OsHMA2 is a plasma membrane-localized transporter for zinc and cadmium in rice that mainly expresses at the phloem region of developed vascular tissues in the nodes and mediates exclusive zinc and cadmium delivery to upper nodes and finally to the panicle.
Abstract
Developing tissues such as meristems and reproductive organs require high zinc, but the molecular mechanisms of how zinc taken up by the roots is preferentially delivered to these tissues with low transpiration are unknown. Here, we report that rice (Oryza sativa) heavy metal ATPase2 (OsHMA2), a member of P-type ATPases, is involved in preferential delivery of zinc to the developing tissues in rice. OsHMA2 was mainly expressed in the mature zone of the roots at the vegetative stage, but higher expression was also found in the nodes at the reproductive stage. The expression was unaffected by either zinc deficiency or zinc excess. OsHMA2 was localized at the pericycle of the roots and at the phloem of enlarged and diffuse vascular bundles in the nodes. Heterologous expression of OsHMA2 in yeast (Saccharomyces cerevisiae) showed influx transport activity for zinc as well as cadmium. Two independent Tos17 insertion lines showed decreased zinc concentration in the crown root tips, decreased concentration of zinc and cadmium in the upper nodes and reproductive organs compared with wild-type rice. Furthermore, a short-term labeling experiment with 67Zn showed that the distribution of zinc to the panicle and uppermost node I was decreased, but that, to the lower nodes, was increased in the two mutants. Taken together, OsHMA2 in the nodes plays an important role in preferential distribution of zinc as well as cadmium through the phloem to the developing tissues.
Zinc is an essential metal for all organisms. In human genome, about 2,800 proteins, which account for 10% of total proteins, require zinc for structural or functional activities (Andreini et al., 2009; Maret and Li, 2009). In plants, zinc is involved in many enzyme activities, maintenance of integrity of biomembranes, RNA and DNA metabolism, carbohydrate metabolism, cell division, protein synthesis, gene expression regulation, and so on (Broadley et al., 2011). Zinc is also required for the metabolism of auxin. Therefore, zinc deficiency in plants causes stunted growth and “little leaf” (Broadley et al., 2011).
Developing tissues with low transpiration especially requires high zinc for the active cell division and growth. For example, in pollen tubes, the zinc concentration at the growing tip was about 150 µg g–1 dry weight compared with about 50 µg g–1 in more basal regions (Ender et al., 1983). In the newly emerged root tips of wheat (Triticum aestivum) plants, zinc concentration is about 220 µg g–1 (Ozturk et al., 2006). In rice (Oryza sativa) shoot meristems, more than 10 times the amount of zinc was found compared with mature leaf blades (Kitagishi and Obata, 1986). However, the molecular mechanisms underlying preferential delivery of zinc to these developing tissues are unknown.
Zinc is taken up by the roots through the ZIP (for ZRT, IRT-related proteins) transporters (Grotz et al., 1998; Guerinot, 2000). Release of zinc into the xylem (xylem loading) is mediated by two P1B-type ATPases, AtHMA2 and AtHMA4, in Arabidopsis (Arabidopsis thaliana), which are localized at the pericycle (Hussain et al., 2004; Verret et al., 2004; Wong and Cobbett, 2009; Wong et al., 2009). Knockout of AtHMA4 and AtHMA2 or AtHMA4 alone resulted in a reduced translocation of zinc from the roots to the shoots. Recently, mutation of OsHMA2 was also reported to cause decreased translocation of zinc and cadmium from the roots to the shoots at the vegetative growth stage in rice (Satoh-Nagasawa et al., 2012; Takahashi et al., 2012); however, the exact mechanism underlying the involvement of this gene in the root-shoot translocation is not yet elucidated. Different from other metals such as manganese, zinc taken up by the roots is preferentially translocated to the shoot meristems and other developing tissues to meet the high demand of zinc in these tissues. In rice, 65Zn taken up by the roots is translocated not to the expanded and active leaves with high transpiration but to the developing tissues, including unexpanded leaf blade, leaf sheath, and young panicles with very low transpiration within 1 h (Obata et al., 1980; Obata and Kitagishi, 1980a). By contrast, manganese taken up by the roots is translocated to the expanded and active leaves (Obata and Kitagishi, 1980a). Higher and faster zinc accumulation was observed at the basal part of elongating leaf sheath, which includes intercalary meristem (Obata et al., 1980; Obata and Kitagishi, 1980a; Kitagishi and Obata, 1986). High zinc accumulation in the meristem was also reported in tomato (Solanum lycopersicum; Langston, 1956) and subterranean clover (Trifolium subterraneum; Riceman and Jones, 1958a, 1958b). These observations suggest that distribution of zinc to the developing tissues does not depend on transpiration-dependent xylem-mediated transport.
At the reproductive growth stage, the preferential accumulation of zinc was also found in the nodes of rice, which probably act as relay points for zinc distribution to the panicles (Obata and Kitagishi, 1980b), which have high zinc requirement for filling the grain. Deficiency of zinc causes low fertility (Obata and Kitagishi, 1980a). However, the molecular mechanisms on how zinc is preferentially delivered to these developing tissues have not been understood. In this study, we found that a member of P1B-type ATPase, OsHMA2, is involved in this process. OsHMA2 was previously implicated in the root-to-shoot translocation of zinc and cadmium at the vegetative stage (Satoh-Nagasawa et al., 2012; Takahashi et al., 2012). However, we found that the major role of OsHMA2 in the nodes is to preferentially deliver zinc to the developing tissues. This is especially important for delivering zinc to the grain at the reproductive growth stage. We also found that this transporter is responsible for cadmium distribution to the grains in rice.
RESULTS
Cloning of Full-Length OsHMA2 cDNA and Phylogenetic Analysis
Because the full-length complementary DNA (cDNA) sequence of OsHMA2 is not available in the Rice Annotation Project Database (http://rapdb.dna.affrc.go.jp/), we first cloned the full-length cDNA of OsHMA2 from the roots of japonica rice ‘Nipponbare’ by reverse transcription (RT)-PCR based on two putative truncated cDNA clones, AK107235/Os06g0700700 and NM_001188052/Os06g0700650, containing putative translational start and stop sites, respectively. The cloned cDNA contained an open reading frame (ORF), with 3,204 bp encoding a 1,067-amino acid peptide (Supplemental Fig. S1A). Like other HMA members, the protein is characterized by eight transmembrane helices, a CPx/SPC motif in the transmembrane domain 6, and putative transition metal-binding domains at the N and/or C termini (Williams and Mills, 2005). Comparison of amino acid core sequences without N- and C-terminal variable regions (Supplemental Fig. S1A) showed that OsHMA2 core shares 66.6% identity with OsHMA3, a tonoplast-localized transporter of cadmium (Ueno et al., 2010). The identity with AtHMA2, AtHMA3, and AtHMA4 core was 62.9%, 59.1%, and 62.1%, respectively (Supplemental Figs. S1A and S2).
Expression Patterns of OsHMA2 at Different Growth Stages
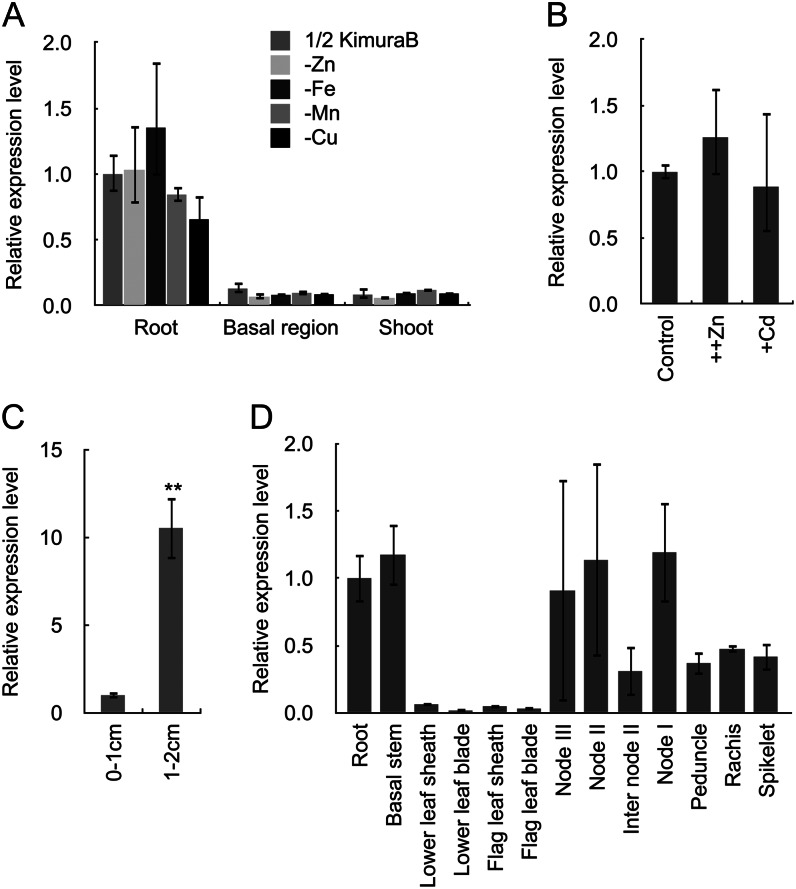
Expression profile of OsHMA2 in different tissues of rice grown hydroponically was investigated by quantitative RT-PCR at the vegetative stage. The expression of OsHMA2 was mainly detected in the roots, with much lower level in the shoots and basal stem (Fig. 1A). To investigate the effect of metal deficiency on the expression of OsHMA2, cDNAs isolated from plants grown in metal-free solution were used. The metal deficiency status has been monitored by respective marker genes (Zheng et al., 2012). Deficiency of zinc, iron, manganese, or copper did not affect the expression of OsHMA2 (Fig. 1A). The expression was also unaffected by excess zinc or cadmium supply (Fig. 1B). Spatial expression analysis showed that the expression was much higher in the mature root region than in the root tip region (Fig. 1C).
Figure 1.
Expression pattern of OsHMA2 gene. A, Tissue-dependent expression of OsHMA2 at vegetative stage. Plants were grown in a nutrient solution in the absence of zinc, iron, manganese, or copper or presence of these metals for 7 d. The roots, shoots, and basal region (2 cm from the root-shoot junction) were sampled for expression analysis. B, Response of OsHMA2 to excess zinc (10 µm) and cadmium (1 µm) for 1 d in vegetative roots. C, Spatial expression in seminal root. Different segments (0–1 cm and 1–2 cm from the apex) were sampled from 5-d-old seedlings. D, Tissue-specific expression at flowering stage. Different tissues samples were taken from rice grown in a paddy field at flowering stage. The expression was determined by quantitative RT-PCR, and expression level of Actin and HistoneH3 were used as an internal control. Error bars represent ± sd of three independent biological replicates. Statistical comparison was performed by one-way ANOVA followed by the Tukey’s multiple comparison test (A and B) or Student’s t test (C). Differences were not significant between different metal deficiency treatment and the control condition (A and B); **P < 0.01 (C).
At reproductive stage, higher expression was found in the basal stem containing unelongated nodes and upper nodes (nodes I–III) at the elongated stem of rice grown in the field, in addition to the roots (Fig. 1D).
Tissue Specificity of Localization of OsHMA2
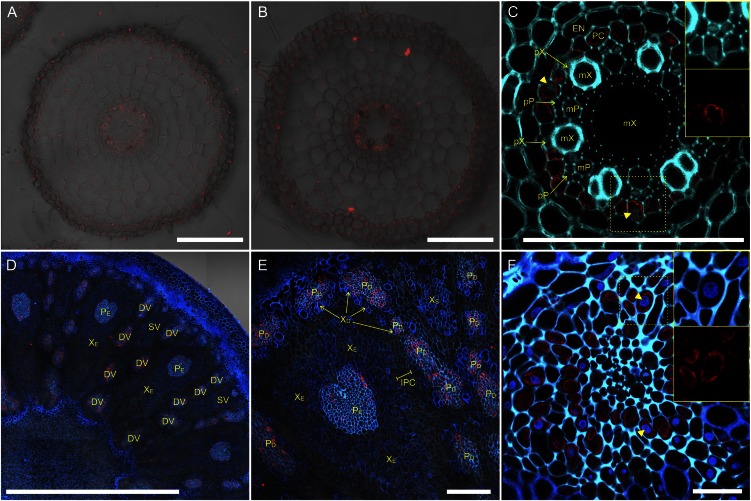
To examine the localization of OsHMA2 protein in different tissues, we performed immunohistochemical staining with a polyclonal antibody against C-terminal peptide of OsHMA2. Fluorescence signal was very weak at the root tip of wild-type rice (5 mm from the apex; Fig. 2A), but strongly detected in the pericycle cell layer of the mature zone (20 mm from the apex; Fig. 2, B and C). No signals in root of two mutant lines (described later) indicate the specificity of the antibody to OsHMA2 (Supplemental Fig. S3, A and B).
Figure 2.
Localization of OsHMA2 in rice root and node. Immunohistochemical staining of OsHMA2 with anti-OsHMA2 polyclonal antibody was performed. A and B, Root cross section at 5 mm (A) or 20 mm (B) from the apex. C, Magnified image of the stele in B. D–F, Cross section of node I at the flowering stage with different magnification. F, Close-up of phloem region of a diffuse vascular bundle. Red color indicates the OsHMA2-specific signal. Blue color indicates cell wall autofluorescence and nucleus stained by DAPI (yellow arrowheads). Insets in C and F are channel-separated magnified images at yellow-dotted areas. Bars = 100 μm (A–C and E), 1 mm (D), and 20 μm (F). Endodermis (EN), pericycle (PC), xylem vessel of proto- and metaxylem (pX and mX, respectively), and sieve tube of proto- and metaphloem (pP and mP, respectively) in the root and diffuse vascular bundle (DV), small vascular bundle (SV), xylem area of enlarged and diffuse vascular bundle (XE and XD, respectively), phloem area of enlarged and diffuse vascular bundle (PE and PD, respectively), and intervening parenchyma cells (IPC) in the node are shown.
In node I (uppermost node connecting to panicle and flag leaf) at the flowering stage, OsHMA2 protein was localized in the phloem region of both the diffuse vascular bundle and the enlarged vascular bundle (Fig. 2, D–F). OsHMA2 protein was also detected in the phloem region at the unelongated basal node and node III (Supplemental Fig. S3, C and D).
To confirm immunostaining results above, we also prepared transgenic rice carrying OsHMA2 promoter-GFP gene. Immunostaining with an anti-GFP antibody also showed the localization at the pericycle layer of the roots and phloem region at the nodes (Supplemental Fig. S3, E and F).
Subcellular Localization of OsHMA2
OsHMA2 was reported to be localized to the plasma membrane in onion (Allium cepa) epidermal cells when a GFP fusion gene was introduced transiently (Satoh-Nagasawa et al., 2012; Takahashi et al., 2012). To examine whether OsHMA2 protein shows similar subcellular localization in rice plants, we first performed immunostaining together with nuclear staining by 4′,6-diamino-2-phenylindole (DAPI). In a magnified image of the root stele, signal from OsHMA2 antibody was detected at cell wall inscribed area of the pericycle cells, which comprehend the nuclei stained by DAPI, but no signal was accompanied with the nuclei (Fig. 2C, inset). Similar subcellular localization was also observed in the phloem parenchyma cells at the upper node (Fig. 2F, inset). These results indicate that OsHMA2 protein was not localized to the endoplasmic organelles, such as the vacuole, Golgi apparatus, endoplasmic reticulum (ER), and nuclei, but most likely localized to the plasma membrane.
To confirm this result, we further performed western-blot analysis with Suc density gradient fractionation of microsome prepared from rice roots. OsHMA2 protein was detected in the same fractionation pattern as H+-ATPase, a plasma membrane marker, but different fractionation patterns from V-type ATPase, Binding immunoglobulin protein, and ADP-ribosylation factor1, which present markers for tonoplast, ER, and Golgi, respectively (Supplemental Fig. S4). Taken together, our results indicate that OsHMA2 protein is localized to the plasma membrane in rice.
Mutation of OsHMA2 Resulted in Impaired Delivery of Zinc to the Developing Tissues at Vegetative Growth Stage
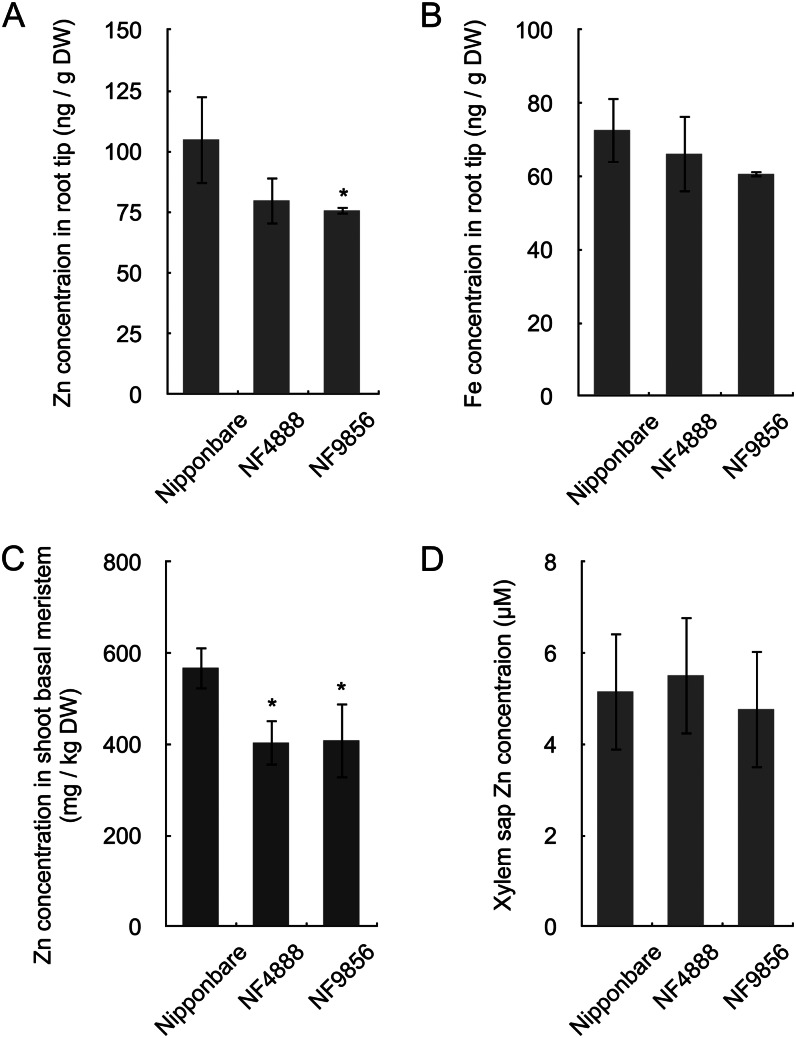
To investigate the role of OsHMA2 in zinc distribution, we obtained two mutants with Tos17 retrotransposon insertion (NF4888 and NF9856). Both mutants have an insertion at the different positions of the 10th exon (Supplemental Fig. S1B). Gene expression analysis showed that the full length of OsHMA2 was not expressed in both lines (Supplemental Fig. S1C). Decreased root-to-shoot translocations of zinc and cadmium in the mutants were observed in our experimental conditions (Supplemental Figs. S5 and S6). This result is consistent with two previous studies (Satoh-Nagasawa et al., 2012; Takahashi et al., 2012). However, a detailed analysis showed that the growth was also affected by mutation of OsHMA2. When the two mutant lines and the wild-type rice were grown hydroponically for 5 weeks in the presence of normal-level zinc (0.4 µm), both the shoot and roots were smaller in the two mutant lines than the wild-type rice (Supplemental Fig. S7). Furthermore, spatial analysis revealed that in contrast to the total zinc concentration in the roots (Supplemental Fig. S6), the zinc concentration in the tip region (0–5 mm from the apex) of the developing crown roots was lower in the mutant lines than that of the wild-type rice (Fig. 3A). As a control, the iron concentration in the same root segment was also determined, but there was no difference between the wild-type rice and mutant lines (Fig. 3B). The shoot basal meristem contained much higher levels of zinc compared with whole shoots in all lines (Fig. 3C; Supplemental Fig. S5), but the zinc concentration in this part was much lower in the two mutant lines than the wild-type rice (Fig. 3C). No difference was found in the concentration of other metals in the shoot meristem region between lines (Supplemental Fig. S8). On the other hand, the zinc concentration of the xylem sap did not differ between the wild-type and mutant lines (Fig. 3D).
Figure 3.
Metal concentration in wild-type rice and two oshma2 mutants. A and B, Concentration of zinc (A) and iron (B) in the root tips. Root tips (0–5 mm) were cut from seedlings with a razor. C, Zinc concentration in the shoot basal meristem. The lowest 2-cm part of leaf sheath of youngest expanded leaf and inner organs was excised. D, Zinc concentration in the xylem sap of 3-week-old seedlings. Error bars represent ± sd of three independent biological replicates. Statistical comparison was performed by one-way ANOVA followed by the Tukey’s multiple comparison test. All data were compared with cv Nipponbare (*P < 0.05).
Because recent studies reported that OsHMA2 may be implicated in cadmium transport (Nocito et al., 2011; Satoh-Nagasawa et al., 2012; Takahashi et al., 2012), we then also compared cadmium concentration between the mutant lines and wild-type rice. Similar to zinc, the mutant lines contained lower levels of cadmium in the shoots and slightly higher cadmium in the roots compared with the wild-type rice (Supplemental Figs. S5, S6, and S8).
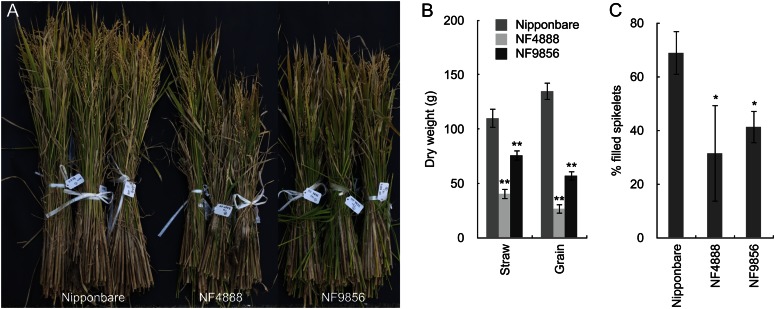
Mutation of OsHMA2 Resulted in Significant Reduction of Grain Yield
Since the expression of OsHMA2 in the nodes was very high at the reproductive growth stage (Fig. 1D), we grew the mutant lines and the wild-type rice in the field until ripening to investigate the physiological role of node-localized OsHMA2 in zinc distribution. A significant difference in the growth and yield was found between the mutant lines and wild-type rice (Fig. 4A). The biomass of the straw was decreased by 35% to 68% in the mutant lines compared with the wild-type rice (Fig. 4B). The grain yield was especially reduced in the mutant lines by 58% to 80%, mainly due to decreased fertility (Fig. 4C).
Figure 4.
Effect of OsHMA2 mutation on rice productivity. A, Growth of wild-type rice and two mutants at harvest. B, Dry weight of straw and grain. C, Fertility of the seeds. The wild-type rice and two oshma2 mutants (NF4888 and NF9856) were grown in a paddy field till ripening. Error bars represent ± sd of three independent biological replicates. Statistical comparison was performed by one-way ANOVA followed by the Tukey’s multiple comparison test. Data were compared with cv Nipponbare (*P < 0.05, **P < 0.01 [B and C]).
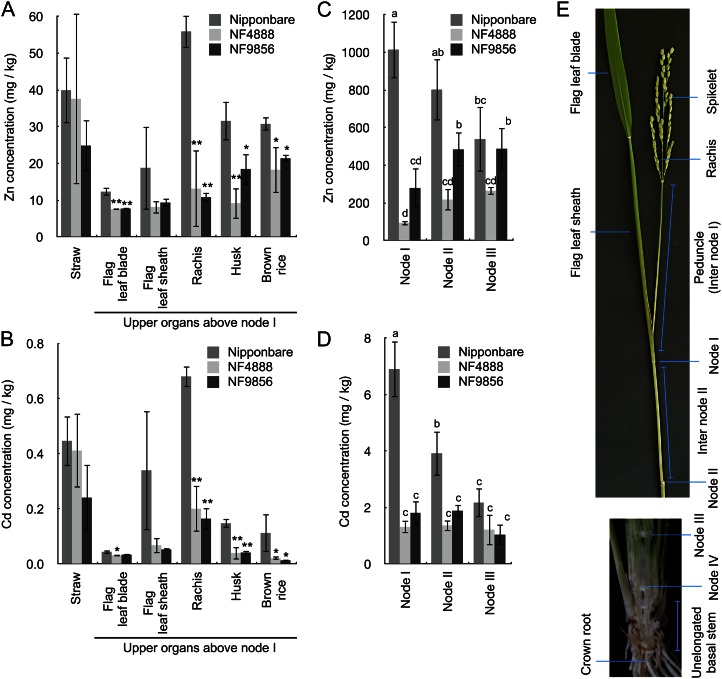
At the harvest, the concentration of zinc and cadmium in the straw (most of the shoot) was not significantly different between the wild-type rice and the mutant lines (Fig. 5, A and B). However, in upper organs above node I, composed of flag leaf, rachis, husk, and brown rice, the concentration of both zinc and cadmium was significantly decreased in the mutant lines (Fig. 5, A and B). The concentration of zinc and cadmium at different nodes was also compared between the mutant lines and the wild-type rice. In the nodes (nodes I–III from the top of the culm), among cations examined, only the concentrations of zinc and cadmium were much lower in the mutant lines than that in the wild type in upper nodes I and II (Fig. 5, C and D; Supplemental Fig. S9). In node III, the concentration of zinc and cadmium was similar between the wild type and the mutants (Fig. 5, C and D). The concentration of other metals was similar or slightly higher in the mutant lines than that in the wild-type rice (Supplemental Fig. S9). Interestingly, in the wild-type rice, the upper node contained more zinc and cadmium than the lower node, whereas each node similarly contained lower levels of zinc and cadmium in the mutant lines (Fig. 5, C and D).
Figure 5.
Metal concentration of wild-type rice and oshma2 mutants at harvest. A and B, Concentration of zinc (A) and cadmium (B) in different tissues. C and D, Concentration of zinc (C) and cadmium (D) in different nodes. The wild-type rice ‘Nipponbare’ and two oshma2 mutants (NF4888 and NF9856) were grown in a paddy field till ripening. Error bars represent ± sd of three independent biological replicates. Statistical comparison was performed by one-way ANOVA followed by the Tukey’s multiple comparison test. Data were compared with cv Nipponbare (*P < 0.05, **P < 0.01 [A and B]). Different letters indicate significance (P < 0.05 [C and D]). E, Different organs sampled used for analysis at the heading stage. Organs above node II (top, leaf attached to node II was removed) and longitudinal section of shoot basal region containing approximately 10 unelongated nodes (bottom) are shown.
Mutation of OsHMA2 Altered the Zinc Distribution at the Reproductive Growth Stage
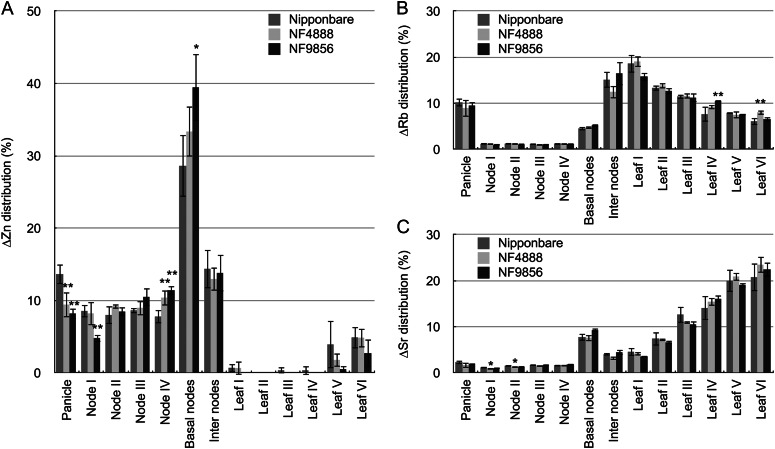
To confirm that the decreased zinc concentration observed only in the upper and reproductive organs of the mutant lines is caused by altered distribution of zinc, we conducted a short-term labeling experiment with a 67Zn stable isotope. Plants were fed with 0.4 µm 67Zn from the roots for 2 d just before heading, and the distribution ratio (ΔZn ) within the above-ground part was calculated. Rubidium and strontium, markers of phloem and xylem transport, respectively (Kuppelwieser and Feller, 1991), were also fed together. Results showed that most 67Zn taken up during 2 d was distributed to the panicles, nodes, and internodes, but hardly to the leaves (Fig. 6A), regardless of the wild-type rice or oshma2 mutants. However, compared with the wild-type rice, the distribution of zinc to the panicle and uppermost node I was lower, but distribution to the lower node IV and basal nodes was higher in the two mutants (Fig. 6A). By contrast, there was almost no difference in the distribution of strontium and rubidium between the wild-type rice and mutant lines (Fig. 6, B and C). A larger part of rubidium was distributed to the panicle, internodes, and upper leaves (Fig. 6B), while most strontium was distributed to the lower leaves with high transpiration (Fig. 6C). These distribution patterns are different from that of zinc (Fig. 6A). These results suggest that OsHMA2 plays an important role in preferential delivery of zinc to the upper parts.
Figure 6.
Short-term distribution of zinc, rubidium, and strontium in wild-type rice and oshma2 mutants. The wild-type rice ‘Nipponbare’ and two oshma2 mutants (NF4888 and NF9856) were grown hydroponically till just before heading. 67Zn stable isotope, rubidium, and strontium were fed from nutrient solution for 2 d. Distribution ratio of ΔZn (A), ΔRb (B), and ΔSr (C) within the above-ground tissues were calculated from net increase of each element in each part. Error bars represent ± sd of four independent biological replicates. Statistical comparison was performed by one-way ANOVA followed by the Tukey’s multiple comparison test. All data were compared with cv Nipponbare (*P < 0.05, **P < 0.01).
Heterologous Expression of OsHMA2 in Yeast
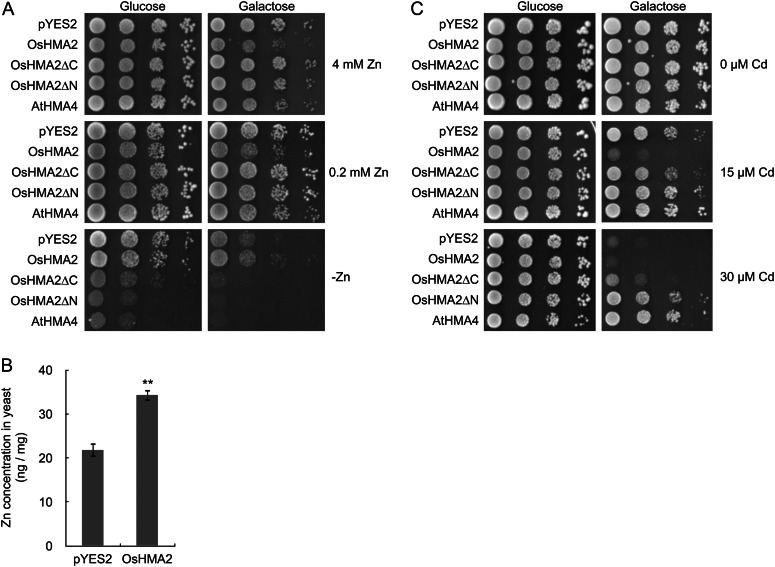
Since only zinc and cadmium concentration in the mutant lines was altered (Figs. 3 and 5), we then tested the transport activity of OsHMA2 for zinc and cadmium in yeast (Saccharomyces cerevisiae). When expressed in a yeast mutant defective in zinc uptake, zinc-regulated transporter1 (zrt1)zrt2, under the control of a Gal-inducible promoter, yeast expressing full-length OsHMA2 showed inhibited growth at high zinc concentrations, but complement the yeast growth on zinc-limited medium in the presence of Gal (Fig. 7A). However, when gene expression was suppressed by the presence of Glc, these differences in the growth were not found between yeast carrying OsHMA2 or an empty vector, indicating an OsHMA2 expression-dependent effect. Furthermore, zinc uptake in liquid medium was also higher in the yeast expressing OsHMA2 than the empty vector control (Fig. 7B). These results indicate that OsHMA2 functions as a transporter for zinc uptake in yeast.
Figure 7.
Yeast transport assay for zinc and cadmium. A, Growth of zrt1zrt2 (zinc uptake-deficient yeast strain) harboring empty vector pYES2, full-length OsHMA2, OsHMA2ΔC(707–1,067), OsHMA2ΔN(2–706), and AtHMA4. The yeast was incubated on a plate containing 4, 0.2, or 0 mm ZnSO4 in the presence of Glc or Gal for 3 d. B, Zinc uptake in yeast carrying OsHMA2 or empty vector. The yeast was exposed to a solution containing 5 µm ZnSO4 for 6 h. Error bars represent ± sd of three independent biological replicates. Statistical comparison was performed by Student’s t test (**P < 0.01). C, Growth of wild-type yeast strain BY4741 transformed with empty vector pYES2, full-length OsHMA2, OsHMA2ΔC(707–1,067), OsHMA2ΔN(2–706), and AtHMA4 in the presence of 0, 15 or 30 µm CdSO4 with Glc or Gal for 3 d.
We also tested the transport activity of OsHMA2 for cadmium in two different yeast strains, BY4741 and INVSc1. Results from both strains showed that in the presence of Glc (no gene induction), the growth was similar between the yeast carrying OsHMA2 and empty vector control on the medium either with or without cadmium (Fig. 7C). However, when the gene expression was induced by Gal, the growth was significantly inhibited in the yeast expressing OsHMA2 compared with empty vector control in the presence of cadmium, although the growth was similar in the absence of cadmium (Fig. 7C; Supplemental Fig. S10). As a control, yeast expressing AtHMA4 was also tested. In contrast to OsHMA2, expression of AtHMA4 enhanced the tolerance to cadmium (Fig. 7C; Supplemental Fig. S10A). These results indicate that, different from AtHMA4, an efflux transporter of zinc/cadmium (Papoyan and Kochian, 2004; Verret et al., 2005), OsHMA2 is an influx transporter for cadmium in yeast.
Because OsHMA2 is characterized by a long C-terminal region (705–1,067 amino acids), which is much longer and has almost no similarity with other HMA members (Supplemental Fig. S1), we tested the role of the C-terminal region in zinc and cadmium sensitivity in yeast. When the C-terminal truncated version of OsHMA2 (ΔC 707–1,067) was expressed, unlike the full-length OsHMA2, it was not only unable to complement the growth, but also showed more severe growth defect in zrt1zrt2 under zinc-limited condition (Fig. 7A). The tolerance to cadmium was slightly increased at 30 µm cadmium (Fig. 7C). On the other hand, when N-terminal truncated OsHMA2 lacking any transmembrane domains (ΔN 2–706) was expressed in yeast, highly increased sensitivity to zinc deficiency and tolerance to cadmium were observed (Fig. 7, A and C). These results consistently suggest that truncation of either C-terminal or N-terminal regions alters the transport activity, further indicating that the full-length OsHMA2 protein functions as an influx transporter of zinc and cadmium in yeast.
In addition, we also compared the growth of yeast expressing OsHMA2 in two other strains, the wild type (BY4741) and a zinc-hypersensitive mutant defective in vacuole zinc sequestration (zinc resistance conferring1 [zrc1] cobalt toxicity1 [cot1]). However, different from zrt1zrt2 (Fig. 7A), the growth did not differ in yeast expressing OsHMA2 and the empty vector in both yeast strains under zinc deficiency and toxicity conditions (Supplemental Fig. S10, B and C), due to competition with endogenous Zrt1 and Zrt2 transporters. These results further suggest that OsHMA2 functions as a transporter for zinc uptake in yeast.
DISCUSSION
OsHMA2 Is a Transporter for Preferentially Delivering Zinc to the Developing Tissues in Rice
Previous characterization of OsHMA2 showed that OsHMA2 is implicated in the root-shoot translocation of zinc and cadmium (Satoh-Nagasawa et al., 2012; Takahashi et al., 2012; Supplemental Figs. S5 and S6). However, we show here that the major role of OsHMA2 in rice is to preferentially deliver zinc (and cadmium) to the developing tissues with high zinc requirement, but low transpiration at both the vegetative and reproductive growth stage through phloem transport.
OsHMA2 is mainly localized to the plasma membrane of pericycle cells of the mature root zone at the vegetative growth stage (Fig. 2, A–C). Pericycle cell layer is adjacent to both xylem vessel and phloem cells, and lateral connections by plasmodesmata between neighbor pericycle cells are very tight (Ma and Peterson, 2001). It is likely that this cell layer contributes to both xylem and phloem transport by control of efflux to the apoplast and influx to the symplast, respectively. Our results suggest that the role of OsHMA2 is to transport zinc from the apoplast of the stele into the phloem for delivering zinc to the developing tissues. This is supported by the following evidence (Fig. 8A). Firstly, when OsHMA2 was mutated, the zinc concentration in the root tip region was decreased (Fig. 3A), although the zinc concentration in the whole roots was increased in the mutant lines (Supplemental Fig. S6). In the root tips, the vascular system has not been developed well; therefore, nutrients should be provided through the symplastic transport. Considering that OsHMA2 is mainly expressed in the mature root zone (Fig. 1C), it is likely that OsHMA2 is involved in translocating zinc from the mature root zone to the root tips via the phloem. Secondly, the zinc concentration in the xylem sap was not affected by the oshma2 mutation (Fig. 3D), but the shoots and basal meristem of the mutant lines contained a lower zinc concentration compared with the wild-type rice (Fig. 3C; Supplemental Fig. S5). This suggests that OsHMA2 is not involved in the xylem loading, but in the phloem loading, which facilitates the translocation of zinc to the developing tissues, including root tips and shoot meristem.
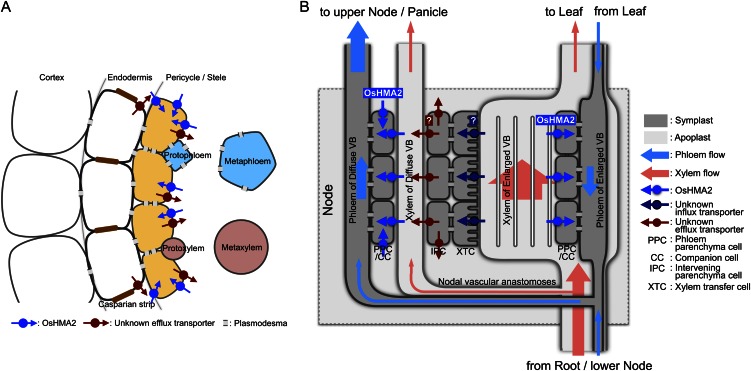
Figure 8.
Schematic diagram of zinc and cadmium transport by OsHMA2 in root and nodes. A, Function of OsHMA2 in the roots. OsHMA2 localized at the pericycle cells transports zinc and cadmium from the apoplast to the symplast to facilitate translocation via the phloem. B, Function of OsHMA2 in nodes. OsHMA2 localized in the phloem parenchyma and companion cells of both enlarged and diffuse vascular bundles in the nodes functions to load zinc and cadmium to the phloem for the preferential distribution to the upper nodes and panicles.
At reproductive growth stage, OsHMA2 was highly expressed in the nodes. Rice nodes have remarkably developed vascular systems composing of an enlarged vascular bundle and a diffuse vascular bundle (Kawahara et al., 1974; Hoshikawa, 1989). Enlarged vascular bundles in a node have a developed huge xylem area and are connected with a vascular bundle in a leaf and a lower node. On the other hand, diffuse vascular bundles are originated at the node surrounding the enlarged vascular bundle and are connected to the upper node (or panicle; Fig. 8B; Kawahara et al., 1974; Hoshikawa, 1989). Therefore, the node is an important place for delivering nutrients to the developing tissues or panicles through intervascular transfer, which transfers nutrients from the transpirational stream in the xylem of enlarged vascular bundles into the xylem or phloem of the diffuse vascular bundles (Kawahara et al., 1974; Hoshikawa, 1989). A previous study showed that at the reproductive growth stage, zinc taken up by the roots was rapidly accumulated at the nodes and then preferentially distributed to the panicles, but hardly to the expanded leaves (Obata and Kitagishi, 1980a, 1980b). In fact, zinc was mainly present in the parenchyma tissues between enlarged vascular bundles and diffuse vascular bundles in rice nodes (Obata and Kitagishi, 1980b; Yamaguchi et al., 2012). OsHMA2 protein was localized in the phloem region of the enlarged and diffuse vascular bundles in the nodes (Fig. 2, D–F; Supplemental Fig. S3). Mutation of OsHMA2 resulted in a significant decrease in the zinc concentration in the upper nodes (Fig. 5C). As a result, zinc concentration of the mutants was decreased in the upper organs connecting with node I, including flag leaf, rachis, husk, and brown rice (Fig. 5A). These results indicate that OsHMA2 in the nodes plays an important role in preferentially delivering zinc to the panicles. This is supported by a short-term labeling experiment with stable isotope 67Zn; mutation of OsHMA2 resulted in a decreased zinc distribution to the panicles (Fig. 6A).
To play a role in preferential distribution of zinc as discussed above, OsHMA2 probably functions as an influx transporter; otherwise, decreased concentration of zinc in the root tips, nodes, and reproductive organs of the mutants could not be explained (Figs. 3 and 5). Similar zinc concentration in the xylem sap could also not be explained (Fig. 3D). Our yeast assay experiment supported that, different from other HMA members, OsHMA2 is an influx zinc transporter, at least in yeast (Fig. 7; Supplemental Fig. S10). This unique feature of OsHMA2 may be associated with its long C-terminal region, which is lacking in other HMA members (Supplemental Fig. S1). In fact, deletion of the C terminus changed the transport activity in yeast (Fig. 7).
Our yeast result is different from two previous studies (Nocito et al., 2011; Satoh-Nagasawa et al., 2012), but consistent with a recent study with HvHMA2, the closest homolog in barley (Hordeum vulgare; Mills et al., 2012). Increased cadmium tolerance by expression of OsHMA2 was very small in the study by Satoh-Nagasawa et al. (2012). Furthermore, expression of truncated cDNAs from mutants showed the same result as the full-length OsHMA2 in yeast, although there was a clear difference in zinc and cadmium accumulation between the wild-type rice and mutants, raising a question as to whether their yeast assay can explain the phenotypic difference. Heterologous expression of plant genes in yeast sometimes yielded different results. For example, expression of AtHMA4 in yeast resulted in hypersensitivity to excess zinc and cadmium in one study (Baekgaard et al., 2010), but resulted in increased tolerance to excess zinc and cadmium in other studies (Papoyan and Kochian, 2004; Mills et al., 2005; Verret et al., 2005). These inconsistencies may be attributed to different yeast strains, experimental conditions, expression vector, medium components, and so on. In this study, we used two different yeast strains (BY4741 and INVSc1) for cadmium sensitivity assay, one of which is the same as that used by Nocito et al. (2011). However, we got the same results in both strains (Fig. 7; Supplemental Fig. S10). Furthermore, we used AtHMA4 as a positive control and found that different from OsHMA2, AtHMA4 enhanced the tolerance to cadmium under the same experimental conditions (Fig. 7; Supplemental Fig. S10), which is consistent with previous results (Papoyan and Kochian, 2004; Mills et al., 2005; Verret et al., 2005). Based on the phenotype observed in the rice mutant lines as discussed above and cell specificity of the localization (Figs. 2, 3, and 5), it is reasonable to consider that wild-type OsHMA2 functions as an influx transporter of cadmium and zinc for phloem loading.
OsHMA2 Is Also Responsible for Cadmium Partition
Cadmium accumulation in rice grain is a serious health problem. Recently, a member of the Nramp family, Nramp5, was identified as a major transporter for cadmium uptake in rice (Sasaki et al., 2012). On the other hand, OsHMA3, a homolog of OsHMA2, is found to be responsible for cadmium sequestration into the vacuoles (Ueno et al., 2010). Different from OsHMA2, OsHMA3 is mainly expressed in the roots and localized to the tonoplast. Overexpression of OsHMA3 resulted in a significant decrease of cadmium in the rice grain (Ueno et al., 2010). However, the molecular mechanism on cadmium distribution to the grain is still poorly understood. In rice shoot, the partition pattern of cadmium is similar to that of zinc, which is characterized by direct translocation, regardless of the transpiration rate, to the youngest leaf, nodes, and developing panicles (Fujimaki et al., 2010). Mutation of OsHMA2 also resulted in decreased root-to-shoot translocation of cadmium at the vegetative growth stage (Supplemental Figs. S5 and S6; Satoh-Nagasawa et al., 2012; Takahashi et al., 2012). At the reproductive stage, the cadmium concentration only in the upper parts, including grain and upper nodes, of the mutants were decreased similar to zinc (Fig. 5). This indicates that, like zinc, partition of cadmium in rice is also mediated by OsHMA2. This is also supported by transport activity for cadmium in yeast; expression of OsHMA2 decreased cadmium tolerance in yeast (Fig. 7; Supplemental Fig. S10).
Recently, a transporter, OsLCT1, was reported to be involved in cadmium distribution in rice (Uraguchi et al., 2011). Rice low-affinity cation transporter1 (OsLCT1) showed efflux transport activity for several cations, including cadmium (but not zinc), and knockdown of OsLCT1 resulted in decreased cadmium (but not zinc) concentration in the grain (Uraguchi et al., 2011). However, in contrast to OsHMA2 expression, which is highly expressed in the nodes throughout the reproductive growth, expression of OsLCT1 in the nodes was only observed at the ripening stage. Higher expression of OsLCT1 was observed in the leaf blade rather than the nodes at each reproductive growth period (Uraguchi et al., 2011). These differences indicate that the cadmium translocation pathway mediated by OsLCT1 is different from that of OsHMA2. OsLCT1 might be involved in retranslocation of cadmium and some other elements from the leaf blades, rather than in the distribution control within the nodes.
In conclusion, OsHMA2 is a transporter for phloem loading of both zinc and cadmium in rice. This transporter plays a key role in preferential distribution of zinc at the roots and nodes (Fig. 8) to developing tissues, which require high zinc. At the nodes, there are two ways to load zinc into the phloem. One is that zinc in the xylem of enlarged vascular bundles coming from the roots or lower nodes is directly loaded to the phloem by OsHMA2 located at the phloem of the same enlarged vascular bundle (Fig. 8B). The remaining zinc in the xylem of enlarged vascular bundles will be taken up into the xylem transfer cells around the enlarged vascular bundles (Kawahara et al., 1974; Hoshikawa, 1989) by an unidentified transporter and then released to the apoplast/xylem of diffuse vascular bundles by another unidentified transporter at the intervening parenchyma cells, followed by loading zinc into the phloem of diffuse vascular bundles by OsHMA2 (Fig. 8B). Our results showed that OsHMA2 is involved in the later step of intervascular transfer of zinc, reloading of zinc from the intervening parenchyma tissues into the phloem of diffuse vascular bundles (Fig. 8B).
MATERIALS AND METHODS
Plant Materials and Growth Condition
Wild-type rice (Oryza sativa ‘Nipponbare’) and two independent Tos17 insertion mutant lines (NF4888 and NF9856) from the Rice Mutant Panel (http://tos.nias.affrc.go.jp/) were used (Supplemental Fig. S1, B and C).
For hydroponic experiments, seeds of wild-type rice and mutant lines were soaked in water overnight at 25°C in the dark and then transferred to a net floating on 0.5 mm CaCl2 solution. On day 7, seedlings were transferred to a 3.5-L plastic pot containing one-half-strength Kimura B solution and grown in a greenhouse at 25°C to 30°C. The nutrient solution contained the macronutrients (NH4)2SO4 (0.18 mm), MgSO4⋅7H2O (0.27 mm), KNO3 (0.09 mm), Ca(NO3)2⋅4H2O (0.18 mm), and KH2PO4 (0.09 mm) and the micronutrients MnCl2⋅4H2O (0.5 μm), H3BO3 (3 μm), (NH4)6Mo7O24⋅4H2O (1 μm), ZnSO4⋅7H2O (0.4 μm), CuSO4⋅5H2O (0.2 μm), and Fe-EDTA (20 μm). The pH of this solution was adjusted to 5.6, and the nutrient solution was renewed every 2 d. Field experiments were carried out at the experimental paddy field of Okayama University. Three-week seedlings of wild-type rice and two mutants precultured hydroponically were transplanted to the field in mid-June and harvested at the end of September. All experiments were repeated with three biological replicates.
Cloning of Full-Length OsHMA2 ORF and Phylogenetic Analysis
To clone the full-length ORF sequence of OsHMA2, total RNA was extracted from rice roots using an RNeasy Plant Mini Kit (Qiagen) and then converted to cDNA using the protocol supplied by the manufacturers of SuperScript II (Invitrogen). The full-length ORF was amplified by PCR using primers 5′-AGATGGCGGCGGAGGGAG-3′ and 5′-CTACTACTCCACTACGATCTCAG-3′, which were designed based on two putative truncated cDNA clones in the Rice Annotation Project Database (http://rapdb.dna.affrc.go.jp/), Os06g0700650 and Os06g0700700. They have a putative translational start and stop site, respectively. The sequence of the amplified cDNA was confirmed by a sequence analyzer (ABI Prism 3100; Applied Biosystems).
For phylogenetic analysis, peptide sequence alignment of rice and Arabidopsis (Arabidopsis thaliana) HMAs (Supplemental Fig. S1) was analyzed by ClustalW using default settings (http://clustalw.ddbj.nig.ac.jp/). The phylogenetic tree was constructed using the neighbor-joining algorithm by MEGA4 software (http://megasoftware.net/) after ClustalW alignment with 1,000 bootstrap trials.
Expression Patterns
For the tissue-specific expression and metal deficiency-response analysis of OsHMA2 at vegetative stage, 2-week-old seedlings were exposed to one-half-strength Kimura B solution with or without zinc, iron, manganese, or copper (Zheng et al., 2012). After 1 week, roots, shoots, and basal region (2 cm above the roots) were sampled and subjected to RNA extraction. The metal deficiency status has been confirmed by marker genes as described in Zheng et al. (2012). For excess zinc- and cadmium-response analysis, 3-week-old seedlings were exposed to a one-half-strength Kimura B solution with 10 µm zinc or 1 µm cadmium for 1 d, and the roots were sampled for RNA extraction. To examine the spatial expression in the roots, the different root segments (0–1 cm and 1–2 cm from the apex) of 7-d-old seedlings were sampled. To investigate the expression pattern of OsHMA2 at reproductive stage, plants were grown in the field till flowering, and then the roots, basal stem, lower leaf sheath, lower leaf blade, flag leaf sheath, flag leaf blade, nodes I to III, internode II, peduncle, rachis, and spikelet were sampled.
Total RNA was extracted by using an RNeasy Plant Mini Kit and then converted to cDNA followed by DNase I treatment using the protocol supplied by the manufacturers of SuperScript II. Specific cDNAs were amplified by SsoFast EvaGreen Supermix (Bio-Rad), and quantitative real-time PCR was performed with the following primer sets for OsHMA2: 5′-CATAGTGAAGCTGCCTGAGATC-3′ and 5′-GATCAAACGCATAGCAGCATCG-3′ on CFX384 (Bio-Rad). HistoneH3 and Actin were used as internal standards, with primer pairs 5′-AGTTTGGTCGCTCTCGATTTCG-3′ and 5′-TCAACAAGTTGACCACGTCACG-3′ for HistoneH3 and 5′-GACTCTGGTGATGGTGTCAGC-3′ and 5′-GGCTGGAAGAGGACCTCAGG-3′ for Actin. The expression data were normalized by these two genes, and relative expression was calculated by the comparative cycle threshold method using CFX Manager software (Bio-Rad).
Transgenic Rice Carrying OsHMA2 Promoter-GFP
To investigate the cellular expression of OsHMA2, we introduced a construct consisting of the promoter (2.13 kb) of OsHMA2 fused with GFP to rice using an Agrobacterium tumefaciens-mediated transformation system (Hiei et al., 1994). The 2.13-kb region upstream of the initiation codon of OsHMA2 was amplified by PCR from cv Nipponbare genomic DNA using primers AAGCTTCAACTCTTTTCTTCCGTTTGTGT and GGATCCCTCTCCTCACTCTCTCCCTCTT. The amplified fragment was cut with HindIII and BamHI and then cloned into pPZP2H-lac (Fuse et al., 2001) carrying GFP and the terminator of the nopaline synthase gene, generating the OsHMA2 promoter-GFP construct. The localization of GFP was observed as described below.
Immunohistological Staining of OsHMA2 and GFP
The synthetic peptide CSHSIVKLPEIVVE (positions 1,054–1,067 of OsHMA2) was used to immunize rabbits to obtain antibodies against OsHMA2. Keyhole limpet hemocyanin was used as carrier protein for the immunization. The obtained antiserum was purified through a peptide affinity column before use. To detect GFP protein in transgenic rice tissues, antibody against GFP (A11122; Molecular Probes) was used. Roots (10-d-old seedlings), basal stem (4 weeks old), node III, and node I (flowering stage) of wild-type rice ‘Nipponbare’, NF4888, NF9856, and OsHMA2 promoter-GFP transgenic rice were used for immunostaining of OsHMA2 protein or GFP as described previously (Yamaji and Ma, 2007). Fluorescence of secondary antibody (Alexa Fluor 555 goat anti-rabbit IgG; Molecular Probes) was observed with confocal laser scanning microscopy (LSM700; Carl Zeiss).
Subcellular Localization
Western-blot analysis with Suc density gradient fractionation of microsomes prepared from rice roots was performed according to a previous study (Ueno et al., 2010). Microsomal fraction was layered on Suc density gradients (20%–60%) with 10 mm Tris-HCl (pH 7.6), 1 mm EDTA, and 1 mm dithiothreitol centrifuged at 100,000g for 2 h. The same quantity of protein samples in each fraction was subjected to SDS-PAGE and immunoblotting. The blots were treated with 1:100 dilutions of OsHMA2 antibody, 1:1,000 H+-ATPase (plasma membrane marker; Agrisera), 1:5,000 of V-type ATPase (tonoplast marker; Agrisera), 1:2,000 of ER luminal binding protein (BiP, ER marker; Cosmo Bio), and 1:1,000 of ADP-ribosylation factor (Golgi marker; Agrisera), respectively. Anti-Rabbit IgG (H+L) HRP Conjugate (1:10,000 dilution; Promega) was used as a secondary antibody, and the ECL Plus Western Blotting Detection System (GE Healthcare) was used for detection via chemiluminescence.
Phenotypic Analysis of Mutant Lines
Shoots, roots, and crown root tip (0–5 mm from the apex) were harvested from 5-week-old seedlings grown hydroponically and subjected to mineral analysis. For xylem sap collection, shoots were excised 2 cm above the roots with a razor, and xylem sap was collected from the cut surface for 30 min using micropipettes. To avoid contamination of symplastic zinc from the damaged cells, initial exudates (1–2 µL) were discarded. For analysis of mineral accumulation, 3-week-old seedlings were exposed to the nutrient solution containing 100 nm cadmium for 1 week, and then the roots, shoots, and shoot basal meristem (lowest 2-cm part of leaf sheath of youngest expanded leaf and inner organs) were harvested separately.
For rice grown in the field, different tissues, including brown rice, husk, rachis, flag leaf blade, flag leaf sheath, nodes I to III, and the remaining part of the shoot (straw) were separately harvested at ripening. The dry weight of the straw and grain was recorded after drying at 40°C for 2 weeks. The fertility was investigated by soaking the seeds in 8.5% (w/v) NaCl solution.
Short-Term Labeling Experiment with 67Zn, Rubidium, and Strontium
Wild-type rice and two mutant lines were grown hydroponically till just before heading as described above. The roots were exposed to a one-half-strength Kimura B solution containing 1 µm rubidium and 1 µm strontium and 0.4 µm zinc labeled with 67Zn (Taiyo Nippon Sanso). The feeding solution was renewed daily. After 2 d, the above-ground part of each plant was separated into panicle, nodes I to IV, unelongated basal nodes, total internodes, and leaves I (flag leaf) to VI. The concentration of 67Zn isotope, rubidium, and strontium in each organ were determined by inductively coupled plasma-mass spectrometry (ICP-MS) after digestion as described below. Distribution ratio within the above-ground part was calculated from net increase of 67Zn (i.e. the total 67Zn amount minus the natural abundance of 67Zn), rubidium, and strontium. For each treatment, four biological replicates were made, and the experiment was repeated twice.
Mineral Determination
Plant samples were dried at 70°C and then digested with concentrated nitric acid (60% [w/v]) at 140°C. The concentrations of zinc, cadmium, potassium, magnesium, calcium, manganese, iron, copper, rubidium, and strontium in plant digests and xylem sap were determined by ICP-MS (Agilent 7700). For 67Zn determination, an isotope mode was applied.
Transport Activity Assay in Yeast
Total RNA was extracted from the roots of rice and Arabidopsis (ecotype Columbia) with an RNeasy Plant Mini Kit. The total RNA was converted to cDNA using the protocol attached to SuperScript II. The cDNA fragment containing an entire ORF for AtHMA4 and OsHMA2 and C- and N-terminal truncated versions of OsHMA2 were amplified by RT-PCR using the primers 5′-GGATCCGAAAATGGCGTTACAAAACAAAG-3′ and 5′- GTCAAGCACTCACATGGTGATGGTG-3′ for AtHMA4, 5′-AGAGCTCAAGATGGCGGCGGAGGGAGG-3′ and 5′-TTCTAGACTACTACTCCACTACGATCTCAG-3′ for OsHMA2, 5′-ATGTGTGCTGCTTCTCATCACGGATCT-3′ and 5′-TTCTAGACTACTACTCCACTACGATCTCAG-3′ for OsHMA2ΔC(707–1,067), and 5′-AAAATGGCGGCGGAGGGAGGGAGGT-3′ and 5′-CTATTTCTTCGCCTTTCGACTGTCCT-3′ for OsHMA2ΔN(2–706). The fragment was directly cloned into the pYES2.1/V5-His-TOPO vector (Invitrogen). After sequence confirmation, the resulting plasmid or the empty vector was introduced into yeast (Saccharomyces cerevisiae) according to the manufacturer’s protocols (S.c. EasyComp Transformation Kit; Invitrogen). The yeast strains used in this study were BY4741 (MATα his2Δ0 met15Δ0 ura3Δ0), INVSc1 (Invitrogen), the zinc uptake-deficient double-mutant zrt1zrt2 (MATα ade6 can1 his3 leu2 trp1 ura3 zrt1∷LEU2 zrt2∷HIS3), and the zrc1cot1 mutant, which is hypersensitive to zinc (MATa;his3Δ1;leu2Δ0;met15Δ0;ura3Δ0;zrc1::natMX;cot1::kanMX4), which was obtained from Ute Kraemer.
Growth of yeast strain BY4741, zrt1zrt2, and zrt1cot1 on different zinc conditions were tested on a synthetic medium containing 2% (w/v) Glc or Gal, 0.67% (w/v) yeast nitrogen base without metals (BIO 101 Systems), 0.2% (w/v) appropriate amino acids, and 2% (w/v) agar buffered at pH 6 with 50 mm MES and supplemented with 0 or 10 mm ZnSO4 for BY4741, 0, 0.2, or 4 mm ZnSO4 for zrt1zrt2, and 0, 200, or 400 µm ZnSO4 for zrc1cot1. After spotting at four yeast cell dilutions (optical densities at 600 nm of 0.2, 0.02, 0.002, and 0.0002), plates were incubated for 3 d at 30°C.
For measurement of Zn2+ uptake, yeast transformants were grown in a synthetic medium containing 2% (w/v) Gal, 0.67% (w/v) yeast nitrogen base without metals, and 0.2% (w/v) appropriate amino acids buffered at pH 6 with 50 mm MES. Cells at midexponential phase were harvested and transferred to liquid synthetic complete-uracil medium containing 2% (w/v) Gal at pH 4.6 for induction of the Gal promoter. After cells were cultured for 2 h, the precultured yeast was adjusted to an optical density at 600 nm value of 3.0 by reducing the amount of liquid. Ten micromolar ZnSO4 was then added to the medium. After 6 h of incubation with gentle shaking, cells were harvested by centrifugation, washed three times with deionized water, and then digested with 2 n HCl. The concentration of zinc in the digest solution was determined by ICP-MS. Three biological replicates for each treatment were made.
For evaluation of cadmium tolerance in yeast (BY4741 and INVSc1), transformants were cultured and spotted on synthetic complete-uracil plates containing Glc or Gal and supplemented with 0, 15, 20, or 30 µm CdSO4. The plate was incubated for 3 d at 30°C.
Statistical Analysis
Data were analyzed using one-way ANOVA followed by Student’s t test or Tukey’s test. Significance was defined as *P < 0.05 or **P < 0.01.
Sequence data from this article can be found in the GenBank/EMBL databases under accession number AB697186 for OsHMA2.
Supplemental Data
The following material is available in the online version of this article.
Supplemental Figure S1. Sequence, structure, and knockout mutants of OsHMA2.
Supplemental Figure S2. Phylogenetic relationship of HMA proteins in rice and Arabidopsis.
Supplemental Figure S3. Immunohistochemical staining of OsHMA2 and GFP.
Supplemental Figure S4. Subcellular localization of OsHMA2 by western-blot analysis.
Supplemental Figure S5. Mineral concentration of the shoots at the vegetative stage.
Supplemental Figure S6. Mineral concentration of the root at the vegetative stage.
Supplemental Figure S7. Growth of wild-type rice and mutants at the vegetative stage.
Supplemental Figure S8. Mineral concentration of the shoot basal meristem at the vegetative stage.
Supplemental Figure S9. Mineral concentration in the nodes.
Supplemental Figure S10. Yeast transport assay for cadmium and zinc.
Acknowledgments
We thank the Rice Genome Resource Center for providing Tos17 insertion lines.
Glossary
- cDNA
complementary DNA
- RT
reverse transcription
- ORF
open reading frame
- DAPI
4',6-diamino-2-phenylindole
- ER
endoplasmic reticulum
- ICP-MS
inductively coupled plasma-mass spectrometry
References
- Andreini C, Bertini I, Rosato A. (2009) Metalloproteomes: a bioinformatic approach. Acc Chem Res 42: 1471–1479 [DOI] [PubMed] [Google Scholar]
- Baekgaard L, Mikkelsen MD, Sørensen DM, Hegelund JN, Persson DP, Mills RF, Yang Z, Husted S, Andersen JP, Buch-Pedersen MJ, et al. (2010) A combined zinc/cadmium sensor and zinc/cadmium export regulator in a heavy metal pump. J Biol Chem 285: 31243–31252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broadley M, Brown P, Cakmak I, Rengel Z, Zhao F. (2011) Function of nutrients: micronutrients. In Marschner P, ed, Marschner’s Mineral Nutrition of Higher Plants. Academic Press, San Diego, pp 212–223 [Google Scholar]
- Ender C, Li MQ, Martin B, Povh B, Nobiling R, Reiss HD, Traxel K. (1983) Demonstration of polar zinc distribution in pollen tubes of Lilium longiflorum with the Heidelberg proton microprobe. Protoplasma 116: 201–203 [Google Scholar]
- Fujimaki S, Suzui N, Ishioka NS, Kawachi N, Ito S, Chino M, Nakamura S. (2010) Tracing cadmium from culture to spikelet: noninvasive imaging and quantitative characterization of absorption, transport, and accumulation of cadmium in an intact rice plant. Plant Physiol 152: 1796–1806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuse T, Sasaki T, Yano M. (2001) Ti-plasmid vectors useful for functional analysis of rice genes. Plant Biotechnol 18: 219–222 [Google Scholar]
- Grotz N, Fox T, Connolly E, Park W, Guerinot ML, Eide D. (1998) Identification of a family of zinc transporter genes from Arabidopsis that respond to zinc deficiency. Proc Natl Acad Sci USA 95: 7220–7224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerinot ML. (2000) The ZIP family of metal transporters. Biochim Biophys Acta 1465: 190–198 [DOI] [PubMed] [Google Scholar]
- Hiei Y, Ohta S, Komari T, Kumashiro T. (1994) Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J 6: 271–282 [DOI] [PubMed] [Google Scholar]
- Hoshikawa K. (1989) The Growing Rice Plant. Nobunkyo, Tokyo [Google Scholar]
- Hussain D, Haydon MJ, Wang Y, Wong E, Sherson SM, Young J, Camakaris J, Harper JF, Cobbett CS. (2004) P-type ATPase heavy metal transporters with roles in essential zinc homeostasis in Arabidopsis. Plant Cell 16: 1327–1339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawahara H, Chonan N, Matsuda T. (1974) Studies on morphogenesis in rice plants 7. The morphology of vascular bundles in the vegetative nodes of the culm. Jpn J Crop Sci 43: 389–401 [Google Scholar]
- Kitagishi K, Obata H. (1986) Effects of zinc deficiency on the nitrogen metabolism of meristematic tissues of rice plants with reference to protein synthesis. Soil Sci Plant Nutr 3: 397–405 [Google Scholar]
- Kuppelwieser H, Feller U. (1991) Transport of Rb and Sr to the ear in mature, excised shoots of wheat: effects of temperature and stem length on Rb removal from the xylem. Plant Soil 132: 281–288 [Google Scholar]
- Langston R. (1956) Distribution patterns of radioisotopes in plants. Proc Am Soc Hortic Sci 68: 370–376 [Google Scholar]
- Ma F, Peterson CA. (2001) Frequencies of plasmodesmata in Allium cepa L. roots: implications for solute transport pathways. J Exp Bot 52: 1051–1061 [DOI] [PubMed] [Google Scholar]
- Maret W, Li Y. (2009) Coordination dynamics of zinc in proteins. Chem Rev 109: 4682–4707 [DOI] [PubMed] [Google Scholar]
- Mills RF, Francini A, Ferreira da Rocha PSC, Baccarini PJ, Aylett M, Krijger GC, Williams LE. (2005) The plant P1B-type ATPase AtHMA4 transports Zn and Cd and plays a role in detoxification of transition metals supplied at elevated levels. FEBS Lett 579: 783–791 [DOI] [PubMed] [Google Scholar]
- Mills RF, Peaston KA, Runions J, Williams LE. (2012) HvHMA2, a P(1B)-ATPase from barley, is highly conserved among cereals and functions in Zn and Cd transport. PLoS ONE 7: e42640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nocito FF, Lancilli C, Dendena B, Lucchini G, Sacchi GA. (2011) Cadmium retention in rice roots is influenced by cadmium availability, chelation and translocation. Plant Cell Environ 34: 994–1008 [DOI] [PubMed] [Google Scholar]
- Obata H, Kitagishi K. (1980a) Longitudinal distribution pattern of Zn and Mn in leaf with special reference to aging: behavior of zinc in rice plants (I). Jpn J Soil Sci Plant Nutri 51: 285–291 [Google Scholar]
- Obata H, Kitagishi K. (1980b) Investigation on pathway of Zn in vegetative node of rice plants by autoradiography: behavior of zinc in rice plants (III). Jpn J Soil Sci Plant Nutri 51: 297–301 [Google Scholar]
- Obata H, Oosawa J, Kitagishi K. (1980) Time course of Zn or Mn accumulation within individual leaves: behavior of zinc in rice plants (II). Jpn J Soil Sci Plant Nutri 51: 292–296 [Google Scholar]
- Ozturk L, Yazici MA, Yucel C, Torun A, Cekic C, Bagci A, Ozkan H, Braun HJ, Sayers Z, Cakmak I. (2006) Concentration and localization of zinc during seed development and germination in wheat. Physiol Plant 128: 144–152 [Google Scholar]
- Papoyan A, Kochian LV. (2004) Identification of Thlaspi caerulescens genes that may be involved in heavy metal hyperaccumulation and tolerance. Characterization of a novel heavy metal transporting ATPase. Plant Physiol 136: 3814–3823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riceman DS, Jones GB. (1958a) Distribution of zinc in subterranean clover (Trifolium subterraneum L.) grown to maturity in a culture solution containing zinc labeled with the radioactive isotope 65Zn. Aust J Agric Res 9: 446–463 [Google Scholar]
- Riceman DS, Jones GB. (1958b) Distribution of dry weight and of zinc and copper among the individual leaves of seedlings of subterranean clover (Trifolium subterraneum L.) grown in complete culture solution and in a culture solution deficient in zinc. Ibid 9: 730–744 [Google Scholar]
- Sasaki A, Yamaji N, Yokosho K, Ma JF. (2012) Nramp5 is a major transporter responsible for manganese and cadmium uptake in rice. Plant Cell 24: 2155–2167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoh-Nagasawa N, Mori M, Nakazawa N, Kawamoto T, Nagato Y, Sakurai K, Takahashi H, Watanabe A, Akagi H. (2012) Mutations in rice (Oryza sativa) heavy metal ATPase 2 (OsHMA2) restrict the translocation of zinc and cadmium. Plant Cell Physiol 53: 213–224 [DOI] [PubMed] [Google Scholar]
- Takahashi R, Ishimaru Y, Shimo H, Ogo Y, Senoura T, Nishizawa NK, Nakanishi H. (2012) The OsHMA2 transporter is involved in root-to-shoot translocation of Zn and Cd in rice. Plant Cell Environ 35: 1948–1957 [DOI] [PubMed] [Google Scholar]
- Ueno D, Yamaji N, Kono I, Huang CF, Ando T, Yano M, Ma JF. (2010) Gene limiting cadmium accumulation in rice. Proc Natl Acad Sci USA 107: 16500–16505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uraguchi S, Kamiya T, Sakamoto T, Kasai K, Sato Y, Nagamura Y, Yoshida A, Kyozuka J, Ishikawa S, Fujiwara T. (2011) Low-affinity cation transporter (OsLCT1) regulates cadmium transport into rice grains. Proc Natl Acad Sci USA 108: 20959–20964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verret F, Gravot A, Auroy P, Leonhardt N, David P, Nussaume L, Vavasseur A, Richaud P. (2004) Overexpression of AtHMA4 enhances root-to-shoot translocation of zinc and cadmium and plant metal tolerance. FEBS Lett 576: 306–312 [DOI] [PubMed] [Google Scholar]
- Verret F, Gravot A, Auroy P, Preveral S, Forestier C, Vavasseur A, Richaud P. (2005) Heavy metal transport by AtHMA4 involves the N-terminal degenerated metal binding domain and the C-terminal His11 stretch. FEBS Lett 579: 1515–1522 [DOI] [PubMed] [Google Scholar]
- Williams LE, Mills RF. (2005) P(1B)-ATPases—an ancient family of transition metal pumps with diverse functions in plants. Trends Plant Sci 10: 491–502 [DOI] [PubMed] [Google Scholar]
- Wong CKE, Cobbett CS. (2009) HMA P-type ATPases are the major mechanism for root-to-shoot Cd translocation in Arabidopsis thaliana. New Phytol 181: 71–78 [DOI] [PubMed] [Google Scholar]
- Wong CKE, Jarvis RS, Sherson SM, Cobbett CS. (2009) Functional analysis of the heavy metal binding domains of the Zn/Cd-transporting ATPase, HMA2, in Arabidopsis thaliana. New Phytol 181: 79–88 [DOI] [PubMed] [Google Scholar]
- Yamaguchi N, Ishikawa S, Abe T, Baba K, Arao T, Terada Y. (2012) Role of the node in controlling traffic of cadmium, zinc, and manganese in rice. J Exp Bot 63: 2729–2737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaji N, Ma JF. (2007) Spatial distribution and temporal variation of the rice silicon transporter Lsi1. Plant Physiol 143: 1306–1313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng L, Yamaji N, Yokosho K, Ma JF. (2012) YSL16 is a phloem-localized transporter of the copper-nicotianamine complex that is responsible for copper distribution in rice. Plant Cell 24: 3767–3782 [DOI] [PMC free article] [PubMed] [Google Scholar]