An exploratory data-analysis method describes changes in gene expression profiles across multiple tissues between species.
Abstract
Developmental differences between species commonly result from changes in the tissue-specific expression of genes. Clustering algorithms are a powerful means to detect coexpression across tissues in single species but are not often applied to multidimensional data sets, such as gene expression across tissues in multiple species. As next-generation sequencing approaches enable interspecific analyses, methods to visualize and explore such data sets will be required. Here, we analyze a data set comprising gene expression profiles across six different tissue types in domesticated tomato (Solanum lycopersicum) and a wild relative (Solanum pennellii). We find that self-organizing maps are a useful means to analyze interspecies data, as orthologs can be assigned to independent levels of a “super self-organizing map.” We compare various clustering approaches using a principal component analysis in which the expression of orthologous pairs is indicated by two points. We leverage the expression profile differences between orthologs to look at tissue-specific changes in gene expression between species. Clustering based on expression differences between species (rather than absolute expression profiles) yields groups of genes with large tissue-by-species interactions. The changes in expression profiles of genes we observe reflect differences in developmental architecture, such as changes in meristematic activity between S. lycopersicum and S. pennellii. Together, our results offer a suite of data-exploration methods that will be important to visualize and make biological sense of next-generation sequencing experiments designed explicitly to discover tissue-by-species interactions in gene expression data.
The hypothesis that cis-regulatory changes in gene expression are sufficient to cause morphological differences between species has been demonstrated multiple times (Britten and Davidson, 1969, 1971; King and Wilson, 1975; for review, see Doebley and Lukens, 1998; Romero et al., 2012). In many of these examples, the changes in regulation do not necessarily affect gene expression levels per se but rather alter the spatiotemporal pattern of expression. One of the most intuitive examples of how changes in the spatial expression of genes can modify form is the Hox genes, the ever-shifting, augmented, and diminished expression of which across modular animal body plans has created a staggering diversity of morphologies (Carroll, 2000). Changes in gene expression profiles across tissues between species are an example of “heterotropy,” describing spatial differences (rather than temporal, as in heterochrony) between species (Carroll, 2008). Both heterotropy and heterochrony are important factors when analyzing differences in gene expression between plant species, which iteratively produce different types of organs (Chitwood et al., 2012b; Chitwood and Sinha, 2013).
Our understanding of how cis-regulatory changes and the modulation of spatial expression affect morphological change has been limited by technology. Whereas previously, forward and reverse genetic approaches were used to study a single gene or a few genes in great detail, microarrays and next-generation sequencing now allow transcriptome-wide assessments of expression levels to be determined (Wang et al., 2009). To date, a number of studies have used these technologies to create expression atlases in plants, albeit in a single species. Cell-type resolution of gene expression in the root (Birnbaum et al., 2003; Brady et al., 2007; Dinneny et al., 2008), temporal and spatial expression patterns in leaves and the meristem (Efroni et al., 2008; Jiao et al., 2009; Li et al., 2010; Park et al., 2012; Takacs et al., 2012), and comparisons of gene expression across mutant genotypes in specific tissues (Eveland et al., 2010) are just a few examples of the unprecedented perspective into the developmental regulation of gene expression that transcriptomic approaches provide.
Nonetheless, transcriptome-wide changes in spatial expression between species remain understudied. Recently, a comparison of gene expression in six different organs of 10 different mammals and birds revealed lineage-specific changes in expression profiles and determined the role of selection in modulating spatial expression profiles (Brawand et al., 2011). In plants, a comparison of gene expression in floral organs among angiosperms demonstrated increasing canalization and organ-specific expression in derived lineages (Chanderbali et al., 2010).
Interspecies studies, such as the above, will undoubtedly become more commonplace as next-generation sequencing obviates the deficit of sequenced genomes/transcriptomes in nonmodel organisms through de novo assembly. Such studies will require exploratory statistical methods to quantitatively analyze the overwhelming number of tissue expression patterns and their possible differences between species. Statistical models that can identify genes with significant tissue-by-species interaction effects (i.e. changes in the tissue-specific expression of genes between species) exist, but they are incapable of informing about patterns across multiple tissues. Additionally, a researcher may be concerned with overall patterns of tissue-by-species changes in the data set and not just those that pass an arbitrary statistical significance threshold. Another problem facing interspecies comparisons is orthology. Beyond the immediate problem of identifying orthologs is whether orthologs should be analyzed independently or somehow comparatively as pairs. Although powerful, commonly used clustering methods, such as hierarchical clustering and k-means clustering, are often used to cluster across a single factor, such as tissue, and are not designed to deal with multidimensional data sets (such as when a gene has multiple expression patterns for each ortholog).
Here, we analyze a data set sampling the transcriptome of six different tissues in two different species: domesticated tomato (Solanum lycopersicum ‘M82’) and a desert-adapted wild relative, Solanum pennellii. We find that self-organizing maps (SOMs), a type of artificial neural network, are convenient for clustering in the context of multiple factors (Kohonen, 1997; Tamayo et al., 1999; Wehrens and Buydens, 2007). Our SOM clusters yield groups of genes with similar expression profiles that have a biological basis, as revealed through Gene Ontology (GO) enrichment analysis and the inclusion of relevant genes with known tissue-specific functions. We also find that the manner in which the expression profile of a gene changes between species depends on overall tissue-specific expression. That is, ortholog pairs assigned to specific clusters, based on their overall expression profile across tissues, exhibit distinct changes in the patterns of their expression profiles between species. Clustering genes based not on their absolute expression profile but rather on their change in expression pattern between species reveals a group of genes that have increased expression in the meristematic tissues of S. lycopersicum, reflective of the increased meristem size in this species relative to S. pennellii. Included in this group of genes are LeT6, WIRY4, and the PIN1 tomato (Solanum spp.) ortholog, all known modulators of tomato shoot apical meristem development (Janssen et al., 1998a, 1998b; Kim et al., 2003; Reinhardt et al., 2003; Pattison and Catalá, 2012; Yifhar et al., 2012). Our results provide a means to explore the tissue-specific expression of genes between species and demonstrate the utility of next-generation sequencing approaches in exploring the molecular changes associated with morphological evolution.
RESULTS
SOMs and superSOMs
A number of methods exist to cluster genes into groups, the members of which possess similar expression profiles over the levels of a factor (e.g. genes with similar expression profiles measured across tissues). The various solutions to this problem carry different limitations (Tamayo et al., 1999). Human-guided clustering has obvious advantages, as nuanced criteria difficult to express algorithmically can be used to discern complex phenomena. Unfortunately this method is subjective, does not scale well, and has rarely been used (Cho et al., 1998). One of the most commonly used approaches is hierarchical clustering, in which genes are placed into a rigid hierarchy of subset groups (Eisen et al., 1998). This may be the ideal way to describe some data sets. However, the numerous patterns of gene expression across the tissues of an organism are not necessarily hierarchically organized. Another popular approach is k-means clustering, in which a set number of clusters (“k”) divides the space describing all expression patterns such that the distance of all gene expression profiles to cluster geometric centers (“centroids”) is minimized (Tavazoie et al., 1999). A disadvantage of k-means clustering is that it does not take into account the topology of clusters to each other; it merely partitions the gene expression space (similar to a Voronoi diagram; Aurenhammer, 1991).
SOMs are another commonly used method to cluster gene expression profiles (Tamayo et al., 1999). SOMs result from a process in which neighboring clusters influence each other, resulting in a network topology reminiscent of biological systems (especially neural networks; Kohonen, 1982, 1997; Kangas et al., 1990; Wehrens and Buydens, 2007). The process begins by randomly assigning data to clusters. That is, a gene expression profile is at random assigned to a cluster, becoming its “codebook vector.” The codebook vector is the gene expression profile that represents the cluster in subsequent training. An expression profile is then randomly selected and assigned to the closest “winning cluster,” based on proximity to the cluster codebook vector using a distance metric. When assigned to a cluster, the data alter the respective cluster’s codebook vector as a weighted average. SOMs are spatially constrained in that the influence of data assigned to a winning cluster extends beyond the winning cluster to neighboring clusters. This is a key difference from k-means clusters: SOMs literally arrange clusters as a map (for an example, see Fig 1, E, F, H, and I) and, therefore, have a topology. This spatial constraint influences the extent to which neighboring clusters alter each other. The process described above is repeated over a specified number of iterations as the clusters adapt to the given data set. At the end of this training period, data are given a final assignment to winning clusters.
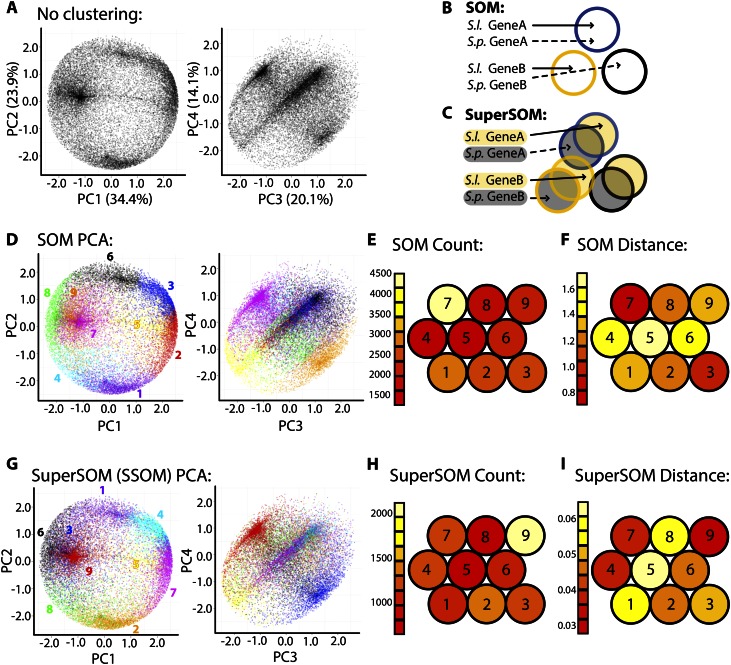
Figure 1.
PCA, SOMs, and superSOMs. A, PCA was performed on gene expression across tissues. The expression profile of each gene is represented twice: one point representing the S. lycopersicum ortholog and the other representing the S. pennellii ortholog. Densities representing overabundant gene expression patterns are easily observed. Variance explained by each PC is indicated. B and C, Diagrams demonstrating the differences between SOMs and superSOMs. Under a regular SOM approach (B), S. lycopersicum (S.l.) and S. pennellii (S.p.) orthologs are assigned to clusters without regard to the species they represent. Using SOM methods, orthologs can be assigned to different clusters. In a superSOM scheme (C), S. lycopersicum orthologs are clustered in a dimension separate from S. pennellii orthologs. Each ortholog in a superSOM is assigned to the same cluster. D to I, PCA and clustering results for SOM (D–F) and superSOM (G–I) approaches. D and G, Genes belonging to different SOM (D) and superSOM (G) clusters are indicated by different colors and projected on the PC space. Note the ability of both methods to explain major densities represented in the PC space and the high correspondence between the expression profiles of genes represented by the clusters in each data set (i.e. SOM/superSOM clusters include genes occupying similar regions and densities in the PC space). Color assignments are arbitrary, and corresponding SOM and superSOM clusters are not the same color. E and H, Number of genes assigned to each SOM cluster (E) and ortholog pairs assigned to each superSOM cluster (H). Note that there are half as many assignments to clusters in a superSOM scheme compared with SOMs. F and I, Mean Euclidean distance of cluster members to codebook vectors in the SOM (F) and superSOM (I). For SOM and superSOM diagrams, the 3 × 3 hexagonal topology is shown. Clusters with similar codebook vectors lie closer to each other than those with disparate codebook vectors. Red indicates low count/Euclidean distance, and white indicates high count/Euclidean distance.
The influence of neighboring clusters on each other is not unlike the Hebbian theory of “neurons that fire together wire together,” and indeed, SOMs are classified as “artificial neural networks” (Hebb, 1949). The principle of clustering using spatial constraints and topology has ramifications in developmental biology and the creation of gene expression atlases. For example, if gene expression is influenced by continuous, spatial factors in an organism (e.g. morphogens and/or hormone gradients) and finite, discrete subsets of the organism are sampled (e.g. cell types, tissues, and/or organs), a properly constructed SOM can reflect the underlying topology and relationships between identified clusters of gene coexpression (Kohonen, 1982; Kangas et al., 1990).
To explore the utility of SOMs in identifying groups of genes with similar expression profiles, we analyzed RNA-Seq data collected from S. lycopersicum and S. pennellii. Gene expression values from the inflorescence, mature leaf, root, seedling, stem, and vegetative apex were mean centered and variance scaled to measure differences attributable to changes in tissue-specific expression rather than magnitude. Importantly, the expression profiles of each ortholog (one from S. lycopersicum, the other from S. pennellii) were scaled across tissues independently. The result is that purely developmental differences in expression profiles of orthologs across tissues are being analyzed. That is, additive species effects (e.g. intrinsically higher expression in one species compared with the other) are effectively eliminated. Genes with differential expression across two species are best analyzed using statistical methods such as fitting generalized linear models and focusing on those genes significant for the species model term (Oshlack et al., 2010). Although we focus on tissue effects rather than species effects in this study, the approaches we describe can accommodate species effects by analyzing ortholog pairs and clustering using the expression of genes across 12 tissues (six for each species). Had the analysis been performed this way, patterns of variance with respect to tissue and species would be confounded, as witnessed by the correlation of principal component (PC) values with fold change expression values between orthologs (Supplemental Fig. S1).
Before clustering, we decided on a cluster number, a problem of variable and feature selection that plagues SOMs and k-means clustering alike (Guyon and Elisseeff, 2003). To help inform our cluster number decision, we visualized the expression profiles of genes through a principal component analysis (PCA; Fig. 1A; Supplemental Fig. S2). In this PCA, each gene is represented twice: one data point representing the tissue expression of the S. lycopersicum ortholog and the other point representing the profile of the S. pennellii ortholog. By observing densities in the PC space (the densest regions representing many genes with similar expression profiles), an appropriate cluster number can be estimated. There should be sufficient clusters to describe distinct, prevalent expression patterns (densities in the PC space) but not so many clusters that a given expression pattern is redundantly covered.
After visualizing the PC space and deciding on a cluster number and SOM topology (3 × 3, hexagonal), we then created a SOM in which the orthologs from each species were independently assigned to clusters. The result of this clustering scheme is that it is possible for orthologs of a gene to be assigned to different clusters (Fig. 1B). Genes belonging to different clusters explain common expression patterns in the data, as visualized by coloring cluster membership in PC space and the correspondence of cluster identity with distinct densities (Fig. 1D). The clusters representing the most distinct densities (e.g. SOM cluster 7, magenta; Fig. 1D) can have high cluster membership (which can contribute to very dense PC regions; Fig. 1E) and lower Euclidean distances of cluster members to the cluster codebook vector (essentially the centroid; Fig. 1F). Clusters representing more diffuse patterns in PC space (e.g. SOM cluster 5, yellow; Fig. 1D) contain members with higher Euclidean distances to their codebook vectors (Fig. 1F).
The robustness of clustering over a single factor (tissue, in this instance) using a SOM is exemplified by the fact that k-means and hierarchical clustering yield comparable results. If the k number of clusters is selected to match that used for the SOM (nine clusters; Fig. 1D), then genes are assigned to k-means clusters that bear a striking resemblance in the expression to genes assigned to SOM clusters (Supplemental Figs. S3 and S4). Similarly, if hierarchical clustering on the genes used for SOM clustering is performed, genes assigned to the same SOM cluster often cluster together (Supplemental Fig. S5). If merely clustering over a single factor (tissue) were our only goal, then any of these three widely used clustering approaches (SOM, k-means clustering, hierarchical clustering) could be used.
However, none of the methods, as presented above, can accommodate clustering when two or more factors are being analyzed. Such a consideration is important in evolutionary and developmental studies, in which species is an additional factor to that of tissue. Allowing orthologs to cluster independently does not satisfy the reality that each is a presumed, derived variant of a common ancestral gene. Is there a way that clustering could proceed considering the relationship of orthologs to each other? SOMs are particularly well suited to this question of dimensionality (in this instance, the additional dimension, or factor, of species identity). In a “superSOM,” clusters have dimensionality and a separate identity associated with each data set, but ultimately, data must be assigned to the same cluster (Wehrens and Buydens, 2007). The distance of data to a cluster is measured as a weighted sum between the identities associated with each data dimension. The dimensionality of superSOMs has obvious relevance to measuring gene expression data between orthologous groups of genes across species.
Using a superSOM, orthologous pairs of genes were assigned to clusters based on their expression profile within each species (Fig. 1C). Compared with a SOM approach, in which all genes, regardless of their species affiliation, were clustered, the results are remarkably similar. Using a 3 × 3 hexagonal topology, both SOM and superSOM clusters explain similar densities in PC space (Fig. 1, D and G; note that colors indicating SOM and superSOM clusters in PC space are arbitrary and not the same between corresponding clusters, so that SOM and superSOM clusters, which are different, are not confused for each other). For example, SOM cluster 7 and superSOM cluster 9 both contain genes residing in a similar region of PC space (Fig. 1, D and G), have relatively high cluster membership counts (Fig. 1, E and H), and have low member Euclidean distances to the codebook vector profile (Fig. 1, F and I). Similarly, SOM cluster 5 and superSOM cluster 5 contain members occupying a sparse region of PC space (Fig. 1, D and G) and have members with high Euclidean distance to the codebook vectors (Fig. 1, F and I).
The correspondence between SOM and superSOM clusters is best realized by comparing codebook vectors (Fig. 2A). The codebook vectors of the resultant superSOM and SOM clusters are highly correlated. Additionally, within the superSOM results, the S. lycopersicum and S. pennellii codebook vectors are highly correlated. For example, SOM cluster 6 and superSOM cluster 1 are both highly expressed in the leaf relative to other tissues (Fig. 2A). Both the S. lycopersicum and corresponding S. pennellii orthologs assigned to superSOM cluster 1 have a similar high leaf expression profile, although there are some differences (S. lycopersicum has higher stem expression relative to S. pennellii). superSOM and SOM results vary from one analysis to another (as do the results from k-means and many other clustering methods) due to the random selection of initial data inputs. However, the similarity between the results in this instance demonstrates the consistency of SOM methods when applied to tissue-by-species gene expression data sets.
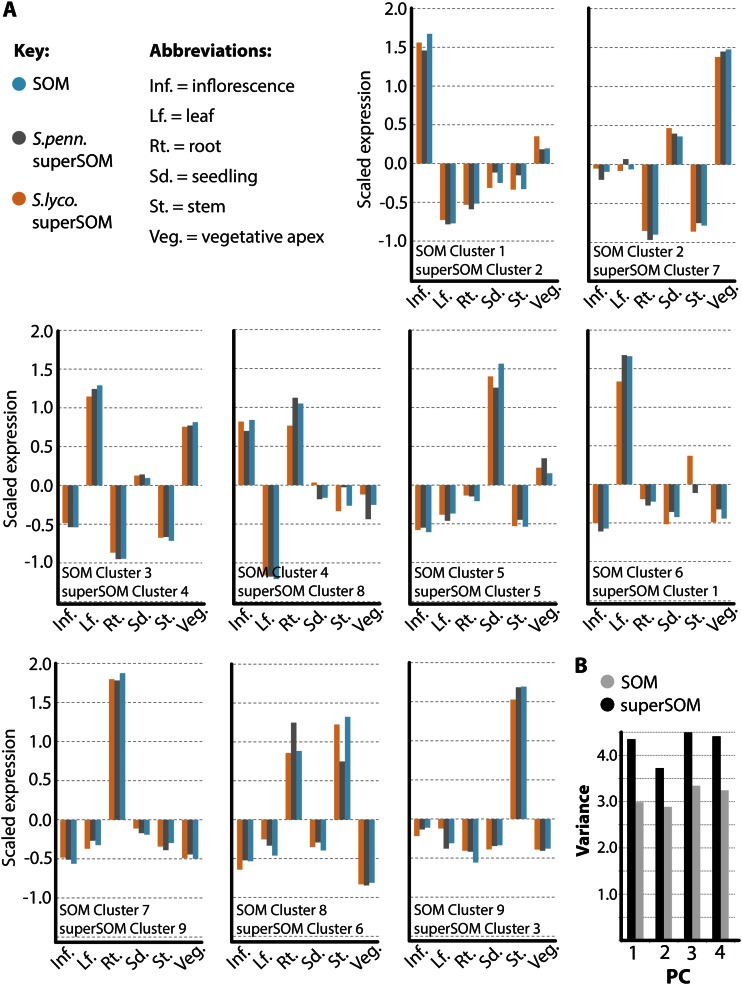
Figure 2.
Correspondence between SOM and superSOM results. A, Codebook vector values representing the identities of SOM and superSOM clusters used in this study. SOM and superSOM clusters correspond based on their overall expression profiles and positions in PC space (Fig. 1). Codebook vectors representing SOM clusters (blue) and superSOM clusters (S. lycopersicum [S.lyco.] in orange and S. pennellii [S.penn.] in gray) exhibit high correlation. B, Comparison of the sum of variances for PCs explaining variation in gene expression across clusters using SOM and superSOM approaches. As expected, because orthologs are forced to occupy the same cluster in a superSOM approach, superSOM cluster members exhibit higher variance in gene expression relative to SOM clusters. Inf., Inflorescence; Lf., leaf; Rt., root; Sd., seedling; St., stem; Veg., vegetative apex. [See online article for color version of this figure.]
To ask if SOM-based clustering yields biologically relevant results, we performed GO enrichment analysis of the clustered genes (Supplemental Tables S1 and S2). We found compelling GO enrichments in many clusters. For example, SOM cluster 1/superSOM cluster 2 genes exhibit high inflorescence expression (Fig. 2A) and are significantly enriched for GO terms relating to cell division, differentiation, embryo and flower development, and epigenetic regulation (Supplemental Table S2). Included in these clusters are CRABS CLAW, ARGONAUTE4, DUO POLLEN1, SQUAMOSA PROMOTER-BINDING PROTEIN-LIKE3/SQUAMOSA PROMOTER-BINDING PROTEIN-LIKE8, ABNORMAL FLORAL ORGANS, STYLISH1, REDUCED VERNALIZATION RESPONSE1, VERNALIZATION INSENSITIVE3, AGAMOUS, UNUSUAL FLORAL ORGANS, APETALA1/APETALA3, SEPALLATA1/SEPALLATA2/SEPALLATA3/SEPALLATA4, CURLY LEAF, INCURVATA2, KRYPTONITE, EXCESS MICROSPOROCYTES1, and PISTILLATA homologs, all known regulators of inflorescence development (Supplemental Table S1). Likewise, genes with high expression in the vegetative apex (SOM cluster 2/superSOM cluster 7; Fig. 2A) are significantly enriched for GO terms associated with development, transcriptional regulation, and translation (Supplemental Table S2). Included among these genes are homologs of the stomatal regulators MUTE, SPEECHLESS, FAMA, TOO MANY MOUTHS, and STOMATAL DENSITY AND DISTRIBUTION and the regulators of shoot apical meristem development SAW1, ASYMMETRIC LEAVES2, TCP DOMAIN PROTEIN4/TCP DOMAIN PROTEIN5/TCP DOMAIN PROTEIN12, YABBY2/YABBY5, MERISTEM LAYER1, and PRESSED FLOWER (Supplemental Table S1). SOM cluster 3/superSOM cluster 4 genes are highly expressed in photosynthetic structures such as leaves, the vegetative apex, and seedlings (Fig. 2A) and are predictably enriched for terms relating to photosynthesis, thykaloids, plastids, and biosynthetic processes (Supplemental Table S2). Genes that possess relatively high expression in tissues with vascular transport and translocation functions, such as the stem and root (SOM clusters 7 and 9/superSOM clusters 9 and 3; Fig. 2A), are enriched for transport, endoplasmic reticulum, Golgi apparatus, and carbohydrate metabolic GO terms (Supplemental Table S2). Transporters and secretion pathway genes, including nitrate, ammonium, potassium, zinc, and sugar transporters; cation efflux proteins; PHOSPHATE1; major facilitator superfamily transporters; multi antimicrobial extrusion protein transporters; sodium-calcium exchangers; general secretory pathway members; proton-dependent oligopeptide transporters; expansins, invertases, pectin esterases, pectin methylesterase inhibitors, hydrolases, transferases, and proteases; nodulin-like proteins; syntaxins; SUCROSE SYNTHASE6; and GNOM are represented in these clusters (Supplemental Table S1).
Despite similarities in the SOM and superSOM results presented above, differences do exist. For example, because of the constraint that orthologs occupy the same cluster in a superSOM, the variance of gene expression patterns for genes assigned to superSOM clusters is higher than that for those assigned to SOM clusters (Fig. 2B). This observation is reflective of the “compromise” made in assigning a pair of orthologs to a cluster, compared with assigning the ortholog from each species to its ideal cluster in a SOM scheme (Fig. 1, B and C). The greatest dissimilarity between SOM and superSOM results, however, lies in comparing ortholog expression across species, which we detail in the next section.
Species Differences in Expression between superSOM- and SOM-Assigned Orthologs
As presented, SOM-based approaches consistently assign genes with similar tissue-specific expression patterns to clusters. Within this context, we wanted to determine the best way to visualize the additional factor of species, and more importantly, tissue-by-species interactions. Such changes in the tissue-specific regulation of gene expression between species are key to understanding the transcriptome-wide changes that track morphological evolution. We began by examining differences between orthologs assigned to superSOM clusters. Assignment of both members of an orthologous pair to the same cluster simplifies matters, as a within-cluster comparison of species differences in tissue expression can be made.
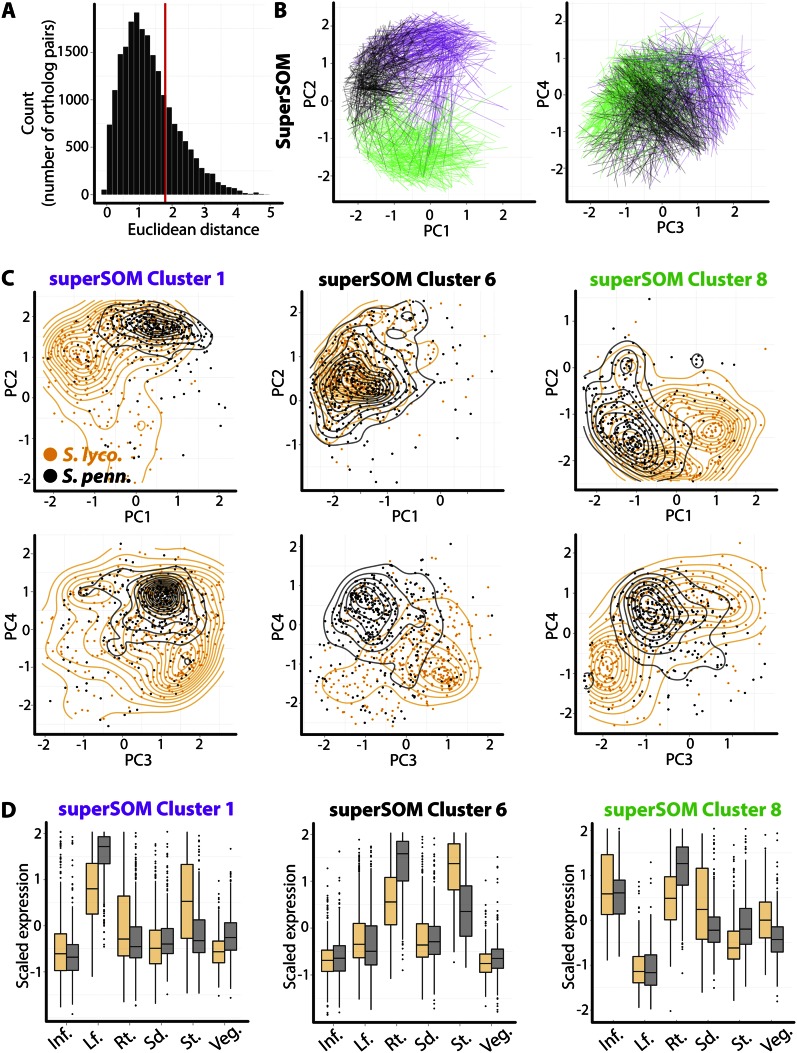
To aid the analysis of gene expression over six tissues, we use PCs as a distance metric. Calculating the Euclidean distance in PC space between orthologs yields a distribution skewed toward longer distances (Fig. 3A). To focus on the most extreme changes in tissue-specific expression, we examined only those orthologs occupying the upper quartile of Euclidean distance from each other. In three examples, superSOM clusters 1 (purple), 6 (black), and 8 (green), the distance between orthologs with the most divergent expression patterns can be visualized as lines connecting their positions in PC space (Fig. 3B).
Figure 3.
Differences in tissue-specific expression of superSOM clustered orthologous pairs. A, Distribution of Euclidean distances between corresponding orthologs in PC space. This metric represents the overall difference in tissue-specific expression between orthologs. To focus on those genes with the greatest tissue-by-species differences, we analyze only those orthologs belonging to the upper quartile of the distribution (indicated by the red line). B, Lines connecting the positions of orthologs in PC space for three select clusters: superSOM cluster 1 (purple), cluster 6 (black), and cluster 8 (green). C, An easier way to analyze the tissue-specific expression of different species is to look at the overall distribution of orthologs in PC space. Shown are the PC positions of genes belonging to each cluster, separated by ortholog identity and layered with contour plots to aid visualization. Note the different regions of PC space that orthologs from S. lycopersicum (S. lyco.) and S. pennellii (S. penn.) occupy. D, Differences in PC space occupied by orthologs translate into tissue-by-species changes in expression patterns: superSOM cluster 1 genes, for example, exhibit lower expression in leaves and higher expression in stems in S. lycopersicum compared with S. pennellii. Inf., Inflorescence; Lf., leaf; Rt., root; Sd., seedling; St., stem; Veg., vegetative apex.
An analysis of the overall distribution of S. lycopersicum and S. pennellii orthologs belonging to each cluster gives a better idea of the tissue-specific differences in expression between species orthologs. Clear shifts in expression profiles can be observed in PC space using this method (Fig. 3C). The S. lycopersicum orthologs in superSOM cluster 1, for example, have lower PC1 and PC2 values and are bimodally distributed around their S. pennellii counterparts in PC3-PC4 space. Of course, these shifts in PC space represent changes in the expression patterns of the genes between these two species (Fig. 3D). superSOM cluster 1 genes are highly expressed in the leaf in both species, but S. pennellii orthologs show higher expression in the leaf and lower expression in the stem compared with S. lycopersicum. Another way to interpret these results is that in S. lycopersicum these genes have more intermediary expression between the stem and leaf but more disparate expression in S. pennellii, a recurring theme in the transcriptome-wide differences between these species that we discuss again later. Similarly, superSOM cluster 6 genes are expressed differentially in the root and stem, with lower S. lycopersicum expression in the root and higher expression in the stem relative to S. pennellii. The shifts in tissue-specific expression between species for superSOM cluster 8 are complex but are distinct when viewed in PC space and demonstrate the importance of using methods such as these to visualize tissue-by-species changes in gene expression.
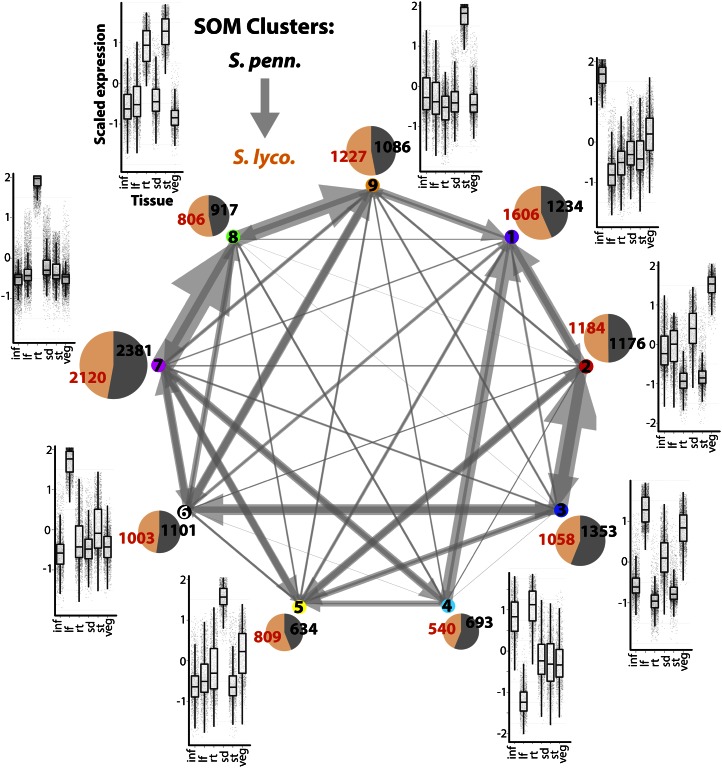
Shifts in tissue-specific expression between orthologs can be interpreted using SOM results as well. Because orthologs are independently assigned to clusters in this scheme, one way to interpret shifts in expression is to focus on those orthologs assigned to different clusters. Approximately 39% of orthologs are assigned to different SOM clusters (4,073 orthologs out of 10,516 with significant tissue terms analyzed in this study). For those orthologs assigned to different clusters, we visualized their assignments as a directional network (arrows point from S. pennellii to S. lycopersicum orthologs), in which edge size is proportional to the number of displaced orthologs (Fig. 4; note that the directionality of an arrow is only used to indicate differences in expression between orthologs and is not meant to convey information about evolutionary direction.). Clear biases in the distribution of orthologs were observed for clusters, many of which reflect the tissue-by-species changes in expression observed in the superSOM data (Fig. 3). Many of the displaced orthologs are assigned to neighboring clusters in PC space. For example, the S. lycopersicum orthologs corresponding to the S. pennellii orthologs assigned to SOM cluster 7 are predominantly assigned to SOM cluster 8 (Fig. 4). There is a similar displacement between SOM clusters 8 and 9, both of which occupy a similar place in PC space and are characterized by genes with high expression in the stem and root (Figs. 1D and 2). The overall tendency of S. lycopersicum orthologs to be displaced in a SOM cluster 7 → cluster 8 → cluster 9 direction is reflective of the differences in expression seen in superSOM cluster 6 (Fig. 3, C and D), in which S. lycopersicum genes have lower expression in the root and higher expression in the stem. Similarly, the displacement of S. lycopersicum orthologs away from SOM cluster 4 to SOM clusters 1 and 5 (Fig. 4) reflects the lower root expression and higher seedling expression observed in S. lycopersicum orthologs occupying superSOM cluster 8 (Fig. 3, C and D).
Figure 4.
Displacement of orthologs to different clusters under a SOM clustering scheme. Using a traditional SOM approach, orthologs can be assigned to different clusters. Shown is a network representation of the assignment of orthologs to different SOM clusters. Arrows represent the displacement of S. lycopersicum (S. lyco.) orthologs (arrow tips) to clusters other than those occupied by their S. pennellii (S. penn.) counterparts (arrow bases). Arrow sizes are proportional to the number of displaced orthologs. Some trends, such as the tendency for S. pennellii → S. lycopersicum displacement in a SOM cluster 7 → cluster 8 → cluster 9 direction, are similar to trends observed in superSOM clusters (Fig. 3). For example, the SOM cluster 7 → cluster 8 → cluster 9 trend reflects lower expression in the root and higher expression in the stem of S. lycopersicum orthologs, a pattern exhibited by superSOM cluster 6. Pie charts adjacent to each cluster are proportional in size to the number of genes to which they are assigned and depict the relative ratio of S. pennellii (black) and S. lycopersicum (orange) members. Box plots next to each cluster show the expression pattern of cluster members. inf, Inflorescence; lf, leaf; rt, root; sd, seedling; st, stem; veg, vegetative apex. [See online article for color version of this figure.]
What is the biological basis of these shifts in the tissue-specific expression of genes between species? As the organs we sampled represent homologous structures, the predominant cause of these shifts represent (1) changes in the expression levels of genes, (2) differences in the overall architecture of the organs sampled (i.e. different morphologies and proportions of specific tissues in organs), or (3) developmental differences between organs at the time of sampling. Each is informative with respect to the nature of the differences between species, the former with respect to gene expression and the latter two with respect to the developmental consequences of changes in gene regulatory networks. An important caveat in the interpretation of results such as these is that the effects on gene expression from the above sources are confounded. Additional measures, such as time series data or modeled correction for growth rate (Chitwood et al., 2012a, 2012b; Chitwood and Sinha, 2013), are required to separate these important contributors to variance in biological data. Furthermore, tissue effects are confounded with environmental effects due to the necessity to grow younger and older tissues in different locations for which different growth conditions were used (as explained in “Materials and Methods”).
Cluster-Specific Changes between Ortholog Expression Profiles
The SOM and superSOM methods described above cluster based on scaled expression across tissues (Figs. 1 and 2). Only after clustering are differences in the expression profiles of orthologs analyzed (Figs. 3 and 4). That is, the differences in expression between orthologs are only being analyzed indirectly, as an after-the-fact comparison. Nonetheless, using the above approaches, distinct changes in gene expression profiles are observable. For example, the genes in superSOM cluster 1 have high expression in the leaf and vary in their leaf and stem expression between S. lycopersicum and S. pennellii (Fig. 3, C and D). However, the orthologs in superSOM cluster 6 have high expression in the root and stem and vary mostly between species in these tissues (compared with the leaf and stem in superSOM cluster 1). The tissue-specific changes in species expression patterns are exhibited by the SOM data as well, and clusters have differing propensities to share displaced orthologs with other clusters (Fig. 4). In both cases, these methods can be used to highlight particular groups of genes that show specific changes in tissue expression patterns between species.
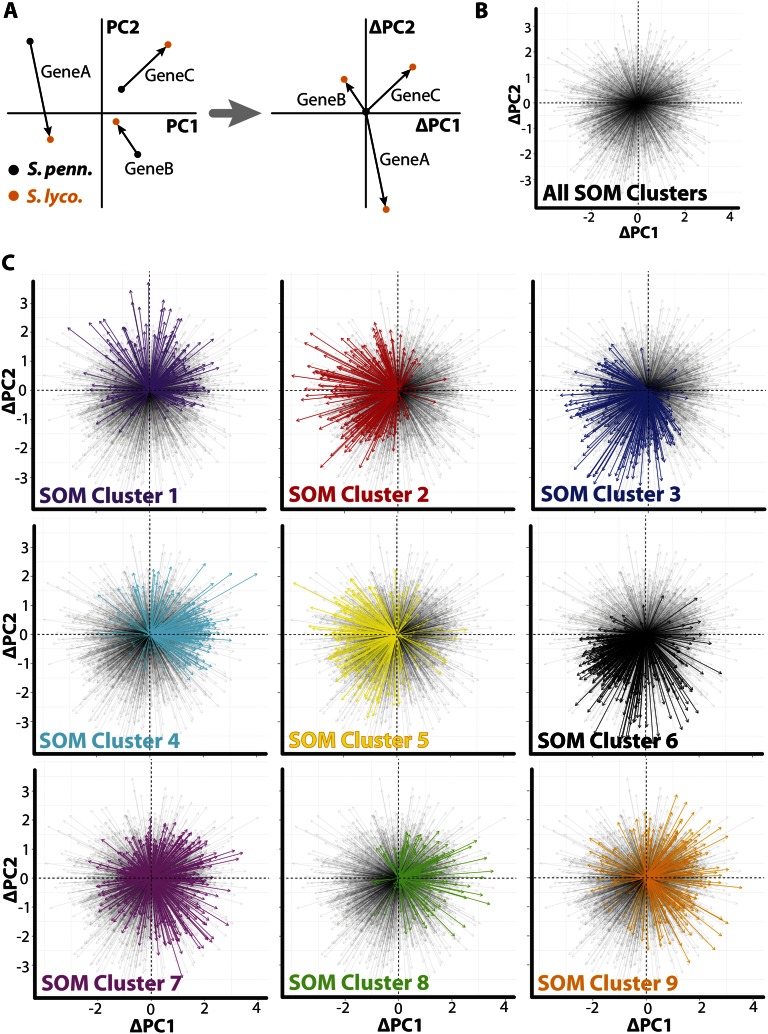
To explore changes in gene expression patterns more directly, in a manner that is not obscured by their innate tissue-specific expression, we developed a vector-based metric based on displacement between orthologs in PC space (Fig. 5A). In this approach, the PC coordinates of both orthologs are recentered such that the S. pennellii ortholog resides at the origin. Each ortholog pair then corresponds to a vector, centered at the origin, the trajectory of which represents the changes in the expression profile that occur between S. pennellii and S. lycopersicum. Collectively, the ortholog vectors emanate without obvious biases in all directions from the origin, demonstrating that most of the possible differences in ortholog expression are represented (Fig. 5B).
Figure 5.
Expression differences between species are biased by tissue context. A, In order to focus on changes in expression profiles, a vector-based approach was used. Ortholog pairs are represented as multidimensional vectors, the magnitude and direction of which represent changes in the expression profiles of genes between species. To focus only on expression changes, vectors were translated in coordinate PC space such that they originate at the origin. Arrow bases represent S. pennellii (S. penn.) orthologs, and arrow tips represent S. lycopersicum (S. lyco.). B, All such vectors represented in the data set originating from the origin. Note that major biases in the change of expression pattern between species are not evident and that most possible changes in expression pattern are represented. C, Vectors representing changes in tissue-specific expression for each SOM cluster are shown. Vectors belonging to orthologs from each cluster are in solid colors overlaid upon all other transparent vectors. Orthologs belonging to different clusters show distinct changes in their tissue-specific expression pattern between species, indicating that genes with particular expression profiles are biased toward a specific set of tissue by species change. Much of this bias is imposed by extreme expression patterns (such as those that occupy the edges of PC space shown in Fig. 1) in S. pennellii and S. lycopersicum expression profiles possessing more generic expression patterns.
To assay the changes in expression associated with different expression profiles, we looked at the SOM clusters to which S. pennellii genes were assigned. If the vectors are examined on a per cluster basis, obvious biases in vector direction are revealed that vary by cluster (Fig. 5C). These biases are connected with the overall innate tissue-specific expression of the cluster. If a comparison is made with SOM cluster position in PC space (Fig. 1D), there is a tendency for vectors to be oriented “inward” toward the origin (Fig. 5C). As the genes used for clustering are those that showed significant differential expression between species, they occupy extreme expression patterns (the periphery of the PC space shown in Fig. 1D). Because ortholog vectors are oriented inward (a region of PC space with more generic, rather than disparate, tissue expression patterns; i.e. the center of PC space represents an expression pattern that is “flat,” with equal expression across all tissues), the implication is that S. lycopersicum orthologs possess more “generic” profiles (i.e. less variable) across tissues compared with S. pennellii.
The propensity for less variable S. lycopersicum expression profiles is transcriptome wide (Fig. 5C). Whether this is a phenomenon specific to tomato or widespread throughout domesticated species and their wild relatives remains to be investigated. Another possibility is that this phenomenon results from environmental effects. For example, perhaps S. pennellii activates tissue-specific stress responses when grown in a greenhouse/chamber compared with S. lycopersicum, which is bred for the cultivated conditions used for this experiment. It remains to be seen if these trends would hold if, instead, the experiment were performed under the xeric conditions to which S. pennellii is adapted.
Clustering by Changes in Expression Profile
When analyzing species-by-tissue interactions, we are most interested in groups of genes that change their tissue-specific expression between species in a distinct manner, regardless of their actual innate expression profile. Take, for example, a group of genes with varied expression patterns (root genes, leaf genes, stem genes, genes expressed in multiple tissues, and so on) that have higher expression in the root in S. pennellii compared with S. lycopersicum. Instead of this diverse group of genes being clustered together by their attribute of higher root expression in S. pennellii, they would instead be categorized into a number of clusters based on their varying expression profiles. Is there a way to cluster genes based on the changes in their expression profile between species?
In order to group genes by changes in expression pattern between species, irrespective of their overall tissue specificity, we first took the difference in PC values between orthologs for each PC, or ΔPC (calculated for each of six PCs, representing the entirety of expression profile variance in the data set). ΔPCs 1 to 6 were assigned to different levels of a six-layer superSOM. The weight of each layer is equal to the variance explained by each PC (Supplemental Fig. S2). The result is that the Euclidean distance of each gene to a cluster is determined as a weighted sum of its ΔPC values 1 to 6, in a manner that is proportional to the amount of variance explained by each PC. By this method, orthologs are clustered by their displacement through PC space rather than their absolute position in PC space. In other words, genes are clustered by the difference in their expression profiles between orthologs.
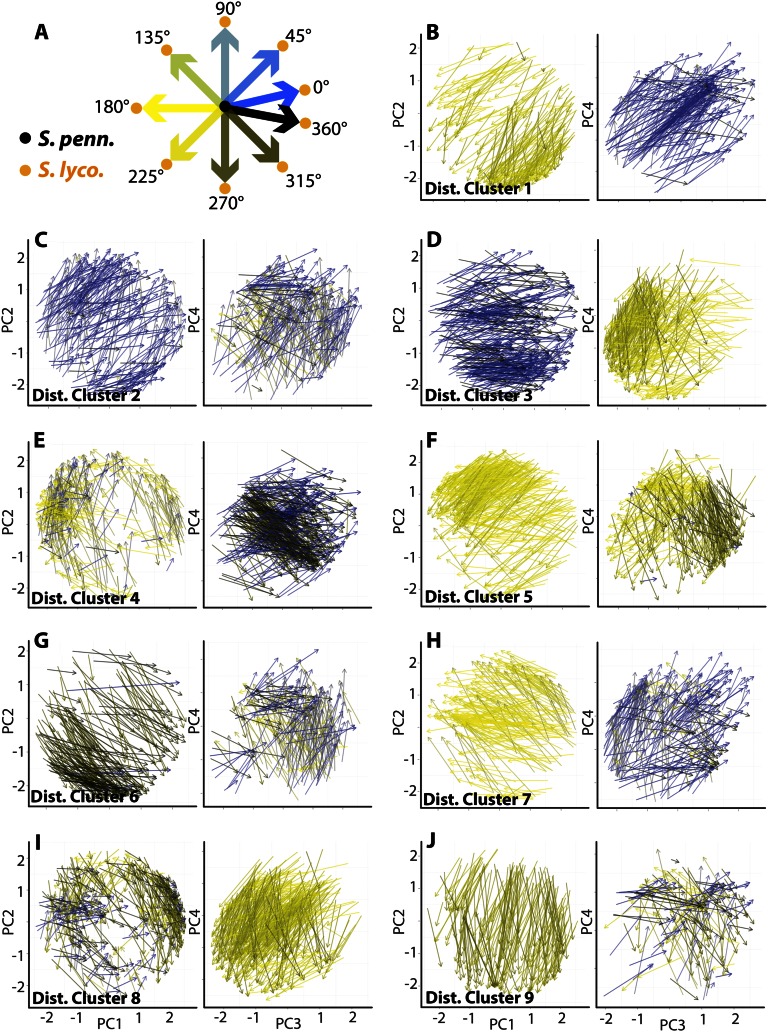
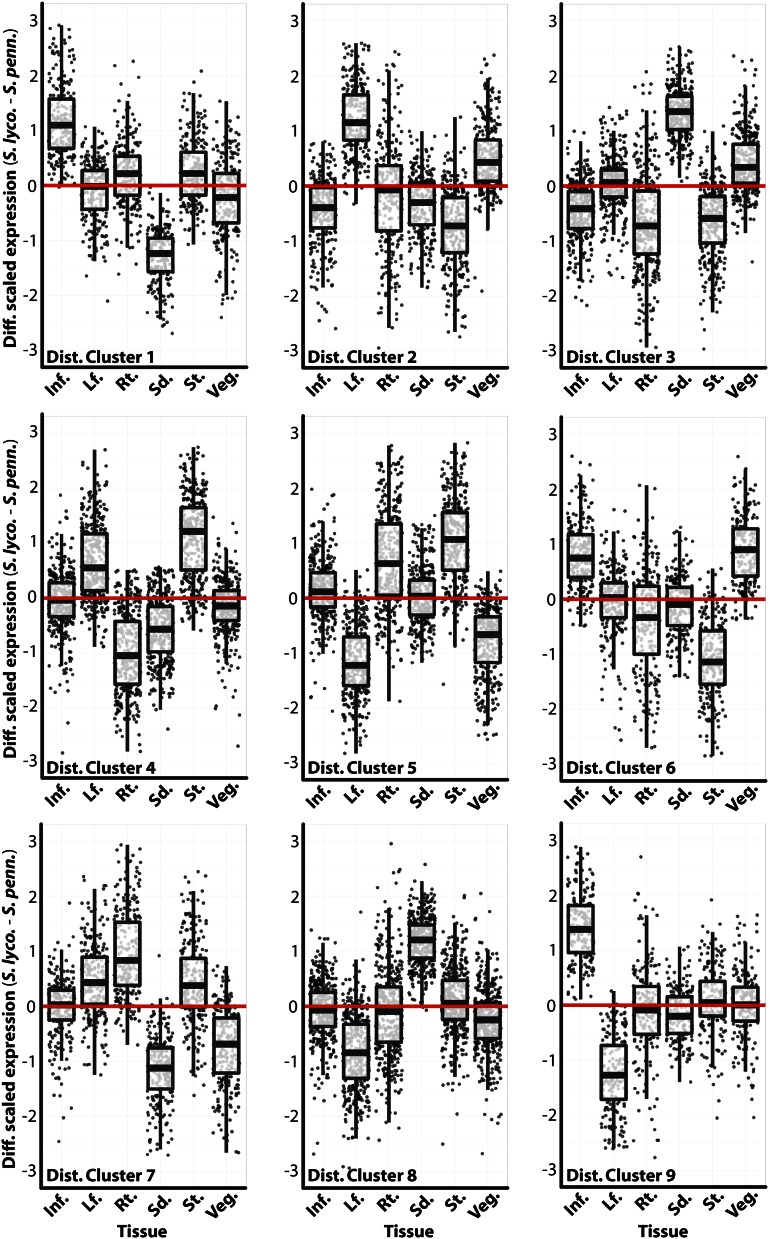
The result of the clustering predictably reveals groups of ortholog vectors, defined by expression profile changes between S. lycopersicum and S. pennellii, that move through PC space in a similar fashion (Fig. 6; Supplemental Table S3). These are groups of genes that exhibit similar changes in expression profile but not necessarily a similar expression profile. The similar changes in expression profile can also be observed as differences in scaled gene expression between S. lycopersicum and S. pennellii orthologs in each tissue (Fig. 7). Just like groups of genes with similar profiles (Fig. 2), clusters of genes with similar changes in expression (“distance clusters”) possess members of functional relevance. For example, distance cluster 7 genes exhibit higher root and stem expression in S. lycopersicum relative to S. pennellii (Fig. 7). Among these genes are SUCROSE SYNTHASE6 and ALTERED PHLOEM DEVELOPMENT homologs, as well as proton-dependent oligopeptide, inorganic phosphate, myoinositol, and multi antimicrobial extrusion protein efflux transporters, all genes associated with the vascular and transport functions of the root and stem (Supplemental Table S3). Distance cluster 9 genes are up-regulated in the S. lycopersicum inflorescence and down-regulated in leaves relative to S. pennellii (Fig. 7). Among these genes is SELF-PRUNING3D (the tomato TFL1/FT homolog) as well as STERILE APETALA and NO TRANSMITTING TRACT homologs, all with inflorescence- and flowering-specific functions (Supplemental Table S3).
Figure 6.
Clustering based on changes in expression profile between species. A, Key for the color scheme used. To aid the visualization of vectors, the angular direction of vectors (within the context of the PCs graphed) is indicated by color. The result is that vectors with similar direction (changes in expression profile between species) are shown in similar colors. B to J, Vectors (arrow base representing S. pennellii [S. penn.] and arrow tip representing S. lycopersicum [S. lyco.]) were clustered based on changes in their component PC values. Changes in PC values were each assigned to an independent layer of a superSOM, the weight of which is proportional to the variance explained by the PC. The result of the cluster analysis is groups of genes with similar changes in their expression profiles between species, regardless of their overall tissue-specific expression. Change in direction in PC space is multidimensional, which must be considered when analyzing the shared properties of vectors belonging to a distance cluster.
Figure 7.
Changes in gene expression between species by tissue, in clusters grouped by change in gene expression profile. For distance clusters (Fig. 6) that possess genes with similar displacement in PC space, the difference in scaled gene expression values between species (S. lycopersicum [S. lyco.]–S. pennellii [S. penn.]) for each tissue is shown. For example, distance cluster 9 genes show higher expression in the inflorescence and lower expression in leaves in S. lycopersicum compared with S. pennellii. Relevant genes in each cluster, the biology of which are reflective of the gene expression pattern, are discussed in the text. Inf., Inflorescence; Lf., leaf; Rt., root; Sd., seedling; St., stem; Veg., vegetative apex. [See online article for color version of this figure.]
Increased Shoot Apical Meristem Activity in S. lycopersicum
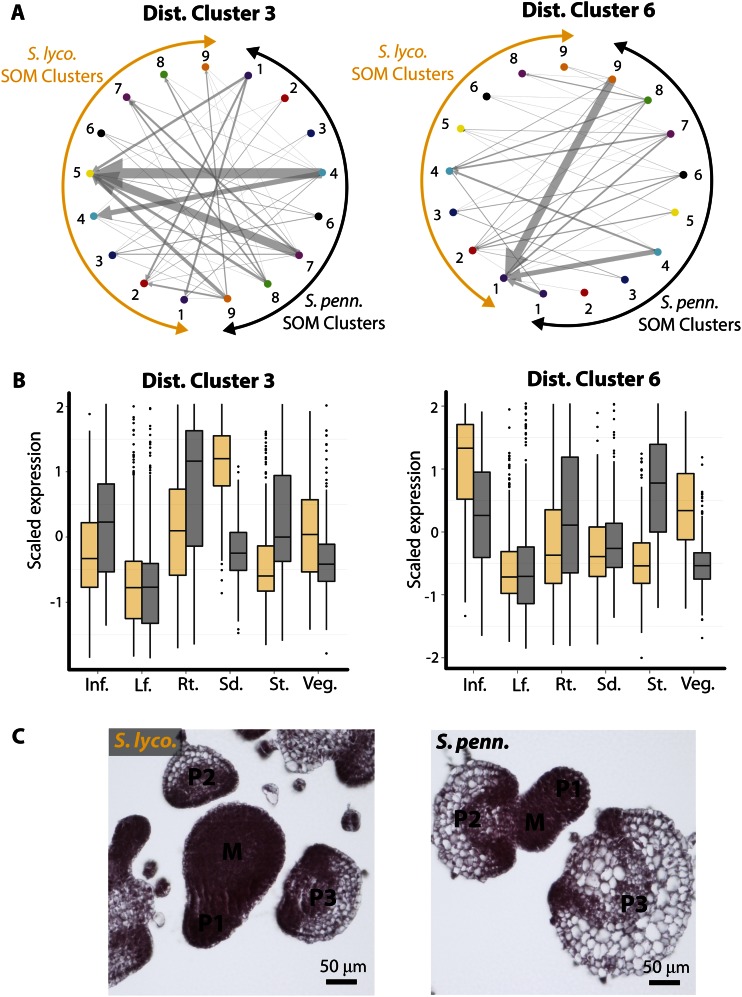
A GO enrichment analysis (Supplemental Table S4) focused our attention to distance clusters 3 and 6, which are enriched for terms similar to those associated with genes with high inflorescence and vegetative meristem expression (Supplemental Table S2). Distance cluster 3 is enriched for terms related to translation (an indication of active cell growth), and distance cluster 6 is enriched for “cell cycle” and “multicellular organismal development” terms. Both clusters exhibit changes in expression such that genes are more highly expressed in meristem-enriched tissues (inflorescence, vegetative meristem, seedling) in S. lycopersicum (Fig. 7). Because we are describing changes in expression profiles, we revisited the displacement of orthologs between SOM clusters (Fig. 4). Distance cluster 3 genes are predominantly displaced to SOM cluster 5, which exhibits high seedling expression (Fig. 8A). This is consistent with the very high seedling expression in S. lycopsersicum relative to S. pennellii in this group of genes (Figs. 7 and 8B). Additionally, distance cluster 3 genes are more highly expressed in the vegetative apex of S. lycopersicum and the stem of S. pennellii. Likewise, distance cluster 6 genes are predominantly displaced from SOM cluster 9 to SOM cluster 1, which contains genes with high vegetative apex expression (Fig. 8A). This group of genes are highly expressed in the inflorescence and vegetative apex of S. lycopersicum, and, like distance cluster 3, show high expression in the stem (a differentiated structure) of S. pennellii (Figs. 7 and 8B).
Figure 8.
Bias toward meristematic expression in S. lycopersicum (S. lyco.) relative to S. pennellii (S. penn.). A, Distance clusters 3 and 6, which are enriched for GO terms associated with meristematic tissues, were examined with respect to the displacement of orthologous genes between different SOM clusters. The orthologs assigned to S. pennellii SOM clusters (black arcs) and the corresponding S. lycopersicum SOM clusters (orange arcs) are indicated by arrows (S. pennellii → S. lycopersicum direction). Note that unlike Figure 4, genes belonging to the same cluster are indicated. There is a trend for S. lycopersicum orthologs to become displaced to clusters with high expression in meristematic tissues. For example, in distance cluster 3, S. lycopersicum orthologs tend to occupy SOM cluster 5, with high seedling expression. In distance cluster 6, S. lycopersicum orthologs become displaced toward SOM cluster 1, with high vegetative apex expression. B, The displacement of distance cluster genes toward meristematic SOM clusters in S. lycopersicum is reflected in their expression profiles. Distance cluster 3 genes show much higher expression in the seedling (and somewhat in the vegetative meristem) in S. lycopersicum relative to S. pennellii. Distance cluster 6 genes show much higher vegetative apex expression (and somewhat higher in the inflorescence) in S. lycopersicum relative to S. pennellii. Both clusters show higher expression in the stem (a differentiated structure) in S. pennellii relative to S. lycopersicum. Inf., Inflorescence; Lf., leaf; Rt., root; Sd., seedling; St., stem; Veg., vegetative apex. C, The higher expression of genes in the meristematic tissues of S. lycopersicum is consistent with species differences relative to S. pennellii. The vegetative meristem of S. lycopersicum is much larger than that of S. pennellii, with more cells. Furthermore, the differentiated primordia of S. pennellii are larger, with more vacuolated cells, relative to the primordia of a similar stage in S. lycopersicum, indicating premature differentiation relative to S. lycopersicum. [See online article for color version of this figure.]
Together, distance clusters 3 and 6 represent a major trend in our data set: a propensity for S. lycopersicum genes to be more highly expressed in the meristem-rich tissues of the seedling, inflorescence, and vegetative meristem and less expressed in the differentiated stem relative to S. pennellii. These genes are enriched for terms related to active cellular growth, translation, and development and are displaced to SOM clusters in S. lycopersicum with high expression in the seedling and vegetative apex.
Are there any morphological features that correspond to these trends in gene expression? One of the most conspicuous differences between S. lycopersicum and its wild relatives is the size of its vegetative meristem and cells (Fig. 8C). The vegetative meristem of S. lycopersicum is two to three times the size of S. pennellii, with many more cells. Additionally, the leaf primordia of S. pennellii are larger than those of a similar developmental stage in S. lycopersicum, and possess larger, more vacuolated cells. Collectively, the active growing region of S. lycopersicum exhibits morphological features consistent with prolonged indeterminacy and delayed differentiation relative to its desert relative S. pennellii (Fig. 8C).
Among the genes included in distance cluster 6 (which exhibits the highest increase in vegetative apex expression in S. lycopersicum; Fig. 7) are known regulators of shoot apical meristem development. LeT6 (a tomato Knotted1 homolog) and PIN1 and CURLY LEAF homologs regulate indeterminacy, the specification of primordia, and epigenetic states of the shoot apical meristem, respectively (Goodrich et al., 1997; Janssen et al., 1998a, 1998b; Reinhardt et al., 2003; Xu and Shen, 2008; Pattison and Catalá, 2012). Additionally included in this cluster are WIRY4 (the SUPPRESSOR OF GENE SILENCING3 homolog involved in specifying adaxial fate) and an ORGAN BOUNDARY1 (or LIGHT-SENSITIVE HOMOLOG3) homolog, which are known regulators of leaf complexity (Kim et al., 2003; Cho and Zambryski, 2011; Yifhar et al., 2012), which dramatically varies between S. lycopersicum and S. pennellii.
Together, the enrichment of GO terms and the inclusion of known shoot apical meristem regulators validates the concept of clustering by differences in gene expression between species as a means to discover those genes closely associated with morphological change during evolution.
DISCUSSION
Finding trends in tissue-by-species gene expression data that are congruent with observed biology is difficult to achieve. Statistical models that fit individual genes with significant interaction terms are a powerful tool but may fail to identify large groups of coexpressed genes or genes expressed in a particular fashion that make sense with respect to a relevant biological question. Exploratory data analysis can help identify trends but can be difficult to implement with multidimensional data sets, such as when studying changes in gene expression across tissues and species. Popular methods, such as k-means and hierarchical clustering, sidestep this issue by either concatenating data sets or not distinguishing between different levels of important factors. With regard to orthologous sets of genes, as encountered in a tissue-by-species data set, clustering using a superSOM approach is particularly powerful (Figs. 1–3). By clustering groups of orthologs rather than genes, and independently taking into account patterns of variance within each species, comparisons between orthologs remain relevant. Clustering without multiple levels can still yield insights, albeit in a more complicated form as a network of displaced orthologs assigned to different clusters (Fig. 4).
Such methods principally cluster on a single factor (tissue) and afterward look for differences in the other (species; Figs. 1–4). Groups of identified genes may be confounded for two different aspects of their differential expression: their overall tissue-specific expression and tissue-by-species differences. Indeed, we show that groups of genes with distinct gene expression profiles exhibit biases in their tissue-by-species changes in expression (Fig. 5). One way to separate these effects is to find groups of genes with similar changes in expression pattern, regardless of their overall tissue-specific expression. A method to achieve this is to cluster genes based on their displacement in PC space, representing the changes in their gene expression profiles between species (Figs. 6 and 7). In our data set, such an approach reveals a trend of higher expression of S. lycopersicum genes in meristem-containing tissue relative to its wild relative S. pennellii, a pattern consistent with observed biology (Fig. 8). Moreover, within such clusters, we identify key components of gene regulatory networks governing indeterminacy and shoot apical meristem development in tomato, including LeT6, WIRY4, and PIN1, validating our approach (Janssen et al., 1998a, 1998b; Kim et al., 2003; Reinhardt et al., 2003; Pattison and Catalá, 2012; Yifhar et al., 2012).
As next-generation sequencing enables transcriptomics to become widespread, multidimensional analyses, similar to the one presented here, will become commonplace. Eventually, in addition to tissue and species factors, developmental time and environmental components will be analyzed to yield comprehensive data sets approaching the totality of gene expression present in evolving populations. Major changes in our thinking about the analysis of such data sets will be required to reveal underlying trends in gene expression consistent with, and eventually explaining, complex phenotypic phenomena. Although the methods outlined here are only a first step, we show how multilevel SOMs and clustering of gene expression changes can begin to reconcile phenotypic differences between species with tissue-by-species interactions in gene expression.
MATERIALS AND METHODS
Plant Materials
Solanum lycopersicum ‘M82’ and Solanum pennellii (LA0716) were donated by Dani Zamir (Hebrew University of Jerusalem). Seeds were germinated on Murashige and Skoog plates kept in the dark for 3 d. Plates were then exposed to light and grown upright at 22°C. After 10 d, seedlings were transferred to soil and kept in chambers until anthesis. After flowering, plants were transferred to the greenhouse. Roots and aerial seedling tissue were collected 10 d after germination. Vegetative apices were collected from plants when the third leaf reached approximately 1 mm. The stem between the fourth and fifth leaves and inflorescences were collected when fully formed (50 d after germination for S. lycopersicum and 56 d after germination for S. pennellii). Total RNA from all tissues was extracted using Trizol (Invitrogen) according to the manufacturer’s standard protocol. Histology was performed using eosin Y staining and standard histology protocols.
Library Preparation and Sequencing
RNA-Seq libraries for the transcriptome experiment were prepared using the Illumina RNA-Seq sample preparation kit (RS-100-0801). Custom paired-end adapters were used to multiplex libraries. Eight paired-end adapters with a unique 3-bp barcode sequence (AAA, AGG, CAC, CGT, GCT, GTC, TCA, and TTG) at the end of the adapter were used for the library preparation. Barcodes were chosen so any one sequencing error in the barcode cannot transform one barcode into another, dramatically reducing the chance of contamination between libraries due to sequencing errors.
PE1 and PE2 primers were mixed in annealing buffer (10 mm Tris-HCl, pH 7.5, 1 mm EDTA, and 50 mm NaCl) and annealed by heating to 95°C and gradually cooling down to 4°C. The complementary DNA (cDNA) libraries were quantified using Bioanalyzer (Agilent), then pooled in random subsets of eight samples and sequenced (paired end, 85 bp each) in the Illumina genome analyzer GAII.
A total of 57 libraries from S. lycopersicum and S. pennellii (as well as Solanum pimpinellifolium LA1589 and Solanum habrochaites LA1777, the data from which are not analyzed here) were sequenced in 14 lanes from seven different 84-cycle runs of the Illumina GAII. These sequencing runs resulted in 406,874,298 85-bp reads and 169,290,821 paired-end and single-end reads. Ninety-five percent of these reads contained the expected barcodes and were separated by library. Illumina adapter sequences and low-quality bases (coded as B in the fastq files) were trimmed from the 3′ ends of the reads. A total of 480,097,244 reads that were 50 bp or longer after trimming were used for our analyses.
Generation of Matched cDNA References
To facilitate mapping and accurate expression analysis of RNA-Seq reads to different species across the tomato complex, we took advantage of a draft S. pennellii genomic sequence (version 0.6.1; S. pennellii Consortium, unpublished data) to build a matched set of reference cDNAs for S. lycopersicum and S. pennellii. The goal was to obtain a matched set of references of equal length containing sequences known to exist in both species and retaining species-specific polymorphisms. The following steps were used for each coding sequence (CDS) defined in the ITAG2.3 set. (1) S. lycopersicum CDSs were used to BLAST against S. pennellii scaffolds using megaBLAST (Zhang et al., 2000; settings: -e 1e-50 -m 7 -N 2 -t 18 -W 11 -A 50) to identify the appropriate scaffold and region. (2) S. pennellii scaffold sequence encompassing the BLAST hit region and an additional 2 kb on either side was retrieved, and GMAP (Wu and Watanabe, 2005) was used to thread the S. lycopersicum CDS onto the S. pennellii scaffold (settings: -n 1 -f 1). (3) GMAP output was parsed to create matching S. lycopersicum and S. pennellii CDSs. Only the matching regions were retained. (4) The matched sets were then filtered to only retain good hits. To accomplish this, the predicted S. pennellii CDSs were BLASTed against the full S. lycopersicum CDS set (a reciprocal BLAST) using megaBLAST (settings as above). To retain matched pairs, we required that the best reciprocal BLAST hit was to the original ITAG CDS, that the best BLAST hit had an e-value at least 103 more significant than the second best hit, and (because we were also interested in obtaining upstream promoter regions not used in this work) that the 5′ high-scoring segment pairs be at least 50 bp, have a 90% identity, and be within 300 bp of the query start. In this way, from the original 34,727 annotated ITAG CDSs (median length of 834), we created 28,801 matched CDS pairs (median length of 849). While a number of gene models are lost using this technique, it is justified for differential expression analysis by the increased short-read mapping accuracy allowed by the matched set.
We used BWA and Samtools to map RNA-Seq reads to the matched reference cDNA set. The parameters for BWA were “-n 0.1 -e 12 -k 1 -l 25”; Samtools was used with -n 1 to select reads that mapped unambiguously to the reference. Read counts from this alignment were used in the analysis of differential expression.
Analysis of Differential Gene Expression
Sequences were filtered, trimmed, and mapped to the appropriate matched cDNA reference. To reduce the loss of counts due to inefficient mapping of paired ends to the shortened matched cDNA sequences, we mapped only the first paired end for our expression analysis. Matched cDNA references were screened out of the analysis if reads from both species showed biased mapping to one of the species-specific matched cDNAs (log fold change > 1 for both S. lycopersicum and S. pennellii reads). Poor samples were identified and removed using a combination of replicate correlation coefficient, correlation plots, and MA plots. The raw count data were then normalized using a modified trimmed mean of M values method (Robinson and Oshlack, 2010). Low expressed genes were filtered on a minimum sum of 20 counts over all samples for further analysis. Genes that did not pass this threshold were considered not expressed. Differential expression was calculated by fitting a quasi-Poisson generalized linear model at the gene level using tissue, species, and tissue-by-species interaction as factors and extracting significance using an F test.
SOMs
Only those genes with a tissue multiple test-corrected P < 0.05 (determined from the model described above) and model-fitted expression values for both S. lycopersicum and S. pennellii were used for subsequent analysis. Thus, only genes that vary significantly in expression across tissue types and can be compared between species are analyzed. In order to remove differences due to the magnitude of gene expression and focus only on gene expression profiles, expression values were mean centered and variance scaled using the scale function (R base package; R Development Core Team, 2012) separately in S. lycopersicum and S. pennellii.
To cluster S. lycopersicum and S. pennellii genes across tissues, a multilevel 3 × 3 hexagonal SOM was used (Kohonen, 1997; Wehrens and Buydens, 2007). For the analysis referred to as superSOM, matching genes between species were assigned to separate superSOM levels of equal weight using the supersom function (R Kohonen package). For the analysis referred to as SOM, all genes, regardless of the species to which they belong, were clustered under a typical SOM scheme (som function). Under both methods, 100 training iterations were used during clustering, over which the α-learning rate decreased from 0.05 to 0.01. The final assignments of genes to winning units form the basis of the gene clusters discussed in this work. Further analyses of SOM and superSOM cluster memberships were carried out using PCA and various graphical and network visualization methods, described below. Codebook vectors were retrieved from the SOM and superSOM analysis to understand the expression patterns represented by each cluster.
Clusters of genes were then analyzed for the enrichment of GO terms at a 0.05 false discovery rate cutoff (Young et al., 2010; goseq Bioconductor package).
PCA
The outcome of both SOM and superSOM methods was visualized in PCA space. Because SOM and superSOM results will vary from simulation to simulation, it is important to compare and visualize results using an invariant method such as PCA (1) to ensure that major variance patterns in the data set are in fact being explained by SOM clusters and (2) to verify the consistency of clustering methods. The results of the PCA were also used to describe changes in the difference between expression profiles between species.
Every gene is represented by two points in PCA space, one representing the tissue expression pattern of the S. lycopersicum member and the other the S. pennellii member. Genes were assigned PC values based on their expression profiles across tissues, regardless of their species identity (R stats package, prcomp function). Details of the variance explained by each PC and the PCA loadings are given in Supplemental Figure S2.
Visualizing Differences in Expression Profiles between Species
The PC space is defined by two points for each gene, one representing the expression profile in S. lycopersicum and the other S. pennellii, providing a means to visualize and explore the differences in the gene expression profiles between these species. To focus on those genes with the most exaggerated expression profile differences between species, we calculated the Euclidean distance between genes in PC space (using all PCs such that more than 99% of variance is accounted for). The distribution is skewed toward higher distances, and we analyzed displacement in PC space for those genes occupying the upper quartile of Euclidean distance from each other.
For superSOM clusters, differences between corresponding S. lycopersicum and S. pennellii genes were analyzed on a per cluster basis. A variety of visualization methods using ggplot2 (Wickham, 2009) in R were used, including lines connecting the points representing expression profiles between species in PC space (geom_segment function), contours overlaid upon scatterplots separated by species identity (geom_point and stat_density2d), and box plots (geom_boxplot). For SOM clusters, the assignment of genes representing different species to different clusters was visualized using network graphics in the program Gephi (Bastian et al., 2009). Results were visualized as a directed, circular layout network. The direction of arrows indicates the assignment of genes to clusters in an S. pennellii → S. lycopersicum direction. Arrow size is proportional to the number of genes represented.
Vector Analysis of Differential Gene Expression
To separate change in expression pattern from the actual expression profile of genes, a vector-based approach was used. To understand the expression profile changes particular to each SOM cluster, vectors in which the base represents the S. pennellii expression profile in PC space and the tip representing S. lycopersicum were translated such that every vector originates from the origin. Vectors were visualized as arrows using the geom_segment function.
In order to cluster multidimensional vectors representing changes in expression profiles across PC space, the vector components represented by each PC were extracted (simply the difference in PC values between species for each gene). Vectors were then clustered using a superSOM, in which each level is assigned a different PC vector component and the weight of the level is equal to the variance explained by the PC. The validity of this method to find groups of genes with similar changes in gene expression profiles between species was verified by visualizing the directional changes of genes belonging to each distance cluster in PC space. Vectors representing the genes in each cluster were visualized as arrows using the geom_segment function in PC space. To help further visualize directional change, arrow color was assigned based on angle with respect to the PC coordinate system being visualized.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. The additional influence of species effects on gene expression profiles.
Supplemental Figure S2. Principal component analysis statistics.
Supplemental Figure S3. Correspondence between k-means clustering and SOMs results.
Supplemental Figure S4. Similarity in expression between genes assigned to corresponding k-means and SOM clusters.
Supplemental Figure S5. Correspondence between hierarchical clustering and SOMs results.
Supplemental Table S1. SOM and superSOM cluster identities, fitted gene expression values, and principal component values.
Supplemental Table S2. GO enrichment analysis of SOM and superSOM clusters.
Supplemental Table S3. Distance cluster identities.
Supplemental Table S4. GO enrichment analysis of distance clusters.
Glossary
- SOM
self-organizing map
- GO
Gene Ontology
- PCA
principal component analysis
- PC
principal component
- superSOM
super self-organizing map
- ΔPC
difference in principal component values between orthologs for each principal component
- CDS
coding sequence
- cDNA
complementary DNA
References
- Aurenhammer F. (1991) Voronoi diagrams: a survey of a fundamental geometric data structure. ACM Comput Surv 23: 345–405 [Google Scholar]
- Bastian M, Heymann S, Jacomy M (2009) Gephi: an open source software for exploring and manipulating networks. In Proceedings of the Third International Conference on Weblogs and Social Media. AAAI Press, Menlo Park, CA, pp 361–362 [Google Scholar]
- Birnbaum K, Shasha DE, Wang JY, Jung JW, Lambert GM, Galbraith DW, Benfey PN. (2003) A gene expression map of the Arabidopsis root. Science 302: 1956–1960 [DOI] [PubMed] [Google Scholar]
- Brady SM, Orlando DA, Lee JY, Wang JY, Koch J, Dinneny JR, Mace D, Ohler U, Benfey PN. (2007) A high-resolution root spatiotemporal map reveals dominant expression patterns. Science 318: 801–806 [DOI] [PubMed] [Google Scholar]
- Brawand D, Soumillon M, Necsulea A, Julien P, Csárdi G, Harrigan P, Weier M, Liechti A, Aximu-Petri A, Kircher M, et al. (2011) The evolution of gene expression levels in mammalian organs. Nature 478: 343–348 [DOI] [PubMed] [Google Scholar]
- Britten RJ, Davidson EH. (1969) Gene regulation for higher cells: a theory. Science 165: 349–357 [DOI] [PubMed] [Google Scholar]
- Britten RJ, Davidson EH. (1971) Repetitive and non-repetitive DNA sequences and a speculation on the origins of evolutionary novelty. Q Rev Biol 46: 111–138 [DOI] [PubMed] [Google Scholar]
- Carroll SB. (2000) Endless forms: the evolution of gene regulation and morphological diversity. Cell 101: 577–580 [DOI] [PubMed] [Google Scholar]
- Carroll SB. (2008) Evo-devo and an expanding evolutionary synthesis: a genetic theory of morphological evolution. Cell 134: 25–36 [DOI] [PubMed] [Google Scholar]
- Chanderbali AS, Yoo MJ, Zahn LM, Brockington SF, Wall PK, Gitzendanner MA, Albert VA, Leebens-Mack J, Altman NS, Ma H, et al. (2010) Conservation and canalization of gene expression during angiosperm diversification accompany the origin and evolution of the flower. Proc Natl Acad Sci USA 107: 22570–22575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chitwood DH, Headland LR, Filiault DL, Kumar R, Jiménez-Gómez JM, Schrager AV, Park DS, Peng J, Sinha NR, Maloof JN. (2012a) Native environment modulates leaf size and response to simulated foliar shade across wild tomato species. PLoS ONE 7: e29570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chitwood DH, Headland LR, Kumar R, Peng J, Maloof JN, Sinha NR. (2012b) The developmental trajectory of leaflet morphology in wild tomato species. Plant Physiol 158: 1230–1240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chitwood DH, Sinha NR. (2013) A census of cells in time: quantitative genetics meets developmental biology. Curr Opin Plant Biol 16: 92–99 [DOI] [PubMed] [Google Scholar]
- Cho E, Zambryski PC. (2011) Organ boundary1 defines a gene expressed at the junction between the shoot apical meristem and lateral organs. Proc Natl Acad Sci USA 108: 2154–2159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho RJ, Campbell MJ, Winzeler EA, Steinmetz L, Conway A, Wodicka L, Wolfsberg TG, Gabrielian AE, Landsman D, Lockhart DJ, et al (1998) A genome-wide transcriptional analysis of the mitotic cell cycle. Mol Cell 2: 65–73 [DOI] [PubMed] [Google Scholar]
- Dinneny JR, Long TA, Wang JY, Jung JW, Mace D, Pointer S, Barron C, Brady SM, Schiefelbein J, Benfey PN. (2008) Cell identity mediates the response of Arabidopsis roots to abiotic stress. Science 320: 942–945 [DOI] [PubMed] [Google Scholar]
- Doebley J, Lukens L. (1998) Transcriptional regulators and the evolution of plant form. Plant Cell 10: 1075–1082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efroni I, Blum E, Goldshmidt A, Eshed Y. (2008) A protracted and dynamic maturation schedule underlies Arabidopsis leaf development. Plant Cell 20: 2293–2306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen MB, Spellman PT, Brown PO, Botstein D. (1998) Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA 95: 14863–14868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eveland AL, Satoh-Nagasawa N, Goldshmidt A, Meyer S, Beatty M, Sakai H, Ware D, Jackson D. (2010) Digital gene expression signatures for maize development. Plant Physiol 154: 1024–1039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodrich J, Puangsomlee P, Martin M, Long D, Meyerowitz EM, Coupland G. (1997) A Polycomb-group gene regulates homeotic gene expression in Arabidopsis. Nature 386: 44–51 [DOI] [PubMed] [Google Scholar]
- Guyon I, Elisseeff A. (2003) An introduction to variable and feature selection. J Mach Learn Res 3: 1157–1182 [Google Scholar]
- Hebb DO (1949) The Organization of Behavior. John Wiley & Sons, New York [Google Scholar]
- Janssen BJ, Lund L, Sinha N. (1998a) Overexpression of a homeobox gene, LeT6, reveals indeterminate features in the tomato compound leaf. Plant Physiol 117: 771–786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssen BJ, Williams A, Chen JJ, Mathern J, Hake S, Sinha N. (1998b) Isolation and characterization of two knotted-like homeobox genes from tomato. Plant Mol Biol 36: 417–425 [DOI] [PubMed] [Google Scholar]
- Jiao Y, Tausta SL, Gandotra N, Sun N, Liu T, Clay NK, Ceserani T, Chen M, Ma L, Holford M, et al. (2009) A transcriptome atlas of rice cell types uncovers cellular, functional and developmental hierarchies. Nat Genet 41: 258–263 [DOI] [PubMed] [Google Scholar]
- Kangas JA, Kohonen TK, Laaksonen JT. (1990) Variants of self-organizing maps. IEEE Transactions on Neural Networks 1: 93–99 [DOI] [PubMed] [Google Scholar]
- Kim M, Pham T, Hamidi A, McCormick S, Kuzoff RK, Sinha N. (2003) Reduced leaf complexity in tomato wiry mutants suggests a role for PHAN and KNOX genes in generating compound leaves. Development 130: 4405–4415 [DOI] [PubMed] [Google Scholar]
- King MC, Wilson AC. (1975) Evolution at two levels in humans and chimpanzees. Science 188: 107–116 [DOI] [PubMed] [Google Scholar]
- Kohonen T. (1982) Self-organized formation of topologically correct feature maps. Biol Cybern 43: 59–69 [Google Scholar]
- Kohonen T (1997) Self-Organizing Maps. Springer, New York [Google Scholar]
- Li P, Ponnala L, Gandotra N, Wang L, Si Y, Tausta SL, Kebrom TH, Provart N, Patel R, Myers CR, et al. (2010) The developmental dynamics of the maize leaf transcriptome. Nat Genet 42: 1060–1067 [DOI] [PubMed] [Google Scholar]
- Oshlack A, Robinson MD, Young MD. (2010) From RNA-seq reads to differential expression results. Genome Biol 11: 220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SJ, Jiang K, Schatz MC, Lippman ZB. (2012) Rate of meristem maturation determines inflorescence architecture in tomato. Proc Natl Acad Sci USA 109: 639–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattison RJ, Catalá C. (2012) Evaluating auxin distribution in tomato (Solanum lycopersicum) through an analysis of the PIN and AUX/LAX gene families. Plant J 70: 585–598 [DOI] [PubMed] [Google Scholar]
- R Development Core Team (2012). R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna, Austria [Google Scholar]
- Reinhardt D, Pesce ER, Stieger P, Mandel T, Baltensperger K, Bennett M, Traas J, Friml J, Kuhlemeier C. (2003) Regulation of phyllotaxis by polar auxin transport. Nature 426: 255–260 [DOI] [PubMed] [Google Scholar]
- Robinson MD, Oshlack A. (2010) A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol 11: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero IG, Ruvinsky I, Gilad Y. (2012) Comparative studies of gene expression and the evolution of gene regulation. Nat Rev Genet 13: 505–516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takacs EM, Li J, Du C, Ponnala L, Janick-Buckner D, Yu J, Muehlbauer GJ, Schnable PS, Timmermans MC, Sun Q, et al (2012) Ontogeny of the maize shoot apical meristem. Plant Cell 24: 3219–3234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamayo P, Slonim D, Mesirov J, Zhu Q, Kitareewan S, Dmitrovsky E, Lander ES, Golub TR. (1999) Interpreting patterns of gene expression with self-organizing maps: methods and application to hematopoietic differentiation. Proc Natl Acad Sci USA 96: 2907–2912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavazoie S, Hughes JD, Campbell MJ, Cho RJ, Church GM. (1999) Systematic determination of genetic network architecture. Nat Genet 22: 281–285 [DOI] [PubMed] [Google Scholar]
- Wang Z, Gerstein M, Snyder M. (2009) RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet 10: 57–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wehrens R, Buydens LMC. (2007) Self- and super-organizing maps in R: the Kohonen package. J Stat Softw 21: 1–19 [Google Scholar]
- Wickham H (2009) ggplot2: Elegant Graphics for Data Analysis. Springer, New York [Google Scholar]
- Wu TD, Watanabe CK. (2005) GMAP: a genomic mapping and alignment program for mRNA and EST sequences. Bioinformatics 21: 1859–1875 [DOI] [PubMed] [Google Scholar]
- Xu L, Shen WH. (2008) Polycomb silencing of KNOX genes confines shoot stem cell niches in Arabidopsis. Curr Biol 18: 1966–1971 [DOI] [PubMed] [Google Scholar]
- Yifhar T, Pekker I, Peled D, Friedlander G, Pistunov A, Sabban M, Wachsman G, Alvarez JP, Amsellem Z, Eshed Y. (2012) Failure of the tomato trans-acting short interfering RNA program to regulate AUXIN RESPONSE FACTOR3 and ARF4 underlies the wiry leaf syndrome. Plant Cell 24: 3575–3589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young MD, Wakefield MJ, Smyth GK, Oshlack A. (2010) Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biol 11: R14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Schwartz S, Wagner L, Miller W. (2000) A greedy algorithm for aligning DNA sequences. J Comput Biol 7: 203–214 [DOI] [PubMed] [Google Scholar]