Abstract
The stress-induced expression of many fission yeast genes is dependent upon the Sty1 MAP kinase (MAPK) and Atf1 transcription factor. Atf1 is phosphorylated by Sty1 yet this phosphorylation is not required for stress-induced gene expression suggesting another mechanism exists whereby Sty1 activates transcription. Here we show that Sty1 associates with Atf1-dependent genes and is recruited to both their promoters and coding regions. This occurs in response to various stress conditions coincident with the kinetics of Sty1’s own activation. Association with promoters is not a consequence of increased nuclear accumulation of Sty1 nor does it require the phosphorylation of Atf1. However, recruitment is completely abolished in a mutant lacking Sty1 kinase activity. Both Atf1 and its binding partner Pcr1 are required for association of Sty1 with Atf1-dependent promoters, suggesting that this heterodimer must be intact for optimal recruitment of the MAPK. However, many Atf1-dependent genes are still expressed in a pcr1Δ mutant but with significantly delayed kinetics thus providing an explanation for the relatively mild stress sensitivity displayed by pcr1Δ. Consistent with this delay, Sty1 and Atf1 cannot be detected at these promoters in this condition suggesting that their association with chromatin is weak or transient in the absence of Pcr1.
Keywords: S. pombe, Sty1, Atf1, Pcr1, Stress response
Introduction
All cells sense and react to changes in their environment. Single-celled organisms are especially vulnerable to such events and must respond to alterations in pH, osmolarity and temperature as well as contending with potentially harmful toxins. One of the major ways in which cells respond to such changes is by radically altering their programme of gene expression, leading to increased levels of proteins with stress protectant functions and a decrease in non-essential activities; this enables resources to be concentrated on adaptation to stress and the repair of damaged macromolecules (reviewed in (1)).
Stress-activated MAP kinase pathways (SAPKs) are central in all eukaryotes to the facilitation of gene expression changes that occur in response to certain environmental cues. The mammalian SAPKs, p38 and JNK, respond to diverse stresses and signals and as a result phosphorylate a number of targets including the ATF2 transcription factor (2-6). In fission yeast, the Sty1 MAPK (also known as Spc1/Phh1 – hereafter referred to as Sty1), is closely related to p38 and like its mammalian counterpart, responds to a variety of stimuli including osmotic, oxidative and heat stress (7-9). Upon exposure to stressors, Sty1 is activated by its MAPK kinase (Wis1), via phosphorylation at Thr171 and Tyr173 and consequently translocates to the nucleus (7,10). Activated Sty1 phosphorylates a number of targets including the b-ZIP transcription factor Atf1, the fission yeast homologue of mammalian ATF2 (11,12). Both Sty1 and Atf1 are required for many of the stress-induced transcriptional changes that occur upon stress; a large subset of these occur in response to several different stresses and as a result have been termed the core environmental stress response or CESR (13). Atf1 binds to its cognate site as a dimer with a second bZIP protein, Pcr1 (14). This heterodimer binds to DNA with much greater affinity than either homodimer (15,16). However, whilst cells lacking atf1 are sensitive to a variety of insults, the stress phenotypes displayed by a pcr1 deletant are much less severe, suggesting a less crucial role in regulating gene expression (15,17,18). Many signalling kinases, including SAPKs, modulate gene expression through regulatory phosphorylation of their target transcription factors or other chromatin-bound proteins (19). Previously we have addressed the function of Atf1 phosphorylation by Sty1. Surprisingly, phosphorylation is not required for stress-induced activation of Atf1-target genes but rather serves to positively regulate the stability of the Atf1 protein. Accordingly, the Atf1-11M phospho-mutant protein, which lacks intact MAPK sites, displays decreased stability compared to its wild type counterpart and accumulates to a lesser extent upon stress. Nevertheless, there is still robust activation of Atf1-dependent gene expression. Consistent with this, atf1-11M does not share the stress sensitivities displayed by an atf1 deletion mutant (18).
These results posed a dilemma: Atf1 is phosphorylated by Sty1; its role in stress-induced activation of gene expression is Sty1-dependent, yet phosphorylation of Atf1 by Sty1 does not appear to be critical for transcriptional activation per se. Thus, what is the essential role of Sty1 in stress-induced gene expression?
Clues may come from studies of Hog1, the budding yeast homologue of Sty1, which is primarily responsible for mediating the osmostress response in this yeast (reviewed in (20)). Hog1 can modulate transcription by phosphorylating its target transcription factors, thereby affecting their ability to activate gene expression (21,22). However, utilizing the chromatin immunoprecipitation technique (ChIP) which allows the mapping of DNA-protein interactions, Hog1 was shown to associate closely with target promoters upon exposure to increased osmolarity but not in response to other stressors such as heat and ethanol (23). The kinase is targeted and anchored to promoters by various translation factors such as Hot1. Once there, Hog1 serves to recruit key components of the general transcriptional machinery such as RNA pol II (24). Indeed, tethering of Hog1 to an artificial promoter by use of a lexA fusion system is sufficient to drive gene expression in a stress-dependent manner (24). In addition, the kinase can affect transcription by directing the association of chromatin-remodelling enzymes, such as the Rpd3 histone deacetylase, to osmoresponsive promoters (25). These findings raised the possibility that the kinase might play a structural role during transcription in addition to functions that require its catalytic activity. Recent studies have shown that Hog1 not only regulates transcriptional initiation but also affects the elongation cycle. The kinase can associate with the ORF as well as the terminator region of osmo-responsive genes where it appears to act as a component of the RNA polII elongation complex (26).
This close association between a protein kinase involved in signal transduction and genes under its control is likely to be a widespread phenomenon. Other budding yeast protein kinases subsequently found to interact directly with chromatin include Tor1, Sch9, Snf1, subunits of PKA and the MAP kinases Fus3 and Kss1, (27-29) whilst in higher eukaryotes, p38 and ERK have been reported to associate with specific promoters during myogenic and adipogenic differentiation respectively (30,31).
We reasoned that the essential role of Sty1 in activating stress-induced gene expression in fission yeast might involve direct association with stress-dependent genes and phosphorylation of chromatin-bound targets other than Atf1. Here we show that Sty1 is indeed recruited to promoters and ORFs of Atf1-dependent genes. Moreover, we have carried out a detailed analysis of the requirements for such association and in particular, have examined the role of the Atf1/Pcr1 heterodimer in this recruitment.
Experimental procedures
Yeast strains and general methods
S. pombe strains used in this study are listed in Table 1. Yeast media and general experimental methods were as described (32).
Table 1.
| Source | |
|---|---|
| NJ2 h− ura4-D18 leu1-32 ade6-M210 his7-366 | C. Hoffman |
| NJ23 h− 972 | Lab stock |
| NJ42 h− pcr1::ura4 ura4-D18 | This study |
| NJ260 h− atf1::kanr leu1-32 ura4-D18 ade6-M210 his7-366 | (18) |
| NJ294 h− sty1::kanr leu1-32 ura4-D18 ade6-M216 his7-366 | This study |
| NJ675 h− sty1-HA6His:ura4 leu1-32 ura4-D18 ade6-M216 his7-366 | This study |
| NJ711 h− sty1-12myc:ura4 leu1-32 ura4-D18 | (10) |
| NJ746 h?sty1-12myc:ura4 atf1-11M-2HA6his:LEU2 leu1-32 ura4-D18 | This study |
| NJ752 h?sty1-12myc:ura4 atf1-2HA6his:LEU2 leu1-32 ura4-D18 | This study |
| NJ757 h− sty1::kanr ura4-D18 | This study |
| NJ760 h?sty1::kanr leu1::nmt41sty1-3Pk:ura4 ura4-D18 (sty1-Pk) | This study |
| NJ765 h?sty1::kanr leu1::nmt41sty1K49R-3Pk:ura4 ura4-D1 (stykd1-Pk) | This study |
| NJ917 h?sty1-12myc:ura4 atf1-2HA6His:LEU2pcr1::kanr | This study |
| NJ922 h−sty1-12myc:ura4pcr1::kanr leu1-32 ura4-D18 | This study |
| NJ959 h? sty1-12myc:ura4 atf1::kanr leu1-32 ura4-D18 | This study |
| NJ961 h− sty1-12myc:ura4 caf1::natr leu1-32 ura4-D18 | This study |
| CW139 h− ura4-D18 | Lab stock |
To construct the sty1 kinase-dead Pk-tagged strain (NJ765) and its control strain (NJ760), first an exact deletion of sty1 was generated in a leu1+ura4-D18 strain (CW139) using standard procedures (33); oligonucleotide sequences available upon request. The sty1 and sty1 kinase-dead (K49R) coding sequences were amplified from plasmids by PCR using primers Sty1_BamH1_fw and Sty1_nostop_re_v2 (oligonucleotide sequences available upon request). The amplified 1kb PCR product was digested with XmaI and BamHI and subcloned into previously SmaI/BamHI cut plasmid pREP41Pkc (34). The obtained plasmids (pREP41sty1-Pkc and pREP41sty1kd-Pkc) were digested with SacI/PstI, the resulting 3kb band isolated and subsequently subcloned into the pINTA vector (35) which had been prepared by digestion with SacI and PstI, giving rise to plasmids pINTAsty1-Pkc and pINTAsty1kd-Pkc. The integration cassettes from plasmids pINTAsty1-Pkc and pINTAsty1kd-Pkc were obtained by a NotI digest, subsequently isolated and used for transformation of strain NJ757. Ura+/leu− colonies were selected and the integration events checked by genomic PCR and sequencing. The strains were verified by assessing cell length and stress sensitivity with the sty1-Pk (NJ760) and sty1kd-Pk (NJ765) behaving as wild type and the sty1Δ mutant respectively. These cells were grown in minimal media to ensure full induction from the nmt41 promoter.
ChIP assays
The ChIP procedure is based on methods described elsewhere (18) with modifications: in the case of Sty1-myc ChIPs, cultures were cross-linked for 30 minutes with 1% Formaldehyde at 24°C. Immunoprecipitation was performed with 15μL Dynal protein A-coated magnetic beads, which were previously incubated over night with 4-5μg of anti-Atf1 polyclonal antibody, anti-Pcr1 rabbit polyclonal antibody (18), anti c-Myc (A-14)X rabbit polyclonal antibody (Santa Cruz Biotechnology), anti-HA (12CA5, CRUK) or mouse anti V5 TAG (Pk) antibody (Serotec), respectively. We used standard PCR as well as real-time PCR to quantify the relative amount of DNA corresponding to stress induced genes isolated from each immunoprecipitate. For standard PCR, gpd1 was compared to the stress independent promoters, hmg1 and cdc2. Primers used are denoted relative to translation start codons and corresponded to −315 to −294 and −148 to −129 for the cdc2 promoter, −494 to −475 and −275 to −256 for the gpd1 promoter and −423 to −404 and −149 to −130 for the hmg1 promoter. For real time PCR quantifications, the relative enrichments of gpd1, hsp9, ctt1 and pka1 were measured over a stress independent promoter (cdc2). The relative enrichments of stress independent genes (cdc2 ORF, pol1 ORF) were measured as negative controls. Primers used are denoted relative to translation start codons and corresponded to −214 to −192 and −177 to −144 for the cdc2 promoter, +471 to +498 and +518 to +541 for cdc2 ORF, −446 to −421 and −388 to −363 for the ctt1 promoter, +1379 to +1399 and +1423 to +1439 for the ctt1 ORF, −402 to −380 and −360 to −340 for gpd1 promoter, +612 to +628 and +646 to +665 for gpd1 ORF, +1213 to +1241 and +1266 to +1286 for gpd1 3′UTR, −264 to −245 and −220 to −199 for the hsp9 promoter, −652 to −630 and −608 to −580 for the pka1 promoter, +622 to +639 and +664 to +684 for the pka1 ORF, +1615 to +1636 and +1657 to +1681 for the pka1 3′UTR, +2520 to +2543 and +2567 to +2588 for pol1 ORF. In addition, the following primers were used for the pka1 promoter (−143 to −121 and −101 to −75); there was no enrichment of Sty1 to these sequences (this data was not shown as part of Figure 7). A fraction of each IP was assayed by western blotting to ensure that equal quantities of Sty1 were being isolated from the samples before and after stress.
Western blotting
Atf1 and Pcr1 were detected as described (18). Activated Sty1 was detected using the anti-phospho-p38 antiserum (Genway). Other antibodies used were anti-HA antiserum (12CA5-Cancer Research UK), anti-myc (Serotec) and anti-Pk (Serotec). All antibodies were used in western blots at a dilution of 1 in 1000.
Northern blots
5μg of total RNA isolated at the time points indicated in the figures were prepared and analyzed as described (18). For the genes shown in Figure 10C, we also assessed their expression in a sty1-myc pcr1Δ mutant and found that the pattern of expression was identical to that shown for pcr1Δ.
Microarray experiments Culture, stress conditions and harvesting
RNA isolation, labelling, microarray hybridization, and data processing were performed as described using our in-house spotted arrays (36). We used the same experimental design as described (13): labelled samples from each stress time point were hybridized with a labelled reference pool containing an equal amount of all the RNA samples from the wild type time points of the corresponding stress. Two independent biological repeats were processed using a dye-swap for labelling. Microarrays were scanned with a Genepix 4000B scanner and analyzed with GenePix software (Axon Instruments). After data acquisition and within-array normalization, the ratios of each gene (time point/reference pool) were divided by the corresponding ratios of untreated wild type cells (0h wild type/reference pool). Thus, the reported ratios represent the expression levels at each time point relative to the expression levels of the untreated wild type cells from the same stress experiment. Expression ratios of biological repeat experiments were averaged. Hierarchical clustering and other data evaluation were performed using GeneSpring (Agilent). In total, 6 microarrays were used in this study and compared to the data from wild type and atf1Δ cells described in a previous study (13). The complete processed data set is available here: http://www.sanger.ac.uk/PostGenomics/S_pombe/.
Immunofluoresence
For immunofluoresence microscopy, cells were fixed as described (37). For visualizing myc-tagged proteins, mouse anti-myc (Cancer Research UK, 9E10) was used at a dilution of 1 to 100. Secondary antibodies were diluted 250-fold for use (Molecular Probes, Alexa Fluor 488 anti-mouse #A-11001). DNA was detected with DAPI (4,6-diamidine- 2-phenyl-indole) in Vectashield mounting medium (Vector, H-1200). Images were viewed using an Olympus BX51 microscope fitted with a 100mW epifluorescence bulb and using analysis software (Soft Imaging System, GmbH). Forscoring phenotypes, a minimum of 500 cells were scored for each genotype.
Results
Sty1 is recruited to the gpd1 promoter upon osmostress with kinetics that are coincident with those of its own activation
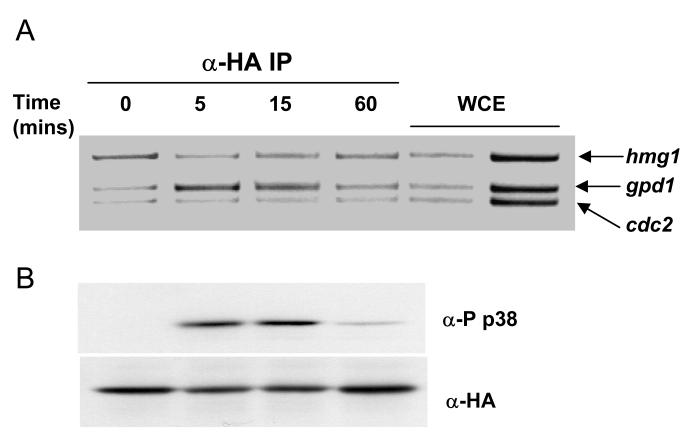
We first sought to examine whether Sty1 could associate with the promoters of stress-induced genes. Previously we have used chromatin immunoprecipitation assays (ChIP) to demonstrate that Atf1 is bound to the promoters of three stress-induced genes that form part of the CESR, namely gpd1, hsp9 and ctt1 (18).These three genes encode proteins with important stress protectant functions and they depend upon both Sty1 and Atf1 for their expression (13). The primer pairs used to amplify the promoters and control regions in this study are indicated in Figure 1 along with their locations relative to the start codons. In order to assess whether Sty1 could associate with chromatin, we examined whether Sty1 was recruited to the gpd1 promoter upon osmotic stress using a ChIP assay analyzed by standard PCR. These experiments were performed using a strain in which the sty1 gene has been fused at its C-terminus with 6 histidine residues and 2 copies of the HA epitope. This strain has previously been verified as behaving as wild type (7). Samples were collected during an hour of osmotic stress; association of Sty1 with the gpd1 promoter was assessed by an increase in the ratio of the PCR product generated by primers specific for the gpd1 promoter compared to the control products which were amplified by primers recognizing the non-stress specific promoters, cdc2 and hmg1. Sty1 recruitment was evident after 5 minutes of stress but had receded after an hour (Figure 2A). This association with the promoter matched the kinetics of Sty1 activation which was assessed by western blotting of Sty1 using antibodies that recognize the phosphorylated activation site. Sty1 phosphorylation was observed at 5 minutes but had almost disappeared by 60 minutes (Figure 2B). This recruitment is consistent with the kinetics of gpd1 transcript accumulation ((13) and Figure 10C). We observed significant amounts of gpd1 mRNA at 15 minutes which had reduced considerably by 60 minutes. These data show that Sty1 is recruited to chromatin of an Atf1-dependent gene upon osmotic stress and that the kinetics of Sty1 activation match those of its association with the gpd1 target gene and those of transcript accumulation. This is consistent with the essential role of Sty1 in transcriptional activation requiring its direct recruitment to stress-induced genes.
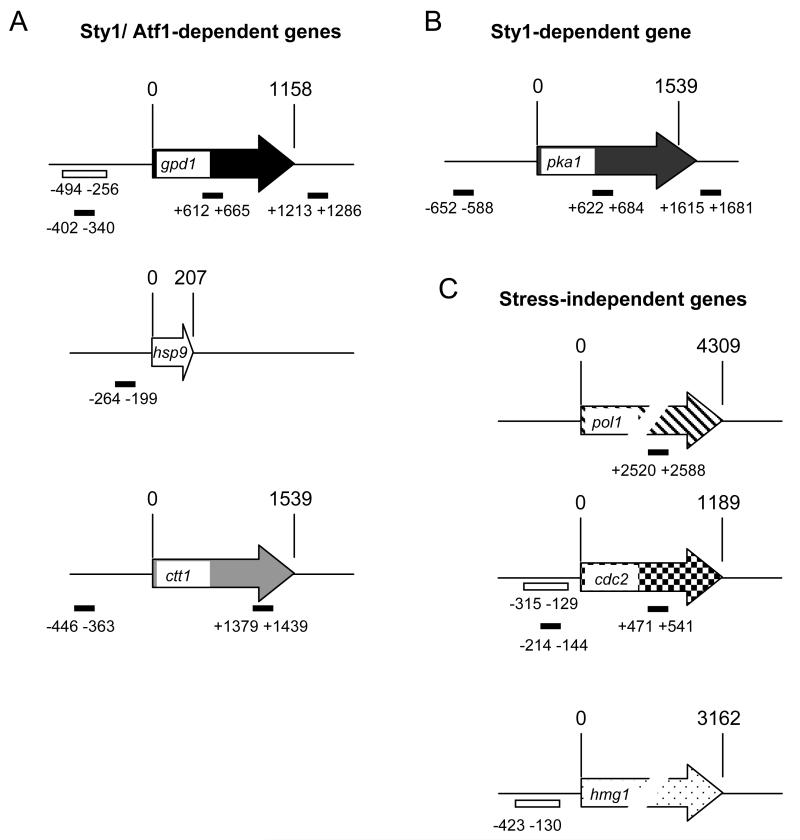
Figure 1.
The locations of primer pairs used in the ChIP analysis.
(A) The stress-induced genes, gpd1, hsp9 and ctt1 are dependent upon Sty1 and Atf1 for their induction.
(B) pka1 is a stress-induced Sty1-dependent, Atf1-independent gene.
(C) pol1, cdc2 and hmg1 are stress and Sty1/Atf1 independent genes and are used as negative controls. The numbers refer to the starting point of each primer relative to the atg (designated as 1). For example, for gpd1, the amplicons −402 to −340, +612 to +666 and +1213 to +1286 represent promoter, ORF and terminator regions respectively. For ctt1, the amplicons −446 to −363 and +1379 to +1439 represent promoter and ORF regions respectively. The amplicons analyzed by real time PCR assays are shown as solid black bars. Those used in standard PCR (Figure 2) are shown as open bars.
Figure 2.
Sty1 is recruited to the gpd1 promoter upon osmotic stress in a manner coincident with its own activation.
(A) ChIP assays showing recruitment of Sty1-6His2HA to the gpd1 promoter upon osmotic stress. Samples were prepared from sty1-6His2HA cells treated with 1M sorbitol for the time points indicated (in minutes). The DNA recovered from the IP was assayed by PCR using primers specific to the gpd1, hmg1 and cdc2 promoters, the latter two of which are not induced upon stress. The control lanes show DNA amplified from two different amounts of whole cell extracts (WCE) prior to performing the IP.
(B) The kinetics of Sty1 activation were assayed by western blotting of protein extracts prepared from the cells exposed to osmotic stress in (A). The blots were probed with antibodies against the activation site of the p38 MAP kinase (anti-phospho p38) which recognize the activation site of Sty1 when dually phosphorylated upon Thr171 and Tyr173. Total levels of Sty1 were assessed by re-probing the blot with anti-HA antiserum.
Sty1 is recruited to Atf1-dependent promoters upon a variety of stress conditions
Next we assessed whether Sty1 could associate with promoters under different types of stress other than osmostress. For these assays we used a strain where the sty1 gene had been C-terminally fused with 12 copies of the myc epitope at its native locus such that the expression of sty1-myc is under the control of the sty1 promoter. In terms of phenotype and gene induction this strain behaves as wild type (10,38) and data not shown. The sty1-myc cells were isolated in the presence and absence of stress and ChIP assays were performed in triplicate. The DNA recovered from the IPs was analyzed by quantitative real-time PCR. Recruitment was determined by comparing the enrichment in the immunoprecipitates of regions corresponding to the gpd1, ctt1 and hsp9 promoters compared with the cdc2 control promoter, which is not expected to be bound by Sty1 (fold over cdc2 promoter). In addition we measured recruitment to regions of DNA that would not be expected to be bound by Sty1 namely the cdc2 ORF and pol1 ORF; (we have previously shown that Atf1 is not bound to these control regions (18)). We found that Sty1 is recruited to the gpd1, ctt1 and hsp9 promoters upon sorbitol-induced osmotic stress, confirming the result observed with the standard PCR assay. Upon stress we observed between 3 and 7.5 fold enrichment of these promoters in the Sty1-containing IPs compared to either the sty1-myc strain in the absence of stress or to an untagged control (wild type) (Figure 3A).
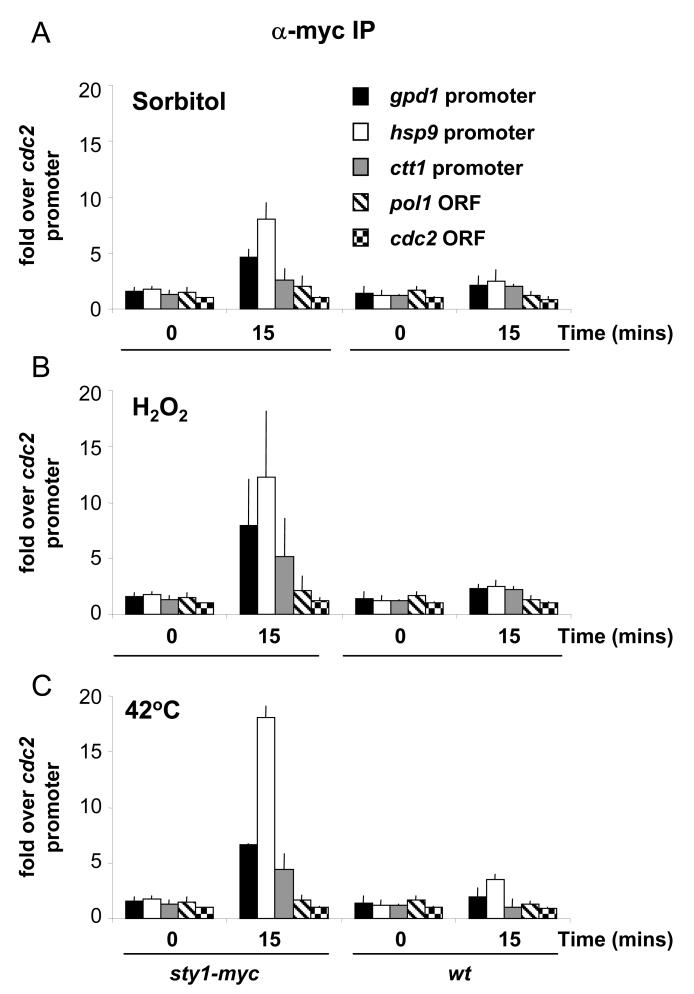
Figure 3.
Recruitment of Sty1 to stress-induced promoters upon a variety of stresses as determined by quantitative ChIP assays.
(A) The sty1-12myc strain and an untagged wild type strain (wt) were sampled before and after exposure to 1M sorbitol for 15 minutes. The experiment was performed in triplicate and results are shown with standard deviation. This is the case for all subsequent ChIP data that has been examined by quantitative PCR. The DNA bound to Sty1 was assayed by real time PCR and the results are expressed as fold enrichment over the cdc2 promoter (which is independent of both Sty1 and Atf1 for its expression and is not induced upon stress).
(B) Recruitment of Sty1 to stress-induced promoters upon oxidative stress. The sty1-12myc strain and an untagged wild type strain (wt) were sampled before and after exposure to 2mM H2O2 for 15 minutes. The experiment was performed in triplicate and the assays were performed as described in (A).
(C) Recruitment of Sty1 to stress-induced promoters upon heat stress. The sty1-12myc strain and an untagged strain (wt) were sampled before and after shifting the cultures to 42°C for 15 minutes. The experiment was performed in triplicate and the assays were performed as described in (A).
The lack of recruitment of Sty1 to promoters in the absence of stress is consistent with the predominantly cytoplasmic localization of Sty1 under basal conditions (10). We also monitored the amount of Sty1 that was recovered in either the absence or presence of stress for these and all other assays in this study and found that equal amounts of Sty1 were precipitated in both conditions (data not shown).
The Hog1 MAP kinase has been shown to be recruited to promoters only upon osmotic stress and not during exposure to heat, ethanol or sorbic acid (23). As Sty1 is required for gene expression upon exposure to a variety of stressors (13), we examined whether it was also recruited to the CESR promoters upon other stress conditions. As shown in Figures 3B and 3C, Sty1 was recruited to the gpd1, hsp9 and ctt1 promoters upon treatment of cells with oxidative stress as well as upon heat shock. Taken together, these data indicate that Sty1 is recruited to a number of Atf1-dependent promoters in response to different stress conditions, consistent with the critical role of Sty1 in controlling transcription upon exposure to a variety of stressors (13,39).
Nuclear localization of Sty1 is not sufficient to induce promoter recruitment in the absence of Sty1 activation
The findings described above are consistent with Sty1 activation being a prerequisite for promoter binding. The simplest way to test this hypothesis would be to assess Sty1 promoter recruitment in a mutant where its activation was abolished such as the wis1Δ mutant. However, in this instance Sty1 would be constitutively cytoplasmic as activation by Wis1 is required for its translocation into the nucleus (10). To resolve this matter we made use of a mutation in the caf1 gene (also known as hba1) which encodes a Ran-binding protein which plays a role in controlling the localization of Sty1. Loss of this gene results in constitutive accumulation of Sty1 in the nucleus (40). Sty1 is not phosphorylated under basal conditions in the caf1Δ mutant ((40), data not shown), and the levels of Sty1 do not change upon stress in this mutant background (data not shown). As Wis1 is predominantly cytoplasmic, the caf1Δ mutation results in the separation of most of the Sty1 MAPK from its upstream MAPKK.
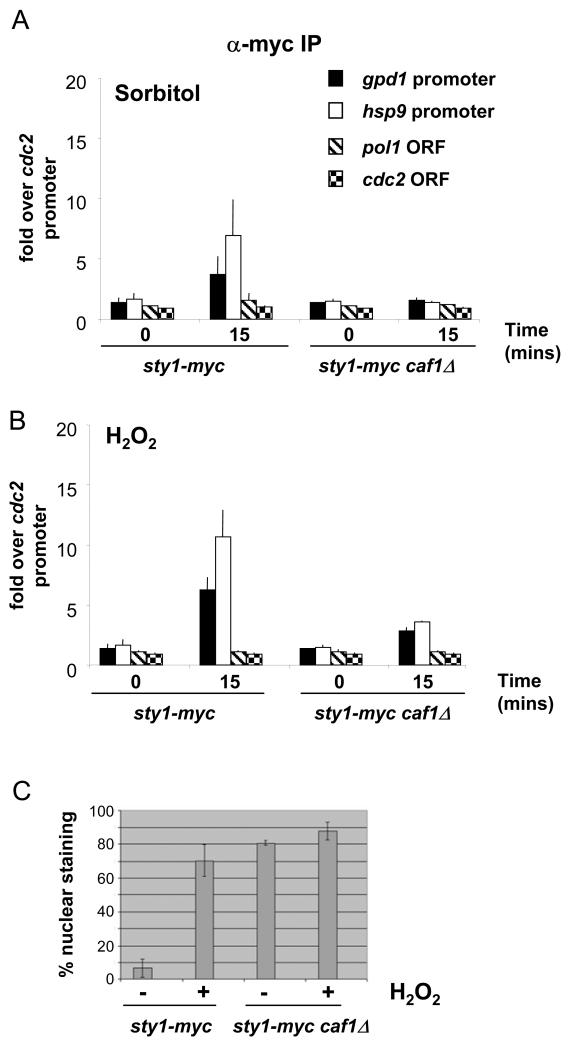
We examined promoter recruitment of Sty1 in a caf1Δ mutant and did not observe any significant promoter association upon either osmotic (Figure 4A) or oxidative stress (Figure 4B). We performed indirect immunofluorescence to check that Sty1-myc was indeed constitutively localized in the nucleus and found that Sty1 displayed a nuclear localization in 80% of sty1-myc caf1Δ cells under basal conditions (Figure 4C), which is in good agreement with the original study (40). Upon oxidative stress, there was a only a very slight recruitment of the kinase to the CESR promoters presumably because the fraction of Sty1 that remains cytoplasmic in the caf1Δ mutant can still be activated; alternatively this could be due to the activation of constitutively nuclear Sty1 by the fraction of Wis1 that shuttles in and out of the nucleus (41). We conclude that Sty1 recruitment to promoters does not arise simply as a consequence of its increased nuclear accumulation that occurs upon stress but rather that Sty1 needs to be activated in order to associate with promoters.
Figure 4.
Nuclear localization of Sty1 is not sufficient to induce promoter recruitment in the absence of Sty1 activation.
(A) ChIP assays to assess the recruitment of Sty1 to stress-induced promoters in caf1Δ cells. Assays were carried out as in Figure 3A using samples prepared from sty1-myc caf1Δ and wild type cells that had been exposed to 1M sorbitol for 15 minutes.
(B) ChIP assays as in (A) but using cells that had been exposed to 2mM H2O2 for 15 minutes.
(C) Results of indirect immunofluoresence to assess the nuclear localization of Sty1 in caf1Δ cells in the absence and presence of stress (2mM H2O2 for 15 minutes). More than 500 cells were counted for each strain in each condition. The results of three independent experiments are shown.
Sty1 kinase activity is required for its recruitment
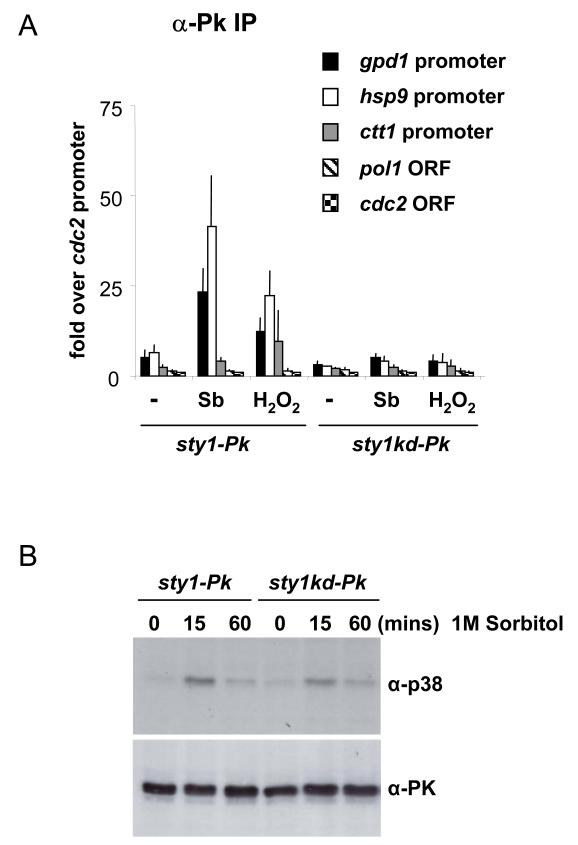
As activation of Sty1 is a prerequisite for recruitment to promoters, we speculated that its own catalytic activity would also be required. To address this issue, we generated a strain that expressed a version of Sty1 that has a mutation in the predicted ATP binding site resulting in a so-called “kinase-dead” form of the enzyme (K49R). This mutated version of the sty1 coding region (sty1kd) was integrated into the genome of a sty1Δ mutant at a heterologous locus and expressed from the nmt41 promoter. In addition, the sty1kd ORF was fused to 3 copies of the Pk epitope at its C-terminus (sty1kd-Pk). As a control, a strain was generated whereby a wild type version of the sty1 gene was integrated into the genome and epitope-tagged in the same way (sty1-Pk). This strain behaved as wild type with respect to cell length and stress resistance suggesting that Sty1 is expressed at adequate levels and that the epitopes do not inhibit Sty1 function. In contrast, cells with the sty1kd mutation displayed stress sensitivities and cell length defects similar to a sty1 deletion mutant suggesting that Sty1 activity had been compromised (data not shown). We assessed recruitment of Sty1kd protein to promoters. As shown in Figure 5A, the mutant kinase did not associate with promoters, in contrast to the control (sty1-Pk), suggesting that the catalytic activity of Sty1 is essential for its recruitment to stress-induced genes. The kinase-dead version of Sty1 was activated to comparable levels as the wild type protein excluding incomplete activation as a reason for its inability to bind to promoters (Figure 5B). The lack of recruitment could not be explained by loss of the Atf1-Pcr1 heterodimer as these factors were both found to be associated with the promoters in question in the sty1kd mutant (data not shown). Taking the above data together, we conclude that Sty1’s catalytic activity is required for its association with stress-responsive promoters.
Figure 5.
Sty1 kinase activity is required for its recruitment to stress-responsive promoters.
(A) ChIP assays to assess the recruitment of Sty1 and Sty1 kinase-dead to stress-induced promoters. Assays were carried out as in Figure 3A using samples prepared from sty1-Pk and sty1kd-Pk cells that had been exposed to 1M sorbitol or 2mM H2O2 for 15 minutes. Cells grown without stress were used as a control (indicated by -).
(B) Western Blot to examine the activation of Sty1 in the sty1-kinase dead (sty1kd-Pk) strain compared to the sty1-Pk strain. Whole cell extracts were prepared before or after 1M sorbitol stress. Sty1 activation was assessed using anti-phospho p38 antiserum. The loading was assessed using anti-Pk antiserum.
Phosphorylation of Atf1 is not required for recruitment of Sty1
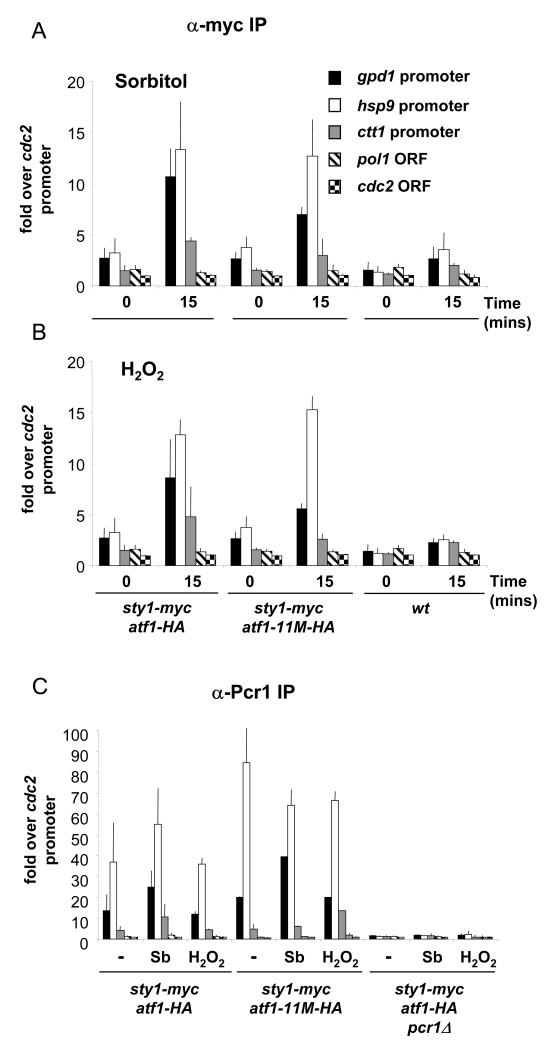
Next we sought to determine what promoter-bound factors are important for Sty1 recruitment. The requirement of Sty1 kinase activity for its association with promoters led us to examine whether phosphorylation of Atf1 was the critical event mediating the recruitment. Sty1 phosphorylates Atf1 which results in the modulation of Atf1’s stability. However, loss of this phosphorylation through mutation of the 11 potential MAPK sites in Atf1 does not abolish stress-induced transcription of Atf1 target genes. Accordingly, this mutated version of Atf1, Atf1-11M, is still bound to Atf1-dependent promoters (18). We asked whether Sty1 recruitment is dependent upon these phosphorylation sites being intact. As shown in Figure 6A and B, Sty1 is still recruited to promoters in the atf1-11M mutant, both upon osmotic and oxidative stress. Furthermore, the levels of recruitment are comparable to those seen in an atf1+ strain. We characterized the status of these promoters further by asking whether Pcr1 is still bound in the atf1-11M mutant. As shown in Figure 6C, Pcr1 is bound to Atf1-dependent promoters in the absence and presence of stress. This is not affected by a lack of Atf1 phosphorylation.
Figure 6.
The atf1-11M allele does not inhibit recruitment of Sty1 to Atf1-dependent promoters.
(A) Samples were processed for ChIP assays as described in Figure 3A. The following strains were used: wild type (wt), sty1-myc atf1-HA and sty1-myc atf1-11M-HA. The stress applied was 1M sorbitol for 15 minutes. As the atf1-11M allele is tagged with the HA epitope, an equivalent version of atf1+ was used as a control (sty1-myc atf1-HA).
(B) Samples of the strains described in (A) were processed for ChIP analysis. Cells were treated with 2mM H2O2 for 15 minutes.
(C) Pcr1 is bound to promoters in the atf1-11M mutant. Samples were processed for ChIP assays as described in Figure 3A. The following strains were used: sty1-myc atf1-HA, sty1-myc atf1-11M-HA and sty1-myc atf1-HA pcr1Δ. The stressors applied were 1M sorbitol or 2mM H2O2 for 15 minutes. The DNA bound to Pcr1 was recovered after immunoprecipitation reactions using antiserum raised against Pcr1.
Taken together, these data indicate that phosphorylation of Atf1 is not the critical event in recruitment of Sty1 to stress-dependent promoters. Moreover, it is consistent with our previous study which demonstrated that transcription is still activated upon stress in the atf1-11M mutant (18).
Sty1 is not recruited to an Atf1-independent gene
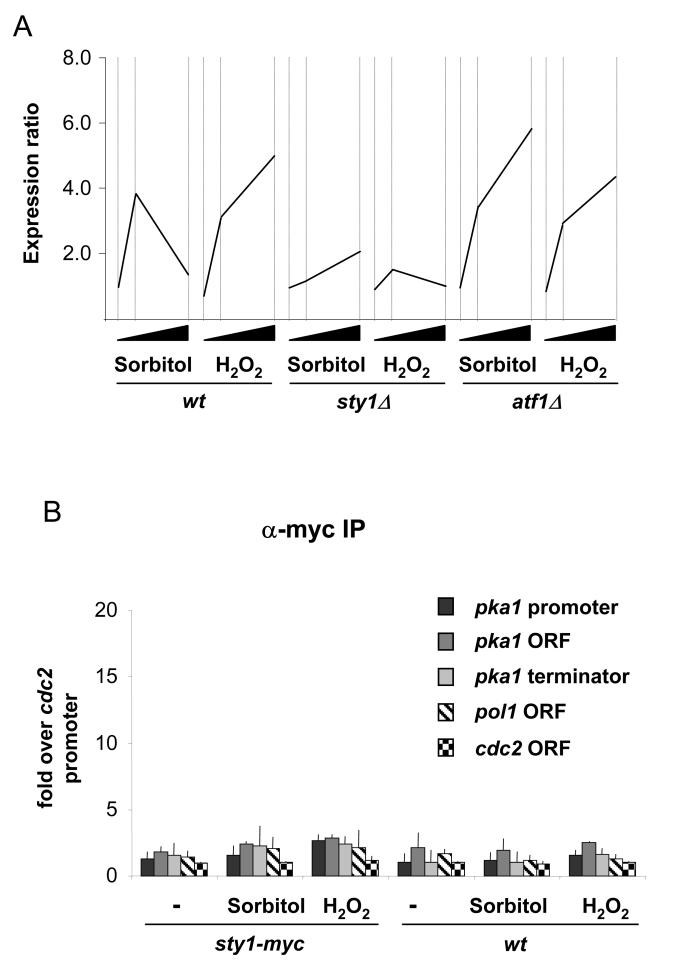
The genes that we had assessed above, namely gpd1, hsp9 and ctt1, all require Atf1 for their expression. However, in our previous analysis of stress induced transcription by whole genome profiling (13), we identified a group of stress-induced CESR genes that are dependent upon Sty1 but not Atf1 (see Figure 5, group 3 genes of Chen et al., (13)). We asked whether Sty1 was recruited to the promoter of one of these Sty1 dependent/Atf1 independent genes: namely pka1. The expression data for pka1 are shown in Figure 7A and the requirement for Sty1 but not Atf1 is evident.
Figure 7.
Sty1 is not recruited to an Atf1-independent gene.
(A) Expression of the pka1 gene upon osmotic and oxidative stress in wild type, sty1Δ and atf1Δ cells. The data has been extracted from our previous microarray study (13). The fold-induction is shown on the y-axis. The time points of stress were 0, 15 and 60 minutes and this is represented by the black wedges on the x-axis.
(B) Samples were processed for ChIP assays as described in Figure 3A using sty1-myc cells. Three different primer sets were used to assess recruitment to the promoter and gave similar results (see Figure 1B). The data for the set corresponding to −652 to −588 are shown here (pka1 promoter). In addition, primer pairs corresponding to the pka1 ORF and terminator were also tested and gave similar results to those designed to amplify the promoter.
The primer pairs used for this ChIP analysis are indicated in Figure 1A. Sty1 was not recruited to the pka1 promoter (or to the body of the gene) upon either osmotic or oxidative stress (Figure 7B). These data suggest that Sty1 controls pka1 and probably other Atf1 independent genes by a mechanism that does not involve intimate association with promoters.
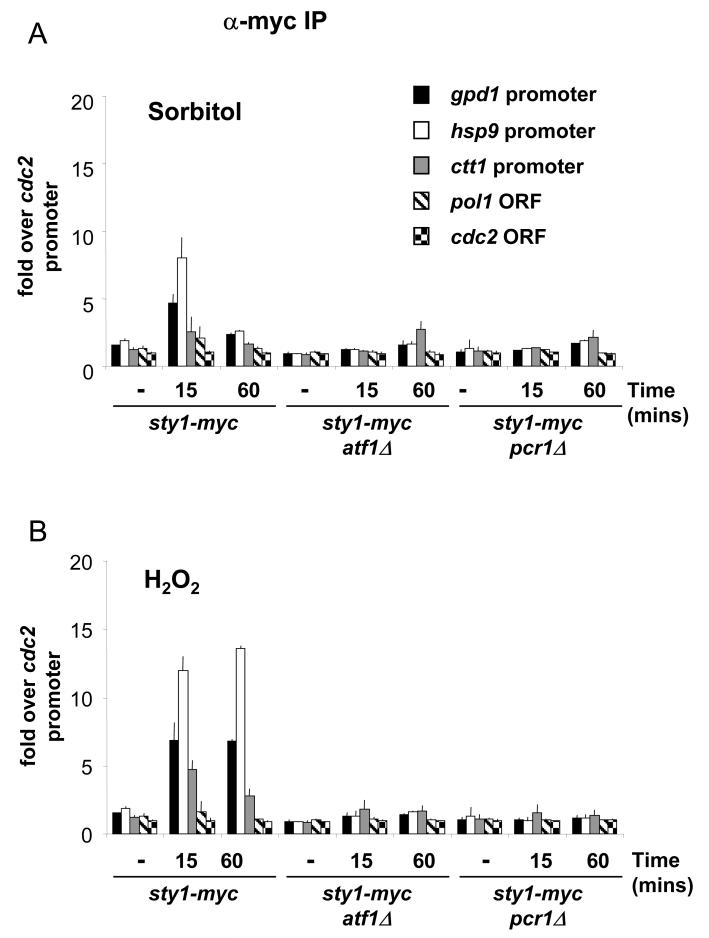
Sty1 recruitment is abolished in cells lacking either Atf1 or Pcr1
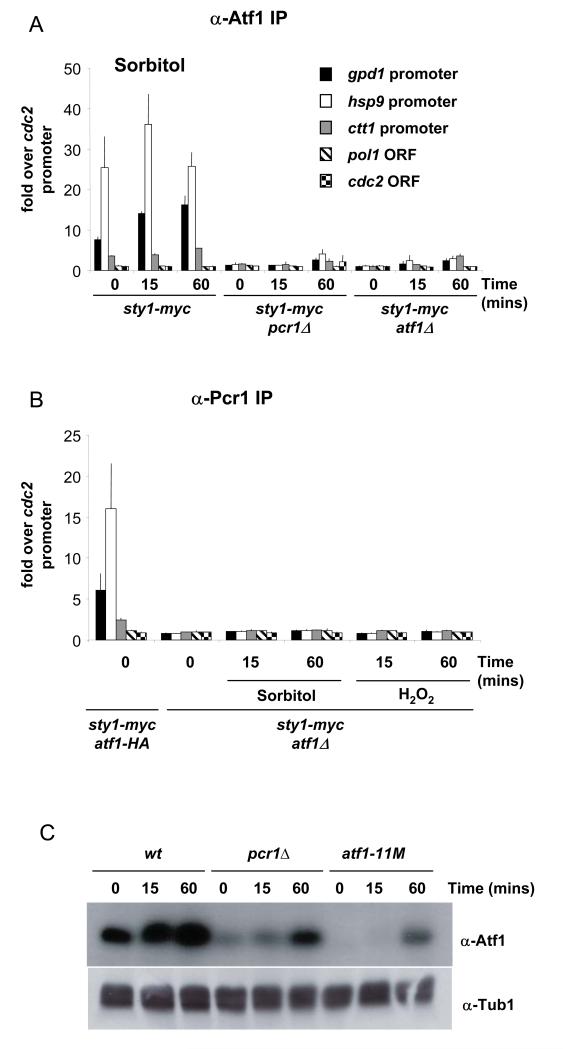
The recruitment of Sty1 to Atf1-dependent genes but not to one whose expression is independent of this factor suggested that Atf1 is required for association of the kinase with chromatin, even though phosphorylation of Atf1 is not. We tested this by examining Sty1 recruitment in an atf1Δ mutant. As shown, there was no association of Sty1 with the promoters of Atf1 dependent CESR genes in atf1Δ upon exposure to either osmotic or oxidative stress (Figures 8A and B respectively). These data are consistent with Atf1 acting as a recruiting factor for Sty1. It is possible however, that recruitment is determined by Pcr1 and that in the absence of Atf1, Pcr1 can no longer bind to promoters as it preferentially forms heterodimers with Atf1 rather than homodimers. To address this possibility, we examined whether Pcr1 could still bind to the CESR promoters in the atf1Δ mutant. As shown in Figure 9B, we were unable to detect binding of Pcr1 to hsp9, ctt1 and gpd1 promoters in the atf1Δ mutant either upon osmotic or oxidative stress.
Figure 8.
Sty1 is not recruited to stress-responsive promoters in the absence of either Atf1 or Pcr1.
(A) Samples were processed for ChIP assays as described in Figure 3A. The following strains were used: sty1-myc, sty1-myc atf1Δ and sty1-myc pcr1Δ. The stress applied was 1M sorbitol for 15 minutes.
(B) Samples of the strains described in (A) were processed for ChIP analysis. Cells were treated with 2mM H2O2 for 15 minutes.
Figure 9.
Atf1 is not detected at promoters in the absence of Pcr1 and vice versa.
(A) Samples were processed for ChIP assays as described in Figure 3A. The following strains were used: sty1-myc, sty1-myc atf1Δ and sty1-myc pcr1Δ. The stress applied was 1M sorbitol for 15 minutes. The DNA bound to Atf1 was recovered after immunoprecipitation reactions using antiserum raised against Atf1.
(B) Samples of the strains described in (A) were processed for ChIP analysis. Cells were treated with 2mM H2O2 for 15 minutes. The DNA bound to Pcr1 was recovered after immunoprecipitation reactions using antiserum raised against Pcr1.
(C) Western Blot to examine the levels of Atf1 protein in pcr1Δ compared to the atf1-11M mutant. The loading was assessed using anti-tubulin antiserum.
If Atf1 alone was determining Sty1 recruitment, it might be possible to observe association of Sty1 with promoters in the absence of Pcr1 and thus we assessed whether we could detect Sty1 recruitment in a pcr1Δ mutant. As shown, Sty1 was not recruited to the promoters of the three CESR genes in the absence of Pcr1, either upon osmotic stress (Figure 8A) or oxidative stress (Figure 8B). This was true at both early and late time points (15 and 60 minutes). Given the dependency of Pcr1 binding upon Atf1 (Figure 9B), we asked whether the converse was true by examining the binding of Atf1 to promoters in the absence of Pcr1. As shown in Figure 9A, Atf1 was no longer associated with the CESR promoters in the pcr1Δ mutant upon either osmotic or oxidative stress. We have previously shown that the levels of Atf1 are regulated by Pcr1 and vice versa (18). Even though there is less Atf1 protein in a pcr1Δ mutant, there is still more than in the atf1-11M mutant (Figure 9C) and we observed binding of both Sty1 and Pcr1 in the latter (Figure 6). Thus we conclude that the lack of binding of Atf1 and Sty1 to promoters in pcr1Δ, is due to loss of Pcr1 and Atf1 at promoters and not caused by reduced levels of Atf1. Taking all the above data together, we conclude that Sty1 cannot be recruited to chromatin in the absence of either Atf1 or Pcr1. Given the interdependency of binding between Atf1 and Pcr1, it was not possible to differentiate between recruitment being dictated by the intact Atf1-Pcr1 heterodimer or by one of these factors alone.
Some Atf1-dependent genes are still expressed in a pcr1Δ mutant
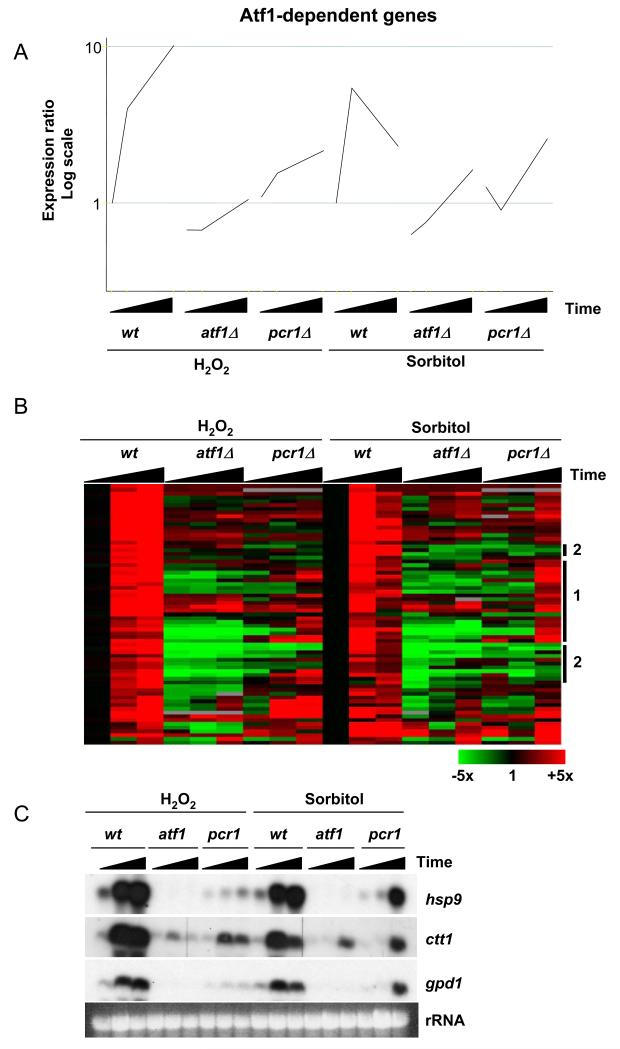
The dependency of Sty1 recruitment upon Pcr1 presented an interesting dilemma: cells lacking Pcr1 grow in a variety of conditions that are stressful enough to render atf1Δ and sty1Δ mutants inviable (15,17,18). These observations suggested that in the absence of Pcr1, cells are still able to produce sufficient mRNA from genes encoding stress protectant functions to ensure survival. Surprisingly, this transcription appears to be independent of the stable recruitment of both Sty1 and Atf1 to promoters (Figures 8 and 9B). In order to address this issue, we performed genome-wide expression analyses of the response to osmotic and oxidative stress in a pcr1Δ mutant and compared the findings to our previous data acquired from wild type and atf1Δ cells (13). For this analysis, we exposed pcr1Δ cells to sorbitol and H2O2 and collected samples at 15 and 60 minutes after stress. The resulting changes in gene expression were analyzed by DNA microarray hybridization. Clearly many of the changes in gene expression that are dependent upon Atf1 (according to a previous study (13)) also require Pcr1. This is illustrated by the average expression patterns of Atf1-dependent genes as shown in Figure 10A. However, the average increase in expression of these genes is higher in pcr1Δ than in atf1Δ upon both osmotic and oxidative stress consistent with the less severe phenotype displayed by the pcr1Δ mutant.
Figure 10.
The expression of Atf1-dependent genes in a pcr1Δ mutant in response to osmotic and oxidative stress.
(A) The pcr1Δ mutant was treated with 1M sorbitol and 0.5mM H2O2 and samples taken before and at 15 and 60 minutes after stress. The data was compared with gene expression changes that occur in wild type (wt) and atf1Δ cells under the same stress conditions (13). Average gene expression profiles of Atf1-dependent genes in wt, atf1Δ and pcr1Δ cells are shown.
(B) A hierarchical cluster analysis of the data described in (A) is shown with rows representing genes and columns representing experimental time points. The mRNA levels at each time point relative to the levels in the same cells before stress treatments are colour-coded as indicated at the bottom with missing data in grey. Specific gene groups are highlighted at the right as discussed in the main text.
(C) Northern blot to show the expression of the CESR genes analyzed by the ChIP assays. Total RNA was isolated from the strains in the absence of stress and upon 15 or 60 minutes after the application of either osmotic or oxidative stress as described in (A) and is indicated by black wedges. The expression of hsp9, ctt1 and gpd1 was assessed by northern blot using probes corresponding to these genes. Equal loading was assessed by total rRNA.
Figure 10B shows a cluster analysis of the Atf1-dependent stress-induced genes (13). Whilst most genes require both factors for optimal expression, many genes remain substantially expressed in the pcr1Δ mutant, although in many cases with altered kinetics such that transcript levels peak at 60 minutes rather than at 15 minutes as observed in wild type (e.g., expression patterns of group 1 genes in Figure 10B). In contrast, there are also some genes whose expression is absolutely dependent upon both Atf1 and Pcr1 for their expression upon stress (e.g., group 2 genes in Figure 10B). The genes that fall into these two categories are listed in Table 2. The two groups of genes cannot be distinguished on the basis of associated GO terms and both groups contain a mixture of well defined stress protectant genes and uncharacterized genes. The CESR genes examined with respect to Sty1 recruitment, namely gpd1, hsp9 and ctt1, are all dependent upon both Atf1 and Pcr1 for optimal transcription, but upon sorbitol stress in particular, display a considerable delay in their expression. We confirmed their patterns of expression by northern blotting. As shown in Figure 10C, the pcr1Δ mutant displayed a greatly reduced level of expression for each of the three genes upon exposure to H2O2 but considerable induction of all three genes after 60 minutes of sorbitol stress. It is likely that some of the remaining expression observed upon H2O2 stress is regulated by Pap1 (42) (our unpublished results). However, this transcription factor is only activated by oxidative stress (43) and thus cannot explain the delayed transcription observed upon osmotic stress. As described earlier, we cannot detect the binding of either Atf1 or Sty1 to promoters in the absence of Pcr1 (Figures 8 and 9). Thus the Atf1-dependent expression in pcr1Δ must be driven either by the binding of Atf1 and Sty1 at levels that are sufficiently low and/or transient enough to evade detection by the ChIP assays or alternatively by other factors whose expression depends upon Atf1 and Sty1.
Table 2.
Atfl-dependent genes that are induced in the pcrlΔ mutant but with altered kinetics (Group 1)
| Gene name | Annotation |
|---|---|
| vipl | Protein with rrm RNA recognition motif |
| mfp2 | Protein kinase inhibitor |
| SPAC9E9.04 | Homologue of Bap31 |
| hsp9 | Heat shock protein |
| gpd1 | Glycerol-3-phoshate dehydrogenase |
| SPCP31B10.06 | Protein containing a C2 domain |
| SPAC2F3.05 | Protein with aldo-keto reductase family domain |
| SPAC22F8.05 | Predicted alpha,alpha-trehalose-phosphate synthase |
| SPCC338.12 | Member of the subtilisin N-terminal region containing family |
| SPBC21C3.19 | Protein of unknown function |
| SPBC11C11.06c | Protein of unknown function |
| git5 | Heterotrimeric G protein beta subunit Git5, Git5, Gpb1 |
| SPAC4D7.02c | Member of the glycerophosphoryl diester phosphodiesterase family |
| zym1 | Putative class I metallothionein |
| SPBC725.10 | Protein with similarity to peripheral-type benzodiazepine receptor |
| SPAC57A 7.02c | Protein of unknown function |
| SPCC16A11.15c | Protein of unknown function |
| pyp2 | Protein tyrosine phosphatase |
| SPBC660.05 | Hypothetical protein |
| SPAC26F1.07 | Protein with high similarity to aldehyde reductase |
| SPBC16A3.02c | Member of the zinc-binding dehydrogenase family |
|
| |
| Atf1-dependent genes that are not induced in atf1Δ or pcr1Δ (Group 2) | |
| Gene name | Annotation |
|
| |
| SPAC15E1.02c | Protein of unknown function |
| SPAC4H3.08 | Protein containing a short chain dehydrogenase domain |
| SPCC757.03c | Member of the DJ-1 or PfpI family |
| SPCC1393.12 | Protein of unknown function |
| gpx1 | glutathione peroxidase Gpx1 |
| rds1 | Stress response protein |
| SPAC19G12.09 | Protein containing an aldo-keto reductase family domain |
| grx1 | Glutaredoxin |
| mrf1;etr1 | Member of the zinc-binding dehydrogenase family |
| SPAC23D3.11 | Protein containing a short chain dehydrogenase domain |
| exg3 | Protein with low similarity to exo-beta-(1,3)-glucanase |
| SPBC2A9.02 | Protein of unknown function |
| SPBC23G7.10c | Protein containing an NADH:flavin oxidoreductase or NADH oxidase family domain |
| SPAC23D3.05c | Pseudogene |
| SPBC1773.06c | Member of the zinc-binding dehydrogenase family |
| SPAC977.13c | Pseudogene |
Annotations are based on S. pombe GeneDB at http://www.genedb.org/genedb/pombe/index.jsp.
Taken together, these data demonstrate that whilst optimal stress-induced gene expression requires both Atf1 and Pcr1, significant but delayed induction of many Atf1-dependent genes still occurs in cells lacking Pcr1, particularly upon osmotic stress. These findings provide an explanation for the less severe stress phenotypes displayed by pcr1Δ compared to atf1Δ.
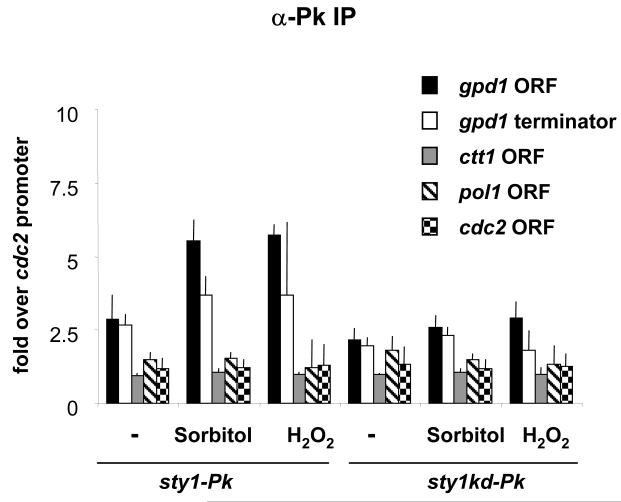
Sty1 is recruited to the open reading frames of stress-induced genes
In addition to its recruitment to promoters, the budding yeast MAPK Hog1 is also associated with the ORF of stress-induced genes where it interacts with components of the transcriptional elongation complex (26). Thus we examined whether Sty1 also associates with the coding regions of stress-induced genes. As the average size of DNA produced during the ChIP analysis was 500bp we chose to examine association of Sty1 with gpd1 and ctt1 as these genes are 1158bp and 1539bp in length, respectively. Primer pairs could therefore be designed sufficiently far apart so as not to amplify both promoter and ORF from the same piece of DNA (primer pairs shown in Figure 1). During initial experiments using myc-tagged Sty1, we observed recruitment of the kinase to both the ORF and terminator regions of gpd1 upon osmotic and oxidative stress, although the fold increase of binding over the control regions was considerably less than we had observed for the promoter regions (data not shown). However, using the untagged control strain we also observed higher levels of non-specific enrichment of these regions than we had observed during the promoter analysis. This problem was prevented by using the Pk-tagged version of Sty1 for the analysis. As a control, we used the kinase-dead mutant of Sty1 which was not recruited to the promoters of stress-dependent genes (Figure 5). Upon both osmotic and oxidative stress, Sty1 was recruited to the ORF and terminator region of the gpd1 gene (Figure 11) but not to the ORFs of the constitutively expressed cdc2 or pol1 genes. Interestingly, we did not observe recruitment to the ctt1 ORF whereas above we showed that Sty1 is associated with the promoter of this gene. The reason for this is not clear but we can eliminate primer design as an explanation as the oligonucleotides in question do amplify this region from whole cell extract (data not shown).
Figure 11.
Sty1 is recruited to the coding and terminator region of a stress-induced gene. ChIP assays perfomed as described in Figure 5 using primer pairs to amplify the gpd1 ORF and terminator (as described in Figure 1A). The cells were exposed to stress for 15 minutes.
We conclude that Sty1 can be recruited not only to promoters but also to the body of at least some stress-induced genes as well as their terminator regions. This suggests that the role of SAPKs in transcriptional elongation control may be evolutionarily conserved.
Discussion
Sty1 is stably associated with Atf1-dependent stress responsive promoters
Many stress-induced transcriptional events in fission yeast depend upon the Sty1 MAPK and its target transcription factor Atf1 (13). However, in contrast to a number of transcription factors that are regulated by MAPKs, the phosphorylation of Atf1 by Sty1 is not required for robust induction of gene expression (18). We postulated that the essential role of Sty1 in transcriptional activation might be a more direct one necessitating its recruitment to chromatin where it could either phosphorylate other targets and/or act as a structural adaptor in transcriptional initiation complexes. Here we have examined the recruitment of Sty1 to promoters using three different epitope-tagged versions of the kinase, assaying the binding using both real-time and standard gel-based PCR as a read-out of association and performing the analysis with a variety of primer pairs to identify the genes in question. We conclude that Sty1 is indeed recruited to stress-dependent promoters.
Recruitment of Sty1 to chromatin by Atf1/Pcr1
Sty1 interacts with Atf1 (11,12), the latter being a good candidate for the factor that recruits Sty1 to chromatin. However, given the loss of detectable binding of Atf1 in pcr1Δ and vice versa, it is hard to assess the contribution of Pcr1 to this process as it is not possible to study Sty1 recruitment in a situation where only one of the two bZIP proteins is bound to DNA. Thus Sty1 recruitment could be mediated exclusively through either Atf1 or Pcr1, or through the heterodimer. Our in vivo binding data confirms previous in vitro assays that demonstrate that Atf1 and Pcr1 bind avidly to DNA as a heterodimer but poorly as homodimers (16). It also confirms genetic data demonstrating that the Atf1-Pcr1 heterodimer (rather than either corresponding homodimer) is required for activation of the ade6 meiotic hot spot for recombination (15).
We observed binding of Atf1 and Pcr1 to stress-responsive genes under basal conditions with just a modest increase in binding upon stress. In contrast, we do not see recruitment of Sty1 to chromatin until the cells are stressed consistent with its change in localization that occurs upon such conditions. Many stress-induced genes are transcribed at a low level in the absence of stress. Our data suggests that this basal transcription is dependent upon both Atf1 and Pcr1 ((13) and Figure 10B and C) and that the stress-induced increase in transcription is mediated by the recruitment of Sty1 to these promoters. We hypothesize that Sty1 recruits additional transcriptional machinery and/or chromatin remodelling activities to mediate the dramatic upregulation in expression of various stress-responsive genes.
Sty1 kinase activity is required for its recruitment to chromatin
We made several observations suggesting that Sty1 kinase activity is required for its association with promoters. Firstly, we found that the kinetics of Sty1 recruitment match those of its own activation (Figure 2). Secondly, forced nuclear localization in the absence of stress-induced activation of Sty1 was not sufficient to result in the association of the kinase with promoters (Figure 4); and finally the catalytically inactive form of Sty1 was not recruited to stress-responsive genes. These findings suggest that phosphorylation of a Sty1 target is necessary for optimal recruitment. However, this target is not Atf1 as recruitment still occurs in the atf1-11M mutant (Figure 6). We also believe that this factor is unlikely to be Pcr1 as it does not appear to be a target for Sty1 phosphorylation (12). In budding yeast, Hog1 is required for the recruitment of a variety of factors to osmo-responsive genes including RNA polII and mediator components although there is currently no evidence to suggest that it phosphorylates any of these factors (24,25). A kinase-dead allele of Hog1 is still recruited to the GPD1 promoter but to a lesser extent than wild type. In contrast, recruitment to other genes, CTT1, STL1 and HSP12 is abolished in the hog1 kinase-dead mutant (23). Our current work is focussed on identifying the critical chromatin bound target(s) for Sty1.
Many Atf1-dependent genes are still induced in a pcr1Δ mutant
Both Atf1 and Pcr1 are required for binding to the meiotic hotspot created by a mutation in the ade6 gene (15) and for chromatin remodelling in this region (44). In contrast, our data shows that in spite of a requirement for the heterodimer to bind to stress responsive genes, there are considerable differences in the transcription profiles of the atf1Δ and pcr1Δ mutants. Our results provide an explanation as to why pcr1Δ cells are not as sensitive to stress as atf1Δ cells (15,18,45); in a pcr1Δ mutant, there is still considerable expression of Atf1-dependent genes, albeit with delayed kinetics. These results raise the question as to what is controlling expression of Atf1-dependent genes in the pcr1Δ mutant, since neither Atf1 nor Pcr1 can be detected on the promoters (Figures 8 and 9). We propose that Atf1 binds weakly and/or transiently to promoters as a homodimer or as a heterodimer with another factor and that the ChIP assay is not sensitive enough to detect such binding. In contrast, Pcr1 may not be able to bind to DNA at all as a homodimer in vivo or as a heterodimer with a factor other than Atf1 thus explaining why there is no significant transcription of Atf1-dependent genes in the atf1Δ mutant. Furthermore, the levels of Pcr1 are greatly reduced in the atf1Δ mutant (18). We have tried to detect the presence of homodimers of either Atf1 or Pcr1 by differentially epitope-tagging the two copies of each protein in a diploid cell and looking for their co-immuoprecipiation but we failed to observe either homodimer (our unpublished results).
What distinguishes the two classes of Atf1-dependent genes such that one group absolutely requires Pcr1 for expression whereas the other group can still be induced to some extent in the pcr1Δ mutant? One possibility is that the promoters of the genes have subtly different sequences representing the Atf1/Pcr1 binding site such that Atf1 homodimers, or heterodimers between Atf1 and another bZIP partner, can still bind to the promoters of genes in group 1 but not group 2. In this regard it is interesting to note that a family of CRE-related sites has been identified to which Atf1/Pcr1 can bind in vitro (46). Future work will characterize the binding sites for Atf1 in the different groups of genes that we have identified (Figure 10) and determine whether they differ in their ability to bind Atf1 homodimers.
Sty1 is recruited to the ORFs of stress-induced genes
We also identified recruitment of Sty1 to the ORF of the gpd1 gene. It remains to be seen whether this association is due to its direct recruitment to these sequences or whether it is there as a consequence of its initial recruitment to promoters and subsequent progression into the body of the gene. For Hog1, its recruitment to ORFs has been uncoupled from promoter recruitment by virtue of constructing chimeric genes with different promoters. Indeed the 3′UTR of the STL1 gene was shown to be important for this (26). The recruitment of MAP kinases to ORFs has not been reported in higher eukaryotes, and thus our observation that Sty1 is targeted to the coding regions of genes makes it more likely that this finding is conserved and raises the possibility that Sty1 may play a role in transcriptional elongation.
Budding and fission yeast are believed to have diverged more than 500 million years ago, yet the recruitment of MAP kinases to stress-activated promoters has been conserved. It remains to be seen whether this aspect of stress signalling is confined to single-celled eukaryotes or if it extends to higher eukaryotes but our results make it more likely that such a mechanism will be universally conserved. The Sty1 pathway is similar to higher eukaryotic stress-activated MAPK pathways and furthermore both fission yeast and mammalian kinases are activated by a number of different signals, hence studying the role of Sty1 at promoters will hopefully provide a useful paradigm for understanding how p38 and JNK might operate.
Acknowledgements
This work was supported by Cancer Research UK and European Union funding (to W.R.) via the “Adaptation to changing nutritional environments” (ACE) project grant HPRN-CT-2002-00249. We thank Gustav Ammerer for helpful discussions, Paul Russell and Elena Hidalgo for strains and reagents and Wolfgang Breitwieser for critically reading the manuscript.
References
- 1.Gasch AP. Yeast. 2007;24(11):961–976. doi: 10.1002/yea.1512. [DOI] [PubMed] [Google Scholar]
- 2.Hibi M, Lin A, Smeal T, Minden A, Karin M. Genes Dev. 1993;7(11):2135–2148. doi: 10.1101/gad.7.11.2135. [DOI] [PubMed] [Google Scholar]
- 3.Derijard B, Hibi M, Wu IH, Barrett T, Su B, Deng T, Karin M, Davis RJ. Cell. 1994;76(6):1025–1037. doi: 10.1016/0092-8674(94)90380-8. [DOI] [PubMed] [Google Scholar]
- 4.Livingstone C, Patel G, Jones N. Embo J. 1995;14(8):1785–1797. doi: 10.1002/j.1460-2075.1995.tb07167.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Raingeaud J, Gupta S, Rogers JS, Dickens M, Han J, Ulevitch RJ, Davis RJ. J Biol Chem. 1995;270(13):7420–7426. doi: 10.1074/jbc.270.13.7420. [DOI] [PubMed] [Google Scholar]
- 6.van Dam H, Wilhelm D, Herr I, Steffen A, Herrlich P, Angel P. Embo J. 1995;14(8):1798–1811. doi: 10.1002/j.1460-2075.1995.tb07168.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shiozaki K, Russell P. Nature. 1995;378(6558):739–743. doi: 10.1038/378739a0. [DOI] [PubMed] [Google Scholar]
- 8.Millar JB, Buck V, Wilkinson MG. Genes Dev. 1995;9(17):2117–2130. doi: 10.1101/gad.9.17.2117. [DOI] [PubMed] [Google Scholar]
- 9.Kato T, Jr., Okazaki K, Murakami H, Stettler S, Fantes PA, Okayama H. FEBS Lett. 1996;378(3):207–212. doi: 10.1016/0014-5793(95)01442-x. [DOI] [PubMed] [Google Scholar]
- 10.Gaits F, Degols G, Shiozaki K, Russell P. Genes Dev. 1998;12(10):1464–1473. doi: 10.1101/gad.12.10.1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shiozaki K, Russell P. Genes Dev. 1996;10(18):2276–2288. doi: 10.1101/gad.10.18.2276. [DOI] [PubMed] [Google Scholar]
- 12.Wilkinson MG, Samuels M, Takeda T, Toone WM, Shieh JC, Toda T, Millar JB, Jones N. Genes Dev. 1996;10(18):2289–2301. doi: 10.1101/gad.10.18.2289. [DOI] [PubMed] [Google Scholar]
- 13.Chen D, Toone WM, Mata J, Lyne R, Burns G, Kivinen K, Brazma A, Jones N, Bahler J. Mol Biol Cell. 2003;14(1):214–229. doi: 10.1091/mbc.E02-08-0499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Watanabe Y, Yamamoto M. Mol Cell Biol. 1996;16(2):704–711. doi: 10.1128/mcb.16.2.704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kon N, Krawchuk MD, Warren BG, Smith GR, Wahls WP. Proc Natl Acad Sci U S A. 1997;94(25):13765–13770. doi: 10.1073/pnas.94.25.13765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wahls WP, Smith GR. Genes Dev. 1994;8(14):1693–1702. doi: 10.1101/gad.8.14.1693. [DOI] [PubMed] [Google Scholar]
- 17.Ohmiya R, Kato C, Yamada H, Aiba H, Mizuno T. Mol Gen Genet. 1999;261(2):297–306. doi: 10.1007/s004380050970. [DOI] [PubMed] [Google Scholar]
- 18.Lawrence CL, Maekawa H, Worthington JL, Reiter W, Wilkinson CR, Jones N. J Biol Chem. 2007;282(8):5160–5170. doi: 10.1074/jbc.M608526200. [DOI] [PubMed] [Google Scholar]
- 19.Edmunds JW, Mahadevan LC. J Cell Sci. 2004;117(Pt 17):3715–3723. doi: 10.1242/jcs.01346. [DOI] [PubMed] [Google Scholar]
- 20.Hohmann S. Microbiol Mol Biol Rev. 2002;66(2):300–372. doi: 10.1128/MMBR.66.2.300-372.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.de Nadal E, Casadome L, Posas F. Mol Cell Biol. 2003;23(1):229–237. doi: 10.1128/MCB.23.1.229-237.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Proft M, Struhl K. Mol Cell. 2002;9(6):1307–1317. doi: 10.1016/s1097-2765(02)00557-9. [DOI] [PubMed] [Google Scholar]
- 23.Alepuz PM, Jovanovic A, Reiser V, Ammerer G. Mol Cell. 2001;7(4):767–777. doi: 10.1016/s1097-2765(01)00221-0. [DOI] [PubMed] [Google Scholar]
- 24.Alepuz PM, de Nadal E, Zapater M, Ammerer G, Posas F. Embo J. 2003;22(10):2433–2442. doi: 10.1093/emboj/cdg243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.De Nadal E, Zapater M, Alepuz PM, Sumoy L, Mas G, Posas F. Nature. 2004;427(6972):370–374. doi: 10.1038/nature02258. [DOI] [PubMed] [Google Scholar]
- 26.Proft M, Mas G, de Nadal E, Vendrell A, Noriega N, Struhl K, Posas F. Mol Cell. 2006;23(2):241–250. doi: 10.1016/j.molcel.2006.05.031. [DOI] [PubMed] [Google Scholar]
- 27.Pokholok DK, Zeitlinger J, Hannett NM, Reynolds DB, Young RA. Science. 2006;313(5786):533–536. doi: 10.1126/science.1127677. [DOI] [PubMed] [Google Scholar]
- 28.Li H, Tsang CK, Watkins M, Bertram PG, Zheng XF. Nature. 2006;442(7106):1058–1061. doi: 10.1038/nature05020. [DOI] [PubMed] [Google Scholar]
- 29.Pascual-Ahuir A, Proft M. Embo J. 2007;26(13):3098–3108. doi: 10.1038/sj.emboj.7601756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Simone C, Forcales SV, Hill DA, Imbalzano AN, Latella L, Puri PL. Nat Genet. 2004;36(7):738–743. doi: 10.1038/ng1378. [DOI] [PubMed] [Google Scholar]
- 31.Vicent GP, Ballare C, Nacht AS, Clausell J, Subtil-Rodriguez A, Quiles I, Jordan A, Beato M. Mol Cell. 2006;24(3):367–381. doi: 10.1016/j.molcel.2006.10.011. [DOI] [PubMed] [Google Scholar]
- 32.Moreno S, Klar A, Nurse P. Methods Enzymol. 1991;194:795–823. doi: 10.1016/0076-6879(91)94059-l. [DOI] [PubMed] [Google Scholar]
- 33.Bahler J, Wu JQ, Longtine MS, Shah NG, McKenzie A, 3rd, Steever AB, Wach A, Philippsen P, Pringle JR. Yeast. 1998;14(10):943–951. doi: 10.1002/(SICI)1097-0061(199807)14:10<943::AID-YEA292>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 34.Craven RA, Griffiths DJ, Sheldrick KS, Randall RE, Hagan IM, Carr AM. Gene. 1998;221(1):59–68. doi: 10.1016/s0378-1119(98)00434-x. [DOI] [PubMed] [Google Scholar]
- 35.Petersen J, Paris J, Willer M, Philippe M, Hagan IM. J Cell Sci. 2001;114(Pt 24):4371–4384. doi: 10.1242/jcs.114.24.4371. [DOI] [PubMed] [Google Scholar]
- 36.Lyne R, Burns G, Mata J, Penkett CJ, Rustici G, Chen D, Langford C, Vetrie D, Bahler J. BMC Genomics. 2003;4(1):27. doi: 10.1186/1471-2164-4-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wilkinson CR, Wallace M, Morphew M, Perry P, Allshire R, Javerzat JP, McIntosh JR, Gordon C. Embo J. 1998;17(22):6465–6476. doi: 10.1093/emboj/17.22.6465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gaits F, Russell P. Mol Biol Cell. 1999;10(5):1395–1407. doi: 10.1091/mbc.10.5.1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Degols G, Shiozaki K, Russell P. Mol Cell Biol. 1996;16(6):2870–2877. doi: 10.1128/mcb.16.6.2870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Castillo EA, Vivancos AP, Jones N, Ayte J, Hidalgo E. J Biol Chem. 2003;278(42):40565–40572. doi: 10.1074/jbc.M305859200. [DOI] [PubMed] [Google Scholar]
- 41.Nguyen AN, Ikner AD, Shiozaki M, Warren SM, Shiozaki K. Mol Biol Cell. 2002;13(8):2651–2663. doi: 10.1091/mbc.02-03-0043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Quinn J, Findlay VJ, Dawson K, Millar JB, Jones N, Morgan BA, Toone WM. Mol Biol Cell. 2002;13(3):805–816. doi: 10.1091/mbc.01-06-0288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Toone WM, Kuge S, Samuels M, Morgan BA, Toda T, Jones N. Genes Dev. 1998;12(10):1453–1463. doi: 10.1101/gad.12.10.1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yamada T, Mizuno K, Hirota K, Kon N, Wahls WP, Hartsuiker E, Murofushi H, Shibata T, Ohta K. Embo J. 2004;23(8):1792–1803. doi: 10.1038/sj.emboj.7600138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kon N, Schroeder SC, Krawchuk MD, Wahls WP. Mol Cell Biol. 1998;18(12):7575–7583. doi: 10.1128/mcb.18.12.7575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fox ME, Yamada T, Ohta K, Smith GR. Genetics. 2000;156(1):59–68. doi: 10.1093/genetics/156.1.59. [DOI] [PMC free article] [PubMed] [Google Scholar]