High-throughput sequencing of Arabidopsis pollen cDNA uncovers novel transcription and alternative splicing.
Abstract
Pollen grains of Arabidopsis (Arabidopsis thaliana) contain two haploid sperm cells enclosed in a haploid vegetative cell. Upon germination, the vegetative cell extrudes a pollen tube that carries the sperm to an ovule for fertilization. Knowing the identity, relative abundance, and splicing patterns of pollen transcripts will improve our understanding of pollen and allow investigation of tissue-specific splicing in plants. Most Arabidopsis pollen transcriptome studies have used the ATH1 microarray, which does not assay splice variants and lacks specific probe sets for many genes. To investigate the pollen transcriptome, we performed high-throughput sequencing (RNA-Seq) of Arabidopsis pollen and seedlings for comparison. Gene expression was more diverse in seedling, and genes involved in cell wall biogenesis were highly expressed in pollen. RNA-Seq detected at least 4,172 protein-coding genes expressed in pollen, including 289 assayed only by nonspecific probe sets. Additional exons and previously unannotated 5′ and 3′ untranslated regions for pollen-expressed genes were revealed. We detected regions in the genome not previously annotated as expressed; 14 were tested and 12 were confirmed by polymerase chain reaction. Gapped read alignments revealed 1,908 high-confidence new splicing events supported by 10 or more spliced read alignments. Alternative splicing patterns in pollen and seedling were highly correlated. For most alternatively spliced genes, the ratio of variants in pollen and seedling was similar, except for some encoding proteins involved in RNA splicing. This study highlights the robustness of splicing patterns in plants and the importance of ongoing annotation and visualization of RNA-Seq data using interactive tools such as Integrated Genome Browser.
Deep sequencing of complementary DNA (cDNA; RNA-Seq) allows researchers to measure RNA abundance on a genome-wide scale and to detect previously unannotated genes and splice variants (for review, see Wang et al., 2009). Prior to RNA-Seq, genome-scale assays of RNA abundance typically employed hybridization of labeled samples onto DNA microarrays. In Arabidopsis (Arabidopsis thaliana), the ATH1 array from Affymetrix has been used most extensively. As of this writing, more than 8,000 individual hybridizations of ATH1 arrays are available through resources such as the Gene Expression Omnibus, NASCArrays, and ArrayExpress. As a relatively mature technology, DNA microarrays have several advantages, including well-established sample preparation and data analysis protocols, rapid turn-around times, and a wealth of archived data and data-mining methodologies. However, expression microarrays are limited, as they require fabrication and thus depend on prior knowledge of genes and gene sequences.
RNA-Seq experiments, because they provide counts of molecules, offer a number of potential advantages for downstream data analysis and interpretation. Following normalization for transcript size and sequencing depth, it is possible to rank genes according to their overall expression levels within the same sample (Mortazavi et al., 2008). By contrast, with microarrays, robust statistical and biophysical methods do not yet exist for deducing the relative abundance of different transcripts in the same sample. This is because the hybridization properties of DNA microarray probes vary from gene to gene, making it difficult to quantify targets based on hybridization intensities alone. Currently, the only comparison that can be made between genes within the same sample is to examine the overlap between genes called as “present” or “absent,” which are qualitative expression measurements from the MAS5 algorithm developed by Affymetrix. This can be done because Affymetrix microarray probe sets include both perfect match and mismatch probes for each intended target. In theory, the mismatch probes detect nonspecific hybridization, while the perfect match probes detect both nonspecific hybridization and bona fide hybridization with the probe set’s intended target. If a statistically significant difference is observed between the perfect match and mismatch probe readings, then the MAS5 algorithm “calls” the probe set target as present. If no such difference is observed, then the statistical test either did not have enough power to detect a real difference (false negative) or the test correctly determined that the target was not present (true negative). Many studies have taken advantage of this aspect of Affymetrix arrays to identify genes that are detectably expressed in pollen or other sample types. However, nothing can be said about the relative “presentness” of two targets based on these readings, because the intensity values are not directly comparable between different probe sets. By contrast, the digital, counts-based nature of RNA-Seq data provides a novel opportunity to quantify RNA abundances and identify RNAs that are the most (or least) abundant in a given tissue or cell type. As a result, we can use RNA-Seq data to revisit ideas about the relative abundance of transcripts encoding signal transduction molecules, transcription factors, structural components of cells, and so on.
Analysis of alternative splicing is another area where RNA-Seq can uncover new information. The most recent (June 2009) Arabidopsis Columbia-0 (Col-0) genome annotation (The Arabidopsis Information Resource 10 [TAIR10]) includes nearly 5,000 protein-coding loci that are annotated as alternatively spliced, a form of posttranscriptional and sometimes cotranscriptional regulation in which the primary product of transcription undergoes diverse splicing patterns, depending on time of day and temperature (Staiger et al., 2003; Sanchez et al., 2010; Seo et al., 2012), stress treatments (Palusa et al., 2007; Gulledge et al., 2012), or tissue type (Li et al., 2010). Differentially spliced regions are often small (English et al., 2010); therefore, it is difficult to design expression microarray probes that can distinguish splicing variants and still have good hybridization properties. RNA-Seq experiments, by contrast, do not require designing probes; with sufficient depth of sequencing and adequate read lengths, it should be possible to obtain enough sequences covering differentially spliced regions to detect when splicing patterns differ across developmental stages, organ types, or treatment regimes. For the same reason, RNA-Seq might uncover new genes and new splice forms when cell types or developmental stages that are not already well represented in EST databases are surveyed.
Pollen grains of higher plants are one such example where RNA-Seq might reveal a wealth of new information about transcription. Pollen grains of higher plants are haploid male gametophytes. A pollen grain contains two cell types: two sperm cells and a vegetative cell that produces the pollen tube that carries the sperm cells to an ovule, where they fertilize the egg and central cell. Several studies that used expression microarrays to survey gene expression in Arabidopsis pollen have been published, including studies of gene expression during pollen development (Honys and Twell, 2003), in mature pollen (Becker et al., 2003; Pina et al., 2005), and during pollen tube growth and maturation (Wang et al., 2008; Qin et al., 2009; Boavida et al., 2011). However, for the reasons noted above, it was likely that some transcripts in pollen remained undiscovered and that relative transcript abundance in pollen was unknown. Therefore, we carried out an RNA-Seq experiment with Arabidopsis pollen and compared pollen RNA-Seq data with two seedling RNA-Seq data sets.
We identified at least 500 genes in the TAIR10 gene annotations that are not represented on the ATH1 microarray but are expressed in pollen. Nearly 1,800 protein-coding genes on the ATH1 array are represented by promiscuous probe sets that cross hybridize to multiple, related targets; we identified at least 239 genes for these probe sets that were expressed in pollen. An analysis of splicing patterns in both pollen and seedling revealed more than 2,000 previously unannotated but well-supported novel splicing events (at least 10 spliced reads per event) corresponding to new introns. Fewer than 20 genes annotated as alternatively spliced in TAIR10 were differentially spliced between pollen and seedling. We also identified pollen-expressed genes with additional exons and genes with previously unannotated 5′ and 3′ untranslated regions (UTRs). Thus, this RNA-Seq data set provides resources that can guide comprehensive assessment of annotation accuracy, alternative splicing, and gene expression in Arabidopsis pollen.
RESULTS
Gene Expression in Pollen
To generate RNA-Seq data sets, we prepared one cDNA library from pollen and two libraries from seedling RNA and sequenced them on two separate Illumina flow cells, generating between 38 and 52 million reads of 75 bases each, per library. The reads were aligned onto the reference Arabidopsis genome assembly (TAIR10) and sorted into two groups: reads designated single-mapping reads that mapped exactly once onto the genome and reads designated multimapping reads that mapped multiple times and therefore could not be unambiguously assigned to a single location. Table I shows the yields and alignment percentages for each library in each category. Because RNA was isolated from a pooled pollen sample, the data should be considered from an “averaged sample” and not from traditional biological replicates, similar to the averaged sample used for a pollen proteomics study (Holmes-Davis et al., 2005).
Table I. Sequencing yields and alignments overview.
Each library was sequenced in two lanes on an Illumina GAIIx sequencer. The number of reads obtained from the three libraries totaled 110.4 million.
| Library | Reads | Aligneda | Uniquely Alignedb |
|---|---|---|---|
| millions | % | ||
| Pollen lane 1 | 26.9 | 85.7 | 81.9 |
| Pollen lane 2 | 25.2 | ||
| Seedling 1, lane 1 | 24.7 | 63.6 | 61.0 |
| Seedling 1, lane 2 | 22.7 | ||
| Seedling 2, lane 1 | 17.8 | 80.9 | 78.8 |
| Seedling 2, lane 2 | 20.4 | ||
Percentage of reads aligned to one or more locations in the genome. bPercentage of reads aligned to only one location in the genome.
We used Integrated Genome Browser (IGB; Nicol et al., 2009) to visualize the read alignments and compare patterns of splicing and gene expression between seedling and pollen samples. For readers to view their own genes of interest in the browser, data sets are available via an IGB QuickLoad data repository site at http://www.igbquickload.org/pollen. Available data sets include read alignments, coverage graphs, and junction features representing splicing events. To view the data, readers can download and launch IGB from bioviz.org and open the RNA-Seq folder listed under the IGBQuickLoad.org data source. Instructions for visualization and analysis of read alignments are available at bioviz.org.
For each gene, we counted the number of single-mapping overlapping reads and used these counts to generate raw count-based and normalized expression values. Normalized expression values calculated for each gene included reads per million (RPM) and reads per kilobase million (RPKM), equal to RPM divided by the largest transcript size per gene in kilobases. Thus, one read per million corresponds to at least 30 reads in total, since each library yielded at least 30 million single-mapping read sequences. All three data sets (read counts, RPM, and RPKM) are useful depending on the analysis: RPKM is particularly useful for within-sample comparisons, while RPM and read counts are useful for assessing overall expression levels between samples. Supplemental Tables containing read counts (Supplemental Table S1), RPM (Supplemental Table S2), and RPKM (Supplemental Table S3) are available for annotated genes in TAIR10, including protein-coding genes, transposable element genes, and noncoding genes where the primary gene product is RNA. Reads that overlapped genes annotated as pseudogenes in TAIR10 were also counted.
To assess expression in the seedling and pollen data sets, we used read counts normalized for sequencing depth (RPM; Supplemental Table S2), requiring at least 5 RPM to designate a gene as expressed, equivalent to around 150 overlapping reads. Note that this threshold is based on the biological intuition that observing at least this many reads is likely due to transcription of the gene rather than due to alignment mistakes or other artifacts, such as contamination from nonpollen material in the case of the pollen library. To evaluate this threshold, we examined pollen read counts and normalized RPM expression values for 24 genes encoding light-harvesting complex proteins (Umate, 2010). Although pollen grains contain plastids, these genes are unlikely to be highly expressed in pollen, so observation of high expression from these genes might indicate contamination from vegetative material that escaped exclusion during vacuum collection of pollen, wherein pollen is collected on a 6-μm mesh (Johnson-Brousseau and McCormick, 2004). For 21 of the 24 light-harvesting complex genes, we observed pollen expression less than 0.5 RPM (i.e. fewer than 20 reads per gene). Three genes (At1g45474, At1g76570, and At5g54270) had higher expression values (2.8, 1.3, and 1.5 RPM), so we examined these in IGB. We found reads overlapping each of the three genes, but in two cases (At1g45474 and At4g54270), the unusually high read counts in pollen were because the 3′ UTR of these two genes overlapped genes on the opposite strand that were highly expressed in pollen. For At1g76570, the pollen reads overlapped with some, but not all, of the exons of the annotated gene. Nonetheless, in all three cases, the normalized read counts were smaller than 5 RPM, suggesting that this is a useful, if highly conservative, threshold for determining whether a gene is expressed in a sample. However, it is important to note that, depending on the gene or the question being asked, other thresholds may be more appropriate. These examples also illustrate that it is important for researchers to examine RNA-Seq reads in a browser such as IGB before drawing conclusions. We note that most genes with RPM > 1 also appear to be bona fide pollen-expressed genes, because the RNA-Seq reads, although fewer, convincingly align to the annotated exon/intron gene models in TAIR10.
The TAIR10 genome annotation includes 27,416 protein-coding genes: using 5 RPM as the threshold, 4,172 (15%) of the protein-coding genes were expressed in pollen. Nearly three times as many protein-coding genes were expressed in seedling, where 14,040 (51%) protein-coding genes had RPM of at least 5. The greater diversity of genes expressed in the seedling data sets, despite the equally strict requirement of 5 RPM per sample type and comparable sequencing depth, is likely due to the more diverse cell types present in seedling than in pollen. Of the protein-coding genes that were expressed in pollen at 5 RPM or higher, 228 (5.5%) had no detectable expression (no overlapping reads) in the seedling data sets.
Comparison of the Pollen RNA-Seq Data Set with Present/Absent Calls from Microarray-Based Expression Analyses
To determine which genes are expressed in a sample, Affymetrix microarray platforms have used a qualitative expression measurement known as a present/absent/marginal call, which relies on the difference between perfect and mismatch probes in a probe set to determine whether the designated target mRNA for the microarray is present in the sample. The equivalent procedure in RNA-Seq analysis involves observing a nonzero or threshold number of aligned reads in a sample library. As discussed previously, identifying the best threshold for determining whether a gene is expressed depends on the experiment, the expected purity of the sample, and the question being addressed. Therefore, we compared RNA-Seq RPM expression values with qualitative present/absent calls from ATH1-based microarray studies of mature, dry pollen (Wang et al., 2008; Qin et al., 2009) that most closely matched the RNA-Seq experiment in terms of sample collection. We obtained the original mature pollen CEL files from each microarray study (GSE6696 and GSE17343), reprocessed the data, and regenerated the present/absent calls. Since 2008, new versions of the Arabidopsis gene annotations and ATH1 probe set-to-gene annotations have been released. Therefore, to ensure comparability with our RNA-Seq data set, we used the most recent mappings provided by The Arabidopsis Information Resource (TAIR) for the probe set to locus identifiers.
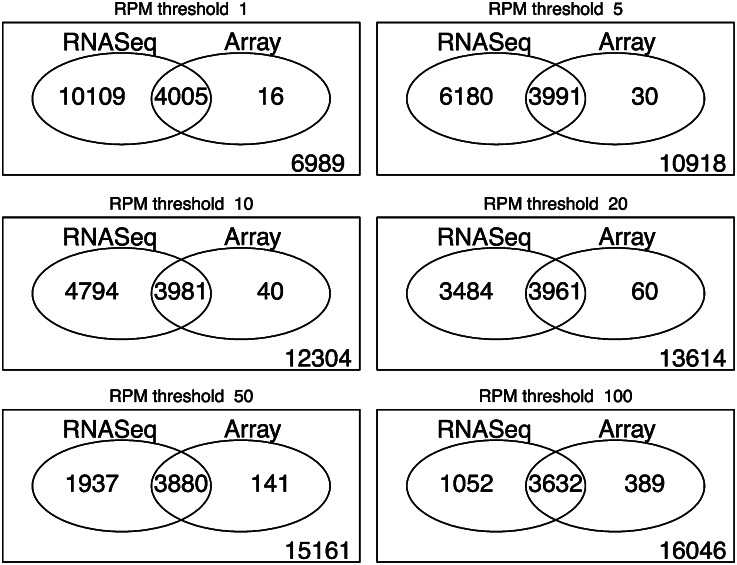
There was a high concordance between read counts in the pollen RNA-Seq and present calls in the microarray data. According to the TAIR probe set annotations, the ATH1 array has 21,119 locus-specific probe sets that interrogate one locus. In the microarray studies, 4,021 of these locus-specific probe sets were called present in at least five of six pollen microarrays. The concordance between the array results and the RNA-Seq results was striking in that nearly all of the genes called present (3,991 of 4,021) had RPM of 5 or larger in the RNA-Seq data set. However, as described above, an RNA-Seq expression level of 5 RPM is equivalent to around 150 overlapping reads per gene and may be overly conservative for some applications. Therefore, we also examined how lower and higher RPM expression thresholds affected the correspondence between RNA-Seq and microarray present calls. As the Venn diagrams in Figure 1 show, the most liberal RPM threshold we tested (1 RPM) had the highest overlap with the present calls from the microarray data. Using this threshold, there were only 16 genes called as present by the microarrays that had no single-mapping overlapping reads in the RNA-Seq data. Increasing the RPM threshold reduced the microarray/RNA-Seq overlap. When the threshold was increased to 100 RPM, the overlap dropped from 4,005 to 3,632, an 11% change. By contrast, the change in the nonintersecting set of genes that were called expressed by RNA-Seq but absent by microarray dropped from 10,109 to 1,052, a more than 10-fold change. Thus, in general, genes called as present by microarray tend to be the most abundantly expressed (higher read counts) when examined in RNA-Seq, illustrating the higher sensitivity of RNA-Seq in terms of qualitative expression values as well as the robustness of the ATH1 microarray present/absent calls.
Figure 1.
Concordance between RNA-Seq and microarray present calls. Each diagram reports the size of different subsets of single-target (no cross hybridization) ATH1 probe set/target pairs. The array subset contains probe sets that were called as present in at least five of six mature, dry pollen ATH1 microarrays from Gene Expression Omnibus expression series GSE6696 and GSE17343, where the sample collection protocol was the same as that used for the RNA-Seq library. The RNA-Seq subset contains probe set target genes with pollen RPM expression values as high or higher than the indicated threshold. The intersections of RNA-Seq and array subsets are genes/probe sets called present by both methods. Values reported in the lower right corner of each diagram are probe set/targets called absent by both methods.
Genes Highly Expressed in Pollen
As discussed in the introduction, the digital, counts-based nature of RNA-Seq data allows ranking genes according to their expression levels, following normalization for transcript size (Mortazavi et al., 2008). Because many studies have already been published that highlight differential gene expression between pollen and other sample types, we focused our analysis on abundances of transcripts within the pollen sample relative to each other in order to identify the most highly expressed genes in pollen. It has been shown that de novo transcription is not strictly required to initiate pollen tube growth (Honys and Twell, 2004), suggesting that the pollen grain is well stocked with RNAs that encode proteins required for early stages of pollen germination and pollen tube growth. To identify the most abundantly expressed genes in pollen, we ranked the genes according to their size-normalized RPKM expression measurements (Supplemental Table S3) and focused on the top 2% most highly expressed genes. This “top-pollen” list included 672 genes with normalized expression values (RPKM) greater than 146, equivalent to 4,000 or more overlapping reads for most of these genes. The functional annotations associated with this top-pollen list were highly enriched for functions related to cell wall modifications, including synthesis, modification, or degradation of polysaccharides. These included COBL10 (At3g20580), two cellulose synthase-like proteins (CSLD1 and At4g07960), expansin A4 (At2g39700), and 16 and 21 genes annotated as putative pectin lyases and pectin methyl esterase inhibitors, respectively. Genes encoding other cell wall-related functions (e.g. glycosyl hydrolases) were also common, as were genes annotated with cell wall degradation functions, such as At1g02790, encoding polygalacturonase 4, and At5g19580, encoding a putative glyoxal oxidase. Several genes encoding SKS family (for SKU5-similar) proteins, a 19-member family of cell wall-associated proteins, were also high on the top-pollen list. The founding member of the SKU family, SKU5 (At4g12420), is required for root growth and is an extracellular protein that attaches to the outer leaflet of the plasma membrane via a glycosylphosphatidylinositol anchor (Sedbrook et al., 2002). SKU5 is only weakly expressed in pollen but abundantly expressed in seedling. Four other members of the SKS family, At3g13400 (SKS13), At1g55560 (SKS14), At3g13390 (SKS11), and At1g55570 (SKS12), are highly expressed in pollen (RPKM > 2,000) and weakly expressed in seedling (RPKM < 1). These genes were previously noted as being expressed in mature pollen (Wang et al., 2008), but until now their high abundance relative to other genes was unknown. The large stockpile of cell wall-related transcripts in the pollen grain helps explain why pollen is capable of germination and tube production even when transcription is inhibited (Honys and Twell, 2003; Wang et al., 2008).
Microarray studies of pollen tube growth have shown that transcripts for genes involved in cellular transport and signal transduction increase in abundance during pollen tube growth (Wang et al., 2008; Qin et al., 2009; Boavida et al., 2011). These observations not only underline the role of the secretory pathway and cell-to-cell signaling in pollen tube growth and guidance; they also highlight a more fundamental fact of pollen physiology, namely, that new transcription takes place during pollen tube growth. Thus, although transcription is not strictly necessary during early stages of pollen tube growth, at some point following rehydration of the pollen grain, synthesis of new RNAs begins. However, the identity of all transcriptional regulators responsible for the activation of RNA synthesis and differential gene expression following pollen germination is unknown. To identify candidate transcription factors that might play a role in these processes, we examined highly expressed genes annotated with the Gene Ontology (GO) term “transcription factor activity” (GO:0003700) in the pollen RNA-Seq data set. There were 16 highly expressed transcription factors (RPKM ≥ 90), including five with average seedling RPKM < 1. These five were a bZIP family transcription factor (At1g35490), two zinc finger (C2H2 type) family proteins (At4g35700 and At4g35610), a myb domain protein101 (At2g32460), and a No Apical Meristem domain transcriptional regulator superfamily protein (At1g60240). Such proteins might help activate and regulate de novo transcription during pollen tube growth.
It is known that interactions between the stylar tissue and the growing pollen tube are involved in guiding the growing pollen tube toward an ovary. Thus, it is interesting to examine categories of genes involved in signal transduction and cell-cell communication in the top-pollen list. We found that transcripts for several genes encoding signal transduction proteins were extremely abundant in pollen. Among the most abundant were transcripts for N-MYC down-regulated-like1 (NDL1; At5g56750), with RPKM of 1,664. Transcripts for this gene were more than 100-fold less abundant in seedling (14 RPKM). The NDL1 protein interacts with the trimeric G-protein β-subunit protein AGB1 (At4g34460) and together with AGB1 mediates auxin transport in the root (Mudgil et al., 2009); to our knowledge, no role for NDL1 in pollen development or pollen tube formation has been identified, but given the extremely high levels of NDL1 transcripts in pollen, such a role now seems highly likely. Interestingly, the mRNA for the NDL1 interactor AGB1 was less abundantly expressed in pollen than in seedling (6.5 versus 89 RPKM in pollen and seedling, respectively), suggesting that signal transduction pathways involving AGB1 and NDL1 differ between seedling and pollen. Annotated protein kinases were also among the most abundantly expressed genes, including 11 with RPKM > 1,000 (At5g12000, At2g43230, At3g02810, At1g17540, At1g61860, At2g07180, At3g20190, At3g18810, At5g18910, At3g08730, and At1g76370). Transcripts for several members of the 34-member Rapid Alkalinization Factor (RALF) family of peptide growth factors (Cao and Shi, 2012), including RALF8 (At1g61563), RALF9 (At1g61566), RALF26 (At3g25170), RALF19 (At2g33775), RALF4 (At1g28270), RALF15 (At2g22055), RALF25 (At3g25165), and RALF30 (At4g13075), were also extremely abundant. Their role in pollen has not been determined, and none was as highly expressed in seedling.
To validate the inspection of the top-pollen list, we used a GO enrichment analysis tool (Eden et al., 2009; http://cbl-gorilla.cs.technion.ac.il) to identify GO terms that were significantly enriched among higher ranking genes with respect to their expression in pollen. The advantage of this tool, in comparison with other GO enrichment methods, is that it uses the ordering of gene rankings to identify enriched terms and does not require preselection of a potentially arbitrary cutoff for differential or high expression. The tool identified several terms as significantly enriched that were consistent with the categories discussed above (P ≤ 10−3). Enriched terms included GO biological process categories related to cell wall organization, modification, and biogenesis. Enriched cellular component terms included categories related to cell wall, pollen tube, and cell projection, while enriched molecular function categories included terms related to protein phosphorylation, including protein kinase activity.
Genes Expressed in Pollen But Not Represented on the ATH1 Array
According to the TAIR10 mappings of probe set to gene annotations, the ATH1 array design lacks probe sets for 5,525 protein-coding loci. Another 2,142 genes annotated as pseudogenes or for which the final gene product is RNA (e.g. tRNA and microRNA genes) also lack a corresponding probe set on the ATH1 array. Of the 5,525 annotated protein-coding loci that have no corresponding probe set, there were 451 that were expressed in pollen with normalized expression values of 5 RPM or greater, roughly corresponding to at least 150 reads, given the depth of sequencing in the pollen sample. Of the non-protein-coding genes with no probe set that were expressed at 5 RPM or more in pollen, two were annotated as microRNAs, 17 as pseudogenes, three as ribosomal RNA genes, one as a pre-tRNA gene, and 55 as other RNA (mostly potential natural antisense genes; Table II). Many of these genes were highly expressed in pollen but not detectably expressed in seedling. Table III presents a list of genes that were highly expressed in pollen, were entirely absent (0 RPM) from the seedling data set, and were not represented on the ATH1 array. These genes represent potential new candidates for functional analyses.
Table II. Probe set-to-target gene annotations for gene types annotated in TAIR10.
Gene type designations are from TAIR. The column labeled No Probe Set gives the number of genes of various types that have no ATH1 probe set. The column labeled Multitarget Probe Set Only reports genes that are interrogated by one or more multitarget probe sets but have no corresponding unique probe set.
| Gene Type (as Annotated in TAIR10) | No Probe Set |
Multitarget Probe Set Only |
||
|---|---|---|---|---|
| Pollen RPM ≥ 5 | Seedling RPM ≥ 5 | Pollen RPM ≥ 5 | Seedling RPM ≥ 5 | |
| protein_coding | 451 | 1,110 | 239 | 766 |
| transposable_element_gene | 19 | 23 | 6 | 2 |
| pseudogene | 17 | 22 | 0 | 2 |
| other_rna | 55 | 145 | 1 | 0 |
| pre_trna | 1 | 1 | 0 | 0 |
| mirna | 2 | 3 | 0 | 0 |
| small_nucleolar_rna | 0 | 1 | 0 | 0 |
| ribosomal_rna | 3 | 3 | 0 | 0 |
| snRNA | 0 | 0 | 0 | 0 |
| Total | 548 | 1,308 | 246 | 770 |
Table III. Protein-coding genes highly expressed in pollen (RPM ≥ 146), undetectable in seedling, and not represented on the ATH1 array.
Gene symbols and descriptions are summarized from TAIR10 annotations.
| Arabidopsis Genome Initiative No. | Symbol | Description |
|---|---|---|
| AT3G01230 | Unknown protein | |
| AT5G28690 | Protein of unknown function (DUF1685) | |
| AT1G08140 | CHX6A | Cation/H+ exchanger6A |
| AT2G22055 | RALFL15 | RALF-like15 |
| AT1G08150 | CHX5 | Cation/hydrogen exchanger family protein |
| AT5G26717 | Putative membrane lipoprotein | |
| AT5G60615 | Defensin-like (DEFL) family protein | |
| AT3G05725 | Protein of unknown function (DUF3511) | |
| AT5G19473 | RPM1-interacting protein4 (RIN4) family protein | |
| AT4G21895 | DNA binding | |
| AT5G02110 | CYCD7 | CYCLIN D7;1 |
| AT4G11485 | LCR11 | Low-molecular-weight Cys-rich11 |
| AT1G80470 | F-box/RNI-like/FBD-like domains-containing protein | |
| AT2G31430 | Plant invertase/pectin methylesterase inhibitor superfamily protein | |
| AT4G19038 | LCR15 | Low-molecular-weight Cys-rich15 |
| AT1G80470 | F-box/RNI-like/FBD-like domains-containing protein | |
| AT2G31430 | Plant invertase/pectin methylesterase inhibitor superfamily protein | |
| AT4G19038 | LCR15 | Low-molecular-weight Cys-rich15 |
| AT4G04078 | Unknown protein |
Quantitative Real-Time PCR Validation of Relative Gene Expression Values in Pollen
A potential benefit of RNA-Seq data are that size-normalized expression values (RPKM) provide a way to rank genes in terms of their relative expression levels. To test this idea and also to confirm the expression of genes not represented on the ATH1 array, we identified nine pollen-expressed genes that varied in expression from 0.54 to 1,375 RPKM in pollen and were not represented on the ATH1 array. We also selected an additional gene (At4g34270) that has performed well as a standard in quantitative real-time PCR (qPCR) experiments but is expressed at very low levels in pollen (0.16 RPKM, seven overlapping single-mapping reads) according to the RNA-Seq data and thus may provide a useful lower bound for detectable expression in pollen. We tested the expression of all 10 genes using qPCR analysis, using four pollen cDNA samples from four separate pollen collections, including the sample used to created the pollen RNA-Seq library. We found that the relationship between normalized quantification cycle and RPKM values was linear when plotted on a semilog scale, with an r2 value of 0.96. Thus, we observed a close correspondence between expression levels detected by RNA-Seq and expression levels measured by qPCR (Supplemental Fig. S1).
Genes Interrogated by Promiscuous Probe Sets on the ATH1 Array
The ATH1 microarray contains 1,065 promiscuous or cross-hybridizing probe sets that recognize more than one target gene. In most cases, these targets have related functions and come from the same gene family. Sequence similarity in their 3′ regions, the part of the transcript used to select ATH1 probes, made designing specific probe sets impossible for these genes. There are 1,795 protein-coding genes on the ATH1 microarray interrogated only by promiscuous probe sets and having no unique probe set. Because of the relatively long read lengths in the pollen RNA-Seq data set, it was feasible to use the read alignments for genes interrogated by promiscuous probe sets to delimit which genes were expressed in pollen. To assess this, we used reads that mapped to exactly one genomic location and focused on genes with normalized expression values of 1 RPM or higher. Sorting Supplemental Table S2 (RPM) on the promiscuous probe set column groups genes that are interrogated by the same probe set. In many cases, we could delimit expression in pollen to one member represented by a given promiscuous probe set. For example, using a threshold of RPM > 5, there are three protein phosphatases represented by a promiscuous probe (266834_s_at); At2g30020 is expressed in pollen (RPM 14.3), while At4g08260 and At3g27140 are not (RPM < 1). A promiscuous probe set (265133_s_at) represents two genes encoding members of the plant self-incompatibility protein S1 family, but only At1g5150 is expressed in pollen (RPM 1000), while At1g51240 is not. A striking example of the usefulness of such analyses is a promiscuous probe set (254473_s_at) that represents 15 genes encoding proteins with DUF1204; of these, only At4g20700 is expressed in pollen (RPM 20.6); none of the others are expressed in seedling either. Thus, the Supplemental Tables published with this article can be used to guide the characterization of the roles of such family members, for example, by limiting the number of transfer DNA insertional mutants that need to be characterized and reducing the need to generate double or higher order mutants. In other cases, the RNA-Seq data showed that both genes corresponding to a promiscuous probe were expressed in pollen but provided quantitative data about their expression levels. For example, ATH1 probe 253898_s_at corresponds to two genes encoding β-subunits of Trp synthase, but At5g54810 (RPM 540) is more highly expressed than At4g27070 (RPM 13).
Regions of Previously Unannotated Transcription
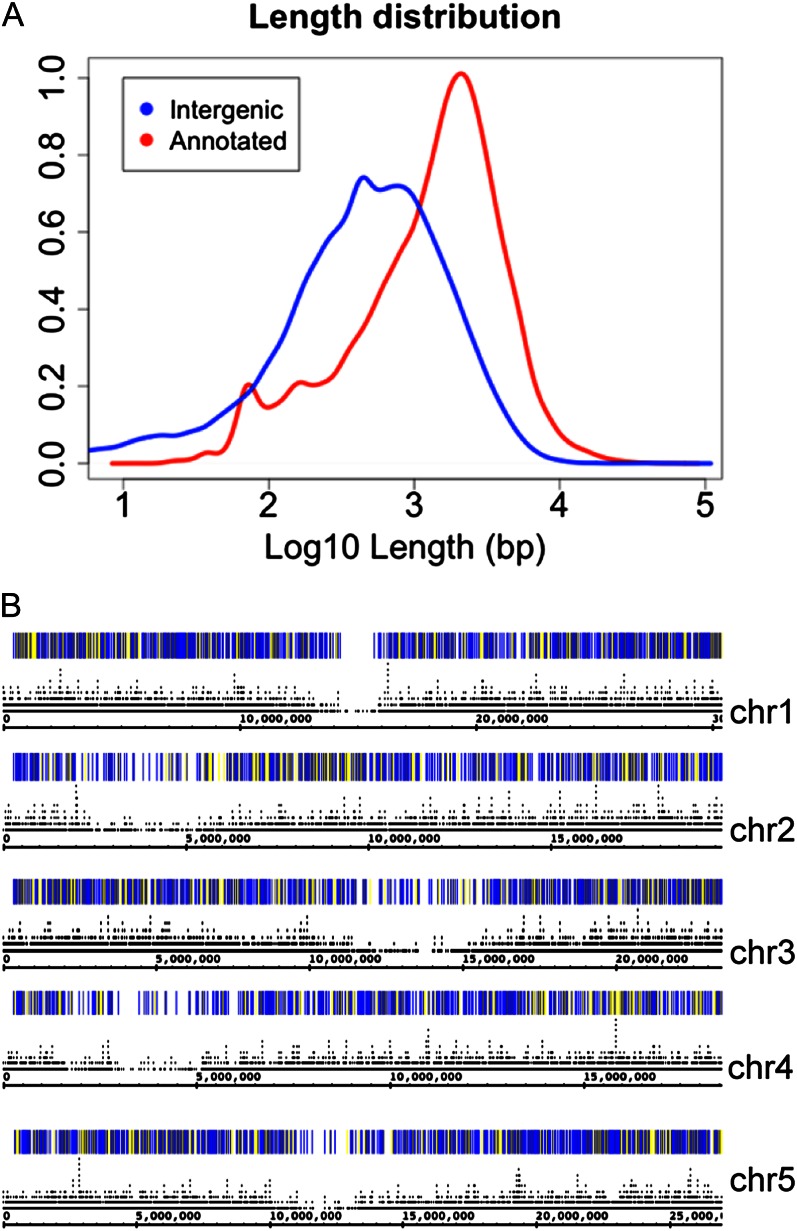
We searched for evidence of previously unannotated transcription in the pollen RNA-Seq data set using utilities from the BedTools suite (Quinlan and Hall, 2010) and manual inspection of read alignments and BedTools outputs in IGB. To start, we identified regions representing “dark matter” of the genome (i.e. regions in the genome outside any annotated gene or transposable element gene). The vast majority (almost 90%) of these intergenic regions were smaller than 2,000 bases and their median size was only 481 bases, reflecting the compactness of the Arabidopsis genome and the depth to which it has been annotated. As shown in Figure 2A, the size distribution of the intergenic regions was approximately log-normally distributed, as has also been observed for gene and transcript sizes (Cui and Loraine, 2006). That is, the distribution was highly skewed with a long right tail but became symmetrical once it was log transformed.
Figure 2.
Reads mapping outside any known gene or transposable element. A, Distribution of log-transformed (base 10) intergenic region (blue) and annotated gene sizes (red), in bp. Values on the y axis indicate the distribution density. B, Heat maps indicate the density of reads mapping to the indicated chromosome regions of the Arabidopsis genome. Darker (blue) hues indicate greater coverage, and yellow hues indicate less coverage. Individual gene models are shown as black marks above each chromosome; base pair positions are indicated. In most cases, stacked gene models that occupy the same horizontal location are annotated transcript variants arising from alternative splicing, alternative promoters, or alternative 3′ end processing.
We next identified all reads whose alignments were entirely contained within the intergenic dark matter regions and did not also extend into nearby annotated genes or transposable element genes. Only 500,714 (1.2%) of the more than 42.6 million single-mapping reads obtained from the pollen library mapped entirely within an intergenic region. As shown in Figure 2B, these reads mapped throughout the genome and were sparsest in regions that were also gene poor. We then identified reads that were either entirely contained within an intergenic region (Fig. 2B) or that crossed over from an intergenic region into a neighboring annotated gene or transposable element. We collapsed these reads into read scaffolds to create a new set of novel annotations representing overlapping, collapsed stacks of reads. Thus, each collapsed read region represents a set of reads whose alignments overlap along the genomic sequence axis, suggesting that they arose from the same gene. Next, we identified the subset of these regions that did not in turn overlap with any known gene or transposable element. This yielded 5,312 distinct, merged regions representing novel transcription.
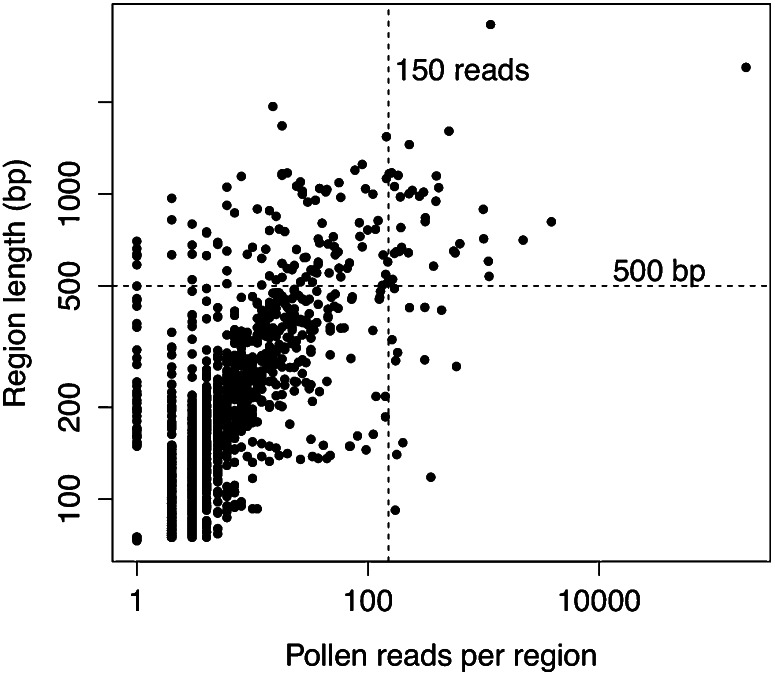
As with the intergenic regions, the distribution of regions of novel transcription was skewed with a long right tail but appeared normal once it was log transformed. Larger regions were associated with a larger numbers of reads (Fig. 3); the Pearson’s correlation coefficient between the logarithm (base 10) of the number of counts and region sizes was 0.8. Around half the regions (3,753) had no coverage (zero reads) from either of the two seedling data sets. Only 232 regions had combined pollen and seedling raw read counts of 50 or larger, and of these, only 74 were 500 bases or larger.
Figure 3.
Relationships between number of reads and region size. The x axis shows the number of reads per region, and the y axis shows the size per region in bp. Both axes are on logarithmic (base 10) scale. There were 35 regions 500 bases or larger with 150 or more reads.
For each novel region that was 500 bases or larger, we used BLASTX to detect potential homologies, since homology to a known protein would suggest that the region encodes a protein or is a pseudogene that once did. We found that 54 of the 135 regions contained significant homologies to known proteins. The list of regions and their expression levels (in reads per region) from pollen and seedling RNA-Seq data sets are in Supplemental Table S4.
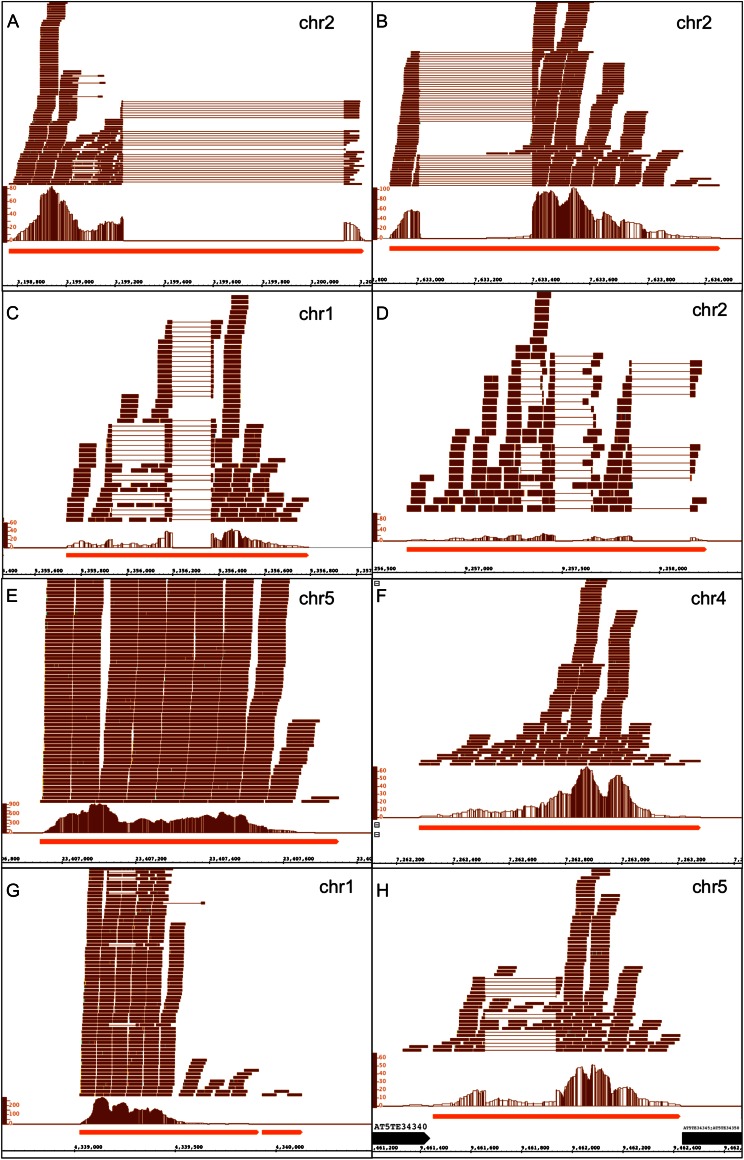
We selected 14 such regions for PCR testing, focusing on larger regions (500 bp or larger) with higher coverage in pollen (100 reads or more). For each, primers internal to putative exons rather than from putative exon-exon junctions were used, so that genomic DNA could be used as a positive control for PCR (Supplemental Table S6). As shown in Figure 4, many regions contained gapped reads suggestive of introns; in all cases, the implied intron boundaries were flanked by the canonical GT and AG splice-site consensus sequences. For each region, we attempted to amplify cDNA prepared from a sample of pollen RNA that was different from the pollen RNA used to generate the RNA-Seq data. Primers for all but two regions amplified a band of the expected size. The primer pairs that failed also failed to amplify genomic DNA, suggesting that the negative results for these regions were due to a primer problem rather than to a lack of expression.
Figure 4.
Putative new genes expressed in pollen. Screen captures from IGB show regions of novel transcription outside previously annotated loci, including annotated transposable elements. Read alignments are in the top track. Thin lines indicate reads mapping across introns. Graphs indicate the number of reads that map at positions indicated in the coordinates track. The bottom track contains TAIR10 gene model annotations, including protein-coding genes, transposable element genes, noncoding genes, and pseudogenes. The chromosome number is indicated in the upper right corner of each image. Regions are as follows: chr2:3198768 to 3200220 (A), chr2:7632907 to 7634053 (B), chr1:5355736 to 5356794 (C), chr2:9256704 to 9258245 (D), chr5:23406941 to 23407752 (E), chr4:7262280 to 7263282 (F), chr1:4339027 to 4339918 (G), and chr5:9461451 to 9462429 (H). [See online article for color version of this figure.]
Revised Annotations for Pollen-Expressed Genes
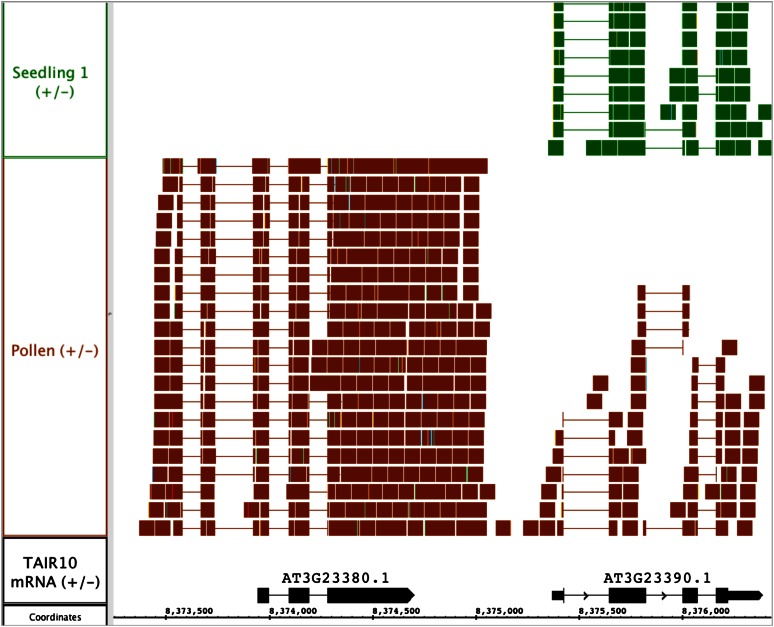
To determine if the annotations for any pollen-expressed genes needed revision, we examined the subpopulation of reads that overlapped the boundaries of known genes but that also crossed over into neighboring intergenic regions. These “cross-over” reads provide evidence for new 5′ or 3′ ends in existing gene models. There were 378,630 such cross-over reads in the pollen RNA-Seq data set. Of these, 370,360 overlapped with protein-coding genes; the rest overlapped with other types of annotated loci, such as pseudogenes or transposable element genes. One example of a protein-coding gene is shown in Figure 5; this protein (At3g23380; RIC5) is annotated in TAIR as playing a role in pollen tube growth and function via its interactions with Rop1 (Wu et al., 2001). Figure 5 presents an image from IGB in which a large number of reads link RIC5 transcription with its flanking intergenic region. As shown in Figure 5, the annotated translation of the protein is not changed. According to the pollen RNA-Seq data set, we found more than 5,000 protein-coding genes that had at least one overlapping read that extended beyond their currently annotated 5′ and 3′ ends. Of these, more than 1,500 had at least 20 reads supporting an extension and nearly 500 genes had 100 or more reads supporting an extension. Supplemental Table S5 reports gene regions and the number of reads from the pollen data that crossed over into intergenic regions. Using Supplemental Table S5 together with visualization in the IGB to assess such 5′ and 3′ extensions can help in the design of promoter constructs or in the manipulation of other regulatory regions that might impact gene expression.
Figure 5.
Visualization from IGB depicting additional exons for RIC5. Single-mapping reads from pollen and seedling RNA-Seq data sets are shown aligned onto the reference Arabidopsis genome. The top row of the pollen reads track contains reads that are drawn on top of each other due to space limitations. [See online article for color version of this figure.]
Novel Splicing Events (New Introns)
We investigated the extent to which the pollen and seedling RNA-Seq data sets revealed new splicing events affecting TAIR10 annotated genes. To assess splicing, we focused on gapped reads, which are reads that aligned across putative introns and are from exon-exon junctions. Our RNA-Seq data sets also contained many so-called “intron reads,” reads that mapped within annotated introns and might represent instances of retained intron alternative splicing events. However, because it is difficult (if not impossible) to determine when intron reads come from partially processed or mature mRNA species, we focused mainly on alternative splicing as detected by gapped reads, since such reads can only arise from spliced transcripts and thus represent bona fide products of splicing.
For the splicing analysis, we only examined single-mapping reads, since many of the multimapping gapped reads aligned to more than one location within the same gene. These reads could not be unambiguously assigned to a single splicing event and so were not useful for this analysis. To reduce false positives due to alignment artifacts, we restricted the analysis to gapped reads that aligned with at least five bases on either side of the putative intron, reasoning that, in most cases, five bases should be enough to anchor a read to a unique location within a gene. The gapped read alignments that met these criteria were used as inputs to a custom program (FindJunctions) that generates scored, exon-exon junction features similar to the “junctions.bed” file output by TopHat. For each junction feature, the score represented the number of gapped reads that supported the junction, and the internal coordinates of the junction feature indicated the 5′ and 3′ ends of a putative intron flanked by the junction feature. BED12 format files containing junction features created by the FindJunctions program and by TopHat are available for download and/or visualization in the IGB from the QuickLoad site.
We then used the output of the FindJunctions program to identify and score new splicing events. Taken together, the seedling and pollen data sets contained gapped reads supporting a total of 117,104 junctions, of which 98,532 were already annotated as part of the TAIR10 annotations and 18,572 were new. Of the new junctions, 1,045 overlapped with but extended beyond the affected genes and the rest (17,527) were internal to the gene models. The 1,045 junction features that overlapped with but extended beyond their respective annotated genes represent potential new exons that extend the 5′ and 3′ boundaries of the gene models. There were 709 genes that could be extended in this way. The remaining 17,527 junctions that were entirely contained within their respective genes affected 8,169 genes. However, nearly half of the novel junction features (46.3%) were supported by only one gapped read, suggesting that many of the novel junctions either represent low-frequency splicing events or alignment artifacts (Table IV). Of the 17,527 novel splicing events that were internal to gene models, there were 1,908 with 10 or more supporting reads, including some that had a very high level of support.
Table IV. Novel splicing events.
The column labeled Majority Pollen Supported reports the number of new splicing events where 80% (four of five) or more of the supporting gapped reads were from the pollen RNA-Seq data set. Internal refers to new junctions that are internal to currently annotated gene models and thus represent likely alternative splicing or cases where the annotated gene model is inaccurate. Extended refers to new junctions that extend 5′ or 3′ of the currently annotated gene models and in most cases represent new UTR exons. NA, Not applicable.
| Minimum Read Support | New Splicing Events |
Majority Pollen Supported | Genes | |
|---|---|---|---|---|
| Internal | Extended | |||
| 1 | 17,527 | 1,045 | NA | 8,547 |
| 2 | 9,407 | 597 | NA | 5,601 |
| 5 | 3,821 | 276 | 998 | 2,757 |
| 10 | 1,908 | 153 | 550 | 1,518 |
| 20 | 872 | 74 | 303 | 724 |
| 50 | 287 | 29 | 128 | 259 |
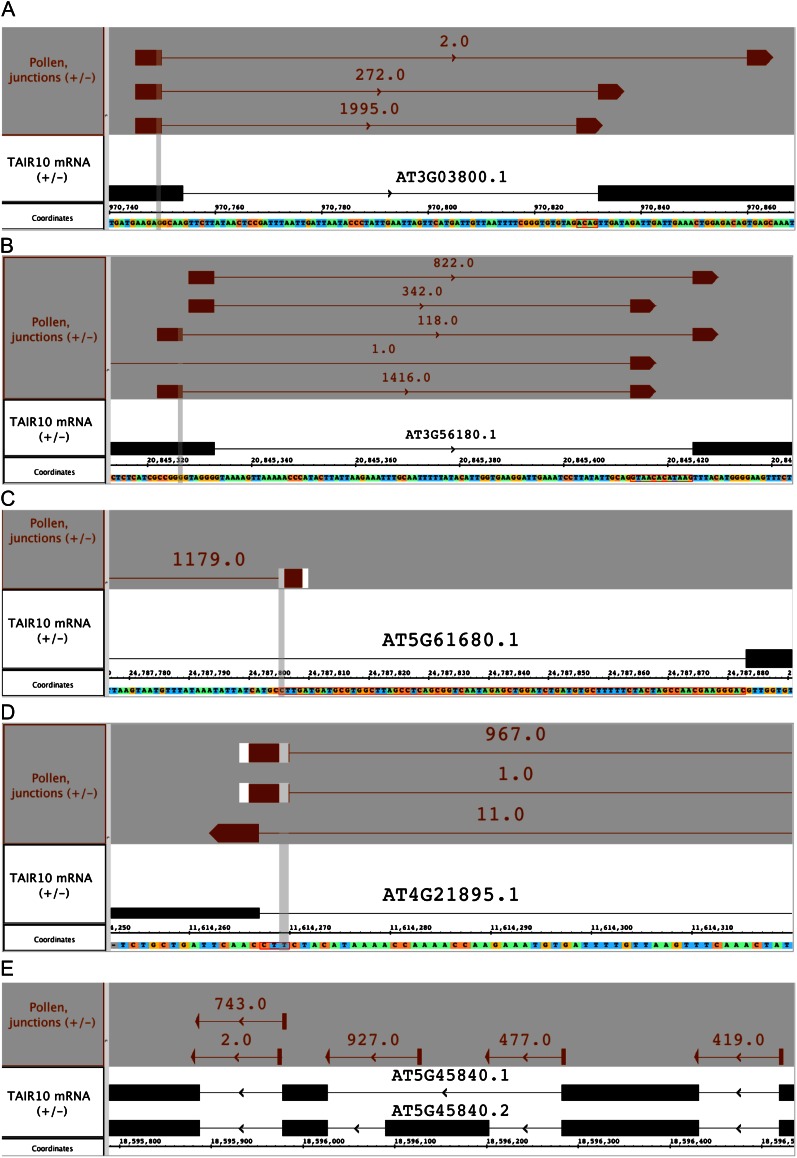
We then sorted the novel, internally located splicing events according to their level of support in the pollen data set and examined the best supported ones in IGB (Table V). In nearly every case, the novel splicing event as detected by the RNA-Seq analysis was the most abundantly expressed. That is, the novel splicing event revealed what appeared to be the bona fide transcript and the annotated gene model was either incorrect or the annotated variant was the minor form (Fig. 6). In every case, the difference affected the conceptual translation, either by adding or deleting one or more amino acids, but did not change the frame of translation or introduce a premature stop codon.
Table V. Highly supported new internal splicing events where most support is from pollen.
Junction Feature denotes genomic coordinates indicating the start and end of the inferred intron, using interbase for the coordinate system. P and S denote the number of gapped reads from the pollen and seedling data sets that support the new introns. Consequence notes changes to the annotated gene model’s conceptual translation.
| Junction Feature | Locus | Description | P | S | Consequence |
|---|---|---|---|---|---|
| J:chr3:970750-970828:+ | AT3G03800 | Syntaxin131 | 1,995 | 0 | Change one amino acid |
| J:chr3:20845327-20845413:+ | AT3G56180 | Unknown function (DUF567) | 1,416 | 0 | Adds one amino acid |
| J:chr5:24787626-24787805:- | AT5G61680 | Pectin-lyase-like | 1,179 | 0 | Adds 26 amino acids |
| J:chr4:11614270-11614908:- | AT4G21895 | DNA binding | 967 | 0 | Adds one amino acid |
| J:chr5:18596030-18596125:- | AT5G45840 | Leu-rich repeat kinase | 927 | 9 | Removes 11 amino acids |
| J:chr3:22391885-22392120:+ | AT3G60570 | β-Expansin | 867 | 0 | Adds 12 amino acids |
| J:chr3:9928426-9928605:-a | AT3G26934 | Unknown | 811 | 0 | Removes two amino acids |
| J:chr4:13927840-13927925:+ | AT4G28000 | P-loop nucleoside triphosphate hydrolase | 711 | 0 | Adds two amino acids |
New junction was the minor form.
Figure 6.
Visualizations from IGB depicting novel junction features. The number of spliced reads supporting junctions predicted from spliced read alignments appear above each junction feature. The annotated gene model is shown in black above the sequence axis. Arrows indicate the direction of transcription. [See online article for color version of this figure.]
Pollen-Specific Splicing Patterns
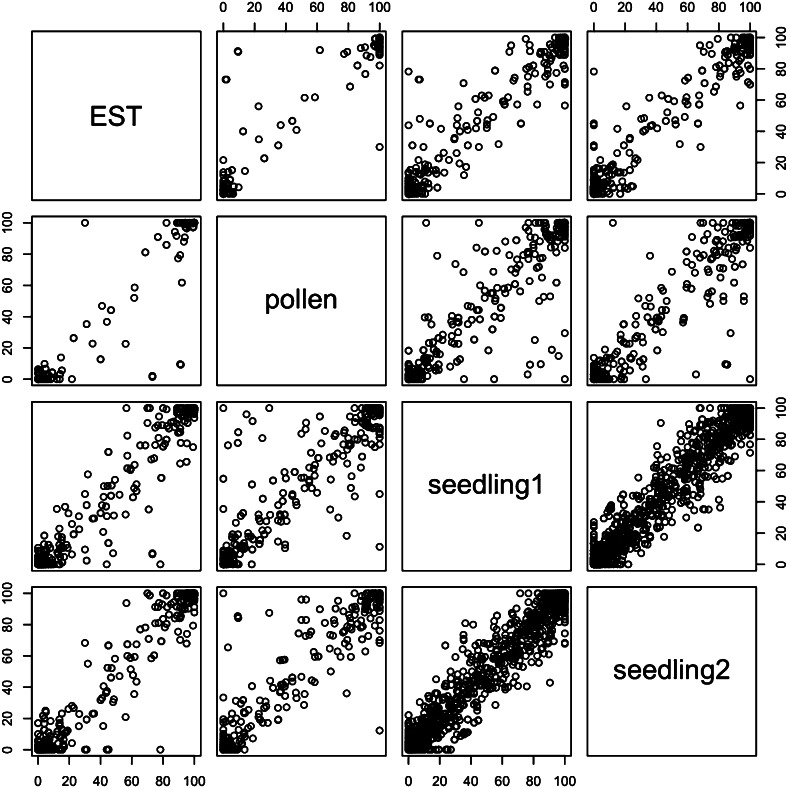
To determine if there were any splicing patterns specific to pollen, we compared patterns of splicing in pollen with patterns of splicing in seedling and with splicing patterns detected in a study of ESTs (English et al., 2010). For this analysis, we used the TAIR10 gene models as a guide, using previously classified alternative splicing events and focusing on alternative donors, alternative acceptors, and exon-skipping events. For each alternative splicing event, we calculated a splicing prevalence score as the percentage of gapped reads supporting the short-form event (called GA) relative to the total number of gapped reads mapping to the differentially spliced region. Many low-frequency alternative splicing choices might be overlooked in cases of low coverage across the differentially spliced region, thus leading to an overestimate or underestimate of the true prevalence score. Therefore, we limited the analysis to alternative splicing events with at least 20 gapped reads crossing the differentially spliced region. Among genes that were expressed in both seedling and pollen, the splicing prevalence score between pollen and seedling was highly correlated, yielding a Pearson’s correlation coefficient of 0.98 (Table VI; Fig. 7). The correlation of the splicing prevalence scores between ESTs, seedling, and pollen was also high. That is, in all three data sets (seedling, ESTs, and pollen), the dominant isoform was usually the same, suggesting that splice-site choice is remarkably stable.
Table VI. Alternative splicing correlation between EST, pollen, and seedling data sets.
Pearson’s correlation coefficients for the percentage of reads (or ESTs) supporting the shorter form (GA) of alternative splicing events are shown. Events with 20 or more reads (or ESTs) supporting either variant form were included in the calculation.
| Sample Type | ESTs | Pollen | Seedling 1 | Seedling 2 |
|---|---|---|---|---|
| ESTs | 1 | 0.9592 | 0.9789 | 0.9800 |
| Pollen | – | 1 | 0.9845 | 0.9876 |
| Seedling 1 | – | – | 1 | 0.9940 |
| Seedling 2 | – | – | – | 1 |
Figure 7.
Splice pattern correlation between ESTs, pollen, and seedling RNA-Seq data sets. The percentages of ESTs or RNA-Seq reads supporting the shorter form (GA) variant for all annotated alternative splicing events in TAIR10 are shown as scatterplots. Events with 20 or more overlapping reads are shown.
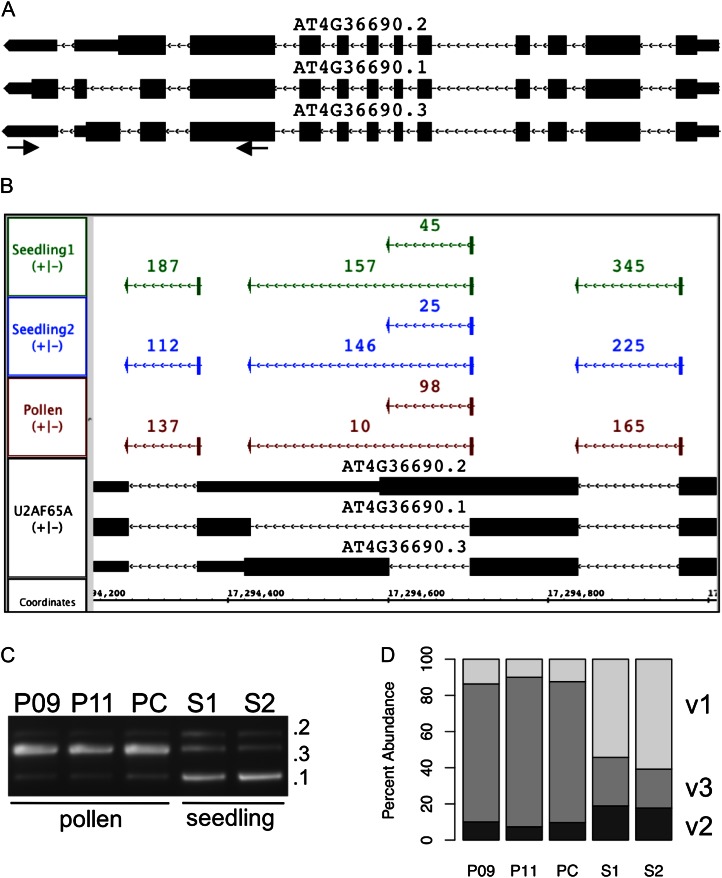
However, there were a few noteworthy exceptions to this general trend (Table VII). These genes included U2AF65A (At4g36690) and other genes with functions related to RNA processing. According to the RNA-Seq data, the U2AF65A variant (At4g36690.3) is the dominant isoform in pollen (Fig. 8A), while in seedling, variant 1 (At4g36690.1) is the most abundant. The main difference between isoforms is that they encode different C termini; the pollen-preferred isoform (0.3) lacks part of a C-terminal RNA recognition motif that is present in the other forms. We used PCR (Fig. 8B) to assess the relative abundance of the isoforms in seedling and pollen, using cDNAs from the originally sequenced pollen sample, two new pollen RNA samples, and two independent seedling RNA samples. The PCR experiment confirmed that isoform 3 was indeed the most prevalent in pollen, whereas in seedling, isoform 1 was most abundant (Fig. 8, C and D). Another pollen RNA-Seq data set from maize (Zea mays; Davidson et al., 2011) is publicly available from the Short Read Archive (SRR189771) and contains around 25 million 35-base reads. We obtained the sequences, aligned them onto the genome, and made the data publicly available for visualization in IGB. Using IGB, we inspected read alignments for six maize genes that were reported on the Phytozome comparative genomics Web site (Goodstein et al., 2012) as putative U2AF65A homologs. Five of the six were annotated as alternatively spliced and had overlapping pollen RNA-Seq reads, but there were no spliced alignments covering the alternatively spliced regions, making it impossible to determine whether the U2AF65A splicing pattern we observed in Arabidopsis is conserved in maize.
Table VII. Genes with pollen-specific splicing patterns.
GA is the gene model encoding the short-form transcript. P and S are the average percentage of reads in pollen and seedling supporting the GA form. Start and End indicate the start and end of the differentially spliced region in interbase genomic coordinates.
| GA | Description | P | S | Start | End |
|---|---|---|---|---|---|
| AT3G17120.1 | Unknown protein | 0 | 100 | 5,841,823 | 5,842,067 |
| AT2G19480.2 | NFA2, nucleosome assembly protein | 100 | 12 | 8,440,871 | 8,440,889 |
| AT3G58510.3 | DEA(D/H) box RNA helicase | 15 | 98 | 21,640,171 | 21,640,277 |
| AT3G06330.3 | RING/U-box superfamily protein | 19 | 96 | 1,917,423 | 1,917,507 |
| AT5G52040.1 | ATRSP41, RNA binding (involved in splicing) | 10 | 84 | 21,131,561 | 21,131,887 |
| AT4G36690.1 | U2AF65A, U2 snRNP auxiliary factor, large subunit, splicing factor | 9 | 82 | 17,294,430 | 17,294,603 |
| AT1G67480.2 | Gal oxidase/Kelch repeat | 3 | 71 | 25,276,345 | 25,276,463 |
| AT4G21720.1 | Unknown protein | 25 | 90 | 11,543,743 | 11,543,795 |
| AT5G65685.1 | UDP-glycosyltransferase | 29 | 94 | 26,275,685 | 26,275,722 |
| AT1G09140.1 | ATSRP30 (involved in splicing) | 79 | 27 | 2,943,226 | 2,943,564 |
Figure 8.
Pollen-specific alternative splicing in U2AF65A. A, Three gene models for U2AF65A. Blocks represent exons, and taller blocks indicate translated regions within an exon. The gene is transcribed from the minus strand of chromosome 4, and transcription is shown as proceeding from right to left. Not shown is variant 4, which is identical to variant 1 in the variably spliced 3′ region. Arrows indicate PCR primer locations. B, Closeup of the 3′ differentially spliced region with scored junction features from pollen and seedling RNA-Seq data sets, where scores represent the number of spliced reads supporting the junction. C, PCR products from amplifying pollen and seedling cDNA samples. Pollen samples included the same pollen RNA used to create the pollen library (P09) and RNA extracted from two independent pollen collections (P11 and PC). S1 and S2 are seedling cDNA samples prepared independently from the RNA-Seq seedling libraries. D, Relative amounts of variants 1, 2, and 3 PCR product amplified from pollen and seedling cDNA. Relative amounts were calculated from C. [See online article for color version of this figure.]
DISCUSSION
We performed ultra-high-throughput sequencing (RNA-Seq) of cDNA prepared from mature, dry pollen and aerial parts of 3-week-old Arabidopsis seedlings for comparison. By aligning sequences onto the reference genome sequence and comparing alignments with annotated genes, we identified protein-coding, microRNA, noncoding RNA, and pseudogenes that were expressed in pollen. Although the pollen collection method we used employs multiple filters to eliminate nonpollen debris, there was the possibility for contamination from nonpollen material. Therefore, we used light-harvesting complex genes, which are not expected to be expressed in pollen, to identify a ballpark expression level threshold of 5 RPM for calling a gene as present in the pollen data set. Based on this threshold, the diversity of gene expression was more than two times higher in seedling than in pollen. The 5-RPM threshold was useful for performing genome-scale analysis, but when analyzing individual genes, this threshold may be too high. We found many genes known to be expressed in pollen that were expressed at levels less than 5 RPM and that also had overlapping reads that matched annotated intron-exon boundaries. Before drawing conclusions about the expression of individual genes, it is important to examine read alignments.
We compared RNA-Seq and microarray present/absent calls from two pollen microarray studies that used the same collection method (Wang et al., 2008; Qin et al., 2009). Genes called present by microarray were nearly always also called present by RNA-Seq, even at high RPM thresholds (Fig. 1). This high correspondence provides independent corroboration of the accuracy and reliability of microarray present/absent calls. We also found that many more genes were detected as expressed in the RNA-Seq data, illustrating the greater ability of high-throughput sequencing to detect expression invisible to the other method. As more RNA-Seq data sets from Arabidopsis become available, it will be valuable to compare present/absent calls from comparable array studies. The Gene Expression Omnibus contains more than 8,000 ATH1 arrays, and many tools are available to visualize and mine these data (for review, see Brady and Provart, 2009; Usadel et al., 2009; Rung and Brazma, 2013). Comparing RNA-Seq and array data will lead to better methods for extracting new value from archived microarray expression data and reveal the strengths and weaknesses of both methods. Previously, high-throughput gene expression studies of pollen used DNA microarrays to call genes as present or absent or to chart gene expression changes during pollen germination, tube growth, and fertilization. However, due to the limitations of the technology, the relative abundance of RNAs present in pollen remained unknown. By comparing RNA-Seq read counts between genes, we assessed the relative abundance of RNAs present in mature, dry pollen. We found that the highest expressed genes in pollen were annotated with cell wall-related functions, including both cell wall biosynthesis and degradation. Cell wall proteins, protein kinases, and several members of the RALF peptide multigene family were also extremely abundant. Many genes in these categories have also been found to be up- and down-regulated during pollen tube growth. Using qPCR, we tested the relative abundance of 10 genes expressed at different levels in pollen, including nine that were not represented on the ATH1 microarray. Expression levels determined by qPCR and RNA-Seq were highly correlated. Altogether, these observations suggest that the pollen grain is well stocked with RNAs encoding proteins involved in tube growth but that new transcription is also important.
The ATH1 microarray was designed nearly 10 years ago (Redman et al., 2004) and lacks probe sets for more than 10,000 annotated genes, including more than 5,000 protein-coding genes. We used the RNA-Seq data to identify pollen-expressed genes that are not represented on the ATH1 array. Even using the relatively stringent expression cutoff of 5 RPM, we identified more than 500 pollen-expressed genes with no ATH1 probe set, including pseudogenes and three microRNA genes. Many genes are interrogated by probe sets that recognized multiple targets, typically members of the same gene family. The RNA-Seq read alignments allowed us to determine which gene family members were expressed in pollen. This finding illustrates the potential value of this data set to pollen researchers and also shows how tissue-specific RNA-Seq data sets may aid gene expression studies in the future. The ability to distinguish the expression of closely related paralogs will accelerate genetic studies by limiting the number of mutants needed to test gene function.
The Arabidopsis genome is among the best-annotated model organism genomes. Until recently, gene structure annotation has been based on computational gene prediction and EST alignments. There are no ESTs from Arabidopsis pollen available from the National Center for Biotechnology Information, suggesting that there might be some unannotated, pollen-expressed genes awaiting discovery. We used the RNA-Seq data to search for transcription outside known genes and transposable elements (Figs. 2 and 3) and found fewer than 50 convincing examples of novel transcription, of which 12 out of 14 tested were verified by PCR (Fig. 4). Given the compactness of the Arabidopsis genome and the thoroughness with which it has been analyzed and annotated, finding so few entirely novel genes is not surprising. However, the RNA-Seq data contained abundant support for new 3′ and 5′ UTRs (Fig. 5), new introns (Fig. 6), and alternative splicing. The TAIR10 annotations used information from an early RNA-Seq study (Filichkin et al., 2010), but the level of coverage and read lengths were much less extensive than those presented here. Other groups have also reported a high degree of novel splicing in wild-type Arabidopsis plants (Marquez et al., 2012), but these new splicing events have not yet been combined and incorporated into existing gene models.
Because pollen consists of two cell types, it was possible to use the pollen RNA-Seq data to address questions about tissue-specific splicing raised by our earlier study of alternative splicing prevalence in Arabidopsis (English et al., 2010). We found that for most annotated alternative splicing events, the minor isoform in one data set (e.g. seedling) was also the minor isoform in other data sets (e.g. pollen or ESTs; Fig. 7). However, we identified a small number of genes where the ratios of major to minor isoforms in the pollen and seedling varied significantly. Interestingly, many of these genes encoded proteins with roles in splicing, including the Arabidopsis homolog of U2AF65A, whose pollen-specific splicing pattern we verified by PCR (Fig. 8). However, the fact that we identified so few differentially spliced genes in pollen should not be taken as evidence that there is not much pollen-specific splicing. The data presented here came from only one pollen library and two seedling libraries, and it is possible that deeper sequencing from more libraries will uncover more examples.
Because Arabidopsis is often used as a reference plant to guide research in other agriculturally important plants, and since pollen is an important model system for plant cell biology, it will be important to systematically combine the RNA-Seq data presented here with data from other studies to improve annotation of the genome. The tools needed to perform this annotation on a genome-wide scale are being developed and will likely be incorporated into the International Arabidopsis Information Portal project (International Arabidopsis Informatics Consortium, 2012). However, the impact of these data are not limited to annotation. The richness, size, and newness of RNA-Seq data mean that, like microarrays, their usefulness can extend beyond their initial publication as new methods for mining and analyzing the data are developed. We found that the one of the most useful tools for analysis of the data was IGB, a visualization tool that supports highly interactive exploration of results. The pollen RNA-Seq data will become an important resource for pollen biologists, and visualization of the RNA-Seq read alignments in the IGB will inform and improve analyses of pollen-expressed genes.
MATERIALS AND METHODS
Sample Collection for RNA-Seq
Arabidopsis (Arabidopsis thaliana) Col-0 seeds were sown on soil, kept at 4°C for 3 d, and then transferred to a temperature-controlled growth room set to 22°C. Seedlings were grown in a 16-h/8-h light/dark cycle under 100 to 120 μmol m−2 s−1. After 21 d of growth, the aerial portions of the seedlings were collected twice per day for 5 d and pooled for RNA extraction. The experiment was then repeated following the same collection scheme, thus providing two distinct biological replicates. Mature, dry pollen was harvested from Col-0 plants using a vacuum collection device as described (Johnson-Brousseau and McCormick, 2004).
RNA Extraction and Illumina Library Preparation
Seedlings or pollen were ground into a fine powder with a mortar and pestle, and RNA was isolated via TRI-reagent extraction with cleanup on Qiagen Plant RNeasy (catalog no. 74104) columns. All RNAs were treated with DNaseI using the Plant RNeasy Kit. A starting amount of 15 μg of total RNA from each sample (seedling and pollen) was used in the library preparation, using the Illumina mRNA-seq Sample Preparation Kit (catalog no. RS-930-1001, part no. 1004898). mRNA was isolated and purified via Sera-Mag Magnetic Oligo(dT) Beads, washed, and then fragmented using divalent cations under elevated temperature. First- and second-strand cDNAs were synthesized, and an end-repair step was performed to convert overhangs into blunt ends. Next, the 3′ ends were adenylated for ligation of the Illumina adapters. cDNA templates were then purified by gel isolation for a size selection of approximately 250 bp and amplified via PCR so that an adequate amount of sample was available for sequencing (10 μL of a 25 ng μL−1 sample). Products were isolated with the Qiagen QIAquick PCR Purification Kit (catalog no. 28104) and added to the flow cell for sequencing.
Preparation of cDNA for PCR Validation Experiments
Two biological replicates of seedling cDNA and four biological replicates of pollen cDNA were prepared as follows. Total RNA from the aerial portions of 24-d-old seedlings grown at 22°C under long-day (16 h of light, 8 h of dark) conditions, or pollen, was extracted using the Qiagen RNeasy Plant Mini Kit (catalog no. 79403). RNAs were treated with DNaseI on the RNeasy Kit column prior to cDNA synthesis. Samples were quantified spectrophotomerically via Nanodrop, and 1 μg of total RNA was used to synthesize first-strand cDNA with an oligo(dT) primer and SMART MMLV reverse transcriptase (Clontech; catalog no. 639522) in a 20-μL reaction. Unless otherwise specified, all PCRs used 2 μL of 1:20 dilutions of the cDNA reaction mixture.
End-Point PCR Validation of Novel Transcription in Pollen
Regions of novel transcription with 50 or more overlapping reads from the pollen RNA-Seq data were selected and checked visually in IGB. Using Primer3, one pair of primers was designed to amplify each of the selected regions (Supplemental Table S7). Where possible, primers were chosen to include intronic regions to distinguish products arising from cDNA rather than from genomic DNA. Each primer pair was used in gradient PCRs containing 0.5 μL of a 1:20 dilution of the first-strand cDNA synthesis reactions described above or 25 ng of genomic DNA as a positive control. Gradient PCR conditions were as follows: denaturation at 94°C for 4 min, cycling at 94°C for 30 s, annealing at varying temperatures for 30 s, and extension at 72.0°C for 30 s. Annealing temperatures ranged from 50°C to 68°C. Reaction products were visualized by agarose gel electrophoresis, and the identities of amplicons were verified by Sanger sequencing.
qPCR Validation of Expression in Pollen
Target amplicons ranging in size from 90 to 110 bases were selected by visualizing gene structures and pollen RNA-Seq reads in IGB. Primers were designed using Primer3 and are listed in Supplemental Table S8. Each primer pair flanked an intron to allow the assessment of possible contamination from genomic DNA. qPCR was performed in 10-μL reaction volumes containing SoFast EvaGreen supermix at 1× concentration (Bio-Rad; catalog no. 172-5203), 500 nm forward and reverse primers, and 2 μL of a 1:20 dilution of pollen cDNA. For each gene, there were four biological replicates of pollen cDNA with two technical replicates per biological replicate. qPCR amplifications were performed on two separate plates, where each plate contained primer pairs for two genes (At5g45740 and At2g39805) to assess plate comparability and allow standardization across plates. qPCR expression values were calculated for each gene as the difference between the quantification cycle of the gene and the reference gene, averaged over technical replicates. The difference values for each gene were averaged across pollen samples and then compared with their corresponding RPKM values from the pollen RNA-Seq data.
PCR Testing of U2AF65A Splicing Pattern
Relative quantification of splice variant products was performed as described (Venables et al., 2012). Primers flanking the alternatively spliced 3′ region of U2AF65A (5′-CCCATCTCTAGCTGCGACTC-3′ and 5′-CAATCACGCAAAAGGGTCTT-3′) were used to amplify all three known splice variants in reactions containing 1 μL of 1:20 dilutions of pollen or seedling cDNA in 20-μL reactions. Three cDNA samples from pollen samples were tested, including the same pollen sample used to create the library and two other samples collected at different times. To assay the relative abundance of splice variant PCR products, 7 μL from each amplification reaction was combined with loading buffer and separated on 1.2% agarose gels containing ethidium bromide and Mr standards. Gels were photographed, and the brightness of each PCR product was measured using VisionWorks LS Image Acquisition and Analysis Software version 7.0.1 (UVP; www.uvp.com/visionworks.html) with background correction. Size-normalized intensities per base pair were determined by dividing band intensity by product size. Size-normalized band intensity values were added to calculate a total transcript level for each lane. The relative amount of each isoform product was calculated as a percentage of the total transcript level per lane by dividing each individual value by the total.
Sequence Processing
Sequences were aligned onto the latest Arabidopsis Col-0 genome assembly (TAIR10; released June 2009) using TopHat version 1.3.3 and BowTie version 0.12.7, with maximum intron size set to 2,000 bases. Maize (Zea mays) sequences were aligned onto the latest maize B73 genome (RefGen_v2; released March 2010) with maximum intron size set to 8,000 bases. The resulting BAM (binary alignment) files were then sorted and indexed using samtools version 0.1.18. The number of alignments or hits (NH) tag for each alignment was used to separate the alignments into BAM files with reads that mapped once (NH:i:1) and reads that mapped more than once (NH:i:n; where n > 1). Files with single-mapping reads have the extension sm.bam, and files with multimapping reads have the extension mm.bam. For analysis of expression, detection of novel genes, novel splicing, and retained introns, only the single-mapping “sm” reads were used. A Java program (FindJunction) was developed that creates junction features from spliced read alignments and counts the number of reads per sample that support each junction. The source code is available from the GenoViz open source project repository hosted at SourceForge.net. RPM expression values were calculated as the number of reads overlapping a gene divided by the total number of single-mapping reads obtained per sample library and multiplied by 1,000,000. RPKM expression values were calculated as RPM divided by the length (in kilobases) of the longest annotated transcript.
Alternative Splicing Analysis
Alternative splicing events were classified using the intron-exon overlap method as described previously (English et al., 2010), in which differentially spliced regions are detected by comparing pairs of gene models annotated to the same locus. To determine support for known splicing choices, reads supporting annotated splicing events were counted using spliced read alignments as support for specific splice-site choices. For each annotated alternative splicing event, the reads per sample type that supported mutually exclusive alternative splicing choices were counted.
Microarray and GO Analyses
Probe set-to-target gene mappings were taken from the TAIR Web site: ftp://ftp.arabidopsis.org/home/tair/Microarrays/Affymetrix/affy_ATH1_array_elements-2010-12-20.txt. Present/absent calls were generated using the MAS5 algorithm as implemented in the Bioconductor “affy” library. The versions of R and Bioconductor were 2.15 and 2.10, respectively. GO terms were from the TAIR annotations file ftp://ftp.arabidopsis.org/home/tair/Ontologies/Gene_Ontology/ATH_GO_GOSLIM.txt.
Data Availability
Illumina sequence data are available from NCBI under Short Read Archive accession SRP022162. Processed alignment, junction, and coverage graph files are available for visualization in Integrated Genome Browser via IGB QuickLoad data source http://www.igbquickload.org/pollen.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Correspondence between qPCR and RNA-Seq expression values.
Supplemental Table S1. RNA-Seq gene expression data in reads per gene.
Supplemental Table S2. RNA-Seq gene expression data in RPM per gene.
Supplemental Table S3. RNA-Seq gene expression data in RPKM per gene.
Supplemental Table S4. Regions containing potential new genes, with counts from pollen and seedling.
Supplemental Table S5. Genes with new 5′ and/or 3′ regions and the number of reads supporting the extended regions.
Supplemental Table S6. New and known splice junctions from seedling and pollen RNA-Seq spliced reads with number of spliced reads from each library.
Supplemental Table S7. Primers used for testing expression of previously unannotated genes.
Supplemental Table S8. Primers used for expression testing by qPCR.
Acknowledgments
We thank Mily Ron for providing pollen RNA, Anuj Puram and Hiral Vora for the FindJunctions program, Alyssa Gulledge for advice on using IGB to assess splicing, Tyler Estrada for assistance with figures, and members of the Integrative Pollen Biology Research Coordination Network for comments on this work.
Glossary
- Col-0
Columbia-0
- TAIR10
The Arabidopsis Information Resource 10
- TAIR
The Arabidopsis Information Resource
- UTR
untranslated region
- IGB
Integrated Genome Browser
- RPM
reads per million
- RPKM
reads per kilobase million
- GO
Gene Ontology
- qPCR
quantitative real-time PCR
- cDNA
complementary DNA
References
- Becker JD, Boavida LC, Carneiro J, Haury M, Feijó JA. (2003) Transcriptional profiling of Arabidopsis tissues reveals the unique characteristics of the pollen transcriptome. Plant Physiol 133: 713–725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boavida LC, Borges F, Becker JD, Feijó JA. (2011) Whole genome analysis of gene expression reveals coordinated activation of signaling and metabolic pathways during pollen-pistil interactions in Arabidopsis. Plant Physiol 155: 2066–2080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brady SM, Provart NJ. (2009) Web-queryable large-scale data sets for hypothesis generation in plant biology. Plant Cell 21: 1034–1051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao J, Shi F. (2012) Evolution of the RALF gene family in plants: gene duplication and selection patterns. Evol Bioinform Online 8: 271–292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui X, Loraine A. (2006) Global correlation analysis between redundant probe sets using a large collection of Arabidopsis ath1 expression profiling data. Comput Syst Bioinformatics Conf 223–226 [PubMed] [Google Scholar]
- Davidson RM, Hansey CN, Gowda M, Childs KL, Lin H, Vaillancourt B, Sekhon RS, de Leon N, Kaeppler S, Jiang N, et al (2011) Utility of RNA-seq for analysis of maize reproductive transcriptomes. Plant Genome 4: 191–203 [Google Scholar]
- Eden E, Navon R, Steinfeld I, Lipson D, Yakhini Z. (2009) GOrilla: a tool for discovery and visualization of enriched GO terms in ranked gene lists. BMC Bioinformatics 10: 48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- English AC, Patel KS, Loraine AE. (2010) Prevalence of alternative splicing choices in Arabidopsis thaliana. BMC Plant Biol 10: 102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filichkin SA, Priest HD, Givan SA, Shen R, Bryant DW, Fox SE, Wong WK, Mockler TC. (2010) Genome-wide mapping of alternative splicing in Arabidopsis thaliana. Genome Res 20: 45–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodstein DM, Shu S, Howson R, Neupane R, Hayes RD, Fazo J, Mitros T, Dirks W, Hellsten U, Putnam N, et al (2012) Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res 40: D1178–D1186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gulledge AA, Roberts AD, Vora H, Patel K, Loraine AE. (2012) Mining Arabidopsis thaliana RNA-seq data with Integrated Genome Browser reveals stress-induced alternative splicing of the putative splicing regulator SR45a. Am J Bot 99: 219–231 [DOI] [PubMed] [Google Scholar]
- Holmes-Davis R, Tanaka CK, Vensel WH, Hurkman WJ, McCormick S. (2005) Proteome mapping of mature pollen of Arabidopsis thaliana. Proteomics 5: 4864–4884 [DOI] [PubMed] [Google Scholar]
- Honys D, Twell D. (2003) Comparative analysis of the Arabidopsis pollen transcriptome. Plant Physiol 132: 640–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honys D, Twell D. (2004) Transcriptome analysis of haploid male gametophyte development in Arabidopsis. Genome Biol 5: R85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Arabidopsis Informatics Consortium (2012) Taking the next step: building an Arabidopsis information portal. Plant Cell 24: 2248–2256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson-Brousseau SA, McCormick S. (2004) A compendium of methods useful for characterizing Arabidopsis pollen mutants and gametophytically-expressed genes. Plant J 39: 761–775 [DOI] [PubMed] [Google Scholar]
- Li P, Ponnala L, Gandotra N, Wang L, Si Y, Tausta SL, Kebrom TH, Provart N, Patel R, Myers CR, et al. (2010) The developmental dynamics of the maize leaf transcriptome. Nat Genet 42: 1060–1067 [DOI] [PubMed] [Google Scholar]
- Marquez Y, Brown JW, Simpson C, Barta A, Kalyna M. (2012) Transcriptome survey reveals increased complexity of the alternative splicing landscape in Arabidopsis. Genome Res 22: 1184–1195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. (2008) Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods 5: 621–628 [DOI] [PubMed] [Google Scholar]
- Mudgil Y, Uhrig JF, Zhou J, Temple B, Jiang K, Jones AM. (2009) Arabidopsis N-MYC DOWNREGULATED-LIKE1, a positive regulator of auxin transport in a G protein-mediated pathway. Plant Cell 21: 3591–3609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicol JW, Helt GA, Blanchard SG, Jr, Raja A, Loraine AE. (2009) The integrated genome browser: free software for distribution and exploration of genome-scale datasets. Bioinformatics 25: 2730–2731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palusa SG, Ali GS, Reddy AS. (2007) Alternative splicing of pre-mRNAs of Arabidopsis serine/arginine-rich proteins: regulation by hormones and stresses. Plant J 49: 1091–1107 [DOI] [PubMed] [Google Scholar]
- Pina C, Pinto F, Feijó JA, Becker JD. (2005) Gene family analysis of the Arabidopsis pollen transcriptome reveals biological implications for cell growth, division control, and gene expression regulation. Plant Physiol 138: 744–756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin Y, Leydon AR, Manziello A, Pandey R, Mount D, Denic S, Vasic B, Johnson MA, Palanivelu R. (2009) Penetration of the stigma and style elicits a novel transcriptome in pollen tubes, pointing to genes critical for growth in a pistil. PLoS Genet 5: e1000621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM. (2010) BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26: 841–842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redman JC, Haas BJ, Tanimoto G, Town CD. (2004) Development and evaluation of an Arabidopsis whole genome Affymetrix probe array. Plant J 38: 545–561 [DOI] [PubMed] [Google Scholar]
- Rung J, Brazma A. (2013) Reuse of public genome-wide gene expression data. Nat Rev Genet 14: 89–99 [DOI] [PubMed] [Google Scholar]
- Sanchez SE, Petrillo E, Beckwith EJ, Zhang X, Rugnone ML, Hernando CE, Cuevas JC, Godoy Herz MA, Depetris-Chauvin A, Simpson CG, et al. (2010) A methyl transferase links the circadian clock to the regulation of alternative splicing. Nature 468: 112–116 [DOI] [PubMed] [Google Scholar]
- Sedbrook JC, Carroll KL, Hung KF, Masson PH, Somerville CR. (2002) The Arabidopsis SKU5 gene encodes an extracellular glycosyl phosphatidylinositol-anchored glycoprotein involved in directional root growth. Plant Cell 14: 1635–1648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo PJ, Park MJ, Lim MH, Kim SG, Lee M, Baldwin IT, Park CM. (2012) A self-regulatory circuit of CIRCADIAN CLOCK-ASSOCIATED1 underlies the circadian clock regulation of temperature responses in Arabidopsis. Plant Cell 24: 2427–2442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staiger D, Zecca L, Wieczorek Kirk DA, Apel K, Eckstein L. (2003) The circadian clock regulated RNA-binding protein AtGRP7 autoregulates its expression by influencing alternative splicing of its own pre-mRNA. Plant J 33: 361–371 [DOI] [PubMed] [Google Scholar]
- Umate P. (2010) Genome-wide analysis of the family of light-harvesting chlorophyll a/b-binding proteins in Arabidopsis and rice. Plant Signal Behav 5: 1537–1542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usadel B, Obayashi T, Mutwil M, Giorgi FM, Bassel GW, Tanimoto M, Chow A, Steinhauser D, Persson S, Provart NJ. (2009) Co-expression tools for plant biology: opportunities for hypothesis generation and caveats. Plant Cell Environ 32: 1633–1651 [DOI] [PubMed] [Google Scholar]
- Venables JP, Vignal E, Baghdiguian S, Fort P, Tazi J. (2012) Tissue-specific alternative splicing of Tak1 is conserved in deuterostomes. Mol Biol Evol 29: 261–269 [DOI] [PubMed] [Google Scholar]
- Wang Y, Zhang WZ, Song LF, Zou JJ, Su Z, Wu WH. (2008) Transcriptome analyses show changes in gene expression to accompany pollen germination and tube growth in Arabidopsis. Plant Physiol 148: 1201–1211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Gerstein M, Snyder M. (2009) RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet 10: 57–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G, Gu Y, Li S, Yang Z. (2001) A genome-wide analysis of Arabidopsis Rop-interactive CRIB motif-containing proteins that act as Rop GTPase targets. Plant Cell 13: 2841–2856 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Illumina sequence data are available from NCBI under Short Read Archive accession SRP022162. Processed alignment, junction, and coverage graph files are available for visualization in Integrated Genome Browser via IGB QuickLoad data source http://www.igbquickload.org/pollen.