KCS9 is involved in the elongation of C22 to C24 fatty acids, which are essential precursors for the biosynthesis of cuticular waxes, aliphatic suberins, and membrane lipids, including sphingolipids and phospholipids.
Abstract
Very-long-chain fatty acids (VLCFAs) with chain lengths from 20 to 34 carbons are involved in diverse biological functions such as membrane constituents, a surface barrier, and seed storage compounds. The first step in VLCFA biosynthesis is the condensation of two carbons to an acyl-coenzyme A, which is catalyzed by 3-ketoacyl-coenzyme A synthase (KCS). In this study, amino acid sequence homology and the messenger RNA expression patterns of 21 Arabidopsis (Arabidopsis thaliana) KCSs were compared. The in planta role of the KCS9 gene, showing higher expression in stem epidermal peels than in stems, was further investigated. The KCS9 gene was ubiquitously expressed in various organs and tissues, including roots, leaves, and stems, including epidermis, silique walls, sepals, the upper portion of the styles, and seed coats, but not in developing embryos. The fluorescent signals of the KCS9::enhanced yellow fluorescent protein construct were merged with those of BrFAD2::monomeric red fluorescent protein, which is an endoplasmic reticulum marker in tobacco (Nicotiana benthamiana) epidermal cells. The kcs9 knockout mutants exhibited a significant reduction in C24 VLCFAs but an accumulation of C20 and C22 VLCFAs in the analysis of membrane and surface lipids. The mutant phenotypes were rescued by the expression of KCS9 under the control of the cauliflower mosaic virus 35S promoter. Taken together, these data demonstrate that KCS9 is involved in the elongation of C22 to C24 fatty acids, which are essential precursors for the biosynthesis of cuticular waxes, aliphatic suberins, and membrane lipids, including sphingolipids and phospholipids. Finally, possible roles of unidentified KCSs are discussed by combining genetic study results and gene expression data from multiple Arabidopsis KCSs.
Very-long-chain fatty acids (VLCFAs) are fatty acids of 20 or more carbons in length and are essential precursors of functionally diverse lipids, cuticular waxes, aliphatic suberins, phospholipids, sphingolipids, and seed oils in the Brassicaceae. These lipids are involved in various functions, such as acting as protective barriers between plants and the environment, impermeable barriers to water and ions, energy-storage compounds in seeds, structural components of membranes, and lipid signaling, which is involved in the hypersensitive response (Pollard et al., 2008; Kunst and Samuels, 2009; Franke et al., 2012). VLCFAs are synthesized by the microsomal fatty acid elongase complex, which catalyzes the cyclic addition of a C2 moiety obtained from malonyl-CoA to C16 or C18 acyl-CoA. The fatty acid elongation process has been shown to proceed through a series of four reactions: condensation of the C2 carbon moiety to acyl-CoA by 3-ketoacyl coenzyme A synthase (KCS), reduction of KCS by 3-ketoacyl coenzyme A reductase (KCR), dehydration of 3-hydroxyacyl-CoA by 3-hydroxyacyl-CoA dehydratase (PAS2), and reduction of trans-2,3-enoyl-CoA by trans-2-enoyl-CoA reductase (ECR). Except for KCS isoforms with redundancy, disruption of KCR1, ECR/ECERIFERUM10 (CER10), or PAS2 exhibited severe morphological abnormalities and embryo lethality, suggesting that VLCFA homeostasis is essential for plant developmental processes (Zheng et al., 2005; Bach et al., 2008; Beaudoin et al., 2009).
Cuticular waxes that cover plant aerial surfaces are known to be involved in limiting nonstomatal water loss and gaseous exchanges (Boyer et al., 1997; Riederer and Schreiber, 2001), repelling lipophilic pathogenic spores and dust (Barthlott and Neinhuis, 1997), and protecting plants from UV light (Reicosky and Hanover, 1978). VLCFAs that are synthesized in the epidermal cells are either directly used or further modified into aldehydes, alkanes, secondary alcohols, ketones, primary alcohols, and wax esters for the synthesis of cuticular waxes. Reverse genetic analysis and Arabidopsis (Arabidopsis thaliana) epidermal peel microarray analysis (Suh et al., 2005) has enabled the research community to identify the functions of many genes involved in cuticular wax biosynthesis (Kunst and Samuels, 2009): CER1 (Bourdenx et al., 2011; Bernard et al., 2012), WAX2/CER3 (Chen et al., 2003; Rowland et al., 2007; Bernard et al., 2012), and MAH1(Greer et al., 2007; Wen and Jetter, 2009) have been shown to be involved in the decarbonylation pathway to form aldehydes, alkanes, secondary alcohols, and ketones, and acyl-coenzyme A reductase (FAR; Aarts et al., 1997; Rowland et al., 2006) and WSD1 (Li et al., 2008) have been shown to be involved in the decarboxylation pathway for the synthesis of primary alcohols and wax esters. The export of wax precursors to the extracellular space is mediated by a heterodimer of the ATP-binding cassette transporters in the plasma membrane (Pighin et al., 2004; Bird et al., 2007; McFarlane et al., 2010). In addition, glycosylphosphatidylinositol-anchored LTP (LTPG1) and LTPG2 contribute either directly or indirectly to the export of cuticular wax (DeBono et al., 2009; Lee et al., 2009; Kim et al., 2012).
VLCFAs that are synthesized in the endodermis of primary roots, seed coats, and the chalaza-micropyle region of seeds are used as precursors for the synthesis of aliphatic suberins. The suberin layer is known to function as a barrier against uncontrolled water, gas, and ion loss and provides protection from environmental stresses and pathogens (Pollard et al., 2008; Franke et al., 2012). For aliphatic suberin biosynthesis, the ω-carbon of the VLCFAs is oxidized by the fatty acyl ω-hydroxylase (Xiao et al., 2004; Li et al., 2007; Höfer et al., 2008; Molina et al., 2008, 2009; Compagnon et al., 2009; Li-Beisson et al., 2009), and the ω-hydroxy VLCFAs are further oxidized into α,ω-dicarboxylic acids by the HOTHEAD-like oxidoreductase (Kurdyukov et al., 2006). α,ω-Dicarboxylic acids are acylated to glycerol-3-P via acyl-CoA:glycerol-3-P acyltransferase (Beisson et al., 2007; Li et al., 2007; Li-Beisson et al., 2009; Yang et al., 2010) or to ferulic acid. In addition, C18, C20, and C22 fatty acids are also reduced by FAR enzymes to primary fatty alcohols, which are a common component in root suberin (Vioque and Kolattukudy, 1997). Finally, the aliphatic suberin precursors are likely to be extensively polymerized and cross linked with the polysaccharides or lignins in the cell wall.
In addition, VLCFAs are found in sphingolipids, including glycosyl inositolphosphoceramides, glycosylceramides, and ceramides and phospholipids, such as phosphatidylethanolamine (PE) and phosphatidyl-Ser (PS), which are present in the extraplastidial membrane (Pata et al., 2010; Yamaoka et al., 2011). For sphingolipid biosynthesis, VLCFA-CoAs and Ser are condensed to form 3-keto-sphinganine, which is subsequently reduced to produce sphinganine, a long chain base (LCB). LCBs are known to be further modified by 4-hydroxylation, 4-desaturation, and 8-desaturation (Lynch and Dunn, 2004; Chen et al., 2006, 2012; Pata et al., 2010). The additional VLCFAs are linked with 4-hydroxy LCBs via an amino group to form ceramides (Chen et al., 2008). The presence of VLCFA in sphingolipids may contribute to an increase of their hydrophobicity, membrane leaflet interdigitation, and the transition from a fluid to a gel phase, which is required for microdomain formation. In plants, PS is synthesized from CDP-diacylglycerol and Ser by PS synthase or through an exchange reaction between a phospholipid head group and Ser by a calcium-dependent base-exchange-type PS synthase (Vincent et al., 1999; Yamaoka et al., 2011). PE biosynthesis proceeds through decarboxylation via PS decarboxylase (Nerlich et al., 2007), the phosphoethanolamine transfer from CDP-ethanolamine to diacylglycerol (Kennedy pathway), and the exchange of the head group of PE with Ser via a base-exchange enzyme (Marshall and Kates, 1973). In particular, PS containing a relatively large amount of VLCFAs is enriched in endoplasmic reticulum (ER)-derived vesicles that may function in stabilizing small (70- to 80-nm-diameter) vesicles (Vincent et al., 2001).
During the fatty acid elongation process, the first committed step is the condensation of C2 units to acyl-CoA by KCS. Arabidopsis harbors a large family containing 21 KCS members (Joubès et al., 2008). Characterization of Arabidopsis KCS mutants with defects in VLCFA synthesis revealed in planta roles and substrate specificities (based on differences in carbon chain length and degree of unsaturation) of KCSs. For example, FAE1, a seed-specific condensing enzyme, was shown to catalyze C20 and C22 VLCFA biosynthesis for seed storage lipids (James et al., 1995). KCS6/CER6/CUT1 and KCS5/CER60 are involved in the elongation of fatty acyl-CoAs longer than C28 VLCFA for cuticular waxes in epidermis and pollen coat lipids (Millar et al., 1999; Fiebig et al., 2000; Hooker et al., 2002). KCS20 and KCS2/DAISY are functionally redundant in the two-carbon elongation to C22 VLCFA, which is required for cuticular wax and root suberin biosynthesis (Franke et al., 2009; Lee et al., 2009). When KCS1 and KCS9 were expressed in yeast (Saccharomyces cerevisiae), KCS1 showed broad substrate specificity for saturated and monounsaturated C16 to C24 acyl-CoAs and KCS9 utilized the C16 to C22 acyl-CoAs (Trenkamp et al., 2004; Blacklock and Jaworski, 2006; Paul et al., 2006). Recently, CER2 encoding putative BAHD acyltransferase was reported to be a fatty acid elongase that was involved in the elongation of C28 fatty acids for the synthesis of wax precursors (Haslam et al., 2012).
In this study, the expression patterns and subcellular localization of KCS9 were examined, and an Arabidopsis kcs9 mutant was isolated to investigate the roles of KCS9 in planta. Diverse classes of lipids, including cuticular waxes, aliphatic suberins, and sphingolipids, as well as fatty acids in various organs were analyzed from the wild type, the kcs9 mutant, and complementation lines. The combined results of this study revealed that KCS9 is involved in the elongation of C22 to C24 fatty acids, which are essential precursors for the biosynthesis of cuticular waxes, aliphatic suberins, and membrane lipids, including sphingolipids. To the best of our knowledge, this is the first study where a KCS9 isoform involved in sphingolipid biosynthesis was identified.
RESULTS
Phylogenetic Tree and Relative Expression of 21 KCS Genes in Arabidopsis
In previous studies, the Arabidopsis genome was shown to contain 21 KCS genes (Joubès et al., 2008). To analyze the relationship among KCS genes, pairwise multiple alignment was carried out using ClustalW Web software (http://www.genome.jp/tools/clustalw) with the BLOSUM matrix form, which is based on amino acid sequences, and then a rooted phylogenetic tree with branch length (Unweighted Pair Group Method with Arithmetic Mean) was constructed (Supplemental Fig. S1A). Based on the Arabidopsis microarray analysis (http://www.arabidopsis.org; Suh et al., 2005), the relative expression patterns of 21 KCS genes were analyzed in various organs or tissues, including seeds, roots, flowers, leaves, stems, and epidermis. KCS genes were divided into eight groups (I–VIII). Group I was first divided from the other KCSs. However, the expression patterns of the KCS genes in group I were very similar to those in groups IV and VI; they were expressed in aerial parts but rarely or not expressed in roots. KCS21 and KCS7 in group II and KCS15 in group III were very rarely or not expressed in Arabidopsis organs. KCS18 (FAE1) and KCS19 displayed seed-specific and seed-predominant expression, respectively. KCS10 (FDH), KCS4, KCS9, KCS11, KCS20, KCS2 (DAISY), and KCS1 genes were ubiquitously expressed in Arabidopsis, suggesting that they may play a role in VLCFA synthesis, which is required for growth and development. KCS14 and KCS13 in group VIII were predominantly expressed in reproductive organs rather than vegetative organs. KCS10 (FDH), KCS6 (CER6/CUT1), KCS5 (CER60), KCS20, KCS2 (DAISY), and KCS1, which are involved in cuticular wax biosynthesis (Millar et al., 1999; Todd et al., 1999; Fiebig et al., 2000; Hooker et al., 2002; Franke et al., 2009; Lee et al., 2009), showed higher expression in stem epidermal peels than in stems (Supplemental Fig. S1B).
Spatial and Temporal Expression of KCS9 in Arabidopsis
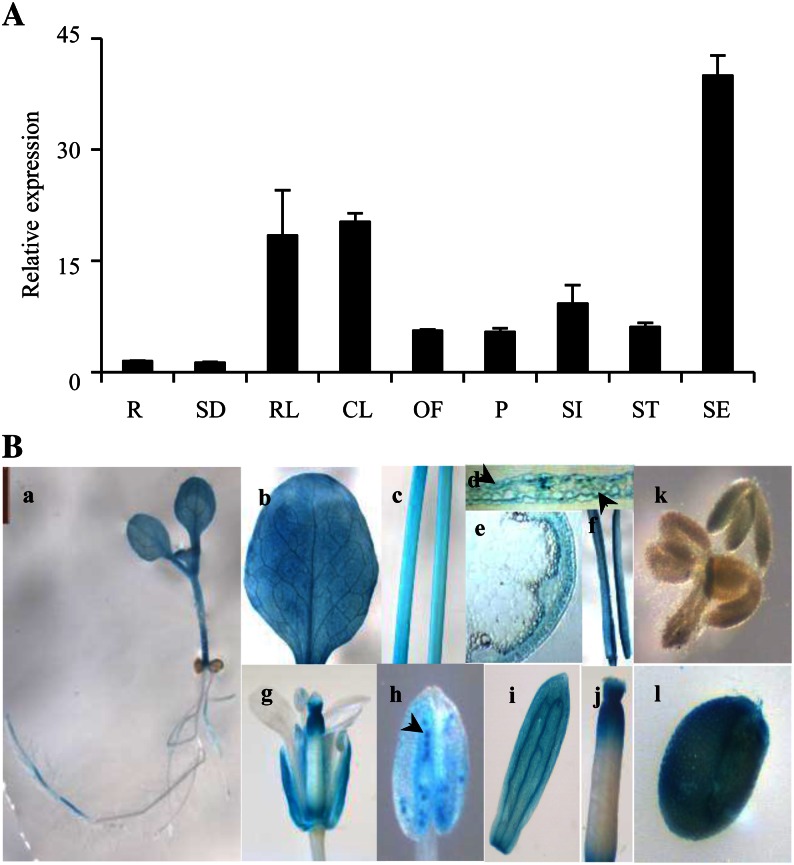
To confirm the relative expression level of the KCS9 gene in different organs or tissues, total RNAs were isolated from 2-week-old roots, 2-week-old seedlings, rosette leaves, cauline leaves, open flowers, pollen grains, and siliques from 6-week-old plants, and stems and stem epidermal peels from 5-week-old plants and subsequently subjected to quantitative reverse transcription (qRT)-PCR analysis. Eukaryotic translation initiation factor4-a (EIF4-a) was used as an internal factor to verify RNA quantity and quality (Gutierrez et al., 2008). As shown in Figure 1A, the level of the KCS9 transcripts was highest in the stem epidermal peels and was moderately detected in aerial parts of plants, including rosette and cauline leaves, open flowers, pollen grains, silique walls, and stems. Also, the expression of the KCS9 gene was relatively very low in roots and young seedlings. This result was very similar to those obtained from the microarray analysis (Fig. 1A; Supplemental Fig. S1B).
Figure 1.
A, qRT-PCR analysis of KCS9 in various Arabidopsis organs including stem epidermis. Total RNAs were isolated from 10-d-old seedlings and various organs of 6-week-old Arabidopsis and subjected to qRT-PCR analysis. EIF4-a was used as an internal standard to verify RNA quantity and quality (Gutierrez et al., 2008). R, Roots; SD, seedlings; RL, rosette leaves; CL, cauline leaves; OF, open flowers; P, pollen grains; SI, siliques harvested at 7 d after flowering; ST, stems; SE, stem epidermis. B, GUS expression under the control of the KCS9 promoter in transgenic Arabidopsis. Panels are as follows: a, young seedling; b, mature leaf; c, stem; d, cross section of leaf; e, cross section of stem; f, silique walls; g, flower; h, anther (arrowhead indicates pollen grain); i, petal; j, style; k, developing embryos; l, developing seed.
To investigate the spatial and temporal expression of the KCS9 gene, a GUS reporter gene under the control of the KCS9 promoter was introduced into Arabidopsis. T2 transgenic plants were histochemically stained with 5-bromo-4-chloro-3-indolyl β-d-glucuronide for GUS expression. After chlorophyll pigments were removed by sequential incubation in 10% to 100% ethanol, the stained organs were imaged using a microscope (Leica L2) or embedded in acrylic resin and cross sectioned. GUS expression was observed in young seedlings, leaves, stems, silique walls, anthers, sepals, upper portion of styles, and seed coats but not in developing embryos. As was observed in the qRT-PCR analysis, GUS expression was also found in leaf and stem epidermal cells (Fig. 1B).
KCS9::Enhanced Yellow Fluorescent Protein Is Localized to the ER
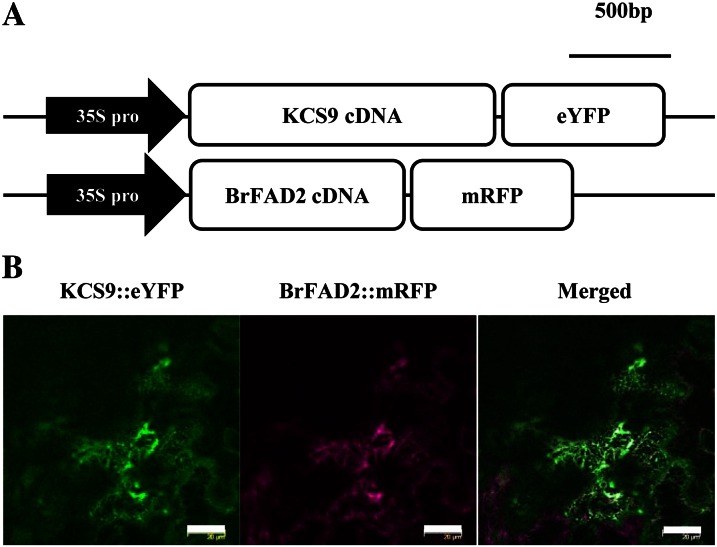
To examine the subcellular localization of KCS9, the coding sequence region of the KCS9 gene was amplified by PCR using the F1/R1 primer set (Supplemental Table S1). The amplified DNA fragments were inserted into the pPZP212 vector containing the cauliflower mosaic virus (CaMV) 35S promoter and enhanced yellow fluorescent protein (eYFP), and the resultant construct was named pKCS9::eYFP (Fig. 2A). In addition, the pBrFAD2::mRFP (for monomeric red fluorescent protein) construct, which harbored the microsomal oleic acid desaturase gene from Brassica rapa under the control of the CaMV 35S promoter, was used as an ER marker (Jung et al., 2011). Agrobacteria transformed with the pKCS9::eYFP or pBrFAD2::mRFP construct were coinfiltrated into tobacco (Nicotiana benthamiana) epidermal cells. After 48 h of incubation, fluorescent signals were observed using an AOBS/Tandem laser confocal scanning microscope (Leica). The green fluorescent signals from KCS9::eYFP were found to be exactly merged with the red fluorescent signals from pBrFAD2::mRFP (Fig. 2B), indicating that KCS9 was localized in the ER.
Figure 2.
Subcellular localization of Arabidopsis KCS9 in tobacco epidermis. A, Schematic diagrams of pKCS9::eYFP and pBrFAD2::mRFP constructs. 35S pro, Promoter of cauliflower mosaic virus 35S RNA. B, Fluorescent signals of KCS9::eYFP and BrFAD2::mRFP in tobacco epidermal cells. Genes encoding fluorescent proteins were translationally fused to KCS9 and BrFAD2 under the control of the CaMV 35S promoter. The constructed vectors were coinfiltrated into tobacco epidermis via A. tumefaciens-mediated transformation, and the fluorescent signals were observed using a laser confocal scanning microscope 48 h after infiltration. BrFAD2::mRFP was used as an ER marker (Jung et al., 2011). Bars = 10 µm.
Isolation of T-DNA Insertion kcs9 Mutant and Complementation Plants
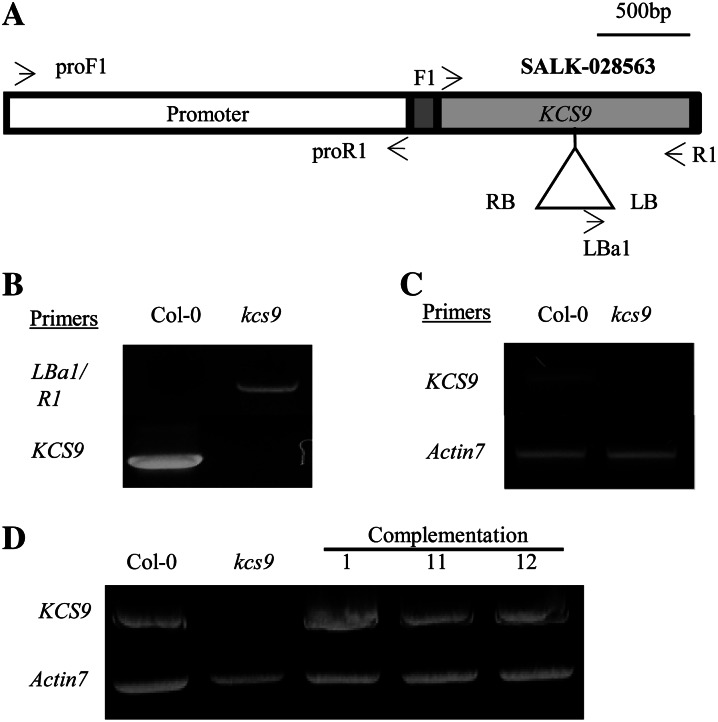
To investigate the function of KCS9 in Arabidopsis, a transfer DNA (T-DNA)-inserted kcs9 mutant (SALK 028563) was isolated by genomic DNA PCR using F1/R1 and Lba1/R1 primer sets (Supplemental Table S1), as shown in Figure 3, A and B. To examine the level of KCS9 transcripts, total RNAs were isolated from the wild type (Columbia-0 [Col-0]) and the kcs9 mutant and subjected to reverse transcription (RT)-PCR. Expression of the KCS9 transcripts was observed in the wild type but not in the kcs9 mutants (Fig. 3C). Under long-day growth conditions for Arabidopsis (16 h of light/8 h of dark), the kcs9 mutants were found to grow and develop normally. No significant alteration in root growth of kcs9 mutants compared with the wild type was observed under room and cold (10°C and 15°C) temperatures (data not shown).
Figure 3.
Isolation of T-DNA-inserted kcs9 mutant (A–C) and generation of complementation lines of the kcs9 mutant (D). A, Genomic organization of the T-DNA-tagged KCS9 gene. T-DNA-inserted kcs9 seeds were obtained from SALK (SALK 028563). The promoter region, 5′ and 3′ untranslated regions, and coding region of the KCS9 gene are shown in the white box, gray boxes, and black box, respectively. LB, Left border; RB, right border. B, Genomic DNA was isolated from the wild type (Col-0) and the kcs9 mutant, and T-DNA insertion was confirmed by genomic DNA PCR using the LBa1 and R1 primers shown in Supplemental Table S1. C, The levels of KCS9 transcripts in 10-d-old seedlings of the wild type (Col-0) and the kcs9 mutant were analyzed by RT-PCR using the F1 and R1 primers shown in Supplemental Table S1. Actin7 was used to determine the quantity and quality of the cDNAs. D, The binary vector harboring KCS9::eYFP under the control of the CaMV 35S promoter was transformed into Arabidopsis for complementation of the kcs9 mutant. The transgenic seedlings were selected on 1/2 MS medium with kanamycin, and leaves of 3-week-old plants were used for RNA isolation to analyze the expression of KCS9 by RT-PCR analysis.
For complementation of the kcs9 mutant, the KCS9::eYFP construct under the control of the CaMV 35S promoter shown in Figure 2A was transformed into the kcs9 mutant via Agrobacterium tumefaciens-mediated transformation. Kanamycin-resistant transgenic kcs9 lines were further selected by identifying the introduced genes using genomic DNA PCR analysis. The level of the KCS9 transcripts was measured by RT-PCR in the leaves of the wild type, kcs9, and complementation lines. The KCS9 transcripts were observed in complementation lines as well as the wild type (Fig. 3D).
KCS9 Is Involved in VLCFA Synthesis in Various Arabidopsis Organs
As shown in Figure 1 and Supplemental Figure S1B, the KCS9 gene was ubiquitously expressed. To investigate if KCS9 is involved in VLCFA synthesis in various Arabidopsis organs and tissues, total fatty acids were extracted from leaves, aerial parts, and roots of young seedlings and stems, flowers, and silique walls of the wild type, kcs9, and three complementation lines and analyzed using gas chromatography (GC).
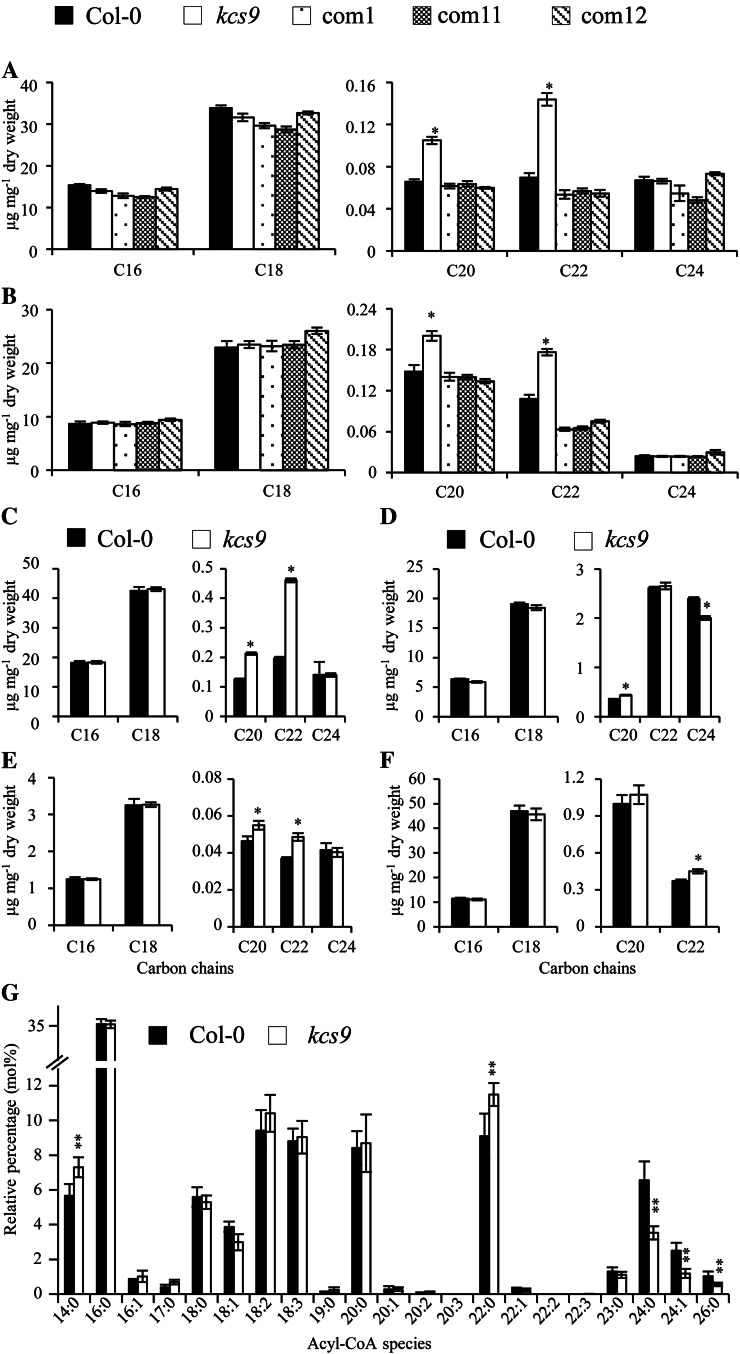
In the fatty acid analysis, we found that the composition of VLCFAs was altered in the kcs9 mutant relative to the wild type, although the VLCFA content was very low in all organs tested except the roots. In leaves and stems, the C20 and C22 fatty acid content was higher in kcs9 than in the wild type, and the chemical mutant phenotype was completely restored in the complementation lines (Fig. 4, A and B). An increase in the amount of C20 and C22 fatty acids was also observed in the aerial parts of young seedlings and flower organs of the kcs9 mutant (Fig. 4, C and E). Similarly, the C22 fatty acid content was higher in kcs9 silique walls than in the wild type (Fig. 4F). In addition, the C20 fatty acid content in the roots of kcs9 was higher, but the C24 fatty acid content was significantly lower (Fig. 4D). However, no significant differences were observed in total amounts of fatty acids from the various organs of the wild type, kcs9, and the three complementation lines, except for the roots (Supplemental Fig. S2A). This difference in VLCFA composition was closely related to the expression pattern of the KCS9 gene observed in the various Arabidopsis organs. This result indicates that KCS9 might function in the elongation of C22 VLCFAs to C24 VLCFAs.
Figure 4.
Fatty acid analysis (A–F) and fatty acyl-CoA profiling (G). A to F, Fatty acids were extracted from lyophilized leaves (A), stems (B), aerial parts (C) and roots (D) of young seedlings, flowers (E), and silique walls (F) of the wild type (Col-0), kcs9, and complementation lines (com1, com11, and com12) and analyzed using GC. G, Fatty acyl-CoA profiling was performed on lyophilized roots of wild-type and kcs9 mutant plants. The x axis represents the carbon chain length of the fatty acids. Values shown are means of four experiments ± sd. Asterisks denote statistical differences with respect to the wild type: *P < 0.05, **P < 0.01.
In addition, the profile of fatty acyl-CoAs was analyzed from roots of kcs9 and wild-type plants to identify the products of KCS9. The contents of C24:0- and C26:0-CoAs were decreased by approximately 50%, but the levels of C14:0- and C22:0-CoAs were significantly increased in the kcs9 mutant relative to the wild type (Fig. 4G), indicating that KCS9 is involved in the elongation of C22:0-CoA to C24:0-CoA. Interestingly, the C24:1-CoA levels were decreased, but the C22:1-CoA contents were not significantly altered in kcs9 compared with the wild type (Fig. 4G).
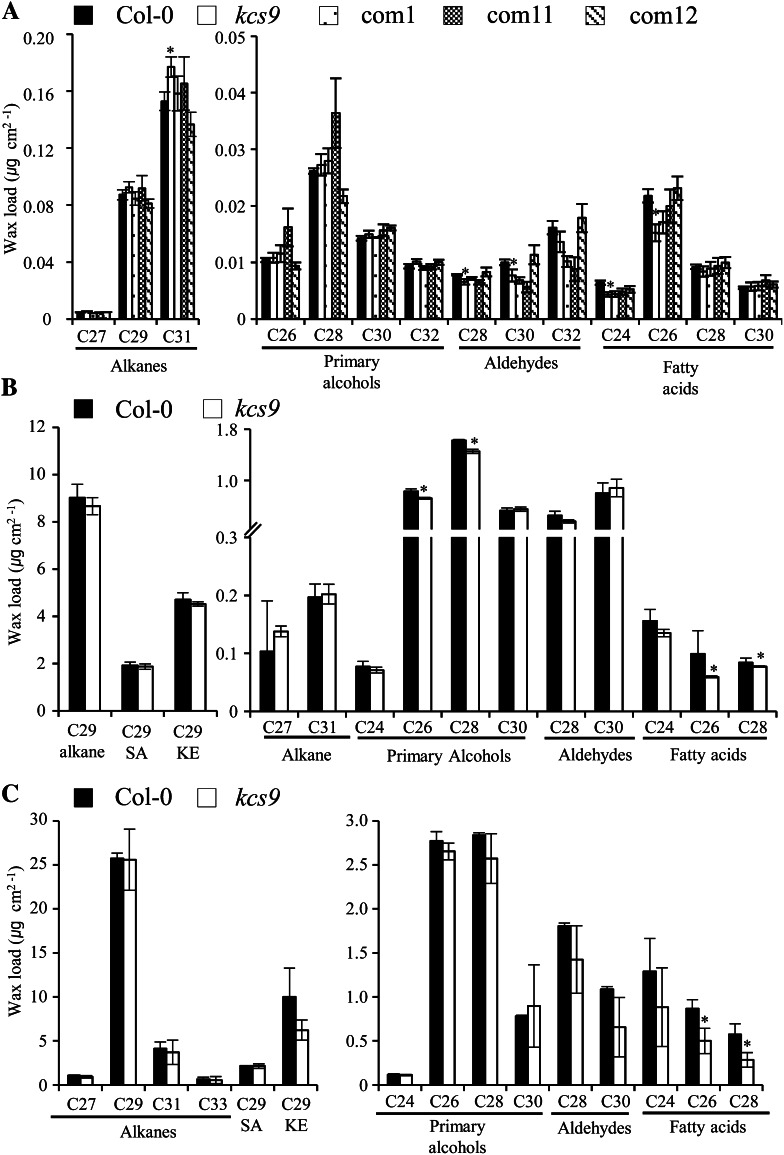
KCS9 Is Involved in Cuticular Wax Biosynthesis
Higher expression of the KCS9 gene in stem epidermal peels than in stems suggests that KCS9 may be involved in cuticular wax biosynthesis. To determine if this was the case, cuticular waxes were extracted from leaves, stems, and seed coats of the wild type, kcs9, and three complementation lines and analyzed using GC. In the leaves, the amounts of C24 and C26 VLCFAs were decreased by 40% and 25% in kcs9 compared with the wild type, respectively. In addition, the chemical mutant phenotype was rescued in the leaves of the complementation lines by the expression of KCS9 under the control of the CaMV 35S promoter (Fig. 5A). The contents of C26 and C28 primary alcohols, C28 aldehydes, and C26 VLCFAs were decreased in kcs9 stems relative to the wild type (Fig. 5B). These decreases in C26 and C28 VLCFAs were observed in kcs9 seed coats (Fig. 5C). However, total wax amounts were not significantly altered in kcs9 relative to the wild type (Supplemental Fig. S2B). This result revealed that KCS9 is involved in cuticular wax biosynthesis.
Figure 5.
Cuticular wax composition and amount in leaves (A), stems (B), and silique walls (C) of the wild type (Col-0), kcs9, and complementation lines (com1, com11, and com12). Cuticular waxes were extracted from 5-week-old plants with chloroform and analyzed using GC. The x axis represents the carbon chain length of VLCFAs and their derivatives. Values shown are means of four experiments ± sd. Asterisks indicate statistical differences with respect to the wild type: *P < 0.05. KE, Ketone; SA, secondary alcohol.
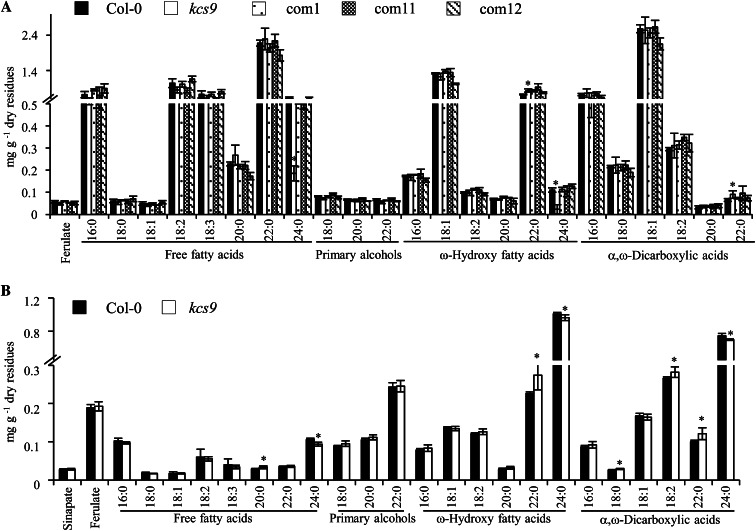
KCS9 Is Involved in Suberin Polyester Biosynthesis
As shown in Figure 1B, GUS expression in the roots and seed coats prompted us to examine the composition and amounts of suberin polyesters. Aliphatic suberin polyesters were extracted from roots and seed coats of the wild type, kcs9, and three complementation lines and hydrolyzed, and lipid-soluble extracts were analyzed using GC-mass spectrometry. The amounts of C24 fatty acids and C24 ω-hydroxy fatty acids were decreased, but the level of C22 fatty acids was increased in kcs9 roots relative to the wild type (Fig. 6A). Increased levels of C20 and C22 ω-hydroxy fatty acids and C18, C18:2, and C22 α,ω-dicarboxylic acids, but decreased levels of C24 VLCFAs and α,ω-dicarboxylic acids, were also observed in the kcs9 seed coat relative to the wild type (Fig. 6B). In addition, the chemical phenotype of the kcs9 mutant was rescued in complementation lines (Fig. 6A). No significant changes were observed in total amounts of aliphatic suberins from the roots of the wild type, kcs9, and three complementation lines (Supplemental Fig. S2C). This result also indicates that KCS9 is involved in the elongation of C24 fatty acids from C22 fatty acids, which are required for the biosynthesis of the suberin polyesters present in roots and seed coats.
Figure 6.
Aliphatic suberin composition and amount in roots (A) and seed coats (B) of the wild type (Col-0), kcs9, and complementation lines (com1, com11, and com12). Two-week-old roots and seed coats were lyophilized, delipidated, and hydrolyzed, and then lipid-soluble extracts were analyzed using GC and GC-mass spectrometry. Values shown are means of three experiments ± sd. Asterisks indicate statistical differences with respect to the wild type: *P < 0.05.
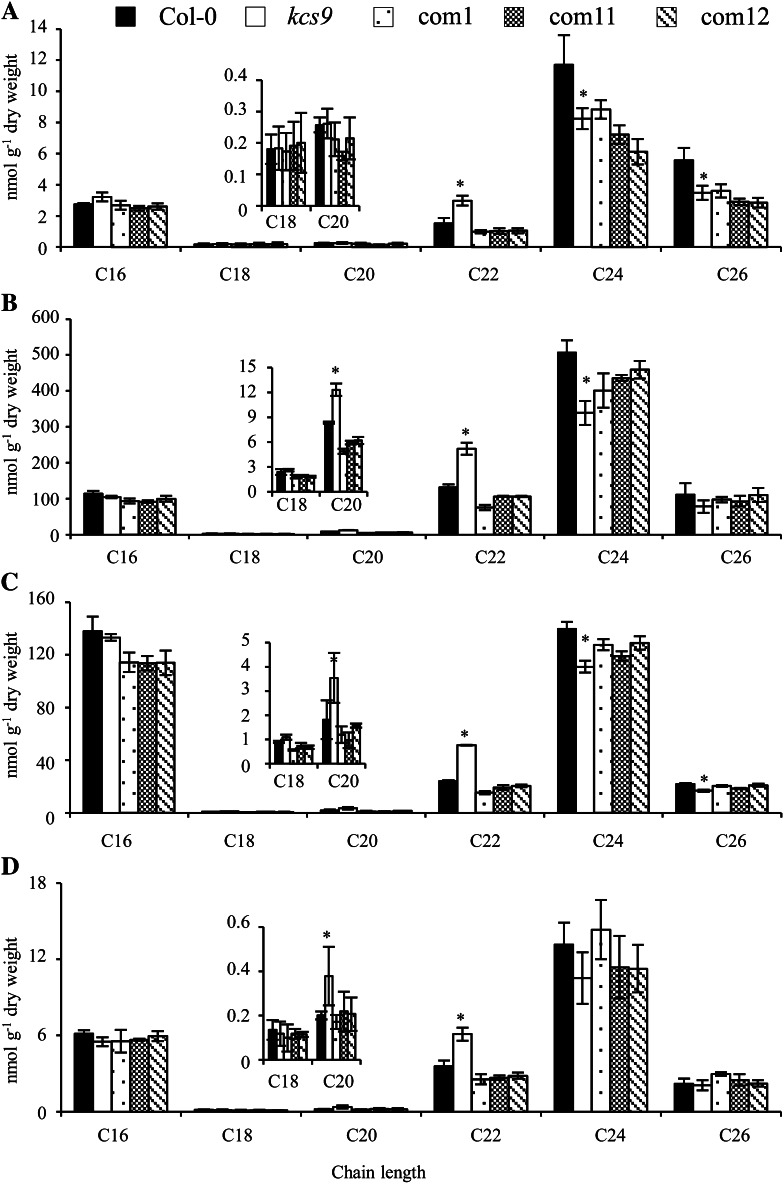
KCS9 Is Involved in Sphingolipid Biosynthesis
Because VLCFA moieties are also essential components in sphingolipids and phospholipids such as PS and PE, the role of KCS9 was investigated in sphingolipid and phospholipid metabolism. Sphingolipid analysis was performed on 2-week-old seedlings of the wild type, kcs9, and three complementation lines. The amounts of C20 and C22 VLCFAs were increased by 20 mol %, but the levels of C24 VLCFAs were decreased by 25% in all sphingolipid species, ceramide, glycosyl inositolphosphoceramide, glucosylceramide, and hydroxyceramide in kcs9 compared with the wild type (Fig. 7). The C26 VLCFA content was also decreased in ceramide and glucosylceramide of the kcs9 mutant relative to the wild type (Fig. 7, A and C). However, the total amount of sphingolipids was not altered in the kcs9 mutant relative to the wild type (Supplemental Fig. S2D). The sphingolipid profile of complementation lines was almost identical to that of the wild type (Fig. 7). Similar results were also observed in the sphingolipid analysis of leaves (Supplemental Fig. S3).
Figure 7.
Sphingolipid composition and amount in young seedling of the wild type, kcs9, and complementation lines (com1, com11, and com12). A, Ceramide. B, Glycosyl inositolphosphoceramide. C, Glucosylceramide. D, Hydroxyceramide. The amounts of all components with equal carbon chain lengths in each class of sphingolipids were combined. Values shown are means of three experiments ± sd. Asterisks indicate statistical differences with respect to the wild type: *P < 0.05.
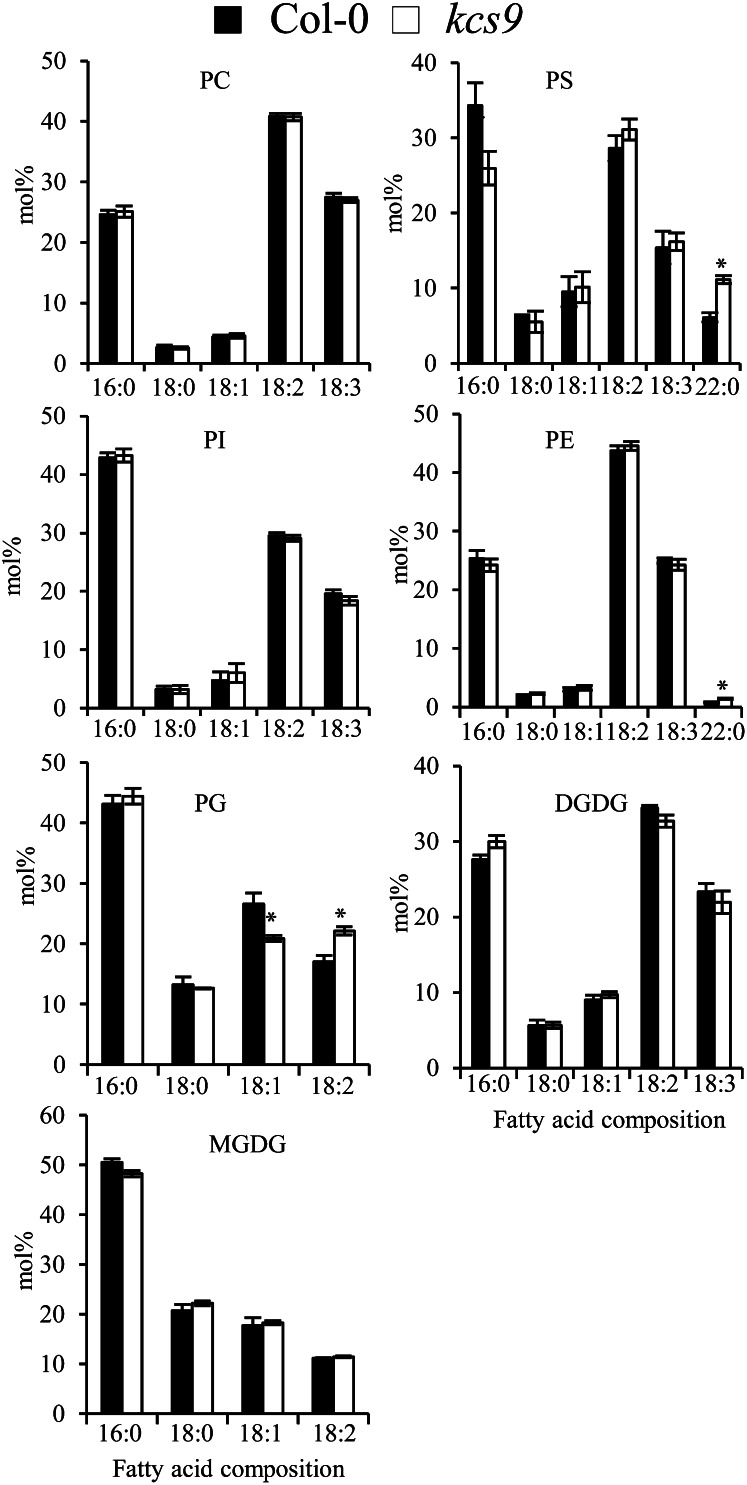
In addition, glycerolipid analysis was performed on 10-d-old roots of wild-type and kcs9 plants. As expected, the amounts of C22 fatty acids were increased in PS and PE, but no changes were observed in phosphatidylcholine, phosphatidylinositol, monogalactosyl diacylglycerol, and digalactosyl diacylglycerol in kcs9 compared with the wild type. Unexpectedly, a decrease in the content of oleic acids and an increase in the content of linoleic acids in phosphatidylglycerol were observed in the kcs9 mutant relative to the wild type (Fig. 8). These results confirmed the role of KCS9 in sphingolipid and phospholipid biosynthesis.
Figure 8.
Glycerolipid analysis in roots of the wild type and the kcs9 mutant. Glycerolipids were extracted from 2-week-old roots and separated by thin-layer chromatography. Fatty acid composition from individual lipids was analyzed by GC. Values shown are means of three experiments ± sd. Asterisks indicate statistical differences with respect to the wild type: *P < 0.05. DGDG, Digalactosyl diacylglycerol; MGDG, monogalactosyl diacylglycerol; PC, phosphatidylcholine; PG, phosphatidylglycerol; PI, phosphatidylinositol.
DISCUSSION
In plants, VLCFAs of up to 34 carbons in length are essential substrates for the biosynthesis of cuticular waxes, sphingolipids and membrane lipids, and suberin polyesters as well as storage triacylglycerols in Brassica spp. seeds. The committed step of VLCFA biosynthesis is processed by KCS. KCSs are known to display substrate specificity that depends on chain length and the presence or the number of double bonds in fatty acids. They exhibit differential expression patterns in specific organs or tissues and under various environmental stresses, which are closely correlated with their functions. In this study, we isolated the KCS9 gene, which is preferentially expressed in stem epidermal cells, based on the result obtained from stem epidermal peel microarray analysis (Suh et al., 2005). The characterization of an Arabidopsis kcs9 knockout mutant revealed that the KCS9 gene is involved in the biosynthesis of C22 to C24 VLCFAs, which are required for the synthesis of cuticular waxes, aliphatic suberins, and membrane lipids, including sphingolipids.
The expression patterns of KCSs in specific or preferential organs and tissues of plants are closely related with their roles in planta. Seed-specific FAE1 is involved in erucic acid synthesis in Brassica spp. seed oils, which is used for the production of lubricants, cosmetics, and pharmaceuticals (James et al., 1995). KCS1, FDH/KCS10, CUT1/KCS6/CER6, CER60/KCS5, KCS2, and KCS20, which are specifically or preferentially expressed in stem epidermal peels compared with stems, function in the elongation of VLCFAs, which are required for cuticular wax biosynthesis (Millar et al., 1999; Todd et al., 1999; Fiebig et al., 2000; Hooker et al., 2002; Suh et al., 2005; Lee et al., 2009). The expression of KCS2/DAISY and KCS20 in the endodermis of roots and the chalaza-micropyle region of seeds has also been implicated in the biosynthesis of aliphatic suberins (Franke et al., 2009; Lee et al., 2009). Similar findings were also observed in this study. (1) The preferential expression of KCS9 in stem and leaf epidermal cells was associated with cuticular wax biosynthesis. (2) The expression of KCS9 in roots and seed coats was related to aliphatic suberin biosynthesis. (3) The ubiquitous expression of KCS9 except embryos at low levels was related to phospholipid and sphingolipid biosynthesis. Therefore, the roles of unidentified KCSs might be suggested based on their expression patterns in plants (Supplemental Fig. S1B). For an example, KCS12 and KCS3, which showed higher expression in stem epidermis than in stem, might be involved in cuticular wax biosynthesis. In addition to tissue- or organ-specific expression patterns of diverse KCSs, the expression of some KCSs could be controlled by various environmental stresses. The level of KCS2/DAISY transcripts was highly increased following applications of osmotic, salt, and drought stresses (Franke et al., 2009; Lee et al., 2009), whereas the expression of KCS9 was not significantly altered under the same stress conditions (Supplemental Fig. S4).
Enzymes consisting of the fatty acid elongase complex have been reported to be localized in the ER membranes. ER localization was observed in the transgenic Arabidopsis leaf expressing the YFP-AtKCR1 (Beaudoin et al., 2009) or the GFP-ECR gene (Zheng et al., 2005). HCD/PAS2-GFP protein fusion was colocalized with the ECR/CER10 mRFP1 associated with the ER. Based on the infiltration of the fluorescent protein fusion constructs into the tobacco epidermis, KCS1, KCS3, KCS5, KCS6, KCS8, KCS10, and KCS12 were identified to be targeted into the ER membranes (Joubès et al., 2008). KCS9 was also found to be localized in the ER network of tobacco epidermal cells in this study. Based on an analysis of membrane topology (http://www.cbs.dtu.dk/services/TMHMM-2.0), it was determined that the KCSs possess two or three membrane-spanning domains. In addition, the protein-protein interaction between YFPN-PAS2 and CER10-YFPC was observed in the ER of Arabidopsis epidermal cells using the bimolecular fluorescence complementation assay (Bach et al., 2008). The protein-protein interactions between PAS1 and PAS2, KCR, or ECR suggest that PAS1 acts as a molecular scaffold for the fatty acid elongase complex in the ER (Roudier et al., 2010), suggesting that the plant fatty acid elongase complex might function in the form of a supramolecular complex. However, if KCSs also interact with PAS1, other members of the fatty acid elongase complex, and/or their own KCS members, have not been reported yet.
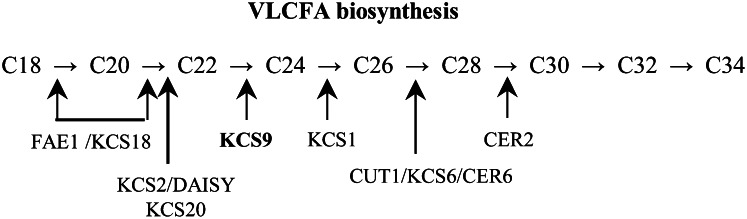
In yeast, a large family of elongase proteins (Elops) has been shown to catalyze the elongation of VLCFAs. Elo1p and Elo2p are responsible for the elongation of C14 to C16 and up to C24, respectively. Elo3p is required for the conversion of C24 to C26 (Oh et al., 1997). By complementation analysis of the yeast elo1Δ elo2Δ elo3Δ triple mutant, several Arabidopsis KCSs, including FAE1, KCS2/DAISY, and KCS20, have been identified (Paul et al., 2006). In vitro elongase activity using microsomes extracted from yeast expressing Arabidopsis KCSs demonstrated that KCSs have substrate specificity for the production of VLCFAs (Trenkamp et al., 2004; Blacklock and Jaworski, 2006; Paul et al., 2006). Additionally, forward and reverse genetic analyses of Arabidopsis have also revealed the substrate specificity of Arabidopsis KCSs, and the results reported to date are summarized in Figure 9. Arabidopsis FAE1/KCS18 is involved in the elongation of fatty acids up to C22 VLCFA in seeds (James et al., 1995). When compared with the wild type, both kcs2/daisy and kcs20 mutants exhibited significant reductions in C22 and C24 VLCFAs but accumulation of C20 VLCFAs, indicating that they function in the elongation of C20 to C22 VLCFAs (Franke et al., 2009; Lee et al., 2009). In this study, KCS9 was clearly shown to have fatty acid elongase activity for the synthesis of C24 VLCFAs from C22. Interestingly, the C24:1-CoA levels were decreased, but the C22:1-CoA contents were not significantly altered, in the profile of fatty acyl-CoAs in kcs9 relative to the wild type. The reduction of C24:1-CoA levels might be caused by the reduction of substrate (C24:0) levels of acyl-CoA desaturase (Smith et al., 2013). Disruption of KCS1 or CER6 resulted in a reduction of all wax monomers longer than C24, suggesting that KCS1 and CUT1/CER6/KCS6 are required for the elongation of C24 VLCFAs (Millar et al., 1999; Todd et al., 1999). Although the structure of CER2 is not related to KCSs, CER2 was reported to be involved in the elongation of C28 fatty acids, perhaps through acyltransferase activity. However, the functional mechanism of CER2 in VLCFA elongation should be further investigated (Haslam et al., 2012). Interestingly, the chain length of VLCFAs is determined by the distance between the active site and the Lys residue of KCS, which is based on the molecular caliper mechanism in yeast (Denic and Weissman, 2007). However it seems that this mechanism is not conserved in Arabidopsis KCSs, which are structurally unrelated to the ELO class of condensing enzymes, although the HXXHH or HXXXH motif that is essential for Elop activity is observed in Arabidopsis KCSs.
Figure 9.
Substrate specificity of KCSs and CER2, which are involved in the production of VLCFAs in Arabidopsis. Numbers correspond to the number of carbon chains of VLCFAs. The elongation steps that are catalyzed by KCSs and CER2 are indicated by arrows.
Several Arabidopsis and rice (Oryza sativa) KCSs that catalyze the condensation of two-carbon to acyl-CoA in VLCFA synthesis have been relatively well identified. Among them, FAE1, CER6, and KCS2/DAISY have been shown to be the major enzymes associated with VLCFA biosynthesis for Brassica spp. seed oil, cuticular wax, and aliphatic suberin synthesis, respectively (James et al., 1995; Millar et al., 1999; Todd et al., 1999; Yephremov et al., 1999; Fiebig et al., 2000; Hooker et al., 2002; Yu et al., 2008; Franke et al., 2009; Lee et al., 2009). KCS2/DAISY and KCS20 are functionally redundant in cuticular wax and root suberin biosynthesis (Lee et al., 2009). However, no KCS involved in VLCFA elongation for sphingolipid synthesis has been reported yet. The LCB1, SBH1 and SBH2, and TSC10A and TSC10B genes encoding the LCB1 subunit of Ser palmitoyltransferase, sphingolipid base hydroxylase1 and sphingolipid base hydroxylase2, and proteins similar to the yeast 3-KDS reductase, which are involved in the sphingolipid biosynthetic pathway, respectively, have been shown to be expressed in whole plants (Chen et al., 2006, 2008; Chao et al., 2011), suggesting that the Arabidopsis KCSs expressed ubiquitously in various organs and tissues may be involved in VLCFA elongation, which is used for sphingolipid biosynthesis. In this study, we found that ubiquitously expressed KCS9 is involved in the elongation of C22 to C24 VLCFAs, which is required for sphingolipid synthesis. This result is also supported by the evidence that the inositolphosphoceramides were synthesized in the yeast elo1Δ elo2Δ elo3Δ triple mutant expressing the KCS9 gene (Paul et al., 2006). Based on previous reports (Zheng et al., 2005; Bach et al., 2008; Beaudoin et al., 2009), a deficiency in sphingolipids results in severe morphological abnormalities and embryo lethality. However, such abnormal phenotypic features were not observed in most Arabidopsis kcs mutants, although fdh and kcs2 kcs20 mutants exhibit floral organ fusion and reduction of root growth, respectively (Yephremov et al., 1999; Lee et al., 2009). These results indicate that more than one KCS enzyme may be involved in VLCFA elongation, which is necessary for sphingolipid synthesis. Therefore, ubiquitously expressed KCS4, KCS11, KCS2, KCS20, and KCS1 as well as KCS9, shown in Supplemental Figure S1B, might be involved in the elongation during sphingolipid synthesis.
In conclusion, genetic and biochemical characterization of the kcs9 mutant provides information for the substrate specificity of the KCS9 enzyme in VLCFA elongation and reveals that VLCFAs synthesized by KCS9 are essential precursors required for the synthesis of cuticular waxes, aliphatic suberins, and membrane lipids, including sphingolipids. Further research is needed to give insight into fundamental questions. What is the proper combination of multiple KCSs for the elongation of a specific VLCFA? And how do plant KCSs determine VLCFA length?
MATERIALS AND METHODS
Plant Materials and Growth Conditions
The Arabidopsis (Arabidopsis thaliana) T-DNA insertion mutant was obtained from SALK (SALK 028563) as ecotype Col-0. Using the genomic DNA PCR method, T-DNA-inserted homozygous plants were isolated due to the loss of antibiotic resistance. The sterilized seeds were germinated on one-half-strength Murashige and Skoog (1/2 MS) medium containing 1% (w/v) Suc and 0.6% (w/v) agar. For the screening of transgenic seeds, kanamycin (25 μg mL−1) was supplemented in the 1/2 MS medium. Plants were grown on soil under a long-day condition (16 h/8 h of light/dark) at 22°C with 60% humidity. For the generation of complementation lines of kcs9 mutants, the KCS9::eYFP construct under the control of the CaMV 35S promoter (for details, see “Subcellular Localization” below) was introduced into the kcs9 mutants via Agrobacterium tumefaciens-mediated transformation using the vacuum infiltration method (Bechtold et al., 1993). In the subcellular localization assay, the third and fourth leaves of 3-week-old tobacco (Nicotiana benthamiana) were used to inoculate A. tumefaciens harboring the pKCS9::eYFP or pBrFAD2::mRFP construct (Jung et al., 2011).
Gene Expression Analysis
Total RNAs were extracted using the RNeasy Plant Mini Kit (Qiagen) and converted into complementary DNAs (cDNAs) by reverse transcriptase following the manufacturer’s protocols (Promega). qRT-PCR was performed using the Q-F1 and Q-R1 primer set shown in Supplemental Table S1. As an internal control, EIF4-a was amplified using EIF4-F1 and EIF4-R1 (Supplemental Table S1). For quantitative analysis of RNA transcript levels, the KAPA SYBR FAST qRT-PCR kit (KAPA Biosystem) and the C1000 thermal cycler (Bio-Rad) were used according to each manufacturer’s instructions.
Subcellular Localization
To certify the subcellular localization, the binary plasmid pPZP212 (Hajdukiewicz et al., 1994) was modified by inserting 35S promoter, eYFP, and Rbcs terminator. The coding region of the KCS9 gene was amplified from Arabidopsis leaf cDNAs using the F1/R1 primer set (Supplemental Table S1). The synthesized DNA fragment was inserted into modified pPZP212 vector under the control of the 35S promoter. The resultant binary plasmid was referred to as pKCS9::eYFP. The BrFAD2::mRFP construct was used as an ER marker (Jung et al., 2011). The binary constructs were transformed into the A. tumefaciens strain GV3101 using the freeze-thaw method (An, 1987). A. tumefaciens harboring each vector was coinfiltrated into the abaxial epidermal cells of tobacco leaves at an optical density of 600 nm = 0.8.
GUS Assay
To examine GUS expression under the control of the KCS9 promoter, the promoter region of KCS9 (approximately 2 kb) was amplified by genomic DNA PCR using proF1 and proR1 (Supplemental Table S1), and the amplified DNA fragments were inserted into the XbaI and SmaI sites in the pBI101 vector. Arabidopsis was transformed by A. tumefaciens harboring the generated vector (Bechtold et al., 1993). After screening on the 1/2 MS medium supplemented with 25 μg mL−1 kanamycin and 100 μg mL−1 carbenicillin, transgenic seedlings and various organs of transgenic plants were stained with the GUS staining solution (100 mm sodium phosphate, pH 7.0, 1 mm 5-bromo-4-chloro-3-indolyl-β-d-glucuronide, 0.5 mm potassium ferrocyanide, 0.5 mm potassium ferricyanide, 10 mm Na2EDTA, and 0.1% [v/v] Triton X-100) using 5-bromo-4-chloro-3-indolyl β-d-glucuronide as a substrate at 37°C for 12 h. The chlorophylls of stained tissues were removed using graded ethanol from 10% to 100%. The images were acquired using a Leica L2 microscope. To visualize the cross sections of stems and leaves, dehydrated GUS samples were embedded in acrylic resin (LR White resin; London Resin Company) and then sliced using a MT990 microtom (RMC) at thicknesses ranging from 10 to 20 μm. The sliced tissues were observed and photographed with a light microscope (Leica L2).
Fatty Acid Analysis
Fatty acids were extracted from the aerial parts and roots of 2-week-old seedlings and leaves, stems, flowers, and silique walls of 4- to 6-week-old plants and transmethylated in 1 mL of methanol containing 5% (v/v) sulfuric acid at 90°C for 1 h. Heptadecanoic acid (17:0) was used as an internal standard. One milliliter of aqueous 0.9% NaCl was added, and the fatty acid methyl esters were recovered by three sequential extractions with 2 mL of hexane. The fatty acid methyl esters were then analyzed using GC (Shimadzu) as described by Jung et al. (2011). The fatty acids were identified by comparing the retention times and mass spectra with standards.
Cuticular Wax Analysis
Cuticular waxes were extracted by immersing the leaves, stems, and silique walls in 5 mL of chloroform. n-Octacosane (200 μg g−1 fresh weight), docosanoic acid (200 μg g−1 fresh weight), and 1-tricosanol (200 μg g−1 fresh weight) were used as internal standards. After the solvent of the wax extracts was removed under nitrogen gas, bis-N,N-trimethylsilyl trifluoroacetamide (Sigma):pyridine (1:1, v/v) was added and incubated at 90% for 30 min. The reaction samples were evaporated again under nitrogen gas and dissolved in heptane:toluene (1:1, v/v). The composition and amount of cuticular waxes were analyzed using GC (Shimadzu) as described by Kim et al. (2012). Single compounds were quantified against internal standards by automatically integrating the peak areas.
Suberin Polyester Analysis
Suberin polyester analysis from roots of 2-week-old seedlings and seed coats of dried seeds was performed using the methods described by Beisson et al. (2007). Methyl heptadecanoate and ω-pentadecalactone (Sigma) were used as internal standards. The lyophilized roots and seed coats were boiled in 25 mL of isopropanol for 10 min and delipidated in CHCl3:CH3OH (2:1, v/v) and CHCl3:CH3OH (1:2, v/v). The dried solvent-extracted residues of the roots and seed coats were methanolyzed using NaOCH3 and then depolymerized by hydrogenolysis. The prepared samples were separated and quantified by GC-mass spectrometry (QP2010; Shimazu) using the method described by Lee et al. (2009).
Sphingolipidomic Analysis
Two-week-old Arabidopsis seedlings were used for sphingolipid analysis as described by Markham and Jaworski (2007). Three milliliters of extraction solvent (isopropanol:water:hexane, 55:25:20, v/v/v) and 10 µL of an internal standard solution were added to approximately 10 to 15 mg of lyophilized tissues and homogenized. GM1 (monosialotetrahexosylganglioside), glucosyl-C12-ceramide, C12-ceramide, C17-sphingosine, and C17-sphingosine-1-P were used as internal standards. Completely homogenized tissues were incubated at 60°C for 15 min and centrifuged at 500g for 10 min. The supernatant was transferred into a new glass tube, and remaining pellets were reextracted with 3 mL of extraction solvent. After heating and spinning, the second supernatant was added to the first supernatant. The extracted solvent was dried under nitrogen gas and dissolved in 2 mL of 33% methylamine in ethanol:water (7:3, v/v). The dissolved mixtures were sonicated and heated at 50°C for 1 h. Subsequently, the solvent was evaporated under nitrogen gas and redissolved in sample solvent (tetrahydrofuran:methanol:water, 2:1:2, v/v/v) containing 0.1% formic acid. Extracted sphingolipids were analyzed by HPLC-electrospray ionization-tandem mass spectrometry (ESI-MS/MS) as described by Markham and Jaworski (2007).
Glycerolipid Analysis
Roots of 2-week-old wild-type and kcs9 mutant plants were ground with liquid nitrogen and quickly immersed in 3 mL of preheated (75°C) isopropanol with 0.01% butylated hydroxytoluene (Sigma) for 15 min. Chloroform (1.5 mL) and 0.6 mL of water were added, vortexed, and agitated for 1 h. Lipid extracts were transferred into a new glass tube with Teflon-lined screw caps. Four milliliters of chloroform:methanol (2:1) with 0.01% butylated hydroxytoluene was added and shaken for 30 min. The extraction procedure was repeated three times. The extracts were washed with 1 mL of 1 m KCl and 2 mL of water. After centrifuging, the upper phase was discarded and the lower phase was used for further experiments. Individual lipids were separated by one-dimensional thin-layer chromatography on (NH4)2SO4-impregnated silica gel G (Miquel and Browse, 1992). The thin-layer chromatography plate was developed with acetone:toluene:water (91 mL:30 mL:7.5 mL) solvent. Glycerophospholipids were located by spraying the plate with 0.01% primuline in 80% acetone and then visualized under UV light. To determine the fatty acid composition of individual lipids, silica gel from each lipid spot was scraped, and fatty acid methyl esters were prepared and analyzed as described above.
Acyl-CoA Profiling
Acyl-CoAs were extracted from pooled, lyophilized dry residues of 2-week-old roots of kcs9 and wild-type plants according to the extraction protocol of Larson and Graham (2001) with modifications. Four technical replicates were prepared using 5 mg of lyophilized tissue per replicate. Freshly prepared extraction buffer (400 µL) containing 10 pmol of pentadecanoyl-CoA (C15:0-CoA) as an internal standard was added to the sample. The extract was washed three times with 400 µL of heptane saturated with isopropanol:water (1:1, v/v). Saturated (NH4)2SO4 (10 µL) was added to the extract and mixed by inversion. Methanol:chloroform (2:1, v/v) at 1.2 mL was added, incubated at room temperature for 20 min, and then centrifuged at 20,000g for 2 min. The supernatant was transferred to a 13- × 100-mm glass tube and evaporated at 42°C under nitrogen to dryness using a Reacti-Vap evaporator (Thermo). The fatty acyl-CoAs were resuspended in 400 µL of 30% acetonitrile in water for liquid chromatography-ESI-MS/MS analysis. Acyl-CoAs were separated by HPLC on a Phenomenex Luna C18 column (3 × 150 mm, 3.5-µm particle size, fitted with a guard cartridge and operated at a flow rate of 300 µL min−1) and quantified by ESI-MS/MS using the method of Han et al. (2010) with modifications. The liquid chromatography-mass spectrometry system consisted of a Prominence ultra-performance liquid chromatography device (Shimadzu) equipped with three pumps and a QTRAP 4000 mass spectrometer (Applied Biosystems). Injection volumes of 30 to 50 µL were used. Solvent A (acetonitrile:water, 10:90 [v/v] containing 15 mm NH4OH), solvent B (acetonitrile:water 90:10 [v/v] containing 15 mm NH4OH), and solvent C (acetonitrile:water 70:30 [v/v] containing 0.1% formic acid) were used the following experiment. The 16-min gradient was initiated at 10% B/90% A (0 min) and increased to 30% B/70% A (at 5 min), further increased to 50% B/50% A (at 10 min), held at 50% B/50% A (at 11 min), returned to 0% B/0% A (12 min), and held at 100% C (from 12 to 13 min) for column regeneration. Column reequilibration at 0% B/100% A was from 14 to 15 min. For fatty acyl-CoA detection by ESI-MS/MS, the QTRAP mass spectrometer was operated in positive mode with the following instrument settings: spray voltage, 5,000 V; source temperature, 350°C; gas 1, 70 pounds per square inch (psi); gas 2, 30 psi; curtain gas, 15 psi; collision gas pressure medium, entrance potential 10. Declustering potentials (ranging from 180 to 210 V) and collision energies (ranging from 50 to 54 V) were optimized using fatty acyl-CoA standards. Data collection was done using Analyst 1.5 software, and data analysis was done using Multiquant 2.0 software (ABSciex).
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession number At2g16280 (KCS9).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Phylogenetic tree and relative expression patterns of the 21 KCS genes in Arabidopsis.
Supplemental Figure S2. Total amounts of fatty acids, cuticular waxes, aliphatic suberins, and sphingolipids from the wild type, kcs9, and complementation lines.
Supplemental Figure S3. Sphingolipid analysis in leaves of the wild type, kcs9, and complementation lines.
Supplemental Figure S4. Expression of the KCS9 gene by application of osmotic and salt stresses and abscisic acid hormone.
Supplemental Table S1. Oligonucleotide sequences of primers used in this study.
Acknowledgments
We thank the Arabidopsis Biological Resource Center (http://www.arabidopsis.org) for providing T-DNA-tagged kcs9 mutants. We also thank Dr. Young-Woo Seo (Korea Basic Science Institute) for confocal microscopy and image analysis.
Glossary
- VLCFA
very-long-chain fatty acid
- PE
phosphatidylethanolamine
- PS
phosphatidyl-Ser
- LCB
long chain base
- ER
endoplasmic reticulum
- qRT
quantitative reverse transcription
- CaMV
cauliflower mosaic virus
- Col-0
Columbia-0
- RT
reverse transcription
- GC
gas chromatography
- 1/2 MS
one-half-strength Murashige and Skoog
- ESI-MS/MS
electrospray ionization tandem mass spectrometry
- cDNA
complementary DNA
- T-DNA
transfer DNA
- mRFP
monomeric red fluorescent protein
- psi
pounds per square inch
References
- Aarts MG, Hodge R, Kalantidis K, Florack D, Wilson ZA, Mulligan BJ, Stiekema WJ, Scott R, Pereira A. (1997) The Arabidopsis MALE STERILITY 2 protein shares similarity with reductases in elongation/condensation complexes. Plant J 12: 615–623 [DOI] [PubMed] [Google Scholar]
- An G. (1987) Binary Ti vectors for plant transformation and promoter analysis. Methods Enzymol 153: 292–305 [Google Scholar]
- Bach L, Michaelson LV, Haslam R, Bellec Y, Gissot L, Marion J, Da Costa M, Boutin JP, Miquel M, Tellier F, et al. (2008) The very-long-chain hydroxy fatty acyl-CoA dehydratase PASTICCINO2 is essential and limiting for plant development. Proc Natl Acad Sci USA 105: 14727–14731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barthlott W, Neinhuis C. (1997) Characterization and distribution of water-repellent, self-cleaning plant surfaces. Ann Bot (Lond) 79: 667–677 [Google Scholar]
- Beaudoin F, Wu X, Li F, Haslam RP, Markham JE, Zheng H, Napier JA, Kunst L. (2009) Functional characterization of the Arabidopsis β-ketoacyl-coenzyme A reductase candidates of the fatty acid elongase. Plant Physiol 150: 1174–1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bechtold N, Ellis J, Pelletier G. (1993) In planta Agrobacterium mediated gene transfer by infiltration of adult Arabidopsis thaliana plants. Comptes Rendus de l’Academie des Sciences Serie III 316: 1194–1199 [Google Scholar]
- Beisson F, Li Y, Bonaventure G, Pollard M, Ohlrogge JB. (2007) The acyltransferase GPAT5 is required for the synthesis of suberin in seed coat and root of Arabidopsis. Plant Cell 19: 351–368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernard A, Domergue F, Pascal S, Jetter R, Renne C, Faure JD, Haslam RP, Napier JA, Lessire R, Joubès J. (2012) Reconstitution of plant alkane biosynthesis in yeast demonstrates that Arabidopsis ECERIFERUM1 and ECERIFERUM3 are core components of a very-long-chain alkane synthesis complex. Plant Cell 24: 3106–3118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird D, Beisson F, Brigham A, Shin J, Greer S, Jetter R, Kunst L, Wu X, Yephremov A, Samuels L. (2007) Characterization of Arabidopsis ABCG11/WBC11, an ATP binding cassette (ABC) transporter that is required for cuticular lipid secretion. Plant J 52: 485–498 [DOI] [PubMed] [Google Scholar]
- Blacklock BJ, Jaworski JG. (2006) Substrate specificity of Arabidopsis 3-ketoacyl-CoA synthases. Biochem Biophys Res Commun 346: 583–590 [DOI] [PubMed] [Google Scholar]
- Bourdenx B, Bernard A, Domergue F, Pascal S, Léger A, Roby D, Pervent M, Vile D, Haslam RP, Napier JA, et al (2011) Overexpression of Arabidopsis ECERIFERUM1 promotes wax very-long-chain alkane biosynthesis and influences plant response to biotic and abiotic stresses. Plant Physiol 156: 29–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer JS, Wong SC, Farquhar GD. (1997) CO2 and water vapor exchange across leaf cuticle (epidermis) at various water potentials. Plant Physiol 114: 185–191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao DY, Gable K, Chen M, Baxter I, Dietrich CR, Cahoon EB, Guerinot ML, Lahner B, Lü S, Markham JE, et al. (2011) Sphingolipids in the root play an important role in regulating the leaf ionome in Arabidopsis thaliana. Plant Cell 23: 1061–1081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M, Han G, Dietrich CR, Dunn TM, Cahoon EB. (2006) The essential nature of sphingolipids in plants as revealed by the functional identification and characterization of the Arabidopsis LCB1 subunit of serine palmitoyltransferase. Plant Cell 18: 3576–3593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M, Markham JE, Cahoon EB. (2012) Sphingolipid Δ8 unsaturation is important for glucosylceramide biosynthesis and low-temperature performance in Arabidopsis. Plant J 69: 769–781 [DOI] [PubMed] [Google Scholar]
- Chen M, Markham JE, Dietrich CR, Jaworski JG, Cahoon EB. (2008) Sphingolipid long-chain base hydroxylation is important for growth and regulation of sphingolipid content and composition in Arabidopsis. Plant Cell 20: 1862–1878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Goodwin SM, Boroff VL, Liu X, Jenks MA. (2003) Cloning and characterization of the WAX2 gene of Arabidopsis involved in cuticle membrane and wax production. Plant Cell 15: 1170–1185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Compagnon V, Diehl P, Benveniste I, Meyer D, Schaller H, Schreiber L, Franke R, Pinot F. (2009) CYP86B1 is required for very long chain ω-hydroxyacid and α,ω-dicarboxylic acid synthesis in root and seed suberin polyester. Plant Physiol 150: 1831–1843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeBono A, Yeats TH, Rose JK, Bird D, Jetter R, Kunst L, Samuels L. (2009) Arabidopsis LTPG is a glycosylphosphatidylinositol-anchored lipid transfer protein required for export of lipids to the plant surface. Plant Cell 21: 1230–1238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denic V, Weissman JS. (2007) A molecular caliper mechanism for determining very long-chain fatty acid length. Cell 130: 663–677 [DOI] [PubMed] [Google Scholar]
- Fiebig A, Mayfield JA, Miley NL, Chau S, Fischer RL, Preuss D. (2000) Alterations in CER6, a gene identical to CUT1, differentially affect long-chain lipid content on the surface of pollen and stems. Plant Cell 12: 2001–2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franke R, Höfer R, Briesen I, Emsermann M, Efremova N, Yephremov A, Schreiber L. (2009) The DAISY gene from Arabidopsis encodes a fatty acid elongase condensing enzyme involved in the biosynthesis of aliphatic suberin in roots and the chalaza-micropyle region of seeds. Plant J 57: 80–95 [DOI] [PubMed] [Google Scholar]
- Franke RB, Dombrink I, Schreiber L. (2012) Suberin goes genomics: use of a short living plant to investigate a long lasting polymer. Front Plant Sci 3: 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greer S, Wen M, Bird D, Wu X, Samuels L, Kunst L, Jetter R. (2007) The cytochrome P450 enzyme CYP96A15 is the midchain alkane hydroxylase responsible for formation of secondary alcohols and ketones in stem cuticular wax of Arabidopsis. Plant Physiol 145: 653–667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez L, Mauriat M, Guénin S, Pelloux J, Lefebvre J-F, Louvet R, Rusterucci C, Moritz T, Guerineau F, Bellini C, et al (2008) The lack of a systematic validation of reference genes: a serious pitfall undervalued in reverse transcription-polymerase chain reaction (RT-PCR) analysis in plants. Plant Biotechnol J 6: 609–618 [DOI] [PubMed] [Google Scholar]
- Hajdukiewicz P, Svab Z, Maliga P. (1994) The small, versatile pPZP family of Agrobacterium binary vectors for plant transformation. Plant Mol Biol 25: 989–994 [DOI] [PubMed] [Google Scholar]
- Han J, Clement JM, Li J, King A, Ng S, Jaworski JG. (2010) The cytochrome P450 CYP86A22 is a fatty acyl-CoA ω-hydroxylase essential for estolide synthesis in the stigma of Petunia hybrida. J Biol Chem 285: 3986–3996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haslam TM, Fernández AM, Zhao L, Kunst L. (2012) Arabidopsis ECERIFERUM2 is a component of the fatty acid elongation machinery required for fatty acid extension to exceptional lengths. Plant Physiol 160: 1164–1174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Höfer R, Briesen I, Beck M, Pinot F, Schreiber L, Franke R. (2008) The Arabidopsis cytochrome P450 CYP86A1 encodes a fatty acid ω-hydroxylase involved in suberin monomer biosynthesis. J Exp Bot 59: 2347–2360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooker TS, Millar AA, Kunst L. (2002) Significance of the expression of the CER6 condensing enzyme for cuticular wax production in Arabidopsis. Plant Physiol 129: 1568–1580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- James DW, Jr, Lim E, Keller J, Plooy I, Ralston E, Dooner HK. (1995) Directed tagging of the Arabidopsis FATTY ACID ELONGATION1 (FAE1) gene with the maize transposon activator. Plant Cell 7: 309–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joubès J, Raffaele S, Bourdenx B, Garcia C, Laroche-Traineau J, Moreau P, Domergue F, Lessire R. (2008) The VLCFA elongase gene family in Arabidopsis thaliana: phylogenetic analysis, 3D modelling and expression profiling. Plant Mol Biol 67: 547–566 [DOI] [PubMed] [Google Scholar]
- Jung JH, Kim H, Go YS, Lee SB, Hur CG, Kim HU, Suh MC. (2011) Identification of functional BrFAD2-1 gene encoding microsomal delta-12 fatty acid desaturase from Brassica rapa and development of Brassica napus containing high oleic acid contents. Plant Cell Rep 30: 1881–1892 [DOI] [PubMed] [Google Scholar]
- Kim H, Lee SB, Kim HJ, Min MK, Hwang I, Suh MC. (2012) Characterization of glycosylphosphatidylinositol-anchored lipid transfer protein 2 (LTPG2) and overlapping function between LTPG/LTPG1 and LTPG2 in cuticular wax export or accumulation in Arabidopsis thaliana. Plant Cell Physiol 53: 1391–1403 [DOI] [PubMed] [Google Scholar]
- Kunst L, Samuels L. (2009) Plant cuticles shine: advances in wax biosynthesis and export. Curr Opin Plant Biol 12: 721–727 [DOI] [PubMed] [Google Scholar]
- Kurdyukov S, Faust A, Trenkamp S, Bär ST, Franke R, Efremova N, Tietjen K, Schreiber L, Saedler H, Yephremov A. (2006) Genetic and biochemical evidence for involvement of HOTHEAD in the biosynthesis of long-chain α-,ω-dicarboxylic fatty acids and formation of extracellular matrix. Planta 224: 315–329 [DOI] [PubMed] [Google Scholar]
- Larson TR, Graham IA. (2001) A novel technique for the sensitive quantification of acyl CoA esters from plant tissues. Plant J 25: 115–125 [DOI] [PubMed] [Google Scholar]
- Lee SB, Jung SJ, Go YS, Kim HU, Kim JK, Cho HJ, Park OK, Suh MC. (2009) Two Arabidopsis 3-ketoacyl CoA synthase genes, KCS20 and KCS2/DAISY, are functionally redundant in cuticular wax and root suberin biosynthesis, but differentially controlled by osmotic stress. Plant J 60: 462–475 [DOI] [PubMed] [Google Scholar]
- Li F, Wu X, Lam P, Bird D, Zheng H, Samuels L, Jetter R, Kunst L. (2008) Identification of the wax ester synthase/acyl-coenzyme A:diacylglycerol acyltransferase WSD1 required for stem wax ester biosynthesis in Arabidopsis. Plant Physiol 148: 97–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Beisson F, Koo AJK, Molina I, Pollard M, Ohlrogge JB. (2007) Identification of acyltransferases required for cutin biosynthesis and production of cutin with suberin-like monomers. Proc Natl Acad Sci USA 104: 18339–18344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li-Beisson Y, Pollard M, Sauveplane V, Pinot F, Ohlrogge JB, Beisson F. (2009) Nanoridges that characterize the surface morphology of flowers require the synthesis of cutin polyester. Proc Natl Acad Sci USA 106: 22008–22013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch DV, Dunn TM. (2004) An introduction to plant sphingolipids and a review of recent advances in understanding their metabolism and function. New Phytol 161: 677–702 [DOI] [PubMed] [Google Scholar]
- Markham JE, Jaworski JG. (2007) Rapid measurement of sphingolipids from Arabidopsis thaliana by reversed-phase high-performance liquid chromatography coupled to electrospray ionization tandem mass spectrometry. Rapid Commun Mass Spectrom 21: 1304–1314 [DOI] [PubMed] [Google Scholar]
- Marshall MO, Kates M. (1973) Biosynthesis of phosphatidyl ethanolamine and phosphatidyl choline in spinach leaves. FEBS Lett 31: 199–202 [DOI] [PubMed] [Google Scholar]
- McFarlane HE, Shin JJ, Bird DA, Samuels AL. (2010) Arabidopsis ABCG transporters, which are required for export of diverse cuticular lipids, dimerize in different combinations. Plant Cell 22: 3066–3075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar AA, Clemens S, Zachgo S, Giblin EM, Taylor DC, Kunst L. (1999) CUT1, an Arabidopsis gene required for cuticular wax biosynthesis and pollen fertility, encodes a very-long-chain fatty acid condensing enzyme. Plant Cell 11: 825–838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miquel M, Browse J. (1992) Arabidopsis mutants deficient in polyunsaturated fatty acid synthesis: biochemical and genetic characterization of a plant oleoyl-phosphatidylcholine desaturase. J Biol Chem 267: 1502–1509 [PubMed] [Google Scholar]
- Molina I, Li-Beisson Y, Beisson F, Ohlrogge JB, Pollard M. (2009) Identification of an Arabidopsis feruloyl-coenzyme A transferase required for suberin synthesis. Plant Physiol 151: 1317–1328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molina I, Ohlrogge JB, Pollard M. (2008) Deposition and localization of lipid polyester in developing seeds of Brassica napus and Arabidopsis thaliana. Plant J 53: 437–449 [DOI] [PubMed] [Google Scholar]
- Nerlich A, von Orlow M, Rontein D, Hanson AD, Dörmann P. (2007) Deficiency in phosphatidylserine decarboxylase activity in the psd1 psd2 psd3 triple mutant of Arabidopsis affects phosphatidylethanolamine accumulation in mitochondria. Plant Physiol 144: 904–914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh CS, Toke DA, Mandala S, Martin CE. (1997) ELO2 and ELO3, homologues of the Saccharomyces cerevisiae ELO1 gene, function in fatty acid elongation and are required for sphingolipid formation. J Biol Chem 272: 17376–17384 [DOI] [PubMed] [Google Scholar]
- Pata MO, Hannun YA, Ng CK. (2010) Plant sphingolipids: decoding the enigma of the sphinx. New Phytol 185: 611–630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul S, Gable K, Beaudoin F, Cahoon E, Jaworski J, Napier JA, Dunn TM. (2006) Members of the Arabidopsis FAE1-like 3-ketoacyl-CoA synthase gene family substitute for the Elop proteins of Saccharomyces cerevisiae. J Biol Chem 281: 9018–9029 [DOI] [PubMed] [Google Scholar]
- Pighin JA, Zheng H, Balakshin LJ, Goodman IP, Western TL, Jetter R, Kunst L, Samuels AL. (2004) Plant cuticular lipid export requires an ABC transporter. Science 306: 702–704 [DOI] [PubMed] [Google Scholar]
- Pollard M, Beisson F, Li Y, Ohlrogge JB. (2008) Building lipid barriers: biosynthesis of cutin and suberin. Trends Plant Sci 13: 236–246 [DOI] [PubMed] [Google Scholar]
- Reicosky DA, Hanover JW. (1978) Physiological effects of surface waxes. I. Light reflectance for glaucous and nonglaucous Picea pungens. Plant Physiol 62: 101–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riederer M, Schreiber L. (2001) Protecting against water loss: analysis of the barrier properties of plant cuticles. J Exp Bot 52: 2023–2032 [DOI] [PubMed] [Google Scholar]
- Roudier F, Gissot L, Beaudoin F, Haslam R, Michaelson L, Marion J, Molino D, Lima A, Bach L, Morin H, et al. (2010) Very-long-chain fatty acids are involved in polar auxin transport and developmental patterning in Arabidopsis. Plant Cell 22: 364–375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowland O, Lee R, Franke R, Schreiber L, Kunst L. (2007) The CER3 wax biosynthetic gene from Arabidopsis thaliana is allelic to WAX2/YRE/FLP1. FEBS Lett 581: 3538–3544 [DOI] [PubMed] [Google Scholar]
- Rowland O, Zheng H, Hepworth SR, Lam P, Jetter R, Kunst L. (2006) CER4 encodes an alcohol-forming fatty acyl-coenzyme A reductase involved in cuticular wax production in Arabidopsis. Plant Physiol 142: 866–877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith MA, Dauk M, Ramadan H, Yang H, Seamons LE, Haslam RP, Beaudoin F, Ramirez-Erosa I, Forseille L. (2013) Involvement of Arabidopsis ACYL-COENZYME A DESATURASE-LIKE2 (At2g31360) in the biosynthesis of the very-long-chain monounsaturated fatty acid components of membrane lipids. Plant Physiol 161: 81–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh MC, Samuels AL, Jetter R, Kunst L, Pollard M, Ohlrogge JB, Beisson F. (2005) Cuticular lipid composition, surface structure, and gene expression in Arabidopsis stem epidermis. Plant Physiol 139: 1649–1665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todd J, Post-Beittenmiller D, Jaworski JG. (1999) KCS1 encodes a fatty acid elongase 3-ketoacyl-CoA synthase affecting wax biosynthesis in Arabidopsis thaliana. Plant J 17: 119–130 [DOI] [PubMed] [Google Scholar]
- Trenkamp S, Martin W, Tietjen K. (2004) Specific and differential inhibition of very-long-chain fatty acid elongases from Arabidopsis thaliana by different herbicides. Proc Natl Acad Sci USA 101: 11903–11908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent P, Maneta-Peyret L, Cassagne C, Moreau P. (2001) Phosphatidylserine delivery to endoplasmic reticulum-derived vesicles of plant cells depends on two biosynthetic pathways. FEBS Lett 498: 32–36 [DOI] [PubMed] [Google Scholar]
- Vincent P, Maneta-Peyret L, Sturbois-Balcerzak B, Duvert M, Cassagne C, Moreau P. (1999) One of the origins of plasma membrane phosphatidylserine in plant cells is a local synthesis by a serine exchange activity. FEBS Lett 464: 80–84 [DOI] [PubMed] [Google Scholar]
- Vioque J, Kolattukudy PE. (1997) Resolution and purification of an aldehyde-generating and an alcohol-generating fatty acyl-CoA reductase from pea leaves (Pisum sativum L.). Arch Biochem Biophys 340: 64–72 [DOI] [PubMed] [Google Scholar]
- Wen M, Jetter R. (2009) Composition of secondary alcohols, ketones, alkanediols, and ketols in Arabidopsis thaliana cuticular waxes. J Exp Bot 60: 1811–1821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao F, Goodwin SM, Xiao Y, Sun Z, Baker D, Tang X, Jenks MA, Zhou JM. (2004) Arabidopsis CYP86A2 represses Pseudomonas syringae type III genes and is required for cuticle development. EMBO J 23: 2903–2913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaoka Y, Yu YB, Mizoi J, Fujiki Y, Saito K, Nishijima M, Lee Y, Nishida I. (2011) PHOSPHATIDYLSERINE SYNTHASE1 is required for microspore development in Arabidopsis thaliana. Plant J 67: 648–661 [DOI] [PubMed] [Google Scholar]
- Yang W, Pollard M, Li-Beisson Y, Beisson F, Feig M, Ohlrogge JB. (2010) A distinct type of glycerol-3-phosphate acyltransferase with sn-2 preference and phosphatase activity producing 2-monoacylglycerol. Proc Natl Acad Sci USA 107: 12040–12045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yephremov A, Wisman E, Huijser P, Huijser C, Wellesen K, Saedler H. (1999) Characterization of the FIDDLEHEAD gene of Arabidopsis reveals a link between adhesion response and cell differentiation in the epidermis. Plant Cell 11: 2187–2201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu D, Ranathunge K, Huang H, Pei Z, Franke R, Schreiber L, He C. (2008) Wax Crystal-Sparse Leaf1 encodes a beta-ketoacyl CoA synthase involved in biosynthesis of cuticular waxes on rice leaf. Planta 228: 675–685 [DOI] [PubMed] [Google Scholar]
- Zheng H, Rowland O, Kunst L. (2005) Disruptions of the Arabidopsis enoyl-CoA reductase gene reveal an essential role for very-long-chain fatty acid synthesis in cell expansion during plant morphogenesis. Plant Cell 17: 1467–1481 [DOI] [PMC free article] [PubMed] [Google Scholar]