GLUP6/GEF is the activator of Rab5 GTPase, and the cycling of GTP- and GDP-bound forms of this regulatory protein is essential for the intracellular transport of proglutelin and α-globulin from the Golgi to PSV and in the maintenance of the general structural organization of the endomembrane system in rice seeds.
Abstract
Rice (Oryza sativa) glutelins are synthesized on the endoplasmic reticulum as a precursor, which are then transported via the Golgi to protein storage vacuoles (PSVs), where they are proteolytically processed into acidic and basic subunits. The glutelin precursor mutant6 (glup6) accumulates abnormally large amounts of proglutelin. Map-base cloning studies showed that glup6 was a loss-of-function mutant of guanine nucleotide exchange factor (GEF), which activates Rab GTPase, a key regulator of membrane trafficking. Immunofluorescence studies showed that the transport of proglutelins and α-globulins to PSV was disrupted in glup6 endosperm. Secreted granules of glutelin and α-globulin were readily observed in young glup6 endosperm, followed by the formation of large dilated paramural bodies (PMBs) containing both proteins as the endosperm matures. The PMBs also contained membrane biomarkers for the Golgi and prevacuolar compartment as well as the cell wall component, β-glucan. Direct evidence was gathered showing that GLUP6/GEF activated in vitro GLUP4/Rab5 as well as several Arabidopsis (Arabidopsis thaliana) Rab5 isoforms to the GTP-bound form. Therefore, loss-of-function mutations in GEF or Rab5 disrupt the normal transport of proglutelin from the Golgi to PSVs, resulting in the initial extracellular secretion of these proteins followed, in turn, by the formation of PMBs. Overall, our results indicate that GLUP6/GEF is the activator of Rab5 GTPase and that the cycling of GTP- and GDP-bound forms of this regulatory protein is essential for the intracellular transport of proglutelin and α-globulin from the Golgi to PSVs and in the maintenance of the general structural organization of the endomembrane system in rice seeds.
Developing rice (Oryza sativa) seeds accumulate three types of storage proteins: glutelin, prolamine, and globulin fractions, which are soluble in dilute acid and alkali, alcohol, and salt solutions, respectively. Rice glutelins are synthesized on the endoplasmic reticulum (ER) as precursors (proglutelins; Yamagata et al., 1982), which are then transported and deposited in the protein storage vacuole (PSV; Krishnan et al., 1986; Yamagata and Tanaka, 1986), where they are proteolytically processed to acidic and basic subunits (Yamagata et al., 1982). α-Globulins are also transported and packaged in the PSV, where together with glutelins they form protein body type II (PB-II; Yamagata et al., 1982; Krishnan et al., 1992). Many storage proteins synthesized on the ER pass through the Golgi apparatus on their way to the PSV (Chrispeels, 1983). These storage proteins are concentrated at the ends of the Golgi cis-cisternae and released as dense vesicles (Chrispeels, 1983; Hohl et al., 1996; Okita and Rogers, 1996). In rice, glutelins and α-globulins are detected within dense vesicles closely associated with the Golgi (Krishnan et al., 1986, 1992), suggesting that the proteins are packaged at the Golgi apparatus and sorted from the trans-Golgi network to the PSV.
We have previously identified eight mutants, endosperm storage protein2 (esp2) and glutelin precursor mutant1 (Glup1) to glup7, which accumulated abnormally high amounts of proglutelin precursors. Genetic analysis showed that these eight mutants can be resolved into four classes (Ueda et al., 2010). Among them, the rice line glup4 was identified as a loss-of-function mutant of Rab5, a small GTPase that regulates membrane vesicle fusion. Intracellular transport of glutelins and α-globulins is disrupted in glup4, where the bulk of these storage proteins are secreted and accumulate in the space formed between the invaginating plasma membrane and the cell wall, forming a paramural body (PMB). Hence, GLUP4/Rab5 is required for the intracellular transport of proglutelins from the Golgi to PSVs in developing rice endosperm (Fukuda et al., 2011). Seeds of glup6, which belongs to the same class as the glup4/rab5 mutant, exhibit a protein pattern similar to glup4 in having elevated proglutelin levels and reduced levels of acidic and basic glutelin subunits, prolamines, and 26-kD α-globulin (Ueda et al., 2010). These observations suggest that glup6 acts in the same pathway responsible for the intracellular transport of proglutelins from the Golgi to the PSV as glup4. As the glup6 gene is unknown, the exact role of GLUP6 protein in the intracellular transport of proglutelins and its possible relationship to Rab5 are unclear.
Rab GTPases are involved in various trafficking pathways, including endocytosis, cytokinesis, and post-Golgi trafficking to the plasma membrane and vacuoles (Woollard and Moore, 2008). Land plants have evolved a unique set of Rab GTPases that presumably reflect the specific demands of plant membrane trafficking. Arabidopsis (Arabidopsis thaliana) contains three Rab5 members, ARA7/RABF2b, RHA1/RABF2a, and ARA6/RABF1. ARA7/RABF2b and RHA1/RABF2a are conventional types of Rab5, while ARA6/RABF1 is unique and found only in plants (Ueda et al., 2001, 2004). These conventional Rab5 isoforms are localized on the Golgi apparatus, the trans-Golgi network, and different populations of multivesicular endosomes/prevacuolar compartments (PVCs) in Arabidopsis and tobacco (Nicotiana tabacum; Sohn et al., 2003; Kotzer et al., 2004; Ueda et al., 2004; Haas et al., 2007; Stierhof and El Kasmi, 2010; Ebine et al., 2011). In rice endosperm, GLUP4/Rab5 is localized on multiple compartments, including the plasma membrane, Golgi apparatus, Golgi-associated dense vesicles, and PSV (Fukuda et al., 2011). These subcellular locations support a role for GLUP4/Rab5 in endocytosis and in trafficking between the Golgi and PSV. In animal cells, the activation of Rab5 is mediated by multiple guanine nucleotide exchange factors (GEFs) such as Rabex5, RIN1, RIN2, RIN3, RME6, Alsin, and ALS2CL, which convert Rab5-GDP to the Rab5-GTP form (Carney et al., 2006). All of the Rab5 GEFs identified to date contain a highly conserved Vps9 catalytic domain, which was originally identified in the yeast (Saccharomyces cerevisiae) GEF, Vps9p (Burd et al., 1996; Horiuchi et al., 1997; Hama et al., 1999). In Arabidopsis, the major GEF, VPS9a, activates all Rab5 members (ARA7, RHA1, and ARA6) to GTP-bound forms. The abnormal PMB structures containing membrane vesicles are more frequently observed in vps9a than in the wild type (Goh et al., 2007). PMBs were first reported as a unique structure that originated from invaginations of the plasma membrane (Marchant and Robards, 1968) and later were proposed to develop by the release of internal vesicles of multivesicular bodies into the paramural space (An et al., 2006b). PMBs are frequently observed in plant tissues in which exocytosis and endocytosis actively occur (Wang et al., 2005; An et al., 2006a, 2006b). This view is consistent with the condition seen in the rice Rab5 glup4 mutant, where glutelin precursors as well as many other components of the endosome system, including β-glucan, assemble to form large PMBs in developing endosperm (Fukuda et al., 2011).
In order to obtain further insights into the intracellular trafficking of storage proteins within the endomembrane system in rice endosperm, we undertook a study to identify the GLUP6 gene as a Rab5-type GEF containing a conserved Vps9 domain using a map-based cloning approach. The phenotypic properties of the glup6/gef mutation were nearly identical to those observed for the loss-of-function mutant of GLUP4/Rab5 and GLUP6/GEF-activated GLUP4/Rab5 by exchanging GDP with GTP. Our results indicate that the GDP/GTP recycling of GLUP4/Rab5 by GLUP6/GEF is a key regulatory process in the trafficking of storage proteins to the PSV and in the general maintenance of the endomembrane system in developing rice endosperm.
RESULTS
The glup6 Mutant Accumulates Large Amounts of Proglutelin
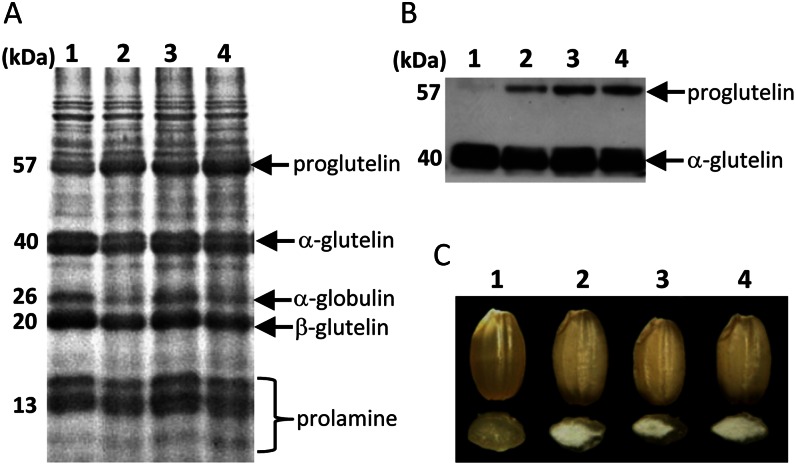
Figure 1A shows the profiles of total seed protein samples from three independent glup6 mutant lines, EM939, EM1327, and EM1484, as well as from the wild type. All three glup6 lines contained elevated amounts of the 57-kD proglutelin polypeptide compared with the wild type. The change in the proglutelin levels between the wild type and the glup6 mutants was readily discernible by immunoblot analysis (Fig. 1B). Figure 1C shows that the grain morphology is also significantly altered in the glup6 mutants. All three mutant lines exhibited a floury endosperm.
Figure 1.
Storage protein composition in seeds of the wild type and glup6 mutant lines. A and B, Seed protein extracts were separated on SDS-PAGE gels (A) and then subjected to immunoblot analysis with anti-glutelin acidic subunit (B). C, The floury endosperm of glup6 mutant lines. Lane 1, the wild type; lane 2, EM939; lane 3, EM1327; lane 4, EM1484. EM939, EM1327, and EM1484 are glup6 allelic mutant lines. [See online article for color version of this figure.]
Supplemental Figure S1 shows the plant phenotypes of the various glup6 mutant lines. Panicle length in EM1327 and primary internode length in EM1327 and EM1484 were reduced significantly compared with the wild type. However, the other mutant lines were similar to the wild type, indicating that the reduction of panicle and primary internode lengths in glup6 lines was not characteristic of the glup6 mutation.
Construction of a Genetic Linkage Map of the GLUP6 Gene
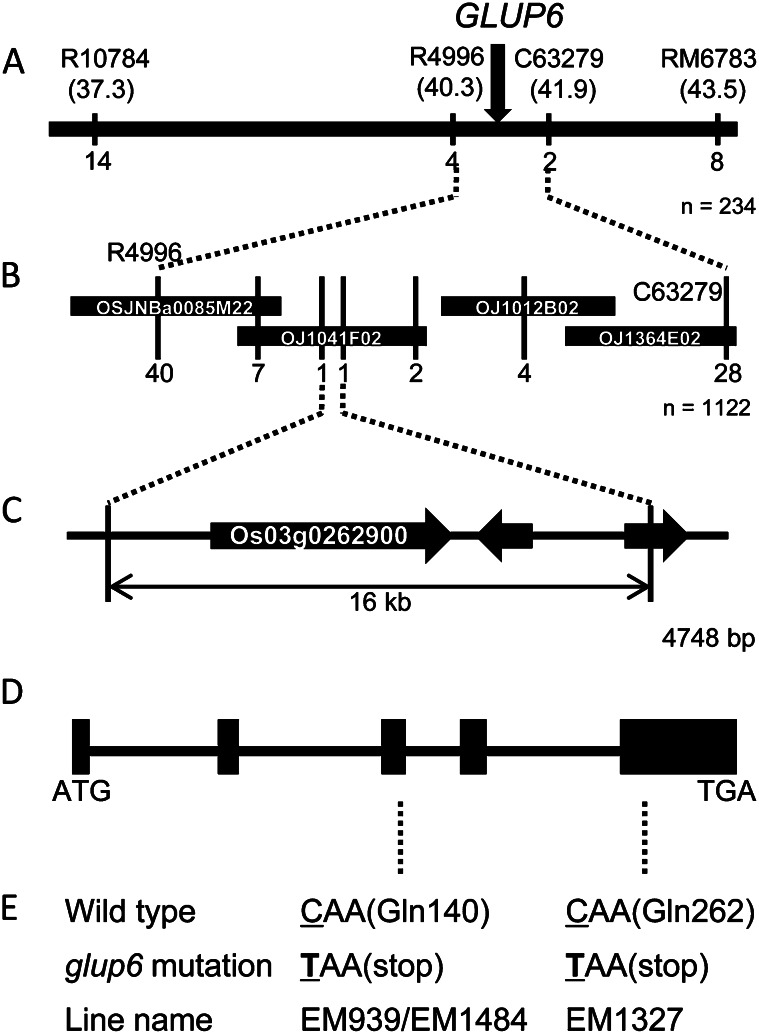
We had previously reported that the GLUP6 gene was located on chromosome 3 (Sato et al., 2003). To identify the GLUP6 gene, a genetic linkage map was constructed (Fig. 2) and then used for linkage analysis using homozygous glup6 plants generated from the F2 progeny of the cross between EM939 and an indica rice cultivar, Kasalath. By studying the linkage analysis of 234 glup6 homozygous plants, the locus was mapped within a 1.6-centimorgan (cM) region between sequence-tagged sites (STS) markers R4996 (40.3 cM from the short arm terminal) and C63279 (41.9 cM) on chromosome 3 (Fig. 2A). A search of the bacterial artificial chromosome (BAC) clone database (International Rice Genome Sequence Project; http://rgp.dna.affrc.go.jp/E/IRGSP/index.html) revealed that the region between R4996 and C63279 corresponded to four overlapping BAC clones. Additional linkage analysis of 1,122 glup6 homozygous plants using STS markers associated with these BAC clones narrowed the GLUP6 locus to within 16 kb of the BAC clone OJ1041F02 (Fig. 2B). A search of the Rice Genome Automated Annotation System (RiceGAAS; http://ricegaas.dna.affrc.go.jp/) revealed the presence of three predicted genes within the 16-kb candidate region of the GLUP6 locus (Fig. 2C).
Figure 2.
Delimiting the candidate genomic region of the GLUP6 gene and the nature of the mutations in various glup6 lines. A, Genetic linkage map of 234 F2 plants showing the relative position of GLUP6 with linkage markers on chromosome 3. The numbers under the linkage map indicate the number of recombinants with the marker. The numbers in parentheses under the linkage marker indicate the genetic distance (cM) from the short arm terminal of the chromosome. B, Fine-scale, high-resolution genetic linkage map of the GLUP6 region developed from the analysis of 1,122 F2 plants and BAC clone contigs spanning the GLUP6 region. The numbers under the BAC clones indicate the number of recombinants in the adjacent marker. C, The open reading frames (thick arrows) in the GLUP6 candidate region predicted by RiceGAAS. D, The structure of the GLUP6 candidate gene, Os03g0262900. ATG and TAA denote the initiation and termination codons, respectively. Black boxes indicate the positions of the five exons. E, Mutation sites in glup6 allelic lines. In EM939, EM1327, and EM1484, a C-to-T mutation resulted in the generation of a nonsense codon and premature termination of the open reading frame.
Based on the genomic sequence data obtained from the Rice Annotation Project Database (http://rapdb.dna.affrc.go.jp/), the genome sequences of the predicted genes in glup6 lines, EM939, EM1327, and EM1484, and the wild type variety, Taichung65, were analyzed. The coding region of the Os03g0262900 gene showed changes in all three EM lines with base substitution C to T evident at nucleotide 418 in EM939 and EM1484 and at nucleotide 784 in EM1327. These base substitutions resulted in Gln-140 in EM939 and EM1484 and Gln-262 in EM1327 being replaced by a premature termination stop codon (Fig. 2E). No other changes in nucleotide sequence were detected in the other two genes that resided within the 16-kb candidate region among the three glup6 lines and Taichung65. Hence, Os03g0262900 was regarded as the GLUP6 candidate gene. A search of Os03g0262900 by RiceGAAS and the Rice Genome Annotation Project Database (http://rice.plantbiology.msu.edu/index.shtml) revealed that the GLUP6 candidate belonged to a gene family containing the Vps9 domain (Supplemental Fig. S2) and was classified as a GEF for the Rab5 group. Analysis of the Rice Expression Profile Database (http://ricexpro.dna.affrc.go.jp/pro/) showed that the GLUP6 gene was expressed throughout plant development (Supplemental Fig. S3).
The full-length open reading frame of Os03g0262900 was amplified with gene-specific primers using complementary DNA (cDNA) reverse transcribed from mRNA extracted from the immature seeds of the wild-type Taichung65 as a template. Sequencing analysis of the amplified single band demonstrated that the GLUP6 candidate gene possessed a coding sequence of 1,443 bp coding for 480 deduced amino acids spanning over five exons (Supplemental Fig. S2).
Complementation Test
In order to demonstrate directly that the GLUP6 candidate gene, Os03g0262900, is the responsible gene of the glup6 mutation, gene complementation studies were conducted. A search of the Rice Genome Resource Center (National Institute of Agrobiological Sciences) for Os03g0262900 cDNA clones resulted in the identification of J033091P13. This cDNA clone contained not only the full-length coding sequence but also an intron between nucleotides Gly-618 located at the 3′ end of exon 4 and Gly-619 located at the 5′ end of exon 5 (Supplemental Fig. S2).
The full-length cDNA with the intron and an approximately 2-kb promoter region of the candidate gene were amplified by PCR from J033091P13 and genomic DNA, respectively, and the products were cloned in an entry vector containing the GFP gene to create a fusion protein (Supplemental Fig. S4A). The constructed chimeric gene was then introduced into the binary vector DV9, which, in turn, was transferred into the glup6 line EM939 via Agrobacterium tumefaciens strain EHA105. Seeds from primary transgenic lines were observed to segregate for normal (denoted by stars) and glup6 elevated levels of proglutelin as viewed by SDS-PAGE analysis (Supplemental Fig. S4B). Immunoblot analysis showed the presence of GFP in seeds that contained normal amounts of proglutelin and its absence in seeds containing elevated levels of proglutelin (Supplemental Fig. S4C). This cosegregation of GFP with normal proglutelin levels indicates that the glup6 mutation is complemented by the introduced J033091P13. These results demonstrate that the phenotype of the elevated proglutelin level observed in glup6 is attributable to the mutations in the gene encoding a putative Rab5 GEF, which we will now call GLUP6/GEF.
Immunoblot analysis using an antibody raised against GLUP6/GEF revealed two reacting polypeptide bands in developing wild-type seeds with the bottom band missing in all glup6 lines (Fig. 3). The theoretical molecular mass of GLUP6/GEF is 53.8 kD, a value in close agreement with the measured molecular size of the bottom polypeptide band. The ubiquitous presence of the top polypeptide band in Figure 3 may be due to another GEF-related protein. As all glup6 lines contain nonsense mutations, even one close to the C terminus (EM1327), the lack of accumulated truncated polypeptides indicates that the polypeptide and the mRNA are unstable, the latter macromolecule likely degraded by nonsense-mediated mRNA decay.
Figure 3.
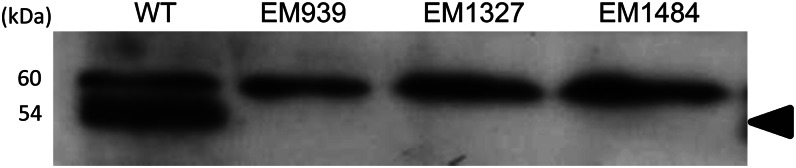
Immunoblot analysis of the seed protein from glup6 lines using anti-GLUP6/GEF antibody. The arrowhead indicates the GLUP6/GEF protein-specific band (53.8 kD). Note that EM939, EM1327, and EM1484 are devoid of GLUP6/GEF protein, indicating that premature termination of translation caused by the nonsense mutation results in general instability of the truncated GEF polypeptide or nonsense-mediated decay of the transcript. WT, Wild type.
Protein Body Formation in glup6
In order to elucidate the function of GLUP6/GEF in the intracellular transport of proglutelins, the endosperm of glup6 lines was analyzed by immunofluorescence microscopy (Fig. 4; Supplemental Fig. S5). In wild-type endosperm, protein bodies containing prolamines (protein body type I [PB-I]) and glutelins and α-globulins (PB-II) increased in size and number as the seeds developed (Fig. 4, A and B; Supplemental Fig. S5, A and B). In glup6 lines EM939 and EM1327, one or more large distended structures reactive to glutelin antibodies were conspicuously observed close to the cell wall (Fig. 4, B and C; Supplemental Fig. S5, C–F). To ensure that the observed protein distribution pattern was not an artifact of the secondary antibodies, these studies were repeated except that fluorescent labels on the secondary mouse and rabbit antibodies were switched from fluorescein isothiocyanate (FITC) to rhodamine (mouse) and from rhodamine to FITC (rabbit; Supplemental Fig. S5, Q–V). The same protein distribution patterns were seen as in Figure 4, indicating that the distribution of storage proteins in glup6 was not an immunofluorescence artifact.
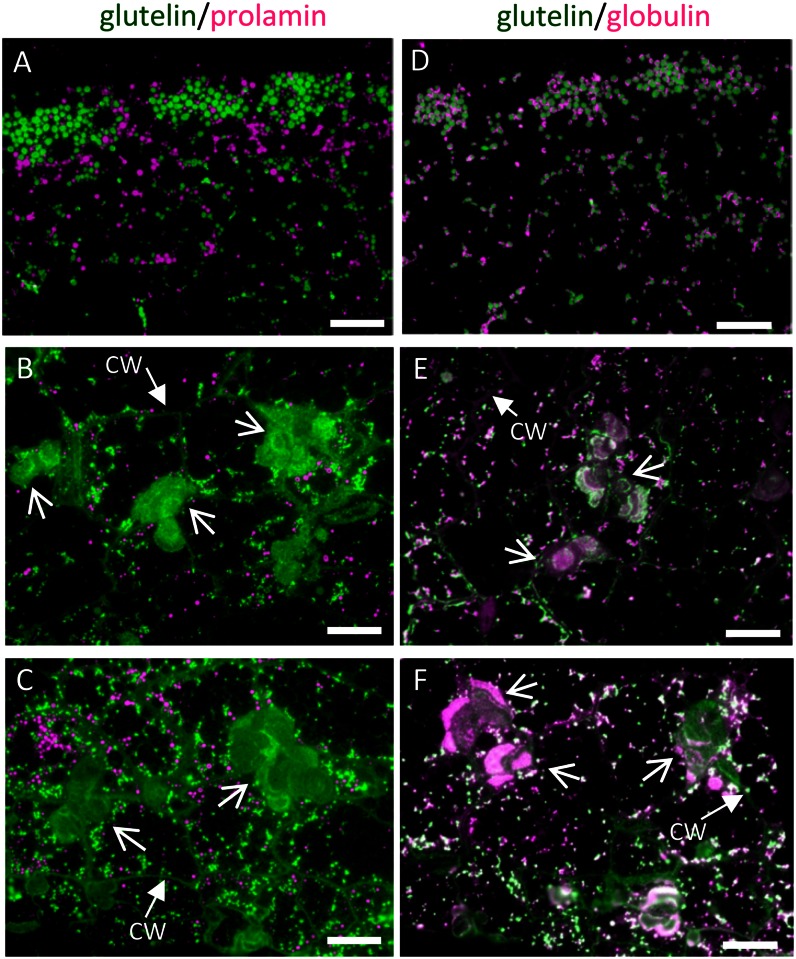
Figure 4.
The distribution of storage proteins in the wild type (A and D) and glup6 mutant lines EM939 (Gln-140 stop; B and E) and EM1327 (Gln-262 stop; C and F) as viewed by immunofluorescence microscopy. A to C depict the distribution of glutelins and prolamines, while D to F depict the distribution of glutelin and α-globulin in 3-WAF seeds. Secondary antibodies against mouse and rabbit IgGs labeled with FITC (green) and rhodamine (magenta), respectively, were used to detect antigens recognized by mouse anti-glutelin and rabbit anti-prolamine, respectively, in A to C and mouse anti-glutelin and rabbit anti-α-globulin, respectively, in D to F. Note that glutelins and globulins, which are normally packaged in PSVs (A and D), are secreted and mislocalized to the large PMBs (arrows) located adjacent to the cell wall (B, C, E, and F). CW, Cell wall. Bars = 20 μm.
The size of the large distended structures in glup6 mutants increased during seed development, unlike PB-II, whose size did not change (Supplemental Fig. S5, C–F and I–L). As this distended structure was common to all three independently derived glup6 lines, its formation can be directly attributed to the loss of function of GLUP6/GEF. These results suggest that the normal transport of proglutelin to the PSV is disrupted in the glup6 mutant and that GLUP6/GEF is an essential factor in this transport process.
In wild-type cells, glutelins and α-globulins are packaged together in PSV, although their distribution is stratified in this organelle, with glutelins located in the bulky crystalloid and α-globulins located at the peripheral, amorphous matrix regions (Fig. 4D; Kumamaru et al., 2010). The novel distended structures in the glup6 endosperm were labeled not only by anti-glutelin but also by anti-α-globulin (Fig. 4, E and F; Supplemental Fig. S5, I–L). In many instances, the distribution of glutelins and α-globulins is stratified, a condition similar to that seen in normal PB-II. The morphology of the large structures in the glup6 mutant resembles the PMBs observed in glup4 mutants (Supplemental Fig. S6, A and B; Fukuda et al., 2011), which are defective in Rab5 GTPase, although the size of the structures in glup6 were typically larger than the PMBs seen in glup4. The cell wall region adjacent to the large structure in glup6 was also labeled by the anti-glutelin antibody, suggesting that the proglutelin was secreted into the extracellular space. By contrast, the development of prolamine protein bodies appeared similar to that seen in the wild-type condition (Fig. 4, B and C).
α-Globulins accumulate in the peripheral regions of PB-II in the wild type (Fig. 4D; Krishnan et al., 1992). In glup6, the distribution of glutelin and α-globulin within PB-IIs appeared normal, although the PB-IIs were much smaller than that seen in the wild type (Fig. 4, D–F). While the size of PB-II in the wild type increased gradually during endosperm development, the size of PB-II in all glup6 lines changed very little during development (Supplemental Fig. S5). These observations indicate that the normal transport of α-globulins to PB-II (PSV) is also disrupted by the loss of function of GLUP6/GEF in glup6 mutants.
Transmission Electron Microscopy of glup6 Mutant Endosperm Indicates That the Large Distended Structures Are PMBs
To elucidate whether the large distended structures in the glup6 endosperm are PMBs similar to those observed in the glup4 endosperm, developing glup6 and wild-type seeds were analyzed by transmission electron microscopy (Fig. 5). In glup6 endosperm at 1 week after flowering (WAF), electron-dense deposits (indicated by arrowheads) were detected in the extracellular space between the cell walls of adjacent cells (Fig. 5, D and E), while electron-dense granules (indicated by arrowheads) were routinely observed in the paramural space between the plasma membrane and the cell wall (Fig. 5F). The electron-dense deposits and granules were not observed in the wild-type endosperm (Fig. 5, A and B). The electron-dense granules in glup6 were labeled by the glutelin antibodies (Fig. 6B), indicating that significant amounts of proglutelins are secreted extracellularly instead of being transported to their normal deposition site, PB-II.
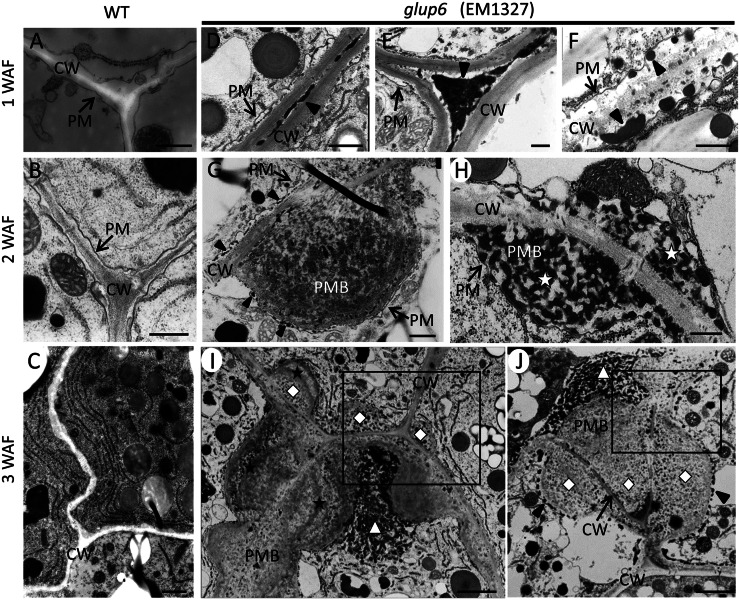
Figure 5.
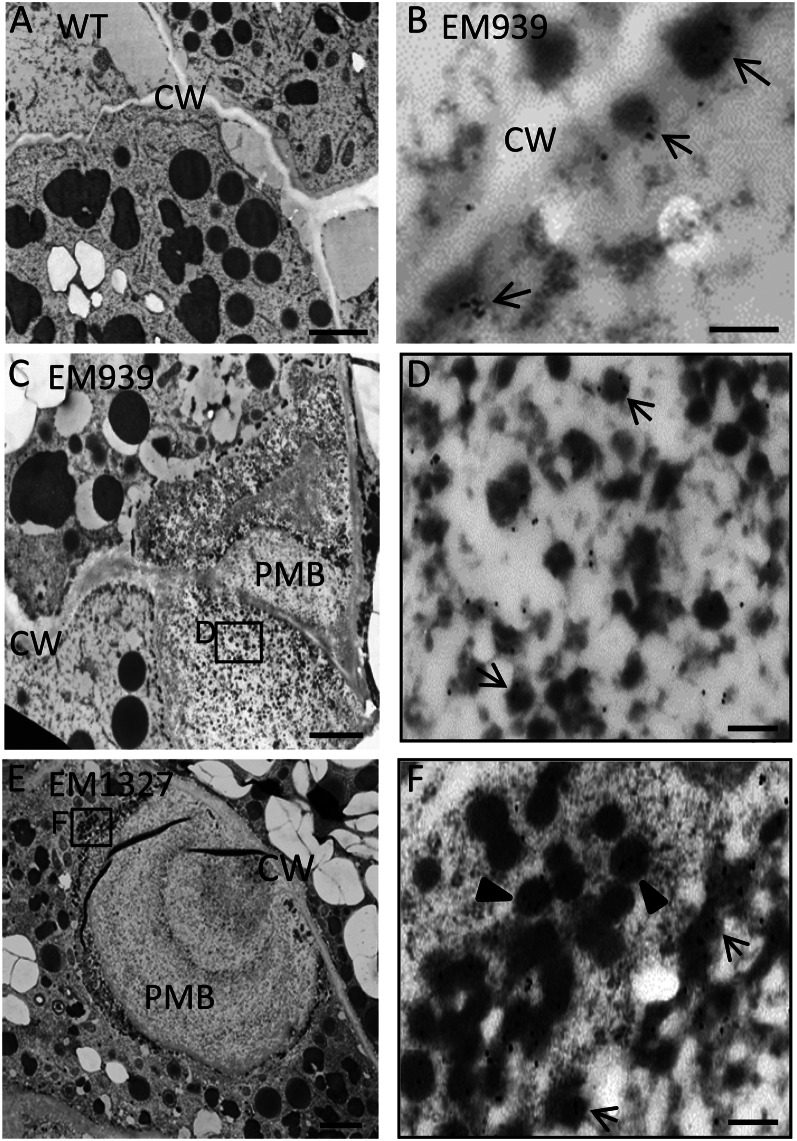
Electron micrographs depicting the secretory granules and ultrastructure of PMBs in developing endosperms of glup6, EM1327. A to C depict sections of developing endosperm in the wild type (WT) at 1, 2, and 3 WAF, respectively. Corresponding sections from glup6, EM1327, are shown in D to J. Arrowheads in D and E denote the electron-dense materials in the extracellular space, while the arrowheads in F indicate electron-dense granules in the space between the plasma membrane (PM) and the cell wall (CW). G and H show the presence of newly formed PMBs located between the plasma membrane and cell wall containing the substance with electron-lucent granules (G) and electron-dense granules (stars in H). The arrowhead in G (top cell) indicates the electron-dense granules within the paramural space (between the cell wall and plasma membrane), while the arrowhead in the bottom cell depicts electron-dense granules between the plasma membrane and PMB. In several instances, internal inclusions of the PMBs are surrounded by cell wall or cell wall-like materials (stars) in I. High-density small granules (diamonds) and high-density unstructured granules (triangles) shows the distinct inclusions within PMBs (I and J). The most outer layer of the PMB was surrounded by electron-dense granules (J; arrowheads). Enlarged images of the boxes in I and J are shown in Supplemental Figure S8. Bars in A, B, and D to H = 500 nm; bar in C = 1 μm; bars in I and J = 2 μm.
Figure 6.
Immunolocalization of glutelin to electron-dense granules and PMBs in glup6 mutants. A, The wild type (WT). B to D, EM939. E and F, EM1327. B, Seed at 1 WAF. A and C to F, Seeds at 2 WAF. D and F, Enlarged images of the areas enclosed by boxes in C and E, respectively. The secreted electron-dense granules, which are reactive to anti-glutelin (arrows), are located between the cell wall (CW) and plasma membrane (B). The granules within PMBs (D) and the dense vesicles close to the PMB in cytoplasm (F) are depicted with arrows and arrowheads, respectively. Gold particles (15 nm) indicate the reaction of anti-glutelin. Bars in A, C, and E = 2 μm; bars in B, D, and F = 200 nm.
In 2-WAF glup6 seeds, the large distended structures containing numerous small granules were readily observed in the paramural space between the plasma membrane and the cell wall (Fig. 5, G and H), indicating that the structures are PMBs. The PMB in glup6 seeds seems to have the same structure as those observed in glup4/rab5a seeds (Supplemental Fig. S6, E and F; Fukuda et al., 2011). The PMBs were very heterogeneous in appearance, with large, dispersed electron-dense granular material. Lumps of PMBs of uniform electron density were observed adjacent to the cell wall. Small electron-dense granules (arrowheads) were observed between the cell wall and the plasma membrane, as seen in the top cell, and between the plasma membrane and the PMBs, as seen in the bottom cell (Fig. 5G). In contrast to the more electron-uniform PMBs seen in Figure 5G, the PMBs in Figure 5H appeared multivesicular in being composed of many irregularly shaped electron-dense granules (Fig. 5H, white stars). The reason for these differences in appearance between the PMBs seen in Figure 5, G and H, is not clear, but a likely possibility is that the former image is a surface view of the electron-uniform PMBs covered with β-glucan (Supplemental Fig. S7) while the latter image is a view of the interior of a PMB containing electron-dense granules.
In 3-WAF glup6 seeds, the PMBs were larger and much more complex in appearance. Several PMBs were observed in areas between two or three neighboring cells (Fig. 5, I and J). A similar spatial distribution of PMBs was also seen in glup4 lines (Supplemental Fig. S6G). Individual PMBs were composed of two or three inclusions (diamonds) separated by cell wall-like materials (stars) and numerous granules with high electron density (triangles; Fig. 5, I and J). These granules associated with the PMB were labeled with anti-glutelin (Fig. 6, C–F). The most outer layer of the PMB was surrounded by electron-dense granules (arrowheads) and, in turn, collectively bounded by the plasma membrane (Fig. 5J, arrowheads; Supplemental Fig. S8).
Interestingly, the electron-dense deposits detected in the extracellular space in 1-WAF seeds disappeared in 3-WAF glup6 seeds, while the electron-dense granules in the paramural space were observed during 1 to 3 WAF, suggesting that the PMBs were formed by the accumulation of the secreted granules and β-glucan. Although the precise relationship between the disappearance of the deposits and the formation of PMBs is not clear, it is highly likely that the secreted protein deposits aggregate together to form PMBs, with continual growth by direct secretion of granules at the cytoplasmic face of the PMB (Fig. 5, G and J). Overall, these results indicate that a significant amount of proglutelin is not transported to its normal deposition site, PB-II, but is secreted extracellularly to form the PMBs and that GLUP6/GEF is required for the intracellular trafficking of proglutelins and α-globulins to the PSV.
GLUP6/GEF Activates GLUP4/Rab5
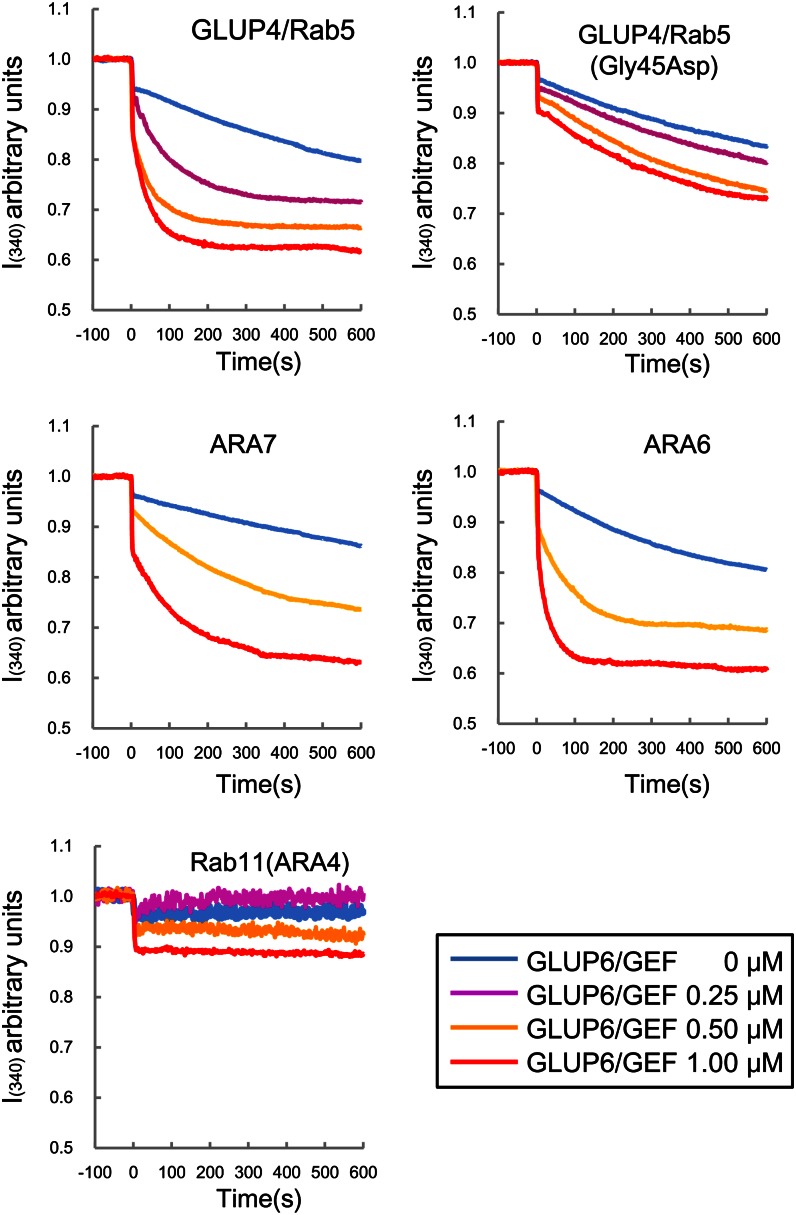
The effect of the glup6/gef mutation was nearly identical to that observed for the glup4/rab5 mutation, although the phenotype of glup6 was much more severe than that of glup4. Considering that Rab5 proteins are activated by GEFs containing the Vps9 domain in yeast, animal, and plant cells (Carney et al., 2006; Goh et al., 2007), the similar phenotypic characteristics displayed by these two mutant lines suggest that the functions of the two genes are closely related and that GLUP4/Rab5 may be activated by GLUP6/GEF. To verify this possibility, in vitro GEF assays, which monitor the change in intrinsic Trp fluorescence upon nucleotide exchange, were conducted using glutathione S-transferase (GST)-tagged recombinant proteins (Fig. 7). Reactions containing GLUP6/GEF and GLUP4/Rab5 readily showed that the Rab5 was activated by GLUP6/GEF in a dose-dependent manner. By contrast, the Rab5 variant containing a mutation (Gly-45Asp) found in the glup4 mutant line, EM960, was not activated efficiently by GLUP6/GEF. As a reference, the Arabidopsis Rab5s ARA6 and ARA7 and Arabidopsis Rab11 were subjected to the GEF assays. ARA6 and ARA7 are known to be activated by the Arabidopsis GEF, VPS9a, while Rab11 is not (Anai et al., 1994; Goh et al., 2007). Hence, ARA6 and ARA7 served as positive controls for the GEF assay, while Rab11 served as a negative control. Dose-dependent activation of Arabidopsis Rab5 members, the conventional type ARA7 and the plant-specific type ARA6, by GLUP6/GEF was observed (Fig. 7). By contrast, Arabidopsis Rab11 was not activated by GLUP6/GEF. The intrinsic nucleotide exchange on GLUP4/Rab5 was lost by the addition of EDTA, while GLUP6/GEF did not show any change in autofluorescence by itself (Supplemental Fig. S9). These results demonstrate that GLUP6/GEF is a Rab5-specific activator.
Figure 7.
In vitro GEF assay of GLUP6/GEF. The conformational changes of Rab5s and Rab11 (as 1 μm GST fusion proteins) upon GDP/guanylyl imidodiphosphate exchange were measured by monitoring Trp autofluorescence when incubated with 0 μm (blue), 0.25 μm (magenta), 0.5 μm (yellow), and 1 μm (red) GST-GLUP6/GEF. GLUP4/Rab5 and GLUP4/Rab5 (Gly-45Asp) indicate the wild type and mutant of rice GLUP4/Rab5 in EM960, respectively. Note that GST-GLUP6/GEF activates GLUP4/Rab5 as well as the Arabidopsis Rab5s, ARA6 and ARA7, but not Arabidopsis Rab11.
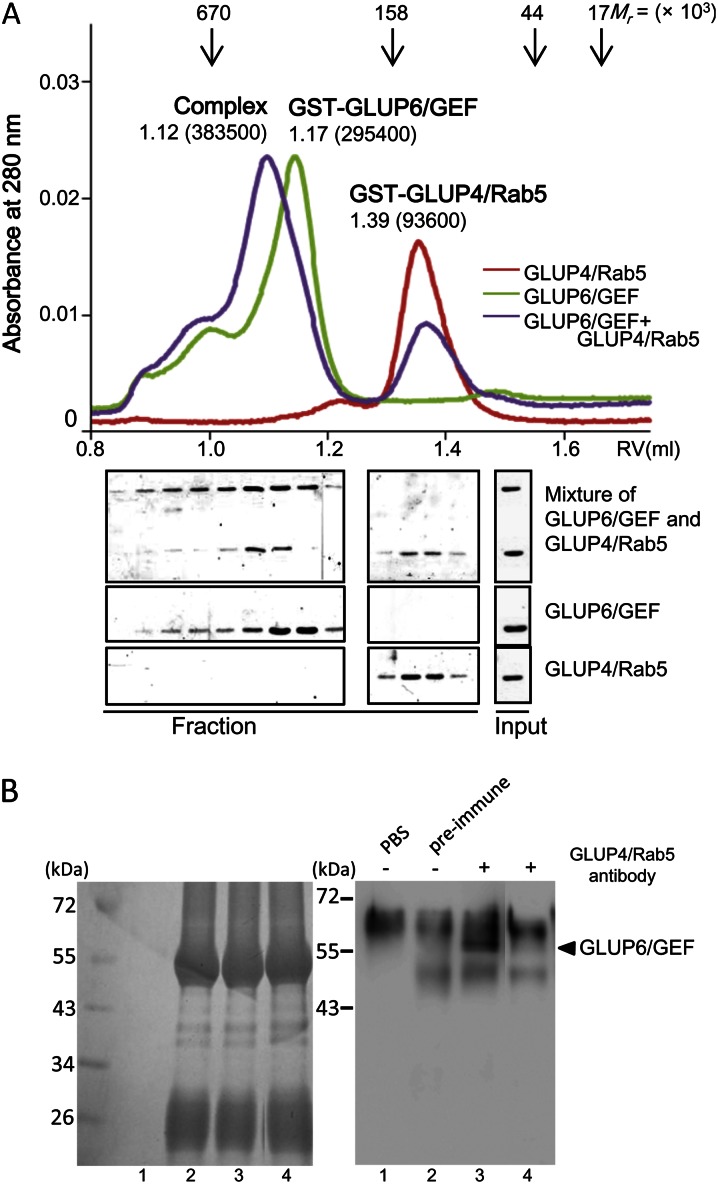
To investigate the possible physical interaction between GLUP6/GEF and GLUP4/Rab5, mixtures of these purified proteins were resolved by gel filtration chromatography (Fig. 8A). Purified GST-GLUP4/Rab5 and GST-GLUP6/GEF eluted as single peaks at estimated molecular masses of 94 and 295 kD, respectively. As the theoretical molecular masses of GST-GLUP4/Rab5 and GST-GLUP6/GEF are calculated to be 48.1 and 79.8 kD, respectively, it is thought that purified GST-GLUP4/Rab5 and GST-GLUP6/GEF eluted from the gel filtration column as an oligomer (93.6 and 295.4 kD, respectively). When both proteins were preincubated together, the peak of GST-GLUP6/GEF corresponding to 295.4 kD shifted to the position at 383.5 kD, suggesting a complex formation of the two proteins by direct interaction in solution. To escape the effect of the GST tag on each protein, thrombin treatment was performed and proteins without the tags were prepared. These proteins were subjected to gel filtration analysis with the same condition, and an elution peak corresponding to the size of 252.6 kD, including both GST-free GLUP4/Rab5 and GST-free GLUP6/GEF, was obtained (Supplemental Fig. S10).
Figure 8.
The interaction between GLUP4/Rab5 and GLUP6/GEF as assessed by gel filtration analysis (A) and coimmunoprecipitation (B). A, The physical interaction between GST-GLUP4/Rab5 and GST-GLUP6/GEF in vitro was analyzed by gel filtration assay. The elution profiles, monitored by the A280, are shown in GST-GLUP4/Rab5 (red), GST-GLUP6/GEF (green), and the mixture of both (blue). The peak positions of the marker proteins are indicated on the top, and the elution volumes and estimated molecular weights in parentheses are indicated above the peaks of each sample. Aliquots (12 μL) of each eluate fraction were subjected to 10% SDS-PAGE followed by Coomassie Brilliant Blue staining. B, Coimmunoprecipitation of GLUP4/Rab5 and GLUP6/GEF. A total protein extract from developing seed was immunoprecipitated using the anti-GLUP4/Rab5 antibody (lanes 3 and 4) prior to analysis of the captured material by SDS-PAGE and western blotting using the anti-GLUP6/GEF antibody. Negative control reactions containing PBS (lane 1) or preimmune antibody (lane 2) in place of anti-GLUP4/Rab5 antibody are shown. Lanes 1 to 3 and 4 show the protein extract from the wild type and glup6 seeds, respectively. The high background seen in lanes 2 to 4 is due to the long exposures required to detect the Rab5-GEF immunoprecipitate. Rab5 is present in small amounts in developing endosperm.
To obtain further evidence for physical interactions between these proteins, coimmunoprecipitation experiments were performed. Proteins extracted from developing rice seeds were incubated with protein A-agarose beads bound with anti-GLUP4/Rab5, and the resulting captured material was then analyzed by western blotting using anti-GLUP6/GEF. Figure 8B shows that anti-GLUP4/Rab5 is able to coimmunoprecipitate protein complexes containing GLUP6/GEF (lane 3), whereas neither protein was detected in the controls using preimmune antibody (lane 2) or when the protein extract from glup6 seeds was used (lane 4). Extraneous bands nonspecifically bound by protein A resin were detected in all lanes (lanes 2–4). This high background is due to the fact that GLUP4/Rab5 was present at very low levels in developing rice endosperm. Hence, to detect the small amounts of GLUP4/Rab5 associated with GLUP6/GEF, prolonged exposures were required, resulting in the visualization of proteins nonspecifically adsorbed to the protein A resin. Overall, results from the GEF assay, gel filtration analysis, and coimmunoprecipitation studies demonstrate that GLUP6/GEF is an activator of GLUP4/Rab5.
DISCUSSION
Factors for the Intracellular Transport of the Proglutelin
Significant insights on the cellular processes involved in the ER-associated synthesis and maturation of proglutelin and its transport to the PSV have been obtained from genetic studies (Ueda et al., 2010). In the rice esp2 line, which contains a deletion mutation in PDIL1-1 encoding a lumenal protein disulfide isomerase, proglutelins accumulate abnormally within the ER instead of being transported to the PSV (Takemoto et al., 2002; Satoh-Cruz et al., 2010a). A second mutant defective in the processes responsible for traffic and the deposition of glutelins in the PSV is glup4 (Ueda et al., 2010), which harbors a defective Rab5, the small GTPase required for endosomal and vacuolar trafficking (Sohn et al., 2003; Kotzer et al., 2004; Ebine et al., 2011). Mutation in Rab5 causes the secretion of glutelins and the formation of large PMB membranous complexes (Supplemental Fig. S6). The PMBs contain glutelins and α-globulins as well as protein markers of the ER, Golgi, PVC, PSV, and plasma membrane as well as the cell wall component, β-glucan (Fukuda et al., 2011). Hence, Rab5 is required not only for the transport of glutelins and α-globulins to the PSV but also for proper maintenance of the endomembrane system during rice endosperm development. The proglutelins are cleaved proteolytically into two subunits by vacuolar processing enzyme and accumulate within the PSV. The proteolytic cleavage of proglutelin by vacuolar processing enzyme plays an important role in the formation of crystalline structures of the glutelin in PSVs (Kumamaru et al., 2010).
This study demonstrates that the GEF encoded by the GLUP6 gene is the putative activator of GLUP4/Rab5. Several lines of evidence support the direct relationship between these two proteins. First, mutant lines for these two genes exhibit nearly identical cellular phenotypic properties. During early endosperm development in both mutations, glutelins are secreted and present as small granules in the extracellular space (Fig. 5; Supplemental Fig. S6). Subsequently, the appearance of glutelin-containing PMBs occurs with the apparent disappearance of extracellular granules. Additionally, proteins normally localized to the Golgi and PVCs are also missorted to the PMB (Supplemental Fig. S11), a condition identical to that seen in the glup4/rab5 mutant (Fukuda et al., 2011). Lastly, we show that GLUP6/GEF interacts directly with GLUP4/Rab5 and activates it by exchanging GDP with GTP (Figs. 7 and 8). These facts indicate that GLUP6/GEF is the guanine nucleotide exchange factor for GLUP4/Rab5 GTPase. Moreover, the recycling of Rab5 from an inactive GDP-bound form to an active GTP-bound form is essential for the efficient sorting of proglutelins from the Golgi to the PSV and for the overall maintenance of the endomembrane system within developing rice endosperm cells.
Activation of the Rab5 Family by GEF
Although glup6 and glup4 mutants share many cellular defects, the PMBs in glup6 are more numerous and larger than those seen in glup4 (Figs. 4 and 5; Supplemental Fig. S6). The exact basis for this more severe phenotype in glup6 is not known, but the presence of multiple Rab5 and GEF activities in rice endosperm may account for the severe phenotype in glup6. Rab5 proteins, like all small GTPases, are negatively regulated by GTPase-activating proteins, which accelerate the intrinsic rate of GTP hydrolysis, and positively regulated by GEFs, which stimulate the release of GDP for the binding of GTP (Carney et al., 2006). The Arabidopsis Rab5 GEF, VPS9a, activates all three Rab5 members but not Rab11 to the GTP-bound form (Goh et al., 2007). The highly conserve Vps9 domain is responsible for the specific activation of Rab5 proteins by Rab5 GEFs (Carney et al., 2006). Here, we showed that GLUP6/GEF was capable of activating GLUP4/Rab5 as well as the two Arabidopsis Rab5 proteins, ARA6 andARA7 (Fig. 7). In addition to GLUP4/Rab5, rice endosperm contains other expressed Rab5 forms (Fukuda et al., 2011), which may partially overlap in function with the GLUP4/Rab5. In fact, genetic evidence is consistent with this view that glup6 and glup4 genes act additively based on the extent of proglutelin accumulated in the double mutant compared with that seen for each mutant line (Ueda et al., 2010). These results lead us to hypothesize that GLUP6/GEF can activate other Rab5 proteins than GLUP4/Rab5 and that GEFs other than GLUP6/GEF can also activate GLUP4/Rab5. These multiple Rab5 and GEF isoforms confer different contributions to the intracellular trafficking of proglutelins and α-globulins from the Golgi to the PSV, with GLUP4/Rab5 and GLUP6/GEF having dominant roles during endosperm development.
Similar to glup4, glup6 seeds exhibit a floury endosperm (Fig. 1C). The glup4 mutant line has a higher Suc content and higher activities of three enzymes involved in converting Suc to starch and cellulose (Suc synthase, UDP-Glc pyrophosphorylase, and cell wall-bound Suc invertase) but lower levels of the starch regulatory enzyme ADP-Glc pyrophosphorylase (Wang et al., 2010). Thus, alterations in the endomembrane system can have far-reaching consequences on other processes, as made evident by its significant changes in starch synthesis in glup4 and glup6 seeds.
Secretion of Proglutelin
In Arabidopsis vps9a-1 mutant plants, the loss of GEF results in the missorting of vacuolar proteins and the formation of PMBs. In addition, incomplete cell plates and abnormal cell wall depositions are frequently observed in embryos at the torpedo stage, suggesting the role of the endocytic pathway in cell plate formation and cell wall biogenesis (Goh et al., 2007). Although similar defects were seen in the rice glup6/gef mutant, there are notable differences in this study. Unlike the small (approximately 0.2 µm) cell wall-free PMBs seen in Arabidopsis vps9a-1 (Goh et al., 2007), the PMBs in glup6 are much larger (approximately 2 µm and larger) and covered with cell wall β-glucan (Supplemental Fig. S7). Moreover, intracellular cell wall depositions observed in the Arabidopsis vps9a are not seen in the rice glup6 lines.
The reason for the secretion of proglutelins due to the loss of function of GLUP4/Rab5 and GLUP6/GEF is not known. In Arabidopsis atvsr1, which contains a transfer DNA insertion in the vacuolar sorting receptor gene, the transport of 12S globulin and 2S albumin storage proteins is redirected from the PSVs and instead secreted where they form large extracellular deposits (Shimada et al., 2003). Mutation in MAG1/AtVPS29, which is involved in recycling a vacuolar sorting receptor between the Golgi and the PSV and required the efficient sorting of seed storage proteins, missorts the precursor form of storage proteins by secreting them from the cells (Shimada et al., 2006). Hence, these genetic studies strongly suggest that mutations in factors involved in membrane vesicular transport to PSV tend to result in the secretion of cargo proteins to the extracellular space. This is consistent with the observations that mutations in the rice Rab5, which is normally associated with the Golgi, PVC, and PSV, result in the secretion of storage proteins.
Lastly, mutations in GLUP4/Rab5 not only affect protein trafficking but also RNA trafficking in developing endosperm. The storage protein RNAs are localized to discrete subdomains of the cortical ER (Li et al., 1993). Prolamine RNAs are localized to the protein body ER that bound PB-I, while glutelin RNAs are sorted to the cisternal-ER (Choi et al., 2000; Hamada et al., 2003; Washida et al., 2009). In situ reverse transcription-PCR studies using fluorescently labeled nucleotides show that glutelin RNAs are missorted in glup4 mutant lines. Instead of their normal distribution on the cisternal-ER, glutelin RNAs are found distributed on PB-I as well as to the PMBs (Doroshenk et al., 2010). Similar to that seen for glup4, glutelin RNAs are also mislocalized in glup6, where they are seen on both PB-I and PMBs (Crofts et al., 2005). The dependence on GLUP4/Rab5 and GLUP6/GEF supports the role of membrane vesicles as the transport vehicles for RNAs as well as for storage proteins. Current efforts are directed at identifying the links between RNA localization and vesicular transport.
MATERIALS AND METHODS
Plant Materials
The rice (Oryza sativa) glup6 mutant lines EM939, EM1327, and EM1484, induced by N-methyl-N-nitrosourea mutagenesis, which were characterized by their accumulation of substantial amounts of the 57-kD proglutelin precursor (Satoh-Cruz et al., 2010b; Ueda et al., 2010), were used in these experiments. The full-length cDNA clone for OsVSR3 (AK072667) was obtained from the National Institute of Agrobiological Sciences in Tsukuba, Japan. pAcGFP1-Golgi and pDsRed-Monomer-N1 were purchased from Clontech. Rice plants were grown in the field or a glasshouse (transgenic lines) at Kyushu University, and developing seeds were harvested and isolated for biochemical and microscopic analyses.
SDS-PAGE and Western-Blot Analysis
Proteins were extracted from seeds using 0.125 m Tris-HCl, 4% SDS, 4 m urea, and 5% β-mercaptoethanol, pH 6.8 (Ushijima et al., 2011). The proteins were resolved by SDS-PAGE, transferred electrically to nitrocellulose membranes, and then incubated for 1 h with Tris-buffered saline (TBS), 10 mm Tris-HCl, 0.15 m NaCl, 5% skim milk, pH 7.5, and primary antibody (1:1,000 dilution). The blot was washed three times with TBS containing 0.05% Tween 20 and then incubated with TBS containing 5% skim milk and the secondary antibody (1:2,500 dilution). The blot was washed three times with TBS containing 0.05% Tween 20 and incubated with the ECL Detection Kit (GE Healthcare). The blot was then exposed to x-ray film for visualization of the bound primary antibody.
Construction of a Genetic Linkage Map
The glup6 line, EM939, was crossed with the indica rice cultivar Kasalath. The F2 seeds were cut in half, and total proteins extracted from the nonembryo half were subjected to SDS-PAGE. Seeds containing abnormally high amounts of 57-kD polypeptide as assessed by SDS-PAGE were judged as homozygous for glup6. Homozygous glup6 plants were cultivated from the embryo-containing half seeds, and genomic DNA was isolated from the seedlings. A high-density genetic linkage map of the GLUP6 gene was constructed using cleaved-amplified polymorphic sequence and STS markers of chromosome 3.
DNA Sequencing Analysis
DNA sequencing analysis was performed as described previously (Kumamaru et al., 2010). Total genomic DNAs from the leaves of glup6 lines and the wild type were obtained using the cetyltrimethylammonium bromide method (Murray and Thompson, 1980). DNA sequence was determined using an ABI PRISM 3100 Genetic Analyzer (Applied Biosystems). DNA sequence analysis was performed using EditView1.0.1 and AutoAssembler 2.1. Comparisons between the wild-type and mutant sequences were performed using ClustalW of the DNA Data Bank of Japan (http://www.ddbj.nig.ac.jp/top-e.html).
Plasmid Construction and Rice Transformation
The AcGFP1-Golgi (Clontech) contains a segment of human β-1,4-galactosyltransferase and localizes to the trans-medial region of the Golgi apparatus (Miao et al., 2006). The GFP sequences were replaced with DsRed-Monomer (Clontech), generating Golgi-DsRed. The C-terminal region of OsVSR3 containing the membrane-spanning domain was amplified by PCR with primers 5′-ATAGGATCCAGCAAAGTTGCTTCTTCGTC-3′ and 5′-TATCTCGAGCCCAGTTAAGCTTGCTGCAA-3′ using a full-length cDNA clone (AK072667) as a template. The PCR product was then inserted downstream of the signal peptide GFP (spGFP) gene (Kawagoe et al., 2005), generating spGFP-VSR3. The two fusion genes were expressed under the control of the rice β--tonoplast intrinsic proteins (Onda et al., 2009). The binary vector containing the Golgi-DsRed and spGFP-VSR3 was constructed by the Gateway system (Invitrogen) as described previously (Onda et al., 2009). Transformation of rice was performed as described previously (Kawagoe et al., 2005).
Microscopic Analysis
For immunofluorescence and immunoelectron microscopic analysis, the samples were fixed as described previously (Takemoto et al., 2002).
Immunofluorescence Microscopic Analysis
Sections were treated with blocking buffer containing 0.8% bovine serum albumin and 0.1% gelatin in phosphate-buffered saline (PBS) and then incubated in the appropriate antibodies diluted in blocking buffer. Nonspecifically bound antibodies were removed by washing the section five times with PBS. The sections were treated with blocking buffer, followed by incubating with secondary antibodies (anti-mouse FITC [green] and anti-rabbit rhodamine [red]; Funakoshi Chemical), and then observed microscopically (BZ-9000; Keyence). Samples were treated with antibodies raised against glutelin basic subunit (1:5,000), 14-kD prolamine (1:1,000), or α-globulin (1:5,000). Red fluorescent images were converted to magenta with Adobe Photoshop.
Transmission Immunoelectron Microscopic Analysis
Ultrathin sections (90–100 nm) were cut with a diamond knife and placed on 200-mesh gilder grids. The grids were treated with blocking buffer (TBS: 10 mm Tris, 500 mm NaCl, and 0.3% Tween, pH 7.2) containing 1% bovine serum albumin for 60 min. The grids were then incubated overnight with the appropriate antibodies diluted with blocking buffer. Unreacted antibodies were removed by washing the section four times in a drop of blocking buffer. The grids were then incubated for 60 min with secondary antibodies (protein A-gold labeled with 5 or 15 nm diameter; Funakoshi Chemical), diluted with blocking buffer as described above, and then sequentially stained with 0.25% KMnO4 and 1% uranylacetate. Samples for cytochemical electron microscopy were embedded in Epon, and sections were stained in Reynolds’ lead staining solution consisting of 3.44% Pb(NO3)2, 4.63% Na2(H6H5O7)2H2O, and 0.21 n NaOH in CO2-free distilled water (Nagamine et al., 2011). The sections were then observed by transmission electron microscopy (JEM-1220) at 80 kV. Laser-scanning confocal microscopic analysis was performed as described previously (Onda et al., 2009).
Expression and Purification of GST Fusion Proteins
GST fusions of GLUP6/GEF, GLUP4/Rab5, and GLUP4/Rab5 (Gly-45Asp) were produced in Escherichia coli BL21 (DE3) using pGEX 4T-1 expression vector (GE Healthcare) and purified according to the procedure described earlier (Goh et al., 2007). The primer sequences for construction of the expression plasmids are shown in Supplemental Table S1, and the experimental procedures are described in the table legend. Rab5s (ARA6 and ARA7) and Rab11 from Arabidopsis (Arabidopsis thaliana), fused with GST, were prepared as described in the previous report (Goh et al., 2007). All of the Rab proteins were purified in the GDP-bound form in the presence of Mg2+ and without EDTA or GDP. After purification of GST-tagged Rabs and GLUP6/GEF using a glutathione-Sepharose 4B column (GE Healthcare), the bound protein fractions were loaded onto a desalting column (GE Healthcare) to remove glutathione and stored frozen at −80°C. The whole purification procedure was finished in 1 d to maximize enzyme activity.
In order to remove the GST tag, the purified GST fusion proteins were reloaded onto GST spin columns (GE Healthcare) and then subjected to thrombin cleavage capture using the manufacturer’s recommendations (Novagen).
Guanine Nucleotide-Exchange Assay
The nucleotide-exchange assay was performed according to the method described previously (Goh et al., 2007). Each purified GST-Rab protein was preloaded with a 25 m excess of GDP at 25°C for 2 h. The excess GDP was removed by a desalting column. For each assay, 1 µm GST-Rab GDP form was preincubated with or without GST-GLUP6/GEF in GEF assay buffer containing 20 mm Tris-HCl, pH 8.0, 150 mm NaCl, and 0.5 mm MgCl2 for 100 s. The nucleotide-exchange reaction was started by the addition of 0.1 mm guanylyl imidodiphosphate. The Trp intrinsic fluorescence of Rab proteins was detected at 340 nm by excitation at 298 nm using a fluorescence spectrophotometer (F-2500; Hitachi). Each experiment was completed within 2 d, as the activity of Rab GTPase decreases rapidly after thawing.
Immunoprecipitation
The extraction from rice seed was performed according to the method described previously (Satoh-Cruz et al., 2010a). Protein A resin, which had previously been incubated overnight with 1× PBS (no antibody control) or preimmune or anti-GLUP4/Rab5 antibody, was washed three times with 1 mL of immunoprecipitation buffer (20 mm Tris-HCl, pH 7.5, 75 mm NaCl, 1 mm EDTA, and 0.1% Nonidet P-40) prior to the addition of 1.5 mL of the cleared supernatant described above. Extracts and antibody were incubated for 2 h prior to washing five times with 1 mL of immunoprecipitation buffer. The protein A was then boiled in SDS-containing sample buffer prior to western blotting with anti-GLUP6/GEF antibody at a dilution of 1:1,000.
Gel Filtration Analysis
A gel filtration binding assay was performed according to the methods described previously (Hamel et al., 2011). Gel filtration chromatography was carried out using the SMART system (GE Healthcare) with a GEF assay buffer as the mobile phase and a flow rate of 40 μL min−1. GST-GLUP6/GEF and the GDP form of GST-GLUP4/Rab5 were separately purified one more time by gel filtration (Superdex 200 3.2/30 column; GE Healthcare). The GDP form of GST-GLUP4/Rab5 and GST-GLUP6/GEF were individually loaded as controls. The mixture of GST-GLUP4/Rab5 GDP and GST-GLUP6/GEF (quantity ratio is 1:1) was loaded on the gel filtration column. To escape the effect of GST attached to each target protein, GST-free GLUP4/Rab5 GDP form, GST-free GLUP6/GEF, and their mixture (2:1 GLUP4/Rab5 GDP:GLUP6/GEF) were directly loaded onto a gel filtration column. Because of the nonspecific cleavage of GLUP6/GEF by the thrombin treatment, each gel filtration fraction was subjected to western-blot analysis to identify the proteins included in each fraction.
All of the manipulations described above were completed in 2 d to minimize the effects of protein instability. The molecular masses of the proteins were estimated from the elution profiles of gel filtration standard marker proteins (Bio-Rad), which included thyroglobulin (670,000), γ-globulin (158,000), ovalbumin (44,000), and myoglobin (17,000).
Antibodies
Seed storage proteins were separated by SDS-PAGE, and individual bands were excised and eluted from the gel by preparative electrophoresis. Antibodies against glutelin acidic subunit and α-globulin were then raised in mice and rabbits, respectively. The specificity of antibody against α-globulin was confirmed by immunoblot analysis (Supplemental Fig. S12), and the antibody reacted with matrix in PSV (Kumamaru et al., 2010). Antibody against 14-kD prolamine was raised in rabbit, and its specificity was confirmed as described previously (Nagamine et al., 2011).
Antibody against the recombinant protein of rice GLUP6/GEF expressed in E. coli was raised in rabbits. Antibody against (1,3;1,4)-β-glucan was purchased from Biosupplies Australia.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Plant phenotypes of glup6 lines.
Supplemental Figure S2. cDNA and the deduced amino acid sequences and EST clones corresponding to Os03g0262900.
Supplemental Figure S3. Expression profile of GLUP6/GEF in rice by the Rice Expression Profile Database.
Supplemental Figure S4. Complementation test for the GEF gene in the glup6 mutant.
Supplemental Figure S5. Immunofluorescence microscopy of protein bodies and the structures in various glup6 allelic lines.
Supplemental Figure S6. Microscopic analysis of protein bodies and the PMBs in glup4 endosperm, EM956.
Supplemental Figure S7. Immunofluorescence microscopy showing the localization of β-glucan and glutelin in developing endosperm of glup6 mutant (3-WAF seeds).
Supplemental Figure S8. Enlarged image of PMBs in glup6 (3-WAF seeds), EM1327.
Supplemental Figure S9. Control of the in vitro GEF assay.
Supplemental Figure S10. Interaction between GLUP4/Rab5 and GLUP6/GEF cleaved GST tag as assessed by gel filtration analysis.
Supplemental Figure S11. Localization of Golgi-DsRed and GFP-PVC (VSR3) in developing endosperm (2 WAF) of the glup6 mutant.
Supplemental Figure S12. Specificity of anti-α-globulin antibody by immunoblot analysis.
Supplemental Table S1. List of PCR primers.
Glossary
- ER
endoplasmic reticulum
- PSV
protein storage vacuole
- PB-II
protein body type II
- PMB
paramural body
- PVC
prevacuolar compartment
- cM
centimorgan
- BAC
bacterial artificial chromosome
- RiceGAAS
Rice Genome Automated Annotation System
- PB-I
protein body type I
- FITC
fluorescein isothiocyanate
- WAF
week after flowering
- GST
glutathione S-transferase
- TBS
Tris-buffered saline
- PBS
phosphate-buffered saline
- cDNA
complementary DNA
- STS:
sequence-tagged sites
- spGFP:
signal peptide GFP
References
- An Q, Ehlers K, Kogel KH, van Bel AJ, Hückelhoven R. (2006a) Multivesicular compartments proliferate in susceptible and resistant MLA12-barley leaves in response to infection by the biotrophic powdery mildew fungus. New Phytol 172: 563–576 [DOI] [PubMed] [Google Scholar]
- An Q, Hückelhoven R, Kogel KH, van Bel AJ. (2006b) Multivesicular bodies participate in a cell wall-associated defence response in barley leaves attacked by the pathogenic powdery mildew fungus. Cell Microbiol 8: 1009–1019 [DOI] [PubMed] [Google Scholar]
- Anai T, Matsui M, Nomura N, Ishizaki R, Uchimiya H. (1994) In vitro mutation analysis of Arabidopsis thaliana small GTP-binding proteins and detection of GAP-like activities in plant cells. FEBS Lett 346: 175–180 [DOI] [PubMed] [Google Scholar]
- Burd CG, Mustol PA, Schu PV, Emr SD. (1996) A yeast protein related to a mammalian Ras-binding protein, Vps9p, is required for localization of vacuolar proteins. Mol Cell Biol 16: 2369–2377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carney DS, Davies BA, Horazdovsky BF. (2006) Vps9 domain-containing proteins: activators of Rab5 GTPases from yeast to neurons. Trends Cell Biol 16: 27–35 [DOI] [PubMed] [Google Scholar]
- Choi SB, Wang C, Muench DG, Ozawa K, Franceschi VR, Wu Y, Okita TW. (2000) Messenger RNA targeting of rice seed storage proteins to specific ER subdomains. Nature 407: 765–767 [DOI] [PubMed] [Google Scholar]
- Chrispeels MJ. (1983) The Golgi apparatus mediates the transport of phytohemagglutinin to the protein bodies in been cotyledon. Planta 158: 140–151 [DOI] [PubMed] [Google Scholar]
- Crofts AJ, Washida H, Okita TW, Satoh M, Ogawa M, Kumamaru T, Satoh H. (2005) The role of mRNA and protein sorting in seed storage protein synthesis, transport, and deposition. Biochem Cell Biol 83: 728–737 [DOI] [PubMed] [Google Scholar]
- Doroshenk KA, Crofts AJ, Washida H, Satoh-Cruz M, Crofts N, Okita TW, Morris RT, Wyrick JJ, Fukuda M, Kumamaru T, et al (2010) Characterization of the rice glup4 mutant suggests a role for the small GTPase Rab5 in the biosynthesis of carbon and nitrogen storage reserves in developing endosperm. Breed Sci 60: 556–567 [Google Scholar]
- Ebine K, Fujimoto M, Okatani Y, Nishiyama T, Goh T, Ito E, Dainobu T, Nishitani A, Uemura T, Sato MH, et al. (2011) A membrane trafficking pathway regulated by the plant-specific RAB GTPase ARA6. Nat Cell Biol 13: 853–859 [DOI] [PubMed] [Google Scholar]
- Fukuda M, Satoh-Cruz M, Wen L, Crofts AJ, Sugino A, Washida H, Okita TW, Ogawa M, Kawagoe Y, Maeshima M, et al (2011) The small GTPase Rab5a is essential for intracellular transport of proglutelin from the Golgi apparatus to the protein storage vacuole and endosomal membrane organization in developing rice endosperm. Plant Physiol 157: 632–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goh T, Uchida W, Arakawa S, Ito E, Dainobu T, Ebine K, Takeuchi M, Sato K, Ueda T, Nakano A. (2007) VPS9a, the common activator for two distinct types of Rab5 GTPases, is essential for the development of Arabidopsis thaliana. Plant Cell 19: 3504–3515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas TJ, Sliwinski MK, Martínez DE, Preuss M, Ebine K, Ueda T, Nielsen E, Odorizzi G, Otegui MS. (2007) The Arabidopsis AAA ATPase SKD1 is involved in multivesicular endosome function and interacts with its positive regulator LYST-INTERACTING PROTEIN5. Plant Cell 19: 1295–1312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hama H, Tall GG, Horazdovsky BF. (1999) Vps9p is a guanine nucleotide exchange factor involved in vesicle-mediated vacuolar protein transport. J Biol Chem 274: 15284–15291 [DOI] [PubMed] [Google Scholar]
- Hamada S, Ishiyama K, Sakulsingharoj C, Choi SB, Wu Y, Wang C, Singh S, Kawai N, Messing J, Okita TW. (2003) Dual regulated RNA transport pathways to the cortical region in developing rice endosperm. Plant Cell 15: 2265–2272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamel B, Monaghan-Benson E, Rojas RJ, Temple BR, Marston DJ, Burridge K, Sondek J. (2011) SmgGDS is a guanine nucleotide exchange factor that specifically activates RhoA and RhoC. J Biol Chem 286: 12141–12148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohl I, Robinson DG, Chrispeels MJ, Hinz G. (1996) Transport of storage proteins to the vacuole is mediated by vesicles without a clathrin coat. J Cell Sci 109: 2539–2550 [DOI] [PubMed] [Google Scholar]
- Horiuchi H, Lippé R, McBride HM, Rubino M, Woodman P, Stenmark H, Rybin V, Wilm M, Ashman K, Mann M, et al (1997) A novel Rab5 GDP/GTP exchange factor complexed to Rabaptin-5 links nucleotide exchange to effector recruitment and function. Cell 90: 1149–1159 [DOI] [PubMed] [Google Scholar]
- Kawagoe Y, Suzuki K, Tasaki M, Yasuda H, Akagi K, Katoh E, Nishizawa NK, Ogawa M, Takaiwa F. (2005) The critical role of disulfide bond formation in protein sorting in the endosperm of rice. Plant Cell 17: 1141–1153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotzer AM, Brandizzi F, Neumann U, Paris N, Moore I, Hawes C. (2004) AtRabF2b (Ara7) acts on the vacuolar trafficking pathway in tobacco leaf epidermal cells. J Cell Sci 117: 6377–6389 [DOI] [PubMed] [Google Scholar]
- Krishnan HB, Franceschi VR, Okita TW. (1986) Immunochemical studies on the role of the Golgi complex in protein-body formation in rice seeds. Planta 169: 471–480 [DOI] [PubMed] [Google Scholar]
- Krishnan HB, White JA, Pueppke SG. (1992) Characterization and location of rice (Oryza sativa L.) seed globulins. Plant Sci 81: 1–11 [Google Scholar]
- Kumamaru T, Uemura Y, Inoue Y, Takemoto Y, Siddiqui SU, Ogawa M, Hara-Nishimura I, Satoh H. (2010) Vacuolar processing enzyme plays an essential role in the crystalline structure of glutelin in rice seed. Plant Cell Physiol 51: 38–46 [DOI] [PubMed] [Google Scholar]
- Li X, Franceschi VR, Okita TW. (1993) Segregation of storage protein mRNAs on the rough endoplasmic reticulum membranes of rice endosperm cells. Cell 72: 869–879 [DOI] [PubMed] [Google Scholar]
- Marchant R, Robards AW. (1968) Membrane systems associated with the plasmalemma of plant cells. Ann Bot (Lond) 32: 457–471 [Google Scholar]
- Miao Y, Yan PK, Kim H, Hwang I, Jiang L. (2006) Localization of green fluorescent protein fusions with the seven Arabidopsis vacuolar sorting receptors to prevacuolar compartments in tobacco BY-2 cells. Plant Physiol 142: 945–962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray MG, Thompson WF. (1980) Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res 8: 4321–4325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagamine A, Matsusaka H, Ushijima T, Kawagoe Y, Ogawa M, Okita TW, Kumamaru T. (2011) A role for the cysteine-rich 10 kDa prolamin in protein body I formation in rice. Plant Cell Physiol 52: 1003–1016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okita TW, Rogers JC. (1996) Compartmentation of proteins in the endomembrane system of plant cells. Annu Rev Plant Physiol Plant Mol Biol 47: 327–350 [DOI] [PubMed] [Google Scholar]
- Onda Y, Kumamaru T, Kawagoe Y. (2009) ER membrane-localized oxidoreductase Ero1 is required for disulfide bond formation in the rice endosperm. Proc Natl Acad Sci USA 106: 14156–14161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato M, Ishihara D, Tian HD, Takemoto Y, Kumamaru T, Satoh H. (2003) Gene mapping and immunocytochemistry of rice glup6 mutant. Rice Genet Newsl 20: 43–45 [Google Scholar]
- Satoh-Cruz M, Crofts AJ, Takemoto-Kuno Y, Sugino A, Washida H, Crofts N, Okita TW, Ogawa M, Satoh H, Kumamaru T. (2010a) Protein disulfide isomerase like 1-1 participates in the maturation of proglutelin within the endoplasmic reticulum in rice endosperm. Plant Cell Physiol 51: 1581–1593 [DOI] [PubMed] [Google Scholar]
- Satoh-Cruz M, Fukuda M, Ogawa M, Satoh H, Kumamaru T. (2010b) Glup4 gene encodes small GTPase, Rab5a in rice. Rice Genet Newsl 25: 48–49 [Google Scholar]
- Shimada T, Fuji K, Tamura K, Kondo M, Nishimura M, Hara-Nishimura I. (2003) Vacuolar sorting receptor for seed storage proteins in Arabidopsis thaliana. Proc Natl Acad Sci USA 100: 16095–16100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada T, Koumoto Y, Li L, Yamazaki M, Kondo M, Nishimura M, Hara-Nishimura I. (2006) AtVPS29, a putative component of a retromer complex, is required for the efficient sorting of seed storage proteins. Plant Cell Physiol 47: 1187–1194 [DOI] [PubMed] [Google Scholar]
- Sohn EJ, Kim ES, Zhao M, Kim SJ, Kim H, Kim YW, Lee YJ, Hillmer S, Sohn U, Jiang L, et al (2003) Rha1, an Arabidopsis Rab5 homolog, plays a critical role in the vacuolar trafficking of soluble cargo proteins. Plant Cell 15: 1057–1070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stierhof YD, El Kasmi F. (2010) Strategies to improve the antigenicity, ultrastructure preservation and visibility of trafficking compartments in Arabidopsis tissue. Eur J Cell Biol 89: 285–297 [DOI] [PubMed] [Google Scholar]
- Takemoto Y, Coughlan SJ, Okita TW, Satoh H, Ogawa M, Kumamaru T. (2002) The rice mutant esp2 greatly accumulates the glutelin precursor and deletes the protein disulfide isomerase. Plant Physiol 128: 1212–1222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda T, Uemura T, Sato MH, Nakano A. (2004) Functional differentiation of endosomes in Arabidopsis cells. Plant J 40: 783–789 [DOI] [PubMed] [Google Scholar]
- Ueda T, Yamaguchi M, Uchimiya H, Nakano A. (2001) Ara6, a plant-unique novel type Rab GTPase, functions in the endocytic pathway of Arabidopsis thaliana. EMBO J 20: 4730–4741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda Y, Satoh-Cruz M, Matsusaka H, Takemoto-Kuno Y, Fukuda M, Okita TW, Ogawa M, Satoh H, Kumamaru T. (2010) Gene-gene interactions between mutants that accumulate abnormally high amounts of proglutelin in rice seed. Breed Sci 60: 568–574 [Google Scholar]
- Ushijima T, Matsusaka H, Jikuya H, Ogawa M, Satoh H, Kumamaru T. (2011) Genetic analysis of cysteine-poor prolamin polypeptides reduced in the endosperm of the rice esp1 mutant. Plant Sci 181: 125–131 [DOI] [PubMed] [Google Scholar]
- Wang Q, Kong L, Hao H, Wang X, Lin J, Samaj J, Baluska F. (2005) Effects of brefeldin A on pollen germination and tube growth: antagonistic effects on endocytosis and secretion. Plant Physiol 139: 1692–1703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Ren Y, Liu X, Jiang L, Chen L, Han X, Jin M, Liu S, Liu F, Lv J, et al. (2010) OsRab5a regulates endomembrane organization and storage protein trafficking in rice endosperm cells. Plant J 64: 812–824 [DOI] [PubMed] [Google Scholar]
- Washida H, Kaneko S, Crofts N, Sugino A, Wang C, Okita TW. (2009) Identification of cis-localization elements that target glutelin RNAs to a specific subdomain of the cortical endoplasmic reticulum in rice endosperm cells. Plant Cell Physiol 50: 1710–1714 [DOI] [PubMed] [Google Scholar]
- Woollard AA, Moore I. (2008) The functions of Rab GTPases in plant membrane traffic. Curr Opin Plant Biol 11: 610–619 [DOI] [PubMed] [Google Scholar]
- Yamagata H, Sugimoto T, Tanaka K, Kasai Z. (1982) Biosynthesis of storage proteins in developing rice seeds. Plant Physiol 70: 1094–1100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamagata H, Tanaka K. (1986) The site of synthesis and accumulation of storage proteins. Plant Cell Physiol 27: 135–145 [Google Scholar]