In MRB-G fruits, the expression of PcMYB10 was reduced by the methylation of regions −604 to −911 bp and −1,218 to −1,649 bp in its promoter. As a result, the expression of PcUFGT, a key gene involved in anthocyanin biosynthesis and regulated by PcMYB10, was also reduced. This might cause the inhibition of anthocyanin biosynthesis and the formation of green-skinned sport.
Abstract
Varieties of the European pear (Pyrus communis) can produce trees with both red- and green-skinned fruits, such as the Max Red Bartlett (MRB) variety, although little is known about the mechanism behind this differential pigmentation. In this study, we investigated the pigmentation of MRB and its green-skinned sport (MRB-G). The results suggest that a reduction in anthocyanin concentration causes the MRB-G sport. Transcript levels of PcUFGT (for UDP-glucose:flavonoid 3-O-glucosyltransferase), the key structural gene in anthocyanin biosynthesis, paralleled the change of anthocyanin concentration in both MRB and MRB-G fruit. We cloned the PcMYB10 gene, a transcription factor associated with the promoter of PcUFGT. An investigation of the 2-kb region upstream of the ATG translation start site of PcMYB10 showed the regions −604 to −911 bp and −1,218 to −1,649 bp to be highly methylated. A comparison of the PcMYB10 promoter methylation level between the MRB and MRB-G forms indicated a correlation between hypermethylation and the green-skin phenotype. An Agrobacterium tumefaciens infiltration assay was conducted on young MRB fruits by using a plasmid constructed to silence endogenous PcMYB10 via DNA methylation. The infiltrated fruits showed blocked anthocyanin biosynthesis, higher methylation of the PcMYB10 promoter, and lower expression of PcMYB10 and PcUFGT. We suggest that the methylation level of PcMYB10 is associated with the formation of the green-skinned sport in the MRB pear. The potential mechanism behind the regulation of anthocyanin biosynthesis is discussed.
Anthocyanin is a water-soluble flavonoid compound. It confers various different colors to plants and is essential in plant resistance, pollen spread, and UV protection (Feild et al., 2001; Regan et al., 2001; Schaefer et al., 2004). In addition, the coloration resulting from anthocyanin is an important agronomic trait, with roles in landscaping, the food industry, and health care (Lila, 2004; He and Giusti, 2010). The biosynthesis of anthocyanin is a complex process involving the regulation of a number of structural and regulatory genes (Koes et al., 2005). Mutations of these genes have led to aberrant anthocyanin biosynthesis and color changes of plant organs. For example, mutations in the structural genes CHS (for chalcone synthase), DFR (for dihydroflavonol 4-reductase), and ANS (for anthocyanidin synthase) involved in anthocyanin biosynthesis cause flower color changes (Stracke et al., 2009; Tanaka et al., 2009; Vanholme et al., 2010). Genetic mutations causing color changes of plant organs also occur in genes regulating anthocyanin biosynthesis, such as transcription factors (Boss et al., 1996; Morita et al., 2006; Park et al., 2007; Allan et al., 2008; Chiu et al., 2010). The transcription factors involved in anthocyanin biosynthesis include R2R3-MYB, bHLH (for basic helix-loop-helix), and WDR (for WD-repeat protein), which regulate the downstream structural genes by forming the MYB-bHLH-WD40 complex, which is involved in the anthocyanin biosynthesis pathway (Holton and Cornish, 1995; Winkel-Shirley, 2001; Schwinn et al., 2006; Gonzalez et al., 2008). Mutations including nucleotide substitutions, deletions, and insertions in the MYB gene result in color changes in plant organs (Boss et al., 1996; Morita et al., 2006). Transposon insertions in the bHLH gene cause white flowers in common morning glory (Ipomoea purpurea; Park et al., 2007), whereas simultaneous mutations of a nucleotide deletion in MYB and a nucleotide insertion in WDR change morning glory flower color from blue to white (Chiu et al., 2010).
Several cis-acting elements in the promoter of key genes regulating anthocyanin biosynthesis are activated by a series of signal transduction processes under the light and temperature receptors (Vitrac et al., 2000; Dela et al., 2003; Zhang et al., 2003). Transposon insertions and deletions in the promoter region of grape (Vitis vinifera) VvMYBA1 lead to a peel color change from green to red (Poudel et al., 2008), a single-nucleotide polymorphism in an apple (Malus domestica) MdMYB1 promoter causes anthocyanin biosynthesis abnormalities (Ban et al., 2007), and a microsatellite insertion in the MdMYB10 promoter results in red flesh of apple (Espley et al., 2009).
The natural occurrence of DNA methylation is another cause of plant mutation. In toadflax (Linaria vulgaris), radially symmetric floral mutants arise from epialleles of the CYCLOIDEA gene, which encodes a transcription activator of the floral asymmetry development process (Cubas et al., 1999). Hypermethylation of the phosphoribosylanthranilate isomerase1-4 (pai1-4) locus in Arabidopsis (Arabidopsis thaliana) is associated with transcriptional repression of PAI (Bender and Fink, 1995). Furthermore, hypermethylation in the promoter of the white pericarp/red cob (P1-wr) gene causes maize (Zea mays) color change, and hypermethylation in the 3′ untranslated region of the allele gene of Pl (Pl-Bh) leads to the formation of red-blotting maize (Cocciolone and Cone, 1993; Sekhon and Chopra, 2009). Hypermethylation in the promoter of MdMYB10 causes a striped pigmentation on Honey Crisp apple fruit (Telias et al., 2011).
There are some investigations of color mutants in pear (Pyrus communis; Thomas et al., 2010). For example, Wu et al. (2012) studied the pigmentation of Early Red Doyenne du Comice, which is a red-skin sport of Doyenne du Comice, and found that the PyMADS18 gene is involved in the early stages of anthocyanin biosynthesis. Pierantoni et al. (2010) reported that the red color trait of Max Red Bartlett (MRB) seems to have no direct relation with the MYB10 gene. Thus far, the mechanism behind pigmentation color mutants in pear is still unclear.
MRB is a red-skinned European pear variety (Abu-Qaoud et al., 1990). We recently found many green-skinned fruits on MRB trees (MRB-G; Supplemental Fig. S1). Based on 4 years of observation, the mutation rate of MRB is 21% to 24% (Supplemental Table S1), and the green-skinned fruits occur stably on a whole branch, or together with red-skinned fruits on a single branch, or appeared in one cluster with red-skinned fruits. Such chimerism is caused by a mutation of meristematic cells during the development of plant tissues (Pohlheim, 2003). The common mutant types causing chimera include chromosomal aberrations, transposon activation, point mutations, and DNA methylation (Pohlheim, 2003). Chromosomal aberrations and point mutations do not occur very often under natural conditions, while DNA methylation and transposon activation can happen frequently in plants (Gong et al., 2008).
In this study, we analyzed the mechanism of the formation of MRB-G and found that the methylation of the PcMYB10 promoter might be the factor causing green-skinned MRB-G. The possible reasons behind the methylation of the PcMYB10 promoter are discussed.
RESULTS
The Concentration of Anthocyanin Is Lower in the Skin of MRB-G Fruits
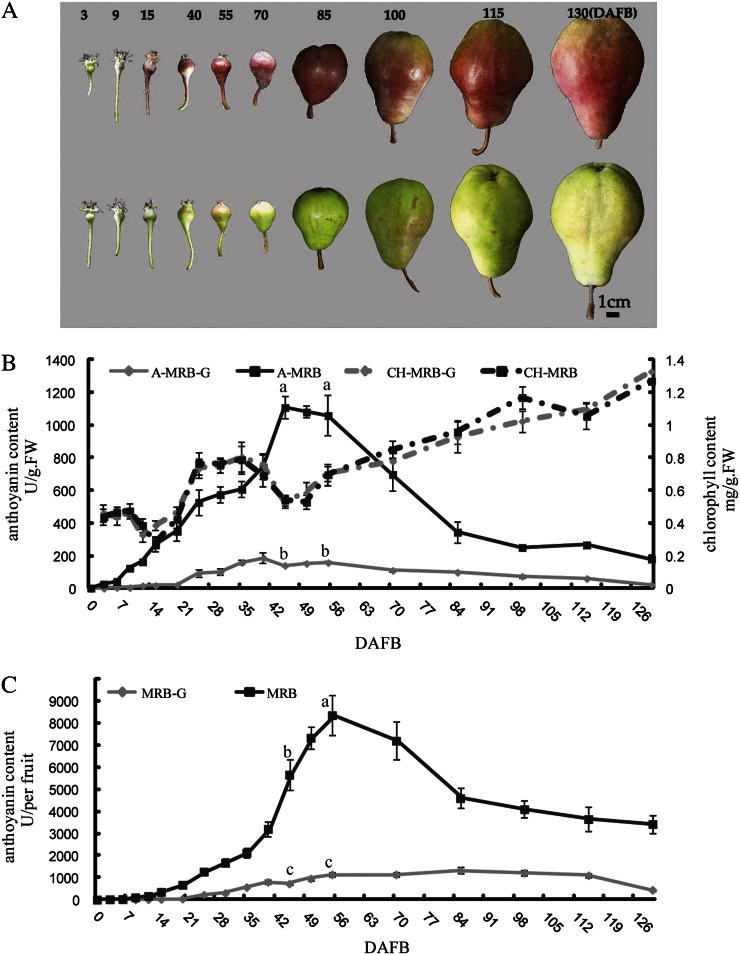
The concentrations of anthocyanin and chlorophyll in the skin of MRB and MRB-G fruits were measured at different development stages (Fig. 1). The concentration of chlorophyll in fruit skin changed similarly in both MRB and MRB-G, and no significant difference was observed between them (Fig. 1B), indicating that chlorophyll was not responsible for MRB-G. In MRB fruit skin, the concentration of anthocyanin started to increase at 9 d after full bloom (DAFB), peaked at 45 to 55 DAFB, and then gradually decreased until 85 DAFB (Fig. 1B). In MRB-G fruit skin, the concentration of anthocyanin increased slightly and then declined during fruit development, and it was significantly lower than that in MRB (Fig. 1B). We also investigated the amount of anthocyanin in the whole fruit. The concentration of fruit skin anthocyanin in whole fruit was paralleled by the concentration by unit weight in MRB and MRB-G (Fig. 1C).
Figure 1.
Change of anthocyanin in MRB and MRB-G. A, Change of fruit pigmentation of MRB (top row) and MRB-G (bottom row). The numbers at top indicate DAFB. Bar = 1 cm. B, Concentrations of anthocyanin and chlorophyll in fruit skin of MRB and MRB-G during fruit development. The numbers under the x axis indicate DAFB. The skin of a whole fruit was collected at the indicated day and mixed. One gram of this mixture was used to measure the concentrations of anthocyanin and chlorophyll (see “Materials and Methods”). A-MRB and A-MRB-G refer to the concentration of anthocyanin in MRB and MRB-G, respectively, and CH-MRB and CH-MRB-G refer to the chlorophyll concentration in MRB and MRB-G. FW, Fresh weight. C, Concentration of anthocyanin in one whole fruit of MRB and MRB-G. The whole fruit was used for the measurement of anthocyanin, and three fruits were used for each measurement. The skin of each fruit was shaved off and mixed. Three replications were performed, and the error bars represent se.
UFGT (for UDP-Glc:flavonoid 3-O-glucosyltransferase) is a key enzyme in anthocyanin biosynthesis. The activity of UFGT was measured in fruit skin at 55 and 115 DAFB of MRB and MRB-G (Supplemental Fig. S2). UFGT enzyme was active in both MRB and MRB-G fruit. Its activity in MRB was 25% higher than that in MRB-G at 55 DAFB and 15% higher at 115 DAFB. From 55 to 115 DAFB, the activity of UFGT did not change in MRB-G, similar to that of anthocyanin concentration.
The Expression of PcUFGT, a Key Structural Gene in Anthocyanin Biosynthesis, Was Much Lower in MRB-G Fruits
To investigate whether structural genes in anthocyanin biosynthesis were involved in the trait differences between MRB and MRB-G, we isolated seven structural genes: PcPAL (for l-Phe ammonia lyase), PcCHS, PcCHI, PcF3H (for flavonone-3-hydroxylase), PcDFR, PcANS, and PcANR (for anthocyanidin reductase). These genes were amplified from 55-DAFB fruit skin of MRB and MRB-G, with primers designed according to the relevant gene sequences as found in GenBank (Supplemental Table S2). Two other structural genes, PcUFGT and PcF3′H (for flavonoid 3′-hydroxylase), were isolated by homology cloning and RACE. The coding regions of PcUFGT and PcF3′H were 1,440 and 1,536 bp, respectively, and showed 89.05% identity with apple MdUFGT (AF117267) and 95.05% identity with MdF3′H (FJ919632). All genes obtained were submitted to GenBank under the accession numbers listed in Supplemental Table S2.
The complementary DNA (cDNA) sequences of the nine structural genes above were compared between MRB and MRB-G; however, no differences were found. We also compared the expression patterns of these structural genes in MRB and MRB-G during fruit development (Supplemental Fig. S4). There was no difference in the expression of PcPAL, PcCHS, PcCHI, and PcF3′H between MRB and MRB-G. The expression of PcF3H, PcDFR1/2, PcANS, and PcANR was higher in MRB than in MRB-G in the early fruit development stage (about 0–55 DAFB) but lower in the middle fruit development stage (about 55–85 DAFB). The expression of PcUFGT in MRB fruit increased from 9 DAFB, reached a maximum at 55 DAFB, and then declined to a very low level. In MRB-G fruits, PcUFGT expression increased slightly and then dropped at 70 DAFB. Moreover, the expression of PcUFGT was substantially higher in MRB fruits than in MRB-G. This expression pattern paralleled anthocyanin accumulation differences between MRB and MRB-G fruits. Interestingly, PcUFGT expression in MRB and MRB-G fruit decreased sharply at 70 and 55 DAFB, respectively, suggesting that anthocyanin biosynthesis may have stopped or slowed after this stage. In addition, the expression of all genes increased sharply at 130 DAFB.
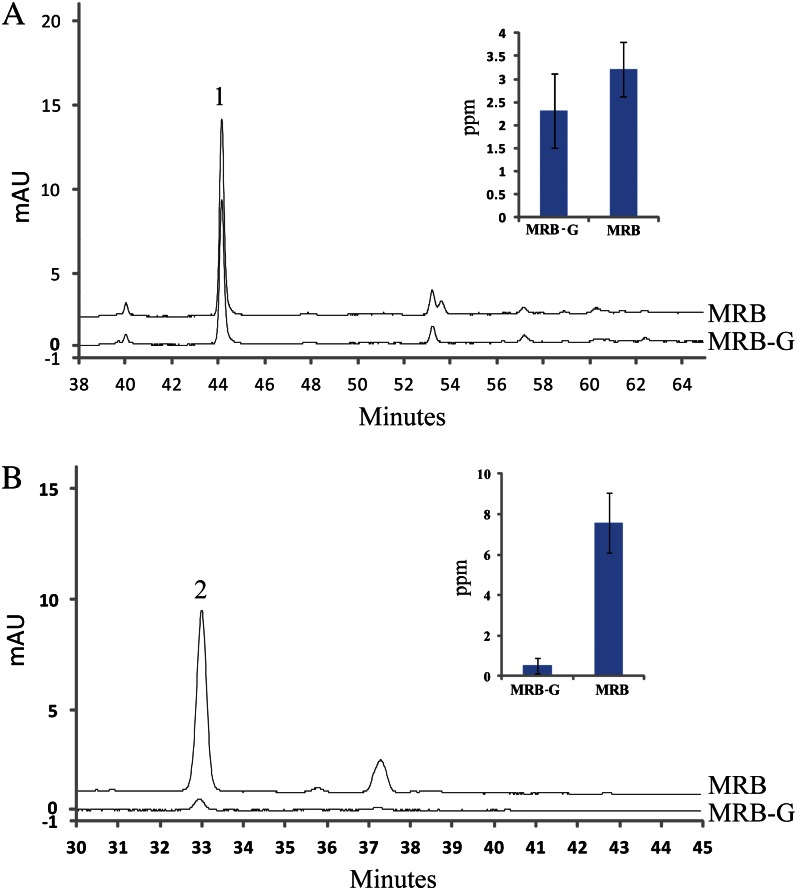
To further assess whether PcUFGT is a key causal factor behind the different colors of MRB and MRB-G, we analyzed the concentrations of the substrate and product of the UFGT enzyme in the fruits of MRB and MRB-G at 55 DAFB. The concentration of the main product of the UFGT enzyme, cyanidin 3-galactoside, showed a clear difference between MRB and MRB-G, while cyanidin, the substrate of the UFGT enzyme, showed no observable difference (Fig. 2).
Figure 2.
HLPC analysis of anthocyanin precursors and anthocyanin. The fruit peel at 55 DAFB was used as the material to analyze the amount of cyanidin 3-galactoside (B) and its precursor cyanidin (A). Peaks 1 and 2 indicate cyanidin and cyanidin 3-galactoside, respectively. All tests were performed with three biological repeats. mAU, Milliabsorbance unit. [See online article for color version of this figure.]
The above results indicated that the reduced concentration of anthocyanin in MRB-G could be the result of the low expression of PcUFGT.
Lower Expression of PcMYB10 Led to Lower Expression of PcUFGT in MRB-G Fruits
To investigate why PcUFGT was expressed at lower levels in MRB-G, we amplified the promoter region of PcUFGT (1,790 bp from the transcription initiation site) from genomic DNA of MRB and MRB-G, but no nucleotide sequence difference was observed.
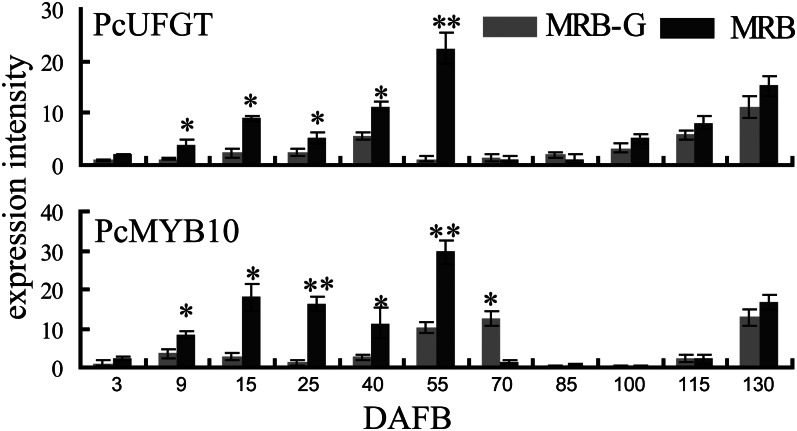
The structural genes of anthocyanin biosynthesis are usually regulated by a protein complex formed from three types of transcription factors, MYB, bHLH, and WD40 (Gonzalez et al., 2008). We isolated three transcription factor genes related to anthocyanin synthesis: PcMYB10 (JX403957), PcbHLH (JX403960), and PcWD40 (JX403961) from MRB. Among these genes, PcMYB10 has the same sequence as MYB10 (EU153575) of pear, and PcbHLH and PcWD40 have higher identity with their respective homologs in apple (Supplemental Table S2). The coding regions of PcMYB10, PcbHLH, and PcWD40 were the same between MRB and MRB-G, but their expression levels showed some differences. In particular, the expression of PcMYB10 exactly paralleled the expression of PcUFGT, both being much lower in MRB-G fruits than in MRB (Fig. 3). Therefore, we focused on the PcMYB10 gene in our study.
Figure 3.
Relative expression of PcUFGT and PcMYB10 during MRB and MRB-G fruit development. Numbers under the x axis indicate DAFB. Total RNA of the fruits collected at each indicated date was used for cDNA synthesis and quantitative reverse transcription-PCR (see “Materials and Methods”). Three replications were performed, and the error bars represent se. Asterisks indicate significantly different values (*P < 0.05, **P < 0.01).
To investigate the regulation of PcUFGT by PcMYB10, we isolated the promoter region of PcUFGT (1,790 bp from the transcription initiation site), which we named proUFGT (JX403963). When proUFGT was ligated to pCAMBIA 1305 vector holding a GFP gene and transformed into tobacco (Nicotiana benthamiana) leaves, it was able to initiate GFP expression (Supplemental Fig. S5), suggesting that it was an active promoter.
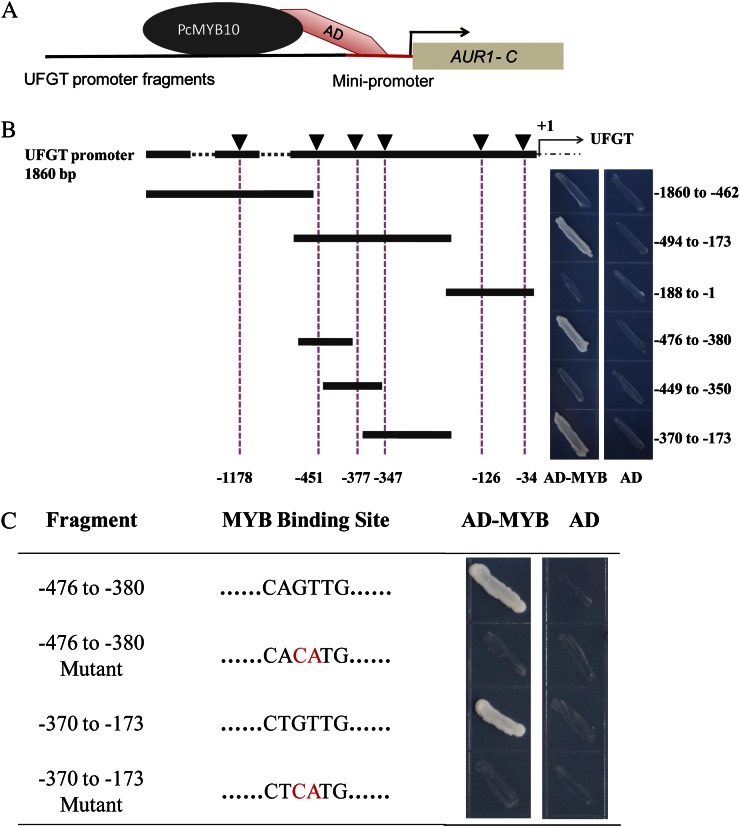
proUFGT contained six MYB-binding sites (MBS) related to anthocyanin biosynthesis (Supplemental Table S3). A yeast one-hybrid (Y1H) assay was performed to test the association of PcMYB10 with proUFGT. The results showed that PcMYB10 could associate with two MBS located at −347 and −451 bp of proUFGT (Fig. 4B), and the mutation in these binding sites could break the association (Fig. 4C), supporting the hypothesis that PcMYB10 was the transcription factor associating with PcUFGT. These results indicated that the lower expression of the transcription factor PcMYB10 resulted in the lower expression of PcUFGT in MRB-G.
Figure 4.
The association of PcMYB10 with the promoter of PcUFGT by Y1H assay. A, The Y1H system was used to determine PcMYB10-binding sites at the UFGT promoter. B, Diagrams of the UFGT promoter deletion series and Y1H analysis of PcMYB10. Six MBS are indicated by black triangles. Numbers under the dotted line indicate the locations of MBS, and numbers on the right side indicate the fragments on the promoter on PcUFGT used for Y1H assays. C, Two MBS were mutated and interacted with the PcMYB10 protein. Each fragment of the UFGT promoter was ligated to pAbAi vector to generate pAbAi-bait plasmids, and AD-MYB was transformed into the Y1HGold strain holding pAbAi-bait and screened again on a selective synthetic dextrose medium lacking uracil/AbA plate. An empty pGADT7 vector (AD) was transformed as a negative control. [See online article for color version of this figure.]
The High Degree of Methylation in the PcMYB10 Promoter Resulted in Its Lower Expression in MRB-G Fruits
To investigate why PcMYB10 was expressed at lower levels in MRB-G, the promoter of PcMYB10 (1,725 bp from the transcription initiation site) was isolated from MRB and MRB-G by chromosome walking and assigned the name proPcMYB10 (JX403962). It contained a core promoter at position −60 bp and an enhancer promoter at position −675 bp (as analyzed by TSSP software on softberry; http://linux1.softberry.com/berry.phtml). Besides the promoter elements, the sequence also contained many cis-acting elements related to light, temperature, and other environmental factors (Supplemental Fig. S6A; Supplemental Table S4). To test the activation of proPcMYB10, the 1,725-bp proPcMYB10 was cloned into a pCAMBIA 1305 vector and transformed into tobacco leaves. The result showed that it could activate the expression of GFP (Supplemental Fig. S5), indicating that proPcMYB10 was an active promoter.
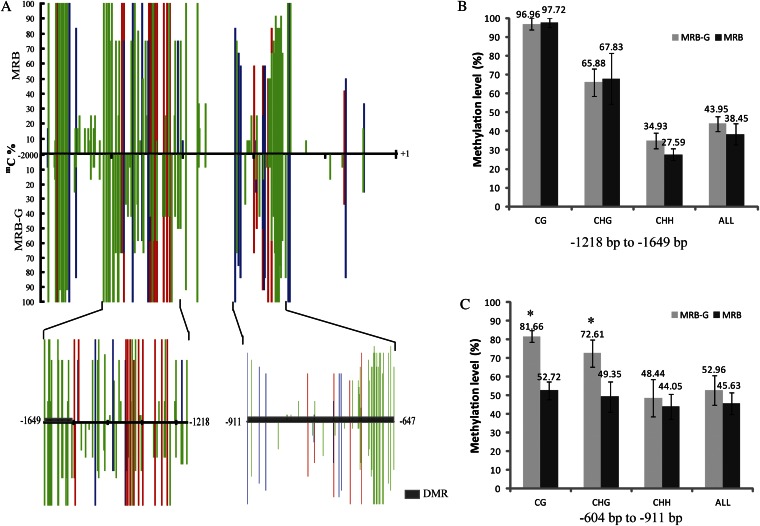
The nucleotide sequence of proPcMYB10 was compared between MRB and MRB-G, but no difference was found. This indicated that the methylation of proPcMYB10 might cause the differential PcMYB10 expression between MRB and MRB-G. Therefore, the methylation of proPcMYB10 was predicted by a methylation-specific endonuclease (McrBC)-PCR. The degree of methylation in the regions +1 to −377 bp and −378 to −707 bp showed no difference between MRB and MRB-G fruit skin. However, the regions −707 to −1,208 bp and −1,209 to −1,847 bp were quite different in methylation (Supplemental Fig. S6B). To verify this result, we also analyzed the methylation level of cytosine in proPcMYB10 of MRB and MRB-G fruit skin by bisulfite sequencing (BSP)-PCR. The regions −604 to −911 bp, −1,218 to −1,649 bp, and −1,804 to −1,965 bp were highly methylated, and the methylation level of regions −604 to −911 bp and −1,218 to −1,649 bp showed the most difference between MRB and MRB-G (Fig. 5A). The methylation level of the region 604 to −911 bp in proPcMYB10 of MRB fruit skin was 45.6% and showed no significant difference with MRB-G. Moreover, the methylation levels of CG and CHG (where H is A, C, or T) cytosines were 28.9% and 23.2% lower in MRB than those in MRB-G (Fig. 5B). Methylation of the −1,218 to −1,649 bp region of proPcMYB10 in MRB fruit skin was 5.5% lower than that in MRB-G. In this region, only the CHH-type cytosine methylation was 7.3% lower in MRB than that in MRB-G (Fig. 5B). Interestingly, the CHH-type cytosine methylation inside this region (−1,557 to 1,628 bp) in MRB was 27% lower than that in MRB-G (Supplemental Fig. S7C).
Figure 5.
Cytosine methylation levels of proPcMYB10 in MRB and MRB-G fruit peel. A, Cytosine methylation levels of proPcMYB10 estimated using BSP-PCR. B, Methylation levels of different types of cytosine in the −1,218 to −1,649 bp region of proPcMYB10. C, Methylation levels of different types of cytosine in the −604 to −911 bp region of proPcMYB10. A total of 500 ng of genomic DNA extracted from 55-DAFB fruit peels of MRB and MRB-G was used to perform bisulfite modification, the product was used as a template to amplify proPcMYB10 by five primer pairs, and 36 independent clones from each PCR were sequenced and analyzed by the software Kismeth. Three biological replications were performed. mC%, Percentage methylation of each cytosine in 36 clones; DMR, differentially methylated region. Red, blue, and green bars indicate CG-, CHG-, and CHH-type cytosine methylation, respectively. +1 indicates the translation start site, and the x axis shows the nucleotide positions relative to the ATG translation start site. H indicates A, C, or T. Asterisks indicate significantly different values (P < 0.05).
We also analyzed the methylation level of the −1,557 to −1,628 bp region of proPcMYB10 in young fruit skin (55 DAFB) and mature fruit skin (115 DAFB) of MRB and MRB-G. The methylation of this region in young and mature MRB fruit skin was 48.7% (Supplemental Fig. S7, A and C) and 46.1% (Supplemental Fig. S7, B and C), respectively, which was 27.0% and 32.4% lower than that in MRB-G. These data indicated that the methylation level of −1,557 to −1,628 bp proPcMYB10 did not change much during fruit development.
Virus-Induced Gene Silencing Led to Abnormal Coloring of Young MRB Fruit Peel
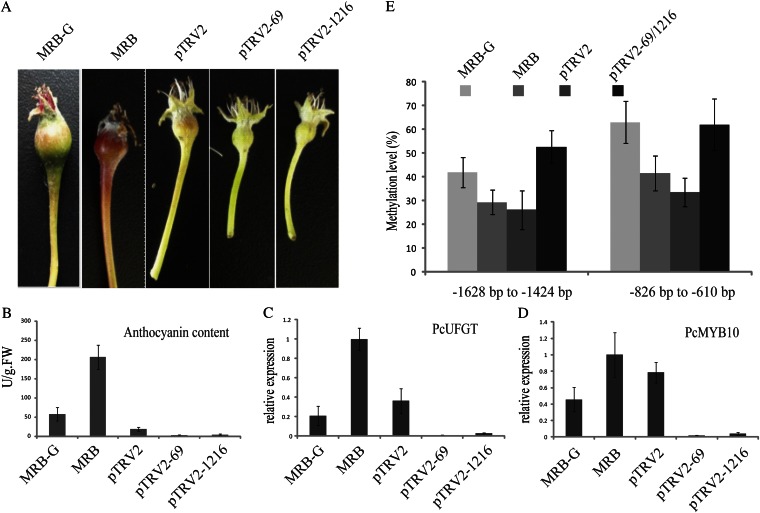
Virus-induced gene silencing (VIGS) potentially induces two types of gene silencing: posttranscriptional gene silencing and transcriptional gene silencing. Viral vectors containing target gene fragments transformed into the plant can inhibit the expression of homologous genes and lead to posttranscriptional gene silencing (Brodersen et al., 2008). If DNA sequences such as promoter sequence are ligated into viral vector and transformed into the plant, this causes transcriptional repression through the cytosine methylation of corresponding DNA sequences, which leads to transcriptional gene silencing (Bai et al., 2011). In this study, we used two VIGS plasmids to induce transcriptional gene silencing of PcMYB10 in young MRB fruit: pTRV2-69 containing the DNA fragment −604 to −911 bp and pTRV2-1216 containing the DNA fragment −1,218 to −1,649 bp of proPcMYB10. MRB inflorescences were bagged through full bloom to 10 DAFB to keep the fruit unpigmented. After removing the bags, the fruits (on trees) were immediately infiltrated with Agrobacterium tumefaciens strain AH105 holding plasmids pTRV2-69 and pTRV2-1216 and exposed to natural light. Fruits infiltrated with empty pTRV2 vector were used as a control. Four days after infiltration, the control fruits turned red, while fruits infiltrated with pTRV2-69 or pTRV2-1216 remained green (Fig. 6A).
Figure 6.
Transcriptional gene silencing of the PcMYB10 gene by VIGS in MRB young fruit. A, The infiltrated fruits were exposed to natural light for 4 d. B, Concentration of anthocyanin in the skin of infiltrated fruits. FW, Fresh weight. C, Expression of PcUFGT in the infiltrated fruits. D, Expression of PcMYB10 in the infiltrated fruits. E, Methylation analysis of proPcMYB10 in the peel of MRB-G, MRB, and MRB fruit infiltrated with pTRV-69 and pTRV-1216. MRB inflorescences were bagged at full bloom, bags of fruitlets at 10 DAFB were removed, and the fruit was used for coinfiltration by pTRV1 and pTRV2 or pTRV2-69 and pTRV2-1216 (see “Materials and Methods”). All tests were performed in three replications. pTRV2, Infiltrated with pTRV2 (empty vector); pTRV2-69, infiltrated with pTRV2-69 (carrying the −604 to −911 bp fragment of PcMYB10); pTRV2-1216, infiltrated with pTRV2-1216 (carrying the −1,218 to −1,649 bp fragment of PcMYB10). pTRV1 was coinfiltrated with the other plasmids in every experiment.
We then measured the concentration of anthocyanin in the peel of infiltrated fruits. It was 7.46 and 3.99 times lower in fruits infiltrated with pTRV2-69 and pTRV2-1216 than in control fruits (Fig. 6B). In fruit infiltrated with pTRV2-69 and pTRV2-1216, the expression of PcUFGT was 37.2 and 12.1 times lower than in control fruits, respectively. And the expression of PcMYB10 was 44.6 and 19.7 times lower than in control fruits (Fig. 6, C and D). Then, we analyzed the methylation level of the −826 to −610 bp and −1,628 to −1,424 bp regions of proPcMYB10 in infiltrated fruits. The methylation level increased 28.5% in the −826 to −610 bp region and 26.7% in the −1,628 to −1,424 bp region after infiltration (Fig. 6E). At the same time, the methylation levels of all three cytosine types (CHH, CHG, and CG) were increased significantly (Supplemental Fig. S10).
These results demonstrated that the methylation of proPcMYB10 repressed the expression of PcMYB10 and PcUFGT and subsequently inhibited the biosynthesis of anthocyanin, which probably caused the formation of a green-skin chimera sport of MRB.
DISCUSSION
Plant diversity is enhanced by the process of mutation. The color change of plant tissue is one of the commonly observed phenotypes of natural mutation. The mechanisms of color change in some plants have been studied extensively (Tegopati, 2001; Li et al., 2005; Yamaguchi et al., 2009; Palai and Rout, 2011), but not in the sport varieties of the European pear. In this paper, we studied the mechanism of the formation of green-skinned MRB-G fruit and demonstrated that hypermethylation of the PcMYB10 promoter was probably the causative factor. To our knowledge, this is the first report on the mechanism of fruit color change in the European pear.
The Change of Anthocyanin Concentration in MRB and MRB-G
Chlorophyll and anthocyanin are the most important pigments in the color formation of plant tissues, including fruit. In this study, we measured the concentrations of chlorophyll and anthocyanin in fruit skin of MRB and MRB-G at different developmental stages (Fig. 1A). The concentration of chlorophyll showed no difference between MRB and MRB-G fruit skin (Fig. 1B), suggesting that the formation of green skin of MRB-G is not related to chlorophyll. A very low concentration of anthocyanin was observed in MRB-G fruit (from 0 to 55 DAFB; Fig. 1B), which was not unexpected due to its green color. In addition, unlike the anthocyanin biosynthesis in apple fruit, which starts immediately prior to fruit maturation (Honda et al., 2002), the concentration of anthocyanin in MRB fruits increased from 9 DAFB and started to decline after 55 DAFB (Fig. 1B). This change paralleled the change of fruit pigmentation (Fig. 1A), indicating that the degradation of anthocyanin might be faster than its biosynthesis after 55 DAFB. On the other hand, UFGT is a key enzyme in plant anthocyanin biosynthesis (Kobayashi et al., 2002; Ubi et al., 2006). In MRB, the UFGT enzyme activity in the peel of mature fruit was lower than that in the peel of young fruit (Supplemental Fig. S2), as was UFGT expression. Therefore, the biosynthesis of anthocyanin declined at the later developmental stages of MRB fruit. Steyn et al. (2004) have reported that the anthocyanin content in Rosemarie pear declines at late development stages, due to an increase of the anthocyanin degradation rate. In the case of MRB fruit, further study is needed to confirm whether the degradation of anthocyanin is increased at later stages.
Expression of Key Genes Involved in the Biosynthesis of Anthocyanin
The biosynthesis of anthocyanin involves at least nine enzymes (Supplemental Fig. S3). We compared the expression of these genes during the development of MRB and MRB-G fruits (Fig. 3; Supplemental Fig. S4). Several genes, such as PcF3H, PcANS, PcANR, and PcUFGT, were expressed differentially, leading us to speculate that these genes contributed to the difference between MRB and MRB-G fruits. Furthermore, the expression of PcUFGT showed obvious consistency with the changing pattern of anthocyanin. This result is consistent with the results in other species, such as apple and grape (Kobayashi et al., 2002; Ubi et al., 2006), suggesting an essential role of UFGT in the biosynthesis of anthocyanin. In our study, we measured the concentration of the UFGT enzyme product, which was significantly higher in MRB than in MRB-G fruit, again suggesting its important role (Supplemental Fig. S2). As a result, our study focused on the function of PcUFGT in the difference of anthocyanin biosynthesis between MRB and MRB-G fruits. However, no difference was observed between MRB and MRB-G by analyzing the coding region and promoter region of PcUFGT, suggesting that other factors upstream may contribute to the difference between MRB and MRB-G.
Transcription factors such as MYB are important regulators in the biosynthesis of anthocyanin in plants (Kobayashi et al., 2002; Ban et al., 2007; Gonzalez et al., 2008; Espley et al., 2009). The promoter region and expression pattern of PcMYB10 were analyzed in developing fruit of MRB and MRB-G. The nucleotide sequence of the PcMYB10 promoter was the same in MRB as in MRB-G, while PcMYB10 expression was different between MRB and MRB-G and showed the same trend as PcUFGT during fruit development (Fig. 3). Interestingly, the expression patterns of both PcUFGT and PcMYB10 paralleled the change of anthocyanin concentration in MRB and MRB-G fruits (Figs. 1B and 3). Our study also showed that PcMYB10 could associate with the promoter of PcUFGT (Fig. 4), indicating that PcMYB10 might be a key gene contributing to the difference between MRB and MRB-G.
In addition, the expression of anthocyanin biosynthesis genes and the regulatory gene PcMYB10 increased rapidly at 130 DAFB in MRB and MRB-G (Fig. 3; Supplemental Fig. S4). Because the fruits at 130 DAFB are fully mature, the contents of secondary metabolites such as a variety of flavonoids have changed greatly (Bain, 1961); we presume that the expression increase in anthocyanin biosynthesis genes at 130 DAFB is an adaptation to secondary metabolite variation at fruit maturity.
Methylation of the PcMYB10 Promoter
The aberrant expression of MYB could be caused by promoter sequence variations, such as insertion of transposons (Kobayashi et al., 2004) and other sequences (Espley et al., 2009) into the promoter region, and additionally by promoter methylation (Cocciolone and Cone, 1993; Telias et al., 2011). In this study, the expression of PcMYB10 was much lower in MRB-G fruit than in MRB fruit (Fig. 3). To ascertain the reason for the expression difference of PcMYB10, we cloned the promoter of PcMYB10 from MRB and MRB-G fruits and compared their sequences, but no differences were observed. This suggests that the decreased expression of PcMYB10 in MRB-G might result from the methylation of its promoter region. Therefore, we analyzed the methylation of the PcMYB10 promoter. In addition to some very small changes in methylation between MRB and MRB-G that may be caused by multiple cell types, methylation of the −604 to −911 bp and −1,218 to −1,649 bp regions in the PcMYB10 promoter was much higher in MRB-G fruit skin than that from MRB fruit skin. A similar result was obtained in apple, in which the methylation of the promoter of MdMYB10 caused a striped phenotype in Honey Crisp (Telias et al., 2011). To better understand the different methylation between MRB and MRB-G fruit, we also analyzed the methylation of three types of cytosine (CHH, CHG, and CG) in the two regions mentioned above. CG and CHG cytosine types in the −604 to −911 bp region of the PcMYB10 promoter have higher methylation levels in MRB-G fruit peel than in MRB. In region −1,218 to −1,649 bp of the PcMYB10 promoter, only CHH-type cytosine showed a higher methylation level in MRB-G fruit peel than in MRB. The type of DNA methylation that induced plant color mutation was largely CG/CHG cytosine methylation, such as the CG/CHG cytosine methylation that occurs in the promoter of the P1-wr gene, inducing color mutation of maize (Sekhon and Chopra, 2009). However, there are no previous reports showing that CHH cytosine hypermethylation and CG/CHG cytosine hypermethylation exist in different regions of a single promoter. We speculate that the DNA methylation-based regulatory mechanism of woody plants (e.g. pear) is more complicated than that occurring in herbaceous plants (e.g. maize), and the existence of both types of CHH and CG/CHG cytosine methylation in the promoter of PcMYB10 increases the stability of the green mutant of MRB through the generations.
In region −1,557 to −1,628 bp, CHH-type cytosine methylation level in MRB fruit peel was 27% lower than in MRB-G. This region may play an important role in the regulation of the PcMYB10 promoter, but with no strong cis-acting elements in this region. We speculate that methylation is not directly blocking the binding of the MYB but acting through an alternative mechanism. We also tested the methylation of −1,557 to −1,628 bp of the PcMYB10 promoter in leaf of MRB and MRB-G and found that methylation level in MRB-G leaves was only 15.6% higher than in MRB (Supplemental Fig. S9), which is different from the results in fruit. We suspect that this variation is due to the different tissues.
To clarify whether PcMYB10 promoter methylation induced the aberrant expression of PcMYB10 in MRB-G, we used a VIGS system in which the endogenous PcMYB10 promoter was methylated. This suppressed its expression, which indicated that the methylation level of the PcMYB10 promoter was increased in fruits infiltrated with plasmid pTRV2-69 and pTRV2-1216 (Fig. 6E; Supplemental Fig. S10), and the anthocyanin synthesized in these fruits decreased to trace levels (Fig. 6B). Meanwhile, the expression of PcUFTG and PcMYB10 also decreased in the filtrated fruits (Fig. 6, C and D). These data indicate that DNA methylation in the promoter of PcMYB10 was associated with reduced transcript accumulation, which repressed the expression of PcUFGT and the biosynthesis of anthocyanin and, consequently, led to green-skinned sport.
Cytosine methylation variations have the potential to play a role in phenotypic plasticity as a response to environmental stress. For example, the mangrove trees (Rhizophoraceae) growing on riverside habitat microenvironments are larger and display more cytosine methylation than stunted trees from nearby salt marshes (Lira-Medeiros et al., 2010). This finding is consistent with the view that epigenetic variation among individuals with similar genotypes can drive phenotypic variation in response to varying physical environments (Richards, 2011). Actually, MRB is a red-peel sport of Bartlett found in the United States, and MRB-G is found in an orchard near Beijing. The difference in methylation level between MRB and MRB-G might be caused by the adaption to new environments. Further study is needed to determine this.
MRB is a red-skin sport of the green-skinned variety Bartlett. In our study, the methylation level of −1,557 to −1,628 bp in the PcMYB10 promoter of MRB fruit peel was not significantly different from that of Bartlett (Supplemental Fig. S8), which indicated that MRB-G was not a reversion of Bartlett.
The expression of F3H, DFR, ANS, and ANR was increased during the mid development stage of MRB-G fruit. While other elements such as transcription factors might be available to regulate their expression, more work needs to be done to examine whether they contribute to the formation of green-skinned sport.
CONCLUSION
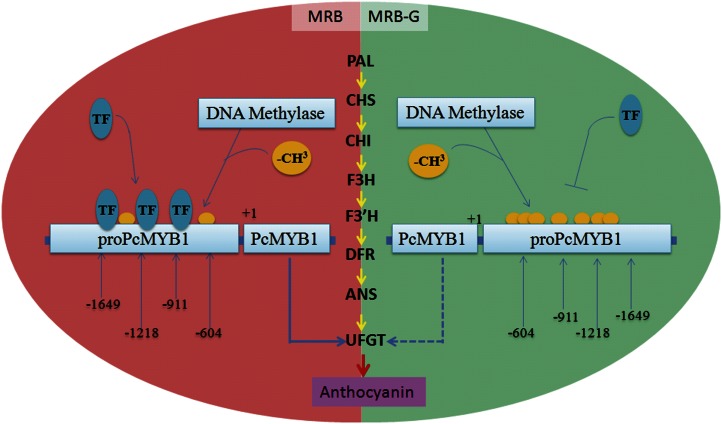
In MRB-G fruits, the expression of PcMYB10 was reduced by the methylation of regions −604 to −911 bp and −1,218 to −1,649 bp in its promoter. As a result, the expression of PcUFGT, a key gene involved in anthocyanin biosynthesis and regulated by PcMYB10, was also reduced. This might be associated with the inhibition of anthocyanin biosynthesis and the formation of green-skinned sport (Fig. 7).
Figure 7.
Model showing the molecular mechanism of the formation of the MRB-G mutant. The methylation of the −604 to −911 bp and −1,218 to −1,649 bp regions in the PcMYB10 promoter inhibits the expression of PcMYB10. PcMYB10 subsequently represses the expression of PcUFGT, which is a key structural gene in anthocyanin biosynthesis. As a result, the biosynthesis of anthocyanin is greatly decreased, causing the green-skinned sport. TF, Transacting factor. The solid and dotted arrows below the PcMYB10 gene indicate normal and repressive expression, respectively. Orange balls indicate −CH3 (methyl). [See online article for color version of this figure.]
MATERIALS AND METHODS
Plant Materials and Nucleic Acid Isolation
MRB and MRB-G branches of pear (Pyrus communis) were found in an orchard located in the suburbs of Beijing, China. Fruits of full bloom and 3, 6, 9, 12, 15, 20, 25, 30, 35, 40, 45, 50, 55, 70, 85, 100, 115, and 130 DAFB were collected from both clones, and the skin was carefully shaved off and collected for the measurement of anthocyanin and chlorophyll contents and nucleic acid isolation. DNA isolation was conducted using the cetyl-trimethyl-ammonium bromide method (Ma and Zhang, 2009). Total RNA was extracted according to Gong et al. (2011). RNA isolated from sample of 55 DAFB was used for gene isolation, and samples of 55 and 115 DAFB were used for methylation analysis of the PcMYB10 gene promoter.
Measurements of Anthocyanin, Chlorophyll, and UFGT Activity
Measurement of total anthocyanin was according to Rabino and Mancinelli (1986), with slight modifications. Briefly, frozen ground peel tissue (0.5 g) was homogenized in 2 mL of methanol with 1% HCl. The homogenate was left 8 h in dark conditions and then centrifuged for 5 min at 8,000g. Absorbance of the supernatant was measured at 530 and 600 nm using a UV-visible spectrophotometer (UNICO UV-2000). Three biological replicates were performed. Relative anthocyanin content was calculated according to the following formula: anthocyanin content (units g−1 fresh weight) = 2 × (OD553nm − OD600nm)/100, where OD equals optical density. Chlorophyll content determination was performed as described by Bruuinsma (1963).
UFGT activity was measured according to the method of Lister et al. (1996) with minor modifications. One gram of fruit skin was ground into fine powder in liquid nitrogen, adding 7.5 mL of extraction buffer (0.2 mol L−1 borate buffer, pH 8.8, 0.005 mol L−1 β-mercaptoethanol, 0.001 mol L−1 EDTA, and 0.001 mol L−1 dithiothreitol) and 10% polyvinylpolypyrrolidone homogenate. After centrifugation at 8,000g for 5 min, the supernatant was collected as enzyme solution and used for UFGT activity measurement. A total of 100 μL of enzyme solution was added to the reaction mixture containing 100 μL of 50 mmol L−1 Gly buffer (pH 8.5), 15 μL of 2 mg mL−1 substrate quercetin (Sigma Chemical), and 10 μL of 15 mg mL−1 UDP-Gal (Sigma Chemical). The mixture was incubated in a 30°C water bath for 30 min, followed by the addition of 75 μL of 20% TCA in methanol to terminate the reaction, then centrifuged at 8,000g for 5 min. The supernatant was collected and used for HPLC (Beckman) analysis. Five microliters of sample was injected into a C18 column, with mobile phase A (acetic acid; 10%, v/v) and mobile phase B (acetonitrile) at A:B = 85:15. The flow rate was 1 mL min−1, with elution for 15 min, 350-nm UV detector assay quercetin content, and UFGT activity was calculated according to a decrease of quercetin with units of mmol g−1 fresh weight h−1. Three biological replicates were performed.
Analysis of the Concentrations of Anthocyanin and Anthocyanin Precursor
Fruit peel was ground into powder in liquid nitrogen. In the direct analysis, samples were dispersed in a 1:1 water:methanol solution containing 2% HCl and sonicated for 30 min. The concentration was approximately 1 mg mL−1 for fruit peel powder. A portion of each sample solution was passed through a 0.22-μm polytetrafluoroethylene filter.
In the hydrolysis analysis, samples were dispersed in 1:1 water:methanol containing 2 n HCl and sonicated for 30 min. A portion of each sample solution was filtered through a 0.22-μm polytetrafluoroethylene filter and transferred to a vial with a Teflon-lined screw cap. The vial was placed in a preheated dry bath and hydrolyzed at 100°C for 60 min. Hydrolyzed samples were immediately cooled to room temperature for analysis.
HPLC analysis was conducted on a Shimadzu SPD-M10A LC-10 HPLC device with a diode-array detector. The separation of anthocyanin and anthocyanin precursor was accomplished on a Welch ultimate LP-C18 (5 μm, 4.6 × 250 mm). Mobile phases were 2% formic acid (A) and acetonitrile (B). The gradient condition was 0 to 30 min, 15% B; 30 to 60 min, 23% B; 60 to 70 min, 40% B. The flow rate was 0.8 mL min−1; column temperature was 40°C; detection was at 520 nm; 10 μL was injected; and postrun time was 15 min.
Isolation of Structure and Regulatory Genes Related to Anthocyanin Biosynthesis
One microgram of total RNA was DNase I treated and used for cDNA synthesis with oligo(dT) primers and the SMARTer PCR cDNA Synthesis Kit (Clontech). Full-length cDNAs of pear genes PAL, CHS, CHI, F3H, DFR, ANS, and ANR were amplified by gene-specific primers based on the available sequences in GenBank (DQ230992, AY786998, EF446163, AF497633, AY227731, DQ230994, and DQ251189). Fragments of pear genes F3′H, UFGT, MYB, bHLH, and WD40 were cloned, using degenerate primers according to the sequence information in grape (Vitis vinifera; DQ786632, AF000372, AB097923, ABM92332, and DQ517914) and apple (Malus domestica; FJ919632, DQ886413, DQ267897, HM122458, and GU173814), and their full-length cDNAs were obtained by using the SMARTer RACE cDNA Amplification Kit (Clontech). All genes obtained were submitted to GenBank with the accession numbers given in Supplemental Table S2. Primer pairs used in this study are listed in Supplemental Table S5.
Real-Time Quantitative PCR Analysis
One microgram of total RNA was used for cDNA synthesis with the primeScript Master Mix Kit (TaKaRa). Quantitative reverse transcription-PCR was performed with the SYBR Premix ExTaq II Kit (TaKaRa), and amplification was real-time monitored on an Option Continuous Fluorescence Detection System (Bio-Rad). Actin (AB190176) was used for normalization. Primer information is given in Supplemental Table S5.
Isolation and Identification of PcUFGT and PcMYB10 Gene Promoters
The promoter regions of PcMYB10 and PcUFGT were isolated using the Genome Walking Kit (TaKaRa) according to the manufacturer’s protocols. The analysis of these two promoters was performed by the online software PlantCARE (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/). The primer sequences used are given in Supplemental Table S5.
Transient expression was employed to test the activity of promoters as described by Bai et al. (2011). Briefly, the promoters of PcMYB10 and PcUFGT were ligated to pCAMBIA 1305 vector with the GFP gene, generating constructs proPcMYB10-GFP and proUFGT-GFP, respectively. The GFP gene was inserted into pCAMBIA 1305 vector as a negative control (GFP control vector). The constructs were introduced into Agrobacterium tumefaciens strain EHA105, and the A. tumefaciens strain was infiltrated into tobacco (Nicotiana benthamiana) leaves. GFP fluorescence was photographed using a stereomicroscope (SZX16-DP72; Olympus).
Y1H Assay
Y1H assays were performed using the Matchmaker Gold Yeast One-Hybrid System Kit (Clontech) according to the manufacturer’s protocols. The PcMYB10 gene was ligated to pGADT7 to generate the construct AD-MYB. The fragments of the promoter of UFGT were ligated to pAbAi vector to generate pAbAi-bait plasmids, which were then linearized and transformed into yeast strain Y1HGold and selected with a selective synthetic dextrose medium lacking uracil plate. The constructs of AD-MYB were transformed into strain Y1HGold holding pAbAi-bait and screened on an SD/-Ura/AbA plate. All transformations and screenings were performed three times.
Methylation Analysis
McrBC-PCR was used to analyze the methylation of relative sequences. A 1-μg DNA sample isolated from 55-DAFB fruit skin was digested with McrBC (New England Biolabs), according to the manufacturer’s instructions, with three biological replicates. For the negative control, GTP was replaced by water. The digested DNA was used as a template for semiquantitative PCR analysis. The PcMYB10 gene promoter sequence was divided into four fragments, +1 to −377 bp, −377 to −707 bp, −707 to −1,208 bp, and −1,208 to −1,847 bp, and amplified with their respective primers (Supplemental Table S5). The amount of amplification product was used to estimate the degree of methylation of the corresponding region.
BSP-PCR analysis was performed as described by Telias et al. (2011). Briefly, 500 ng of genomic DNA from 55- and 115-DAFB fruit skin was treated by the EZ DNA Methylation-Gold Kit (Zymo Research). Using the treated DNA as template, PcMYB10 promoter fragments were amplified by Zymo Taq DNA Polymerase (Zymo Research), ligated to PMD18-T vector (TaKaRa), and then sequenced. Sequences of 36 independent clones of each fragment were analyzed with the online software Kismeth (Gruntman et al., 2008), and the methylation level of each fragment was calculated. PcMYB10 promoter was amplified by five pairs of degenerate primers (Supplemental Table S5).
VIGS in Pear
Regions −604 to −911 bp and −1,218 to −1,649 bp in the PcMYB10 promoter were amplified by HS Taq DNA polymerase (TaKaRa) and ligated to the pMD18-T vector. The plasmids were digested by EcoRI and XhoI and ligated to the EcoRI-XhoI-cut pTRV2 vector to obtain two constructs: pTRV2-69 (containing the −604 to −911 bp fragment) and pTRV2-1216 (containing the −1,218 to −1,649 bp fragment). pTRV1, pTRV2 (empty plasmid), pTRV2-69, and pTRV2-1216 were transformed into A. tumefaciens strain EHA105.
The inflorescences of MRB were bagged at the full-bloom stage. The bags were removed from fruits at 10 DAFB, and these fruits (on the tree) were subjected to A. tumefaciens-mediated tobacco rattle virus vector infiltration. The A. tumefaciens-mediated tobacco rattle virus infiltration was performed according to Fu et al. (2005) and Bai et al. (2011) with some modifications. A. tumefaciens strains containing pTRV1 and pTRV2 or their derivative vectors, pTRV2-69 and pTRV2-1216, were mixed in a 1:1 ratio and infiltrated into 10-DAFB pear fruit flesh near the petiole. After 4 d of exposure to natural light on the tree, anthocyanin content, PcUFGT and PcMYB10 gene expression, and cytosine methylation of the corresponding regions in the PcMYB10 promoter were tested by the method described above. Fruits coinfiltrated with pTRV1 and pTRV2 were used as controls. Primer sequences used for methylation analysis are given in Supplemental Table S5.
Sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: PcPAL, JX403947; PcCHS, JX403948; PcCHI, JX403949; PcF3H, JX403950; PcF3'H, JX403951; PcDFR1/2, JX403952/3; PcANS, JX403954; PcANR, JX403955; PcUFGT, JX403956; PcMYB10, JX403957; PcbHLH, JX403960; and PcWD40, JX403961.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Phenotypes of MRB (left) and its green-peel chimera mutant MRB-G (right).
Supplemental Figure S2. Activity of PcUFGT in MRB and MRB-G fruit.
Supplemental Figure S3. The pathway of anthocyanin biosynthesis in plants.
Supplemental Figure S4. Relative expression of genes related to anthocyanin biosynthesis during MRB and MRB-G fruit development.
Supplemental Figure S5. Identification of proPcMYB10 and proPcUFGT.
Supplemental Figure S6. Identification of the regions of proPcMYB10 with different methylation levels between MRB and MRB-G by McrBC-PCR.
Supplemental Figure S7. Methylation levels of the −1,628 to −1,557 bp region of proPcMYB10 in 55-DAFB and 115-DAFB fruit peel of MRB and MRB-G.
Supplemental Figure S8. Methylation levels of the proPcMYB10 −1,628 to −1,557 bp region in fruit peel of MRB and Bartlett.
Supplemental Figure S9. Methylation levels of the proPcMYB10 −1,628 to −1,557 bp region in leaves of MRB and MRB-G.
Supplemental Figure S10. Methylation analysis of proPcMYB10 in the peel of MRB fruit infiltrated with pTRV-69 and pTRV-1216 by BSP-PCR.
Supplemental Table S1. Mutant rate of MRB to MRB-G.
Supplemental Table S2. Information of genes isolated.
Supplemental Table S3. Parts of cis-acting elements in proPcUFGT of MRB fruit peel.
Supplemental Table S4. Parts of cis-acting elements in proPcMYB10 of MRB fruit peel.
Supplemental Table S5. Sequence information of PCR primers used in this study.
Acknowledgments
We thank Dr. Susan Brown from the Department of Horticulture, Cornell University, for giving helpful comments on the manuscript.
Glossary
- MRB
Max Red Bartlett
- MRB-G
green-skinned sport of Max Red Bartlett
- DAFB
d after full bloom
- MBS
MYB-binding sites
- BSP
bisulfite sequencing
- VIGS
virus-induced gene silencing
- Y1H
yeast one-hybrid
- cDNA
complementary DNA
- McrBc
methylation-specific endonuclease
References
- Abu-Qaoud H, Skirvin RM, Chevreau E. (1990) In vitro separation of chimeral pears into their component genotypes. Euphytica 48: 189–196 [Google Scholar]
- Allan AC, Hellens RP, Laing WA. (2008) MYB transcription factors that colour our fruit. Trends Plant Sci 13: 99–102 [DOI] [PubMed] [Google Scholar]
- Bai S, Kasai A, Yamada K, Li T, Harada T. (2011) A mobile signal transported over a long distance induces systemic transcriptional gene silencing in a grafted partner. J Exp Bot 62: 4561–4570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bain JM. (1961) Some morphological, anatomical, and physiological changes in the pear fruit (Pyrus communis var. Williams Bon Chrétien) during development and following harvest. Aust J Bot 9: 99–123 [Google Scholar]
- Ban Y, Honda C, Hatsuyama Y, Igarashi M, Bessho H, Moriguchi T. (2007) Isolation and functional analysis of a MYB transcription factor gene that is a key regulator for the development of red coloration in apple skin. Plant Cell Physiol 48: 958–970 [DOI] [PubMed] [Google Scholar]
- Bender J, Fink GR. (1995) Epigenetic control of an endogenous gene family is revealed by a novel blue fluorescent mutant of Arabidopsis. Cell 83: 725–734 [DOI] [PubMed] [Google Scholar]
- Boss PK, Davies C, Robinson SP. (1996) Analysis of the expression of anthocyanin pathway genes in developing Vitis vinifera L. cv Shiraz grape berries and the implications for pathway regulation. Plant Physiol 111: 1059–1066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodersen P, Sakvarelidze-Achard L, Bruun-Rasmussen M, Dunoyer P, Yamamoto YY, Sieburth L, Voinnet O. (2008) Widespread translational inhibition by plant miRNAs and siRNAs. Science 320: 1185–1190 [DOI] [PubMed] [Google Scholar]
- Bruuinsma J. (1963) The quantitative analysis of chlorophylls a and b in plant extracts. Photochem Photobiol 2: 241–249 [Google Scholar]
- Chiu LW, Zhou X, Burke S, Wu X, Prior RL, Li L. (2010) The purple cauliflower arises from activation of a MYB transcription factor. Plant Physiol 154: 1470–1480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cocciolone SM, Cone KC. (1993) Pl-Bh, an anthocyanin regulatory gene of maize that leads to variegated pigmentation. Genetics 135: 575–588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cubas P, Vincent C, Coen E. (1999) An epigenetic mutation responsible for natural variation in floral symmetry. Nature 401: 157–161 [DOI] [PubMed] [Google Scholar]
- Dela G, Or E, Ovadia R, Nissim-Levi A, Weiss D, Oren-Shamir M. (2003) Changes in anthocyanin concentration and composition in Jaguarrose flowers due to transient high-temperature conditions. Plant Sci 164: 333–340 [Google Scholar]
- Espley R, Brendolise C, Chagné D, Kutty-Amma S, Green S, Volz R, Putterill J, Schouten HJ, Gardiner SE, Hellens RP, et al (2009) Multiple repeats of a promoter segment causes transcription factor autoregulation in red apple. Plant Cell 21: 168–183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feild TS, Lee DW, Holbrook NM. (2001) Why leaves turn red in autumn: the role of anthocyanins in senescing leaves of red-osier dogwood. Plant Physiol 127: 566–574 [PMC free article] [PubMed] [Google Scholar]
- Fu DQ, Zhu BZ, Zhu HL, Jiang WB, Luo YB. (2005) Virus-induced gene silencing in tomato fruit. Plant J 43: 299–308 [DOI] [PubMed] [Google Scholar]
- Gong YM, Cao HN, Zong CW, Jin C, Xue XU. (2011) Three methods for extracting total RNA from leaves of pear varieties. Hubei Agricultural Sciences 1: 1–3 [Google Scholar]
- Gong ZY, Yu HX, Yi CD. (2008) Process of somaclone variation in plant. Chinese Agricultural Science Bulletin 24: 65–68 [Google Scholar]
- Gonzalez A, Zhao M, Leavitt JM, Lloyd AM. (2008) Regulation of the anthocyanin biosynthetic pathway by the TTG1/bHLH/Myb transcriptional complex in Arabidopsis seedlings. Plant J 53: 814–827 [DOI] [PubMed] [Google Scholar]
- Gruntman E, Qi Y, Slotkin RK, Roeder T, Martienssen RA, Sachidanandam R. (2008) Kismeth: analyzer of plant methylation states through bisulfite sequencing. BMC Bioinformatics 9: 371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He J, Giusti MM. (2010) Anthocyanins: natural colorants with health-promoting properties. Annu Rev Food Sci Technol 1: 163–187 [DOI] [PubMed] [Google Scholar]
- Holton TA, Cornish EC. (1995) Genetics and biochemistry of anthocyanin biosynthesis. Plant Cell 7: 1071–1083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda C, Kotoda N, Wada M, Kondo S, Kobayashi S, Soejima J, Zhang Z, Tsuda T, Moriguchi T. (2002) Anthocyanin biosynthetic genes are coordinately expressed during red coloration in apple skin. Plant Physiol Biochem 40: 955–962 [Google Scholar]
- Kobayashi S, Goto-Yamamoto N, Hirochika H. (2004) Retrotransposon-induced mutations in grape skin color. Science 304: 982. [DOI] [PubMed] [Google Scholar]
- Kobayashi S, Ishimaru M, Hiraoka K, Honda C. (2002) Myb-related genes of the Kyoho grape (Vitis labruscana) regulate anthocyanin biosynthesis. Planta 215: 924–933 [DOI] [PubMed] [Google Scholar]
- Koes R, Verweij W, Quattrocchio F. (2005) Flavonoids: a colorful model for the regulation and evolution of biochemical pathways. Trends Plant Sci 10: 236–242 [DOI] [PubMed] [Google Scholar]
- Li MT, Yu LJ, Wang LM, Liu JM, Lei C. (2005) [The heredity of flower colors and the discovery of flower color chimera in Chrysanthemum species]. Yi Chuan 27: 948–952 [PubMed] [Google Scholar]
- Lila MA. (2004) Anthocyanins and human health: an in vitro investigative approach. J Biomed Biotechnol 2004: 306–313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lira-Medeiros CF, Parisod C, Fernandes RA, Mata CS, Cardoso MA, Ferreira PC. (2010) Epigenetic variation in mangrove plants occurring in contrasting natural environment. PLoS ONE 5: e10326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister CE, Lancaster JE, Walker JRL. (1996) Developmental changes in enzymes of flavonoid biosynthesis in the skins of red and green apple cultivars. J Sci Food Agric 71: 313–320 [Google Scholar]
- Ma YZ, Zhang YX. (2009) Effect of different tissues in Pyrus L. for genome DNA extract on RAPD. Southwest China J Agr Sci 22: 1042–1045 [Google Scholar]
- Morita Y, Saitoh M, Hoshino A, Nitasaka E, Iida S. (2006) Isolation of cDNAs for R2R3-MYB, bHLH and WDR transcriptional regulators and identification of c and ca mutations conferring white flowers in the Japanese morning glory. Plant Cell Physiol 47: 457–470 [DOI] [PubMed] [Google Scholar]
- Palai SK, Rout GR. (2011) Characterization of new variety of chrysanthemum by using ISSR markers. Hortic Bras 29: 613–617 [Google Scholar]
- Park KI, Ishikawa N, Morita Y, Choi JD, Hoshino A, Iida S. (2007) A bHLH regulatory gene in the common morning glory, Ipomoea purpurea, controls anthocyanin biosynthesis in flowers, proanthocyanidin and phytomelanin pigmentation in seeds, and seed trichome formation. Plant J 49: 641–654 [DOI] [PubMed] [Google Scholar]
- Pierantoni L, Dondini L, De Franceschi P, Musacchi S, Winkel BSJ, Sansavini S. (2010) Mapping of an anthocyanin-regulating MYB transcription factor and its expression in red and green pear, Pyrus communis. Plant Physiol Biochem 48: 1020–1026 [DOI] [PubMed] [Google Scholar]
- Pohlheim F. (2003) Vergleichende Untersuchungen zur Sprossvariation bei Plectranthus L'Herit. (Lamiaceae). Feddes Repert 114: 488–496 [Google Scholar]
- Poudel PR, Goto-yamamoto N, Mochioka R, Kataoka I, Beppuet K. (2008) Expression analysis of UDP-glucose:flavonoid 3-O-glucosyltransferase (UFGT) gene in an interspecific hybrid grape between Vitis ficifolia var. Ganebu and Vitis vinifera cv. Muscat of Alexandria. Plant Biotechnol Rep 2: 233–238 [Google Scholar]
- Rabino I, Mancinelli AL. (1986) Light, temperature, and anthocyanin production. Plant Physiol 81: 922–924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regan BC, Julliot C, Simmen B, Viénot F, Charles-Dominique P, Mollon JD. (2001) Fruits, foliage and the evolution of primate colour vision. Philos Trans R Soc Lond B Biol Sci 356: 229–283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards EJ. (2011) Natural epigenetic variation in plant species: a view from the field. Curr Opin Plant Biol 14: 204–209 [DOI] [PubMed] [Google Scholar]
- Schaefer HM, Schaefer V, Levey DJ. (2004) How plant-animal interactions signal new insights in communication. Trends Ecol Evol 19: 577–584 [Google Scholar]
- Schwinn K, Venail J, Shang Y, Mackay S, Alm V, Butelli E, Oyama R, Bailey P, Davies K, Martin C. (2006) A small family of MYB-regulatory genes controls floral pigmentation intensity and patterning in the genus Antirrhinum. Plant Cell 18: 831–851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekhon RS, Chopra S. (2009) Progressive loss of DNA methylation releases epigenetic gene silencing from a tandemly repeated maize Myb gene. Genetics 181: 81–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steyn WJ, Holcroft DM, Wand SJE, Jacobs G. (2004) Anthocyanin degradation in detached pome fruit with reference to preharvest red color loss and pigmentation patterns of blushed and fully red pears. J Am Soc Hortic Sci 129: 13–19 [Google Scholar]
- Stracke R, De Vos RCH, Bartelniewoehner L, Ishihara H, Sagasser M, Martens S, Weisshaar B. (2009) Metabolomic and genetic analyses of flavonol synthesis in Arabidopsis thaliana support the in vivo involvement of leucoanthocyanidin dioxygenase. Planta 229: 427–445 [DOI] [PubMed] [Google Scholar]
- Tanaka Y, Brugliera F, Chandler S. (2009) Recent progress of flower colour modification by biotechnology. Int J Mol Sci 10: 5350–5369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tegopati B. (2001) The study on growth and development of mango flower. Chimera 6: 1–11 [Google Scholar]
- Telias A, Lin-Wang K, Stevenson DE, Cooney JM, Hellens RP, Allan AC, Hoover EE, Bradeen JM. (2011) Apple skin patterning is associated with differential expression of MYB10. BMC Plant Biol 11: 93–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas LA, Sehata MJ, du Preez MG, Rees JG, Ndimba BK. (2010) Establishment of proteome spot profiles and comparative analysis of the red and green phenotypes of ‘Bon Rouge’ pear (Pyrus communis L.) leaves. Afr J Biotechnol 9: 4334–4341 [Google Scholar]
- Ubi BE, Honda C, Bessho H, Kondo S, Wada M, Kobayashi S, Moriguchi T. (2006) Expression analysis of anthocyanin biosynthetic genes in apple skin: effect of UV-B and temperature. Plant Sci 170: 571–578 [Google Scholar]
- Vanholme R, Ralph J, Akiyama T, Lu F, Pazo JR, Kim H, Christensen JH, Van Reusel B, Storme V, De Rycke R, et al. (2010) Engineering traditional monolignols out of lignin by concomitant up-regulation of F5H1 and down-regulation of COMT in Arabidopsis. Plant J 64: 885–897 [DOI] [PubMed] [Google Scholar]
- Vitrac X, Larronde F, Krisa S, Decendit A, Deffieux G, Mérillon JM. (2000) Sugar sensing and Ca2+-calmodulin requirement in Vitis vinifera cells producing anthocyanins. Phytochemistry 53: 659–665 [DOI] [PubMed] [Google Scholar]
- Winkel-Shirley B. (2001) Flavonoid biosynthesis: a colorful model for genetics, biochemistry, cell biology, and biotechnology. Plant Physiol 126: 485–493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Zhao G, Yang YN, Le WQ, Khan MA, Zhang SL, Gu C, Huang WJ. (2012) Identification of differentially expressed genes related to coloration in red/green mutant pear (Pyrus communis L.). Tree Genet Genomes 1: 1–9 [Google Scholar]
- Yamaguchi H, Shimizu A, Hase Y, Degi K, Tanaka A, Morishita T. (2009) Mutation induction with ion beam irradiation of lateral buds of chrysanthemum and analysis of chimeric structure of induced mutants. Euphytica 165: 97–103 [Google Scholar]
- Zhang F, Gonzalez A, Zhao MZ, Payne CT, Lloyd A. (2003) A network of redundant bHLH proteins functions in all TTG1-dependent pathways of Arabidopsis. Development 130: 4859–4869 [DOI] [PubMed] [Google Scholar]