Abstract
Drosophila melanogaster is a cosmopolitan species that colonizes a great variety of environments. One trait that shows abundant evidence for naturally segregating genetic variance in different populations of D. melanogaster is cold tolerance. Previous work has found quantitative trait loci (QTL) exclusively on the second and the third chromosomes. To gain insight into the genetic architecture of cold tolerance on the X chromosome and to compare the results with our analyses of selective sweeps, a mapping population was derived from a cross between substitution lines that solely differed in the origin of their X chromosome: one originates from a European inbred line and the other one from an African inbred line. We found a total of six QTL for cold tolerance factors on the X chromosome of D. melanogaster. Although the composite interval mapping revealed slightly different QTL profiles between sexes, a coherent model suggests that most QTL overlapped between sexes, and each explained around 5–14% of the genetic variance (which may be slightly overestimated). The allelic effects were largely additive, but we also detected two significant interactions. Taken together, this provides evidence for multiple QTL that are spread along the entire X chromosome and whose effects range from low to intermediate. One detected transgressive QTL influences cold tolerance in different ways for the two sexes. While females benefit from the European allele increasing their cold tolerance, males tend to do better with the African allele. Finally, using selective sweep mapping, the candidate gene CG16700 for cold tolerance colocalizing with a QTL was identified.
Keywords: cold stress, QTL analysis, selective sweep mapping, thermal adaptation
The search for adaptive signatures in genomic data is essential to unravel the evolutionary history of a species (Bersaglieri et al. 2004; Palaisa et al. 2004; Beisswanger et al. 2006; Kane & Rieseberg 2008). Comprehensive studies of genome-wide patterns of variation provided insight into allele distributions and ecologically favoured genotypes in different species (Hinds et al. 2005; Nordborg et al. 2005; Ometto et al. 2005; Borevitz et al. 2007; Hutter et al. 2007). However, these studies were based on patterns of genetic variation that can be altered by nonselective forces like population demography. Mapping methods for quantitative trait loci (QTL) overcome this problem (Mackay 2001). Once the QTL are discovered, one can quantify their impact by estimating allelic effects and interactions and find plausible candidate genes that control the trait.
Traditionally, QTL studies start out with parental lines, such as inbred lines of certain plant or animal species, artificially selected for a divergent phenotype (Long et al. 1995; Moehring & Mackay 2004; Norry et al. 2008). Likewise, natural populations used for QTL analysis should exhibit divergent traits that have been driven apart by natural or sexual selection. Drosophila melanogaster is a cosmopolitan species with locally adapted populations, which makes it a suitable model for naturally based QTL studies (David & Capy 1988). The ancestral population in central equatorial Africa split off from the melanogaster-simulans ancestor around 2–3 Myr ago (Cariou 1987; Lachaise & Silvain 2004). By contrast, the non-African populations have a by far younger history. The habitat expansion to the Eurasian continent started in sub-Saharan Africa around 15 000 BP (David & Capy 1988; Baudry et al. 2004; Stephan & Li 2007). Stable derived populations in major parts of Europe and Asia as well as new populations in North America and Australia suggest recent adaptation to new environmental conditions. The occurrence of geographical patterns and latitudinal clines in populations on different continents for several phenotypic traits, such as developmental time (James & Partridge 1995), temperature tolerance (James et al. 1997; Hoffmann et al. 2002), egg size (Azevedo et al. 1996), diapause (Mitrovski & Hoffmann 2001) and body size (David & Capy 1988; Van ‘t Land et al. 1999), suggest climatic factors as important selective pressures. Thus, one of the first important adaptive steps for a tropical species, like D. melanogaster, is an increase in cold tolerance while colonizing temperate regions.
Indeed, numerous studies suggest that D. melanogaster steadily adapts to new temperature conditions (Mckenzie 1975; Azevedo et al. 1996; James et al. 1997; Gibert et al. 2001; Hoffmann et al. 2003; Anderson et al. 2005). It generally maintains a stable population within the thermal limits of 12–32 °C, even though far colder temperatures can temporarily be accepted (David & Clavel 1967; Stanley et al. 1980). All developmental stages of D. melanogaster have the capacity for cold tolerance adaptation; nonetheless, the adult stage is the strongest candidate to survive drops in temperature on a seasonal scale (Mckenzie 1975; Tucić 1979; Izquierdo 1991). The larval phase apparently runs through critical periods of development, which are highly susceptible to thermal changes and tend to collapse when exposed to cold. After eclosion, the flies overcome these critical periods of development and selection might favour adults for overwintering.
Outside the tropical regions, however, D. melanogaster is in any case closely dependent on human activity (Lachaise & Silvain 2004). When temperature drops below 6 °C, they locally hide in human settlements and enter quiescence, a resting stage that might have serious consequences on sex-specific physical constraints (Izquierdo 1991). With rising temperature in springtime, populations might regenerate and the capacity for reconstructing a population is mostly attributable to inseminated females (Izquierdo 1991). Certainly, the most critical factor influencing the survival of Drosophila populations in temperate regions is wintertime.
Tucić (1979) first suggested that cold tolerance in D. melanogaster is controlled by various groups of genes at particular developmental stages. He proposed that the genetic factors controlling cold tolerance are spread along the entire genome and that the relative importance of genes varies between chromosomes, such that most genes are located on chromosomes 2 and 3, and only relatively few are present on the X chromosome. However, in contrast to the autosomes, QTL on the X chromosome for cold tolerance have not been detected in previous studies (Morgan & Mackay 2006; Norry et al. 2008).
Here, we present the results of a QTL analysis of cold tolerance in D. melanogaster concentrating on the X chromosome. In this context, cold tolerance is referred to as a short time response to a cold shock that is induced by a temporary and severe cold treatment (0 °C) (for the definition of cold shock, see Loeschcke & Sørensen 2005). The individual recovery time of such a cold shock was shown to be a reliable measure of the organismal defence abilities against low temperatures in Drosophila species (Gibert et al. 2001). We chose the X chromosome for two reasons: (i) to revisit earlier studies that did not find QTL on the X and (ii) to compare the QTL analysis with our map of selective sweep regions (that is most detailed for the X chromosome; Li & Stephan 2006) to search for candidate genes of cold tolerance. In other words, the selective sweep approach is employed here to fine-map QTL by testing which genes under a QTL were subjected to recent strong positive selection and may therefore be used for further analysis.
For the QTL study, we used two highly inbred parental lines with homogenized genetic background and unique X chromosomes that were each derived from wild populations of two very different thermal environments: sub-Saharan Africa (Lake Kariba, Zimbabwe) and Western Europe (Leiden, The Netherlands). The parental lines exhibit significantly different phenotypes for cold tolerance, such that the European line is much better adapted to cold than the African one. Reciprocal crosses of the parental lines were used to establish a panel of X-chromosomal recombinant (XR) lines. With these XR lines, we addressed the following questions: (i) Do we find a major QTL with large effect or several QTL with intermediate effects? (ii) Which genomic regions of the X chromosome are associated with cold tolerance? (iii) Are there interactions between QTL? (iv) Do QTL–sex interactions play a role for cold tolerance? For the selective sweep analysis, we used samples that have been collected from the same two local populations from Africa and Europe. In fact, the two lines that led to the construction of the XR lines were derived from these samples.
Materials and methods
Drosophila stocks
Experiments were carried out using highly inbred isofemale lines of D. melanogaster. All stocks were kept at 23 °C, 45% humidity and under constant light conditions. Development took place on a high-nutrient killed yeast food medium (12 mL) in glass vials of 200 mL. Two unrelated lines of wild origin served as starting point for the QTL analysis: line A157 was sampled in Africa (Lake Kariba, Zimbabwe; Begun & Aquadro 1993) and line E14 was collected in Europe (Leiden, Netherlands 1999). The demographic and selective history of the populations from which these two lines were sampled has been studied in detail using genome scans (Glinka et al. 2003; Ometto et al. 2005; Beisswanger et al. 2006; Li & Stephan 2006; Hutter et al. 2007). The cold tolerance profiles between lines A157 and E14 differ significantly (Welch’s t-test, P < 0.001).
By constructing substitution lines, we assessed the presence of cold tolerance factors on the X chromosome. Using the balancer line 6418 (FM7j balancer X chromosome obtained from Bloomington Stock Center; see legend to Supplementary Fig. S1), we introgressed a wild-type (wt) X chromosome into the balancer line background. First, we isolated a single female offspring from a wt-balancer cross and then back-crossed it for seven generations to males of the balancer line (Supplementary Figs S1 and S2). The resulting lines carry the genetic background of the balancer line (mitochondria, 2nd, 3rd and 4th chromosome) and the X chromosome of either the A157 or the E14 wild-type. These lines will be referred to as A* and E*, respectively.
We checked subsequently whether these lines carry the same autosomal background by sequencing several short DNA fragments of the autosomes (data not shown). This confirmed that A* and E* were identical except for their X chromosomes. Moreover, both lines still show significantly different cold tolerance profiles (Welch’s t-test, P < 0.001), which made them suitable parental lines to establish an array of XR lines by directed crossings. The offspring of two reciprocal crosses between A* and E* was allowed to mate freely for five generations to accumulate recombination events. During this phase, generations were strictly separated by removing the adults just before their offspring hatched. Male flies of the F5 generation were then crossed and back-crossed to the balancer line to establish a population of 186 XR lines each carrying a unique recombinant X chromosome. Once the XR lines were created, they were maintained in the laboratory by full-sib mating and monthly mass transfer.
Cold tolerance phenotype
As a measurement of cold tolerance, we used a modified protocol for the chill coma recovery test of David et al. (1998). At temperatures below 0 °C, D. melanogaster adults lose their mobility and feeding capacity experiencing a knockdown that is called chill coma. When they are brought back to a higher temperature, they progressively recover from chill coma and regain normal activity. A fly is considered as having recovered when it is able to stand on its legs (David et al. 1998). The time they take for doing so is called the chill coma recovery time (CCRT). The CCRT is tightly linked to cold tolerance as the recovery from a cold treatment is strongly correlated with the duration of cold treatment (David et al. 1998). The test is nonlethal, and the time metric corresponds to a quantitative variable. To remove any influence of breeding conditions, all tested flies were maintained at low densities (Peters & Barbosa 1977). As thermotolerance is highly correlated with maturity (David et al. 1998), male and female flies were all tested at the age of 5 days.
For the chill coma recovery test, flies were individually placed into empty 8-ml vials without anaesthesia (Milton & Partridge 2008), fixed in racks and placed in an ice-water bath of 0 °C. After 7 h of exposure, flies were brought back to room temperature (23 ± 1 °C) and CCRT was recorded in minutes (min). Dead individuals were not included into the analysis. They represented less than 1% of the total tested flies and were randomly distributed among lines. As controls, flies from the lines A* and E* were tested at the same time as the XR lines.
Marker selection and genotyping
A previously conducted genome scan of the X chromosome revealed that lines A157 and E14 differ by numerous single nucleotide differences (SNPs) (Glinka et al. 2003; Ometto et al. 2005). These SNPs are thus reliable markers for the African and European genotypes. A total of 23 SNPs from neutrally evolving fragments that were uniformly distributed along the chromosome were selected to serve as genetic markers for the QTL analysis. To maximize the mapping resolution, the marker density was adjusted to the recombination rate: marker density was thus increased in regions of higher recombination rate (for further information on markers, see Supplementary Table S1).
By extracting genomic DNA from pools of 20 females with the Puregene DNA isolation kit (Gentra System, Minneapolis, USA), we genotyped each XR line. Custom Taqman SNP genotyping assays from Applied Bio-systems (ABI, Foster City, USA) were performed on the DNA samples and ran on an ABI 7500 Real Time thermocycler PCR machine. The presence of either genotype at each marker position was determined with the 7500 Fast Software version 2.0 provided by ABI. To infer marker linkage, we used the frequency of recombination events between marker pairs (RecombRate software, Comeron et al. 1999) and applied Haldane’s map function to obtain a marker map in centimorgan (cM) (Haldane 1919). Markers show significant segregation distortion (SD) with the European alleles being overrepresented. As this phenomenon affects the whole X chromosome, it is not likely to result from either genotyping errors or linked segregation distortion loci. One possibility that would lead to the chromosome-wide overrepresentation of the European genotype would be the reduced inheritance of African alleles in an early step of XR line establishment. The reciprocal crosses of the A* and E* lines (see Material and Methods, Drosophila stocks) might be a crucial point at which such a shift could happen by chance. A sensible reduction in offspring number of males carrying the African X chromosome can reduce the amount of African alleles in the XR line population. The SD is, however, not expected to produce a higher false positive rate, nor will it significantly change the position or the effect of QTL, particularly in additive models of a large XR line population (Xu 2008; Zhang et al. 2010).
Quantitative genetic analysis
The statistical analysis was performed using the R software (version 2.8.0). To determine the differences in cold resistance between the parental lines A* and E*, we log-transformed the CCRT measurements to improve normality of the residuals (ln(CCRT)). A two-way ANOVA was used to infer the effects of line (A* or E*), sex (male or female) and their interactions.
The data obtained from the XR line population were analysed using different statistical approaches. We used a linear regression model to reveal the genotype-by-sex interactions (GSI). According to the model y = μ + aL + bS + γ(aL × bS) + εL,S,i, we quantified the influence of the line aL, the sex bS and their interactions γ on the variance of the ln(CCRT) y. The parameter μ is the population mean; the residuals εL,S,i are assumed to follow a normal distribution with constant variance. Each XR line represents a different multi-locus genotype for the X chromosome, and within-line phenotypic variation can exclusively be traced back to trait plasticity as there is no within-line genetic variation. We estimated the sex-specific X-chromosomal heritability in a broad sense: , where is the between-line variance and is the within-line variance for ln(CCRT). Another measurement of GSI is the cross-sex genetic correlation coefficient of mean ln(CCRT) of males and females between the XR lines rGS = covMF/(σM × σF).
Interval mapping
To investigate the QTL positions, their contributions to variation among lines and to estimate their effect, we used the Windows QTL cartographer version 2.5 (Wang et al. 2007) and R/qtl (Broman et al. 2003). The input for the analysis is the line-dependent mean values of ln(CCRT) and the vector of marker genotypes (0 = African genotype, 1 = European genotype). Within an interval between two markers, we used a fixed number of grid positions with a density of 1 cM to estimate the probabilities of the underlying genotypes conditioned on the observed marker data using the Haley–Knott regression (Haley & Knott 1992). The results are expressed as LOD-score (Sen and Churchill 2001). This score is plotted at every 1 cM grid position of the X-chromosome to give a likelihood profile. A QTL is more likely to be located at the region with high LOD values. We performed a hierarchical interval mapping for males and females separately, starting with a one-QTL analysis, which was then expanded to a more advanced two-QTL analysis and the composite interval mapping (CIM) to handle the nature of more complex QTL. The number of marker cofactors for the CIM should not exceed 2 n where n is the number of available markers (Jansen & Stam 1994). Thus, we used up to 10 putative cofactors performing a forward and backward selection of the CIM model with a window size of 10 cM. Each significant threshold was established by randomly permuting the trait values among the marker genotypes 1000 times. The empirical critical LOD value corresponding to α = 0.05 was determined according to the 5% cut-off of the distribution of maximal LOD values obtained by each permutation. To test whether there is evidence for a QTL and whether the two-QTL analysis is a significant improvement over a one-QTL analysis, the LOD score had to exceed its threshold.
Multiple-QTL model
The multiple-QTL model was used to regress over the main candidate QTL positions to identify the most important elements that shape the genetic architecture of cold tolerance on the X chromosome of D. melanogaster. As there is a continuum of potential covariates, we used the knowledge of the interval mapping to pick candidate QTL regions for model fitting. Again, the likelihood ratio test statistic was used to infer the model fit expressed in LOD scores. Model search was conducted by backward elimination: we started with the largest model comprising all candidate loci that were found in both the male and female data sets and stepwise removed the covariate that had the smallest LOD value. Hence, the starting point of the analysis was identical for males and females. By backward selection, we constructed a nested sequence of models of decreasing size all the way to the null model. For model selection, we chose the one with the largest model LOD score. By including pairwise interactions, we enforced a hierarchy under which the inclusion of the interaction of certain markers requires the inclusion of main effects for each marker individually.
Mapping of QTL–sex interactions
Loci that influence the cold tolerance differently for males and females underlie QTL–sex interactions. These interactions might preserve genetic variation within the population as males and females lack a consistent response to natural selection. To localize such regions, we used the average difference of ln(CCRT) between males and females of each XR line as the phenotype in a QTL analysis. If there is no variation in the differences between males and females, there can be no QTL–sex interactions, but if the differences vary between the XR lines, we might find associations of this variation and a chromosomal region. Again, a significance threshold was derived by 1000 permutations.
Analysis of a selective sweep region
Of the selective sweep map of the X chromosome inferred by Li & Stephan (2006) that consisted of 54 and 55 putative 100-kb long fragments in the African and European populations, respectively, where sweeps were identified, we investigated one region in detail (window 55 of the European population, cytological position 15E3). This region was chosen because it fulfils necessary conditions for the occurrence of a (complete) sweep in a local population influencing a quantitative trait: (i) the sweep colocalizes with a QTL, namely the QTL at position 56 cM (cytological position 13E-20E, Fig. 3), which is significant in both males and females and is lacking QTL–sex interactions; (ii) the sweep is specific to a population. Because the sweep occurred in the European, but not in the African population, allelic differences at the gene(s) affecting the trait may therefore be found. In contrast, if a recent sweep was shared between these populations (such as for the D. melanogaster polyhomeotic locus, Beisswanger & Stephan 2008), the gene targeted by positive selection would not contribute to the quantitative trait.
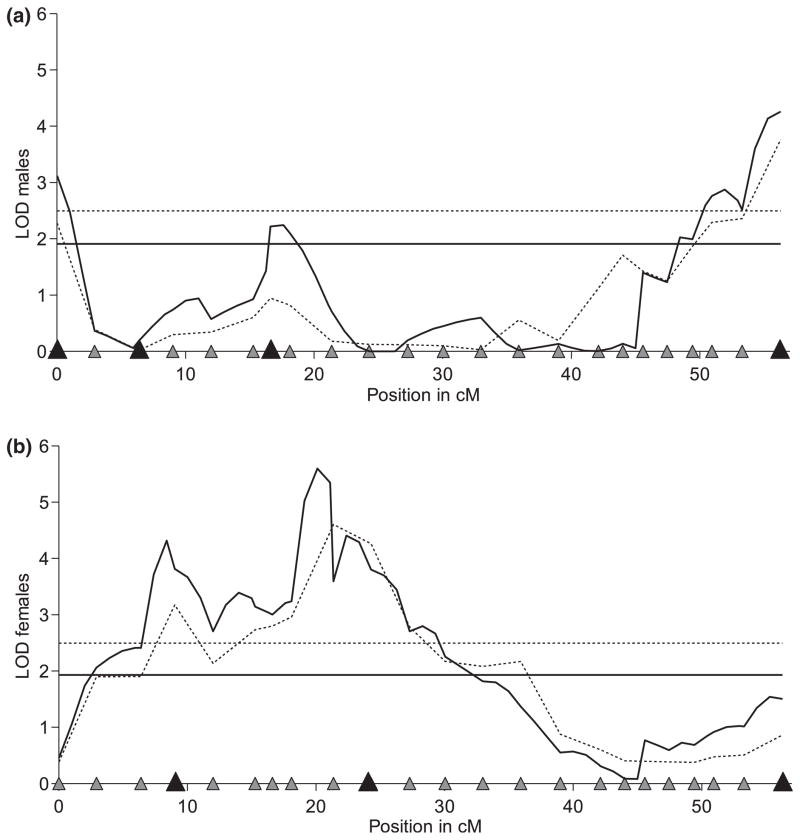
Fig. 3.
Results of interval-mapping methods for ln[chill coma recovery time (CCRT)] in males (a) and females (b) of Drosophila melanogaster. The one-quantitative trait loci (QTL) analysis (dotted lines) and the composite interval mapping (CIM) (solid lines) for males and females give consistent results, but the CIM better resolved the genetic architecture of multiple QTL for cold tolerance on the X chromosome. The horizontal lines correspond to the threshold values (dotted line for the one-QTL model and the solid one for the CIM). The distribution of the 23 genetic markers used is shown on the x-axis by triangles, where larger black triangles indicate the locations of QTL cofactors. Each position at which the LOD score exceeds the threshold value is a genomic region showing significant association with CCRT—i.e. a QTL.
While the analysis by Li & Stephan (2006) was carried out using the site frequency spectrum of SNPs averaged over a limited number of 500-bp long fragments (of at least three in a 100-kb window), we increased the amount of sequence data in window 55 in two steps as described by Svetec et al. (2009): first, 12 additional fragments of 500-bp length were PCR-amplified and re-sequenced in both the African and European samples used by Li & Stephan (2006) (11 lines for the African sample and 12 for the European one), and second, a region of 6.4 kb (between absolute positions 16 992 569 and 16 998 925; release 5.29 of Flybase, http://flybase.org) was completely re-sequenced in these 23 lines. This fine-scale analysis allowed us to determine the target of selection very precisely (i.e. down to the level of individual genes). The new sequence data have been deposited in GenBank (accession nos HQ005309 to HQ005331).
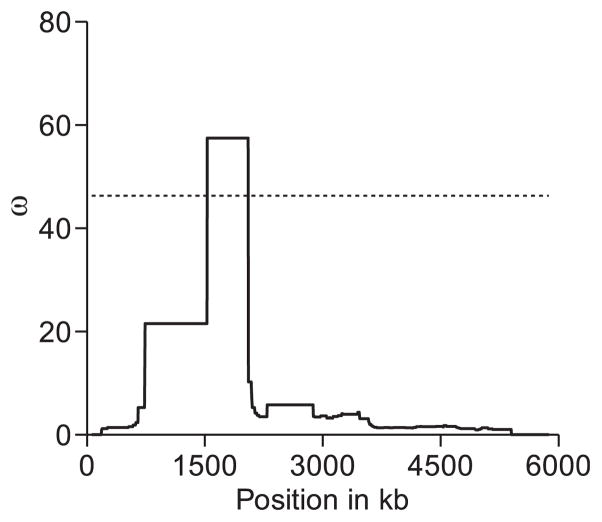
This data set was then subjected to an analysis of linkage disequilibrium (LD) using the ω statistic (Kim & Nielsen 2004). Elevated values of ω provide evidence of a selective sweep, and the peak of this statistic (ωMAX) indicates the location of the target of selection in the genome (Pavlidis et al. 2010). Positions containing insertions or deletions (indels) were excluded.
The study by Li & Stephan (2006) provided evidence for recent positive selection in this genomic region based on the site frequency spectrum. In this study, we confirm their results using a LD-based statistic. In the statistical hypothesis testing, we did not use the ascertainment bias correction of Thornton & Jensen (2007). Their correction is applied when a genomic region is chosen for selective sweep analysis based on a priori information (e.g. reduced polymorphism levels). In the current analysis, we already know that this genomic region deviates from neutrality (Li & Stephan 2006). A low P-value for the ωMAX is therefore expected because there is evidence that the genomic region is an outlier when the null (neutral) hypothesis is assumed to be correct.
To assess the statistical significance of the maximum value ωMAX, we ran 10 000 neutral simulations with the ms software (Hudson 2002). The demographic scenario of the African and European populations of D. melanogaster (Li & Stephan 2006) represented the null hypothesis. The mutation rate (1.47 × 10−9) was estimated from the observed number of polymorphisms in the African population using the method of Zivkovic & Wiehe (2008). Thus, the African population was used as a proxy for selective neutrality. The recombination rate (3.6 × 10−8) was obtained from the D. melanogaster recombination rate calculator (Fiston-Lavier et al. 2010). Only the European subset of each simulation was used to assess the significance of ωMAX.
Results
Thermotolerance in the parental lines
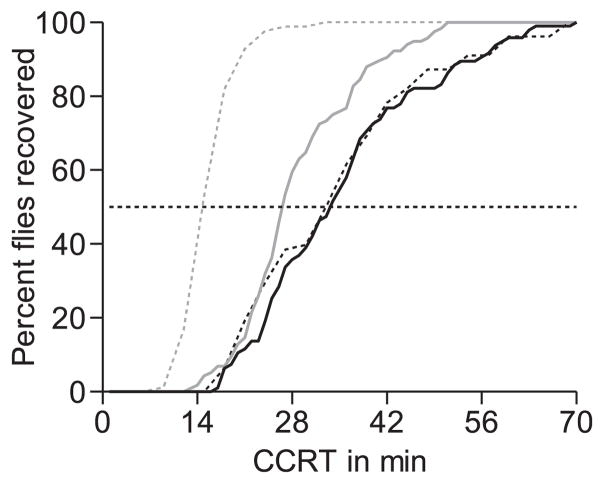
The CCRT of the original lines A157 and E14 differed significantly; i.e. A157 males: 62.1 ± 1.3 (SE) min, females: 66.4 ± 3.8 (SE) min; E14 males: 28.6 ± 0.5 (SE), females: 23.5 ± 1.3 (SE). The CCRT of the parental lines A* and E* were also significantly different: 39.4 ± 2.3 (SE) and 35.5 ± 1.3 (SE) min for males and females with the African X chromosome, respectively, and 27.5 ± 1.0 (SE) and 29.1 ± 0.8 (SE) min for males and females with the European X chromosome (Fig. 1, Table 1, two-way ANOVA for log-transformed data). However, there was no significant variation in CCRT for sexes among the parental lines as well as no detectable line–sex interaction (Table 1).
Fig. 1.
Chill coma recovery curves of the parental lines (A* in black, E* in grey) separated for males (doted lines) and females (solid lines). Both males and females of the European line recovered significantly earlier from the chill coma than those of the African line (Welch’s t-test, P < 0.001). The black dotted horizontal line shows the time at which 50% of the line-specific flies recovered.
Table 1.
Results of the two-way ANOVA of log-transformed chill coma recovery time of the parental lines A* and E*
| d.f. | Sum Sq | Mean Sq | F value | Pr(>F) | |
|---|---|---|---|---|---|
| Line | 1 | 3.0128 | 3.0128 | 31.7842 | 4.511e–08*** |
| Sex | 1 | 0.0936 | 0.0936 | 0.9879 | 0.3212 ns |
| Line × sex | 1 | 0.2334 | 0.2334 | 2.4626 | 0.1178 ns |
| Residuals | 258 | 24.4556 | 0.0948 |
ns, not significant;
significant with P < 0.001.
Thermotolerance in the XR lines
Mean values ranged from 22.9 ± 1.3 (SE) min to 42.2 ± 3.5 (SE) min for males and from 22.7 ± 1.2 (SE) min to 46.7 ± 4.1 (SE) min for the females. The results of the two-way ANOVA of the XR lines revealed highly significant line-specific variation for ln(CCRT) (Table 2). The main effect of sex is only slightly significant as it is not constant across lines; the line–sex interaction is highly significant (Table 2). Thus, genotype-by-sex interactions are present in our XR line population. The increase in mean ln(CCRT) between the XR lines is smooth, suggesting multiple QTL with small to intermediate effects (Fig. 2a,b). In females, one XR line significantly exceeded the cold tolerance of the parental E* line (Welch’s t-test and Holm–Bonferroni correction, P < 0.05), showing a transgressive phenotype.
Table 2.
Results of the ANOVA of log-transformed chill coma recovery time of X-chromosomal recombinant lines
| d.f. | Sum Sq | Mean Sq | F value | Pr(>F) | |
|---|---|---|---|---|---|
| Line | 189 | 64.64 | 0.34200 | 3.9779 | 2.2e–16*** |
| Sex | 1 | 0.53 | 0.53241 | 6.1925 | 0.01286* |
| Line × sex | 182 | 25.32 | 0.13910 | 1.6179 | 4.411e–07*** |
| Residuals | 5867 | 504.42 | 0.08598 |
significant with P < 0.05,
P < 0.001
Fig. 2.
Mean ln[chill coma recovery time (CCRT)] in males (a) and females (b) of the X-chromosomal recombinant (XR) lines of Drosophila melanogaster. The smooth increase indicates multiple quantitative trait loci (QTL) with small to intermediate effects. Paternal cold tolerance (E*, grey triangle on the x-axis) as well as cold sensitivity (A*, black triangle) was not significantly higher in the males of the XR lines, whereas the females of one XR line showed a significantly shorter CCRT than the E* line (Welch’s t-test and Holm–Bonferroni correction, P < 0.05).
Sex-specific X-chromosomal heritability in the broad sense, , varied between sexes ranging from 0.2 for males to 0.13 for females. The reduced linear regression model among lines within sexes was highly significant for males and females, jointly indicating an X-linked genetic basis for cold tolerance in both sexes (P < 0.001). The coefficient of cross-sex genetic correlation rGS was low (0.35) but highly significant (Pearson’s product–moment correlation, P < 0.001), displaying a weak correlated response of both sexes under selection. These estimates support the hypothesis that X-linked cold tolerance is low but present, and a mixture of commonly inherited and sex-specific loci affects males and females of D. melanogaster.
One-QTL and two-QTL analyses
The one-QTL interval mapping revealed one significant QTL position for cold tolerance on the X chromosome of D. melanogaster for both males and females. Although the LOD curve of males shows at least three peaks, only one exceeds the threshold of the 5% significance level (Fig. 3a, dotted line). This QTL position is located towards the right tip of the chromosome close to the centromere between the last two markers at 56 cM. Thus, for the one-QTL analysis, this is the strongest candidate in males. The LOD curve of females shows a double peak with the highest LOD value at position 22 cM between markers six and eleven (Fig. 3b, dotted line). The second peak at around 9 cM also exceeds the threshold but cannot be reliably decoupled from the main candidate at 22 cM. The thresholds of 2.5 LOD were established by 1000 permutations for males and females separately.
The two-QTL analysis revealed insight into a more complex QTL structure. The first model extension comprehended the possibility for a second QTL with additive effects (additive model, H1a), while the second model extension allowed for a second QTL with epi-static effects (full model, H1f). For males of the XR line population, the additive and the full models reach their maxima at the same positions, 0 cM and 56 cM, showing very highly significant LOD scores for both models (additive model: lod.add = 7.45; full model: lod.full = 8.32). By comparing the two-QTL analysis to the one-QTL analysis, we exclusively quantified the support of the second QTL position. Under the additive model as well as under the full model, there is significant evidence for a second QTL in the male data set, concerning the two candidate regions at 0 cM and 56 cM (lod.av1 = 3.09, lod.fv1 = 3.97). There is no clear hint for an interaction between them (lod.int = 0.87).
The females of the XR line population show similar consistent results for the two-QTL genome scan as the males. The additive and the full models maximize at analogous positions (additive model positions at 8 cM and 22 cM, full model positions at 10 cM and 23 cM), thus overlapping with the maxima of the one-QTL analysis. The scores are high, giving strong support for at least one QTL at either of these positions (lod.add = 7.17, lod.full = 8.23). The evidence for the second QTL is congruent in both models. The implementations of a second QTL in the additive model and the full model with the interaction term exceed the thresholds (lod.av1 = 1.85, lod.fv1 = 2.90), giving significant support for the second QTL position at 22–23 cM.
Composite interval mapping
For the males of the XR line population, four cofactors were included in the CIM model (marker 1, 3, 7 and 23, Fig. 3a, black triangles), leading to one more candidate region that exceeded the threshold at position 18 cM (Fig. 3a, solid line). The formerly detected QTL positions at 0 cM and 56 cM remained significant. For females of the XR line population, the QTL positions remained unchanged at around 9 cM and 23 cM although the candidate region slightly increased. Overall, three markers were included as cofactors (Fig. 3b, black triangles, marker 4, 10 and 23).
Multiple-QTL model
The most significant QTL positions defined by the two-QTL analysis and CIM entered the multiple linear regression analysis to estimate the main QTL and their interaction effects. For both males and females, an additive model with interactions was favoured to summarize the main QTL and their effects. After the backward elimination procedure, overall four main effects and two interactions remained important in the multiple-QTL model for the males of the XR line population. In descending order, these are position 56 cM (LOD = 6.64), 0 cM (LOD = 4.82), 18 cM (LOD = 4.08), 24 cM (LOD = 2.66), interaction 24 × 18 cM (LOD = 2.17) and 0 × 56 cM (LOD = 1.16). The general model fit is good (LOD = 12.40) and explains around 26% of the trait variance (Table 3). For the females of the XR line population, overall four main effects and one interaction remained important in the model fit after the elimination procedure. These are position 24 cM (LOD = 6.95), 17 cM (LOD = 4.31), 9 cM (LOD = 2.99), 56 cM (LOD = 2.68) and the interaction 24 × 17 cM (LOD = 4.18). The general model fit is good (LOD = 12.89) and explains around 28% of the trait variance (Table 4).
Table 3.
Multiple-QTL model for males of the XR line population of Drosophila melanogaster
| Model formula: y = Q1 + Q3 + Q5 + Q2 + Q2:Q5 + Q3:Q1
| |||||
|---|---|---|---|---|---|
| d.f. | SS | MS | LOD | %Var | |
| Model | 6 | 0.70 | 0.12 | 12.40 | 26.31 |
| Error | 180 | 1.95 | 0.01 | ||
| Total | 186 | 2.65 | |||
| Drop one QTL at a time ANOVA table
| ||||||
|---|---|---|---|---|---|---|
| Cytologic position | d.f. | Type III SS | LOD | %Var | Estimated allele effect (SE) | |
|
|
||||||
| Intercept | 3.51 ± 0.02 | |||||
| X@56 | 13E-20E | 2 | 0.35 | 6.64 | 13.09 | −0.16 ± 0.03 |
| X@0 | 1A-3F | 2 | 0.25 | 4.82 | 9.30 | −0.13 ± 0.03 |
| X@18 | 7B-8E | 2 | 0.21 | 4.08 | 7.79 | −0.06 ± 0.03 |
| X@24 | 8E-11D | 2 | 0.13 | 2.66 | 4.99 | 0.06 ± 0.03 |
| X@24:X@18 | 1 | 0.11 | 2.17 | 4.04 | −0.16 ± 0.05 | |
| X@0:X@56 | 1 | 0.06 | 1.16 | 2.13 | 0.13 ± 0.06 | |
QTL, quantitative trait loci; XR, X-chromosomal recombinant.
Table 4.
Multiple QTL model for females of the XR line population of Drosophila melanogaster
| Model formula: y = Q2 + Q6 + Q4 + Q1 + Q2:Q6
| |||||
|---|---|---|---|---|---|
| d.f. | SS | MS | LOD | %Var | |
| Model | 5 | 0.77 | 0.15 | 12.89 | 27.58 |
| Error | 178 | 2.02 | 0.01 | ||
| Total | 183 | 2.79 | |||
| Drop one QTL at a time ANOVA table
| ||||||
|---|---|---|---|---|---|---|
| Cytologic position | d.f. | Type III SS | LOD | %Var | Estimated allele effect (SE) | |
|
|
||||||
| Intercept | 3.52 ± 0.02 | |||||
| X@24 | 8E-11D | 2 | 0.38 | 6.95 | 13.75 | −0.05 ± 0.03 |
| X@17 | 6C-10B | 2 | 0.23 | 4.31 | 8.25 | 0.02 ± 0.03 |
| X@9 | 4C14-5C7 | 1 | 0.16 | 2.99 | 5.63 | −0.11 ± 0.03 |
| X@56 | 13E-20E | 1 | 0.14 | 2.68 | 5.02 | −0.08 ± 0.02 |
| X@24:X@17 | 1 | 0.22 | 4.18 | 7.96 | −0.25 ± 0.06 | |
QTL, quantitative trait loci; XR, X-chromosomal recombinant.
Overall, males and females of D. melanogaster show similar genetic architecture for cold tolerance on the X chromosome. They share most of the detected QTL, such as positions 17 cM, 18 cM, 24 cM and 56 cM, although it needed a coherent multiple-QTL model to show this. Two QTL, however, remained unique to one of both data sets: position 0 cM to males and position 9 cM to females. The estimated additive effects have mainly a negative sign, which indicates that the African alleles generally reduce cold tolerance. Three of the studied QTL positions, however, have a positive sign: the QTL at position 24 cM as well as the interaction 0 × 56 cM in males and the QTL at position 17 cM in females. Whereas the effect of the QTL at position 17 cM in females is low, the effect of the QTL at position 24 cM in males is a transgressive QTL. Males benefit from the African allele to increase their cold tolerance. The interaction effect 0 × 56 cM is based on the product of the genotypes at position 0 cM and 56 cM. The effect of position 0 cM depends on whether a male fly has the African or European allele at position 56 cM. Thus, the positive sign of the interaction effect indicates an increased cold tolerance when both loci exhibit the African genotype, although the QTL at positions 0 cM and 56 cM individually show negative effects.
QTL–sex interactions
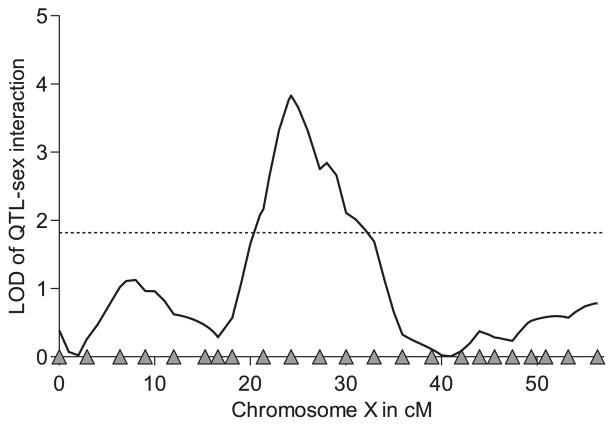
Consistent with the highly significant genotype-by-sex interactions of the XR line population, the differences between males and females show considerable variation. The result of the one-QTL analysis shows a strong association between the differences in cold tolerance between sexes and the position at 24 cM (Fig. 4). The LOD value of 3.83 clearly exceeds the threshold of 1.79. At this position, females substantially increase their cold tolerance by the European allele (Welch’s t-test, P < 0.001), but in males, both genotypes perform equally well (Welch’s t-test ns, Fig. 5). The difference of genotype-based cold tolerance of males and females assumes its maximum at position 24 cM and gives reliable evidence for QTL–sex interactions on the X chromosome.
Fig. 4.
Localization of quantitative trait loci (QTL)–sex interactions on the X chromosome of Drosophila melanogaster. The one-QTL analysis was sufficient to reveal a peak at 24.3 cM, which exceeds the 5% threshold of LOD = 1.79 (dotted line).
Fig. 5.
ln[chill coma recovery time (CCRT)] results in relation to the X-chromosomal recombinant (XR) line genotype at marker position 24 cM for males and females. While females significantly decrease their CCRT when having the European genotype at this marker position (Welch’s t-test, P < 0.001), males tend to increase their CCRT. However, the difference in CCRT between the males with European and African genotype was not significant. Such contrasting patterns for males and females are called quantitative trait loci (QTL)–sex interactions.
A selective sweep associated with CCRT colocalizes with a QTL
The profile of the ω statistic in the fully re-sequenced region is shown in Fig. 6. The dashed line depicts the 95th percentile of ωMAX obtained by the neutral simulations (see Materials and Methods). The P-value for the observed ωMAX is 0.037. The maximum of the ω distribution is located in the intergenic region between the computer-predicted genes CG16700 and CG4991. In a very recent study, the expression variation observed at CG16700 has been associated with CCRT in another derived population (from North America; Ayroles et al. 2009). This suggests that CG16700 (but not CG4991) is a candidate gene of cold tolerance.
Fig. 6.
Profile of the ω statistic across the completely re-sequenced region of the European population. This 6.4-kb region consists of the intergenic fragment between the genes CG16700 and CG4991. The region shown on the x-axis is shorter than 6.4 kb because the genomic segment at the right end, between positions 5921 and 6379, was not included in the analysis because of smaller sample size (missing data). These positions correspond to the 3′UTR of CG4991. Likewise, positions containing indels were excluded. The dotted line represents the 5% significance level.
Discussion
Rationale
Numerous QTL studies used artificially selected parental lines (Tucić 1979; Heyen et al. 1999; Tuiskula-Haavisto et al. 2002; Norry et al. 2004, 2007; Morgan & Mackay 2006) to maximize the initial genetic differences. Strongly divergent genetic effects increase the chances to detect significant QTL. With the recent advances in statistics and molecular biology, it is possible to detect QTL that are based on much lower levels of phenotypic variation. We thus decided to investigate naturally segregating variance for cold tolerance occurring between two wild lines of D. melanogaster (A157 and E14) that were sampled in sub-Saharan Africa and Western Europe. These two lines exhibit a divergent cold tolerance phenotype: line A157 is less tolerant to cold than line E14. This is an expected result as previous studies showed that cold tolerance follows a geographical pattern and latitudinal clines (Bubliy et al. 2002; Hoffmann et al. 2002).
The presence of cold tolerance factors on the X chromosome of D. melanogaster is demonstrated by the fact that the two substitution lines A* and E* exhibit divergent cold tolerance phenotypes. To map these factors on the X chromosome, an XR line population was constructed and studied. The broad sense heritability of cold tolerance estimated for the XR line population con-firmed that 13–20% of the phenotypic variance has a genetic basis on the X chromosome. We thus used a QTL mapping approach to locate the X-linked factors influencing cold tolerance. Furthermore, using selective sweep analysis, we fine-mapped the genomic region colocalizing with one of the QTL.
QTL analysis
As we detected a significant effect of sex as well as line–sex interactions on cold tolerance, we decided to treat the male and female data sets separately to study sex antagonistic effects, which are referred to as QTL–sex interactions. Splitting the data set decreased the sample size and could thus have reduced the power to detect the cold tolerance factors that are shared between sexes. However, our results clearly show that each of the two data sets remained sufficiently large to detect significant QTL effects. Moreover, the high number of scored individuals (about 10 individuals per XR line), the high marker density (23 along the X chromosome) and the use of a homogenized genetic background certainly raised our chance to detect QTL.
The basic one-QTL analysis was not sufficient to explain the trait variance in our XR line population. Therefore, we extended the one-QTL analysis and considered multiple QTL to distribute residual variance to several loci with putative low to intermediate effects. The two-QTL analysis allows for the possibility of two QTL positions, while the CIM method relaxes the limitation of two QTL using a number of marker cofactors. The selection of cofactors, however, is critical and somehow arbitrary, which can result in conspicuous randomness in the choice of cofactors and misleading conclusions of QTL positions. Using the candidates of the two-QTL analysis and the CIM, comprising QTL positions, cofactors and interactions, we constructed a coherent model for both sexes, based on a backward selection process to quantify the impact of main factors as well as interactions.
Overall, we found six QTL that were significantly associated with cold tolerance on the X chromosome, explaining about 26–28% of the total phenotypic variance observed in males and females. Consistently, the distribution of mean CCRT in the XR line population (Fig. 2) does not show a biphasic response as expected in case of one major QTL. Indeed, each of the detected QTL accounts for 5–14% of the total phenotypic variation. This also explains the failure of the one-QTL analysis as it lacks the possibility to sufficiently distribute the residual variance of the trait and dissolve its complexity. It is important to notice that on average, the QTL effects are likely to be slightly overestimated because the XR lines have a homogenous autosomal background. In other words, in the present study, we focused on phenotypic variation in cold tolerance generated by naturally occurring alleles of the X chromosome only.
It is a common finding that genes contribute differently to complex traits in males and females. Among the six QTL, two were exclusively detected in either males (0 cM) or females (9 cM), two are partially overlapping (17 cM and 18 cM), and two were found at the same position in males and females (24 cM, 56 cM). But sex specificity has to be taken with caution. Detecting a QTL in one sex and not in the other might be attributable to a lack of power in the analysis. Precise investigations into the sex-specific effects of the different alleles must be performed to assess the sex-specific role of any of these QTL. The QTL positions 17 cM and 18 cM do not perfectly colocalize but partially overlap and are both involved in interactions with the QTL position at 24 cM. This could suggest that female position 17 cM and male position 18 cM are confounded. Only further fine-mapping experiments would confirm whether the same sets of genes contribute to cold tolerance in both males and females.
One CIM cofactor was found in the male data set at position 6 cM, but it does not appear as main effect in the multiple-QTL analysis, neither in males nor in females. Thus, it is the sole cofactor whose effect in the CIM could not be verified with the multiple-QTL model. However, one has to keep in mind that a classical drawback of association mapping studies also applies to the present study: we simply might miss some additional QTL because their effects are below the detection level of the interval mapping.
In Fig. 2, the ranges of phenotypic values of the parental lines and XR lines are different for the two sexes. More precisely, the parental values are closer, and the XR line values are more spread out for the females than for the males. In other words, transgressive segregation (i.e. the existence of hybrid offspring with phenotypes exceeding the E* line) is present when looking at females. One female line significantly exceeds the parental cold tolerance. As dominance is excluded (the XR lines are homozygotes), additive effects between loci are the most likely explanation for the female transgressive segregation observed in Fig. 2.
Interactions between QTL
We detected several QTL that showed significant levels of interactions (in males: 0 cM × 56 cM and 18 cM × 24 cM, in females 17 cM × 24 cM). Interactions, also called epistasis, are referred to as departure from additivity, mainly multiplicativity of allelic effects. QTL mapping allows one to detect such interactions, but it does not provide any information about the nature of these interactions: protein–protein (for example, cofactors), DNA–protein (like gene expression regulation) or physiological pathway interactions (for example, enzyme cascades in a metabolic pathway).
Generally, interactions might not necessarily be the consequence of gene action but reflect the way how selection operates at these loci (Crow 2010). Genes with small additive effects might accumulate slightly deleterious mutations until a certain threshold. Individuals that reached the threshold will be subject to truncation selection and are eliminated from the population. This means deleterious mutations in multiple QTL are jointly removed, creating the signal of interactions. Crow (2010) called this ‘quasi-epistasis’ that is attributable to selection’s grouping of alleles with similar effect.
QTL–sex interactions
The QTL at position 24 cM exhibits two interesting features: it interacts with both QTL positions at 17 cM and 18 cM, but it also generates a QTL–sex interaction. We thus included sex differentiation factors as candidate genes for this region only (Supplementary Table S2). Moreover, for the QTL at position 24 cM (marker 10), the female cold tolerance pattern is consistent with what is observed all along the chromosome: the European allele confers a significantly higher cold tolerance than the African allele. Males, however, exhibit a transgressive QTL and reduce their CCRT by having the African allele. This finding agrees with the highly significant but low cross-sex genetic correlation coefficient of males and females, indicating a weak correlated response of both sexes under selection. Sex antagonistic selection maintains genetic variation within a population and is thus not suitable for a selective sweep analysis in the context of cold tolerance. Although the coherent multiple-QTL model revealed similar architectures for males and females, there are some noticeable differences, one of which can be quantified by means of a one-QTL analysis (Fig. 4).
Several scenarios could explain that cold tolerance genes are present in the African population. First, in the past, the African population might have experienced colder periods and, because of its large population size, some cold tolerance alleles could still segregate. A second explanation would be that some positively selected traits in Africa, such as desiccation resistance or heat tolerance, could confer higher cold tolerance. However, the reason why the African QTL allele at position 24 cM generates such strong divergent effects on males and females requires more investigations.
Candidate genes and selective sweep mapping
Thermal adaptation in Drosophila is a complex topic that comprises a variety of physiological processes (whether considering a short-term or long-term treatment, Loeschcke & Sørensen 2005). In the case of chill coma recovery, adaptation might occur by a rapid cold response triggered by multiple genes. Besides the direct temperature resistance, further mechanisms such as energy allocation shifts and metabolic regulation could increase cold tolerance (see Supplementary Data S1).
Thus far, the X chromosome was thought to be rather poor in cold tolerance factors as all major genes (ppk, trap1, Fst, Sas, desat2, Catsup, Ddc, nompA, hsp 70, Dca and hsr-omega) were mapped on the autosomes (Goto 2000, 2001; Morgan & Mackay 2006; Norry et al. 2007, 2008; Sinclair et al. 2007). Recent expression analyses, however, produced many candidates on the X chromosome that are associated with the cold tolerance phenotype measured by cold shock treatments. Telonis-Scott et al. (2009) created lines of D. melanogaster that were artificially selected for cold resistance by breeding individuals with elevated chill coma recovery abilities (protocol described in Anderson et al. 2005). By comparing gene expression levels of selected and nonselected lines, they found ten genes on the X chromosome that significantly altered their expression level, eight of which are located within our QTL. Another expression study (Ayroles et al. 2009) revealed 144 genes with expression levels correlated with the ability to recover from chill coma. Out of this set, 91 genes are located within our QTL.
Nonetheless, it remains challenging to narrow down the list of candidate genes that are promising for further study. Here, we used selective sweep analysis for fine-mapping the genomic region colocalizing with a QTL to search for suitable candidate genes. Selective sweep mapping under the QTL most proximal to the centromere revealed evidence for recent positive selection in the European population. The target of selection was located in the intergenic region between two adjacent genes, CG16700 and CG4991 (Fig. 6). In combination with the recent results of Ayroles et al. (2009) that showed an association of CG16700 with CCRT in another derived population (from North America), our sweep analysis suggests that CG16700 is a candidate gene for cold tolerance. Functional investigations, however, are needed to validate this result.
This example shows that fine mapping based on selective sweeps may provide a powerful approach to identify candidate genes that colocalize with QTL. A similar strategy can be used in cases in which other fine-scale analyses of QTL, such as deletion mapping, are not applicable (Rubin et al. 2010).
Additional genes on the X chromosome that potentially affect CCRT and colocalize with the identified QTL have been obtained in database searches (see Supplementary Table S2).
Supplementary Material
Data S1 Candidate genes.
Fig. S1 Construction of the substitution lines A* and E*.
Fig. S2 Homogenization of maternally inherited factors in A* and E*.
Table S1 Marker information and related accession nos of sequenced fragments of isofemale fly lines E14 and A157 of D. melanogaster (Glinka et al. 2003; Ometto et al. 2005).
Table S2 Candidate gene list for all six QTL positions.
Acknowledgments
We thank David De Lorenzo for discussion, Aurélie Bonin and her team of reviewers for valuable suggestions on previous versions of the manuscript, and Anne Wilken and Susanne Voigt for excellent technical assistance. This research was funded by grant STE 325/12 from the DFG Research Unit 1078 to WS, National Institutes of Health grant GM074244 to KWB, and doctoral fellowships from the Volkswagen-Foundation to PP and RW. S.
Footnotes
N.S. has special interest in the behavior, the genetics, and the life history of insects. A.W. works on QTL and selective sweep mapping in model organisms. R.W. works on adaptive consequences of standing genetic variation in natural populations, including humans. P.P. develops algorithms to detect selective sweeps in populations that have experienced past demographic changes. J.C. is focusing on theoretical models of genetic effects for improving quantitative analyses in general and QTL analyses in particular. K.B’s research interests include the characterization of meiotic recombination and the development of improved methods and software for QTL mapping. The main focus of D.M.’s research is on the development of model-based methods for the analysis of genetic data. W.S. works on various questions of theoretical and empirical population genetics (in particular adaptation).
Data accessibility:
data deposited at Dryad: doi:10.5061/dryad.8083.
References
- Anderson AR, Hoffmann AA, McKechnie SW. Response to selection for rapid chill-coma recovery in Drosophila melanogaster: physiology and life-history traits. Genetical Research. 2005;85:15–22. doi: 10.1017/s0016672304007281. [DOI] [PubMed] [Google Scholar]
- Ayroles JF, Carbone MA, Stone EA, et al. Systems genetics of complex traits in Drosophila melanogaster. Nature Genetics. 2009;41:299–307. doi: 10.1038/ng.332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azevedo RBR, Vernon F, Partridge L. Thermal evolution of egg size in Drosophila melanogaster. Evolution. 1996;50:2338–2345. doi: 10.1111/j.1558-5646.1996.tb03621.x. [DOI] [PubMed] [Google Scholar]
- Baudry E, Viginier B, Veuille M. Non-African populations of Drosophila melanogaster have a unique origin. Molecular Biology and Evolution. 2004;21:1482–1491. doi: 10.1093/molbev/msh089. [DOI] [PubMed] [Google Scholar]
- Begun DJ, Aquadro CF. African and North American populations of Drosophila melanogaster are very different at the DNA level. Nature. 1993;365:548–550. doi: 10.1038/365548a0. [DOI] [PubMed] [Google Scholar]
- Beisswanger S, Stephan W. Evidence that strong positive selection drives neofunctionalization in the tandemly duplicated polyhomeotic genes in Drosophila. Proceedings of the National Academy of Sciences, USA. 2008;105:5447–5452. doi: 10.1073/pnas.0710892105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beisswanger S, Stephan W, De Lorenzo D. Evidence for a selective sweep in the wapl region of Drosophila melanogaster. Genetics. 2006;172:265–274. doi: 10.1534/genetics.105.049346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bersaglieri T, Sabeti PC, Patterson N, et al. Genetic signatures of strong recent positive selection at the lactase gene. American Journal of Human Genetics. 2004;74:1111–1120. doi: 10.1086/421051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borevitz JO, Hazen SP, Michael TP, et al. Genome-wide patterns of single-feature polymorphism in Arabidopsis thaliana. Proceedings of the National Academy of Sciences, USA. 2007;104:12057–12062. doi: 10.1073/pnas.0705323104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broman KW, Wu H, Sen S, Churchill GA. R/qtl: QTL mapping in experimental crosses. Bioinformatics. 2003;19:889–890. doi: 10.1093/bioinformatics/btg112. [DOI] [PubMed] [Google Scholar]
- Bubliy OA, Riihimaa A, Norry FM, Loeschcke V. Variation in resistance and acclimation to low-temperature stress among three geographical strains of Drosophila melanogaster. Journal of Thermal Biology. 2002;27:337–344. [Google Scholar]
- Cariou ML. Biochemical phylogeny of the eight species in the Drosophila melanogaster subgroup, including D. sechellia and D. orena. Genetical Research. 1987;50:181–185. doi: 10.1017/s0016672300023673. [DOI] [PubMed] [Google Scholar]
- Comeron JM, Kreitman M, Aguadé M. Natural selection on synonymous sites is correlated with gene length and recombination in Drosophila. Genetics. 1999;151:239–249. doi: 10.1093/genetics/151.1.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crow JF. On epistasis: why it is unimportant in polygenic directional selection. Philosophical Transactions of the Royal Society B: Biological Sciences. 2010;365:1241–1244. doi: 10.1098/rstb.2009.0275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David JR, Capy P. Genetic variation of Drosophila melanogaster natural populations. Trends in Genetics. 1988;4:106–111. doi: 10.1016/0168-9525(88)90098-4. [DOI] [PubMed] [Google Scholar]
- David J, Clavel MF. Influence of temperature during the course of development on various biometric characteristics of adult Drosophila melanogaster Meigen. Journal of Insect Physiology. 1967;13:717–729. doi: 10.1016/0022-1910(67)90121-7. [DOI] [PubMed] [Google Scholar]
- David JR, Gibert P, Pla E, et al. Cold stress tolerance in Drosophila: analysis of chill coma recovery in D. melanogaster. Journal of Thermal Biology. 1998;23:291–299. [Google Scholar]
- Fiston-Lavier AS, Singh ND, Lipatov M, Petrov DA. Drosophila melanogaster recombination rate calculator. Gene. 2010;463:18–20. doi: 10.1016/j.gene.2010.04.015. [DOI] [PubMed] [Google Scholar]
- Gibert P, Moreteau B, Petavy G, Karan D, David JR. Chill-coma tolerance, a major climatic adaptation among Drosophila species. Evolution. 2001;55:1063–1068. doi: 10.1554/0014-3820(2001)055[1063:cctamc]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Glinka S, Ometto L, Mousset S, Stephan W, De Lorenzo D. Demography and natural selection have shaped genetic variation in Drosophila melanogaster: a multi-locus approach. Genetics. 2003;165:1269–1278. doi: 10.1093/genetics/165.3.1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto SG. Expression of Drosophila homologue of senescence marker protein-30 during cold acclimation. Journal of Insect Physiology. 2000;46:1111–1120. doi: 10.1016/s0022-1910(99)00221-8. [DOI] [PubMed] [Google Scholar]
- Goto SG. A novel gene that is up-regulated during recovery from cold shock in Drosophila melanogaster. Gene. 2001;270:259–264. doi: 10.1016/s0378-1119(01)00465-6. [DOI] [PubMed] [Google Scholar]
- Haldane JBS. The combination of linkage values, and the calculation of distance between the loci of linked factors. Journal of Genetics. 1919;8:299–309. [Google Scholar]
- Haley CS, Knott SA. A simple regression method for mapping quantitative trait loci in line crosses using flanking markers. Heredity. 1992;69:315–324. doi: 10.1038/hdy.1992.131. [DOI] [PubMed] [Google Scholar]
- Heyen DW, Weller JI, Ron M, et al. A genome scan for QTL influencing milk production and health traits in dairy cattle. Physiological Genomics. 1999;1:165–175. doi: 10.1152/physiolgenomics.1999.1.3.165. [DOI] [PubMed] [Google Scholar]
- Hinds DA, Stuve LL, Nilsen GB, et al. Whole-genome patterns of common DNA variation in three human populations. Science. 2005;307:1072–1079. doi: 10.1126/science.1105436. [DOI] [PubMed] [Google Scholar]
- Hoffmann AA, Anderson A, Hallas R. Opposing clines for high and low temperature resistance in Drosophila melanogaster. Ecology Letters. 2002;5:614–618. [Google Scholar]
- Hoffmann AA, Sørensen JG, Loeschcke V. Adaptation of Drosophila to temperature extremes: bringing together quantitative and molecular approaches. Journal of Thermal Biology. 2003;28:175–216. [Google Scholar]
- Hudson RR. Generating samples under a Wright-Fisher neutral model of genetic variation. Bioinformatics. 2002;18:337–338. doi: 10.1093/bioinformatics/18.2.337. [DOI] [PubMed] [Google Scholar]
- Hutter S, Li H, Beisswanger S, De Lorenzo D, Stephan W. Distinctly different sex ratios in African and European populations of Drosophila melanogaster inferred from chromosomewide single nucleotide polymorphism data. Genetics. 2007;177:469–480. doi: 10.1534/genetics.107.074922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izquierdo JI. How does Drosophila melanogaster overwinter? Entomologia Experimentalis et Applicata. 1991;59:51–58. [Google Scholar]
- James AC, Partridge L. Thermal evolution of rate of larval development in Drosophila melanogaster in laboratory and field populations. Journal of Evolutionary Biology. 1995;8:315–330. [Google Scholar]
- James AC, Azevedo RB, Partridge L. Genetic and environmental responses to temperature of Drosophila melanogaster from a latitudinal cline. Genetics. 1997;146:881–890. doi: 10.1093/genetics/146.3.881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen RC, Stam P. High resolution of quantitative traits into multiple loci via interval mapping. Genetics. 1994;136:1447–1455. doi: 10.1093/genetics/136.4.1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kane NC, Rieseberg LH. Genetics and evolution of weedy Helianthus annuus populations: adaptation of an agricultural weed. Molecular Ecology. 2008;17:384–394. doi: 10.1111/j.1365-294X.2007.03467.x. [DOI] [PubMed] [Google Scholar]
- Kim Y, Nielsen R. Linkage disequilibrium as a signature of selective sweeps. Genetics. 2004;167:1513–1524. doi: 10.1534/genetics.103.025387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lachaise D, Silvain JF. How two Afrotropical endemics made two cosmopolitan human commensals: the Drosophila melanogaster-D. simulans palaeogeographic riddle. Genetica. 2004;120:17–39. doi: 10.1023/b:gene.0000017627.27537.ef. [DOI] [PubMed] [Google Scholar]
- Li H, Stephan W. Inferring the demographic history and rate of adaptive substitution in Drosophila. PLoS Genetics. 2006;2:e166. doi: 10.1371/journal.pgen.0020166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loeschcke V, Sørensen JG. Acclimation, heat shock and hardening—a response from evolutionary biology. Journal of Thermal Biology. 2005;30:255–257. [Google Scholar]
- Long AD, Mullaney SL, Reid LA, et al. High resolution mapping of genetic factors affecting abdominal bristle number in Drosophila melanogaster. Genetics. 1995;139:1273–1291. doi: 10.1093/genetics/139.3.1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay TFC. Quantitative trait loci in Drosophila. Nature Reviews Genetics. 2001;2:11–20. doi: 10.1038/35047544. [DOI] [PubMed] [Google Scholar]
- Mckenzie J. The influence of low temperature on survival and reproduction in populations of Drosophila melanogaster. Australian Journal of Zoology. 1975;23:237–247. [Google Scholar]
- Milton CC, Partridge L. Brief carbon dioxide exposure blocks heat hardening but not cold acclimation in Drosophila melanogaster. Journal of Insect Physiology. 2008;54:32–40. doi: 10.1016/j.jinsphys.2007.08.001. [DOI] [PubMed] [Google Scholar]
- Mitrovski P, Hoffmann AA. Postponed reproduction as an adaptation to winter conditions in Drosophila melanogaster: evidence for clinal variation under semi-natural conditions. Proceedings of the Royal Society in London. Series B, Biological Sciences. 2001;268:2163–2168. doi: 10.1098/rspb.2001.1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moehring AJ, Mackay TFC. The quantitative genetic basis of male mating behavior in Drosophila melanogaster. Genetics. 2004;167:1249–1263. doi: 10.1534/genetics.103.024372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan TJ, Mackay TFC. Quantitative trait loci for thermotolerance phenotypes in Drosophila melanogaster. Heredity. 2006;96:232–242. doi: 10.1038/sj.hdy.6800786. [DOI] [PubMed] [Google Scholar]
- Nordborg M, Hu TT, Ishino Y, et al. The pattern of polymorphism in Arabidopsis thaliana. PLoS Biology. 2005;3:e196. doi: 10.1371/journal.pbio.0030196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norry FM, Dahlgaard J, Loeschcke V. Quantitative trait loci affecting knockdown resistance to high temperature in Drosophila melanogaster. Molecular Ecology. 2004;13:3585–3594. doi: 10.1111/j.1365-294X.2004.02323.x. [DOI] [PubMed] [Google Scholar]
- Norry FM, Gomez FH, Loeschcke V. Knockdown resistance to heat stress and slow recovery from chill coma are genetically associated in a quantitative trait locus region of chromosome 2 in Drosophila melanogaster. Molecular Ecology. 2007;16:3274–3284. doi: 10.1111/j.1365-294X.2007.03335.x. [DOI] [PubMed] [Google Scholar]
- Norry FM, Scannapieco AC, Sambucetti P, Bertoli CI, Loeschcke V. QTL for the thermotolerance effect of heat hardening, knockdown resistance to heat and chill-coma recovery in an intercontinental set of recombinant inbred lines of Drosophila melanogaster. Molecular Ecology. 2008;17:4570–4581. doi: 10.1111/j.1365-294X.2008.03945.x. [DOI] [PubMed] [Google Scholar]
- Ometto L, Glinka S, De Lorenzo D, Stephan W. Inferring the effects of demography and selection on Drosophila melanogaster populations from a chromosome-wide scan of DNA variation. Molecular Biology and Evolution. 2005;22:2119–2130. doi: 10.1093/molbev/msi207. [DOI] [PubMed] [Google Scholar]
- Palaisa K, Morgante M, Tingey S, Rafalski A. Long-range patterns of diversity and linkage disequilibrium surrounding the maize Y1 gene are indicative of an asymmetric selective sweep. Proceedings of the National Academy of Sciences, USA. 2004;101:9885–9890. doi: 10.1073/pnas.0307839101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlidis P, Jensen JD, Stephan W. Searching for footprints of positive selection in whole-genome SNP data from nonequilibrium populations. Genetics. 2010;185:907–922. doi: 10.1534/genetics.110.116459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters TM, Barbosa P. Influence of population density on size, fecundity, and developmental rate of insects in culture. Annual Review of Entomology. 1977;22:431–450. [Google Scholar]
- Rubin CJ, Zody MC, Eriksson J, et al. Whole-genome resequencing reveals loci under selection during chicken domestication. Nature. 2010;464:587–591. doi: 10.1038/nature08832. [DOI] [PubMed] [Google Scholar]
- Sen S, Churchill GA. A statistical framework for quantitative trait mapping. Genetics. 2001;159:371–387. doi: 10.1093/genetics/159.1.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinclair BJ, Gibbs AG, Roberts SP. Gene transcription during exposure to, and recovery from, cold and desiccation stress in Drosophila melanogaster. Insect Molecular Biology. 2007;16:435–443. doi: 10.1111/j.1365-2583.2007.00739.x. [DOI] [PubMed] [Google Scholar]
- Stanley S, Parsons P, Spence G, Weber L. Resistance of species of the Drosophila melanogaster subgroup to environmental extremes. Australian Journal of Zoology. 1980;28:413–421. [Google Scholar]
- Stephan W, Li H. The recent demographic and adaptive history of Drosophila melanogaster. Heredity. 2007;98:65–68. doi: 10.1038/sj.hdy.6800901. [DOI] [PubMed] [Google Scholar]
- Svetec N, Pavlidis P, Stephan W. Recent strong positive selection on Drosophila melanogaster HDAC6, a gene encoding a stress surveillance factor, as revealed by population genomic analysis. Molecular Biology and Evolution. 2009;26:1549–1556. doi: 10.1093/molbev/msp065. [DOI] [PubMed] [Google Scholar]
- Telonis-Scott M, Hallas R, McKechnie SW, Wee CW, Hoffmann AA. Selection for cold resistance alters gene transcript levels in Drosophila melanogaster. Journal of Insect Physiology. 2009;55:549–555. doi: 10.1016/j.jinsphys.2009.01.010. [DOI] [PubMed] [Google Scholar]
- Thornton KR, Jensen JD. Controlling the false-positive rate in multilocus genome scans for selection. Genetics. 2007;175:737–750. doi: 10.1534/genetics.106.064642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucić N. Genetic capacity for adaptation to cold resistance at different developmental stages of Drosophila melanogaster. Evolution. 1979;33:350–358. doi: 10.1111/j.1558-5646.1979.tb04688.x. [DOI] [PubMed] [Google Scholar]
- Tuiskula-Haavisto M, Honkatukia M, Vilkki J, et al. Mapping of quantitative trait loci affecting quality and production traits in egg layers. Poultry Science. 2002;81:919–927. doi: 10.1093/ps/81.7.919. [DOI] [PubMed] [Google Scholar]
- Van ‘t Land J, Putten PV, Zwaan B, Kamping A, Delden WV. Latitudinal variation in wild populations of Drosophila melanogaster: heritabilities and reaction norms. Journal of Evolutionary Biology. 1999;12:222–232. [Google Scholar]
- Wang S, Basten CJ, Zeng Z-B. Windows QTL Cartographer 2.5. Department of Statistics, North Carolina State University; Raleigh, NC: 2007. [Google Scholar]
- Xu S. Quantitative trait locus mapping can benefit from segregation distortion. Genetics. 2008;180:2201–2208. doi: 10.1534/genetics.108.090688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Wang S, Li H, et al. Effects of missing marker and segregation distortion on QTL mapping in F2 populations. Theoretical and Applied Genetics. 2010;121:1071–1082. doi: 10.1007/s00122-010-1372-z. [DOI] [PubMed] [Google Scholar]
- Zivkovic D, Wiehe T. Second-order moments of segregating sites under variable population size. Genetics. 2008;180:341–357. doi: 10.1534/genetics.108.091231. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data S1 Candidate genes.
Fig. S1 Construction of the substitution lines A* and E*.
Fig. S2 Homogenization of maternally inherited factors in A* and E*.
Table S1 Marker information and related accession nos of sequenced fragments of isofemale fly lines E14 and A157 of D. melanogaster (Glinka et al. 2003; Ometto et al. 2005).
Table S2 Candidate gene list for all six QTL positions.