Identification of novel loci that regulate root cellular and morphological development that may be useful for breeding programs.
Abstract
While the Arabidopsis (Arabidopsis thaliana) root has been elegantly characterized with respect to specification of cell identity, its development is missing a number of cellular features present in other species. We have characterized the root development of a wild and a domesticated tomato species, Solanum pennellii and Solanum lycopersicum ‘M82.’ We found extensive differences between these species for root morphology and cellular development including root length, a novel gravity set point angle, differences in cortical cell layer patterning, stem cell niche structure, and radial cell division. Using an introgression line population between these two species, we identified numerous loci that regulate these distinct aspects of development. Specifically we comprehensively identified loci that regulate (1) root length by distinct mechanisms including regulation of cell production within the meristem and the balance between cell division and expansion, (2) the gravity set point angle, and (3) radial cell division or expansion either in specific cell types or generally across multiple cell types. Our findings provide a novel perspective on the regulation of root growth and development between species. These loci have exciting implications with respect to regulation of drought resistance or salinity tolerance and regulation of root development in a family that has undergone domestication.
The root system is of vital importance to plants because it anchors the plant and its cells absorb and transport water, nutrients, and solutes to the shoot. The root system has a complex branching architecture with numerous cell types whose development must be dynamic, plastic, and highly responsive to the environment to maximize plant fitness and yield. To optimize root system architecture for the specific environment in which the plant is growing, developmental programs associated with distinct developmental stages, and cell types are specifically and precisely regulated by both local and global signals. For instance, local nitrogen sources induce root hair tip growth and can regulate lateral root initiation (Malamy and Ryan, 2001; Bloch et al., 2011) while the search for water regulates primary root growth (Saucedo et al., 2012). This complex architecture and plasticity complicate the ability to enumerate and study root architecture concomitantly at all cellular and tissue levels. Because of this limitation, population-level studies are typically limited to measuring architecture-level variables such as root length, number, and branching, without focusing on the development of specific cell types that give rise to this architecture.
Root cell type specification and development has been extensively studied using classical genetic methods in the model plant Arabidopsis (Arabidopsis thaliana). These studies revealed the elegant simplicity of the Arabidopsis root at the cellular level (Dolan et al., 1993). In Arabidopsis there is an invariant number of cells within the single cortical and endodermal layers of the primary root but variable cell numbers within lateral roots. The core of the root stem cell niche is formed by a set of four quiescent center (QC) cells, with a set of initial cells that give rise to all cell types in the root surrounding the QC. Developmental genetic studies in Arabidopsis have identified a variety of genes that regulate root length, lateral root number, and radial patterning (Benfey and Scheres, 2000; Mähönen et al., 2000; Schiefelbein et al., 2009). This includes the identification of genes that regulate vascular cell proliferation, endodermis and epidermis cell identity, and the asymmetric division of the cortex-endodermis initial (CEI).
Arabidopsis has provided an excellent base model for root cellular development, yet as with any species there are unique cellular aspects that are present and/or missing within Arabidopsis that necessitate the study of other species. For instance, Arabidopsis is unusual as it contains only four QC cells, whereas most monocot and dicot species contain a greater number of QC cells (Jiang et al., 2003). To date, the regulatory mechanisms controlling this diversity in QC cell number are completely unknown. Additionally, most monocot and dicot species contain numerous cortex layers that are the product of repeated divisions of a CEI cell, whereas Arabidopsis only contains a single cortex layer (Dolan et al., 1993). The cell number in the cortex and the endodermis is invariant in the Arabidopsis primary root, but variable in many other plant species. Regulation of radial cell number variability in these cell types as well as the pericycle has never been addressed in any plant species. Furthermore, in 80% of flowering plant species, the outer layer of the root’s cortex, or exodermis, contains a suberinized cell wall to restrict passage of solutes from the outside of the root to the inside, but Arabidopsis does not contain a suberinized exodermis. The exodermis has been reported to be derived from an independent cortical initial, suggesting it is an independent specialized cell type whose genetics are not addressable within Arabidopsis (Heimsch and Seago, 2008). Genes regulating the specification of the exodermis and the production of multiple cortical layers have not been identified in monocots or dicots. Thus, classical genetic approaches have not addressed the genetic mechanisms regulating cell proliferation and patterning decisions within many cell types not present in Arabidopsis.
One approach with significant potential to identify these unresolved genetic mechanisms and integrate them into the broader control of root system architecture is the use of natural variation within and between species (Shindo et al., 2007). The use of stable mapping populations such as a homozygous introgression line (IL) between two different species provides a stable genetic pool from which to repeatedly phenotype different cellular and morphological aspects of root architecture and integrate them into a common model. This quantitative genetic analysis is typically conducted using quantitative trait locus (QTL) mapping, which has identified loci or, in a small number of cases, genes that regulate root length in monocots and dicots (Bettey et al., 2000; Mouchel et al., 2004; Loudet et al., 2005; Fitz Gerald et al., 2006; Reymond et al., 2006; Fita et al., 2008; Khan et al., 2012). These studies however have typically been limited to the analysis of large-effect loci (Loudet et al., 2005; Reymond et al., 2006) and have not coordinately dissected root architecture at both the morphological and cellular levels.
To determine how tomato (Solanum spp.) root morphogenesis is determined by cellular features including radial patterning, radial cell proliferation, radial cell expansion, and compensatory changes in cell expansion when cell proliferation is altered (Hemerly et al., 1995), we performed a detailed characterization of root development in two Solanum species. We used Solanum pennellii, a wild tomato species, and cv M82 of the domesticated species Solanum lycopersicum and their derived IL population. A wild species, S. pennellii is found in coastal deserts and rocky, arid soil and exhibits drought and salt tolerance and pathogen resistance in comparison with the domesticated cv M82 (Dehan and Tal, 1978; Koca et al., 2006; Easlon and Richards, 2009). In this study, we identified significant developmental differences between the two species by measuring a large range of root traits including the cell number within individual cell types, CEI spatiotemporal patterning differences, variability in cortex cell layer and QC cell number, root growth, and a novel gravity set point angle. To explore the link between the whole organ phenotype and cellular level using interspecific genetic variation we used the IL population derived from a cross between cv M82 and S. pennellii (Eshed and Zamir, 1995). This population comprises 76 segmental ILs with marker-defined genomic regions of S. pennellii substituting for homologous intervals of the cultivated variety cv M82 that partition the tomato genome into 107 bins. Measuring the above cellular and morphological phenotypes in these lines identified numerous major- and minor-effect loci for each phenotype, showing that interspecific variation in root development involves a complex suite of genetic changes, many of which display cell type-specific effects.
RESULTS
Root Growth and Development Differ at the Morphological and Cellular Level between the Wild (S. pennellii) and Domesticated (cv M82) Tomato Species
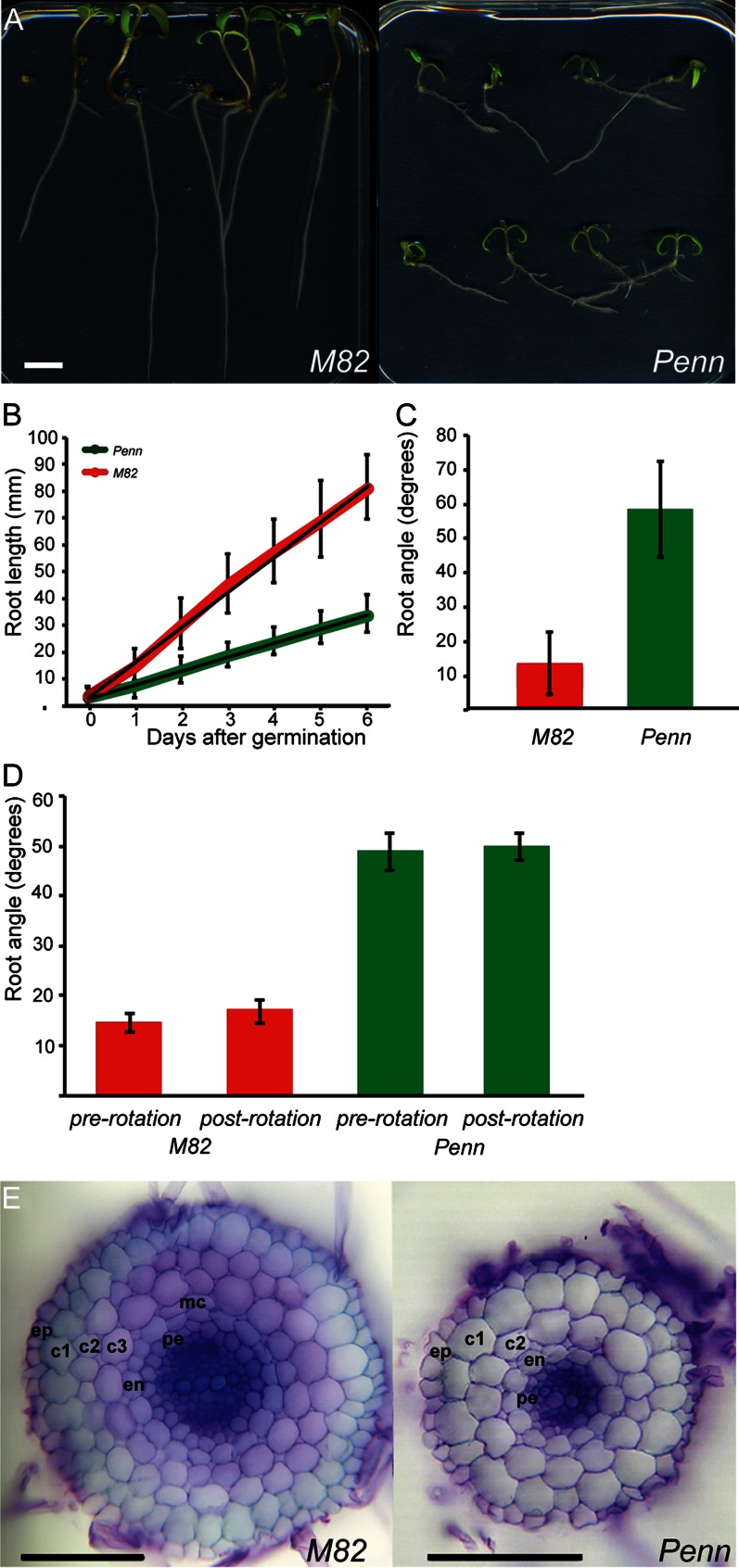
Roots of domesticated tomato were significantly longer than those of the wild species for each day up to 6 d after germination and had an increased growth rate for the domesticated cv M82 compared with S. pennellii (12.8 mm d–1 versus 5.1 mm d–1, respectively; Fig. 1, A and B). Primary roots of S. pennellii grew at a significantly different angle relative to the gravity vector than those of cv M82 (Fig. 1, A and C). The roots of both species were exposed to a change in the gravity vector by rotating the plates 90° (Fig. 1D). Both species responded to this rotation with a change in growth direction equivalent to that observed before the shift (Fig. 1D).
Figure 1.
Root morphology and cellular anatomy of S. lycopersicum ‘M82’ and S. pennellii (Penn). In all cases, error bars represent sd. A, cv M82 and S. pennellii root length and growth angle. Bar = 1 cm. B, Root length. All points at and following 1 d post germination are significant at P < 0.001; n = 24–60 (cv M82), n = 73–79 (S. pennellii). C, Root growth angle. The angle is significantly different at P < 0.001; n = 59 (cv M82), n = 76 (S. pennellii). D, Mean root angles before (pre) and after (post) turning growing roots 90°. Angles are significantly different between genotypes (P < 0.001) but not between treatments (P = 0.14 and 0.08 for cv M82 and S. pennellii, respectively); n = 13 (cv M82), n = 7 (S. pennellii). E, Cross sections of cv M82 and S. pennellii roots. Bar = 100 µm. Ep, Epidermis; c1, outer cortex layer; c2, S cortex layer; c3, inner cortex layer; mc, middle cortex; en, endodermis; pe, pericycle. S. pennellii is missing the c3 layer.
S. pennellii roots had one less cell layer compared with cv M82 (Fig. 1E). The extra, complete layer was found throughout the root and was derived from a CEI in contrast to a middle cortex layer. Endodermis identity was assessed by locating the Casparian strip. A single endodermal layer was identified in both S. pennellii and cv M82 (Supplemental Fig. S1; Fig. 2G). Additionally, a suberinized outer cortical layer, or exodermis, was identified in both species (Supplemental Fig. S1; Fig. 2G).
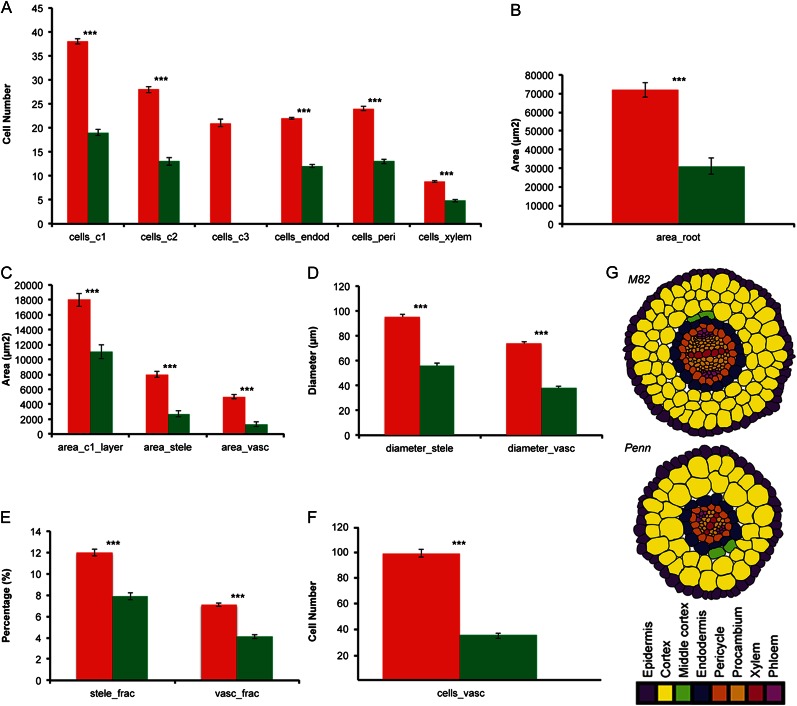
Figure 2.
Quantification of cellular anatomy in cv M82 and S. pennellii (Penn). The least squares mean was calculated from all the measurements for both species across all experiments. In all cases, *** = P < 0.001 as determined by a Student’s t test, and cells were measured from a 1-cm section flanking the middle of the root. A–F, cv M82 (red columns) and S. pennellii (green columns). cells_c1, Cells in the first cortex layer; cells_c2, cells in the second cortex layer; cells_c3, cells in the third cortex layer; cells_endod, cells in the endodermis; cells_peri, cells in the pericycle; cells_xylem, cells in the xylem; area_c1_layer, area of the outer cortex layer; area_stele, area of the stele; area_vasc, area of the vasculature; diameter_stele, diameter of stele across the median xylem axis; diameter_vasc, diameter of vasculature across the median xylem axis; stele_frac, fraction of root area occupied by stele area; vasc_frac, fraction of root area occupied by vascular cylinder; cells_vasc, total number of cells in the vascular cylinder. G, Cartoon of radial cell patterning in cv M82 and S. pennellii. Images are traced from the representative cross sections in Figure 1E.
Root area and radial cell number for all cell types measured (cortex, endodermis, pericycle, and xylem) were reduced in S. pennellii as compared with cv M82 (Fig. 2). Both species were found to have diarch vascular architecture, but the vascular cylinder of S. pennellii had significantly fewer cells than cv M82 (Fig. 2, F and G).
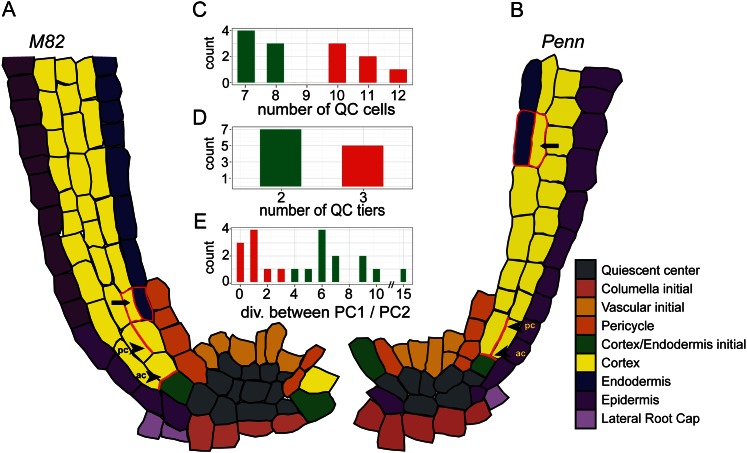
The stem cell niche structure in both species was identified by locating replicating cells and the columella stem cell niche (Supplemental Fig. S2). The number of QC cells and their arrangement differed between the two species, with seven to eight cells (sd = 0.53) in S. pennellii in two tiers and 10 to 12 cells (sd = 0.82) in cv M82 in three tiers (Fig. 3, A–D; Supplemental Fig. S2).
Figure 3.
Characterization of the stem cell niche of cv M82 and S. pennellii (Penn). A–B, Cellular architecture of cv M82 (A) and S. pennellii (B) stem cell niche as seen in a representative median longitudinal optical section. Pc, Periclinal division; ac, anticlinal division. C–D, Number of QC cells (C) and QC tiers (D) of cv M82 (red) and S. pennellii (green). The difference in QC cell number was significant at P = 2.48e–5 using a Student’s t test. E, The number of anticlinal divisions between the first periclinal division and the second periclinal division in cv M82 (red) and S. pennellii (green). The last periclinal division in cv M82 and S. pennellii that gives rise to the endodermis is indicated with an arrow. PC1, Periclinal division 1; PC2, periclinal division 2.
All cortex layers and the endodermis in cv M82 and S. pennellii were derived from periclinal divisions of the CEI or its daughters (Fig. 3, A–B and E). Without a molecular marker to specifically identify the CEI, we defined cells surrounding the stem cell niche that give rise to either cortex or endodermis cells a CEI. The third cortex layer in cv M82 was derived from an additional periclinal division of a CEI daughter cell. A difference in the timing of these periclinal divisions was also observed between the two species. In cv M82 only one to three anticlinal divisions (sd = 1) were observed, while in S. pennellii four to 15 divisions were observed (sd = 2.9) before the second periclinal division (Fig. 3E).
Root Length Is Controlled by Multiple Loci Influencing Cell Division and Expansion
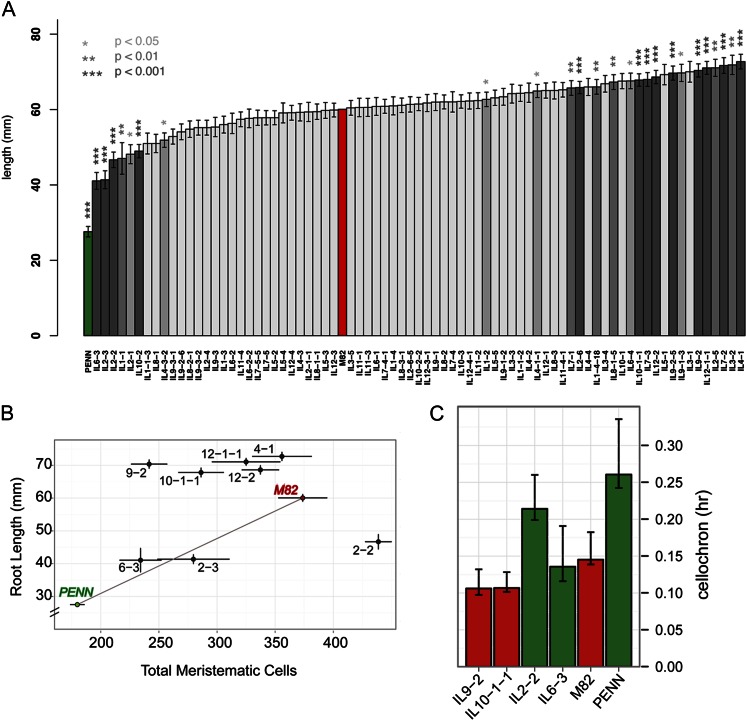
QTL analysis was performed for root length across the IL population. Many genotypes showed a transgressive increase in root length compared to the cv M82 parent (Fig. 4A). While several genotypes displayed significantly shorter roots, none displayed a transgressive decrease in root length relative to S. pennellii (Fig. 4A). However, a number of S. pennellii loci provided a long root phenotype compared with the corresponding cv M82 locus.
Figure 4.
Identification of QTLs regulating root length. A, Mean root length values across ILs. Asterisks and bar shading indicate corrected significance as determined using the Dunnett multiple hypothesis testing correction method. Error bars represent se. Parent’s values are shown in red (cv M82) and green (S. pennellii). B, Correlation of root length and meristematic zone cell number in parents and a subset of ILs. Parents are colored red (cv M82) and green (S. pennellii). Error bars represent se. Total meristematic zone cell number is calculated as the number of cells in the outer and inner cortex layer and in the endodermis. C, Cellochron analysis to determine the dynamic rate of addition of a cell to the root’s elongation zone. PENN, S. pennellii.
The short-rooted S. pennellii displayed fewer cells circumferentially in the outer and inner cortex layers (Fig. 2A) and fewer cells within the root meristem (longitudinally) relative to cv M82 (Fig. 4B). The length of meristem cells within the two species was slightly different. Differences in cell expansion coupled with differences in cell production rate within the meristem or differences in exit from the root’s meristematic zone to elongation zone control variation in root length (Supplemental Fig. S3).
We measured the number of cells in the root’s meristematic zone (cortex layers and endodermis) and plotted it relative to root length, for a subset of ILs that displayed the longest (9-2, 10-1-1, 12-1-1, 4-1) and shortest (6-3, 2-3, 2-2) root phenotypes (Fig. 4B). In contrast to the parents, changes in root length among the ILs were not always explicable by differences in the number of cells within the root’s meristematic zone. Four ILs stood out in this regard (Fig. 4B). IL 6-3 showed a decrease in the number of cells within the root meristem that was consistent with its shorter root length similar to S. pennellii. In stark contrast, IL 2-2 roots were essentially as short as IL 6-3 and showed the greatest number of cells within the root meristem (approximately 438; Fig. 4B). Conversely, the long roots of ILs 9-2 and 10-1-1 showed a decrease in cell number within the root’s meristematic zone, relative to cv M82 (Fig. 4B).
The dynamics of a cell’s transition from the meristematic zone to the elongation was calculated. A cellochron (c) represents the time interval during which a new cell is added to a file within the elongation zone (Fig. 4C; Silk et al., 1989; Beemster and Baskin, 1998). The short root/high meristem cell number IL 2-2 had a cellochron that was much longer relative to cv M82, consistent with its greatly increased meristem size. IL 2-2 had a short root due to fewer cells entering the elongation zone each hour (Fig. 4, B and C). Similarly, the long root/low meristem cell number ILs 9-2 and 10-1-1 had a cellochron that was shorter relative to cv M82, leading to fewer cells within the root meristem and more cells that entered the elongation zone each hour (Fig. 4C).
Multiple Loci Govern Developmental Control of Root Angle
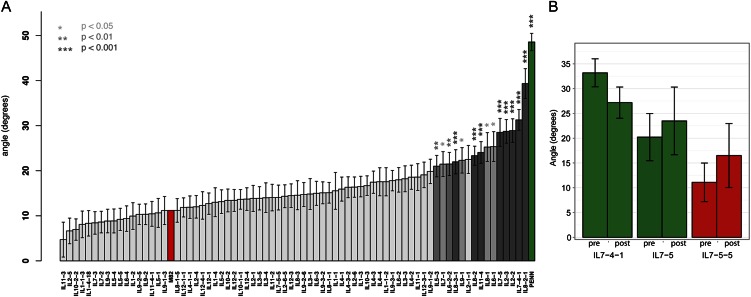
One striking developmental difference between the roots of S. pennellii and cv M82 was the differential establishment of a growth angle away from the gravity vector in S. pennellii (Figs. 1A and 5A). Several ILs had angles significantly less vertical than cv M82 (Fig. 5A). Transgressive individuals showing a root angle closer to vertical than cv M82’s were seen, but none passed Dunnett corrected significance thresholds (Fig. 5A). Growth angle was found to be a polygenic trait with numerous loci of moderate effect.
Figure 5.
Identification of QTLs regulating root growth angle. A, Mean root angle values across ILs. Asterisks and bar shading indicate corrected significance. Error bars show se. Parent’s values are shown in red (cv M82) and green (S. pennellii). B, Analysis of gravity response in a subset of ILs. Mean root angles were measured before (pre) and after (post) turning growing roots 90°. Angles are not significantly different between treatments (IL 7-4-1: P = 0.15, n = 11; IL 7-5-5: P = 0.68, n = 6; IL 7-5: P = 0.53, n = 8). Red represents roots that grow more like cv M82, and green represents ILs that grow more like S. pennellii.
To determine if these loci were affecting the growth angle relative to gravity rather than causing insensitivity to gravity, we changed the gravity vector for two lines with an S. pennellii-like growth angle phenotype and one like cv M82 (Fig. 5B). In each case the ILs responded to the change in gravity, just like cv M82 and S. pennellii (Fig. 5B), indicating that these loci affected the growth set angle, not response to the gravity vector.
Identification of Loci Regulating Root Radial Cell Division, Cell Expansion, and Their Proportions
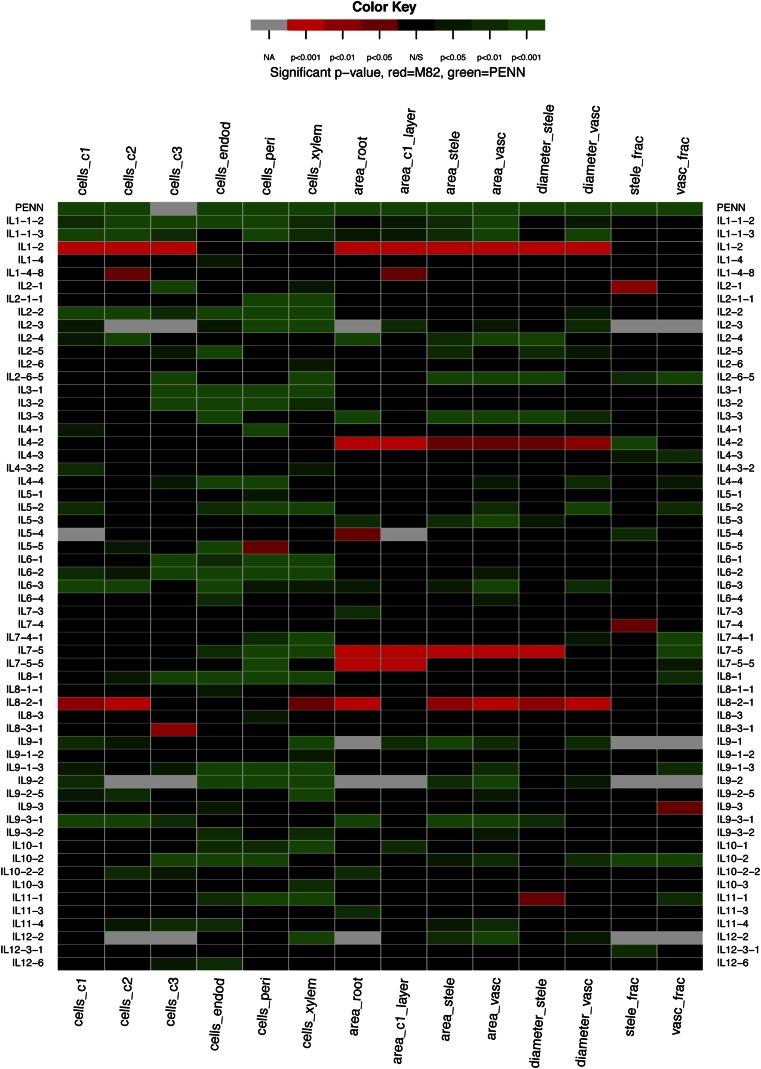
As previously shown, there were numerous cellular differences in the development of the cv M82 and S. pennellii roots (Fig. 2). We measured 14 different cellular traits in cv M82, S. pennellii, and the ILs (Fig. 6; Supplemental Table S3). Many genetic loci contributed to each of the cellular traits and were distributed throughout the genome. The most striking cellular difference between cv M82 and S. pennellii was the additional cortex layer present in cv M82 (Fig. 2G). However, no IL contained only two cortex layers, suggesting that it is a complex trait requiring multiple loci. As with the root length and root angle traits, no ILs showed a stronger phenotype than S. pennellii for any of the cellular traits (Supplemental Data Set S1). Contrary to the angle trait, several ILs showed transgressive phenotypes that were higher than cv M82 (Supplemental Data Set S1). Relative to the root length and root angle traits, a large number of QTLs had the capacity to control cellular architecture traits (Fig. 6). Many of the ILs contained QTLs contributing to multiple traits (Fig. 6). Furthermore, the majority of these loci resulted in cell numbers or dimensions that were less than cv M82 (Fig. 6).
Figure 6.
Identification of QTLs regulating cellular morphology and root development. Significance of a trait is represented relative to cv M82 in heat map format with * = P < 0.05, ** = P < 0.01, *** = P < 0.001 as determined by ANOVA with Dunnett multiple hypothesis testing correction. Only ILs with at least one significant trait are displayed. If a trait is transgressive to cv M82 (i.e. increases in cell number or other parameters) then the trait is represented in shades of red, whereas if there are fewer cells or other parameters, then the trait is represented in shades of green. Abbreviations are as for Figure 2.
Independence of Genetic Loci Controlling Root Cellular Patterning
Thirty-nine of the 53 ILs that showed a change in radial cell number showed a similar change in at least one other cell type (Fig. 6; Supplemental Fig. S4). This argued for the existence of loci that acted as general regulators of radial cell number, but there was also evidence for loci that functioned as cell type-specific radial division regulators. For example, IL 1-4-8 had a positive effect only on cell number in the second cortex layer while IL 8-3-1 positively regulated only the cell number in the third cortex layer. IL 1-4 had a decrease in cell number in the endodermis only while IL 5-1, IL 7-5-5, and IL 8-3 all decreased cell number only in the pericycle.
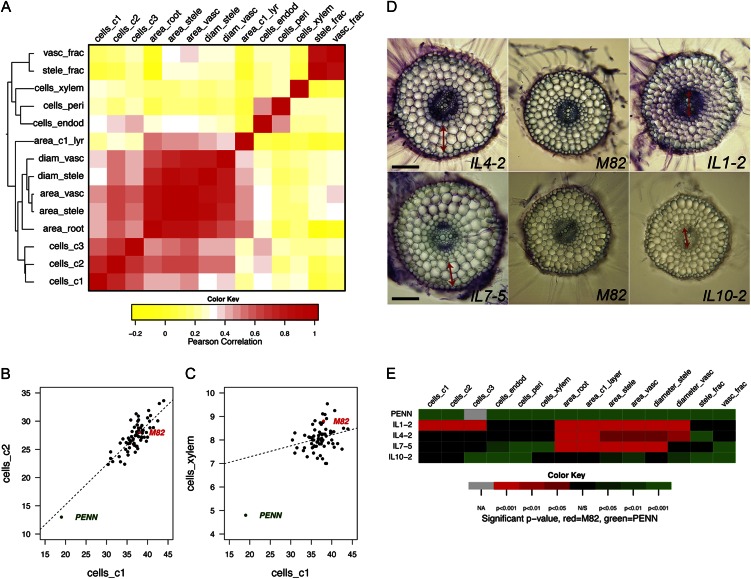
A relatively high correlation between cell number in the first and second cortex layer was observed, with a more modest correlation between these layers and the innermost layer (Fig. 7, A–C; Supplemental Table S4). Additionally, endodermis and pericycle cell number closely correlated with each other and were less correlated with xylem cell number. Finally, cell number in the cortex layers formed a separate cluster from endodermal, pericycle, and xylem cell number (Fig. 7A). This suggested that genetic control of radial cell number variability became gradually uncoupled from the outer cell layers to the stele.
Figure 7.
The correlation of cellular traits. A, Correlation plot showing overall similarity in cell number and other traits. B–C, Plots showing partial correlation between cell number in cells_c1 and cells_c2 and in cells_c1 and cells_xylem. D, Cross sections illustrating growth constraints. The double-headed arrow indicates the tissue layer, which has expanded presumably to compensate for a change in division in another cell layer. Bar = 100 µm. E, A subset of the heat map in Figure 6 corresponding to the ILs shown in D. Abbreviations are as for Figure 2.
Several ILs showed expected changes in division with area. These included IL 1-2, where cell number was increased across the cortex layers and, accordingly, root area and area of the first cortex layer were increased (Fig. 7, D–E). In the other direction, ILs 8-2-1, 9-1, 9-3-1, 10-2-2, and 11-3 had a decrease in cell number in the cortex, and root area and area of the first cortex layer were decreased (Fig. 6).
Two ILs were identified where expansion of the outer cortex layer appeared to compensate for changes in inner cell layers. In IL 7-5, cell number was decreased across several cell layers including the endodermis, pericycle, and xylem, but root area and area of the first cortex layer were increased (Fig. 7, D–E). In IL 4-2, the total root and stele areas were significantly larger than in cv M82, but no significant difference in the cell number of any layer compared with cv M82 was observed (Fig. 7, D–E). Furthermore, the fraction of the stele and vasculature relative to whole root area was significantly smaller than cv M82, indicating that the outer radial layers expanded in a different ratio than the inner vascular cylinder.
In contrast, we also identified ILs where there was expansion in the vasculature relative to the outer cortex layer. In IL 1-2 there was an increase in the number of cells in the outer cortex layers but no increase in endodermis, pericycle, and xylem cell numbers. However, the diameter and area of the stele and vascular cylinder were increased, in addition to whole root area. This suggested increased expansion of xylem cells and also potentially expansion of other cell types within the stele. IL 10-2 showed a decreased fraction of stele area relative to whole root area with no change in the number of cells within the xylem (Fig. 7, D–E). There must therefore be decreased expansion of cells within the xylem axis or phloem and procambial cells.
DISCUSSION
Our study uses interspecific variation between S. lycopersicum ‘M82’ and S. pennellii inbred accession LA716 to identify numerous genetic loci that regulate a diversity of root traits. We will discuss the significance of several main findings of this work. First, the results here provide a framework for developmental genetic studies of roots with respect to traits not present in Arabidopsis. Second, we highlight our comprehensive dissection of the S. pennellii genome and its ability to regulate root length by coordinating cell division, elongation, and their dynamics. Third, we discuss examples of constrained regulation of growth to maintain morphology, and cases where growth between different layers was uncoupled. Finally, we discuss the adaptive significance of several of these root traits.
The detailed characterization of the developmental ontogeny of root development in both species establishes a framework for developmental genetic analysis of this rising model organism. Genetic variation in novel traits that have never, to our knowledge, been described with respect to tomato or Arabidopsis root development were characterized. These include consistent root growth away from the gravity vector that is distinct from agravitropic growth responses (Michniewicz et al., 2007). A “gravitropic set angle” had been previously theoretically proposed (Digby and Firn, 1995), and our data supports the existence of such a growth response that is genetically variable. The presence of genetic variation leading to an additional cortex layer, an increase in QC cell number, and repeated periclinal divisions of the CEI daughters provide an excellent system to study these features that are common in both dicot and monocot plant crop species, but not in Arabidopsis. In fact, two potentially genetically separable components of QC development were observed—cell number and niche organization. In cv M82, there are three tiers of cells, whereas in S. pennellii there are two tiers with fewer cells. In contrast, only a single tier of two cells in the medial, longitudinal axis of Arabidopsis has been described, while an amorphous set of hundreds of cells in the maize (Zea mays) QC has been observed (Jiang et al., 2003). Further analysis of this IL population can facilitate the eventual identification of genes that regulate proliferation within the QC as well as the tiered organization of the QC.
The spatiotemporal division patterns by which the CEI regulates the formation of multiple cortex layers is also quite striking. In the case of this CEI in cv M82, it appears that there are three distinct formative divisions of a stem cell that give rise to three cortex layers and one endodermis layer. Therefore, the third cortex layer in cv M82 is not the same as the middle cortex, although it should be noted that the middle cortex was identified in similar proportions in both species. Interestingly, the periclinal divisions of the CEI daughters are all morphologically symmetric as opposed to the morphologically asymmetric division, which is characteristic of the Arabidopsis CEI. Thus it appears that there are two transit amplifying populations (proliferative stem cell progeny) within these cell layers—the first is the anticlinal divisions that occur between division of the CEI and its daughters that will undergo periclinal divisions, and the second is within the root meristematic zone (Scheres, 2007). In Arabidopsis and rice (Oryza sativa), the SHORT-ROOT (SHR) transcription factor, in conjunction with its downstream target and physically interacting partner SCARECROW (SCR), regulates the asymmetric division of the CEI. In Arabidopsis this is accomplished via several feedforward loops involving SHR, SCR, RETINOBLASTOMA-RELATED, CYCLIN D6;1, and auxin (Cui et al., 2007; Sozzani et al., 2010). It will be interesting to determine whether these molecules will play a conserved role in these divisions in tomato.
Our detailed analysis of root growth provides some interesting perspective on the mechanisms controlling root length between these two species. There are many different S. pennellii loci that decrease root length and that increase root length transgressively from cv M82. Many of these are major-effect loci ranging from 24% to 58% of the variation observed, and at least from the four ILs characterized in increased detail, regulation of root length is achieved by multiple mechanisms including control of cell division within the root meristem as well as by controlling the rate of exit from the root meristem into the elongation zone and by root expansion. Likely, many of the QTLs identified also regulate additional aspects of root growth including limits on cell expansion and control of QC size—precocious differentiation of the root’s QC can result in determinate root growth and a short root phenotype (Sena et al., 2004; Moubayidin et al., 2010; Sozzani et al., 2010).
Regulation of morphology or shape by tightly controlled division and expansion has been closely studied in both the Arabidopsis leaf and the Drosophila spp. wing. In both of these cases, reduction of cell division by down-regulation of a Cell Division Cycle2 kinase resulted in increased expansion of underlying cells to maintain overall morphology and an increase in cell division by overexpression of a Drosophila E2F transcription factor resulted in a decrease of cell expansion again to maintain morphology (Hemerly et al., 1995; Weigmann et al., 1997; Neufeld et al., 1998). In the Drosophila spp. wing, targeted disruption of growth in defined tissues resulted in changes in growth and cell proliferation in neighboring tissues likely by unidentified cell nonautonomous mechanisms (Mesquita et al., 2010). Further support for the strong regulation of overall morphology came from a quantitative genetic study of variation in wing shape and cell number showing that the genetic correlation of wing shape across sexes and environments is considerably higher than that of cell division for individual cell types, indicating that cell division is more variable than the compensation mechanisms (Birdsall et al., 2000). In our study we identified significant genetic variation in cell division and size and some examples of conserved compensation between outer tissue layers and the inner vascular cylinder. However, we also found ILs that had no apparent compensation to maintain overall root morphology and its proportions. The ability to compensate between layers is therefore genetically programmed and variable between species. This observation suggests several possible mechanisms: (1) a cell nonautonomous mechanism that is genetically variable, or (2) an altered set point for compensation. Thus, these loci likely contain genes that are responsible for regulating compensation in expansion between neighboring tissues, presumably also by cell nonautonomous mechanisms. The extensive amount of genetic variability in tissue compensation and cell numbers and cell expansion suggests that these traits are partially independent and potentially have arisen through the different selection pressures experienced leading to wild and domesticated tomato.
The ecology of the tomato species provides some perspective as to the potential adaptive significance of the gravity and root length traits. S. pennellii inbred accession LA716 is derived from the coastal desert with rocky soils (Moyle, 2008). The short root phenotype with a growth angle away from the gravity vector may have developed to facilitate growth in an environment with thin top soil and rocky terrain, as well as to enable efficient capture of water in the desert environment. S. pennellii is drought resistant (Pillay and Beyl, 1990; Easlon and Richards, 2009; Ziaf et al., 2011), and water restriction experiments with short root or angled ILs can certainly test this hypothesis. Furthermore, the identification of lines that were able to uncouple the root length phenotype from the growth angle phenotype can help determine the contribution of each trait to such a drought resistant phenotype. Three remaining questions exist with respect to the growth angle trait: Was this trait selected against during domestication? What is the molecular mechanism by which it occurs? What would the adaptive advantage of this trait be? The first question can be answered by determining whether this trait is found within other tomato species at a variety of positions within the Solanum section of the Lycopersicon phylogeny (Moyle, 2008) and particularly whether it is found in Solanum pimpinellifolium, which is likely the most recent wild ancestor of the domesticated tomato species (Tomato Genome Consortium, 2012). Identification of the genes underlying the loci that regulate this growth response will allow us to test for molecular signatures of past selection such as elevated nonsynonymous/synonymous substitution ratios or allele frequency changes (Nielsen, 2005; Oleksyk et al., 2010). Finally, identifying the genes underlying these loci will determine the molecular mechanism underlying this response and provide insight into what degree it involves manipulation of classic growth response hormone pathways, like those involved in auxin signaling. Transcriptional regulation has been shown to play a major role in distinct aspects of root growth and development (Lee and Schiefelbein, 1999; Nakajima et al., 2001; Bonke et al., 2003; Sena et al., 2004; Nawy et al., 2005; Levesque et al., 2006), so identification of expression QTL regulating these responses may facilitate such endeavors.
In summary, the work presented here will be of great importance in exploring the molecular mechanisms by which root development has evolved with respect to root morphology and cellular anatomy, as well as the adaptive significance of these developmental traits.
MATERIALS AND METHODS
Plant Material and Growth Conditions
The IL population comprises 76 lines, each containing a single chromosomal segment of the wild tomato Solanum pennellii inbred accession LA716 in the genetic background of the cultivated tomato Solanum lycopersicum ‘M82’. The IL population provides a complete coverage of the wild species genome. The development and genetic characterization of the ILs have been described by Eshed and Zamir (1995).
Tomato roots were grown on Murashige and Skoog (MS) plates containing 4.3 g L–1 MS (Caisson, catalog no. MSP01–50LT), 0.5 g L−1 2-(N-morpholino)ethanesulfonic, 10 g L−1 Suc, pH = 5.8, and 8 g L−1 Agar (Difco, catalog no. 214530). Seeds were placed in tissue-embedding cassettes and surface sterilized in 70% ethanol for 2 min, then in 3% hypochlorite for 20 min, followed by three washes with distilled water. Seven sterilized seeds were placed on each MS plate in a row 2.5 cm from the top of a square 12- × 12-cm plate, and then sealed with a 3M surgical tape. In each experiment at least three plates for each line were grown. Plates were placed vertically in a rack in a randomized block design, and the experiments were carried out in a growth chamber with a 16:8 light:dark cycle at 22°C and 50%–75% humidity with a light intensity of 100 μE. The germination date was monitored for each seed. Roots were phenotyped at 5 d after germination for root length and growth angle experiments, at 7 d after germination for cellular anatomy experiments, and 3 and 6 d post germination for the gravitropic response experiment. Plates were opened only once to ensure same growth conditions.
Microscopy
Bright field microscopy
Imaging was performed using an Olympus Vanox microscope (root cross sections). Images were captured with a PIXERA Pro-600ES camera or a Zeiss Axioplan Imaging 2 microscope (amyloplast staining, characterization of the stem cell niche). Axioplan images were captured with a CCD Zeiss camera using the AxioVision 4.8 software.
Differential interference contrast optics
Differential interference contrast (DIC) optics were used with a Zeiss Axioplan imaging 2 microscope (amyloplast staining, characterization of the stem cell niche).
Confocal laser scanning microscopy
Optical sections through whole root tips and root transverse sections were obtained using an Olympus FV1000 confocal laser scanning microscope (Olympus America) which was equipped with a second, photokinetic scan head (405-nm laser line) and two spectral scan emission detectors. Objective lenses of 40× with optical zoom of 1.3× or 60× with no optical zoom were used; images and image stacks were captured at an 800 × 800 pixel resolution using Nyquist settings, resulting in 0.70- to 1.0-micron-thick optical sections (suberin staining, cell wall imaging, root stem cell niche).
High-Throughput Histological Analysis
Tissue embedding and sectioning
A section 1 cm long flanking the midlength of 7-d-old roots (for all cellular trait experiments) was cut, and 1 cm of the root tip (to determine whether the extra cortex layer was found throughout the root for the parents) was obtained. Three to five root segments were bundled together and placed in a 1.5-ml conical tube containing 3% molten agarose at 60°C with the apical end facing down. After the agarose solidified, the bottom of the tube was cut with a hot scalpel and the agarose plugs (approximately 2 cm) containing the root segments were placed in glass vials containing formaldehyde-acetic acid fixative (70% ethanol, 5% acetic acid, 5% formaldehyde) for at least 48 h at 4°C followed by dehydration in sequential ethanol dilutions (75%, 50%, 30%, 10%) for 30 min each then placed in dH2O. Sections (approximately 120 µm) were made using a Vibratom Series 1000 sectioning system. Approximately 20 sections from each plug were placed in a 12-well plate and stained with 0.05% toluidine blue (Ruzin, 1999) for 10 s followed by destaining with distilled water. Images were observed under light microscopy (see above for microscope type) and were taken in 20× magnification using a PIXERA Pro-600ES camera.
Suberin Staining to Identify the Endodermis and Exodermis
Staining of root sections was done according to Brundrett et al. (1988). In short, root agarose cross sections (300 µm) were placed in a small weighing boat and incubated overnight at room temperature with 0.1% (w/v) berberine hemisulfate (Sigma) dissolved in distilled water. The sections were rinsed three times with distilled water and counterstained with 0.5% (w/v) aniline blue in distilled water for 10 min at room temperature. The sections were rinsed again as above and transferred into 0.1% (w/v) FeCl3 in 50% (v/v) glycerine. After 10 min in this solution the sections were transferred to a microscope slide and mounted in the same solution. Roots were viewed using confocal laser scanning microscopy with the Olympus FV1000. Root transverse sections for suberin staining were imaged using the fluorescein isothiocyanate preset (excitation: 488 nm) with emission capture to all wavelengths greater than 510 nm. This setting was sufficient to detect the berberine staining on lignified and suberized cell walls.
Amyloplast Staining to Define the Base of the QC
For visualizing starch in whole roots, the roots (3 to 4 cm from the tip) were fixed for 1 to 2 min in 3:1 ethanol:acetic acid solution, then only the root tip was dipped for 3 s in Lugol staining solution (1.5 g potassium iodide, 0.5 g iodine in 30 mL distilled water). Roots were mounted in chloral-hydrate solution (8:3:1 chloral-hydrate:distilled water:glycerol). Roots were observed under a Zeiss Axioplan imaging 2 microscope, and DIC was used. Images were captured with a CCD Zeiss camera using the AxioVision 4.8 software. Cells above the first tier of cells with amyloplasts are the columella stem cell initials. Cells above the columella stem cells are the base of the QC.
Root Cell Wall Imaging Using Calcofluor
Tips (1 to 1.5 cm) of 5- to 7-d-old roots were fixed with 3.7% formaldehyde, 0.1% Triton-X in 1× phosphate-buffered saline (PBS) followed by staining with 0.1% calcofluor in 1× PBS for 1 h then rinsed with PBS and mounted with ProLongGold antifade reagent for clearing (Invitrogen) and examined with confocal imaging as described with the Olympus FV1000. To detect cell walls, roots stained with calcofluor were imaged using the 4′,6-diamino-phenylindole preset (excitation: 405 nm, emission: 450 nm).
Identification of Cells Undergoing S-Phase and All Root Nuclei
Click-iT EdU Alexa Fluor488 cell proliferation assay (Invitrogen) was used according to Kotogány et al. (2010). In short, tips (1 to 1.5 cm) of 5- to 7-d-old roots were submerged in 20 µm EdU in distilled water for 3 h then fixed for 30 min (3.7% formaldehyde, 0.1% Triton-X in 1× PBS) at room temperature. Material was rinsed with PBS (3 × 10 min), and root tips were incubated in the EdU detection cocktail for 30 min followed by PBS washes (3 × 10 min). To visualize all nuclei 0.1% (v/v) of To-Pro3 dye (Invitrogen) was added to the last wash, and roots were incubated for 1 h to overnight. To detect cells undergoing the S-phase of the cell cycle in addition to the cell wall being stained with calcofluor using confocal laser scanning microscopy on the Olympus FV1000, the 4′,6-diamino-phenylindole setting was employed to image cell walls (calcofluor), the Alexa Fluor488 setting (excitation: 488 nm, emission: 519 nm) was employed to image EdU (nuclei in S-phase), and the Cyanine 5 setting (excitation: 673 nm, emission: 707 nm) was used to image all nuclei.
QCs were identified by a lack of EdU staining and being present above the tier of the columella initial cells.
Characterization of the Stem Cell Niche
To characterize the root QC and CEI cell division patterns, whole roots were scanned using confocal laser scanning microscopy with the Olympus FV1000 in order to determine the median plane through the root tip. In addition, at least 10 to 30 optical slices per stack were captured around the median plane in order to ensure that the center of the root apical meristem was imaged. The median plane through the root apex was further assessed based on the symmetry of the locations of CEIs relative to the root tip. Image stacks were processed in the Olympus FluoView software (version 2.04), and images were exported to ImageJ (version 1.45s, National Institutes of Health [http://rsbweb.nih.gov/ij/]) or Adobe Photoshop CS5. In all cases, images were modified uniformly using the γ setting in FluoView, and/or by adjusting the location of the bottom and/or top of the histogram in the look-up table in FluoView or the equivalent levels of modification in Photoshop and ImageJ. To determine the number of anticlinal divisions between periclinal division 1 and 2 in Figure 3E, we counted nine roots (cv M82) and 12 roots (S. pennellii).
Root Length, Growth Angle, and Cellular Architecture Data Acquisition from Images
Plates containing the roots were scanned with Epson Perfection 4490 scanner at 300 dots per inch resolution next to a ruler. Phenotyping of roots was performed on the root images using ImageJ software (http://rsbweb.nih.gov/ij/index.html). Root lengths and diameters were traced using the segmented line tool. The angle of root growth was calculated using the angle tool, cell number was calculated using the cell counter plug-in, and area was calculated using the polygon selection tool.
Meristem Cell Length Measurements
For all meristem measurements, n = 13 roots for cv M82 and n = 16 roots for S. pennellii. For meristematic cell lengths, n = 34 cells for cv M82 and n = 33 cells for S. pennellii. For mature cell lengths, n = 22 cells for cv M82 and n = 34 for S. pennellii. Roots were mounted in chloral-hydrate solution (8:3:1 chloral-hydrate:distilled water:glycerol). Images were obtained with DIC optics using a Zeiss Axioskop 2 microscope.
Cellochron Analysis
We carried out cellochron analysis by calculating local cell length (mm) in the root maturation zone relative to local cell velocity (mm h–1; Silk et al., 1989; Beemster and Baskin, 1998). A cellochron (c) represents the time interval during which a new cell is added to a file within the elongation zone. The ratio of mature cell length to root extension rate was used to evaluate c at the base of the elongation zone, the point at which the rate of cell expansion surpasses the rate of cell division. Roots were mounted in chloral-hydrate solution (8:3:1 chloral-hydrate:distilled water:glycerol). Images were obtained with DIC optics using a Zeiss Axioskop 2 microscope. Mature cell length was acquired from the images of the root above the elongation zone. Individual cells of each genotype (range: n = 10, n = 34) were measured using the line tool in ImageJ. Cell length values were then divided by organ extension velocity, determined by dividing root lengths for each genotype by the number of hours from germination (mm h–1). Error bars represent 95% bootstrap confidence intervals of error distributions created by sampling ratios of mature cell length and growth velocity data.
Statistical Data Analysis
Experimental design: root length and angle across all ILs and parents
Root length and angle were measured across five independent experiments. In each experiment a minimum of three individual roots were sampled. Each root can be considered as a biological replicate. For root length, we had a minimum of eight biological replicates and a maximum of 79 biological replicates with a mean of 40 ± 14 replicates. For root angle, a minimum of eight biological replicates and a maximum of 42 biological replicates with a mean of 23 ± 8 biological replicates were sampled. The angle was defined as that of the gravity vector to the line from the center of the root tip to the center of the root halfway back to the first obvious bend.
Experimental design: root angle in response to gravity stimulus
The angle was measured just prior to plate rotation and 3 d after plate rotation.
Experimental design: cellular measurements of the parents
For cellular measurements to assess whether there were differences in the parents, a minimum of 12 biological replicates and a maximum of 14 biological replicates for cv M82 were analyzed, and for S. pennellii, a minimum of nine biological replicates and a maximum of 11 biological replicates were analyzed (Fig. 2). For analysis of cell number, area, and circumferences, the experimental effect was assessed using a linear fixed-effect model via the lm and ANOVA functions in the stats package within the R statistical programming language (R Development Core Team, 2012). The specific model that was fit to the data were TRT = GEN + EXP + GEN*EXP + e (where TRT is the trait value being fitted, GEN is the genotype fitted as a fixed effect, EXP is the experiment fitted as a random effect, and e is the error; Supplemental Table S1). For this analysis we considered EXP a fixed effect to maximize our ability to test whether there was a significant impact of experiment on the traits. Genotype contributes to the majority of the variation observed, as determined by the sum of squares of GEN relative to those for EXP or GEN × EXP. From Supplemental Table S1, the average sum of squares for GEN × EXP was 3.5% of that for genotype. To assess the amount of variation attributable to each factor, a linear random-effect model was fit to the same data using the lme function in the lme4 package (Bates et al., 2012) with the same model formula (Supplemental Table S2). We also modeled genotype and experiment as random effects and determined the proportion of variance attributable to each and to the GEN × EXP interaction (Supplemental Table S2). Here the GEN × EXP variance was on average 7.8% of the variance contributed by genotype alone. Both of these modeling approaches suggest that experiment is not an influential effect on the ability to detect QTLs for these phenotypes.
Statistical analysis: identification of significant length and growth angle QTLs
Only 72 ILs had a minimum of three biological replicates (Supplemental Data Set S1). S. pennellii and cv M82 were included as controls in each experiment. Where cv M82 and S. pennellii were represented in multiple experiments, cv M82 and S. pennellii least squares means (means across experiments of the predicted values for each genotype) were computed by fitting data to the linear fixed-effect model TRT = EXP + GEN using the lm function and predicting means with the predict function in the stats package. For QTL detection purposes, the data for a specific IL is only compared with the cv M82 controls present in the same experiment within which that IL was measured. Phenotype data were analyzed using the R statistical programming language (R Development Core Team, 2012) and mixed linear models with the ANOVA function in the stats package and the lme function in the nmle package (Pinheiro et al., 2013) were fit to the data. The final model used was TRT = GEN + EXP + e. Multiple testing correction was done post hoc using Dunnett’s test with the glht and mcp functions in the multcomp package (Hothorn et al., 2008) across all the IL/cv M82 comparisons. A bar plot was made for each trait, showing means across all sampled roots for each genotype including cv M82 and S. pennellii and using bar color to indicate significance of difference of trait value compared with cv M82, and those significance values were also depicted with colors in heat maps whose axes were genotype and trait.
Statistical analysis: identification of significant cellular QTLs
In order to measure the cellular traits, the introgression lines were divided across three different experiments. To identify significant cellular QTLs, data from each experiment were analyzed independently, and the cellular trait of an IL in one experiment was compared to that trait in cv M82 within the same experiment. For each IL there were a minimum of three biological replicates and a maximum of 14 biological replicates. Appropriate statistical analysis of cellular phenotypes required testing for potential effects of the situational variables “plug” (several roots bundled together in a group), “section” (a thin section of a plug), and “individual” (an individual root within the plug, or a single biological replicate) and a derived variable “genoplug” (genotype and plug). These were analyzed using linear mixed-effect models with the lme function and the models then compared using the ANOVA function. The three models were TRT = GEN + GPG/IND/SEC, TRT = GEN + GPG/IND, and TRT = GEN + IND. The models all used genotype (GEN) as a fixed effect while the other variables (GPG for genoplug, IND for individual, SEC for section) were nested random effects. No significant random effects of GPG, IND, or SEC were found, and as such we utilized the simplest model, TRT = GEN + e, to test whether a specific IL was significantly different from cv M82. Again, multiple testing correction was done post hoc using Dunnett’s test with the glht and mcp functions in the multcomp package (Hothorn et al., 2008) across all 76 IL/cv M82 comparisons.
To assess the genetic correlation between cellular phenotypes across the ILs, we conducted a partial Pearson correlation of each trait pair while accounting for the different experiments in which the ILs were measured, using the pcor.test function in the ggm package (Marchetti et al., 2012). S. pennellii was not included as it was an outlier for all measurements. The resulting partial Pearson correlation moments were then used to generate a heat map of all pairwise phenotypic correlations. R scripts are available in the software section of the lab Web site at http://www-plb.ucdavis.edu/labs/brady/software.html.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. cv M82 and S. pennellii
Supplemental Figure S2. Characterization of QC niche.
Supplemental Figure S3. DIC image of root tips.
Supplemental Figure S4. Number of ILs that show a change in cell number.
Supplemental Table S1. ANOVA table output.
Supplemental Table S2. lm model output.
Supplemental Table S3. Trait means and P values.
Supplemental Table S4. Partial Pearson correlations of trait means.
Supplemental Data Set S1. Bar graphs of cellular trait means.
Acknowledgments
We thank Neelima Sinha, Dan Chitwood, and Lauren Hughes for supplying valuable plant material and good discussion; Gloria Muday and Greg Maloney for good discussion; Yasunori Ichihashi for advice on the EdU assay; and the Tomato Genetics Resource Center at the University of California, Davis, for supplying seeds. We thank Jaimie Van Norman and Terri Long for critical review of the manuscript. We thank Pamela Riley for excellent greenhouse assistance, and Mallorie Taylor-Teeples, Nell Laub, Marla McPherson, Natasha Worden, Kristine Santos, Quynh Dinh, Duy Nguyen, Adi Chatow, Vasuda Chauhan, and Demelsa Menendez for excellent technical assistance. We also thank Jason Corwin for valuable statistical discussions and assistance with R. We thank Wendy Silk for assistance in the kinematic analysis.
Glossary
- QTL
quantitative trait locus
- IL
introgression lines
- MS
Murashige and Skoog medium
- QC
quiescent center
- DIC
differential interference contrast
- PBS
phosphate-buffered saline
- CEI
cortex-endodermis initial
References
- Bates D, Maechler M, Bolker B (2012). lme4: Linear mixed-effects models using S4 classes. R package version 0.999999-0. http://CRAN.R-project.org/package=lme4 (November 13, 2012)
- Beemster GT, Baskin TI. (1998) Analysis of cell division and elongation underlying the developmental acceleration of root growth in Arabidopsis thaliana. Plant Physiol 116: 1515–1526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benfey PN, Scheres B. (2000). Root development. Curr Biol 10: R813–815 [DOI] [PubMed] [Google Scholar]
- Bettey M, Finch-Savage WE, King GJ, Lynn JR. (2000) Quantitative genetic analysis of seed vigour and pre-emergence seedling growth traits in Brassica oleracea. New Phytol 148: 277–286 [Google Scholar]
- Birdsall K, Zimmerman E, Teeter K, Gibson G. (2000) Genetic variation for the positioning of wing veins in Drosophila melanogaster. Evol Dev 2: 16–24 [DOI] [PubMed] [Google Scholar]
- Bloch D, Monshausen G, Singer M, Gilroy S, Yalovsky S. (2011) Nitrogen source interacts with ROP signalling in root hair tip-growth. Plant Cell Environ 34: 76–88 [DOI] [PubMed] [Google Scholar]
- Bonke M, Thitamadee S, Mähönen AP, Hauser MT, Helariutta Y. (2003) APL regulates vascular tissue identity in Arabidopsis. Nature 426: 181–186 [DOI] [PubMed] [Google Scholar]
- Brundrett MC, Enstone DE, Peterson CA. (1988) A berberine-aniline blue fluorescent staining procedure for suberin, lignin, and callose in plant-tissue. Protoplasma 146: 133–142 [Google Scholar]
- Cui H, Levesque MP, Vernoux T, Jung JW, Paquette AJ, Gallagher KL, Wang JY, Blilou I, Scheres B, Benfey PN. (2007) An evolutionarily conserved mechanism delimiting SHR movement defines a single layer of endodermis in plants. Science 316: 421–425 [DOI] [PubMed] [Google Scholar]
- Dehan K, Tal M. (1978) Salt tolerance in the wild relatives of the cultivated tomato: Responses of Solanum pennellii to high salinity. Irrig Sci 1: 71–76 [Google Scholar]
- Digby J, Firn RD. (1995) The gravitropic set-point angle (GSA): the identification of an important developmentally controlled variable governing plant architecture. Plant Cell Environ 18: 1434–1440 [DOI] [PubMed] [Google Scholar]
- Dolan L, Janmaat K, Willemsen V, Linstead P, Poethig S, Roberts K, Scheres B. (1993) Cellular organisation of the Arabidopsis thaliana root. Development 119: 71–84 [DOI] [PubMed] [Google Scholar]
- Easlon HM, Richards JH. (2009) Drought response in self-compatible species of tomato (Solanaceae). Am J Bot 96: 605–611 [DOI] [PubMed] [Google Scholar]
- Eshed Y, Zamir D. (1995) An introgression line population of Lycopersicon pennellii in the cultivated Tomato enables the identification and fine mapping of yield-associated QTL. Genetics 141: 1147–1162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fita A, Picó B, Monforte AJ, Nuez F. (2008) Genetics of root system architecture using near-isogenic lines of melon. J Am Soc Hortic Sci 133: 448–458 [Google Scholar]
- Fitz Gerald JN, Lehti-Shiu MD, Ingram PA, Deak KI, Biesiada T, Malamy JE. (2006) Identification of quantitative trait loci that regulate Arabidopsis root system size and plasticity. Genetics 172: 485–498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heimsch C, Seago JL., Jr (2008) Organization of the root apical meristem in angiosperms. Am J Bot 95: 1–21 [DOI] [PubMed] [Google Scholar]
- Hemerly A, Engler JdeA, Bergounioux C, Van Montagu M, Engler G, Inzé D, Ferreira P. (1995) Dominant negative mutants of the Cdc2 kinase uncouple cell division from iterative plant development. EMBO J 14: 3925–3936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hothorn T, Bretz F, Westfall P. (2008) Simultaneous inference in general parametric models. Biom J 50: 346–363 [DOI] [PubMed] [Google Scholar]
- Jiang K, Meng YL, Feldman LJ. (2003) Quiescent center formation in maize roots is associated with an auxin-regulated oxidizing environment. Development 130: 1429–1438 [DOI] [PubMed] [Google Scholar]
- Khan N, Kazmi RH, Willems LAJ, van Heusden AW, Ligterink W, Hilhorst HWM. (2012) Exploring the natural variation for seedling traits and their link with seed dimensions in tomato. PLoS ONE 7: e43991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koca H, Ozdemir F, Turkan I. (2006) Effect of salt stress on lipid peroxidation and superoxide dismutase and peroxidase activity of Lycopersicon esculentum and L. pennellii. Biol Plant 50: 745–748 [Google Scholar]
- Kotogány E, Dudits D, Horváth GV, Ayaydin F. (2010) A rapid and robust assay for detection of S-phase cell cycle progression in plant cells and tissues by using ethynyl deoxyuridine. Plant Methods 6: 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MM, Schiefelbein J. (1999) WEREWOLF, a MYB-related protein in Arabidopsis, is a position-dependent regulator of epidermal cell patterning. Cell 99: 473–483 [DOI] [PubMed] [Google Scholar]
- Levesque MP, Vernoux T, Busch W, Cui H, Wang JY, Blilou I, Hassan H, Nakajima K, Matsumoto N, Lohmann JU, et al. (2006) Whole-genome analysis of the SHORT-ROOT developmental pathway in Arabidopsis. PLoS Biol 4: e143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loudet O, Gaudon V, Trubuil A, Daniel-Vedele F. (2005) Quantitative trait loci controlling root growth and architecture in Arabidopsis thaliana confirmed by heterogeneous inbred family. Theor Appl Genet 110: 742–753 [DOI] [PubMed] [Google Scholar]
- Mähönen AP, Bonke M, Kauppinen L, Riikonen M, Benfey PN, Helariutta Y. (2000) A novel two-component hybrid molecule regulates vascular morphogenesis of the Arabidopsis root. Genes Dev 14: 2938–2943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malamy JE, Ryan KS. (2001) Environmental regulation of lateral root initiation in Arabidopsis. Plant Physiol 127: 899–909 [PMC free article] [PubMed] [Google Scholar]
- Marchetti GM, Drton M, Sadeghi K (2012). ggm: A package for Graphical Markov Models. R package version 1.995-3. http://CRAN.R-project.org/package=ggm (March 7, 2013)
- Mesquita D, Dekanty A, Milán M. (2010) A dp53-dependent mechanism involved in coordinating tissue growth in Drosophila. PLoS Biol 8: e1000566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michniewicz M, Brewer PB, Friml JI. (2007). Polar auxin transport and asymmetric auxin distribution. The Arabidopsis Book 5: e0108, doi/10.1199/tab.0108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moubayidin L, Perilli S, Dello Ioio R, Di Mambro R, Costantino P, Sabatini S. (2010). The rate of cell differentiation controls the Arabidopsis root meristem growth phase. Curr Biol 20: 1138–1143 [DOI] [PubMed] [Google Scholar]
- Mouchel CF, Briggs GC, Hardtke CS. (2004) Natural genetic variation in Arabidopsis identifies BREVIS RADIX, a novel regulator of cell proliferation and elongation in the root. Genes Dev 18: 700–714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moyle LC. (2008) Ecological and evolutionary genomics in the wild tomatoes (Solanum sect. Lycopersicon). Evolution 62: 2995–3013 [DOI] [PubMed] [Google Scholar]
- Nakajima K, Sena G, Nawy T, Benfey PN. (2001) Intercellular movement of the putative transcription factor SHR in root patterning. Nature 413: 307–311 [DOI] [PubMed] [Google Scholar]
- Nawy T, Lee J-Y, Colinas J, Wang JY, Thongrod SC, Malamy JE, Birnbaum K, Benfey PN. (2005) Transcriptional profile of the Arabidopsis root quiescent center. Plant Cell 17: 1908–1925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neufeld TP, de la Cruz AF, Johnston LA, Edgar BA. (1998) Coordination of growth and cell division in the Drosophila wing. Cell 93: 1183–1193 [DOI] [PubMed] [Google Scholar]
- Nielsen R. (2005) Molecular signatures of natural selection. Annu Rev Genet 39: 197–218 [DOI] [PubMed] [Google Scholar]
- Oleksyk TK, Smith MW, O’Brien SJ. (2010) Genome-wide scans for footprints of natural selection. Philos Trans R Soc Lond B Biol Sci 365: 185–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillay I, Beyl C. (1990) Early responses of drought-resistant and -susceptible tomato plants subjected to water stress. J Plant Growth Regul 9: 213–219 [Google Scholar]
- Pinheiro J, Bates D, DebRoy S, Sarkar D, R Development Core Team (2013). nlme: Linear and Nonlinear Mixed Effects Models. R package version 3.1-108.
- R Development Core Team (2012). R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. http://www.R-project.org/ (November 13, 2012)
- Reymond M, Svistoonoff S, Loudet O, Nussaume L, Desnos T. (2006) Identification of QTL controlling root growth response to phosphate starvation in Arabidopsis thaliana. Plant Cell Environ 29: 115–125 [DOI] [PubMed] [Google Scholar]
- Ruzin SE (1999). Plant Microtechnique and Microscopy. Oxford University Press, New York [Google Scholar]
- Saucedo M, Ponce G, Campos ME, Eapen D, García E, Luján R, Sánchez Y, Cassab GI. (2012) An altered hydrotropic response (ahr1) mutant of Arabidopsis recovers root hydrotropism with cytokinin. J Exp Bot 63: 3587–3601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheres B. (2007) Stem-cell niches: nursery rhymes across kingdoms. Nat Rev Mol Cell Biol 8: 345–354 [DOI] [PubMed] [Google Scholar]
- Schiefelbein J, Kwak SH, Wieckowski Y, Barron C, Bruex A. (2009) The gene regulatory network for root epidermal cell-type pattern formation in Arabidopsis. J Exp Bot 60: 1515–1521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sena G, Jung JW, Benfey PN. (2004) A broad competence to respond to SHORT ROOT revealed by tissue-specific ectopic expression. Development 131: 2817–2826 [DOI] [PubMed] [Google Scholar]
- Shindo C, Bernasconi G, Hardtke CS. (2007) Natural genetic variation in Arabidopsis: tools, traits and prospects for evolutionary ecology. Ann Bot (Lond) 99: 1043–1054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silk WK, Lord EM, Eckard KJ. (1989) Growth patterns inferred from anatomical records: empirical tests using longisections of roots of Zea mays L. Plant Physiol 90: 708–713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sozzani R, Cui H, Moreno-Risueno MA, Busch W, Van Norman JM, Vernoux T, Brady SM, Dewitte W, Murray JA, Benfey PN. (2010) Spatiotemporal regulation of cell-cycle genes by SHORTROOT links patterning and growth. Nature 466: 128–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomato Genome Consortium (2012) The tomato genome sequence provides insights into fleshy fruit evolution. Nature 485: 635–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigmann K, Cohen SM, Lehner CF. (1997) Cell cycle progression, growth and patterning in imaginal discs despite inhibition of cell division after inactivation of Drosophila Cdc2 kinase. Development 124: 3555–3563 [DOI] [PubMed] [Google Scholar]
- Ziaf K, Loukehaich R, Gong P, Liu H, Han Q, Wang T, Li H, Ye Z. (2011) A multiple stress-responsive gene ERD15 from Solanum pennellii confers stress tolerance in tobacco. Plant Cell Physiol 52: 1055–1067 [DOI] [PubMed] [Google Scholar]