Abstract
The first step during bacillithiol (BSH) biosynthesis involves the formation of N-acetylglucosaminylmalate from UDP-N-acetylglucosamine and L-malate and is catalyzed by a GT4 class glycosyltransferase enzyme (BshA). Recombinant Staphylococcus aureus and Bacillus subtilis BshA were highly specific and active with L-malate but the former showed low activity with D-glyceric acid and the latter with D-malate. We show that BshA is inhibited by BSH and similarly that MshA (first enzyme of mycothiol biosynthesis) is inhibited by the final product MSH.
Keywords: mycothiol, bacillithiol, Staphylococcus aureus, glycosyltransferase, mycobacteria, thiol
Introduction
In nearly all eukaryotes, glutathione is the major low-molecular weight thiol that serves as a cofactor for a wide range of enzymes involved in protecting against reactive oxygen species, reactive nitrogen species, and many other toxins. GSH is found in most Gram-negative bacteria, including cyanobacteria, but only rarely in Gram-positive bacteria, which include a wide range of human pathogens. In the high GC Actinobacteria, a low-molecular-weight cysteine derivative called mycothiol (MSH) is the major thiol [1] while in the low GC Gram-positive bacteria, a similar thiol, bacillithiol (BSH), is present [2]. Both MSH and BSH contain the cysteinylglucosamine core moiety but in BSH L-malic acid is substituted for myo-inositol and the cysteine amino group is not acylated (Fig. 1a and 1b).
Fig. 1.
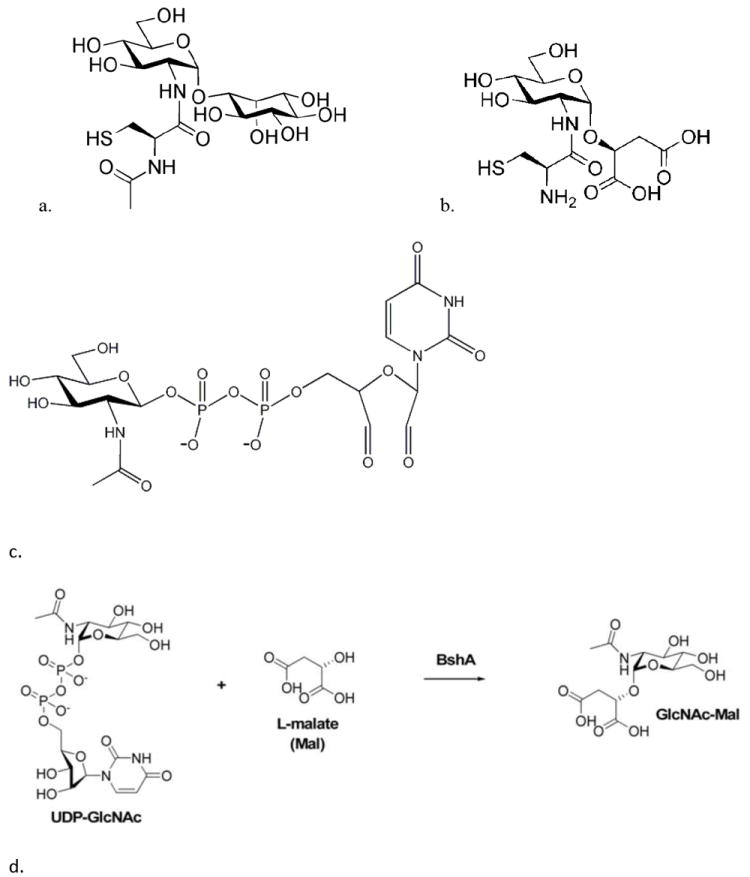
Structure of a) mycothiol, b) bacillithiol, c) O-UDP-N-acetylglucosamine (oxidized UDP-N-acetylglucosamine); note that the ribose moiety in UDP-N-acetylglucosamine has been oxidized to a dialdehydo- moiety, and d) BshA glycosyl transferase reaction
The first step in MSH biosynthesis involves the formation of N-acetylglucosaminylinositol-3- phosphate from UDP-N-acetyl glucosamine (UDP-GlcNAc) and 1-L-inositol-1-phosphate, a process catalyzed by a GT4 class glycosyltransferase (MshA) [3]. A similar first step occurs during BSH biosynthesis where a B. subtilis GT4 class glycosyltransferase enzyme (BshA) catalyzes the formation of N-acetylglucosaminylmalate (GlcNAc-Mal) from UDP-GlcNAc and L-malate (Fig. 1d) [4]. In 2008, Ruane et al. reported the crystal structure of a putative glycosyltransferase, ORF BA1558, which is the B. anthracis homolog of BshA [5]. Parsonage et al. (2010) reported the structure of this B. anthracis BshA with UDP-malate ternary complex and described the phenotype of the mutant disrupted in this gene [6]. Herein, we report on the characterization of B. subtilis BshA and S. aureus BshA and show that BSH is able to inhibit BshA. As a dominant thiol in significant pathogens, BSH biochemistry, in particular enzymes like BshA that are involved in the biosynthesis of BSH, may prove to be good targets for antibiotics directed against these pathogens.
Materials and Methods
Cloning and expression of B. subtilis and S. aureus BshA
BshA was PCR amplified from B. subtilis JH642 using primers BshAN5 (5’-CACCATGAGAAAACTAAA AATAGGA) and BshAN3 (5’-TCACTCCGGTTCTGCTAAATCGGC) and proofreading Pfu DNA polymerase (Stratagene) [7]. The amplicon was cloned into pet151 TOPO vector (Invitrogen) and expressed in BL-21 Star (DE3) pLysS E. coli strain as the N-terminal His-6 tagged protein. The protein was purified to homogeneity on a chelate chromatography nickel column (Clontech). The eluted proteins were dialyzed against the BshA assay buffer, 100 mM NaCl, 10 mM MgCl2, and 25 mM HEPES pH 7.5, to which 1 mM β-mercaptoethanol was added and concentrated with Centricon 30 ultrafilter (Amicon) and snap frozen in 10% glycerol. Similarly, the S. aureus BshA was PCR amplified using SBshAN5 (5’-CACCATGAAGATAGGTATAAC) and SBshAN3 (5’- TTACTCGCCTTTACTTTTGTT), expressed and purified using PrepEase His tag protein purification Prep (USB Corporation). The BshA protein fraction was loaded onto a Sephadex G25 column to remove the imidazole and eluted with BshA assay buffer to which 2 mM dithiothreitol (DTT) was added. S. aureus BshA was snap frozen in individual aliquots in glycerol as it eluted off the column.
Assay for glycosyltransferase activity
S. aureus and B. subtilis BshA were assayed in 100 mM NaCl, 10 mM MgCl2, and 25 mM HEPES pH 7.5 at 37°C with 2 mM DTT and 1 mM β-mercaptoethanol added to the buffer respectively. The consumption of UDP-GlcNAc and release of UDP were analyzed by HPLC with detection of peaks spectrophotometrically at 260 nm as previously described for MshA with minor modifications [3]. The following HPLC conditions were used: Buffer A: 2 mM tetrabutylammonium phosphate, pH 5.4; Buffer B: 20 mM KH2PO4, 50% methanol, 10 mM tetrabutylammonium phosphate, pH 5.4; elution program: 0-1 min, 15%B; 1 to 31 min, linear 15-100% Buffer B; 31-32 min, linear 100%-15% Buffer B; 45 min reinjection. The retention times for the standards, uridine, UMP, UDP-GlcNAc, UDP, and UTP were 4.9, 14.2, 20.5, 21.9, and 25.6 min, respectively. S. aureus BshA substrate specificity and B. subtilis BshA substrate specificity was determined.
Inhibition of BshA and MshA glycosyltransferase with BSH, MSH and O-UDP-GlcNAc
B. subtilis BshA was preincubated for 15 min at 37°C with various concentrations of BSH, diluted into the BshA reaction mixture, and glycosyltransferase activity was measured as the release of UDP as described above. The dose-response curves for BSH were determined in duplicate and half maximal inhibitory concentration (IC50) calculated from these curves. Likewise, M. smegmatis cell lysate was preincubated with various concentrations of MSH, diluted into the MshA reaction mixture containing 1 mM 1-L-inositol-1-phosphate and 1 mM UDP-GlcNAc. To determine if 2’,3’-dialdehydo-UDP-N-acetylglucosamine (O-UDP-GlcNAc; Fig. 1c) from oxidation of UDP-GlcNAc inhibits BshA and/or MshA activity, both BshA and M. smegmatis cell lysate were preincubated with various concentrations of O-UDP-GlcNAc and activity assays performed.
Size determination of BshA glycosyl transferase
Gel filtration using Sephacryl 200 was used to determine the native molecular weight for both S. aureus and B. subtilis BshA. The following proteins were used to calibrate the column: cytochrome C, ovalbumin, bovine serum albumin, phophorylase B, aldehyde dehydrogenase, β amylase, and apoferritin.
Thiol determination
Briefly, 60 μg of purified protein was treated with 10 mM DTT, 10 mM diamide, or water at room temperature for 15 min. To denature the protein and stop the reaction, acetonitrile was added to the reaction followed by centrifugation to collect the pelleted protein. The pelleted protein was washed, brought up in 100 μl of 6 M guanidine HCl, and incubated at 37°C for 25 min. Protein concentration was determined by measuring absorbance at 280 nm. To measure the thiol content, the samples were treated with 0.16 mM 5,5’-dithiobis(2-nitrobenzoic acid) (DTNB) for 15 min and the absorbance at 412 nm was measured.
Molecular Modeling and Data analysis
To determine basic kinetic parameters for each substrate, initial velocity plots at saturating concentrations of one substrate were fit to the Michaelis-Menten equation. All data were analyzed using KaleidaGraph (Synergy Software).
Protein threading on B. subtilis and S. aureus BshA was performed using B. anthracis BA1558 X-Ray crystal structure (PDB accession no.: 2JJM) with IP, integer programming-based threading engine of RAPTOR [8] 3D structure modeling tool OWL and displayed with MolScript [9].
Results and Discussion
We recently identified the gene, bshA, responsible for the first step in the biosynthesis of BSH in Bacillus subtilis [4]. This gene codes for a retaining glycosyltransferase which uses L-malate as the acceptor substrate and UDP-GlcNAc as the donor substrate. Disruption of this gene in B. subtilis results in sensitivity to alkylating agents, fosfomycin and methylglyoxal, indicating involvement of BSH in detoxification of toxins, to environmental stresses such as osmotic stress and acid stress, protection against toxins and a decrease in sporulation [4].
In this report, we present a characterization of B. subtilis BshA and the medically relevant S. aureus BshA. When the two genes were expressed in E. coli, the resulting recombinant proteins behaved very differently during the purification process. B. subtilis BshA purification resulted in a good yield and the protein remained soluble even after concentration to 7mg ml-1; however, S. aureus BshA started precipitating as the protein eluted off the Ni+ affinity column. Thus, we switched to commercially prepared Prep Ease His Tag Protein Purification columns for rapid purification of the protein followed by Sephadex G25 gel filtration column to rid eluent of imidazole. As the fractions were eluted, they were frozen in 10% glycerol to prevent further precipitation.
Both recombinant S. aureus and B. subtilis BshA exhibited marked activity when assayed by HPLC for UDP production using UDP-GlcNAc and L-malate as substrates. Both substrates exhibited Michaelis-Menten kinetics in the initial velocity plots for L-malate and UDP-GlcNAc, and no curvature was seen in the Lineweaver-Burk linear replots (data not shown). The kinetic parameters for each substrate were determined after fitting to the Michaelis-Menten Equation. The apparent Km for UDP-GlcNAc at 3 mM malate is within the significant error range (250 ± 110 μM for S. aureus and 370 ± 94 μM for B. subtilis) (Table 1). The apparent Km for UDP-GlcNAc reported for B. anthracis BshA is equal to that of B. subtilis at 370 μM determined with the HPLC assay [6] and the apparent Km for UDP-GlcNAc for C. diptheraie MshA is 210 ± 20 μM [10]. In contrast, the apparent Km for L-malate is not within the significant error range between the S. aureus and B. subtilis enzymes (710 ± 110 μM for S. aureus BshA and 410 ± 70 μM for B. subtilis BshA) and the Vmax for the S. aureus enzyme is also 3 to 4 fold higher than the B. subtilis enzyme. Substrate specificity studies further highlight the differences between the two enzymes with S. aureus BshA showing measurable specific activity with not only L-malate but D-glyceric acid and B. subtilis BshA showing measurable specific activity with L-malate and the entantiomer D-malate (Table 2). The kcat calculated from the UDP-GlcNAc Vmax is 24-25 sec-1 and 6.1-7.8 sec-1 for S. aureus and B. subtilis BshA, respectively. In comparison, the kcat for B. anthracis BshA [6] and Corynebacterium diptheraie MshA [10] are 28 sec-1 and 12.5 sec-1 respectively.
Table 1.
Apparent Km and Vmax of Staphylococcus aureus and Bacillus subtilis BshA. For L-malate concentration dependence, 10 mM UDP-N-acetylglucosamine (UDP-GlcNAc) fixed and L-malate concentration varied from 0.1mM to 3.2 mM; for UDP-GlcNAc concentration dependence, 3 mM of L-malate fixed and UDP-GlcNAc varied from 1 mM to 15 mM; and, 0.35 μg BshA added to the reactions.
| Substrate | Km [μM] | Vmax [nmol min-1mg-1] |
|---|---|---|
| L-malate | ||
| S. aureus | 710 ± 110 | 35 600 ± 1800 |
| B. subtilis | 410 ± 70 | 11 200 ± 100 |
| UDP-GlcNAc | ||
| S. aureus | 250 ± 110 | 34 200 ± 3700 |
| B. subtilis | 370 ± 94 | 8700 ± 100 |
Table 2.
Substrate specificity of S. aureus and B. subtilis BshA. For B. subtilis BshA, reaction conditions consisted of 0.3 mM acceptor substrate, 1.0 mM UDP-GlcNAc and 0.16 μg BshA. For S. aureus BshA, reaction conditions consisted of 0.7 mM acceptor substrate, 3.0 mM UDP-GlcNAc and 0.048 μg BshA.
| S. aureus | B. subtilis | ||||
|---|---|---|---|---|---|
|
| |||||
| Substrate | Specific Activity (nmoles min-1 mg-1) | Relative Rate (%) | Specific Activity (nmoles min-1 mg-1) | Relative Rate (%) | |
| L-malic acid | 18,600 ± 6,300 | 100 | 6,600 ± 150 | 100 | |
| D-malic acid | <100 | <0.5 | 230 ± 8 | 3.4 | |
| inositol-1-L-phosphate | <100 | <0.5 | <100 | <0.5 | |
| glycolic acid | <100 | <0.5 | <100 | <0.5 | |
| D-lactic acid | <100 | <0.5 | <100 | <0.5 | |
| D, L-isocitric acid | <100 | <0.5 | <100 | <0.5 | |
| D-glyceric acid | 430 ± 180 | 2.3 | <100 | <0.5 | |
| citric acid | <100 | <0.5 | <100 | <0.5 | |
In 2008, Ruane and colleagues [5] first reported the structure of Bacillus anthracis BshA (BA1558). Parsonage et al (2010) further crystallized this enzyme in complex with UDP and malate [6]. B. anthracis BshA has 63.1% sequence identity and 77.6% sequence similarity to B. subtilis BshA and 53.2% sequence identity and 72.5% sequence similarity to S. aureus BshA. We thus modeled the S. aureus and B. subtilis BshA on B. anthracis BshA. Molecular modeling indicated that all three proteins contain the typical GT-B fold of glycosyltransferases, which consists of two “Rossmann-like” beta/alpha/beta domains separated by a deep crevice in the inter-domain region and a kinked C-terminal α-helix that crosses over from the C-terminal domain to contact the N-terminal domain [5]. However there are some differences, most notably in the position of active site residues among the three proteins (Fig. 2). The active site Lys211 (Lys 212 in B. subtilis BshA, Lys 209 in S. aureus BshA), which binds to one of the phosphate oxygens of UDP, is present in an α-helix in approximately the same position in all three structures. On the other hand, the B. anthracis BshA His120 and B. subtilis BshA His121, which binds to malate, is present at the end of the β-sheet while S. aureus BshA His118 is in the middle of a β sheet. Also, the positions of the B. anthracis BshA Glu282 and Glu290 binding sites and the relevant amino acids in B. subtilis BshA (Glu283, Glu291) and S. aureus BshA (Glu280, Glu288) are very different among the three protein structures. Both residues are involved in substrate recognition of UDP, Glu282 binding to one of the phosphate oxygens of UDP and Glu290 binding to the ribose-2’-OH and ribose-3’-OH. These variations in the structure of the proteins may account for the difference in the behavior of the enzymes.
Fig. 2.
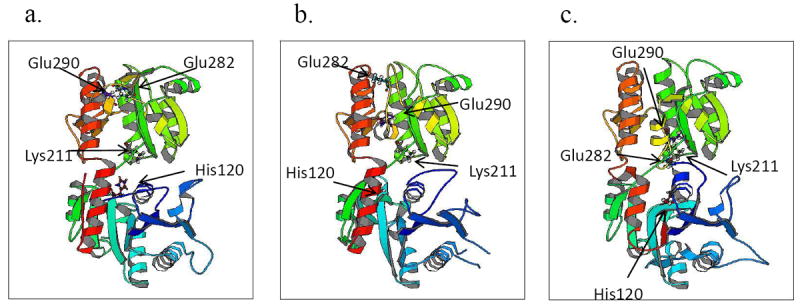
3D structure of B. subtilis and S. aureus BshA, a) B. anthracis BshA (BA1558), b) B. subtilis BshA, and c) S. aureus BshA. Residues identified in S. aureus and B. subtilis are equivalent to B. anthracis residue numbers.
Ruane et al. (2008) [5] reported that B. anthracis monomers are arranged into a tetramer and Parsonage et al. (2010) [6] confirmed that B. anthracis BshA consists of eight monomers arranged as a dimer of tetramers. To determine if B. subtilis BshA and S. aureus BshA are also tetramers, gel filtration chromatography of purified recombinant proteins was conducted. B. subtilis BshA is composed of 377 amino acid residues with a monomer molecular weight of 42 kDa respectively. Analysis by gel filtration chromatography yielded apparent molecular weights of 160,000 kDa, suggesting that B. subtilis BshA is a tetramer. However, we were unable to determine the size of S. aureus BshA even after treatment with reducing agents, DTT and Tris(2-carboxyethyl) phosphine (TCEP) and alkylating agent, iodoacetamide, which resulted in precipitation of protein during gel filtration chromatography. To examine the oligomeric state of S. aureus BshA, thiol titration was performed in guanidine HCl using DTNB. In S. aureus BshA, three out of the four cysteines were free upon treatment with DTT and pretreatment with diamide, which oxidized the cysteines, did not yield any free thiols. These results suggest that either one of the cysteine in the native protein is buried and inaccessible to DTT or is in the disulfide form. S. aureus BshA is thus also likely to form a disulfide linked dimer.
Regulation of BSH synthesis is likely to be multifactorial, including transcriptional control and feedback inhibition. GshA, γ-Glutamate-cysteine ligase, which catalyzes the first of two steps in the pathway for the biosynthesis of glutathione, is feedback-inhibited by glutathione [11] and CoA inhibits pantethonate kinase, the first enzyme in the CoA biosynthesis pathway [12]. To determine whether MSH and BSH biosynthesis are regulated by feedback inhibition, B. subtilis BshA and M. smegmatis cell lysate were pre-incubated with BSH and MSH respectively followed by the enzymatic assay. The IC50 for BSH inhibition of BshA is 0.7 mM, within the biological range for this thiol (Fig. 3), and the IC50 for MSH inhibition of MshA is 3.6 mM, also within the biological range for this thiol (Fig. 4a). Thus like glutathione and CoA, the levels of BSH of MSH are modulated by feedback inhibition of the first enzyme in the biosynthetic pathway.
Fig. 3.

Inhibiton of B. subtilis BshA by BSH. a) B. subtilis BshA (0.71 μg) preincubated with BSH and assayed for BshA activity with 0.5mM UDP-GlcNAc and 0.5 mM L-malate
Fig. 4.
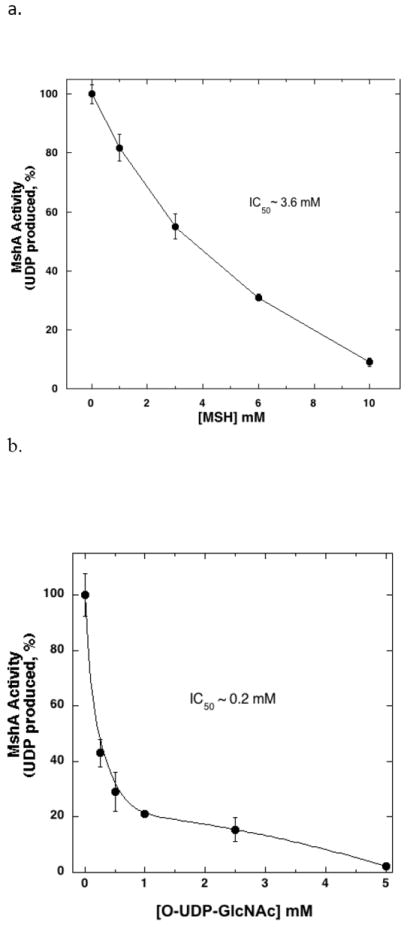
a) M. smegmatis cell free extract (2.7 mg total protein) preincubated with MSH and assayed for MshA activity with 1mM UDP-GlcNAc and 1mM 1-L-inositiol-1-phosphate. b) M. smegmatis cell free extract preincubated with O-UDP-GlcNAc and assayed for MshA activity with 1 mM UDP-GlcNAc and 1 mM 1-L-inositiol-1-phosphate. Control MshA rates, without inhibitor, were 0.4 nmoles min-1 mg protein-1.
BshA and MshA catalyze the first committed step in BSH and MSH biosynthesis respectively and thus these enzymes are considered to be potential drug targets. Recently, Frantom et al. (2011) reported that UDP-(5F)-GlcNAc, a sugar nucleotide with an electron withdrawing substituent, acts as a slow-binding, competitive inhibitor of C. glutamicum MshA (Ki ~1.6 μM) [13]. O-UDP-GlcNAc (Fig. 1c) inhibits UDP-N-acetylglucosamine 2-epimerase by binding to the UDP-GlcNAc binding site irreversibly [14]. As UDP-GlcNAc is a substrate for both BshA and MshA, O-UDP-GlcNAc was assayed for its ability to inhibit these enzymes. Preincubation of the inhibitor with the M. smegmatis cell lysate followed by the MshA assay resulted in an IC50 of 0.2 mM for MshA (Fig. 4b). Similar inhibition studies of BshA with O-UDP-GlcNAc did not show consistent inhibition. This inhibitor may serve as a basis for the chemical synthesis of further inhibitors for MshA.
Highlights.
Staphylococcus aureus and Bacillus subtilis BshA, which catalyze the first step in bacillithiol biosynthesis, are highly specific and active with L-malate.
S. aureus BshA has low activity with D-glyceric acid while B. subtilis BshA has low activity with D-malic acid.
Feedback inhibition of BshA and MshA occurs with bacillithiol and mycothiol, respectively.
Acknowledgments
This work was supported by NIH grants GM061223 to MR, AI072133 to RCF, and a MBRS RISE student stipend to HU.
Abbreviations
- BSH
bacillithiol
- MSH
mycothio
- UDP-GlcNAc
UDP-N-acetyl glucosamine
- GlcNAc-Mal
N-acetylglucosaminylmalate
- DTT
dithiothreitol
- DTNB
5,5’-dithiobis(2-nitrobenzoic acid)
- TCEP
Tris(2-carboxyethyl) phosphine
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Newton GL, Arnold K, Price MS, Sherrill C, delCardayre SB, Aharonowitz Y, Cohen G, Davies J, Fahey RC, Davis C. Distribution of thiols in microorganisms: mycothiol is a major thiol in most actinomycetes. J Bacteriol. 1996;178:1990–1995. doi: 10.1128/jb.178.7.1990-1995.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Newton GL, Rawat M, La Clair JJ, Jothivasan VK, Budiarto T, Hamilton CJ, Claiborne A, Helmann JD, Fahey RC. Bacillithiol is an antioxidant thiol produced in Bacilli. Nat Chem Biol. 2009;5:625–627. doi: 10.1038/nchembio.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Newton GL, Ta P, Bzymek KP, Fahey RC. Biochemistry of the initial steps of mycothiol biosynthesis. J Biol Chem. 2006;281:33910–20. doi: 10.1074/jbc.M604724200. [DOI] [PubMed] [Google Scholar]
- 4.Gaballa A, Newton GL, Antelmann H, Parsonage D, Upton H, Rawat M, Claiborne A, Fahey RC, Helmann JD. Biosynthesis and functions of bacillithiol, a major low-molecular-weight thiol in Bacilli. Proc Natl Acad Sci USA. 2010;107:6482–6. doi: 10.1073/pnas.1000928107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ruane KM, Davies GJ, Martinez-Fleites C. Crystal structure of a family GT4 glycosyltransferase from Bacillus anthracis ORF BA1558. Proteins. 2008;73:784–7. doi: 10.1002/prot.22171. [DOI] [PubMed] [Google Scholar]
- 6.Parsonage D, Newton GL, Holder RC, Wallace BD, Paige C, Hamilton CJ, Dos Santos PC, Redinbo MR, Reid SD, Claiborne A. Characterization of the N-acetyl-α-D-glucosaminyl l-malate synthase and deacetylase functions for bacillithiol biosynthesis in Bacillus anthracis. Biochemistry. 2010;49:8398–414. doi: 10.1021/bi100698n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: a laboratory manual. 2 Cold Spring Harbor Laboratory Press; Plainview, NY: 1989. [Google Scholar]
- 8.Xu J, Li M, Kim D, Xu Y. RAPTOR: Optimal Protein Threading by Linear Programming. J Bioinform Comput Bio. 2003;1:95–117. doi: 10.1142/s0219720003000186. [DOI] [PubMed] [Google Scholar]
- 9.Kraulis PJ. MOLSCRIPT: a program to produce both detailed and schematic plots of protein structures. J Appl Cryst. 1991;24:946–950. [Google Scholar]
- 10.Vetting MW, Frantom PA, Blanchard JS. Structural and enzymatic analysis of MshA from Corynebacterium glutamicum: substrate assisted catalysis. J Biol Chem. 2008;283:15834–15844. doi: 10.1074/jbc.M801017200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Apontoweil P, Berends W. Glutathione biosynthesis in Escherichia coli K 12. Properties of the enzymes and regulation. Biochim Biophys Acta. 1975;399:1–9. doi: 10.1016/0304-4165(75)90205-6. [DOI] [PubMed] [Google Scholar]
- 12.Karasawa T, Yoshida K, Furukawa K, Hosoki K. Feedback Inhibition of Pantothenate Kinase by Coenzyme A and Possible Role of the Enzyme for the Regulation of Cellular Coenzyme A Level. J Biochem. 1972;71:1065–1067. doi: 10.1093/oxfordjournals.jbchem.a129854. [DOI] [PubMed] [Google Scholar]
- 13.Frantom PA, Coward JK, Blanchard JS. UDP-(5F)-GlcNAc acts as a slow-binding inhibitor of MshA, a retaining glycosyltransferase. J Am Chem Soc. 2010;132:6626–7. doi: 10.1021/ja101231a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Blume A, Chen H, Reutter W, Schmidt RR, Hinderlich S. 2’, 3’-Dialdehydo-UDP-N-acetylglucosamine inhibits UDP-N-acetylglucosamine 2-epimerase, the key enzyme of sialic acid biosynthesis. FEBS Lett. 2002;521:127–32. doi: 10.1016/s0014-5793(02)02856-9. [DOI] [PubMed] [Google Scholar]


