as a consequence of changes in the overall structure of JAZ10.4. Nevertheless, our findings suggest that JAZ-JAZ and JAZ-NINJA interactions involve common sequence determinants and raise the question of how JAZ homomeric and heteromeric interactions affect JAZ repressor activity (Howe, 2010). JAZ8 is unique in this context because, despite the presence of a conserved TIFY motif, it interacts very weakly with NINJA but retains the ability to heterodimerize with other JAZs (Pauwels et al., 2010; Shyu et al., 2012; this study). The differential interaction of JAZ8 and JAZ10 with NINJA indicates that unidentified sequence determinants within the ZIM domain play an important role in JAZ-NINJA coupling.
Negative Feedback Regulation by JAZ10.4
In comparison with most other JAZ proteins in Arabidopsis, a distinguishing feature of JAZ10.4 is its enhanced stability in cells containing high levels of JA. The increased stability of JAZ10.4 is a direct consequence of an alternative splicing event that results in loss of the COI1-interacting degron and, as a consequence, resistance to JA-induced degradation (Chung and Howe, 2009). In contrast to labile JAZs, the strength of repression by JAZ10.4 is expected to increase as the expression of JAZ10 increases. Consistent with this idea, overexpression of either the JAZ10.4 complementary DNA (cDNA) or a JAZ10 genomic clone from the constitutive cauliflower mosaic virus 35S promoter results in strong JA-insensitive root phenotypes (Chung and Howe, 2009; Chung et al., 2010). Given that JAZ10 expression is tightly controlled by the JA pathway (Yan et al., 2007; Chung et al., 2008; Fernández-Calvo et al., 2011; Demianski et al., 2012), an unresolved question has been whether JAZ10.4 accumulates in JA-stimulated wild-type plants and, if so, whether its induced expression is sufficient to attenuate JA responses. Here, we demonstrate that the expression of JAZ10.4 from the native JAZ10 promoter does in fact complement the JA-hypersensitive phenotype of jaz10-1 seedlings. We also show that the reduced sensitivity of pJAZ10:HA-JAZ10.4 lines to exogenous JA correlates with the level of JAZ10.4 protein in JA-stimulated seedlings. These collective findings strongly support a role for JAZ10.4 as an endogenous negative regulator of JA signaling.
Because the expression of JAZ10.4 in pJAZ10:HA-JAZ10.4 lines does not depend on alternative splicing of JAZ10 pre-mRNA, the level of JAZ10.4 in these lines may be greater than that produced in wild-type plants. We attempted to account for this possibility by characterizing multiple independent transgenic lines that exhibit a range of sensitivity to JA. When grown on relatively low levels of exogenous JA (10 μm), all lines tested were less sensitive to JA than the JA-hypersensitive jaz10-1 mutant and, for the most part, resembled wild-type seedlings. At higher concentrations of JA, some lines were significantly less sensitive to JA than the wild type. Based on the level of JAZ10.4 accumulation in representative low-expressing (i.e. 4.1) and high-expressing (e.g. 14.4) lines, we attribute variation in hormone sensitivity to differences in the strength of JAZ10.4 expression. Regardless of this variation, the fact that all pJAZ10:HA-JAZ10.4 lines were less sensitive to JA than jaz10-1 seedlings suggests that the hypersensitive phenotype of jaz10-1 results in part from the loss of production of JAZ10.4.
Our results support a model of negative feedback inhibition by JAZ10.4. At low JA levels, one or more JAZ repressors interact with MYC TFs to inhibit the expression of JAZ10 and shut off the production of JAZ10 splice variants. This is supported by the fact that JAZ10 expression is nearly abolished in a myc2/myc3/myc4 triple mutant (Fernández-Calvo et al., 2011) and by the existence of putative MYC2/MYC3/MYC4-binding G-box elements in the promoter of JAZ10 (Chini et al., 2007). Additionally, cycloheximide induction experiments showed that JAZ10 is a primary rather than secondary JA-response gene (Chung et al., 2008). In response to environmental or developmental cues that result in increased JA-Ile levels, we propose that JAZ repressors bound to the promoter of JAZ10 are degraded, thus allowing transcriptional activation of JAZ10 by MYC2/MYC3/MYC4. The identities of specific JAZ proteins that silence the expression of JAZ10 and other primary response genes under low-JA conditions remain to be determined. Following JA-induced transcription of JAZ10, JAZ10 pre-mRNA is subject to alternative splicing events that give rise to JAZ10.1, JAZ10.3, and JAZ10.4. The relative abundance of each JAZ10 isoform will depend not only on the efficiency of the respective alternative splicing events but also the differential stability of the proteins in JA-stimulated cells (Chung and Howe, 2009). Indeed, we found that JAZ10.4 rapidly accumulates in response to JA treatment and that accumulation of the protein correlates with the reduced sensitivity of seedlings to JA. Based on the ability of JAZ10.4 to repress JA responses and to bind MYC2/MYC3/MYC4, JAZ10.4 likely restrains the expression of genes controlled by these TFs. Additional experiments are needed to test this hypothesis. Given that JA-induced expression of JAZ10 is dependent on MYC2/MYC3/MYC4 (Fernández-Calvo et al., 2011), it is possible that JAZ10.4 negatively regulates its own production by binding to MYC2/MYC3/MYC4 and inhibiting JAZ10 transcription. This function of JAZ10 in the feedback control of JA signaling supports the emerging view that alternative splicing plays a fundamental role in plant adaptation to environmental stress (Reddy, 2007; Barbazuk et al., 2008).
It is well established that plants, like animals, have evolved various mechanisms to desensitize cells to the presence of a hormone, thereby restraining the duration and amplitude of hormone-induced responses. Among the proposed mechanisms involved in the attenuation of JA responses are JA-induced removal of JA-Ile (Kitaoka et al., 2011; Koo et al., 2011; Heitz et al., 2012; Koo and Howe, 2012; Woldemariam et al., 2012), JA-induced expression of MYC2-related TFs that negatively regulate JA responses (Nakata et al., 2013), and, as described here, JA-induced synthesis of stable JAZ repressors (Chung and Howe, 2009; Chung et al., 2010; Shyu et al., 2012). Negative regulatory feedback by JAZ10.4 and other stable JAZs may provide stability to the JA network by limiting the range over which the concentrations of signaling components, including MYC-related TFs, JAZs, and JA-Ile, fluctuate during the JA response. These JAZs may be important for curtailing JA-related defense responses that are energetically demanding or toxic to the cell or for maintaining appropriate growth rates in fluctuating environmental conditions (Moreno et al., 2009; Cerrudo et al., 2012; Yang et al., 2012). The ability of JAZ10 to physically interact with Arabidopsis DELLA proteins (Yang et al., 2012), which mediate growth repression, raises the interesting possibility that stable JAZ10 splice variants may act synergistically to promote growth in the presence of high JA levels. Tissue- or cell-specific accumulation of JA-resistant JAZs may provide a mechanism to generate spatial heterogeneity in growth and defense responses (Kessler and Baldwin, 2002; Melotto et al., 2006). A better understanding of JAZ protein expression patterns promises to provide insight into how these key regulators enhance plant fitness in challenging and continuously changing environments.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Arabidopsis (Arabidopsis thaliana) plants were grown at 21°C under long-day conditions as described by Chung and Howe (2009). The Columbia ecotype was used as the wild-type genetic background for all experiments. The jaz10-1 mutant (SAIL_92_D08) was obtained from the Arabidopsis Biological Resource Center. The 35S:JAZ8ΔEAR line was described previously by Shyu et al. (2012). JA-mediated root growth inhibition assays were performed as described previously (Chung and Howe, 2009; Shyu et al., 2012).
Y2H Analysis
JAZ10.4 was used as the bait in a Y2H screen performed by Hybrigenics. A cDNA encoding JAZ10.4 was cloned into pB27 as a C-terminal fusion to LexA (N-LexA-JAZ10.4-C) and subsequently used to screen a random-primed cDNA library prepared from 1-week-old Arabidopsis seedlings. Thirty-two million clones (3.2-fold coverage of the library) were screened using a mating approach with yeast (Saccharomyces cerevisiae) strains HGX13 and L40ΔGal4 as described previously (Fromont-Racine et al., 1997). His+ colonies (295 total) were selected on a medium lacking Trp, Leu, and His and supplemented with 0.5 mm 3-aminotriazole to reduce autoactivation by the “bait” protein. cDNA fragments corresponding to positive “prey” clones were amplified by PCR and sequenced at their 5′ and 3′ junctions. The resulting sequences were searched against GenBank and assigned a quality score indicative of the confidence of interaction (Formstecher et al., 2005). Only those interactions with the highest confidence values are reported in this paper.
All other Y2H assays were performed with the Matchmaker LexA system (Clontech) as described previously (Melotto et al., 2008; Chung and Howe, 2009). JAZ10.4 constructs were subcloned into the pGILDA vector to generate translational fusions with the LexA DNA-binding domain. Full-length cDNAs encoding MYC2 and NINJA were subcloned into the pB42AD vector to generate fusions with the B42 activation domain. Various bait-prey pairs were cotransformed into yeast (Saccharomyces cerevisiae) strain EGY48 using the Frozen-EZ Yeast Transformation II kit (Zymo Research). Photographic images of Y2H plates were taken after 1 or 2 d of incubation at 30°C (Chung and Howe, 2009). Y2H assays were replicated at least three times. A complete list of primers and constructs used for cloning into pGILDA and pB42AD are described in Supplemental Table S1.
Transgene Constructs
All PCRs were performed with KAPA HIFI Polymerase (Kapa Biosystems) following the manufacturer’s instructions. All primer sets used for PCR are listed in Supplemental Table S1. For cloning with Gateway vectors (Invitrogen), PCR amplicons were purified and cloned into pEntr-D-Topo using the pENTR Directional TOPO Cloning Kit (Invitrogen). Sequence-verified clones were transferred to the corresponding destination vector (Supplemental Table S1) using the Gateway LR Clonase II Enzyme Mix (Invitrogen). JAZ10.4 and JAZ10.4RRR→AAA constructs were cloned by Gateway reaction into the pEarley101 destination vector to generate a C-terminal fusion protein with YFP (Earley et al., 2006). We followed a two-step selection process to obtain lines that express these fusion proteins at relatively high levels. First, transgenic seedlings were screened on Murashige and Skoog agar plates for resistance to kanamycin (50 μg mL−1). Second, a Leica M165FC Fluorescence Stereomicroscope was used to screen the resulting kanamycin-resistant lines for seedlings that accumulate the JAZ-YFP fusion protein in the nucleus.
Expression of the N-terminal HA-tagged JAZ10.4 protein from the native JAZ10 promoter was achieved as follows. A 2.0-kb JAZ10 promoter sequence, similar to that described by Sehr et al. (2010), was amplified by PCR using XhoI-predigested T31B5 bacterial artificial chromosome DNA as a template. The JAZ10 promoter was cloned directionally into pEntr-D-Topo (to generate pEntr-JAZ10promoter) using forward and reverse primers carrying NotI restriction sites (Supplemental Table S1). JAZ10.4 cDNA was amplified with a primer set designed to add an HA epitope tag to the N terminus of the protein, and the resulting amplicon was cloned into pEntr-D-Topo (to give pEntr-HA-JAZ10.4). The JAZ10 promoter was released from pEntr-JAZ10promoter using the NotI restriction enzyme and ligated to the NotI-linearized pEntr-HA-JAZ10.4 using the Rapid Ligation Kit (Roche). An LR Clonase (Invitrogen) reaction was used to transfer this construct into the final destination vector, pGWB401 (Nakagawa et al., 2007). The final construct was confirmed by sequencing and transformed into Agrobacterium tumefaciens strain C58C1 for the generation of transgenic Arabidopsis plants (Chung and Howe, 2009). Kanamycin-resistant T1 seedlings were selected on agar plates containing 50 μg mL−1 antibiotic. T2 seedlings were used to select lines containing a single transfer DNA insertion based on a 3:1 segregation ratio of the antibiotic resistance marker. T3 seedlings were used to screen for lines that were homozygous for the transgene.
The MYC2 cDNA was PCR amplified using cDNA synthesized from total RNA isolated from rosette leaves of 5-week-old Arabidopsis plants. Oligonucleotide primer sequences are described in Supplemental Table S1. The resulting MYC2 amplicon was cloned into pENTR/D-TOPO (Invitrogen) and transferred into the pJYP006 binary vector to create a vector (35S:Myc-MYC2) in which the N terminus of MYC2 is fused to nine repeats of the c-Myc epitope. This vector was introduced into A. tumefaciens strain GV3100 for transformation of the jin1-9 mutant. T1 plants were grown on soil for 7 d and sprayed with a solution containing 0.3 µm glufosinate (Finale; AgrEvo Environmental Health). T2 plants containing a single transfer DNA insertion (as determined by 3:1 segregation of glufosinate resistance) were used for western-blot analysis to confirm the expression of Myc-MYC2. A homozygous T3 line was used as a source of leaf extracts for in vitro pull-down assays.
We used four PCRs to generate the JAZ8ΔEAR-ZIM10 construct. In one reaction, JAZ8ΔEAR cDNA was used as a template with primer set JAZ8-pENTR-FP and JAZ8-ZIM10-N-RP to amplify cDNA encoding the first 44 amino acids of JAZ8ΔEAR. Another reaction also used JAZ8ΔEAR as a template with primer set JAZ8-ZIM10-C-FP and JAZ8-pENTR-RP to amplify cDNA encoding the last 59 amino acids of JAZ8ΔEAR (amino acids 73–131). In the third PCR, the amplicon from the first PCR (JAZ8ΔEAR N-terminal 44 amino acids) and JAZ10.1 cDNA were both used as templates with primer set JAZ8-pENTR-FP and JAZ10-KpnI-RP to generate a JAZ8ΔEAR-JAZ10 chimera. Finally, the JAZ8ΔEAR-JAZ10 chimera and cDNA encoding the last 59 amino acids of JAZ8ΔEAR (amplicon from the second PCR) were used as templates with primer set JAZ8-pENTR-FP and JAZ8-pENTR-RP to generate the final JAZ8ΔEAR-ZIM10 construct. JAZ8ΔEAR-ZIM10 was cloned into vector pENTR-TOPO for sequencing and subcloning. Following A. tumefaciens-mediated transformation, 32 independent T1 plants were transferred to soil and RNA was extracted from leaf tissue. JAZ8ΔEAR-ZIM10 gene-specific primers (JAZ8-pENTR-FP and JAZ8-pENTR-RP) were used in reverse transcription-PCR to confirm expression of the transgene. Ten lines were selected and further propagated for the identification of homozygous T3 lines.
Site-Directed Mutagenesis
Single and multiple amino acid residues within JAZ proteins were substituted to Ala with the Quick-Change II site-directed mutagenesis kit (Stratagene), as described previously (Chung and Howe, 2009; Shyu et al., 2012). PCR was performed with Pfu Turbo DNA Polymerase to generate the constructs listed in Supplemental Table S1. pGEM-T vector harboring the wild-type JAZ cDNA was used as a template for mutagenesis. All constructs were sequenced to confirm the corresponding mutation.
In Vitro Pull-Down Assays
Cloning, expression, and purification of recombinant JAZ proteins as maltose-binding protein- and hexa-His-tagged fusions (referred to as JAZ-His) were done as described previously (Thines et al., 2007; Chung et al., 2010). Primers used for cloning are described in Supplemental Table S1. JAZ-MYC2 interactions were analyzed in pull-down assays employing purified JAZ-His proteins and an epitope-tagged (9× c-Myc) derivative of MYC2, which was expressed under the control of the cauliflower mosaic virus 35S promoter in the jin1-9 mutant background of Arabidopsis as described above. Leaf tissue from this transgenic line (35S:Myc-MYC2) was ground to a fine powder in liquid nitrogen and extracted in 2 mL of homogenization buffer (Chung et al., 2010) per gram of ground tissue with the addition of Complete Mini protease inhibitor tablet-EDTA free (Roche) and 50 µm MG132 (Sigma-Aldrich). The extract was centrifuged at 20,200g for 15 min at 4°C, and the resulting supernatant was subjected to a second centrifugation round under the same conditions. In vitro pull-down assays were done as described previously (Chung et al., 2010) using 25 µg of recombinant JAZ-His and 0.5 mg of protein of 35S:Myc-MYC2 leaf extract. MYC2 binding was assessed by western-blot analysis using an anti-cMyc antibody (Covance). All in vitro pull-down assays were repeated at least three times.
Analysis of JAZ10 Protein Complexes Isolated from Arabidopsis T87 Cells
Arabidopsis T87 cells (Axelos et al., 1992) were obtained from the RIKEN BioResource Center. Cells were transformed with A. tumefaciens strain EHA105 as described by Held et al. (2012). All transgenic Arabidopsis cell lines were generated by transformation with vector pVKgw-N-YFP, which fuses YFP to the N terminus of JAZ10 isoforms (Held et al., 2012). The expression of YFP-JAZ10 proteins in T87 cells was assessed by western-blot analysis using an anti-GFP antibody (Molecular Probes). Selected T87 cell lines were inoculated into 50 mL of fresh medium and grown for 4 d to exponential phase (Held et al., 2012). For experiments involving JA treatment, a solution of MeJA (Sigma-Aldrich) was added to a final concentration of 50 µm to exponentially growing cells. Cells were harvested 2 h later on Whatman filter paper using a vacuum trap connected to a ceramic funnel. The fresh cell mass was immediately estimated and frozen in liquid nitrogen. A small aliquot of cells in the original flask was set aside for imaging by confocal microscopy (Olympus FluoView 1000 Laser Scanning Confocal Microscope). Frozen cells were ground with a mortar and pestle to a fine powder in liquid nitrogen. Protein was extracted by the addition of 2 mL of lysis buffer (25 mm Tris-HCl, pH 7.5, 15 mm MgCl, 150 mm NaCl, 0.01% Tween 20, 50 µm MG132, 1 mm phenylmethylsulfonyl fluoride, 14.6 µm β-mercaptoethanol, and one tablet of MiniProtean cocktail [Roche] per 10 mL of lysis buffer) per gram of liquid nitrogen-ground cell powder. The resulting mixture was thawed with gentle rocking at 4°C for 30 min and then centrifuged at 26,890g at 4°C for 30 min. The supernatant was decanted to a prechilled tube, and GFP-Trap_A resin (ChromoTek) was added (10 µL of prewashed resin per 10 g of cell powder). Prior to this step, GFP-Trap resin was washed three times with 500 µL of dilution buffer (identical to lysis buffer but lacking Tween 20). The crude cell extract/GFP-Trap resin mixture was incubated for 2 h at 4°C with gentle rocking and then centrifuged at 900g for 5 min in a swinging-bucket centrifuge (GS-6R; Beckman) at 4°C. The resulting supernatant was transferred to a prechilled tube for a second round of affinity purification using the conditions described above. Protein-bound resin from both rounds of purification was pooled and washed three times with 500 µL of ice-cold dilution buffer.
Antibody-bound proteins were digested on bead by incubation for 6 h at 37°C in 10 µL of a solution containing trypsin (5 ng µL−1) and 50 mm ammonium bicarbonate. The solution was acidified by the addition of 5% formic acid and then centrifuged at 14,000g. Peptide-containing supernatant was removed and concentrated by solid-phase extraction with OMIX tips (www.varian.com). Purified peptides were then resuspended in 20 µL of a solution containing 2% acetonitrile and 0.1% trifluoroacetic acid. An aliquot (10 µL) of this fraction was injected by a Waters nanoAcquity Sample Manager (www.waters.com) and loaded for 5 min onto a Waters Symmetry C18 peptide trap (5 µm, 180 µm × 20 mm) at 4 µL min−1 in 5% acetonitrile/0.1% formic acid. The bound peptides were eluted onto a Waters BEH C18 nanoAcquity column (1.7 µm, 100 µm × 100 mm) over 35 min with a gradient of 5% B to 30% B in 21 min, ramped up to 90% B at 23 min and held for 1 min, then dropped back to 5% B at 24.1 min using a Waters nanoAcquity ultra-performance liquid chromatography device (buffer A = 99.9% water/0.1% formic acid, buffer B = 99.9% acetonitrile/0.1% formic acid) with an initial flow rate of 0.8 µL min−1.
Eluted peptides were sprayed into a ThermoFisher LTQ Linear Ion Trap mass spectrometer outfitted with a MICHROM Bioresources ADVANCE nano-spray source. The top five ions in each survey scan were then subjected to data-dependent zoom scans followed by low-energy collision-induced dissociation, and the resulting MS/MS spectra were converted to peptide lists with BioWorks Browser version 3.3.1 (ThermoFisher) using the default LTQ instrument parameters to filter out nonpeptide signals recorded during data acquisition. Peptide lists were searched against a custom protein sequence database consisting of Arabidopsis (The Arabidopsis Information Resource 10; www.arabidopsis.org) combined with common laboratory contaminants (downloaded from www.ncbi.nlm.nih.gov) using the Mascot searching algorithm, version 2.4 (www.matrixscience.com). The Mascot output was then analyzed with Scaffold version 3.6.0 (www.proteomesoftware.com) to probabilistically validate protein identifications using the ProteinProphet computer algorithm (Nesvizhskii et al., 2003). Only those peptides satisfying the Scaffold 95% confidence filter were reported.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Expression of JAZ10 splice variants in Arabidopsis T87 cell cultures.
Supplemental Figure S2. The RRR motif is required for the interaction of MYC2 with JAZ10.4 but not JAZ10.1.
Supplemental Figure S3. Subcellular localization of JAZ10.4 and JAZ10.4RRR→AAA-YFP.
Supplemental Table S1. Description of oligonucleotide primers used in this study.
Acknowledgments
We gratefully acknowledge Linda Danhof for performing T87 Arabidopsis cell transformations and for the maintenance of transformed cultures as well as the RIKEN BioResource Center for providing T87 cells. We thank Yuki Yoshida for providing seeds of the jaz10-1 mutant. The SAIL_92_D08 line and the T31B5 bacterial artificial chromosome were obtained from the Arabidopsis Biological Resource Center. Michael Held and Federica Brandizzi are acknowledged for providing the pVKgw-N-YFP vector. We also thank Doug Whitten and the MSU Proteomics Facility for assistance with proteomic analyses, and Ian Major for helpful comments on the manuscript. Ben MacNeille, Muhammad Nuraiman Rosme, and Li Deng are acknowledged for technical assistance throughout the project.
Glossary
- JA
jasmonate
- JA-Ile
jasmonoyl-l-Ile
- TF
transcription factor
- EAR
ERF-associated amphiphilic repression
- Y2H
yeast two-hybrid
- MS
mass spectrometry
- YFP
yellow fluorescent protein
- CMID
cryptic MYC2-interacting domain
- HA
hemagglutinin
- MeJA
methyl jasmonate
- cDNA
complementary DNA
References
- Arabidopsis Interactome Mapping Consortium (2011) Evidence for network evolution in an Arabidopsis interactome map. Science 333: 601–607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axelos M, Curie C, Mazzolini L, Bardet C, Lescure B. (1992) A protocol for transient gene expression in Arabidopsis thaliana protoplasts isolated from cell-suspension cultures. Plant Physiol Biochem 30: 123–128 [Google Scholar]
- Barbazuk WB, Fu Y, McGinnis KM. (2008) Genome-wide analyses of alternative splicing in plants: opportunities and challenges. Genome Res 18: 1381–1392 [DOI] [PubMed] [Google Scholar]
- Browse J. (2009) Jasmonate passes muster: a receptor and targets for the defense hormone. Annu Rev Plant Biol 60: 183–205 [DOI] [PubMed] [Google Scholar]
- Causier B, Ashworth M, Guo WJ, Davies B. (2012) The TOPLESS interactome: a framework for gene repression in Arabidopsis. Plant Physiol 158: 423–438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerrudo I, Keller MM, Cargnel MD, Demkura PV, de Wit M, Patitucci MS, Pierik R, Pieterse CMJ, Ballaré CL. (2012) Low red/far-red ratios reduce Arabidopsis resistance to Botrytis cinerea and jasmonate responses via a COI1-JAZ10-dependent, salicylic acid-independent mechanism. Plant Physiol 158: 2042–2052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q, Sun J, Zhai Q, Zhou W, Qi L, Xu L, Wang B, Chen R, Jiang H, Qi J, et al. (2011) The basic helix-loop-helix transcription factor MYC2 directly represses PLETHORA expression during jasmonate-mediated modulation of the root stem cell niche in Arabidopsis. Plant Cell 23: 3335–3352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Z, Sun L, Qi T, Zhang B, Peng W, Liu Y, Xie D. (2011) The bHLH transcription factor MYC3 interacts with the jasmonate ZIM-domain proteins to mediate jasmonate response in Arabidopsis. Mol Plant 4: 279–288 [DOI] [PubMed] [Google Scholar]
- Chini A, Fonseca S, Chico JM, Fernández-Calvo P, Solano R. (2009) The ZIM domain mediates homo- and heteromeric interactions between Arabidopsis JAZ proteins. Plant J 59: 77–87 [DOI] [PubMed] [Google Scholar]
- Chini A, Fonseca S, Fernández G, Adie B, Chico JM, Lorenzo O, García-Casado G, López-Vidriero I, Lozano FM, Ponce MR, et al (2007) The JAZ family of repressors is the missing link in jasmonate signalling. Nature 448: 666–671 [DOI] [PubMed] [Google Scholar]
- Chung HS, Cooke TF, Depew CL, Patel LC, Ogawa N, Kobayashi Y, Howe GA. (2010) Alternative splicing expands the repertoire of dominant JAZ repressors of jasmonate signaling. Plant J 63: 613–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung HS, Howe GA. (2009) A critical role for the TIFY motif in repression of jasmonate signaling by a stabilized splice variant of the JASMONATE ZIM-domain protein JAZ10 in Arabidopsis. Plant Cell 21: 131–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung HS, Koo AJK, Gao X, Jayanty S, Thines B, Jones AD, Howe GA. (2008) Regulation and function of Arabidopsis JASMONATE ZIM-domain genes in response to wounding and herbivory. Plant Physiol 146: 952–964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung HS, Niu Y, Browse J, Howe GA. (2009) Top hits in contemporary JAZ: an update on jasmonate signaling. Phytochemistry 70: 1547–1559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demianski AJ, Chung KM, Kunkel BN. (2012) Analysis of Arabidopsis JAZ gene expression during Pseudomonas syringae pathogenesis. Mol Plant Pathol 13: 46–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earley KW, Haag JR, Pontes O, Opper K, Juehne T, Song KM, Pikaard CS. (2006) Gateway-compatible vectors for plant functional genomics and proteomics. Plant J 45: 616–629 [DOI] [PubMed] [Google Scholar]
- Fernández-Calvo P, Chini A, Fernández-Barbero G, Chico JM, Gimenez-Ibanez S, Geerinck J, Eeckhout D, Schweizer F, Godoy M, Franco-Zorrilla JM, et al. (2011) The Arabidopsis bHLH transcription factors MYC3 and MYC4 are targets of JAZ repressors and act additively with MYC2 in the activation of jasmonate responses. Plant Cell 23: 701–715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonseca S, Chini A, Hamberg M, Adie B, Porzel A, Kramell R, Miersch O, Wasternack C, Solano R. (2009) (+)-7-iso-Jasmonoyl-L-isoleucine is the endogenous bioactive jasmonate. Nat Chem Biol 5: 344–350 [DOI] [PubMed] [Google Scholar]
- Formstecher E, Aresta S, Collura V, Hamburger A, Meil A, Trehin A, Reverdy C, Betin V, Maire S, Brun C, et al. (2005) Protein interaction mapping: a Drosophila case study. Genome Res 15: 376–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fromont-Racine M, Rain JC, Legrain P. (1997) Toward a functional analysis of the yeast genome through exhaustive two-hybrid screens. Nat Genet 16: 277–282 [DOI] [PubMed] [Google Scholar]
- Glazebrook J. (2005) Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu Rev Phytopathol 43: 205–227 [DOI] [PubMed] [Google Scholar]
- Heitz T, Widemann E, Lugan R, Miesch L, Ullmann P, Désaubry L, Holder E, Grausem B, Kandel S, Miesch M, et al (2012) Cytochromes P450 CYP94C1 and CYP94B3 catalyze two successive oxidation steps of plant hormone jasmonoyl-isoleucine for catabolic turnover. J Biol Chem 287: 6296–6306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Held MA, Danhof L, Brandizzi F (2013) Identifying protein-protein interactions in Arabidopsis by co-immunoprecipitation of GFP-tagged proteins and proteomic detection. In JA Rothnagel, ed, Fluorescent Proteins. Humana Press, Totawa, NJ (in press) [Google Scholar]
- Howe GA. (2010) Ubiquitin ligase-coupled receptors extend their reach to jasmonate. Plant Physiol 154: 471–474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe GA, Jander G. (2008) Plant immunity to insect herbivores. Annu Rev Plant Biol 59: 41–66 [DOI] [PubMed] [Google Scholar]
- Howe GA, Lightner J, Browse J, Ryan CA. (1996) An octadecanoid pathway mutant (JL5) of tomato is compromised in signaling for defense against insect attack. Plant Cell 8: 2067–2077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagale S, Links MG, Rozwadowski K. (2010) Genome-wide analysis of ethylene-responsive element binding factor-associated amphiphilic repression motif-containing transcriptional regulators in Arabidopsis. Plant Physiol 152: 1109–1134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsir L, Schilmiller AL, Staswick PE, He SY, Howe GA. (2008) COI1 is a critical component of a receptor for jasmonate and the bacterial virulence factor coronatine. Proc Natl Acad Sci USA 105: 7100–7105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazan K, Manners JM. (2013) MYC2: the master in action. Mol Plant (in press) [DOI] [PubMed] [Google Scholar]
- Kessler A, Baldwin IT. (2002) Plant responses to insect herbivory: the emerging molecular analysis. Annu Rev Plant Biol 53: 299–328 [DOI] [PubMed] [Google Scholar]
- Kessler A, Halitschke R, Baldwin IT. (2004) Silencing the jasmonate cascade: induced plant defenses and insect populations. Science 305: 665–668 [DOI] [PubMed] [Google Scholar]
- Kitaoka N, Matsubara T, Sato M, Takahashi K, Wakuta S, Kawaide H, Matsui H, Nabeta K, Matsuura H. (2011) Arabidopsis CYP94B3 encodes jasmonyl-L-isoleucine 12-hydroxylase, a key enzyme in the oxidative catabolism of jasmonate. Plant Cell Physiol 52: 1757–1765 [DOI] [PubMed] [Google Scholar]
- Koo AJ, Cooke TF, Howe GA. (2011) Cytochrome P450 CYP94B3 mediates catabolism and inactivation of the plant hormone jasmonoyl-L-isoleucine. Proc Natl Acad Sci USA 108: 9298–9303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koo AJ, Howe GA. (2009) The wound hormone jasmonate. Phytochemistry 70: 1571–1580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koo AJ, Howe GA. (2012) Catabolism and deactivation of the lipid-derived hormone jasmonoyl-isoleucine. Front Plant Sci 3: 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Zhao Y, McCaig BC, Wingerd BA, Wang J, Whalon ME, Pichersky E, Howe GA. (2004) The tomato homolog of CORONATINE-INSENSITIVE1 is required for the maternal control of seed maturation, jasmonate-signaled defense responses, and glandular trichome development. Plant Cell 16: 126–143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenzo O, Chico JM, Sánchez-Serrano JJ, Solano R. (2004) JASMONATE-INSENSITIVE1 encodes a MYC transcription factor essential to discriminate between different jasmonate-regulated defense responses in Arabidopsis. Plant Cell 16: 1938–1950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConn M, Browse J. (1996) The critical requirement for linolenic acid is pollen development, not photosynthesis, in an Arabidopsis mutant. Plant Cell 8: 403–416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConn M, Creelman RA, Bell E, Mullet JE, Browse J. (1997) Jasmonate is essential for insect defense in Arabidopsis. Proc Natl Acad Sci USA 94: 5473–5477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melotto M, Mecey C, Niu Y, Chung HS, Katsir L, Yao J, Zeng W, Thines B, Staswick P, Browse J, et al (2008) A critical role of two positively charged amino acids in the Jas motif of Arabidopsis JAZ proteins in mediating coronatine- and jasmonoyl isoleucine-dependent interactions with the COI1 F-box protein. Plant J 55: 979–988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melotto M, Underwood W, Koczan J, Nomura K, He SY. (2006) Plant stomata function in innate immunity against bacterial invasion. Cell 126: 969–980 [DOI] [PubMed] [Google Scholar]
- Moreno JE, Tao Y, Chory J, Ballaré CL. (2009) Ecological modulation of plant defense via phytochrome control of jasmonate sensitivity. Proc Natl Acad Sci USA 106: 4935–4940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagpal P, Ellis CM, Weber H, Ploense SE, Barkawi LS, Guilfoyle TJ, Hagen G, Alonso JM, Cohen JD, Farmer EE, et al (2005) Auxin response factors ARF6 and ARF8 promote jasmonic acid production and flower maturation. Development 132: 4107–4118 [DOI] [PubMed] [Google Scholar]
- Nakagawa T, Kurose T, Hino T, Tanaka K, Kawamukai M, Niwa Y, Toyooka K, Matsuoka K, Jinbo T, Kimura T. (2007) Development of series of Gateway binary vectors, pGWBs, for realizing efficient construction of fusion genes for plant transformation. J Biosci Bioeng 104: 34–41 [DOI] [PubMed] [Google Scholar]
- Nakata M, Mitsuda N, Herde M, Koo AJ, Moreno JE, Suzuki K, Howe GA, Ohme-Takagi M. (2013) A bHLH-type transcription factor, JA-ASSOCIATED MYC2-LIKE1, acts as repressor to negatively regulate jasmonate signaling in Arabidopsis thaliana. Plant Cell (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nesvizhskii AI, Keller A, Kolker E, Aebersold R. (2003) A statistical model for identifying proteins by tandem mass spectrometry. Anal Chem 75: 4646–4658 [DOI] [PubMed] [Google Scholar]
- Niu YJ, Figueroa P, Browse J. (2011) Characterization of JAZ-interacting bHLH transcription factors that regulate jasmonate responses in Arabidopsis. J Exp Bot 62: 2143–2154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauwels L, Barbero GF, Geerinck J, Tilleman S, Grunewald W, Pérez AC, Chico JM, Bossche RV, Sewell J, Gil E, et al. (2010) NINJA connects the co-repressor TOPLESS to jasmonate signalling. Nature 464: 788–791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauwels L, Goossens A. (2011) The JAZ proteins: a crucial interface in the jasmonate signaling cascade. Plant Cell 23: 3089–3100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy ASN. (2007) Alternative splicing of pre-messenger RNAs in plants in the genomic era. Annu Rev Plant Biol 58: 267–294 [DOI] [PubMed] [Google Scholar]
- Sehr EM, Agusti J, Lehner R, Farmer EE, Schwarz M, Greb T. (2010) Analysis of secondary growth in the Arabidopsis shoot reveals a positive role of jasmonate signalling in cambium formation. Plant J 63: 811–822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheard LB, Tan X, Mao HB, Withers J, Ben-Nissan G, Hinds TR, Kobayashi Y, Hsu FF, Sharon M, Browse J, et al. (2010) Jasmonate perception by inositol-phosphate-potentiated COI1-JAZ co-receptor. Nature 468: 400–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shyu C, Figueroa P, Depew CL, Cooke TF, Sheard LB, Moreno JE, Katsir L, Zheng N, Browse J, Howe GA. (2012) JAZ8 lacks a canonical degron and has an EAR motif that mediates transcriptional repression of jasmonate responses in Arabidopsis. Plant Cell 24: 536–550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song S, Qi T, Huang H, Ren Q, Wu D, Chang C, Peng W, Liu Y, Peng J, Xie D. (2011) The jasmonate-ZIM domain proteins interact with the R2R3-MYB transcription factors MYB21 and MYB24 to affect jasmonate-regulated stamen development in Arabidopsis. Plant Cell 23: 1000–1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staswick PE, Su W, Howell SH. (1992) Methyl jasmonate inhibition of root growth and induction of a leaf protein are decreased in an Arabidopsis thaliana mutant. Proc Natl Acad Sci USA 89: 6837–6840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szemenyei H, Hannon M, Long JA. (2008) TOPLESS mediates auxin-dependent transcriptional repression during Arabidopsis embryogenesis. Science 319: 1384–1386 [DOI] [PubMed] [Google Scholar]
- Thines B, Katsir L, Melotto M, Niu Y, Mandaokar A, Liu G, Nomura K, He SY, Howe GA, Browse J. (2007) JAZ repressor proteins are targets of the SCF(COI1) complex during jasmonate signalling. Nature 448: 661–665 [DOI] [PubMed] [Google Scholar]
- Vanholme B, Grunewald W, Bateman A, Kohchi T, Gheysen G. (2007) The tify family previously known as ZIM. Trends Plant Sci 12: 239–244 [DOI] [PubMed] [Google Scholar]
- Wager A, Browse J. (2012) Social network: JAZ protein interactions expand our knowledge of jasmonate signaling. Front Plant Sci 3: 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Withers J, Yao J, Mecey C, Howe GA, Melotto M, He SY. (2012) Transcription factor-dependent nuclear localization of a transcriptional repressor in jasmonate hormone signaling. Proc Natl Acad Sci USA 109: 20148–20153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woldemariam MG, Onkokesung N, Baldwin IT, Galis I. (2012) Jasmonoyl-L-isoleucine hydrolase 1 (JIH1) regulates jasmonoyl-L-isoleucine levels and attenuates plant defenses against herbivores. Plant J 72: 758–767 [DOI] [PubMed] [Google Scholar]
- Yan JB, Zhang C, Gu M, Bai ZY, Zhang WG, Qi TC, Cheng ZW, Peng W, Luo HB, Nan FJ, et al (2009) The Arabidopsis CORONATINE INSENSITIVE1 protein is a jasmonate receptor. Plant Cell 21: 2220–2236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Y, Stolz S, Chételat A, Reymond P, Pagni M, Dubugnon L, Farmer EE. (2007) A downstream mediator in the growth repression limb of the jasmonate pathway. Plant Cell 19: 2470–2483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang DL, Yao J, Mei CS, Tong XH, Zeng LJ, Li Q, Xiao LT, Sun TP, Li JG, Deng XW, et al. (2012) Plant hormone jasmonate prioritizes defense over growth by interfering with gibberellin signaling cascade. Proc Natl Acad Sci USA 109: E1192–E1200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida Y, Sano R, Wada T, Takabayashi J, Okada K. (2009) Jasmonic acid control of GLABRA3 links inducible defense and trichome patterning in Arabidopsis. Development 136: 1039–1048 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Turner JG. (2008) Wound-induced endogenous jasmonates stunt plant growth by inhibiting mitosis. PLoS ONE 3: e3699. [DOI] [PMC free article] [PubMed] [Google Scholar]