EMF1 and ULT1 counteract each other in the regulation of flowering and stress tolerance.
Abstract
Epigenetic regulation of gene expression is of fundamental importance for eukaryotic development. EMBRYONIC FLOWER1 (EMF1) is a plant-specific gene that participates in Polycomb group-mediated transcriptional repression of target genes such as the flower MADS box genes AGAMOUS, APETALA3, and PISTILLATA. Here, we investigated the molecular mechanism underlying the curly leaf and early flowering phenotypes caused by reducing EMF1 activity in the leaf primordia of LFYasEMF1 transgenic plants and propose a combined effect of multiple flower MADS box gene activities on these phenotypes. ULTRAPETALA1 (ULT1) functions as a trithorax group factor that counteracts Polycomb group action in Arabidopsis (Arabidopsis thaliana). Removing ULT1 activity rescues both the abnormal developmental phenotypes and most of the misregulated gene expression of LFYasEMF1 plants. Reducing EMF1 activity increases salt tolerance, an effect that is diminished by introducing the ult1-3 mutation into the LFYasEMF1 background. EMF1 is required for trimethylating lysine-27 on histone 3 (H3K27me3), and ULT1 associates with ARABIDOPSIS TRITHORAX1 (ATX1) for trimethylating lysine-3 on histone 4 (H3K4me3) at flower MADS box gene loci. Reducing EMF1 activity decreases H3K27me3 marks and increases H3K4me3 marks on target gene loci. Removing ULT1 activity has the opposite effect on the two histone marks. Removing both gene activities restores the active and repressive marks to near wild-type levels. Thus, ULT1 acts as an antirepressor that counteracts EMF1 action through modulation of histone marks on target genes. Our analysis indicates that, instead of acting as off and on switches, EMF1 and ULT1 mediate histone mark deposition and modulate transcriptional activities of the target genes.
The life cycle of eukaryotes is marked by developmental phases specified by spatial and temporal patterns of gene expression. Polycomb group (PcG) and trithorax group (trxG) proteins function as epigenetic repressors and activators, respectively. They act dynamically in regulating gene expression patterns to specify cell fates and maintain differentiated cell states (Schuettengruber et al., 2007; Hennig and Derkacheva, 2009; Holec and Berger, 2012).
PcG genes were first discovered in Drosophila melanogaster as repressors of the homeotic Hox genes involved in embryo segmentation (Lewis, 1978; Jurgens, 1985). The two D. melanogaster PcG protein complexes, Polycomb Repressive Complex1 (PRC1) and PRC2, the components of which are highly conserved among eukaryotic organisms, cooperate to maintain silencing through trimethylation of histone 3 Lys-27 (H3K27me3) of their target genes (Schwartz and Pirrotta, 2007). D. melanogaster trxG proteins were identified as suppressors of PcG mutations that cause trimethylation of histone 3 Lys-4 (H3K4me3) and activate gene expression (Poux et al., 2002; Klymenko and Müller, 2004; Schwartz and Pirrotta, 2007). Subtle regulation of histone methylation homeostasis by PcG and trxG on target genes determines gene expression status; however, the molecular mechanisms that translate histone modification patterns to specific transcriptional states remain unclear (Poux et al., 2002; Klymenko and Müller, 2004).
In Arabidopsis (Arabidopsis thaliana), proteins homologous to core components of D. melanogaster PRC2 are required for three developmental functions: FERTILIZATION INDEPENDENT SEED2-harboring PRC2 regulates seed development (Chaudhury et al., 2001; Köhler et al., 2003); VERNALIZATION2 (VRN2)-PRC2 is involved in vernalization-mediated flowering via the regulation of FLOWERING LOCUS C (FLC; Gendall et al., 2001; Wood et al., 2006; De Lucia et al., 2008); and EMBRYONIC FLOWER2 (EMF2)-PRC2 is required to maintain the vegetative phase by repressing the expression of the flower homeotic MADS box genes AGAMOUS (AG), APETALA3 (AP3), and PISTILLATA (PI; Kinoshita et al., 2001; Yoshida et al., 2001; Chanvivattana et al., 2004). Plants impaired in EMF2-PRC2 complex members display pleiotropic phenotypes, including early flowering, curly leaves, terminal flower, and abnormal flower organs, caused by the widespread misexpression of multiple flower homeotic genes (Sánchez et al., 2009). In Arabidopsis, the PRC1 RING-finger homologs, AtRING1A/B and AtBMI1A/B, are essential for maintaining cell identity (Sanchez-Pulido et al., 2008; Xu and Shen, 2008; Bratzel et al., 2010). Several other proteins are involved in AtPRC1 functions, namely LIKE HETEROCHROMATIN PROTEIN1/TERMINAL FLOWER2 (TFL2), VRN1, and EMF1 (Mylne et al., 2006; Turck et al., 2007; Zhang et al., 2007; Calonje et al., 2008). EMF1 binds four RING-finger proteins and is required for H2A Lys-119 monoubiquitination activity on select EMF1 target genes (Bratzel et al., 2010).
EMF1 is a plant-specific protein that encodes a transcriptional regulator (Aubert et al., 2001; Calonje et al., 2008). Although EMF1 shares no sequence homology with animal PcG protein genes, it is involved in plant PcG-mediated gene repression and targets flower homeotic genes directly, as does EMF2 (Kim et al., 2010). Mutations in EMF1 and EMF2 cause plants to skip the vegetative phase, transforming the embryonic meristem from an indeterminate to a determinate state and producing a terminal flower (Sung et al., 1992; Yang et al., 1995; Chen et al., 1997; Yoshida et al., 2001). Global gene expression analysis revealed that EMF1 regulates a remarkably high number of genes involved in transcription factor activity, developmental pathways, microRNA gene silencing, and stress responses in Arabidopsis (Kim et al., 2010). Tissue-specific removal of EMF1 activity from leaf primordia in transgenic plants expressing antisenseEMF1 under the control of the LEAFY (LFY) promoter (LFYasEMF1) permits vegetative growth but leads to early flowering plants with upward-curled leaves (Sánchez et al., 2009) similar to those of curly leaf (clf), a PcG mutant (Goodrich et al., 1997). Genome-wide investigation of EMF1 targets has revealed that EMF1 mediates gene expression via diverse mechanisms. It acts along with PRC2 to repress the flower MADS box genes as well as with PRC1 to regulate the expression of many other genes (Kim et al., 2012).
The Arabidopsis genome encodes nine proteins related to the D. melanogaster TRITHORAX proteins (Alvarez-Venegas and Avramova, 2001; Baumbusch et al., 2001; Ng et al., 2007). Among these, the Arabidopsis homolog of trxG, ARABIDOPSIS TRITHORAX1 (ATX1), harbors a SET domain with weak methylating activity in vitro (Alvarez-Venegas et al., 2003). ATX1 and ARABIDOPSIS TRITHORAX-RELATED7 are required for expression of the MADS box genes involved in flower organogenesis and of the floral repressor FLC (Tamada et al., 2009). atx1 mutants have minor defects in leaf and flower organs (Pien et al., 2008). The ULTRAPETALA1 (ULT1) gene encodes a protein containing a putative DNA-binding SAND (for Sp100, AIRE-1, NucP41/75, DEAF-1) domain and a B box-like motif that may mediate protein-protein interactions (Carles et al., 2005). Although ULT1 has no homology with animal trxG components, ult1 mutations suppress all of the phenotypes caused by mutations in CLF, indicating that ULT1 meets the genetic definition of a trxG gene (Carles and Fletcher, 2009). In addition, ULT1 physically interacts with ATX1 and acts as an antirepressor to CLF-mediated gene repression in Arabidopsis by limiting the deposition of repressive H3K27me3 marks on target gene chromatin (Carles and Fletcher, 2009).
In this study, we investigated the impact of EMF1 in every stage of Arabidopsis development through a detailed characterization of the LFYasEMF1 defects. Comparing the gene expression pattern of LFYasEMF1 and plants with similar phenotypes led to the conclusion that it is the combined transcriptional activity from multiple flower MADS box genes, rather than the ectopic expression of a single MADS box gene, that regulates flowering time and leaf differentiation in these plants. Our investigation of the effect of ULT1 on EMF1-mediated gene repression in Arabidopsis showed that ULT1 functions as an antirepressor that counteracts EMF1-mediated repression. Furthermore, our analysis revealed a close link between transcriptional activity and differences in the active and repressive histone marks on the target genes. We propose that, in addition to maintaining off/on states, these histone marks play a role in modulating transcriptional activity.
RESULTS
The Phenotypes of ult1-3 emf1-2 Double Mutants Are emf1 Like
Plants impaired in EMF1 activity (e.g. emf1-2 mutants) flower upon germination (Supplemental Fig. S1A). Plants that lack ULT1 function, such as ult1-3 null mutants, flower late and produce extra floral organs, predominantly sepals and petals, hence the name ULTRAPETALA (Fletcher, 2001). ult1 mutations rescue the clf phenotypes (Carles and Fletcher, 2009). Because EMF1 shows a close functional relationship to EMF2, which forms a putative PRC2 with CLF, we investigated whether ULT1 could counteract EMF1 gene action. emf1-2 ult1-3 double mutants were generated as described in “Materials and Methods” to determine whether ult1 can rescue the emf1 phenotype (Supplemental Fig. S1, B and C). ult1-3 seedlings resemble those of the wild type (Supplemental Fig. S1A), whereas emf1-2 ult1-3 plants look liked the emf1-2 plants, producing oval-shaped, petioleless cotyledons that later become carpelloid, extremely short hypocotyls, and no leaf primordia (Supplemental Fig. S1A; Chen et al., 1997; Moon et al., 2003). Since the emf1-2 ult1-3 double mutants are similar to emf1-2 single mutants, we concluded that the ult1-3 mutation cannot rescue the emf1-2 phenotypes.
The ult1 Mutation Rescued LFYasEMF1 Phenotypes
The ult1 mutant has no visible phenotype in the seedling and early vegetative phases (Fletcher, 2001; Carles et al., 2004). Hence, we reasoned that ULT1 probably does not play a major role in development until the adult phase of Arabidopsis. Therefore, ult1 cannot counteract the loss of EMF1 function during the seedling growth phase. However, ult1 mutations delay flowering and can restore the curly leaf and early flowering phenotypes of the clf mutant (Chanvivattana et al., 2004; Carles and Fletcher, 2009). We have previously generated transgenic LFYasEMF1 plants (Sánchez et al., 2009). The LFY promoter is known to be active in leaf primordia by 4 d after germination (DAG) and is gradually up-regulated during vegetative development, peaking in the floral meristem (Blázquez et al., 1997). Similar to clf mutants (Goodrich et al., 1997), these plants showed curly leaves and early flowering phenotypes in both long-day (LD) and short-day (SD) conditions, implying a functional relationship between EMF1 and CLF.
To study whether ult1 mutations can rescue the LFYasEMF1 phenotypes, we crossed homozygous ult1-3 plants with homozygous LFYasEMF1 transgenic plants and selected F2 plants homozygous for LFYasEMF1 (see “Materials and Methods”). We then genotyped these plants and their progeny to identify those homozygous for the ult1-3 allele (Supplemental Fig. S1, B and C) and characterized plants homozygous for both LFYasEMF1 and ult1-3, as described below.
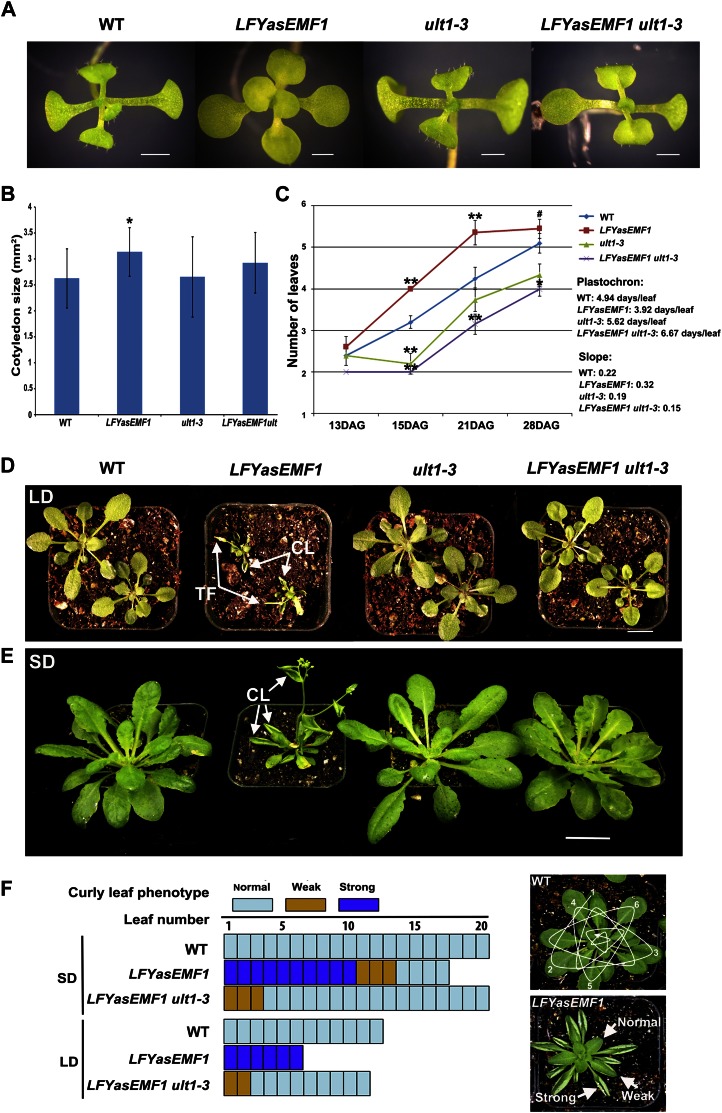
In early development, the cotyledons of LFYasEMF1 plants had shorter petioles and larger blades than wild-type cotyledons (Fig. 1, A and B). The blades were round but did not curl downward like wild-type cotyledon blades (Fig. 1A). Introduction of the ult1-3 mutation into the LFYasEMF1 plants rescued the altered cotyledon growth patterns (Fig. 1, A and B). LFYasEMF1 plants produced rosette leaves at a faster pace or displayed heterochronic changes when grown under SD conditions (Fig. 1C). Wild-type plants produced a new leaf every 4.9 d, ult1-3 plants every 5.6 d, LFYasEMF1 plants every 3.9 d, and LFYasEMF1 ult1-3 plants every 6.7 d. The double mutant leaf production rate is closer to the wild type and ult1-3 than to LFYasEMF1. These results showed that EMF1 and ULT1 act in an opposite manner on the rate of leaf production, with EMF1 impeding it and ULT1 promoting it. The ult1 mutation also seems to be epistatic to LFYasEMF1, indicating that normal ULT1 activity is required to condition the LFYasEMF1 phenotype.
Figure 1.
ult1-3 restores seedling and curly leaf phenotypes of LFYasEMF1. A, Three-week-old wild-type (WT), LFYasEMF1, ult1-3, and LFYasEMF1 ult1-3 plants grown on agar plates under SD conditions. The LFYasEMF1 cotyledons are flat and round, while those of the other three genotypes are slightly downward curled. B, Adaxial surface areas of the cotyledons of the four genotypes grown on agar plates under SD conditions. Significant difference from the wild type (Student’s t test) is marked with the asterisk (P < 0.05). C, Plastochron of the four plants grown on agar plates under SD conditions. The plastochron was calculated based on the slope of the number of days per leaf produced from 13 through 21 DAG. Significant differences from the wild type (Student’s t test) are marked with asterisks (* P < 0.05, **P < 0.01). The number sign indicates that the plants have bolted. D and E, Phenotypes of soil-grown LFYasEMF1 plants rescued by the ult1-3 mutation. Plants shown are 4 weeks old grown in LD (D) or 8 weeks old grown in SD in a greenhouse (E) after 2 weeks of growth on agar plates under SD conditions. CL, Curly leaf; TF, terminal flower. F, Curly leaf phenotype of LFYasEMF1 and LFYasEMF1 ult1-3 plants grown under LD and SD conditions. Blue areas, normal, not curly; brown areas, weak, mild curliness; purple areas, strong curliness. The top panel at right shows the strategy for counting leaf number from the bottom up, and the bottom panel at right shows variations in the extent of curliness. Bars = 1 mm (A), 1 cm (D), and 2 cm (E).
Later in development, upward-curling leaves is the most prominent phenotype when EMF1 activity is reduced in the leaf primordia of LFYasEMF1 plants. Soil-grown LFYasEMF1 plants produced small rosette leaves that were curled upward along the proximal distal axis, whereas wild-type and ult1-3 leaves had a broad and flat lamina (Fig. 1, D and E). The increasing curvature as the leaf enlarged was classified as strong, weak, or normal. The oldest 10 leaves had the most pronounced curvature with leaf margins curled upward, and the 11th to 15th leaves were weakly curled (Fig. 1F). Newly emerging LFYasEMF1 leaves were flat and similar to wild-type leaves but curled upward as the leaf expanded and matured. Introduction of the ult1 mutation rescued the LFYasEMF1 curly rosette leaf phenotypes (Fig. 1, D–F). As shown in Figure 1F, only the first three largest leaves in LFYasEMF1 ult1-3 plants displayed mild curliness, demonstrating a nearly full rescue of the LFYasEMF1 curly leaf phenotype by the ult1-3 mutation. We likewise observed that the upward-curled cauline leaves of LFYasEMF1 plants (Fig. 1E) were rescued by the introduction of the ult-3 mutation.
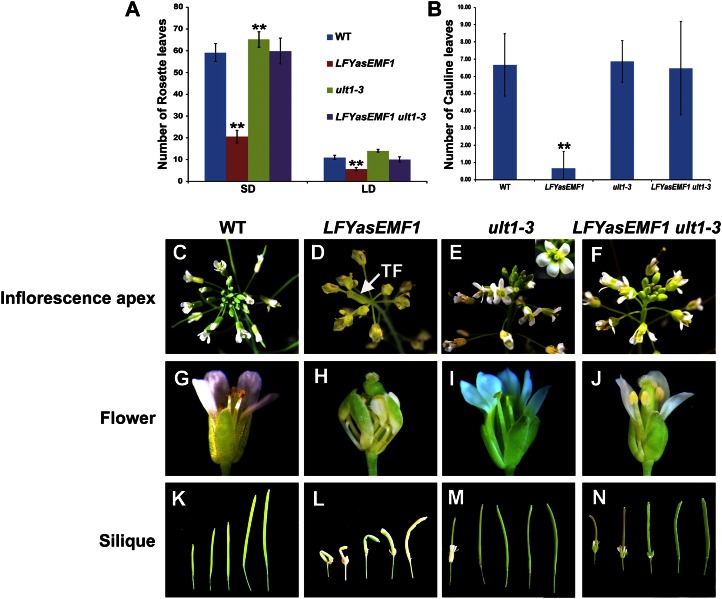
The vegetative-to-reproductive transition takes place prematurely in LFYasEMF1 plants (Sánchez et al., 2009). These plants produced fewer rosette leaves than wild-type plants (LFYasEMF1, 20.6 ± 2.8 at SD and 5.7 ± 0.8 at LD; wild type, 59.1 ± 4.1 at SD and 11 ± 1 at LD; Fig. 2A). ult1-3 mutants flowered late (Carles et al., 2005) and produced more rosette leaves than wild-type plants (ult1-3, 65.2 ± 3.5 at SD and 14.1 ± 0.7 at LD; Fig. 2A). Introduction of ult1-3 into LFYasEMF1 restored rosette leaf numbers to nearly those of wild-type plants (LFYasEMF1 ult1-3, 59.9 ± 6 at SD and 10.1 ± 1.1 at LD; Fig. 2A). The early flowering phenotype of LFYasEMF1 plants was rescued by ult1-3 under both SD and LD conditions. LFYasEMF1 plants also produced far fewer cauline leaves and inflorescence nodes than wild-type or ult1-3 plants (Fig. 2B). Consequently, they often produced a short inflorescence with a terminal flower phenotype like that of the emf1 mutants (Figs. 1, D and E, and 2D). The inflorescence defects of LFYasEMF1 were rescued by the ult1-3 mutation, with the numbers of cauline leaf nodes (Fig. 2B) and flower buds (Fig. 2, C–F) in LFYasEMF1 ult1-3 plants similar to those of wild-type plants.
Figure 2.
ult1-3 represses LFYasEMF1 early flowering and abnormal flower phenotypes. A and B, Flowering times of wild-type (WT), LFYasEMF1, ult1-3, and LFYasEMF1 ult1-3 plants, measured as rosette leaf number (A) during LD and SD growing conditions in soil and cauline leaf number (B) during LD growing conditions in soil after 15 d on agar plates grown under SD conditions. Error bars represent se. Asterisks indicate values that are significantly different from the wild type (**P < 0.01 using Student’s t test). C, A wild-type inflorescence apex. D, A LFYasEMF1 inflorescence apex bearing abnormal flowers and a terminal flower (TF). E, An ult1-3 inflorescence apex that generated flowers containing additional floral organs. The inset shows the extra petal in an ult1-3 flower. F, A LFYasEMF1 ult1-3 inflorescence apex that is normal. G, A wild-type flower with four petals, four sepals, six stamens, and two carpels. H, A LFYasEMF1 flower with four curly petals, four curly sepals, four curly stamens, and two carpels forming a twisted pistil. I, An ult1-3 flower with six sepals, six petals, six stamens, and two carpels. J, A LFYasEMF1 ult1-3 flower with four normal petals and sepals, six stamens, and two carpels. K to N, Siliques from wild-type (K), LFYasEMF1 (L), ult1-3 (M), and LFYasEMF1 ult1-3 (N) plants.
LFYasEMF1 flowers displayed a variety of phenotypes (Fig. 2, D and H). In severely affected flowers, usually terminal flowers, the first and second whorl organs were missing, the number of stamens was reduced, and multiple carpelloid organs were fused together (Supplemental Fig. S2, A–C). We also observed the “flower-in-flower” phenotype, where one flower arose inside another in LFYasEMF1 plants (Supplemental Fig. S2, D–F). In flowers with milder phenotypes, the sepals and petals were smaller and narrower, and all flower organs became curled like the rosette and cauline leaves. As a result, the sepals could not completely enclose the inner organs (Fig. 2H). The flower buds often opened later than normal (e.g. after silique elongation). Consequently, silique growth was constrained compared with wild-type and ult1-3 siliques (Fig. 2, H and K–M). Abnormal flower development was followed by poor seed set in LFYasEMF1 plants (Fig. 2L). Thus, reducing EMF1 activity affected flowering time and floral organ development as well as vegetative growth. ult1-3 flowers often contained more floral organs that frequently included extra petals (Fig. 2, E and I; Fletcher, 2001). The introduction of ult1-3 restored LFYasEMF1 to an almost normal phenotype that included sepals, petals, stamens, pistils, and siliques (Fig. 2, J and N). LFYasEMF1 ult1-3 plants did not have extra petals. Occasionally, the floral organs of LFYasEMF1 ult1-3 plants were slightly abnormal, with smaller and narrower sepals and petals. In summary, eliminating ULT1 function via the ult1-3 mutation rescued the LFYasEMF1 defects in almost every aspect of seedling and adult development.
Global Gene Expression Pattern in Wild-Type, LFYasEMF1, ult1-3, and LFYasEMF1 ult1-3 Plants
To investigate the molecular basis of the rescued phenotype, we compared global gene expression patterns in 7- and 15-DAG LFYasEMF1, ult1-3, LFYasEMF1 ult1-3, and wild-type seedlings grown under SD conditions using a custom-designed Agilent GeneChip containing probe sets representing 29,104 unique Arabidopsis genes (see “Materials and Methods”). We analyzed the 22,770 Arabidopsis genes that met the criterion of a hybridization signal greater than or equal to 50 in at least one of the four plant samples. Genes showing more than a 2-fold change in the hybridization signals were designated as up-regulated or down-regulated if those signals were higher or lower in the mutants than in the wild type (Supplemental Table S1). GeneChip results were confirmed by reverse transcription (RT)-PCR (Supplemental Fig. S3).
We first analyzed gene expression in the single mutant parent plants LFYasEMF1 and ult1-3. In LFYasEMF1 plants, 2,415 genes (10.6%) were up-regulated and 1,746 genes (7.7%) were down-regulated at 7 DAG. Thus, 18.3% of genes analyzed had altered expression in 7-DAG LFYasEMF1 plants (Table I). In 15-DAG LFYasEMF1 plants, 1,438 genes (6.3%) were up-regulated and 1,706 genes (7.5%) were down-regulated: 13.8% of the genes differed in expression (Table I). The ult1-3 mutants showed a lower percentage of misregulated genes than the LFYasEMF1 plants (Table I).
Table I. Gene expression changes in LFYasEMF1 and ult1-3.
Up and Down denote the number (percentage) of genes up-regulated and down-regulated, respectively, more than 2-fold in mutant or transgenic plants compared with the wild type.
ult1-3 Restored the Misregulated Gene Expression Pattern of LFYasEMF1
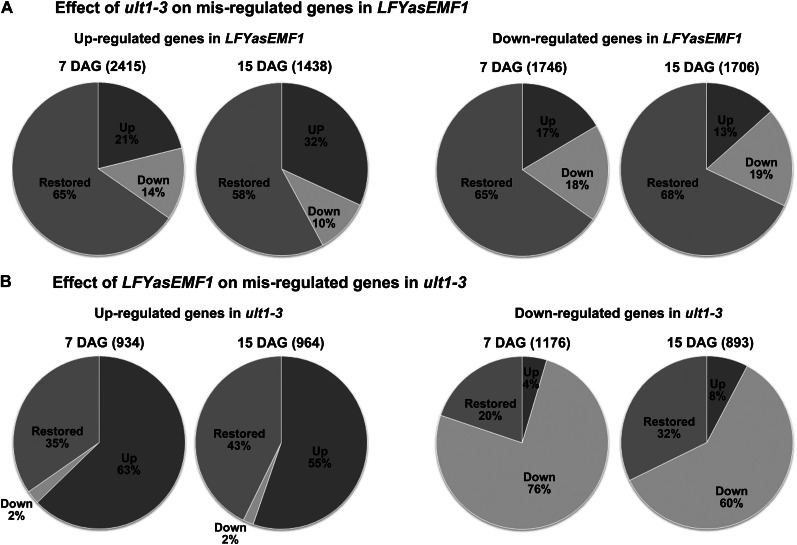
We then investigated whether ult1-3 affected the LFYasEMF1 gene expression pattern and found that the majority of the misregulated genes in LFYasEMF1 were restored to wild-type levels by the ult1-3 mutation (Fig. 3A; Supplemental Table S2). We found that 1,905 (79%) of 2,415 up-regulated genes in the 7-DAG LFYasEMF1 seedlings were no longer up-regulated in LFYasEMF1 ult1-3 seedlings. Among these, 1,577 genes (65%) were restored to wild-type expression levels and 328 genes (14%) were down-regulated (Fig. 3A; Supplemental Table S3). Similarly, 68% of the up-regulated genes in 15-DAG LFYasEMF1 were no longer up-regulated in 15-DAG LFYasEMF1 ult1-3 plants, with 832 genes (58%) restored to wild-type levels and 148 genes (10%) being down-regulated (Fig. 3A; Supplemental Table S3). Also, of the down-regulated genes in LFYasEMF1, 65% were restored to wild-type levels at 7 DAG and 68% were restored to wild-type levels at 15 DAG in LFYasEMF1 ult1-3 seedlings (Fig. 3A; Supplemental Table S3). The high percentage of genes with restored expression levels corresponds well with the phenotypic rescue of LFYasEMF1 by the ult1-3 mutation.
Figure 3.
Gene expression pattern in LFYasEMF1 ult1-3 seedlings. A, Pie charts of gene expression pattern affected by ult1-3 on 2-fold or greater up-regulated (left) and down-regulated (right) genes in LFYasEMF1 plants at 7 or 15 DAG. The number reflects the percentage of misregulated genes in LFYasEMF1 restored to wild-type expression levels or 2-fold or greater up-regulated (blue) or down-regulated levels in LFYasEMF1 ult1-3 plants. B, Pie charts of gene expression patterns affected by LFYasEMF1 on 2-fold or greater up-regulated (left) and down-regulated (right) genes in ult1-3 plants at 7 or 15 DAG. The number reflects the percentage of misregulated genes in ult1-3 restored to wild-type expression levels or 2-fold or greater up-regulated or down-regulated levels in LFYasEMF1 ult1-3 plants.
EMF1 interacts with several thousand target genes, 3,227 (58%) of which are also marked with H3K27me3 (EMF1_K27) in Arabidopsis seedlings (Kim et al., 2012). These EMF1_K27 genes are most likely regulated via PcG mechanisms. We investigated the impact of LFYasEMF1 on the EMF1_K27 genes and found that 326 (22.7%) of the 1,438 up-regulated genes in 15-DAG LFYasEMF1 plants were EMF1_K27 genes (Table II). Similarly, 295 (17.3%) down-regulated genes were EMF1_K27 genes (Table II). Both classes are greater than 11.4%, the fraction of EMF1_K27 genes in the Arabidopsis genome (Kim et al., 2012). Of the 326 up-regulated EMF1_K27 genes in LFYasEMF1 seedlings, 171 (52.5%) were not up-regulated in LFYasEMF1 ult1-3 seedlings. These included 150 (46.1%) restored to wild-type expression levels and 21 (6.4%) that became down-regulated (Table II). Of the 295 down-regulated EMF1_K27 genes in LFYasEMF1 plants, 250 (84.8%) were not down-regulated in LFYasEMF1 ult1-3 plants, including 194 (65.8%) restored to wild-type expression levels and 56 (19%) that became up-regulated (Table II). The majority of the genes for which their misregulation in LFYasEMF1 plants was restored by the ult1-3 mutation are EMF1_K27 genes, suggesting that ULT1 acts on many EMF1 target genes and antagonizes EMF1 action on gene expression.
Table II. Effects of ult1-3 on expression patterns of misregulated EMF1_K27 genes in LFYasEMF1 plants.
| Gene | Genome | LFYasEMF1 (1,438)a |
LFYasEMF1 ult1-3 |
LFYasEMF1 (1,706)a |
LFYasEMF1 ult1-3 |
||||
|---|---|---|---|---|---|---|---|---|---|
| Up | Up | Down | Restored | Down | Up | Down | Restored | ||
| EMF1_K27 (3,227)b | 11.4% | 326 (22.7%)c | 155 (47.5%) | 21 (6.4%) | 150 (46.1%) | 295 (17.3%)c | 56 (19%) | 45 (15.3%) | 194 (65.8%) |
Up- or down-regulated genes in LFYasEMF1 plants by at least 2-fold at 15 DAG. bGenes that are bound by EMF1 and marked by H3K27me3, 3,227 of 29,336, in the Arabidopsis genome (Kim et al., 2012). cNumber (percentage) of EMF1_K27 genes up- or down-regulated in LFYasEMF1 plants at 15 DAG.
To understand the function of the LFYasEMF1 misregulated genes restored by ult1-3, we considered the 15 functional categories described previously (Kim et al., 2010), with appropriate modifications of the gene list. These categories include genes involved in flower organ identity, flowering time, seed development, photoreceptor and photosynthesis system, cell expansion, auxin, GA, ethylene, and abscisic acid synthesis and signaling, stress, cold, and heat responses, and histone and transcription factor genes (Supplemental Table S1). Analysis of the genes in these 15 categories showed that ult1-3 can restore the expression of approximately 60% of the up- and down-regulated genes in LFYasEMF1 seedlings to wild-type levels (Supplemental Fig. S4A). In every one of the 15 categories, expression of a substantial number of genes was restored (Table III; Supplemental Table S4). Restored genes that affect flower organ identity, flowering time, seed formation, and stress are described further.
Table III. ult1-3 restoration of misregulated genes in LFYasEMF1 plants at 7 DAG.
| Category | No. of Genes Investigated | Change in Gene Expression | No. of Genes Misregulated in LFYasEMF1a | No. of Genes with Expression Restored to Wild-Type Level in LFYasEMF1 ult1-3b | Example of Genes with Expression Restored to Wild-Type Level in LFYasEMF1 ult1-3c |
|---|---|---|---|---|---|
| Abscisic acid | 12 | Up | 2 | 1 | RNA-binding protein |
| Down | 0 | 0 | |||
| Auxin | 107 | Up | 8 | 4 | Auxin-responsive family protein |
| Down | 13 | 11 | SAUR24, GH3.17, IAA14, SAUR19 | ||
| Cold | 19 | Up | 3 | 0 | |
| Down | 0 | 0 | |||
| Ethylene | 56 | Up | 14 | 8 | RRTF1, ERF5, ERF73, ERF-1, ORA59, MBF1C |
| Down | 0 | 0 | |||
| Expansin | 20 | Up | 0 | 0 | |
| Down | 5 | 3 | EXPA1, EXPA5, EXPA10 | ||
| Flower organ identity | 14 | Up | 10 | 10d | AG, SEP1, SEP2, SEP3, AP1, AP3, PI, SHP1, SHP2, STK |
| Down | 1 | 0 | |||
| Flowering time | 98 | Up | 12 | 9 | AGL24, TFL1, TEM1, ATC, MAF3, CO, SPL3, CPL3, FPF1 |
| Down | 11 | 8 | SPL1, SPL5, FLD, DNF, TOE1, FRI, GID1B, GA1 | ||
| GA | 13 | Up | 1 | 1 | GASA6 |
| Down | 1 | 0 | |||
| Heat | 50 | Up | 9 | 3 | HSFB2A, HSFA7A |
| Down | 4 | 2 | HSFC1, HSP23.6-MITO | ||
| Histone | 36 | Up | 5 | 4 | HTA12 |
| Down | 0 | 0 | |||
| Photoreceptor | 28 | Up | 2 | 1 | ELIP1 |
| Down | 3 | 2 | PAT1, IOS1 | ||
| Photosynthesis | 54 | Up | 1 | 1 | PSII 5-kD protein |
| Down | 2 | 1 | Chlorophyll a/b-binding protein | ||
| Seed | 119 | Up | 25 | 12 | Protease inhibitor genes, XYP1, M17, LEC1 |
| Down | 5 | 3 | SIP1, SIP2, EM1 | ||
| Stress | 46 | Up | 2 | 2 | STZ |
| Down | 2 | 0 | |||
| Transcription factor | 588 | Up | 77 | 48 | No apical meristem (NAM) family genes |
| Down | 46 | 22 | |||
| Total | 1,260 | 264 | 156 |
Genes more than 2-fold up- or down-regulated in LFYasEMF1 plants at 7 DAG. Fold changes are calculated using the signal intensity, which measures the change in expression level for each probe set between wild-type and mutant or transgenic plants. bGenes up- or down-regulated in LFYasEMF1 plants that were restored to wild-type expression levels (less than 2-fold) by ult1-3 at 7 DAG. cFor a complete list, see Supplemental Table S1. dEight of the 10 flower organ identity genes remained up-regulated by more than 2-fold in LFYasEMF1 ult1-3 plants but are included here because their up-regulation is greatly reduced (Supplemental Table S1).
Flower Organ Identity Genes
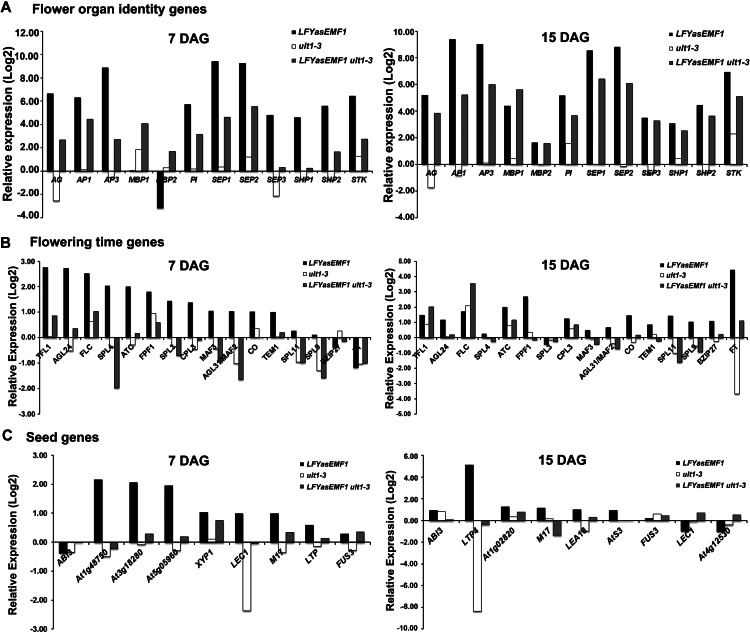
The 10 flower-specific MADS box genes AG, PI, APETALA1 (AP1), AP3, SHATTERPROOF1 (SHP1), SHP2, SEPALLATA1 (SEP1), SEP2, SEP3, and SEEDSTICK (STK) are all EMF1_K27 genes. In LFYasEMF1 plants, they showed significant ectopic expression at 7 DAG that increased at 15 DAG (Fig. 4A; Supplemental Table S1). This suggests that more antisenseEMF1, and hence less EMF1, is present in the plants at 15 DAG than at 7 DAG, consistent with the increasing LFY promoter activity as plants mature (Blázquez et al., 1997; Winter et al., 2011). In contrast, the mRNA levels of the 10 flower MADS box genes in ult1-3 plants were unchanged except for AG and SEP3, which were lower, and SEP2, which was higher, than in wild-type plants (Supplemental Table S1). Introduction of the ult1-3 mutation significantly reduced the expression level of these 10 highly up-regulated genes in LFYasEMF1 plants, although eight of the 10 genes still showed more than 2-fold up-regulation at 7 DAG (Fig. 4A; Supplemental Table S1). For example, AG had a 110-fold up-regulation in LFYasEMF1 but only 6-fold in LFYasEMF1 ult1-3. The highly up-regulated genes are not completely restored to wild-type levels (Supplemental Table S1).
Figure 4.
Opposite regulation of gene expression by EMF1 and ULT1. A, Relative expression levels of flower MADS box genes in plants at 7 DAG (left panel) and 15 DAG (right panel) using data based on GeneChip analysis. The log2 values of the fold change were calculated for comparison of the changes in gene expression levels in LFYasEMF1, ult1-3, and LFYasEMF1 ult1-3 plants relative to wild-type plants. A log2 ratio of 1 is the same as a 2-fold change in gene expression. B, Relative expression changes of flowering-time genes. C, Relative expression changes of seed genes.
Flowering-Time Genes
Among the 98 flowering-time genes investigated (Supplemental Table S1), 12 genes are up-regulated in LFYasEMF1 plants at 7 DAG and 11 genes are up-regulated at 15 DAG (Table III; Supplemental Table S4). Eight of these are directly regulated by EMF1: AGAMOUS-LIKE24 (AGL24), FLC, FLOWERING PROMOTING FACTOR1 (FPF1), MADS AFFECTING FLOWERING2 (MAF2)/AGL31, MAF3, SQUAMOSA PROMOTER BINDING PROTEIN-LIKE3 (SPL9), TEMPRANILLO1 (TEM1), and TFL1 (Kim et al., 2012). AGL24, FPF1, and SPL9 promote flowering (Kania et al., 1997; Melzer et al., 1999; Michaels et al., 2003; Schwarz et al., 2008; Wang et al., 2009), whereas FLC, TEM1, TFL1, MAF2, and MAF3 repress flowering (Michaels and Amasino, 1999; Ratcliffe et al., 2003; Castillejo and Pelaz, 2008).
Genes misregulated in 7- and/or 15-DAG LFYasEMF1 plants include 17 that are not EMF1 targets, such as CENTRORADIALIS (ATC), CONSTANS (CO), SPL3, SPL4, SPL5, and FLOWERING LOCUS T (FT; Supplemental Table S1). EMF1 may regulate these genes indirectly through the activities of genes it does interact with and regulate directly. For example, EMF1 interacts directly with FLC but does not interact with FT chromatin (Kim et al., 2010, 2012). FT mRNA levels increase as plants mature (Farrona et al., 2011), but FT is not up-regulated in LFYasEMF1 plants until 15 DAG. Ectopic expression of FLC is lower at 15 DAG than at 7 DAG in LFYasEMF1 plants (Fig. 4B; Supplemental Table S1), which would reduce FLC-mediated FT repression at 15 DAG. Thus, FT mRNA levels are much higher at 15 DAG than at 7 DAG in wild-type plants. The age-dependent increase in FT mRNA levels is enhanced by the reduced expression of FLC in LFYasEMF1 plants (Fig. 4B).
In LFYasEMF1 ult1-3 plants, nine of 12 flowering-time genes up-regulated in LFYasEMF1 plants at 7 DAG and six of 11 genes up-regulated at 15 DAG were restored to wild-type expression levels by the introduction of the ult1-3 mutation (Table III; Supplemental Table S4). Similarly, eight of 11 down-regulated genes in LFYasEMF1 plants at 7 DAG and three of seven down-regulated genes at 15 DAG were restored to wild-type expression levels (Table III; Supplemental Table S4). Together, the expression levels of 73% of the genes directly or indirectly regulated by EMF1 were restored in LFYasEMF1 ult1-3 plants at 7 and/or 15 DAG. The restoration of the misregulated flowering-time genes is consistent with the normal flowering time of the LFYasEMF1 ult1-3 plants.
Seed Genes
We examined 119 genes involved in seed development. The 25 genes up-regulated at 7 DAG and the nine genes up-regulated at 15 DAG in LFYasEMF1 plants include the LATE EMBRYOGENESIS ABUNDANT, OLEOSIN2, SEED STORAGE PROTEIN/LIPID TRANSFER PROTEIN (LTP), ARABIDOPSIS THALIANA SEED GENE3, and LEAFY COTYLEDON1 (LEC1) genes (Fig. 4C; Supplemental Table S1). Sixteen of the 7-DAG up-regulated genes and six of the 15-DAG up-regulated genes showed reduced ectopic expression in LFYasEMF1 ult1-3 plants. Among these, 12 of the 7-DAG genes and three of the 15-DAG genes were restored to wild-type expression levels in LFYasEMF1 ult1-3 plants (Table III; Supplemental Table S4). Three major seed regulator genes, ABSCISIC ACID INSENSITIVE3 (ABI3), FUSCA3, and LEC2, to which EMF1 binds (To et al., 2006; Kim et al., 2012), did not show increased expression in LFYasEMF1 plants by GeneChip analysis. ABI3 mRNA levels were confirmed by RT-PCR (Supplemental Fig. S3). Seed storage protein and lipid transfer protein genes (e.g. LTP4), which are downstream genes controlled by these major seed regulators, did show ectopic expression in LFYasEMF1 plants that was restored by the ult1-3 mutation (Supplemental Table S1). We also found that five seed development genes were down-regulated in the 7-DAG LFYasEMF1 seedlings and 13 seed development genes were down-regulated in the 15-DAG LFYasEMF1 seedlings. More than 50% of these were restored to wild-type expression levels in LFYasEMF1 ult1-3 seedlings (Table III; Supplemental Table S4). The ectopic expression and restoration of downstream seed storage gene expression suggest that there are subtle changes in the expression of the seed regulatory genes that are not readily detectable in LFYasEMF1 plants.
EMF1 and ULT1 Modulate the Salt Stress Response
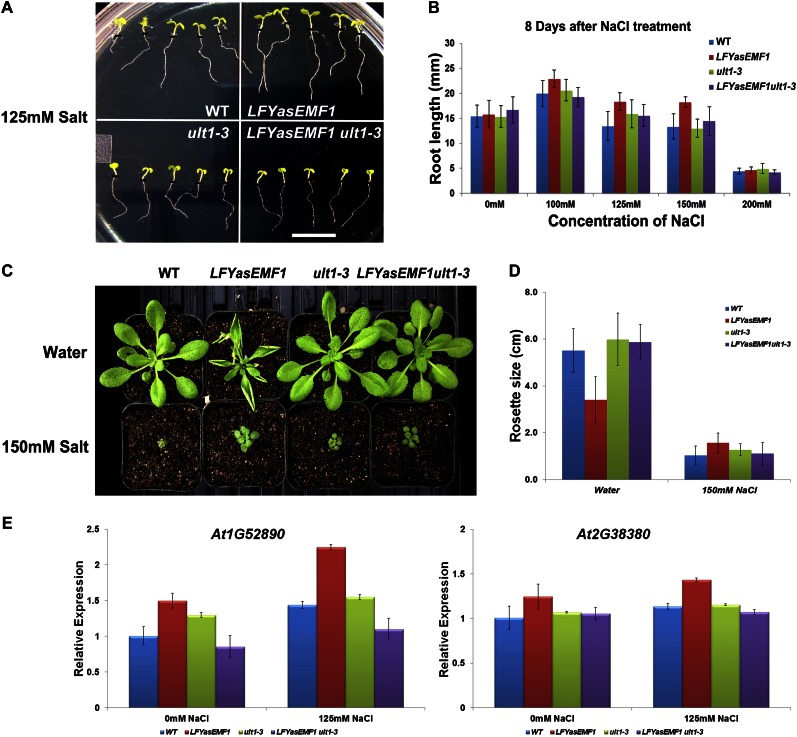
Our previous study showed that EMF1 binds preferentially to genes involved in biotic and abiotic stress responses (Kim et al., 2012). To investigate whether the EMF1 interaction with stress genes would affect the stress response, we studied salt tolerance in wild-type, LFYasEMF1, ult1-3, and LFYasEMF1 ult1-3 seedlings and adult plants. To avoid adverse effects of salt treatment on germination, we transferred 3-DAG wild-type, LFYasEMF1, ult1-3, and LFYasEMF1 ult1-3 seedlings from Murashige and Skoog (MS) plates to MS medium containing different concentrations of salt (0, 100, 125, 150, and 200 mm) and grew them for an additional 8 d under SD conditions. The LFYasEMF1 roots grew longer than the wild-type and ult1-3 roots on 100, 125, and 150 mm salt plates (Fig. 5, A and B), suggesting that LFYasEMF1 seedlings are more salt tolerant than the wild-type and ult1-3 seedlings.
Figure 5.
ult1-3 restores LFYasEMF1 enhanced salt tolerance and salt gene expression. A, Salt tolerance test of seedling growth. Wild-type (WT), LFYasEMF1, ult1-3, and LFYasEMF1 ult1-3 seedlings at 3 DAG were transferred from MS medium to MS medium containing 125 mm salt and grown for an additional 8 d. Short black lines indicate the junction of hypocotyl and root. B, Root length of seedlings grown on medium with and without salt, showing an enhanced salt tolerance in LFYasEMF1 that was restored by the introduction of ult1 into LFYasEMF1. Root length was measured after 8 d of growth on MS or MS with different concentrations of salt (n = 20). C, Salt tolerance test of plant growth in soil. Wild-type, LFYasEMF1, ult1-3, and LFYasEMF1 ult1-3 plants at 36 DAG were watered with or without 150 mm salt. D, Effect of salt on shoot growth, showing an enhanced salt tolerance in LFYasEMF1 that was restored in LFYasEMF1 ult1-3 plants. Rosette size was measured by rosette diameter. While wild-type, ult1-3, and LFYasEMF1 ult1-3 plants displayed about 80% inhibition of shoot growth by the 150 mm salt treatment, LFYasEMF1 growth inhibition was 50% at 36 DAG (n = 10). E, Quantitative RT-PCR analysis of the transcript levels of two salt-responsive genes, AT2G38380 and AT1G52890, in 6-DAG seedlings grown with or without salt.
The growth advantage of LFYasEMF1 seedlings on salt was diminished by the introduction of the ult1-3 mutation, as shown by a modest change in LFYasEMF1 ult1-3 root length compared with wild-type and ult1-3 root length. At 200 mm salt, the seedling root growth of all four genotypes was equally inhibited (Fig. 5, A and B). We then transferred these plants to soil and continued the same salt treatment as on the agar plates and the same SD condition. As mentioned above (Fig. 1, D and E), LFYasEMF1 plants watered with 0 mm salt exhibited curly leaves, early flowering, and smaller rosette size. However, they were more tolerant to salt treatment, as shown by their larger rosette size compared with wild-type, ult1-3, and LFYasEMF1 ult1-3 plants when watered with 150 mm salt (Fig. 5, C and D). Whereas the wild-type, ult1-3, and LFYasEMF1 ult1-3 plants displayed about 80% inhibition of shoot growth by the 150 mm salt treatment, LFYasEMF1’s growth inhibition was 50% (Fig. 5D). Thus, both seedling and adult growth of LFYasEMF1 plants was more salt tolerant, and this tolerance was reduced by the introduction of the ult1-3 mutation.
To elucidate the molecular mechanism of the EMF1-mediated salt response, we studied the expression levels of two salt-responsive genes, a gene belonging to the peroxidase superfamily, AT2G38380, and a NAC domain protein gene, AT1G52890 (Tran et al., 2004; Jiang et al., 2007), in wild-type, LFYasEMF1, ult1-3, and LFYasEMF1 ult1-3 seedlings. As expected, salt increased, albeit to varying degrees, the transcript levels of these two genes in all four types of seedlings (Fig. 5E; Supplemental Fig. S5). Importantly, LFYasEMF1 plants accumulated more salt-inducible transcripts, especially AT1G52890, than wild-type plants when grown in the absence of salt. Salt treatment further increased its transcript level in LFYasEMF1 more than it did in wild-type plants (Fig. 5E). The AT1G52890 transcript level in ult1-3 was higher than in wild-type plants (Fig. 5E). However, a comparison of the transcript levels between LFYasEMF1 and LFYasEMF1 ult1-3 plants showed that the introduction of ult1-3 greatly reduced the transcript levels of these two genes in LFYasEMF1 plants. These results indicate that the transcripts of salt-inducible genes are more readily accumulated in plants with reduced EMF1 activity, confirming a role for EMF1 in regulating the stress response. Removing ULT1 activity restored the misregulation of these genes in LFYasEMF1 plants, consistent with the reduced salt tolerance in LFYasEMF1 ult1 plants to a level closer to that of wild-type and ult1-3 plants (Fig. 5, B and C).
The Effect of LFYasEMF1 on the ult1-3 Misregulated Gene Expression Pattern
Since reducing ULT1 activity restored gene expression patterns in LFYasEMF1, we also analyzed the impact of the LFYasEMF1 transgene on the misregulation of genes in ult1-3 plants. Interestingly, LFYasEMF1 had a limited impact on misregulated genes in the ult1-3 background. The majority of genes up-regulated (63% at 7 DAG, 55% at 15 DAG) or down-regulated (60% at 7 DAG, 76% at 15 DAG) in ult1-3 plants remained up- and down-regulated in LFYasEMF1 ult1-3 plants (Fig. 3B; Supplemental Tables S2 and S5). Similarly, analysis of the genes in 15 categories showed that the majority of the up- and down-regulated genes in ult1-3 plants remained up- and down-regulated in LFYasEMF1 ult1-3 plants (Supplemental Fig. S4B). Nevertheless, LFYasEMF1 had a stronger impact on gene expression pattern in 15-DAG than in 7-DAG ult1-3 plants, suggesting that ULT1 and EMF1 share more target genes at 15 DAG than at 7 DAG. This is consistent with the observation that the ult1 mutation does not cause visible phenotypes in early development and cannot rescue emf1 mutations that obliterated seedling and rosette development.
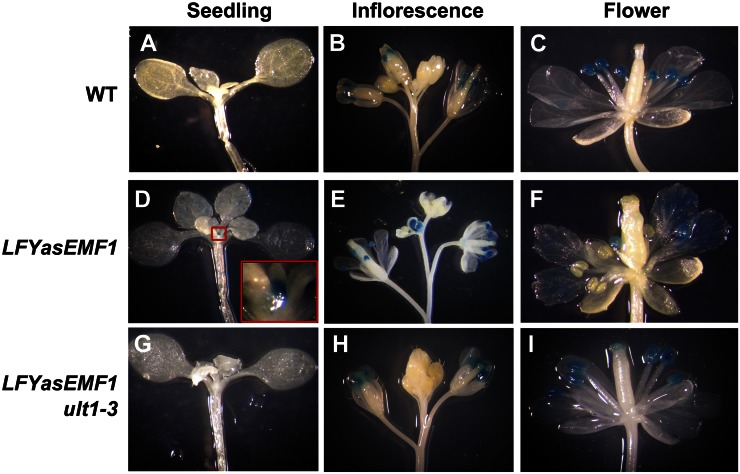
Temporal and Spatial AP3::GUS Expression Patterns in LFYasEMF1 and LFYasEMF1 ult1-3
To investigate the role of the misexpressed MADS box genes on the aberrant development of LFYasEMF1, we studied the temporal and spatial expression patterns of AP3 in wild-type, LFYasEMF1, and LFYasEMF1 ult1-3 plants harboring an AP3::GLUCURONIDASE (AP3::GUS) reporter construct. AP3 is normally activated in stage 3 flower primordia to specify petal and stamen identity (Krizek and Fletcher, 2005). The LFYasEMF1 plants at 21 DAG grown under SD conditions looked like wild-type plants but showed ectopic AP3::GUS activity in the shoot apex (Fig. 6, A and D). As expected, wild-type and LFYasEMF1 ult1-3 shoot apices did not show GUS activity at this developmental stage (Fig. 6, A and G). After flowering, AP3::GUS activity in wild-type flowers was restricted to the stamens (Fig. 6, B and C). However, the LFYasEMF1 plants exhibited ectopic AP3::GUS activity in buds, petals, receptacles, and even some carpels (Fig. 5, E and F). Furthermore, the six stamens varied in the extent of GUS activity (Fig. 6F). The LFYasEMF1 ult1-3 flowers had wild-type-like AP3::GUS patterns (Fig. 6, H and I). Ectopic AP3 activity in LFYasEMF1 shoot apices at 21 DAG is consistent with its early flowering phenotype. Disturbed EMF1 activity in the flower corresponds with the misexpression of AP3 that resulted in abnormal flower organ development in severe cases. This indicates that tight regulation of EMF1 is necessary for normal floral organ differentiation. The absence of ULT1 activity can counteract the effects of LFYasEMF1 and restore normal AP3 expression patterns in the shoot apex and in the flower.
Figure 6.
AP3::GUS activity in wild-type (WT), LFYasEMF1, and LFYasEMF1 ult1-3 plants. A, Wild-type plants at 21 DAG, grown under SD conditions, showing no AP3::GUS activity. B and C, Wild-type plants showing AP3::GUS activity in the stamens but not in the flower buds or petals. D, LFYasEMF1 plants at 21 DAG showing strong AP3::GUS activity at the shoot apex. The inset shows a magnified shoot apex. E and F, LFYasEMF1 plants showing ectopic AP3::GUS activity in flower buds and petals but not all the stamens. Some stamens display no GUS activity. G, LFYasEMF1 ult1-3 plants at 21 DAG showing no AP3::GUS activity. H and I, LFYasEMF1 ult1-3 plants showing AP3::GUS activity in the stamens but not in the flower buds or petals.
Effects of LFYasEMF1 and ult1-3 on the Histone Methylation Patterns of the Flower Homeotic Genes
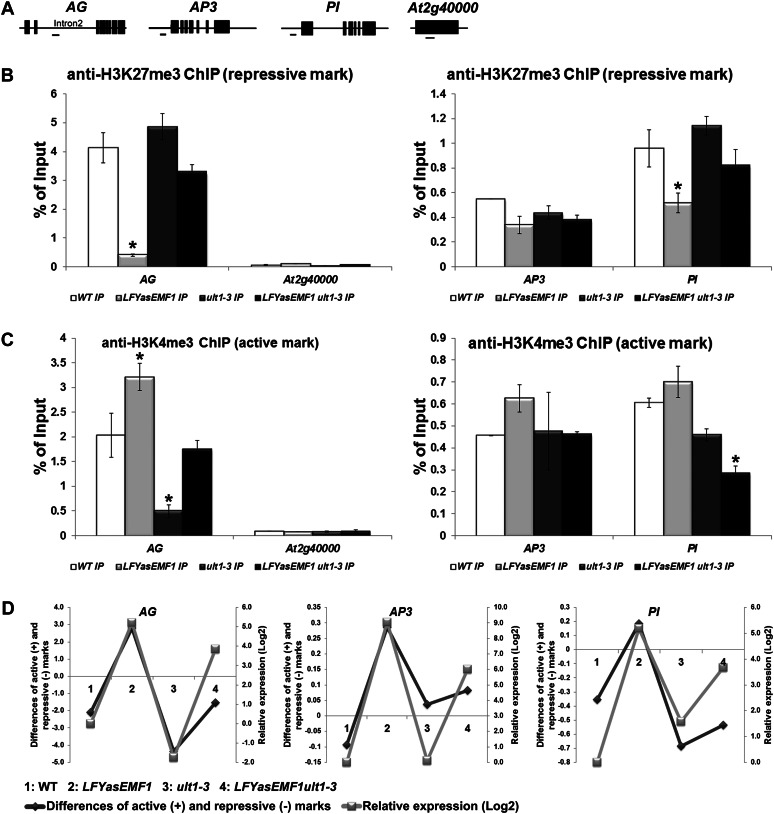
Genome-wide analysis of histone methylation patterns revealed that the 5′ end sequences in the promoter and gene body are highly enriched in H3K4me3 or H3K27me3 marks (Zhang et al., 2009; Kim et al., 2012). Therefore, we performed chromatin immunoprecipitation (ChIP)-quantitative PCR (qPCR) analysis in these regions of the three EMF1_K27 genes, AG, AP3, and PI, using AT2G4000, which is not trimethylated, as a negative control for H3K27me3 (Kim et al., 2012; Fig. 7A).
Figure 7.
Influence of EMF1 and ULT1 on AG, AP3, and PI histone methylation patterns and transcriptional activities. A, Schematic representation of AG, AP3, PI, and AT2G40000 gene structures. The exon/intron structures are depicted as black boxes/black lines. The regions amplified by qPCR are depicted as horizontal lines below the schemes. B, ChIP analysis of H3K27me3 levels at the AG, AP3, and PI loci in wild-type (WT), LFYasEMF1, ult1-3, and LFYasEMF1 ult1-3 seedlings at 15 DAG. Anti-H3K27me3 antibody was used to immunoprecipitate (IP) nuclear proteins from plants. Input is preimmunoprecipitated DNA after sonication. ChIP products were analyzed by qPCR using the primers corresponding to the gene region shown in A. ChIP results are expressed as a percentage of input DNA, with error bars representing sd of three replicates. C, ChIP analysis of H3K4me3 levels at the AG, AP3, and PI loci in wild-type, LFYasEMF1, ult1-3, and LFYasEMF1 ult1-3 seedlings at 15 DAG. ChIP and qPCR were carried out as described in B. Asterisks in B and C indicate values that are significantly different from the wild type (P < 0.05 using Student’s t test). D, Correlation between the relative mRNA expression levels (log2) and the difference between the active H3K4me3 (+) and repressive H3K27me3 (−) marks on the AG, AP3, and PI genes calculated by subtracting the enrichment level of the repressive H3K27me3 signal from that of the active H3K4me3 signal based on qPCR results in B and C. The relative expression levels of AG, AP3, and PI in plants at 15 DAG were based on GeneChip data (Supplemental Table S1). The log2 values of the fold changes were calculated for comparisons of the changes in expression level for each gene in the four plant samples. A log2 ratio of 1 is the same as a fold change of 2.
ChIP results showed that the H3K27me3-repressive marks are greatly reduced at the AG, AP3, and PI loci in LFYasEMF1 seedlings at 15 DAG relative to wild-type seedlings (Fig. 7B). Conversely, ult1-3 seedlings were slightly enriched in H3K27me3 at the AG and PI loci compared with wild-type seedlings, and both loci showed significant gains compared with LFYasEMF1 seedlings (Fig. 7B). LFYasEMF1 ult1-3 seedlings had more H3K27me3 marks at these loci than LFYasEMF1 seedlings but fewer than wild-type and ult1-3 seedlings (Fig. 7B). These results are consistent with reduced expression of these genes in LFYasEMF1 ult1-3 compared with LFYasEMF1 plants and slightly increased expression compared with wild-type plants (Fig. 4A). Despite the similar phenotype rescue of clf mutants and LFYasEMF1 plants by the ult1 mutation, there is a difference between the interaction of ULT1 with EMF1 and that of ULT1 with CLF. In clf-2 plants, AG was depleted of H3K27me3 regardless of the presence or absence of ULT1 (Carles and Fletcher, 2009). In LFYasEMF1 plants, however, removing ULT1 restored H3K27me3 to a nearly wild-type level. One reason may be that CLF is the methyltransferase while EMF1 participates in its enzyme activity via yet unknown mechanisms.
H3K4me3 is a histone mark for active chromatin. However, it is also found on inactive genes in mammalian cells and on the AG loci in Arabidopsis seedlings (Bernstein et al., 2006; Saleh et al., 2007). The AG locus was more enriched for the H3K4me3 activation marks in LFYasEMF1 seedlings than in wild-type seedlings (Fig. 7C). That the AG locus in LFYasEMF1 plants has more active H3K4me3 marks and fewer repressive H3K27me3 marks (Fig. 7, B and C) indicates that the locus has shifted toward a more active state. This is consistent with ectopic AG expression in LFYasEMF1 plants before flowering. A previous study reported minor effects of ult1 on H3K4me3 deposition in the wild type, although ult1-3 did reduce its deposition in clf-2 (Carles and Fletcher, 2009). Here, we found that H3K4me3 marks on the AG locus were greatly reduced in ult1-3 seedlings compared with wild-type and LFYasEMF1 seedlings (Fig. 7C). In the LFYasEMF1 ult1-3 plants, the level of H3K4me3 marks on the AG locus recovered to a level near that of wild-type plants (Fig. 7C). These results indicate that, besides restricting H3K27me3 on PRC2 target loci, ULT1 plays an important role in H3K4 trimethylation. The other two MADS box genes, AP3 and PI, also displayed varying levels of H3K4me3 enrichment in LFYasEMF1, ult1-3, and LFYasEMF1 ult1-3 plants (Fig. 7C). Importantly, introduction of the ult1-3 mutation reduced the H3K4me3 marks on all three genes in the LFYasEMF1 background. The finding that removing ULT1 function from LFYasEMF1 plants reduced active marks but increased repressive marks on target genes is consistent with ULT1 and EMF1 having antagonistic functions. H3K4 trimethylation in the absence of ULT1 may result from gene redundancy (i.e. ULT2 may substitute for ULT1 functions in ult1-3 mutants; Carles et al., 2005). While EMF1 is a single gene, it may not be the core component of the PRC2 necessary for the H3K27 trimethylase activity of CLF.
The coexistence of the active and repressive histone marks on the target genes led us to examine the data further. We subtracted the signals corresponding to the repressive H3K27me3 histone marks on the target gene chromatin from those of the active H3K4me3 marks and plotted this relative value for each genotype against the relative mRNA transcription levels (Fig. 7D; Supplemental Table S6). This revealed that the more active (+) or less repressive (−) the differences between the histone marks, the higher the transcriptional activities of the three genes, as the relative AG transcript levels among the four genotypes are LFYasEMF1 > LFYasEMF1 ult1 > the wild type > ult1-3 (Fig. 7D). A similar pattern was observed with AP3 and PI (Fig. 7D). These data indicate a close correlation between the difference of the active and repressive histone marks and transcriptional activity.
DISCUSSION
Since vegetative growth is largely eliminated in emf1 mutants, we employed tissue-specific removal of EMF1 activity to study the effects of EMF1 during the vegetative phase. Analyses of the LFYasEMF1 phenotype and expression patterns showed that EMF1 is required for lateral organ differentiation and stable phase transitions in Arabidopsis. We found that ectopic expression of not one but multiple MADS box genes underlies the curly leaf and early flowering phenotypes of LFYasEMF1 plants. These plants were also useful in investigating the epigenetic mechanisms of gene regulation by EMF1 and ULT1. In this study, we discovered that ult1 can suppress the defects caused by partial removal of EMF1 activity but not the defects at germination caused by the emf1 mutation. Our results indicate also that the difference in repressive and active histone marks on target genes modulates transcriptional activity in Arabidopsis.
Role of EMF1 in Arabidopsis Seedling Development
Reducing EMF1 activity at the onset of leaf development did not prevent rosette leaf growth. This indicates that EMF1 is not required for organ initiation, although it is required for subsequent leaf differentiation. LFYasEMF1 plants produced bigger and rounder cotyledons than wild-type plants and also formed new rosette leaves at a faster pace. SPL9, an SBP box transcriptional factor involved in the juvenile-to-adult phase transition, is up-regulated in LFYasEMF1 plants (Supplemental Table S1). spl9 mutants produce leaves at a slower pace and are delayed in flowering (Schwarz et al., 2008). Thus, the SPL9 up-regulation in LFYasEMF1 plants is consistent with their faster pace of leaf production and earlier flowering. The rosette leaves of LFYasEMF1 plants were small and curled upward along the proximal distal axis. In fact, all lateral organs, including cauline leaves and flower organs, are curled in LFYasEMF1 plants. The molecular mechanisms underlying these phenotypes are discussed below.
Role of EMF1 in Flower Meristem Determination and Flower Organ Development
EMF1 involvement in flower organ development is evidenced by the fact that LFYasEMF1 plants displayed a variety of phenotypes, ranging from reduced flower organ size and missing flower organs to terminal flower, floral homeotic defects, and floral reversion in severe cases. This is counter to the expectation that epigenetic repressors would play a limited role in flower organ development when the flower MADS box genes they target are active. Flower patterning depends on PRC2 activity (Wu et al., 2012). The abnormal flower morphology (Fig. 2; Supplemental Fig. S2) and irregular AP3::GUS expression patterns in LFYasEMF1 plants (Fig. 6) indicate that a fine-tuning of flower MADS box gene expression during successive flower organ transition also depends on EMF1-mediated gene silencing.
The flower-in-flower phenotype observed in LFYasEMF1 plants (Supplemental Fig. S2, D–F) is a form of floral reversion, in which the flower meristem reverts back to the inflorescence meristem. Floral reversion occurs in plants impaired in the floral meristem identity genes LFY and AP1 (Weigel et al., 1992; Wagner et al., 1999; Parcy et al., 2002). Ectopic expression of AGL24, an EMF1_K27 gene, also led to floral reversion (Yu et al., 2004). Reduced EMF1 activity in LFYasEMF1 flowers would allow ectopic expression of AP1 and AGL24 (Supplemental Table S1), resulting in flower formation within a flower. Before bolting, AP1 and its downstream flower MADS box genes are ectopically expressed in the shoot apex of LFYasEMF1 plants (Fig. 6D; Supplemental Table S1; Sánchez et al., 2009). This may be sufficient to initiate terminal flower formation in LFYasEMF1 plants (Parcy et al., 2002).
Role of EMF1 in the Salt Stress Response
EMF1 preferentially binds genes involved in biotic and abiotic stress responses (Kim et al., 2012), suggesting its involvement in stress regulation. To investigate the function of EMF1 interaction with stress gene chromatin, we investigated the salt stress response and found that LFYasEMF1 plants were more, albeit mildly, salt tolerant than wild-type plants. The LFY promoter is known to be active in leaf primordia by 4 DAG and is gradually up-regulated during vegetative development, peaking in the floral meristem but not in the rest of the plant (Blázquez et al., 1997). Hence, only leaf primordia would experience reduced EMF1 due to the expression of asEMF1 driven by the LFY promoter, whereas EMF1 would be expressed at normal levels in mature organs. Localized reduction of EMF1 activity in the shoot apex region may not lead to high stress tolerance. Inducible expression of asEMF1 in response to stress in whole plants would potentially increase the extent of salt tolerance.
Because LFYasEMF1 plants have more EMF1 activity and display weaker phenotypes than emf1 mutants, LFYasEMF1 plants would not display as many misregulated genes as emf1 mutants. Thus, two stress-related genes are up-regulated in 7-DAG LFYasEMF1 seedlings (Table III), whereas 21 genes are up-regulated in 7-DAG emf1-2 seedlings (Kim et al., 2010). Nevertheless, we identified two salt-inducible genes that are EMF1 bound and trimethylated on H3K27 (EMF1_K27), At2G38380 and At1G52890. Both of these genes are expressed at a higher level in LFYasEMF1 than in wild-type plants when grown in the absence or presence of salt, consistent with the ability of LFYasEMF1 plants to endure higher salt stress conditions. In the LFYasEMF1 ult1-3 plants, the transcript levels of these two genes and the salt tolerance of the plants were reduced to near the wild-type level. Reduction of salt tolerance in LFYasEMF1 plants by removing ULT1 activity indicates that EMF1 regulation of the two salt-inducible genes is counteracted by ULT1 activity, as is the case for the flower MADS box genes (see below). Should future investigations of the tissue-specific expression pattern of stress genes indicate an advantage of expressing salt-inducible genes in roots (or shoots) only, it will be possible to express asEMF1 under the control of a root-specific promoter in generating plants with high salt tolerance.
Mechanisms Underlying the Curly Leaf and Early Flowering Phenotypes
LFYasEMF1 and clf plants, as well as transgenic plants constitutively expressing AG, FT, and SEP3 under the control of the 35S promoter (35S::AG, 35S::FT, and 35S::SEP3; Mizukami and Ma, 1992; Honma and Goto, 2001; Lopez-Vernaza et al., 2012), all have curly leaf and early flowering phenotypes. Besides the similarity in phenotypes, these plants share one other thing in common: ectopic expression of multiple flower MADS box genes (Table IV). The aberrant phenotypes can be rescued by removing the ectopic activity of just one of the flower MADS box genes without altering the ectopic expression of the others. For example, AG, SEP3, FT, and FLC are ectopically expressed in clf (Goodrich et al., 1997; Lopez-Vernaza et al., 2012). Mutations in SEP3, FT, or FLC were able to suppress the clf phenotype, while AG remained highly expressed (Lopez-Vernaza et al., 2012). Thus, AG misexpression is not the only reason for the curly leaf phenotype.
Table IV. Ectopic expression of MADS box and flowering-time genes in plants displaying early flowering and curly leaf phenotypes.
+ indicates genes up-regulated as reported in the references provided.
| Genotype | Phenotypes |
MADS Box and Flowering-Time Genes Up-Regulated |
References | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Flowering | Leaf | AG | SEP3 | AP3 | PI | AP1 | FUL | FT | FLC | ||
| 35S::AG | Early | Curly | + | + | + | + | Mizukami and Ma (1992); Gómez-Mena et al. (2005) | ||||
| 35S::FT | Early | Curly | +a | + | +a | +a | + | + | + | Teper-Bamnolker and Samach (2005) | |
| 35S::SEP3 | Early | Curly | + | + | + | + | Honma and Goto (2001); Castillejo et al. (2005) | ||||
| 35S::AP3 35S::PI | Early | Curly | + | + | Krizek and Meyerowitz (1996); Honma and Goto (2001) | ||||||
| clf | Early | Curly | + | + | + | + | + | Goodrich et al. (1997); Lopez-Vernaza et al. (2012) | |||
| LFYasEMF1 | Early | Curly | + | + | + | + | + | + | + | + | Sánchez et al. (2009); this study |
Genes up-regulated as predicted by the up-regulation of SEP3 in the 35S::FT plants.
Many MADS box genes are ectopically expressed in LFYasEMF1 plants, including the 10 flower homeotic genes and the flowering-time genes FLC, SUPPRESSOR OF OVEREXPRESSION OF CONSTANS1, and AGL24. In the LFYasEMF1 ult1-3 plants, ectopic expression of these genes is greatly reduced. Flowering-time genes regulate each other and the downstream flower homeotic genes, which also regulate each other, in a complex network. Some regulatory actions are positive and some are negative. MADS domain proteins form homodimers and heterodimers, ternary, quaternary, and multimers to regulate downstream gene expression (Honma and Goto, 2001; de Folter et al., 2005; Liu et al., 2008). The end result of the ectopic expression of one or more flower homeotic or flowering-time genes would be the excessive accumulation of multiple flower MADS domain proteins. This ectopic expression of one or more MADS domain proteins could create new complexes, displacing or substituting for other MADS domain proteins in stage-specific multiprotein complexes. Nevertheless, protein complexes of diverse components seem to act redundantly by regulating similar downstream genes, presumably those regulated by MADS domain protein complexes at that developmental stage. Although LFYasEMF1, clf, 35S::AG, 35S::FT, and 35S::SEP3 plants are not identical in their gene expression patterns, each shows ectopic expression of multiple flower MADS box genes (Table IV). This is sufficient to cause the common aberrant phenotypes.
Epigenetic Regulation of Flower MADS Box Genes, AG, AP3, and PI
ULT1 harbors the SAND domain with DNA-binding activity; thus, it may function as a transcriptional coactivator to regulate a few specific genes. As a result, the ult1 mutation may rescue the curly leaf and early flowering phenotypes of LFYasEMF1 through the reduced expression of a few genes such as the MADS box genes. However, 2,110 genes in diverse functional groups are misregulated in ult1 mutants (Table I), so ULT1 is not likely to act on the relatively few floral homeotic genes. Based on the ability of ULT1 to act as an antirepressor of PcG, a fundamental property of a trxG factor, we propose that ULT1 is an epigenetic regulator rather than a transcriptional activator. First, the ult1 mutation completely suppresses the phenotypes caused by the mutations in CLF, a well-characterized PcG gene (Carles and Fletcher, 2009). Second, besides the flower organ and flowering-time genes, the ult1-3 mutation restores the expression of many classes of genes in LFYasEMF1 plants, including seed genes, histone genes, stress genes, expansion genes, and hormone response genes (Table III; Supplemental Table S4). In the case of the stress genes, we established a direct link between the misregulation and restoration of the EMF1-targeted stress genes with the LFYasEMF1 and LFYasEMF1 ult1-3 plants, respectively, to show a separation of the EMF1-mediated flower program from that of the stress program. Third, the ult1-3 mutation not only affects the transcript levels of its target genes but also their chromatin marks, H3K4me3, functioning to limit the deposition of repressive histone H3K27me3 marks. Fourth, ULT1 can physically interact with the Arabidopsis trxG factor ATX1 (Carles and Fletcher, 2009).
The ult1 mutation counteracted the impact of the LFYasEMF1 transgene at nearly every stage of development. ult1-3 restored the expression of the majority (68%–82%) of misregulated genes in LFYasEMF1 seedlings to wild-type levels. This suggests a molecular basis for the phenotype rescue and a mechanism for the restoration of gene expression. PcG and trxG factors trimethylate a different Lys on the same histone of the common target genes. The presence of both active H3K4me3 and repressive H3K27me3 at silent loci, termed the “bivalent mark,” was proposed to mark silent genes poised for activation during differentiation in mouse embryonic stem cells (Bernstein et al., 2006). Thus, the existence of both H3K27me3 and H3K4me3 marks on AG, AP3, and PI silent loci might represent a plastic state of these genes in seedling cells that are subject to activation by changing developmental and environmental cues. However, how PcG and trxG act antagonistically in regulating gene activities remains unclear.
Recent studies showed that H3K27me3 stimulates PRC2 activity, whereas H3K4me3 inhibits it through allosteric inhibition of H3K27me3 deposition (Schmitges et al., 2011). A genome-wide study showed that EMF1 is required for H3K27me3 on a fraction of its target gene loci in Arabidopsis seedlings, indicating EMF1 acting along with PRC2 for the trimethylation of H3K27 on these loci (Kim et al., 2012). Consistent with a feedback mechanism of H3K27me3 stimulation of PRC2, reduced EMF1 in LFYasEMF1 plants would reduce H3K27me3-repressive mark deposition, in turn reducing PRC2 activity on the flower MADS box genes. In ult1-3 mutants, we found reduced H3K4me3 active marks, which would remove the allosteric inhibition of the PRC2 deposition of the repressive marks, resulting in high levels of repressive marks, on the target gene histones. The restoration of the repressive marks in LFYasEMF1 ult1-3 seedlings shows that PRC2 is able to integrate opposing H3K4me3 and H3K27me3 modifications into an intermediary H3K27 methylation activity (Schmitges et al., 2011).
Although H3K4me3 and H3K27me3 are strongly associated with active and inactive chromatin, respectively, the debate continues whether these histone marks, H3K4me3 in particular, are the cause or consequence of transcriptional change (Henikoff and Shilatifard, 2011). In the case of EMF1 and ULT1, their molecular and biochemical functions have not been fully characterized. The ULT1 protein contains the SAND domain, known to function in chromatin-dependent transcriptional control, and can be associated with other modules, including the PHD finger (Bottomley et al., 2001). EMF1 is a plant-specific protein that contains a few known motifs, such as the ATP/GTP-binding motif (P loop) and the LXXLL motif, which is thought to be a transcriptional regulatory motif (Aubert et al., 2001) and is implicated in chromatin compaction (Calonje et al., 2008; Beh et al., 2012). If these two proteins are required for histone methylation, as our data indicate, the simple explanation would be that histone methylation change is causal to the transcriptional change seen in the plants impaired in their expression. On the other hand, should these two proteins affect transcription directly, the change in histone methylation pattern may be a consequence of the transcription change in the LFYasEMF1 and ult1 plants. Regardless, our analysis revealed a close relationship between histone methylation levels and transcriptional activity. Mutually exclusive PcG and trxG domains are known to maintain stable off and on developmental states, respectively (Papp and Müller, 2006); nevertheless, the inhibitory circuitry present in PRC2 does not function as a binary on/off switch (Schmitges et al., 2011). Our study further demonstrates a quantitative correlation rather than an on/off relationship between the histone methylation pattern and the transcript levels of the three flower MADS box genes. There is an additive effect of the repressive (−) and active (+) marks on the chromatin, such that the more (+) histone marks are found on a given gene the higher the transcript levels, whereas the presence of fewer (+) or more (−) histone marks leads to lower transcript levels. We propose that a balance of the H3K27 and H3K4 methyl marks on the gene locus mediates its transcriptional activity. This indicates that, besides maintaining active versus repressive chromatin states, histone methylation marks may function as modulators of target gene expression levels.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Wild-type, mutant, and transgenic plants used in this study are all in the Arabidopsis (Arabidopsis thaliana) Columbia background. The LFYasEMF1 transgenic plants harbor an AP3::GUS construct, kindly provided by Dr. Vivian Irish (Yale University), and a construct that expresses asEMF1 under the control of the LFY promoter (Sánchez et al., 2009). For all experiments, Arabidopsis seeds were surface sterilized and plated on agar plates containing two-fifth-strength MS Basal Salt Mixture (Sigma), 1.5% Suc, and 0.8% agar. The plates were placed at 4°C for 2 d and moved to an SD growth room (8 h of light/16 h of dark) at 21°C for 15 d. Seedlings were then transferred to soil to be grown under SD or LD conditions (16 h of light/8 h of dark) at 21°C in the greenhouse.
Genetic Crosses, Cotyledon Measurements, and GUS Activity Assays
emf1 ult1 double mutants were generated by crossing homozygous ult1-3 plants with heterozygous emf1-2 plants and identifying homozygous ult1-3 progeny in the F2 population based on ult1-3’s extra-petal phenotype. The F3 segregating progeny displaying the emf1-2 phenotype were subjected to PCR-based genotyping to identify ult1-3 alleles using the primers shown in Supplemental Table S7.
To introduce the ult1-3 mutation into the LFYasEMF1 background, we crossed homozygous LFYasEMF1 plants harboring AP3::GUS (Sánchez et al., 2009) with ult1-3 plants. F2 progeny were grown on two-fifth-strength MS medium containing 40 mg L−1 hygromycin B to select for homozygous LFYasEMF1 individuals, which would segregate 100% hygromycin-resistant progeny. Progeny that showed AP3::GUS activity in stamens were subsequently identified. Homozygous LFYasEMF1 ult1-3 plants were screened by allele-specific polymorphism genotyping, using the primers listed in Supplemental Table S7 to genotype ult1-3 and LFYasEMF1.
To measure cotyledon sizes, we took photographs of the cotyledons of 2-week-old seedlings and obtained the surface area of the cotyledon blades using the ImageJ software program (http://rsb.info.nih.gov/ij/).
GUS activity in transgenic plants was assayed as described (Pu et al., 2008) with slight modifications. Briefly, seedlings or tissues were incubated in 2 mm 5-bromo-4-chloro-3-indolyl-β-d-GlcA in 50 mm phosphate buffer, pH 7.0, containing 0.5 mm K3Fe(CN)6 and 0.5 mm K4Fe(CN)6 for 12 h at 37C, rinsed with 70% ethanol (v/v) at room temperature, and observed and photographed with a dissecting microscope. All subsequent image manipulation and figure preparation were performed with Adobe Photoshop.
Salt Treatment, and Root Length and Rosette Diameter Measurements
Wild-type, LFYasEMF1, ult1-3, and LFYasEMF1 ult1-3 seeds were sown on MS agar medium. The plates were incubated at 4°C for 2 d before being transferred to 21°C under SD conditions. The 3-DAG seedlings of the wild type, LFYasEMF1, ult1-3, and LFYasEMF1 ult1-3 were transferred from MS plates to MS medium supplemented with different concentrations of salt (0, 100, 125, 150, and 200 mm) and then grown for an additional 8 d under SD conditions. The root length was measured daily for 8 d post transfer (n = 20). The 15-DAG seedlings were transferred to soil and continued with the same salt treatment as when they were grown on agar plates. The rosette diameter of plants grown in soil was measured at 36 DAG to obtain average rosette size (n = 10).
Microarray Experiments and Data Analysis
Total RNA was extracted by TRIzol Reagent (Invitrogen) following instructions in the user’s manual. DNA was eliminated by the use of the TURBO DNA-free kit (Ambion). Microarray experiments were performed in the DNA microarray core laboratory (Institute of Plant and Microbial Biology, Academia Sinica; http//ipmb.sinica.edu.tw/affy/). One microgram of total RNA from each sample was used as a template to generate amino allyl-modified RNA (aRNA). The amplified aRNA was labeled with Alexa Fluor 555 reactive dyes (Invitrogen) and fragmented according to the Amino Allyl MessageAmp aRNA Amplification Kit (Ambion) instruction manual before hybridization to the Arabidopsis Gene Expression Microarray chip version 4 with 43,803 Arabidopsis probes represented (G2519F; Agilent Technologies; http://www.genomics.agilent.com). The chips were hybridized with equal amounts (1.65 μg) of Alexa Fluor 555-labeled aRNA for 17 h at 65°C and washed according to the manufacturer’s instructions in the Agilent Gene Expression Hybridization Kit. Chip images were scanned and analyzed by the use of Feature Extraction software (version 10.7.1.1) following the GE1-107_Sep09 protocol (Agilent Technologies). Microarray hybridization was repeated two times with independent biological samples. The results presented were based on one set of microarray analyses. Genes with hybridization signals of 50 or greater in at least one of four plant samples were analyzed. The fold change of gene expression was based on the ratio of mutant or transgenic plants to the wild type, with the ratio greater than 2-fold used to identify up- or down-regulated genes.
RT-PCR and Real-Time Quantitative RT-PCR
Total RNA was isolated from 6-DAG (Fig. 5) or 15-DAG (Supplemental Fig. S3) seedlings using the RNeasy plant kit (Qiagen) as described previously (Pu et al., 2008). RNA samples were treated with RNase-free DNaseI (Invitrogen) for 15 min at room temperature, and then DNaseI was inactivated by treatment with 25 mm EDTA solution. First-strand complementary DNA (cDNA) synthesis was performed on 1 μg of total RNA using SuperScript II reverse transcriptase (Invitrogen) and 12mer to 18mer oligo(dT) according to the manufacturer’s instructions. For RT-PCR, samples of the first-strand cDNA were then used in PCR with Phusion high-fidelity DNA polymerase according to the recommendations of the supplier. At least two independent experiments employing biological replicates were performed. Three technical replicates of RT-PCR were performed for each biological sample with similar results. Thirty-one to 36 cycles of PCR were used for all genes with the gene-specific primers listed in Supplemental Table S7. The intensity of DNA bands in each image (Supplemental Fig. S5) was measured using ImageJ software. The relative DNA level of each gene was then normalized to each UBQ control and the wild type and is represented as an Arabic number beneath each gel image. For quantitative RT-PCR, samples of the first-strand cDNA were then analyzed (Thermocycler ABI 7300) using SYBR Green and standard settings. Quantitative RT-PCR was performed in a final volume of 20 μL. Three technical replicates were done for each sample. Amplification of UBQ was used as an internal control to normalize all data. Quantification was determined by applying the comparative cycle threshold formula (Pu et al., 2008). All gene-specific primers are listed in Supplemental Table S7. The intensity of DNA bands in each image (Supplemental Fig. S5) was measured using ImageJ software. The relative DNA level of each gene was then normalized to each UBQ control and the wild type and is represented as an Arabic number beneath each gel image.
ChIP and ChIP-qPCR
ChIP experiments were performed as described previously (Kim et al., 2010). Briefly, fresh tissues of 15-DAG seedlings were infiltrated in 1% formaldehyde solution under a vacuum for 30 min to cross link the chromatin. The reaction was stopped by adding 0.1 m Gly. Fixed tissues were ground in liquid nitrogen, nuclei were isolated, and chromatin was extracted and sheared by sonication (Microson MS-50; 10 s on and 10 s off for 10 times) to generate 0.5- to 2-kb fragments (Bowler et al., 2004). Anti-H3K27me3 (Millipore) and anti-H3K4me3 (Abcam) antibodies were used to immunoprecipitate the fragmented chromatin. Cross linking of immunoprecipitated chromatin was reversed with 5 m salt, and DNA was precipitated with 100% ethanol. For the input control, 5 m salt was added to 0.5% total chromatin before immunoprecipitation to reverse the cross linking, and DNA was isolated by 100% ethanol. The relative amount of DNA was determined by spectrophotometry (NanoDrop ND1000). Immunoprecipitated DNA was analyzed by qPCR (Thermocycler ABI 7300) using SYBR Green and standard settings. PCR was performed in a final volume of 20 μL, and dilutions of purified input DNA were measured together with the immunoprecipitated DNA samples. Three technical replicates were done for each sample. Quantification was determined by applying the 2-∆Ct formula (SuperArray ChIP-qPCR user manual; Bioscience Corporation). Average immunoprecipitates from chromatin isolated independently are expressed on graphs as percentages of corresponding input DNA with error bars representing the sd. Primers used for the detection of AG, AP3, PI, and AT2G40000 are listed in Supplemental Table S7.
The microarray data from this article are deposited in the Gene Expression Omnibus with accession number GSE39947. Sequence data can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: At5G11530 (EMF1), At4G28190 (ULT1), At3G54340 (AP3), At4G18960 (AG), and At5G20240 (PI).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Phenotype and genotype of emf1-2 ult1-3 double mutants.
Supplemental Figure S2. Homeotic defects of LFYasEMF1 flowers.
Supplemental Figure S3. Expression of the flower MADS box and seed genes in plants impaired in EMF1 and/or ULT1.
Supplemental Figure S4. Restoration of misregulated genes of 15 functional categories in LFYasEMF1 ult1-3 seedlings.
Supplemental Figure S5. Effects of emf1 and ult1 on salt-responsive gene expression.
Supplemental Table S1. Expression pattern of the 15 categories of genes in wild-type, LFYasEMF1, ult1-3, and LFYasEMF1 ult1-3 Arabidopsis at 7 and 15 DAG.
Supplemental Table S2. Significantly misregulated genes in LFYasEMF1 and ult1-3 and restored in LFYasEMF1 ult1-3.
Supplemental Table S3. Expression change by ult1-3 on genes misregulated in LFYasEMF1 plants.
Supplemental Table S4. ult1-3 restoration of misregulated genes in LFYasEMF1 plants at 15 DAG.
Supplemental Table S5. Expression change by LFYasEMF1 on misregulated genes in ult1-3 plants.
Supplemental Table S6. Differences of active H3K4me3 (+) and repressive H3K27me3 (−) marks on the flower MADS box genes AG, AP3, and PI.
Supplemental Table S7. Primers.
Acknowledgments
We thank the following people from the Department of Plant and Microbial Biology, University of California, Berkeley: Yvonne Kim, Nathalie Quintero, Tiffany Wang, and Gilbert Garcia for assistance and Drs. Andy Jackson and Russell Jones for critical reading of the manuscript and their valuable comments.
Glossary
- PcG
Polycomb group
- trxG
trithorax group
- DAG
days after germination
- LD
long-day
- SD
short-day
- RT
reverse transcription
- MS
Murashige and Skoog
- qPCR
quantitative PCR
- ChIP
chromatin immunoprecipitation
- aRNA
amino allyl-modified RNA
- cDNA
complementary DNA
References
- Alvarez-Venegas R, Avramova Z. (2001) Two Arabidopsis homologs of the animal trithorax genes: a new structural domain is a signature feature of the trithorax gene family. Gene 271: 215–221 [DOI] [PubMed] [Google Scholar]
- Alvarez-Venegas R, Pien S, Sadder M, Witmer X, Grossniklaus U, Avramova Z. (2003) ATX-1, an Arabidopsis homolog of trithorax, activates flower homeotic genes. Curr Biol 13: 627–637 [DOI] [PubMed] [Google Scholar]
- Aubert D, Chen LJ, Moon YH, Martin D, Castle LA, Yang CH, Sung ZR. (2001) EMF1, a novel protein involved in the control of shoot architecture and flowering in Arabidopsis. Plant Cell 13: 1865–1875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumbusch LO, Thorstensen T, Krauss V, Fischer A, Naumann K, Assalkhou R, Schulz I, Reuter G, Aalen RB. (2001) The Arabidopsis thaliana genome contains at least 29 active genes encoding SET domain proteins that can be assigned to four evolutionarily conserved classes. Nucleic Acids Res 29: 4319–4333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beh LY, Colwell LJ, Francis NJ. (2012) A core subunit of Polycomb repressive complex 1 is broadly conserved in function but not primary sequence. Proc Natl Acad Sci USA 109: E1063–E1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, et al. (2006) A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell 125: 315–326 [DOI] [PubMed] [Google Scholar]
- Blázquez MA, Soowal LN, Lee I, Weigel D. (1997) LEAFY expression and flower initiation in Arabidopsis. Development 124: 3835–3844 [DOI] [PubMed] [Google Scholar]
- Bottomley MJ, Collard MW, Huggenvik JI, Liu ZH, Gibson TJ, Sattler M. (2001) The SAND domain structure defines a novel DNA-binding fold in transcriptional regulation. Nat Struct Biol 8: 626–633 [DOI] [PubMed] [Google Scholar]
- Bowler C, Benvenuto G, Laflamme P, Molino D, Probst AV, Tariq M, Paszkowski J. (2004) Chromatin techniques for plant cells. Plant J 39: 776–789 [DOI] [PubMed] [Google Scholar]
- Bratzel F, López-Torrejón G, Koch M, Del Pozo JC, Calonje M. (2010) Keeping cell identity in Arabidopsis requires PRC1 RING-finger homologs that catalyze H2A monoubiquitination. Curr Biol 20: 1853–1859 [DOI] [PubMed] [Google Scholar]
- Calonje M, Sanchez R, Chen LJ, Sung ZR. (2008) EMBRYONIC FLOWER1 participates in polycomb group-mediated AG gene silencing in Arabidopsis. Plant Cell 20: 277–291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carles CC, Choffnes-Inada D, Reville K, Lertpiriyapong K, Fletcher JC. (2005) ULTRAPETALA1 encodes a SAND domain putative transcriptional regulator that controls shoot and floral meristem activity in Arabidopsis. Development 132: 897–911 [DOI] [PubMed] [Google Scholar]
- Carles CC, Fletcher JC. (2009) The SAND domain protein ULTRAPETALA1 acts as a trithorax group factor to regulate cell fate in plants. Genes Dev 23: 2723–2728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carles CC, Lertpiriyapong K, Reville K, Fletcher JC. (2004) The ULTRAPETALA1 gene functions early in Arabidopsis development to restrict shoot apical meristem activity and acts through WUSCHEL to regulate floral meristem determinacy. Genetics 167: 1893–1903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castillejo C, Romera-Branchat M, Pelaz S. (2005) A new role of the Arabidopsis SEPALLATA3 gene revealed by its constitutive expression. Plant Journal 43: 586–596 [DOI] [PubMed] [Google Scholar]
- Castillejo C, Pelaz S. (2008) The balance between CONSTANS and TEMPRANILLO activities determines FT expression to trigger flowering. Curr Biol 18: 1338–1343 [DOI] [PubMed] [Google Scholar]
- Chanvivattana Y, Bishopp A, Schubert D, Stock C, Moon YH, Sung ZR, Goodrich J. (2004) Interaction of Polycomb-group proteins controlling flowering in Arabidopsis. Development 131: 5263–5276 [DOI] [PubMed] [Google Scholar]
- Chaudhury AM, Koltunow A, Payne T, Luo M, Tucker MR, Dennis ES, Peacock WJ. (2001) Control of early seed development. Annu Rev Cell Dev Biol 17: 677–699 [DOI] [PubMed] [Google Scholar]
- Chen L, Cheng JC, Castle L, Sung ZR. (1997) EMF genes regulate Arabidopsis inflorescence development. Plant Cell 9: 2011–2024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Folter S, Immink RG, Kieffer M, Parenicová L, Henz SR, Weigel D, Busscher M, Kooiker M, Colombo L, Kater MM, et al (2005) Comprehensive interaction map of the Arabidopsis MADS box transcription factors. Plant Cell 17: 1424–1433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Lucia F, Crevillen P, Jones AME, Greb T, Dean C. (2008) A PHD-polycomb repressive complex 2 triggers the epigenetic silencing of FLC during vernalization. Proc Natl Acad Sci USA 105: 16831–16836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrona S, Thorpe FL, Engelhorn J, Adrian J, Dong X, Sarid-Krebs L, Goodrich J, Turck F. (2011) Tissue-specific expression of FLOWERING LOCUS T in Arabidopsis is maintained independently of polycomb group protein repression. Plant Cell 23: 3204–3214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher JC. (2001) The ULTRAPETALA gene controls shoot and floral meristem size in Arabidopsis. Development 128: 1323–1333 [DOI] [PubMed] [Google Scholar]
- Gendall AR, Levy YY, Wilson A, Dean C. (2001) The VERNALIZATION 2 gene mediates the epigenetic regulation of vernalization in Arabidopsis. Cell 107: 525–535 [DOI] [PubMed] [Google Scholar]
- Gomez-Mena C, de Folter S, Costa MM, Angenent GC, Sablowski R. (2005) Transcriptional program controlled by the floral homeotic gene AGAMOUS during early organogenesis. Development 132: 429–438 [DOI] [PubMed] [Google Scholar]
- Goodrich J, Puangsomlee P, Martin M, Long D, Meyerowitz EM, Coupland G. (1997) A Polycomb-group gene regulates homeotic gene expression in Arabidopsis. Nature 386: 44–51 [DOI] [PubMed] [Google Scholar]
- Henikoff S, Shilatifard A. (2011) Histone modification: cause or cog? Trends Genet 27: 389–396 [DOI] [PubMed] [Google Scholar]
- Hennig L, Derkacheva M. (2009) Diversity of Polycomb group complexes in plants: same rules, different players? Trends Genet 25: 414–423 [DOI] [PubMed] [Google Scholar]
- Holec S, Berger F. (2012) Polycomb group complexes mediate developmental transitions in plants. Plant Physiol 158: 35–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honma T, Goto K. (2001) Complexes of MADS-box proteins are sufficient to convert leaves into floral organs. Nature 409: 525–529 [DOI] [PubMed] [Google Scholar]
- Jiang Y, Yang B, Harris NS, Deyholos MK. (2007) Comparative proteomic analysis of NaCl stress-responsive proteins in Arabidopsis roots. J Exp Bot 58: 3591–3607 [DOI] [PubMed] [Google Scholar]
- Jurgens G. (1985) A group of genes controlling the spatial expression of the bithorax complex in Drosophila. Nature 316: 153–155 [Google Scholar]
- Kania T, Russenberger D, Peng S, Apel K, Melzer S. (1997) FPF1 promotes flowering in Arabidopsis. Plant Cell 9: 1327–1338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SY, Lee J, Eshed-Williams L, Zilberman D, Sung ZR. (2012) EMF1 and PRC2 cooperate to repress key regulators of Arabidopsis development. PLoS Genet 8: e1002512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SY, Zhu T, Sung ZR. (2010) Epigenetic regulation of gene programs by EMF1 and EMF2 in Arabidopsis. Plant Physiol 152: 516–528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita T, Harada JJ, Goldberg RB, Fischer RL. (2001) Polycomb repression of flowering during early plant development. Proc Natl Acad Sci USA 98: 14156–14161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klymenko T, Müller J. (2004) The histone methyltransferases Trithorax and Ash1 prevent transcriptional silencing by Polycomb group proteins. EMBO Rep 5: 373–377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köhler C, Hennig L, Bouveret R, Gheyselinck J, Grossniklaus U, Gruissem W. (2003) Arabidopsis MSI1 is a component of the MEA/FIE Polycomb group complex and required for seed development. EMBO J 22: 4804–4814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krizek BA, Meyerowitz EM. (1996) The Arabidopsis homeotic genes APETALA3 and PISTILLATA are sufficient to provide the B class organ identity function. Development 122: 11–22 [DOI] [PubMed] [Google Scholar]
- Krizek BA, Fletcher JC. (2005) Molecular mechanisms of flower development: an armchair guide. Nat Rev Genet 6: 688–698 [DOI] [PubMed] [Google Scholar]
- Lewis EB. (1978) A gene complex controlling segmentation in Drosophila. Nature 276: 565–570 [DOI] [PubMed] [Google Scholar]
- Liu C, Chen H, Er HL, Soo HM, Kumar PP, Han JH, Liou YC, Yu H. (2008) Direct interaction of AGL24 and SOC1 integrates flowering signals in Arabidopsis. Development 135: 1481–1491 [DOI] [PubMed] [Google Scholar]
- Lopez-Vernaza M, Yang S, Müller R, Thorpe F, de Leau E, Goodrich J. (2012) Antagonistic roles of SEPALLATA3, FT and FLC genes as targets of the polycomb group gene CURLY LEAF. PLoS ONE 7: e30715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melzer S, Kampmann G, Chandler J, Apel K. (1999) FPF1 modulates the competence to flowering in Arabidopsis. Plant J 18: 395–405 [DOI] [PubMed] [Google Scholar]
- Michaels SD, Amasino RM. (1999) FLOWERING LOCUS C encodes a novel MADS domain protein that acts as a repressor of flowering. Plant Cell 11: 949–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaels SD, Ditta G, Gustafson-Brown C, Pelaz S, Yanofsky M, Amasino RM. (2003) AGL24 acts as a promoter of flowering in Arabidopsis and is positively regulated by vernalization. Plant J 33: 867–874 [DOI] [PubMed] [Google Scholar]
- Mizukami Y, Ma H. (1992) Ectopic expression of the floral homeotic gene AGAMOUS in transgenic Arabidopsis plants alters floral organ identity. Cell 71: 119–131 [DOI] [PubMed] [Google Scholar]
- Moon YH, Chen L, Pan RL, Chang HS, Zhu T, Maffeo DM, Sung ZR. (2003) EMF genes maintain vegetative development by repressing the flower program in Arabidopsis. Plant Cell 15: 681–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mylne JS, Barrett L, Tessadori F, Mesnage S, Johnson L, Bernatavichute YV, Jacobsen SE, Fransz P, Dean C. (2006) LHP1, the Arabidopsis homologue of HETEROCHROMATIN PROTEIN1, is required for epigenetic silencing of FLC. Proc Natl Acad Sci USA 103: 5012–5017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng DW, Wang T, Chandrasekharan MB, Aramayo R, Kertbundit S, Hall TC. (2007) Plant SET domain-containing proteins: structure, function and regulation. Biochim Biophys Acta 1769: 316–329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papp B, Müller J. (2006) Histone trimethylation and the maintenance of transcriptional ON and OFF states by trxG and PcG proteins. Genes Dev 20: 2041–2054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parcy F, Bomblies K, Weigel D. (2002) Interaction of LEAFY, AGAMOUS and TERMINAL FLOWER1 in maintaining floral meristem identity in Arabidopsis. Development 129: 2519–2527 [DOI] [PubMed] [Google Scholar]
- Pien S, Fleury D, Mylne JS, Crevillen P, Inzé D, Avramova Z, Dean C, Grossniklaus U. (2008) ARABIDOPSIS TRITHORAX1 dynamically regulates FLOWERING LOCUS C activation via histone 3 lysine 4 trimethylation. Plant Cell 20: 580–588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poux S, Horard B, Sigrist CJA, Pirrotta V. (2002) The Drosophila trithorax protein is a coactivator required to prevent re-establishment of polycomb silencing. Development 129: 2483–2493 [DOI] [PubMed] [Google Scholar]
- Pu L, Li Q, Fan XP, Yang WC, Xue YB. (2008) The R2R3 MYB transcription factor GhMYB109 is required for cotton fiber development. Genetics 180: 811–820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratcliffe OJ, Kumimoto RW, Wong BJ, Riechmann JL. (2003) Analysis of the Arabidopsis MADS AFFECTING FLOWERING gene family: MAF2 prevents vernalization by short periods of cold. Plant Cell 15: 1159–1169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saleh A, Al-Abdallat A, Ndamukong I, Alvarez-Venegas R, Avramova Z. (2007) The Arabidopsis homologs of trithorax (ATX1) and enhancer of zeste (CLF) establish ‘bivalent chromatin marks’ at the silent AGAMOUS locus. Nucleic Acids Res 35: 6290–6296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez R, Kim MY, Calonje M, Moon YH, Sung ZR. (2009) Temporal and spatial requirement of EMF1 activity for Arabidopsis vegetative and reproductive development. Mol Plant 2: 643–653 [DOI] [PubMed] [Google Scholar]
- Sanchez-Pulido L, Devos D, Sung ZR, Calonje M. (2008) RAWUL: a new ubiquitin-like domain in PRC1 ring finger proteins that unveils putative plant and worm PRC1 orthologs. BMC Genomics 9: 308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitges FW, Prusty AB, Faty M, Stützer A, Lingaraju GM, Aiwazian J, Sack R, Hess D, Li L, Zhou SL, et al. (2011) Histone methylation by PRC2 is inhibited by active chromatin marks. Mol Cell 42: 330–341 [DOI] [PubMed] [Google Scholar]
- Schuettengruber B, Chourrout D, Vervoort M, Leblanc B, Cavalli G. (2007) Genome regulation by polycomb and trithorax proteins. Cell 128: 735–745 [DOI] [PubMed] [Google Scholar]
- Schwartz YB, Pirrotta V. (2007) Polycomb silencing mechanisms and the management of genomic programmes. Nat Rev Genet 8: 9–22 [DOI] [PubMed] [Google Scholar]
- Schwarz S, Grande AV, Bujdoso N, Saedler H, Huijser P. (2008) The microRNA regulated SBP-box genes SPL9 and SPL15 control shoot maturation in Arabidopsis. Plant Mol Biol 67: 183–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung ZR, Belachew A, Shunong B, Bertrand-Garcia R. (1992) EMF, an Arabidopsis gene required for vegetative shoot development. Science 258: 1645–1647 [DOI] [PubMed] [Google Scholar]
- Tamada Y, Yun JY, Woo SC, Amasino RM. (2009) ARABIDOPSIS TRITHORAX-RELATED7 is required for methylation of lysine 4 of histone H3 and for transcriptional activation of FLOWERING LOCUS C. Plant Cell 21: 3257–3269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teper-Bamnolker P, Samach A. (2005) The flowering integrator FT regulates SEPALLATA3 and FRUITFULL accumulation in Arabidopsis leaves. Plant Cell 17: 2661–2675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- To A, Valon C, Savino G, Guilleminot J, Devic M, Giraudat J, Parcy F. (2006) A network of local and redundant gene regulation governs Arabidopsis seed maturation. Plant Cell 18: 1642–1651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran LS, Nakashima K, Sakuma Y, Simpson SD, Fujita Y, Maruyama K, Fujita M, Seki M, Shinozaki K, Yamaguchi-Shinozaki K. (2004) Isolation and functional analysis of Arabidopsis stress-inducible NAC transcription factors that bind to a drought-responsive cis-element in the early responsive to dehydration stress 1 promoter. Plant Cell 16: 2481–2498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turck F, Roudier F, Farrona S, Martin-Magniette ML, Guillaume E, Buisine N, Gagnot S, Martienssen RA, Coupland G, Colot V. (2007) Arabidopsis TFL2/LHP1 specifically associates with genes marked by trimethylation of histone H3 lysine 27. PLoS Genet 3: 855–866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner D, Sablowski RW, Meyerowitz EM. (1999) Transcriptional activation of APETALA1 by LEAFY. Science 285: 582–584 [DOI] [PubMed] [Google Scholar]
- Wang JW, Czech B, Weigel D. (2009) miR156-regulated SPL transcription factors define an endogenous flowering pathway in Arabidopsis thaliana. Cell 138: 738–749 [DOI] [PubMed] [Google Scholar]
- Weigel D, Alvarez J, Smyth DR, Yanofsky MF, Meyerowitz EM. (1992) LEAFY controls floral meristem identity in Arabidopsis. Cell 69: 843–859 [DOI] [PubMed] [Google Scholar]
- Winter CM, Austin RS, Blanvillain-Baufumé S, Reback MA, Monniaux M, Wu MF, Sang Y, Yamaguchi A, Yamaguchi N, Parker JE, et al. (2011) LEAFY target genes reveal floral regulatory logic, cis motifs, and a link to biotic stimulus response. Dev Cell 20: 430–443 [DOI] [PubMed] [Google Scholar]
- Wood CC, Robertson M, Tanner G, Peacock WJ, Dennis ES, Helliwell CA. (2006) The Arabidopsis thaliana vernalization response requires a polycomb-like protein complex that also includes VERNALIZATION INSENSITIVE 3. Proc Natl Acad Sci USA 103: 14631–14636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu MF, Sang Y, Bezhani S, Yamaguchi N, Han SK, Li Z, Su Y, Slewinski TL, Wagner D. (2012) SWI2/SNF2 chromatin remodeling ATPases overcome polycomb repression and control floral organ identity with the LEAFY and SEPALLATA3 transcription factors. Proc Natl Acad Sci USA 109: 3576–3581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L, Shen WH. (2008) Polycomb silencing of KNOX genes confines shoot stem cell niches in Arabidopsis. Curr Biol 18: 1966–1971 [DOI] [PubMed] [Google Scholar]
- Yang CH, Chen LJ, Sung ZR. (1995) Genetic regulation of shoot development in Arabidopsis: role of the EMF genes. Dev Biol 169: 421–435 [DOI] [PubMed] [Google Scholar]
- Yoshida N, Yanai Y, Chen LJ, Kato Y, Hiratsuka J, Miwa T, Sung ZR, Takahashi S. (2001) EMBRYONIC FLOWER2, a novel polycomb group protein homolog, mediates shoot development and flowering in Arabidopsis. Plant Cell 13: 2471–2481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H, Ito T, Wellmer F, Meyerowitz EM. (2004) Repression of AGAMOUS-LIKE 24 is a crucial step in promoting flower development. Nat Genet 36: 157–161 [DOI] [PubMed] [Google Scholar]
- Zhang X, Bernatavichute YV, Cokus S, Pellegrini M, Jacobsen SE. (2009) Genome-wide analysis of mono-, di- and trimethylation of histone H3 lysine 4 in Arabidopsis thaliana. Genome Biol 10: R62.1–62.14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Germann S, Blus BJ, Khorasanizadeh S, Gaudin V, Jacobsen SE. (2007) The Arabidopsis LHP1 protein colocalizes with histone H3 Lys27 trimethylation. Nat Struct Mol Biol 14: 869–871 [DOI] [PubMed] [Google Scholar]