Complex transcriptional response of AtALMT1 malate transporter could account for its contribution to pleiotropic traits.
Abstract
In Arabidopsis (Arabidopsis thaliana), malate released into the rhizosphere has various roles, such as detoxifying rhizotoxic aluminum (Al) and recruiting beneficial rhizobacteria that induce plant immunity. ALUMINUM-ACTIVATED MALATE TRANSPORTER1 (AtALMT1) is a critical gene in these responses, but its regulatory mechanisms remain unclear. To explore the mechanism of the multiple responses of AtALMT1, we profiled its expression patterns in wild-type plants, in transgenic plants harboring various deleted promoter constructs, and in mutant plants with defects in signal transduction in response to various inducers. AtALMT1 transcription was clearly induced by indole-3-acetic acid (IAA), abscisic acid (ABA), low pH, and hydrogen peroxide, indicating that it was able to respond to multiple signals, while it was not induced by methyl jasmonate and salicylic acid. The IAA-signaling double mutant nonphototropic hypocotyls4-1; auxin-responsive factor19-1 and the ABA-signaling mutant aba insensitive1-1 did not respond to auxin and ABA, respectively, but both showed an Al response comparable to that of the wild type. A synthetic microbe-associated molecular pattern peptide, flagellin22 (flg22), induced AtALMT1 transcription but did not induce the transcription of IAA- and ABA-responsive biomarker genes, indicating that both Al and flg22 responses of AtALMT1 were independent of IAA and ABA signaling. An in planta β-glucuronidase reporter assay identified that the ABA response was regulated by a region upstream (−317 bp) from the first ATG codon, but other stress responses may share critical regulatory element(s) located between −292 and −317 bp. These results illustrate the complex regulation of AtALMT1 expression during the adaptation to abiotic and biotic stresses.
Organic acid (OA) excretion/uptake plays various roles in many plant tissues. For example, it regulates stomatal closure in guard cells (Vahisalu et al., 2008), and in root tissues, it plays roles in nutrient uptake (e.g. iron [Durrett et al., 2007] and phosphorus [Neumann et al., 1999]) and in the detoxification of toxic ions (e.g. aluminum [Al; Pellet et al., 1995] and copper [Murphy et al., 1999]) in the rhizosphere. Excretion of OA from the roots can also recruit beneficial bacteria that enhance defense mechanisms through induced systemic resistance (Rudrappa et al., 2008; Lakshmanan et al., 2012). These events are regulated in a complex manner by a system involving OA transporters and OA synthesis (Delhaize et al., 1993; López-Bucio et al., 2000) at both the transcriptional and posttranslational levels (e.g. the SLOW ANION CHANNEL-ASSOCIATED1 [SLAC1] response to CO2; Negi et al., 2008). It is important to explore these mechanisms at the molecular level to understand the complex roles of OA transport in biological processes and its contribution to pleiotropic traits.
Arabidopsis (Arabidopsis thaliana) ALUMINUM-ACTIVATED MALATE TRANSPORTER1 (AtALMT1; Hoekenga et al., 2006) was first identified as an ortholog of TaALMT1, which encodes a root-localized malate transporter in wheat (Triticum aestivum). This protein plays a critical role in Al tolerance by detoxifying Al rhizotoxicity (Sasaki et al., 2004). Excretion of malate via AtALMT1 is induced by infection of aerial tissues by pathogenic bacteria (Rudrappa et al., 2008). In such cases, malate recruits beneficial bacteria to form a biofilm at the root surface, activating induced systemic reactions to protect the plant against bacterial infection. Thus, AtALMT1 has pleiotropic effects in both abiotic (i.e. Al resistance) and biotic (i.e. beneficial bacteria recruitment) stress resistance through its role in exuding malate from the roots. Its encoding gene, AtALMT1, is a good target for studies aimed at understanding the complex nature of regulation related to the multiple roles of OA transport.
The AtALMT1 protein transports malate into the rhizosphere, where it detoxifies Al3+ by converting it to the much less phytotoxic Al-malate chelating complex (Hoekenga et al., 2006). A knockout mutation of the gene resulted in hypersensitivity to Al, suggesting that this protein is essential for the survival of Arabidopsis in acid soils. In our previous study, we showed that malate excretion as a mechanism of Al tolerance is likely to be optimized to minimize carbon loss via both transcriptional and posttranslational regulation (Kobayashi et al., 2007). In that study, AtALMT1 expression was limited to the root tip, the tissue most sensitive to Al rhizotoxicity, under Al treatment, but it was barely expressed in response to other rhizotoxic ions. In addition, Al activation was identified in the activation process of malate transport. Pharmacological analyses suggested that both processes involve protein phosphorylation/dephosphorylation. Although the mechanisms of protein activation have not been clarified yet, bacterial infection of aerial tissues induced AtALMT1 transcription in the roots (Rudrappa et al., 2008). Treatment of aerial tissues with the elicitors coronatine and microbe-associated molecular patterns such as flagellin22 (flg22 [QRLSTGSRINSAKDDAAGLQIA]; Felix et al., 1999) also induced transcription of AtALMT1 in the roots (Lakshmanan et al., 2012). This finding indicated that the transcriptional regulation of AtALMT1 responds to multiple signals.
Multisignal regulation has been reported for some transporters such as the inorganic anion transporter SLAC1, which regulates stomatal closure (Vahisalu et al., 2008). The SLAC1 malate transporter plays important roles in photosynthesis and the drought response (Geiger et al., 2009; Kusumi et al., 2012), both of which are regulated by complex systems that are responsive to multiple signal inducers such as abscisic acid (ABA) and reactive oxygen species. Some of the signaling pathways in those systems involve protein phosphorylation, such as type 2C protein phosphatase (PP2C)/SNF1-related protein kinase2 (SnRK2) in the ABA response (Umezawa et al., 2010). The multiple biological roles of AtALMT1 suggest that it is regulated by such a complex system. In this study, we profiled AtALMT1 expression in response to various phytohormones and other chemicals. The aim of this study was to explore the complex transcriptional regulation of this gene, which encodes a protein that plays roles in various stress responses.
RESULTS
Profiling of ALMT1 Transcription in Response to Rhizotoxins and Chemical Treatments
AtALMT1 transcription was profiled using transgenic plants carrying an AtALMT1promoter::GUS (−1,110 from the first ATG; the full promoter region) fusion construct after short-term (6-h) treatments with phytohormones and chemical inducers (i.e. salicylic acid [SA] and methyl jasmonate [MeJA]). Other than benzylaminopurine (BAP), all phytohormones and chemical inducers resulted in GUS activity (visualized as blue staining) in the root apices, but there were differences in the density and tissue localization of the staining among treatments (Fig. 1A). The indole-3-acetic acid (IAA)-treated roots were stained in the elongation zone and vascular tissues in the upper parts. ABA treatment induced dense staining in the elongation zone, while the density depended on the ABA concentration. Roots treated with GA3 showed blue staining throughout the whole root apex, although the staining was much lighter than that in the IAA- and ABA-treated roots. The ethylene precursor 1-aminocyclopropane-1-carboxylic acid (ACC) resulted in slight staining at the root apex, which was similar to the root tips treated with SA and MeJA, which are major inducers of the biotic stress response (Fig. 1A). This indicated that some phytohormones can induce AtALMT1 expression within the short term, but IAA and ABA have stronger activity to induce expression. Among the abiotic stressors, Al, low pH, and hydrogen peroxide (H2O2) treatments induced stronger GUS staining than did NaCl, copper, and cadmium treatments. Transcript analysis confirmed that there were higher transcript levels of AtALMT1 in densely stained treatments than in lightly stained treatments (Fig. 1B).
Figure 1.
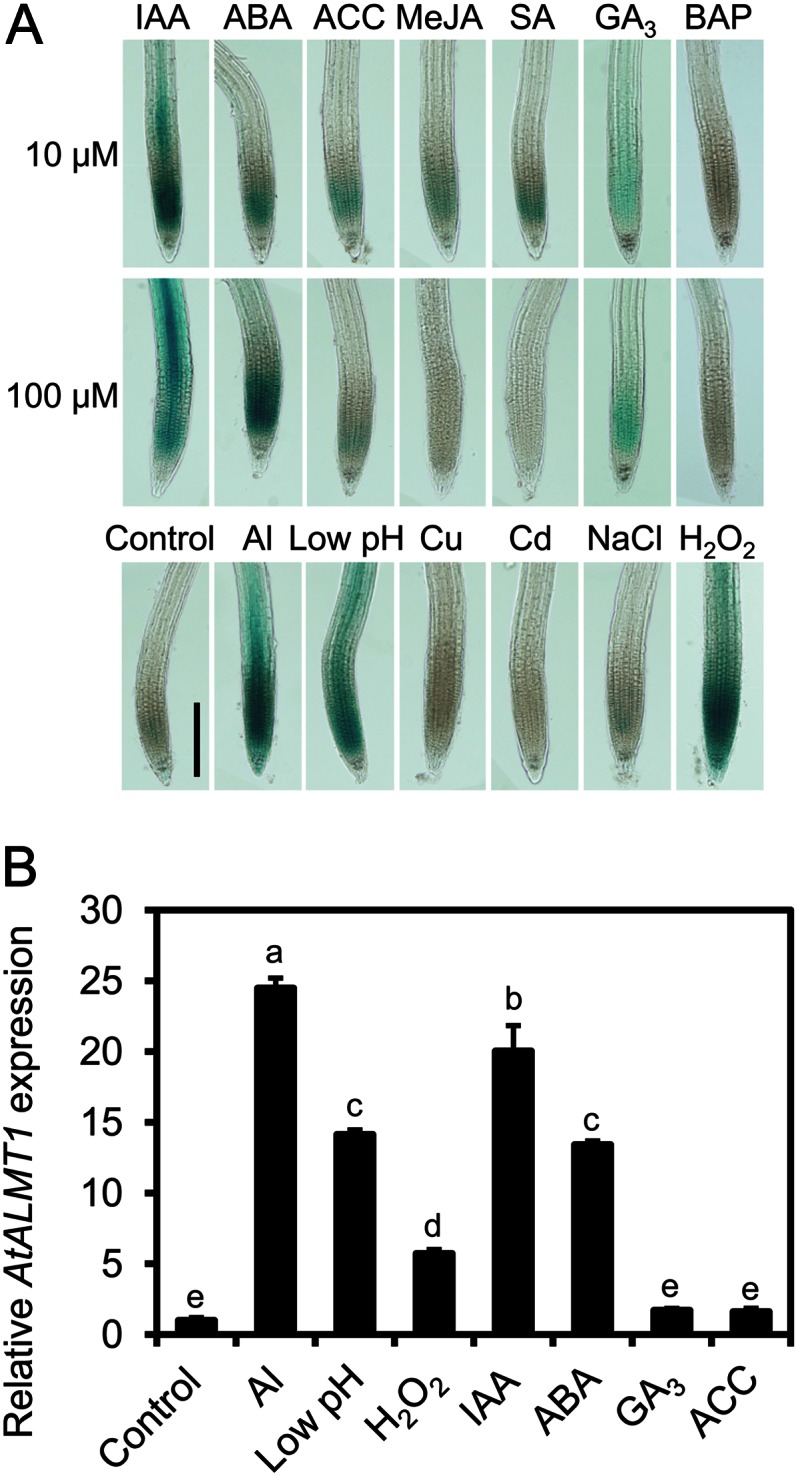
GUS expression and AtALMT1 transcription in roots of Arabidopsis under various treatments. Transgenic Col-0 harboring AtALMT1promoter::GUS (the GUS reporter gene fused with the full promoter sequence of −1,110 bp from the first ATG) and wild-type Col-0 were analyzed after exposure to various phytohormones (10 or 100 μm IAA, ABA, ACC, MeJA, SA, GA3, and BAP), H2O2(300 μm; pH 5.5), or rhizotoxic ions (10 μm AlCl3, 1.6 μm CuSO4, 15 μm CdCl2, and 50 mm NaCl at pH 5.5 or pH 4.7 for low pH). Activation of the AtALMT1 promoter was observed by GUS staining (blue). A, GUS staining patterns in a transgenic line. Bar = 20 μm. B, AtALMT1 transcript levels in wild-type Col-0 in response to various inducers determined by real-time RT-PCR (using UBQ1 as an internal control). Values shown are means ± sd (n = 3). Different letters indicate statistically significant differences (P < 0.05, Tukey’s test). Seedlings were incubated to various solutions for 6 h before histochemical and transcripts analyses.
There were differences in GUS expression patterns in root tissues among the various treatments (Fig. 1A). Only Al and H2O2 induced GUS expression in the whole root tip, including in root cap cells. Al, low pH, and IAA treatments induced GUS expression in the inner and upper parts of roots. Roots treated with ABA showed dense blue staining that was limited to the elongation zone and the meristem. These results confirmed that AtALMT1 transcription is induced by various signal inducers, while the position in the root tissues and the expression level are variable among the treatments.
AtALMT1 Expression in IAA- and ABA-Signaling Mutants
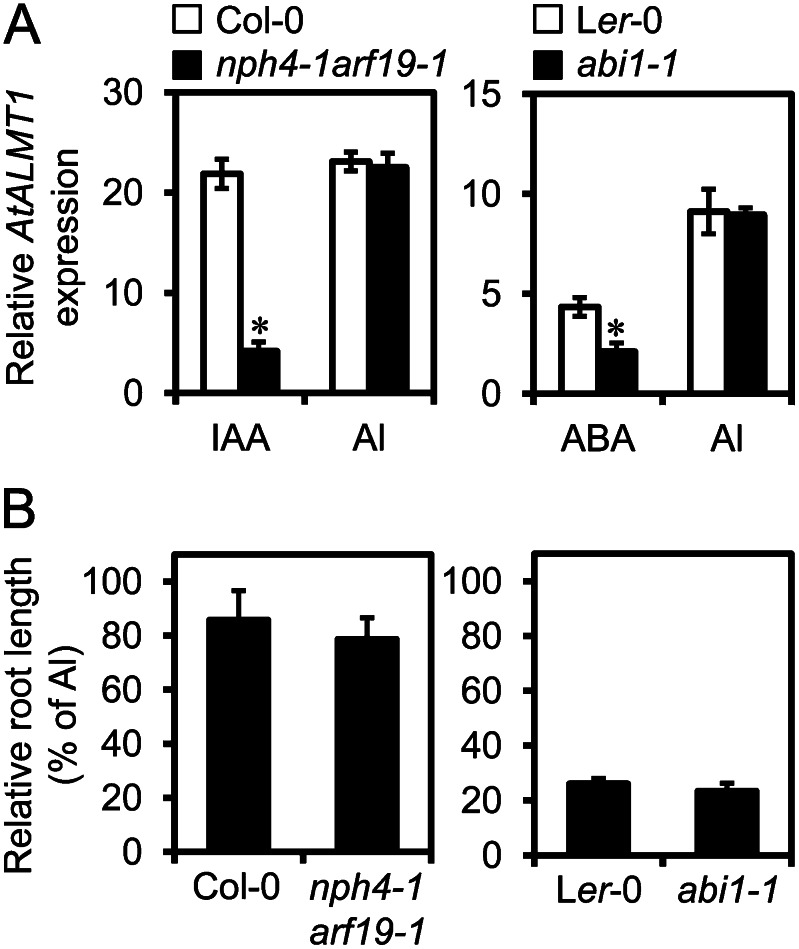
To characterize the induction of AtALMT1 mediated by IAA- and ABA-signaling pathways, we analyzed transcript levels of AtALMT1 in an IAA-signaling double mutant, nonphototropic hypocotyls4-1; auxin-responsive factor19-1 (nph4-1arf19-1; Okushima et al., 2005, 2007), and an ABA-signaling mutant, aba insensitive1-1 (abi1-1; Leung et al., 1994; Meyer et al., 1994). Both mutants defective in the ability to activate transcription responded to ABA and IAA (see “Discussion”). Transcript levels of AtALMT1 in the IAA- and ABA-signaling mutants were analyzed by real-time reverse transcription (RT)-PCR. After 6 h of IAA treatment, AtALMT1 transcript levels were higher in the wild type than in nph4-arf19-1 (Fig. 2A). Compared with that in the wild type, ABA-induced AtALMT1 transcription was significantly decreased in the ABA-signaling mutant abi1-1 (Fig. 2A). The fact that ABA- and IAA-induced AtALMT1 transcription was decreased in these mutants suggested that IAA and ABA signaling are involved in the activation of AtALMT1 transcription.
Figure 2.
AtALMT1 expression and Al tolerance in ABA- and IAA-signaling mutants. A, Roots of the IAA-signaling mutant nph4-1arf19-1 (Col-0 background) and the ABA-signaling mutant abi1-1 (Ler-0 background) were immersed in solutions containing 10 μm IAA and 100 μm ABA, or in Al (10 μm AlCl3) rhizotoxic solution, for 6 h. AtALMT1 transcripts were quantified by real-time RT-PCR using UBQ1 as an internal control. Transcript levels in parental accessions, Col-0 and Ler-0, were also quantified. Asterisks indicate significant differences from Col-0 (P < 0.05, Student’s t test). B, Al tolerance of mutants and parental accessions, as determined by root growth assay (length of the primary root after 7 d in 4 μm AlCl3 solution, pH 5.0). Values are means ± sd (n = 5).
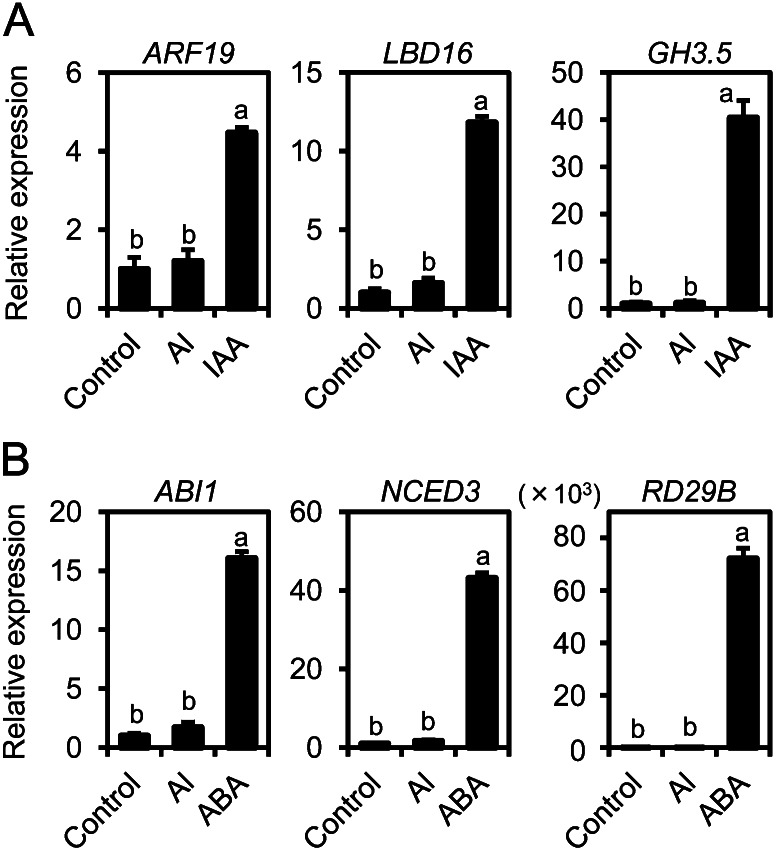
To evaluate cross talk among these phytohormones and Al-inducible AtALMT1 expression, the transcript levels of AtALMT1 and IAA- and ABA-responsive genes were compared between the mutants and wild-type parental accessions and between the control and an Al treatment in the wild type. In the Al treatment, transcript levels of AtALMT1 in the mutants were comparable to those in the parental accessions (Fig. 2A), suggesting that AtALMT1 transcription in response to Al treatment was not solely regulated by ABA and IAA. The transcript levels of some IAA-responsive and ABA-responsive genes were far lower in the Al treatment than in the IAA and ABA treatments (Fig. 3). Such genes included the IAA-responsive genes ARF19, LATERAL ORGAN BOUNDARIES DOMAIN16 (LBD16), and GH3.5 and the ABA-responsive genes ABI1, NINE-CIS-EPOXYCAROTENOID DIOXYGENASE3 (NCED3), and RESPONSIVE TO DESSICATION29B (RD29B). Primary root growth of the IAA- and ABA-signaling mutants was comparable to that of the wild-type parental accessions in Al-toxic solution (Fig. 2B). These findings suggested that ABA and IAA signaling were not required to maintain the Al tolerance of the primary root via AtALMT1 expression.
Figure 3.
Transcription of IAA- and ABA-responsive genes under Al treatment. Transcript levels of IAA-responsive genes (ARF19, LBD16, and GH3.5; A) and ABA-responsive genes (ABI1, NCED3, and RD29B; B) are shown in the wild type (Col-0) under Al and phytohormone treatments. Seedlings were grown for 6 d in control nutrient solution, and then roots were incubated in 10 μm IAA, 100 μm ABA, or 10 μm AlCl3 at pH 5.5 for 6 h. Transcript levels were analyzed by real-time RT-PCR. Different letters indicate significant differences compared with the control (P < 0.05, Tukey’s test).
Taken together, these results suggested that Al-responsive AtALMT1 transcription, at least after a short-term (less than 6 h) Al treatment, is regulated by a pathway other than the ABA and IAA signaling pathways, which may not have a critical role in Al tolerance (as measured by primary root growth) mediated by malate excretion.
Promoter Analysis to Characterize AtALMT1 Responses to Inducers
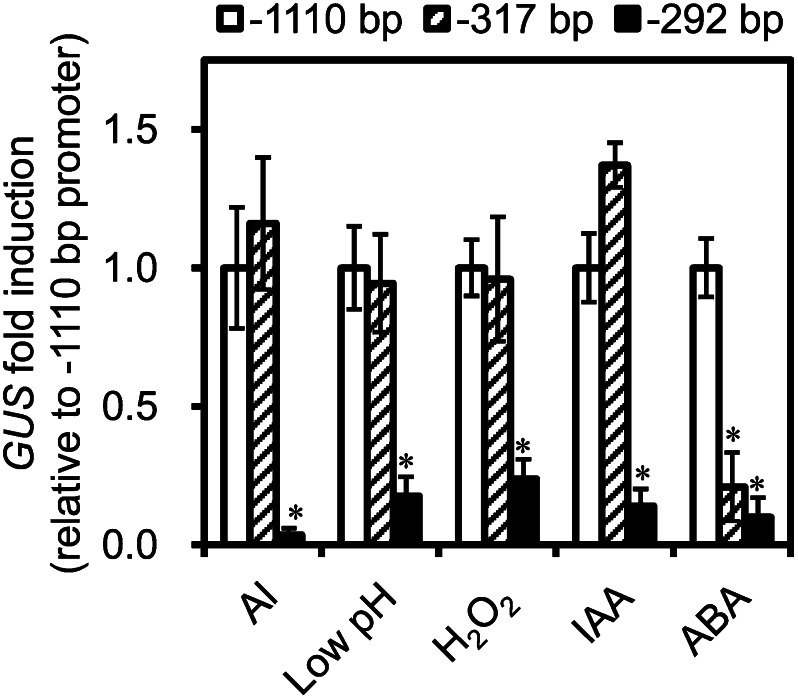
To characterize in detail the regulation of AtALMT1 transcription by Al and other inducers, we analyzed GUS expression in AtALMT1promoter::GUS transgenic lines carrying different lengths of the 5′ region of the AtALMT1 promoter. Using the full promoter construct (−1,110bp::GUS) as the reference, we compared GUS expression driven by deleted promoter regions (−317 bp [-317bp::GUS] and −292 bp [−292bp::GUS]) in response to various signal inducers. GUS expression was lower in the −292bp-GUS line than in the −1,110bp::GUS line in response to Al, low pH, IAA, ABA, and H2O2. GUS expression in response to the various inducers was similar between the −1,110bp::GUS line and the −317bp::GUS line, except when ABA was used as the signal inducer, where the GUS expression in the −317bp::GUS line was markedly lower than that in the −1,110bp::GUS line (Fig. 4). This result indicated that the major ABA-regulating element was localized farther upstream of the AtALMT1 promoter than −317 bp. To find cis-acting elements in the promoter, we searched the AtALMT1 promoter sequence between −317 and −1,110 bp using the PLACE and PlantCARE databases (Higo et al., 1999; Lescot et al., 2002). Both databases predicted cis-acting elements related to drought-inducible elements, CAAT box (CCAAT) and Myb binding site (CAACTG), in that region (Supplemental Table S1). In the Al, low-pH, IAA, and H2O2 treatments, there was lower GUS expression in the −292bp::GUS line than in the −1110bp::GUS line and the −317bp::GUS line (Fig. 4). This indicated that the critical regulatory element(s) common to those treatments would be in the region from −292 to −317 bp, while no consensus sequence was predicted by the same databases.
Figure 4.
GUS transcription in transgenic plants transformed with different AtALMT1 promoter deletion constructs in response to various signaling factors. Transcript levels of GUS in −1,110, −317, and −292 bp AtALMT1promoter:GUS transgenic plants were analyzed by real-time quantitative RT-PCR. Seedling roots were treated with 10 μm AlCl3, 300 μm H2O2, 10 μm IAA, or 100 μm ABA at pH 5.5 or pH 4.7 (low pH) for 6 h. Transcript levels of GUS were normalized to that of UBQ1, and then fold change (treatment/control) was calculated for each line. Relative fold change (the relative value of each line to mean fold change of the −1,110 bp AtALMT1 promoter construct) was calculated. Values are means ± se (n = 3). Asterisks in each treatment represent significant differences compared with the −1,110 bp promoter construct (P < 0.05, Student’s t test).
Activation of AtALMT1 in Roots of Arabidopsis by flg22
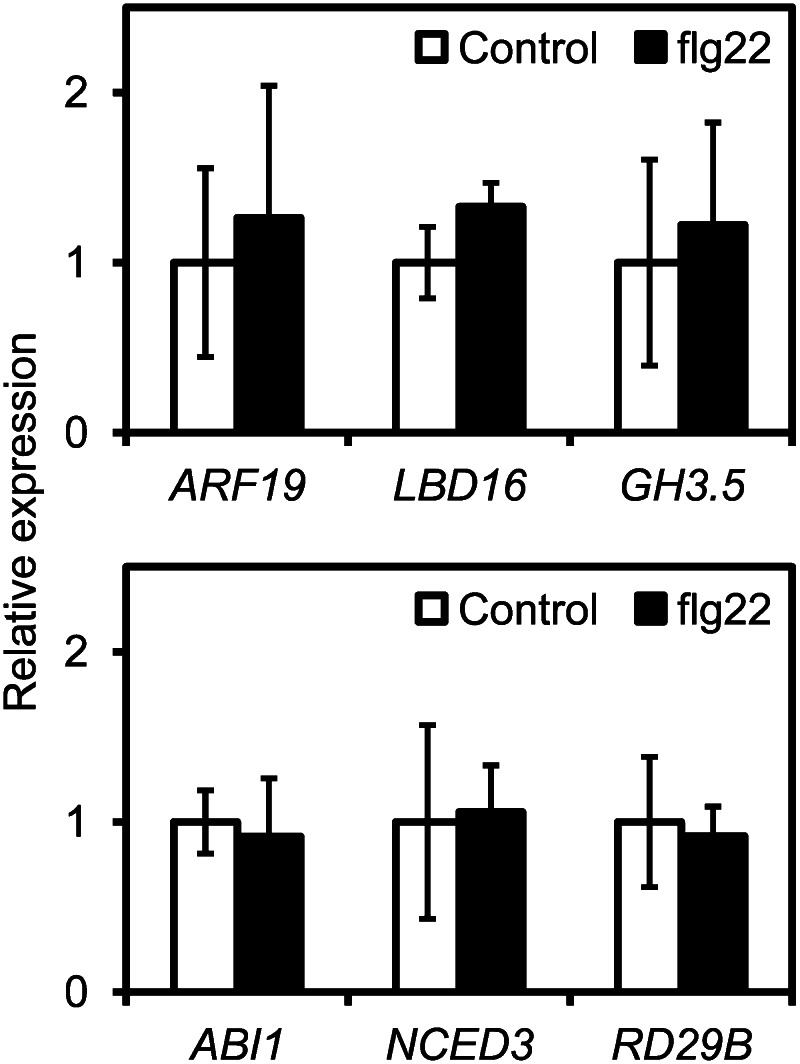
Some bacteria induce AtALMT1 expression through shoot-root signaling (Lakshmanan et al., 2012); however, the direct activation of AtALMT1 transcription in the roots had not been analyzed. To determine whether AtALMT1 expression could be induced by bacterial challenge to the roots, we treated the roots with flg22, a biological inducer of the plant-bacteria response, and performed histochemical analysis and expression analysis of GUS expression from AtALMT1promoter::GUS fusion constructs. In the −1,110bp::GUS line, GUS was expressed throughout the whole root tip in response to flg22, similar to the pattern of GUS transcription in response to Al treatment (Fig. 5A) Compared with that in the wild type, flg22-induced AtALMT1 transcription was significantly decreased in the flg22-signaling mutant flagellin-sensitive2 (fls2), which is defective in the flg22 receptor for activating transcription in response to flg22 (Fig. 5B). The fls2 mutant showed AtALMT1 expression by Al treatment, which was comparable to Columbia (Col-0; Fig. 5B). It suggests that flg22 signaling is involved in the activation of AtALMT1 expression but is not affected by Al-responsive induction. In addition, real-time RT-PCR analyses showed that flg22-induced GUS transcription in the −317bp::GUS line was comparable to that in the −1,110bp::GUS line. However, the fold change in GUS transcript levels was significantly reduced in the −292bp::GUS line compared with those in the other two lines (Fig. 5C), the same pattern observed in response to other inducers. Using inducible biomarker genes, we evaluated IAA and ABA signaling in flg22-induced AtALMT1 expression. All of the IAA-responsive genes (ARF19, LBD16, and GH3.5) and the ABA-responsive genes (ABI1, NCED3, and RD29B) did not respond to flg22 treatment of the roots (Fig. 6), suggesting that the short-term response of AtALMT1 expression induced by flg22 differs from that induced by IAA and ABA and that it is independent from the Al response.
Figure 5.
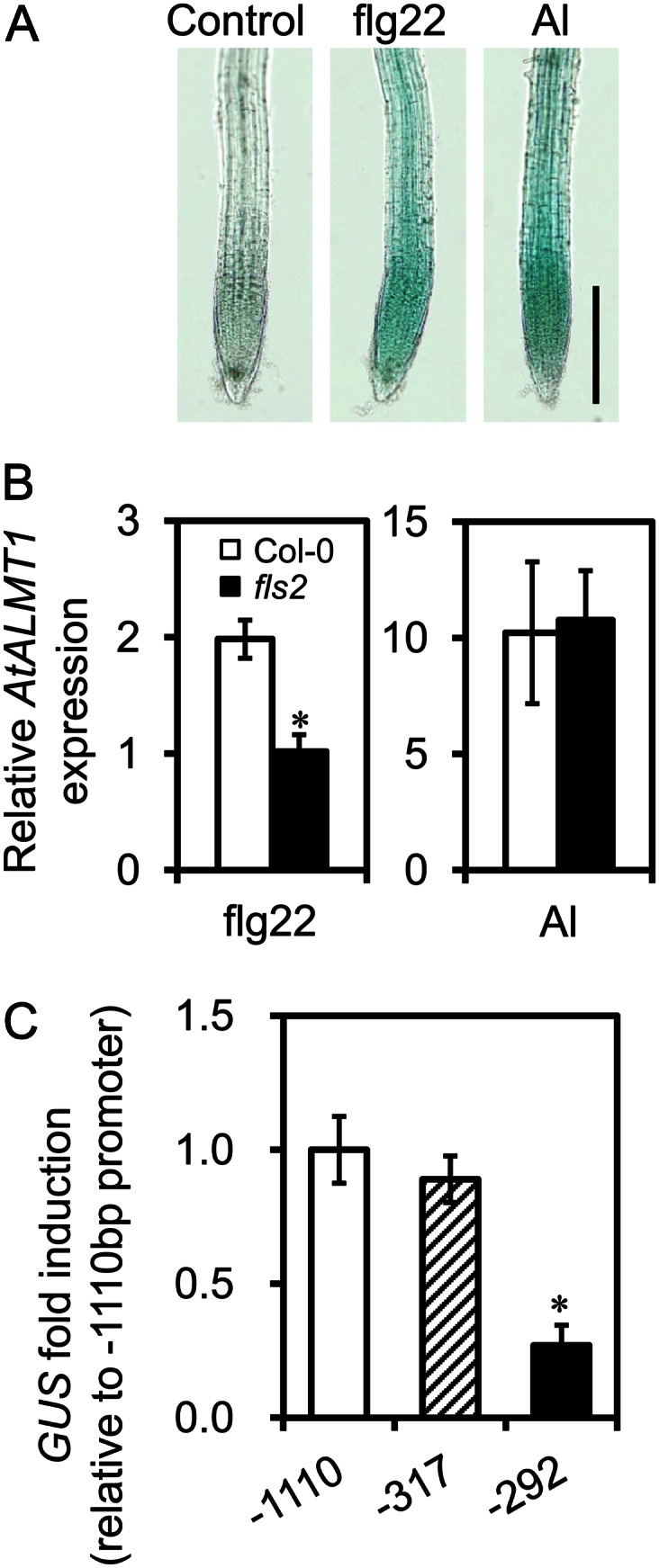
AtALMT1 transcription and GUS expression in roots treated with flg22. A, Induction of AtALMT1 by flg22 in transgenic Arabidopsis carrying AtALMT1promoter::GUS constructs. The images show GUS staining in roots after 6 h of treatment with flg22 (10 μm) and AlCl3 (10 μm). Bar = 20 μm. B, AtALMT1 transcript levels in roots of the flg22 signaling mutant fls2 and the wild type (Col-0) incubated for 6 h with or without flg22 (10 μm) or AlCl3 (10 μm). Values are means ± sd (n = 3). The asterisk indicates a significant difference from the Col-0 transcript level (P < 0.05, Student’s t test). C, Transcript levels of GUS in −1,110, −317, and −292 bp AtALMT1promoter:GUS transgenic plants analyzed by real-time quantitative RT-PCR. Roots of seedlings were treated with 10 μm flg22 for 6 h. GUS transcript levels were normalized to that of UBQ1, and then fold change (treatment/control) was calculated for each line. Relative fold change (the relative value of each line to mean fold change of the −1,110 bp AtALMT1 promoter construct) was calculated. Values are means ± se (n = 3). The asterisk represents a significant difference from the −1,110 bp promoter construct (P < 0.05, Student’s t test).
Figure 6.
Transcription of IAA- and ABA-responsive genes in response to flg22 treatment of roots. Transcript levels of IAA-responsive genes (ARF19, LBD16, and GH3.5) and ABA-responsive genes (ABI1, NCED3, and RD29B) are shown in roots of the wild type (Col-0) treated with flg22. All seedlings were grown for 6 d in control nutrient solution, and then roots were incubated in 10 μm flg22 for 6 h. Transcript levels were analyzed by real-time RT-PCR and normalized to that of UBQ1.
Transcript Profiling of AtMATE and ALS3
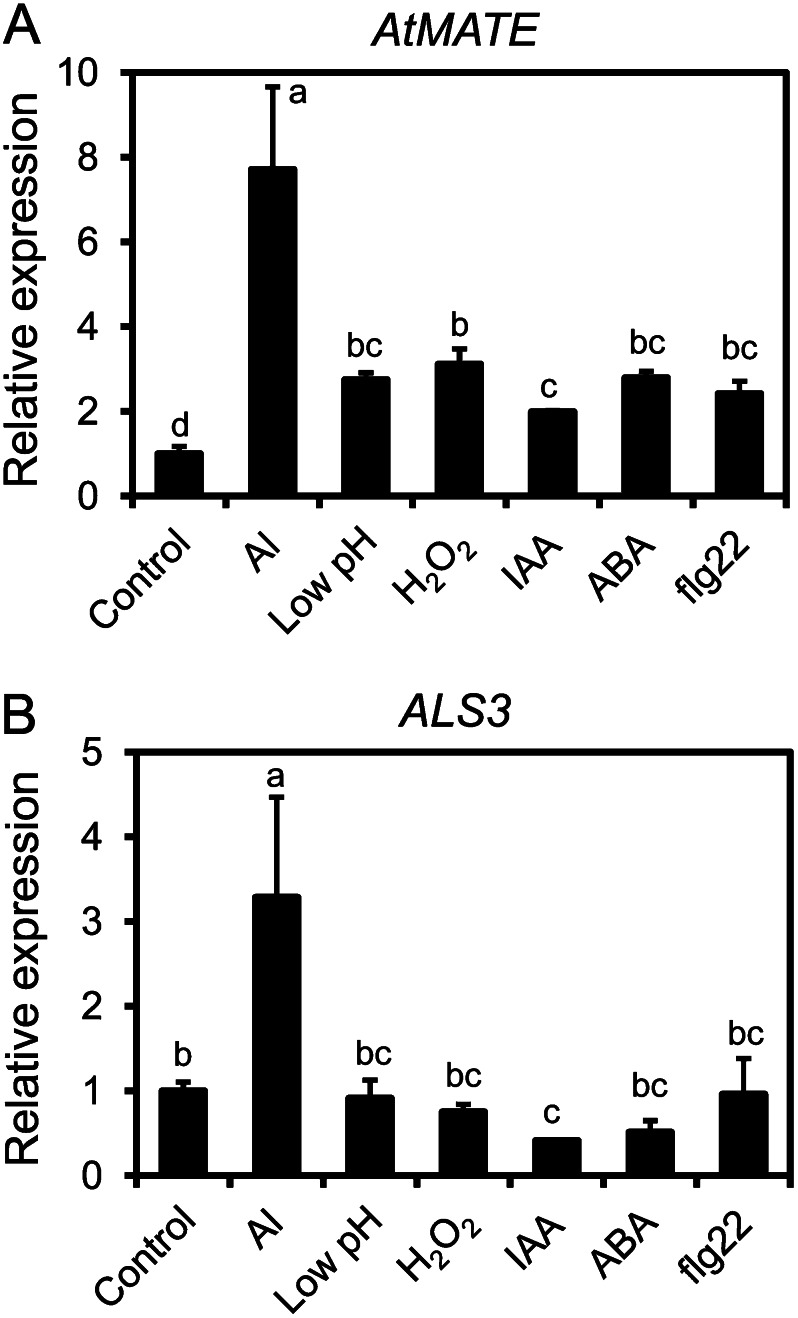
We analyzed the transcript levels of some other Al tolerance genes in response to inducers that activate AtALMT1 expression. AtMATE encodes an Al-responsive citrate transporter that belongs to the multidrug and toxic compound extrusion (MATE) efflux protein family (Liu et al., 2009), while ALUMINUM SENSITIVE3 (ALS3; Larsen et al., 2005) encodes a putative homolog of rice (Oryza sativa) SENSITIVE TO ALUMINUM RHIZOTOXICITY2, which encodes a subunit of a UDP-Glc transporter in rice (Huang et al., 2009). The transcription pattern of AtMATE was similar to that of AtALMT1. Although there were lower transcript levels of AtMATE (Fig. 7A) than AtALMT1 (Fig. 1B), all of the inducers of AtALMT1 transcription also induced AtMATE transcription (Fig. 7A). However, only the Al treatment was able to induce ALS3 transcription (Fig. 7B). This indicated that similar complex regulation of transcription would be shared by some other Al tolerance genes.
Figure 7.
Transcription of AtMATE and ALS3 in Arabidopsis roots in response to various treatments. Transcript levels of Al-responsive genes (AtMATE [A] and ALS3 [B]) were determined by real-time PCR in wild-type (Col-0) seedling roots after 6-h treatments with IAA (10 μm), ABA (100 μm), H2O2 (300 μm), AlCl3 (10 μm), flg22 (10 μm), or low pH (pH 4.7). Transcript levels were normalized to that of UBQ1, and relative expression (treatment/control) was calculated. Values are means ± sd (n = 3). Different letters indicate significant differences from the transcript level in the control (P < 0.05, Tukey’s test).
DISCUSSION
In this study, we identified that some signal inducers can trigger AtALMT1 expression within the short term (less than 6 h). Phytohormones (IAA and ABA) and other inducers (H2O2, low pH, and flg22 peptide; Fig. 5) all induced AtALMT1 transcription. This finding showed that AtALMT1 is regulated by multiple stressors that result in the production of these signal inducers. Our analyses using IAA- and ABA-signaling mutants, nph4-1arf19-1 and abi1-1, respectively, identified that AtALMT1 expression in response to each inducer is mediated by particular pathways linked to their genotypes. In the nph4-1arf19-1 double mutant, IAA signaling regulated by ARF-Aux/IAA proteins and SCFTIR1/AFB-mediated Aux/IAA proteolysis is blocked (Quint and Gray, 2006). The ABA-insensitive mutant abi1-1 shows a block in ABA signaling through the PP2C/SnRK2 pathway (Umezawa et al., 2009). In our ongoing research, we have not found any experimental evidence that ABA and IAA signaling are directly involved in the short-term activation of AtALMT1 expression by Al. Al treatments induced AtALMT1 transcription in these mutants (Fig. 2A) but did not induce ABA- and IAA-inducible biomarker genes (Fig. 3). This finding confirmed that Al activation of AtALMT1 transcription is not simply regulated by one of these phytohormones.
ABA- and IAA-signaling pathways did not directly contribute to the short-term expression of AtALMT1 induced by flg22 (Figs. 2 and 6). In addition, SA and MeJA, which are major inducers of the biotic stress response, could not induce AtALMT1 transcription (Fig. 1). This finding showed that these signal inducers do not directly contribute to AtALMT1 expression in response to flg22, similar to the response to Al. However, H2O2 and low-pH stress could be involved in flg22 and Al activation of AtALMT1 transcription. A transcriptome analysis showed that gene expression patterns during the bacterial response resembled those under low-pH stress in Arabidopsis (Lager et al., 2010), while the microbe-associated molecular pattern response results in H2O2 production, which is coupled with flg22 recognition (Torres et al., 2006). In this study, H2O2 and low pH activated AtALMT1 transcription. Consistent with this, Al treatments trigger H2O2 accumulation in the root tip (Kobayashi et al., 2005) and decrease cellular pH (Moseyko and Feldman, 2001). Further research is required to clarify the interactions among these factors in Al- and flg22-responsive AtALMT1 transcription.
Although the IAA- and ABA-signaling mutants did not show enhanced Al sensitivity in our experimental conditions (Fig. 2B), the responses involving IAA and ABA might have roles in Al tolerance in the natural environment. We used the length of the primary roots in hydroponic culture as the indicator of Al tolerance. The architecture of whole roots affects water acquisition and drought resistance in plants in the natural environment (Xiong et al., 2006). For example, IAA accumulation is an essential step for lateral root development, and thus, nph4-1arf19-1 cannot form lateral roots (Okushima et al., 2007). We observed that the transgenic line carrying AtALMT1promoter::GFP expressed GFP at that site without any exogenous inducer treatments (Supplemental Fig. S1). Inducible expression mediated by IAA could result in an accumulation of AtALMT1 proteins in the tip of lateral roots before they come into contact with rhizotoxic Al. Dehydrated roots in dried soils produce ABA, which can induce stomatal closure via long-distance signaling, thus increasing drought tolerance (Zhang et al., 2006). We observed that ABA treatment induced AtALMT1 transcription. In a drought situation, it is reasonable to expect that AtALMT1 transcription induced by ABA (i.e. like that in the root tip in Fig. 1A) could protect the roots from Al rhizotoxicity, which, if unchecked, could further exacerbate drought sensitivity by inhibiting root development. In addition, ABA accumulation in response to Al occurs in some crop plants such as barley (Hordeum vulgare) and soybean (Glycine max; Kasai et al., 1993; Hou et al., 2010). Together, these findings suggest that responses involving IAA and ABA play a role in Al tolerance in Arabidopsis in the natural soil environment.
Lakshmanan et al. (2012) reported that application of flg22 to the aerial parts induced AtALMT1 expression in the roots, which is concomitant with ABA accumulation and stomatal closure in the shoots. This suggested that AtALMT1 expression mediated by ABA signaling would have a role in biotic stress resistance, in particular, in the shoot-root interaction. On the other hand, in this study, flg22 treatment of the roots induced AtALMT1 transcription within 6 h (Fig. 5). This pattern of AtALMT1 expression could be reasonable for recruiting beneficial rhizobacteria similar to Bacillus subtilis FB17 (Rudrappa et al., 2008). FB17 was attracted to the root surface by malate chemotaxis, and then it induced systemic resistance to protect aerial tissues from infection by pathogenic bacteria. AtALMT1-mediated malate excretion is a critical step for the attraction of beneficial bacteria to the root surface. Interestingly, FB17-derived elicitors (i.e. autoclaved cells) did not induce AtALMT1, possibly because of differences of its flagella that are not highly conserved in the structure of flg22 (Lakshmanan et al., 2012). The significant induction of AtALMT1 by flg22 to the roots suggests that other members of the rhizobacterial community conserving flg22 structure may support the recruitment of beneficial bacteria that require AtALMT1-dependent malate excretion.
Promoter-GUS reporter analyses revealed some of the mechanisms underlying the complex regulation of AtALMT1 expression in response to various inducers. These analyses indicated that an ABA-responsive element is located in the upstream (−317 to −1,110 bp) promoter region, while the responsive element for other signal inducers (Fig. 4) was located closer to the start codon, between −317 and −292 bp. Several drought responsive cis-element motifs were predicted by the PlantCARE and PLACE databases (Supplemental Table S1). A common region (−317 to −292 bp) contained a putative cis-acting site, GGN(T/g/a/C)V(C/A/g)S(C/G) (Tsutsui et al., 2011), that binds the ALUMINUM RESISTANCE TRANSCRIPTION FACTOR1 (ART1) zinc finger transcription factor (Yamaji et al., 2009), while we cannot show further experimental evidence confirming the regulatory element(s) in the region. AtALMT1 expression requires Arabidopsis SENSITIVE TO PROTON RHIZOTOXICITY1 (AtSTOP1; Iuchi et al., 2007), which contains sequences highly homologous to those of the zinc finger domains of ART1. It is possible that an interaction with AtSTOP1 is critical for regulating AtALMT1 expression via the common region. This possibility requires further research, but AtMATE, which is regulated by AtSTOP1, showed multiple responses to the same signal inducers, suggesting that some of the genes regulated by AtSTOP1 may share similar complex regulatory mechanisms, allowing responses to various stressors.
In conclusion, our results illustrate the complex patterns of AtALMT1 transcription in response to various signal inducers. However, for successful malate excretion, the AtALMT1 protein must be activated. This would also be regulated in a complex manner and would be another critical factor in regulating malate excretion from the roots. Previous research on Al tolerance indicated that protein phosphorylation/dephosphorylation is involved in Al activation of AtALMT1 (Kobayashi et al., 2007). Although this process has yet to be characterized in detail, the increase in malate excretion induced by beneficial bacteria (Rudrappa et al., 2008) would require a similar mechanism. Among the tested signal inducers, ABA and H2O2 induced malate excretion during 24 h of treatment (Supplemental Fig. S2). This suggests that some signal inducers may activate the AtALMT1 protein. Further research on the activation process of the protein is required to understand how plants regulate malate excretion from the roots for acquiring stress resistance.
MATERIALS AND METHODS
Plant Materials
Arabidopsis (Arabidopsis thaliana) accessions Col-0 and Landsberg erecta (Ler-0) were obtained from the RIKEN BioResource Center (BRC) and the Nottingham Arabidopsis Stock Centre. The IAA-signaling double mutant nph4-larf19-1 was kindly provided by Dr. Hidehiro Fukaki (Kobe University). The ABA-signaling mutant abi1-1 was obtained from the RIKEN BRC. The flg22-signaling mutant fls2 was kindly provided by Dr. Ken Shirasu (RIKEN Plant Science Center). The mutant abi1-1 was in the Ler-0 background, and other lines/mutants were in the Col-0 background.
Construction of Transgenic Lines for AtALMT1 Promoter Analysis
A series of transgenic plants carrying AtALMT1promoter::GUS fusion constructs in Col-0 were prepared by Agrobacterium tumefaciens-mediated transformation using the following procedures. All PCRs were carried out using PrimeSTAR max (Takara) high-fidelity Taq polymerase, and the sequences of the amplified products were checked with an ABI PRISM 3100 Genetic Analyzer using the BigDye Terminator version 3.1 Cycle Sequencing Kit (Applied Biosystems) according to the manufacturer’s protocol. Details of primer sequences are shown in Supplemental Table S2. A series of 5′ deleted promoters, −1,110, −317, and −292 bp from A of the start codon of AtALMT1, were amplified by PCR. Each sequence was attached to the 5′ end of the open reading frame of the GUS gene connected to the nopaline synthase terminator by overlapping extension PCR (Horton et al., 1989). The amplicon was digested with SfiI and then introduced into the transfer DNA region of pBE2113 (Mitsuhara et al., 1996), which contains a kanamycin resistance cassette as the selection marker. We used the hypervirulent A. tumefaciens strain GV3101 to transform plant tissues using the floral dip method (Clough and Bent, 1998). The transgenic seeds that were obtained were screened using kanamycin (50 g mL−1 in Murashige and Skoog medium containing 1% agar; Murashige and Skoog, 1962) as the selection marker.
Growth Conditions and Root Growth Test
Arabidopsis seedlings were grown hydroponically according to the method of Kobayashi et al. (2007) in modified MGRL solution diluted to 1:50 (with inorganic phosphate and pH 5.6 for transcript analyses; without phosphorus and pH 5.0 for growth tests to determine Al tolerance). Seedlings were grown at 24°C ± 2°C under a 12-h-light/12-h-dark photoperiod, with light supplied at a photosynthetic photon flux density of 37 μmol m−2 s−1. The culture solutions were renewed every 2 d. Seedlings were grown for 6 d for transcript analyses and GUS staining and for 7 d for the root growth test to assess Al tolerance, unless mentioned otherwise. Root length was measured at day 7 in control and Al-toxic solutions (4 μm AlCl3). Five of the 10 seedlings with the longest roots in Al-toxic solution were used to calculate mean values and sd to assess the Al tolerance of the lines, as described previously (Kobayashi et al., 2007).
Stress, Phytohormone, and Chemical Treatments
The roots of seedlings pregrown as described above were transferred to solutions containing various rhizotoxins (10 μm AlCl3, 1.6 μm CuSO4, 15 μm CdCl2, or 50 mm NaCl) or chemicals (10 or 100 μm IAA, ABA, ACC, BAP, GA3, MeJA, or SA, or 300 mm H2O2) and then incubated for 6 h. All rhizotoxic ions and chemicals were added to solution containing 1/50 MGRL nutrients with extra CaCl2 added to make a final concentration of 200 μm, pH 5.5 (solution did not contain phosphorus).
Histochemical Analysis of Reporter Expression
GUS staining was performed as described previously (Kobayashi et al., 2007) with −1,110bp AtALMT1promoter::GUS transgenic lines. Briefly, the roots of seedlings were treated with rhizotoxins and chemicals as described above and stained with staining solution (1.0 mm X-glucuronide, 0.1 m sodium phosphate buffer [pH 7.0], 10 mm EDTA [pH 8.0], 0.5 mm potassium ferricyanide [pH 7.0], 0.5 mm potassium ferrocyanide [pH 7.0], 0.3% Triton X-100, and 20% methanol) for 15 min at 37°C. The samples were observed and photographed with an Olympus BX51 microscope equipped with an Olympus DP70 camera system.
RNA Extraction and Expression Analysis
Total RNA was extracted as described (Suzuki et al., 2003). First-strand complementary DNA was synthesized from total RNA with ReverTra Ace (Toyobo) and oligo(dT)18 primers. Quantitative real-time RT-PCR was performed using gene-specific primer pairs (Supplemental Table S3) with SYBR Premix Ex Taq II (Takara Bio) and the Thermal Cycler Dice Real Time System II (Takara Bio) following the manufacturer’s instructions. Reactions were performed with three biological replicates for each sample. Transcript levels of the target gene were normalized to that of UBQ1.
Promoter Motif Search
PlantCARE (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) and PLACE/single scan (http://www.dna.affrc.go.jp/PLACE/) software were used to scan for cis-elements in the AtALMT1 promoter sequence (−1,110 bp from A of the first ATG). The cis-elements identified using PlantCARE were evaluated by scanning at the PLACE database. Only cis-elements related to the drought response in Arabidopsis are shown in Supplemental Table S3.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Histochemical analysis of AtALMT1 expression during lateral root formation in Arabidopsis.
Supplemental Figure S2. Malate excretion from Arabidopsis roots after treatment with Al and other signal inducers, inducing AtALMT1 expression.
Supplemental Table S1. List of cis-acting elements of the AtALMT1 promoter.
Supplemental Table S2. List of primer sequences for the AtALMT1promoter::GUS construct of the transgenic plant.
Supplemental Table S3. Sequences of primers used for quantitative RT-PCR.
Acknowledgments
We are grateful for the technical support provided by Atsuko Iuchi, Fumie Mori, and Setsuko Kawamura of the RIKEN BRC. We thank the RIKEN BRC, the Arabidopsis Biological Resource Center, and the Nottingham Arabidopsis Stock Centre for providing Arabidopsis seeds. We are grateful to Dr. Hidehiro Fukaki and Dr. Ken Shirasu for providing Arabidopsis mutants.
Glossary
- OA
organic acid
- Al
aluminum
- ABA
abscisic acid
- SA
salicylic acid
- MeJA
methyl jasmonate
- BAP
benzylaminopurine
- IAA
indole-3-acetic acid
- ACC
1-aminocyclopropane-1-carboxylic acid
- H2O2
hydrogen peroxide
- RT
reverse transcription
- Col-0
Columbia
- Ler-0
Landsberg erecta
- BRC
BioResource Center
References
- Clough SJ, Bent AF. (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Delhaize E, Ryan PR, Randall PJ. (1993) Aluminum tolerance in wheat (Triticum aestivum L.). II. Aluminum-stimulated excretion of malic acid from root apices. Plant Physiol 103: 695–702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durrett TP, Gassmann W, Rogers EE. (2007) The FRD3-mediated efflux of citrate into the root vasculature is necessary for efficient iron translocation. Plant Physiol 144: 197–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felix G, Duran JD, Volko S, Boller T. (1999) Plants have a sensitive perception system for the most conserved domain of bacterial flagellin. Plant J 18: 265–276 [DOI] [PubMed] [Google Scholar]
- Geiger D, Scherzer S, Mumm P, Stange A, Marten I, Bauer H, Ache P, Matschi S, Liese A, Al-Rasheid KA, et al (2009) Activity of guard cell anion channel SLAC1 is controlled by drought-stress signaling kinase-phosphatase pair. Proc Natl Acad Sci USA 106: 21425–21430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higo K, Ugawa Y, Iwamoto M, Korenaga T. (1999) Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Res 27: 297–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoekenga OA, Maron LG, Piñeros MA, Cançado GM, Shaff J, Kobayashi Y, Ryan PR, Dong B, Delhaize E, Sasaki T, et al. (2006) AtALMT1, which encodes a malate transporter, is identified as one of several genes critical for aluminum tolerance in Arabidopsis. Proc Natl Acad Sci USA 103: 9738–9743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton RM, Hunt HD, Ho SN, Pullen JK, Pease LR. (1989) Engineering hybrid genes without the use of restriction enzymes: gene splicing by overlap extension. Gene 77: 61–68 [DOI] [PubMed] [Google Scholar]
- Hou N, You J, Pang J, Xu M, Chen G, Yang Z. (2010) The accumulation and transport of abscisic acid in soybean (Glycine max L.) under aluminum stress. Plant Soil 330: 127–137 [Google Scholar]
- Huang CF, Yamaji N, Mitani N, Yano M, Nagamura Y, Ma JF. (2009) A bacterial-type ABC transporter is involved in aluminum tolerance in rice. Plant Cell 21: 655–667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iuchi S, Koyama H, Iuchi A, Kobayashi Y, Kitabayashi S, Kobayashi Y, Ikka T, Hirayama T, Shinozaki K, Kobayashi M. (2007) Zinc finger protein STOP1 is critical for proton tolerance in Arabidopsis and coregulates a key gene in aluminum tolerance. Proc Natl Acad Sci USA 104: 9900–9905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasai M, Sasaki M, Tanakamaru S, Yamamoto Y, Matsumoto H. (1993) Possible involvement of abscisic acid in increases in activities of two vacuolar H+-pumps in barley roots under aluminum stress. Plant Cell Physiol 34: 1335–1338 [Google Scholar]
- Kobayashi Y, Furuta Y, Ohno T, Hara T, Koyama H. (2005) Quantitative trait loci controlling aluminium tolerance in two accessions of Arabidopsis thaliana (Landsberg erecta and Cape Verde Islands). Plant Cell Environ 28: 1516–1524 [Google Scholar]
- Kobayashi Y, Hoekenga OA, Itoh H, Nakashima M, Saito S, Shaff JE, Maron LG, Piñeros MA, Kochian LV, Koyama H. (2007) Characterization of AtALMT1 expression in aluminum-inducible malate release and its role for rhizotoxic stress tolerance in Arabidopsis. Plant Physiol 145: 843–852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusumi K, Hirotsuka S, Kumamaru T, Iba K. (2012) Increased leaf photosynthesis caused by elevated stomatal conductance in a rice mutant deficient in SLAC1, a guard cell anion channel protein. J Exp Bot 63: 5635–5644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lager I, Andréasson O, Dunbar TL, Andreasson E, Escobar MA, Rasmusson AG. (2010) Changes in external pH rapidly alter plant gene expression and modulate auxin and elicitor responses. Plant Cell Environ 33: 1513–1528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakshmanan V, Kitto SL, Caplan JL, Hsueh YH, Kearns DB, Wu YS, Bais HP. (2012) Microbe-associated molecular patterns-triggered root responses mediate beneficial rhizobacterial recruitment in Arabidopsis. Plant Physiol 160: 1642–1661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen PB, Geisler MJ, Jones CA, Williams KM, Cancel JD. (2005) ALS3 encodes a phloem-localized ABC transporter-like protein that is required for aluminum tolerance in Arabidopsis. Plant J 41: 353–363 [DOI] [PubMed] [Google Scholar]
- Lescot M, Déhais P, Thijs G, Marchal K, Moreau Y, Van de Peer Y, Rouzé P, Rombauts S. (2002) PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res 30: 325–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung J, Bouvier-Durand M, Morris PC, Guerrier D, Chefdor F, Giraudat J. (1994) Arabidopsis ABA response gene ABI1: features of a calcium-modulated protein phosphatase. Science 264: 1448–1452 [DOI] [PubMed] [Google Scholar]
- Liu J, Magalhaes JV, Shaff J, Kochian LV. (2009) Aluminum-activated citrate and malate transporters from the MATE and ALMT families function independently to confer Arabidopsis aluminum tolerance. Plant J 57: 389–399 [DOI] [PubMed] [Google Scholar]
- López-Bucio J, Nieto-Jacobo MF, Ramírez-Rodríguez VV, Herrera-Estrella L. (2000) Organic acid metabolism in plants: from adaptive physiology to transgenic varieties for cultivation in extreme soils. Plant Sci 160: 1–13 [DOI] [PubMed] [Google Scholar]
- Meyer K, Leube MP, Grill E. (1994) A protein phosphatase 2C involved in ABA signal transduction in Arabidopsis thaliana. Science 264: 1452–1455 [DOI] [PubMed] [Google Scholar]
- Mitsuhara I, Ugaki M, Hirochika H, Ohshima M, Murakami T, Gotoh Y, Katayose Y, Nakamura S, Honkura R, Nishimiya S, et al. (1996) Efficient promoter cassettes for enhanced expression of foreign genes in dicotyledonous and monocotyledonous plants. Plant Cell Physiol 37: 49–59 [DOI] [PubMed] [Google Scholar]
- Moseyko N, Feldman LJ. (2001) Expression of pH-sensitive green fluorescent protein in Arabidopsis thaliana. Plant Cell Environ 24: 557–563 [DOI] [PubMed] [Google Scholar]
- Murashige T, Skoog F. (1962) A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol Plant 15: 473–497 [Google Scholar]
- Murphy AS, Eisinger WR, Shaff JE, Kochian LV, Taiz L. (1999) Early copper-induced leakage of K+ from Arabidopsis seedlings is mediated by ion channels and coupled to citrate efflux. Plant Physiol 121: 1375–1382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negi J, Matsuda O, Nagasawa T, Oba Y, Takahashi H, Kawai-Yamada M, Uchimiya H, Hashimoto M, Iba K. (2008) CO2 regulator SLAC1 and its homologues are essential for anion homeostasis in plant cells. Nature 452: 483–486 [DOI] [PubMed] [Google Scholar]
- Neumann G, Massonneau A, Martinoia E, Römheld V. (1999) Physiological adaptations to phosphorus deficiency during proteoid root development in white lupin. Planta 208: 373–382 [Google Scholar]
- Okushima Y, Fukaki H, Onoda M, Theologis A, Tasaka M. (2007) ARF7 and ARF19 regulate lateral root formation via direct activation of LBD/ASL genes in Arabidopsis. Plant Cell 19: 118–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okushima Y, Overvoorde PJ, Arima K, Alonso JM, Chan A, Chang C, Ecker JR, Hughes B, Lui A, Nguyen D, et al. (2005) Functional genomic analysis of the AUXIN RESPONSE FACTOR gene family members in Arabidopsis thaliana: unique and overlapping functions of ARF7 and ARF19. Plant Cell 17: 444–463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellet DM, Grunes DL, Kochian LV. (1995) Organic acid exudation as an aluminum-tolerance mechanism in maize (Zea mays L.). Planta 196: 788–795 [Google Scholar]
- Quint M, Gray WM. (2006) Auxin signaling. Curr Opin Plant Biol 9: 448–453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudrappa T, Czymmek KJ, Paré PW, Bais HP. (2008) Root-secreted malic acid recruits beneficial soil bacteria. Plant Physiol 148: 1547–1556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki T, Yamamoto Y, Ezaki B, Katsuhara M, Ahn SJ, Ryan PR, Delhaize E, Matsumoto H. (2004) A wheat gene encoding an aluminum-activated malate transporter. Plant J 37: 645–653 [DOI] [PubMed] [Google Scholar]
- Suzuki Y, Hibino T, Kawazu T, Wada T, Kihara T, Koyama H. (2003) Extraction of total RNA from leaves of Eucalyptus and other woody and herbaceous plants using sodium isoascorbate. Biotechniques 34: 988–990, 992–993 [DOI] [PubMed] [Google Scholar]
- Torres MA, Jones JD, Dangl JL. (2006) Reactive oxygen species signaling in response to pathogens. Plant Physiol 141: 373–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsutsui T, Yamaji N, Feng Ma J. (2011) Identification of a cis-acting element of ART1, a C2H2-type zinc-finger transcription factor for aluminum tolerance in rice. Plant Physiol 156: 925–931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umezawa T, Nakashima K, Miyakawa T, Kuromori T, Tanokura M, Shinozaki K, Yamaguchi-Shinozaki K. (2010) Molecular basis of the core regulatory network in ABA responses: sensing, signaling and transport. Plant Cell Physiol 51: 1821–1839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umezawa T, Sugiyama N, Mizoguchi M, Hayashi S, Myouga F, Yamaguchi-Shinozaki K, Ishihama Y, Hirayama T, Shinozaki K. (2009) Type 2C protein phosphatases directly regulate abscisic acid-activated protein kinases in Arabidopsis. Proc Natl Acad Sci USA 106: 17588–17593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vahisalu T, Kollist H, Wang YF, Nishimura N, Chan WY, Valerio G, Lamminmäki A, Brosché M, Moldau H, Desikan R, et al (2008) SLAC1 is required for plant guard cell S-type anion channel function in stomatal signalling. Nature 452: 487–491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong L, Wang RG, Mao G, Koczan JM. (2006) Identification of drought tolerance determinants by genetic analysis of root response to drought stress and abscisic acid. Plant Physiol 142: 1065–1074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaji N, Huang CF, Nagao S, Yano M, Sato Y, Nagamura Y, Ma JF. (2009) A zinc finger transcription factor ART1 regulates multiple genes implicated in aluminum tolerance in rice. Plant Cell 21: 3339–3349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Jia W, Yang J, Ismail AM. (2006) Role of ABA in integrating plant responses to drought and salt stresses. Field Crops Res 97: 111–119 [Google Scholar]