The proteasome-mediated regulation of class II ERF transcriptional repressors is involved in the progression of leaf senescence.
Abstract
Leaf senescence is the final process of leaf development that involves the mobilization of nutrients from old leaves to newly growing tissues. Despite the identification of several transcription factors involved in the regulation of this process, the mechanisms underlying the progression of leaf senescence are largely unknown. Herein, we describe the proteasome-mediated regulation of class II ETHYLENE RESPONSE FACTOR (ERF) transcriptional repressors and involvement of these factors in the progression of leaf senescence in Arabidopsis (Arabidopsis thaliana). Based on previous results showing that the tobacco (Nicotiana tabacum) ERF3 (NtERF3) specifically interacts with a ubiquitin-conjugating enzyme, we examined the stability of NtERF3 in vitro and confirmed its rapid degradation by plant protein extracts. Furthermore, NtERF3 accumulated in plants treated with a proteasome inhibitor. The Arabidopsis class II ERFs AtERF4 and AtERF8 were also regulated by the proteasome and increased with plant aging. Transgenic Arabidopsis plants with enhanced expression of NtERF3, AtERF4, or AtERF8 showed precocious leaf senescence. Our gene expression and chromatin immunoprecipitation analyses suggest that AtERF4 and AtERF8 targeted the EPITHIOSPECIFIER PROTEIN/EPITHIOSPECIFYING SENESCENCE REGULATOR gene and regulated the expression of many genes involved in the progression of leaf senescence. By contrast, an aterf4 aterf8 double mutant exhibited delayed leaf senescence. Our results provide insight into the important role of class II ERFs in the progression of leaf senescence.
Leaf senescence, which is characterized by progressive yellowing, is the final stage of leaf development and involves the mobilization of nutrients from old leaves to newly growing tissues. The progression of leaf senescence requires programmed cell death combined with the cessation of photosynthesis, organelle breakdown, and protein degradation. The regulation of leaf senescence depends largely on the developmental age of plants, although it is also influenced by various external stimuli (Gan and Amasino, 1997). The detection of internal and external signals activates various processes mediated by signaling molecules such as plant hormones and reactive oxygen species (ROS; Buchanan-Wollaston et al., 2003; Lim et al., 2007).
The progression of leaf senescence is associated with the down-regulation of genes involved in chlorophyll biosynthesis, carbon metabolism, and photosynthesis and the up-regulation of genes involved in responses to hormones, ROS, and various stresses (Gepstein, 2004; Lin and Wu, 2004; Buchanan-Wollaston et al., 2005; van der Graaff et al., 2006; Balazadeh et al., 2008; Breeze et al., 2011). The coordinated regulation of gene expression during leaf senescence depends on the combined action of several families of transcription factors (TFs). In particular, genes coding for NO APICAL MERISTEM/ARABIDOPSIS TRANSLATION ACTIVATION FACTOR/CUP-SHAPED COTYLEDON (NAC), zinc finger, WRKY, MYB, and APETALA2/ETHYLENE RESPONSE FACTOR (AP2/ERF) TFs are transcriptionally up-regulated during leaf senescence (Lin and Wu, 2004; Buchanan-Wollaston et al., 2005; Breeze et al., 2011). Functional analyses using Arabidopsis (Arabidopsis thaliana) have revealed that NAC and WRKY TFs, including NAC-like ACTIVATED BY AP3/PI, ANAC092/ORESARA1 (ORE1), ORE1-SISTER1, VND-INTERACTING2, NAC WITH TRANSMEMBRANE MOTIF-LIKE4, JUNGBRUNNEN1, WRKY6, WRKY53, WRKY54, and WRKY75, regulate leaf senescence (Robatzek and Somssich, 2002; Miao et al., 2004; Guo and Gan, 2006; Kim et al., 2009; Balazadeh et al., 2011; Besseau et al., 2012; Lee et al., 2012; Wu et al., 2012). By contrast, the role of AP2/ERF TFs in leaf senescence remains unclear.
AP2/ERF TFs consist of 146 members in Arabidopsis and are involved in responses to various external and internal stimuli. Several AP2/ERFs modulate responses to leaf senescence-associated signaling molecules such as ROS, ethylene, jasmonic acid (JA), abscisic acid (ABA), and cytokinin (Nakano et al., 2006; Mizoi et al., 2012). Gain-of-function studies suggest that RELATED TO ABI3/VP1 and C-REPEAT BINDING FACTOR/DEHYDRATION RESPONSIBLE ELEMENT BINDING1 positively and negatively regulate leaf senescence, respectively (Sharabi-Schwager et al., 2010; Woo et al., 2010). However, knowledge of the molecular mechanisms of these AP2/ERFs in the regulation of leaf senescence is lacking.
Class II ERFs are characterized by the ERF-associated amphiphilic repression (EAR) motif (Ohta et al., 2001). Because class II ERFs repress target gene transcription in the presence of ERF activators in transient gene expression assays (Fujimoto et al., 2000; Ohta et al., 2000, 2001), understanding the regulation of class II ERFs is key to elucidating the complex mechanisms underlying AP2/ERF-mediated gene regulation. In addition to studying the stress response-associated transcriptional control of class II ERF genes, (Suzuki et al., 1998; Yamamoto et al., 1999; Fujimoto et al., 2000; Kitajima et al., 2000; Nishiuchi et al., 2004), we previously analyzed the posttranslational regulation of class II ERFs and showed that a class II ERF from tobacco (Nicotiana tabacum), NtERF3, physically interacts with the ubiquitin-conjugating (UBC) enzyme NtUBC2 (Koyama et al., 2003). Coexpression of the dominant-negative form of NtUBC2 enhances the repressive activity of NtERF3 in transient assays, suggesting that NtUBC2 may regulate NtERF3 activity. By contrast, NtUBC2 does not interact with NtERF2, a transcriptional activator. The function of NtUBC2 in the ubiquitin-proteasome system (Dreher and Callis, 2007; Vierstra, 2009) suggests that NtERF3 may be regulated by proteolysis.
In this study, we demonstrate the involvement of proteasomes in the control of NtERF3 and two homologous Arabidopsis class II ERFs, AtERF4 and AtERF8. We also performed functional and gene expression analyses to clarify the role of these class II ERFs in the progression of leaf senescence.
RESULTS
Rapid Degradation of NtERF3 in Vitro
Because NtERF3 physically and functionally interacts with NtUBC2 (Koyama et al., 2003), we first examined the regulation of NtERF3 by proteolysis in vitro (Fig. 1). NtERF3, NtERF2, and GFP, as a stable protein model, were individually produced in Escherichia coli and purified with affinity resin (Supplemental Fig. S1). Each protein was incubated for 0, 15, 30, and 60 min in a cultured tobacco XD6S cell extract, and protein stability was analyzed with immunoblotting. Whereas NtERF3 was extremely unstable in the tobacco cell extract solution (Fig. 1A), NtERF2 and GFP were stable for 60 min (Fig. 1, B and C). NtERF3 levels remained stable in a bovine serum albumin solution (Fig. 1D), suggesting that NtERF3 is specifically degraded by plant proteins.
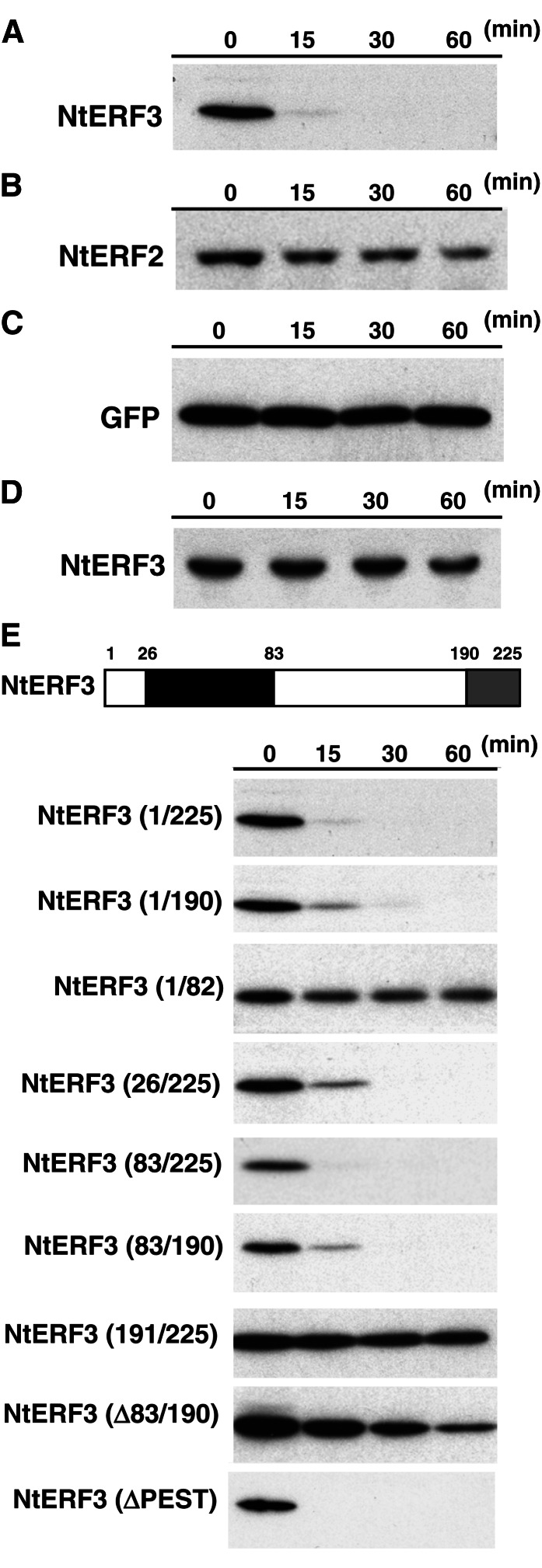
Figure 1.
Rapid degradation of NtERF3 in vitro. A to D, Levels of recombinant NtERF3 (A and D), NtERF2 (B), and GFP (C) incubated in the cell extract (A–C) or bovine serum albumin (D) solutions for the indicated times. Recombinant NtERF3, NtERF2, and GFP were detected via immunoblotting using anti-NtERF3 (A and D), anti-NtERF2 (B), and anti-GFP (C) antibodies, respectively. E, Mapping the domain responsible for NtERF3 instability in vitro. The top section shows a schematic representation of NtERF3. Black and gray boxes indicate the DBD and EAR repression domain, respectively. Numbers show the position of amino acid residues from the first Met. The bottom sections show the levels of the respective regions of NtERF3 at the time points indicated in the cell extract solution. These regions of NtERF3 were detected via immunoblotting using an anti-NtERF3 antibody. The molecular size of each protein is provided in Supplemental Fig. S1.
To identify the amino acid residues involved in the stability of NtERF3, we produced truncated NtERF3 proteins in E. coli (Fig. 1E; Supplemental Fig. S1). NtERF3 contains a conserved DNA-binding domain, an EAR domain, and an NtUBC2-interacting region consisting of amino acid residues 83 to 190 (NtERF3 [83/190]; Fig. 1E; Koyama et al., 2003). Incubating NtERF3 (1/225), NtERF3 (1/190), NtERF3 (26/225), NtERF3 (83/190), and NtERF3 (83/225) in the tobacco cell extract solution caused their rapid degradation, whereas NtERF3 (∆83/190), in which amino acid residues 83 to 190 were deleted, was degraded at a much slower rate. By contrast, NtERF3 (1/82) and NtERF3 (191/225) remained undegraded. These results suggested that NtERF3 (83/190) was essential and sufficient for NtERF3 degradation, whereas the repression and ERF domains were not involved in the stabilization of the protein. NtERF3 amino acid residues 83 to 95 contain a PEST motif, which is involved in the instability of proteins (Asher et al., 2006); however, deletion of the PEST motif did not affect the stability of NtERF3 in vitro (Fig. 1E).
Low Accumulation of NtERF3 in Plant Cells
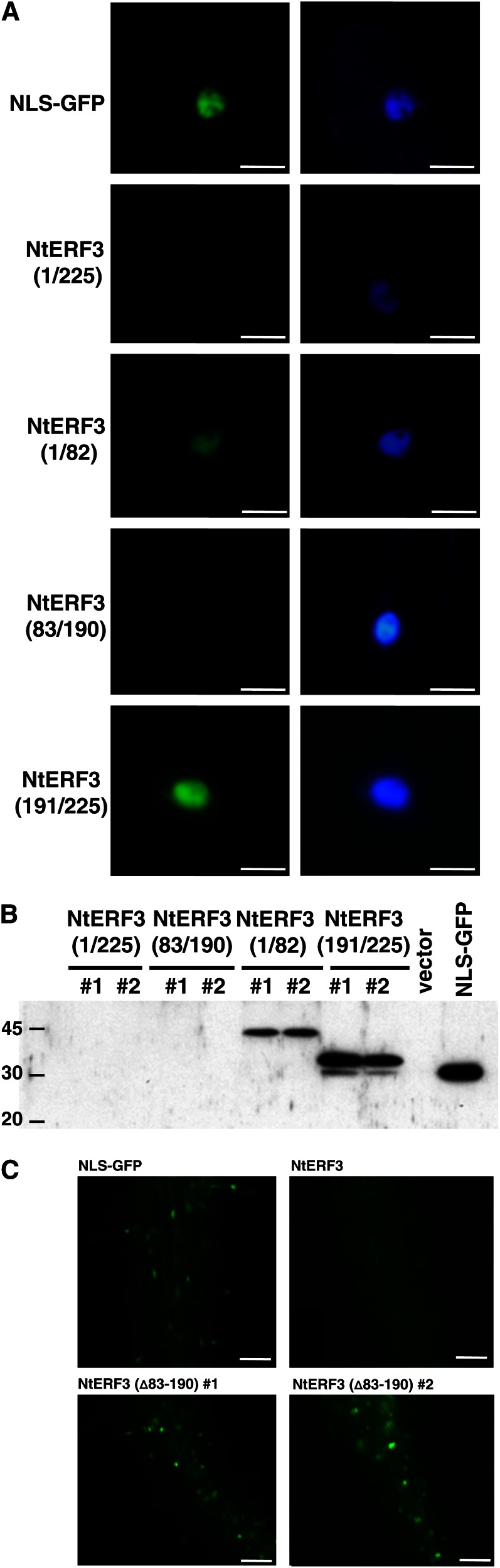
To examine the stability of NtERF3 in plants, we individually fused the coding sequences (CDSs) for the respective regions of NtERF3 with the nuclear localization signal (NLS)-GFP sequence under the control of the Cauliflower mosaic virus 35S promoter (Pro-35S:NLS-GFP), and the resulting fusion genes were transformed into cultured tobacco XD6S cells (Supplemental Fig. S2A). More than 100 kanamycin-resistant tobacco calluses for each construct were examined for GFP florescence. We detected no GFP signal in the Pro-35S:NLS-GFP-NtERF3 and Pro-35S:NLS-GFP-NtERF3 (83/190) tobacco calluses, whereas the Pro-35S:NLS-GFP-NtERF3 (1/82) and Pro-35S:NLS-GFP-NtERF3 (191/225) tobacco calluses showed strong nuclear GFP fluorescence (Fig. 2A). Consistent with these results, immunoblot analysis of protein extracts from Pro-35S:NLS-GFP-NtERF3 (1/82) and Pro-35S:NLS-GFP-NtERF3 (191/225) tobacco calluses showed positive reactivity against an anti-GFP antibody, whereas extracts from Pro-35S:NLS-GFP-NtERF3 and Pro-35S:NLS-GFP-NtERF3 (83/190) tobacco calluses were negative (Fig. 2B).
Figure 2.
Low accumulation of NtERF3 in plant cells. A, GFP florescence in the nucleus of transgenic tobacco cells in which the individual fused gene was introduced (left). 4′,6-Diamidino-2-phenylindole staining (right) shows the location of the nucleus. B, Immunoblot analysis of transgenic tobacco cells after introduction of the fusion genes. Two independent lines per construct were subjected to immunoblot analysis using an anti-GFP antibody. The molecular size markers are shown on the left. C, GFP florescence of Pro-35S:NLS-GFP-NtERF3 and Pro-35S:NLS-GFP-NtERF3 (∆83/190) roots of 3-d-old Arabidopsis seedlings. NLS, Nuclear localization signal. Bars = 10 µm (A) and 50 µm (C).
No kanamycin-resistant Pro-35S:NLS-GFP-NtERF3 (∆83/190) tobacco calluses were recovered, suggesting that the fusion gene may confer a lethal phenotype on tobacco cells and this lethal gene may have a less severe effect in a heterologous system. As expected, two lines of Pro-35S:NLS-GFP-NtERF3 (∆83/190) Arabidopsis plants grew and produced small amounts of seed (Supplemental Fig. S2B; see Fig. 4). Because Pro-35S:NLS-GFP-NtERF3 (∆83/190) Arabidopsis plants exhibited a severe defective phenotype, 3-d-old seedlings were analyzed by florescence microscopy. The cells of Pro-35S:NLS-GFP-NtERF3 (∆83/190) Arabidopsis plants showed strong GFP florescence similar to that of Pro-35S:NLS-GFP Arabidopsis plants; the florescence was absent in Pro-35S:NLS-GFP-NtERF3 Arabidopsis plants (Fig. 2C). These results suggest that NtERF3 levels were low in plant cells and that the NtERF3 (83/190) region may determine protein instability.
Figure 4.
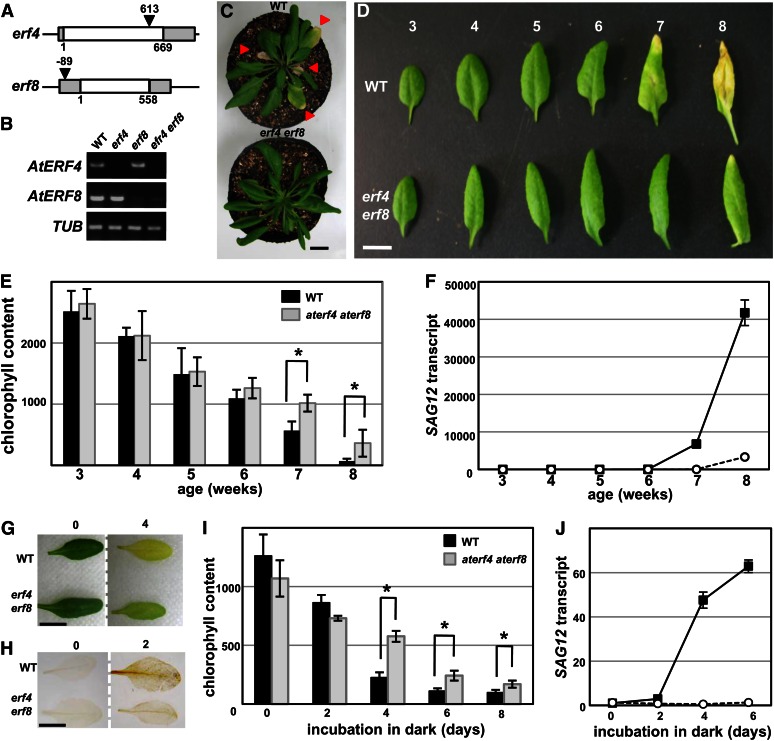
Ectopic expression of NtERF3, AtERF4, and AtERF8 genes induced cell death and precocious leaf senescence. A and B, Wild-type (A) and Pro-35S:NLS-GFP-NtERF3 (∆83/190; B) Arabidopsis plants grown on Murashige and Skoog plates for 3 weeks. C, Wild-type (left) and Pro-35S:NLS-GFP-NtERF3 (right) Arabidopsis plants grown on soil for 5 weeks. Inflorescences were detached for a detailed view. D to F, Wild-type (D), Pro-35S:AtERF4-HA (E), and Pro-35S:AtERF8-HA (F) Arabidopsis plants grown for 4 weeks. G and H, Trypan blue (G) and DAB (H) staining of wild-type, Pro-35S:NLS-GFP-NtERF3 (∆83/190), Pro-35S:AtERF4-HA, and Pro-35S:AtERF8-HA leaves of 3-week-old Arabidopsis plants (from left to right). Bars = 1 cm (A–F) and 0.5 mm (G and H).
Proteasomal Degradation of Class II ERF Repressors
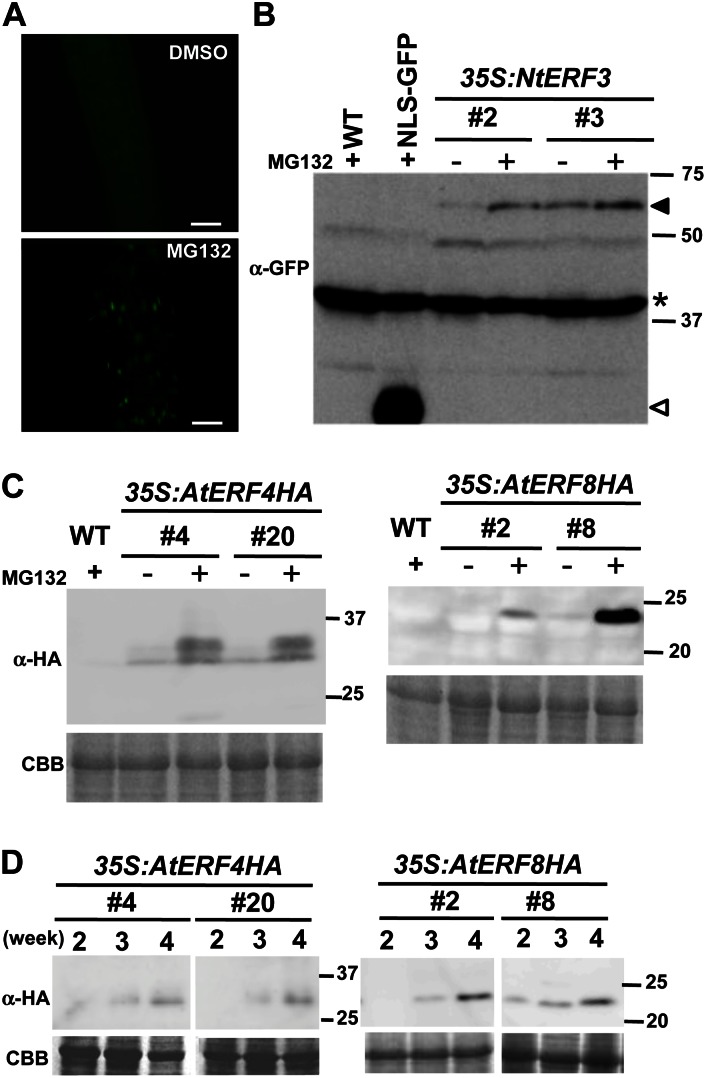
To characterize the mechanism underlying the proteolytic degradation of NtERF3, we treated Pro-35S:NLS-GFP-NtERF3 Arabidopsis plants with MG132, a proteasome inhibitor. Fluorescence microscopy and immunoblot analyses showed that MG132 treatment enhanced the accumulation of NLS-GFP-NtERF3 (Fig. 3A).
Figure 3.
NtERF3, AtERF4, and AtERF8 levels in Arabidopsis plants increased by MG132 and aging. A, GFP florescence of the roots of Pro-35S:NLS-GFP-NtERF3 Arabidopsis seedlings treated with DMSO and MG132. Bars = 50 µm. B and C, Accumulation of NLS-GFP-NtERF3 (B), AtERF4-HA, and AtERF8-HA (C) induced by MG132 treatment. Two independent lines of 3-week-old Pro-35S:NLS-GFP-NtERF3 (35S:NtERF3), Pro-35S:AtERF4-HA, and Pro-35S:AtERF8-HA Arabidopsis plants treated with DMSO (–) and MG132 (+) were subjected to immunoblot analysis using anti-GFP (B) and anti-HA antibodies (C). In B, the black and white triangles indicate NLS-GFP-NtERF3 and NLS-GFP signals, respectively. Signals due to the nonspecific cross reactivity with an unknown protein (B, asterisk) and Coomassie Brilliant Blue (CBB) staining (C) serve as loading controls. Molecular size markers are shown on the right. D, Accumulation of AtERF4-HA and AtERF8-HA induced by plant aging. Pro-35S:AtERF4-HA and Pro-35S:AtERF8-HA Arabidopsis plants were grown on plates for 2, 3, and 4 weeks and subjected to immunoblot using anti-HA antibody. WT, Wild type. [See online article for color version of this figure.]
We further investigated the role of proteasomes in the stability of the NtERF3 homologs AtERF4 and AtERF8 (Ohta et al., 2001). Hemaglutinin (HA)-tagged AtERF4 and AtERF8 under the control of the 35S promoter (Pro-35S:AtERF4-HA and Pro-35S:AtERF8-HA) were separately introduced into Arabidopsis plants (Supplemental Fig. S2B). These transgenic plants were grown for 3 weeks and treated with MG132. Immunoblot analysis showed that MG132 treatment increased AtERF4-HA and AtERF8-HA levels, whereas dimethyl sulfoxide (DMSO) treatment had no effect (Fig. 3, B and C). Furthermore, an upper-shifted band corresponding to AtERF4-HA suggested posttranslational modification of AtERF4 (Fig. 3C). These results provided evidence of the involvement of proteasome in the control of NtERF3, AtERF4, and AtERF8 stabilities.
Interestingly, AtERF4-HA and AtERF8-HA accumulated in aging plants in the absence of MG132 treatment (Fig. 3D). However, unlike MG132-treated plants, no shift of AtERF4-HA was detected in older plants. These results indicated that aging also stabilized AtERF4 and AtERF8.
Cell Death and Precocious Leaf Senescence Induced by Class II ERFs
Our study of the stability of ERF indicated that ectopic expression of class II ERF repressor genes induced a phenotype characterized by cell death and precocious leaf senescence (Fig. 4). We therefore attempted to characterize the functions of class II ERF repressors in Arabidopsis plants. Two Pro-35S:NLS-GFP-NtERF3 (∆83/190) lines of Arabidopsis plants established through several independent transformation trials were fertile but severely deformed (Fig. 4, A and B), indicating the lethality of high NtERF3 accumulation. Transformation of Pro-35S:NLS-GFP-NtERF3, Pro-35S:AtERF4-HA, and Pro-35S:AtERF8-HA genes into Arabidopsis plants induced significant but relatively moderate phenotypes associated with cell death. A significant proportion of Pro-35S:AtERF4-HA (15 of 40 lines) and Pro-35S:AtERF8-HA (13 of 46 lines) Arabidopsis plants exhibited moderate phenotypes with lesions in both cotyledons and leaves, and they often died before flowering. Certain Pro-35S:NLS-GFP-NtERF3 (two lines), Pro-35S:AtERF4-HA (11 lines), and Pro-35S:AtERF8-HA (eight lines) Arabidopsis plants exhibited mild phenotypes with smaller leaves in the seedling stage and precocious senescence in rosette leaves (Fig. 4, C–F).
The Pro-35S:NLS-GFP-NtERF3 (∆83/190), Pro-35S:AtERF4-HA, and Pro-35S:AtERF8-HA leaves with the moderate phenotypes showed strong signals of trypan blue pigment, a dead cell marker (Fig. 4G). These plants also displayed a brown precipitate owing to the reaction of hydrogen peroxide (H2O2) and diaminobenzidine (DAB; Fig. 4H). These results suggested that ectopic expression of NtERF3, AtERF4, and AtERF8 caused cell death and precocious leaf senescence associated with H2O2 production.
Regulation of Genes Involved in Leaf Senescence by AtERF4
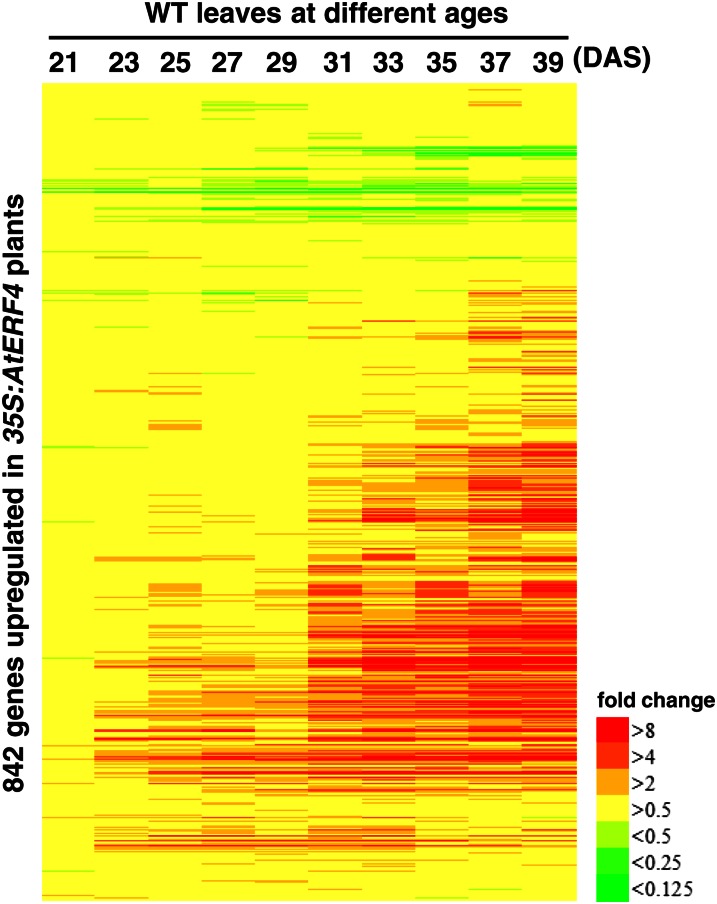
Because ectopic expression of the class II ERF genes examined had a similar effect on inducing cell death and precocious leaf senescence, these ERFs might regulate expression of a common set of downstream genes. Previous studies revealed the induction of AtERF4 expression by ethylene, ABA, and JA, which regulate the progression of leaf senescence (Fujimoto et al., 2000; McGrath et al., 2005; Yang et al., 2005), and then AtERF4 was used as a model for further characterization of downstream genes. Microarray analysis detected 929 genes transcriptionally increased more than 2-fold (P value of dependent Student’s t test < 0.05; false discovery rate [FDR] < 0.04225; see “Materials and Methods”) and 687 genes decreased less than one-half (P < 0.05) in 2-week-old Pro-35S:AtERF4-HA plants compared with Pro-35S:NLS-GFP-HA plants, which showed normal morphology. Comparative analysis of public databases revealed that a considerable number of the genes up-regulated in Pro-35S:AtERF4-HA plants were also up-regulated in the older leaves of the wild type (Fig. 5). The heat map included 842 genes recorded in the data sets (Breeze et al., 2011) of the 929 genes up-regulated in Pro-35S:AtERF4-HA plants and showed that their transcripts increased according to the age of plants (Fig. 5). In addition, reverse transcription (RT)-PCR analysis revealed the up-regulation of senescence-associated genes SAG12 and SAG13 in Pro-35S:AtERF4-HA plants (Supplemental Fig. S3). Whereas the phenotype of Pro-35S:AtERF4-HA plants could be the result of senescence, hypersensitive reaction-like cell death, or other necrosis-related events (Fig. 4), our results suggested that ectopic expression of AtERF4 induced the expression of genes associated with leaf senescence.
Figure 5.
Expression of the genes regulated by AtERF4 in response to plant aging. Genes up-regulated by AtERF4 are aligned along the vertical axis, and the ages of wild-type (WT) Arabidopsis plants are given along the horizontal axis. The bar represents the transcript level of each gene and is colored according to the extent of fold change in the data set (Breeze et al., 2011).
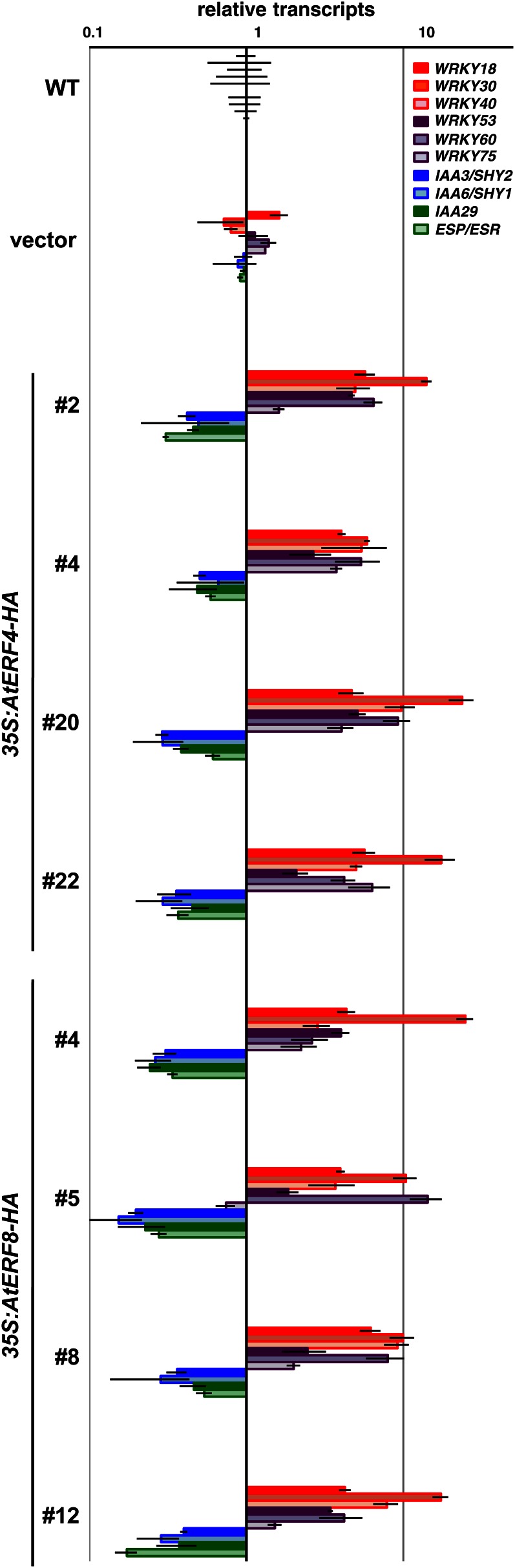
We searched for TF binding sites in the 1,000-bp region upstream of the transcription initiation site of the genes up-regulated in Pro-35S:AtERF4-HA plants and found that the group of genes containing a W-box, which is a binding motif of WRKY TFs, was overrepresented (Supplemental Table S1). Consistently, 17 of 71 WRKY genes were significantly up-regulated in Pro-35S:AtERF4-HA plants (Supplemental Tables S2 and S3). Among them, WRKY30, WRKY53, and WRKY75 were positive regulators of leaf senescence, whereas WRKY18, WRKY40, and WRKY60 were involved in the basal defense response against pathogen attack (Xu et al., 2006; Miao and Zentgraf, 2007; Besseau et al., 2012; Li et al., 2012). In our RT-PCR analysis, WRKY18, WRKY30, WRKY40, WRKY53, WRKY60, and WRKY75 transcripts were increased in Pro-35S:AtERF4-HA and Pro-35S:AtERF8-HA plants (Fig. 6), indicating that AtERF4 and AtERF8 regulated the expression of these genes.
Figure 6.
Expression of the genes downstream of the class II ERFs in 35S:AtERF4-HA and 35S:AtERF8-HA Arabidopsis plants. Transcript levels were determined with RT-PCR of aliquots of total RNAs from wild-type (WT), Pro-35S:NLS-GFP-HA (vector), and four independent lines of Pro-35S:AtERF4-HA and Pro-35S:AtERF8-HA Arabidopsis plants. The wild-type values were set at 1. Error bars indicate sd.
Conversely, Pro-35S:AtERF4-HA plants showed reduced expression of the genes down-regulated in aging plants. Our comparative analysis revealed that AUXIN/INDOLE-3-ACETIC ACID (AUX/IAA) genes, which are negative regulators of auxin responses and down-regulated in senescent leaves (van der Graaff et al., 2006), were overrepresented in the genes down-regulated in Pro-35S:AtERF4-HA plants (Supplemental Tables S2 and S4). Moreover, EPITHIOSPECIFIER PROTEIN/EPITHIOSPECIFYING SENESCENCE REGULATOR (ESP/ESR), a negative regulator of leaf senescence via the antagonistic modulation of WRKY53 activity at the transcriptional and posttranslational levels (Miao and Zentgraf, 2007), was down-regulated in Pro-35S:AtERF4-HA plants. RT-PCR analysis showed that transcripts for IAA3/SHORT HYPOCOTYL2 (SHY2), IAA6/SHY1, IAA29, and ESP/ESR were reduced in Pro-35S:AtERF4-HA and Pro-35S:AtERF8-HA plants (Fig. 6). The gene expression profiles of these plants were consistent with the precocious leaf senescence phenotype.
Delayed Leaf Senescence in an aterf4 aterf8 Double Mutant
To clarify the physiological role of class II ERFs in plants, we established aterf4 and aterf8 single and aterf4 aterf8 double mutants (Fig. 7, A and B). Whereas the single mutants had apparently normal leaves, the aterf4 aterf8 double mutant had irregular, downward curled leaves and delayed leaf yellowing (Fig. 7, C and D; Supplemental Fig. S4). The age-dependent reduction of chlorophyll content and activation of senescence marker gene SAG12 in wild-type leaves was delayed in the leaves of the aterf4 aterf8 mutant (Fig. 7, E and F). A dark-induced senescence assay demonstrated that mature leaves of the wild type incubated in the dark displayed yellowing (Oh et al., 1997; Weaver and Amasino, 2001), whereas those of the aterf4 aterf8 double mutants showed delayed yellowing (Fig. 7G). DAB staining demonstrated that H2O2 accumulated significantly in dark-induced senescing leaves of wild-type plants but to a lesser degree in the aterf4 aterf8 mutant (Fig. 7H). The dark-induced decline of chlorophyll content was also delayed in aterf4 aterf8 mutant leaves, and activation of SAG12 expression was inhibited (Fig. 7, I and J). These results indicated that aterf4 aterf8 delayed leaf senescence.
Figure 7.
Delayed leaf senescence in an aterf4 aterf8 double mutant. A, Schematic diagrams of the AtERF4 and AtERF8 structures and the transfer DNA tag insertions. The white and gray boxes represent coding and untranslated regions, respectively. The triangles show the transfer DNA insertion site. Numbers indicate positions of the transfer DNA tag insertion relative to the translational initiation site. B, AtERF4 and AtERF8 transcripts in aterf4 and aterf8 mutants and aterf4 aterf8 double mutants. The CDSs of AtERF4 and AtERF8 were amplified by RT-PCR of aliquots of total RNAs from 3-week-old wild type (WT) and erf mutants. An extraordinarily low level of AtERF8 transcript was detected in aterf8 mutants and aterf4 aterf8 double mutants. C, Rosettes of 6-week-old wild-type (left) and aterf4 aterf8 (right) plants. Triangles indicate senescing leaves. Inflorescences were detached for detailed view. D, The sixth leaf of wild-type (top) and aterf4 aterf8 (bottom) plants of different ages. E, Chlorophyll content of the sixth leaf of wild-type and aterf4 aterf8 plants at indicated ages. The error bars and asterisks indicate sd (n = 12) and sds at P < 0.001 by Student’s t test. F, SAG12 transcript was determined by RT-PCR of aliquots of total RNAs from the sixth leaves of wild-type (black squares) and aterf4 aterf8 (white circles) plants at the indicated ages. The values of 3-week-old wild-type leaves were set at 1. Error bars indicate sd of technical triplicates. G, Dark-induced senescence in wild-type and aterf4 aterf8 leaves. The detached leaves were photographed on days 0 (left) and 4 (right) after incubation in the dark. H, DAB staining of the detached leaves at 0 (left) and 2 (right) d after incubation in the dark. I, Chlorophyll content in wild-type and aterf4 aterf8 leaves at indicated days after incubation in the dark. The error bars and asterisks indicate sd (n = 7) and sds at P < 0.001 by Student’s t test. J, The SAG12 transcript was determined with RT-PCR using aliquots of total RNAs from wild-type (black squares) and aterf4 aterf8 (white circles) leaves incubated for the indicated days in dark. The values of wild-type leaves at day 0 were set at 1. Error bars indicate sd of technical triplicates. Bars = 1 cm.
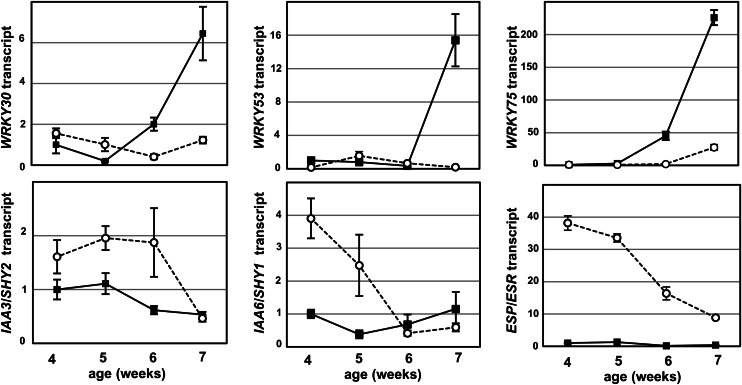
In the leaves of the aterf4 aterf8 mutant, the expression of genes involved in the positive regulation of leaf senescence was down-regulated, whereas that of the negative regulator genes was increased (Fig. 8). WRKY30, WRKY53, and WRKY75 transcripts were low in the mature leaves of the wild type and subsequently increased in older leaves, but were maintained at low levels in mature and old leaves of aterf4 aterf8 mutants. Conversely, IAA3/SHY2, IAA6/SHY6, and ESP/ESR transcripts in mature leaves of the aterf4 aterf8 mutant were increased compared with those of the wild type, whereas these transcripts were gradually reduced in older leaves (Fig. 8; Supplemental Fig. S5). Taken together, these results suggest that the delayed leaf senescence phenotype of the aterf4 aterf8 mutant was responsible for changes in the expression profile.
Figure 8.
Expression of leaf senescence-related genes during the aging of wild-type and aterf4 aterf8 plants. Transcript levels were determined with RT-PCR using aliquots of total RNAs from the sixth leaves of wild-type (black squares) and aterf4 aterf8 (white circles) plants at the indicated ages. The values of 4-week-old wild-type leaves were set at 1. Error bars indicate sd of technical triplicates.
Delayed leaf senescence in aterf4 aterf8 plants was unlikely to have resulted from general growth retardation because flowering time in this mutant was comparable to that of wild-type plants (Supplemental Fig. S6). Furthermore, no defects in the general components of the ethylene and JA response pathways in the aterf4 aterf8 mutant was detected; it progressed at a pace similar to that of the wild type in the presence of 1-aminocyclopropanecarboxylic acid, a precursor of ethylene, and methyl jasmonate (Supplemental Fig. S7).
Binding of AtERF4 and AtERF8 to ESP/ESR
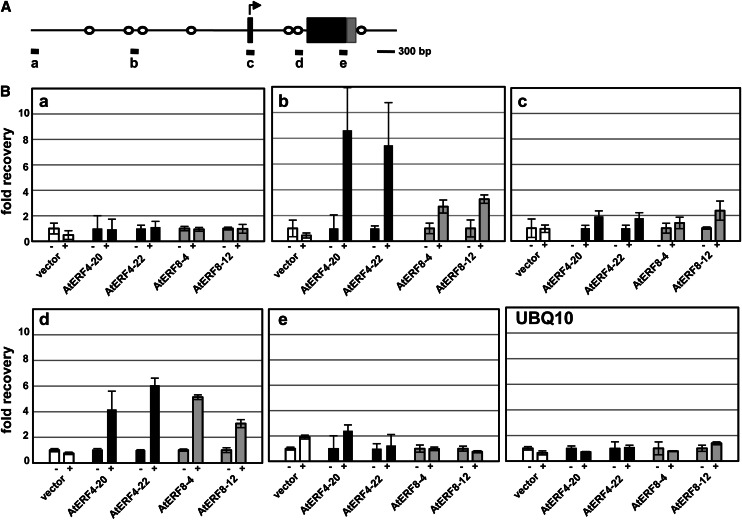
Because AtERF4 and AtERF8 function primarily as transcriptional repressors (Ohta et al., 2001), we presumed that their target genes might be down-regulated in Pro-35S:AtERF4-HA and Pro-35S:AtERF8-HA Arabidopsis plants and up-regulated in aterf4 aterf8 mutants. As described above, the pattern of ESP/ESR expression suggested that AtERF4 and AtERF8 targeted ESP/ESR. A chromatin immunoprecipitation (ChIP) assay of Pro-35S:AtERF4-HA and Pro-35S:AtERF4-HA Arabidopsis plants using anti-HA antibody detected the enrichment of the promoter and intron sequences of ESP/ESR that contain CCGnC motifs, the target sequence of AtERF4 (Fig. 9; Yang et al., 2009). By contrast, the ChIP assay detected no enrichment of the ESP/ESR CDS or the UBIQUITIN10 (UBQ10) sequence (Fig. 9). In addition, the ChIP assay using Pro-35S:NLS-GFP-HA Arabidopsis plants did not detect enrichment of the ESP/ESR sequences (Fig. 9). These results suggested specific binding of AtERF4 and AtERF8 to ESP/ESR in plant cells.
Figure 9.
Binding of AtERF4 and AtERF8 to the genomic region of ESP/ESR. A, A diagram of the genomic region of ESP/ESR. Black and gray boxes indicate the coding and untranslated regions, respectively. White circles show the position of the CCGnC motif, a target sequence of AtERF4. Thick bars below the diagram indicate the regions amplified with sets of primers in the ChIP analysis in B. B, Enrichment of the ESP/ESR sequences in the ChIP analysis. Chromatins were prepared from Pro-35S:NLS-GFP-HA (vector) and two independent lines of Pro-35S:AtERF4-HA (AtERF4) and Pro-35S:AtERF8-HA (AtERF8) Arabidopsis plants, immunoprecipitated in the absence (–) and presence (+) of antibodies, and subjected to quantitative PCR. The fold recovery was relative to the value processed in the absence of antibodies. Error bars indicate sd of technical triplicates.
Induction of AtERF4 and AtERF8 Expression during Leaf Senescence
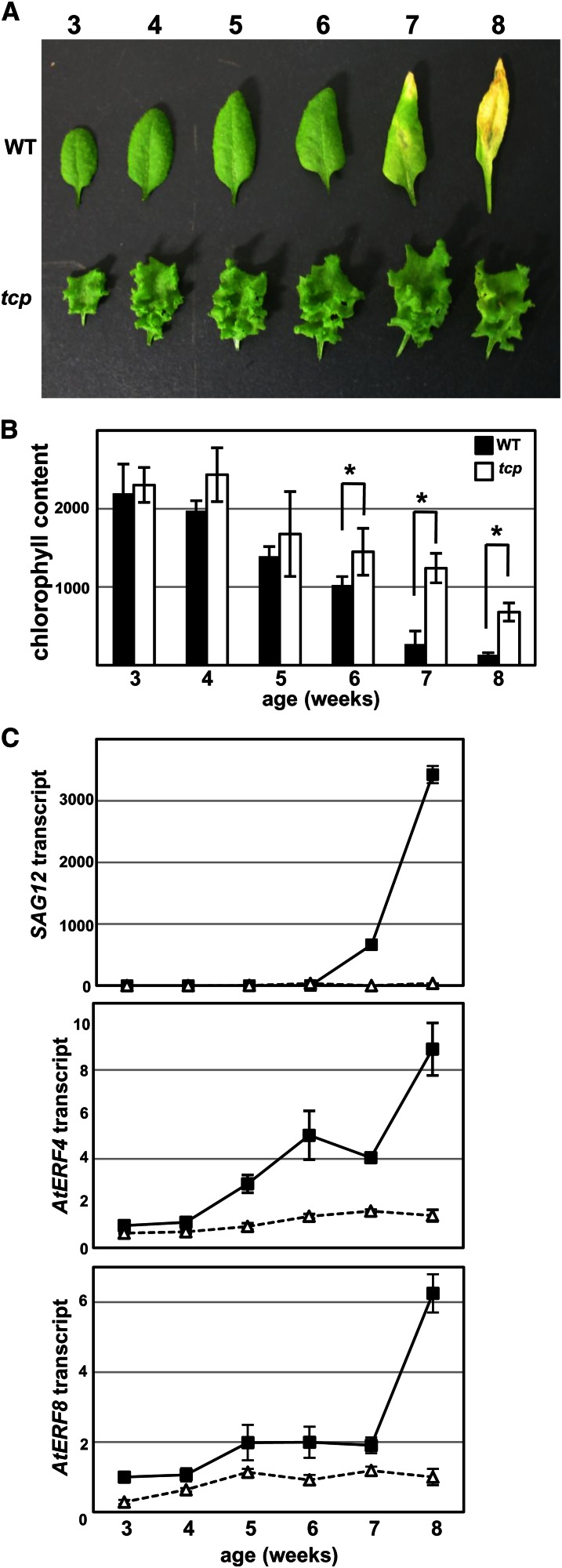
We investigated the transcriptional regulation of AtERF4 and AtERF8 in leaf senescence. Because the TEOSINTE BRANCHED1/CYCLOIDEA/PROLIFERATING CELL NUCLEAR ANTIGEN BINDING FACTOR (TCP) genes are reportedly involved in the progression of leaf senescence (Schommer et al., 2008), we confirmed that compared with the wild type, a tcp3 tcp4 tcp5 tcp10 tcp13 (quintuple tcp) mutant displayed delayed yellowing of leaves, decline in chlorophyll content, and SAG12 activation (Koyama et al., 2010; Fig. 10, A–C), and we used the quintuple tcp mutant as a control for plants with delayed senescence. Our RT-PCR analysis detected basal levels of AtERF4 and AtERF8 transcripts in leaves of 3-old-week wild-type plants (Figs. 7B and 10C; Yang et al., 2005). The increase in AtERF4 and AtERF8 transcripts began 5 weeks after germination and preceded leaf yellowing, but this increase was delayed in the quintuple tcp mutant (Fig. 10C). These results suggested that AtERF4 and AtERF8 expression was induced during leaf senescence.
Figure 10.
Expression of AtERF4 and AtERF8 during leaf senescence of wild-type (WT) and quintuple tcp plants. A, The delayed yellowing of leaves in quintuple tcp mutant. The sixth leaf of wild-type (top) and quintuple tcp (bottom) plants of different ages was photographed. B, Chlorophyll content in the sixth leaf of the wild type and quintuple tcp mutant at indicated ages. The error bars and asterisks indicate sd (n = 12) and sds at P < 0.001 by Student’s t test. C, Expression of SAG12, AtERF4, and AtERF8 in the wild type and the quintuple tcp mutant. Transcript levels were determined with RT-PCR using aliquots of total RNAs from the sixth leaves of the wild type (black squares) and quintuple tcp mutants (white triangles) at the indicated ages. Each value of 3-week-old leaves was set at 1. Error bars indicate sd of technical triplicates.
DISCUSSION
Class II ERF Regulatory Cascade for the Progression of Leaf Senescence
The involvement of class II ERFs in the responses to pathogens, salt, ethylene, JA, and ABA has been reported previously (McGrath et al., 2005; Nasir et al., 2005; Song et al., 2005; Yang et al., 2005; Li et al., 2011). In particular, NbCD1, a Nicotiana benthamiana class II ERF, positively regulates cell death in the defense response of tobacco and Arabidopsis (Nasir et al., 2005). Herein, we expand these findings on the proteasome-mediated control of class II ERFs and describe a novel gene regulatory cascade involving AtERF4 and AtERF8 that progresses leaf senescence.
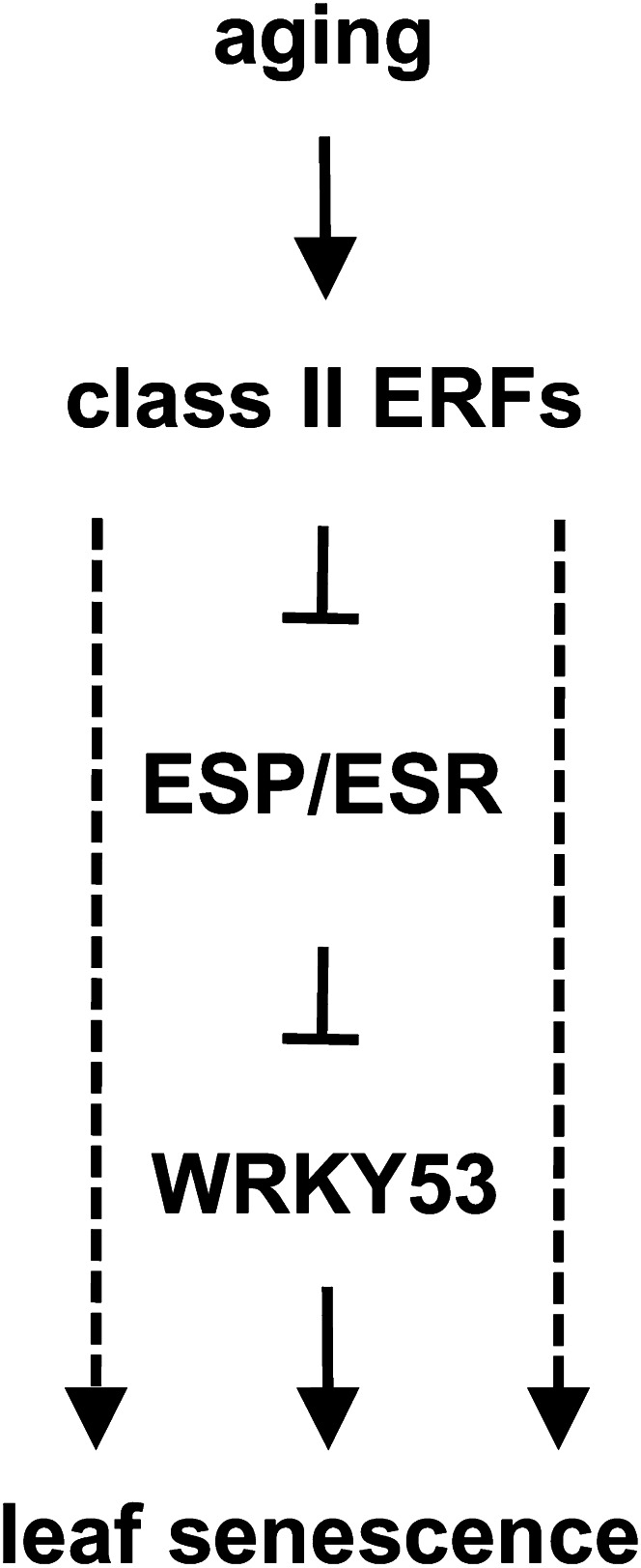
Figure 11A presents our working model of a regulatory cascade involving the class II ERFs that progresses leaf senescence. Our gene expression and ChIP analyses suggest ESP/ESR as a direct target of AtERF4 and AtERF8. The ectopic expression of AtERF4 and AtERF8 transcriptionally repressed ESP/ESR, whereas loss of function of these ERF genes derepressed its expression. The inhibitory effect of ESP/ESR on the progression of leaf senescence depends largely on the activity of WRKY53, a positive regulator of leaf senescence (Miao and Zentgraf, 2007). ESP/ESR inhibits the activity of WRKY53 through physical interaction that may prevent WRKY53 DNA-binding activity and then retards leaf senescence. In addition, ESP/ESR transcriptionally suppresses WRKY53 expression. Because the primary biochemical function of AtERF4 and AtERF8 is the repression of transcription, the repression of ESP/ESR by AtERF4 and AtERF8 may activate WRKY53 and, as a consequent, initiate the progression of leaf senescence. The JA-inducible and ecotype-dependent expression of ESP/ESR has been reported (Lambrix et al., 2001; Miao et al., 2007), but the molecular mechanisms of the regulation are unclear. Our findings of the gradual reduction of ESP/ESR expression during aging (Supplemental Fig. S5) and its regulation by class II ERFs provide novel insights into the regulation of ESP/ESR.
Figure 11.
Model showing the roles of the class II ERFs in the progression of leaf senescence. Schematic representation of a regulatory cascade involving the class II ERFs for the progression of leaf senescence. Aging activates AtERF4 and AtERF8 at both the transcriptional and the posttranslational levels. These ERFs directly repress transcription of ESP/ESR to derepress the activity of WRKY53. WRKY53 and uncharacterized factors downstream of ERFs are involved in the progression of leaf senescence, as indicated by solid and dashed arrows, respectively.
The ectopic expression of AtERF4 and AtERF8 induced expression of WRKY30, WRKY53, and WRKY75 genes, which positively regulated leaf senescence, whereas loss of function of these ERF genes inhibited their expression. By contrast, AtERF4 did not change the expression of WRKY6, another regulator of the progression of leaf senescence, in our microarray analysis (Supplemental Table S3). These results suggest that the expression of WRKY30, WRKY53, and WRKY75 genes are positively regulated by AtERF4 and AtERF8 but not the general effects of the leaf senescence-associated phenotype. In addition to aging and ESP/ESR, salicylate, H2O2, pathogens, a mitogen-activated protein kinase kinase kinase, and histone methylation are involved in the regulation of WRKY53 (Miao et al., 2007; Miao and Zentgraf, 2007; Ay et al., 2009). Interaction between WRKY30 and WRKY53 in yeast (Saccharomyces cerevisiae) two-hybrid system suggests the possible involvement of these WRKYs in the same signaling pathway (Besseau et al., 2012). By contrast, the regulation of WRKY30 and WRKY75 is almost unknown. Our results characterize AtERF4 and AtERF8 as novel regulators of these WRKY genes. Involvement of ESP/ESR and WRKY53 in the regulation of expression of WRKY30 and WRKY75 remains unclear.
Moreover, AtERF4 and AtERF8 might regulate these WRKY genes directly and indirectly in uncharacterized pathways. In addition to ESP/ESR, AtERF4 and AtERF8 negatively regulate the expression of many genes, including AUX/IAA genes. Several studies have reported on both the positive and the negative roles of auxin in the progression of leaf senescence (Ellis et al., 2005; van der Graaff et al., 2006; Hou et al., 2013). Because IAA3/SHY2 suppresses expression of auxin-inducible genes through physical interactions with AUXIN RESPONSE FACTORs (ARFs) and several ARFs act as positive regulators of leaf senescence (Ellis et al., 2005; Weijers et al., 2005), the negative effects on IAA3/SHY2 by AtERF4 and AtERF8 may be involved in the progression of leaf senescence. The repression of AUX/IAA transcriptions by AtERF4 and AtERF8 may be responsible for a specific, rather than a general, role of auxin in the progression of leaf senescence.
Because plants stimulate leaf senescence prematurely under unfavorable conditions (Buchanan-Wollaston et al., 2003; Lim et al., 2007), stress responsive genes downstream of AtERF4 and AtERF8 likely contribute to signaling pathway integration for the regulation of leaf senescence. ESP/ESR, the target of AtERF4 and AtERF8, has important roles in defense against herbivores and pathogens (Lambrix et al., 2001; Miao et al., 2007). WRKY genes are involved in leaf senescence and various stress responses (Rushton et al., 2010). WRKY53 and WRKY75 are involved in responses to biotic stress and nutrient starvation, respectively (Devaiah et al., 2007; Miao and Zentgraf, 2007). WRKY18, WRKY40, and WRKY60 enhance the basal defense system against pathogens (Xu et al., 2006). In addition, our microarray analysis suggested that AtERF4 up-regulates ORE1, ERF1, JASMONATE ZIM-DOMAIN1, ZINC FINGER OF ARABIDOPSIS THALIANA7 (ZAT7), ZAT10, ZAT12, MYB2, and MYB44, which regulate responses to ethylene, JA, ABA, H2O2, and dehydration (Solano et al., 1998; Sakamoto et al., 2004; Davletova et al., 2005; Chini et al., 2007; Ciftci-Yilmaz et al., 2007; Thines et al., 2007; Jung et al., 2008; Kim et al., 2009; Guo and Gan, 2011). Because these stresses affect the progression of leaf senescence, we postulate that these class II ERFs may integrate aging and stress signaling pathways. Whereas the similar phenotype of Pro-35S:NLS-GFP-NtERF3 Arabidopsis plants to those of Pro-35S:AtERF4-HA and Pro-35S:AtERF8-HA Arabidopsis plants suggests the involvement of NtERF3 in the progression of cell death and leaf senescence, the downstream genes of NtERF3 in tobacco plants remain to be clarified.
Proteolytic Control of Class II ERFs by Proteasomes
We demonstrate that aging stimulates expression of AtERF4 and AtERF8 in leaves and increases ERF accumulation. In addition to elucidating the stress response-associated transcriptional control of class II ERFs (Suzuki et al., 1998; Yamamoto et al., 1999; Fujimoto et al., 2000; Kitajima et al., 2000; Nishiuchi et al., 2004), our study adds some insights to the developmental regulation of these ERFs. The induction of AtERF4 and AtERF8 expression through aging is consistent with data from several transcriptome analyses (Breeze et al., 2011), suggesting robust developmental control of AtERF4 and AtERF8 during leaf senescence. The delayed induction of AtERF4 and AtERF8 expression in quintuple tcp mutant leaves suggests the involvement of the TCP genes in the regulation of class II ERFs, although the detailed mechanisms of this induction remain unclear. The progression of leaf senescence requires the coordination of many signals, and the regulation of class II ERFs during senescence is considered to be under the control of a complex process.
Because the class II ERFs act as positive regulators of leaf senescence, our results suggest that both transcriptional activation and posttranslational stabilization of AtERF4 and AtERF8 are important for the progression of senescence. The fine-tuning of their accumulation may set the pace of the progression of leaf senescence. In younger plants, proteolytic regulation maintains low levels of class II ERFs (Fig. 3). The severe cell death observed in the transgenic Arabidopsis plants with constitutive accumulation of NtERF3 (∆83/190) demonstrates that the enhanced and continuous accumulation of class II ERFs causes premature death. AtERF4 and AtERF8 transcripts are maintained at basal levels in younger leaves (Figs. 7B and 10C; Yang et al., 2005), but rapidly activated to cope with various environmental stimuli (McGrath et al., 2005; Nasir et al., 2005; Song et al., 2005; Yang et al., 2005). Therefore, proteolytic regulation may prevent their excess and prolonged accumulation to protect cells against premature death.
Whereas selective protein turnover is regulated by proteasomes during leaf senescence, bulk protein degradation is processed by autophagy (Vierstra, 2009). Prior studies have described the role of several components of the ubiquitin-proteasome system (Woo et al., 2001; Gepstein, 2004; Lin and Wu, 2004; Buchanan-Wollaston et al., 2005; van der Graaff et al., 2006; Peng et al., 2007; Raab et al., 2009; Miao and Zentgraf, 2010; Breeze et al., 2011). In particular, WRKY53 is degraded by a homologous to the E6-AP carboxyl terminus domain ubiquitin ligase (Miao and Zentgraf, 2010). Our present study suggests that proteolytic control of class II ERFs by proteasomes occurs and these factors are involved in the progression of leaf senescence. In addition to the characterization of NtUBC2 as an interaction partner of NtERF3 (Koyama et al., 2003), investigations for identification of components that degrade class II ERFs remain to be performed. These class II ERFs interact with the corepressor TOPLESS to negatively regulate target gene transcription (Causier et al., 2012). The posttranslational control of class II ERFs by proteasomes suggests that a sophisticated mechanism may underlie the progression of leaf senescence.
In summary, we showed the proteasomal regulation, age-dependent accumulation, and regulatory cascade of class II ERFs during leaf development. Our results provide cues of a sophisticated mechanism for the progression of leaf senescence involving class II ERFs.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Growth conditions and the transformation of tobacco (Nicotiana tabacum) XD6S cells and Arabidopsis (Arabidopsis thaliana) plants have been described previously (Yamamoto et al., 1999; Koyama et al., 2010). Arabidopsis ecotype Columbia was used throughout this study unless otherwise indicated. erf4-1 (SALK_073394; Alonso et al., 2003; McGrath et al., 2005) and erf8-1 (FLAG157D10; Wassilewskija; Samson et al., 2002) were crossed three times to Columbia plants before use. The tcp quintuple mutant (tcp3 tcp4 tcp5 tcp10 tcp13) has been described previously (Koyama et al., 2010).
Plasmids
For recombinant protein production in Escherichia coli, the CDSs of NtERF2, NtERF3, GFP, and various regions of NtERF3 were cloned into pET32 (Novagen). For the expression of GFP fusion genes in plant cells, the NLS of the SV-40 T antigen from pGAD424 (Clontech) was inserted upstream of GFP, and the entire or partial CDSs of NtERF3 were cloned downstream of GFP. The resulting fusion GFP genes were individually inserted into pBI101 for the transformation of tobacco XD6S cells and Arabidopsis plants. For the overexpression of AtERF4 and AtERF8, the CDSs without the stop codon of AtERF4 and AtERF8 were respectively ligated into p35SHAG, in which the LDLDLELRLGFA sequence of p35SSRDXG was replaced by the HA sequence and transferred into pBCKH (Mitsuda et al., 2006). The primers used for plasmid construction are listed in Supplemental Table S5.
In Vitro Degradation Assay
The 6× His-tagged NtERFs and GFP were produced in E. coli, purified using a nickel-chelating resin according to manufacturer instructions (Amersham Pharmacia), and dialyzed against a solution containing 20 mm Tris (pH 8.0) and 100 mm NaCl. The cell-free degradation assay was modified from previous reports as described below (Osterlund et al., 2000). For preparation of the extract solution, cultured tobacco XD6S cells were ground in liquid nitrogen, suspended in a buffer containing 50 mm Tris (pH 7.5), 10 mm NaCl, 10 mm MgCl2, 5 mm dithiothreitol, 2 mm ATP, and COMPLETE EDTA-free Protease Inhibitor Cocktail (Promega), and centrifuged at 15,000 rpm for 10 min at 4°C. The supernatant was diluted to 4 mg mL–1 to make the cell extract solution. Aliquots containing 100 ng recombinant NtERFs and GFP were added into 200 µL cell extract solution, incubated at room temperature for 0, 15, 30, or 60 min, mixed in an equal volume of 2× SDS loading buffer (Tris, pH 6.8, 2% SDS (w/v), 1 mm EDTA, and 2-mercaptoethanol), and boiled to terminate the reaction. The proteins were separated by SDS-PAGE, blotted onto Immobilon-P membranes (Millipore), and subjected to immunoblot analysis.
Preparation of Tobacco and Arabidopsis Proteins
For preparation of plant protein samples, tobacco XD6S callus lines at 2 weeks after the inoculation on new plates and Arabidopsis seedlings grown on Murashige and Skoog plates were ground in liquid nitrogen, suspended in extraction buffer (Tris, pH 6.8, 8 m urea, 0.5% SDS (w/v), 1 mm EDTA, and 2-mercaptoethanol), incubated for 2 min at 95°C, and mixed with one-third volume of SDS/urea loading buffer (Tris, pH 6.8, 8 m urea, 0.5% SDS (w/v), 1 mm EDTA, and 2-mercaptoethanol). Aliquots containing 10 and 30 µg of protein from tobacco XD6S cells and Arabidopsis plants, respectively, were separated using SDS-PAGE, blotted onto Immobilon-P membranes, and subjected to immunoblot analysis.
Antibodies and Immunoblot Analysis
For production of anti-NtERF2 and anti-NtERF3 antibodies, the recombinant NtERF2 and NtERF3 proteins fused with maltose-binding protein were produced in E. coli, purified using an amylose resin (New England Laboratory) according to manufacturer instructions, and injected into rabbits. For immunoblot analysis, membranes were incubated with the appropriate primary and secondary antibodies. The antibodies used were as follows: anti-NtERF2 and anti-NtERF3 antibodies (1:2,000), anti-GFP antibody (1:5,000; Medical and Biological Laboratory), peroxidase-conjugated anti-HA antibody (1:500; Roche), and peroxidase-conjugated anti-rabbit IgG (1:10,000; Medical and Biological Laboratory). Immunoreactive proteins were detected using the ECL Plus Western Blotting Kit (GE Healthcare).
Microscopy
The fluorescence images in Figure 2, A and C were, respectively, monitored with an Olympus BHS-RFC as described previously (Ohta et al., 2000) and a Keyence BZ-9000 with a GFP Bandpass filter at a fixed one-third second exposure. Bright field images were obtained with a Leica MZ FL III.
Gene Expression Analysis
Total RNA was prepared from tobacco XD6S cells and Arabidopsis plants using TRIzol reagent, FastRNA Green (Qbiogene), or RNeasy Plant Mini (Qiagen) kits. RNA blot analysis was performed as described previously (Koyama et al., 2003). For RT, templates were reverse transcribed from total RNA using SuperScript II first-strand complementary DNA synthesis (Invitrogen). Real-time PCR was performed using first-strand complementary DNA, a pair of primers (Supplemental Table S5), and iQTM CYBR Green PCR Supermix (Bio-Rad) with a CFX96 real-time PCR system (Bio-Rad). Transcript levels were detected in triplicate using a standard curve derived from the reference sample. The relative values of the transcripts were normalized to the UBQ1 level.
Microarray analysis was performed with the two-color method using aliquots of total RNA from four biological replicates of 2-week-old Pro-35S:AtERF4-HA (line no. 20) and Pro-35S:NLS-GFP-HA Arabidopsis plants with an Agilent Arabidopsis V3 (4x44k) microarray. We used previously described preparation and statistical analyses (Koyama et al., 2010) with the following exceptions. Only genes with average detection values of at least 1.5 in both test and reference samples were analyzed. The P value for each gene was calculated using a dependent Student’s t test. To estimate the FDR, we calculated the Q value from the P value using QVALUE software with the default settings (Storey and Tibshirani, 2003) and selected up- and down-regulated (>2-fold and <0.5-fold) genes with a P value of 0.05 or less (FDR < 0.04225). In Figure 5, the transcript level of each gene at different ages was calculated as relative to that at 19 d after sowing (Breeze et al., 2011), and a clustered heat map was prepared using Cluster3 software (Eisen et al., 1998). To evaluate overrepresentation of some gene lists among up- and down-regulated genes, we performed a binomial test using R (http://www.r-project.org/).
ChIP
Nuclear extract was prepared from 3-week-old Arabidopsis plants, subjected to sonication, and immunoprecipitated with 500 ng anti-HA antibody (clone 3F10; Roche) as described previously (Koyama et al., 2010). The chromatin precipitated was reverse cross linked and purified with ethanol and served as a template for PCR (CFX96 real-time PCR system; Bio-Rad) using an appropriate set of primers (Supplemental Table S5). The values were calculated with a standard curve generated from the input sample and normalized using a background value determined from the eukaryotic INITIATION FACTOR4A sequence. Figure 10 shows a representative from three biological replicates with similar trends.
Chemical Treatment of Plants
For MG132 treatment, Arabidopsis plants were immersed in a solution containing 50 mm MES (pH 5.7), 0.05% (v/v) Tween 20, and 50 µm MG132 (Calbiochem) for 6 h in light. For trypan blue staining, Arabidopsis plants were immersed in trypan blue solution (25% (v/v) lactic acid, 23% (v/v) phenol, and 2.5 mg mL–1 trypan blue [Sigma]) for 1 h and rinsed several times in chloral hydrate solution (25 g chloral hydrate in 10 mL water). For DAB staining, Arabidopsis plants were incubated in DAB buffer (50 mm Tris-acetate, pH 5.0, 0.05% (v/v) Tween 20, 1 mg mL–1 DAB [Serva]) in the dark for 24 h, cleared in boiling ethanol, and rendered transparent in chloral hydrate solution.
Dark-Induced Senescence Assay
The sixth leaves were detached from 5-week-old Arabidopsis plants, floated on water, and incubated in the dark. For chlorophyll extraction, the leaves were soaked in 80% (v/v) acetone. The chlorophyll content was determined via spectrophotometric analysis (Lichtenthaler and Buschmann, 2001) and normalized to the fresh weight of the leaves.
Sequence data from this article can be found in the GenBank or Arabidopsis Genome Initiative data libraries under accession numbers NtERF3 (D38124), AtERF4 (AT3G15210), AtERF8 (AT1G53170), WRKY18 (AT4G31800), WRKY30 (AT5G24110), WRKY40 (AT1G80840), WRKY53 (AT4G23810), WRKY60 (AT2G25000), WRKY75 (AT5G13080), ESP/ESR (AT1G54040), IAA3/SHY2 (AT1G04240), IAA6/SHY1 (AT1G52830), IAA29 (AT4G32280), SAG12 (AT5G45890), SAG13 (AT2G29350), UBQ1 (AT3G52590), UBQ10 (AT4G05320), and eukaryotic INITIATION FACTOR4A (AT3G13920). Microarray data from this article can be found in the National Center for Biotechnology Information Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE41053.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Production and purification of NtERF proteins in E. coli.
Supplemental Figure S2. Expression of NtERF3, AtERF4, AtERF8, and GFP genes in tobacco XD6S callus lines and Arabidopsis plants.
Supplemental Figure S3. Expression of SAG genes in Pro-35S:AtERF4-HA Arabidopsis plants.
Supplemental Figure S4. Leaf senescence of aterf4 and aterf8 single mutants.
Supplemental Figure S5. Expression of ESP/ESR during leaf senescence in wild-type leaves.
Supplemental Figure S6. Timing of flowering in wild-type and aterf4 aterf8 double mutant plants.
Supplemental Figure S7. An ethylene precursor- and methyl jasmonate-stimulated dark-induced senescence of aterf4 aterf8 double mutant leaves.
Supplemental Table S1. Cis-elements enriched in the genes regulated in 35S:AtERF4-HA plants.
Supplemental Table S2. Families containing the genes up- and down-regulated in 35S:AtERF4-HA plants.
Supplemental Table S3. Expression of WRKY genes in 35S:AtERF4-HA plants.
Supplemental Table S4. Expression of AUX/IAA genes in 35S:AtERF4-HA plants.
Supplemental Table S5. Primers used in this study.
Acknowledgments
We thank the Arabidopsis Biological Resource Center and the French National Institute of Agricultural Research for providing seeds, Yuko Takiguchi for her technical contribution regarding the microarray experiment, and Ryouhei Terauchi, Hiroyuki Kanzaki, Hideaki Shinshi, Kaoru Suzuki, Keiichiro Hiratsu, and Kyoko Matsui for helpful discussions.
Glossary
- ROS
reactive oxygen species
- TF
transcription factor
- JA
jasmonic acid
- ABA
abscisic acid
- CDSs
coding sequences
- DMSO
dimethyl sulfoxide
- H2O2
hydrogen peroxide
- DAB
diaminobenzidine
- FDR
false discovery rate
- RT
reverse transcription
- ChIP
chromatin immunoprecipitation
References
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al. (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Asher G, Reuven N, Shaul Y. (2006) 20S proteasomes and protein degradation “by default.” Bioessays 28: 844–849 [DOI] [PubMed] [Google Scholar]
- Ay N, Irmler K, Fischer A, Uhlemann R, Reuter G, Humbeck K. (2009) Epigenetic programming via histone methylation at WRKY53 controls leaf senescence in Arabidopsis thaliana. Plant J 58: 333–346 [DOI] [PubMed] [Google Scholar]
- Balazadeh S, Riaño-Pachón DM, Mueller-Roeber B. (2008) Transcription factors regulating leaf senescence in Arabidopsis thaliana. Plant Biol (Stuttg) 10: 63–75 [DOI] [PubMed] [Google Scholar]
- Balazadeh S, Kwasniewski M, Caldana C, Mehrnia M, Zanor MI, Xue GP, Mueller-Roeber B. (2011) ORS1, an H2O2-responsive NAC transcription factor, controls senescence in Arabidopsis thaliana. Mol Plant 4: 346–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besseau S, Li J, Palva ET. (2012) WRKY54 and WRKY70 co-operate as negative regulators of leaf senescence in Arabidopsis thaliana. J Exp Bot 63: 2667–2679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breeze E, Harrison E, McHattie S, Hughes L, Hickman R, Hill C, Kiddle S, Kim YS, Penfold CA, Jenkins D, et al. (2011) High-resolution temporal profiling of transcripts during Arabidopsis leaf senescence reveals a distinct chronology of processes and regulation. Plant Cell 23: 873–894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchanan-Wollaston V, Earl S, Harrison E, Mathas E, Navabpour S, Page T, Pink D. (2003) The molecular analysis of leaf senescence—a genomics approach. Plant Biotechnol J 1: 3–22 [DOI] [PubMed] [Google Scholar]
- Buchanan-Wollaston V, Page T, Harrison E, Breeze E, Lim PO, Nam HG, Lin JF, Wu SH, Swidzinski J, Ishizaki K, et al. (2005) Comparative transcriptome analysis reveals significant differences in gene expression and signalling pathways between developmental and dark/starvation-induced senescence in Arabidopsis. Plant J 42: 567–585 [DOI] [PubMed] [Google Scholar]
- Causier B, Ashworth M, Guo W, Davies B. (2012) The TOPLESS interactome: a framework for gene repression in Arabidopsis. Plant Physiol 158: 423–438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chini A, Fonseca S, Fernández G, Adie B, Chico JM, Lorenzo O, García-Casado G, López-Vidriero I, Lozano FM, Ponce MR, et al. (2007) The JAZ family of repressors is the missing link in jasmonate signalling. Nature 448: 666–671 [DOI] [PubMed] [Google Scholar]
- Ciftci-Yilmaz S, Morsy MR, Song L, Coutu A, Krizek BA, Lewis MW, Warren D, Cushman J, Connolly EL, Mittler R. (2007) The EAR-motif of the Cys2/His2-type zinc finger protein Zat7 plays a key role in the defense response of Arabidopsis to salinity stress. J Biol Chem 282: 9260–9268 [DOI] [PubMed] [Google Scholar]
- Davletova S, Schlauch K, Coutu J, Mittler R. (2005) The zinc-finger protein Zat12 plays a central role in reactive oxygen and abiotic stress signaling in Arabidopsis. Plant Physiol 139: 847–856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devaiah BN, Karthikeyan AS, Raghothama KG. (2007) WRKY75 transcription factor is a modulator of phosphate acquisition and root development in Arabidopsis. Plant Physiol 143: 1789–1801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreher K, Callis J. (2007) Ubiquitin, hormones and biotic stress in plants. Ann Bot (Lond) 99: 787–822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen MB, Spellman PT, Brown PO, Botstein D. (1998) Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA 95: 14863–14868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis CM, Nagpal P, Young JC, Hagen G, Guilfoyle TJ, Reed JW. (2005) AUXIN RESPONSE FACTOR1 and AUXIN RESPONSE FACTOR2 regulate senescence and floral organ abscission in Arabidopsis thaliana. Development 132: 4563–4574 [DOI] [PubMed] [Google Scholar]
- Fujimoto SY, Ohta M, Usui A, Shinshi H, Ohme-Takagi M. (2000) Arabidopsis ethylene-responsive element binding factors act as transcriptional activators or repressors of GCC box-mediated gene expression. Plant Cell 12: 393–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan S, Amasino RM. (1997) Making sense of senescence (molecular genetic regulation and manipulation of leaf senescence). Plant Physiol 113: 313–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gepstein S. (2004) Leaf senescence—not just a ‘wear and tear’ phenomenon. Genome Biol 5: 212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, Gan S. (2006) AtNAP, a NAC family transcription factor, has an important role in leaf senescence. Plant J 46: 601–612 [DOI] [PubMed] [Google Scholar]
- Guo Y, Gan S. (2011) AtMYB2 regulates whole plant senescence by inhibiting cytokinin-mediated branching at late stages of development in Arabidopsis. Plant Physiol 156: 1612–1619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou K, Wu W, Gan SS. (2013) SAUR36, a small auxin up RNA gene, is involved in the promotion of leaf senescence in Arabidopsis. Plant Physiol 161: 1002–1009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung C, Seo JS, Han SW, Koo YJ, Kim CH, Song SI, Nahm BH, Choi YD, Cheong JJ. (2008) Overexpression of AtMYB44 enhances stomatal closure to confer abiotic stress tolerance in transgenic Arabidopsis. Plant Physiol 146: 623–635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JH, Woo HR, Kim J, Lim PO, Lee IC, Choi SH, Hwang D, Nam HG. (2009) Trifurcate feed-forward regulation of age-dependent cell death involving miR164 in Arabidopsis. Science 323: 1053–1057 [DOI] [PubMed] [Google Scholar]
- Kitajima S, Koyama T, Ohme-Takagi M, Shinshi H, Sato F. (2000) Characterization of gene expression of NsERFs, transcription factors of basic PR genes from Nicotiana sylvestris. Plant Cell Physiol 41: 817–824 [DOI] [PubMed] [Google Scholar]
- Koyama T, Mitsuda N, Seki M, Shinozaki K, Ohme-Takagi M. (2010) TCP transcription factors regulate the activities of ASYMMETRIC LEAVES1 and miR164, as well as the auxin response, during differentiation of leaves in Arabidopsis. Plant Cell 22: 3574–3588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koyama T, Okada T, Kitajima S, Ohme-Takagi M, Shinshi H, Sato F. (2003) Isolation of tobacco ubiquitin-conjugating enzyme cDNA in a yeast two-hybrid system with tobacco ERF3 as bait and its characterization of specific interaction. J Exp Bot 54: 1175–1181 [DOI] [PubMed] [Google Scholar]
- Lambrix V, Reichelt M, Mitchell-Olds T, Kliebenstein DJ, Gershenzon J. (2001) The Arabidopsis epithiospecifier protein promotes the hydrolysis of glucosinolates to nitriles and influences Trichoplusia ni herbivory. Plant Cell 13: 2793–2807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Seo PJ, Lee HJ, Park CM. (2012) A NAC transcription factor NTL4 promotes reactive oxygen species production during drought-induced leaf senescence in Arabidopsis. Plant J 70: 831–844 [DOI] [PubMed] [Google Scholar]
- Li Z, Zhang L, Yu Y, Quan R, Zhang Z, Zhang H, Huang R. (2011) The ethylene response factor AtERF11 that is transcriptionally modulated by the bZIP transcription factor HY5 is a crucial repressor for ethylene biosynthesis in Arabidopsis. Plant J 68: 88–99 [DOI] [PubMed] [Google Scholar]
- Li Z, Peng J, Wen X, Guo H. (2012) Gene network analysis and functional studies of senescence-associated genes reveal novel regulators of Arabidopsis leaf senescence. J Integr Plant Biol 54: 526–539 [DOI] [PubMed] [Google Scholar]
- Lichtenthaler HK, Buschmann C. (2001) Chlorophylls and carotenoids: measurement and characterization by UV-VIS spectroscopy. In Wrolstad RE, Acree TE, An H, Decker EA, Penner MH, Reid DS, Schwartz SJ, Shoemaker CF, Sporns P, eds, Current Protocols in Food Analytical Chemistry. Wiley & Sons, New York [Google Scholar]
- Lim PO, Kim HJ, Nam HG. (2007) Leaf senescence. Annu Rev Plant Biol 58: 115–136 [DOI] [PubMed] [Google Scholar]
- Lin JF, Wu SH. (2004) Molecular events in senescing Arabidopsis leaves. Plant J 39: 612–628 [DOI] [PubMed] [Google Scholar]
- McGrath KC, Dombrecht B, Manners JM, Schenk PM, Edgar CI, Maclean DJ, Scheible WR, Udvardi MK, Kazan K. (2005) Repressor- and activator-type ethylene response factors functioning in jasmonate signaling and disease resistance identified via a genome-wide screen of Arabidopsis transcription factor gene expression. Plant Physiol 139: 949–959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao Y, Laun T, Zimmermann P, Zentgraf U. (2004) Targets of the WRKY53 transcription factor and its role during leaf senescence in Arabidopsis. Plant Mol Biol 55: 853–867 [DOI] [PubMed] [Google Scholar]
- Miao Y, Laun TM, Smykowski A, Zentgraf U. (2007) Arabidopsis MEKK1 can take a short cut: it can directly interact with senescence-related WRKY53 transcription factor on the protein level and can bind to its promoter. Plant Mol Biol 65: 63–76 [DOI] [PubMed] [Google Scholar]
- Miao Y, Zentgraf U. (2007) The antagonist function of Arabidopsis WRKY53 and ESR/ESP in leaf senescence is modulated by the jasmonic and salicylic acid equilibrium. Plant Cell 19: 819–830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao Y, Zentgraf U. (2010) A HECT E3 ubiquitin ligase negatively regulates Arabidopsis leaf senescence through degradation of the transcription factor WRKY53. Plant J 63: 179–188 [DOI] [PubMed] [Google Scholar]
- Mitsuda N, Hiratsu K, Todaka D, Nakashima K, Yamaguchi-Shinozaki K, Ohme-Takagi M. (2006) Efficient production of male and female sterile plants by expression of a chimeric repressor in Arabidopsis and rice. Plant Biotechnol J 4: 325–332 [DOI] [PubMed] [Google Scholar]
- Mizoi J, Shinozaki K, Yamaguchi-Shinozaki K. (2012) AP2/ERF family transcription factors in plant abiotic stress responses. Biochim Biophys Acta 1819: 86–96 [DOI] [PubMed] [Google Scholar]
- Nakano T, Suzuki K, Fujimura T, Shinshi H. (2006) Genome-wide analysis of the ERF gene family in Arabidopsis and rice. Plant Physiol 140: 411–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasir KH, Takahashi Y, Ito A, Saitoh H, Matsumura H, Kanzaki H, Shimizu T, Ito M, Fujisawa S, Sharma PC, et al. (2005) High-throughput in planta expression screening identifies a class II ethylene-responsive element binding factor-like protein that regulates plant cell death and non-host resistance. Plant J 43: 491–505 [DOI] [PubMed] [Google Scholar]
- Nishiuchi T, Shinshi H, Suzuki K. (2004) Rapid and transient activation of transcription of the ERF3 gene by wounding in tobacco leaves: possible involvement of NtWRKYs and autorepression. J Biol Chem 279: 55355–55361 [DOI] [PubMed] [Google Scholar]
- Oh SA, Park JH, Lee GI, Paek KH, Park SK, Nam HG. (1997) Identification of three genetic loci controlling leaf senescence in Arabidopsis thaliana. Plant J 12: 527–535 [DOI] [PubMed] [Google Scholar]
- Ohta M, Matsui K, Hiratsu K, Shinshi H, Ohme-Takagi M. (2001) Repression domains of class II ERF transcriptional repressors share an essential motif for active repression. Plant Cell 13: 1959–1968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta M, Ohme-Takagi M, Shinshi H. (2000) Three ethylene-responsive transcription factors in tobacco with distinct transactivation functions. Plant J 22: 29–38 [DOI] [PubMed] [Google Scholar]
- Osterlund MT, Hardtke CS, Wei N, Deng XW. (2000) Targeted destabilization of HY5 during light-regulated development of Arabidopsis. Nature 405: 462–466 [DOI] [PubMed] [Google Scholar]
- Peng M, Hannam C, Gu H, Bi YM, Rothstein SJ. (2007) A mutation in NLA, which encodes a RING-type ubiquitin ligase, disrupts the adaptability of Arabidopsis to nitrogen limitation. Plant J 50: 320–337 [DOI] [PubMed] [Google Scholar]
- Raab S, Drechsel G, Zarepour M, Hartung W, Koshiba T, Bittner F, Hoth S. (2009) Identification of a novel E3 ubiquitin ligase that is required for suppression of premature senescence in Arabidopsis. Plant J 59: 39–51 [DOI] [PubMed] [Google Scholar]
- Robatzek S, Somssich IE. (2002) Targets of AtWRKY6 regulation during plant senescence and pathogen defense. Genes Dev 16: 1139–1149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rushton PJ, Somssich IE, Ringler P, Shen QJ. (2010) WRKY transcription factors. Trends Plant Sci 15: 247–258 [DOI] [PubMed] [Google Scholar]
- Sakamoto H, Maruyama K, Sakuma Y, Meshi T, Iwabuchi M, Shinozaki K, Yamaguchi-Shinozaki K. (2004) Arabidopsis Cys2/His2-type zinc-finger proteins function as transcription repressors under drought, cold, and high-salinity stress conditions. Plant Physiol 136: 2734–2746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samson F, Brunaud V, Balzergue S, Dubreucq B, Lepiniec L, Pelletier G, Caboche M, Lecharny A. (2002) FLAGdb/FST: a database of mapped flanking insertion sites (FSTs) of Arabidopsis thaliana T-DNA transformants. Nucleic Acids Res 30: 94–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schommer C, Palatnik JF, Aggarwal P, Chételat A, Cubas P, Farmer EE, Nath U, Weigel D. (2008) Control of jasmonate biosynthesis and senescence by miR319 targets. PLoS Biol 6: e230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharabi-Schwager M, Lers A, Samach A, Guy CL, Porat R. (2010) Overexpression of the CBF2 transcriptional activator in Arabidopsis delays leaf senescence and extends plant longevity. J Exp Bot 61: 261–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solano R, Stepanova A, Chao Q, Ecker JR. (1998) Nuclear events in ethylene signaling: a transcriptional cascade mediated by ETHYLENE-INSENSITIVE3 and ETHYLENE-RESPONSE-FACTOR1. Genes Dev 12: 3703–3714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song CP, Agarwal M, Ohta M, Guo Y, Halfter U, Wang P, Zhu JK. (2005) Role of an Arabidopsis AP2/EREBP-type transcriptional repressor in abscisic acid and drought stress responses. Plant Cell 17: 2384–2396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storey JD, Tibshirani R. (2003) Statistical significance for genomewide studies. Proc Natl Acad Sci USA 100: 9440–9445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki K, Suzuki N, Ohme-Takagi M, Shinshi H. (1998) Immediate early induction of mRNAs for ethylene-responsive transcription factors in tobacco leaf strips after cutting. Plant J 15: 657–665 [DOI] [PubMed] [Google Scholar]
- Thines B, Katsir L, Melotto M, Niu Y, Mandaokar A, Liu G, Nomura K, He SY, Howe GA, Browse J. (2007) JAZ repressor proteins are targets of the SCF(COI1) complex during jasmonate signalling. Nature 448: 661–665 [DOI] [PubMed] [Google Scholar]
- van der Graaff E, Schwacke R, Schneider A, Desimone M, Flügge UI, Kunze R. (2006) Transcription analysis of Arabidopsis membrane transporters and hormone pathways during developmental and induced leaf senescence. Plant Physiol 141: 776–792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vierstra RD. (2009) The ubiquitin-26S proteasome system at the nexus of plant biology. Nat Rev Mol Cell Biol 10: 385–397 [DOI] [PubMed] [Google Scholar]
- Weaver LM, Amasino RM. (2001) Senescence is induced in individually darkened Arabidopsis leaves, but inhibited in whole darkened plants. Plant Physiol 127: 876–886 [PMC free article] [PubMed] [Google Scholar]
- Weijers D, Benkova E, Jäger KE, Schlereth A, Hamann T, Kientz M, Wilmoth JC, Reed JW, Jürgens G. (2005) Developmental specificity of auxin response by pairs of ARF and Aux/IAA transcriptional regulators. EMBO J 24: 1874–1885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo HR, Chung KM, Park JH, Oh SA, Ahn T, Hong SH, Jang SK, Nam HG. (2001) ORE9, an F-box protein that regulates leaf senescence in Arabidopsis. Plant Cell 13: 1779–1790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo HR, Kim JH, Kim J, Kim J, Lee U, Song IJ, Kim JH, Lee HY, Nam HG, Lim PO. (2010) The RAV1 transcription factor positively regulates leaf senescence in Arabidopsis. J Exp Bot 61: 3947–3957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu A, Allu AD, Garapati P, Siddiqui H, Dortay H, Zanor MI, Asensi-Fabado MA, Munné-Bosch S, Antonio C, Tohge T, et al. (2012) JUNGBRUNNEN1, a reactive oxygen species-responsive NAC transcription factor, regulates longevity in Arabidopsis. Plant Cell 24: 482–506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Chen C, Fan B, Chen Z. (2006) Physical and functional interactions between pathogen-induced Arabidopsis WRKY18, WRKY40, and WRKY60 transcription factors. Plant Cell 18: 1310–1326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto S, Suzuki K, Shinshi H. (1999) Elicitor-responsive, ethylene-independent activation of GCC box-mediated transcription that is regulated by both protein phosphorylation and dephosphorylation in cultured tobacco cells. Plant J 20: 571–579 [DOI] [PubMed] [Google Scholar]
- Yang Z, Tian L, Latoszek-Green M, Brown D, Wu K. (2005) Arabidopsis ERF4 is a transcriptional repressor capable of modulating ethylene and abscisic acid responses. Plant Mol Biol 58: 585–596 [DOI] [PubMed] [Google Scholar]
- Yang S, Wang S, Liu X, Yu Y, Yue L, Wang X, Hao D. (2009) Four divergent Arabidopsis ethylene-responsive element-binding factor domains bind to a target DNA motif with a universal CG step core recognition and different flanking bases preference. FEBS J 276: 7177–7186 [DOI] [PubMed] [Google Scholar]