DNA Pol λ participates in the repair of double strand breaks induced by high salinity and DNA cross-linking agent in Arabidopsis seedlings, demonstrating the importance of Pol λ in the double strand break repair signaling network in higher plant genome.
Abstract
DNA polymerase λ (Pol λ) is the sole member of family X DNA polymerase in plants and plays a crucial role in nuclear DNA damage repair. Here, we report the transcriptional up-regulation of Arabidopsis (Arabidopsis thaliana) AtPolλ in response to abiotic and genotoxic stress, including salinity and the DNA cross-linking agent mitomycin C (MMC). The increased sensitivity of atpolλ knockout mutants toward high salinity and MMC treatments, with higher levels of accumulation of double strand breaks (DSBs) than wild-type plants and delayed repair of DSBs, has suggested the requirement of Pol λ in DSB repair in plants. AtPolλ overexpression moderately complemented the deficiency of DSB repair capacity in atpolλ mutants. Transcriptional up-regulation of major nonhomologous end joining (NHEJ) pathway genes KU80, X-RAY CROSS COMPLEMENTATION PROTEIN4 (XRCC4), and DNA Ligase4 (Lig4) along with AtPolλ in Arabidopsis seedlings, and the increased sensitivity of atpolλ-2/atxrcc4 and atpolλ-2/atlig4 double mutants toward high salinity and MMC treatments, indicated the involvement of NHEJ-mediated repair of salinity- and MMC-induced DSBs. The suppressed expression of NHEJ genes in atpolλ mutants suggested complex transcriptional regulation of NHEJ genes. Pol λ interacted directly with XRCC4 and Lig4 via its N-terminal breast cancer-associated C terminus (BRCT) domain in a yeast two-hybrid system, while increased sensitivity of BRCT-deficient Pol λ-expressing transgenic atpolλ-2 mutants toward genotoxins indicated the importance of the BRCT domain of AtPolλ in mediating the interactions for processing DSBs. Our findings provide evidence for the direct involvement of DNA Pol λ in the repair of DSBs in a plant genome.
Plants, with their inherent immobility and obligatory dependence on sunlight for energy, face tremendous challenges in maintaining the integrity of the genome, which is under continuous assault from environmental factors like UV and ionizing radiation, high salinity, chemical mutagens, and free radicals or alkylating agents generated by endogenous processes (Britt, 1999; Tuteja et al., 2009). Lesions in the DNA, contributed by various damaging agents, may result in changes in both the chemical and physical structures of DNA (Bray and West, 2005) and thus generate both cytotoxic and genotoxic effects (Amoroso et al., 2011). Therefore, plant cells have evolved with highly efficient and wide-ranging mechanisms for the detection and repair of DNA damage to ensure genome stability (Tuteja et al., 2009).
Family X DNA polymerases (Pols) are mainly involved in DNA repair and recombination pathways (Ramadan et al., 2004; Roy et al., 2009). Mammalian DNA polymerase λ (Pol λ), a relatively newly identified X family member, is widespread among higher eukaryotes, both in animals and plants. Human DNA Pol λ is an exonuclease-deficient single-polypeptide DNA polymerase that shares a high degree of sequence homology with mammalian DNA Pol β (32% amino acid identity with human Pol β). In vitro and in vivo studies have indicated that both enzymes share many of their biochemical properties, including a 5′-deoxyribose phosphate lyase activity required to complement the DNA synthesis step associated with base excision repair (García-Díaz et al., 2000; Braithwaite et al., 2005). The involvement of Pol λ has also been strongly suggested in filling gaps in DNA during nonhomologous end joining (NHEJ) of double strand breaks (DSBs; Lee et al., 2004; Ma et al., 2004; Moon et al., 2007; Garcia-Diaz et al., 2009). Studies in mammalian cells have demonstrated a protective role of DNA Pol λ against oxidative DNA damage (Braithwaite et al., 2005). A potential role of this enzyme as a “mismatch extender” during NHEJ and its possible involvement in translesion synthesis have been suggested earlier (Picher et al., 2006). Recent studies have indicated a role of Pol λ in the repair of smoking-induced DNA damage in respiratory epithelium (Ohba et al., 2009) and microhomology-mediated DNA strand annealing and elongation (Crespan et al., 2012).
Genome-wide sequence analyses have suggested that DNA Pol λ represents the only member of family X DNA Pols in plants and is possibly involved in base excision repair (Uchiyama et al., 2004). Recently, we have reported the requirement of Arabidopsis (Arabidopsis thaliana) DNA Pol λ (AtPolλ) in the repair of UV-B-induced DNA damage via the nucleotide excision repair pathway (Roy et al., 2011). In addition, the role of DNA Pol λ in high-fidelity translesion DNA synthesis in response to oxidative DNA damage has been demonstrated in Arabidopsis (Amoroso et al., 2011). However, the involvement of DNA Pol λ in DSB repair in plants has not been characterized so far.
In this study, we provide evidence for the involvement of AtPolλ in the repair of DSBs, induced by high salinity and DNA cross-linking agent, via the NHEJ pathway in Arabidopsis. We have further demonstrated direct interactions of AtPolλ with the Arabidopsis homolog of X-RAY CROSS COMPLEMENTATION PROTEIN4 (AtXRCC4; AT3G23100) and DNA Ligase4 (AtLig4; AT5G57160), the two major components of the NHEJ pathway of DSB repair in plants, and the importance of the N-terminal breast cancer-associated C terminus (BRCT) domain of AtPolλ in mediating the interactions for processing DSBs.
RESULTS
Conservation of the AtPolλ Gene Sequence in Higher Plant Genomes
AtPolλ (AT1G10520) belongs to family X DNA Pol based on its sequence homology with mammalian DNA Pol β, DNA Pol μ, and terminal deoxynucleotidyl transferase and appears to be the single representative of family X DNA Pols in plants to carry out nuclear DNA repair functions (Amoroso et al., 2011; Roy et al., 2011). The full-length AtPolλ, expressed in Escherichia coli cells, has been found as a single polypeptide protein with an intrinsic DNA polymerase activity (Supplemental Fig. S1) and the biochemical properties closely related to human DNA Pol λ and other members of X family DNA Pols (Supplemental Fig. S2). Sequential and phylogenetic analyses have revealed close similarity of AtPolλ with mammalian DNA Pol β, Pol λ, as well other members of family X DNA Pols from animal and plant sources (Supplemental Figs. S3–S5; Supplemental Table S2).
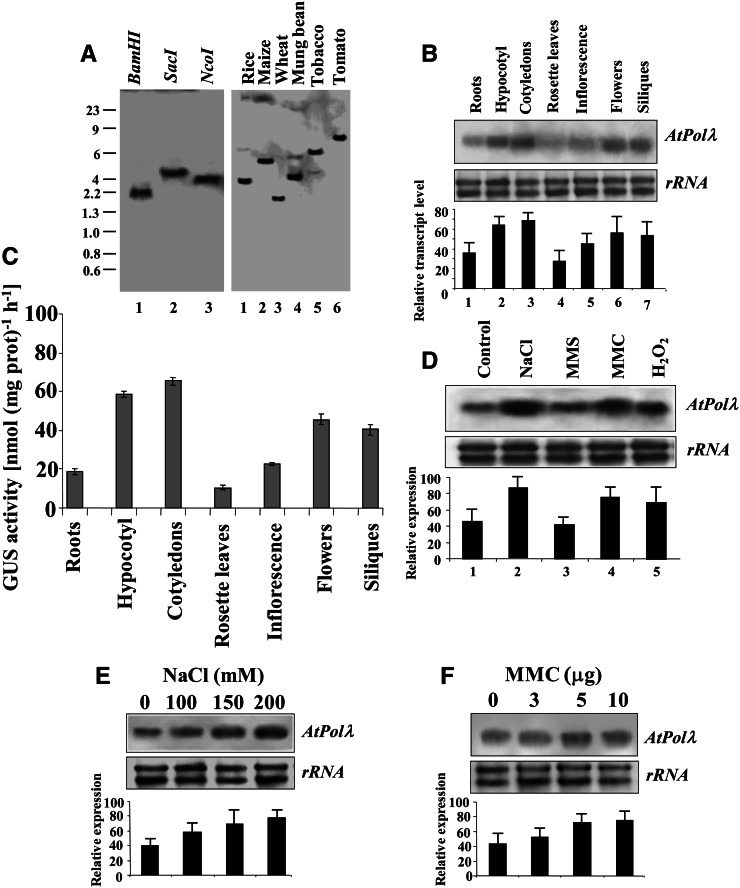
The members of family X DNA Pols have been found to be evolutionarily conserved across different phyla. In addition to eukaryotes, a number of family X DNA Pols have been characterized in all forms of life, including archaea, eubacteria, and viruses (Fan and Wu, 2004). To analyze the organization of the AtPolλ gene (3.4 kb) in the Arabidopsis genome, [α32P]dCTP-labeled full-length complementary DNA (cDNA) of the gene (1.59 kb) was used as a probe in genomic Southern-blot studies. A single strong band of hybridization signal was detected with each of the three different restriction endonuclease-digested genomic DNA samples from Arabidopsis (Fig. 1A, left panel), suggesting that AtPolλ exists as a single copy in the Arabidopsis genome. Furthermore, Southern hybridization analysis using restriction-digested genomic DNA samples from various other plant species with labeled 1.59-kb AtPolλ cDNA probe have indicated positive genomic hybridization signals (Fig. 1A, right panel), indicating evolutionary conservation of the Pol λ gene sequence among higher plant genomes. In silico analysis for the presence of Pol λ homologs in different plant lineages has revealed the presence of a single copy of the gene among a majority of plant species. Interestingly, we have found the existence of two copies of a hypothetical Pol λ gene in Medicago domestica and Arabidopsis lyrata and three copies in Carica papaya and Selaginella moellendorfi (Supplemental Fig. S6).
Figure 1.
Conservation of the DNA Pol λ gene in plant genomes. A, Genomic hybridization analysis of the Pol λ gene using genomic DNA samples from Arabidopsis (left panel) and various other plant species (right panel). DNA size markers (in kb) are indicated on the left. B, The accumulation pattern of the AtPolλ transcript in different tissues of Arabidopsis was analyzed by RNA gel blotting using 20 μg of total RNA (top panel). Ethidium bromide-stained ribosomal RNA (rRNA) is shown as a loading control (middle panel). Quantification of the data by densitometry (Bio-Rad Image Densitometer G700) is shown in the bottom panel. Data points represent mean values from three independent trials. C, Measurement of GUS activity in various tissues of transgenic Arabidopsis plants carrying the AtPolλ promoter-GUS transgene. Bars represent mean values from three independent observations. D, DNA damage regulated the expression of AtPolλ. RNA gel-blot analysis shows total RNA (20 μg) from 7-d-old wild-type Arabidopsis seedlings exposed to high salinity (200 mm NaCl) and various other genotoxic agents (80 µL L−1 MMS, 5 μg mL−1 MMC, and 20 mm H2O2; top panel). Ethidium bromide-stained rRNA is shown as a loading control (middle panel). Quantification of the data is shown in the bottom panel. Data points represent mean values from three independent trials. E and F, RNA gel-blot analysis using total RNA (20 μg) from 7-d-old wild-type Arabidopsis seedlings exposed to increasing concentrations of NaCl (E) or MMC (F) for 8 h (top panels). Ethidium bromide-stained rRNA is shown as a loading control in each case (middle panels). Quantification of the data is shown in the bottom panels. Data points represent mean values from three independent trials. Representative gel images from at least three independent trials are shown for A, B, D, E, and F. A 1.59-kb AtPolλ cDNA fragment was used as a probe in DNA and RNA gel blotting.
The AtPolλ Transcript Accumulates in Meristematic and Meiotic Tissues
To investigate the tissue-specific expression patterns of AtPolλ transcript in Arabidopsis, we next performed RNA gel-blot analysis using total RNA isolated from various tissues of 7-d-old seedlings and 4-week-old Arabidopsis plants. AtPolλ transcript expression levels were relatively higher in hypocotyls and cotyledons from 7-d-old seedlings, and detectable expression was also obtained in roots from 7-d-old seedlings. In adult plants, the AtPolλ transcript accumulation level was low in rosette leaves and inflorescence axes, while appreciable levels of expression were detected in flowers and siliques (Fig. 1B). We further used transgenic Arabidopsis lines harboring the 789-bp upstream promoter sequence of AtPolλ in fusion with the GUS reporter to assess tissue-specific expression of the gene. A representative transgenic line, revealed by GUS stain, was used to test the expression levels of AtPolλ in various tissues by GUS activity assay (Roy Choudhury et al., 2008). GUS activity levels in the various tissues in Arabidopsis plants were consistent with RNA gel-blot results (Fig. 1C). The accumulation of AtPolλ transcript in actively growing tissues indicates a possible function of the gene in DNA replication and repair during seedling growth, while AtPolλ transcript expression in flower supports the proposed role of Pol λ in meiotic DNA repair (García-Díaz et al., 2000).
Abiotic and Genotoxic Stress Induces AtPolλ Expression
Recent studies have demonstrated roles of DNA Pol λ in UV-B-induced DNA damage repair and translesion DNA synthesis in the repair of oxidative DNA damage (Amoroso et al., 2011; Roy et al., 2011), suggesting the requirement of Pol λ in different repair pathways in plants. To further characterize the transcriptional induction of the Pol λ gene in response to abiotic and genotoxic agent treatments, we next examined AtPolλ message levels in 7-d-old wild-type Arabidopsis seedlings subjected to treatment with various stress factors, as described in “Materials and Methods.” As shown in Figure 1D, 200 mm NaCl treatment strongly induced AtPolλ transcript expression (approximately 2.2-fold higher than the control). In addition, mitomycin C (MMC) and hydrogen peroxide (H2O2; an oxidative agent) treatments also caused transcriptional up-regulation of AtPolλ. Interestingly, methyl methane sulfonate (MMS; an alkylating agent) treatment did not cause any considerable increase in AtPolλ mRNA levels as compared with the control condition.
Earlier reports have demonstrated a role of DNA Pol λ in DSB repair (Ma et al., 2004), while high salinity and MMC are known to induce DSBs in DNA (Dmitrieva and Burg, 2005; Watanabe et al., 2009). Therefore, we next specifically examined the induction pattern of AtPolλ message levels, particularly in response to high salinity and MMC treatments, in 7-d-old wild-type Arabidopsis seedlings. As shown in Figure 1, E and F, a dose-dependent increase in AtPolλ transcript accumulation was detected when 7-d-old wild-type Arabidopsis seedlings were treated with increasing concentrations of NaCl and MMC, respectively. Overall, these results have indicated transcriptional up-regulation of AtPolλ in wild-type Arabidopsis seedlings in response to high salinity and genotoxic agents, including MMC and H2O2. The increased message level of AtPolλ following H2O2 treatment was consistent with the role of Pol λ in the repair of oxidative DNA damage indicated in earlier studies (Amoroso et al., 2011).
We have also compared the transcript expression data with the gene expression data sets related to AtPolλ (AT1G10520) available in the Arabidopsis eFP browser (http://bar.utoronto.ca/efp/cgi-bin/efpWeb.cgi; Winter et al., 2007). The data available in this browser regarding transcript expression levels of AtPolλ in roots, hypocotyls, cotyledons, and leaves for Columbia (Col-0) plants were found to be consistent with our results (Fig. 1B). The expression levels found in whole stem (top of stem), stigma, ovary and dry pollen, and seed also supported our results regarding AtPolλ expression patterns detected in inflorescences, flowers (including stigma, ovary, and pollen), and siliques (including seeds; Fig. 1B). Moreover, the induction patterns of AtPolλ in roots and shoots in 18-d-old wild-type Arabidopsis (Col-0), found in the eFP browser, after treatments with 150 mm NaCl and genotoxic agents (1.5 μg mL−1 bleomycin plus 22 μg mL−1 MMC for up to 24 h) were in relative agreement with our findings of high salinity- and MMC-mediated induction of AtPolλ transcripts in 7-d-old Arabidopsis seedlings (Fig. 1D).
AtPolλ Loss-of-Function Mutants Display Increased Sensitivity to High Salinity and DNA Cross-Linking Agent
To investigate the biological function of Pol λ in the repair of DNA damage caused by high salinity and MMC treatments in Arabidopsis seedlings, we next analyzed the phenotypes of three independent transfer DNA (T-DNA) insertion mutant lines of AtPolλ, atpolλ-1 (Salk_075391C), atpolλ-2 (Salk_070973), and atpolλ-3 (Salk_061978), with T-DNA insertions at three different sites in the AtPolλ gene (At1G10520; Supplemental Fig. S7A). The T-DNA insertion mutant lines were found to be homozygous. AtPolλ transcript and protein expression analyses have indicated that atpolλ-1 and atpolλ-2 represent true null mutation lines (Supplemental Fig. S7, B and C).
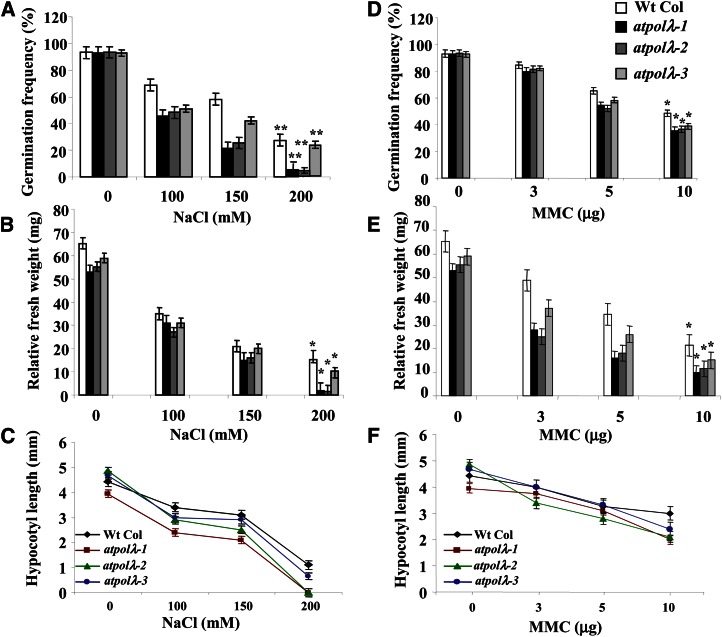
The atpolλ-1, atpolλ-2, and atpolλ-3 homozygous mutants were viable and did not display any significant morphological deviation from the wild-type phenotype under standard growth conditions except for the early flowering phenotype in the atpolλ-1 mutant line (Supplemental Fig. S8). On the other hand, our results indicate transcriptional up-regulation of AtPolλ in 7-d-old wild-type Arabidopsis seedlings following high salinity and MMC treatments. Therefore, we then examined the relative sensitivity of atpolλ mutant seedlings toward high salinity and a DNA cross-linking agent by subjecting the wild type and the atpolλ mutant lines to high salinity and other genotoxic agents (see “Materials and Methods”). Growth responses in 7-d-old seedlings were affected by high NaCl and MMC in both the wild type and atpolλ mutants. However, the effects were relatively more pronounced in atpolλ-1 and atpolλ-2 seedlings. The germination rates and seedling fresh weights were considerably affected by high salinity in atpolλ knockout lines as compared with untreated seedlings (P < 0.001–0.01; Fig. 2, A and B). Prominent inhibition in hypocotyl length elongation was also detected in atpolλ-1 and atpolλ-2 mutant seedlings due to salt treatment (P < 0.001; Fig. 2C; Supplemental Table S1).
Figure 2.
Analysis of the growth response of atpolλ mutants in the presence of high salinity and DNA cross-linking agent. Arabidopsis seeds from the wild type (Wt Col) and atpolλ mutants were plated on MS-agar medium containing different concentrations of NaCl or MMC. One-week-old plants were tested for their sensitivity. A and D, Analysis of germination rates in 4-d-old wild-type and atpolλ mutant plants grown in the absence or presence of increasing concentrations of NaCl (A) or MMC (D). B and E, Determination of relative fresh weights of 1-week-old wild-type and atpolλ mutant seedlings grown in the absence or presence of increasing concentrations of NaCl (B) or MMC (E). C and F, Measurement of hypocotyl length in 1-week-old wild-type and atpolλ mutant seedlings grown in the absence or presence of increasing concentrations of NaCl (C) or MMC (F). The bars represent mean values from three independent observations. *P < 0.05, **P < 0.01 relative to respective controls (n = 3). [See online article for color version of this figure.]
As compared with untreated seedlings, the atpolλ mutant lines also displayed a hypersensitive growth response in the presence of the DNA cross link-inducing agent MMC at 10 μg mL−1, as evidenced in germination rates, fresh weights of seedlings, and hypocotyl length elongation (P < 0.01–0.04; Fig. 2, D–F; Supplemental Table S1). However, compared with salinity treatment, the responses were relatively less severe. Interestingly, the sensitivity of 7-d-old seedlings of the atpolλ-1/atpolλ-2 mutant line (having both alleles, one on each homologous chromosome) toward high NaCl or MMC did not differ significantly from the atpolλ-1 mutant line, indicating a dominant effect of the atpolλ-1 mutant allele over atpolλ-2 (data not shown). However, none of the atpolλ mutants showed any significant sensitivity after treatment with the alkylating agent MMS (data not shown). Furthermore, 3-week-old atpolλ knockout mutant lines also displayed increased sensitivity toward high salinity and MMC but not in the presence of MMS (data not shown). Together, these results indicated the role of AtPolλ in the repair of DNA damage like DSBs induced in the presence of high salinity and a DNA cross-linking agent. Conversely, the function of this protein in the repair of methylated bases seemed to be dispensable.
AtPolλ Protein Accumulates in Wild-Type Arabidopsis Seedlings following High Salinity and MMC Treatments
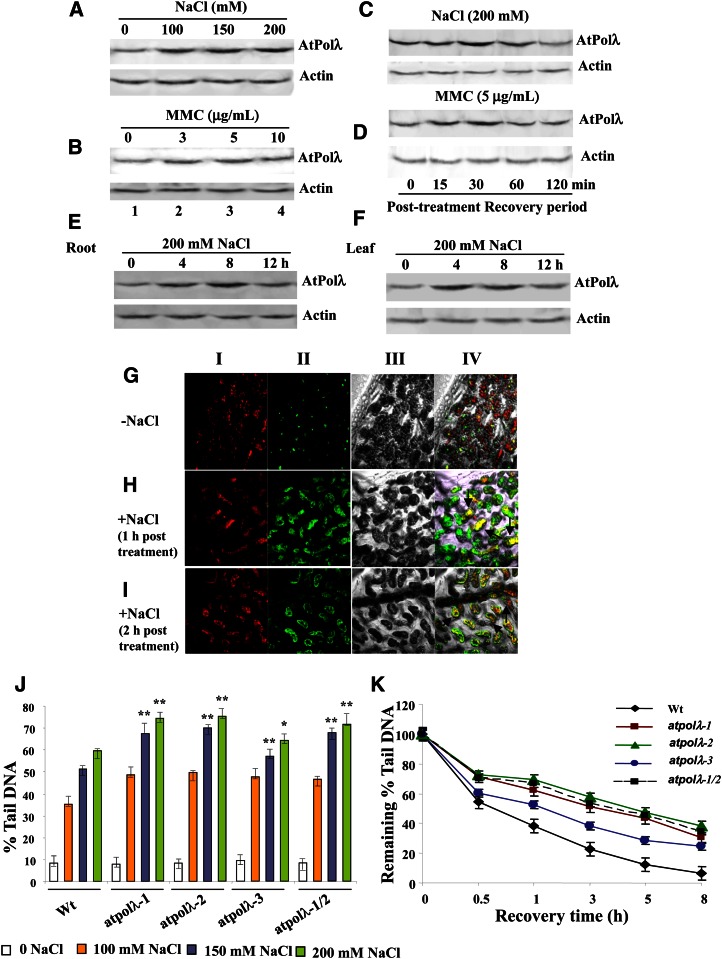
Our results indicate increased sensitivity of atpolλ mutants toward high salinity and a DNA cross link-inducing agent (Fig. 2). Therefore, to further understand the functional relevance of DNA Pol λ in the repair of high salinity- and MMC-induced DNA damage, we next analyzed the accumulation pattern of AtPolλ protein in wild-type Arabidopsis seedlings after high salinity and MMC treatments. AtPolλ protein accumulation increased steadily in the presence of increasing NaCl concentrations (Fig. 3A). A similar response was also noticed in the presence of increasing concentrations of MMC (Fig. 3B), supporting the idea of a requirement of DNA Pol λ in the repair of DNA damage generated by high salinity and MMC. To further verify these observations, we next analyzed changes in AtPolλ protein level in 7-d-old wild-type Arabidopsis seedlings during the posttreatment recovery periods in the absence of genotoxins. AtPolλ protein level increased appreciably within 30 to 60 min of recovery after salinity treatment (Fig. 3C) and also during 15 to 60 min of recovery following MMC treatment (Fig. 3D). Untreated control seedlings were recovered under identical time points and showed no significant level of variation in AtPolλ protein during the recovery periods (data not shown). MMS treatment did not cause any notable change in AtPolλ protein level during the recovery periods (data not shown). Together, these results further indicated the involvement of Pol λ in the repair of high salt- and MMC-induced DNA damage in Arabidopsis seedlings.
Figure 3.
AtPolλ protein accumulation in wild-type Arabidopsis seedlings after high salt and MMC treatment. A and B, Immunodetection of AtPolλ protein using anti-AtPolλ polyclonal antibody in total protein extracts (40 μg) from 7-d-old wild-type seedlings exposed to increasing concentrations of NaCl (0–200 mm) or MMC (0–10 μg mL−1) for 8 h (top panels). C and D, Immunoblot analysis of protein extracts from 7-d-old wild-type seedlings transferred to one-half-strength MS liquid medium for recovery at various time points after 200 mm NaCl or 5 μg mL−1 MMC treatment for 8 h (top panels). E and F, Accumulation of AtPolλ protein in the roots (E) and leaves (F) of 2-week-old wild-type Arabidopsis seedlings exposed to 200 mm NaCl for various time points analyzed by immunoblotting (top panels). The level of actin protein, detected using anti-actin antibody, is shown as a loading control (bottom panels in A–F). The 0-h time point served as the control. Representative gel images from at least three independent experiments are shown. G to I, Immunolocalization of AtPolλ in leaf cells of 7-d-old wild-type Arabidopsis seedlings from untreated control (G) and after 1 h (H) and 2 h (I) of recovery following 200 mm NaCl treatment for 6 h. Nuclei were stained with 4′,6-diamino-phenylindole (red fluorescence in panel I). AtPolλ was detected using anti-AtPolλ IgG coupled to fluorescein isothiocyanate (green fluorescence in panel II). Panel III shows differential interference contrast images, and nuclear localization of AtPolλ is shown in the merged images of panels I and II (panel IV; yellow fluorescence zones indicated by arrows). J, Relative accumulation levels of DSBs in 7-d-old wild-type (Wt) and atpolλ mutant seedlings exposed to various concentrations of NaCl for 6 h. The extent of DNA damage is represented by the fraction of DNA migrating in the comet tail. Bars represent mean values from three independent trials. K, Mean percentage of DNA in the comet tails of 7-d-old wild-type and atpolλ mutant seedlings at various recovery time points. Maximum damage is normalized as 100% at 0 h for all lines. Data shown are means ± sd of three independent replications. *P < 0.05, **P < 0.01 relative to respective controls (n = 3). [See online article for color version of this figure.]
Studies in mammalian systems have indicated that high NaCl induces DSBs in DNA and interferes with the activation of several components of the classical DNA damage response, including meiotic recombination11 (Mre11), H2AX, and Chk1, resulting in an inhibition of DNA repair (Dmitrieva and Burg, 2005; Irarrazabal et al., 2006). High salinity also induces DSBs in plant genomes and inhibits plant growth and fertility (Boyko et al., 2010). Plants repair DSBs mainly via the NHEJ pathway. Although earlier studies in mammalian systems have revealed a role of DNA Pol λ in NHEJ-mediated DSB repair (Ma et al., 2004), the involvement of Pol λ in DSB repair in plants remains uncharacterized (Waterworth et al., 2011). Our results indicate high NaCl-mediated induction of AtPolλ expression both at transcript and protein levels in Arabidopsis seedlings (Figs. 1 and 3) and increased sensitivity of atpolλ knockout mutants toward high salinity. In addition, the effects of salinity stress on atpolλ mutant lines were more prominent than MMC treatment (Fig. 2). Therefore, based on these observations, we further specifically investigated the effect of salt stress on the expression of AtPolλ protein in Arabidopsis seedlings. Two-week-old wild-type Arabidopsis seedlings exposed to 200 mm NaCl for increasing time points showed enhanced accumulation of AtPolλ protein within 4 to 8 h of salt stress in root and leaf tissues (Fig. 3, E and F). Although AtPolλ protein accumulation did not increase further after 8 h of treatment, the accumulation level appeared to be relatively faster in leaf tissues than in roots, indicating that the salt stress-mediated DNA repair signaling is probably perceived faster in leaves than in roots. Seedlings were also treated with 100 mm LiCl to examine whether enhanced accumulation of AtPolλ protein was specific to NaCl. LiCl treatment did not induce AtPolλ transcript or protein expression to any significant level (data not shown). However, severe growth retardation of seedlings was observed in the presence of 200 to 300 mm LiCl.
Nuclear presence would be required for Pol λ to participate in the DNA damage repair pathway. Therefore, we next studied the changes in the subcellular localization of AtPolλ protein in leaf tissues of 7-d-old wild-type Arabidopsis seedlings after salt treatment. In contrast to untreated controls and in addition to cytosolic staining, a notable increase in the nuclear localization signal of AtPolλ was detected after high NaCl treatment (Fig. 3, G–I, panel IV). Protein gel blot studies using cytosolic and nuclear protein fractions from control and salt-treated leaf samples also indicated increased AtPolλ in the nuclear protein fraction from salt-treated leaf tissues (data not shown). Taken together, these findings demonstrate enhanced nuclear localization of AtPolλ after high NaCl treatment, consistent with a possible role of DNA Pol λ in nuclear DNA repair in response to salinity-induced DNA damage.
atpolλ Mutants Are Impaired in Their Capacity to Repair DNA DSBs
Our results indicate that loss of AtPolλ function leads to enhanced sensitivity of atpolλ knockout mutants toward high salinity (Fig. 2, A–C), while salt treatment causes increased accumulation of AtPolλ in wild-type Arabidopsis seedlings (Fig. 3A). Since high salinity is known to induce DSBs in DNA and Pol λ has been implicated in NHEJ-mediated DSB repair (Capp et al., 2006; Moon et al., 2007; Garcia-Diaz et al., 2009), we next analyzed the extent of DSB accumulation and repair in the wild type and atpolλ mutants following the exposure of seedlings to high salinity (200 mm NaCl). A comet assay was carried out under neutral conditions (Kozak et al., 2009) using a nuclear suspension prepared from control and salt-treated 7-d-old seedlings to measure the relative accumulation and repair of DSBs (Supplemental Fig. S9, A–F). The atpolλ-1, atpolλ-2, atpolλ-3, and atpolλ-1/atpolλ-2 mutant lines showed approximately 26%, 29%, 16%, and 27% higher DSBs, respectively, than wild-type seedlings after exposure to 150 mm NaCl (P < 0.002–0.006; Fig. 3J). Seedlings treated with 200 mm NaCl caused approximately 23%, 25%, 11%, and 22% higher DSB accumulation in atpolλ-1, atpolλ-2, atpolλ-3, and atpolλ-1/atpolλ-2 mutants than wild-type seedlings (P < 0.001–0.004; Fig. 3J).
To further investigate the relative efficiencies of DSB repair in the wild type and atpolλ mutant lines, we performed DSB repair kinetics through a time course of recovery for 0.5 to 8 h of incubation of seedlings in one-half-strength liquid Murashige and Skoog (MS) medium after salt treatment (Supplemental Fig. S9, B–F). In both the wild type and atpolλ mutants, DSB repair rates were relatively similar during the early phase of recovery (0–0.5 h). However, wild-type seedlings showed better DSB repair rates in the later phases of recovery. Whereas approximately 82% DSB repair could be detected in wild-type seedlings after 8 h of recovery, atpolλ-1, atpolλ-2, atpolλ-3, and atpolλ-1/atpolλ-2 mutant lines showed approximately 65%, 59%, 70%, and 61% DSB repair under similar recovery periods (P < 0.0001–0.001; Fig. 3K). Together, these observations indicate that AtPolλ loss of function impairs the DSB repair efficiency, suggesting a requirement of DNA Pol λ for the repair of DSBs in plants.
AtPolλ Overexpression Enhances DSB Repair Efficiency in atpolλ Mutants
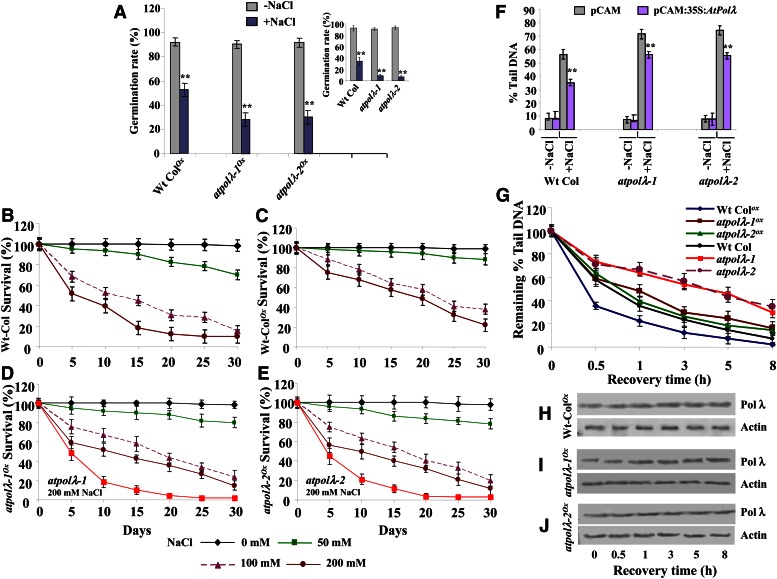
The increased sensitivity of atpolλ mutants, particularly atpolλ-1 and atpolλ-2, toward higher salinity seemed to result from the loss of AtPolλ function (Fig. 2, A–C). Therefore, we reasoned that if DNA Pol λ acts in DSB repair, the DSB repair efficiency in atpolλ knockout mutants could be enhanced by overexpressing AtPolλ. Transgenic Arabidopsis plants of the wild type and atpolλ mutants (atpolλ-1 and atpolλ-2) overexpressing full-length AtPolλ cDNA driven by the constitutive cauliflower mosaic virus (CaMV) 35S promoter were used to address this possibility. The growth responses of AtPolλ-overexpressing transgenic lines of the wild type and atpolλ mutants (Wt-ColOx and atpolλOx) were compared with those of nonoverexpressor (empty vector-transformed line used as a control) seedlings in the absence and presence of 200 mm NaCl (Supplemental Fig. S10). The salt-induced inhibition in seed germination rates was minimized in the Wt-ColOx and atpolλOx lines as compared with nonoverexpressor lines (Fig. 4A; P < 0.001–0.004).
Figure 4.
Effect of the overexpression of AtPolλ in the wild type (Wt Col) and atpolλ mutants. A, Seed germination rates measured in 4-d-old wild-type and atpolλ mutant plants overexpressing AtPolλ and in empty vector-transformed nonoverexpressor lines (inset) in the absence or presence of 200 mm NaCl. Data points represent mean values from three independent experiments. B to E, Survival rates of nonoverexpressor (empty vector transformed) and AtPolλ overexpressor lines of wild-type and atpolλ mutant plants in the presence of high salinity. Three-week-old seedlings were transferred in soil pots and supplied with 50, 100, and 200 mm NaCl solution. Data points represent mean values of three independent observations. Survival rates of empty vector-transformed control (without transgene) atpolλ-1 and atpolλ-2 mutant lines in the presence of 200 mm NaCl are shown in red traces in D and E, respectively. F, Evaluation of the extent of the accumulation of DSBs by comet assay in nonoverexpressor control and AtPolλ-overexpressing transgenic 7-d-old wild-type and atpolλ mutant seedlings in the absence or presence of 200 mm NaCl for 6 h. G, Repair of DSBs in nonoverexpressor control and AtPolλ-overexpressing transgenic 7-d-old wild-type and atpolλ mutant seedlings during the post-salt treatment incubation in one-half-strength liquid MS medium for various time points. The data represent mean values ± se of triplicate assays. **P < 0.01 relative to respective controls (n = 3). H to J, Immunoblots showing the expression of DNA Pol λ in protein extracts from AtPolλ-overexpressing transgenic 7-d-old wild-type and atpolλ mutant seedlings during post-salt treatment incubation periods. Expression levels of actin are shown as a loading control. Images are representative of three trials with similar results. [See online article for color version of this figure.]
To analyze the extent of tolerance to salinity stress due to Pol λ overexpression, we compared the survival rates of nonoverexpressor plants with Wt-ColOx and atpolλOx lines following exposure to different concentrations of NaCl (Fig. 4, B–E). The percentage of survival was scored at 5-d intervals following the transfer of 3-week-old plants under controlled growth conditions. After 30 d, approximately 20% to 22% of Wt-ColOx plants survived in the presence of 200 mm NaCl, as compared with less than 10% survival of nonoverexpressor wild-type plants (Fig. 4, B and C). Under similar conditions, atpolλOx lines showed approximately 10% to 12% survival in the presence of 200 mm NaCl, as compared with 2% to 5% survival of the atpolλ nonoverexpressor mutant lines (Fig. 4, D and E, red traces).
It was further analyzed whether AtPolλ overexpression affects the DNA repair efficiency in the wild type and atpolλ mutant lines in response to salinity treatment. The induction of DSBs in Wt-ColOx, atpolλ-1Ox, and atpolλ-2Ox seedlings was approximately 36%, 22%, and 27% less in comparison with the respective nonoverexpressor lines (P < 0.002–0.005; Fig. 4F). AtPolλ overexpression was found to improve the DSB repair efficiency in atpolλ-1 and atpolλ-2 mutants over nonoverexpressor lines (P < 0.01–0.02; Fig. 4G). The accumulation pattern of DNA Pol λ in overexpressor lines during the post-salt treatment recovery periods (Fig. 4, H–J) was also consistent with the proposed function of this protein in the repair of high salt-induced DSBs in Arabidopsis seedlings. Taken together, these observations demonstrate that DNA Pol λ overexpression helps to enhance the repair efficiency of DSBs generated by high salinity and confers a better growth response under salt stress. We additionally tested the possibility of whether AtPolλ overproduction produces any salt tolerance effect in other organisms, such as the model prokaryotic E. coli and the unicellular eukaryote yeast (Saccharomyces cerevisiae). Relatively better cell survival rates were detected in E. coli (Supplemental Fig. S11) and yeast cells (data not shown) overexpressing full-length AtPolλ protein.
High NaCl and MMC Treatments Induce the Expression of NHEJ Genes in Arabidopsis Seedlings
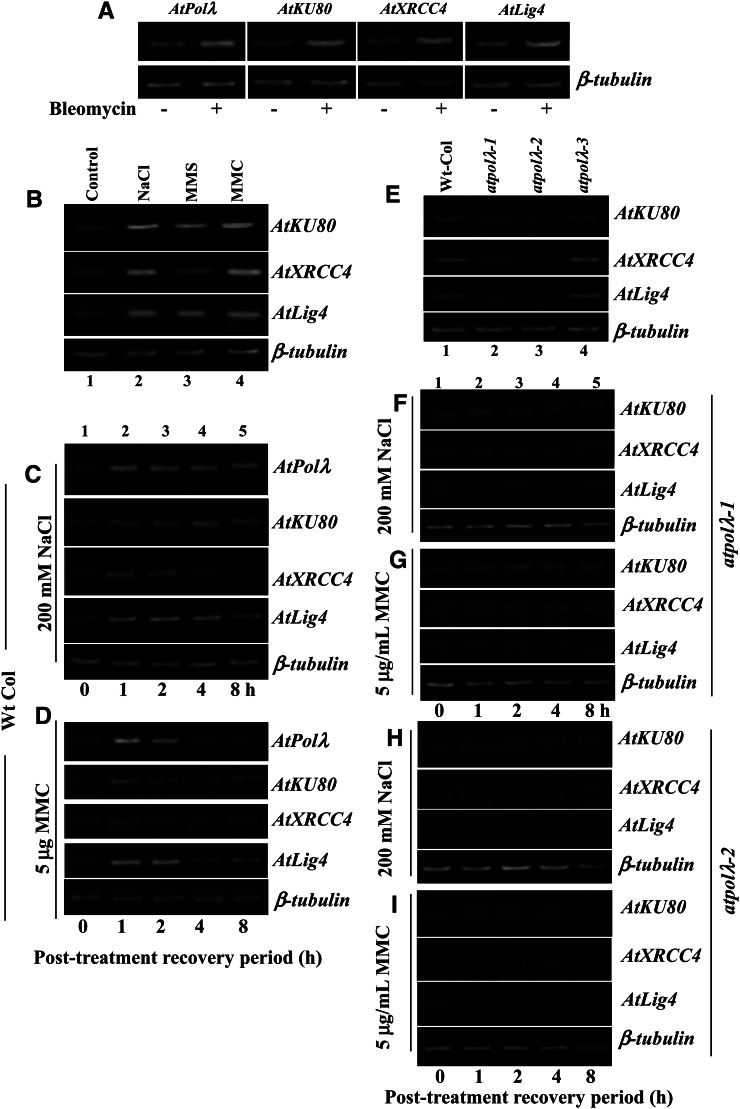
In plants, DSBs are mainly repaired by the NHEJ mechanism, which requires the step-wise participation of proteins including KU70-KU80 heterodimer, a family X DNA Pol for gap-filling DNA synthesis, and the XRCC4-DNA ligase IV complex (Fan and Wu, 2004; Lee et al., 2004; Crespan et al., 2012). In plants, a role of DNA Pol λ in DSB repair and its interaction with other components of the NHEJ pathway have not been characterized. Our results involving comet analysis in the wild type and atpolλ knockout mutants (Fig. 3, J and K) and the phenotypes of AtPolλ-overexpressing atpolλ transgenic lines (Fig. 4) indicate a possible function of DNA Pol λ in the repair of DSBs generated by high salinity. To further study AtPolλ function in DSB repair and its interaction with the other components of the NHEJ pathway in plants, we first analyzed the steady-state transcript levels of three major NHEJ-related genes in Arabidopsis, AtKU80 (AT1G48050), AtXRCC4 (AT3G23100), and AtLig4 (AT5G57160), along with AtPolλ in 7-d-old wild-type Arabidopsis seedlings after exposure to bleomycin, a well-known chemical agent that induces DSBs in DNA (Supplemental Table S3). In comparison with the untreated control, endogenous message levels of the NHEJ genes including AtPolλ increased significantly in bleomycin-treated seedlings (Fig. 5A), suggesting transcriptional induction of these genes in response to DSBs induced by bleomycin, consistent with the notion of the involvement of NHEJ genes in DSB repair. NaCl and MMC treatments also caused transcriptional up-regulation of the NHEJ genes, although different magnitudes of induction were evident (Fig. 5B; Supplemental Fig. S12, A–C). In addition, increased transcript expression of AtKU80 and AtLig4 was also detected following MMS treatment (Fig. 5B, first and third panels, lane 3). These results indicate that DSBs induced by high NaCl and MMC treatments are repaired via the “classic” NHEJ pathway. Induction of AtKU80 and AtLig4 transcripts after MMS treatment may suggest the possible involvement of these genes in the repair of DNA base methylation.
Figure 5.
Expression of DSB repair genes in response to DNA-damaging agents. A, Changes in the transcript abundance of AtPolλ, AtKU80, AtXRCC4, and AtLig4 analyzed by semiquantitative reverse transcription-PCR from 7-d-old wild-type Arabidopsis seedlings without treatment and following exposure to bleomycin (10 μg mL−1) for 8 h (top panels). The − and + symbols indicate the absence and presence of bleomycin. B, Expression of core NHEJ-related DSB repair genes AtKU80, AtXRCC4, and AtLig4 in 7-d-old wild-type Arabidopsis seedlings after treatments with DNA-damaging agents. Lane 1 in each panel indicates mRNA levels of the respective genes in untreated control seedlings, while lanes 2 to 4 indicate expression of the genes after exposure of seedlings to 200 mm NaCl, 80 µL L−1 MMS, and 5 μg mL−1 MMC, respectively, for 8 h. C and D, Changes in mRNA levels of NHEJ genes in 7-d-old wild-type seedlings in various recovery time periods after exposure to 200 mm NaCl (C) and 5 μg mL−1 MMC (D) for 8 h. The 0-h time point served as the control (lane 1). The AtPolλ loss-of-function mutation reduces the expression of core DSB repair genes. E, Expression of NHEJ genes in 7-d-old wild-type and atpolλ mutant seedlings. F and G, Changes in mRNA levels of NHEJ genes in 7-d-old atpolλ-1 mutant seedlings during various recovery time periods after treatment with 200 mm NaCl (F) and 5 μg mL−1 MMC (G) for 8 h. H and I, Expression of NHEJ genes in 7-d-old atpolλ-2 mutant seedlings during posttreatment recovery periods after exposure to 200 mm NaCl (H) and 5 μg mL−1 MMC (I) for 8 h. The 0-h time point served as the control (lane 1). Expression of β-tubulin is shown as a control (bottom panels). Quantification of the changes in transcript levels is shown in Supplemental Figure S12 for B to D and in Supplemental Figure S13 for E to I. Specificity of the amplification of transcripts was confirmed by direct sequencing of the gel-purified PCR products.
Analysis of transcript expression profiles of NHEJ-related genes in salt-treated wild-type seedlings revealed an increase in AtPolλ mRNA level (more than 2-fold) during the 1- to 2-h posttreatment incubation periods over the control. AtKU80, AtXRCC4, and AtLig4 transcript levels increased approximately 1.6-, 2-, and 2.6-fold under similar conditions (Fig. 5C; Supplemental Fig. S12, D–G). MMC treatment resulted in approximately 2.4-, 2.2-, 1.9-, and 3.1-fold increases in AtPolλ, AtKU80, AtXRCC4, and AtLig4 mRNA levels during the 1- to 2-h recovery periods over the control (Fig. 5D; Supplemental Fig. S12, H–K). Together, these results indicate transcriptional up-regulation of NHEJ genes during the early recovery periods after induction of DSBs by high salt or MMC treatment.
AtPolλ Loss of Function Leads to Decreased Expression of NHEJ Genes
Based on the above results of transcriptional activation of the NHEJ genes along with AtPolλ in wild-type Arabidopsis seedlings following high NaCl and MMC treatments, we further tested the possible interaction of AtPolλ with other NHEJ genes by investigating whether AtPolλ loss of function affects the transcriptional response of the NHEJ genes. As compared with wild-type plants, the mRNA levels of the NHEJ genes were found to be noticeably reduced in atpolλ-1 and atpolλ-2 mutant lines (Fig. 5E; Supplemental Fig. S13, A–C). In addition, NaCl or MMC treatment did not cause any significant induction of the NHEJ genes in atpolλ-1 and atpolλ-2 mutants (Supplemental Fig. S14, A and B). In both atpolλ-1 and atpolλ-2 mutants, the expression levels of the NHEJ genes remained largely unaffected during the recovery periods after high salt or MMC treatment (Fig. 5, F–I; Supplemental Fig. S13). Interestingly, we have also detected relatively decreased transcript levels of AtPolλ in the homozygous T-DNA insertion mutant lines of both AtXRCC4 (SALK_052736C) and AtLig4 (SALK_044027C) compared with wild-type Arabidopsis seedlings (Supplemental Fig. S14C). These observations have indicated that a defect in one NHEJ component may affect the expression of the other components of the NHEJ pathway and possibly indicate a complex transcriptional regulation of NHEJ genes.
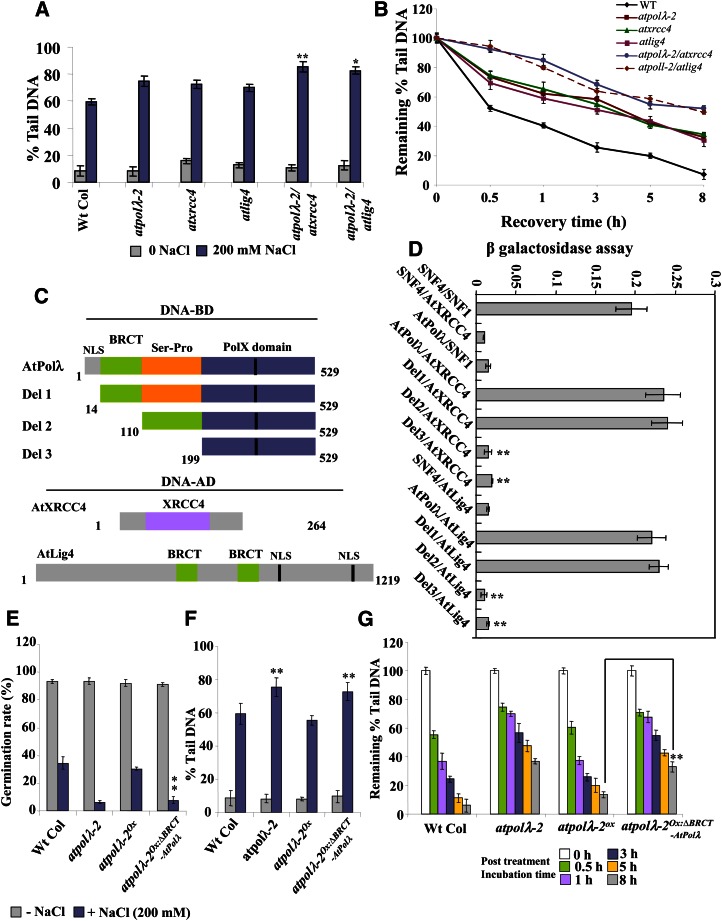
Similar to atpolλ mutants, the atxrcc4 and atlig4 T-DNA insertion mutants showed increased sensitivity toward high salinity and MMC treatments (data not shown). Based on these observations, we next analyzed the sensitivity of atpolλ-2/atxrcc4 and atpolλ-2/atlig4 double mutants toward genotoxin treatments to further understand the possible interactions of AtPolλ with AtXRCC4 and AtLig4. After high salt treatment, atpolλ-2/atxrcc4 and atpolλ-2/atlig4 double mutants showed higher DSB accumulation than single mutant lines (P < 0.01–0.007; Fig. 6A) and displayed slower DSB repair rates. During post-salt treatment recovery phases, the atpolλ-2/atxrcc4 and atpolλ-2/atlig4 double mutants showed approximately 45% to 48% and approximately 42% to 51% DSB repair, in contrast to approximately 58% to 68%, approximately 59% to 66%, and approximately 57% to 70% DSB repair in atpolλ-2, atxrcc4, and atlig4 single mutant lines after 5 to 8 h of recovery (P < 0.007–0.01; Fig. 6B). The atpolλ-2/atxrcc4 and atpolλ-2/atlig4 double mutant lines also displayed delayed DSB repair rates compared with the single mutant lines after MMC treatment (data not shown). Together, these results have indicated the involvement of DNA Pol λ and other NHEJ components, including XRCC4 and DNA Lig4, in the repair of high salinity- and MMC-induced DSBs.
Figure 6.
The atpolλ-2/atxrcc4 and atpolλ-2/atlig4 double mutants show delayed repair of DSBs. A, Seven-day-old wild-type (Wt Col), atpolλ-2, atxrcc4, atlig4, atpolλ-2/atxrcc4, and atpolλ-2/atlig4 seedlings were exposed to 200 mm NaCl for 8 h, and the extent of accumulation of DSBs was measured by comet assay under neutral conditions. B, Reduction in DSBs when seedlings were maintained in one-half-strength liquid MS medium for various time points after salt treatment. The results represent mean values ± se of triplicate assays. WT, Wild type. C, Characterization of the interaction of AtPolλ with AtXRCC4 and AtLig4 by yeast two-hybrid assay. Different constructs of AtPolλ were expressed as fusion proteins with the GAL4 DNA-binding domain (GAL4-DB), while AtXRCC4 and AtLig4 full-length proteins were expressed as fusion proteins with the GAL4 transcriptional activation domain (GAL4-AD). NLS, Nuclear localization signal. D, Interactions between proteins were measured by the induction of β-galactosidase expression in yeast following the manufacturer’s protocol. Error bars indicate se from triplicate assays. BRCT-mediated interactions of AtPolλ with AtXRCC4 and AtLig4 are important for DSB repair. E to G, Germination rates, accumulation of DSBs after salt treatment, and kinetics of DSB repair during post-salt treatment recovery periods in the wild type, the atpolλ-2 mutant line, AtPolλ-overexpressing transgenic atpolλ-2 mutants, and BRCT domain-deficient AtPolλ-overexpressing transgenic atpolλ-2 mutant lines. Data shown are means ± sd of three independent replications. *P < 0.05, **P < 0.01 relative to respective controls (n = 3). [See online article for color version of this figure.]
AtPolλ Interacts with AtXRCC4 and AtLig4
The N-terminal region of AtPolλ contains a conserved BRCT domain (Supplemental Fig. S3A), which is shared among a wide range of plant DNA repair proteins and required for protein-protein interactions. Previous reports in mammalian systems indicated interaction of Pol λ with the XRCC4-DNA Lig4 complex via its N-terminal BRCT domain, resulting in activation of the DNA synthesis activity of Pol λ (Fan and Wu, 2004). We next performed yeast-two hybrid interaction tests to establish the involvement of DNA Pol λ with XRCC4-Lig4 for NHEJ-mediated DSB repair (Fig. 6C). We confirmed that AtPolλ interacts sufficiently with AtXRCC4 and AtLig4 (Fig. 6D). However, deletion of the N-terminal BRCT domain of AtPolλ (Del2 and Del3) strongly diminished its interaction with AtXRCC4 and AtLig4, demonstrating the necessity of the BRCT domain of AtPolλ for the interaction. Based on these observations, we then determined the in vivo significance of the BRCT domain-mediated interactions of AtPolλ with XRCC4 and Lig4 by expressing the BRCT domain-deficient DNA Pol λ in the atpolλ-2 knockout line (atpolλ-2Ox:∆BRCT-AtPolλ; Supplemental Fig. S15, A–C). The atpolλ-2Ox:∆BRCT-AtPolλ transgenic seedlings did not display any significant morphological difference from the atpolλ-2 mutant line under standard growth conditions (Supplemental Fig. S15D), while it showed increased sensitivity in the presence of high salinity (Supplemental Fig. S15, E–H). The inhibition in seed germination and the extent of accumulation of DSBs in atpolλ-2Ox:∆BRCT-AtPolλ transgenic seedlings were comparable to the atpolλ-2 mutant line (Fig. 6, E and F). Furthermore, similar to the atpolλ-2 mutant line, atpolλ-2Ox:∆BRCT-AtPolλ transgenic seedlings showed delayed DSB repair rates compared with the AtPolλ-overexpressing atpolλ-2 transgenic line after salt treatment (Fig. 6G). These results were consistent with the yeast two-hybrid data and demonstrate that the BRCT-mediated interaction of DNA Pol λ with XRCC4-DNA Lig4 is essential in processing DNA ends in the classic NHEJ pathway of DSB repair.
DISCUSSION
The involvement of family X DNA Pol in DSB repair in plants remains mostly unclear. In this study, we bring further arguments to support the role of DNA Pol λ in the repair of high salinity- and DNA cross linking agent-induced DSBs in Arabidopsis. Analysis of AtPolλ endogenous message levels in wild-type Arabidopsis seedlings in the presence of high salinity and DNA cross linking agent (Fig. 1D) and the increased sensitivity of atpolλ T-DNA knockout mutants toward these genotoxic agents (Fig. 2) have indicated a possible function of Pol λ in the repair of DNA damage induced by high NaCl and MMC. Furthermore, enhanced accumulation of the AtPolλ protein in salt- and MMC-treated wild-type Arabidopsis seedlings (Fig. 3, A–F) and slower DSB repair rates in wild-type and atpolλ mutant seedlings following exposure to high salinity, as detected by comet assays (Fig. 3, J and K), have provided important evidence to suggest the involvement of Pol λ in the repair of DSBs induced by high salinity or MMC treatment in Arabidopsis.
In plants, the majority of DSBs in somatic cells are repaired via the NHEJ pathway (West et al., 2004; Puchta, 2005). However, as compared with plants, knowledge on the NHEJ-mediated DSB repair mechanism is more advanced in mammals, where the KU complex has been shown to recruit nuclease activities, DNA polymerases, and the end-processing factors, including the XRCC4-Lig4 complex, at the sites of DSBs (Lieber, 2010). The homologs of many of these factors, including the involvement of DNA Pol λ in DSB repair, have not been characterized in plants (Waterworth et al., 2011). Furthermore, information on high NaCl- or genotoxins like DNA cross linking agent-induced DNA damage and the repair mechanism is still limited in plants. Previous studies have reported an up-regulation of pea (Pisum sativum) Topoisomerase2 (TOP2) and DNA Helicase45, which serve as important components of the DNA replication and repair machinery, in response to various abiotic stresses, including high salinity (Hettiarachchi et al., 2005; Sanan-Mishra et al., 2005). It has been suggested that TOP2 may play a role in chromatin remodeling and the maintenance of proper DNA topology, thus affecting the expression of several stress-regulated genes (Hettiarachchi et al., 2005). The pea helicase PDH45 mRNA has been shown to be induced in pea seedlings in response to high salt, while overexpression of the gene under the control of a constitutive CaMV 35S promoter conferred salinity tolerance in tobacco (Nicotiana tabacum) plants, indicating PDH45, a DEAD box helicase, as a possible genetic determinant of salt tolerance and yield stability. It has been proposed that PDH45 is probably active at the translation level to facilitate stable protein synthesis, or it may act as part of multisubunit protein complexes to regulate gene expression under stress (Sanan-Mishra et al., 2005).
In our study, under salt stress, the overexpressor lines (both the wild type and atpolλ mutants) showed relatively reduced sensitivity toward high salinity, as observed in seed germination percentage, relative survival rates of seedlings, and improved DSB repair, than controls in the presence of high NaCl (Fig. 4, A and C–G), suggesting that a salt tolerance trait was functional and stable in transgenic plants. These observations were consistent with our assumption regarding the involvement of Pol λ in the repair of high NaCl-induced DSBs, thus providing better protection to overexpressor lines in the presence of high salinity. Studies from mammalian systems indicated that DNA Pol λ is recruited by the KU complex via its BRCT domain at the site of DSBs, where it participates in the processing of DNA ends in association with the XRCC4-Lig4 complex (Ma et al., 2004; Fan and Wu, 2004). In plants, as the only family X DNA Pol, the gap-filling DNA synthesis during NHEJ-mediated DSB repair is presumably carried out by Pol λ for processing DNA ends with the XRCC4-Lig4 complex. Therefore, it is likely that overexpression of AtPolλ in atpolλ knockout lines enhances the DSB repair efficiency and thus confers an improved response in the presence of high salinity. However, the enhanced DSB repair efficiency in transgenic wild-type Arabidopsis overexpressing AtPolλ was interesting. Recent studies have suggested functional redundancy of the components of other repair pathways during the initial phases of DSB repair. Extensive analyses of DSB repair kinetics in mutant lines have revealed the involvement of plant single strand break repair pathway proteins like DNA Lig1 and XRCC1 in DSB repair pathways in higher plants. KU80 and XRCC1 have been shown to act redundantly in the initial phases of DSB repair (Charbonnel et al., 2010). Based on this, we assume that the classic NHEJ components may interact with the components of other repair pathways, which also participate in DSB repair depending on the damage response. Therefore, studies on the possible interactions of Pol λ with other “nonclassic” NHEJ components of DSB repair will probably help to further uncover the role of Pol λ in plant DSB repair.
Transcriptional changes are an integral part of the DNA damage response in most organisms, including plants. However, information on the transcriptional regulation of DSB repair genes is rather limited. In plants, Ataxia Telangiectasia Mutated Kinase regulates the transcriptional DNA damage response, which is highly specific to DSB-inducing genotoxins rather than a general response to DNA damage (Molinier et al., 2005). In our study, transcriptional up-regulation of the NHEJ-related genes AtKU80, AtXRCC4, and AtLig4 in addition to AtPolλ in wild-type Arabidopsis seedlings after high NaCl and MMC treatments (Fig. 5B) have provided evidence for the involvement of classic NHEJ components in the repair of DSBs generated by high salt or MMC treatment, with DNA Pol λ as the probable gap-filling enzyme. Interestingly, expression levels of NHEJ genes, particularly AtXRCC4 and AtLig4, were found to be compromised in atpolλ knockout mutants (Fig. 5E), and even genotoxin treatments did not induce the expression of NHEJ genes in atpolλ mutants (Supplemental Fig. S14, A and B). These observations probably indicate the existence of a complex feedback/feed-forward transcriptional regulation of the components acting downstream of the NHEJ pathway after the initial binding of the Ku70-Ku80 heterodimer at the DSB termini. In the absence of gap-filling repair synthesis activity, a feed-forward inhibitory signal was probably activated to reduce the expression of genes encoding components of the ligation complex. On the other hand, the reduced expression of AtPolλ in atxrcc4 and atlig4 mutants (Supplemental Fig. S14C) may indicate the feedback inhibitory loop.
Studies from the mammalian system have indicated that, depending on the damage response, the homologous recombination (HR) and NHEJ may either compete or cooperate to repair DSBs (Hartlerode and Scully, 2009). However, we were unable to detect any significant change in the endogenous message levels of AtRAD51 (AT5G20850) and AtRAD54 (AT3G19210), two important members of Arabidopsis HR pathway, in atpolλ knockout lines (Supplemental Fig. S14D) and other NHEJ mutants, including atxrcc4 and atlig4 (data not shown), as compared with wild-type plants. In addition, RAD51 and RAD54 mRNA levels did not display any notable difference during the posttreatment recovery period in one-half-strength MS medium following treatments of atpolλ-1 and atpolλ-2 seedlings with high NaCl or MMC (Supplemental Fig. S14, E–H). These primary observations indicate that distinct regulations probably exist for NHEJ- and HR-related transcriptional DNA damage responses. However, further studies at the genetic and molecular levels are required to shed new light on the regulation and interactions of NHEJ and HR genes in higher plant genomes.
The molecular components of the HR and NHEJ pathways have been shown to be highly conserved among eukaryotes, and previous studies have indicated requirements of both these pathways for DSB repair in plants (Bray and West, 2005). The major components of DSB detection in plants include the KU70-KU80 complex, which has high affinity for broken DNA ends and also acts as a core component of the NHEJ pathway (West et al., 2004). In addition, the MRN complex, comprising of Mre11, RAD50, and NBS1, has also been implicated in DSB detection and is involved in both NHEJ- and HR-mediated DSB repair pathways (Amiard et al., 2010). In mammals, the involvement of DNA Pol λ in NHEJ as a gap-filling DNA Pol and its interaction with the XRCC4-DNA Lig4 complex via the N-terminal BRCT domain have been demonstrated (Fan and Wu, 2004; Lee et al., 2004). However, in plants, the interactions among the components of the DSB repair pathway remain mostly unclear. Previous reports have demonstrated interaction between Arabidopsis DNA Lig4 (AtLig4) and XRCC4 (AtXRCC4) via the tandem BRCT domains of the AtLig4 protein (West et al., 2000). In our study, yeast two-hybrid interaction tests confirmed direct interactions of AtPolλ with AtXRCC4 and AtLig4 (Fig. 6D). Studies using BRCT domain-deficient AtPolλ have demonstrated the key role of this domain in mediating the interactions. This finding was consistent with the DSB repair phenotype of the atpolλ-2 knockout mutant line expressing BRCT domain-deficient AtPolλ (atpolλ-2Ox:∆BRCT-AtPolλ), which displayed increased sensitivity toward high salinity and delayed DSB repair rate similar to atpolλ knockout mutant lines, suggesting the requirement of the N-terminal BRCT domain of AtPolλ in mediating the interaction with XRCC4 and Lig4 for processing broken DNA ends.
Based on the observed interaction of AtPolλ with AtXRCC4 and AtLig4, the enhanced sensitivity of atpolλ-2/atxrcc4 and atpolλ-2/atlig4 double mutants toward the genotoxic agents was puzzling, as we expected a similar level of sensitivity to the single mutants. However, studies involving plants carrying mutations for known DSB repair factors, like KU80, XRCC1 (a single strand break repair protein involved in DSB repair), XRCC2 (functions in HR pathway), and XPF (involved in single strand annealing of DSBs), both singly or in combinations, have suggested robust mechanisms for the DSB repair pathway in higher plants. In addition to KU-dependent end joining as the major pathway, an additional uncharacterized DSB repair pathway has been found to be active in plants (Waterworth et al., 2011). Moreover, the biological functions of many of the DNA Pols in plants have not been well characterized. Therefore, we anticipate the possibility of interactions of classic NHEJ components with the components of backup DSB repair pathways active in plants. In the absence of Pol λ activity, XRCC4-DNA Lig4 may participate in DSB repair, where components of a “backup” DSB repair pathway or additional DNA Pol may substitute for the tailoring activity (gap filling) of DNA Pol λ, which is consistent with the delayed DSB repair phenotype in atpolλ knockout mutants after genotoxin treatment. A similar situation probably arises for DNA Pol λ in xrcc4 and atlig4 mutants, where the involvement of components of a backup DSB repair pathway may replace the gap-sealing function of classic NHEJ components for repairing DSBs. However, when two major NHEJ components are simultaneously inactive, as in the case of double mutants, DSB repair even via utilization of the backup components probably becomes more severely affected, resulting in an enhanced phenotype of double mutants. Therefore, functional characterization of several additional components related to DSB repair but with overlapping or coordinating functions is necessary to further understand the DSB repair mechanisms in higher plant genomes.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
The Arabidopsis (Arabidopsis thaliana) mutants atpolλ-1 (Salk_075391C), atpolλ-2 (Salk_070973), atpolλ-3 (Salk_061978), atxrcc4 (Salk_052736C), and atlig4 (Salk_044027C) are in the Col-0 accession. The atpolλ-1, atpolλ-2, atpolλ-1, atxrcc4, and atlig4 T-DNA insertion lines were obtained from the T-DNA knockout collections (Alonso et al., 2003) at the Arabidopsis Biological Resource Center. Arabidopsis Col-0 seeds were surface sterilized and grown in potting soil (Soilrite) or growth medium supplemented with 0.8% Bactoagar and 1% Suc. Unless stated otherwise, the seeds in the potting soil or on growth medium-agar plates were cold treated at 4°C for 3 to 5 d and then transferred to light chambers maintained at 22°C with 16-h-light/8-h-dark periods. The intensity of white fluorescent light (Philips) was 100 µmol m−2 s−1. To obtain the homozygous mutant lines, plants heterozygous or homozygous for the atpolλ-1, atpolλ-2, or atpolλ-3 mutation were subjected to PCR genotyping analyses. Individual plants were examined by PCR using the left border-specific primer LBb1.3 (5′-ATTTTGCCGATTTCGGAAC-3′) and the AtPolλ-specific primers LP1 (5′-CAATGACCGAACTGGAGCTAG-3′) and RP1 (5′-ATACGTGTCAACGCCTGAATC-3′) for atpolλ-1, LP2 (5′-TTGGTTGGTGTCAGGTAAAGC-3′) and RP2 (5′-TCCGATTTCAACCATGAAAAC-3′) for atpolλ-2, and LP3 (5′-GACATTGCTGAGAAATTTCC-3′) and RP3 (5′-CTCGCTACCACTTATTGCAGG-3′) for atpolλ-3. The wild-type (Col-0) line was used to compare the phenotypic and genotypic differences of atpolλ mutant lines.
To test for the existence of any dominance between atpolλ-1 and atpolλ-2 mutant alleles, the homozygous atpolλ-1 (Col-0) mutant line was genetically crossed with the atpolλ-2 (Col-0) homozygous mutant line to bring together the two mutant alleles in the same plant with both alleles present, one on each homologous chromosome. The genotype of the atpolλ-1/atpolλ-2 mutant line was tested from about 35 seedlings by genomic PCR in F3 progeny. Homozygous mutant lines for atxrcc4 and atlig4 were obtained by PCR genotyping analyses using the left border-specific primer (LBb1.3) and LP4 (5′-GATCCCTCTAGAGCATCGGAG-3′) and RP4 (5′-TATAAAATCCACTAAGGCGCG-3′) for atxrcc4 and LP5 (5′-GCTTCAAGTGAGAACAGGTGC-3′) and RP5 (5′-CTGATTCGAACCAAACTCAGC-3′) for atlig. For the generation of atpolλ-2/atxrcc4 and atpolλ-2/atlig4 double mutant lines, homozygous atpolλ-2 (Col-0) mutant plants were genetically crossed with atxrcc4 and atlig4 (Col-0) homozygous mutant lines. Homozygous double mutant lines for atpolλ-2/atxrcc4 and atpolλ-2/atlig4 were screened and confirmed by the absence of the corresponding wild-type alleles in F2 progeny by genotyping.
For the generation of AtPolλ overexpressor lines, the 1.59-kb full-length AtPolλ cDNA was amplified by PCR using the gene-specific primers (forward) 5′-AGCTGGATCCATGGCGGCAAAGCGAGG-3′ and (reverse) 5′-GTTGAGCTCTCAGAGATTCCTCTCTCGTGT-3′ and cloned into the BamHI-SacI sites (underlined) of pCAMBIA 1201 binary vector under the control of the CaMV 35S constitutive promoter. Plants were transformed by Agrobacterium-mediated floral dip method. Wild-type Col-0 and atpolλ-1 and atpolλ-2 transgenic lines overexpressing the 1.59-kb full-length cDNA of AtPolλ under the control of the CaMV 35S constitutive promoter were described previously (Roy et al., 2011). Arabidopsis transgenic lines containing the 789-bp upstream promoter sequence of AtPolλ in fusion with the GUS reporter have been described previously (Roy et al., 2012). Generation of the transgenic atpolλ-2 mutant line expressing the BRCT domain-deficient AtPolλ cDNA has been described in Supplemental Materials and Methods S1.
Treatment of Seedlings and Quantitative Plant Growth Measurement
To examine the expression pattern of AtPolλ in wild-type (Col-0) Arabidopsis seedlings following treatments with salinity and other genotoxic agents during the early seedling stage, surface-sterilized seeds were plated on growth medium (containing 0.8% Bactoagar and 1% Suc), transferred to a growth chamber with 16-h-light/8-h-dark photocycles, and then 7-d-old seedlings were transferred one-half-strength liquid MS medium containing specific or different concentrations of NaCl, MMS, MMC, and H2O2. Seedlings were incubated for 8 h, and then whole seedling samples or the shoots and roots were collected separately for RNA and protein isolation. To examine the growth response of the wild type (Col-0) and atpolλ mutants toward salinity and other genotoxic stress factors at the early seedling stage, surface-sterilized seeds of the wild type and atpolλ mutants were spread on MS agar (containing 0.8% Bactoagar and 1% Suc) supplemented with different concentrations of NaCl, MMS, and MMC. After cold treatment, plates were transferred to growth chambers with 16-h-light/8-h-dark conditions. One week later, plants were screened for their sensitivity. For recovery, after high salinity or MMC treatment, seedlings were incubated in one-half-strength MS medium in the absence of genotoxic agents for various time points. It is also important to indicate here that during the initial hours of posttreatment recovery or incubation periods, plant tissues were presumably still under the influence of the genotoxic agents used for treatment, such as NaCl or MMC, which might not have completely washed out during the initial recovery time points. Quantitative plant growth measurements for the hypocotyl length of seedlings, length of the primary roots, and numbers of lateral roots were made using ImageJ software.
Southern Blotting and RNA Gel-Blot Analysis
Genomic DNA was isolated from leaves of 12-d Arabidopsis (wild-type Col-0) plants by following the method of Murray and Thompson (1980). Approximately 10 μg of restriction endonuclease-digested genomic DNA was separated by electrophoresis on a 0.8% agarose gel, samples were blotted onto a Hybond-N nylon membrane filter (Amersham), and the filter was hybridized to 32P-labeled AtPolλ cDNA as a probe (1.59 kb) under normal hybridization and washing conditions following Sambrook et al. (1989).
Total RNA was isolated from approximately 100 mg of plant tissue samples by using the RNeasy plant mini kit (Qiagen). Northern-blot analysis was carried out using 20 μg of total RNA samples following Sambrook et al. (1989). AtPolλ cDNA (1.59 kb), labeled using the Megaprime DNA labeling system (Amersham Biosciences), was used as a probe. 28S and 18S ribosomal RNA (rRNA) were used as loading controls.
Preparation of Total Protein Extracts, Immunoblotting, and Immunolocalization Assay
Total protein extracts were prepared from approximately 100 mg of plant tissues by following the method described previously (Roy et al., 2011). Protein concentration in each sample was determined by the Bradford assay (Bradford, 1976) using bovine serum albumin as a standard (Fraction V; Sigma). Immunoblot analysis was carried out using the Super Signal West Pico Chemiluminescent substrate kit (Pierce, Thermo Scientific) by following the manufacturer’s instructions. Affinity-purified rabbit anti-AtPolλ polyclonal antibody was used as the primary antibody, while goat anti-rabbit IgG (horseradish peroxidase conjugated; Thermo Scientific) was used as the secondary antibody. The immunolocalization assay was performed essentially by following the protocol of Paciorek et al. (2006). Fluorescein isothiocyanate-coupled affinity-purified anti-AtPolλ IgG was used to detect AtPolλ protein in leaf cells of 7-d-old wild-type seedlings before and after 200 mm NaCl treatment for 6 h followed by incubation of seedlings in one-half-strength liquid MS medium for 1 and 2 h. Leaf tissue sections were used for in situ localization of AtPolλ protein as described previously (Roy et al., 2011).
Analysis of the Induction and Repair of DSBs by Comet Assay
The extent of the accumulation of nuclear DNA damage in the wild type and mutant lines in response to high salinity and other chemical agents was analyzed by comet assay under neutral conditions to detect the DSBs as described previously Kozak et al. (2009).
Yeast Two-Hybrid Assay
The full-length and N-terminal deleted versions of AtPolλ cDNA fragments were cloned into the plasmid vector pAS.1 to create GAL4-DNA-binding domain fusion proteins (GAL4-DB). AtXRCC1 and AtLig4 cDNAs were cloned into the plasmid vector pGAD-C1 to create GAL4-activation domain (GAL4-AD) fusion proteins (Fig. 6C). All fusions were confirmed by sequencing. The plasmids pSE1111 and pSE1112, expressing the SNF1-GAL4-activation domain and SNF4-GAL4-DNA-binding domain, respectively, were used as controls. Yeast (Saccharomyces cerevisiae) strain Y190 was used for interaction studies (Durfee et al., 1993), and the quantitative strength of the interaction was determined by β-galactosidase expression assay using o-nitrophenyl-β-d-galactopyranoside as a substrate following the manufacturer’s instructions.
Statistical Analysis
The ANOVA (Statstica 6.0) software package was used for statistical analysis for comparing the results of germination percentage, various morphological parameters of wild-type and mutant seedlings, and extent of the accumulation of DNA damage and recovery following exposure of seedlings to high salinity and other chemical agents. P ≤ 0.05 was considered significant.
Sequence data can be found in The Arabidopsis Information Resource under the following accession numbers: AtPolλ (AT1G10520), AtKU80 (AT1G48050), AtXRCC4 (AT3G23100), AtLig4 (AT5G57160), AtRAD51 (AT5G20850), AtRAD54 (AT3G19210), and β-tubulin (AT5G44340).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Expression and purification of recombinant AtPolλ.
Supplemental Figure S2. Biochemical characterization of recombinant AtPolλ.
Supplemental Figure S3. Structural features of AtPolλ.
Supplemental Figure S4. Base compositional bias differences among Pol λ sequences.
Supplemental Figure S5. Sequence alignment of AtPolλ PolX domain.
Supplemental Figure S6. Pol λ homologs in different plant lineages.
Supplemental Figure S7. Structure of AtPolλ gene and its T-DNA insertions.
Supplemental Figure S8. Morphological features of atpolλ mutants.
Supplemental Figure S9. Salinity induced DSB accumulation and repair in atpolλ mutants.
Supplemental Figure S10. AtPolλ overexpression in atpolλ mutants.
Supplemental Figure S11. AtPolλ overexpression and salinity tolerance in E. coli.
Supplemental Figure S12. Quantification of NHEJ gene expression in Col-0 plants.
Supplemental Figure S13. Quantification of NHEJ genes in atpolλ mutants.
Supplemental Figure S14. Expression of NHEJ genes in atpolλ mutants.
Supplemental Figure S15. Phenotype of atpolλ-2 plants expressing ▵BRCT:AtPolλ.
Supplemental Table S1. Growth responses of atpolλ mutants in presence of genotoxins.
Supplemental Table S2. Genes encoding family X DNA Pols from plants and mammals.
Supplemental Table S3. Primers used for semi-quantitative RT-PCR.
Supplemental Materials and Methods S1. Supplemental Results, Materials, and Methods.
Acknowledgments
We thank Dr. Samuel H. Wilson and Dr. Rajendra Prasad (National Institute of Environmental Health Services) for generously sharing the anti-rat DNA Pol β antibody. We also thank Prof. John Doonan (Institute of Biological, Environmental, and Rural Sciences, Aberystwyth University) for providing Agrobacterium tumefaciens strain GV3101. We are thankful to Jadav Kumar Ghosh, Ashim Poddar, and Dipak C. Konar (Bose Institute) for proving necessary technical support.
Glossary
- NHEJ
nonhomologous end joining
- DSB
double strand break
- cDNA
complementary DNA
- MMC
mitomycin C
- H2O2
hydrogen peroxide
- MMS
methyl methane sulfonate
- Col-0
Columbia
- T-DNA
transfer DNA
- MS
Murashige and Skoog
- CaMV
cauliflower mosaic virus
- BRCT
breast cancer-associated C terminus
- HR
hypersensitive response
- rRNA
ribosomal RNA
References
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al. (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Amiard S, Charbonnel C, Allain E, Depeiges A, White CI, Gallego ME. (2010) Distinct roles of the ATR kinase and the Mre11-Rad50-Nbs1 complex in the maintenance of chromosomal stability in Arabidopsis. Plant Cell 22: 3020–3033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amoroso A, Concia L, Maggio C, Raynaud C, Bergounioux C, Crespan E, Cella R, Maga G. (2011) Oxidative DNA damage bypass in Arabidopsis thaliana requires DNA polymerase λ and proliferating cell nuclear antigen 2. Plant Cell 23: 806–822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyko A, Golubov A, Bilichak A, Kovalchuk I. (2010) Chlorine ions but not sodium ions alter genome stability of Arabidopsis thaliana. Plant Cell Physiol 51: 1066–1078 [DOI] [PubMed] [Google Scholar]
- Bradford MM. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72: 248–254 [DOI] [PubMed] [Google Scholar]
- Braithwaite EK, Kedar PS, Lan L, Polosina YY, Asagoshi K, Poltoratsky VP, Horton JK, Miller H, Teebor GW, Yasui A, et al (2005) DNA polymerase λ protects mouse fibroblasts against oxidative DNA damage and is recruited to sites of DNA damage/repair. J Biol Chem 280: 31641–31647 [DOI] [PubMed] [Google Scholar]
- Bray CM, West CE. (2005) DNA repair mechanisms in plants: crucial sensors and effectors for the maintenance of genome integrity. New Phytol 168: 511–528 [DOI] [PubMed] [Google Scholar]
- Britt AB. (1999) Molecular genetics of DNA repair in higher plants. Trends Plant Sci 4: 20–25 [DOI] [PubMed] [Google Scholar]
- Capp JP, Boudsocq F, Bertrand P, Laroche-Clary A, Pourquier P, Lopez BS, Cazaux C, Hoffmann JS, Canitrot Y. (2006) The DNA polymerase lambda is required for the repair of non-compatible DNA double strand breaks by NHEJ in mammalian cells. Nucleic Acids Res 34: 2998–3007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charbonnel C, Gallego ME, White CI. (2010) Xrcc1-dependent and Ku-dependent DNA double-strand break repair kinetics in Arabidopsis plants. Plant J 64: 280–290 [DOI] [PubMed] [Google Scholar]
- Crespan E, Czabany T, Maga G, Hübscher U. (2012) Microhomology-mediated DNA strand annealing and elongation by human DNA polymerases λ and β on normal and repetitive DNA sequences. Nucleic Acids Res 40: 5577–5590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dmitrieva NI, Burg MB. (2005) Hypertonic stress response. Mutat Res 569: 65–74 [DOI] [PubMed] [Google Scholar]
- Durfee T, Becherer K, Chen PL, Yeh SH, Yang Y, Kilburn AE, Lee WH, Elledge SJ. (1993) The retinoblastoma protein associates with the protein phosphatase type 1 catalytic subunit. Genes Dev 7: 555–569 [DOI] [PubMed] [Google Scholar]
- Fan W, Wu X. (2004) DNA polymerase lambda can elongate on DNA substrates mimicking non-homologous end joining and interact with XRCC4-ligase IV complex. Biochem Biophys Res Commun 323: 1328–1333 [DOI] [PubMed] [Google Scholar]
- Garcia-Diaz M, Bebenek K, Larrea AA, Havener JM, Perera L, Krahn JM, Pedersen LC, Ramsden DA, Kunkel TA. (2009) Template strand scrunching during DNA gap repair synthesis by human polymerase lambda. Nat Struct Mol Biol 16: 967–972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Díaz M, Domínguez O, López-Fernández LA, de Lera LT, Saníger ML, Ruiz JF, Párraga M, García-Ortiz MJ, Kirchhoff T, del Mazo J, et al (2000) DNA polymerase lambda (Pol lambda), a novel eukaryotic DNA polymerase with a potential role in meiosis. J Mol Biol 301: 851–867 [DOI] [PubMed] [Google Scholar]
- Hartlerode AJ, Scully R. (2009) Mechanisms of double-strand break repair in somatic mammalian cells. Biochem J 423: 157–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hettiarachchi GH, Reddy MK, Sopory SK, Chattopadhyay S. (2005) Regulation of TOP2 by various abiotic stresses including cold and salinity in pea and transgenic tobacco plants. Plant Cell Physiol 46: 1154–1160 [DOI] [PubMed] [Google Scholar]
- Irarrazabal CE, Burg MB, Ward SG, Ferraris JD. (2006) Phosphatidylinositol 3-kinase mediates activation of ATM by high NaCl and by ionizing radiation: role in osmoprotective transcriptional regulation. Proc Natl Acad Sci USA 103: 8882–8887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak J, West CE, White C, da Costa-Nunes JA, Angelis KJ. (2009) Rapid repair of DNA double strand breaks in Arabidopsis thaliana is dependent on proteins involved in chromosome structure maintenance. DNA Repair (Amst) 8: 413–419 [DOI] [PubMed] [Google Scholar]
- Lee JW, Blanco L, Zhou T, Garcia-Diaz M, Bebenek K, Kunkel TA, Wang Z, Povirk LF. (2004) Implication of DNA polymerase lambda in alignment-based gap filling for nonhomologous DNA end joining in human nuclear extracts. J Biol Chem 279: 805–811 [DOI] [PubMed] [Google Scholar]
- Lieber MR. (2010) The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annu Rev Biochem 79: 181–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y, Lu H, Tippin B, Goodman MF, Shimazaki N, Koiwai O, Hsieh CL, Schwarz K, Lieber MR. (2004) A biochemically defined system for mammalian nonhomologous DNA end joining. Mol Cell 16: 701–713 [DOI] [PubMed] [Google Scholar]
- Molinier J, Oakeley EJ, Niederhauser O, Kovalchuk I, Hohn B. (2005) Dynamic response of plant genome to ultraviolet radiation and other genotoxic stresses. Mutat Res 571: 235–247 [DOI] [PubMed] [Google Scholar]
- Moon AF, Garcia-Diaz M, Batra VK, Beard WA, Bebenek K, Kunkel TA, Wilson SH, Pedersen LC. (2007) The X family portrait: structural insights into biological functions of X family polymerases. DNA Repair (Amst) 6: 1709–1725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray MG, Thompson WF. (1980) Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res 8: 4321–4325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohba T, Kometani T, Shoji F, Yano T, Yoshino I, Taguchi K, Kuraoka I, Oda S, Maehara Y. (2009) Expression of an X-family DNA polymerase, pol lambda, in the respiratory epithelium of non-small cell lung cancer patients with habitual smoking. Mutat Res 677: 66–71 [DOI] [PubMed] [Google Scholar]
- Paciorek T, Sauer M, Balla J, Wiśniewska J, Friml J. (2006) Immunocytochemical technique for protein localization in sections of plant tissues. Nat Protoc 1: 104–107 [DOI] [PubMed] [Google Scholar]
- Picher AJ, García-Díaz M, Bebenek K, Pedersen LC, Kunkel TA, Blanco L. (2006) Promiscuous mismatch extension by human DNA polymerase lambda. Nucleic Acids Res 34: 3259–3266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puchta H. (2005) The repair of double-strand breaks in plants: mechanisms and consequences for genome evolution. J Exp Bot 56: 1–14 [DOI] [PubMed] [Google Scholar]
- Ramadan K, Shevelev I, Hübscher U. (2004) The DNA-polymerase-X family: controllers of DNA quality? Nat Rev Mol Cell Biol 5: 1038–1043 [DOI] [PubMed] [Google Scholar]
- Roy S, Choudhury SR, Singh SK, Das KP. (2011) AtPolλ, a homolog of mammalian DNA polymerase λ in Arabidopsis thaliana, is involved in the repair of UV-B induced DNA damage through the dark repair pathway. Plant Cell Physiol 52: 448–467 [DOI] [PubMed] [Google Scholar]
- Roy S, Choudhury SR, Singh SK, Das KP. (2012) Functional analysis of light-regulated promoter region of AtPolλ gene. Planta 235: 411–432 [DOI] [PubMed] [Google Scholar]
- Roy S, Singh SK, Choudhury SR, Sengupta DN. (2009) An insight into the biological functions of family X-DNA polymerase in DNA replication and repair of plant genome. Plant Signal Behav 4: 678–681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy Choudhury S, Roy S, Das R, Sengupta DN. (2008) Differential transcriptional regulation of banana sucrose phosphate synthase gene in response to ethylene, auxin, wounding, low temperature and different photoperiods during fruit ripening and functional analysis of banana SPS gene promoter. Planta 229: 207–223 [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T (1989) Molecular Cloning: A Laboratory Manual, Ed 2. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- Sanan-Mishra N, Pham XH, Sopory SK, Tuteja N. (2005) Pea DNA helicase 45 overexpression in tobacco confers high salinity tolerance without affecting yield. Proc Natl Acad Sci USA 102: 509–514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuteja N, Ahmad P, Panda BB, Tuteja R. (2009) Genotoxic stress in plants: shedding light on DNA damage, repair and DNA repair helicases. Mutat Res 681: 134–149 [DOI] [PubMed] [Google Scholar]
- Uchiyama Y, Kimura S, Yamamoto T, Ishibashi T, Sakaguchi K. (2004) Plant DNA polymerase lambda, a DNA repair enzyme that functions in plant meristematic and meiotic tissues. Eur J Biochem 271: 2799–2807 [DOI] [PubMed] [Google Scholar]
- Watanabe K, Pacher M, Dukowic S, Schubert V, Puchta H, Schubert I. (2009) The STRUCTURAL MAINTENANCE OF CHROMOSOMES 5/6 complex promotes sister chromatid alignment and homologous recombination after DNA damage in Arabidopsis thaliana. Plant Cell 21: 2688–2699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterworth WM, Drury GE, Bray CM, West CE. (2011) Repairing breaks in the plant genome: the importance of keeping it together. New Phytol 192: 805–822 [DOI] [PubMed] [Google Scholar]
- West CE, Waterworth WM, Jiang Q, Bray CM. (2000) Arabidopsis DNA ligase IV is induced by γ-irradiation and interacts with an Arabidopsis homologue of the double strand break repair protein XRCC4. Plant J 24: 67–78 [DOI] [PubMed] [Google Scholar]
- West CE, Waterworth WM, Sunderland PA, Bray CM. (2004) Arabidopsis DNA double-strand break repair pathways. Biochem Soc Trans 32: 964–966 [DOI] [PubMed] [Google Scholar]
- Winter D, Vinegar B, Nahal H, Ammar R, Wilson GV, Provart NJ. (2007) An “Electronic Fluorescent Pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS ONE 2: e718. [DOI] [PMC free article] [PubMed] [Google Scholar]