There is a phase of diversifying selection of duplicated genes following function relaxation after whole-genome duplication.
Abstract
Episodes of whole-genome duplication (WGD) followed by gene loss dominate the evolutionary history of flowering plants. Despite the importance of understanding gene evolution following WGD, little is known about the evolutionary dynamics of this process. In this study, we analyzed duplicated genes from three WGD events in the Arabidopsis (Arabidopsis thaliana) lineage using multiple data types. Most duplicated genes that have survived from the most recent WGD (α) are under purifying selection in modern Arabidopsis populations. Using the number of identified protein-protein interactions as a proxy for functional divergence, approximately 92.7% of α-duplicated genes were diverged in function from one another in modern Arabidopsis populations, indicating that their preservation is no longer explicable by dosage balance. Dosage-balanced retention declines with antiquity of duplication: 24.1% of α-duplicated gene pairs in Arabidopsis remain in dosage balance with interacting partners, versus 12.9% and 9.4% for the earlier β-duplication and γ-triplication. GO-slim (a cut-down version of gene ontologies) terms reinforce evidence from protein-protein interactions, showing that the putatively diverged gene pairs are adapted to different cellular components. We identified a group of α-duplicated genes that show higher than average single-nucleotide polymorphism density, indicating that a period of positive selection, potentially driving functional divergence, may have preceded the current phase of purifying selection. We propose three possible paths for the evolution of duplicated genes following WGD.
With a growing number of genome sequences available, whole-genome duplication (WGD) has been found to be widespread in the evolutionary history of many species (Wolfe and Shields, 1997; Lynch and Conery, 2000; Bowers et al., 2003; Dehal and Boore, 2005). WGD survives more frequently in plants than in animals (Li, 1997), and most evidence comes from the study of angiosperm (flowering plant) genomes (Tang et al., 2008). In the eudicot lineage, an ancient whole-genome triplication event predates the split of Arabidopsis (Arabidopsis thaliana; Bowers et al., 2003), papaya (Carica papaya; Ming et al., 2008), soybean (Glycine max; Schmutz et al., 2010), poplar (Populus trichocarpa; Tuskan et al., 2006), and grape (Vitis vinifera; Jaillon et al., 2007). Following this ancient hexaploidy, two additional WGDs occurred independently in both the Arabidopsis and soybean lineages, and one in the poplar lineage, but none in grape or papaya. In the monocot lineage, an ancient WGD estimated at approximately 70 million years ago predated the divergence of the cereals (Paterson et al., 2004), with a more ancient duplication also evident (Tang et al., 2010). Besides the two ancient WGDs, a more recent WGD (approximately 11.9 million years ago) was found in maize (Zea mays; Blanc and Wolfe, 2004a; Swigonová et al., 2004), and many additional grasses are neopolyploids or recent paleopolyploids. Two additional ancestral WGDs may have contributed to the rise of seed plants and flowering plants (Jiao et al., 2011).
Gene duplication has been widely accepted as an important factor in evolution. Many models have been proposed to account for this process, including neofunctionalization (Ohno, 1970), subfunctionalization (Force et al., 1999), adaptive conflict (Hughes, 1994; Des Marais and Rausher, 2008), dosage balance (Papp et al., 2003; Birchler and Veitia, 2007; Liang et al., 2008), benefit of increasing dosage (Romero and Palacios, 1997), paralogous heterozygote advantage (Spofford, 1969; Proulx and Phillips, 2006), and adaptive radiation (Francino, 2005), which have been reviewed in detail (Zhang, 2003; Van de Peer, 2004; Conant and Wolfe, 2008; Ponting, 2008; Innan and Kondrashov, 2010). Early study of gene duplications rarely discriminated between duplication types and treated all duplicated genes as homologous gene sets. More recent studies generally differentiate between genes derived from small-scale duplication and WGD (Davis and Petrov, 2005; Hakes et al., 2007).
Genes duplicated by WGD experience a variety of fates, with the most frequent being nonfunctionalization and/or the loss of one member of such gene pairs. Genes retained in duplicate for long periods after WGD show greater than average representation of members of signal transduction and transcription factor gene families, and genes involved in DNA repair have been preferentially lost in Arabidopsis and rice (Oryza sativa; Blanc and Wolfe, 2004b; Maere et al., 2005; Freeling and Thomas, 2006; Paterson et al., 2006). By one estimate, approximately 57% of the retained duplicated genes are diverged in function following the most recent WGD in Arabidopsis (Blanc and Wolfe, 2004b). Analysis of very recent WGD gene pairs from Arabidopsis reveals reduced levels of nucleotide polymorphism, indicating a possible selective sweep soon after duplication (Moore and Purugganan, 2003). In modern populations, the coding regions of WGD duplicated genes appear to be more evolutionarily constrained than those of singleton genes, perhaps suggesting that some paralogous genes may buffer essential functions at an early stage of duplication (Chapman et al., 2006).
A particularly important model underlying gene evolution following WGD is the dosage-balance model (Birchler and Veitia, 2007), based on the hypothesis that retained duplicated genes are dosage sensitive and depend on each other. Under this model, gene pairs that are retained tend to be balanced in dosage with each other. Loss of only one such gene causes imbalance and decreases fitness. Gene pairs may be duplicated or lost synchronously.
The primary assumption of the dosage-balance model, that the functions of retained genes should remain unchanged throughout evolutionary time, is problematic. For example, this contradicts the finding that 57% of the recent WGD gene pairs have diverged in function in Arabidopsis (Blanc and Wolfe, 2004b). Under the dosage-balance model, retained duplicated genes might also be predicted to have protein interaction partners that are also retained as duplicated genes from the same WGD event, a prediction that can now be tested. Although the dosage-balance hypothesis applies well in gene family evolution in yeast (Saccharomyces cerevisiae; Papp et al., 2003), it has been used widely to account for the preservation of WGD genes in plants (Freeling and Thomas, 2006; Birchler and Veitia, 2010). The much smaller effective population size of plants than yeast, and the associated greater tolerance of mutations that are initially slightly deleterious, may lead to profound differences in the fates of genes and mutations in yeast and higher plants, respectively (Lynch et al., 1995). Furthermore, the dynamics of gene evolution following WGD is not well understood.
Could the dosage-balance hypothesis explain the pattern of retention of duplicated genes following WGD? Do the functions of genes diverge after duplication? Does this process follow genetic drift or is it driven by selection? In this study, we used multiple types of genome-wide data in Arabidopsis to (1) further evaluate the dosage-balance hypothesis; (2) reevaluate gene functional divergence following WGD using protein-protein interaction data; and (3) analyze selection pressure in gene pairs formed by WGD. We propose a model that reconciles these findings with prior theory and results in three general evolutionary paths that may be taken by duplicated genes following WGD.
RESULTS
Gene Function Divergence following WGD
The functional relationship between gene pairs is an important factor in studying gene evolution following WGD. A total of 13,568 nonredundant protein-protein physical interaction pairs involving 5,531 genes to date are verified by various experiments in Arabidopsis (Stark et al., 2006; Arabidopsis Interactome Mapping Consortium, 2011). Based on these data, there are 464 (12.8%), 162 (11.2%), and 76 (14.6%) duplicated gene pairs that are known to have interaction partners for α-, β-, and γ-WGD events, respectively (Table I). To investigate functional divergence between members of duplicated gene pairs, we define a function index based on the genes that they interact with (see “Materials and Methods”). The function of a gene could be indicated by the genes it interacts with. We assume that two duplicated genes share the same group of interaction partners immediately following duplication (Zhang, 2003). As function diverges, we expect the members of a duplicated gene pair to acquire new interaction partners and/or lose original partners. The number of different interaction partners would thus be a measure of the function divergence between duplicated gene pairs (Supplemental Fig. S1).
Table I. Statistics of protein-protein interaction data in different WGD categories.
| WGD | Total No. of Gene Pairs | No. of Genes That Have Interactors | No. of Gene Pairs in Which Only a Single Gene Has Interactors | No. of Gene Pairs in Which Both Genes Have Interactors | No. of Duplicate Gene Pairs Interacting with Each Other | No. of Gene Pairs Showing Dosage Balancea |
|---|---|---|---|---|---|---|
| α | 3,614 | 1,822 | 894 | 464 | 69 | 871 |
| β | 1,451 | 721 | 397 | 162 | 24 | 187 |
| γ | 521 | 285 | 133 | 76 | 17 | 49 |
The number of gene pairs whose interaction partner maintains the duplication status from the same WGD event.
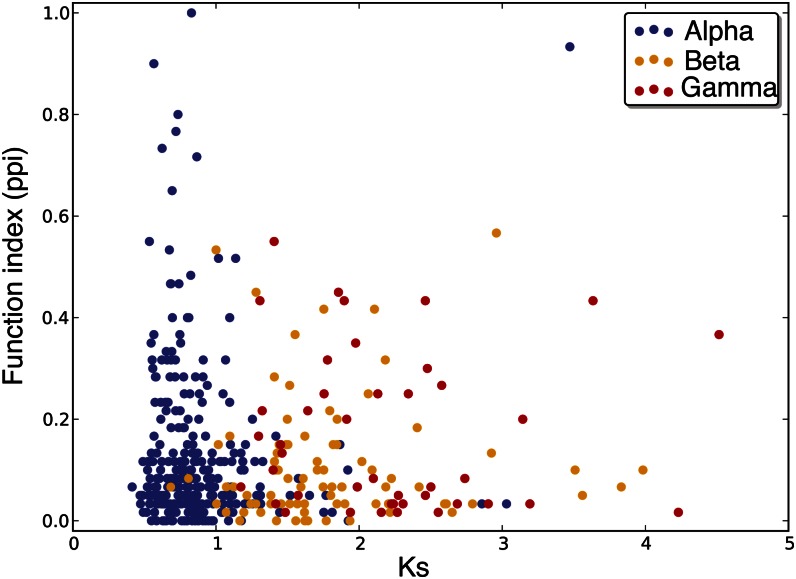
Using protein-protein interaction as a proxy for function, we studied functional divergence between duplicated genes. Figure 1 shows the degree of functional divergence of gene pairs derived from three WGD events in Arabidopsis plotted against putatively neutral sequence divergence measured by the synonymous substitution rate (Ks). Gene pairs from γ-WGD show significantly higher function index than α- and β-WGD gene pairs (Student’s t test, P = 0.0278 and P = 0.0116). Almost all gene pairs are diverged in function at Ks > 2. This trend is also exemplified in that most duplicated genes from the γ-WGD event are diverged in function (Supplemental Fig. S2). Random sampling of functionally nonrelated gene pairs (Blanc and Wolfe, 2004b) found 57% of recent duplicated gene pairs to show function divergence with a correlation coefficient cutoff of r = 0.52, with a 5% false-positive rate. Our findings suggest that their estimation might be conservative: of 464 gene pairs derived from the most recent WGD, 92.7% had different interaction partners, indicating the divergence of function, compared with 97.3% for the more ancient γ-WGD. There is a subset of duplicated gene pairs that are completely diverged in function (i.e. with no shared interaction partners), unexpectedly faster than other gene pairs produced at the same time, which indicates selection driving them to diverge in function.
Figure 1.
Function divergence between duplicated Arabidopsis genes on the time scale measured by Ks. Blue dots represent duplicated gene pairs from the α-WGD event (most recent), green dots from the β-WGD event, and red dots from the γ-WGD event (most ancient). ppi, Protein-protein interaction.
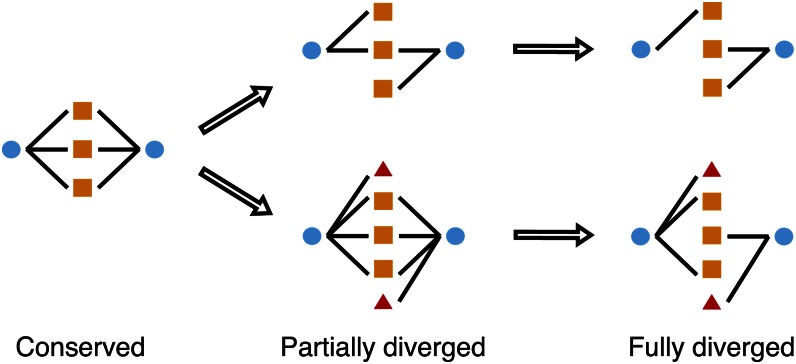
Strong evidence of a trend toward function divergence contrasts with the central assumption of the dosage-balance hypothesis as a mechanism to explain duplicate retention for these genes. In this respect, we further divided duplicated gene pairs from WGDs into three groups according to their function-divergence status: conserved gene pairs (numbering 52) that share all interaction partners, partially diverged gene pairs (166) that share some interaction partners, and fully diverged gene pairs (211) that share no interaction partners (Fig. 2).
Figure 2.
Categories of WGD-duplicated genes based on the divergence of protein interaction partners. Blue circles represent WGD-duplicated paralogous gene pairs. Orange squares are the original interacting partners shared by both copies. Red triangles are newly gained interacting partners indicating function divergence. Arrows suggest the process of function divergence of WGD-duplicated genes. The top and bottom paths refer to divergence by loss and gain of interaction partners, respectively.
Conserved, partially diverged, and fully diverged gene pairs differ in several important ways. First, the three groups differ by the types and proportions of protein domains that they contain (Supplemental Fig. S3). We searched the Pfam protein domain database for each pair of genes in all three groups (Finn et al., 2010). Genes in the fully diverged group show significantly higher numbers of protein domain types than genes in the other two groups (Mann-Whitney U test, P = 0.000175 and P = 0.0072). Second, there is a difference of gene function enriched in each group, as indicated by GO-slim terms, a subset of the terms in the whole gene ontology. Genes functioning in mitochondria and involved in electron transport or energy pathways are significantly enriched in the conserved and partially diverged groups (Supplemental Table S1). The enriched gene functions also differ in significance between partially and fully diverged groups, although they overlap. Genes responsive to abiotic or biotic stimulus and stress and involved in signal transduction are enriched in partially diverged groups at a higher significance level. Genes functioning as protein binding and working in the cytosol and plasma membrane are most enriched in the fully diverged group. Third, gene coexpression correlation also differs between the three functional groups (Supplemental Fig. S4), with the conserved group showing a higher level of correlation in expression profiles and the partially and fully diverged groups showing progressively lower correlations. The difference between the fully diverged group and the other two groups reaches statistical significance (Mann-Whitney U test, P = 0.0357 and P = 0.0123), but this trend clearly parallels protein-protein interaction data, suggesting that the three function-divergence groups reflect different evolutionary dynamics.
Gene Function Divergence According to GO-slim Terms
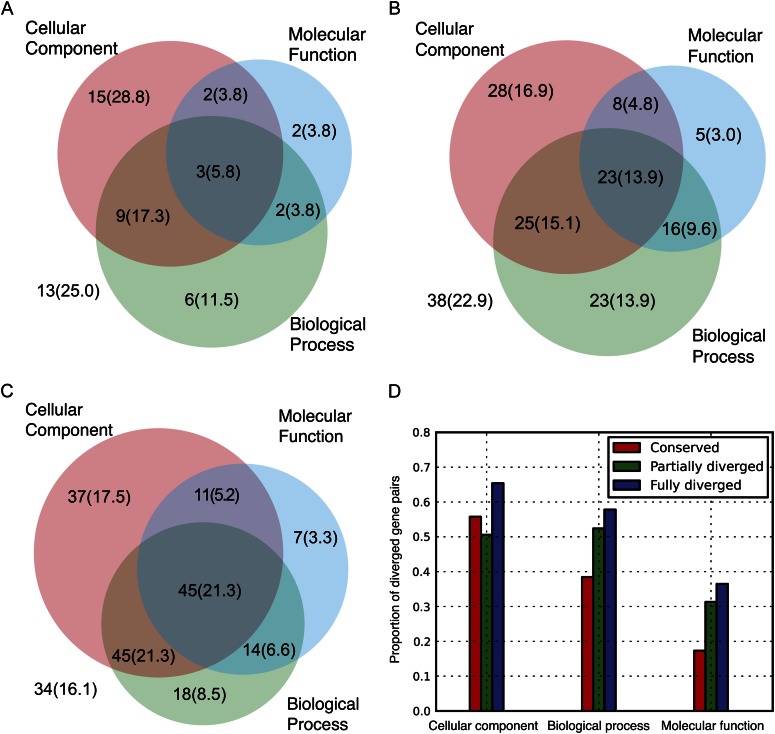
To further investigate the functional divergence between the three groups, we studied the gene annotation of duplicated gene pairs. We classified gene annotation divergence in each of three function categories defining different aspects of the function of a gene (Harris et al., 2004), namely cell component, molecular function, and biological process, for each function-divergence group (Fig. 3). If we consider members of duplicated gene pairs differing in at least one function category to have experienced function divergence, then 73% of the most recent WGD genes show function divergence compared with 77% for β and 82% for γ.
Figure 3.
Function divergence indicated by GO-slim terms. Venn diagrams show the numbers of diverged gene pairs in three Gene Ontology (GO) function categories. The integer indicates the number of gene pairs that shows diverged GO annotations in the function category. The number in parentheses shows the percentage. The numbers outside all three circles represents the number (percentage) of nondiverged gene pairs in all three GO function categories. A, Conserved. B, Partially diverged. C, Fully diverged. D, Proportion of diverged gene pairs in each GO category.
A total of 84% of genes differ by at least one of three function categories in the fully diverged group, 77% for the partially diverged group, and 75% for the conserved group. A higher proportion of duplicated gene pairs in the fully diverged group show divergence in the cell component function category than the other two groups (Fig. 3D). For example, AT2G16600 and AT4G34870 both function in chloroplast, cytosol, and plasma membrane, but AT4G34870 also functions extracellularly. Likewise, genes AT1G16030 and AT1G79930 derived from WGD both function in cytosol and plasma membrane. However, AT1G16030 also functions in cell wall and chloroplast, while AT1G79930 functions in the nucleus. Many such cases are identified in the fully diverged group (Supplemental Table S2). It is important to note that protein interactions that happen within plasma membranes are harder to recover in screens like yeast two-hybrid analysis, perhaps contributing to the enrichment of these gene pairs in the fully diverged category.
Dosage Balance and Retention of Duplicated Genes
According to the dosage-balance hypothesis, interacting genes should remain at the same level of duplication; that is, if one gene retains a duplicated copy, then its interacting gene(s) should also be retained in duplicate. To test this, we searched all the interactive gene pairs (based on 13,568 interactions available) in each of three WGDs in Arabidopsis and found 871, 187, and 49 duplicated pairs from α-, β-, and γ-WGDs that have the same copy number as their interaction partners from the same WGD event, consistent with the dosage-balance hypothesis (Table I). In contrast, 1,046, 436, and 170 duplicated gene pairs have lost/gained an interacting partner following WGD, in conflict with the dosage-balance hypothesis. Dosage-balanced retention declines steadily with the antiquity of duplication: 24.1% of duplicated gene pairs from the most recent WGD in Arabidopsis have remained in dosage balance with interacting partners, versus 12.9% and 9.4% for the earlier β-duplication and γ-triplication.
Selection Underlying the Divergence of Functions of Duplicated Genes
The study of protein-protein interaction data above suggests that 92.7% of the paralogous gene pairs may have diverged in function since the most recent WGD event in Arabidopsis. Evidence from an analysis of protein functional domains and GO-slim annotation terms shows that patterns of functional divergence have not been random but have differentially affected gene functional groups, implying the action of selection.
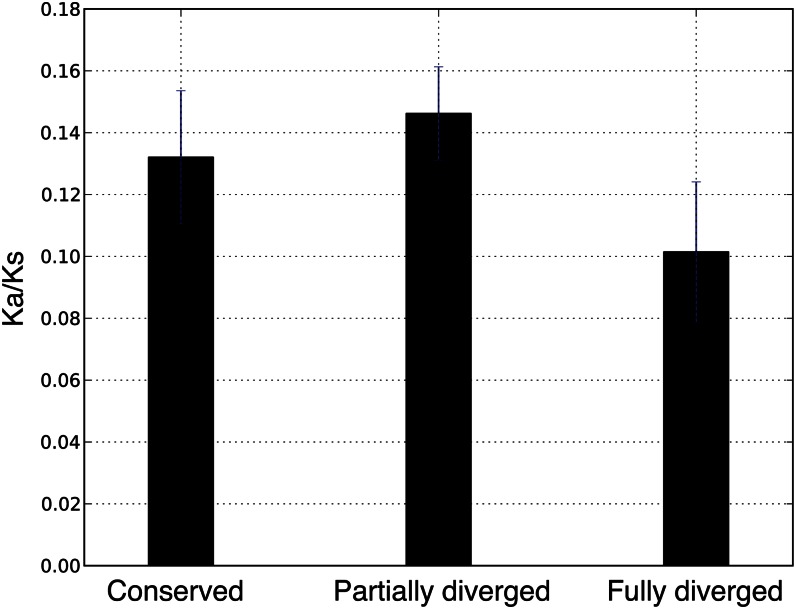
To investigate the nature of the selection pressure contributing to functional divergence between duplicated gene pairs, we first compared the ratio of the number of nonsynonymous substitutions per nonsynonymous site (Ka) to the number of synonymous substitutions per synonymous site (Ks) of duplicated gene pairs among the three function-divergence groups (Fig. 4). Duplicated genes from all three groups have low Ka/Ks ratios, indicating that they are generally under purifying selection. The fully diverged group shows the lowest Ka/Ks ratio (i.e. strongest evidence of purifying selection), although it could not be distinguished statistically from that of the conserved group. Curiously, the Ka/Ks ratio of the partially diverged group is marginally larger than that of the fully diverged group (Mann-Whitney U test, P = 0.0934), suggesting that the genes in this group are experiencing weaker purifying selection or an averaging of stronger positive and negative selection.
Figure 4.
Comparison of Ka/Ks ratios among function-divergence groups. The mean Ka/Ks ratio in each group is shown with a black bar, with error bars indicating se. Two-sample independent t tests show that there is significant difference between partially and fully diverged groups (P = 0.0371). [See online article for color version of this figure.]
Single-Nucleotide Polymorphism
Selection commonly affects the density of single-nucleotide polymorphism (SNP) near loci under selection. Low SNP frequency in a population indicates loci that have recently experienced strong positive selection (Hartl and Clark, 2007). Indeed, loci that are under diversifying selection usually exhibit lower SNP density than the average across the genome (Moore and Purugganan, 2003).
We compared the coding SNP density of duplicated genes from the three function-divergence groups (Fig. 5). The SNP density of all three groups is significantly lower than genome-wide coding SNP density. This is not readily explained by traditional evolutionary theory, which predicts an increase in polymorphism between duplicated genes due to function redundancy.
Figure 5.
Comparison of coding SNP density in modern Arabidopsis populations among different function-divergence groups. Black bars indicate means of coding SNP density, with error bars indicating se. Mann-Whitney U tests show that the SNP density of gene pairs in the partially diverged group is significantly lower than that of fully diverged and conserved groups (P = 2.619e-11 and P = 0.000349). There is no significant difference between the fully diverged and conserved groups. The red line indicates the genome-wide average of coding SNP density. [See online article for color version of this figure.]
A recent study shows that nonreciprocal DNA exchanges, such as “illegitimate recombination” or “gene conversion,” might be a major driver of concerted evolution, reducing the polymorphism rate between duplicated genes (Wang et al., 2009). To test this, we performed a phylogenetic comparison of gene quartets from Arabidopsis and Arabidopsis lyrata, which diverged more recently than the α-WGD. Among 39, 125, and 141 gene quartets from conserved, partially diverged, and fully diverged groups that could be tested, only one case (from the partially diverged group) shows evidence of gene conversion, reflected as greater similarity of α-WGD paralogs within a nucleus than orthologs between the species (Fig. 5). Therefore, we conclude that the low SNP rate in WGD-duplicated genes could not be explained by gene conversion. Instead, the data indicate that positive selection might have affected most of the α-WGD-duplicated genes. However, the significantly lower SNP density of duplicated gene pairs of the partially diverged group than the fully diverged and conserved groups (Mann-Whitney U test, P = 2.619e-11 and P = 0.000349) suggests that selective sweeps may have affected the partially diverged group more recently than the other groups.
BLOSUM80 Score
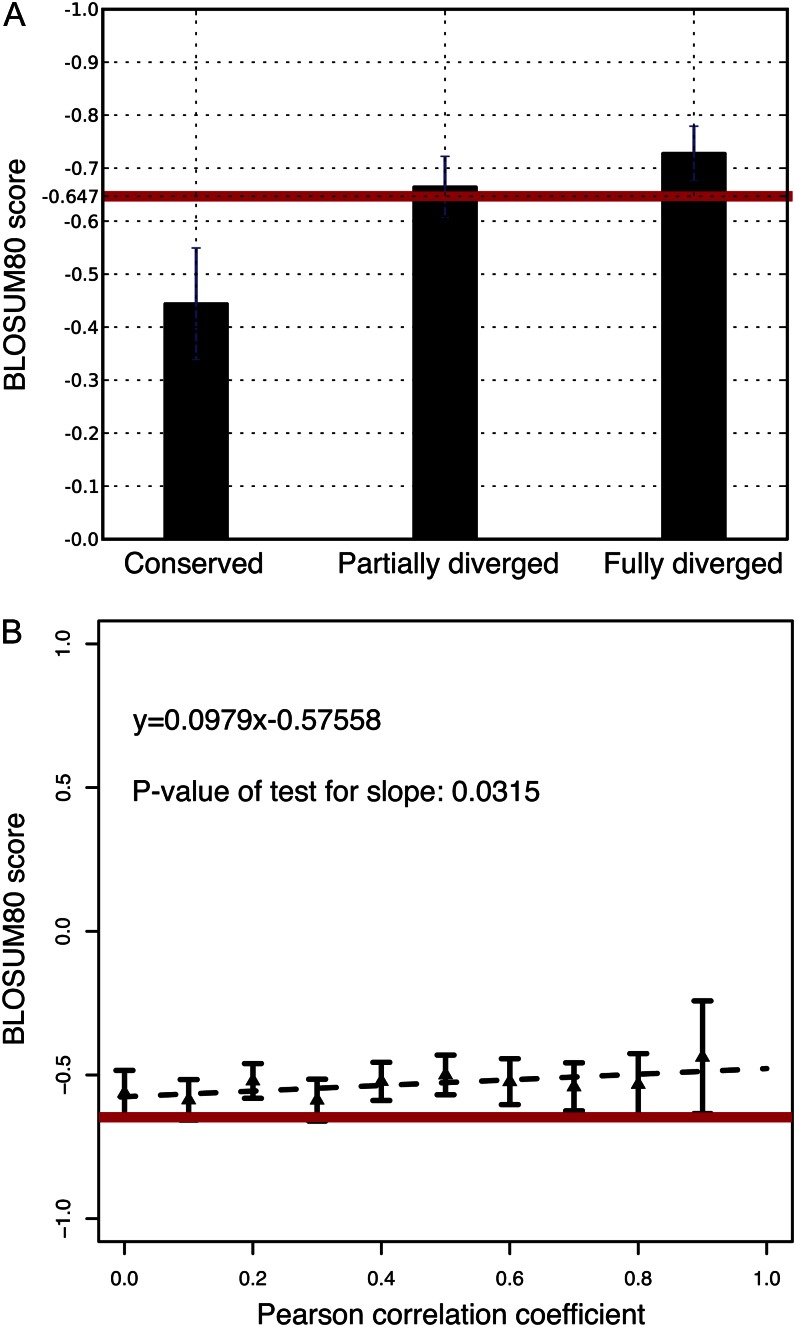
To further study the nature of selection pressure acting on duplicated gene pairs, we compared the severity of nonsynonymous SNPs among the three function-divergence groups. We used the BLOSUM80 amino acid substitution matrix, which assigns each amino acid substitution a score according to its frequency of occurrence in protein sequence alignments that are more than 80% identical. Less frequent substitutions are inferred to represent more severe changes to protein function. Loci that have experienced positive selection would be expected to have increased average severity of nonsynonymous SNPs relative to others in the population. Figure 6A shows that in the modern Arabidopsis population, there are significantly larger numbers of severe nonsynonymous substitutions in the fully and partially diversified groups than in the conserved group (Mann-Whitney U test, P = 0.00183 and P = 0.00233). It also shows that nonsynonymous SNPs cause more severe changes to protein functions in the fully diversified group than the genome-wide average. In contrast, most nonsynonymous SNPs in genes of the conserved group have relatively minor effects on protein function, lower than the genome-wide average. These observations provide further evidence that duplicated gene pairs from the fully diversified groups have been or are under diversifying selection.
Figure 6.
Comparison of the severity of nonsynonymous nucleotide polymorphism in modern Arabidopsis populations among function-divergence groups. Average BLOSUM80 score over all nonsynonymous SNPs in each duplicated gene is calculated as a measure of the severity of amino acid changes, with smaller BLOSUM80 scores indicating greater severity. The red lines indicate the genome-wide average of BLOSUM80 scores. A, Comparison of the mean BLOSUM80 scores among function-divergence groups, with error bars indicating se. Mann-Whitney U tests show that there are significantly larger numbers of severe nonsynonymous substitutions in the fully and partially diversified groups than in the conserved group (P = 0.00183 and P = 0.00233). B, Comparison of coding SNP severity and degrees of expression divergence for each duplicated gene pair. Linear regression of the mean BLOSUM80 score is shown as a broken line with the function of the regression line. [See online article for color version of this figure.]
SNP severity is directly related to gene expression divergence. We plotted the BLOSUM80 score against coexpression correlations between duplicated gene pairs from the most recent WGD (Fig. 6B). Duplicated gene pairs with lower expression correlation contain more severe amino acid substitutions than those with higher expression correlation, consistent with the inference that function divergence between duplicated genes has been driven by diversifying selection. The most recent WGD duplicated genes show less severe amino acid substitutions than the average of all genes in the genome. This indicates that most beneficial nonsynonymous SNPs have been driven to high frequency in the population sometime in the past and that purifying selection is now generally purging severe nonsynonymous SNPs.
DISCUSSION
Interrelationships among several different data types relating to gene function, considered in the context of three different episodes of duplication separated by tens of millions of years, illustrate divergent paths for the retention of duplicated genes following WGD. While paleoduplicated genes still present in Arabidopsis are generally under purifying selection, consistent with prior findings (Chapman et al., 2006), many show evidence of having experienced a diversifying selection phase before returning to purifying selection in modern populations. Populations of duplicated genes that formed at the same time now range from being “fully diverged” in protein-protein interaction partners and expression patterns across cell components to being “conserved,” retaining similar protein-protein interaction partners and closely correlated expression patterns.
The Dosage-Balance Hypothesis
Dosage balance has been used to explain the preferential retention of duplicated genes after WGD (Birchler and Veitia, 2007). Some authors suggest that gene-balanced duplications provide a sufficient explanation for the morphological complexity of both plant and animal eukaryotic lineages (Freeling and Thomas, 2006). However, a key assumption of the dosage-balance hypothesis is that both members of duplicated gene pairs should retain original gene functions. Many studies show that the functions of genes retained in duplicate for long time periods are generally diverged (Gu et al., 2002; Wagner, 2002; Makova and Li, 2003; Blanc and Wolfe, 2004b; He and Zhang, 2005). This study suggests that approximately 90% of these genes have diverged in protein interaction partners. The dosage-balance hypothesis and its assumption of conserved gene function are quite reasonable immediately after WGD, particularly under autopolyploidy, in which two duplicated genes are nearly if not fully identical. However, it is problematic to apply the dosage-balance hypothesis to explain retained duplicated genes that have substantially diverged in function, as appears to be the case for most genes after a long evolutionary time. Another prediction of the dosage-balance hypothesis is that both members of protein interaction pairs should have the same duplication status, which now only holds true for approximately 24% of duplicated gene pairs from the most recent Arabidopsis WGD (i.e. the majority of retained duplicated genes are no longer in dosage balance with their interaction partners). These observations might be reconciled if dosage balance is a major player in duplicate retention soon after WGD, with its effect gradually declining in the long term.
Our findings are essentially consistent with the recently proposed “two-phase adaptation process” of WGD gene evolution (Bekaert et al., 2011). The dosage balance of regulatory genes might contribute to many biological processes (Birchler et al., 2005), a hypothesis also supported by our analysis (Supplemental Table S1).
We note that a small but distinctive group of genes show no functional divergence (function index = 0) even after extensive neutral sequence divergence. Genes with kinase and transcription factor activity are among the most significantly enriched functions in this group, although they are also enriched in partially and fully diverged groups. Genes functioning in mitochondria and involved in electron transport or energy pathways are enriched only in the conserved group (Supplemental Table S1), suggesting that genes involved in energy production are particularly unlikely to experience subfunctionalization/neofunctionalization when compared with all duplicated genes in the Arabidopsis genome.
Function Divergence by Microadaptation
Through the analysis of protein-protein interaction data, we identified a group of genes that are fully diverged in interaction partners identified to date between members of duplicated gene pairs. Population SNP patterns indicate that these genes might have been affected by ancient selective sweeps (Figs. 4–6). Analysis of GO-slim annotations shows that a higher proportion of fully diverged than partially diverged or conserved duplicated genes are diverged in the cell component category (Fig. 3D). For most diverged gene pairs, one member is adapted to at least one unique compartment (Supplemental Table S2).
These lines of evidence support the notion that purifying selection on both genes is relaxed following WGD, allowing either member to accumulate functional mutations that sometimes result in the exploration of new biochemical niches. Higher SNP density in gene pairs of the fully diverged group may indicate that they experienced positive selection earlier than the partially diverged duplicated genes, with the population SNP density now restored and purifying selection (observed by Ka/Ks ratio) acting to preserve the new functions indicated by GO-slim annotation. This inference is consistent with their greater divergence in protein interaction partners. If the new biochemical niche enhances the fitness of an organism in its environment, the causal mutation should spread rapidly in the population. We call this process microadaptation, due to its being vividly similar to conventional adaptation selection.
What forces drive microadaptation? In most cases, WGD causes the instability of neopolyploids and reduces the fitness of the host (Comai, 2005; Mayrose et al., 2011). However, genome sequence data show that all flowering plant species can be traced to a polyploid ancestor. Appropriate molecular mechanisms that contribute to the occasional survival of polyploid plants are not clear yet. Our discovery of a group of duplicated genes that show extensive function divergence driven by microadaptation might have contributed to this process. For example, many fully diverged gene pairs now working in different cell components (i.e. with different GO-slim annotations; Fig. 3D; Supplemental Table S2) may reflect greater fluidity of cellular membrane systems and may have conferred increased survival at low temperatures. Indeed, polyploid plants are more abundant in cold than in warm environments, for example, increasing from 31% in Sicily to 54.5% in Iceland (Tischler, 1935) and showing similar patterns in other areas (Johnson and Packer, 1965; Soltis and Soltis, 1999; Brochmann et al., 2004). The Cretaceous-Paleogene extinction event, suggested by some to be associated with a selective advantage for polyploids (Fawcett et al., 2009), may have been due not only to asteroid impact but to climate change, having been preceded by several million years of global cooling (Raup, 1986).
Gene Evolution after WGD in Plants
The dosage-balance hypothesis stresses that purifying selection on dosage balance determines the retention of duplicated genes. The dosage-balance model per se does not address the mechanisms of evolutionary innovation, although the preservation of some genes in duplicate is an essential first step toward permitting such mechanisms to operate. Many WGD events in plants have been closely related to species diversity and complexity (Swigonová et al., 2004; Jiao et al., 2011). Freeling and Thomas (2006) noted that, “In order to recruit a diverged, duplicate functional module to a new boundary, gene dosage sensitivity must be avoided or mitigated.” In this sense, dosage balance may be regarded as a source of purifying selection on the copy numbers of genes that are involved in protein complexes rather than a general model for post-WGD gene evolution. Moreover, dosage balance applies with less efficiency in plants than in single-celled organisms, where the hypothesis was first proposed. Microbes have vastly larger effective population sizes and more intense selection on genome size, so only the few rare events that confer strong advantages are likely to be maintained very long (Lynch et al., 1995). Plant species usually have relatively small effective population sizes, and duplications that are neutral or even slightly deleterious may very well persist for some time, allowing subsequent changes that either make them advantageous or purge them.
Both the level of functional divergence between gene pairs and the percentage of gene pairs that have diverged increase with time since duplication. However, rather than being a continuous process, our findings suggest that divergence occurs in a more episodic manner, by one of three evolutionary paths. In path I, purifying selection protects both gene copies from functional divergence immediately following a WGD until purifying selection is reduced or removed. The source of purifying selection could be, for example, dosage balance. When purifying selection is removed, one or both duplicated genes may follow path II, with relaxed selection. The speed of functional divergence may depend on the rates of single-nucleotide mutation, unequal crossover, transposable element visitations, gene conversion, and other factors. These factors may vary widely among lineages or even among locations within a genome; for example, euchromatin is substantially different from heterochromatin. Once mutagenesis alters the function of one copy, purifying selection may immediately act on the other copy. The divergence of function between the two copies may continue to grow unless/until the new function becomes fixed. In path III, purifying selection on each member of a duplicated gene pair is relaxed immediately following a WGD. Many such genes may simply be lost, as are the vast majority of duplicated genes, but a few may now follow path II. If an altered function of one copy is beneficial, becoming fixed and then subjected to purifying selection, microadaptation has taken place. In partial summary, different sets of WGD-duplicated genes may follow different evolutionary paths following a WGD. In the context of our three proposed paths, it important to note that purifying selection is not a binary trait but can occur to varying degrees; indeed, any nonpseudogene experiences some level of purifying selection.
Genetic drift and diversifying selection are the two ways by which polyploidy can be fixed in a population. Plant species usually have relatively small effective population size (e.g. compared with microbes), which makes genetic drift relatively more effective. However, some evidence suggests that the survival of polyploid plants may often coincide with the occurrence of extreme environments (Raup, 1986; Crow and Wagner, 2006; Fawcett et al., 2009), suggesting that adaptive selection might play a key role in fixing paleopolyploidy in the population. In conclusion, we suggest that it may be of singularly high importance to the evolution of natural polyploid plants that at least a subset of genes experience path III.
MATERIALS AND METHODS
Duplicated Genes
Duplicated genes from three WGDs in Arabidopsis (Arabidopsis thaliana) were defined as described (Bowers et al., 2003).
Protein-Protein Interaction Analysis
To investigate gene functions, we used empirical protein-protein interaction data for Arabidopsis from BioGRID version 3.1.76 (Stark et al., 2006) and an interactome study (Arabidopsis Interactome Mapping Consortium, 2011). All nonphysical interactions are excluded. A total of 13,568 physical interaction gene pairs are used after further removing redundant pairs. Data are derived from one of the following experiments: affinity-capture (mass spectrometry, RNA, and western), biochemical activity, cocrystal structure, cofractionation, colocalization, copurification, far western, fluorescence resonance energy transfer, protein-fragment complementation assay, protein-peptide, protein-RNA, reconstituted complex and two-hybrid analyses.
Function index is defined by proportions of unique interactions divided by the total number of interactions of duplicated gene pairs. For example, for duplicated gene pair A and B: a, as the number of interactions of gene A; b, as the number of interactions of gene B; s, as the number of common interaction partners shared by A and B; u, as the number of unique interaction partners of gene A; v, as the number of unique interaction partners of gene.
The function index of gene pair A and B is given by
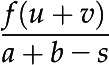 |
where f is a weighting factor defined as (a + b)/max(t) and t is a vector of the number of interaction partners for all gene pairs from three WGDs. f gives different weight to gene pairs, as it is more informative for gene pairs with a larger number of interaction partners than for those with fewer partners.
Gene Coexpression Analysis
Arabidopsis gene expression data obtained with the Affymetrix ATH1 Genome Array (GPL198) were downloaded from the National Center for Biotechnology Information Gene Expression Omnibus database (Suyama et al., 2006). There are 65 data sets and 7,421 samples. Data were normalized using the robust multiarray average algorithm implemented in the Bioconductor packages of statistical computing language R (Gentleman et al., 2004). We further filtered the data set to remove outlier data points using arrayQualityMetrics in the Bioconductor packages (Kauffmann et al., 2009). Each gene expression profile was compared with all other genes using the standard Pearson correlation coefficient.
SNP Analysis
Genome-wide SNP data from Arabidopsis were downloaded from Biomart (release 5) in the Ensembl plants database (http://plants.ensembl.org/index.html). To assess the severity of nonsynonymous SNPs, an average BLOSUM80 score over all nonsynonymous polymorphisms for each gene is calculated based on the BLOSUM80 protein substitution matrix (Henikoff and Henikoff, 1992).
Gene Ontology Analysis
In order to get a functional overview of the genes, the GO-slim classification of a gene was determined (Larkin et al., 2007). All records are derived from literature-based annotations and protein domain-based electronic annotations. Gene function divergence in each Gene Ontology function category is defined by the number of differential annotation terms between duplicated gene pairs in each category.
Calculation of Ka and Ks Values
Protein sequences were aligned using ClustalW (Larkin et al., 2007). Coding sequence alignments were guided by protein sequence alignment using PAL2NAL (Suyama et al., 2006). Ka and Ks values were calculated using the Nei-Gojobori method implemented in the yn00 program in the PAML package (Yang, 2007).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Distribution of unique protein-protein interaction partners in duplicated gene pairs.
Supplemental Figure S2. Comparison of the distribution of protein-protein interaction categories in duplicated gene pairs.
Supplemental Figure S3. The number of unique protein domain in three functional groups.
Supplemental Figure S4. Gene expression correlations among three functional groups.
Supplemental Figure S5. A gene quartet from Arabidopsis and Arabidopsis lyrata suggests gene conversion.
Supplemental Table S1. Function enrichment of three functional groups indicated by GO-slim terms.
Supplemental Table S2. Cell component divergence between duplicated gene pairs in the fully diverged group.
Glossary
- WGD
whole-genome duplication
- Ks
synonymous substitutions per synonymous site
- SNP
single-nucleotide polymorphism
- Ka
nonsynonymous substitutions per nonsynonymous site
References
- Arabidopsis Interactome Mapping Consortium (2011) Evidence for network evolution in an Arabidopsis interactome map. Science 333: 601–607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bekaert M, Edger PP, Pires JC, Conant GC. (2011) Two-phase resolution of polyploidy in the Arabidopsis metabolic network gives rise to relative and absolute dosage constraints. Plant Cell 23: 1719–1728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birchler JA, Riddle NC, Auger DL, Veitia RA. (2005) Dosage balance in gene regulation: biological implications. Trends Genet 21: 219–226 [DOI] [PubMed] [Google Scholar]
- Birchler JA, Veitia RA. (2007) The gene balance hypothesis: from classical genetics to modern genomics. Plant Cell 19: 395–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birchler JA, Veitia RA. (2010) The gene balance hypothesis: implications for gene regulation, quantitative traits and evolution. New Phytol 186: 54–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanc G, Wolfe KH. (2004a) Widespread paleopolyploidy in model plant species inferred from age distributions of duplicate genes. Plant Cell 16: 1667–1678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanc G, Wolfe KH. (2004b) Functional divergence of duplicated genes formed by polyploidy during Arabidopsis evolution. Plant Cell 16: 1679–1691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowers JE, Chapman BA, Rong J, Paterson AH. (2003) Unravelling angiosperm genome evolution by phylogenetic analysis of chromosomal duplication events. Nature 422: 433–438 [DOI] [PubMed] [Google Scholar]
- Brochmann C, Brysting AK, Alsos IG, Borgen L, Grundt HH, Scheen AC, Elven R. (2004) Polyploidy in arctic plants. Biol J Linn Soc Lond 82: 521–536 [Google Scholar]
- Chapman BA, Bowers JE, Feltus FA, Paterson AH. (2006) Buffering of crucial functions by paleologous duplicated genes may contribute cyclicality to angiosperm genome duplication. Proc Natl Acad Sci USA 103: 2730–2735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comai L. (2005) The advantages and disadvantages of being polyploid. Nat Rev Genet 6: 836–846 [DOI] [PubMed] [Google Scholar]
- Conant GC, Wolfe KH. (2008) Turning a hobby into a job: how duplicated genes find new functions. Nat Rev Genet 9: 938–950 [DOI] [PubMed] [Google Scholar]
- Crow KD, Wagner GP. (2006) What is the role of genome duplication in the evolution of complexity and diversity? Mol Biol Evol 23: 887–892 [DOI] [PubMed] [Google Scholar]
- Davis JC, Petrov DA. (2005) Do disparate mechanisms of duplication add similar genes to the genome? Trends Genet 21: 548–551 [DOI] [PubMed] [Google Scholar]
- Dehal P, Boore JL. (2005) Two rounds of whole genome duplication in the ancestral vertebrate. PLoS Biol 3: e314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Des Marais DL, Rausher MD. (2008) Escape from adaptive conflict after duplication in an anthocyanin pathway gene. Nature 454: 762–765 [DOI] [PubMed] [Google Scholar]
- Fawcett JA, Maere S, Van de Peer Y. (2009) Plants with double genomes might have had a better chance to survive the Cretaceous-Tertiary extinction event. Proc Natl Acad Sci USA 106: 5737–5742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn RD, Mistry J, Tate J, Coggill P, Heger A, Pollington JE, Gavin OL, Gunasekaran P, Ceric G, Forslund K, et al. (2010) The Pfam protein families database. Nucleic Acids Res 38: D211–D222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Force A, Lynch M, Pickett FB, Amores A, Yan YL, Postlethwait J. (1999) Preservation of duplicate genes by complementary, degenerative mutations. Genetics 151: 1531–1545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francino MP. (2005) An adaptive radiation model for the origin of new gene functions. Nat Genet 37: 573–577 [DOI] [PubMed] [Google Scholar]
- Freeling M, Thomas BC. (2006) Gene-balanced duplications, like tetraploidy, provide predictable drive to increase morphological complexity. Genome Res 16: 805–814 [DOI] [PubMed] [Google Scholar]
- Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, et al. (2004) Bioconductor: open software development for computational biology and bioinformatics. Genome Biol 5: R80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu ZL, Nicolae D, Lu HHS, Li WH. (2002) Rapid divergence in expression between duplicate genes inferred from microarray data. Trends Genet 18: 609–613 [DOI] [PubMed] [Google Scholar]
- Hakes L, Pinney JW, Lovell SC, Oliver SG, Robertson DL. (2007) All duplicates are not equal: the difference between small-scale and genome duplication. Genome Biol 8: R209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris MA, Clark J, Ireland A, Lomax J, Ashburner M, Foulger R, Eilbeck K, Lewis S, Marshall B, Mungall C, et al. (2004) The Gene Ontology (GO) database and informatics resource. Nucleic Acids Res 32: D258–D261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartl DL, Clark AG (2007) Principles of Population Genetics, Ed 4. Sinauer Associates, Sunderland, MA [Google Scholar]
- He X, Zhang J. (2005) Rapid subfunctionalization accompanied by prolonged and substantial neofunctionalization in duplicate gene evolution. Genetics 169: 1157–1164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S, Henikoff JG. (1992) Amino acid substitution matrices from protein blocks. Proc Natl Acad Sci USA 89: 10915–10919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes AL. (1994) The evolution of functionally novel proteins after gene duplication. Proc Biol Sci 256: 119–124 [DOI] [PubMed] [Google Scholar]
- Innan H, Kondrashov F. (2010) The evolution of gene duplications: classifying and distinguishing between models. Nat Rev Genet 11: 97–108 [DOI] [PubMed] [Google Scholar]
- Jaillon O, Aury JM, Noel B, Policriti A, Clepet C, Casagrande A, Choisne N, Aubourg S, Vitulo N, Jubin C, et al. (2007) The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature 449: 463–467 [DOI] [PubMed] [Google Scholar]
- Jiao Y, Wickett NJ, Ayyampalayam S, Chanderbali AS, Landherr L, Ralph PE, Tomsho LP, Hu Y, Liang H, Soltis PS, et al. (2011) Ancestral polyploidy in seed plants and angiosperms. Nature 473: 97–100 [DOI] [PubMed] [Google Scholar]
- Johnson AW, Packer JG. (1965) Polyploidy and environment in arctic Alaska. Science 148: 237–239 [DOI] [PubMed] [Google Scholar]
- Kauffmann A, Gentleman R, Huber W. (2009) arrayQualityMetrics: a Bioconductor package for quality assessment of microarray data. Bioinformatics 25: 415–416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, et al. (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23: 2947–2948 [DOI] [PubMed] [Google Scholar]
- Li WH (1997) Molecular Evolution. Sinauer Associates, Sunderland, MA [Google Scholar]
- Liang H, Plazonic KR, Chen J, Li WH, Fernández A. (2008) Protein under-wrapping causes dosage sensitivity and decreases gene duplicability. PLoS Genet 4: e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M, Conery J, Burger R. (1995) Mutation accumulation and the extinction of small populations. Am Nat 146: 489–518 [Google Scholar]
- Lynch M, Conery JS. (2000) The evolutionary fate and consequences of duplicate genes. Science 290: 1151–1155 [DOI] [PubMed] [Google Scholar]
- Maere S, De Bodt S, Raes J, Casneuf T, Van Montagu M, Kuiper M, Van de Peer Y. (2005) Modeling gene and genome duplications in eukaryotes. Proc Natl Acad Sci USA 102: 5454–5459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makova KD, Li WH. (2003) Divergence in the spatial pattern of gene expression between human duplicate genes. Genome Res 13: 1638–1645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayrose I, Zhan SH, Rothfels CJ, Magnuson-Ford K, Barker MS, Rieseberg LH, Otto SP. (2011) Recently formed polyploid plants diversify at lower rates. Science 333: 1257. [DOI] [PubMed] [Google Scholar]
- Ming R, Hou S, Feng Y, Yu Q, Dionne-Laporte A, Saw JH, Senin P, Wang W, Ly BV, Lewis KL, et al. (2008) The draft genome of the transgenic tropical fruit tree papaya (Carica papaya Linnaeus). Nature 452: 991–996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore RC, Purugganan MD. (2003) The early stages of duplicate gene evolution. Proc Natl Acad Sci USA 100: 15682–15687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno S (1970) Evolution by Gene Duplication. Springer-Verlag, Berlin [Google Scholar]
- Papp B, Pál C, Hurst LD. (2003) Dosage sensitivity and the evolution of gene families in yeast. Nature 424: 194–197 [DOI] [PubMed] [Google Scholar]
- Paterson AH, Bowers JE, Chapman BA. (2004) Ancient polyploidization predating divergence of the cereals, and its consequences for comparative genomics. Proc Natl Acad Sci USA 101: 9903–9908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paterson AH, Chapman BA, Kissinger JC, Bowers JE, Feltus FA, Estill JC. (2006) Many gene and domain families have convergent fates following independent whole-genome duplication events in Arabidopsis, Oryza, Saccharomyces and Tetraodon. Trends Genet 22: 597–602 [DOI] [PubMed] [Google Scholar]
- Ponting CP. (2008) The functional repertoires of metazoan genomes. Nat Rev Genet 9: 689–698 [DOI] [PubMed] [Google Scholar]
- Proulx SR, Phillips PC. (2006) Allelic divergence precedes and promotes gene duplication. Evolution 60: 881–892 [PubMed] [Google Scholar]
- Raup DM. (1986) Biological extinction in Earth history. Science 231: 1528–1533 [DOI] [PubMed] [Google Scholar]
- Romero D, Palacios R. (1997) Gene amplification and genomic plasticity in prokaryotes. Annu Rev Genet 31: 91–111 [DOI] [PubMed] [Google Scholar]
- Schmutz J, Cannon SB, Schlueter J, Ma J, Mitros T, Nelson W, Hyten DL, Song Q, Thelen JJ, Cheng J, et al. (2010) Genome sequence of the palaeopolyploid soybean. Nature 463: 178–183 [DOI] [PubMed] [Google Scholar]
- Soltis DE, Soltis PS. (1999) Polyploidy: recurrent formation and genome evolution. Trends Ecol Evol 14: 348–352 [DOI] [PubMed] [Google Scholar]
- Spofford JB. (1969) Heterosis and evolution of duplications. Am Nat 103: 407–432 [Google Scholar]
- Stark C, Breitkreutz BJ, Reguly T, Boucher L, Breitkreutz A, Tyers M. (2006) BioGRID: a general repository for interaction datasets. Nucleic Acids Res 34: D535–D539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suyama M, Torrents D, Bork P. (2006) PAL2NAL: robust conversion of protein sequence alignments into the corresponding codon alignments. Nucleic Acids Res 34: W609–W612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swigonová Z, Lai J, Ma J, Ramakrishna W, Llaca V, Bennetzen JL, Messing J. (2004) Close split of sorghum and maize genome progenitors. Genome Res 14: 1916–1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang H, Bowers JE, Wang X, Ming R, Alam M, Paterson AH. (2008) Synteny and collinearity in plant genomes. Science 320: 486–488 [DOI] [PubMed] [Google Scholar]
- Tang H, Bowers JE, Wang X, Paterson AH. (2010) Angiosperm genome comparisons reveal early polyploidy in the monocot lineage. Proc Natl Acad Sci USA 107: 472–477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tischler G. (1935) Die Bedeutung der Polyploidie für die Verbreitung der Angiospermen. Bot Jahrb 47: 1–36 [Google Scholar]
- Tuskan GA, Difazio S, Jansson S, Bohlmann J, Grigoriev I, Hellsten U, Putnam N, Ralph S, Rombauts S, Salamov A, et al. (2006) The genome of black cottonwood, Populus trichocarpa (Torr. & Gray). Science 313: 1596–1604 [DOI] [PubMed] [Google Scholar]
- Van de Peer Y. (2004) Computational approaches to unveiling ancient genome duplications. Nat Rev Genet 5: 752–763 [DOI] [PubMed] [Google Scholar]
- Wagner A. (2002) Asymmetric functional divergence of duplicate genes in yeast. Mol Biol Evol 19: 1760–1768 [DOI] [PubMed] [Google Scholar]
- Wang X, Tang H, Bowers JE, Paterson AH. (2009) Comparative inference of illegitimate recombination between rice and sorghum duplicated genes produced by polyploidization. Genome Res 19: 1026–1032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe KH, Shields DC. (1997) Molecular evidence for an ancient duplication of the entire yeast genome. Nature 387: 708–713 [DOI] [PubMed] [Google Scholar]
- Yang ZH. (2007) PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol 24: 1586–1591 [DOI] [PubMed] [Google Scholar]
- Zhang JZ. (2003) Evolution by gene duplication: an update. Trends Ecol Evol 18: 292–298 [Google Scholar]