A novel genetical genomics analysis of metabolites in four developmental stages of Arabidopsis germination unravels both genetic and genetic × environment loci controlling these metabolites.
Abstract
A complex phenotype such as seed germination is the result of several genetic and environmental cues and requires the concerted action of many genes. The use of well-structured recombinant inbred lines in combination with “omics” analysis can help to disentangle the genetic basis of such quantitative traits. This so-called genetical genomics approach can effectively capture both genetic and epistatic interactions. However, to understand how the environment interacts with genomic-encoded information, a better understanding of the perception and processing of environmental signals is needed. In a classical genetical genomics setup, this requires replication of the whole experiment in different environmental conditions. A novel generalized setup overcomes this limitation and includes environmental perturbation within a single experimental design. We developed a dedicated quantitative trait loci mapping procedure to implement this approach and used existing phenotypical data to demonstrate its power. In addition, we studied the genetic regulation of primary metabolism in dry and imbibed Arabidopsis (Arabidopsis thaliana) seeds. In the metabolome, many changes were observed that were under both environmental and genetic controls and their interaction. This concept offers unique reduction of experimental load with minimal compromise of statistical power and is of great potential in the field of systems genetics, which requires a broad understanding of both plasticity and dynamic regulation.
The use of natural variation to disentangle the genetic (G) mechanisms underlying phenotypic differences has been very successful both in crop plants and in the model plant Arabidopsis (Arabidopsis thaliana; Alonso-Blanco et al., 2009). Most of the variation within wild or domesticated plant species is of quantitative nature determined by G polymorphisms at multiple loci. Such quantitative trait loci (QTL) can be analyzed efficiently using experimental mapping populations such as recombinant inbred lines (RILs) derived from directed crosses. Nowadays, many well-structured RIL populations are available, often accompanied with detailed studies of phenotypic variation (Mitchell-Olds and Schmitt, 2006). The complexity of quantitative traits is further determined by the interactions between genomic loci (i.e. epistasis) and between the genotype and the environment (genetic × environmental [G:E]). While epistasis can be effectively identified in QTL analyses, albeit with lower power than main effects, the detection of G:E interactions requires experimentation in multiple conditions of interest. Because of the large population sizes often needed to obtain sufficient statistical power for QTL detection, G:E interactions are usually ignored in experimental setups. However, a better understanding of the perception and processing of environmental (E) signals is greatly needed, because interactions provide important insights in adaptation mechanisms and evolutionary constraints such as balancing and disruptive selection. To obtain a more detailed view of the molecular mechanisms underlying phenotypic variation, genetical genomics studies, in which molecular traits are genetically analyzed, have been successfully applied to enhance a directed strategy to identify causal relationships (Kliebenstein et al., 2006; Keurentjes et al., 2007a; van Leeuwen et al., 2007; Wentzell et al., 2007; West et al., 2007; Rowe et al., 2008). The observed phenotype is often the resultant of a functional cascade of gene transcription followed by protein translation and modification, which finally leads to a highly dynamic metabolome underlying emergent properties (Kooke and Keurentjes, 2011). With the technological advances made in genomic analytical platforms, such as transcriptomics, proteomics, and metabolomics, the large-scale, high-throughput analyses needed for quantitative G approaches have become feasible (Jansen and Nap, 2001; Keurentjes et al., 2008). To incorporate developmental and E perturbation in the often expensive and laborious omic analyses, an alternative experimental setup, coined generalized genetical genomics (GGG), using balanced fractions of a RIL population has been proposed (Li et al., 2008). It provides a cost-effective experimental setup for hypothesis-generating research in multiple environments. Such an approach aims for the creation of subpopulations of RILs, one for each environment to be tested, with an optimal distribution of parental alleles over all available markers (Li et al., 2009). When these subpopulations are subjected to E perturbation, the emerging phenotypes can be explained by several sources of variation: G variation, E variation, and G:E variation. Whenever the resulting phenotype is not or only mildly affected by E interactions (G:E), the analysis of the different subpopulations can be combined, gaining the full power of a complete population. However, when a trait shows strong G:E interaction (e.g. those that only express G variation in specific environments), the power to detect QTL is dependent on those subpopulations expressing the G variation. Although G:E interactions have been detected previously in genetical genomics studies for expression (Li et al., 2006; Smith and Kruglyak, 2008; Gerrits et al., 2009; Yeung et al., 2011) and metabolite content (Zhu et al., 2012) by analyzing all lines in a population under different environments, the GGG concept offers an effective way of studying a combination of G and E perturbations and is of great potential in the field of systems genetics, in which a broad understanding of both plasticity and dynamics is required (Li et al., 2008). The fundamental basis of the experimental design and data analysis using a full model (Y = E + G + G:E + e), where Y is observed phenotype and e is residual error, is generally valid and frequently used (Churchill, 2002; Li et al., 2006; Gerrits et al., 2009). As a proof of principle, we present experimental data on the G regulation of primary metabolism in dry and imbibed Arabidopsis seeds using a GGG design and discuss the application and implications of such a strategy.
Plants are extremely rich in biochemical compounds, and major roles in plant development, adaptation, and defense have been identified for biosynthesis pathways and their products (Binder, 2010). The biosynthetic pathways of primary metabolites are well studied and often well conserved between different taxa (Peregrín-Alvarez et al., 2009). Nonetheless, quantitative variation for many of these compounds can be observed between natural variants, which might be reflected in their different growth characteristics. The analysis of single-gene mutants, for example, has unraveled many key components in biochemical pathways and has demonstrated their role in phenotypic traits (Fiehn et al., 2000). In Arabidopsis, G variation for many of its metabolic compounds has been observed (Kliebenstein et al., 2001a; Keurentjes et al., 2006; Rowe et al., 2008), but G:E interactions were ignored in these studies and only addressed by Chan et al. (2011). Metabolic profiling at different growth stages has further revealed important fluxes that regulate plant development and adaptation (de Oliveira Dal’Molin et al., 2010). Using the accumulated historical mutations that occur in natural variants in combination with metabolic profiling in a generalized design offers the unique possibility of identifying G effects over a series of developmental stages. Here, we report on the interaction of four different physiological environments (i.e. developmental stages) in dry and imbibed seeds with two founder genotypes in a RIL population. To detect the majority of the most prominent primary metabolites, we used gas chromatography-mass spectrometry of polar extracts (Roessner et al., 2000; Lisec et al., 2008). These include essential metabolites such as sugars, amino acids, and organic acids, which are key compounds in reserve storage and catabolism, growth, and energy metabolism.
The switch from a dry seed, which is equipped for optimal survival and storage of reserves, toward an imbibed seed, in which energy needed for germination is released and which prepares for autotrophic production, is remarkable. Reserves that have been stored during seed maturation are degraded and remobilized during germination (Bewley, 1997; Shu et al., 2008), a process that is heavily influenced by the capacity of carbon/nitrogen partitioning of a maturing seed (Dowdle et al., 2007). Arabidopsis mutants affected in their oil reserve content or its mobilization show delayed but not full inhibition of germination (Kinnersley and Turano, 2000; Bouché and Fromm, 2004; Shu et al., 2008; Kelly et al., 2011). This suggests an additional metabolic switch that occurs during seed desiccation after seed maturation involving a change from accumulation of oil and storage proteins to the synthesis of free amino acids, sugars, fatty acids, and their degradation products functioning to prepare for rapid metabolic recovery during imbibition (Fait et al., 2006; Angelovici et al., 2010). Imbibition of mature seeds specifically shows reduction of the metabolites that accumulate during the desiccation period. Upon germination, an increase of many metabolites, including amino acids, sugars, and organic acids, can be observed again, which reflects the increase of autotrophic activity (Fait et al., 2006). Profiling the primary metabolome over different developmental stages in a mapping population is therefore expected to reveal the dynamics of G regulation of many of these important processes. We will demonstrate here that much of the observed variation in biochemical profiles can be attributed to genotype-by-environment interactions, which can be effectively identified in a GGG approach.
RESULTS AND DISCUSSION
Experimental Design
Previous studies that focused on the comparative analysis of developmental and metabolic variation suggest a link between central metabolism and plant physiology, but G coregulation is not frequently observed (Keurentjes et al., 2006; Meyer et al., 2007). That said, in several studies in Arabidopsis, a major metabolite QTL cluster is associated with the ERECTA locus, representing a strong regulator of development, which is known for its pleiotropic effects (Fu et al., 2009). To circumvent this strong bias, we used two natural variants, Bayreuth (Bay-0) and Shahdara (Sha), which are not polymorphic for the ERECTA locus. The Bay-0 × Sha RIL population (Loudet et al., 2002) has previously been shown to contain G variation for seed germination (Joosen et al., 2012) and other physiological traits (Loudet et al., 2003b, 2005, 2008; Barriere et al., 2005; Diaz et al., 2006; Reymond et al., 2006; Meng et al., 2008), anion strength (Loudet et al., 2003a), carbohydrate content (Calenge et al., 2006), gene expression (West et al., 2007), and primary (Rowe et al., 2008) and secondary metabolite levels (Wentzell et al., 2007).
Powerful mapping of G variation in a RIL population is dependent on the size of the population, the level of recombination, and an evenly genome-wide distribution of the parental alleles. In this study, a core set of the Bay-0 × Sha RIL population (Loudet et al., 2002) consisting of 165 lines and optimized for the aforementioned factors was used. This core population was divided in four subpopulations optimized for the distribution of parental alleles using the R package DesignGG, aiming at the most accurate estimate of G and G:E effects (Li et al., 2009; Supplemental Fig. S1).
Comparison of Different Designs Using Classic Phenotypes
Standard QTL mapping procedures can efficiently capture G variation and epistasis, but do not take E perturbation into consideration. Appropriate modeling of the G variance-covariance in the data is of great importance when combining information from different environments in QTL analysis (Churchill, 2002). Linear models are particularly well suited for this. Here, E differences are incorporated as an additional variable in a generalized design (GGG design). To enable mapping of the observed trait variation and taking the four developmental stages into consideration, an R script was developed, which uses functions and data structures from the R/qtl package (Broman et al., 2003; Arends et al., 2010; Supplemental File S3). The R script uses a linear model to calculate the likelihood of genotype-to-phenotype linkage for each marker with the following formula:
where yi is the ith observation of the studied phenotype, variable gi is the genotype, ei is a vector with seed conditions, and gi:ei the interaction term. The values βj represent parameters to be estimated, and εi is the error term. The simplified description (Y = E + G + G:E + ε) of this linear model will be used henceforward. Separate likelihood estimates (–log probability, henceforth log of the odds [LOD] scores) are generated for the E, G, and G:E effects.
To validate the use of a GGG design, we studied the G and interacting effects between G and E on phenotypes in four different E conditions. These phenotypes were obtained by studying different germination parameters under different E conditions (Joosen et al., 2012). In total, we compared the power of different designs by performing QTL analysis for 96 classic phenotypes under four different environments (Joosen et al., 2012; Table I). Furthermore, we also investigate the interacting effect between G and E. The full-model mapping (Y = E + G + G:E + ε) was applied to a full-block design, random design, and GGG design. Single-marker mapping (Y = G + ε) was applied to a single-block design. The number of detected QTL and interacting QTL (false discovery rate = 0.05, based on >10,000 runs permutation) with the different designs are shown in Table I. In the full-block design, all samples were allocated to the four conditions. Obviously, this is the most expensive way of performing the experiment, as the required resources and effort are quadrupled. As a consequence of the size of the experiment, the power of detecting G effects is the best for this design. Unfortunately, we cannot afford such expensive experiments in many situations due to limited resources and time. The single-block design only focuses on one of the four conditions, as in most published genetical genomics studies to date. In this way, the samples size for the selected condition is n, and we will have equal power, as in the full-block design, for detecting the G effects for this particular condition. Clearly, this design will miss the information from the other three conditions, and interacting effects between G and E factors cannot be investigated. To study both G and interacting effects with a limited budget, the random and the GGG design allocate the n different samples to the four environments evenly, measuring n/4 samples in each condition. Although the possibility to detect G effects is only slightly better for the GGG design, the detection of interacting QTL is clearly improved in the GGG design compared with the random design. These results show that the optimal allocation of samples in the GGG design clearly improves the ability to detect both G and interacting effects and that the GGG design results in the maximization of detected variation in relation to the necessary resources, with only a minimal compromise of statistical power compared with the full-block design.
Table I. Comparing different experimental designs.
Comparison of different experimental designs to study G and G:E effects on classic phenotypes in four different conditions. Each E condition is indicated with different gradient of gray in the blocks. In total there are n (164) genetically different RILs, and the data were analyzed in four different ways. The last two rows compare the number of QTLs for main G effect and G:E interacting effect detected using different design strategies. The numbers in parentheses indicate the QTLs that share confidence intervals (1.5 drop-off) with the full-block design.
| Design | QTL | Interacting QTL |
|---|---|---|
| Full-Block | 96 | 30 |
| Best power for G effect | ||
| Most expensive | ||
| Best power for G:E effect | ||
| Single-Block | 93 | 0 |
| Same power for G effect in the selected condition | ||
| Less expensive | ||
| Missing G:E effect | ||
| Random | 78(75) | 17(5) |
| Limited power for G effect | ||
| Less expensive | ||
| Limited power for G:E effect | ||
| GGG | 81(67) | 27(12) |
| Optimal power for G effect | ||
| Less expensive | ||
| Optimal power for G:E effect |
Metabolic Analyses
To study the metabolic status of Arabidopsis seeds during germination, four biologically important developmental stages of seed germination with expected variation in metabolite levels to different extent were selected. The first two stages, being freshly harvested primary dormant (PD) and after-ripened (AR) nondormant dry seeds, respectively, are expected to comprise a very similar metabolome, as most, if not all, metabolic fluxes are arrested in the dry seed. The oil-rich (approximately 40%) Arabidopsis seeds (Hobbs et al., 2004) typically desiccate to moisture contents below 5%, which results in an arrest of all enzymatic reactions due to the lack of free water. The other two stages represented 6-h-imbibed (6H) seeds and seeds at radicle protrusion (RP), respectively. Full rehydration of dry seeds typically completes in less than 2 h, and although developmental differences are not yet expected, many metabolic processes will have started after 6 h of imbibition (Nakabayashi et al., 2005; Howell et al., 2009). RP marks the end point of germination sensu stricto and is known to be accompanied by a major switch of both the transcriptome and metabolome (Nakabayashi et al., 2005; Fait et al., 2006). These four developmental stages are anticipated to vary to different degrees in their metabolic profiles, with hardly any difference between dry seed samples, some differences between dry and imbibed seeds, and very pronounced differences between dry seeds and seeds at RP.
To determine the metabolic status of G variants in these different developmental stages, all individuals in the four subpopulations and their parental accessions were subjected to gas chromatography-time of flight-mass spectrometry. Each sample consists of the polar fraction of a methanol extract of a bulk of approximately 700 to 1,000 seeds (20 mg). Samples were analyzed in random order and interspersed with pooled sample controls to control for experimental errors. The metabolic profiling of the segregating RILs was performed, and the use of segregation population provided an intrinsic replication for each genotypic marker (Jansen and Nap, 2001). In total, 7,537 mass peaks were detected, representing 161 metabolites, according to centrotyping based on retention time and correlation structure (Tikunov et al., 2011). In total, 63 metabolites could be annotated using an in-house-constructed library and a publicly available mass spectra library (Schauer et al., 2005; Supplemental File S1).
The parental accessions Bay-0 and Sha were measured in duplicate for all four developmental stages, allowing us to model the influence of condition and accession using a multifactor univariate ANOVA:
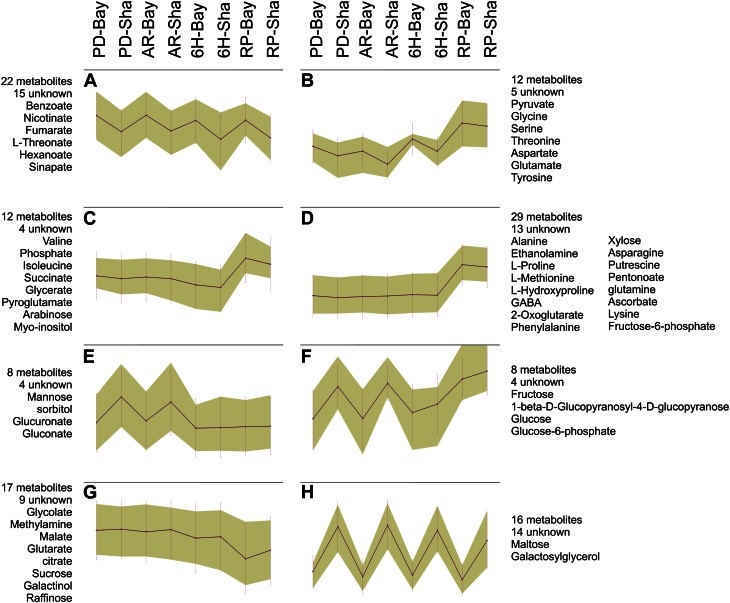
ANOVA for the parental samples identified 108 metabolites showing significant variation (false discovery rate < 0.05) between developmental stages (E) and 85 showing variation between the parents (G), with an overlap of 54 metabolites showing variation between both variables in an interactive way (G:E; Supplemental File S2). For 37 metabolites, no significant variation was detected between the parental accessions or in any of the developmental stages. A self-organizing map, created from the metabolites showing significant variation between the parents, groups different metabolites according to their accumulation pattern over different genotypes and developmental stages (Fig. 1). Clearly different patterns of variation can be observed, namely G in Figure 1, A and H, E in Figure 1, C and D, G + E in Figure 1, B and G, and G:E in Figure 1, E and F, illustrating the complex regulation of metabolic processes and the need for sophisticated analysis methods, such as principle component analysis or multiple QTL mapping (Arends et al., 2010).
Figure 1.
Self-organizing map, grouping different metabolites according to their accumulation pattern over different genotypes, and developmental stages of significantly variable metabolites (ANOVA F, P < 0.05) measured in the parental lines Bay-0 and Sha in four developmental stages. Two independent biological replicates were measured for each combination of parent and developmental stage. [See online article for color version of this figure.]
Because metabolite levels are varying between both parents and between the chosen seed germination stages, a segregation of metabolic accumulation can be expected in the RIL population of 164 lines. A principle component analysis of the metabolic profiles, revealing the internal structure in the data, shows that the first component clearly separates 6H seeds and seeds at RP from both PD and AR seeds, explaining 37% of the total variation (Supplemental Fig. S2). This confirms the large metabolic changes accompanying the transition from dry arrested seeds to the imbibed and germinating developmental stages. As expected, no obvious differences could be detected between the metabolomes of PD and AR dry seeds. The second component, explaining 11% of the total variation, separates the parental accessions, indicating that this component explains a lot of the G variation in metabolic profiles. These results demonstrate that Bay-0 and Sha possess G variation for the accumulation of primary metabolites, which segregates in their recombinant offspring and which is strongly influenced by the developmental stage used for profiling.
Transgressive segregation was visualized by comparing parental and RIL metabolite level distributions (Supplemental Fig. S3). Some positive and negative transgression is observed for most of the metabolites in which the metabolite accumulation in a RIL is respectively higher or lower compared with the respectively highest or lowest parent. In addition, 15 metabolites were detected in RILs that were not present in either parent. This suggests that new allele combinations in the RIL population resulted in enhanced accumulation or even novel formation of metabolites, although it could also be that those metabolites were missed in the parents because of the limits of the technology and methodology used in this study.
G Mapping of Metabolites in a GGG Design
In the experimental setup of this study, the E variation is defined as variation observed between the four developmental stages (PD, AR, 6H, and RP). Significance thresholds, determined by permutation analysis (n = 1,000, P < 0.01) for each metabolite, ranged from LOD 3.43 to LOD 3.50 and was stringently set to LOD 4 for all analyses. Mapping resulted in 120 significant QTLs in the G component for 83 metabolites and 31 G:E QTLs for 27 metabolites, ranging from one to four QTLs per metabolite. Thirteen of the G:E QTLs are significant in the G component as well. For 66 metabolites, no significant QTL was detected. Clustered heat maps for both the G and the G:E QTL profiles were created (Supplemental Figs. S4 and S5).
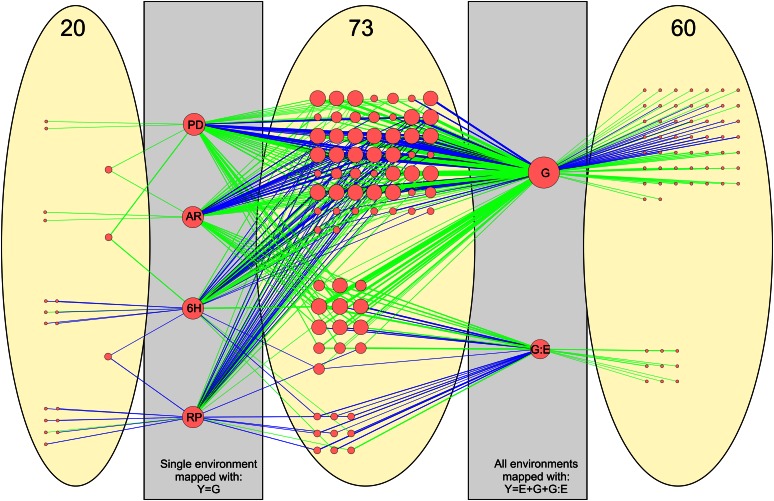
To test the performance of the generalized mapping procedure, QTLs detected in individual environments (using the linear model yi = β0 + β1gi + εi, henceforth Y = G + ε) were compared with QTLs detected in the combined mapping approach (using the linear model Y = E + G + G:E + ε; Fig. 2; Supplemental Table S1). QTLs were binned in upper or lower chromosome arms to reduce the effects of small positional shifts. Results were plotted in a network, with nodes representing QTLs connected with edges to nodes representing the mapping populations in which they were detected (Fig. 2). QTLs are grouped in three sections according to their detection in the different mapping procedures. The middle section shows 73 QTLs that were detected in both the Y = E + G + G:E + ε model and in one or more single-environment mappings using the Y = G + ε model. This shows that most of the G variation present in the single environments can effectively be captured by using the generalized model. The presence of 60 QTLs that were only significantly detected in the Y = E + G + G:E + ε model (right section) shows the combined power of the generalized approach and the usage of more genotypes. These QTLs are not detected in the single-environment mapping in which only 41 individuals were used. Combining all data across all environments in the linear model increases power to detect QTLs, but it should be noted that there are also 20 minor QTLs (left section) that are only significant in the single-environment mapping with the Y = G + ε model. These QTLs are not detected in the Y = E + G + G:E + ε model. This can be explained by two factors: (1) environments in which the G variation is not expressed introduce noise in the experimental data and thereby decrease mapping power, and (2) deviations from a balanced allele distribution in the different subpopulations can introduce some stochasticity around the threshold level, although this is not the case in our data.
Figure 2.
Comparison of QTLs detected within single environments (PD, AR, 6H, and RP) by using the simple Y = G + ε model with QTLs detected when combining environments via the full Y = E + G + G:E + ε model. QTLs were binned to two regions per chromosome (i.e. top and bottom region). When comparing QTLs of a single trait from two models, they are considered as shared ones if QTLs fall in the same region. In total, we found 73 QTLs shared between the two models, as shown in the middle ellipse. There are 20 and 60 QTLs that are only detected in the simple and full model, respectively. Nodes indicate metabolite QTLs, and node size shows the degree of connectivity. Nodes are connected by edges, which show the link between a QTL and a mapping population (single environments versus multiple environments). Separate nodes are created for the G component and the G:E component. Edge line color represents direction of the QTLs (green for higher levels in Sha and blue for higher levels in Bay-0). Line width indicates LOD scores. Detailed results comparing overlapping QTLs based on 95% confidence interval between models are shown in Supplemental Table S1.
Importantly, all major-to-moderate-effect-size QTLs could be detected using the generalized model, even when these QTLs were not detected in the separate environment models. Although it is difficult to compare power with the latter models, because population sizes differ, the generalized design efficiently identifies all relevant QTLs, which were detected by the four separate models, and in addition, it detects G:E interactions. In a general exploratory study, the reduction in experimental burden therefore amply outweighs the incidental failure to detect the limited number of small-effect QTLs. The application of a GGG design can thus be an important advancement in evolutionary and ecological studies assessing the contribution of G and E effects to natural variation in life history traits.
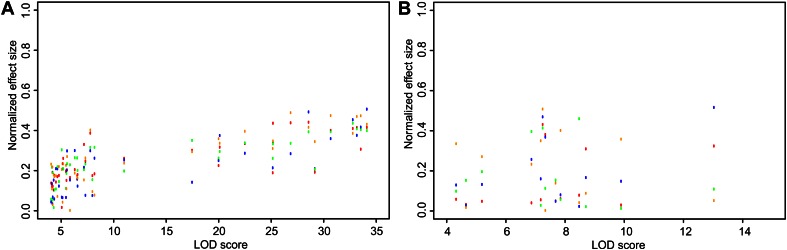
For breeding purposes, the allelic effect size is an important measure, and differentiation of the environment in which the allelic effect is expressed can be very useful. In the generalized setup, the allelic effect size of those metabolites with significant QTLs is separated per environment (Supplemental Files S4 and S5). For every QTL that is consistently detected in all four conditions, a LOD score for G effect (Fig. 3, x axis) is obtained from full-model mapping. For these QTLs, normalized allelic effect sizes are calculated by Z-score transformations for each environment (Fig. 3, y axis). QTLs detected in the G component of the linear model (Fig. 3A) show an expected linear relationship between LOD score and effect size in all measured environments. This correlation is much weaker for QTLs detected in the G:E component of the linear model (Fig. 3B) because the G variation is not expressed in all environments. QTLs of metabolites with strong G:E interaction, therefore, display larger effect sizes in fewer environments compared with G-component QTLs of similar significance levels.
Figure 3.
Effect sizes for each individual developmental stage are plotted against the derived LOD score. A, Normalized allelic effect size per environment against LOD scores from the G component. B, Normalized allelic effect size per environment against LOD scores from the G:E interaction component. Colors indicate the developmental stages (red for PD, blue for AR, green for 6H, and orange for RP).
Clearly, the choice of environments used in such study is crucial (Li et al., 2008). Limited power can be expected when environments vary too much and no overlapping G variation is present, and contrarily, there is hardly any additive value of the design when using very similar environments. In this study, we carefully selected four biologically relevant developmental stages of seed germination with expected variation in metabolite levels to different extent and consider them as an E factor in the follow-up statistical analysis. The selected developmental stages start from PD dry seeds to seeds at the point of RP. The first two stages, being freshly harvested PD and AR nondormant dry seeds, respectively, are expected to comprise a very similar metabolome, as most, if not all, metabolic fluxes are arrested in the dry seed. The other two stages represent 6H seeds and seeds at RP, respectively. Different levels of E variation were obtained and could be mapped by the G and/or G:E component of the linear model.
G Regulation of Metabolic Traits
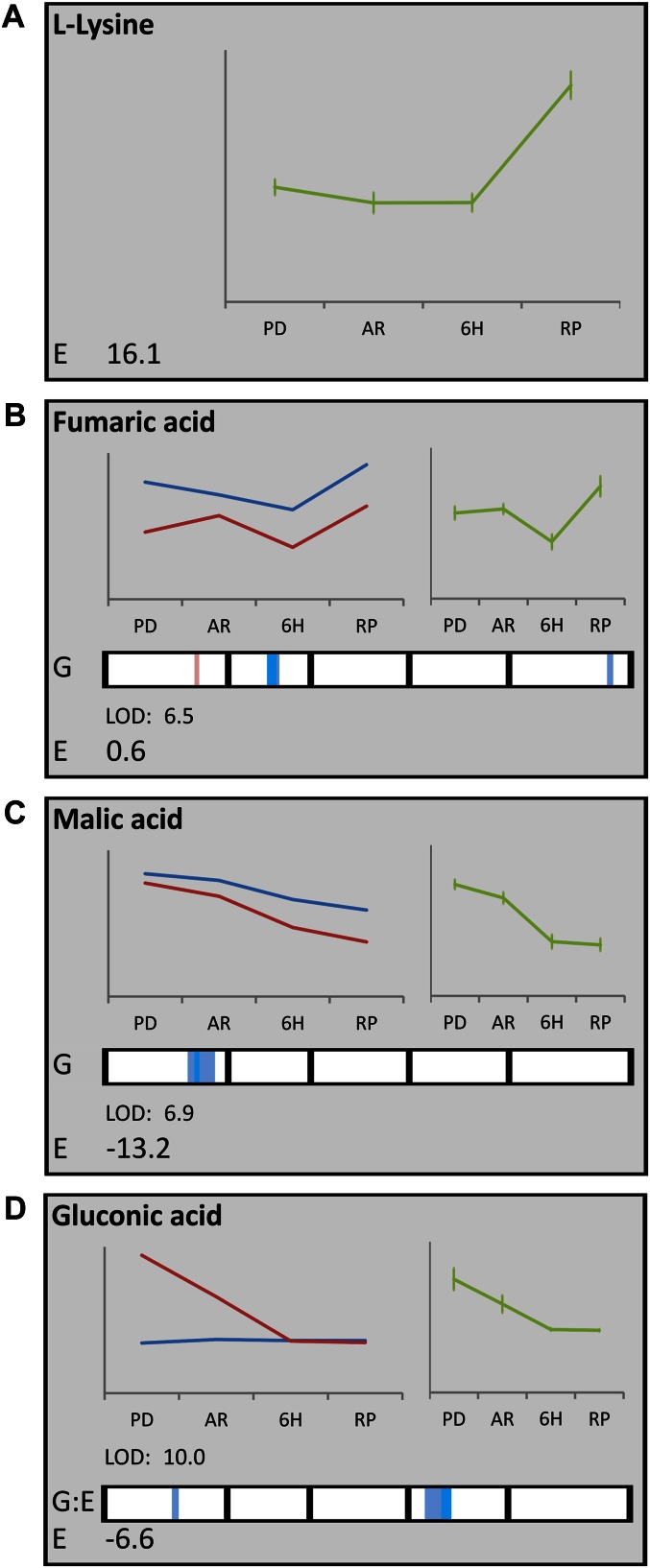
One of the most rewarding benefits of the generalized approach is the possibility to analyze metabolic fluxes over different environments or developmental stages in addition to the effect of G variation. The acquired information of both sources of variation can be effectively displayed in so-called flash cards, in which line graphs illustrate the G and E effect and detected QTLs are plotted in heat bars (Fig. 4; Supplemental Fig. S6). The individual components of the linear model Y = E + G + G:E + ε provide the valuable measures for the various sources of variation. For example, Lys content strongly increases in germinating seeds, indicated by a significant LOD score of 16.1 for the E effect, but no G variation for Lys could be detected (Fig. 4A). For this metabolite, G variants vary indistinguishably from each other over different environments. By contrast, fumaric acid shows little variation between the developmental stages (LOD 0.6), but displays strong G variation explained by a highly significant QTL (LOD 6.5) for the G effect at the center of chromosome 2. Higher levels for fumaric acid are detected in all developmental stages for those lines harboring the Bay-0 allele (Fig. 4B). An example of the additive effect of E and G factors is the decrease in levels of malic acid in imbibed seeds. Here, a strong E effect (LOD 13.2) is accompanied with an additional G effect, explained by a G QTL (LOD 6.9) at the bottom of chromosome 1. Note that the G effect here is similar in all environments (Fig. 4C). This is not the case for gluconic acid, levels of which are strongly affected by the G:E interaction. A strong G:E QTL (LOD 10) is detected at the top of chromosome 4. The Sha allele at this position causes higher levels of gluconic acid in dry seeds but not in imbibed seeds (Fig. 4D). This strong negative E effect (LOD 6.6) is also responsible for the apparent directional shift of the G:E QTL effect.
Figure 4.
Normalized metabolite changes during four developmental stages (PD, AR, 6H, and RP). Each section represents a single metabolite and contains information about E variation (green line plot represents the average over all lines within a single developmental stage) and G variation (blue lines represent the metabolite levels for lines carrying the Bay-0 allele for the most significant QTL, and red lines represent those for the Sha allele-carrying lines). QTL profiles for metabolites with either G or G:E variation are indicated at the bottom of each section by a heat bar representing the five chromosomes, and a false-color scale is used to indicate the QTL significance. For G QTLs, positive values (light and dark blue) represent a larger effect on the metabolite content for the Bay-0 allele, and negative values (light and dark red) represent a larger effect on the metabolite content for the Sha allele. Interpretation of the color scale for G:E QTLs is less intuitive because strong negative E effects can result in inversion of the QTL LOD score (e.g. gluconic acid). The presented effect plot (left line plot) shows the true allele effect. E variation is expressed as LOD score in the lower left corner. Depending on the most significant variation, either G or G:E interaction effects are also indicated with LOD scores in the lower left corner below or above the heat bar, respectively. A, l-Lys, showing only E variation. B, Fumaric acid, showing G variation. C, Malic acid, showing both G and E variation. D, Gluconic acid, showing interaction between G:E variation.
Similar to the self-organizing maps in Figure 1, flashcards can be instrumental in the identification of metabolic relationships, with the added value of G regulatory information. This is illustrated by integrating flashcards of all metabolites that were identified in this study with a general Arabidopsis metabolic pathway diagram (http://www.KEGG.jp; Supplemental Fig. S7). For instance, several pathways in carbohydrate metabolism, such as the biosynthesis routes for Gal, pentose phosphate, starch/Suc, and amino and nucleotide sugars, are highly interconnected and are therefore subject to coregulation mechanisms. A number of compounds involved in different subparts of the carbohydrate network module (e.g. Glc-6-P, maltose, Man, GlcA, and gluconic acid) share a strong QTL at the top of chromosome 4. This suggests that the observed variation for these compounds has a single G basis, possibly affecting competition for a general precursor or directing feedback loops. In addition, many of these compounds show strong positive or negative correlation due to E control. G coregulation was also observed for amino acid metabolism. Amino acids are substrate for the synthesis of aminoacyl-tRNAs, which, in turn, are essential substrates for translation (Sheppard et al., 2008). A single G:E QTL at the bottom of chromosome 1 was detected for eight amino acids, explaining a large part of the observed G variation. The joined analysis of environmentally and genetically induced variation in metabolic profiles can thus identify causal relationships between different modular parts of metabolic networks and associate these connections with relevant biological processes.
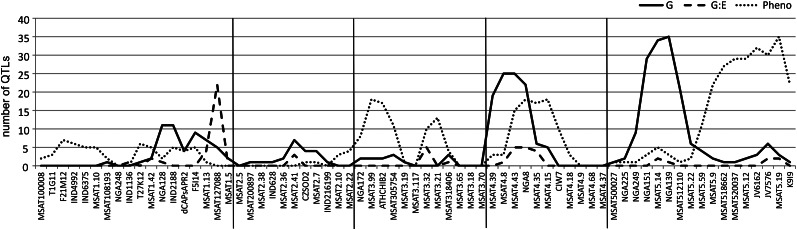
Regulatory Hotspots and Physiological Coregulation
As noted, the accumulation of several metabolites maps to identical positions, suggesting that these might be regulated by a common G factor. Although colocating QTLs can be the result of independent closely linked G factors, such coinciding QTLs are expected to occur more or less randomly by chance. Any deviation from expected frequency distributions along the genome thus hints at G coregulation (Breitling et al., 2008). When plotted against their genomic position, eight of such suggestive QTL hotspots can be seen (Fig. 5), of which, the two major ones (chromosomes 4-MSAT4.8 and 5-NGA139) colocate with previously identified hotspots for metabolic regulation (Kliebenstein et al., 2001b; Keurentjes et al., 2006; Wentzell et al., 2007; Rowe et al., 2008). Interestingly, both these loci have been shown to play a role in glucosinolate biosynthesis. The AOP locus at chromosome 4 regulates side-chain modification, while the MAM locus at chromosome 5 determines chain elongation, but these compounds are not targeted for in gas chromatography-mass spectrometry analysis, which predominantly detects primary metabolites. As for many glucosinolates, for some metabolites, including γ-aminobutyric acid (GABA) and maltose, QTLs were detected at both positions. In other cases, a single QTL was detected at chromosome 4 or 5, e.g. Glc-6-P and Tyr, respectively. Although the identified primary metabolites are not directly connected with the glucosinolate biosynthesis pathway, such associations have been reported before (Rowe et al., 2008). These results might suggest alternative functions for AOP and MAM or a role in resource competition and allocation in central metabolism. This suggestion is further supported by the fact that these loci link to flowering time and the circadian clock regulation in the Bay-0 × Sha population (Chan et al., 2011). It also cannot be ruled out that other genes overlapping the AOP or MAM regions are causal for the observed variation.
Figure 5.
Number of significant QTLs plotted against the G location. Metabolic QTLs are represented by the black (G) and dashed (G:E) lines. Germination-related QTLs (Joosen et al., 2012) are shown by the dotted line.
Because many metabolites appear to be coregulated, the strong impact of some loci on central metabolism might also exert its effect on physiological traits. Recently, the G landscape of seed germination in the same population has been described, for which seed germination parameters were acquired under a wide range of E conditions (Joosen et al., 2012). A comparison between variation in germination characteristics and metabolite levels might reveal compounds involved in the process of germination. Although no clear colocation of hotspots for germination and metabolite QTLs could be observed, incidental coincidence between isolated QTLs of both types of traits did occur. For instance, G variation for seed size colocates with a large metabolic QTL cluster on the lower arm of chromosome 1 (approximately 75 centimorgans). This cluster contains many QTLs for amino acids, but also many QTLs for components of the TCA cycle (e.g. fumarate and malate). In plants, Leu, Ile, and Val can be broken down, and the end products of their catabolic pathways enter the TCA cycle to generate energy. It has been shown that these amino acids promote their own degradation, but only during seed germination or senescence or under sugar starvation (Binder, 2010). This suggests that the degradation pathways provide alternative carbon sources for the plant in extreme conditions. In addition, branched-chain amino acids and their derived α-keto acids are cytotoxic, and preventing accumulation through degradation may be an important detoxification mechanism (Fujiki et al., 2000). Higher levels of both fumarate and malate, as a result of the degradation of a surplus of amino acids, might thus be indicative for larger seed sizes. A second QTL for seed size on chromosome 5 colocates with a QTL of opposite effect for GABA accumulation. Interestingly, Bay-0 alleles at both QTLs confer larger seed size, suggesting that there was selection pressure for large seed size in the environment where Bay-0 was collected, as was also observed in a different population (Alonso-Blanco et al., 1999). However, where levels of fumarate and malate are increased in larger seeds, the accumulation of GABA is decreased. GABA is known to be involved in a range of cellular processes (Palanivelu et al., 2003) and is rapidly accumulated in response to biotic and abiotic stresses (Kinnersley and Turano, 2000). It has been postulated that it has roles in herbivore deterrence, pH and redox regulation, energy production, and maintenance of carbon/nitrogen balance (Bouché and Fromm, 2004). In a recent study, GABA levels in seeds were shown to increase by expressing Glu decarboxylase under a seed maturation-specific phaseolin promoter (Fait et al., 2011). In accordance with our findings, this resulted in smaller seed size and reduced seed vigor in T3 plants. No opposite seed size effect could be detected at a GABA QTL with increased levels due to the Bay-0 allele at the top of chromosome 4, but colocating G variation for germination on abscisic acid, heat sensitivity, and dormancy was observed at this position. These cases illustrate the power of joined G analyses of metabolic and physiological traits for generation of hypotheses that can help in the functional annotation of plant metabolites and their possible role in the regulation of important physiological processes.
Confirmation of Metabolic QTLs
To independently confirm the effect of a single locus, it must be isolated and tested in an isogenic background. Several methods can be followed to perform such an independent confirmation of QTLs. A powerful approach is the use of residual heterozygosity in early generations of RILs. The Bay-0 × Sha RIL population (420 lines in total) was genotyped at F6, in which approximately 97% homozygosity is reached in each line. This resulted in the presence of residual heterozygosity in at least a single RIL at almost all genome positions. Those heterozygous regions are segregating in a Mendelian fashion in the next generation and can be used to confirm QTL positions, as it provides a possibility to study both parental alleles at the locus of interest in an otherwise homozygous background (Tuinstra et al., 1997). In a heterogeneous inbred family (HIF), those heterozygous regions are fixed, and two separate lines containing the alleles of both parents, respectively, are maintained.
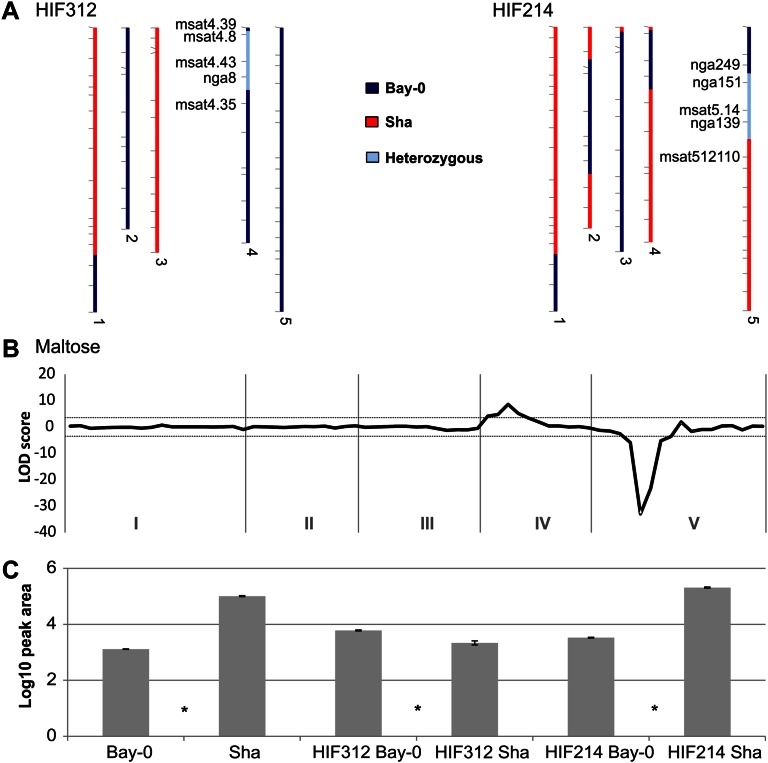
HIF312 and HIF214 are segregating for regions at the top of chromosomes 4 and 5 (Fig. 6A), respectively, and cover the region in which the two major metabolite hotspots were detected. AR dry seeds were used to profile the HIFs for metabolic content because many of the QTLs detected in this region showed a large-effect size at the dry seed stages. Significant differences between parental alleles using four replicates were defined by a two-tailed Student’s t test (P < 0.05). In total, 34 out of 64 QTLs could be confirmed using this approach (Supplemental Fig. S8). For maltose, for instance, two QTLs with opposite direction were found (Fig. 6B), which could both be confirmed using the two distinct HIFs (Fig. 6C). In a number of cases, a HIF effect was observed that was not detected significantly in the RIL population (e.g. digalactosylglycerol). This might be the result from the higher power in near isogenic lines due to the absence of epistatic interactions (Keurentjes et al., 2007b). Nonetheless, a substantial number of QTLs could not be confirmed by the HIF lines. The enrichment for small-effect QTLs in the unconfirmed class suggests that four replicates generate insufficient power to identify significant differences for these metabolites in the HIF experiments, although we cannot rule out that they are false positives from the QTL analysis. Furthermore, QTLs depending on epistatic interactions cannot be detected in some near-isogenic lines. In addition, a number of QTL support intervals are broader than the region covered by the HIF, and thus, the causal G polymorphism within the QTL interval, but outside the region covered by the HIF, would have been missed.
Figure 6.
QTL confirmation for maltose using the HIF approach. Two QTL regions (top chromosome 4 and top chromosome 5) were analyzed using AR seeds of lines HIF312 and HIF214 (A). The QTL profile for maltose (B) shows two significant QTLs (dashed line indicates the LOD 4 significance threshold). The lower section (C) shows the parental levels for maltose and the confirmation for both QTLs by the segregating HIF lines (either fixed for Bay-0 or Sha alleles at the heterozygous interval). Significant differences (Student’s t test, P < 0.05) are indicated with an asterisk in between the two contrasting samples.
The analyses of the HIF lines indicate that most of the large-effect QTLs can be accurately detected using a generalized genomics approach. Although an underestimation of small-effect QTLs can be expected, this is largely compensated by the higher power of detecting G and E interactions.
CONCLUSION
The use of natural variation is a valuable tool to dissect the genetics of complex traits, and the addition of powerful omics analysis provides a great resource to disentangle molecular mechanisms. However, the expensive nature of many omics experiments limits researchers to deploy perturbation of either environment or development. New strategies are needed to enable the switch from genetical genomics to system genetics. Here, we have reported on a strategy to divide a RIL population in well-defined subpopulations and to use those to perturb the environment or developmental stage. To this end, a novel R script has been created to enable QTL mapping using a linear model that includes the possibility to account for G and E variation. This R script is fast enough to analyze hundreds to thousands of traits and creates possibilities to extend the GGG strategy to whole-genome gene expression analysis by either microarray or next-generation sequence approaches (Joosen et al., 2009; Ligterink et al., 2012).
Efficient QTL mapping is strongly dependent on the population size and recombination frequency. Keurentjes et al. (2007b) studied the effect of the population size and showed a linear relationship between the number of individuals used for mapping and the smallest detectable G effect. In this light, it might seem undesirable to split a RIL population in smaller subpopulations. This is true when G variation is only detectable in a single unique environment or developmental stage leading to a strong G:E interaction. More often, variation is subject to the environment, without a complete abolishment of the G variation. In those cases, the E effects can be normalized, and the power of detecting a QTL is increased to the total number of lines used in the different subpopulations. The availability of a genome-wide set of HIF lines of the Bay-0 × Sha RIL population provides a solid and fast way to confirm QTLs. By using this approach, we tested two of the observed QTL hotspots and were able to confirm many of the detected QTLs. When resources are limited, this can be regarded as a good alternative for replicating the whole experiment during, for example, different growth seasons.
Many studies have shown the highly dynamic nature of molecular mechanisms leading toward seed germination (for review, see Catusse et al., 2008; Daszkowska-Golec, 2011; Weitbrecht et al., 2011). Performing expensive genetical genomic experiments without any perturbation of the environment will therefore always raise questions about the possible extrapolation of the results when slightly different conditions are used. Information about the flux of a metabolite within a range of developmental stages or within a range of environments allows a much more precise interpretation of the molecular effects. By using the generalized strategy, we showed that it is possible to deduct the metabolic fluxes (Fig. 4). This extra level of information is a very valuable addition and helps to interpret the effect of G variation in the context of a dynamic and constantly changing metabolome.
Metabolite hotspots can reveal important loci involved in major metabolic pathway differences between two natural variants. In several studies, the detected omics hotspots did not colocate more than expected by chance with phenotypic hotspots (Keurentjes et al., 2006; Meyer et al., 2007). However, in this study, we detected some colocating QTLs, which might be explained by the narrow developmental window in which both metabolite and phenotypic QTLs (Joosen et al., 2012) were gathered. We detected overlapping QTLs for amino acid synthesis, TCA cycle compounds, and seed size at the bottom of chromosome 1 and also colocation between QTLs for GABA, seed size, and germination under stress conditions at the top of chromosome 5 (Joosen et al., 2012). These colocating QTLs are interesting leads for further research, which is necessary to elucidate the true causal molecular mechanisms.
In conclusion, in the era of large systems genetics initiatives, we propose to consider the use of a generalized design for genetical genomics studies. The simultaneous acquisition of both G variation and developmental fluxes is a cost-effective approach, enabling a much better understanding of the processes involved. We see great potential in further exploration of the generalized design for transcriptome or other omics-related studies.
MATERIALS AND METHODS
Plant Material
Seeds from the core population (165 lines) of the Arabidopsis (Arabidopsis thaliana) Bay-0 × Sha RIL population (Loudet et al., 2002) and HIF lines were obtained from the Versailles Biological Resource Centre for Arabidopsis (http://dbsgap.versailles.inra.fr/vnat). The population is mapped with 69 markers, with an average distance between the markers of 6.1 centimorgans (Loudet et al., 2002). Maternal plants were grown in a fully randomized setup and, seeds from four to seven plants per RIL were bulk harvested. Plants were grown on 4- × 4-cm rockwool plugs (MM40/40, Grodan B.V.) and watered with 1 g L–1 Hyponex fertilizer (nitrogen:phosphorus:potassium, 7:6:19; http://www.hyponex.co.jp) in a climate chamber (20°C day, 18°C night) with 16 h of light (35 W m–2) at a relative humidity of 70%. Seeds were either stored at –80°C 1 week after harvest (PD) or AR at room temperature and ambient relative humidity until maximum germination potential after 5 d of imbibition was reached. AR seeds were imbibed on water-saturated filter paper at 20°C for 6 h and quickly transferred to a dry filter paper for 1 min to remove excess of water (6H). Manual selection with the help of a binocular was carried out to harvest seeds, with the radicle at the point of protrusion (RP). Three RP lines failed the metabolite analysis and were replaced by dry PD samples.
Metabolite Analysis
The metabolite extraction was performed based on a previously described method (Roessner et al., 2000) with some modifications. Seeds (20 mg) were homogenized using a microdismembrator (Sartorius) in 2-mL tubes with two iron balls (2 and 5 mm) precooled in liquid nitrogen. Seven microliters of methanol:chloroform (4:3) was added together with the standard (0.2 mg mL–1 ribitol) and mixed thoroughly. After 10 min of sonication, 200 µL Milli-Q was added to the mixture, followed by vortexing and centrifugation (5 min, 13,500 rpm). The methanol phase was collected in a glass vial. Five hundred microliters of methanol/chloroform was added to the remaining organic phase and kept on ice for 10 min. Two hundred microliters of MQ was added followed by vortexing and centrifugation (5 min, 13,500 rpm). Again, the methanol phase was collected and mixed with the other collected phase. One hundred microliters was dried overnight using a SpeedVac (35°C, Savant SPD121).
A GC-TOF-MS method (Carreno-Quintero et al., 2012) was used with some minor modifications. Detector voltage was set at 1,600 V. Raw data were processed using the ChromaTOF software 2.0 (Leco Instruments) and further processed using the MetAlign software (Lommen, 2009) to extract and align the mass signals. A signal-to-noise ratio of 2 was used. The output was further processed by the MetAlign Output Transformer (Plant Research International), and mass signals that were present in less than three RILs were discarded. Centrotypes were created using the MSClust program (Tikunov et al., 2011). The mass spectra of these centrotypes were used for the identification by matching to an in-house-constructed library and the National Institute of Standards and Technology NIST05 (http://www.nist.gov/srd/mslist.cfm) libraries. This identification is based on spectra similarity and comparison of retention indices calculated by using a third-order polynomial function (Strehmel et al., 2008).
QTL Mapping
Data were preprocessed using a log transformation, and per-phenotype outliers were removed after Z transformation (Z-score > 3). With the open-source statistical package R (version 2.14.1), we fitted a basic linear model (yi = β0 + β1gi + εi) on the four conditions separately. This was followed by a combined mapping allowing for a developmental covariate and interaction term between the G marker and developmental stage (yi = β0 + β1ei + β2gi + β3ei:gi + εi). P values from all mappings are transformed into LOD scores by taking the –log. In addition, raw and normalized effects were calculated for each individual environment. Normalized effects were calculated by dividing the difference between the maximum and the minimum value for that trait by the mean effect at the marker. LOD significance was determined using permutations for the combined mapping of the four environments; a LOD score of 4 was found to be significant (Breitling et al., 2008). Supplemental File S3 contains the R script used for the data analysis.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Allele distribution within the Bay-0 × Sha RIL population and the four selected subpopulations.
Supplemental Figure S2. Principal component analysis plot showing the first two principal components of the metabolite analysis in the Bay-0 × Sha RIL population.
Supplemental Figure S3. Transgression plot.
Supplemental Figure S4. Clustered heat map from the G component showing the LOD profiles of all metabolites.
Supplemental Figure S5. Clustered heat map from the G:E component showing the LOD profiles of all metabolites.
Supplemental Figure S6. Flashcards of all identified metabolites.
Supplemental Figure S7. Kyoto Encyclopedia of Genes and Genomes metabolic pathway with flashcards overlay of the metabolites identified in this study.
Supplemental Figure S8. Overview from HIF analysis with all metabolites with significant QTL confirmation.
Supplemental Table S1. Overview of QTLs shared between different models based on 95% confidence intervals.
Supplemental File S1. Metabolite centrotype data.
Supplemental File S2. ANOVA results from metabolic profiling of the parental lines Bay-0 and Sha.
Supplemental File S3. R script with original data files allowing reanalysis of all data provided in this paper.
Supplemental File S4. Summary of all detected metabolic G QTLs.
Supplemental File S5. Summary of all detected metabolic G:E QTLs.
Acknowledgments
We thank Linus van der Plas for critical reading of the article.
Glossary
- QTL
quantitative trait loci
- RIL
recombinant inbred line
- G:E
genetic × environment
- GGG
generalized genetical genomics
- G
genetic
- E
environmental
- Bay-0
Bayreuth
- Sha
Shahdara
- PD
primary dormant
- AR
after-ripened
- 6H
6-h-imbibed
- RP
radicle protrusion
- HIF
heterogeneous inbred family
- LOD
log of the odds
- GABA
γ-aminobutyric acid
References
- Alonso-Blanco C, Aarts MG, Bentsink L, Keurentjes JJ, Reymond M, Vreugdenhil D, Koornneef M. (2009) What has natural variation taught us about plant development, physiology, and adaptation? Plant Cell 21: 1877–1896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso-Blanco C, Blankestijn-de Vries H, Hanhart CJ, Koornneef M. (1999) Natural allelic variation at seed size loci in relation to other life history traits of Arabidopsis thaliana. Proc Natl Acad Sci USA 96: 4710–4717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angelovici R, Galili G, Fernie AR, Fait A. (2010) Seed desiccation: a bridge between maturation and germination. Trends Plant Sci 15: 211–218 [DOI] [PubMed] [Google Scholar]
- Arends D, Prins P, Jansen RC, Broman KW. (2010) R/qtl: high-throughput multiple QTL mapping. Bioinformatics 26: 2990–2992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barriere Y, Laperche A, Barrot L, Aurel G, Briand M, Jouanin L. (2005) QTL analysis of lignification and cell wall digestibility in the Bay-0 × Shahdara RIL progeny of Arabidopsis thaliana as a model system for forage plant. Plant Sci 168: 1235–1245 [Google Scholar]
- Bewley JD. (1997) Seed germination and dormancy. Plant Cell 9: 1055–1066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binder S. (2010) Branched-chain amino acid metabolism in Arabidopsis thaliana. The Arabidopsis Book 8: e0137, doi/10.1199/tab.0137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouché N, Fromm H. (2004) GABA in plants: just a metabolite? Trends Plant Sci 9: 110–115 [DOI] [PubMed] [Google Scholar]
- Breitling R, Li Y, Tesson BM, Fu J, Wu C, Wiltshire T, Gerrits A, Bystrykh LV, de Haan G, Su AI, et al. (2008) Genetical genomics: spotlight on QTL hotspots. PLoS Genet 4: e1000232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broman KW, Wu H, Sen Ś, Churchill GA. (2003) R/qtl: QTL mapping in experimental crosses. Bioinformatics 19: 889–890 [DOI] [PubMed] [Google Scholar]
- Calenge F, Saliba-Colombani V, Mahieu S, Loudet O, Daniel-Vedele F, Krapp A. (2006) Natural variation for carbohydrate content in Arabidopsis. Interaction with complex traits dissected by quantitative genetics. Plant Physiol 141: 1630–1643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carreno-Quintero N, Acharjee A, Maliepaard C, Bachem CW, Mumm R, Bouwmeester H, Visser RG, Keurentjes JJ. (2012) Untargeted metabolic quantitative trait loci analyses reveal a relationship between primary metabolism and potato tuber quality. Plant Physiol 158: 1306–1318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catusse J, Job C, Job D. (2008) Transcriptome- and proteome-wide analyses of seed germination. C R Biol 331: 815–822 [DOI] [PubMed] [Google Scholar]
- Chan EKF, Rowe HC, Corwin JA, Joseph B, Kliebenstein DJ. (2011) Combining genome-wide association mapping and transcriptional networks to identify novel genes controlling glucosinolates in Arabidopsis thaliana. PLoS Biol 9: e1001125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churchill GA. (2002) Fundamentals of experimental design for cDNA microarrays. Nat Genet 32: 490–495 [DOI] [PubMed] [Google Scholar]
- Daszkowska-Golec A. (2011) Arabidopsis seed germination under abiotic stress as a concert of action of phytohormones. OMICS 15: 763–774 [DOI] [PubMed] [Google Scholar]
- de Oliveira Dal’Molin CG, Quek LE, Palfreyman RW, Brumbley SM, Nielsen LK. (2010) AraGEM, a genome-scale reconstruction of the primary metabolic network in Arabidopsis. Plant Physiol 152: 579–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz C, Saliba-Colombani V, Loudet O, Belluomo P, Moreau L, Daniel-Vedele F, Morot-Gaudry JF, Masclaux-Daubresse C. (2006) Leaf yellowing and anthocyanin accumulation are two genetically independent strategies in response to nitrogen limitation in Arabidopsis thaliana. Plant Cell Physiol 47: 74–83 [DOI] [PubMed] [Google Scholar]
- Dowdle J, Ishikawa T, Gatzek S, Rolinski S, Smirnoff N. (2007) Two genes in Arabidopsis thaliana encoding GDP-L-galactose phosphorylase are required for ascorbate biosynthesis and seedling viability. Plant J 52: 673–689 [DOI] [PubMed] [Google Scholar]
- Fait A, Angelovici R, Less H, Ohad I, Urbanczyk-Wochniak E, Fernie AR, Galili G. (2006) Arabidopsis seed development and germination is associated with temporally distinct metabolic switches. Plant Physiol 142: 839–854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fait A, Nesi AN, Angelovici R, Lehmann M, Pham PA, Song L, Haslam RP, Napier JA, Galili G, Fernie AR. (2011) Targeted enhancement of glutamate-to-γ-aminobutyrate conversion in Arabidopsis seeds affects carbon-nitrogen balance and storage reserves in a development-dependent manner. Plant Physiol 157: 1026–1042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiehn O, Kopka J, Dörmann P, Altmann T, Trethewey RN, Willmitzer L. (2000) Metabolite profiling for plant functional genomics. Nat Biotechnol 18: 1157–1161 [DOI] [PubMed] [Google Scholar]
- Fu J, Keurentjes JJ, Bouwmeester H, America T, Verstappen FW, Ward JL, Beale MH, de Vos RC, Dijkstra M, Scheltema RA, et al. (2009) System-wide molecular evidence for phenotypic buffering in Arabidopsis. Nat Genet 41: 166–167 [DOI] [PubMed] [Google Scholar]
- Fujiki Y, Sato T, Ito M, Watanabe A. (2000) Isolation and characterization of cDNA clones for the e1β and E2 subunits of the branched-chain α-ketoacid dehydrogenase complex in Arabidopsis. J Biol Chem 275: 6007–6013 [DOI] [PubMed] [Google Scholar]
- Gerrits A, Li Y, Tesson BM, Bystrykh LV, Weersing E, Ausema A, Dontje B, Wang X, Breitling R, Jansen RC, et al. (2009) Expression quantitative trait loci are highly sensitive to cellular differentiation state. PLoS Genet 5: e1000692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobbs DH, Flintham JE, Hills MJ. (2004) Genetic control of storage oil synthesis in seeds of Arabidopsis. Plant Physiol 136: 3341–3349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell KA, Narsai R, Carroll A, Ivanova A, Lohse M, Usadel B, Millar AH, Whelan J. (2009) Mapping metabolic and transcript temporal switches during germination in rice highlights specific transcription factors and the role of RNA instability in the germination process. Plant Physiol 149: 961–980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen RC, Nap JP. (2001) Genetical genomics: the added value from segregation. Trends Genet 17: 388–391 [DOI] [PubMed] [Google Scholar]
- Joosen RV, Arends D, Willems LA, Ligterink W, Jansen RC, Hilhorst HW. (2012) Visualizing the genetic landscape of Arabidopsis seed performance. Plant Physiol 158: 570–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joosen RV, Ligterink W, Hilhorst HW, Keurentjes JJ. (2009) Advances in genetical genomics of plants. Curr Genomics 10: 540–549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly AA, Quettier AL, Shaw E, Eastmond PJ. (2011) Seed storage oil mobilization is important but not essential for germination or seedling establishment in Arabidopsis. Plant Physiol 157: 866–875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keurentjes JJ, Fu J, Terpstra IR, Garcia JM, van den Ackerveken G, Snoek LB, Peeters AJ, Vreugdenhil D, Koornneef M, Jansen RC. (2007a) Regulatory network construction in Arabidopsis by using genome-wide gene expression quantitative trait loci. Proc Natl Acad Sci USA 104: 1708–1713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keurentjes JJ, Koornneef M, Vreugdenhil D. (2008) Quantitative genetics in the age of omics. Curr Opin Plant Biol 11: 123–128 [DOI] [PubMed] [Google Scholar]
- Keurentjes JJB, Bentsink L, Alonso-Blanco C, Hanhart CJ, Blankestijn-De Vries H, Effgen S, Vreugdenhil D, Koornneef M. (2007b) Development of a near-isogenic line population of Arabidopsis thaliana and comparison of mapping power with a recombinant inbred line population. Genetics 175: 891–905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keurentjes JJB, Fu J, de Vos CHR, Lommen A, Hall RD, Bino RJ, van der Plas LHW, Jansen RC, Vreugdenhil D, Koornneef M. (2006) The genetics of plant metabolism. Nat Genet 38: 842–849 [DOI] [PubMed] [Google Scholar]
- Kinnersley AM, Turano FJ. (2000) Gamma aminobutyric acid (GABA) and plant responses to stress. Crit Rev Plant Sci 19: 479–509 [Google Scholar]
- Kliebenstein DJ, Gershenzon J, Mitchell-Olds T. (2001a) Comparative quantitative trait loci mapping of aliphatic, indolic and benzylic glucosinolate production in Arabidopsis thaliana leaves and seeds. Genetics 159: 359–370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kliebenstein DJ, Lambrix VM, Reichelt M, Gershenzon J, Mitchell-Olds T. (2001b) Gene duplication in the diversification of secondary metabolism: tandem 2-oxoglutarate-dependent dioxygenases control glucosinolate biosynthesis in Arabidopsis. Plant Cell 13: 681–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kliebenstein DJ, West MAL, van Leeuwen H, Loudet O, Doerge RW, St. Clair DA. (2006) Identification of QTLs controlling gene expression networks defined a priori. BMC Bioinformatics 7: 308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kooke R, Keurentjes JJ. (2012) Multi-dimensional regulation of metabolic networks shaping plant development and performance. J Exp Bot 63: 3353–3365 [DOI] [PubMed] [Google Scholar]
- Li Y, Álvarez OA, Gutteling EW, Tijsterman M, Fu J, Riksen JAG, Hazendonk E, Prins P, Plasterk RHA, Jansen RC, et al. (2006) Mapping determinants of gene expression plasticity by genetical genomics in C. elegans. PLoS Genet 2: e222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Breitling R, Jansen RC. (2008) Generalizing genetical genomics: getting added value from environmental perturbation. Trends Genet 24: 518–524 [DOI] [PubMed] [Google Scholar]
- Li Y, Swertz MA, Vera G, Fu J, Breitling R, Jansen RC. (2009) DesignGG: an R-package and web tool for the optimal design of genetical genomics experiments. BMC Bioinformatics 10: 188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ligterink W, Joosen RVL, Hilhorst HWM. (2012) Unravelling the complex trait of seed quality: using natural variation through a combination of physiology, genetics and -omics technologies. Seed Sci Res 22: S45–S52 [Google Scholar]
- Lisec J, Meyer RC, Steinfath M, Redestig H, Becher M, Witucka-Wall H, Fiehn O, Törjék O, Selbig J, Altmann T, et al. (2008) Identification of metabolic and biomass QTL in Arabidopsis thaliana in a parallel analysis of RIL and IL populations. Plant J 53: 960–972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lommen A. (2009) MetAlign: interface-driven, versatile metabolomics tool for hyphenated full-scan mass spectrometry data preprocessing. Anal Chem 81: 3079–3086 [DOI] [PubMed] [Google Scholar]
- Loudet O, Chaillou S, Camilleri C, Bouchez D, Daniel-Vedele F. (2002) Bay-0 × Shahdara recombinant inbred line population: a powerful tool for the genetic dissection of complex traits in Arabidopsis. Theor Appl Genet 104: 1173–1184 [DOI] [PubMed] [Google Scholar]
- Loudet O, Chaillou S, Krapp A, Daniel-Vedele F. (2003a) Quantitative trait loci analysis of water and anion contents in interaction with nitrogen availability in Arabidopsis thaliana. Genetics 163: 711–722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loudet O, Chaillou S, Merigout P, Talbotec J, Daniel-Vedele F. (2003b) Quantitative trait loci analysis of nitrogen use efficiency in Arabidopsis. Plant Physiol 131: 345–358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loudet O, Gaudon V, Trubuil A, Daniel-Vedele F. (2005) Quantitative trait loci controlling root growth and architecture in Arabidopsis thaliana confirmed by heterogeneous inbred family. Theor Appl Genet 110: 742–753 [DOI] [PubMed] [Google Scholar]
- Loudet O, Michael TP, Burger BT, Le Metté C, Mockler TC, Weigel D, Chory J. (2008) A zinc knuckle protein that negatively controls morning-specific growth in Arabidopsis thaliana. Proc Natl Acad Sci USA 105: 17193–17198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng P-H, Macquet A, Loudet O, Marion-Poll A, North HM. (2008) Analysis of natural allelic variation controlling Arabidopsis thaliana seed germinability in response to cold and dark: identification of three major quantitative trait loci. Mol Plant 1: 145–154 [DOI] [PubMed] [Google Scholar]
- Meyer RC, Steinfath M, Lisec J, Becher M, Witucka-Wall H, Törjék O, Fiehn O, Eckardt Ä, Willmitzer L, Selbig J, et al. (2007) The metabolic signature related to high plant growth rate in Arabidopsis thaliana. Proc Natl Acad Sci USA 104: 4759–4764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell-Olds T, Schmitt J. (2006) Genetic mechanisms and evolutionary significance of natural variation in Arabidopsis. Nature 441: 947–952 [DOI] [PubMed] [Google Scholar]
- Nakabayashi K, Okamoto M, Koshiba T, Kamiya Y, Nambara E. (2005) Genome-wide profiling of stored mRNA in Arabidopsis thaliana seed germination: epigenetic and genetic regulation of transcription in seed. Plant J 41: 697–709 [DOI] [PubMed] [Google Scholar]
- Palanivelu R, Brass L, Edlund AF, Preuss D. (2003) Pollen tube growth and guidance is regulated by POP2, an Arabidopsis gene that controls GABA levels. Cell 114: 47–59 [DOI] [PubMed] [Google Scholar]
- Peregrín-Alvarez JM, Sanford C, Parkinson J. (2009) The conservation and evolutionary modularity of metabolism. Genome Biol 10: R63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reymond M, Svistoonoff S, Loudet O, Nussaume L, Desnos T. (2006) Identification of QTL controlling root growth response to phosphate starvation in Arabidopsis thaliana. Plant Cell Environ 29: 115–125 [DOI] [PubMed] [Google Scholar]
- Roessner U, Wagner C, Kopka J, Trethewey RN, Willmitzer L. (2000) Technical advance: simultaneous analysis of metabolites in potato tuber by gas chromatography-mass spectrometry. Plant J 23: 131–142 [DOI] [PubMed] [Google Scholar]
- Rowe HC, Hansen BG, Halkier BA, Kliebenstein DJ. (2008) Biochemical networks and epistasis shape the Arabidopsis thaliana metabolome. Plant Cell 20: 1199–1216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schauer N, Steinhauser D, Strelkov S, Schomburg D, Allison G, Moritz T, Lundgren K, Roessner-Tunali U, Forbes MG, Willmitzer L, et al. (2005) GC-MS libraries for the rapid identification of metabolites in complex biological samples. FEBS Lett 579: 1332–1337 [DOI] [PubMed] [Google Scholar]
- Sheppard K, Yuan J, Hohn MJ, Jester B, Devine KM, Söll D. (2008) From one amino acid to another: tRNA-dependent amino acid biosynthesis. Nucleic Acids Res 36: 1813–1825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu XL, Frank T, Shu QY, Engel KH. (2008) Metabolite profiling of germinating rice seeds. J Agric Food Chem 56: 11612–11620 [DOI] [PubMed] [Google Scholar]
- Smith EN, Kruglyak L. (2008) Gene-environment interaction in yeast gene expression. PLoS Biol 6: e83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strehmel N, Hummel J, Erban A, Strassburg K, Kopka J. (2008) Retention index thresholds for compound matching in GC-MS metabolite profiling. J Chromatogr B Analyt Technol Biomed Life Sci 871: 182–190 [DOI] [PubMed] [Google Scholar]
- Tikunov YM, Laptenok S, Hall RD, Bovy A, de Vos RC. (2012) MSClust: a tool for unsupervised mass spectra extraction of chromatography-mass spectrometry ion-wise aligned data. Metabolomics 8: 714–718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuinstra MR, Ejeta G, Goldsbrough PB. (1997) Heterogeneous inbred family (HIF) analysis: a method for developing near-isogenic lines that differ at quantitative trait loci. Theor Appl Genet 95: 1005–1011 [Google Scholar]
- van Leeuwen H, Kliebenstein DJ, West MAL, Kim K, van Poecke R, Katagiri F, Michelmore RW, Doerge RW, St Clair DA. (2007) Natural variation among Arabidopsis thaliana accessions for transcriptome response to exogenous salicylic acid. Plant Cell 19: 2099–2110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weitbrecht K, Müller K, Leubner-Metzger G. (2011) First off the mark: early seed germination. J Exp Bot 62: 3289–3309 [DOI] [PubMed] [Google Scholar]
- Wentzell AM, Rowe HC, Hansen BG, Ticconi C, Halkier BA, Kliebenstein DJ. (2007) Linking metabolic QTLs with network and cis-eQTLs controlling biosynthetic pathways. PLoS Genet 3: 1687–1701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- West MAL, Kim K, Kliebenstein DJ, van Leeuwen H, Michelmore RW, Doerge RW, St Clair DA. (2007) Global eQTL mapping reveals the complex genetic architecture of transcript-level variation in Arabidopsis. Genetics 175: 1441–1450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeung KY, Dombek KM, Lo K, Mittler JE, Zhu J, Schadt EE, Bumgarner RE, Raftery AE. (2011) Construction of regulatory networks using expression time-series data of a genotyped population. Proc Natl Acad Sci USA 108: 19436–19441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Sova P, Xu Q, Dombek KM, Xu EY, Vu H, Tu Z, Brem RB, Bumgarner RE, Schadt EE. (2012) Stitching together multiple data dimensions reveals interacting metabolomic and transcriptomic networks that modulate cell regulation. PLoS Biol 10: e1001301. [DOI] [PMC free article] [PubMed] [Google Scholar]