A bHLH gene (PtrbHLH) confers cold tolerance and modulates peroxidase-mediated scavenging of H2O2.
Abstract
The basic helix-loop-helix (bHLH) transcription factors are involved in a variety of physiological processes. However, plant bHLHs functioning in cold tolerance and the underlying mechanisms remain poorly understood. Here, we report the identification and functional characterization of PtrbHLH isolated from trifoliate orange (Poncirus trifoliata). The transcript levels of PtrbHLH were up-regulated under various abiotic stresses, particularly cold. PtrbHLH was localized in the nucleus with transactivation activity. Overexpression of PtrbHLH in tobacco (Nicotiana tabacum) or lemon (Citrus limon) conferred enhanced tolerance to cold under chilling or freezing temperatures, whereas down-regulation of PtrbHLH in trifoliate orange by RNA interference (RNAi) resulted in elevated cold sensitivity. A range of stress-responsive genes was up-regulated or down-regulated in the transgenic lemon. Of special note, several peroxidase (POD) genes were induced after cold treatment. Compared with the wild type, POD activity was increased in the overexpression plants but decreased in the RNAi plants, which was inversely correlated with the hydrogen peroxide (H2O2) levels in the tested lines. Treatment of the transgenic tobacco plants with POD inhibitors elevated the H2O2 levels and greatly compromised their cold tolerance, while exogenous replenishment of POD enhanced cold tolerance of the RNAi line. In addition, transgenic tobacco and lemon plants were more tolerant to oxidative stresses. Yeast one-hybrid assay and transient expression analysis demonstrated that PtrbHLH could bind to the E-box elements in the promoter region of a POD gene. Taken together, these results demonstrate that PtrbHLH plays an important role in cold tolerance, at least in part, by positively regulating POD-mediated reactive oxygen species removal.
As sessile organisms, plants are constantly exposed to an array of environmental stresses, among which cold has been shown to be a key factor affecting growth and development, crop productivity, and geographic distribution. Over a long evolutionary period, plants have developed a set of sophisticated mechanisms to adapt to or survive under adverse conditions. Significant advancements, made during the past decades, have revealed some of the mechanisms underlying plant responses to various abiotic stresses (Liu et al., 1998; Thomashow, 1999; Cook et al., 2004; Kaplan et al., 2004; Vogel et al., 2005; Shulaev et al., 2008; Miller et al., 2009; Qin et al., 2011; Delhaize et al., 2012). It is now clear that the stress response is a highly complex process regulated by multiple signaling pathways. A generic stress signal transduction pathway starts with signal perception through known or unknown sensors, followed by the activation of second messengers, such as Ca2+, reactive oxygen species (ROS), and inositol phosphates. The signal is then transduced and relayed by various signaling components to switch on adaptive responses, leading to a range of physiological and metabolic alterations that impart stress tolerance.
Emerging evidence has begun to show that the stress signaling cascade encompasses a large number of stress-responsive genes, which can be categorized into two major groups based on the functions of their products. The first group is composed of functional proteins that act directly in protecting the cells from stress-associated damage, while the second group consists of regulatory proteins that function in signal transduction and gene expression regulation (Agarwal et al., 2006; Chinnusamy et al., 2006; Shinozaki and Yamaguchi-Shinozaki, 2007; Lata and Prasad, 2011). Physiological functions of many stress-responsive genes have been characterized, although the detailed molecular mechanisms underlying the abiotic stress responses are far from being completely elucidated. Exploitation of these genes cannot only improve our understanding of the stress response mechanisms but also make it possible to create transgenic plants with enhanced stress tolerance through transgene technology (Wang et al., 2003; Lata and Prasad, 2011; Qin et al., 2011). For the latter aspect, many studies have shown that overexpression of stress-responsive genes could enhance the stress tolerance of plants. However, it is important to point out that the constitutive overexpression of different stress-responsive genes may give rise to various outcomes (Agarwal et al., 2006). In general, genetic manipulation of regulatory genes is thought to be a more powerful approach for elevating stress tolerance in plants when compared with similar efforts on downstream individual stress-responsive genes. Transcription factors (TFs) constitute a group of important regulatory genes that play pivotal roles in the control of expression of stress-responsive genes. Overexpression of a TF may activate a group of target genes that function in a concerted manner to counteract the adverse effects of abiotic stresses. Therefore, genetic engineering of TFs has been proposed to be a robust strategy for improving the stress tolerance of crop plants (Thomashow et al., 2001; Sreenivasulu et al., 2007; Nakashima et al., 2009; Golldack et al., 2011).
Plants have a wide range of TFs; for instance, the Arabidopsis (Arabidopsis thaliana) genome contains more than 1,500 TFs, accounting for nearly 6% of its total genes (Riechmann et al., 2000). Among the TFs, the basic helix-loop-helix (bHLH) motif-containing TFs are important regulatory components of the transcriptional networks. As suggested by the name, the bHLH motif consists of conserved amino acids with two functionally distinct regions, the N-terminal basic region and the helix-loop-helix region. The basic region, which is composed of approximately 15 amino acids, including several basic residues, determines the specificity of the DNA-protein interactions. The helix-loop-helix region contains two amphipathic α-helices connected by a loop region of variable length and is responsible for the formation of homodimers or heterodimers (Buck and Atchley, 2003; Toledo-Ortiz et al., 2003; Li et al., 2006). The bHLH TFs are widely distributed in eukaryotes and have been extensively studied in animal systems. Plant bHLH TFs are relatively less characterized, albeit more and more relevant studies have been reported in recent years since the first plant bHLH-containing protein was characterized (Ludwig et al., 1989). To date, plant bHLH proteins have been shown to function in the transcriptional regulation of a diversity of biological processes, including flowering (Ito et al., 2012), trichome or root hair development (Bernhardt et al., 2003; Karas et al., 2009; Tominaga-Wada et al., 2012), chloroplast development (Monte et al., 2004), biosynthesis of flavonoid, isoquinoline alkaloid, and anthocyanin (Nesi et al., 2000; Ohno et al., 2011; Yamada et al., 2011; Xie et al., 2012), and nodule vascular patterning (Godiard et al., 2011). In addition, several plant bHLH proteins, such as PHYTOCHROME-INTERACTING FACTOR3 (PIF3), PIF4, and LONG HYPOCOTYL IN FAR-RED1, participate in the photoinduced signal transduction (Ni et al., 1998; Fairchild et al., 2000; Huq and Quail, 2002). Furthermore, some plant bHLH TFs are responsive to abiotic stresses. For example, INDUCER OF CBF EXPRESSION1 (ICE1) and ICE2 of Arabidopsis and MdCIbHLH1 of apple (Malus domestica) were suggested to be involved in the cold stress response (Chinnusamy et al., 2003; Fursova et al., 2009; Feng et al., 2012). OsbHLH148, a rice (Oryza sativa) bHLH gene, functioned in drought tolerance as a component of the jasmonate signaling module (Seo et al., 2011). Other plant bHLH genes, such as AtbHLH38, AtbHLH39, and FER-LIKE IRON DEFICIENCY-INDUCED TRANSCRIPTION FACTOR (FIT), were induced by iron deficiency; their roles in iron acquisition and heavy metal detoxification have been supported by transgenic manipulation (Yuan et al., 2008; Lingam et al., 2011; Wu et al., 2012). Recently, another two bHLH genes, AtbHLH100 and AtbHLH101, have also been shown to be key regulators of iron-deficiency responses via a FIT-independent pathway (Sivitz et al., 2012). Collectively, these findings suggest that plant bHLH TFs are involved in the regulation of plant responses to various abiotic stresses.
The bHLH TFs are a large gene family in the plant genome; there are 167 genes in Arabidopsis and 162 in rice (Toledo-Ortiz et al., 2003; Li et al., 2006). Although some bHLH TFs have been characterized, the biological functions of most of the plant bHLH TFs remain unclear, especially in nonmodel plants, such as trifoliate orange (Poncirus trifoliata), in a genus closely related to Citrus. Trifoliate orange plants are extremely cold hardy when fully acclimated (Peng et al., 2012), making it a good source of valuable genes involved in cold stress tolerance. However, no information is available on the functional identification of bHLH genes in trifoliate orange. Recently, we identified a cold-inducible homolog of Arabidopsis ICE1 in trifoliate orange, which could enhance cold tolerance when overexpressed in tobacco (Nicotiana tabacum; X.S. Huang and J.H. Liu, unpublished data). Thus, we wondered if other bHLH genes of trifoliate orange might also be involved in cold stress response. In this study, we report the identification of another bHLH gene from trifoliate orange (designated as PtrbHLH) and demonstrate that PtrbHLH plays a positive role in cold tolerance. We also show that expression levels of a number of stress-associated genes were modified in the transgenic lemon (Citrus limon), including those encoding peroxidase (POD). The PtrbHLH-overexpressing lines possessed higher POD activity, accumulated less hydrogen peroxide (H2O2), and displayed better tolerance to oxidative stresses, whereas lower POD activity and more H2O2 were detected in the RNA interference (RNAi) lines under cold stress. Exogenous application of POD inhibitors to transgenic tobacco plants elevated H2O2 accumulation and impaired cold tolerance, whereas replenishment of POD to the RNAi plants suppressed H2O2 accumulation and enhanced cold tolerance. Furthermore, PtrbHLH was shown to bind to E-box cis-elements in the promoter region of the POD gene. Overall, our data suggest that PtrbHLH might act as a positive regulator of cold tolerance, which is partially ascribed to regulating POD-mediated H2O2 scavenging.
RESULTS
Isolation and Sequence Analysis of PtrbHLH
Increasing evidence indicates that plant bHLH TFs play an important role in abiotic stress responses, but bHLH TFs have not yet been characterized in trifoliate orange. To determine whether bHLH genes from trifoliate orange function in cold tolerance, we employed an approach that combined bioinformatics database search and reverse transcription (RT)-PCR to isolate bHLH genes. RT-PCR of trifoliate orange complementary DNA (cDNA) was carried out with a pair of primers designed according to the contig merged from the retrieved sequences. The RT-PCR yielded a single fragment of 1,921 bp, which contains an intact open reading frame (ORF) of 1,464 bp. Motif scanning against MyHits (http://myhits.isb-sib.ch/cgi-bin/motif_scan) showed that the predicted product of the ORF possesses a typical bHLH domain of 48 amino acids, consisting of a basic region of 15 amino acids and two helices (14 amino acids each) that were collected by a loop of five amino acids. For the convenience of description, the gene was designated as PtrbHLH. The bHLH domain of PtrbHLH displayed a significant degree of identity with those from other plant species (Supplemental Fig. S1). The predicted PtrbHLH contains 487 amino acid residues with a calculated molecular mass of 53.6 kD and a pI of 5.30. BLAST analysis showed that at the protein level, PtrbHLH had 62% sequence identity to GmICE2 of soybean (Glycine max) and 61% to ICE2 of Arabidopsis but shared less than 46% identity with ICE1. A phylogenetic tree constructed based on the sequences of PtrbHLH and the Arabidopsis bHLHs indicates that PtrbHLH is most closely related to AtbHLH33 (ICE2; Supplemental Fig. S2).
Expression Profiles of PtrbHLH in Response to Abiotic Stresses
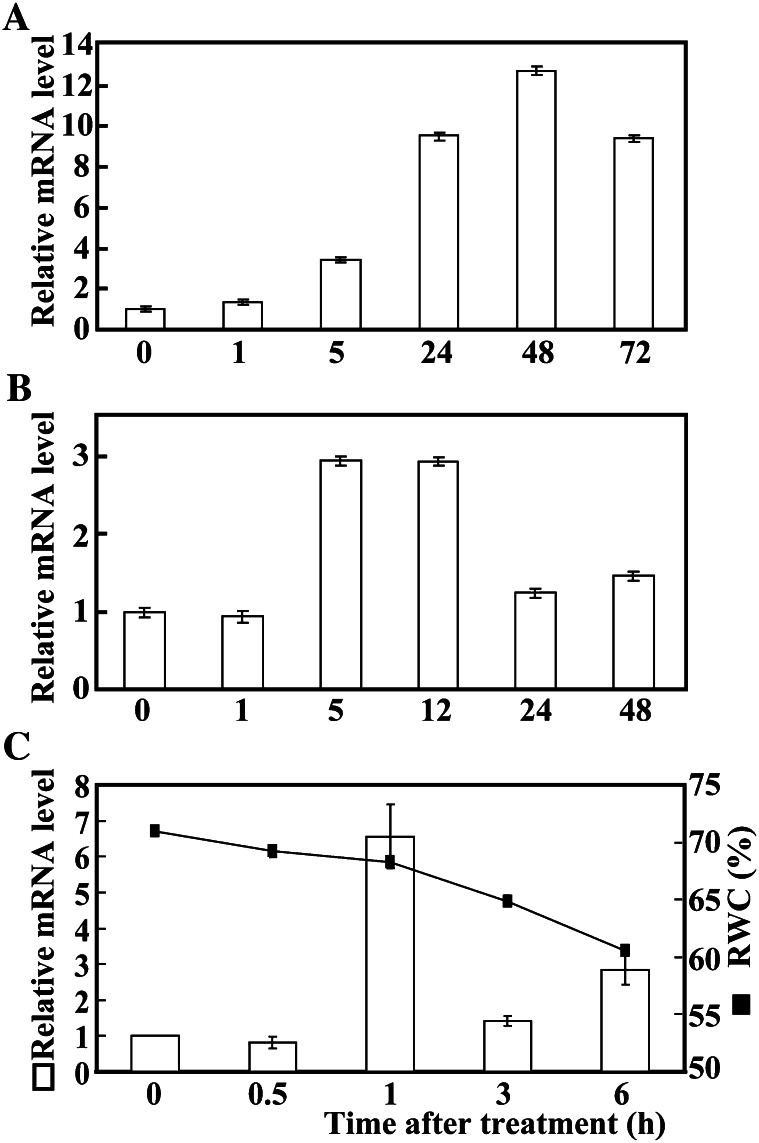
Real-time quantitative PCR (qPCR) was used to examine the expression profiles of PtrbHLH under various abiotic stresses, including cold, dehydration, and salt. Steady-state mRNA levels of PtrbHLH were elevated under all tested stresses, while the expression patterns varied. PtrbHLH transcript level increased progressively under cold stress until reaching the highest level at 48 h (greater than 12-fold induction), which exhibited a slight decrease at the last time point (Fig. 1A). Upon exposure to salt, PtrbHLH was induced by nearly 3-fold at 5 and 12 h, followed by reduction to a level slightly higher than that at the onset of the salt treatment (Fig. 1B). The decrease of relative water content in the leaves was slow within 1 h of dehydration in an ambient environment but accelerated at 3 and 6 h. Under such circumstances, PtrbHLH mRNA abundance underwent minor change at 0.5 h, increased to the highest level at 1 h, followed by reduction at the last two time points (Fig. 1C). A stronger induction of PtrbHLH transcript level was observed under cold treatment when compared with salt and dehydration.
Figure 1.
Time-course expression analysis of PtrbHLH under various stress treatments. A, Expression of PtrbHLH under cold stress for 0, 1, 5, 24, 48, and 72 h. B, Expression of PtrbHLH under salt stress for 0, 1, 5, 12, 24, and 48 h. C, Expression of PtrbHLH and relative water content (RWC) under dehydration stress (25°C and relative room humidity of 44%) for 0, 0.5, 1, 3, and 6 h. Error bars for qPCR analysis stand for sd based on four replicates. Relative water content analysis was repeated twice with three replicates.
PtrbHLH Is Localized in the Nucleus
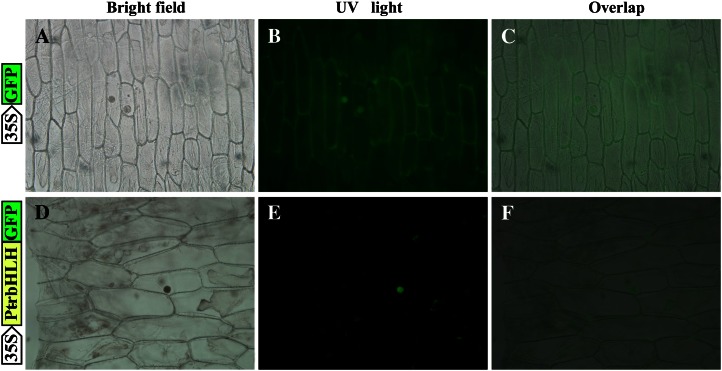
Sequence analysis showed that PtrbHLH possesses a nuclear localization signal (positions 298–315), implying that it may be a nuclear protein. To test this, PtrbHLH was in-frame translationally fused to the N terminus of the GFP reporter gene under the control of the cauliflower mosaic virus 35S promoter (CaMV 35S). The fusion plasmid and the control (GFP) were then separately transferred into onion (Allium cepa) epidermal cells, followed by monitoring of green fluorescence with a microscope. Transient expression assays indicated that GFP alone was detected in the cytoplasm and nucleus, whereas the PtrbHLH-GFP fusion protein was exclusively restricted in the nucleus (Fig. 2). Nuclear localization of PtrbHLH was further confirmed by rice protoplast transformation (data not shown). These observations suggest that PtrbHLH is subcellularly localized in the nucleus, consistent with its nature as a TF.
Figure 2.
Subcellular localization analysis of PtrbHLH. The PtrbHLH-GFP fusion protein and GFP (used as a control), driven by CaMV 35S, were separately transformed into onion epidermal cells and visualized by fluorescence microscopy. Images were taken in the representative cells expressing GFP (A and B) or the PtrbHLH-GFP fusion protein (D and E) under bright field (A and D) or dark field (B and E). The merged images are shown in C and F, respectively. [See online article for color version of this figure.]
Transactivation Analysis and Determination of the Transactivation Region
The presence of transactivation activity is an important feature for TFs. A yeast two-hybrid assay was used to determine if PtrbHLH acts as a transactivator. To this end, PtrbHLH was fused downstream of the yeast GAL4 DNA-binding domain in the pDEST32 vector to form recombinant vector pDEST32-PtrbHLH, using pDEST32 as a control vector. The two vectors were transformed into yeast cells, which were then screened on the selection medium synthetic dropout (SD)/−Leu/−His. The yeast cells transformed with either vector could grow on the synthetic dropout medium (SD/−Leu), whereas only the cells transformed with the recombinant vector survived on the selection medium alone or supplemented with 30 mm 3-amino-1,2,4-triazole (3-AT; Supplemental Fig. S3, A and B), suggesting that PtrbHLH possesses transactivation activity.
To determine which region of PtrbHLH is involved in transcriptional transactivation, yeast two-hybrid assays were carried out using intact or truncated PtrbHLH as an effector (Supplemental Fig. S3C). The transfected yeast cells harboring either the full-length PtrbHLH (FL) or the truncated version D1 (deletion of the first 43 amino acids at the N terminus) grew well on the selection medium, suggesting that the N-terminal 43 residues are dispensable for PtrbHLH’s transactivation activity. On the contrary, when the 108 (D2), 151 (D3), 198 (D4), 273 (D5), or 379 (D6) amino acids at the N terminus were deleted, no interaction was detected, which was further supported by the colony-lift filter assay (Supplemental Fig. S3, D and E). Taken together, these results demonstrate that the amino acids from positions 43 to 108 in PtrbHLH are critical for the transactivation activity of PtrbHLH.
Overexpression of PtrbHLH Increases the Cold Tolerance of Transgenic Plants
Given that cold stress resulted in the strongest accumulation of PtrbHLH transcript (Fig. 1), we speculated that PtrbHLH may play a critical role in the regulation of cold stress response. To test this hypothesis, we generated transgenic tobacco and lemon plants expressing PtrbHLH via Agrobacterium tumefaciens-mediated transformation (Supplemental Figs. S4 and S5). Two independent overexpression lines of tobacco (OE8 and OE10) and lemon (#2 and #8) with high transcript levels of PtrbHLH were used for further analysis, along with the corresponding untransformed wild type.
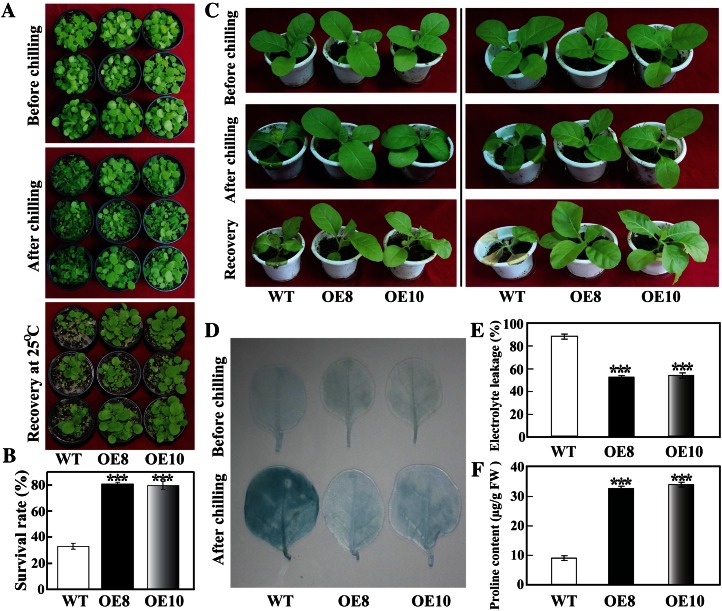
To investigate whether PtrbHLH overexpression could enhance the cold tolerance of the transgenic plants, we first analyzed the tolerance of transgenic tobacco to chilling stress (0°C) using 30-d-old seedlings. Under normal growth conditions, there was no apparent difference in plant morphology between the transgenic plants and the wild type. When treated for 23 h at 0°C, chilling injury was observed in the leaves of the wild-type plants, but the transgenic plants were affected to a lesser extent (Fig. 3A). After recovery for 10 d at ambient temperature, 80.7% of OE8 plants and 79.7% of OE10 plants survived, while the survival rate of the wild type was only 33.0% (Fig. 3B). The difference in the survival rates between the transgenic lines and the wild type was statistically significant.
Figure 3.
Cold tolerance assay of transgenic tobacco plants overexpressing PtrbHLH. A, Phenotypes of 30-d-old transgenic plants (OE8 and OE10) and the wild type (WT) before and after chilling treatment (0°C for 23 h), followed by recovery in an ambient environment for 10 d. B, Survival rates of the transgenic and wild-type plants after recovery from chilling treatment. The survival rate is the ratio of the number of live plants and the total number of plants tested under stress. C, Phenotypes of 60-d-old transgenic and wild-type plants before/after chilling treatment and after recovery. D, Cell death analysis of the leaves via trypan blue staining before and after chilling treatment. E and F, Electrolyte leakage (E) and Pro content (F) in wild-type and transgenic plants measured after chilling treatment. FW, Fresh weight. Asterisks indicate significant differences between the transgenic lines and the wild type (***P < 0.001). [See online article for color version of this figure.]
When 60-d-old tobacco plants were used for the chilling tolerance experiment, similar results were obtained: the transgenic lines were more resistant to chilling stress and displayed less damage after recovery compared with the wild type (Fig. 3C). Cell death and electrolyte leakage are reliable indicators of cell injuries caused by abiotic stresses. The leaves were comparably stained with trypan blue between the two transgenic lines and the wild type without stress treatment. However, deeper staining was visualized in the wild type after exposure to the chilling treatment as compared with the transgenic plants (Fig. 3D), indicating that cell death was more serious in the wild type. Furthermore, the transgenic plants had lower electrolyte leakage relative to the wild type (Fig. 3E). Pro, a stress-related metabolite, has been thought of as an important compatible solute contributing to osmotic adjustment, enabling plants to better tolerate the adverse impacts of abiotic stresses (Kaplan et al., 2007; Nounjan et al., 2012). In addition, Pro may function in stabilization and protection of the membranes, proteins, and enzymes (Hoque et al., 2008). Thus, Pro contents in the tested plants were measured. As can be seen in Figure 3F, the Pro contents of the two transgenic lines were significantly higher than that of the wild type after chilling stress. These results indicate that overexpression of PtrbHLH conferred enhanced chilling tolerance on the transgenic tobacco plants.
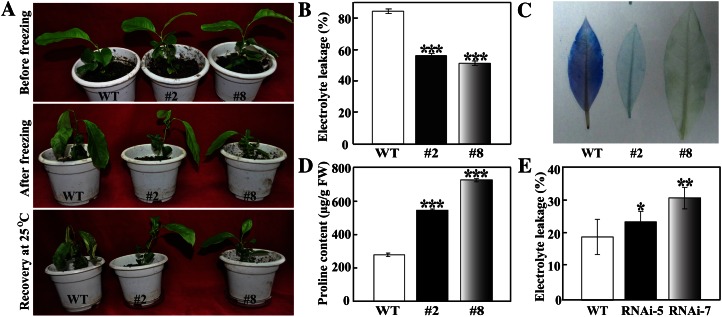
The transgenic lemon plants were morphologically indistinguishable from the untransformed control in the absence of cold stress. However, exposure to −3°C for 3 h following 48 h at 0°C caused more serious leaf drooping in the wild type than in the transgenic lines (Fig. 4A). After recovery for 5 d at 25°C, the survival rates of #2 and #8 were 72.2% and 88.9%, respectively, significantly higher than the 27.8% of the wild type. Consistent with the enhanced cold tolerance phenotype, lower levels of electrolyte leakage and weaker trypan blue staining were detected in the transgenic plants compared with the wild type (Fig. 4, B and C), indicating that the former suffered from less severe freezing-associated cell damage. Meanwhile, the Pro contents of #2 and #8 were significantly higher than that of the wild type (Fig. 4D). These results demonstrate that overexpression of PtrbHLH could enhance freezing tolerance in lemon.
Figure 4.
Cold tolerance assay of transgenic lemon or trifoliate orange RNAi plants. A, Phenotypes of lemon transgenic plants (#2 and #8) and the wild type (WT) before and after freezing treatment (−3°C for 3 h), followed by recovery in an ambient environment for 5 d. B to D, Electrolyte leakage (B), cell death (C), and Pro content (D) of wild-type lemon and transgenic plants analyzed after freezing treatment. FW, Fresh weight. E, Electrolyte leakage of wild-type trifoliate orange and RNAi lines after chilling treatment. Asterisks indicate significant differences between the transgenic lines and the wild type (*P < 0.05, **P < 0.01, ***P < 0.001). [See online article for color version of this figure.]
Suppression of PtrbHLH in Trifoliate Orange Confers Sensitivity to Cold Stress
To further elucidate the role of PtrbHLH in cold tolerance, RNAi was used to suppress the expression of PtrbHLH in trifoliate orange. Six independent transgenic lines were produced using the PtrbHLH-RNAi construct. The expression levels of PtrbHLH were dramatically decreased in two lines, implying that PtrbHLH has been successfully suppressed (Supplemental Fig. S6). These two RNAi lines, which were designated as RNAi-5 and RNAi-7, showed no difference in phenotype and growth performance under normal conditions but displayed more serious injury in the top-most leaves compared with the wild type under chilling conditions (data not shown). Electrolyte leakage in the RNAi lines (24.0% for RNAi-5 and 31.7% for RNAi-7) were significantly higher than the 19.8% of the wild type (Fig. 4E), which suggests that suppression of PtrbHLH expression may result in cold susceptibility. Taken together, the above findings demonstrate that PtrbHLH acts as a positive regulator of cold tolerance.
Overexpression of PtrbHLH in Lemon Leads to Dramatic Transcriptomic Alterations
As a TF, PtrbHLH possibly regulates an array of downstream target genes, which may account for the noticeable enhancement of chilling or freezing tolerance in the overexpression lines. To test this speculation, we carried out a preliminary microarray analysis to compare the expression profiles between wild-type lemon and one overexpression line (#8) before and after cold treatment using the Affymetrix GeneChip Citrus Genome Array, which contains 30,171 probe sets representing up to 33,879 citrus transcripts.
Using 2-fold change as a selection threshold, a total of 107 and 338 genes were up- and down-regulated, respectively, in #8 compared with the wild type without cold stress (Supplemental Fig. S7A; Supplemental Tables S1 and S2). However, 711 and 546 genes were up- and down-regulated, respectively, in line #8 compared with the wild type under cold conditions (Supplemental Fig. S7B; Supplemental Tables S3 and S4), indicating a noticeable alteration of the gene expression profiling in the transgenic plant. Among the differentially expressed genes (DEGs), 13 and 10 genes were commonly up- or down-regulated in #8 before and after cold treatment (Supplemental Fig. S7C). To validate the results from this preliminary microarray analysis, the expression of six up-regulated genes was analyzed by qPCR (Supplemental Fig. S7D). It is evident that the expression patterns of the selected genes by qPCR were largely consistent with those of the microarray data despite the difference in the absolute fold change between the two methods, suggesting that results from this preliminary microarray analysis were reliable.
Gene Ontology (GO) analysis of the DEGs in the transgenic line showed that GO terms under the category of “biological process” before stress treatment were primarily related to “metabolic process,” “cellular process,” and “response to stimulus,” while “cellular process,” “biological regulation,” and “metabolic process” predominated after cold treatment. Molecular functions of the DEGs were mainly related to “catalytic activity,” “binding,” and “transporter activity,” with the largest distribution on “catalytic activity” before and after cold stress. Cellular component included the GO terms “cell part,” “membrane-bounded organelle,” and “vesicle” with or without stress, in which “cell part” had the highest count (Supplemental Figs. S8 and S9).
It is worth mentioning that many DEGs identified from the preliminary microarray analysis are stress-responsive functional proteins that have been demonstrated to play a direct role in stress responses, such as low-temperature protein, late embryogenesis abundant protein, miraculin-like protein2, water channel-like protein, Pro-rich protein, dehydration-responsive protein, and POD. In addition, several regulatory proteins associated with stress signal transduction, such as protein phosphatase 2C and mitogen-activated protein kinase, and TFs, including NAC (for NAM, ATAF, and CUC), WRKY, and APETALA2/ethylene-responsive factor (AP2/ERF), were also activated in the overexpression line. Interestingly, nine POD genes were shown to be up-regulated in the transgenic plant after cold stress, suggesting that they may be closely associated with stress tolerance.
Analysis of POD Activity and ROS Levels in the Transgenic Plants
It is known that POD plays a key role in scavenging H2O2, one of the major ROS (Gill and Tuteja, 2010). Since several POD genes were up-regulated in the PtrbHLH overexpression line after cold stress, according to the preliminary microarray analysis, it is possible that the enhanced cold tolerance of the transgenic line might be associated with more robust detoxification of ROS. In order to test this hypothesis, we performed a series of experiments to determine whether overexpression of PtrbHLH affects POD-mediated ROS scavenging and how this regulation influences the cold tolerance of the transgenic plants.
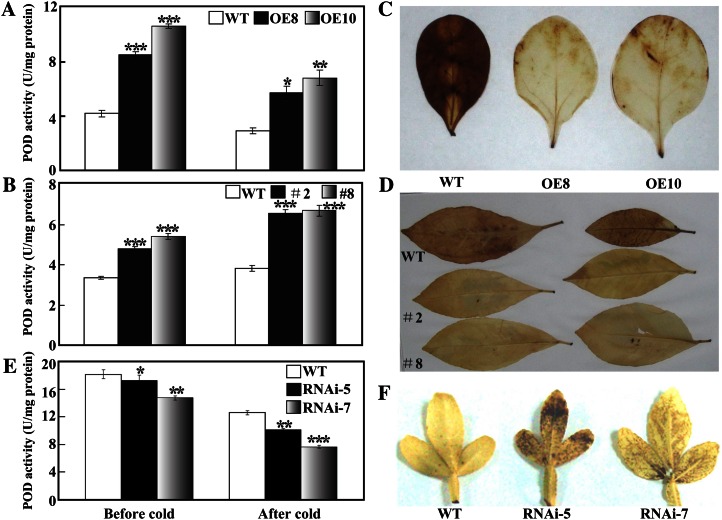
First, POD activities were measured in transgenic and wild-type plants of tobacco and lemon treated with or without cold stress (0°C for tobacco and −3°C for lemon). The transgenic lemon and tobacco plants possessed higher POD activity than their corresponding wild type under normal conditions. After the cold treatment, POD activity was increased in lemon but decreased in tobacco; however, the activities of the transgenic plants were clearly higher than those of the corresponding wild type (Fig. 5, A and B). Accumulation of H2O2 in the leaves after cold stress was examined by histochemical staining with 3,3′-diaminobenzidine (DAB). Brown-colored polymeric oxidation products were visualized in lemon and tobacco, whereas the intensity of the wild type was markedly stronger than that of the transgenic plants, suggesting that the accumulation of H2O2 was alleviated in the latter (Fig. 5, C and D). In addition, POD activity and H2O2 accumulation were examined in the trifoliate orange RNAi lines and the wild type. POD activity in the RNAi lines was lower than in the wild type before and after cold treatment (Fig. 5E). Conversely, stronger DAB staining was detected in the RNAi lines as compared with the wild type, suggesting that the accumulation of H2O2 was elevated when PtrbHLH was down-regulated (Fig. 5F). Taken together, these results demonstrate that PtrbHLH regulates the POD-mediated ROS detoxification in the transgenic plants.
Figure 5.
Analysis of POD activity and H2O2 accumulation in the transgenic plants. A to D, POD activity (A and B) and H2O2 accumulation (C and D) in transgenic tobacco (A and C) and lemon (B and D) plants overexpressing PtrbHLH before and after cold treatment, in comparison with the corresponding wild type (WT). E and F, POD activity (E) and H2O2 accumulation (F) in cold-treated leaves collected from the wild type and trifoliate orange RNAi lines. Asterisks indicate significant differences between the transgenic lines and the wild type under the same conditions (*P < 0.05, **P < 0.01, ***P < 0.001). [See online article for color version of this figure.]
Inhibition of POD Compromises the Chilling Tolerance of Transgenic Tobacco
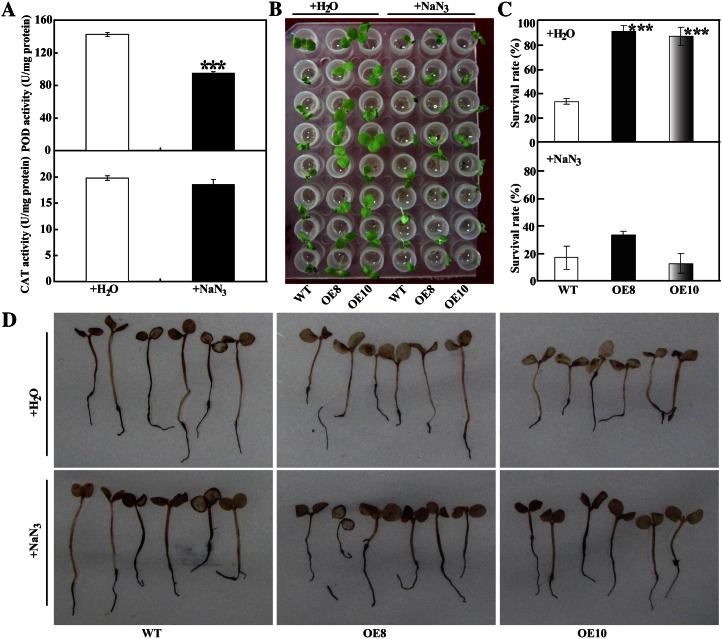
As alluded to above, the transgenic plants showed higher POD activity and conferred enhanced chilling or freezing tolerance. This raised the question of whether the POD-mediated ROS scavenging is associated with cold tolerance of the overexpression lines. To answer this question, transgenic tobacco plants were treated with 5 mm NaN3, a potential inhibitor that has been reported to suppress POD (Duarte-Vázquez et al., 2001; Zhan et al., 2003). To determine how NaN3 affected endogenous antioxidant enzymes, the activities of POD and CAT, two enzymes involved in H2O2 scavenging, were analyzed. Treatment with NaN3 led to significant reduction of POD activity relative to water treatment (from 142.9 to 95.3 units mg−1 protein), whereas CAT activity was negligibly altered (Fig. 6A). When the seedlings treated with water were exposed to cold, the transgenic plants showed remarkably better growth as compared with the wild type. NaN3 treatment stunted the growth of transgenic and wild-type plants during chilling treatment. The dramatic phenotypic difference between the transgenic lines and the wild type observed with the water treatment was abolished when the inhibitor was used, since their phenotypes were comparable to each other (Fig. 6B). Without the inhibitor application, more than 80% of the transgenic seedlings survived after cold treatment, whereas only 33.3% of the wild-type plants survived. However, when NaN3 was applied, the survival rate of the two transgenic lines was sharply decreased, being only slightly higher (OE8, 33.3%) or even lower (OE10, 12.5%) than the wild type (16.7%; Fig. 6C). This result was also reflected by the DAB staining for H2O2. When treated with water, the transgenic seedlings showed weaker DAB staining compared with the wild type; however, when NaN3 was used, the transgenic lines and the wild type displayed similar DAB staining intensity (Fig. 6D).
Figure 6.
Treatment with NaN3 compromised the chilling tolerance of transgenic tobacco plants. A, POD and CAT activities in plants treated with water or 5 mm NaN3 for 12 h. B, Phenotypes of chilling-treated transgenic and wild-type (WT) plants that were pretreated with water or NaN3. The treatment was repeated three times with eight replicates for different lines at each repetition. C and D, Survival rates (C) and H2O2 accumulation (D) of transgenic and wild-type plants analyzed at the end of the chilling treatment. Asterisks (***P < 0.001) indicate significant differences between water and NaN3 treatment (A) or between the transgenic lines and the wild type (C). [See online article for color version of this figure.]
To further consolidate the role of POD role in chilling tolerance, another two inhibitors were tested, p-chloromercurisulfonic acid (p-CMPSA) and potassium periodate, which have been shown to inhibit POD activity (Mittler and Zilinskas, 1992; Rudrappa et al., 2007). The transgenic plants were treated with either p-CMPSA or potassium periodate for 3 h before exposure to chilling stress at 0°C. After 24 h of chilling treatment, growth of the transgenic plants pretreated with the two inhibitors was worse compared with that of the plants treated with water, as more damage was observed in the former (Supplemental Fig. S10, A and D). Under chilling conditions, the transgenic plants pretreated with the inhibitors had lower survival rates and higher electrolyte leakage than those pretreated with water (Supplemental Fig. S10, B and E). Histochemical staining with DAB showed that after chilling treatment, more H2O2 accumulated, reflected by the deeper staining, in the plants pretreated with the two inhibitors when compared with water treatment (Supplemental Fig. S10, C and F). The above results together showed that the POD inhibitors impaired the capacity of ROS scavenging and substantially compromised the chilling tolerance of the transgenic plants, which implies that POD-mediated ROS homeostasis is tightly involved in the cold tolerance.
Exogenous Replenishment of POD to RNAi Plants Increased Chilling Tolerance
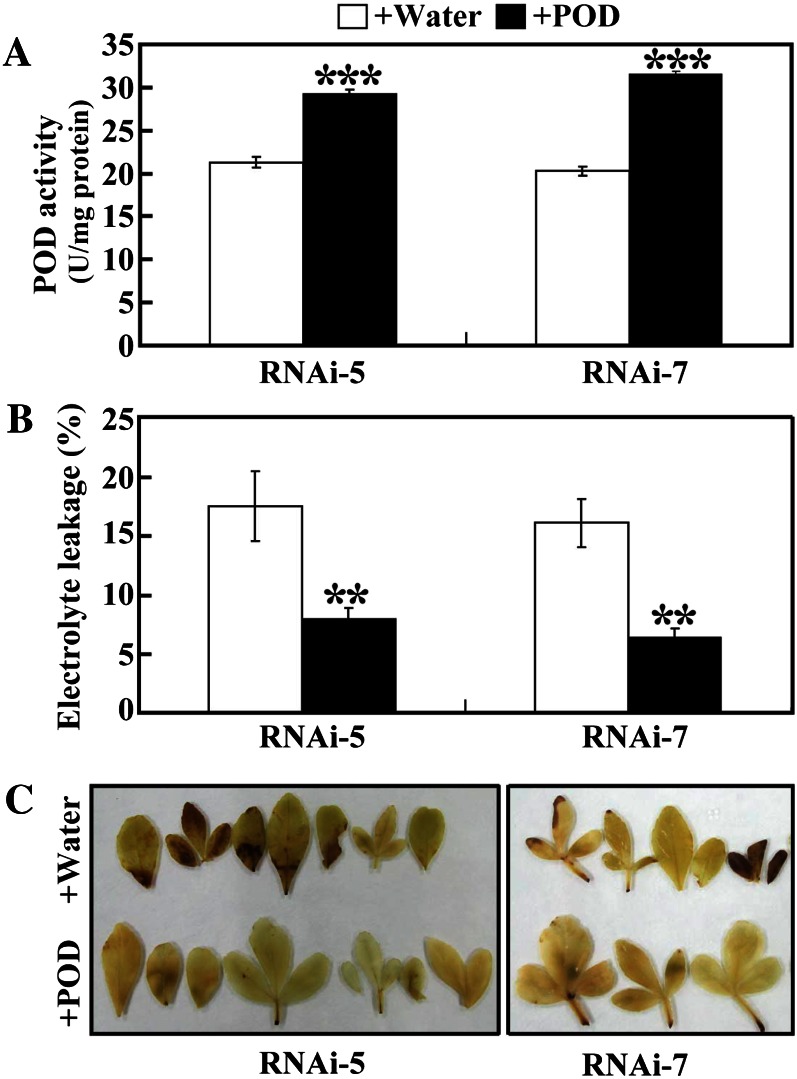
To further examine the role of POD in cold tolerance, POD was exogenously supplied to the RNAi lines (RNAi-5 and RNAi-7) before chilling stress, using water incubation as a control. Incubation of the RNAi plants in the POD solution for 12 h increased the endogenous POD activities of RNAi-5 and RNAi-7 by 38.0% and 54.7%, respectively, which was significantly higher than that of the same line incubated with water (Fig. 7A). Supplementation of POD to the RNAi plants significantly decreased the electrolyte leakage levels (Fig. 7B) and alleviated H2O2 accumulation (Fig. 7C) after chilling stress when compared with the water incubation. These data demonstrate that exogenous application of POD to the RNAi plants could enhance the chilling tolerance.
Figure 7.
Replenishment of POD to the RNAi lines (RNAi-5 and RNAi-7) enhanced chilling tolerance. A, Endogenous POD activity of the RNAi-5 and RNAi-7 plants incubated with water or POD solution for 12 h. B, Electrolyte leakage of chilling-treated RNAi-5 and RNAi-7 plants preincubated with water or POD solution. Asterisks indicate significant differences between the water treatment and POD treatment (**P < 0.01, ***P < 0.001). C, H2O2 accumulation in leaves of chilling-treated RNAi-5 and RNAi-7 plants preincubated with water or POD solution. [See online article for color version of this figure.]
PtrbHLH-Overexpressing Plants Are More Tolerant to Oxidative Stress
The above results (Fig. 5) suggest that the endogenous H2O2 was detoxified in a more efficient manner in the PtrbHLH-overexpressing lines under cold stress, leading to a lower level of oxidative stress. As POD activity was increased in the overexpression plants, in-depth work was performed to test whether they were more tolerant to oxidative stresses caused by two inducers, methyl viologen (MV) and H2O2.
When 30-d-old tobacco seedlings were treated with MV for 17 h, most of the wild-type plants died, whereas the transgenic lines exhibited better growth phenotypes (Supplemental Fig. S11A). Survival rates of OE8 and OE10 were 71.8% and 65.6%, respectively, significantly higher than the 18.8% of the wild type (Supplemental Fig. S11B). DAB staining revealed that the wild type accumulated more H2O2 in comparison with the two transgenic lines (Supplemental Fig. S11C). In addition, oxidative stress tolerance was also examined using tobacco leaf discs. Incubation of the leaf discs in water for 2 d did not cause any conspicuous difference among the tested materials. However, when the leaf discs were floated on 2% H2O2 solution for the same period, the wild-type discs became browned and necrotic, whereas some discs of the transgenic lines still remained green (Supplemental Fig. S11D). No difference in H2O2 accumulation was observed among the water-treated discs, but much deeper staining occurred in the wild type compared with the two transgenic lines after H2O2 treatment (Supplemental Fig. S11E).
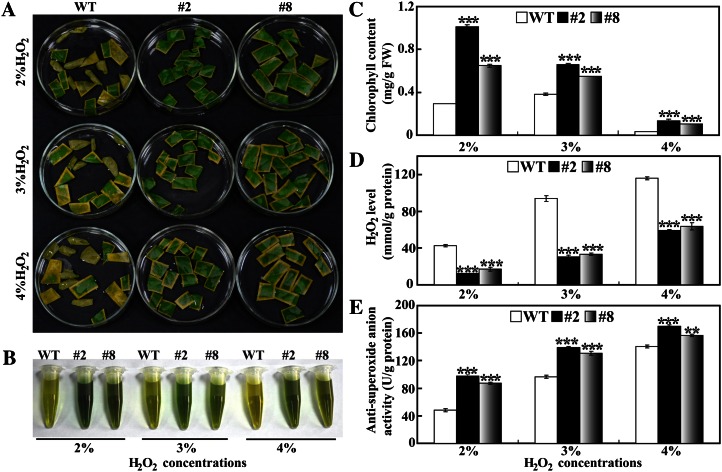
To assess the oxidative stress tolerance of the transgenic lemon, the leaves were cut into small pieces and treated for 32 h with H2O2 at different concentrations (2%, 3%, and 4%). A large proportion of the wild-type pieces were bleached and turned browned, while the color of the transgenic lines was better retained irrespective of the H2O2 concentration. Increase in the H2O2 concentration led to more serious color loss for all lines, but the transgenic lines displayed better leaf color compared with the wild type at the same concentration (Fig. 8A). Higher chlorophyll content was measured in the transgenic lines than in the wild type (Fig. 8, B and C). Endogenous H2O2 content in the leaf pieces was enhanced with the increase of exogenous H2O2 concentration, whereas the transgenic lines contained significantly lower levels of H2O2 than the wild type (Fig. 8D). On the contrary, higher antisuperoxide anion activity was observed in the transgenic lines, indicating that the levels of superoxide anion in these lines might be lower than in the wild type (Fig. 8E). These results indicated that the transgenic plants were more tolerant to the oxidative stresses.
Figure 8.
Oxidative stress tolerance assay of transgenic lemon. A, Representative photographs showing leaf pieces of transgenic (#2 and #8) and wild-type (WT) plants after H2O2 treatment. B and C, Chlorophyll extraction solutions (B) and chlorophyll contents (C) in leaf pieces of transgenic and wild-type plants. FW, Fresh weight. D and E, Endogenous H2O2 contents (D) and antisuperoxide anion activity (E) in leaf pieces of transgenic and wild-type plants. Asterisks indicate significant differences between the transgenic lines and the wild type under the same treatment (**P < 0.01, ***P < 0.001). [See online article for color version of this figure.]
PtrbHLH Binds to E-Box Elements in the Promoter Region of the POD Gene
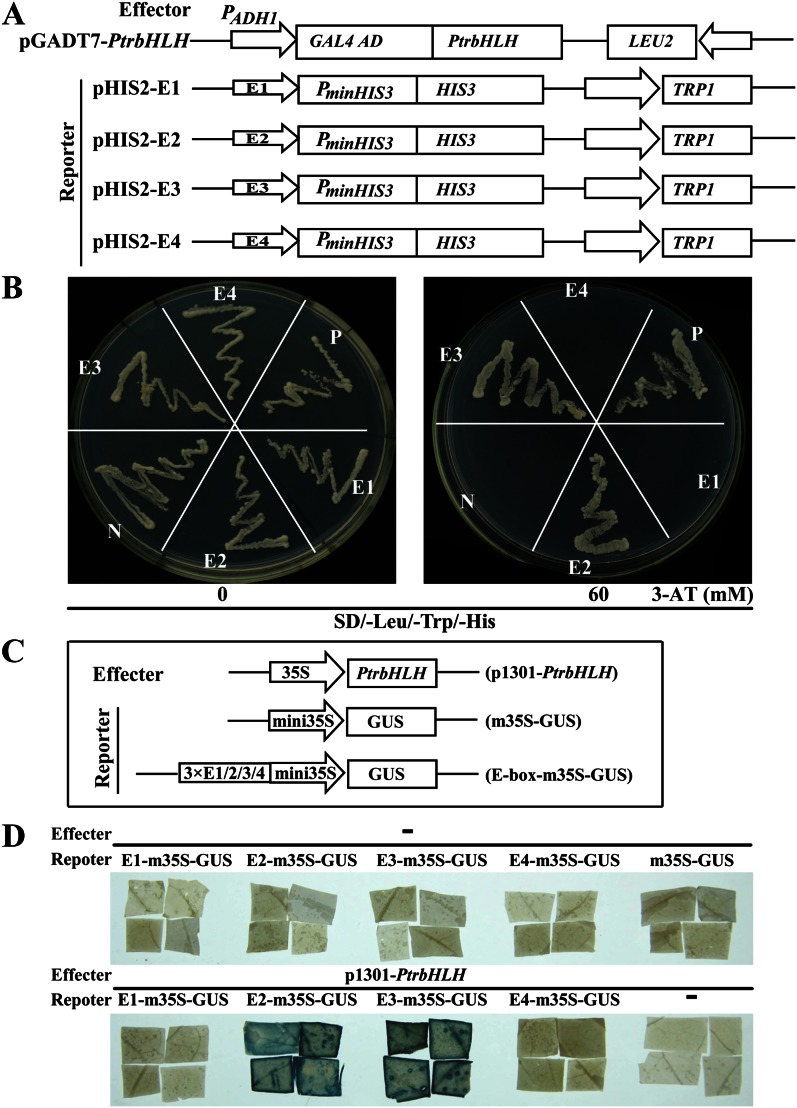
The microarray data and stress tolerance analysis suggest that POD might be one of the potential target genes that are regulated by PtrbHLH. It is known that bHLH protein can regulate gene expression by binding to the E-box (5′-CANNTG-3′) cis-element, as defined by Fisher and Goding (1992) and Meshi and Iwabuchi (1995). Bioinformatics analysis showed that the 2,320-bp promoter sequence of the POD gene (Cit.8519.1.S1_x_at) contains four potential E-box sequences at the distal upstream region. They are designated as E1 (−2,121 to −2,110; 5′-GGGCAGATGTTG-3′), E2 (−1,895 to −1,884; 5′-AAGCAAATGTAA-3′), E3 (−1,863 to −1,852; 5′-CATCAAATGGCT-3′), and E4 (−1,405 to −1,394; 5′-CATCAATTGTTT-3′), in which the putative E-box cis-elements are underlined. Therefore, we investigated whether PtrbHLH could bind to these elements using a yeast one-hybrid assay. The PtrbHLH ORF was fused to the GAL4 activation domain in the vector pGADT7-Rec2 to generate fusion plasmid pGADT7-PtrbHLH. Four synthetic oligonucleotide sequences containing triple repetitions of each fragment were separately cloned into pHIS2, giving rise to the pHIS2-E1/E2/E3/E4 reporter construct (Fig. 9A). The effector pGADT7-PtrbHLH and reporter containing any one of the four E-box elements (pHIS2-E1/E2/E3/E4) were cotransformed into yeast strain Y187, which was then plated on SD/−Leu/−Trp/−His medium supplemented with or without 60 mm 3-AT. When the transformants were selected on the medium without 3-AT, all of the yeast cells showed normal growth. However, when 3-AT was added, only the cells cotransformed with pGADT7-PtrbHLH and pHIS2-E2 or pHIS2-E3 could grow (Fig. 9B). These results indicated that the PtrbHLH protein could bind to the E-box elements in the POD promoter.
Figure 9.
Analysis of PtrbHLH binding to E-box elements in the promoter region of a POD gene. A, Schematic structures of the yeast one-hybrid effector (pGADT7-PtrbHLH) and reporter vector pHIS2-E1/E2/E3/E4 (E1, E2, E3, and E4 indicate the four putative E-box elements). B, Growth of yeast (strain Y187) cells transformed with the effector plasmid and the reporter plasmid on SD/−Leu/−Trp/−His supplemented with or without 60 mm 3-AT. N, Negative control (p53HIS2 plus pGAD-PtrbHLH); P, positive control (p53HIS2 plus pGAD-53). C, Schematic structures of the effector (p1301-PtrbHLH) and reporter vector (E-box-m35S-GUS or m35S-GUS) used for transient expression analysis. D, GUS staining of representative leaf pieces infiltrated with only reporters (top panel) or those coinfiltrated with the effector and the reporters (E-box-m35S-GUS; bottom panel). − indicates the absence of effector (top panel) or reporters (bottom panel). [See online article for color version of this figure.]
To confirm the results of the yeast one-hybrid assay, transient expression analysis was carried out using PtrbHLH as an effector. The E-box sequences were repeated three times and separately fused upstream of the minimal 35S GUS (m35S-GUS) to generate four different reporters (E-box-m35S-GUS), using m35S-GUS as a control reporter (Fig. 9C). The leaf pieces infiltrated with only the reporters (either E-box-m35S-GUS or m35S-GUS) or the effector did not show blue color after the GUS staining assay. Likewise, leaf pieces cotransformed with the effector and two reporters (E1-m35S-GUS and E4-m35S-GUS) were not stained blue, whereas blue color was observed on the leaf pieces coinfiltrated with the effector and E2-m35S-GUS or E3-m35S-GUS (Fig. 9D). A transient GUS assay, consistent with the yeast one-hybrid results, further supported the interaction between PtrbHLH and the E-box elements in the promoter region of the POD gene.
DISCUSSION
TFs regulate target genes by interacting with the relevant cis-acting elements in the promoter region, which reinforces the notion that they are desirable candidate genes for genetic transformation (Thomashow et al., 2001; Yamaguchi-Shinozaki and Shinozaki, 2005; Shinozaki and Yamaguchi-Shinozaki, 2007; Lata and Prasad, 2011; Osakabe et al., 2011; Qin et al., 2011). Plant genomes contain different families of TFs that are implicated in stress responses, and many of them have been thoroughly studied, including CBF, NAC, MYB, AP2/EREBP, bZIP, and WRKY. Conversely, the plant bHLH proteins, another important group of TFs, have been less well characterized. So far, only a few bHLH genes have been reported to play essential roles in stress tolerance (Chinnusamy et al., 2003; Fursova et al., 2009; Seo et al., 2011). As bHLH TFs constitute a large family, exploration of more stress-responsive bHLH genes will provide a better understanding of the roles of individual members of this family in the stress signaling network.
Here, we report the identification of a bHLH gene (PtrbHLH) in trifoliate orange. Although more than 100 bHLH genes are present in the Arabidopsis and rice genomes, the exact number of bHLH family members in trifoliate orange is still unknown, because the whole genome sequence is not available at present. Multiple sequence alignment suggests that the bHLH domain and zipper region of PtrbHLH share striking sequence similarities with those of the bHLH proteins from other plants; however, the N-terminal sequences flanking the bHLH domains were extensively distinct from each other. As the bHLH domain functions in DNA binding and dimerization, the presence of the highly conserved bHLH motif indicates that PtrbHLH might have biological functions similar to other bHLH genes. At the whole-polypeptide level, PtrbHLH shares approximately 46% and 62% homology with ICE1 and ICE2, respectively, suggesting that PtrbHLH is more closely related to ICE2 than ICE1. This is supported by the phylogenetic analysis, where PtrbHLH is clustered with GmICE2 and ICE2, and by the absence of two characteristic sequences of ICE1, GAQPTLFQFKA and LPPT (Badawi et al., 2008). These observations seem to suggest that PtrbHLH might be a novel bHLH gene in trifoliate orange.
When the expression patterns of stress-responsive bHLH genes were analyzed, we found that they exhibited different expression dynamics under various stresses. For example, the transcript level of ICE1 was up-regulated by cold and salt but not by dehydration (Chinnusamy et al., 2003). The mRNA abundance of AtMYC2, a bHLH gene, was up-regulated at the early stage of drought and showed a steady increase under salt stress (Abe et al., 2003). The OsbHLH148 transcript level was rapidly and transiently induced by dehydration and salt, while low temperature resulted in progressive activation (Seo et al., 2011). In our work, the PtrbHLH transcript was continuously induced by cold but only transiently by salt. Dehydration also caused a prominent up-regulation of PtrbHLH, with the largest induction at the early stage. The expression patterns of PtrbHLH are comparable to those of OsbHLH148. Although several bHLH genes were up-regulated by cold, the transcript level of CsICE1 in Camellia sinensis was not altered by chilling treatment (Wang et al., 2012). These results seem to indicate that the bHLH genes were diversely modulated under various abiotic stresses, implying that they possibly play different roles in mediating the response to specific stresses. This is not unique, since different members in a certain TF family have been demonstrated to display various responses under the same stresses (Wang et al., 2003). Disparity in the expression profiles may be ascribed to differences in genotypes, the tissues used for stress treatment, or the stress conditions. Another explanation is that the analyzed bHLH genes are different members in the large family, although they share a high degree of sequence homology in the bHLH domain. In addition, they may be controlled by various regulators upstream of the stress signaling network.
Because cold stress resulted in a stronger induction of PtrbHLH mRNA levels than salt and drought, we made efforts to elucidate the roles of PtrbHLH in cold tolerance by generating transgenic plants transformed with either overexpression or RNAi vector. PtrbHLH was transformed into tobacco, a model plant that has been extensively used for functional analysis of genes from many plants, and into lemon, a cold-sensitive perennial plant of significant agronomic value, to ensure the reliability and stability of PtrbHLH’s function in stress tolerance. It is evident that alteration of the PtrbHLH transcript amount resulted in a notable change of cold tolerance in the transgenic plants. Overexpression of PtrbHLH in both tobacco and lemon pronouncedly conferred enhanced tolerance to cold stress under chilling or freezing temperature, as measured by electrolyte leakage, survival rate, and chlorophyll content, along with phenotypic observation. However, when PtrbHLH was knocked down in trifoliate orange, the RNAi lines showed an increased sensitivity to the chilling stress compared with the wild type. These data demonstrate that PtrbHLH plays a positive regulatory role in cold tolerance.
In order to elucidate the molecular mechanisms underlying the enhanced cold tolerance, transcriptional profiles were compared between wild-type lemon and the transgenic line (#8) before and after cold stress. This preliminary microarray analysis showed that overexpression of PtrbHLH resulted in a comprehensive transcriptomic modification in the transgenic line. Of special note, PtrbHLH not only induces but also suppresses the transcript levels of many genes, implying both positive and negative impacts on the expression atlas. Such a phenomenon is not an exception, as extensive transcriptional reprogramming has been demonstrated in many reports comparing global transcript profiles between transgenic plants overexpressing a TF and their wild-type counterparts (Xiang et al., 2008; Tang et al., 2012). It is worth mentioning that the number of genes with altered expression levels under cold (711 up-regulated and 546 down-regulated) is remarkably larger than that under normal conditions (107 and 338, respectively). An explanation for this is that PtrbHLH undergoes certain unidentified modifications upon exposure to cold, which alters its regulatory mode in activation or suppression of the genes involved in the stress signaling network. Although we did not figure out the modifications in this study, earlier work on ICE1, a bHLH gene, may provide supporting evidence for this assumption. It has been shown that cold promoted the ubiquitination of ICE1 mediated by the RING finger gene, facilitating the degradation of this protein (Dong et al., 2006; Miura et al., 2011). In contrast, SIZ1 (a SUMO E3 ligase)-dependent sumoylation was induced by cold, leading to a reduction of ICE1 polyubiquination and subsequent activation/stabilization of the protein (Miura et al., 2007). Therefore, under cold stress, PtrbHLH might experience the posttranslational modification that is associated with the regulation of a wider spectrum of stress-responsive genes. Among the differentially expressed genes in the transgenic line, many have been annotated or confirmed to be involved in stress tolerance, either regulatory genes or functional ones. This suggests that PtrbHLH overexpression may extensively activate the stress-related genes, promoting the synthesis of diverse functional proteins or molecules that impart stress adaptation or tolerance, leading to enhanced cold tolerance in the transgenic plants.
ROS are highly reactive and toxic molecules that cause damage to proteins, lipids, and nucleic acids (Gill and Tuteja, 2010). ROS accumulation relies on the balance between production and scavenging. It is well known that ROS in plants are maintained at a low level under optimal growth conditions but are dramatically stimulated upon exposure to abiotic stresses (Miller et al., 2010). Overproduction of ROS results in oxidative stress, causing lipid peroxidation and membrane damage (Mittler, 2002; Suzuki et al., 2012). Plants have established a set of antioxidant defense machinery, including both enzymatic and nonenzymatic antioxidants, which works in concert to scavenge ROS and protect plant cells from oxidative stress (Gill and Tuteja, 2010; Miller et al., 2010). POD, one of the important antioxidant enzymes, plays a critical role in scavenging H2O2. In the preliminary microarray analysis, the genes encoding POD were overrepresented in the transgenic line compared with the wild type under cold stress. Meanwhile, higher POD activity was detected in the overexpression plants, implying that they might possess a more efficient antioxidant network than the wild type. This is corroborated by the accumulation of lower amounts of H2O2 in the transgenic lines. As an excess of H2O2 in plant cells is extremely detrimental to cellular functions, these findings indicate that the transgenic lines suffered from less severe oxidative stress under cold conditions, consistent with the reduced levels of electrolyte leakage, a parameter for determining the magnitude of membrane damage. Earlier studies have shown that ROS accumulation is closely associated with cold stress (Suzuki and Mittler, 2006); it is thus conceivable that a robust ROS scavenging system will be effective for removing the ROS produced under cold stress. In this regard, the enhanced cold tolerance of the transgenic plants overexpressing PtrbHLH might be ascribed, at least in part, to maintaining intracellular ROS pools at low levels. This notion is supported by the following arguments. First, treatment with the three inhibitors that are reported to suppress POD (Mittler and Zilinskas, 1992; Duarte-Vázquez et al., 2001; Zhan et al., 2003; Rudrappa et al., 2007) led to an elevation of H2O2 levels and compromised the chilling tolerance of transgenic tobacco. The difference in stress tolerance between the transgenic lines and the wild type was largely diminished compared with that under water treatment. Second, the trifoliate orange PtrbHLH-RNAi lines exhibited lower POD activity, higher H2O2 levels, and larger electrolyte leakage under chilling stress, indicating that suppression of PtrbHLH rendered the RNAi plants more susceptible to the cold stress. The plant phenotype or parameters of the RNAi lines were opposite to what was seen with the overexpression lines. In contrast, exogenous supplementation of POD to the RNAi line alleviated the H2O2 accumulation and elevated the chilling tolerance. These results indicate that PtrbHLH functions to confer cold tolerance through regulating POD-mediated ROS scavenging in the transgenic plants.
It is known that oxidative stress arises from either excessive ROS accumulation under abiotic stresses or specific treatment with chemicals like MV or H2O2 (Gill and Tuteja, 2010; Miller et al., 2010; Ning et al., 2010). As the transgenic plants had higher POD activity, we were interested in finding out their performance under treatment with MV or H2O2. The transgenic plants of tobacco and lemon were damaged to a less serious degree by the two agents, suggesting that they were more resistant to the oxidative stresses than the wild type. Lower levels of H2O2 were accumulated in the transgenic plants treated with these agents, indicating that the H2O2 produced under the oxidative stress was detoxified faster, which may be ascribed to the presence of higher POD activities. This experiment provided a line of convincing evidence supporting the enhanced oxidative stress tolerance in the transgenic plants overexpressing PtrbHLH.
The bHLH TFs have been shown to regulate gene expression through binding to the conserved cis-element E-box or G-box (5′-CANNTG-3′; Meshi and Iwabuchi, 1995) in the promoters of related genes involved in various physiological processes. For example, ICE1 was shown to bind to the consensus sequence of the CBF3 promoter region and regulated CBF3 expression (Chinnusamy et al., 2003). Recently, FLOWERING BHLH1 was shown to preferentially bind to the E-box of the CONSTANS gene and regulated flowering in Arabidopsis (Ito et al., 2012). In this study, four putative E-box variants were predicted in the promoter region of the POD gene, but PtrbHLH only interacted with two (E2 and E3) of them. The presence of the E-box variants in the promoter region has been assumed to be a strategy for offering appropriate binding sites for different bHLH proteins (Badawi et al., 2008; Carretero-Paulet et al., 2010). This proposition is comprehensible, as bHLH proteins exist as a large family with numerous members; each member in the family may have its own or a preferential binding target, although common binding motifs have been reported for different TFs in the same family. As a matter of fact, the specific interaction between bHLH protein and its target elements has been reported previously. For example, TaICE87 and TaICE41, two bHLH genes in wheat (Triticum aestivum), were shown to interact with different cis-elements in the promoter region of TaCBFIVd-B9 (Badawi et al., 2008). Binding specificity may be caused by variation of the flanking sequences surrounding the cis-element or by specific sequences in the TF (Fisher and Goding, 1992). Although we did not decipher the mechanisms of the binding preference in this study, our survey of the interaction between PtrbHLH and the putative E-box elements revealed that PtrbHLH acts as a transcription activator and regulates the expression of the POD gene through binding to E-box elements. This presumably explains the up-regulation of POD transcript levels and the higher POD activity in the transgenic plants overexpressing the PtrbHLH gene.
CONCLUSION
Our data demonstrate that PtrbHLH is a stress-responsive TF and plays a positive role in cold and oxidative stress tolerance. Activation of the POD-mediated scavenging of H2O2 produced under stress is possibly responsible for the enhanced cold tolerance rendered by PtrbHLH. It should be mentioned that there might be other target genes under the control of PtrbHLH. We noticed that some genes involved in ROS scavenging, such as APX, were also up-regulated in the overexpression line. Efforts are ongoing to investigate whether these genes are also regulated by PtrbHLH to gain a better understanding of the regulatory mechanisms of the TF.
MATERIALS AND METHODS
Plant Materials and Stress Treatments
Eight-month-old uniform and healthy shoots detached from 2-year-old trifoliate orange (Poncirus trifoliata) seedlings grown in a nursery at Huazhong Agricultural University were used to determine the expression patterns of PtrbHLH under cold and salt stresses. For cold stress, the shoots were put in the chamber set at 4°C for 0, 1, 5, 24, 48, and 72 h. Salinity treatment was applied by placing the shoots in 200 mm NaCl solution for 0, 1, 5, 12, 24, and 48 h. For dehydration stress, the aerial parts of 7-month-old seedlings were put on filter paper above a work bench and dried at 25°C (in autumn, with relative humidity of 44.0%) for 0, 0.5, 1, 3, and 6 h. Relative water content in the leaves was measured at each time point of the dehydration treatment using a Moisture Balance (Mettler; HG63) according to the manufacturer’s instructions. Cold treatment was carried out in a growth chamber in the dark, while salt and dehydration treatments were performed in an ambient environment with natural daylight. The stress treatments were initiated at 9 am of the first day of the experiment, and the fully expanded leaves on the top of the shoots were collected at the indicated time points of each treatment, immediately frozen in liquid nitrogen, and stored at −80°C for further analysis.
Gene Isolation and Sequence Analysis
The citrus database HarvEST (http://harvest.ucr.edu) was searched using a keyword “ICE1,” which yielded two outputs. The first is composed of a full-length sequence with high identity to the ICE1 gene, while the second contains a total of 17 EST sequences that could be merged into one contig with a complete ORF. Based on the contig sequence, GSP1 primers (unless otherwise stated, all primers are listed in Supplemental Table S5) were designed for RT-RCR amplification of trifoliate orange cDNA prepared from leaves treated for 48 h at 4°C. PCR, in a total volume of 50 µL, consisted of 250 ng of cDNA, 1× TransStart FastPfu buffer, 0.25 mm deoxyribonucleotide triphosphate, 2.5 units of TransStart FastPfu DNA polymerase, and 0.4 µm of each primer. RT-PCR was performed with the following protocol: one cycle at 95°C for 2 min, 40 cycles of 95°C for 20 s, 55°C for 20 s, 72°C for 60 s, and 72°C for 5 min. The PCR product was recovered, cloned into pMD18-T vector (TaKaRa), and sequenced (United Gene). Sequence analysis was done by the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/), and prediction of the bHLH domain was performed on Motif scan (http://myhits.isb-sib.ch/cgi-bin/motif_scan). The sequence alignment was carried out using ClustalW, and the phylogenetic tree was constructed by the neighbor-joining method using MEGA 4.0.
Gene Expression Analysis by Real-Time qPCR
Real-time qPCR was applied to evaluate transcription levels of PtrbHLH under different treatments. Total RNA was extracted from the samples using Trizol reagent (Invitrogen) according to the manufacturer’s instructions. After treatment with RNase-free DNase I, 1 µg of RNA was synthesized into cDNA with the RevertAid First Strand cDNA Synthesis Kit (MBI). The 10-µL qPCR solutions contained 5 µL of SYBR-Green PCR Master Mix (Applied Biosystems), 0.25 µm forward and 0.25 µm reverse primers (GSP2), and 50 ng of cDNA template. Quadruple qPCR was performed on an ABI 7500 Real-Time PCR System (Applied Biosystems) using the following cycling regime: 50°C for 2 min, 95°C for 10 min, followed by 40 cycles of 95°C for 15 s and 58°C for 1 min. The Actin gene (BQ623464.1) was amplified in parallel as an internal reference gene. The relative expression levels of the amplified products were calculated based on the comparative threshold cycle method (Livak and Schmittgen, 2001). Transcript abundance of the examined gene was normalized against those of the reference gene.
Subcellular Localization Analysis
To determine the subcellular localization of PtrbHLH, pMD18-T containing PtrbHLH was amplified with GSP4; the amplicon was digested with NcoI and SpeI and subcloned into the pCAMBIA1302 vector containing the GFP reporter gene to produce fusion construct pCAMBIA1302-PtrbLHLH-GFP under the control of the CaMV 35S. The fusion construct and the control vector (pCAMBIA1302) were separately transferred into Agrobacterium tumefaciens strain EHA105 by heat shock. A. tumefaciens-mediated transformation of onion (Allium cepa) epidermal cells was done as described by Huang et al. (2011). The transformed onion cells were observed with a universal fluorescence microscope (Olympus; BX61).
Transcriptional Activation Assay
For the transactivation assay, intact or deleted (D1–D6) PtrbHLH ORFs were amplified with corresponding primers (GSPF1–GSPF7) and inserted into the EcoRI and XhoI sites of pENTR3C (Invitrogen). The recombinant constructs pENTR3C-PtrbHLH(FL) or pENTR3C-PtrbHLH(D1) to pENTR3C-PtrbHLH(D6) were then fused in frame downstream of the yeast GAL4 DNA-binding domain in pDEST32 (Invitrogen). The yeast strain MaV203 (Invitrogen) containing the HIS3 and LacZ reporter genes was independently transformed with the plasmids (recombinant constructs or pDEST32) according to the manufacturer’s instructions. The transformants were plated on SD/−Leu or SD/−Leu/−His supplemented with 3-AT (0 or 30 mm) to test the expression of the reporter gene HIS3. The colony-lift filter assay using 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside was carried out based on the instruction manual (Invitrogen) to examine expression of the reporter gene LacZ.
Generation of Transgenic Plants by A. tumefaciens-Mediated Transformation
PtrbHLH cDNA was amplified with specific primers (GSP1) containing SalI or KpnI restriction sites. The PCR product was confirmed, and digested by SalI and KpnI, before ligation into the XhoI/KpnI-linearized binary vector pBI121 driven by the CaMV 35S. For the construction of PtrbHLH-RNAi, a fragment of the PtrbHLH ORF (432 bp, 37–468 bp) was amplified by PCR using primers of GSP3, which was then introduced into the pHGRV vector. All of the constructs were transferred into A. tumefaciens strain EHA105. The overexpression vector was used to transform tobacco (Nicotiana tabacum) and lemon (Citrus limon), and the RNAi vector was transformed into trifoliate orange. Tobacco transformation was performed based on a leaf disc method (Horsch et al., 1985; Huang et al., 2010), while lemon and trifoliate orange were transformed using shoot segments as explants (Fu et al., 2011). Kanamycin-resistant plants were identified by PCR using two pairs of primers (NPTII and CaMV 35S-PtrbHLH); only those yielding the expected PCR fragments by both primers were regarded as positive. In addition, overexpression of PtrbHLH in two of the putative lines was examined by semiquantitative RT-PCR using specific primers (GSP5) using the protocol described by Huang et al. (2010). The reference genes, Actin and Ubiquitin, were used as internal controls for lemon/trifoliate orange and tobacco, respectively. Positive tobacco T0 plants were transplanted to soil and grown in the greenhouse for the collection of T1 and T2 seeds, while lemon and trifoliate orange transgenic plants were vegetatively multiplied using stem segments of in vitro seedlings.
Cold Tolerance Assays of the Transgenic Plants
Seeds of tobacco transgenic lines and the wild type were sown in plastic pots filled with a 1:1 mixture of vermiculite and soil under a photoperiod of 16 h of light/8 h of dark at 25°C. To test the cold tolerance, 30- or 60-d-old tobacco plants were directly exposed to 0°C for 23 h without cold acclimation (Hegedüs et al., 2004), followed by recovery at 25°C for 10 or 5 d, respectively. Electrolyte leakage, cell death, Pro content, H2O2 accumulation, and POD activity were assayed after the chilling treatment was stopped, while survival rate was evaluated after the recovery. In another experiment, two- or three-leaf-stage seedlings were hydroponically grown in inhibitor-containing solutions (5 mm NaN3 for 12 h, 0.5 mm p-CMPSA for 3 h, or 5 mm potassium periodate for 3 h), using water as a control, before cold treatment at 0°C for 24 h, followed by examination of survival rate and analysis of H2O2 accumulation. Tobacco leaves were harvested after NaN3 treatment to analyze its effect on the activities of endogenous POD and CAT.
Lemon plants were transplanted to soil pots and grown for 60 d. Transgenic and wild-type plants were first acclimated at 0°C for 48 h and then treated at −3°C for 3 h (Mohammadian et al., 2012), followed by recovery at 25°C for 5 d. Electrolyte leakage, cell death, POD activity, and Pro content were assessed after freezing treatment. Uniform seedlings from 150-d-old wild-type and RNAi lines of trifoliate orange were exposed to 0°C for 5 d, followed by analysis of electrolyte leakage, POD activity, and H2O2 accumulation. In order to test the role of POD in cold tolerance, the RNAi lines (RNAi-5 and RNAi-7) were incubated in either water or 50 mg L−1 POD for 12 h and then subjected to chilling treatment at 0°C for 24 h. POD activity of the RNAi lines was measured before chilling treatment, while the leaves collected after cold stress were subjected to analysis of electrolyte leakage and in situ H2O2 accumulation.
Oxidative Stress Tolerance Assays of the Transgenic Plants
For the oxidative stress test, tobacco seedlings at the two-leaf stage were treated with 8 µm MV for 17 h in an ambient environment (28°C). In addition, leaf discs prepared from fully expanded leaves of 60-d-old plants using a cork borer were incubated in 2% H2O2 or water for 2 d. After the treatment, photographs were taken and H2O2 accumulation was determined, while survival rate was measured for the MV treatment. As for lemon, leaf pieces of wild-type and transgenic plants were incubated in H2O2 (2%, 3%, and 4%) for 32 h. The leaf pieces collected at the end of the treatment were used for measurement of total chlorophyll content, H2O2 level, and antisuperoxide anion activity (an index indicating superoxide anion levels).
Physiological Measurement and Histochemical Staining
Electrolyte leakage and chlorophyll content were measured based on the procedures described in earlier studies (Huang et al., 2010; Wang et al., 2011). Pro content was spectrophotometrically determined according to Zhao et al. (2009). Antioxidant enzyme (POD and CAT) activity, H2O2 level, and antisuperoxide anion activity were quantified using the relevant detection kits (Nanjing Jiancheng Bioengineering Institute) based on the manufacturer’s instructions. In situ H2O2 accumulation and cell death were examined via histochemical staining by DAB (Shi et al., 2010; Huang et al., 2011) and trypan blue (Pogány et al., 2009), respectively.
Preliminary Microarray Analysis
Transcriptional profiling of wild-type lemon and transgenic line #8 was carried out by microarray analysis using the Affymetrix GeneChip Citrus Genome Array. Two biological replicates were used for each of the genotypes, the wild type and #8, under normal growth conditions, while three biological replicates were used for each of them after cold treatment (0°C for 6 h). Each biological replicate consisted of three to four plants, which were mixed after collection to generate one independent sample pool. One biological replicate was used to hybridize with one chip. Total RNA was extracted from every biological replicate using Trizol reagent (Invitrogen). Synthesis of cDNA, probe labeling, chip hybridization, washing, staining, and scanning were performed according to the Affymetrix GeneChip Expression Analysis Technical Manual (BGI). Data analysis was carried out as described by Zhu et al. (2011). In brief, the scanned images from the arrays were processed using GeneChip Operating Software (GCOS 1.2; Affymetrix) with default settings to generate raw data intensity files (CEL files). The CEL files were then imported into the Bioconductor system (R software) using the Affy package, and robust multiarray analysis (Bolstad et al., 2003) was applied to preprocess and normalize the raw data to calculate the expression values. The signal ratios of each gene were compared between the wild type and #8 under the same growth conditions; the probe sets with 2-fold change (either up- or down-regulation) in #8 were considered as DEGs (Ning et al., 2010; Tang et al., 2012). Function analysis of the DEGs and GO term annotation were carried out with the Blast2go software. The microarray results were verified by qPCR analysis of the expression levels of six genes with the protocol mentioned above except using primers specific to the examined genes (Supplemental Table S5).
Yeast One-Hybrid Assays in Yeast
Putative E-box sequences were identified in the promoter region of the POD gene (Cit.8519.1.S1_x_at) based on the citrus genome sequence. Yeast one-hybrid assay was performed to investigate whether PtrbHLH could interact with the E-box elements. The full-length ORF of PtrbHLH was amplified by PCR using primers containing NdeI and BamHI restriction sites (GSP6) and fused to the GAL4 activation domain in the vector pGADT7-Rec2 (Clontech) to create the fusion protein pGADT7-PtrbHLH (effector vector). A 42-bp oligonucleotide sequence containing triple tandem repeats of each E-box sequence was cloned into pHIS2 to generate pHIS2-E-box (reporter vector). Both the effector vector and reporter vector were cotransformed into yeast strain Y187 following the manufacturer’s instructions (Clontech). The transformed cells were selected on SD/−Leu/−Trp/−His medium supplemented with or without 60 mm 3-AT to examine protein-DNA interaction.
Transient Expression Analysis
The transient expression assay was carried out to confirm the interaction between PtrbHLH and the E-box elements using a method modified from that of Li et al. (2010). To this end, the minimal-100 CaMV 35S (m35S) was PCR amplified from the pBI221 vector using primers containing HindIII or BamHI restriction sites (GSP7), inserted upstream of the GUS gene of pCAMBIA1391 to generate the m35S-GUS recombinant construct. Four sequences containing three repetitions of each E-box element, added with PstI and EcoRV restriction sites, were inserted upstream of the m35S sequence of pBI221 to generate E-box-m35S-GUS-pBI221, which was double digested by PstI and XbaI and subcloned into pCAMBIA1301 to form a recombinant construct of E-box-m35S-GUS-pCAMBIA1301. Then, the constructs were inserted into the pCAMBIA1391 vector at the HindIII and BamHI sites to produce the reporter constructs (E-box-m35S-GUS), which were mobilized into A. tumefaciens (GV3101). The PtrbHLH overexpression construct was used as the effector. A. tumefaciens-mediated transformation of tobacco leaf pieces excised from 70-d-old seedlings was performed as described previously (Huang et al., 2010) with the exception of using the A. tumefaciens cells harboring the effector and the reporter at a 1:1 ratio, while the infection with m35S-GUS or the effector alone was used as the control. After coculture in the dark for 3 d at 25°C, the leaf pieces were subjected to GUS activity determination via the histochemical method, as described previously (Jefferson et al., 1987). The transient expression assay was repeated three times, giving the same results each time.
Statistical Analysis
All experimental data are averages of at least three independent replicates. The data were statistically evaluated by applying Fisher’s lsd test, in the ANOVA program of SAS (SAS Institute), taking P < 0.05 (*), P < 0.01 (**), and P < 0.001 (***) as significantly different.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession number JX512645.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Amino acid sequence alignment of bHLH domains from plants.
Supplemental Figure S2. Phylogenetic tree constructed using PtrbHLH and Arabidopsis bHLHs.
Supplemental Figure S3. Transactivation assay of PtrbHLH in yeast.
Supplemental Figure S4. Generation and molecular identification of transgenic tobacco plants overexpressing PtrbHLH.
Supplemental Figure S5. Generation and molecular identification of transgenic lemon plants overexpressing PtrbHLH.
Supplemental Figure S6. Generation and molecular identification of trifoliate orange RNAi plants.
Supplemental Figure S7. Preliminary microarray analysis of wild-type lemon and transgenic line #8.
Supplemental Figure S8. GO analysis of the differentially expressed genes before cold treatment.
Supplemental Figure S9. GO analysis of the differentially expressed genes after cold treatment.
Supplemental Figure S10. Treatment with p-CMPSA and potassium periodate compromised the chilling tolerance of transgenic tobacco plants.
Supplemental Figure S11. Oxidative stress tolerance assay of the transgenic tobacco plants.
Supplemental Table S1. List of up-regulated genes in the transgenic line under normal conditions.
Supplemental Table S2. List of down-regulated genes in the transgenic line under normal conditions.
Supplemental Table S3. List of up-regulated genes in the transgenic line under cold stress.
Supplemental Table S4. List of down-regulated genes in the transgenic line under cold stress.
Supplemental Table S5. Primers used in this study.
Acknowledgments
We are grateful to Dr. Yan Xu (Northwest A&F University) for the generous gift of pBI221 used for transient expression analysis. Thanks are extended to Dr. Kevin Folta (University of Florida), Dr. Shunyuan Xiao (University of Maryland), and Dr. Randall Niedz (U.S. Department of Agriculture) for their valuable suggestions on the improvement of the manuscript.
Glossary
- ROS
reactive oxygen species
- TF
transcription factor
- H2O2
hydrogen peroxide
- RNAi
RNA interference
- RT
reverse transcription
- ORF
open reading frame
- qPCR
quantitative PCR
- CaMV 35S
cauliflower mosaic virus 35S promoter
- SD
synthetic dropout
- DEGs
differentially expressed genes
- GO
Gene Ontology
- POD
peroxidase
- DAB
3,3′-diaminobenzidine
- p-CMPSA
p-chloromercurisulfonic acid
- MV
methyl viologen
- cDNA
complementary DNA
- 3-AT
3-amino-1,2,4-triazole
References
- Abe H, Urao T, Ito T, Seki M, Shinozaki K, Yamaguchi-Shinozaki K. (2003) Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) function as transcriptional activators in abscisic acid signaling. Plant Cell 15: 63–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agarwal PK, Agarwal P, Reddy MK, Sopory SK. (2006) Role of DREB transcription factors in abiotic and biotic stress tolerance in plants. Plant Cell Rep 25: 1263–1274 [DOI] [PubMed] [Google Scholar]
- Badawi M, Reddy YV, Agharbaoui Z, Tominaga Y, Danyluk J, Sarhan F, Houde M. (2008) Structure and functional analysis of wheat ICE (inducer of CBF expression) genes. Plant Cell Physiol 49: 1237–1249 [DOI] [PubMed] [Google Scholar]
- Bernhardt C, Lee MM, Gonzalez A, Zhang F, Lloyd A, Schiefelbein J. (2003) The bHLH genes GLABRA3 (GL3) and ENHANCER OF GLABRA3 (EGL3) specify epidermal cell fate in the Arabidopsis root. Development 130: 6431–6439 [DOI] [PubMed] [Google Scholar]
- Bolstad BM, Irizarry RA, Astrand M, Speed TP. (2003) A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics 19: 185–193 [DOI] [PubMed] [Google Scholar]
- Buck MJ, Atchley WR. (2003) Phylogenetic analysis of plant basic helix-loop-helix proteins. J Mol Evol 56: 742–750 [DOI] [PubMed] [Google Scholar]
- Carretero-Paulet L, Galstyan A, Roig-Villanova I, Martínez-García JF, Bilbao-Castro JR, Robertson DL. (2010) Genome-wide classification and evolutionary analysis of the bHLH family of transcription factors in Arabidopsis, poplar, rice, moss, and algae. Plant Physiol 153: 1398–1412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinnusamy V, Ohta M, Kanrar S, Lee BH, Hong X, Agarwal M, Zhu JK. (2003) ICE1: a regulator of cold-induced transcriptome and freezing tolerance in Arabidopsis. Genes Dev 17: 1043–1054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinnusamy V, Zhu J, Zhu JK. (2006) Salt stress signaling and mechanisms of plant salt tolerance. Genet Eng (N Y) 27: 141–177 [DOI] [PubMed] [Google Scholar]
- Cook D, Fowler S, Fiehn O, Thomashow MF. (2004) A prominent role for the CBF cold response pathway in configuring the low-temperature metabolome of Arabidopsis. Proc Natl Acad Sci USA 101: 15243–15248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delhaize E, Ma JF, Ryan PR. (2012) Transcriptional regulation of aluminum tolerance genes. Trends Plant Sci 7: 341–348 [DOI] [PubMed] [Google Scholar]
- Dong CH, Agarwal M, Zhang YY, Xie Q, Zhu JK. (2006) The negative regulator of plant cold responses, HOS1, is a RING E3 ligase that mediates the ubiquitination and degradation of ICE1. Proc Natl Acad Sci USA 103: 8281–8286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duarte-Vázquez MA, García-Almendárez BE, Regalado C, Whitaker JR. (2001) Purification and properties of a neutral peroxidase isozyme from turnip (Brassica napus L. var. Purple Top White Globe) roots. J Agric Food Chem 49: 4450–4456 [DOI] [PubMed] [Google Scholar]
- Fairchild CD, Schumaker MA, Quail PH. (2000) HFR1 encodes an atypical bHLH protein that acts in phytochrome A signal transduction. Genes Dev 14: 2377–2391 [PMC free article] [PubMed] [Google Scholar]
- Feng XM, Zhao Q, Zhao LL, Qiao Y, Xie XB, Li HF, Yao YX, You CX, Hao YJ. (2012) The cold-induced basic helix-loop-helix transcription factor gene MdCIbHLH1 encodes an ICE-like protein in apple. BMC Plant Biol 12: 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher F, Goding CR. (1992) Single amino acid substitutions alter helix-loop-helix protein specificity for bases flanking the core CANNTG motif. EMBO J 11: 4103–4109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu XZ, Khan EU, Hu SS, Fan QJ, Liu JH. (2011) Overexpression of the betaine aldehyde dehydrogenase gene from Atriplex hortensis enhances salt tolerance in the transgenic trifoliate orange (Poncirus trifoliata L. Raf.). Environ Exp Bot 74: 106–113 [Google Scholar]
- Fursova OV, Pogorelko GV, Tarasov VA. (2009) Identification of ICE2, a gene involved in cold acclimation which determines freezing tolerance in Arabidopsis thaliana. Gene 429: 98–103 [DOI] [PubMed] [Google Scholar]
- Gill SS, Tuteja N. (2010) Reactive oxygen species and antioxidant machinery in abiotic stress tolerance in crop plants. Plant Physiol Biochem 48: 909–930 [DOI] [PubMed] [Google Scholar]
- Godiard L, Lepage A, Moreau S, Laporte D, Verdenaud M, Timmers T, Gamas P. (2011) MtbHLH1, a bHLH transcription factor involved in Medicago truncatula nodule vascular patterning and nodule to plant metabolic exchanges. New Phytol 191: 391–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golldack D, Lüking I, Yang O. (2011) Plant tolerance to drought and salinity: stress regulating transcription factors and their functional significance in the cellular transcriptional network. Plant Cell Rep 30: 1383–1391 [DOI] [PubMed] [Google Scholar]
- Hegedüs A, Erdei S, Janda T, Tóth E, Horváth G, Dudits D. (2004) Transgenic tobacco plants overproducing alfalfa aldose/aldehyde reductase show higher tolerance to low temperature and cadmium stress. Plant Sci 166: 1329–1333 [Google Scholar]
- Hoque MA, Banu MNA, Nakamura Y, Shimoishi Y, Murata Y. (2008) Proline and glycinebetaine enhance antioxidant defense and methylglyoxal detoxification systems and reduce NaCl-induced damage in cultured tobacco cells. J Plant Physiol 165: 813–824 [DOI] [PubMed] [Google Scholar]
- Horsch RB, Fry JE, Hoffmann NL, Eichholtz D, Rogers SG, Fraley RT. (1985) A simple and general method for transferring genes into plants. Science 227: 1229–1231 [DOI] [PubMed] [Google Scholar]
- Huang XS, Liu JH, Chen XJ. (2010) Overexpression of PtrABF gene, a bZIP transcription factor isolated from Poncirus trifoliata, enhances dehydration and drought tolerance in tobacco via scavenging ROS and modulating expression of stress-responsive genes. BMC Plant Biol 10: 230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang XS, Luo T, Fu XZ, Fan QJ, Liu JH. (2011) Cloning and molecular characterization of a mitogen-activated protein kinase gene from Poncirus trifoliata whose ectopic expression confers dehydration/drought tolerance in tobacco. J Exp Bot 62: 5191–5206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huq E, Quail PH. (2002) PIF4, a phytochrome-interacting bHLH factor, functions as a negative regulator of phytochrome B signaling in Arabidopsis. EMBO J 21: 2441–2450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito S, Song YH, Josephson-Day AR, Miller RJ, Breton G, Olmstead RG, Imaizumi T. (2012) FLOWERING BHLH transcriptional activators control expression of the photoperiodic flowering regulator CONSTANS in Arabidopsis. Proc Natl Acad Sci USA 109: 3582–3587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW. (1987) GUS fusions: β-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J 6: 3901–3907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan F, Kopka J, Haskell DW, Zhao W, Schiller KC, Gatzke N, Sung DY, Guy CL. (2004) Exploring the temperature-stress metabolome of Arabidopsis. Plant Physiol 136: 4159–4168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan F, Kopka J, Sung DY, Zhao W, Popp M, Porat R, Guy CL. (2007) Transcript and metabolite profiling during cold acclimation of Arabidopsis reveals an intricate relationship of cold-regulated gene expression with modifications in metabolite content. Plant J 50: 967–981 [DOI] [PubMed] [Google Scholar]
- Karas B, Amyot L, Johansen C, Sato S, Tabata S, Kawaguchi M, Szczyglowski K. (2009) Conservation of Lotus and Arabidopsis basic helix-loop-helix proteins reveals new players in root hair development. Plant Physiol 151: 1175–1185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lata C, Prasad M. (2011) Role of DREBs in regulation of abiotic stress responses in plants. J Exp Bot 62: 4731–4748 [DOI] [PubMed] [Google Scholar]
- Li H, Xu Y, Xiao Y, Zhu Z, Xie X, Zhao H, Wang Y. (2010) Expression and functional analysis of two genes encoding transcription factors, VpWRKY1 and VpWRKY2, isolated from Chinese wild Vitis pseudoreticulata. Planta 232: 1325–1337 [DOI] [PubMed] [Google Scholar]
- Li XX, Duan XP, Jiang HX, Sun YJ, Tang YP, Yuan Z, Guo JK, Liang WQ, Chen L, Yin JY, et al. (2006) Genome-wide analysis of basic/helix-loop-helix transcription factor family in rice and Arabidopsis. Plant Physiol 141: 1167–1184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingam S, Mohrbacher J, Brumbarova T, Potuschak T, Fink-Straube C, Blondet E, Genschik P, Bauer P. (2011) Interaction between the bHLH transcription factor FIT and ETHYLENE INSENSITIVE3/ETHYLENE INSENSITIVE3-LIKE1 reveals molecular linkage between the regulation of iron acquisition and ethylene signaling in Arabidopsis. Plant Cell 23: 1815–1829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Kasuga M, Sakuma Y, Abe H, Miura S, Yamaguchi-Shinozaki K, Shinozaki K. (1998) Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought- and low-temperature-responsive gene expression, respectively, in Arabidopsis. Plant Cell 10: 1391–1406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2-Δ Δ C(T) method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Ludwig SR, Habera LF, Dellaporta SL, Wessler SR. (1989) Lc, a member of the maize R gene family responsible for tissue-specific anthocyanin production, encodes a protein similar to transcriptional activators and contains the myc-homology region. Proc Natl Acad Sci USA 86: 7092–7096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meshi T, Iwabuchi M. (1995) Plant transcription factors. Plant Cell Physiol 36: 1405–1420 [PubMed] [Google Scholar]
- Miller G, Schlauch K, Tam R, Cortes D, Torres MA, Shulaev V, Dangl JL, Mittler R. (2009) The plant NADPH oxidase RBOHD mediates rapid systemic signaling in response to diverse stimuli. Sci Signal 2: ra45. [DOI] [PubMed] [Google Scholar]
- Miller G, Suzuki N, Ciftci-Yilmaz S, Mittler R. (2010) Reactive oxygen species homeostasis and signalling during drought and salinity stresses. Plant Cell Environ 33: 453–467 [DOI] [PubMed] [Google Scholar]
- Mittler R. (2002) Oxidative stress, antioxidants and stress tolerance. Trends Plant Sci 7: 405–410 [DOI] [PubMed] [Google Scholar]
- Mittler R, Zilinskas BA. (1992) Molecular cloning and characterization of a gene encoding pea cytosolic ascorbate peroxidase. J Biol Chem 267: 21802–21807 [PubMed] [Google Scholar]
- Miura K, Jin JB, Lee J, Yoo CY, Stirm V, Miura T, Ashworth EN, Bressan RA, Yun DJ, Hasegawa PM. (2007) SIZ1-mediated sumoylation of ICE1 controls CBF3/DREB1A expression and freezing tolerance in Arabidopsis. Plant Cell 19: 1403–1414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura K, Ohta M, Nakazawa M, Ono M, Hasegawa PM. (2011) ICE1 Ser403 is necessary for protein stabilization and regulation of cold signaling and tolerance. Plant J 67: 269–279 [DOI] [PubMed] [Google Scholar]
- Mohammadian MA, Largani ZK, Sajedi RH. (2012) Quantitative and qualitative comparison of antioxidant activity in the flavedo tissue of three cultivars of citrus fruit under cold stress. Aust J Crop Sci 6: 402–406 [Google Scholar]
- Monte E, Tepperman JM, Al-Sady B, Kaczorowski KA, Alonso JM, Ecker JR, Li X, Zhang YL, Quail PH. (2004) The phytochrome-interacting transcription factor, PIF3, acts early, selectively, and positively in light-induced chloroplast development. Proc Natl Acad Sci USA 101: 16091–16098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakashima K, Ito Y, Yamaguchi-Shinozaki K. (2009) Transcriptional regulatory networks in response to abiotic stresses in Arabidopsis and grasses. Plant Physiol 149: 88–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nesi N, Debeaujon I, Jond C, Pelletier G, Caboche M, Lepiniec L. (2000) The TT8 gene encodes a basic helix-loop-helix domain protein required for expression of DFR and BAN genes in Arabidopsis siliques. Plant Cell 12: 1863–1878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni M, Tepperman JM, Quail PH. (1998) PIF3, a phytochrome-interacting factor necessary for normal photoinduced signal transduction, is a novel basic helix-loop-helix protein. Cell 95: 657–667 [DOI] [PubMed] [Google Scholar]
- Ning J, Li X, Hicks LM, Xiong L. (2010) A Raf-like MAPKKK gene DSM1 mediates drought resistance through reactive oxygen species scavenging in rice. Plant Physiol 152: 876–890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nounjan N, Nghia PT, Theerakulpisut P. (2012) Exogenous proline and trehalose promote recovery of rice seedlings from salt-stress and differentially modulate antioxidant enzymes and expression of related genes. J Plant Physiol 169: 596–604 [DOI] [PubMed] [Google Scholar]
- Ohno S, Hosokawa M, Hoshino A, Kitamura Y, Morita Y, Park KI, Nakashima A, Deguchi A, Tatsuzawa F, Doi M, et al (2011) A bHLH transcription factor, DvIVS, is involved in regulation of anthocyanin synthesis in dahlia (Dahlia variabilis). J Exp Bot 62: 5105–5116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osakabe Y, Kajita S, Osakabe K. (2011) Genetic engineering of woody plants: current and future targets in a stressful environment. Physiol Plant 142: 105–117 [DOI] [PubMed] [Google Scholar]
- Peng T, Zhu XF, Fan QJ, Sun PP, Liu JH. (2012) Identification and characterization of low temperature stress responsive genes in Poncirus trifoliata by suppression subtractive hybridization. Gene 492: 220–228 [DOI] [PubMed] [Google Scholar]
- Pogány M, von Rad U, Grün S, Dongó A, Pintye A, Simoneau P, Bahnweg G, Kiss L, Barna B, Durner J. (2009) Dual roles of reactive oxygen species and NADPH oxidase RBOHD in an Arabidopsis-Alternaria pathosystem. Plant Physiol 151: 1459–1475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin F, Shinozaki K, Yamaguchi-Shinozaki K. (2011) Achievements and challenges in understanding plant abiotic stress responses and tolerance. Plant Cell Physiol 52: 1569–1582 [DOI] [PubMed] [Google Scholar]
- Riechmann JL, Heard J, Martin G, Reuber L, Jiang CZ, Keddie J, Adam L, Pineda O, Ratcliffe OJ, Samaha RR, et al. (2000) Arabidopsis transcription factors: genome-wide comparative analysis among eukaryotes. Science 290: 2105–2110 [DOI] [PubMed] [Google Scholar]
- Rudrappa T, Lakshmanan V, Kaunain R, Mandayam Singara NM, Neelwarne B. (2007) Purification and characterization of an intracellular peroxidase from genetically transformed roots of red beet (Beta vulgaris L.). Food Chem 105: 1312–1320 [Google Scholar]
- Seo JS, Joo J, Kim MJ, Kim YK, Nahm BH, Song SI, Cheong JJ, Lee JS, Kim JK, Choi YD. (2011) OsbHLH148, a basic helix-loop-helix protein, interacts with OsJAZ proteins in a jasmonate signaling pathway leading to drought tolerance in rice. Plant J 65: 907–921 [DOI] [PubMed] [Google Scholar]
- Shi J, Fu XZ, Peng T, Huang XS, Fan QJ, Liu JH. (2010) Spermine pretreatment confers dehydration tolerance of citrus in vitro plants via modulation of antioxidative capacity and stomatal response. Tree Physiol 30: 914–922 [DOI] [PubMed] [Google Scholar]
- Shinozaki K, Yamaguchi-Shinozaki K. (2007) Gene networks involved in drought stress response and tolerance. J Exp Bot 58: 221–227 [DOI] [PubMed] [Google Scholar]
- Shulaev V, Cortes D, Miller G, Mittler R. (2008) Metabolomics for plant stress response. Physiol Plant 132: 199–208 [DOI] [PubMed] [Google Scholar]
- Sivitz AB, Hermand V, Curie C, Vert G. (2012) Arabidopsis bHLH100 and bHLH101 control iron homeostasis via a FIT-independent pathway. PLoS ONE 7: e44843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sreenivasulu N, Sopory SK, Kavi Kishor PB. (2007) Deciphering the regulatory mechanisms of abiotic stress tolerance in plants by genomic approaches. Gene 388: 1–13 [DOI] [PubMed] [Google Scholar]
- Suzuki N, Koussevitzky S, Mittler R, Miller G. (2012) ROS and redox signalling in the response of plants to abiotic stress. Plant Cell Environ 35: 259–270 [DOI] [PubMed] [Google Scholar]
- Suzuki N, Mittler R. (2006) Reactive oxygen species and temperature stresses: a delicate balance between signaling and destruction. Physiol Plant 126: 45–51 [Google Scholar]
- Tang N, Zhang H, Li X, Xiao J, Xiong LZ. (2012) Constitutive activation of transcription factor OsbZIP46 improves drought tolerance in rice. Plant Physiol 158: 1755–1768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toledo-Ortiz G, Huq E, Quail PH. (2003) The Arabidopsis basic/helix-loop-helix transcription factor family. Plant Cell 15: 1749–1770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomashow MF. (1999) Plant cold acclimation: freezing tolerance genes and regulatory mechanisms. Annu Rev Plant Physiol Plant Mol Biol 50: 571–599 [DOI] [PubMed] [Google Scholar]
- Thomashow MF, Gilmour SJ, Stockinger EJ, Jaglo-Ottosen KR, Zarka DG. (2001) Role of the Arabidopsis CBF transcriptional activators in cold acclimation. Physiol Plant 112: 171–175 [Google Scholar]
- Tominaga-Wada R, Iwata M, Nukumizu Y, Sano R, Wada T. (2012) A full-length R-like basic-helix-loop-helix transcription factor is required for anthocyanin upregulation whereas the N-terminal region regulates epidermal hair formation. Plant Sci 183: 115–122 [DOI] [PubMed] [Google Scholar]
- Vogel JT, Zarka DG, Van Buskirk HA, Fowler SG, Thomashow MF. (2005) Roles of the CBF2 and ZAT12 transcription factors in configuring the low temperature transcriptome of Arabidopsis. Plant J 41: 195–211 [DOI] [PubMed] [Google Scholar]
- Wang J, Sun PP, Chen CL, Wang Y, Fu XZ, Liu JH. (2011) An arginine decarboxylase gene PtADC from Poncirus trifoliata confers abiotic stress tolerance and promotes primary root growth in Arabidopsis. J Exp Bot 62: 2899–2914 [DOI] [PubMed] [Google Scholar]
- Wang WX, Vinocur B, Altman A. (2003) Plant responses to drought, salinity and extreme temperatures: towards genetic engineering for stress tolerance. Planta 218: 1–14 [DOI] [PubMed] [Google Scholar]
- Wang Y, Jiang CJ, Li YY, Wei CL, Deng WW. (2012) CsICE1 and CsCBF1: two transcription factors involved in cold responses in Camellia sinensis. Plant Cell Rep 31: 27–34 [DOI] [PubMed] [Google Scholar]
- Wu H, Chen C, Du J, Liu H, Cui Y, Zhang Y, He Y, Wang Y, Chu C, Feng Z, et al (2012) Co-overexpression FIT with AtbHLH38 or AtbHLH39 in Arabidopsis-enhanced cadmium tolerance via increased cadmium sequestration in roots and improved iron homeostasis of shoots. Plant Physiol 158: 790–800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang Y, Tang N, Du H, Ye H, Xiong LZ. (2008) Characterization of OsbZIP23 as a key player of the basic leucine zipper transcription factor family for conferring abscisic acid sensitivity and salinity and drought tolerance in rice. Plant Physiol 148: 1938–1952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie XB, Li S, Zhang RF, Zhao J, Chen YC, Zhao Q, Yao YX, You CX, Zhang XS, Hao YJ. (2012) The bHLH transcription factor MdbHLH3 promotes anthocyanin accumulation and fruit colouration in response to low temperature in apples. Plant Cell Environ 35: 1884–1897 [DOI] [PubMed] [Google Scholar]
- Yamada Y, Kokabu Y, Chaki K, Yoshimoto T, Ohgaki M, Yoshida S, Kato N, Koyama T, Sato F. (2011) Isoquinoline alkaloid biosynthesis is regulated by a unique bHLH-type transcription factor in Coptis japonica. Plant Cell Physiol 52: 1131–1141 [DOI] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki K, Shinozaki K. (2005) Organization of cis-acting regulatory elements in osmotic- and cold-stress-responsive promoters. Trends Plant Sci 10: 88–94 [DOI] [PubMed] [Google Scholar]
- Yuan YX, Wu HL, Wang N, Li J, Zhao WN, Du J, Wang DW, Ling HQ. (2008) FIT interacts with AtbHLH38 and AtbHLH39 in regulating iron uptake gene expression for iron homeostasis in Arabidopsis. Cell Res 18: 385–397 [DOI] [PubMed] [Google Scholar]
- Zhan WB, Chen J, Zhang ZD, Zhou L, Fukuda H. (2003) Elimination of shrimp endogenous peroxidase background in immunodot blot assays to detect white spot syndrome virus (WSSV). Dis Aquat Organ 53: 263–265 [DOI] [PubMed] [Google Scholar]
- Zhao L, Liu FX, Xu WY, Di C, Zhou SX, Xue YB, Yu JJ, Su Z. (2009) Increased expression of OsSPX1 enhances cold/subfreezing tolerance in tobacco and Arabidopsis thaliana. Plant Biotechnol J 7: 550–561 [DOI] [PubMed] [Google Scholar]
- Zhu A, Li W, Ye J, Sun X, Ding Y, Cheng Y, Deng X. (2011) Microarray expression profiling of postharvest Ponkan mandarin (Citrus reticulata) fruit under cold storage reveals regulatory gene candidates and implications on soluble sugars metabolism. J Integr Plant Biol 53: 358–374 [DOI] [PubMed] [Google Scholar]