In Arabidopsis leaf pavement cells and trichomes, the ER is a reservoir for W/SRC signaling and may have a key role in the early steps of W/SRC activation.
Abstract
During plant cell morphogenesis, signal transduction and cytoskeletal dynamics interact to locally organize the cytoplasm and define the geometry of cell expansion. The WAVE/SCAR (for WASP family verprolin homologous/suppressor of cyclic AMP receptor) regulatory complex (W/SRC) is an evolutionarily conserved heteromeric protein complex. Within the plant kingdom W/SRC is a broadly used effector that converts Rho-of-Plants (ROP)/Rac small GTPase signals into Actin-Related Protein2/3 and actin-dependent growth responses. Although the components and biochemistry of the W/SRC pathway are well understood, a basic understanding of how cells partition W/SRC into active and inactive pools is lacking. In this paper, we report that the endoplasmic reticulum (ER) is an important organelle for W/SRC regulation. We determined that a large intracellular pool of the core W/SRC subunit NAP1, like the known positive regulator of W/SRC, the DOCK family guanine nucleotide-exchange factor SPIKE1 (SPK1), localizes to the surface of the ER. The ER-associated NAP1 is inactive because it displays little colocalization with the actin network, and ER localization requires neither activating signals from SPK1 nor a physical association with its W/SRC-binding partner, SRA1. Our results indicate that in Arabidopsis (Arabidopsis thaliana) leaf pavement cells and trichomes, the ER is a reservoir for W/SRC signaling and may have a key role in the early steps of W/SRC assembly and/or activation.
The W/SRC (for WASP family verprolin homologous/suppressor of cAMP receptor regulatory complex) and Actin-Related Protein (ARP)2/3 complex are part of an evolutionarily conserved Rho-of-Plants (ROP)/Rac small GTPase signal transduction cascade that controls actin-dependent morphogenesis in a wide variety of tissues and developmental contexts (Smith and Oppenheimer, 2005; Szymanski, 2005; Yalovsky et al., 2008). Many of the components and regulatory relationships among the complexes were discovered based on the stage-specific cell-swelling and -twisting phenotypes of the distorted class of Arabidopsis (Arabidopsis thaliana) trichome mutants (Szymanski et al., 1999; Zhang et al., 2005, 2008; Djakovic et al., 2006; Le et al., 2006; Uhrig et al., 2007). However, in both maize (Zea mays) and Arabidopsis, W/SRC and/or ARP2/3 are required for normal pavement cell morphogenesis (Frank and Smith, 2002; Mathur et al., 2003b; Brembu et al., 2004). Compared with other Arabidopsis pavement cell mutants, the shape defects of the distorted group are relatively mild. However, the distorted mutants and spike1 (spk1) differ from most other morphology mutants in that they display gaps in the shoot epidermis, most frequently at the interface of pavement cells and stomata (Qiu et al., 2002; Le et al., 2003; Li et al., 2003; Mathur et al., 2003b; Zhang et al., 2005; Djakovic et al., 2006). The cell gaps may reflect either uncoordinated growth between neighboring cells or defective cortical actin-dependent secretion of polysaccharides and/or proteins that promote cell-cell adhesion (Smith and Oppenheimer, 2005; Hussey et al., 2006; Leucci et al., 2007).
In tip-growing cells, there is a strict requirement for actin to organize the trafficking and secretion activities of the cell to restrict growth to the apex. In Arabidopsis, the W/SRC-ARP2/3 pathway is not an essential tip growth component, because null alleles of both W/SRC and ARP2/3 subunits do not cause noticeable pollen tube or root hair phenotypes (Le et al., 2003; Djakovic et al., 2006). However, reverse genetic analysis of the W/SRC subunit BRK1 and ARP2/3 in the tip-growing protonemal cells of Physcomitrella patens revealed the obvious importance of this pathway (Harries et al., 2005; Perroud and Quatrano, 2008). Along similar lines, in two different legume species, W/SRC subunits are required for a normal root nodulation response to symbiotic bacteria (Yokota et al., 2009; Miyahara et al., 2010), indicating a conditional importance for this pathway in root hair growth. These genetic studies centered on the W/SRC and ARP2/3 pathways, in addition to those that involve a broader collection of actin-based morphology mutants (Smith and Oppenheimer, 2005; Blanchoin et al., 2010), are defining important cytoskeletal proteins and new interactions with the endomembrane system during morphogenesis. However, it is not completely clear how unstable actin filaments and actin bundle networks dictate the growth patterns of cells (Staiger et al., 2009).
The difficulty of understanding the functions of specific actin arrays can be explained, in part, by the fact that plant cells that employ a diffuse growth mechanism have highly unstable cortical actin filaments and large actin bundles that do not have a geometry that obviously relates to the direction of growth or a specific subcellular activity (Blanchoin et al., 2010). This is in contrast to the cortical endocytic actin patches in yeast (Saccharomyces cerevisiae; Evangelista et al., 2002; Kaksonen et al., 2003) and cortical meshworks in the lamellipodia of crawling cells (Pollard and Borisy, 2003) that reveal subcellular locations where actin works to locally control membrane dynamics. In thick-walled plant cells, the magnitude of the forces that accompany turgor-driven cell expansion exceed those that could be generated by actin polymerization by orders of magnitude (Szymanski and Cosgrove, 2009). Localized cell wall loosening or the assembly of an anisotropic cell wall generates asymmetric yielding responses to turgor-induced stress (Baskin, 2005; Cosgrove, 2005). Therefore, the actin-based control of cell boundary dynamics is indirect, and the actin cytoskeleton influences cell shape change, in part, by actin and/or myosin-dependent trafficking of hormone transporters (Geldner et al., 2001) and organelles (Prokhnevsky et al., 2008), including those that control the localized delivery of protein complexes and polysaccharides that pattern the cell wall (Leucci et al., 2007; Gutierrez et al., 2009). In this scheme for actin-based growth control, the actin network dynamically rearranges at spatial scales that span from approximately 1- to 10-µm subcellular domains that may locally position organelles (Cleary, 1995; Gibbon et al., 1999; Szymanski et al., 1999) to the more than 100-µm actin bundle networks that operate at the spatial scales of entire cells (Gutierrez et al., 2009; Dyachok et al., 2011). It is clear from the work of several laboratories that the W/SRC and ARP2/3 protein complexes are required to organize cortical actin and actin bundle networks in trichomes (Szymanski et al., 1999; Le et al., 2003; Deeks et al., 2004; Zhang et al., 2005) and cylindrical epidermal cells (Mathur et al., 2003b; Dyachok et al., 2008, 2011). A key challenge now is to understand how plant cells deploy these approximately 10- to 20-nm heteromeric protein complexes to influence the patterns of growth at cellular scales.
The genetic and biochemical control of ARP2/3 is complicated, but this is a tractable problem in plants, because the pathway is relatively simple compared with most other species in which it has been characterized. For example, in organisms ranging from yeast to humans, there are multiple types of ARP2/3 activators, protein complexes, and pathways that activate ARP2/3 (Welch and Mullins, 2002; Derivery and Gautreau, 2010). However, the maize and Arabidopsis genomes encode only WAVE/SCAR homologous proteins that can potently activate ARP2/3 (Frank et al., 2004; Basu et al., 2005). Detailed genetic and biochemical analyses of the WAVE/SCAR gene family in Arabidopsis demonstrated that the plant activators function interchangeably within the context of the W/SRC and define the lone pathway for ARP2/3 activation (Zhang et al., 2008). Bioinformatic analyses are consistent with a prominent role for W/SRC in the angiosperms, because in general, WASH complex subunits, which are structurally similar to WAVE/SCAR proteins, are largely absent from the higher plant genomes, while WAVE/SCAR genes are highly conserved (Kollmar et al., 2012).
The components and regulatory schemes of the W/SRC-ARP2/3 pathway in Arabidopsis and P. patens are conserved compared with vertebrate species that employ these same protein complexes (Szymanski, 2005). For example, mutant complementation tests indicate that human W/SRC and ARP2/3 complex subunits can substitute for the Arabidopsis proteins (Mathur et al., 2003b). Furthermore, biochemical assays of Arabidopsis W/SRC (Basu et al., 2004; El-Assal et al., 2004; Frank et al., 2004; Le et al., 2006; Uhrig et al., 2007) and ARP2/3 assembly (Kotchoni et al., 2009) have shown that the binary interactions among W/SRC subunits and ARP2/3 complex assembly mechanisms are indistinguishable from those that have been observed for human W/SRC (Gautreau et al., 2004) and yeast ARP2/3 (Winter et al., 1999). After an initial period of controversy concerning the biochemical control of W/SRC, it is now apparent that vertebrate W/SRC (Derivery et al., 2009; Ismail et al., 2009), like the ARP2/3 complex (Machesky et al., 1999), is intrinsically inactive and requires positive regulation by Rac and other factors to fully activate ARP2/3 (Ismail et al., 2009; Lebensohn and Kirschner, 2009; Chen et al., 2010). Although overexpression of dominant negative ROP mutants causes trichome swelling and a reduced trichome branch number (Fu et al., 2002), the involvement of ROPs in trichome morphogenesis has been difficult to prove with a loss-of-function ROP allele because so many ROPs are expressed in this cell type (Marks et al., 2009). Existing reports on ROP loss-of-function mutants demonstrate the importance of pavement cell morphogenesis but do not document a trichome phenotype (Fu et al., 2005; Xu et al., 2010). A recent report describes a clever strategy to generate ROP loss-of-function lines that used the ectopic expression of ROP-specific bacterial toxins. There was a strong association between inducible expression of the toxins and the appearance of trichomes with severe trichome swelling and reduced branch number phenotypes (Singh et al., 2012). Although the exact mechanism of ROP-dependent control of W/SRC remains to be determined, the results described above in combination with the detection of direct interactions between the ROPGEF SPK1, active forms of ROP, and W/SRC subunits (Basu et al., 2004, 2008; Uhrig et al., 2007) strongly suggest that W/SRC is a ROP effector complex.
The major challenge in the field now is to better understand the cellular control of W/SRC and how the complex is partitioned into active and inactive pools. In mammalian cells that crawl on a solid substrate, current models propose that a cytosolic pool of inactive WAVE/SCAR proteins and W/SRC is locally recruited and activated at specific plasma membrane surfaces in response to signals from some unknown Rac guanine nucleotide-exchange factor (GEF), protein kinase, and/or lipid kinase (Oikawa et al., 2004; Lebensohn and Kirschner, 2009; Chen et al., 2010). However, in Drosophila melanogaster neurons (Bogdan and Klämbt, 2003) and cultured human melanoma cells (Steffen et al., 2004), there are large pools of W/SRC with a perinuclear or organelle-like punctate localization that has no obvious relationship to cell shape or motility, raising uncertainty about the cellular mechanisms of W/SRC activation and the importance of different subcellular pools of the complex.
In plants, cell fractionation experiments indicate that SCAR1 and ARP2/3 have an increased association with membranes compared with their animal counterparts (Dyachok et al., 2008; Kotchoni et al., 2009). In tip-growing moss protonemal cells, both the W/SRC subunit BRK1 and ARP2/3 localize to a population of unidentified organelles within the apical zone (Perroud and Quatrano, 2008). Similar live-cell imaging experiments in Arabidopsis reported a plasma membrane localization for SCAR1 and BRK1 in a variety of shoot epidermal and root cortex, and their accumulation at young trichome branch tips and at three-way cell wall junctions may define subcellular domains for W/SRC-ARP2/3-dependent actin filament nucleation at the plasma membrane (Dyachok et al., 2008). However, to our knowledge, active W/SRC, defined here as the fraction of W/SRC that colocalizes with ARP2/3 or actin, has not been reported in plants, and the plasma membrane is not necessarily the only organelle involved in W/SRC regulation. For example, the reported accumulation of BRK1 and SCAR1 at three-way cell wall junctions has a punctate appearance at the cell cortex that may not simply correspond to the plasma membrane (Dyachok et al., 2008). Also, in young stage 4 trichomes, there was an uncharacterized pool of intracellular SCAR1, but not BRK1, that localized to relatively large punctate structures (Dyachok et al., 2008). The endoplasmic reticulum (ER) may also be involved in W/SRC regulation. The ER-localized DOCK family ROPGEF SPK1 (Zhang et al., 2010) physically associates with multiple W/SRC proteins (Uhrig et al., 2007; Basu et al., 2008) and, based on genetic criteria, is an upstream, positive regulator of the W/SRC-ARP2/3 pathway (Basu et al., 2008). In the leaf, one function of SPK1 is to promote normal trafficking between the ER and Golgi; however, arp2/3 mutants do not share ER-stress phenotypes with spk1 (Zhang et al., 2010), making it unclear if SPK1 and the ER are directly involved in W/SRC signaling.
This paper focuses on the localization and control of the W/SRC subunit NAP1/GNARLED/NAPP/HEM1/2. Arabidopsis NAP1 directly interacts with the ROP/Rac effector subunit SRA1/PIROGI/KLUNKER/PIRP (Basu et al., 2004; El-Assal et al., 2004; Uhrig et al., 2007). Based on the equally severe syndrome of nap1 and arp2/3 null phenotypes, and double mutant analyses, the only known function of NAP1 is to positively regulate ARP2/3 (Brembu et al., 2004; Deeks et al., 2004; El-Din El-Assal et al., 2004; Li et al., 2004). The vertebrate SRA1-NAP1 dimer is important for W/SRC assembly (Gautreau et al., 2004) and forms an extended physical surface that trans-inhibits the C-terminal ARP2/3-activating domain of WAVE/SCAR (Chen et al., 2010). The plant NAP1 and SRA1 proteins share end-to-end amino acid conservation with their vertebrate homologs and may form a heterodimer with similar functions (Basu et al., 2004; El-Assal et al., 2004; Uhrig et al., 2007). We report here that Arabidopsis NAP1 is strongly associated with ER membranes. In a detailed series of localization experiments, we detect a complicated intracellular distribution of NAP1 among the ER, the nucleus, and unidentified submicrometer punctae. A large pool of ER-associated NAP1 is inactive, based on the low level of colocalization with actin.
Its accumulation on the ER does not require activating signals from either SPK1 or SRA1. These data indicate that the ER is a reservoir for W/SRC signaling and suggest that early steps in the positive regulation of NAP1 and the W/SRC occur on the ER surface.
RESULTS
NAP1 Strongly Associates with Membranes and Cofractionates with ER Membranes
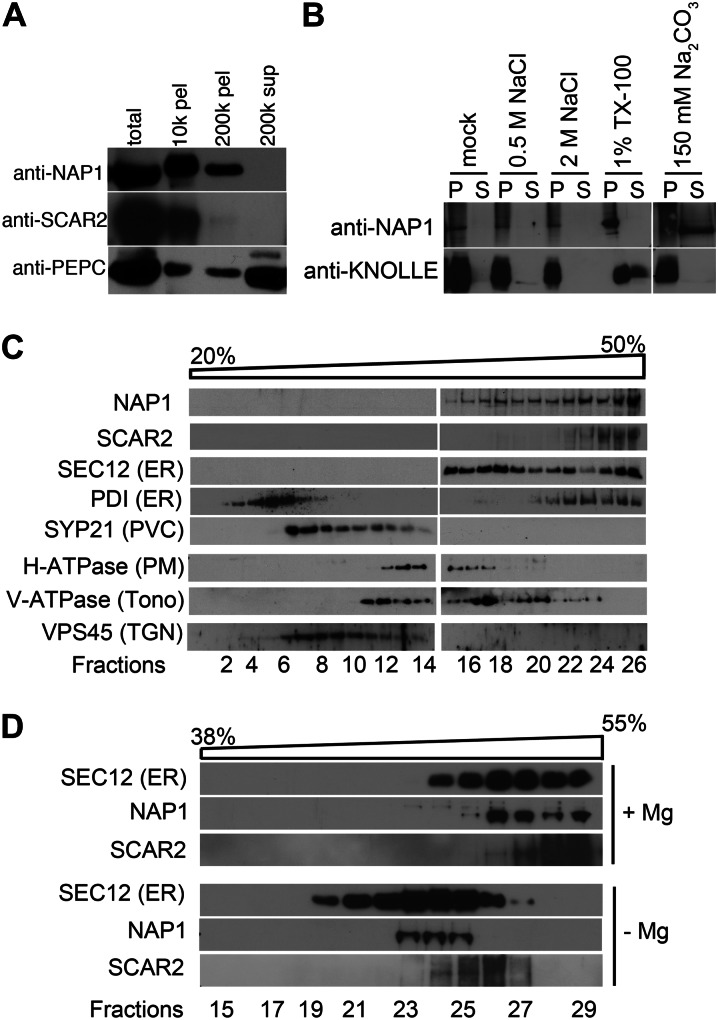
Because NAP1 is widely expressed in the shoots (El-Assal et al., 2004), we were able to examine the subcellular distribution of NAP1 extracts prepared from shoots at 16 to 18 d after germination (DAG). Cell extracts were separated by differential centrifugation, and the supernatant and pellet fractions were loaded by equal proportions of cytosolic and particulate fractions and subjected to western-blot analysis (Fig. 1A). The specificity of the NAP1 antibody that had been previously reported for western blotting (Le et al., 2006) was reconfirmed (Supplemental Fig. S1A). We consistently detected NAP1 among the pellet fractions, with the majority of the protein present in the 10,000g pellet fraction. It proved difficult to detect any NAP1 in the cytosolic 200,000g supernatant fraction, and only a small proportion of NAP1 was detected in the cytosol in overexposed blots (Supplemental Fig. S2). Consistent with previous reports of SCAR1 being present in the particulate fractions (Dyachok et al., 2008, 2011), the vast majority of SCAR2 was in the 10,000g pellet organelle fraction and was detected in soluble fraction only upon overexposure of western blots (Supplemental Fig. S2). The cytosolic enzyme phosphoenolpyruvate carboxylase served as a control for the quality of the cell fractionation and was detected primarily in the cytosolic fraction (Fig. 1A). Because the S/WRC subunits lack any signal sequences or strongly predicted membrane-spanning segments, these results suggested that NAP1 was a peripheral membrane-associated protein. The association of NAP1 with membranes was relatively strong, because it could not be dissociated from crude microsomes following treatment with high concentrations of salt or the nonionic detergent Triton X-100 (Fig. 1B). The membrane-associated NAP1 was resistant to most commonly used detergents but could be solubilized with deoxycholate (Supplemental Fig. S3). As expected, NAP1 had the properties of a peripheral membrane protein, because it was efficiently solubilized by treatment with Na2CO3 (Fujiki et al., 1982), while the integral membrane protein KNOLLE was not (Fig. 1B).
Figure 1.
Arabidopsis NAP1 strongly associates with membranes and cofractionates with the ER marker protein SEC12. A, Cell fractionation of NAP1 and SCAR2 in wild-type leaf extracts. Different fractions of cell extracts were loaded by equal proportion and blotted with anti-NAP1 and anti-SCAR2 antibodies, and the NAP1 blot was reprobed with anti-phosphoenolpyruvate carboxylase (PEPC) antibody. The centrifugation speeds and fraction type are indicated. B, NAP1 is a peripheral membrane protein. Assays determined the NAP1 membrane association for the different buffer conditions indicated at the top of the panel. Membrane-binding assays under different binding conditions were recentrifuged, and the partitioning of NAP1 between the pellet (P) and supernatant (S) fractions was analyzed by western blotting. C, Western-blot data of Suc velocity gradient fractions indicating the sedimentation behavior of NAP1 and SCAR2 relative to a panel of organelle marker proteins. SEC12 and PDI were used as ER markers. SYP21, H-ATPase, V-ATPase subunit E, and VPS45 were used as markers for prevacuolar compartment (PVC), plasma membrane (PM), tonoplast (Tono), and trans-Golgi network (TGN), respectively. D, Disassembly of ER membrane-associated ribosomes by chelating Mg2+ with EDTA shifts the distribution of NAP1, SCAR2, and SEC12 to lower density fractions.
To determine if NAP1 and SCAR2 associated preferentially with subsets of organelle membranes, crude microsomes were separated by Suc velocity gradient centrifugation. Three independent Suc gradient assays revealed a clear specificity for NAP1 membrane association, because NAP1 positive membranes were consistently distributed among the most dense fractions (Fig. 1C). In leaf microsomes, vesicles that are positive for ER marker proteins are relatively dense and are distributed near the bottom of the gradient (Ueda et al., 2010; Zhang et al., 2010). The NAP1-positive membranes overlapped to the greatest degree with the subset of dense membranes that contained known ER marker proteins: the ER-specific GEF SEC12 and the luminal ER protein PDI (Fig. 1C). Although NAP1, SCAR2, PDI, and SEC12 were all present in the most dense membrane fractions, there was variability among the marker proteins in their distribution on the gradient, perhaps reflecting the heterogeneity of the composition of the ER within and/or between cell types. The S/WRC subunits SCAR1 and BRK1 have been reported to have a plasma membrane localization based on live-cell imaging using fluorescent protein-tagged proteins (Dyachok et al., 2008, 2011). The microsome-associated pool of NAP1 that we detected in fractionated cells did not appear to be solely plasma membrane associated, because the plasma membrane H+-ATPase is resolved from the ER-containing fractions (Fig. 1C). However, there was some overlap of NAP1 signal with the plasma membrane marker, and there may be a minor pool of NAP1 that stably associates with the plasma membrane in this assay. When the Suc gradient fractions were probed with the SCAR2 antibody, it was clear that SCAR2, like the dense NAP1-, PDI-, and SEC12-positive vesicles, was also resolved from all of the other marker proteins that were analyzed (Fig. 1C). These results do not necessarily contradict previous imaging data obtained from pavement cells and trichomes (Dyachok et al., 2008, 2011). First, mesophyll cells are the predominant source of organelles for these experiments, and NAP1 and SCAR2 may have a different localization in this cell type. Second, these experiments are biased to detect proteins with stable associations with organelles that survive for approximately 20 h during the fractionation protocols. A small or unstable pool of membrane-associated NAP1 or SCAR2 that was independent of the ER might not be detected in these experiments. To test further for a possible ER association of NAP1 and SCAR2, we treated crude microsomes with EDTA to dissociate ribosomes and to specifically decrease the buoyant density of ER membranes. In this experiment, NAP1-, SCAR2-, and SEC12-positive membranes were now shifted to less dense fractions relative to the untreated control membranes (Fig. 1D). This result was consistent with an ER localization of NAP1 and SCAR2, but in this assay, we again detected subtle differences in the properties of NAP1- and SCAR2-positive membranes (Fig. 1D) that could reflect differing interactions of the two W/SRC subunits with distinct peripheral ER proteins or different subdomains of the ER. Collectively, these results indicate that the vast majority of NAP1 and SCAR2 in cell extracts is membrane associated and may be enriched in ER membranes.
NAP1 Localizes to Multiple Organelles in Developing Pavement Cells and Trichomes
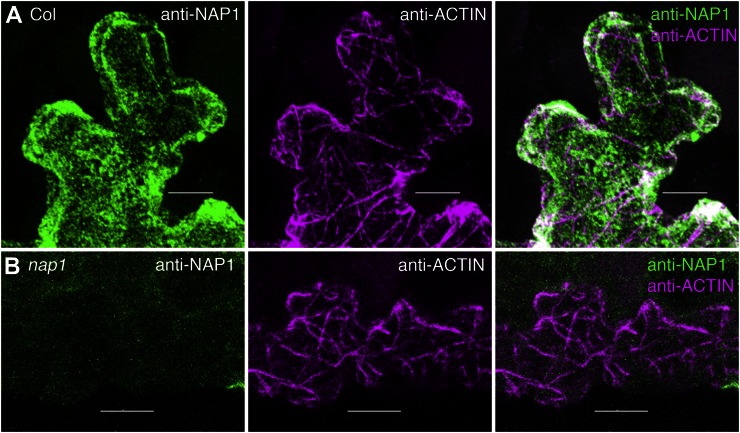
To learn more about the precise localization of NAP1 in epidermal cells, we conducted a series of immunolocalization experiments using the same custom NAP1 antibody (Le et al., 2006). The utility of the NAP1 antiserum for localization was demonstrated by comparing the fluorescence signal of the crude serum with that of the preimmune control serum from the same rabbit. A maximum projection of the entire confocal volume containing the epidermis and a layer of underlying mesophyll cells revealed a dramatically enhanced signal for the crude NAP1 serum compared with the preimmune control under the same imaging conditions (Supplemental Fig. S1, B and C). Affinity purification of the NAP1 antibody using a peptide antigen affinity column helped to reduce the frequency of random spots of NAP1 signal. In freeze-shattering immunolabeling experiments, the affinity-purified NAP1 antibody generated strong and specific signals. Efficiently fixed and well-labeled wild-type pavement cells were identified based on filamentous anti-actin antibody in one channel and strong NAP1 in a second channel (Fig. 2A). In this whole-mount labeling technique, the antibody cannot penetrate adjacent cells that are not fractured, and these cells defined the level of autofluorescence. The anti-NAP1 signal was specific, because in repeated identical double-labeling experiments conducted using nap1 leaves, actin signal was obvious, but the NAP1 channel had only background levels of signal (Fig. 2B).
Figure 2.
Whole-mount freeze shattering of the epidermis detects highly specific signal for the affinity-purified anti-NAP1 antibody. A, Confocal image projection of a freeze-shattered region of the epidermis including a cracked labeled cell and an unlabeled neighboring cell. Immunolabeling is shown for wild-type leaf pavement cells with anti-NAP1 (left) and anti-actin (middle) antibodies. Right, Overlay image of the two channels. Genotypes, antibodies, and keys to the false coloring of the signals are indicated in this and all other similarly organized image panels. B, Anti-NAP1 signal is absent in the nap1/grl-6 null mutant. Immunolabeling is shown for nap1 leaf pavement cells with anti-NAP1 (left) and anti-actin (middle) antibodies. Right, Overlay image of the two channels. The image settings in B were the same as in A. Bars = 5 μm in A and 10 μm in B.
The initial series of NAP1 localization experiments focused on epidermal pavement cells in cotyledons and leaves. We immediately noticed that NAP1 was sensitive to detergent extraction, because its pattern was very diffuse in samples processed using standard cytoskeleton localization protocols (Szymanski et al., 1999), but when detergent extraction times were reduced (see “Materials and Methods”), approximately 40% of the cotyledon pavement cells from 2- and 10-DAG seedlings and leaf pavement cells consistently displayed a reticulate network of NAP1 signal and/or punctate NAP1-positive structures that were 0.7 ± 0.2 µm (n = 126) in diameter (Fig. 3). Reducing detergent concentrations from 1% to 0.1% Triton X-100 yielded an identical pattern of reticulate and/or punctate NAP1 and increased the percentage of cells with this pattern to approximately 80%. However, this level of detergent precluded double-labeling experiments with actin, because actin filaments could only rarely be detected in these mildly extracted cells; therefore, 1% Triton X-100 was used with a reduced incubation time. The image planes displayed in the left panels of Figure 3 illustrate the reticulate pattern, and those on the right include image planes that highlight examples of the small NAP1 punctae in pavement cells from cotyledons and leaves. Using fluorescent protein tagging and live-cell imaging, the W/SRC complex subunits BRK1 and SCAR1 commonly display increased signal at three-way cell wall junctions (Dyachok et al., 2008, 2011). In our immunolocalization experiments, bright NAP1 spots could be detected at three-way cell wall junctions (Fig. 4A); however, clusters of relatively high NAP1 signal (Fig. 2A) and NAP1 punctae (Fig. 3) did not have an obviously restricted localization to a particular cortical location. The NAP1-positive structures could be observed at different cortical locations, independent of cell shape. In the future, it will be important to generate fluorescent protein fusions to NAP1 to enable direct comparisons of its localization with those of BRK1 and SCARs. However, to our knowledge, a functional fluorescent protein-tagged NAP1 subunit has not been reported in a genetic system. In 2-DAG, but not 10-DAG, cotyledon pavement cells, NAP1 was also localized to larger elliptical structures that were present at moderate abundance of approximately 10 to 20 per cell (Fig. 3A). These elongated NAP1-positive structures, which were approximately parallel to the image plane, had a mean length of 3.9 ± 0.6 µm and a mean width of 1.7 ± 0.3 µm (n = 51). The sizes of these relatively large NAP1-positive structures and their presence only in early-stage cotyledons are similar to what has been previously reported for ER bodies (Matsushima et al., 2003).
Figure 3.
NAP1 has a complex punctate and reticulate pattern in cotyledon and leaf pavement cells at different developmental stages. A, Confocal image projection of NAP1 localization in 2-DAG wild-type cotyledon pavement cells. NAP1-positive reticulate and cylindrical structures are shown (left). NAP1-positive small punctae are highlighted (right). B, NAP1 localization patterns in 10-DAG wild-type cotyledon pavement cells. Localization patterns are displayed as in A. Image planes include a cortical subregion of the apical surface of a highly lobed, fully expanded cell. C, NAP1 localization patterns in 14-DAG wild-type leaf pavement cells. Localizations patterns are displayed as in A. Bars = 5 μm.
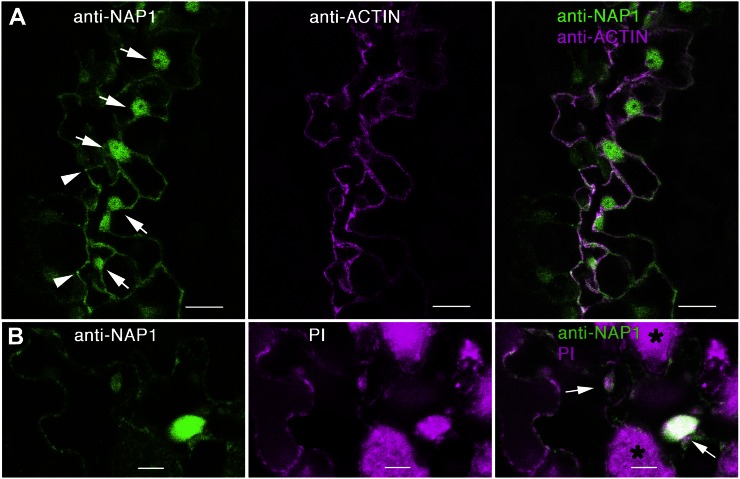
Figure 4.
NAP1 is localized to the nucleus in pavement cells. A, Wild-type leaf pavement cells immunolabeled with anti-NAP1 (left) and anti-actin (middle) showing NAP1 localization in the nuclei of cells along the freeze fracture on the leaf tissue, as indicated by arrows (left). Right, Overlay image of the two channels. Left, Arrowheads indicate relatively bright NAP1 signal at three-way cell wall junctions of three different pavement cells. Images are projections of confocal images that include most of the pavement cell volume. B, Double labeling with leaf pavement cells labeled with anti-NAP1 antibody (left) and propidium iodide (PI; middle). Right, Overlay image of the two channels. Arrows (right) indicate double-labeled nuclei. The asterisks (right) show mesophyll cells that have chlorophyll autofluorescence under image settings for propidium iodide. Bars = 5 μm.
We also consistently detected a nuclear pool of NAP1 in pavement cells. In Figure 4A, a freeze-shattered crack in the adaxial epidermis that occurred along a series of adjacent cells is shown. Each of these cells had clearly labeled nuclei that were present in optical sections that included the nucleus and the lumen of the large central vacuole that did not contain either F-actin or NAP1 signal. The identity of these NAP1-positive compartments as nuclei was inferred based on the presence of a nucleolus-like structure within the organelle that was not NAP1 positive and the common occurrence of actin meshworks and bundles that surround the organelle. To conclusively prove that these organelles are nuclei, samples were double labeled with anti-NAP1 and propidium iodide, which binds to nucleic acids and labels nuclei. NAP1-positive nuclei in pavement cells were also labeled with propidium iodide (Fig. 4B). Most of the background signal in the propidium iodide channel is due to chlorophyll autofluorescence from the underlying mesophyll cell layer that is unavoidable, since the nuclei are usually positioned along the basal surface of the pavement cells. The presence or absence of NAP1 in pavement cell nuclei was unpredictable, with no obvious correlations between cell size or shape. Prominent nuclear labeling of NAP1 was also observed in trichomes (Fig. 5D). Close inspection of trichomes at different stages detected NAP1 signal above background levels at all stages of morphogenesis. For example, of the 117 NAP1- and GFP-HDEL-labeled trichomes that were part of this study, 37 cells had a distinctive nuclear envelope that could be visualized with the GFP-HDEL signal (five stage 1/2 cells, three stage 3/4 cells, 26 stage 5 cells, and three stage 6 cells). NAP1 was present in the nucleus in every instance; therefore, NAP1 localization to the nucleus can occur throughout trichome development.
Figure 5.
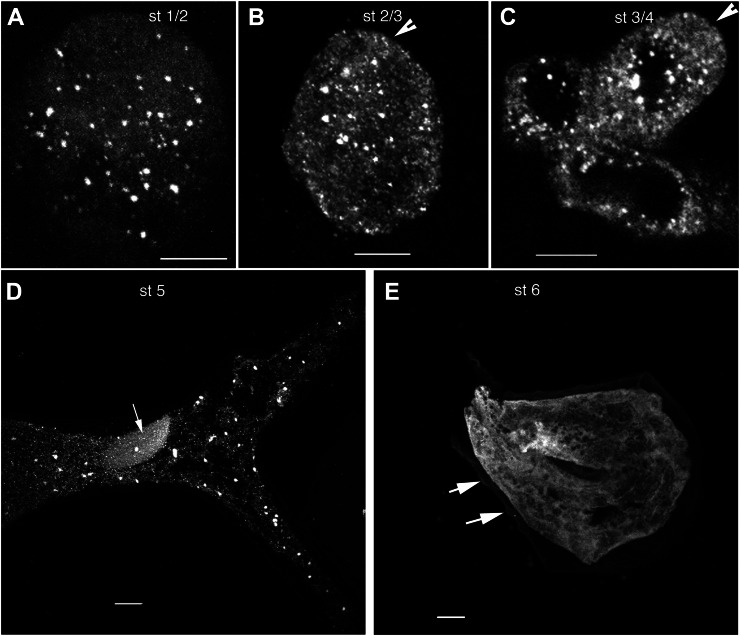
In trichomes, NAP1 transitions from a distributed population of punctae in growing cells from stages 1 through 5 to a reticulate pattern in fully expanded stage 6 cells. A, Immunolocalized NAP1 is concentrated at punctate structures that are distributed throughout a spherical stage 1 (st 1/2) trichome cell. The image shown is the maximum Z-projection of a confocal image stack of anti-NAP1 signal from nearly all of the cell volume. B, NAP1 localization in a cylindrical unbranched stage 2/3 (st 2/3) trichome that is at the transition to branch initiation. The arrowhead indicates the position of a small bulge that defines the position of a newly initiated branch. The confocal image is a Z-projection containing nearly all of the cell volume. C, NAP1 localization in a stage 4 (st 4) trichome that contains a short branch with a blunt tip. NAP1 is localized to punctate structures. The arrowhead indicates a blunt trichome branch tip at the stage when NAP1 is essential for polarized growth. The image is the maximum Z-projection of part of a confocal image stack that contains nearly all of the upper trichome. D, NAP1 localization in a stage 5 (st 5) trichome. NAP1 is localized to punctate structures and diffuse membrane structures. The large elliptical structure in the trichome’s stalk is the nucleus, as indicated by the arrow. The image is the maximum Z-projection of a confocal image stack. E, NAP1 localization in a stage 6 (st 6) trichome. NAP1 shows a reticulate structure and diffuse sheets of membrane. Punctate NAP1 structures were rarely found in this stage 6 cell. The arrows indicate the cell wall tubercles that define stage 6 cells that have ceased cell expansion. Bars = 5 μm in A and B and 10 μm in C to E.
In addition to the nuclear pool of NAP1 in trichomes, there was an interesting punctate distribution that varied depending on the developmental stage. In stage 6 trichomes, NAP1 had the appearance of fenestrated sheets and a reticulate meshwork that was similar to what has been described previously for the ER using ER-targeted GFP-HDEL fusion proteins (Haseloff et al., 1997; Ridge et al., 1999). NAP1 localization was also determined in actively growing trichomes from stage 1 through stage 5 (Szymanski et al., 1998). The distorted mutants are defined by a stage 4-dependent requirement for actin and ARP2/3 as the trichome branch buds transition from a symmetrical bulge (stage 3) to an elongated structure (Szymanski et al., 1999). At these early stages, including stage 3/4 cells (Fig. 5), NAP1 had a distributed punctate signal. Although the punctate signal in projected images of stage 3/4 and stage 5 trichomes appeared to be contained within developing vacuole-like organelles with a diameter of approximately 10 µm (Fig. 5, C and D), in single image planes, the punctae were always adjacent to, but not within, the putative vacuole compartments. The size and shape of the NAP1 punctae were quantified from thresholded images of stage 3/4 and stage 5 cells. The mean maximum diameter of NAP1 punctae increased from 1.1 ± 0.4 µm (n = 151) in stage 3/4 cells to 2.3 ± 1.5 µm (n = 162) in stage 5 cells (Supplemental Table S1). In stage 3/4 cells, BRK1 was previously localized to the plasma membrane along the tip of an expanding branch, while SCAR1 had a more complex localization that included both tip localization and a putative localization to intracellular organelles that was distinct from BRK1 (Dyachok et al., 2008). Although our freeze-shattering experiments frequently detected NAP1 punctae at or near the apex of branches (Fig. 5C), in 12 stage 4 cells in which the branch tip was well labeled, we did not observe a strong concentration of NAP1 at this distal location.
NAP1 Colocalizes with the ER in Pavement Cells and Trichomes
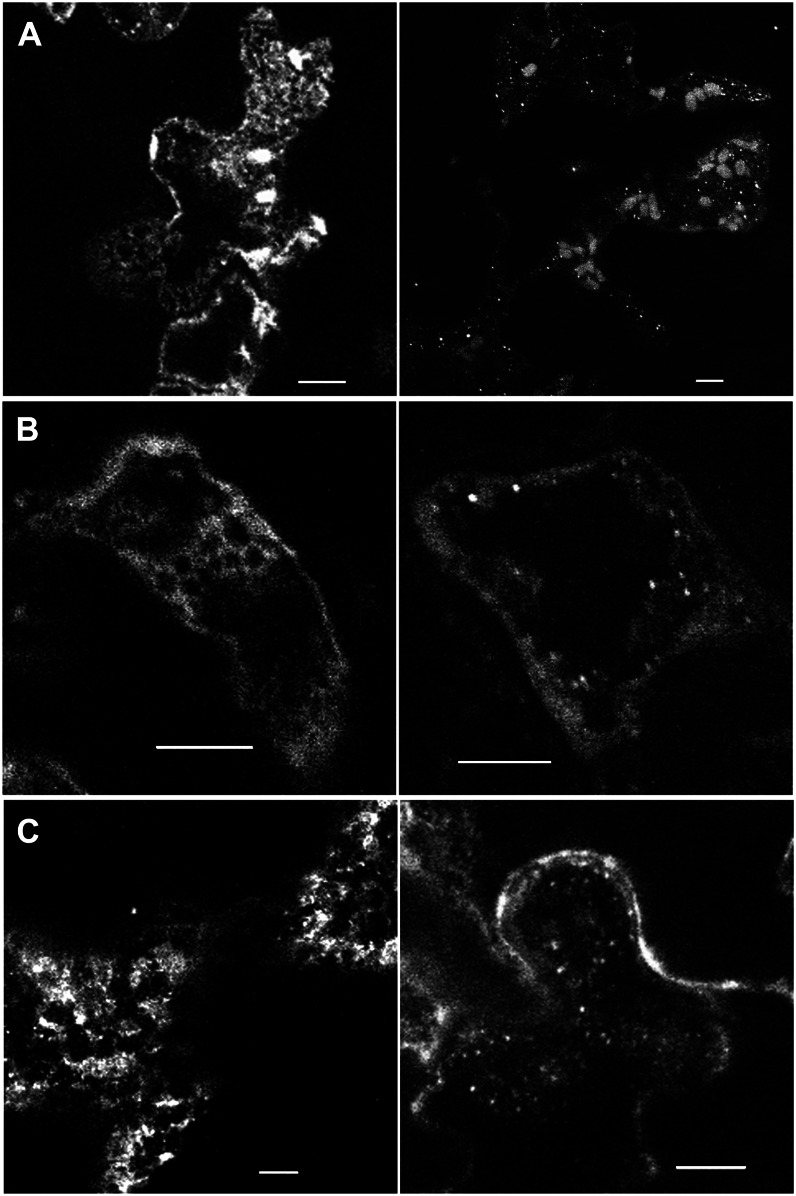
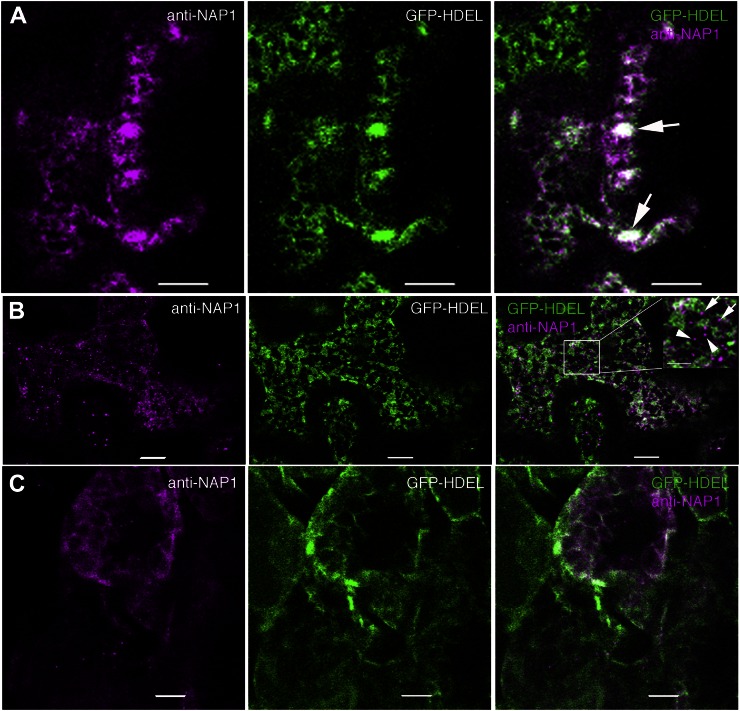
To determine if the NAP1-positive structures detected in pavement cells corresponded to the ER, we localized NAP1 in the Arabidopsis Biological Resource Center transgenic line CS16251 that expressed a luminal GFP-HDEL (Nelson et al., 2007) reporter that specifically labels the ER (Fig. 6). In living cells, GFP-HDEL displays the characteristic reticulate network of ER in vacuolated pavement cells (Zhang et al., 2010). Our immunolocalization protocols are known to preserve the native patterns of the plasma membrane and ER in pavement cells (Zhang et al., 2010). In 2-DAG cotyledon pavement cells, the relatively large punctae strongly colocalized with bright GFP-HDEL-positive structures that, based on their size, shape, and number, corresponded to ER bodies (Hara-Nishimura and Matsushima, 2003; Ogasawara et al., 2009). The colocalization can be visualized qualitatively as the white pixels in the merged panel in Figure 6A that reflect overlapping false-colored magenta NAP1 pixels with the green GFP-HDEL pixels. In freeze-shattered GFP-HDEL-expressing pavement cells that had a reticulate NAP1 localization, there was a substantial overlap of the NAP1 and GFP-HDEL signals (Fig. 6, A and B). To quantify the colocalization between NAP1 and GFP-HDEL, we calculated Pearson correlation coefficients from single image planes that contained at least 100 µm2 of the cortical cytoplasm. The Pearson correlations coefficient was 0.59 ± 0.07 (n = 15). This was a moderately high correlation coefficient, only slightly lower than the value of 0.73 that was reported for two ER proteins in a two-color live-cell imaging experiment (Sroubek and McDonald, 2011). The colocalization of NAP1 and GFP-HDEL was not limited to pavement cells and was also observed in mesophyll cells that were also labeled in freeze-shattering experiments (Fig. 6C). In double-labeled pavement cells, most of the NAP1 signal had the overall appearance of a general ER marker. However, we also consistently detected smaller, approximately 0.7-µm spherical NAP1-positive structures that did not obviously colocalize with the GFP-HDEL ER marker. Rather, they were often adjacent to the ER (Fig. 6B). After a close inspection of image stacks that contained a high density of NAP1 punctae, it was found that 80% (n = 242) of the punctae were just touching or partially overlapping with GFP-HDEL-positive ER tubules and dilated cisternae. This pattern has been previously described for ER exit site (ERES) marker proteins and SPK1 in immunolabeled cells (Yang et al., 2005; Zhang et al., 2010) and may reflect a specialized domain of the ER that is not labeled with GFP-HDEL. The remaining 20% of the NAP1 punctae were physically isolated from the ER, which is quite well preserved (Fig. 6B, inset), and could reflect the existence of a small pool of NAP1 that is physically separated from GFP-HDEL and the ER on some other organelle surface or as part of a cluster of cytosolic protein complexes. As shown previously (Zhang et al., 2010), our fixation methods retain the plasma membrane localization pattern of PHOT1-GFP, and in double-labeled cells, some of the NAP1 signal clearly does not overlap with the plasma membrane marker (Supplemental Fig. S4), but due to the thinness of the cortical cytoplasm in vacuolated plant cells, there are also many instances in which NAP1 signal overlapped significantly with PHOT1-GFP, and we cannot rule out the existence of a plasma membrane pool of NAP1 in this cell type.
Figure 6.
NAP1 colocalizes with the ER marker GFP-HDEL in ER bodies and/or reticulate structures in leaf pavement cells and mesophyll cells. Wild-type cells expressing transgenic GFP-HDEL (middle) were labeled with anti-NAP1 antibody that was detected with an Alexa546-conjugated secondary antibody (left). Right, Overlay images of the two channels. A, In pavement cells, NAP1 colocalizes with GFP-HDEL-positive ER cisternae, ER tubules, and ER bodies. The arrows point to ER bodies that contain both GFP-HDEL and NAP1. B, In pavement cells, punctate NAP1 signals do not completely overlap with GFP-HDEL but, instead, are either adjacent to GFP-HDEL-labeled ER or in seemingly independent structures. Right, The inset image is the enlarged boxed region in the same panel. The arrows indicate NAP1 punctae that are in physical contact with GFP-HDEL-labeled ER. The arrowheads indicate NAP1 punctae that were not associated with the GFP-HDEL ER. C, NAP1 colocalizes with GFP-HDEL in mesophyll cells. Bars = 5 μm in A, 10 μm in B and C, and 5 μm in the inset in B.
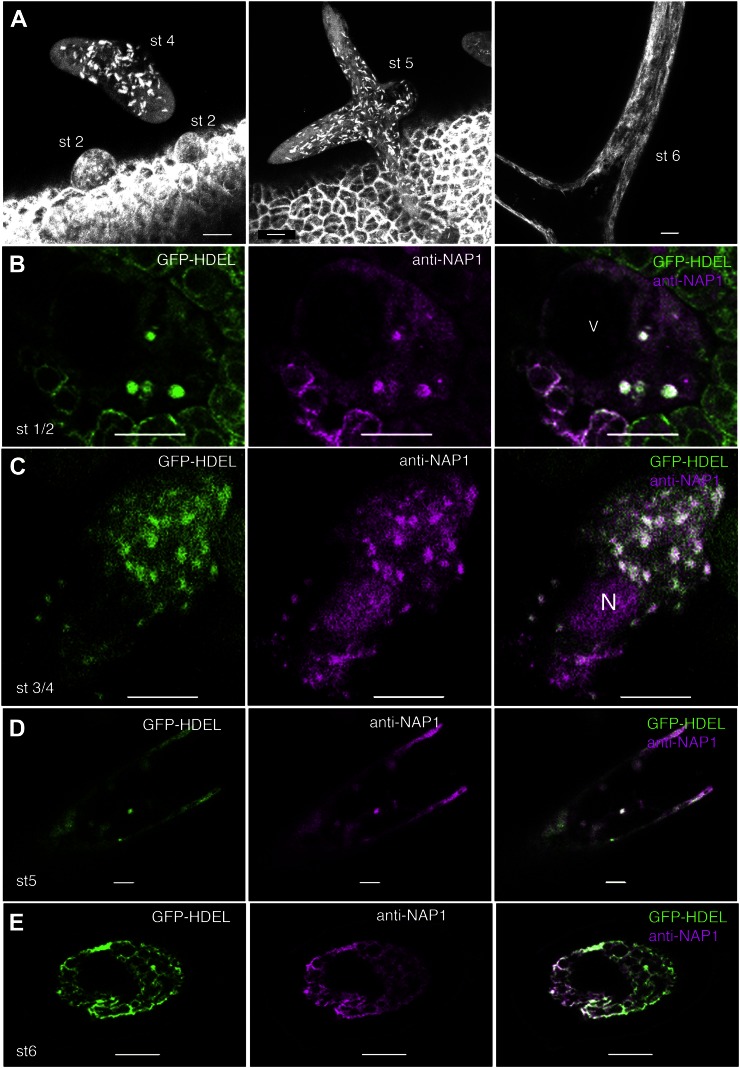
To determine if the punctate pattern of NAP1 in trichomes also corresponded to the ER, living 10- to 12-DAG GFP-HDEL plants were mounted on microscope slides and scanned to locate trichomes at specific developmental stages. Interestingly, the GFP-HDEL localization in developing trichomes included both diffuse meshworks and discrete punctae, the latter of which resembled the NAP1 localization pattern (Fig. 7A). In spherical stage 1 and unbranched cylindrical stage 2 trichomes, a subpool of the GFP-HDEL reporter localized to numerous, bright, nodule-like structures that were scattered throughout a more uniform diffuse fluorescence signal. This localization pattern persisted through stage 5 of trichome development. The overall shape of NAP1 and GFP-HDEL-positive structures appeared slightly elongated, and this was reflected in the measured mean circularity shape factors that were slightly less than 1, which is the circularity value of a perfect circle (Supplemental Table S1). An elongated nodule-like ER morphology is observed in rapidly growing pollen tubes (Lovy-Wheeler et al., 2007) and is a documented altered morphology of the rough ER-containing nodules that occur in response to oryzalin treatment (Langhans et al., 2009). The mean maximum diameter of the GFP-HDEL organelles in live cells was slightly larger than the NAP1 punctae in fixed samples and increased from 1.1 ± 0.4 µm in stage 3/4 cells to 2.1 ± 0.8 µm in stage 5 trichomes (Supplemental Table S1). The smaller size of NAP1 punctae in fixed wild-type cells compared with GFP-HDEL in living cells was probably an artifact of fixation and/or incomplete labeling or a potential effect of GFP-HDEL on the size of the structure. Although it is clear that the punctate appearance of NAP1 in trichomes did not require GFP-HDEL overexpression, because that same pattern was detected in untransformed plants (Fig. 5, A–D), it is possible that GFP-HDEL overexpression artificially forms or alters these ER nodule-like structures. The exact morphological changes in the ER during trichome development are not well characterized, and additional analyses of ER dynamics in trichomes are needed. However, the transition of the ER from nodules to a diffuse reticulate network lacking GFP-HDEL-positive nodules occurred in this transgenic line (Fig. 7A, right) and resembled the pattern of NAP1 localization in mature trichomes in freeze-shattered cells from untransformed lines (Fig. 5E).
Figure 7.
In expanding trichomes, the ER consists of a distributed population of nodule-like structures that strongly colocalize with NAP1. A, Localization patterns of GFP-HDEL in wild-type live-cell trichomes. GFP-HDEL is localized to punctate structures in early stage 2 (st 2) trichomes and slightly larger elliptical structures in stage 4 (st 4) trichomes, as shown by the maximum Z-projection of multiple confocal images. GFP-HDEL is localized to elliptical structures in stage 5 (st 5) trichomes, as shown by the maximum Z-projection of a confocal image stack containing most of the stage 5 trichome that is transitioning to a pointed branch tip morphology (middle). GFP-HDEL is localized to membrane sheets and extended tubules in stage 6 (st 6) trichomes, as shown by the maximum Z-projection of confocal images containing part of a stage 6 trichome (right). B, Single confocal image slice showing the colocalization between NAP1 (middle) and GFP-HDEL (left) in a stage 2 trichome at punctate structures, as shown by the white pixels (right). V, Lumen of the central vacuole. C, Single confocal image slice showing the colocalization between NAP1 (middle) and GFP-HDEL (left) in a stage 3/4 trichome at punctate structures, as shown by the white pixels (right). N, Nucleus. D, Single confocal image slice showing the colocalization between NAP1 (middle) and GFP-HDEL (left) in a stage 5 trichome containing punctate structures, as shown by the white pixels (right). E, Single confocal image slice showing the colocalization between NAP1 (middle) and GFP-HDEL (left) in a stage 6 trichome with a reticulate membrane organization, as shown by the white pixels (right). Bars = 10 μm in A to C and 5 μm in D and E.
As a direct test for the colocalization of NAP1 to ER nodule-like structures in trichomes, we conducted a series of NAP1 freeze-shattering experiments in GFP-HDEL-expressing lines. A punctate distribution of NAP1 was again detected in stage 1/2 (Fig. 7B), stage 3/4 (Fig. 7C), and stage 5 (Fig. 7D) trichomes, and in each case, there was an obvious colocalization of NAP1 and the GFP-HDEL-positive nodules. As expected, nuclear pools of NAP1 (Fig. 7C) did not colocalize with GFP-HDEL. In mature stage 6 trichomes, both NAP1 and GFP-HDEL consistently displayed a reticulate network appearance (Fig. 7E).
To test the possibility that NAP1 and the S/WRC-ARP2/3 pathway promoted the formation or maintenance of these specialized ER structures, the GFP-HDEL reporter was crossed into nap1 and dis2 plants and analyzed for the presence of the GFP-HDEL-positive nodules as a function of trichome development (Supplemental Fig. S5). Upon examining the marker in hundreds of trichomes from the wild type and distorted mutants at each developmental stage, we detected no apparent difference in the size or density of the GFP-HDEL-positive nodules. We conclude that this specialized GFP-HDEL-positive structure does not require W/SRC or ARP2/3 activity for its formation. Instead, the nodules likely reflect the general structure of the ER in the trichomes of these transgenic lines, and probably many ER-localized proteins have this localization pattern.
Inactive NAP1 Is ER Localized
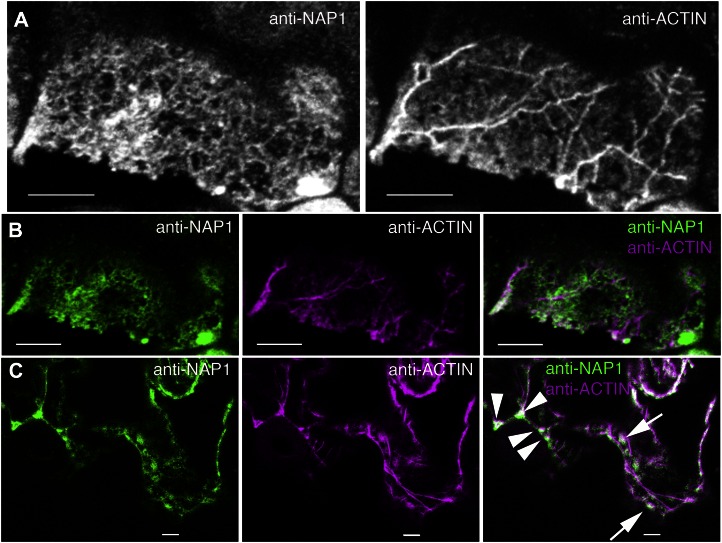
Based on the existing literature, the general morphology and localization of the ER and the actin cytoskeleton in leaf epidermal cells are quite distinct (Qiu et al., 2002; Saint-Jore et al., 2002; Djakovic et al., 2006; Ueda et al., 2010). Therefore, it seemed unlikely that the general ER-associated pool of NAP1 was active in terms of localized ARP2/3 activation. Nonetheless, colocalization of NAP1 with actin is a useful method to define and localize an active pool. To our knowledge, double labeling of W/SRC subunits and actin has not been reported previously in a plant system. In practice, we found double labeling of NAP1 and actin using whole-mount fixed samples to be technically challenging, because the long-term detergent extraction conditions needed to efficiently label actin bundle networks would either partially or completely solubilize NAP1 in most cells. The problem was most severe in young-stage trichomes with a dense cytoplasm, because one could find cells that were either labeled for actin or NAP1, but it was nearly impossible to find cells that were well labeled for both. In highly vacuolated pavement cells, it was possible to double label NAP1 and actin, but the labeling efficiency was still relatively low. For example, three freeze-shattered samples were screened to identify 13 cells that had the typical reticulate and punctate NAP1 signal and a well-labeled actin cytoskeleton. Manual inspection of the images revealed very little overlap of the actin and NAP1 signals in most cases (Fig. 8A). Pearson correlation coefficients of double-labeled cells had a mean value of 0.19 ± 0.16 (n = 26), reflecting an overall low and variable level of signal overlap (Fig. 8, A and B). The vast majority of ER-associated NAP1 appeared to be inactive, either because it was not actively engaged in ARP2/3 activation at a particular instant in time or because it was engaged in some other unknown activity that was independent of ARP2/3. It is also apparent that we did not detect all of the actin in these cells; therefore, the measured correlation coefficient probably underestimated the true level of colocalization, and it is possible that subpools of ER-associated NAP1 are fully active. There was qualitative evidence for spatial overlap between the NAP1 and actin signals in these double-labeled cells. In many cases, NAP-positive punctae were often detected adjacent to actin bundles (Fig. 8C). The proximity of NAP1 to actin bundles does not necessarily reflect a subcellular pool of fully active NAP1 that causes ARP2/3-dependent actin polymerization but, instead, could reflect the existence of NAP1-positive ER domains or organelles with latent ARP2/3 activation potential that are anchored to, or moving on, actin bundles. At a low frequency, we also detected clusters of NAP1 (Fig. 2A) or NAP1 punctae (Fig. 8C) that obviously overlapped with the actin signal. It was clear from the above experiments that much of the NAP1 signal was independent of the actin network; however, additional experimental tools are needed to accurately identify and quantify actin-associated NAP1 in cells.
Figure 8.
ER-localized NAP1 has a low level of colocalization with the actin cytoskeleton. A, Maximum Z-projection of seven confocal image slices from a wild-type leaf pavement cell double labeled for NAP1 (left) and actin (right). B, Single confocal image slices contained within the Z-projection in A. An example of the type of region of interest used for Pearson correlation coefficient is shown. Left, Single confocal image slice from the anti-NAP1 channel. Middle, Corresponding image slice from the actin channel. Right, Overlay image of two channels. C, Single confocal optical section showing the double labeling of NAP1 (left) and actin (middle) in wild-type pavement cells. Colocalization of NAP1 and actin was observed at three-way cell wall junctions (arrowheads) and punctate ER structures (arrows). Bars = 5 μm.
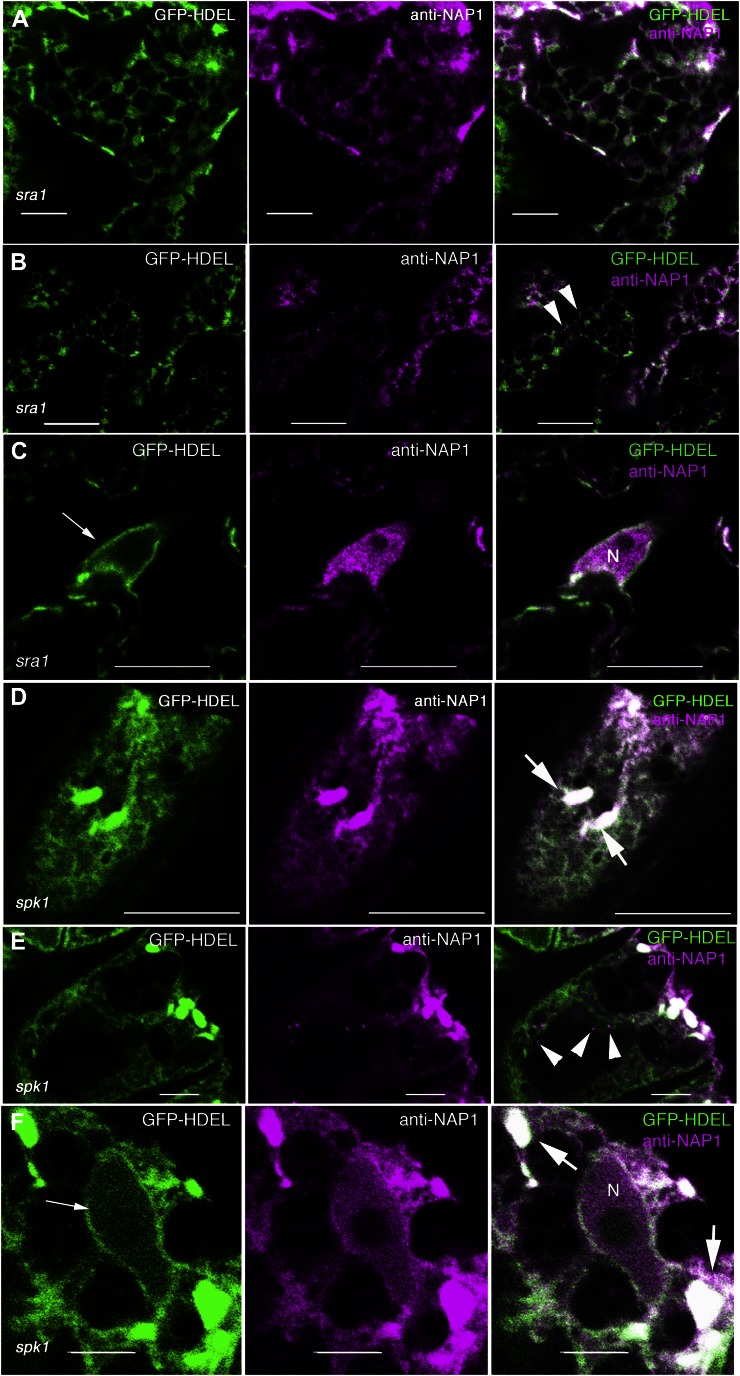
As an independent test to determine if the ER-localized NAP1 required an assembled or active W/SRC complex, we conducted a series of NAP1 and GFP-HDEL colocalization experiments in the mutant backgrounds that contain inactive NAP1. Arabidopsis SRA1 physically interacts with NAP1 (Basu et al., 2004; El-Assal et al., 2004; Uhrig et al., 2007). Null alleles of sra1 have a strong distorted trichome phenotype, and based on the array and severity of its shoot phenotypes, it functions solely within the W/SRC pathway (Basu et al., 2004; Brembu et al., 2004). In many systems, the stability of the NAP1 protein requires SRA1 (Derivery et al., 2009). However, in Arabidopsis, NAP1 protein levels are only slightly reduced in sra1 (Le et al., 2006), and there have been other reports using human cell lines in which W/SRC subunits had protein stabilities that were largely independent of the other subunits of the complex (Steffen et al., 2004; Zipfel et al., 2006). Interestingly the reticulate and punctate localization of NAP1 and GFP-HDEL in sra1 pavement cells was virtually indistinguishable from the wild type (Fig. 9A). The Pearson correlation coefficient of NAP1 and GFP-HDEL in sra1 (0.56 ± 0.10; n = 13) was not significantly different from the wild type. In sra1 pavement cells, NAP1 punctae were present (Fig. 9B), and a nuclear pool of NAP1 was also consistently detected (Fig. 9C). SPK1 is an ER-localized ROPGEF (Zhang et al., 2010) and a known positive regulator of the W/SRC pathway (Basu et al., 2008; Zhang et al., 2010). In spk1 pavement cells, the ER has a reduced amount of ER tubulation and a more diffuse appearance compared with the wild type (Zhang et al., 2010), but it still contained normal GFP-HDEL-positive ER bodies (Fig. 9, D and E). In spk1 cells, NAP1 strongly colocalized with the ER bodies and had a similar overall localization pattern compared with GFP-HDEL (Fig. 9, D and E). The approximately 0.7-µm-diameter NAP1 punctae (Fig. 9E) and a nuclear pool of NAP1 (Fig. 9F) were also present in spk1 cells. Collectively, these genetic data clearly indicated that inactive NAP1 associates with the ER in vivo.
Figure 9.
The general ER localization of NAP1 is independent of the integrity of W/SRC and SPK1-dependent positive regulation upstream SPK1 signaling. A to C, Localization of the GFP-HDEL ER marker (left) and NAP1 (middle) in the pavement cells of sra1. Single image planes are shown in all panels. Right, Overlay of the two false-colored image channels. A, NAP1 strongly colocalizes with GFP-HDEL in sra1 pavement cells, as shown by the white pixels (right). B, Punctate NAP1 adjacent to GFP-HDEL-positive ER was present in sra1, as shown by the arrowheads (right). C, NAP1 is present in the nucleus in sra1. GFP-HDEL labels the nuclear envelope, as shown by the arrow (left). D to F, Localization of the GFP-HDEL ER marker (left) and NAP1 (middle) in the pavement cells of spk1. Single image planes are shown in all panels. Right, Overlay of the two false-colored image channels. D, NAP1 strongly colocalizes with ER bodies and reticulate ER marked by GFP-HDEL in spk1 pavement cells, as shown by the white pixels (right). E, Punctate NAP1 adjacent to GFP-HDEL-positive ER was present in spk1, as shown by the arrowheads (right). F, NAP1 is present in the nucleus in spk1. Arrowheads indicate discrete NAP1 punctae, thick arrows indicate ER bodies, and thin arrows indicate nuclear membrane. N, Nucleus. Bars = 5 μm in A, E, and F and 10 μm in B to D.
DISCUSSION
In diverse plant species, the W/SRC and ARP2/3 complexes mediate actin-based cellular organization and growth control in response to endogenous morphogenesis signals (Szymanski et al., 1999; Mathur et al., 2003a; Harries et al., 2005), light (Dyachok et al., 2011), and symbiotic interactions with nitrogen-fixing bacteria (Yokota et al., 2009; Miyahara et al., 2010). In angiosperms, ARP2/3 activation pathways are relatively simple compared with other organisms, because the ROP effector W/SRC complex acts as the sole source of activating signals (Szymanski, 2005; Zhang et al., 2008). In Arabidopsis, multiple activities, including the assembly of WAVE/SCAR proteins into a functional W/SRC through physical interactions with BRK1 (Frank et al., 2004; Zhang et al., 2005) and Abl-interactor-like proteins (Basu et al., 2005; Jörgens et al., 2010), and positive regulation by the ROPGEF SPK1 (Basu et al., 2008) are required to properly activate ARP2/3. However, the subcellular localization of pathway components is poorly understood, and a major challenge in the field is to determine the cellular mechanisms that partition W/SRC between inactive and fully activated pools.
Previous studies suggest that the plasma membrane is an important location for W/SRC activity (Dyachok et al., 2008, 2011). However, SPK1 localizes to the ER and ERES (Zhang et al., 2010), creating uncertainty about which organelles participate in W/SRC regulation. In this paper, we discover the importance of the ER in the cell biology of W/SRC regulation, because a large fraction of the cellular pool of NAP1 and SCAR2 localizes to the ER surface. The ER-associated pool of NAP1 is not necessarily active because little of it colocalizes with actin, and its localization to this organelle surface is independent of either the normal assembly of the W/SRC or its activation by SPK1. These new results indicate that multiple organelles participate in the activation and recycling of W/SRC subunits and suggest that activating signals from the ROPGEF SPK1 that positively regulate the entire W/SRC-ARP2/3 pathway originate from the ER surface.
NAP1 was localized to the ER in two independent sets of experiments. First, we identified a pool of NAP1 in crude leaf microsomes based on the comigration of NAP1-positive vesicles with the ER marker protein SEC12 in Suc velocity gradient centrifugation experiments (Fig. 1). SEC12-, SCAR2-, and NAP1-positive membranes were shifted to less dense Suc fractions when ER-associated ribosomes were destabilized by chelating free Mg2+. Immunolocalization experiments also detected a reticulate pattern for NAP1, which is a well-known and specific pattern that is specific to the ER (Haseloff et al., 1997; Ridge et al., 1999). The identity of the NAP1-positive compartment as the ER was conclusively proved by double labeling NAP1 and the known ER marker GFP-HDEL (Nelson et al., 2007) in pavement cells, trichomes, and mesophyll cells. In each of these cell types, there was an obvious overlap of NAP1 signals and the ER, and NAP1 localized to the ER regardless of the morphology of the organelle or the developmental stage of the cell (Figs. 6 and 7). For example, in pavement cells of 2- or 10-DAG cotyledons and leaves, NAP1 localized to ER cisternae, tubules, and, when present, ER bodies. Expanding trichomes appear to have an unusual ER domain as defined by GFP-HDEL, which consists of independent approximately 2.1- to 2.7-µm nodule-like structures that transition into a more typical reticulate appearance in mature stage 6 cells (Fig. 7A; Supplemental Table S1). In trichomes, NAP1 colocalizes with GFP-HDEL independent of cell stage or ER morphology (Fig. 7, B–E). In developing trichomes, we found no correlation between the presence of GFP-HDEL-positive nodules and the activity state of NAP1 or ARPC2, because neither gene product is required for their formation (Supplemental Fig. S5), and there is no correlation between the timing and persistence of GFP-HDEL-positive nodule formation and the strict stage-specific requirements for W/SRC and ARP2/3 during the transition to branch elongation during stage 4.
A key result from this work is that the bulk of the ER-associated NAP1 is inactive with respect to W/SRC activity and ARP2/3 activation. Although double labeling of a W/SRC subunit and actin has not been reported previously in plants, colocalization with actin is one useful assay to identify putatively active pools of NAP1. In pavement cells, we find that a large fraction of NAP1 colocalizes with the ER marker, GFP-HDEL (Pearson correlation coefficient = 0.59), but in the same cell type, there is very little overlap of the NAP1 and actin signals (Pearson correlation coefficient = 0.19). The low level of colocalization of NAP1 and actin indicates that most of the ER-associated NAP1 is inactive. This conclusion is consistent with several published observations. First, the spatial patterns of the ER and actin in pavement cells are distinct, as shown using a variety of methods to localize actin and ER either independently (Haseloff et al., 1997; Scott et al., 1999; Qiu et al., 2002; Djakovic et al., 2006) or simultaneously in two-color live-cell imaging studies (Ueda et al., 2010). Nonoverlapping patterns of NAP1 and the actin network would not be expected if all of the ER-associated NAP1 was engaged in ARP2/3 activation and local actin polymerization. Stable colocalization of actin and the ER may be restricted to prominent actin bundles that support myosin-dependent directional transport (Avisar et al., 2009; Sparkes et al., 2009; Ueda et al., 2010). NAP1 does not obviously mediate the interaction between actin bundles in the ER, because NAP1 did not colocalize with actin bundles. Second, W/SRC and ARP2/3 mutants have an extremely subtle pavement cell actin phenotype (Mathur et al., 2003a; Djakovic et al., 2006). This would not be the case if the ER was coated with active NAP1 and ARP2/3. We need to emphasize that, although this work clearly demonstrates large pools of inactive NAP1 on the ER surface, it does not exclude the ER as a location where NAP1 and W/SRC could activate ARP2/3 and cause actin polymerization. For example, despite technical limitations in which actin and NAP1 could be double labeled only in particular cell types, we detected examples in which clusters of NAP1 signal (Fig. 2A) or individual NAP1 punctae (Fig. 8C) colocalized with actin. These may correspond to specialized domains of the ER where NAP1 and the W/SRC locally activate ARP2/3-dependent nucleation. Additional experimental tools and approaches are needed to determine where W/SRC is fully activated and the physiological significance of ARP2/3-generated filaments during pavement cell growth.
In trichomes, ARP2/3 is required to maintain a net longitudinal actin bundle alignment in developing branches and to maintain the polarized growth of the branch (Szymanski et al., 1999; Le et al., 2003; Deeks et al., 2004). Live-cell imaging of BRK1 and ectopically expressed SCAR1 reported a branch tip localization in a stage 4 cell (Dyachok et al., 2008), implying that the apical plasma membrane is a domain for localized ARP2/3 activation. Although the question of tip-localized actin in leaf hairs has not been directly addressed, published images clearly show elevated actin signal at this location in some cells (Le et al., 2003; Djakovic et al., 2006). In crawling cells, a fully assembled W/SRC relocalizes and promotes ARP2/3 activation near the leading edge (Steffen et al., 2004). If only fully assembled W/SRC has activity in plant cells, then NAP1 should also accumulate at the apical plasma membrane in stage 4 trichomes. It could be that the putative pool of apical NAP1 at or near the distal plasma membrane was either selectively extracted by detergents during permeabilization or obscured by the relatively large pools of intracellular, organelle-associated NAP1. The nonoverlapping localizations of BRK1 and SCAR1 (Dyachok et al., 2008) and reports in metazoan cells that NAP1 and BRK1 homologs have important activities that are independent of fully assembled W/SRC (Weiner et al., 2006; Derivery et al., 2008) suggest that W/SRC assembly is a point of regulation (Derivery and Gautreau, 2010). In this scenario, the punctate localization of SCAR1 and NAP1 in trichomes may correspond to an ER domain where W/SRC assembly and positive regulation by SPK1 serves as one level of regulation that enables full W/SRC assembly and its relocalization to another membrane surface, such as the plasma membrane. Although our data suggest that signaling for W/SRC is initiated on the ER surface, the potential importance of regulated W/SRC assembly and/or spatially staged small GTPase activation merits further research.
NAP1 signal in trichomes is highly concentrated in GFP-HDEL-positive nodules that are distributed throughout the cell (Figs. 5C and 7C); however, this pool of NAP1 is unlikely to promote organelle-associated actin polymerization in all instances. Published analyses of the actin cytoskeleton in trichomes using four different probes for actin in living and fixed cells (Le et al., 2003; Mathur et al., 2003a; Deeks et al., 2004; Basu et al., 2005) failed to detect any indication of a distributed population of punctate actin signal that would be expected if the organelle-associated pool of NAP1 was fully active.
The presence of inactive NAP1 on the ER surface may be a general feature of leaf epidermal cells. Interestingly, the ER localization of NAP1 in pavement cells is not affected either in spk1, in which W/SRC is inactive (Basu et al., 2008), or in sra1 cells, in which W/SRC is incompletely assembled and has little or no activity (Basu et al., 2004; Brembu et al., 2004; Saedler et al., 2004). These genetic data clearly demonstrate that inactive NAP1 associates with ER membranes and that its binding partner, SRA1, is not necessary for either the stability of the NAP1 protein or its association with the ER. The ER localization of NAP1 could be due either to the direct binding of NAP1 to a specific lipid(s) or proteins on the ER surface or to the similar binding activity of an inactive NAP1 complex that includes a subset of known W/SRC subunits and/or other, to be identified, proteins.
The bulk ER is not the sole subcellular location for NAP1. In pavement cells, we consistently detect NAP1 in approximately 0.6-µm-diameter punctate structures that are found both adjacent to the ER and independent from the ER network (Fig. 6B). Therefore, there are subpools of NAP1 that may segregate into specialized subdomains of the ER or perhaps even relocalize or traffic to organelles that are distinct from the ER system. Live cell probes for NAP1 will be needed to unambiguously define these potentially specialized protein pools. However, our experiments here demonstrate that these 0.6-µm NAP1 punctae are not, by definition, active, because they are commonly observed in both the spk1 and sra1 mutants (Fig. 9, B and E). We find that, like SPK1 (Zhang et al., 2010), there is an obvious nuclear pool of NAP1 in pavement cells and trichomes (Figs. 4 and 5D). In dark-grown roots, the nuclear pool of SCAR1 is specifically degraded (Dyachok et al., 2011). We find that in aerial tissues, nuclear NAP1 is stable and not necessarily active, because its localization is also unaltered in sra1 and spk1 (Fig. 9, C and F). The presence of NAP1 in the nucleoplasm indicates that, despite the extremely low levels of soluble NAP1 detected in cell fractionation assays (Fig. 1A; Supplemental Fig. S2), a cytosolic pool must exist in order for nuclear import to occur. In animal cells, the recruitment of soluble W/SRC and its clustering on specific membrane surfaces is a prominent feature of proposed models for W/SRC regulation (Lebensohn and Kirschner, 2009; Chen et al., 2010). This is because, in nonplant species, a significant fraction of cellular W/SRC is cytosolic (Eden et al., 2002; Bogdan and Klämbt, 2003). In addition, biochemical studies that employ soluble vertebrate WAVE/SCAR proteins or fully assembled W/SRC detect specific binding to phospholipids that is consistent with a cytosolic recruitment model (Oikawa et al., 2004; Steffen et al., 2004; Lebensohn and Kirschner, 2009). However, in many motile cell types, including neurons, NAP1 has a striking punctate or perinuclear localization pattern that may reflect the general importance of multiple organelles in the cellular cycles of W/SRC activation and use (Bogdan and Klämbt, 2003; Steffen et al., 2004; Millius et al., 2009).
To conclude, our results indicate that in the shoot epidermis the ER serves a reservoir of inactive NAP1, and perhaps SCAR2, that has the potential to be either assembled into an intact W/SRC or activated to elicit ARP2/3-dependent actin polymerization in response to cellular signals. This is an important discovery because it suggests that the endogenous signals and protein machineries that mediate the initial steps of NAP1- and W/SRC-positive regulation are located at the ER surface, rather than residing exclusively at the plasma membrane. Given that SPK1 and ROP localize to the ER and accumulate at ERES (Bracha et al., 2002; Zhang et al., 2010), it is possible that SPK1 and ROP-GTP are direct positive regulators of NAP1-containing complexes at the specialized domains of the ER. Key questions now are centered on the precise nature of NAP1-positive regulation, how it relates to W/SRC assembly, and its full activation at specific cellular locations that have importance during ARP2/3-dependent growth. The ERES seems unlikely to be a primary location for ARP2/3 activation, because it is not a known site of actin accumulation, and the normal export of cargo from ERES to the Golgi does not require either an intact actin cytoskeleton or ARP2/3 (Brandizzi et al., 2002; Zhang et al., 2010). If the clustering of SCAR1 and BRK1 at the apex of stage 4 trichomes (Dyachok et al., 2008) reflects a fully activated pool of W/SRC at the plasma membrane, it is possible that the cellular control of ARP2/3 activation occurs in steps at multiple organelle surfaces in the secretory pathway. The further use of the distorted mutants and the development of new live-cell probes and localization tools for W/SRC subunits and ARP2/3 should enable future experiments that systematically dissect the cellular control of W/SRC in the context of actin-based morphogenesis.
MATERIALS AND METHODS
Plant Strains and Growth Conditions
The Arabidopsis (Arabidopsis thaliana) ecotype Columbia (Col-0) was used as the wild type. The GFP-HDEL transgenic line was obtained from the Arabidopsis Biological Resource Center with the stock number CS16251. The construct contains the AtWAK2 signal sequence and a C-terminal fusion between GFP and the HDEL ER-retention signal as described previously (Nelson et al., 2007). The mutant alleles of nap1 (grl-6), spk1-1, and sra1 (pir-3) are as described previously (Qiu et al., 2002; Basu et al., 2004; El-Assal et al., 2004). The plant materials for immunolocalization and live-cell imaging were grown on one-half-strength Murashige and Skoog medium with 0.8% Bacto agar and 1% Suc at 22°C with constant illumination. Cell fractionation experiments employed soil-grown plants with constant illumination.
Cell Fractionation, NAP1 Solubilization, and Suc Velocity Gradients
Two grams of leaf material of 14- to 16-DAG plants was dissected and quickly transferred to the ice-cold homogenization buffer, which contains 20 mm HEPES-KCl, pH 7.2, 50 mm KOAc, 2 mm Mg(OAc)2, 250 mm sorbitol, 1 mm EDTA, 1 mm EGTA, 1 mm dithiothreitol, 1 mm phenylmethylsulfonyl fluoride, and 1% (v/v) protease inhibitor cocktail (Sigma), and then homogenized with a Polytron (Brinkman Scientific). The homogenate was filtered through two layers of Miracloth, and the flow through was spun at 1,000g for 10 min at 4°C in a Beckman F0850 rotor. The resulting supernatant was defined as total protein. For cell fractionation experiments, the total protein was sequentially centrifuged at the g forces shown in the figures at 4°C for 30 min. The pellet fractions were resuspended in the same volume of homogenization buffer as the supernatant. The fractions were then loaded by equal proportion, in which the equal cell fractions by volume were loaded for SDS-PAGE and western blotting. In the representative blots shown in Figure 1A, the absolute amounts of proteins loaded in the total, 10k pellet, 200k pellet, and 200k supernatant fractions are 73, 13, 12, and 33 µg, respectively.
For the NAP1 solubilization experiment, the total protein was centrifuged at 200,000g for 1 h at 4°C in a Beckman Optima MAX tabletop ultracentrifuge. The pellet fraction after 200,000g spin was resuspended in a total volume of 500 µL with resuspension buffer (10 mm HEPES, pH 7.2, 150 mm NaCl, 1 mm EDTA, 10% glycerol, 2 mm dithiothreitol, 1 mm phenylmethylsulfonyl fluoride, and 1% protease inhibitor cocktail). Twenty microliters of resuspended material was used for each trial, with the final volume being 200 µL. Reactions containing buffer controls and the salts and detergents listed in the figure were rocked at room temperature for 30 min and then centrifuged at 200,000g for 1 h. Equal proportions of the resulting supernatants and insoluble material were separated on SDS-PAGE gels and blotted against anti-NAP1 and anti-KNOLLE primary antibodies.
For Suc density gradient separations of membrane proteins, the filtrate from 2 g of shoot tissue was centrifuged at 200,000g at 4°C for 1 h. The resulting pellet fraction was resuspended in 200 μL of 10 mm Tris-HCl, pH 7.5, 150 mm NaCl, 1 mm EDTA, 10% (v/v) glycerol, phenylmethylsulfonyl fluoride, and 1 mm protease inhibitors and directly loaded on Suc solution that had a 20% to 50% (w/v) continuous density gradient. Suc density gradients were centrifuged at 100,000g for 16 h at 4°C. Fractions in different concentrations of Suc were loaded on 12% SDS-PAGE for western blotting. For western blotting, SDS-PAGE gels were cast and run using a Bio-Rad Mini-Protein3 Cell and transferred to 0.2-µm reinforced nitrocellulose membranes (Whatman Optitran BA-S 83) in 48 mm Tris, 39 mm Gly, 20% (v/v) methanol, and 0.037% SDS buffer using an Idea Scientific Genie Blotter or a Bio-Rad Mini Trans-Blot Electrophoretic transfer cell. Membranes were probed using rabbit antibodies diluted to 1:1,000, except anti-KNOLLE, which was used at 1:4,000, and incubated overnight at 4°C, washed three or four times for 10 min at room temperature with chilled TTBS, and then incubated with a horseradish peroxidase-conjugated goat anti-rabbit secondary antibody (Pierce) at 1:50,000 for 1 to 2 h. Signal was detected on x-ray film with SuperSignal West Pico Chemiluminescent Substrate (Pierce). The V-ATPase subunit E antibody was purchased from Agrisera. The NAP1 and SCAR2 antibodies have been described previously (Le et al., 2006).
Immunolocalization by Freeze Shattering and Confocal Microscopy
The immunolocalization protocol was the same as reported previously (Zhang et al., 2010), except that the permeabilization step after freeze shattering was 2 h. The same immunolocalization protocol was applied to all samples, including the wild type (Col-0), nap1 (grl-6), spike1, and GFP-HDEL. A previously described NAP1 peptide antibody was affinity purified by coupling 2 mg of each of the two antigenic peptides to a SulfoLink Kit (Pierce) column according to the manufacturer’s protocol. A total of 25 mL of anti-NAP1 final bleed was applied to this column and then eluted with 100 mm Gly, pH 2.5. One-milliliter fractions were collected into equal volumes of 50 μL of 1 m Tris-HCl, pH 9.5. Fractions enriched with the purified antibody were pooled, dialyzed, and concentrated with a centrifugal filter device (Amicon Centriprep YM-30; Millipore). The crude NAP1 serum, the NAP1 preimmune serum, and the affinity-purified anti-NAP1 antibodies were used at 1:400 dilution. The mouse monoclonal anti-actin (JLA20) antibody (Calbiochem) was used at 1:400. Alexa488-conjugated goat anti-rabbit (IgG), Alexa546-conjugated goat anti-rabbit (IgG), FITC-conjugated goat anti-mouse (IgM), and rhodamine-conjugated goat anti-mouse (IgM) secondary antibodies (Life Technologies) were used at 1:400 dilutions. A Zeiss 710 laser scanning confocal microscope (Carl Zeiss) was used for imaging. System-optimized settings for GFP and Alexa546 dual-channel imaging were used for the samples that have GFP and Alexa546 fluorescence. System-optimized settings for Alexa488 and Rhodamine Red dual-channel imaging were used for samples that have Alexa488 and Rhodamine Red fluorescence. GFP single-channel imaging settings were used for GFP-HDEL live-cell imaging. A 63× oil-immersion objective (1.4 numerical aperture) was used for fixed samples, and a 63× water-immersion objective (1.3 numerical aperture) was used for GFP-HDEL live-cell imaging.
Particle Size and Pearson Correlation Coefficient Measurements
Confocal image stacks from NAP1-labeled trichome cells in Col-0 and GFP-HDEL-expressing lines were used for measuring the size of GFP-HDEL- and NAP1-positive nodules. To measure a single nodule, the image stacks were manually inspected for selected image planes that contained this structure, and then a maximum Z-projection of these image planes was made. The resulting image was thresholded to eliminate background fluorescence and generate a binary image. A circular region of interest was created for each nodule using ImageJ (http://rsb.info.nih.gov/ij/), and the descriptive parameters of this structure were measured using the Analyze Particles function of ImageJ. For image planes that had multiple ER GFP-HDEL-positive nodules, a larger region of interest that included all nodules was drawn to get the descriptive parameters for each of the structures. Small particles that were less than four pixels in size were discarded. NAP1 punctae size quantification followed a similar procedure. To quantify the colocalization between GFP-HDEL and NAP1, confocal image stacks that contained GFP-HDEL in the green channel and NAP1 in the red channel were merged to an RGB stack file. The stacks were manually inspected to identify cortical regions that had both GFP-HDEL and NAP1 signal. A free hand drawing tool was used to select the region in the image field that had both GFP-HDEL and NAP1. The PSC colocalization plugin in ImageJ was used to quantify the Pearson correlation coefficient between signals in two channels in the selected region. The default threshold value of 40 for eight-bit images was above the background fluorescence and was used for both channels for quantification. Two to three image planes were used for colocalization analysis in each image stack. Similar procedures were used for NAP1 and actin colocalization quantification.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. NAP1 antibody specificity.
Supplemental Figure S2. Overexposure of the NAP1 and SCAR2 western blots.
Supplemental Figure S3. Detergent solubilization of NAP.
Supplemental Figure S4. Double labeling of the plasma membrane marker PHOT1 and NAP1.
Supplemental Figure S5. GFP-HDEL-positive ER nodules in nap1 and dis2 trichomes.
Supplementary Table 1. Quantification of the size of NAP1-positive ER nodules.
Acknowledgments
We would like to thank Makoto Yanagisawa for helpful comments on the manuscript. We are grateful to Natasha Raikhel for the gifts of the SEC12, SYP21, and VPS45 antibodies. Thank you to Gerd Juergens for the KNOLLE antibody. Thank you to Mike Sussman for the H+-ATPase antibody.
Glossary
- W/SRC
WASP family verprolin homologous/suppressor of cAMP receptor regulatory complex
- DAG
d after germination
- Col-0
Columbia
- ER
endoplasmic reticulum
- ROP
Rho-of-Plants
- ERES
ER exit site
References
- Avisar D, Abu-Abied M, Belausov E, Sadot E, Hawes C, Sparkes IA. (2009) A comparative study of the involvement of 17 Arabidopsis myosin family members on the motility of Golgi and other organelles. Plant Physiol 150: 700–709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baskin TI. (2005) Anisotropic expansion of the plant cell wall. Annu Rev Cell Dev Biol 21: 203–222 [DOI] [PubMed] [Google Scholar]
- Basu D, El-Assal Sel-D, Le J, Mallery EL, Szymanski DB. (2004) Interchangeable functions of Arabidopsis PIROGI and the human WAVE complex subunit SRA1 during leaf epidermal development. Development 131: 4345–4355 [DOI] [PubMed] [Google Scholar]
- Basu D, Le J, El-Essal Sel-D, Huang S, Zhang C, Mallery EL, Koliantz G, Staiger CJ, Szymanski DB. (2005) DISTORTED3/SCAR2 is a putative Arabidopsis WAVE complex subunit that activates the Arp2/3 complex and is required for epidermal morphogenesis. Plant Cell 17: 502–524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basu D, Le J, Zakharova T, Mallery EL, Szymanski DB. (2008) A SPIKE1 signaling complex controls actin-dependent cell morphogenesis through the heteromeric WAVE and ARP2/3 complexes. Proc Natl Acad Sci USA 105: 4044–4049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchoin L, Boujemaa-Paterski R, Henty JL, Khurana P, Staiger CJ. (2010) Actin dynamics in plant cells: a team effort from multiple proteins orchestrates this very fast-paced game. Curr Opin Plant Biol 13: 714–723 [DOI] [PubMed] [Google Scholar]
- Bogdan S, Klämbt C. (2003) Kette regulates actin dynamics and genetically interacts with Wave and Wasp. Development 130: 4427–4437 [DOI] [PubMed] [Google Scholar]
- Bracha K, Lavy M, Yalovsky S. (2002) The Arabidopsis AtSTE24 is a CAAX protease with broad substrate specificity. J Biol Chem 277: 29856–29864 [DOI] [PubMed] [Google Scholar]
- Brandizzi F, Snapp EL, Roberts AG, Lippincott-Schwartz J, Hawes C. (2002) Membrane protein transport between the endoplasmic reticulum and the Golgi in tobacco leaves is energy dependent but cytoskeleton independent: evidence from selective photobleaching. Plant Cell 14: 1293–1309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brembu T, Winge P, Seem M, Bones AM. (2004) NAPP and PIRP encode subunits of a putative wave regulatory protein complex involved in plant cell morphogenesis. Plant Cell 16: 2335–2349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Borek D, Padrick SB, Gomez TS, Metlagel Z, Ismail AM, Umetani J, Billadeau DD, Otwinowski Z, Rosen MK. (2010) Structure and control of the actin regulatory WAVE complex. Nature 468: 533–538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleary AL. (1995) F-actin redistributions at the division site in living Tradescantia stomatal complexes as revealed by microinjection of rhodamine-phalloidin. Protoplasma 185: 152–165 [Google Scholar]
- Cosgrove DJ. (2005) Growth of the plant cell wall. Nat Rev Mol Cell Biol 6: 850–861 [DOI] [PubMed] [Google Scholar]
- Deeks MJ, Kaloriti D, Davies B, Malhó R, Hussey PJ. (2004) Arabidopsis NAP1 is essential for Arp2/3-dependent trichome morphogenesis. Curr Biol 14: 1410–1414 [DOI] [PubMed] [Google Scholar]
- Derivery E, Fink J, Martin D, Houdusse A, Piel M, Stradal TE, Louvard D, Gautreau A. (2008) Free Brick1 is a trimeric precursor in the assembly of a functional wave complex. PLoS ONE 3: e2462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derivery E, Gautreau A. (2010) Generation of branched actin networks: assembly and regulation of the N-WASP and WAVE molecular machines. Bioessays 32: 119–131 [DOI] [PubMed] [Google Scholar]
- Derivery E, Lombard B, Loew D, Gautreau A. (2009) The Wave complex is intrinsically inactive. Cell Motil Cytoskeleton 66: 777–790 [DOI] [PubMed] [Google Scholar]
- Djakovic S, Dyachok J, Burke M, Frank MJ, Smith LG. (2006) BRICK1/HSPC300 functions with SCAR and the ARP2/3 complex to regulate epidermal cell shape in Arabidopsis. Development 133: 1091–1100 [DOI] [PubMed] [Google Scholar]
- Dyachok J, Shao M-R, Vaughn K, Bowling A, Facette M, Djakovic S, Clark L, Smith L. (2008) Plasma membrane-associated SCAR complex subunits promote cortical F-actin accumulation and normal growth characteristics in Arabidopsis roots. Mol Plant 1: 990–1006 [DOI] [PubMed] [Google Scholar]
- Dyachok J, Zhu L, Liao F, He J, Huq E, Blancaflor EB. (2011) SCAR mediates light-induced root elongation in Arabidopsis through photoreceptors and proteasomes. Plant Cell 23: 3610–3626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eden S, Rohatgi R, Podtelejnikov AV, Mann M, Kirschner MW. (2002) Mechanism of regulation of WAVE1-induced actin nucleation by Rac1 and Nck. Nature 418: 790–793 [DOI] [PubMed] [Google Scholar]
- El-Assal Sel-D, Le J, Basu D, Mallery EL, Szymanski DB. (2004) Arabidopsis GNARLED encodes a NAP125 homolog that positively regulates ARP2/3. Curr Biol 14: 1405–1409 [DOI] [PubMed] [Google Scholar]
- El-Din El-Assal S, Le J, Basu D, Mallery EL, Szymanski DB. (2004) DISTORTED2 encodes an ARPC2 subunit of the putative Arabidopsis ARP2/3 complex. Plant J 38: 526–538 [DOI] [PubMed] [Google Scholar]
- Evangelista M, Pruyne D, Amberg DC, Boone C, Bretscher A. (2002) Formins direct Arp2/3-independent actin filament assembly to polarize cell growth in yeast. Nat Cell Biol 4: 260–269 [DOI] [PubMed] [Google Scholar]
- Frank M, Egile C, Dyachok J, Djakovic S, Nolasco M, Li R, Smith LG. (2004) Activation of Arp2/3 complex-dependent actin polymerization by plant proteins distantly related to Scar/WAVE. Proc Natl Acad Sci USA 101: 16379–16384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank MJ, Smith LG. (2002) A small, novel protein highly conserved in plants and animals promotes the polarized growth and division of maize leaf epidermal cells. Curr Biol 12: 849–853 [DOI] [PubMed] [Google Scholar]
- Fu Y, Gu Y, Zheng Z, Wasteneys GO, Yang Z. (2005) Arabidopsis interdigitating cell growth requires two antagonistic pathways with opposing action on cell morphogenesis. Cell 120: 687–700 [DOI] [PubMed] [Google Scholar]
- Fu Y, Li H, Yang Z. (2002) The ROP2 GTPase controls the formation of cortical fine F-actin and the early phase of directional cell expansion during Arabidopsis organogenesis. Plant Cell 14: 777–794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiki Y, Hubbard AL, Fowler S, Lazarow PB. (1982) Isolation of intracellular membranes by means of sodium carbonate treatment: application to endoplasmic reticulum. J Cell Biol 93: 97–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautreau A, Ho HY, Li J, Steen H, Gygi SP, Kirschner MW. (2004) Purification and architecture of the ubiquitous Wave complex. Proc Natl Acad Sci USA 101: 4379–4383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geldner N, Friml J, Stierhof Y-D, Jürgens G, Palme K. (2001) Auxin transport inhibitors block PIN1 cycling and vesicle trafficking. Nature 413: 425–428 [DOI] [PubMed] [Google Scholar]
- Gibbon BC, Kovar DR, Staiger CJ. (1999) Latrunculin B has different effects on pollen germination and tube growth. Plant Cell 11: 2349–2363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez R, Lindeboom JJ, Paredez AR, Emons AM, Ehrhardt DW. (2009) Arabidopsis cortical microtubules position cellulose synthase delivery to the plasma membrane and interact with cellulose synthase trafficking compartments. Nat Cell Biol 11: 797–806 [DOI] [PubMed] [Google Scholar]
- Hara-Nishimura I, Matsushima R. (2003) A wound-inducible organelle derived from endoplasmic reticulum: a plant strategy against environmental stresses? Curr Opin Plant Biol 6: 583–588 [DOI] [PubMed] [Google Scholar]
- Harries PA, Pan A, Quatrano RS. (2005) Actin-related protein2/3 complex component ARPC1 is required for proper cell morphogenesis and polarized cell growth in Physcomitrella patens. Plant Cell 17: 2327–2339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haseloff J, Siemering KR, Prasher DC, Hodge S. (1997) Removal of a cryptic intron and subcellular localization of green fluorescent protein are required to mark transgenic Arabidopsis plants brightly. Proc Natl Acad Sci USA 94: 2122–2127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussey PJ, Ketelaar T, Deeks MJ. (2006) Control of the actin cytoskeleton in plant cell growth. Annu Rev Plant Biol 57: 109–125 [DOI] [PubMed] [Google Scholar]
- Ismail AM, Padrick SB, Chen B, Umetani J, Rosen MK. (2009) The WAVE regulatory complex is inhibited. Nat Struct Mol Biol 16: 561–563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jörgens CI, Grünewald N, Hülskamp M, Uhrig JF. (2010) A role for ABIL3 in plant cell morphogenesis. Plant J 62: 925–935 [DOI] [PubMed] [Google Scholar]
- Kaksonen M, Sun Y, Drubin DG. (2003) A pathway for association of receptors, adaptors, and actin during endocytic internalization. Cell 115: 475–487 [DOI] [PubMed] [Google Scholar]
- Kollmar M, Lbik D, Enge S. (2012) Evolution of the eukaryotic ARP2/3 activators of the WASP family: WASP, WAVE, WASH, and WHAMM, and the proposed new family members WAWH and WAML. BMC Res Notes 5: 88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotchoni SO, Zakharova T, Mallery EL, Le J, El-Assal Sel-D, Szymanski DB. (2009) The association of the Arabidopsis actin-related protein2/3 complex with cell membranes is linked to its assembly status but not its activation. Plant Physiol 151: 2095–2109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langhans M, Niemes S, Pimpl P, Robinson DG. (2009) Oryzalin bodies: in addition to its anti-microtubule properties, the dinitroaniline herbicide oryzalin causes nodulation of the endoplasmic reticulum. Protoplasma 236: 73–84 [DOI] [PubMed] [Google Scholar]
- Le J, El-Assal Sel-D, Basu D, Saad ME, Szymanski DB. (2003) Requirements for Arabidopsis ATARP2 and ATARP3 during epidermal development. Curr Biol 13: 1341–1347 [DOI] [PubMed] [Google Scholar]
- Le J, Mallery EL, Zhang C, Brankle S, Szymanski DB. (2006) Arabidopsis BRICK1/HSPC300 is an essential WAVE-complex subunit that selectively stabilizes the Arp2/3 activator SCAR2. Curr Biol 16: 895–901 [DOI] [PubMed] [Google Scholar]
- Lebensohn AM, Kirschner MW. (2009) Activation of the WAVE complex by coincident signals controls actin assembly. Mol Cell 36: 512–524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leucci MR, Di Sansebastiano G-P, Gigante M, Dalessandro G, Piro G. (2007) Secretion marker proteins and cell-wall polysaccharides move through different secretory pathways. Planta 225: 1001–1017 [DOI] [PubMed] [Google Scholar]
- Li S, Blanchoin L, Yang Z, Lord EM. (2003) The putative Arabidopsis arp2/3 complex controls leaf cell morphogenesis. Plant Physiol 132: 2034–2044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Sorefan K, Hemmann G, Bevan MW. (2004) Arabidopsis NAP and PIR regulate actin-based cell morphogenesis and multiple developmental processes. Plant Physiol 136: 3616–3627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovy-Wheeler A, Cárdenas L, Kunkel JG, Hepler PK. (2007) Differential organelle movement on the actin cytoskeleton in lily pollen tubes. Cell Motil Cytoskeleton 64: 217–232 [DOI] [PubMed] [Google Scholar]
- Machesky LM, Mullins RD, Higgs HN, Kaiser DA, Blanchoin L, May RC, Hall ME, Pollard TD. (1999) Scar, a WASp-related protein, activates nucleation of actin filaments by the Arp2/3 complex. Proc Natl Acad Sci USA 96: 3739–3744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marks MD, Wenger JP, Gilding E, Jilk R, Dixon RA. (2009) Transcriptome analysis of Arabidopsis wild-type and gl3-sst sim trichomes identifies four additional genes required for trichome development. Mol Plant 2: 803–822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathur J, Mathur N, Kernebeck B, Hülskamp M. (2003a) Mutations in actin-related proteins 2 and 3 affect cell shape development in Arabidopsis. Plant Cell 15: 1632–1645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathur J, Mathur N, Kirik V, Kernebeck B, Srinivas BP, Hülskamp M. (2003b) Arabidopsis CROOKED encodes for the smallest subunit of the ARP2/3 complex and controls cell shape by region specific fine F-actin formation. Development 130: 3137–3146 [DOI] [PubMed] [Google Scholar]
- Matsushima R, Kondo M, Nishimura M, Hara-Nishimura I. (2003) A novel ER-derived compartment, the ER body, selectively accumulates a beta-glucosidase with an ER-retention signal in Arabidopsis. Plant J 33: 493–502 [DOI] [PubMed] [Google Scholar]
- Millius A, Dandekar SN, Houk AR, Weiner OD. (2009) Neutrophils establish rapid and robust WAVE complex polarity in an actin-dependent fashion. Curr Biol 19: 253–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyahara A, Richens J, Starker C, Morieri G, Smith L, Long S, Downie JA, Oldroyd GE. (2010) Conservation in function of a SCAR/WAVE component during infection thread and root hair growth in Medicago truncatula. Mol Plant Microbe Interact 23: 1553–1562 [DOI] [PubMed] [Google Scholar]
- Nelson BK, Cai X, Nebenführ A. (2007) A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. Plant J 51: 1126–1136 [DOI] [PubMed] [Google Scholar]
- Ogasawara K, Yamada K, Christeller JT, Kondo M, Hatugai N, Hara-Nishimura I, Nishimura M. (2009) Constitutive and inducible ER bodies of Arabidopsis thaliana accumulate distinct β-glucosidases. Plant Cell Physiol 50: 480–488 [DOI] [PubMed] [Google Scholar]
- Oikawa T, Yamaguchi H, Itoh T, Kato M, Ijuin T, Yamazaki D, Suetsugu S, Takenawa T. (2004) PtdIns(3,4,5)P3 binding is necessary for WAVE2-induced formation of lamellipodia. Nat Cell Biol 6: 420–426 [DOI] [PubMed] [Google Scholar]
- Perroud P-F, Quatrano RS. (2008) BRICK1 is required for apical cell growth in filaments of the moss Physcomitrella patens but not for gametophore morphology. Plant Cell 20: 411–422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard TD, Borisy GG. (2003) Cellular motility driven by assembly and disassembly of actin filaments. Cell 112: 453–465 [DOI] [PubMed] [Google Scholar]
- Prokhnevsky AI, Peremyslov VV, Dolja VV. (2008) Overlapping functions of the four class XI myosins in Arabidopsis growth, root hair elongation, and organelle motility. Proc Natl Acad Sci USA 105: 19744–19749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu JL, Jilk R, Marks MD, Szymanski DB. (2002) The Arabidopsis SPIKE1 gene is required for normal cell shape control and tissue development. Plant Cell 14: 101–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridge RW, Uozumi Y, Plazinski J, Hurley UA, Williamson RE. (1999) Developmental transitions and dynamics of the cortical ER of Arabidopsis cells seen with green fluorescent protein. Plant Cell Physiol 40: 1253–1261 [DOI] [PubMed] [Google Scholar]
- Saedler R, Zimmermann I, Mutondo M, Hülskamp M. (2004) The Arabidopsis KLUNKER gene controls cell shape changes and encodes the AtSRA1 homolog. Plant Mol Biol 56: 775–782 [DOI] [PubMed] [Google Scholar]
- Saint-Jore CM, Evins J, Batoko H, Brandizzi F, Moore I, Hawes C. (2002) Redistribution of membrane proteins between the Golgi apparatus and endoplasmic reticulum in plants is reversible and not dependent on cytoskeletal networks. Plant J 29: 661–678 [DOI] [PubMed] [Google Scholar]
- Scott A, Wyatt S, Tsou PL, Robertson D, Allen NS. (1999) Model system for plant cell biology: GFP imaging in living onion epidermal cells. Biotechniques 26: 1125–, 1128–1132. [DOI] [PubMed] [Google Scholar]
- Singh MK, Ren F, Giesemann T, Bosco CD, Pasternak TP, Blein T, Ruperti B, Schmidt G, Aktories K, Molendijk AJ, et al (2012) Modification of plant Rac/Rop GTPase signalling using bacterial toxin transgenes. Plant J 73: 314–324 [DOI] [PubMed] [Google Scholar]
- Smith LG, Oppenheimer DG. (2005) Spatial control of cell expansion by the plant cytoskeleton. Annu Rev Cell Dev Biol 21: 271–295 [DOI] [PubMed] [Google Scholar]
- Sparkes I, Runions J, Hawes C, Griffing L. (2009) Movement and remodeling of the endoplasmic reticulum in nondividing cells of tobacco leaves. Plant Cell 21: 3937–3949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sroubek J, McDonald TV. (2011) Protein kinase A activity at the endoplasmic reticulum surface is responsible for augmentation of human ether-a-go-go-related gene product (HERG). J Biol Chem 286: 21927–21936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staiger CJ, Sheahan MB, Khurana P, Wang X, McCurdy DW, Blanchoin L. (2009) Actin filament dynamics are dominated by rapid growth and severing activity in the Arabidopsis cortical array. J Cell Biol 184: 269–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffen A, Rottner K, Ehinger J, Innocenti M, Scita G, Wehland J, Stradal TEB. (2004) Sra-1 and Nap1 link Rac to actin assembly driving lamellipodia formation. EMBO J 23: 749–759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szymanski DB. (2005) Breaking the WAVE complex: the point of Arabidopsis trichomes. Curr Opin Plant Biol 8: 103–112 [DOI] [PubMed] [Google Scholar]
- Szymanski DB, Cosgrove DJ. (2009) Dynamic coordination of cytoskeletal and cell wall systems during plant cell morphogenesis. Curr Biol 19: R800–R811 [DOI] [PubMed] [Google Scholar]
- Szymanski DB, Jilk RA, Pollock SM, Marks MD. (1998) Control of GL2 expression in Arabidopsis leaves and trichomes. Development 125: 1161–1171 [DOI] [PubMed] [Google Scholar]
- Szymanski DB, Marks MD, Wick SM. (1999) Organized F-actin is essential for normal trichome morphogenesis in Arabidopsis. Plant Cell 11: 2331–2347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda H, Yokota E, Kutsuna N, Shimada T, Tamura K, Shimmen T, Hasezawa S, Dolja VV, Hara-Nishimura I. (2010) Myosin-dependent endoplasmic reticulum motility and F-actin organization in plant cells. Proc Natl Acad Sci USA 107: 6894–6899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhrig JF, Mutondo M, Zimmermann I, Deeks MJ, Machesky LM, Thomas P, Uhrig S, Rambke C, Hussey PJ, Hülskamp M. (2007) The role of Arabidopsis SCAR genes in ARP2-ARP3-dependent cell morphogenesis. Development 134: 967–977 [DOI] [PubMed] [Google Scholar]
- Weiner OD, Rentel MC, Ott A, Brown GE, Jedrychowski M, Yaffe MB, Gygi SP, Cantley LC, Bourne HR, Kirschner MW. (2006) Hem-1 complexes are essential for Rac activation, actin polymerization, and myosin regulation during neutrophil chemotaxis. PLoS Biol 4: e38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch MD, Mullins RD. (2002) Cellular control of actin nucleation. Annu Rev Cell Dev Biol 18: 247–288 [DOI] [PubMed] [Google Scholar]
- Winter DC, Choe EY, Li R. (1999) Genetic dissection of the budding yeast Arp2/3 complex: a comparison of the in vivo and structural roles of individual subunits. Proc Natl Acad Sci USA 96: 7288–7293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu T, Wen M, Nagawa S, Fu Y, Chen JG, Wu MJ, Perrot-Rechenmann C, Friml J, Jones AM, Yang Z. (2010) Cell surface- and rho GTPase-based auxin signaling controls cellular interdigitation in Arabidopsis. Cell 143: 99–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yalovsky S, Bloch D, Sorek N, Kost B. (2008) Regulation of membrane trafficking, cytoskeleton dynamics, and cell polarity by ROP/RAC GTPases. Plant Physiol 147: 1527–1543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y-D, Elamawi R, Bubeck J, Pepperkok R, Ritzenthaler C, Robinson DG. (2005) Dynamics of COPII vesicles and the Golgi apparatus in cultured Nicotiana tabacum BY-2 cells provides evidence for transient association of Golgi stacks with endoplasmic reticulum exit sites. Plant Cell 17: 1513–1531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokota K, Fukai E, Madsen LH, Jurkiewicz A, Rueda P, Radutoiu S, Held M, Hossain MS, Szczyglowski K, Morieri G, et al. (2009) Rearrangement of actin cytoskeleton mediates invasion of Lotus japonicus roots by Mesorhizobium loti. Plant Cell 21: 267–284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Kotchoni SO, Samuels AL, Szymanski DB. (2010) SPIKE1 signals originate from and assemble specialized domains of the endoplasmic reticulum. Curr Biol 20: 2144–2149 [DOI] [PubMed] [Google Scholar]
- Zhang C, Mallery EL, Schlueter J, Huang S, Fan Y, Brankle S, Staiger CJ, Szymanski DB. (2008) Arabidopsis SCARs function interchangeably to meet actin-related protein 2/3 activation thresholds during morphogenesis. Plant Cell 20: 995–1011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Dyachok J, Krishnakumar S, Smith LG, Oppenheimer DG. (2005) IRREGULAR TRICHOME BRANCH1 in Arabidopsis encodes a plant homolog of the actin-related protein2/3 complex activator Scar/WAVE that regulates actin and microtubule organization. Plant Cell 17: 2314–2326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zipfel PA, Bunnell SC, Witherow DS, Gu JJ, Chislock EM, Ring C, Pendergast AM. (2006) Role for the Abi/wave protein complex in T cell receptor-mediated proliferation and cytoskeletal remodeling. Curr Biol 16: 35–46 [DOI] [PubMed] [Google Scholar]