Genetic and candidate gene approaches are combined to identify an O-methyltransferase with a major role in methoxypyrazine biosynthesis in grapevine.
Abstract
Methoxypyrazines (MPs) are strongly odorant volatile molecules with vegetable-like fragrances that are widespread in plants. Some grapevine (Vitis vinifera) varieties accumulate significant amounts of MPs, including 2-methoxy-3-isobutylpyrazine (IBMP), which is the major MP in grape berries. MPs are of particular importance in white Sauvignon Blanc wines. The typicality of these wines relies on a fine balance between the pea pod, capsicum character of MPs and the passion fruit/grapefruit character due to volatile thiols. Although MPs play a crucial role in Sauvignon varietal aromas, excessive concentrations of these powerful odorants alter wine quality and reduce consumer acceptance, particularly in red wines. The last step of IBMP biosynthesis has been proposed to involve the methoxylation of the nonvolatile precursor 2-hydroxy-3-isobutylpyrazine to give rise to the highly volatile IBMP. In this work, we have used a quantitative trait loci approach to investigate the genetic bases of IBMP biosynthesis. This has led to the identification of two previously uncharacterized S-adenosyl-methionine-dependent O-methyltransferase genes, termed VvOMT3 and VvOMT4. Functional characterization of these two O-methyltransferases showed that the VvOMT3 protein was highly specific and efficient for 2-hydroxy-3-isobutylpyrazine methylation. Based on its differential expression in high- and low-MP-producing grapevine varieties, we propose that VvOMT3 is a key gene for IBMP biosynthesis in grapevine.
The pleasure experienced while enjoying a glass of wine is the result of sophisticated sensory, neurophysiological, and psychological processes triggered by wine aroma. Wine flavor is the result of a complex mixture of volatile compounds in the headspace of the glass that induces feelings of pleasure at the brain level (Shepherd, 2006). During the last 40 years, over 800 volatile molecules have been formally identified in wines, in concentrations ranging from hundreds of milligrams per liter down to a few picograms per liter (Ebeler and Thorngate, 2009; Styger et al., 2011). Among all of them, a relatively limited number of compounds, called varietal (or primary) aromas, play a crucial role in wine flavor and typicality. These aromas, which are related to the grape variety, belong to a limited number of chemical families, including monoterpenes, C13 norisoprenoids, volatile sulfur compounds, and methoxypyrazines (MPs; Ebeler and Thorngate, 2009). Quite frequently, they exist mostly in the grape (Vitis vinifera) berry as nonvolatile, odorless, “bound” forms that can be released by chemical and enzymatic reactions occurring during the winemaking and wine aging processes, thus enhancing wine’s varietal expression (Styger et al., 2011). Two classical examples are the glycoside precursors of the monoterpenols (Strauss et al., 1986) and the cysteinylated or glutathionylated precursors of the volatile thiols (Tominaga et al., 1998; Peña-Gallego et al., 2012). Noticeable exceptions are the MPs, which are found in grape berries exclusively as free, volatile molecules.
MPs are strongly odorant volatile heterocycles, with vegetable-like fragrances, that are widely occurring in the plant kingdom (Maga, 1982). In grape, they can be detected in fruits, leaves, shoots, and roots (Dunlevy et al., 2010). They are found in different grape varieties and are particularly abundant in the so-called Bordeaux cultivars (i.e. cv Cabernet Franc, Cabernet Sauvignon [CS], Sauvignon Blanc, Merlot, and Carménère [Car]; Bayonove et al., 1975; Lacey et al., 1991; Roujou de Boubée et al., 2002; Belancic and Agosin, 2007), whereas they are rarely detected in other cultivars, such as cv Pinot Noir (PN), Chardonnay, or Petit Verdot (PV). This finding indicates a strong genotype dependency of MP biosynthesis (Koch et al., 2010). MPs are accumulated in berries until bunch closure or véraison, and then their level declines after véraison (Hashizume and Samuta, 1999; Ryona et al., 2008). MP concentration in wine is highly correlated with the grape berry content at harvest (Roujou de Boubée et al., 2002). Three MPs are found in grape berries: 2-methoxy-3-isobutylpyrazine (IBMP), which is the most abundant, and two others, 2-methoxy-3-isopropylpyrazine (IPMP) and 2-methoxy-3-sec-butylpyrazine (SBMP; Ebeler and Thorngate, 2009). Both IBMP and IPMP display very low sensory detection thresholds in the wine matrix, ranging from 1 to 16 ng L–1.
MPs are of particular importance in white Sauvignon Blanc wines. The typicality of these wines relies on a fine balance between the pea pod, capsicum character of MPs and the passion fruit/grapefruit character due to volatile thiols (Dubourdieu et al., 2006; Lund et al., 2009). Although MPs play a crucial role in Sauvignon varietal aromas, excessive concentrations of these extremely powerful odorants will reduce consumer acceptance (Parr et al., 2007). In red wine, MPs are considered as off-flavor, and red wines can be depreciated by concentrations above 10 ng L–1 (Allen et al., 1991; Roujou de Boubée et al., 2000; Belancic and Agosin, 2007). Given the importance of MPs, either as typical varietal aromas or as detrimental off-flavors, deciphering the genetic and molecular determinism of their accumulation is of high interest for viticulture.
In spite of this, until recently little was known about the MP biosynthesis pathway or the MP biosynthetic genes, either in grapevine or other plant species. Theoretical biosynthesis pathways have been proposed since the mid-1970s. They all start by the addition of an α-dicarbonyl on a branched amino acid (Leu for IBMP, Val for IPMP) to form a 2-hydroxy-3-alkylpyrazine, which is subsequently transformed into the corresponding MP, by a methoxylation reaction (Murray and Whitfield 1975; Gallois et al., 1988). While the initial addition step remains to be demonstrated in plants, an S-adenosyl-l-Met (SAM)-dependent O-methyltransferase (OMT), capable of converting 2-hydroxy-3-isobutylpyrazine (IBHP) into IBMP, has been detected in CS shoots, partially purified and sequenced (Hashizume et al., 2001a, 2001b; Fig. 1). Recently, Dunlevy et al. (2010) characterized two OMTs, VvOMT1 and VvOMT2, capable of methylating IBHP in vitro, albeit with high apparent Km values. To investigate the genetic bases of MP biosynthesis in grape berries, we performed a quantitative trait loci (QTL) analysis, which has led to the identification of two previously uncharacterized OMTs termed VvOMT3 and VvOMT4. Functional characterization of these two OMTs showed that VvOMT3 was highly specific and efficient for IBHP methylation. Based on its differential expression in high-MP and low-MP grapevine varieties, we propose that VvOMT3 and, to a lesser extent, VvOMT4 are key genes for MP biosynthesis in grapevine berries.
Figure 1.
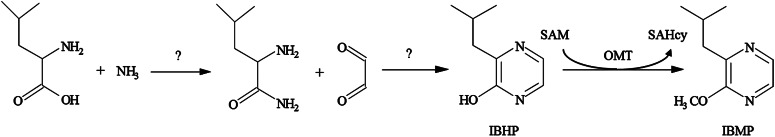
Putative biosynthesis pathway for IBMP adapted from Hashizume et al. (2001a). SAHcy, S-Adenosyl-l-homo-Cys.
RESULTS
QTL Analysis of MP Biosynthesis in Grapevine
In grapevine, MPs have been shown to accumulate both in leaves and berries (Roujou de Boubée, 2000; Dunlevy et al., 2010). To investigate the genetic bases of IBMP biosynthesis, a CS × Vitis riparia ‘Gloire de Montpellier’ (RGM) F1 progeny comprised of 130 genotypes was analyzed for IBMP content using solid-phase extraction (SPE)-gas chromatography-mass spectrometry (GC-MS). CS and RGM are known to produce contrasted MP levels; CS is a high producer unlike RGM, which does not accumulate significant levels of MPs.
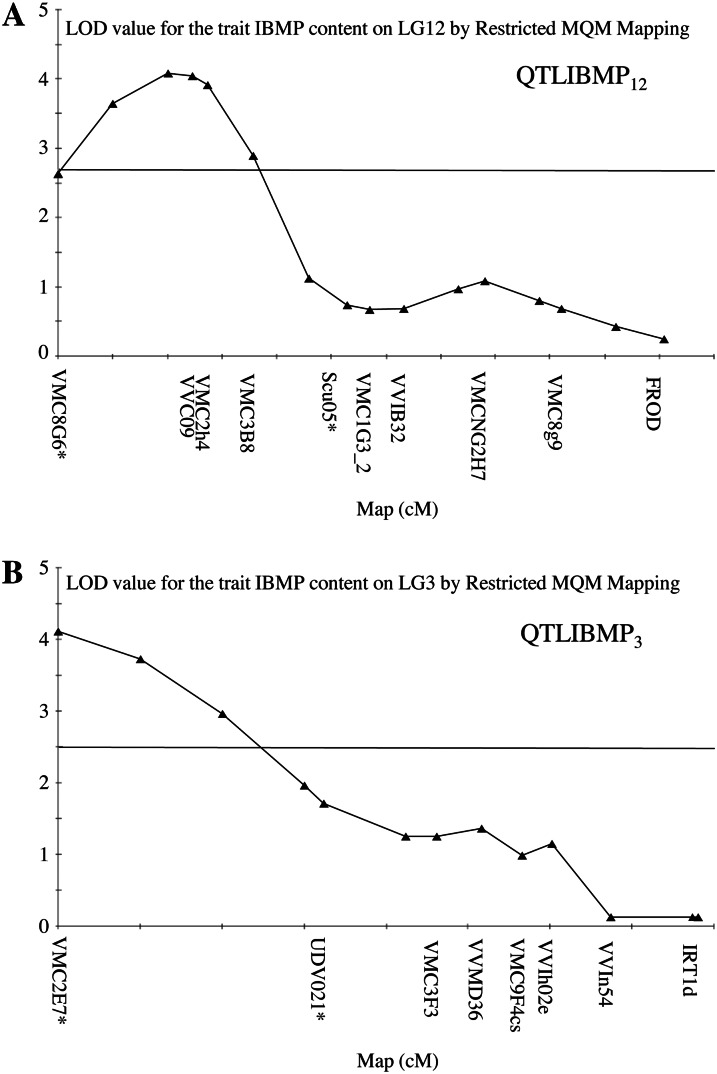
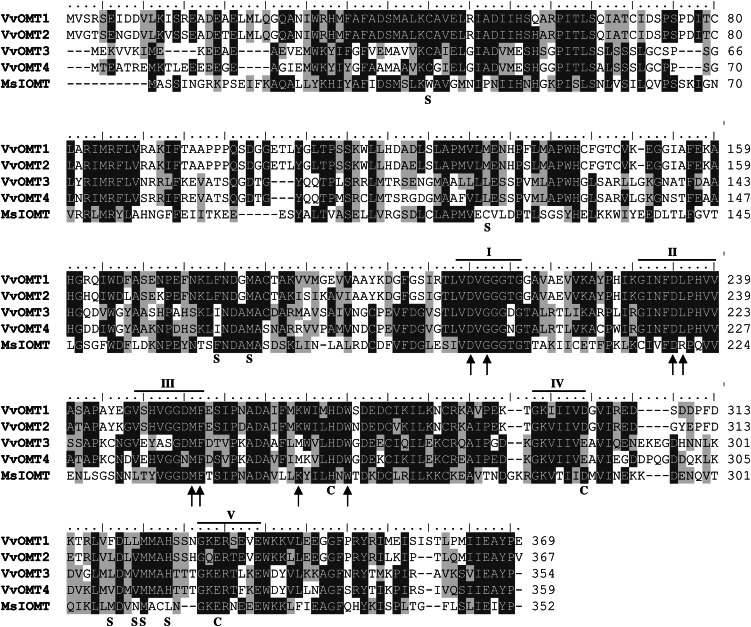
As a first approach, IBMP was quantified in basal leaves of the F1 CS × RGM progeny. The distribution of IBMP contents segregating among the F1 population is presented in Supplemental Figure S1. IBMP contents measured in CS and RGM leaves were 50 and 1.5 ng kg–1, respectively. In the F1 CS × RGM population, IBMP contents were highly variable, ranging between 0 and 1,946 ng kg–1 of fresh leaf weight, with a mean value of 50.4 ng kg–1. IBMP content did not follow a normal distribution, and 29.2% of the genotypes produced no IBMP. Log-transformed values were used to detect QTL related to IBMP content (Supplemental Fig. S1B). For QTL analysis, an F1 CS × RGM consensus map was used. This map consisted of a total of 206 genetic markers ordered into 19 linkage groups (LGs) depicting the 19 Vitis spp. chromosomes, with 186 mapped markers and an average of 9.8 markers per LG (Marguerit et al., 2009). Using a restricted multiple QTL mapping (MQM) analysis, five QTLs, significant at P = 0.05 at the LG level, were identified. They explained 41% of the total variance for IBMP content (Table I). Each QTL explained separately around 10% of IBMP content variance. Three of them were significant at the whole-genome level (P = 0.05; Table I). The markers flanking these QTLs were used to identify the corresponding genomic regions. Analysis of these regions based on the grapevine reference genome sequence (Jaillon et al., 2007) showed that several hundred genes were present in each interval (Table I). Because the biosynthetic pathway leading to IBMP is still hypothetical except for the last O-methylation step, we decided to focus on candidate OMT genes. Such candidate OMTs were identified within the flanking marker interval of two of the three genome-wide significant QTLs (Fig. 2; Table I). The first QTL, explaining 11.4% of the phenotypic variance for IBMP content, was located on LG12 between the markers VMC8G6 and scu05 (Fig. 2A; Table I). This QTL, QTLIBMP12, was supported by a maximum log of the odds (LOD) of 4.49 in restricted MQM analysis. The second QTL, QTLIBMP3, was identified on LG3 between markers VMC2E7 and UDV021, explaining 11% of the variance with a maximum LOD of 4.66 (Fig. 2B; Table I). Among the OMT genes present within the flanking marker interval of QTLIBMP12, we identified the VvOMT1 and VvOMT2 genes, which have been previously proposed to be involved in IBMP biosynthesis (Dunlevy et al., 2010). The physical interval surrounding the maximum LOD of QTLIBMP3 encompassed 211 predicted unigenes. Among these unigenes, a cluster of two genes encoding putative OMTs within 7 kb was identified, referred to hereafter as VvOMT3 (VIT_03s0038g03090) and VvOMT4 (VIT_03s0038g03080). These two genes had similar structures, with two exons and one intron, and their corresponding transcripts were 1,062 and 1,080 bp, respectively. VvOMT3 and VvOMT4 proteins shared 74% identity/86% similarity (Supplemental Fig. S2; Fig. 3) and contained the five characteristic domains of plant OMTs. An additional gene, VIT_03s0038g03070 (Fig. 3), very similar to VvOMT4 was located close to this gene. However, this putative pseudogene was predicted to encode a truncated OMT protein, which was very unlikely to be functional. Therefore, these four genes, VvOMT1, VvOMT2, VvOMT3, and VvOMT4, were chosen for further characterization to investigate their potential involvement in the final O-methylation step of MP biosynthesis (Murray and Whitfield, 1975).
Table I. QTLs for IBMP content in leaves of the F1 population CS × RGM1995-1.
| QTL | LG/Chr | Peak LOD | Confidence Interval | Markers Flanking QTL | LOD Threshold P = 0.05 on the LG | P-Value Threshold at the LG Level | P-Value Threshold at the Whole-Genome Level | % Expl | Distance between Flanking Markers on Chr Sequence | Predicted Genes between Flanking Markers on Chr Sequence |
|---|---|---|---|---|---|---|---|---|---|---|
| cM | Mbp | |||||||||
| QTLIBMP12 | 12 | 4.49 | 0–21 | VMC8G6-scu05 | 2.7 | 0.001 | 0.03 | 11.4 | 6.73 | 606 |
| QTLIBMP3 | 3 | 4.66 | 0–15 | VMC2E7-UDV021 | 2.5 | 0.001 | 0.02 | 11 | 1.93 | 211 |
| QTLIBMP5 | 5 | 4.33 | 43–60 | VMC9B5-VMC4C6 | 2.7 | 0.001 | 0.04 | 10.8 | 5.26 | 383 |
| QTLIBMP9 | 9 | 3.37 | 53–74 | VMC4H6-VMC2E11 | 3.3 | 0.05 | 0.24 | 8.8 | 13.02 | 444 |
| QTLIBMP18 | 18 | 3.32 | 76–91 | VMC6F11-VMC7F2 | 2.9 | 0.02 | 0.24 | 9.4 | 3.70 | 206 |
Figure 2.
LOD score plots for IBMP QTLs on LG3 and LG12. A, QTLIBMP12, QTL located on LG12. B, QTLIBMP3, QTL identified on LG3. QTLs were detected on the F1 CS × RGM mapping population on the consensus map. Asterisks indicate flanking markers used to identify the corresponding genomic regions on chromosome 3 and 12 sequences. The horizontal axis is graduated every 5 centimorgans (cM). The horizontal line indicates the minimum LOD score retained for QTLs detection at the LG level (P = 0.05).
Figure 3.
Alignment of the predicted amino acid sequences of VvOMT1, VvOMT2, VvOMT3, and VvOMT4. I to V, amino acid domains conserved among plant OMTs. ↑, SAM-binding residues; C, catalytic residues; S, substrate-binding residues of MsIOMT (for M. sativa Isoflavone OMT) as determined by Zubieta et al. (2001). The black and gray shading represents identical and similar amino acids, respectively.
Analysis of Candidate VvOMT Gene Expression
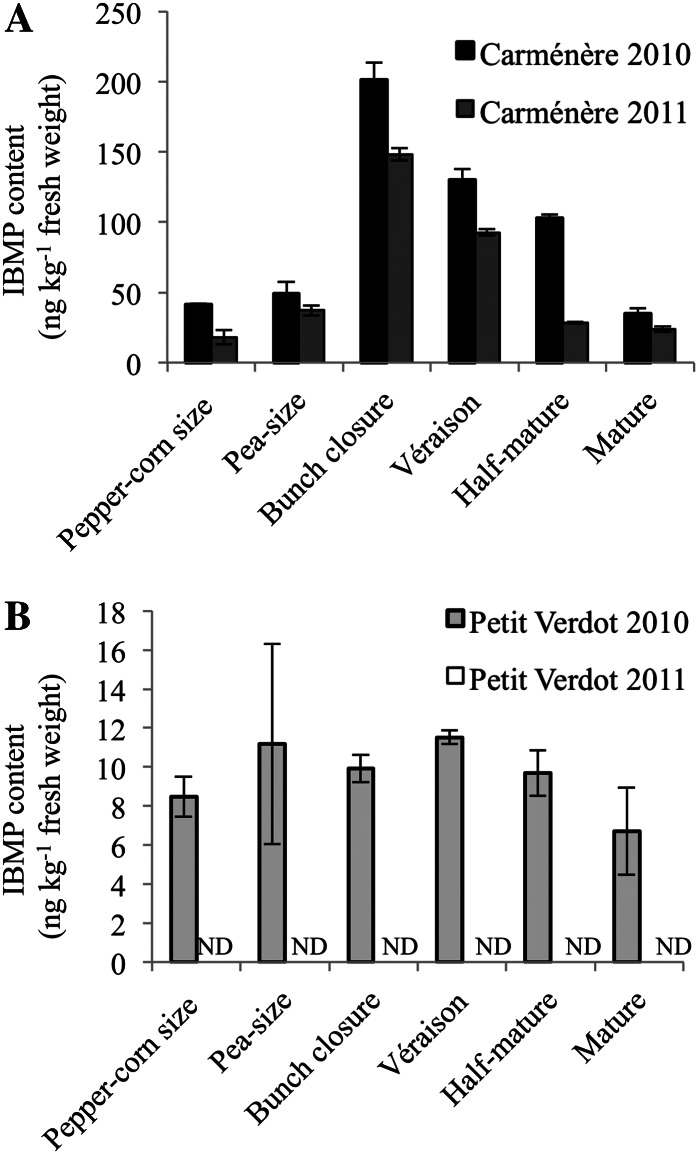
The QTL analysis was performed on the F1 CS × RGM model population, which is well suited to investigate the genetic bases of IBMP biosynthesis, but does not have any enological application. To confirm the potential role of VvOMT genes in IBMP biosynthesis in wine-relevant varieties, Car and PV were selected for the next experiments. Car and PV are used worldwide in various winemaking areas, including Bordeaux, France, and are known to produce highly contrasted IBMP amounts. Using solid-phase microextraction (SPME)-GC-MS, IBMP was quantified in Car and PV berries harvested at six different time points throughout berry development during the 2010 and 2011 growing seasons (Supplemental Fig. S3; Fig. 4). As expected, IBMP levels were always much higher in Car berries than in PV berries. For both cultivars, the IBMP accumulation profiles obtained in 2010 and 2011 were very similar. However, IBMP contents in Car berries were slightly higher in 2010 than in 2011. In 2010, the amount of IBMP in Car berries increased from 41.5 ng kg–1 fresh weight at peppercorn-size stage to a peak of 201 ng kg–1 at bunch closure stage and then declined to 34.6 ng kg–1 at mature stage (Fig. 4A). IBMP levels in PV berries were extremely low in 2010 and did not change throughout berry development. PV berries did not accumulate any detectable amounts of IBMP in 2011 (Fig. 4B).
Figure 4.
IBMP levels throughout Car and PV berry development. IBMP content was investigated throughout the 2010 and 2011 growing seasons and is expressed as nanograms per kilogram of fresh berry weight. IBMP levels are reported as means of three independent experiments ± sd (bars). ND, Not detectable.
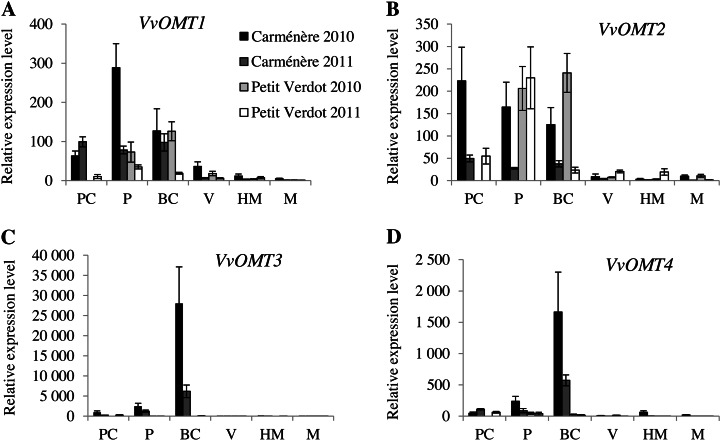
The expression patterns of VvOMT1, VvOMT2, VvOMT3, and VvOMT4 were investigated by real-time PCR using berries harvested during the 2010 and 2011 growing seasons (Fig. 5). For each cultivar, similar expression profiles of VvOMT genes were obtained in 2010 and 2011. However, the expression profiles obtained for OMT genes related to QTLIBMP12 (i.e. VvOMT1 and VvOMT2) and those related to QTLIBMP3 (i.e. VvOMT3 and VvOMT4) were very different. The expression levels of all VvOMT genes were low from véraison to mature stages. Expression of VvOMT1 and VvOMT2 did not differ significantly at bunch closure stage between Car and PV berries (Fig. 5, A and B). By contrast, the expression levels of VvOMT3 and VvOMT4 clearly differed between Car and PV berries (Fig. 5, C and D). In Car berries, VvOMT3 and VvOMT4 expression increased from peppercorn stage, reached a peak in expression at bunch closure stage, and was low from véraison to mature stages during both growing seasons (Fig. 5C). Expression levels of VvOMT genes were generally lower in 2011 than in 2010. The VvOMT3 gene was highly expressed in the high-MP Car berries at bunch closure stage, at the peak of IBMP production, whereas the expression of this gene was very low in PV berries at the same stage.
Figure 5.
Expression profiles of VvOMT1, VvOMT2, VvOMT3, and VvOMT4 genes throughout Car and PV berry development. The expression profiles of VvOMT1 (A), VvOMT2 (B), VvOMT3 (C), and VvOMT4 (D) were investigated throughout the 2010 and 2011 growing seasons from peppercorn size to mature stages. Gene expression was normalized with VvGAPDH and VvEF1γ. All results were expressed as means ± sd (bars) for three independent experiments and relative to the lowest level of expression detected. PC, Peppercorn size (EL-29); P, pea size (EL-31); BC, bunch closure (EL-32); V, véraison (EL-35); HM, half mature (10-potential alcohol degree, EL-36); M, mature (EL-38).
Expression of Recombinant VvOMT Proteins in Escherichia coli
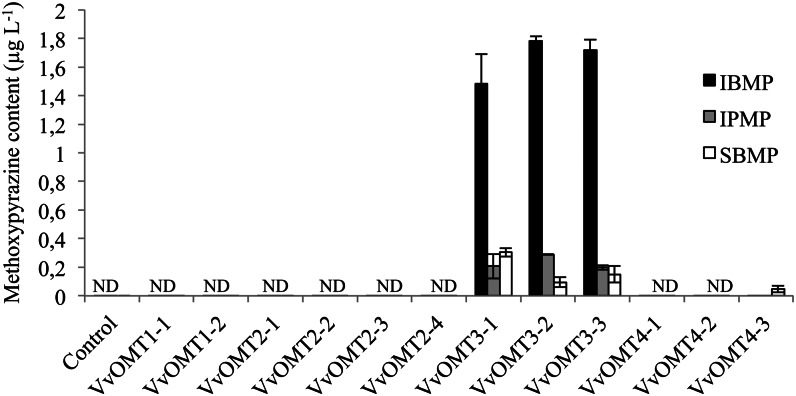
To further investigate the potential role of VvOMT3 and VvOMT4 genes in IBMP biosynthesis and to characterize their allelic diversity, several VvOMT3 and VvOMT4 alleles were cloned from Car, CS, PN, and PV, together with VvOMT1 and VvOMT2 alleles for comparison. IBMP is known to be abundant in Car and CS (Roujou de Boubée et al., 2002; Belancic and Agosin, 2007), whereas PN and PV berries produce very low amounts of IBMP (Koch et al., 2010). A detailed comparison between all alleles is presented in Supplemental Table S1. The VvOMT1-2 and VvOMT2-2 alleles were identical, respectively, to VvOMT1 and VvOMT2 characterized previously (Dunlevy et al., 2010; Vallarino et al., 2011). No amino acid polymorphism between the VvOMT alleles was predicted to alter the conserved domains or binding residues common to all plant OMTs. All complementary DNAs (cDNAs) corresponding to the cloned VvOMT alleles were cloned into the Gateway-compatible vector pHNGWA and expressed in E. coli as NusA fusion proteins (Busso et al., 2005). Unexpectedly, some VvOMT-expressing E. coli cultures exhibited a distinctive pepper-like odor, prompting us to analyze the culture media. GC-MS analyses of culture supernatants revealed that bacterial expression of some recombinant VvOMT alleles resulted in the release of significant amounts of MPs (IBMP, IPMP, and SBMP), indicating that odorless nonmethylated precursor molecules were present in E. coli (Fig. 6). The strongly pepper-scented VvOMT3-1-, VvOMT3-2-, and VvOMT3-3-expressing cultures produced high amounts of MPs, with levels of IBMP being significantly higher than those of IPMP and SBMP (Fig. 6). Expression of VvOMT4-3 allele resulted in the accumulation of low levels of SBMP. Conversely, cultures expressing VvOMT1 and VvOMT2 alleles, as well as control E. coli cultures, did not accumulate any detectable amounts of MPs.
Figure 6.
Analysis of MP contents in culture media of E. coli expressing recombinant VvOMT alleles. MPs were extracted from the culture supernatants using SBSE and quantified by GC-MS. MP amounts are means ± ses (bars) of three independent experiments. ND, Not detectable.
Only VvOMT3 alleles led to significant amounts of MPs in E. coli culture media. This could reflect a particular activity of VvOMT3 for MP biosynthesis, but it could also be due to differences in expression or solubility compared with other VvOMT proteins. However, SDS-PAGE analysis of E. coli protein extracts showed that all VvOMT proteins were efficiently expressed as soluble fusion proteins (Supplemental Fig. S4A). A selection of fusion proteins were purified and incubated with IBHP in the presence of SAM as a preliminary activity screen. Incubation of most purified NusA-VvOMT fusion proteins with IBHP resulted in the biosynthesis of detectable amounts of IBMP. However, VvOMT3-1, VvOMT3-2, and VvOMT3-3 proteins catalyzed the formation of much larger amounts of IBMP than those observed with other VvOMT proteins (Supplemental Fig. S4B).
Detailed Characterization of VvOMT3 Proteins
Gene expression analyses, MP accumulation in E. coli culture supernatants, and preliminary activity tests were consistent with VvOMT3 being the most important VvOMT gene for IBMP biosynthesis. Therefore, VvOMT3 alleles from grapevine varieties producing high and low amounts of MPs were selected for detailed biochemical characterization. The proteins corresponding to the VvOMT3-1 allele present in Car and CS and to the VvOMT3-2 allele present in both PN and PV were expressed in E. coli as His-tagged NusA fusion proteins and purified on a metal affinity resin. Determination of the kinetic parameters of NusA-VvOMT3-1 and NusA-VvOMT3-2 showed that both enzymes exhibit similar affinity for their IBHP substrate, the turnover rate of NusA-VvOMT3-2 being slightly higher than that of NusA-VvOMT3-1 (Table II). The product of both VvOMT3 proteins incubated with IBHP in the presence of SAM was unambiguously identified as IBMP, compared with the corresponding authentic standard (Supplemental Fig. S5). Incubation of VvOMT3 proteins with a selection of potential substrates, including 5-methoxyresorcinol, resveratrol, and quercetin, did not lead to the formation of significant amounts of methylation products (Supplemental Fig. S4C).
Table II. Kinetic parameters of recombinant VvOMT3-1 and VvOMT3-2 using IBHP as a substrate.
Data are expressed as means of triplicate assays using the NusA-VvOMT3 fusion proteins, and se values are given in parentheses.
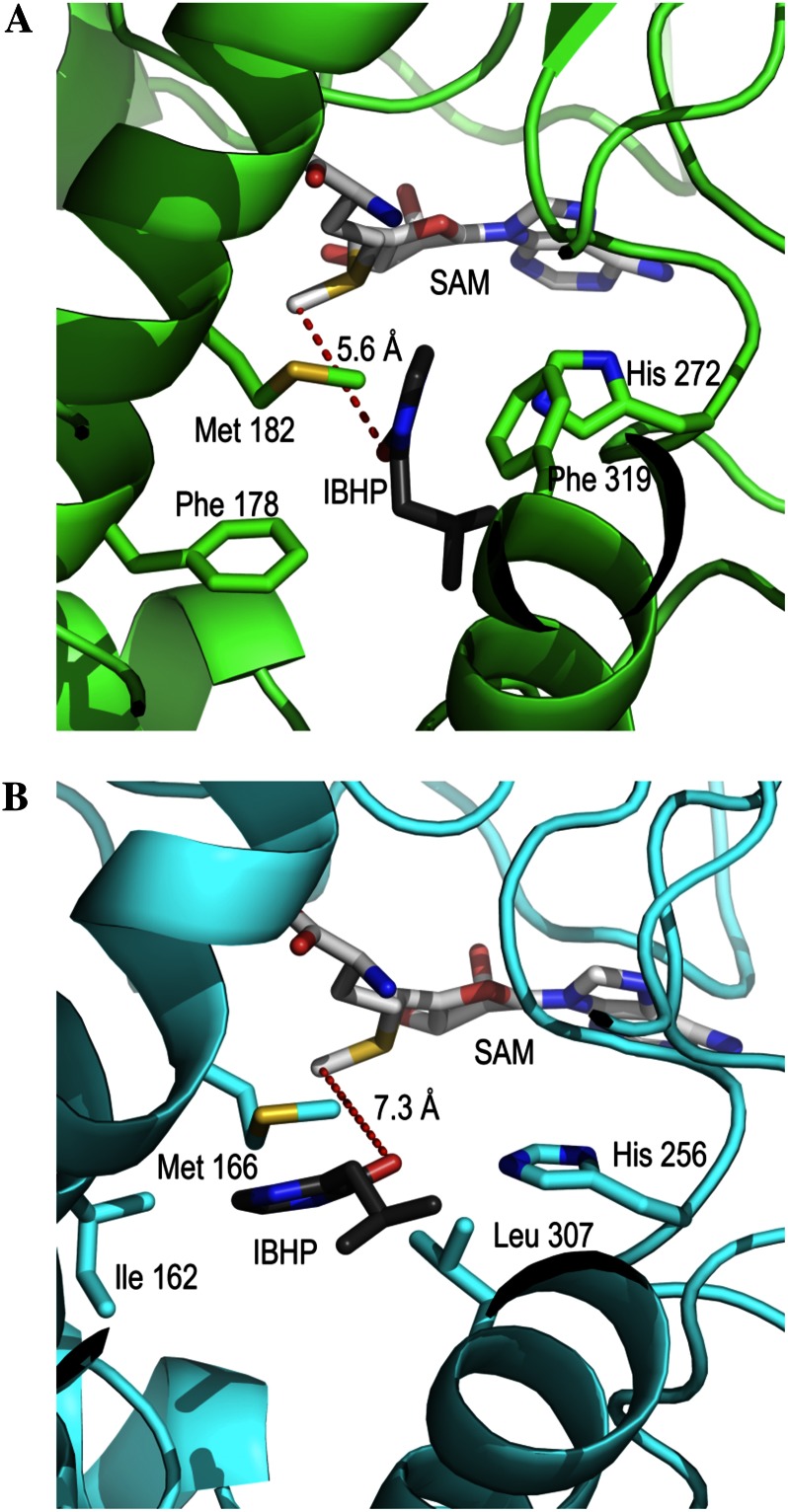
VvOMT1 and VvOMT2 proteins have previously been shown to methylate several substrates, including IBHP, in vitro (Dunlevy et al., 2010). The structural bases for their activity have recently been investigated (Vallarino et al., 2011). However, VvOMT3 exhibited much higher methylation efficiencies and a very narrow specificity for IBHP, which prompted us to investigate the structural basis for these remarkable characteristics. For comparison, VvOMT1-2 (identical to VvOMT1; Vallarino et al., 2011) and VvOMT3-2 alleles were selected for structure modeling based on available plant OMT structures. VvOMT1 shared a high homology with other OMTs (isoflavone OMT from Medicago sativa 1FP2, 39% identity; monolignol OMT from Clarkia breweri 3REO, 37% identity; and isoflavanone 4′-OMT from Medicago truncatula 1ZGA, 39% identity). VvOMT3 has less homology with templates retained for homology modeling (caffeic acid OMT from Lolium perenne 3P9C, 31% identity; chalcone OMT from M. sativa 1FP1, 30% identity; and isoflavanone 4′-OMT from M. truncatula 1ZGA, 34% identity). Because dimerization has been reported to be critical for catalytic activity (Zubieta et al., 2001; Noel et al., 2003), VvOMT1 and VvOMT3 were modeled as homodimers. Molecular dynamics was performed after homology modeling to remove the bias from the template structures. Comparison of the trajectories of several independent molecular dynamics runs shows the convergence of the system to a stable low-energy model. After about 100-picosecond equilibration, the IBHP molecule and the α-carbons of the protein remained in stable positions, with minor fluctuations. In the VvOMT1 model, the IBHP substrate in the active site is close to His 272 and Met 182, and the distance between the oxygen atom of IBHP and the methyl group of SAM is 5.6 Å. In the VvOMT3-2 model, the general structure of the active site is similar to VvOMT1. The SAM is at the same position, and the distance between the oxygen atom of IBHP and the methyl group of SAM is 7.3 Å. However, the large and hydrophobic residues Phe 178 and Phe 319 present in VvOMT1 are replaced by Ile 162 and Leu 307 in VvOMT3 (Fig. 7).
Figure 7.
Molecular modeling of VvOMT1 and VvOMT3. Models shown are three-dimensional structures of the active sites of VvOMT1 (A) and VvOMT3 (B). The models incorporate SAM plus IBHP substrate.
DISCUSSION
VvOMT3 and VvOMT4 Gene Expression Profiles Mirror IBMP Accumulation in Grape Berries
The availability of the sequence of the grapevine reference genome (Jaillon et al., 2007) has allowed considerable progress in grapevine genetics. In particular, quantitative genetics approaches have proven very useful to identify genes involved in agronomically relevant traits (Costantini et al., 2008; Battilana et al., 2009; Duchêne et al., 2009, 2012). As a first approach, a CS × RGM progeny has been used to analyze the genetic bases of IBMP biosynthesis in leaves, allowing the detection of five significant QTLs, explaining 41% of the variance in IBMP content. The last step of MP biosynthesis involves an O-methylation reaction, and candidate OMTs have been proposed to catalyze this reaction (Murray and Whitfield, 1975; Hashizume et al., 2001b; Dunlevy et al., 2010). Therefore, we first focused on candidate OMTs present between the markers flanking the detected QTLs. In particular, four candidate OMT genes colocalized with QTLIBMP3 and QTLIBMP12. These two QTLs were significant, with a false discovery probability of P = 0.03 at the whole-genome level and of P = 0.001 at the LG level. We did not identify any obvious candidate genes potentially involved in IBMP biosynthesis under the three other QTLs. The VvOMT1 and VvOMT2 genes potentially associated to QTLIBMP12 have been characterized previously, unlike the VvOMT3 and VvOMT4 genes within the interval of QTLIBMP3 that were identified in this study.
To investigate further these candidates in a winemaking-relevant context, transcript levels of all four OMT genes were monitored by quantitative PCR during berry development in both Car and PV for two consecutive years. Several major conclusions can be drawn from the comparison of the normalized expression data. First, the expression levels of VvOMT3 and VvOMT4 closely mirror IBMP accumulation patterns in berries, with the peak of transcript accumulation for these genes at bunch closure matching the maximum of IBMP levels in the fruits. By contrast, transcript levels for VvOMT1 and VvOMT2 start to accumulate earlier at peppercorn size and peak at pea size, when low amounts of IBMP are detected even in the strong MP producer Car. Such discrepancy between VvOMT1 and VvOMT2 transcript levels and MP content have already been reported in CS berries by Dunlevy et al. (2010), who showed that the maximum mRNA accumulation occurred 15 d before the peak of IBMP. Second, expression levels of VvOMT3 and VvOMT4 are extremely contrasted between the high IBMP producer Car and the low producer PV, particularly at bunch closure stage. At this stage, depending on the year, VvOMT3 and VvOMT4 expression levels are 100 to 4,000 times higher and 40 to 128 times higher, respectively, in Car than in PV. Conversely, VvOMT1 and VvOMT2 expression patterns do not significantly differ in Car and PV berries at bunch closure. Taken together, these expression data are consistent with VvOMT3 and, to a lesser extent, VvOMT4 being key genes in IBMP biosynthesis in grape berries. This prompted us to investigate the catalytic properties of the corresponding proteins after heterologous expression in E. coli.
VvOMT3 Catalyzes the Formation of IBMP in Vivo and in Vitro
Pyrazines are produced by a vast diversity of organisms, including bacteria, fungi, plants, and animals, indicating that their biosynthesis may rely on ubiquitous metabolic pathways (Müller and Rappert, 2010). A relatively small number of bacteria have been shown to produce MPs. These include Pseudomonas perolens, which releases IPMP and SBMP (Cheng et al., 1991). Although E. coli has not been reported to normally produce MPs, expression of VvOMT3 alleles resulted in the release of large amounts of MPs in the growth medium, amounting to several micrograms per liter. This corresponds to 100 to 1,000 times the perception thresholds of these molecules, resulting in a very strong and typical pepper-like odor. This simple experiment provides important information about MP biosynthesis. First, these results show that E. coli produces significant amounts of MP precursors, including IBHP, whose potential functions are not known. The reason why E. coli does not normally produce volatile MPs seems to be the lack of an appropriate OMT, which would efficiently methylate IBHP and other MP precursors. Therefore, E. coli provides a very simple and efficient system for the functional characterization of OMTs involved in MP biosynthesis in other organisms. Second, analysis of in vivo MP production in E. coli following expression of VvOMT genes unambiguously identified VvOMT3 alleles as the most efficient for the methylation of MP precursors. Although VvOMT1 and VvOMT2 were efficiently expressed in E. coli, they did not lead to the accumulation of detectable amounts of MPs. VvOMT1 and VvOMT2 proteins have been shown to methylate IBHP in vitro, albeit with poor affinity (Dunlevy et al., 2010). These results illustrate the fact that OMT proteins may be promiscuous with respect to substrate acceptance in vitro, making it difficult to define a physiological function from such in vitro data.
The VvOMT3-induced MP biosynthesis in E. coli and the high level of expression of this gene in grape berries provide strong evidence in favor of a major role of VvOMT3 in IBMP biosynthesis. Therefore, two VvOMT3 alleles from high-MP and low-MP grapevine varieties were selected for detailed characterization. Both VvOMT3 proteins exhibited high affinity for the IBHP substrate, with Km values below 2 µm. These Km values are lower than most of those of other plant OMTs for their corresponding substrates (Lavid et al., 2002; Zubieta et al., 2002), but low Km values are expected for low-abundance substrates such as IBHP (Harris et al., 2012). Conversely, both VvOMT3 alleles exhibited very low turnover rates, although catalytic constant values in the same order of magnitude have already been reported for other plant OMTs (Baerson et al., 2008). However, this apparently low catalytic efficiency did not prevent the VvOMT3-induced accumulation of large amounts of MPs in E. coli. Furthermore, MPs are low-abundance compounds in grape berries, generally amounting to less than a few hundred nanograms per kilogram of fresh weight (Bayonove et al., 1975; Lacey et al., 1991; Roujou de Boubée et al., 2002; Belancic and Agosin, 2007). These low amounts are sufficient to impact wine quality, due to the extremely low perception thresholds of these compounds. However, the production of such amounts of MPs may not require a large global enzyme activity and is likely to be compatible with relatively inefficient enzymes. Alternatively, recombinant VvOMT3 proteins may be unstable when purified from E. coli. This may result in the presence of a large proportion of inactive proteins in the purified fractions, which would be responsible for the low apparent efficiency of VvOMT3 enzymes. The VvOMT3-2 allele present in the low-MP varieties PN and PV was slightly more active than the VvOMT3-1 allele from the high-MP varieties Car and CS. This confirms that high-MP and low-MP phenotypes are due to differential expression rather than differential activity of particular VvOMT3 alleles. Unlike VvOMT1 and VvOMT2, which were found to be active with a broad range of substrates (Dunlevy et al., 2010), VvOMT3 exhibited a remarkable specificity for IBHP. No significant methylation activity could be obtained using several other potential substrates belonging to the phenol, stilbene, and flavonol families. Although VvOMT3 activity was tested only with the IBHP substrate in vitro, VvOMT3-induced accumulation of MPs in E. coli suggests that VvOMT3 can methylate several MP precursors, including 2-hydroxy-3-isopropylpyrazine and 2-hydroxy-3-S-butylpyrazine in addition to IBHP.
To investigate the structural basis for the remarkable specificity of VvOMT3, both VvOMT3 and VvOMT1 proteins were modeled and compared with the model of VvOMT1 reported recently (Vallarino et al., 2011). Following homology modeling based on known OMT structures, stable low-energy models were obtained after convergence of several independent molecular dynamics runs. This modeling approach allowed an optimized docking of the SAM and IBHP ligands in flexible enzymes. The VvOMT1 model obtained by this protocol is very close to the result already obtained by Vallarino et al. (2011), which validates the modeling approach used. Although the general structure of the VvOMT3 model is very similar to that of VvOMT1, the active site of VvOMT3 is larger and the hydrophobic residues surrounding the substrate-binding pocket have more flexible side chains. VvOMT3 exhibits much higher affinity (Km = 1.9 µm) than VvOMT1 (Km = 539 µm) for the IBHP substrate. In contrast to the rigid active site of VvOMT1, the more flexible environment provided by the VvOMT3 substrate-binding pocket may be partly responsible for its greatly improved efficiency for IBHP methylation.
CONCLUSION
Despite its capital interest for grapevine enological potential and wine quality, the genetic and molecular basis of aroma biosynthesis has just started to be deciphered. In this work, a combination of genetic and candidate gene approaches has led to the characterization of VvOMT3, a novel OMT with remarkable specificity and efficiency for IBMP biosynthesis. The VvOMT3 gene is highly expressed in the berries of MP-rich grapevine varieties, consistent with a major role of this gene in the biosynthesis of MPs in grape berries. The reason for differential VvOMT3 expression in high-MP and low-MP varieties is not known. Careful examination of the PN40024 grapevine reference genome sequence (Jaillon et al., 2007) indicates the presence of numerous relicts of transposable elements around VvOMT3 and VvOMT4 genes, which may impact their expression. Moreover, it should be noted that allelic variations for VvOMT3 and VvOMT4 exist between high- and low-IBMP-producing grapevine cultivars (Supplemental Table S1), suggesting that these genes could be used as molecular markers for breeding purposes. There is currently great interest in breeding new grapevine varieties resistant to diseases of major economic importance in viticulture (Peressotti et al., 2010). These breeding programs based primarily on resistance characters may also offer the opportunity to improve quality-related traits. The identification of major genes of MP biosynthesis will provide useful tools for current and future breeding programs to obtain grapevine cultivars with appropriate levels of MPs.
MATERIALS AND METHODS
Plant Material
F1 Population
The mapping pedigree used in this study consisted of 138 F1 individuals derived from the interspecific cross of grapevine (Vitis vinifera) CS × Vitis riparia RGM. This F1 population, named CS × RGM1995-1, was developed at Institut National de la Recherche Agronomique, Bordeaux, France. The consensus genetic map of the F1 population CS × RGM1995-1 was previously obtained (Marguerit et al., 2009). Basal leaves of 130 F1 individual plants, grown in a greenhouse, were harvested at the end of August during the 2009 growing season. Leaves were stored at –20°C until biochemical analysis.
Samples from Vineyard
Experimental material was harvested during the 2010 and 2011 growing seasons from Car and PV grapevines grown at the Château Dillon vineyards of the Lycée Agricole de Bordeaux-Blanquefort in Blanquefort, France. Berries were collected at six different time points corresponding to peppercorn-size (EL-29), pea-size (EL-31), bunch closure (EL-32), véraison (EL-35), half-mature (10-potential alcohol degree, EL-36), and mature (EL-38) stages as defined by the viticulturists of Château Dillon and the modified E-L system dedicated to identify grapevine growth stages.
For each date, bunch samples were collected separately along grapevine rows located in the middle of the plot. Whole bunches were collected and pooled along rows, except on the three to four first vine stocks on both sides of rows, to minimize differences brought about by phytosanitary treatments, sun exposure, or bunch size.
Extra berries were harvested at the bunch closure stage during the 2009 growing seasons from CS and PN grapevines grown at the grapevine collection of the Bordeaux Institut National de la Recherche Agronomique research station.
All samples were frozen immediately in liquid nitrogen and stored at –80°C until further use.
GC-MS Quantification of IBMP
IBMP has been quantified in grape berries using a stable isotope dilution assay modified from that of Dunlevy et al. (2010). Frozen samples were ground using a conventional blender then in a hydroalcoholic solution using a FastPrep protocol (MP Biomedicals). Typically, to 10 g of ground sample transferred to a 50-mL tube, 30 g of 6.35-mm ceramic spheres (MP Biomedicals) and 10 mL of 70% (v/v) hydroalcoholic solution were added. The tubes were shaken in the FastPrep Instrument (MP Biomedicals) for 20 s at 6 m s–1 and then removed and centrifuged at 7,500g for 15 min at 4°C. The upper phase, containing IBMP, was handled carefully to avoid transferring debris. Then, depending on the material, IBMP extraction before gas chromatography analysis was performed either by SPE for ground leaf samples (40 g) or by SPME for ground berry samples (10 g). Analyses of MPs in Escherichia coli growth media was performed using stir bar sorptive extraction (SBSE).
IBMP Analysis by SPE Protocol
A 20-µL aliquot of deuterated 2H2-IBMP (Orga-Link), at 10 mg L–1 in ethanol, was supplemented as an internal standard to the upper layer of the leaf extract. After dilution in osmosed water (4 volumes), the extract was kept at 4°C for 12 h and filtered (1-µm porosity, Millipore). A purification step on a C18 reversed-phase SPE column (6 mL, 20-μm porosity; Superclean, Supelco) was then performed after rinsing of the column successively by ethyl acetate, ethanol, and ethanol 10% (v/v) volume in water (3 mL at each step, respectively). IBMP was then eluted with 3 mL of CH2CL2:ether mixture (1:2, v/v). The obtained purified solution was subsequently dried over NaSO4, concentrated under a nitrogen flux (at 100 mL min–1) to 100 µL, and then stored at –20°C before analysis by GC-MS (3 µL injected; splitless injection purge time, 1 min). In the range of concentrations from 20 ng L–1 to 2 µg L–1 of IBMP supplemented in leaf extract, the regression equation was linear (IBMP [ng L–1] = 66.28x + 47.49 [x = ratio of peak surface of IBMP on the peak surface of the internal standard]; R2 = 0.993). The repeatability for IBMP (determined five times on the same sample) was 12%. Limit of quantification was 20 ng L–1.
IBMP Analysis by SPME Protocol
Two milliliters of the berry extract were diluted in 12 mL of osmosed water and transferred to a glass headspace vial. A 20-µL aliquot of 1 mg L–1 2H2-IBMP and 3 g of NaCl (99%, Sigma-Aldrich) were added to the vial to increase partitioning into the headspace. SPME extraction was done with a divinylbenzene-carboxen-polydimethylsiloxane fiber (50/30 µm, Supelco). The samples were submitted to agitation (500 rpm for 5 s, stop for 2 s) for 5 min at 60°C and then to the extraction with the SPME fiber for 20 min at 60°C. SPME injection was then realized in splitless mode for 10 min with a desorption temperature of 230°C. In the range of concentrations from 20 ng L–1 to 2 µg L–1 of IBMP supplemented in berry extract, the regression equation was linear (IBMP [ng L–1] = 0.0002x – 0.0017 [x = ratio of peak surface of IBMP on the peak surface of the internal standard]; R2 = 0.999). The repeatability for IBMP (determined five times on the same sample) was 15%. Limit of quantification was 30 ng L–1.
MP Analysis Using SBSE
Following E. coli growth in 2× yeast tripticase medium, cells were harvested by centrifugation (10 min at 6,000g), and MPs were extracted from 10 mL of culture supernatant by SBSE (Gerstel) for 2 h at room temperature according to the protocol of Coelho et al. (2009). MPs were subsequently analyzed by GC-MS.
Gas Chromatography Analysis Coupled to Mass Spectrometry
For both SPE and SPME samples, a 6890 N Gas Chromatograph (Agilent Technologies) equipped with a Combi PAL autosampler (CTC Analytics) with a liquid injection (SPE extracts) or SPME injection mode was used. The gas chromatograph was coupled to an HP 5973N mass selective detector (Agilent Technologies) functioning in electron impact mode at 70 eV. The analyses were performed on a Carbowax 20 m capillary column (BP20, 50 m, 0.25-mm internal diameter, 0.2-µm film thickness, Scientific Glass Engineering). Helium N60 (Air Liquide) was used as a carrier gas. Column head pressure was at 155 kPa (purge flow at 50 mL min–1). The temperature program was as follows: initial hold for 1 min at 45°C, followed by a 3°C min–1 ramp to 120°C and then 50°C min–1 to 240°C. The final temperature was held for 8 min. The injector port was at 230°C, and the injection mode was splitless. The mass selective detector transfer line temperature was 240°C. The quantification was performed in selected-ion monitoring mode after evidencing the retention time of the quantified compound with that of the internal standard in full-scan mode. Data processing was carried out by Chemstation software (Agilent Technologies). The qualifier ions were mass-to-charge ration (m/z) 94, 124, and 151 for IBMP and m/z 95, 127, and 154 for 2H2-IBMP. The quantifier ions were m/z 124 and 127 for IBMP and m/z 151 and 154 for 2H2-IBMP. The IBMP quantification was carried out by comparing ion peak areas of IBMP with that of the 2H2-IBMP internal standard. All SPE-GC-MS and SPME-GC-MS assays were performed in triplicate for each sample.
Liquid Chromatography Coupled to Mass Spectrometry Analysis
The analyses were carried out on an UltiMate 3000 ultra-HPLC-Diode Array Detector system (Dionex) coupled to an Exactive mass spectrometer equipped with an electrospray ionization source (Thermo Fisher Scientific). The column (C18 PolarTec 50 × 2 mm i.d., 1.8 µm; Macherey-Nagel) was developed at 20°C with eluent B (water with 0.1% [v/v] formic acid) and eluent A (acetonitrile with 0.1% [v/v] formic acid) at a flow rate of 0.4 mL min–1 as follows: conditions of 95% B were maintained for 1 min, followed by a gradient over 1 min to 80% B, which was followed again by a gradient over 1 min to 35% B and an additional 0.6 min to 5% B, and these final conditions were maintained for 0.8 min prior to reequilibration to 95% B. The electrospray ionization source was operating in positive mode. The ion transfer capillary temperature was 315°C, the needle voltage was 3.4 kV, and the capillary voltage was 60 V. Nitrogen was used as sheath gas and auxiliary gas, which were maintained at 50 and 15 arbitrary units, respectively. Mass spectra were acquired within a range of mass-to-charge ratio of 100 to 700 atomic mass units, using a resolution of 50,000 (full width at half maximum). Data processing was carried out using the Xcalibur software (Thermo Fisher Scientific). Enzymatically formed IBMP was identified according to its mass spectrum and retention time compared with those of an authentic IBMP standard (Sigma). Amounts were quantified by mass peak integration and calculated using a calibration curve of an authentic IBMP standard in a range of 1 to 330 nm. Other reaction products were identified by the specific m/z of their molecule ions and their retention times.
QTL Analysis
To reduce the positive skewness of IBMP content distribution, square root and log transformations were tested. A natural logarithmic transformation of the data approximated a normal distribution. QTL analysis has been done with both nontransformed and transformed data, without modifying the QTL detection (data not shown). QTL detection was performed with MapQTL 4.0 software (Kyazma) on the consensus map of F1 population CS × RGM1995-1 using interval mapping and MQM. Information on molecular marker characteristics, genotyping conditions, and map construction parameters were the same as those described previously (Marguerit et al., 2009). The maximum number of cofactors retained was five. The significant LOD threshold was calculated with P = 0.05 for the LG and with P = 0.05 for the whole genome through 1,000 permutations. The maximum LOD value was retained for QTL position and a ±2-LOD interval for the confidence interval. The percentage of variance explained by each QTL and the total variance explained by all the QTLs affecting IBMP trait were obtained using restricted MQM implemented with MapQTL 4.0 software.
Genetic markers flanking the confidence interval of QTLs were positioned on the grape genome using a BLASTN procedure against the 12× grape genome assembly (http://genomes.cribi.unipd.it/grape/). Gene lists between flanking markers were extracted from whole-genomic Genome Flat File and used to estimate the number of genes in each genomic region of interest.
RNA Extraction and cDNA Synthesis
Total RNAs from Car, CS, PN, and PV samples were isolated as previously described by Reid et al. (2006). Pedicel and seeds of berries were removed before grinding samples in liquid nitrogen. Total RNAs were subjected to DNA digestion using Ambion TURBO DNA-free DNase (Life Technologies) according to the manufacturer’s instructions. RNA content was measured at 260 nm with a Nanodrop 2000C spectrophotometer (Labtech) and visualized by electrophoresis on 1.5% (w/v) agarose gels. cDNAs were synthesized from DNA-free RNA using SuperScript III reverse transcriptase (Invitrogen) according to the manufacturer’s instructions. The cDNA obtained was diluted (1:10) in distilled water.
Quantitative Real-Time PCR
Real-time PCR was carried out using a CFX96 real-time system (Bio-Rad). Ten-microliter reaction mixes were prepared, which included 5 µL of iQSYBRGreen Supermix (Applied Biosystems), 0.2 µm of each primer, and 2 µL of diluted cDNA. All biological samples were tested in triplicate. Negative controls were included in each run. PCR conditions were initial denaturation at 95°C for 3 min, followed by 40 cycles of 95°C for 10 s and 57°C for 15 s. Amplification was followed by melting curve analysis to check the specificity of primers in each reaction. Primer sequences were designed with Beacon Designer 7 software (Premier Biosoft International) and are presented in Supplemental Table S2. The efficiency of the primer pairs was measured on a cDNA serial dilution.
Results were normalized with the internal reference genes VvEF1γ (for Elongation Factor1γ; AF176496) and VvGAPDH (for Glyceraldehyde 3-P Dehydrogenase; VIT_17s0000g10430). The expression stability measurement of these two housekeeping genes was performed using geNorm software (Vandesompele et al., 2002). Normalized expression of the VvOMT genes was calculated using the Bio-Rad CFX Manager software with a method derived from the algorithms outlined by Vandesompele et al. (2002). Gene transcripts were quantified upon normalization to VvEF1γ and VvGAPDH by using the comparative cycle threshold method (i.e. comparing the cycle threshold of the target gene with that of VvEF1γ and VvGAPDH). Gene expressions were expressed as mean ± sd calculated for three independent experiments.
Cloning of Full-Length VvOMT cDNAs
The full-length cDNA of VvOMTs (VvOMT1, VvOMT2, VvOMT3, and VvOMT4) were amplified from cDNA of bunch closure berries (Car, CS, PN, and PV) with high-fidelity Taq polymerase (Phusion DNA Polymerase; Finnzymes). Primer sequences are presented in Supplemental Table S3. The amplified cDNA for each VvOMT was cloned into the pGEM-T Easy Vector (Promega), and the resulting plasmid was sequenced. VvOMT1 and VvOMT2 sequences were obtained from GenBank. VvOMT3 and VvOMT4 putative sequences were identified from the PN40024 genomic sequence (Jaillon et al., 2007). VvOMT proteins were aligned using ClustalW (Thompson et al., 1994).
Characterization of Recombinant VvOMT Proteins
Full-length VvOMT cDNAs were cloned into pDONR201 Gateway-compatible vector (Invitrogen) to generate entry clones. These entry clones were sequenced to verify that no mutation had been introduced. VvOMT cDNAs were then transferred into the pHNGWA destination vector (accession no. EU680842; Busso et al., 2005) by using LR clonase (Invitrogen) according to the manufacturer’s protocol. pHNGWA-VvOMT plasmids were transformed into ArcticExpress BL21 (DE3) E. coli cells (Agilent) harboring a plasmid, which encodes two cold-adapted chaperonins under constitutive expression. Overnight cultures (2 mL) were inoculated into 50 mL of 2× yeast tripticase medium containing 1% (v/v) glycerol and grown at 28°C to an optical density at 600 nm of 1.0 prior to induction with 1 mm isopropylthio-β-d-galactoside. Cultures were then grown further for 12 h at 10°C. After harvesting the cells by centrifugation (10 min at 6,000g), the pellet was resuspended in 10 mL of purification buffer (25 mm HEPES, pH 7, and 1 mm β-mercaptoethanol), and cell lysis was carried out by sonication. Nonsoluble parts were removed by centrifugation (5 min at 15,000g), and NusA fusion proteins were purified using Protino Ni Ted 1000 packed columns (Macherey-Nagel) according to the manufacturer’s instructions. The NusA-VvOMT fusion proteins were eluted with purification buffer containing 200 mm imidazol. Protein expression and purification steps were monitored by SDS/PAGE. Protein concentrations were determined with the Bio-Rad protein assay kit.
Enzyme Assays
For the determination of kinetic parameters, purified NusA-VvOMT (2.5 µg) was incubated with IBHP ranging from 0.25 to 50 µm in the presence of 500 µm SAM in incubation buffer (25 mm HEPES, pH 7, 1 mm MgCl2, 1 mm β-mercaptoethanol, and 1 µg µL–1 bovine serum albumin) in a final volume of 100 µL for 2.5 h at 28°C. The reactions were stopped by addition of 100 µL of ethyl acetate, followed by vigorous vortexing and centrifugation (5 min at 15,000g). The organic phase containing reaction products and the nontransformed substrate was transferred into a new tube and mixed with one volume of methanol prior to liquid chromatography-mass spectrometry analyses. Km and Vmax values were calculated from Lineweaver-Burk plots. For substrate specificity studies, purified NusA-VvOMT (approximately 2.5 µg) was incubated with different substrates at a final concentration of 20 µm (IBHP, 5-methoxyresorcinol, resveratrol, and quercetin) in the presence of 500 µm SAM in incubation buffer in a final volume of 100 µL for 2.5 h at 28°C. Reaction products were extracted as described above and analyzed by liquid chromatography-mass spectrometry.
Homology Modeling
Structural models of OMTs of VvOMT1-2 (Uniprot A5AD94_VITVI) and VvOMT3-2 (this work) were generated by homology modeling. Sequence and structure alignments were done using @TOME-2 pipeline (Pons and Labesse, 2009). For VvOMT1, structural alignment was performed with Protein Data Bank entries 1FP2, 3REO, and 1ZGA. For VvOMT3, structural alignment was performed with Protein Data Bank entries 3P9C, 1FP1, and 1ZGA. SAM and IBHP structures and topologies were automatically generated with PRODRG (Schüttelkopf and van Aalten, 2004) and manually corrected using recommendations for small molecule topologies (Lemkul et al., 2010). SAM and the methylated hydroxyl group of IBHP were placed at the same positions as in the template structures and were introduced in the model at the beginning of homology modeling using MODELER (Sali and Blundell, 1993). VvOMT1-2 and VvOMT3-2 were generated as homodimers. The models with the lowest Discrete Optimized Protein Energy score (Shen and Sali, 2006) among 100 generated models were selected for energy minimization and molecular dynamics.
Molecular Dynamics Simulations
Simulations were performed using the GROMACS package (Van Der Spoel et al., 2005) in cubic boxes filled with SPC216 water molecules and GROMOS43a1 as force field. Before molecular dynamics, the protein-ligands complex was subjected to energy minimization and positional restraints cycles. The simulation was carried out with periodic boundary conditions by adding sodium ions to have a value of zero as the net electrostatic charge of the system. The bond lengths were constrained by the all atoms LINCS algorithm. Particle mesh Ewald algorithm was used for the electrostatic interactions with a cutoff of 0.9 nm. The simulations were performed at neutral pH with runs of 10 ns at 300 K coupling the system to an external bath. GROMACS routines were utilized to check the trajectories and the quality of the simulations. The structure of the final model was checked using MOLPROBITY (Chen et al., 2010).
All sequence data from this article can be found in the GenBank/EMBL data libraries under the accession numbers listed in Supplemental Table S1.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Distribution of the IBMP content in leaves of the F1 CS × RGM population used in QTL mapping.
Supplemental Figure S2. Genomic structure of the open reading frames of the VvOMT3 and VvOMT4 genes cluster on chromosome 3.
Supplemental Figure S3. Harvested stages throughout cv Carménère and Petit Verdot berry development.
Supplemental Figure S4. Preliminary characterization of VvOMT enzymatic activity.
Supplemental Figure S5. Identification of VvOMT3-1 and VvOMT3-2 reaction product.
Supplemental Table S1. VvOM1, VvOMT2, VvOMT3, and VvOMT4 alleles identified in Carménère, Cabernet Sauvignon, Pinot Noir and Petit Verdot cultivars.
Supplemental Table S2. Quantitative PCR primer sequences.
Supplemental Table S3. Sequences of primers used throughout the cloning of VvOMT cDNA.
Acknowledgments
We thank Bernard Douens, Jean-Pierre Petit, and Jean-Paul Robert for taking care of the F1 CS × RGM population, Claude Bonnet, Cyril Hévin (Unité Mixte de Recherche 1287 Ecophysiologie et Génomique Fonctionnelle de la Vigne), and Mr. Dominique Foucault (Château Dillon) for their help in berry collection, Adline Delcamp and Mélanie Massonet (Unité Mixte de Recherche 1287 Ecophysiologie et Génomique Fonctionnelle de la Vigne), and Monique Pons (Equipe d′Accueil 4577 nologie) for their help in IBMP content analysis and in VvOMT characterization, and Bernard Jost (Microarray and Sequencing Platform, IGBMC) for helpful discussions.
Glossary
- MP
methoxypyrazine
- IBMP
2-methoxy-3-isobutylpyrazine
- IPMP
2-methoxy-3-isopropylpyrazine
- SBMP
2-methoxy-3-sec-butylpyrazine
- SAM
S-adenosyl-l-Met
- IBHP
2-hydroxy-3-isobutylpyrazine
- QTL
quantitative trait loci
- CS
cv Cabernet Sauvignon
- RGM
cv Gloire de Montpellier
- SPE
solid-phase extraction
- GC-MS
gas chromatography-mass spectrometry
- SPME
solid-phase microextraction
- MQM
multiple quantitative trait loci mapping
- LOD
log of the odds
- Car
cv Carménère
- PV
cv Petit Verdot
- PN
cv Pinot Noir
- cDNA
complementary DNA
- SBSE
stir bar sorptive extraction
- m/z
mass-to-charge ratio
- LG
linkage group
- Kcat
catalytic constant
References
- Allen MS, Lacey MJ, Harris RLN, Brown WV. (1991) Contribution of methoxypyrazines to Sauvignon Blanc wine aroma. Am J Enol Vitic 42: 109–112 [Google Scholar]
- Baerson SR, Dayan FE, Rimando AM, Nanayakkara NPD, Liu CJ, Schroerder J, Fishbein M, Pan Z, Kagan IA, Pratt LH, et al. (2008) A functional genomics investigation of allelochemical biosynthesis in Sorghum bicolor root hairs. J Biol Chem 283: 3231–3247 [DOI] [PubMed] [Google Scholar]
- Battilana J, Costantini L, Emanuelli F, Sevini F, Segala C, Moser S, Velasco R, Versini G, Stella Grando M. (2009) The 1-deoxy-D: -xylulose 5-phosphate synthase gene co-localizes with a major QTL affecting monoterpene content in grapevine. Theor Appl Genet 118: 653–669 [DOI] [PubMed] [Google Scholar]
- Bayonove C, Cordonnier RE, Dubois P. (1975) Etude d'une fraction caractéristique de l'arôme du raisin de la variété Cabernet-Sauvignon: mise en évidence de la 2-méthoxy-3-isobutylpyrazine. CR Acad Sci 60: 1321–1329 [Google Scholar]
- Belancic A, Agosin E. (2007) Methoxypyrazines in grapes and wines of Vitis vinifera cv. Carmenere. Am J Enol Vitic 58: 462–469 [Google Scholar]
- Busso D, Delagoutte-Busso B, Moras D. (2005) Construction of a set Gateway-based destination vectors for high-throughput cloning and expression screening in Escherichia coli. Anal Biochem 343: 313–321 [DOI] [PubMed] [Google Scholar]
- Chen VB, Arendall WB, III, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC. (2010) MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr 66: 12–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng TB, Reineccius GA, Bjorklund JA, Leete E. (1991) Biosynthesis of 2-methoxy-3-isopropylpyrazine in Pseudomonas perolens. J Agric Food Chem 39: 1009–1012 [Google Scholar]
- Coelho E, Coimbra MA, Nogueira JMF, Rocha SM. (2009) Quantification approach for assessment of sparkling wine volatiles from different soils, ripening stages, and varieties by stir bar sorptive extraction with liquid desorption. Anal Chim Acta 635: 214–221 [DOI] [PubMed] [Google Scholar]
- Costantini L, Battilana J, Lamaj F, Fanizza G, Grando MS. (2008) Berry and phenology-related traits in grapevine (Vitis vinifera L.): from quantitative trait loci to underlying genes. BMC Plant Biol 8: 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubourdieu D, Tominaga T, Masneuf I, Peyrot Des Gachons C, Murat ML. (2006) The role of yeasts in grape flavor development during fermentation: the example of Sauvignon blanc. Am J Enol Vitic 57: 81–88 [Google Scholar]
- Duchêne E, Butterlin G, Claudel P, Dumas V, Jaegli N, Merdinoglu D. (2009) A grapevine (Vitis vinifera L.) deoxy-D: -xylulose synthase gene colocates with a major quantitative trait loci for terpenol content. Theor Appl Genet 118: 541–552 [DOI] [PubMed] [Google Scholar]
- Duchêne E, Butterlin G, Dumas V, Merdinoglu D. (2012) Towards the adaptation of grapevine varieties to climate change: QTLs and candidate genes for developmental stages. Theor Appl Genet 124: 623–635 [DOI] [PubMed] [Google Scholar]
- Dunlevy JD, Soole KL, Perkins MV, Dennis EG, Keyzers RA, Kalua CM, Boss PK. (2010) Two O-methyltransferases involved in the biosynthesis of methoxypyrazines: grape-derived aroma compounds important to wine flavour. Plant Mol Biol 74: 77–89; erratumDunlevy JD, Soole KL, Perkins MV, Dennis EG, Keyzers RA, Kalua CM, Boss PK. (2013) Plant Mol Biol 81: 523 [DOI] [PubMed] [Google Scholar]
- Ebeler SE, Thorngate JH. (2009) Wine chemistry and flavor: looking into the crystal glass. J Agric Food Chem 57: 8098–8108 [DOI] [PubMed] [Google Scholar]
- Gallois A, Kergomard A, Adda J. (1988) Study of the biosynthesis of 3-isopropyl2-methoxypyrazine produced by Pseudomonas taetrolens. Food Chem 28: 299–309 [Google Scholar]
- Harris SA, Ryona I, Sacks GL. (2012) Behavior of 3-isobutyl-2-hydroxypyrazine (IBHP), a key intermediate in 3-isobutyl-2-methoxypyrazine (IBMP) metabolism, in ripening wine grapes. J Agric Food Chem 60: 11901–11908 [DOI] [PubMed] [Google Scholar]
- Hashizume K, Samuta T. (1999) Grape maturity and light exposure affect berry methoxypyrazine concentration. Am J Enol Vitic 50: 194–198 [Google Scholar]
- Hashizume K, Tozawa K, Endo M, Aramaki I. (2001a) S-Adenosyl-l-methionine-dependent O-methylation of 2-hydroxy-3-alkylpyrazine in wine grapes: a putative final step of methoxypyrazine biosynthesis. Biosci Biotechnol Biochem 65: 795–801 [DOI] [PubMed] [Google Scholar]
- Hashizume K, Tozawa K, Hiraga Y, Aramaki I. (2001b) Purification and characterization of a O-methyltransferase capable of methylating 2-hydroxy-3-alkylpyrazine from Vitis vinifera L. (cv. Cabernet Sauvignon). Biosci Biotechnol Biochem 65: 2213–2219 [DOI] [PubMed] [Google Scholar]
- Jaillon O, Aury J-M, Noel B, Policriti A, Clepet C, Casagrande A, Choisne N, Aubourg S, Vitulo N, Jubin C, et al. (2007) The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature 449: 463–467 [DOI] [PubMed] [Google Scholar]
- Koch A, Doyle CL, Matthews MA, Williams LE, Ebeler SE. (2010) 2-Methoxy-3-isobutylpyrazine in grape berries and its dependence on genotype. Phytochemistry 71: 2190–2198 [DOI] [PubMed] [Google Scholar]
- Lacey MJ, Allen MS, Harris RLN, Brown WV. (1991) Methoxypyrazines in Sauvignon blanc grapes and wines. Am J Enol Vitic 42: 103–108 [Google Scholar]
- Lavid N, Schwab W, Kafkas E, Koch-Dean M, Bar E, Larkov O, Ravid U, Lewinsohn E. (2002) Aroma biosynthesis in strawberry: S-adenosylmethionine:furaneol O-methyltransferase activity in ripening fruits. J Agric Food Chem 50: 4025–4030 [DOI] [PubMed] [Google Scholar]
- Lemkul JA, Allen WJ, Bevan DR. (2010) Practical considerations for building GROMOS-compatible small-molecule topologies. J Chem Inf Model 50: 2221–2235 [DOI] [PubMed] [Google Scholar]
- Lund C, Thompson MK, Benkwitz F, Wohler MW, Triggs CM, Gardner R, Heymann H, Nicolau L. (2009) New Zealand Sauvignon blanc distinct flavor characteristics: sensory, chemical and consumer aspects. Am J Enol Vitic 60: 1–12 [Google Scholar]
- Maga JA. (1982) Pyrazines in foods: an update. Crit Rev Food Sci Nutr 16: 1–48 [DOI] [PubMed] [Google Scholar]
- Marguerit E, Boury C, Manicki A, Donnart M, Butterlin G, Némorin A, Wiedemann-Merdinoglu S, Merdinoglu D, Ollat N, Decroocq S. (2009) Genetic dissection of sex determinism, inflorescence morphology and downy mildew resistance in grapevine. Theor Appl Genet 118: 1261–1278 [DOI] [PubMed] [Google Scholar]
- Müller R, Rappert S. (2010) Pyrazines: occurrence, formation and biodegradation. Appl Microbiol Biotechnol 85: 1315–1320 [DOI] [PubMed] [Google Scholar]
- Murray KE, Whitfield FB. (1975) Occurrence of 3-alkyl-2-methoxypyrazines in raw vegetables. J Sci Food Agric 26: 973–986 [Google Scholar]
- Noel JP, Dixon RA, Pichersky E, Zubieta C, Ferrer J-L. (2003) Structural, functional, and evolutionary basis for methylation of plant small molecules. Recent Adv Phytochem 37: 37–58 [Google Scholar]
- Parr WV, Green JA, White KG, Sherlock RR. (2007) The distinctive flavour of New Zealand Sauvignon blanc: sensory characterization by wine professionals. Food Qual Prefer 18: 849–861 [Google Scholar]
- Peña-Gallego A, Hernández-Orte P, Cacho J, Ferreira V. (2012) S-Cysteinylated and S-glutathionylated thiol precursors in grapes. A review. Food Chem 131: 1–13 [Google Scholar]
- Peressotti E, Wiedemann-Merdinoglu S, Delmotte F, Bellin D, Di Gaspero G, Testolin R, Merdinoglu D, Mestre P. (2010) Breakdown of resistance to grapevine downy mildew upon limited deployment of a resistant variety. BMC Plant Biol 10: 147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pons JL, Labesse G. (2009) @TOME-2: a new pipeline for comparative modeling of protein-ligand complexes. Nucleic Acids Res 37: W485–W491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid KE, Olsson N, Schlosser J, Peng F, Lund ST. (2006) An optimized grapevine RNA isolation procedure and statistical determination of reference genes for real-time RT-PCR during berry development. BMC Plant Biol 6: 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roujou de Boubée D (2000) Research on 2-methoxy-3-isobutylpyrazine in grapes and wines. Analytical, biological and agronomical approaches. PhD thesis. Bordeaux 2 University, Bordeaux, France [Google Scholar]
- Roujou de Boubée D, Cumsille AM, Pons M, Dubourdieu D. (2002) Location of 2-methoxy-3-isobutylpyrazine in Cabernet-Sauvignon grape bunches and its extractability during vinification. Am J Enol Vitic 53: 1–5 [Google Scholar]
- Roujou de Boubée D, Van Leeuwen C, Dubourdieu D. (2000) Organoleptic impact of 2-methoxy-3-isobutylpyrazine on red bordeaux and loire wines. Effect of environmental conditions on concentrations in grapes during ripening. J Agric Food Chem 48: 4830–4834 [DOI] [PubMed] [Google Scholar]
- Ryona I, Pan BS, Intrigliolo DS, Lakso AN, Sacks GL. (2008) Effects of cluster light exposure on 3-isobutyl-2-methoxypyrazine accumulation and degradation patterns in red wine grapes (Vitis vinifera L. Cv. Cabernet Franc). J Agric Food Chem 56: 10838–10846 [DOI] [PubMed] [Google Scholar]
- Sali A, Blundell TL. (1993) Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol 234: 779–815 [DOI] [PubMed] [Google Scholar]
- Schüttelkopf AW, van Aalten DM. (2004) PRODRG: a tool for high-throughput crystallography of protein-ligand complexes. Acta Crystallogr D Biol Crystallogr 60: 1355–1363 [DOI] [PubMed] [Google Scholar]
- Shen MY, Sali A. (2006) Statistical potential for assessment and prediction of protein structures. Protein Sci 15: 2507–2524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepherd GM. (2006) Smell images and the flavour system in the human brain. Nature 444: 316–321 [DOI] [PubMed] [Google Scholar]
- Strauss CR, Wilson B, Gooley PR, Williams PJ. (1986) Role of monoterpenes in grape and wine flavor. In Parliament C, ed, Biogeneration of Aromas. American Chemical Society, Washington, DC, pp 222–242 [Google Scholar]
- Styger G, Prior B, Bauer FF. (2011) Wine flavor and aroma. J Ind Microbiol Biotechnol 38: 1145–1159 [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TG. (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22: 4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tominaga T, Peyrot des Gachons C, Dubourdieu D. (1998) New type of flavor precursors in Vitis vinifera L. cv. Sauvignon Blanc: S-cysteine conjugates. J Agric Food Chem 46: 5215–5219 [DOI] [PubMed] [Google Scholar]
- Vallarino JG, Lopez-Cortes XA, Dunlevy JD, Boss PK, Gonzalez-Nilo FD, Moreno YM. (2011) Biosynthesis of methoxypyrazines: elucidating the structural/functional relationship of two Vitis vinifera O-methyltransferases capable of catalyzing the putative final step of the biosynthesis of 3-alkyl-2-methoxypyrazine. J Agric Food Chem 59: 7310–7316 [DOI] [PubMed] [Google Scholar]
- Van Der Spoel D, Lindahl E, Hess B, Groenhof G, Mark AE, Berendsen HJ. (2005) GROMACS: fast, flexible, and free. J Comput Chem 26: 1701–1718 [DOI] [PubMed] [Google Scholar]
- Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F. (2002) Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 3: 1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zubieta C, He XZ, Dixon RA, Noel JP. (2001) Structures of two natural product methyltransferases reveal the basis for substrate specificity in plant O-methyltransferases. Nat Struct Biol 8: 271–279 [DOI] [PubMed] [Google Scholar]
- Zubieta C, Kota P, Ferrer J-L, Dixon RA, Noel JP. (2002) Structural basis for the modulation of lignin monomer methylation by caffeic acid/5-hydroxyferulic acid 3/5-O-methyltransferase. Plant Cell 14: 1265–1277 [DOI] [PMC free article] [PubMed] [Google Scholar]