The catalytic mechanism and exact specificity for hydroxycinnamoyltransferase from sorghum were determined by comprehensive approaches with crystal structures of apo-form and ternary product complex, site-directed mutagenesis, and kinetic and thermodynamic analyses.
Abstract
Hydroxycinnamoyltransferase (HCT) from sorghum (Sorghum bicolor) participates in an early step of the phenylpropanoid pathway, exchanging coenzyme A (CoA) esterified to p-coumaric acid with shikimic or quinic acid as intermediates in the biosynthesis of the monolignols coniferyl alcohol and sinapyl alcohol. In order to elucidate the mode of action of this enzyme, we have determined the crystal structures of SbHCT in its apo-form and ternary complex with shikimate and p-coumaroyl-CoA, which was converted to its product during crystal soaking. The structure revealed the roles of threonine-36, serine-38, tyrosine-40, histidine-162, arginine-371, and threonine-384 in catalysis and specificity. Based on the exact chemistry of p-coumaroyl-CoA and shikimic acid in the active site and an analysis of kinetic and thermodynamic data of the wild type and mutants, we propose a role for histidine-162 and threonine-36 in the catalytic mechanism of HCT. Considering the calorimetric data, substrate binding of SbHCT should occur sequentially, with p-coumaroyl-CoA binding prior to the acyl acceptor molecule. While some HCTs can use both shikimate and quinate as an acyl acceptor, SbHCT displays low activity toward quinate. Comparison of the structure of sorghum HCT with the HCT involved in chlorogenic acid synthesis in coffee (Coffea canephora) revealed many shared features. Taken together, these observations explain how CoA-dependent transferases with similar structural features can participate in different biochemical pathways across species.
Lignin is a major structural and protective component of plant cell walls. Lignin exists as a polymer of mainly three hydroxycinnamyl alcohols and related compounds, referred to as monolignols. The most common monolignols are coniferyl, sinapyl, and p-coumaryl alcohol (Ralph et al., 2004; Vanholme et al., 2010). After polymerization, structures derived from those compounds are referred to as guaiacyl, syringyl, and p-hydroxyphenyl subunits, respectively. The specific composition of lignin subunits varies among species, tissues, and developmental stages. Gymnosperm trees produce lignin that is primarily made of guaiacyl subunits, angiosperm trees contain guaiacyl and syringyl subunits, whereas grasses contain guaiacyl and syringyl subunits with small amounts (approximately 5%) of p-hydroxyphenyl residues. This observed variation in subunit composition across species may reflect the heterogeneity in substrate specificity and kinetic parameters among various monolignol biosynthetic enzymes (Weng et al., 2008).
Biosynthesis of the monolignols occurs via the phenylpropanoid pathway using Phe precursors (Vanholme et al., 2010). Phe ammonia lyase, cinnamate-4-hydroxylase, and 4-coumarate coenzyme A (CoA) ligase (4CL) generate p-coumaroyl-CoA from Phe (Vanholme et al., 2010). Grasses can bypass cinnamate-4-hydroxylase by using Tyr as a substrate for Phe ammonia lyase (Neish, 1961; Rösler et al., 1997). The hydroxycinnamoyltransferase (HCT) enzymes exchange the CoA functionality esterified to p-coumaric acid with shikimic or quinic acid to allow for the subsequent conversion of the p-coumaroyl moiety to a caffeoyl moiety by p-coumarate-3′-hydroxylase (C3′H). The hydroxycinnamoyl-CoA shikimate hydroxycinnamoyltransferases (HSTs) exhibit preference for shikimate, whereas the hydroxycinnamoyl-CoA quinate hydroxycinnamoyltransferases prefer quinate as a substrate (Sander and Petersen, 2011). Subsequent reactions ultimately lead to coniferyl and sinapyl alcohol via reduction of the γ-carbon on the propane side chain and substitution of the C3 and C5 positions of the phenol ring (Boerjan et al., 2003).
Sorghum (Sorghum bicolor) is an attractive bioenergy crop with typical dry biomass yields between 20 and 25 Mg ha−1 and yields as high as 40 Mg ha−1 possible under optimal conditions (Venuto and Kindiger, 2008). Moreover, sorghum utilizes nitrogen-based fertilizer more efficiently than maize (Zea mays) and sugarcane (Saccharum officinarum), leading to less groundwater contamination and lower CO2 emission (Propheter and Staggenborg, 2010; Wortmann and Regassa, 2011). Overall, sorghum has a higher sugar yield potential per land area and requires less water for growth than maize, allowing it to grow in a more diverse range of environments (Saballos, 2008). The sorghum genome sequence has been released (Paterson et al., 2009), and Targeting Induced Local Lesions in Genomes populations exist (Xin et al., 2008) in which various cell wall mutants have been identified (Sattler et al., 2012; Vermerris and Saballos, 2012).
A detailed understanding of the catalytic mechanism of phenylpropanoid-related enzymes will enable the targeted modification of lignin subunit composition. The presence of lignin poses a major obstacle to the production of biofuels and chemicals from lignocellulosic biomass, because of its ability to hinder the activity of enzymes required to degrade cellulose to sugars that can be fermented for ethanol production (Yang and Wyman, 2004; Berlin et al., 2006). Genetic modification of plant cell wall composition, especially lignin content and subunit composition, has been shown to improve biomass conversion to fermentable sugars (Chen and Dixon, 2007; Vermerris et al., 2007; Jung et al., 2012). In particular, HCT silencing in Arabidopsis (Arabidopsis thaliana) causes an accumulation of p-hydroxyphenyl residues in the lignin and decreased content of guaiacyl and syringyl residues, leading to a dwarf phenotype (Li et al., 2010). Down-regulation of HCT has also been shown to result in decreased plant growth in alfalfa (Medicago sativa; Shadle et al., 2007). Concomitantly, ruminant digestibility and the yield of fermentable sugars following enzymatic saccharification increased (Chen and Dixon, 2007; Shadle et al., 2007). Reduced HCT activity may alter cell wall polymer interactions and allow better access of cellulolytic enzymes to the cellulose. Therefore, it has the potential to reduce the energy and processing costs associated with the conversion of biomass to fuels and chemicals. However fine-tuning will be necessary to limit the negative impacts on plant growth, which will require a detailed understanding of the catalytic mechanism of HCT.
Given the difference in lignin subunit composition among different species and the prominence of grasses among dedicated bioenergy crops, we have focused on elucidating the crystal structure and activity of monolignol-related enzymes of sorghum, starting with the HST-like HCT. HCT belongs to the BAHD superfamily of plant-specific acyl-CoA-dependent acyltransferases (Ma et al., 2005; D’Auria, 2006). However, the BAHD superfamily has functionally and structurally diverse members that frequently possess little (as low as 10%) sequence identity among them (St-Pierre and Luca, 2000). Recent studies led to the crystal structure of the HST-like HCT from robusta coffee (Coffea canephora), an angiosperm dicot with a binding pocket elucidated by molecular docking and mutagenesis (Lallemand et al., 2012). In this report, we present the three-dimensional structures of HCT in its apo-form and ternary complex, supplemented by mutagenic studies to elucidate its reaction mechanism and structural relationship to other members in this growing functional class.
RESULTS
Determination of the Three-Dimensional Structure of SbHCT
The structures of the recombinant SbHCT were determined in their apo-form and ternary complex with p-coumaroyl-CoA and shikimate at 2.0 and 2.4 Å resolution, respectively. The structure of apo-form SbHCT was determined by molecular replacement using the coordinates of apo-form HCT from coffee (Protein Data Bank [PDB] identifier 4G0B).
Static light scattering was employed to determine the exact oligomeric status of SbHCT (2 mg mL–1) in solution. The single peak identified with this analysis (Supplemental Fig. S1) revealed the monomeric (49 kD) nature of SbHCT. Consistently, the SbHCT crystal contained one molecule per asymmetric unit, and the crystal lattice did not show any extensive packing interactions.
Global Structure of SbHCT
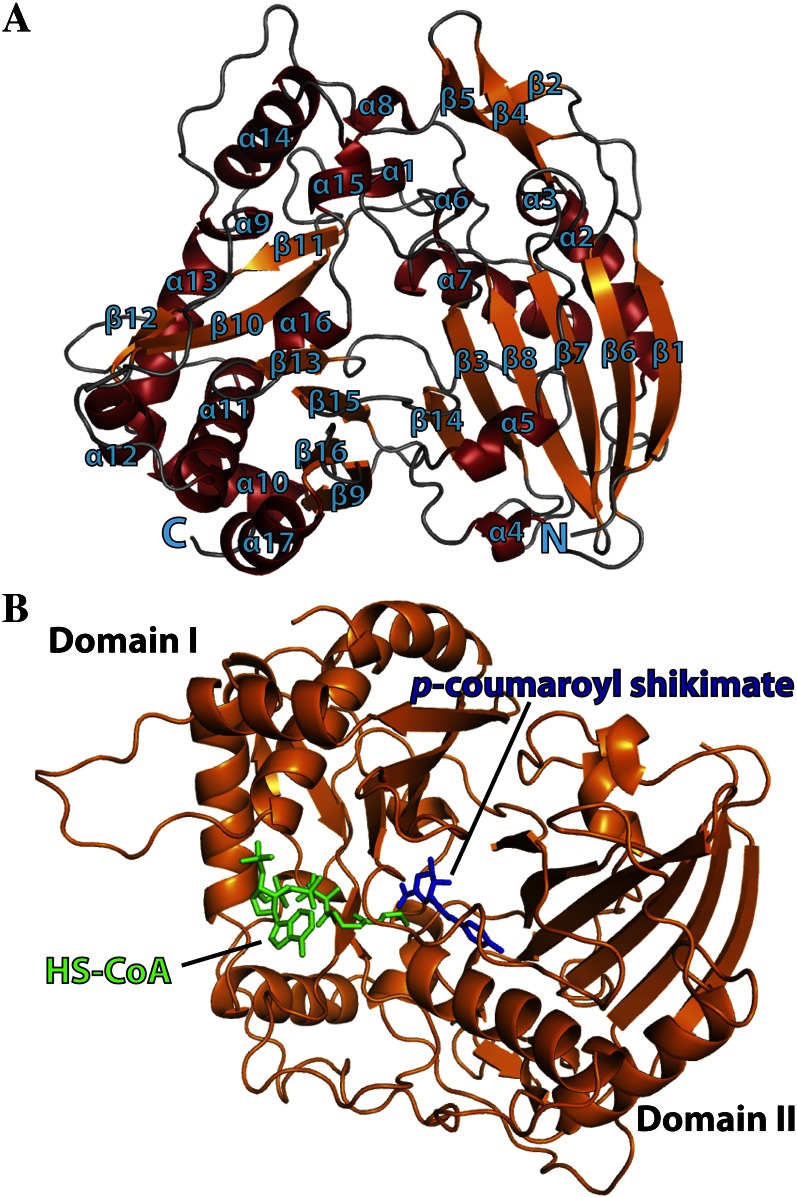
The global structure of the SbHCT molecule contained two domains (domain I and domain II) consisting of 16 β-strands and 17 α-helices (Fig. 1). Domain I included residues 1 to 193 and 389 to 409, while domain II included residues 194 to 388 and 410 to 448. Both domains displayed a mixed β-sheet in their cores flanked by α-helices on both sides of the sheet. The two β-sheets in the core of each domain had a similar shape, even though some of the β-strands in each sheet had different directionality. Specifically, the β-sheet constituting the core of domain I was arranged β1-β6-β7-β8-β3-β14, while the β-sheet of domain II was ordered as β9-β16-β15-β13-β10-β11-β12. In addition, domain I has another small protruded, antiparallel β-sheet in the order β2-β5-β4, which was oriented nearly perpendicular to the mixed β-sheet core of domain I. Although the domain structures were superimposable, sequence identity between the β-sheet amino acids of each domain was relatively low (approximately 10%–12%). The residues of the two core β-sheets were of hydrophobic nature. Consequently, the hydrophobic residues interacted with the hydrophobic side of the amphiphilic α-helices, whose hydrophilic faces were solvent exposed. Overall, the surface residues of SbHCT were very hydrophilic, explaining its high solubility.
Figure 1.
Ribbon diagram representing the crystal structure of SbHCT with bound p-coumaroyl shikimate and HS-CoA. A, Secondary structure elements have been numbered sequentially as α1 to α17 and β1 to β16. N and C refer to the N- and C-terminal regions, respectively. α-Helices are shown in red and β-strands are shown in orange. B, The p-coumaroyl shikimate is in blue and HS-CoA is in green. The glycosidic bond dihedral angles of the adenosine unit of p-coumaroyl-CoA in both binary and ternary complexes adopt the syn conformation. This figure was generated using Open-Source PyMOL (version 1.4).
The number of peripheral α-helices of each domain was different. Some of these observed α-helices had only one turn, while others displayed a severely bent conformation due to the insertion of Pro or Gly residues in the middle of the corresponding helices. In particular, the five helices connecting β12 and β13 were interrupted several times by Gly and Pro residues. Significantly, some of these helix-breaking sites were mapped at the point of amino acid insertions and deletions when SbHCT was compared with other known structures of acyltransferases, as discussed in detail below.
The Cα positions of the ternary complex were superimposable to those of apo-form SbHCT, with a root mean square deviation value of 0.8 Å excluding the disordered area (residues 219–231). Notably, the backbone of several areas, which included residues 30 to 37, 130 to 140, 369 to 380, and 407 to 413, were shifted more than 2 Å upon complex formation, resulting in a significant closure of the substrate-binding pockets. Some of those shifted regions contained the residues involved in the coordination of substrates.
There were two regions of high-temperature factors in the SbHCT molecule. One region was part of the long crossover (residues 182–236) that linked domain I and domain II. The electron density corresponding to those residues (216PAMLSSEPPQAALTAKPATPP236) was not visible. The other high-temperature factor region was the C-terminal end of α10 and the following loop (residues 257–267), which contained three Arg residues located in the p-coumaroyl-CoA entry site.
Substrate-Binding Site
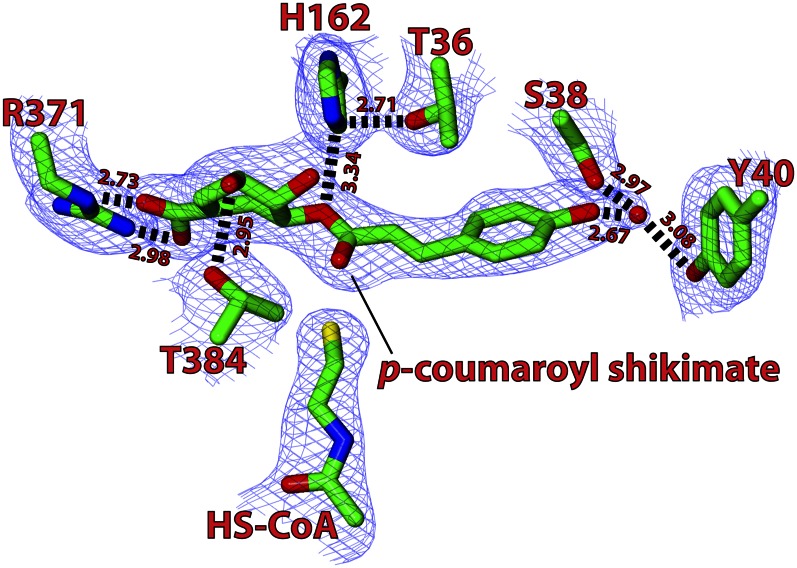
By generating difference Fourier maps (|Fo| − |Fc|; Fo and Fc are observed and calculated structure factors, respectively) of the ternary complex data, it was possible to visualize the position of the bound p-coumaroyl-CoA and shikimate, both of which were located between domains I and II (Figs. 1 and 2) and were clear enough to show the conformation of each molecule. In addition, the corresponding densities indicated that the two molecules had reacted and existed in the product state as free HS-CoA and p-coumaroyl shikimate (Fig. 2).
Figure 2.
Structure of the substrate-binding pocket of SbHCT in the ternary complex. The difference Fourier map contoured at 1.5 σ clearly shows the p-coumaroyl shikimate and HS-CoA reaction products in the ternary complex, with residues important for binding and catalysis highlighted and their residue numbers indicated. The water molecules are shown as red spheres. This figure was generated using CCP4 Molecular Graphics (version 2.5.0).
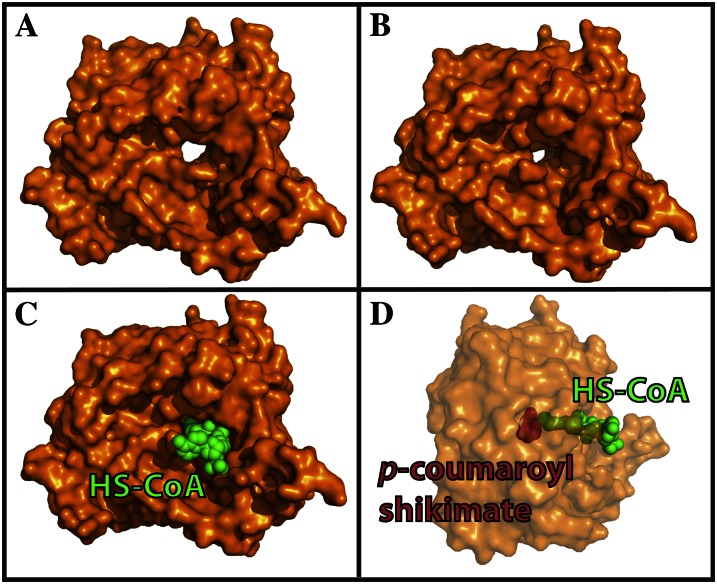
In the apo-structure of SbHCT, the area corresponding to the binding pocket was part of a well-defined tunnel that pierces through the entire body of the protein (Fig. 3). One side of this tunnel was formed by the amino end of α7 and the carboxy ends of both β10 and β13. The other side of the tunnel was formed by the amino end of β3, α15, and α16 and the loop connecting β14 and β15. The residues lining the wall of this tunnel were predominantly hydrophobic. Three polyethylene glycol, one glycerol, and several ordered solvent molecules were located along the path of the tunnel in the apo-form crystal structure. Those small organic molecules from the crystallization buffer, such as polyethelene glycol, were visualized in the p-coumaroyl-CoA tunnel, indicating that the tunnel had a preference for molecules capable of adopting a linear chain formation.
Figure 3.
Surface representation of SbHCT. The channel running through the whole enzyme is clearly visible in apo-form (A), which is significantly constricted in ternary complex form (B). The central cavity piercing through the enzyme and the active site are occupied by a p-coumaroyl shikimate and HS-CoA molecules, shown in space-filling model (C and D). The transparency of the protein surface in D was increased to improve the distinction between p-coumaroyl shikimate (blue) and HS-CoA (green), which is a 90° rotated view from the view of C. [See online article for color version of this figure.]
The pantothenate unit of p-coumaroyl-CoA was located between β10 and β13 of domain II, separating the carboxy ends of those two β-strands and accommodating its fully extended conformation (Figs. 1 and 3). The carbonyl oxygen of the pantothenate unit interacted with the backbone amide of Asp-300. The glycosidic dihedral angle of the adenosine unit of HS-CoA in the ternary complexes adopted the “anti” conformation. Although their distances and angular orientations were not optimal, the guanidinium side chains of Arg-252 and Arg-304 weakly interacted with the phosphate groups of HS-CoA. In addition, the 3′ and 5′ phosphates attached to the adenosine moiety were positioned within positive dipoles contributed by main-chain N-H ends of the α11 helix. The adenine ring was located at the surface of the enzyme, exposed to solvents, and without any noticeably strong interaction with enzyme.
Contrary to the weak coordination for HS-CoA, p-coumaroyl shikimate was in noticeably significant interaction with the enzyme. The phenolic moiety of p-coumaroyl shikimate was indirectly anchored to both hydroxyl side chains of Ser-38 and Tyr-40 by an ordered solvent molecule. In addition, the carbonyl oxygen of p-coumaroyl shikimate forms a hydrogen bond with the Nε1 of the Trp-386 indole ring (Fig. 2).
The shikimate moiety in p-coumaroyl shikimate is also coordinated by the neighboring residues. The hydroxyl group on the C3 atom forms a hydrogen bond with the hydroxyl side chains of Thr-384, and its carboxyl group forms a salt bridge with the guanido group of Arg-371. The side chain of Arg-371 is somewhat disordered in the apo-form, but its temperature factors are substantially reduced upon complex formation. The hydroxyl group on the C4 atom forms a hydrogen bond with a water molecule that is in turn connected to a solvent area at the entrance site.
Isothermal Titration Calorimetry
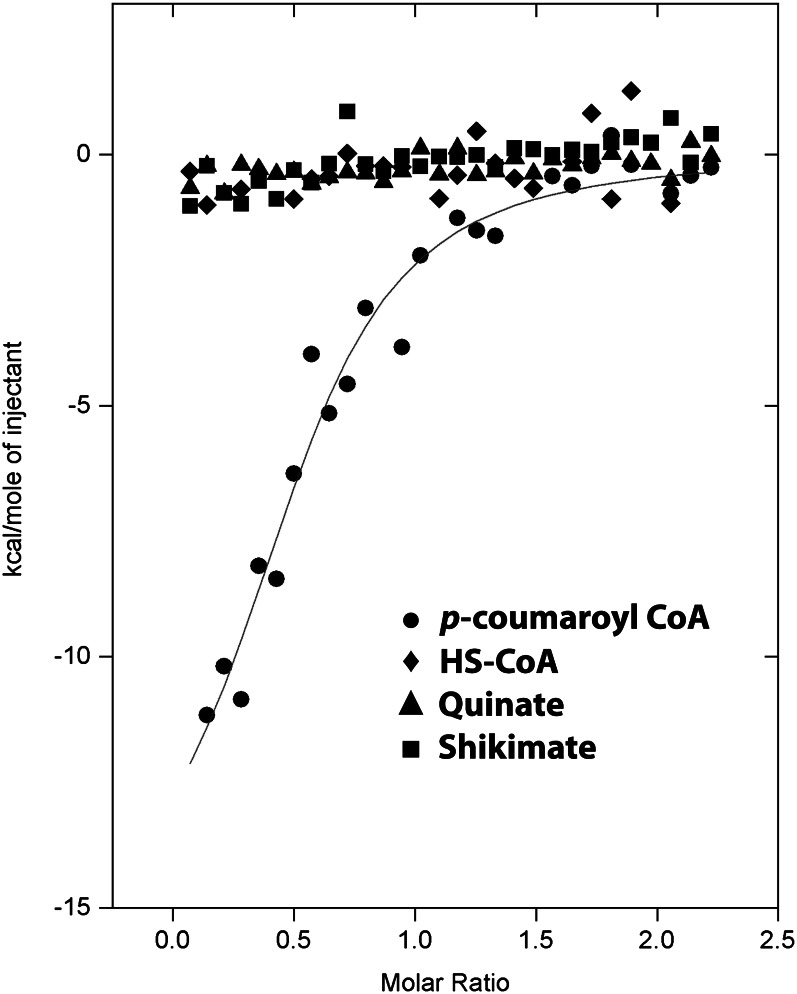
To monitor the differential binding affinities among p-coumaroyl-CoA, HS-CoA, shikimate, and quinate, thermodynamic characterizations were performed with isothermal titration calorimetry (ITC). As shown in Figure 4, a significant amount of heat was released only when SbHCT associated with p-coumaroyl-CoA. The binding interactions with p-coumaroyl-CoA had significant enthalpic contributions, −15.9 kcal mol−1. In addition, ITC data revealed a slightly unfavorable entropic contribution, −26.8 cal mol−1 degree−1, which may mean that the active site of the enzyme was slightly stabilized upon binding to the p-coumaroyl moiety and that only a few solvent molecules were freed from the pocket. Based on the ITC data analysis, the dissociation constant value for p-coumaroyl-CoA was calculated to be 1.6 µm for SbHCT.
Figure 4.
Measurement of substrate binding through ITC experiments. The trends of heat released by serial injections of p-coumaroyl-CoA (circles), HS-CoA (diamonds), quinate (triangles), and shikimate (squares) into the SbHCT solution were monitored. Only p-coumaroyl-CoA showed the typical heat-releasing pattern. Solid lines represent the least square fits of the data.
Characterization of Steady-State Kinetics for SbHCT and Its Mutant Forms
In order to further examine the catalytic roles of the residues implicated by the crystal structures, a set of mutant SbHCT complementary DNAs (cDNAs) were generated via site-directed mutagenesis and expressed in Escherichia coli. The recombinant proteins in which residues of interest had been replaced with Ala were purified via affinity chromatography and characterized. The six mutants were T36A, S38A, Y40A, H162A, R371A, and T384A. The similar shapes and intensities for the far UV-circular dichroism spectra indicate that the wild type and all six mutants contain a similar amount of secondary structures (Supplemental Fig. S1).
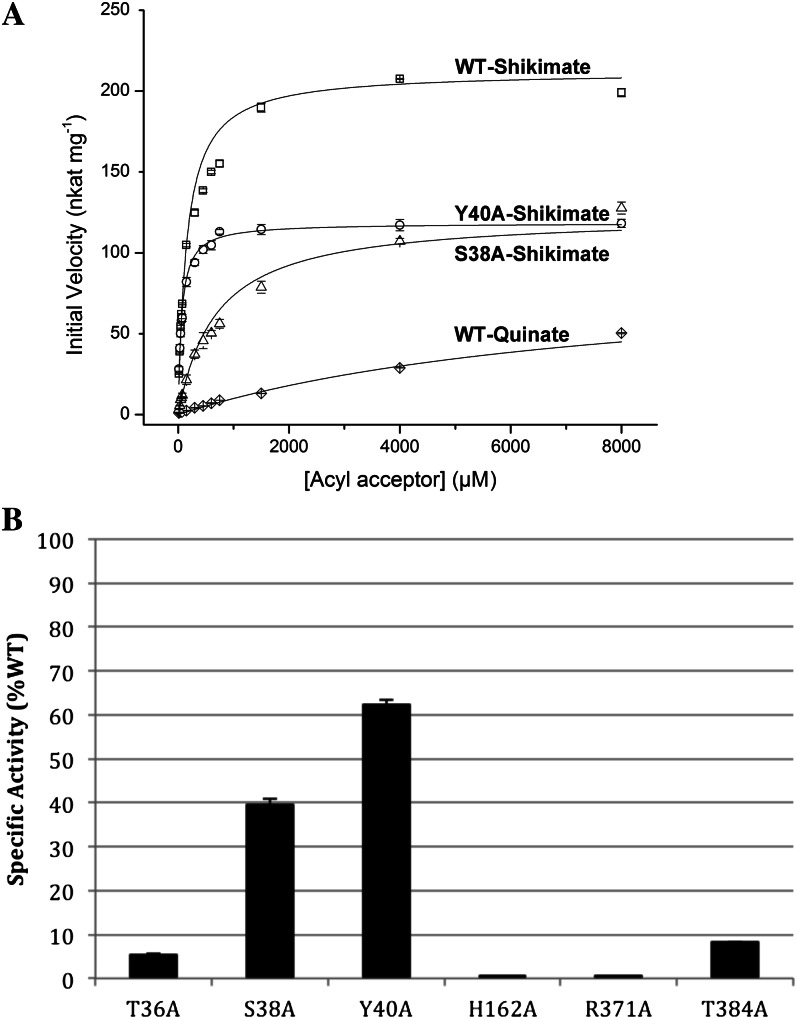
The steady-state kinetics experiments were performed by holding the concentration of wild-type and mutant enzymes, and p-coumaroyl-CoA, constant at 80.5 nm and 0.05 mm, respectively, while shikimate concentrations were varied from 15 μm to 8 mm (Fig. 5A). The calculated Km value for shikimate of wild-type SbHCT was 153 ± 12.4 μm. The Vmax value of wild-type SbHCT was 212 ± 3.96 nkat mg−1 (nmol s−1 mg−1). The Km values for shikimate of S38A and Y40A mutant SbHCT were 678 ± 62.1 and 61.3 ± 3.7 μm, respectively. The Vmax values of these two mutant SbHCTs were 123 ± 5.57 and 118 ± 1.91 nkat mg−1, respectively. The specific activity of wild-type SbHCT was 558 ± 6.75 nkat mg−1 at 750 μm shikimate, while the specific activities of T36A, S38A, Y40A, and T384A were 6%, 40%, 62%, and 9% of that of the wild type, respectively. H162A and R371A displayed no detectable activity (Fig. 5B). Although S38A and Y40A still displayed some residual activity, the reduction in activity in all six mutants confirms the importance of these residues for enzyme function.
Figure 5.
Activity of SbHCT mutants. Mutation of the active-site residues identified in the crystal structure results in the reduction of SbHCT kinetic activity. A, Michaelis-Menten kinetics for the wild type (WT) and S38A and Y40A mutants of SbHCT. B, Relative specific activities of mutants compared with that of wild-type SbHCT. The activities were measured at 750 µm shikimate and compared with the wild type in percentage terms.
In addition, by holding the same concentration of wild-type HCT and p-coumaroyl-CoA constant at 80.5 nm and 0.05 mm, respectively, quinate concentrations were varied from 15 μm to 8 mm (Fig. 5A). As shown in Figure 5A, it has low activity and a continuously rising curve; thus, meaningful kinetic parameters for wild-type SbHCT with quinate could not be determined. The specific activity of wild-type SbHCT at 750 μm quinate was 8.95 ± 0.06 nkat mg−1.
Identification of Related BAHD Family Members via Distance Matrix Alignment and BLAST Searches
All BAHD family members contain an HXXXD motif, and some of them also share a DFGWG motif with minor degeneracy in the sequence (St-Pierre and Luca, 2000; Burhenne et al., 2003). The HXXXD motif contains a catalytic His residue, offering an explanation for its conservation across the members of the BAHD superfamily, whereas the DFGWG motif is usually located distal to the active site (Ma et al., 2005; Lallemand et al., 2012). Despite the presence of these two conserved motifs, members of the BAHD superfamily have varying acyl donor specificities that are difficult to predict by sequence alignment and other conventional bioinformatics tools (Luo et al., 2007).
In order to establish the structural classification for SbHCT and identify its closely related structural homologs in the PDB, a Distance Matrix Alignment search was performed (Holm and Sander, 1993). The results showed that the highest match was to HCT from coffee (4G0B), with a Z-score of 58.5, which was followed by vinorine synthase from Rauvolfia serpentina (2BGH), with a Z-score of 35.3, trichothecene 3-O-acetyltransferase from Fusarium sporotrichioides (2ZBA; Z-score = 34.8), malonyltransferase from tobacco (Nicotiana tabacum; 2XR7; Z-score = 33.8), and malonyltransferase from Chrysanthemum × morifolium (2E1T; Z-score = 33.5). Subsequent proteins had much lower Z-scores; for example, choline acetyltransferase from rat (1T1U) was 10.7 and carnitine acetyltransferase from mouse (1NDB) was 5.8. Significantly, all the enzymes with high Z-scores were CoA-dependent transferases and synthases.
Additionally, a search for proteins with similar amino acid sequences in the PDB using BLAST (Altschul et al., 1990) revealed that HCT from coffee (4G0B) showed the highest similarity (75%) to SbHCT, followed by vinorine synthase (2BGH; 43%), two malonyltransferases (2XR7 [42%] and 2E1T [46%]), and trichothecene 3-O-acetyltransferase (2ZBA; 39%). Like HCT, vinorine synthase also belongs to the BAHD superfamily (Ma et al., 2005). Except for those four enzymes, the corresponding levels of sequence similarity for the following enzymes in the BLAST list were quite low.
In general, the CoA-dependent enzymes with the highest scores of similarity had two mixed β-sheets at the core, which resembled SbHCT in terms of size and relative orientation between two domains. However, the structural heterogeneity was apparent among those CoA-dependent enzymes in terms of backbone topology and number of secondary structural elements, which correlated with their great diversity of function.
In SbHCT, the conserved HXXXD motif (162HHVAD166) is located on α6, which is the interface between the two domains (Fig. 1). The catalytic His-162 in this motif is located in the middle of the tunnel and is accessible from both sides of the SbHCT molecule through the tunnel. On one side of the His-162 residue, one-half of the tunnel was established by splitting two antiparallel β-strands of domain I, the amino ends of β3 and the carboxy end of β14. The other side was established by splitting the carboxy ends of two parallel β-strands, β10 and β13 of domain II.
The carboxyl side chain of conserved residue Asp-166 in the HXXXD motif established a salt bridge with Arg-302, which is completely conserved among the other enzymes with high Z-scores. These two salt bridge-forming residues, Asp-166 and Arg-302, were located at the apex positions of domains I and II, respectively, which could lock the interdomain motion. Therefore, this interaction could serve a critically important role in maintaining the proper interdomain distance and conformation rather than being directly involved in the reaction mechanism. On the other hand, the two animal transferases, carnitine acetyltransferase from mouse (1NDB) and choline transferase from rat (1T1U), showed an identical conformation of the carboxyl side chain of the Asp (or Glu in the case of carnitine acetyltransferase) in the HXXXD motif, such that it can interact simultaneously with Arg and His that are in the corresponding positions of Arg-302 and His-162 in SbHCT. In SbHCT, however, the distance between the carboxyl oxygen atom of Asp-162 and the Nδ1 atom of the His-162 observed in SbHCT is longer than a normal hydrogen bond in those structures.
The 395DFGWG399 motif is located between β13 and β14, which belong to the core β-sheets of domain I and domain II, respectively (Fig. 1). Considering its distance from the active site, the direct involvement of the 395DFGWG399 motif in the enzymatic reaction is very unlikely. Instead, by inserting two hydrophobic side chains of Phe-396 and Trp-398 in the DFGWG motif into the hydrophobic core of domain II, this highly conserved motif may instead be involved in stabilizing the overall two-domain structure of SbHCT and other enzymes in the BAHD superfamily.
DISCUSSION
Substrate Binding and Catalytic Mechanism
Based on the positions of p-coumaroyl shikimate and HS-CoA molecules, it is likely that shikimate and p-coumaroyl-CoA enter the active site through their unique entry sites on opposite faces of the enzyme instead of sharing both entry sites. A dual-port system as observed in SbHCT has also been proposed for other BAHD members based on crystal structures and kinetic data (Keating et al., 2002; Ma et al., 2005). Notably, the size and arrangement of the secondary structural elements constituting the entrance site for the CoA are very similar among the above-mentioned CoA-dependent transferases of high similarity (Supplemental Fig. S2). On the other hand, the entry sites for the proposed substrates all have different sizes and arrangements of secondary structural elements, probably dictating the substrate preference for an individual enzyme supporting two independent entry sites. Considering the substantial closure of the binding pockets, the significant affinity of SbHCT for p-coumaroyl-CoA (1.6 µm), and the negligible affinity for shikimate in our ITC data, it is likely that p-coumaroyl-CoA preconditions the binding of shikimate.
The binding pocket for shikimate is predominantly hydrophobic, being surrounded by Val-31, Pro-32, Ala-298, Ile-318, Phe-376, Leu-414, Phe-416, and Leu-418. In general, those residues were not conserved among the compared enzymes, other than being hydrophobic in nature (Supplemental Fig. S2). In the midst of those apolar residues were two polar residues, Arg-371 and Thr-384, at strategic locations for binding a shikimate molecule (Fig. 2) that diffused in through this short, solvent-exposed channel. However, our ITC data showed a negligible affinity of apo-SbHCT for shikimate, reflecting the weak nature of the interaction between the two without p-coumaroyl-CoA.
In the ternary complex of SbHCT, the imidazole ring of His-162 was in close proximity to the C5 atom of the shikimate moiety. Thus, the attached hydroxyl oxygen that couples shikimate to the p-coumaroyl moiety were positioned close (3.3 Å) to the Nε2 atom of imidazole (Fig. 2). The same Nε2 atom also formed a hydrogen bond with the hydroxyl side chain of Thr-36, possibly leading to proper orientation or modulation of pKa for catalysis. In addition, the Nδ1 atom of the imidazole ring in the ternary complex forms a hydrogen bond with both carbonyl oxygens of Pro-32 and Phe-34 (Fig. 6). Such coordinating interaction for Pro-32, Phe-34, and Thr-36 was not observed in the apo-form. Thus, the side chain of His-162 showed significant conformational differences between the apo-form and ternary complex structures of SbHCT. Upon superimposing the backbones of the compared CoA-dependent transferases in their apo-form, the flexible nature of this His residue was obvious.
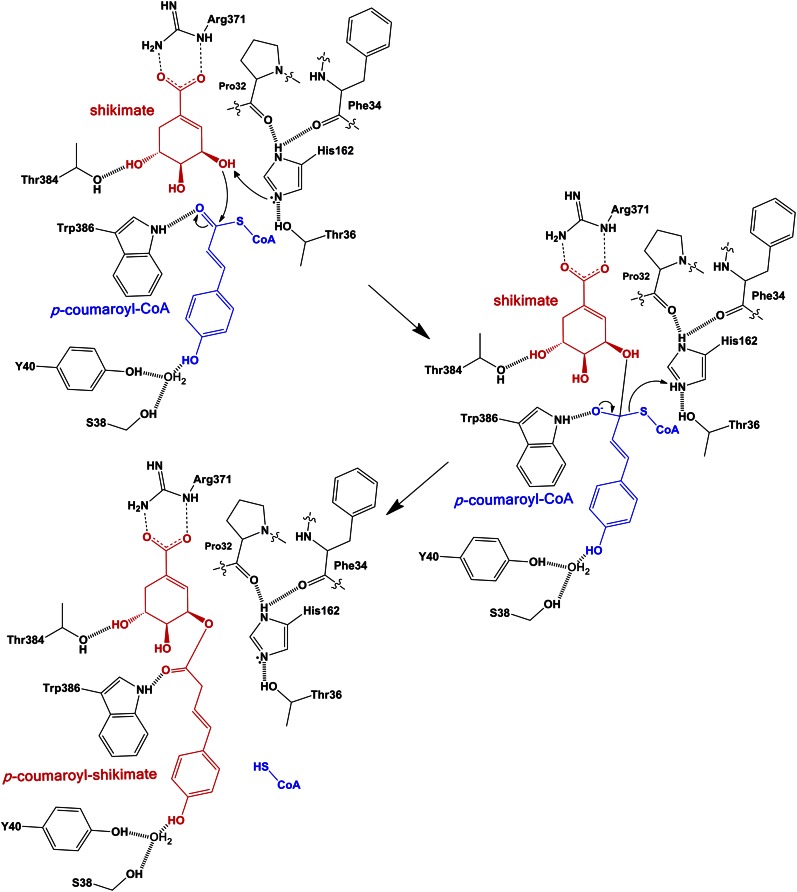
Figure 6.
Proposed catalytic mechanism of SbHCT. His-162 serves as a general base by abstracting a proton from shikimic acid during catalysis. The oxyanion in the tetrahedral transition state is stabilized by forming hydrogen bonds with both side chains of Thr-384 and Trp-386. The arrows indicate the movement of two electrons. This figure was drawn with ChemDraw 12.0. [See online article for color version of this figure.]
In the catalytic mechanism (Fig. 6), based on the combined structural and kinetic data, the imidazole Nε2 atom of His-162 with the aid of Thr-36 can be properly oriented for abstracting a proton from the hydroxyl group at the C3 atom of shikimate, acting as a general base and enabling a nucleophilic attack of the resulting oxyanion on the γ-carbon of p-coumaroyl-CoA. Consistent with this model, the H162A mutant did not have any significant activity and the T36A mutant had significantly reduced activity (Fig. 5).
The oxyanion of the tetrahedral intermediate is in a hydrophobic environment and greatly stabilized by forming a hydrogen bond with the properly positioned side chains of the above-mentioned Trp-386 (Fig. 6). Therefore, SbHCT has a well-adopted active site for preferential binding to a transition state. In the apo-form, the corresponding position for the oxyanion is occupied by a water molecule. The formation of p-coumaroyl shikimate is completed with the release of HS-CoA, with the proton being donated by His-162 (Fig. 6), which showed very weak affinity to the enzyme (Fig. 4).
Substrate Specificity of SbHCT and Other Members of the BAHD Family
The BAHD family comprises a diverse group of enzymes that share structural features despite limited sequence similarity. These features include the two-domain structure with distinct α-helices and β-sheets as well as a central tunnel that pierces the entire enzyme. The proposed reaction mechanism whereby the different molecules participating in the reaction enter and exit from opposite sides of the tunnel would enable the conservation of structural features adapted to one compound (e.g. hydroxycinnamoyl-CoA esters), while the structural features adapted to the other compound can be varied, ultimately leading to the ability to synthesize a wide array of compounds using a similar design. In SbHCT, most of the residues in contact with p-coumaroyl shikimate displayed a flexible nature in its apo-form, but upon complex formation, they establish a tight fit together with domain motion. Therefore, more detailed knowledge of the structure of this class of enzymes and elucidation of the reaction mechanisms may lead to the ability to synthesize novel compounds with, for example, medicinal properties.
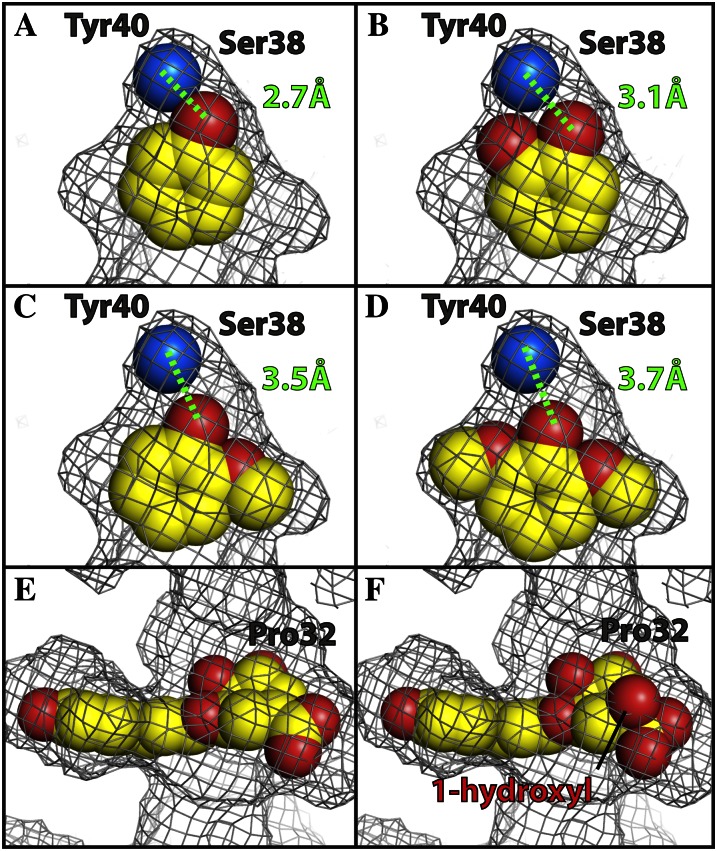
The proposed reaction mechanism is well supported by the structural and kinetic analyses as well as by observations with the mutant SbHCTs. The residual activity in S38A and Y40A is consistent with the subtle role of these residues in substrate binding. The phenolic moiety of p-coumaroyl shikimate was indirectly anchored to both hydroxyl side chains of Ser-38 and Tyr-40 by an ordered solvent molecule (Fig. 7A). Due to the tightness of the corresponding binding pocket, the modeled caffeoyl-CoA needs to be shifted in order to accommodate its 3- and 4-hydroxyl groups, which would lead to a slightly weaker interaction (Fig. 7B). The binding of feruloyl-CoA (Fig. 7C) or sinapoyl-CoA (Fig. 7D), which contain methoxy groups, one (on C3) and two (on C3 and C5), respectively, would not fit without a serious adjustment of backbone residues constituting the pocket. In addition, their hydrophobic methoxy group(s) will significantly slow the diffusion of the corresponding CoA adduct along the tunnel of the same hydrophobic nature, affecting the turnover rate of the enzyme.
Figure 7.
Steric fit of p-coumaroyl-CoA, shikimate, quinate, and other modeled CoA compounds into the binding pocket of SbHCT. A, p-Coumaroyl-CoA:4-hydroxyl group (red) forms a hydrogen bond (2.7 Å) with the water molecule (blue), coordinated by hydroxyl side chains of Ser-38 and Tyr-40. B to F, Caffeoyl-CoA (B), feruloyl-CoA (C), sinapoyl-CoA (D), shikimate (E), and modeled quinate (F), of which the 1-hydroxyl group (highlighted) makes a severe steric clash with Pro-32. The gray grid represents the boundary of the van der Waals surface of the substrate-binding pocket. As indicated with the black line, the 1-hydroxyl group of quinate is bulging through this boundary specifically at the location of Pro-32. This figure was drawn with Open-Source PyMOL (version 1.4). [See online article for color version of this figure.]
The binding pocket of shikimate (Fig. 7E) was also very tight. Thus, accommodating quinate, which has a 1-hydroxyl group and a 1-carboxyl group in a tetrahedral geometry compared with the trigonal one observed in shikimate, requires a substantial cost for both entropy and enthalpy, significantly affecting its dissociation constant. The 1-hydroxyl group of the modeled quinate illustrates a large steric clash with Pro-32 and can be seen bulging out of the calculated van der Waals surface of the pocket (Fig. 7F). Consequently, the activity of SbHCT for quinate was quite low (Fig. 5A). Considering the fact that most of the residues constituting the binding pockets of SbHCT and CcHCT are identical (Supplemental Fig. S2), the reported increased Km value of quinate (430 μm) for CcHCT compared with that of shikimate (75 μm; Lallemand et al., 2012) is also likely due to steric clash. Therefore, the production of compounds other than p-coumaroyl shikimate by wild-type SbHCT should be very minimal. The availability of both apo-form and ternary complex structures of SbHCT allows for a sophisticated engineering of binding pockets for various CoA esters and quinate/shikimate in order to derive the desired products, which could prove to be more effective than previous attempts based on the apo-form CcHCT structure (Lallemand et al., 2012).
The Potential Role of HCT in Determining Lignin Subunit Composition
As stated in the introduction, grasses contain a higher proportion of p-hydroxyphenyl residues in their lignin than angiosperm dicots. As transgenic Arabidopsis and alfalfa plants in which HCT has been down-regulated accumulate higher proportions of p-hydroxyphenyl residues in their lignin, a possible explanation for the higher p-hydroxyphenyl content in grass lignin could be a less efficient conversion of p-coumaroyl-CoA to caffeoyl-CoA, generating a larger substrate pool of p-coumaroyl-CoA for conversion to p-coumaraldehyde by cinnamoyl-CoA reductase. This may be due to grasses having lower activity of HCT, C3′H, or both. Kinetic parameters of HCT from tobacco (NtHCT) and coffee (CcHCT), both angiosperm dicots, have been reported by Hoffmann et al. (2004) and Lallemand et al. (2012), respectively, and can thus serve as the basis for comparison with SbHCT, assuming that CcHCT is involved in the biosynthesis of chlorogenic acid and monolignols and that the experimental conditions used to determine the kinetic parameters were similar to those used for SbHCT. Km values for shikimate were 75 μm for CcHCT, 750 μm for NtHCT, and 153 μm for SbHCT. The Vmax values were 13.55 nkat mg−1 for CcHCT, 0.14 nkat mg−1 for NtHCT, and 212 nkat mg−1 for SbHCT, suggesting that NtHCT has the lowest affinity for shikimate, the lowest catalytic activity, and that SbHCT is intermediate both in terms of substrate affinity and catalytic activity. Based on these kinetic data, we have no clear evidence of HCT acting as an inherent control point of coniferyl and sinapyl alcohol synthesis. Combined with the suggestion that alfalfa HCT may carry out a reversible (Lee et al., 2011) conversion of p-coumaroyl shikimate and HS-CoA to p-coumaroyl-CoA and shikimate, it would be worth comparing the kinetic parameters of C3′H in grasses versus angiosperm dicots as a possible cause for the higher proportion of p-hydroxyphenyl residues in grasses.
Balancing the Impact on Lignin Subunit Composition with Plant Growth
The down-regulation of HCT in Arabidopsis and alfalfa leads to smaller plants as a direct consequence of changes to their lignin (Shadle et al., 2007; Li et al., 2010). While the biomass from the resulting transgenic plants can be more easily converted to fermentable sugars, the overall yield will be reduced due to the small plant size. A reduction, rather than an elimination of HCT activity, may offer a way to maximize sugar yield. Site-directed mutagenesis based on the crystal structures of apo-form and ternary complex of SbHCT and other members of the BAHD superfamily allowed us to generate several SbHCT mutants with reduced activity, namely T36A, T384A, S38A, and Y40A, which have 6%, 9%, 40%, and 62% activity compared with that of the wild type, respectively. These mutants, which affect substrate affinity and specificity, are prime candidates for engineering HCT, which could modify the cell wall composition of sorghum without any significant negative effect on plant growth. However, those mutations affecting catalytic residues, such as H162A and T384A, or the residues critical for the substrate affinity, such as R371A, completely abolished the enzymatic activity and, thus, cannot be candidates for any gene manipulation. The use of Targeting Induced Local Lesions in Genomes populations to identify mutants in which these key residues have been altered, or the use of novel allele-editing tools such as zinc finger nucleases (Wright et al., 2005; Shukla et al., 2009) or transcription activator-like effector nucleases (Christian et al., 2010; Miller et al., 2011), can be explored to further investigate this in sorghum.
MATERIALS AND METHODS
Chemicals
Chemicals were obtained from Sigma-Aldrich or Fisher Scientific. Crystallization screens were obtained from Hampton Research.
Cloning and Subcloning of SbHCT
Sorghum (Sorghum bicolor) HCT (Sb04g025760.1) cDNA was amplified by reverse transcription-PCR and cloned. Total RNA isolated from 6-week-old stalks was reverse transcribed using the First Strand cDNA Synthesis Kit and an oligo(dT) primer (Roche). HCT cDNA was PCR amplified using the following primers (SbHCT-F, 5′-GCTACCTGCTTTTACACCGC-3′; SbHCT-R, 5′-TAAACCAAACGGCACAAAAA-3′) and Turbo Pfu polymerase (Stratagene) following the manufacturer’s protocol and cloned into Zero-Blunt plasmid vector (Invitrogen) following the manufacturer’s protocol. The HCT coding region was PCR amplified from the vector with the following primers (SbHCT_PciI-F, 5′-CAAACATGTATATCACGGTGCGGGGT-3′; SbHCT_EcoRI-R, 5′-TTCGAATTCAGAAGTCGTAGATGAG-3′) for cloning into the NcoI and EcoRI restrictions sites of pET30a (Novagen; now EMD Millipore) following PciI and EcoRI digestion of the PCR fragment. The digested vector and PCR fragment were ligated overnight at 16°C with T4 ligase. The DNA sequence of the inserted fragment was verified by DNA sequencing. Rosetta (DE3) pLysS competent cells (EMD Millipore) were used for transformation to enhance the expression of eukaryotic protein.
The site-directed mutations described within this article were created in the HCT coding region through PCR-based plasmid amplification using the QuikChange II Site-Directed Mutagenesis protocol and Turbo Pfu polymerase (Stratagene; now Agilent). Complementary plus- and minus-strand oligonucleotides containing corresponding amino acid changes were used to introduce the nucleotide changes into the pET30 plasmid, followed by DpnI (New England Biolabs) digestion to remove the template prior to the transformation of Escherichia coli. The presence of these point mutations was confirmed by automated DNA sequencing. The oligonucleotides were used for each mutagenesis (Supplemental Table S3).
Expression and Purification of SbHCT
For SbHCT expression, 100 mL of Luria-Bertani medium supplemented with kanamycin (100 µg mL−1) and chloramphenicol (34 µg mL−1) was inoculated with a freezer stock of Rosetta (DE3) cells containing pET30a-SbHCT. After growing overnight at 37°C, this culture was then used to inoculate 1.5 L of Luria-Bertani medium. Cells were grown at 37°C with constant shaking until A600 reached 0.6. At that point, the temperature of the incubator was reduced to 20°C and 0.1 mm isopropyl β-d-thiogalactopyranoside was added to induce SbHCT expression. After incubating for 12 h, the induced cells were harvested by centrifugation at 5,000 rpm for 10 min at 4°C. The resulting cell pellet was resuspended with 25 mL of lysis buffer (20 mm Tris, 300 mm NaCl, and 20 mm imidazole, pH 8.0) and was sonicated six times for 10 s each (model 450 sonifier; Branson Ultrasonics). Lysates were cleared by centrifugation at a speed of 15,000 rpm for 30 min.
The resulting supernatant was mixed with 30 mL of nickel-nitrilotriacetic acid agarose (Qiagen) that had been preequilibrated with the same lysis buffer and placed into a column. After washing the column with 400 mL of wash buffer (20 mm Tris, 300 mm NaCl, and 20 mm imidazole, pH 8.0), the SbHCT was then eluted with elution buffer (20 mm Tris, 300 mm NaCl, and 200 mm imidazole, pH 8.0). The corresponding fractions containing SbHCT (200 mm imidazole) were desalted and concentrated into 20 mm Tris (pH 8.5) by ultrafiltration with an Amicon 8050 cell with a 30-kD cutoff membrane (Millipore). The concentrated SbHCT solution, without His-tag cleavage, was applied to a Mono-Q column (GE Healthcare) that was preequilibrated with 20 mm Tris buffer (pH 8.5) at a flow rate of 2.0 cm3 min–1. The catalytically active SbHCT was eluted at approximately 200 mm NaCl with a linear gradient.
All the purification steps were analyzed by electrophoresis on a 12% Tris-Gly SDS-PAGE gel stained with Coomassie blue. Corresponding protein concentrations were determined using the Bio-Rad protein assay kit, with bovine serum albumin as a standard. The overall yield for SbHCT was 10.4 mg L−1.
Synthesis and Purification of p-Coumaroyl-CoA
Sb4CL was expressed and purified (Saballos et al., 2012) in an identical manner to SbHCT, with a final concentration of approximately 8 mg mL−1. p-Coumaroyl-CoA was generated by ligation of p-coumaric acid with CoA catalyzed by the purified Sb4CL in a 40-mL reaction containing 50 mm Na2HPO4 (pH 7.1), 5 mm MgCl2, 2 mm p-coumaric acid, 2.5 mm ATP, 800 µm HS-CoA, and 410 µg of 4CL. The reaction was incubated at room temperature in the dark for 4 h and heat inactivated by boiling for 10 min. Formation of the reaction product, p-coumaroyl-CoA, was monitored at 333 nm using the previously described extinction coefficient (ε = 21 mm−1 cm−1; Rautengarten et al., 2010). Reaction products were then cleared of Sb4CL protein by ethanol precipitation for 6 h at −20°C. The resulting solution was HPLC purified with a semipreparative reverse-phase C18 column (XTerra MS C18, 2.5 µm, 10 × 50 mm; Waters) running at 4 mL min−1. A flow of 0.3 mL min−1 and a gradient of solvent A (0.5% trifluoroacetic acid) and solvent B (acetonitrile) were applied (0–1.3 min, 10% B isocratic; 1.3–6.3 min, 10%–30% B linear; 6.3–10.1 min, 30% B isocratic; 10.1–11.4 min, 30%–35% B linear; 11.4–13.9 min, 35%–100% B linear; 13.9–15.2 min, 100% B isocratic; 15.2–15.7 min, 100%–10% B linear; 15.7–17.5 min, 10% B isocratic).
Multiangle Laser Light Scattering
The weight-average molecular mass of SbHCT was measured by combined size-exclusion chromatography and multiangle laser light scattering. A total of 100 μg of SbHCT was loaded onto a BioSep-SEC-S 2000 column (Phenomenex) and eluted isocratically with a flow rate of 0.5 mL min−1. The eluate was passed through tandem UV detector (Gilson), Optilab interferometric refractometer (Wyatt Technology), and Dawn laser light-scattering detector (Wyatt Technology). Scattering data were analyzed using the Zimm fitting method with software (ASTRA) provided by the instrument manufacturer.
Crystallization and Data Collection
Apo-form crystals of recombinant SbHCT were grown at 4°C using hanging-drop vapor diffusion. The solution of SbHCT (10 mg mL−1) in 20 mm Tris (pH 8.5) was mixed with an equal volume of reservoir (0.1 m MES monohydrate, pH 6.0, and 22% [v/v] polyethylene glycol 400) and equilibrated against the reservoir solution. Diffraction-quality crystals appeared after 14 to 20 d. The apo-form crystals of SbHCT belonged to hexagonal space group P65 with unit cell dimensions of a = b = 135.9 Å, c = 64.0 Å, α = β = 90°, γ = 120°. There is one SbHCT molecule in the asymmetric unit. Diffraction data up to 2.0 Å resolution were collected at the Berkeley Advanced Light Source (ALS; beam line 8.2.1). For data collection at 100 K, crystals were transferred stepwise to a cryoprotection solution containing all components of the reservoir and an additional 20% glycerol. Apo-form crystals were soaked with a reservoir solution containing approximately 5 mm p-coumaroyl-CoA and 5 mm shikimic acid to obtain ternary complex with substrate and cofactor. The crystal-substrate-cofactor mixture was incubated for 1.5 h at 4°C prior to looping. Diffraction data were collected up to 2.4 Å. The ternary data were collected at the Berkeley ALS (beam line 8.2.2). Diffraction data were processed and scaled with the HKL2000 package (Otwinowski and Minor, 1997). The statistics for the diffraction data are listed in Table I.
Table I. Crystallographic data and refinement statistics of SbHCT.
| Parameter | Apo-Form | Complex |
|---|---|---|
| Data | ||
| Beam line | ALS 8.2.1 | ALS 8.2.1 |
| Wavelength (Å) | 1.0 | 1.0 |
| Resolution (Å) | 50–2.0 | 50–2.4 |
| Space group | P65 | P65 |
| Cell dimensions (Å) | a = 135.9 | a = 137.2 |
| b = 135.9 | b = 137.2 | |
| c = 64.0 | c = 63.6 | |
| Asymmetric unit | 1 | 1 |
| Total observations | 491,611 | 465,320 |
| Unique reflections | 44,714 | 44,241 |
| Completeness (%) | 99.9 (94.0) | 97.0 (90.0) |
| Rsyma,b | 0.076 (0.627) | 0.086 (0.479) |
| Refinement | ||
| Resolution (Å) | 44.5–2.0 | 43.4–2.4 |
| No. of reflections | 43,938 | 25,964 |
| Rcrystc (%) | 18.3 | 20.4 |
| Rfreed (%) | 20.7 | 25.4 |
| r.m.s.d.e bonds (Å) | 0.007 | 0.008 |
| r.m.s.d.e angles (°) | 1.012 | 1.119 |
| No. of atoms | ||
| Protein and ligand | 3,409 | 3,421 |
| Water | 334 | 127 |
Numbers in parentheses refer to the highest shell. bRsym = Σ|Ih − < Ih>|/ΣIh, where < Ih> is the average intensity over symmetry equivalent reflections. cRcryst = Σ|Fobs − Fcalc|/Fobs, where summation is over the data used for refinement. dRfree was calculated as for Rcryst using 5% of the data that was excluded from refinement. eRoot mean square deviation.
Phasing and Refinement
Initial phases of the SbHCT diffraction data were obtained by the molecular replacement method with CcHCT from coffee (Coffea canephora; Lallemand et al., 2012) as a search model and using Phaser (McCoy et al., 2007) as implemented in PHENIX (Adams et al., 2002), which resulted in a clearly interpretable electron density map with many well-defined secondary structural elements. The corresponding amino acids were assigned and manually fitted into this map using the software COOT (Jones et al., 1991; Emsley et al., 2010) and refined using PHENIX (Adams et al., 2002), with the simulated annealing protocol resulting in Rwork 18.3% and Rfree 20.7% for random 5% set. The ternary complex of SbHCT was refined using the apo-form model as input coordinates. The final refinement statistics for the apo-form and ternary complex of SbHCT are provided in Table I. The root mean square deviations from ideal geometry of the final coordinates corresponding to the apo-form are 0.007 Å for bonds and 1.01° for angles. The values for the ternary complex are 0.008 Å for bonds and 1.12° for angles. All coordinates and diffraction data have been deposited in the PDB: 4KE4 (apo-form SbHCT) and 4KEC (ternary complex form SbHCT).
ITC
ITC reactions were carried out with a VP-ITC instrument (MicroCal). SbHCT was prepared for ITC by dialyzing into titration buffer (20 mm Tris, pH 8.5) for 24 h at 4°C. The concentration of protein in the calorimetric reaction cell was diluted to 10 μm. All titrations were performed at 25°C with a stirring speed of 300 rpm and 30 injections (10 μL each). Ligands, p-coumaroyl-CoA, HS-CoA, shikimate, and quinate were diluted to 100 μm into the same titration buffer and injected into the cell containing SbHCT solution, and the heat of binding was observed. Ligands were also titrated against buffer to account for the heat of dilution. Ligand concentrations were adjusted to obtain significant heat of binding, and the time intervals between injections were also adjusted to ensure proper baseline equilibration. All samples were degassed prior to titration.
Enzyme Assays of SbHCT
Kinetic parameters of SbHCT were determined by enzymatic assay. Reaction conditions were 40 mm KH2PO4, pH 6.5, 50 mm p-coumaroyl-CoA, and increasing concentrations of shikimate. The reaction was monitored by the decrease in A342 using the previously determined extinction coefficient (ε = 13.6 mm−1 cm−1; Ulbrich and Zenk, 1980). The reaction was initiated by the addition of 2.4 µg of purified SbHCT. Specific activities of mutant versions of SbHCT were assessed using identical reaction conditions at 750 µm shikimate.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Molecular mass determination/circular dichroism spectra for SbHCT wild type and mutants.
Supplemental Figure S2. Multiple sequence alignment of SbHCT and homologs.
Supplemental Table S1. Oligonucleotides used for site-directed mutagenesis.
Acknowledgments
We thank Tammy Gries for her technical assistance on experiments presented in this paper.
Glossary
- HCT
hydroxycinnamoyltransferase
- C3′H
p-coumarate-3′-hydroxylase
- HST
hydroxycinnamoyl-coenzyme A shikimate hydroxycinnamoyltransferase
- 4CL
4-coumarate coenzyme A ligase
- PDB
Protein Data Bank
- ITC
isothermal titration calorimetry
- ALS
Advanced Light Source
- cDNA
complementary DNA
References
- Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC. (2002) PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr 58: 1948–1954 [DOI] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. (1990) Basic local alignment search tool. J Mol Biol 215: 403–410 [DOI] [PubMed] [Google Scholar]
- Berlin A, Balakshin M, Gilkes N, Kadla J, Maximenko V, Kubo S, Saddler J. (2006) Inhibition of cellulase, xylanase and beta-glucosidase activities by softwood lignin preparations. J Biotechnol 125: 198–209 [DOI] [PubMed] [Google Scholar]
- Boerjan W, Ralph J, Baucher M. (2003) Lignin biosynthesis. Annu Rev Plant Biol 54: 519–546 [DOI] [PubMed] [Google Scholar]
- Burhenne K, Kristensen BK, Rasmussen SK. (2003) A new class of N-hydroxycinnamoyltransferases: purification, cloning, and expression of a barley agmatine coumaroyltransferase (EC 2.3.1.64). J Biol Chem 278: 13919–13927 [DOI] [PubMed] [Google Scholar]
- Chen F, Dixon RA. (2007) Lignin modification improves fermentable sugar yields for biofuel production. Nat Biotechnol 25: 759–761 [DOI] [PubMed] [Google Scholar]
- Christian M, Cermak T, Doyle EL, Schmidt C, Zhang F, Hummel A, Bogdanove AJ, Voytas DF. (2010) Targeting DNA double-strand breaks with TAL effector nucleases. Genetics 186: 757–761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Auria JC. (2006) Acyltransferases in plants: a good time to be BAHD. Curr Opin Plant Biol 9: 331–340 [DOI] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, Cowtan K. (2010) Features and development of Coot. Acta Crystallogr D Biol Crystallogr 66: 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann L, Besseau S, Geoffroy P, Ritzenthaler C, Meyer D, Lapierre C, Pollet B, Legrand M. (2004) Silencing of hydroxycinnamoyl-coenzyme A shikimate/quinate hydroxycinnamoyltransferase affects phenylpropanoid biosynthesis. Plant Cell 16: 1446–1465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm L, Sander C. (1993) Protein structure comparison by alignment of distance matrices. J Mol Biol 233: 123–138 [DOI] [PubMed] [Google Scholar]
- Jones TA, Zou JY, Cowan SW, Kjeldgaard M. (1991) Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr A 47: 110–119 [DOI] [PubMed] [Google Scholar]
- Jung JH, Fouad WM, Vermerris W, Gallo M, Altpeter F. (2012) RNAi suppression of lignin biosynthesis in sugarcane reduces recalcitrance for biofuel production from lignocellulosic biomass. Plant Biotechnol J 10: 1067–1076 [DOI] [PubMed] [Google Scholar]
- Keating TA, Marshall CG, Walsh CT, Keating AE. (2002) The structure of VibH represents nonribosomal peptide synthetase condensation, cyclization and epimerization domains. Nat Struct Biol 9: 522–526 [DOI] [PubMed] [Google Scholar]
- Lallemand LA, Zubieta C, Lee SG, Wang Y, Acajjaoui S, Timmins J, McSweeney S, Jez JM, McCarthy JG, McCarthy AA. (2012) A structural basis for the biosynthesis of the major chlorogenic acids found in coffee. Plant Physiol 160: 249–260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y, Chen F, Gallego-Giraldo L, Dixon RA, Voit EO. (2011) Integrative analysis of transgenic alfalfa (Medicago sativa L.) suggests new metabolic control mechanisms for monolignol biosynthesis. PLoS Comput Biol 7: e1002047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Bonawitz ND, Weng JK, Chapple C. (2010) The growth reduction associated with repressed lignin biosynthesis in Arabidopsis thaliana is independent of flavonoids. Plant Cell 22: 1620–1632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo J, Nishiyama Y, Fuell C, Taguchi G, Elliott K, Hill L, Tanaka Y, Kitayama M, Yamazaki M, Bailey P, et al. (2007) Convergent evolution in the BAHD family of acyl transferases: identification and characterization of anthocyanin acyl transferases from Arabidopsis thaliana. Plant J 50: 678–695 [DOI] [PubMed] [Google Scholar]
- Ma X, Koepke J, Panjikar S, Fritzsch G, Stöckigt J. (2005) Crystal structure of vinorine synthase, the first representative of the BAHD superfamily. J Biol Chem 280: 13576–13583 [DOI] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. (2007) Phaser crystallographic software. J Appl Cryst 40: 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JC, Tan S, Qiao G, Barlow KA, Wang J, Xia DF, Meng X, Paschon DE, Leung E, Hinkley SJ, et al. (2011) A TALE nuclease architecture for efficient genome editing. Nat Biotechnol 29: 143–148 [DOI] [PubMed] [Google Scholar]
- Neish A. (1961) Formation of m- and p-coumaric acids by enzymatic deamination of the corresponding isomers of tyrosine. Phytochemistry 1: 1–24 [Google Scholar]
- Otwinowski Z, Minor W. (1997) Processing of x-ray diffraction data collected in oscillation mode. Methods Enzymol 276: 307–326 [DOI] [PubMed] [Google Scholar]
- Paterson AH, Bowers JE, Bruggmann R, Dubchak I, Grimwood J, Gundlach H, Haberer G, Hellsten U, Mitros T, Poliakov A, et al. (2009) The Sorghum bicolor genome and the diversification of grasses. Nature 457: 551–556 [DOI] [PubMed] [Google Scholar]
- Propheter J, Staggenborg S. (2010) Performance of annual and perennial biofuel crops: nutrient removal during the first two years. Agron J 102: 798–805 [Google Scholar]
- Ralph J, Lundquist K, Brunow G, Lu F, Kim H, Schatz P, Marita J, Hatfield R, Ralph S, Christensen J, et al. (2004) Lignins: natural polymers from oxidative coupling of 4-hydroxyphenylpropanoids. Phytochem Rev 3: 29–60 [Google Scholar]
- Rautengarten C, Baidoo E, Keasling J, Scheller H. (2010) A simple method for enzymatic synthesis of unlabeled and radiolabeled hydroxycinnamate-CoA. Bioenerg Res 3: 115–122 [Google Scholar]
- Rösler J, Krekel F, Amrhein N, Schmid J. (1997) Maize phenylalanine ammonia-lyase has tyrosine ammonia-lyase activity. Plant Physiol 113: 175–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saballos A (2008) Development and utilization of sorghum as a bioenergy crop. In W Vermerris, ed, Genetic Improvement of Bioenergy Crops. Springer, New York, pp 211–248 [Google Scholar]
- Saballos A, Sattler SE, Sanchez E, Foster TP, Xin Z, Kang C, Pedersen JF, Vermerris W. (2012) Brown midrib2 (Bmr2) encodes the major 4-coumarate:coenzyme A ligase involved in lignin biosynthesis in sorghum (Sorghum bicolor (L.) Moench). Plant J 70: 818–830 [DOI] [PubMed] [Google Scholar]
- Sander M, Petersen M. (2011) Distinct substrate specificities and unusual substrate flexibilities of two hydroxycinnamoyltransferases, rosmarinic acid synthase and hydroxycinnamoyl-CoA:shikimate hydroxycinnamoyl-transferase, from Coleus blumei Benth. Planta 233: 1157–1171 [DOI] [PubMed] [Google Scholar]
- Sattler SE, Palmer NA, Saballos A, Greene AM, Xin Z, Sarath G, Vermerris W, Pedersen JF (2012) Identification and characterization of four missense mutations in Brown midrib12 (Bmr12), the caffeic acid O-methyltransferase (COMT) of sorghum. Bioenerg Res 5: 855–865 [Google Scholar]
- Shadle G, Chen F, Srinivasa Reddy MS, Jackson L, Nakashima J, Dixon RA. (2007) Down-regulation of hydroxycinnamoyl CoA:shikimate hydroxycinnamoyl transferase in transgenic alfalfa affects lignification, development and forage quality. Phytochemistry 68: 1521–1529 [DOI] [PubMed] [Google Scholar]
- Shukla VK, Doyon Y, Miller JC, DeKelver RC, Moehle EA, Worden SE, Mitchell JC, Arnold NL, Gopalan S, Meng X, et al. (2009) Precise genome modification in the crop species Zea mays using zinc-finger nucleases. Nature 459: 437–441 [DOI] [PubMed] [Google Scholar]
- St-Pierre B, Luca V (2000) Evolution of acyltransferase genes: origin and diversification of the BAHD superfamily of acyltransferases involved in secondary metabolism. In JT Romeo, R Ibrahi, L Varin, V De Luca, eds, Evolution of Metabolic Pathways. Recent Advances in Phytochemistry, Vol 34. Elsevier Science Ltd, Oxford, pp 285–315 [Google Scholar]
- Ulbrich B, Zenk M. (1980) Partial purification and properties of p-hydroxycinnamoyl-CoA:shikimate-p-hydroxycinnamoyl transferase from higher plants. Phytochemistry 19: 1625–1629 [Google Scholar]
- Vanholme R, Demedts B, Morreel K, Ralph J, Boerjan W. (2010) Lignin biosynthesis and structure. Plant Physiol 153: 895–905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venuto B, Kindiger B. (2008) Forage and bioenergy feedstock production from hybrid forage sorghum and sorghum-sudangrass hybrids. Grassl Sci 54: 188–196 [Google Scholar]
- Vermerris W, Saballos A (2012) Genetic enhancement of sorghum for biomass utilization. In A Paterson, ed, Genetics and Genomics of the Saccharinae. Springer, New York, pp 391–428 [Google Scholar]
- Vermerris W, Saballos A, Ejeta G, Mosier N, Ladisch M, Carpita N. (2007) Molecular breeding to enhance ethanol production from corn and sorghum stover. Crop Sci 47: S142–S153 [Google Scholar]
- Weng JK, Li X, Stout J, Chapple C. (2008) Independent origins of syringyl lignin in vascular plants. Proc Natl Acad Sci USA 105: 7887–7892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wortmann C, Regassa T. (2011) Sweet sorghum as a bioenergy crop for the US Great Plains. In MA dos Santos Bernardes, ed, Economic Effects of Biofuel Production. InTech, Rijeka, Croatia, pp 225–240 [Google Scholar]
- Wright DA, Townsend JA, Winfrey RJ, Jr, Irwin PA, Rajagopal J, Lonosky PM, Hall BD, Jondle MD, Voytas DF. (2005) High-frequency homologous recombination in plants mediated by zinc-finger nucleases. Plant J 44: 693–705 [DOI] [PubMed] [Google Scholar]
- Xin Z, Wang ML, Barkley NA, Burow G, Franks C, Pederson G, Burke J. (2008) Applying genotyping (TILLING) and phenotyping analyses to elucidate gene function in a chemically induced sorghum mutant population. BMC Plant Biol 8: 103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang B, Wyman CE. (2004) Effect of xylan and lignin removal by batch and flowthrough pretreatment on the enzymatic digestibility of corn stover cellulose. Biotechnol Bioeng 86: 88–95 [DOI] [PubMed] [Google Scholar]