Pollen-expressed arabinokinase-like protein gene CAP1 is important for rice pollen development, and its related genes are conserved in both monocotyledonous and dicotyledonous plants.
Abstract
We isolated a pollen-defective mutant, collapsed abnormal pollen1 (cap1), from Tos17 insertional mutant lines of rice (Oryza sativa). The cap1 heterozygous plant produced equal numbers of normal and collapsed abnormal grains. The abnormal pollen grains lacked almost all cytoplasmic materials, nuclei, and intine cell walls and did not germinate. Genetic analysis of crosses revealed that the cap1 mutation did not affect female reproduction or vegetative growth. CAP1 encodes a protein consisting of 996 amino acids that showed high similarity to Arabidopsis (Arabidopsis thaliana) l-arabinokinase, which catalyzes the conversion of l-arabinose to l-arabinose 1-phosphate. A wild-type genomic DNA segment containing CAP1 restored mutants to normal pollen grains. During rice pollen development, CAP1 was preferentially expressed in anthers at the bicellular pollen stage, and the effects of the cap1 mutation were mainly detected at this stage. Based on the metabolic pathway of l-arabinose, cap1 pollen phenotype may have been caused by toxic accumulation of l-arabinose or by inhibition of cell wall metabolism due to the lack of UDP-l-arabinose derived from l-arabinose 1-phosphate. The expression pattern of CAP1 was very similar to that of another Arabidopsis homolog that showed 71% amino acid identity with CAP1. Our results suggested that CAP1 and related genes are critical for pollen development in both monocotyledonous and dicotyledonous plants.
The life cycles of plants and eukaryotic algae include alternating haploid gametophytic and diploid sporophytic phases. The gametophytic phase begins with the completion of meiosis. In angiosperms, microspores derived from meiotic tetrads undergo haploid mitosis. The asymmetrical division yields a large vegetative cell and a small generative cell. Soon, the generative cell migrates into the cytoplasm of the vegetative cell, where it eventually divides into two sperm cells. The haploid phase ends with the completion of double fertilization (Raghavan, 1989; Itoh et al., 2005; Wilson and Zhang, 2009; Tian et al., 2010; Twell, 2011). The timing of sperm cell formation depends on the plant species. For example, the generative cells of Cruciferae and Gramineae plants, with tricellular pollen, divide into two sperm cells within the pollen grains at flowering, while the division in Solanaceae and Liliaceae plants, with bicellular pollen, occurs within the pollen tube after pollination (Twell, 1994; McCormick, 2004). In mature pollen, the chromatin of generative and sperm cell nuclei is highly condensed, whereas the vegetative nucleus contains diffuse chromatin (Tanaka, 1997; Borg et al., 2009). In addition, for example in rice (Oryza sativa) and maize (Zea mays), the cytoplasm of vegetative cells in mature pollen grains is full of large starch granules to supply carbon skeletons and energy for pollen tube growth (Datta et al., 2002; Zhang et al., 2011).
Several recent studies using microarrays from rice (Wang et al., 2005; Hobo et al., 2008; Suwabe et al., 2008; Deveshwar et al., 2011; Sato et al., 2011) and Arabidopsis (Arabidopsis thaliana; Becker et al., 2003; Honys and Twell, 2003; Pina et al., 2005; Grennan, 2007) showed that more than 10,000 genes are expressed in anthers or pollen grains during pollen development. In addition to these comprehensive gene expression profiles, the functions of at least 37 genes expressed in male gametophyte involved in pollen development in Arabidopsis have been identified by mutant analysis (Twell et al., 2006; Twell, 2010). For example, loss of function of the GLC-6-P TRANSLOCATOR1, the reversibly glycosylated polypeptides RPG1 and RPG2 for polysaccharide biosynthesis, the cellulose synthases CESA1 to CESA3, CESA6, and CESA9, the long-chain bases LCB1, LCB2A, and LCB2C for sphingolipid synthesis, or the glucan synthases GSL8 and GSL10 cause male gametophytic lethality during pollen development (Niewiadomski et al., 2005; Drakakaki et al., 2006; Persson et al., 2007; Teng et al., 2008; Töller et al., 2008). Other genes affect gametophyte cell division and patterning. Mutations in the germline-specific R2R3 MYB transcription factor DUO POLLEN1 results in defective generative cell division (Borg et al., 2011), and disruption of the microtubule-associated protein GEMINI POLLEN1, the FUSED-kinase TWO-IN-ONE, or γ tubulins TUBG1 and TUBG2 affects asymmetric microspore mitosis (Twell et al., 2002; Oh et al., 2005; Pastuglia et al., 2006). Thus, although the functions of many genes in Arabidopsis male gametophytes have been elucidated, information on rice genes expressed after the differentiation of archesporial cells is mainly restricted to the gene products of the sporophyte in microsporocytes or anther tissues (Wilson and Zhang, 2009).
Meiosis is a first crucial event in pollen development. The meiotic proteins MEIOSIS ARRESTED AT LEPTOTENE1 (MEL1), of the ARGONAUTE family of RNA interference (RNAi) proteins, HOMOLOGOUS PAIRING ABERRATION IN RICE MEIOSIS1-3, associated with synapsis of homologous chromosomes, and MEL2, a novel RNA recognition motif protein, are essential for viable pollen production (Nonomura et al., 2004a, 2004b, 2007, 2011; Yuan et al., 2009). The innermost layer (tapetum) of the anther wall is also important for early pollen stages, such as microsporocytes and microspores. Mutant analyses of defective tapetums have revealed several genes critical for pollen development, such as the gibberellin-regulated R2R3 MYB transcription factor GAMYB; the C2-GRAM domain protein gene ORYZA SATIVA NO POLLEN; UNDEVELOPED TAPETUM1, which encodes a basic helix-loop-helix transcription factor; WAX-DEFICIENT ANTHER1, encoding an enzyme involved in the synthesis of very-long-chain fatty acids; basic helix-loop-helix protein genes TAPETUM DEGENERATION RETARDATION and ETERNAL TAPETUM1 (EAT1); UDP GLC PYROPHOSPHORYLASE1 (UGP1); the lipid transfer protein gene named OsC6; the R2R3 MYB transcription factor CARBON STARVED ANTHER (CSA); a cytochrome P450 family gene, CYP704B2; a C-class MADS box gene, MADS3; APOPTOSIS INHIBITOR5 (API5); and MICROSPORE AND TAPETUM REGULATOR1. In these mutants, cells within anthers began to degenerate at the meiosis or microspore stages, eventually resulting in no pollen or complete pollen collapse (Kaneko et al., 2004; Jiang et al., 2005; Jung et al., 2005, 2006; Li et al., 2006, 2010a, 2011; Chen et al., 2007; Chhun et al., 2007; Aya et al., 2009; Zhang et al., 2010a, 2010b, 2013; Hu et al., 2011; Tan et al., 2012; Niu et al., 2013).
Unlike information on anther- and meiosis-related genes in rice, only a few genes expressed in male gametophytes have been identified by loss-of-function analyses. RICE IMMATURE POLLEN1 (RIP1) encodes a conserved protein with five domains of WD40 repeats, which are thought to be involved in protein-protein interactions, and its transcript was abundant in the late stages of pollen development (Han et al., 2006). The rip1 mutation delayed pollen maturation, and the pollen grains did not germinate, resulting in male sterility (Han et al., 2006). OsRAD21-3, one of four homologs of radiation-sensitive (RAD) mutant 21 of the fission yeast in the rice genome, is preferentially expressed in microspores and pollen grains and is required for postmeiotic pollen development (Tao et al., 2007). The RA68 gene, with unknown function, is expressed preferentially in shoots and flowers, and RNAi plants of RA68 are defective in microspore mitosis and starch accumulation (Li et al., 2010b). The mutant pollen grains of SUC TRANSPORTER1 and rice IMPORTIN β1, which encodes an importin-β protein involved in the import of nuclear proteins, matured normally, but reciprocal cross experiments between heterozygous mutants and wild-type plants clearly showed that mutant alleles could not be transmitted through the male gametophyte, suggesting that the pollen grains of the mutant plants were dysfunctional (Hirose et al., 2010; Han et al., 2011). The RICE GLYCOSYLTRANSFERASE1 (OsGT1) gene is highly expressed in mature pollen and is essential for intine construction (Moon et al., 2013). Thus, our knowledge about the functions of male gametophyte-expressed genes in rice is still fragmentary, and further identification and characterization of these genes are necessary for a comprehensive understanding of pollen differentiation in angiosperms.
The endogenous retrotransposon Tos17 of rice is highly activated during tissue culture but inactive in regenerated plants (Hirochika et al., 1996). Insertional mutagenesis with Tos17 has been shown to be a useful tool for functional analysis of rice genes associated with many agronomic and biological traits (Miyao et al., 2003; Hirochika, 2010). We investigated genes important to pollen development in rice using pollen semisterility mutants tagged by insertional mutagenesis with Tos17. Here, we showed that the ARABINOKINASE-like gene, which is expressed preferentially in anthers, was required for rice pollen development, but affected neither female genetic transmission nor vegetative tissue development, and that highly homologous proteins were present in various plant species.
RESULTS
Isolation of Mutants Defective in Pollen Formation
Wild-type mature rice pollen grains are spherical in shape (approximately 40 µm in diameter), have a single aperture (germination pore), and are rich in starch granules in the vegetative cell. They contain two highly condensed sperm cell nuclei and one vegetative nucleus with dispersed chromatin. To search for mutants defective in these phenotypic traits, long-term in vitro-cultured calli were regenerated to mature plants and allowed to set pollen. We expected insertion-inactivation of putative target genes as a result of Tos17-mediated mutagenesis. We searched for plant lines defective in sperm and/or vegetative cells by examining starch accumulation and nuclear morphology under light microscopy. We expected to isolate mutant lines showing approximately 50% pollen fertility and high seed fertility compared with wild-type cv Nipponbare. Such mutants are hereafter referred to as pollen semisterile (PSS). In the first screening, we randomly chose 500 independent plants (M0 line) from individually cultured calli. Ten seeds (M1 line) from each plant were sown and grown to maturity. If at least one of the 10 plants showed a PSS phenotype, we considered the line a candidate for pollen mutants. We identified 95 candidate lines, more than one-half of which showed collapsed pollen with staining of neither starch nor nuclei. From these 95 lines, we selected 48 candidates based on the severity of abnormal starch and nuclear staining and examined their phenotypes in the M2 generation as a second screening. Sixty plants from each candidate line were grown to maturity and examined for their PSS phenotype. Twenty-nine lines contained at least one plant with the same PSS phenotype as the parental M1 line. Matured pollen of these mutant plants contained either one or two nuclei with full starch, two nuclei with little starch, or neither nucleus nor starch granules. To investigate whether the PSS phenotype was linked to Tos17 insertion events, we performed Southern hybridization analysis on 60 plants in each of the 29 mutant lines using a Tos17 DNA segment as a probe. In the ND2104 line, an approximately 3-kb fragment was always associated with the PSS phenotype (Supplemental Fig. S1).
We further examined the mutationally altered phenotype of mature pollen of the ND2104 line. In wild-type plants, almost all pollen grains were uniformly round in shape and contained normal levels of starch, as revealed by complete staining with iodine-potassium iodide (IKI) solution (Fig. 1A). By contrast, the ND2104 line produced 50% abnormal pollen; the remaining 50% were indistinguishable from wild-type pollen. Abnormal pollen grains were smaller in size (approximately 30 µm in diameter) than wild-type grains (40 µm), and many were collapsed. Most of the collapsed grains contained no starch, but in a few cases, a limited number of starch granules were observed (Fig. 1B). Mature pollen grains were stained with hematoxylin to visualize nuclei. Two identical sperm cell nuclei and one vegetative nucleus were clearly visible within individual wild-type pollen grains (Fig. 1C). In the mutant ND2104 line, the pollen grains with normal levels of starch showed the wild-type pattern of nuclear staining, whereas none of the collapsed pollen grains contained nuclei (Fig. 1D). Viable grains were stained purple by Alexander’s stain (Fig. 1E), while aborted pollen grains from mutant plants were blue (Fig. 1F). In addition, no cell wall fluorescence was detectable from collapsed pollen grains stained with calcofluor white solution, whereas all normal pollen grains emitted blue-white fluorescence (Fig. 1, G and H). Thus, these mutant pollen grains had lost almost all cytoplasm and comprised only exine, i.e. they were empty pollen grains. To test pollen germination, grains were sown on pollen germination medium; after 2 h, many normal pollen grains from the mutant plant had rehydrated and germinated, but almost no collapsed pollen grains had rehydrated (Fig. 1I). More than 70% of cv Nipponbare pollen grains germinated on the germination medium, but only about 38% of ND2104 pollen grains germinated (Fig. 1J).
Figure 1.
Phenotype and germinability of pollen grains affected by a cap1 mutation. Pollen grains were isolated from wild-type (A, C, and E) and heterozygous (+/–) cap1 (B, D, and F–H) mutant plants, and pollen germinability was examined in vitro (I and J). Pollen grains were stained with IKI solution to visualize starch granules (A and B), with hematoxylin solution to identify nuclei (C and D), with Alexander’s solution to test viability (E and F), and with calcofluor white solution to visualize cell wall (G and H). Wild-type pollen grains fully accumulated starch granules (A) and contained two sperm nuclei with condensed chromatin (C, arrowheads) and one vegetative nucleus with diffused chromatin (C, arrow). In Alexander staining, viable pollen grains were purple (E), whereas mutant grains of the cap1 heterozygous (+/–) plant were stained blue (F, arrows). No/few starch granules (B) or cytoplasmic materials (F) were detected, and neither nucleus nor cell wall was observed (G and H) in mutant pollen grains (arrows). Pollen germination of cap1 on the germination medium is shown in I. Arrowheads in I indicate germinating pollen tubes. Arrows in B, D, F–H, and I indicate collapsed mutant pollen grains. The pollen germination rates of cap1 (+/–) and cv Nipponbare are shown in J. Error bars indicate sd. Bars = 100 µm (A, B, and E–H), 50 µm (C and D), and 200 µm (I).
Molecular Cloning
To determine the chromosomal locus where Tos17 was inserted, we cloned a part of the 3-kb fragment containing the Tos17 responsible for the PSS phenotype (Supplemental Fig. S1). Genomic DNAs from PSS and wild-type plants were digested with XbaI, a linker was ligated to these fragments, and then PCR was performed using Tos17- and linker-specific primers. Several PSS-specific fragments were amplified and cloned into the vector pCRII. Sequence analysis identified an XbaI segment composed of a 2,716-bp portion of Tos17 and 111 bp of rice genome (Fig. 2A). The total size of 2,827 bp agreed well with the 3-kb length identified by Southern blotting (Supplemental Fig. S1). We further performed PCR genotyping (Supplemental Fig. S2) using primers designed to amplify both the genomic segment and the composite segment comprising Tos17 (Supplemental Table S1). All mutants generated the composite 532-bp fragment, whereas wild-type rice plants produced only the 762-bp genomic fragment (Supplemental Fig. S2). These results strongly suggested that the insertion of Tos17 was responsible for the mutant phenotype.
Figure 2.
Genomic structure of the CAP1 locus and pollen phenotypes of allelic mutant lines. A, The CAP1 genomic segment (long black bar, top) was approximately 8.8 kb in length. CAP1 comprised 28 exons (thick bars, bottom). The 3′-end part of another gene of unknown function (dotted and short black bar, top) was upstream of CAP1. The ApaI-SalI fragment (approximately 15.0 kb) was used for the complementation test in Figure 5. Arrowheads with numbers indicate the position of primers used in this study (Supplemental Table S1). The insertion positions and directions of Tos17 in ND2104 and in five allelic lines are shown with white boxes with arrowheads. In ND2104, an XbaI fragment derived from the XbaI sites in exon 10 of CAP1 and in Tos17 corresponded to the approximately 3-kb band in the Southern analysis (Supplemental Fig. S1). B, In the NF1037 and GN4590 lines, Tos17 was inserted into introns, as shown above. Genotyping was performed by PCR using the primers shown here and in Supplemental Table S1. Mature pollen grains were stained with IKI. Each line segregated to wild-type (W) and heterozygous (H) plants. Bars = 100 µm. C, In NC3090, NG3477, and NG0590 lines, Tos17 was inserted into exons, as shown above. Each line segregated to wild-type and heterozygous plants. A mutant homozygous for the Tos17 insertion (T17) also segregated in the NG0590 line. Bars = 100 µm.
The gene Os02g0141300, in which Tos17 inserted, is composed of 28 exons that encode a protein of 996 amino acids and a molecular mass of 109 kD. It appears to be a member of the galactokinase, homo-Ser kinase, mevalonate kinase, and phosphomevalonate kinase (GHMP) superfamily. The organization of exons and introns was verified against the full-length complementary DNA (cDNA) sequence (Kikuchi et al., 2003). Tos17 was inserted in exon 10 in ND2104 (Fig. 2A). Because its functional assignment is still inconclusive (see below), the gene was tentatively called COLLAPSED ABNORMAL POLLEN1 (CAP1). Detailed analysis allowed us to determine the arrangement of three prominent structural domains. The N-terminal half of CAP1 contained a glycosyltransferase family 1 domain (30–338 amino acids), while the C-terminal half contained both a Gal-binding (GB) signature (496–540 amino acids) and a GHMP N-terminal (GHMP-N) domain (638–704 amino acids; Fig. 3A). The GHMP-N domain is involved in ATP binding (Tsay and Robinson, 1991; Lee and Leustek, 1999).
Figure 3.
Domain structure and multiple alignments of CAP1 and related proteins. A, The domain structures of CAP1 (Os02g0141300, 996 amino acids), OsARA1 (Os06g0702500, 994 amino acids), AtARA1 (identified by Sherson et al. [1999]; At4g16130, 1,039 amino acids), AtAra2 (At3g42850, 964 amino acids), Sel1 (S. moellendorffii, 964 amino acids), and Phy1 (P. patens, 991 amino acids) are listed. Black triangles indicate the Tos17 insertion positions in cap1 mutant alleles. Accession numbers were NP_001045858 (CAP1), NP_001058491 (OsARA1), NP_193348 (AtARA1), NP_189871 (AtARA2), XP_002976688 (Sel1), and XP_001784003 (Phy1). B, Multiple alignment of amino acid sequences of the GT1 domain. Identical amino acids are highlighted in gray. Numbers indicate the positions of amino acid residues. The thick bar indicates the position of the GT1 domain of CAP1. C, Multiple alignment of amino acid sequences of the GB and C-terminal domains of CAP1. Identical amino acids are highlighted in gray. Numbers indicate the positions of amino acid residues. The thick bar indicates the position of the GB domain, and the double lines indicate the N-terminal domain. GT1, Glycosyltransferase domain 1 of the glycosyltransferase family; N, N-terminal domain; C, C-terminal domain of GHMP superfamily.
A closely related protein, Os06g0702500, was also found in the rice genome. This polypeptide comprised 994 amino acids with 79% amino acid sequence identity to CAP1 (Fig. 3B). Similar proteins were found in many higher plants, including Gramineae and Arabidopsis (between 90% and 71% amino acid identity), the fern Selaginella moellendorffii (67%), and the moss Physcomitrella patens (63%; Figs. 3 and 4). In Gramineae, there were at least two similar proteins in a genome that were divided into two phylogenetically distinct clades, the CAP1 clade (with about 90% identity to CAP1) and the Os06g0702500 protein clade (about 80% identity; Fig. 4). The Arabidopsis genome also contained two genes similar to CAP1; AtARA1 (At4g16130) had 75% amino acid identity to CAP1 and encoded arabinokinase (Dolezal and Cobbett, 1991; Gy et al., 1998; Sherson et al., 1999), and At3g42850, tentatively referred to as AtARA2, had 71% identity. Similarly, Os06g0702500 was tentatively termed OsARA1. CAP1 and the other proteins exhibited similar domain structures to AtARA1 (Fig. 3A). No highly homologous proteins were found in bacteria or animal genomes. Therefore, these results suggest that CAP1 is a highly conserved plant-specific gene whose product appears to act as an arabinokinase.
Figure 4.
Phylogenetic tree of CAP1 and related proteins in plants. Twenty protein sequences (Bra1 and Bra2 in Brachypodium distachyon, Sor1 and Sor2 in Sorghum bicolor, Hor1 and Hor2 in Hordeum vulgare, Pop1 and Pop2 in Populus trichocarpa, Vit1 and Vit2 in Vitis vinifera, Gly1 in Glycine max, Med1 in Medicago truncatula, Ric1 in Ricinus communis, and Phy2 in P. patens) in addition to the six proteins in Figure 3 were selected by National Center for Biotechnology Information/BLAST homology search using the amino acid sequence of CAP1. Their accession numbers were XP_003560519 (Bra1), XP_003574519 (Bra2), XP_002437544 (Sor1), XP_002453285 (Sor2), BAJ85666 (Hor1), BAJ94139 (Hor2), XP_002332102 (Pop1), XP_002331441 (Pop2), CBI20799 (Vit1), XP_002266644 (Vit2), XP_003550127 (Gy1), XP_003588615 (Med1), XP_002527993 (Ric1), and XP_001754993 (Phy2). Gramineae, dicotyledon, and lower-plant proteins are highlighted in blue, red, and yellow, respectively.
Genetic Characterization
In the initial screening described above, approximately one-half of the pollen had the collapsed phenotype. Thus, all of the male gametophytes carrying the Tos17 insertion were expected to die. To further investigate this possibility, we obtained seeds via self-pollination and grew them to maturity. Pollen fertility was evaluated with IKI staining, and Tos17 insertion was genotyped via PCR of leaf DNA. Unexpectedly, plants homozygous (–/–) for the Tos17 insertion were obtained; they comprised 13.3% (43 of 324) of the progeny (Table I). This segregation distortion suggested that a small fraction of the pollen with the Tos17 insertion was active. To confirm this observation, reciprocal crosses were performed using mutant heterozygous (+/–) and wild-type cv Nipponbare plants. When maternal cv Nipponbare plants were crossed with pollen from heterozygous PSS plants, heterozygotes plants should never be obtained if the PSS phenotype due to Tos17 insertion was complete. In fact, however, we obtained a few PSS heterozygous progeny plants (Table I). Therefore, some of the mutant pollen grains were viable. By contrast, when the PSS heterozygotes were used as female recipients, fertile wild-type and PSS heterozygous plants segregated in a 1:1 ratio (wild-type fertile:PSS heterozygous = 60:58, χ2 = 0.034; Table I), demonstrating that female fertility of the PSS plants was normal, irrespective of the presence or absence of the Tos17 insertion. The mutant homozygous plants for disrupted CAP1 could produce seeds using wild-type pollen grains (Table I). This further showed normal female transmission.
Table I. Transmission efficiency of the CAP1 allele in rice.
Numerals indicate the number of plants in each genotype class. Numbers in parentheses are the percentages of each pollen phenotype and PCR genotyping in F1 plants.
| Parental Genotypes (Female × Male) | CAP1 +/+ | cap1 +/– | cap1 –/– |
|---|---|---|---|
| cap1 +/– × cap1 +/– | 132 (40.7) | 149 (46.0) | 43 (13.3) |
| CAP1 +/+ × cap1 +/– | 18 (75.0) | 6 (25.0) | − |
| cap1 +/– × CAP1 +/+ | 60 (50.8) | 58 (49.2) | − |
| cap1 –/– × CAP1 +/+ | 0 (0) | 34 (100) | − |
The mutant homozygous plants had vegetative developments, flowering times, panicle morphologies, and tiller numbers that were indistinguishable from those of wild-type or heterozygous plants (Supplemental Fig. S3A). In addition, anthers of the homozygous cap1 plants were very similar in size and form to those of heterozygous and wild-type plants, but almost all pollen grains were collapsed (Supplemental Fig. S3, B and C). The numbers of abnormal pollen grains varied among both spikelets and individual plants. On average, only 4.2% of the pollen grains were normal. Close examination of the pollen revealed that a small fraction appeared to be normal (Supplemental Fig. S3C). As expected, very few (three) seeds were obtained from crosses of 831 spikelets (seed fertility, 0.36%) of homozygous plants (Table II). One of the three seeds did not germinate, while the remaining two grew normally. Genotyping showed that these plants were homozygous for the Tos17 insertion, and they had the collapsed pollen phenotype of their parent plants (data not shown).
Table II. Analysis of seeds in mature rice panicles.
Developed seeds and unfertilized spikelets were counted in 10 to 20 mature panicles from two plants of each genotype.
| Genotype | Developed Seeds | Unfertilized Spikelets | Developed Seeds |
|---|---|---|---|
| % | |||
| Wild type | 847 | 925 | 91.6 |
| cap1 +/– | 660 | 838 | 78.8 |
| cap1 –/– | 3 | 831 | 0.36 |
A database search for the allelic lines of the CAP1 gene using the Tos17 mutant panel (http://tos.nias.affrc.go.jp/) identified 39 lines in addition to ND2104. Tos17 mainly inserted in the C-terminal half of the protein (data not shown). We chose lines in which Tos17 inserted in introns (lines NF1037 and NG4590) and exons (lines NC3090, NG3477, and NG0590) for additional analysis (Fig. 2A). Nine to 19 seeds (M1 line) from individual lines were grown, genotyped by PCR analysis, and characterized for pollen morphology by IKI staining. In NF1037 and NG4590 (intron insertion lines), the heterozygous plants produced normal pollen, similar to the noninsertional wild-type (+/+) plants (Fig. 2B). By contrast, in NC3090, NG3477, and NG0590 (exon insertion lines), all seven heterozygous plants had PSS phenotypes very similar to that of ND2104, and all 29 noninsertional wild-type plants derived from these three lines produced normal pollen grains (Fig. 2C). As shown in Figure 3A, Tos17 was inserted in the glycosyltransferase family 1 domain in ND2104, between the GB and GHMP-N domains in NC3090, and the downstream of the GHMP-N domain in NG3477 and NG0590, generating in-frame stop codons (data not shown). Moreover, one mutant homozygous plant was obtained from NG0590; it produced many abnormal pollen grains in addition to a few normal ones (Fig. 2C). The number of abnormal grains varied among spikelets, as described above for ND2104. Thus, insertions into introns of the CAP1 gene did not affect pollen development, but insertion events into exons caused aberrant pollen maturation.
Complementation Analysis
To determine whether aberrant pollen resulted from a mutation in the CAP1 gene, we obtained the 15.0-kb ApaI-SalI genome fragment containing the entire CAP1 gene, as well as the 2.4-kb upstream and 3.8-kb downstream regions (Fig. 2A). The 15.0-kb segment also contained truncated genes of unknown function; they included only the C-terminal regions and were unlikely to contribute to complementation (Fig. 2A). The 15.0-kb fragment was introduced into ND2104 homozygotes. Complementation was evaluated by the number of normal pollen grains with fully produced starch granules. When pollen grains from 13 regenerated T0 plants (g1–g13) were stained with IKI, more than 10% were normal (Fig. 5A). Among them, g5, g7, and g9 plants each yielded more than 30% normal pollen (Fig. 5, A and B), while five nontransgenic homozygous plants (n1–n5) with the Tos17 insertion produced an average of only 4.2% normal pollen grains. No recovery was shown in 10 transformants (v1–v10) using the empty vector (pPZP2H-lac) as a negative control; these plants produced only 3.0% normal pollen grains (Fig. 5A). These results suggested that the pollen phenotype of ND2104 was caused by the insertion of Tos17 in the CAP1 gene.
Figure 5.
Complementation analysis of cap1 mutants. A, Complementation of the cap1 mutant was evaluated as the percentage of pollen grains fully stained with IKI. All plants were homozygous for the Tos17 insertion. Five nontransgenic plants (T17ho, n1–n5), 10 transgenic lines with control vector (vector, v1–v10), and 13 transgenic lines with 15.0-kb (ApaI-SalI) segment vector (15-kb [ApaI-SalI] segment, g1–g13) were examined for pollen fertility using 10 (n1–n5) or three (v1–v10 and g1–g13) spikelets. Error bars indicate sd. B, Pollen grains of transgenic g5, g7, and g9 plants stained with IKI. Bar = 200 µm. [See online article for color version of this figure.]
Observations of Mutant Pollen during Development
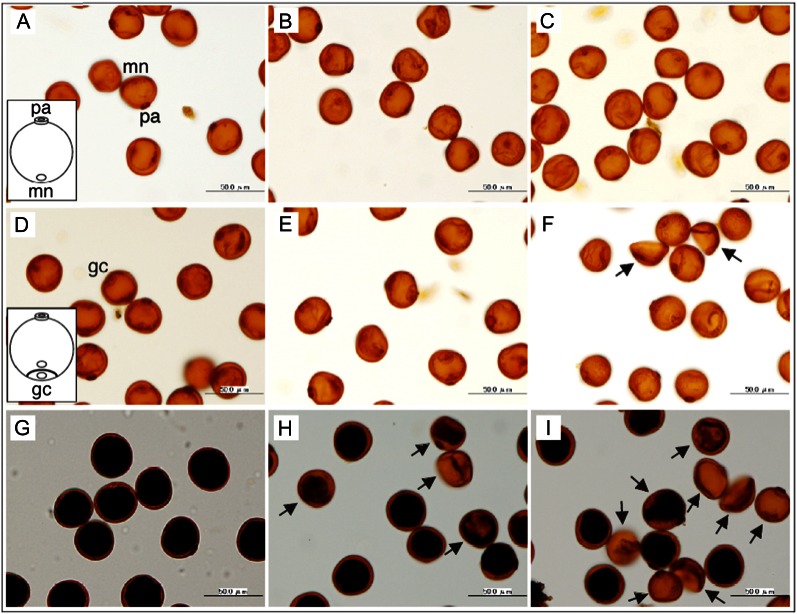
To elucidate the developmental defects in cap1 mutants, we examined pollen morphology and starch accumulation (Fig. 6). In wild-type rice, the microspore nucleus migrated from the central region toward the generative pole, which is opposite the single pollen aperture (Fig. 6A). This stage corresponds to the anther stage 10 (Zhang et al., 2011). Then, the polarized microspore underwent asymmetric cell division and cell plate formation to generate bicellular pollen grains (stage 11a; Fig. 6D). Subsequently, vegetative cells synthesized starch granules in the midbicellular stage (stage 11b; Fig. 6G), and full starch accumulation was observed in mature pollen (stage 13; Fig. 1A). In cap1 mutant lines, microspore morphology and development were indistinguishable from those of wild-type plants until the polarized stage (Fig. 6, A–C). However, after microspore mitosis, a few bicellular pollen grains in homozygotes were collapsed (Fig. 6F). In the midbicellular stage, the number of collapsed pollen grains with poor starch accumulation increased (Fig. 6I). By the end of pollen development, almost all grains were collapsed, and starch accumulation was rarely observed (Supplemental Fig. S3). In heterozygous plants, the aberrant pollen grains that were similar to those of homozygous mutants were conspicuous at the midbicellular stage (Fig. 6H). Because almost no starch accumulation was observed in the final stage in homozygous mutants, starch that was synthesized in the middle stage was apparently degraded during subsequent pollen development. In contrast to abnormality of the developing pollen grains, the anther walls of homozygous cap1 plants were very similar to those of wild-type plants during anther development (Supplemental Fig. S3, D–I; stages 10–13). We further examined the developing pollen of homozygous cap1 plants by nuclear staining (Supplemental Fig. S3, J–P). When microspores of homozygous mutant and wild-type plants at stage 10 were stained with 4′,6-diamidino-2-phenylindole (DAPI), almost all cells contained single nuclei (Supplemental Fig. S3, J, M, and P). At bicellular pollen stage (stage 11), approximately 80% of the pollen grains in wild-type plants contained two nuclei, but only 33.3% of the pollen grains of cap1 plants were bicellular stage. No nuclei were detectable in more than one-half of pollen grains (61.1%) from the homozygous cap1 plants (Supplemental Fig. S3, K, N, and P). The numbers of binucleate (bicellular) pollen grains in cap1 varied among spikelet. This variation in number of normal pollen grains probably reflects progressive cell death of cap1 pollen at this stage. At tricellular pollen stage (stage 12), two sperm nuclei and one vegetative nucleus were detected in each pollen grain of wild-type plants. By contrast, no nuclei were detected in almost all pollen grains of homozygous cap1 mutants (Supplemental Fig. S3, L, O, and P). These results suggested that the cap1 mutation affected not anther formation but pollen development in rice.
Figure 6.
Pollen development of cap1 mutants. Microspores (A, B, and C; stage 10 in Zhang et al. [2011]) and early (D, E, and F; stage 11a) and middle (G, H, and I; stage 11b) stages of bicellular pollen grains were isolated from wild-type (A, D, and G), Tos17 heterozygous (B, E, and H), and Tos17 homozygous (C, F, and I) plants. Microspores and pollen grains were stained with IKI. Microspore nuclei (mn; A), pollen apertures (pa; A), and generative cells (gc) in early bicellular pollen grains (D) were easily observed because starch accumulation occurred after the middle bicellular pollen stage. Arrows (F, H, and I) indicate collapsed pollen grains. Bars = 50 µm.
Gene Expression Analysis of CAP1
To examine the expression profiles of CAP1 in various rice tissues, we performed a semiquantitative reverse transcription (RT)-PCR analysis (Fig. 7A). After 25 cycles of RT-PCR, a very weak signal was detectable in meiotic-stage spikelets (stages 7 and 8) and in microspore stage anthers (stages 9 and 10), and no signal was detected in leaf blades, roots, lemmas/paleas, or flowering-stage pistils, nor in tricellular pollen stage anthers (stages 12 and 13), while a prominent signal was detected in anthers at the bicellular pollen stage (stage 11). After 35 cycles, a prominent or weak signal was detected in all tissues investigated. The RT-PCR data of CAP1 (Os02g0141300) agreed with the microarray expression profile in the RiceXpro database (Supplemental Fig. S4A; http://ricexpro.dna.affrc.go.jp/). We next examined the temporal and spatial expression pattern of CAP1 during anther development by in situ hybridization (Fig. 7, B–G). No signal was detected in the anther of microspore (stage 10), bicellular pollen (stage 11), and tricellular pollen (stage 13) stages using a digoxigenin (DIG)-labeled CAP1 sense probe as a control. By contrast, the hybridization signals by a DIG-labeled CAP1 antisense probe were present not only in developing pollen, but also in tapetum and endothecium (anther wall). Thus, CAP1 is preferentially expressed in anthers during pollen development. The developmental stage, when increased CAP1 expression was observed, coincided with the timing of morphological and biochemical alterations in cap1 mutants (Fig. 6; Supplemental Fig. S3), suggesting that CAP1 is closely associated with bicellular-stage pollen at stage 11.
Figure 7.
Gene expression pattern of CAP1. A, RT-PCR analysis of CAP1 and ubiquitin in vegetative and reproductive tissues. Total RNA was extracted from leaf blades of 2-month-old plants (Le), roots of 2-week-old plants (Ro), lemmas/paleas (L/P), and pistils (Pi) from spikelets before anthesis, spikelets at the meiotic stage (Me; stages 7 and 8 in Zhang et al. [2011]), and anthers at the microspore (Mi; stage 10), bicellular (Bi; stage 11), and tricellular (Tr; stages 12 and 13) stages and subjected to RT-PCR analysis. Amplifications of CAP1 were performed with 25 (top) and 35 (middle) cycles; the ubiquitin gene was amplified with 25 cycles (bottom). B–G, In situ localization of CAP1 transcript within wild-type anther locules at microspore (B and E; stage 10), bicellular pollen (C and F; stage 11), and tricellular pollen (D and G; stage 13) stages. Probed with the CAP1 antisense (B–D) and sense (E–G) probes. BP, Bicellular pollen; Ep, epidermis; En, endothecium; MS, microspore; Ta, tapetum; TP, tricellular pollen. Bar = 50 µm.
To analyze the expression profiles of three CAP1-related genes (OsARA1, Os06g0702500; AtARA1, At4g16130; and AtARA2, At3g42850) in the rice and Arabidopsis genomes, we extracted information from the RiceXpro and Arabidopsis electronic Fluorescent Pictograph browser (http://bar.utoronto.ca/efp/cgi-bin/efpWeb.cgi; Winter et al., 2007) expression databases. The OsARA1 gene was expressed in a temporally and spatially independent manner (Supplemental Fig. S4B). Similarly, the AtARA1 gene was expressed in a wide variety of tissues during development (Supplemental Fig. S5A). By contrast, the AtARA2 gene was preferentially expressed in pollen and at especially high levels during the bicellular stage (Supplemental Fig. S5). These results suggested that AtARA2 in Arabidopsis is an ortholog of CAP1 and that the putative arabinokinase is important for pollen development in both monocotyledonous and dicotyledonous plants.
DISCUSSION
In this study, we successfully isolated 29 pollen-defective mutants from Tos17-induced mutant rice lines. We examined, in detail, one pollen mutant line, ND2104, with abnormal collapsed pollen grains. The mutation affected only pollen development; vegetative growth and female function appeared to be normal. The Tos17 insertion in the ND2104 line occurred in exon 10 of the gene Os02g0141300, whose product was annotated as a member of the GHMP superfamily. Phenotype analysis using mutant alleles and a complementation test using the normal rice genome segment revealed that the Tos17 insertion in ND2104 disrupted Os02g0141300, which was designated as CAP1. If the cap1 mutation affected sporophytic processes, no pollen-defective phenotypes should be detected among heterozygous plants. The cap1 heterozygotes produced approximately 50% normal and 50% abnormal pollen grains. Thus, cap1 seemed to affect gametophyte pollen development. Hobo et al. (2008) showed that CAP1 is expressed in both microspore/pollen and tapetum tissues by laser microdissection-mediated microarray. In situ hybridization signals were detected in both the pollen grains and the anther wall at stages 10 to 13 of pollen development. However, the function of CAP1 in the tapetum is probably limited, because the homozygous cap1 plants formed normal anther wall and could produce a few pollen grains and seeds, suggesting the preservation of normal tapetum function. Almost all cap1 pollen grains were collapsed, lacked cytoplasmic components and cell walls, and could not germinate on the medium. During pollen development, microspores with the cap1 mutation gave rise to normal vegetative and generative cells, but abnormalities were evident at the bicellular pollen/anther stage 11. The number of abnormal pollen grains with no visible nuclei increased in this stage, and almost all grains were collapsed at flowering. The diameter of pollen grains at the bicellular stage was nearly identical (approximately 30 µm) between normal and mutant pollen grains. However, at the flowering stage, wild-type pollen grains were approximately 40 µm in diameter, whereas mutant pollen grains had not grown. Thus, cap1 pollen grain size was fixed at the bicellular stage. In addition, transcripts of CAP1 were preferentially accumulated at the bicellular pollen/anther stage 11. Therefore, the CAP1 protein is very likely important for bicellular pollen development.
A homology database search identified CAP1-related proteins in several plant species, including Gramineae and dicots. CAP1 had high (71%–79%) amino acid sequence identity and shared domain organization with another rice OsARA1 protein and with two Arabidopsis proteins, AtARA1 and AtARA2. Because AtARA1 was previously identified as an arabinokinase, CAP1 and its homologs probably act as arabinokinases as well. The expression patterns of these genes varied. CAP1 and AtARA2 transcripts accumulated preferentially in anthers, whereas OsARA1 and AtARA1 transcripts occurred ubiquitously in virtually all tissues. Because CAP1 was required for normal pollen development and its expression pattern was similar to that of AtARA2, AtARA2 is likely the ortholog of CAP1. However, neither AtARA1 nor OsARA1 seems to be directly involved in pollen development. This assumption is supported by the finding that the AtARA1 mutation ara1 segregates in a Mendelian fashion (Dolezal and Cobbett, 1991), suggesting that ara1 does not affect normal pollen development. We proposed that CAP1 would behave in a male gametophyte-specific way. Nevertheless, the self-fertilized progeny of heterozygous cap1 plants generated homozygous cap1 plants at 13.3% of the total progeny, suggesting that a few pollen grains were viable, even if cap1 was defective. Both CAP1 and OsARA1 transcripts were detected in bicellular and tricellular pollen grains by our in situ hybridization analysis and by microarray analysis of laser microdissection (Hobo et al., 2008). Thus, CAP1 and OsARA1 are probably partially redundant in male gametophytes of rice. A small number of cap1 pollen grain for producing 13.3% homozygous plants might be viable by OsARA1 protein. Further inspection of structural similarities among CAP1-related genes in Gramineae revealed that these genes belong to CAP1 and OsARA1 clades; the former group may play an important role in pollen development in Gramineae.
l-Arabinokinase (EC 2.7.1.46) phosphorylates l-arabinose to generate l-arabinose 1-P (Ara-1-P), which is converted to UDP-l-arabinose by UDP-sugar pyrophosphorylase (USP; Reiter, 2008; Kotake et al., 2010). UDP-l-arabinose is subsequently incorporated into cell wall polysaccharides (Reiter, 2008; Kotake et al., 2010). Mutants of AtUSP are recognized as gametophytic mutants with abnormal collapsed pollen grains, cytoplasm degradation, and no intine (cell wall; Schnurr et al., 2006; Kotake et al., 2007). Because the phenotype of usp mutants resembles that of cap1 mutants and arabinokinase is situated upstream of USP, its mutation might have affected normal pollen intine development. Alternatively, the cap1 mutant may accumulate toxic levels of l-arabinose due to failure to convert arabinose to Ara-1-P. Consistent with this prediction, Arabidopsis ara1 mutants do not effectively convert arabinose to Ara-1-P and are sensitive to exogenous arabinose application (Dolezal and Cobbett, 1991). Thus, l-arabinose metabolism appears to be required for normal pollen development.
The functional domains of CAP1 protein were predicted, as in Arabidopsis ARA1s. Three allelic mutant lines, NC3090, NG3477, and NG0590, produced very similar collapsed pollen grains to cap1. Although NG3477 and NG0590 mutants maintained all three potential functional domains intact, the plants exhibited the collapsed pollen phenotype. Thus, the C-terminal portion of CAP1 seems to be important for enzymatic function, in addition to the putative functional domains. This hypothesis should be confirmed by biochemical analysis of the enzymatic properties of CAP1.
Several sporophyte-expressed mutations of GAMYB, Ugp1, CSA, API5, and EAT1 affect normal anther formation and result in collapsed pollen (Chen et al., 2007; Aya et al., 2009; Zhang et al., 2010b; Li et al., 2011; Niu et al., 2013), resembling that of the cap1 mutant. By contrast, RA68 gene, which encodes a protein with unknown function in rice, is expressed in the tapetum and in developing spores. Pollen grains of RA68 RNAi plants were arrested in the mid/late uninucleate microspore stage, or microspore mitosis (Li et al., 2010b). OsRAD21-3, one of four rice RAD21-related genes, is expressed in pollen grains. Some pollen grains of RNAi plants of the gene were also arrested at the microspore stage (Tao et al., 2007). Another example is RIP1, which encodes a WD-repeat protein. The rip1 pollen development began to delay at the early bicellular pollen stage (at late stage 11b; Han et al., 2006). Although the sperm cells appeared to form normally in mature rip1 pollen grains, the vegetative cells were immature at flowering, resulting in deficient pollen germination (Han et al., 2006). Recently, the OsGT1 gene has been shown to be essential for intine formation, and OsGT1 acts at the early bicellular pollen stage (stage 11b; Moon et al., 2013). By contrast, pollen grains of the cap1 mutant began collapse at the early bicellular pollen stage (stage 11b). Consequently, CAP1 appears to play a role in the bicellular pollen stage (stage 11), as does both RIP1 and OsGT1, while RA68 and RAD21-3 act in the late uninucleate microspore at stage 10. The cap1 pollen grains exhibited a striking phenotype compared with these other pollen mutations. Identification of CAP1 orthologs in other plant species will enhance our understanding of the relationship between arabinokinase and pollen development in flowering plants.
MATERIALS AND METHODS
Plant Materials and Mutant Screening
Rice (Oryza sativa) ‘Nipponbare’ was used for Tos17-induced mutagenesis. Calli induced from rice seeds were propagated for 5 months in a liquid culture (http://tos.nias.affrc.go.jp/; Hirochika et al., 1996). They were then allowed to regenerate to maturity. During this culturing process, an endogenous retrotransposon of rice Tos17 is activated and inserted to cause gene mutations (Hirochika et al., 1996). Starch and nuclei of pollen grains were stained according to the procedures of Yamagata et al. (2010) and Kindiger and Beckett (1985), respectively. Panicles 4 to 6 d after heading were first fixed in 70% (v/v) ethanol. One preflowering spikelet was cut out from the fixed panicle, and six anthers from the spikelet were stained. For starch staining, two of six anthers were excised, placed onto a slide glass, squashed in a drop of 1% (w/v) IKI solution, and cleared of debris with forceps. After incubation at room temperature for a few minutes, more than 300 pollen grains were examined by light microscopy (Olympus BX41). To stain nuclei, the remaining four anthers were squashed on a glass slide in three to five drops of solution containing 35% (w/v) chloral hydrate and 45% (v/v) acetic acid. The specimens were gently heated over an alcohol lamp for 5 to 7 s until the pollen grains became transparent, and pollen nuclei were then stained for a few minutes with a solution of 1% (w/v) hematoxylin, 50% (v/v) propionic acid, and 0.25% (w/v) FeNH4(SO4)2. More than 300 pollen grains of each specimen were examined by microscopy, as above.
Pollen Viability, Cell Wall Staining, and in Vitro Pollen Germination
To test for pollen viability, anthers before anthesis were directly immersed and squashed with forceps in Alexander’s solution (Alexander, 1969), and released pollen grains were incubated at 25°C for 2 d. To stain cell walls, anthers before anthesis were fixed in solution containing 2.5% (v/v) formaldehyde and 70% (v/v) ethanol. Cells were stained with 0.02% (w/v) calcofluor white (fluorescent brightener 28, Sigma) in 0.1 m Tris-HCl buffer (pH 9.0) for a few minutes. Pollen grains were examined under a light microscope (Alexander’s staining) or an epifluorescence microscope fitted with U-MWU2 filters (BP330-385, BA420, and DM400, Olympus; calcofluor white staining). In vitro pollen germination was tested according to Mizuta et al. (2010) with minor modifications. Pollen grains from dehisced anthers were immediately placed on a pollen germination medium containing 17.5% (w/v) Suc, 0.01% (w/v) H3BO3, 0.05% (w/v) CaCl2⋅2H2O, 0.01% (w/v) KH2PO4, and 0.7% (w/v) agarose and incubated at 30°C for 2 h in a humid chamber. More than 300 pollen grains were observed in three independent examinations of three to five spikelets each.
Southern Hybridization, Molecular Cloning, and Genotyping of Plants by PCR
Genomic DNA was extracted from leaf blades of 2-month-old plants according to the method described by Rogers and Bendich (1988). For Southern hybridization, genomic DNA (approximately 5 µg) was digested with XbaI at 37°C for 6 h and electrophoresed on a 0.8% (w/v) agarose gel at 4°C for 24 h. DNA fragments were transferred onto a nylon membrane (Pall BioSupport Division). A 900-bp fragment of Tos17 (Hirochika et al., 1996) was amplified by PCR and used as a probe. The probe was labeled with a horseradish peroxidase using an enhanced chemiluminescence system (GE Healthcare). Hybridization and signal detection on chemiluminescence films were performed according to manufacture’s instructions.
Amplification of the Tos17-flanking region was performed by an adapter ligation-PCR protocol (Siebert et al., 1995) with modifications. Briefly, genomic DNA from a mutant plant was digested with XbaI, and the 5′-protruding DNA ends were filled in using the Klenow fragment. Both blunt-ended XbaI fragments were ligated to asymmetric double-stranded adapters consisting of a 48-bp oligonucleotide (forward strand, 5′-GTA ATA CGA CTC ACT ATA GGG CAC GCG TGG TCG ACG GCC CGG GCT GGT-3′) and an 8-bp oligonucleotide (reverse strand, 5′-PO4-ACC AGC CC-NH2-3′) and used as template for PCR. A composite fragment of the Tos17 segment and flanking genomic region was amplified by primary PCR using the Tos17-specific gwTos1 primer and the adapter-specific AP1 primer. A subsequent secondary nested PCR used the Tos17-specific gwTos2 primer and the adapter-specific AP2 primer (Supplemental Table S1). The first PCR mixture (40 μL) consisted of 1× PCR buffer, approximately 25 ng DNA with adapter, 0.32 mm deoxynucleoside triphosphate (dNTP) mixture, 0.5 µm AP1 primer, 1.0 µm gwTos1 primer, and 1× Advantage 2 polymerase mix (Takara Bio). The PCR program was 30 cycles of 95°C per 10 s and 68°C per 2 min. In the second PCR, 1.0 µL of a 1:100 dilution of the first PCR mixture was used as template with the AP2 and gwTos2 primers. This PCR involved 22 cycles of 95°C per 10 s and 68°C per 2 min. Amplified product that was specific to the mutant plant was subcloned into pCRII (Invitrogen Life Technologies), and the nucleotide sequence of the insert was determined.
To determine the position of the Tos17 insertion in ND2104, the nucleotide sequence was searched using BLAST in the Rice Annotation Project Database (http://rapdb.dna.affrc.go.jp/; Ohyanagi et al., 2006). Additional allelic mutants were identified using the Rice Tos17 Insertion Mutant Database (http://tos.nias.affrc.go.jp/). The domain structure of the polypeptides was examined using the Pfam database (http://pfam.sanger.ac.uk/; Punta et al., 2012) and with SMART, a simple modular architecture research tool database (http://smart.embl-heidelberg.de/; Letunic et al., 2012).
For PCR genotyping of the ND2104 mutant line and additional mutant alleles, genomic DNA (approximately 0.1 µg) was used as a template for PCR. The PCR mixture (20 µL) consisted of 1× GoTaq master mix (0.2 mm dNTP mixture, 1.5 mm MgCl2, 1× GoTaq PCR buffer, and 1 unit GoTaq DNA polymerase; Promega KK), 0.4 µm of a gene-specific primer pair (Supplemental Table S1), and 0.4 µm of the T17L primer (Supplemental Table S1). Amplification was performed with 35 cycles of 94°C per 1 min, 60°C per 1 min, and 72°C per 1 min.
Multiple Sequence Alignment and Phylogenetic Analysis
The multiple sequence alignment of amino acid sequences of CAP1 and related proteins was performed using the online ClustalW tool (http://clustalw.ddbj.nig.ac.jp/) using default parameters, and a phylogenetic tree was generated using the neighbor-joining method (Saitou and Nei, 1987) in GENETYX-Tree 2.1.0 (Software Development).
Genetic Transmission of cap1 Gene
Reciprocal crosses of wild-type and heterozygous plants were performed and examined as described previously (Nonomura et al., 2003). In brief, plants were grown to flowering stages, flowered spikelets were removed, and panicles were immersed in 42°C hot water for 7 min for emasculation. Young unopened spikelets were removed, and the opened mature flowers were pollinated with foreign pollen grains. To examine self-fertilization in the heterozygous and homozygous mutants, the spikelets that had flowered were removed from the panicles, the remaining panicles were covered with paper bags, and the eventual seeds were counted.
Observation of Cells during Pollen Development
Developmental stages of pollen grains were estimated based on visual inspection of the plants and microscopic observation. When the distances between the auricles of the flag and penultimate leaves were between –5 and 0 cm and between 3 and 6 cm, the spikelets of panicles contained microsporocytes (stages 7 and 8) and microspores (stages 9 and 10), respectively. Subsequently, when the distances between the auricle of the penultimate leaf and the top of the panicle were 8 to 10 cm and greater than 10 cm, the spikelets of panicles contained bicellular (stage 11) and tricellular pollen grains (stages 12–14), respectively. The panicles containing developing pollen grains were harvested from wild-type cv Nipponbare and mutant plants and fixed in solution containing 2.5% (v/v) formaldehyde and 70% (v/v) ethanol. Cells were incubated with the 1% (w/v) IKI solution for pollen starch staining, as described above. On the other hand, to visualize nuclei, cells were incubated with 2 μg mL–1 DAPI in phosphate-buffered saline for 1 h at 60°C as described by Han et al. (2006). More than 300 pollen grains of three to five spikelets at each developmental stage were examined using epifluorescence microscope fitted with U-MWU2 filters after DAPI staining. Detailed developmental stages were determined based on nuclear position, nuclear number, or accumulation of starch granules.
RT-PCR Analysis
Total RNA was extracted from leaf blade, root, lemma, palea, pistil, spikelet, and anther tissues using the RNeasy Plant Mini Kit (Qiagen). To avoid amplifying genomic DNA, the RNA fractions were treated with RNase-Free DNase I (RQ1; Promega KK). A total of 0.5 µg RNA was reverse transcribed using oligo(dT)18 primer for the first-strand cDNA synthesis in a 20-µL reaction mixture according to the manufacturer’s instruction (Transcriptor First Strand cDNA Synthesis Kit; Roche Diagnostics KK). The PCR mixture (20 μL) consisted of 1× PCR buffer, 0.5 µL cDNA product, 0.2 mm dNTP mixture, 0.4 µm gene-specific primers (CP-8328 and CP-8783 for CAP1 or 5-RUbi and 3-RUbi for rice ubiquitin; Supplemental Table S1), and 1× Advantage 2 polymerase mix. Amplification was performed by 25 or 35 cycles of 94°C per 1 min, 60°C per 1 min, and 72°C per 1 min.
Preparation of Anther Sections and in Situ Hybridization
For observation of anther walls, anthers in various stages of pollen development were fixed in solution containing 2.5% (v/v) formaldehyde and 70% (v/v) ethanol. After fixation, the tissues were dehydrated in an ethanol series and embedded in paraffin (Paraplast Plus, Oxford Labware). Sections of 8 µm in thickness were hydrated and stained with 0.1% (w/v) toluidine blue O (Kanto Chemical) and destained in distilled water.
For in situ hybridization, anthers were fixed with formaldehyde-acetic acid solution (10% [v/v] formalin, 5% [v/v] acetic acid, and 50% [v/v] ethanol) for 24 h at 4°C, dehydrated, and embedded in paraffin. Sections (8 µm) were hydrated and treated with 10 µg mL–1 of proteinase K (Roche Diagnostics KK) at 37°C for 40 min. The RT-PCR product of CAP1 was subcloned into pCRII vector, and the nucleotide sequence of the insert was confirmed by sequencing. DIG-labeled antisense and sense RNA probes were synthesized according to the manufacturer’s instructions using T7 and SP6 RNA polymerase (Digoxigenin Labeling Kit, Roche Diagnostics KK), after linearization of the plasmid. Specimens were incubated in the hybridization buffer (50% [v/v] deionized formamide, 300 mm NaCl, 1 mm EDTA, 1× Denhardt’s solution, 100 μg mL–1 denatured salmon sperm DNA, 50 µg mL–1 yeast [Saccharomyces cerevisiae] tRNA, 10% [w/v] dextran sulfate, and 10 mm Tris-HCl, pH 8.0) for 1 h and were then hybridized with the DIG-labeled probes overnight at 45°C in a moist chamber. Slides were washed in 2× SSC and then reacted with RNase A (10 μg mL–1) at 37°C for 30 min. After several washes with 2× and 0.2× SSC, detection of labeled probes was performed using alkaline phosphatase-conjugated anti-DIG Fab fragments and nitroblue tetrazolium/5-bromo-4-chloro-3-indolyl phosphate as substrate (Roche Diagnostics KK).
Genetic Complementation
The bacterial artificial chromosome clones including the CAP1 gene were searched in the Rice Genome Sequencing Consortium database (http://www.genome.arizona.edu/shotgun/rice/). One clone, OSJNBa0014I14, contained the full-length CAP1 gene. The 15.0-kb ApaI-SalI segment of the clone was composed of full-length CAP1 together with 2.4-kb upstream and 3.8-kb downstream regions (Fig. 2A). The fragment was subcloned to the pPZP2H-lac binary vector (Fuse et al., 2001) to generate pKU61. Either pKU61 or an empty vector plasmid (as control) was introduced into homozygous Tos17 insertion mutants by Agrobacterium tumefaciens-mediated transformation. Plants were regenerated as described by Hiei et al. (1994).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Southern-blot linkage analysis of the PSS phenotype (cap1 mutation) and Tos17 insertions.
Supplemental Figure S2. Genotyping of ND2104-line plants by PCR.
Supplemental Figure S3. Phenotype of plants homozygous for the Tos17 insertion.
Supplemental Figure S4. Tissue- and development-specific expression data of CAP1 and closely related genes in rice.
Supplemental Figure S5. Tissue- and development-specific expression data of CAP1-related genes in Arabidopsis.
Supplemental Table S1. PCR primers and oligonucleotides used in this study.
Acknowledgments
We thank M. Eiguchi (National Institute of Genetics) and S. Takahashi, K. Toyosawa, and N. Watanabe (Akita Prefectural University) for excellent technical assistance, Dr. A. Yoshimura and his laboratory members (Kyushu University) for kindly supporting us in the first screening of pollen mutants, Dr. M. Yano (National Institute of Agrobiological Sciences) for kindly providing the pPZP2H-lac vector, and Dr. N. Kurata (National Institute of Genetics) for helpful discussions and comments.
Glossary
- RNAi
RNA interference
- PSS
pollen semisterile
- IKI
iodine-potassium iodide
- GHMP
galactokinase, homo-Ser kinase, mevalonate kinase, and phosphomevalonate kinase
- cDNA
complementary DNA
- GB
Gal-binding
- GHMP-N
galactokinase, homo-Ser kinase, mevalonate kinase, and phosphomevalonate kinase N-terminal
- DAPI
4′,6-diamidino-2-phenylindole
- RT
reverse transcription
- DIG
digoxigenin
- Ara-1-P
l-arabinose 1-P
- dNTP
deoxynucleoside triphosphate
References
- Alexander MP. (1969) Differential staining of aborted and nonaborted pollen. Stain Technol 44: 117–122 [DOI] [PubMed] [Google Scholar]
- Aya K, Ueguchi-Tanaka M, Kondo M, Hamada K, Yano K, Nishimura M, Matsuoka M. (2009) Gibberellin modulates anther development in rice via the transcriptional regulation of GAMYB. Plant Cell 21: 1453–1472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker JD, Boavida LC, Carneiro J, Haury M, Feijó JA. (2003) Transcriptional profiling of Arabidopsis tissues reveals the unique characteristics of the pollen transcriptome. Plant Physiol 133: 713–725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borg M, Brownfield L, Khatab H, Sidorova A, Lingaya M, Twell D. (2011) The R2R3 MYB transcription factor DUO1 activates a male germline-specific regulon essential for sperm cell differentiation in Arabidopsis. Plant Cell 23: 534–549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borg M, Brownfield L, Twell D. (2009) Male gametophyte development: a molecular perspective. J Exp Bot 60: 1465–1478 [DOI] [PubMed] [Google Scholar]
- Chen R, Zhao X, Shao Z, Wei Z, Wang Y, Zhu L, Zhao J, Sun M, He R, He G. (2007) Rice UDP-glucose pyrophosphorylase1 is essential for pollen callose deposition and its cosuppression results in a new type of thermosensitive genic male sterility. Plant Cell 19: 847–861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chhun T, Aya K, Asano K, Yamamoto E, Morinaka Y, Watanabe M, Kitano H, Ashikari M, Matsuoka M, Ueguchi-Tanaka M. (2007) Gibberellin regulates pollen viability and pollen tube growth in rice. Plant Cell 19: 3876–3888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta R, Chamusco KC, Chourey PS. (2002) Starch biosynthesis during pollen maturation is associated with altered patterns of gene expression in maize. Plant Physiol 130: 1645–1656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deveshwar P, Bovill WD, Sharma R, Able JA, Kapoor S. (2011) Analysis of anther transcriptomes to identify genes contributing to meiosis and male gametophyte development in rice. BMC Plant Biol 11: 78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolezal O, Cobbett CS. (1991) Arabinose kinase-deficient mutant of Arabidopsis thaliana. Plant Physiol 96: 1255–1260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drakakaki G, Zabotina O, Delgado I, Robert S, Keegstra K, Raikhel N. (2006) Arabidopsis reversibly glycosylated polypeptides 1 and 2 are essential for pollen development. Plant Physiol 142: 1480–1492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuse T, Sasaki T, Yano M. (2001) Ti-plasmid vectors useful for functional analysis of rice genes. Plant Biotechnol 18: 219–222 [Google Scholar]
- Grennan AK. (2007) An analysis of the Arabidopsis pollen transcriptome. Plant Physiol 145: 3–4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gy I, Aubourg S, Sherson S, Cobbett CS, Cheron A, Kreis M, Lecharny A. (1998) Analysis of a 14-kb fragment containing a putative cell wall gene and a candidate for the ARA1, arabinose kinase, gene from chromosome IV of Arabidopsis thaliana. Gene 209: 201–210 [DOI] [PubMed] [Google Scholar]
- Han MJ, Jung KH, Yi G, An G. (2011) Rice Importin β1 gene affects pollen tube elongation. Mol Cells 31: 523–530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han MJ, Jung KH, Yi G, Lee DY, An G. (2006) Rice Immature Pollen 1 (RIP1) is a regulator of late pollen development. Plant Cell Physiol 47: 1457–1472 [DOI] [PubMed] [Google Scholar]
- Hiei Y, Ohta S, Komari T, Kumashiro T. (1994) Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J 6: 271–282 [DOI] [PubMed] [Google Scholar]
- Hirochika H. (2010) Insertional mutagenesis with Tos17 for functional analysis of rice genes. Breed Sci 60: 486–492 [Google Scholar]
- Hirochika H, Sugimoto K, Otsuki Y, Tsugawa H, Kanda M. (1996) Retrotransposons of rice involved in mutations induced by tissue culture. Proc Natl Acad Sci USA 93: 7783–7788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirose T, Zhang Z, Miyao A, Hirochika H, Ohsugi R, Terao T. (2010) Disruption of a gene for rice sucrose transporter, OsSUT1, impairs pollen function but pollen maturation is unaffected. J Exp Bot 61: 3639–3646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobo T, Suwabe K, Aya K, Suzuki G, Yano K, Ishimizu T, Fujita M, Kikuchi S, Hamada K, Miyano M, et al. (2008) Various spatiotemporal expression profiles of anther-expressed genes in rice. Plant Cell Physiol 49: 1417–1428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honys D, Twell D. (2003) Comparative analysis of the Arabidopsis pollen transcriptome. Plant Physiol 132: 640–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu L, Liang W, Yin C, Cui X, Zong J, Wang X, Hu J, Zhang D. (2011) Rice MADS3 regulates ROS homeostasis during late anther development. Plant Cell 23: 515–533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh J, Nonomura K, Ikeda K, Yamaki S, Inukai Y, Yamagishi H, Kitano H, Nagato Y. (2005) Rice plant development: from zygote to spikelet. Plant Cell Physiol 46: 23–47 [DOI] [PubMed] [Google Scholar]
- Jiang SY, Cai M, Ramachandran S. (2005) The Oryza sativa no pollen (Osnop) gene plays a role in male gametophyte development and most likely encodes a C2-GRAM domain-containing protein. Plant Mol Biol 57: 835–853 [DOI] [PubMed] [Google Scholar]
- Jung KH, Han MJ, Lee DY, Lee YS, Schreiber L, Franke R, Faust A, Yephremov A, Saedler H, Kim YW, et al. (2006) Wax-deficient anther1 is involved in cuticle and wax production in rice anther walls and is required for pollen development. Plant Cell 18: 3015–3032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung KH, Han MJ, Lee YS, Kim YW, Hwang I, Kim MJ, Kim YK, Nahm BH, An G. (2005) Rice Undeveloped Tapetum1 is a major regulator of early tapetum development. Plant Cell 17: 2705–2722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko M, Inukai Y, Ueguchi-Tanaka M, Itoh H, Izawa T, Kobayashi Y, Hattori T, Miyao A, Hirochika H, Ashikari M, et al. (2004) Loss-of-function mutations of the rice GAMYB gene impair α-amylase expression in aleurone and flower development. Plant Cell 16: 33–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi S, Satoh K, Nagata T, Kawagashira N, Doi K, Kishimoto N, Yazaki J, Ishikawa M, Yamada H, Ooka H, et al. (2003) Collection, mapping, and annotation of over 28,000 cDNA clones from japonica rice. Science 301: 376–379 [DOI] [PubMed] [Google Scholar]
- Kindiger B, Beckett JB. (1985) A hematoxylin staining procedure for maize pollen grain chromosomes. Stain Technol 60: 265–269 [DOI] [PubMed] [Google Scholar]
- Kotake T, Hirosawa C, Ando Y, Tsumuraya Y. (2010) Generation of nucleotide sugars for biomass formation in plants. Plant Biotechnol 27: 231–236 [Google Scholar]
- Kotake T, Hojo S, Yamaguchi D, Aohara T, Konishi T, Tsumuraya Y. (2007) Properties and physiological functions of UDP-sugar pyrophosphorylase in Arabidopsis. Biosci Biotechnol Biochem 71: 761–771 [DOI] [PubMed] [Google Scholar]
- Lee M, Leustek T. (1999) Identification of the gene encoding homoserine kinase from Arabidopsis thaliana and characterization of the recombinant enzyme derived from the gene. Arch Biochem Biophys 372: 135–142 [DOI] [PubMed] [Google Scholar]
- Letunic I, Doerks T, Bork P. (2012) SMART 7: recent updates to the protein domain annotation resource. Nucleic Acids Res 40: D302–D305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Pinot F, Sauveplane V, Werck-Reichhart D, Diehl P, Schreiber L, Franke R, Zhang P, Chen L, Gao Y, et al. (2010a) Cytochrome P450 family member CYP704B2 catalyzes the ω-hydroxylation of fatty acids and is required for anther cutin biosynthesis and pollen exine formation in rice. Plant Cell 22: 173–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N, Zhang DS, Liu HS, Yin CS, Li XX, Liang WQ, Yuan Z, Xu B, Chu HW, Wang J, et al. (2006) The rice tapetum degeneration retardation gene is required for tapetum degradation and anther development. Plant Cell 18: 2999–3014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T, Gong C, Wang T. (2010b) RA68 is required for postmeiotic pollen development in Oryza sativa. Plant Mol Biol 72: 265–277 [DOI] [PubMed] [Google Scholar]
- Li X, Gao X, Wei Y, Deng L, Ouyang Y, Chen G, Li X, Zhang QF, Wu C. (2011) Rice APOPTOSIS INHIBITOR5 coupled with two DEAD-box adenosine 5′-triphosphate-dependent RNA helicases regulates tapetum degeneration. Plant Cell 23: 1416–1434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormick S. (2004) Control of male gametophyte development. Plant Cell 16: S142–S153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyao A, Tanaka K, Murata K, Sawaki H, Takeda S, Abe K, Shinozuka Y, Onosato K, Hirochika H. (2003) Target site specificity of the Tos17 retrotransposon shows a preference for insertion within genes and against insertion in retrotransposon-rich regions of the genome. Plant Cell 15: 1771–1780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuta Y, Harushima Y, Kurata N. (2010) Rice pollen hybrid incompatibility caused by reciprocal gene loss of duplicated genes. Proc Natl Acad Sci USA 107: 20417–20422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon S, Kim SR, Zhao G, Yi J, Yoo Y, Jin P, Lee SW, Jung KH, Zhang D, An G. (2013) Rice glycosyltransferase1 encodes a glycosyltransferase essential for pollen wall formation. Plant Physiol 161: 663–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niewiadomski P, Knappe S, Geimer S, Fischer K, Schulz B, Unte US, Rosso MG, Ache P, Flügge UI, Schneider A. (2005) The Arabidopsis plastidic glucose 6-phosphate/phosphate translocator GPT1 is essential for pollen maturation and embryo sac development. Plant Cell 17: 760–775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu N, Liang W, Yang X, Jin W, Wilson ZA, Hu J, Zhang D. (2013) EAT1 promotes tapetal cell death by regulating aspartic proteases during male reproductive development in rice. Nat Commun 4: 1445. [DOI] [PubMed] [Google Scholar]
- Nonomura KI, Eiguchi M, Nakano M, Takashima K, Komeda N, Fukuchi S, Miyazaki S, Miyao A, Hirochika H, Kurata N. (2011) A novel RNA-recognition-motif protein is required for premeiotic G1/S-phase transition in rice (Oryza sativa L.). PLoS Genet 7: e1001265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonomura KI, Miyoshi K, Eiguchi M, Suzuki T, Miyao A, Hirochika H, Kurata N. (2003) The MSP1 gene is necessary to restrict the number of cells entering into male and female sporogenesis and to initiate anther wall formation in rice. Plant Cell 15: 1728–1739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonomura KI, Morohoshi A, Nakano M, Eiguchi M, Miyao A, Hirochika H, Kurata N. (2007) A germ cell specific gene of the ARGONAUTE family is essential for the progression of premeiotic mitosis and meiosis during sporogenesis in rice. Plant Cell 19: 2583–2594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonomura KI, Nakano M, Fukuda T, Eiguchi M, Miyao A, Hirochika H, Kurata N. (2004a) The novel gene HOMOLOGOUS PAIRING ABERRATION IN RICE MEIOSIS1 of rice encodes a putative coiled-coil protein required for homologous chromosome pairing in meiosis. Plant Cell 16: 1008–1020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonomura KI, Nakano M, Murata K, Miyoshi K, Eiguchi M, Miyao A, Hirochika H, Kurata N. (2004b) An insertional mutation in the rice PAIR2 gene, the ortholog of Arabidopsis ASY1, results in a defect in homologous chromosome pairing during meiosis. Mol Genet Genomics 271: 121–129 [DOI] [PubMed] [Google Scholar]
- Oh SA, Johnson A, Smertenko A, Rahman D, Park SK, Hussey PJ, Twell D. (2005) A divergent cellular role for the FUSED kinase family in the plant-specific cytokinetic phragmoplast. Curr Biol 15: 2107–2111 [DOI] [PubMed] [Google Scholar]
- Ohyanagi H, Tanaka T, Sakai H, Shigemoto Y, Yamaguchi K, Habara T, Fujii Y, Antonio BA, Nagamura Y, Imanishi T, et al. (2006) The Rice Annotation Project Database (RAP-DB): hub for Oryza sativa ssp. japonica genome information. Nucleic Acids Res 34: D741–D744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastuglia M, Azimzadeh J, Goussot M, Camilleri C, Belcram K, Evrard JL, Schmit AC, Guerche P, Bouchez D. (2006) γ-Tubulin is essential for microtubule organization and development in Arabidopsis. Plant Cell 18: 1412–1425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persson S, Paredez A, Carroll A, Palsdottir H, Doblin M, Poindexter P, Khitrov N, Auer M, Somerville CR. (2007) Genetic evidence for three unique components in primary cell-wall cellulose synthase complexes in Arabidopsis. Proc Natl Acad Sci USA 104: 15566–15571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pina C, Pinto F, Feijó JA, Becker JD. (2005) Gene family analysis of the Arabidopsis pollen transcriptome reveals biological implications for cell growth, division control, and gene expression regulation. Plant Physiol 138: 744–756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Punta M, Coggill PC, Eberhardt RY, Mistry J, Tate J, Boursnell C, Pang N, Forslund K, Ceric G, Clements J, et al. (2012) The Pfam protein families database. Nucleic Acids Res 40: D290–D301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghavan V. (1989) mRNAs and a cloned histone gene are differentially expressed during anther and pollen development in rice (Oryza sativa L.). J Cell Sci 92: 217–229 [DOI] [PubMed] [Google Scholar]
- Reiter WD. (2008) Biochemical genetics of nucleotide sugar interconversion reactions. Curr Opin Plant Biol 11: 236–243 [DOI] [PubMed] [Google Scholar]
- Rogers SO, Bendich AJ. (1988) Extraction of DNA from plant tissue. In Gelvin SB, Schilperoort RA, eds, Plant Molecular Biology Manual. Kluwer Academic Publishers, Dordrecht, The Netherlands, pp 1–10 [Google Scholar]
- Saitou N, Nei M. (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4: 406–425 [DOI] [PubMed] [Google Scholar]
- Sato Y, Antonio BA, Namiki N, Takehisa H, Minami H, Kamatsuki K, Sugimoto K, Shimizu Y, Hirochika H, Nagamura Y. (2011) RiceXPro: a platform for monitoring gene expression in japonica rice grown under natural field conditions. Nucleic Acids Res 39: D1141–D1148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnurr JA, Storey KK, Jung HJ, Somers DA, Gronwald JW. (2006) UDP-sugar pyrophosphorylase is essential for pollen development in Arabidopsis. Planta 224: 520–532 [DOI] [PubMed] [Google Scholar]
- Sherson S, Gy I, Medd J, Schmidt R, Dean C, Kreis M, Lecharny A, Cobbett C. (1999) The arabinose kinase, ARA1, gene of Arabidopsis is a novel member of the galactose kinase gene family. Plant Mol Biol 39: 1003–1012 [DOI] [PubMed] [Google Scholar]
- Siebert PD, Chenchik A, Kellogg DE, Lukyanov KA, Lukyanov SA. (1995) An improved PCR method for walking in uncloned genomic DNA. Nucleic Acids Res 23: 1087–1088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suwabe K, Suzuki G, Takahashi H, Shiono K, Endo M, Yano K, Fujita M, Masuko H, Saito H, Fujioka T, et al. (2008) Separated transcriptomes of male gametophyte and tapetum in rice: validity of a laser microdissection (LM) microarray. Plant Cell Physiol 49: 1407–1416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan H, Liang W, Hu J, Zhang D. (2012) MTR1 encodes a secretory fasciclin glycoprotein required for male reproductive development in rice. Dev Cell 22: 1127–1137 [DOI] [PubMed] [Google Scholar]
- Tanaka I. (1997) Differentiation of generative and vegetative cells in angiosperm pollen. Sex Plant Reprod 10: 1–7 [Google Scholar]
- Tao J, Zhang L, Chong K, Wang T. (2007) OsRAD21-3, an orthologue of yeast RAD21, is required for pollen development in Oryza sativa. Plant J 51: 919–930 [DOI] [PubMed] [Google Scholar]
- Teng C, Dong H, Shi L, Deng Y, Mu J, Zhang J, Yang X, Zuo J. (2008) Serine palmitoyltransferase, a key enzyme for de novo synthesis of sphingolipids, is essential for male gametophyte development in Arabidopsis. Plant Physiol 146: 1322–1332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian Y, Yang H, Zhang H, Dai Q, Fang J, Qing X, Lu X. (2010) The molecular mechanisms of male reproductive organogenesis in rice (Oryza sativa L.). Plant Growth Regul 61: 11–20 [Google Scholar]
- Töller A, Brownfield L, Neu C, Twell D, Schulze-Lefert P. (2008) Dual function of Arabidopsis glucan synthase-like genes GSL8 and GSL10 in male gametophyte development and plant growth. Plant J 54: 911–923 [DOI] [PubMed] [Google Scholar]
- Tsay YH, Robinson GW. (1991) Cloning and characterization of ERG8, an essential gene of Saccharomyces cerevisiae that encodes phosphomevalonate kinase. Mol Cell Biol 11: 620–631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twell D. (1994) The diversity and regulation of gene expression in the pathway of male gametophyte development. In Scott RJ, Stead AD, eds, Molecular and Cellular Aspects of Plant Reproduction, Vol 55 Cambridge University Press, Cambridge, UK, pp 83–135 [Google Scholar]
- Twell D. (2010) Male gametophyte development. In Pua EC, Davey MR, eds, Plant Developmental Biology: Biotechnological Perspectives, Vol 1 Springer-Verlag, Berlin, pp 225–244 [Google Scholar]
- Twell D. (2011) Male gametogenesis and germline specification in flowering plants. Sex Plant Reprod 24: 149–160 [DOI] [PubMed] [Google Scholar]
- Twell D, Oh SA, Honys D. (2006) Pollen development, a genetic and transcriptomic view. In Malhó R, ed, The Pollen Tube: A Cellular and Molecular Perspective. Plant Cell Monographs, Vol 3 Springer-Verlag, Berlin, pp 15–45 [Google Scholar]
- Twell D, Park SK, Hawkins TJ, Schubert D, Schmidt R, Smertenko A, Hussey PJ. (2002) MOR1/GEM1 has an essential role in the plant-specific cytokinetic phragmoplast. Nat Cell Biol 4: 711–714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Liang Y, Li C, Xu Y, Lan L, Zhao D, Chen C, Xu Z, Xue Y, Chong K. (2005) Microarray analysis of gene expression involved in anther development in rice (Oryza sativa L.). Plant Mol Biol 58: 721–737 [DOI] [PubMed] [Google Scholar]
- Wilson ZA, Zhang DB. (2009) From Arabidopsis to rice: pathways in pollen development. J Exp Bot 60: 1479–1492 [DOI] [PubMed] [Google Scholar]
- Winter D, Vinegar B, Nahal H, Ammar R, Wilson GV, Provart NJ. (2007) An “Electronic Fluorescent Pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS ONE 2: e718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamagata Y, Yamamoto E, Aya K, Win KT, Doi K, Sobrizal, Ito T, Kanamori H, Wu J, Matsumoto T, et al. (2010) Mitochondrial gene in the nuclear genome induces reproductive barrier in rice. Proc Natl Acad Sci USA 107: 1494–1499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan W, Li X, Chang Y, Wen R, Chen G, Zhang QF, Wu C. (2009) Mutation of the rice gene PAIR3 results in lack of bivalent formation in meiosis. Plant J 59: 303–315 [DOI] [PubMed] [Google Scholar]
- Zhang D, Liang W, Yin C, Zong J, Gu F, Zhang D. (2010a) OsC6, encoding a lipid transfer protein, is required for postmeiotic anther development in rice. Plant Physiol 154: 149–162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Luo X, Zhu L. (2011) Cytological analysis and genetic control of rice anther development. J Genet Genomics 38: 379–390 [DOI] [PubMed] [Google Scholar]
- Zhang H, Liang W, Yang X, Luo X, Jiang N, Ma H, Zhang D. (2010b) Carbon starved anther encodes a MYB domain protein that regulates sugar partitioning required for rice pollen development. Plant Cell 22: 672–689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Xu C, He Y, Zong J, Yang X, Si H, Sun Z, Hu J, Liang W, Zhang D. (2013) Mutation in CSA creates a new photoperiod-sensitive genic male sterile line applicable for hybrid rice seed production. Proc Natl Acad Sci USA 110: 76–81 [DOI] [PMC free article] [PubMed] [Google Scholar]