The cyp78a8 cyp78a9 loss-of-function mutant has reduced seed set due to outer integument development arrest leading to female sterility.
Abstract
Synchronized communication between gametophytic and sporophytic tissue is crucial for successful reproduction, and hormones seem to have a prominent role in it. Here, we studied the role of the Arabidopsis (Arabidopsis thaliana) cytochrome P450 CYP78A9 enzyme during reproductive development. First, controlled pollination experiments indicate that CYP78A9 responds to fertilization. Second, while CYP78A9 overexpression can uncouple fruit development from fertilization, the cyp78a8 cyp78a9 loss-of-function mutant has reduced seed set due to outer ovule integument development arrest, leading to female sterility. Moreover, CYP78A9 has a specific expression pattern in inner integuments in early steps of ovule development as well as in the funiculus, embryo, and integuments of developing seeds. CYP78A9 overexpression did not change the response to the known hormones involved in flower development and fruit set, and it did not seem to have much effect on the major known hormonal pathways. Furthermore, according to previous predictions, perturbations in the flavonol biosynthesis pathway were detected in cyp78a9, cyp78a8 cyp78a9, and empty siliques (es1-D) mutants. However, it appeared that they do not cause the observed phenotypes. In summary, these results add new insights into the role of CYP78A9 in plant reproduction and present, to our knowledge, the first characterization of metabolite differences between mutants in this gene family.
Angiosperms have evolved the processes of double fertilization and fruit development as pivotal steps of their survival and dispersal strategies. Pollination and fertilization are essential for fruit initiation, considering that the angiosperm flower initiates terminal senescence and abscission programs if pollination has not taken place (Vivian-Smith et al., 2001; Fuentes and Vivian-Smith, 2009). For centuries, humans endeavored to prevent this association in order to develop seedless fruits. Most research has concentrated on the role of endogenous phytohormones as triggers for fruit initiation after fertilization, and different strategies such as exogenous application or artificial overproduction of plant hormones (Fuentes and Vivian-Smith, 2009), mutation, and misexpression of specific genes have been tested. The principal lines of evidence suggest that increased auxin and GA content in ovules and ovary leads to parthenocarpic fruits in Arabidopsis (Arabidopsis thaliana; Varoquaux et al., 2000; Goetz et al., 2007; Alabadí et al., 2009; Dorcey et al., 2009; Fuentes and Vivian-Smith, 2009; Pandolfini et al., 2009; Carbonell-Bejerano et al., 2010). But even with this increasing insight, the available knowledge of the mechanisms that underlie parthenocarpy is still limited.
The Arabidopsis transposon activation-tagged mutant empty siliques (es1-D) overexpresses the cytochrome P450 CYP78A9 gene and is characterized by the development of parthenocarpic fruits (Marsch-Martinez et al., 2002; de Folter et al., 2004). When crossed to wild-type plants, it results in the formation of viable seed. However, its fertility is reduced compared with wild-type plants. Moreover, siliques that result from the pollination of mutant es1-D plants with wild-type pollen grow larger than unpollinated mutant siliques and wider than wild-type siliques (Marsch-Martinez et al., 2002). A transfer DNA (T-DNA) activation-tagging screen also identified a mutant overexpressing CYP78A9 with a similar phenotype (Ito and Meyerowitz, 2000).
P450 cytochromes represent a family of heme-containing enzymes belonging to the monooxygenase group, which are found in all kingdoms and show extraordinary diversity in their chemical reactions (Schuler et al., 2006; Mizutani and Ohta, 2010). In plants, they are involved in the metabolism of most phytohormones, including auxins, GAs, cytokinins, brassinosteroids, abscisic acid, and jasmonic acid, as well as many secondary metabolites (defensive compounds and fatty acids; Werck-Reichhart et al., 2002; Bak et al., 2011).
Arabidopsis has six CYP78A genes (Bak et al., 2011). The overexpression of KLUH/CYP78A5, ENHANCER OF DA1-1 (EOD3)/CYP78A6, and CYP78A9 genes all produced a general growth phenotype. This phenotype included large siliques and short stamens, a delay in bud opening and organ abscission, reduced fertility, a more severe phenotype in basipetal flowers, and longevity. This shared phenotype between the different genes indicates that they might act on a related metabolic network (Zondlo and Irish, 1999; Ito and Meyerowitz, 2000; Marsch-Martinez et al., 2002; Fang et al., 2012). An insertional knockout line for KLUH/CYP78A5 showed a higher rate of leaf initiation (Wang et al., 2008). The smaller size of leaves, sepals, and petals in homozygous cyp78a5/klu plants was found to be due to a decreased cell number and not caused by a decreased cell size (Anastasiou et al., 2007). The use of cyp78a5/klu plants, in which KLUH/CYP78A5 expression can be induced, showed that CYP78A5 prevents a premature arrest of growth by maintaining cell proliferation. Comparison of regions of organ proliferation and CYP78A5 petal expression domains indicated that CYP78A5 acts non cell autonomously (Anastasiou et al., 2007). The same mode of action occurs in seeds. KLUH/CYP78A5 is expressed in the inner integument of developing ovules and stimulates cell proliferation, thereby determining seed size (Adamski et al., 2009). A loss-of-function mutant for the closely related gene CYP78A7 has also been isolated, although it does not show an evident phenotype (Wang et al., 2008). However, embryos of the double knockout mutant cyp78a5/klu cyp78a7 did not develop correctly. Their size was small and their shoot apical meristem continued to grow, resulting in supernumerary cotyledons. When some seedlings survived, they gave rise to small plants with compacted rosette leaves and an increased leaf initiation rate. It seems, therefore, that KLUH/CYP78A5 and CYP78A7 play redundant roles in regulating the relative growth of the shoot apical meristem and the rest of the plant (Wang et al., 2008). Adamski et al. (2009) reported that the homozygous insertional mutants of CYP78A9 showed a reduction in seed size; however, they did not detect any genetic interaction between CYP78A9 and KLUH/CYP78A5. On the contrary, Fang and coworkers (2012) proposed that CYP78A9 and EOD3/CYP78A6 have an overlapping function in the control of seed size, as noted by the synergistic enhancement of the cyp78a6 seed size phenotype produced in the cyp78a9 background.
Although the catalytic function of the Arabidopsis CYP78A enzymes remains unknown, the expression pattern and the in vivo effect of the related genes bring up their possible engagement in the biosynthesis of some unknown type of plant growth regulator (Zondlo and Irish, 1999; Ito and Meyerowitz, 2000; Anastasiou et al., 2007; Kai et al., 2009).
Hormones seem to have a prominent role in synchronizing fertilization and fruit growth. Dorcey and coworkers (2009) have proposed that a fertilization-dependent auxin response in ovules triggers fruit development through the stimulation of GA metabolism in Arabidopsis. However, this does not explain the phenotype of the empty siliques activation tagging Arabidopsis mutant (es1-D), in which fruit set can be uncoupled from fertilization.
In this work, we provide a detailed description of CYP78A9 function during reproductive development. The genetic relation of CYP78A9 with its closest paralogs, CYP78A6 and CYP78A8, shows a redundant function controlling floral organ growth and integument development, which in turn affect fertility. All the defects observed in the cyp78a8 cyp78a9 double homozygous mutant are related to the sporophyte before fertilization, which supports the idea that this family of genes acts maternally during reproductive development. Moreover, the expression pattern of CYP78A9 studied in detail added a new insight into the possible function of this gene during reproductive development. CYP78A9 expression was observed in integuments during ovule development and highlights the possible communication role of the CYP78A9-produced signal between sporophytic and gametophytic tissue. During seed development, CYP78A9 expression was also present in embryo and seed coat. In addition, the pattern of pCYP78A9::GUS visible in the funiculus 12 h after hand pollination, together with the fact that CYP78A9 overexpression uncoupled fruit development from fertilization, supports the possible role of a CYP78A9-produced signal in synchronizing fertilization and fruit development. Furthermore, metabolic profiling of mutants compared with the wild type showed perturbation in flavonoid content. However, the differences in kaempferol and quercetin content did not explain the observed phenotypes of the mutants. In summary, the results add new insights to the role of CYP78A9 in plant reproduction and present, to our knowledge, the first metabolic characterization of mutants in this family of genes.
RESULTS
Characterization of the Arabidopsis Activation-Tagging Line es1-D
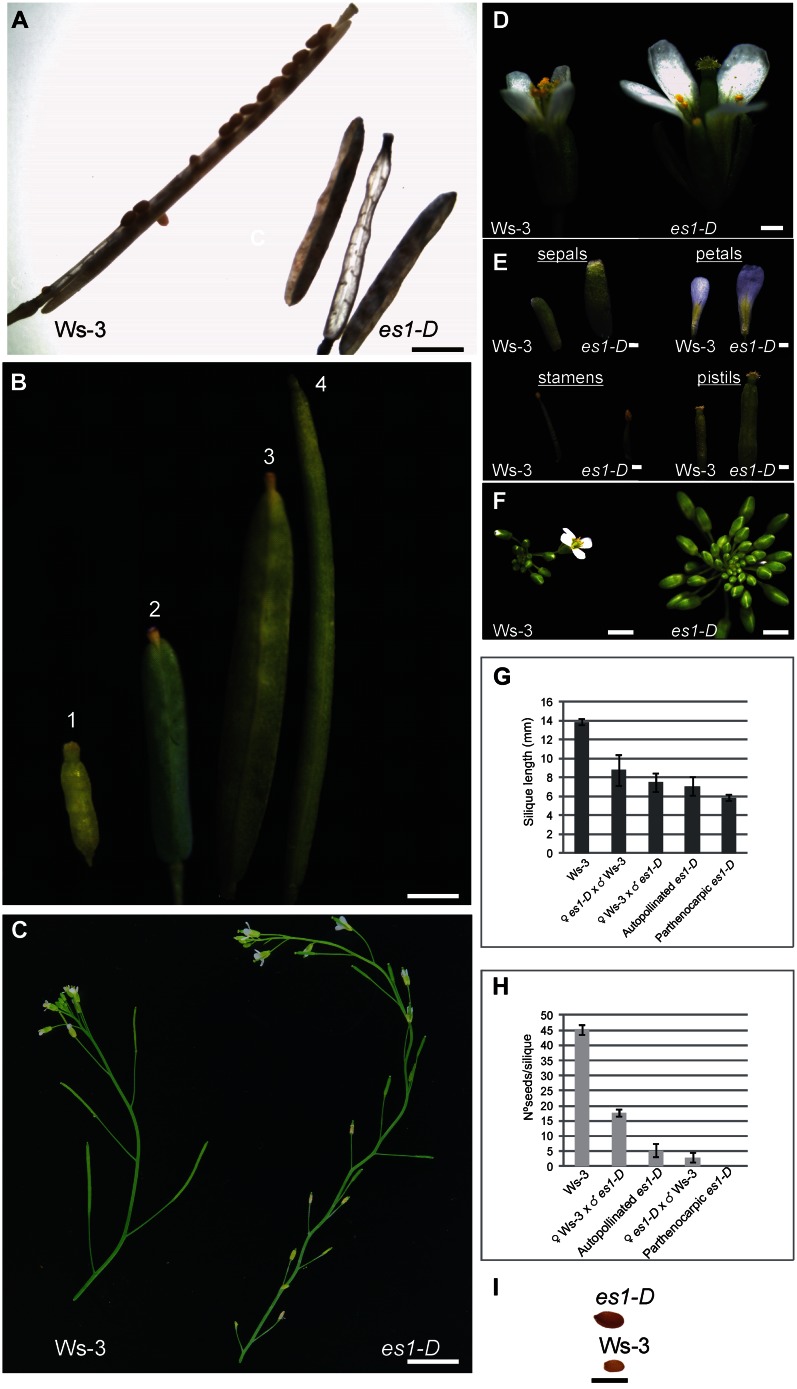
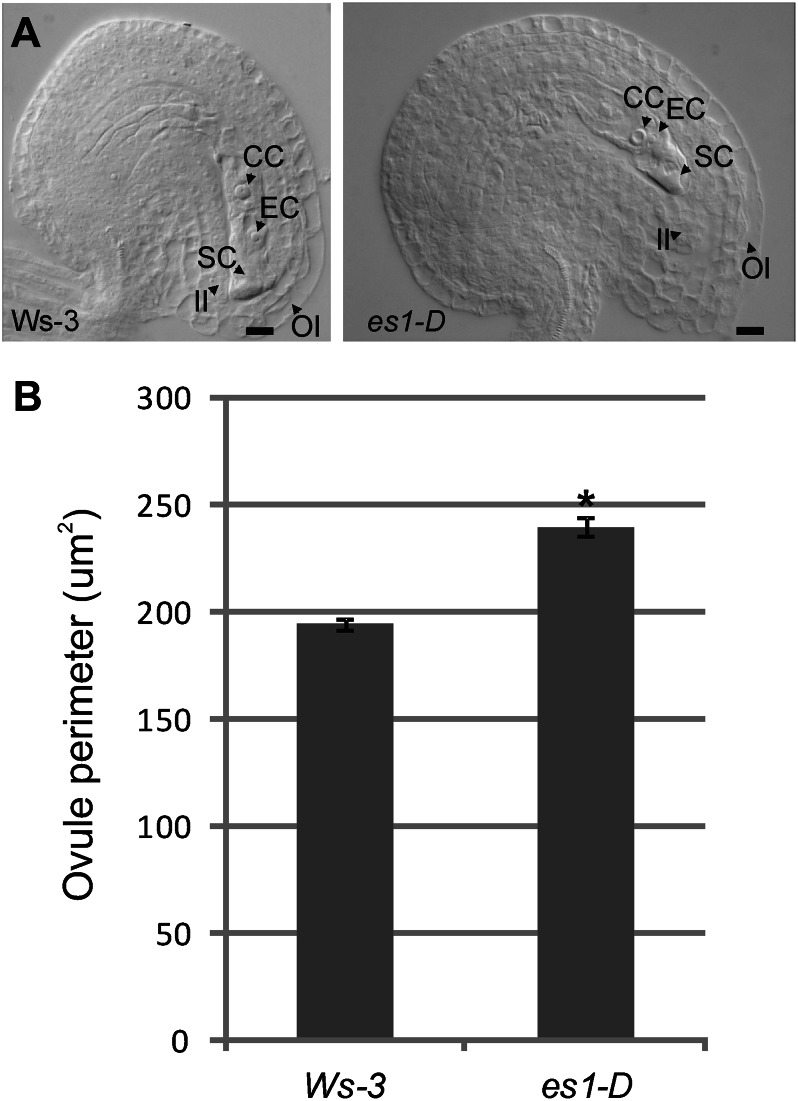
In the activation-tagged es1-D mutant (Supplemental Fig. S1; Marsch-Martinez et al., 2002; de Folter et al., 2004), androecia and gynoecia developed in an uncoordinated manner; anthers dehisce later than in wild-type Arabidopsis plants, resulting in a lack of pollination (Fig. 1). Interestingly, fruits developed even without pollination, which never happens in wild-type plants (Vivian-Smith et al., 2001). Despite the apparently normal carpel morphology, the majority of the mature fruits were empty (Fig. 1A). These parthenocarpic fruits were wider and shorter than the wild type (Fig. 1G), and when developing flowers were emasculated, the produced empty fruits reached a longer and wider size (Fig. 1B). At the end of the es1-D life cycle, pollination occurs but seed yield and fruit length are still reduced (Fig. 1, G and H). Reciprocal crosses were made to analyze whether the male and/or the female reproductive side was affected. The reduced fertility could not be rescued with wild-type pollen (Fig. 1H), suggesting that es1-D has defects during ovule development. Ovule number was not changed in es1-D but had larger ovules and displayed outer integuments with more and larger cells than the wild type (45–50 compared with 29–35 cells in wild-type Wassilewskija [Ws-3]; Fig. 2A). The ovule perimeter was significantly (P < 0.001, Student’s t test) larger in es1-D compared with wild-type Ws-3 (Fig. 2B). However, es1-D pollen also failed to produce normal fruit set when crossed to the wild type: only 17 to 20 seeds per fruit were observed (Fig. 1H), compared with normally 40 to 50 seeds found in wild-type fruits. Although the fruits of this cross (♀ Ws-3 × ♂ es1-D) had more seeds, they did not reach the length of a wild-type fruit (Fig. 1G). These results indicate that both male and female reproductive development is affected in the es1-D mutant.
Figure 1.
Flower and fruit phenotypes of es1-D and wild-type plants. A, Open mature Ws-3 and es1-D fruits. B, es1-D fruit development: 1, es1-D pistil at stage 13; 2, es1-D nonemasculated developed fruit; 3, es1-D developed fruit from a pistil emasculated at stage 13; 4, wild-type developed fruit. C, Detail of the apical part of the main inflorescence stem from wild-type and es1-D plants. Note the zigzag pattern in the es1-D mutant. D, Stage 14 wild-type and es1-D flowers. The uncoordinated development of androecium and gynoecium in the es1-D mutant is visible. E, Sepals, petals, stamens, and pistils of the flowers shown in D. F, Closeup view of wild-type and es1-D inflorescences showing a strong delay in bud opening in es1-D compared with the wild type. G, Silique length comparison between es1-D parthenocarpic, self-pollinated, and wild-type or es1-D cross-pollinated fruits. H, Seed set comparison between es1-D parthenocarpic, self-pollinated, and wild-type or es1-D cross-pollinated fruits. I, Representative es1-D and wild-type seeds. Values represent means ± se (n = 10 for autopollinated Ws-3, autopollinated es1-D, and parthenocarpic es1-D; n = 6 for reciprocal crosses). Bars = 0.2 cm in A, B, and F; 2 cm in C; 0.2 mm in D and E; and 1 mm in I.
Figure 2.
The es1-D mutant has larger ovules and develops outer integuments with more and larger cells. A, Mature ovules from Ws-3 and es1-D plants, showing bigger es1-D ovules with more cells in the outer integument. CC, Central cell; EC egg cell; II, inner integument; OI, outer integument; SC, synergid cell. Bars = 10 µm. B, Ovule perimeter comparison between Ws-3 and es1-D, showing that ovule perimeter was significantly larger in es1-D than in Ws-3 plants (*P < 0.001). Values represent means ± se (n = 40).
Furthermore, we observed that es1-D sepals, petals, pistils, and seeds were larger in comparison with the wild type (Fig. 1, D, E, and I). However, es1-D stamen filaments failed to elongate (Fig. 1E). Moreover, basipetal floral buds were slow to open, and stamen filament elongation was more severely affected. The more acropetal flowers were less severely affected, although the sepals and petals of those flowers did not abscise normally. In addition to the flower phenotypes, the es1-D plants showed larger dark green leaves and stout stems with a zigzag pattern (Fig. 1C), a phenotype that has also been observed when KLUH/CYP78A5 and EOD3/CYP78A6, CYP78A subfamily members, are overexpressed (Zondlo and Irish, 1999; Ito and Meyerowitz, 2000; Marsch-Martinez et al., 2002; Fang et al., 2012).
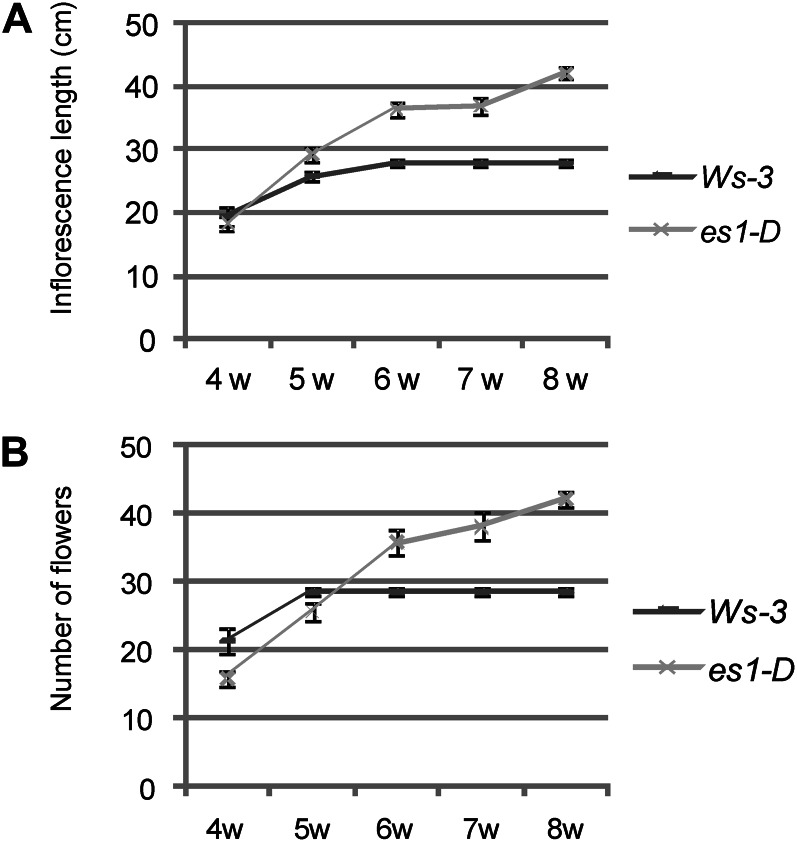
The es1-D mutant flowered later (21 d) than the wild-type plants (16 d), but no difference in the number of leaves produced at the moment of flowering was observed (Supplemental Fig. S2). Furthermore, the period of flower production and the total number of flowers produced in the main inflorescence of mature es1-D and wild-type plants were compared. At week 5 after flower initiation, wild-type plants stopped producing flowers, while at week 8, the main inflorescence of a still growing es1-D plant was producing flowers (Fig. 3B). Data presented in Figure 3 show that es1-D clearly produces flowers over a longer period and that the growth of its main inflorescence is extended by several weeks (Fig. 3A); therefore, it produced more flowers, but not because of faster growth. This phenotype was also described by Hensel et al. (1994) for mutants that have reduced fertility, whereby mutations that reduced the number of seeds per silique by more than 50% were associated with an increased proliferative capacity of the main inflorescence stem.
Figure 3.
Increased longevity of es1-D plants. A, Inflorescence length of wild-type Ws-3 and es1-D plants. The es1-D mutant lives longer and produces longer inflorescence stems than the wild type. B, Number of flowers per main inflorescence stem in wild-type and es1-D plants. es1-D produces more flowers. Values represent means ± se (n = 10). Numbers at bottom represent weeks (w) of growth.
To confirm that the observed phenotypes in es1-D were due to CYP78A9 overexpression, the entire genomic region was cloned and expressed constitutively under the control of the 35S promoter (35S::gCYP78A9). We obtained 21 independent lines, and 17 of them displayed the es1-D phenotype (Supplemental Fig. S3).
The phenotypes observed in the activation-tagging mutant es1-D as well as in the transgenic lines constitutively overexpressing CYP78A9 suggest that CYP78A9 has a function controlling floral organ size and, moreover, that it is involved in producing a signal that can overcome the pistil senescence program that initiates when in wild-type plants fertilization does not occur (Vivian-Smith et al., 2001; Fuentes and Vivian-Smith, 2009).
Characterization of Arabidopsis cyp78a9, cyp78a6, and cyp78a8 Single and Double Mutants
To better understand the function of CYP78A9, T-DNA insertion loss-of-function mutants for CYP78A9 and for its closest paralogs, CYP78A6 and CYP78A8, were analyzed (Supplemental Fig. S1, B–E). Homozygous plants were identified by PCR genotyping, and reverse transcription (RT)-PCR experiments confirmed that they were true knockout mutants (Supplemental Fig. S4).
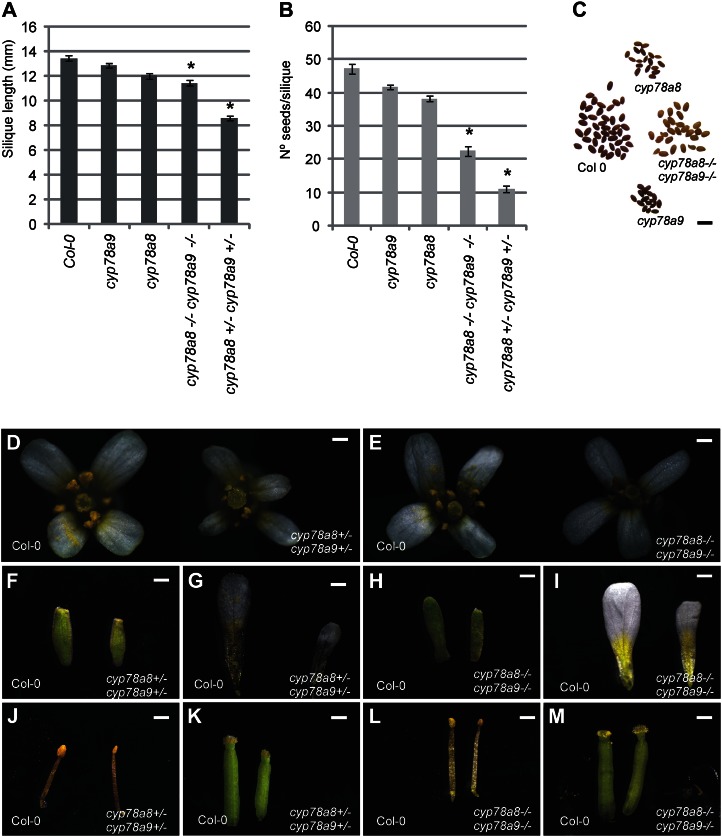
Regarding fruit development, no evident loss-of-function phenotype was observed in cyp78a9, cyp78a6, and cyp78a8 single mutants compared with wild-type Columbia (Col-0). All single mutants showed no difference in fruit length and seed number per fruit compared with each other and with the wild type (P < 0.001; Fig. 4, A and B; Supplemental Fig. S5, A and B). To test whether CYP78A6 or CYP78A8 acts redundantly with CYP78A9 in regulating fruit length and seed set, we crossed cyp78a9 to cyp78a6 and to cyp78a8. A reduction in both fruit length and seed set was already evident in F1 plants cyp78a8 (+/−) cyp78a9 (+/−) compared with the wild type and single mutants (P ≤ 0.001). These results were also confirmed in F2 double heterozygous plants (Fig. 4, A and B).
Figure 4.
Flower and fruit phenotypes of cyp78a9, cyp78a8, and double mutants. A, Mature silique length for the wild type and cyp78a9 and cyp78a8 single and double mutants. Silique length is reduced in the cyp78a8 (−/−) cyp78a9 (−/−) and cyp78a8 (+/−) cyp78a9 (+/−) double mutants. B, Seed yield. The number of seeds per silique is severely reduced in cyp78a8 (−/−) cyp78a9 (−/−) and cyp78a8 (+/−) cyp78a9 (+/−) double mutants. C, Mature seeds of the wild type and cyp78a9 and cyp78a8 single and double mutants. Note the pale testa color of the double mutant. D, Flower phenotypes of the cyp78a8 (+/−) cyp78a9 (+/−) double heterozygous mutant compared with the Col-0 wild type. E, Flower phenotypes of the cyp78a8 (−/−) cyp78a9 (−/−) double homozygous mutant compared with the Col-0 wild type. F to M, Sepals, petals, stamens, and pistils of the flowers presented in D and E. Note the reduction of organ size in the double homozygous and double heterozygous mutants. Values represent means ± se (n = 20). *Significantly different from Col-0 at P < 0.001. Bars = 0.2 mm in D to M and 1 mm in C.
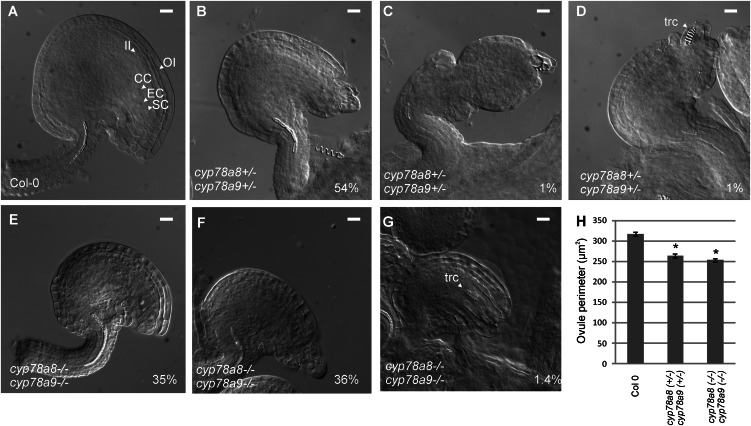
This reduction in seed set was related to defects in ovule development. From a total of 194 cyp78a8 (+/−) cyp78a9 (+/−) ovules analyzed, 109 (56%) presented different levels of integument development arrest. The most abundant phenotype, 105 of 194 ovules (54%), was one that still conserved to some extent the asymmetry of a wild-type ovule but presented short integuments that failed to accommodate the developing embryo sac, resulting in physical restriction of the gametophyte leading to female sterility (Fig. 5B). The affected ovules (56%) were smaller, and the reduction of the cyp78a8 (+/−) cyp78a9 (+/−) ovule perimeter was significantly different (at P < 0.001 by Student’s t test) compared with wild-type Col-0 (Fig. 5H). Furthermore, in severe cases (around 1% of ovules), the outer integument interrupted its growth and the ovule developed a somehow orthotropic morphology (Fig. 5C). In other cases (around 1% of ovules), tracheid-like cells appeared at the position of the embryo sac (Fig. 5D).
Figure 5.
Ovule phenotypes of cyp78a9, cyp78a8, and double mutants. A, Mature ovules from the wild type (Col-0). B to D, Mature ovules from the cyp78a8 (+/−) cyp78a9 (+/−) double heterozygous mutant. From 194 ovules analyzed, 105 (54%) have shorter outer integuments that failed to accommodate the embryo sac but still conserve ovule characteristic asymmetry (B), around 1% have severe arrest of outer integument growth (C), and in some cases (1%), tracheid-like cells appear at the position of the embryo sac (D). E to G, Mature ovules from the cyp78a8 (−/−) cyp78a9 (−/−) double homozygous mutant. From 213 ovules analyzed, 74 (35%) have shorter outer integuments than the wild type but still have a normal embryo sac (E), 77 (36%) present growth arrest of the outer integument and embryo sac abortion (F), and three (1.4%) present tracheid-like cells in the position of the embryo sac (G). CC, Central cell; EC, egg cell; II, inner integument; OI, outer integument; SC, synergid cell; trc, tracheid-like cell. Bars = 10 µm. H, Comparison of ovule perimeter between Col-0 and double homozygous and heterozygous mutants. *Significantly different from Col-0 at P < 0.001. Values represent means ± se (n = 40).
Interestingly, the cyp78a8 (−/−) cyp78a9 (−/−) double homozygous mutant showed alterations in silique length and seed set, but to a lesser extent than in the cyp78a8 (+/−) cyp78a9 (+/−) double mutant (Fig. 4, A and B). In the double homozygous mutant, ovules were also affected in outer integument development. From 213 analyzed ovules, 74 (35%) were significantly (P < 0.001 by Student’s t test) reduced in size (Fig. 5H) due to fewer cells in the outer integument (15–20 compared with 29–33 cells in wild-type Col-0) but still presented a normal embryo sac (Fig. 5E). Seventy-seven (36%) developed short integuments that failed to accommodate the developing embryo sac (Fig. 5F), and three (1.4%) presented tracheid-like cells at the position of the embryo sac (Fig. 5G). The frequency of the phenotype severity correlates well with the observed seed set.
In addition, the cyp78a8 (+/−) cyp78a9 (+/−) and cyp78a8 (−/−) cyp78a9 (−/−) double mutants presented a reduction in floral organ size compared with the wild type (Fig. 4, D–M), which is the opposite effect seen from when CYP78A9 is overexpressed (Fig. 1). This phenomenon is also reported for mutants with outer integument defects, as for AINTEGUMENTA (ANT), in which strong mutants presented random reduction of organ size in the outer whorls (Klucher et al., 1996). Furthermore, a change in color of the seed testa was observed in cyp78a8 (−/−) cyp78a9 (+/−) and cyp78a8 (−/−) cyp78a9 (−/−) (pale brown; Fig. 4C), which is normally seen in mutants related to the phenylpropanoid pathway (Buer et al., 2010).
An analysis of the cross between cyp78a6 and cyp78a9 showed no reduction in silique length, although the number of seeds produced per silique was slightly diminished in F1 generation plants (Supplemental Fig. S5, A and B). However, Fang and coworkers (2012) showed that cyp78a6 cyp78a9 had no reduced seed set but reduced seed size due to smaller cells in the integuments of the developing seeds, and they proposed that cyp78a9 synergistically enhanced the seed-size phenotype of cyp78a6.
In our work, an analysis of the cyp78a8 (−/−) cyp78a9 (−/−) and cyp78a8 (+/−) cyp78a9 (+/−) double mutants showed a clear genetic interaction between these genes controlling seed set. Moreover, the defects observed in cyp78a8 (−/−) cyp78a9 (−/−) and cyp78a8 (+/−) cyp78a9 (+/−) double mutants are linked to the sporophyte before fertilization, which supports the idea that this family of genes acts maternally during reproductive development.
CYP78A9 Expression during Reproductive Development
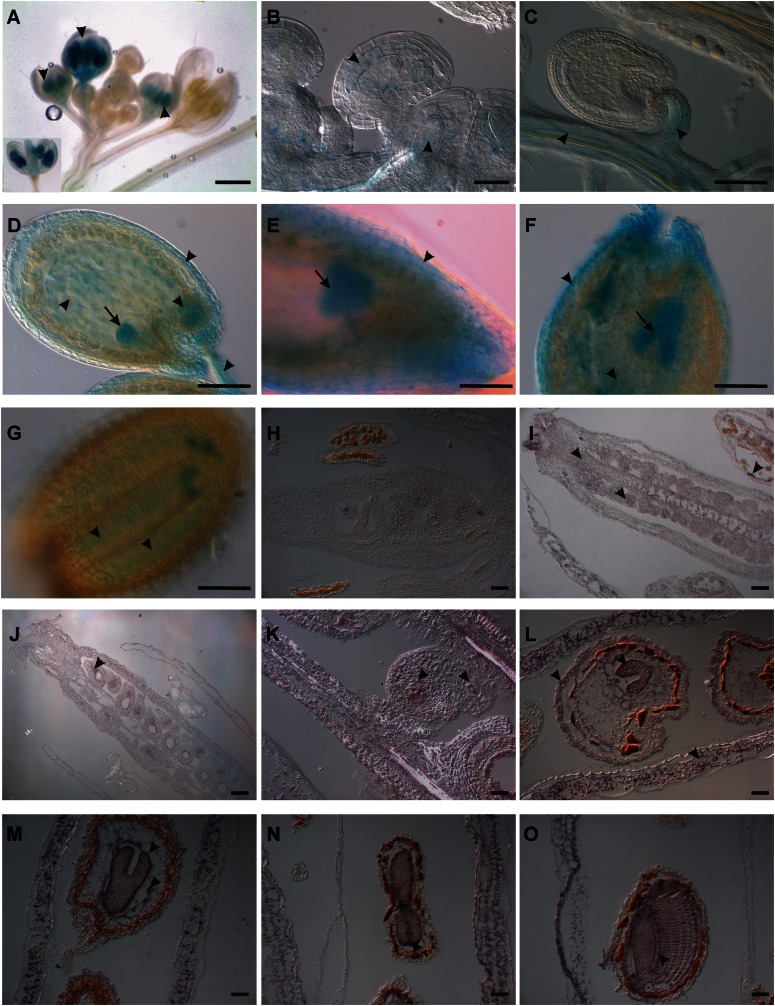
In order to learn more about the CYP78A9 gene, we first made a transcriptional fusion with the GUS reporter under the control of the putative 3-kb CYP78A9 promoter region (CYP78A9::GFP:GUS; Fig. 6, A–G). Transgenic plants were analyzed during reproductive development, and GUS staining was observed first at floral developmental stage 11 according to Smyth et al. (1990). At stage 11, GUS staining was localized in anthers, and around stage 12, staining was also observed at the gynoecium (Fig. 6A) and the inner integuments of the ovules (Fig. 6B). At stage 13, when flowers open and fertilization occurs, the staining was localized at the placenta and the funiculi (Fig. 6C). During seed development, strong staining was observed in the embryo, from the globular stage until the curly leaf stage of the embryo (Fig. 6, C–G, arrows), with a peak at the torpedo stage (Fig. 6F, arrow). Furthermore, signal was also evident at the maternal chalazal region, the endosperm, and the developing testa (Fig. 6, D–F, arrowheads).
Figure 6.
Expression patterns of CYP78A9 in flower, fruit, and seed tissues using promoter::GUS analysis (A–G) and in situ hybridization (H–O). A, pCYP78A9::GUS flowers showing GUS signal in anthers at stages 11 and 12 (arrowheads). At stage 12, the signal is also localized in pistils. B, Ovules of a stage 10 bud showing expression in the inner integuments (arrowheads). C, Ovule 24 h after pollination showing signal in the funiculus and placenta (arrowheads). D, Developing seed showing CYP78A9 expression in integuments, funiculus, endosperm, and embryo at the globular stage (arrowheads). E, Seed with heart-stage embryo. The GUS signal is localized in the embryo (arrow) and in the integuments (arrowhead). F, Seed with torpedo-stage embryo. At this stage, the signal is stronger in the embryo (arrow) and endosperm (arrowhead) but still localized in the integuments (arrowhead). G, Seed with curly leaf-stage embryo. The signal is localized in the cotyledons and the radicle (arrowheads). H, CYP78A9 sense probe hybridized to longitudinal sections of wild-type gynoecium from a closed bud. No signal is observed. I, CYP78A9 antisense probe hybridized to longitudinal sections of a wild-type closed bud. CYP78A9 is expressed in ovules, stigma, marginal tissue, and tapetum (arrowheads). J, CYP78A9 antisense probe hybridized to a wild-type pollinated flower showing expression in developing seeds and valves (arrowheads). K, CYP78A9 antisense probe hybridized to an early developing seed showing expression in chalaza and micropylar regions (arrowheads). L to O, CYP78A9 antisense probe hybridized to wild-type fruits showing seeds with embryo at heart, torpedo, and curly stage. CYP78A9 is expressed in the embryo and strongly in the epidermis of wild-type embryo (arrowheads), with the strongest signal observed in torpedo-stage embryos (arrowhead; O). Expression can also be seen in the integuments of the developing seeds and in the valves of the fruit (arrowheads). Bars = 1 mm in A to G and 10 µm in H to O.
A more detailed expression analysis by in situ hybridization confirmed the observed GUS patterns and gave additional information on CYP78A9 expression (Fig. 6, H–O). Floral buds at stage 10 showed that CYP78A9 was expressed in the tapetum cells of the anthers (Fig. 6I). Moreover, at this stage, CYP78A9 mRNA was localized in ovules, placenta, and also in the valves and stigma. After fertilization, signal in the developing seeds was stronger compared with mature ovules and was localized to the chalazal and micropylar regions (Fig. 6, J–K). During seed development, mRNA localization was observed in the embryo in the same manner as seen in transgenic plants with CYP78A9::GFP:GUS; however, the highest in situ hybridization signal was localized at the epidermis of developing embryos (Fig. 6, L–O). Furthermore, signal was detected in endosperm, in the testa of the seed, and in valves of developing fruits.
Notably, mRNA expression analysis in es1-D mutant floral buds presented the same expression pattern compared with the wild type, although with an approximately 2-fold increase in signal intensity, confirming that the es1-D phenotype is due to the altered amount of mRNA and not to ectopic expression of CYP78A9 (Supplemental Fig. S6).
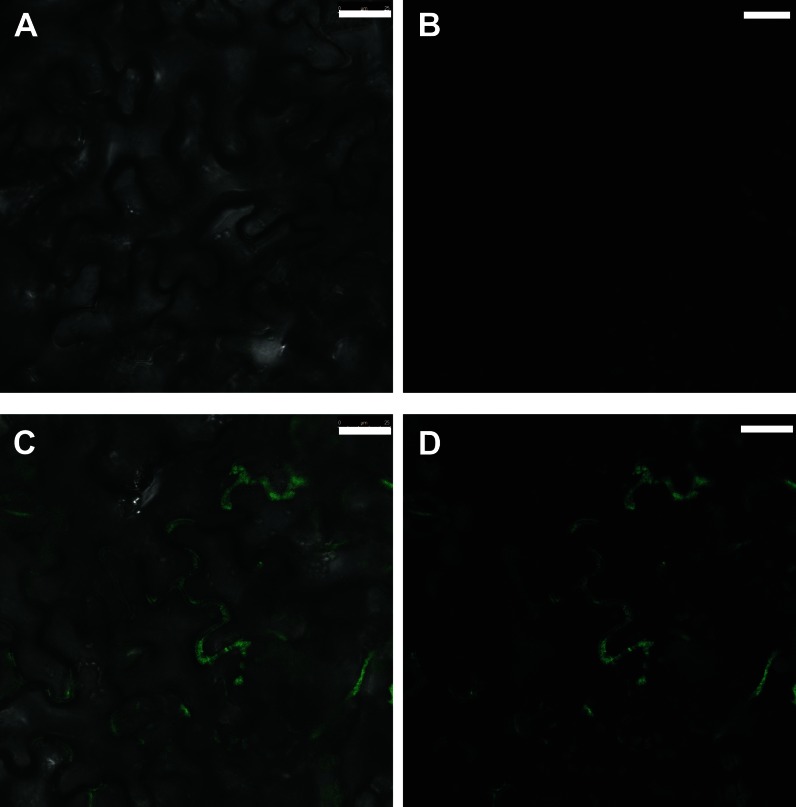
Furthermore, we analyzed the protein localization for CYP78A9. For this, a transient expression assay was performed by infecting tobacco (Nicotiana tabacum) leaves with Agrobacterium tumefaciens containing a 35S::gCYP78A9:GFP translational GFP fusion construct. GFP fluorescence signal was detected by confocal microscopy at both sides of the cellular wall of consecutive cells, suggesting that CYP78A9 could be a plasma membrane-associated protein (Fig. 7).
Figure 7.
Cellular CYP78A9 protein localization. A and B, Background fluorescence signal of noninfected tobacco leaves. A, Bright-field view. B, Dark-field view. C and D, Fluorescence GFP signal of gCYP78A9:GFP in infected tobacco leaves with 35S::gCYP78A9:GFP. C, Bright-field view. D, Dark-field view. The fluorescence signal detected suggests that the CYP78A9 protein is associated with the plasma membrane. Bars = 25 µm.
CYP78A9 Responds to the Fertilization Event
Because of the fertility-related phenotypes observed, we investigated what happened with the expression of CYP78A9 during fertilization. In situ and CYP78A9::GFP:GUS analyses (Fig. 6) demonstrated signal in mature ovules and in the septum before fertilization. After fertilization, signal was observed in developing seed and septum, but also in the funiculus and placental tissue. The funiculus is the connection between the mother plant, via the placenta, and the developing offspring and is important for nutrition. Stadler and coworkers (2005) demonstrated that the release of photoassimilates and macromolecules to the developing seed is mediated by the outer integument, which represents a symplastic extension of the funicular phloem.
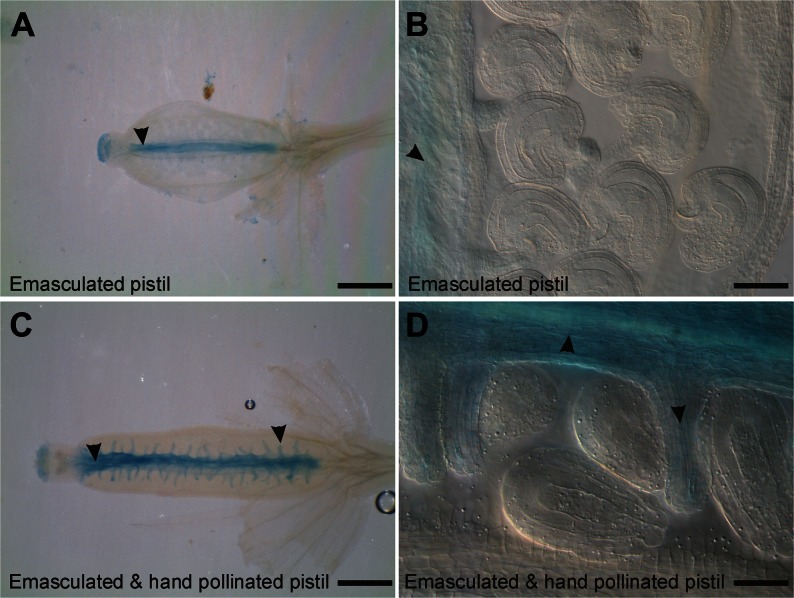
Developing flowers of transgenic CYP78A9::GFP:GUS plants were emasculated and observed 12 h after hand pollination. At this time point, fertilization has normally taken place (Faure et al., 2002). As a control, CYP78A9::GFP:GUS plants were emasculated and stained after 12 h. In these control plants, GUS signal in the ovary was observed only in the septum (Fig. 8, A and B). Interestingly, in emasculated and afterward hand-pollinated pistils, GUS signal was observed also in the funiculus and in the placental tissue (Fig. 8, C and D). These results indicate that the CYP78A9 promoter responds to the fertilization event.
Figure 8.
CYP78A9 promoter (pCYP78A9::GUS) responds to the fertilization event. A, Pistil 12 h after emasculation showing expression in the stigma and septum (arrowhead) but not in the funiculus. B, Closeup view of ovules from the emasculated pistil (in C). Signal is only present in the septum but not in ovules. C, Pistil 12 h after hand pollination showing expression in the stigma, placenta, and funiculus (arrowheads). D, Closeup view of ovules of the pollinated pistil (in A) showing clear GUS signal localization in the funiculus and placenta (arrowheads) but not in ovules. Bars = 1 mm.
The CYP78A9-Produced Signal Seems To Be Different from the Known Hormones That Trigger Fruit Growth after Fertilization
To determine whether CYP78A9 was involved in known pathways regulating fruit growth, the response patterns of two of the major hormones involved in this process, auxins and GAs, were analyzed in the es1-D mutant.
A cross between es1-D and the auxin-response marker DR5::GUS (Ulmasov et al., 1997) showed a similar expression pattern to wild-type DR5::GUS plants throughout flower development (Supplemental Fig. S7). The only noted difference was observed in stage 14 flowers. In wild-type DR5::GUS plants, where normal fertilization occurs at floral stage 13, GUS signal was seen in fertilized ovules (developing seeds) and in funiculi, while no signal was observed in this tissue in es1-D DR5::GUS plants due to the absence of pollination (Supplemental Fig. S7). However, it is possible that the auxin increase required to produce parthenocarpic fruit development occurs earlier in the es1-D mutant.
We analyzed the expression of the GA biosynthesis marker line GA20ox1::GUS (Desgagné-Penix et al., 2005) in the es1-D background. The same expression pattern was observed during flower and fruit development in the wild-type background as in the es1-D background (Supplemental Fig. S7). These data suggest that GAs cannot be the cause of the uncoupled growth of es1-D fruit from fertilization.
On the other hand, we evaluated the transcriptional effect of the application of different hormones, as well as hormone inhibitors in seedlings, on the expression of CYP78A9. For this, publicly available microarray data present in the Electronic Fluorescent Pictograph browser (Winter et al., 2007) was used. We observed that CYP78A9 responded transcriptionally to the application of many hormones (Supplemental Fig. S8) as well as hormone inhibitors (Supplemental Fig. S9). The CYP78A9 expression changes observed did not allow us to identify if CYP78A9 might be involved in one of these hormone pathways; however, a hypothesis about CYP78A9 expression being regulated by inputs from multiple hormone signals could be made.
Microarray Expression Analysis in the es1-D Mutant
In order to increase our knowledge about the transcriptional responses to CYP78A9 activity, a microarray experiment comparing es1-D and wild-type closed floral buds from young primary inflorescences was performed. CYP78A9-responsive genes were defined as genes that were significantly differentially expressed in es1-D (z score > 2 sd) compared with the wild type, which resulted in 409 up-regulated and 825 down-regulated genes (Supplemental Table S1).
Some hormone and secondary metabolite enzymes were identified. Among the up-regulated genes, we can mention CYP724A1, encoding a brassinosteroid C-22 hydroxylase (Zhang et al., 2012), FMO GS-OX1, a glucosinolate oxygenase (Hansen et al., 2007), and MIOX1, a MYOINOSITOL OXYGENASE1 involved as an entry point of ascorbate biosynthesis (Endres and Tenhaken, 2009), converting myoinositol into d-GlcA, which is an important precursor for cell walls and regulates the flux of inositol into the phosphoinositol pathway (Alimohammadi et al., 2012).
The down-regulated genes included YUC1 (Cheng et al., 2006, 2007), AT3G51730 (SAUR-like response factor), IAA3/SHY2 (Tian et al., 2002), and IAA27/PAP2 (Liscum and Reed, 2002), involved in auxin metabolism and signaling; Jasmonate-Regulated Gene21 and a lipooxygenase, involved in jasmonic acid biosynthesis (Melan et al., 1993); BRI1, associated with brassinosteroid signaling (Noguchi et al., 1999); and AT2G39540, which encodes a GA-regulated protein (Roxrud et al., 2007).
Interestingly, CYP78A8 was down-regulated in response to the overexpression of CYP78A9 as well as chalcone isomerase, responsible of converting chalcones into flavonones (Buer et al., 2010), and CYP71B16, reported as coexpressed with the phenylpropanoid pathway (Ehlting et al., 2006). A relationship between the phenylpropanoid pathway and parthenocarpy was suggested also by the gene expression profiles of highly and weakly parthenocarpic pear (Pyrus spp.) cultivars. The highly parthenocarpic group showed changes in genes located in the junctions of the metabolic routes leading to the production of flavonoids or monolignols and sinapoylmalate (Nishitani et al., 2012).
Several genes related to ovule and stamen development were changed. Concerning ovule integument development, we found HUELLENLOS up-regulated (Skinner et al., 2001) and ANT down-regulated (Elliott et al., 1996).
In relation to stamen development and to the early defects observed in es1-D, short filaments and delay in anther dehiscence, MYB21 and MYB65 were down-regulated (Millar and Gubler, 2005; Song et al., 2011). With regard to pollen development and pollen tube growth, TCP16 (Takeda et al., 2006), AT1G10770 (pectin methylesterase/invertase inhibitor; Zhang et al., 2010), phospholipase (PLA2-γ; Kim et al., 2011), and the arabinogalactan proteins (AGP7, AGP11, AGP22, and AGP23) were found to be down-regulated (Pereira et al., 2006; Seifert and Roberts, 2007; Coimbra et al., 2009; Ellis et al., 2010).
Furthermore, a comparison was made between the data obtained in es1-D and the described sets of hormone-regulated genes (Nemhauser et al., 2006), but no clear overlap was found (Supplemental Table S2).
Previous information on transcriptional responses to CYP78A9 was obtained by de Folter et al. (2004) in fruits, using a macroarray containing probes to analyze the expression of over 1,100 transcription factors. By means of a comparison between the microarray (this study) and the macroarray (de Folter et al., 2004) data, 28 down-regulated and 16 up-regulated genes were down- and up-regulated in both data sets, respectively (Supplemental Table S3). Among these, we found a series of transcription factors that could be related to the altered phenotypes seen in the es1-D mutant. For instance, ANT and BEL1-like (SAWTOOTH1 [BLH2], SAWTOOTH2 [BLH4], and BEL1-LIKE HOMEODOMAIN5 [BLH5]) transcription factors were related to integument development (Elliott et al., 1996). Furthermore, MYB3, down-regulated in both experiments, is a transcriptional repressor of proanthocyanidin biosynthesis (Feller et al., 2011). Interestingly, two enzymes involved in flavonol production were identified in the common gene list, a family 1 glycosyl transferase (up-regulated) and chalcone flavonone isomerase (TT5; down-regulated).
In summary, the microarray experiment showed an altered transcriptional pattern of genes associated with male and female reproductive development. Related to hormones, some genes were found to be down-regulated, although no clear trends could be observed. A set of 16 P450 enzymes changed their levels of expression in the es1-D mutant. Some of them were not annotated as being part of a specific biosynthetic pathway, and of the ones that were annotated, each participates in a different pathway (CYP79F1, glucosinolate biosynthesis; CYP710A2, brassinosteroid biosynthesis; CYP86A7, fatty acid metabolic process). Interestingly, CYP78A8, one of CYP78A9’s closest paralogs, was down-regulated in response to CYP78A9 overexpression. Furthermore, enzymes involved in the phenylpropanoid pathway, such as chalcone isomerase (TT5), and other P450 enzymes, such as CYP71B16 and CYP716A1, reported as coexpressed with this pathway, responded to the overexpression of CYP78A9.
CYP78A9 Function Prediction Using Public Database Analyses
Another strategy undertaken for the identification of the role of CYP78A9 was the analysis of genes coexpressed with CYP78A9 based on floral tissue public microarray data. A total of 88 genes (Supplemental Table S4) were found to be coexpressed with CYP78A9 using the Algorithm for the Reconstruction of Accurate Cellular Networks (Margolin et al., 2006a, 2006b). These genes were then annotated and grouped as belonging to one of five functional categories using the gene functional classification tool DAVID (Huang et al., 2008, 2009). Group 1 was enriched for genes linked to cyanoamino acid metabolism, phenylpropanoid, starch and Suc biosynthesis, and lipid metabolism processes. Group 2 included genes involved in fatty acid metabolism, alkaloid, and flavonoid biosynthetic processes. Protein kinase signaling pathway and ATP-binding cassette transporter G (in animals related to eye pigment transport; Schmitz et al., 2001) formed part of group 3. Flavonoid and methylglyoxal catabolic process, protein kinase related to fatty acid signaling pathway, triacylglycerol and ketone body metabolism, and Phe, Tyr, and Trp biosynthesis genes formed group 4. Finally, genes belonging to group 5 were implicated in calcium-dependent lipid-binding signaling involved in cell death.
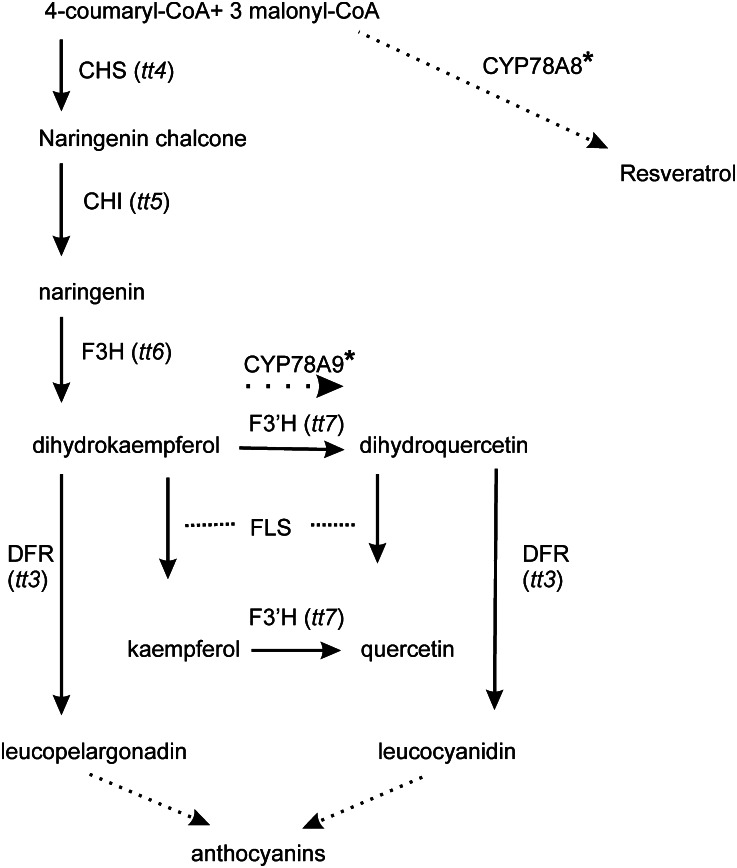
Predictions made by AraCyc, a metabolic pathway reference database (Zhang et al., 2005; Rhee et al., 2006), positioned the CYP78A9 protein in the phenylpropanoid pathway, more specifically in the flavonol biosynthesis pathway, at the conversion of dihydrokaempferol to dihydroquercetin, and/or in luteolin biosynthesis and in eriodictyol formation from naringenin, steps known to be catalyzed by the enzyme flavonoid 3′-monooxygenase (F3′H; Fig. 9). In the case of CYP78A8, the Cytochrome P450 Expression Database (Ehlting et al., 2006) showed that the top-scoring coexpressed pathway was monoterpene/sesquiterpene/diterpene biosynthesis, and the Kyoto Encyclopedia of Genes and Genomes (Kanehisa and Goto, 2000; Kanehisa et al., 2012) predicted its function in stilbenoid, diarylheptanoid, and gingerol biosynthesis. For CYP78A6, there are no predictions available.
Figure 9.
The flavonoid branch of the phenylpropanoid pathway. Known locations of Arabidopsis tt mutations are indicated in parentheses on the phenylpropanoid pathway. CYP78A9 and CYP78A8 are located in the pathway based on the AraCyc and Kyoto Encyclopedia of Genes and Genomes bioinformatic predictions, respectively.
All these data, together with bioinformatics predictions reported in the Cytochrome P450 Expression Database (Ehlting et al., 2006), suggest that CYP78A9 could be an enzyme in the core phenylpropanoid pathway (Fig. 9). However, we cannot discard its function in other predicted pathways, like cell wall carbohydrate metabolism or lipid, fatty acid, and isoprenoid biosynthesis.
Kaempferol and Quercetin Contents in Leaves and Inflorescences of cyp78a9, cyp78a8, cyp78a8 cyp78a9, and es1-D Mutants
The response patterns of DR5::GUS and GA20ox::GUS as well as the transcriptional responses to the overexpression of CYP78A9 indicate that the CYP78A9-produced metabolite seems to be different from the known hormones that trigger fruit growth after fertilization. The group of genes found to be coexpressed with CYP78A9 in public flower microarray data showed that this gene could be involved in many processes, including secondary metabolism and signaling pathways. Moreover, AraCyc predicted the function of CYP78A9 at the same point in the phenylpropanoid pathway as F3′H. These data, together with the changes observed in the testa color of the double mutant that are similar to that observed in the transparent testa7 (tt7) mutant (mutated in F3′H), motivated us to explore whether CYP78A9 could act in the flavonol biosynthesis pathway.
Metabolic profiling was performed using liquid chromatography-mass spectrometry (LC-MS) and multiple fragmentation. Acetone crude extracts from leaves and inflorescences were screened by LC-MS (ACQUITY UPLC-LCT Premier XE equipment; Waters). The analyses of the spectra obtained both from the mutant and its respective wild-type ecotype were performed using MarkerLynx (Waters) and an integrated package comparing and discriminating data sets using multivariable statistics such as principal component analysis (PCA). Selected metabolites were further fragmented using the SYNAPT HDMS system (Waters), and fragmentation spectra of the highlighted metabolites were compared with authentic kaempferol and quercetin standards.
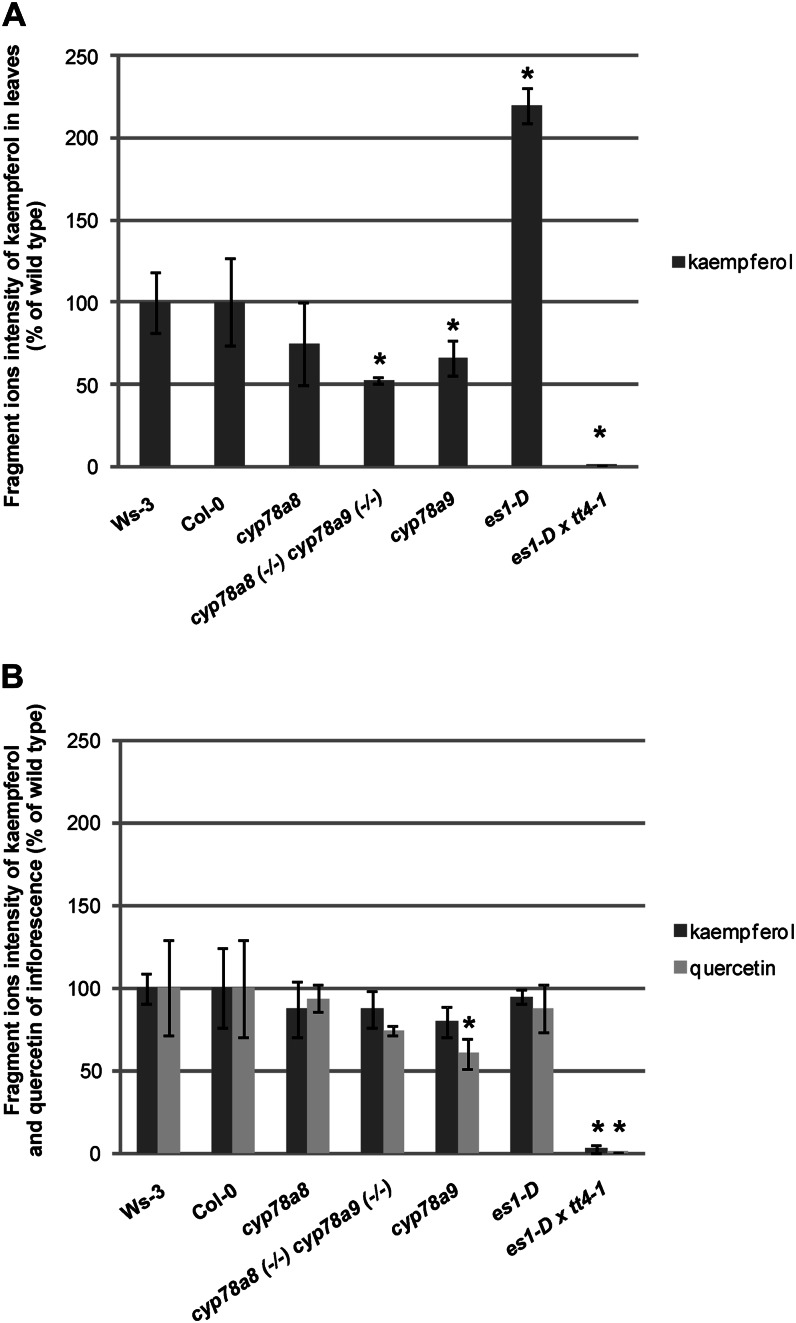
In order to find out if metabolic fluxes were guided toward a specific direction in the phenylpropanoid pathway, we analyzed hydrolyzed acetone extracts, which reveals the total pool of kaempferol and quercetin in the samples. cyp78a9 and cyp78a8 (−/−) cyp78a9 (−/−) leaves have 40% and 50% kaempferol content reductions, respectively, compared with wild-type Col-0 (P < 0.05), in contrast to the 120% increase of kaempferol content that presented in es1-D leaves with respect to wild-type Ws-3 (P < 0.05; Fig. 10A). Even though cyp78a8 leaves showed a tendency of kaempferol content reduction compared with the wild type, this difference was not statistically significant (Fig. 10A). It has been reported that Arabidopsis leaves have a reduced flux through the flavonoid biosynthetic pathway and accumulate a higher ratio of kaempferol to quercetin derivatives than flowers and seedlings (Sheahan and Cheong, 1998). In line with this, in our study, hardly any quercetin intensities in leaves were detected, and no difference by PCA analysis was found.
Figure 10.
Kaempferol and quercetin perturbations were found in cyp78a9, cyp78a8 cyp78a9, and es1-D mutants. A, Fragment ion intensities of kaempferol from hydrolyzed samples from leaves of Ws-3, Col-0, cyp78a8, cyp78a9, cyp78a8 (−/−) cyp78a9 (−/−), es1-D, and tt4-1 es1-D. B, Fragment ion intensities of kaempferol and quercetin from hydrolyzed samples from inflorescences of Ws-3, Col-0, cyp78a8, cyp78a9, cyp78a8 (−/−) cyp78a9 (−/−), es1-D, and tt4-1 es1-D. *Significantly different from the wild type at P < 0.05. Values represent means ± sd in percentage of the wild type (n = 9).
Inflorescences presented less difference than leaves with respect to kaempferol content. However, cyp78a9 inflorescences showed a significant reduction in quercetin content with respect to the wild type (Fig. 10B).
In summary, although we observed alterations in the kaempferol and quercetin contents in cyp78a9, cyp78a8 (−/−) cyp78a9 (−/−), and es1-D mutants, which supports the predictions of CYP78A9 having a certain function in this pathway, the phenotypes of the mutants in terms of seed set are not in accordance with those observed for the known tt7 and tt4 mutants, which do not show these levels of seed set impairment (Ylstra et al., 1996).
Genetic Interaction between CYP78A9 and Chalcone Synthase (tt4)?
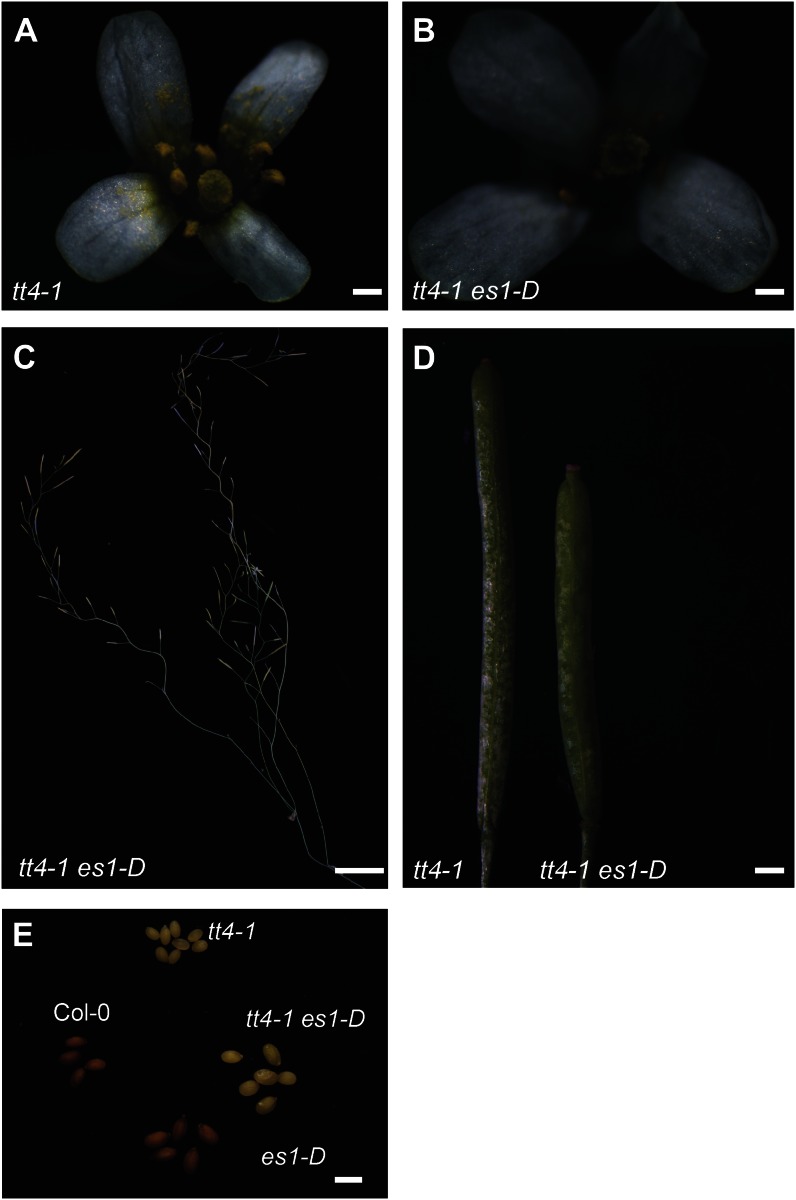
Given the prediction of AraCyc that positioned CYP78A9 in the flavonol biosynthesis pathway, at the conversion of dihydrokaempferol to dihydroquercetin, and the perturbations in kaempferol levels observed in es1-D (CYP78A9 overexpression), cyp78a9, and cyp78a8 (−/−) cyp78a9 (−/−) mutants, we wanted to further clarify the relationship between CYP78A9 and the flavonoid branch of the phenylpropanoid pathway. To test this, we examined the cross between es1-D (CYP78A9 overexpression) and tt4-1 (a null mutant that has a lesion in chalcone synthase [CHS] resulting in no flavonoid production and known for its yellowish seed color; Peer et al., 2001). The phenotype of the tt4-1 es1-D double mutant plants, found in the F2 generation, presented the characteristic yellow testa seed color of the tt4-1 mutant but also the reduction in fertility, the larger seed, the zigzag stem, and the floral organ size phenotypes of the es1-D mutant (Fig. 11). This means that there is no genetic interaction between CYP78A9 and tt4-1, because the double mutant showed a purely additive phenotype. Metabolic analysis of this double mutant showed neither kaempferol nor quercetin accumulation (Fig. 10), suggesting that the alterations in kaempferol observed in es1-D cannot be responsible for the CYP78A9 overexpression phenotypes. Moreover, previous studies showed that the Arabidopsis tt4-1 mutant, which lacks all downstream compounds of the flavonoid pathway, has no impairment in seed set (Burbulis et al., 1996; Ylstra et al., 1996).
Figure 11.
Phenotype of the tt4-1 es1-D double mutant. A and B, tt4-1 (A) and tt4-1 es1-D (B) flowers. Note the tt4-1 es1-D larger organ size. C, tt4-1 es1-D plant showing the characteristic zigzag pattern as seen in the es1-D stem. D, tt4-1 and tt4-1 es1-D fruits. Note the characteristic phenotype from es1-D, in which fruits are wider and shorter than tt4-1 fruits. E, Seeds from Col-0 and single and double mutants, showing the additive phenotype present in the tt4-1 es1-D double mutant. Bars = 0.2 mm in A and B; 2 cm in C; 0.2 cm in D; and 1 mm in E.
Preliminary Metabolic Screen
A preliminary metabolic screen was performed in the es1-D mutant using LC-MS and multiple fragmentations (Table I). Acetone crude extracts from leaves and inflorescences were screened by LC-MS (ACQUITY UPLC-LCT Premier XE equipment; Waters). Following the same procedure as described before, after PCA, selected metabolites were further fragmented using the SYNAPT HDMS system (Waters), and fragmentation spectra of highlighted metabolites were compared with authentic standards (kaempferol and quercetin), public databases, and recently published data (von Roepenack-Lahaye et al., 2004; Beekwilder et al., 2008; Kachlicki et al., 2008; McNab et al., 2009; Bollinger et al., 2010; Gouveia and Castilho, 2010). A distinct pattern of metabolite markers was observed by PCA for the mutants when compared with wild-type plants (Table I), indicating that the genetic alterations gave rise to changes in metabolite composition and concentration. Among the variations in metabolite concentration, the data highlighted a difference in flavonol content between es1-D and its wild-type ecotype. Kaempferol-glycosylated derivatives were the most abundant markers during the MarkerLynx PCA (Table I). However, the presence of quercetin derivatives was also notable, even if found at a lower proportion in the es1-D overexpressing mutant compared with the wild type. A unique accumulation profile was observed in different tissues of es1-D and the wild type (Table I). Inflorescence tissue showed more diversity in the nature of the flavonol aglycones than did the leaves. Finally, compounds not assigned as part of the flavonol pathway were found in varying concentrations in the es1-D mutant and the wild type. Within the data collected in the electrospray ionization negative mode of inflorescence tissue, the presence of lysophosphatidic acids (LPAs) was confirmed by the typical fragmentation of LPA showing peaks at mass-to-charge ratio (m/z) 153 and 79. As an example, the concentration of LPA (m/z 734.29) was higher in the es1-D mutant than in the wild type (Table I). Also, the presence of glucohirsutin (m/z 492.10), a glucosinolate, was confirmed by its fragmentation pattern (m/z 428 and 311) and reported as being more concentrated in es1-D inflorescences (Table I).
Table I. Relative amounts of metabolites identified in crude extracts of Arabidopsis leaves and inflorescences by ACQUITY UPLC-LCT Premier XE (Waters).
The values shown are averages of the relative peak response area in three replicates ± se. The up-regulation (↑) or down-regulation (↓) ratio is shown in boldface when there is a statistically significant difference (P < 0.05). The fragmentation spectra of metabolites were compared with authentic standards (for kaempferol and quercetin), with public databases, and with recently published data for the other metabolites found (von Roepenack-Lahaye et al., 2004; Beekwilder et al., 2008; Kachlicki et al., 2008; McNab et al., 2009; Bollinger et al., 2010; Gouveia and Castilho, 2010). ESI, Electrospray ionization; NS, not significant.
| Tissue/ESI | m/z | MS2 | Identification | Ws-3 | es1-D | Ratio | P |
|---|---|---|---|---|---|---|---|
| Leaves | |||||||
| Negative | 624.16 | 539, 503, 341, 313 | Unknown flavonoid aglycone | 122.1 ± 17.5 | 258.9 ± 12.4 | ↑2.12 | 1,277E-05 |
| 819.16 | 749, 477, 455, 277 | Unknown | 106.6 ± 42.7 | 172.9 ± 9.4 | ↑1.62 | 0.0494 | |
| 820.46 | 477, 461, 313 | Iso-rhamnoetin 3-O-glucoside-7-O-rhamnoside [M-H]− | 241.3 ± 11.1 | 161.1 ± 57. | NS | ||
| 431.15 | 341, 285, 153 | Kaempferol rhamnoside [M-H]− | 66.8 ± 4.9 | 86.9 ± 6.0 | ↑1.3 | 0.0023 | |
| 785.21 | 447, 341 | Unknown | 84.8 ± 6.6 | 44.8 ± 1.6 | ↓0.53 | 0.0008 | |
| Positive | 797.45 | 519, 230, 112 | Unknown | 235.1 ± 10.9 | 247.9 ± 12.0 | NS | |
| 327.16 | [Quercetin +Na + 2]2+ | 67.9 ± 28.9 | 75.4 ± 15.2 | NS | |||
| 593.28 | 533, 460 | Unknown | 140.7 ± 3.6 | 128.2 ± 7.7 | ↓0.87 | 0.0537 | |
| 329.15 | Unknown | 140.2 ± 7.1 | 92.9 ± 15.2 | ↓0.66 | 0.0041 | ||
| Inflorescence | |||||||
| Negative | 624.16 | 477, 461, 313 | Iso-rhamnoetin 3-O-glucoside-7-O-rhamnoside | 407.6 ± 17.4 | 382.2 ± 20.5 | ↓0.94 | 0.0439 |
| 786.21 | 609,447, 421,341 | Unknown | 228.5 ± 12.4 | 222.9 ± 13.3 | NS | ||
| 492.10 | 428, 234,97,80 | Glucohirsutin | 24.7 ± 7.3 | 165.6 ± 78.1 | ↑6.71 | 0.0067 | |
| 593.11 | 447, 285 | Kaempferol 3-O-glucoside-7-O-rhamnoside | 139.3 ± 7.7 | 115.5 ± 31.3 | NS | ||
| 669.41 | 381, 255, 153, 79 | LPA | 114.4 ± 6.3 | 105.0 ± 11.9 | NS | ||
| 639.16 | 607, 461, 422, 341, 297 | Unknown | 109.1 ± 5.3 | 99.3 ± 9.4 | NS | ||
| 734.29 | 422, 377, 277, 153, 79 | LPA | 91.5 ± 8.5 | 103.8 ± 9.8 | ↑1.13 | 0.0433 | |
| 663.17 | 477, 341, 311, 153, 79 | LPA | 101.9 ± 6.7 | 86.6 ± 20.4 | NS | ||
| 786.22 | 609, 447, 341, 153, 79 | LPA | 82.7 ± 4.3 | 83.7 ± 4.9 | NS | ||
| Positive | 579.25 | 433, 381, 287 | Kaempferol 3-O-rhamnopyranoside-7-O-rhamnopyranoside | 235.1 ± 10.9 | 247.9 ± 12.0 | NS | |
| 741.23 | 595, 433, 381, 287 | Kaempferol O-rhamnoside O-hexosyl-rhamnoside hexoside (K-Rha-Gly-3-Rha-7) | 179.9 ± 12.0 | 182.1 ± 11.9 | NS | ||
| 599.21 | 433, 365, 287 | Kaempferol-3-O-glycosyl-7-O-rhamnoside | 90.1 ± 14.9 | 135.3 ± 47.0 | NS | ||
| 625.18 | 463, 381, 365, 316 | Rhamnosyl hexosyl methyl quercetin | 109.8 ± 6.9 | 98.9 ± 9.9 | NS | ||
| 580.15 | 433, 287 | Neoeriocitrin | 79.6 ± 5.8 | 96.5 ± 17.5 | NS | ||
| 742.23 | 570, 433, 287 | Kaempferol rhamnoside-3-O-glycoside-7-O-rhamnoside | 66.6 ± 4.3 | 67.1 ± 5.3 | NS | ||
| 611.16 | 449, 381, 355, 303, 221 | Quercetin conjugate | 67.9 ± 4.6 | 53.9 ± 5.7 | NS | ||
| 323.19 | 223, 192, 138, 130, 84 | Unknown | 63.8 ± 12.2 | 64.4 ± 12.6 | NS | ||
| 609.18 | 355, 303, 281, 221 | Quercetin conjugate | 58.2 ± 2.9 | 53.9 ± 5.7 | NS |
In summary, this work provides, to our knowledge, the first characterization of metabolite differences between mutants in this gene family, giving interesting indicators for future investigations of the reaction(s) they catalyze.
DISCUSSION
Parthenocarpic Fruit Development in a CYP78A9 Overexpression Line, and the Hormonal Context
Overexpression of the cytochrome P450 CYP78A9 gene allows fruit growth to be uncoupled from fertilization. In the es1-D activation-tagging mutant, where the CYP78A9 gene is overexpressed, androecia and gynoecia develop in an uncoordinated manner. Anthers present a dehiscence delay and the filaments never reach the stigma, so pollination does not occur. In wild-type plants during the maturation and receptive periods, specific molecular pathways restrict the growth of the pistil and accessory tissues and stop them from developing into fruits (Vivian-Smith et al., 2001; Goetz et al., 2006). Apparently, though, CYP78A9 overexpression can overcome this restriction, allowing the pistil to grow before the androecium is mature. Dorcey and coworkers (2009) further showed that a fertilization-dependent auxin signal induces GA biosynthesis (via GA20ox1, GA20ox2, and GA3ox1) that, in turn, triggers fruit growth. However, es1-D DR5::GUS plants did not show the characteristic expression pattern in ovules and funiculus as the wild type, but fruits were developed. Moreover, es1-D GA20ox1::GUS plants showed the same expression pattern as wild-type plants, suggesting that GAs cannot be attributed as the cause for the uncoupled growth of es1-D fruit from fertilization. However, both auxin and GA probably act in conjunction with the CYP78A9-produced signal in the fruit growth program.
CYP78A6, CYP78A8, and CYP78A9 Are Involved in Reproductive Development
In the more acropetal flowers of es1-D plants, at the end of its life cycle, pollination and fertilization occur, although fertility is still reduced. Cross-pollination experiments showed that both pollen and ovules of es1-D were affected. The most relevant altered feature in es1-D ovules was integument size, with more and larger cells than in the wild type. This fact also has been recently reported for eod31-D, the activation-tagging mutant for CYP78A6 (the closest paralog of CYP78A9), in which the ovules have more and larger integument cells (Fang et al., 2012), and has also been reported for plants with directed expression of KLUH/CYP78A5 to the outer integuments (Adamski et al., 2009). In contrast, when cyp78a6 cyp78a9 and cyp78a8 cyp78a9 double mutants were analyzed, a reduction in fertility was detected and was more severe in the latter. The reduction of fertility in the cyp78a8 cyp78a9 double mutant was correlated with the growth arrest of outer integuments that fail to accommodate the embryo sac.
Arabidopsis mutants with impaired integument initiation and outgrowth, such as ant, inner no outer (a YABBY transcription factor), and bel1, are associated with aborted embryo sac development and reduced fertility (Ray et al., 1994; Elliott et al., 1996; Baker et al., 1997; Villanueva et al., 1999). Other mutants with altered integuments have been proven to present abnormalities during seed development, as is the case of the abs stk double mutant, which completely lacks endothelium (Mizzotti et al., 2012), or the ttg2 mutation, which affects primarily cell elongation in the integuments by altered proanthocyanidin synthesis (Garcia et al., 2005). The gametophytic mutants characterized until now all have normal development of sporophytic tissues. Accordingly, a hierarchical communication between the two generations has been suggested, assigning a higher order to the sporophytic maternal tissues (Bencivenga et al., 2011). The fruit without fertilization (fwf/arf8) Arabidopsis parthenocarpic mutant showed extended outer integuments (Vivian-Smith et al., 2001), as observed in es1-D. Interestingly, two integument-defective mutants have been reported to affect the parthenocarpic fruit development of the Arabidopsis fwf/arf8 mutant. The ABERRANT TESTA SHAPE (ats1/kan4-1) and bel1 mutants enhanced parthenocarpic fruit development. The need of emasculation is negated when the double mutant of 3-KETOACYL-COENZYME A SYNTHASE6 (pop1) and fwf1 is combined with the ats1 mutant, which only develops one integument (Vivian-Smith et al., 2001). Based on this evidence, the third floral whorl prevents fruit initiation, possibly by a shared pathway with the integuments mediating this communication. Nevertheless, which class of messengers are involved (e.g. metabolites, small peptides, or hormones) has not been deciphered yet (Vivian-Smith et al., 2001; Bencivenga et al., 2011; Mizzotti et al., 2012).
Previous work using cyp78a mutants and CYP78A-overexpressing plants proposed that CYP78A family members are involved in the generation of novel signaling compounds that are mainly related to the control of organ size and shape as well as plant architecture (Ito and Meyerowitz, 2000; Miyoshi et al., 2004; Anastasiou et al., 2007; Adamski et al., 2009). A KLUH/CYP78A5-dependent signal has been demonstrated to act in a non-cell-autonomous manner to shape and size floral organs and to maternally limit the size of the outer integuments to control seed size (Adamski et al., 2009). While klu/cyp78a5 had reduced petal and integument cell numbers, transgenic plants that overexpressed KLUH/CYP78A5 had increased numbers of petal and integument cells, indicating that this gene regulates organ size through cell proliferation (Zondlo and Irish, 1999; Anastasiou et al., 2007; Adamski et al., 2009). The maternal control of seed size has also been demonstrated for CYP78A6- and CYP78A9-dependent signals (Adamski et al., 2009; Fang et al., 2012). There is also a report of the biological role of members of this family in Physcomitrella patens, where the two CYP78A members act redundantly during protonemal growth and gametophore development. In that study, the cyp78a27 cyp78a28 double mutant as well as both overexpression lines resulted in impaired gametophore development and affected the endogenous levels of several plant hormones (Lohmann et al., 2010; Katsumata et al., 2011). So it seems that the CYP78A family of P450 monoxygenases has a conserved role across the plant kingdom. In this work, we confirmed the role of CYP78A9 in controlling floral organ size and its role during integument development. The analysis of the cyp78a8 cyp78a9 double mutant showed a clear genetic interaction between these genes controlling seed set via outer integument development. The defects observed in cyp78a8 cyp78a9 double mutants are linked to the sporophyte before fertilization, which supports the idea that this family of genes act maternally during reproductive development. Although, the defects detected in the cyp78a8 cyp78a9 mutant were linked to the sporophyte before fertilization, the expression pattern detected during seed and embryo development suggests that CYP78A9 could have a possible communication role between the embryo and the seed coat while they develop. Furthermore, the fact that CYP78A9::GFP:GUS responds to the fertilization event indicates the possible communication role of CYP78A9 between the placenta, funiculus, and ovule during the fertilization process.
CYP78A8 and CYP78A9 in Metabolic Processes
Many studies suggest that members of the CYP78A family produce a novel kind of signal. Anastasiou and colleagues (2007) demonstrated that there is no clear overlap between the KLUH/CYP78A5-regulated genes and the genes controlling the known plant hormones, and the klu/cyp78a5 phenotype could not be rescued by the application of the known hormones. Microarray data obtained here also showed no clear changes in hormonal pathways due to the overexpression of CYP78A9. Imaishi and colleagues (2000) reported that the recombinant CYP78A1 protein in yeast is able to catalyze the 12-hydroxylation of lauric acid. KLUH/CYP78A5, CYP78A7, and CYP78A10 produced in insect cells are also able to catalyze the ω-hydroxylation of short-chain fatty acids, with lauric acid as the preferred substrate (Kai et al., 2009). However, no rescue of the klu/cyp78a5 phenotype was obtained with the application of 12-hydroxyl-lauric acid, pointing out that this might not be the only substrate in planta. Su and Hsu (2010) showed that transient expression in petals in Phalaenopsis spp. of CYP78A2 cloned from the same species increased anthocyanin content in that organ. CYP78A2 boosts the pathway without the biosynthesis of any new anthocyanin, and the effect is not limited to Phalaenopsis spp. but also occurs in rose (Rosa spp.) and carnation (Dianthus caryophyllus; Su and Hsu, 2010). However, the authors could not prove if CYP78A2 acts directly in this pathway or indirectly via the production of another hormone or secondary metabolite (Su and Hsu, 2010).
In an effort to confirm the prediction that CYP78A9 has an overlapping function with F3′H in the conversion of dihydrokaempferol to dihydroquercetin, we performed a metabolic exploration of the flavonoid pathway. Here, one would expect a partial block of the pathway to result in an increase of dihydrokaempferol or kaempferol. Still, quercetin can be present, as the double mutant is in an F3′H-intact background. Interestingly, hydrolyzed extracts showed that cyp78a9 and cyp78a8 (−/−) cyp78a9 (−/−) leaves have 40% and 50% kaempferol content reductions, respectively, compared with wild-type Col-0 (P < 0.05), in contrast to the 120% increase of kaempferol content that presented in es1-D leaves with respect to wild-type Ws-3 (P < 0.05). However, cyp78a9 inflorescences showed a significant reduction in quercetin content with respect to the wild type. Flavonoids are plant secondary metabolites that comprise pigments such as chalcones and anthocyanins as well as colorless molecules such as flavonones, flavones, and flavonols (Buer et al., 2010). The presence of flavonoids has been reported in pollen and pistils of many plant species. In petunia (Petunia hybrida) and maize (Zea mays), chs mutants are blocked in the first step of flavonoid biosynthesis and are defective in pollen tube growth. Kaempferol was identified as the pollen germination-inducing factor when applied to mutant stigma (Mo et al., 1992). Biochemical experiments and analyses of auxin fluxes in flavonoid-deficient mutants suggest that flavonols negatively regulate auxin transport processes, which are increased in tt4 mutants (Jacobs and Rubery, 1988; Brown et al., 2001; Buer and Muday, 2004; Peer et al., 2004).
Recently, experiments in Arabidopsis have shown the ability of unglycosylated kaempferol and quercetin to compete with the auxin transport inhibitor 1-N-naphthylphthalamic acid for a high-affinity binding site found in a protein complex containing PGP1, PGP2, and MDR1/PGP19, proteins that belong to the ATP-binding cassette transporters (Noh et al., 2001; Murphy et al., 2002; Geisler et al., 2005). Flavonols are believed to directly modulate auxin transport, a process that is reduced in the kaempferol-overaccumulating mutant tt7 defective in F3′H (Peer et al., 2004; Santelia et al., 2008). Alterations in seed size and development were observed in mutants defective in proanthocyanidin synthesis or accumulation, as is the case for ttg2, which is affected in integument cell elongation (Garcia et al., 2005), and for the abs stk double mutant, which has severely reduced fertility and completely lacks endothelium (Mizzotti et al., 2012). A link between parthenocarpy and the flavonoid pathway has been established in tomato (Solanum lycopersicum) by Schijlen and coworkers (2007). Down-regulation of the flavonoid pathway using RNA interference-mediated suppression of CHS rendered plants with impaired pollen tube growth and pollination-dependent parthenocarpic fruit development (Schijlen et al., 2007). Parthenocarpy can also be achieved by overexpression of the grape (Vitis vinifera) Stilbene Synthase gene (Ingrosso et al., 2011). This enzyme competes for the same substrates as CHS, and its overexpression produced a decrease in flavonoid content that leads to male-sterile pollen in tobacco and a decreased seed set in tomato fruits. This connection could not be made in Arabidopsis, as the tt4 (CHS) mutant did not present male sterility and parthenocarpic development, although a slight reduction in seed set was observed (Burbulis et al., 1996; Ylstra et al., 1996). Recently, Mahajan and colleagues (2011) have proposed a new strategy to generate fruits with reduced seed set in tobacco. By means of posttranscriptional gene silencing of Flavonol Synthase (FLS), they obtained plants reduced in quercetin and anthocyanidin contents but increased in catechin, epicatechin, and epigallocatechin. Interestingly, FLS-silenced lines were significantly reduced in seed number (Mahajan et al., 2011). In summary, flavonoids are important metabolites that have diverse biological functions. Within the past few years, increasing interest in these metabolites has been reported because of their possible role during reproductive development and the implied potential of biotechnologically controlled seed set by manipulating this pathway (Taylor and Grotewold, 2005; Falcone Ferreyra et al., 2012).
Although the metabolic data showed differential accumulation of flavonoids between the wild type and mutants, some contradictions did not support these alterations being in line with the CYP78A9 catalytic bioinformatics prediction or CYP78A9 as being the direct cause of the phenotypes observed in the mutants. First, kaempferol levels of cyp78a9 and cyp78a8 cyp78a9 mutants were not in accordance with the data reported for the tt7 mutant, deficient in F3′H activity, which overaccumulates kaempferol and does not produce quercetin (Shirley et al., 1995; Peer et al., 2001). In this sense, the cyp78a8 cyp78a9 kaempferol profile resembles more the tt5 mutant, which shows a drastically reduced flux through the biosynthetic pathway in relation to the wild type (Sheahan and Cheong, 1998); however, it does not have the same change in testa color: cyp78a8 cyp78a9 has pale brown testa and tt5 has yellowish testa. The profiles reported for tt3 and tt6 mutants did not resemble the situation of our mutant, as tt3 has been reported to accumulate excess quercetin and kaempferol (Peer et al., 2001) and tt6 has reduced kaempferol content and has no quercetin (Shirley et al., 1995). Moreover, the tt7 (kaempferol overaccumulator) and tt4 (devoid of kaempferol) mutants did not show the levels of seed set impairment seen in the cyp78a8 cyp78a9 double mutant (Ylstra et al., 1996). Second, the tt4-1 es1-D double mutant showed a purely additive phenotype and did not accumulate either kaempferol or quercetin, suggesting that the alterations in flavonoids present in the overexpression mutant are not responsible for the observed phenotypes. Third, variations in other metabolites, like glucohirsutin (a glucosinolate), LPAs, unknown flavonoid aglycones, and unidentified compounds, were found in the analyses. And fourth, Kai and coworkers (2009) showed that CYP78A5/KLUH, CYP78A7, and CYP78A10 catalyze the ω-hydroxylation of short-chain fatty acids and proposed that this family of enzymes modifies a fatty acid-related molecule that could participate in another biosynthetic pathway.
Although CYP78A9 was predicted to possibly be chloroplast localized (Schuler et al., 2006), our work suggest its localization to the plasma membrane. Based on this information, together with experimental evidence that CYP78A9 forms a protein-protein interaction with CALMODULIN7 (At3g43810) and with the protein kinase superfamily protein (At1g48210), localized also in the plasma membrane (Popescu et al., 2007), and with predictions made by the Algorithm for the Reconstruction of Accurate Cellular Networks that this protein could be implicated with calcium-dependent lipid-binding signaling involved in cell death, makes this an interesting case to explore.
Furthermore, future studies should elucidate whether a specific metabolite causes all the observed morphological phenotypes or different metabolites cause particular phenotypes.
CONCLUSION
Our findings suggest that CYP78A9 has a function during reproductive development. The genetic evidence supports the idea that CYP78A9 and its closest paralogs participate in a pathway that controls floral organ size and ovule integument development, as denoted by the phenotypes of es1-D overexpression and cyp78a8 cyp78a9 double mutants.
The CYP78A9-specific expression pattern suggests that the produced signal coordinates growth between sporophytic and gametophytic tissue and between the structures that protect the ovules and the seed while they develop. Studies with the CYP78A9 promoter line suggest an interesting function of the gene in communication between the placenta, funiculus, and ovule during the fertilization process. Metabolic analyses of the mutants showed the existence of alterations in flavonoid content with respect to the wild type. However, these alterations seem not to cause the observed phenotypes, as the tt4-1 es1-D double mutant presents purely additive phenotypes without having kaempferol and quercetin contents. Despite this, the work presented here contributes to the characterization of metabolite differences between mutants in this gene family, giving interesting indicators for future investigation of the reaction(s) they catalyze.
MATERIALS AND METHODS
Plant Growth and Plant Material
Arabidopsis (Arabidopsis thaliana) plants were germinated in soil (3:1:1, peat moss:perlite:vermiculite) in a growth chamber at 22°C under long-day conditions (8 h of dark/16 h of light) and transferred to standard greenhouse conditions (22°C–27°C, natural light). The flowering-time assay was performed in a growth chamber at 22°C under long-day conditions. The es1-D mutant was identified by activation tagging in the Arabidopsis Ws-3 background (Marsch-Martinez et al., 2002). The SALK line (cyp78a9; SALK_0665880), SAIL lines (cyp78a6, CS833552; cyp78a8, CS823514), the marker line DR5::GUS (Ulmasov et al., 1997), and GA20ox1::GUS (CS57942; Desgagné-Penix et al., 2005) are all in the Col-0 background.
Constructs and Transformation
For the promoter analysis, 3,020 bp directly upstream of the CYP78A9 (At3g61880) translational start was amplified and cloned into the pENTR-D vector (Invitrogen) using specific primers (forward, 5′-GGTGGGATACCGGTCAAGTG-3′; reverse, 5′-GGATGCAGAGGAACAAGAGAGAG-3′). The resulting construct was sequence verified and recombined with the binary vector pBGWFS7.0 (Karimi et al., 2002), resulting in the vector CYP78A9::GFP:GUS. Wild-type Arabidopsis plants (ecotype Ws-3) were transformed using the floral dip method (Clough and Bent, 1998), and transformants were identified through BASTA selection.
To confirm that the phenotype of es1-D was due to the overexpression of the CYP78A9 gene, the entire genomic region was cloned using the forward primer 5′-ATGGCCACCAAGCTCGACAC-3′ and the reverse primer 5′-TCATACACTAAAACTGCGCCTGG-3′ into the pENTR-D vector (Invitrogen), verified by sequencing, and recombined with the Gateway plasmid pK7WG2D (Karimi et al., 2002), resulting in the vector 35S::gCYP78A9. Wild-type Arabidopsis plants (ecotype Ws-3) were transformed using the floral dip method (Clough and Bent, 1998), and transformants were identified through kanamycin selection.
For protein localization, the pENTR-D + gCYP78A9 (entire genomic region) plasmid was recombined with the Gateway binary vector pB7FWG2 (Karimi et al., 2002), resulting in the vector 35S::gCYP78A9:GFP. Agrobacterium tumefaciens containing this construct was grown to an optical density greater than 1 in 5 mL of Luria-Bertani medium with appropriate antibiotics. The culture was centrifuged at 4,000 rpm for 10 min at room temperature, and the pellet was resuspended in 3 mL of a solution of 10 mm MgCl2, 10 mm MES, pH 5.6, and acetosyringone to a final concentration of 200 µm was added. This suspension was left for 3 h at room temperature with weak shaking. Tobacco (Nicotiana tabacum) leaves were infiltrated with a syringe of 10 mL on the abaxial face. Fluorescence signal was observed with confocal microscopy (TCS SPE; Leica) after 3 d of incubation in greenhouse conditions. A 488-nm argon laser line was used for excitation, and emission was detected between 505 and 550 nm.
Histology
For GUS analysis, Arabidopsis tissues were incubated overnight at 37°C with a 5-bromo-4-chloro-3-indolyl-β-glucuronic acid solution (Gold Biotechnology; Jefferson et al., 1987). For clearings, tissue was treated overnight with Hoyer’s solution (Anderson, 1954) plus 20% lactic acid. Light images from GUS-stained tissues and from plant phenotypes were obtained using a Leica EZ4 D stereomicroscope. Images of ovules were obtained using a Leica CTR6000 device equipped with differential interference contrast (Nomarski) optics.
RNA Extraction and RT-PCR
RNA was isolated using LiCl (Verwoerd et al., 1989). Around 1 µg of RNA was treated with DNase I (Invitrogen), and one-tenth of the treated RNA was used for complementary DNA (cDNA) synthesis with Moloney murine leukemia virus reverse transcriptase or SuperScript II RNase H reverse transcriptase (both from Invitrogen), following the supplier’s instructions. The obtained cDNA was used for gene expression analyses. PCR experiments were performed using cDNA from wild-type and mutant tissues. A PCR using ACTIN (forward, 5′-GTGTTGGACTCTGGAGATGGTGTG-3′; reverse, 5′-GCCAAAGCAGTGATCTCTTTGCTC-3′) primers for all the samples was used as a control. The reactions were performed as follows: 95°C for 3 min, 30 cycles of 95°C for 30 s, 55°C for 40 s, and 72°C for 2 min, followed by 72°C for 5 min.
In Situ Hybridization
Arabidopsis tissue was collected, and for siliques, transverse cuts were made in order to remove the apical and basal tips (to allow better infiltration), fixed, and embedded in Paraplast. In summary, the samples were placed in 10-mL vials, and fixation was performed with 50% ethanol, 5% glacial acetic acid, and 3.7% formaldehyde solution, vacuum was applied twice for 15 min followed by replacement of fixative, and then the samples were left overnight at 4°C. Before imbibing the tissue in Paraplast, the samples were dehydrated in an ethanol series (50%, 60%, 70%, 80%, 90%, and 95%), 30 min between each step, followed by incubation overnight in 95% ethanol at 4°C. Afterward, samples were incubated twice in 100% ethanol, 30 min between steps, and then left overnight at 4°C. Next, the tissue samples were taken through a HISTO-CLEAR series of 1 h each of 25% HISTO-CLEAR:75% ethanol, 50% HISTO-CLEAR:50% ethanol, 75% HISTO-CLEAR:25% ethanol, and finally, 100% HISTO-CLEAR. Infiltration was made with 100% fresh HISTO-CLEAR and 10 to 15 chips of Paraplast and incubated overnight at room temperature. The next day, the solution was replaced with 100% Paraplast and repeated five times during a time span of 6 h at 60°C, followed by making the molds. After this, sections (10 μm) were made on a rotary microtome (Leica RM 2025) and mounted on slides.
A CYP78A9 PCR fragment corresponding to nucleotides 1,755 to 1,960 (using forward primer 5′-CACTATAGGGCAACACGTATCAAGATGTTAGTTTA-3′ and reverse primer 5′-CCAGGCGCAGTTTTAGTGTA-3′) was cloned into PGEM-T easy vector (Promega). The sense and antisense probes were synthesized by an in vitro transcription reaction using T7 and SP6 polymerase (Metabion), respectively. Prior to hybridization, the slides were dewaxed in HISTO-CLEAR twice for 10 min, 100% ethanol for 2 min, and an ethanol series (95%–30%; 1 min each), transferred to saline solution (8.5 g L−1 NaCl) for 15 min, and then for 5 min in 1× phosphate-buffered saline (PBS). To improve probe penetration into the tissue, slides were incubated with proteinase K (1 µg mL−1) at 37°C for 30 min. The proteinase K digestion was stopped by keeping the slides for 2 min in 2 mg mL−1 Gly-1× PBS. For postfixation, the slides were transferred to fresh fixation solution and kept for 10 min at room temperature. Next, the slides were washed twice with 1× PBS for 5 min and transferred to 0.1 m triethanolamine plus 1 mL of acetic anhydride for 10 min, followed by a 5-min wash in 1× PBS and a reverse ethanol series (30%, 50%, 85%, 95%, 100%) of 2 min each, a step in 0.5% NaCl for 2 min, and 2 min with 1× PBS. Hybridization was done in a humidified box with a digoxigenin-labeled probe at 52°C overnight. Immunological detection was performed with a 1:1,250 antibody final concentration (Anti-Digoxigenin-AP; Roche) in bovine serum albumin (10 g L−1) solution for 2 h and an overnight incubation in nitroblue tetrazolium/5-bromo-4-chloro-3-indolyl phosphate-containing solution as described previously (Coen et al., 1990).
Expression Analysis (Microarray)
RNA was extracted (Verwoerd et al., 1989) from pooled inflorescences of 20 plants containing close buds only, comparing es1-D with wild-type Arabidopsis plants (Ws-3). Labeled cDNA (Alexa555 and Alexa647) was used for hybridization (in duplicate) to the 70-mer oligo Arabidopsis V.3.0.3 microarrays, which was performed at the Unidad de Microarreglos de DNA, Instituto de Fisiologia, Universidad Nacional Autónoma de México (http://microarrays.ifc.unam.mx/).
Microarray data analysis was performed with free software genArise, developed in the Computing Unit of the Cellular Physiology Institute of UNAM (http://www.ifc.unam.mx/genarise/). genArise carries out a number of transformations: background correction, Lowess normalization, intensity filter, analysis of replicates, and selection of differentially expressed genes. The software identifies differentially expressed genes by calculating an intensity-dependent z score using a sliding-window algorithm to calculate the mean and sd within a window surrounding each data point and then defines a z score where z measures the number of sd a data point has from the mean. zi = (Ri – mean(R))/sd(R), where zi is the z score for each element, Ri is the log ratio for each element, and sd(R) is the sd of the log ratio. Ratio calculations of significant changes in gene expression derived from globally normalized data are performed by simply computing the ratio of the average of all the measurements from one condition or sample to another. With this criterion, the elements with a z score greater than 2 sd are considered significantly differentially expressed genes (Cheadle et al., 2003).
PCR-Based Genotyping
Identification of the cyp78a9 mutant allele was performed by PCR analysis using the primer LBb1 (5′-GCGTGGACCGCTTGCTGCAACT-3′) on the T-DNA left border and the primer 5′-TCATACACTAAAACTGCGCCTGG-3′. The CYP78A9 wild-type allele was identified using the primer 5′-ATGGCCACCAAGCTCGACAC-3′ in combination with the primer 5′-TCATACACTAAAACTGCGCCTGG-3′. The cyp78a6 mutant allele was identified using primers LB3 (5′-ATTTTGCCGATTTCGGAAC-3′) and 5′-CCGGTTAAAGAATCGGCTTAC-3′, and the CYP78A6 wild-type allele was identified using the primer combination 5′-AACTCCAAAGGATCAACCCAC-3′ and 5′-CCGGTTAAAGAATCGGCTTAC-3′. For the identification of the cyp78a8 mutant allele, LB3 in combination with 5′-CTGAGATGAGTAACGCCAAGC-3′ were used, and for the CYP78A8 wild-type allele, the primers 5′-ATAGCCCACATGTTGACCATC-3′ and 5′-CTGAGATGAGTAACGCCAAGC-3′ were used.
Metabolite Analysis by LC-MS and Tandem Mass Spectrometry Using ACQUITY UPLC-LCT Premier XE and the SYNAPT HDMS System, Respectively
Sample Preparation
Frozen plant material (fully expanded leaves after flowering and whole flowers) was ground in liquid nitrogen. For each 100 mg of fresh tissue, 300 µL of cold acetone was added, and the mixture was vortexed, sonicated for 5 min, and then centrifuged at 16,100g to separate the crude extract from the tissue. The supernatant was used for analysis (crude extracts) and for the hydrolysis of aglycones from the glycosylated flavonoid compounds. The hydrolysis was performed following the protocol of Burbulis et al. (1996) by heating equal volumes of crude extract and 2 n HCl at 70°C for 40 min. The flavonoids were separated from the aqueous volume with an equal volume of ethyl acetate by vortexing and centrifuging at 16,100g for 10 min. The upper organic layer was removed, placed in new Eppendorf tubes, and lyophilized. The lyophilized samples were dissolved in 1,000 μL of 100% methanol and filtered through a 0.22-μm filter before injection onto the chromatographic column. For each sample, three biological replicates were included. Every sample was injected in triplicate.
Chromatography
Chromatographic separation was performed on an ACQUITY BEH C-18 column (2.1 × 50 mm i.d., 1.7 μm; Waters) using an ACQUITY UPLC-LCT Premier XE (Waters). The column was maintained at 50°C and eluted with a 10-min linear gradient for hydrolyzed samples (method 1) or was maintained at 35°C and eluted with a 30-min gradient applied to the crude extracts (method 2). For method 1, the mobile phase, at a flow rate of 0.5 mL min−1, consisted of water:methanol (75:25; + 0.125% formic acid) and was maintained for 10 min. The volume of sample injected onto the column was 5 μL. For method 2, the mobile phase, at a flow rate of 0.2 mL min−1, consisted of a starting mixture of solvents A and B (methanol:water, 1:9; A, 100% methanol; B, water + 0.1% formic acid). A decrease of solvent B up to 20% over 15 min was then performed. Solvent B was returned to its initial composition over 1 min, and the initial condition was maintained for 15 min in order to equilibrate the column. The volume of sample injected onto the column was 5 μL.
Mass Spectrometry
The eluent was introduced into the UPLC-LCT Premier XE mass spectrometer (Waters) by electrospray ionization, with capillary and cone voltages set in the positive ion mode to 3,100 and 70 V and in the negative mode to 3,300 and 40 V. The desolvation gas was set to 850 L h−1 at a temperature of 350°C for the positive mode and to 650 L h−1 and 200°C for the negative mode. The cone gas was set to 10 L h−1 and the source temperature was set to 80°C for the positive mode and to 10 L h−1 and 100°C for the negative mode. Continuum data were acquired from m/z 50 to 1,000 using an accumulation time of 0.2 s per spectrum. All spectra were mass corrected in real time by reference to Leu enkephalin (2 μg mL−1), infused at 5 μL min−1 through an independent reference electrospray. The resolution of the system was of 11,000 for the positive mode and 10,500 for the negative mode.
Data processing was performed by MarkerLynx, an application manager of the MassLynx 4.1 software (Waters) with the following parameters: retention time range of 0.1 to 9.80 min for the hydrolyzed samples and 0.64 to 30 min for the crude extract; mass tolerance of 0.30 D; peak width and baseline noise were automatically calculated by the program; mass window at 0.05 D; and retention time window at 0.2 min. Automatic smoothing was applied, and isotopic peaks were removed from the data. The data were analyzed by PCA within the MarkerLynx application manager on the mean center of the peak area intensities, taking scaling into account. Fragmentation analysis was performed on MarkerLynx selected metabolites using the SYNAPT HDMS system (Waters). The mass spectra were obtained applying a cone voltage ranging from 15 to 80 V, leading to the fragmentation of the base peak of interest. The fragmentation spectra of the highlighted metabolites were compared with authentic standards (for kaempferol and quercetin), with public databases, and with recently published data for the other metabolites found (von Roepenack-Lahaye et al., 2004; Beekwilder et al., 2008; Kachlicki et al., 2008; McNab et al., 2009; Bollinger et al., 2010; Gouveia and Castilho, 2010). The collected peak lists with m/z and peak area intensities were further processed with Excel software, and the statistically significant differences in individual markers between the wild type and the mutants were demonstrated by pairwise t test (two tailed, two sample, unequal variance).
Sequence data for this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: CYP78A9 (At3g61880), CYP78A6 (At2g46660), and CYP78A8 (At1g01190). The ArrayExpress Database microarray data submission number is E-MEXP-3770.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Position of Activating I Element affecting the CYP78A9 gene.
Supplemental Figure S2. es1-D flowering time.
Supplemental Figure S3. 35S::gCYP78A9 phenotype.
Supplemental Figure S4. T-DNA insertional line genotyping and RT-PCR data.
Supplemental Figure S5. Fruit size and seed yield from the wild type and cyp78a9 and cyp78a6 single and double mutants.
Supplemental Figure S6. Comparison of in situ hybridization between Ws-3 and es1-D buds.
Supplemental Figure S7. Expression pattern of auxin and GA in es1-D compared with the wild type (Ws-3) during flower development.
Supplemental Figure S8. CYP78A9 transcriptional effect upon the application of different hormones to seedlings.
Supplemental Figure S9. CYP78A9 transcriptional effect upon the application of different hormone inhibitors to seedlings.
Supplemental Table S1. Microarray up-regulated and down-regulated genes at 2 sd.
Supplemental Table S2. Comparison of CYP78A9-regulated and phythormone-responsive genes.
Supplemental Table S3. Gene list derived for the comparison between macroarray and microarray data.
Supplemental Table S4. CYP78A9-coexpressed genes in flower tissue.
Supplemental Table S5. Gene functional classification derived from the genes coexpressed with CYP78A9 in flower tissue.
Acknowledgments
We thank the SALK Institute, the Arabidopsis Biological Resource Center, and the Nottingham Arabidopsis Stock Centre for seeds. We thank Jorge Ramirez for the microarray service at the Unidad de Microareglos, Instituto de Fisiologia, Universidad Nacional Autónoma de México. Furthermore, we thank Gerardo Acosta García for discussions, Paulo Cázares-Flores for the 35S::gCYP78A9 construct, Daniela Ramos-Cruz for the flowering-time assay, and Edmundo Lozoya-Gloria and Yolanda Rodriguez-Aza for HPLC standards.
Glossary
- Ws-3
Wassilewskija
- Col-0
Columbia
- RT
reverse transcription
- F3′H
flavonoid 3′-monooxygenase
- LC-MS
liquid chromatography-mass spectrometry
- PCA
principal component analysis
- LPA
lysophosphatidic acid
- m/z
mass-to-charge ratio
- T-DNA
transfer DNA
- cDNA
complementary DNA
- PBS
phosphate-buffered saline
References
- Adamski NM, Anastasiou E, Eriksson S, O’Neill CM, Lenhard M. (2009) Local maternal control of seed size by KLUH/CYP78A5-dependent growth signaling. Proc Natl Acad Sci USA 106: 20115–20120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alabadí D, Blázquez MA, Carbonell J, Ferrándiz C, Pérez-Amador MA. (2009) Instructive roles for hormones in plant development. Int J Dev Biol 53: 1597–1608 [DOI] [PubMed] [Google Scholar]
- Alimohammadi M, de Silva K, Ballu C, Ali N, Khodakovskaya MV. (2012) Reduction of inositol (1,4,5)-trisphosphate affects the overall phosphoinositol pathway and leads to modifications in light signalling and secondary metabolism in tomato plants. J Exp Bot 63: 825–835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anastasiou E, Kenz S, Gerstung M, MacLean D, Timmer J, Fleck C, Lenhard M. (2007) Control of plant organ size by KLUH/CYP78A5-dependent intercellular signaling. Dev Cell 13: 843–856 [DOI] [PubMed] [Google Scholar]
- Anderson LE. (1954) Hoyer's solution as a rapid permanent mounting medium for bryophytes. Bryologist 57: 242–244 [Google Scholar]
- Bak S, Beisson F, Bishop G, Hamberger B, Höfer R, Paquette S, Werck-Reichhart D. (2011) Cytochromes p450. The Arabidopsis Book 9: e0144, [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker SC, Robinson-Beers K, Villanueva JM, Gaiser JC, Gasser CS. (1997) Interactions among genes regulating ovule development in Arabidopsis thaliana. Genetics 145: 1109–1124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beekwilder J, van Leeuwen W, van Dam NM, Bertossi M, Grandi V, Mizzi L, Soloviev M, Szabados L, Molthoff JW, Schipper B, et al. (2008) The impact of the absence of aliphatic glucosinolates on insect herbivory in Arabidopsis. PLoS ONE 3: e2068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bencivenga S, Colombo L, Masiero S. (2011) Cross talk between the sporophyte and the megagametophyte during ovule development. Sex Plant Reprod 24: 113–121 [DOI] [PubMed] [Google Scholar]
- Bollinger JG, Ii H, Sadilek M, Gelb MH. (2010) Improved method for the quantification of lysophospholipids including enol ether species by liquid chromatography-tandem mass spectrometry. J Lipid Res 51: 440–447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown DE, Rashotte AM, Murphy AS, Normanly J, Tague BW, Peer WA, Taiz L, Muday GK. (2001) Flavonoids act as negative regulators of auxin transport in vivo in Arabidopsis. Plant Physiol 126: 524–535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buer CS, Imin N, Djordjevic MA. (2010) Flavonoids: new roles for old molecules. J Integr Plant Biol 52: 98–111 [DOI] [PubMed] [Google Scholar]
- Buer CS, Muday GK. (2004) The transparent testa4 mutation prevents flavonoid synthesis and alters auxin transport and the response of Arabidopsis roots to gravity and light. Plant Cell 16: 1191–1205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burbulis IE, Iacobucci M, Shirley BW. (1996) A null mutation in the first enzyme of flavonoid biosynthesis does not affect male fertility in Arabidopsis. Plant Cell 8: 1013–1025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbonell-Bejerano P, Urbez C, Carbonell J, Granell A, Perez-Amador MA. (2010) A fertilization-independent developmental program triggers partial fruit development and senescence processes in pistils of Arabidopsis. Plant Physiol 154: 163–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheadle C, Vawter MP, Freed WJ, Becker KG. (2003) Analysis of microarray data using Z score transformation. J Mol Diagn 5: 73–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y, Dai X, Zhao Y. (2006) Auxin biosynthesis by the YUCCA flavin monooxygenases controls the formation of floral organs and vascular tissues in Arabidopsis. Genes Dev 20: 1790–1799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y, Dai X, Zhao Y. (2007) Auxin synthesized by the YUCCA flavin monooxygenases is essential for embryogenesis and leaf formation in Arabidopsis. Plant Cell 19: 2430–2439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Coen ES, Romero JM, Doyle S, Elliott R, Murphy G, Carpenter R. (1990) floricaula: a homeotic gene required for flower development in Antirrhinum majus. Cell 63: 1311–1322 [DOI] [PubMed] [Google Scholar]
- Coimbra S, Costa M, Jones B, Mendes MA, Pereira LG. (2009) Pollen grain development is compromised in Arabidopsis agp6 agp11 null mutants. J Exp Bot 60: 3133–3142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Folter S, Busscher J, Colombo L, Losa A, Angenent GC. (2004) Transcript profiling of transcription factor genes during silique development in Arabidopsis. Plant Mol Biol 56: 351–366 [DOI] [PubMed] [Google Scholar]
- Desgagné-Penix I, Eakanunkul S, Coles JP, Phillips AL, Hedden P, Sponsel VM. (2005) The auxin transport inhibitor response 3 (tir3) allele of BIG and auxin transport inhibitors affect the gibberellin status of Arabidopsis. Plant J 41: 231–242 [DOI] [PubMed] [Google Scholar]
- Dorcey E, Urbez C, Blázquez MA, Carbonell J, Perez-Amador MA. (2009) Fertilization-dependent auxin response in ovules triggers fruit development through the modulation of gibberellin metabolism in Arabidopsis. Plant J 58: 318–332 [DOI] [PubMed] [Google Scholar]
- Ehlting J, Provart NJ, Werck-Reichhart D. (2006) Functional annotation of the Arabidopsis P450 superfamily based on large-scale co-expression analysis. Biochem Soc Trans 34: 1192–1198 [DOI] [PubMed] [Google Scholar]
- Elliott RC, Betzner AS, Huttner E, Oakes MP, Tucker WQ, Gerentes D, Perez P, Smyth DR. (1996) AINTEGUMENTA, an APETALA2-like gene of Arabidopsis with pleiotropic roles in ovule development and floral organ growth. Plant Cell 8: 155–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis M, Egelund J, Schultz CJ, Bacic A. (2010) Arabinogalactan-proteins: key regulators at the cell surface? Plant Physiol 153: 403–419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endres S, Tenhaken R. (2009) Myoinositol oxygenase controls the level of myoinositol in Arabidopsis, but does not increase ascorbic acid. Plant Physiol 149: 1042–1049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falcone Ferreyra ML, Rius S, Casati P. (2012) Flavonoids: biosynthesis, biological functions and biotechnological applications. Front Plant Sci 3: 222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang W, Wang Z, Cui R, Li J, Li Y. (2012) Maternal control of seed size by EOD3/CYP78A6 in Arabidopsis thaliana. Plant J 70: 929–939 [DOI] [PubMed] [Google Scholar]
- Faure JE, Rotman N, Fortuné P, Dumas C. (2002) Fertilization in Arabidopsis thaliana wild type: developmental stages and time course. Plant J 30: 481–488 [DOI] [PubMed] [Google Scholar]
- Feller A, Machemer K, Braun EL, Grotewold E. (2011) Evolutionary and comparative analysis of MYB and bHLH plant transcription factors. Plant J 66: 94–116 [DOI] [PubMed] [Google Scholar]
- Fuentes S, Vivian-Smith A (2009) Fertilisation and fruit initiation. In L Østergaard, ed, Fruit Development and Seed Dispersal. Annual Plant Reviews, Vol 38. Wiley-Blackwell, Hoboken, NJ, pp 107–171 [Google Scholar]
- Garcia D, Fitz Gerald JN, Berger F. (2005) Maternal control of integument cell elongation and zygotic control of endosperm growth are coordinated to determine seed size in Arabidopsis. Plant Cell 17: 52–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisler M, Blakeslee JJ, Bouchard R, Lee OR, Vincenzetti V, Bandyopadhyay A, Titapiwatanakun B, Peer WA, Bailly A, Richards EL, et al. (2005) Cellular efflux of auxin catalyzed by the Arabidopsis MDR/PGP transporter AtPGP1. Plant J 44: 179–194 [DOI] [PubMed] [Google Scholar]
- Goetz M, Hooper LC, Johnson SD, Rodrigues JCM, Vivian-Smith A, Koltunow AM. (2007) Expression of aberrant forms of AUXIN RESPONSE FACTOR8 stimulates parthenocarpy in Arabidopsis and tomato. Plant Physiol 145: 351–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goetz M, Vivian-Smith A, Johnson SD, Koltunow AM. (2006) AUXIN RESPONSE FACTOR8 is a negative regulator of fruit initiation in Arabidopsis. Plant Cell 18: 1873–1886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouveia SC, Castilho PC. (2010) Characterization of phenolic compounds in Helichrysum melaleucum by high-performance liquid chromatography with on-line ultraviolet and mass spectrometry detection. Rapid Commun Mass Spectrom 24: 1851–1868 [DOI] [PubMed] [Google Scholar]
- Hansen BG, Kliebenstein DJ, Halkier BA. (2007) Identification of a flavin-monooxygenase as the S-oxygenating enzyme in aliphatic glucosinolate biosynthesis in Arabidopsis. Plant J 50: 902–910 [DOI] [PubMed] [Google Scholar]
- Hensel LL, Nelson MA, Richmond TA, Bleecker AB. (1994) The fate of inflorescence meristems is controlled by developing fruits in Arabidopsis. Plant Physiol 106: 863–876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA. (2008) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4: 44–57 [DOI] [PubMed] [Google Scholar]
- Huang W, Sherman BT, Lempicki RA. (2009) Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res 37: 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imaishi H, Matsuo S, Swai E, Ohkawa H. (2000) CYP78A1 preferentially expressed in developing inflorescences of Zea mays encoded a cytochrome P450-dependent lauric acid 12-monooxygenase. Biosci Biotechnol Biochem 64: 1696–1701 [DOI] [PubMed] [Google Scholar]
- Ingrosso I, Bonsegna S, De Domenico S, Laddomada B, Blando F, Santino A, Giovinazzo G. (2011) Over-expression of a grape stilbene synthase gene in tomato induces parthenocarpy and causes abnormal pollen development. Plant Physiol Biochem 49: 1092–1099 [DOI] [PubMed] [Google Scholar]
- Ito T, Meyerowitz EM. (2000) Overexpression of a gene encoding a cytochrome P450, CYP78A9, induces large and seedless fruit in Arabidopsis. Plant Cell 12: 1541–1550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs M, Rubery PH. (1988) Naturally occurring auxin transport regulators. Science 241: 346–349 [DOI] [PubMed] [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW. (1987) GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J 6: 3901–3907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kachlicki P, Einhorn J, Muth D, Kerhoas L, Stobiecki M. (2008) Evaluation of glycosylation and malonylation patterns in flavonoid glycosides during LC/MS/MS metabolite profiling. J Mass Spectrom 43: 572–586 [DOI] [PubMed] [Google Scholar]
- Kai K, Hashidzume H, Yoshimura K, Suzuki H, Sakurai N, Shibata D, Ohta D. (2009) Metabolomics for the characterization of cytochromes P450-dependent fatty acid hydroxylation reactions in Arabidopsis. Plant Biotechnol 26: 175–182 [Google Scholar]
- Kanehisa M, Goto S. (2000) KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res 28: 27–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Goto S, Sato Y, Furumichi M, Tanabe M. (2012) KEGG for integration and interpretation of large-scale molecular data sets. Nucleic Acids Res 40: D109–D114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi M, Inzé D, Depicker A. (2002) Gateway vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci 7: 193–195 [DOI] [PubMed] [Google Scholar]
- Katsumata T, Fukazawa J, Magome H, Jikumaru Y, Kamiya Y, Natsume M, Kawaide H, Yamaguchi S. (2011) Involvement of the CYP78A subfamily of cytochrome P450 monooxygenases in protonema growth and gametophore formation in the moss Physcomitrella patens. Biosci Biotechnol Biochem 75: 331–336 [DOI] [PubMed] [Google Scholar]
- Kim HJ, Ok SH, Bahn SC, Jang J, Oh SA, Park SK, Twell D, Ryu SB, Shin JS. (2011) Endoplasmic reticulum- and Golgi-localized phospholipase A2 plays critical roles in Arabidopsis pollen development and germination. Plant Cell 23: 94–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klucher KM, Chow H, Reiser L, Fischer RL. (1996) The AINTEGUMENTA gene of Arabidopsis required for ovule and female gametophyte development is related to the floral homeotic gene APETALA2. Plant Cell 8: 137–153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liscum E, Reed JW. (2002) Genetics of Aux/IAA and ARF action in plant growth and development. Plant Mol Biol 49: 387–400 [PubMed] [Google Scholar]
- Lohmann D, Stacey N, Breuninger H, Jikumaru Y, Müller D, Sicard A, Leyser O, Yamaguchi S, Lenhard M. (2010) SLOW MOTION is required for within-plant auxin homeostasis and normal timing of lateral organ initiation at the shoot meristem in Arabidopsis. Plant Cell 22: 335–348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahajan M, Ahuja PS, Yadav SK. (2011) Post-transcriptional silencing of flavonol synthase mRNA in tobacco leads to fruits with arrested seed set. PLoS ONE 6: e28315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margolin AA, Nemenman I, Basso K, Wiggins C, Stolovitzky G, Dalla Favera R, Califano A. (2006a) ARACNE: an algorithm for the reconstruction of gene regulatory networks in a mammalian cellular context. BMC Bioinformatics (Suppl 1) 7: S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margolin AA, Wang K, Lim WK, Kustagi M, Nemenman I, Califano A. (2006b) Reverse engineering cellular networks. Nat Protoc 1: 662–671 [DOI] [PubMed] [Google Scholar]
- Marsch-Martinez N, Greco R, Van Arkel G, Herrera-Estrella L, Pereira A. (2002) Activation tagging using the En-I maize transposon system in Arabidopsis. Plant Physiol 129: 1544–1556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNab H, Ferreira ESB, Hulme AN, Quye A. (2009) Negative ion ESI-MS analysis of natural yellow dye flavonoids: an isotopic labelling study. Int J Mass Spectrom 284: 57–65 [Google Scholar]
- Melan MA, Dong X, Endara ME, Davis KR, Ausubel FM, Peterman TK. (1993) An Arabidopsis thaliana lipoxygenase gene can be induced by pathogens, abscisic acid, and methyl jasmonate. Plant Physiol 101: 441–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar AA, Gubler F. (2005) The Arabidopsis GAMYB-like genes, MYB33 and MYB65, are microRNA-regulated genes that redundantly facilitate anther development. Plant Cell 17: 705–721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyoshi K, Ahn B-O, Kawakatsu T, Ito Y, Itoh J-I, Nagato Y, Kurata N. (2004) PLASTOCHRON1, a timekeeper of leaf initiation in rice, encodes cytochrome P450. Proc Natl Acad Sci USA 101: 875–880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizutani M, Ohta D. (2010) Diversification of P450 genes during land plant evolution. Annu Rev Plant Biol 61: 291–315 [DOI] [PubMed] [Google Scholar]
- Mizzotti C, Mendes MA, Caporali E, Schnittger A, Kater MM, Battaglia R, Colombo L. (2012) The MADS box genes SEEDSTICK and ARABIDOPSIS Bsister play a maternal role in fertilization and seed development. Plant J 70: 409–420 [DOI] [PubMed] [Google Scholar]
- Mo Y, Nagel C, Taylor LP. (1992) Biochemical complementation of chalcone synthase mutants defines a role for flavonols in functional pollen. Proc Natl Acad Sci USA 89: 7213–7217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy AS, Hoogner KR, Peer WA, Taiz L. (2002) Identification, purification, and molecular cloning of N-1-naphthylphthalmic acid-binding plasma membrane-associated aminopeptidases from Arabidopsis. Plant Physiol 128: 935–950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemhauser JL, Hong F, Chory J. (2006) Different plant hormones regulate similar processes through largely nonoverlapping transcriptional responses. Cell 126: 467–475 [DOI] [PubMed] [Google Scholar]
- Nishitani C, Yamaguchi-Nakamura A, Hosaka F, Terakami S, Shimizu T, Yano K, Itai A, Saito T, Yamamoto T. (2012) Parthenocarpic genetic resources and gene expression related to parthenocarpy among four species in pear (Pyrus spp.). Sci Hortic (Amsterdam) 136: 101–109 [Google Scholar]
- Noguchi T, Fujioka S, Choe S, Takatsuto S, Yoshida S, Yuan H, Feldmann KA, Tax FE. (1999) Brassinosteroid-insensitive dwarf mutants of Arabidopsis accumulate brassinosteroids. Plant Physiol 121: 743–752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noh B, Murphy AS, Spalding EP. (2001) Multidrug resistance-like genes of Arabidopsis required for auxin transport and auxin-mediated development. Plant Cell 13: 2441–2454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandolfini T, Molesini B, Spena A (2009) Parthenocarpy in crop plants. In L Østergaard, ed, Fruit Development and Seed Dispersal. Annual Plant Reviews, Vol 38. Wiley-Blackwell, Hoboken, NJ, pp 326–345 [Google Scholar]
- Peer WA, Bandyopadhyay A, Blakeslee JJ, Makam SN, Chen RJ, Masson PH, Murphy AS. (2004) Variation in expression and protein localization of the PIN family of auxin efflux facilitator proteins in flavonoid mutants with altered auxin transport in Arabidopsis thaliana. Plant Cell 16: 1898–1911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peer WA, Brown DE, Tague BW, Muday GK, Taiz L, Murphy AS. (2001) Flavonoid accumulation patterns of transparent testa mutants of Arabidopsis. Plant Physiol 126: 536–548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira LG, Coimbra S, Oliveira H, Monteiro L, Sottomayor M. (2006) Expression of arabinogalactan protein genes in pollen tubes of Arabidopsis thaliana. Planta 223: 374–380 [DOI] [PubMed] [Google Scholar]
- Popescu SC, Popescu GV, Bachan S, Zhang Z, Seay M, Gerstein M, Snyder M, Dinesh-Kumar SP. (2007) Differential binding of calmodulin-related proteins to their targets revealed through high-density Arabidopsis protein microarrays. Proc Natl Acad Sci USA 104: 4730–4735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray A, Robinson-Beers K, Ray S, Baker SC, Lang JD, Preuss D, Milligan SB, Gasser CS. (1994) Arabidopsis floral homeotic gene BELL (BEL1) controls ovule development through negative regulation of AGAMOUS gene (AG). Proc Natl Acad Sci USA 91: 5761–5765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee SY, Zhang P, Foerster H, Tissier C (2006) AraCyc: overview of an Arabidopsis metabolism database and its applications for plant research. In K Saito, R Dixon, L Willmitzer, eds, Plant Metabolomics. Biotechnology in Agriculture and Forestry, Vol 57. Springer, Berlin, pp 141–154 [Google Scholar]
- Roxrud I, Lid SE, Fletcher JC, Schmidt ED, Opsahl-Sorteberg HG. (2007) GASA4, one of the 14-member Arabidopsis GASA family of small polypeptides, regulates flowering and seed development. Plant Cell Physiol 48: 471–483 [DOI] [PubMed] [Google Scholar]
- Santelia D, Henrichs S, Vincenzetti V, Sauer M, Bigler L, Klein M, Bailly A, Lee Y, Friml J, Geisler M, et al. (2008) Flavonoids redirect PIN-mediated polar auxin fluxes during root gravitropic responses. J Biol Chem 283: 31218–31226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schijlen EGWM, de Vos CHR, Martens S, Jonker HH, Rosin FM, Molthoff JW, Tikunov YM, Angenent GC, van Tunen AJ, Bovy AG. (2007) RNA interference silencing of chalcone synthase, the first step in the flavonoid biosynthesis pathway, leads to parthenocarpic tomato fruits. Plant Physiol 144: 1520–1530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz G, Langmann T, Heimerl S. (2001) Role of ABCG1 and other ABCG family members in lipid metabolism. J Lipid Res 42: 1513–1520 [PubMed] [Google Scholar]
- Schuler M, Duan H, Bilgin M, Ali S. (2006) Arabidopsis cytochrome P450s through the looking glass: a window on plant biochemistry. Phytochem Rev 5: 205–237 [Google Scholar]
- Seifert GJ, Roberts K. (2007) The biology of arabinogalactan proteins. Annu Rev Plant Biol 58: 137–161 [DOI] [PubMed] [Google Scholar]
- Sheahan J, Cheong H. (1998) The colorless flavonoids of Arabidopsis thaliana (Brassicaceae). II. Flavonoid 3′ hydroxylation and lipid peroxidation. Am J Bot 85: 476–480 [PubMed] [Google Scholar]
- Shirley BW, Kubasek WL, Storz G, Bruggemann E, Koornneef M, Ausubel FM, Goodman HM. (1995) Analysis of Arabidopsis mutants deficient in flavonoid biosynthesis. Plant J 8: 659–671 [DOI] [PubMed] [Google Scholar]
- Skinner DJ, Baker SC, Meister RJ, Broadhvest J, Schneitz K, Gasser CS. (2001) The Arabidopsis HUELLENLOS gene, which is essential for normal ovule development, encodes a mitochondrial ribosomal protein. Plant Cell 13: 2719–2730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth DR, Bowman JL, Meyerowitz EM. (1990) Early flower development in Arabidopsis. Plant Cell 2: 755–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song S, Qi T, Huang H, Ren Q, Wu D, Chang C, Peng W, Liu Y, Peng J, Xie D. (2011) The jasmonate-ZIM domain proteins interact with the R2R3-MYB transcription factors MYB21 and MYB24 to affect jasmonate-regulated stamen development in Arabidopsis. Plant Cell 23: 1000–1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadler R, Lauterbach C, Sauer N. (2005) Cell-to-cell movement of green fluorescent protein reveals post-phloem transport in the outer integument and identifies symplastic domains in Arabidopsis seeds and embryos. Plant Physiol 139: 701–712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su V, Hsu B-D. (2010) Transient expression of the cytochrome p450 CYP78A2 enhances anthocyanin production in flowers. Plant Mol Biol Rep 28: 302–308 [Google Scholar]
- Takeda T, Amano K, Ohto MA, Nakamura K, Sato S, Kato T, Tabata S, Ueguchi C. (2006) RNA interference of the Arabidopsis putative transcription factor TCP16 gene results in abortion of early pollen development. Plant Mol Biol 61: 165–177 [DOI] [PubMed] [Google Scholar]
- Taylor LP, Grotewold E. (2005) Flavonoids as developmental regulators. Curr Opin Plant Biol 8: 317–323 [DOI] [PubMed] [Google Scholar]
- Tian Q, Uhlir NJ, Reed JW. (2002) Arabidopsis SHY2/IAA3 inhibits auxin-regulated gene expression. Plant Cell 14: 301–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulmasov T, Murfett J, Hagen G, Guilfoyle TJ. (1997) Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. Plant Cell 9: 1963–1971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varoquaux F, Blanvillain R, Delseny M, Gallois P. (2000) Less is better: new approaches for seedless fruit production. Trends Biotechnol 18: 233–242 [DOI] [PubMed] [Google Scholar]
- Verwoerd TC, Dekker BM, Hoekema A. (1989) A small-scale procedure for the rapid isolation of plant RNAs. Nucleic Acids Res 17: 2362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villanueva JM, Broadhvest J, Hauser BA, Meister RJ, Schneitz K, Gasser CS. (1999) INNER NO OUTER regulates abaxial- adaxial patterning in Arabidopsis ovules. Genes Dev 13: 3160–3169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vivian-Smith A, Luo M, Chaudhury A, Koltunow A. (2001) Fruit development is actively restricted in the absence of fertilization in Arabidopsis. Development 128: 2321–2331 [DOI] [PubMed] [Google Scholar]
- von Roepenack-Lahaye E, Degenkolb T, Zerjeski M, Franz M, Roth U, Wessjohann L, Schmidt J, Scheel D, Clemens S. (2004) Profiling of Arabidopsis secondary metabolites by capillary liquid chromatography coupled to electrospray ionization quadrupole time-of-flight mass spectrometry. Plant Physiol 134: 548–559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J-W, Schwab R, Czech B, Mica E, Weigel D. (2008) Dual effects of miR156-targeted SPL genes and CYP78A5/KLUH on plastochron length and organ size in Arabidopsis thaliana. Plant Cell 20: 1231–1243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werck-Reichhart D, Bak S, Paquette S. (2002) Cytochromes p450. The Arabidopsis Book 1: e0028, [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter D, Vinegar B, Nahal H, Ammar R, Wilson GV, Provart NJ. (2007) An “Electronic Fluorescent Pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS ONE 2: e718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ylstra B, Muskens M, Van Tunen AJ. (1996) Flavonols are not essential for fertilization in Arabidopsis thaliana. Plant Mol Biol 32: 1155–1158 [DOI] [PubMed] [Google Scholar]
- Zhang GY, Feng J, Wu J, Wang XW. (2010) BoPMEI1, a pollen-specific pectin methylesterase inhibitor, has an essential role in pollen tube growth. Planta 231: 1323–1334 [DOI] [PubMed] [Google Scholar]
- Zhang P, Foerster H, Tissier CP, Mueller L, Paley S, Karp PD, Rhee SY. (2005) MetaCyc and AraCyc: metabolic pathway databases for plant research. Plant Physiol 138: 27–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R, Xia X, Lindsey K, da Rocha PS. (2012) Functional complementation of dwf4 mutants of Arabidopsis by overexpression of CYP724A1. J Plant Physiol 169: 421–428 [DOI] [PubMed] [Google Scholar]
- Zondlo SC, Irish VF. (1999) CYP78A5 encodes a cytochrome P450 that marks the shoot apical meristem boundary in Arabidopsis. Plant J 19: 259–268 [DOI] [PubMed] [Google Scholar]