Knowledge and assays from Arabidopsis axillary meristem biology can be successfully applied to Salix spp. and can increase the understanding of a fundamental aspect of SRC biomass production, allowing more targeted breeding.
Abstract
The success of the short-rotation coppice system in biomass willow (Salix spp.) relies on the activity of the shoot-producing meristems found on the coppice stool. However, the regulation of the activity of these meristems is poorly understood. In contrast, our knowledge of the mechanisms behind axillary meristem regulation in Arabidopsis (Arabidopsis thaliana) has grown rapidly in the past few years through the exploitation of integrated physiological, genetic, and molecular assays. Here, we demonstrate that these assays can be directly transferred to study the control of bud activation in biomass willow and to assess similarities with the known hormone regulatory system in Arabidopsis. Bud hormone response was found to be qualitatively remarkably similar in Salix spp. and Arabidopsis. These similarities led us to test whether Arabidopsis hormone mutants could be used to assess allelic variation in the cognate Salix spp. hormone genes. Allelic differences in Salix spp. strigolactone genes were observed using this approach. These results demonstrate that both knowledge and assays from Arabidopsis axillary meristem biology can be successfully applied to Salix spp. and can increase our understanding of a fundamental aspect of short-rotation coppice biomass production, allowing more targeted breeding.
Willow (Salix spp.) is widely distributed in temperate regions, where some species are particularly well adapted to being grown as a biomass energy crop in short-rotation coppice (SRC) cycles, providing a renewable source of nearly carbon-neutral energy. Important SRC characteristics include the potential for high yields in short time periods, ease of vegetative propagation, and the ability to resprout after multiple harvests (Keoleian and Volk, 2005). Biomass willows, which are derived from basket-making varieties, are initially established by planting 25- to 30-cm-long stem cuttings in spring at densities of 10,000 to 20,000 per hectare. In the establishment year, growth largely occurs as single stems, which can reach heights of up to 2.5 m between spring and late autumn. These stems are coppiced just above ground level in winter (December to January) after leaf shedding. Once conditions are favorable in spring, the cut stools resprout to produce multiple stems. The stems will continue growing for a further 3 years, at which point they may be 7 to 8 m high, before being coppiced again to produce the first biomass harvest. Short-rotation coppicing continues on this 3-year cycle for 25 to 30 years before the willow stools are killed off and plowed into the ground. Coppicing enables farmers to grow the crop in shorter cycles than is possible in conventional forestry. In addition, harvesting or decapitation causes reinvigoration (the coppicing phenomenon) and, in some species, can even accelerate growth toward the theoretical maximum (Cannell et al., 1987). Previous studies have shown that the development and structure of the resprouted shoots depend on the position, abundance, and activity of dormant axillary buds on the stool, which can differ among willow species (Sennerby-Forsse and Zsuffa, 1995). However, although the success of the whole SRC system relies on the reactivation of these buds in response to coppicing, surprisingly little is known about the regulation of this process.
In contrast to willow, the mechanisms underlying axillary bud activation in Arabidopsis (Arabidopsis thaliana) have been investigated in some detail over the past few years, resulting in the identification of a network of interacting plant hormones that move systemically through the plant to control bud growth (Leyser, 2009). Central to this network is auxin, synthesized in the growing shoot apex and transported basipetally in the polar auxin transport stream (PATS), from where it acts indirectly to inhibit bud growth (Thimann and Skoog, 1933; Morris, 1977; Booker et al., 2003).
The indirect mode of action of auxin can be achieved by two mechanisms. First, if it is assumed that for active growth, buds must establish their own PATS into the main stem, then high auxin concentrations in the main stem can prevent bud activation by reducing the sink strength of the main stem for auxin, thereby preventing the canalization of auxin transport out of the bud. Auxin transport canalization involves an initial flow of auxin from a source to a sink, which both up-regulates and polarizes auxin transport in the direction of this initial flow, gradually “canalizing” it into files of cells with high auxin transport capacity toward the sink (Sachs, 1981). This process strongly correlates with bud activation (Morris, 1977; Li and Bangerth, 1999; Prusinkiewicz et al., 2009; Balla et al., 2011).
Second, auxin in the main stem has been shown to regulate the synthesis of two other hormones that regulate branching. Auxin can down-regulate the synthesis of cytokinin (Li et al., 1995; Nordström et al.., 2004; Tanaka et al., 2006), a known positive regulator of branching (for review, see Müller and Leyser, 2011). Furthermore, auxin can up-regulate the transcription of strigolactone biosynthetic genes (Hayward et al., 2009). Strigolactones or a derivative (SL) were recently identified (Gomez-Roldan et al., 2008; Umehara et al., 2008) as the upwardly mobile branch-inhibiting signal proposed to exist through studies of increased branching mutants, including more axillary growth (max) mutants of Arabidopsis, ramosus (rms) mutants of pea (Pisum sativum), decreased apical dominance (dad) mutants of petunia (Petunia hybrida), and dwarf (d) or high tillering dwarf mutants of rice (Oryza sativa; for review, see Domagalska and Leyser, 2011). The branchiness of a subset of these mutants can be rescued by grafting to wild-type roots or by SL addition (Napoli, 1996; Beveridge et al., 1997; Morris et al., 2001; Turnbull et al., 2002; Sorefan et al., 2003; Booker et al., 2005; Simons et al., 2007; Gomez-Roldan et al., 2008; Umehara et al., 2008; Crawford et al., 2010). Genes implicated in SL biosynthesis include MAX3/RMS5/DAD3/D17 and MAX4/RMS1/DAD1/D10, which encode divergent plastidic carotenoid cleavage dioxygenases (Sorefan et al., 2003; Booker et al., 2004; Snowden et al., 2005; Johnson et al., 2006; Zou et al., 2006; Arite et al., 2007), MAX1, a member of the cytochrome P450 family, and CYP711A1, which is predicted to work downstream of MAX3 and MAX4 (Booker et al., 2005). Mutants defective in SL signaling cannot be rescued by grafting to wild-type roots or by SL addition and include MAX2/RMS4/D3, which encodes an F-box protein (Stirnberg et al., 2002; Ishikawa et al., 2005; Johnson et al., 2006).
Since both cytokinin and SL are transported up the plant in the transpiration stream (Li et al., 1995; Kohlen et al., 2011), they can be carried directly into axillary buds to regulate their growth. This could be achieved by influencing the transcription of growth-regulating genes in the bud (Aguilar-Martínez et al., 2007; Brewer et al., 2009; Braun et al., 2012; Dun et al., 2012) and/or by modulating auxin transport properties, both locally and systemically, thereby affecting the ability of buds to establish auxin transport canalization into the main stem (Bennett et al., 2006; Lazar and Goodman, 2006; Lin et al., 2009; Prusinkiewicz et al., 2009; Crawford et al., 2010; Marhavý et al., 2011; Shinohara et al., 2013). Activation of buds will in turn affect the amount of auxin transported out into the main stem. Thus, auxin, cytokinin, and SL interact in multiple feedback loops to regulate branching.
Increasing concerns over climate change and energy security have heightened interest in the development of biorenewables for heat, power, and transport fuels, and willow has a recognized contribution to make. Salix spp. varieties initially used for biomass plantations were selected for basket-making qualities, and optimization of the crop is needed to realize fully the potential of SRC willow for bioenergy and biofuels (Karp et al., 2011). Bud outgrowth is a trait that shows great variability in the U.K. National Willow Collection and would be an excellent target for breeders. However, while the developmental biology of buds has been well characterized, bud physiology has not been investigated in any detail. In this study, techniques developed to dissect the regulation of bud outgrowth in Arabidopsis are tested on Salix spp. with a long-term aim of using them to understand branching control in willow and thus to assess the branching potential of future elite lines. The impressive similarity between the regulatory systems in willow and Arabidopsis prompted us to assess the possibility of using Arabidopsis hormone mutants as a platform to assess allelic variation in cognate Salix spp. genes, since routine transformation of willow is currently not possible. Our results suggest that this is a highly promising strategy.
RESULTS
Bud Hormone Responses
The activation of Arabidopsis buds on decapitated nodal stem segments can be inhibited by the application of auxin to the apical stump of the stem (Chatfield et al., 2000). To assess whether the same is true for Salix spp., nodal segments were excised from the secondary branches produced when stem cuttings from field-grown plants were sprouted in water. The field-grown stem segments had been harvested during the previous dormant cycle, cut into 30-cm lengths, and stored at −4°C. On removal from cold storage, they were soaked in water for 24 h and then stood in a beaker with just the basal end of the sticks in water. Bud break on the stem segments was observed nearly synchronously within 7 d, producing the side branches. On these freshly sprouted secondary branches, the axillary meristems remained dormant indefinitely unless the apex was removed, which resulted in activation of the axillary meristems in an apical-basal gradient similar to that observed in Arabidopsis (Hempel and Feldmann, 1994; Grbić and Bleecker, 1996). This suggested that the nodes on these branches were suitable candidates for the assay, so nodal segments were taken from them. Unlike the Arabidopsis assay, the tissue used was not grown in asceptic conditions, so a greater number of nodes were initially set up in each experiment to allow for losses due to contamination.
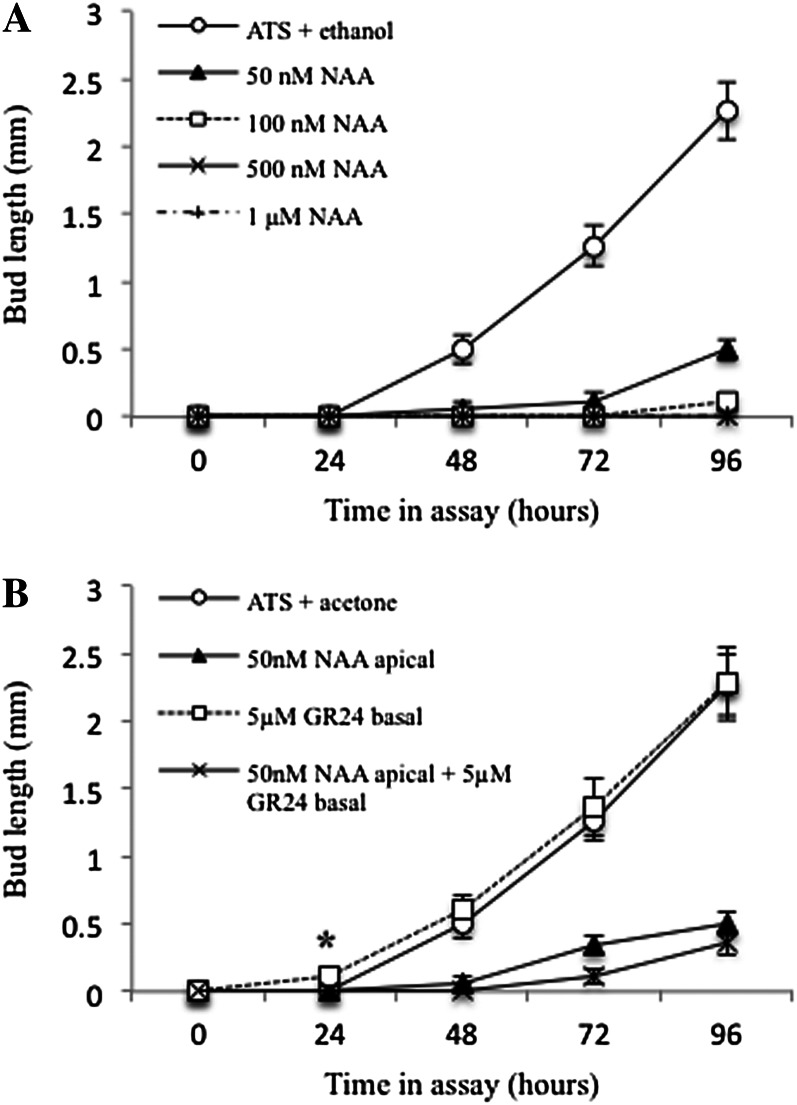
Application of 50 nm 1-naphthaleneacetic acid (NAA) apically to the Salix spp. stem was able to inhibit the outgrowth of Salix spp. buds for between 2 and 4 d, while 1 µm inhibited them beyond the duration of the experiment (Fig. 1A), indicating much greater sensitivity to auxin than that typically observed in Arabidopsis, where 1 µm NAA inhibits buds for approximately 4 d (Chatfield et al., 2000).
Figure 1.
Effects of apical auxin (NAA) and basal strigolactone (GR24) treatments on Salix spp. bud outgrowth. Excised nodes were inserted between two agar-solidified media blocks in a petri dish such that treatments could be applied apically or basally to the node. A, The apical blocks contained a range of NAA concentrations (50 nm–1 µm) or no NAA (plus an equal volume of carrier ethanol). B, The apical block contained either 50 nm NAA or no NAA (plus an equal volume of ethanol carrier), and the basal block contained 5 µm GR24 or no GR24 (plus an equal volume of acetone carrier). The length of the bud was measured every 24 h. Data represent means ± se (n = 15–20). The experiment was replicated four times. *Basal GR24 and ATS control means are significantly different at P < 0.05 (Student’s t test).
In Arabidopsis one-node assays of this type, basal application of GR24, a synthetic SL, has no effect on bud growth, but it can enhance the ability of apical auxin to inhibit bud activity. Similarly, basal 5 µm GR24 combined with apical 50 nm NAA prolonged Salix spp. bud inhibition beyond the effect of apical NAA alone, particularly in the first 96 h (Fig. 1B). Interestingly, with basal 5 µm GR24 treatment alone, bud swelling and subsequent outgrowth were observed 24 h earlier than in the no-treatment control. This effect was consistent through four replicates of the experiment and was statistically significant (P < 0.05, Student’s t test). After only 24 h of treatment, the GR24-treated buds began to swell, which was not observed until 48 h in the control buds, indicating that GR24 stimulates bud outgrowth in this assay.
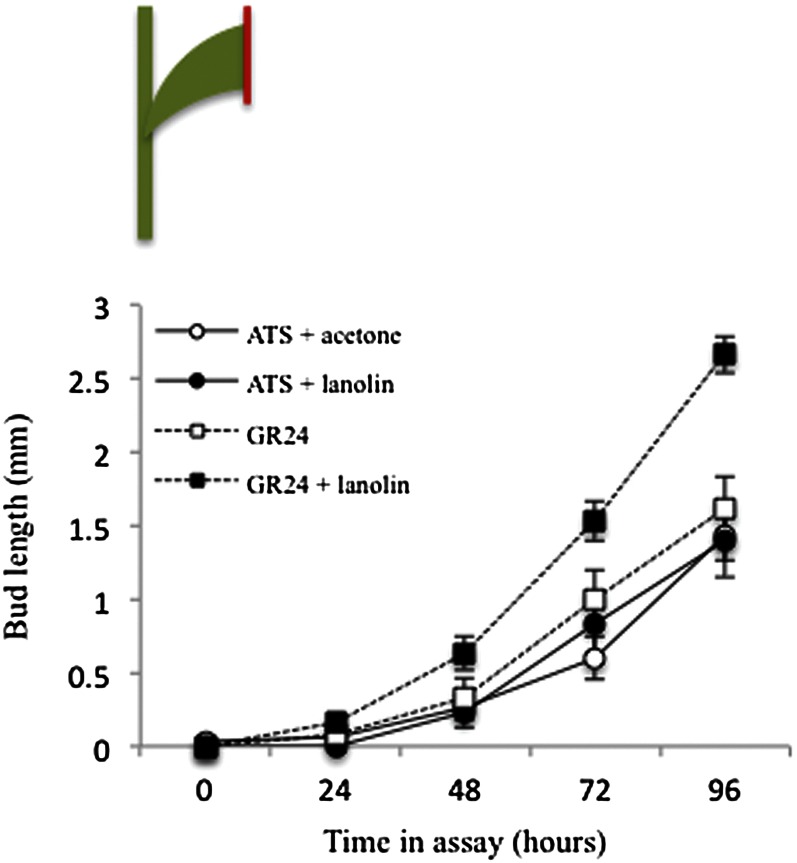
We had not previously observed such stimulation of bud activity by GR24 treatments in similar assays in Arabidopsis. However, by comparing activation times for solitary buds on isolated nodes versus the two buds on two-node segments, we had observed that faster bud activation can occur in the absence of competition from another bud (Crawford et al., 2010). Furthermore, GR24 treatment can promote bud activation when supplied to whole plants with compromised auxin transport (Shinohara et al., 2013). In contrast to Arabidopsis, where there is only one bud per node, in Salix spp., each node carries a group of three buds with a large central bud and two smaller buds (Brunkener, 1984, 1988; Paukkonen et al., 1992). Although these cited references studied much older tissue than the juvenile tissue used in these studies, we were able to confirm the presence of the three buds within the bud scale (Supplemental Fig. S1). Therefore, we hypothesized that in untreated nodal segments, activation of the large central buds may be slightly delayed by competition with the smaller adjacent buds. GR24 may be promoting Salix spp. bud outgrowth by shutting down competition between the three Salix spp. buds, allowing the central bud to establish PATS into the main stem more rapidly. Apart from competition between the three buds, the rapidity for PATS establishment is likely further compromised by the presence of two competing auxin sinks near the buds, namely the leaf trace and the PATS in the main stem (Ongaro et al., 2008). To reduce the complexity of this system and increase the importance of bud-bud competition, GR24 delivery to the leaf trace was reduced by sealing it with lanolin, inhibiting transpiration (Fig. 2). Under these circumstances, we predicted that SL treatment would have a more dramatic effect in promoting activation of the central bud. Slightly earlier bud outgrowth was observed again with basal GR24 treatment, and this was considerably enhanced in the nodes that had their leaves sealed with lanolin (Fig. 2). No differences were observed between the no-GR24 nodes with and without lanolin.
Figure 2.
Strigolactone (GR24) enhances competition between Salix spp. buds. Excised nodes were inserted between two agar-solidified media blocks in a petri dish such that treatments could be applied basally to the node. The nodal segments had their subtending leaf trimmed to 1 cm and the edge either left unsealed or sealed with lanolin (as in the cartoon above, with lanolin shown in red). Basal treatment was either 5 µm GR24 or no GR24 (plus an equal volume of acetone carrier). The length of the bud was measured every 24 h. Data represent means ± se (n = 15–20). [See online article for color version of this figure.]
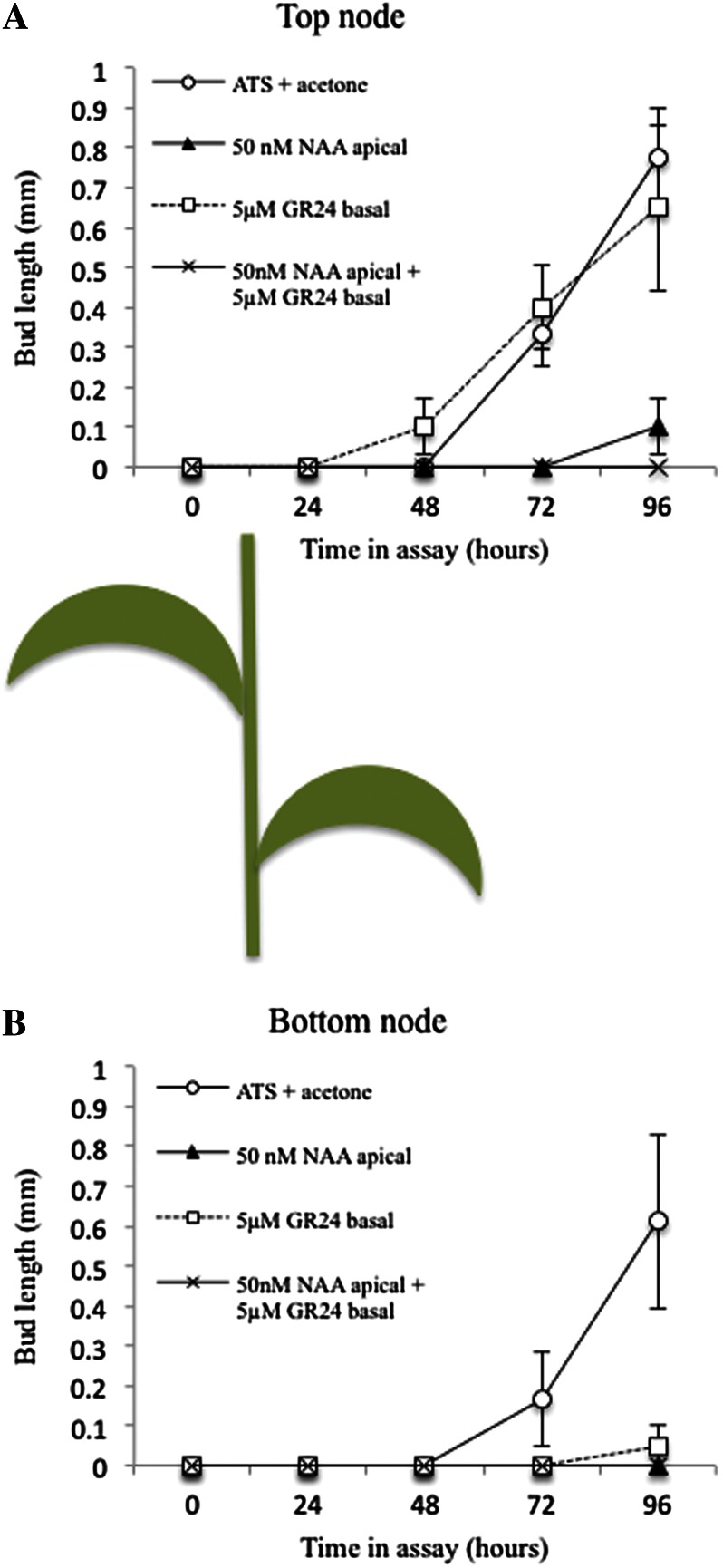
In the one-node assays, basal GR24 was able to inhibit bud outgrowth only in the presence of a strong auxin source. To investigate this further, a two-node system was used, with the idea that activation of one bud would contribute auxin to the stem, providing an auxin source. With two nodes, the behavior of the apical node was similar to the single-node experiments (Fig. 3A), although bud activation was slowed in all treatments, consistent with balanced competition between the central buds at the two nodes (compare Figs. 1B and 3). There was stronger inhibition in the combined GR24 and NAA treatment than with NAA alone. The apical bud always dominated the basal bud, which showed considerably delayed and slower outgrowth than the apical bud in all cases, including the untreated control (Fig. 3B). The basal bud was strongly inhibited by GR24 alone, similar to the combined GR24 and NAA treatment. These data show that, as in Arabidopsis, GR24 is able to inhibit bud outgrowth only in the presence of a competing auxin source.
Figure 3.
Effects of synthetic strigolactone (GR24) and auxin (NAA) on bud outgrowth in two-node stem segments. Excised segments, encompassing two nodes, with their associated buds (as in the cartoon below) were inserted between two agar-solidified media blocks in a petri dish such that treatments could be applied apically or basally to the section. The apical block contained either 50 nm NAA or no NAA (plus an equal volume of ethanol carrier), and the basal block contained 5 µm GR24 or no GR24 (plus an equal volume of acetone carrier). The lengths of the more apical (A) and basal (B) buds were measured every 24 h. Data represent means ± se (n = 15–20). [See online article for color version of this figure.]
Testing Salix spp. MAX Allelic Variation through Transformation
Given the similarities between Salix spp. and Arabidopsis bud activation, we decided to assess the possibility of using Arabidopsis mutants as a platform to test for functional allelic variation in the cognate willow genes. SxMAX1, SxMAX2, and SxMAX4 genes were amplified from willow lines as indicated in Table I, including the parents of the K8 mapping population (siblings S3 and R13), Salix viminalis × (S. viminalis × [S. viminalis × Salix schwerinii]), which has been used to identify a number of yield quantitative trait loci (QTLs; Hanley et al., 2006). All of the willow lines used in this study were confirmed diploids, but there is still the possibility that the amplified sequences are not alleles of the targeted genes but rather paralogs or duplicates. To assess this possibility, all the amplified sequences were directly sequenced in order to survey all polymorphisms expected for that primer pair before multiple individual clones were sequenced. For plants where polymorphisms were detected within the genomic DNA, these were accounted for in the sequences of the two parent clone sequences per plant/gene. Each primer set identified a maximum of only two sequences per targeted gene per plant genome (Table I). Where two sequences were detected, support that they were indeed alleles at a single locus came from linkage studies in which single-nucleotide polymorphisms between the two sequences were used to test for segregation in appropriate mapping family progeny. Only SxMAX4E could not be tested in this way, because no mapping family is available at present. In all cases, the polymorphisms segregated, which would not be the case if the single-nucleotide polymorphism was detected as a result of the amplification of two homozygous, but unlinked, gene homologs. The cloned sequences for each of the SxMAX genes, except SxMAX4E, were mapped to comparable positions in mapping populations (Supplemental Table S1), in agreement with the poplar (Populus trichocarpa) orthologs targeted at the outset. Unrooted trees also supported orthology between all SxMAX alleles and the original poplar targets. Supplemental Figure S2 illustrates that all Salix spp. alleles are more similar to the original poplar target than they are to any other poplar homolog, supporting the case that only true Salix spp. alleles were identified.
Table I. Information on the cloning of the Salix spp. MAX alleles.
F, Forward, R, reverse.
| Allele | Primersa | GenBank Accession No. | Clonal Name | DNA Source Species | No. of Sequences Recovered | Closest Poplar Homolog |
|---|---|---|---|---|---|---|
| SxMAX1B | F: 5′-ATGGATTTACAGGTTTTGTTTACAG-3′ | JN613462 | S3 | S. viminalis × [S. viminalis × (S. viminalis × S. schwerinii)] | 1 | 0006s24320 |
| R: 5′-TCAAGTTCGTTTTATGATTCTAAGC-3′ | ||||||
| SxMAX2A | F: 5′-ATGGCTGCTACCATGAACGATC-3′ | JN613463 | S3 | S. viminalis × [S. viminalis × (S. viminalis × S. schwerinii)] | 1 | 0014s13910 |
| R: 5′-TCAGTCGAGGATCGGACGCC-3′ | ||||||
| SxMAX2B | F: 5′-ATGGCTGCTACCATGAACGATC-3′ | JN613464 | R13 | S. viminalis × [S. viminalis × (S. viminalis × S. schwerinii)] | 2 | 0014s13910 |
| R: 5′-TCAGTCGAGGATCGGACGCC-3′ | ||||||
| SxMAX4B | F: 5′-ATGGCTTCCTTGGCATTTTCC-3′ | JN613465 | S3 | S. viminalis × [S. viminalis × (S. viminalis × S. schwerinii)] | 1 | 0006s25490 |
| R: 5′-TTATTTCTTTGGCACCCAGCATC-3′ | ||||||
| SxMAX4E | F: 5′-CTCCAACTTGGTATGTCTCCC-3′ | JX391953 | RES0453 | Salix aurita | 2 | 0006s25490 |
| R: 5′-GATAGCTAAATCACACAACCCC-3′ | ||||||
| SxMAX4G | F: 5′-CTCCAACTTGGTATGTCTCCC-3′ | JN613468 | Ulbricht-weide | Salix purpurea × S. viminalis | 1 | 0006s25490 |
| R: 5′-GATAGCTAAATCACACAACCCC-3′ |
The motif CACC was added to the 5′ end of all forward primers for directional cloning purposes.
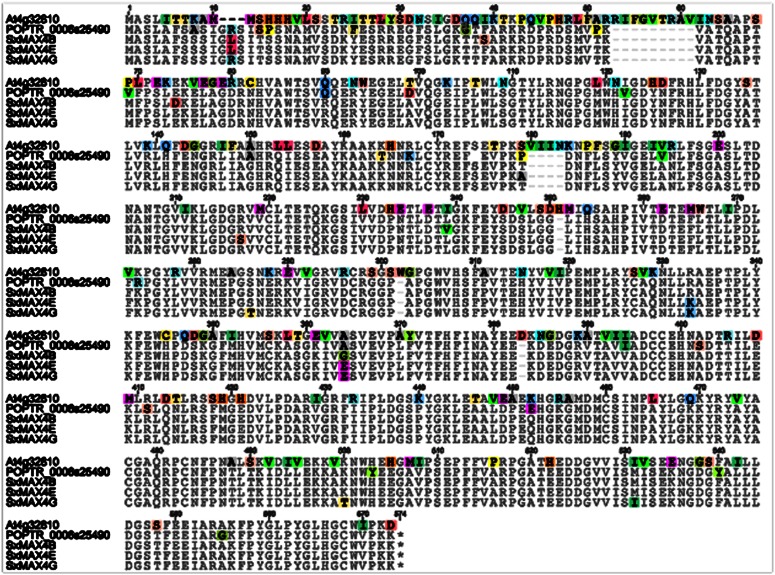
The SxMAX alleles were transformed into well-characterized loss-of-function Arabidopsis max mutants to assess the functional significance of the observed allelic variation in willow. Multiple independent lines were generated for each construct to determine whether the alleles were capable of rescuing fully the max mutant phenotypes in at least some lines. In a MUSCLE (Drummond et al., 2011) alignment of Arabidopsis MAX4, PtMAX4, and the three Salix spp. MAX4 alleles (Fig. 4), amino acid differences were observed in all three SxMAX alleles as compared with each other. In SxMAX4E, for example, a highly conserved Arg at position 217 was replaced by a Ser residue, and a Thr present in SxMAX4B and Gly at position 179 were replaced by an Ala.
Figure 4.
MUSCLE alignment of the predicted protein sequences of Arabidopsis MAX4 (At4g32810), poplar MAX4 (POPTR_0006s25490), and the three Salix spp. MAX4 alleles B, E, and G. Nonconsensus residues were highlighted according to Geneious Pro 5.5.6 default settings.
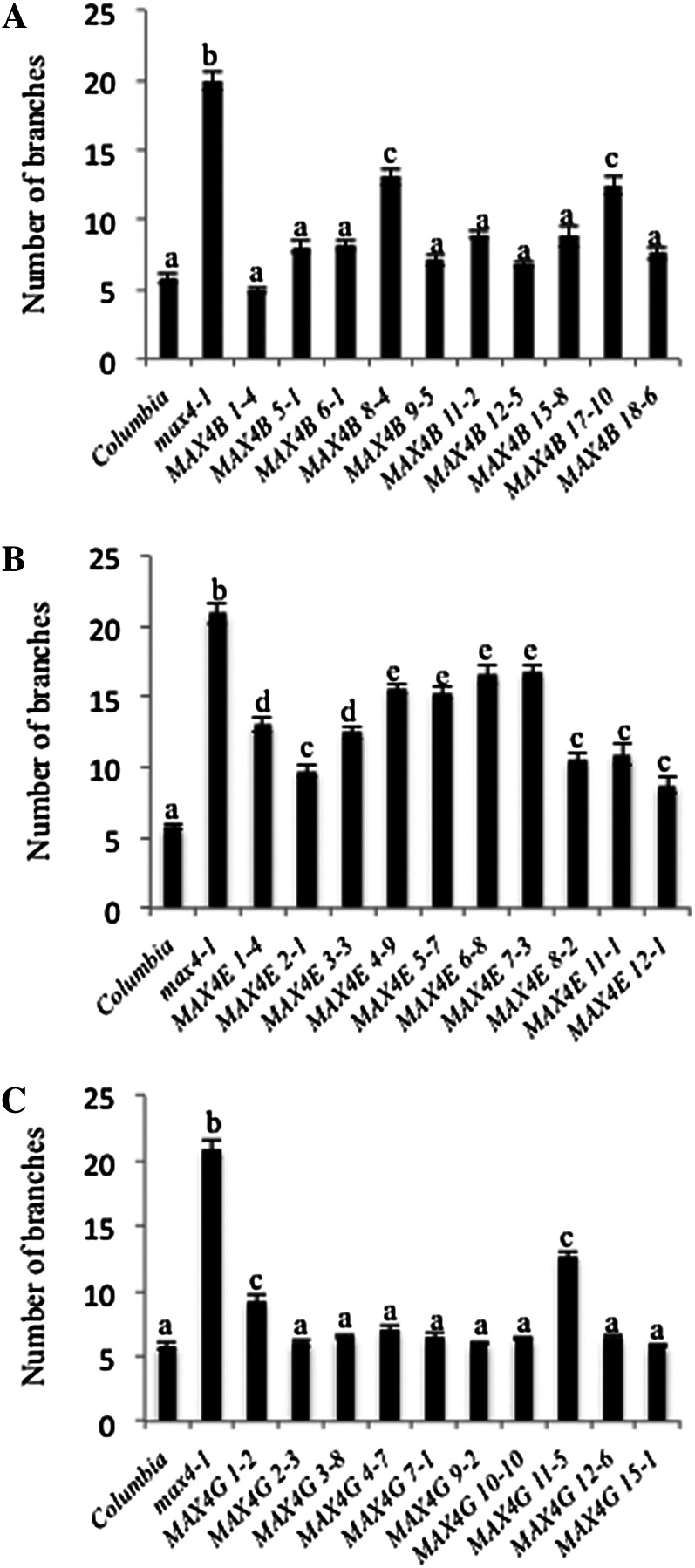
In branching assays, the majority of the independent transgenic lines (eight of 10) expressing either SxMAX4B or SxMAX4G were able to restore the Arabidopsis max4-1 branching phenotype to the wild type (Fig. 5). With both constructs, a small number of lines did not show full rescue, which could be partly due to differences in the expression levels of the transgene (Supplemental Fig. S3). In contrast, SxMAX4E was not able to rescue fully the max4-1 branching defect in any of the independent lines studied but only reduced branching by around one-third in the majority of the transgenic lines. The amino acid polymorphisms identified in SxMAX4E, therefore, are candidates for this inability to rescue fully the max4 branching defect.
Figure 5.
Testing allelic variation in SxMAX4 alleles through the rescue of branching in max4. The branching assay was as described by Greb et al. (2003). Branch numbers were determined for each of the three Salix spp. alleles tested in 10 independent transgenic lines, with wild-type Columbia and max4-1 as controls. Data represent mean numbers of branches ± se (n = 15–20). The different letters in A, B, and C denote significant differences at P < 0.05 in mean values as determined using Tukey’s test. Values with the same lowercase letter are not significantly different from one another.
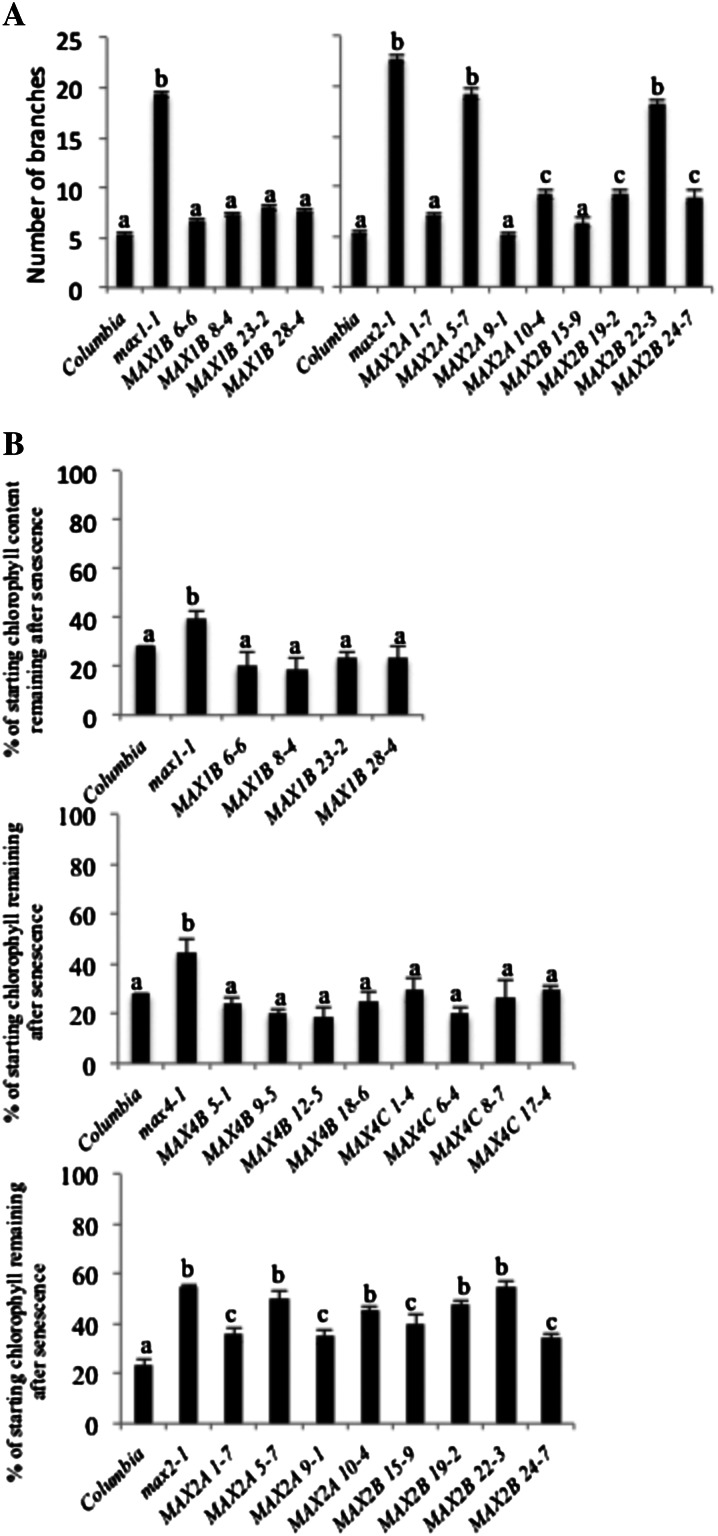
SxMAX1B fully rescued the max1 branching defect in all of the transgenic lines studied (data for a subset of lines are shown in Fig. 6A). A MUSCLE (Drummond et al., 2011) alignment of Arabidopsis MAX1, PtMAX1, and SxMAX1B (Supplemental Fig. S4) showed that there were amino acid differences, but these clearly did not affect the ability of the Salix spp. sequence to complement max1.
Figure 6.
The delayed senescence defect of max mutants could be rescued by their cognate Salix spp. MAX SL biosynthesis genes but not by the SL signaling gene SxMAX2. A, The branching assay was as described by Greb et al. (2003). Branch numbers were determined in 10 independent transgenic lines per Salix spp. allele plus wild-type Columbia, max1-1, and max2-1. Data represent mean numbers of branches ± se (n = 15–20). B, Cotyledons of 7-d-old seedlings were excised and placed in darkness for 8 d to induce senescence. The percentage of the starting chlorophyll remaining after senescence was calculated from the mean chlorophyll a + b content in control (unsenesced) and senesced cotyledons. Data represent mean percentage of remaining chlorophyll in three independent experiments ± se (n = 3; each sample = 40 cotyledons). The different letters denote significant differences at P < 0.05 in mean values as determined using Tukey’s test. Values with the same lowercase letter are not significantly different from one another.
Mutants in max2 differ from the other Arabidopsis max mutants by being nongraft or SL rescuable; thus, MAX2 is implicated in SL signaling (Stirnberg et al., 2002; Booker et al., 2005; Crawford et al., 2010). A MUSCLE (Drummond et al., 2011) alignment of Arabidopsis MAX2, PtMAX2, SxMAX2A, and SxMAX2B showed a number of amino acid differences between the Arabidopsis, poplar, and Salix spp. sequences but only two amino acid differences between the two Salix spp. sequences (Supplemental Fig. S4). SxMAX2A and SxMAX2B were both capable of rescuing the branching defect of max2 (Fig. 6A), with some of the independent transgenic lines showing wild-type branch numbers, but there was substantial variation between independent transgenic lines, conspicuously more than with either the SxMAX4 or SxMAX1 construct. For both Salix spp. MAX2 constructs, it was not possible to make any link between the degree of branching rescue and the expression levels of the transgene, suggesting that there may be posttranscriptional effects (Supplemental Fig. S5).
Bud Auxin Responses
To investigate whether the restored shoot-branching phenotypes correlated with restored bud auxin responses, the ability of apically applied auxin to inhibit bud outgrowth was tested using isolated nodal segments (Chatfield et al., 2000). The max mutants are auxin resistant in this assay (Bennett et al., 2006). Bud outgrowth was tested in the presence and absence of 1 μm apical auxin (NAA) in three of the SxMAX2A transgenic lines displaying full (SxMAX2A 1-7), partial (SxMAX2A 10-4), or no (SxMAX2A 5-7) rescue. The fully rescuing line, namely SxMAX2A 1-7, showed wild-type sensitivity to apical auxin, with little bud activation after 144 h of incubation, similar to wild-type plants, whereas buds on excised nodes from lines SxMAX2A 10-4 and 5-7 behaved similarly to max2-1, with significant elongation by 144 h (Supplemental Fig. S5).
Senescence
Although relatively unpleiotropic, the SL mutants do confer additional phenotypes. In particular, max mutants have delayed leaf senescence, a phenotype that is particularly strong in max2 mutants (Woo et al., 2001). To determine whether this SL function was also conserved between Salix spp. and Arabidopsis, we tested whether the willow genes could also complement the delayed senescence of the max mutants using cotyledon senescence assays. SxMAX1B, SxMAX4B, and SxMAX4C were able to rescue the delayed-senescence phenotype of their respective max mutants (Fig. 6B). However, for SxMAX2A or SxMAX2B, regardless of the degree of branching rescue, only a slight rescue of the delayed-senescence phenotype was observed. For example, SxMAX2A transgenic lines 1-7 and 9-1, which could fully rescue the max2 branching defect, only halved the difference between max2 and wild-type leaf senescence. Thus, the SL biosynthesis genes (SxMAX4 and SxMAX1) were able to complement delayed senescence and branching (Figs. 5 and 6A, respectively), while the SL signaling gene SxMAX2 was only fully able to rescue branching.
DISCUSSION
The genus Salix contains a number of species of great value as biomass crops (Gullberg, 1993; Åhman and Larsson, 1994; Lindegaard and Barker, 1997). Over the past few years, significant advances have been made in the breeding of biomass willows (Zsuffa, 1990; Gullberg, 1993; Åhman and Larsson, 1994; Lindegaard and Barker, 1997), with cultivars such as Tora and Torhild (bred in Sweden) and Resolution (bred in the United Kingdom) giving average biomass yields of 12 to 14 oven-dried tons per hectare per year in trials. Previous studies have shown that willows of quite different architecture (e.g. many thin stems or few thick stems) can achieve high biomass yields, suggesting that it may be necessary to breed for multiple ideotypes (Tharakan et al., 2005). Having the ability to select specific coppice types would assist the breeder enormously in developing genotypes for different environments and/or end uses. Our studies demonstrate that both knowledge and assays from Arabidopsis axillary meristem biology can be successfully applied to Salix spp. to increase our understanding of this fundamental aspect of SRC biomass production, thus allowing more targeted breeding.
Salix spp. Bud Response to Strigolactone and Auxin
The inhibition of bud outgrowth by an actively growing shoot apex, apical dominance, is mediated by auxin synthesized in the young expanding leaves at the apex (Ljung et al., 2001) and transported basipetally down the main stem in the PATS (Thimann and Skoog, 1933). The action of auxin in inhibiting buds is still not fully understood, but it is clear that it acts indirectly without entering the bud (Morris, 1977; Booker et al., 2003). SLs, when directly applied to buds, can inhibit them (Gomez-Roldan et al., 2008; Brewer et al., 2009), evidence that contributed to the proposal that they may be acting as second messengers for auxin, relaying the inhibitory signal from the stem to buds. Alternatively, it has been proposed that PAT in the main stem can inhibit bud activity through influencing stem sink strength for auxin, thereby affecting the crucial establishment of auxin export from the bud and thus bud activation (Li and Bangerth, 1999; Bennett et al., 2006; Prusinkiewicz et al., 2009; Balla et al., 2011). SL action in this case would be through systemically dampening auxin transport canalization. We found that, as with other species (Crawford et al., 2010; Liang et al., 2010), SL was only effective at inhibiting isolated willow buds in the presence of an apical auxin source. Indeed, when an isolated Salix spp. bud was treated with SL alone, it activated slightly earlier than untreated buds. This does not support a direct role for SL acting locally in the bud to inhibit growth (Brewer et al., 2009; Dun et al., 2012); rather, it argues for the auxin transport canalization model for the regulation of bud outgrowth and its modulation by SLs (Shinohara et al., 2013).
Crawford et al. (2010) proposed that SLs were acting to set the global context by which buds compete for auxin export into the main stem. In a low-SL situation, many buds can activate and contribute auxin to the main stem. In a high-SL situation, the first bud to activate can dominate and prevent the activation of other buds. Such competition could also be an extremely important factor locally and be influenced by developmental and environmental factors. In Salix spp., the buds initiate and differentiate in groups of three (Supplemental Fig. S1; Brunkener, 1984, 1988; Paukkonen et al., 1992). The main bud in the center lies between two flanking buds that have their own bud scales, and all three buds share a common bud scale. At an early stage, the two flanking buds are formed in the axils of the two bud scales in the primary bud, in close connection to the main vascular system of the primary shoot (Sennerby-Forsse et al., 1992). Competition, therefore, is not just between different buds on the stem but also among the three buds in the cluster. SL may be promoting Salix spp. bud outgrowth by shutting down competition between the three Salix spp. buds and allowing the central bud rapidly to dominate and activate. Ongaro et al. (2008) showed that buds could form vascular connections with either the leaf trace or the main stem vascular bundles. If GR24 treatment is enhancing the competitive advantage of the central Salix spp. bud and the path for vascularization is simplified by reducing GR24 delivery to the leaf trace, then the central bud may be able to activate even more rapidly. This may explain why lanolin treatment at the leaf excision site had no effect by itself but could enhance the effect of GR24 treatment in stimulating the activation of the central bud.
Bud outgrowth in Salix spp. was inhibited at lower concentrations of applied auxin than in Arabidopsis and chrysanthemum (Dendranthema grandiflorum; Chatfield et al., 2000; Liang et al., 2010). This suggests a stronger inhibitory effect of an active apex upon the subtending branches or generally lower levels of endogenous auxin. Stronger apical dominance on each twig may reduce self-shading and thus contribute to optimal light structure in the canopy.
Two-node assays in Arabidopsis and chrysanthemum gave somewhat different results, suggesting that there may be different relative bud-bud competitiveness in these two species. In chrysanthemum, similar to Salix spp., the apical bud was always favored over the basal bud. This is expected because auxin exported from the apical bud and transported down the stem in the PATS can easily influence the auxin sink strength of the stem at the position of the basal bud, whereas the influence of auxin exported from the basal bud on the apical bud is likely to be less strong. In Arabidopsis, the basal bud in two-node assays is able to dominate, possibly because it is typically larger than the apical bud at the start of the assay and, therefore, at a competitive advantage (Ongaro et al., 2008; Crawford et al., 2010; Liang et al., 2010). The apical Salix spp. bud behaved similarly to buds isolated on single-node segments; however, the basal bud was inhibited, especially if basal GR24 and/or apical auxin were added. Presumably, this was due to the inhibitory effect of auxin exported from the active apical bud. However, bud outgrowth in both buds was delayed, as compared with single nodes, suggesting some effective competition from the basal bud.
Uncoupling the S. viminalis MAX2 Senescence Response from Branching
In the transformation-rescue experiments, alleles of the SxMAX genes involved in strigolactone biosynthesis (SxMAX1 and SxMAX4) were able to rescue both the delayed senescence as well as the highly branched phenotypes of their cognate mutants, while alleles of the SxMAX2 strigolactone signaling gene were only able to rescue fully the branching phenotypes. Bud outgrowth sensitivity to auxin was also rescuable by Salix spp. MAX2. MAX2/ORE9/PPS encodes an F-box protein involved in inflorescence architecture, senescence, early seedling light response, and karrikin signaling (Woo et al., 2001; Stirnberg et al., 2002; Shen et al., 2007; Nelson et al., 2011). MAX2 is a component of an SCF (for SKP, Cullin, and F-box protein) complex (Stirnberg et al., 2007) and, therefore, is predicted to be involved in targeting specific proteins for ubiquitination, probably marking them for proteolysis. The relationship between the different roles for MAX2 is poorly understood. It is possible that these diverse roles are mediated by multiple targets for SCFMAX2. Therefore, the different abilities of the willow alleles to rescue Arabidopsis senescence and branching could be because willow MAX2 can recognize the branching-specific targets but not the senescence-specific targets. This could be because of divergent coevolution of willow MAX2 and its senescence-specific targets or because the regulation of senescence is a newer role gained by AtMAX2 or one that has been lost by SxMAX2. Studying more divergent Salix spp. MAX2 alleles and comparing branching and senescence QTLs in Salix spp. may provide more insight into the evolution of these MAX2 roles.
Future Prospects for Breeding
The identification of QTLs and an increased number of markers now available in willow have increased the likelihood of uncovering the genetic basis for developmental processes (Tsarouhas et al., 2002; Hanley, 2003, Hanley et al., 2006). To be able to select more effectively for high biomass yield, a better understanding of how the relevant growth processes important for high biomass are regulated is crucial, together with the identification of useful variation in the genes that underpin them in different willow lines and species. The MAX genes were chosen for further study because there is already a body of evidence around them and previously developed assays to test their involvement in shoot branching in Arabidopsis, which could be used to study yield and architecture variation in biomass willow. Transformation of the study species provides the ultimate test for QTL candidate gene confirmation; however, this is not currently possible in willow. Cognate Arabidopsis null mutants, therefore, are an attractive option as a platform for the assessment of functional allelic variation. SxMAX4B and SxMAX4G were able to rescue fully the max4-1 branching defect. SxMAX4E, however, was less effective. Polymorphisms identified in SxMAX4E may be responsible for the inability of this allele to show full MAX4 activity. These include changes in residues highly conserved among MAX4 orthologs, such as R217S, but no specific function has been proposed for this residue (Messing et al., 2010; Delaux et al., 2012). These results provide support for the efficacy of using an Arabidopsis transformation system to identify functionally relevant allelic variation in Salix spp. Analysis of SxMAX4E in mapping populations will help determine whether these polymorphisms are of future interest to willow breeders.
Complementary to using transformation as a means of assessing allelic variation, physiological assays can be used as signatures to determine the underlying causes of branching variation. For example, max mutants and auxin signaling mutants both have increased branching and auxin-resistant bud activation, but max mutants uniquely have increased auxin transport and the overaccumulation of PIN proteins at the plasma membrane (Sorefan et al., 2003; Bennett et al., 2006; Lazar and Goodman, 2006). We have shown that the relevant assays from Arabidopsis can be readily transferred to willow and, therefore, can be used in a combinatorial way, along with candidate gene selection and QTL mapping, to assist breeders in their efforts to improve yield in biomass Salix spp.
CONCLUSION
In willow, bioenergy-relevant traits, such as coppicing response and shoot number, are difficult to study due to the size of the plant, the length of the SRC cycle, and the complex nature of many of the traits. For example, shoot numbers measured at the time of harvest are not only the result of the number of buds that sprouted after coppicing but also of the extent to which stems were subsequently self-thinned. In order to dissect these traits, tractable experimental systems are needed where hypotheses concerning the roles of different genes can be challenged. We have used Arabidopsis to identify functional differences in willow alleles in biological assays that help to formulate hypotheses about what these alleles may be doing in willow. In the context of a program of work aimed at testing hypotheses about causal loci underlying QTLs, our results show that this could be a useful tool. Once Salix spp. lines carrying representative alleles have been identified, bud physiology can be analyzed to identify further similarities with strigolactone biology beyond branch numbers. In combination, these results can inform genetic studies in willow, allowing crosses to be set up to test associations between specific alleles of the gene in question and the strigolactone-related traits.
MATERIALS AND METHODS
Plant Lines and Plant Growth
Unless otherwise stated, plants were grown in a growth room at 21°C with 16 h of daylight, 8 h of night, and a light intensity of 100 to 120 μmol m−2 s−1. All Arabidopsis (Arabidopsis thaliana) max lines are in a Columbia-0 background. For transformation, max1-1, max2-1, and max4-1 plants were grown in 8-cm-square pots containing F2 compost treated with Intercept 70WG (both from Levington Horticulture). Plants grown for the shoot-branching assays were sown in P40 multitrays (Desch Plantpak) in F2 compost as above.
Salix viminalis ‘Bowles Hybrid’ stem cuttings (25–30 cm long), harvested during the dormant cycle, were removed from cold storage (−4°C) and soaked in water for 24 h. Growth of shoots from these stems was activated by standing them in a beaker of water in the growth room. By activating the buds in this way, we observed nearly synchronous activation of the axillary buds to give secondary branches of equivalent age and length.
Hormone Stocks
NAA (Sigma) was dissolved in 70% ethanol, and GR24 (LeadGen Labs) was dissolved in acetone.
Bud Activity Assays
Bud hormone response assays were performed as described by Chatfield et al. (2000) with the exceptions that the subtending leaf was trimmed to around 1 cm to fit into the square petri dish and the tissue used was not sterile. Nodal segments from the freshly sprouted secondary branches produced from the willow (Salix spp.) stem segments, containing either one or two axillary nodes, were placed between two agar-solidified Arabidopsis Salt (ATS; Wilson et al., 1990) media slabs to which hormone treatments could be applied. All experiments were repeated a minimum of four times.
Isolation of the Salix spp. MAX Genes
Putative orthologs of the Arabidopsis MAX1, MAX2, and MAX4 genes were identified in the poplar (Populus trichocarpa) database version 1.1 using TBLASTN on http://genome.jgi-psf.org/Poptr1_1.home.html. For both MAX1 and MAX4, two highly homologous gene sequences were detected on poplar linkage groups VI and XVIII in accordance with the known Salicoid whole-genome duplication event (Tuskan et al., 2006). However, only one clear homolog was identified in poplar for MAX2, located on linkage group XIV. For all three genes, the most homologous gene was used in downstream functional studies. Putative Salix spp. orthologs were amplified initially from genotype R13 of Salix viminalis × [S. viminalis × (S. viminalis × Salix schwerinii)] (Hanley et al., 2006) using primers designed to predicted poplar coding sequence. Resulting PCR products were gel purified (QIAquick Gel Extraction Kit; Qiagen), sequenced, and mapped on a poplar-anchored Salix spp. map (K8; Hanley et al., 2006) to confirm synteny. Salix spp. sequence was assembled using ContigExpress Vector NTI 10.1.1 (Invitrogen). RACE was used to confirm transcript ends using complementary DNA synthesized from R13 RNA (extracted according to Chang et al., 1993). For this, the GeneRacer Kit (Invitrogen) was used according to the manufacturer’s instructions.
To identify different alleles for functional testing, SxMAX alleles were amplified from several Salix spp. genomic DNA samples as part of an ongoing screen (for details, see Table I). Amplification was performed using AccuPrime Pfx SuperMix (Invitrogen), and the products were gel purified, cloned into the pENTR/D-TOPO vector (Invitrogen), and sequenced.
Poplar gene models (version 2.2; http://www.phytozome.net) were used to predict SxMAX protein-coding sequences, which were then translated and aligned with Arabidopsis and poplar protein sequences using the MUSCLE algorithm within Geneious Pro 5.5.6 software (Drummond et al., 2011) using default settings. Only SxMAX2 required minor manual adjustment. Nonconsensus residues were highlighted according to Geneious Pro 5.5.6 default settings.
To confirm the orthology of the SxMAX alleles with the initial poplar target, and to limit the possibility that Salix spp. paralogs had been amplified, which is expected in the willow genome as a consequence of the Salicoid duplication event (Hanley et al., 2006; Tuskan et al., 2006), unrooted trees were generated from coding sequences of the Salix spp. alleles and poplar homologs (Supplemental Fig. S2). Poplar coding sequences were retrieved from http://www.phytozome.net via BLAST searches to the poplar genome using the SxMAX1B, SxMAX2A, or SxMAX4B coding sequence as the query sequence. The BLAST hits concurred with the original Arabidopsis TBLASTN searches performed to identify putative poplar orthologs. SxMAX1B retrieved POPTR_0006s24320.1 and its homeolog POPTR_0018s07540, and SxMAX4B retrieved POPTR_0006s25490.1 and its homeolog POPTR_0018s08010. SxMAX2A BLAST results included a single candidate ortholog, POPTR_14s13910, and two more distant, nonhomeologous homologs, POPTR_0011s07320 and POPTR_0002s21260. SxMAX1C, an allele of Salix spp. genotype R13, was included in the MAX1 tree for the purpose of constructing an unrooted tree using the neighbor-joining tree-build method. Coding sequences were aligned using the MUSCLE algorithm within Geneious Pro 5.5.6 software set to default settings. These alignments were used to generate unrooted trees using Geneious Tree Builder set to default settings (genetic distance model, Jukes-Cantor; tree-build method, neighbor joining; bootstrap resampling, 100 replicates) also available in Geneious Pro 5.5.6.
Generation of Transgenic Plants
The SxMAX alleles were transferred from pENTR/D-TOPO to the Gateway-compatible binary destination vector pK7WG2 (Karimi et al., 2002) using LR Clonase II Enzyme Mix (Invitrogen) and transformed into the corresponding Arabidopsis max mutant background via Agrobacterium tumefaciens strain GV3101 using the floral dip method (Clough and Bent, 1998). Transformants were selected on agar-solidified ATS medium containing 50 μg mL−1 kanamycin (Sigma). For each construct, at least 10 independent single insertion lines were taken to homozygosity.
Shoot-Branching Assay
To quantify branching, a decapitation assay was used (Greb et al., 2003), in which the total number of rosette branches produced following decapitation was scored.
Cotyledon Senescence Assays
Seeds were sown in 8-cm pots and then stratified at 4°C for 7 d before being transferred to the growth room for a further 7 d. The cotyledons were then carefully excised, and six tubes of 40 cotyledons each were collected for each genotype. Three of the tubes were flash frozen in liquid nitrogen before storage at −80°C. Senescence was induced in the other three tubes by floating the cotyledons on 3 mm MES buffer (pH 5.8) at 21°C for 8 d in the dark (Woo et al., 2001).
Chlorophyll was extracted by boiling the cotyledon samples in 1 mL of 96% ethanol at 80°C for 4 h. Chlorophyll content was calculated as described by Lichtenthaler and Wellburn (1983) using absorbance measurements taken at wavelengths of 665 and 649. The percentage of starting chlorophyll content remaining after senescence was calculated as the mean chlorophyll a + b in senesced leaves divided by the mean chlorophyll a + b in unsenesced leaves, then expressed as a percentage. The mean and se of at least three separate experiments were calculated.
Gene Expression
Using the Qiagen RNeasy plant mini kit (Qiagen), total RNA was isolated from duplicate 7-d-old Arabidopsis seedlings and cDNA synthesized from 500 ng of RNA using SuperScript II (Invitrogen) as per the manufacturer’s instructions. This was then diluted 20-fold and used in quantitative PCR in an Illumina Lightcycler with SYBR Green for detection. Reactions were set up in triplicate, using wild-type Columbia and the corresponding max mutant as controls, and the cycle threshold values were calculated from the accompanying software. The obtained values were calculated against those of UBQ5, which was used as an internal standard. Fold difference was calculated from the means of the cycle threshold values from the triplicate assays. Salix spp. MAX4 primers used were SxMAX4F (5′-CCGGCACCTTTTCGATGGCTATG-3′) and SxMAX4R (5′-GCCGCCTTGTAAGCCTCCGATT-3′). Salix spp. MAX2 primers used were SxMAX2F (5′-CCTGTTTCAGATCGGTGACTC-3′) and SxMAX2R (5′-GTACCCATCTTCAGGTTC-3′).
The following sequences have been deposited into GenBank (http://www.ncbi.nlm.nih.gov): SxMAX1B, accession number JN613462; SxMAX2A, JN613463; SxMAX2B, JN613464; SxMAX4B, JN613465; SxMAX4E, JX391953; and SxMAX4G, JN613468.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Juvenile S. viminalis ‘Bowles Hybrid’ nodes have both a central bud and two flanking buds.
Supplemental Figure S2. Unrooted trees with consensus support (%) labels, illustrating the homology between Salix spp. alleles of MAX genes MAX1, MAX2, and MAX4 and poplar homologs.
Supplemental Figure S3. Quantitative reverse transcription PCR analysis of transgene expression levels in SxMAX4 alleles B, E, and G.
Supplemental Figure S4. MUSCLE alignments of Salix spp., poplar, and Arabidopsis MAX1 and MAX2 sequences.
Supplemental Figure S5. Expression and bud outgrowth analysis of SxMAX2 lines.
Supplemental Table S1. Map positions of the Salix spp. MAX genes.
Acknowledgments
We thank Rachel Williams who worked on the SxMAX2 senescence assays as part of her undergraduate project, Harry Whitwell who produced the scanning electron micrographs of developing willow buds during his undergraduate project, and the University of York horticulture staff for their expertise in looking after our plants.
Glossary
- SRC
short-rotation coppice
- PATS
polar auxin transport stream
- SL
strigolactone derivative
- NAA
1-naphthaleneacetic acid
- QTL
quantitative trait locus
- ATS
Arabidopsis Salt
References
- Aguilar-Martínez JA, Poza-Carrión C, Cubas P. (2007) Arabidopsis BRANCHED1 acts as an integrator of branching signals within axillary buds. Plant Cell 19: 458–472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Åhman I, Larsson S. (1994) Genetic improvement of willow (Salix) as a source of bioenergy. Nor J Agric Sci (Suppl) 18: 47–56 [Google Scholar]
- Arite T, Iwata H, Ohshima K, Maekawa M, Nakajima M, Kojima M, Sakakibara H, Kyozuka J. (2007) DWARF10, an RMS1/MAX4/DAD1 ortholog, controls lateral bud outgrowth in rice. Plant J 51: 1019–1029 [DOI] [PubMed] [Google Scholar]
- Balla J, Kalousek P, Reinöhl V, Friml J, Procházka S. (2011) Competitive canalization of PIN-dependent auxin flow from axillary buds controls pea bud outgrowth. Plant J 65: 571–577 [DOI] [PubMed] [Google Scholar]
- Bennett T, Sieberer T, Willett B, Booker J, Luschnig C, Leyser O. (2006) The Arabidopsis MAX pathway controls shoot branching by regulating auxin transport. Curr Biol 16: 553–563 [DOI] [PubMed] [Google Scholar]
- Beveridge CA, Symons GM, Murfet IC, Ross JJ, Rameau C. (1997) The rms1 mutant of pea has elevated indole-3-acetic acid levels and reduced root-sap zeatin riboside content but increased branching controlled by graft-transmissable signal(s). Plant Physiol 115: 1251–1258 [Google Scholar]
- Booker J, Auldridge M, Wills S, McCarty D, Klee H, Leyser O. (2004) MAX3/CCD7 is a carotenoid cleavage dioxygenase required for the synthesis of a novel plant signaling molecule. Curr Biol 14: 1232–1238 [DOI] [PubMed] [Google Scholar]
- Booker J, Chatfield S, Leyser O. (2003) Auxin acts in xylem-associated or medullary cells to mediate apical dominance. Plant Cell 15: 495–507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booker J, Sieberer T, Wright W, Williamson L, Willett B, Stirnberg P, Turnbull C, Srinivasan M, Goddard P, Leyser O. (2005) MAX1 encodes a cytochrome P450 family member that acts downstream of MAX3/4 to produce a carotenoid-derived branch-inhibiting hormone. Dev Cell 8: 443–449 [DOI] [PubMed] [Google Scholar]
- Braun N, de Saint Germain A, Pillot J-P, Boutet-Mercey S, Dalmais M, Antoniadi I, Li X, Maia-Grondard A, Le Signor C, Bouteiller N, et al. (2012) The pea TCP transcription factor PsBRC1 acts downstream of strigolactones to control shoot branching. Plant Physiol 158: 225–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brewer PB, Dun EA, Ferguson BJ, Rameau C, Beveridge CA. (2009) Strigolactone acts downstream of auxin to regulate bud outgrowth in pea and Arabidopsis. Plant Physiol 150: 482–493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunkener L (1984) Gross Morphology and Anatomy of Current Shoots of Salix. Energy Forestry Project Report 34. Swedish University of Agricultural Science, Uppsala, Sweden
- Brunkener L (1988) Gross Morphological and Anatomical Aspects of Shoot Growth in Salix. Energy Forestry Project Report 45. Swedish University of Agricultural Science, Uppsala, Sweden
- Cannell MGR, Milne R, Sheppard LJ, Unsworth MH. (1987) Radiation interception and productivity of willow. J Appl Ecol 24: 261–278 [Google Scholar]
- Chang S, Puryear J, Cairney J. (1993) A simple and efficient method for extracting RNA from pine trees. Plant Mol Biol Rep 11: 113–116 [Google Scholar]
- Chatfield SP, Stirnberg P, Forde BG, Leyser O. (2000) The hormonal regulation of axillary bud growth in Arabidopsis. Plant J 24: 159–169 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Crawford S, Shinohara N, Sieberer T, Williamson L, George G, Hepworth J, Müller D, Domagalska MA, Leyser O. (2010) Strigolactones enhance competition between shoot branches by dampening auxin transport. Development 137: 2905–2913 [DOI] [PubMed] [Google Scholar]
- Delaux P-M, Xie X, Timme RE, Puech-Pages V, Dunand C, Lecompte E, Delwiche CF, Yoneyama K, Bécard G, Séjalon-Delmas N. (2012) Origin of strigolactones in the green lineage. New Phytol 195: 857–871 [DOI] [PubMed] [Google Scholar]
- Domagalska MA, Leyser O. (2011) Signal integration in the control of shoot branching. Nat Rev Mol Cell Biol 12: 211–221 [DOI] [PubMed] [Google Scholar]
- Drummond AJ, Ashton B, Buxton S, Cheung M, Cooper A, Duran C, Field M, Heled J, Kearse M, Markowitz S, et al (2011) Geneious version 5.4. http://www.geneious.com/ (June 19, 2012)
- Dun EA, de Saint Germain A, Rameau C, Beveridge CA. (2012) Antagonistic action of strigolactone and cytokinin in bud outgrowth control. Plant Physiol 158: 487–498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Roldan V, Fermas S, Brewer PB, Puech-Pagès V, Dun EA, Pillot JP, Letisse F, Matusova R, Danoun S, Portais JC, et al. (2008) Strigolactone inhibition of shoot branching. Nature 455: 189–194 [DOI] [PubMed] [Google Scholar]
- Grbić B, Bleecker AB. (1996) An altered body plan is conferred on Arabidopsis plants carrying dominant alleles of two genes. Development 122: 2395–2403 [DOI] [PubMed] [Google Scholar]
- Greb T, Clarenz O, Schafer E, Muller D, Herrero R, Schmitz G, Theres K. (2003) Molecular analysis of the LATERAL SUPPRESSOR gene in Arabidopsis reveals a conserved control mechanism for axillary meristem formation. Genes Dev 17: 1175–1187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gullberg U. (1993) Towards making willows pilot species for coppicing production. For Chron 69: 721–726 [Google Scholar]
- Hanley SJ (2003) Genetic mapping of important agronomic traits in biomass willow. PhD thesis. University of Bristol, Bristol, UK [Google Scholar]
- Hanley SJ, Mallott MD, Karp A. (2006) Alignment of a Salix linkage map to the Populus genomic sequence reveals macrosynteny between willow and poplar genomes. Tree Genet Genomes 3: 35–48 [Google Scholar]
- Hayward A, Stirnberg P, Beveridge C, Leyser O. (2009) Interactions between auxin and strigolactone in shoot branching control. Plant Physiol 151: 400–412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hempel FD, Feldmann LJ. (1994) Bi-directional inflorescence development in Arabidopsis thaliana: acropetal initiation of flowers and basipetal initiation of paraclades. Planta 192: 276–286 [Google Scholar]
- Ishikawa S, Maekawa M, Arite T, Onishi K, Takamure I, Kyozuka J. (2005) Suppression of tiller bud activity in tillering dwarf mutants of rice. Plant Cell Physiol 46: 79–86 [DOI] [PubMed] [Google Scholar]
- Johnson X, Brcich T, Dun EA, Goussot M, Haurogné K, Beveridge CA, Rameau C. (2006) Branching genes are conserved across species: genes controlling a novel signal in pea are coregulated by other long-distance signals. Plant Physiol 142: 1014–1026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi M, Inzé D, Depicker A. (2002) Gateway vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci 7: 193–195 [DOI] [PubMed] [Google Scholar]
- Karp A, Hanley SJ, Trybush SO, Macalpine W, Pei M, Shield I. (2011) Genetic improvement of willow for bioenergy and biofuels. J Integr Plant Biol 53: 151–165 [DOI] [PubMed] [Google Scholar]
- Keoleian GA, Volk TA. (2005) Renewable energy from willow biomass crops: life cycle energy, environmental and economic performance. Crit Rev Plant Sci 24: 385–406 [Google Scholar]
- Kohlen W, Charnikhova T, Qing L, Bours R, Domagalska MA, Beguerie S, Verstappen F, Leyser O, Bouwmeester H, Ruyter-Spira C. (2011) Strigolactones are transported through the xylem and play a key role in shoot architectural response to phosphate deficiency in nonarbuscular mycorrhizal host Arabidopsis thaliana. Plant Physiol 155: 679–686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazar G, Goodman HM. (2006) MAX1, a regulator of the flavonoid pathway, controls vegetative axillary bud outgrowth in Arabidopsis. Proc Natl Acad Sci USA 103: 472–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leyser O. (2009) The control of shoot branching: an example of plant information processing. Plant Cell Environ 32: 694–703 [DOI] [PubMed] [Google Scholar]
- Li C, Bangerth F. (1999) Autoinhibition of indoleacetic acid transport in the shoot of two-branched pea (Pisum sativum) plants and its relationship to correlative dominance. Physiol Plant 106: 415–420 [Google Scholar]
- Li CJ, Herrera GJ, Bangerth F. (1995) Effect of apex excision and replacement by 1-naphthylacetic acid on cytokinin concentration and apical dominance in pea plants. Physiol Plant 94: 465–469 [Google Scholar]
- Liang J, Zhao L, Challis R, Leyser O. (2010) Strigolactone regulation of shoot branching in chrysanthemum (Dendranthema grandiflorum). J Exp Bot 61: 3069–3078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichtenthaler HK, Wellburn AR (1983) Determinations of total carotenoids and chlorophylls a and b of leaf extracts in different solvents. Biochem Soc Trans 11: 591–592 [Google Scholar]
- Lin H, Wang R, Qian Q, Yan M, Meng X, Fu Z, Yan C, Jiang B, Su Z, Li J, et al. (2009) DWARF27, an iron-containing protein required for the biosynthesis of strigolactones, regulates rice tiller bud outgrowth. Plant Cell 21: 1512–1525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindegaard KN, Barker JHA. (1997) Breeding willows for biomass. Asp Appl Biol 49: 155–162 [Google Scholar]
- Ljung K, Bhalerao RP, Sandberg G. (2001) Sites and homeostatic control of auxin biosynthesis in Arabidopsis during vegetative growth. Plant J 28: 465–474 [DOI] [PubMed] [Google Scholar]
- Marhavý P, Bielach A, Abas L, Abuzeineh A, Duclercq J, Tanaka H, Pařezová M, Petrášek J, Friml J, Kleine-Vehn J, et al. (2011) Cytokinin modulates endocytic trafficking of PIN1 auxin efflux carrier to control plant organogenesis. Dev Cell 21: 796–804 [DOI] [PubMed] [Google Scholar]
- Messing SAJ, Gabelli SB, Echeverria I, Vogel JT, Guan JC, Tan BC, Klee HJ, McCarty DR, Amzel LM. (2010) Structural insights into maize viviparous14, a key enzyme in the biosynthesis of the phytohormone abscisic acid. Plant Cell 22: 2970–2980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris DA. (1977) Transport of exogenous auxin in two-branched pea seedlings (Pisum sativum L.). Planta 136: 91–96 [DOI] [PubMed] [Google Scholar]
- Morris SE, Turnbull CG, Murfet IC, Beveridge CA. (2001) Mutational analysis of branching in pea: evidence that Rms1 and Rms5 regulate the same novel signal. Plant Physiol 126: 1205–1213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller D, Leyser O. (2011) Auxin, cytokinin and the control of shoot branching. Ann Bot (Lond) 107: 1203–1212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Napoli C. (1996) Highly branched phenotype of the petunia dad1-1 mutant is reversed by grafting. Plant Physiol 111: 27–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson DC, Scaffidi A, Dun EA, Waters MT, Flematti GR, Dixon KW, Beveridge CA, Ghisalberti EL, Smith SM. (2011) F-box protein MAX2 has dual roles in karrikin and strigolactone signaling in Arabidopsis thaliana. Proc Natl Acad Sci USA 108: 8897–8902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordström A, Tarkowski P, Tarkowska D, Norbaek R, Åstot C, Dolezal K, Sandberg G. (2004) Auxin regulation of cytokinin biosynthesis in Arabidopsis thaliana: a factor of potential importance for auxin-cytokinin-regulated development. Proc Natl Acad Sci USA 101: 8039–8044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ongaro V, Bainbridge K, Williamson L, Leyser O. (2008) Interactions between axillary branches of Arabidopsis. Mol Plant 1: 388–400 [DOI] [PubMed] [Google Scholar]
- Paukkonen K, Kauppi A, Ferm A. (1992) Origin, structure and shoot-formation ability of buds in cutting-origin stools of Salix ‘Aquatica’. Flora 186: 53–65 [Google Scholar]
- Prusinkiewicz P, Crawford S, Smith RS, Ljung K, Bennett T, Ongaro V, Leyser O. (2009) Control of bud activation by an auxin transport switch. Proc Natl Acad Sci USA 106: 17431–17436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachs T. (1981) The control of patterned differentiation of vascular tissues. Adv Bot Res 9: 151–162 [Google Scholar]
- Sennerby-Forsse L, Ferm A, Kauppi A (1992) Coppicing ability and sustainability. In CP Mitchell, JB Ford-Robertson, T Hinckley, L Sennerby-Forsse, eds, Ecophysiology of Short Forest Crops. Elsevier Applied Science, London, pp 146–184 [Google Scholar]
- Sennerby-Forsse L, Zsuffa L. (1995) Bud structure and resprouting in coppiced stools of Salix viminalis L., S. eriocephala Michx., and S. amygdaloides Anders. Trees Struct Funct 9: 224–234 [Google Scholar]
- Shen H, Luong P, Huq E. (2007) The F-box protein MAX2 functions as a positive regulator of photomorphogenesis in Arabidopsis. Plant Physiol 145: 1471–1483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinohara N, Taylor C, Leyser O. (2013) Strigolactone can promote or inhibit shoot branching by triggering rapid depletion of the auxin efflux protein PIN1 from the plasma membrane. PLoS Biol 11: e1001474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons JL, Napoli CA, Janssen BJ, Plummer KM, Snowden KC. (2007) Analysis of the DECREASED APICAL DOMINANCE genes of petunia in the control of axillary branching. Plant Physiol 143: 697–706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snowden KC, Simkin AJ, Janssen BJ, Templeton KR, Loucas HM, Simons JL, Karunairetnam S, Gleave AP, Clark DG, Klee HJ. (2005) The Decreased apical dominance1/Petunia hybrida CAROTENOID CLEAVAGE DIOXYGENASE8 gene affects branch production and plays a role in leaf senescence, root growth, and flower development. Plant Cell 17: 746–759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorefan K, Booker J, Haurogné K, Goussot M, Bainbridge K, Foo E, Chatfield S, Ward S, Beveridge C, Rameau C, et al. (2003) MAX4 and RMS1 are orthologous dioxygenase-like genes that regulate shoot branching in Arabidopsis and pea. Genes Dev 17: 1469–1474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stirnberg P, Furner IJ, Ottoline Leyser HM. (2007) MAX2 participates in an SCF complex which acts locally at the node to suppress shoot branching. Plant J 50: 80–94 [DOI] [PubMed] [Google Scholar]
- Stirnberg P, van De Sande K, Leyser HM. (2002) MAX1 and MAX2 control shoot lateral branching in Arabidopsis. Development 129: 1131–1141 [DOI] [PubMed] [Google Scholar]
- Tanaka M, Takei K, Kojima M, Sakakibara H, Mori H. (2006) Auxin controls local cytokinin biosynthesis in the nodal stem in apical dominance. Plant J 45: 1028–1036 [DOI] [PubMed] [Google Scholar]
- Tharakan PJ, Volk TA, Nowak CA, Abrahamson LP. (2005) Morphological traits of 30 willow clones and their relationship to biomass production. Can J For Res 35: 421–431 [Google Scholar]
- Thimann KV, Skoog F. (1933) Studies on the growth hormone of plants. III. The inhibiting action of the growth substance on bud development. Proc Natl Acad Sci USA 19: 714–716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsarouhas V, Gullberg U, Lagercrantz U. (2002) An AFLP and RFLP linkage map and quantitative trait locus (QTL) analysis of growth traits in Salix. Theor Appl Genet 105: 277–288 [DOI] [PubMed] [Google Scholar]
- Turnbull CG, Booker JP, Leyser HM. (2002) Micrografting techniques for testing long-distance signalling in Arabidopsis. Plant J 32: 255–262 [DOI] [PubMed] [Google Scholar]
- Tuskan GA, Difazio S, Jansson S, Bohlmann J, Grigoriev I, Hellsten U, Putnam N, Ralph S, Rombauts S, Salamov A, et al. (2006) The genome of black cottonwood, Populus trichocarpa (Torr. & Gray). Science 313: 1596–1604 [DOI] [PubMed] [Google Scholar]
- Umehara M, Hanada A, Yoshida S, Akiyama K, Arite T, Takeda-Kamiya N, Magome H, Kamiya Y, Shirasu K, Yoneyama K, et al. (2008) Inhibition of shoot branching by new terpenoid plant hormones. Nature 455: 195–200 [DOI] [PubMed] [Google Scholar]
- Wilson AK, Pickett FB, Turner JC, Estelle M. (1990) A dominant mutation in Arabidopsis confers resistance to auxin, ethylene and abscisic acid. Mol Gen Genet 222: 377–383 [DOI] [PubMed] [Google Scholar]
- Woo HR, Chung KM, Park JH, Oh SA, Ahn T, Hong SH, Jang SK, Nam HG. (2001) ORE9, an F-box protein that regulates leaf senescence in Arabidopsis. Plant Cell 13: 1779–1790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou J, Zhang S, Zhang W, Li G, Chen Z, Zhai W, Zhao X, Pan X, Xie Q, Zhu L. (2006) The rice HIGH-TILLERING DWARF1 encoding an ortholog of Arabidopsis MAX3 is required for negative regulation of the outgrowth of axillary buds. Plant J 48: 687–698 [DOI] [PubMed] [Google Scholar]
- Zsuffa L. (1990) Genetic improvement of willows for energy plantations. Biomass 22: 35–47 [Google Scholar]