NBS-LRR genes, which constitute the major class of plant innate immune receptors, are massively degraded through microRNA-guided generation of secondary small interfering RNAs in several perennial species.
Abstract
Small RNAs (sRNAs), including microRNA (miRNA) and short-interfering RNA (siRNA), are important in the regulation of diverse biological processes. Comparative studies of sRNAs from plants have mainly focused on miRNA, even though they constitute a mere fraction of the total sRNA diversity. In this study, we report results from an in-depth analysis of the sRNA population from the conifer spruce (Picea abies) and compared the results with those of a range of plant species. The vast majority of sRNA sequences in spruce can be assigned to 21-nucleotide-long siRNA sequences, of which a large fraction originate from the degradation of transcribed sequences related to nucleotide-binding site-leucine-rich repeat-type resistance genes. Over 90% of all genes predicted to contain either a Toll/interleukin-1 receptor or nucleotide-binding site domain showed evidence of siRNA degradation. The data further suggest that this phased degradation of resistance-related genes is initiated from miRNA-guided cleavage, often by an abundant 22-nucleotide miRNA. Comparative analysis over a range of plant species revealed a huge variation in the abundance of this phenomenon. The process seemed to be virtually absent in several species, including Arabidopsis (Arabidopsis thaliana), rice (Oryza sativa), and nonvascular plants, while particularly high frequencies were observed in spruce, grape (Vitis vinifera), and poplar (Populus trichocarpa). This divergent pattern might reflect a mechanism to limit runaway transcription of these genes in species with rapidly expanding nucleotide-binding site-leucine-rich repeat gene families. Alternatively, it might reflect variation in a counter-counter defense mechanism between plant species.
Small RNAs (sRNAs) are abundant in plants, and a large portion of them have important roles in the regulation of diverse biological processes, including regulation of patterning and development, response to the environment, defense against pathogens, and silencing of endogenous transposable elements (Vaucheret, 2006; Chapman and Carrington, 2007; Chen, 2009; Ruiz-Ferrer and Voinnet, 2009; Voinnet, 2009). The common origin of all sRNAs is double-stranded RNA (dsRNA) that can be derived from a diverse set of sources, including transcription of inverted repeat structures, convergent transcription or virus replication, as well as transcription of genes containing a hairpin structure by RNA polymerase II (microRNA [miRNA]; Voinnet, 2009). dsRNA can also arise from the action of specific RNA-dependent RNA polymerases. The resulting dsRNA is cleaved into short RNA duplexes typically between 18 and 24 nucleotides (nt) long by a dicer-like protein (DCL). After cleavage, a specific strand is incorporated into an RNA-induced silencing complex (RISC) that, upon identification of a complementary sequence, might inactive the target by cleavage or translational repression of mRNA. Alternatively, RISCs can target DNA for methylation of DNA or histones (Ramachandran and Chen, 2008).
Numerous studies of both model and nonmodel plant species have revealed that the most abundant class of plant sRNAs are the short-interfering RNAs (siRNAs; Rajagopalan et al., 2006). These siRNAs mediate the silencing of both endogenous and exogenous genes. To date, the majority of endogenously produced siRNAs have been shown to act on transposable elements and repetitive DNA through DNA and chromatin modification (Ramachandran and Chen, 2008; Jin and Zhu, 2010). In angiosperms, these siRNAs are typically 24 nt long and are processed by a specific dicer protein, DCL3 (Xie et al., 2004). These 24-nt siRNAs are dependent on RNA-dependent RNA polymerase2 for their generation and on Argonaute4 for their function in DNA or histone methylation (Xie et al., 2004; Kasschau et al., 2007). This pathway also requires the plant-specific RNA polymerases Pol IV and Pol V (Haag and Pikaard, 2011). Pol IV has a major role to initiate the generating 24-nt siRNAs, which are guided to sites transcribed by Pol V.
The second most abundant class of sRNAs is miRNAs, which regulate target transcripts by cleavage or translational repression (Brodersen et al., 2008). miRNAs originate from DNA-encoded transcripts that form imperfect fold-back structures. miRNA genes produce primary miRNA transcripts that are cleaved, mainly by DCL1, to produce miRNA duplexes. Thereafter, one strand of the miRNA duplex is selectively incorporated into the RISC. Plant genomes typically contain hundreds of MIR genes (i.e. regions encoding primary miRNAs; Cuperus et al., 2011).
Besides these general and shared features of sRNA populations among plant species, there are also major differences, but as many of the available analysis tools to study sRNA rely on already identified sRNA sequences in other species, novelties in individual species have been, until recently, largely overlooked. Furthermore, we have a much better understanding of the biogenesis and function of specific groups of sRNAs (e.g. miRNAs), and as a result, the vast majority of sRNA studies report putative miRNA sequences and their gene targets and more seldom discuss other groups of sRNA sequences. In two recent studies of short RNAs from Medicago truncatula and tomato (Solanum lycopersicum; Zhai et al., 2011; Shivaprasad et al., 2012), several novel aspects were identified and revealed that a substantial fraction of the total population of sRNAs in these species can be classified as siRNAs originating from nucleotide-binding site (NBS)-Leu-rich repeat (LRR) genes. Similar findings have not been reported in the model plants Arabidopsis (Arabidopsis thaliana) and rice (Oryza sativa). Furthermore, these siRNAs were often observed in 21-nt spaced (phased) intervals and initiated through cleavage by 22-nt-long miRNAs, analogous to the generation of trans-acting short-interfering RNA (tasiRNA; Zhai et al., 2011). As NBS-LRR genes are key genes in plant defense systems, this observation has posed an intriguing hypothesis that such miRNAs could be part of a counter-counter defense system. Support for this hypothesis was recently found in a detailed study in tomato, where miRNA-mediated silencing of NBS-LRR genes was reduced in plants infected by viruses and bacteria (Shivaprasad et al., 2012). An alternative explanation could be that the mechanism evolved to constrain the expression of a rapidly expanding and evolving gene family in a similar fashion that has been proposed for the pentatricopeptide (PPR) gene family in Arabidopsis (Sunkar and Zhu, 2004; Cuperus et al., 2011). These differences in siRNA-mediated degradation of NBS-LRR genes between plant species are intriguing and warrant further studies of plant sRNAs in additional species.
In this study, we report a detailed characterization of the sRNA population from the gymnosperm species spruce (Picea abies). In line with previous reports from Pinaceae spp., we found a very low abundance of 24-nt sRNAs (Dolgosheina et al., 2008; Morin et al., 2008), and the most abundant group of sRNAs in spruce is 21 nt long. Among these 21-nt sRNAs, we could identify and validate several miRNA sequences, but the majority of spruce 21-nt sRNAs could be characterized as siRNAs. More than 25% of all 21-nt short reads collected originated from phased degradation of NBS-LRR genes. In an attempt to better understand this massive production of short reads from NBS-LRR genes, we broadened our study and reanalyzed short-read data from a diverse set of plant species with an emphasis on reads that originate from this type of gene.
RESULTS
Spruce sRNAs
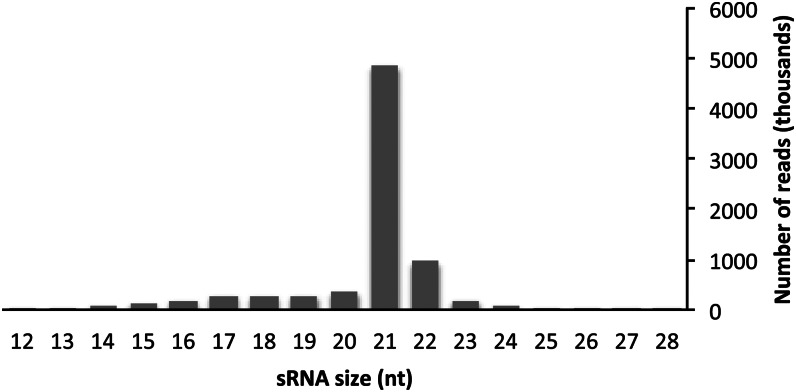
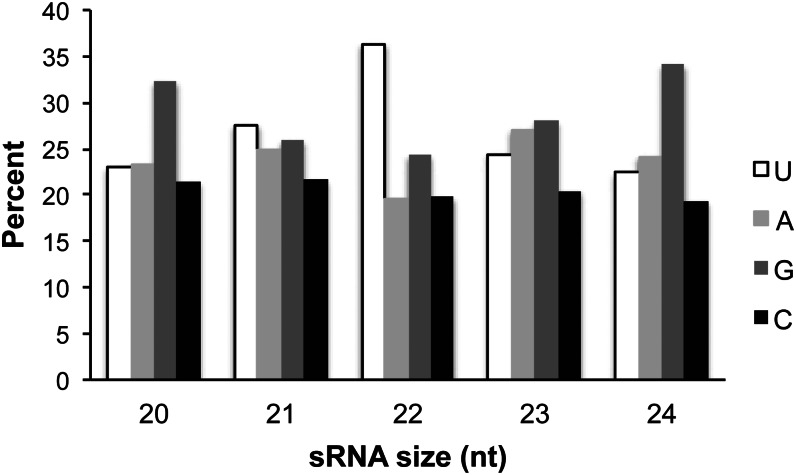
sRNA was isolated from newly flushed buds of spruce, and more than 10 million sequence reads were generated from the resulting libraries. After quality filtering, 8.3 million reads were retained for further analysis. Constraining the reads to a length between 18 and 29 nt (7 million reads) and collapsing them into unique sequences yielded 59,616 entities. The copy number of each sequence was highly variable, and the most abundant sequence occurred in 459,908 copies, and 1,160,718 sequences occurred less than 10 times. The clearly most abundant size class was 21 nt, constituting 72% of all reads with a size between 18 and 24 nt (Fig. 1). The 22-nt class was the second most abundant class (12% of the reads), while the 24-nt class constituted only 1%. The 5′-terminal nucleotide is important for sorting of sRNAs to Argonaute complexes in angiosperm plants. In Arabidopsis, the majority of miRNAs possess a 5′-uridine, which directs those to Argonaute1 (Mi et al., 2008). Among all spruce sRNA sequences, no bias for a particular 5′ nucleotide was evident. Calculating the percentage of each 5′-terminal nucleotide for sRNAs of different lengths indicated an overrepresentation of uridine for 22-nt sRNAs only (36% of all 22-nt sRNAs start with a 5′-uridine; Fig. 2).
Figure 1.
Observed length distribution of sRNA sequences from spruce. The total number of reads in each size class is plotted.
Figure 2.
Percentage of each 5′-terminal nucleotide of spruce sRNA sequence. Estimates were calculated separately for sRNAs of different length (20–24 nt).
The 24-nt siRNAs are known to guide the DNA methylation and heterochromatin formation of repetitive and transposable elements in angiosperms (Herr, 2005; Morin et al., 2008). As such, 24-nt siRNAs seems to be rare or absent in spruce, and we wanted to investigate to what extent other sizes of Pinaceae spp. sRNAs might target repetitive DNA, so we mapped spruce sRNAs to four spruce and two Picea glauca bacterial artificial chromosomes (BACs; in total, 657 kb; De Paoli 2006; Hamberger et al., 2009). Allowing one mismatch and multiple hits resulted in 1% of the sRNA sequences mapping to these references. We also mapped Pinus contorta sRNAs to genome sequence data from 10 annotated Pinus taeda BACs. Despite an estimated high repeat content (Kovach et al., 2010), a low number of sRNAs mapped to the genomic sequence of the 10 BACs (in total, 1 Mb). Mapping 134,580 sRNAs and allowing for two mismatches resulted in only 0.5% of the reads mapping, and no clustering of reads on repetitive elements was discernible. These results suggest that the role of 24-nt sRNAs in angiosperms, to silence repetitive and transposable elements, is not to a large extent performed by other types of sRNAs in Pinaceae spp.
Spruce miRNAs
The 59,616 unique sRNA sequences and 166,685 Picea spp. EST clusters were used to identify potential miRNA and miRNA* and their potential precursor sequences. This identified a large number of potential miRNA/miRNA* that, based on sequence similarity, could be grouped into 137 families. Fifty of these showed high similarity (mismatches of 2 bp or less) to the plant miRNAs in miRBase (Kozomara and Griffiths-Jones, 2011), including 26 miRNAs previously reported for spruce by Yakovlev et al. (2010). Among the 87 novel putative families, reads corresponding to both the miRNA and miRNA* sequences were identified in our short-read data for 17 of them (Supplemental Table S1). While the vast majority of miRNA families also identified in angiosperms were associated with a most abundant read of 21 nt in our data (17 out of 18), a larger fraction of those identified previously only in Pinaceae spp. were associated with a most abundant read of 22 nt (18 out of 33).
Target prediction for the 137 potential miRNA/miRNA* sequences identified a large number of putative targets (Supplemental Table S1). Without additional data, these predictions should not be taken as strong evidence of true targets, but we noted that in almost one-third of the miRNA families with a predicted target, at least one of the targets is annotated as a resistance gene. This targeting of resistance genes is even more pronounced in the families that seem specific to Pinaceae, whereas we found no example of resistance targets among the families that are shared between angiosperms and gymnosperms. Furthermore, among the predicted 87 novel miRNAs identified in this study, 22 were predicted to target resistance genes (Supplemental Table S1).
Degradation of NBS-LRR Genes Is a Main Source of sRNAs in Spruce
To further characterize the sRNA sequences, the collapsed reads were mapped to the Picea spp. EST clusters from the Gene Index Project (Spruce Gene Index [SGI]; in total, 79,409 clusters). Allowing for multiple hits and one mismatch resulted in mapping of 57% of all sRNA reads (39% of all unique sRNA sequences) to 48,242 clusters. Out of those clusters, 14,231 contained a single mapped sRNA sequence, while 10 or more different sRNA sequences mapped to each of 11,189 clusters (Supplemental Table S2).
As the majority of EST clusters are likely to code for proteins, a surprisingly large number of EST clusters were characterized by a substantial number of reads mapping to each of them. The maximum number of unique sequences mapping to an individual EST cluster was 33,862, which corresponds to 544,261 reads including redundant identical sequences. The mean and median numbers of reads (including redundant ones) per EST cluster were 468 and three, respectively. As we allowed for multiple hits, these numbers do not truly reflect the number of reads originating from each EST cluster but at least suggest that a multitude of sRNA sequences originate from each of a substantial number of different genes corresponding to these EST clusters. To obtain a rough minimum estimate of the number of genes spawning these massive numbers of sRNAs, we aligned the sRNA sequences without mismatch and retained only reads with a single hit in the reference. With these parameters, 11,806 EST clusters from the SGI Picea spp. gene catalog obtained at least one hit, and 1,605 of those were hit by 10 or more sequences. The pattern seen for sRNAs from newly flushed buds was further compared with data from three other tissues (needles, immature female buds, and lateral bud meristem), and the correlation of the number of mapped reads per EST cluster was in all cases above 0.9 (data not shown), showing that the observed pattern is not specific for flushing buds.
Examining the annotation of the EST clusters to which high numbers of sRNA reads mapped identified a large proportion of genes annotated as NBS-LRR resistance genes. Domain prediction and comparison with known conifer NBS-LRR genes further supported a very large number of them as putative NBS-LRR sequences (Supplemental Table S2). Out of the 100 EST clusters with the highest number of different sRNA sequences mapping to them, 73 were classified as NBS-LRR sequences, and of the 4.7 million reads that in total mapped to SGI Picea spp. EST clusters, 2.1 million (44%) mapped to clusters for which the sequence showed high similarity to NBS-LRR genes (Supplemental Table S2). Restricting sRNA reads to those with a length of 21 nt reinforced this observation further: 90 out of the 100 genes with highest read counts were classified as NBS-LRR-type sequences, and out of 3.3 million 21-nt reads that mapped to SGI EST clusters, 1.6 million (48%) mapped to clusters classified as NBS-LRR-type sequences.
Figure 3 shows the distribution of the number of 21-nt reads per EST cluster, illustrating the preponderance of NBS-LRR type among those with high read counts. As NBS-LRR genes constitute around 1% of all EST clusters, these data reveal a strong enrichment of NBS-LRR genes among those with many sRNA reads mapping to them.
Figure 3.
Distribution of the number of sRNA reads mapping to individual EST clusters. The x axis shows log2-transformed length-normalized read counts, where the numbers of sRNA reads mapping to each EST cluster are normalized by dividing by the length of the cluster and multiplying by the average read length (21 nt). Mean and median of nontransformed data for the two classes of genes are 4.4 and 0.07 for non-NBS-LRR genes and 241 and 10.0 for NBS-LRR genes, respectively.
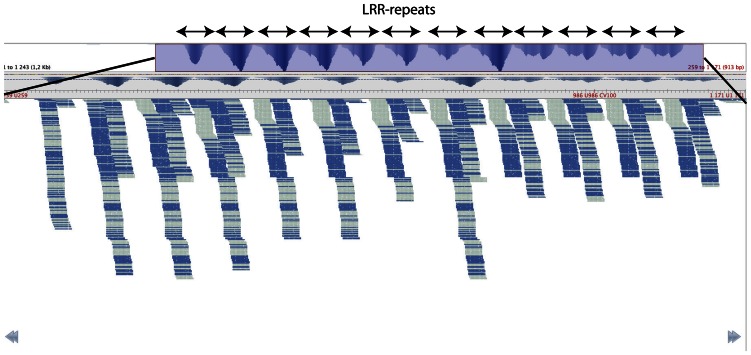
As many of the EST clusters in our reference are most likely not full length, we also mapped our sRNA reads to the newly released P. glauca gene catalog (which contains more than 23,000 mRNA sequences that are annotated as full length; Rigault et al., 2011) and a full-length Toll/interleukin-1 receptor (TIR)-NBS-LRR gene from Picea sitchensis (ACN49932; Liu and Ekramoddoullah, 2011). Mapping also to these references confirms the observed pattern, and genes with similarity to NBS-LRR-type resistance genes typically had unique reads mapping to them at multiple positions. An example is shown in Figure 4, where the EST cluster from the P. glauca gene catalog with the largest number of mapped unique reads is shown. In total, 3,547 different sRNA sequences mapped to this cluster, and on average, each sequence mapped seven times with a range from one to 12. This pattern is partly explained by the highly repeated structure of the reference sequence, with 12 tandem repeats of 72 nt in length potentially coding for LRRs (Fig. 4).
Figure 4.
Coverage plot of sRNA reads mapping to TC142020. Allowing for one mismatch and multiple hits resulted in 3,547 unique sRNA sequences mapping to this EST cluster. Each sRNA sequence is represented by one read. The top panel shows a coverage plot of the complete sequence, and the blue shaded area is shown in the bottom panel displaying individual reads. Read orientation is depicted by light or dark blue color in the bottom panel. The plot was produced with Tablet (Milne et al., 2010).
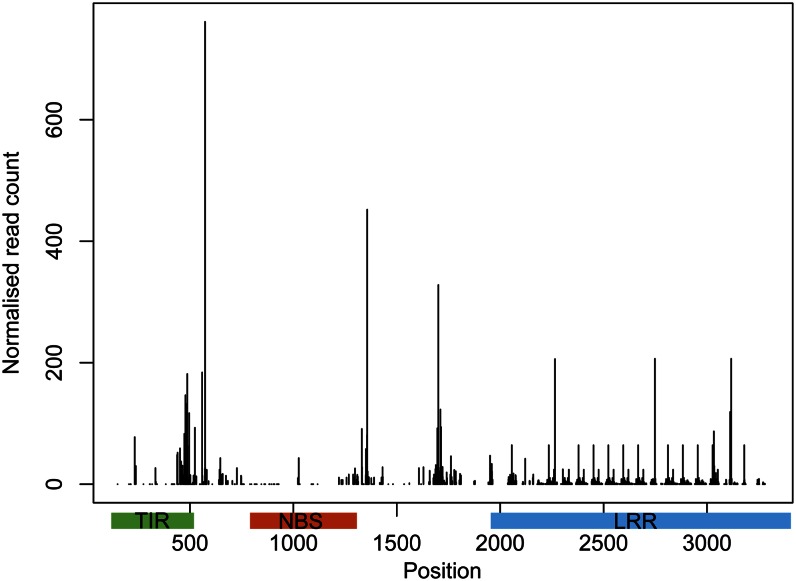
Figure 5 shows the pattern observed for the P. sitchensis full-length TIR-NBS-LRR gene (after compensation for the fact that many reads map to multiple positions by dividing the short read counts by the number of places they mapped to). The obtained coverage distribution suggests that the sRNA sequences are derived from a large portion of the transcript, but some areas might spawn more sRNAs than others.
Figure 5.
Coverage plot of sRNA reads mapped to a full-length spruce TIR-NBS-LRR gene (ACN40032). Normalized read count is the number of reads mapping divided by the number of matches for each read in the whole reference. [See online article for color version of this figure.]
The high numbers of sRNAs mapping to NBS-LRR-type genes suggest that such genes are generally more subjected to siRNA degradation. If this were the case, we might expect low levels of intact mRNA from such genes. To compare estimates of mRNA levels, we mapped six Illumina Digital Gene Expression (DGE) libraries to the SGI EST clusters (Supplemental Table S2). Clusters with more than 100 mapped sRNA sequences had a mean of 93 DGE tags per 1,000 bp, while those with fewer than 100 sRNA sequences had a mean of 11,931 DGE tags per 1,000 bp. Thus, in general, abundant sRNA mapping is associated with low mRNA levels. For genes with similarity to NBS-LRR-type resistance genes, the corresponding numbers were 91 and 313 DGE tags per 1,000 bp for those with more and fewer than 100 mapped sRNA reads, respectively. Thus, NBS-LRR genes seem to have generally low mRNA levels, but those spawning high numbers of sRNAs do not seem to show strikingly lower mRNA levels.
The reports on phased degradation of NBS-LRR genes in angiosperms prompted us to search for phased 21-nt siRNAs originating from spruce NBS-LRR loci, applying the phasing algorithm used by Zhai et al. (2011). A total of 208 loci had a phase score larger than 15, and more than 50% of these loci contained either NBS or LRR domains, and another significant portion lacked a significant BLASTX hit against protein databases, some of which might constitute tasiRNA loci (Supplemental Table S2).
As repetitive structures (e.g. LRRs) in the reference sequence might obscure phasing signals, we also focused on genes comprising TIR or NBS but no LRR domains and with more than 100 unique sequences mapping to them. For these, the average phase scores were 15 and 12, respectively, compared with the average of nine observed for all genes (with more than 100 unique sRNA sequences mapping to them; Supplemental Table S2).
A few highly abundant 22-nt miRNA families that triggered siRNA generation from multiple NBS-LRR genes were identified in Medicago and Solanaceae spp. (Zhai et al., 2011; Li et al., 2012; Shivaprasad et al., 2012). Many of these belong to the related families miR482 and miR2118. Family miR482 was identified also in spruce, where sRNA sequences matching both miR482 and miR2118 were identified and were by our pipeline included in the miR482 family. Of the 38 predicted target ESTs for this family, only two were annotated as NBS-LRR genes (Supplemental Table S1).
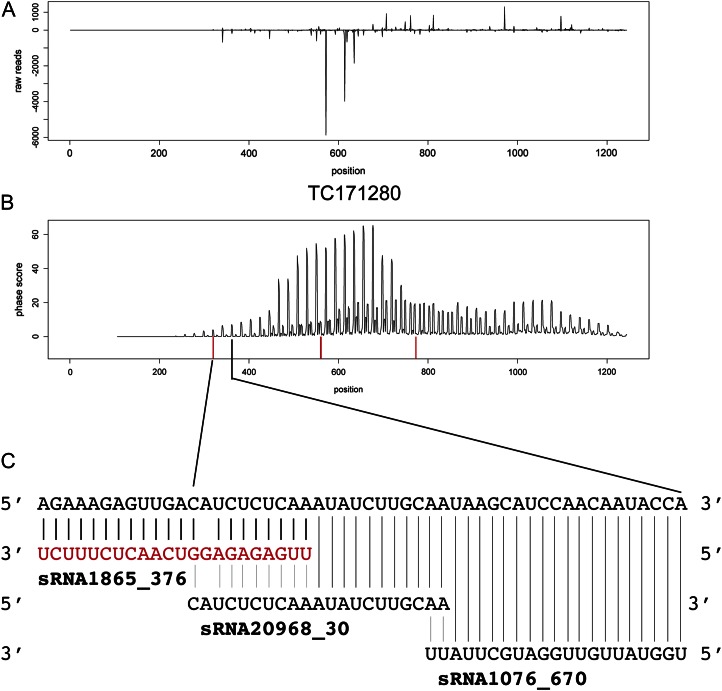
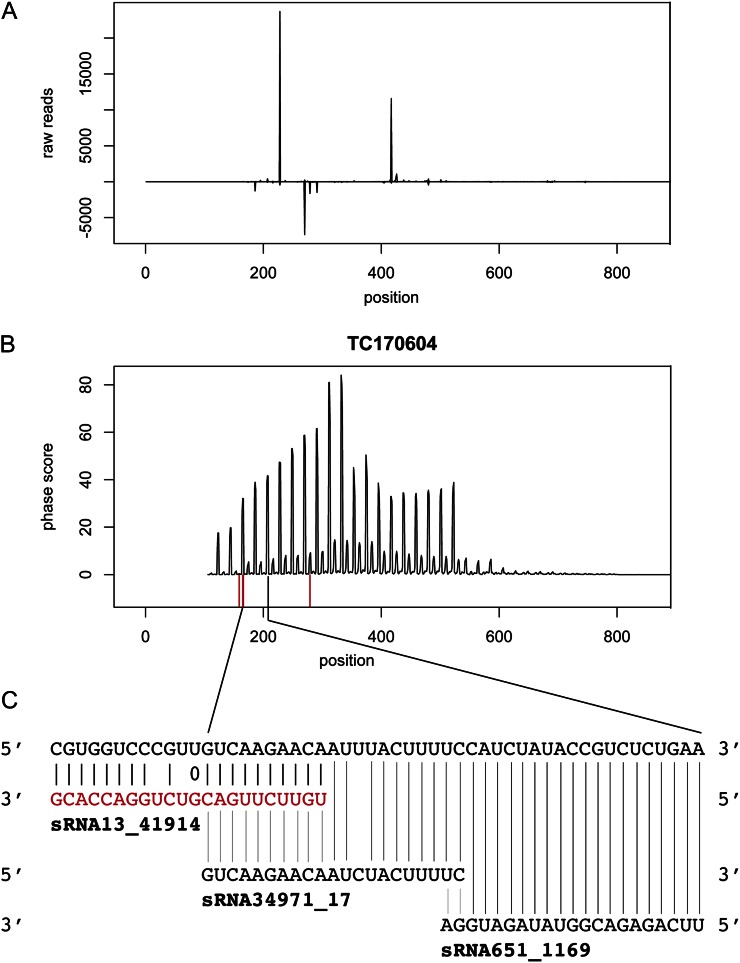
Focusing on genes with TIR or NBS domains, several examples of genes with a significant phase score and 22-nt sRNAs mapping in positions consistent with them triggering phased 21-nt sRNAs were found. Figure 6 shows data from TC171280, which is predicted to contain an NBS domain but no LRRs (Supplemental Table S2). A high phase score was observed with the major phase, consistent with the mapping of 22-nt sRNA1865. Additional 22-nt sRNAs map to downstream positions that indicate the triggering of phased 21-nt sRNAs in alternative phases. Figure 7 shows data for transcript TC170604, which is predicted to contain a TIR domain but no NBS or LRR domains (Supplemental Table S2). Again, a 22-nt sRNA, sRNA13, maps in a position consistent with the major phase for 21-nt sRNAs.
Figure 6.
Phasing pattern observed after mapping spruce 21-nt sRNAs to TC171280. A, sRNA abundance plot (reads mapping in forward and reverse orientation are plotted above and below the horizontal line, respectively). B, Phasing score distribution is plotted above the horizontal line, and predicted cleavage positions for 22-nt miRNAs are shown in red below. C, A magnification around the predicted miRNA cleavage sites, with the predicted 22-nt miRNA in red and two phased 21-nt sRNAs in black. [See online article for color version of this figure.]
Figure 7.
Phasing pattern observed after mapping spruce 21-nt sRNAs to TC170604. A, sRNA abundance plot (reads mapping in forward and reverse orientation are plotted above and below the horizontal line, respectively). B, Phasing score distribution is plotted above the horizontal line, and predicted cleavage positions for 22-nt miRNAs are shown in red below. C, A magnification around the predicted miRNA cleavage sites, with the predicted 22-nt miRNA in red and two phased 21-nt sRNAs in black. [See online article for color version of this figure.]
Among the 22-nt sRNA sequences, a clear majority seem to be specific to Picea spp., and of the 52 most abundant 22-nt sRNA sequences (those represented by more than 1,000 reads), 19 were predicted by our stringent pipeline to be miRNAs. Furthermore, 20 of them mapped to one or more EST clusters classified as NBS-LRR sequences (in the expected orientation, and with a maximum of three mismatches). Among the 10 most abundant 22-nt sRNAs, seven were predicted miRNAs and nine were predicted to target NBS-LRR-like sequences (Table I). Out of those nine, six target regions were predicted to code for TIR domains, whereas none were predicted to target an NBS domain. Another set (14) of the 52 most abundant 22-nt sRNA sequences mapped to EST clusters that displayed high phase scores but were not classified as NBS-LRR sequences. Only one of those showed significant homology to plant proteins, and the rest might thus represent tasiRNAs.
Table I. Ten most abundant 22-nt sRNAs in spruce.
| sRNA | Reads | Target 1 | Phmaxa | Target 2 | Phmaxa | NBS-LRRb |
|---|---|---|---|---|---|---|
| 2* | 148,890 | TC161536 | 7.3 | TC130897 | 6.1 | 3 |
| 3* | 133,356 | TC139913 | 25.1 | CO219226 | 19.4 | 14 |
| 5* | 62,691 | TC143748 | 23.8 | 1 | ||
| 7 | 55,848 | TC143748 | 23.8 | CO234506 | 15.3 | 8 |
| 9 | 53,201 | TC143748 | 23.8 | CO234506 | 15.3 | 7 |
| 13* | 41,914 | DR540978 | 45.7 | TC143748 | 23.8 | 4 |
| 14* | 40,208 | TC128174 | 35.4 | TC167323 | 21.2 | 0 |
| 25 | 25,571 | TC137450 | 6.6 | 1 | ||
| 35* | 20,410 | TC160176 | 13.7 | TC158558 | 4.9 | 3 |
| 41* | 17,814 | TC143748 | 23.8 | TC161536 | 7.3 | 9 |
As one particular TAS gene (TAS3) has been show to be conserved from mosses to angiosperms, we specifically searched for evidence of a conserved miR390-TAS3-ARF pathway in spruce. Among the 14 predicted targets of spruce miR390, one EST cluster displayed a high phase score suggesting siRNA generation (TC161203; Supplemental Table S2). Alignment of TC161203 with TAS3 sequences from Arabidopsis and tobacco (Nicotiana tabacum) revealed a conservation of two miR390 target sites flanking a conserved area of predicted tasiRNA production (Fig. 8). This pattern has previously been reported as strong evidence for conservation of the miR390-TAS3-ARF pathway in multiple plant species, including P. taeda (Axtell et al., 2006; Krasnikova et al., 2009).
Figure 8.

Nucleotide sequence alignment of TAS3 loci from Arabidopsis and tobacco and a putative TAS3 locus from spruce (TC161203). Boxes indicate miR390 complementary sites and predicted tasiARF-encoding sequence regions. Shading shows residues identical for each given position. GenBank accession numbers of the sequences included in the alignment are as follows: Arabidopsis, BX838290; tobacco, FJ804751.
Amounts of sRNA Originating from NBS-LRR Genes Vary Largely between Species
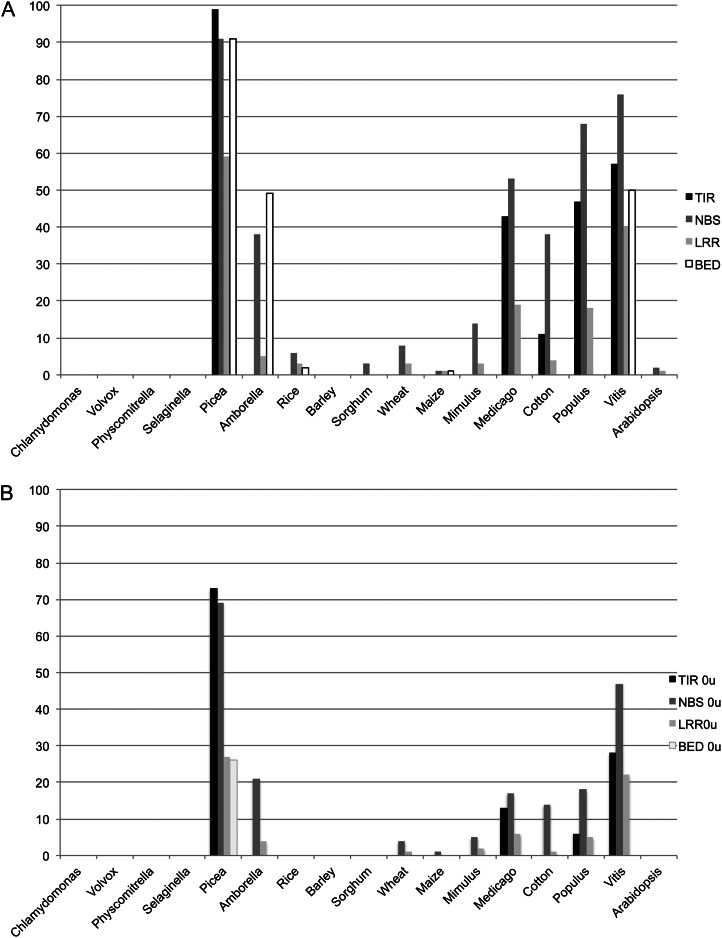
The high abundance of 21-nt sRNAs mapping to NBS-LRR genes in spruce, and recent reports of similar findings in Medicago and Solanum spp. (Zhai et al., 2011; Shivaprasad et al., 2012), prompted us to survey this phenomenon in additional plant species. To facilitate comparisons across species, we first chose to map 21-nt sRNA data from 16 plant species to databases derived from the Gene Index Project. Focusing on NBS-LRR-type resistance genes (i.e. those EST clusters containing one or more of the following domains: TIR, BED, NBS, LRR), the patterns were highly variable across species. When allowing for one mismatch and multiple hits for each sRNA, around 90% of all spruce genes containing TIR, NBS, or BED domains were hit by more than 10 unique sRNA reads. Focusing on NBS domain-containing genes, high percentages were also obtained in Amborella, Medicago, Gossypium, Populus, and Vitis spp., while low percentages of gene hits by 21-nt sRNAs were generally found in monocots and nonangiosperm species (except Picea spp.; Fig. 9A). Excluding Picea spp., the highest percentages were obtained for the NBS domain, followed by TIR in most species. It should be noted that NBS-LRR genes with TIR domains are rare in monocots (Tarr and Alexander, 2009). Mapping 21-nt reads with zero mismatches and retaining only unique hits resulted in a similar pattern across species, albeit a generally lower percentage was observed (Fig. 9).
Figure 9.
Percentage of NBS-LRR-type genes hit by more than 10 unique sRNA reads in different plant species. For each species, genes that contained any of TIR, NBS, LRR, or BED domains were selected, and for each class, the percentage of those genes that were hit by more than 10 different unique 21-nt sRNA sequences was calculated. In the top graph, sRNA reads were mapped allowing one mismatch, and reads mapping at multiple positions in the whole reference were retained. In the bottom graph, mapping was performed with only perfect matches and discarding reads mapping to multiple positions.
To further compare the sRNA populations of different species, we used sRNA data from four angiosperm species for which genome sequence data are available, two perennials (poplar [Populus trichocarpa] and grape [Vitis vinifera]) and two annuals (Arabidopsis and rice).
Arabidopsis
For the Arabidopsis data, containing close to 300,000 unique sRNA sequences, 36% mapped to 11,859 complementary DNA (cDNA) models in The Arabidopsis Information Resource. A single sRNA mapped to 4,040 cDNA models, while 4,356 models spawned more than 10 different sRNA sequences. The maximum number of unique sRNA sequences mapping to an individual cDNA model was 8,157. Among the 500 annotated cDNA models with the highest read counts, 489 were classified as transposable element genes, while none of those cDNA models were annotated as any type of resistance gene. Mapping only sRNAs of 21 nt still resulted in a predominance of transposable elements among cDNA models with high sRNA read counts (Supplemental Table S3). In addition, a few PPR and tetratricopeptide repeat proteins as well as TAS genes were identified with high 21-nt sRNA read counts. Calculating the ratio of 21- to 24-nt reads mapping to each cDNA model identified PPRs and tetratricopeptide repeats as those with the highest ratios (Supplemental Fig. S1; Supplemental Table S3). High ratios were also typically obtained for TAS genes and miRNA genes. This group also contained a single TIR-NBS-LRR class disease resistance gene.
Rice
Seventy-five percent of 1,751,364 reads representing 31,474 unique sRNA sequences originating from rice seedlings mapped to rice transcripts (V6.1). The maximum number of unique sRNA sequences mapping to a single transcript was 6,424, and 5,502 transcripts spawned more than 10 unique sRNA sequences. Although many transcripts were annotated as ordinary protein-coding genes, a vast majority of transcripts spawning high numbers of sRNAs showed similarity to sequences in The Institute for Genomic Research Oryza Repeat Database (Supplemental Table S4). The clearly most prominent type of repeat identified was miniature inverted-repeat transposable element (171 out of 200 with the highest read counts; Supplemental Table S4). This suggests that many putative protein-coding genes contain short miniature inverted-repeat transposable elements that spawn high numbers of sRNAs. Additional annotations among transcripts spawning high numbers of sRNAs included ribosomal DNA-like sequences and retrotransposons. For transcripts with high read counts, the ratio of 21- to 24-nt sRNAs was close to 1 (0.89 for transcripts with more than 100 sRNA sequences mapping). The few NBS-LRR types with a significant number of sRNAs mapping (e.g. LOC_Os11g12330; Supplemental Table S4) also show similarity to sequences in The Institute for Genomic Research Oryza Repeat Database, and 24-nt sRNAs generally dominated over 21-nt sRNAs at these loci.
Poplar
Mapping 2.1 million sRNA sequences from xylem tissue to poplar transcripts (version 2.0) allowing one mismatch and multiple hits resulted in 10% mapped reads (27% of all 21-nt sRNAs and 5% of all 24-nt sRNAs). A single sRNA read mapped to 8,723 transcripts, 4,452 transcripts spawned more than 10 sRNA reads, and the maximum number of reads mapped to a single transcript was 17,157. Among the top-ranking genes were those with annotations relating to NBS-LRR resistance genes and retroelements (e.g. ORF V protein, AP endonuclease/reverse transcriptase, non-LTR retroelement reverse transcriptase, polyprotein, etc.; Supplemental Table S5). For 21-nt sRNAs, there was a predominance of NBS-LRR-type resistance genes, while for 24-nt sRNAs, a majority of genes were annotated as transposable elements. In particular, sorting the genes on the ratio of 21- to 24-nt sRNAs mapping to them, the vast majority of genes with high ratios were NBS-LRR-type resistance genes (among the top 100 genes, 91 were annotated as NBS-LRR resistance genes). Table II shows the ratio of 21- to 24-nt sRNAs mapping to the different categories of genes that were found among those with high read counts. Except for NBS-LRR resistance genes, a couple of other gene classes (GRAS family transcription factors and xylem Ser proteinase1) showed high ratios, while those with annotation relating to transposable elements showed ratios below 1.
Table II. Ratio of 21-nt sRNAs to 24-nt sRNAs for selected gene classes in poplar.
The ratio was calculated for those annotation classes containing five or more genes among those 500 with the highest sRNA counts.
| Annotation | No. of Genes | Ratio | Total Reads |
|---|---|---|---|
| NBS-LRR resistance protein | 512 | 15.8 | 183,882 |
| Unknown function | 472 | 1.9 | 74,508 |
| Reverse transcriptase | 64 | 0.2 | 15,173 |
| Polyprotein | 21 | 0.5 | 10,647 |
| ORF V protein | 5 | 0.4 | 7,429 |
| Retroelement | 14 | 0.4 | 6,796 |
| GRAS family transcription factor | 22 | 16.6 | 2,987 |
| Retrotransposon | 11 | 0.3 | 1,590 |
| Xylem Ser proteinase1 | 5 | 12.9 | 1,006 |
To study the genomic sRNA distribution, the 21- and 24-nt sRNA reads were also mapped to scaffold 1 of the poplar genome version 2.0 (length of 48.4 Mb). For both types of sRNAs, 27% mapped to scaffold 1. In general most reads mapped to intergenic regions (93% of 21-nt sRNAs and 97% of 24-nt sRNAs), and distributions for 21- and 24-nt sRNAs were largely similar (Supplemental Fig. S2). Focusing on NBS-LRR-type resistance genes, the pattern observed for mapping sRNA to transcripts was evident also when mapping to scaffold 1. Supplemental Figure S3 shows a region with a cluster of resistance genes, where a clear pattern of 21-nt sRNAs as opposed to 24-nt sRNAs mapping to those genes is observed. This cluster also contains a gene annotated as a transposase (POPTR_0001s42600.1) for which the opposite pattern is observed, with mainly 24-nt sRNAs mapping. For the NBS-LRR resistance genes, the majority of 21-nt sRNAs mapped to exons, indicating that most sRNAs are derived from processed transcripts (Supplemental Fig. S4).
Grape
sRNAs isolated from leaf and stem, constituting 114,688 unique sequences, were mapped against the grape mRNA (version 2). Allowing one mismatch and multiple hits resulted in 10% mapped sRNA sequences (17% of 21-nt sRNAs and 4% of 24-nt sRNAs). A single sRNA read mapped to 3,887 transcripts, 552 transcripts spawned more than 10 sRNA sequences, and the maximum number of sequences mapped to a single transcript was 919. The majority of mRNA sequences with large numbers of mapping sRNAs showed similarity to NBS-LRR resistance genes. Among the top 200 genes with the highest number of sRNAs mapping, at least 121 (60%) were identified as NBS-LRR-type resistance genes (Supplemental Table S6). In contrast, among those with from one to 10 hits, 74 out of 6,671 were identified as NBS-LRR-type resistance genes.
DISCUSSION
Studies of sRNAs in plants in general, and conifers in particular, have focused on the identification of miRNA sequences. In this study, we aimed at a more general characterization of sRNAs in spruce. To set the data into perspective, we also analyzed some aspects of sRNA patterns in other plant species. The most striking result relates to the very large proportion of sRNAs that are derived from NBS-LRR resistance gene sequences in spruce, a pattern that to a lesser extent is observed only in some other plant species.
In contrast to most angiosperm species, the vast majority of sRNAs were 21 nt long in our spruce sRNA libraries, and the frequency of 24-nt sRNAs was very low (estimated at 1% in spruce). This pattern is consistent with data from several other Pinaceae conifer spp.: Araucaria araucana, P. glauca, P. contorta, Pinus strobus, Pseudotsuga menziesii, and Thuja plicata (Dolgosheina et al., 2008), and has also been observed previously in spruce (Yakovlev et al., 2010), suggesting that it might be general for conifers in the Pinaceae family. Concordant with these findings, Dolgosheina et al. (2008) found no evidence for a DCL3 enzyme in this group of conifers, which in angiosperms is crucial for the production of 24-nt sRNAs. Until a comprehensive survey of siRNAs in multiple tissues is available, it cannot be ruled out that the DLC3-dependent 24 nt are present in higher abundance in some tissues also in species from the Pinaceae family. This study included actively growing needles, dormant vegetative buds, and female reproductive structures at an early stage of development. It is conceivable that 24-nt siRNAs are produced during later stages of reproductive development in spruce, but the frequency observed in vegetative tissues is clearly lower than that in the angiosperms studied so far.
The apparent lack of DCL3 and the low frequency of 24-nt sRNAs are not features common to all conifers, as it was recently shown that Cunninghamia lanceolata has an sRNA distribution more similar to angiosperm species, including a large fraction of 24-nt sRNAs (Wan et al., 2012). They also reported the presence of sequences predicted to code for proteins necessary for the biogenesis of 24-nt sRNAs, including DCL3.
An intriguing question stemming from the apparent lack of a pathway producing 24-nt siRNAs in Pinaceae spp. is whether the role of 24-nt sRNAs, in methylation and the silencing of transposable elements, is conducted by some other sRNA species or perhaps by an alternative mechanism. As species in the Pinaceae family tend to have exceptionally large genomes compared with both angiosperms and other gymnosperm families (mean 1C in Pinaceae = 23.76 pg, Cypressaceae = 12.43 pg, and Taxaceae = 11.05 pg, while that in angiosperms = 5.94 pg), it has been speculated that the large size may in part be explained by less efficient mechanisms for the silencing of transposable elements (Dolgosheina et al., 2008; Burleigh et al., 2012). Only 1% of the short reads obtained from spruce mapped to the six available BAC sequences from Picea spp. In an attempt to compare this estimate with a species with a high abundance of 24-nt sRNAs mainly associated with repetitive DNA (poplar; Klevebring et al., 2009), we mapped poplar sRNAs against scaffold 1 of the poplar genome assembly (48.4 Mb). Dividing the reference in 74 windows each with a size corresponding to the sum of the six Picea spp. BACs, we obtained an average of 2% of sRNAs mapping per window. The estimate of 1% obtained with Picea spp. BACs lies in the lower 10% of the distribution obtained in poplar, indicating a somewhat, but not substantially, lower frequency of sRNAs mapping to genomic DNA. As the majority of the analyzed Picea spp. BACs contain repetitive DNA (mainly retroelements; De Paoli, 2006; Hamberger et al., 2009), the estimated fraction of sRNAs mapping to these regions seems to be low. However, it is clear that more data including a reference genome are needed to test if mechanisms involving sRNAs are important in the silencing of retroelements in conifers. As most retroelements in conifers seem to be old (De Paoli, 2006), the majority are likely silenced by mutations, thus reducing the need for a mechanism silencing their transcription. However, based on comparisons of repetitive elements between Taxodium distichum and P. taeda (Liu et al., 2011; Magbanua et al., 2011), it was suggested that Pinaceae spp. might contain more recently expanded repetitive element families, which in turn could explain part of the larger genome sizes observed in Pinaceae spp. Thus, a less efficient siRNA-guided silencing of transposable elements in Pinaceae spp. seems plausible.
miRNAs in Spruce
The identification of novel miRNAs in spruce and their targets was not the main focus of this study, and we can conclude that the vast majority of 21-nt sRNAs in spruce can be classified as siRNAs and that only a very small part of the spruce 21-nt sRNA population is composed of miRNAs. The exceptionally large fraction of 21-nt siRNAs complicates the identification and analysis of miRNAs and their targets. Still, a number of the novel miRNA families have support by both the large number of reads and the presence of both miRNA and miRNA* sequences and a potential precursor, suggesting that they are true miRNAs.
We found that seven of the predicted miRNA families had predicted target genes with high sequence similarities to genes with characterized functions in other plant species: MIR156, MIR160, MIR164, MIR166, MIR172, MIR395, and MIR858 (Rhoades et al., 2002; Takuno and Innan, 2011; Khraiwesh et al., 2012). Even if our data do not allow us to conclusively identify correct targets of the identified miRNA families, we identified a substantial number of putative miRNA families that may target resistance genes of the NBS-LRR class (Supplemental Table S1). In particular, several predicted highly abundant conifer-specific 22-nt miRNAs were identified that are likely to trigger siRNA production from the target loci (see below).
Generation of siRNAs from NBS-LRR Genes in Spruce
We found that a surprisingly high portion of sRNAs in spruce are derived from NBS-LRR-type resistance genes. Out of the close to 6 million 21-nt sRNA reads included in the analysis, 1.6 million (27%) mapped to sequences related to NBS-LRR genes, and these reads constitute 48% of all reads mapping to transcribed sequences. Considering the small fraction of the transcriptome that these NBS-LRR sequences represent, these estimates are compelling. Recent studies have highlighted a new role for miRNAs and siRNAs in plant defense (Zhai et al., 2011; Li et al., 2012; Shivaprasad et al., 2012). The 22-nt miRNAs specifically targeting NBS-LRR genes trigger siRNAs at these loci, indicating an important role in the regulation of resistance. In this study, we found evidence that this mechanism is a main source generating 21-nt sRNAs from spruce NBS-LRR genes.
Although high phase scores that indicate secondary siRNA production in a phased pattern were rare in our data, we identified several loci with high phase scores and with a 22-nt sRNA mapping in a position consistent with it acting as a trigger of the phased 21-nt sRNA. A generally low frequency of significant phase scores could, even in the presence of phased degradation, be explained by the presence of many partial sequences in the reference database, the repetitive nature of LRRs, and that triggering of siRNAs occurs at several positions in a gene. In the partial sequences, the region where siRNA generation is initiated might be lacking, and strong phasing is only expected close to the initiation site. Studies of NBS-LRR genes in angiosperms have so far only identified NBS and TIR domains as targets for the miRNA-mediated initiation of siRNAs (Zhai et al., 2011; Shivaprasad et al., 2012). In our data, we identified many EST clusters with only LRR domains that spawned large numbers of siRNAs. These generally had low phase scores, probably also as an effect of their structure with short highly similar repetitive regions, which may obscure a phasing signal. In this study, we also found indications that more than one 22-nt miRNA might initiate siRNAs in different phases in one gene, which would significantly reduce the phase score statistic, despite a phased degradation (Figs. 6 and 7).
Available data in angiosperms have implicated one diverse miRNA superfamily (including miR482 and miR2118) as initiators of secondary siRNAs on NBS-LRR mRNA (Zhai et al., 2011; Shivaprasad et al., 2012). In contrast, our data set contained a substantial fraction of highly abundant and diverse 22-nt sRNAs, several of which were predicted miRNAs and predicted to initiate secondary siRNAs from NBS-LRR transcripts. Most of those miRNAs were predicted to target TIR domains, but due to the lack of a comprehensive full-length transcript database for spruce, it is premature to evaluate the target specificity of the spruce miRNA initiating secondary siRNAs. Based on the available data, we find no support for a major role for the miR482/miR2118 superfamily as regulators of NBS-LRR genes in spruce. Rather, a large number of other highly abundant miRNAs seem to specifically target such genes in this species.
An alternative mechanism that could potentially generate siRNAs is natural antisense transcription, resulting in dsRNAs and subsequent siRNA production. Due to the nature of NBS-LRR gene evolution, natural antisense transcription might be more prevalent among such genes (Yi and Richards, 2009). Our DGE tag data do not support natural antisense transcription as a general mechanism for sRNAs derived from NBS-LRR-type resistance genes. Although EST clusters with high ratios of antisense to sense DGE tags were identified only rarely (Supplemental Table S2), there was no general tendency for such genes to show high ratios.
siRNA-Mediated Degradation of NBS-LRR Genes Varies Widely between Species
Our survey of sRNAs mapping to NBS-LRR genes revealed striking differences in the prevalence of this phenomenon. On the basis of the available data, we can only speculate on the reason for this large diversity. Clearly, spruce stands out as extreme in terms of the amount of sRNA that is derived from NBS-LRR genes as well as the percentage of such genes that spawn siRNAs. The mechanism seems to reach a significant level only in seed plants. This increase is correlated with a drastic expansion of the number of NBS-LRR genes occurring in seed plants (Yue et al., 2012). However, a very low frequency of NBS-LRR genes from monocots spawn siRNAs, and a high frequency was observed only in some dicot species, with very low frequencies observed in Arabidopsis.
The low frequency observed in monocots refutes the hypothesis that the mechanism is directly related to the number of NBS-LRR genes. However, estimates of sequence divergence between NBS-LRR genes in Arabidopsis, rice, poplar, and grape suggest that recent tandem duplication has been more frequent in the two perennial species (Yang et al., 2008). These data also indicate increased numbers of gene conversion events and unequal crossing over among NBS-LRR genes in the perennial species, and it was suggested that the excess of recent duplications and a higher homologous recombination rate in grape and poplar could compensate for the long generation time coupled to a low nucleotide substitution rate as compared with the pathogens (Yang et al., 2008). A reliable estimate of the number of NBS-LRR genes in conifers and their pattern of evolution is premature and has to await a conifer genome sequence, but recent expansion of the NBS-LRR gene family in some species might explain part of the divergent patterns of siRNA degradation of NBS-LRR genes observed between species.
Recently, it was suggested that the miRNA-mediated mechanism generating siRNAs from NBS-LRR genes is part of counter-counter defense against pathogens (Shivaprasad et al., 2012). A silencing cascade mediated by miR482 that targets NBS-LRR genes was suppressed in plants infected with viruses or bacteria, leading to an increase in mRNA levels of NBS-LRR target genes, likely contributing to increased defense reactions. To what extent such a counter-counter defense mechanism is present in different species is currently unknown, as is the extent to which it could explain the large variation in siRNA production observed between species. An interesting question is whether the evolution of such a counter-counter defense mechanism is affected by life history, such as perenniality. A common feature of several species with high numbers of siRNAs mapping to NBS-LRR genes is that they are long-lived perennial species (Picea, Populus, Vitis, and Gossypium spp. but not Medicago spp.). A major difference between annuals and perennials, affecting the evolution of resistance, is that perennials will almost certainly encounter many different pathogens before reproduction, and the long generation time confers problems in matching the evolutionary rates of the pathogens. Besides higher duplication and recombination rates among NBS-LRR genes, additional layers of defense might be beneficial.
Recent data from legumes suggest that the repression of NBS-LRR genes by specific miRNAs may have evolved to avoid plant defense responses during symbiotic interactions (Bazin et al., 2012). Such a mechanism could potentially explain the high incidence of miRNA-guided siRNA-mediated degradation of NBS-LRR genes observed in Medicago spp. Such an explanation seems less likely for the high incidence of siRNA degradation of resistance genes seen in perennials. Even though symbiotic mycorrhizal interactions are common among those species, a massive degradation of NBS-LRR genes in needles, as observed in Picea spp., seems costly and detrimental to pathogen resistance.
MATERIALS AND METHODS
Sequencing of sRNAs from Spruce
Tissue was collected from newly flushed buds of spruce (Picea abies) and frozen in liquid nitrogen. Total RNA was isolated using a modified version of the procedure of Azevedo et al. (2003), where precipitation was done in isopropanol instead of LiCl in order to retain sRNAs. sRNA libraries were constructed using the Illumina Small RNA kit version A, where PAGE gels were used to select sRNAs with a size between 18 and 30 nt. Each library was based on a pool of RNA from six trees. Two libraries from separate pools were sequenced using the Illumina platform.
After removal of the adaptor sequence, about 10.6 million reads were obtained. These reads were filtered by removing those matching structural RNAs (ribosomal RNA, transfer RNA) using BLASTN searches against Rfam and mononucleotide repeats. After these filtering steps, 8,310,409 reads were kept for further analysis.
DGE Tag Data
To estimate gene expression, we used six digital gene expression libraries constructed from individual needle tissue samples of spruce seedlings. The libraries were sequenced on the Illumina platform each to a depth of around 4 million reads. After filtering, the reads were mapped against the Picea spp. EST clusters from the Gene Index Project. Expression levels were estimated by the number of reads mapping to each transcript divided by the length of each transcript.
Identification of miRNAs and Their Targets
Short reads were searched against the Rfam (Gardner et al., 2009) database, and matches to other types of known sRNAs were removed. The remaining short reads were compared with miRBase using the National Center for Biotechnology Information “blast-short” program with an e-value = 1,000 and word size = 7. Sequences with no more than two mismatches (including overhangs, but no gaps allowed) were identified as miRNAs conserved between species. Precursors were identified in three steps. First, we searched a spruce transcriptome database that included the spruce putative unique transcripts assembly of Chen et al. (2012), the Picea glauca gene catalog of Rigault et al. (2011), and the SGI EST clusters (Supplemental Table S7) with the filtered short reads. Sequences with a maximum of two mismatches to a short read (no gaps allowed) and to which the short read mapped less than six times were used to predict primary precursors using the miRDeep pipeline (Friedländer et al., 2008). The predicted primary precursors were then filtered to remove protein-coding mRNAs by searching against the UniProtKB plant protein database. In addition, to minimize the number of false positives in the prediction, we used a classification method, PlantMiRNAPred (Xuan et al., 2011), for the further classification of valid plant pre-miRNA sequences. We used the SGI data set for the prediction of miRNA targets using psRNATarget (Dai and Zhao, 2011).
Analysis of siRNAs
sRNA data for the various species were mainly downloaded as Gene Expression Omnibus data sets, series GSE28755 (for details, see Supplemental Table S7). sRNA was mapped against various reference sequences using Bowtie (version 0.12.7; Langmead 2010). To facilitate comparisons among the different species, we used EST clusters from the Plant Gene Index (Supplemental Table S7), as most species lack a reference genome. For Arabidopsis (Arabidopsis thaliana), rice (Oryza sativa), poplar (Populus trichocarpa), and grape (Vitis vinifera), we also mapped sRNAs against annotated full-length transcripts (Supplemental Table S7). For spruce, we also used the Arborea P. glauca gene catalog that contains more than 23,000 mRNA sequences that are annotated as full length (Rigault et al., 2011). In order to predict protein domains and identify putative resistance genes, we translated all EST clusters to protein sequences (all six frames) and used PFAM protein families (Punta et al., 2012) and the program hmmsearch with default settings (http://hmmer.janelia.org). The following hmm profiles were used: LRR_1, PF00560; LRR_2, PF07723; LRR_3, PF07725; LRR_4, PF12799; LRR_5, PF13306; LRR_6, PF13516; LRR_7, PF13504; LRR_8, PF13855; NBS, PF00931; TIR, PF01582.
In addition, spruce sRNA data were mapped against six Picea spp. BACs (four spruce ones supplied by E. De Paoli and two from P. glauca; Hamberger et al., 2009). Finally, Pinus contorta sRNA (ftp://ftp03.bcgsc.ca/public/pine_rice_smRNA/pine_all_454_reads.fna) was also mapped to 10 Pinus taeda BACs (Kovach et al., 2010).
Analysis of sRNA Phasing
Phasing analysis was conducted based on the calculation described by De Paoli et al. (2009), with some modification, essentially as described by Zhai et al. (2011). Reads on the sense and antisense strands were offset by −1 nt and +1 nt, respectively, to account for a 2-nt overhang occurring during dsRNA cleavage. The number of phase cycle positions occupied by at least one read in the current 210-nt window must be greater than 3, and the most abundant read at those positions should not constitute more than 90% of the sum of all reads at phase cycle positions. The calculations were performed in R (R Development Core Team, 2008).
All sequence data generated in this study are available from GenBank under accession numbers SRR813801, SRR815091, SRR824098 to SRR824103, SRR824149, and SRR824150.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Mapping of 21-nt sRNA to a region of Arabidopsis chromosome 1.
Supplemental Figure S2. Distribution of 21- and 24-nt sRNA mapping to scaffold 1 of poplar.
Supplemental Figure S3. sRNA distribution in an NBS-LRR region on poplar scaffold 1.
Supplemental Figure S4. sRNA abundance at a poplar NBS-LRR gene locus.
Supplemental Table S1. Identified P. abies miRNA.
Supplemental Table S2. Spruce sRNA and gene expression data.
Supplemental Table S3. Arabidopsis sRNA data.
Supplemental Table S4. Rice sRNA data.
Supplemental Table S5. Poplar sRNA data.
Supplemental Table S6. Grape sRNA data.
Supplemental Table S7. sRNA data sets used in this study.
Acknowledgments
We thank Kerstin Jeppson for sRNA extractions and Illumina library preparations and the Uppsala SciLifeLab for generating the sequence data. We are also in debt to Emanuele De Paoli and Jixian Zhai for answering questions regarding the calculation of phase scores from Illumina data.
Glossary
- sRNA
small RNA
- dsRNA
double-stranded RNA
- miRNA
microRNA
- RISC
RNA-induced silencing complex
- siRNA
short-interfering RNA
- NBS
nucleotide-binding site
- LRR
Leu-rich repeat
- tasiRNA
trans-acting short-interfering RNA
- PPR
pentatricopeptide
- BAC
bacterial artificial chromosome
- nt
nucleotide
- cDNA
complementary DNA
- SGI
Spruce Gene Index
- DGE
Digital Gene Expression
References
- Axtell MJ, Jan C, Rajagopalan R, Bartel DP. (2006) A two-hit trigger for siRNA biogenesis in plants. Cell 127: 565–577 [DOI] [PubMed] [Google Scholar]
- Azevedo H, Lino-Neto T, Tavares RM. (2003) An improved method for high-quality RNA isolation from needles of adult Maritime pine trees. Plant Mol Biol Rep 21: 333–338 [Google Scholar]
- Bazin J, Bustos-Sanmamed P, Hartmann C, Lelandais-Brière C, Crespi M. (2012) Complexity of miRNA-dependent regulation in root symbiosis. Philos Trans R Soc Lond B Biol Sci 367: 1570–1579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodersen P, Sakvarelidze-Achard L, Bruun-Rasmussen M, Dunoyer P, Yamamoto YY, Sieburth L, Voinnet O. (2008) Widespread translational inhibition by plant miRNAs and siRNAs. Science 320: 1185–1190 [DOI] [PubMed] [Google Scholar]
- Burleigh JG, Barbazuk WB, Davis JM, Morse AM, Soltis PS. (2012) Exploring diversification and genome size evolution in extant gymnosperms through phylogenetic synthesis. J Bot 2012: 1–6 [Google Scholar]
- Chapman EJ, Carrington JC. (2007) Specialization and evolution of endogenous small RNA pathways. Nat Rev Genet 8: 884–896 [DOI] [PubMed] [Google Scholar]
- Chen J, Uebbing S, Gyllenstrand N, Lagercrantz U, Lascoux M, Källman T. (2012) Sequencing of the needle transcriptome from Norway spruce (Picea abies Karst L.) reveals lower substitution rates, but similar selective constraints in gymnosperms and angiosperms. BMC Genomics 13: 589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X. (2009) Small RNAs and their roles in plant development. Annu Rev Cell Dev Biol 25: 21–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuperus JT, Fahlgren N, Carrington JC. (2011) Evolution and functional diversification of MIRNA genes. Plant Cell 23: 431–442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai X, Zhao PX. (2011) psRNATarget: a plant small RNA target analysis server. Nucleic Acids Res 39: W155–W159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Paoli E (2006) Genetic diversity, linkage disequilibrium and repetitive component of the genome in Norway spruce (Picea abies (L.) Karst.). PhD thesis. Universita Degli Studi Di Udine, Udine, Italy [Google Scholar]
- De Paoli E, Dorantes-Acosta A, Zhai J, Accerbi M, Jeong DH, Park S, Meyers BC, Jorgensen RA, Green PJ. (2009) Distinct extremely abundant siRNAs associated with cosuppression in petunia. RNA 15: 1965–1970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolgosheina EV, Morin RD, Aksay G, Sahinalp SC, Magrini V, Mardis ER, Mattsson J, Unrau PJ. (2008) Conifers have a unique small RNA silencing signature. RNA 14: 1508–1515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedländer MR, Chen W, Adamidi C, Maaskola J, Einspanier R, Knespel S, Rajewsky N. (2008) Discovering microRNAs from deep sequencing data using miRDeep. Nat Biotechnol 26: 407–415 [DOI] [PubMed] [Google Scholar]
- Gardner PP, Daub J, Tate JG, Nawrocki EP, Kolbe DL, Lindgreen S, Wilkinson AC, Finn RD, Griffiths-Jones S, Eddy SR, et al (2009) Rfam: updates to the RNA families database. Nucleic Acids Res 37: D136–D140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haag JR, Pikaard CS. (2011) Multisubunit RNA polymerases IV and V: purveyors of non-coding RNA for plant gene silencing. Nat Rev Mol Cell Biol 12: 483–492 [DOI] [PubMed] [Google Scholar]
- Hamberger B, Hall D, Yuen M, Oddy C, Hamberger B, Keeling CI, Ritland C, Ritland K, Bohlmann J. (2009) Targeted isolation, sequence assembly and characterization of two white spruce (Picea glauca) BAC clones for terpenoid synthase and cytochrome P450 genes involved in conifer defence reveal insights into a conifer genome. BMC Plant Biol 9: 106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herr AJ. (2005) Pathways through the small RNA world of plants. FEBS Lett 579: 5879–5888 [DOI] [PubMed] [Google Scholar]
- Jin H, Zhu JK. (2010) A viral suppressor protein inhibits host RNA silencing by hooking up with Argonautes. Genes Dev 24: 853–856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasschau KD, Fahlgren N, Chapman EJ, Sullivan CM, Cumbie JS, Givan SA, Carrington JC. (2007) Genome-wide profiling and analysis of Arabidopsis siRNAs. PLoS Biol 5: e57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khraiwesh B, Zhu JK, Zhu J. (2012) Role of miRNAs and siRNAs in biotic and abiotic stress responses of plants. Biochim Biophys Acta 1819: 137–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klevebring D, Street NR, Fahlgren N, Kasschau KD, Carrington JC, Lundeberg J, Jansson S. (2009) Genome-wide profiling of Populus small RNAs. BMC Genomics 10: 620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovach A, Wegrzyn JL, Parra G, Holt C, Bruening GE, Loopstra CA, Hartigan J, Yandell M, Langley CH, Korf I, et al (2010) The Pinus taeda genome is characterized by diverse and highly diverged repetitive sequences. BMC Genomics 11: 420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozomara A, Griffiths-Jones S. (2011) miRBase: integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res 39: D152–D157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krasnikova MS, Milyutina IA, Bobrova VK, Ozerova LV, Troitsky AV, Solovyev AG, Morozov SY. (2009) Novel miR390-dependent transacting siRNA precursors in plants revealed by a PCR-based experimental approach and database analysis. J Biomed Biotechnol 2009: 952304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B. (2010) Aligning short sequencing reads with Bowtie. Curr Protoc Bioinformatics 32: 11.7.1–11.7.14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F, Pignatta D, Bendix C, Brunkard JO, Cohn MM, Tung J, Sun H, Kumar P, Baker B. (2012) MicroRNA regulation of plant innate immune receptors. Proc Natl Acad Sci USA 109: 1790–1795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu JJ, Ekramoddoullah AKM. (2011) Genomic organization, induced expression and promoter activity of a resistance gene analog (PmTNL1) in western white pine (Pinus monticola). Planta 233: 1041–1053 [DOI] [PubMed] [Google Scholar]
- Liu W, Thummasuwan S, Sehgal SK, Chouvarine P, Peterson DG. (2011) Characterization of the genome of bald cypress. BMC Genomics 12: 553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magbanua ZV, Ozkan S, Bartlett BD, Chouvarine P, Saski CA, Liston A, Cronn RC, Nelson CD, Peterson DG. (2011) Adventures in the enormous: a 1.8 million clone BAC library for the 21.7 Gb genome of loblolly pine. PLoS ONE 6: e16214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi S, Cai T, Hu Y, Chen Y, Hodges E, Ni F, Wu L, Li S, Zhou H, Long C, et al. (2008) Sorting of small RNAs into Arabidopsis argonaute complexes is directed by the 5′ terminal nucleotide. Cell 133: 116–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milne I, Bayer M, Cardle L, Shaw P, Stephen G, Wright F, Marshall D. (2010) Tablet: next generation sequence assembly visualization. Bioinformatics 26: 401–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morin RD, Aksay G, Dolgosheina E, Ebhardt HA, Magrini V, Mardis ER, Sahinalp SC, Unrau PJ. (2008) Comparative analysis of the small RNA transcriptomes of Pinus contorta and Oryza sativa. Genome Res 18: 571–584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Punta M, Coggill PC, Eberhardt RY, Mistry J, Tate J, Boursnell C, Pang N, Forslund K, Ceric G, Clements J, et al. (2012) The Pfam protein families database. Nucleic Acids Res 40: D290–D301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team (2008) R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna [Google Scholar]
- Rajagopalan R, Vaucheret H, Trejo J, Bartel DP. (2006) A diverse and evolutionarily fluid set of microRNAs in Arabidopsis thaliana. Genes Dev 20: 3407–3425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramachandran V, Chen X. (2008) Small RNA metabolism in Arabidopsis. Trends Plant Sci 13: 368–374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhoades MW, Reinhart BJ, Lim LP, Burge CB, Bartel B, Bartel DP. (2002) Prediction of plant microRNA targets. Cell 110: 513–520 [DOI] [PubMed] [Google Scholar]
- Rigault P, Boyle B, Lepage P, Cooke JEK, Bousquet J, MacKay JJ. (2011) A white spruce gene catalog for conifer genome analyses. Plant Physiol 157: 14–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Ferrer V, Voinnet O. (2009) Roles of plant small RNAs in biotic stress responses. Annu Rev Plant Biol 60: 485–510 [DOI] [PubMed] [Google Scholar]
- Shivaprasad PV, Chen HM, Patel K, Bond DM, Santos BACM, Baulcombe DC. (2012) A microRNA superfamily regulates nucleotide binding site-leucine-rich repeats and other mRNAs. Plant Cell 24: 859–874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunkar R, Zhu JK. (2004) Novel and stress-regulated microRNAs and other small RNAs from Arabidopsis. Plant Cell 16: 2001–2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takuno S, Innan H. (2011) Selection fine-tunes the expression of microRNA target genes in Arabidopsis thaliana. Mol Biol Evol 28: 2429–2434 [DOI] [PubMed] [Google Scholar]
- Tarr DEK, Alexander HM. (2009) TIR-NBS-LRR genes are rare in monocots: evidence from diverse monocot orders. BMC Res Notes 2: 197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaucheret H. (2006) Post-transcriptional small RNA pathways in plants: mechanisms and regulations. Genes Dev 20: 759–771 [DOI] [PubMed] [Google Scholar]
- Voinnet O. (2009) Origin, biogenesis, and activity of plant microRNAs. Cell 136: 669–687 [DOI] [PubMed] [Google Scholar]
- Wan LC, Wang F, Guo X, Lu S, Qiu Z, Zhao Y, Zhang H, Lin J. (2012) Identification and characterization of small non-coding RNAs from Chinese fir by high throughput sequencing. BMC Plant Biol 12: 146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z, Johansen LK, Gustafson AM, Kasschau KD, Lellis AD, Zilberman D, Jacobsen SE, Carrington JC. (2004) Genetic and functional diversification of small RNA pathways in plants. PLoS Biol 2: E104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xuan P, Guo M, Liu X, Huang Y, Li W, Huang Y. (2011) PlantMiRNAPred: efficient classification of real and pseudo plant pre-miRNAs. Bioinformatics 27: 1368–1376 [DOI] [PubMed] [Google Scholar]
- Yakovlev IA, Fossdal CG, Johnsen OY. (2010) MicroRNAs, the epigenetic memory and climatic adaptation in Norway spruce. New Phytol 187: 1154–1169 [DOI] [PubMed] [Google Scholar]
- Yang S, Zhang X, Yue JX, Tian D, Chen JQ. (2008) Recent duplications dominate NBS-encoding gene expansion in two woody species. Mol Genet Genomics 280: 187–198 [DOI] [PubMed] [Google Scholar]
- Yi H, Richards EJ. (2009) Gene duplication and hypermutation of the pathogen Resistance gene SNC1 in the Arabidopsis bal variant. Genetics 183: 1227–1234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue JX, Meyers BC, Chen JQ, Tian D, Yang S. (2012) Tracing the origin and evolutionary history of plant nucleotide-binding site-leucine-rich repeat (NBS-LRR) genes. New Phytol 193: 1049–1063 [DOI] [PubMed] [Google Scholar]
- Zhai J, Jeong DH, De Paoli E, Park S, Rosen BD, Li Y, González AJ, Yan Z, Kitto SL, Grusak MA, et al. (2011) MicroRNAs as master regulators of the plant NB-LRR defense gene family via the production of phased, trans-acting siRNAs. Genes Dev 25: 2540–2553 [DOI] [PMC free article] [PubMed] [Google Scholar]