The growth phenotype of the rus1/wxr3 mutant is related to a defect in auxin transport caused by decreased accumulation of auxin transporters.
Abstract
The phytohormone auxin regulates virtually every aspect of plant development. To identify new genes involved in auxin activity, a genetic screen was performed for Arabidopsis (Arabidopsis thaliana) mutants with altered expression of the auxin-responsive reporter DR5rev:GFP. One of the mutants recovered in the screen, designated as weak auxin response3 (wxr3), exhibits much lower DR5rev:GFP expression when treated with the synthetic auxin 2,4-dichlorophenoxyacetic acid and displays severe defects in root development. The wxr3 mutant decreases polar auxin transport and results in a disruption of the asymmetric auxin distribution. The levels of the auxin transporters AUXIN1 and PIN-FORMED are dramatically reduced in the wxr3 root tip. Molecular analyses demonstrate that WXR3 is ROOT ULTRAVIOLET B-SENSITIVE1 (RUS1), a member of the conserved Domain of Unknown Function647 protein family found in diverse eukaryotic organisms. Our data suggest that RUS1/WXR3 plays an essential role in the regulation of polar auxin transport by maintaining the proper level of auxin transporters on the plasma membrane.
The plant hormone indole-3-acetic acid (IAA) is the most important natural auxin. It regulates virtually every aspect of plant development, including embryogenesis, root initiation, lateral root development, tropic responses, leaf formation, stem elongation, and fruit development (Möller and Weijers, 2009; Sundberg and Østergaard, 2009; Takahashi et al., 2009; Overvoorde et al., 2010; Scarpella et al., 2010; Vernoux et al., 2010). Auxin is synthesized in young aerial tissues and actively transported to other parts of the plant in a polar fashion to form and maintain auxin gradients (Grieneisen et al., 2007; Grunewald and Friml, 2010; Zhao, 2010). Polar auxin transport is mediated by plasma membrane-localized transporters, including the PIN-FORMED (PIN) and P-glycoprotein (PGP) auxin transporters, and the AUXIN RESISTANT1/LIKE AUXIN RESISTANT1 (AUX1/LAX) auxin permeases (Okada et al., 1991; Bennett et al., 1996; Müller et al., 1998; Marchant et al., 1999; Friml et al., 2002a, 2002b, 2003; Bouchard et al., 2006; Blakeslee et al., 2007; Cho et al., 2007).
The AUX1 and PIN proteins display tissue-specific expression patterns and regulated subcellular localization on the plasma membrane, which in the case of the PIN proteins determines the direction of auxin flow (Teale et al., 2006; Wisniewska et al., 2006; Grunewald and Friml, 2010). For example, in the root, PIN1 localizes at the basal (root apex-facing) side of the root vasculature; meanwhile PIN2 is at the basal side of the root cortical cells and the apical (shoot apex-facing) side of the epidermal and root cap cells (Gälweiler et al., 1998; Müller et al., 1998). AUX1 is expressed in the stele, columella, epidermis, and lateral root cap and localizes on the apical side of root protophloem cells (Bennett et al., 1996). The localization of the PINs is dynamic and changes rapidly through vesicle endocytic recycling (Grunewald and Friml, 2010). The fungal toxin brefeldin A (BFA) is a recycling inhibitor that is widely used to study this process. After treatment of Arabidopsis (Arabidopsis thaliana) roots with BFA, the plasma membrane-localized PINs are rapidly internalized and accumulate in so-called BFA compartments in the cytosol in a reversible manner (Geldner et al., 2001).
Several factors that are important for the polar localization of PINs have been identified. PIN polarity requires the appropriate sterol composition in the plasma membrane and cellulose-based extracellular matrix in the cell wall (Willemsen et al., 2003; Men et al., 2008; Feraru et al., 2011). Recycling of PIN proteins is a dynamic process that involves clathrin-dependent endocytosis, guanine-nucleotide exchange factor for ADP-ribosylation factor GTPase (ARF-GEF)-dependent movement to the plasma membrane, and retromer-dependent vacuolar targeting for degradation (Feraru and Friml, 2008). Mutants defective in any of these processes exhibit altered localization or expression of PIN proteins in the plant (Steinmann et al., 1999; Jaillais et al., 2006; Dhonukshe et al., 2007; Jaillais and Gaude, 2007; Kleine-Vehn et al., 2008; Naramoto et al., 2010; Dhonukshe, 2011). In addition, PIN polarity is required for its phosphorylation status. PINOID, a Ser/Thr kinase, phosphorylates PIN proteins and is crucial for apical PIN delivery; while protein phosphatase 2A (PP2A) functions antagonistically to PINOID (Friml et al., 2004; Michniewicz et al., 2007; Zhang et al., 2010).
Once auxin is transported into a specific cell, it promotes the recruitment of the Aux/IAA transcriptional repressors to the SCFTIR1/AFB ubiquitin E3 complex. The Aux/IAAs are ubiquitinylated and degraded by the 26S proteasome, thus allowing the activation of ARF-dependent transcription (Tiwari et al., 2001; Dharmasiri et al., 2005a, 2005b; Kepinski and Leyser, 2005; Weijers et al., 2005). Previous studies showed that the auxin receptors TRANSPORT INHIBITOR RESPONSE/AUXIN SIGNALING F-BOX protein (TIR1/AFB) are required for the establishment of the root meristem and postembryonic root growth in Arabidopsis (Dharmasiri et al., 2005b). In order to identify new genes that function in auxin signaling in the root, we used a well-characterized auxin reporter, DR5rev:GFP. Several mutants with shorter primary roots and decreased DR5rev:GFP expression upon auxin treatment were isolated. Here, we report the characterization of weak auxin response3 (wxr3), an allele of the ROOT ULTRAVIOLET B-SENSITIVE1 (RUS1) gene, which encodes a Domain of Unknown Function647 (DUF647) protein (Tong et al., 2008). We present data showing that the wxr3 mutant exhibits dramatically reduced levels of auxin transporters, which leads to a reduction in polar auxin transport and defects in the auxin response.
RESULTS
The wxr3 Mutant Displays Severe Defects in Root Development and 2,4-D Response
To identify new genes affecting auxin response, approximately 5,000 transgenic seeds (DR5rev:GFP background) were mutagenized with ethyl methanesulfonate, and the M2 population was screened for mutants with altered expression of GFP in the root (Ge et al., 2010). A number of mutants with a shorter primary root and reduced GFP signal upon auxin treatment were isolated. One of these mutants, called wxr3, was further characterized. Segregation analysis revealed that wxr3 behaves as a recessive mutation.
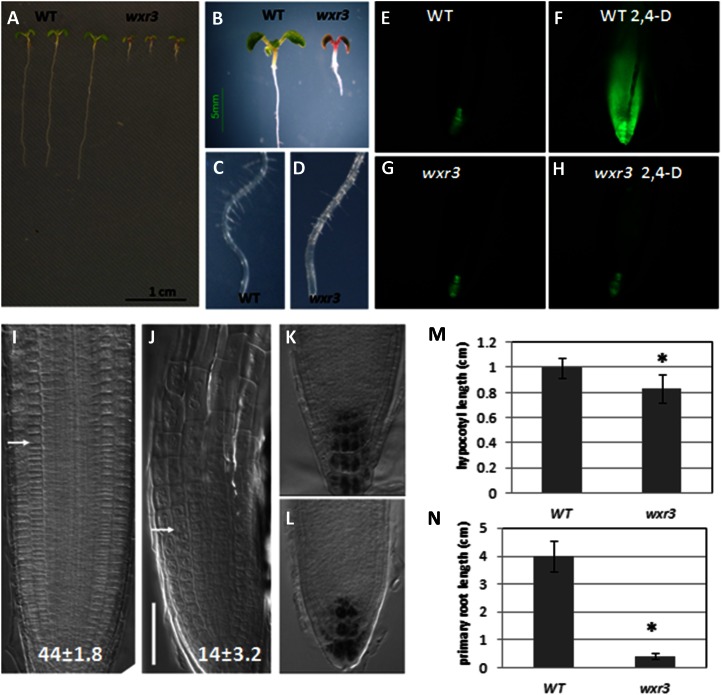
The wxr3 mutant displays much lower levels of DR5rev:GFP expression after treatment with the synthetic auxin 2,4-dichlorophenoxyacetic acid (2,4-D). Six-day-old wxr3 seedlings, treated with 80 nm 2,4-D overnight, do not exhibit an increase in DR5rev:GFP signal in the root apex (Fig. 1, G and H). In contrast, GFP signal increases dramatically in the control line (Fig. 1, E and F). Meanwhile, the wxr3 mutant has severe defects in root development. Ten-day-old wxr3 seedlings display extremely shorter primary roots, shorter hypocotyl length, smaller cotyledons, and anthocyanin accumulation in the shoot meristem (Fig. 1, A, B, M, and N). The primary root length of wild-type seedlings is about 4.0 ± 0.5 cm (mean ± se; n = 14) after 7 d of growth, while the wxr3 primary root length is only 0.4 ± 0.1 cm (n = 16). The root hairs of the wxr3 mutant initiate normally but are deficient in elongation (Fig. 1, C and D). Lugol staining shows that the wxr3 mutant has fewer and disorganized columella cells (Fig. 1, K and L). In addition, we found that the overall organization of the wxr3 root is altered. Mutant roots display an irregular cell pattern with a much shorter elongation zone consisting of fewer but larger cells (Fig. 1, I and J). The wild-type roots have 44 ± 1.8 (n = 10) meristem cortex cells, whereas the wxr3 mutant has only 14 ± 3.2 (n = 10) cells.
Figure 1.
The wxr3 mutant displays severe defects in root development and 2,4-D response. A and B, Phenotypes of 10-d-old wxr3 seedlings compared with the wild type (WT). C and D, Root hair development in 10-d-old wild-type (C) and wxr3 (D) seedlings. E to H, The wxr3 mutant has dramatically reduced DR5rev:GFP level after 2,4-D treatment. Without 2,4-D, the wxr3 mutant (G) has a similar level of GFP signal to the control line (E); after 80 nm 2,4-D treatment for 12 h, the GFP level is dramatically increased in the control line (F) but not in the wxr3 mutant (H). I and J, The structure of the root apex of wild-type (I) and wxr3 (J) seedlings. The arrows denote the boundary between the meristem and the elongation zone. Numbers indicate cortical cell numbers in the meristem zone. Bar = 50 μm. K and L, Lugol staining of columella cells of wild-type (K) and wxr3 (L) roots. M, The wxr3 mutant displays a severe defect in primary root elongation compared with the wild type. N, The wxr3 mutant has a shorter hypocotyl length compared with the wild type. An ANOVA followed by Fisher’s lsd mean separation test (SPSS) were performed on the data. The asterisks indicate statistically significant differences between the wild type and the wxr3 mutant (ANOVA, P < 0.01). Error bars represent se. All seedlings in E to N were 7 d old.
To further characterize root developmental defects, the wxr3 mutant was crossed with transgenic lines expressing a cell division marker (CYCB1;1) and root development markers (SCARECROW [SCR] and SHORTROOT [SHR]; Di Laurenzio et al., 1996; Doerner et al., 1996; Helariutta et al., 2000). The wxr3 mutant displays lower expression of the cyclin-dependent kinase CYCB1;1 in the root tip compared with the control, indicating that fewer cells are actively dividing (Supplemental Fig. S1, A and B). Also, the wxr3 mutant exhibits reduced expression of the root stem cell identity markers SCR and SHR, suggesting that root development is abnormal in the mutant (Supplemental Fig. S1, C–F). Despite the severity of the root defects, the wxr3 mutant does not exhibit strong defects in rosette and inflorescence development (Supplemental Fig. S2).
The wxr3 Mutation Does Not Affect SCFTIR1/AFB-Dependent Auxin Signaling
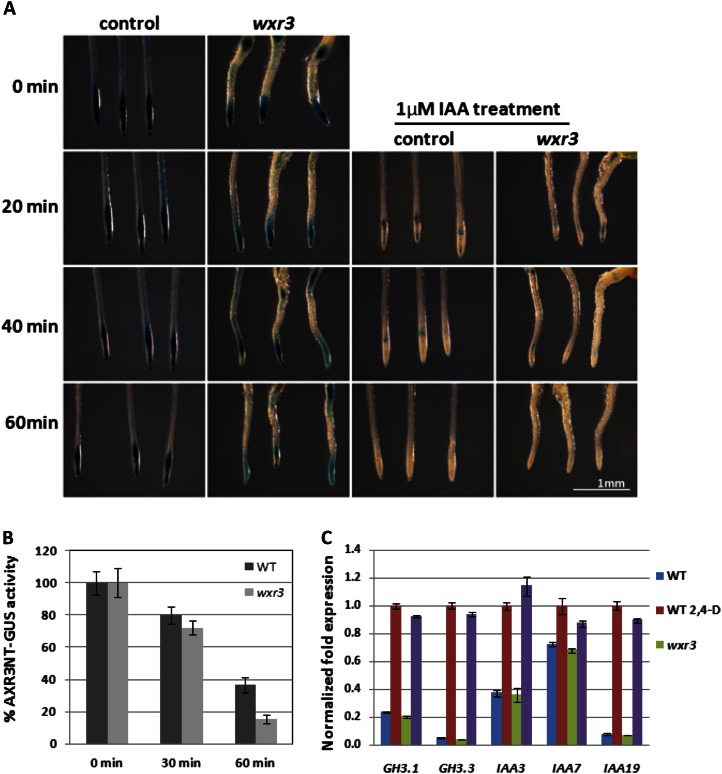
To determine if WXR3 is required for the activity of the SCFTIR1/AFB complex, the wxr3 mutant was crossed to the pHS:AXR3NT-GUS transgenic line. This line expresses domains I and II of AXR3/IAA17 (AXR3NT) upon heat shock and is used as a reporter for the auxin-dependent degradation of the Aux/IAA proteins (Gray et al., 2001). In the F2 generation, homozygous pHS:AXR3NT-GUS wxr3 plants were identified and GUS staining was performed at intervals after a 2-h heat-shock treatment. The wxr3 mutant exhibits less GUS staining after the initial heat shock compared with the control. However, GUS staining is rapidly reduced by IAA treatment in both lines (Fig. 2, A and B). GUS activity was then measured by 4-methylumbelliferyl-β-d-glucuronide (MUG) assay. We found that loss of GUS activity after IAA treatment occurred at a similar rate in the mutant and control lines, suggesting that the degradation of the AXR3NT-GUS protein is not defective in wxr3 (Fig. 2, A and B). Thus, the wxr3 mutation does not appear to affect the function of SCFTIR1/AFB in the plant.
Figure 2.
The wxr3 mutation does not directly affect SCFTIR1/AFB-dependent auxin signaling. A, GUS staining of pHS:AXR3NT-GUS in the wxr3 mutant background at different time points with or without IAA treatment after 2 h at 37°C temperature treatment. B, GUS activity determined by MUG assay of the seedlings in A. C, Levels of Aux/IAA and GH3 transcripts in wild-type (WT) and wxr3 seedlings treated with 20 μm IAA for 1 h. Error bars represent se.
To determine whether the wxr3 mutant is deficient in the induction of auxin-responsive gene expression, the levels of Aux/IAA and GH3 transcripts were determined after auxin treatment using real-time quantitative PCR. The results show that after treatment with 20 µm IAA for 1 h, the induction of Aux/IAA and GH3 expression is similar in the wild type and mutant, indicating that the wxr3 mutation does not directly affect auxin-dependent gene expression (Fig. 2C). Taken together, these results suggest that the wxr3 mutant does not have a defect in SCFTIR1/AFB-dependent auxin signaling.
The wxr3 Mutant Has a Defect in Polar Auxin Transport
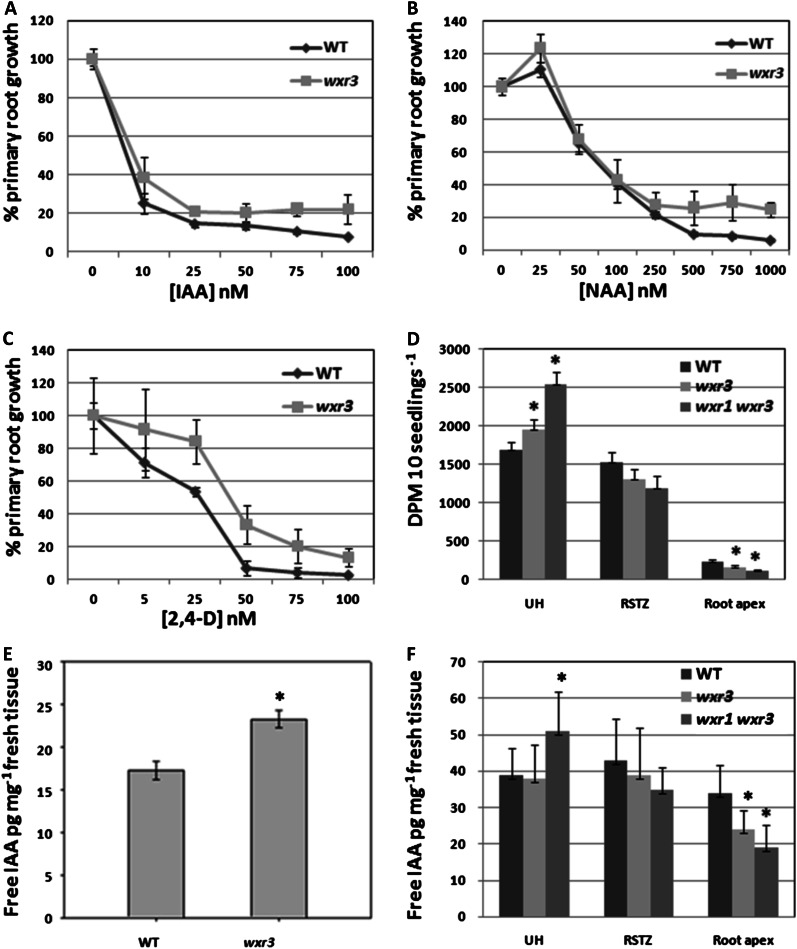
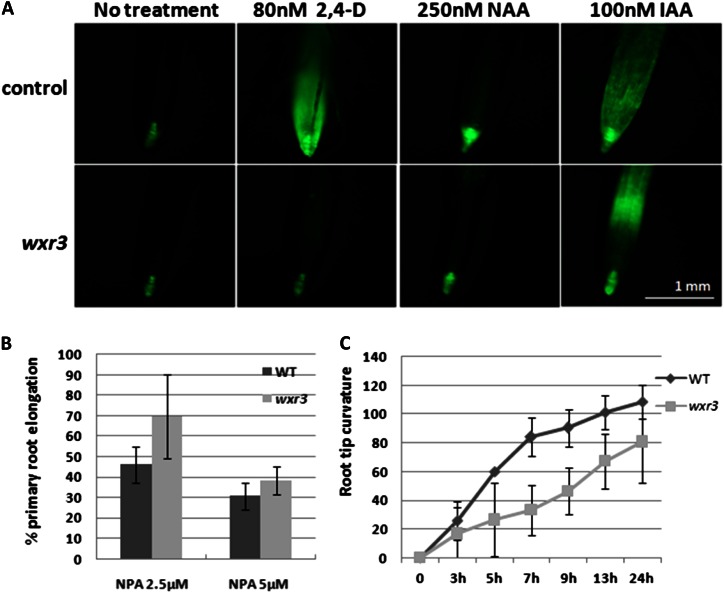
To determine if auxin transport is affected in the wxr3 mutant, we took advantage of the fact that the synthetic auxins 2,4-D and napthaleneacetic acid (NAA) are not substrates for the auxin efflux and influx carriers, respectively (Marchant et al., 1999). Five-day-old wxr3 seedlings were transferred onto fresh Arabidopsis + Suc (ATS) plates supplemented with different concentrations of IAA, NAA, or 2,4-D. After another 5 d of growth, the length of newly grown primary root was measured and expressed relative to growth on control plates. The results show that the wxr3 DR5rev:GFP line exhibits a differential response to IAA and NAA compared with 2,4-D. The mutant has a similar response to the wild type at low concentrations of IAA and NAA treatment and is slightly resistant at higher concentrations (Fig. 3, A and B). However, the mutant is strongly resistant to 2,4-D (Fig. 3C) throughout the concentration range. This suggests that the mutant may be affected in auxin influx. Similar results were obtained when we examined DR5rev:GFP expression after auxin treatment. In the mutant, 2,4-D does not induce DR5 activity after overnight treatment. NAA treatment results in a slight increase in GFP signal, while IAA clearly enhances DR5rev:GFP expression (Fig. 4A). However, the spatial distribution of DR5rev:GFP expression is altered in the mutant, with reduced signal in the cell division zone but enhanced expression in the elongation zone, compared with IAA-treated wild-type roots. Moreover, the wxr3 mutant displays a clear resistance to the auxin efflux inhibitor naphthylphthalamic acid with respect to primary root elongation and has a delayed response during root gravitropism (Fig. 4, B and C). These results indicate that polar auxin transport, possibly both auxin efflux and influx components, may be defective in the wxr3 mutant.
Figure 3.
The wxr3 mutant exhibits decreased polar auxin transport resulting in a disruption in auxin distribution. A to C, Primary root elongation of the wxr3 mutant is strongly resistant to 2,4-D (C) but not to IAA (A) or NAA (B) at low concentrations. D, Transport of [3H]IAA is strongly reduced at the wxr3 root apex compared with the control. E and F, The level of free IAA is increased in the shoot (E) but decreased in root tips (F) of the wxr3 mutant compared with the control. UH, Upper hypocotyl; RSTZ, root-shoot transition zone. Asterisks indicate statistically significant differences between the wild type (WT) and the wxr3 mutant (ANOVA, P < 0.05). Error bars represent se. Seedlings in E were 7 d old. For D and F, seedlings were grown until the hypocotyls were 5 mm, 5 d old for the wild type and 6 d old for wxr3-1.
Figure 4.
The wxr3 mutant displays defects in auxin transport. A, DR5rev:GFP expression in the wxr3 mutant and the control line treated with various auxins. Seedlings were 7 d old. B, Root elongation of wxr3 seedlings is resistant to naphthylphthalamic acid (NPA) treatment. C, The roots of wxr3 seedlings show a delayed response to gravity compared with the wild type (WT). Error bars represent se.
To further characterize this defect, auxin transport was directly measured. The results show that the wxr3 mutant significantly reduces the transport of applied [3H]IAA at the root apex region, indicating that the mutant has a defect in polar auxin transport (Fig. 3D). To determine whether the defect in auxin transport affects the distribution of auxin in the seedling, the level of free IAA was measured in the wxr3 mutant. Compared with the wild type, wxr3 seedlings have a higher level of free IAA in aerial tissues but a reduction of IAA in the root tip region, suggesting that auxin synthesized in the shoot is not efficiently transported into the root tip (Fig. 3, E and F). These results indicate that the wxr3 mutation interferes with IAA distribution by altering polar auxin transport.
The wxr3 Mutation Affects the Accumulation of PIN Proteins on the Plasma Membrane
Auxin transporters, such as AUX1 and the PINs, play a critical role in polar auxin transport. Because of the auxin transport defect in wxr3 plants, the expression and distribution of AUX1 and PINs were determined in the mutant. To visualize AUX1, the pAUX1:AUX1-YFP transgene was crossed into wxr3 seedlings (Swarup et al., 2004). The distribution and level of AUX1-YFP were studied by confocal microscopy. The results show that the AUX1-YFP level is much lower in the wxr3 mutant compared with the control line (Fig. 5, A and B). To examine PIN1 and PIN2 levels, an in situ immunodetection assay was performed using anti-PIN1 and anti-PIN2 antisera. Like AUX1, the levels of PIN1 and PIN2 are dramatically reduced in the mutant (Fig. 5, C–F). Finally, we used a pPIN3:PIN3-GFP transgene to show a similar defect in PIN3 level in the mutant as well (Fig. 5, G and H; Kleine-Vehn et al., 2010). However, the subcellular localization of PINs does not exhibit any obvious difference in the wxr3 mutant either with or without BFA treatment compared with the control (Fig. 5, A–H; Supplemental Fig. S3).
Figure 5.
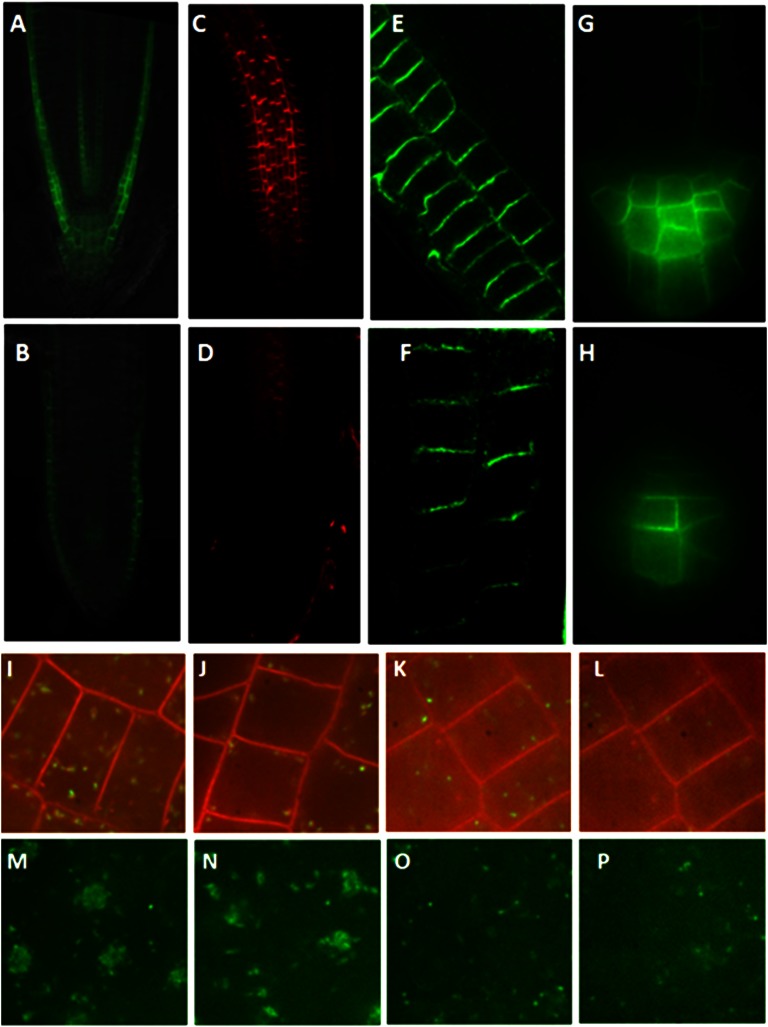
The wxr3 mutant has reduced levels of auxin transporters and endosomal markers in the cell. A and B, Expression of pAUX1:AUX1-YFP in the root tip of the control line (A) and the wxr3 mutant (B). C and D, PIN1 protein levels in the root tip of the wild type (C) and the wxr3 mutant (D). E and F, PIN2 protein levels in the root tip of the wild type (E) and the wxr3 mutant (F). G and H, Expression of pPIN3:PIN3-GFP in the root tip of the control line (G) and the wxr3 mutant (H). I and J, The wxr3 mutant (J) has lower levels of VHA-a1-GFP compared with control seedlings (I). K and L, The wxr3 mutant (L) exhibits a lower level of ARA7-GFP compared with control seedlings (K). M and N, Subcellular localization of VHA-a1-GFP in the wxr3 mutant (N) and control seedlings (M) after 50 μm BFA treatment for 2 h. O and P, Subcellular localization of ARA7-GFP in the wxr3 mutant (P) and control seedlings (O) after 50 μm BFA treatment for 2 h.
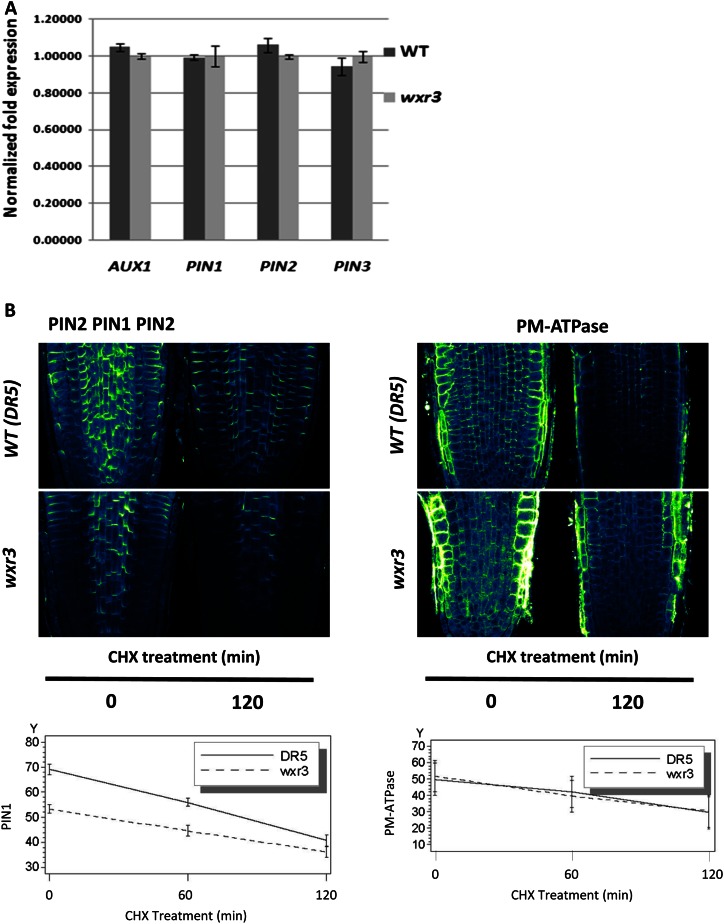
The results of real-time quantitative PCR experiments indicate that the RNA levels of AUX1, PIN1, PIN2, and PIN3 are similar between the mutant and the wild type, suggesting that the reduction of PIN levels in wxr3 is not related to their transcription (Fig. 6A). To test whether the wxr3 mutation affects the stability of the PINs, cycloheximide (CHX), an inhibitor of protein biosynthesis, was used to treat seedlings. We also examined the level of the plasma membrane (PM)-ATPase to determine if wxr3 has a global effect on the accumulation of plasma membrane proteins. After treatment with CHX, the protein level of PIN1/PIN2 was determined through in situ immunodetection assay. The results confirm that PIN1/PIN2 levels are reduced in the mutant but that this effect is not due to increased degradation (Fig. 6B). The levels of PM-ATPase are similar in the mutant and the wild type, indicating that WXR3 is not universally required for the accumulation of membrane proteins.
Figure 6.
The wxr3 mutation does not affect the stability of PIN proteins on the plasma membrane. A, RNA levels of AUX1 and PINs in wild-type (WT) and wxr3 seedlings as measured by quantitative RT-PCR. Error bars represent se. B, Disappearance of proteins from the plasma membrane after treatment with 50 μm CHX is not affected by the wxr3 mutation. Roots were treated with antibody against PIN1/PIN2 (left) or PM-ATPase (right). PIN1 and PM-ATPase levels were quantified and are expressed as arbitrary units over time.
A recent study demonstrated that the shr mutant exhibits a progressive reduction in the levels of the PIN proteins in the root (Lucas et al., 2011). This fact raises the possibility that in wxr3-1, reduced auxin transport is a consequence of reduced SHR levels. However, the root tips of 6-d-old wxr3 seedlings have lower IAA levels than the wild type, while shr root tips have higher IAA levels at this age (Lucas et al., 2011). For this reason, we think that the reduction in SHR levels is probably not the primary cause of decreased PIN levels in wxr3. Nevertheless, in the future, it will be interesting to explore the potential relations between WXR3 and SHR during the development of root tissues.
Lower PIN Levels May Be Related to Endosome Trafficking
To determine whether the reduction in auxin transporter levels is related to endosome recycling, the wxr3 mutant was crossed to transgenic lines expressing the VACUOLAR PROTON ATPASE (VHA)-a1-GFP and ARABIDOPSIS RAB GTPASE HOMOLOG F2B-GFP (ARA7-GFP), markers for trans-Golgi network and prevacuolar compartment localization, respectively (Lee et al., 2004; Dettmer et al., 2006). The trans-Golgi network and prevacuolar compartment function as early and late endosome, respectively, in plant cells. Examination of these lines shows that both markers are decreased in the wxr3 mutant, indicating that the wxr3 mutant has fewer early and late endosomes (Fig. 5, I–L). However, recycling of these endosomes is still sensitive to BFA, similar to the control (Fig. 5, M–P).
To further explore the effects of BFA on PIN1/PIN2 accumulation in the mutant, we performed a BFA washout experiment (Supplemental Fig. S3). Seedlings were treated with 50 μm BFA for 60 min and then the BFA was washed out for 120 min. The fraction of PIN1/PIN2 containing BFA bodies was similar in wxr3 and wild-type roots throughout the experiment, indicating that the mutant is not affected in a BFA-sensitive aspect of endosome recycling.
WXR3 Is a Member of the DUF647 Family
Genetic analyses indicate that the phenotype of the wxr3 mutant is induced by a single recessive mutation. To gain insight into the function of WXR3, the mutation was cloned by a map-based strategy. The wxr3 mutant (ecotype Columbia [Col-0]) was crossed to Landsberg erecta, and mutant plants were recovered from the F2 population. After analysis of 421 plants, the wxr3 mutation was mapped to bacterial artificial chromosome F16L2 on chromosome 3 (Supplemental Fig. S4). Based on sequencing data, a G-to-A mutation is located at the 3′ end of the second exon of the At3g45890 gene (Supplemental Fig. S4). The mutation causes an RNA splicing error that retains the second intron in the mature mRNA. This splice product, identified by reverse transcription (RT)-PCR and sequencing, introduces a premature stop codon, which may result in a truncated protein. To verify that this mutation is responsible for the phenotype conferred by the wxr3 mutant, a complementation assay was performed. Wild-type At3g45890 genomic DNA including 2 kb of DNA sequence upstream of At3g45890 was introduced into the wxr3 DR5rev:GFP mutant, and homozygous transgenic lines were isolated. These lines display a wild-type phenotype both with respect to primary root growth and the DR5rev:GFP response to 2,4-D, confirming that At3g45890 is WXR3 (Supplemental Fig. S4). At3g45890 was previously identified as RUS1, functioning in UV-B light response (Tong et al., 2008).
RUS1/WXR3 belongs to the DUF647 protein family, so named for the presence of the DUF647 domain in the C terminus. In the Arabidopsis genome, there are six DUF647 family members. RUS1/WXR3 protein has a unique N-terminal extension before a Gly-rich region (10 Gly residues in a 12-amino acid region) compared with other DUF647 proteins (Supplemental Fig. S4). Another DUF647 protein, WXR1/RUS2, was identified in the same mutant screen (Ge et al., 2010). The wxr1 mutant displays similar root morphology to the wxr3 mutant and defects in polar auxin transport. We also generated the wxr1 wxr3 double mutant. These plants exhibit more severe defects in auxin transport and plant developmental growth (Fig. 3, D and F; Supplemental Fig. S2). This suggests that WXR1/RUS2 and WXR3/RUS1 have a related and overlapping function in the plant.
To analyze the expression pattern of the RUS1/WXR3 gene, the pWXR3:WXR3-GUS construct was created and introduced into the wxr3 mutant. Analysis of the transgenic plants reveals that the RUS1/WXR3 protein is most abundant in the cotyledons, roots, and hypocotyls (Supplemental Fig. S5, A–I). Particularly strong GUS staining is observed in the leaf veins, root vascular tissues, root tip, and lateral root primordia. Previous work showed that RUS3/WXR1 is localized in plastids (Ge et al., 2010). To investigate the subcellular localization of RUS1/WXR3, a p35S:WXR3-GFP construct was introduced into a transgenic line carrying the plastid mCherry-based marker pt-rb (Nelson et al., 2007). Examination of this line shows that WXR3-GFP colocalizes with pt-rb, suggesting that RUS1/WXR3 is also localized to the plastid (Supplemental Fig. S5, J–L).
Since overexpression of RUS1/WXR3 by the 35S promoter does not display a strong effect on root development, RUS1/WXR3 was placed under the control of an estradiol-inducible promoter and transformed into wild-type plants (Curtis and Grossniklaus, 2003). Without estradiol treatment, the root morphology of the transgenic line is similar to the wild type. However, after 4 μm estradiol treatment for 2 d, the line displays many more root hairs, suggesting that overexpression of RUS1/WXR3 can enhance root hair initiation and elongation, consistent with a role for RUS1/WXR3 in polar auxin transport (Supplemental Fig. S6).
DISCUSSION
Root Development in the wxr3 Mutant
Auxin plays a key role in embryogenesis and postembryonic development in plants. Many genes involved in auxin signaling have been shown to function in aspects of root development, including primary root elongation, root hair initiation, lateral root development, and root gravitropic response (Müller et al., 1998; Hamann et al., 1999, 2002; Marchant et al., 1999, 2002; Nagpal et al., 2000; Reed, 2001). The wxr3 mutant exhibits severe defects in root meristem maintenance, root hair elongation, and gravitropic response. In addition, the expression of the auxin reporter DR5rev:GFP is not induced by 2,4-D treatment. In contrast, DR5rev:GFP expression is strongly induced by IAA, suggesting that wxr3 does not affect SCFTIR1/AFB-dependent auxin signaling. This is consistent with our observation that the mutation does not stabilize the AXR3NT-GUS protein in the plant. However, the wxr3 mutation does alter the spatial distribution of DR5rev:GFP expression. This difference may reflect a change in auxin transport. Indeed, we found that wxr3 seedlings accumulate auxin in the apical region. In young seedlings, auxin is synthesized in the cotyledons and developing leaves and is actively transported through the hypocotyl to the root, which enables primary root elongation and lateral root development. Direct measurement of auxin transport in the wxr3 mutant demonstrates a defect in this process. We propose that the defects in root development observed in wxr3 seedlings are related to reduced auxin transport.
Function of the WXR3 Protein
The PINs and AUX1/LAX are crucial for auxin transport. Loss-of-function mutants in auxin transporter genes display severe defects in plant development. For example, the pin1 mutant is defective in organ initiation and exhibits pin-shaped inflorescences devoid of flowers (Okada et al., 1991). In the wxr3 mutant, the protein levels of the auxin transporters PIN1, PIN2, PIN3 and AUX1 are dramatically reduced on the plasma membrane. This suggests that the defect in polar auxin transport in the wxr3 mutant is caused by decreased levels of the transporters. The wxr3 mutant exhibits fewer early and late endosomes recycling in the cell compared with the control. It is possible that the wxr3 mutation causes a nonspecific defect in plasma membrane protein turnover.
RUS1/WXR3 localizes at plastids. In a previous study, RUS1/WXR3 was identified in a screen for plants that are hypersensitive to very-low-fluence UVB (less than 0.1 μmol m−2 s−1) as well, indicating that RUS1/WXR3 functions in this light response (Tong et al., 2008). Light plays an essential role in many plant developmental processes, including the regulation of polar auxin transport (Lau and Deng, 2010). Light affects the expression and subcellular localization of auxin transporters (Laxmi et al., 2008; Keuskamp et al., 2010; Ding et al., 2011; Liu et al., 2011; Wan et al., 2012). Future studies will determine whether these light effects involve RUS1/WXR3 function.
The DUF647 Proteins and Plant Development
The Arabidopsis genome encodes six DUF647 family members. Two of these, RUS1/WXR3 and RUS2/WXR1, were isolated in the same two mutant screens for either auxin response defects or hypersensitivity to UV-B light (Tong et al., 2008; Leasure et al., 2009; Ge et al., 2010). Both of the mutants have very similar phenotypes with defects in root development, polar auxin transport, and response to UV-B. The wxr1 wxr3 double mutant displays much more severe developmental defects throughout plant growth, indicating a crucial function of the DUF647 family in plant growth. RUS1/WXR3 and RUS2/WXR1 have the same subcellular localization. A recent study shows that they interact with each other through the yeast (Saccharomyces cerevisiae) two-hybrid system (Leasure et al., 2009). The fact that the double mutant has a more severe phenotype than either single mutant might suggest that the two proteins are acting at least partly independently. However, it is important to note that it is not clear if either the rus1/wxr3 or rus2/wxr1 mutations are nulls. Thus, the double mutant phenotype may represent the additive effect of two hypomorphic mutations. All of these data suggest that they may form a complex in vivo. In the future, it will be important to determine how these proteins impact on PIN protein trafficking and homeostasis.
MATERIALS AND METHODS
Plant Materials and Conditions
The Arabidopsis (Arabidopsis thaliana) mutants and transgenic lines used in this study were generated in Col-0. Seeds were surface sterilized for 20 min in 30% commercial bleach, plated on ATS medium supplemented with 0.8% agar, and stratified for 2 to 4 d at 4°C. ATS medium consists of 1% Suc, 5 mm KNO3, 2.5 mm KPO4, 2 mm MgSO4, 2 mm Ca(NO3)2, 50 μm Fe-EDTA, and 1 mL L−1 micronutrients. All seedling experiments were performed under long-day conditions (16 h of light/8 h of dark) in the growth chamber (80 μmol m−2 s−1, 22°C), unless otherwise stated. Plants in soil were grown in long-day conditions at 22°C.
For immunolocalization studies, DR5rev::GFP and wxr3-3 seeds were gas sterilized, sown on solid AM+ medium (one-half-strength Murashige and Skoog [MS] with 1% Suc and 0.8% agar, pH 5.9), and incubated at 4°C for 2 d. Seedlings were grown on vertical plates at 21°C under continuous light conditions for 6 d.
Ethyl Methanesulfonate Mutagenesis and Mutant Screen
Ethyl methanesulfonate mutagenesis and the mutant screen were as described by Ge et al. (2010). Briefly, approximately 5,000 DR5rev:GFP transgenic seeds were treated with 0.3% ethyl methanesulfonate for 10 h. After several rounds of washing, the seeds were sown in soil. To screen, about 40,000 M2 seeds were germinated on ATS medium. Seedlings with a short root compared with the control line were selected and treated with 75 nm 2,4-D for 12 h. Seedlings with reduced GFP levels were recovered for further study. Each candidate mutant was backcrossed with DR5rev:GFP (Col-0) plants three times before characterization.
Root Inhibition Assay and Gravitropism Response
To assess the effect of auxin on root growth, seedlings were grown in yellow light for 5 d to enhance the primary root elongation of the wxr3 mutant and then transferred to fresh ATS medium with different concentrations of NAA, 2,4-D, or IAA. After an additional 5 d, root growth after transfer was measured. To determine the gravitropic response of roots, 5-d-old seedlings were rotated 90° clockwise and root tip bending was recorded by a Nikon SMZ1500 dissecting microscope every 2 h for 12 h. All measurements were performed using ImageJ software (http://rsbweb.nih.gov/ij/index.html).
Plasmid Constructs and Plant Transformation
The pWXR3:WXR3-GUS construct was made by introducing WXR3 genomic DNA with the 2-kb 5′ upstream region into PMDC163 vector using the Gateway LR Reaction Kit (Invitrogen). The p35S:WXR3-GFP construct was generated by cloning full-length WXR3 complementary DNA into the PMDC83 vector. The primers can be found in Supplemental Table S1. The binary plasmids were introduced into Agrobacterium tumefaciens strain GV3101. Transformation of Col-0 plants was performed by floral dip (Clough and Bent, 1998), and the transgenic plants were screened on ATS medium supplemented with 50 μg L−1 hygromycin.
GUS Staining
Six-day-old pHS:AXR3NT-GUS and wxr3 pHS:AXR3NT-GUS plants were heat shocked at 37°C for 2 h in ATS solution and then treated with or without IAA. At time intervals, whole seedlings were collected in GUS staining solution [100 mm Na2PO4, pH 7.0, 10 mm EDTA, 0.1% Triton X-100, 1.0 mm K3Fe(CN)6, and 2 mm 5-bromo-4-chloro-3-indolyl-β-glucuronic acid], vacuum infiltrated for 20 min, and stained overnight at 37°C. The seedlings were cleared in 70% ethanol and imaged with a Nikon SMZ1500 dissecting microscope. To measure GUS activity, protein was harvested by grinding whole seedlings in liquid nitrogen and vortexing vigorously in extraction buffer (50 mm Na2PO4, pH 7.0, 10 mm dithiothreitol, 1 mm EDTA, 0.1% sodium lauryl sarcosine, and 0.1% Triton X-100). Cellular debris was removed by centrifugation at maximum speed for 10 min at 4°C, and protein concentration was determined by the Bradford assay. Twenty microliters of extract was added to 180 μL of MUG assay buffer (1 mm MUG in extraction buffer). The reaction was incubated for 4 h at 37°C in the dark and then added to an equal volume of 0.2 m Na2CO3 to stop the reaction. Enzyme activity was measured using a spectrofluorometer.
Auxin Transport Assay and Quantification of Free IAA Levels
The auxin transport assay and the quantification of free IAA levels were described by Ge et al. (2010). Seedlings were grown in light on 1% phytagar plates containing 0.25× MS (pH 5.2) and 1% Suc until the hypocotyl reached 5 mm in length. Before the assay, 10 seedlings were transferred to vertically discontinuous filter paper strips saturated in 0.25× MS and allowed to equilibrate for 1.5 h. Auxin solutions were made up in 0.25% agarose containing 2% dimethyl sulfoxide and 25 mm MES (pH 5.2). A 0.1-μL microdroplet containing 500 nm unlabeled IAA and 500 nm [3H]IAA (specific activity, 25 Ci mmol−1; American Radiochemical) was placed on the apical tip of the seedlings. Seedlings were then incubated in the dark for 5 h. After incubation, the cotyledons were removed, and sections were harvested. The upper hypocotyl section is below the excised apex where auxin is applied, the root-shoot transition zone is a 2-mm section centered on this zone, and the root apex is a 2-mm section starting from the root tip. [3H]IAA was counted in a low-range scintillation counter. To measure free auxin levels, seedlings were grown as described for the auxin transport assay. Sampled sections were homogenized in liquid nitrogen, diluted with 800 μL of 0.05 m sodium phosphate, pH 7.0, and 0.02% (w/v) sodium diethyldithiocarbamate, combined with 4 ng [13C]IAA working standard, shaken for 15 min at 4°C, and combined with 1 m HCl to a final pH of 2.7. Samples were passed through a 0.45-μm syringe filter and applied to an Isolute C8-EC (500 mg per 3 mL; no. 291-0050-B) solid-phase extraction column preconditioned by methanol/acetic acid. Then, samples were analyzed by gas chromatography-mass spectrometry. Quantifications are based on comparisons of IAA peaks with [13C]IAA standards normalized to the fresh weight of the original sample. The results were repeated in three independent experiments.
RNA Extraction and Real-Time Quantitative PCR
Total RNA was extracted from 7-d-old seedlings using the RNeasy Plant Mini Kit (Qiagen). Yield was quantified by the Thermo Scientific NanoDrop 2000. A total of 1 μg of RNA was used for complementary DNA synthesis through the SuperScript III First-Strand Synthesis Kit (Invitrogen). Quantitative RT-PCR was performed as described previously (Greenham et al., 2011). The primers can be found in Supplemental Table S1. Data were normalized to the reference gene PP2AA3 according to the comparative cycle threshold method (Czechowski et al., 2005).
Pharmacological Treatments and Immunolocalization
For pharmacological treatments, 6-d-old seedlings were used. Stock solutions of CHX and/or BFA in dimethyl sulfoxide were diluted in liquid AM+ medium to a final concentration of 50 μm. Seedlings were treated on cell culture plates for the indicated times.
In situ immunolocalization and detection were performed on 6-d-old seedlings using the In Situ Pro robot (Intavis). Antibodies and dilutions were as follows: rabbit anti-PIN1 (1:1,000), rabbit anti-PIN2 (1:1,000), rabbit anti-ATPase (1:1,000; Paciorek et al., 2005), and anti-rabbit IgG (whole molecule) F(ab′)2 fragment Cy3 (Sigma; 1:600).
Quantitative Microscopy and Statistical Analysis
Confocal microscopy and imaging were done on a Zeiss 710 confocal microscope. ImageJ was used to measure signal intensity. The effect of CHX on PIN1 and ATPase signal was determined in stele cells using 10 to 20 images per treatment and seed line for three experiments. A three-way ANOVA model was fit to the data with treatment, line, and experiment as main effect terms and including all two-way and three-way interaction terms using the SAS PROC MIXED procedure. To determine the effects of BFA, the intracellular and plasma membrane signal intensities were measured in 12 to 15 cells from three to five images for each treatment and seed line for two experiments. Graphs represent the percentage of cells containing intracellular aggregates after BFA treatment and washouts or the average of ratios of intracellular to plasma membrane for each cell. A three-way ANOVA model was fit to the data. An extra nested term was added to the model as a random term (plant nested in line). The significance of the random term was tested with a log-likelihood ratio test and was found not to be significant at the 0.05 significance level.
The Arabidopsis Genome Initiative locus numbers for the major genes discussed in this article are as follows: RUS1/WXR3 (At3g45890), RUS2/WXR1 (At2g31190), TIR1 (At3g62980), ABCB1/PGP1 (At2g36910), ABCB19/PGP19 (At3g28860), PIN1 (At1g73590), PIN2 (At5g57090), PIN3 (At1g70940), AUX1 (At2g38120), VHA-A1 (At2g28520), and ARA7 (At4g19640).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. The wxr3 mutant exhibits decreased expression of CYCB1;1, SCR, and SHR compared with the control.
Supplemental Figure S2. Phenotypic characterization of the wxr1, wxr3 mutant and wxr1 wxr3 double mutant.
Supplemental Figure S3. The wxr3 mutation does not affect response to BFA.
Supplemental Figure S4. Positional cloning of the wxr3 mutation.
Supplemental Figure S5. Expression pattern and subcellular localization of RUS1/WXR3 protein.
Supplemental Figure S6. Inducible overexpression of WXR3 enhances root hair initiation and elongation.
Supplemental Table S1. Primer sequences.
Acknowledgments
The transgenic lines VHA-a1-GFP and ARA7-GFP were kindly provided by Gerd Jurgens. The pAUX1:AUX1-YFP transgenic line was provided by Malcolm Bennett. M.K. thanks Veronique Storme for assistance with statistical analysis.
Glossary
- IAA
indole-3-acetic acid
- BFA
brefeldin A
- 2,4-D
2,4-dichlorophenoxyacetic acid
- MUG
4-methylumbelliferyl-β-d-glucuronide
- NAA
naphthylacetic acid
- CHX
cycloheximide
- Col-0
ecotype Columbia
- RT
reverse transcription
- MS
Murashige and Skoog
- NAA
napthaleneacetic acid
- ATS
Arabidopsis + Suc
- PM
plasma membrane
References
- Bennett MJ, Marchant A, Green HG, May ST, Ward SP, Millner PA, Walker AR, Schulz B, Feldmann KA. (1996) Arabidopsis AUX1 gene: a permease-like regulator of root gravitropism. Science 273: 948–950 [DOI] [PubMed] [Google Scholar]
- Blakeslee JJ, Bandyopadhyay A, Lee OR, Mravec J, Titapiwatanakun B, Sauer M, Makam SN, Cheng Y, Bouchard R, Adamec J, et al. (2007) Interactions among PIN-FORMED and P-glycoprotein auxin transporters in Arabidopsis. Plant Cell 19: 131–147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouchard R, Bailly A, Blakeslee JJ, Oehring SC, Vincenzetti V, Lee OR, Paponov I, Palme K, Mancuso S, Murphy AS, et al (2006) Immunophilin-like TWISTED DWARF1 modulates auxin efflux activities of Arabidopsis P-glycoproteins. J Biol Chem 281: 30603–30612 [DOI] [PubMed] [Google Scholar]
- Cho M, Lee SH, Cho HT. (2007) P-glycoprotein4 displays auxin efflux transporter-like action in Arabidopsis root hair cells and tobacco cells. Plant Cell 19: 3930–3943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Curtis MD, Grossniklaus U. (2003) A Gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol 133: 462–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czechowski T, Stitt M, Altmann T, Udvardi MK, Scheible WR. (2005) Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol 139: 5–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dettmer J, Hong-Hermesdorf A, Stierhof YD, Schumacher K. (2006) Vacuolar H+-ATPase activity is required for endocytic and secretory trafficking in Arabidopsis. Plant Cell 18: 715–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dharmasiri N, Dharmasiri S, Estelle M. (2005a) The F-box protein TIR1 is an auxin receptor. Nature 435: 441–445 [DOI] [PubMed] [Google Scholar]
- Dharmasiri N, Dharmasiri S, Weijers D, Lechner E, Yamada M, Hobbie L, Ehrismann JS, Jürgens G, Estelle M. (2005b) Plant development is regulated by a family of auxin receptor F box proteins. Dev Cell 9: 109–119 [DOI] [PubMed] [Google Scholar]
- Dhonukshe P. (2011) PIN polarity regulation by AGC-3 kinases and ARF-GEF: a recurrent theme with context dependent modifications for plant development and response. Plant Signal Behav 6: 1333–1337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhonukshe P, Aniento F, Hwang I, Robinson DG, Mravec J, Stierhof YD, Friml J. (2007) Clathrin-mediated constitutive endocytosis of PIN auxin efflux carriers in Arabidopsis. Curr Biol 17: 520–527 [DOI] [PubMed] [Google Scholar]
- Di Laurenzio L, Wysocka-Diller J, Malamy JE, Pysh L, Helariutta Y, Freshour G, Hahn MG, Feldmann KA, Benfey PN. (1996) The SCARECROW gene regulates an asymmetric cell division that is essential for generating the radial organization of the Arabidopsis root. Cell 86: 423–433 [DOI] [PubMed] [Google Scholar]
- Ding Z, Galván-Ampudia CS, Demarsy E, Łangowski L, Kleine-Vehn J, Fan Y, Morita MT, Tasaka M, Fankhauser C, Offringa R, et al (2011) Light-mediated polarization of the PIN3 auxin transporter for the phototropic response in Arabidopsis. Nat Cell Biol 13: 447–452 [DOI] [PubMed] [Google Scholar]
- Doerner P, Jørgensen JE, You R, Steppuhn J, Lamb C. (1996) Control of root growth and development by cyclin expression. Nature 380: 520–523 [DOI] [PubMed] [Google Scholar]
- Feraru E, Feraru MI, Kleine-Vehn J, Martinière A, Mouille G, Vanneste S, Vernhettes S, Runions J, Friml J. (2011) PIN polarity maintenance by the cell wall in Arabidopsis. Curr Biol 21: 338–343 [DOI] [PubMed] [Google Scholar]
- Feraru E, Friml J. (2008) PIN polar targeting. Plant Physiol 147: 1553–1559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friml J, Benková E, Blilou I, Wisniewska J, Hamann T, Ljung K, Woody S, Sandberg G, Scheres B, Jürgens G, et al (2002a) AtPIN4 mediates sink-driven auxin gradients and root patterning in Arabidopsis. Cell 108: 661–673 [DOI] [PubMed] [Google Scholar]
- Friml J, Vieten A, Sauer M, Weijers D, Schwarz H, Hamann T, Offringa R, Jürgens G. (2003) Efflux-dependent auxin gradients establish the apical-basal axis of Arabidopsis. Nature 426: 147–153 [DOI] [PubMed] [Google Scholar]
- Friml J, Wiśniewska J, Benková E, Mendgen K, Palme K. (2002b) Lateral relocation of auxin efflux regulator PIN3 mediates tropism in Arabidopsis. Nature 415: 806–809 [DOI] [PubMed] [Google Scholar]
- Friml J, Yang X, Michniewicz M, Weijers D, Quint A, Tietz O, Benjamins R, Ouwerkerk PB, Ljung K, Sandberg G, et al. (2004) A PINOID-dependent binary switch in apical-basal PIN polar targeting directs auxin efflux. Science 306: 862–865 [DOI] [PubMed] [Google Scholar]
- Gälweiler L, Guan C, Müller A, Wisman E, Mendgen K, Yephremov A, Palme K. (1998) Regulation of polar auxin transport by AtPIN1 in Arabidopsis vascular tissue. Science 282: 2226–2230 [DOI] [PubMed] [Google Scholar]
- Ge L, Peer W, Robert S, Swarup R, Ye S, Prigge M, Cohen JD, Friml J, Murphy A, Tang D, et al (2010) Arabidopsis ROOT UVB SENSITIVE2/WEAK AUXIN RESPONSE1 is required for polar auxin transport. Plant Cell 22: 1749–1761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geldner N, Friml J, Stierhof YD, Jürgens G, Palme K. (2001) Auxin transport inhibitors block PIN1 cycling and vesicle trafficking. Nature 413: 425–428 [DOI] [PubMed] [Google Scholar]
- Gray WM, Kepinski S, Rouse D, Leyser O, Estelle M. (2001) Auxin regulates SCF(TIR1)-dependent degradation of AUX/IAA proteins. Nature 414: 271–276 [DOI] [PubMed] [Google Scholar]
- Greenham K, Santner A, Castillejo C, Mooney S, Sairanen I, Ljung K, Estelle M. (2011) The AFB4 auxin receptor is a negative regulator of auxin signaling in seedlings. Curr Biol 21: 520–525 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Grieneisen VA, Xu J, Marée AF, Hogeweg P, Scheres B. (2007) Auxin transport is sufficient to generate a maximum and gradient guiding root growth. Nature 449: 1008–1013 [DOI] [PubMed] [Google Scholar]
- Grunewald W, Friml J. (2010) The march of the PINs: developmental plasticity by dynamic polar targeting in plant cells. EMBO J 29: 2700–2714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamann T, Benkova E, Bäurle I, Kientz M, Jürgens G. (2002) The Arabidopsis BODENLOS gene encodes an auxin response protein inhibiting MONOPTEROS-mediated embryo patterning. Genes Dev 16: 1610–1615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamann T, Mayer U, Jürgens G. (1999) The auxin-insensitive bodenlos mutation affects primary root formation and apical-basal patterning in the Arabidopsis embryo. Development 126: 1387–1395 [DOI] [PubMed] [Google Scholar]
- Helariutta Y, Fukaki H, Wysocka-Diller J, Nakajima K, Jung J, Sena G, Hauser MT, Benfey PN. (2000) The SHORT-ROOT gene controls radial patterning of the Arabidopsis root through radial signaling. Cell 101: 555–567 [DOI] [PubMed] [Google Scholar]
- Jaillais Y, Fobis-Loisy I, Miège C, Rollin C, Gaude T. (2006) AtSNX1 defines an endosome for auxin-carrier trafficking in Arabidopsis. Nature 443: 106–109 [DOI] [PubMed] [Google Scholar]
- Jaillais Y, Gaude T. (2007) Sorting out the sorting functions of endosomes in Arabidopsis. Plant Signal Behav 2: 556–558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kepinski S, Leyser O. (2005) The Arabidopsis F-box protein TIR1 is an auxin receptor. Nature 435: 446–451 [DOI] [PubMed] [Google Scholar]
- Keuskamp DH, Pollmann S, Voesenek LA, Peeters AJ, Pierik R. (2010) Auxin transport through PIN-FORMED 3 (PIN3) controls shade avoidance and fitness during competition. Proc Natl Acad Sci USA 107: 22740–22744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleine-Vehn J, Ding Z, Jones AR, Tasaka M, Morita MT, Friml J. (2010) Gravity-induced PIN transcytosis for polarization of auxin fluxes in gravity-sensing root cells. Proc Natl Acad Sci USA 107: 22344–22349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleine-Vehn J, Langowski L, Wisniewska J, Dhonukshe P, Brewer PB, Friml J. (2008) Cellular and molecular requirements for polar PIN targeting and transcytosis in plants. Mol Plant 1: 1056–1066 [DOI] [PubMed] [Google Scholar]
- Lau OS, Deng XW. (2010) Plant hormone signaling lightens up: integrators of light and hormones. Curr Opin Plant Biol 13: 571–577 [DOI] [PubMed] [Google Scholar]
- Laxmi A, Pan J, Morsy M, Chen R. (2008) Light plays an essential role in intracellular distribution of auxin efflux carrier PIN2 in Arabidopsis thaliana. PLoS ONE 3: e1510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leasure CD, Tong H, Yuen G, Hou X, Sun X, He ZH. (2009) ROOT UV-B SENSITIVE2 acts with ROOT UV-B SENSITIVE1 in a root ultraviolet B-sensing pathway. Plant Physiol 150: 1902–1915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee GJ, Sohn EJ, Lee MH, Hwang I. (2004) The Arabidopsis rab5 homologs rha1 and ara7 localize to the prevacuolar compartment. Plant Cell Physiol 45: 1211–1220 [DOI] [PubMed] [Google Scholar]
- Liu X, Cohen JD, Gardner G. (2011) Low-fluence red light increases the transport and biosynthesis of auxin. Plant Physiol 157: 891–904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas M, Swarup R, Paponov IA, Swarup K, Casimiro I, Lake D, Peret B, Zappala S, Mairhofer S, Whitworth M, et al. (2011) Short-Root regulates primary, lateral, and adventitious root development in Arabidopsis. Plant Physiol 155: 384–398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchant A, Bhalerao R, Casimiro I, Eklöf J, Casero PJ, Bennett M, Sandberg G. (2002) AUX1 promotes lateral root formation by facilitating indole-3-acetic acid distribution between sink and source tissues in the Arabidopsis seedling. Plant Cell 14: 589–597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchant A, Kargul J, May ST, Muller P, Delbarre A, Perrot-Rechenmann C, Bennett MJ. (1999) AUX1 regulates root gravitropism in Arabidopsis by facilitating auxin uptake within root apical tissues. EMBO J 18: 2066–2073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Men S, Boutté Y, Ikeda Y, Li X, Palme K, Stierhof YD, Hartmann MA, Moritz T, Grebe M. (2008) Sterol-dependent endocytosis mediates post-cytokinetic acquisition of PIN2 auxin efflux carrier polarity. Nat Cell Biol 10: 237–244 [DOI] [PubMed] [Google Scholar]
- Michniewicz M, Zago MK, Abas L, Weijers D, Schweighofer A, Meskiene I, Heisler MG, Ohno C, Zhang J, Huang F, et al. (2007) Antagonistic regulation of PIN phosphorylation by PP2A and PINOID directs auxin flux. Cell 130: 1044–1056 [DOI] [PubMed] [Google Scholar]
- Möller B, Weijers D. (2009) Auxin control of embryo patterning. Cold Spring Harb Perspect Biol 1: a001545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller A, Guan C, Gälweiler L, Tänzler P, Huijser P, Marchant A, Parry G, Bennett M, Wisman E, Palme K. (1998) AtPIN2 defines a locus of Arabidopsis for root gravitropism control. EMBO J 17: 6903–6911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagpal P, Walker LM, Young JC, Sonawala A, Timpte C, Estelle M, Reed JW. (2000) AXR2 encodes a member of the Aux/IAA protein family. Plant Physiol 123: 563–574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naramoto S, Kleine-Vehn J, Robert S, Fujimoto M, Dainobu T, Paciorek T, Ueda T, Nakano A, Van Montagu MC, Fukuda H, et al (2010) ADP-ribosylation factor machinery mediates endocytosis in plant cells. Proc Natl Acad Sci USA 107: 21890–21895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson BK, Cai X, Nebenführ A. (2007) A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. Plant J 51: 1126–1136 [DOI] [PubMed] [Google Scholar]
- Okada K, Ueda J, Komaki MK, Bell CJ, Shimura Y. (1991) Requirement of the auxin polar transport system in early stages of Arabidopsis floral bud formation. Plant Cell 3: 677–684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overvoorde P, Fukaki H, Beeckman T. (2010) Auxin control of root development. Cold Spring Harb Perspect Biol 2: a001537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paciorek T, Zazímalová E, Ruthardt N, Petrásek J, Stierhof YD, Kleine-Vehn J, Morris DA, Emans N, Jürgens G, Geldner N, et al (2005) Auxin inhibits endocytosis and promotes its own efflux from cells. Nature 435: 1251–1256 [DOI] [PubMed] [Google Scholar]
- Reed JW. (2001) Roles and activities of Aux/IAA proteins in Arabidopsis. Trends Plant Sci 6: 420–425 [DOI] [PubMed] [Google Scholar]
- Scarpella E, Barkoulas M, Tsiantis M. (2010) Control of leaf and vein development by auxin. Cold Spring Harb Perspect Biol 2: a001511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinmann T, Geldner N, Grebe M, Mangold S, Jackson CL, Paris S, Gälweiler L, Palme K, Jürgens G. (1999) Coordinated polar localization of auxin efflux carrier PIN1 by GNOM ARF GEF. Science 286: 316–318 [DOI] [PubMed] [Google Scholar]
- Sundberg E, Østergaard L. (2009) Distinct and dynamic auxin activities during reproductive development. Cold Spring Harb Perspect Biol 1: a001628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swarup R, Kargul J, Marchant A, Zadik D, Rahman A, Mills R, Yemm A, May S, Williams L, Millner P, et al (2004) Structure-function analysis of the presumptive Arabidopsis auxin permease AUX1. Plant Cell 16: 3069–3083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi H, Miyazawa Y, Fujii N. (2009) Hormonal interactions during root tropic growth: hydrotropism versus gravitropism. Plant Mol Biol 69: 489–502 [DOI] [PubMed] [Google Scholar]
- Teale WD, Paponov IA, Palme K. (2006) Auxin in action: signalling, transport and the control of plant growth and development. Nat Rev Mol Cell Biol 7: 847–859 [DOI] [PubMed] [Google Scholar]
- Tiwari SB, Wang XJ, Hagen G, Guilfoyle TJ. (2001) AUX/IAA proteins are active repressors, and their stability and activity are modulated by auxin. Plant Cell 13: 2809–2822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong H, Leasure CD, Hou X, Yuen G, Briggs W, He ZH. (2008) Role of root UV-B sensing in Arabidopsis early seedling development. Proc Natl Acad Sci USA 105: 21039–21044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernoux T, Besnard F, Traas J. (2010) Auxin at the shoot apical meristem. Cold Spring Harb Perspect Biol 2: a001487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan Y, Jasik J, Wang L, Hao H, Volkmann D, Menzel D, Mancuso S, Baluška F, Lin J. (2012) The signal transducer NPH3 integrates the phototropin1 photosensor with PIN2-based polar auxin transport in Arabidopsis root phototropism. Plant Cell 24: 551–565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weijers D, Benkova E, Jäger KE, Schlereth A, Hamann T, Kientz M, Wilmoth JC, Reed JW, Jürgens G. (2005) Developmental specificity of auxin response by pairs of ARF and Aux/IAA transcriptional regulators. EMBO J 24: 1874–1885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willemsen V, Friml J, Grebe M, van den Toorn A, Palme K, Scheres B. (2003) Cell polarity and PIN protein positioning in Arabidopsis require STEROL METHYLTRANSFERASE1 function. Plant Cell 15: 612–625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisniewska J, Xu J, Seifertová D, Brewer PB, Ruzicka K, Blilou I, Rouquié D, Benková E, Scheres B, Friml J. (2006) Polar PIN localization directs auxin flow in plants. Science 312: 883. [DOI] [PubMed] [Google Scholar]
- Zhang J, Nodzynski T, Pencík A, Rolcík J, Friml J. (2010) PIN phosphorylation is sufficient to mediate PIN polarity and direct auxin transport. Proc Natl Acad Sci USA 107: 918–922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y. (2010) Auxin biosynthesis and its role in plant development. Annu Rev Plant Biol 61: 49–64 [DOI] [PMC free article] [PubMed] [Google Scholar]