Abstract
Targeting protein–protein interactions (PPI) is an emerging field in drug discovery. Dimerization and PPI are essential properties of human immunodeficiency virus (HIV)-1 proteins, their mediated functions, and virus biology. Additionally, dimerization is required for the functional interaction of HIV-1 proteins with many host cellular components. In this study, a bimolecular fluorescence complementation (BiFC)-based screening assay was developed that can quantify changes in dimerization, using HIV-1 viral protein R (Vpr) dimerization as a “proof of concept.” Results demonstrated that Venus Vpr (generated by BiFC Vpr constructs) could be competed off in a dose-dependent manner using untagged, full-length Vpr as a competitor molecule. The change in signal intensity was measured quantitatively through flow cytometry and fluorescence microscopy in a high content screening assay. High content imaging was used to screen a library of small molecules for an effect on Vpr dimerization. Among the tested molecules, a few of the small molecules demonstrate an effect on Vpr dimerization in a dose-dependent manner.
Keywords: BiFC, protein-protein interaction, HIV-1 Vpr, dimerization, drug targets
Introduction
Protein–protein interactions (PPI) are essential for many cellular functions, including cancer and host pathogen interactions.1 Thus, a variety of methods have been developed to evaluate direct PPI in vitro and in vivo for future targeting strategies.2–4 In vitro methods such as copurification and affinity precipitation assays require the removal of proteins from their native environment and are not compatible with high throughput screening (HTS). A number of cell-based assay technologies that are compatible with HTS have been applied to PPI targets including yeast and mammalian 2 hybrid assays, fluorescence resonance energy transfer, bioluminescence resonance energy transfer (BRET), positional biosensors, and protein–fragment complementation assays (PCA).5 The visualization and quantification of direct protein interactions within living cells provide two important advantages over other methods – the protein partners are expressed in their normal cellular environment, and their subcellular localization can be determined. Bimolecular fluorescence complementation (BiFC) approaches use protein interaction partners expressed as chimeras with fragments of a fluorescent protein. BiFC is based on the formation of a fluorescent complex by fragments of fluorescent proteins for which association is facilitated by the interaction between the proteins fused to these fragments. BiFC analysis has been used successfully to study the direct interaction of many different proteins in different cell types and organisms.6–9 Recent studies have also shown that BiFC analysis can be used for HTS to assess the effects of small molecules on protein complexes, providing a method to measure spatial and temporal changes in protein complexes that are a response to drugs.10
Dramatic improvements in treating human immunodeficiency virus-type 1 (HIV-1) infected individuals have been attained with highly active antiretroviral therapy. Most antiretroviral treatment regimes, however, fail to provide long-term suppression of viral replication and therefore do little to control disease progression.11–14 This has prompted investigators to explore small molecule inhibitors that target conserved functions of other viral proteins as well as viral-host interactions. Most of the HIV-1 viral proteins (Gag, Nef, viral protein R [Vpr], reverse transcriptase, integrase, Rev, Env and protease) form dimers and/or oligomers that are critical to their functions in the viral life cycle.15–20 Darunavir, a protease inhibitor, was found to also inhibit protease dimerization; it has also shown decreased escape mutant formation compared to other protease inhibitors.21 Thus, viral protein dimerization is a viable target in HIV-1 and can provide additional potent antivirals. To test this hypothesis and to develop a high-throughput cell-based screening platform, we used HIV-1 Vpr as a “proof of concept” molecule and evaluated a method to block protein dimerization and/or PPI. Furthermore, there are no Food and Drug Administration-approved antivirals that target this protein, despite evidence that HIV-1 Vpr is implicated in pathogenesis;22,23 however, studies are in progress to target HIV-1 Vpr by many investigators.24–28
HIV-1 encoded Vpr, a nonstructural protein, is incorporated into the virus particle and possesses several characteristic features that are known to play important roles in HIV-1 replication. Biochemical and nuclear magnetic resonance studies suggest that Vpr has three alpha helices connected by loops that interact with each other to form dimers and oligomers upon expression.29–32 Many of the functions of Vpr in the cell are carried out by virion-associated Vpr (similar to de novo synthesized Vpr), suggesting that the incorporation of Vpr into virus particles is important in HIV-1 biology.33–35 In infected cells, Vpr is primarily present as dimers, though at higher concentrations it is known to form trimers and oligomers.36,37 Preliminary studies on Vpr dimerization using a BiFC assay system show that dimerization-defective Vpr molecules fail to incorporate into virus particles, suggesting that Vpr dimerization is essential for virion incorporation, interaction with host cellular proteins, and downstream functions of Vpr.38 The focus of this report is first to develop an assay to measure changes in Vpr dimerization and, secondly, to screen two libraries for inhibitors of Vpr dimerization. Results indicate that BiFC-based high content cell-based screening provides a measureable readout that will be useful to screen compound libraries. Two small libraries were screened, one of overlapping Vpr peptides, and another of leucine rotamers designed to mimic alpha helices. Together, these studies validate the usefulness of this assay and potential for future drug screening.
Materials and methods
Cell lines and plasmids
HeLa cell lines were grown in Dulbecco’s modified Eagle medium (Gibco®; Life Technologies, Carlsbad, CA, USA) supplemented with 10% fetal bovine serum (HyClone; Thermo Fisher Scientific, Inc, Waltham, MA, USA), 1% penicillin–streptomycin (Invitrogen; Life Technologies), and 1% L-glutamine (Invitrogen; Life Technologies). The Venus-Vpr chimeric constructs were generated as described.39 Briefly, sequences encoding the amino (residues 1–173; referred to as VN) or carboxyl (residues 155–238; referred to as VC) fragments of Venus fluorescence protein were fused to the N terminus of HIV-1 Vpr via a six alanine linker. Venus-Vpr refers to cells expressing both VC-Vpr and VN-Vpr via transient transfection. HIV-1 Vpr containing Flag-tag was used as “untagged” Vpr in competition assays.
Vpr peptides and leucine rotamers
HIV-1 consensus B VPR peptides (15 amino acids in length, with eleven amino acid overlaps) were obtained from the National Institutes of Health (NIH) AIDS Reagent repository and dissolved according to the datasheet for these peptides. Vpr peptides were dissolved either in phosphate buffered saline (PBS) (at 10 μg/mL) or in dimethyl sulfoxide (DMSO) (at 1 μg/mL) based on the information provided in the Vpr peptide datasheets from NIH, ARRP (AIDS Research and Reagent program). Leucine rotamers were synthesized as small molecular weight mimics of α-helices of proteins and dissolved in DMSO for further use.
Transfection
Cells were transfected with equal quantities of VN-Vpr and VC-Vpr using the PolyJet reagent as suggested (SignaGen Laboratories, Rockville, MD, USA). Five hours post-transfection in screening assays, cells were trypsinized and replated into a 96-well plate at a density of 45,000 cells/mL and treated with Vpr peptides or compounds diluted in an appropriate solvent in triplicate. Twenty-four hours posttreatment, cells were fixed and analyzed.
Western blot and immunofluorescence
Cells were seeded on six-well plates and transfected with a total of 2 μg of plasmid (combinations of Venus-Vpr, Vpr-flag, and empty vector). Cells were lysed, and protein levels were quantitated with a bicinchoninic assay (Pierce Biotechnology, Inc, Rockford, IL, USA). Samples were transferred to a polyvinylidene fluoride membrane (EMD Millipore, Billerica, MA, USA) and probed for Flag, HA-tag, or tubulin using monoclonal antibodies. Membranes were developed using enhanced chemiluminescence substrate (Pierce Biotechnology, Inc). For immunofluorescence, cells were transfected with a total of 2 μg of plasmid (combinations of Venus Vpr, Vpr-Flag, and empty vector) using PolyJet. Eighteen hours post-transfection, cells were fixed and stained with anti-HA and anti-Flag antibody.40 Cell nuclei were stained with Hoescht 33342 (Life Technologies, Grand Island, NY, USA). Fluorescence was detected using an Olympus Fluoview 500 upright microscope (Olympus, Center Valley, PA, USA) with appropriate filters. Spot intensity quantification was performed using MetaMorph II software (Molecular Devices, LLC, Sunnyvale, CA, USA).
High content imaging analysis
Cells were transfected as described above, fixed, and cell nuclei were stained with Hoescht 33342. Cells were stored in PBS until automated fluorescence microscopy analysis with the ArrayScan VTI HCS Reader imaging cytometer (Thermo Scientific Cellomics; Thermo Fisher Scientific, Inc, Waltham, MA, USA). Data were collected in the fluorescein isothiocyanate, tetramethylrhodamine-5-(and-6)-isothiocyanate (TRITC), and 4′,6-diamidino-2-phenylindole dihydrochloride channels, and analyzed using the BioApplications platform (Thermo Fisher Scientific, Inc). Background fluorescence was eliminated through comparison of fluorescein isothiocyanate (FITC) and TRITC channel intensities, and dying cells were excluded based on small nuclear area using the standard protocols developed by the Drug Discovery Institute of University of Pittsburgh. The number of viable cells per field was recorded for each well and used to assess comparative cytotoxicity. Viable cells were analyzed for mean nuclear BiFC intensity. The signal-to-noise ratio (S/N) was calculated as:
| (1) |
The Z′ factor measures the separation band using a combination of the variability and the difference in mean signal between the positive and negative controls.41 The Z factor (Z′) was calculated as:
| (2) |
Data normalization
All Vpr peptides and small molecules were tested in triplicate on each plate. The replicate wells were averaged, and the average between these three wells was used to normalize the data between plates. Based on criteria published by the University of Pittsburgh Drug Discovery Institute, Z-scores were used to normalize the data across multiple plates.42 The Z-score was calculated as follows:
| (3) |
where x is the value to be normalized, μ is the mean, and σ is the SD of the mean. The mean was defined as the average across all replicates of the appropriate solvent-treated, transfected control on a single plate. To compare between plates, the Z-scores from individual plates were averaged for each small molecule. Due to the exploratory nature of the assay, the hit threshold was set at one standard deviation from the mean.
Results
Kinetics of protein expression
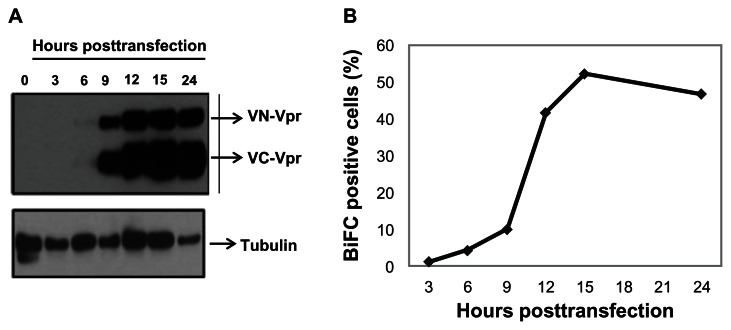
BiFC-based reconstitution of the Venus fluorophore is irreversible. Therefore, it was necessary to understand the kinetics of protein expression in order to time the addition of dimerization inhibitors before the expressed proteins form dimers and/or oligomers. To determine the speed of transfected protein expression, cells were cotransfected with equal amounts of VN-Vpr and VC-Vpr plasmid (referred as Venus-Vpr), collected at various times post-transfection, and assessed by Western blot (Figure 1A). Vpr expression was detected as early as 6 hours post-transfection in HeLa cells; by 12 hours post-transfection, the proteins levels were saturated in cell lysate (Figure 1A). Simultaneous measurement of BiFC within the same culture indicates that only 10% of cells are BiFC-positive and the percentage increases drastically to >40% by 12 to 15 hours post-transfection, remaining steady thereafter (Figure 1B). The delay between protein expression and BiFC fluorescence detection is likely due to the maturation time needed for the reconstitution of the Venus molecule.43
Figure 1.
Vpr expression kinetics and generation of BiFC in transfected cells.
Notes: Cells were cotransfected with equal amounts of VN-Vpr and VC-Vpr plasmids. Hours post-transfection, cells were collected, split into two halves, and used for further analysis. (A) Cells were lysed and analyzed via Western blot using anti-HA antibody to detect Vpr expression. Tubulin was used as loading control. (B) Cells were fixed and analyzed via flow cytometry to detect the percentage of BiFC-positive cells. Results represent one of three independent experiments.
Abbreviations: Vpr, viral protein R; BiFC, bimolecular fluorescence complementation; VN-Vpr, Vpr fused to N-terminus of Venus protein; VC-Vpr, Vpr fused to C-terminus of Venus protein; HA, HA-tagged Vpr.
Competition assay
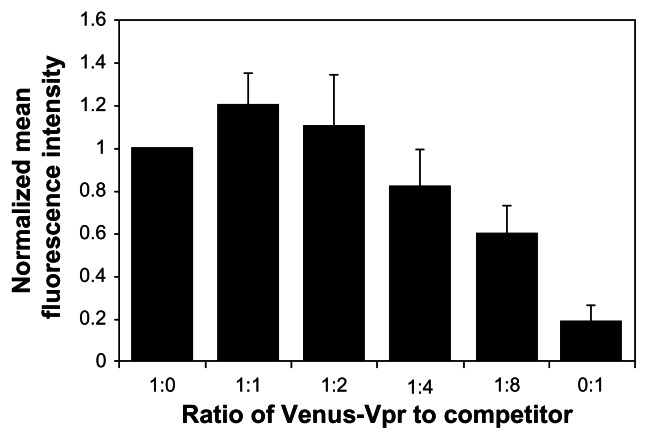
In order to function as a screening tool, the BiFC system must be able to quantitatively detect changes in dimerization levels. In the absence of a known positive control, we used a competition assay to detect a decrease in Vpr dimerization through the BiFC signal. Reconstitution of the fluorophore by dimerization of the Venus-Vpr fusion proteins was competed off by cotransfection of increasing quantities of untagged, full-length Vpr (Vpr-Flag), and BiFC signal was measured. The input of Venus-Vpr was held constant to allow for comparison among the different ratios of Venus-Vpr to competitor. Increasing amounts of untagged Vpr, up to a maximum ratio of 1:8 Venus-Vpr to Vpr-Flag, were assessed at 18 hours post-transfection for their effect on BiFC fluorescence. Results indicate the mean fluorescence intensity (MFI) exhibits a linear decrease as levels of competitor Vpr-flag increase. At the 1:8 ratio, the MFI decreased by 40% compared to Venus-Vpr alone (Figure 2). To verify the level of Venus-Vpr protein expression in these samples, we performed a Western blot on cell lysates from these cotransfected cells, and the results indicated no change in Vpr fusion protein expression (data not shown). Flow cytometry gates were set to capture all BiFC positive cells, independent of intensity. No significant change was observed in the percentage of BiFC positive cells (data not shown), which suggests that transfection efficiency was not affected by the addition of competitor Vpr-flag plasmid.
Figure 2.
Competition assay to detect the loss of BiFC signal generated by Vpr dimerization.
Notes: Cells were transfected with a constant amount of Venus-Vpr plasmid with increasing concentrations of untagged Vpr plasmid. DNA concentration and volume were normalized using empty vector DNA. Eighteen hours post-transfection, cells were collected and analyzed by flow cytometry. Results represent the mean of four independent experiments, and data from each experiment were normalized to the 1:0 (Venus Vpr: Untagged Vpr) sample.
Abbreviations: BiFC, bimolecular fluorescence complementation; Vpr, viral protein R; DNA, deoxyribonucleic acid.
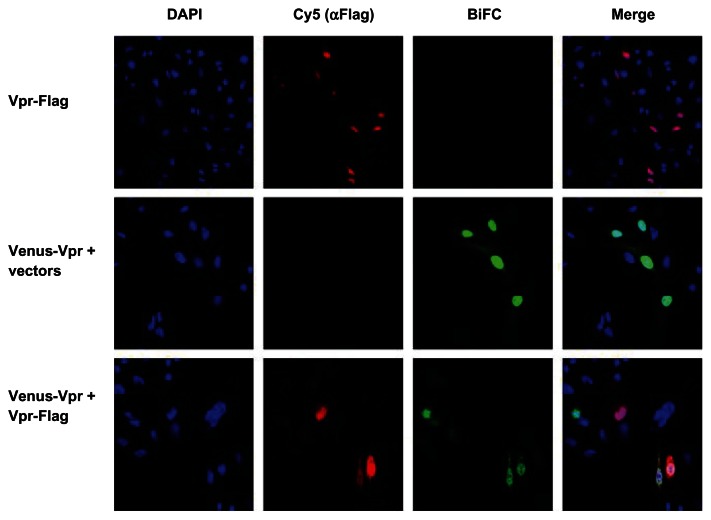
We next assessed the decrease in BiFC signal intensity using immunostaining followed by imaging using a confocal microscope. HeLa cells cotransfected with Venus-Vpr alone or with Vpr-Flag at a 1:8 ratio were fixed and stained with anti-Flag antibody (Figure 3). A visible decrease in BiFC fluorescence intensity was observed in cells cotransfected with Venus-Vpr and Vpr-flag (bottom panel) compared with Venus-Vpr and vector deoxyribonucleic acid (DNA) (middle panel). The average intensity of the BiFC signal in the nucleus for Venus-Vpr transfection without Vpr-flag was 223.34 ± 47.09. For Vpr-flag cotransfected cells, five separate images containing twelve cotransfected cells were analyzed to generate an average pixel intensity of 141.10 ± 22.85. When normalized to the Venus-Vpr transfection, there is a 37% decrease in pixel intensity. These results show that a decrease in dimerization can be quantified at similar levels by both flow cytometry through MFI of BiFC positive cells, and fluorescence microscopy through average pixel intensity.
Figure 3.
Fluorescence intensity of BiFC generated by Venus-Vpr in the presence of competitor untagged Vpr.
Notes: Cells were seeded onto cover slips and transfected with Venus-Vpr plasmids, Vpr-Flag, or Venus-Vpr and Vpr-Flag. Cells were fixed 18 hours post-transfection, stained with Cy5-conjugated Flag M5 antibody to detect the Vpr competitor molecule. Cells were viewed under confocal microscope at 60X magnification. Blue, DAPI to stain the nucleus; green, BiFC signal to visualize Venus-Vpr; red, Flag-Cy5 signal to detect the competitor Vpr molecule.
Abbreviations: BiFC, bimolecular fluorescence complementation; Vpr, viral protein R; DAPI, 4′,6-diamidino-2-phenylindole, dihydrochloride.
Analysis of peptide library
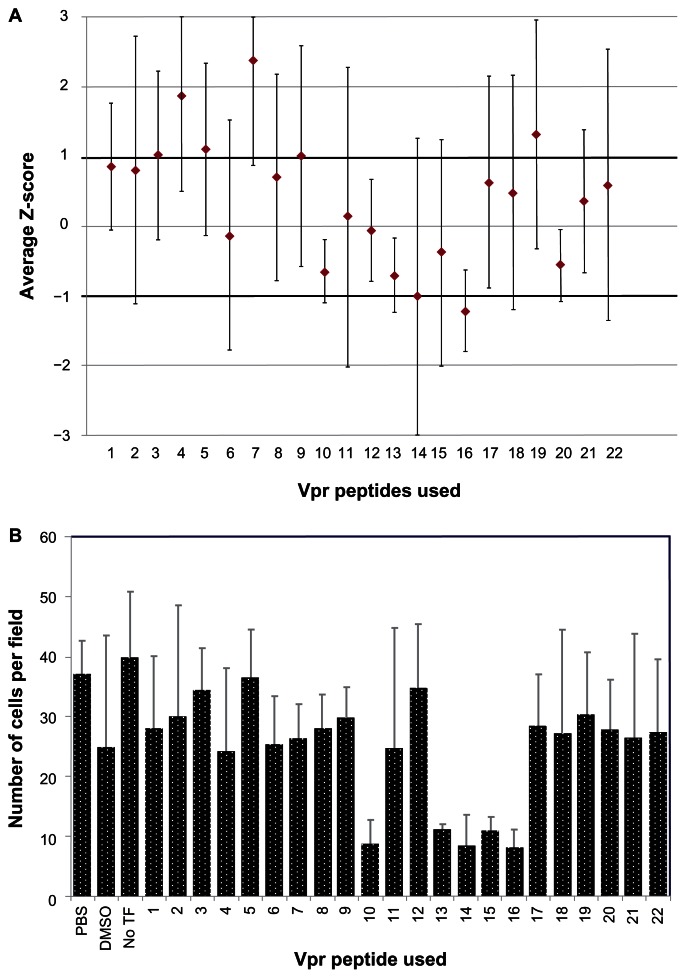
A library of overlapping Vpr peptides was screened by high content imaging analysis for its ability to block Vpr dimerization. Z-scores were calculated and the average scores from four repetitions were plotted (Figure 4A). While six peptides (p4, p5, p7, p14, p16, and p19) showed greater than one SD from the mean BiFC signal intensity, all of the SDs overlapped the variability window and thus could not definitely be considered a deviation from the mean. It is interesting to note that four of these peptides (p4, p5, p7 and p19) showed an increase, suggesting enhanced dimerization/oligomerization, whereas p14 and p16 showed a negative impact on dimerization. Next, we assessed the toxicity of Vpr peptides, as Vpr peptides in certain helices are known to induce apoptosis and cell death.38 Results presented in Figure 4B show the average number of cells per field for each peptide treatment measured using the HCS tool-based cell viability. The peptides dissolved in PBS (p1–9, p11, p17–22) were well tolerated when compared to the PBS vehicle control; however, with the exception of p12, the peptides dissolved in DMSO exhibited significant cytotoxicity compared to the DMSO vehicle control. The DMSO vehicle control averaged 25 cells per field, whereas peptides 10, 13, 14, 15, and 16 averaged between eight and twelve cells per field, suggesting high toxicity. Further analysis indicates that these peptides are part of the third helical domain and the C-terminal domain of Vpr, both of which have been implicated in Vpr-mediated cell death and apoptosis.44,45 Together, these results did not identify a Vpr inhibitory peptide, but rather they identified positive regulators of Vpr dimerization, suggesting these regions of the Vpr molecule might be involved in forming higher orders of oligomerization.
Figure 4.
Ability of Vpr peptides to interfere with Vpr dimerization.
Note: (A) Vpr peptides were screened for an effect on nuclear BiFC fluorescence using high content imaging. Cells transfected with Venus-Vpr plasmids were treated with Vpr peptides or vehicle controls (DMSO or PBS) and assessed for BiFC signal as described in methods. Data were normalized to the solvent-appropriate control cells. The average and standard deviation across four independent experiments were calculated and plotted. (B) The average number of viable cells per field, after excluding cells for morphology and nuclear size using the image software, were recorded for each well (based on the average of >20 fields per well). The figure represents one of four independent experiments.
Abbreviations: Vpr, viral protein R; BiFC, bimolecular fluorescence complementation; PBS, phosphate buffered saline; DMSO, dimethyl sulfoxide; TF, transfection.
Analysis of leucine rotamer library
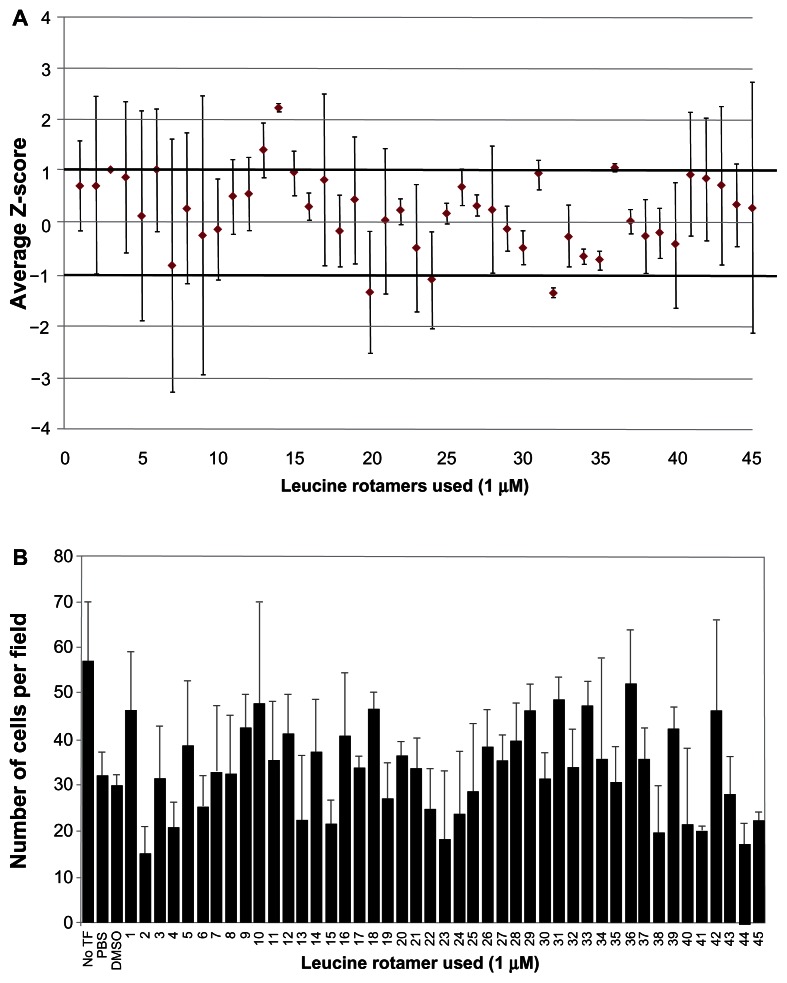
A leucine rotamer library, containing 45 small molecules, was screened using high content image analysis for effects on Vpr dimerization measured by BiFC signal (Figure 5A). Of the 45 leucine rotamers screened, three had greater than one SD separation from the mean. Rotamer 14 had a separation of +2.23 SDs from the transfected control, indicating that it increased the mean BiFC intensity in the nucleus. Rotamer 36 also increased nuclear BiFC signal, but was less potent (separation of +1 SDs from the transfected control) than rotamer 14. Although three rotamers (20, 24, and 32) decreased the nuclear BiFC intensity by more than 1 SD from the mean, the level of variation for 20 and 24 are high; in contrast, rotamer 32 decreased the nuclear BiFC signal by greater than 1 SD. Rotamer 32 showed a separation of −1.37 SDs from the mean. Based on the initial screen, rotamers 14, 32, and 36 were rescreened at higher concentrations (10 μM). The negative effect of rotamer 32 remained the same at both concentrations (data not shown). The positive effect of rotamer 14 on BiFC signal was magnified to greater than 3 SDs at 10 μM. At a 1 μM concentration, rotamer 36 treatment resulted in a slight increase in nuclear BiFC fluorescence; however, at a 10 μM concentration, a negative effect on nuclear BiFC fluorescence was observed. Next, the cytotoxicity of the leucine rotamers was assessed at a 1 μM concentration. Rotamers 2, 4, 15, 41, 44, and 45 had cell counts 20% lower than the DMSO-treated control (Figure 5B). Interestingly, eleven rotamer treatments, including rotamer 36, displayed cell counts over 20% higher than the control. Overall, transfected cells tolerated leucine rotamers well at the concentration used for high content screening analysis.
Figure 5.
Ability of leucine rotamer library to inhibit Vpr dimerization measured by BiFC signal.
Note: (A) Small molecules (leucine rotamers) were screened for their ability to interfere with Vpr dimerization measured by BiFC fluorescence using high content imaging. Cells transfected with Venus-Vpr plasmids were treated with leucine rotamer library (1 μM) or vehicle control (DMSO) and assessed for BiFC signal. Data was normalized to vehicle exposed transfected cells using Z-score methods. The average and standard deviation from two independent experiments were calculated and plotted. (B) The average number of viable cells per field, after excluding cells for morphology and nuclear size, were recorded for each well (based on the average of >20 fields per well) using the image software. Figure represents one of two independent experiments.
Abbreviations: Vpr, viral protein R; BiFC, bimolecular fluorescence complementation; TF, transfection; PBS, phosphate buffered saline; DMSO, dimethyl sulfoxide.
Discussion
Advances in the study of PPI in the last 10 years have opened up promising new lines of research in the field of therapeutics.26,46–48 One specific type of PPI, the formation of dimers, has been identified as a drug target in almost all HIV proteins.49–55 Our laboratory had previously shown that a pair of BiFC plasmids for Vpr produce Vpr dimers that restore the fluorescent molecule when cotransfected.39 Here we assessed whether this BiFC system could be used to detect a decrease in Vpr dimerization using a competition assay; a similar strategy has been used to discover high affinity binders of transcription factors.56 Using a triple plasmid transfection strategy, we cotransfected the two Venus fragment-tagged Vpr plasmids and an untagged Vpr expression plasmid or vector control. At higher input levels of untagged Vpr, a decrease in the MFI, but not the percentage of BiFC-positive cells, was observed. This indicated that the transfection efficiency was the same among the samples, but that triple plasmid transfected cells fluoresce less brightly than cells transfected with DNA vector control. This suggests that untagged Vpr can compete for the formation of dimers in a dose-dependent manner. The MFI decreases as the concentration of untagged Vpr increases, indicating the feasibility of this system as a screening tool. Surprisingly, a slight increase in both the percentage of BiFC-positive cells and the MFI was seen between the 1:1 ratio of Venus-Vpr to competitor and the Venus-Vpr alone. Vpr is known to form dimers, trimers, and hexamers in a concentration-dependent manner;31 therefore, one possible explanation for the increased MFI is the formation of higher-order oligomers.
Imaging-based high content screening using automated fluorescence microscopy has several advantages over other screening methods. The major advantage is the lack of manipulation of the cells, which reduces the background signal in addition to giving us the ability to incorporate other parameters such as toxicity, mitochondrial function, and subcellular distribution of the target protein(s). Another important feature of a high content screen is the Z-score. Z-scores present the difference between a value and the control in terms of SDs from the control mean. The hit threshold of greater than 1 SD from the mean was chosen for three reasons. First, this is a cell-based screen, and the library members may be impermeable to cells. Secondly, the final concentrations of small molecules that we are applying to cells are in the low micromolar range. In vitro HTS assays typically discover hits in the high micromolar or millimolar range. The low concentration of the small molecule treatment may reduce the magnitude of the result. Thirdly, there is no known inhibitor of Vpr dimerization to use as a positive control, and thus no effective dose is available for reference and/or calibration.
The screen of the peptide library yielded five compounds whose Z-scores were greater than 1 SD away from the mean, but all five had large margins of error that extended into the hit threshold. The screen of the leucine rotamer library yielded three compounds with Z-scores greater than 1 SD from the mean whose error bars did not cross the hit threshold. These rotamers (14, 32, and 36) were assessed at a tenfold higher dilution (10 μM) to further evaluate the dose-dependent effect. Rotamers 32 and 36 displayed 67% toxicity at this level, but rotamer 14 remained relatively nontoxic. Rotamer 32 had a negative effect on BiFC intensity, but increasing the concentration of 32 did not increase the magnitude of the effect. Interestingly, 14 showed increased BiFC fluorescence compared to the control, and it responded in a dose-dependent manner when tested at a higher concentration. While this was not the desired outcome of the screen, a compound that increases dimerization of Vpr could have laboratory relevance. Vpr tagged with enhanced green fluorescent protein is used to create fluorescently tagged virus particles for studies on viral entry and uncoating. If an increase in Vpr dimerization/oligomerization results in increased incorporation into the viral particle, this small molecule could be used to increase the overall intensity of fluorescent virions, which would aid in imaging studies.
Taken together, a system to detect the dimerization of HIV-1 Vpr was developed and assessed. It is capable of measuring changes in fluorescence intensity through both flow cytometry and high content imaging. We concluded that BiFC is a valid system for detecting interference with dimerization and moved to a small-scale high content screen. Though we selected Vpr dimerization as a “proof of concept” molecule in this report, BiFC-based HCS can be applied to other HIV-1 viral proteins such as protease and Gag, as well as viral–host protein interactions. Most HIV viral proteins (Gag, Nef, RT, Vpr, and protease) form dimers and/or oligomers, and this unique feature is important for their functions in virus biology.15–19,31 Furthermore, drug resistance mutations within the oligomeric domains of HIV-1 proteins are lethal to their functions, thus disrupting such protein–protein interactions with small molecules will provide probe compounds with strong antiviral effects that are less susceptible to resistance development.
Acknowledgments
This work was supported in part by R01 MH087247 to VA from the NIMH, NIH. The following reagent was obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH: HIV-1 consensus B Vpr (15-mer); peptides – complete set. This project used the UPCI cytometry facility that is supported in part by award P30CA047904.
Footnotes
Disclosure
The authors report no conflicts of interest in this work. The authors have no financial conflicts of interest.
References
- 1.Tafforeau L, Rabourdin-Combe C, Lotteau V. Virus-human cell interactomes. Methods Mol Biol. 2012;812:103–120. doi: 10.1007/978-1-61779-455-1_6. [DOI] [PubMed] [Google Scholar]
- 2.de Chassey B, Meyniel-Schicklin L, Aublin-Gex A, André P, Lotteau V. New horizons for antiviral drug discovery from virus-host protein interaction networks. Curr Opin Virol. 2012;2(5):606–613. doi: 10.1016/j.coviro.2012.09.001. [DOI] [PubMed] [Google Scholar]
- 3.Kar G, Kuzu G, Keskin O, Gursoy A. Protein-protein interfaces integrated into interaction networks: implications on drug design. Curr Pharm Des. 2012;18(30):4697–4705. doi: 10.2174/138161212802651643. [DOI] [PubMed] [Google Scholar]
- 4.Bultinck J, Lievens S, Tavernier J. Protein-protein interactions: network analysis and applications in drug discovery. Curr Pharm Des. 2012;18(30):4619–4629. doi: 10.2174/138161212802651562. [DOI] [PubMed] [Google Scholar]
- 5.Gell DA, Grant RP, Mackay JP. The detection and quantitation of protein oligomerization. Adv Exp Med Biol. 2012;747:19–41. doi: 10.1007/978-1-4614-3229-6_2. [DOI] [PubMed] [Google Scholar]
- 6.Kerppola TK. Design and implementation of bimolecular fluorescence complementation (BiFC) assays for the visualization of protein interactions in living cells. Nat Protoc. 2006;1(3):1278–1286. doi: 10.1038/nprot.2006.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zal T. Visualization of protein interactions in living cells. Self Nonself. 2011;2(2):98–107. doi: 10.4161/self.2.2.17932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ventura S. Bimolecular fluorescence complementation: illuminating cellular protein interactions. Curr Mol Med. 2011;11(7):582–598. doi: 10.2174/156652411800615117. [DOI] [PubMed] [Google Scholar]
- 9.Hollender CA, Liu Z. Bimolecular fluorescence complementation (BiFC) assay for protein-protein interaction in onion cells using the helios gene gun. J Vis Exp. 2010;(40):1963. doi: 10.3791/1963. pii. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.MacDonald ML, Lamerdin J, Owens S, et al. Identifying off-target effects and hidden phenotypes of drugs in human cells. Nat Chem Biol. 2006;2(6):329–337. doi: 10.1038/nchembio790. [DOI] [PubMed] [Google Scholar]
- 11.Yerly S, Rickenbach M, Popescu M, Taffe P, Craig C, Perrin L for Swiss HIV Cohort Study. Drug resistance mutations in HIV-1-infected subjects during protease inhibitor-containing highly active antiretroviral therapy with nelfinavir or indinavir. Antivir Ther. 2001;6(3):185–189. [PubMed] [Google Scholar]
- 12.Hirsch MS, Conway B, D’Aquila RT, et al. Antiretroviral drug resistance testing in adults with HIV infection: implications for clinical management. International AIDS Society – USA Panel. JAMA. 1998;279(24):1984–1991. doi: 10.1001/jama.279.24.1984. [DOI] [PubMed] [Google Scholar]
- 13.Sarmati L, Nicastri E, Parisi SG, et al. Failure of stavudine-lamivudine combination therapy in antiretroviral-naive patients with AZT-like HIV-1 resistance mutations. J Med Virol. 2001;65(4):631–636. doi: 10.1002/jmv.2083. [DOI] [PubMed] [Google Scholar]
- 14.Aleman S, Söderbärg K, Visco-Comandini U, Sitbon G, Sönnerborg A. Drug resistance at low viraemia in HIV-1-infected patients with antiretroviral combination therapy. AIDS. 2002;16(7):1039–1044. doi: 10.1097/00002030-200205030-00010. [DOI] [PubMed] [Google Scholar]
- 15.Hübner W, Chen P, Del Portillo A, Liu Y, Gordon RE, Chen BK. Sequence of human immunodeficiency virus type 1 (HIV-1) Gag localization and oligomerization monitored with live confocal imaging of a replication-competent, fluorescently tagged HIV-1. J Virol. 2007;81(22):12596–12607. doi: 10.1128/JVI.01088-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ye H, Choi HJ, Poe J, Smithgall TE. Oligomerization is required for HIV-1 Nef-induced activation of the Src family protein-tyrosine kinase, Hck. Biochemistry. 2004;43(50):15775–15784. doi: 10.1021/bi048712f. [DOI] [PubMed] [Google Scholar]
- 17.Liu LX, Heveker N, Fackler OT, et al. Mutation of a conserved residue (D123) required for oligomerization of human immunodeficiency virus type 1 Nef protein abolishes interaction with human thioesterase and results in impairment of Nef biological functions. J Virol. 2000;74(11):5310–5319. doi: 10.1128/jvi.74.11.5310-5319.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Srivastava S, Sluis-Cremer N, Tachedjian G. Dimerization of human immunodeficiency virus type 1 reverse transcriptase as an antiviral target. Curr Pharm Des. 2006;12(15):1879–1894. doi: 10.2174/138161206776873590. [DOI] [PubMed] [Google Scholar]
- 19.Wapling J, Moore KL, Sonza S, Mak J, Tachedjian G. Mutations that abrogate human immunodeficiency virus type 1 reverse transcriptase dimerization affect maturation of the reverse transcriptase heterodimer. J Virol. 2005;79(16):10247–10257. doi: 10.1128/JVI.79.16.10247-10257.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Koh Y, Matreyek KA, Engelman A. Differential sensitivities of retroviruses to integrase strand transfer inhibitors. J Virol. 2011;85(7):3677–3682. doi: 10.1128/JVI.02541-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang Y, Liu Z, Brunzelle JS, et al. The higher barrier of darunavir and tipranavir resistance for HIV-1 protease. Biochem Biophys Res Commun. 2011;412(4):737–742. doi: 10.1016/j.bbrc.2011.08.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kogan M, Rappaport J. HIV-1 accessory protein Vpr: relevance in the pathogenesis of HIV and potential for therapeutic intervention. Retrovirology. 2011;8:25. doi: 10.1186/1742-4690-8-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Majumder B, Venkatachari NJ, Srinivasan A, Ayyavoo V. HIV-1 mediated immune pathogenesis: spotlight on the role of viral protein R (Vpr) Curr HIV Res. 2009;7(2):169–177. doi: 10.2174/157016209787581445. [DOI] [PubMed] [Google Scholar]
- 24.Ong EB, Watanabe N, Saito A, et al. Vipirinin, a coumarin-based HIV-1 Vpr inhibitor, interacts with a hydrophobic region of VPR. J Biol Chem. 2011;286(16):14049–14056. doi: 10.1074/jbc.M110.185397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hagiwara K, Murakami T, Xue G, et al. Identification of a novel Vpr-binding compound that inhibits HIV-1 multiplication in macrophages by chemical array. Biochem Biophys Res Commun. 2010;403(1):40–45. doi: 10.1016/j.bbrc.2010.10.107. [DOI] [PubMed] [Google Scholar]
- 26.Suzuki T, Yamamoto N, Nonaka M, et al. Inhibition of human immunodeficiency virus type 1 (HIV-1) nuclear import via Vpr-Importin alpha interactions as a novel HIV-1 therapy. Biochem Biophys Res Commun. 2009;380(4):838–843. doi: 10.1016/j.bbrc.2009.01.180. [DOI] [PubMed] [Google Scholar]
- 27.Schafer EA, Venkatachari NJ, Ayyavoo V. Antiviral effects of mifepristone on human immunodeficiency virus type-1 (HIV-1): targeting Vpr and its cellular partner, the glucocorticoid receptor (GR) Antiviral Res. 2006;72(3):224–232. doi: 10.1016/j.antiviral.2006.06.008. [DOI] [PubMed] [Google Scholar]
- 28.Kamata M, Wu RP, An DS, et al. Cell-based chemical genetic screen identifies damnacanthal as an inhibitor of HIV-1 Vpr induced cell death. Biochem Biophys Res Commun. 2006;348(3):1101–1106. doi: 10.1016/j.bbrc.2006.07.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Morellet N, Bouaziz S, Petitjean P, Roques BP. NMR structure of the HIV-1 regulatory protein VPR. J Mol Biol. 2003;327(1):215–227. doi: 10.1016/s0022-2836(03)00060-3. [DOI] [PubMed] [Google Scholar]
- 30.Wecker K, Morellet N, Bouaziz S, Roques BP. NMR structure of the HIV-1 regulatory protein Vpr in H2O/trifluoroethanol. Comparison with the Vpr N-terminal (1–51) and C-terminal (52–96) domains. Eur J Biochem. 2002;269(15):3779–3788. doi: 10.1046/j.1432-1033.2002.03067.x. [DOI] [PubMed] [Google Scholar]
- 31.Zhao LJ, Wang L, Mukherjee S, Narayan O. Biochemical mechanism of HIV-1 Vpr function. Oligomerization mediated by the N-terminal domain. J Biol Chem. 1994;269(51):32131–32137. [PubMed] [Google Scholar]
- 32.Singh SP, Tomkowicz B, Lai D, et al. Functional role of residues corresponding to helical domain II (amino acids 35 to 46) of human immunodeficiency virus type 1 Vpr. J Virol. 2000;74(22):10650–10657. doi: 10.1128/jvi.74.22.10650-10657.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Poon B, Chang MA, Chen IS. Vpr is required for efficient Nef expression from unintegrated human immunodeficiency virus type 1 DNA. J Virol. 2007;81(19):10515–10523. doi: 10.1128/JVI.00947-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Majumder B, Janket ML, Schafer EA, et al. Human immunodeficiency virus type 1 Vpr impairs dendritic cell maturation and T-cell activation: implications for viral immune escape. J Virol. 2005;79(13):7990–8003. doi: 10.1128/JVI.79.13.7990-8003.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Arokium H, Kamata M, Chen I. Virion-associated Vpr of human immunodeficiency virus type 1 triggers activation of apoptotic events and enhances fas-induced apoptosis in human T cells. J Virol. 2009;83(21):11283–11297. doi: 10.1128/JVI.00756-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fritz JV, Didier P, Clamme JP, et al. Direct Vpr-Vpr interaction in cells monitored by two photon fluorescence correlation spectroscopy and fluorescence lifetime imaging. Retrovirology. 2008;5:87. doi: 10.1186/1742-4690-5-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jenkins Y, Pornillos O, Rich RL, Myszka DG, Sundquist WI, Malim MH. Biochemical analyses of the interactions between human immunodeficiency virus type 1 Vpr and p6(Gag) J Virol. 2001;75(21):10537–10542. doi: 10.1128/JVI.75.21.10537-10542.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pandey RC, Datta D, Mukerjee R, Srinivasan A, Mahalingam S, Sawaya BE. HIV-1 Vpr: a closer look at the multifunctional protein from the structural perspective. Curr HIV Res. 2009;7(2):114–128. doi: 10.2174/157016209787581508. [DOI] [PubMed] [Google Scholar]
- 39.Venkatachari NJ, Walker LA, Tastan O, et al. Human immunodeficiency virus type 1 Vpr: oligomerization is an essential feature for its incorporation into virus particles. Virol J. 2010;7:119. doi: 10.1186/1743-422X-7-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Thotala D, Schafer EA, Tungaturthi PK, et al. Structure-functional analysis of human immunodeficiency virus type 1 (HIV-1) Vpr: role of leucine residues on Vpr-mediated transactivation and virus replication. Virology. 2004;328(1):89–100. doi: 10.1016/j.virol.2004.07.013. [DOI] [PubMed] [Google Scholar]
- 41.Zhang JH, Chung TD, Oldenburg KR. A simple statistical parameter for use in evaluation and validation of high throughput screening assays. J Biomol Screen. 1999;4(2):67–73. doi: 10.1177/108705719900400206. [DOI] [PubMed] [Google Scholar]
- 42.Shun TY, Lazo JS, Sharlow ER, Johnston PA. Identifying actives from HTS data sets: practical approaches for the selection of an appropriate HTS data-processing method and quality control review. J Biomol Screen. 2011;16(1):1–14. doi: 10.1177/1087057110389039. [DOI] [PubMed] [Google Scholar]
- 43.Kerppola TK. Bimolecular fluorescence complementation (BiFC) analysis as a probe of protein interactions in living cells. Annu Rev Biophys. 2008;37:465–487. doi: 10.1146/annurev.biophys.37.032807.125842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Roumier T, Vieira HL, Castedo M, et al. The C-terminal moiety of HIV-1 Vpr induces cell death via a caspase-independent mitochondrial pathway. Cell Death Differ. 2002;9(11):1212–1219. doi: 10.1038/sj.cdd.4401089. [DOI] [PubMed] [Google Scholar]
- 45.Arunagiri C, Macreadie I, Hewish D, Azad A. A C-terminal domain of HIV-1 accessory protein Vpr is involved in penetration, mitochondrial dysfunction and apoptosis of human CD4+ lymphocytes. Apoptosis. 1997;2(1):69–76. doi: 10.1023/a:1026487609215. [DOI] [PubMed] [Google Scholar]
- 46.Christ F, Voet A, Marchand A, et al. Rational design of small-molecule inhibitors of the LEDGF/p75-integrase interaction and HIV replication. Nat Chem Biol. 2010;6(6):442–448. doi: 10.1038/nchembio.370. [DOI] [PubMed] [Google Scholar]
- 47.Cen S, Peng ZG, Li XY, et al. Small molecular compounds inhibit HIV-1 replication through specifically stabilizing APOBEC3G. J Biol Chem. 2010;285(22):16546–16552. doi: 10.1074/jbc.M109.085308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kodama Y, Hu CD. Bimolecular fluorescence complementation (BiFC): a 5-year update and future perspectives. Biotechniques. 2012;53(5):285–298. doi: 10.2144/000113943. [DOI] [PubMed] [Google Scholar]
- 49.Bannwarth L, Rose T, Dufau L, et al. Dimer disruption and monomer sequestration by alkyl tripeptides are successful strategies for inhibiting wild-type and multidrug-resistant mutated HIV-1 proteases. Biochemistry. 2009;48(2):379–387. doi: 10.1021/bi801422u. [DOI] [PubMed] [Google Scholar]
- 50.Camarasa MJ, Velázquez S, San-Félix A, Pérez-Pérez MJ, Gago F. Dimerization inhibitors of HIV-1 reverse transcriptase, protease and integrase: a single mode of inhibition for the three HIV enzymes? Antiviral Res. 2006;71(2–3):260–267. doi: 10.1016/j.antiviral.2006.05.021. [DOI] [PubMed] [Google Scholar]
- 51.DiMattia MA, Watts NR, Stahl SJ, et al. Implications of the HIV-1 Rev dimer structure at 3.2 A resolution for multimeric binding to the Rev response element. Proc Natl Acad Sci U S A. 2010;107(13):5810–5814. doi: 10.1073/pnas.0914946107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Doménech R, Abian O, Bocanegra R, et al. Dendrimers as potential inhibitors of the dimerization of the capsid protein of HIV-1. Biomacromolecules. 2010;11(8):2069–2078. doi: 10.1021/bm100432x. [DOI] [PubMed] [Google Scholar]
- 53.Frankel AD, Chen L, Cotter RJ, Pabo CO. Dimerization of the tat protein from human immunodeficiency virus: a cysteine-rich peptide mimics the normal metal-linked dimer interface. Proc Natl Acad Sci U S A. 1988;85(17):6297–6300. doi: 10.1073/pnas.85.17.6297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hayouka Z, Rosenbluh J, Levin A, et al. Inhibiting HIV-1 integrase by shifting its oligomerization equilibrium. Proc Natl Acad Sci U S A. 2007;104(20):8316–8321. doi: 10.1073/pnas.0700781104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Miller JH, Presnyak V, Smith HC. The dimerization domain of HIV-1 viral infectivity factor Vif is required to block virion incorporation of APOBEC3G. Retrovirology. 2007;4:81. doi: 10.1186/1742-4690-4-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mason JM, Müller KM, Arndt KM. Positive aspects of negative design: simultaneous selection of specificity and interaction stability. Biochemistry. 2007;46(16):4804–4814. doi: 10.1021/bi602506p. [DOI] [PubMed] [Google Scholar]