Deubiquitination plays a role in daylength measurement to regulate flowering time.
Abstract
Protein ubiquitination is involved in most cellular processes. In Arabidopsis (Arabidopsis thaliana), ubiquitin-mediated protein degradation regulates the stability of key components of the circadian clock feedback loops and the photoperiodic flowering pathway. Here, we identified two ubiquitin-specific proteases, UBP12 and UBP13, involved in circadian clock and photoperiodic flowering regulation. Double mutants of ubp12 and ubp13 display pleiotropic phenotypes, including early flowering and short periodicity of circadian rhythms. In ubp12 ubp13 double mutants, CONSTANS (CO) transcript rises earlier than that of wild-type plants during the day, which leads to increased expression of FLOWERING LOCUS T. This, and analysis of ubp12 co mutants, indicates that UBP12 and UBP13 regulate photoperiodic flowering through a CO-dependent pathway. In addition, UBP12 and UBP13 regulate the circadian rhythm of clock genes, including LATE ELONGATED HYPOCOTYL, CIRCADIAN CLOCK ASSOCIATED1, and TIMING OF CAB EXPRESSION1. Furthermore, UBP12 and UBP13 are circadian controlled. Therefore, our work reveals a role for two deubiquitinases, UBP12 and UBP13, in the control of the circadian clock and photoperiodic flowering, which extends our understanding of ubiquitin in daylength measurement in higher plants.
Protein ubiquitination is a critical posttranslational mechanism regulating diverse cellular processes and signal transduction pathways in eukaryotes. Ubiquitin protein is a 76-amino-acid-long polypeptide conserved throughout all eukaryotic organisms. Attachment of ubiquitin to a Lys residue in the substrate protein requires multiple steps catalysis by E1 activating, E2 conjugating, and E3 ligating enzymes (Finley, 2009). Among these enzymes, E3 ligases are responsible for specific substrate recognition. According to their mechanisms of action and subunit composition, four main types of E3s have been identified in plants, including E3-associated protein carboxyl terminus, Really Interesting New Gene (RING), U-box, and cullin-RING ligases (Vierstra, 2009). In Arabidopsis (Arabidopsis thaliana), more than 1,400 genes encode components of the ubiquitin-proteasome pathway, and 90% of these genes encode subunits of E3 ligases (Moon et al., 2004). In higher plants, E3 ligases play important roles in hormone responses, photomorphogenesis, senescence, circadian rhythm, and floral development (Moon et al., 2004). For example, CONSTITUTIVE PHOTOMORPHOGENESIS1 (COP1), a RING domain E3 ligase, plays extensive roles in light response and photomorphogenesis by targeting multiple proteins, such as LONG HYPOCOTYL5, Phytochrome A, and LONG AFTER FAR-RED LIGHT1 (Lau and Deng, 2012). Moreover, COP1 also regulates the circadian clock and flowering time by destabilizing GIGANTEA (GI) and CONSTANS (CO; Jang et al., 2008; Liu et al., 2008a; Yu et al., 2008).
Ubiquitination is dynamic and reversible; the enzymatic reaction that opposes ubiquitin conjugation is deubiquitination. In human genome, 79 deubiquitinating enzymes (DUBs) were predicted (Nijman et al., 2005). Most DUBs, roughly 80%, belong to four subfamilies with Cys active sites containing a highly conserved catalytic triad; these families are ubiquitin C-terminal hydrolases, ubiquitin-specific proteases (UBPs/USPs), ovarian tumor proteases, and Machado-Josephin domain proteins (Katz et al., 2010). A minor subfamily is the JAB1/MPN/MOV34 metalloenzyme, members of which have a zinc active site (Wing, 2003; Reyes-Turcu et al., 2009). The USP family includes more than 50 members in humans and is the largest family of DUBs. USPs are involved in tumor suppression, DNA repair, neural stem cell progenitor maintenance, immune response, viral replication, and epigenetic control (Katz et al., 2010; Nicholson and Suresh Kumar, 2011; Neutzner and Neutzner, 2012).
Compared with the large numbers of E3 ligases, DUBs in Arabidopsis comprise a relatively smaller group. USPs, the largest subfamily, include 27 proteins with Cys- and His-box signature motifs, which have been predicted to confer deubiquitination activities (Yan et al., 2000). Among these proteins, UBP2 (Yan et al., 2000), UBP3, UBP4 (Chandler et al., 1997), UBP12 (Ewan et al., 2011), UBP14 (Doelling et al., 2001), UBP15 (Liu et al., 2008b), and UBP26 (Sridhar et al., 2007) were shown to be active enzymes in vitro. These UBPs are involved in different signaling pathways and cellular processes. For example, UBP1 and UBP2 are required for the resistance to the amino acid analog Canavanine, but the single and double mutants have no obvious phenotype under normal growth conditions (Yan et al., 2000). UBP3 and UBP4 are homologs and share 93% amino acid sequence identity; they redundantly affect pollen development and transmission (Chandler et al., 1997; Doelling et al., 2007). The ubp14 mutant shows embryonic lethality at the globular stage (Doelling et al., 2001). The ubp15 mutant displays leaf developmental defects and other phenotypes, such as early flowering, weak apical dominance, and reduced fertility (Liu et al., 2008b). UBP26 can remove the monoubiquitin at Lys-143 on H2B and controls heterochromatic silencing (Sridhar et al., 2007). The ubp26 mutant also causes early flowering by repression of FLOWERING LOCUS C (FLC) transcription and seed developmental defects by activation of the imprinted gene PHERES1 (Luo et al., 2008; Schmitz et al., 2009). UBP12 in Arabidopsis or NtUBP12 in tobacco (Nicotiana tabacum) acts as a negative regulator in plant immunity (Ewan et al., 2011). Therefore, unraveling the biological functions of UBPs and their substrates in Arabidopsis will add another layer to our understanding of the ubiquitination dynamics in plant development.
In this work, we report a novel role for two DUBs, UBP12 and UBP13. These two enzymes are themselves circadian regulated, and the corresponding hypomorphic alleles display a short period of the circadian clock. Study of their roles in flowering time indicates that they repress premature flowering through the photoperiod pathway. Thus, we demonstrate that deubiquitination is also important for circadian clock and photoperiodic flowering regulation.
RESULTS
UBP12 and UBP13 Confer Deubiquitination Activities in Vitro
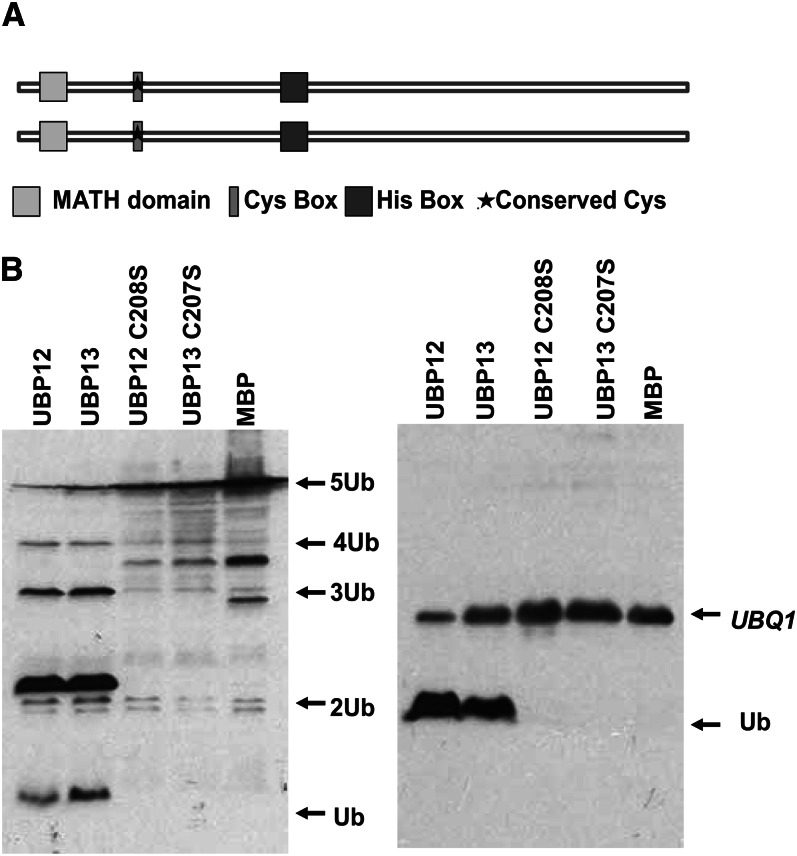
UBP12 (At5g06600) and UBP13 (At3g11910) are two Arabidopsis homologs of human ubiquitin-specific protease USP7/HAUSP (first identified as a herpes virus-associated cellular factor; Everett et al., 1997). UBP12 and UBP13 share high amino acid sequence similarity, with 91% sequence identity, suggesting that their function may be redundant. They also share 34% amino acid sequence identity with USP7 (Supplemental Fig. S1) and have the conserved Cys- and His-box signature motifs, indicating that they have potential deubiquitination activity. At the N termini, these three proteins contain a meprin and tumor necrosis factor receptor-associated factor homology (MATH) domain, which is not found in other Arabidopsis UBP proteins (Fig. 1A).
Figure 1.
UBP12 and UBP13 have deubiquitination activities. A, Schematic diagram of the UBP12 and UBP13 protein structures. The MATH domain, conserved Cys-box, and His-box are shown as squares or rectangles. The star indicates the Cys residue required for enzymatic activity. B, UBP12 and UBP13 are active DUBs. In vivo cleavage of hexameric polyubiquitin (UBQ10, left) and ubiquitin extension protein (UBQ1, right). UBP12, UBP13, and their mutants UBP12C208S and UBP13C207S were coexpressed with the substrates UBQ1 and UBQ10 in E. coli; the cleavage products were detected by immunoblot analyses with anti-ubiquitin antibodies. The positions of the substrates and cleaved products are indicated by arrows.
To determine whether UBP12 and UBP13 have deubiquitination activity in vitro, we performed enzymatic activity assays using the hexameric polyubiquitin protein UBQ10 and ubiquitin extension protein UBQ1, which bears the 52 amino acid ribosomal protein appended to a single ubiquitin moiety as substrates. When wild-type UBP12 or UBP13 and their mutant forms UBP12C208S and UBP13C207S, in which conserved Cys of the enzymatic active sites were substituted by Ser, were coexpressed with UBQ10 and UBQ1 in Escherichia coli as described (Yan et al., 2000), we can detect the cleaved products by immunoblotting analysis with ubiquitin antibody. The wild-type UBP12 and UBP13 were capable of cleaving ubiquitin from both UBQ10 and UBQ1 (Fig. 1B). However, neither of the mutants, UBP12C208S or UBP13C207S, showed any enzymatic activity, indicating that activities of UBP12 and UBP13 were dependent on the conserved Cys residue. This result is consistent with previous findings that UBP12 can remove ubiquitin from Lys-48-linked ubiquitin chain (Ewan et al., 2011). All of these results demonstrate that UBP12 and UBP13 are bona fide DUBs in Arabidopsis.
UBP12 and UBP13 Are Ubiquitously Expressed and Localize to Both Cytoplasm and Nucleus
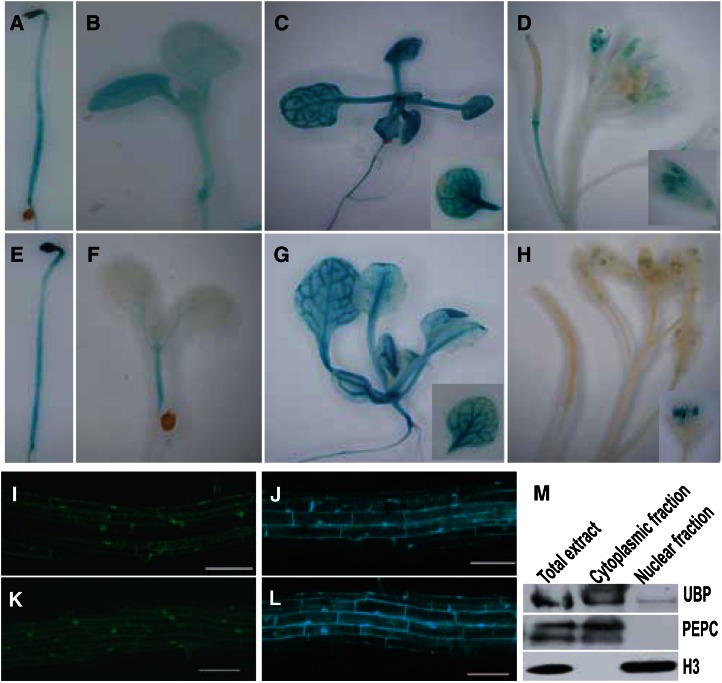
To figure out the biological functions of UBP12 and UBP13 in plant development, we first determined UBP12 and UBP13 expression patterns by examining the GUS signal in transgenic plants with GUS expressed under the control of the UBP12 or UBP13 promoter. UBP12 and UBP13 were both expressed in the hypocotyl, cotyledon, leaf, root, and inflorescence, especially in the vascular part of these tissues (Fig. 2, A–H). However, a few differences were observed. First, UBP12 was expressed in hypocotyl and cotyledon of 4-d-old plants (Fig. 2B), whereas UBP13 was mainly expressed in hypocotyl, but not cotyledon, of 4-d-old seedlings (Fig. 2F). Second, in flowers, UBP12 was expressed in carpel, sepal, and pollen (Fig. 2D), whereas UBP13 was mainly expressed only in the pollen (Fig. 2H). The highly overlapping expression patterns of UBP12 and UBP13 in Arabidopsis suggest that they may be functionally redundant in regulating plant development.
Figure 2.
UBP12 and UBP13 have similar expression pattern and protein localization. A to D, GUS staining of dark-grown seedlings (A), 4-d-old seedlings under LD condition (B), 14-d-old seedlings under LD condition (C), and inflorescences (D) of the transformants containing UBP12pro:GUS. E to H, GUS staining of dark-grown seedlings (E), 4-d-old seedlings under LD condition (F), 14-d-old seedlings under LD condition (G), and inflorescences (H) of the transformants containing UBP13pro:GUS. I to L, UBP12 fused to GFP (K) and UBP13 fused to CFP (L) localize to the nuclei and cytoplasm; 35Spro:GFP (I) and 35Spro:CFP (J) were used as control. Bar = 200 µm. UBP12/UBP13 protein was detected in both cytoplasmic and nuclear fractions. Histone H3 and phosphoenolpyruvate carboxylase (PEPC) were used as control for nuclear or cytoplasmic fraction, respectively.
To examine the subcellular localizations of UBP12 and UBP13 proteins, we generated green fluorescent protein (GFP)- and cyan fluorescent protein (CFP)-tagged UBP12 and UBP13. In UBP12-GFP and UBP13-CFP transgenic plants, we observed that UBP12 and UBP13 were located in both cytoplasm and nucleus (Fig. 2, K and L), which was similar to the GFP (Fig. 2I) and CFP (Fig. 2J) alone in 35S:GFP and 35S:CFP transgenic plants. Moreover, we detected the UBP12/UBP13 protein in separated cytoplasmic or nuclear fractions using UBP antibodies. Consistent with our observation in these transgenic plants, the UBP12/UBP13 can be detected in both cytoplasm and nucleus, though more UBP12/UBP13 can be detected in the cytoplasm (Fig. 2M). These results suggest that UBP12 and UBP13 might affect substrates in both the cytoplasmic and nucleic compartments.
Mutations of UBP12 and UBP13 Exhibit Pleiotropic Phenotypes
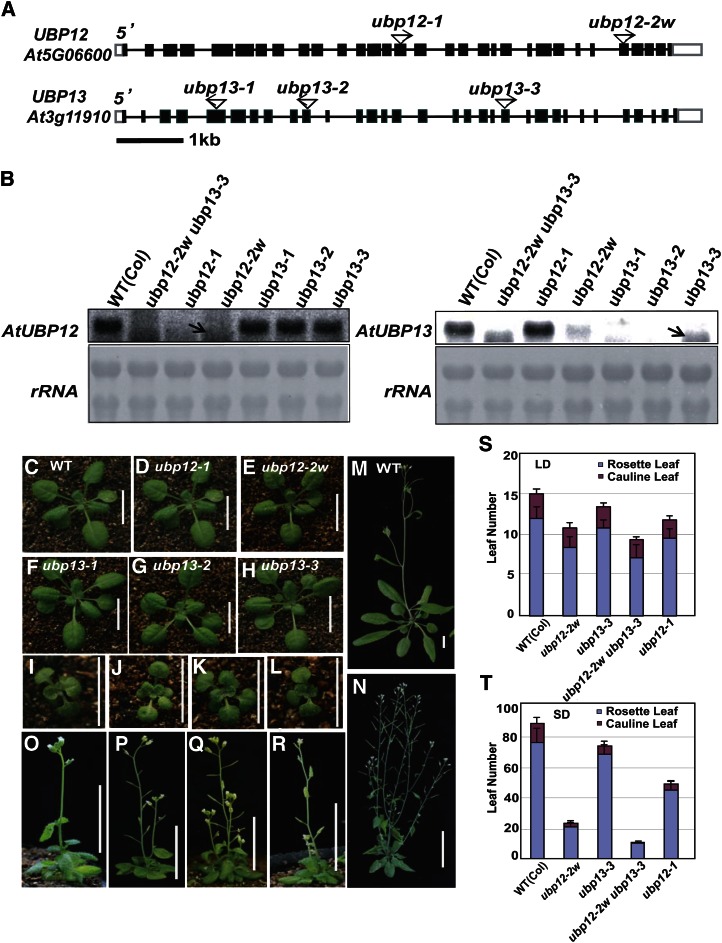
To investigate the biological functions of UBP12 and UBP13, we isolated mutants from transfer DNA (T-DNA) insertion populations of Arabidopsis. Two insertional mutants of UBP12 were identified, and the alleles were named as ubp12-1 (GABI_244E11) and ubp12-2w (GABI_742C10; Fig. 3A, top); the alleles contain T-DNA insertions in exons 15 and 28, respectively. Three mutant alleles of UBP13 were identified and designated as ubp13-1 (SALK_128312), ubp13-2 (SALK_024054), and ubp13-3 (SALK_132368; Fig. 3A, bottom). T-DNAs were inserted in the fifth, 10th, and 21st exons of these three mutants, respectively.
Figure 3.
Mutations of UBP12 and UBP13 affect plant development and flowering time. A, Schematic diagrams of the UBP12 and UBP13 gene structures, with the T-DNA insertion sites indicated. Black boxes indicate exons, white boxes indicate untranslated regions, and lines indicate introns. B, Northern blots showing the expression levels of UBP12 and UBP13 in the T-DNA insertion mutants (top). Ribosomal RNA stained with methylene blue was used as loading control (bottom). C to L, Phenotypes of 24-d-old seedlings of the wild type (C), ubp12-1 (D), ubp12-2w (E), ubp13-1 (F), ubp13-2 (G), ubp13-3 (H), ubp12-2w ubp13-1 (I), ubp12-2w ubp13-2 (J), ubp12-2w ubp13-3 (K), and ubp12-1 ubp13-3 (L). Bar = 1 cm. M to R, Phenotypes of the wild type (M), ubp12-2w (N), ubp12-2w ubp13-1 (O), ubp12-2w ubp13-2 (P), ubp12-2w ubp13-3 (Q), and ubp12-1 ubp13-3 (R) after bolting. Bar = 1 cm. S and T, Statistical analysis of leaf numbers of ubp12-2w, ubp13-3, ubp12-2w ubp13-3 double mutants, and ubp12-1 under LD (S) and SD (T) conditions compared with wild-type plants. Values are means ± sd of at least 20 plants. WT, Wild type.
By northern-blot analysis, no accumulation of full-length UBP12 mRNA was detected in ubp12-1 and ubp12-2w mutant plants, and no full-length UBP13 mRNA was detected in ubp13-1, ubp13-2, and ubp13-3 (Fig. 3B). However, one smaller segment was found in ubp12-2w and ubp13-3 mutant plants, suggesting that ubp12-2w and ubp13-3 are not null alleles for UBP12 or UBP13. In the ubp12-2w mutant, surprisingly, the mRNA level of UBP13 was also decreased (Fig. 3B), which might result from high transcription of the 3′ primer region of UBP12 in ubp12-2w causing suppression of UBP13 in trans (Supplemental Fig. S2), indicating that ubp12-2w is a weak double mutant, although there is only one T-DNA insertion in the genome (Supplemental Fig. S3). Therefore, we named it as ubp12-2w. Different from other single mutants (Fig. 3, C, D, F, G, and H) with no obvious developmental phenotypes, the ubp12-2w exhibited distinct phenotypes, including small plants, round leaves, short petioles, dwarfism, and more branches after bolting (Fig. 3, E and N).
The similar expression patterns of UBP12 and UBP13 (Fig. 2, A–H) indicate that they could have redundant biological functions in regulating plant development. To test this, we generated double mutants and obtained ubp12-2w ubp13-1 (Fig. 3, I and O), ubp12-2w ubp13-2 (Fig. 3, J and P), ubp12-2w ubp13-3 (Fig. 3, K and Q), and ubp12-1 ubp13-3 (Fig. 3, L and R). All these double mutants displayed similar but much more severe phenotypes than ubp12-2w (Fig. 3, E and N), including smaller plants, rounder leaves, shorter petioles at seedling stage, more severe dwarf statures, and more bushy plants at mature stage. Among these viable double mutants, ubp12-2w ubp13-3 showed weakest developmental phenotypes in every aspect we observed, which is consistent with the fact that partial transcripts can be detected in ubp12-2w or ubp13-3. Fertility of all these double mutants was dramatically decreased. Only ubp12-2w ubp13-3 set enough seeds for further research, whereas ubp12-1 ubp13-3 was completely infertile. The homozygous ubp12-1 ubp13-1 and ubp12-1 ubp13-2 double mutants could not be obtained, suggesting that these two genes are important for Arabidopsis embryo development and/or male/female gametophyte function. Taken together, we conclude that UBP12 and UBP13 are involved in diverse developmental processes.
In addition to the developmental patterning defects of ubp12-2w and ubp12-2w ubp13-3, the single mutants of ubp12-1 and ubp12-2w and the double mutant ubp12-2w ubp13-3 also showed early-flowering phenotype under both long-day (LD; 16-h light/8-h dark; Fig. 3S) and short-day (SD; 8-h-light/16-h-dark) conditions (Fig. 3T) compared with wild-type plants. The phenotypes were profounder under SD. The ubp12-2w ubp13-3 mutant flowered after forming only about 10 leaves under SD, which is similar to the mutants under LD condition, indicating that the double mutant is insensitive to photoperiod. The ubp12-1 ubp13-3 double mutant also showed early flowering, if only according to leaf numbers (Supplemental Fig. S4, A and B). However, ubp12-1 ubp13-3 displayed drastic developmental retardation under LD and SD, suggesting that it is not reasonable for us to analyze the flowering time (Supplemental Fig. S4C). Taken together, these results indicate that DUBs UBP12 and UBP13 are involved in the photoperiodic floral transition pathway.
Role of UBP12 and UBP13 in Photoperiodic Flowering Requires CO
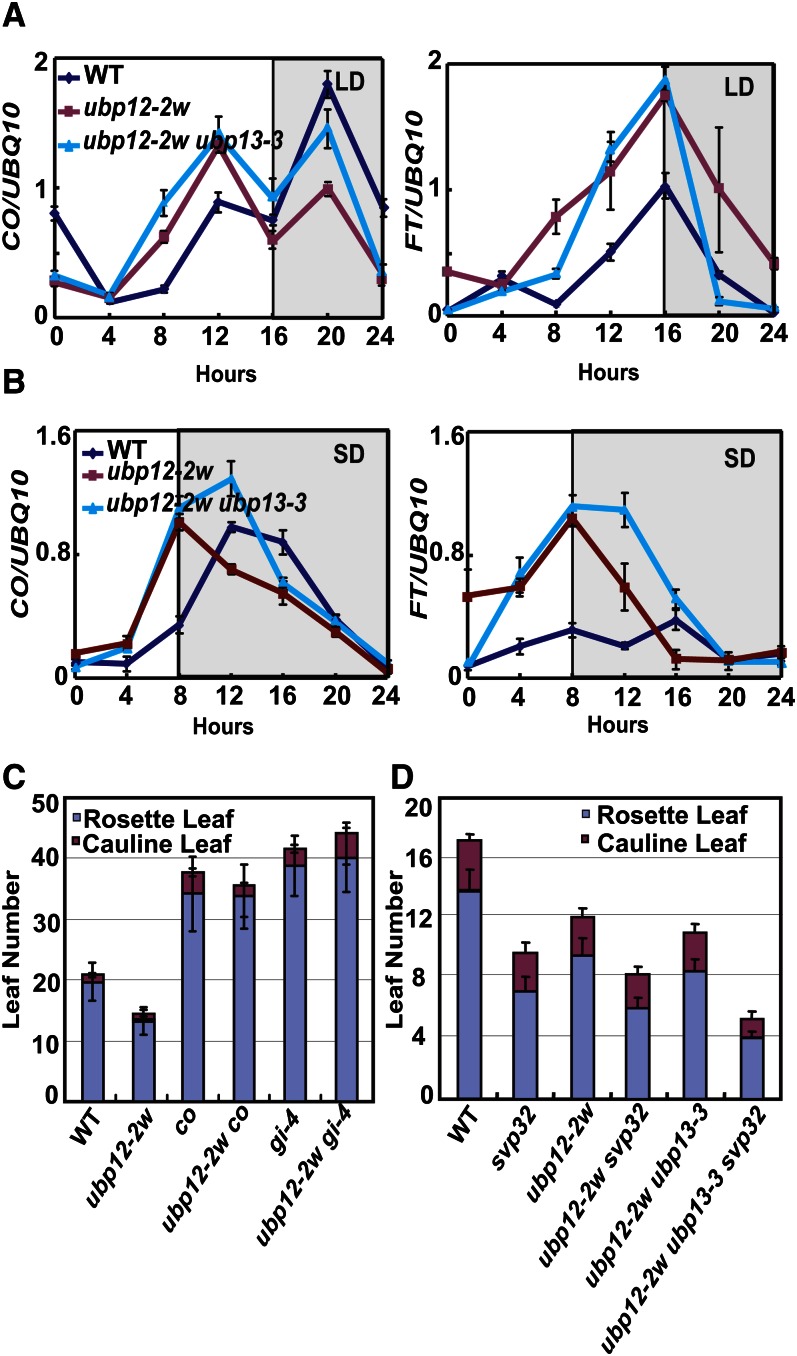
In Arabidopsis, transcriptional regulation of CO is crucial for daylength measurement. CO is activated under proper daylength and subsequently up-regulates the expression of FLOWERING LOCUS T (FT) to promote flowering. Changes in CO transcription are the key of many daylength-insensitive mutants (Yanovsky and Kay, 2002). To determine if the early-flowering phenotype caused by mutations of UBP12 and UBP13 depends on CO and FT, we measured CO and FT transcripts by quantitative reverse transcription PCR (qRT-PCR) at 4-h intervals for 24 h under both LD and SD conditions. In ubp12-2w and ubp12-2w ubp13-3 double mutants, the expression level of CO started to increase at 4 or 8 h after dawn, which was earlier than that in wild-type plants under both LD (Fig. 4A, left) and SD (Fig. 4B, left) conditions. This led to elevated CO expression during the day and then activated FT expression (Fig. 4A, right), which was more evident in the SD condition (Fig. 4B). By contrast, the transcription of FLC was not affected in ubp12 or ubp13 single and double mutants (Supplemental Fig. S5). These results indicate that UBP12 and UBP13 act in the photoperiodic flowering pathway by regulating CO and FT transcriptions.
Figure 4.
UBP12 and UBP13 regulate photoperiodic flowering. A, Expression patterns and transcript levels of CO (left) and FT (right) under LD condition in the wild type, ubp12-2w, ubp13-3, and ubp12-2w ubp13-3 double mutant. Values are means ± sd of three independent experiments. B, Expression patterns and transcript levels of CO (left) and FT (right) under SD condition in the wild type, ubp12-2w, ubp13-3, and ubp12-2w ubp13-3 double mutant. Values are means ± sd of three independent experiments. C, Statistical analysis of leaf numbers of ubp12-2w co and ubp12-2w gi-4 double mutants under LD condition. Values are means ± sd of at least 20 plants. D, Statistical analysis of leaf numbers of ubp12-2w svp32 and ubp12-2w ubp13-3 svp32 plants under LD condition. Values are means ± sd of at least 20 plants. WT, Wild type.
Genetic analysis of ubp12-2w co double mutants further supported our results. Both the ubp12-2w co double mutants and co single mutants flowered after forming around 30 leaves, but the ubp12-2w mutant flowered with only around 10 leaves under LD condition (Fig. 4C; Supplemental Fig. S6). This indicates that UBP12 and UBP13 act upstream of CO. GI is an upstream regulator of CO and positively regulates CO transcription. So we also analyzed ubp12-2w gi-4 mutants and found that they flowered as late as gi-4 plants (Fig. 4C; Supplemental Fig. S6), indicating that GI is also downstream of UBP12 and UBP13 in regulating photoperiodic flowering. We also tested one MADS box protein involved in photoperiodic flowering regulation, SHORT VEGETATIVE PHASE (SVP), which negatively regulates FT expression in a CO-independent manner (Kim et al., 2005; Fujiwara et al., 2008; Li et al., 2008). ubp12-2w svp32 and ubp12-2w ubp13-3 svp32 showed earlier flowering than svp32 and ubp12-2w or ubp12-2w ubp13-3 under LD condition (Fig. 4D), indicating that UBP12/UBP13 and SVP regulate photoperiodic flowering in parallel. These genetic interactions suggest that UBP12 and UBP13 regulate flowering time through GI and CO.
Mutations of UBP12 and UBP13 Result in Altered Circadian Rhythm
In the photoperiodic flowering pathway, the circadian oscillators take part in the daylength measurement by regulating CO expression (Imaizumi and Kay, 2006). The plant circadian clock is composed of multiple feedback loops (Harmer, 2009). In Arabidopsis, two MYB transcription factors, CIRCADIAN CLOCK-ASSOCIATED1 (CCA1) and LATE ELONGATED HYPOCOTYL (LHY), are expressed in the morning. They can bind to the promoter of the evening gene TIMING OF CAB EXPRESSION1 (TOC1) to repress its transcription (Alabadí et al., 2001; Carré and Kim, 2002). In turn, TOC1 regulates the expressions of CCA1 and LHY as a repressor (Gendron et al., 2012; Huang et al., 2012; Pokhilko et al., 2012).
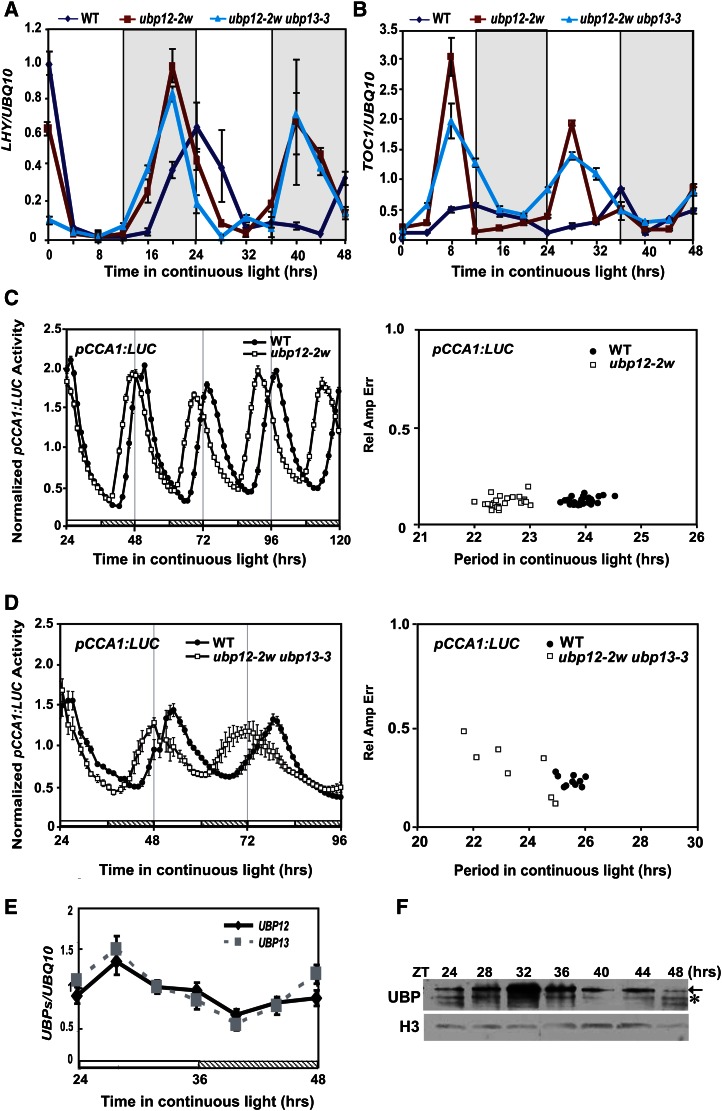
We examined whether mutation of UBP12 and UBP13 affects circadian rhythm and the expression patterns of these core clock genes. The rhythmic accumulations of LHY and TOC1 were tested by qRT-PCR in ubp12-2w and ubp12-2w ubp13-3 mutants. Under constant white light (LL) free-running condition, the periods of LHY (Fig. 5A) and TOC1 (Fig. 5B) expression were shortened by 4 h in ubp12-2w and ubp12-2w ubp13-3 double mutants compared with that of wild-type plants. In addition, the transcription level of TOC1 was obviously increased in ubp12-2w and ubp12-2w ubp13-3 mutants (Fig. 5B). Moreover, the period of CCA1 promoter:LUCIFERASE (pCCA1:LUC) circadian rhythm was also shortened by 4 h in ubp12-2w (n = 23) and ubp12-2w ubp13-3 mutants (n = 7) compared with the wild type (Fig. 5, C and D). Taken together, these results indicate that UBP12 and UBP13 function in the periodic control of the expression of core clock oscillators in Arabidopsis. Then we tested whether UBP12 and UBP13 are themselves circadianly regulated. mRNA levels of UBP12 and UBP13 oscillated under LL condition (Fig. 5E). The transcripts of UBP12 and UBP13 increased and reached their highest level at 4 h (Zeitgeber time [ZT] 28) and then decreased and reached their lowest level around 16 h (ZT 40). Accumulation of UBP12 and UBP13 proteins was also highest at 8 h (ZT 32) and lowest at 16 h (ZT 40), which is similar to their mRNA levels (Fig. 5F). These results demonstrate that UBP12 and UBP13 are under circadian control. Taken together, we show that UBP12 and UBP13 are essential for proper circadian rhythm.
Figure 5.
Mutation of UBP12 and UBP13 shortens the circadian period under LL. The expression patterns of LHY (A) and TOC1 (B) under LL condition were analyzed by qRT-PCR. Wild-type, ubp12-2w, and ubp12-2w ubp13-3 plants were grown for 10 d in 12-h-light/12-h-dark cycles before released to LL and sampled every 4 h from ZT0. Circadian rhythm of pCCA1:LUC was detected in transgenic seedlings in ubp12-2w (n = 23; C) or ubp12-2w ubp13-3 (n = 7; D). The pCCA1:LUC transgenic plants in ubp12-2w and ubp12-2w ubp13-3 were grown for 7 or 15 d, respectively, in 12-h-light/12-h-dark cycles and transferred to LL at ZT 0. Relative amplitude error is a measure of the strength of the oscillation. Statistical analysis of period length showed that in each case, the periods of ubp12-2w and ubp12-2w ubp13-3 were shorter than that of the wild type (P < 0.001). UBP12 and UBP13 expression levels were tested from ZT 24 to 48 by qRT-PCR in wild-type plants (E). Values are means ± sd of three independent experiments. The protein level of UBP12/UBP13 under LL was tested by immunoblot using UBP12/UBP13 antibodies from ZT 24 to 48 (F). The arrow indicates the UBP protein position. The asterisk indicates the nonspecific band. H3 was used as the loading control. WT, Wild type.
DISCUSSION
The circadian clock coordinates diverse aspects of plant development with daily cycles and promotes their adaption to the environment (McClung, 2011; Nagel and Kay, 2012). In Arabidopsis, proteasomal degradation pathway functions in the circadian clock and photoperiodic flowering by regulating the stability of key components in these pathways. Our research identified two circadian-regulated ubiquitin-specific proteases, UBP12 and UBP13, functioning in photoperiodic flowering and the circadian clock, which broadened our understanding of the proteasomal degradation mechanism in these processes.
UBP12 and UBP13 in Regulating Diverse Aspects of Plant Development
The ubiquitin-proteasome pathway contributes significantly to various aspects of development in Arabidopsis (Moon et al., 2004). As a counterbalance to ubiquitination, DUBs should also regulate diverse developmental processes. UBP12 and UBP13 affect plant development extensively. In addition to their role in the circadian clock and flowering, UBP12 and UBP13 are required for immunity against virulent Pseudomonas syringae in tomato (Solanum lycopersicum; Ewan et al., 2011). It is very likely that UBP12 and UBP13 target key factors in these processes and regulate protein levels by counteracting ubiquitin-mediated degradation.
Phylogenetic analysis shows that UBP12 and UBP13 are similar to human USP7, which plays crucial roles in diverse cellular processes by deubiquitinating different substrates to regulate protein stability and subcellular localization (Li et al., 2002; van der Knaap et al., 2005; van der Horst et al., 2006; Song et al., 2008; Daubeuf et al., 2009; Maertens et al., 2010; Huang et al., 2011; Khoronenkova et al., 2012). Therefore, like USP7, UBP12 and UBP13 might have a wide range of targets in different developmental processes. Identifying the substrates of UBP12 and UBP13 in the future will help us to understand how they contribute to various aspects of plant development.
UBP12/UBP13 in Circadian Clock and Photoperiodic Flowering Time Regulation
Daylength measurement plays an essential role in plant growth and development. Plants use endogenous clocks to adjust their physiological and developmental stages according to the change of environment (Harmer, 2009). In Arabidopsis, transcriptional regulation of CO is crucial in the daylength measurement. Under SD, the expression of CO is repressed before dusk, and FT can only express at a low level, which is insufficient to induce flowering (Imaizumi and Kay, 2006). On the contrary, the ubp12 ubp13 double mutant shows reduced sensitivity to daylength and results in advance expression of CO before dusk. As a result, FT is highly expressed in ubp12 ubp13 double mutants, which leads to the early-flowering phenotype. Moreover, our results indicate that both UBP12 and UBP13 are required to maintain the appropriate expression phases of the circadian genes.
UBP12 and UBP13 are DUBs and might act in circadian and photoperiodic flowering by altering the proteasomal degradation pathway. By now, there are four E3 ligases, FLAVIN-BINDING, KELCH REPEAT AND F-BOX1 (FKF1; Nelson et al., 2000; Imaizumi et al., 2005; Sawa et al., 2007; Fornara et al., 2009), ZEITLUPE (ZTL; Más et al., 2003; Kiba et al., 2007; Kim et al., 2007), LOV KELCH PROTEIN2 (Baudry et al., 2010; Takase et al., 2011; Ito et al., 2012), and COP1 (Jang et al., 2008; Liu et al., 2008a), shown to function in the plant circadian rhythm and photoperiodic flowering pathway.
The mutation of FKF1 displays late flowering under LD, and the day peak of CO appears 3 h later in fkf1 mutants than in wild-type plants. In contrast with fkf1, the ubp12 ubp13 double mutant is early flowering, and CO expression rises earlier in ubp12 ubp13 double mutants than in the wild type under SD and LD. The ztl mutant exhibits long periodicity in LL and decreased amplitude of the circadian genes. By contrast, ubp12 ubp13 double mutants have short periodicity, and the expression level of the circadian gene TOC1 is increased. These observations suggest that UBP12 and UBP13 might counteract the functions of these F-box proteins. It would be interesting to test if UPB12 and UBP13 can regulate the ubiquitination and stability of the substrates of these F-box proteins in the future.
The cop1 mutant shows short periodicity and early flowering under LD and SD conditions by affecting CO protein stability and altering the CO and LHY expression through impacting on GI degradation (Jang et al., 2008; Liu et al., 2008a; Yu et al., 2008). Mutation of UBP12 and UBP13 resulted in similar phenotypes as cop1 regarding flowering time, CO expression, and changes in circadian rhythm. These similarities suggest that UBP12 and UBP13 are not likely to function antagonistically to COP1. It would be interesting to see how these biochemically opposite enzymes contribute the same way to the response to daylength measurement. Thus, understanding the relationship between UBP12/UBP13 and COP1 in the future could help us to understand the circadian clock and photoperiodic flowering regulations better.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Arabidopsis (Arabidopsis thaliana) ecotype Columbia wild-type and mutant plants were grown on vermiculite saturated with water under either LD (16-h-light/8-h-dark) or SD (8-h-light/16-h-dark) conditions with an intensity of 80 to 120 μE m–2 s–1 of white light at 23°C as described previously (Lu et al., 2011). ubp12-1 (GABI_244E11), ubp12-2w (GABI_742C10), ubp13-1 (SALK_128312), ubp13-2 (SALK_024054), ubp13-3 (SALK_132368), co (Liu et al., 2008a), gi-4, and svp32 (Fujiwara et al., 2008) were used for genetics analysis. The primers used for genotyping are listed in Supplemental Table S1.
Alignment Analysis
Amino acid sequences of UBP12, UBP13, and mammalian USP7 were aligned using Jalview software through ClustalW. The domains were analyzed by the Pfam protein families database (http://pfam.sanger.ac.uk/).
GUS Staining and GFP Location
UBP12pro::GUS and UBP13pro::GUS transgenic (T3) lines were used to determine the expression pattern of UBP12/UBP13 via histochemical GUS staining as described (Niu et al., 2008). The promoters of UBP12 and UBP13 were amplified by primers CX3019 and CX2973 and CX2971 and CX2972, respectively, and cloned into p1391Z (XF388) vector. The complementary DNAs (cDNAs) of UBP12 and UBP13 were amplified by primers CX2969 and CX2970 and CX2967 and CX2968, respectively, and cloned into vectors pCAMBIA1300-35S-GFP (XF215) or pCAMBIA1300-35S-CFP (XF953). The subcellular localization of UBP12 and UBP13 was performed by observing the roots of transgenic (T3) plants carrying C-terminal fusion of GFP to UBP12 and CFP to UBP13 driven by the 35S promoter and analyzed by confocal microscopy (Leica TCS SP5).
Deubiquitin Activity Assay
The full-length UBP12 or UBP13 coding sequences were amplified by primers CX3090 and CX3092 or CX3090 and CX3091, respectively. Their mutant forms, UBP12C208S and UBP13C207S, were generated by QuikChange II Site-Directed Mutagenesis Kit (Stratagene) using primers CX3287 and CX3288 and CX3285 and CX3286, respectively. These products were cloned into MBP-LIC (XF510) vector and then coexpressed with the substrates UBQ1 and UBQ10 in Escherichia coli as described (Yan et al., 2000). Lysates were subjected to SDS-PAGE, transferred to polyvinylidene difluoride membranes, and detected by immunoblot with anti-ubiquitin antibodies. The primers used for these constructions are listed in Supplemental Table S1.
RNA Gel-Blot Analysis, DNA Gel-Blot Analysis, and Quantitative PCR
Total RNA was extracted using Trizol reagent (Invitrogen), 10 d after germination from whole seedlings grown under the indicated conditions (LD, SD, and LL conditions) on Murashige and Skoog plates. RNA (20 µg per lane) was separated in an agarose gel containing 1% (v/v) formaldehyde, blotted onto Hybond N+ membrane (GE Healthcare), and probed with the PCR-amplified DNA fragments using specific primer pairs (CX3977 and CX3118 for UBP12 and CX3976 and CX3120 for UBP13). Total DNA was extracted using cetyl-trimethyl-ammonium bromide reagent and digested by the restriction enzyme EcoRI or XhoI, and then digested DNA was separated in an agarose gel, blotted onto a Hybond N+ membrane, and probed with the PCR-amplified 35S promoter using specific primer pairs CX2532 and CX2533. For qRT-PCR, 2.0 μg total RNA was treated with DNaseI (Ambion), and then the first-strand cDNA was synthesized by using a cDNA synthesis kit (Transgen). qRT-PCR was performed using a CFX96 Real-Time PCR Instrument (Bio-Rad) with the SYBR Green reaction mix (Kangwei S-7567). Primers for qRT-PCR can be found in Supplemental Table S1.
Bioluminescence Measurement
For luciferase measurement, the pCCA1:LUC transgenic seedlings were entrained for 7 d or 15 d in 12-h-light/12-h-dark cycles at 22°C before transfer to continuous light. Since the first day in continuous light condition, seedlings were transferred to 96-well microplates (Perkin-Elmer) containing 200 μL Murashige and Skoog medium plus 2% (w/v) Suc and 30 μL 2.5 mm luciferin. The bioluminescence production was record with a Packard TopCount luminometer (Xu et al., 2010). Data were assayed using the Biological Rhythms Analysis Software System 2.1.4, which integrates the fast Fourier transform nonlinear least squares analysis for circadian rhythms (Plautz et al., 1997).
UBP Antibody and UBP Abundance in Arabidopsis
UBP12/UBP13-specific antibody was produced using a fragment of the N-terminal 315 amino acids of UBP13 expressed in E. coli. Polyclonal antisera was raised in mouse and affinity purified by UBP13 antigen. The specificity of UBP antibody was confirmed by Western blot using the wild type and the double mutant of ubp12-2wubp13-2. As showed in Supplemental Figure S7, the full-length band of UBP is present in the wild type but not in the double mutant, whereas the partial UBP12 is present in the mutant only. Total extracts were prepared from wild-type Columbia grown under LL condition every 4 h, and the nuclear and cytoplasmic fractions were separated as described (Weigel and Glazebrook, 2002). Immunoblot assay was performed as described (Lu et al., 2011). Histone H3 was used as control for loading and nuclear fraction. Phosphoenolpyruvate carboxylase was used as control for cytoplasmic fraction; antibodies included anti-H3:ab1791 (Abcam) and anti-PEPC:1004163 (Rockland).
Generation of Double Mutants
The double mutants were generated from the cross of homozygous mutants and identified from the F2 progeny grown on soil by comparing with their parental phenotypes and PCR-based characterization. The primers used for genotyping are listed in Supplemental Table S1.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Amino acid sequence alignment of UBP12, UBP13, and HsUSP7.
Supplemental Figure S2. Schematic diagram of raw unique reads mapped onto the region around the 3′ primer end of UBP12 in the WT and the ubp12-2w ubp13-3 double mutant.
Supplemental Figure S3. The number of T-DNA insertions in ubp12-2w.
Supplemental Figure S4. Statistical analysis of leaf numbers of ubp12-2w, ubp13-3, ubp12-2w ubp13-3, ubp12-1, and ubp12-1 ubp13-3 mutants under LD and SD conditions compared with wild-type plants.
Supplemental Figure S5. FLC expression level in ubp12 and ubp13 single and double mutants of 12 DAG seedlings.
Supplemental Figure S6. The phenotypes of ubp12-2w co and ubp12-2w gi-4 double mutants.
Supplemental Figure S7. UBP antibody specificity assay by western blot.
Supplemental Table S1. Primers used for qRT-PCR, constructions, and genotyping.
Acknowledgments
We thank Qingbao Zhu for technical help, Richard D. Vierstra (University of Wisconsin-Madison, Madison, WI) for providing UBQ1 and UBQ10 plasmids, Hongquan Yang (Shanghai Jiaotong University) for providing co (SAIL_H_024) seed, and Chentao Lin (University of California, Los Angeles) for providing gi-4 mutant seed.
Glossary
- DUB
deubiquitinating enzyme
- CFP
cyan fluorescent protein
- T-DNA
transfer DNA
- LD
long-day
- SD
short-day
- qRT-PCR
quantitative reverse transcriptase PCR
- LL
constant white light
- cDNA
complementary DNA
- ZT
Zeitgeber time
References
- Alabadí D, Oyama T, Yanovsky MJ, Harmon FG, Más P, Kay SA. (2001) Reciprocal regulation between TOC1 and LHY/CCA1 within the Arabidopsis circadian clock. Science 293: 880–883 [DOI] [PubMed] [Google Scholar]
- Baudry A, Ito S, Song YH, Strait AA, Kiba T, Lu S, Henriques R, Pruneda-Paz JL, Chua NH, Tobin EM, et al. (2010) F-box proteins FKF1 and LKP2 act in concert with ZEITLUPE to control Arabidopsis clock progression. Plant Cell 22: 606–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carré IA, Kim JY. (2002) MYB transcription factors in the Arabidopsis circadian clock. J Exp Bot 53: 1551–1557 [DOI] [PubMed] [Google Scholar]
- Chandler JS, McArdle B, Callis J. (1997) AtUBP3 and AtUBP4 are two closely related Arabidopsis thaliana ubiquitin-specific proteases present in the nucleus. Mol Gen Genet 255: 302–310 [DOI] [PubMed] [Google Scholar]
- Daubeuf S, Singh D, Tan Y, Liu H, Federoff HJ, Bowers WJ, Tolba K. (2009) HSV ICP0 recruits USP7 to modulate TLR-mediated innate response. Blood 113: 3264–3275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doelling JH, Phillips AR, Soyler-Ogretim G, Wise J, Chandler J, Callis J, Otegui MS, Vierstra RD. (2007) The ubiquitin-specific protease subfamily UBP3/UBP4 is essential for pollen development and transmission in Arabidopsis. Plant Physiol 145: 801–813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doelling JH, Yan N, Kurepa J, Walker J, Vierstra RD. (2001) The ubiquitin-specific protease UBP14 is essential for early embryo development in Arabidopsis thaliana. Plant J 27: 393–405 [DOI] [PubMed] [Google Scholar]
- Everett RD, Meredith M, Orr A, Cross A, Kathoria M, Parkinson J. (1997) A novel ubiquitin-specific protease is dynamically associated with the PML nuclear domain and binds to a herpesvirus regulatory protein. EMBO J 16: 566–577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewan R, Pangestuti R, Thornber S, Craig A, Carr C, O’Donnell L, Zhang C, Sadanandom A. (2011) Deubiquitinating enzymes AtUBP12 and AtUBP13 and their tobacco homologue NtUBP12 are negative regulators of plant immunity. New Phytol 191: 92–106 [DOI] [PubMed] [Google Scholar]
- Finley D. (2009) Recognition and processing of ubiquitin-protein conjugates by the proteasome. Annu Rev Biochem 78: 477–513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fornara F, Panigrahi KC, Gissot L, Sauerbrunn N, Rühl M, Jarillo JA, Coupland G. (2009) Arabidopsis DOF transcription factors act redundantly to reduce CONSTANS expression and are essential for a photoperiodic flowering response. Dev Cell 17: 75–86 [DOI] [PubMed] [Google Scholar]
- Fujiwara S, Oda A, Yoshida R, Niinuma K, Miyata K, Tomozoe Y, Tajima T, Nakagawa M, Hayashi K, Coupland G, et al. (2008) Circadian clock proteins LHY and CCA1 regulate SVP protein accumulation to control flowering in Arabidopsis. Plant Cell 20: 2960–2971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendron JM, Pruneda-Paz JL, Doherty CJ, Gross AM, Kang SE, Kay SA. (2012) Arabidopsis circadian clock protein, TOC1, is a DNA-binding transcription factor. Proc Natl Acad Sci USA 109: 3167–3172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmer SL. (2009) The circadian system in higher plants. Annu Rev Plant Biol 60: 357–377 [DOI] [PubMed] [Google Scholar]
- Huang W, Pérez-García P, Pokhilko A, Millar AJ, Antoshechkin I, Riechmann JL, Mas P. (2012) Mapping the core of the Arabidopsis circadian clock defines the network structure of the oscillator. Science 336: 75–79 [DOI] [PubMed] [Google Scholar]
- Huang Z, Wu Q, Guryanova OA, Cheng L, Shou W, Rich JN, Bao S. (2011) Deubiquitylase HAUSP stabilizes REST and promotes maintenance of neural progenitor cells. Nat Cell Biol 13: 142–152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imaizumi T, Kay SA. (2006) Photoperiodic control of flowering: not only by coincidence. Trends Plant Sci 11: 550–558 [DOI] [PubMed] [Google Scholar]
- Imaizumi T, Schultz TF, Harmon FG, Ho LA, Kay SA. (2005) FKF1 F-box protein mediates cyclic degradation of a repressor of CONSTANS in Arabidopsis. Science 309: 293–297 [DOI] [PubMed] [Google Scholar]
- Ito S, Song YH, Imaizumi T. (2012) LOV domain-containing F-box proteins: light-dependent protein degradation modules in Arabidopsis. Mol Plant 5: 573–582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang S, Marchal V, Panigrahi KC, Wenkel S, Soppe W, Deng XW, Valverde F, Coupland G. (2008) Arabidopsis COP1 shapes the temporal pattern of CO accumulation conferring a photoperiodic flowering response. EMBO J 27: 1277–1288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz EJ, Isasa M, Crosas B. (2010) A new map to understand deubiquitination. Biochem Soc Trans 38: 21–28 [DOI] [PubMed] [Google Scholar]
- Khoronenkova SV, Dianova II, Ternette N, Kessler BM, Parsons JL, Dianov GL. (2012) ATM-dependent downregulation of USP7/HAUSP by PPM1G activates p53 response to DNA damage. Mol Cell 45: 801–813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiba T, Henriques R, Sakakibara H, Chua NH. (2007) Targeted degradation of PSEUDO-RESPONSE REGULATOR5 by an SCFZTL complex regulates clock function and photomorphogenesis in Arabidopsis thaliana. Plant Cell 19: 2516–2530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim WY, Fujiwara S, Suh SS, Kim J, Kim Y, Han L, David K, Putterill J, Nam HG, Somers DE. (2007) ZEITLUPE is a circadian photoreceptor stabilized by GIGANTEA in blue light. Nature 449: 356–360 [DOI] [PubMed] [Google Scholar]
- Kim WY, Hicks KA, Somers DE. (2005) Independent roles for EARLY FLOWERING 3 and ZEITLUPE in the control of circadian timing, hypocotyl length, and flowering time. Plant Physiol 139: 1557–1569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau OS, Deng XW. (2012) The photomorphogenic repressors COP1 and DET1: 20 years later. Trends Plant Sci 17: 584–593 [DOI] [PubMed] [Google Scholar]
- Li D, Liu C, Shen L, Wu Y, Chen H, Robertson M, Helliwell CA, Ito T, Meyerowitz E, Yu H. (2008) A repressor complex governs the integration of flowering signals in Arabidopsis. Dev Cell 15: 110–120 [DOI] [PubMed] [Google Scholar]
- Li M, Chen D, Shiloh A, Luo J, Nikolaev AY, Qin J, Gu W. (2002) Deubiquitination of p53 by HAUSP is an important pathway for p53 stabilization. Nature 416: 648–653 [DOI] [PubMed] [Google Scholar]
- Liu LJ, Zhang YC, Li QH, Sang Y, Mao J, Lian HL, Wang L, Yang HQ. (2008a) COP1-mediated ubiquitination of CONSTANS is implicated in cryptochrome regulation of flowering in Arabidopsis. Plant Cell 20: 292–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Wang F, Zhang H, He H, Ma L, Deng XW. (2008b) Functional characterization of the Arabidopsis ubiquitin-specific protease gene family reveals specific role and redundancy of individual members in development. Plant J 55: 844–856 [DOI] [PubMed] [Google Scholar]
- Lu F, Cui X, Zhang S, Jenuwein T, Cao X. (2011) Arabidopsis REF6 is a histone H3 lysine 27 demethylase. Nat Genet 43: 715–719 [DOI] [PubMed] [Google Scholar]
- Luo M, Luo MZ, Buzas D, Finnegan J, Helliwell C, Dennis ES, Peacock WJ, Chaudhury A. (2008) UBIQUITIN-SPECIFIC PROTEASE 26 is required for seed development and the repression of PHERES1 in Arabidopsis. Genetics 180: 229–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maertens GN, El Messaoudi-Aubert S, Elderkin S, Hiom K, Peters G. (2010) Ubiquitin-specific proteases 7 and 11 modulate Polycomb regulation of the INK4a tumour suppressor. EMBO J 29: 2553–2565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Más P, Kim WY, Somers DE, Kay SA. (2003) Targeted degradation of TOC1 by ZTL modulates circadian function in Arabidopsis thaliana. Nature 426: 567–570 [DOI] [PubMed] [Google Scholar]
- McClung CR. (2011) The genetics of plant clocks. Adv Genet 74: 105–139 [DOI] [PubMed] [Google Scholar]
- Moon J, Parry G, Estelle M. (2004) The ubiquitin-proteasome pathway and plant development. Plant Cell 16: 3181–3195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagel DH, Kay SA. (2012) Complexity in the wiring and regulation of plant circadian networks. Curr Biol 22: R648–R657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson DC, Lasswell J, Rogg LE, Cohen MA, Bartel B. (2000) FKF1, a clock-controlled gene that regulates the transition to flowering in Arabidopsis. Cell 101: 331–340 [DOI] [PubMed] [Google Scholar]
- Neutzner M, Neutzner A. (2012) Enzymes of ubiquitination and deubiquitination. Essays Biochem 52: 37–50 [DOI] [PubMed] [Google Scholar]
- Nicholson B, Suresh Kumar KG. (2011) The multifaceted roles of USP7: new therapeutic opportunities. Cell Biochem Biophys 60: 61–68 [DOI] [PubMed] [Google Scholar]
- Nijman SM, Luna-Vargas MP, Velds A, Brummelkamp TR, Dirac AM, Sixma TK, Bernards R. (2005) A genomic and functional inventory of deubiquitinating enzymes. Cell 123: 773–786 [DOI] [PubMed] [Google Scholar]
- Niu L, Zhang Y, Pei Y, Liu C, Cao X. (2008) Redundant requirement for a pair of PROTEIN ARGININE METHYLTRANSFERASE4 homologs for the proper regulation of Arabidopsis flowering time. Plant Physiol 148: 490–503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plautz JD, Straume M, Stanewsky R, Jamison CF, Brandes C, Dowse HB, Hall JC, Kay SA. (1997) Quantitative analysis of Drosophila period gene transcription in living animals. J Biol Rhythms 12: 204–217 [DOI] [PubMed] [Google Scholar]
- Pokhilko A, Fernández AP, Edwards KD, Southern MM, Halliday KJ, Millar AJ. (2012) The clock gene circuit in Arabidopsis includes a repressilator with additional feedback loops. Mol Syst Biol 8: 574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes-Turcu FE, Ventii KH, Wilkinson KD. (2009) Regulation and cellular roles of ubiquitin-specific deubiquitinating enzymes. Annu Rev Biochem 78: 363–397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawa M, Nusinow DA, Kay SA, Imaizumi T. (2007) FKF1 and GIGANTEA complex formation is required for day-length measurement in Arabidopsis. Science 318: 261–265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz RJ, Tamada Y, Doyle MR, Zhang X, Amasino RM. (2009) Histone H2B deubiquitination is required for transcriptional activation of FLOWERING LOCUS C and for proper control of flowering in Arabidopsis. Plant Physiol 149: 1196–1204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song MS, Salmena L, Carracedo A, Egia A, Lo-Coco F, Teruya-Feldstein J, Pandolfi PP. (2008) The deubiquitinylation and localization of PTEN are regulated by a HAUSP-PML network. Nature 455: 813–817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sridhar VV, Kapoor A, Zhang K, Zhu J, Zhou T, Hasegawa PM, Bressan RA, Zhu JK. (2007) Control of DNA methylation and heterochromatic silencing by histone H2B deubiquitination. Nature 447: 735–738 [DOI] [PubMed] [Google Scholar]
- Takase T, Nishiyama Y, Tanihigashi H, Ogura Y, Miyazaki Y, Yamada Y, Kiyosue T. (2011) LOV KELCH PROTEIN2 and ZEITLUPE repress Arabidopsis photoperiodic flowering under non-inductive conditions, dependent on FLAVIN-BINDING KELCH REPEAT F-BOX1. Plant J 67: 608–621 [DOI] [PubMed] [Google Scholar]
- van der Horst A, de Vries-Smits AM, Brenkman AB, van Triest MH, van den Broek N, Colland F, Maurice MM, Burgering BM. (2006) FOXO4 transcriptional activity is regulated by monoubiquitination and USP7/HAUSP. Nat Cell Biol 8: 1064–1073 [DOI] [PubMed] [Google Scholar]
- van der Knaap JA, Kumar BR, Moshkin YM, Langenberg K, Krijgsveld J, Heck AJ, Karch F, Verrijzer CP. (2005) GMP synthetase stimulates histone H2B deubiquitylation by the epigenetic silencer USP7. Mol Cell 17: 695–707 [DOI] [PubMed] [Google Scholar]
- Vierstra RD. (2009) The ubiquitin-26S proteasome system at the nexus of plant biology. Nat Rev Mol Cell Biol 10: 385–397 [DOI] [PubMed] [Google Scholar]
- Weigel D, Glazebrook J. (2002) Arabidopsis: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- Wing SS. (2003) Deubiquitinating enzymes—the importance of driving in reverse along the ubiquitin-proteasome pathway. Int J Biochem Cell Biol 35: 590–605 [DOI] [PubMed] [Google Scholar]
- Xu X, Xie Q, McClung CR. (2010) Robust circadian rhythms of gene expression in Brassica rapa tissue culture. Plant Physiol 153: 841–850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan N, Doelling JH, Falbel TG, Durski AM, Vierstra RD. (2000) The ubiquitin-specific protease family from Arabidopsis. AtUBP1 and 2 are required for the resistance to the amino acid analog canavanine. Plant Physiol 124: 1828–1843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanovsky MJ, Kay SA. (2002) Molecular basis of seasonal time measurement in Arabidopsis. Nature 419: 308–312 [DOI] [PubMed] [Google Scholar]
- Yu JW, Rubio V, Lee NY, Bai S, Lee SY, Kim SS, Liu L, Zhang Y, Irigoyen ML, Sullivan JA, et al. (2008) COP1 and ELF3 control circadian function and photoperiodic flowering by regulating GI stability. Mol Cell 32: 617–630 [DOI] [PMC free article] [PubMed] [Google Scholar]