Abstract
Objective
To study the effect of familial Alzheimer disease (FAD) mutations and APOE genotype on plasma signaling protein levels.
Design
Cross-sectional comparison of plasma levels of 77 proteins measured using multiplex immune assays.
Setting
A tertiary referral dementia research center.
Participants
Thirty-three persons from families harboring PSEN1 or APP mutations, aged 19 to 59 years.
Main Outcome Measures
Protein levels were compared between FAD mutation carriers (MCs) and non-carriers (NCs) and among APOE genotype groups, using multiple linear regression models.
Results
Twenty-one participants were FAD MCs and 12 were NCs. Six had the APOE ε2/3, 6 had the ε3/4, and 21 had the ε3/3 genotype. Levels of 17 proteins differed among APOE genotype groups, and there were significant interactions between age and APOE genotype for 12 proteins. Plasma levels of apolipoprotein E and superoxide dismutase 1 were highest in the ε2 carriers, lowest in ε4 carriers, and intermediate in the ε3 carriers. Levels of multiple interleukins showed the opposite pattern and, among the ε4 carriers, demonstrated significant negative correlations with age. Although there were no significant differences between FAD MCs and NCs, there were interactions between mutation status and APOE genotype for 13 proteins.
Conclusions
We found different patterns of inflammatory markers in young and middle-aged persons among APOE genotype groups. The APOE ε4 carriers had the lowest levels of apolipoprotein E. Young ε4 carriers have increased inflammatory markers that diminish with age. We demonstrated altered inflammatory responses in young and middle adulthood in ε4 carriers that may relate to AD risk later in life.
The prevailing theory of the etiologic factors associated with Alzheimer disease (AD) is that increased relative production or aggregation and/or decreased removal of the 42 amino acid–length cleavage product (Aβ42) of amyloid precursor protein (APP) are key events in initiating the illness.1 There is much evidence supporting this “amyloid hypothesis,” including that APP degradation products are the principal constituents of the plaques that characterize AD brains2 and that the mutations in the PSEN1, PSEN2, and APP genes causing familial AD (FAD) lead to increased relative or absolute production of the Aβ42 peptide.3 The risk-conferring allele of the gene encoding for apolipoprotein E (APOE ε4)4 has also been shown5 to negatively influence transport and aggregation of the Aβ peptide. Apolipoprotein E (ApoE), however, influences many physiological processes and the function most relevant to AD pathogenesis is not clear.
In addition to the hallmark amyloid plaques and neurofibrillary tangles that characterize AD brains, inflammatory changes are also well described. Upregulation of complement, cytokines, and acute-phase reactants occurs near amyloid plaques6 and appears to be an early event. Higher levels of such inflammatory markers have been reported in the cerebrospinal fluid of persons affected by AD7 and in persons carrying FAD mutations.8 Although it is unclear to what degree inflammation is causative of or reactive to more critical pathogenic events in AD, the demonstration that polymorphisms in the gene encoding for complement receptor 1 (CR1),9 complement factor H (CFH),10 and possibly variants in certain interleukins11 as risk factors for AD argue for a key role.
The primary pathology of AD is in the central nervous system, but there is evidence that chemical changes measurable in plasma, including inflammatory markers, may reflect central nervous system changes. Plasma markers that predict the development of AD would be useful in elucidating the presymptomatic stage of the illness and might provide targets for interventions to prevent the development of or hamper progression of the disease. In both human disease12 and animal models of neurodegeneration,13 systemic inflammation is associated with more rapid disease progression. Ray et al14 identified a panel of 18 plasma markers that were useful in distinguishing patients with AD from individuals serving as controls and in predicting which persons with mild cognitive impairment went on to develop AD.
Persons at risk for familial AD due to PSEN1 and APP mutations, in whom the ultimate development of disease can be predicted with essentially 100% certainty, allow us to sensitively detect biochemical changes occurring during the presymptomatic period.15 The APOE genotype is also a risk factor for the development of AD; its variants have differential effects on inflammation, and its relationship to various biological markers may be more relevant to late-onset AD. The goal of the present study was to assess the influence of FAD mutations, APOE genotype, and age on plasma levels of molecules involved in intercellular communication that are potentially relevant to neurodegenerative disease as identified by Ray et al14 and were successfully adapted to a multiplex immunoassay platform.
METHODS
STUDY POPULATION
Thirty-five persons from families harboring PSEN1 or APP mutations who were free of acute illness were enrolled in the present study. Four persons had dementia and the remaining 31 were at 50% risk of inheriting these FAD mutations. All participants underwent in-depth clinical, imaging, and biochemical assessments. Twenty-six participants were from families with PSEN1 mutations, and 9 were from families harboring APP mutations. Participants were from 14 distinct families, of which 12 had a proband with a proven PSEN1 mutation (A431E substitution in 9,16,17 L235V substitution in 1,18 G206A substitution in 1,18 and S212Y substitution in 1), and 2 had a proband with the V717I substitution in APP.
The Clinical Dementia Rating Scale (CDR)19 was performed with an unrelated informant and the participant, with scores of 0.5, 1, 2, and 3 representing questionable, mild, moderate, and severe stages of dementia, respectively. In all but 4 individuals with dementia and 2 who had undergone clinical presymptomatic testing, clinical assessments were performed with the rater blinded to the participant’s genetic status. Participants were informed that they would be tested for APOE genotype and the FAD mutation for which they were at risk but in the context of the research protocol would not be told the result. All participants provided written informed consent. All study procedures were approved by the institutional review board at University of California at Los Angeles.
Blood was drawn in the morning with participants in a fasting state. Thirty milliliters of plasma was centrifuged, aliquotted into 0.5-mL siliconized polypropylene Eppendorf tubes, and stored at −80°C within 2 hours of being drawn. Plasma samples were coded using unique identifiers and stored until being forwarded to Rules-Based Medicine, Inc, which measured the levels of 77 molecules using Luminex platform–based mutiplex immunoassays (Table 1 lists the analytes measured).
Table 1.
List of Proteins Measured
| Proteins Different Among APOE Genotype Groupsa | Proteins With Interaction Between APOE Genotype and FAD Mutation Status | Proteins Measured in >50% of Casesb | ||
|---|---|---|---|---|
| Apolipoprotein E | Apolipoprotein E | Adiponectin | GRO-α | MIP-1α |
| AXL | AXL receptor tyrosine kinase | Agouti-related protein | HCC-4 | MIP-1β |
| BDNF | IGF-1 | Angiopoietin 2 | Hepatocyte growth factor | NrCAM |
| CD5L | Thrombopoietin | Apolipoprotein B | ICAM-1 | Protein S |
| CgA | Fas ligand | Apolipoprotein D | IGF BP-2 | Pulmonary and activation-regulated chemokine |
| EGF | MDC | Apolipoprotein J (clusterin) | IGF-1 | S100β |
| Basic FGF | IL-1α | β-Lymphocyte chemoattractant | IL-16 | SGOT |
| I-309 (CCL1) | IL-7 | Betacellulin | IL-1α | Sortilin |
| IL-12p40 | IL-12p40 | BMP-6 | IL-1β | sRAGE |
| IL-13 | IL-13 | Ciliary neurotrophic factor | IL-7 | Stem cell factor |
| IL-3 | IL-3 | Complement factor H | IL-8 | Thrombopoietin |
| IL-4 | IL-4 | ENA-78 | Insulin | Thymus-expressed chemokine |
| IL-5 | IL-5 | Eotaxin-3 | M-CSF | TIMP-2 |
| IL-15 | Fas | MCP-1 | TRAIL-R3 | |
| RANTES | Fas ligand | MCP-3 | VEGF | |
| Superoxide dismutase 1 | Fetuin A | MDC | Vitronectin | |
| TIMP-1 | FGF-4 | MIF | ||
Abbreviations: AXL, AXL receptor tyrosine kinase; BDNF, brain-derived neurotrophic factor; BMP-6, bone morphogenetic protein 6; CCL1, CC chemokine 1; CD5L, CD5 antigen–like; CgA, chromogranin A; EGF, epidermal growth factor; FAD, familial Alzheimer disease; FGF, fibroblast growth factor; ICAM-1, intracellular adhesion molecule 1; IGF-1, insulinlike growth factor 1; IGF BP-2, insulinlike growth factor binding protein 2; IL, interleukin; M-CSF, macrophage colony-stimulating factor; MDC, macrophage-derived chemokine; MIF, macrophage migration inhibitory factor; MIP-1, macrophage inflammatory protein 1; NrCAM, neuronal cell adhesion molecule; SGOT, serum glutamic oxaloacetic transaminase; sRAGE, receptor for advanced glycosylation end products; TIMP-1/-2, tissue inhibitor of metalloproteinase 1 and 2; TRAIL-R3, TNF-related apoptosis-inducing ligand receptor 3; VEGF, vascular endothelial growth factor.
Differences analyzed by Kruskal-Wallis test (P < .05, false discovery rate < 0.2).
There was no differentiation between FAD mutation carriers and noncarriers, nor among APOE genotype group.
Microspheres were color-coded by varying the ratio of a pair of dyes impregnated into beads. Each bead set was coated with a reagent specific to the molecules of interest, allowing the capture and detection of specific analytes. Following sample incubation and washes, a fluorescently labeled detection antibody was bound. Complexes were analyzed in the Luminex instrument in which beads passed a pair of lasers that detect the reporter dye on the detection antibody and the dye ratio simultaneously. Eight multiplex assays, comprising all 77 target molecules, were run on each plasma sample. Each multiplex run included analyte-specific standards and controls. Further details regarding the methods are provided in the eAppendix (http://www.archneurol.com). All measures were performed without clinical and genetic information.
GENETIC TESTING
The DNA was extracted and APOE genotyping was performed using standard techniques. The presence of A431E and L235V substitutions in PSEN1 were assessed using restriction fragment length polymorphism analyses. The presence or absence of the G206A substitution in PSEN1 (n = 1) was assessed directly with bidirectional sequencing. The presence of the S212Y mutation in an affected person was ascertained using a technique in which the open reading frame of the coding region of the PSEN1 gene was sequenced (Athena Diagnostics). The presence of the V717I substitution in APP was assessed with direct sequencing.
STATISTICAL ANALYSIS
Demographic factors were compared between FAD MCs and NCs and among APOE genotype groups using 1-way analysis of variance or Fisher exact tests. Measurable levels were unobtainable in 50% or more of the participants for 10 of the 77 proteins, and these were therefore excluded from further analyses. For concentrations that were below the limit of detection for the remaining analytes, values were imputed by providing a value 1% below the lowest detected value for that measure. Levels were compared between FAD MCs and NCs and among APOE genotype groups using nonparametric statistical tests (Wilcoxon and Kruskal-Wallis). Next, proteins were transformed as appropriate (eg, log, power transformations based on quantile-quantile plots for each protein) and we constructed multiple linear regression models with covariates for age, sex, APOE genotype, and FAD mutation status. We also individually tested interaction terms between FAD mutation status and APOE genotype, FAD mutation status and age, FAD gene with risk for mutation (PSEN1 vs APP) and APOE genotype, and APOE genotype and age.
Finally, in cases in which we found significant APOE × age interaction effects, we separately computed correlations between analyte levels and age within APOE genotype groups. To control for false positives due to multiple comparisons, the false discovery rate (FDR) was calculated. The R package QVALUE was used to compute estimated FDRs.20 Protein levels with FDR differences of less than 0.2 were considered significant. Statistical analyses were performed in R version 2.6.1 as well as commercial software (PASW Statistics 18.0; SPSS Inc).
RESULTS
Two individuals were excluded because of extreme outlying measures of multiple protein levels. Twenty-five of the 33 remaining participants were women, with ages ranging from 19 to 59 years. Twenty-one participants were FAD MCs and 12 were NCs (Table 2). Six had the APOE ε2/3, 6 had the ε3/4, and 21 had the ε3/3 genotype. Among FAD MCs, 14 were presymptomatic (CDR, 0), 5 had questionable impairment (CDR, 0.5), and 2 had dementia (CDR, >0.5). There were no significant differences in age, adjusted age, sex distribution, or APOE genotype between FAD MCs and NCs (Table 2). Among the 21 FAD MCs, 16 had PSEN1 mutations and 5 carried the V717I APP mutation. Similarly, there were no significant differences in age or distribution of sex or FAD mutation status among APOE genotype groups (Table 3).
Table 2.
Demographics With Regard to FAD Mutation Status
| Characteristic | No. (%)
|
P Value | |
|---|---|---|---|
| FAD Mutation Carriers (n = 21) | FAD Mutation Noncarriers (n = 12) | ||
| Age, median (range), y | 32.8 (19–55) | 38.8 (19–59) | .11 |
| Adjusted age, mean (SD), ya | −12.4 (11.0) | −6.8 (15.9) | .25 |
| Female sex | 16 (76) | 9 (75) | .99 |
| APOE allele | |||
| 2/3 | 5 (24) | 1 (8) | .38 |
| 3/3 | 14 (67) | 7 (58) | .72 |
| 3/4 | 2 (10) | 4 (33) | .16 |
Abbreviation: FAD, familial Alzheimer disease.
Adjusted age is the participants’ ages in relationship to the median age of dementia diagnosis in the family.
Table 3.
Demographics With Regard to APOE Genotype
| Characteristic | No. (%)
|
P Value | ||
|---|---|---|---|---|
| APOE 2/3 (n = 6) | APOE 3/3 (n = 21) | APOE 3/4 (n = 6) | ||
| Age, mean (SD), y | 34.0 (5.8) | 33.3 (11.1) | 41.8 (9.2) | .20 |
| Female sex | 5 (83) | 15 (71) | 5 (83) | .99 |
| FAD MCs | 5 (83) | 14 (67) | 2 (33) | .22 |
Abbreviations: FAD, familial Alzheimer disease; MCs, mutation carriers.
Plasma levels of CFH (3619 vs 2876 μg/mL; P = .02) were elevated in FAD MCs compared with NCs. This difference did not survive correction for the FDR.
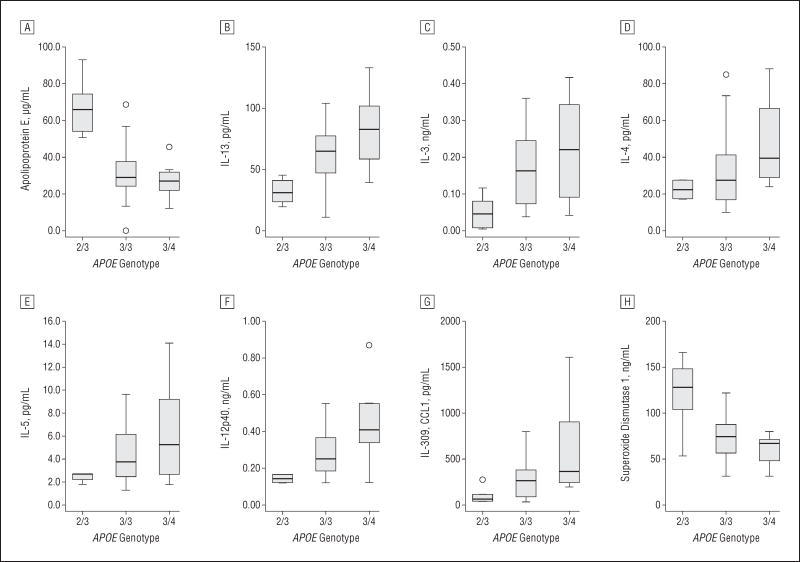
When plasma levels were compared across APOE genotype groups, significant differences were seen in the levels of 17 proteins (in descending order of significance): ApoE, interleukin (IL) 13, epidermal growth factor, IL-15, IL-3, tissue inhibitor of metalloproteinase 1, I-309 (CC chemokine 1), basic fibroblast growth factor, superoxide dismutase 1 (SOD1), chromogranin A, RANTES, CD5 antigen–like, IL-5, brain-derived neurotrophic factor, AXL receptor tyrosine kinase, IL-12p40, and IL-4 (Table 1). Total levels of ApoE were significantly elevated in persons with the APOE ε2/3 genotype relative to the ε3/3 genotype (67.4 vs 34.1 μg/mL; P = .002) and higher in carriers of the ε3/3 relative to the ε3/4 genotype (34.1 vs 27.3 μg/mL; P = .02) (Figure 1A). Levels of IL-13 were significantly elevated in persons with the ε3/3 relative to the ε2/3 genotype (65.6 vs 31.9 pg/mL, P = .005) and higher in persons with the ε3/4 relative to the ε3/3 genotype (85.9 vs 65.6 pg/mL; P = .005) (Figure 1B). The trend for higher plasma interleukin levels in APOE ε3/4 carriers relative to APOE ε3/3 carriers and APOE ε3/3 carriers relative to APOE ε2/3 carriers held for IL-3, IL-4, IL-5, and IL-12p40 (Figure 1C, D, E, and F, respectively). Levels of I-309 were significantly elevated in persons with the APOE ε3/3 relative to ε2/3 genotype (291.7 vs 108.1 pg/mL; P = .02) and in carriers of the APOE ε3/4 relative to the ε3/3 genotype (614.8 vs 291.7 pg/mL; P = .02) (Figure 1G). The SOD1 levels were significantly higher in APOE ε2/3 carriers than in ε3/3 carriers (121.2 vs 73.5 ng/mL; P = .02) and in ε3/3 carriers than in ε3/4 carriers (73.5 vs 60.8 ng/mL; P = .02 (Figure 1H). When covariance analysis was performed for the gene with risk for mutation, the levels of 13 proteins were still significantly related to APOE genotype, although 13 proteins also showed a relationship to the gene containing the mutation. There were significant interactions between APOE genotype and FAD mutation status in the levels of ApoE, IGF-1, IL-13, IL-1α, thrombopoietin, AXL receptor tyrosine kinase, Fas ligand, IL-12p40, IL-3, IL-4, IL-5, IL-7, and MDC (Table 1).
Figure 1.
Mean levels of analytes that differed significantly among APOE genotype groups. The horizontal line in the middle of each box indicates the median, while the top and bottom borders of the box mark the 75th and 25th percentiles, respectively. Whiskers represent outliers between the 1.5 and 3 interquartile ranges, and individual data points indicate extreme values beyond 3 interquartile ranges. IL indicates interleukin.
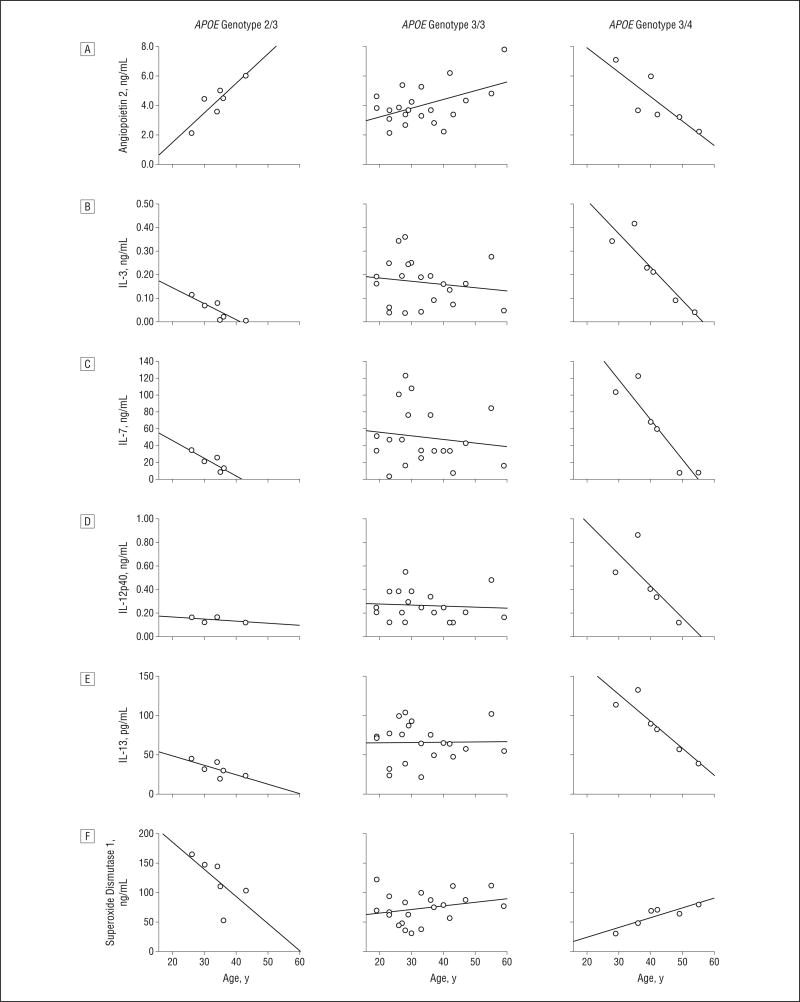
From the multiple regression models we found that angiopoietin 2, stem cell factor, IL-12p40, IL-1α, ENA-78, CNTF, M-CSF, IL-15, IL-13, IL-7, IL-3, and SOD1 showed interactions of APOE genotype with age (Table 4 and Figure 2) (P < .05, FDR < 0.2). The structure of the interaction for angiopoietin 2 showed an increase with age in ε2/3 carriers, a less steep increase with age in ε3/3 carriers, and a decrease with age in ε3/4 carriers. Among the interleukins, levels were highest in younger ε3/4 carriers and decreased with age. With regard to SOD1, levels were highest in young ε2/3 carriers and decreased with age. Although statistically significant, the trends with age for the other analytes were less pronounced.
Table 4.
Spearman Rank Correlations Between Age and Untransformed Analyte Levels
| Characteristic | APOE ε2/3 | P Value | APOE ε3/3 | P Value | APOE ε3/4 | P Value |
|---|---|---|---|---|---|---|
| Angiopoietin 2, ng/mL | 0.89 | .02 | 0.25 | .27 | −0.94 | .005 |
| Stem cell factor, pg/mL | −0.84 | .04 | 0.02 | .94 | 0.97 | .01 |
| IL-1α, ng/mL | 0.50 | .67 | 0.04 | .89 | −0.90 | .04 |
| IL-3, ng/mL | −0.89 | .02 | −0.11 | .65 | −0.94 | .005 |
| IL-7, pg/mL | −0.80 | .10 | −0.18 | .45 | −0.93 | .008 |
| IL-12p40, ng/mL | −0.45 | .22 | −0.14 | .56 | −0.90 | .04 |
| IL-13, pg/mL | −0.77 | .07 | −0.10 | .66 | −0.94 | .005 |
| IL-15, ng/mL | NA | NA | 0.39 | .12 | −1.0 | <.001 |
| ENA-78, ng/mL | −0.77 | .07 | 0.03 | .89 | 0.94 | .005 |
| CNTF, ng/mL | −0.09 | .87 | −0.10 | .67 | 0.90 | .04 |
| M-CSF, ng/mL | 0.37 | .47 | −0.06 | .79 | 0.49 | .33 |
| SOD1, ng/mL | −0.94 | .005 | 0.22 | .33 | 0.83 | .04 |
Abbreviations: CNTF, ciliary neurotrophic factor; IL, interleukin; M-CSF, macrophage colony-stimulating factor; NA, not available; SOD1, superoxide dismutase 1.
Figure 2.
Levels of analytes with significant interactions of APOE genotype and age in which discernible patterns with age were observed. The solid line in each graph represents the linear regression fit across all subjects. The Spearman rank correlations are given in Table 4. IL indicates interleukin.
COMMENT
The levels of plasma proteins involved in intercellular communication may relate to AD pathologic characteristics.14,21 In the current study, we ascertained the levels of many such proteins in persons from FAD families, most of whom were asymptomatic. No significant differences were found between FAD MCs and NCs, although differences among APOE genotype groups suggested an elevation of inflammatory mediators in carriers of the APOE ε4 and ε3 alleles relative to the ε2 allele, with trends toward higher levels in ε4 carriers relative to ε3 carriers. Furthermore, for some markers, there was a decrease with age in ε4 carriers that was not present or less pronounced with other APOE genotypes.
Despite the certainty with which FAD MCs develop the disease and the systemic nature of such mutations, we found minimal differences in plasma protein levels between MCs and NCs. Although CFH was nonsignificantly elevated in FAD MCs, plasma levels of CFH have been found22 to be elevated in patients with AD, using an unbiased proteomic approach, and polymorphisms in the gene for CFH have been linked to AD risk.10 Although FAD mutation status in itself did not have a strong influence on protein plasma levels, there were significant interactions between FAD mutation status and APOE genotype for several proteins, suggesting convergent influences on disease pathogenesis.
In prior studies comparing plasma ApoE concentration between persons with dementia and those serving as controls, elevated,23 decreased,24 and equivalent25 levels have been reported. However, studies in persons without dementia have consistently shown an effect of APOE genotype on plasma ApoE levels, with carriers of the ε2 allele having higher levels than carriers of the ε3 allele, who, in turn, have higher levels than ε4 allele carriers.26 A study in transgenic mice suggested that this was due to increased degradation of the ε4 form of the protein.27 Our finding of elevated ApoE levels in APOE ε2 carriers is consistent with these observations. The mechanisms by which the APOE ε4 allele confers an increased risk for AD are controversial; one possibility is mediation by lower levels of total ApoE protein in addition to disparate functionality conferred by polymorphisms.
In our population, the APOE genotype was related to the levels of the inflammatory markers I-309, IL-1α, IL-3, IL-7, IL-12p40, IL-13, and IL-15, with the ε2 allele being associated with the lowest levels and the ε4 allele with the highest. The interleukins can have either proinflammatory or regulatory roles in immune response, and some are overexpressed in the AD brain.6 Other studies14,21,28 have found plasma levels of interleukins to be inconsistently associated with AD and incipient AD, possibly related to frequent comorbidity in aged populations. Of the 18 markers found to predict AD by Ray et al,14 only 10 could be successfully adapted to the current multiplex immunoassay panel and were therefore included in the present study. Of these, one (IL-3) for which decreased levels were found to predict AD by Ray et al was also found to be elevated in APOE ε4 carriers in our study. Prior studies have focused on populations older than ours and generally have looked at AD or mild cognitive impairment diagnosis without regard to APOE genotype. Our data indicate that some of the variability in prior studies may be the result of age and APOE genotype and suggest that differences in the inflammatory response occur in APOE ε4 carriers and diminish before the age at which AD symptoms begin. The APOE ε4 genotype may therefore exert its influence on AD risk in early adulthood and mid-adulthood, times at which other manifestations of APOE genotype are also evident.29,30
Apolipoprotein E is a pleiotropic protein, and many different mechanisms have been invoked in explaining how the APOE ε4 variant contributes to AD risk. Although effects on Aβ metabolism are most commonly cited,5 many other effects,31–34 including influences on inflammation, have been observed. Microglial activation by APP was blocked by the presence of APOE ε3 but not by APOE ε435 and microglia derived from transgenic mice with the human APOE ε4 allele secrete higher levels of proinflammatory cytokines than do microglia from APOE ε3 mice.36 In humans it has been demonstrated that higher plasma levels of IL-6 are associated with poorer cognition in the elderly and predict steeper decline in memory, an effect greatest in APOE ε4 carriers.37 Both an observational study of incident AD38 and a randomized, prospective controlled study of persons with AD39 suggested that anti-inflammatory interventions have greater benefit in APOE ε4 carriers relative to ε4 noncarriers. If the increased inflammatory response observed in young APOE ε4 carriers is related to the AD process observed later in life, intervention with anti-inflammatory medications at a young age might serve to ameliorate disease pathogenesis later.
An important limitation of this study is the variable degree of relatedness between the participants. Nine of the 14 families had the same PSEN1 mutation (A431E), which has been demonstrated to represent a founder effect,16 and the 2 families with the V717I APP mutation may also be related. Indeed, we found that the gene with risk for mutation was related to the levels of 13 proteins in plasma, and, considering the relatively close genetic relationships among persons at risk for PSEN1 and APP mutation in our cohort, this may represent the effects of non-FAD, non-APOE genetic influences on plasma protein levels. The fact that effects of APOE were still seen when the risk for APP or PSEN1 mutations was covaried, however, indicates that APOE genotype has additional effects.
Another limitation of this study is the small number of participants relative to the large number of variables analyzed. Control for the FDR, the consistency of the pattern of interleukin levels, and the strong interactions between age and APOE genotype for many markers, however, diminishes the likelihood of our results being spurious. Also, considering the small size of the APOE ε2/3 and ε3/4 groups, the Spearman rank correlations should be interpreted with caution. Because we analyzed 67 proteins, it is likely that several would have significant correlations by chance (~3.4 = 0.05 × the number of evaluable proteins); therefore, our findings of 11 protein/age correlations in the ε4 subgroup, although of interest, should be considered pilot data.
Our finding of elevated inflammatory mediators in young adult APOE ε4 carriers is consistent with ApoE ε4 being associated with an increased propensity toward inflammation in the periphery that decreases with aging. Whether this is causative of, reactive to, or incidental to incipient AD pathology is unclear, but the effect was seen in young persons presumed to have few abnormalities. Determining whether interventions influencing inflammatory mechanisms can affect disease in a prospective fashion would allow us to better establish the chain of events leading to AD.
Supplementary Material
Acknowledgments
Funding/Support: This study was supported by Public Health Services K08 AG-22228, California Department of Health Services 04-35522, Alzheimer’s Disease Research Center Grant P50 AG-16570 from the National Institute on Aging, the Easton Consortium for Alzheimer’s Disease Drug Discovery and Biomarkers, the General Clinical Research Centers Program M01-RR00865, the Sidell Kagan Foundation, and the Shirley and Jack Goldberg Trust.
Footnotes
Financial Disclosure: None reported.
Online-Only Material: The eAppendix is available at http://www.archneurol.com.
Author Contributions: Study concept and design: Ringman, Cummings, and Cole. Acquisition of data: Ringman and Geschwind. Analysis and interpretation of data: Ringman, Elashoff, Geschwind, Welsh, Gylys, Lee, and Cole. Drafting of the manuscript: Ringman, Elashoff, and Cummings. Critical revision of the manuscript for important intellectual content: Ringman, Elashoff, Geschwind, Welsh, Gylys, Lee, and Cole. Statistical analysis: Elashoff and Lee. Obtained funding: Cummings. Administrative, technical, and material support: Ringman, Welsh, Gylys, and Cole. Study supervision: Geschwind and Gylys.
References
- 1.Hardy J, Selkoe DJ. The amyloid hypothesis of Alzheimer’s disease: progress and problems on the road to therapeutics. Science. 2002;297(5580):353–356. doi: 10.1126/science.1072994. [DOI] [PubMed] [Google Scholar]
- 2.Wong CW, Quaranta V, Glenner GG. Neuritic plaques and cerebrovascular amyloid in Alzheimer disease are antigenically related. Proc Natl Acad Sci U S A. 1985;82(24):8729–8732. doi: 10.1073/pnas.82.24.8729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Scheuner D, Eckman C, Jensen M, et al. Secreted amyloid beta-protein similar to that in the senile plaques of Alzheimer’s disease is increased in vivo by the presenilin 1 and 2 and APP mutations linked to familial Alzheimer’s disease. Nat Med. 1996;2(8):864–870. doi: 10.1038/nm0896-864. [DOI] [PubMed] [Google Scholar]
- 4.Corder EH, Saunders AM, Strittmatter WJ, et al. Gene dose of apolipoprotein E type 4 allele and the risk of Alzheimer’s disease in late onset families. Science. 1993;261(5123):921–923. doi: 10.1126/science.8346443. [DOI] [PubMed] [Google Scholar]
- 5.Kim J, Basak JM, Holtzman DM. The role of apolipoprotein E in Alzheimer’s disease. Neuron. 2009;63(3):287–303. doi: 10.1016/j.neuron.2009.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Akiyama H, Barger S, Barnum S, et al. Inflammation and Alzheimer’s disease. Neurobiol Aging. 2000;21(3):383–421. doi: 10.1016/s0197-4580(00)00124-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pan S, Rush J, Peskind ER, et al. Application of targeted quantitative proteomics analysis in human cerebrospinal fluid using a liquid chromatography matrix-assisted laser desorption/ionization time-of-flight tandem mass spectrometer (LC MALDI TOF/TOF) platform. J Proteome Res. 2008;7(2):720–730. doi: 10.1021/pr700630x. [DOI] [PubMed] [Google Scholar]
- 8.Ringman JM, Schulman H, Becker C, et al. Proteomic changes in cerebrospinal fluid of presymptomatic and affected persons carrying familial Alzheimer disease mutations. Arch Neurol. 2012;69(1):96–104. doi: 10.1001/archneurol.2011.642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lambert JC, Heath S, Even G, et al. European Alzheimer’s Disease Initiative Investigators. Genome-wide association study identifies variants at CLU and CR1 associated with Alzheimer’s disease. Nat Genet. 2009;41(10):1094–1099. doi: 10.1038/ng.439. [DOI] [PubMed] [Google Scholar]
- 10.Zetterberg M, Landgren S, Andersson ME, et al. Association of complement factor H Y402H gene polymorphism with Alzheimer’s disease. Am J Med Genet B Neuropsychiatr Genet. 2008;147(6):720–726. doi: 10.1002/ajmg.b.30668. [DOI] [PubMed] [Google Scholar]
- 11.Nicoll JA, Mrak RE, Graham DI, et al. Association of interleukin-1 gene polymorphisms with Alzheimer’s disease. Ann Neurol. 2000;47(3):365–368. [PMC free article] [PubMed] [Google Scholar]
- 12.Holmes C, Cunningham C, Zotova E, et al. Systemic inflammation and disease progression in Alzheimer disease. Neurology. 2009;73(10):768–774. doi: 10.1212/WNL.0b013e3181b6bb95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cunningham C, Wilcockson DC, Campion S, Lunnon K, Perry VH. Central and systemic endotoxin challenges exacerbate the local inflammatory response and increase neuronal death during chronic neurodegeneration. J Neurosci. 2005;25(40):9275–9284. doi: 10.1523/JNEUROSCI.2614-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ray S, Britschgi M, Herbert C, et al. Classification and prediction of clinical Alzheimer’s diagnosis based on plasma signaling proteins. Nat Med. 2007;13 (11):1359–1362. doi: 10.1038/nm1653. [DOI] [PubMed] [Google Scholar]
- 15.Ringman JM. What the study of persons at risk for familial Alzheimer’s disease can tell us about the earliest stages of the disorder: a review. J Geriatr Psychiatry Neurol. 2005;18(4):228–233. doi: 10.1177/0891988705281878. [DOI] [PubMed] [Google Scholar]
- 16.Murrell J, Ghetti B, Cochran E, et al. The A431E mutation in PSEN1 causing familial Alzheimer’s disease originating in Jalisco State, Mexico: an additional fifteen families. Neurogenetics. 2006;7(4):277–279. doi: 10.1007/s10048-006-0053-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yescas P, Huertas-Vazquez A, Villarreal-Molina MT, et al. Founder effect for the Ala431Glu mutation of the presenilin 1 gene causing early-onset Alzheimer’s disease in Mexican families. Neurogenetics. 2006;7(3):195–200. doi: 10.1007/s10048-006-0043-3. [DOI] [PubMed] [Google Scholar]
- 18.Athan ES, Williamson J, Ciappa A, et al. A founder mutation in presenilin 1 causing early-onset Alzheimer disease in unrelated Caribbean Hispanic families. JAMA. 2001;286(18):2257–2263. doi: 10.1001/jama.286.18.2257. [DOI] [PubMed] [Google Scholar]
- 19.Morris JC. Clinical dementia rating: a reliable and valid diagnostic and staging measure for dementia of the Alzheimer type. Int Psychogeriatr. 1997;9(suppl 1):173–176. 177–178. doi: 10.1017/s1041610297004870. [DOI] [PubMed] [Google Scholar]
- 20.Storey JD. The positive false discovery rate: a Bayesian interpretation of the q-value. Ann Stat. 2003;31(6):2013–2035. doi: 10.1214/aos/1074290335. [DOI] [Google Scholar]
- 21.Soares HD, Chen Y, Sabbagh M, Roher A, Schrijvers E, Breteler M. Identifying early markers of Alzheimer’s disease using quantitative multiplex proteomic immunoassay panels. Ann N Y Acad Sci. 2009;1180:56–67. doi: 10.1111/j.1749-6632.2009.05066.x. [DOI] [PubMed] [Google Scholar]
- 22.Hye A, Lynham S, Thambisetty M, et al. Proteome-based plasma biomarkers for Alzheimer’s disease. Brain. 2006;129(pt 11):3042–3050. doi: 10.1093/brain/awl279. [DOI] [PubMed] [Google Scholar]
- 23.Taddei K, Clarnette R, Gandy SE, Martins RN. Increased plasma apolipoprotein E (apoE) levels in Alzheimer’s disease. Neurosci Lett. 1997;223(1):29–32. doi: 10.1016/s0304-3940(97)13394-8. [DOI] [PubMed] [Google Scholar]
- 24.Siest G, Bertrand P, Qin B, et al. Apolipoprotein E polymorphism and serum concentration in Alzheimer’s disease in nine European centres: the ApoEurope study. Clin Chem Lab Med. 2000;38(8):721–730. doi: 10.1515/CCLM.2000.102. [DOI] [PubMed] [Google Scholar]
- 25.Slooter AJ, de Knijff P, Hofman A, et al. Serum apolipoprotein E level is not increased in Alzheimer’s disease: the Rotterdam study. Neurosci Lett. 1998;248 (1):21–24. doi: 10.1016/s0304-3940(98)00339-5. [DOI] [PubMed] [Google Scholar]
- 26.van Vliet P, Westendorp RG, Eikelenboom P, et al. Parental history of Alzheimer disease associated with lower plasma apolipoprotein E levels. Neurology. 2009;73(9):681–687. doi: 10.1212/WNL.0b013e3181b59c2e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Riddell DR, Zhou H, Atchison K, et al. Impact of apolipoprotein E (ApoE) polymorphism on brain ApoE levels. J Neurosci. 2008;28(45):11445–11453. doi: 10.1523/JNEUROSCI.1972-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.O’Bryant SE, Xiao G, Barber R, et al. A serum protein-based algorithm for the detection of Alzheimer disease. Arch Neurol. 2010;67(9):1077–1081. doi: 10.1001/archneurol.2010.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reiman EM, Chen K, Alexander GE, et al. Functional brain abnormalities in young adults at genetic risk for late-onset Alzheimer’s dementia. Proc Natl Acad Sci U S A. 2004;101(1):284–289. doi: 10.1073/pnas.2635903100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ringman JM, Medina LD, Braskie M, et al. Effects of risk genes on BOLD activation in presymptomatic carriers of familial Alzheimer’s disease mutations during a novelty encoding task. Cereb Cortex. 2011;21(4):877–883. doi: 10.1093/cercor/bhq158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Trommer BL, Shah C, Yun SH, et al. ApoE isoform-specific effects on LTP: blockade by oligomeric amyloid-31–42. Neurobiol Dis. 2005;18(1):75–82. doi: 10.1016/j.nbd.2004.08.011. [DOI] [PubMed] [Google Scholar]
- 32.Tesseur I, Van Dorpe J, Spittaels K, Van den Haute C, Moechars D, Van Leuven F. Expression of human apolipoprotein E4 in neurons causes hyperphosphorylation of protein tau in the brains of transgenic mice. Am J Pathol. 2000;156 (3):951–964. doi: 10.1016/S0002-9440(10)64963-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nathan BP, Chang KC, Bellosta S, et al. The inhibitory effect of apolipoprotein E4 on neurite outgrowth is associated with microtubule depolymerization. J Biol Chem. 1995;270(34):19791–19799. doi: 10.1074/jbc.270.34.19791. [DOI] [PubMed] [Google Scholar]
- 34.Buttini M, Orth M, Bellosta S, et al. Expression of human apolipoprotein E3 or E4 in the brains of ApoE−/− mice: isoform-specific effects on neurodegeneration. J Neurosci. 1999;19(12):4867–4880. doi: 10.1523/JNEUROSCI.19-12-04867.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Barger SW, Harmon AD. Microglial activation by Alzheimer amyloid precursor protein and modulation by apolipoprotein E. Nature. 1997;388(6645):878–881. doi: 10.1038/42257. [DOI] [PubMed] [Google Scholar]
- 36.Vitek MP, Brown CM, Colton CA. APOE genotype-specific differences in the innate immune response. Neurobiol Aging. 2009;30(9):1350–1360. doi: 10.1016/j.neurobiolaging.2007.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schram MT, Euser SM, de Craen AJ, et al. Systemic markers of inflammation and cognitive decline in old age. J Am Geriatr Soc. 2007;55(5):708–716. doi: 10.1111/j.1532-5415.2007.01159.x. [DOI] [PubMed] [Google Scholar]
- 38.Szekely CA, Breitner JC, Fitzpatrick AL, et al. NSAID use and dementia risk in the Cardiovascular Health Study: role of APOE and NSAID type. Neurology. 2008;70(1):17–24. doi: 10.1212/01.wnl.0000284596.95156.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pasqualetti P, Bonomini C, Dal Forno G, et al. A randomized controlled study on effects of ibuprofen on cognitive progression of Alzheimer’s disease. Aging Clin Exp Res. 2009;21(2):102–110. doi: 10.1007/BF03325217. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.