A transcription factor is expressed in proliferating cells and affects shoot architecture.
Abstract
We report about ERF BUD ENHANCER (EBE; At5g61890), a transcription factor that affects cell proliferation as well as axillary bud outgrowth and shoot branching in Arabidopsis (Arabidopsis thaliana). EBE encodes a member of the APETALA2/ETHYLENE RESPONSE FACTOR (AP2/ERF) transcription factor superfamily; the gene is strongly expressed in proliferating cells and is rapidly and transiently up-regulated in axillary meristems upon main stem decapitation. Overexpression of EBE promotes cell proliferation in growing calli, while the opposite is observed in EBE-RNAi lines. EBE overexpression also stimulates axillary bud formation and outgrowth, while repressing it results in inhibition of bud growth. Global transcriptome analysis of estradiol-inducible EBE overexpression lines revealed 48 EBE early-responsive genes, of which 14 were up-regulated and 34 were down-regulated. EBE activates several genes involved in cell cycle regulation and dormancy breaking, including D-type cyclin CYCD3;3, transcription regulator DPa, and BRCA1-ASSOCIATED RING DOMAIN1. Among the down-regulated genes were DORMANCY-ASSOCIATED PROTEIN1 (AtDRM1), AtDRM1 homolog, MEDIATOR OF ABA-REGULATED DORMANCY1, and ZINC FINGER HOMEODOMAIN5. Our data indicate that the effect of EBE on shoot branching likely results from an activation of genes involved in cell cycle regulation and dormancy breaking.
The aerial shoot system and in particular the branching habit are central elements of the three-dimensional plant structure (Sussex and Kerk, 2001; McSteen and Leyser, 2005). Branch patterns vary widely within and between species; they contribute to the diversity of plant life forms and their ability to populate different ecological niches (Bonser and Aarssen, 2003). Plant architecture and, hence, the branching pattern is also an important attribute to modern crop species; changing the branching habit can help to optimize the use of resources for enhanced growth, biomass accumulation, and seed or fruit yield (García del Moral and García del Moral, 1995; Zhao et al., 2006; Doust, 2007; Boe and Beck, 2008).
The branching habit shows remarkable developmental plasticity controlled by an intricate interplay of various hormones, including auxin, strigolactone, and cytokinin (Wang and Li, 2006, 2008; Beveridge and Kyozuka, 2010; Domagalska and Leyser, 2011), and is also affected by many environmental factors, including light, temperature, humidity, and nutrient availability (Reinhardt and Kuhlemeier, 2002). Axillary shoot development begins with the initiation of an axillary meristem in the axil of a leaf to form a bud through subsequent cell proliferation. Depending on internal and environmental signals, the bud may then grow out farther to form an axillary shoot or (transiently) stop further cell division and remain in a dormant state before cell proliferation may resume later (Shimizu-Sato and Mori, 2001; Kebrom et al., 2006). Thus, molecular mechanisms coordinating progression through the cell cycle are expected to be of great relevance for bud formation and outgrowth (Horvath et al., 2003; Tatematsu et al., 2005).
Various experimental approaches have been used to study the regulation of axillary shoot development. Classical decapitation studies and application of phytohormones showed that auxin plays a central role in controlling shoot branching. Recent findings demonstrate that auxin moving down the plant not only promotes the production of strigolactone by stimulating the expression of strigolactone biosynthesis genes (Foo et al., 2005; Hayward et al., 2009) but also negatively regulates cytokinin content and import (to the bud) to suppress bud outgrowth (second messenger model; Li et al., 1995; Tanaka et al., 2006). Moreover, auxin export from the bud appears to be critical for its outgrowth (auxin transport canalization model; Domagalska and Leyser, 2011). Nevertheless, recent studies have proposed the existence of a rapid, indole acetic acid-independent signal acting as the trigger for bud outgrowth after decapitation. The signal might be a physical response to a change in turgor pressure or an electrical potential, rather than a chemical compound (Ferguson and Beveridge, 2009; Renton et al., 2012).
Although much has been learned about the hormonal control of shoot branching, little is currently known about regulatory processes acting in the bud itself. One of the most prominent bud-specific regulators is TEOSINTE BRANCHED1 (TB1) from maize (Zea mays; Doebley et al., 1997), a member of the plant-specific TCP (for TB1, CYCLOIDEA, and PCF domain) family of transcription factors (Martín-Trillo and Cubas, 2010); similarly, TB1 homologs from other species, including BRANCHED1 (BRC1 [At3g18550]; also called TEOSINTE BRANCHED1-LIKE1 [TBL1] or TCP18; Aguilar-Martínez et al., 2007; Finlayson, 2007) and BRC2 (TCP12) in Arabidopsis (Arabidopsis thaliana) and SlBRC1b in tomato (Solanum lycopersicum; Martín-Trillo et al., 2011; for review, see Domagalska and Leyser, 2011), control shoot branching. Generally, expression of these genes is largely restricted to axillary meristems and buds, and loss of functional TB1, BRC1, or BRC2 genes leads to increased branching, while overexpressing TB1 homologs inhibits branching (Aguilar-Martínez et al., 2007; Finlayson, 2007; Martín-Trillo et al., 2011).
Most cells in dormant axillary buds are arrested at the G1 phase of the cell cycle (Devitt and Stafstrom, 1995; Shimizu and Mori, 1998), and experimental evidence suggests that TCP transcription factors regulate the transition from the G1 to the S phase (Kosugi and Ohashi, 1997). In dormant axillary buds of pea (Pisum sativum), transcript levels of various cell cycle-related genes, such as PROLIFERATING CELL NUCLEAR ANTIGEN, CYCB1;2 (B-type cyclin), CYCD3;1 (D-type cyclin), and CELL DIVISION CYCLE2, were very low, but expression of all genes increased after decapitation in a cell cycle-dependent fashion (Shimizu and Mori, 1998; Shimizu-Sato and Mori, 2001). Global expression profiling confirmed the up-regulation of many cell cycle-related genes in axillary shoots upon main stem decapitation, in addition to genes encoding ribosomal proteins (Tatematsu et al., 2005). In addition, other genes, often of vaguely defined molecular or biochemical functions, are affected in axillary buds by main stem decapitation. In Arabidopsis, expression of the dormancy-associated genes AtDRM1 (At1g28330) and AtDRM1 homolog (At2g33830) is strongly down-regulated within the first 12 to 24 h after main shoot decapitation, while expression of both genes increases again thereafter (Tatematsu et al., 2005). Similarly, expression of the DRM1 ortholog from pea (PsDRM1) strongly declines within several hours of decapitation, while it rapidly resumes when buds become dormant again, revealing it as a good dormancy marker (Stafstrom et al., 1998).
Here, we report about ERF BUD ENHANCER (EBE; At5g61890), a new transcriptional regulator of -cell proliferation and axillary bud outgrowth in Arabidopsis. EBE is a member of the APETALA2/ETHYLENE RESPONSE FACTOR (AP2/ERF) transcription factor superfamily that encompasses 147 members in Arabidopsis (Nakano et al., 2006). The EBE gene is strongly expressed in proliferating cells, with preferential expression during the S phase of the cell cycle. Overexpression of EBE promotes cell proliferation, leading to enhanced callus growth, while the opposite is observed in EBE-RNAi lines. In addition, EBE overexpression in transgenic plants stimulates axillary bud formation and outgrowth, while its repression inhibits bud growth. The effect of EBE on shoot branching likely results from affecting genes involved in cell cycle regulation and dormancy breaking.
RESULTS
EBE Is Prominently Expressed in Proliferating Cells
To discover transcription factors controlling vegetative development, we screened transgenic Arabidopsis lines ectopically expressing transcription factors under the control of the cauliflower mosaic virus (CaMV) 35S promoter. One of the lines that attracted our attention overexpressed an uncharacterized AP2/ERF transcription factor encoded by gene locus At5g61890. As we show below, At5g61890 promotes axillary bud growth; therefore, we named it EBE. EBE belongs to group Xa of the ERF family of the AP2/ERF superfamily; group Xa includes six genes in Arabidopsis and seven genes in rice (Oryza sativa; Nakano et al., 2006). Like other members of ERF group Xa, EBE harbors a characteristic CMX-1 motif of unknown function in its N-terminal region.
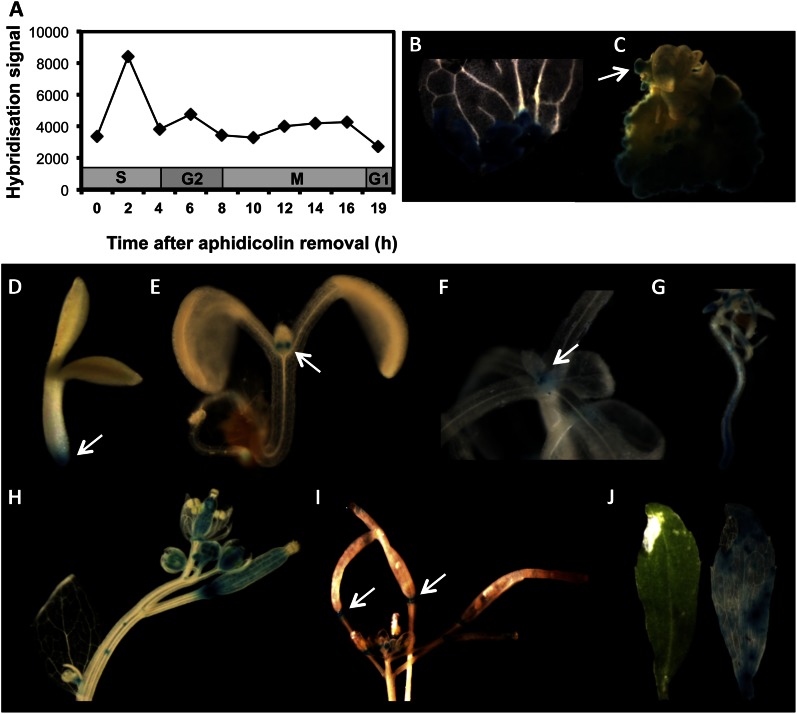
By analyzing public transcriptome data (Genevestigator; Zimmermann et al., 2004), we found that expression of EBE is particularly strong in undifferentiated cells of suspension cultures and in calli. In a global transcriptome data set of Menges et al. (2003), we observed preferential EBE expression during the S phase of the cell cycle, whereas expression was low in other phases (Fig. 1A). To test whether EBE expression in proliferating cells is controlled at the promoter level, we fused its approximately 2-kb promoter to the GUS reporter gene and tested GUS activity in transgenic PromEBE:GUS Arabidopsis plants. Cotyledon segments of three independent PromEBE:GUS lines were incubated on callus-induction medium (CIM). Strong GUS activity was observed in callus (Fig. 1B), while GUS activity was weaker in regenerating shoots (Fig. 1C), and no GUS activity was detected in young regenerating roots. Similarly, in a global transcriptome analysis using Affymetrix ATH1 microarrays, EBE was up-regulated during callus formation 4 and 7 d after incubation in CIM (Che et al., 2006), and it was approximately 3.5-fold up-regulated in Agrobacterium tumefaciens-induced Arabidopsis tumors when compared with tumor-free inflorescence stalk tissue (Deeken et al., 2006). Rapid cell proliferation in tumors is stimulated by high concentrations of cytokinin and auxin synthesized by transfer DNA (T-DNA)-encoded bacterial enzymes. Indeed, tumor growth in plants is reminiscent of callus formation in CIM, supporting our above results. Similarly, EBE expression increased by approximately 10-fold in regenerating stumps of root tips within 5 h after removal of the tip (Sena et al., 2009), and it increased to a similar extent in callus derived from cotyledons and petals (Sugimoto et al., 2010). Using PromEBE:GUS plants, we observed strong GUS staining several hours after wounding of leaves, when callus formation resumed at wound sites (data not shown). Collectively, these data demonstrate that EBE expression is particularly prominent in proliferating cells.
Figure 1.
EBE expression. A, EBE expression during cell cycle progression. Data were extracted from Menges et al. (2003), using Genevestigator. B to J, EBE promoter-driven GUS activity. B and C, GUS activity in calli and regenerating shoot (arrow in C). D and E, Three- and 9-d-old seedlings, respectively. Note GUS staining in the root tip and stipules (arrows). F, Shoot, with staining at the apex (arrow). G, Roots. H, Flowers and young siliques. I, Old siliques, with GUS staining in abscission zones (arrows). J, Senescent leaf: left, unstained; right, GUS stained. Incubation in GUS staining solution was done overnight.
We also analyzed the expression pattern of EBE in whole plants. In 3- to 8-d-old seedlings, GUS activity was detected in root tips (Fig. 1D), stipules (Fig. 1E), and the shoot apex (Fig. 1F). Prominent EBE expression was also observed in tips of lateral roots (Fig. 1G) and various floral tissues (Fig. 1H). Comparatively, strong EBE expression was observed in young siliques (Fig. 1H), while expression in old siliques was largely restricted to abscission zones (Fig. 1I). Weak EBE expression was normally observed during early leaf development, whereas it was virtually absent in bigger leaves but resumed upon leaf senescence (Fig. 1J). We also measured EBE transcript abundance by quantitative reverse transcription (qRT)-PCR in immature and partially (approximately 20%) senescent leaves and found approximately 10-fold higher expression in the latter (data not shown). In some plant lines, hypocotyls and a small area of the cotyledon tip and petioles also showed PromEBE-driven GUS activity.
EBE Overexpression Promotes Callus Growth and Triggers Neoplastic Activity
To assess the in vivo function of EBE, we generated 35S:EBE overexpression lines and chose three representative lines (#11, #21, and #28) for detailed studies. Additionally, we inhibited EBE expression by RNA interference (RNAi) and selected lines (including EBE-RNAi #10, #22 and #17) with strongly reduced EBE transcript abundance in rosette leaves (see below) for analysis.
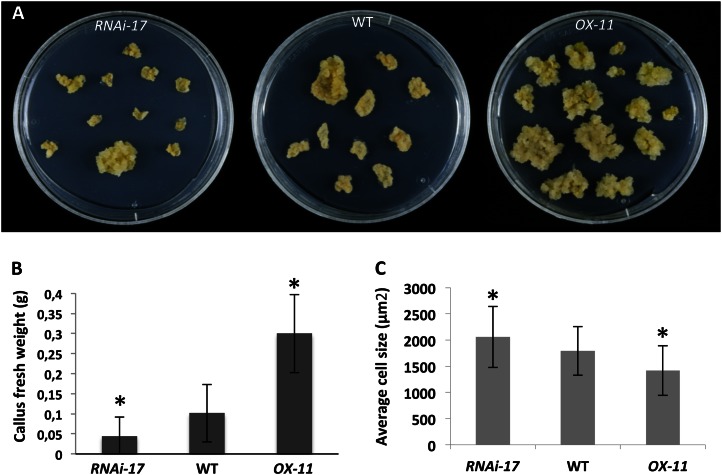
The prominent expression of EBE in proliferating cells and calli prompted us to analyze the effect of EBE overexpression and inhibition on callus formation and shoot and root development. First, cotyledon or young leaf segments obtained from the transgenic 35S:EBE and EBE-RNAi lines as well as from Arabidopsis ecotype Columbia (Col-0) plants were precultured on CIM for 4 d and then transferred to shoot-induction medium, root-induction medium, or fresh CIM for 4 weeks. The sizes and number of the calli formed, the number of adventitious shoots, and root formation were analyzed weekly. With respect to the capacities for shoot or root regeneration, we did not detect significant differences between 35S:EBE, EBE-RNAi, and wild-type plants. However, calli formed from the 35S:EBE lines were considerably larger than those of the wild type, and EBE-RNAi calli were smaller (Fig. 2, A and B). We analyzed cell sizes in 35S:EBE, EBE-RNAi, and wild-type calli and observed smaller cells in EBE overexpressors than in RNAi lines (Fig. 2C); thus, larger calli observed in EBE overexpressors appear to result from more pronounced cell proliferation leading to an increased cell number.
Figure 2.
Effects of altered EBE expression on callus formation. A and B, Overexpression of EBE increases callus size. Data in B represent mean fresh weight ± sd of at least 30 calli, grown in three independent biological experiments. C, Size of cells in calli. Data are means of at least 120 cells each ± sd. In B, asterisks indicate statistically significant differences between the wild type (WT) and either EBE-RNAi-17 or 35S:EBE (OX-11) calli (Student’s t test after false discovery rate correction, P < 0.01). In C, asterisks indicate statistically significant differences between EBE-RNAi-17 and 35S:EBE (OX-11) calli (Student’s t test, P < 0.05).
A prominent feature observed in 35S:EBE plants was neoplasia; tissue similar to green callus often produced “organ-like” structures at wounded sites (or partially senesced tissue) in the absence of external phytohormones (Supplemental Fig. S1, A–C). Upon extended growth, such calli regularly differentiated into organs such as roots, leaves, or shoots (data not shown), while neoplasia was very rarely observed in wild-type plants kept under identical conditions. Cell proliferation and callus formation can be induced from differentiated plant tissues by phytohormone treatment (Mizukami and Fischer, 2000). Apparently, ectopic overexpression of EBE reduces the dependence on external hormones for reentry into the cell cycle and cell proliferation.
EBE Affects Shoot Branching
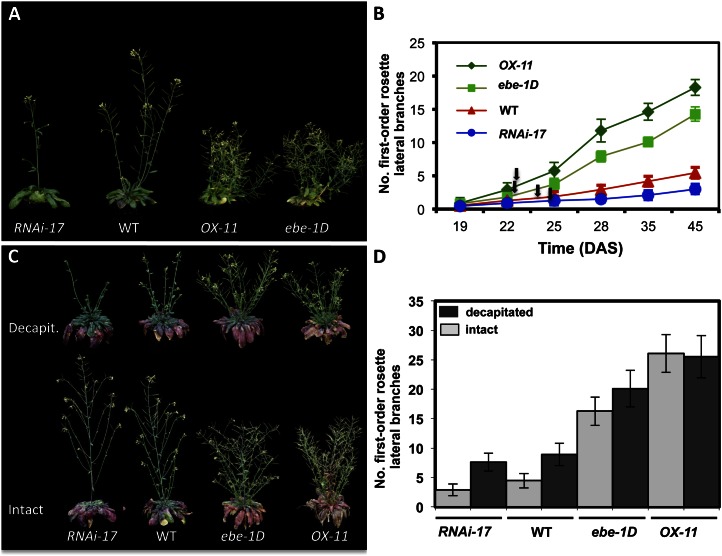
35S:EBE plants remained smaller and produced more first-order lateral rosette branches than the parent wild type (Fig. 3, A and B). After 28 d of growth, wild-type plants displayed around six lateral branches while 35S:EBE lines #21, #11, and #28 displayed about nine, 15, and 20 lateral branches, respectively (Supplemental Fig. S2A). qRT-PCR revealed low EBE expression in leaves of 4-week-old wild-type plants. In 35S:EBE transgenic plants, EBE transcript abundance was positively correlated with the strength of the observed phenotype. EBE expression level determined by qRT-PCR was approximately 10 times lower in line 35S:EBE #21, which displayed a weaker phenotype, than in the strong-phenotype lines #11 and #28 (Supplemental Fig. S2B).
Figure 3.
Phenotypic effects of altered EBE transcript level. A, Phenotypes of 33-d-old transgenic EBE lines compared with wild-type (WT) plants, grown in long-day conditions. B, Number of first-order rosette lateral branches in 35S:EBE (OX-11), ebe-1D, wild-type, and EBE-RNAi-17 plants. Given are means ± sd of 11 different plants of each line at each stage. At time points 28, 35, and 45 d after sowing (DAS), the differences between wild-type and all transgenic plants are statistically significant (Student’s t test after false discovery rate correction, P < 0.01). Arrows indicate flowering time for each line. Flowering time of the transgenic plants did not significantly differ from that of the wild type under our experimental conditions (Student’s t test after false discovery rate correction, P > 0.05; Supplemental Table S5). C, Phenotype of transgenic plants 12 d after decapitation (top) compared with intact plants that were not decapitated (bottom). Plants were 3 months old. D, Number of first-order rosette lateral branches of wild-type and transgenic plants at 12 d after removal of the main shoot, compared with intact plants. In wild-type plants, approximately 50% of the buds in primary rosette branches grew out, whereas in RNAi, ebe-1D, and OX-11 lines, approximately 38%, 80%, and 100%, respectively, of the buds did. Data represent means ± sd (n = 11). Data for intact versus decapitated plants were significantly different for all lines except the strong EBE overexpressor line OX-11 (Student’s t test, P < 0.01).
We next analyzed an Arabidopsis mutant (ebe-1D; SALK_000833) carrying a T-DNA insertion approximately 400 bp upstream of the EBE translation initiation codon (ATG). Homozygous ebe-1D insertion lines, identified by PCR on genomic DNA (Supplemental Fig. S3A), phenotypically resembled 35S:EBE lines (first-order lateral branches were more abundant than in the wild type; Fig. 3, A and B; Supplemental Fig. S3, B and C). In contrast, heterozygous T-DNA insertion lines showed normal growth indistinguishable from the wild type (data not shown). Segregation analysis indicated that ebe-1D had a single T-DNA insertion (data not shown). Expression analysis by qRT-PCR revealed approximately 6-fold elevated EBE transcript abundance in the homozygous ebe-1D mutant compared with the wild type, while EBE expression level was enhanced by up to approximately 10-fold in the 35S:EBE-21 line (Supplemental Fig. S3D). Thus, as in 35S:EBE lines, elevated EBE expression coincided with enhanced formation of lateral branches.
In approximately 4-week-old RNAi lines #10, #22, and #17, EBE transcript level was approximately six, 15, and 27 times, respectively, lower than in Col-0 wild-type plants (Supplemental Fig. S4A). All RNAi lines produced fewer first-order rosette lateral branches than Col-0, and the inhibition level of EBE expression followed the strength of the observed phenotype (Supplemental Fig. S4B). However, the number of first-order (and second-order) cauline branches did not change in EBE transgenic plants compared with the wild type (Supplemental Table S1). Final plant height was reduced in 35S:EBE lines compared with the wild type; a similar observation was made for ebe-1D (Supplemental Table S2), consistent with its elevated EBE expression.
EBE Promotes Axillary Bud Formation and Outgrowth
A well-known developmental phenomenon in plants is apical dominance controlled by the main shoot apical meristem (SAM). Upon decapitation, the inhibitory effect of auxin on axillary buds is removed, resulting in cell proliferation and bud outgrowth. To examine whether the altered number of axillary (i.e. first-order rosette lateral) branches in transgenic plants results from a general effect on lateral bud outgrowth, axillary shoot formation was analyzed in wild-type and transgenic plants before and after main shoot decapitation. In intact (i.e. nondecapitated) plants, the number of axillary shoots was approximately three, five, 16, and 26 in EBE-RNAi-17, the wild type, ebe-1D, and 35S:EBE #11, respectively (Fig. 3, C and D), following the level of EBE expression. To test the effect of decapitation, plants were first grown under short-day conditions for 30 d and then shifted to long-day conditions to induce flowering. Primary shoots were removed at a size of approximately 10 cm. Twelve days after decapitation, wild-type plants had developed approximately nine axillary shoots, whereas EBE-RNAi-17, ebe-1D, and 35S:EBE #11 had produced approximately seven, 20, and 25 axillary shoots, respectively (Fig. 3, C and D). Thus, while additional axillary shoots were formed in wild-type and RNAi plants (about twice as many under decapitation than intact conditions), no additional axillary shoots formed upon decapitation in plants overexpressing EBE (which already had many more axillary shoots in intact plants). The stimulatory effect of decapitation on axillary shoot formation was slightly more pronounced in the RNAi line than in the wild type, indicating that the RNAi-mediated inhibitory effect is partly overcome when expression of the endogenous EBE gene is induced by decapitation (see below). In nondecapitated wild-type plants, more axillary shoots are produced than in RNAi lines, reflecting the higher basal (“decapitation-independent”) level of EBE expression in the wild type. The decapitation-induced effect on axillary shoot formation is then less pronounced in the wild type than in RNAi lines.
To further substantiate the conclusion that EBE affects axillary bud formation, we analyzed the presence of vegetative buds in wild-type and transgenic plants. In accordance with published reports (Grbic, 2005), wild-type plants did not form buds in their cotyledonary nodes. Similarly, buds were absent in cotyledonary nodes of RNAi lines. In contrast, both 35S:EBE and ebe-1D lines produced cotyledonary buds (data not shown).
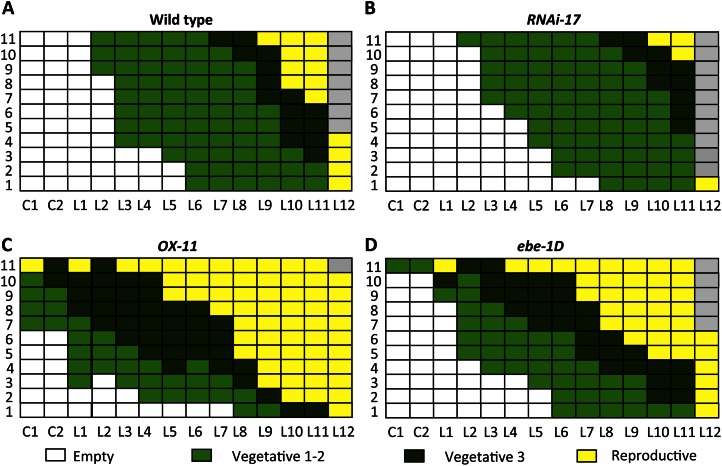
Four weeks after sowing, wild-type and transgenic plants grown under long-day conditions had started flowering, and axillary meristems were initiated in a basipetal order. In wild-type plants, buds closest to the shoot apex were developmentally more advanced than more distant ones; cotyledons and most of the L1 and L2 leaves (which developed after the cotyledons) had no axillary buds, while leaves L3 to L9 generally had buds in vegetative stage 1 or 2 (i.e. did not yet have flowers). One-half of the L10 to L12 leaves contained flowering buds in their axils (Fig. 4A). In contrast, in EBE-RNAi lines, only a few L10, L11, and L12 leaves had flowering buds (Fig. 4B), while in 35S:EBE and ebe-1D plants, flowering buds developed in a high percentage of young leaves (Fig. 4, C and D).
Figure 4.
Developmental stages of buds in the axils of cotyledons (C1 and C2) and rosette leaves (L1–L12) shown on the x axis. The experiment was performed with 11 4-week-old plants grown under long-day conditions, indicated by numbers on the y axis. As EBE-RNAi-17 lines had developed approximately 12 leaves at the analysis time point, data of 12 rosette leaves (L1–L12) are presented for all lines. A, The wild type. B, EBE-RNAi-17. C, 35S:EBE (OX-11). D, ebe-1D. The bud developmental gradient was found to be more pronounced in EBE-RNAi lines than in the wild type but to be less obvious in 35S:EBE and ebe-1D lines. Developmental stages are as follows: vegetative 1, buds with two or more leaf primordia formed, no trichomes, 150 to 250 µm; vegetative 2, mid vegetative stage, buds with differentiating trichome-bearing leaf primordia, less than 400 µm; vegetative 3, late vegetative stage, buds with expanding trichome-bearing leaf primordia, more than 400 µm; reproductive, flower meristems visible within the bud.
In conclusion, our data indicate that EBE influences cellular processes, most likely by affecting cell cycle progression and cell proliferation, during axillary shoot formation in both intact and decapitated plants.
Main Stem Decapitation Stimulates EBE Expression in Axillary Buds
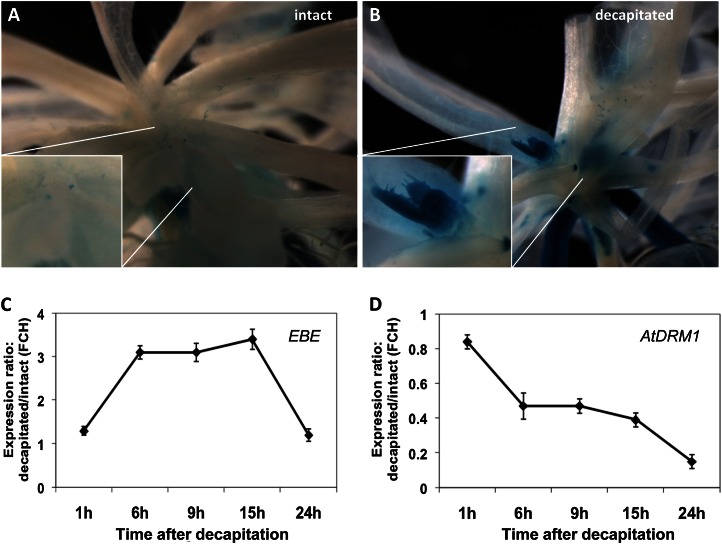
As main stem decapitation promotes axillary bud outgrowth, we tested decapitation-dependent expression of EBE in the PromEBE:GUS lines. Main stems (2–5 cm long) were decapitated when axillary buds were visible. Low EBE promoter-dependent GUS activity was visible in axillary nodes of intact plants, but this activity remarkably increased 6 h after decapitation (Fig. 5, A and B). Decapitation-controlled expression of EBE was confirmed by qRT-PCR in wild-type Arabidopsis plants. EBE transcript abundance was measured in the five uppermost axillary branches developing from either decapitated or intact main stems (control). EBE expression level increased transiently (by up to approximately 3-fold) 6 to 15 h after decapitation of the main stem (Fig. 5C). As a control, we included AtDRM1 (At1g28330), the transcript abundance of which decreased upon decapitation (Fig. 5D), as reported (Tatematsu et al., 2005; Aguilar-Martínez et al., 2007).
Figure 5.
Altered EBE expression before and after decapitation. A and B, GUS staining of axillary shoots before (A) and 6 h after (B) main stem decapitation. C and D, qRT-PCR analysis of EBE (C) and AtDRM1 (D) expression in the five uppermost axillary shoots after main stem decapitation. Symbols in both panels represent means ± sd of three biological experiments. FCH, Fold change.
EBE Contributes to the Maintenance of the SAM and Affects Plastochron
One week after germination, when grown in short-day conditions, we did not observe macroscopic differences between transgenic and wild-type plants, while at 2 weeks, a change in the leaf plastochron (the time interval between the initiation of successive leaves) was evident: leaf initiation occurred in shorter time intervals in 35S:EBE and ebe-1D plants compared with the wild type, reminiscent of a shorter plastochron, whereas plastochron was longer in EBE-RNAi lines (Fig. 6, A and B). Additionally, leaf size remained generally smaller in 35S:EBE #28 and ebe-1D, while it was increased in EBE-RNAi plants compared with the wild type (Fig. 3A; Supplemental Fig. S5A). In accordance with this observation, cell sizes were smaller in 35S:EBE than in wild-type plants (similar to cells in calli, as reported above) and bigger in RNAi plants (Supplemental Fig. S5, B and C).
Figure 6.
Effects of altered EBE expression on plant phenotype. A, EBE up-regulation causes an increase and its down-regulation causes a decrease of the leaf initiation rate as compared with the wild type (WT). Threads of the following colors were used for labeling rosette leaves: orange for leaves L1 and L2, yellow for leaves L3 and L4, and white for leaves L5 and L6. B, Number of rosette leaves of transgenic and wild-type plants during a period of 4 weeks. The symbols represent means ± sd of 11 plants at each time point. At 30 d after sowing (DAS), there is a statistically significant difference between the wild type and all transgenic plants (Student’s t test after false discovery rate correction, P < 0.05). C, Altered phyllotaxy and SAM size in transgenic plants compared with the wild type. The left panel shows an enlarged SAM in a 35S:EBE plant (OX-11) that had arrested without developing further. The right panel shows an arrested SAM in the EBE-RNAi-50 line that occurred after cotyledon development. The three middle panels show the changed phyllotaxy of transgenic plants in comparison with the wild type. In some of the 35S:EBE plants, the angle between the two first true leaves was only about 120°, while in EBE-RNAi plants, the angle was more than 150°C.
It is well established in Arabidopsis that cotyledons and the first pair of true leaves emerge in an approximately decussate (180°) arrangement (Mündermann et al., 2005). We observed that phyllotaxy of the first pair of true leaves was altered in plants strongly overexpressing EBE (Fig. 6C): of 38 35S:EBE-21 plants analyzed, the majority (22 plants) had an angle of 120° to 160°, five plants showed an angle of about 100° to 120°, and 11 plants showed an angle close to 180°C. In contrast, of 55 wild-type plants analyzed, only four displayed an angle of less than 180°. Similarly, of 19 ebe-1D plants tested, the majority displayed an angle of 180°, and five showed an angle between 120° and 160°. Thirty-six of 43 EBE-RNAi-17 plants tested had a normal angle of approximately 180°, and seven lines showed an angle of 140° to 160°.
In addition, in some cases, the size of the SAM was abnormally enlarged in 35S:EBE plants (Fig. 6C), so that SAM development arrested after the formation of a few leaves. Development also arrested in some EBE-RNAi lines after cotyledon growth (Fig. 6C). Analysis by scanning electron microscopy revealed that in some 35S:EBE plants, a leaf primordium was initiated but then moved away from the meristem without developing further (Supplemental Fig. S6A). Such abortive leaf primordia were never observed in wild-type plants under our experimental conditions.
We also analyzed GUS staining in the shoot apex of PromEBE:GUS plants. We observed low but still detectable GUS activity in early leaf primordia (Supplemental Fig. S6B). Taken together, these results suggest a likely involvement of EBE in maintaining the structure of the SAM as well as plastochron and phyllotaxy.
EBE-Triggered Changes in Gene Expression
To identify genes downstream of EBE, we generated transgenic Arabidopsis lines expressing EBE under the control of an estradiol-inducible promoter (EBE-XVE lines) using the system of Zuo et al. (2000). Transgenic seedlings precultured on agar-solidified Murashige and Skoog (MS) medium were transferred to liquid MS medium supplemented with 2 μm β-estradiol (EST). Wild-type control plants (Col-0) were treated in the same way. Leaves of the EST-treated plants were harvested after different time points (from 30 min to 20 h) and tested for EBE expression by qRT-PCR; enhanced EBE expression was already detected at the earliest time point. From four different lines analyzed, we selected EBE-XVE #14 (XVE-14) for further experiments (Supplemental Fig. S7A). To test the effect of chemically induced EBE overexpression on early seedling development, we cultured XVE-14 (and Col-0) seedlings for 4 d on MS agar plates and then transferred them to liquid MS medium containing 10 μm EST. As seen in Supplemental Figure S7B, XVE-14 seedlings developed primary leaves more rapidly than wild-type seedlings and produced more lateral roots, further supporting a role of EBE in plant development.
To identify genes affected by EBE, we studied the transcriptomes in shoots of XVE-14 and wild-type seedlings after 45 min of EST (2 μm) treatment using Affymetrix ATH1 arrays (three biological replications each). Genes showing an expression change of greater than 1.5-fold in all three experiments in XVE-14 but not in wild-type plants were considered to be downstream of EBE. We identified nine up-regulated and 26 down-regulated genes in the EBE-inducible line. Expression of some additional genes was significantly altered in two of the three experiments. We tested the expression of these genes in EST-induced XVE-14 and wild-type plants by qRT-PCR (three biological replicates), confirming differential expression of an additional five up-regulated and eight down-regulated genes. Table I lists all 48 genes robustly affected by inducible EBE overexpression.
Table I. List of genes differentially expressed in EST-inducible EBE overexpression line 14 compared with wild-type plants at 45 min after EST induction.
Three independent biological replications were performed. Numbers represent the expression ratio (fold change). Average values and sd are given as well. Genes are ordered according to their fold change. Asterisks are as follows: *transcription factors up-regulated by EBE; **transcription factors down-regulated by EBE. Bio1, Bio2, and Bio3 represent the first, second, and third biological replicates, respectively.
| Affy Identifier | Arabidopsis Genome Initiative Identifier | Annotation | Bio1 | Bio2 | Bio3 | Average | sd |
|---|---|---|---|---|---|---|---|
| 266743_at | At2g02990a | RNS1 (RNase1) | 9.46 | 13.1 | 5.45 | 9.34 | 3.83 |
| 257673_at | At3g20370 | TRAF-like family protein | 2.78 | 13.3 | 2.10 | 6.07 | 6.29 |
| 266294_at | At2g29500a | HSP20-like chaperone superfamily protein | 5.74 | 3.66 | 3.48 | 4.29 | 1.25 |
| 252189_at | At3g50070a | CYCD3;3 (cyclin D3;3) | 3.02 | 2.50 | 2.80 | 2.77 | 0.26 |
| 262680_at | At1g75880 | SGNH hydrolase-type esterase superfamily protein | 3.01 | 2.99 | 2.30 | 2.77 | 0.41 |
| 259173_at | At3g03640 | BGLU25 (β-glucosidase25) | 1.65 | 4.73 | 1.55 | 2.64 | 1.81 |
| 265097_at | at1g04020a | ATBARD1 (BRCA1-associated RING domain1) | 2.83 | 1.94 | 2.76 | 2.51 | 0.49 |
| 251052_at | At5g02470a | DPa; transcription factor* | 2.97 | 2.65 | 1.68 | 2.43 | 0.67 |
| 251181_at | At3g62820 | Invertase/pectin methylesterase inhibitor family protein | 1.75 | 1.99 | 2.20 | 1.98 | 0.23 |
| 264782_at | At1g08810 | AtMYB60; transcription factor* | 2.54 | 1.86 | 1.54 | 1.98 | 0.51 |
| 267628_at | At2g42280 | bHLH130; transcription factor* | 1.72 | 1.70 | 2.10 | 1.84 | 0.23 |
| 249112_at | At5g43780 | APS4 | 1.57 | 1.81 | 1.96 | 1.78 | 0.20 |
| 267305_at | At2g30070 | ATKT1-ATKUP1 (K+ uptake1) | 1.74 | 1.59 | 1.57 | 1.64 | 0.09 |
| 251556_at | At3g58840 | Tropomyosin related | 1.52 | 1.75 | 1.55 | 1.61 | 0.13 |
| 267092_at | At2g38120a | AUX1 (auxin resistant1) | 0.53 | 0.59 | 0.61 | 0.58 | 0.04 |
| 255433_at | At4g03210 | AtXTH9 | 0.54 | 0.61 | 0.57 | 0.57 | 0.04 |
| 246996_at | At5g67420a | LBD37; transcription factor** | 0.65 | 0.57 | 0.50 | 0.57 | 0.08 |
| 246002_at | At5g20740 | Invertase/pectin methylesterase inhibitor family protein | 0.6 | 0.63 | 0.47 | 0.57 | 0.08 |
| 266391_at | At2g41290 | Strictosidine synthase like | 0.53 | 0.50 | 0.61 | 0.55 | 0.06 |
| 261375_at | At1g53160a | SPL4; transcription factor** | 0.65 | 0.44 | 0.56 | 0.55 | 0.10 |
| 253608_at | At4g30290 | AtXTH19 | 0.64 | 0.52 | 0.47 | 0.54 | 0.09 |
| 260560_at | At2g43590 | Chitinase, putative | 0.60 | 0.45 | 0.55 | 0.53 | 0.08 |
| 246906_at | At5g25475 | AP2/B3-like transcription factor** | 0.60 | 0.43 | 0.56 | 0.53 | 0.09 |
| 253815_at | At4g28250a | ATEXPB3 (expansin B3) | 0.62 | 0.31 | 0.64 | 0.52 | 0.19 |
| 255872_at | At2g30360 | CIPK11 | 0.52 | 0.39 | 0.62 | 0.51 | 0.11 |
| 256452_at | At1g75240 | AtHB33; transcription factor** | 0.58 | 0.43 | 0.46 | 0.49 | 0.08 |
| 262986_at | At1g23390 | Kelch repeat-containing F-box family protein | 0.60 | 0.24 | 0.63 | 0.49 | 0.22 |
| 263951_at | At2g35960 | NHL12 (NDR1/HIN1-like12) | 0.55 | 0.36 | 0.55 | 0.48 | 0.11 |
| 266300_at | At2g01420a | PIN4 (pin-formed4) | 0.37 | 0.4 | 0.63 | 0.47 | 0.14 |
| 252063_at | At3g51590 | LTP12 (lipid transfer protein12) | 0.30 | 0.5 | 0.57 | 0.46 | 0.14 |
| 267460_at | At2g33810 | SPL3; transcription factor** | 0.60 | 0.31 | 0.46 | 0.46 | 0.15 |
| 252549_at | At3g45860 | CRK4; Cys-rich RLK | 0.34 | 0.37 | 0.66 | 0.46 | 0.18 |
| 266223_at | At2g28790 | Thaumatin superfamily protein | 0.64 | 0.29 | 0.35 | 0.43 | 0.19 |
| 245265_at | At4g14400 | ACD6 (accelerated cell death6) | 0.57 | 0.09 | 0.56 | 0.41 | 0.27 |
| 248062_at | At5g55450 | Protease inhibitor/seed storage/lipid transfer protein family protein | 0.30 | 0.38 | 0.48 | 0.39 | 0.09 |
| 245668_at | At1g28330a | AtDRM1 (dormancy-associated protein1) | 0.41 | 0.10 | 0.55 | 0.35 | 0.23 |
| 247718_at | At5g59310 | LTP4 (lipid transfer protein4) | 0.20 | 0.42 | 0.41 | 0.34 | 0.12 |
| 251169_at | At3g63210a | MARD1 (mediator of abscisic acid-regulated dormancy1) | 0.30 | 0.27 | 0.40 | 0.32 | 0.07 |
| 253061_at | At4g37610 | BT5 (BTB and TAZ domain protein5); transcription regulator** | 0.24 | 0.44 | 0.23 | 0.30 | 0.12 |
| 252269_at | At3g49580 | LSU1; response to low sulfur1 | 0.35 | 0.27 | 0.27 | 0.29 | 0.05 |
| 260038_at | At1g68875 | Unknown protein | 0.38 | 0.26 | 0.17 | 0.27 | 0.10 |
| 265117_at | At1g62500 | Protease inhibitor/seed storage/lipid transfer protein family protein | 0.30 | 0.26 | 0.19 | 0.25 | 0.05 |
| 256597_at | At3g28500 | 60S acidic ribosomal protein P2 | 0.12 | 0.35 | 0.29 | 0.25 | 0.12 |
| 257421_at | At1g12030 | Similar to unknown protein | 0.19 | 0.14 | 0.44 | 0.25 | 0.16 |
| 267461_at | At2g33830a | AtDRM1 homolog | 0.22 | 0.12 | 0.39 | 0.24 | 0.14 |
| 258675_at | At3g08770 | LTP6 (lipid transfer protein6) | 0.42 | 0.09 | 0.16 | 0.22 | 0.18 |
| 247717_at | At5g59320 | LTP3 (lipid transfer protein3) | 0.13 | 0.15 | 0.23 | 0.17 | 0.05 |
| 259802_at | At1g72260 | THI2.1 (thionin2.1) | 0.10 | 0.02 | 0.10 | 0.07 | 0.04 |
Genes gained from qRT-PCR experiments; other genes were obtained from microarray analyses.
Notably, of the 14 genes rapidly up-regulated by EBE, three were cell cycle related, including D-type cyclin CYCD3;3 (At3g50070; 2.8-fold), transcription regulator DPa (At5g02470; 2.4-fold), and BRCA1-ASSOCIATED RING DOMAIN1 (BARD1; At1g04020; 2.5-fold). CYCD3;3 transcripts accumulate in the early G1 phase during cell cycle reentry (Menges et al., 2005), and dormancy breaking is generally accompanied by increased expression of CYCD genes (Horvath et al., 2003). DPa, along with E2Fa, is expressed in actively dividing tissues of the SAM, young leaf primordia, vascular tissues of maturing leaf primordia, and axillary buds. DPa interacts with the E2Fa transcription factor to form a complex that stimulates the expression of S phase-specific genes, leading to cell division and a delay of cell differentiation (De Veylder et al., 2002). BARD1 has been reported to control SAM organization and maintenance by restricting WUSCHEL (WUS) expression to the organizing center (Han et al., 2008). WUS encodes a homeodomain protein important for stem cell regeneration in the SAM (Mayer et al., 1998). Two further up-regulated genes encode the transcription factors bHLH130 (At2g42280) and AtMYB60 (At1g08810); however, their potential roles in shoot branching are not known.
AtDRM1 (At1g28330), AtDRM1 homolog (At2g33830), and MEDIATOR OF ABA-REGULATED DORMANCY1 (MARD1/SAG102; At3g63210) were strongly down-regulated after EBE induction. Notably, AtDRM1 and AtDRM1 homolog are also remarkably down-regulated upon initiation of bud growth (Tatematsu et al., 2005), while MARD1 has so far only been shown to affect seed dormancy control (He and Gan, 2004).
Several of the down-regulated genes encode lipid transfer proteins (AtLTP3, At5g59320; AtLTP4, At5g59310; AtLTP6, At3g08770; AtLTP12, At3g51590). LTPs are ubiquitous plant lipid-binding proteins proposed to function in various developmental and stress responses, signaling, and cell wall loosening (Kader, 1996; Bakan et al., 2006; Yeats and Rose, 2008). Furthermore, expression of the cell wall-loosening gene AtEXPB3 (At4g28250; encoding expansin) and the xyloglucan endotransglucosylase/hydrolase genes AtXTH9 (At4g03210) and AtXTH19 (At4g30290) was markedly reduced upon EBE induction. Finally, a thaumatin-like protein (At2g28790) located in the cell wall (Bayer et al., 2006) and a putative chitinase (At2g43590) potentially involved in modulating cell expansion (Kwon et al., 2005) were also down-regulated. Collectively, EBE appears to exert part of its function by affecting the expression of genes encoding different types of cell wall-remodeling proteins.
AUX1 (At2g38120), which encodes an auxin influx carrier, was also down-regulated. During leaf initiation, AUX1 contributes to controlling a regular angle of 137.5° between successive primordia (Bainbridge et al., 2008).
Six of the 34 down-regulated genes encode transcription factors (Table I). ZINC-FINGER HOMEODOMAIN5 (ZHD5; At1g75240), also called ARABIDOPSIS THALIANA HOMEOBOX PROTEIN33, has recently been shown to stimulate shoot growth and the formation of larger leaves upon overexpression. Conversely, inhibition of ZHD5 at the protein level by overexpressing an interacting inhibitor peptide (mini zinc finger 1) resulted in dwarfish plants with small leaves (Hong et al., 2011). These phenotypes are consistent with growth phenotypes observed in our experiments, where overexpression of EBE, thereby lowering ZHD5 expression, similarly caused dwarfish plants with small leaves and multiple shoots, and reducing EBE expression by RNAi inhibits axillary bud outgrowth and promotes the formation of larger leaves. LATERAL ORGAN BOUNDARY DOMAIN37 (LBD37; At5g67420) is another down-regulated transcription factor that has been shown recently to affect nitrogen-dependent basal shoot branching, although its mode of action in this process has not been reported (Rubin et al., 2009). The transcriptional regulators SPL3 (At2g33810) and SPL4 (At1g53160) stimulate vegetative phase change and flowering and are suppressed by miR156 (Wu and Poethig, 2006), but their role in bud outgrowth has not been studied in detail yet. In general, however, members of the SPL family are implicated in controlling plastochron setting (Schwarz et al., 2008; Wang et al., 2008; Leyser, 2009). BT5 (At4g37610), which encodes a cytosolic protein of the BTB/TAZ family (Robert et al., 2009), was also one of the down-regulated genes; its function is unknown. MapMan analysis (Usadel et al., 2006) confirmed that genes controlled by EBE are mainly involved in lipid metabolism (lipid transfer proteins), cell wall modification, RNA regulation of transcription, and auxin metabolism (signal transduction; Supplemental Table S3).
Potential Regulatory Elements in Promoters of EBE Downstream Genes
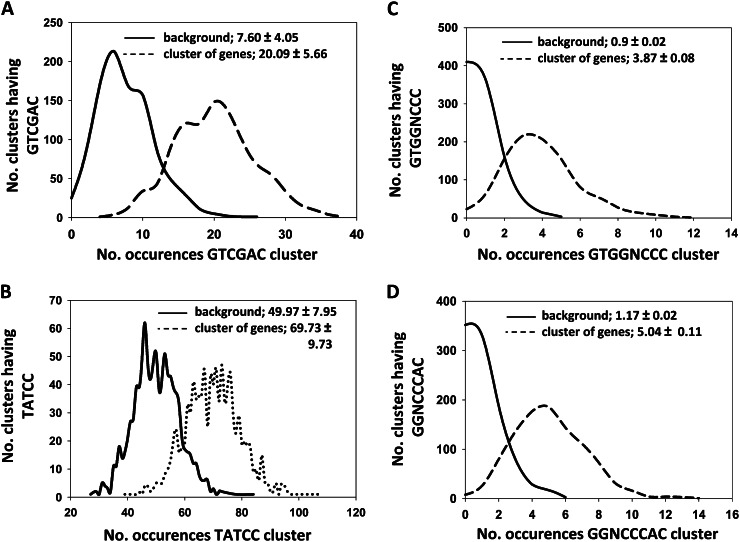
Since EBE is an AP2/ERF transcription factor, we analyzed the 1-kb promoters of the 48 EBE-affected genes for the presence of cis-elements known to be targeted by AP2/ERFs (compare with Supplemental Table S4), using POBO (http://ekhidna.biocenter.helsinki.fi/poxo/pobo) and Expression Angler (http://bar.utoronto.ca/ntools/cgi-bin/BAR_Promomer.cgi; Kankainen and Holm, 2004; Toufighi et al., 2005), revealing a significant overrepresentation of the HvCBF2-binding site GTCGAC (Fig. 7A; Supplemental Table S4). While several promoters contain ERF-like binding sequences (such as CIPK11, AtLTP6, and AtLTP12), some others (AtMYB60 and AtXTH9) did not and, therefore, might represent secondary downstream targets of EBE (Supplemental Table S4).
Figure 7.
Overrepresentation analysis of cis-regulatory motifs in the promoters of early EBE target genes. A, Overrepresentation of the GTCGAC motif in the promoters of the 48 early target genes compared with the background. B, The TATCC motif (SRE) is more abundant in the promoters of the 34 down-regulated EBE early target genes than in the background data set. C and D, Overrepresentation of TCP target sites in the promoters of the 48 EBE downstream targets. All analyses were performed using the POBO program. Dashed lines indicate the occurrence of motifs in EBE downstream genes, and solid lines indicate the occurrence of motifs in the background data set.
Tatematsu et al. (2005) previously searched for sequence motifs overrepresented in promoters of genes up- or down-regulated after main shoot decapitation and observed that promoters of down-regulated genes were enriched for the TATCC motif that resembles the sugar-repressive element (SRE), whereas promoters of up-regulated genes were enriched for GGCCCAWW (named Up1) and AAACCCTA (Up2) motifs. As EBE affects axillary bud outgrowth, we searched for SRE, Up1, and Up2 regulatory elements in 1-kb upstream regions of EBE-affected genes. We observed a significant overrepresentation of the SRE motif in promoters of the 34 down-regulated genes when compared with the Arabidopsis background data set (Fig. 7B). In Arabidopsis, the two TCP transcription factors BRC1 and BRC2 control shoot branching (Aguilar-Martínez et al., 2007; Finlayson, 2007; Martín-Trillo et al., 2011). POBO analysis indicated that the TCP-binding sites GGNCCCAC and G(T/C)GGNCCC (Kosugi and Ohashi, 2002; Welchen and Gonzalez, 2006; Martín-Trillo and Cubas, 2010) are overrepresented in the 1-kb promoters of the 48 EBE downstream genes (Fig. 7, C and D), suggesting that members of the TCP family, including BRC1, may act together with EBE to jointly regulate their expression.
DISCUSSION
Shoot branching is a finely regulated developmental process tuned by both endogenous and environmental cues (Wang and Li, 2008; Finlayson et al., 2010; Domagalska and Leyser, 2011). In many cases, newly formed axillary buds remain dormant for some time before growing out to form a branch. The shoot apex exerts an inhibitory effect on bud outgrowth, a phenomenon known as apical dominance (Shimizu-Sato and Mori, 2001). Apical dominance can be released by environmental and developmental signals (Shimizu-Sato and Mori, 2001; Dun et al., 2006).
Although various genes controlling shoot architecture have been reported, the underlying regulatory mechanisms, in particular those acting in the axillary bud itself, are currently not well understood. Here, we report that EBE, an ERF transcription factor of the AP2/ERF superfamily, has an impact on shoot architecture. As we show, the number of outgrowing buds is decreased in EBE-RNAi lines and increased in EBE overexpression lines compared with the wild type (Fig. 3, A and B). These differences cannot be attributed to potential differences in developmental speed, as RNAi lines never produced as many lateral branches as wild-type plants, even if they were grown until complete senescence. Similarly, EBE overexpressors consistently had higher numbers of lateral branches than wild-type plants even upon full senescence. Moreover, main stem decapitation rapidly (within 1 h) enhances EBE expression in axillary buds of wild-type plants (Fig. 5C). Of note, in estradiol-inducible EBE overexpression lines, AtDRM1, AtDRM1 homolog (At2g33830), and MARD1/SAG102 (At3g63210) are remarkably down-regulated after EBE induction. These genes are also strongly down-regulated in buds that start to grow (AtDRM1 and AtDRM1 homolog; Tatematsu et al., 2005). On the other hand, dormancy release occurs concomitant with the up-regulation of genes that function during G1-to-S-phase transition, such as D-type cyclins (CYCD; Shimizu-Sato and Mori, 2001; Horvath et al., 2003; Tatematsu et al., 2005). Of note, CYCD3;3 is remarkably up-regulated after EBE induction. Furthermore, the promoters of AtDRM1 homolog and MARD1 both contain the GTCGAC motif (HvCBF2-binding sequence), which is overrepresented in the promoters of the 48 EBE target genes (Fig. 7A). Similarly, the AtDRM1 and CYCD3;3 promoters contain GCCGAC and ATCGAC sequences, respectively, both of which are alternative sequences to GTCGAC with lower binding affinities (Xue, 2003). This result indicates that the above genes might be direct targets of EBE. In addition, we observed an overrepresentation of the SRE element, which was previously implicated in axillary bud outgrowth (Tatematsu et al., 2005), in the promoters of the 34 EBE down-regulated genes (Fig. 7B). Although binding of EBE to these motifs has not been tested experimentally yet, this result further supports the model that EBE is an important regulator of axillary bud outgrowth.
Although the altered branching pattern observed in EBE-modified plants may primarily be caused by direct effects on regulatory processes in axillary buds, the alternative possibility, that the developmental phenotype is at least partly caused by perturbed apical dominance, cannot be disregarded at present. Notably, EBE expression is widely detected in various plant tissues rather than being restricted to axillary buds only (Fig. 1). Thus, EBE might actually exert its role in different plant organs by affecting, for example, the function of phytohormones such as auxin, cytokinin, or strigolactones, or other unidentified long-distance signals. In agreement with this hypothesis, the primary shoot apex of 35S:EBE and EBE:RNAi plants is often disorganized (Fig. 6C). Furthermore, ectopic overexpression of EBE triggers neoplastic capacity (Supplemental Fig. S1, A–C), indicating that auxin/cytokinin balance or signaling may be perturbed. The phenotype observed for EBE overexpression plants to some extent resembles that of the altered meristem program1 mutant, which lacks apical dominance and has an elevated cytokinin level (Chaudhury et al., 1993; Nogué et al., 2000). However, in 35S:EBE plants, the branching phenotype may not be causally linked to an elevated cytokinin level. We observed a slightly earlier senescence in 35S:EBE than in wild-type plants (data not shown), indicating that cytokinin level may be reduced rather than increased.
Most plant cells are totipotent. Totipotency requires the acquisition of stem cell potential (dedifferentiation) in response to proper stimuli and expressing this potential during subsequent morphogenesis (regeneration). During dedifferentiation, genome reprogramming occurs to establish a stem cell status, and subsequently, gene activity patterns change in a systematic manner while entering the regeneration phase (Sugiyama, 1999; Grafi, 2004). EBE is preferentially expressed during the S phase of the cell cycle (Fig. 1A) and is highly active in dividing cells (Fig. 1, A and B; Menges et al., 2003; Deeken et al., 2006; Sugimoto et al., 2010), indicating that it regulates the expression of genes important for cell proliferation. Thus, the pleiotropic phenotype induced by the ectopic expression of EBE may to some extent be caused by disconnecting the expression of its target genes from the regular developmental program. In addition, our experimental data suggest that elevated EBE expression in wounded and senescent leaf areas may affect stem cell potentiality and subsequent morphogenesis (regeneration). Notably, transcriptome profiling previously showed remarkable similarities between senescing and dedifferentiating protoplast cells, indicating that both cell states share common molecular characteristics (Damri et al., 2009; Grafi et al., 2011). Those authors proposed a model in which the response of plant cells to stress (which often triggers precocious senescence) converges on cellular dedifferentiation, which leads to a stem cell-like fate before cells adopt a new fate (Grafi et al., 2011). In accordance with this model, tissues phenotypically resembling green calli, or organ-like structures, regenerated from wounded sites or partially senesced tissues of 35S:EBE plants (Supplemental Fig. S1, A–C). Worth mentioning is that EBE resembles the WOUND-INDUCED DEDIFFERENTIATION1 (WIND1) gene and its close homologs WIND2, WIND3, and WIND4, all of which are induced by wounding and promote cell dedifferentiation in Arabidopsis (Iwase et al., 2011). However, a role in bud outgrowth and shoot branching was not reported for these other AP2/ERF transcription factors. Of note, the WIND genes encode group Ib AP2/ERF transcription factors, while EBE is a member of group Xa (Nakano et al., 2006).
It is well established that the SAM, along its radial axis, is organized into a central zone that includes a source of pluripotent stem cells as well as a peripheral zone in which appendages, including leaves and flowers, are generated (Dodsworth, 2009; Barton, 2010; Ha et al., 2010). Transgenic EBE plants showed altered SAM size, leaf initiation rate, phyllotaxy, and axillary bud formation and outgrowth. As shown in Figure 6C, the apical apex of 35S:EBE plants can massively enlarge, indicating that EBE overexpression affects meristem maintenance. Moreover, 35S:EBE plants develop buds in their cotyledonary nodes, which is normally not observed in wild-type plants. On the other hand, leaf primordia were initiated in some cases in 35S:EBE plants but failed to develop further (Supplemental Fig. S6A), indicating that EBE overexpression perturbs peripheral organ development.
Previously, transcription factors of the TCP family, including BRC1 and BRC2 in Arabidopsis, were shown to play a critical role in shoot branching (Aguilar-Martínez et al., 2007; Finlayson, 2007; Martín-Trillo et al., 2011; Braun et al., 2012). Generally, loss of function of these transcription factors leads to increased branching, while overexpression leads to inhibition of bud outgrowth, identifying them as negative regulators of branching. In contrast, EBE, a member of the AP2/ERF transcription factor superfamily (Nakano et al., 2006), acts as a positive regulator of bud outgrowth and shoot branching (Fig. 3). Currently, the regulatory interplay between BRC1 (or BRC2) and EBE in the control of branch growth remains unknown. BRC1 has been suggested to integrate endogenous and environmental signals to control bud outgrowth (Aguilar-Martínez et al., 2007). Class II TCPs, to which BRC1 belongs, have been proposed to repress organ growth by inhibiting cell proliferation at the G1-to-S transition (Martín-Trillo and Cubas, 2010; Aggarwal et al., 2011). EBE shows elevated expression during the S phase of the cell cycle and lower expression levels in the other phases (Fig. 1A; Menges et al., 2003). As TCP-binding sites are overrepresented in promoters of EBE downstream genes, their expression may be jointly regulated by both types of transcription factors. Future experiments will have to unravel the intricacies of the transcriptional control acting during dormancy breaking and bud outgrowth in more detail; the BRC1/BRC2 and EBE transcription factors are excellent starting points for achieving this goal.
MATERIALS AND METHODS
General
Standard molecular biological techniques were performed as described by Sambrook et al. (2001). DNA sequencing was achieved by AGOWA. Oligonucleotides were obtained from MWG Biotech. Unless indicated otherwise, chemicals were purchased from Roche, Merck, Invitrogen, Sigma-Aldrich, and Fluka. Molecular biology kits were obtained from Qiagen and Macherey-Nagel.
Databases and Biocomputational Tools
For sequence and expression analyses, the following databases and tools were used: eFP browser (http://bbc.botany.utoronto.ca/efp/cgi-bin/efpWeb.cgi); ExPASy (http://us.expasy.org/); Expression Angler (http://bar.utoronto.ca/ntools/cgi-bin/BAR_Promomer.cgi); Genevestigator (https://www.genevestigator.ethz.ch); National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/); PLACE (http://www.dna.affrc.go.jp/PLACE/); PlnTFDB (http://plntfdb.bio.uni-potsdam.de/v3.0/); and The Arabidopsis Information Resource (http://www.arabidopsis.org/). Frequencies of motifs in EBE downstream genes were calculated using POBO (http://ekhidna.biocenter.helsinki.fi/poxo/pobo) on 1-kb promoter sequences of all 48 EBE-responsive genes (Table I) extracted from the repository of The Arabidopsis Information Resource. For bootstrap analysis, the following parameters were used: number of pseudoclusters, 1,000; number of sequences in the pseudocluster, 48; promoter length, 1,000 bp.
Biological Material
All experiments were performed using Arabidopsis (Arabidopsis thaliana) ecotype Col-0. Agrobacterium tumefaciens strain GV3101 (pMP90) was used for Arabidopsis transformations. Escherichia coli strain DH5α (Stratagene) was employed for standard DNA work.
Growth Conditions
For tissue culture, Arabidopsis seeds were surface sterilized in 70% ethanol for 2 min and then in sterilization solution (20% sodium hypochlorite) for 20 min. After sterilization, the seeds were sown on one-half-strength MS medium (Murashige and Skoog, 1962) supplemented with 1% (w/v) Suc and appropriate antibiotics and solidified with 0.7% (w/v) phytoagar. After imbibition, the seeds were stratified at 4°C for 3 d. The seeds were germinated at 22°C under a 16-h-day (140 µmol m−2 s−1)/8-h-night regime. Two-week-old seedlings were carefully removed from plates and transplanted to soil in 6-cm pots (Einheitserde GS90; Gebrüder Patzer) or, if necessary, directly subjected to various treatments. Unless otherwise indicated, Arabidopsis plants were grown in controlled conditions in a growth chamber with a 16-h daylength provided by fluorescent light at 80 or 120 µmol m−2 s−1, day/night temperatures of 20°C/16°C, and relative humidity of 60%/75% (long-day conditions). Short-day conditions were as follows: 8-h day (150 µmol m−2 s−1), day/night temperatures of 20°C/18°C, and relative humidity of 60%/75%.
Constructs
35S:EBE
The EBE coding region (Arabidopsis Genome Initiative code At5g61890) was PCR amplified from Arabidopsis. Col-0 complementary DNA (cDNA) was inserted into pUni/V5-His-TOPO (Invitrogen) and, after sequence confirmation, cloned via the PmeI/PacI sites into a modified pGreen0229 plant transformation vector (www.pgreen.ac.uk) containing a CaMV 35S promoter (Skirycz et al., 2006).
EBE-XVE
The EBE coding region was PCR amplified from Arabidopsis Col-0 cDNA using primers EBE-fwd-1 (5′-CTCGAGATGTATGGGAAGAGG-3′) and EBE-rev-1 (5′-ACTAGTTTAATATCCCGAATGAGG-3′). The amplified fragment was inserted into plasmid pCR-2.1-TOPO (Invitrogen) and subsequently fused via XhoI/SpeI sites into the pER8 vector (Zuo et al., 2000).
EBE-RNAi
An approximately 200-bp fragment of the EBE coding region (exon 2) was PCR amplified from Col-0 cDNA using primers EBE-fwd (5′-CACCGGAGAGGGTTCAGCTTGG-3′) and EBE-rev (5′-GTTGTAGCAGCAGTCGTAGTAC-3′). The PCR product was cloned into vector pENTR/D-TOPO of the Gateway system (Invitrogen). After sequencing, the EBE fragment was transferred to Gateway-compatible binary vector pBINAR-RNAi (kindly provided by Dr. Fernando Carrari, Max-Planck Institute of Molecular Plant Physiology) by attL × attR recombination, resulting in plasmid EBE-RNAi. The vector carries the CaMV 35S promoter upstream of the EBE silencing construct. After transformation into Arabidopsis, 25 independent kanamycin-resistant seedlings were chosen to test for reduced EBE transcript level using qRT-PCR.
PromEBE:GUS
An approximately 2-kb fragment upstream of the translation start codon of the EBE gene was PCR amplified from Arabidopsis Col-0 genomic DNA using primers EBE-GUS-fwd (5′-AAGCTTGACATATGGAGAAAGTGTCC-3′) and EBE-GUS-rev (5′-TCTAGATCCGAGGATCTACTTTTGC-3′). The promoter fragment was inserted into plasmid pCR-2.1-TOPO and subsequently fused via SpeI/XhoI (vector sites) to the GUS reporter gene in pGPTV-Kan (Becker et al., 1992), previously cut with XbaI/SalI, resulting in plasmid PromEBE:GUS.
EBE T-DNA Insertion Line
An Arabidopsis line (SALK_000833) carrying a T-DNA insertion approximately 400 bp upstream of the EBE translation start codon (ATG) was obtained from the Nottingham Arabidopsis Stock Centre (http://arabidopsis.info/). Homozygous insertion lines were identified by PCR on genomic DNA. The approximate location of the T-DNA within the EBE gene was confirmed by PCR (Supplemental Fig. S3, A and B). The presence of the EBE wild-type allele was tested using primers EBE-F-promoter (5′-AAGCTTACCTGTCAACTTAGTTAGC-3′) and EBE-R-promoter (5′-TCTAGATCCGAGGATCTACTTTTGC-3′). The presence of the T-DNA insertion allele was tested using T-DNA left border primers (5′-GGCAATCAGCTGTTGCCCGTCTCACTGGTG-3′ and 5′-TGGTTCACGTAGTGGGCCATCG-3′) in combination with the EBE-R-promoter primer. EBE expression in the wild type and T-DNA insertion lines was tested by qRT-PCR.
Estradiol Induction Experiments
EBE-XVE and wild-type (control) seeds were cultured on MS agar plates at 22°C under a 16-h-day (140 µmol m−2 s−1)/8-h-night regime. After 3 weeks, seedlings were transferred to MS liquid medium while shaking. Sixteen hours later, 17-EST (Sigma-Aldrich) was added to the medium to a final concentration of 2 μm. Subsequently, whole shoots or rosette leaves were harvested at different time points after EST addition (30 min, 45 min, and 1, 4, 6, and 20 h).
GUS Assays
GUS activity was determined histochemically as described (Plesch et al., 2001). T2 and T3 plants were used for all studies. Infiltrated plants were incubated in GUS staining solution for 1 to 20 h at 37°C. For wounding experiments, mature leaves were injured with a needle. GUS staining was started 1 to 48 h after application of the wound stress. Plant material for microscopy was infiltrated and embedded in Technovit 7100 (Kulzer) as instructed by the manufacturer. Sections of 4-µm thickness were made on a Leica RM 2155 rotary microtome carrying a disposable Adams steel knife.
Expression Profiling by qRT-PCR
Total RNA extraction, cDNA synthesis, and qRT-PCR were performed as described previously (Caldana et al., 2007; Balazadeh et al., 2008). PCR was run on an ABI PRISM 7900HT sequence detection system (Applied Biosystems Applera).
Microarray Analysis
Three micrograms of quality-checked total RNA obtained from leaves of three biological replicates of EBE-XVE plants at time point 45 min were processed for use in Affymetrix ATH1 hybridizations. Labeling, hybridization, washing, staining, and scanning procedures were performed by an Affymetrix-authorized service provider (ATLAS Biolabs) as described in the Affymetrix technical manual. Raw data (CEL files) obtained from RNA hybridization experiments were normalized with the affyPLM package from the Bioconductor software project (Gentleman et al., 2004) using GCRMA, which employs the GC content of probes in normalization with robust multiple array average and gives one value for each probe set instead of keeping probe-level information (Wu et al., 2004). The transcriptional changes were determined by subtracting the normalized signal intensity of the control samples from that of induced samples. Expression data were submitted to the National Center for Biotechnology Information Gene Expression Omnibus repository (www.ncbi.nlm.nih.gov/geo/) under accession number GSE27855.
Callus Formation and Shoot and Root Regeneration
Leaf segments from seedlings were precultured on CIM (containing 2% Suc, 1 mg L−1 2,4-dichlorophenoxyacetic acid, and 0.2 mg L−1 kinetin) for 4 d and then transferred to shoot-induction medium (containing 2% Suc, 1 mg L−1 benzylaminopurine, and 0.4 mg L−1 naphthalene acetic acid), root-induction medium (containing 2% Suc and 0.4 mg L−1 naphthalene acetic acid), or new CIM. Media were refreshed every 2 weeks.
Analysis of Shoot Branching
Shoot branches or axillary buds were analyzed using a binocular. Rosette leaf axils were examined individually beginning with the oldest one. Leaves were removed successively after study so that the younger leaf axils became accessible. Lateral buds with at least 0.5 cm length were considered as lateral branches. Ten to 20 plants were analyzed in each experiment, and each experiment was performed at least twice.
Main Stem Decapitation
PromEBE:GUS seeds were cultured in one-half-strength MS medium in glass pots. The seeds were germinated at 22°C under a 16-h-day (140 µmol m−2 s−1)/8-h-night regime. Main stems were decapitated 4 to 7 d after bolting, at stem height of 2 to 5 cm, and when axillary buds were visible. GUS activity was analyzed 6 and 14 h after decapitation.
Decapitation-controlled expression of EBE was confirmed in wild-type Arabidopsis plants by qRT-PCR. Plants were grown in long-day conditions, and decapitation was done when the main stem was approximately 10 cm high. The five top axillary shoots were harvested at different time points (1, 6, 9, 15, and 24 h) after decapitation and mixed before analysis. The same procedure was applied for intact plants.
Scanning Electron Microscopy
Tissues were fixed in 4% formaldehyde in 0.07 m sodium phosphate/potassium phosphate buffer (pH 7.2) at 4°C for 4 to 16 h. Then, tissues were rinsed in sodium phosphate/potassium phosphate buffer for 1 h, dehydrated in a graded ethanol series, and critical point dried in vacuum. Samples were coated with gold/palladium and examined at an accelerating voltage of 10 kV with a high-resolution SEM LEO 1550 microscope (Carl Zeiss).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Neoplastic activity in 35S:EBE plants.
Supplemental Figure S2. Phenotypic effects of enhanced EBE transcript level.
Supplemental Figure S3. Analysis of the ebe-1D mutant.
Supplemental Figure S4. Phenotype of EBE-RNAi lines.
Supplemental Figure S5. Leaf and cell sizes.
Supplemental Figure S6. Phenotype of SAM and EBE expression shown by GUS staining.
Supplemental Figure S7. Phenotype induced by estradiol-induced EBE overexpression.
Supplemental Table S1. Number of cauline leaf branches in transgenic lines compared with the wild type.
Supplemental Table S2. Length of primary inflorescences in transgenic and wild-type plants.
Supplemental Table S3. Overrepresentation analysis of the 48 genes rapidly responding to enhanced EBE expression.
Supplemental Table S4. Target DNA-binding sequences of AP2/ERF domain proteins in the promoters of EBE downstream genes.
Supplemental Table S5. Flowering time points in EBE-modified plants.
Acknowledgments
We thank Eugenia Maximova for excellent microscopic assistance; Nooshin Omranian for help with statistical analyses; Josef Bergstein for photographic work; Karin Koehl and her colleagues for expert plant care; Fernando Carrari for providing pBINAR-RNAi vector; Erwan Michard for introducing M.M. to EBE; the anonymous reviewers for helpful comments on the manuscript; and the Nottingham Arabidopsis Stock Centre for providing seeds of the ebe-1D T-DNA insertion line.
Glossary
- CaMV
cauliflower mosaic virus
- CIM
callus-induction medium
- qRT
quantitative reverse transcription
- RNAi
RNA interference
- Col-0
ecotype Columbia
- SAM
shoot apical meristem
- MS
Murashige and Skoog
- EST
β-estradiol
- SRE
sugar-repressive element
- cDNA
complementary DNA
- T-DNA
transfer DNA
References
- Aggarwal P, Padmanabhan B, Bhat A, Sarvepalli K, Sadhale PP, Nath U. (2011) The TCP4 transcription factor of Arabidopsis blocks cell division in yeast at G1→S transition. Biochem Biophys Res Commun 410: 276–281 [DOI] [PubMed] [Google Scholar]
- Aguilar-Martínez JA, Poza-Carrión C, Cubas P. (2007) Arabidopsis BRANCHED1 acts as an integrator of branching signals within axillary buds. Plant Cell 19: 458–472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bainbridge K, Guyomarc’h S, Bayer E, Swarup R, Bennett M, Mandel T, Kuhlemeier C. (2008) Auxin influx carriers stabilize phyllotactic patterning. Genes Dev 22: 810–823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakan B, Hamberg M, Perrocheau L, Maume D, Rogniaux H, Tranquet O, Rondeau C, Blein J-P, Ponchet M, Marion D. (2006) Specific adduction of plant lipid transfer protein by an allene oxide generated by 9-lipoxygenase and allene oxide synthase. J Biol Chem 281: 38981–38988 [DOI] [PubMed] [Google Scholar]
- Balazadeh S, Riaño-Pachón DM, Mueller-Roeber B. (2008) Transcription factors regulating leaf senescence in Arabidopsis thaliana. Plant Biol (Stuttg) (Suppl 1) 10: 63–75 [DOI] [PubMed] [Google Scholar]
- Barton MK. (2010) Twenty years on: the inner workings of the shoot apical meristem, a developmental dynamo. Dev Biol 341: 95–113 [DOI] [PubMed] [Google Scholar]
- Bayer EM, Bottrill AR, Walshaw J, Vigouroux M, Naldrett MJ, Thomas CL, Maule AJ. (2006) Arabidopsis cell wall proteome defined using multidimensional protein identification technology. Proteomics 6: 301–311 [DOI] [PubMed] [Google Scholar]
- Becker D, Kemper E, Schell J, Masterson R. (1992) New plant binary vectors with selectable markers located proximal to the left T-DNA border. Plant Mol Biol 20: 1195–1197 [DOI] [PubMed] [Google Scholar]
- Beveridge CA, Kyozuka J. (2010) New genes in the strigolactone-related shoot branching pathway. Curr Opin Plant Biol 13: 34–39 [DOI] [PubMed] [Google Scholar]
- Boe A, Beck DL. (2008) Yield components of biomass in switchgrass. Crop Sci 48: 1306–1311 [Google Scholar]
- Bonser SP, Aarssen LW. (2003) Allometry and development in herbaceous plants: functional responses of meristem allocation to light and nutrient availability. Am J Bot 90: 404–412 [DOI] [PubMed] [Google Scholar]
- Braun N, de Saint Germain A, Pillot JP, Boutet-Mercey S, Dalmais M, Antoniadi I, Li X, Maia-Grondard A, Le Signor C, Bouteiller N, et al. (2012) The pea TCP transcription factor PsBRC1 acts downstream of strigolactones to control shoot branching. Plant Physiol 158: 225–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caldana C, Scheible WR, Mueller-Roeber B, Ruzicic S. (2007) A quantitative RT-PCR platform for high-throughput expression profiling of 2500 rice transcription factors. Plant Methods 3: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhury AM, Letham S, Craig S, Dennis ES. (1993) amp1: a mutant with high cytokinin levels and altered embryonic pattern, faster vegetative growth, constitutive photomorphogenesis and precocious flowering. Plant J 4: 907–916 [Google Scholar]
- Che P, Lall S, Nettleton D, Howell SH. (2006) Gene expression programs during shoot, root, and callus development in Arabidopsis tissue culture. Plant Physiol 141: 620–637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damri M, Granot G, Ben-Meir H, Avivi Y, Plaschkes I, Chalifa-Caspi V, Wolfson M, Fraifeld V, Grafi G. (2009) Senescing cells share common features with dedifferentiating cells. Rejuvenation Res 12: 435–443 [DOI] [PubMed] [Google Scholar]
- Deeken R, Engelmann JC, Efetova M, Czirjak T, Müller T, Kaiser WM, Tietz O, Krischke M, Mueller MJ, Palme K, et al (2006) An integrated view of gene expression and solute profiles of Arabidopsis tumors: a genome-wide approach. Plant Cell 18: 3617–3634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Veylder L, Beeckman T, Beemster GTS, de Almeida Engler J, Ormenese S, Maes S, Naudts M, Van Der Schueren E, Jacqmard A, Engler G, et al (2002) Control of proliferation, endoreduplication and differentiation by the Arabidopsis E2Fa-DPa transcription factor. EMBO J 21: 1360–1368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devitt ML, Stafstrom JP. (1995) Cell cycle regulation during growth-dormancy cycles in pea axillary buds. Plant Mol Biol 29: 255–265 [DOI] [PubMed] [Google Scholar]
- Dodsworth S. (2009) A diverse and intricate signalling network regulates stem cell fate in the shoot apical meristem. Dev Biol 336: 1–9 [DOI] [PubMed] [Google Scholar]
- Doebley J, Stec A, Hubbard L. (1997) The evolution of apical dominance in maize. Nature 386: 485–488 [DOI] [PubMed] [Google Scholar]
- Domagalska MA, Leyser O. (2011) Signal integration in the control of shoot branching. Nat Rev Mol Cell Biol 12: 211–221 [DOI] [PubMed] [Google Scholar]
- Doust A. (2007) Architectural evolution and its implications for domestication in grasses. Ann Bot (Lond) 100: 941–950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dun EA, Ferguson BJ, Beveridge CA. (2006) Apical dominance and shoot branching: divergent opinions or divergent mechanisms? Plant Physiol 142: 812–819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson BJ, Beveridge CA. (2009) Roles for auxin, cytokinin, and strigolactone in regulating shoot branching. Plant Physiol 149: 1929–1944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finlayson SA. (2007) Arabidopsis Teosinte Branched1-like 1 regulates axillary bud outgrowth and is homologous to monocot Teosinte Branched1. Plant Cell Physiol 48: 667–677 [DOI] [PubMed] [Google Scholar]
- Finlayson SA, Krishnareddy SR, Kebrom TH, Casal JJ. (2010) Phytochrome regulation of branching in Arabidopsis. Plant Physiol 152: 1914–1927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foo E, Bullier E, Goussot M, Foucher F, Rameau C, Beveridge CA. (2005) The branching gene RAMOSUS1 mediates interactions among two novel signals and auxin in pea. Plant Cell 17: 464–474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- García del Moral MB, García del Moral LF. (1995) Tiller production and survival in relation to grain yield in winter and spring barley. Field Crops Res 44: 85–93 [Google Scholar]
- Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, et al. (2004) Bioconductor: open software development for computational biology and bioinformatics. Genome Biol 5: R80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grafi G. (2004) How cells dedifferentiate: a lesson from plants. Dev Biol 268: 1–6 [DOI] [PubMed] [Google Scholar]
- Grafi G, Chalifa-C aspi V, Nagar T, Plaschkes I, Barak S, Ransbotyn V. (2011) Plant response to stress meets dedifferentiation. Planta 233: 433–438 [DOI] [PubMed] [Google Scholar]
- Grbic V. (2005) Comparative analysis of axillary and floral meristem development. Can J Bot 83: 343–349 [Google Scholar]
- Ha CM, Jun JH, Fletcher JC. (2010) Shoot apical meristem form and function. Curr Top Dev Biol 91: 103–140 [DOI] [PubMed] [Google Scholar]
- Han P, Li Q, Zhu Y-X. (2008) Mutation of Arabidopsis BARD1 causes meristem defects by failing to confine WUSCHEL expression to the organizing center. Plant Cell 20: 1482–1493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayward A, Stirnberg P, Beveridge C, Leyser O. (2009) Interactions between auxin and strigolactone in shoot branching control. Plant Physiol 151: 400–412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y, Gan S. (2004) A novel zinc-finger protein with a proline-rich domain mediates ABA-regulated seed dormancy in Arabidopsis. Plant Mol Biol 54: 1–9 [DOI] [PubMed] [Google Scholar]
- Hong SY, Kim OK, Kim SG, Yang MS, Park CM. (2011) Nuclear import and DNA binding of the ZHD5 transcription factor is modulated by a competitive peptide inhibitor in Arabidopsis. J Biol Chem 286: 1659–1668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath DP, Anderson JV, Chao WS, Foley ME. (2003) Knowing when to grow: signals regulating bud dormancy. Trends Plant Sci 8: 534–540 [DOI] [PubMed] [Google Scholar]
- Iwase A, Mitsuda N, Koyama T, Hiratsu K, Kojima M, Arai T, Inoue Y, Seki M, Sakakibara H, Sugimoto K, et al (2011) The AP2/ERF transcription factor WIND1 controls cell dedifferentiation in Arabidopsis. Curr Biol 21: 508–514 [DOI] [PubMed] [Google Scholar]
- Kader J-C. (1996) Lipid transfer proteins in plants. Annu Rev Plant Physiol Plant Mol Biol 47: 627–654 [DOI] [PubMed] [Google Scholar]
- Kankainen M, Holm L. (2004) POBO, transcription factor binding site verification with bootstrapping. Nucleic Acids Res 32: W222–W229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kebrom TH, Burson BL, Finlayson SA. (2006) Phytochrome B represses Teosinte Branched1 expression and induces sorghum axillary bud outgrowth in response to light signals. Plant Physiol 140: 1109–1117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosugi S, Ohashi Y. (1997) PCF1 and PCF2 specifically bind to cis elements in the rice proliferating cell nuclear antigen gene. Plant Cell 9: 1607–1619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosugi S, Ohashi Y. (2002) DNA binding and dimerization specificity and potential targets for the TCP protein family. Plant J 30: 337–348 [DOI] [PubMed] [Google Scholar]
- Kwon H-K, Yokoyama R, Nishitani K. (2005) A proteomic approach to apoplastic proteins involved in cell wall regeneration in protoplasts of Arabidopsis suspension-cultured cells. Plant Cell Physiol 46: 843–857 [DOI] [PubMed] [Google Scholar]
- Leyser O. (2009) The control of shoot branching: an example of plant information processing. Plant Cell Environ 32: 694–703 [DOI] [PubMed] [Google Scholar]
- Li CJ, Guevara E, Herrera J, Bangerth F. (1995) Effect of apex excision and replacement by 1-naphthylacetic acid on cytokinin concentration and apical dominance in pea plants. Physiol Plant 94: 465–469 [Google Scholar]
- Martín-Trillo M, Cubas P. (2010) TCP genes: a family snapshot ten years later. Trends Plant Sci 15: 31–39 [DOI] [PubMed] [Google Scholar]
- Martín-Trillo M, Grandío EG, Serra F, Marcel F, Rodríguez-Buey ML, Schmitz G, Theres K, Bendahmane A, Dopazo H, Cubas P. (2011) Role of tomato BRANCHED1-like genes in the control of shoot branching. Plant J 67: 701–714 [DOI] [PubMed] [Google Scholar]
- Mayer KF, Schoof H, Haecker A, Lenhard M, Jürgens G, Laux T. (1998) Role of WUSCHEL in regulating stem cell fate in the Arabidopsis shoot meristem. Cell 95: 805–815 [DOI] [PubMed] [Google Scholar]
- McSteen P, Leyser O. (2005) Shoot branching. Annu Rev Plant Biol 56: 353–374 [DOI] [PubMed] [Google Scholar]
- Menges M, de Jager SM, Gruissem W, Murray JAH. (2005) Global analysis of the core cell cycle regulators of Arabidopsis identifies novel genes, reveals multiple and highly specific profiles of expression and provides a coherent model for plant cell cycle control. Plant J 41: 546–566 [DOI] [PubMed] [Google Scholar]
- Menges M, Hennig L, Gruissem W, Murray JAH. (2003) Genome-wide gene expression in an Arabidopsis cell suspension. Plant Mol Biol 53: 423–442 [DOI] [PubMed] [Google Scholar]
- Mizukami Y, Fischer RL. (2000) Plant organ size control: AINTEGUMENTA regulates growth and cell numbers during organogenesis. Proc Natl Acad Sci USA 97: 942–947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mündermann L, Erasmus Y, Lane B, Coen E, Prusinkiewicz P. (2005) Quantitative modeling of Arabidopsis development. Plant Physiol 139: 960–968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige T, Skoog F. (1962) A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol Plant 15: 473–497 [Google Scholar]
- Nakano T, Suzuki K, Fujimura T, Shinshi H. (2006) Genome-wide analysis of the ERF gene family in Arabidopsis and rice. Plant Physiol 140: 411–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogué F, Grandjean O, Craig S, Dennis S, Chaudhury M. (2000) Higher levels of cell proliferation rate and cyclin CycD3 expression in the Arabidopsis amp1 mutant. Plant Growth Regul 32: 267–273 [Google Scholar]
- Plesch G, Ehrhardt T, Mueller-Roeber B. (2001) Involvement of TAAAG elements suggests a role for Dof transcription factors in guard cell-specific gene expression. Plant J 28: 455–464 [DOI] [PubMed] [Google Scholar]
- Reinhardt D, Kuhlemeier C. (2002) Plant architecture. EMBO Rep 3: 846–851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renton M, Hanan J, Ferguson BJ, Beveridge CA. (2012) Models of long-distance transport: how is carrier-dependent auxin transport regulated in the stem? New Phytol 194: 704–715 [DOI] [PubMed] [Google Scholar]
- Robert HS, Quint A, Brand D, Vivian-Smith A, Offringa R. (2009) BTB and TAZ domain scaffold proteins perform a crucial function in Arabidopsis development. Plant J 58: 109–121 [DOI] [PubMed] [Google Scholar]
- Rubin G, Tohge T, Matsuda F, Saito K, Scheible WR. (2009) Members of the LBD family of transcription factors repress anthocyanin synthesis and affect additional nitrogen responses in Arabidopsis. Plant Cell 21: 3567–3584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsche EF, Maniatis T (2001) Molecular Cloning: A Laboratory Manual, Ed 3. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- Schwarz S, Grande AV, Bujdoso N, Saedler H, Huijser P. (2008) The microRNA regulated SBP-box genes SPL9 and SPL15 control shoot maturation in Arabidopsis. Plant Mol Biol 67: 183–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sena G, Wang X, Liu HY, Hofhuis H, Birnbaum KD. (2009) Organ regeneration does not require a functional stem cell niche in plants. Nature 457: 1150–1153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu S, Mori H. (1998) Analysis of cycles of dormancy and growth in pea axillary buds based on mRNA accumulation patterns of cell cycle-related genes. Plant Cell Physiol 39: 255–262 [DOI] [PubMed] [Google Scholar]
- Shimizu-Sato S, Mori H. (2001) Control of outgrowth and dormancy in axillary buds. Plant Physiol 127: 1405–1413 [PMC free article] [PubMed] [Google Scholar]
- Skirycz A, Reichelt M, Burow M, Birkemeyer C, Rolcik J, Kopka J, Zanor MI, Gershenzon J, Strnad M, Szopa J, et al (2006) DOF transcription factor AtDof1.1 (OBP2) is part of a regulatory network controlling glucosinolate biosynthesis in Arabidopsis. Plant J 47: 10–24 [DOI] [PubMed] [Google Scholar]
- Stafstrom JP, Ripley BD, Devitt ML, Drake B. (1998) Dormancy-associated gene expression in pea axillary buds: cloning and expression of PsDRM1 and PsDRM2. Planta 205: 547–552 [DOI] [PubMed] [Google Scholar]
- Sugimoto K, Jiao Y, Meyerowitz EM. (2010) Arabidopsis regeneration from multiple tissues occurs via a root development pathway. Dev Cell 18: 463–471 [DOI] [PubMed] [Google Scholar]
- Sugiyama M. (1999) Organogenesis in vitro. Curr Opin Plant Biol 2: 61–64 [DOI] [PubMed] [Google Scholar]
- Sussex IM, Kerk NM. (2001) The evolution of plant architecture. Curr Opin Plant Biol 4: 33–37 [DOI] [PubMed] [Google Scholar]
- Tanaka M, Takei K, Kojima M, Sakakibara H, Mori H. (2006) Auxin controls local cytokinin biosynthesis in the nodal stem in apical dominance. Plant J 45: 1028–1036 [DOI] [PubMed] [Google Scholar]
- Tatematsu K, Ward S, Leyser O, Kamiya Y, Nambara E. (2005) Identification of cis-elements that regulate gene expression during initiation of axillary bud outgrowth in Arabidopsis. Plant Physiol 138: 757–766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toufighi K, Brady SM, Austin R, Ly E, Provart NJ. (2005) The Botany Array Resource: e-northerns, expression angling, and promoter analyses. Plant J 43: 153–163 [DOI] [PubMed] [Google Scholar]
- Usadel B, Nagel A, Steinhauser D, Gibon Y, Bläsing OE, Redestig H, Sreenivasulu N, Krall L, Hannah MA, Poree F, et al (2006) PageMan: an interactive ontology tool to generate, display, and annotate overview graphs for profiling experiments. BMC Bioinformatics 7: 535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J-W, Schwab R, Czech B, Mica E, Weigel D. (2008) Dual effects of miR156-targeted SPL genes and CYP78A5/KLUH on plastochron length and organ size in Arabidopsis thaliana. Plant Cell 20: 1231–1243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Li J. (2006) Genes controlling plant architecture. Curr Opin Biotechnol 17: 123–129 [DOI] [PubMed] [Google Scholar]
- Wang Y, Li J. (2008) Molecular basis of plant architecture. Annu Rev Plant Biol 59: 253–279 [DOI] [PubMed] [Google Scholar]
- Welchen E, Gonzalez DH. (2006) Overrepresentation of elements recognized by TCP-domain transcription factors in the upstream regions of nuclear genes encoding components of the mitochondrial oxidative phosphorylation machinery. Plant Physiol 141: 540–545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G, Poethig RS. (2006) Temporal regulation of shoot development in Arabidopsis thaliana by miR156 and its target SPL3. Development 133: 3539–3547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z, Irizarry RA, Gentleman R, Martinez-Murillo F, Spencer F. (2004) A model-based background adjustment for oligonucleotide expression arrays. J Am Stat Assoc 99: 909–917 [Google Scholar]
- Xue GP. (2003) The DNA-binding activity of an AP2 transcriptional activator HvCBF2 involved in regulation of low-temperature responsive genes in barley is modulated by temperature. Plant J 33: 373–383 [DOI] [PubMed] [Google Scholar]
- Yeats TH, Rose JKC. (2008) The biochemistry and biology of extracellular plant lipid-transfer proteins (LTPs). Protein Sci 17: 191–198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao DL, Atlin GN, Bastiaans L, Spiertz JHJ. (2006) Developing selection protocols for weed competitiveness in aerobic rice. Field Crops Res 97: 272–285 [Google Scholar]
- Zimmermann P, Hirsch-Hoffmann M, Hennig L, Gruissem W. (2004) GENEVESTIGATOR: Arabidopsis microarray database and analysis toolbox. Plant Physiol 136: 2621–2632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo J, Niu QW, Chua NH. (2000) An estrogen receptor-based transactivator XVE mediates highly inducible gene expression in transgenic plants. Plant J 24: 265–273 [DOI] [PubMed] [Google Scholar]