The bacterial effector AvrRpt2 promotes pathogen virulence via stimulating the turnover of Arabidopsis auxin regulators AXR2 and AXR3.
Abstract
To accomplish successful infection, pathogens deploy complex strategies to interfere with host defense systems and subvert host physiology to favor pathogen survival and multiplication. Modulation of plant auxin physiology and signaling is emerging as a common virulence strategy for phytobacteria to cause diseases. However, the underlying mechanisms remain largely elusive. We have previously shown that the Pseudomonas syringae type III effector AvrRpt2 alters Arabidopsis (Arabidopsis thaliana) auxin physiology. Here, we report that AvrRpt2 promotes auxin response by stimulating the turnover of auxin/indole acetic acid (Aux/IAA) proteins, the key negative regulators in auxin signaling. AvrRpt2 acts additively with auxin to stimulate Aux/IAA turnover, suggesting distinct, yet proteasome-dependent, mechanisms operated by AvrRpt2 and auxin to control Aux/IAA stability. Cysteine protease activity is required for AvrRpt2-stimulated auxin signaling and Aux/IAA degradation. Importantly, transgenic plants expressing the dominant axr2-1 mutation recalcitrant to AvrRpt2-mediated degradation ameliorated the virulence functions of AvrRpt2 but did not alter the avirulent function mediated by the corresponding RPS2 resistance protein. Thus, promoting auxin response via modulating the stability of the key transcription repressors Aux/IAA is a mechanism used by the bacterial type III effector AvrRpt2 to promote pathogenicity.
Being sessile and lacking an adaptive immune system, plants possess a sophisticated innate immune system to cope with pathogen attack (Chisholm et al., 2006; Jones and Dangl, 2006; Boller and He, 2009; Dodds and Rathjen, 2010; Spoel and Dong, 2012). The first line of plant inducible defense is triggered by the recognition of conserved microbial components, namely pathogen- or microbe-associated molecular patterns, via cell surface pattern recognition receptors and elicits pathogen-associated molecular pattern-triggered immunity (PTI) or basal defense (Boller and Felix, 2009; Monaghan and Zipfel, 2012; Schwessinger and Ronald, 2012). The perception of microbe-associated molecular patterns induces a series of immune signaling events. The signaling events activated by well-studied bacterial flagellin and elongation factor Tu include rapid receptor dimerization, phosphorylation, and ubiquitination, which lead to oxidative burst, stomatal closure, induction of defense-related genes, and accumulation of antimicrobial compounds. In addition to PTI, plants have also evolved resistance proteins that directly or indirectly recognize cognate pathogen-encoded effectors and result in cultivar-specific effector-triggered immunity often accompanied by localized programmed cell death or hypersensitive response (HR) and systemic defense signaling (DeYoung and Innes, 2006; Bent and Mackey, 2007; Elmore et al., 2011; Maekawa et al., 2011; Bonardi et al., 2012). In order to successfully colonize plant tissue and cause disease, pathogens need to avoid and/or suppress host immune systems and obtain nutrients before they can multiply to high levels.
Pseudomonas syringae is a gram-negative plant pathogen that causes a wide variety of diseases, including blights, leaf spots, and galls, in different plant species and is a model system in molecular plant pathology (Preston, 2000). Sequencing and genetic studies have identified many key pathogenicity and virulence determinants in P. syringae, including global virulence regulators, the type III secretion system, phytotoxins, and exopolysaccharides (Buell et al., 2003). The type III secretion system mediates the delivery of a plethora of effector proteins into host cells, where they sabotage host immune responses and physiology to favor pathogen survival and multiplication in susceptible plants and trigger defense responses in resistant plants (Mudgett, 2005; Göhre and Robatzek, 2008; Büttner and He, 2009; Block and Alfano, 2011; Feng and Zhou, 2012; Lindeberg et al., 2012). Much progress has been made toward understanding the virulence functions of individual effectors in promoting pathogenicity. In particular, many type III effectors are able to interfere with or suppress host immunity by directly hijacking key components in PTI signaling (Mudgett, 2005; Göhre and Robatzek, 2008; Büttner and He, 2009; Block and Alfano, 2011; Feng and Zhou, 2012; Lindeberg et al., 2012). However, it is much less understood how these effectors modulate host physiology to favor infection.
Plant physiological and cellular processes are deliberately orchestrated by the collective action of plant growth hormones (Santner and Estelle, 2009). In addition to their roles in plant growth and development, the homeostasis of individual hormones and their cross talk influence the outcomes of plant-pathogen interactions (Spoel and Dong, 2008; Grant and Jones, 2009; Kazan and Manners, 2009; Robert-Seilaniantz et al., 2011). It is well established that the phytohormones salicylic acid (SA), jasmonic acid, and ethylene play distinct and overlapping roles in plant defense against biotrophic and necrotrophic pathogens and in the establishment of plant systemic defense (Spoel and Dong, 2008; Grant and Jones, 2009; Pieterse et al., 2009). Recent progress has revealed the cross talk of signaling pathways between classical phytohormones, such as auxin, abscisic acid (ABA), cytokinins, and brassinosteroids, and plant disease resistance or susceptibility (Spoel and Dong, 2008; Grant and Jones, 2009; Kazan and Manners, 2009; Robert-Seilaniantz et al., 2011). Free auxin levels increase in plants infected with P. syringae or Xanthomonas spp., and application of exogenous auxin promotes disease susceptibility (O’Donnell et al., 2003; Chen et al., 2007; Wang et al., 2007). Inappropriate activation of ABA biosynthesis and signaling disrupts plant resistance to several pathogens (de Torres Zabala et al., 2009; Fan et al., 2009). Brassinosteroids unidirectionally suppress plant PTI signaling at multiple levels (Albrecht et al., 2012; Belkhadir et al., 2012). Thus, the perturbation of hormonal homeostasis by pathogens constitutes a general virulence strategy for infection. However, the underlying molecular mechanisms remain largely elusive.
Auxin plays important roles in many aspects of plant growth and development, including cell division, expansion, and differentiation (Benjamins and Scheres, 2008; Santner and Estelle, 2009). The direct binding of auxin to its receptor F-box protein TIR1, a subunit of the ubiquitin ligase complex SCFTIR1, stabilizes the interaction with its substrates, members of the auxin/indole acetic acid (Aux/IAA) family of transcription repressors. This promotes SCFTIR1 ubiquitin complex-catalyzed degradation of Aux/IAA proteins, which relieves Aux/IAA-mediated repression of auxin response factors and results in the transcription of auxin-responsive genes (Benjamins and Scheres, 2008; Mockaitis and Estelle, 2008; Santner and Estelle, 2009). Interestingly, many plant pathogenic microorganisms produce auxin during their interactions with plants (Glickmann et al., 1998; Spaepen et al., 2007). Microbially produced auxin is an important pathogenicity determinant in certain plant-pathogen interactions. For example, auxin stimulates cell growth and gall formation in plants infected with Agrobacterium tumefaciens (Lee et al., 2009). Some P. syringae strains are also capable of producing auxin, although its role in pathogenesis is not understood (Glickmann et al., 1998). A recent study suggested that bacterial flagellin-mediated immunity in Arabidopsis (Arabidopsis thaliana) is associated with the suppression of auxin signaling via microRNA-mediated transcriptional regulation of the auxin receptors TIR1, AFB2, and AFB3 (Navarro et al., 2006). The defense signal molecule SA provokes a profound global repression of auxin-related genes and inhibits auxin responses by stabilizing Aux/IAA proteins (Wang et al., 2007). Thus, auxin is associated with disease susceptibility, and the suppression of auxin signaling appears to constitute a part of plant defense mechanisms.
Consistent with auxin’s role in plant disease resistance, we have found that the P. syringae effector AvrRpt2 augments host auxin biosynthesis and signaling and alters auxin physiology (Chen et al., 2007). Transgenic Arabidopsis plants expressing AvrRpt2 exhibit phenotypes reminiscent of auxin mutants with altered auxin physiology, including elongated primary roots and increased numbers of lateral roots, increased response to exogenously applied auxin, and enhanced accumulation of free indole acetic acid during P. syringae infection (Chen et al., 2007). Transient expression of AvrRpt2 in protoplasts promotes auxin-responsive gene expression (Chen et al., 2007). AvrRpt2 is a Cys protease that is active in plant cells, and several host targets of AvrRpt2 have been identified (Axtell et al., 2003; Chisholm et al., 2005). To understand the underlying mechanism through which AvrRpt2 alters auxin physiology, we tested whether AvrRpt2 could modulate the key components in auxin signaling. Here, we report that AvrRpt2 promotes auxin signaling by stimulating the protein turnover of Aux/IAA negative regulators in a proteasome-dependent manner. Apparently, the Cys protease activity of AvrRpt2 is required to exert this function. The additive effect of AvrRpt2 and auxin on Aux/IAA degradation suggests distinct mechanisms operated by AvrRpt2 and auxin to control Aux/IAA stability. A dominant mutation, axr2-1, which is resistant to auxin-mediated degradation, also blocks AvrRpt2-mediated degradation. Importantly, the transgenic plants expressing the dominant axr2-1 mutant abolished the virulence functions of AvrRpt2 but did not affect the RPS2-mediated avirulent function. Taken together, these data indicate that stimulating Aux/IAA protein turnover serves as a virulence mechanism for AvrRpt2 to promote pathogenicity.
RESULTS
AvrRpt2 Stimulates Aux/IAA Turnover
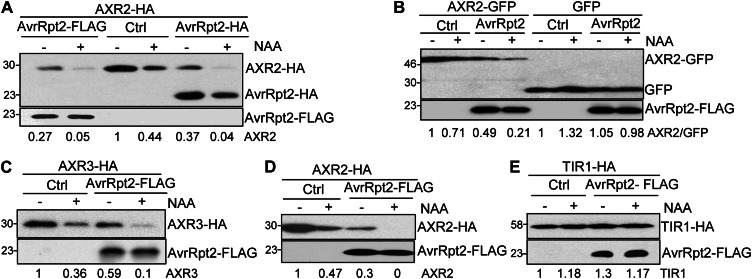
Recent studies demonstrated that auxin binds TIR1 and stabilizes the interaction between TIR1 and its Aux/IAA substrates. This interaction results in the rapid ubiquitination and subsequent degradation of Aux/IAA transcription repressors, thereby activating the expression of a vast array of auxin-responsive genes (Benjamins and Scheres, 2008; Mockaitis and Estelle, 2008; Santner and Estelle, 2009). Apparently, regulated protein degradation is central to most aspects of the auxin response. Thus, we tested whether AvrRpt2 could alter the stability or activity of one or more auxin receptors (e.g. TIR1) or Aux/IAA transcription repressor proteins (e.g. AXR2 and AXR3). We coexpressed epitope-tagged TIR1 or AXR with AvrRpt2 under the control of the cauliflower mosaic virus 35S promoter in Arabidopsis wild-type Columbia (Col-0) protoplasts. Protein expression was detected 6 h after transfection, prior to the occurrence of AvrRpt2-induced cell death in wild-type protoplasts (Gao et al., 2013). As shown in Figure 1A, the protein level of AXR2 was significantly reduced upon treatment with 1 µm 1-naphthalacetic acid (NAA), a synthetic auxin, which is consistent with the previous finding that perception of auxin leads to the degradation of AXR2 (Gray et al., 2001). Interestingly, we repeatedly observed that AvrRpt2 tagged with either hemagglutinin (HA) or FLAG epitope stimulated the degradation of AXR2 (Fig. 1A). Apparently, in the presence of exogenous auxin, AvrRpt2 further accelerated AXR2 protein degradation (Fig. 1A), suggesting an additive effect of AvrRpt2 and auxin on AXR2 protein stability. Similarly, the protein level of AXR2 fused with GFP was diminished, whereas the GFP protein itself remained unaltered upon auxin treatment or in the presence of AvrRpt2 (Fig. 1B), indicating the specificity of the AvrRpt2 effect. Moreover, the stability of AXR3, a close homolog of AXR2, exhibited a similar pattern to AXR2 upon auxin treatment and in the presence of AvrRpt2, suggesting that AvrRpt2 promotes an auxin response by stimulating the degradation of this family of auxin transcription repressor proteins (Fig. 1C). The observed AXR2 and AXR3 protein reduction is unlikely to be caused by the change at the transcript level, since AXR genes were under the control of the constitutively active 35S promoter. Moreover, it has been reported that auxin treatment often stimulates a prolonged transcriptional increase of AXR family genes (Abel et al., 1995).
Figure 1.
AvrRpt2 promotes the turnover of AXR2 and AXR3 but not TIR1. A, Both FLAG and HA epitope-tagged AvrRpt2 stimulate AXR2-HA protein turnover in wild-type Col-0 protoplasts. The control plasmid (Ctrl), AvrRpt2-FLAG, or AvrRpt2-HA was cotransfected with AXR2-HA into wild-type protoplasts. The protoplasts were incubated for 4 h before treatment with1 µm NAA for an additional 2 h, and western-blot analysis was carried out using an α-HA or α-FLAG antibody. The band intensity of AXR2 was quantified by ImageJ software and is labeled on the bottom. B, AvrRpt2 promotes AXR2-GFP but not GFP protein turnover in wild-type protoplasts. C, AvrRpt2-FLAG promotes AXR3-HA protein turnover in wild-type protoplasts. D, AvrRpt2-FLAG promotes AXR2-HA protein turnover in rps2 protoplasts. E, AvrRpt2 does not affect TIR1 protein expression in wild-type protoplasts. All the above experiments were repeated three to five times, and representative results are shown.
While AvrRpt2 plays a role in promoting pathogen virulence in susceptible plants, it is recognized by disease-resistant protein RPS2 in resistant plants to trigger defense responses (Axtell and Staskawicz, 2003; Mackey et al., 2003). To test whether AvrRpt2-induced AXR protein degradation is a result of RPS2-mediated defense responses, we examined AXR2 protein levels in rps2 mutant protoplasts. Similar to what we observed in wild-type protoplasts, AvrRpt2 induced AXR2 protein turnover, which was further enhanced with auxin treatment in rps2 mutant protoplasts (Fig. 1D). These data suggest that AvrRpt2-mediated AXR protein turnover is independent of its cognate RPS2 resistance protein. To elucidate the specificity of AvrRpt2-mediated AXR2/AXR3 degradation, we tested the effect of AvrRpt2 on the stability of the auxin receptor TIR1 in the protoplast transient assay. As reported previously (Gray et al., 2001), the protein level of TIR1 was not affected by treatment with NAA (Fig. 1E). The expression of AvrRpt2 in protoplasts did not cause any detectable changes in the protein level of TIR1 (Fig. 1E). Taken together, these data indicate that AvrRpt2 specifically reduces the protein stability of the negative regulators AXR2 and AXR3 in auxin signaling.
AvrRpt2 Modulates AXR2 Stability during Bacterial Infection
To confirm AvrRpt2-mediated AXR2 protein turnover in plants, we transformed FLAG epitope-tagged AXR2 under the control of the 35S regulatory element into transgenic rps2 mutant plants carrying a dexamethasone (Dex)-inducible AvrRpt2 construct (AXR2-FLAG/Dex::AvrRpt2/rps2). Upon Dex treatment, the expression of AvrRpt2 led to the significant diminution of AXR2 protein in two independent transgenic lines (Fig. 2A). To exclude the possibility of a direct effect of Dex on AXR2 stability, we generated AXR2-FLAG transgenic plants in the wild-type background (AXR2-FLAG/WT). The Dex treatment did not affect AXR2 protein expression in two independent transgenic lines (Supplemental Fig. S1).
Figure 2.
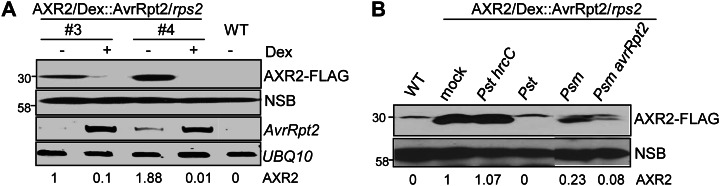
AvrRpt2 promotes AXR2 turnover during bacterial infection. A, Inducible expression of AvrRpt2 in transgenic plants leads to AXR2-FLAG protein turnover. Leaves from two independent lines of 4-week-old 35S::AXR2-FLAG/Dex::AvrRpt2/rps2 transgenic plants were hand inoculated with either 5 µm Dex or water control and harvested 24 h after inoculation. The AXR2-FLAG protein was detected by western blot with an α-FLAG antibody. A nonspecific band (NSB) on the same blot is shown as the protein-loading control. AvrRpt2 expression was detected by RT-PCR analysis, and the UBQ10 gene was used as a control. The band intensity of AXR2 was quantified by ImageJ software and is labeled on the bottom. B, Bacterium-delivered AvrRpt2 stimulates AXR2 protein turnover. Four-week-old 35S::AXR2-FLAG/Dex::AvrRpt2/rps2 transgenic plants were hand inoculated with the indicated bacteria at a concentration of 1 × 108 cfu mL−1 or water as a mock control. Samples were harvested 30 h after inoculation for AXR2-FLAG protein detection with an α-FLAG antibody. The above experiments were repeated three times, and representative results are shown. WT, Wild type.
In a natural infection, it is likely that only minute amounts of type III effector proteins are delivered into host cells by pathogens. To rule out the possibility that AvrRpt2-mediated diminution of AXR2 is attributed to the overexpression of AvrRpt2 protein in protoplasts or transgenic plants, we inoculated AXR2-FLAG transgenic plants with different bacteria and examined the AXR2 protein levels. As shown in Figure 2B, compared with mock treatment, inoculation with virulent P. syringae pv tomato DC3000 (Pst) diminished AXR2 protein level, whereas the corresponding type III secretion mutant Pst hrcC exhibited little effect on AXR2 protein stability. It is well known that a suite of type III effectors from Pst are delivered into host cells to interfere with host responses. Apparently, one or more type III effectors in Pst may possess the ability to regulate AXR2 protein stability. Alternatively, Pst may produce auxin (Glickmann et al., 1998), which could induce AXR2 degradation. The strong diminution of AXR2 protein by Pst prevented us from further investigating the effect of AvrRpt2 delivered from Pst. Interestingly, P. syringae pv maculicola (Psm) triggered a moderate diminution of AXR2 protein compared with mock inoculation (Fig. 2B). Significantly, AXR2 protein level was further reduced with the infection of Psm avrRpt2. These data further support that AvrRpt2 modulates plant auxin physiology and signaling via stimulating Aux/IAA protein turnover during plant-pathogen interaction.
The AvrRpt2-Mediated Auxin Response Depends on Its Cys Protease Activity
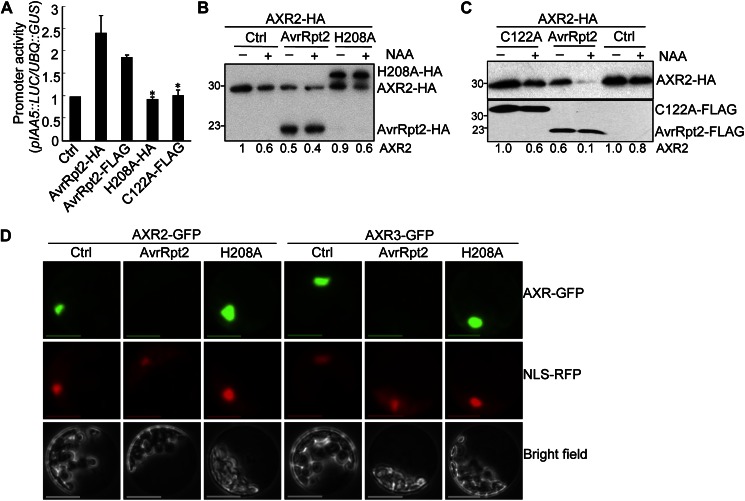
Protein structural prediction and functional analyses indicated that AvrRpt2 is a Cys protease that is autoprocessed inside the plant cell, where it further cleaves several host proteins (Axtell et al., 2003; Chisholm et al., 2005). The conserved residues comprising the catalytic triad of the Cys protease (Cys-122, His-208, and Asp-226) are essential for AvrRpt2 to trigger defense responses mediated by the RPS2 disease resistance gene (Axtell et al., 2003). However, it is not clear whether the Cys protease activity is also required for RPS2-independent AvrRpt2 functions (Lim and Kunkel, 2004a, 2004b). We tested AvrRpt2 mutants, in which the conserved residues comprising the catalytic triad of the Cys protease have been mutated, for the ability to promote auxin response and AXR2 protein stability. IAA5 (for indole-3-acetic acid inducible5) is an early auxin-responsive gene and quickly activated by auxin treatment (Abel et al., 1995). As shown in Figure 3A, the promoter of IAA5 fused with the luciferase reporter (pIAA5::LUC) was activated by AvrRpt2. However, the H208A and C122A mutants of AvrRpt2 lost the ability to activate pIAA5::LUC expression. Similarly, the H208A mutant was unable to activate the expression of another auxin-responsive reporter, pGH3::LUC (Supplemental Fig. S2A). These data indicate that Cys protease activity is required for AvrRpt2 to stimulate the auxin response. Consistent with previous reports (Axtell and Staskawicz, 2003; Chisholm et al., 2005), H208A and C122A mutants accumulated as unprocessed proteins (due to their proteolytic deficiency) to a level comparable to that of wild-type AvrRpt2 (Fig. 3, B and C; Supplemental Fig. S2A). Notably, the AvrRpt2-mediated AXR2 protein diminution was abolished by either the H208A or C122A mutation (Fig. 3, B and C), suggesting the requirement of its Cys protease activity.
Figure 3.
The AvrRpt2-mediated auxin response depends on its Cys protease activity. A, The Cys protease catalytic mutants H208A and C122A of AvrRpt2 are no longer able to activate auxin reporter pIAA5::LUC expression. Protoplasts were cotransfected with a control vector (Ctrl), wild-type AvrRpt2 or its mutants, and the pIAA5::LUC reporter. pUBQ::GUS was included in the transfection as an internal control. The luciferase and GUS activities were detected 6 h after transfection. The promoter activity is shown as the LUC-GUS ratio. Data are shown as means ± sd, and the asterisks indicate significant differences between wild-type AvrRpt2 and its mutants (P < 0.05). B, The H208A mutant blocks AvrRpt2-mediated AXR2 turnover. The control plasmid, AvrRpt2-HA, or AvrRpt2 H208A-HA was cotransfected with AXR2-HA into wild-type protoplasts. The protoplasts were incubated for 4 h before treatment with 1 µm NAA for an additional 2 h. Note that the H208A mutant accumulated as unprocessed protein because of its proteolytic deficiency. The band intensity of AXR2 was quantified by ImageJ software and is labeled on the bottom. C, The C122A mutant blocks AvrRpt2-mediated AXR2 turnover. D, The wild type but not the H208A mutant of AvrRpt2 diminishes the AXR2-GFP and AXR3-GFP fluorescence signals in the nucleus. The control plasmid, AvrRpt2-HA, or AvrRpt2H208A-HA was cotransfected with AXR2-GFP or AXR3-GFP into wild-type protoplasts. An NLS-RFP construct was included in the transfection for nuclear localization control. The samples were collected 10 h after transfection for microscope observation. All the above experiments were repeated three times, and representative results are shown.
We further visualized AXR2-GFP and AXR3-GFP expression with a microscope in the absence or presence of AvrRpt2 or its Cys protease mutant H208A. The clear and strong nuclear fluorescence signal could be easily detected in Arabidopsis cells transfected with AXR2-GFP or AXR3-GFP (Fig. 3D). However, the overall intensity of the green fluorescence signal of AXR2-GFP or AXR3-GFP was significantly reduced, and its nuclear localization was not clearly observed in the cells cotransfected with AvrRpt2. AvrRpt2 did not affect the signal intensity and nuclear localization of cotransfected NLS-RFP, a nucleus-localized red fluorescent protein, in the same cells (Fig. 3D). These results further substantiate our conclusion that AvrRpt2 stimulates the turnover of AXR2 and AXR3. However, the AvrRpt2-induced disappearance of AXR2-GFP or AXR3-GFP signal was no longer observed with the H208A mutant (Fig. 3D). The Cys protease activity-dependent AvrRpt2-induced AXR2-GFP or AXR3-GFP disappearance was also observed in rps2 mutant protoplasts (Supplemental Fig. S2B), reinforcing the RPS2 independence of AvrRpt2-mediated AXR diminution (Fig. 1D). Taken together, these data indicate that promotion of the auxin response and AXR protein turnover by AvrRpt2 likely depends upon its Cys protease activity.
AvrRpt2-Mediated AXR Protein Turnover Is Proteasome Dependent
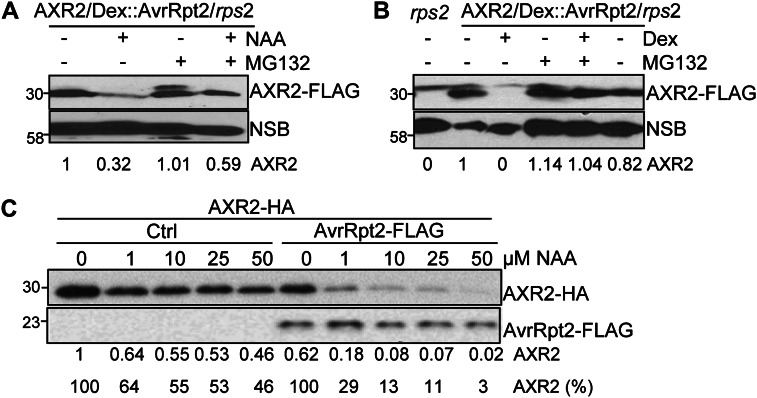
Auxin stimulates Aux/IAA protein degradation via the ubiquitin/proteasome-mediated pathway (Benjamins and Scheres, 2008; Mockaitis and Estelle, 2008; Santner and Estelle, 2009). We addressed the potential mechanism underlying AvrRpt2-mediated AXR2 degradation. Consistent with previous reports (Gray et al., 2001), treatment with MG132, a proteasome inhibitor, blocked NAA-mediated AXR2 protein degradation in AXR2-FLAG/Dex::AvrRpt2/rps2 transgenic plants (Fig. 4A). Interestingly, AvrRpt2-stimulated AXR2 protein turnover was also largely diminished by MG132 treatment (Fig. 4B), suggesting that AvrRpt2 stimulates AXR2 protein degradation via a mechanism that involves the 26S proteasome. Our results raise the possibility that the effect of AvrRpt2 on AXR2 protein stability might be the result of increased auxin level in AvrRpt2-transfected protoplasts or transgenic plants. We have previously detected an increase of auxin level in AvrRpt2 transgenic plants (Chen et al., 2007). To test this hypothesis, we treated AXR2-HA-transfected cells with gradually increased concentrations of NAA and examined AXR2 protein levels. Auxin-mediated AXR2 protein degradation was induced at the concentration of 1 µm NAA, and further increase of NAA concentration up to 50 µm did not dramatically enhance AXR2 protein degradation (Fig. 4C). In contrast, the presence of AvrRpt2 in combination with NAA treatment even at the concentration of 1 µm resulted in a pronounced reduction of AXR2 protein, consistent with the additive effect of AvrRpt2 and auxin in the control of AXR protein stability (Fig. 4C). This result argues against the possibility that AvrRpt2-mediated AXR2 protein turnover is due simply to elevated auxin levels stimulated by AvrRpt2. Apparently, AvrRpt2 and auxin stimulate AXR protein turnover via distinct, yet proteasome-dependent, protein degradation pathways.
Figure 4.
AvrRpt2-mediated AXR2 turnover is proteasome dependent. A, MG132 suppresses auxin-mediated AXR2 turnover. Four-week-old 35S::AXR2-FLAG/Dex-AvrRpt2/rps2 transgenic plants were hand inoculated with 4 µm NAA with or without 5 µm MG132, and the samples were collected 24 h after inoculation for western blot with an α-FLAG antibody. A nonspecific band (NSB) on the same blot is shown as the protein-loading control. B, MG132 suppresses AvrRpt2-mediated AXR2 turnover. Four-week-old 35S::AXR2-FLAG/Dex-AvrRpt2/rps2 transgenic plants were hand inoculated with 5 µm Dex with or without 5 µm MG132, and samples were collected 24 h after inoculation for western blot. C, AvrRpt2 possesses an additive effect with auxin to stimulate AXR2 turnover. Wild-type protoplasts were transfected with a control plasmid or AvrRpt2-FLAG together with AXR2-HA. The protoplasts were incubated for 4 h before treatment with different concentrations of NAA for an additional 2 h. The band intensity of AXR2 was quantified by ImageJ software and is labeled underneath the gel. The numbers in the top row represent the relative value of AXR2 protein level when the sample without NAA treatment or AvrRpt2 transfection was set as 1. The numbers in the bottom row show the relative percentage of AXR2 protein compared with samples without NAA treatment in the absence (control [Ctrl]) or presence of AvrRpt2. All the above experiments were repeated three times, and representative results are shown.
The axr2-1 Mutant Is Resistant to AvrRpt2-Mediated Degradation
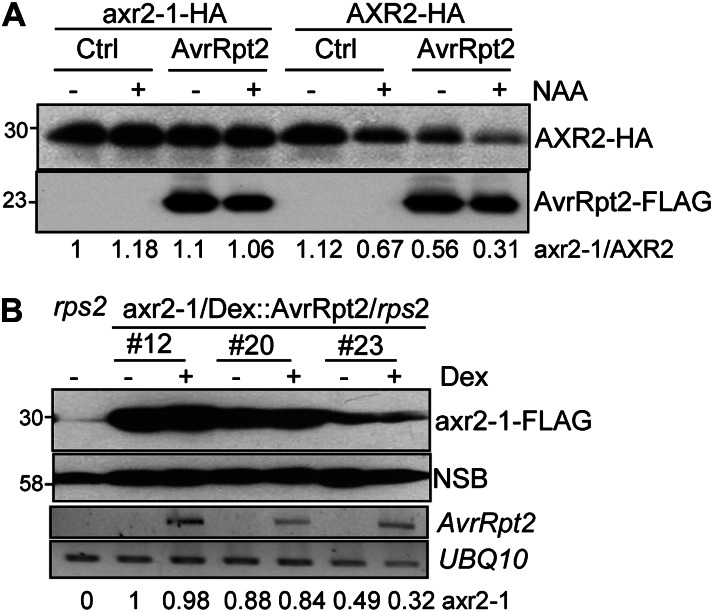
The dominant axr2-1 mutant, with an amino acid substitution in the domain II motif, is known to be resistant to SCFTIR1-dependent degradation upon auxin perception (Gray et al., 2001). We generated the equivalent P87S mutation in AXR2 (AXR2P87S) corresponding to the axr2-1 mutant in our transgenic constructs (Gray et al., 2001). In contrast to wild-type AXR2 protein, axr2-1 mutant protein was no longer degraded upon NAA treatment when transiently expressed in protoplasts (Fig. 5A). This recapitulates the previous report that the axr2-1 mutant is resistant to auxin-mediated protein degradation (Gray et al., 2001). Importantly, the protein level of axr2-1 remained the same in the presence or absence of AvrRpt2, indicating that the axr2-1 mutant was also resistant to AvrRpt2-mediated protein degradation (Fig. 5A). We also generated transgenic plants carrying FLAG epitope-tagged axr2-1 under the control of the 35S promoter in the Dex::AvrRpt2/rps2 background. The phenotype of the axr2-1 transgenic plants with strong protein expression resembled that of dominant axr2-1 genetic mutants (Gray et al., 2001; Supplemental Fig. S3). Consistent with the protoplast transient assay, the axr2-1 protein level was not affected by the expression of AvrRpt2 upon Dex treatment (Fig. 5B).
Figure 5.
axr2-1 is resistant to AvrRpt2-mediated degradation. A, The axr2-1 (AXR2P87S) mutant is insensitive to auxin- and AvrRpt2-mediated degradation in protoplasts. Ctrl, Control. B, The axr2-1 protein is insensitive to AvrRpt2-mediated degradation in transgenic plants. Leaves from three independent lines of 4-week-old axr2-1-FLAG/Dex::AvrRpt2/rps2 transgenic plants were hand inoculated with either 5 µm Dex or water control and harvested 24 h after inoculation. The axr2-1 proteins were detected by western blot with an α-FLAG antibody. A nonspecific band (NSB) on the same blot is shown as the protein-loading control. The expression of AvrRpt2 after Dex treatment is shown by RT-PCR, with UBQ10 as a control. The band intensity of axr2-1 was quantified by ImageJ software and is labeled on the bottom. The above experiments were repeated three times, and representative results are shown.
AvrRpt2-Mediated AXR2 Turnover Is Associated with Its Virulence Function
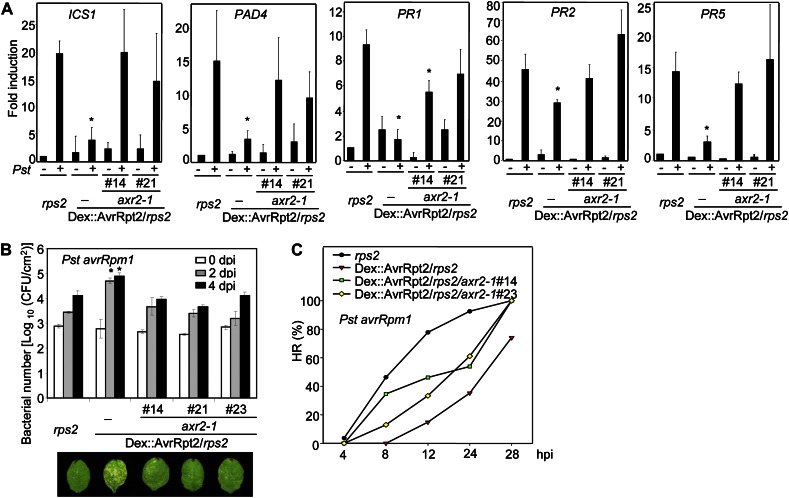
Our results indicate that AvrRpt2 augments the auxin response by promoting Aux/IAA protein turnover and that this is independent of its cognate resistance protein RPS2. Conversely, suppression of auxin signaling is associated with bacterial flagellin-induced and SA-mediated defense (Navarro et al., 2006; Wang et al., 2007). We asked whether stimulating Aux/IAA protein turnover and auxin signaling could be a mechanism underlying the virulence activity for the AvrRpt2 effector. To test this hypothesis, we assayed AvrRpt2 virulence function in dominant axr2-1 transgenic plants in which some aspects of the AvrRpt2-mediated promotion of the auxin response and Aux/IAA protein degradation were abrogated. The axr2-1/Dex::AvrRpt2/rps2 transgenic plants with moderate axr2-1 protein expression were selected for the assays, since these plants did not have obvious growth defects (Supplemental Fig. S3). Pst infection induced the expression of several defense-related genes, such as ICS1, PAD4, PR1, PR2, and PR5, which are involved in SA-mediated defense (Chen et al., 2004; Lim and Kunkel, 2004b). The expression of AvrRpt2 in planta suppressed the induction of these genes by Pst (Chen et al., 2004; Lim and Kunkel, 2004b). The suppression function of AvrRpt2 was largely ameliorated in the axr2-1/Dex::AvrRpt2/rps2 transgenic plants (Fig. 6A).
Figure 6.
The axr2-1 mutant suppresses AvrRpt2 virulence. A, The AvrRpt2-mediated suppression of Pst-induced defense gene expression is abolished in axr2-1 transgenic plants. Leaves from 4-week-old two independent lines (#14 and #21) of axr2-1-FLAG/Dex::AvrRpt2/rps2, Dex::AvrRpt2/rps2, or rps2 plants were syringe inoculated with Pst at a concentration of 1 × 108 cfu mL−1. Samples were harvested at 6 hpi for ICS1, PAD4, and PR1 and at 3 dpi for PR2 and PR5 gene expression with quantitative RT-PCR analysis. UBQ10 was used as an internal control. The data are shown as means ± se from three independent biological replicates. Asterisks indicate significant differences at P < 0.05 when compared with data from rps2 plants with Pst inoculation. B, AvrRpt2-mediated suppression of AvrRpm1 disease resistance is blocked in axr2-1 transgenic plants. Leaves from 4-week-old three independent lines of axr2-1-FLAG/Dex::AvrRpt2/rps2, Dex::AvrRpt2/rps2, or rps2 plants were hand inoculated with Pst avrRpm1 at a concentration of 5 × 105 cfu mL−1. Bacterial growth was measured 0, 2, and 4 dpi. The data are shown as means ± se of three repeats, and asterisks indicate significant differences at P < 0.05 when compared with data from rps2 plants. Disease symptoms at 5 dpi are shown at the bottom. C, AvrRpt2-mediated suppression of AvrRpm1 HR is blocked in axr2-1 transgenic plants. Leaves from 4-week-old two independent lines of axr2-1-FLAG/Dex::AvrRpt2/rps2, Dex::AvrRpt2/rps2, or rps2 plants were hand inoculated with Pst avrRpm1 at a concentration of 1 × 108 cfu mL−1. The HR was observed at the indicated time points and calculated as the percentage of leaves exhibiting the wilting phenotype of total inoculated leaves (n > 25). All the above experiments were repeated three times, and representative results are shown. [See online article for color version of this figure.]
It has been shown that AvrRpt2 inhibits AvrRpm1-mediated disease resistance responses, including restriction of bacterial growth and induction of HR (Ritter and Dangl, 1996; Chen et al., 2000). As shown in Figure 6B, AvrRpm1-mediated restriction of bacterial growth and disease symptom development were substantially reduced upon AvrRpt2 expression in Dex::AvrRpt2/rps2 plants. The bacterial growth of Pst avrRpm1 increased about 10-fold at 2 and 4 d post infection (dpi) in Dex::AvrRpt2/rps2 plants compared with that in rps2 plants. However, the axr2-1 transgenic plants largely ameliorated AvrRpt2’s ability to interfere with AvrRpm1-mediated disease resistance. The disease symptom development and in planta bacterial multiplication in axr2-1/Dex::AvrRpt2/rps2 plants exhibited a similar pattern to those in rps2 plants (Fig. 6B). The infection of Pst avrRpm1 induces a rapid and robust HR, which can be observed by the massive tissue collapse of the infected region 5 to 12 h post inoculation (hpi). The ratio of HR was quantified as the percentage of leaves exhibiting the wilting phenotype of total inoculated leaves (Axtell et al., 2003). The expression of AvrRpt2 substantially delayed the onset of AvrRpm1-mediated tissue collapse. However, this suppression function of AvrRpt2 was largely abolished in transgenic plants with expression of axr2-1 (Fig. 6C).
Stimulation of AXR2 Turnover Is Not Associated with the AvrRpt2 Avirulence Function
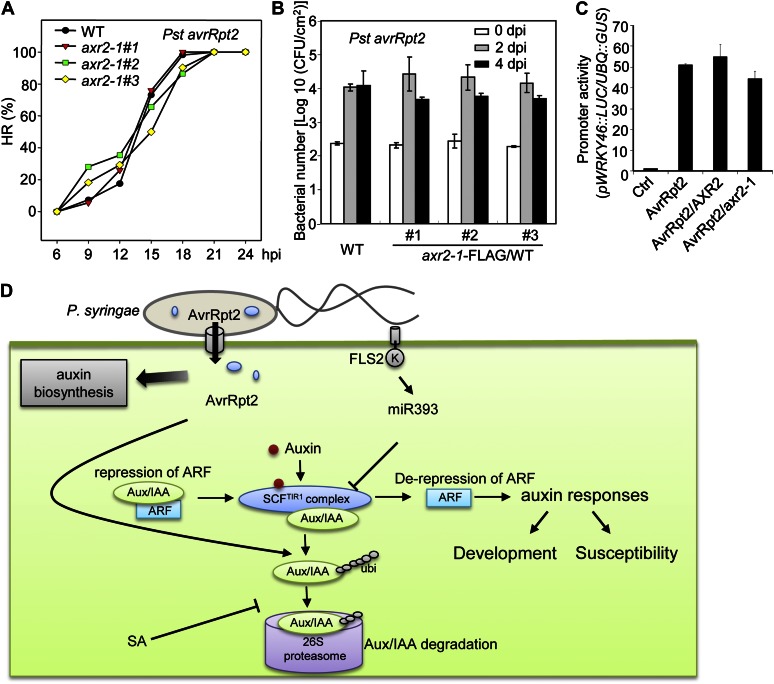
In the presence of the corresponding disease resistance RPS2 gene, AvrRpt2 triggers resistance responses in the host, including restriction of in planta bacterial multiplication and activation of a localized HR to prevent the further spread of pathogens (Axtell and Staskawicz, 2003; Mackey et al., 2003). To investigate whether AvrRpt2-mediated Aux/IAA protein degradation is required for its avirulence function, we generated axr2-1 transgenic plants in the wild-type (RPS2) background. The inoculation of Pst avrRpt2 at a relatively high inoculum concentration elicited an HR in wild-type Arabidopsis plants, with 80% to 100% of inoculated leaves showing tissue collapse at about 15 to 21 hpi (Fig. 7A). The progression of AvrRpt2-mediated HR exhibited a similar pattern in three independent lines of axr2-1 transgenic plants and wild-type Col-0 plants (Fig. 7A). Similarly, the expression of axr2-1 did not change AvrRpt2-mediated disease resistance in wild-type plants. The in planta bacterial multiplication of Pst avrRpt2 was similar in wild-type and axr2-1 transgenic plants (Fig. 7B). Elicitation of AvrRpt2-RPS2 signaling is often accompanied by the transcriptional activation of an array of defense-associated genes, including WRKY46 (Gao et al., 2013). Consistently, expression of either AXR2 or axr2-1 in protoplasts did not alter AvrRpt2-mediated pWRKY46::LUC reporter activity. Together, these data suggest that AvrRpt2-mediated AXR2 protein degradation is not required for AvrRpt2-RPS2 disease resistance.
Figure 7.
The axr2-1 mutant does not affect AvrRpt2 avirulence. A, The axr2-1 mutant does not affect AvrRpt2-mediated HR. Leaves from 4-week-old three independent lines of axr2-1-FLAG/WT or wild-type (WT) plants were hand inoculated with Pst avrRpt2 at a concentration of 1 × 108 cfu mL−1. The HR was observed at the indicated time points and calculated as the percentage of leaves exhibiting the wilting phenotype of total inoculated leaves (n > 25). B, The axr2-1 mutant does not affect AvrRpt2-mediated disease resistance. Leaves from 4-week-old three independent lines of axr2-1-FLAG/WT or wild-type plants were hand inoculated with Pst avrRpt2 at a concentration of 5 × 105 cfu mL−1. Bacterial growth was measured 0, 2, and 4 dpi. The data are shown as means ± se of three repeats. C, The axr2-1 mutant does not affect AvrRpt2-mediated WRKY46 activation. Protoplasts were cotransfected with the control plasmid, AvrRpt2, AXR2, or the axr2-1 mutant together with the pWRKY46::LUC reporter. pUBQ::GUS was included in the transfection as an internal control. The luciferase and GUS activities were detected 6 h after transfection. The promoter activity is shown as the LUC-GUS ratio with means ± sd. D, A model of the AvrRpt2 virulence mechanism via stimulating Arabidopsis Aux/IAA protein turnover. All the above experiments were repeated three times, and representative results are shown. [See online article for color version of this figure.]
DISCUSSION
As one of the earliest identified phytohormones, auxin plays pivotal roles in almost every aspect of plant growth and development (Benjamins and Scheres, 2008; Mockaitis and Estelle, 2008; Santner and Estelle, 2009). Not surprisingly, auxin has been implicated in intimate association with plant resistance and disease susceptibility, although the molecular mechanisms involved still remain largely unknown (Kazan and Manners, 2009; Robert-Seilaniantz et al., 2011). Similar to the plant defense hormones SA and jasmonic acid, auxin differentially regulates the outcomes of plant-pathogen interactions depending on the life styles of the pathogens (Kazan and Manners, 2009; Robert-Seilaniantz et al., 2011). During infection by Pst, the free indole acetic acid level increased and application of auxin promoted disease symptom development (O’Donnell et al., 2003; Chen et al., 2007; Wang et al., 2007). Consistently, the type III effector AvrRpt2 modulates host auxin physiology and signaling as a potential virulence strategy (Chen et al., 2007). Here, we demonstrated that AvrRpt2 promotes the auxin response by stimulating turnover of the negative regulator AXR2 and AXR3 proteins (Fig. 7D). The diminution of these Aux/IAA proteins was observed when AvrRpt2 was expressed in protoplasts, transgenic plants, or delivered by bacteria. AvrRpt2-mediated auxin response and AXR2 degradation depend on the Cys protease activity of AvrRpt2, suggesting that this activity is crucial for AvrRpt2 virulence function. Similar to auxin-stimulated degradation of the Aux/IAA proteins, AvrRpt2-mediated AXR2 degradation is proteasome dependent (Fig. 7D). The virulence, but not avirulence, function of AvrRpt2 was significantly blocked in Arabidopsis plants carrying the dominant axr2-1 mutation, which renders AXR2 resistant to auxin- and AvrRpt2-mediated degradation, suggesting that degradation of Aux/IAA proteins is a key virulence mechanism of AvrRpt2. Interestingly, additional P. syringae effectors also appear to possess the ability to modulate Aux/IAA stability, implicating this as a general virulence mechanism used by pathogenic bacteria (Fig. 2B).
Although the mechanisms are not well understood, it has long been observed that AvrRpt2 promotes pathogen virulence on host plants lacking a functional RPS2 gene (Chen et al., 2000). RIN4, which is cleaved by AvrRpt2 to elicit RPS2 resistance signaling, is predicted to be a virulent target of AvrRpt2 (Axtell and Staskawicz, 2003; Mackey et al., 2003). However, AvrRpt2 promotes P. syringae virulence independent of RIN4, suggesting that AvrRpt2 has virulence mechanisms and targets other than RIN4 (Lim and Kunkel, 2004b). In addition, AvrRpt2 virulence activity likely acts independent or downstream of the defense hormone SA (Chen et al., 2004). This accumulating evidence suggests that AvrRpt2 may promote pathogenesis by modulating some other aspect of host cell physiology or signaling. Here, we provide a mechanism of AvrRpt2 virulence by promoting the degradation of key negative regulators in auxin signaling, thereby increasing auxin response. In addition to AvrRpt2, the Xanthomonas campestris pv vesicatoria effector AvrBs3 induced the expression of several auxin-responsive genes in susceptible pepper (Capsicum annuum) plants (Marois et al., 2002). The ABA signaling pathway is also a target for P. syringae type III effectors (de Torres-Zabala et al., 2007). In addition to type III effectors, other virulence factors, such as the phytotoxin coronatine, could also modulate auxin or other hormone levels or signaling (Thilmony et al., 2006). Thus, the modulation of host hormone physiology is a common virulence strategy among plant pathogens.
Aux/IAA proteins bind and inactivate auxin response factors, many of which are positive regulators of auxin-responsive genes. Auxin acts as a molecular glue to stabilize the interaction between its receptor F-box protein TIR1 and Aux/IAA proteins and to promote Aux/IAA degradation through the ubiquitin-proteasome-dependent pathway (Benjamins and Scheres, 2008; Mockaitis and Estelle, 2008; Santner and Estelle, 2009). It is likely that AvrRpt2-stimulated AXR2/AXR3 degradation is also mediated through the SCFTIR1 ubiquitin complex-proteasome pathway, since MG132 inhibits AXR2/AXR3 degradation by AvrRpt2 and the axr2-1 mutant protein is resistant to AvrRpt2-mediated degradation. However, AvrRpt2-mediated AXR2/AXR3 degradation is not simply due to the elevated auxin level in plants expressing AvrRpt2. Exogenous application of high concentrations of auxin could not achieve the pronounced diminution of Aux/IAA proteins stimulated by the additive effects of AvrRpt2 and auxin, suggesting that AvrRpt2 likely possesses a distinct mechanism to regulate Aux/IAA protein stability (Fig. 4C). It is also possible that AvrRpt2 stabilizes TIR1 and Aux/IAA protein interaction to promote Aux/IAA degradation. Strikingly, Aux/IAA proteins appear to be common targets for pathogen virulence strategies. The plant defense hormone SA stabilizes Aux/IAA to repress auxin signaling as a part of SA-mediated defense mechanisms (Wang et al., 2007). Tobacco mosaic virus infection of Arabidopsis plants results in transcriptional reprogramming of a large number of auxin-responsive genes, which is achieved by the interaction between the tobacco mosaic virus replicase protein and the Arabidopsis Aux/IAA26 protein, thereby preventing the localization of Aux/IAA to the nucleus (Padmanabhan et al., 2008). A spiroketal-macrolide, yokonolide B from Streptomyces diastatochromogenes B59, inhibits the expression of auxin-responsive genes by blocking Aux/IAA protein degradation upstream of AXR and TIR1 (Hayashi et al., 2003).
AvrRpt2 encodes a Cys protease that cleaves itself and several host proteins, and this activity is essential for AvrRpt2 to trigger defense responses mediated by RPS2 (Axtell et al., 2003; Chisholm et al., 2005). Our results indicate that the Cys protease activity is also required for the AvrRpt2-mediated auxin response, suggesting that full AvrRpt2 virulence activity depends on its Cys protease activity. Interestingly, Cys protease activity is also required for AvrRpt2-mediated AXR2 degradation. We tested whether AvrRpt2 was able to directly cleave AXR2 in vitro. Although AvrRpt2 could cleave RIN4 in the presence of cyclophilin ROC1 (Coaker et al., 2005), we failed to detect the cleavage of AXR2 by AvrRpt2 in several attempts. AXR2 and AXR3 are mainly localized in the nucleus (Gray et al., 2001; Fig. 3D). Notably, AvrRpt2 has been observed in the nucleus in addition to localizing at the plasma membrane (Jin et al., 2003). Nevertheless, we cannot rule out the possibility that AvrRpt2 directly degrades/cleaves AXR2/AXR3 to promote the auxin response. Alternatively, AvrRpt2 regulates Aux/IAA protein stability indirectly and cleaves other host protein(s), which are required to stabilize Aux/IAA proteins or inhibit SCFTIR1 complex-mediated Aux/IAA degradation. The identification of additional AvrRpt2 host proteins mediating the auxin response may provide insights into auxin signaling mechanisms in plants.
MATERIALS AND METHODS
Plant Growth and Pathogen Assays
Arabidopsis (Arabidopsis thaliana) wild-type plants (accession Col-0) were grown in pots containing soil (Metro Mix 366) in a growth room at 23°C, 60% relative humidity, and 75 μE m−2 s−1 light with a 12-h photoperiod for approximately 4 weeks before protoplast isolation or bacterial inoculation. Different Pseudomonas syringae strains (P. syringae pv tomato DC3000 [Pst], Pst hrcC, Pst avrRpt2, P. syringae pv maculicola [Psm], and Psm avrRpt2) were grown overnight at 28°C in King’s B medium with appropriate antibiotics. Bacteria were pelleted by 4,000 rpm centrifugation, washed, and diluted to the desired density with water. Arabidopsis leaves were infiltrated with bacteria using a needleless syringe. To measure bacterial growth, leaves were inoculated with P. syringae at a concentration of 5 × 105 colony-forming units (cfu) mL−1, two leaf discs were ground in 100 μL of water, and serial dilutions were plated on King’s B medium with appropriate antibiotics. Bacterial cfu were counted 2 d after incubation at 28°C. Each data point is shown as an average of three biological replicates. At least three independent repeats were performed for all experiments, and representative data are shown. For HR assays, the leaves of 4-week-old plants were syringe infiltrated with bacteria at a concentration of 1 × 108 cfu mL−1, and the tissue collapse was monitored at the indicated time points. The severity of the HR was calculated as the percentage of inoculated leaves showing tissue collapse.
Generation of AXR2 and AXR2P87S Transgenic Plants
Dex-inducible AvrRpt2 (in the rps2-101C mutant background) transgenic plants were obtained from Dr. Fred Ausubel. AXR2-FLAG and axr2-1-FLAG transgenic plants were generated by Agrobacterium tumefaciens-mediated transformation with a binary vector (pCB302) carrying the AXR2 or AXR2P87S mutant gene under the control of the cauliflower mosaic virus 35S promoter with a C-terminal FLAG epitope tag. To screen transgenic plants, the primary transformants were sprayed with Basta and the surviving plants were subjected to western-blot analysis for protein expression. To induce AvrRpt2 expression, the transgenic plants were hand inoculated with 5 μm Dex.
Plasmid Constructs and Point Mutations
The AvrRpt2 in planta expression vector construct, pIAA5::LUC, and pGH3::LUC and pWRKY46::LUC reporter constructs were reported previously (Chen et al., 2007; Gao et al., 2013). AXR2, AXR3, or TIR1 was amplified from complementary DNA of wild-type Col-0 with the following primers (the restriction enzyme sites are underlined, and the start codon is italicized) to clone into the protoplast expression vectors: AXR2-F-BamH1, 5′-CGGGATCCATGATCGGCCAACTTATGAA-3′; AXR2-R-Stu1, 5′-GAAGGCCTAGATCTGTTCTTGCAGTACT-3′; AXR3-F-BamH1, 5′-CGGGATCCATGATGGGCAGTGTCGAG-3′; AXR3-R-Stu1, 5′-GAAGGCCTAGCTCTGCTCTTGCACT-3′; TIR1-BamH1-F, 5′-CGGGATCCATGCAGAAGCGAATAGCCTT-3′; TIR1-Stu1-R, 5′-GAAGGCCTTAATCCGTTAGTAGTAATGA-3′.
Point mutations were generated using the site-specific mutagenesis kit (Stratagene Quick Change site-directed mutagenesis kit) using the following primers for AXR2P87S, AvrRpt2 H208A, and AvrRpt2 C122A mutants (the mutated sites are underlined): AXR2P87S-F, 5′-GTGGTGGGATGG-TCACCTGTGAGGAAC-3′; AXR2P87S-R, 5′-GTTCCTCACAGGTGACCATCCC-ACCAC-3′; AvrRpt2H208A-F, 5′-CCGAATGACAGCTGGGCCATGTCGGT-CCT-3′; AvrRpt2H208A-R, 5′-CCAGTGAGGACCGACATGGCCCAGCT-GTC-3′; AvrRpt2C122A-F, 5′-TGAGCGAATGGGAGCTTGGTATGCCTG-CGC-3′; AvrRpt2C122A-R, 5′-GCGCAGGCATACCAAGCTCCCATTCGC-TCA-3′.
Arabidopsis Protoplast Transient Assays and Western-Blot Analysis
Protoplast transient expression and western-blot analysis were carried out as described previously (Chen et al., 2007). Briefly, 0.2-mL protoplasts at a density of 2 × 105 mL−1 were transfected with 40 μg of total DNA including AvrRpt2 and AXR for western-blot analysis. The ratio of AvrRpt2 and AXR DNA was 1:1. The protoplasts were incubated at room temperature for 4 h before treatment with 1 µm NAA for an additional 2 h. The samples were collected and added to 4× SDS loading buffer for western-blot analysis. For reporter assays, 0.1-mL protoplasts were transfected with a luciferase reporter and AvrRpt2. UBQ10-GUS was cotransfected as an internal control, and the promoter activity was presented as the LUC-GUS ratio. Protoplasts were collected at 6 h after transfection for luciferase and GUS activity assays.
Real-Time Reverse Transcription-PCR Analysis
Total RNA was isolated from leaves inoculated with water or Pst and treated with TRIzol Reagent (Invitrogen). Complementary DNA was synthesized from 1 μg of total RNA with 0.1 μg of oligo(dT) primer and reverse transcriptase (New England Biolabs). Real-time reverse transcription (RT)-PCR analysis was carried out using iTaq SYBR Green Supermix (Bio-Rad) supplemented with ROX in the ABI GeneAmp PCR System 9700. The expression of defense-related genes was normalized to the expression of UBQ10. The primer sequences of different genes are as follows: UBQ10-F, 5′-AGATCCAGGACAAGGAAGGTATTC-3′; UBQ10-R, 5′-CGCAGGACCAAGTGAAGAGTAG-3′; ICS1-F, 5′-CCCTTAACAAGGTTGTTCTTGC-3′; ICS1-R, 5′-CCTTCACGCTGTAACTGTGC-3′; PAD4-F, 5′-TCTTCAGTTAAAGATCAAGGAAGG-3′; PAD4-R, 5′-GGTTGAATGGCCGGTTATC-3′; PR1-F, 5′-CGTTCACATAATTCCCACGAG-3′; PR1-R, 5′-TCAGTGAGACTCGGATGTGC-3′; PR2-F, 5′-GCATTCGCTGGATGTTTTG-3′; PR2-R, 5′-CTTCAACCACACAGCTGGAC-3′; PR5-F, 5′-GTTCATCACAAGCGGCATT-3′; PR5-R, 5′-GTCAATTCAAATCCTCCATCG-3′.
Microscope Observation
GFP fusions of AXR2 or AXR3 were cotransfected with a vector control or avrRpt2. Protein localization was observed 10 h post transfection with a fluorescence microscope (Nikon Eclipse Ti). The nucleus was labeled by cotransfected NLS-RFP.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Dex treatment did not affect AXR2 protein expression in wild-type plants.
Supplemental Figure S2. The conserved catalytic residue His-208 of the Cys protease is essential for AvrRpt2-mediated auxin response and AXR2/AXR3-GFP degradation.
Supplemental Figure S3. Growth phenotype of 35S::axr2-1-FLAG/Dex::AvrRpt2/rps2 transgenic plants.
Acknowledgments
We thank Dr. F. Ausubel for the Dex-inducible AvrRpt2 transgenic plants, Dr. H. Koiwa for the NLS-RFP construct, and Dr. P. de Figueiredo for his generosity in sharing his microscope.
Glossary
- PTI
pathogen-associated molecular pattern-triggered immunity
- HR
hypersensitive response
- SA
salicylic acid
- ABA
abscisic acid
- Aux/IAA
auxin/indole acetic acid
- Col-0
Columbia
- NAA
1-naphthalacetic acid
- HA
hemagglutinin
- Dex
dexamethasone
- Pst
Pseudomonas syringae pv tomato DC3000
- Psm
Pseudomonas syringae pv maculicola
- dpi
d post infection
- hpi
h post inoculation
- cfu
colony-forming units
- RT
reverse transcription
References
- Abel S, Nguyen MD, Theologis A. (1995) The PS-IAA4/5-like family of early auxin-inducible mRNAs in Arabidopsis thaliana. J Mol Biol 251: 533–549 [DOI] [PubMed] [Google Scholar]
- Albrecht C, Boutrot F, Segonzac C, Schwessinger B, Gimenez-Ibanez S, Chinchilla D, Rathjen JP, de Vries SC, Zipfel C. (2012) Brassinosteroids inhibit pathogen-associated molecular pattern-triggered immune signaling independent of the receptor kinase BAK1. Proc Natl Acad Sci USA 109: 303–308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axtell MJ, Chisholm ST, Dahlbeck D, Staskawicz BJ. (2003) Genetic and molecular evidence that the Pseudomonas syringae type III effector protein AvrRpt2 is a cysteine protease. Mol Microbiol 49: 1537–1546 [DOI] [PubMed] [Google Scholar]
- Axtell MJ, Staskawicz BJ. (2003) Initiation of RPS2-specified disease resistance in Arabidopsis is coupled to the AvrRpt2-directed elimination of RIN4. Cell 112: 369–377 [DOI] [PubMed] [Google Scholar]
- Belkhadir Y, Jaillais Y, Epple P, Balsemão-Pires E, Dangl JL, Chory J. (2012) Brassinosteroids modulate the efficiency of plant immune responses to microbe-associated molecular patterns. Proc Natl Acad Sci USA 109: 297–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamins R, Scheres B. (2008) Auxin: the looping star in plant development. Annu Rev Plant Biol 59: 443–465 [DOI] [PubMed] [Google Scholar]
- Bent AF, Mackey D. (2007) Elicitors, effectors, and R genes: the new paradigm and a lifetime supply of questions. Annu Rev Phytopathol 45: 399–436 [DOI] [PubMed] [Google Scholar]
- Block A, Alfano JR. (2011) Plant targets for Pseudomonas syringae type III effectors: virulence targets or guarded decoys? Curr Opin Microbiol 14: 39–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boller T, Felix G. (2009) A renaissance of elicitors: perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu Rev Plant Biol 60: 379–406 [DOI] [PubMed] [Google Scholar]
- Boller T, He SY. (2009) Innate immunity in plants: an arms race between pattern recognition receptors in plants and effectors in microbial pathogens. Science 324: 742–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonardi V, Cherkis K, Nishimura MT, Dangl JL. (2012) A new eye on NLR proteins: focused on clarity or diffused by complexity? Curr Opin Immunol 24: 41–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buell CR, Joardar V, Lindeberg M, Selengut J, Paulsen IT, Gwinn ML, Dodson RJ, Deboy RT, Durkin AS, Kolonay JF, et al. (2003) The complete genome sequence of the Arabidopsis and tomato pathogen Pseudomonas syringae pv. tomato DC3000. Proc Natl Acad Sci USA 100: 10181–10186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Büttner D, He SY. (2009) Type III protein secretion in plant pathogenic bacteria. Plant Physiol 150: 1656–1664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Agnew JL, Cohen JD, He P, Shan L, Sheen J, Kunkel BN. (2007) Pseudomonas syringae type III effector AvrRpt2 alters Arabidopsis thaliana auxin physiology. Proc Natl Acad Sci USA 104: 20131–20136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Kloek AP, Boch J, Katagiri F, Kunkel BN. (2000) The Pseudomonas syringae avrRpt2 gene product promotes pathogen virulence from inside plant cells. Mol Plant Microbe Interact 13: 1312–1321 [DOI] [PubMed] [Google Scholar]
- Chen Z, Kloek AP, Cuzick A, Moeder W, Tang D, Innes RW, Klessig DF, McDowell JM, Kunkel BN. (2004) The Pseudomonas syringae type III effector AvrRpt2 functions downstream or independently of SA to promote virulence on Arabidopsis thaliana. Plant J 37: 494–504 [DOI] [PubMed] [Google Scholar]
- Chisholm ST, Coaker G, Day B, Staskawicz BJ. (2006) Host-microbe interactions: shaping the evolution of the plant immune response. Cell 124: 803–814 [DOI] [PubMed] [Google Scholar]
- Chisholm ST, Dahlbeck D, Krishnamurthy N, Day B, Sjolander K, Staskawicz BJ. (2005) Molecular characterization of proteolytic cleavage sites of the Pseudomonas syringae effector AvrRpt2. Proc Natl Acad Sci USA 102: 2087–2092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coaker G, Falick A, Staskawicz B. (2005) Activation of a phytopathogenic bacterial effector protein by a eukaryotic cyclophilin. Science 308: 548–550 [DOI] [PubMed] [Google Scholar]
- de Torres Zabala M, Bennett MH, Truman WH, Grant MR. (2009) Antagonism between salicylic and abscisic acid reflects early host-pathogen conflict and moulds plant defence responses. Plant J 59: 375–386 [DOI] [PubMed] [Google Scholar]
- de Torres-Zabala M, Truman W, Bennett MH, Lafforgue G, Mansfield JW, Rodriguez Egea P, Bögre L, Grant M. (2007) Pseudomonas syringae pv. tomato hijacks the Arabidopsis abscisic acid signalling pathway to cause disease. EMBO J 26: 1434–1443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeYoung BJ, Innes RW. (2006) Plant NBS-LRR proteins in pathogen sensing and host defense. Nat Immunol 7: 1243–1249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodds PN, Rathjen JP. (2010) Plant immunity: towards an integrated view of plant-pathogen interactions. Nat Rev Genet 11: 539–548 [DOI] [PubMed] [Google Scholar]
- Elmore JM, Lin ZJ, Coaker G. (2011) Plant NB-LRR signaling: upstreams and downstreams. Curr Opin Plant Biol 14: 365–371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan J, Hill L, Crooks C, Doerner P, Lamb C. (2009) Abscisic acid has a key role in modulating diverse plant-pathogen interactions. Plant Physiol 150: 1750–1761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng F, Zhou JM. (2012) Plant-bacterial pathogen interactions mediated by type III effectors. Curr Opin Plant Biol 15: 469–476 [DOI] [PubMed] [Google Scholar]
- Gao X, Chen X, Lin W, Chen S, Lu D, Niu Y, Li L, Cheng C, McCormack M, Sheen J, et al (2013) Bifurcation of Arabidopsis NLR immune signaling via Ca2+-dependent protein kinases. PLoS Pathog 9: e1003127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glickmann E, Gardan L, Jacquet S, Hussain S, Elasri M, Petit A, Dessaux Y. (1998) Auxin production is a common feature of most pathovars of Pseudomonas syringae. Mol Plant Microbe Interact 11: 156–162 [DOI] [PubMed] [Google Scholar]
- Göhre V, Robatzek S. (2008) Breaking the barriers: microbial effector molecules subvert plant immunity. Annu Rev Phytopathol 46: 189–215 [DOI] [PubMed] [Google Scholar]
- Grant MR, Jones JD. (2009) Hormone (dis)harmony moulds plant health and disease. Science 324: 750–752 [DOI] [PubMed] [Google Scholar]
- Gray WM, Kepinski S, Rouse D, Leyser O, Estelle M. (2001) Auxin regulates SCF(TIR1)-dependent degradation of AUX/IAA proteins. Nature 414: 271–276 [DOI] [PubMed] [Google Scholar]
- Hayashi K, Jones AM, Ogino K, Yamazoe A, Oono Y, Inoguchi M, Kondo H, Nozaki H. (2003) Yokonolide B, a novel inhibitor of auxin action, blocks degradation of AUX/IAA factors. J Biol Chem 278: 23797–23806 [DOI] [PubMed] [Google Scholar]
- Jin P, Wood MD, Wu Y, Xie Z, Katagiri F. (2003) Cleavage of the Pseudomonas syringae type III effector AvrRpt2 requires a host factor(s) common among eukaryotes and is important for AvrRpt2 localization in the host cell. Plant Physiol 133: 1072–1082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones JD, Dangl JL. (2006) The plant immune system. Nature 444: 323–329 [DOI] [PubMed] [Google Scholar]
- Kazan K, Manners JM. (2009) Linking development to defense: auxin in plant-pathogen interactions. Trends Plant Sci 14: 373–382 [DOI] [PubMed] [Google Scholar]
- Lee CW, Efetova M, Engelmann JC, Kramell R, Wasternack C, Ludwig-Müller J, Hedrich R, Deeken R. (2009) Agrobacterium tumefaciens promotes tumor induction by modulating pathogen defense in Arabidopsis thaliana. Plant Cell 21: 2948–2962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim MT, Kunkel BN. (2004a) Mutations in the Pseudomonas syringae avrRpt2 gene that dissociate its virulence and avirulence activities lead to decreased efficiency in AvrRpt2-induced disappearance of RIN4. Mol Plant Microbe Interact 17: 313–321 [DOI] [PubMed] [Google Scholar]
- Lim MT, Kunkel BN. (2004b) The Pseudomonas syringae type III effector AvrRpt2 promotes virulence independently of RIN4, a predicted virulence target in Arabidopsis thaliana. Plant J 40: 790–798 [DOI] [PubMed] [Google Scholar]
- Lindeberg M, Cunnac S, Collmer A. (2012) Pseudomonas syringae type III effector repertoires: last words in endless arguments. Trends Microbiol 20: 199–208 [DOI] [PubMed] [Google Scholar]
- Mackey D, Belkhadir Y, Alonso JM, Ecker JR, Dangl JL. (2003) Arabidopsis RIN4 is a target of the type III virulence effector AvrRpt2 and modulates RPS2-mediated resistance. Cell 112: 379–389 [DOI] [PubMed] [Google Scholar]
- Maekawa T, Kufer TA, Schulze-Lefert P. (2011) NLR functions in plant and animal immune systems: so far and yet so close. Nat Immunol 12: 817–826 [DOI] [PubMed] [Google Scholar]
- Marois E, Van den Ackerveken G, Bonas U. (2002) The Xanthomonas type III effector protein AvrBs3 modulates plant gene expression and induces cell hypertrophy in the susceptible host. Mol Plant Microbe Interact 15: 637–646 [DOI] [PubMed] [Google Scholar]
- Mockaitis K, Estelle M. (2008) Auxin receptors and plant development: a new signaling paradigm. Annu Rev Cell Dev Biol 24: 55–80 [DOI] [PubMed] [Google Scholar]
- Monaghan J, Zipfel C. (2012) Plant pattern recognition receptor complexes at the plasma membrane. Curr Opin Plant Biol 15: 349–357 [DOI] [PubMed] [Google Scholar]
- Mudgett MB. (2005) New insights to the function of phytopathogenic bacterial type III effectors in plants. Annu Rev Plant Biol 56: 509–531 [DOI] [PubMed] [Google Scholar]
- Navarro L, Dunoyer P, Jay F, Arnold B, Dharmasiri N, Estelle M, Voinnet O, Jones JD. (2006) A plant miRNA contributes to antibacterial resistance by repressing auxin signaling. Science 312: 436–439 [DOI] [PubMed] [Google Scholar]
- O’Donnell PJ, Schmelz EA, Moussatche P, Lund ST, Jones JB, Klee HJ. (2003) Susceptible to intolerance: a range of hormonal actions in a susceptible Arabidopsis pathogen response. Plant J 33: 245–257 [DOI] [PubMed] [Google Scholar]
- Padmanabhan MS, Kramer SR, Wang X, Culver JN. (2008) Tobacco mosaic virus replicase-auxin/indole acetic acid protein interactions: reprogramming the auxin response pathway to enhance virus infection. J Virol 82: 2477–2485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pieterse CM, Leon-Reyes A, Van der Ent S, Van Wees SC. (2009) Networking by small-molecule hormones in plant immunity. Nat Chem Biol 5: 308–316 [DOI] [PubMed] [Google Scholar]
- Preston GM. (2000) Pseudomonas syringae pv. tomato: the right pathogen, of the right plant, at the right time. Mol Plant Pathol 1: 263–275 [DOI] [PubMed] [Google Scholar]
- Ritter C, Dangl JL. (1996) Interference between two specific pathogen recognition events mediated by distinct plant disease resistance genes. Plant Cell 8: 251–257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert-Seilaniantz A, Grant M, Jones JD. (2011) Hormone crosstalk in plant disease and defense: more than just jasmonate-salicylate antagonism. Annu Rev Phytopathol 49: 317–343 [DOI] [PubMed] [Google Scholar]
- Santner A, Estelle M. (2009) Recent advances and emerging trends in plant hormone signalling. Nature 459: 1071–1078 [DOI] [PubMed] [Google Scholar]
- Schwessinger B, Ronald PC. (2012) Plant innate immunity: perception of conserved microbial signatures. Annu Rev Plant Biol 63: 451–482 [DOI] [PubMed] [Google Scholar]
- Spaepen S, Vanderleyden J, Remans R. (2007) Indole-3-acetic acid in microbial and microorganism-plant signaling. FEMS Microbiol Rev 31: 425–448 [DOI] [PubMed] [Google Scholar]
- Spoel SH, Dong X. (2008) Making sense of hormone crosstalk during plant immune responses. Cell Host Microbe 3: 348–351 [DOI] [PubMed] [Google Scholar]
- Spoel SH, Dong X. (2012) How do plants achieve immunity? Defence without specialized immune cells. Nat Rev Immunol 12: 89–100 [DOI] [PubMed] [Google Scholar]
- Thilmony R, Underwood W, He SY. (2006) Genome-wide transcriptional analysis of the Arabidopsis thaliana interaction with the plant pathogen Pseudomonas syringae pv. tomato DC3000 and the human pathogen Escherichia coli O157:H7. Plant J 46: 34–53 [DOI] [PubMed] [Google Scholar]
- Wang D, Pajerowska-Mukhtar K, Culler AH, Dong X. (2007) Salicylic acid inhibits pathogen growth in plants through repression of the auxin signaling pathway. Curr Biol 17: 1784–1790 [DOI] [PubMed] [Google Scholar]